- 1Department of Infectious Diseases and Immunology, College of Veterinary Medicine, University of Florida, Gainesville, FL, United States
- 2Faculty of Veterinary Science, Prince of Songkla University, Hatyai, Songkhla, Thailand
- 3Faculty of Medical Technology, Prince of Songkla University, Hatyai, Songkhla, Thailand
- 4Emerging Pathogens Institute, University of Florida, Gainesville, FL, United States
- 5The Pathogen and Microbiome Institute, Department of Biological Sciences, Northern Arizona University, Flagstaff, AZ, United States
Introduction: Burkholderia pseudomallei, a Gram-negative bacterium inhabiting soil and fresh water, is the causative agent of melioidosis, a formidable disease in the tropics. The emergence of antibiotic resistance and the extended duration of treatment, up to 20 weeks, have posed significant challenges in combatting melioidosis. As an alternative approach, bacteriophage therapy is being explored.
Methods: To identify the most promising bacteriophage for future therapeutic applications, we designed a screening process to address the barrier of phage specificity due to the O-antigen receptor diversity. By using two biosafe strains, Bp82 (O-antigen type A) and 576mn (O-antigen type B), to represent the major serotype A and B, we screened 145 phage samples collected from soil and water in southern Thailand.
Results: Ten of them demonstrated the ability to overcome differences in O-antigen types, yielding positive plaques formed on culture of both bacterial strains. Subsequently, we isolated 22 bacteriophages from these samples, one was adaptively mutated during the screening process, named ΦPK23V1, which had the ability to infect up to 83.3% (115/138) of tested B. pseudomallei strains, spanning both serogroups. Employing a panel of surface polysaccharide antigen mutant strains, we explored the role of capsular polysaccharide (CPS) and O-antigens as essential components for phage infection. All isolated phages were classified into the P2-like myophage group. Additionally, our research revealed a point mutation in the phage tail fiber gene (gpH), expanding the host range of ΦPK23V1, even in the absence of CPS and O-antigens.
Discussion: However, it was evident that ΦPK23V1 is a lysogenic phage, which cannot be readily applied for therapeutic use. This discovery sheds light on the receptor binding domain of P2-like bacteriophages in B. pseudomallei. Collectively, our study has identified bacteriophages with a broad host range within B. pseudomallei strains, enhancing our understanding of phage–host interactions and offering insights into the role of the phage tail fiber gene in host cell entry.
Introduction
Melioidosis is a severe and life-threatening disease caused by the bacterium Burkholderia pseudomallei (B. pseudomallei), which can be transmitted from the environment to animals and humans. It is predominantly found in the soil and water of tropical regions between latitude 20°N and 20°S, with highly endemic areas including Thailand, Malaysia, India and northern Australia (Currie et al., 2008; Limmathurotsakul et al., 2016; Chewapreecha et al., 2017). The presence of B. pseudomallei in the environments extremely complicates decontamination as it can survive in soil at least 3 meters in depth (Pongmala et al., 2022). This allows the bacterium to persist, and results in annual disease outbreaks in humans and animals in affected regions (Shaw et al., 2022). Treating melioidosis in humans is also difficult, resulting in high mortality rate of approximately 56% in Thailand (Chetchotisakd et al., 2014). The inherent resistance of B. pseudomallei to many commonly used antibiotics, coupled with its ability to survive within host cells, necessitates prolonged antibiotic therapy extending up to 20 weeks (Schweizer, 2013; Dance, 2014; Keragala et al., 2023). This extended treatment duration raises concerns over potential treatment failures due to the capability of the bacterium to develop antibiotic resistance.
The disease gained attention again in 2021 due to the multistate outbreak of a household aromatherapy product contamination with B. pseudomallei, which was sold in four states in the United States (Gee et al., 2022; Petras et al., 2022). This outbreak led to four non–travel-associated cases in Georgia, Kansas, Minnesota, and Texas, with fatalities reported in Kansas and Georgia. In addition, the CDC has reported that B. pseudomallei was isolated from environments in Mississippi in July 2022, and is now considered locally endemic to the Gulf Coast of Mississippi (CDC Health Alert Network, 2022; Petras et al., 2023; Torres, 2023). To date, B. pseudomallei and B. mallei, a clonal related species, have been classified by the Centers for Disease Control and Prevention (CDC) and other government agencies as a Tier 1 (top tier) bio-threat agent of mass destruction (Wiersinga et al., 2018). Unfortunately, an effective vaccine is unavailable for either disease. Moreover, conventional decontamination efforts in areas such as fields or animal farms have proven to be ineffective (Na-ngam et al., 2004; Pongmala et al., 2022). Furthermore, the requirement for a biosafety level (BSL)-3 laboratory when working with B. pseudomallei and B. mallei pose considerable challenges in the development of novel therapeutic agents.
In light of the limited treatment options and the pressing need to combat this highly resistant bacterium, researchers are now exploring phage therapy as a potential avenue for decontaminating affected areas and facilitating diagnosis and treatments. However, the highly specific nature of phages to their host bacteria presents a significant obstacle in finding phages capable of infecting multiple strains of B. pseudomallei, thereby hindering progress in phage research for this bacterium. To address this challenge, a meticulous screening process is essential to identify suitable bacteriophages capable of infecting the diverse strains of B. pseudomallei. The bacterium has become widely distributed in tropical regions, particularly in Southeast Asia, South Asia, northern Australia, and South America. Global phylogenetic analysis has classified B. pseudomallei into two main population groups: the Australian and the Asian groups (Chewapreecha et al., 2017). Notably, discernible variation in the O-antigen component of the lipopolysaccharide (LPS), a known phage receptor, has been observed among B. pseudomallei strains originating from these distinct geographical regions (Tuanyok et al., 2012; Limmathurotsakul et al., 2016). The majority of strains prevalent in Southeast Asia exhibit O-antigen type A, also known as serotype A, while O-antigen type B is relatively infrequent. Conversely, approximately 15% of Australian strains possess O-antigen type B (Tuanyok et al., 2012; Norris et al., 2017b). The O-antigen component of LPS has been reported to serve as a significant receptor for bacteriophages targeting B. pseudomallei, including Burkholderia phage Φ1026b, ΦE202, ΦE125, and ΦX216 (Woods et al., 2002; DeShazer, 2004; Ronning et al., 2010; Kvitko et al., 2012). Evaluation of the specificity of these phages has involved testing them with B. mallei, a genetically closely related species responsible for glanders. The experiments revealed that phage infection was impeded in B. mallei strains harboring mutations in the O-antigen biosynthesis genes, wbiE or wbiG, highlighting the essential role of the O-antigen in phage attachment (Woods et al., 2002; Ronning et al., 2010). Based on multiple studies of phage receptors in B. pseudomallei, the O-antigen component of LPS emerged as the most significant receptor for phage attachment. Therefore, genetic differences between B. pseudomallei serotypes may limit the ability of a single phage to infect both major serotypes.
To identify a board-host range phage for future applications, we conducted a screening process to explore potential phage receptors beyond the O-antigen receptor. The objective was to identify a phage capable of infecting most serotypes of B. pseudomallei strains from different geographic locations. This initiative sought to broaden our understanding of phage–host interactions and potentially expand board-host range bacteriophage isolates available for future studies aimed at combating B. pseudomallei infections.
Materials and methods
Bacterial strains and growth conditions
Burkholderia pseudomallei strain CAM5, a wild-type strain with O-antigen type A and sequence type (ST) 3, a common genotype in southern Thailand, was used as the host strain for phage isolation from soil and water samples as described in the following section. This strain was originally isolated from the lung of a deceased camel during a melioidosis outbreak in a local zoo in Songkhla Province. Under laboratory conditions, strain CAM5 exhibits normal growth in LB medium.
To facilitate research on B. pseudomallei in laboratories with a biosafety level 2 (BSL-2) designation, two virulence-attenuated biosafe strains of B. pseudomallei, Bp82 (serotype A) (Propst et al., 2010) and 576mn (serotype B) (Norris et al., 2017a), were used. The attenuated B. pseudomallei Bp82, 576mn, and their isogenic derivatives Bp82 △wcbI-R (CPS knocked out) from Norris et al., 2017b, Bp82 △wbiD (O-antigen knocked out), Bp82 △wcbI-R △wbiD (double CPS and O-antigen knocked out), and 576mn △wcbB (CPS knocked out) were grown on Luria Bertani (LB) medium supplemented with 80 µg/mL adenine with aeration at 37°C. The creation of these mutant strains and their phenotypic verifications are described in Supplementary Data Sheet 1. Procedures with wildtype B. pseudomallei and B. mallei strains were performed in the select agent approved BSL-3 facilities at the University of Florida. All strains of wildtype B. pseudomallei and the bacterial strains used in this study included B. thailandensis TXDOH, B. thailandensis E555, B. thailandensis E264, B. oklahomensis, B. vietnamensis, B. territorii, B. latens-like spp., B. cenocepacia-like spp., B. cenocepacia, B. multivarans were grown on LB agar or in LB broth. B. mallei strains were grown with LB agar or LB broth supplemented with 4% glycerol. Vibrio vulnificus MLT403 was a gift from Dr. Paul A. Gulig, Department of Molecular Genetics and Microbiology, College of Medicine, University of Florida and used as a host for ΦCK-2 (Cerveny et al., 2002). V. vulnificus MLT403 was cultured in LB supplemented with 0.85% (w/v) NaCl, at 30°C. All bacterial strains used in this study are summarized in Supplementary Table 1.
Phage isolation
A total of 145 soil and water samples were collected from various locations in Songkhla Province and Nakhon Si Thammarat Province, the two largest provinces of southern Thailand. The phage isolation technique utilized in this study was adapted from Clokie and colleagues (Clokie and Kropinski, 2009). In brief, stagnant surface water or a clear soil solution (prepared by mixing 10 g of soil with 10 mL of sterile distilled water and allowing it to precipitate overnight at room temperature) was subjected to centrifugation at 4,000 × g for 15 minutes to eliminate particulate matter. Subsequently, 8 mL of the supernatant was combined with 1 mL of an overnight culture of B. pseudomallei CAM5 and 1 mL of a 10X concentrated LB broth. The resultant solution was then incubated at 37°C for a minimum of 5 hours. Following incubation, the culture was centrifuged at 9,000 × g for 15 minutes and filtered through a 0.22µm Polyvinyl Difluoride (PVDF) syringe filter. To ensure sterility, 10% of the filtered solution was cultured on LB agar at 37°C for 48 hours.
A spot test technique was subsequently employed by applying the filtered supernatant from the previous step onto a double-layer soft agar of the host bacterial strain to assess the presence of phages. Specifically, 100 µL of B. pseudomallei CAM5 culture, harvested at an OD600 of 0.3, was mixed with 4 mL of 0.35% (w/v) molten LB agar (also known as LB soft agar) and promptly poured onto an LB agar plate. The plate was rotated swiftly and let to solidify for 5 minutes. Following this, a 10 µL volume of the phage solution was spotted onto the plate. After allowing the plate to dry for 15 minutes, it was incubated overnight at 37°C. The presence of bacteriophages was confirmed by a large clear zone, or multiple plaques formed on the bacterial lawn (Daubie et al., 2022). These phage isolation steps were conducted in a BSL-2 enhanced laboratory registered with Thailand’s the Ministry of Public Health – Department of Medical Science for use with B. pseudomallei research at Prince of Songkla University. Once the presence of phages was confirmed, the filtered phage solution stocks were shipped to our laboratory at the University of Florida for further analysis. These steps are illustrated in Figure 1.
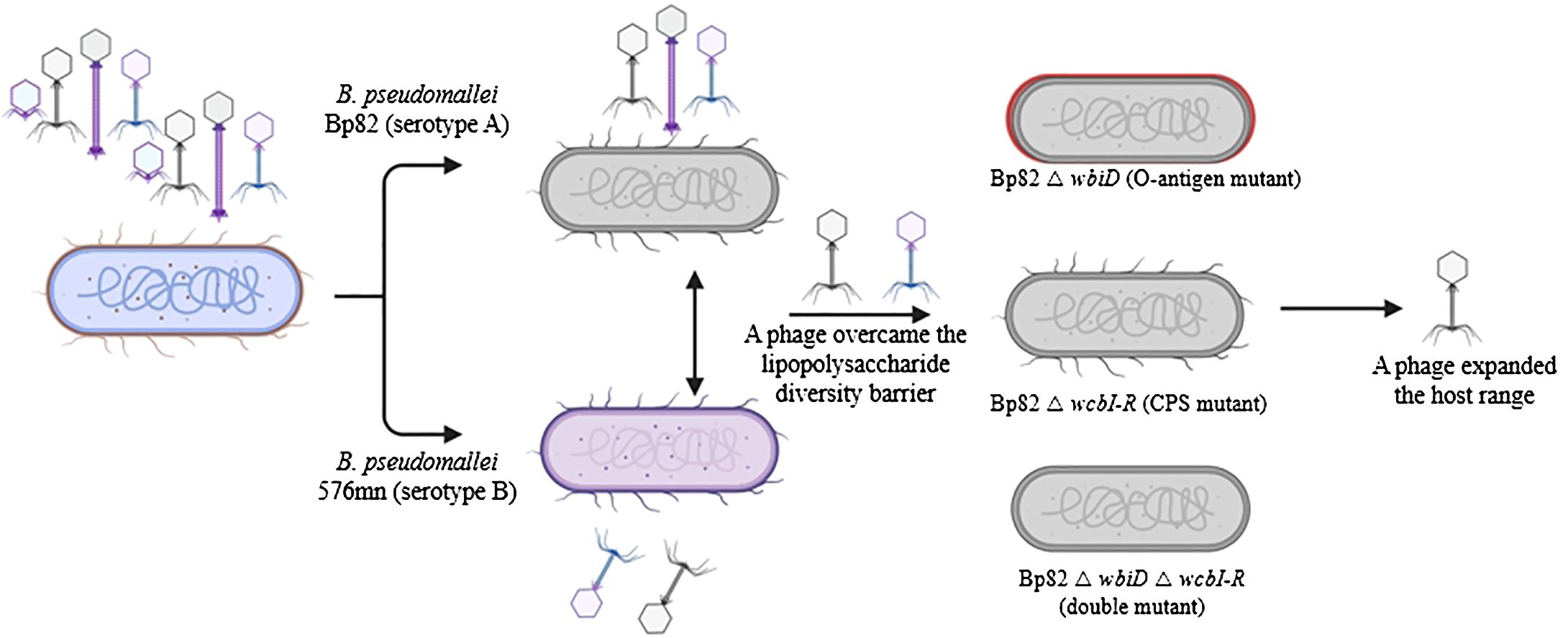
Figure 1. An overview of the phage screening process designed to identify phages capable of overcoming lipopolysaccharide (LPS) variations within the predominant Burkholderia pseudomallei population. The phage library from Thailand was cultivated using attenuated B. pseudomallei strains Bp82 (representing serotype A) and 576mn (representing serotype B). Subsequently, phages exhibiting the ability to infect both serotype A and B were subjected to further testing using capsule polysaccharide mutant strains to determine their potential receptor targets.
Phage screening and purification
Each log-phase bacterial culture of B. pseudomallei Bp82 (serotype A) and 576mn (serotype B), was mixed with 10 µL of the corresponding phage sample. Subsequently, 4 mL of LB soft agar was mixed into the mixture before being poured on top of an LB agar plate. These plates were then incubated overnight at 37°C to facilitate the screening of plaque formation. Samples that exhibited plaque formation in both bacterial strains underwent further screening. Plaques formed on the bacterial lawn were purified through three consecutive rounds of isolation, with a sterile pipet tip being used to select and transfer each plaque. Each isolated plaque was resuspended in 1 mL of storage media (SM) buffer, composed of Tris-HCl (10 mM), NaCl (100 mM), MgSO4 (50 mM), and gelatin (0.01%), adjusted to a pH of 7.5. This step was conducted with a minimum of three repetitions to purify the phage strain and ensure its consistency in plaque morphology. Importantly, throughout this process, all phages were propagated using the same host to prevent potential contamination by temperate phages of host origin. We noted that Bp 82 possessed three different temperate phages but could not form plaques on its own lysogenic host (Khrongsee et al., 2024).
Phage propagation and quantification
The plate lysate technique was used for phage propagation. Initially, 100 µL of the filtered phage solution in SM medium was combined with 100 µL of log-phase bacterial culture at OD600 of 0.3, allowing 15 minutes for phage absorption into the bacteria. Subsequently, a double-layer plaque assay was prepared and incubated overnight at 37°C. To harvest the phages, 10 mL of SM buffer were poured onto the plate, followed by careful removal of the soft agar. The resulting suspension was transferred to a new tube and vigorously vortexed. Afterward, the phage mixture underwent centrifugation at 9,000 × g for 10 minutes, followed by filtration through a 0.22 µm PVDF syringe filter. For phage quantification, ten-fold serial dilutions were prepared in a 96-well plate using SM buffer as the diluent. A 5 µL volume of each dilution was spotted onto bacterial soft agar and incubated overnight at 37°C as described previously. This quantification process was conducted in triplicate, and plaque numbers were subsequently counted.
Host range determination
The spot test technique was used to evaluate host susceptibility for each phage strain. This technique was performed by dropping a volume of 10 µL of 1×107 pfu/mL of each bacteriophage solution on top of the bacterial soft agar as described above. The phage was recorded as positive if clear or turbid plaques were observed. The result was negative if the phage failed to form plaques on the lawn of bacteria. Phage specificity was tested with a panel of B. pseudomallei strains (N=138) from different sources (e.g., humans, animals, and environment) and an exclusive strain panel of other genetically closely related Burkholderia species strains (N =17). All bacterial strains used in this study are listed in Supplementary Table 1. For the growth curve analysis of the host challenge test, a 100 µL volume of phage at a concentration of 106 pfu/mL was combined with 100 µL of log-phase bacteria of approximately 108 cfu/mL in 500 µL of LB broth, all within a 48-well plate. The plate was then read using a Biotek Synergy HDTM (Agilent Technologies Inc., Santa Clara, CA, USA) with triplicate samples for each measurement. To identify phage receptors, two common surface antigens including O-antigens and CPS were tested. B. pseudomallei Bp82 △wbiD (O-antigen knocked out), Bp82 △wcbI-R (CPS knocked out), Bp82 △wcbI-R wbiD (O-antigen and CPS knocked out), and 576mn △wcbB (CPS knocked out) generated in our previous study (Norris et al., 2017a) were spot-tested with phages as described above. Phage adsorption was performed by mixing the phage ~105 pfu with ~108 bacterial cells in 37°C for 15 minutes, then plated following the techniques as described previously (Le et al., 2013).
Transmission electron microscopy
A volume of 100 µL containing at least 108 pfu/mL of phages was diluted with 900 µL of SM buffer. Subsequently, 10 µL of the phage solution was deposited onto a glow-discharged, carbon-stabilized copper grid and allowed to sit for 5 minutes. The grid was then briefly stained with 1% (w/v) uranyl acetate solution for 1 minute. The prepared grids were visualized using a TEM (FEI Tecnai G2 Spirit Twin TEM; Nanoscience Initiative; USA) at the Interdisciplinary Center for Biotechnology Research, University of Florida. Phage size was determined based on the average measurements from five independent phages using DigitalMicrograph® software (Gatan Inc., CA, USA).
Phage DNA extraction
Phage DNA extraction was performed as previously described (Kvitko et al., 2012) with minor modifications as described below. Briefly, 100 mL phage suspension was filtered with a 0.22µm syringe filter to remove possible remaining bacteria. The mixture was then centrifuged at 25,000 × g for 60 minutes, the supernatant was discarded, and a yellow spot phage pellet was observed. One mL of LB broth was added to resuspend the pellet. To extract the phage DNA, 500 µL of concentrated lysate was mixed with 5 μL DNase I (1000 U/mL) and 5 μL RNase A (20 mg/mL) were added and incubated at 37°C for 60 minutes. The phage capsid destabilization and the DNase I inactivation was performed by adding 50 μL of 10% SDS, 40 μL of 0.5 M EDTA (pH 8.0), and 2.5 µL of proteinase K (20 mg/mL). Next, the mixture was incubated at 55°C for 60 minutes. DNA extraction was performed by adding an equal volume of phenol-chloroform-isoamyl alcohol (PCI) in a ratio of 25:24:1, followed by vertexing and centrifugation at 13,000 × g for 5 minutes. The aqueous phase was transferred to a new tube. The DNA extraction was performed twice with PCI and once with chloroform. The DNA was precipitated by adding an equal volume of absolute ethanol containing 50 µL of 3 M sodium acetate pH 5.2 at −20°C overnight. The DNA pellet was retained from centrifugation at 13,000 × g for 10 minutes. The pellet was washed twice with 1 mL of 70% ethanol and air-dried in a biosafety cabinet. Next, the pellet was resuspended in 50 µL of DNase free sterile water.
Phage DNA sequencing and genome comparison
A DNA sequencing library was prepared using a NEBNext® Ultra ™ II DNA Library Prep kit according to the manufacturer’s instructions, and sequencing was performed on an Illumina MiSeq sequencer using a v3 chemistry 600-cycle kit. De novo assembly of the paired-end reads was performed using SPAdes 3.13.0 (Bankevich et al., 2012) using default parameters. The assembled genome was annotated using Genome Annotation services via the Bacterial and Viral Bioinformatics Resource Center (BV-BRC; http://bv-brc.org) (Olson et al., 2023). Comparison of conserved genomic regions in the phage and prophage of Bp K96243 (chromosome II) was performed using, the Artemis Comparison Tool (Carver et al., 2005) and Easyfig 2.2.2 OSX (Sullivan et al., 2011).
Gene comparison and protein structure prediction
The gpH gene sequences of Burkholderia P2-like phages were retrieved from GenBank and subsequently aligned using Multalin (Corpet, 1988) and SnapGene viewer 7.0.2 software. To model the structure of the GpH monomer, we employed SWISS-MODEL (Waterhouse et al., 2018) utilizing the phage tail fiber protein A0A7U4SUZ9_9BURK from Burkholderia sp. Bp5365 as a template. This allowed us to compare the structural differences between GpH protein and those from closely related species. Further insights into the trimeric structure of the GpH protein tip were obtained using the AlphaFold Colab notebook (Evans et al., 2021; Jumper et al., 2021) and visually represented using ChimeraX 1.6.1 software (Pettersen et al., 2021).
Statistical analysis
The adsorption data were analyzed using GraphPad Prism version 9 (GraphPad Software Inc., San Diego, CA, USA). A two-way ANOVA was employed to assess the statistical differences between groups, followed by Tukey’s multiple comparison tests to evaluate the significance of differences between means. Differences were deemed statistically significant at a threshold of p < 0.05.
Results
Phage selection against the O-antigen diversity of B. pseudomallei
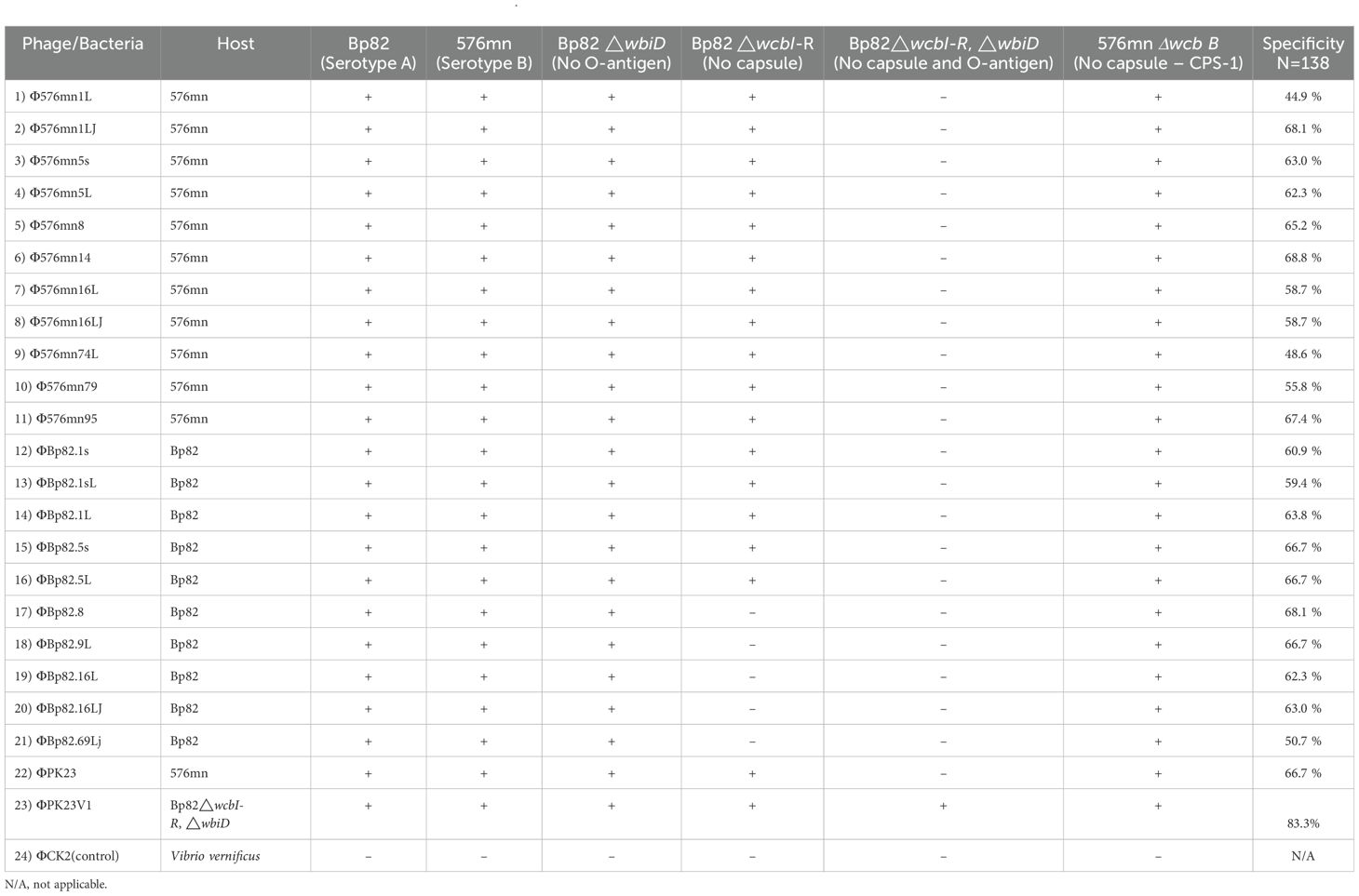
In order to identify phages capable of overcoming the diversity of the O-antigen moiety of B. pseudomallei LPS, we tested 145 phage samples collected from Thailand against the two different serotypic host strains, Bp82 and 576mn. We have found that only 10 out of 145 phage samples were able to form plaques on culture of both B. pseudomallei strains. The phages were further purified from each host strain and tested with one another to ensure that the phage can infect both. After rounds of phage purification, we isolated 22 phage strains based on the differences of their plaque morphology. These phages are listed in Table 1.
Host range and receptor determination
We further conducted a comprehensive assessment of the host range and receptor specificity. Initially, these phages were tested against attenuated B. pseudomallei strains, specifically 576mn and Bp82. As expected, our findings confirmed that these phages indeed had the ability to effectively infect both Bp82 and 576mn strains suggesting the phage could overcome the O-antigen differences. Subsequently, we expanded our investigation to a diverse panel of B. pseudomallei strains and other closely related species within the Burkholderia genus, including representatives from both the B. pseudomallei complex and the B. cepacia complex. While none of the tested phages exhibited infectivity towards species outside the Burkholderia pseudomallei, two distinct exceptions were identified: B. mallei and atypical B. thailandensis TXDOH (Supplementary Table 1).
To gain further insight into phage–host interactions, we conducted additional investigations regarding their infectivity against B. pseudomallei strains with distinct genetic modifications of CPS mutants, O-antigen mutants, and both CPS and O-antigen mutant strains. All 22 phage strains were able to form plaques on the culture lawn of Bp82 △wbiD (O-antigen type A knockout), and demonstrated the ability to maintain infectivity levels comparable to that in Bp82. Conversely, our observations revealed a diminished capacity of most phages to infect the Bp82 △wcbI-R strain (CPS knockout), while displaying complete inability to infect the Bp82 double △wcbI-R△wbiD mutant strain. A comprehensive list of these strains is provided in Table 1. One particularly intriguing discovery was the identification of a mutant phage designated as ΦPK23V1, originating from ΦPK23. This variant exhibited a remarkable ability to infect the Bp82 double △wcbI-R△wbiD mutant strain (Table 1). It is important to note that these phage strains demonstrated specificity to B. pseudomallei between 44.3 to 68.3% (N=142), while ΦPK23V1 was able to infect up to 83.3% (115/138) of the tested B. pseudomallei strains. In the 15-minute adsorption test, ΦPK23V1 exhibited better adsorption compared to the wild type phage ΦPK23, while both phages demonstrated a significant reduction in adsorption when encountering the Bp82 CPS mutant strain. However, there was no significant difference in phage adsorption observed in Bp82 and its double mutant, suggesting a low binding affinity of the phages to the new receptor within 15 minutes. This observation aligns with the results from the growth curve of the host challenge test, where ΦPK23V1 displayed better suppression of the bacterial growth compared to the wild type phage. Importantly, only ΦPK23V1 exhibited the capability to infect the Bp82 double mutant strain, although it took approximately 7 hours for the suppression to become evident (see Figure 2).
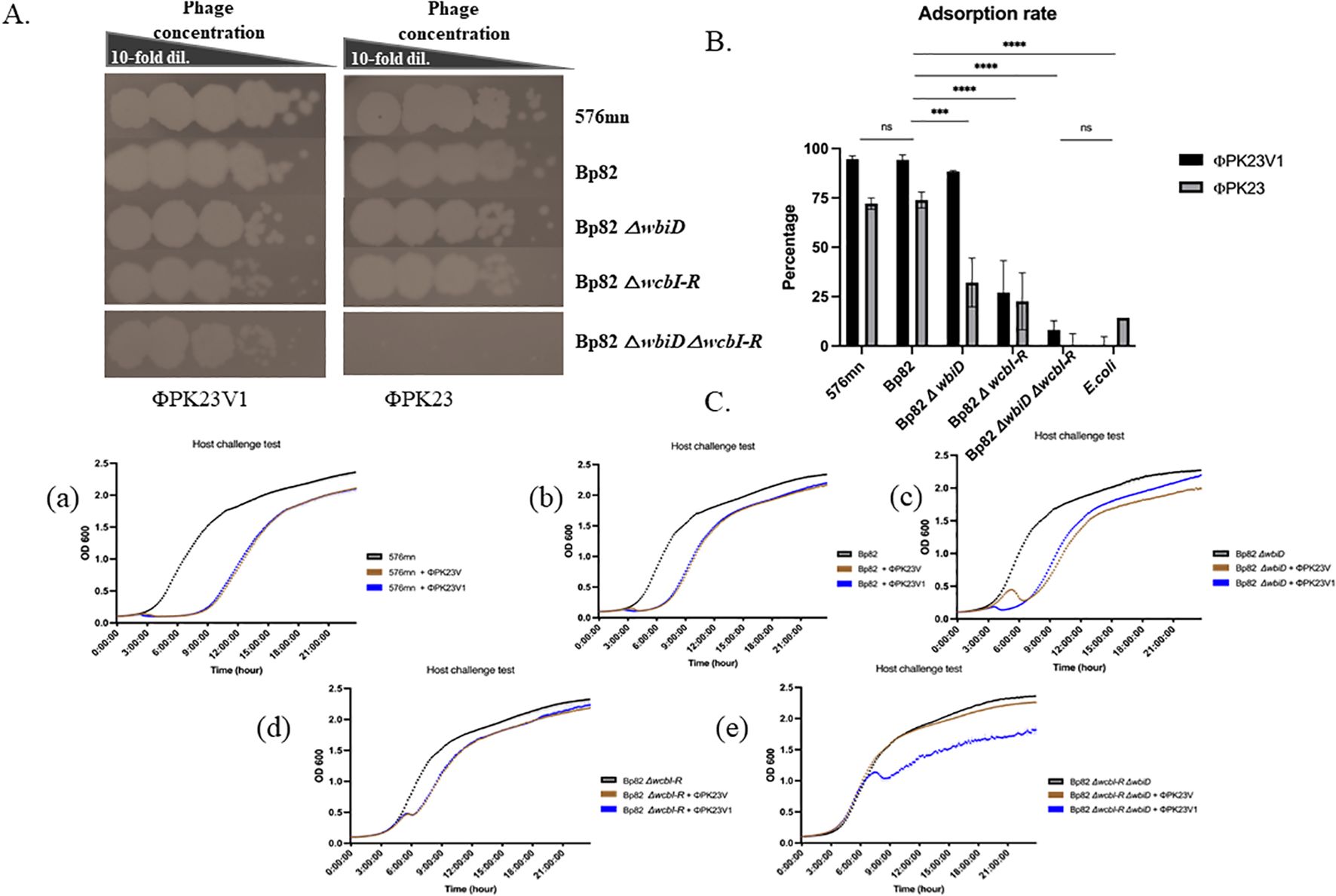
Figure 2. Growth and adsorption of ΦPK23 and ΦPK23V1; (A) spot tests reveal interactions with various surface antigen mutant hosts; (B) the percentage of phage adsorption capacity within 15 minutes when exposed to different surface antigen mutant hosts; and (C), the outcomes of phage–host challenge experiments using diverse surface antigen mutant strains of B. pseudomallei. In panel (C), these experiments allow for a comparison of the killing activity of ΦPK23 (depicted in brown) and ΦPK23V1 phage (shown in blue) at a multiplicity of infection (MOI) of 0.01. Graphs, a) 576mn, representing O-antigen type B; b) Bp82, representing O-antigen type A; c) Bp82 O-antigen mutant strain; d) Bp82 capsule polysaccharide (CPS) mutant strain; and e) Bp82 O-antigen and CPS mutant strains. This figure provides crucial insights into the growth, adsorption, and host interactions of ΦPK23 and ΦPK23V1 across various mutant strains, shedding light on their distinct capabilities and potential applications. ns (non-significant), P > 0.05, ***, P ≤ 0.001, ****, P ≤ 0.0001.
Phage morphology
The phages: Φ576mn8, ΦPK23, Φ576mn95, and ΦBp82.16L were selected for TEM examination. TEM revealed that all phage strains had an icosahedral head, average 50 nm in diameter, with a 150 nm long tail encased in with a sheath. Based on their morphology, we have classified the phages as members of myophages (Figure 3).
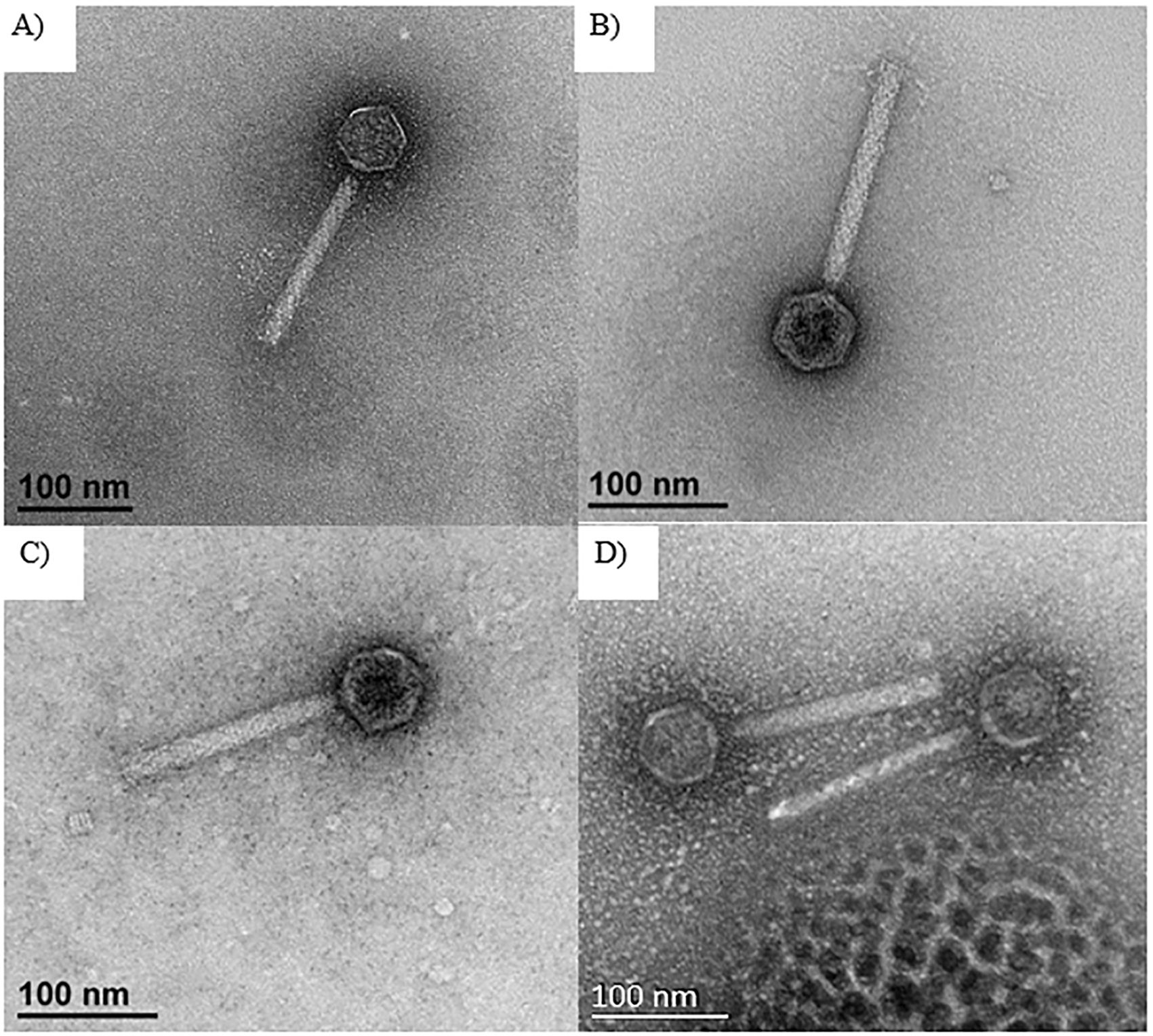
Figure 3. TEM images of selected phages including: (A) Φ576mn8, (B) ΦPK23, (C) Φ576mn95, and (D) ΦBp82.16L. The scale bar in each image represents a length of 100 nanometers, with these images captured at a magnification of ×40,000. All of these phages exhibit an icosahedral head with a long tail encased in a contractile sheath.
Sequencing and genome analysis
The sequencing analysis unveiled that the genome of ΦPK23 spans a linear genome of 35,343 base pairs and encodes 49 predicted proteins (GenBank accession# MZ375754.1). Its cohesive (Cos) sequence was identified as 5’- GGCGAGGAGGGGGAC-3’ based on the identification using PhageTerm software (Garneau et al., 2017) and Li’s method (Li et al., 2014). This phage belongs to the Peduoviridae family, that includes the well-studied Enterobacteria phage P2 (Accession# NC_001895.1), which primarily infects E. coli. Notably, the ΦPK23 genome encompasses an integrase gene and an attachment site (attP). Furthermore, our investigation identified a remarkable similarity between the phage genome and one of the prophages located within genomic island 15 (GI15) on chromosome 2 of B. pseudomallei K96243 (see Figure 4A). When aligning the phage and prophage genomes, it became evident that the genome orientation was disrupted at attP and subsequently reconfigured within the attL and attR regions of the bacterial genome. This reconfiguration resulted in the emergence of short-directed repeat sequences of the attachment sequences (attL and attR). It is essential to note that Bp82 and 576mn, in contrast, do not contain any prophage presence within the GI15 location, and their attP sequences exhibit two mismatches (as depicted in Figure 4B). These findings underscore the complex dynamics of lysogenic phage integration and genomic rearrangement in B. pseudomallei. A constructed phylogenetic tree, based on whole-genome sequences of Burkholderia myophages, delineates a distinct clade closely related to other P2 phages, divergent from Burkholderia siphophages (Figure 5). P2-like myophages and the siphophages are common prophages within B. pseudomallei.
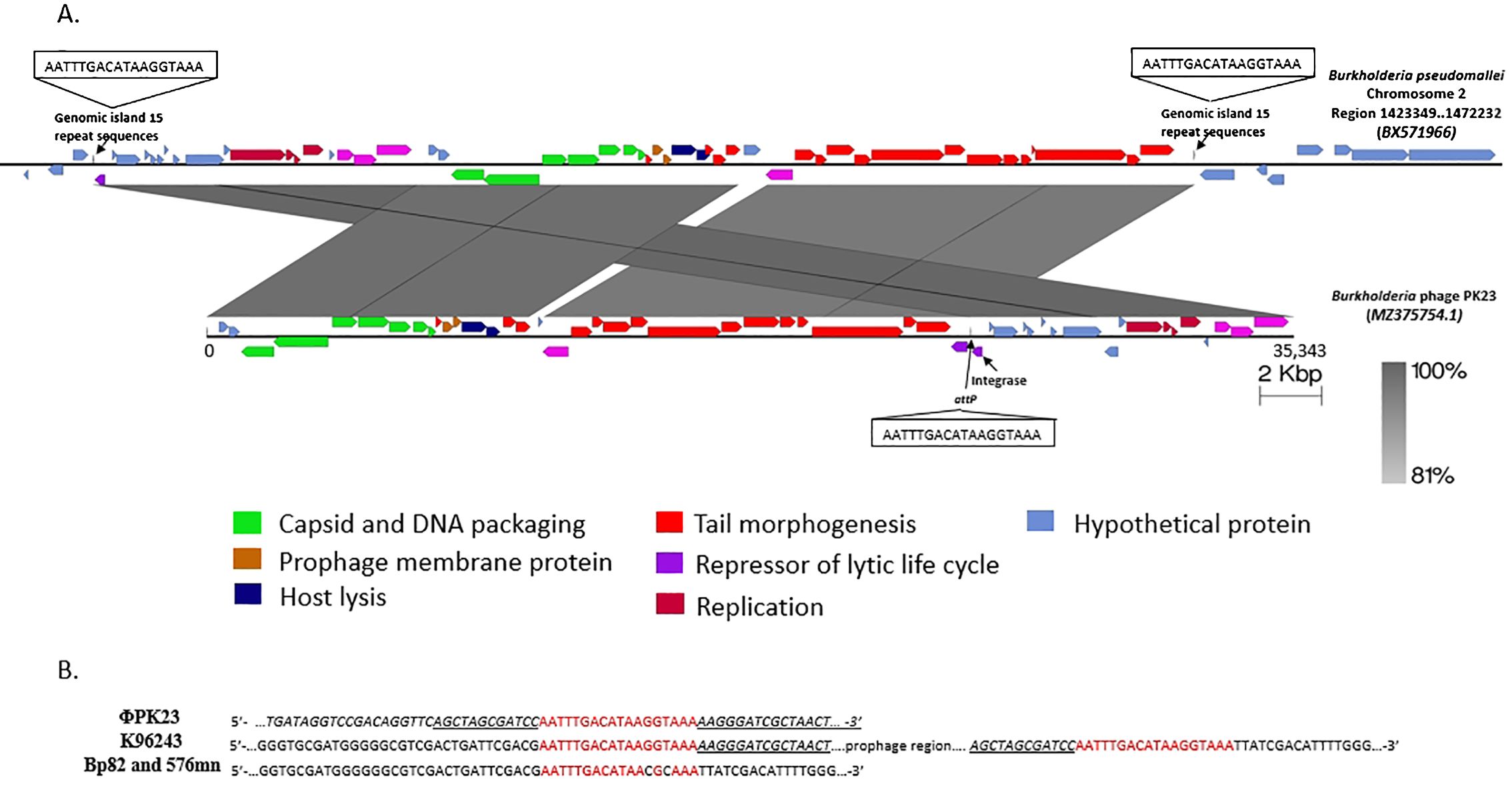
Figure 4. The ΦPK23 genome and a potential integration site. (A) is the genomic representation of Burkholderia phage ΦPK23 and its alignment with one of the prophages situated within genomic island 15 (GI15) on chromosome 2 of B. pseudomallei K96243; distinct gene functions within the phage genome are color-coded. (B) shows the 18 bp attP sequence of ΦPK23, the identical genomic island repeats, attL and attR, in B. pseudomallei K96243, and the two mismatched bases of attB sequence in Bp82 and 576mn.
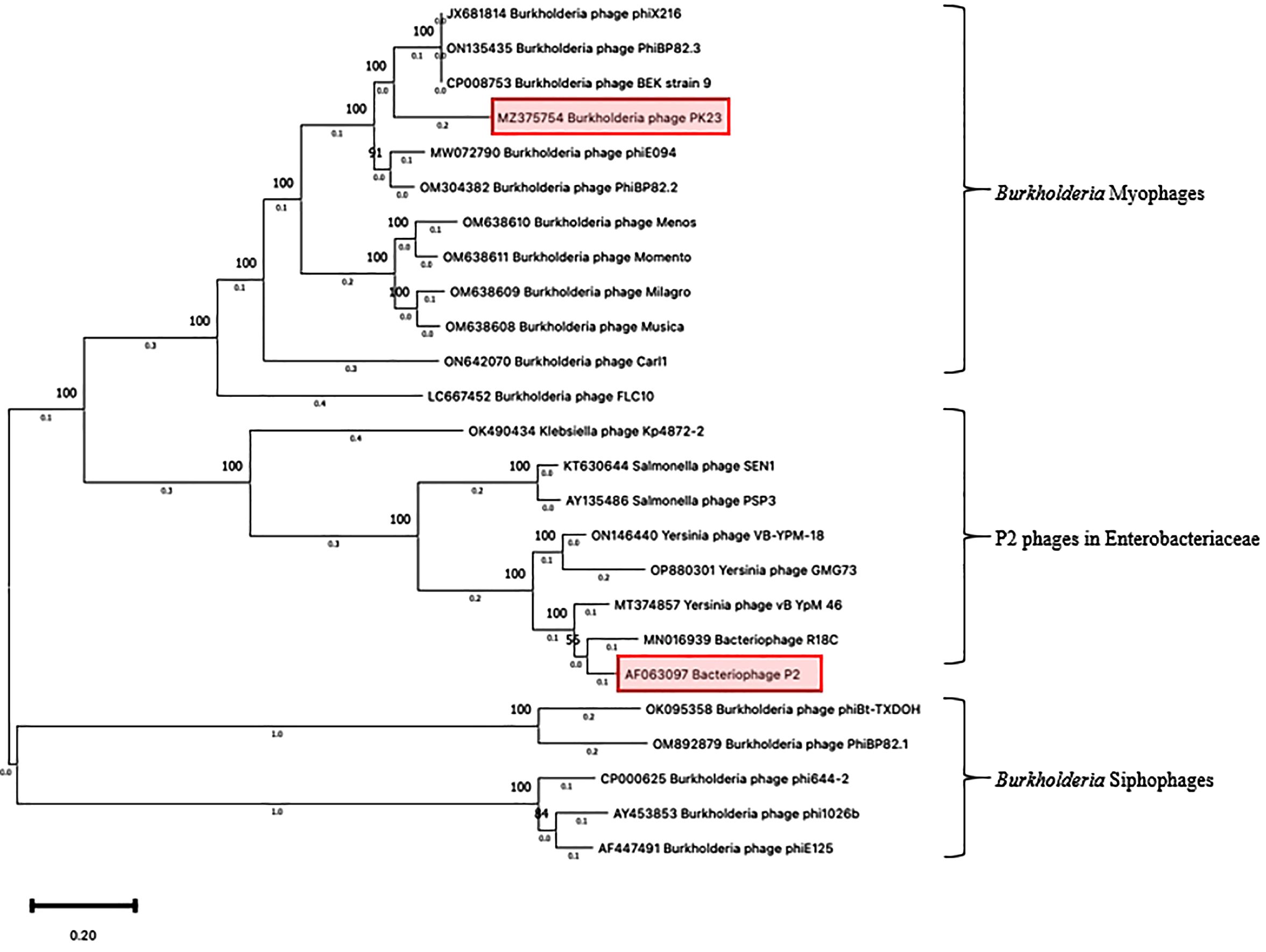
Figure 5. Phylogenetic tree of Burkholderia phage ΦPK23. A phylogenetic tree based on the alignment of the whole genome sequences of Burkholderia phages was generated by the Maximum Likelihood method using the Tamura-Nei model within MEGA 11 (The Pennsylvania State University – Institute of Molecular Evolutionary Genetics, PA, USA). The numbers above nodes represent the bootstrap values (500 replicates).
ΦPK23V1 shares an identical sequence with ΦPK23, except for a single nucleotide mutation at position 2148 within the tail fiber protein gene (gpH), where cytosine is substituted with adenine. This genetic alteration results in a missense mutation in the protein, converting asparagine (N) at amino acid position 716 to lysine (K), as depicted in Figure 6. Utilizing SWISS-MODEL, we conducted a structural analysis of the ΦPK23 GpH, revealing its most probable structural similarity with the phage tail fiber protein A0A7U4SUZ9_9BURK found in Burkholderia sp. strain Bp5365 (Supplementary Figure 1). Interestingly, this structural similarity is consistent across the entire protein, except for a distinct region encompassing amino acid positions 630–750. This region corresponds to the tip of the tail fiber protein, potentially serving as the receptor binding domain. The variations observed in this particular region between these two phages indicate potential differences in their host specificity, implying binding to distinct host species.
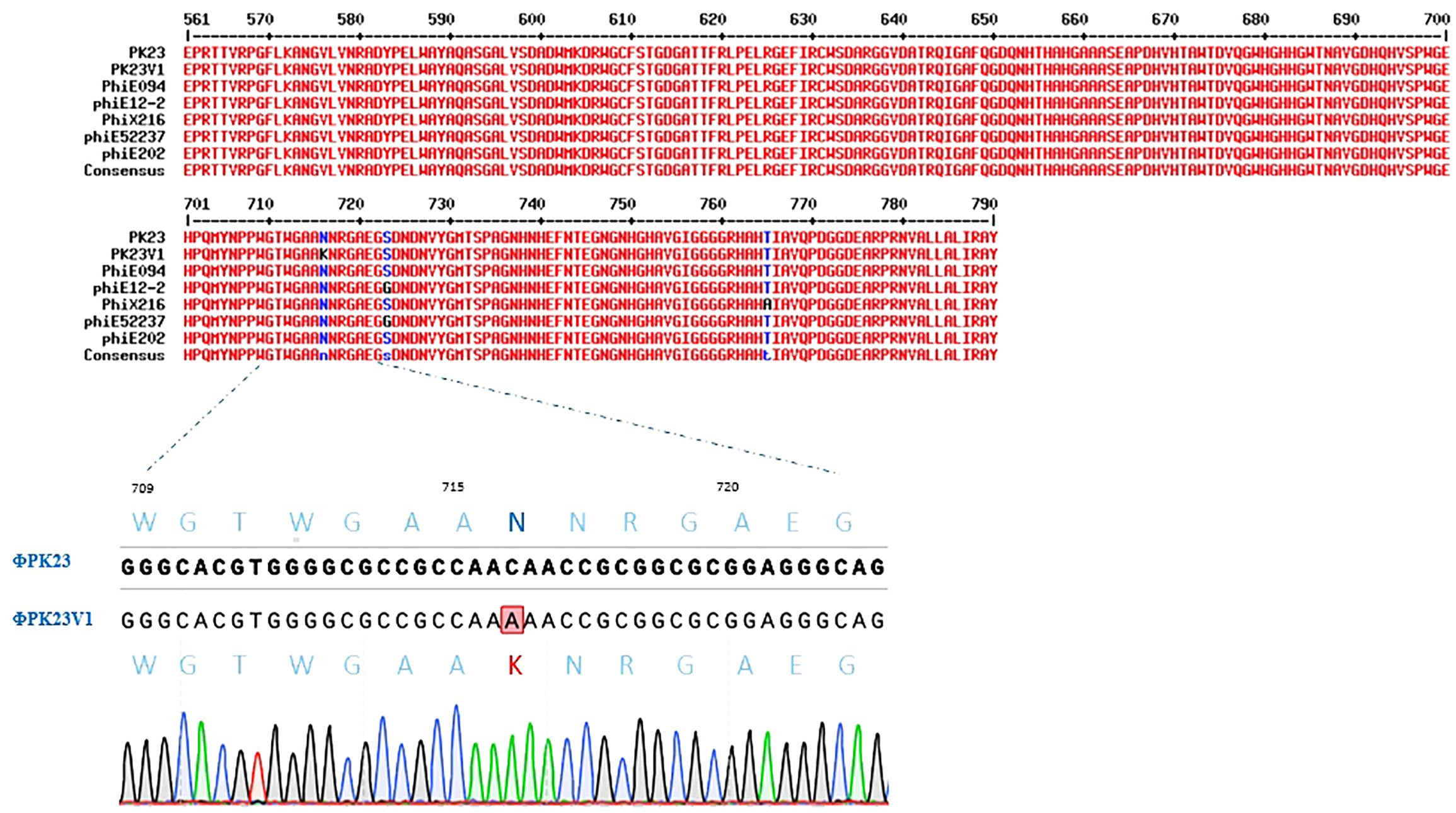
Figure 6. Protein sequence alignment of the P2-like phages known to infect B. pseudomallei, focusing on the amino acid positions spanning 561–790 at the end of the tail fiber protein. The alignment highlights conserved regions shared among all the phages, with a noteworthy exception being the unique lysine substitution at amino acid position 716 in ΦPK23V1. This mutation was confirmed by PCR and Sanger sequencing.
To delve deeper into the implications of the host interactions stemming from the missense mutation in gpH gene we constructed an Alphafold model (Figure 7). This model elucidated the trimeric ribbon structure spanning amino acid positions 561 to the end of the gene at position 790, offering insights into the tip structure of GpH protein. Notably, a majority of amino acids within this region exhibit a charge range from neutral to negative. This negative charge distribution is abundant in the inner core of the tip base, a critical site for binding to the bacterial surface receptor. Remarkably, our investigation also observed an amino acid substitution mutation in gpH with lysine substitutions located on the side of the tip base, resulting in an expansion of its size. Importantly, lysine residues bear positive charges, thereby enhancing the overall positive charge of the protein. These findings shed light on the structural and charge characteristics of the GpH tip, suggesting the potential impact of the lysine substitutions on expanding host receptor interactions.
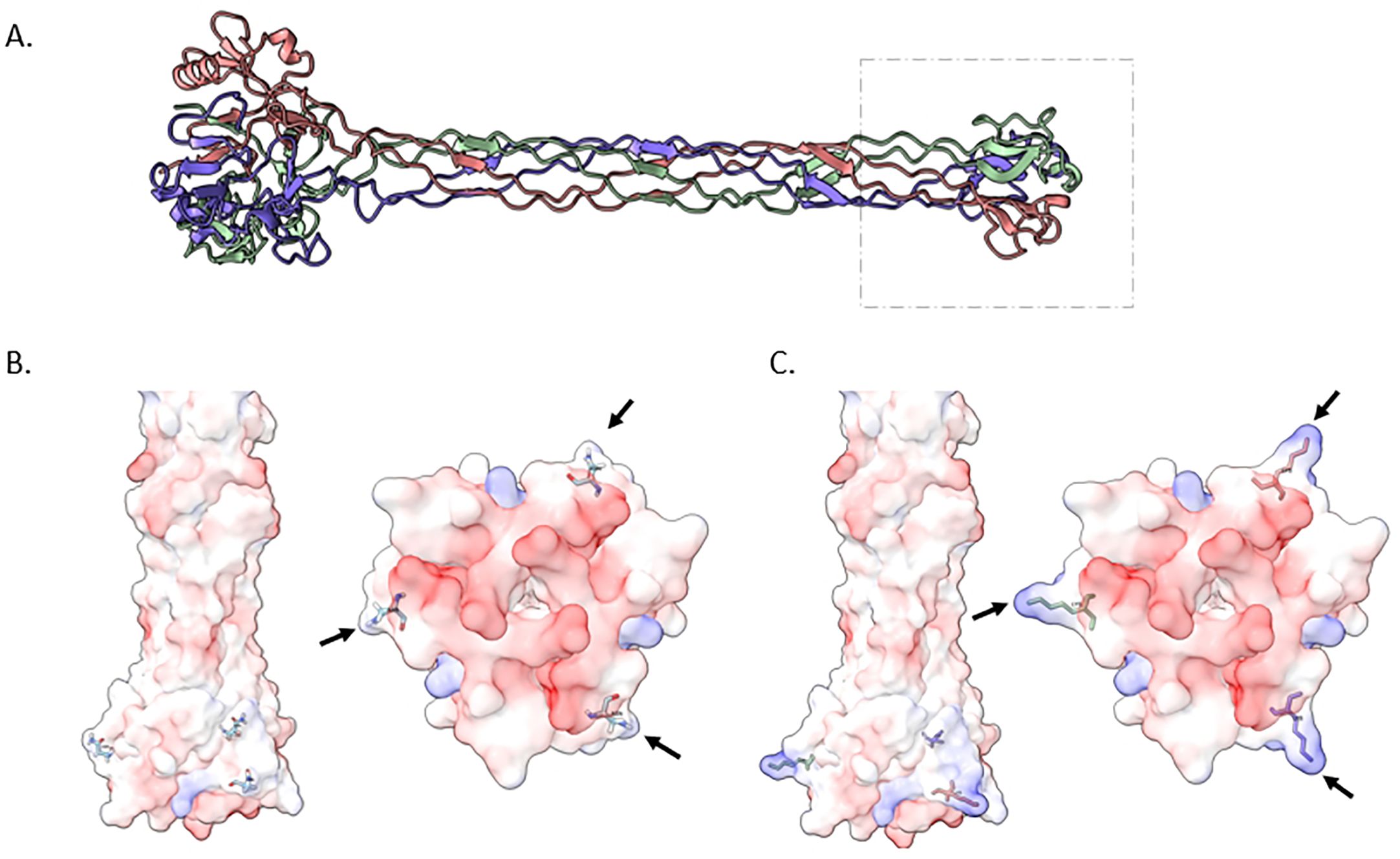
Figure 7. Structural insights into the ΦPK23V1 Tail Fiber Protein (GpH). (A) depicts a trimeric ribbon model constructed from amino acid positions 561 to the end of the protein at position 790. This visualization highlights the tip of the protein, suggesting its potential as the receptor binding domain. (B) offers a 3D view of the wildtype GpH structure, specifically focusing on the tip region. This view is presented from both lateral (left) and ventral (right) perspectives, providing insights into the electrostatic charge distribution of the protein. The majority of amino acids in this region exhibit a range from neutral charge (white) to negative charge (red). This negative charge distribution is especially prominent in the inner core of the tip base, which plays a critical role in binding to the bacterial surface receptor. (C) illustrates a mutant GpH featuring lysine substitutions. These lysine residues are positioned on the side of the tip base, expanding its size. Notably, lysine residues are positively charged, resulting in an overall increase in the positive charge of the protein.
Discussion
We designed a screening procedure to obtain the optimized phages that can infect both B. pseudomallei serotypes. In this study, we applied the B. pseudomallei pangenome analysis studies from Chewapreecha and colleagues revealing that the major populations of B. pseudomallei were Australian and Asian groups (Chewapreecha et al., 2017). In these two populations, we previously established that the surface O-antigens conferred two major serotypes, and that up to around 98% of Southeast Asian strains were serotype A. In contrast, approximately 15% of serotype B was found among Australian strains (Tuanyok et al., 2012). It is important to highlight that most of the publications had previously indicated that B. pseudomallei phages relied on the O-antigen moiety of the lipopolysaccharide (LPS) as a receptor to enter bacterial cell (Woods et al., 2002; DeShazer, 2004; Ronning et al., 2010; Kvitko et al., 2012). Thus, the O-antigen diversity among B. pseudomallei strains could potentially impact phage entry. In this case, we hypothesized that O-antigen differences would limit phage specificity, which could be the obstacle for phage to infect the major populations of B. pseudomallei. To investigate this, we used Bp82 and 576mn, two biosafe strains, to screen phages that overcome the serotypic diversity of B. pseudomallei. These attenuated strains were also suitable for work with phages in a BSL-2 laboratory.
We were able to isolate 23 bacteriophages including the host-adapted mutant ΦPK23V1 which was highly specific to B. pseudomallei. These phages could not infect other non-pathogenic Burkholderia species, even B. thailandensis E555, an atypical strain, which expresses O-antigen and CPS antigen identical to those of B. pseudomallei. However, these phages can infect B. thailandensis TXDOH, an atypical pathogenic strain isolated from a child who developed pneumonia after being submerged in water from a car accident (Glass et al., 2006; Gee et al., 2018). This atypical B. thailandensis strain produces O-antigen similar to that of B. mallei and CPS similar to both B. mallei and B. pseudomallei (Glass et al., 2006; Tuanyok et al., 2012),whereas typical B. thailandensis strains lack the pathogenic CPS. However, we still do not know what receptor these phages used to enter this atypical B. thailandensis strain. As expected, the pathogenic B. mallei strains were infected by these phages similar to the other B. pseudomallei phages (Woods et al., 2002; Kvitko et al., 2012). B. mallei is a clone of B. pseudomallei that has reduced its genome to become a host-adapted pathogen. B. mallei has lost all genomic islands including prophages (Tuanyok et al., 2008) suggesting that B. mallei may have also lost immunity for preventing phage infection. B. mallei is commonly used as a susceptible host to propagate phages from B. pseudomallei and B. thailandensis (Tumapa et al., 2008; Kvitko et al., 2012). Most of our phage strains infected up to 65% of B. pseudomallei strains from both serotypes tested in this study, while ΦPK23V1 had the highest specificity up to 83.3% of the tested B. pseudomallei strains, which was higher than the broad host range ΦST79 and ΦX216 reported previously (Yordpratum et al., 2011; Kvitko et al., 2012). We have learned that some phages capable of infecting the O-antigen type A strains might recognize the O-antigen type B strains as well, while they failed to form a plaque since their genomic DNA was digested by the restriction modification system after injection, or it was just a consequence of abortive infection.
We selected ΦPK23V1, Φ576mn8, Φ576mn95, and ΦBp82.16L to be examined by TEM. The TEM revealed that these phages possessed long tails with contractile sheaths, the virion morphology expected for structure of the myophages (Figure 3). This morphotype is prevalent among B. pseudomallei phages, including Φ52237, ΦE12-2, ΦE202, ΦK96243, ΦX216, ΦE094, ΦvB_HM795, ΦvB_HM387, ΦBP82.2, and ΦBP82.3; all of which are closely related to well-characterized P2 myophages (Holden et al., 2004; Ronning et al., 2010; Kvitko et al., 2012; Muangsombut et al., 2021; Withatanung et al., 2024; Khrongsee et al., 2024). P2-like myophages are commonly identified as prophages within Gram-negative bacteria (Christie and Calendar, 2016). Other prophages in B. pseudomallei, include the siphophages Φ664-2, Φ1026b, ΦE125, ΦBP82.1 (Woods et al., 2002; DeShazer, 2004; Ronning et al., 2010; Khrongsee et al., 2024). Our findings clearly demonstrate that these phages belong to a phylogenetic lineage separate from that of the P2-like myophages. In addition, to date, only two different lytic phage strains have been identified as potential candidates for phage therapy: ΦST79, a myophage isolated in Northeast Thailand (Yordpratum et al., 2011) and ΦvB_BpP_HN01, a podophage, isolated in Hainan, China (Wang et al., 2022). We further tested if the isolated phage strains were affected by the deletion of other surface polysaccharide antigens, such as CPS, to narrow down the receptor of these phages. We have found that while all phages could infect B. pseudomallei Bp82 △wbiD (O-antigen type A knocked out), five could not infect the CPS mutant (△wcbI-R) strain. This suggested that these five phages might have used the CPS as a sole receptor to infect the serotype A strains. However, when these phages were able to infect the CPS mutant of the serotype B B. pseudomallei 576mn △wcbB, suggesting they were able to use another receptor for entry. (e.g., the O-antigen type B). However, future work would be needed to test this hypothesis following the generation knocked-out O-antigen type B strain.
It has been known that O-antigen type A is comprised of a repeating subunit of glucose and talose, while O-antigen type B is comprised of a repeating subunit of xylose-rhamnose-rhamnose-rhamnose-rhamnose-galactose (Norris et al., 2017b). The CPS is comprised of 2-O-acetyl-6-deoxy-β-d-manno-heptopyranose (Reckseidler-Zenteno et al., 2005). Based on the growth curve analysis and adsorption tests, the CPS might play a role as a main phage receptor. This is novel and has not been described in B. pseudomallei phages previously. However, bacteriophages that use CPS receptors have been reported in several bacteria such as E. coli K29 phage (Fehmel et al., 1975), Clostridium perfringen CPS1 phage (Ha et al., 2019), Klebsiella pneumoniae KN2 phages (Hsu et al., 2013), Acinetobacter baumannii AB900 phages (Altamirano et al., 2021). These phages were reported to bind with a sugar subunit of the CPS. Interestingly, CPS1 phage bound to a glucosamine in the CPS subunit was able to bind to other monosaccharides such as galactosamine and acetyl galactosamine. The phage uses difference sugar subunits as receptors depended on the similarity and polarity of the sugar structures. To the best of our knowledge, there are no common chemical structures between the CPS and the LPS molecules in B. pseudomallei. Our study simply identifies that both structures are involved in phage attachment, but the part of the sugar subunit the phage uses as a receptor is still unclear. Future work is needed to develop an appropriate monosaccharide binding assay to confirm this observation.
In a serendipitous discovery, we identified ΦPK23V1, which was isolated from a plaque formed on a lawn of the double CPS and LPS mutant strain by chance when tested with the ΦPK23 phage. Our sequencing analyses of ΦPK23 and ΦPK23V1 revealed their classification within the Peduoviridae family, alongside the well-studied P2 phages. Notably, ΦPK23 exhibited genomic similarity to a prophage identified in B. pseudomallei K96243, located on chromosome 2 within the GI15 region (Holden et al., 2004; Tuanyok et al., 2008). Despite its broad host range, it was evident that ΦPK23 is a lysogenic phage which may not be used directly for phage applications. A particularly intriguing observation pertained to the ability of the phage to induce lytic activity in two of our attenuated strains, seemingly contrary to its lysogenic nature. This may be explained by the presence of two mismatches within the bacterial attachment site (attB) sequence of both B. pseudomallei Bp82 and 576mn, describing its ability to recombine into the bacterial genome. ΦPK23 encodes a short 18-bp attachment sequence (attP), while the stability of its targeting attB sequence across B. pseudomallei strains varies. This stands in contrast to Φ1026b, which employs a 45-base pair segment from the 3’ end of the tRNA-proline sequence (attB) as its attachment site, a feature conserved across B. pseudomallei strains (DeShazer, 2004). It is worth noting that whereas most prophages in B. pseudomallei are lysogenic and often associated with tRNA regions, ΦPK23 or ΦPK23V1 exhibit a distinct genomic profile, encoding a notably short attP sequence of only 18 base pairs. This peculiarity may underlie their capacity to induce lytic activity in our attenuated strains. Given the genome similarity between ΦPK23 and the prophage island GI15 of B. pseudomallei K96243, we were concerned that this phage strain might be a temperate phage of B. pseudomallei CAM5, the phage-propagating host strain, rather than from an environmental source. To investigate this, we sequenced B. pseudomallei CAM5 using Illumina technology (data not shown). As suspected, B. pseudomallei CAM5 harbored a prophage of approximately 35.3 kb, similar to the genome of ΦPK23, but had at least 45 SNPs. We then questioned whether this prophage was functional and could be the source of ΦPK23. To explore this possibility, we attempted to isolate the temperate phage from B. pseudomallei CAM5 using the technique described by Khrongsee et al. (2024). It was observed that B. pseudomallei CAM5 produced temperate phages on the lawn of both B. pseudomallei Bp82 and 576mn, and B. mallei ATCC 23344, but no plaques were observed on its own culture lawn. Since all 145 phage samples from Thailand, including ΦPK23, were isolated from plaques formed on the lawn of B. pseudomallei CAM5, these phage samples, are likely to be phages from environmental sources, presumably temperate phages from other environmental B. pseudomallei strains or other species, rather than from B. pseudomallei CAM5’s own temperate phages.
Recently, Muangsombut and colleagues conducted a study characterizing the utility of the E094 phage tail fiber (GpH) in capturing bacteria for diagnostic purposes (Muangsombut et al., 2021). In our investigation, we identified a single nucleotide mutation (C2148A) leading to a missense mutation at position N716K in the tail fiber protein of ΦPK23V1. This mutation enables ΦPK23V1 to infect B. pseudomallei in the absence of the O-antigen and CPS. Upon analyzing the GpH structure and comparing it to that of the same phage in other Burkholderia species, we observed a high degree of conservation in the protein sequence, except for a specific region around position 640 at the C terminus of the protein (Supplementary Figure 1). Our predicted Alphafold 3D structure revealed that this region constitutes the tip of the tail fiber, and the ΦPK23V1 mutation occurred at the base of this tip, suggesting its potential as a receptor binding domain. We further conducted a trimeric 3D structure analysis of both ΦPK23 and ΦPK23V1, which demonstrated that the mutation results in an amino acid change from Asparagine (N), characterized by non-polar charge, to Lysine (K), a positively polar charge (Figure 7), potentially increasing the phage’s binding affinity due to the altered charge at a critical interaction site. Importantly, this mutation does not seem to affect the tail fiber’s inner core structure, implying that the primary receptor, identified as CPS, remains the same. Our results indicate that ΦPK23V1 exhibits better binding to host cells compared to ΦPK23. Given that the phage possesses multiple receptors for host cell entry, identifying new receptors based solely on susceptible and resistant bacterial sequencing remains challenging. Consequently, the precise targets to which ΦPK23V1 binds, beyond LPS and CPS, remain uncertain and warrant further investigation.
In previous studies, the E. coli P2 phage has been documented to possess two receptor binding proteins, namely the tail fiber protein (GpH) and the spike protein (GpV). These proteins are known to bind to the lipopolysaccharide (LPS) and specific ion molecules, respectively (Yamashita et al., 2011; Cunliffe et al., 2022). However, it remains unclear whether the ΦPK23 GpV protein plays a role in the protein binding domain of B. pseudomallei. But, our investigation clearly demonstrates that a point mutation in gpH within ΦPK23V1 can lead to an expansion of its host range.
In summary, the method developed herein holds the potential for the selection of B. pseudomallei-specific phages, each employing entry mechanisms distinct from O-antigens. Evidently, these phages may offer a solution to overcome the O-antigen diversity and serotype variation within B. pseudomallei strains. As anticipated, our investigation revealed a broad host range among these phages, positioning them favorably for various downstream applications. However, the lysogenic nature of these phages necessitates manipulation prior to practical use. Additionally, our study has facilitated the prediction of a receptor binding domain within the phage tail fiber protein, based on a point mutation within our phage. This discovery sheds light on the mechanisms by which the phage interacts with bacteria during host cell entry, offering valuable insights into this intricate process.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
PK: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. JK: Formal analysis, Investigation, Writing – review & editing. MA: Investigation, Writing – review & editing. KS: Investigation, Writing – review & editing, Methodology. TW: Investigation, Writing – review & editing, Resources. HS: Resources, Writing – review & editing, Funding acquisition. AT: Funding acquisition, Writing – review & editing, Conceptualization, Data curation, Formal analysis, Investigation, Project administration, Supervision.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported in part to HS and AT by the Defense Threat Reduction Agency (DTRA) Grant no. HDTRA1-21-1-0029.
Acknowledgments
We express our profound gratitude to Drs. Paul A Gulig, Mary M Brown, Sarah M Doore, Andrew B Alison, Rosanna Marsella, and Henry Heine for their invaluable guidance and supervision of PK throughout his graduate programs at the University of Florida. We thank Puttawat Suphaprueksapong and Jason J. Thornton for their technical assistances. AT is a member of the University of Florida Phage Research Consortium.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbrio.2024.1433593/full#supplementary-material
Supplementary Figure 1 | An AlphaFold monomeric model, a SWISS-MODEL, of the phage tail fiber protein A0A7U4SUZ9_9BURK from Burkholderia sp Bp5365, which exhibits a remarkable structural similarity to the phage tail fiber protein (GpH) found in ΦPK23 and ΦPK23V1. In this representation, the blue color highlight regions with similar protein structure to ΦPK23 GpH, while orange signifies distinct structural features. Notably, the differences were observed particularly at the tip of the phage tail fiber protein.
References
Altamirano F. L. G., Forsyth J. H., Patwa R., Kostoulias X., Trim M., Subedi D., et al. (2021). Bacteriophages targeting Acinetobacter baumannii capsule induce antimicrobial resensitization. Nat Microbiol 6, 157–161. doi: 10.1038/s41564-020-00830-7
Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. J. Comput. Mol. Cell Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Carver T. J., Rutherford K. M., Berriman M., Rajandream M.-A., Barrell B. G., Parkhill J. (2005). ACT: the artemis comparison tool. Bioinformatics 21, 3422–3423. doi: 10.1093/bioinformatics/bti553
CDC Health Alert Network (2022). Melioidosis Locally Endemic in Areas of the Mississippi Gulf Coast after Burkholderia pseudomallei Isolated in Soil and Water and Linked to Two Cases – Mississippi 2020 and 2022. Available online at: https://www.emergency.cdc.gov/han/2022/pdf/CDC_HAN_470.pdf (accessed July 27, 2022).
Cerveny K. E., DePaola A., Duckworth D. H., Gulig P. A. (2002). Phage therapy of local and systemic disease caused by Vibrio vulnificus in iron-dextran-treated mice. Infect. Immun. 70, 6251–6262. doi: 10.1128/iai.70.11.6251-6262.2002
Chetchotisakd P., Chierakul W., Chaowagul W., Anunnatsiri S., Phimda K., Mootsikapun P., et al. (2014). Trimethoprim-sulfamethoxazole versus trimethoprimsulfamethoxazole plus doxycycline as oral eradicative treatment for melioidosis (MERTH): A multicentre, double-blind, non-inferiority, randomised controlled trial. Lancet 383, 807–814. doi: 10.1016/S0140-6736(13)61951-0
Chewapreecha C., Holden M. T. G., Vehkala M., Välimäki N., Yang Z., Harris S. R., et al. (2017). Global and regional dissemination and evolution of Burkholderia pseudomallei. Nat. Microbiol. 2, 16263. doi: 10.1038/nmicrobiol.2016.263
Christie G. E., Calendar R. (2016). Bacteriophage P2. Bacteriophage 6, e1145782. doi: 10.1080/21597081.2016.1145782
Clokie M. R. J., Kropinski A. (2009). Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions. New York, NY, USA: Humana Press, c/o Springer Science+Business Media, LLC.
Corpet F. (1988). Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890. doi: 10.1093/nar/16.22.10881
Cunliffe T. G., Parker A. L., Jaramillo A. (2022). Pseudotyping bacteriophage P2 tail fibers to extend the host range for biomedical applications. ACS Synth. Biol. 11, 3207–3215. doi: 10.1021/acssynbio.1c00629
Currie B. J., Dance D. A. B., Cheng A. C. (2008). The global distribution of Burkholderia pseudomallei and melioidosis: an update. Trans. R. Soc Trop. Med. Hyg. 102 Suppl, S1–S4. doi: 10.1016/S0035-9203(08)70002-6
Dance D. (2014). Treatment and prophylaxis of melioidosis. Int. J. Antimicrob. Agents 43, 310–318. doi: 10.1016/j.ijantimicag.2014.01.005
Daubie V., Chalhoub H., Blasdel B., Dahma H., Merabishvili M., Glonti T., et al. (2022). Determination of phage susceptibility as a clinical diagnostic tool: A routine perspective. Front. Cell. Infect. Microbiol. 12, 1000721. doi: 10.3389/fcimb.2022.1000721
DeShazer D. (2004). Genomic diversity of Burkholderia pseudomallei clinical isolates: subtractive hybridization reveals a Burkholderia mallei-specific prophage in B. pseudomallei 1026b. J. Bacteriol. 186, 3938–3950. doi: 10.1128/JB.186.12.3938-3950.2004
Evans R., O’Neill M., Pritzel A., Antropova N., Senior A., Green T., et al. (2021). Protein complex prediction with AlphaFold-Multimer. bioRxiv. doi: 10.1101/2021.10.04.463034
Fehmel F., Feige U., Niemann H., Stirm S. (1975). Escherichia coli capsule bacteriophages. VII. Bacteriophage 29-host capsular polysaccharide interactions. J. Virol. 16, 591 LP–591601. doi: 10.1128/JVI.16.3.591-601.1975
Garneau J. R., Depardieu F., Fortier L. C., Bikard D., Monol M. (2017). PhageTerm: a tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci. Rep. 7, 8292. doi: 10.1038/s41598-017-07910-5
Gee J. E., Bower W. A., Kunkel A., Petras J., Gettings J., Bye M., et al. (2022). Multistate outbreak of melioidosis associated with imported aromatherapy spray. N. Engl. J. Med. 386, 861–868. doi: 10.1056/NEJMoa2116130
Gee J. E., Elrod M. G., Gulvik C. A., Haselow D. T., Waters C., Liu L., et al. (2018). Burkholderia Thailandensis isolated from infected wound, Arkansas, USA. Emerg. Infect. Dis. 24, 2091–2094. doi: 10.3201/eid2411.180821
Glass M. B., Gee J. E., Steigerwalt A. G., Cavuoti D., Barton T., Hardy R. D., et al. (2006). Pneumonia and septicemia caused by Burkholderia Thailandensis in the United States. J. Clin. Microbiol. 44, 4601–4604. doi: 10.1128/JCM.01585-06
Ha E., Chun J., Kim M., Ryu S. (2019). Capsular polysaccharide is a receptor of a Clostridium perfringens bacteriophage CPS1. Viruses 11, 1002. doi: 10.3390/v11111002
Holden M. T. G., Titball R. W., Peacock S. J., Cerdeño-Tárraga A. M., Atkins T., Crossman L. C., et al. (2004). Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc. Natl. Acad. Sci. 101, 14240–14245. doi: 10.1073/pnas.0403302101
Hsu C.-R., Lin T.-L., Pan Y.-J., Hsieh P.-F., Wang J.-T. (2013). Isolation of a bacteriophage specific for a new capsular type of Klebsiella pneumoniae and characterization of its polysaccharide depolymerase. PloS One 8, 1–9. doi: 10.1371/journal.pone.0070092
Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Keragala K. A. R. K., Gunathilaka M. G. R. S. S., Senevirathna R. M. I. S. K., Jayaweera J. A. A. S. (2023). Efficacy and safety of co-trimoxazole in eradication phase of melioidosis; systematic review. Ann. Clin. Microbiol. Antimicrob. 22, 74. doi: 10.1186/s12941-023-00620-z
Khrongsee P., Irby I., Akaphan P., Alami-Rose M. A., Kaewrakmuk J., Tuanyok A. (2024). A comprehensive study of prophage islands in Burkholderia pseudomallei complex (BPC). Front. Bacteriol. 3, 1339809. doi: 10.3389/fbrio.2024.1339809
Kvitko B. H., Cox C. R., DeShazer D., Johnson S. L., Voorhees K. J., Schweizer H. P. (2012). [amp]]phiv;X216, a P2-like bacteriophage with broad Burkholderia pseudomallei and B. mallei strain infectivity. BMC Microbiol. 12, 289. doi: 10.1186/1471-2180-12-289
Le S., He X., Tan Y., Huang G., Zhang L., Lux R., et al. (2013). Mapping the tail fiber as the receptor binding protein responsible for differential host specificity of Pseudomonas aeruginosa bacteriophages PaP1 and JG004. PloS One 8, e68562. doi: 10.1371/journal.pone.0068562
Li S., Fan H., An X., Fan H., Jiang H., Chen Y., et al. (2014). Scrutinizing virus genome termini by high-throughput sequencing. PloS One 9, e85806. doi: 10.1371/journal.pone.0085806
Limmathurotsakul D., Golding N., Dance D. A. A. B. B., Messina J. P., David M., Pigott D. M., et al. (2016). Predicted global distribution of Burkholderia pseudomallei and burden of melioidosis. Nat. Microbiol. 1, 1–13. doi: 10.1038/nmicrobiol.2015.8.Predicted
Muangsombut V., Withatanung P., Chantratita N., Chareonsudjai S., Lim J., Galyov E. E., et al. (2021). Rapid clinical screening of Burkholderia pseudomallei colonies by a bacteriophage tail fiber-based latex agglutination assay. Appl. Environ. Microbiol. 87, e0301920. doi: 10.1128/AEM.03019-20
Na-ngam N., Angkititakul S., Noimay P., Thamlikitkul V. (2004). The effect of quicklime (calcium oxide) as an inhibitor of Burkholderia pseudomallei. Trans. R. Soc Trop. Med. Hyg. 98, 337–341. doi: 10.1016/j.trstmh.2003.10.003
Norris M. H., Rahman Khan M. S., Schweizer H. P., Tuanyok A. (2017a). An avirulent Burkholderia pseudomallei ΔpurM strain with atypical type B LPS: expansion of the toolkit for biosafe studies of melioidosis. BMC Microbiol. 17, 132. doi: 10.1186/s12866-017-1040-4
Norris M. H., Schweizer H. P., Tuanyok A. (2017b). Structural diversity of Burkholderia pseudomallei lipopolysaccharides affects innate immune signaling. PloS Negl. Trop. Dis. 11, e0005571. doi: 10.1371/journal.pntd.0005571
Olson R. D., Assaf R., Brettin T., Conrad N., Cucinell C., Davis J. J., et al. (2023). Introducing the Bacterial and Viral Bioinformatics Resource Center (BV-BRC): a resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 51, D678–D689. doi: 10.1093/nar/gkac1003
Petras J. K., Elrod M. G., Ty M., Adams P., Zahner D., Adams A., et al. (2022). Notes from the field: Burkholderia pseudomallei detected in a raccoon carcass linked to a multistate aromatherapy-associated melioidosis outbreak - Texas 2022. MMWR. Morb. Mortal. Wkly. Rep. 71, 1597–1598. doi: 10.15585/mmwr.mm7150a5
Petras J. K., Elrod M. G., Ty M. C., Dawson P., O’Laughlin K., Gee J. E., et al. (2023). Locally acquired melioidosis linked to environment - Mississippi 2020-2023. N. Engl. J. Med. 389, 2355–2362. doi: 10.1056/NEJMoa2306448
Pettersen E. F., Goddard T. D., Huang C. C., Meng E. C., Couch G. S., Croll T. I., et al. (2021). UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82. doi: 10.1002/pro.3943
Pongmala K., Pierret A., Oliva P., Pando A., Davong V., Rattanavong S., et al. (2022). Distribution of Burkholderia pseudomallei within a 300-cm deep soil profile: implications for environmental sampling. Sci. Rep. 12, 8674. doi: 10.1038/s41598-022-12795-0
Propst K. L., Mima T., Choi K.-H., Dow S. W., Schweizer H. P. (2010). A Burkholderia pseudomallei deltapurM mutant is avirulent in immunocompetent and immunodeficient animals: candidate strain for exclusion from select-agent lists. Infect. Immun. 78, 3136–3143. doi: 10.1128/IAI.01313-09
Reckseidler-Zenteno S. L., DeVinney R., Woods D. E. (2005). The capsular polysaccharide of Burkholderia pseudomallei contributes to survival in serum by reducing complement factor C3b deposition. Infect. Immun. 73, 1106–1115. doi: 10.1128/IAI.73.2.1106-1115.2005
Ronning C. M., Losada L., Brinkac L., Inman J., Ulrich R. L., Schell M., et al. (2010). Genetic and phenotypic diversity in Burkholderia: contributions by prophage and phage-like elements. BMC Microbiol. 10, 202. doi: 10.1186/1471-2180-10-202
Schweizer H. P. (2013). Mechanisms of antibiotic resistance in Burkholderia pseudomallei: implications for treatment of melioidosis Herbert. Futur. Microbiol. 7, 1389–1399. doi: 10.2217/fmb.12.116.Mechanisms
Shaw T., Assig K., Tellapragada C., Wagner G. E., Choudhary M., Göhler A., et al. (2022). Environmental factors associated with soil prevalence of the melioidosis pathogen Burkholderia pseudomallei: A longitudinal seasonal study from South West India. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.902996
Sullivan M. J., Petty N. K., Beatson S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Torres A. G. (2023). The public health significance of finding autochthonous melioidosis cases in the continental United States. PloS Negl. Trop. Dis. 17, e0011550. doi: 10.1371/journal.pntd.0011550
Tuanyok A., Leadem B. R., Auerbach R. K., Beckstrom-Sternberg S. M., Beckstrom-Sternberg J. S., Mayo M., et al. (2008). Genomic islands from five strains of Burkholderia pseudomallei. BMC Genomics 9, 566. doi: 10.1186/1471-2164-9-566
Tuanyok A., Stone J. K., Mayo M., Kaestli M., Gruendike J., Georgia S., et al. (2012). The genetic and molecular basis of O-antigenic diversity in Burkholderia pseudomallei lipopolysaccharide. PloS Negl. Trop. Dis. 6, e1453. doi: 10.1371/journal.pntd.0001453
Tumapa S., Holden M. T. G., Vesaratchavest M., Wuthiekanun V., Limmathurotsakul D., Chierakul W., et al. (2008). Burkholderia pseudomallei genome plasticity associated with genomic island variation. BMC Genomics 9, 190. doi: 10.1186/1471-2164-9-190
Wang Y., Li X., Dance D. A. B., Xia H., Chen C., Luo N., et al. (2022). A novel lytic phage potentially effective for phage therapy against Burkholderia pseudomallei in the tropics. Infect. Dis. Poverty. 11, 87. doi: 10.1186/s40249-022-01012-9
Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Wiersinga W. J., Virk H. S., Torres A. G., Currie B. J., Peacock S. J., Dance D. A. B. B., et al. (2018). Melioidosis. Nat. Rev. Dis. Prim. 4, 17107. doi: 10.1038/nrdp.2017.107
Withatanung P., Janesomboon S., Vanaporn M., Muangsombut V., Charoensudjai S., Baker D. J., et al. (2024). Induced Burkholderia prophages detected from the hemoculture: a biomarker for Burkholderia pseudomallei infection. Front. Microbiol. 15. doi: 10.3389/fmicb.2024.1361121
Woods D., Jeddeloh J., Fritz D., DeShazer D. (2002). Burkholderia Thailandensis E125 Harbors a Temperate Bacteriophage Specific for Burkholderia mallei. J. Bacteriol. 184, 4003–4017. doi: 10.1128/JB.184.14.4003-4017.2002
Yamashita E., Nakagawa A., Takahashi J., Tsunoda K., Yamada S., Takeda S. (2011). The host-binding domain of the P2 phage tail spike reveals a trimeric iron-binding structure. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67, 837–841. doi: 10.1107/S1744309111005999
Keywords: melioidosis, Burkholderia pseudomallei, bacteriophage, phage-tail fiber, GpH
Citation: Khrongsee P, Kaewrakmuk J, Alami-Rose M, Subramaniam K, Waltzek TB, Schweizer HP and Tuanyok A (2024) Exploring Burkholderia pseudomallei-specific bacteriophages: overcoming O-antigen specificity and adaptive mutation in phage tail fiber. Front. Bacteriol. 3:1433593. doi: 10.3389/fbrio.2024.1433593
Received: 16 May 2024; Accepted: 27 August 2024;
Published: 18 September 2024.
Edited by:
Daniel Yero, Autonomous University of Barcelona, SpainReviewed by:
Sohinee Sarkar, Royal Children’s Hospital, AustraliaQianfeng Xia, Hainan Medical University, China
Copyright © 2024 Khrongsee, Kaewrakmuk, Alami-Rose, Subramaniam, Waltzek, Schweizer and Tuanyok. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Apichai Tuanyok, dHVhbnlva0B1ZmwuZWR1
†Present address: Thomas B. Waltzek, Washington State University, Pullman, WA, United States
 Pacharapong Khrongsee
Pacharapong Khrongsee Jedsada Kaewrakmuk
Jedsada Kaewrakmuk Mariam Alami-Rose
Mariam Alami-Rose Kuttichantran Subramaniam
Kuttichantran Subramaniam Thomas B. Waltzek
Thomas B. Waltzek Herbert P. Schweizer
Herbert P. Schweizer Apichai Tuanyok
Apichai Tuanyok