- 1Department of Obstetrics and Gynecology, Institute of Medical Science, Banaras Hindu University (BHU), Varanasi, India
- 2Department of Neurology, All India Institute of Medical Sciences (AIIMS), Rishikesh, India
- 3Department of General Medicine, Institute of Medical Science, Banaras Hindu University (BHU), Varanasi, India
- 4Centre of Experimental Medicine and Surgery, Institute of Medical Sciences, Banaras Hindu University (BHU), Varanasi, India
Introduction: Enteric fever is widespread in many regions of developing countries. Despite low sensitivity, blood culture remains the gold standard diagnostic test for enteric fever. Diagnostic tests like Widal lack the desired specificity; hence, patients are overtreated many times. Inaccessibility to proper medical care in developing countries further poses a challenge to diagnosis by these conventional methods, promoting the needless intake of over-the-counter drugs by people. Although rapid kit-based tests are available, the reliability of these diagnostic tests in terms of specificity and sensitivity is quite variable. We aimed to validate the reliability of Typhipoint EIA (ELISA-based test) against blood clot nested PCR for enteric fever, as a gold standard, in view of the reported variable culture yield by calculating the sensitivity, specificity, and likelihood ratio.
Methods: A total of 100 patients were included in the study out of 152 patients screened, based on the inclusion criteria. The clinical profile of provisional enteric fever was recorded along with the amplification of the DNA fragment of flagellin (H1-d), and the stkG gene of Salmonella typhi and Salmonella paratyphi A, respectively, by nested PCR performed on blood clots, urine, and stool samples. Further validation of the ELISA-based test, i.e., Typhipoint EIA, was done considering nested PCR as a gold standard. The control group consisted of 40 healthy subjects.
Results: Nested PCR of the blood clots showed 84% positivity. Total culture positivity was found in 89 samples (combined), and among all samples for culture, clot culture was positive in 52 (52%), urine culture in 5 (5%), and stool culture in 32 (32%) cases. The total number of Typhipoint EIA IgM-positive cases was 83 (83%). The validation of Typhipoint EIA IgM showed 92.9% sensitivity and 68.8% specificity against blood clot PCR for Salmonella typhi.
Discussion: The Typhipoint EIA test for the diagnosis of enteric fever is quite sensitive as well as specific. It may be advised that two to three specific antigens of S. typhi should be spotted on the test kit for a satisfactory level of diagnosis of enteric fever in field conditions. This will help achieve the desired accuracy of the rapid test to avoid unnecessary antimicrobial therapy and costly investigations.
Introduction
Enteric fever is endemic in most developing countries. Incidence is 14 million cases every year, killing almost 0.13 to 0.2 million people, with India accounting for more than half of the total cases, i.e., 8.3 million cases and 72,000 fatalities, as per the global burden of disease 2017 (John et al., 2016; Stanaway et al., 2019), and it continues to maintain its pace like many past years. In 2019, the mean typhoid fever incidences in India accounted for 81–499 person-year per Lakh population as per the United Nations World Population Prospects database (https://population.un.org/wpp/). Severe enteric fever surveillance has also been conducted in India in hospital and laboratory-based settings for accurately assessing the incidence of enteric disease (John et al., 2018). The prevalence for laboratory-diagnosed cases of typhoid is 9.7% and paratyphoid is 0.9% (John et al., 2016). However, this figure is quite heterogeneous. The global case fatality rate (higher among children and the elderly) is approximately 0.95%. The bacteremia caused by different serovars of Salmonella enterica (Salmonella typhi and Paratyphi A, B, and C) has been studied in various contexts. Studies have shown that the virulence of different Salmonella serovars in chickens can be assessed using a bacteremia model (Millemann et al., 2005). FTIR spectroscopy has been used to identify typhoidal and para-typhoidal Salmonella serovars associated with bacteremia (Cordovana et al., 2021). Research on Malawian strains provides insights into genetic variations and host adaptation (Bronowski et al., 2013). The various diagnostic methods, including culture-based methods, immunomagnetic separation assays, whole-genome sequencing, Fourier-transform infrared spectroscopy, CRISPRs, and polymerase chain reaction (PCR), offer faster and more sensitive results but have limitations like high costs, specialized equipment, and potential non-vertical transmission events (Corrêa et al., 2018; Saha et al., 2023). It will be advantageous to make additional attempts to identify early infection biomarkers and distinguish between chronic illness from persistent active infection (Saha et al., 2023). A recent study developed a molecular classification assay for clade typing of Salmonella enterica serovar Enteritidis, responsible for gastroenteritis and nontyphoidal Salmonellosis (Gallichan et al., 2022). Not only this, but the diagnostic tool’s accuracy of antibody tests like TUBEX, Typhidot, and Test-iT is also quite heterogeneous in terms of sensitivity and specificity, as concluded in many meta-analyses (Arora et al., 2019). The Widal test has already lost its diagnostic value in developed countries in light of its low prevalence, availability of safe drinking water, and better laboratory bacterial isolation facilities (Mawazo et al., 2019). In developing countries, the relevance of the Widal test is being debated for its poor performance when compared with blood culture. Although bone marrow/blood culture has already been established as a gold standard for the diagnosis of typhoid and paratyphoid fever, its sensitivity is quite variable as reported in many studies taking laboratory technique as an important confounding factor (Stanaway et al., 2019). The epidemiology of enteric fevers in India is poorly understood due to the substantial correlation between enteric fevers, mortality, and morbidity in the country and the absence of reliable surveillance tests. However, surveillance studies have been conducted such as the “National Surveillance System for Enteric Fever in India” to find patients (especially children 6 months to 15 years of age) who might get neglected with facility-based monitoring systems; the study employs active case-seeking techniques in addition to surveillance program initiated for monitoring enteric fever from hospitals called “The Surveillance of Enteric Fever in Asia Project” (Saha et al., 2023). Generally, bone marrow culture/blood culture for the isolation of Salmonella typhi is a sensitive method and is suggested as the gold standard. However, bone marrow collection is a very painful procedure and demands technical expertise. Peripheral blood culture is relatively more practical but unfortunately lacks the desired sensitivity (Wain and Hosoglu, 2008). Furthermore, blood culture facilities are available only in tertiary-level hospitals in the majority of typhoid-endemic developing countries. Moreover, the culture isolation procedure takes at least 3–5 days to reach the final diagnosis for positive bacterial growth (Wain and Hosoglu, 2008). This delay in diagnosis may lead to the unnecessary administration of antibiotics or, sometimes, non-initiation of specific therapy. That is why there is an urgent need for reliable and rapid antibody-based tests to facilitate an early diagnosis of typhoid fever. There are many antibody-based tests (semiquantitative slide agglutination test, single-tube Widal test, TUBEX, Typhipoint EIA test, Typhidot test, etc.) that have been evaluated in typhoid-endemic areas with blood culture as the gold standard (Choo et al., 1994; Sánchez-Jiménez and Cardona-Castro, 2004). The nested PCR-based methods have been well established as the most sensitive and specific in the diagnosis of typhoid fever (Hatta and Smits, 2007; Pratap et al., 2013). Considering the aforementioned facts, we decided to conduct the current study to validate ELISA-based rapid diagnostic test Typhipoint EIA results against nested PCR for Salmonella typhi and paratyphi, considering nested PCR as a gold standard and compared it with the culture results.
Materials and methods
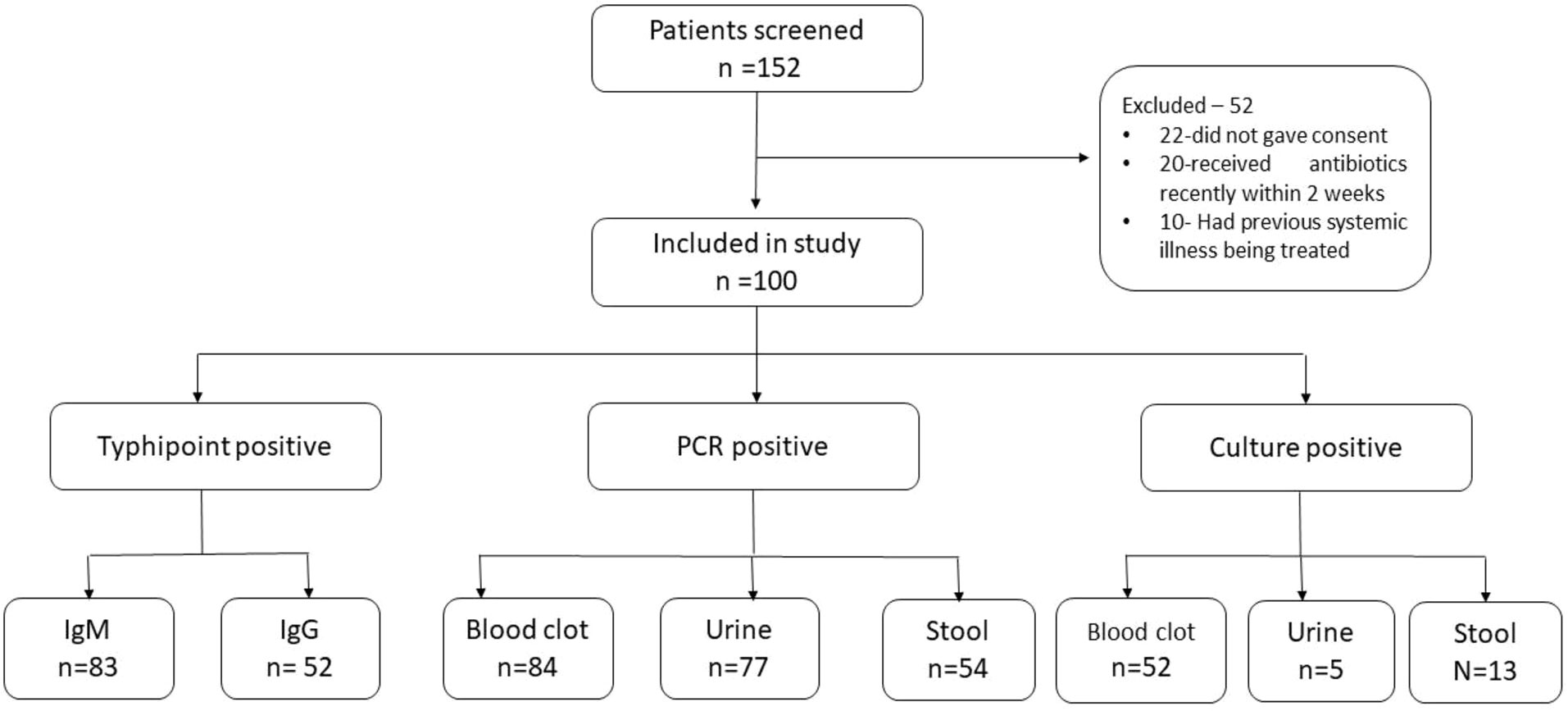
A total of 152 patients were screened, out of which 52 were excluded as per the pre-decided exclusion criterion (Figure 1). The final study population comprised 100 suspected cases of typhoid fever and 40 healthy non-febrile subjects from the same area belonging to the age group of 13–80 years. This study included patients who visited the outpatient department of medicine and also patients who were admitted to the ward of the department of medicine of the concerned hospital. Those who had received antibiotic treatment within 2 weeks before coming to the hospital or had a history of significant organ/system disease that interfered with the diagnosis were excluded. This tertiary-level hospital is situated in the eastern part of North India, on the basin of the river Ganga. The Ganga basin is known for being a typhoid-endemic region.
It was a diagnostic test accuracy (DTA) cross-sectional study. One part (blood clot culture) of the study was conducted as a part of the dissertation, and this part is still unpublished. It was conducted in collaboration with the Department of General Medicine and the Department of Microbiology, Institute of Medical Sciences, Banaras Hindu University, Varanasi-221005. Well-informed consent was obtained from each of the participants, and the study was approved by the Institute’s Ethics Committee (approval number: 291).
Collection of specimens
For evaluations, around 10 ml of blood sample was collected by venipuncture in a sterile tube with aseptic precaution, 5 g–10 g of stool specimen was collected in a sterile wide-mouth container, and around 100 ml of urine sample was collected in a sterile container from study subjects. Blood was allowed to clot at room temperature for 30 min, and the separated serum was stored at −20°C. Instead of the standard laboratory method for the culture of specimens, we followed the method previously reported in the same laboratory for a better culture yield (Ahirwar et al., 2014). To start with, the reference strain of S. typhi (MTCC 3216) was made in saline (0.85% NaCl) at a concentration of 3 × 108 CFU/ml. Around 10 ml of brain heart infusion broth was distributed in 19 different tubes with different pH values (i.e., 2.0, 2.5, 3.0, 3.5, 4.0, 4.5, 5.0, 5.5, 6.0, 6.5, 7.0, 7.5, 8.0, 8.5, 9.0, 9.5, 10.0, 10.5, and 11.0). Then, bacterial suspension (0.5 ml) was added to each of the above tubes, and subcultures were made at different time intervals (5, 10, 20, 30, 60, 120, and 180 min).
Extraction of genomic DNA from blood and stool specimens
Around 3 ml–5 ml of blood clot was subjected to DNA extraction by using the phenol-chloroform and proteinase-K method (Joseph and Russell David, 2006), whereas 3 g–5 g of stool was subjected to DNA extraction by a modified method to minimize carryover of PCR inhibitors (Hart et al., 2015). The additional steps in this protocol were to add 3 g–5 g of stool or deposit of 100-ml centrifuged urine to 10 ml of 10% formal saline (formaldehyde 40% W/V and 0.85% W/V NaCl) to make the suspension and addition of 3 ml of ether before centrifugation at 1,800 g for 5 min.
PCR and nested PCR
For detecting the presence of pathogen-specific gene sequences, 100 ng of each of the extracted DNA from blood, stool, and urine specimens were subjected separately to PCR amplification, targeting S. typhi-specific flagellin (fliC) gene sequences. Primers used were those designed by Song et al. (1993), which were further modified by Frankel et al. (1994) (Frankel et al., 1989; Song et al., 1993). For primary PCR amplification, forward primer 5'-ACTGCTAAAACCACTACT-3' (ST1) and reverse primer 5'-TTAACGCAGTAAAGAGAG-3' (ST2) were used, whereas for nested PCR amplification, forward primer 5'-AGATGGTACTGGCGTTGCTC-3' (ST3) and reverse primer 5'-TGGAGACTTCGGTCGCGTAG-3' (ST4) were used to amplify 343-bp and 229-bp nucleotide sequences, respectively. The amplification protocol was as described previously (Kumar et al., 2012).
Typhipoint ELISA for IgM and IgG
Typhipoint is an enzyme immunoassay (EIA) for the detection of IgM and IgG antibodies against the outer membrane protein of S. typhi. This test was performed strictly following the instructions given by the manufacturer (AB Diagnopath Manufacturing Pvt., Ltd., New Delhi, India). In brief: All the reagents were shaken well before the test procedure. On each of the test strips, 300 µl of sample diluent was put into the test well. To each well, 3 µl of serum sample or control was added. Incubation was done on the rocker platform for 15 min. The sample was aspirated, and 1× diluent was used for washing three times. Anti-human IgM and IgG antibodies were added to the respective wells. The strip was covered and incubated at room temperature for 30 min on the rocker platform. Washing was done again with the wash buffer three times for 5 min. Now 300 µl of color-development solution was added into each well, and it was incubated for 10 min. The reaction was stopped by aspirating the color-development solution. The strip was examined for the color developed by the naked eye (Choo et al., 1999; Olsen et al., 2004).
Statistical analysis
The prevalence of Typhoid cases, i.e., 20% in our hospital settings with a 10% margin of error, n = 64 (sample size), was taken. With the increase in implementing design effect, the resized sample size comes to 96, and considering 5% non-responsive patients, n = 100. The parameters of bio-statistics used to validate the test in relation to the most appropriate gold standard were sensitivity and specificity. Positive predictive value (PPV+), negative predictive value (NPV−), likelihood ratio for a positive test result (LR+), and likelihood ratio for a negative test result (LR−) were also calculated using “MedCalc,” a diagnostic test evaluation calculator from MedCalc Software Ltd. (https://www.medcalc.org/calc/diagnostic_test.php (Version 20.110).
Results
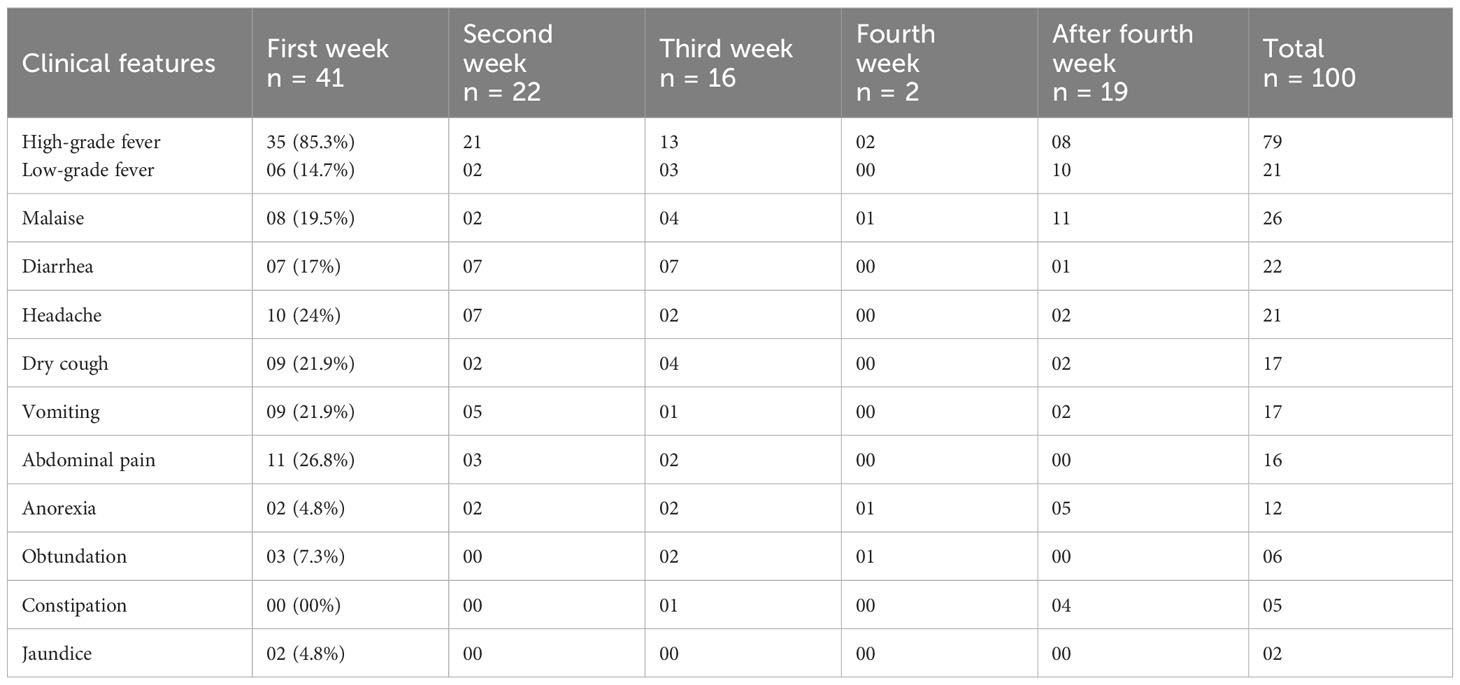
Out of the 152 patients screened, 100 were included in the study as per the inclusion criteria enumerated in Figure 1 without any regard for age or sex. The selected 100 patients were clinically suspected of having enteric fever. Fever, malaise, diarrhea, and headache were the most common presentations. The age range of the study population was 13 to 75 years; the mean age was 28.69 years (SD ±1.49), with the majority 59 (59%) falling into the age group of 13 to 25 years, followed by 31 (31%) between 26 and 50 years, and 10 (10%) were more than 50 years. The patient population had men and women in a ratio of 1.78:1 (64%:36%). With regard to the time of presentation (visit), 41 (41%) patients presented in the first week of illness, followed by 22 (22%), 16 (16%), 2 (2%), and 19 (19%) in the second, third, fourth, and after fourth weeks of illness, respectively (Table 1).
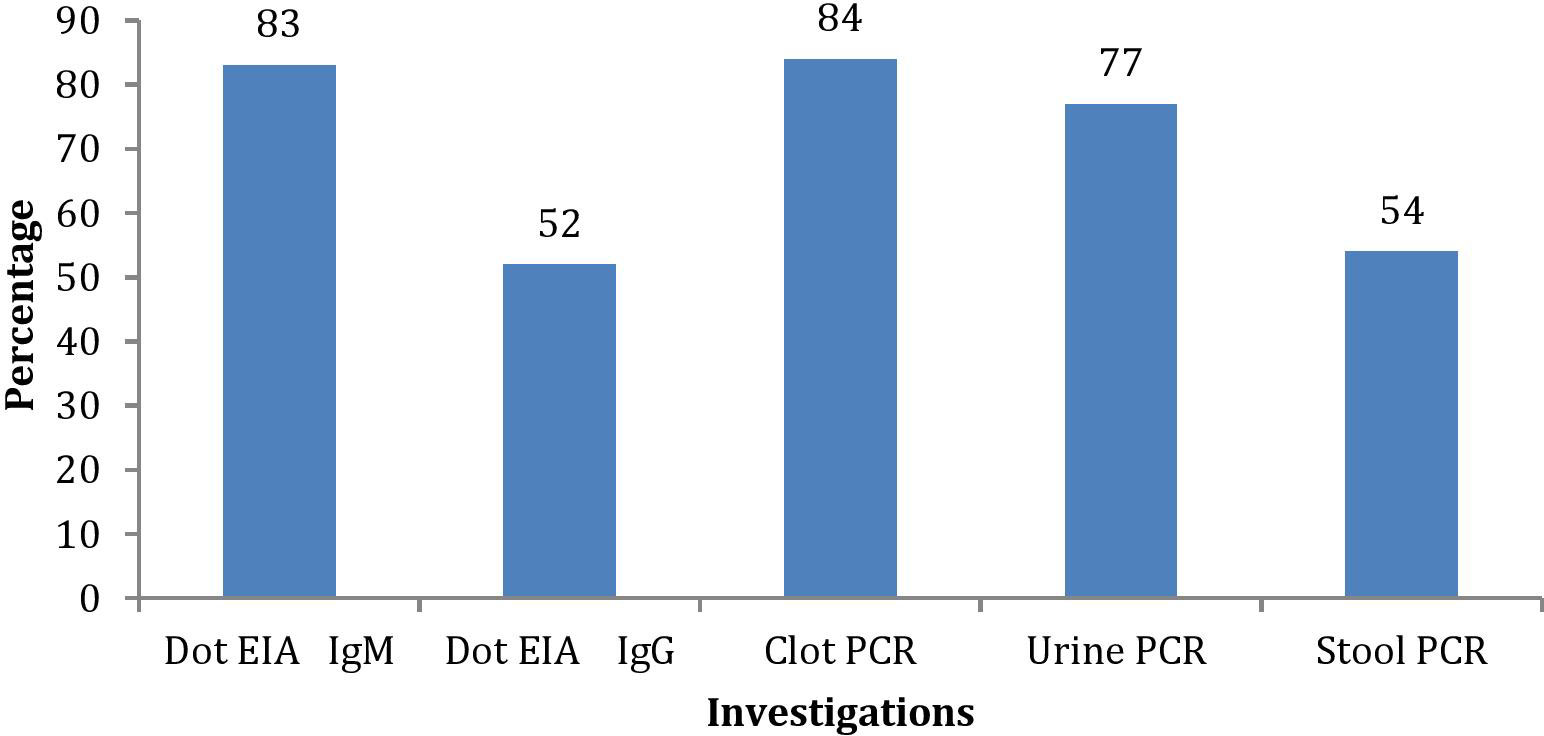
Among the investigations performed for the diagnosis of typhoid in suspected enteric fever patients, a total of 91 patients were either IgM or IgG positive (83 IgM positive and 8 only IgG positive). Blood clot culture (n = 52, 52%), stool culture (n = 32, 32%), and urine culture (n = 5, 5%) were positive in suspected cases of enteric fever (Figures 2, 3). Out of these, patient stool culture alone was positive in 13 (13%) and urine culture alone was positive in 5 (5%); hence, a total of 67 patients were detected positive by either clot, stool, or urine culture.
The nested polymerase chain reactions (nPCR) were done in blood clot samples of all suspected cases, and after the nested PCR, the amplified products were run on gel electrophoresis. The nPCR for Salmonella typhi specifically amplified the fli-C flagellin gene fragment of 343 bp (Figure 4), whereas the nPCR for Salmonella paratyphi A specifically amplified the stkG fimbrial gene fragment of 229 bp (Figure 5). Nested PCR was positive in 85 suspected cases of enteric fever (n = 85; 85%). We also did nPCR in urine and stool samples of all the suspected enteric fever cases, which showed positive results in 37 cases (37%) and 54 (54%) cases, respectively. It is pertinent to mention here that none of the urine, stool, and blood clot samples collected from afebrile healthy controls were found positive for S. typhi-specific amplification in nested PCR (Figures 4, 5). The Typhipoint EIA was performed on all the serum samples collected from suspected typhoid fever cases. A total of 83 (83%) were found to be positive for S. typhi-specific IgM antibodies. Among 17 antibody-negative samples, six patients were found to be positive upon S. typhi-specific blood clot nPCR amplification. On the other hand, five patients were observed to be positive for S. typhi IgM antibodies by the Typhipoint EIA test, and their specific amplification by nPCR was negative. Thus, 78 patients were found to be true positives when blood clot nPCR was taken as the gold standard.
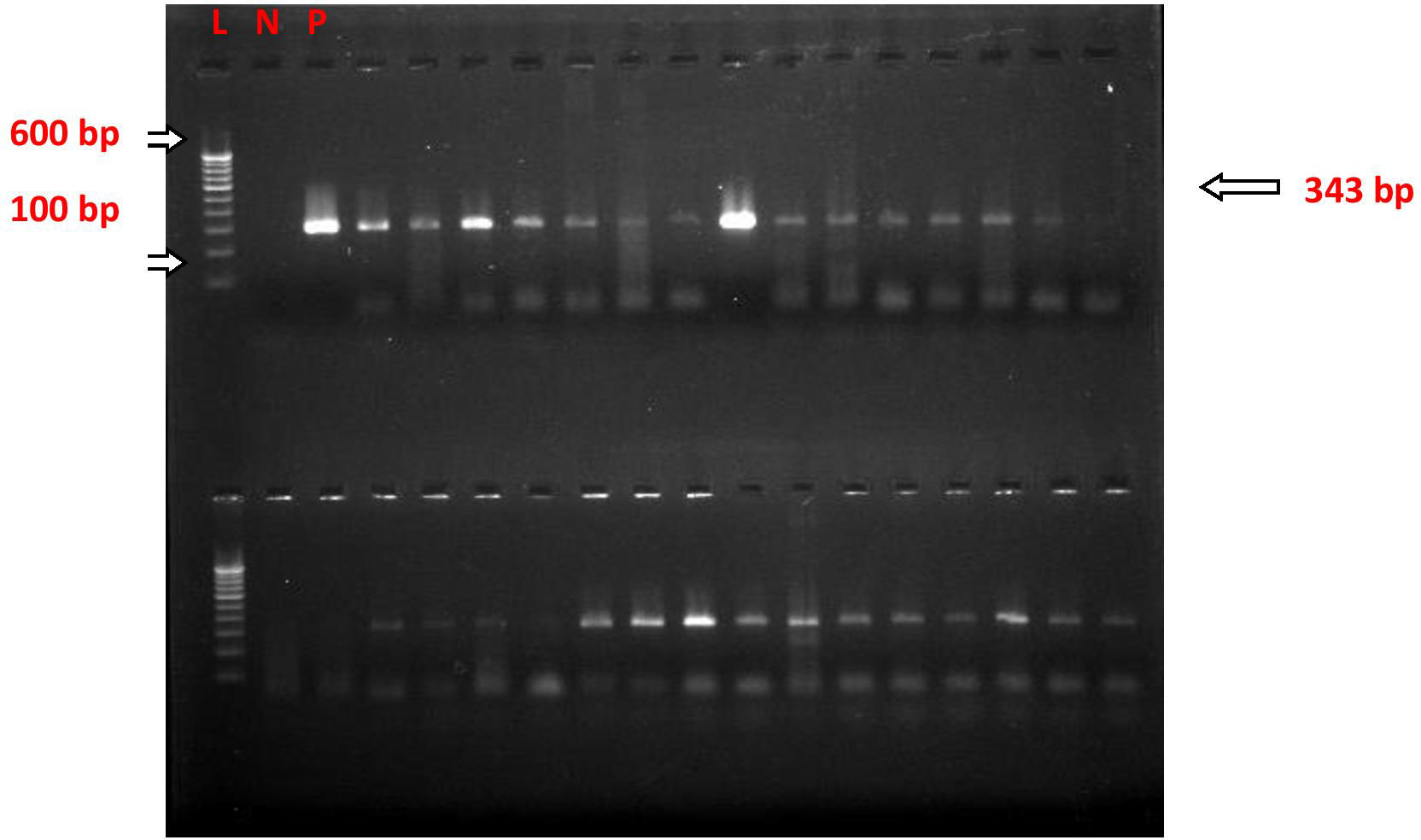
Figure 4 nPCR for Salmonella typhi fli-C gene (flagellin) Blood Clot Samples; after nested PCR, the PCR products were run on gel electrophoresis, and the image was taken in the multi-image light cabinet. L, ladder (molecular marker); N, negative control; P, positive control.
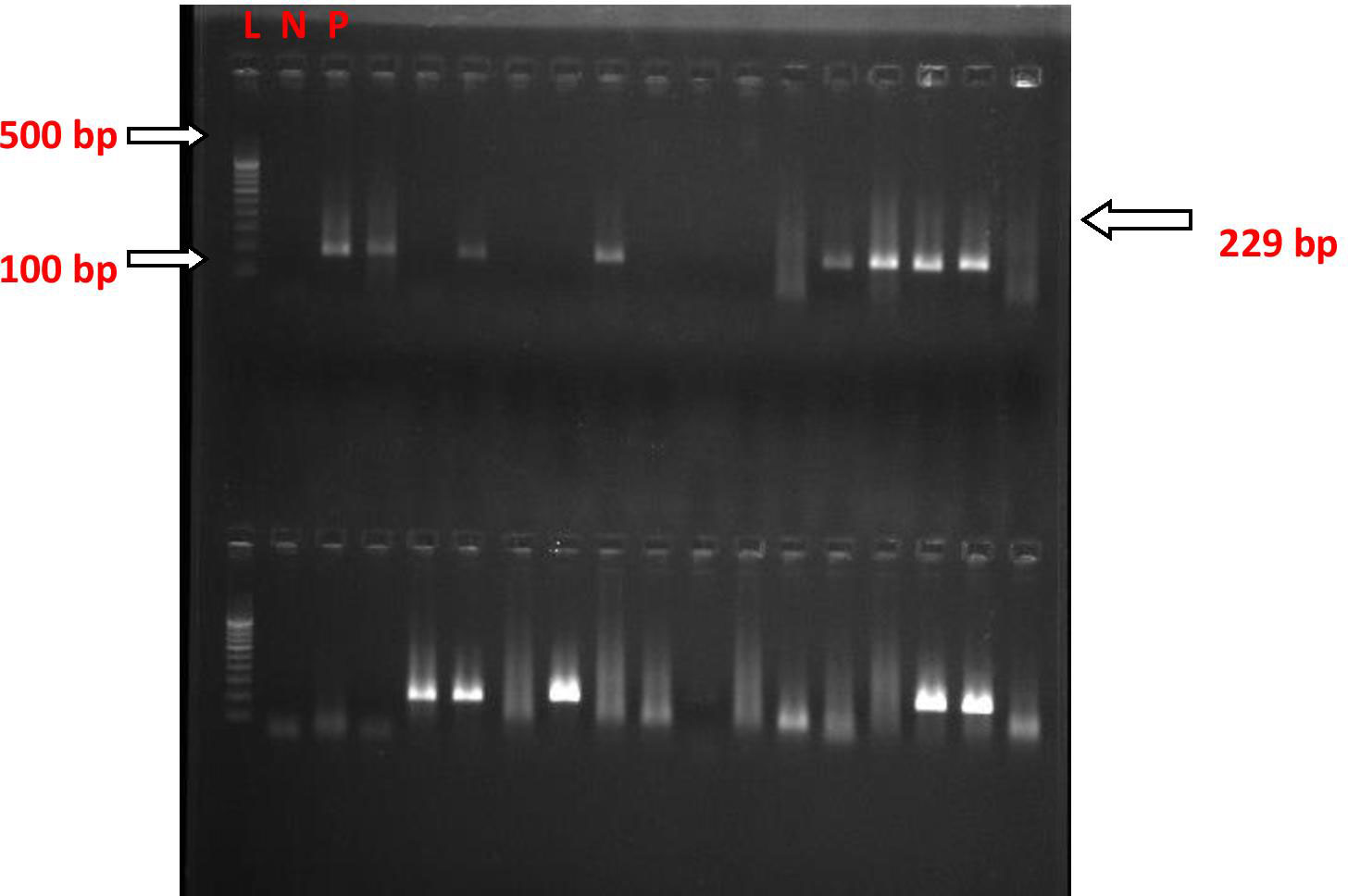
Figure 5 nPCR for the Salmonella paratyphi A stkG (fimbrial) gene in blood clot culture; after nested PCR, the PCR products were run on gel electrophoresis, and the image was taken in the multi-image light cabinet. L, ladder (molecular marker); N, negative control; P, positive control.
The Typhipoint EIA IgM antibody-positive results alone with blood clot nPCR for Salmonella typhi were validated (Tables 2, 3). The total number of either Typhipoint EIA IgM or IgG antibody-positive cases was 91%. The validation of Typhipoint EIA for either IgM or IgG antibody-positive cases for the diagnosis of enteric fever against clot nPCR Salmonella typhi as the gold standard showed a sensitivity of 97.62% (95% CI, 92% to 99%), but the specificity was reduced to 43.75% (95% CI, 20% to 70%). Since enteric fever also includes infection with Salmonella paratyphi, we validated Typhipoint EIA result against the total number of either S. typhi nPCR-positive cases or Salmonella paratyphi nPCR-positive cases (Figure 6). The Typhipoint EIA IgM antibody positivity as diagnostic indices showed a sensitivity of 91.76% (95% CI, 84% to 97%), and a specificity of 66.67% (95% CI, 38% to 88%), a positive predictive value of 93.9% (95% CI, 88% to 97%), and a negative predictive value of 58.8% (95% CI, 39% to 76%) (LR + 2.75, LR − 0.12).
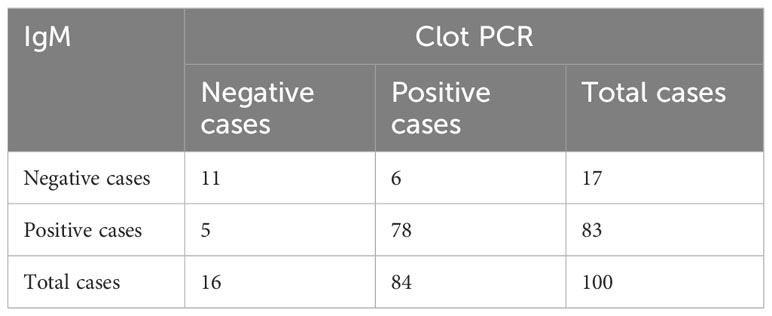
Table 2 Results of Typhipoint EIA IgM and blood clot PCR for S. typhi in clinically suspected cases.
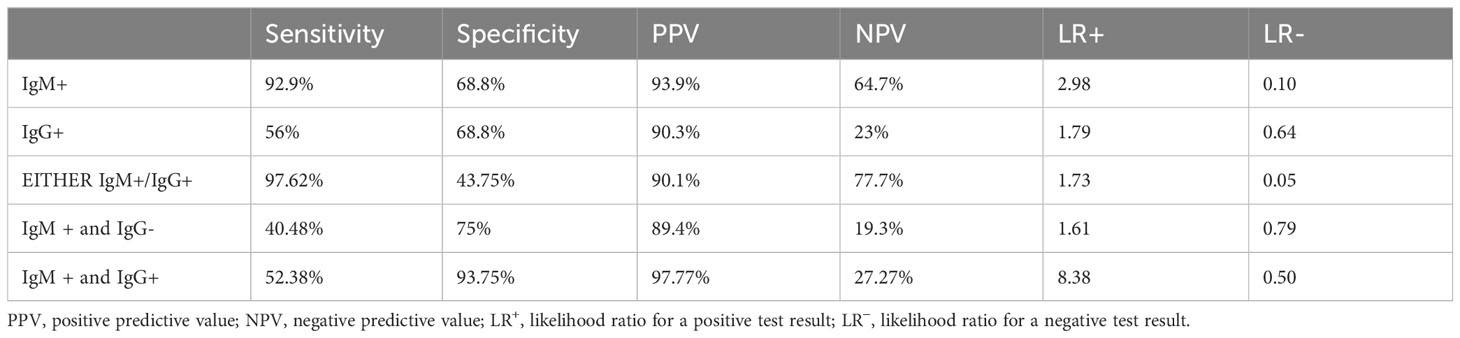
Table 3 Diagnostic validity of various diagnostic tests for detection of Salmonella typhi against clot PCR for S. typhi.
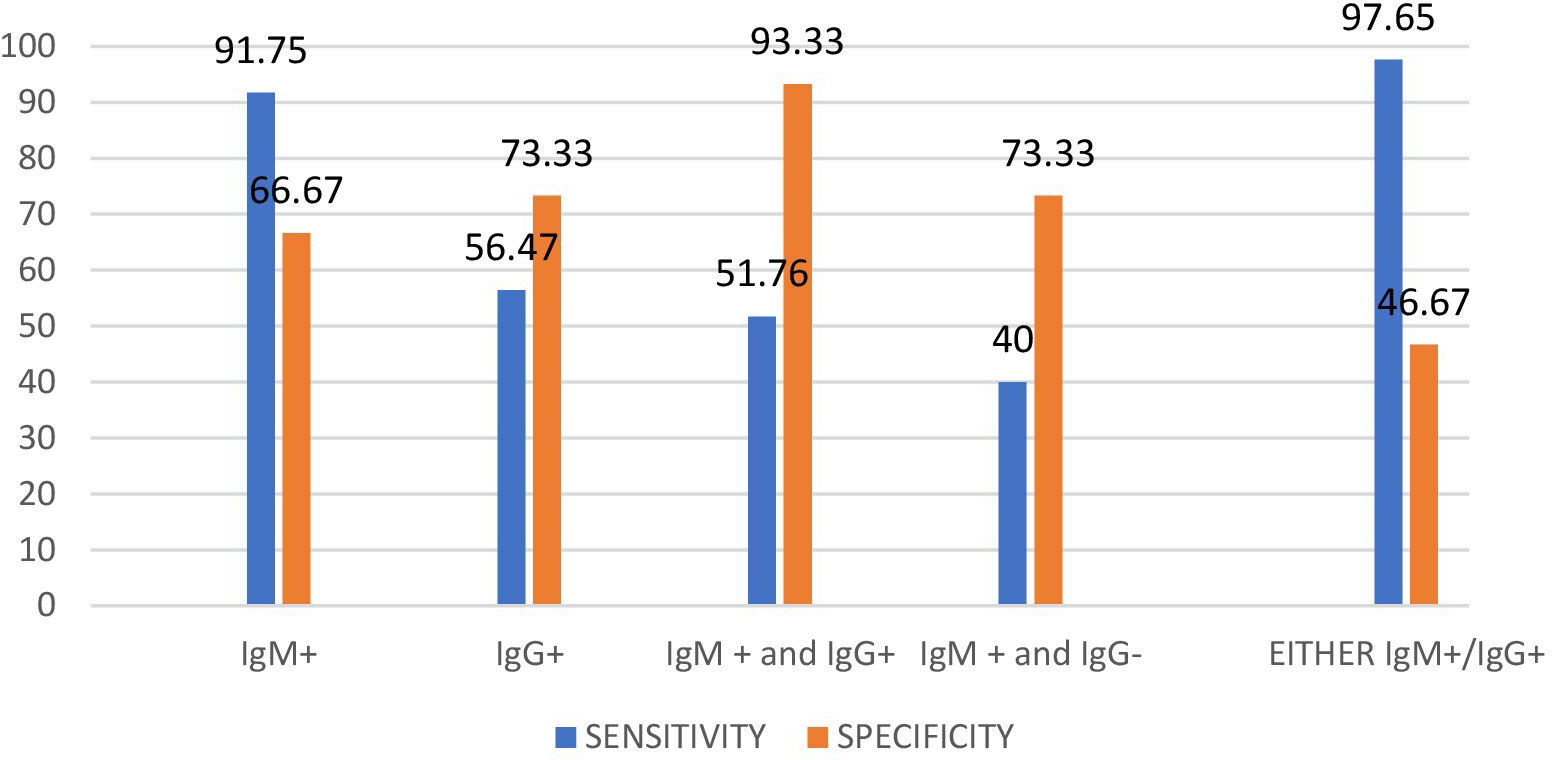
Figure 6 Diagnostic validity of Typhipoint EIA test with respect to Clot PCR for S. typhi and paratyphi together as gold standard.
Since the illness in our study lasted longer than 2 weeks, we used either Typhipoint EIA IgM or IgG antibody-positive result validation as a diagnostic test, with Clot Nested PCR for S. typhi and S. paratyphi together as the gold standard test, to determine the presence of both new and chronic infections. The sensitivity was found to be 97.65% (95% CI, 92% to 99%), and specificity was reduced to 46.67% (95% CI, 21% to 73%) with PPV-91.2% (95% CI, 87% to 94%) and NPV-77.7% (95% CI, 45% to 94%) (Tables 2–4). We did blood clot, urine, and stool cultures for Salmonella in all of the specimens for the diagnosis of enteric fever, as the bone marrow aspirate culture, the gold standard for investigations, is an invasive and painful procedure. A total of 52% of cases were blood clot culture positive, 13% were stool culture positive alone, and 5 cases were urine culture positive alone. A total of 70 samples comprising blood clots, urine, or stool cultures from suspected enteric fever patients were evaluated. The cumulative result from all three specimens (blood, urine, and stool) after acid exposure was 77.7% positive for the isolation of the S. typhi serotype. Acid exposure cultures also succeeded in isolating bacteria from urine samples (5.5%) and stool samples (40%).
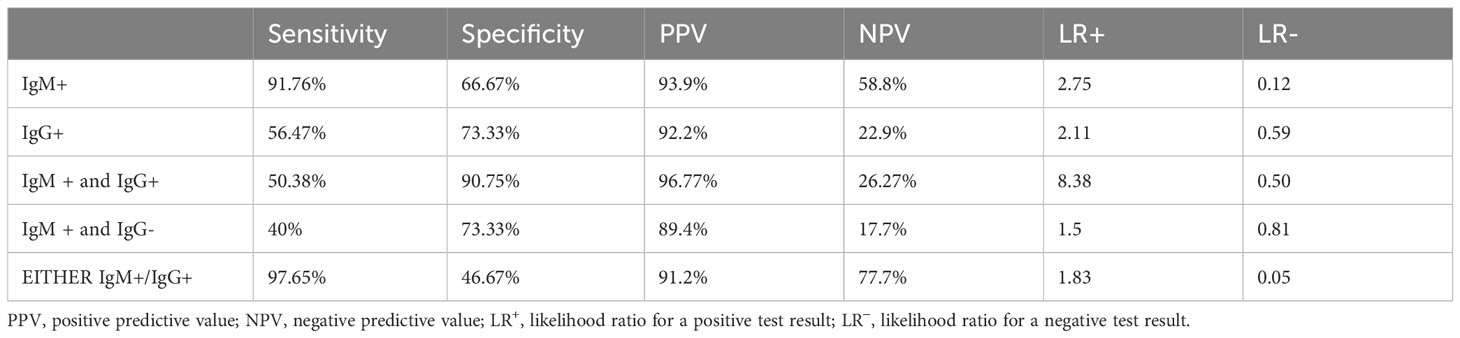
Table 4 Diagnostic validity of various diagnostic tests with respect to clot PCR for S. typhi and S. paratyphi together as gold standard.
We tried to find out the validation of the Typhipoint EIA test against blood clot culture in our study subjects (Table 5). Validation of IgM-positive cases against either clot/stool/urine culture for Salmonella typhi as a gold standard showed desired sensitivity and specificity (Table 6). Hence, we can see that Typhipoint EIA IgM antibody alone or IgM/IgG antibody positivity-based tests are quite sensitive, but specificity is quite low once we take culture for Salmonella typhi as the gold standard. At the same time, if blood clot nPCR for Salmonella typhi is considered as gold standard, then a higher sensitivity (reaching 90%) and specificity (reaching almost 70%) with rational PPV and NPV were achieved (Tables 5, 6).
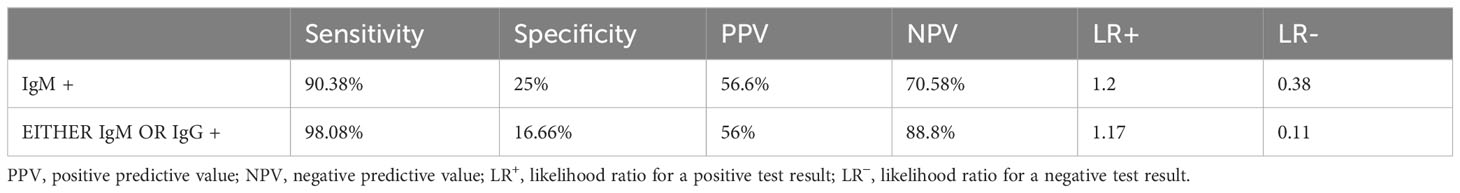
Table 5 Diagnostic validity of various diagnostic tests with respect to clot culture for Salmonella as gold standard.
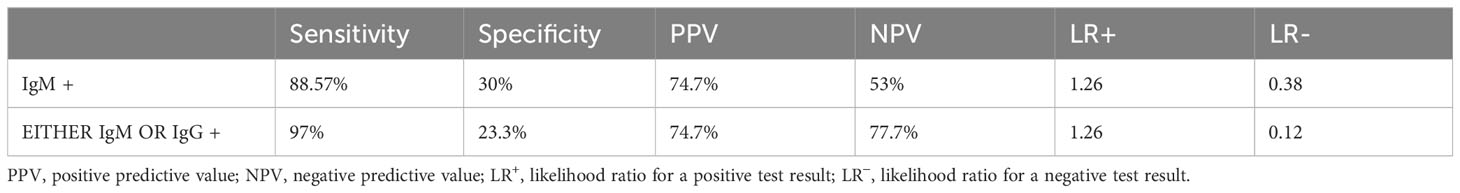
Table 6 Diagnostic validity of various diagnostic tests with respect to either culture(clot/stool/urine) for Salmonella as gold standard positive.
Discussion
The Widal test quantifies the agglutinating antibodies for the antigen of S. typhi, in particular lipopolysaccharide and flagella in the serum sample of suspected enteric fever (Olapoenia and King, 2000). Typhipoint EIA detects IgM and IgG antibodies against 50 kDa, the outer membrane protein of Salmonella typhi. The early humoral response of the immune system against the pathogen is the formation of IgM, whereas the delayed response is the formation of IgG antibodies; detection of IgG and IgM together predicts a middle phase of enteric fever (Iványi et al., 1966). A study by Mawazo et al. concluded that the Widal test is not a reliable method for the diagnosis of enteric fever, as the false positive and negative results are quite frequent (sn-81.5%, sp-18.3%, PPV-10.1%, NPV-89.7%); moreover, enteric fever seldom co-relates to the blood culture (kappa = 0.014, p < 0.05) and stool culture (kappa = 0.22, p < 0.05) (Mawazo et al., 2019). The threshold of antibody titer is considered as 1:80 for TO and 1:160 for anti-TH to diagnose enteric fever cases(11. In contrast, studies from African countries still conclude that this test holds clinical importance for the diagnosis of enteric fever (Mengist and Tilahun, 2017). Therefore, it is necessary to develop and adopt a new rapid diagnostic method that would show higher specificity than Widal and be cost-effective for developing countries.
In the present study, the rapid typhoid antibody test (Typhipoint ELISA) appears to have better sensitivity (92.9%) (95% CI, 85% to 97%) and specificity (68.8%) (95% CI, 41% to 89%), when nested PCR-based S. typhi detection was carried out in clinically suspected typhoid fever patients as gold standard of investigation against the single-tube Widal test as per a previous study (Prakash et al., 2005). In addition, a cross-sectional comparative study conducted in the West region of Cameroon also suggested Typhidot immunoassay (sn-80.56%, sp-94.03%, PPV-66.67%, NPV-97.03%) to be a better diagnostic test as compared with Widal (sn-94.44%, sp-48.35%, PPV-21.32%, NPV-98.33%) (Ousenu et al., 2021). The expected LR− value for any satisfactory diagnostic test is ≤0.1 (Darton et al., 2017); in our study, the LR− value for IgM antibody detection by Typhipoint EIA was 0.10 and it seems to be satisfactory, but for the IgG antibody, the value was quite high, i.e., 0.64. The sensitivity and specificity of the Typhipoint EIA IgM test with respect to clot culture were 90.38% and 25%, respectively (LR + 1.2; LR − 0.31). In many previous studies, the high false negative culture results were correlated with the laboratory differences in the methods, prior antibiotic therapy, and cumbersome procedures (Olapoenia and King, 2000; Darton et al., 2017; Srinivasan et al., 2021). We got a similar diagnostic yield with the culture results, and Typhipoint EIA (either IgM or IgG positive) validation with respect to clot culture apparently scored poor performance in terms of statistical numbers (sensitivity—98.08%; specificity—16.66%; LR + 1.17; LR − 0.11). The yield of blood culture varies between 40% and 60% in many studies; thus, it indicates a high possibility of false negative results (Crump et al., 2015; Mehmood et al., 2015). A meta‐analysis by Wijedoru et al. revealed an average sensitivity of 69% (95% CI 59% to 78%) and a specificity of 90% (95% CI 78% to 93%) for the Typhipoint EIA test and other prototype tests (kit-based test); for the Widal method, sensitivity was 78% (95% CI, 71% to 85%) and specificity was 87% (95% CI, 82% to 91%) (Wijedoru et al., 2017). On the other hand, the TUBEX method has its disadvantage of the coloring score system (Bakr et al., 2010). In another study, validation of the ELISA-based Typhoid test (kit-based test) showed a sensitivity of 84% (95%, CI 73% to 91%) and a specificity of 79% (95% CI 70% to 87%) (Fadeel et al., 2011). Analyzing the 13 different studies which also included the Typhidot test with indeterminate test results, the sensitivity was 78% (95% CI 65% to 87%) and the specificity was 77% (95% CI 66% to 86%). No significant difference was found in either sensitivity or specificity when the results of TUBEX, Typhidot, and Test‐IT Typhoid tests were compared (Ismail, 2000; Fadeel et al., 2011).
The mainstay of diagnosis in clinical practice is a positive blood culture, although the test is only positive in 40% to 80% of cases. This low sensitivity could be related to many confounding factors like the low number of bacteria circulating in the first week of the disease, prior antimicrobial therapy, the type of culture medium used, the ratio of blood to broth, the stage of illness at the time of presentation, and the duration of incubation (Wain et al., 1998; Mogasale et al., 2016). There are certain modified tests available nowadays like the Typhidot-M test. In this modified test, inactivation of IgG allows access to the IgM, and hence it is more specific for diagnosis in the early stage of enteric fever; it has a sensitivity ranging from 68% to 95% and a specificity ranging from 75% to 95% as shown in various studies. The high negative predictive value (NPV) of this test would be useful in areas of high endemicity (Wijedoru et al., 2017). Thus, this test may be more appropriate than the Widal test in the diagnosis of enteric fever. However, this test may be false negative during the second week of illness due to falling levels of IgM. Although this ELISA-based method is not very satisfactory, it is better than the Widal test, as shown by earlier studies (Keddy et al., 2011). The explanation for better sensitivity resulted in the way that using the ELISA-based method, the enzymatic reaction is amplified by activating several substrate molecules, whereas in the Widal test, antibodies are binding resulting in agglutination of the bacteria.
In our study, the patients presented with early or late symptoms; hence, the culture results were combined for all of the three samples (clot, stool, and urine) to detect S. typhi. The sensitivity and specificity of the Typhipoint EIA test were not supportive to confirm the diagnosis of enteric fever (Table 6), and these values did not improve much when they were compared, in terms of specificity, with clot culture alone. Traditionally blood clot culture has been described as the gold standard to validate the diagnostic test accuracy of a new diagnostic method. Hence, in our study, we did the DTA of Typhipoint EIA considering PCR as the gold standard and also compared the DTA data of Typhipoint EIA considering blood clot culture for diagnosis of typhoid fever from samples drawn at one instance to find out which results suit better to be called as the gold standard diagnostic test for validation of any newer or improvised Typhipoint EIA test in future. Thus, to go ahead with a definitive treatment is not possible with such data. Although most of the previous studies have been done considering blood culture or bone marrow aspirate culture as the gold standard, our findings are well in agreement with those that have taken the study subjects which were exclusively culture positive (Escamilla et al., 1986; Mogasale et al., 2016). The sensitivity and specificity of blood culture were found to be 61% and 100%, respectively, against a bone marrow culture comparator (Mogasale et al., 2016). Even the blood culture and clot culture had shown the same rate of isolation (Mogasale et al., 2016). Isolation of organisms from blood is the gold standard for diagnosis of enteric fever. The results of culture and isolation for S. typhi in our study have similarities with the studies reported earlier (Escamilla et al., 1986; Mogasale et al., 2016). However, the clot culture was more sensitive for isolating Salmonella than the whole blood culture and Salmonella growth was faster in the culture of blood clots compared with the whole blood (Escamilla et al., 1986). In our study, the false positives for the antibodies against S. typhi may not be true false positives; rather, the most sensitive nested PCR-based method may miss those enteric fever cases which are caused by paratyphoid bacteria. The positivity in healthy volunteers for the S. typhi-specific antibody may not necessarily be false positive. However, in an endemic area, a subclinical infection, chronic carrier state, or convalescent state cannot be denied. Similarly, false negative cases may occur because of a variable host response against bacteria in enteric fever. To overcome this, targeting more than one antigen of S. typhi may give better results if we have to rely upon the ELISA-based antibody detection diagnostic tools at all. A similar suggestion has also been given by Fadeel et al. (2011) from Egypt because masking of IgM by IgG antibody may occur due to the robust response of IgG in case of reinfection or presence of IgG in patients from endemic areas (Fadeel et al., 2011). Earlier, a study from the same laboratory showed that blood clot yield after brief acid exposure was 53.3% (Ahirwar et al., 2014). This was much higher than the conventional method (8.8%); in addition, acid exposure cultures also succeeded in isolating bacteria from urine samples (5.5%) and stool samples (40%), which was not the case with the conventional method. The cumulative result from all the three specimens (blood, urine, stool) after acid exposure was 77.7% positive for the isolation of the S. typhi serotype as compared with 8.8% by the conventional method. This observation was reported for the first time. The gold standard is bone marrow culture/blood clot culture; however, it is a painful procedure, and the yield is poor in terms of sensitivity especially for a chronic carrier, whereas Typhipoint immunoassay can provide a rapid and easy investigation along with high sensitivity and specificity. The nested PCR technique detects salmonella with high accuracy and should be used as a preferred diagnostic test for culture-negative patients (Escamilla et al., 1986; Khan et al., 2012). In this regard, when we compared the yield of Typhipoint EIA IgM-positive test results against nested PCR as a gold standard for the investigation, we got a high sensitivity and comparable specificity, high positive predictive value, and negative predictive value (Table 3). When either IgM/IgG positive test results were validated, we found a significant drop in specificity but increased sensitivity (Table 3); this may be explained by few subjects having previous infection. We also checked for diagnostic accuracy of Typhipoint EIA tests against the results of nested PCR for S. typhi and S. paratyphi together. A gold standard for investigation and the results of IgM and either IgM/IgG were not much different (Table 4). This was done to find out the missing cases of typhoid due to Salmonella paratyphi alone. The specificity of nested PCR is almost equal to bone marrow culture, and sensitivity is equally comparable with the readily available rapid diagnostic immunoassay Typhipoint test which can be used for screening or surveillance purposes for a population.
Asymptomatic carriers represent an important reservoir that helps to transmit the disease and is responsible for the outbreaks of enteric fever in endemic regions. Identifying this carrier state has always been an epidemiological challenge (Divyashree et al., 2016; Kurtz et al., 2017; Voysey et al., 2020). The current gold standard investigation used for the diagnosis of carriers is the stool culture, but it is quite tedious and also has a low sensitivity. To overcome this problem, a combination of stool, urine, and clot samples could be used in nested PCR for diagnosis of the chronic carrier state. When we evaluated the accuracy of Typhipoint EIA test results against the nested PCR results of all three samples (blood clot, urine, stool), it showed good sensitivity of 88.57% and low specificity of 30%, and no difference in diagnostic validity was found when these samples were checked with PCR for both S. typhi and S. paratyphi.
The lacunae in the clinical application of Typhipoint EIA are its false negativity in the case of patients contracting enteric fever in the early phase of illness and its lack of specificity owing to not being able to detect the MDR strain of salmonella; thereby, these cases would remain undetected (Ismail, 2000; Divyashree et al., 2016). Due to the masking effect of IgG over IgM, this test would give false negative results of a given high IgG concentration. The low sensitivity of blood culture yield in Salmonella typhi and paratyphi is due to a lack of reference standards, which may lead to erroneous results depending on lab techniques and self-treatment with the widely available over-the-counter antibiotics intake in India before sampling for the blood culture (Wain etal., 1998; Bakr et al., 2010; Mogasale et al., 2016).
The choice of antibiotics and the appropriate time to start the treatment of enteric fever are lacking due to poor sensitivity in the diagnosis via the previously established gold standard, i.e., blood culture. False negative or false positive detection of S. typhi and S. paratyphi leads to either undertreatment or overtreatment, respectively, which is responsible for the occurrence of antibiotic-resistant patterns. AMR genes were carried via the pHCM1, pK91, and pPRJEB21992 plasmids, which together with the SGI11 variations were responsible for administering the resistance phenotypes, pK91 (contain qnr genes) conferred high ciprofloxacin resistance (Lima et al., 2019). Target gene mutations such as DNA gyrase and Topoisomerase IV, efflux system, quorum sensing, and biofilm formation are known to contribute to increased resistance; overexpression of these systems results in therapeutic failure (Martins et al., 2011).
Overuse and misuse of antibiotics can lead to antibiotic resistance in S. typhi and S. paratyphi, as subtherapeutic drug levels cannot eliminate the growth of bacteria but rather only suppress them. Similar concerns are raised by the study conducted in Ethiopia about patients being falsely diagnosed via the Widal test and further inappropriately treated leading to high multidrug resistance of Salmonella typhi and Salmonella paratyphi isolates to commonly available antimicrobials (Deksissa and Gebremedhin, 2019). This is also supported by a finding of a study conducted at Cornell University, Ithaca, New York, USA, which emphasizes that although PCR-based nucleic acid detection is reasonably rapid it requires technical labor; hence, there is a need to identify a more accessible and inexpensive procedure (Deksissa and Gebremedhin, 2019; Neupane et al., 2021). Non-typhoidal serotypes of Salmonella cause gastroenteritis, and non-typhoidal salmonellosis is self-limiting (Hannemann and Galán, 2017) and resolves without antibiotics; however, certain complications occur in immunocompromised patients. Therefore, the acute condition needs to be treated effectively. The chronic carrier of enteric fever is generally associated with gall bladder and gastrointestinal tract diseases and malignancy. The chronic carrier is required to be identified with an appropriate diagnostic method that shows high specificity and sensitivity. In the given situation, our data suggest that validation of the ELISA-based rapid test should be done considering nested PCR of blood clot as a gold standard of investigations for the diagnosis of enteric fever as it can reflect the appropriate benchmark for a future improved ELISA-based rapid test. The typhi antigens HIyE and YncE have been reported to be beneficial biomarkers for acute typhoid and chronic carriers, respectively (Franklin et al., 2020). We recommend rapid detection of enteric fever by targeting two or more such antigens via Typhipoint EIA for diagnosis and surveillance in developing countries. However, IgG antibodies are detectable even after the third week of successful treatment; therefore, nested PCR is suggested for detecting such chronic carriers of enteric fever as being capable of detecting the drug-resistant bacteria.
Conclusion
Single antigen-targeted ELISA-based methods seem to reach a satisfactory level of sensitivity for their use in resource-poor settings. Contrastingly, the blood culture method is not a very sensitive tool and is time-consuming whereas the PCR-based investigations are, although very sensitive and specific, quite costly for developing countries and are currently only available at tertiary healthcare centers. Therefore, a strong push may be needed for standardizing antibody-based investigation. It may be advised that two to three specific antigens of S. typhi should be spotted on the membrane for the ELISA-based diagnostic kits to allow it to reach the desired level of diagnostic accuracy for the detection of enteric fever in terms of acceptable diagnostic yield for field conditions, especially in developing countries and endemic areas. The validation of such diagnostic tools should be done with DNA-based investigations instead of inconsistent culture-based investigations. We detect antibodies against S. typhi (Vi), 9,12:d, the H=d flagellar, and ‘Vi’ capsular antigen in the Widal agglutination test. HIyE and YncE typhi antigens are considered to be useful biomarkers for acute typhoid or chronic carriers, respectively (Franklin et al., 2020). For Typhipoint EIA, an antibody against outer membrane protein is detected. In the future, an evaluation of the presence of antibodies against the antigens identified in the current study for their potential application in diagnosing enteric fever could be attempted for their possible inclusion in the next-generation Typhipoint EIA kit, which would then have substantially improved levels of diagnostic test accuracy for their use in field conditions.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
This study received approval from the Ethical Committee Institute of Medical Sciences, Banaras Hindu University, Varanasi, Uttar Pradesh- 221005, India (Approval number: 291). All methods were performed in accordance with the ethical standards of the Declaration of Helsinki and its later amendments.
Author contributions
AD: Conceptualization, Formal analysis, Writing – original draft. AT: Conceptualization, Formal analysis, Writing – original draft. SJ: Writing – review & editing. KR: Writing – review & editing. JR: Writing – review & editing. SS: Writing – review & editing. DK: Conceptualization, Formal analysis, Writing – original draft.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
We are grateful to Prof. Gopal Nath for his support and coordination to accomplish the study. We express our sincere gratitude to all the patients for their involvement in the study. The authors alone are responsible for the content and writing of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
EIA, Enzyme immunoassay; PCR, Polymerase chain reaction; Sn, Sensitivity; Sp, Specificity; PPV, Positive predictive value; NPV, Negative predictive value; LR +, likelihood ratio for positive test results; LR, Likelihood ratio for negative test results; DTA, Diagnostic Test Accuracy.
References
Ahirwar S. K., Pratap C. B., Patel S. K., Shukla V. K., Singh I. G., Mishra O. P., et al. (2014). Acid exposure induces multiplication of Salmonella enterica serovar Typhi. J. Clin. Microbiol. 52 (12), 4330–4333. doi: 10.1128/JCM.02275-14
Arora P., Thorlund K., Brenner D. R., Andrews J. R. (2019). Comparative accuracy of typhoid diagnostic tools: a Bayesian latent-class network analysis. PloS Negl. Trop. Dis. 13 (5), e0007303. doi: 10.1371/journal.pntd.0007303
Bakr W. M., El Attar L. A., Ashour M. S., El Tokhy A. M. (2010). Tubex test versus Widal test in the diagnosis of typhoid fever in Kafr El-Shekh, Egypt. J. Egyptian Public Health Assoc. 85 (5-6), 285–296.
Bronowski C., Fookes M. C., Gilderthorp R., Ashelford K. E., Harris S. R., Phiri A., et al. (2013). Genomic haracterization of invasive non-typhoidal Salmonella enterica Subspecies enterica Serovar Bovismorbificans isolates from Malawi. PloS Negl. Trop. Dis. 7 (11), e2557. doi: 10.1371/journal.pntd.0002557
Choo K. E., Davis T. M., Ismail A., Ibrahim T. T., Ghazali W. N. (1999). Rapid and reliable serological diagnosis of enteric fever: comparative sensitivity and specificity of Typhidot and Typhidot-M tests in febrile Malaysian children. Acta Tropica 72 (2), 175–183. doi: 10.1016/S0001-706X(98)00095-3
Choo K. E., Oppenheimer S. J., Ismail A. B., Ong K. H. (1994). Rapid serodiagnosis of typhoid fever by dot enzyme immunoassay in an endemic area. Clin. Infect. Dis. 19 (1), 172–176. doi: 10.1093/clinids/19.1.172
Cordovana M., Mauder N., Kostrzewa M., Wille A., Rojak S., Hagen R. M., et al. (2021). Classification of Salmonella enterica of the (Para-) typhoid fever group by Fourier-transform infrared (FTIR) spectroscopy. Microorganisms 9 (4), 853. doi: 10.3390/microorganisms9040853
Corrêa I. M., Pereira L. Q., Silva I. G., Altarugio R., Smaniotto B. D., Silva T. M., et al. (2018). Comparison of three diagnostic methods for Salmonella enterica serovars detection in chicken rinse. Pesquisa Veterinária Brasileira 38, 1300–1306. doi: 10.1590/1678-5150-pvb-5211
Crump J. A., Sjölund-Karlsson M., Gordon M. A., Parry C. M. (2015). Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin. Microbiol. Rev. 28 (4), 901–937. doi: 10.1128/CMR.00002-15
Darton T. C., Zhou L., Blohmke C. J., Jones C., Waddington C. S., Baker S., et al. (2017). Blood culture-PCR to optimise typhoid fever diagnosis after controlled human infection identifies frequent asymptomatic cases and evidence of primary bacteraemia. J. Infect. 74 (4), 358–366. doi: 10.1016/j.jinf.2017.01.006
Deksissa T., Gebremedhin E. Z. (2019). A cross-sectional study of enteric fever among febrile patients at Ambo hospital: prevalence, risk factors, comparison of Widal test and stool culture and antimicrobials susceptibility pattern of isolates. BMC Infect. Dis. 19 (1), 1–2. doi: 10.1186/s12879-019-3917-3
Divyashree S., Nabarro E. B., Veeraraghavan B., Rupali P. (2016). Enteric fever in India: current scenario and future directions. Trop. Med. Int. Health 21 (10), 1255–1262. doi: 10.1111/tmi.12762
Escamilla J. O., Florez-Ugarte H. E., Kilpatrick M. E. (1986). Evaluation of blood clot cultures for isolation of Salmonella typhi, Salmonella paratyphi-A, and Brucella melitensis. J. Clin. Microbiol. 24 (3), 388–390. doi: 10.1128/jcm.24.3.388-390.1986
Fadeel M. A., House B. L., Wasfy M. M., Klena J. D., Habashy E. E., Said M. M., et al. (2011). Evaluation of a newly developed ELISA against Widal, TUBEX-TF and Typhidot for typhoid fever surveillance. J. Infect. Dev. Ctries. 5 (03), 169–175. doi: 10.3855/jidc.1339
Frankel G., Newton S. M., Schoolnik G. K., Stocker B. A. (1989). Intragenic recombination in a flagellin gene: characterization of the H1-j gene of Salmonella typhi. EMBO J. 8 (10), 3149–3152. doi: 10.1002/j.1460-2075.1989.tb08468.x
Franklin F., Chong C. W., Chua L. H., Anthony A. A., Liew M. W., Aziah I., et al. (2020). Evaluation of Salmonella Typhi antigen YncE alongside HlyE for the detection of typhoid fever and its carriers. Med. Microbiol. Immunol. 209, 593–601. doi: 10.1007/s00430-020-00667-1
Gallichan S., Perez-Sepulveda B. M., Feasey N. A., Hinton J. C., Thomas J., Smith A. M. (2022). Multiplex PCR assay for clade typing of Salmonella enterica serovar enteritidis. Microbiol. Spectrum 10 (6), e03182–e03122. doi: 10.1128/spectrum.03182-22
Hannemann S., Galán J. E. (2017). Salmonella enterica serovar-specific transcriptional reprogramming of infected cells. PloS Pathog 13 (7), e1006532. doi: 10.1371/journal.ppat.1006532
Hart M. L., Meyer A., Johnson P. J., Ericsson A. C. (2015). Comparative evaluation of DNA extraction methods from feces of multiple host species for downstream next-generation sequencing. PloS One 10 (11), e0143334. doi: 10.1371/journal.pone.0143334
Hatta M., Smits H. L. (2007). Detection of Salmonella typhi by nested polymerase chain reaction in blood, urine, and stool samples. Am. J. Trop. Med. hygiene. 76 (1), 139–143. doi: 10.4269/ajtmh.2007.76.139
Ismail A. (2000). New advances in the diagnosis of typhoid and detection of typhoid carriers. Malaysian J. Med. Sciences: MJMS. 7 (2), 3.
Iványi J., Valentová V., Cerný J. (1966). The dose of antigen required for the suppression of the IgM and IgG antibody response in chickens. I. The kinetics and characterization of serum antibodies. Folia Biologica. 12 (3), 157–167.
John J., Bavdekar A., Rongsen-Chandola T., Dutta S., Kang G. (2018). Estimating the incidence of enteric fever in children in India: a multi-site, active fever surveillance of pediatric cohorts. BMC Public Health 18, 1–6. doi: 10.1186/s12889-018-5498-2
John J., Van Aart C. J., Grassly N. C. (2016). The burden of typhoid and paratyphoid in India: systematic review and meta-analysis. PloS Negl. Trop. Dis. 10 (4), e0004616. doi: 10.1371/journal.pntd.0004616
Joseph S., Russell David W. (2006). Purification of nucleic acids by extraction with phenol: chloroform. Cold Spring Harb. Protoc. 1, 4455. doi: 10.1101/pdb.prot4455
Keddy K. H., Sooka A., Letsoalo M. E., Hoyland G., Chaignat C. L., Morrissey A. B., et al. (2011). Sensitivity and specificity of typhoid fever rapid antibody tests for laboratory diagnosis at two sub-Saharan African sites. Bull. World Health Organization. 89 (9), 640–647. doi: 10.2471/BLT.11.087627
Khan S., Harish B. N., Menezes G. A., Acharya N. S., Parija S. C. (2012). Early diagnosis of typhoid fever by nested PCR for flagellin gene of Salmonella enterica serotype Typhi. Indian J. Med. Res. 136 (5), 850.
Kumar G., Pratap C. B., Mishra O. P., Kumar K., Nath G. (2012). Urine in the diagnosis of typhoid fever by using nested PCR targeting flagellin gene (fliC). J. Clin. Microbiol. 50 (6), 1964–1967. doi: 10.1128/JCM.00031-12
Kurtz J. R., Goggins J. A., McLachlan J. B. (2017). Salmonella infection: Interplay between the bacteria and host immune system. Immunol. Lett. 190, 42–50. doi: 10.1016/j.imlet.2017.07.006
Lima N. C., Tanmoy A. M., Westeel E., De Almeida L. G., Rajoharison A., Islam M., et al. (2019). Analysis of isolates from Bangladesh highlights multiple ways to carry resistance genes in Salmonella Typhi. BMC Genomics 20 (1), 1–5. doi: 10.1186/s12864-019-5916-6
Martins M., McCusker M., Amaral L., Fanning S. (2011). Mechanisms of antibiotic resistance in Salmonella: efflux pumps, genetics, quorum sensing and biofilm formation. Lett. Drug Design Discov. 8 (2), 114–123. doi: 10.2174/157018011794183770
Mawazo A., Bwire G. M., Matee M. I. (2019). Performance of Widal test and stool culture in the diagnosis of typhoid fever among suspected patients in Dar es Salaam, Tanzania. BMC Res. Notes 12 (1), 1–5. doi: 10.1186/s13104-019-4340-y
Mehmood K., Sundus A., Naqvi I. H., Ibrahim M. F., Siddique O., Ibrahim N. F. (2015). Typhidot-A blessing or a menace. Pakistan J. Med. Sci. 31 (2), 439. doi: 10.12669/pjms.312.5934
Mengist H. M., Tilahun K. (2017). Diagnostic value of Widal test in the diagnosis of typhoid fever: a systematic review. J. Med. Microbiol. Diagn. 6 (01), 1–4. doi: 10.4172/2161-0703.1000248
Millemann Y., Mouline C., Lafont J.-P., Chaslus-Dancla E. (2005). Bacteraemia assays in chickens as a model for the evaluation of the virulence of Salmonella enterica serovars Typhimurium and Enteritidis strains. Rev. médecine vétérinaire. 156 (2), 70–76.
Mogasale V., Ramani E., Mogasale V. V., Park J. (2016). What proportion of Salmonella Typhi cases are detected by blood culture? A systematic literature review. Ann. Clin. Microbiol. Antimicrobials 15, 1–8. doi: 10.1186/s12941-016-0147-z
Neupane D. P., Dulal H. P., Song J. (2021). Enteric fever diagnosis: current challenges and future directions. Pathogens 10 (4), 410. doi: 10.3390/pathogens10040410
Olapoenia L. A., King A. L. (2000). Widal agglutination test-100 years later: still plagued by controversy. Prostgrad. Med. J. 76, 80–84. doi: 10.1136/pmj.76.892.80
Olsen S. J., Pruckler J., Bibb W., Thanh N. T., Trinh T. M., Minh N. T., et al. (2004). Evaluation of rapid diagnostic tests for typhoid fever. J. Clin. Microbiol. 42 (5), 1885–1889. doi: 10.1128/JCM.42.5.1885-1889.2004
Ousenu K., Ali I. M., Sama L. F., Ndam M. N., Tchouangueu T. F., Tume C. B. (2021). A cross-sectional comparative study of the performance of the Widal test and the typhidot immunoassay for typhoid fever diagnosis in the west region of Cameroon. Can. J. Infect. Dis. Med. Microbiol. doi: 10.1155/2021/8279122
Prakash P., Mishra O. P., Singh A. K., Gulati A. K., Nath G. (2005). Evaluation of nested PCR in diagnosis of typhoid fever. J. Clin. Microbiol. 43 (1), 431–432. doi: 10.1128/JCM.43.1.431-432.2005
Pratap C. B., Kumar G., Patel S. K., Verma A. K., Shukla V. K., Kumar K., et al. (2013). Targeting of putative fimbrial gene for detection of S. Typhi in typhoid fever and chronic typhoid carriers by nested PCR. J. Infect. Dev. Ctries. 7 (07), 520–527. doi: 10.3855/jidc.2561
Saha T., Arisoyin A. E., Bollu B., Ashok T., Babu A., Issani A., et al. (2023). Enteric fever: diagnostic challenges and the importance of early intervention. Cureus 15 (7). doi: 10.7759/cureus.41831
Sánchez-Jiménez M. M., Cardona-Castro N. (2004). Validation of a PCR for diagnosis of typhoid fever and salmonellosis by amplification of the hilA gene in clinical samples from Colombian patients. J. Med. Microbiol. 53 (9), 875–878. doi: 10.1099/jmm.0.45630-0
Song J. H., Cho H., Park M. Y., Na D. S., Moon H. B., Pai C. H. (1993). Detection of Salmonella typhi in the blood of patients with typhoid fever by polymerase chain reaction. J. Clin. Microbiol. 31 (6), 1439–1443. doi: 10.1128/jcm.31.6.1439-1443.1993
Srinivasan M., Sindhu K. N., Ramanujam K., Ramasamy R. K., Subramaniam S., Ganesan S. K., et al. (2021). Factors predicting blood culture positivity in children with enteric fever. J. Infect. Dis. 224 (Supplement_5), S484–S493. doi: 10.1093/infdis/jiab357
Stanaway J. D., Reiner R. C., Blacker B. F., Goldberg E. M., Khalil I. A., Troeger C. E., et al. (2019). The global burden of typhoid and paratyphoid fevers: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect. Dis. 19 (4), 369–381. doi: 10.1016/S1473-3099(18)30685-6
Voysey M., Pant D., Shakya M., Liu X., Colin-Jones R., Theiss-Nyland K., et al. (2020). Under-detection of blood culture-positive enteric fever cases: the impact of missing and methods for adjusting incidence estimates. PloS Negl. Trop. Dis. 14 (1), e0007805. doi: 10.1371/journal.pntd.0007805
Wain J., Diep T. S., Ho V. A., Walsh A. M., Hoa N. T., Parry C. M., et al. (1998). Quantitation of bacteria in blood of typhoid fever patients and relationship between counts and clinical features, transmissibility, and antibiotic resistance. J. Clin. Microbiol. 36 (6), 1683–1687. doi: 10.1128/JCM.36.6.1683-1687.1998
Wain J., Hosoglu S. (2008). The laboratory diagnosis of enteric fever. J. Infect. Dev. Ctries. 2 (06), 421–425. doi: 10.3855/jidc.155
Keywords: enteric fever, nested PCR, Typhipoint EIA, Salmonella typhi, Salmonella paratyphi
Citation: Diwaker A, Tiwari A, Jain S, Rupali KA, Ram J, Singh S and Kishore D (2024) Enteric fever and the diagnostic tools: defining the accuracy. Front. Bacteriol. 3:1332180. doi: 10.3389/fbrio.2024.1332180
Received: 02 November 2023; Accepted: 08 January 2024;
Published: 29 January 2024.
Edited by:
Rajan P. Adhikari, Integrated BioTherapeutics, Inc., United StatesReviewed by:
Bilal Ahmad Tantry, Government Medical College, IndiaMuhammad Asif Zahoor, Government College University, Faisalabad, Pakistan
Copyright © 2024 Diwaker, Tiwari, Jain, Rupali, Ram, Singh and Kishore. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dhiraj Kishore, ZGhpcmFqa2lzaG9yZUBnbWFpbC5jb20=
 Amita Diwaker
Amita Diwaker Ashutosh Tiwari2
Ashutosh Tiwari2 Shubham Jain
Shubham Jain Kumari Astha Rupali
Kumari Astha Rupali Jitendra Ram
Jitendra Ram Samer Singh
Samer Singh Dhiraj Kishore
Dhiraj Kishore