
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Audiol. Otol., 15 June 2023
Sec. Auditory Science
Volume 1 - 2023 | https://doi.org/10.3389/fauot.2023.1191143
 Maria-Pia Tuset1†
Maria-Pia Tuset1† Matthew D. Wiefels1†
Matthew D. Wiefels1† Keelin McKenna1,2
Keelin McKenna1,2 Jeenu Mittal1
Jeenu Mittal1 Chirag Gowda1
Chirag Gowda1 Rahul Mittal1
Rahul Mittal1 Adrien A. Eshraghi1,3,4,5*
Adrien A. Eshraghi1,3,4,5*Deciphering the inner ear transcriptome has allowed major breakthroughs in therapeutic development for auditory disorders. Single-cell sequencing (SCS) technologies, through the analysis of cellular heterogeneity and the identification of novel gene markers, can significantly enhance our understanding about auditory disorders. The application of SCS has allowed the identification of gene targets specific to the inner ear cellular subtypes such as supporting cells, hair cells, stria vascularis cells, and spiral ganglion neuron cells. SCS has provided new insights into the molecular mechanisms underlying the pathophysiology of auditory disorders such as noise-trauma induced hearing loss, ototoxicity, sudden sensorineural hearing loss (SSNHL), vestibular schwannomas and acute otitis media, leading to the discovery of novel therapeutic targets. The objective of this perspective article is to discuss recent advancements regarding the utilization of SCS for inner ear research. With global initiatives striving to overcome the increasing burden of hearing loss, the SCS technique holds a great potential in revolutionizing the management of auditory disorders, paving the way to develop effective treatment modalities.
Single-cell sequencing (SCS) has emerged as a powerful technique leading to key discoveries in the medical field (Allen et al., 2023; Krämer et al., 2023; Yu et al., 2023; Zhang et al., 2023). Previous studies focused on analyzing bulk tissue samples and determined the average expression of gene transcripts (Pijuan-Sala et al., 2019). SCS analyzes the transcriptome of individual cells and can help in understanding the specific function of a single cell (Chang et al., 2023). Understanding tissue heterogeneity has proven impactful in cancer research, where tumor heterogeneity and rare cell types have allowed a better understanding of the disease (Pyle and Hoa, 2020; van der Leun et al., 2020; Lei et al., 2021; Liu et al., 2023; Xia et al., 2023).
Besides cancer, auditory research has also been greatly impacted by SCS. There has been an increased interest in the auditory community to study inner ear tissue heterogeneity, auditory disorders and their underlying molecular mechanisms using SCS (Ranum et al., 2019; Pyle and Hoa, 2020; Petitpré et al., 2022). SCS has allowed the identification of unknown cell subtypes in the inner ear and novel cellular markers through the study of cellular heterogeneity (Yizhar-Barnea and Avraham, 2017).
The aim of this brief perspective article is to discuss the recent advancements regarding the utilization of SCS for inner ear research (Table 1). SCS holds a great potential to better understand the function of the heterogenous cell population in the inner ear as well as to decipher the molecular mechanisms underlying auditory disorders, thus paving the way to develop novel therapeutic modalities.
Since the launch of the human genome project in the year 2000 and the development of next generation sequencing (NGS) technologies, the tools available to study the genetic basis of disease pathophysiology have continued to transform. NGS offers high-throughput sequencing, but requires an important pool of cells and provides bulk sample gene expression (van Dijk et al., 2014). However, transcript expression depends on gene expression level and cell populations. To address this issue, the development of SCS has allowed genomic, epigenomic, and whole transcriptome information to be obtained from single cells (Wu et al., 2021). Using this revolutionary technique, novel discoveries in tissue heterogeneity and biology at the single-cell level have been achieved.
First described by Tang et al. (2009), who used individual blastomeres, SCS involves cell isolation, capture, and lysis followed by nucleic acid extraction, amplification, and sequencing using NGS (Figure 1; Wu et al., 2021). A recent study compared the different techniques used in cell type-specific expression analysis of the inner ear. It was concluded that a combined approach using fluorescent-activated cell sorting (FACS) and SCS was best to overcome the specific challenges of inner ear single-cell transcriptome analysis (Hertzano et al., 2021).
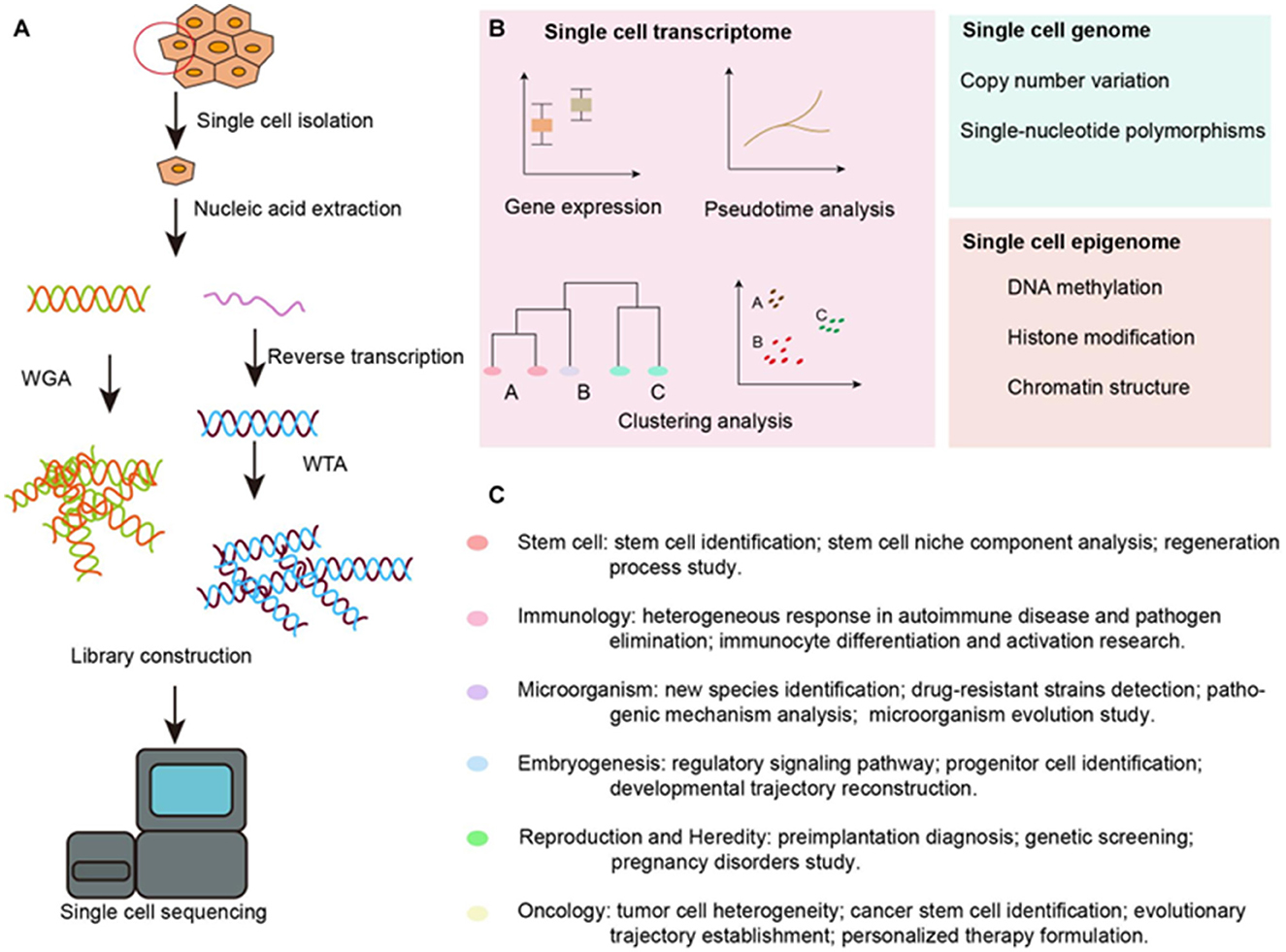
Figure 1. Overview of SCS workflow and applications in the inner ear. (A) Schematic strategy of SCS workflow. After dissociation of the organ or tissue of interest to live single cell, cells are then captured by various methods and lysed to release RNA and DNA fragment, the former is reversed transcribed to synthesized cDNA. DNA fragment or cDNA must be amplified to generate sequencing library. Next-generation sequencing is subsequently performed to generate the readouts that can be assigned to single cells via cell-specific barcodes. (B) Analysis of single cell transcriptome, genome and epigenome data. (C) Diverse fields of basic research that have been impacted by SCS technologies. WGA, whole genome amplification; WTA, whole transcriptome amplification. Taken from Wu et al. (2021) distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited.
To date, SCS of the inner ear has been performed primarily on animal models, especially mice, focusing on inner ear development and hair cell regeneration (Durruthy-Durruthy and Heller, 2015; Pyle and Hoa, 2020; Qian et al., 2022). Given the global burden of hearing loss, SCS can be used to identify the different signaling cascades involved in the auditory pathway. Through the application of SCS, differential expression of transcript isoforms of deafness genes can be analyzed in single inner ear hair cells (HCs). Identifying these isoforms is a powerful tool for gene therapy development, allowing cell-type specificity and limiting off-target effects. A study analyzed 12 deafness genes and led to the identification of 20 highly conserved exons as therapeutic targets (Ranum et al., 2019). As cochlear tissue is underrepresented in annotation databases, these studies are essential to support targeted and personalized treatment of auditory disorders.
Advances in SCS have made unprecedented strides in understanding the genetic etiology of auditory disorders. Several genes involved in the auditory pathways have been identified using SCS. The organ of Corti (OC), composed of HCs and spiral ganglion neurons (SGNs), has been largely investigated for its role in neural sound transmission. Conversely, the stria vascularis (SV), well-known for its role in providing the necessary blood supply to the OC, has been found to play an important sensory role using SCS technique. The SV is composed of three main cell types: marginal cells, intermediate cells, and basal cells, along with other cell types such as spindle cells and macrophages. Single-cell RNA sequencing has led to the discovery of several genes in the SV implicated in hearing that were challenging to identify previously. These genes include Abcg1 and Heyl in marginal cells, P2rx2 and Kcnj16 in spindle cells, Nrp2 and Kcnj13 in intermediate cells, and Sox8 and Nr2f2 in basal cells (Korrapati et al., 2019). The information derived from the identification of genes implicated in sensorineural hearing loss (SNHL) using SCS can be used to develop cell-targeted therapeutic strategies.
Single-cell RNA sequencing has also been used to analyze the sensory epithelial cells of the inner ear leading to the identification of several new genes. These genes include Rasd2, Anxa4, and Pcp4 on HCs and Mia1 and Cdh4 on medial supporting cells and Cntn1 within lateral supporting cells (Burns et al., 2015). In the inner ear, HCs and supporting cells are both found in epithelial tissues involved in balance and hearing. Damaged or dysfunctional genes in these cells may contribute to SNHL and/or vestibular pathologies. Research into the regeneration of HCs and supporting cells in the cochlea may show promise for patients with SNHL due to a variety of factors including loud noise exposure, chemotherapy, and aging. Specifically, researchers have investigated the potential of using supporting cells to act as precursors for generating HCs (Walters et al., 2017). The expression of genes associated with epithelial tissue in the cochlea identified through SCS can be manipulated to attenuate hearing loss. For instance, Walters et al. discovered that deleting the p27 gene while upregulating the expression of the Atoh1 gene in adult mouse cochleae triggered the conversion of supporting cells to HCs following damage to HCs resulting from loud noise. Similarly, upregulation of Pou4f3 or Gata3 with Atoh1 bolstered the transformation of supporting cells to HCs in adult mice (Walters et al., 2017). These findings demonstrate the potential for gene therapy to be an important tool in attenuating SNHL.
SGNs are neural cells that transmit information from HCs of the cochlea to the central cochlear nucleus in the brainstem (Shrestha et al., 2018). Type I SGNs make up the majority of SGNs and innervate inner HCs (IHCs) while Type II SGNs innervate OHCs. Type I SGNs function primarily to transmit sound information to the brain while Type II SGNs play a role in pain sensing and damage perception; these differences in function are mainly due to differences in structure and molecular signaling (Liu et al., 2015). Outer hair cells (OHCs) amplify sounds that enter the cochlea. These amplified sound-induced vibrations are transmitted to IHCs, which trigger the opening of transduction channels at the apical surfaces of IHCs, thus causing the excitation of SGNs (Sun et al., 2018). SCS has been used to identify 3 subtypes of Type I SGNs: subtypes Ia, Ib, and Ic (Shrestha et al., 2018). These subtypes differ by their expression of multiple genes, such as Pou4f1, Lypd1, Calb2, and Ntng1, that affect neuronal differentiation and function. For instance, many of these genes encode proteins related to calcium and potassium channel proteins in the plasma membranes of Type I SGNs. Variation in gene expression among the Type I SGN subtypes correlates with differences in expression of certain proteins, most notably membrane proteins, which can result in differences in physiology and synaptic transmission among SGN subtypes (Shrestha et al., 2018). Using SCS, it may be possible to identify different gene isoforms expressed in SGN cellular subtypes so as to specifically address the cells with pathologic ionic homeostasis through cell-specific gene targeting.
SCS is now being increasingly used to better understand the molecular mechanisms underlying drug-induced ototoxicity such as due to cisplatin or aminoglycosides. Cisplatin is a chemotherapeutic drug that is used in the treatment of many malignant tumors. However, it has been found that cisplatin accumulates preferentially in the basal turn of the cochlea, causing loss of OHCs in the OC, leading to high-frequency hearing loss (Prayuenyong et al., 2021). Cisplatin enters the inner ear by crossing the blood-labyrinthine barrier specifically, entering the SV marginal cells, using CTR1 and OCT2 (Wang et al., 2023). This damages the Na+/K+/2Cl- channel, which is integral in maintaining a healthy endolymph. Aminoglycosides, broad-spectrum antibiotics, are also known to cause ototoxicity in a similar manner but are thought to utilize TRPV1 and TRPV4 (Steyger, 2021). A pharmacokinetic study demonstrated that cisplatin continues to accumulate in the inner ear following each dose, and the cells show limited ability to eliminate it. The highest level of accumulation was identified in the SV. Additionally, they analyzed postmortem cochleae from patients who were treated with cisplatin and identified that the drug was still present up to 18 months after treatment (Breglio et al., 2017).
Researchers have used SCS to discover the role of soluble resistance-related calcium binding protein (sorcin) in cisplatin-induced ototoxicity (Wang et al., 2021). Sorcin is a calcium-binding protein that is known for its involvement in calcium homeostasis, apoptosis, and tumor progression (Shabnam et al., 2018). In particular, it can reduce apoptosis by regulating apoptotic proteins, including Bax, Bcl-2, and caspase-3, and is involved in signaling pathways, including ERK, MAPK/ERK, STAT3, and P13K/Akt (Shabnam et al., 2018). Sorcin is expressed in cochlear HCs and auditory HEI-OC-1 cells (Wang et al., 2021). It has been shown that with increasing levels of cisplatin, sorcin decreased, and the damage was most pronounced in the basal turn of the cochlea (Wang et al., 2021). They also found that when sorcin was knocked out using siRNA, there was a higher level of pro-apoptotic proteins, such as caspase-3 and Bax, and a decreased amount of anti-apoptotic proteins, such as Bcl-2, in response to cisplatin exposure (Wang et al., 2021). Additionally, higher levels of reactive oxygen species (ROS) and matrix metalloproteases (MMP) were observed, indicating mitochondrial dysfunction (Wang et al., 2021). There was activation of the ERK1/2 signaling pathway, previously implicated in apoptosis of HCs following knockdown of sorcin. SCS can be utilized to further investigate the molecular mechanisms underlying the otoprotective effects of sorcin and its cell-specific expression following cisplatin exposure paving the way for developing treatment modalities to prevent hearing loss.
SCS is also being utilized to study how noise affects the cochlear sensory compartment and leads to SNHL. Milon et al. (2021) sequenced cell-type specific immunoprecipitated RNA (RiboTag-IP) to isolate cells along with SCS to study specific transcriptional cell responses to stress (Milon et al., 2021). Mice were exposed to 8 to 16 kHz noise bands at 105 dB for 2 h, causing a permanent threshold shift across all frequencies. OHCs and support cells were analyzed and a difference in 1,947 and 1,796 expressed genes, respectively, compared to controls was observed. There was an increase in genes implicated in apoptosis (Jun, Bcl10, and Tgfb2) as well as immune-related genes (Ifr7, Ccl5, and Tlr3). Additionally, some were specific for OHCs, which included genes that function in mRNA metabolic processes (Sfpq, Hnrnpu, and Snrmp48), and some were specific for supporting cells, including genes for the transmission of nerve impulses (Grm3, Asic2, and Cacna1e). It was also observed that type 1A SGN elicit a strong transcriptional response to noise. The promoters of genes expressed by these cells were highly interactive with transcription factors from the activating transcription factor (ATF) family, specifically Atf3, Atf4, and Atf5. These factors may be protective after noise exposure because they upregulate pro-survival genes, such as Fgf21, Cntfr, Hspa5, and Gas6. Next, they studied lateral wall (LW). A total of 408 differentially expressed genes were downregulated in these cells. There was a downregulation of potassium ion transport genes suggesting decreased functioning of LW cells in transporting potassium into the endolymph following noise exposure (Milon et al., 2021). While there are cell-type specific responses to noise trauma, the cochlea was also found to mount a response that was shared amongst the cell types. About 36 genes, including Stat3 and Irf7, were found to be upregulated across all cell types and were specifically responsive to transcription factors IRF7 and STAT3 (Milon et al., 2021). Using this gene expression data, metformin was identified as the top candidate for potential pharmacologic therapy for noise-induced hearing loss (NIHL). These findings suggest the potential of SCS to identify novel therapeutic agents based on cell-specific gene expression following auditory insults such as noise exposure.
Sudden sensorineural hearing loss (SSNHL) is defined as a hearing loss of at least 30 dB that spans a minimum of three contiguous audiometric frequencies and develops within 72 h (Nelson et al., 2021). SCS holds great potential to understand the genetic etiology of SSNHL. However, so far only one study has used this technique. A study reviewed SCS data that had been performed on particular cell types within the cochlea, including the SV, Pillar cells and Deiter cells, as well as IHCs and OHCs, to determine if there were certain genes implicated in SSNHL (Nelson et al., 2021). It was observed that the SV showed increased expression of Sod1, Gpx3, Mif, Gjb2, Nr3c1, Clock, Gpx1, Sod2, and Nr3c2. Pillar and Deiters cells show increased expression of Apoe, Gjb2, and Pon2. Finally, IHCs, OHCs, and Deiters cells show increased expression of Sod1, Gpx1, Mif , Gjb2, Nr3c1, Nr3c2, and Gpx3. Superoxide dismutase (Sod1/2) and macrophage migration inhibitory factor (Mif) may be particularly important and have been seen to be implicated in other types of hearing loss as well, including noise-induced and age-related (Nelson et al., 2021). Additionally, they evaluated the different expression of glutathione peroxidase isoforms. Notably, in the SV, Gpx3 was expressed at a higher rate than Gpx1 in the marginal, intermediate, and basal cells. This is in contrast to other cells in the OC, which preferentially expressed Gpx1 (Nelson et al., 2021). An important limitation of this study was that they used a previously published mouse whole transcriptome dataset with poor resolution, impeding any definitive conclusion concerning gene expression patterns. Additionally, candidate genes were identified based on previously identified pathways, such as inflammation or oxidative stress. Finally, the data addressed was purely observational, and the functional significance of the identified genes was not determined (Nelson et al., 2021). However, results suggest that utilizing SCS methods can enhance our understanding regarding the underlying molecular mechanisms involved in SSNHL.
Vestibular schwannomas (VS) are benign tumors arising from the vestibulocochlear nerve's Schwann cells and are the most common neoplastic cause of hearing loss. Current treatment options comprise surgery or radiotherapy, and monoclonal antibody angiogenesis inhibitors for VS associated to neurofibromatosis type 2 (NF2). Although bulk analysis of VS tissue has been performed (de Vries et al., 2019; Huang et al., 2019), no information on the cellular microenvironnement is available. By applying SCS to VS tissue samples, the different cell types and factors in these tumors as well as candidate biomarkers of VS can be identified leading to the discovery of potential drug targets. A study performed SCS on three human tissue samples of sporadic VS and was able to identify six cell types involved in the tumor microenvironment: Schwann cells, myeloid cells, fibroblasts, lymphocytes (B and T cells), and endothelial cells (Yidian et al., 2022). Six Schwann cell subgroups were identified based on antigen presenting genes and HLA phenotypes. A particular subgroup SC-C3 was enriched in hematopoiesis and in the MAPK signaling pathway, typically activated after peripheral nerve injury, and initiating cell differentiation. Cellular subgroup SC-C4 was correlated to chemokine activation and SC-C5 to myelination, suggesting the high heterogeneity of the expression profiles and function of this cell type in VS. Analysis of transcription factors revealed SC-C5 expressed TEAD4, involved in the Hippo signaling pathway which induces apoptosis and limits proliferation (Yidian et al., 2022). Analysis of the myeloid cell population, and more particularly a subtype of monocytes, activated PI3K a transduction pathway involved in chemotherapy resistance. Through SCS, cellular interactions were also studied, and allowed to identify specific cellular links, especially those between fibroblasts and immune cells. Two different fibroblast clusters were found to be mainly involved in the CXCL signaling pathway, promoting angiogenesis in tumor tissue (Yidian et al., 2022). The application of SCS in sporadic VS revealed fundamental characteristics on cellular heterogeneity, microenvironment, and molecular pathways, paving the way for specific drug development.
Acute otitis media (AOM) refers to middle ear infection leading to inflammation and is a global healthcare burden. Studies on middle ear mucosa (MEM) have been performed using flow cytometry, bulk DNA analysis (Forséni Flodin, 2001; Jecker et al., 2001), and more recently SCS (Ryan et al., 2020). An animal study focusing on MEM, used SCS to clarify the alterations induced by inflammation and multicellular interactions resulting in the identification of new cell subtypes (Rao et al., 2021). Eight major cell subtypes were identified by SCS. Macrophage subtype M2 specifically expressed mrc1, and communicated with all other cellular subtypes, thus playing a key role in the intercellular crosstalk during inflammation. Colony-stimulating factor 1 (CSF1) and IL-34 interaction with CSF1-receptor were both increased in the M2 subpopulation of inflamed and normal mucosa, suggesting the important role of these macrophages in the middle ear. This study identified dual-feature cells and described their specific roles by comparing SCS data to the Gene Ontology database (Rao et al., 2021). Key discoveries such as the crosstalk through IGF-1 between macrophages and the dual-feature epithelial-phagocytic cells were described, such as the increase in IGF1 in normal mucosa and its decrease AOM. SCS holds a great potential in elucidating precise molecular pathways underlying pathophysiology of AOM that needs to be harnessed in future studies.
The advent of SCS has significantly enhanced our understanding of inner ear cellular heterogeneity as well as the genes and molecular pathways involved in hearing loss. Studies using SCS have identified novel marker genes involved in auditory disorders such as ototoxicity, NIHL, and SSHNL, thus providing the key targets for gene therapy. Nevertheless, the limits of viral transfections, their efficacy on the human ear, and the acceptability of genetic treatment by patients, will determine the clinical translation of gene therapy.
Although SCS has revolutionized genetic research, current methods still need some improvement. The cell isolation for SCS is challenging, as the inner ear lies nested at the base of the skull in the robust petrous part of the temporal bone, making it difficult to access and reach the desired cell types. In addition, cells are scarce, with hair cells representing <3% of an intact sensory epithelium, as well as difficult to dissect from the labyrinthine structures. The cell dissociation of SCS can also alter the gene expression profiles. The other important limitation of SCS is that it requires a large number of intact viable cells to correctly profile auditory disorders. However, current techniques impact the spatial resolution necessary to identify cells and their function (Ranum et al., 2019). With the concurrent development of spatial transcriptomics, linking gene function to the anatomical location of cells, it is feasible to overcome some of these challenges (Srivatsan et al., 2021; Vahid et al., 2023). Given the overwhelming amount of data collected through SCS, data storage and data analysis software development is another major limiting factor for this technology. Efforts aimed toward creating and continuously updating universal database platforms will determine the optimal use of the entirety of the complex information discovered through SCS.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
We are grateful to Valerie Gramling for critical reading of manuscript.
RM and AAE declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Allen, W. E., Blosser, T. R., Sullivan, Z. A., Dulac, C., and Zhuang, X. (2023). Molecular and spatial signatures of mouse brain aging at single-cell resolution. Cell 186, 194–208.e18. doi: 10.1016/j.cell.2022.12.010
Breglio, A. M., Rusheen, A. E., Shide, E. D., Fernandez, K. A., Spielbauer, K. K., McLachlin, K. M., et al. (2017). Cisplatin is retained in the cochlea indefinitely following chemotherapy. Nat. Commun. 8, 1654. doi: 10.1038/s41467-017-01837-1
Burns, J. C., Kelly, M. C., Hoa, M., Morell, R. J., and Kelley, M. W. (2015). Single-cell RNA-Seq resolves cellular complexity in sensory organs from the neonatal inner ear. Nat. Commun. 6, 8557. doi: 10.1038/ncomms9557
Chang, P. -C., Lin, Y. -C., Yen, H. -J., Hueng, D. -Y., Huang, S. -M., and Li, Y. -F. (2023). Ancient ubiquitous protein 1 (AUP1) is a prognostic biomarker connected with TP53 mutation and the inflamed microenvironments in glioma. Cancer Cell Int. 23, 62. doi: 10.1186/s12935-023-02912-y
de Vries, W. M., Briaire-de Bruijn, I. H., van Benthem, P. P. G., van der Mey, A. G. L., and Hogendoorn, P. C. W. (2019). M-CSF and IL-34 expression as indicators for growth in sporadic vestibular schwannoma. Virchows Arch. 474, 375–381. doi: 10.1007/s00428-018-2503-1
Durruthy-Durruthy, R., and Heller, S. (2015). Applications for single cell trajectory analysis in inner ear development and regeneration. Cell Tissue Res. 361, 49–57. doi: 10.1007/s00441-014-2079-2
Forséni Flodin, M. (2001). Macrophages and possible osteoclast differentiation in the rat bullar bone during experimental acute otitis media, with reference to tympanosclerosis. Otol. Neurotol. 22, 771–775. doi: 10.1097/00129492-200111000-00010
Hertzano, R., Gwilliam, K., Rose, K., Milon, B., and Matern, M. S. (2021). Cell type-specific expression analysis of the inner ear: A technical report. Laryngoscope 131(Suppl.5), S1–S16. doi: 10.1002/lary.28765
Huang, X., Xu, J., Shen, Y., Zhang, L., Xu, M., Chen, M., et al. (2019). Protein profiling of cerebrospinal fluid from patients undergoing vestibular schwannoma surgery and clinical significance. Biomed. Pharmacother. 116, 108985. doi: 10.1016/j.biopha.2019.108985
Jecker, P., Pabst, R., and Westermann, J. (2001). Proliferating macrophages, dendritic cells, natural killer cells, T and B lymphocytes in the middle ear and Eustachian tube mucosa during experimental acute otitis media in the rat. Clin. Exp. Immunol. 126, 421–425. doi: 10.1046/j.1365-2249.2001.01543.x
Korrapati, S., Taukulis, I., Olszewski, R., Pyle, M., Gu, S., Singh, R., et al. (2019). Single cell and single nucleus RNA-seq reveal cellular heterogeneity and homeostatic regulatory networks in adult mouse stria vascularis. Front. Mol. Neurosci. 12, 316. doi: 10.3389/fnmol.2019.00316
Krämer, B., Nalin, A. P., Ma, F., Eickhoff, S., Lutz, P., Leonardelli, S., et al. (2023). Single-cell RNA sequencing identifies a population of human liver-type ILC1s. Cell Rep. 42, 111937. doi: 10.1016/j.celrep.2022.111937
Lei, Y., Tang, R., Xu, J., Wang, W., Zhang, B., Liu, J., et al. (2021). Applications of single-cell sequencing in cancer research: progress and perspectives. J. Hematol. Oncol. 14, 91. doi: 10.1186/s13045-021-01105-2
Liu, C., Glowatzki, E., and Fuchs, P. A. (2015). Unmyelinated type II afferent neurons report cochlear damage. Proc. Natl. Acad. Sci. U. S. A. 112, 14723–14727. doi: 10.1073/pnas.1515228112
Liu, C., Li, X., Huang, Q., Zhang, M., Lei, T., Wang, F., et al. (2023). Single-cell RNA-sequencing reveals radiochemotherapy-induced innate immune activation and MHC-II upregulation in cervical cancer. Signal Transduct. Target Ther. 8, 44. doi: 10.1038/s41392-022-01264-9
Milon, B., Shulman, E. D., So, K. S., Cederroth, C. R., Lipford, E. L., Sperber, M., et al. (2021). A cell-type-specific atlas of the inner ear transcriptional response to acoustic trauma. Cell Rep. 36, 109758. doi: 10.1016/j.celrep.2021.109758
Nelson, L., Johns, J. D., Gu, S., and Hoa, M. (2021). Utilizing single cell RNA-sequencing to implicate cell types and therapeutic targets for SSNHL in the adult cochlea. Otol. Neurotol. 42, e1410–e1421. doi: 10.1097/MAO.0000000000003356
Petitpré, C., Faure, L., Uhl, P., Fontanet, P., Filova, I., Pavlinkova, G., et al. (2022). Single-cell RNA-sequencing analysis of the developing mouse inner ear identifies molecular logic of auditory neuron diversification. Nat. Commun. 13, 3878. doi: 10.1038/s41467-022-31580-1
Pijuan-Sala, B., Griffiths, J. A., Guibentif, C., Hiscock, T. W., Jawaid, W., Calero-Nieto, F. J., et al. (2019). A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566, 490–495. doi: 10.1038/s41586-019-0933-9
Prayuenyong, P., Baguley, D. M., Kros, C. J., and Steyger, P. S. (2021). Preferential cochleotoxicity of cisplatin. Front. Neurosci. 15, 695268. doi: 10.3389/fnins.2021.695268
Pyle, M. P., and Hoa, M. (2020). Applications of single-cell sequencing for the field of otolaryngology: A contemporary review. Laryngoscope Investig. Otolaryngol. 5, 404–431. doi: 10.1002/lio2.388
Qian, F., Wei, G., Gao, Y., Wang, X., Gong, J., Guo, C., et al. (2022). Single-cell RNA-sequencing of zebrafish hair cells reveals novel genes potentially involved in hearing loss. Cell Mol. Life Sci. 79, 385. doi: 10.1007/s00018-022-04410-2
Ranum, P. T., Goodwin, A. T., Yoshimura, H., Kolbe, D. L., Walls, W. D., Koh, J.-Y., et al. (2019). Insights into the biology of hearing and deafness revealed by single-cell RNA sequencing. Cell Rep. 26, 3160–3171.e3. doi: 10.1016/j.celrep.2019.02.053
Rao, Y., Zhong, D., Qiu, K., Cheng, D., Li, L., Zhang, Y., et al. (2021). Single-cell transcriptome profiling identifies phagocytosis-related dual-feature cells in a model of acute otitis media in rats. Front. Immunol. 12, 760954. doi: 10.3389/fimmu.2021.760954
Ryan, A. F., Nasamran, C. A., Pak, K., Draf, C., Fisch, K. M., Webster, N., et al. (2020). Single-cell transcriptomes reveal a complex cellular landscape in the middle ear and differential capacities for acute response to infection. Front. Genet. 11, 358. doi: 10.3389/fgene.2020.00358
Shabnam, B., Padmavathi, G., Banik, K., Girisa, S., Monisha, J., Sethi, G., et al. (2018). Sorcin a potential molecular target for cancer therapy. Transl. Oncol. 11, 1379–1389. doi: 10.1016/j.tranon.2018.08.015
Shrestha, B. R., Chia, C., Wu, L., Kujawa, S. G., Liberman, M. C., and Goodrich, L. V. (2018). Sensory neuron diversity in the inner ear is shaped by activity. Cell 174, 1229–1246.e17. doi: 10.1016/j.cell.2018.07.007
Srivatsan, S. R., Regier, M. C., Barkan, E., Franks, J. M., Packer, J. S., Grosjean, P., et al. (2021). Embryo-scale, single-cell spatial transcriptomics. Science. 373, 111–117. doi: 10.1126/science.abb9536
Steyger, P. S. (2021). Mechanisms of aminoglycoside- and cisplatin-induced ototoxicity. Am. J. Audiol. 30, 887–900. doi: 10.1044/2021_AJA-21-00006
Sun, S., Babola, T., Pregernig, G., So, K., Nguyen, M., Su, M., et al. (2018). Hair cell mechanotransduction regulates spontaneous activity and spiral ganglion subtype specification in the auditory system. Cell 174, 1247–1263.e15. doi: 10.1016/j.cell.2018.07.008
Tang, F., Barbacioru, C., Wang, Y., Nordman, E., Lee, C., Xu, N., et al. (2009). mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6, 377–382. doi: 10.1038/nmeth.1315
Vahid, M. R., Brown, E. L., Steen, C. B., Zhang, W., Jeon, H. S., Kang, M., et al. (2023). High-resolution alignment of single-cell and spatial transcriptomes with CytoSPACE. Nat. Biotechnol. doi: 10.1038/s41587-023-01697-9. [Epub ahead of print].
van der Leun, A. M., Thommen, D. S., and Schumacher, T. N. (2020). CD8+ T cell states in human cancer: Insights from single-cell analysis. Nat. Rev. Cancer 20, 218–232. doi: 10.1038/s41568-019-0235-4
van Dijk, E. L., Auger, H., Jaszczyszyn, Y., and Thermes, C. (2014). Ten years of next-generation sequencing technology. Trends Genet. 30, 418–426. doi: 10.1016/j.tig.2014.07.001
Walters, B. J., Coak, E., Dearman, J., Bailey, G., Yamashita, T., Kuo, B., et al. (2017). In vivo Interplay between p27Kip1, GATA3, ATOH1, and POU4F3 converts non-sensory cells to hair cells in adult mice. Cell Rep. 19, 307–320. doi: 10.1016/j.celrep.2017.03.044
Wang, D., Shi, S., Hsieh, Y.-L., Wang, J., Wang, H., and Wang, W. (2021). Knockdown of sorcin increases HEI-OC1 cell damage induced by cisplatin in vitro. Arch. Biochem. Biophys. 701, 108752. doi: 10.1016/j.abb.2021.108752
Wang, X., Zhou, Y., Wang, D., Wang, Y., Zhou, Z., Ma, X., et al. (2023). Cisplatin-induced ototoxicity: From signaling network to therapeutic targets. Biomed. Pharmacother. 157, 114045. doi: 10.1016/j.biopha.2022.114045
Wu, M., Xia, M., Li, W., and Li, H. (2021). Single-cell sequencing applications in the inner ear. Front. Cell Dev. Biol. 9, 637779. doi: 10.3389/fcell.2021.637779
Xia, J., Xie, Z., Niu, G., Lu, Z., Wang, Z., Xing, Y., et al. (2023). Single-cell landscape and clinical outcomes of infiltrating B cells in colorectal cancer. Immunology 168, 135–151. doi: 10.1111/imm.13568
Yidian, C., Chen, L., Hongxia, D., Yanguo, L., and Zhisen, S. (2022). Single-cell sequencing reveals the cell map and transcriptional network of sporadic vestibular schwannoma. Front. Mol. Neurosci. 15, 984529. doi: 10.3389/fnmol.2022.984529
Yizhar-Barnea, O., and Avraham, K. B. (2017). Single cell analysis of the inner ear sensory organs. Int. J. Dev. Biol. 61, 205–213. doi: 10.1387/ijdb.160453ka
Yu, L., Zhang, Y., Liu, C., Wu, X., Wang, S., Sui, W., et al. (2023). Heterogeneity of macrophages in atherosclerosis revealed by single-cell RNA sequencing. FASEB J. 37, e22810. doi: 10.1096/fj.202201932RR
Keywords: auditory disorders, single cell sequencing, inner ear, molecular mechanisms, cellular diversity, cochlea
Citation: Tuset MP, Wiefels MD, McKenna K, Mittal J, Gowda C, Mittal R and Eshraghi AA (2023) Single-cell sequencing: a powerful technique to understand the pathophysiology of auditory disorders. Front. Audiol. Otol. 1:1191143. doi: 10.3389/fauot.2023.1191143
Received: 21 March 2023; Accepted: 11 May 2023;
Published: 15 June 2023.
Edited by:
Achintya K. Bhowmik, Stanford University, United StatesReviewed by:
Shiming Yang, PLA General Hospital, ChinaCopyright © 2023 Tuset, Wiefels, McKenna, Mittal, Gowda, Mittal and Eshraghi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adrien A. Eshraghi, YWVzaHJhZ2hpQG1lZC5taWFtaS5lZHU=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.