
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Bacteriol., 26 July 2023
Sec. Molecular Bacteriology and Microbiome
Volume 2 - 2023 | https://doi.org/10.3389/fbrio.2023.1229077
This article is part of the Research TopicEditors' Showcase: Molecular Bacteriology and MicrobiomeView all 7 articles
Colorectal cancer (CRC) is a leading cause of cancer-related deaths globally. Incidence rates among individuals under 50 years are rising, which has led to the lowering of the recommended screening age from 50 to 45 years for those at an average risk. While numerous risk factors are associated with the development of CRC, most cases contain microbial signatures representative of dysbiosis, indicating a role for the gut microbiome in disease pathogenesis. To date, most research has investigated individual members of the gut microbiota independently; however, it is widely established that microbes interact with each other in the gut. More recently, two specific species of the microbiota have revealed a pro-carcinogenic synergism in vivo. Strains of both Bacteroides fragilis and Escherichia coli have been linked to CRC in clinical studies and been shown to induce carcinogenesis in mouse models through B. fragilis toxin and colibactin, respectively. The link between these two bacteria is found within their spatial association: biofilms, or mucosal-associated microbial aggregates. In this review, we discuss the roles of B. fragilis and E. coli in healthy and diseased guts, current evidence associating each bacterium with CRC individually, and their synergistic contributions to the pathogenesis of CRC. Future investigation of CRC should focus on bacterial biofilms and additional potential pro-carcinogenic synergisms between other species of the gut microbiota to improve prevention and screening measures.
Colorectal cancer (CRC) is the third most common cancer among men and women, surpassed only by prostate and breast cancer, for men and women, respectively, and lung cancer (Siegel et al., 2022). In 2022, there were an estimated 151,030 new cases of CRC and 52,980 deaths in the United States alone (Siegel et al., 2022). Interestingly, from 2014 to 2018, annual CRC incidence rates declined by approximately 2% in individuals 50 years and older but increased by approximately 1.5% in individuals younger than 50. As a result of this alarming increase, the American Cancer Society has recommended that CRC screenings begin at age 45 for those at an average risk (Siegel et al., 2022).
The etiology of CRC can be influenced by a multitude of factors, including sporadic mutations, pre-existing chronic inflammation, and hereditary factors (Mármol et al., 2017). However, if these factors are closely examined, a common denominator may be identified in all individuals regardless of the age of onset and origin: the gut microbiome. Under conditions of dysbiosis, a change in the microbiome potentially induced by these factors, members of the microbiota can invade the intestinal epithelial cell (IEC) barrier, inducing damage, inflammation, and the formation of biofilms (Chew et al., 2020). Recently, Dejea et al. (2018) found evidence of a spatial association of B. fragilis and E. coli in biofilms, as well as a synergistic pro-carcinogenic involvement between these predominant bacterial species of the gut microbiota (Dejea et al., 2018). While the exact mechanism of microbial-induced CRC has yet to be elucidated, the Alpha bug model (Sears and Pardoll, 2011) and driver-passenger model (Tjalsma et al., 2012) provide some explanation of how the microbiota may induce carcinogenesis in general. This review focuses on the roles of B. fragilis and E. coli in the presence and absence of disease, and the current evidence associating each bacterium with CRC individually and in synergism with one another.
Microbiota are extensive communities of microorganisms distributed throughout the human body, with the gastrointestinal tract being one of the most densely populated environments. The microbiome includes the genes harbored by these microorganisms (Turnbaugh et al., 2007). With its array of functions and critical roles in many different aspects of human health, the microbiome is sometimes regarded as its own organ (Baquero and Nombela, 2012). The human gut microbiome can house over 35,000 bacterial species and constitutes approximately 70% of the total microorganisms present in humans (Sekirov et al., 2010). Under normal healthy conditions, gut microbiota exist in a commensal relationship with the host, aiding in digestion, metabolism, and immunity. However, in conditions of dysbiosis, disease can quickly ensue from alterations or imbalances in the composition of the microbiota. Opportunistic and/or pathogenic bacteria can outcompete the commensal bacteria and subsequently invade the IEC barrier, causing inflammation and the development of a possible pre-malignant environment.
A vast majority of the microorganisms present in the gut exist in the intestinal lumen; however, some are also found in the mucosal layer of the epithelium (Van den Abbeele et al., 2011). When bacteria interact and invade the mucin layer, making contact with the epithelium, they can induce structural changes to IECs, resulting in intestinal inflammation and changes to the surrounding microbial community, including the formation of biofilms (Tytgat et al., 2019; Chew et al., 2020). Biofilms, bacterial aggregates or polymicrobial communities, originate as small aggregates of adhered bacteria that evolve into mature biofilms upon enclosure in a matrix of secreted polysaccharides, proteins, and nucleotides, collectively known as the extracellular polymeric substances (EPS) (Flemming and Wingender, 2010; Tytgat et al., 2019). The EPS matrix functions to promote adhesion and aggregation of bacteria, serves as a protective layer for the biofilm community, and acts as a nutrient source (Flemming and Wingender, 2010). While gut biofilms can include pathogenic or nonpathogenic bacterial species with a propensity for beneficial or detrimental roles, they appear to be potential markers for disease development (Tytgat et al., 2019), and it has been suggested that microbial-induced CRC is dependent on the formation of biofilms in the gut (Dejea et al., 2014; Chew et al., 2020). The pathogenicity of biofilms can be attributed to their ability to localize bacteria to mucosal surfaces, promote interactions and cooperation between bacterial species, and protect bacteria from harm (Tytgat et al., 2019).
Biofilms are present in both healthy individuals and CRC patients, although more frequently seen in the latter (13% in healthy and 50% in CRC populations) (Dejea et al., 2014; Chew et al., 2020). Biofilms associated with CRC have been shown to harbor different bacterial species, including those from Veillonellaceae, Lachnospiraceae, Coriobacteriaceae, Bacteroidetes, and Firmicutes (Dejea et al., 2014; Drewes et al., 2017). The transfer of microbiota from biofilm-positive mucosa, obtained from both healthy individuals and patients with CRC, to germ-free mice led to the development of CRC (Tomkovich et al., 2020). Both tumor and non-tumor tissues harboring biofilms revealed reduced expression of E-cadherin in crypt cells, increased polyamine production, increased interleukin 6 (IL-6) expression and activation of signal transducer and activator of transcription (STAT3) in IECs, and increased IEC proliferation (Figure 1) (Dejea et al., 2014; Chew et al., 2020). E-cadherin is a tumor suppressor protein that forms adherens junctions, and loss of E-cadherin correlates with increased tumoral invasiveness and metastasis (Vleminckx et al., 1991). Interleukin-6 and its pro-inflammatory effects contribute to immunosuppression, angiogenesis, and increased tumor cell proliferation, survival, and metastasis. Interleukin-6 also activates STAT3 signaling, further promoting tumor proliferation and carcinogenesis. Biofilm organization may also show spatial preference, as biofilms have been associated with 89% of right-sided tumors versus only 12% of left-sided tumors (Dejea et al., 2014).
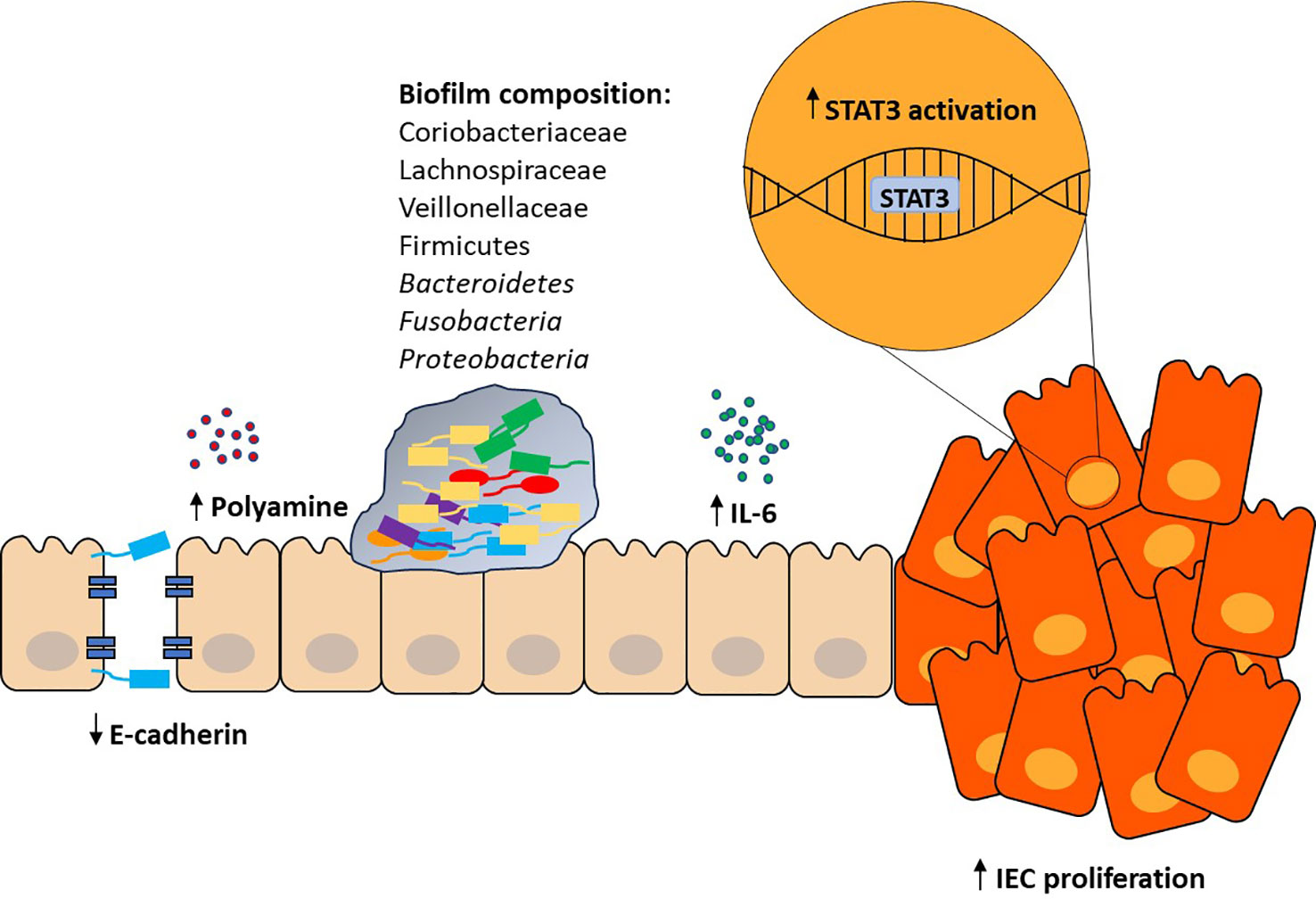
Figure 1 Proposed role of gut biofilm in CRC. Investigation of gut biofilms has revealed numerous changes to the IEC barrier and local inflammatory environment. Increased polyamine production leads to a myriad of functions regulated by the gut microbiota. Loss of E-cadherin results in a breach in the tight junction integrity of the IEC barrier. Increased IL-6 expression and STAT3 activation results in IEC proliferation, invasiveness, and angiogenesis. Combined, these effects reveal the pro-carcinogenic potential of gut biofilms in the development of CRC.
With an increase in research focused on the role of the gut microbiome in disease development, an association between specific members of the microbiota and CRC has been revealed (Cheng et al., 2020; Abdulla et al., 2021). For example, B. fragilis has been shown to induce epithelial-mesenchymal transition (EMT) in the colon; cleave E-cadherin through B. fragilis toxin (BFT); activate the Wnt, nuclear factor-κB (NF-κB), STAT3, and mitogen-activated protein kinase (MAPK) signaling pathways; activate production of pro-inflammatory cytokines; and enhance expression of oncogenes (Abdulla et al., 2021). Escherichia coli alters cell cycle progression through induction of DNA damage by its virulence factor, colibactin (Abdulla et al., 2021). Fusobacterium nucleatum has also been shown to trigger EMT in the colon, activate NF-κB and the production of pro-inflammatory cytokines, and inhibit anti-tumor immunity, all contributing to inflammation and tumorigenesis (Abdulla et al., 2021). Peptostreptococcus anaerobius alters Toll-like receptors 2 and 4 (TLR2 and TLR4) signaling, leading to accumulation of reactive oxygen species (Abdulla et al., 2021). Streptococcus gallolyticus has been implicated in activation of Wnt signaling and upregulation of oncogenes (Abdulla et al., 2021). While these associations have been influential in further elucidating the relationship between microbiota and CRC, how these bacteria interact with each other specifically in the context of inflammation and CRC development must be examined to fully uncover the mechanisms underlying microbial-induced CRC. B. fragilis and E. coli are the first of these bacterial species discovered to exist in a synergistic interaction affecting the development of CRC. Hence, we will focus our discussion on B. fragilis and E. coli.
The Bacteroides species are Gram-negative, obligate anaerobic bacteria that represent approximately 25% of all anaerobes in the gut microbiota (Wexler, 2007). Bacteroides spp. mostly exist in a commensal relationship with their host and serve key roles in nutrition and immunity (Wexler, 2007; Sears, 2009; Zafar and Saier, 2021). However, some strains can be opportunistic pathogens, causing widespread disease if they escape the gut. B. fragilis is one of the most widely studied of these strains due to its opportunistic nature and high virulence. Furthermore, B. fragilis is the most commonly isolated anaerobe from clinical cases of diarrhea, sepsis, and extra-intestinal infections (Haghi et al., 2019).
Strains of B. fragilis are characterized based on the presence or absence of a 20-kDa protein toxin called B. fragilis toxin (BFT), or fragilysin (Moncrief et al., 1995; Wu et al., 1998; Wu et al., 2004). Enterotoxigenic B. fragilis (ETBF) contain this toxin, while non-enterotoxigenic B. fragilis (NTBF) do not. BFT is transcribed from the B. fragilis toxin gene (bft) and is one of three different isotypes, BFT-1, BFT-2, and BFT-3 (Franco et al., 2002). BFT functions as a zinc-dependent metalloprotease that cleaves E-cadherin and causes its degradation (Wu et al., 1998; Wu et al., 2004). E-cadherin is necessary for maintaining the mechanical integrity of the IEC barrier and proper maturation of Paneth and goblet cells, and reduced levels of E-cadherin has been linked to both ulcerative colitis and Crohn’s disease (Schneider et al., 2010). BFT-induced cleavage of E-cadherin stimulates the opportunistic nature of ETBF and promotes disease pathogenesis. ETBF and its toxic properties were initially described in lamb diarrheal disease and later spread to other livestock, such as piglets, foals, and calves (Myers et al., 1984; Myers et al., 1985; Myers and Shoop, 1987; Myers et al., 1987a; Collins et al., 1989). It was further described in cases of diarrhea in children (Myers et al., 1987b; Sack et al., 1992; Sack et al., 1994; Durmaz et al., 2005). It has since been established as a global cause of diarrhea (Sears, 2009).
Bacteroides fragilis toxin induces morphological changes in IECs including loss of cell-cell adhesions, rounding, swelling, and alterations to the actin cytoskeleton (Weikel et al., 1992; Donelli et al., 1996). It enhances expression of chemokines (interleukin 8 (IL-8), transforming growth factor beta (TGF-β), epithelial neutrophil-activating protein 78 (ENA-78), and growth-regulated alpha protein (GRO-α)) (Sanfilippo et al., 2000; Kim et al., 2001); secretory response factors in the gut (cyclooxygenase-2 (COX-2) and prostaglandin E2) (Kim et al., 2006); leukocyte recruitment proteins (intercellular adhesion molecule 1 (ICAM-1)) (Roh et al., 2011); and anti-inflammatory mediators (heme oxygenase-1 (HO-1)) (Ko et al., 2016; Ko et al., 2020). BFT activates NF-κB, β-catenin, and MAPK signaling (KIM et al., 2002; Kim et al., 2005) and plays a role in the upregulation of the DNA damage marker, phosphorylated H2AX (γ-H2AX), and production of ROS through spermine oxidase (SMO) (Goodwin et al., 2011). Recently, Allen et al. (2022) utilized whole-genome and whole-exome sequencing on mouse tumors after colonization with ETBF and found that ETBF did not produce a unique mutational profile (Allen et al., 2022). ETBF-induced tumors followed a similar mutational pattern as spontaneous tumors, resulting from errors in DNA mismatch repair machinery and homologous recombination. Interestingly, B. fragilis was significantly enriched in mismatch repair-deficient (dMMR) CRC but not mismatch repair-proficient (pMMR) CRC (Hale et al., 2018).
B. fragilis has also been shown to modulate the anti-tumor immune response to immunotherapy (Vétizou et al., 2015). Mouse tumors grown in germ-free conditions or with broad-spectrum antibiotics were unresponsive to cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4) blockade; however, when B. fragilis was recolonized in mice, these tumors regained responsiveness to CTLA-4 treatment in a manner consistent with an elevated T helper 1 (Th1) response and maturation of intratumoral dendritic cells (DCs) (Vétizou et al., 2015). These findings not only suggest the need of gut microbiota for the efficacy of some immunotherapies, but also the protective, anti-cancer properties of B. fragilis in bolstering immunotherapy.
It should be noted that Kordahi et al. (2021) recently discovered a possible role of NTBF in CRC development, despite being the so-called non-enterotoxigenic form. Researchers found that B. fragilis isolates from patients with colonic polyps were colonized mostly with NTBF, not ETBF, and NTBF was positively correlated with levels of interleukin 12p40 (IL-12p40) and polyp size (Kordahi et al., 2021). These NTBF isolates were enriched in genes related to lipopolysaccharide (LPS) synthesis and were able to induce TLR4 signaling and a pro-inflammatory response (Kordahi et al., 2021). Based on the findings by Kordahi et al. (2021), NTBF should not be excluded as a plausible contributor to CRC development solely due to its lack of BFT.
Escherichia coli is a Gram-negative, facultative anaerobe present in the gut microbiome of over 90% of people (Martinson and Walk, 2020). Aside from a few select pathogenic strains, E. coli has a commensal relationship with its host and maintains a myriad of critical roles in the human gut, such as vitamin K production (Suvarna et al., 1998) and protection against pathogenic microbes in the gut (Richter et al., 2018). Furthermore, E. coli uses up oxygen present in the gut, thus bolstering an anaerobic environment for obligate anaerobes such as B. fragilis to thrive (Mueller et al., 2015). The essential physiological roles and relative abundance of E. coli in the human gut make it a great candidate for investigating microbial-induced CRC.
Escherichia coli strains are divided into four phylotypes, A, B1, B2, and D (Friesen, 1988; Herzer et al., 1990; Clermont et al., 2000). Most commensal strains belong to phylotype A, while most pathogenic strains belong to phylotype B2 and D (Bingen et al., 1998; Boyd and Hartl, 1998; Picard et al., 1999; Johnson and Stell, 2000). Pathogenic strains of E. coli are common causes of urinary tract infections, sepsis, meningitis, etc. (Johnson and Russo, 2002a; Johnson and Russo, 2002b). The pathogenicity of certain E. coli strains can be attributed to the presence of different virulence factors and the ability to induce a genotoxic effect in cells. The bacterial toxins that stimulate this genotoxic effect are termed cyclomodulins (Nougayrède et al., 2005). Cyclomodulins interfere with the eukaryotic cell cycle through induction of DNA damage and genomic instability, both of which can contribute to carcinogenesis (Gagnière et al., 2016).
The B2 phylotype of E. coli possesses a genomic island called the polyketide synthetase (pks) island (Nougayrède et al., 2006; Nowrouzian and Oswald, 2012) which carries the information to produce a polyketide-peptide toxin called colibactin (Nougayrède et al., 2006; Nowrouzian and Oswald, 2012). The structure of colibactin has recently been confirmed as containing an α-dicarbonyl group and two electrophilic spirocyclopropyldihydro-2-pyrrolone groups that can undergo ring-opening to attach to DNA (Xue et al., 2019). Although the exact signaling mechanism of colibactin has yet to be uncovered, its genotoxic activity in eukaryotic cells has been examined. Like the other cyclomodulins of E. coli, colibactin translocates to the nucleus where it induces interstrand crosslinks and double-stranded DNA breaks (Nougayrède et al., 2006; Dougherty and Jobin, 2021). Colibactin has also been shown to trigger alkylation and formation of adenine adducts in vitro and in vivo through the electrophilic cyclopropane groups (Wilson et al., 2019).
It has been established that pks+ E. coli can induce a unique mutational profile, providing evidence of the causality between colibactin and mutations in IECs (Pleguezuelos-Manzano et al., 2020). Whole genome sequencing (WGS) revealed a specific mutational signature of colibactin characterized by single base substitutions (SBS-pks), specifically T>N substitutions at ATA, ATT, and TTT sites, as well as a small indel signature (ID-pks) characterized by single T deletions within T homopolymers (Pleguezuelos-Manzano et al., 2020). These same SBS-pks and ID-pks mutational signatures were found enriched in CRC-derived metastases compared to other solid cancer metastases (Pleguezuelos-Manzano et al., 2020). Although further research is necessary, taken altogether, these data may suggest that in respect to the proposed models of the relationship between the microbiota and CRC, pks+ E. coli may be the driver that initiates CRC development while ETBF further promotes it via extra-genomic mechanisms.
Colibactin has also been shown to exert its genotoxic effects on the local microbial community. Infection of pregnant mice with the genotoxic B2 phylotype of E. coli revealed microbial changes in the guts of mouse offspring at two different time points: 15 and 35 days. At day 15, the mice displayed increased abundance of Lachnospiraceae, decreased abundance of Proteobacteria, and significant alterations to the overall microbial profile despite no change in overall diversity (Tronnet et al., 2020). At day 35, the mice displayed decreased abundance of Firmicutes and related taxa, increased abundance of Deferribacteres and Tenericutes, increased overall diversity, and enrichment in microbial DNA replication and repair pathways (Tronnet et al., 2020). The findings from Tronnet et al. (2020) reveal a significant role for colibactin in influencing and modifying the surrounding microbial community. Further findings from Chen et al. (2022) revealed that the genotoxic B2 phylotype of E. coli reduced populations of Vibrio cholerae by three to five orders of magnitude, inhibited the growth of other Vibrio species, of Enterobacter aerogenes and Staphylococcus, and provided direct evidence of colibactin-mediated reduction of B. fragilis (Chen et al., 2022), thus emphasizing the role of colibactin in shaping the gut microbiota and influencing the interactions with invading pathogens.
The precise mechanism(s) of how the gut microbiome contributes to carcinogenesis and the development of CRC is still not completely understood, perhaps due to the vast array of microorganisms present, extreme interpersonal heterogeneity, or simply due to microbiome research being a somewhat newly growing field. Previous studies have proposed two explanations that attempt to shed light on the relationship between gut microbiota and CRC: 1) the Alpha-bug model and 2) the driver-passenger model (Figure 2).
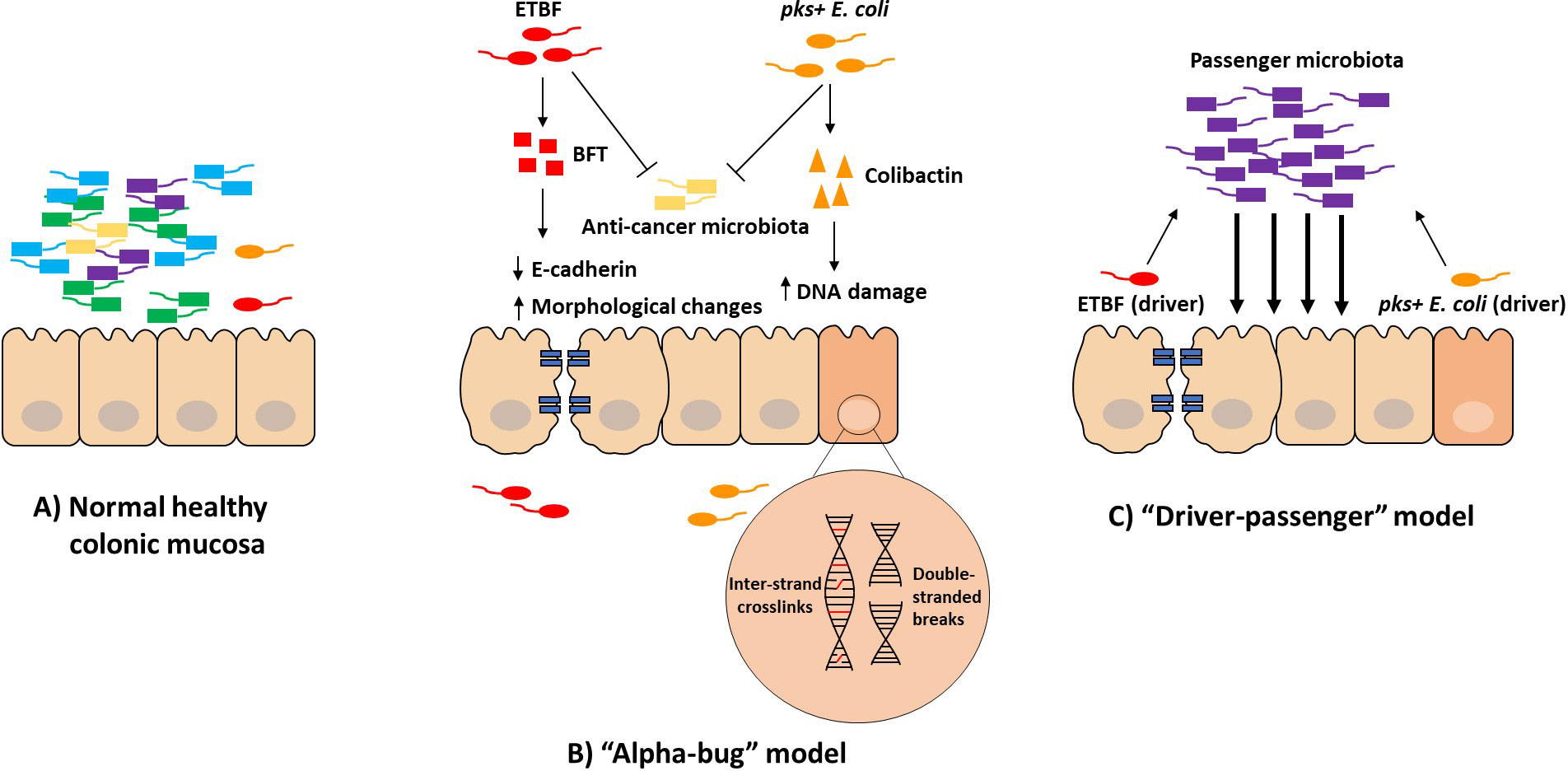
Figure 2 Proposed models of the relationship between gut microbiota and CRC. Under normal healthy conditions, gut microbiota exist in a commensal relationship with the host (A). Under conditions of dysbiosis, opportunistic and/or pathogenic microbiota can invade the IEC barrier and promote an inflammatory and pro-carcinogenic environment. The “Alpha-bug” model (B) describes the ability of the “Alpha-bug” (e.g., ETBF and pks+ E. coli) to induce carcinogenesis directly by its respective genotoxins and indirectly by inhibiting anti-cancer microorganisms and altering the local microbiome. The “driver-passenger” model (C) describes the ability of the “driver” to initiate carcinogenesis while the “passengers” maintain and bolster the diseased state.
The “Alpha-bug” model, proposed by Sears and Pardoll (2011), refers to the pro-carcinogenic bacterium as an “Alpha-bug” that demonstrates a three-fold potential in stimulating CRC pathogenesis (Sears and Pardoll, 2011). First, the Alpha-bug can induce carcinogenesis directly through its virulence or genotoxic metabolites. Second, the Alpha-bug can alter its local microbial community to further promote the transition to an oncogenic state and the genotoxic activity of the Alpha-bug. Third, the Alpha-bug can inhibit anti-cancer microorganisms by “crowding out” these protective species, thus further enhancing carcinogenesis (Sears and Pardoll, 2011). This combination of inducing tumorigenesis directly, remodeling the microbial community, and removing anti-cancer bacteria is a potential model of microbial-involvement in the development of CRC by an Alpha-bug. Both B. fragilis and E. coli have the potential to be Alpha-bugs (Figure 2B).
The “driver-passenger” model, first proposed by Tjalsma et al. (2012), distinguishes bacteria into two types based on their temporal association with the development of CRC (Tjalsma et al., 2012). First, the “driver” bacterium utilizes its genotoxins to induce damage to the IEC barrier. This begins a cascade of changes that eventually lead to CRC pathogenesis. The genotoxin-induced epithelial changes cause alterations in the local microbiome, allowing for opportunistic pathogens, the “passengers”, to thrive and further promote carcinogenesis. Thus, in this model, two key bacterial players contribute to disease pathogenesis (Figure 2C). Although the driver bacterium initiates CRC development, it is the opportunistic pathogens that maintain the diseased state.
Before discussing the synergistic involvement of B. fragilis and E. coli in the development of CRC, it is important to first discuss the evidence for involvement of each bacterium in CRC individually. B. fragilis has been extensively linked to CRC in vivo. Gnotobiotic mice developed intestinal ulcerations, edema, and inflammatory infiltration after infection with BFT-producing ETBF, but not NTBF (Nakano et al., 2006). Furthermore, ETBF was shown to cause colitis independently (Rhee et al., 2009; Goodwin et al., 2011; Wick et al., 2014; Hwang et al., 2020), worsen colitis induced by dextran sodium sulfate (DSS) (Rabizadeh et al., 2007), induce colon tumorigenesis in a Stat3/T helper 17 cell (Th17)-mediated manner (Wu et al., 2009; Chung et al., 2018; Liu et al., 2020), increase median amounts of colon tumors with increased duration of ETBF colonization (DeStefano Shields et al., 2016), and promote polyp formation and tumorigenesis in an azoxymethane (AOM)/DSS mouse model (Hwang et al., 2020).
Several human observational studies have also demonstrated a link between B. fragilis and CRC. A study by Toprak et al. (2006) first provided the evidence for this association, with bft being detected at a higher rate in stool samples of CRC patients compared to the control group (Toprak et al., 2006). A later study by Boleij et al. (2015) also detected bft at higher rates in colon mucosal samples of CRC patients compared to the control group (Boleij et al., 2015) and more so in late- versus early-stage CRC, a finding that was supported by another study (Viljoen et al., 2015). Other studies have shown an association between ETBF and CRC; ETBF was detected at higher rates in tumors compared to normal tissues and controls in 97 CRC patients from South China (Zhou et al., 2016). Also, an association was found between ETBF positivity and low-grade dysplasia, tubular adenomas, and serrated polyps, indicating a possible role of ETBF in CRC development (Purcell et al., 2017). Further studies have successfully reproduced these findings of increased detection of B. fragilis in CRC patients. Despite this, some studies have shown results contradictory to the proposition that B. fragilis expression is enhanced in CRC (Dutilh et al., 2013; Zeller et al., 2014; Lennard et al., 2016; Wirbel et al., 2019; Piciocchi et al., 2021; Shariati et al., 2021). Whether due to method design, confounding variables, etc., more research is needed to compare studies and truly uncover the causative role of B. fragilis in CRC development.
Escherichia coli has also demonstrated a role in colon tumorigenesis in vivo. Germ-free Il10-/- mice colonized with pks+ or pks- E. coli developed severe colitis, and AOM-treated germ free Il10-/- mice colonized with pks+ E. coli developed increased colonic tumorigenesis (Arthur et al., 2012). Interestingly, AOM-treated germ-free Il10-/-Rag2-/- mice, lacking functional T and B cells, did not develop colonic inflammation or tumorigenesis when colonized with pks+ E. coli, suggesting that pks+ E. coli requires inflammation to promote tumorigenesis initiated by AOM. AOM/DSS-treated mice also developed enhanced colonic tumorigenesis when colonized with pks+ E. coli (Cougnoux et al., 2014). Furthermore, multiple intestinal neoplasia (Min) mice, harboring a mutant allele in the APC gene, colonized with pks+ E. coli developed increased colonic polyps compared to uncolonized controls (Bonnet et al., 2014).
The role of E. coli has also been reported by human observational studies of CRC. In a study by Arthur et al. (2012), the abundance of pks+ E. coli was significantly increased in 35 IBD and 21 CRC patients (Arthur et al., 2012). Other studies investigating the abundance of pks+ E. coli in patients with CRC or diverticulosis found higher percentages of E. coli in CRC patients compared to patients with diverticulitis, although the percentages varied slightly between studies (Buc et al., 2013; Bonnet et al., 2014; Raisch et al., 2014). Elevated levels of pks+ E. coli markers have also been detected in stool samples of patients diagnosed with CRC (Eklöf et al., 2017). Also, higher levels of pks+ E. coli were detected in CRC patients compared to healthy controls (Prorok-Hamon et al., 2014; Iyadorai et al., 2020). Some studies suggest E. coli B2 strains heavily associate with cancerous lesions more so due to a result of tumorigenesis as opposed to a cause of tumorigenesis (Brader et al., 2008; Stritzker et al., 2010; Kocijancic et al., 2016). Again, more research and longitudinal studies are needed to uncover the causative roles of pks+ E. coli in the development of CRC.
The synergistic potential of B. fragilis and E. coli was initially described in intra-abdominal infection (Rotstein et al., 1985). Bacteroides fragilis-E. coli coinfection of intraperitoneal fibrin clots in rats resulted in increased mortality compared to either bacterium alone. Similarly, B. fragilis enhanced the severity and persistence of E. coli-induced abscesses and vice versa (Rotstein et al., 1989).
Based on the proposed models of the relationship between the gut microbiota and CRC, it is unlikely for one microorganism to individually affect the colon in the transition to an oncogenic state. Considering both B. fragilis and E. coli are plausible “Alpha-bugs,” the possibility that these two bacteria work together to contribute to the development of CRC should not be excluded. Furthermore, biofilms found in the colons of patients diagnosed with CRC are commonly composed of both ETBF and E. coli, providing further evidence of this profound interaction (Dejea et al., 2018).
Dejea et al. (2018) provided the first evidence of a possible synergistic role between B. fragilis and E. coli in the development of CRC (Dejea et al., 2018). Examination of the colonic mucosa of five patients with familial adenomatous polyposis (FAP) revealed the presence of biofilms. These FAP biofilms were found throughout the colon and consisted of mainly B. fragilis and E. coli. Moreover, the mucosal samples from 25 patients with FAP were significantly associated with ETBF and pks+ E. coli compared to the mucosal samples from healthy controls.
To investigate whether ETBF and pks+ E. coli coinfection enhances tumorigenesis compared to either bacterium alone, Dejea et al. utilized ApcMinΔ716/+ and AOM-injected wild-type (WT) mice. Compared to infection with either bacterium alone, AOM-treated WT mice developed more tumors when coinfected with ETBF and pks+ E. coli, and coinfected ApcMinΔ716/+ mice had shortened survival times. Interestingly, AOM-treated WT mice developed few to no tumors when infected with ETBF or pks+ E. coli alone, indicating the necessity for both bacteria to synergistically induce carcinogenesis (Dejea et al., 2018). Coinfected mice also displayed significantly increased colonic hyperplasia and micro-adenomas compared to monoinfection with either bacterium, and significantly increased inflammation compared to monoinfection with pks+ E. coli but not monoinfection with ETBF (Dejea et al., 2018).
Dejea et al. also discovered that coinfected mice had a significantly increased fecal IgA response to pks+ E. coli compared to mice infected with pks+ E. coli alone, and displayed an increase in mucosal-associated pks+ E. coli in the distal colon compared to only the colonic lumen in monoinfection with pks+ E. coli (Dejea et al., 2018). This indicates that increased tumorigenesis may be a result of a shift in the spatial arrangement of pks+ E. coli from the lumen to the colonic mucosa. ETBF alone or with pks+ E. coli significantly reduced the mucous depth like the well-known colonic mucin-degrading bacterium, Akkermansia muciniphila. However, coinfection of AOM-treated WT mice with A. muciniphila and pks+ E. coli resulted in reduced tumorigenesis, suggesting that alterations to the mucosa alone is insufficient in promoting carcinogenesis and hinting at a potential additive effect of B. fragilis in carcinogenesis. Additionally, DNA damage in IECs of coinfected mice was significantly increased compared to mice infected with each bacterium alone. Combined, these results indicate that ETBF-induced mucosal degradation is necessary for enhanced pks+ E. coli colonization, possibly allowing colibactin to further damage IECs. The data from Dejea et al. (2018) suggest a role for ETBF and pks+ E. coli interactions in promoting carcinogenesis during coinfection of the gut, and the direct evidence provided by Chen et al. (2022) for colibactin-mediated reduction of B. fragilis further emphasize the complex interactions between ETBF and E. coli and their potential influence on gut pathogenesis. Further investigation into the targeted killing of B. fragilis by colibactin-producing E. coli is needed to elucidate these interactions and their possible effects on the gut microbiome.
Colorectal cancer is one of the most common and deadly cancer types globally, and despite advancements in our understanding, detection, and treatment, the incident risk for individuals under 50 years is increasing. While numerous risk factors are associated with the development of CRC, more than 80% of cases contain microbial signatures representative of dysbiosis (Drewes et al., 2017). However, the role of the gut microbiome remains elusive. Although several studies have examined the composition of the gut microbiome and particular microbiota that may have a role in the development of CRC, a definitive, direct causal role has yet to be uncovered. Pathogenic strains of two bacteria that are normal inhabitants of the gut microbiome, B. fragilis and E. coli, have been shown to induce carcinogenesis in vivo and have been associated with CRC in human observational studies. More recently, however, a synergistic role of B. fragilis and E. coli has been shown in mouse models, suggesting that analysis of both may be of importance in future screening and preventive measures.
The synergistic activity of B. fragilis and E. coli in vivo presents an interesting and exciting area for potential future research. But how do we expand our understanding of potential pro-carcinogenic synergisms between other species of the microbiota? The answer to this question requires a retrospective view. The synergism between ETBF and pks+ E. coli was investigated, in-part, due to their high association in bacterial-biofilms of CRC. So, to uncover additional microbial associations and possible synergistic contributions with CRC, biofilms must be at the forefront of investigation. Biofilms of CRC patients frequently house Bacteroidetes, Lachnospiraceae, Fusobacterium and Proteobacteria taxa (Dejea et al., 2014; Drewes et al., 2017), so investigating biofilms may allow us to identify possible, future synergistic candidates. Biofilms serve as one route to further uncover the role of the microbiome in the development of CRC.
So, how do we fill in this knowledge gap to further analyze the gut microbiome to improve and develop new cancer prevention and screening measures? First, and most importantly, more longitudinal studies are needed on the gut microbiome and CRC to allow for cross-study comparisons, which has been a limiting factor thus far. Slight differences between in vivo studies, mostly methodology related, have made cross-study comparisons challenging and unclear, and human observational studies show mixed results. Second, advanced sequencing technology has proved influential in the discovery and analysis of microbiota when traditional culture methods present problems. This technology needs to be further employed in early-life studies of microbiota, as well as in identifying additional biofilms and associated microbiota that may play a role in CRC development. With that, we may be able to utilize these CRC-associated biofilms and microbiota to predict disease risk and clinical outcomes. Third, further studies and research should focus on the development of therapeutics and treatments geared towards the microbiome for improved CRC treatment and prevention. As each individual has a unique, personalized gut microbiome, personalized treatment strategies will be ideal in ensuring the best clinical results. Overall, the gut microbiome has a wide range of potential benefits in managing CRC from screening and predictive biomarkers to prevention and treatments. As once stated by the wise Dutch philosopher Desiderius Erasmus, “prevention is better than cure” and this holds even more true in the present day and the current state of the gut microbiome in CRC.
CL and RL-K conceived and designed the study topic, conducted the review, and wrote the paper. CL created the figures. Both authors contributed to the article and approved the submitted version.
The authors thank the Department of Biomedical Education, California Health Sciences University-College of Osteopathic Medicine for their support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdulla M.-H., Agarwal D., Singh J. K., Traiki T. B., Pandey M. K., Ahmad R., et al. (2021). Association of the microbiome with colorectal cancer development (Review). Int. J. Oncol. 58, 1–12. doi: 10.3892/ijo.2021.5197
Allen J., Rosendahl Huber A., Pleguezuelos-Manzano C., Puschhof J., Wu S., Wu X., et al. (2022). Colon Tumors in Enterotoxigenic Bacteroides fragilis (ETBF)-Colonized Mice Do Not Display a Unique Mutational Signature but Instead Possess Host-Dependent Alterations in the APC Gene. Microbiol. Spectr. 10, e01055–e01022. doi: 10.1128/spectrum.01055-22
Arthur J. C., Perez-Chanona E., Mühlbauer M., Tomkovich S., Uronis J. M., Fan T.-J., et al. (2012). Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123. doi: 10.1126/science.1224820
Baquero F., Nombela C. (2012). The microbiome as a human organ. Clin. Microbiol. Infection 18, 2–4. doi: 10.1111/j.1469-0691.2012.03916.x
Bingen E., Picard B., Brahimi N., Mathy S., Desjardins P., Elion J., et al. (1998). Phylogenetic analysis of escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 group strains. J. Infect. Dis. 177, 642–650. doi: 10.1086/514217
Boleij A., Hechenbleikner E. M., Goodwin A. C., Badani R., Stein E. M., Lazarev M. G., et al. (2015). The bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin. Infect. Dis. 60, 208–215. doi: 10.1093/cid/ciu787
Bonnet M., Buc E., Sauvanet P., Darcha C., Dubois D., Pereira B., et al. (2014). Colonization of the human gut by E. coli and colorectal cancer risk. Clin. Cancer Res. 20, 859–867. doi: 10.1158/1078-0432.CCR-13-1343
Boyd E. F., Hartl D. L. (1998). Chromosomal regions specific to pathogenic isolates of escherichia coli have a phylogenetically clustered distribution. J. Bacteriol 180, 1159–1165. doi: 10.1128/JB.180.5.1159-1165.1998
Brader P., Stritzker J., Riedl C. C., Zanzonico P., Cai S., Burnazi E. M., et al. (2008). Escherichia coli nissle 1917 facilitates tumor detection by positron emission tomography and optical imaging. Clin. Cancer Res. 14, 2295–2302. doi: 10.1158/1078-0432.CCR-07-4254
Buc E., Dubois D., Sauvanet P., Raisch J., Delmas J., Darfeuille-Michaud A., et al. (2013). High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PloS One 8, e56964. doi: 10.1371/journal.pone.0056964
Chen J., Byun H., Liu R., Jung I.-J., Pu Q., Zhu C. Y., et al. (2022). A commensal-encoded genotoxin drives restriction of Vibrio cholerae colonization and host gut microbiome remodeling. Proc. Natl. Acad. Sci. 119, e2121180119. doi: 10.1073/pnas.2121180119
Cheng Y., Ling Z., Li L. (2020). The intestinal microbiota and colorectal cancer. Front. Immunol. 11. doi: 10.3389/fimmu.2020.615056
Chew S.-S., Tan L. T.-H., Law J. W.-F., Pusparajah P., Goh B.-H., Ab Mutalib N. S., et al. (2020). Targeting gut microbial biofilms—A key to hinder colon carcinogenesis? Cancers (Basel) 12, 2272. doi: 10.3390/cancers12082272
Chung L., Orberg E. T., Geis A. L., Chan J. L., Fu K., DeStefano Shields C. E., et al. (2018). Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host Microbe 23, 203–214.e5. doi: 10.1016/j.chom.2018.01.007
Clermont O., Bonacorsi S., Bingen E. (2000). Rapid and simple determination of the escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66, 4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000
Collins J. E., Bergeland M. E., Myers L. L., Shoop D. S. (1989). Exfoliating colitis associated with enterotoxigenic bacteroides fragilis in a piglet. J. Vet. Diagn. Invest. 1, 349–351. doi: 10.1177/104063878900100413
Cougnoux A., Dalmasso G., Martinez R., Buc E., Delmas J., Gibold L., et al. (2014). Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut 63, 1–11. doi: 10.1136/gutjnl-2013-305257
Dejea C. M., Fathi P., Craig J. M., Boleij A., Taddese R., Geis A. L., et al. (2018). Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 359, 592–597. doi: 10.1126/science.aah3648
Dejea C. M., Wick E. C., Hechenbleikner E. M., White J. R., Mark Welch J. L., Rossetti B. J., et al. (2014). Microbiota organization is a distinct feature of proximal colorectal cancers. Proc. Natl. Acad. Sci. U.S.A. 111, 18321–18326. doi: 10.1073/pnas.1406199111
DeStefano Shields C. E., Van Meerbeke S. W., Housseau F., Wang H., Huso D. L., Casero R. A., et al. (2016). Reduction of murine colon tumorigenesis driven by enterotoxigenic bacteroides fragilis using cefoxitin treatment. J. Infect. Dis. 214, 122–129. doi: 10.1093/infdis/jiw069
Donelli G., Fabbri A., Fiorentini C. (1996). Bacteroides fragilis enterotoxin induces cytoskeletal changes and surface blebbing in HT-29 cells. Infect. Immun. 64, 113–119. doi: 10.1128/iai.64.1.113-119.1996
Dougherty M. W., Jobin C. (2021). Shining a light on colibactin biology. Toxins (Basel) 13, 346. doi: 10.3390/toxins13050346
Drewes J. L., White J. R., Dejea C. M., Fathi P., Iyadorai T., Vadivelu J., et al. (2017). High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes 3, 34. doi: 10.1038/s41522-017-0040-3
Durmaz B., Dalgalar M., Durmaz R. (2005). Prevalence of Enterotoxigenic Bacteroides fragilis in patients with diarrhea: A controlled study. Anaerobe 11, 318–321. doi: 10.1016/j.anaerobe.2005.06.001
Dutilh B. E., Backus L., van Hijum S. A. F. T., Tjalsma H. (2013). Screening metatranscriptomes for toxin genes as functional drivers of human colorectal cancer. Best Pract. Res. Clin. Gastroenterol. 27, 85–99. doi: 10.1016/j.bpg.2013.03.008
Eklöf V., Löfgren-Burström A., Zingmark C., Edin S., Larsson P., Karling P., et al. (2017). Cancer-associated fecal microbial markers in colorectal cancer detection. Int. J. Cancer 141, 2528–2536. doi: 10.1002/ijc.31011
Flemming H.-C., Wingender J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633. doi: 10.1038/nrmicro2415
Franco A. A., Cheng R. K., Goodman A., Sears C. L. (2002). Modulation of bft expression by the Bacteroides fragilis pathogenicity island and its flanking region. Mol. Microbiol. 45, 1067–1077. doi: 10.1046/j.1365-2958.2002.03077.x
Friesen J. D. (1988). Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Science 240, 1678+. doi: 10.1126/science.240.4859.1678
Gagnière J., Raisch J., Veziant J., Barnich N., Bonnet R., Buc E., et al. (2016). Gut microbiota imbalance and colorectal cancer. World J. Gastroenterol. 22, 501–518. doi: 10.3748/wjg.v22.i2.501
Goodwin A. C., Shields C. E. D., Wu S., Huso D. L., Wu X., Murray-Stewart T. R., et al. (2011). Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 108, 15354–15359. doi: 10.1073/pnas.1010203108
Haghi F., Goli E., Mirzaei B., Zeighami H. (2019). The association between fecal enterotoxigenic B. fragilis with colorectal cancer. BMC Cancer 19, 879. doi: 10.1186/s12885-019-6115-1
Hale V. L., Jeraldo P., Chen J., Mundy M., Yao J., Priya S., et al. (2018). Distinct microbes, metabolites, and ecologies define the microbiome in deficient and proficient mismatch repair colorectal cancers. Genome Med. 10, 78. doi: 10.1186/s13073-018-0586-6
Herzer P. J., Inouye S., Inouye M., Whittam T. S. (1990). Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriology 172, 6175–6181. doi: 10.1128/jb.172.11.6175-6181.1990
Hwang S., Lee C. G., Jo M., Park C. O., Gwon S.-Y., Hwang S., et al. (2020). Enterotoxigenic Bacteroides fragilis infection exacerbates tumorigenesis in AOM/DSS mouse model. Int. J. Med. Sci. 17, 145–152. doi: 10.7150/ijms.38371
Iyadorai T., Mariappan V., Vellasamy K. M., Wanyiri J. W., Roslani A. C., Lee G. K., et al. (2020). Prevalence and association of pks+ Escherichia coli with colorectal cancer in patients at the University Malaya Medical Centre, Malaysia. PloS One 15, e0228217. doi: 10.1371/journal.pone.0228217
Johnson J. R., Russo T. A. (2002a). Extraintestinal pathogenic Escherichia coli: the other bad E coli. J. Lab. Clin. Med. 139, 155–162. doi: 10.1067/mlc.2002.121550
Johnson J. R., Russo T. A. (2002b). Uropathogenic Escherichia coli as agents of diverse non-urinary tract extraintestinal infections. J. Infect. Dis. 186, 859–864. doi: 10.1086/342490
Johnson J. R., Stell A. L. (2000). Extended virulence genotypes of escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181, 261–272. doi: 10.1086/315217
KIM J. M., CHO S. J., OH Y.-K., JUNG H.-Y., KIM Y.-J., KIM N. (2002). Nuclear factor-kappa B activation pathway in intestinal epithelial cells is a major regulator of chemokine gene expression and neutrophil migration induced by Bacteroides fragilis enterotoxin. Clin. Exp. Immunol. 130, 59–66. doi: 10.1046/j.1365-2249.2002.01921.x
Kim J. M., Jung H. Y., Lee J. Y., Youn J., Lee C.-H., Kim K.-H. (2005). Mitogen-activated protein kinase and activator protein-1 dependent signals are essential for Bacteroides fragilis enterotoxin-induced enteritis. Eur. J. Immunol. 35, 2648–2657. doi: 10.1002/eji.200526321
Kim J. M., Lee J. Y., Yoon Y. M., Oh Y.-K., Kang J. S., Kim Y.-J., et al. (2006). Bacteroides fragilis enterotoxin induces cyclooxygenase-2 and fluid secretion in intestinal epithelial cells through NF-κB activation. Eur. J. Immunol. 36, 2446–2456. doi: 10.1002/eji.200535808
Kim J. M., Oh Y. K., Kim Y. J., Oh H. B., Cho Y. J. (2001). Polarized secretion of CXC chemokines by human intestinal epithelial cells in response to Bacteroides fragilis enterotoxin: NF-κB plays a major role in the regulation of IL-8 expression. Clin. Exp. Immunol. 123, 421–427. doi: 10.1046/j.1365-2249.2001.01462.x
Ko S. H., Jeon J. I., Woo H. A., Kim J. M. (2020). Bacteroides fragilis enterotoxin upregulates heme oxygenase-1 in dendritic cells via reactive oxygen species-, mitogen-activated protein kinase-, and Nrf2-dependent pathway. World J. Gastroenterol. 26, 291–306. doi: 10.3748/wjg.v26.i3.291
Ko S. H., Rho D. J., Jeon J. I., Kim Y.-J., Woo H. A., Lee Y. K., et al. (2016). Bacteroides fragilis Enterotoxin Upregulates Heme Oxygenase-1 in Intestinal Epithelial Cells via a Mitogen-Activated Protein Kinase- and NF-κB-Dependent Pathway, Leading to Modulation of Apoptosis. Infect. Immun. 84, 2541–2554. doi: 10.1128/IAI.00191-16
Kocijancic D., Felgner S., Frahm M., Komoll R.-M., Iljazovic A., Pawar V., et al. (2016). Therapy of solid tumors using probiotic Symbioflor-2 – restraints and potential. Oncotarget 7, 22605–22622. doi: 10.18632/oncotarget.8027
Kordahi M. C., Stanaway I. B., Avril M., Chac D., Blanc M.-P., Ross B., et al. (2021). Genomic and functional characterization of a mucosal symbiont involved in early-stage colorectal cancer. Cell Host Microbe 29, 1589–1598.e6. doi: 10.1016/j.chom.2021.08.013
Lennard K. S., Goosen R. W., Blackburn J. M. (2016). Bacterially-associated transcriptional remodelling in a distinct genomic subtype of colorectal cancer provides a plausible molecular basis for disease development. PloS One 11, e0166282. doi: 10.1371/journal.pone.0166282
Liu Q.-Q., Li C.-M., Fu L.-N., Wang H.-L., Tan J., Wang Y.-Q., et al. (2020). Enterotoxigenic Bacteroides fragilis induces the stemness in colorectal cancer via upregulating histone demethylase JMJD2B. Gut Microbes 12, 1788900. doi: 10.1080/19490976.2020.1788900
Mármol I., Sánchez-de-Diego C., Pradilla Dieste A., Cerrada E., Yoldi M. J. R. (2017). Colorectal carcinoma: A general overview and future perspectives in colorectal cancer. Int. J. Mol. Sci. 18, 197. doi: 10.3390/ijms18010197
Martinson J. N. V., Walk S. T. (2020). Escherichia coli residency in the gut of healthy human adults. EcoSal Plus 9, 1–27. doi: 10.1128/ecosalplus.ESP-0003-2020
Moncrief J. S., Obiso R., Barroso L. A., Kling J. J., Wright R. L., Van Tassell R. L., et al. (1995). The enterotoxin of Bacteroides fragilis is a metalloprotease. Infect. Immun. 63, 175–181. doi: 10.1128/iai.63.1.175-181.1995
Mueller N. T., Bakacs E., Combellick J., Grigoryan Z., Dominguez-Bello M. G. (2015). The infant microbiome development: mom matters. Trends Mol. Med. 21, 109–117. doi: 10.1016/j.molmed.2014.12.002
Myers L. L., Firehammer B. D., Shoop D. S., Border M. M. (1984). Bacteroides fragilis: a possible cause of acute diarrheal disease in newborn lambs. Infect. Immun. 44, 241–244. doi: 10.1128/iai.44.2.241-244.1984
Myers L. L., Shoop D. S. (1987). Association of enterotoxigenic Bacteroides fragilis with diarrheal disease in young pigs. Am. J. Vet. Res. 48, 774–775.
Myers L. L., Shoop D. S., Byars T. D. (1987a). Diarrhea associated with enterotoxigenic Bacteroides fragilis in foals. Am. J. Vet. Res. 48, 1565–1567.
Myers L. L., Shoop D. S., Firehammer B. D., Border M. M. (1985). Association of enterotoxigenic bacteroides fragilis with diarrheal disease in calves. J. Infect. Dis. 152, 1344–1347. doi: 10.1093/infdis/152.6.1344
Myers L. L., Shoop D. S., Stackhouse L. L., Newman F. S., Flaherty R. J., Letson G. W., et al. (1987b). Isolation of enterotoxigenic Bacteroides fragilis from humans with diarrhea. J. Clin. Microbiol. 25, 2330–2333. doi: 10.1128/jcm.25.12.2330-2333.1987
Nakano V., Gomes D. A., Arantes R. M. E., Nicoli J. R., Avila-Campos M. J. (2006). Evaluation of the pathogenicity of the bacteroides fragilis toxin gene subtypes in gnotobiotic mice. Curr. Microbiol. 53, 113–117. doi: 10.1007/s00284-005-0321-6
Nougayrède J.-P., Homburg S., Taieb F., Boury M., Brzuszkiewicz E., Gottschalk G., et al. (2006). Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313, 848–851. doi: 10.1126/science.1127059
Nougayrède J.-P., Taieb F., Rycke J. D., Oswald E. (2005). Cyclomodulins: bacterial effectors that modulate the eukaryotic cell cycle. Trends Microbiol. 13, 103–110. doi: 10.1016/j.tim.2005.01.002
Nowrouzian F. L., Oswald E. (2012). Escherichia coli strains with the capacity for long-term persistence in the bowel microbiota carry the potentially genotoxic pks island. Microbial Pathogenesis 53, 180–182. doi: 10.1016/j.micpath.2012.05.011
Picard B., Garcia J. S., Gouriou S., Duriez P., Brahimi N., Bingen E., et al. (1999). The Link between Phylogeny and Virulence in Escherichia coli Extraintestinal Infection. Infect. Immun. 67, 546–553. doi: 10.1128/IAI.67.2.546-553.1999
Piciocchi A., Germinario E. A. P., Garcia Etxebarria K., Rossi S., Sanchez-Mete L., Porowska B., et al. (2021). Association of polygenic risk score and bacterial toxins at screening colonoscopy with colorectal cancer progression: A multicenter case-control study. Toxins (Basel) 13, 569. doi: 10.3390/toxins13080569
Pleguezuelos-Manzano C., Puschhof J., Huber A. R., van Hoeck A., Wood H. M., Nomburg J., et al. (2020). Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature 580, 269–273. doi: 10.1038/s41586-020-2080-8
Prorok-Hamon M., Friswell M. K., Alswied A., Roberts C. L., Song F., Flanagan P. K., et al. (2014). Colonic mucosa-associated diffusely adherent afaC+ Escherichia coli expressing lpfA and pks are increased in inflammatory bowel disease and colon cancer. Gut 63, 761–770. doi: 10.1136/gutjnl-2013-304739
Purcell R. V., Pearson J., Aitchison A., Dixon L., Frizelle F. A., Keenan J. I. (2017). Colonization with enterotoxigenic Bacteroides fragilis is associated with early-stage colorectal neoplasia. PloS One 12, e0171602. doi: 10.1371/journal.pone.0171602
Rabizadeh S., Rhee K.-J., Wu S., Huso D., Gan C. M., Golub J. E., et al. (2007). Enterotoxigenic bacteroides fragilis: A potential instigator of colitis. Inflammation Bowel Dis. 13, 1475–1483. doi: 10.1002/ibd.20265
Raisch J., Buc E., Bonnet M., Sauvanet P., Vazeille E., de Vallée A., et al. (2014). Colon cancer-associated B2 Escherichia coli colonize gut mucosa and promote cell proliferation. World J. Gastroenterol. 20, 6560–6572. doi: 10.3748/wjg.v20.i21.6560
Rhee K.-J., Wu S., Wu X., Huso D. L., Karim B., Franco A. A., et al. (2009). Induction of persistent colitis by a human commensal, enterotoxigenic bacteroides fragilis, in wild-type C57BL/6 mice. Infect. Immun. 77, 1708–1718. doi: 10.1128/IAI.00814-08
Richter T. K. S., Michalski J. M., Zanetti L., Tennant S. M., Chen W. H., Rasko D. A. (2018). Responses of the human gut escherichia coli population to pathogen and antibiotic disturbances. mSystems 3, e00047–e00018. doi: 10.1128/mSystems.00047-18
Roh H. C., Yoo D. Y., Ko S. H., Kim Y.-J., Kim J. M. (2011). Bacteroides fragilis Enterotoxin Upregulates Intercellular Adhesion Molecule-1 in Endothelial Cells via an Aldose Reductase-, MAPK-, and NF-κB–Dependent Pathway, Leading to Monocyte Adhesion to Endothelial Cells. J. Immunol. 187, 1931–1941. doi: 10.4049/jimmunol.1101226
Rotstein D., Kao J., Houston K. (1989). Reciprocalsynergy between Escherichia co/i and Bacteroides fragdis in an intra-abdominal infection model. (Microbiology Society) J. Med. Microbiol. doi: 10.1099/00222615-29-4-269
Rotstein O. D., Pruett T. L., Simmons R. L. (1985). Lethal Microbial Synergism in Intra-abdominal Infections: Escherichia coli and Bacteroides fragilis. Arch. Surg. 120, 146–151. doi: 10.1001/archsurg.1985.01390260016003
Sack R. B., Albert M. J., Alam K., Neogi P. K., Akbar M. S. (1994). Isolation of enterotoxigenic Bacteroides fragilis from Bangladeshi children with diarrhea: a controlled study. J. Clin. Microbiol. 32, 960–963. doi: 10.1128/jcm.32.4.960-963.1994
Sack R. B., Myers L. L., Almeido - Hill J., Shoop D. S., Bradbury W. C., Reid R., et al. (1992). Enterotoxigenic bacteroides fragilis: epidemiologic studies of its role as a human diarrhoeal pathogen. J. Diarrhoeal Dis. Res. 10, 4–9.
Sanfilippo L., Li C. K. F., Seth R., Balwin T. J., Menozzi M. G., Mahida Y. R. (2000). Bacteroides fragilis enterotoxin induces the expression of IL-8 and transforming growth factor-beta (TGF-β) by human colonic epithelial cells. Clin. Exp. Immunol. 119, 456–463. doi: 10.1046/j.1365-2249.2000.01155.x
Schneider M. R., Dahlhoff M., Horst D., Hirschi B., Trülzsch K., Müller-Höcker J., et al. (2010). A key role for E-cadherin in intestinal homeostasis and paneth cell maturation. PloS One 5, e14325. doi: 10.1371/journal.pone.0014325
Sears C. L. (2009). Enterotoxigenic Bacteroides fragilis: a Rogue among Symbiotes. Clin. Microbiol. Rev. 22, 349–369. doi: 10.1128/CMR.00053-08
Sears C. L., Pardoll D. M. (2011). Perspective: alpha-bugs, their microbial partners, and the link to colon cancer. J. Infect. Dis. 203, 306–311. doi: 10.1093/jinfdis/jiq061
Sekirov I., Russell S. L., Antunes L. C. M., Finlay B. B. (2010). Gut microbiota in health and disease. Physiol. Rev. 90, 859–904. doi: 10.1152/physrev.00045.2009
Shariati A., Razavi S., Ghaznavi-Rad E., Jahanbin B., Akbari A., Norzaee S., et al. (2021). Association between colorectal cancer and Fusobacterium nucleatum and Bacteroides fragilis bacteria in Iranian patients: a preliminary study. Infect. Agent Cancer 16, 41. doi: 10.1186/s13027-021-00381-4
Siegel R. L., Miller K. D., Fuchs H. E., Jemal A. (2022). Cancer statistics. CA: A Cancer J. Clin. 72, 7–33. doi: 10.3322/caac.21708
Stritzker J., Weibel S., Seubert C., Götz A., Tresch A., van Rooijen N., et al. (2010). Enterobacterial tumor colonization in mice depends on bacterial metabolism and macrophages but is independent of chemotaxis and motility. Int. J. Med. Microbiol. 300, 449–456. doi: 10.1016/j.ijmm.2010.02.004
Suvarna K., Stevenson D., Meganathan R., Hudspeth M. E. S. (1998). Menaquinone (Vitamin K2) Biosynthesis: Localization and Characterization of the menA Gene from Escherichia coli. J. Bacteriol 180, 2782–2787. doi: 10.1128/JB.180.10.2782-2787.1998
Tjalsma H., Boleij A., Marchesi J. R., Dutilh B. E. (2012). A bacterial driver–passenger model for colorectal cancer: beyond the usual suspects. Nat. Rev. Microbiol. 10, 575–582. doi: 10.1038/nrmicro2819
Tomkovich S., Dejea C. M., Winglee K., Drewes J. L., Chung L., Housseau F., et al. (2020). Human colon mucosal biofilms from healthy or colon cancer hosts are carcinogenic. J. Clin. Invest. 129, 1699–1712. doi: 10.1172/JCI124196
Toprak N. U., Yagci A., Gulluoglu B. M., Akin M. L., Demirkalem P., Celenk T., et al. (2006). A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin. Microbiol. Infection 12, 782–786. doi: 10.1111/j.1469-0691.2006.01494.x
Tronnet S., Floch P., Lucarelli L., Gaillard D., Martin P., Serino M., et al. (2020). The genotoxin colibactin shapes gut microbiota in mice. mSphere 5, 1–11. doi: 10.1128/msphere.00589-20
Turnbaugh P. J., Ley R. E., Hamady M., Fraser-Liggett C., Knight R., Gordon J. I. (2007). The human microbiome project: exploring the microbial part of ourselves in a changing world. Nature 449, 804–810. doi: 10.1038/nature06244
Tytgat H. L. P., Nobrega F. L., van der Oost J., de Vos W. M. (2019). Bowel biofilms: tipping points between a healthy and compromised gut? Trends Microbiol. 27, 17–25. doi: 10.1016/j.tim.2018.08.009
Van den Abbeele P., Van de Wiele T., Verstraete W., Possemiers S. (2011). The host selects mucosal and luminal associations of coevolved gut microorganisms: a novel concept. FEMS Microbiol. Rev. 35, 681–704. doi: 10.1111/j.1574-6976.2011.00270.x
Vétizou M., Pitt J. M., Daillère R., Lepage P., Waldschmitt N., Flament C., et al. (2015). Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084. doi: 10.1126/science.aad1329
Viljoen K. S., Dakshinamurthy A., Goldberg P., Blackburn J. M. (2015). Quantitative Profiling of Colorectal Cancer-Associated Bacteria Reveals Associations between Fusobacterium spp., Enterotoxigenic Bacteroides fragilis (ETBF) and Clinicopathological Features of Colorectal Cancer. PloS One 10, e0119462. doi: 10.1371/journal.pone.0119462
Vleminckx K., Vakaet L., Mareel M., Fiers W., van Roy F. (1991). Genetic manipulation of E-cadherin expression by epithelial tumor cells reveals an invasion suppressor role. Cell 66, 107–119. doi: 10.1016/0092-8674(91)90143-m
Weikel C. S., Grieco F. D., Reuben J., Myers L. L., Sack R. B. (1992). Human colonic epithelial cells, HT29/C1, treated with crude Bacteroides fragilis enterotoxin dramatically alter their morphology. Infect. Immun. 60, 321–327. doi: 10.1128/iai.60.2.321-327.1992
Wexler H. M. (2007). Bacteroides: the good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 20, 593–621. doi: 10.1128/CMR.00008-07
Wick E. C., Rabizadeh S., Albesiano E., Wu X., Wu S., Chan J., et al. (2014). Stat3 activation in murine colitis induced by enterotoxigenic. Bacteroides fragilis. Inflammation Bowel Dis. 20, 821–834. doi: 10.1097/MIB.0000000000000019
Wilson M. R., Jiang Y., Villalta P. W., Stornetta A., Boudreau P. D., Carrá A., et al. (2019). The human gut bacterial genotoxin colibactin alkylates DNA. Science 363, eaar7785. doi: 10.1126/science.aar7785
Wirbel J., Pyl P. T., Kartal E., Zych K., Kashani A., Milanese A., et al. (2019). Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 25, 679–689. doi: 10.1038/s41591-019-0406-6
Wu S., Lim K.-C., Huang J., Saidi R. F., Sears C. L. (1998). Bacteroides fragilis enterotoxin cleaves the zonula adherens protein, E-cadherin. Proc. Natl. Acad. Sci. U.S.A. 95, 14979–14984. doi: 10.1073/pnas.95.25.14979
Wu S., Powell J., Mathioudakis N., Kane S., Fernandez E., Sears C. L. (2004). Bacteroides fragilis Enterotoxin Induces Intestinal Epithelial Cell Secretion of Interleukin-8 through Mitogen-Activated Protein Kinases and a Tyrosine Kinase-Regulated Nuclear Factor-κB Pathway. Infect. Immun. 72, 5832–5839. doi: 10.1128/IAI.72.10.5832-5839.2004
Wu S., Rhee K.-J., Albesiano E., Rabizadeh S., Wu X., Yen H.-R., et al. (2009). A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 15, 1016–1022. doi: 10.1038/nm.2015
Xue M., Kim C., Healy A., Wernke K., Wang Z., Frischling M., et al. (2019). Structure elucidation of colibactin and its DNA cross-links. Science 365, eaax2685. doi: 10.1126/science.aax2685
Zafar H., Saier M. H. (2021). Gut Bacteroides species in health and disease. Gut Microbes 13, 1848158. doi: 10.1080/19490976.2020.1848158
Zeller G., Tap J., Voigt A. Y., Sunagawa S., Kultima J. R., Costea P. I., et al. (2014). Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 10, 766. doi: 10.15252/msb.20145645
Keywords: colorectal cancer, gut microbiota, microbiome, Escherichia coli, Bacteroides fragilis, biofilm
Citation: Lichtenstern CR and Lamichhane-Khadka R (2023) A tale of two bacteria – Bacteroides fragilis, Escherichia coli, and colorectal cancer. Front. Bacteriol. 2:1229077. doi: 10.3389/fbrio.2023.1229077
Received: 25 May 2023; Accepted: 11 July 2023;
Published: 26 July 2023.
Edited by:
Ansel Hsiao, University of California, Riverside, United StatesReviewed by:
Qinqin Pu, University of Pennsylvania, United StatesCopyright © 2023 Lichtenstern and Lamichhane-Khadka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Reena Lamichhane-Khadka, cmxhbWljaGhhbmVraGFka2FAY2hzdS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.