- 1National Key Laboratory of Intelligent Tracking and Forecasting for Infectious Diseases, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China
- 2Department of Respiratory Medicine, Peking University People’s Hospital, Beijing, China
We report a case of Legionnaires’ disease caused by Legionella bozemanae, which is the first time that L. bozemanae has been isolated from a bronchoalveolar lavage fluid sample from an immunocompromised patient in China. The findings highlight the susceptibility of immunocompromised patients to infections caused by the rare but highly pathogenic L. bozemanae.
Introduction
Legionella, a bacterial genus that is widespread in aquatic environments, enters human alveolar macrophages via the respiratory tract and causes Legionnaires’ disease (Mondino et al., 2020). Legionella bozemanae, which was first isolated in 1959 from the lung tissue of a navy diver, is a rare cause of Legionnaires’ disease, accounting for 3%–5% cases of pneumonia caused by Legionella (Bozeman et al., 1968; Mitchell et al., 1984; Reingold et al., 1984). The associated mortality rate is up to 40% (Taylor and Albrecht, 1995). Diagnosis is difficult because this species is insensitive to serological tests and urinary antigen tests for L. pneumophila serogroup 1 (Guyard and Low, 2011; Widmer et al., 2007). Here we report a case involving an immunocompromised patient in China who developed Legionnaires’ disease caused by L. bozemanae. The clinical manifestations included cough, expectoration, lung infiltration, fever, and dyspnea. Second-generation sequencing of plasma and bronchoalveolar lavage fluid (BALF) showed positivity for L. bozemanae. After anti-infective treatment with rifampicin, moxifloxacin, azithromycin, piperacillin/tazobactam, the patient gradually recovered. To our knowledge, this is the first time that L. bozemanae has been isolated from patient sample in China, and the findings are expected to provide a reference for clinical diagnosis.
Case presentation
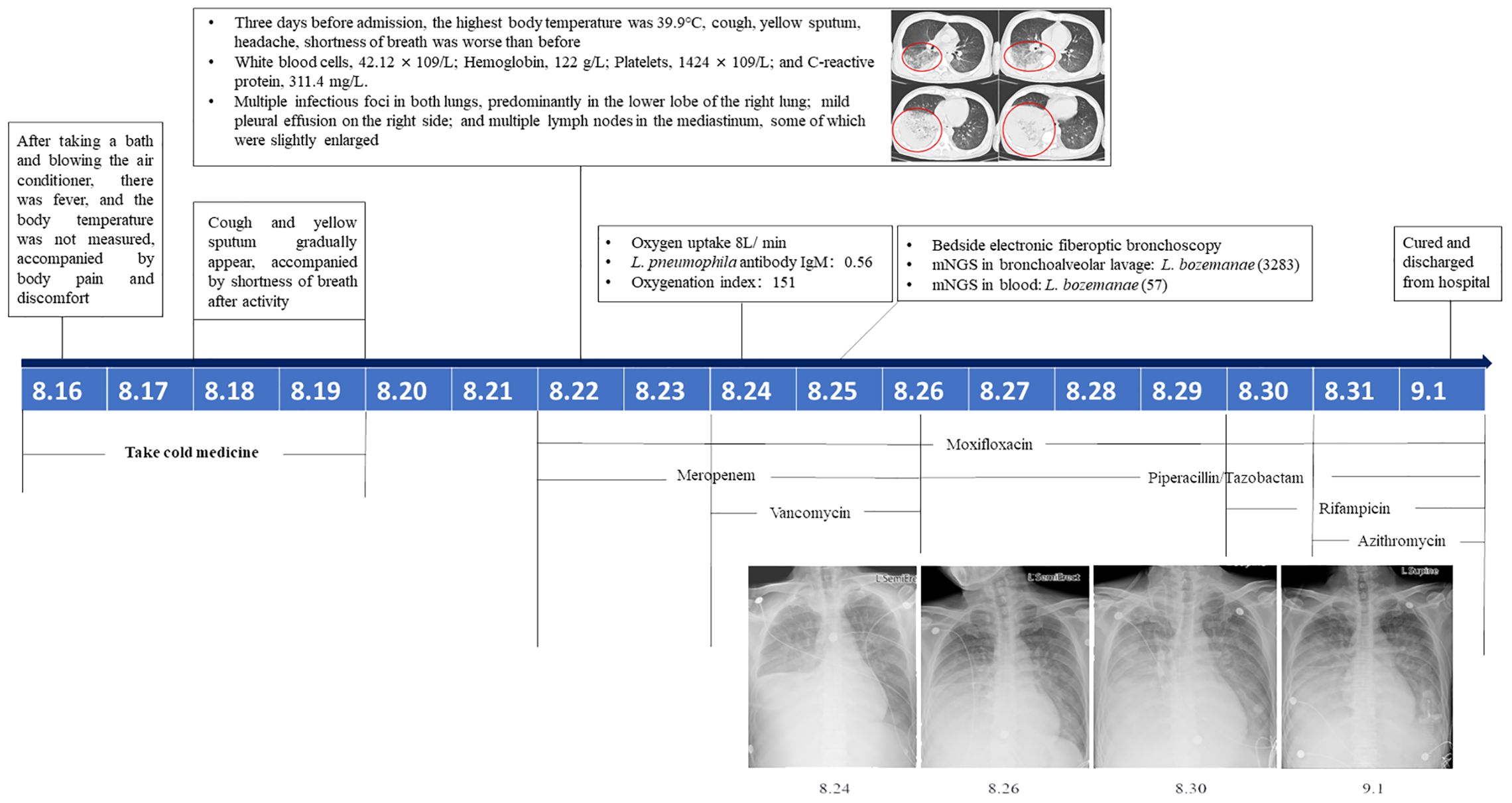
The patient was a 50-year-old male farmer with a 30-year history of smoking 5–20 cigarettes per day and social drinking. On August 24, 2022, he was admitted to a hospital in Beijing with fever, cough, yellow sputum, and progressively worsening dyspnea for 9 days. Eight days before admission, he developed a fever after taking a bath and turning on the air conditioner. The fever was accompanied by peripheral aches and pains followed by the gradual development of a cough and yellow sputum with dyspnea on exertion. The highest recorded temperature was 39.9°C 3 days before admission, with worsening cough and sputum; therefore, he visited an outpatient clinic. Test results showed the following: white blood cells, 42.12 × 109/L; hemoglobin, 122 g/L; platelets, 1424 × 109/L; and C-reactive protein, 311.4 mg/L. Chest computed tomography suggested multiple infectious foci in both lungs, predominantly in the lower lobe of the right lung; mild pleural effusion on the right side; and multiple lymph nodes in the mediastinum, some of which were slightly enlarged. Following admission, L. pneumophila IgM antibody titers were 0.56, 1.24, and 1.31, respectively, and the oxygenation index was 151. Right lower lung palpation showed fibrillation was slightly diminished, and percussion revealed murmurs. The breath sounds were low, with coarse breath sounds heard over the left lung, and no dry rales were heard. The patient’s dyspnea worsened, and he received an oxygen mask (8 L/min) with meropenem (1.0g q8h), moxifloxacin (0.4g qd), and vancomycin (1.0 g q12h) for treating the infection. Bedside fiberoptic bronchoscopy showed some golden-yellow secretions in the lumen, congestion of the tracheal mucosa, and a sharp, central bulge. Second-generation sequencing of plasma and bronchoalveolar lavage fluid (BALF) showed positivity for L. bozemanae. The patient gradually recovered following anti-infective treatment with rifampicin (0.45 g qd), moxifloxacin (0.4 g qd), azithromycin (0.5 g qd) and piperacillin/tazobactam (4.5 g q8h). Eventually, the patient was cured and discharged (Figure 1).
Methods
Bioinformatics
The DNA was extracted using a DNA extraction kit (QIAamp DNA Mini Kit, QIAGEN, Hilden, Germany) following the manufacturer’s instructions. The quality qualified DNA was analysed by Next-generation sequencing (NGS) at Novogene Co., Ltd. (Beijing, China). This DNA sample was fragmented to 350bp size by ultrasonic crushing to construct a small fragment gene library, and double-end sequencing was performed based on Illumina NovaSeq PE150 to remove a certain proportion of low-quality data in the sequenced raw data to ensure the accuracy and reliability of the results of the subsequent information analysis. After obtaining valid data, SOAP denovo software and CISA software were used for assembly and integration.
Single-copy orthologous genes were identified based on gene family clustering, and single-copy core genes were identified based on core-pan analysis. MUSCLE software was used to compare the genes, and the phylogenetic tree was constructed by TreeBeST software using the NJ method and the maximum likelihood method of PHYML.
Intracellular growth assay
Mouse macrophages J774 were cultured in DMEM-H (Gibco, NY, USA) medium supplemented with 10% fetal bovine serum at 37 °C with 5% CO2. Logarithmic growth phase suspensions of L. bozemanae, L. pneumophila ATCC 33152, and L. pneumophila philadelphia1 (JR32) (approximately 1×108 CFU/mL) were diluted 10-fold in DMEM-H medium to bring the infected suspensions to 1×107 CFU/mL. Add 1 mL of infected bacterial suspension to J774 mouse macrophages (1×105 cells/mL) pre-spread in a 24-well plate so that the Multiplicity Of Infection (MOI) is 100. After the infected cells were incubated in a 5% CO2 incubator at 37 °C for 1.5 h, the supernatant solution containing the bacteria was discarded and the cells were washed three times with 500 μL of 10 mM phosphate buffered saline (PBS; Gibco) to remove extracellular bacteria. Then 0.5 mL DMEM-H medium containing 10% fetal bovine serum (FBS; Gibco) was added to each well. To determine the number of proliferating bacteria in J774 cells, 0.5 mL sterile distilled water was added to each well every 24 h to completely suspend the infected cells in the culture medium, and then transferred to a 1.5 mL sterile centrifuge tube. The number of proliferating bacteria was counted by spreading the gradient bacterial solution on buffered charcoal yeast extract (BCYE; Oxoid) plates by serial dilution, and each experiment was repeated three times.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed according to EUCAST recommendations and manufacturer’s instructions (https://www.eucast.org/eucastguidancedocuments/). Epidemiological cut-off (ECOFF) of resistance determined by EUCAST guidelines was used to interpret the results. The minimum inhibitory concentration (MIC) of antibiotics for routine clinical treatment against each strain was evaluated using the E-test strip method, and L. pneumophila ATCC 33152 was used as a control for drug susceptibility testing. Fresh colonies were taken and resuspended in PBS and the concentration of the bacterial solution was adjusted to 0.5 McFarland standard. The surface of BCYE-α plate was uniformly wiped with a sterile cotton swab dipped in bacterial solution, and the E test strip was placed in the center of the plate. After incubation at 35°C for 48 h, the MIC value of the E test strip was determined. The experiment was repeated for three times.
Results and discussion
The patient’s sputum sample was spread on glycine–vancomycin–polymyxin–cycloheximide plates (OXOID, UK) for pathogen isolation. The isolated strain was identified as L. bozemanae by 16S rRNA gene sequencing and next-generation sequencing. Evaluation of phylogenetic trees based on gene families (Figure 2A) and core genes (Figure 2B) showed that L. bozemanae BOZE (ICDC) was most closely related to L. bozemanae 94163278 (Supplementary Data Sheets S1 and S2). Whole genome sequencing data have been deposited in the NCBI BioProject repository (accession no. PRJNA992381) and the BioSample database (accession no. SAMN36348289).
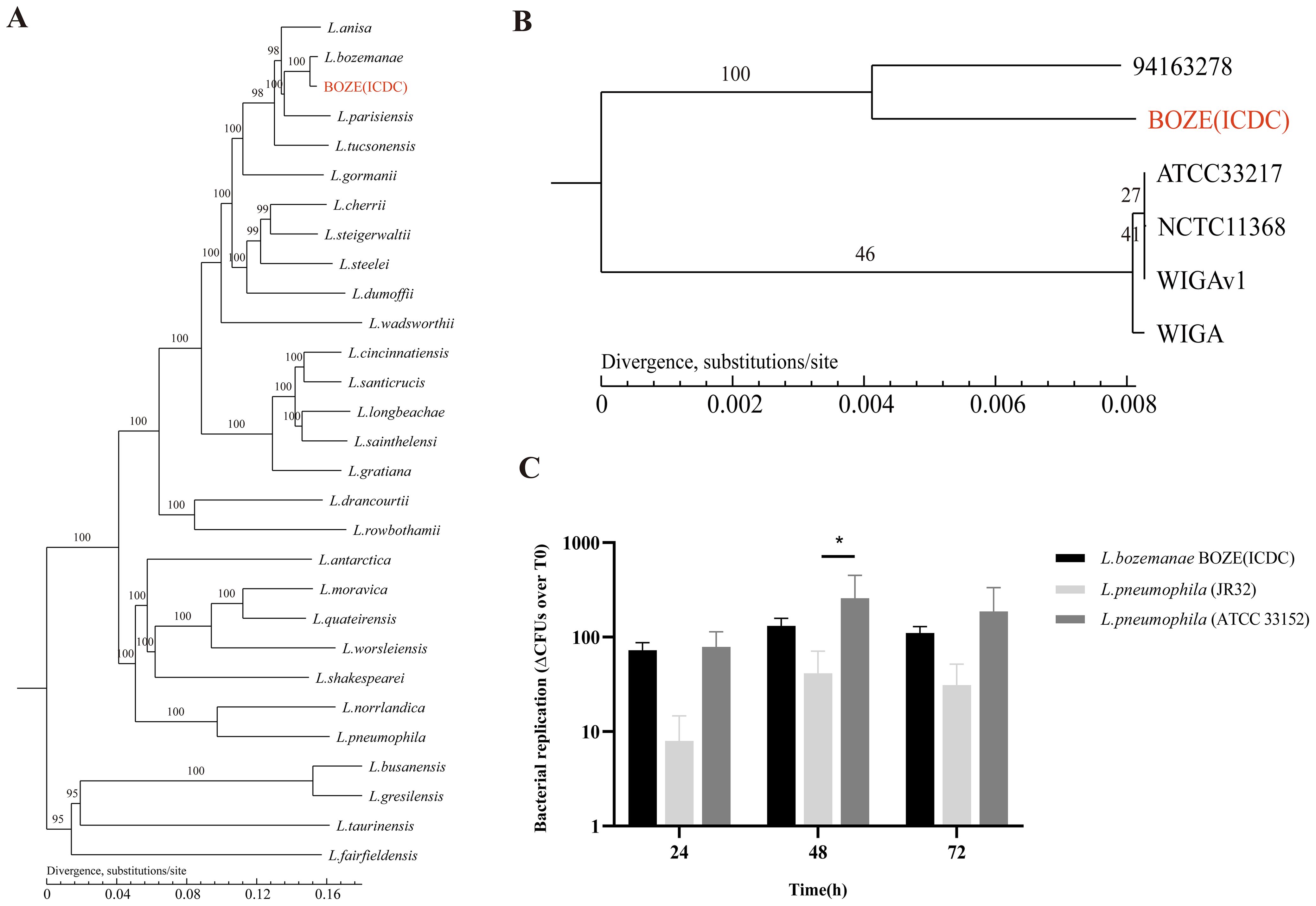
Figure 2. Genetic evolution and pathogenicity of the L. bozemanae strain BOZE (ICDC). (A) Phylogenetic tree based on the gene family for Legionella species. (B) Phylogenetic tree based on the core genes of L. bozemanae. (C) Comparison of intracellular proliferation in J774 mouse macrophages between L. bozemanae strain BOZE (ICDC) and control strains L. pneumophila Philadelphia 1 (JR32) and L. pneumophila ATCC 33152 at T0, T24, T48, and T72 hours. The results are presented as means ± SD from three independent experiments. Significance was assessed at a 95% confidence interval (*p<0.05) by two-way ANOVA with Bonferroni’s multiple-comparison test.
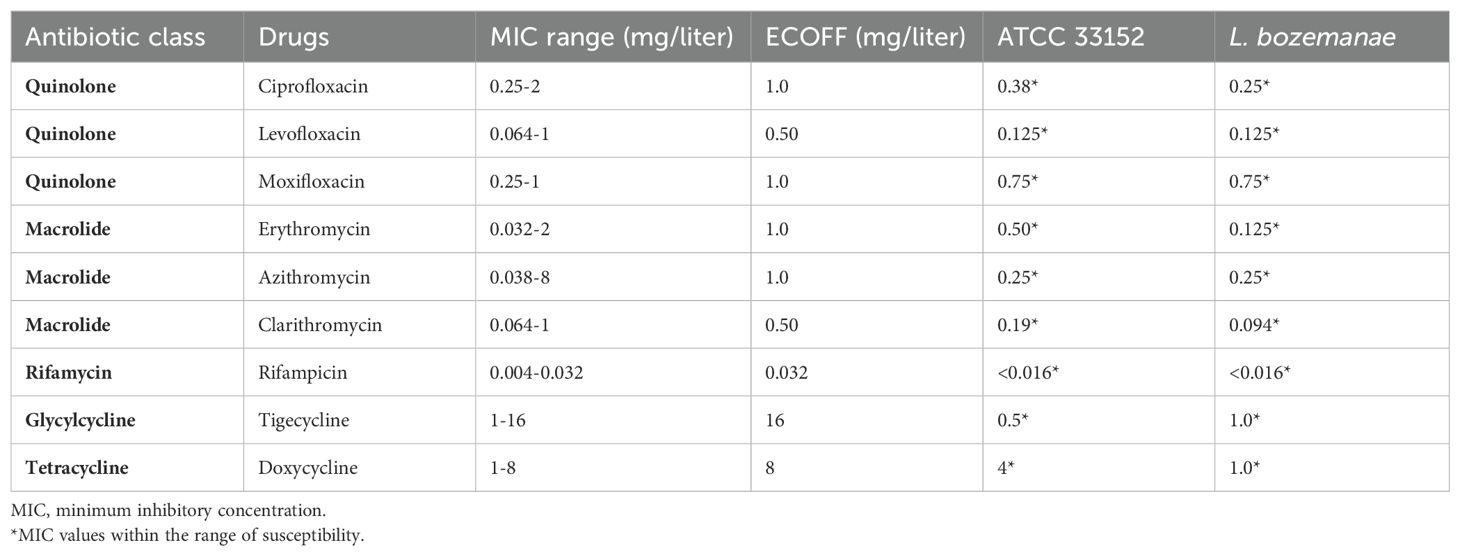
An understanding of the biological features of bacteria can provide a solid basis for infection treatment. At present, 17% environmental and clinical Legionella species in China have been found to exhibit azithromycin resistance (Jia et al., 2019). Therefore, the sensitivity of L. bozemanae BOZE (ICDC) to nine representative antibiotics (ciprofloxacin, levofloxacin, moxifloxacin, erythromycin, azithromycin, clarithromycin, rifampicin, tigecycline and doxycycline) was tested, and it was found that the minimum inhibitory concentration values were significantly lower than the epidemiological cut-off values (Table 1). Thus, routine clinical medications can be used to treat Legionnaires’ disease caused by L. bozemanae BOZE (ICDC). Cytologic experiments showed that the fold change in growth over T0 of L. pneumophila ATCC 33152 and L. bozemanae BOZE(ICDC) was overall higher than that of JR32 at T24, T48, and T72 hours. And similar intracellular proliferative capacity compared to the highly pathogenic and virulent L. pneumophila ATCC 33152 suggests the need to be vigilant about the infection of L. pneumophila BOZE (ICDC) (Figure 2C).
In summary, we reported a rare case of Legionnaires’ disease caused by the rare L. bozemanae in China. Our findings highlight that clinicians should be aware that infection with this species may go undetected in patients with clinical pneumonia, and they should particularly consider Legionella infection in immunocompromised patients. In addition, the surveillance and investigation of L. bozemanae in aquatic environments should be strengthened to prevent and control the outbreak of Legionnaires’ disease caused by L. bozemanae.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI, PRJNA992381, SAMN36348289.
Ethics statement
The study was approved by the Ethical Committee of the Chinese Center for Disease Control and Prevention (No. ICDC-202115). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
PX: Writing – original draft, Writing – review & editing. FW: Software, Writing – original draft. HR: Methodology, Writing – original draft. WN: Validation, Writing – review & editing. NZ: Investigation, Writing – original draft. RL: Investigation, Writing – original draft. YC: Data curation, Writing – original draft. ZG: Methodology, Writing – original draft. TQ: Conceptualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the National Natural Science Foundation of China [grant number 81671985], and the Science Foundation for the State Key Laboratory for Infectious Disease Prevention and Control of China [grant numbers 2022SKLID209, 2019SKLID403], and the Public Health Service Capability Improvement Project of the National Health Commission of the People’s Republic of China [grant numbers 2100409034, 0104205].
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbrio.2024.1476727/full#supplementary-material
Supplementary DATA SHEET S1 | Gene family phylogenetic tree related genes for Legionella.
Supplementary DATA SHEET S2 | Core genes phylogenetic tree related genes for L. bozemanae.
References
Bozeman F. M., Humphries J. W., Campbell. J. M. (1968). A new group of rickettsia-like agents recovered from Guinea pigs. Acta Virol. 12, 87–93. doi: 10.1146/annurev-pathmechdis-012419-032742
Guyard C., Low D. E. (2011). Legionella infections and travel associated legionellosis. Travel Med. Infect. Dis. 9, 176–186. doi: 10.1016/s0163-4453(84)94027-1
Jia X., Ren H., Nie X., Li Y., Li J., Qin T. (2019). Antibiotic resistance and azithromycin resistance mechanism of legionella pneumophila serogroup 1 in China. Antimicrob. Agents Chemother. 63 (10), e00768-19. doi: 10.1093/infdis/149.5.819
Mitchell R. G., Pasvol G., Newnham. R. S. (1984). Pneumonia due to Legionella bozemanii: first report of a case in Europe. J. Infect. 8, 251–255. doi: 10.1093/clinids/20.2.329
Mondino S., Schmidt S., Rolando M., Escoll P., Gomez-Valero L., Buchrieser C. (2020). Legionnaires’ Disease: state of the art knowledge of pathogenesis mechanisms of legionella. Annu. Rev. Pathol. 15, 439–466. doi: 10.1016/j.tmaid.2010.05.006
Reingold A. L., Thomason B. M., Brake B. J., Thacker L., Wilkinson H. W., Kuritsky J. N. (1984). Legionella pneumonia in the United States: the distribution of serogroups and species causing human illness. J. Infect. Dis. 149 (5), 819. doi: 10.1093/infdis/149.5.819
Taylor T. H., Albrecht M. A. (1995). Legionella bozemanii cavitary pneumonia poorly responsive to erythromycin: case report and review. Clin. Infect. Dis. 20, 329–334. doi: 10.1128/aac.00768-19
Keywords: Legionella bozemanae, Legionnaires’ disease, pneumonia, non-pneumophila legionella, antibiotic susceptibility
Citation: Xu P, Wang F, Ren H, Ni W, Zhao N, Li R, Chen Y, Gao Z and Qin T (2024) A case report of severe pulmonary legionellosis caused by Legionella bozemanae. Front. Bacteriol. 3:1476727. doi: 10.3389/fbrio.2024.1476727
Received: 06 August 2024; Accepted: 30 September 2024;
Published: 16 October 2024.
Edited by:
Weng C. Chan, University of Nottingham, United KingdomReviewed by:
Stanimir S. Ivanov, Louisiana State University Health Shreveport, United StatesThorsten Kuczius, University Hospital Münster, Germany
Copyright © 2024 Xu, Wang, Ren, Ni, Zhao, Li, Chen, Gao and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tian Qin, cWludGlhbkBpY2RjLmNu
†These authors have contributed equally to this work and share first authorship
 Peixing Xu
Peixing Xu Fang Wang2†
Fang Wang2† Wentao Ni
Wentao Ni Na Zhao
Na Zhao Ran Li
Ran Li Zhancheng Gao
Zhancheng Gao Tian Qin
Tian Qin