
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Astron. Space Sci., 26 October 2023
Sec. Astrobiology
Volume 10 - 2023 | https://doi.org/10.3389/fspas.2023.1244682
 Korine A. Duval1†
Korine A. Duval1† Tessa B. Van Volkenburg1†
Tessa B. Van Volkenburg1† Kathleen L. Craft2*
Kathleen L. Craft2* Chanel M. Person1
Chanel M. Person1 John S. Harshman3
John S. Harshman3 Diarny O. Fernandes1
Diarny O. Fernandes1 Jennifer S. Benzing3
Jennifer S. Benzing3 Emil G. McDowell1
Emil G. McDowell1 Tyler W. Nelson2
Tyler W. Nelson2 Gautham S. Divakar2
Gautham S. Divakar2 Owen M. Pochettino2
Owen M. Pochettino2 Mark E. Perry2
Mark E. Perry2 Christopher E. Bradburne3*
Christopher E. Bradburne3*In situ sampling missions to detect biosignatures on ocean worlds requires thorough sample preparation to manage the expected chemical complexity of such environments. Proposed instruments must be capable of automatic liquid sample handling to ensure sensitive and accurate detections of biosignatures, regardless of the initial chemical composition. Herein, we outline the design, build, and test of the integrated Biosignature Preparation for Ocean Worlds (BioPOW) system capable of purifying amino acids from icy samples. This four step modular instrument 1) melts ice samples, 2) purifies amino acids via cation exchange chromatography, 3) concentrates via vacuum drying, and 4) derivatizes amino acids to volatilize and enable detection with downstream analytical instruments. Initial experiments validated the thermal performance of the system by melting ice in the sample cup (1 mL sample, 3°C–28°C, <5 min, 1.4 kJ) and heating the derivatization tank past the concentration temperature (20°C–60°C, 12 min, 3.6 kJ) to the derivatization temperature (60°C–90°C, 25 min, 7.5 kJ). Later experiments investigated important factors for automatic cation exchange using a design of experiments approach, and found that initial salt concentration, sample and eluate flow rates, and water wash volumes all play significant roles in reducing conductivity (1.1 x–6.7 x) while maintaining phenylalanine yields between 31% and 94%. The modules were then integrated into a 12 cm × 20 cm × 20 cm fieldable platform for analysis, and the maturation of this design for future spaceflight is discussed.
“Can life have originated elsewhere in our Solar System?” and “How do we best search for it?”; these are a few of the motivating questions behind our exploration of other worlds. The recent Origins, Worlds, and Life (OWL) 2023–2032 Decadal Survey calls out similar questions which shows support in the planetary community for life detection (Committee on the Planetary Science and Astrobiology Decadal Survey et al., 2022). Ocean worlds including Europa, a moon of Jupiter, and Enceladus, a moon of Saturn, likely harbor global oceans beneath their icy surfaces (e.g., Carr et al., 1998; Iess et al., 2014). At Enceladus, salty-ice erupts in geysers from its south polar terrain “tiger stripes” (e.g., Porco et al., 2006; Spencer and Nimmo, 2013 and references within), and researchers have observed putative plumes at Europa (i.e., Roth et al., 2014; Sparks et al., 2016; Jia et al., 2018). These ocean worlds may be excellent places to search for life as their oceans are kept liquid due to the tidal forcing from their eccentric orbits. This causes frictional heating within the interiors that can provide the energy and chemistry (water-rock interactions) needed to make the oceans habitable (Greenberg et al., 2000; Chyba and Phillips, 2001). Additionally, water may be able to make its way through the ice shell via fractures and cryovolcanic activity, potentially producing plumes and surface deposits that can be collected and analyzed (e.g., Manga and Wang, 2007; Quick et al., 2017).
Although promising for life, these environments present challenges for biosignature detection. Samples likely contain low biomass concentrations, have high salt levels, are acidic or basic in pH, and contain complex chemical compositions. For example, at Enceladus the Cassini spacecraft made observations of possible molecular nitrogen (N2), carbon dioxide (CO2), methane (CH4), propane (C3H8), acetylene (C2H2), and several others in the plume material (Waite et al., 2017; Postberg et al., 2018), as well as measurements of sodium and potassium chloride salts in Saturn’s E-ring (Postberg et al., 2009), which is believed to be fed by the Enceladus plume. Additionally, at Europa detections have been made of sodium chloride (NaCl), magnesium chloride (MgCl2), and magnesium sulfate (MgSO4) salts, hydrogen peroxide (H2O2), and possibly radiation processed sulfuric acid (H2SO4) (McCord et al., 1999; Kargel et al., 2000; Dalton, 2007; Carlson et al., 2009; Brown and Hand, 2013; Trumbo et al., 2022). Salts and other interferent chemistries have been shown to have degrading effects on analyses (e.g., Stockton et al., 2009; Gehrke and Leimer, 1970). In addition, introduction of acids and bases into a Gas-Chromatography separation system can degrade the column stationary phase and lead to column bleed, peak tailing, and reduction in chromatogram resolution (Agilent Technologies, 2007). Accordingly, there is a persistent need for low size, mass, and power instrumentation for biosignature sample preparation and analyses in space.
Numerous biosignature detection instruments capable of analyzing signals of interest are currently under development including those through the 2019 NASA ICEE-2 (Instrument Concepts for Europa Exploration) grants, and NASA’s Concepts for Ocean worlds Life Detection Technology (COLDTech) program focused on advancing the technology readiness level (TRL) of biochemical, analytical, and sample handling instruments for a potential future ocean world landed mission (Hand et al., 2017; Hand et al., 2022; Mercer et al., 2023). These include the Sample Processor for Life on Icy Worlds (SPLIce) (Chinn et al., 2017), Microfluidic Organic Analyzer for Biosignatures (MOAB) (Golozar et al., 2022), the Mass Spectrometer for Planetary Exploration-Organic Composition Analyzer (MASPEX-ORCA) (Blase et al., 2020), the Europan Molecular Indicators of Life Investigation (EMILI) (Brinckerhoff et al., 2022), MICA: Icy-World Chemistry Analyzer (Ricco et al., 2022), and others. Additionally, the Enceladus Orbilander mission, which was recently prioritized by the OWL decadal survey (Committee on the Planetary Science and Astrobiology Decadal Survey et al., 2022), describes in situ instrumentation needed to search for signals of life in plume material collected in orbit and from samples on the surface (MacKenzie et al., 2022). Although multiple groups have contributed significant advancements towards the development of biosignature detection, there is a lack of development for sample preparation devices. It is clear, however, that in order to meet the expected environmental challenges in ocean world environments, and improve the sensitivity and efficiency of bio-molecule detection instruments for a wide range of complex mixtures, samples must undergo appropriate pre-processing. This includes concentration, desalination, and purification.
We present here the development to date of an agnostic, readily-integrated sample preparation system for amino acids, the Biosignature Preparation for Ocean Worlds (BioPOW) instrument, developed partially under a 2019 ICEE-2 grant. Building off of our previous work (Volkenburg et al., 2022), this platform is a novel, low size (approximately 12 cm × 20 cm × 20 cm) and mass (590 g) automated device capable of purifying, concentrating, and derivatizing a range of amino acids from complex (e.g., high salinity, high/low pH) ocean world analog mixtures. In this paper, we overview the system operating procedures, demonstrate its utility in purifying a model amino acid without significant loss, and describe its maturation in test readiness level for flight. We also discuss how, beyond our immediate interest in amino acid purification, our approach creates a pathway to purify numerous biosignatures of interest using solid phase microscale columns for in situ space flight investigations.
The automatic BioPOW instrument prototype was constructed as three distinct modules that were later integrated for testing (Figure 1). The fluidic manifold with reservoirs and adaptors was 3D-printed using Stereolithography (SLA) (Figure 1C) using a rigid transparent resin (Somos® WaterShed® XC 11122) allowing for visual inspection of the manifold during testing. It was populated with a micro-metering pump (LPVA1050050L PUMP-VV-50μL-PEEK-TZP CERAMIC) and solenoid valves (LFNA1250125H MIV-FLANGE MOUNT-300 SERIES) manufactured by Lee Company. All channels in the manifold have a 1/32-inch diameter. The sample cup (Figure 1B) and derivatization tank (Figure 1D) both include custom embedded resistive heaters made from 21-gauge nichrome wire (Temco Industrial) and a commercial strip (HAP6945, Minco), respectively. The sample cup (1 mL sample volume) has a titanium outer sleeve with a high temperature custom-machined aluminum nitride-based ceramic inner lining (Shapal) encapsulated with Cerabond 571 ceramic epoxy (Aremco). Additionally, we designed a separate chamber for the drying (concentration) and derivatization steps due to the need to have additional ports for consumables and to provide temperatures in excess of 100°C in a confined region. The derivatization chamber was milled from stainless steel and has three ports, one input for inserting the sample and the derivatization fluid, a second input for an inert carrier gas, and one output for vapor exhaust during the drying stage. For lab testing, we used nitrogen as the carrier gas. Metal, 1/8-inch 304 stainless steel tubing connects the chamber to input sources and the output to accommodate the high temperatures required during derivatization. Otherwise, the remaining manifold elements with inlet and outlet ports that do not directly mount to the manifold are operationally coupled using 1/8-inch outer diameter, 1/16-inch inner diameter tubing.
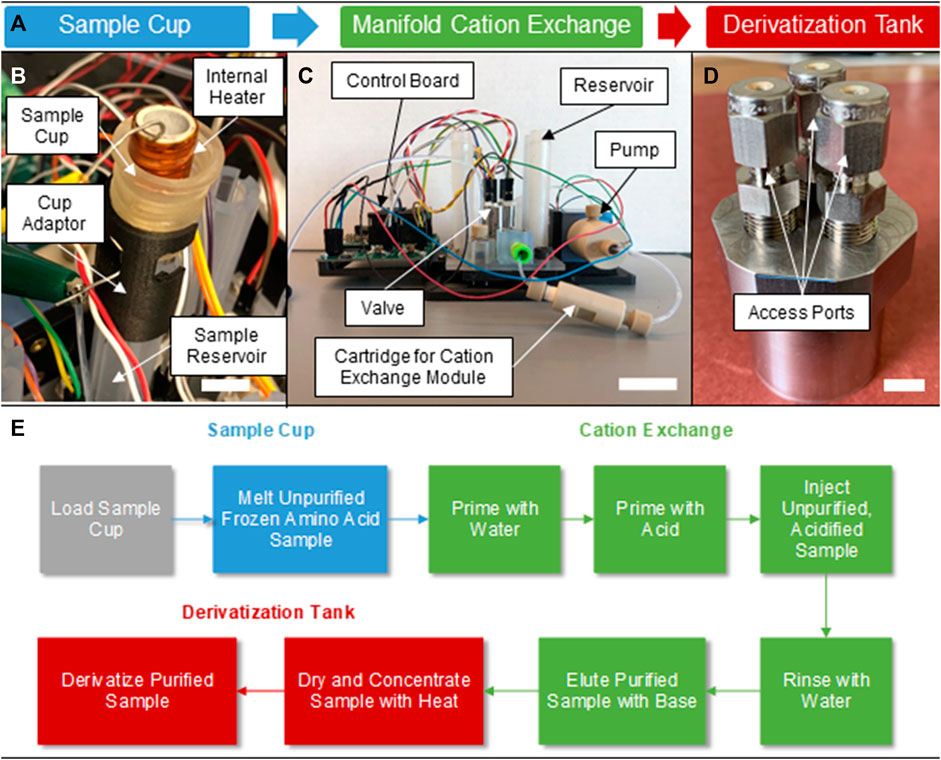
FIGURE 1. Conceptual workflow and components of the BioPOW system. (A) Standalone prototypes and process flow for the three subcomponents of the sample handling and preparation device, BioPOW, for amino acids. (B) Version 2 (v2) of the sample cup showing the internal nichrome heater and ceramic shell, scale bar is 1 cm. (C) The cation exchange manifold used for the DOE and showing mounted passive and active components, scale bar is 4 cm. (D) The derivatization tank showing the three access ports for sample introduction, venting, and gas injection, scale bar is 1 cm. (E) The operating procedure for sample processing. Here, sample loading is performed by an external system.
Control electronics and software were designed, built, and tested to automatically control the processing of a sample, including melting, purification, drying, and derivatization. The electronics breadboard also controlled the linear actuator that seals the sample cup on delivery from a system such as the Collaborative Acceptance and Distribution for Measuring Europan Samples (CADMES) Sample Delivery System, as specified during the NASA ICEE-2 grant, and manages the heater that melts the ice sample. The custom-printed circuit board has an ATmega328 microcontroller to run the experiment and interface with a host computer. The board also has a Texas Instruments DRV8824 micro-stepping motor driver, a MAX6675 K-Type thermocouple amplifier, nine solenoid drivers for controlling the valves, a proportional valve driver circuit, and an LTM4625DC/DC converter for driving the ice cup heaters.
The BioPOW employs an ion exchange chromatography technique to purify and desalinate proteinogenic amino acids, independent of the pH, salinity, and type of salt in the sample. The system works by leveraging three sub-components (Figure 1A): Figure 1B a sample cup for melting the solution, Figure 1C a cation exchange module for purifying and concentrating the amino acids, and Figure 1D a derivatization tank for tagging the amino acids for downstream analysis.
To receive a sample, we designed the sample cup to interface seamlessly with the CADMES Sample Delivery System developed in the ICEE-2 program. This is a mechanical arm proposed for Europa sample collection and transfer to internal processing (Malespin et al., 2020). We explored two versions of the cup design, one complete with a custom receiving and sealing jig that will automatically generate a fluid tight seal (v1, Supplementary Figure S1), and another that can be directly mounted to the sample reservoir (v2, Figure 1B) of the fluidic manifold. For simplicity, we used the direct mount version of the cup in this study. Both cups include a resistive heater that elevates the temperature of the ice above 0°C, causing it to melt for facile transfer to the sample reservoir. For v2, the sample cup melts a packed ice bed in the 1 mL reservoir at an initial wall temperature of 3°C–28°C in less than 5 min with a 5 W power draw (∼1.4 kJ, Supplementary Figure S2). Future designs would also accommodate higher temperatures (>100°C) and incorporate tubing to transfer volatiles released directly to an MS and/or GC-MS for analysis.
The fluidic manifold has embedded microchannels that enable sample and reagent transfer between passive components (i.e., mounted reservoirs, tubing adaptors, and a microscale solid phase cartridge to contain the cation exchange resin) and programmable electromechanical elements (solenoid valves and a micro-metering pump) as shown in Figure 1C. Thus, this standalone manifold is capable of performing the priming conditioning, sample introduction, washing, and elution steps required for cation exchange purification in an automated fashion.
Cation exchange was performed on biotechnology grade AG50W-X8 resin with hydrogen form, 8% crosslink density, 200–400 dry mesh size, 63–150 μm wet bead size, and ∼1,000 molecular weight limit (purchased from Bio-Rad Laboratories). Deionized (DI) water was purchased as NERL™ reagent-grade water (Fisher Scientific). American Chemical Society (ACS)-grade reagents (Millipore Sigma), hydrochloric acid (HCl, 37 w/v %), and ammonium hydroxide (NH4OH, 28–30 w/v %) were diluted in DI water to 50 mM and 1.0 M, respectively. For design of experiments (DOE) optimization, the introduced sample consisted of L-phenylalanine (phe) hydrochloride (100 mM phe in 0.1 M HCl to provide an acidified sample with pH < 2; see cation exchange procedure details in Van Volkenburg et al., 2022) spiked into a NaCl solution (5 M in water). The phe and NaCl solutions were purchased from Millipore Sigma and diluted with DI water to final concentrations of 40 mM phe into 50 mM or 500 mM NaCl. Phe was selected because it has a high binding affinity to the resin in the range of NaCl concentrations tested, enabling initial system optimizations (described below). All buffers and samples were prepared in Biosafety-Level 2 (BSL-2) hoods to reduce the possibility of contamination. Before separation experiments, cation exchange resin was purified with three 15 min spins at 1500 RPM in 1 M NH4OH, discarding the supernatant in between each spin.
Finally, following purification, the eluted sample is transferred to the derivatization tank for drying and resuspension in the derivatization agent (Figure 1D). Distinct from the sample cup, this module has three ports for liquid injection, vacuum drying, and carrier gas injection. The drying process is paramount for concentrating the sample and removing the 1 M NH4OH solution, which is used to elute the amino acids from the cation exchange resin and interferes with the derivatization process. Due to the lower boiling point of NH4OH compared to ice, ∼38°C for a 25% solution at atmospheric pressure (National Center for Biotechnology Information, 2023), and lower temperature needs for the derivatization process, the derivatization tank has less stringent heating requirements than the sample cup. Therefore, the derivatization tank was programmed for two operational modes. During drying, the sample is slowly heated to 60°C and maintained until only a powder remains. The gradual temperature ramp in this step prevents excessive boiling and bubbling of the sample amino acid and NH4OH solution. Following removal of the NH4OH solution, the derivatization agent reacts with the amino acids at 90°C for 1 h. Specifically, starting from room temperature this module takes approximately 12 min to achieve a temperature of 60°C for concentration (∼3.6 kJ) and 25 min to increase from 60°C to 90°C for derivatization (∼7.5 kJ) with a power dissipation of 5 W while empty (Supplementary Figure S3).
The derivatized sample enables analysis via Gas Chromatography-Mass Spectrometry (GC-MS) or GC-GC-MS. Although this study focuses on GC-MS analysis, the BioPOW purification technique is agile and can be readily integrated with alternative downstream techniques, such as capillary electrophoresis-laser induced fluorescence (CE-LIF) (Oborny et al., 2021), by modifying the tagging solution and derivatization protocol. A summary of the operating procedure for the sample preparation system is shown in Figure 1E.
We used a DOE approach (Design of Experiments, 2023) to identify the most significant input variables in the purification of amino acids using the stand-alone cation exchange module (Figure 1C, and green boxes in Figure 1E). By manipulating multiple inputs at the same time, the DOE approach can efficiently identify important interactions between parameters that are not readily evident during single-factor experiments in an expedient manner. To simplify the binding interactions, the introduced sample was a binary system comprised of one amino acid, phe, and one salt, NaCl. We used UV-vis absorbance to determine phe concentration and conductivity to correlate to salinity. Cation exchange efficiency is dependent on numerous kinetic factors including sample and buffer pH, flow rates and residence time, and temperature of the system (Wang et al., 1989; Melis et al., 1996). Based on our previous experiments regarding the most influential factors on cation exchange efficiency, we down-selected the input variables to five factors at two levels. These included sample salt (NaCl) concentrations (50 and 500 mM), and manifold operation values: sample introduction flow rate (5 and 20 μL/s), NH4OH eluate flow rate (5 and 20 μL/s), water wash volume (5x and 10x resin volume, where 5x and 10x indicate the multiplier value to the total resin volume of 50 μL), and eluate buffer volume (5x and 10x resin volume). Here, the maximum flow rate for our chosen pump is 50 μL/s. Thus we chose the volumetric flow rates to be near the lower bound of this range (i.e., 5 μL/s) and towards the center of this range (i.e., 20 μL/s). Additionally, we chose the 5x multiplier as our lower bound for the eluate as advised by the manufacturer to optimize elution (personal communication with BioRad). For the eluate wash an additional 500 µL of buffer was added to each separation to compensate for the dead volume within fluidic lines and ensure all amino acids were collected in the eluate reservoir. Finally, the manifold performance was evaluated by 1) percent phe recovery (Eq. 1) where concentrations and volumes are denoted C and V, respectively, and 2) conductivity decrease factor (Eq. 2), in units of x times decrease characterized by the ratio of conductivity (σ) in the eluate and sample. Parameter values for all nine DOE runs are included in Table 1.
Following separation on the fluidic manifold, 200 µL from both eluate and waste reservoirs was collected in a 96-well plate (UV-STAR) and analyzed with a Spark (TECAN) or ClarioStar Plus (BMG Labtech) plate reader. The absorbance was measured at 258 nm, and converted to concentration using a standard curve of phe in 1 M NH4OH. An additional 300 µL was placed into a low recovery vial and analyzed with a Mettler Toledo conductivity meter with low volume probe attachment (FiveEasy Benchtop F30 Meter). The probe was calibrated by using a 1,413 μS/cm potassium chloride conductivity standard (Mettler Toledo) before each use.
Analysis of the purified sample was performed through manual derivatization and GC-MS in the laboratory to evaluate separation efficiencies. This procedure used previously developed methods at the Southwest Research Institute (SwRI) for amino acid interrogation with N-tert-Butyldimethylsilyl-N-methyltrifluoroacetamide (MTBSTFA) (Blase et al., 2022). MTBSTFA is a silylating agent that rapidly reacts with amines and can be used to convert amino acids to more volatile products, enabling analysis via GC-MS (Stalport et al., 2012). To derivatize the amino acids, 200 µL from both the eluate and waste reservoirs of the manifold was dried in a vacuum oven at 60°C and 80 kPa in low recovery vials (VWR). Samples were then re-hydrated in 100 µL of MTBSTFA, capped, and heated in a vacuum oven at 90°C for 1 h. Samples were cooled to room temperature before analysis with the Agilent Technologies 7890B GC System coupled with a 5977A Mass Spectrometry Detector and 7693A Autosampler. The GC column used was a 30 m × 0.25 mm × 0.25 µm DB-5MS. For measurement, a liquid sample injection (1.0 µL) was performed using the autosampler. The GC inlet was set to 200°C while the flow rate was set to 1.0 mL/min. The GC method was run using the following parameters: 70°C for 1 min, ramp at 30°C/min to 280°C (hold at 280°C for 1 min), then ramp at 40°C/min to 325°C.
Following validation of the sample cup and derivatization tank heating capabilities (Supplementary Figures S2, S3), we performed a series of purification experiments to compare the performance of the stand-alone automated cation exchange manifold with manual introduction of reagents using a syringe pump (Figure 2). Similar to our previous manual study (Volkenburg et al., 2022), we selected 62.5 mM phe in a 1.6 M NaCl solution as the amino acid sample to facilitate rapid quantitation with an ultraviolet-visible (UV-vis) absorbance-based plate reader (Figure 2A). We intentionally overloaded the columns by introducing excess phe in order to ensure there would be a detectable signal in the waste and eluate reservoirs for both UV-vis and GC-MS detection. We kept flow conditions constant between both experiments, adding 800 µL of 10 mM HCl, 1 mL of sample, 800 µL of DI water, and 1.6 mL of 1 M NH4OH sequentially to the cartridge at a flow rate of 50 μL/s. Notably, while the null hypothesis was not rejected for the difference between the phe recovery rate for the manifold and benchtop experiments for the waste reservoir, there was a clear statistical significance for the eluate reservoirs (
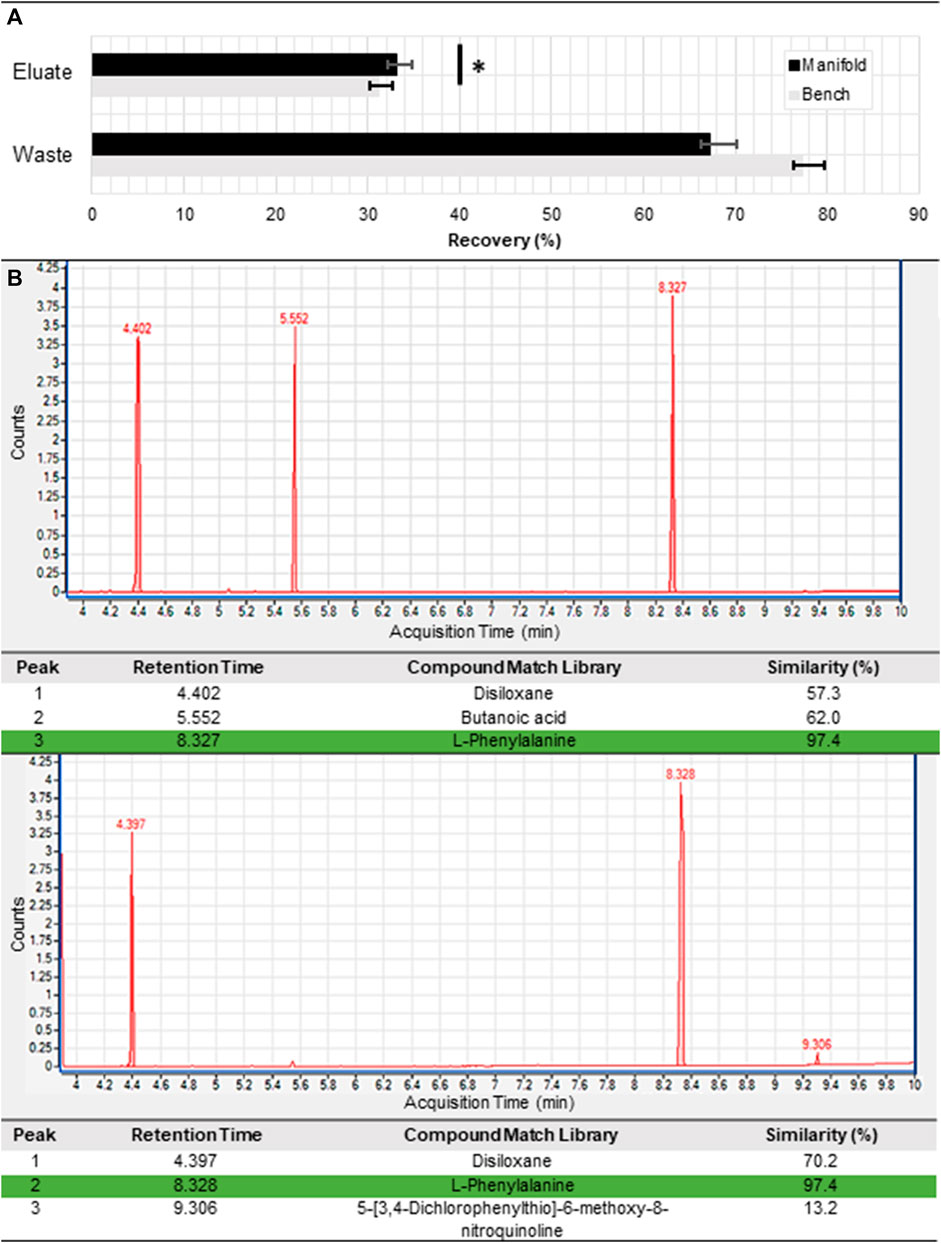
FIGURE 2. Purification of an exemplar amino acid. (A) Phenylalanine recovery from overloaded samples based on fluorescence measurements at the eluate and waste reservoirs of the standalone cation exchange module for manifold and benchtop tests (n = 3). Here, the p-value (determined using a t-test) is 0.04 and 0.364 for the eluate and waste reservoirs, respectively. Detection of a phenylalanine peak from the purified eluted sample by the GC-MS for both the (B) benchtop and (C) manifold processed samples. Phenylalanine detection is highlighted in green.
With these set parameters, we chose nine runs in a fractional factorial design within the bounds of pre-determined high and low values using John’s Macintosh Project (JMP) statistical software (Figure 3A) (Design of Experiments, 2023). A fractional factorial, or screening experiment, allows for detection of the greatest effects. For each experiment, we measured the resulting phe recovery rates and NaCl reduction in each eluate. Next, we used a linear regression fit to identify effects and dependencies amongst the parameters. Leverage plots with the actual values against the predicted values for both conductivity decrease (Figure 3B) and phe recovery (Figure 3C) show a good fit (R2 = 0.99) and a poor fit (R2 = 0.77), respectively. We additionally show the p-value (significance value) for each input, where values less than 0.05 indicate statistical significance [denoted with (*), Figure 3D]. Here, Figure 3D shows that salt concentration, sample flow rate, eluate flow rate, and water wash volume are all statistically significant variables for the salt reduction, but none of the variables are statistically significant for the phe recovery. Finally, these results allowed us to perform a desirability analysis where our response goal was equally weighted between both phe recovery percentage and conductivity decrease factor to determine the optimal parameters for separation. For this analysis we linearly weighted our response goals between 0%–100% phe recovery for the lower and higher bounds, and between ×1 and ×7 for the conductivity decrease factor. Our selected parameters allowed us to compute the optimized values and corresponding implications for each parameter of the DOE (Figure 3E).
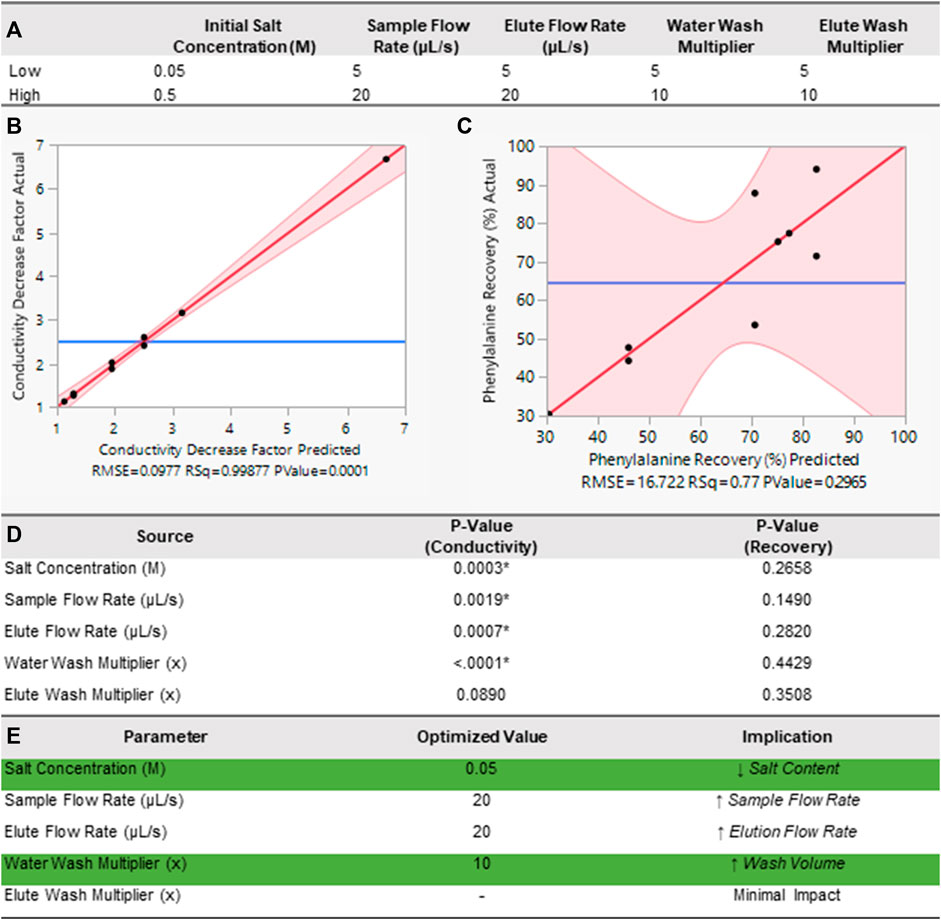
FIGURE 3. DOE analysis of module functionality. (A) The two levels of input variables for the DOE. Actual values plotted against predicted values (leverage plot) from the DOE with a linear regression fit for (B) conductivity decrease and (C) phenylalanine recovery that compares the null hypothesis (blue line) to the alternative hypothesis (red slanted line) with 95% confidence intervals (red shaded region). (D) Table of p-Values for selected input values, where (*) denotes statistical significance (
In our recent study, we explored the operational limits of the cation exchange module (Van Volkenburg et al., 2022). In this work, we mature the process by demonstrating its utility on an automated platform. The automated platform enabled higher throughput screening of the optimal operation parameters for sample purification with significantly reduced human intervention. From our comparison tests, we found that the standard deviations between triplicate runs were small, and well within the error of the measurement method. Despite being statistically different, average phe recovery between the two experiments was very close with 33% for the automated manifold and 31% for the manual benchtop Figure 2A. Because the columns were overloaded, the loss in each experiment was higher than the recovery, but both tests were within 10% of each other (67% for the automated manifold and 77% for the manual benchtop). Notably, the automated platform yielded slightly higher phe recovery rates compared to benchtop experiments with a p-value of 0.04. This slight performance improvement was likely due to the variations in the flow profile between both experiments. While the benchtop experiments were performed manually with a continuous flow syringe pump, the automated manifold employed the use of a micro-metering pump. In contrast to syringe continuous flow, micro-metered fluid injection occurs in a cyclic manner whereby reagents are sequentially withdrawn and dispensed in 50 µL volumes. This improved performance is thus consistent with preliminary findings from our previous studies, where we found that introducing soaks during elution (i.e., intentionally pausing flow in between solvent exchange steps) increased the purification efficiency of our process. Nevertheless, this agreement between experiments validated the performance of our automated setup and indicated we could transition to manifold experiments moving forward.
With performance consistency confirmation, our final tests focused on developing a better understanding of the interaction between operational parameters on the cation exchange manifold. During optimization, we particularly focused on salt reduction as these non-volatile contaminating ions lead to reduced performance of downstream derivatization and amino acid analyses. Thus, our primary goal was to reduce the overall salt content in the purified samples without significant (>50%) loss of amino acids. While we were able to discern clear trends from the impact of varying experimental operational parameters on the conductivity decrease, no statistically significant relationships with phe recovery emerged. The poorer fit for phe recovery could be due to several factors; the main reason is likely bubbles in the lines causing volume variability in the manifold collection reservoirs. Collected eluate volumes had between 3% and 19% differences between the input volumes and actual collected volumes. Automated end-to-end sequential runs enabled by our integrated system will remove the need for an operator to collect samples. Removing human interference during measurement and handling steps should reduce variability of phe concentration measurements. Additionally, bubble injection into the standalone cation exchange manifold may have occurred at our tubing connections made from materials with poor mechanical cycling performance such as the inlet/outlet ports connected to PEEK ¼ −28 fluidic fittings. To eliminate these potential leak points, future manifold designs will transition away from inexpensive, rapid prototyping based materials to high fidelity space rated materials that are permanently mated by brazed or epoxied connections.
After characterizing the manifold performance, we built the fully integrated BioPOW system complete with fluid routing between sample cup and derivatization tank (Figure 4A computer-aided design; Figure 4B assembled prototype). The updated manifold consists of a set of five reagent reservoirs, eleven solenoid valves, a liquid pump, a cation exchange cartridge, and two collection reservoirs (mechanical schematic, Figure 4C). Here, the reagents include cation exchange chemicals (i.e., liquefied sample, acidic solution, basic solution, and water) along with the derivatization agent (MTBSTFA). Additionally, the collection reservoirs divide the sample into the purified eluate and salty “waste”. Importantly, the waste can be captured here for optional downstream analysis. Despite these multiple functionalities, the entire instrument, including the mounted sample cup, side mounted metering pump, and derivatization tank, has a volume of only 12 cm × 20 cm × 20 cm. As designed, this new integrated manifold is about 12.7 cm long × 10.2 cm wide × 10.2 cm tall and 590 g (sample cup is 22 g, manifold with associate valves, pump, and reservoirs is 389 g, and the derivatization tank is 179 g). The associated control board is roughly 12.7 cm long × 12.7 cm wide × 2.5 cm thick and its mass will depend on final flight system requirements. Once in the fluidic manifold, the melted sample is automatically pumped through an in-line, microscale (i.e., 50 μL) packed bed filled with cation exchange resin. Building off our previous work, the now integrated BioPOW system captures, purifies, and elutes amino acids in suspension by performing sequential rinsing steps of an acidic solution, water, and a basic solution with the added capability of pushing eluted samples directly to the derivatization tank for solvent exchange to the derivatization agent.
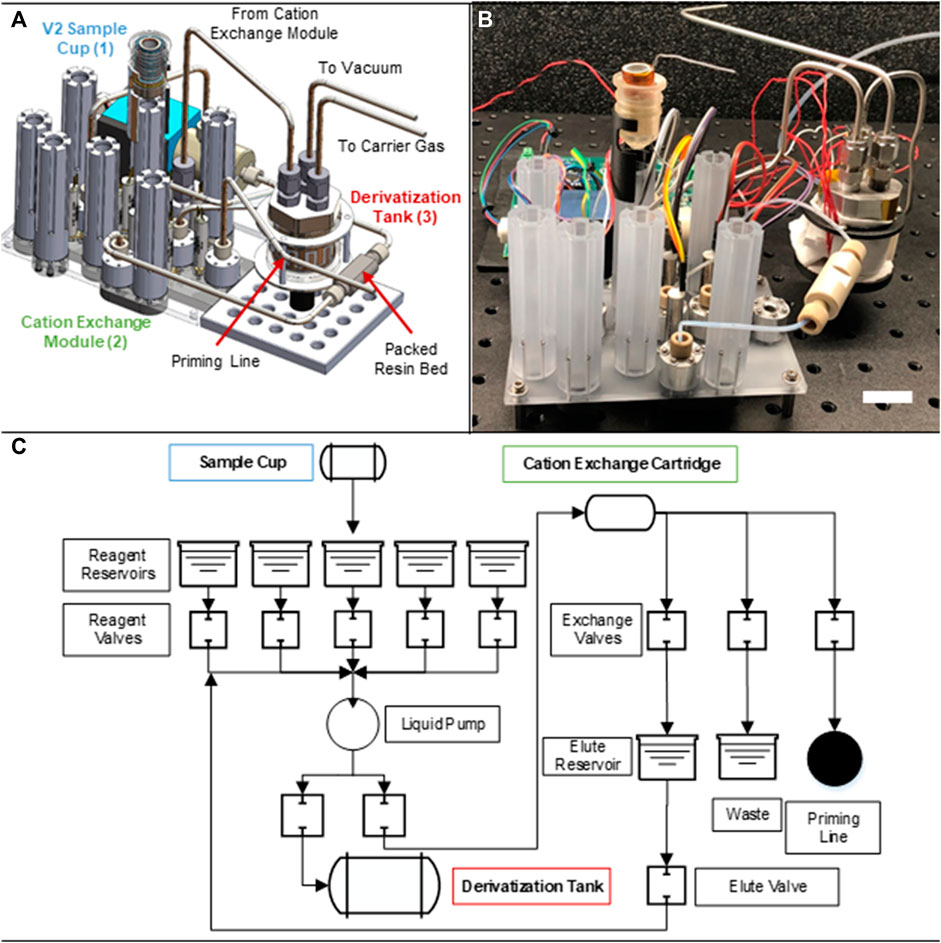
FIGURE 4. Integrated Biosignature Preparation for Ocean Worlds (BioPOW) instrument prototype. This figure shown is a (A) labeled schematic, (B) image of the assembled prototype, scale bar is 2 cm, and (C) mechanical routing of the BioPOW system showing the overall integration scheme.
Currently BioPOW is comprised of a combination of commercial off the shelf (COTS) electronics, rapid prototyping materials (e.g., 3D printed manifold), durable materials (e.g., metals, ceramics), and medical grade electromechanical components. As a result, the present prototype is not yet mature enough for space flight (TRL 4), but a facile route to increasing its technology readiness is available. In this prototype design, all electronics are controlled through an Arduino interface. This open source electronic prototyping platform is responsible for controlling the solenoid valves, a stepper motor to drive the liquid dispense pump, and thermocouples to measure the temperature of the sample cup and derivatization tank during heating. To adapt this design for flight, the Arduino can be replaced with a higher TRL electronic component such as a field programmable gate array (FPGA) and the COTS integrated circuit for motor control can be replaced by high TRL discrete H-bridges. For example, the electronics for the Europa Imaging System (EIS) instrument on the Europa Clipper mission use radiation tolerant FPGAs and H-bridge circuits for motor control which are rated to 300 krad total dose to survive that mission’s expected 150 krad environment inside the spacecraft radiation shield vault. This tolerance level would be sufficient for an Enceladus mission profile and could be supplemented for a Europa mission profile with the addition of physical shielding, as implemented on Europa Clipper and Juno missions (Spaulding and Eremenko, 2015; Bozovich et al., 2018).
Additionally, the sample cup and derivatization tank are already made from space rated materials such as high temperature aluminum nitride, stainless steel, and nichrome. As a result, the manifold and associated microfluidic active and passive mounted components require the most maturation. Depending on the materials selected, radiation can cause embrittlement and lead to leaks and malfunctions. There is a growing body of work surrounding microfluidic manipulation in space using similar electromechanical components including promising results from radiation testing in 50 rad of 300 MeV/n Fe-56 (Ricco et al., 2020; Padgen et al., 2021), and between 0.15–1.2 krad in high-inclination Earth orbit (Kitts et al., 2011). However, the radiation requirements for ocean world missions are much higher. Radiation tests will be performed to upper levels of 300 krad total ionizing dose (TID) to simulate the radiation under shielding that is expected at Europa with a 2-fold factor of safety (150 krad nominal level for design) based on the Europa Lander Report’s design for Europa’s surface environment (Hand et al., 2017; Bozovich et al., 2018). Additionally, the Europa Clipper mission instrument payload has shielding, electromechanical hardware, spacecraft electronics, and materials tested for operation within this harsh environment, of which the BioPOW design will draw from (Bozovich et al., 2018; Centurelli et al., 2018). Future prototypes will focus on replacing the 3D-printed materials with polymer, glass, or metal-based manifolds, switching to reagent introduction via direct injection into the manifold, and performing environmental testing in thermal vacuum chambers and on reduced gravity flights to continue optimizing performance and raising the TRL of the system for space flight. Gas-tight fittings will likely reduce bubble generation, but other mitigation strategies like bubble traps, which have been demonstrated in microgravity, will also be investigated (Sun et al., 2019; Padgen et al., 2020). Additionally, integrated flow sensors and pressure sensors at the inlets and outlet of the derivatization tank respectively will be necessary to ensure operation in microgravity or unexpected tilt regimes. Similarly, the sample cup could use a pressurized neutral gas entering from the top seal to pump the melted fluid into the next chamber. Higher TRL iterations will also investigate cation exchange resin and buffer degradation routes in relevant environmental conditions to see the impact on purification capability in a mission-like scenario, and will explore mitigation strategies such as alternate resin formulations or environmental controls if necessary.
Adequate preparation of the expected complex aqueous sample matrices at ocean worlds is essential to enable detection of amino acids with both high sensitivity and high confidence. This work represents an important step towards a coherent sample preparation system: transitioning from benchtop experiments to an automated platform that can purify and separate bio-molecules for downstream analyses. The BioPOW design encompasses a sample cup to melt icy samples, a cation exchange manifold to purify amino acids, and a dry-down and derivatization tank to concentrate and chemically tag amino acids for detection and characterization. Preliminary studies allowed us to characterize the thermal performance of the stand-alone module operational modes (i.e., melting, drying, and derivatization), and determine the most important operational parameters for reducing salt concentration with the cation exchange module without significantly impacting amino acid recovery. With this improved understanding of the system performance, the sample purification can be optimized for a range of samples types, including more complex sample compositions containing potentially corrosive (e.g., H2SO4 at Europa) and/or rocky materials (e.g., Mars or Ceres rock-ice mixtures). Additionally, the surrounding hardware and connected modules can be used to target other biomolecules in different relevant environments by changing the resin and reagents used. Similarly, the BioPOW modular system was designed with integration flexibility in mind, and can interact both with the CADMES Sample Delivery System to intake a sample (Malespin et al., 2020) and can provide purified and concentrated amino acid solutions with or without derivatization to downstream analytical instruments such as heritage GC-MS systems or proposed CE-LIF microfluidic devices. Through future iterations, the low size and mass BioPOW integrated platform will raise the system’s TRL for flight through electronic and material developments identified in this project, and in doing so, will provide a sample purification platform for improved detections of amino acids at ocean worlds.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
KD, TV, KC, CP, JB, JH, CB, and MP contributed to the conception and design of the chemical experiments. KD, JH, DF, EM, TN, GD, OP, and MP contributed to the mechanical and electrical design of the BioPOW system. TV, CP, and JB, produced and analyzed the experimental results. KD and TV wrote the first draft of the manuscript. All authors contributed to the article and approved the submitted version.
This work was funded using Johns Hopkins University Applied Physics Laboratory (JHU/APL) internal funding and a NASA ICEE-2 grant 80NSSC19K0611 through a subcontract with Southwest Research Institute (SwRI).
The authors appreciate support from Dr. Chris Glein, Dr. Mark Libardoni, and Ryan Blase from SwRI in developing the amino acid derivatization strategy.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fspas.2023.1244682/full#supplementary-material
Agilent Technologies (2007). Major causes of column performance degradation. Available at: https://www.agilent.com/cs/library/support/documents/col%20degrd.pdf (Accessed October 3, 2022).
Blase, R. C., Libardoni, M. J., Miller, G. P., Miller, K. E., Phillips-Lander, C. M., Glein, C. R., et al. (2022). MEMS GC column performance for analyzing organics and biological molecules for future landed planetary missions. Front. Astronomy Space Sci. 9. doi:10.3389/fspas.2022.828103
Blase, R. C., Libardoni, M. J., Miller, G. P., Miller, K. E., Phillips-Lander, C. M., Waite, J. H., et al. (2020). Experimental coupling of a MEMS gas chromatograph and a mass spectrometer for organic analysis in space environments. ACS Earth Space Chem. 4, 1718–1729. doi:10.1021/acsearthspacechem.0c00131
Bozovich, A. N., Rax, B. G., Davila, J., Nguyen, D., Kenna, A. J., Zajac, S. A., et al. (2018). “Compendium of total ionizing dose (TID) test results for the Europa clipper mission,” in 2018 IEEE Radiation Effects Data Workshop (REDW), Waikoloa Village, HI, USA.
Brinckerhoff, W. B., Willis, P. A., Ricco, A. J., Kaplan, D. A., Danell, R. M., Grubisic, A., et al. (2022). Europan molecular Indicators of life investigation (EMILI) for a future Europa lander mission. Front. Space Technol. 2. doi:10.3389/frspt.2021.760927
Brown, M. E., and Hand, K. P. (2013). Salts and radiation products on the surface of Europa. AJ 145, 110. doi:10.1088/0004-6256/145/4/110
Carlson, R. W., Calvin, W. M., Dalton, J. B., Hansen, G. B., Hudson, R. L., Johnson, R. E., et al. (2009). “Europa’s surface composition,” in Europa (Arizona, United States: University of Arizona Press), 283–387.
Carr, M. H., Belton, M. J. S., Chapman, C. R., Davies, M. E., Geissler, P., Greenberg, R., et al. (1998). Evidence for a subsurface ocean on Europa. Nature 391, 363–365. doi:10.1038/34857
Centurelli, J., Turtle, Z. P., Slack, K. A., Osterman, S. N., and Roth, D. R. (2018). “Europa imaging system wide angle camera: the effect of gamma radiation on the refractive index and transmission of radiation resistant glasses,” in Space telescopes and instrumentation 2018: optical, infrared, and millimeter wave. Editors H. A. MacEwen, M. Lystrup, G. G. Fazio, N. Batalha, E. C. Tong, and N. Siegler (Austin, United States: SPIE), 150. doi:10.1117/12.2313596
Chinn, T. N., Lee, A. K., Boone, T. D., Tan, M. X., Chin, M. M., McCutcheon, G. C., et al. (2017). Sample processor for life on icy worlds (SPLIce): design and test results. 3.
Chyba, C. F., and Phillips, C. B. (2001). Possible ecosystems and the search for life on Europa. Proc. Natl. Acad. Sci. 98, 801–804. doi:10.1073/pnas.98.3.801
Committee on the Planetary Science and Astrobiology Decadal SurveySpace Studies Board, Division on Engineering and Physical SciencesNational Academies of Sciences, Engineering, and Medicine (2022). Origins, worlds, and life: a decadal strategy for planetary science and Astrobiology 2023-2032. Washington, D.C.: National Academies Press. doi:10.17226/26522
Dalton, J. B. (2007). Linear mixture modeling of Europa’s non-ice material based on cryogenic laboratory spectroscopy. Geophys. Res. Lett. 34, L21205. doi:10.1029/2007GL031497
Design of Experiments (2023). Design of experiments. Available at: https://www.jmp.com/en_us/applications/design-of-experiments.html (Accessed October 3, 2022).
Golozar, M., Casto, L., Estlack, Z., Kim, J., Butterworth, A. L., and Mathies, R. A. (2022). “Microfluidic organic analyzer for biosignatures (MOAB): a flight-design instrument with the capabilities required to detect trace biosignatures on icy moons,” in 2022 Astrobiology Science Conference (AGU), Atlanta, Georgia, May 15-20, 2022.
Greenberg, R., Geissler, P., Tufts, B. R., and Hoppa, G. V. (2000). Habitability of Europa’s crust: the role of tidal-tectonic processes. J. Geophys. Res. Planets 105, 17551–17562. doi:10.1029/1999JE001147
Hand, K. P., Phillips, C. B., Murray, A., Garvin, J. B., Maize, E. H., Gibbs, R. G., et al. (2022). Science goals and mission architecture of the Europa lander mission concept. Planet. Sci. J. 3, 22. doi:10.3847/PSJ/ac4493
Hand, K. P., Murray, A. E., Garvin, J. B., Brinckerhoff, W. B., Christner, B. C., Edgett, K. S., et al. (2017). Europa lander study 2016 Report the Europa lander science definition team Report. Available at: https://europa.nasa.gov/resources/58/europa-lander-study-2016-report (Accessed March 18, 2022).
Iess, L., Stevenson, D. J., Parisi, M., Hemingway, D., Jacobson, R. A., Lunine, J. I., et al. (2014). The gravity field and interior structure of Enceladus. Science 344, 78–80. doi:10.1126/science.1250551
Jia, X., Kivelson, M. G., Khurana, K. K., and Kurth, W. S. (2018). Evidence of a plume on Europa from Galileo magnetic and plasma wave signatures. Nat. Astron 2, 459–464. doi:10.1038/s41550-018-0450-z
Kargel, J. S., Kaye, J. Z., Head, J. W., Marion, G. M., Sassen, R., Crowley, J. K., et al. (2000). Europa’s Crust and Ocean: origin, composition, and the prospects for life. Icarus 148, 226–265. doi:10.1006/icar.2000.6471
Kitts, C., Rasay, M., Bica, L., Mas, I., Neumann, M., Young, A., et al. (2011). Initial on-orbit engineering results from the O/OREOS nanosatellite.
MacKenzie, S. M., Neveu, M., Davila, A. F., Lunine, J. I., Cable, M. L., Phillips-Lander, C. M., et al. (2022). Science objectives for flagship-class mission Concepts for the search for evidence of life at Enceladus. Astrobiology 22, 685–712. doi:10.1089/ast.2020.2425
Malespin, C. A., Chu, P., Pinnick, V., Grossman, A., Barfknecht, P., Francom, M., et al. (2020). The collaborative acceptance and distribution for measuring europan samples (CADMES) sample delivery system for a future Europa lander. 1943.
Manga, M., and Wang, C.-Y. (2007). Pressurized oceans and the eruption of liquid water on Europa and Enceladus. Geophys. Res. Lett. 34, L07202. doi:10.1029/2007GL029297
McCord, T. B., Hansen, G. B., Matson, D. L., Johnson, T. V., Crowley, J. K., Fanale, F. P., et al. (1999). Hydrated salt minerals on Europa’s surface from the Galileo near-infrared mapping spectrometer (NIMS) investigation. J. Geophys. Res. Planets 104, 11827–11851. doi:10.1029/1999JE900005
Melis, S., Markos, J., Cao, G., and Morbidelli, M. (1996). Ion-exchange equilibria of amino acids on a strong acid resin. Ind. Eng. Chem. Res. 35, 1912–1920. doi:10.1021/ie950569m
Mercer, C. R., Stephan, R. A., Voytek, M. A., Niebur, C., Paganini, L., Schulte, M. D., et al. (2023). NASA science technology development programs for Ocean Worlds exploration, 308–320. doi:10.1061/9780784484470.029
National Center for Biotechnology Information (2023). Ammonium hydroxide PubChem compound summary for CID 14923, ammonium hydroxide. Available at: https://pubchem.ncbi.nlm.nih.gov/compound/14923 (Accessed March 9, 2023).
Oborny, N. J., Kehl, F., Cretu, V., Noell, A. C., and Willis, P. A. (2021). A radiation tolerant laser-induced fluorescence detection system for a potential Europa Lander mission. Acta Astronaut. 186, 465–472. doi:10.1016/j.actaastro.2021.06.012
Padgen, M. R., Chinn, T. N., Friedericks, C. R., Lera, M. P., Chin, M., Parra, M. P., et al. (2020). The EcAMSat fluidic system to study antibiotic resistance in low earth orbit: development and lessons learned from space flight. Acta Astronaut. 173, 449–459. doi:10.1016/j.actaastro.2020.02.031
Padgen, M. R., Liddell, L. C., Bhardwaj, S. R., Gentry, D., Marina, D., Parra, M., et al. (2021). BioSentinel: a biofluidic nanosatellite monitoring microbial growth and activity in deep space. Astrobiology 23, 637–647. ast. 2020. doi:10.1089/ast.2020.2305
Porco, C. C., Helfenstein, P., Thomas, P. C., Ingersoll, A. P., Wisdom, J., West, R., et al. (2006). Cassini observes the active south Pole of Enceladus. Science 311, 1393–1401. doi:10.1126/science.1123013
Postberg, F., Clark, R. N., Hansen, C. J., Coates, A. J., Dalle Ore, C. M., Scipioni, F., et al. (2018). “Plume and surface composition of Enceladus,” in Enceladus and the icy moons of Saturn (Arizona: University of Arizona Press), 129–162.
Postberg, F., Kempf, S., Schmidt, J., Brilliantov, N., Beinsen, A., Abel, B., et al. (2009). Sodium salts in E-ring ice grains from an ocean below the surface of Enceladus. Nature 459, 1098–1101. doi:10.1038/nature08046
Quick, L. C., Glaze, L. S., and Baloga, S. M. (2017). Cryovolcanic emplacement of domes on Europa. Icarus 284, 477–488. doi:10.1016/j.icarus.2016.06.029
Ricco, A. J., Maria, S. R. S., Hanel, R. P., and Bhattacharya, S. (2020). BioSentinel: a 6U nanosatellite for deep-space biological science. IEEE Aerosp. Electron. Syst. Mag. 35, 6–18. doi:10.1109/MAES.2019.2953760
Ricco, A., Quinn, R., Shimada, J., Radosevich, L., Boone, T., Rademacher, A., et al. (2022). “The Microfluidic Icy-world Chemical Analyzer (MICA): measuring soluble chemistry context to understand habitability and the potential for life,” in 2022 Astrobiology Scienc Conference, AGU, Atlanta, Georgia, May 15-20, 2022.
Roth, L., Saur, J., Retherford, K. D., Strobel, D. F., Feldman, P. D., McGrath, M. A., et al. (2014). Transient water vapor at Europa’s south Pole. Science 343, 171–174. doi:10.1126/science.1247051
Sparks, W. B., Hand, K. P., McGrath, M. A., Bergeron, E., Cracraft, M., and Deustua, S. E. (2016). PROBING FOR EVIDENCE OF PLUMES ON EUROPA WITH HST/STIS. ApJ 829, 121. doi:10.3847/0004-637X/829/2/121
Spaulding, M., and Eremenko, A. (2015). “Europa clipper vault shielding optimization approach,” in 2015 IEEE Aerospace Conference, Big Sky, Montana, 7-14 March 2015, 1–6.
Spencer, J. R., and Nimmo, F. (2013). Enceladus: an active ice world in the saturn system. Annu. Rev. Earth Planet. Sci. 41, 693–717. doi:10.1146/annurev-earth-050212-124025
Stalport, F., Glavin, D. P., Eigenbrode, J. L., Bish, D., Blake, D., Coll, P., et al. (2012). The influence of mineralogy on recovering organic acids from Mars analogue materials using the “one-pot” derivatization experiment on the Sample Analysis at Mars (SAM) instrument suite. Planet. Space Sci. 67, 1–13. doi:10.1016/j.pss.2012.02.010
Sun, S., Wang, C., Bi, Y., Li, N., Lü, D., Chen, Q., et al. (2019). An integration design of gas exchange, bubble separation, and flow control in a space cell culture system on board the SJ-10 satellite. Rev. Sci. Instrum. 90, 075114. doi:10.1063/1.5087770
Trumbo, S. K., Becker, T. M., Brown, M. E., Denman, W. T. P., Molyneux, P., Hendrix, A., et al. (2022). A New UV Spectral Feature on Europa: confirmation of nacl in leading-hemisphere chaos terrain. Planet. Sci. J. 3, 27. doi:10.3847/PSJ/ac4580
Volkenburg, T. V., Benzing, J. S., Craft, K. L., Ohiri, K., Kilhefner, A., Irons, K., et al. (2022). Microfluidic chromatography for enhanced amino acid detection at Ocean Worlds. Astrobiology 2021, 0182. doi:10.1089/ast.2021.0182
Waite, J. H., Glein, C. R., Perryman, R. S., Teolis, B. D., Magee, B. A., Miller, G., et al. (2017). Cassini finds molecular hydrogen in the Enceladus plume: evidence for hydrothermal processes. Science 356, 155–159. doi:10.1126/science.aai8703
Keywords: ocean world, amino acids, microfluidics, design of experiments, GC-MS
Citation: Duval KA, Van Volkenburg TB, Craft KL, Person CM, Harshman JS, Fernandes DO, Benzing JS, McDowell EG, Nelson TW, Divakar GS, Pochettino OM, Perry ME and Bradburne CE (2023) Biosignature preparation for ocean worlds (BioPOW) instrument prototype. Front. Astron. Space Sci. 10:1244682. doi: 10.3389/fspas.2023.1244682
Received: 22 June 2023; Accepted: 02 October 2023;
Published: 26 October 2023.
Edited by:
Jennifer Claire Stern, National Aeronautics and Space Administration, United StatesReviewed by:
Antonio J. Ricco, National Aeronautics and Space Administration, United StatesCopyright © 2023 Duval, Van Volkenburg, Craft, Person, Harshman, Fernandes, Benzing, McDowell, Nelson, Divakar, Pochettino, Perry and Bradburne. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kathleen L. Craft, a2F0ZS5jcmFmdEBqaHVhcGwuZWR1; Christopher E. Bradburne, Y2hyaXMuYnJhZGJ1cm5lQGpodWFwbC5lZHU=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.