
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Astron. Space Sci. , 12 August 2022
Sec. Planetary Science
Volume 9 - 2022 | https://doi.org/10.3389/fspas.2022.964657
This article is part of the Research Topic Space Resources and Planetary Sustainability: Challenges and Opportunities View all 5 articles
Researchers have been studying transcriptomic and proteomic responses of plants to ranges of reduced gravitational conditions. These include blue and red light in microgravity, circadian rhythms in microgravity, microgravity in different ecotypes, microgravity on suborbital flights, and they have using a variety of experimental equipment. Recent findings have linked microgravity and transcriptomic changes in genes relating to cell wall synthesis and modification, oxidative stress, abiotic stressors, phytohormones, sugar synthesis and metabolism, ribosomal biogenesis, and plant defense to other organisms. Although we have a better-established profile of the transcriptomic response of plants to reduced gravity, some areas of study have not yet been thoroughly investigated. The initial stages and progression of transcriptional responses to microgravity, the responses of additional plant species, and tissue-specific transcriptional responses to microgravity should all be further investigated in order to better develop our understanding of how plants react to a reduced gravity environment. In the near future, advancing technology, rapidly growing databases, and an increasing number of spaceflight opportunities will allow for more research to be conducted to address these and many other related questions in plant space biology.
The feasibility of long-term space travel is contingent upon humans’ ability to grow plants in space, as they provide oxygen, food, and a psychological benefit for astronauts and future long-term space travelers (Wolverton and Kiss, 2009). In order to make deeper forays into space, we need to ensure that plants can thrive in a reduced gravity environment. We can begin working towards this by studying the ways in which plants respond to microgravity and reduced gravity (e.g., the gravity levels of the Moon and Mars; Kiss, 2014).
Because plants evolved in the conditions on Earth, their growth, development, and responses to stimuli have been refined to be effective in this environment. On Earth, gravity and light are two stimuli which, among other factors, allow plants to determine directionality; sunlight comes from above and gravity pulls downward (Izzo et al., 2022). When plants are grown in the microgravity of space, they do so in the absence of the gravity, which is not only downward directing stimulus but also the force that necessitated the development of plant structural reinforcement.
These deviations from the qualities of their native environment (e.g., altered gravity) are stressors. Plants experience stress from many stimuli: herbivory, wounding and pathogens, drought or flood conditions, deprivation of oxygen or carbon dioxide, excessive or insufficient light, temperature extremes, etc. Plant development and growth are altered when grown in the presence of stressors. Large scale changes in response to stress include reduced growth, biosynthesis of additional pigments, delayed blooming time, thinner leaves, reduced fruit size and number, etc. which are all the result of transcriptomic and proteomic changes (Argueso et al., 2009; Duque et al., 2013; Zhang et al., 2020).
Understanding changes in the transcriptome and proteome that allow plants to react to survive in stressful environments is crucial to our working towards developing crops that will be minimally affected by microgravity. Recently, great strides have been made in pursuit of that goal; the studies in transcriptomic and proteomic response to microgravity have been invaluable steps towards comprehending plant response to microgravity, and many other reviews of plants in space have been published, e.g. Wolverton and Kiss (2009), Ferl et al. (2002), Paul et al. (2013a), and Vandenbrink and Kiss (2016). Here, we reviewed studies published about the -omic response of plants to microgravity, focusing on the last 5 years (but, in some case, looking as far back as 9 years). Studies have been conducted in microgravity, various levels of reduced gravity, different light wavelengths, different lighting cycles, in different life stages, using tissues from different organs and in different species.
To date, there have been numerous studies of the transcriptomic and proteomic effects of microgravity and spaceflight on plants. One such study endeavored to determine the way in which plants respond to altered gravitational conditions by analyzing and comparing transcriptomes of Arabidopsis thaliana after 5 days in microgravity and in Earth gravity (Fengler et al., 2015). By comparing the transcriptomes of spaceflight plants to those of controls, the authors identified differentially expressed genes (DEGs) and then analyzed them using Gene Ontology analysis (GO analysis) to determine any statistically significant patterns in the functions of the DEGs. This study showed a massive impact on the transcriptional frequency of genes involved in translation. DEGs from the chloroplastic genome were found to all be upregulated, including transcripts encoding ribosomal subunits, which is supported by the upregulation of a nuclear coded translational initiation factor. It was also found that many genes involved in the mitochondrial electron transport chain were differentially upregulated in spaceflight, including subunits of enzymes in the respiratory chain (NADH dehydrogenase), and genes relating to ATP biosynthesis. Genes involved in cell wall modification were also found to be differentially upregulated. The only theme identified amongst the downregulated DEGs was a large group of heat shock proteins (HSPs), which are involved in stress responses, and some specifically function as protein folding chaperones. The study notes that other researchers have found HSPs to be upregulated in microgravity, contrary to their observed results. The authors highlighted the need for an onboard centrifuge to serve as a control in spaceflight (as centrifuges can create a gravitational environment through centrifugal force), in addition to a ground control, due to the significant transcriptomic changes that were observed in the onboard 1g control as compared to the 1g ground control (Figure 1). A 1g control run alongside a reduced gravity experimental group in spaceflight has been made possible by several types of hardware currently available to researchers. These spaceflight hardwares contain centrifuges capable of creating 1g gravity to serve as a control in flight (Figure 1). The value of a control group run in parallel with the experimental group is immeasurable, and is vital in future investigations into the -omic response of plants to reduced gravity (Shymanovich and Kiss, 2022).
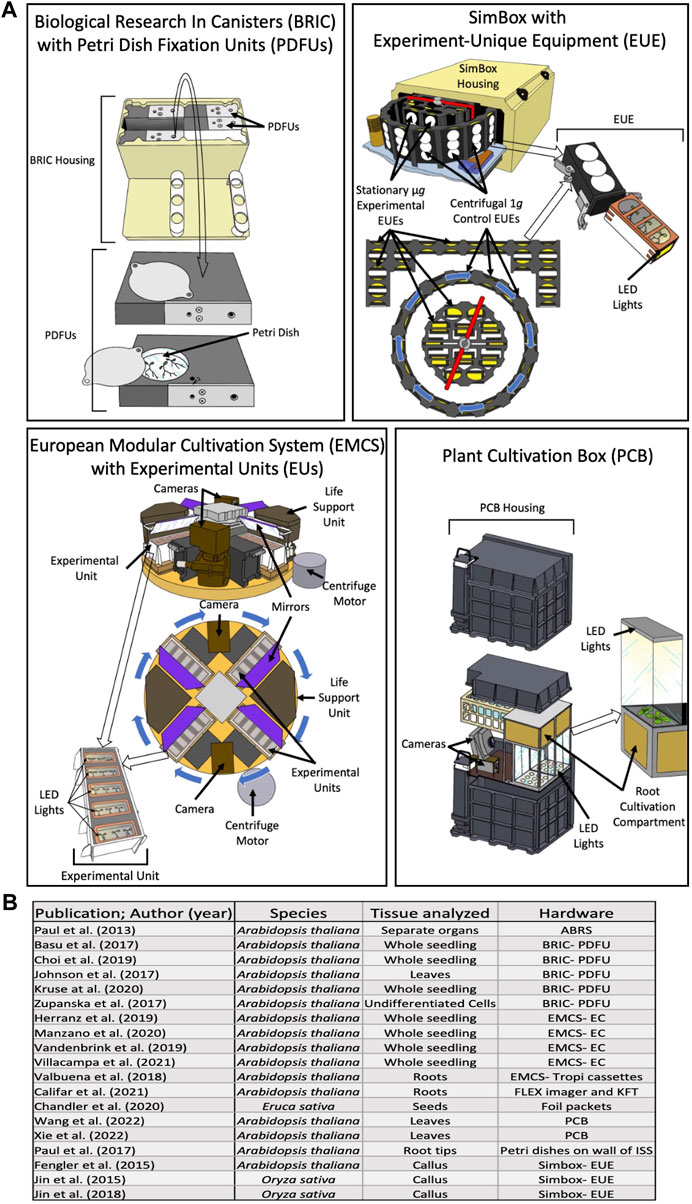
FIGURE 1. (A) Diagrams of the spaceflight hardware used in more than one of the plant biology studies in this review. Circles of arrows indicate centrifuge rotation capability. (B) Summary of the hardware, species and sampled tissues utilized in recent spaceflight experiments. LED light locations are indicated with arrows. EUEs in the center of the SimBox centrifuge and in the back arranged in a bracket shape do not spin, and therefore act as the microgravity experimental group in the same space as the 1g control EUEs loaded into the centrifugal ring. Abbreviations are: Biological Research In Canisters with Petri Dish Fixation Unit (BRIC-PDFU), Kennedy Fixation Tubes (KFT), Experiment-unique equipment (EUE), European Modular Cultivation System with Experimental Containers (EMCS-EC), Advanced Biological Research System (ABRS), and Plant Culture Boxes (PCB).
The transcriptomic response of A. thaliana was further investigated by Johnson et al. (2017). By comparing the transcriptomes from spaceflight and ground control plants, DEGs were identified and analyzed via GO analysis. One subset of these DEGs was involved in cell wall and cytoskeletal structures, including genes that support the cell wall of particularly elongate cells, genes involved in cell wall remodeling, and genes involved in cleaving molecules that reinforce the structure of the cell wall (Johnson et al., 2017). GO analysis further revealed that a number of the DEGs were HSPs (which are involved in general stress responses), genes involved in signal transduction, transporters, and developmental processes. An additional set of DEGs was identified as being related to hypoxia. The researchers suspected that the lack of gaseous convection inside the hardware may contribute to seedlings experiencing oxygen deprivation, however oxygen deprivation appeared to be less of a stressor in this study than it had been in previously conducted experiments. It appears that rather than air, it was the lack of convection of water (caused by the nature of microgravity) that proved to be a strong stressor in this study; many DEGs were identified as being involved in plant stress response to water deprivation. This research found that reduced gravitational stimulus affected the growth and development of plants both directly, as seen in the DEGs relating to cell wall modification, and indirectly through changing the nature of other stimuli, as with the DEGs involved in hypoxia and water deprivation.
The effects of spaceflight on the transcriptome of undifferentiated A. thaliana cells were investigated by Zupanska et al. (2017). These researchers sought to determine the role of Altered Response to Gravity 1 (ARG1), a gene that has been shown to be involved in the process by which plants sense gravity (Zupanska et al., 2017). To determine how ARG1 contributes to the gravitropic response as a whole, they studied undifferentiated cells of Columbia (COL) wildtype and ARG1 knockout plants were grown in darkness on Earth and in space, after which their transcriptomes were compared. By comparing the transcriptomes of the two lines of cells in space and the transcriptomes of each on the ground with those in space, genes that were unusually regulated (regulated in a manner that diverged from what was observed in wildtype) without a functional copy of ARG1 were identified and examined further. Of the 78 genes that were found to be differentially regulated in wildtype cells grown on Earth versus in space, the expression of 75 was different in the ARG1 knockout cells, and were therefore identified as genes dependent upon ARG1 for a normal transcriptional response to spaceflight. The most strongly induced of these genes were transcription factors; one involved in responding to light, gibberellic acid, and abscisic acid, the other in pollen development, pathogen defense, and root development. Within the 75 ARG1 dependent genes were 12 genes that were improperly regulated in ARG1 knockout cells both on Earth and in space. These genes were deemed ARG1 required, and many had roles in pathogen defense, with some linked to cell wall processing and auxin signaling. Two further categories of genes were identified: 1) corrected genes, which were dysregulated in the mutant background on Earth but normalized in space, and 2) compensated genes, which may compensate for the lack of ARG1 in the mutant cells. Many of the corrected genes were found to be linked to transport and vesicle transit, and many of the compensating genes were linked to cell wall metabolism, the endomembrane system, and auxin signaling and regulation of cell polarity. The results of this study indicate that the gene ARG1 is necessary for the adaptation of undifferentiated cells to spaceflight, and reinforces the prevalence of the regulation of cell wall metabolic processes which is a function that has been noted in many transcriptomic analyses of spaceflight plants (e.g., Johnson et al., 2017).
The transcriptomes of Oryza sativa, rice, seedlings were also analyzed after spaceflight (Jin et al., 2015). The transcriptome of plants grown in microgravity revealed a number of roles common to groups DEGs, including a set of transcription factors. Over 70% of the differentially expressed transcription factors were downregulated in response to spaceflight. An additional group of identified DEGs were found to be related to hormone metabolism; four DEGs related to auxin metabolism, one DEG involved in gibberellic acid synthesis was downregulated 4-fold, a negative regulator of cytokinin signaling was upregulated 4-fold, and one DEG was related to ethylene metabolism. A significant number of auxin transporters were identified as being differentially regulated, including ABCs (an auxin transporter-like protein), and potassium transporters which have been linked to auxin transport in A. thaliana. An additional group of microgravity-induced DEGs were involved in cell signaling; eight DEGs related to calcium signaling (four up and four downregulated), receptor kinases (all but four upregulated DEGs were downregulated), and G-proteins which are involved in signal transduction (three downregulated and two upregulated). Further investigation revealed DEGs related to the cell wall; two downregulated secondary cell wall DEGs, four upregulated primary cell-wall-modifying DEGs, two cell wall reorganization DEGs, and four upregulated genes involved in cell wall structure. DEGs related to sugars were identified; two upregulated DEGs involved in starch synthesis, and six involved in the TCA cycle (three up and three downregulated). A group of DEGs was shown to be involved in secondary metabolism, which influences auxin transport and, thereby, alters gravitropism. The final investigated set of DEGs were involved in protein degradation (all but three were downregulated). The results of this study indicated that transcriptional responses to microgravity included modified transcription of genes involved in regulating the structure of the cell wall, the TCA cycle, metabolism, signal transduction, protein modification and degradation, metabolism of phytohormones, and calcium regulation and signaling. Several of these DEG functions have been observed in the response of A. thalina to microgravity.
The previous study investigating the transcriptomic effects of microgravity on rice seedlings was expanded upon by Jin et al. (2018). Transcriptomic analysis of seedlings in microgravity revealed DEGs involved in several different processes and roles, including those in cell wall, stress and water related functions (Jin et al., 2018). Within the DEGs related to the cell wall, three downregulated genes were involved in cell wall degradation, and five upregulated genes were involved in cell wall modification (through loosening the bonds in the cell wall and allowing for expansion and other modifications). The water related DEGs were those of the aquaporin family, which create channels in the cell membrane through which water can flow in and out of cells, facilitating water transport. Of these 17 DEGs, all but two were upregulated when comparing the spaceflight transcriptome with that of the spaceflight 1g control. This study identified water transport and cell wall modifications as the primary ways in which plants respond to the stress of microgravity. They propose that changes in the cell wall and water transport are working together to maintain the structure of an entire plant in microgravity through the changed cell wall configuration and turgor pressure within the cells.
Most plant research is conducted using the model organism Arabidopsis thaliana, however, the wildtype line used in research varies between labs. The most commonly used wildtype lines include Columbia (COL), Wassilewskija (WS), Landsberg (LER), and Cape Verde Islands (CVI). Though these lines are all wildtypes, they differ genetically, as well as in their phenotypes and responses to stimuli (Kiss et al., 2000). To determine which responses to altered gravitational stimulus were unique and which were common to these ecotypes, Choi et al. (2019) analyzed and compared the transcriptomic responses of COL, WS, LER and CVI grown in microgravity. Analysis of the transcriptomes of the four wildtype lines showed that several genes encoding Heat Shock Proteins (HSPs) involved in oxidative stress response and temperature stress response were upregulated in microgravity conditions (Choi et al., 2019). Differentially expressed peroxidase genes were found to be downregulated across the different ecotypes in microgravity, though the specific genes and number of genes affected differed between the lines. Using GO analysis, the researchers learned that genes involved in the response to high light, to temperature and high heat, and to ROS and hydrogen peroxide were significantly downregulated across all wildtype ecotypes. They determined that all four lines exhibited unique transcriptional responses to exposure to microgravity, including responses to drought and flood conditions, to cell wall remodeling, to high light and plastid-related responses.
The difference between the wildtype lines Columbia (COL) and Wassilewskija (WS) wildtypes has been attributed to the gene phyD, of which, unlike COL, WS does not have a functional copy (Paul et al., 2017). In order to determine if the photosensor PhyD is the cause of differing responses to microgravity observed in COL and WS, a line with the gene phyD mutated in the COL background was grown alongside WS and COL in microgravity and their transcriptomes were compared. The genes that were differentially expressed consistently across the ecotypes were involved in light signaling, defense and in cell wall modification, but most genes were not commonly differentially expressed. DEGs that were expressed uniquely among the ecotypes included defense, response to ROS, cell wall metabolism, hormone metabolism and signaling (auxin, gibberellic acid, and abscisic acid), responses to biotic and abiotic stresses and the pathogen defense response. These researchers determined that the COL transcriptome was altered to a greater degree in microgravity than were those of phyD and WS, but that the latter two did still differ in their transcriptomic responses. The results of this study show that the differences between COL and WS are not solely due to the mutation of the PhyD gene, and that the COL transcriptome is more reactive to altered gravitational conditions than was that of WS.
Most research in microgravity is conducted on spacecrafts and the International Space Station (ISS), however, there are alternative means of attaining microgravity environments. Suborbital flights (those which do not reach escape velocity or become artificial satellites around Earth) can provide a reduced gravitational environment for a short time (i.e., a few minutes). Plants were flown on several different types of suborbital flights to determine how their transcriptomes were affected in each environment as compared to those on Earth and in microgravity (Califar et al., 2021). The flights tested included parabolic flights, the Virgin Galactic and Blue Origin suborbital flights, and the F-104 Starfighter flight, all of which create different cycles of hypergravity and microgravity along their flightpath. Transcriptomic analyses showed that there were unique links between specific types of suborbital flight and DEGs with roles in: response to stimuli, light reactions and photosynthesis, reactions to toxins, reactions to dugs/antibiotics, response to iron ions, metal sequestering and homeostasis, regulation of ubiquitination, oligomerization, cell wall organization or biogenesis, callose deposition, sugar metabolism, response to sulfur, and response to nitrogen containing compound. Because these roles were only associated with one type of suborbital flight, they are considered responses to unique characteristics of those flights. Analysis showed that, although specific genes were rarely similarly differentially regulated across them, GO analysis showed that there were DEGs with roles common to all flights, such as hypoxia, oxidative stress, regulation of central carbon and nitrogen metabolism, and regulation of amino acid degradation. These responses were proposed to be the result of suborbital flight and as the authors noted, some do overlap (oxidative stress and hypoxia) with those transcriptomic effects found in microgravity.
Light and gravity are closely linked, as on Earth, they serve as orienting stimuli, indicating directionality for plants. Because reactions to these stimuli are intertwined in plants, the connections between their signals and responses have been investigated. Vandenbrink et al. (2019) investigated how light and gravity stimuli interact by exposing A. thaliana to either 1g or microgravity in blue light. By comparing the transcriptomes of plants grown in microgravity and those grown at 1g, they found that DEGs that were upregulated in microgravity were related to ribosomes, oxidative phosphorylation, and to pathways previously associated with responses to light (light perception and response, starch metabolism, and carotenoid synthesis; (Vandenbrink et al., 2019). Other biological processes in which DEGs are involved include response to an abiotic stimulus, the respiratory electron chain, and generation of energy and metabolites. Further investigation was conducted using pathway analysis, by which the researchers discovered that many downregulated DEGs were related to photosynthesis and photosynthetic machinery. These include DEGs encoding subunits of both PSI and PSII, photosynthetic electron transport, a subunit of APT-ase, and all but one of the subunits of the light-harvesting chlorophyll protein complex (Figure 2). These downregulated DEGs suggest that plant responses to microgravity influence the plant response to blue light. Using pathway analysis provided great insight into the ways plants respond to microgravity and blue light.
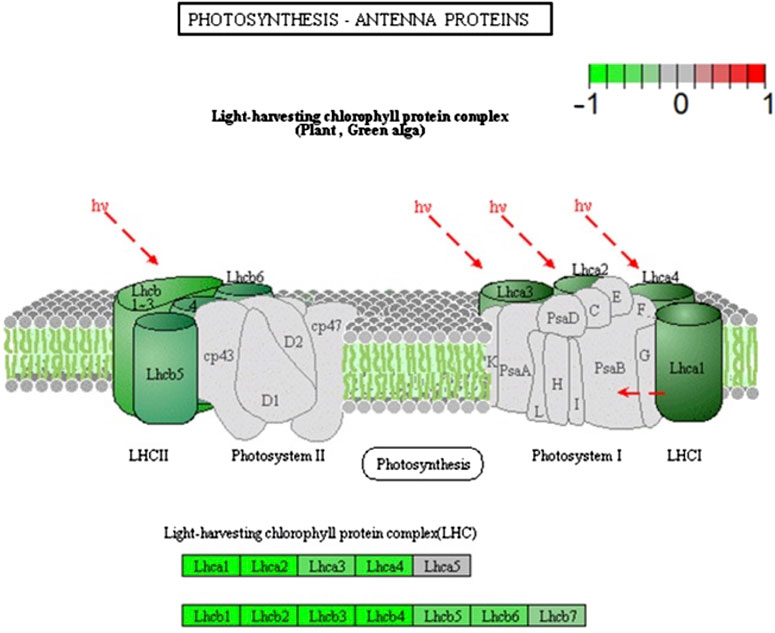
FIGURE 2. A diagram of PSI and PSII indicating the differential repression observed in microgravity and blue light. The brighter the green coloration, the more strongly the gene encoding the subunit was repressed. Modified from Vandenbrink et al. (2019).
Microgravity has been shown to affect the cell cycle, altering cellular growth and proliferation as well as ribosomal biosynthesis. In order to investigate the effects of microgravity on the cell cycle without the confounding ribosomal effect, Manzano et al. (2020) studied the transcriptomes of nucleolin mutants nuc1 and nuc2 grown in microgravity. Nucleolin is crucial to ribosomal biogenesis and is the most common type of protein in the nucleolus, which allowed for the individual study of effect of microgravity on the cellular cycle in red light (Manzano et al., 2020). Nucleolin mutants experience delayed development and reduced growth as well as disorganization of the nucleolus. Transcriptomic analysis showed that in red light and microgravity, the upregulation of DEGs involved in photosynthesis, drug catabolism, pathogenesis, cell wall modification and the biotic stress response as well as the downregulation of genes involved in the response to temperature, hypoxia, oxidative stress, and abscisic acid were similar between the nucleolin mutants and wildtype. These observations suggest that these nucleolins are not necessary for the proper regulation of these responses to red light and microgravity. These researchers showed, however, that in microgravity and red light, wildtype and nuc1-2, but not nuc2-2, genes involved in cold acclimation, hormone metabolism and karrikin, were downregulated. These observations indicate that NUC2 is necessary for the normal regulation of these processes in red light and microgravity. Conversely, nuc2-2 and wildtype similarly regulated genes involved in response to drugs, antibiotics, salicylic acid, and wounding, however, nuc1-1 mutant did not regulate these genes comparably, indicating that NUC1 is necessary for the proper regulation of these responses in these conditions. In dark conditions, it was found that NUC2 could compensate for the loss of NUC1 in regard to regulation of responses to red light, UV, ionizing radiation, gravity, and auxins, but that NUC2 could not do so in regard to developmental processes, mRNA quality regulation, response to temperature and the immune system. In red light conditions, it was concluded that NUC2 has a larger role to play in response to lighting and gravitational conditions. The results of this study illustrate the dynamic interaction of plant responses to light, light quality, and microgravity.
Understanding the -omic changes that plants experience when grown in microgravity could be elucidated by examining their responses to gravity conditions between microgravity (µg) and 1g. By studying the various levels of gravity, the responses of transcriptomes and proteomes could expose how the response to microgravity progresses in a plant. While we know a great deal about how plants function in microgravity, we have little knowledge of how plants function in reduced gravity conditions (those between µg and 1g), such as those found on the surface of the Moon and Mars (Kiss, 2014).
As gravity and light are closely linked stimuli, several studies investigating a range of gravitational conditions (between µg and 1g) have paired gravitational levels and light wavelengths as experimental variables (Valbuena et al., 2018; Herranz et al., 2019; Villacampa et al., 2021). The interaction of blue light and gravity was further explored by analyzing the transcriptomes of blue light-grown plants which were grown in 1 g (Earth), reduced Earth gravity (0.57 g), Mars gravity (0.36 g), gravity of the Moon (0.18 g), low gravity (<0.1 g) and microgravity (µg) (Herranz et al., 2019). These researchers determined that in microgravity, low gravity, and lunar gravity, the alterations to transcriptomes of A. thaliana were similar, which they hypothesized was due to the strength of the photostimulus overwhelming the gravitational stimulus. A stronger gravitational stimulus, that of Mars and reduced Earth gravity, resulted transcriptomes very similar to that of the plants grown at Earth gravity, which suggests that the gravitational stimulus was great enough to overwhelm the effects of the photostimulus. GO analysis showed that DEGs of plants grown in the gravitational conditions of the microgravity, low gravity, and the gravity of the Moon, were related to responses to chitin, to organonitrogen compounds, to a nitrogen compound, to organic and substance, to drugs and to stress. The analysis showed that microgravity and low gravity also shared downregulated DEGs related to response to oxygen containing compounds, to stimuli, and to chemicals. However, these were not present in the transcriptome of plants grown in lunar gravity. GO analysis of the upregulated DEGs in microgravity, low gravity and lunar gravity showed no overlapping DEG roles, but that in microgravity, DEGs were related to photosynthesis and biosynthetic processes, in low gravity, DEGs were related to defense responses to other organisms, and in lunar gravity DEGs were related to responses to chemical substances. Results of this study indicate that there is a complex and dynamic relationship between plant responses to light and gravity.
The Herranz et al. (2019) study complements that conducted by Valbuena et al. (2018), which compared the transcriptomic responses of plants grown in gravity measuring 1, 0.5, 0.3, 0.1 g, µg and red light (Valbuena et al., 2018). In this study, six genes were selected and tracked as indicators of three traits: polar auxin transport and perception, ribosomal biogenesis, and cell cycle regulation. These processes are significantly altered in plants when grown in reduced gravity, but this study showed that red light diminished the effect reduced gravity exerted on them. The return of cellular proliferation and growth to the patterns and levels observed at 1 g was proposed to be the result of auxin transport and perception returning to the patterns and levels observed at 1 g, as auxin regulates the cellular cycle. This hypothesis is supported in the data; gravitational conditions that caused larger degrees of differential expression in one of the selected 6 genes also showed a larger degree of differential expression in the other five. The transcriptome of wildtype grown in reduced gravity showed minor deviation from that grown in 1 g, except in microgravity, where the cellular cycle, ribosome biogenesis and auxin transport and perception are all strongly upregulated. In phyA and phyB mutants, the reduced gravity conditions resulted in similar regulation or downregulation of the six genes in all reduced gravity conditions, with the exception of phyB in microgravity. In microgravity, phyB followed the pattern observed in wildtype; an upregulation of the six indicator genes. Though wildtype and phyB differed in their responses, phyA was the least like the other two lines, which suggests that phytochrome A is necessary to the responses to reduced gravity observed in wildtype and phyB.
The influence of red light in microgravity was further investigated by Villacampa et al. (2021). Transcriptomes of plants grown in red light in the following gravity conditions were analyzed and compared: Earth gravity (1 g), Mars gravity (0.36 g), lunar gravity (0.18 g), low gravity (<0.1 g) and microgravity (µg). Transcriptomes of plants grown in all gravity conditions showed an upregulation in transcription of genes involved in photosynthesis, light responses, pigment biosynthesis, and light harvesting, which is in accordance with previous studies. Additional other categories include genes involved in the response to karrikin, secondary metabolic processes, phenylpropanoid metabolism and the Calvin Cycle (Villacampa et al., 2021). The overlap between results of this study and the established response to microgravity extends to the genes that were upregulated in microgravity alone, which are involved in photosynthesis, cell wall reorganization and biogenesis and the cell cycle. Genes that were downregulated in all gravity conditions were involved in the response to far red light signaling and response, and in proteolysis. In Mars gravity conditions, 13 members of a group of transcription factors, WRKY transcription factors (named after a segment of their highly conserved amino acid sequence), were upregulated in both red light and darkness as compared to transcription levels at 1 g. These transcription factors are involved in the response to hypoxia, plant defense, salt stress, immune processes, osmotic stress, senescence, response to touch, phosphate transport and the response to and homeostasis of hormones, salicylic acid, jasmonic acid, auxin, abscisic acid, and gibberellic acid. The transcriptomic data also showed upregulation of DEGs from the mitochondrial and chloroplastic genomes. Upregulated DEGs from the mitochondrial genome were involved in biosynthesis of ribosomal subunits, of cytochrome oxidase, of NADH dehydrogenase subunits, of ATP binding cassette and of cytochrome C. In the chloroplastic genome, upregulated DEGs were involved in biosynthesis of PSI and PSII subunits, NAD (P) H dehydrogenase, and parts of the photosynthetic electron transport chain. These findings aligns with the established transcriptomic profile of plants in microgravity established in previous studies.
The circadian rhythm of A. thaliana regulates the daily oscillation of about one third of the genome (Covington et al., 2008). One of the many processes regulated by the circadian clock is gravitropism (Tolsma et al., 2021). Many experiments conducted in reduced gravity are done so in a constant light and/or darkness, which disrupts the circadian rhythm (Figure 3). Without one of the most important regulating elements of growth and development, the entirety of a plant response to a stimulus is not discernable. Because of this, several studies have been conducted to determine how A. thaliana responds to microgravity in circadian lighting conditions.
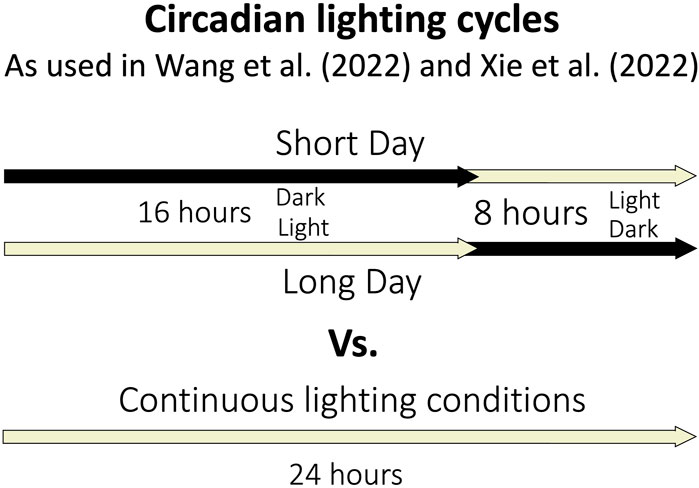
FIGURE 3. Depiction of the lighting timing for circadian and continuous lighting conditions in selected spaceflight experiments.
To determine how microgravity affects blooming time in long day (16 h light, 8 h dark) and short day (8 h light, 16 h dark) conditions Wang et al. (2022) recently conducted transcriptomic analyses were conducted in microgravity (Figure 3). The genes involved regulating the flowing time in long day and short day conditions in Earth gravity have been explored and include Flowering locus T (FT) which is crucial for the day-length related regulation of flowering time (Wang et al., 2022). When plants in microgravity were forced to express FT (in a line called FG), analysis revealed DEGs in both short and long days had roles in cellular component biogenesis, rRNA and ncRNA processing and metabolism, and ribosome biogenesis and assembly, where as wildtype DEGs were involved in amino acid biosynthesis, cytoplasmic translation, maturation, processing and metabolism of various types of RNA, ribosomal biosynthesis and assembly, protein folding and peptide biosynthesis and metabolism. The difference indicates that FT regulates the wildtype regulation of the identified DEGs. Comparing plants grown in long and short days in microgravity revealed DEGs involved in the response to chemical stress, to oxygen containing compound, to ROS and the oxidative stress response, and in the regulation of transcription of abscisic acid, of ribosomal biogenesis, of translation, and in the defense response and organelle organization.
Further comparisons revealed that plants grown in long and short days in microgravity differed in their transcriptomic responses. Analysis revealed that plants grown in microgravity in long day conditions were more reactive on the transcriptomic level (they experienced a greater degree of transcriptomic change with a much larger number of DEGs) than were their short day counterparts. The daylength variable affected circadian clock-controlled regulation of flowering timing in microgravity, as the circadian genes responsible for regulation in both morning and evening were shown to be differentially regulated in microgravity as compared to 1 g, and were differentially regulated between the long and short day conditions.
These findings from Wang et al. (2022) were further expanded upon by Xie et al. (2022). Flowering locus T (FT) is further investigated along with Suppressor of Overexpressor of CO1 (SOC1) in the context of spaceflight. FT expression is influenced by circadian rhythm, day length, temperature, and vernalization, and peak expression occurs just before flowering (Xie et al., 2022). The authors found that FT peaked earlier on Earth than it did in microgravity and that SOC1 may function in concert with FT as hubs for transcriptional changes that occur before flowering. This study showed that, contrary to findings from continuously illuminated experiments conducted in microgravity, the long and short day plants flowered later than their Earth counterparts, though researchers hypothesized that the dense growing conditions in those experiments conducted under continuous illumination may have contributed to the earlier blooming time. Genes found differentially regulated between long and short days were involved in sugar metabolism, responding to chemical stimuli, light, the hormones gibberellic acid (which is crucial to the flowing process), abscisic acid, brassinosteroids, and jasmonic acid. Xie et al. (2022) also found that SOC1 downregulation in leaves was accompanied by the downregulation of genes involved in response to photoperiod, the circadian clock, vernalization, ambient temperature, gibberellic acid, and aging, which indicates that both SOC1 and FT are important for the integration of spaceflight conditions into the flowering regulation pathways. The complex system of transcriptional circadian regulation should be investigated.
Nearly all of the experiments conducted in reduced gravity are done so using seedlings, but plants of different developmental stages utilize different regulation processes and pathways. To fully understand the responses of plants to reduced gravity, the effect of the stimulus must be studied at different life stages. The previously addressed Xie et al. (2022) and Wang et al. (2022) studies were both conducted using mature blooming plants, but it is important to study the response to microgravity at various life stages.
Chandler et al. (2020) analyzed how Eruca sativa (salad rocket) seeds were affected by microgravity and red light. This study aimed to determine the molecular and physiological effects of spaceflight on dry seeds. Previously it was found that seeds that had experienced long-term spaceflight suffered accelerated aging, manifesting as reduced seedling germination and reduced seedling growth, possibly due to microgravity, the different types of radiation in space, short wave UV, mechanical vibrations of the flight, and/or pressure and atmospheric conditions and fluctuations (Musgrave, 2002; Visscher et al., 2016; Chandler et al., 2020). After storage aboard the ISS for 6 months, the authors found that the seeds in this study did not experience any developmental abnormalities. The authors attributed this to the dried state of the seeds, in which condition the seeds are not subject to water-related processes, hormone signaling or gravitational stimulus. Despite the seeds growing into phenotypically normal plants, the seeds that had been stored in space were delayed in germination as compared to Earth control seeds. Using the Earth-based controls to eliminate other variables, it was hypothesized that the vibrations from extreme acceleration and sound levels to which the seeds were exposed in spaceflight may be a contributing cause behind the accelerated aging of seeds in spaceflight. Researchers concluded that the vibrations worked in concert with various types of space radiation causing the accelerated aging observed in the seeds of this study. Transcriptomic analysis of the spaceflight seeds showed that the upregulated DEGs were related to DNA repair, molecular chaperones, response to radiation, and response to light and downregulated DEGs were related to translation and ribosomal assembly.
One study analyzed and compared the transcriptome of different organs from spaceflight plants (Paul et al., 2013b). Using a 60:40 ratio of WS:COL wildtype plants, the authors analyzed the transcriptomic effects of microgravity in distinct tissues. The transcriptomes of the leaves, roots and hypocotyls were independently analyzed, and it was found that different genes are differentially expressed in the three different tissues (Figure 4). Analysis revealed that roles of DEGs found exclusively in the leaves are involved in hormone signaling, particularly auxin signaling, and cell wall-related proteins are downregulated, and those involved in defense to herbivory and pathogens, calcium, and phosphate metabolism genes (which may be associated with wounding and gravity sensing) are upregulated. In the hypocotyl, DEGs associated with cell wall expansion were repressed, and with auxin signaling were upregulated. In the roots, DEGs related to cell wall remodeling were highly upregulated, as were genes associated with light and tactile stimuli. Interestingly, it was found that in the leaf tissue, a gene (PGP18), associated with light perception and cell wall remodeling, was highly upregulated, but the same gene was strongly downregulated in the roots. Ultimately this study indicated that there are significant transcriptomic differences among different organs and that responses are lost in analyzing one tissue alone or the seedling as a whole.
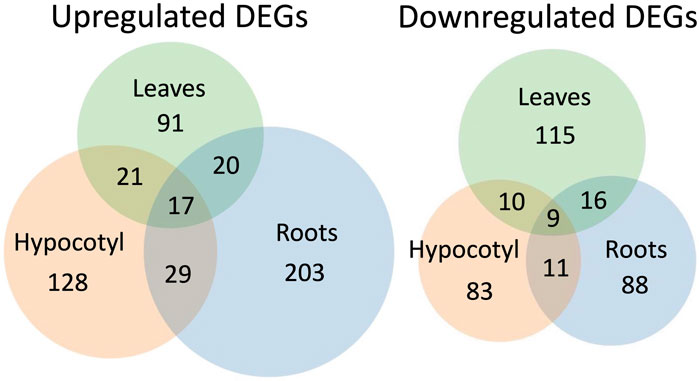
FIGURE 4. Venn diagrams proportionally depicting the DEGs identified in different tissues. Modified from Paul et al. (2013). These data were generated from samples of different wildtype lines. 40% of the tissue analyzed was collected from WS plants, and the remaining 60% was taken from COL plants.
The equipment and spaceflight hardware that allows for microgravity studies are environments in and of themselves, and as such, affect the growth and development of the plants that grow within them.
Currently, there as several types of hardware available to plant space researchers, several of which are depicted in Figure 1. The Biological Research in Canisters with Petri Dish Fixation units (BRIC- PDFU), which are designed by the National Aeronautics and Space Administration (NASA), are each capable of containing 5 PDFUs, with several environmental monitoring devices available to exchange for one of the PDFUs. The BRIC-PDFU is limited in two ways: firstly, until recently (and in all of the studies addressed in this review), the experiments using BRIC had to be conducted in darkness, and secondly, the centrifuges aboard the ISS are not compatible with the BRIC unit, and, therefore, researchers using the BRIC-PDFUs cannot have an inflight 1 g control. The Plant Cultivation Box (PCB), designed by the China National Space Administration, is similar in the regard, as it is also without a self-contained centrifuge, and so, does not have an inflight 1 g control. However, it contains programable LED lights, as well as two cameras which can monitor plant growth and development as the experiment progresses without increasing the necessary crew time. Another highlight of the PCB is its size; larger plants can be grown inside the PCB, which allows for longer experiments and a wider array of plant species which can be grown within it. Both the European Modular Cultivation System with Experimental Units or TROPI cassettes (EMCS-EU or EMCS-TROPI), designed by the European Space Agency, and the SimBox with Experiment-Unique Equipment (SimBox-EUE) have a self-contained centrifuge, and each of the EUs, TROPI cassettes, and EUEs have their own programable LED lighting. Additionally (Shymanovich et al., 2022), the EMCS contains two cameras, like the PCB, which allows researchers to monitor plant growth and development. Of the experimental equipment used by studies in this review, only the BRIC-PDFU, and NASA’s Kennedy Fixation Tube (A hallow cylinder less than 3 cm in diameter, with a growth space around 6 cm long and room enough to hold 25 ml of a fixative agent to preserve the samples at the conclusion of the experiment) are still in use aboard the ISS, and the SimBox aboard the Shenzhou-8 spacecraft; the rest have been retired.
The effects of the hardware BRIC-PDFU (Biological Research In Canisters with Petri Dish Fixation Unit) (Figure 1) on the plants grown inside were studied by Basu et al. (2017). Using the hardware and a controlled environment, seeds were germinated and grown, then their proteomes and transcriptomes were analyzed from seedlings in both conditions (Basu et al., 2017). The study showed that even in Earth gravity, the BRIC hardware caused wide ranging differential regulation of genes, including those related to aerobic respiration, responses to chitin, high light, wounding, drought, toxins, karrikin (a regulator of plant development), heat, cold, salt stress, and hydrogen peroxide, as well as the responses to and signaling pathways of the plant hormones jasmonic acid, abscisic acid, and ethylene. The proteomic analysis showed differing levels of cytoplasmic proteins related to salt stress, oxidative stress and hydrogen peroxide catabolism, temperature stimuli, defense response to a bacterium, translation and ribosome biosynthesis, and the response to the hormone cytokinin. Analysis of the proteome revealed divergent concentrations of membrane-bound proteins in the BRIC plants relating to photosystem repair, actin filament depolymerization, chlorophyll biosynthesis, response to cadmium ions, defense response, cell growth, oxidation-reduction, lipid biosynthesis, camalexin biosynthesis, defense response to a bacterium and wounding, response to cold, salt stress, morphogenesis development, metabolism of hexose, of ADP and ATP, of pyruvate, and of coenzymes and response to the hormone cytokinin. The authors determined that these transcriptional and proteomic changes were determined to be stress-induced responses to the BRIC hardware environment. The dense plating of the seeds (hundreds of seeds on a relatively small petri dish) typically employed in this hardware may have also contributed to the observed influence of the BRIC environment on the transcriptome and proteome of plants. Results of this study indicate that the effects of experimental hardware itself and the ways in which researchers utilize it, should be carefully considered when researchers analyze spaceflight data.
Transcriptomic data from spaceflights are collected primarily using RNAlater, a solution that is meant to prevent further transcription or any degradation of RNA thereby preserving the transcriptome. Because there is a limit to the speed at which the solution can permeate plant cells, the transcriptome and proteome may be altered by using RNAlater. Researchers studied the efficacy of RNAlater through comparison of the transcriptomes and proteomes of plants fixed with RNAlater and liquid nitrogen (Kruse et al., 2017). By comparing samples fixed with RNAlater with those immediately flash frozen in liquid nitrogen, this group found that genes involved in responding to water, water deprivation, desiccation, cuticle development, DNA transcription (elongation), salt stress metabolism of acetyl-CoA, phytosteroid, and brassinosteroids, and biosynthesis of sterols, phytosteroids, and brassinosteroids were differentially upregulated in the transcriptomes of samples fixed with RNAlater. They also determined that RNAlater also affected the levels of post-translational modifications. The RNAlater solution itself was proposed to be the cause of the DEGs related to salt stress, water deprivation and desiccation, as the influx of the solution would elicit these responses to combat the osmotic stress during the time it took to achieve full permeation of all tissue. The protein content of the analyzed samples was mostly unchanged, which the authors attributed to the additional time it takes to alter the proteome via translation as compared to the time to change the transcriptome.
As found by Kruse et al. (2017) in their investigation of RNAlater and Basu et al. (2017) in their investigation of hardware, researchers use significant differences between the proteomes and transcriptomes to determine how plants respond to stimuli, by examining both types of-omic analysis in parallel, researchers are able to look beyond transcriptional regulation to examine post-transcriptional and post-translational regulatory processes. The regulation of protein levels is not determined solely by transcription of genes; between transcription and translation and translation and a functioning protein, a great many regulating processes affect the expression of genes. Investigating how transcription alone is affected does not provide a full assessment of a response to microgravity, or other stimuli.
The regulation of the response to microgravity that occurred between translation and transcription as well as post-translational modifications was investigated by Kruse et al. (2020). The transcriptomic analysis showed that DEGs upregulated in microgravity are plastid localized and plastid produced (Kruse et al., 2020). Some of the plastid related DEGs are photosynthesis-related, despite the unlit growing environment, however the differentially transcribed genes did not ultimately result in differentially abundant proteins, indicating the occurrence of post-transcriptional regulation. Other DEGs were found to be involved in the oxidative stress response, including Reactive Oxygen Species (ROS) quenchers, which, in combination with the activation of ROS responding protein and oxidation of the membrane, the results indicate an oxidative extracellular environment with resistance to the oxidative stress in the cytosol. The oxidative stress response has potentially been amplified by the generalized response to the stress of microgravity. This stress also manifested in the changes to microtubule activity (synthesis, assembly and use within the cell), including those related to responses to ROS. Increased levels of proteins involved in auxin transport were observed, contributing to the changes in plant growth, as did several genes involved in cellulose synthase. Transcriptomic and proteomic results also revealed a connection between microgravity and auxin transport and signaling. Several types of auxin transporters, Pin-formed (PIN), Like Auxin resistant (LAX), and ATP-Binding Cassette (ABC), were upregulated in microgravity, and a downstream target of auxin was post-translationally modified to a lesser degree in space than on Earth, which indicates that the protein is less active. This target, H+ ATP-ase2 (AHA2) is involved in acidification of the cell wall allowing for expansion and is related to the increase in observed increased levels of proteins that negatively regulate cellulose synthesis. These transcriptional and proteomic regulation of cell wall, auxin, oxidative stress, and plastid responses all contribute to the overall reaction of plants to reduced gravity.
Many different studies have been conducted, using many different lighting conditions, hardware, analytical methods, and experimental time periods. The volume of the data accumulated by these studies is immense and diverse but assembling and synthesizing the data into compatible groups for study is invaluable. Some studies conducted in space are compared to the results from one experiment for confirmation as well as to further isolate the contribution of a single variable to the transcriptomic and proteomic responses (e.g., Johnson et al., 2017 and Vandenbrink et al., 2019), but larger scale analyses are more difficult and less common.
In a recent study, Manian et al. (2021) were able to analyze two distinct data sets together and assembled a foundation for the network of genes involved in the plant response to gravity. GLDS-7 and GLDS-120 data sets, made available by NASA’s open source database, GeneLab (https://genelab.nasa.gov), were analyzed. Data included transcriptomic data from light and dark-grown plants in spaceflight and ground control groups, and from two wildtype lines, COL and WS, and one phytochromeD (phyD) mutant (a light-sensing mutant). From these data sets, researchers identified gene networks that are involved in regulating root growth, catabolic processes, and cell wall biosynthesis in response to microgravity (Manian et al., 2021). The genes that serve as hubs were identified and the connections between genes were determined. The researchers then compared the hubs of associated genes identified in the wildtypes and the phyD mutant, in microgravity, and on the ground, and the in the light and dark conditions. Their analysis showed that PhyD plays a role in red-light sensing and stress response signaling, and that hubs involved in RNA-dependent DNA biosynthesis, DNA metabolism, DNA interaction, cell wall biosynthesis, protein transport/regulation, auxin signaling, catabolic processes and stress responses were affected in microgravity. This study provided insight into the means by which genes interact and create the various established responses to microgravity.
The accumulation and conversion of previously collected data sets to one comparable format was a critical step taken to allow for meta-analysis. This step was taken by Barker et al. (2020) to provide researchers with a database that can be used to further elucidate the transcriptomic and proteomic responses to spaceflight (Barker et al., 2020). The database established is named “TOAST”, Test Of Arabidopsis Space Transcriptome, and has organized a huge number of datasets collected in reduced gravity. In TOAST, datasets have been converted (names of genes/profiles translated into the same naming system, data organized in the same manner, etc.) and made compatible with one another for analysis, regardless of the type of assay originally performed. Proteomic, transcriptomic and other studies are available in the database. TOAST is a researcher-friendly database, searchable by a huge number of factors; type of data (e.g., RNAseq, proteome, etc.), species, flight type (e.g., the ISS, the Space Shuttle, the Shenzhou Spaceship, etc.), experimental environment (the hardware used, lighting conditions etc.), tissues sampled and ecotypes used. Through its conversion of datasets, organization and user interface, this database has made large-scale meta-analysis an achievable objective, and will allow researchers to answer any number of questions that were not asked in the original research.
Results from the multitude of recent investigations into the -omic responses of plants to microgravity sometimes conflict, but where the results overlap can certainly advance our understanding of how plants function in microgravity. As findings of any one study are consistently and repeatedly supported by subsequent investigations, these results become an accepted piece of the profile of plants’ transcriptional response to reduced gravity. As far as research has heretofore determined, the profile of transcriptional changes in response to microgravity includes several themes, the first of which is cell wall synthesis and/or modification. Of the studies examined in this review, Califar et al. (2021), Choi et al. (2019), Jin et al. (2018), Jin et al. (2015), Johnson et al. (2017), Kruse et al. (2020), Manian at al. (2021), Manzano et al. (2020), Paul et al. (2017), Paul et al. (2013b), and Villacampa et al. (2021) all found a significant number of DEGs associated with cell wall synthesis and/or modification (Supplementary Table S1). This may be the result of the unexpected gravity stimulus creating unusual structural demands for the plant. Other trends observed trends in the functions of DEGs found in many of these examined experiments include: metabolism/catabolism of sugars (in eight studies), oxidative stress (13 studies), ribosome biogenesis (seven studies), and photosynthesis (12 studies). Many studies also found DEGs with links to phytohormones (17 studies), though there was not an overrepresentation of DEGs related to a particular hormone in most of those studies, genes relating to cytokinin (four studies), abscisic acid (ten studies), gibberellin (five studies), auxin (ten studies), ethylene (seven studies), and salicylic acid (seven studies) were each found repeatedly in these publications. The association of microgravity and the transcriptional responses to abiotic stressors (17 studies) and defense against wounding and infection (13 studies) may similarly be manifestations of a stressed condition. It was proposed that the inclusion of sugar metabolism/catabolism in the themes of DEGs associated with microgravity may be caused by the dark conditions in which many plants were grown (Supplementary Table S1) or by the lack of proper convection within experimental hardware (Johnson et al., 2017). At this stage in our microgravity research, it is difficult to be certain which effects beyond cell wall modification are caused by microgravity rather than other environmental conditions of the experiment.
Plant research is still primarily conducted using the model plant Arabidopsis thaliana. While there are immense benefits to conducting -omics research to this plant (e.g., a well annotated genome, a greater number of established gene and protein interactions/networks, an extensive collection of mutant lines available, etc.), the more that we investigate the -omics of other plant species, the more we will understand about those plants and better we can hypothesize about the -omics of species we have yet to closely study (Vandenbrink et al., 2016). Only three of the studies examined here were from different species; two from Oryza sativa (rice), Jin et al. (2015) and Jin et al. (2018), and one from Eruca sativa (salad rocket), Chandler et al. (2020; Figure 1). The genes, transcriptomes and proteomes of these species are not as well researched as those of A. thaliana, but this is true of those of all other plants, and our understanding of the -omic response of plants cannot be limited to A. thaliana alone. Further developing our knowledge of the genes and proteins of other plant species will allow us to better predict the functions of orthologs, gene networks and protein interactions in unstudied species. The most compelling reason to stray from our beloved A. thaliana is a simple one; it is not a crop plant, and the more tangible benefits from plant research may occur through research in crops.
Despite the problems with RNAlater found by Kruse et al. (2017), (i.e. changes in the transcriptome) for the moment, we have no alternatives and therefore must continue using RNAlater to examine the transcriptomes of spaceflight specimen. Until technology is created that specifically allows for specimen preservation (be it with liquid nitrogen or a method/tool that has yet to be developed) in microgravity, using RNAlater is the best instrument we have for belatedly examining transcriptomes from spaceflight samples. The transcriptomic effects that arise during the delay of RNAlater permeating the cells of a sample could potentially be mitigated by speeding the process; if negative pressure could be created via a vacuum, the solution could infiltrate the plant cells at a greater rate and thereby reduce the time cells would have to react to being introduced to the solution. Augmenting the current hardware with vacuum pumps would be too difficult at this moment, but technology is constantly improving, and this feature could potentially be included in the future.
A reoccurring issue in space -omics research is the sample type upon which analyses are conducted. Most studies use the entire seedling to determine the transcriptome and proteome, however three were conducted using tissue of calli, three used leaves, three used roots, one used root tips, and one used seeds (Figure 5). Only one study conducted distinct analyses on different organs: Paul et al. (2013b). This study was critical to microgravity research; differentiated cell types, e.g., those found in the roots, shoots, and hypocotyl, fulfill distinct roles, and as such, react to stressors like microgravity in different manners. Their results showed that over 84% of DEGs in different organs were differentially regulated exclusively in one of the three tissues, that under 16% of DEGs were commonly differentially expressed in at least two of the three tissues and that a miniscule 0.03% were differentially expressed in all three tissues (Figure 4). Notably, one DEG was opposingly regulated in the roots and shoots; P-Glycoprotein18/ATP-Binding Cassette B 18 (PGP18/ABCB18), a gene associated with light perception and cell wall remodeling. It was strongly upregulated in the leaves but strongly downregulated in the roots. When the tissues of the roots, shoots and hypocotyls are taken as a whole, the protein and transcript expression profiles are averaged together, and this can lead to DEGs or differentially expressed proteins of one tissue being drowned out by the expression in the other two. The individual -omics profile of each tissue is an important part of how plants respond to microgravity, and deserves further investigation. It is worth noting that the tissues analyzed in this study were collected from a mix of COL and WS. These two ecotypes have distinct responses to the stress of microgravity, as demonstrated in Paul et al. (2017), which makes it likely that the differing transcriptional responses to microgravity may be obfuscating one another, much the way that the different tissues would when analyzing entire seedlings.
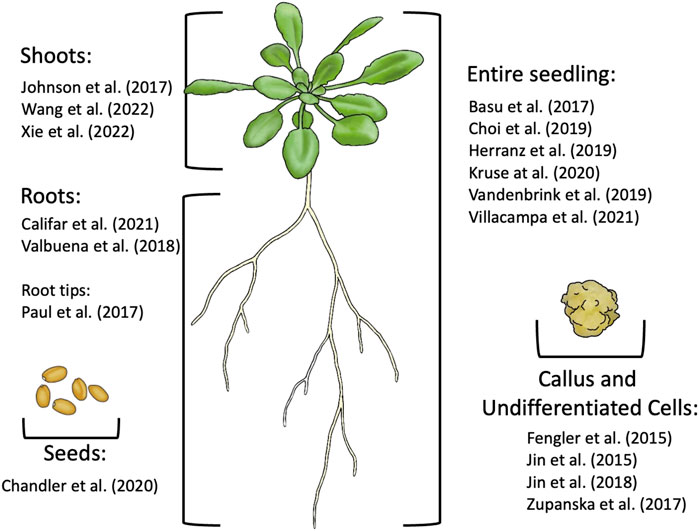
FIGURE 5. Summary of the tissue types used for -omic analysis from recent spaceflight experiments with plants.
The treatment of seedlings can also contribute to the loss of important data. When seedlings are grown in different light strengths, atmospheric conditions and within different hardware, the differing environments can act as confounding variables. Different light levels and qualities were used in the examined studies, ranging from dark conditions, all the way to full spectrum lighting and full spectrum light to red or blue illumination (Figure 6). The lighting schedules among the studies also varied, from constant illumination to long and short day illuminations (Figure 3). There is immense value in testing seedlings in different qualities of light, as well as different lighting schedules, just as there is value in using different hardware, because when the studies are taken as a whole, the varying growing conditions can ameliorate the effect any single confounding variable can have on the results. However, there are drawbacks to the divergent environmental conditions; as Basu et al. (2017) discovered that hardware itself can affect the results of an -omics assay. This was noted by Zupanska et al. (2017), where the researchers pointed out the stressful effects of the BRIC hardware on the plant cells. It was further supported by Johnson et al. (2017), in which researchers compared their data with that of two other experiments and found that many of the DEGs which had been previously identified were not differentially regulated in their results, and that some genes, like HSPs, were actually opposingly differentially regulated within the studies.
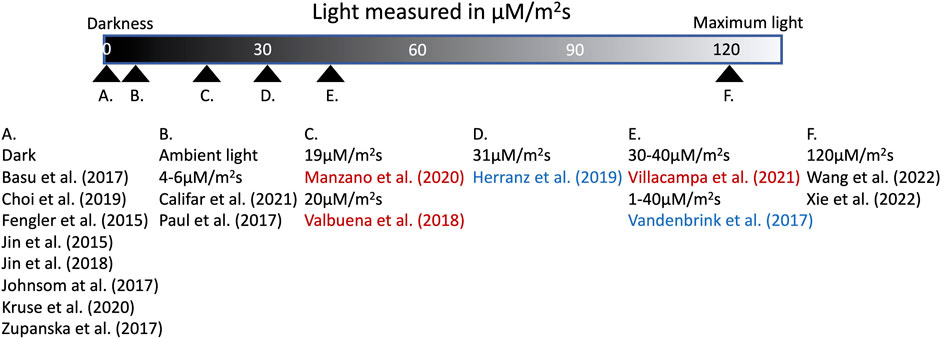
FIGURE 6. Summary of the lighting conditions from recent spaceflight experiments with plants. Experiments conducted using red or blue light are colored accordingly.
Some of these contradictions were hypothesized to be caused by the environmental conditions of the experiments. These hypotheses are supported by the data collected by the controls of the study. The presence of an inflight and ground control, as found in Valbuena et al. (2018) and Fengler et al. (2015), is vital, as the ground and flight controls can help to determine the changes that result from the hardware itself rather than those truly from spaceflight or microgravity. It is especially important to have an inflight control, which, unfortunately, many experiments lack (Shymanovich and Kiss, 2022). However, in our spaceflight experiments, we were able to utilize the centrifuge within the EMCS (shown in Figure 1) to provide a 1g inflight control, e.g., Vandenbrink et al. (2016), Shymanovich et al. (2022).
An additional confounding factor in these analyses is the exposure of root tissue to light. Roots exhibit phototropic effects when they are exposed to direct light (Kiss et al., 2003), however, in the practical application of spaceflight plant research, i.e., cultivating crops in space, the roots of plants will most likely be submerged or covered in some water-conducting material/hardware. The phototropic effects roots experience may not be limited to the proteome and transcriptome of the roots. The downstream effects of growing plants with roots exposed to light could very well be experienced in the leaves as well, as many signaling molecules (e.g., phytohormones, with which several of the examined studies found DEGs involved) carry signals throughout the body of a plant. This means that the experiments that have been conducted are skewed in a way that are not applicable to cultivation in the future. To correct this effect, the hardware used should be augmented to include some light-shielding material over the area in which roots grow, so that the overall effects will be truer to the conditions crops will experience in space, as will the transcriptomic and proteomic profiles.
A missing piece of the microgravity -omics puzzle is the developmental progression of the plant response to microgravity. The existing studies examine the transcriptome and proteome after the plants have grown for at least 2 days in microgravity, but the longer-term transcriptomic and proteomic responses observed in these studies are created by a sequence of microgravity-induced responses, an initial signal leading to another which creates a third, one falling into the next in their specific order. Knowing the steps that occur to cause the observed transcriptional and proteomic changes would provide insight into the effects of each gene and which genes might induce or repress others. By using RNAlater to fix samples at a series of early timepoints (e.g., 10 min, 30 min, 1 h, 2 h, 4 h, etc.) and analyzing the transcriptomes and proteomes, the development of the transcriptomic and proteomic responses to microgravity could be elucidated. The early transcriptional developments could be evaluated using samples collected from suborbital flights, as microgravity is achieved for three to 4 minutes aboard Virgin Galactic and Blue Origin and these flights do cause many of the same transcriptional changes as are observed in spaceflight (Califar et al., 2021).
We are confident that because of the increased availability of commercial spaceflights and suborbital possibilities that there will be more opportunities to overcome the limitations of plant space experiments to date. In addition, the Artemis program will allow us to better assess the biology of plants on the lunar surface. Resolving these issues will be critical for the use of plants in bioregenerative life support systems required for long-range space travel and developing colonies on Mars and other distant planets.
Both authors have participated in the writing of the manuscript and approve the final version.
This work was supported by the National Aeronautics and Space Administration (NASA) via grant 80NSSC17K0546 to JK.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fspas.2022.964657/full#supplementary-material
Argueso, C. T., Ferreira, F. J., and Kieber, J. J. (2009). Environmental perception avenues: The interaction of cytokinin and environmental response pathways. Plant, Cell Environ. 32, 1147–1160. doi:10.1111/j.1365-3040.2009.01940.x
Barker, R., Lombardino, J., Rasmussen, K., and Gilroy, S. (2020). Test of arabidopsis space transcriptome: A discovery environment to explore multiple plant biology spaceflight experiments. Frontiers in Plant Science 11 (2020), 147. doi:10.3389/fpls.2020.00147
Basu, P., Kruse, C. P. S., Luesse, D. R., and Wyatt, S. E. (2017). Growth in spaceflight hardware results in alterations to the transcriptome and proteome. Life Sci. Space Res. (Amst). 15, 88–96. doi:10.1016/j.lssr.2017.09.001
Califar, B., Zupanska, A., Callaham, J. A., Bamsey, M. T., Graham, T., Paul, A.-L., et al. (2021). Shared metabolic remodeling processes characterize the transcriptome of Arabidopsis thaliana within various suborbital flight environments. Gravit. Space Res. 9, 13–29. doi:10.2478/gsr-2021-0002
Chandler, J. O., Haas, F. B., Khan, S., Bowden, L., Ignatz, M., Enfissi, E. M. A., et al. (2020). Rocket science: The effect of spaceflight on germination physiology, ageing, and transcriptome of Eruca sativa seeds. Life 10, 49–25. doi:10.3390/life10040049
Choi, W. G., Barker, R. J., Kim, S. H., Swanson, S. J., and Gilroy, S. (2019). Variation in the transcriptome of different ecotypes of Arabidopsis thaliana reveals signatures of oxidative stress in plant responses to spaceflight. Am. J. Bot. 106, 123–136. doi:10.1002/ajb2.1223
Covington, M. F., Maloof, J. N., Straume, M., Kay, S. A., and Harmer, S. L. (2008). Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 9, R130. doi:10.1186/gb-2008-9-8-r130
Duque, A. S., Martinho de Almeida, A., Bernardes da Silva, A., Marques da Silva, J., Farinha, A. P., Santos, D., et al. (2013). Abiotic stress responses in plants: Unraveling the complexity of genes and networks to survive. London, United Kingdom: Intech Open. doi:10.5772/45842
Fengler, S., Spirer, I., Neef, M., Ecke, M., Nieselt, K., and Hampp, R. (2015). A whole-genome microarray study of Arabidopsis thaliana semisolid callus cultures exposed to microgravity and nonmicrogravity related spaceflight conditions for 5 days on board of Shenzhou 8. Biomed. Res. Int. 2015, 1–15. doi:10.1155/2015/547495
Ferl, R., Wheeler, R., Levine, H. G., and Paul, A. L. (2002). Plants in space. Curr. Opin. Plant Biol. 5, 258–263. doi:10.1016/S1369-5266(02)00254-6
Herranz, R., Vandenbrink, J. P., Villacampa, A., Manzano, A., Poehlman, W. L., Feltus, F. A., et al. (2019). RNAseq analysis of the response of Arabidopsis thaliana to fractional gravity under blue-light stimulation during spaceflight. Front. Plant Sci. 10, 1529–1611. doi:10.3389/fpls.2019.01529
Izzo, L. G., Romano, L. E., Muthert, L. W. F., Iovane, M., Capozzi, F., Manzano, A., et al. (2022). Interaction of gravitropism and phototropism in roots of Brassica oleracea. Environ. Exp. Bot. 193, 104700. doi:10.1016/j.envexpbot.2021.104700
Jin, J., Chen, H., and Cai, W. (2015). Transcriptome analysis of Oryza sativa calli under microgravity. Microgravity Sci. Technol. 27, 437–453. doi:10.1007/s12217-015-9432-2
Jin, J., Chen, H., and Cai, W. (2018). Transcriptomic analysis reveals the effects of microgravity on rice calli on board the Chinese Spaceship Shenzhou 8. Microgravity Sci. Technol. 30, 807–816. doi:10.1007/s12217-018-9633-6
Johnson, C. M., Subramanian, A., Pattathil, S., Correll, M. J., and Kiss, J. Z. (2017). Comparative transcriptomics indicate changes in cell wall organization and stress response in seedlings during spaceflight. Am. J. Bot. 104, 1219–1231. doi:10.3732/ajb.1700079
Kiss, J. Z., Brinckmann, E., and Brillouet, C. (2000). Development and growth of several strains of Arabidopsis seedlings in microgravity. Int. J. Plant Sci. 161, 55–62. doi:10.1086/314223
Kiss, J. Z., Mullen, J. L., Correll, M. J., and Hangarter, R. P. (2003). Phytochromes A and B mediate red-light-induced positive phototropism in roots. Plant Physiol. 131, 1411–1417. doi:10.1104/pp.013847
Kiss, J. Z. (2014). Plant biology in reduced gravity on the Moon and Mars. Plant Biol. (Stuttg). 16, 12–17. doi:10.1111/plb.12031
Kruse, C. P. S., Basu, P., Luesse, D. R., and Wyatt, S. E. (2017). Transcriptome and proteome responses in RNAlater preserved tissue of Arabidopsis thaliana. PLoS One 12, e0175943–10. doi:10.1371/journal.pone.0175943
Kruse, C. P. S., Meyers, A. D., Basu, P., Hutchinson, S., Luesse, D. R., and Wyatt, S. E. (2020). Spaceflight induces novel regulatory responses in Arabidopsis seedling as revealed by combined proteomic and transcriptomic analyses. BMC Plant Biol. 20, 237–316. doi:10.1186/s12870-020-02392-6
Manian, V., Orozco, J., Gangapuram, H., Janwa, H., and Agrinsoni, C. (2021). Network analysis of gene transcriptions of Arabidopsis thaliana in spaceflight microgravity. Genes (Basel) 12, 337. doi:10.3390/genes12030337
Manzano, A., Villacampa, A., Sáez-Vásquez, J., Kiss, J. Z., Medina, F. J., and Herranz, R. (2020). The importance of Earth reference controls in spaceflight -omics research: Characterization of nucleolin mutants from the seedling growth experiments. iScience 23, 101686. doi:10.1016/j.isci.2020.101686
Paul, A. L., Sng, N. J., Zupanska, A. K., Krishnamurthy, A., Schultz, E. R., and Ferl, R. J. (2017). Genetic dissection of the Arabidopsis spaceflight transcriptome: Are some responses dispensable for the physiological adaptation of plants to spaceflight? PLoS One 12, e0180186–24. doi:10.1371/journal.pone.0180186
Paul, A. L., Wheeler, R. M., Levine, H. G., and Ferl, R. J. (2013a). Fundamental plant biology enabled by the space shuttle. Am. J. Bot. 100, 226–234. doi:10.3732/ajb.1200338
Paul, A. L., Zupanska, A. K., Schultz, E. R., and Ferl, R. J. (2013b). Organ-specific remodeling of the Arabidopsis transcriptome in response to spaceflight. BMC Plant Biol. 13, 112. doi:10.1186/1471-2229-13-112
Shymanovich, T., and Kiss, J. Z. (2022). Conducting plant experiments in space and on the Moon. Methods Mol. Biol. 2368, 165–198. doi:10.1007/978-1-0716-1677-2_12
Shymanovich, T., Vandenbrink, J. P., Herranz, R., Medina, F. J., and Kiss, J. Z. (2022). Spaceflight studies identify a gene encoding an intermediate filament involved in tropism pathways. Plant Physiol. biochem. 171, 191–200. doi:10.1016/j.plaphy.2021.12.039
Tolsma, J. S., Ryan, K. T., Torres, J. J., Richards, J. T., Richardson, Z., Land, E. S., et al. (2021). The circadian-clock regulates the Arabidopsis gravitropic response. Gravit. Space Res. 9, 171–186. doi:10.2478/gsr-2021-0014
Valbuena, M. A., Manzano, A., Vandenbrink, J. P., Pereda-Loth, V., Carnero-Diaz, E., Edelmann, R. E., et al. (2018). The combined effects of real or simulated microgravity and red-light photoactivation on plant root meristematic cells. Planta 248, 691–704. doi:10.1007/s00425-018-2930-x
Vandenbrink, J. P., Herranz, R., Medina, F. J., Edelmann, R. E., and Kiss, J. Z. (2016). A novel blue-light phototropic response is revealed in roots of Arabidopsis thaliana in microgravity. Planta 244, 1201–1215. doi:10.1007/s00425-016-2581-8
Vandenbrink, J. P., Herranz, R., Poehlman, W. L., Alex Feltus, F., Villacampa, A., Ciska, M., et al. (2019). RNA-seq analyses of Arabidopsis thaliana seedlings after exposure to blue-light phototropic stimuli in microgravity. Am. J. Bot. 106, 1466–1476. doi:10.1002/ajb2.1384
Vandenbrink, J. P., and Kiss, J. Z. (2016). Space, the final frontier: A critical review of recent experiments performed in microgravity. Plant Sci. 243, 115–119. doi:10.1016/j.plantsci.2015.11.004
Villacampa, A., Ciska, M., Manzano, A., Vandenbrink, J. P., Kiss, J. Z., Herranz, R., et al. (2021). From spaceflight to MARS G-levels: Adaptive response of A. thaliana seedlings in a reduced gravity environment is enhanced by red-light photostimulation. Int. J. Mol. Sci. 22, 899–927. doi:10.3390/ijms22020899
Visscher, A. M., Seal, C. E., Newton, R. J., Frances, A. L., and Pritchard, H. W. (2016). Dry seeds and environmental extremes: Consequences for seed lifespan and germination. Funct. Plant Biol. 43, 656–668. doi:10.1071/FP15275
Wang, L., Xie, J., Mou, C., Jiao, Y., Dou, Y., and Zheng, H. (2022). Transcriptomic analysis of the interaction between Flowering Locus T induction and photoperiodic signaling in response to spaceflight. Front. Cell Dev. Biol. 9, 813246–813318. doi:10.3389/fcell.2021.813246
Wolverton, C., and Kiss, J. Z. (2009). An update on plant space biology. Gravit. Space Res. 22, 13–20.
Xie, J., Wang, L., and Zheng, H. (2022). Molecular basis to integrate microgravity signals into the photoperiodic flowering pathway in Arabidopsis thaliana under spaceflight condition. Int. J. Mol. Sci. 23, 63. doi:10.3390/ijms23010063
Zhang, H., Zhao, Y., and Zhu, J. K. (2020). Thriving under stress: How plants balance growth and the stress response. Dev. Cell 55, 529–543. doi:10.1016/j.devcel.2020.10.012
Keywords: gene profiling, microgravity, plant biology, reduced gravity, spaceflight, transcriptomics
Citation: Hughes AM and Kiss JZ (2022) -Omics studies of plant biology in spaceflight: A critical review of recent experiments. Front. Astron. Space Sci. 9:964657. doi: 10.3389/fspas.2022.964657
Received: 08 June 2022; Accepted: 18 July 2022;
Published: 12 August 2022.
Edited by:
Oliver Ullrich, University of Zurich, SwitzerlandReviewed by:
Richard John Barker, University of Wisconsin-Madison, United StatesCopyright © 2022 Hughes and Kiss. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John Z. Kiss, anpraXNzQHVuY2cuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.