
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bacteriol. , 16 July 2024
Sec. One Health in Bacteriology
Volume 3 - 2024 | https://doi.org/10.3389/fbrio.2024.1432292
Escherichia coli is found in diverse environmental niches, including meat and meat products, and is known for its significance in both food safety and public health. In South Africa, whole genomic sequencing (WGS) efforts for E. coli are overwhelmingly skewed toward human isolates with limited studies conducted on non-human isolates. Therefore, the aim of this study was to use WGS to characterise generic strains of E. coli isolated from animal specimens, meat, and meat-based products in South Africa. Based on WGS analysis, a total of 35 E. coli strains were grouped into five phylogroups (A, B1, B2, C, and E), with A (46%) being the most predominant. Virulence-associated genes identified the isolates as either extra-intestinal pathogenic E. coli (69%) or intestinal pathogenic E. coli (31%) pathotypes. Twenty-three different serotypes were identified, with O101:H37 (17%), O2:H4 (17%), O6:H5 (13%), and O64:H19 (9%) being the predominant ones. Among the 19 different sequence types (STs), ST1858, ST975, and ST10 were the most prevalent (11% each). Various virulence genes, antimicrobial resistance genes, and genetic mobile elements carrying Tn2, IS26, and Tn6196 elements were detected, with the disinfectant resistance sitABCD being the most predominant. The type 1 CRISPR system which functions by storing records of previous invasions to provide immunological memory for a rapid and robust response upon subsequent viral infections was detected in all isolates, consisting of subtypes I-E (86%), I-A (57%), and I-F (11%). The findings of this study provide an insight into the genetic diversity of generic E. coli isolates from animal species, meat, and meat-based products in South Africa.
Food-producing animal slaughter establishments and processors test for E. coli Biotype I (generic E. coli) to verify the effectiveness of their process controls and sanitation practices. Faecal contamination, one of the primary sources of pathogenic and non-pathogenic organisms that contaminate animal protein foods, is commonly indicated by generic E. coli (Gekenidis et al., 2018). This bacterium is prevalent since it inhabits the gastrointestinal tract of humans and warm-blooded animals (Murphy et al., 2021). The performance criteria for generic E. coli are not enforceable, as the bacteria numbers simply represent microbial loads used to monitor and verify whether the slaughter and/or production process was adequately controlled. These criteria provide guidance to livestock slaughter establishments on the effectiveness of their processes in preventing faecal contamination. Test results serve as evidence that the slaughter or production maintained sufficient process controls for hygienic dressing. E. coli is ubiquitous, inhabiting diverse environments such as water sources, animals, and food (Lupindu, 2017; Galindo-Méndez, 2020). However, the characterisation of generic E. coli and its potential pathogenicity to humans is often overlooked, despite E. coli being a significant cause of serious diseases in humans and animals.
In sub-Saharan Africa, E. coli diarrheal infections represent a significant public health challenge, with a high incidence attributed to factors such as limited access to clean water, sanitation and inadequate hygiene (Robert et al., 2021). E. coli strains causing intestinal infections are known as intestinal pathogenic E. coli (InPEC), while those responsible for extraintestinal infections are termed extraintestinal pathogenic E. coli (ExPEC). These infections encompass various pathotypes, each characterised by specific traits (Johnson and Russo, 2002; Meena et al., 2021, 2023). InPEC is linked to pathotypes such as enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), enteroaggregative E. coli (EAEC), shiga toxin-producing E. coli (STEC), diffusely adherent E. coli (DAEC), adherent-invasive E. coli (AIEC), and enteroinvasive E. coli (EIEC). ExPEC infections are associated with pathotypes such as avian pathogenic E. coli (APEC), uropathogenic E. coli (UPEC), neonatal meningitis E. coli (NMEC), and sepsis-associated E. coli (SEPEC), primarily affecting humans (Martinez-Medina, 2021; Abdulabbas et al., 2023).
There is a compelling need to closely monitor the spread of E. coli in animals and food derived from animal sources (Ramos et al., 2020). Furthermore, there’s a growing apprehension regarding the potential for E. coli to acquire antimicrobial resistance (AMR) traits within livestock environments. This evolution could not only complicate but also escalate the cost of treating infections in both humans and animals (Palma et al., 2020). Such antimicrobial resistance poses a significant public health concern, as it reduces the effectiveness of antibiotics, thereby limiting treatment options and potentially increasing the severity and duration of illnesses caused by E. coli infections.
Studies have increasingly highlighted the intriguing link between the virulence of E. coli strains and their Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR-Cas) systems. CRISPR-Cas systems, originally identified as adaptive immune mechanisms in bacteria and archaea, play a crucial role in defending against foreign genetic elements such as bacteriophages and plasmids (Koonin et al., 2017; Murugan et al., 2017). Several studies have suggested that certain E. coli strains with more robust CRISPR-Cas systems may exhibit decreased virulence due to their enhanced ability to fend off invading genetic elements (García-Gutiérrez et al., 2015). Conversely, strains with compromised or less effective CRISPR-Cas systems might be associated with higher virulence as they struggle to combat invasive genetic elements, potentially including virulence factors (Louwen et al., 2014). Furthermore, the interplay between CRISPR and virulence in E. coli is complex and multifaceted. It involves various factors such as the specific composition and activity of CRISPR systems, the presence of virulence genes, and the environmental context in which the bacteria reside. Understanding this interrelationship can offer insights into the evolution of E. coli pathogenicity and potentially inform strategies for combating E. coli-related infections (Kang and Lee, 2022).
Cases and outbreaks of E. coli in both animals and humans are well-documented in South Africa (Gambushe et al., 2022; Khabo-Mmekoa et al., 2022; Manyi-Loh and Lues, 2023). However, the primary focus of whole-genome sequencing (WGS) endeavours has predominantly targeted human clinical strains associated with outbreaks (Muloi et al., 2018; Massella et al., 2020). Additionally, attention has been devoted to analysing environmental samples (Igwaran et al., 2018; Bolukaoto et al., 2021). Nonetheless, genomic data regarding the complete diversity of E. coli colonising food and animals, their array of antimicrobial resistance genes (ARGs), associated mobile genetic elements (MGEs), virulence-associated genes (VAGs), and the possible presence of ExPEC is lacking in South Africa. This study utilised WGS to analyse 35 E. coli isolates collected from animals, meat, and meat products in South Africa in order to highlight the significance and requirement of such studies within South Africa over extended period of time, few of these samples are international samples which were collected from ports of entry in South Africa.
The isolates utilised in this study were obtained from samples processed between 1988 and 2018 at the Bacteriology Laboratory of the Onderstepoort Veterinary Research Institute in South Africa, as part of their routine diagnostic testing. Hence, for this research, a selection of 35 E. coli isolates was made, encompassing diverse geographical regions within the country (Gauteng, Free State, North-West, Limpopo, and Mpumalanga), as well as international locales, different isolation sources (animal, meat and meat products such as a traditional sausage called wors), and a range of animal species (poultry, porcine, bovine and ovine) (Table 1), a water sample was included in this analysis in order to trace the source of E. coli contamination. These international samples are isolates from imported meat samples intended for local consumption. The isolates were stored in lyophilised form and then reconstituted by inoculating them into brain heart infusion (BHI) broth, followed by an incubation period at 37°C for 18 to 24 hours.
Genomic DNA was extracted from overnight cultures using the High Pure PCR template preparation kit (Roche, Germany) in accordance with the manufacturer’s instructions. Purity and concentration of the DNA were assessed using a Nanodrop 1000 spectrophotometer. Subsequently, WGS was performed at the Biotechnology Platform Agricultural Research Council, Onderstepoort, South Africa using a HiSeq 2500 instrument (Illumina, San Diego, CA, USA). The construction of DNA libraries was accomplished using TruSeq DNA library preparation kits (Illumina, San Diego, CA, USA).
The raw read quality was assessed with FastQC v.0.11.9 (Andrews, 2010) and the adapters and low-quality reads were trimmed using Trimmomatic v.0.39 (Bolger et al., 2014). SPAdes (v3.15.3) was used for assembly of each isolate (Prjibelski et al., 2020) and assembly quality was analysed using Quast v4.4 (Gurevich et al., 2013). Genome quality along with contamination levels were assessed using CheckM v1.0.18 (Parks et al., 2015). The isolates were annotated using Prokka v1.13.7 (Seemann, 2014). To determine the strains taxonomic classification, a portion of the complete nucleotide sequences assemblies in this study were aligned using the Basic local alignment tool (BLASTN) against the nucleotide sequences on the NCBI database. Isolate with a percentage identity of >90% to E. coli on the NCBI database were accepted (Peker et al., 2019).
Clermont quadruplex phylo-group assignment technique was employed to determine E. coli phylogroups. Fasta files containing E. coli contigs were uploaded on the Clermont Typing website (http://clermontyping.iame-research.center/) and sequences were analysed using the default settings (Clermont et al., 2019).
The assembled genomes of E. coli isolates were used to perform in silico serotyping of the O and H antigens, employing SerotypeFinder gene database hosted at the Center for Genomic Epidemiology (CGE), accessible at https://cge.food.dtu.dk/services/SerotypeFinder/. Specifically, for the O antigen, the database analysed the wzx, wzy, wzm, and wzt genes, while for flagellin H-antigen, it processed the fliC, flkA, flmA, flnA, and fllA genes (Joensen et al., 2014). The analysis was conducted with the default parameters specified on the website, including a minimum sequence length of 60% and a threshold of 85%.
Escherichia coli pathotypes in this study were classified based on their virulence factor characteristics as seen in literature (Robins-Browne et al., 2016; Jesser and Levy, 2020; Riley, 2020; Enciso-Martínez et al., 2022; Geurtsen et al., 2022) with isolates classified as either intestinal-pathogenic E. coli (InPEC) or extraintestinal E. coli (ExPEC) (Supplementary Material Table S1). Each isolate’s virulence characteristics were analysed and assigned a pathotype based on the presence of the target gene. Identification was based on a combination of the main virulence factor genes which are capable of causing disease and these included genes responsible for attachment, production of toxins or hemolysis. In order to classify InPEC according to the corresponding pathotype, ETEC must contain either LT or ST enterotoxin, STEC must contain any stx gene and the EPEC must contain the intimin eae. On the ExPEC classification, the UPEC, APEC and NMEC pathotype assignment depends on presence of any of the two target genes listed on Supplementary Table S1.
Sequence types were identified through multi-locus sequence typing (MLST) using version 2.0 of the CGE tool, which is accessible at https://cge.food.dtu.dk/services/MLST/. On the CGE tool, select the MLST configuration (Escherichia coli #1), select the minimum depth for allele of 5X, input the assembled genomes (Larsen et al., 2012). Retrieve and analyse the results based on the sequence types identified, confidence scores as well the allele profiles.
As part of the publicly accessible web-based tools for WGS analysis offered by the CGE, virulence factors within this study were determined using this web-based platform. The assembled genomes of E. coli isolates were submitted on the VirulenceFinder database (https://cge.cbs.dtu.dk/services/VirulenceFinder/) to detect virulence genes, E. coli species was selected, a threshold of over 90% identity with a minimum length of 60% was selected and assembled genomes were analysed (Joensen et al., 2014). In order to select significant genes which, encode virulence for InPEC and ExPEC, Chapman et al. (2006) and Badi et al. (2018) were used as a reference guide to select specific genes (Supplementary Data Table S1).
The isolates underwent analysis using PathogenFinder v. 1.1 (https://cge.food.dtu.dk/services/PathogenFinder/) to assess their potential human-associated pathogenicity. Assembled genomes were uploaded, and the phylum selection included all the classes of bacteria that can be detectable by the system. The system predicts the number of pathogenic and non-pathogenic bacterial families (Cosentino et al., 2013).
The presence and characteristics of a cluster of regularly spaced short palindromic repeats (CRISPR) were determined using the CRISPRone online tool (https://omics.informatics.indiana.edu/CRISPRone/). This tool allows searching for CRISPR-Cas system genes and proteins, as well as class types and subtypes of the system. It also returns the number of loci, length, and nucleotide sequences of repeat spacers (Zhang and Ye, 2017).
Resistance genes as well antibiotic phenotypes were obtained from ResFinder v4.1 (https://cge.food.dtu.dk/services/ResFinder/). Plasmid-associated genes and mobile genetic elements genes were obtained from PlasmidFinder v2.1 (https://cge.food.dtu.dk/services/PlasmidFinder/ and MobileElementFinder v1.0.3 (https://cge.food.dtu.dk/services/MobileElementFinder/), respectively. Plasmid Inc types hosted by pathogenwatch (https://pathogen.watch/) was used to verify the result of PlasmidFinder (Argimón et al., 2021).
Assembled genomes of isolates within this study were analysed for presence of antimicrobial resistance phenotypes using Resfinder v4 tool with default parameters, this tool is able analyse antibiograms in silico. Using Resfinder, a threshold percentage identity of 90% with a minimum length of 60% was used for antimicrobial resistance genes (Bortolaia et al., 2020). Presence of antimicrobial resistance genes within the isolates and the antimicrobials identified by the tool were used to infer presence of antimicrobial resistance phenotypes.
A total of 35 isolates were obtained from animal specimens, meat and meat products in South Africa and some are international samples from port of entries into the country. Supplementary Table S5 contains an overall summary of the results obtained grouped according to species. These isolates underwent WGS, and all were identified as E. coli using NCBI BLASTN tool. The BLASTN results showed that all the genomes from this study clustered among publicly available E. coli genomes with a percentage identity of over 95% to those on the NCBI database (Supplementary Table S3) (Peker et al., 2019).
Upon phylogenetic classification revealed phylogroup A as the most prevalent among poultry isolates, accounting for 46% of the total, followed by phylogroup B1 (20%), B2 (6%), with phylogroups C and E contributing 3%, these other phylogroups were widely distributed among various animal species (bovine, ovine and porcine). Phylogroup C was detected in both poultry and porcine samples, while phylogroup E was exclusively isolated from poultry samples. Notably, all phylogroups were observed across different provinces in South Africa. Twenty-three different serotypes were identified with O101:H37 (17%), O2:H4 (17%), O6:H5 (13%) and O64:H19 (9%) being the predominant (Supplementary Tables S2, S5). The distribution of serotypes across provinces showed that majority of isolates originated from the Free State (43%) and Gauteng province (36%), followed by some from Northwest (14%) and only one (7.%) from a port of entry.
In this study, virulence-associated genes of E. coli pathotypes were utilised to characterise isolates based on the pathotype they belong to, thereby classifying them as either InPEC or ExPEC. Majority of the isolates belonged to the ExPEC pathotype which comprised of 69%, with APEC accounting for 37% and UPEC for 31%. Among the APEC pathotype, poultry contributed 20%, while the UPEC pathotype, contributed 9% for bovine, and poultry each and 3% for porcine. In contrast, InPEC pathotypes comprised only 31% of the isolates, with ETEC representing 23%. ETEC pathotype in poultry, and porcine contributed 6% in each, in bovine and ovine contributed 3%. STEC contributed 3% and was found in poultry, EPEC was only found in porcine and contributed 6% of the isolates, Supplementary Table S5 and Figure 1 lists the pathotype assignment for each isolate.
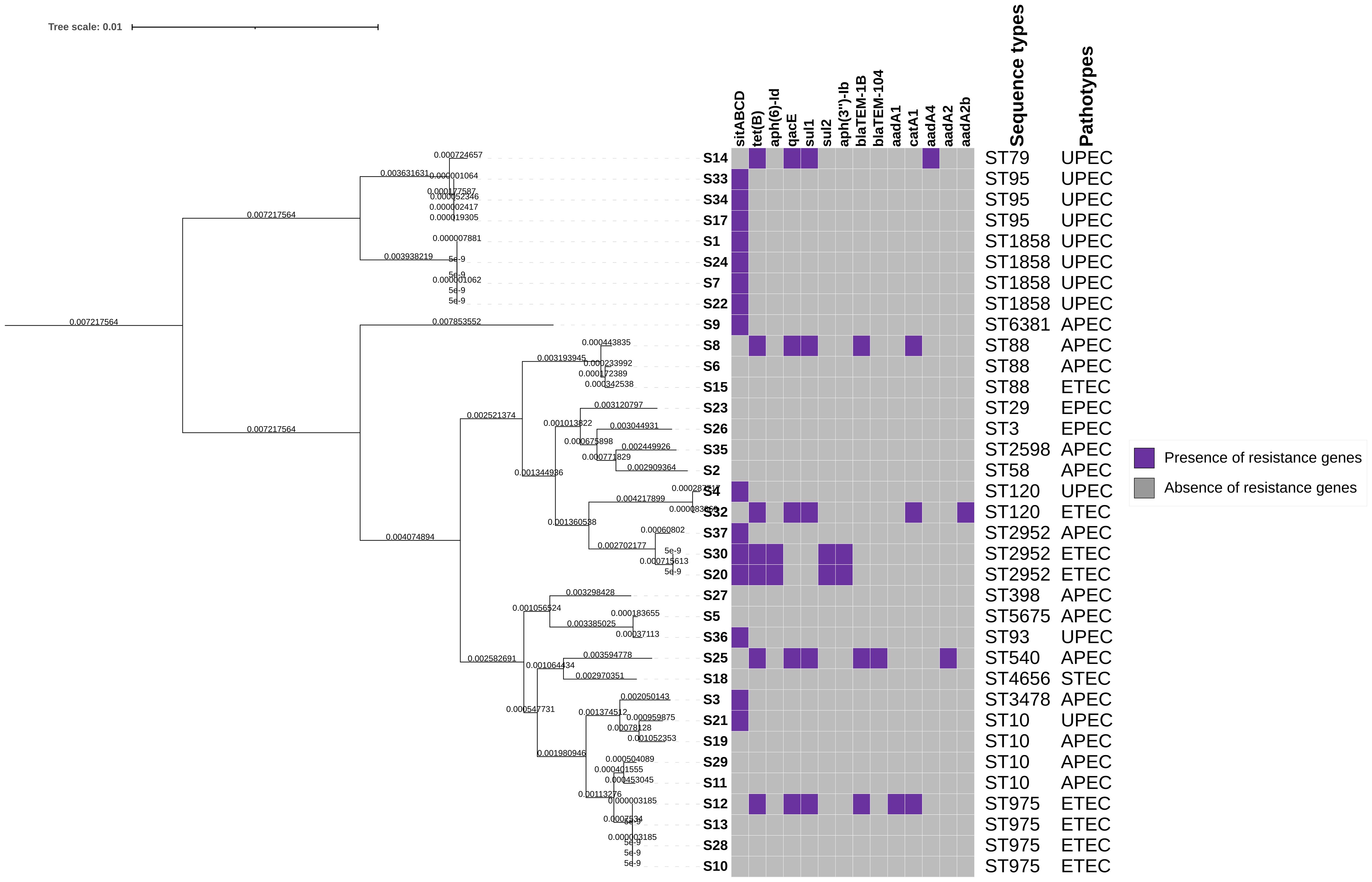
Figure 1 Heat map showing the distribution of E. coli antimicrobial resistance genes, violet colour indicates presence of resistance genes and grey indicates absence of resistance genes. The top panel of the x-axis indicates the antimicrobial resistance genes detected. The right panel of the map indicates the sequence types and pathotypes associated with these isolates.
The isolates were classified into a total of 19 distinct STs. Among these, ST1858, ST975, and ST10 emerged as the most prevalent, each accounting for 11% of the isolates. ST95, ST88, and ST2952, constituted 9%, while ST120 was 6% of the total isolates, respectively. All the identified STs are presented in Figure 2; Supplementary Figure S1 and Supplementary Table S2.
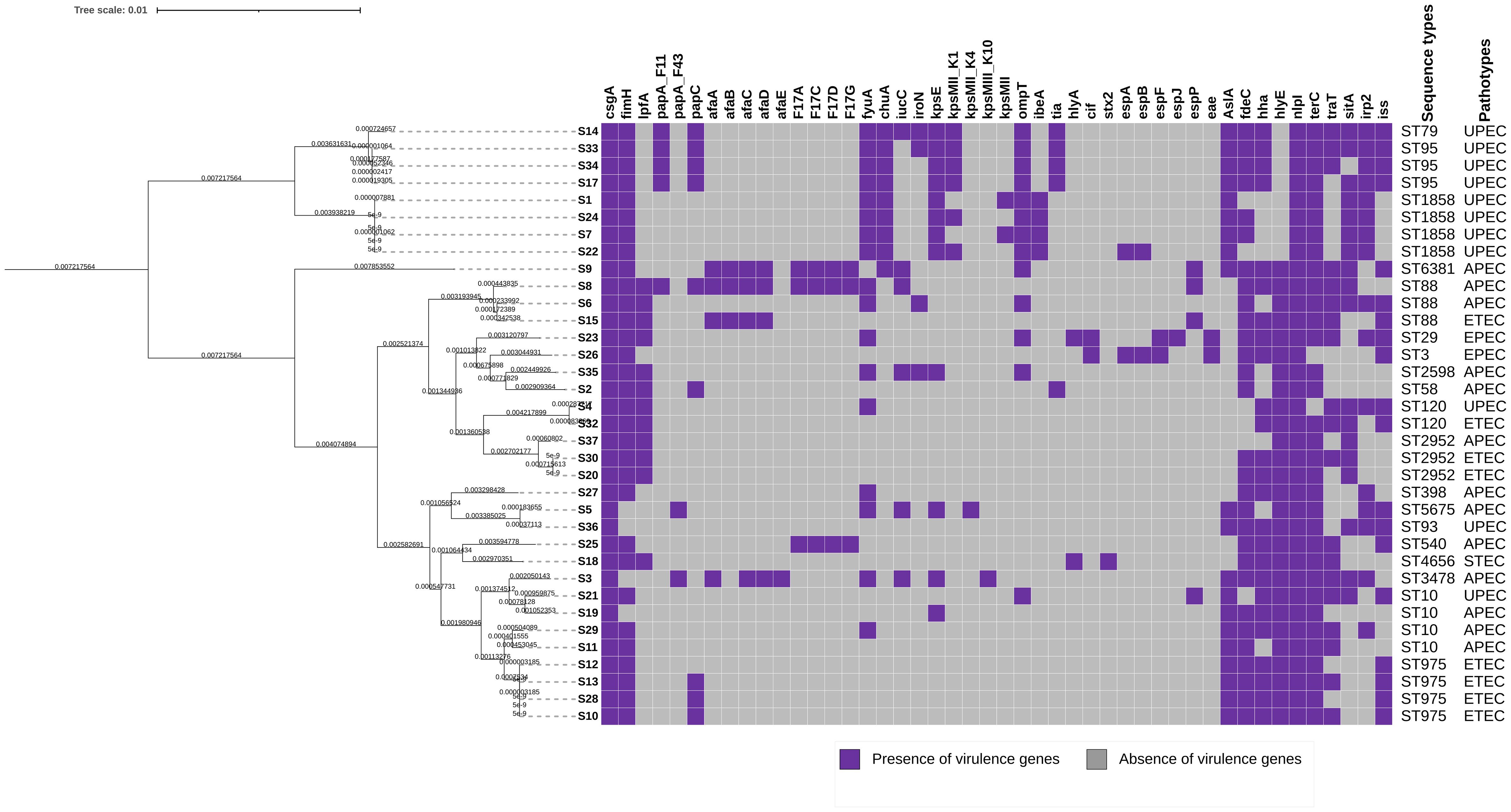
Figure 2 A heatmap of E. coli virulence genes, the map shows presence/absence of genes. Violet colour indicates presence of genes and grey indicates absence of genes in isolates from animal specimen, meat and meat products in South Africa. The genes are listed on the top panel of the x-axis. The right panel indicates the sequence types and pathotypes of each of the isolates represented on the phylogenetic tress.
Figure 2 shows distribution of virulence genes detected in all the isolates. The predominant virulence genes detected in the isolates encompassed a variety of crucial factors. Notably, csgA (100%), encoding the curlin major subunit, and nlpI (100%), responsible for encoding the lipoprotein NlpI precursor, were universally present. This was followed closely by terC (associated with tellurium ion resistance) detected at 94%, and fimH (involved in type 1 fimbriae formation) at 89%. The YHD fimbrial clusters were also prevalent, with 86% for yehA encoding the outer membrane lipoprotein, 88.6% for yehB encoding the usher, yehC (89%) encoding the chaperone, and yehD (86%) encoding the major pilin subunit. Among other notable genes were fdeC (86%), an intimin-like adhesion, and hlyE (77%), encoding avian E. coli haemolysin. The iss gene (54%), associated with increased serum survival, and irp2 (46%), encoding a high molecular weight protein 2 non-ribosomal peptide synthetase, were also detected in the isolates. Less frequently observed genes included stx2 (3%), a Shiga-toxin gene, and various type three secretion effector proteins such as espA, espB, espF, espJ, and cif, collectively found in 6% of the isolates.
Major genes clusters were observed within the study. Genes contributing to E. coli pathogenicity were identified which included adherence genes (csgA, fimH, lpfA, pap, afa), iron-uptake genes (fyuA, chuA, iucC, iroN), the capsule synthesis genes (kpsE, kpsMII, kpsMIII), genes responsible for invasion and survival (ompT, ibeA, tia) and those responsible for the secretion system (esp genes). Other virulence genes which could not be classified into a group were also identified.
All the isolates within this study were confirmed as human pathogens that belong to the Gammaproteobacteria class.
The type 1 CRISPR system was found across all isolates, with a noteworthy distribution of subtypes. Specifically, 86% exhibited subtype I-E, 57% subtype I-A, and 11% subtype I-F. Among the subtypes, 80% featured a singular CRISPR loci, while the remaining 20% exhibited two loci. Subtype I-E demonstrated distinctive characteristics, including the presence of cas3 (82%), cas6e (51%), and cas8e (77%). Subtype I-A was characterised by cas5 (51%) and cas7 (51%), whereas Subtype I-F exhibited cas5f, cas6f, cas7f, and cas8f, each detected in 11% of the isolates. Within the CRISPR system, Cas genes featuring a nuclease with the DEDDh motif were identified in 66% of the isolates. Additionally, the universal cas1 and cas2 elements were present in 63% of the isolates, adding a foundational element to the diversity observed within the CRISPR loci across the studied isolates.
The distribution of AMR gene presence among the isolates is presented in Supplemntary Table S4, Figure 1, Supplementary Figure S1. The disinfectant resistance gene sitABCD (43%) emerged as the most detected resistance gene among these E. coli isolates. Within the subset of isolates exhibiting sitABCD resistance, additional genes were identified, including the tetracycline resistance gene tet(B) (23%), and the disinfectant resistance gene qacE (14%). Sulfonamide resistance genes sul1 (9%) and sul2 (6%) were also identified, along with the chloramphenicol resistance gene catA1 (6%). Beta-lactam resistance genes blaTEM-1B (9%) and blaTEM-104 (3%) were detected. Furthermore, aminoglycoside resistance genes aadA1 (6%), aadA2b (6%), aph(6)-Id, and aph(3’’)-Ib (6%) were observed. Notably, at least fifteen isolates, comprising 43% of the total, did not exhibit any resistance genes.
The putative AMR phenotypes of these isolates were determined in silico using Resfinder tool, this was employed due to lack of the minimum inhibitory concentration (MIC) results and also putting into consideration that these isolates are primarily historic samples. The results revealed that all isolates exhibited resistance to cotrimoxazole (100%), while 97% were resistant to penicillin, Supplementary Table S5. Additionally, resistance to the chemical disinfectant hydrogen peroxide was observed in 37% of the isolates. Furthermore, resistance to amoxicillin (26%), sulfamethazine (17%), tetracycline (14%), chloramphenicol (11%), ceftazidime (14%), aztreonam (9%), piperacillin (9%), ampicillin (6%), and azithromycin (3%) was also identified (Supplementary Table S5). Multidrug resistance (MDR) was observed seven isolates (20%), MDR was in this study is defined as those isolates were each isolate conferred resistance to five or seven antibiotics. The most common class of antibiotics among these multi-drug resistant isolates were beta-lactams, sulphonamides, tetracycline, and chloramphenicol.
Mobile elements detected in the isolates from this study exhibited a diverse array, showcasing various transposable elements (IS elements), ISEc elements, transposons, and the presence of a specific Miniature Inverted-repeat Transposable Element (MITEEc1) of 123bp in each isolate. Among the detected IS elements, a spectrum of types was identified, including IS4, IS5, IS100, IS609, IS682, IS911, and IS45. The ISEc elements displayed similar diversity, encompassing ISEc1, ISEc42, ISEc39, ISEc38, ISEc10, ISEc45, ISEc30, and ISEc52. Notably, transposons such as Tn2 and Tn6196 were also identified, further highlighting the complexity of mobile genetic elements in these isolates.
Antimicrobial resistance genes detected within the multidrug resistant isolate were found to be in different contig positions. Sample S8 isolated from poultry meat was found to have resistance genes on node 245, resistance genes were sul1, aadA1, qacE with IS26, blaTEM-1B was found on node 252 with MGE Tn2 and tet(B) was found on node 222 with no MGE. Sample S12 from porcine meat had resistance on node 121 which carried tet(B) and blaTEM-1B with MGE Tn2, node 143 carried aadA1, qacE and sul1 with no MGE, node 146 carried catA1 with no MGE. Sample S14 from porcine meat had resistance on node 24 which carried aadA4, sul1 and qacE, on the same contig Tn6196 was detected. Node 170 carried tet(B). Sample S20 isolated from animal faeces had a resistance gene aph(6)-Id, aph(3’’)-Ib and sul2 on node 117 and tet(B) was on node 112, MGE were not detected in this isolate. Sample S25 from animal faeces had no MGE, node 180 carried tet(B), node 187 had qacE, sul1 and aadA2, node 295 had blaTEM-1B and blaTEM-104 and node 128 carried catA1. Sample S30 isolated from animal faeces had no MGE, while node 113 had sul2, aph(3’’)-Ib and aph(6)-Id and node 121 carried tet(B). Sample S32 from animal faeces node 228 carried tet(B), node 225 qacE, sul1 and aadA2b, node 185 carried catA1. Node 150 carried blaTEM-1B with Tn2.
As illustrated in Figure 3, the analysis of plasmid replicons using in silico WGS revealed a predominant presence of incompatibility groups, particularly IncFII (n=23, 66%) and IncFIB (n=21, 60%), across various isolates. Additionally, Col plasmid groups were also detected, with Col440I (n=2, 6%), Col156 (n=5, 14%), ColRNAI (n=2, 6%), ColE10 (n=1, 3%), and Col440II (n=2, 6%) exhibiting distribution among the isolates. Other plasmid types, such as IncFIA (n=7, 20%), IncB/O/K/Z (n=1, 3%), IncI2 (Delta) (n=1, 3%), and IncHI2A (n=1, 3%), were also identified, contributing to the overall plasmid landscape. Furthermore, the study identified isolates displaying multidrug resistance that harboured the class 1 integron (intl1), characterised by cassette arrays including aadA1 and aadA2b, thereby adding another layer of genetic complexity to the mobile elements within this E. coli population. These isolates were isolated from porcine and bovine.
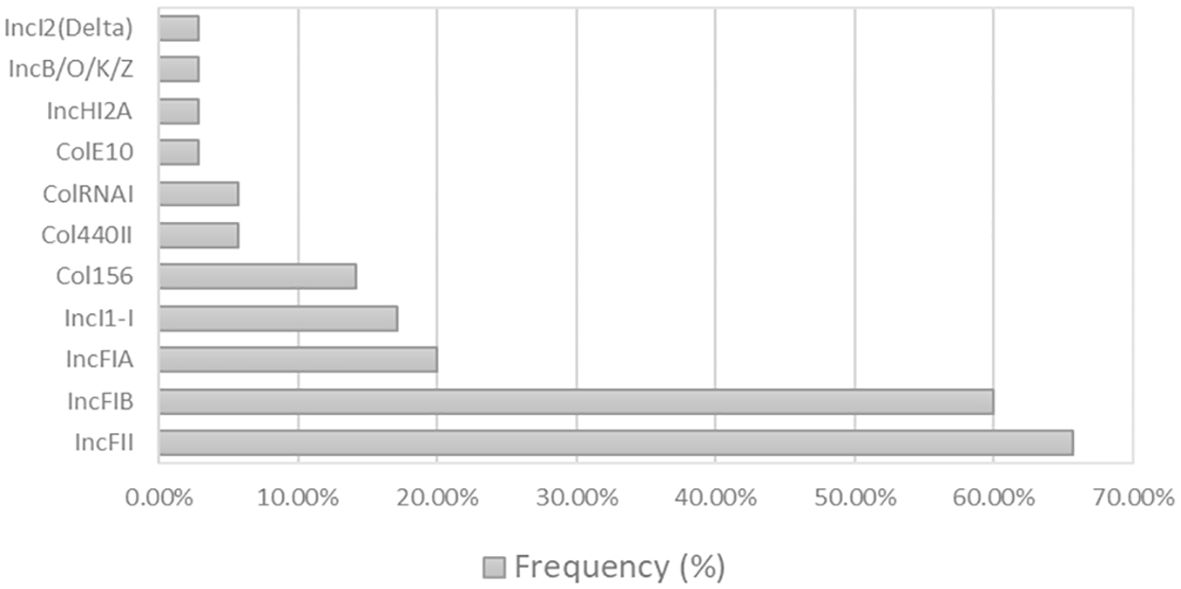
Figure 3 Bar graph of the plasmid Inc Types detected. Eleven plasmids were detected within the thirty-five E. coli isolates. On the left side of the chart, the y-axis indicates the percentage of each plasmid.
A total of 35 generic E. coli isolates from various sources underwent characterisation using WGS. The highly discriminative nature of WGS enables comparison of genetic relatedness among bacteria, even at the sub-species level, thus establishing it as the gold standard for typing bacterial isolates (Uelze et al., 2020). Moreover, WGS facilitates monitoring of antimicrobial resistance, virulence, and pathogenicity profiling, as well as source tracing, root cause analysis of contamination events, and enforcement of quality checks for bacterial analysis (Allard et al., 2018). The application of WGS in this study revealed that the generic isolates of E. coli belonged to five phylogroups: A, B1, B2, C, and E, representing 2, 23 and 19 different pathotypes, serotypes and STs, respectively. Therefore, the information population structure of E. coli circulating in the country of non-human clinical isolates provide critical information for epidemiological purposes.
Understanding the dynamics of the E. coli population is crucial for various reasons, including public health management and food safety. Phylogrouping, a method used to categorise E. coli strains based on genetic relatedness, plays a pivotal role in this regard. By classifying strains into different phylogroups, researchers can gain insights into how these strains are associated with the diseases they cause (Halaji et al., 2022). In the current study, the most prevalent phylogroups identified were A, B2, and B1, with the less common groups being C and E. This finding aligns with previous research indicating that phylogroups A, B1, and B2 are often the most predominant among E. coli strains found in various contexts, including clinical human cases, as well as in food and animal products (Pakbin et al., 2021; Aguirre-Sánchez et al., 2022; Zhao et al., 2022). Of particular note, phylogroup B2 has garnered attention for its association with extra-intestinal infections, indicating its potential role in causing a majority of such infections. This highlights the importance of understanding the distribution and prevalence of different phylogroups in various environments, as it can inform strategies for disease prevention and control.
In the current study, ExPEC strains were identified as the most predominant, collectively contributing at least 69% of the isolates. These strains were primarily classified into two pathotypes: APEC at 37% and UPEC at 31%. ExPEC strains are recognised for their propensity to cause diseases beyond the intestinal tract, including meningitis, urinary tract infections (UTIs), and sepsis (Rocha et al., 2021). In human populations, UPEC is infamous for its association with urinary tract infections, a prevalent and often recurrent medical issue (Whelan et al., 2023). Conversely, APEC is predominantly linked with avian colibacillosis, a significant bacterial infection affecting poultry industries worldwide with serious economic losses and welfare concerns in poultry farming (Kathayat et al., 2021). Most APEC strains identified in this study were found to exhibit diverse phylogenetic origins, with group A being the most prevalent. In contrast, UPEC strains primarily grouped within phylogenetic group B2. This pattern of phylogenetic distribution among APEC and UPEC strains has been consistently observed across multiple research studies (Malema et al., 2018; Rocha et al., 2021; Ghorbani et al., 2022). This phenomenon underscores the significance of phylogenetic characterisation in understanding the epidemiology and pathogenesis of different pathotypes of E. coli, shedding light on potential evolutionary and ecological dynamics shaping their distribution and virulence.
Interestingly, InPEC strains were found in lower numbers (31.3%) compared to ExPEC, with ETEC (23%), STEC (3%), and EPEC (6%) pathotypes being the most predominant. Intestinal diseases caused by InPEC play a significant role in burdening low-income countries with infections (Rojas-Lopez et al., 2018). ETEC, EPEC, and STEC are known to be found in various animal species, including bovine, porcine, ovine or caprine, cats, and dogs, with bovine recognised as the major reservoir of these pathotypes. These three pathotypes are members of the diarrheagenic E. coli (DEC) group, majority of the samples were these pathotypes were identified were from animal meat that is used as a source of protein. It has been observed that ETEC and EPEC pathotype tend to be the most frequently isolated pathotypes which are known to cause severe diarrhoea, this has been observed in South Africa were farming of domestic animals occurs (García et al., 2018; Abdalla et al., 2022). Presence of these pathotype constitutes as a major public health risk since majority of these pathotypes displayed resistance to various antibiotics including tetracycline, cotrimoxazole, chrloramphenicol, penicillin, sulfamethazine and amoxycillin (Eagar and Naidoo, 2017). It has been observed that high prevalence of these pathotypes is due to contamination of food products during processing (Tanih et al., 2015).
The occurrence of both InPEC and ExPEC pathotypes has been described in other studies (Masters et al., 2011; Omolajaiye et al., 2020). In this study, a higher prevalence of ExPEC may indicate that pathogenic E. coli is currently persisting in various sources of food and animals, suggesting a higher potential for health risks associated with foods consumed by humans. In South Africa, the presence of both InPEC and ExPEC has been detected in treated effluents (Omolajaiye et al., 2020), highlighting the importance of comprehensive surveillance and control measures to mitigate the spread of pathogenic E. coli strains in both environmental and food contexts.
In this study, all the E. coli strains were subjected to serotyping using an in silico technique called serotype finder. In the past, O (somatic) and H (flagellar) antigens were utilised for serotyping E. coli isolates as either pathogenic or non-pathogenic (Joensen et al., 2015). Presently, there are 187 O antigens and 53 H antigens (Mare et al., 2021). The O antigen constitutes part of the lipopolysaccharide layer, which forms the outer membrane of E. coli, while the H antigen is the flagellar antigen responsible for E. coli’s motility. Both are considered major surface antigens (Royer et al., 2021). In this study, 21 O antigens were identified: O2, O6, O9, O50, and O101 each contributed to 19% of the total O antigens, making them the most prevalent. This was followed by O8, O11, and O64, which accounted for 10%. Nineteen H antigens were detected, with H4 being the most prevalent, followed by H37 (21%), H16 (16%), and H9, H10, H19, and H27 each contributing 10.5% of the total H antigen. Predominant combinations of serotypes in this study included O6:H5, O9:H37 and O50:H4. Serotype O6:H5 has been linked to causing UTI in humans and domestic animals such as cats and dogs were O6 is found to be most predominant antigen (Johnson et al., 2001; Ksiezarek et al., 2021). There are not that many reports of serotype O9:H37 and O50:H4, but both have been isolated from poultry farms in China that keep goose and ducks (Shawa et al., 2021; Hu et al., 2022).
Among the predominant STs, all detected ST1858 strains were found to belong to the UPEC pathotype. One study conducted in Germany detected ST1858 from a catheter in a hospital with patients treated for UTI (Toval et al., 2014). ST975 belonged to the ETEC pathotype and was found to belong to phylogroup A. ST2952 belonged to phylogroup B1 and belonged to the ETEC pathotype, while ST10 belonged to phylogroup A and was associated with both APEC and UPEC. ST95, a group B2, was associated with the UPEC pathotype. ST88 belonged to phylogroups A and C and was associated with both APEC and ETEC. ST120 belonged to groups A and B1 and was associated with both UPEC and ETEC.
In porcine, ST88 belonging to phylogroup C has been observed, while phylogroup A associated with ST88 has been isolated in avian and are often commensal strains (Abraham et al., 2014; Maluta et al., 2014). Isolates which belonging to A and B1 are often associated with commensal E. coli which is often found in the gut of animals and humans, ST120 is considered a commensal that belongs to phylogroup A (Bahgat et al., 2023). In immunocompromised humans, phylogroup B1 has been associated with resistance to antibiotics and some clones of ST120 have been found to be susceptible (Das et al., 2013; Liu et al., 2015).
This study also detected various virulence factors associated with E. coli pathogenesis. Important virulence factor genes such as fimH, papA/papG, hlyA, usp, cnf1, iutA, fyuA, afa, ompT, sfa, chuA, vat, and fycV were detected in these isolates, and these genes are associated with the UPEC pathotype (Khairy et al., 2019). Virulence genes associated with the APEC pathotype were also detected, including iroN, iss, ompT, tsh, hlyF, cvaC, iutA, sfa, papGII, fimH, and nlpI. The ETEC pathotype presented genes such as elt and est genes, which are associated with diarrhoea in children in low-income countries. The ETEC isolates in this study possessed only the est gene (Singh et al., 2019; Higginson et al., 2022).
The STEC pathotype was detected in very low proportions and presented with the stx2 gene; stx1 was not detected in any of the isolates. The presence of the stx2 gene is often associated with undercooked food or contaminated water, and the disease caused by STEC is characterised by haemolytic uremic syndrome, bloody diarrhoea, and haemorrhagic colitis (Alfinete et al., 2022). The EPEC pathotype presented with the eae gene, which is known for causing severe diarrhoea in children in developing countries, and mortalities in children have also been observed (Bolukaoto et al., 2021).
PathogenFinder, a web-server that can identify genetic characteristics linked to pathogenic and non-pathogenic isolates was used to determine if the isolates in this study have the potential to be pathogenic to humans. It is noted that some isolates may appear to be non-pathogenic, when introduced into favourable environments they may become opportunistic and become pathogenic (Cosentino et al., 2013). All the isolates in this study matched to various pathogenic of E. coli strains with various protein families identified which are linked to E. coli pathogenicity. This led to the prediction that all the isolates in this study have a very high probability (>0.90) of being human pathogens even though only a few indicated to be resistant to multiple antibiotics. In addition, these findings were supported by the virulence characteristics identified using virulencefinder (Montso et al., 2022; Wang et al., 2024).
Four CRISPR loci were found in E. coli: CRISPR 1, 2, 3, and 4. Depending on the presence of the corresponding cas genes, these loci are categorised as Type I-E (CRISPR 1 and 2) or Type I-F (CRISPR 3 and 4) (Xue and Sashital, 2019). In this study, CRISPR 1 was the most prevalent (86%) system, and it had a variable number of CRISPR arrays. CRISPR 3 loci were detected in very low numbers (11%). CRISPR2 and CRISPR4 systems were not detected. These arrays provide RNA molecules with a pattern to follow, enabling CRISPR-associated (Cas) proteins to precisely destroy viruses or bacteria upon re-infection. Only Cas1 and Cas2 are needed for the genetic recording of infections by obtaining spacers from DNA invaders. Despite the diversity observed between mobile genetic elements and CRISPR systems, nearly all known CRISPR-Cas systems share significant conservation in Cas1 and Cas2. Currently, CRISPR-Cas systems are classified into class 1 and 2, which are further subdivided into six types, type I-VI, and thirty-three subtypes (Makarova et al., 2015; Koonin et al., 2017; Murugan et al., 2017).
CRISPR-Cas systems play a crucial role in restricting phage infection and proliferation, serving as a crucial component of bacteriophage resistance mechanisms. These mechanisms enable bacteria to detect and remove phage DNA upon reinfection by gathering and preserving genetic material from previous phage interactions (Oluwarinde et al., 2023). The CRISPR-cas3 protein was the most predominant (83%) of all the CRISPR-cas proteins. Due to the helicase and nuclease activity offered by the CRISPR-cas3 protein, bacteria are well protected against phage attacks, increasing bacteria’s chances of survival and prolonging its viability (Montso et al., 2022).
All the isolates presented with either the CRISPR or cas genes, there was no significant difference observed within the CRISPR-cas system between isolates which showed MDR (20%) and those that did not present with MDR. In this study, it was observed that 80% of the isolates were resistant to fewer antibiotics, while 20% of the were deemed to be multidrug resistant isolates, but no clear difference could be identified. It has been investigated in several studies with E. coli that there is no significant correlation observed between the CRISPR-cas system as well as antibiotic resistance (Touchon et al., 2012; Toro et al., 2014). Studies have shown that the type 1 CRISPR present in E. coli isolates has the ability to prevent a pathogen from acquiring plasmids that are resistant to antibiotics (Tao et al., 2022).
The type CRISPR type I-F constitutes 11% of the strains in the study, isolates belonging to this type were found to group with phylogroup B2 and the isolates were resistant to three or more antibiotics, other B2 phylogroups were found in CRISPR type I-E. Strains within type I-F are often associated with UPEC pathotype and are associated with causing urinary tract infections. Only one isolate within the type I-F which forms part of the MDR strains had plasmid IncFII, other isolates within this type did not present with any plasmid. This may support the finding that the type I-F CRISPR system does interfere with survival of plasmids responsible for antimicrobial resistance (Almendros et al., 2012; Aydin et al., 2017). The type I-E CRISPR system is the most common type to be found in E. coli, this system is considered to be inactive in E. coli since its spacers are not suitable for attacking the viruses that bacteria encounters (Dion et al., 2024). In this study, only four isolates were found to contain both type I-E and I-F CRISPR, this is very uncommon, this may be attributed to by gene flow or ecological diversity of these bacteria (García-Gutiérrez et al., 2015).
Twelve resistance genes were detected in this study, with the most predominant resistance gene being the sitABCD disinfectant gene, which builds the ABC transporter system responsible for causing resistance against hydrogen peroxide (Sabri et al., 2006; Al-Mustapha et al., 2022). The disinfectant qacE was found in 14% of the isolates on IS26, and it is responsible for resistance to quaternary ammonium compounds. Resistance against qacE has been observed in the food industry, where such chemical agents are used (Zou et al., 2014). The resistance gene tet(B) was detected in 23% of the isolates, with no MGE surrounding it. This gene confers resistance to tetracycline and encodes an efflux pump that plays a crucial role in the ability to cause resistance to antibiotics (Arredondo et al., 2019).
The high occurrence of resistance against tetracycline in these isolates may suggest overuse of antibiotics (Jaja et al., 2020). Sulphonamides resistance genes sul1 (9%) and sul2 (6%) were also detected, along with beta-lactam resistance genes blaTEM-1B (9%) and blaTEM-104 (3%). Aminoglycoside resistance genes aadA1 (6%), aadA2b (6%), aph(6)-Id, and aph(3’’)-Ib (6%) were also observed. All the blaTEM-1B genes were found on the transposable element Tn2. Any form of TEM-1 is encoded by either Tn1, Tn2, or Tn3 resistance transposons (Partridge and Hall, 2005; Stephens et al., 2020). Resistance gene sul1, aadA1, qacE were found either on insertion elements IS26 or Tn6196. Tn6196 were found on plasmid IncHI2A (Johansson et al., 2021), IS26 is one of the elements known to speed up transmission of antimicrobial resistance genes in various communities of microorganisms (Behera et al., 2023).
Sulfonamides resistance genes such as the sul1and sul2 are known to confer resistance to sulphonamides antibiotics, these genes can be transferred from commensal bacteria through mobile genetic elements to humans by consumption of meat-based products and in turn persists to more virulence bacteria in the human gut (Soufi et al., 2011). Sulfonamide antibiotics are known to inhibit the enzyme dihydropteroate synthase (DHPS) which is important for synthesis of folate in bacteria. Through the process of horizontal gene transfer, resistance to sulphonamides often occurs (Capasso and Supuran, 2014). Though in this study there was less prevalence of these genes, they are commonly known to be found in high prevalence in domesticated animals, humans and in environments were aquatic animals are cultivated (Jiang et al., 2019). Aminoglycosides are important antibiotics used for various treatments of bacterial infections in humans and animals. Resistance to this antibiotic occurs when the Aminoglycoside nucleotidyltransferases (ANTs) enzymes inactivate this antibiotics. Aminoglycosides molecules have different adenylation positions and there are five classes of ANT enzymes which are responsible for targeting these positions for inactivation. There are also Aminoglycoside phosphotransferases (APHs) enzyme with a specific focus on disabling the antibiotics ability to bind to bacteria (Van Duijkeren et al., 2019).
The mobile genetic elements and plasmids detected in these isolates exhibited significant diversity. Among them, the IncFII, IncFIB, Col440, and Col156 plasmid replicons were the most common. Notably, the IncFIB plasmid replicon is known to carry genes conferring resistance to cephalosporin, a phenomenon observed particularly among poultry farmers in Nigeria (Al-Mustapha et al., 2023), in silico analysis of antibiograms within this study did not detect any isolates resistance to cephalosporins or carbapenems. Additionally, the IS5 element is associated with mcr-9 and blaCTX-M55 resistance mechanisms (Mbanga et al., 2021).
The need for heightened surveillance is underscored by the emergence of antibiotic resistance in diseases affecting both humans and animals. E. coli has long been recognised as a gram-negative indicator bacterium for antibiotic resistance due to its abundance in humans and various animal species, making it a potential vehicle for the spread of resistance genes between organisms (Chantziaras et al., 2014; Jaja et al., 2020). Significant phenotypic resistance was observed, particularly for penicillin, cotrimoxazole, amoxicillin, hydrogen peroxide, and sulfamethazine. The fact that these antibiotics are easily accessible could explain the degree of resistance observed especially in penicillin especially in settings where it is used as prophylactics to treat diseases or as a growth promoter (Kazemnia et al., 2014; Deekshit and Srikumar, 2022). Among the list of priority pathogens that the World health organisation has, E. coli forms part of the pathogens that are known to harbour cryptic resistance genes, these genes are present on the bacteria but do not exhibit the corresponding phenotypic resistance. This suggests that under certain conditions some genes remain silent but when transferred to a new host they become activated (Deekshit and Srikumar, 2022). This phenomena of silent resistance genes has been observed in a number of studies relating E. coli (Zhao et al., 2001; Enne et al., 2006; Li et al., 2014). Antibiotic resistance patterns may vary between animal populations and regions. Studies conducted in Africa have highlighted high levels of antimicrobial resistance in foods derived from animal products, particularly against antibiotics such as tetracycline and sulfamethazine, which are crucial in both human and veterinary medicine (Wesonga et al., 2010; Donkor et al., 2012; Alonso et al., 2017; Jaja et al., 2020).
This study focused on a small subset of generic E. coli; thus, its findings may not be generalised for all E. coli pathotypes. However, this study adds to the knowledge that pathogenic E. coli can survive and be disseminated in the animal specimen, meat and meat products, which is a public health concern. The findings on the mobile genetic elements detected suggests a potential for horizontal gene transfer and spread of resistance genes of E. coli of which some may encode most of the significant virulence factors which affects humans. Some of the virulence factors identified include genes associated with adhesion, invasion and survival and they mainly belong to the ExPEC pathotype which are indicative of a pathogenic potential. The study also identified prevalence of antimicrobial resistance genes which encode for beta-lactams, tetracycline, sulphonamides and disinfectants, these findings suggests that an active surveillance on drug usage is required, and new strategies for control measures for E. coli infections are required. Results presented on this study were mainly from domestic animals that are used as a source of protein, these results demonstrated that there is a high prevalence rate of E. coli isolates, and it is mainly from animal-based food products, mainly poultry, porcine and bovine. More comprehensive studies are required to characterise resistant E. coli from animal-based food products with specific focus on monitoring virulence traits and the genetic traits associated with pathogenicity as well the risk factors posed by this bacterium.
The original contributions presented in the study are publicly available. This data can be found here: NCBI BioProject,PRJNA1126085.
The animal study was approved by University of South Africa, Department of Life and Consumer Sciences Research Ethics Committee (2023/CAES_HREC/010). The study was conducted in accordance with the local legislation and institutional requirements.
RM: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. RP: Conceptualization, Supervision, Writing – review & editing. KM: Conceptualization, Funding acquisition, Writing – review & editing. TM: Writing – review & editing, Conceptualization. IM: Conceptualization, Data curation, Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors are grateful to the Agricultural Research Council-Onderstepoort Veterinary Research Feed and Food laboratory staff that processed the samples.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The views expressed by the authors do not necessarily reflect those of the South African Department of Agriculture, Land Reform and Rural Development.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbrio.2024.1432292/full#supplementary-material
Abdalla S. E., Abia A. L., Amoako D. G., Perrett K., Bester L. A., Essack S. Y. (2022). Food animals as reservoirs and potential sources of multidrug-resistant diarrheagenic E. coli pathotypes: Focus on intensive pig farming in South Africa. Onderstepoort J. Vet. Res. 89 (1), 1963.
Abdulabbas H. S., Al-Khafaji N., Abed S. Y., Al-Dahmoshi H., Al-Baroody H. N. (2023). Phylotypes and Pathotypes of Diarrheagenic Escherichia coli of Gastroenteritis. doi: 10.5772/intechopen.109860
Abraham S., Trott D. J., Jordan D., Gordon D. M., Groves M. D., Fairbrother J. M., et al. (2014). Phylogenetic and molecular insights into the evolution of multidrug-resistant porcine enterotoxigenic Escherichia coli in Australia. Int. J. Antimicrob. Agents. 44 (2), 105–111.
Aguirre-Sánchez J. R., Valdez-Torres J. B., Del Campo N. C., Martínez-Urtaza J., Del Campo N. C., Lee B. G., et al. (2022). Phylogenetic group and virulence profile classification in Escherichia coli from distinct isolation sources in Mexico. Infection Genet. Evolution 106, 105380. doi: 10.1016/j.meegid.2022.105380
Alfinete N. W., Bolukaoto J. Y., Heine L., Potgieter N., Barnard T. G. (2022). Virulence and phylogenetic analysis of enteric pathogenic Escherichia coli isolated from children with diarrhoea in South Africa. Int. J. Infect. Dis. 114, 226–232. doi: 10.1016/j.ijid.2021.11.017
Allard M. W., Bell R., Ferreira C. M., Gonzalez-Escalona N., Hoffmann M., Muruvanda T., et al. (2018). Genomics of foodborne pathogens for microbial food safety. Curr. Opin. Biotechnol. 49, 224–229.
Almendros C., Guzman N. M., Diez-Villasenor C., García-Martínez J., Mojica F. J. (2012). Target motifs affecting natural immunity by a constitutive CRISPR-Cas system in Escherichia coli. PloS One 7 (11), e50797.
Al-Mustapha A. I., Alada S. A., Raufu I. A., Lawal A. N., Eskola K., Brouwer M. S., et al. (2022). Co-occurrence of antibiotic and disinfectant resistance genes in extensively drug-resistant Escherichia coli isolated from broilers in Ilorin, North Central Nigeria. J. Global antimicrobial. resistance 31, 337–344. doi: 10.1016/j.jgar.2022.11.002
Al-Mustapha A. I., Raufu I. A., Ogundijo O. A., Odetokun I. A., Tiwari A., Brouwer M. S., et al. (2023). Antibiotic resistance genes, mobile elements, virulence genes, and phages in cultivated ESBL-producing Escherichia coli of poultry origin in Kwara State, North Central Nigeria. Int. J. Food Microbiol. 389, 110086.
Alonso C. A., Zarazaga M., Ben Sallem R., Jouini A., Ben Slama K., Torres C. (2017). Antibiotic resistance in Escherichia coli in husbandry animals: the African perspective. Lett. Appl. Microbiol. 64, 318–334. doi: 10.1111/lam.12724
Andrews S. (2010). FastQC, a Quality Control tool for High Throughput Sequence Data (Cambridge: Babraham Institute).
Argimón S., David S., Underwood A., Abrudan M., Wheeler N. E., Kekre M., et al. (2021). Rapid genomic characterization and global surveillance of Klebsiella using Pathogenwatch. Clin. Infect. Dis. 73 (Supplement_4), S325–S335.
Arredondo A., Àlvarez G., Nart J., Mor C., Blanc V., León R. (2019). Detection and expression analysis of tet(B) in Streptococcus oralis. J. Oral. Microbiol. 11, 1643204. doi: 10.1080/20002297.2019.1643204
Aydin S., Personne Y., Newire E., Laverick R., Russell O., Roberts A. P., et al. (2017). Presence of Type IF CRISPR/Cas systems is associated with antimicrobial susceptibility in Escherichia coli. J. Antimicrob. Chemother. 72 (8), 2213–2218.
Badi S., Cremonesi P., Abbassi M. S., Ibrahim C., Snoussi M., Bignoli G., et al. (2018). Antibiotic resistance phenotypes and virulence-associated genes in Escherichia coli isolated from animals and animal food products in Tunisia. FEMS Microbiol. letters 365, fny088. doi: 10.1093/femsle/fny088
Bahgat O. T., Rizk D. E., Kenawy H. I., Barwa R. F. (2023). Characterization of Non-O157 Enterohemorrhagic Escherichia coli isolates from clinical and environmental sources.
Behera M., Parmanand, Roshan M., Rajput S., Gautam D., Vats A., et al. (2023). Novel aadA5 and dfrA17 variants of class 1 integron in multidrug-resistant Escherichia coli causing bovine mastitis. Appl. Microbiol. Biotechnol. 107, 433–446. doi: 10.1007/s00253-022-12304-3
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bolukaoto J. Y., Singh A., Alfinete N., Barnard T. G. (2021). Occurrence of hybrid diarrhoeagenic Escherichia coli associated with multidrug resistance in environmental water, Johannesburg, South Africa. Microorganisms 9, 2163. doi: 10.3390/microorganisms9102163
Bortolaia V., Kaas R. S., Ruppe E., Roberts M. C., Schwarz S., Cattoir V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrobial. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Capasso C., Supuran C. T. (2014). Sulfa and trimethoprim-like drugs–antimetabolites acting as carbonic anhydrase, dihydropteroate synthase and dihydrofolate reductase inhibitors. J. Enzyme inhibition medicinal Chem. 29, 379–387. doi: 10.3109/14756366.2013.787422
Chantziaras I., Boyen F., Callens B., Dewulf J. (2014). Correlation between veterinary antimicrobial use and antimicrobial resistance in food-producing animals: a report on seven countries. J. Antimicrobial. Chemother. 69, 827–834. doi: 10.1093/jac/dkt443
Chapman T. A., Wu X. Y., Barchia I., Bettelheim K. A., Driesen S., Trott D., et al. (2006). Comparison of virulence gene profiles of Escherichia coli strains isolated from healthy and diarrheic swine. Appl. Environ. Microbiol. 72, 4782–4795. doi: 10.1128/AEM.02885-05
Clermont O., Dixit O. V., Vangchhia B., Condamine B., Dion S., Bridier-Nahmias A., et al. (2019). Characterization and rapid identification of phylogroup G in Escherichia coli, a lineage with high virulence and antibiotic resistance potential. Environ. Microbiol. 21 (8), 3107–3117.
Cosentino S., Voldby Larsen M., Møller Aarestrup F., Lund O. (2013). PathogenFinder-distinguishing friend from foe using bacterial whole genome sequence data. PloS One 8, e77302. doi: 10.1371/annotation/b84e1af7-c127-45c3-be22-76abd977600f
Das P., Singh A. K., Mukherjee S., Rajendran K., Saha D. R., Koley H., et al. (2013). Composition of Escherichia coli population in the neonatal gut: phylogroups and virulence determinants. J. Med. Microbiol. 62 (11), 1680–1687.
Deekshit V. K., Srikumar S. (2022). ‘To be, or not to be’—The dilemma of ‘silent’antimicrobial resistance genes in bacteria. J. Appl. Microbiol. 133, 2902–2914. doi: 10.1111/jam.15738
Dion M. B., Shah S. A., Deng L., Thorsen J., Stokholm J., Krogfelt K. A., et al. (2024). Escherichia coli CRISPR arrays from early life fecal samples preferentially target prophages. ISME J. 18 (1), wrae005.
Donkor E. S., Newman M. J., Yeboah-Manu D. (2012). Epidemiological aspects of non-human antibiotic usage and resistance: implications for the control of antibiotic resistance in Ghana. Trop. Med. Int. Health 17, 462–468. doi: 10.1111/j.1365-3156.2012.02955.x
Eagar H., Naidoo V. (2017). Veterinary antimicrobial stewardship in South Africa. Int. Biol. Rev. 1 (2).
Enciso-Martínez Y., González-Aguilar G. A., Martínez-Téllez M. A., González-Pérez C. J., Valencia-Rivera D. E., Barrios-Villa E., et al. (2022). Relevance of tracking the diversity of Escherichia coli pathotypes to reinforce food safety. Int. J. Food Microbiol. 374, p.109736. doi: 10.1016/j.ijfoodmicro.2022.109736
Enne V. I., Delsol A. A., Roe J. M., Bennett P. M. (2006). Evidence of antibiotic resistance gene silencing in Escherichia coli. Antimicrobial. Agents chemother. 50, 3003–3010. doi: 10.1128/AAC.00137-06
Galindo-Méndez M. (2020). “Antimicrobial resistance in Escherichia coli,” in E. Coli Infections-Importance of Early Diagnosis and Efficient Treatment, 1–20. doi: 10.5772/intechopen.93115
Gambushe S. M., Zishiri O. T., El Zowalaty M. E. (2022). Review of Escherichia coli O157: H7 prevalence, pathogenicity, heavy metal and antimicrobial resistance, African perspective. Infection Drug Resistance 15, 4645–4673. doi: 10.2147/IDR.S365269
García V., García-Meniño I., Mora A., Flament-Simon S. C., Díaz-Jiménez D., Blanco J. E., et al. (2018). Co-occurrence of mcr-1, mcr-4 and mcr-5 genes in multidrug-resistant ST10 Enterotoxigenic and Shiga toxin-producing Escherichia coli in Spain (2006-2017). Int. J. Antimicrob. Agents 52 (1), 104–108.
García-Gutiérrez E., Almendros C., Mojica F. J., Guzmán N. M., García-Martínez J. (2015). CRISPR content correlates with the pathogenic potential of Escherichia coli. PloS One 10, e0131935. doi: 10.1371/journal.pone.0131935
Gekenidis M. T., Qi W., Hummerjohann J., Zbinden R., Walsh F., Drissner D. (2018). Antibiotic-resistant indicator bacteria in irrigation water: high prevalence of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli. PloS One 13, e0207857. doi: 10.1371/journal.pone.0207857
Geurtsen J., de Been M., Weerdenburg E., Zomer A., McNally A., Poolman J. (2022). Genomics and pathotypes of the many faces of Escherichia coli. FEMS Microbiol. Rev. 46, fuac031. doi: 10.1093/femsre/fuac031
Ghorbani A., Khoshbakht R., Kaboosi H., Shirzad-Aski H., Ghadikolaii F. P. (2022). Study on antibiotic resistance and phylogenetic comparison of avian-pathogenic Escherichia coli (APEC) and uropathogenic Escherichia coli (UPEC) isolates. Veterinary Res. Forum 13, 569. doi: 10.30466/vrf.2021.527563.3160
Gurevich A., Saveliev V., Vyahhi N., Tesler G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Halaji M., Fayyazi A., Rajabnia M., Zare D., Pournajaf A., Ranjbar R. (2022). Phylogenetic group distribution of uropathogenic Escherichia coli and related antimicrobial resistance pattern: a meta-analysis and systematic review. Front. Cell. Infection Microbiol. 12, 790184. doi: 10.3389/fcimb.2022.790184
Higginson E. E., Sayeed M. A., Pereira Dias J., Shetty V., Ballal M., Srivastava S. K., et al. (2022). Microbiome profiling of enterotoxigenic Escherichia coli (ETEC) carriers highlights signature differences between symptomatic and asymptomatic individuals. MBio 13, e00157–e00122. doi: 10.1128/mbio.00157-22
Hu J., Afayibo D. J. A., Zhang B., Zhu H., Yao L., Guo W., et al. (2022). Characteristics, pathogenic mechanism, zoonotic potential, drug resistance, and prevention of avian pathogenic Escherichia coli (APEC). Front. Microbiol. 13, 1049391.
Igwaran A., Iweriebor B. C., Okoh A. I. (2018). Molecular characterization and antimicrobial resistance pattern of Escherichia coli recovered from wastewater treatment plants in Eastern Cape South Africa. Int. J. Environ. Res. Public Health 15, 1237. doi: 10.3390/ijerph15061237
Jaja I. F., Oguttu J., Jaja C. J. I., Green E. (2020). Prevalence and distribution of antimicrobial resistance determinants of Escherichia coli isolates obtained from meat in South Africa. PloS One 15, e0216914. doi: 10.1371/journal.pone.0216914
Jesser K. J., Levy K. (2020). Updates on defining and detecting diarrheagenic Escherichia coli pathotypes. Curr. Opin. Infect. diseases 33, 372. doi: 10.1097/QCO.0000000000000665
Jiang H., Cheng H., Liang Y., Yu S., Yu T., Fang J., et al. (2019). Diverse mobile genetic elements and conjugal transferability of sulfonamide resistance genes (sul1, sul2, and sul3) in Escherichia coli isolates from Penaeus vannamei and pork from large markets in Zhejiang, China. Front. Microbiol. 10, 1787. doi: 10.3389/fmicb.2019.01787
Joensen K. G., Scheutz F., Lund O., Hasman H., Kaas R. S., Nielsen E. M., et al. (2014). Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 52, 1501–1510. doi: 10.1128/JCM.03617-13
Joensen K. G., Tetzschner A. M., Iguchi A., Aarestrup F. M., Scheutz F. (2015). Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 53, 2410–2426. doi: 10.1128/JCM.00008-15
Johansson M. H., Bortolaia V., Tansirichaiya S., Aarestrup F. M., Roberts A. P., Petersen T. N. (2021). Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrobial. Chemother. 76, 101–109. doi: 10.1093/jac/dkaa390
Johnson J. R., Delavari P., Kuskowski M., Stell A. L. (2001). Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J. Infect. Dis. 183 (1), 78–88.
Johnson J. R., Russo T. A. (2002). Extraintestinal pathogenic Escherichia coli:”the other bad E coli. J. Lab. Clin. Med. 139, 155–162. doi: 10.1067/mlc.2002.121550
Kang H. J., Lee Y. J. (2022). Distribution of CRISPR in Escherichia coli isolated from bulk tank milk and its potential relationship with virulence. Animals 12, 503. doi: 10.3390/ani12040503
Kathayat D., Lokesh D., Ranjit S., Rajashekara G. (2021). Avian pathogenic Escherichia coli (APEC): an overview of virulence and pathogenesis factors, zoonotic potential, and control strategies. Pathogens. 10 (4), 467.
Kazemnia A., Ahmadi M., Dilmaghani M. (2014). Antibiotic resistance pattern of different Escherichia coli phylogenetic groups isolated from human urinary tract infection and avian colibacillosis. Iranian Biomed. J. 18, 219. doi: 10.6091/ibj.1394.2014
Khabo-Mmekoa C. M., Genthe B., Momba M. N. B. (2022). Enteric pathogens risk factors associated with household drinking water: A case study in Ugu District Kwa-Zulu Natal Province, South Africa. Int. J. Environ. Res. Public Health 19, 4431. doi: 10.3390/ijerph19084431
Khairy R. M., Mohamed E. S., Abdel Ghany H. M., Abdelrahim S. S. (2019). Phylogenic classification and virulence genes profiles of uropathogenic E. coli and diarrhegenic E. coli strains isolated from community acquired infections. PloS One 14, e0222441. doi: 10.1371/journal.pone.0222441
Koonin E. V., Makarova K. S., Zhang F. (2017). Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 37, 67–78. doi: 10.1016/j.mib.2017.05.008
Ksiezarek M., Novais A., Peixe L. (2021). The darkest place is under the candlestick-healthy urogenital tract as a source of UTI-related Escherichia coli lineages. bioRxiv, 2021–2008.
Larsen M. V., Cosentino S., Rasmussen S., Friis C., Hasman H., Marvig R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50 (4), 1355–1361.
Li X., Yao Z., Yuan L., Qi W., Shuchang A., Jichao C., et al. (2014). Identification of Klebsiella pneumoniae strains harboring inactive extended-spectrum beta-lactamase antibiotic-resistance genes. Chin. Med. J. 127, 3051–3057. doi: 10.3760/cma.j.issn.0366-6999.20140628
Liu C., Qin S., Xu H., Xu L., Zhao D., Liu X., et al. (2015). New Delhi metallo-β-lactamase 1 (NDM-1), the dominant carbapenemase detected in carbapenem-resistant Enterobacter cloacae from Henan Province, China. PloS One 10 (8), e0135044.
Louwen R., Staals R. H., Endtz H. P., van Baarlen P., van der Oost J. (2014). The role of CRISPR-Cas systems in virulence of pathogenic bacteria. Microbiol. Mol. Biol. Rev. 78, 74–88. doi: 10.1128/MMBR.00039-13
Lupindu A. M. (2017). “Isolation and characterization of Escherichia coli from animals, humans, and environment,” in Escherichia Coli-Recent Advances on Physiology, Pathogenesis and Biotechnological Applications (IntechOpen Limited, London, United Kingdom), 187–206.
Makarova K. S., Wolf Y. I., Alkhnbashi O. S., Costa F., Shah S. A., Saunders S. J., et al. (2015). An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 13, 722–736. doi: 10.1038/nrmicro3569
Malema M. S., Abia A. L. K., Tandlich R., Zuma B., Mwenge Kahinda J. M., Ubomba-Jaswa E. (2018). Antibiotic-resistant pathogenic Escherichia coli isolated from rooftop rainwater-harvesting tanks in the Eastern Cape, South Africa. Int. J. Environ. Res. Public Health 15, 892. doi: 10.3390/ijerph15050892
Maluta R. P., Logue C. M., Casas M. R. T., Meng T., Guastalli E. A. L., Rojas T. C. G., et al. (2014). Overlapped sequence types (STs) and serogroups of avian pathogenic (APEC) and human extra-intestinal pathogenic (ExPEC) Escherichia coli isolated in Brazil. PloS One 9 (8), e105016.
Manyi-Loh C. E., Lues R. (2023). A South African perspective on the microbiological and chemical quality of meat: plausible public health implications. Microorganisms 11, 2484. doi: 10.3390/microorganisms11102484
Mare A. D., Ciurea C. N., Man A., Tudor B., Moldovan V., Decean L., et al. (2021). Enteropathogenic Escherichia coli–a summary of the literature. Gastroenterol. Insights 12 (1), 28–40.
Martinez-Medina M. (2021). Pathogenic Escherichia coli: Infections and therapies. Antibiotics 10, 112. doi: 10.3390/antibiotics10020112
Massella E., Reid C. J., Cummins M. L., Anantanawat K., Zingali T., Serraino A., et al. (2020). Snapshot study of whole genome sequences of Escherichia coli from healthy companion animals, livestock, wildlife, humans and food in Italy. Antibiotics 9, 782. doi: 10.3390/antibiotics9110782
Masters N., Wiegand A., Ahmed W., Katouli M. (2011). Escherichia coli virulence genes profile of surface waters as an indicator of water quality. Water Res. 45, 6321–6333. doi: 10.1016/j.watres.2011.09.018
Mbanga J., Amoako D. G., Abia A. L., Allam M., Ismail A., Essack S. Y. (2021). Genomic insights of multidrug-resistant Escherichia coli from wastewater sources and their association with clinical pathogens in South Africa. Front. Veterinary Sci. 8, 636715. doi: 10.3389/fvets.2021.636715
Meena P. R., Priyanka P., Singh A. P. (2023). Extraintestinal pathogenic Escherichia coli (ExPEC) reservoirs, and antibiotics resistance trends: a one-health surveillance for risk analysis from “farm-to-fork. Lett. Appl. Microbiol. 76, ovac016. doi: 10.1093/lambio/ovac016
Meena P. R., Yadav P., Hemlata H., Tejavath K. K., Singh A. P. (2021). Poultry-origin extraintestinal Escherichia coli strains carrying the traits associated with urinary tract infection, sepsis, meningitis and avian colibacillosis in India. J. Appl. Microbiol. 130, 2087–2101. doi: 10.1111/jam.14905
Montso P. K., Bezuidenhout C. C., Mienie C., Somorin Y. M., Odeyemi O. A., Mlambo V., et al. (2022). Genetic diversity and whole genome sequence analysis data of multidrug resistant atypical enteropathogenic Escherichia coli O177 strains: An assessment of food safety and public health implications. Int. J. Food Microbiol. 365, 109555. doi: 10.1016/j.ijfoodmicro.2022.109555
Muloi D., Ward M. J., Pedersen A. B., Fevre E. M., Woolhouse M. E., van Bunnik B. A. (2018). Are food animals responsible for transfer of antimicrobial-resistant Escherichia coli or their resistance determinants to human populations? A systematic review. Foodborne Pathog. Dis. 15, 467–474. doi: 10.1089/fpd.2017.2411
Murphy R., Palm M., Mustonen V., Warringer J., Farewell A., Parts L., et al. (2021). Genomic epidemiology and evolution of Escherichia coli in wild animals in Mexico. Msphere 6, 10–1128. doi: 10.1128/mSphere.00738-20
Murugan K., Babu K., Sundaresan R., Rajan R., Sashital D. G. (2017). The revolution continues: newly discovered systems expand the CRISPR-Cas toolkit. Mol. Cell. 68, 15–25. doi: 10.1016/j.molcel.2017.09.007
Oluwarinde B. O., Ajose D. J., Abolarinwa T. O., Montso P. K., Du Preez I., Njom H. A., et al. (2023). Safety properties of escherichia coli O157: H7 specific bacteriophages: recent advances for food safety. Foods 12, 3989. doi: 10.3390/foods12213989
Omolajaiye S. A., Afolabi K. O., Iweriebor B. C. (2020). Pathotyping and antibiotic resistance profiling of Escherichia coli isolates from children with acute diarrhea in amatole district municipality of Eastern Cape, South Africa. BioMed. Res. Int. 2020 (1), 4250165. doi: 10.1155/2020/4250165
Pakbin B., Allahyari S., Amani Z., Brück W. M., Mahmoudi R., Peymani A. (2021). Prevalence, phylogroups and antimicrobial susceptibility of Escherichia coli isolates from food products. Antibiotics 10, 1291. doi: 10.3390/antibiotics10111291
Palma E., Tilocca B., Roncada P. (2020). Antimicrobial resistance in veterinary medicine: An overview. Int. J. Mol. Sci. 21, 1914. doi: 10.3390/ijms21061914
Parks D. H., Imelfort M., Skennerton C. T., Hugenholtz P., Tyson G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Partridge S. R., Hall R. M. (2005). Evolution of transposons containing bla TEM genes. Antimicrobial. Agents chemother. 49, 1267–1268. doi: 10.1128/AAC.49.3.1267-1268.2005
Peker N., Garcia-Croes S., Dijkhuizen B., Wiersma H. H., van Zanten E., Wisselink G., et al. (2019). A comparison of three different bioinformatics analyses of the 16S–23S rRNA encoding region for bacterial identification. Front. Microbiol. 10, 620. doi: 10.3389/fmicb.2019.00620
Prjibelski A. D., Puglia G. D., Antipov D., Bushmanova E., Giordano D., Mikheenko A., et al. (2020). Extending rnaSPAdes functionality for hybrid transcriptome assembly. BMC Bioinf. 21, 1–9. doi: 10.1186/s12859-020-03614-2
Ramos S., Silva V., Dapkevicius M. D. L. E., Caniça M., Tejedor-Junco M. T., Igrejas G., et al. (2020). Escherichia coli as commensal and pathogenic bacteria among food-producing animals: Health implications of extended spectrum β-lactamase (ESBL) production. Animals 10, 2239. doi: 10.3390/ani10122239
Riley L. W. (2020). Distinguishing pathovars from nonpathovars: Escherichia coli. Microbiol. spectrum 8, 8–4. doi: 10.1128/microbiolspec.AME-0014-2020
Robert E., Grippa M., Nikiema D. E., Kergoat L., Koudougou H., Auda Y., et al. (2021). Environmental determinants of E. coli, link with the diarrheal diseases, and indication of vulnerability criteria in tropical West Africa (Kapore, Burkina Faso). PloS Negl. Trop. Diseases 15, e0009634. doi: 10.1371/journal.pntd.0009634
Robins-Browne R. M., Holt K. E., Ingle D. J., Hocking D. M., Yang J., Tauschek M. (2016). Are Escherichia coli pathotypes still relevant in the era of whole-genome sequencing? Front. Cell. infection Microbiol. 6, 141. doi: 10.3389/fcimb.2016.00141
Rocha D. T. D., Salle F. D. O., Borges K. A., Furian T. Q., Nascimento V. P. D., Moraes H. L. D. S., et al. (2021). Avian pathogenic Escherichia coli (APEC) and uropathogenic Escherichia coli (UPEC): characterization and comparison. J. Infection Developing Countries 15, 962–971. doi: 10.3855/jidc.14217
Rojas-Lopez M., Monterio R., Desvaux M., Rosini R. (2018). Intestinal pathogenic Escherichia coli: Insights for vaccine development. Front. Microbiol. 9, 333650. doi: 10.3389/fmicb.2018.00440
Royer G., Darty M. M., Clermont O., Condamine B., Laouenan C., Decousser J. W., et al. (2021). Phylogroup stability contrasts with high within sequence type complex dynamics of Escherichia coli bloodstream infection isolates over a 12-year period. Genome Med. 13 (1), 77.
Sabri M., Léveillé S., Dozois C. M. (2006). A SitABCD homologue from an avian pathogenic Escherichia coli strain mediates transport of iron and manganese and resistance to hydrogen peroxide. Microbiology 152, 745–758. doi: 10.1099/mic.0.28682-0
Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shawa M., Furuta Y., Paudel A., Kabunda O. B., Mulenga E., Mubanga M., et al. (2021). Clonal relationship between multidrug-resistant Escherichia coli ST69 from poultry and humans in Lusaka, Zambia. FEMS Microbiol. Lett. 368 (21-24), fnac004.
Singh P., Metgud S. C., Roy S., Purwar S. (2019). Evolution of diarrheagenic Escherichia coli pathotypes in India. J. Lab. physicians 11, 346–351. doi: 10.4103/JLP.JLP_58_19
Soufi L., Sáenz Y., Vinué L., Abbassi M. S., Ruiz E., Zarazaga M., et al. (2011). Escherichia coli of poultry food origin as reservoir of sulphonamide resistance genes and integrons. Int. J. Food Microbiol. 144, 497–502. doi: 10.1016/j.ijfoodmicro.2010.11.008
Stephens C., Arismendi T., Wright M., Hartman A., Gonzalez A., Gill M., et al. (2020). F plasmids are the major carriers of antibiotic resistance genes in human-associated commensal Escherichia coli. Msphere 5, 10–1128. doi: 10.1128/mSphere.00709-20
Tanih N. F., Sekwadi E., Ndip R. N., Bessong P. O. (2015). Detection of pathogenic Escherichia coli and Staphylococcus aureus from cattle and pigs slaughtered in abattoirs in Vhembe District, South Africa. Sci. World J. 2015 (1), 195972.
Tao S., Chen H., Li N., Liang W. (2022). The application of the CRISPR-Cas system in antibiotic resistance. Infect. Drug Resist., 4155–4168.
Toro M., Cao G., Ju W., Allard M., Barrangou R., Zhao S., et al. (2014). Association of clustered regularly interspaced short palindromic repeat (CRISPR) elements with specific serotypes and virulence potential of Shiga toxin-producing Escherichia coli. Appl. Environ. Microbiol. 80 (4), 1411–1420.
Touchon M., Charpentier S., Pognard D., Picard B., Arlet G., Rocha E. P., et al. (2012). Antibiotic resistance plasmids spread among natural isolates of Escherichia coli in spite of CRISPR elements. Microbiology 158 (12), 2997–3004.
Toval F., Köhler C. D., Vogel U., Wagenlehner F., Mellmann A., Fruth A., et al. (2014). Characterization of Escherichia coli isolates from hospital inpatients or outpatients with urinary tract infection. J. Clin. Microbiol. 52, 407–418. doi: 10.1128/JCM.02069-13
Uelze L., Grützke J., Borowiak M., Hammerl J. A., Juraschek K., Deneke C., et al. (2020). Typing methods based on whole genome sequencing data. One Health Outlook 2, 1–19. doi: 10.1186/s42522-020-0010-1
Van Duijkeren E., Schwarz C., Bouchard D., Catry B., Pomba C., Baptiste K. E., et al. (2019). The use of aminoglycosides in animals within the EU: development of resistance in animals and possible impact on human and animal health: a review. J. Antimicrobial. Chemother. 74, 2480–2496. doi: 10.1093/jac/dkz161
Wang D., Ji X., Jiang B., Yuan Y., Liang B., Sun S., et al. (2024). Prevalence of Antibiotic Resistance and Virulence Genes in Escherichia coli Carried by Migratory Birds on the Inner Mongolia Plateau of Northern China from 2018 to 2023. Microorganisms 12 (6), 1076.
Wesonga S. M., Muluvi G. M., Okemo P. O., Kariuki S. (2010). Antibiotic resistant Salmonella and Escherichia coli isolated from indiginous Gallus domesticus in Nairobi, Kenya. East Afr. Med. J. 87, 205–210. doi: 10.4314/eamj.v87i5.63075
Whelan S., Lucey B., Finn K. (2023). Uropathogenic Escherichia coli (UPEC)-associated urinary tract infections: the molecular basis for challenges to effective treatment. Microorganisms 11 (9), 2169.
Xue C., Sashital D. G. (2019). Mechanisms of type IE and IF CRISPR-Cas systems in Enterobacteriaceae. EcoSal. plus 8, 10–1128. doi: 10.1128/ecosalplus.esp-0008-2018
Zhang Q., Ye Y. (2017). Not all predicted CRISPR–Cas systems are equal: isolated cas genes and classes of CRISPR like elements. BMC Bioinf. 18, 1–12. doi: 10.1186/s12859-017-1512-4
Zhao S., White D. G., Ge B., Ayers S., Friedman S., English L., et al. (2001). Identification and characterization of integron-mediated antibiotic resistance among Shiga toxin-producing Escherichia coli isolates. Appl. Environ. Microbiol. 67, 1558–1564. doi: 10.1128/AEM.67.4.1558-1564.2001
Zhao X., Zhao H., Zhou Z., Miao Y., Li R., Yang B., et al. (2022). Characterization of extended-spectrum β-lactamase-producing escherichia coli isolates that cause diarrhea in sheep in Northwest China. Microbiol. Spectrum 10, e01595–e01522. doi: 10.1128/spectrum.01595-22
Keywords: phylogroups, extra-intestinal pathogenic E. coli, intestinal pathogenic E. coli, sequence type, virulence genes, antimicrobial resistance genes, CRISPR system
Citation: Malesa R, Pierneef R, Magwedere K, Mafuna T and Matle I (2024) Genomic characterisation of generic Escherichia coli from food-producing animals and products of animal origin in South Africa. Front. Bacteriol. 3:1432292. doi: 10.3389/fbrio.2024.1432292
Received: 13 May 2024; Accepted: 01 July 2024;
Published: 16 July 2024.
Edited by:
Kumaragurubaran Karthik, Tamil Nadu Veterinary and Animal Sciences University, IndiaReviewed by:
Anca Farkas, Babeş-Bolyai University, RomaniaCopyright © 2024 Malesa, Pierneef, Magwedere, Mafuna and Matle. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Refiloe Malesa, bWFsb2tvdHNhckBhcmMuYWdyaWMuemE=; Itumeleng Matle, bWF0bGVpQGFyYy5hZ3JpYy56YQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.