
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Astron. Space Sci., 28 January 2022
Sec. Astrobiology
Volume 9 - 2022 | https://doi.org/10.3389/fspas.2022.817900
This article is part of the Research TopicMars analogs: Environment, Habitability and BiodiversityView all 11 articles
Past acid-sulfate hydrothermal systems on Mars have promise in their ability to have hosted life for billions of years. One method for analyzing these systems is to study analog environments on Earth. To assess the astrobiological potential of Martian acid-sulfate hydrothermal systems, the crater lake of the active Poás Volcano, Laguna Caliente, was sampled in 2013 and 2017. Laguna Caliente presents an extremely dynamic terrestrial environment with near-ambient to boiling temperatures, pH fluctuations from −0.87 to 1.5, a wide range of chemistries and redox potential, and frequent phreatic-to-phreatomagmatic eruptions. Samples of lake fluid, sulfur clumps, and lake bottom sediment underwent 16S rRNA gene sequencing and metagenomic “shotgun” sequencing, which revealed this lake hosts an extremely low biodiversity of microorganisms dominated by Acidiphilium spp. Shotgun metagenomics of the samples suggests this community has numerous genetic adaptations that confer survival, including functional pathways to reduce the effects of toxic metals and numerous metabolic pathways utilizing a variety of simple and complex sugar molecules. The identification of these various metabolic pathways suggests adaptations related to carbon limited environments, fulfillment of high energy requirements, and survival in a hostile volcanic setting. The perseverance of life in Laguna Caliente indicates life on Mars could have thrived in analogous environments, stressing the need for the search for life in relict Martian acid-sulfate hydrothermal systems.
The science discipline of Astrobiology is rather broad and leads to research ranging from studying the origins of life on Earth to searching for extraterrestrial life on alien worlds (Domagal-Goldman et al., 2016; Davila, 2021). To prepare for space missions with key goals to find signs of life on these extraterrestrial planets and moons, we must better understand how life in these environments may look like (Ehrenfreund et al., 2011). The search for signs of past life on Mars is a key NASA priority (National Research Council, 2011; Changela et al., 2021), demonstrated in 2021 by the arrival of the Perseverance rover on the Red Planet (Mangold et al., 2021). As of the time of this writing, neither extant life nor definitive signs of past life on Mars have been found; however, the habitability of Mars has been assessed to great detail (e.g., Cockell et al., 2012). For example, it has been shown that early Mars had persistent liquid surface water, biogenic elements, and environments conducive to life (e.g., Grotzinger et al., 2015). While recent landing sites for habitability studies have focused on sedimentary environments (Kereszturi et al., 2016; Vago et al., 2018), relict hydrothermal settings may provide a better setting to investigate the astrobiological potential of early Mars (e.g., Michalski et al., 2018).
During the early period of Mars’ history, volcanic/magmatic processes were widespread and liquid water was present in surface and near surface environments. The surface heat flux on early Mars was roughly an order of magnitude higher than at present (Hauck Ii and Phillips, 2002), and enormous amounts of crust were emplaced via volcanic and magmatic processes until ∼3 billion years ago (e.g., Phillips et al., 2001; Craddock and Greeley, 2009; Carr and Head, 2010). Moreover, widespread liquid water was present at or near the surface during this time period (e.g., Hynek and Phillips, 2003; Fairén et al., 2003). Mars had an active, globally integrated, hydrologic cycle from ∼3.8 to 3.6 Ga, based on the valley networks, which were incised at this time (Fassett and Head, 2008a; Hoke and Hynek, 2009; Hynek et al., 2010). Hundreds of paleolake basins have also been identified (e.g., Fassett and Head, 2008b; Goudge et al., 2016) and a number of these host fan delta deposits (e.g., Di Achille and Hynek, 2010; Wilson et al., 2021). After this time, significant water reserves remained in Mars’ crust, as evidenced by the enormous outflow channels, which are thought to have been caused by confined aquifers bursting onto the surface through a cryosphere (e.g., Rotto and Tanaka, 1995; Andrews-Hanna and Phillips, 2007).
Given the abundant crustal/surface waters and rampant volcanism and magmatism, it is thought that large-scale hydrothermal alteration was common on early Mars, leading to extensive deposits of acid-sulfate alteration minerals (Bibring et al., 2005; Gendrin et al., 2005; Solomon et al., 2005; Ehlmann et al., 2011; Hynek et al., 2013). While surface waters may have been limited in time, volcanic activity likely occurred for much longer. With the lack of plate tectonics on Mars, the preserved geological record suggests intermittent summit eruptions occurred on individual volcanoes spanning 4 billion years (Robbins et al., 2011). Surface volcanic activity may have continued to <50,000 years ago (Horvath et al., 2021), and recent results from the InSight mission support a recently active volcano-tectonic system at Cerberus Fossae (Banerdt et al., 2020), which is believed to host an ice-saturated regolith (e.g., Moitra et al., 2021). Collectively, these observations suggest that subsurface hydrothermal activity may be ongoing within Mars’ crust. In addition to volcanism, hydrothermal systems on Mars may also have been initiated from large impacts and could have provided a volumetrically large, long duration, cradle for the origin of life (Osinski et al., 2020). There are at least 78 large impact basins recorded to have formed prior to ∼3.5 Ga (Robbins et al., 2013) with some likely hosting hydrothermal systems for up to 10 million years (Abramov and Kring, 2005).
Relict hydrothermal systems on Mars have been identified extensively on the surface via orbiters and landers obtaining mineralogy and geomorphology data. For example, the Spirit rover in Gusev crater found sulfate and silica accumulations suggesting acid-sulfate alteration occurred in fumarolic and/or hot spring environments next to Home Plate (Squyres et al., 2007; Squyres et al., 2008; Yen et al., 2008; Schmidt et al., 2009; Ruff et al., 2011). Hydrothermal processes have also been proposed for the MER Opportunity landing site (McCollom and Hynek, 2005) and at Gale crater, the home of the Curiosity rover (Yen et al., 2021). High temperature acid-sulfate alteration has also been hypothesized to have altered mineralogies present in Valles Marineris (Gendrin et al., 2005; Chojnacki and Hynek, 2008; Weitz et al., 2011; Thollot et al., 2012; Marcucci et al., 2013), the volcanic cone on the Nili Patera volcano (e.g., Skok et al., 2010), and numerous other volcanic edifices (Gulick, 1998; Tanaka et al., 1998; Dohm and Tanaka, 1999; Gulick, 2001; Hynek et al., 2010; El Maarry et al., 2012).
On Earth, hydrothermal environments often provide sources of heat, energy, and water for life (Walter and Des Marais, 1993; Schulze-Makuch et al., 2007; Hays et al., 2017). It is also hypothesized that the last universal common ancestor of life (LUCA) on Earth was likely a thermophile or hyperthermophile (Nisbet and Sleep, 2001; Giulio, 2003; Gaucher et al., 2008) potentially utilizing iron-sulfur redox chemistry in a hydrothermal setting (Martin and Russell, 2003; Wächtershäuser, 2006; Martin et al., 2008; Weiss et al., 2016). It is conceivable that life on Mars could have formed in a homologous manner in Martian hydrothermal environments and been sustained for billions of years through volcanism or impact-related hydrothermal activity. The condensation of volcanic vapors (fumaroles) and crustal hydrothermal circulation could have provided warm and moist habitats for life even while cold and dry conditions persisted globally. Understanding terrestrial life in environments analogous to Martian hydrothermal systems is crucial in order to characterize how life may have existed on Mars, which conditions extraterrestrial life could adapt to, and to better search for life on the Red Planet. Here, we characterize the taxonomic and functional diversity of microbial communities in the Poás Volcano in Costa Rica; arguably one of the most extreme Martian analog environments on Earth.
The volcanic crater of Poás Volcano, Costa Rica (Figure 1) has been characterized as a Martian analog due to its mineralogical consistencies with relict Martian hydrothermal systems (Hynek et al., 2014; Black et al., 2015; Black et al., 2016; Rodríguez and van Bergen, 2017; Black and Hynek, 2018). The Poás Volcano is a basaltic andesite stratovolcano in the Central Cordillera of Costa Rica (Prosser and Carr, 1987). Poás has been active throughout the Holocene; phreatic to phreatomagmatic eruptions are common even in times of quiescence, such as between 1955 and 2017. These eruptions mostly consist of unpredictable steam-driven expulsions from the crater lake, Laguna Caliente (Figure 1C). A magmatic episode occurred from April to November 2017, and phreatomagmatic events resumed in 2019 (Salvage et al., 2018; Wang et al., 2020).
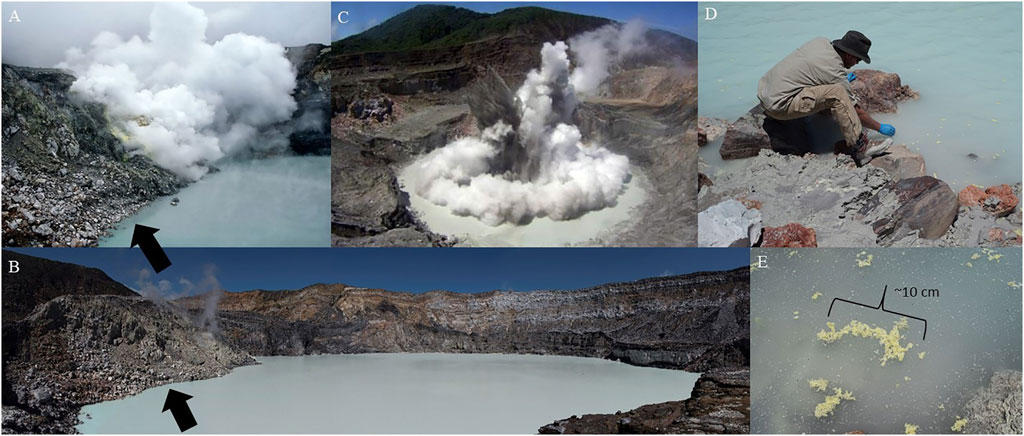
FIGURE 1. (A) The active Poás Crater and Laguna Caliente during the November 20, 2013 field campaign, and (B) reduced activity during the March 23, 2017 field campaign. The arrows point to the same sampling site adjacent to fumarolic activity (10°11′48.09″N, 84°13′48.30″W). (C) A phreatic eruption in Poás Volcano, June 2014 (Courtesy of OVISCORI). (D) Sampling clumps of elemental sulfur floating on Laguna Caliente during the 2017 field campaign where fluids and lake bottom sediments were collected 2 m offshore. (E) A close-up of the sulfur clumps with dead insects.
Poás Volcano hosts Laguna Caliente, an ultra-acidic sulfurous crater lake that is primarily sourced through a subsurface groundwater system (Sanford et al., 1995). Acid-sulfate alteration of the parent rocks has led to an abundance of a variety of sulfates, Fe-oxides, and clays akin to those found on Mars in relict hydrothermal settings (Hynek et al., 2014; Black et al., 2015; Black et al., 2016; Rodríguez and van Bergen, 2017; Rodríguez and van Bergen, 2017; Black and Hynek, 2018). Poás is an extremely dynamic terrestrial environment with recorded temperatures ranging from near-ambient to boiling and pH values from −0.87 to 1.5; sometimes spanning these ranges over the course of days to weeks (Rowe et al., 1992b). Negative pH values are attributed to high normality (>1 M) of hydrogen ions in solution. Temperature and pH changes are reflective of an underlying magma body (<500 m depth to zero depth during eruptions) (Rymer and Brown, 1989; Rymer et al., 2000), fluctuating as a result of rising magma dendrites and magmatic gases interacting with meteoric and lake water (Rymer et al.,2000). Volcanic gases (HCl and H2S and SO2, which hydrolyze to H2SO4) contribute to the extreme acidity of the lake. Times of lower pH are a result of increased contribution from HCl and SO2 relative to H2S (Rowe et al., 1992a). The lake has historically exhibited a wide range of chemistries as well as redox potential (Supplementary Tables S1, S2 in the Supplemental Information), including a variety of S and Fe redox chemistry. The large range of sulfur oxidation states (from 0 with elemental sulfur to +6 with sulfate; sulfite and thiosulfate were also detected, Martínez-Cruz et al., 2019) in this lake comprises a complex sulfur cycle which could provide metabolic options for chemolithotrophic archaea and bacteria (Friedrich, 1997; Hynek et al., 2018).
On November 20, 2013, Laguna Caliente was sampled in order to characterize its microbiology for the first time (Figure 1A). This study was described in Hynek et al. (2018), which reported a “veritable monoculture” of a bacterium in the Acidiphilium genus in the phylum Proteobacteria. Nearly 98% of amplicons from 16S rRNA sequencing fell within a single Operational Taxonomic Unit (OTU), whose taxonomy was most closely related to Acidiphilium angustum strain KLB (at least 97% similarity).
Members of the Acidiphilium genus have been shown to grow in temperatures ranging from ∼17°C–45°C, and in a pH range of ∼1.5–7.5 with optimal growth at pH ∼3 (Hiraishi et al., 1998; Küsel et al., 1999; Schippers, 2007; Li et al., 2020)—conditions less extreme than that of Laguna Caliente. Members of this genus have also been identified in, and isolated from, acid mine drainages, hydrothermal environments, a coal mine drainage, and another Mars analog in Rio Tinto, Spain (Harrison, 1981; Wichlacz et al., 1986; Lane et al., 1992; Kishimoto et al., 1995; González-Toril et al., 2003; Amils et al., 2007; San Martin-Uriz et al., 2011).
All members of the Acidiphilium genus are aerobic acidophiles, and these members have exhibited a wide range of environmental adaptations (Hiraishi et al., 1998; Dopson and Johnson, 2012). For example, some members of the Acidiphilium genus have been identified to oxidize elemental sulfur for growth (Rohwerder et al., 2003; Rohwerder and Sand, 2003; Ghosh and Dam, 2009) with some utilizing the bacterial Sox (sulfur oxidation) metabolic pathway (Liu et al., 2016). Other members of this genus are able to reduce Fe(III) (Lane et al., 1992; Küsel et al., 1999), reduce Cu(II) in the presence of iron (Johnson et al., 2017), and reduce Cr(VI) (Cummings et al., 2007). Genes conferring nickel and arsenic resistance have also been identified in Acidiphilium spp. isolated from Rio Tinto, Spain (Mirete et al., 2017). The ability of acidophiles, like Acidiphilium spp., to live and grow in low pH has been attributed to highly impermeable cell membranes, reversed membrane potential, a predominance of secondary transporters, and methods to alleviate lowered pH for protons that cross through the cytoplasm (Baker-Austin and Dopson, 2007). Overall, members of the Acidiphilium genus have included obligate heterotrophs, facultative chemolithotrophs, and autotrophs (Hiraishi et al., 1998; Dopson and Johnson, 2012).
Environmental systems with extremely low microbial biodiversity, as described in Hynek et al. (2018), are rare on Earth. For comparison, the Iron Mountain Mine in California hosts an acid mine drainage with pH as low as −3.6, and a diverse microbial community that includes Bacteria, Archaea, and Eukarya was identified in a sample of pH ∼0.5 (Nordstrom and Alpers, 1999; Edwards et al., 2000). It seems possible that in Laguna Caliente the combination of an extremely low pH, fluctuating temperatures (38°C–90°C, Rowe et al., 1992b), volcanic dynamics, and a high concentration of dissolved ions and toxic metals has restricted life in a way that only highly specialized bacteria can survive in this environment (Hynek et al., 2018; Wang et al., 2020).
To make predictions about how the organisms identified by Hynek et al. (2018) are able to survive in this challenging environment, samples were collected again by our team on March 28, 2017 (Figures 1B,D). In addition to describing the community makeup for temporal comparison to Hynek et al. (2018), we wanted to look specifically at the metabolic pathways and adaptations that could allow organisms to survive in such harsh conditions. Hydrothermal systems have the potential to be sterilizing by increasing temperatures, lowering pH, or inserting toxic chemicals to the environment. On Mars, the surface environment can also be sterilizing from incoming surface radiation and low atmospheric pressure. Thus, groundwater systems connected to hydrothermal systems are likely safe havens for single-celled organisms during volcanism-induced sterilization events. Because of this, if microorganisms were to exist in Martian hydrothermal systems, they likely would have required biological mechanisms to survive. An ability to survive on the fringes, such as in connected groundwater systems, would increase the chances for microorganisms to survive for extended periods of time. As a Martian analog, the Poás Volcano serves as a proxy for studying the adaptations that may have allowed any organisms to survive in hydrothermal systems on Mars. To accomplish the goals of this study, samples from both field campaigns were further scrutinized with shotgun metagenomics.
The first field campaign occurred November 20, 2013, as described in Hynek et al. (2018), and the second field campaign occurred March 28, 2017. For both field campaigns, Laguna Caliente exhibited convective activity as evidenced by roiling of the lake’s surface and adjacent active fumaroles. Sampling both years occurred at the same locale (Figures 1A,B).
Sampling in 2013 is described in more detail in Hynek et al. (2018), but to summarize, fluids from Laguna Caliente were collected aseptically in duplicate in sterile 50 ml Falcon tubes (Corning Incorporated, Corning, New York, United States) 2 m offshore, with the use of a sterile aluminum pole. In the 2017 sampling expedition, lake bottom sediment samples entrenched with lake fluids were collected using the same methods. Floating sulfur clump samples, also taken in the 2017 sampling expedition, were collected by hand closer to shore (Figures 1D,E) aseptically in duplicate in sterile 50 ml Falcon tubes with lake fluids entrenched. All samples were immediately stored on water ice. Within hours the samples were frozen to −4°C and within days were transferred to a −80°C freezer for later DNA extraction.
Temperature and pH were read by field instruments as well as pH paper for fidelity. Lake fluids were collected in sterile Falcon tubes and then filtered through 0.2 µm Sterivex filters (Cole-Parmer, Vernon Hills, Illinois, United States) and stored in the dark until analysis by ICP-MS and ICP-OES at Activation Laboratories Ltd. (Actlabs, Ancaster, Ontario, Canada) for cations, anions, and a full elemental scan.
Mineralogical and fluid chemistry samples of Laguna Caliente were taken in both 2013 and 2017 (Supplementary Tables S1, S2 in the Supplemental Information). For the 2017 field campaign, the temperature was 10°C lower than in the 2013 sampling and the pH was measured at 1.45 (compared to 0.29 in 2013). Compared to March 2013, the fumarolic activity was much less and there was no indication of significant HCl or H2S in the gases, only SO2 (Figure 1). The higher pH in the 2017 sampling is likely due to two factors; 1) lower observed volcanic activity, and 2) sampling after significant rainfall in the prior weeks (unlike our 2013 campaign), which could have diluted the lake fluids. The lower lake acidity in 2017 coincided with decreased concentrations of sulfate and chlorine (and many other elements). Nevertheless, the environmental conditions from 2017 still remained extreme for most mesophilic organisms.
Supplementary Table S1 in the Supplemental Information reports concentrations of elements in Laguna Caliente that have potential to be utilized in microbial metabolism (S and Fe). Additionally, concentrations of potentially hazardous elements to microorganisms that are discussed in the metagenomic results (Ag, As, Co, Cd, Cr, Cu, Ni, and Zn) are also included in Supplementary Table S2 in the Supplemental Information.
DNA was extracted from the −80°C frozen samples using the MoBio PowerMax Soil Kit (Qiagen, Germantown, MD, United States) following manufacturer’s instructions. Duplicate extractions were performed using material from each sample (lake fluid, lake bottom sediment, sulfur clumps). The DNA from the replicate extractions from each sample was combined and concentrated with Amicon Ultra-4 spin filters (MilliporeSigma, Burlington, MA, United States) three times for each sample to increase DNA concentration enough for downstream analysis.
For sequence-based analyses of 16S rRNA genes of the 2013 Laguna Caliente sample, we use the approaches described by Carini et al. (2016). To summarize, the V4 region of the 16S rRNA gene was PCR-amplified with barcoded primers (515F, 5′-GTGCCAGCMGCCGCGGTAA-3′ and 806R, 5′-GGACTACVSGGGTATCTAAT-3′ (Caporaso et al., 2010). Products from duplicate PCR reactions for each sample, as well as “no template” and “DNA extraction” negative controls, were cleaned, normalized, and pooled with the SequalPrepTM Normalization Plates (Thermo Fisher Scientific, Carlsbad, CA, United States) in the Fierer Lab at the University of Colorado (CU) Boulder, and sequenced on an Illumina MiSeq platform (Illumina, San Diego, CA, United States) by using v2 500-cycle paired-end kits at the CU Boulder Biofrontiers Sequencing Center. These sequence reads can be found in the supplementary material in Hynek et al. (2018).
16S rRNA gene sequences were processed using the DADA2 pipeline (Callahan et al., 2017). Sequences were quality filtered and clustered into amplicon sequence variants (ASVs). Taxonomic information was assigned to ASVs using a naive Bayesian classifier method (Wang et al., 2007), which takes the set of ASVs generated and compares them to a training set of reference sequences from the 16S rRNA bacterial and archaeal SILVA database (Quast et al., 2013; Yilmaz et al., 2014). A minimum bootstrapping threshold required to return a taxonomic classification of 50% similarity was used for analysis. ASVs associated with chloroplast, mitochondria, and eukaryotes were removed prior to downstream analyses.
For the metagenomic “shotgun” sequencing, three samples in total were sequenced. The same sample from Laguna Caliente in 2013 that underwent 16S rRNA sequencing was sequenced as well as two additional samples from the 2017 field campaign: a sulfur clump floating on the lake and bottom sediment from the shore of the lake - both mixed with lake fluid. These three samples were prepared using the Nextera XT DNA Library Prep Kit (Illumina, San Diego, CA, United States) by the University of Colorado BioFrontiers Institute. Afterwards, the samples were sequenced using a NovaSEQ 6000 Sequencing System (Illumina, San Diego, CA, United States) with paired-end 150 cycle 2 × 150 bp format by the Genomics and Microarray Core at the University of Colorado Denver Cancer Center with an targeted insert size of 250 bp. The sequencing data generated from these samples can be accessed in the NCBI Sequence Read Database, project accession number PRJNA760478.
Samples had between 2.4 × 107 and 5.2 × 107 reads for a total of 1.1 × 108 reads total across the three samples. Removal of adapter sequences from raw sequence data was done with Cutadapt (Martin, 2011). Quality filtering and trimming was performed with Sickle v.1.33 (-q 20 -I 50) (Joshi and Fass, 2011). After this trimming and filtering a total of 7.3 × 106 reads, just 7% of the total raw reads recovered from sequencing, remained for use in downstream analysis (average of 2.4 × 106 reads per sample). The relative abundance and diversity of bacteria and archaea in the metagenomic samples were classified by identifying 16S rRNA gene reads from the metagenomic sequence data using Metaxa2 (Bengtsson-Palme et al., 2015).
Assembly-free analysis on the adapter trimmed and quality filtered data was performed with SqueezeMeta on Reads v0.1.0, Sept 2019 with the alternative analysis mode sqm_reads.pl script (Tamames and Puente-Sánchez, 2019). With these settings paired end reads were grouped for annotation. Similarity searches for eggnog predictions (Huerta-Cepas et al., 2015) and KEGG classifications (Kanehisa and Goto, 2000) were done using Diamond (Buchfink et al., 2015). Annotated reads were rarified to 8917733, the lowest number of reads across the three samples using the “Vegan” package in R. The rarified table was normalized using MUSiCC to obtain accurate and biologically meaningful measures of gene abundance based on universal single copy genes (Manor and Borenstein, 2015).
Contigs were also generated from the trimmed and filtered sequence data using MEGAHIT, a NGS de novo assembler (Li et al., 2015). To identify and categorize specific iron genes and iron gene operons in these contigs, we used FeGenie (Braun, 2003; Garber et al., 2020).
As described above, 16S rRNA genes sequences were recovered from both 16S rRNA sequencing and by extracting the 16S rRNA gene reads from the metagenomic sequence data using Metaxa2. In contrast with Hynek et al. (2018) who used OTUs, sequences were grouped with 100% similarity as Amplicon Sequence Variants (ASV) to categorize the microbes in the sample [see Callahan et al. (2017) for more explanation of why we chose to use ASVs instead of OTUs].
Re-processing the data from Hynek et al. (2018) as ASVs, instead of OTUs, produced similar results. Instead of ∼98% of OTUs being assigned to Acidiphilium angustum, ∼92% of ASVs were assigned to the Acidiphilum spp. (Figure 2). This slight difference is expected, as the adoption of ASVs occurred to help preserve the presence of rare taxa in sequencing data that were often hidden by OTU clustering (Callahan et al., 2017). This data clearly shows that Acidiphilium spp. is the overwhelmingly dominant genus in this system in 2013, supporting what was described by Hynek et al. (2018).
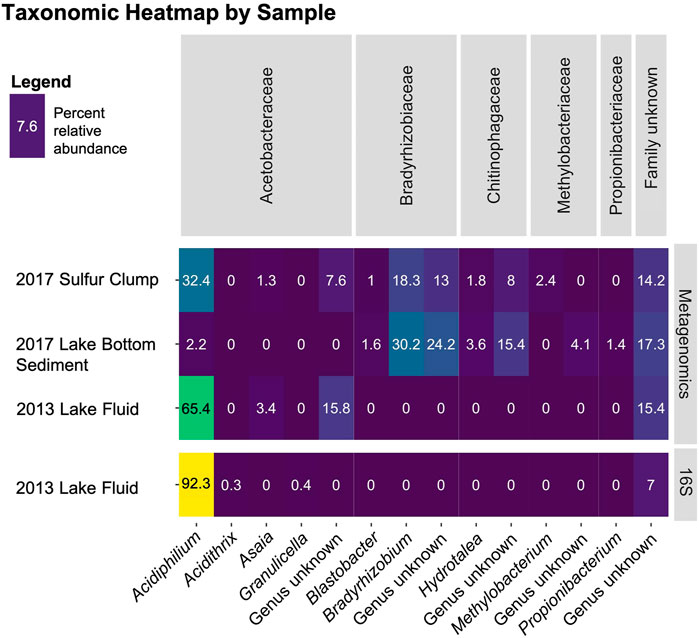
FIGURE 2. Microbial communities of Laguna Caliente. The relative abundance of microbial communities in each of the four samples displayed were determined either by 16S rRNA marker gene sequencing or were recovered from metagenomic “shotgun” sequencing by Metaxa2 as indicated in the right panel. Identified bacterial families are shown across the top of the heatmap while genera are displayed across the bottom.
The taxonomic data recovered from the shotgun metagenomics shows an even wider spread of microbial diversity. The data from the 2013 Laguna Caliente fluid sample shows that ∼64% of reads converge on Acidiphilium spp. ∼3% of this sequence was assigned to Asaia spp., which is another acidophile that was first identified in a fluid of pH 3.5 (Moore et al., 2002). The 2017 sulfur clump data shows only ∼32% of reads assigned to Acidiphilium spp. and the 2017 lake bottom sediment data shows that only ∼2% of identified 16S sequences assigned to Acidiphilium spp. (Figure 2).
As Laguna Caliente is an environment with high concentrations of sulfur in a variety of oxidation states, we investigated what sulfur metabolisms were present in the metagenome. Additionally, alternative types of metabolisms were investigated including complex carbon metabolisms, photosynthesis, and iron and arsenic oxidation. Many environmental adaptation genes were also investigated, such as heat shock operons, proton exporters, and heavy metal detoxifiers that are commonly found in other organisms of the genus Acidiphilium (Supplementary Table S3 in the Supplemental Information). Reads associated with these pathways were found in all three samples, unless noted otherwise. We acknowledge that while the metagenomics analyses used here can identify the presence/absence of genes and thus provide insights into potential functions, we cannot say whether or not these genes are active without metatranscriptomics.
The metagenomic sequencing identified a wide range of sulfur metabolism genes in our samples (Figure 3). These genes indicate the ability to reduce inorganic sulfur from sulfate to sulfide (+6 to −2) using assimilatory sulfate reduction. Assimilatory sulfate reduction is a pathway that allows cells to further incorporate sulfide into carbon skeletons of amino acids to form cysteine or homocysteine (Brunold, 1993).
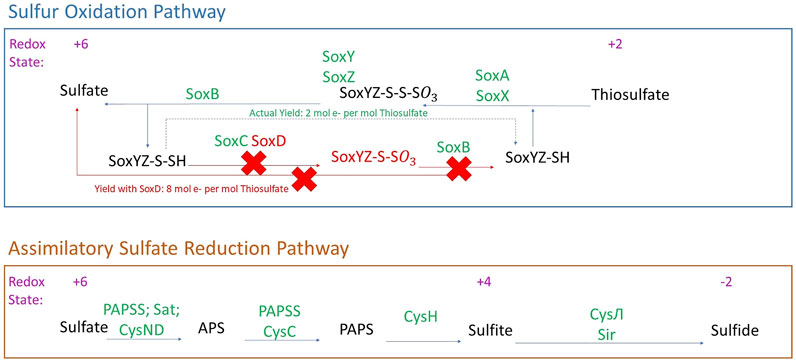
FIGURE 3. Sulfur metabolism in Laguna Caliente metagenomic samples. Genes identified in the metagenomes are presented with green text. Genes and products not identified in the metagenomes are presented with red text. (Modified from Kanehisa and Goto, 2000).
The metagenomes also host a nearly complete Sox system including soxA, soxB, soxC, soxX, soxY, and soxZ. However, we did not find soxD in the assembly-free analysis, and we did not search for it in the assembled data since that was used solely for the FeGenie analysis. With soxD absent, the heterotetrameric SoxCD complex would not be formed and sulfur-dependent cytochrome c reduction cannot be accomplished. Nevertheless, oxidation from thiosulfate to sulfate might still be accomplished; the SoxCD heterotetramer may be bypassed to allow for sulfur oxidation. In a full soxXYZABCD system, the yield is 8 mol electrons produced per mol of thiosulfate; however, with the SoxCD heterotetramer absent, the yield is reduced to 2 mol of electrons per mol of thiosulfate (Friedrich et al., 2001).
The annotated reads also contained various pathways central to carbon metabolism in microorganisms. Genes related to the metabolism of simple sugars are present; gene regions related to glycolysis, gluconeogenesis, lactate fermentation, the pentose phosphate pathway, the citric acid cycle, and oxidative phosphorylation were all identified. Additionally, genes whose presence may indicate the utilization of more complex sugar molecules were also identified, including those assigned to the glyoxylate cycle, the ethylmalonyl-CoA pathway, the polyhydroxybutyrate cycle (Figure 4), the serine glyoxylate pathway, the methylglyoxal pathway, and the Entner-Doudoroff pathway (Yamauchi and Yamada, 1981; Ackermann et al., 1995; Okubo et al., 2010; Schneider et al., 2012). Identified genes and their abundance in the Laguna Caliente samples of the Entner-Doudoroff, methylglyoxal, and serine-glyoxylate pathways are reported in Supplementary Table S4 in the Supplemental Information. Note that only the 2017 sulfur clump sample included genes for methylglyoxal synthase, which would allow for the full methylglyoxal pathway.
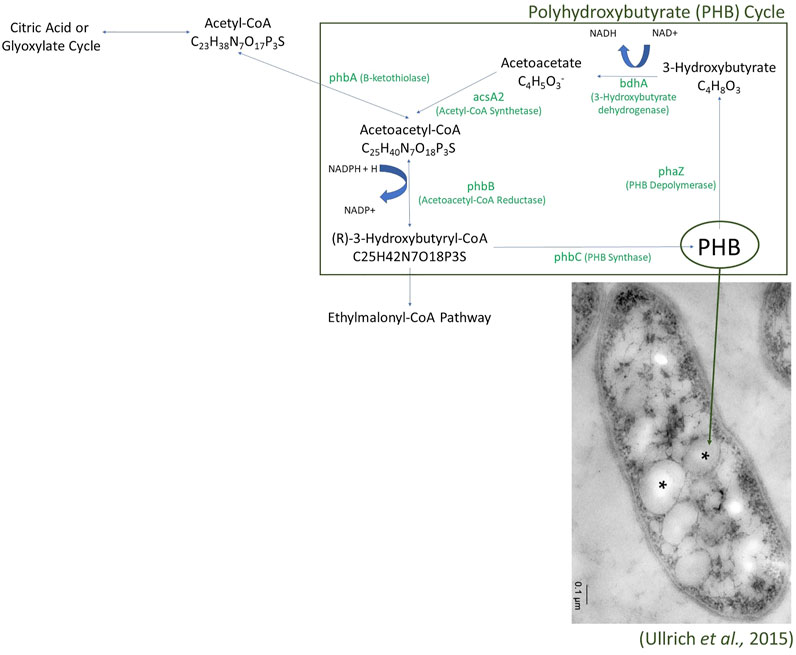
FIGURE 4. The Polyhydroxybutyrate (PHB) Cycle as found in the Laguna Caliente metagenomic samples. Enzymes identified in the metagenomes are presented with green text.
It has been suggested that the glyoxylate cycle and the ethylmalonyl-CoA pathway are anabolic pathways (Schneider et al., 2012; Chew et al., 2019), and the polyhydroxybutyrate cycle results in the creation of microbial plastic granules utilized in times of energy and carbon limitations (Ackermann et al., 1995). Peña-Ocaña et al. (2022) report metabolic maps and networks constructed from genes identified in mesophilic and hyperthermophilic samples from the El Chichón volcano crater lake that include some of the genes identified in the Laguna Caliente metagenomic samples, such as the Entner-Doudoroff pathway and sulfur oxidation.
In addition to sugar metabolism genes, genes commonly utilized in photosynthesis were identified, including genes related to the Calvin Cycle and photorespiration. These included RuBisCO, phosphoribulokinase, fructose-1,6-bisphosphatase, photosystems I and II, light-harvesting complex I and II, cytochrome b6f, and photosynthetic reaction center L and M subunits. Note that only the 2013 metagenome contained sequences encoding for photosystems I and II, light-harvesting complex I and II, and cytochrome b6f.
While the trademark for acid-resistance in bacteria is an impermeable cell membrane characterized by a complex lipid bilayer (Baker-Austin and Dopson, 2007), metagenomic analysis can identify the presence of genes that encode for proton export that might confer acid resistance. All samples contained the full genetic pathways to encode for nuo, cyo, and F1Fe-ATPase that when up-regulated confer acid resistance by proton efflux (Krulwich et al., 2011).
Genes related to the decarboxylase/antiporter-dependent acid-resistance systems (AR2 and AR3) were identified. These genes include glutamate decarboxylase isozymes (GadA and GadB), arginine decarboxylase (AdiA), and cognate antiporters (GadC for glutamate and AdiC for arginine). Briefly, this system promotes intracellular conversion of glutamate to GABA and arginine to agmatine for efflux out of the cell. This system expels 1 mol of H+ per mol of glutamate or arginine promoting pH homeostasis and acid resistance (Foster, 2004).
The metagenome also contained many genes that confer heat resistance, including genes that are typically found in the heat-shock operons hrcA-grpE-dnaJ, groES-groEL, and dnaK-sHSP. The upregulation of these operons is known to confer heat resistance in thermophiles such as Thermotoga maritima (Wang et al., 2015). Additionally, increased dnaK mRNA and DnaK protein have been correlated in cells experiencing acid shock as well as in acid ‘adapted’ cells (Jayaraman et al., 1997).
Laguna Caliente hosts a high concentration of iron ions (276–631 ppm, Supplementary Table S1 in the Supplemental Information). The metagenome contained numerous genes involving the transport and metabolism of iron. Iron uptake was found in the 2013 fluid and 2017 lake bottom sediment samples, marked by the presence of genes found in the fbpABC operon. Genes conferring the ability to reduce Fe(III) were not identified in the metagenome; however, the Cyc2 gene was present in the 2013 fluid sample and the 2017 lake bottom sediment sample, which is a gene utilized in iron oxidation (Castelle et al., 2008). Specifically, the metagenome contained the gene sequence for cluster 2 of Cyc2 that was found in an isolate of the acidophilic iron-oxidizing bacterium, Acidithiobacillus ferrooxidans. This cytochrome c is characterized by a relatively large redox potential (560 mV at pH 4.8) during the oxidation of Fe(II) to Fe(III) (Castelle et al., 2008).
Genes that code for the Ton system (TonB-ExbB-ExbD) protein complex were also present in all the samples. TonB can transport siderophores, hemes, and iron from transferrin/lactoferrin. It is understood to transport iron complexes the majority of the time; however, it is also known to uptake vitamin B12, nickel chelates, and carbohydrates (Noinaj et al., 2010; Garber et al., 2020).
Chromate is a strong oxidant and a potential mutagen to cells (Cummings et al., 2007). Genes encoding for NAD(P)H-dependent chromate reductase were found in the metagenome. NAD(P)H-dependent chromate reductase reduces Cr(VI) to Cr(III) thereby conferring resistance to chromate.
Arsenic, which is very toxic to cells, was also found in high concentrations in Laguna Caliente (Supplementary Table S2 in the Supplemental Information). Arsenic resistance is likely conferred by multiple arsenic resistance genes that were found in the metagenomes (Supplementary Table S3 in the Supplemental Information). As(V) and As(III) enter cells utilizing the Pst/Pit and GlpF transporters, respectively (Kalia and Joshi, 2009); genes encoding for both transporters are found in the metagenomes. Once in the cell, arsenic can be detoxified and transported through multiple mechanisms. The ars operon, arsRDABC, reduces As(V) into As(III) (via ArsC) and exports As(III) out of the cell (via ArsAB) (Kalia and Joshi, 2009). The metagenome contained arsA, arsB, arsC, and arsR; while arsD was missing in the samples; arsD is often absent in bacteria. arsD encodes a negative regulatory protein and arsR encodes an arsenite inducible repressor (Kalia and Joshi, 2009). Genes encoding for the arsenic methyltransferase (ArsM) protein are also present in the metagenome; ArsM detoxifies As(III) by converting As(III) to trimethylarsine, which is then exported out of the cell via passive diffusion (Bentley and Chasteen, 2002; Kalia and Joshi, 2009). The samples also contained genes encoding for arsenite oxidase (Aox) protein. Aox allows cells to oxidize extracellular As(III) to As(V), thereby gaining energy from this process (Kalia and Joshi, 2009).
The metagenomes also contained genes that encode the cusCBA operon (Supplementary Table S3 in the Supplemental Information). The cusCBA operon allows for metal efflux of Cu(I) and Ag(I). Additionally, genes encoding the Cue system (CopA and CueO) were also present. The Cue system detoxifies intracellular Cu(I) by exporting it to the periplasm (via CopA) and oxidizing the periplasmic Cu(I) to Cu(II) (via CueO) (Gudipaty et al., 2012). Note that genes conferring copper reduction were not found.
Genes encoding for the cusCBA operon were identified twice in the metagenome using the KEGG classification. The second identification of cus operon genes contains sequences that are homologous with two other metal efflux systems, Cnr and Czc. The KEGG classification system does not differentiate between these three systems using this sequence; i.e., they all have the same sequence ID’s for each component (Cnr/Czc/Cus: C-K15725, A-K15726, B-K15727). While the data just presented on the genes encoding the cusCBA operon are likely to be unique to cus, the additional set of sequences may represent the cnr operon, the czc operon, the cus operon again, or up to all three operons. cnrCBA is the structural locus of the cnr operon, and the cnrCBA encodes an efflux pump that exports Co(II) and Ni(II) (Tibazarwa et al., 2000). The czcCBA operon encodes another homologous efflux pump protein complex that has the ability to export Co(II), Zn(II), and Cd(II) (Rensing et al., 1997). Ubiquitous in all microorganisms, multidrug efflux pumps may also export heavy metals in addition to other substrates (Blanco et al., 2016).
Both the metagenomic and 16S rRNA gene sequencing efforts show that the 2013 Laguna Caliente fluid sample contains >90% Acidiphilium (Figure 2). The 2017 samples, on the other hand, contain a larger diversity of bacteria including Acidiphilium spp. and Asaia spp., known acidophiles (Figure 2). However, members of the Bradyrhizobiaceae, Chitinophagaceae, and Methylobacteriaceae families were also represented. Some of these could be laboratory contaminants, but all are commonly found in soils and none of which are acidophiles (Kämpfer et al., 2011; Kovaleva et al., 2014; Cao et al., 2017; Ormeño-Orrillo and Martínez-Romero, 2019), so it is unlikely that they inhabited the harsh conditions of Laguna Caliente. More likely, since the 2017 campaign occurred after rainfall, these soil biome microbes were probably washed into the lake from the surrounding vegetated crater walls. It is also possible that a higher pH allowed for more biodiversity. However, considering the samples from 2017 contained more than just lake fluids (i.e., sulfur clumps, that also hosted dead insects (Figure 1E), or lake bottom sediment) and that the additional bacteria discovered are commonly found in soils, it is likely that these additional bacteria were sourced from the surrounding soils in the Poás Volcano crater. This is corroborated by the result that the 2017 lake bottom sediment sample had the lowest percentage of Acidiphilium spp. and the highest percentage of these soil taxa. Moving forward, samples from the lake fluid should be scrutinized more, and field campaigns should prioritize collection of samples farther away from the lake shore.
The samples all contained surprisingly high potential functional diversity for remarkably low biodiversity, especially for the 2013 Laguna Caliente fluid sample which was largely dominated by the genus Acidiphilium. Many of the genes related to carbon cycling pathways we identified utilize glyoxylate and the breaking down of it into malate to produce energy. It is more profound that there are many ways for this system of bacteria to create energy including sulfur oxidation, iron oxidation, arsenic oxidation, utilization of simple sugars, utilization of complex sugars, and carbon fixation. The finding of genes consistent with polyhydroxybutyrate (PHB) granule formation and utilization suggests that microorganisms in these communities may be creating energy reserves to use in times of energy limitations (Ackermann et al., 1995). For example, PHB granules have been found in Acidiphilium spp. JA12-A1 (Ullrich et al., 2015).
The chief question resulting from these findings is “why would a microbial community need so many ways to produce energy when there is an essentially unlimited source of energy (sulfur, in many oxidation states) present?” While sulfur oxidation through the Sox system yields less energy in the absence of soxD gene, there still remains an effectively unlimited supply of sulfur in various redox states present in Laguna Caliente to make up for the absence of soxD gene. We provide three potential explanations to this question.
Our first hypothesis is that since Laguna Caliente is limiting in organic and inorganic carbon (Barry, 2019); bacteria living in the lake would have to utilize carbon fixation, carbon storage (PHB granules), and carbon anabolism (glyoxylate cycle, ethylmalonyl-CoA pathway) to make up for this lack of organics.
Our second hypothesis for why we have identified genes related to so many metabolic pathways in the samples is that the bacteria would need a sufficiently high yield of ATP to survive in Laguna Caliente. We have identified various adaptations this sample may utilize to deal with the harsh conditions of the lake. Of these, the ATP-dependent molecular chaperone proteins utilize ATP directly to ensure proper protein folding in times of elevated heat, and in some cases, acidity (Jayaraman et al., 1997). Transport proteins also utilize a high amount of ATP for active transport; of the efflux pumps described in this paper, the multidrug efflux pump, F1Fe-ATPase, CopA, arsAB, and the Ton system all use ATP to transport substrates across the cell membrane (Kalia and Joshi, 2009; Noinaj et al., 2010; Krulwich et al., 2011; Gudipaty et al., 2012; Blanco et al., 2016). Other environmental adaptation genes, such as those in the cusCBA, cnrCBA, czcCBA operons rely on secondary active transport, utilizing concentration gradients facilitated by ATP (Rensing et al., 1997; Tibazarwa et al., 2000; Long et al., 2012). The upregulation of glycolysis is also noted to occur in thermophiles to provide immediate energy to cope with heat stress (Wang et al., 2015).
Note that many of the genes and adaptations we discuss are also very common in microorganisms found in less extreme environments (e.g., the Ton system, the heat-shock operons, the multidrug efflux pumps, and acid-resistance genes). The confirmation of the ability for our bacteria to actually use these systems in times of environmental stress would have to be measured in the laboratory using other methods like transcriptomics. It has proven difficult to replicate the extreme conditions of Laguna Caliente in vitro, and to date, bacterial culturing and isolation experiments have been unsuccessful. While the metagenomic data does not prove that our samples utilize these systems and adaptations to survive, we show the capabilities of these bacteria. And while the presence of these genes in the metagenome do not prove these bacteria use these genes to survive, it is highly likely that they would use the pathways available to them to survive in Laguna Caliente; an environment that is severely extreme and hostile to life. Thus, it is conceivable that an exceptionally high yield of ATP would be needed for the upkeep of various mechanisms to ensure survival in Laguna Caliente.
Our third hypothesis for the presence of numerous metabolic pathways in our sample is the most eccentric. We start with asking the question, “what if the presence of multiple metabolic pathways is the defining feature that allows this community to survive in a volcanically active environment?” Recall that the Poás Volcano exhibited occasional-to-frequent phreatic-to-phreatomagmatic eruptions in recent years (Figure 1C). These eruptions have the ability to make Laguna Caliente extremely hot, decrease the pH exponentially, and influx large amounts of environmental toxins. While life has proven persistent in a host of hostile conditions, it is probable that these eruptions could make Laguna Caliente too hostile for even these resolute microorganisms.
It is conceivable that bacteria in Laguna Caliente could be transported away from the lake and into the surrounding soils via water level fluctuations, groundwater diffusion, or phreatic eruptions. So, prior to a phreatic eruption there would exist microbes both in Laguna Caliente and in the surrounding soils and connected groundwater system. Then later, a dramatic eruption could sterilize life in Laguna Caliente potentially draining the lake, which is what occurred in April 2017. Nevertheless, bacteria in the surrounding soils would be able to survive farther away from the eruption using complex carbon metabolism such as multiple carbon anabolism pathways, photosynthesis, and PHB granules formed in times of higher energy availability. The combination of these complex carbon metabolisms, in addition to simple carbon pathways and sulfur, iron, and arsenic oxidation would allow microbes to survive in ever-changing conditions and environments in the Poás Volcano crater. Then, when milder conditions return in Laguna Caliente and the lake reforms and expands, certain bacteria would be returned to their native environment: either washed in from the surrounding surface or via a connected groundwater system associated with the lake. This could help explain the presence of so many energy metabolism pathways in a sample of such low biodiversity. This could also help explain the relative persistence of the identified microbes, particularly Acidiphilium spp., which is known to be able to tolerate a wide variety of challenging conditions (Hiraishi et al., 1998; Dopson and Johnson 2012; Ullrich et al., 2015), between the November 2013 and March 2017 field campaigns.
In April 2017 after the second sampling, Poás Volcano entered an eruptive phase for over 2 years. Laguna Caliente was drained during explosive eruptions that added tephra and lavas to the crater floor, effectively sterilizing it. In October 2019, surface volcanic activity waned enough for the crater lake to reform and we were able to sample it in November 2019. While the results of that expedition are ongoing, future expeditions to the Poás Volcano will continue to characterize both fluid samples of Laguna Caliente and surrounding soil samples to further scrutinize this theory of microbial persistence through volcanic eruptions.
All landed missions to Mars that have focused on water, habitability, and astrobiology this millennium have had landing sites in sedimentary environments. While the conditions on Mars were more habitable early in its history, with an active water cycle for up to millions of years (e.g., Hoke et al., 2011), it is likely that surface waters did not persist for much longer. The Gale crater lake might have lasted 10,000 years (Grotzinger et al., 2015). Formation timescales of deltas on Mars suggest 1,000s of years (Hoke et al., 2014). Hoke and Hynek (2009) argued that there was a ∼200-million-year time period when fluvial valleys formed, albeit intermittently. Others argue that all the geomorphic fluviolacustrine features on Mars could have been formed from the melting of ice during punctuated events (e.g., Fastook and Head, 2015; Palumbo et al., 2018). To us, the geologically short duration of surface waters (e.g., Kereszturi, 2012) combined with harsh surface conditions (e.g., radiation, aridity, low temperature) imply a low probability for the origin and sustainment of life on Mars in sedimentary environments. Conversely, protracted groundwater and hydrothermal systems within the crust (e.g., Solomon et al., 2005; Ehlmann et al., 2011), and at individual volcanic centers active for most of Mars’ history (Robbins et al., 2011) provide a much higher probability for Martian life. Additionally, even if life existed in the brief episodes of surface waters, it is likely that when Mars dried up, life would have condensed onto smaller habitable niches such as acid-sulfate hydrothermal systems, which were likely in existence for billions of years.
Hydrothermal environments are suitable habitats as they provide all the ingredients for life (Walter and Des Marais, 1993; Schulze-Makuch et al., 2007; Hays et al., 2017), and they were likely one of the most stable habitats on Mars due to persistent volcanism and continuous groundwater (Hynek et al., 2013), as well as within long-lived impact-generated hydrothermal systems (e.g., Abramov and Kring, 2005; Osinski et al., 2020). Relict Martian hydrothermal systems were likely as dynamic and at times as hostile to life as Laguna Caliente is at present. Hence, this environment might be key to understanding if and how life on the Red Planet existed.
While hydrothermal environments could provide warm and moist locales for life to thrive, a drastic environmental change, such as an influx of high temperature- or high acidity fluids, has the potential to sterilize any residential microorganisms. Outside of these habitable niches, the Martian surface can also be sterilizing with incoming surface radiation and hostile atmospheric conditions such as cold temperatures, low humidity, and low pressure. In the event of microbe-killing events in Martian hydrothermal environments, the most clement environment for refuge would be connected groundwater systems. So, in order to maximize survival in Martian hydrothermal environments, microbes would need to be able to live in a variety of conditions such as hot, hydrothermal environments and connected groundwater systems. Thus, microbes on Mars would likely have to have had multiple adaptations to survive over millions or billions of years among hydrothermal features.
Our study is of considerable astrobiological interest because per our microbial community dominated by Acidiphilium spp., there appears to be various metabolic pathways and numerous adaptations that may possibly allow the community to survive in the ever-changing conditions and on the fringes of Laguna Caliente. The ability for Acidiphilium spp., in particular, to survive in a diverse set of extreme environments has been attributed to an abundant repertoire of genes acquired through horizontal gene transfer (HGT) (Li et al., 2020).
Like the microbial community in Laguna Caliente, any life in Martian hydrothermal environments would likely have developed considerable flexibility and tolerances to live in dynamic and extreme conditions. While our study suggests this bacterial community consists of highly evolved organisms suited for survival in the Poás Volcano, its rather low biodiversity suggests they live more independently compared to most described natural ecosystems. This makes it a rather simplistic system for maintaining survival in a hydrothermal environment. On the extreme end of this situation, single-species ecosystems are rare in nature, as very seldom can a singular species survive without symbiotic interactions with neighboring microbes (Chivian et al., 2008).
If life originated in the Noachian epoch (>3.7 bya) when Mars exhibited more widespread habitable conditions, we hypothesize that HGT events in the beginnings of Martian life, as was proposed to occur on Earth in the time of LUCA (e.g., Fournier et al., 2015), would benefit the development of functional diversity necessary to survive in extreme environments. Then in later times when global conditions became more hostile to life (aridification and acidification), microorganisms that live in communities with high functional diversity and low alpha diversity, like our microbial community dominated by Acidiphilium spp., would have the best chance of surviving in acid-sulfate hydrothermal systems. Thus, an independent single-celled organism (e.g., Chivian et al., 2008; Hynek et al., 2018) or a low biodiversity microbial community (described herein) that can survive in both Martian hydrothermal environments and in connected groundwater systems in times of volcanic dynamics may functionally resemble early Martian life.
One limitation of our study is that our microbial community likely utilizes aerobic metabolisms, and oxygen is scarce in the Martian atmosphere (Trainer et al., 2019), thereby making aerobic metabolism difficult. Nevertheless, aerobic methanotrophic metabolism has been proposed to be functional in the low temperatures, pressures, and oxygen concentrations present on Mars (Seto et al., 2019). Additionally, it has been suggested that sufficient oxygen for aerobic metabolisms would be present in briny near surface waters (Stamenković et al., 2018), such as hydrothermal systems. Although driven by abiotic processes, it has also been proposed that Mars underwent a large oxygenation event in the Late Noachian (Liu et al., 2021), which could have shaped metabolisms of future life. Oxygen may also be outgassed in hydrothermal environments or exist as free oxygen in fluids. Even if aerobic metabolisms could not be supported on Mars, it is probable that Martian microbes could develop analogous anaerobic metabolisms and environmental adaptations akin to our model community allowing for survival in extreme conditions and fringe environments. On Earth, obligate and facultative anaerobic acidophilic/acidotolerant thermophiles were cultivated from terrestrial hot springs and deep-sea hydrothermal vent chimneys, although their metabolic strategies were undetermined (Prokofeva et al., 2005).
While the low biodiversity and low biomass of our isolates from Laguna Caliente may suggest life on Mars is a difficult proposition, we argue that these findings show extreme perseverance of life in hostile and dynamic Mars-like conditions. Microorganisms on Earth have evolved to survive and thrive in a great host of conditions. If life on Mars began in a similar manner as life on Earth, it is conceivable that microorganisms on Mars could have evolved in an analogous manner to extremophiles on Earth, developing some of the adaptations found in this study.
Hydrothermal systems were likely the longest-lived habitable environment through Mars’ history. This environment does not come without its challenges for life, which include potential sterilization events as a result of volcanic activity. The key solution for this challenge would be a self-sufficient microorganism or low biodiversity ecosystem that contains numerous adaptations and metabolisms capable of withstanding minor environmental changes and capable of surviving in connected groundwater systems during more extreme environmental fluctuations. This study addresses the challenges for life in Martian hydrothermal systems, using an analog system from Costa Rica, to show that life can in fact adapt to such environmental extremes. In the Poás Volcano, a microbial community largely dominated by Acidiphilium spp. appears to be a suitable model community that can even withstand phreatic eruptions. Our study shows that life contains the genetic potential and functional flexibility to survive in such extreme environments. Appropriately, we propose that acid-sulfate hydrothermal systems be prioritized in the search for life on Mars.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Written informed consent was obtained from BH for the publication of any potentially identifiable images or data included in this article.
BH is the academic advisor of JW and ND, and he conceived this project. BH performed field sampling with GA. JW performed extractions of the samples. ND performed post-sequence processing of all the samples and submitted it to the NCBI Sequence Read Database. JW investigated the genes of interest from the sequence reads and wrote the first draft of the manuscript. ND, BH, and JW refined sections of the manuscript from the original draft. All authors contributed to manuscript revision, read, and approved the submitted version.
The Leadership Education for Advancement and Promotion (LEAP) program at the University of Colorado Boulder and NASA’s Early Career Fellowship #NNX12AF20G helped fund this research.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors would like to thank the Leadership Education for Advancement and Promotion (LEAP) program at the University of Colorado Boulder and NASA’s Early Career Fellowship #NNX12AF20G for financial support. We sincerely appreciate the in-country support of Alvarado Induni Guillermo, Instituto Costarricense de Electricidad (ICE), and Observatorio Vulcanológico y Sismológico de Costa Rica (OVISCORI). We also are indebted to Alexis Templeton and Noah Fierer for use of their laboratory facilities to complete this work.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fspas.2022.817900/full#supplementary-material
Abramov, O., and Kring, D. A. (2005). Impact-induced Hydrothermal Activity on Early Mars. J. Geophys. Res. 110 (E12). doi:10.1029/2005JE002453
Ackermann, J.-u., Müller, S., Lösche, A., Bley, T., and Babel, w. (1995). Methylobacterium Rhodesianum Cells Tend to Double the DNA Content under Growth Limitations and Accumulate PHB. J. Biotechnol. 39 (1), 9–20. doi:10.1016/0168-1656(94)00138-3
Amils, R., González-Toril, E., Fernández-Remolar, D., Gómez, F., Aguilera, Á., and Rodríguez, N. (2007). Extreme Environments as Mars Terrestrial Analogs: The Rio Tinto Case. Planet. Space Sci. 55(3), 370–381. doi:10.1016/j.pss.2006.02.006
Andrews-Hanna, J. C., and Phillips, R. J. (2007). Hydrological Modeling of Outflow Channels and Chaos Regions on Mars. Journal of Geophysical Research. Planets 112 (E8). doi:10.1029/2006JE002881
Baker-Austin, C., and Dopson, M. (2007). Life in Acid: pH Homeostasis in Acidophiles. Trends Microbiol. 15 (4), 165–171. doi:10.1016/j.tim.2007.02.005
Banerdt, W. B., Smrekar, S. E., Banfield, D., Giardini, D., Golombek, M., Johnson, C. L., et al. (2020). Initial Results from the InSight mission on Mars. Nat. Geosci. 13 (3), 183–189. doi:10.1038/s41561-020-0544-y
Barry, P. (2019). Carbon and noble Gas Data from Costa Rica Seeps, Version 1.0. Interdiscip. Earth Data Alliance (Ieda). Accessed 2020-10-18. doi:10.1594/IEDA/111271
Bengtsson-Palme, J., Hartmann, M., Eriksson, K. M., Pal, C., Thorell, K., Larsson, D. G. J., et al. (2015). metaxa2: Improved Identification and Taxonomic Classification of Small and Large Subunit rRNA in Metagenomic Data. Mol. Ecol. Resour. 15 (6), 1403–1414. doi:10.1111/1755-0998.12399
Bentley, R., and Chasteen, T. G. (2002). Microbial Methylation of Metalloids: Arsenic, Antimony, and Bismuth. Microbiology and Molecular Biology Reviews. MMBR 66 (2), 250–271. doi:10.1128/MMBR.66.2.250-271.2002
Bibring, J. P., Langevin, Y., Gendrin, A., Gondet, B., Poulet, F., Berthé, M., et al. (2005). Mars Surface Diversity as Revealed by the OMEGA/Mars Express Observations. Science 307 (5715), 1576. doi:10.1126/science.1108806
Black, S. R., and Hynek, B. M. (2018). Characterization of Terrestrial Hydrothermal Alteration Products with Mars Analog Instrumentation: Implications for Current and Future Rover Investigations. Icarus 307, 235–259. doi:10.1016/j.icarus.2017.10.032
Black, S. R., Hynek, B. M., and Alvarado, G. E. (2015). “Spectral Identification of Acid Sulfate Alteration Products in Costa Rica Volcanoes: Implications for Early Mars [abstract 2260],” in 46th Lunar and Planetary Science Conference, Houston (Lunar and Planetary Institute). LPI Contribution No. 1832.
Black, S. R., Hynek, B. M., Hoover, R., Beckerman, L. G., and Alvarado, G. E. (2016). “Characterization of Hydrothermal Alteration in Costa Rica: Mineralogy, Methodology, and Implications for Mars [abstract 2546],” in 47th Lunar and Planetary Science Conference, Houston (Lunar and Planetary Institute). LPI Contribution No. 1903.
Blanco, P., Hernando-Amado, S., Reales-Calderon, J. A., Corona, F., Lira, F., Alcalde-Rico, M., et al. (2016). Bacterial Multidrug Efflux Pumps: Much More Than Antibiotic Resistance Determinants. Microorganisms 4 (1). doi:10.3390/microorganisms4010014
Braun, V. (2003). Iron Uptake by Escherichia coli. Front. Bioscience-Landmark 8 (6), 1409–1421. doi:10.2741/1232
Brunold, C. (1993). “Regulatory Interactions between Sulfate and Nitrate Assimilation,” in Sulfur Nutrition and Assimilation in Higher PlantsRegulatory Agricultural and Environmental Aspects. Editors L. J. de Kok, I. Stulen, H. Rennenberg, C. Brunold, Rauseret al. (The Hague: SPB Academic Publishing), 61–76.
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 12 (1), 59–60. doi:10.1038/nmeth.3176
Callahan, B. J., McMurdie, P. J., and Holmes, S. P. (2017). Exact Sequence Variants Should Replace Operational Taxonomic Units in Marker-Gene Data Analysis. ISME J. 11 (12), 2639–2643. doi:10.1038/ismej.2017.119
Cao, M., Huang, J., Li, J., Qiao, Z., and Wang, G. (2017). Edaphobaculum Flavum Gen. nov., Sp. nov., a Member of Family Chitinophagaceae, Isolated from Grassland Soil. Int. J. Syst. Evol. Microbiol. 67 (11), 4475–4481. doi:10.1099/ijsem.0.002316
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 7 (5), 335–336. doi:10.1038/nmeth.f.303
Carini, P., Marsden, P. J., Leff, J. W., Morgan, E. E., Strickland, M. S., and Fierer, N. (2016). Relic DNA Is Abundant in Soil and Obscures Estimates of Soil Microbial Diversity. Nat. Microbiol. 2 (3), 16242. doi:10.1038/nmicrobiol.2016.242
Carr, M. H., and Head, J. W. (2010). Geologic History of Mars. Earth Planet. Sci. Lett. 294 (3), 185–203. doi:10.1016/j.epsl.2009.06.042
Castelle, C., Guiral, M., Malarte, G., Ledgham, F., Leroy, G., Brugna, M., et al. (2008). A New Iron-oxidizing/O2-Reducing Supercomplex Spanning Both Inner and Outer Membranes, Isolated from the Extreme Acidophile Acidithiobacillus Ferrooxidans. J. Biol. Chem. 283 (38), 25803–25811. doi:10.1074/jbc.M802496200
Changela, H. G., Chatzitheodoridis, E., Antunes, A., Beaty, D., Bouw, K., Bridges, J. C., et al. (2021). Mars: New Insights and Unresolved Questions. Int. J. Astrobiology 20 (6), 394–426. doi:10.1017/S1473550421000276
Chew, S. Y., Chee, W. J. Y., and Than, L. T. L. (2019). The Glyoxylate Cycle and Alternative Carbon Metabolism as Metabolic Adaptation Strategies of Candida Glabrata: Perspectives from Candida Albicans and Saccharomyces cerevisiae. J. Biomed. Sci. 26 (1), 52. doi:10.1186/s12929-019-0546-5
Chivian, D., Brodie, E. L., Alm, E. J., Culley, D. E., Dehal, P. S., DeSantis, T. Z., et al. (2008). Environmental Genomics Reveals a Single-Species Ecosystem Deep within Earth. Science 322 (5899), 275. doi:10.1126/science.1155495
Chojnacki, M., and Hynek, B. M. (2008). Geological Context of Water-Altered Minerals in Valles Marineris, Mars. Journal of Geophysical Research. Planets 113 (E12). doi:10.1029/2007JE003070
Cockell, C. S., Balme, M., Bridges, J. C., Davila, A., and Schwenzer, S. P. (2012). Uninhabited Habitats on Mars. Icarus 217 (1), 184–193. doi:10.1016/j.icarus.2011.10.025
Craddock, R. A., and Greeley, R. (2009). Minimum Estimates of the Amount and Timing of Gases Released into the Martian Atmosphere from Volcanic Eruptions. Icarus 204 (2), 512–526. doi:10.1016/j.icarus.2009.07.026
Cummings, D. E., Fendorf, S., Singh, N., Sani, R. K., Peyton, B. M., and Magnuson, T. S. (2007). Reduction of Cr(VI) under Acidic Conditions by the Facultative Fe(III)-Reducing Bacterium Acidiphilium Cryptum. Environ. Sci. Tech. 41 (1), 146–152. doi:10.1021/es061333k
Davila, A. F. (2021). Life on Mars: Independent Genesis or Common Ancestor. Astrobiology 21 (7), 802–812. doi:10.1089/ast.2020.2397
Di Achille, G., and Hynek, B. M. (2010). Ancient Ocean on Mars Supported by Global Distribution of Deltas and Valleys. Nat. Geosci. 3 (7), 459–463. doi:10.1038/ngeo891
Dohm, J. M., and Tanaka, K. L. (1999). Geology of the Thaumasia Region, Mars: Plateau Development, valley Origins, and Magmatic Evolution. Planet. Space Sci. 47 (3), 411–431. doi:10.1016/S0032-0633(98)00141-X
Domagal-Goldman, S. D., Wright, K. E., Domagal-Goldman, S. D., Wright, K. E., Adamala, K., Arina de la Rubia, L., et al. (2016). The Astrobiology Primer v2.0. Astrobiology 16 (8), 561–653. doi:10.1089/ast.2015.1460
Dopson, M., and Johnson, D. B. (2012). Biodiversity, Metabolism and Applications of Acidophilic Sulfur-Metabolizing Microorganisms. Environ. Microbiol. 14 (10), 2620–2631. doi:10.1111/j.1462-2920.2012.02749.x
Edwards, K. J., Bond, P. L., Druschel, G. K., McGuire, M. M., Hamers, R. J., and Banfield, J. F. (2000). Geochemical and Biological Aspects of Sulfide mineral Dissolution: Lessons from Iron Mountain, California. Chem. Geology. 169 (3), 383–397. doi:10.1016/S0009-2541(00)00216-3
Ehlmann, B. L., Mustard, J. F., Clark, R. N., Swayze, G. A., and Murchie, S. L. (2011). Evidence for Low-Grade Metamorphism, Hydrothermal Alteration, and Diagenesis on Mars from Phyllosilicate Mineral Assemblages. Clays and Clay Minerals 59 (4), 359–377. doi:10.1346/CCMN.2011.0590402
Ehrenfreund, P., Röling, W. F. M., Thiel, C. S., Quinn, R., Sephton, M. A., Stoker, C., et al. (2011). Astrobiology and Habitability Studies in Preparation for Future Mars Missions: Trends from Investigating Minerals, Organics and Biota. Int. J. Astrobiology 10 (3), 239–253. doi:10.1017/S1473550411000140
El Maarry, M. R., Dohm, J. M., Marzo, G. A., Fergason, R., Goetz, W., Heggy, E., et al. (2012). Searching for Evidence of Hydrothermal Activity at Apollinaris Mons, Mars. Icarus 217 (1), 297–314. doi:10.1016/j.icarus.2011.10.022
Fairén, A. G., Dohm, J. M., Baker, V. R., de Pablo, M. A., Ruiz, J., Ferris, J. C., et al. (2003). Episodic Flood Inundations of the Northern plains of Mars. Icarus 165 (1), 53–67. doi:10.1016/S0019-1035(03)00144-1
Fassett, C. I., and Head, J. W. (2008a). The Timing of Martian valley Network Activity: Constraints from Buffered Crater Counting. Icarus 195 (1), 61–89. doi:10.1016/j.icarus.2007.12.009
Fassett, C. I., and Head, J. W. (2008b). Valley Network-Fed, Open-basin Lakes on Mars: Distribution and Implications for Noachian Surface and Subsurface Hydrology. Icarus 198 (1), 37–56. doi:10.1016/j.icarus.2008.06.016
Fastook, J. L., and Head, J. W. (2015). Glaciation in the Late Noachian Icy Highlands: Ice Accumulation, Distribution, Flow Rates, Basal Melting, and Top-Down Melting Rates and Patterns. Sci. Planet. Space Sci. 106, 82–98. doi:10.1016/j.pss.2014.11.028
Foster, J. W. (2004). Escherichia coli Acid Resistance: Tales of an Amateur Acidophile. Nat. Rev. Microbiol. 2 (11), 898–907. doi:10.1038/nrmicro1021
Fournier, G. P., Andam, C. P., and Gogarten, J. P. (2015). Ancient Horizontal Gene Transfer and the Last Common Ancestors. BMC Evol. Biol. 15, 70. doi:10.1186/s12862-015-0350-0
Friedrich, C. G. (1997). “Physiology and Genetics of Sulfur-Oxidizing Bacteria,” in Advances in Microbial Physiology. Editor R. K. Poole (Academic Press), 235–289. doi:10.1016/s0065-2911(08)60018-1
Friedrich, C. G., Rother, D., Bardischewsky, F., Quentmeier, A., and Fischer, J. (2001). Oxidation of Reduced Inorganic Sulfur Compounds by Bacteria: Emergence of a Common Mechanism. Appl. Environ. Microbiol. 67 (7), 2873. doi:10.1128/AEM.67.7.2873-2882.2001
Garber, A. I., Nealson, K. H., Okamoto, A., McAllister, S. M., Chan, C. S., Barco, R. A., et al. (2020). FeGenie: A Comprehensive Tool for the Identification of Iron Genes and Iron Gene Neighborhoods in Genome and Metagenome Assemblies. Front. Microbiol. 11 (37). doi:10.3389/fmicb.2020.00037
Gaucher, E. A., Govindarajan, S., and Ganesh, O. K. (2008). Palaeotemperature Trend for Precambrian Life Inferred from Resurrected Proteins. Nature 451 (7179), 704–707. doi:10.1038/nature06510
Gendrin, A., Mangold, N., Bibring, J. P., Langevin, Y., Gondet, B., Poulet, F., et al. (2005). Sulfates in Martian Layered Terrains: The OMEGA/Mars Express View. Science 307 (5715), 1587. doi:10.1126/science.1109087
Ghosh, W., and Dam, B. (2009). Biochemistry and Molecular Biology of Lithotrophic Sulfur Oxidation by Taxonomically and Ecologically Diverse Bacteria and Archaea. FEMS Microbiol. Rev. 33 (6), 999–1043. doi:10.1111/j.1574-6976.2009.00187.x
Giulio, M. D. (2003). The Universal Ancestor Was a Thermophile or a Hyperthermophile: Tests and Further Evidence. J. Theor. Biol. 221 (3), 425–436. doi:10.1006/jtbi.2003.3197
González-Toril, E., Llobet-Brossa, E., Casamayor, E. O., Amann, R., and Amils, R. (2003). Microbial Ecology of an Extreme Acidic Environment, the Tinto River. Appl. Environ. Microbiol. 69 (8), 4853. doi:10.1128/AEM.69.8.4853-4865.2003
Goudge, T. A., Fassett, C. I., Head, J. W., Mustard, J. F., and Aureli, K. L. (2016). Insights into Surface Runoff on Early Mars from Paleolake basin Morphology and Stratigraphy. Geology 44 (6), 419–422. doi:10.1130/G37734.1
Grotzinger, J. P., Gupta, S., Malin, M. C., Rubin, D. M., Schieber, J., Siebach, K., et al. (2015). Deposition, Exhumation, and Paleoclimate of an Ancient lake deposit, Gale Crater. Mars. Sci. 350 (6257), aac7575. doi:10.1126/science.aac7575
Gudipaty, S. A., Larsen, A. S., Rensing, C., and McEvoy, M. M. (2012). Regulation of Cu(I)/Ag(I) Efflux Genes in Escherichia coli by the Sensor Kinase CusS. FEMS Microbiol. Lett. 330 (1), 30–37. doi:10.1111/j.1574-6968.2012.02529.x
Gulick, V. C. (1998). Magmatic Intrusions and a Hydrothermal Origin for Fluvial Valleys on Mars. J. Geophys. Res. Planets 103 (E8), 19365–19387. doi:10.1029/98JE01321
Gulick, V. C. (2001). Origin of the valley Networks on Mars: a Hydrological Perspective. Geomorphology 37 (3), 241–268. doi:10.1016/S0169-555X(00)00086-6
Harrison, A. P. (1981). Acidiphilium Cryptum Gen. nov., Sp. nov., Heterotrophic Bacterium from Acidic Mineral Environments. Int. J. Syst. Evol. Microbiol. 31 (3), 327–332. doi:10.1099/00207713-31-3-327
Hauck Ii, S. A., and Phillips, R. J. (2002). Thermal and Crustal Evolution of Mars. J. Geophys. Res. Planets 107 (E7), 6–1–6-19. doi:10.1029/2001JE001801
Hays, L. E., Graham, H. V., Des Marais, D. J., Hausrath, E. M., Horgan, B., McCollom, T. M., et al. (2017). Biosignature Preservation and Detection in Mars Analog Environments. Astrobiology 17 (4), 363–400. doi:10.1089/ast.2016.1627
Hiraishi, A., Nagashima, K. V. P., Matsuura, K., Shimada, K., Takaichi, S., Wakao, N., et al. (1998). Phylogeny and Photosynthetic Features of Thiobacillus Acidophilus and Related Acidophilic Bacteria: its Transfer to the Genus Acidiphilium as Acidiphilium Acidophilum Comb. Nov. Int. J. Syst. Evol. Microbiol. 48 (4), 1389–1398. doi:10.1099/00207713-48-4-1389
Hoke, M. R. T., and Hynek, B. M. (2009). Roaming Zones of Precipitation on Ancient Mars as Recorded in valley Networks. Journal of Geophysical Research. Planets 114 (E8). doi:10.1029/2008JE003247
Hoke, M. R. T., Hynek, B. M., Di Achille, G., and Hutton, E. W. H. (2014). The Effects of Sediment Supply and Concentrations on the Formation Timescale of Martian Deltas. Icarus 228, 1–12. doi:10.1016/j.icarus.2013.09.017
Hoke, M. R. T., Hynek, B. M., and Tucker, G. E. (2011). Formation Timescales of Large Martian valley Networks. Earth Planet. Sci. Lett. 312 (1), 1–12. doi:10.1016/j.epsl.2011.09.053
Horvath, D. G., Moitra, P., Hamilton, C. W., Craddock, R. A., and Andrews-Hanna, J. C. (2021). Evidence for Geologically Recent Explosive Volcanism in Elysium Planitia, Mars. Icarus 365, 114499. doi:10.1016/j.icarus.2021.114499
Huerta-Cepas, J., Szklarczyk, D., Forslund, K., Cook, H., Heller, D., Walter, M. C., et al. (2015). eggNOG 4.5: a Hierarchical Orthology Framework with Improved Functional Annotations for Eukaryotic, Prokaryotic and Viral Sequences. Nucleic Acids Res. 44 (D1), D286–D293. doi:10.1093/nar/gkv1248
Hynek, B. M., Beach, M., and Hoke, M. R. T. (2010). Updated Global Map of Martian valley Networks and Implications for Climate and Hydrologic Processes. Journal of Geophysical Research. Planets 115 (E9). doi:10.1029/2009JE003548
Hynek, B. M., McCollom, T. M., Marcucci, E. C., Brugman, K., and Rogers, K. L. (2013). Assessment of Environmental Controls on Acid-Sulfate Alteration at Active Volcanoes in Nicaragua: Applications to Relic Hydrothermal Systems on Mars. J. Geophys. Res. Planets 118 (10), 2083–2104. doi:10.1002/jgre.20140
Hynek, B. M., McCollom, T. M., McHenry, L. J., and Alvarado, G. E. (2014). “Assessing Hydrothermal Alteration on Early Mars through Analog Environments in Nicaragua, Costa Rica, Ice-Land, and Hawaii [abstract 2172],” in 45thLunar and Planetary Science Conference, Houston (Lunar and Planetary Institute). LPI Contribution No. 1777.
Hynek, B. M., and Phillips, R. J. (2003). New Data Reveal Mature, Integrated Drainage Systems on Mars Indicative of Past Precipitation. Geology 31 (9), 757–760. doi:10.1130/G19607.1
Hynek, B. M., Rogers, K. L., Antunovich, M., Avard, G., and Alvarado, G. E. (2018). Lack of Microbial Diversity in an Extreme Mars Analog Setting. Astrobiology 18 (7), 923–933. Poás Volcano, Costa Rica. doi:10.1089/ast.2017.1719
Jayaraman, G. C., Penders, J. E., and Burne, R. A. (1997). Transcriptional Analysis of the Streptococcus Mutans hrcA, grpE and dnaK Genes and Regulation of Expression in Response to Heat Shock and Environmental Acidification. Mol. Microbiol. 25 (2), 329–341. doi:10.1046/j.1365-2958.1997.4671835.x
Johnson, D. B., Hedrich, S., and Pakostova, E. (2017). Indirect Redox Transformations of Iron, Copper, and Chromium Catalyzed by Extremely Acidophilic Bacteria. Front. Microbiol. 8, 211. doi:10.3389/fmicb.2017.00211
Kalia, K., and Joshi, D. N. (2009). “CHAPTER 72-Detoxification of Arsenic,” in Handbook of Toxicology of Chemical Warfare Agents. Editor R. C. Gupta. (San Diego: Academic Press), 1083–1100. doi:10.1016/b978-012374484-5.00072-9:
Kämpfer, P., Lodders, N., and Falsen, E. (2011). Hydrotalea Flava Gen. nov., Sp. nov., a New Member of the Phylum Bacteroidetes and Allocation of the Genera Chitinophaga, Sediminibacterium, Lacibacter, Flavihumibacter, Flavisolibacter, Niabella, Niastella, Segetibacter, Parasegetibacter, Terrimonas, Ferruginibacter, Filimonas and Hydrotalea to the Family Chitinophagaceae Fam. Nov. Int. J. Syst. Evol. Microbiol. 61 (3), 518–523. doi:10.1099/ijs.0.023002-0
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28 (1), 27–30. doi:10.1093/nar/28.1.27
Kereszturi, A. (2012). Review of Wet Environment Types on Mars with Focus on Duration and Volumetric Issues. Astrobiology 12 (6), 586–600. doi:10.1089/ast.2011.0686
Kereszturi, A., Bradak, B., Chatzitheodoridis, E., and Ujvari, G. (2016). Indicators and Methods to Understand Past Environments from ExoMars Rover Drills. Origins Life Evol. Biospheres 46 (4), 435–454. doi:10.1007/s11084-016-9492-3
Kishimoto, N., Fukaya, F., Inagaki, K., Sugio, T., Tanaka, H., and Tano, T. (1995). Distribution of Bacteriochlorophyll a Among Aerobic and Acidophilic Bacteria and Light-Enhanced CO2-incorporation in Acidiphilium Rubrum. FEMS Microbiol. Ecol. 16 (4), 291–296. doi:10.1111/j.1574-6941.1995.tb00293.x
Kovaleva, J., Degener, J. E., and van der Mei, H. C. (2014). <span Class="named-Content Genus-Species" Id="named-Content-1">Methylobacterium</span> and its Role in Health Care-Associated Infection. J. Clin. Microbiol. 52 (5), 1317. doi:10.1128/JCM.03561-13
Krulwich, T. A., Sachs, G., and Padan, E. (2011). Molecular Aspects of Bacterial pH Sensing and Homeostasis. Nat. Rev. Microbiol. 9 (5), 330–343. doi:10.1038/nrmicro2549
Küsel, K., Dorsch, T., Acker, G., and Stackebrandt, E. (1999). Microbial Reduction of Fe(III) in Acidic Sediments: Isolation of <em>Acidiphilium Cryptum</em> JF-5 Capable of Coupling the Reduction of Fe(III) to the Oxidation of Glucose. Appl. Environ. Microbiol. 65 (8), 3633. doi:10.1128/AEM.65.8.3633-3640.1999
Lane, D. J., Harrison, A. P., Stahl, D., Pace, B., Giovannoni, S. J., Olsen, G. J., et al. (1992). Evolutionary Relationships Among Sulfur- and Iron-Oxidizing Eubacteria. J. Bacteriol. 174 (1), 269–278. doi:10.1128/jb.174.1.269-278.1992
Li, D., Liu, C.-M., Luo, R., Sadakane, K., and Lam, T.-W. (2015). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31 (10), 1674–1676. doi:10.1093/bioinformatics/btv033
Li, L., Liu, Z., Zhang, M., Meng, D., Liu, X., Wang, P., et al. (2020). Insights into the Metabolism and Evolution of the Genus Acidiphilium, a Typical Acidophile in Acid Mine Drainage. mSystems 5 (6), e00867–00820. doi:10.1128/mSystems.00867-20
Liu, J., Michalski, J. R., Tan, W., He, H., Ye, B., and Xiao, L. (2021). Anoxic Chemical Weathering under a Reducing Greenhouse on Early Mars. Nat. Astron. 5 (5), 503–509. doi:10.1038/s41550-021-01303-5
Liu, Y., Yang, H., Zhang, X., Xiao, Y., Guo, X., and Liu, X. (2016). Genomic Analysis Unravels Reduced Inorganic Sulfur Compound Oxidation of Heterotrophic Acidophilic Acidicaldus Sp. Strain DX-1. Biomed. Res. Int. 2016, 8137012. doi:10.1155/2016/8137012
Long, F., Su, C.-C., Lei, H.-T., Bolla, J. R., Do, S. V., and Yu, E. W. (2012). Structure and Mechanism of the Tripartite CusCBA Heavy-Metal Efflux Complex. Philosophical Transactions of the Royal Society of London. Series B. Biol. Sci. 367 (1592), 1047–1058. doi:10.1098/rstb.2011.0203
Mangold, N., Gupta, S., Gasnault, O., Dromart, G., Tarnas, J. D., Sholes, S. F., et al. (2021). Perseverance Rover Reveals an Ancient delta-lake System and Flood Deposits at Jezero Crater. Mars. Sci. 374 (6568), 711–717. doi:10.1126/science.abl4051
Manor, O., and Borenstein, E. (2015). MUSiCC: a Marker Genes Based Framework for Metagenomic Normalization and Accurate Profiling of Gene Abundances in the Microbiome. Genome Biol. 16 (1), 53. doi:10.1186/s13059-015-0610-8
Marcucci, E. C., Hynek, B. M., Kierein-Young, K. S., and Rogers, K. L. (2013). Visible-near-infrared Reflectance Spectroscopy of Volcanic Acid-Sulfate Alteration in Nicaragua: Analogs for Early Mars. J. Geophys. Res. Planets 118 (10), 2213–2233. doi:10.1002/jgre.20159
Martin, M. (2011). Cutadapt Removes Adapter Sequences from High-Throughput Sequencing readsNext Generation Sequencing Data AnalysisDO - 10.14806/ej.17.1.200. EMBnet.journal Vol. 17 (No 1). doi:10.14806/ej.17.1.200
Martin, W., Baross, J., Kelley, D., and Russell, M. J. (2008). Hydrothermal Vents and the Origin of Life. Nat. Rev. Microbiol. 6 (11), 805–814. doi:10.1038/nrmicro1991
Martin, W., and Russell, M. J. (2003). On the Origins of Cells: a Hypothesis for the Evolutionary Transitions from Abiotic Geochemistry to Chemoautotrophic Prokaryotes, and from Prokaryotes to Nucleated Cells. Philosophical Transactions of the Royal Society of London. Series B. Biol. Sci. 358 (1429), 59–85. doi:10.1098/rstb.2002.1183
Martínez-Cruz, M., van Bergen, M. J., Takano, B., Fernández-Soto, E., and Barquero-Hernández, J. (2019). “Behaviour of Polythionates in the Acid Lake of Poás Volcano: Insights into Changes in the Magmatic-Hydrothermal Regime and Subaqueous Input of Volatiles,” in Poás Volcano: The Pulsing Heart of Central America Volcanic Zone. Editors F. Tassi, O. Vaselli, and R. A. Mora Amador (Cham: Springer International Publishing), 155–202. doi:10.1007/978-3-319-02156-0_7
McCollom, T. M., and Hynek, B. M. (2005). A Volcanic Environment for Bedrock Diagenesis at Meridiani Planum on Mars. Nature 438 (7071), 1129–1131. doi:10.1038/nature04390
Michalski, J. R., Onstott, T. C., Mojzsis, S. J., Mustard, J., Chan, Q. H. S., Niles, P. B., et al. (2018). The Martian Subsurface as a Potential Window into the Origin of Life. Nat. Geosci. 11 (1), 21–26. doi:10.1038/s41561-017-0015-2
Mirete, S., Morgante, V., and González-Pastor, J. E. (2017). “Acidophiles: Diversity and Mechanisms of Adaptation to Acidic Environments,” in Adaption of Microbial Life to Environmental Extremes: Novel Research Results and Application. Editors H. Stan-Lotter, and S. Fendrihan (Cham: Springer International Publishing), 227–251. doi:10.1007/978-3-319-48327-6_9
Moitra, P., Horvath, D. G., and Andrews-Hanna, J. C. (2021). Investigating the Roles of Magmatic Volatiles, Ground Ice and Impact-Triggering on a Very Recent and Highly Explosive Volcanic Eruption on Mars. Earth Planet. Sci. Lett. 567, 116986. doi:10.1016/j.epsl.2021.116986
Moore, J. E., McCalmont, M., Xu, J., Millar, B. C., and Heaney, N. (2002). Asaia sp., an Unusual Spoilage Organism of Fruit-Flavored Bottled Water. Appl. Environ. Microbiol. 68 (8), 4130–4131. doi:10.1128/AEM.68.8.4130-4131.2002
National Research Council (2011). Vision and Voyages for Planetary Science in the Decade 2013-2022. Washington, DC: The National Academies Press.
Nisbet, E. G., and Sleep, N. H. (2001). The Habitat and Nature of Early Life. Nature 409 (6823), 1083–1091. doi:10.1038/35059210
Noinaj, N., Guillier, M., Barnard, T. J., and Buchanan, S. K. (2010). TonB-Dependent Transporters: Regulation, Structure, and Function. Annu. Rev. Microbiol. 64 (1), 43–60. doi:10.1146/annurev.micro.112408.134247
Nordstrom, D. K., and Alpers, C. N. (1999). Negative pH, Efflorescent Mineralogy, and Consequences for Environmental Restoration at the Iron Mountain Superfund Site, California. Proc. Natl. Acad. Sci. 96 (7), 3455. doi:10.1073/pnas.96.7.3455
Okubo, Y., Yang, S., Chistoserdova, L., and Lidstrom, M. E. (2010). Alternative Route for Glyoxylate Consumption during Growth on Two-Carbon Compounds by Methylobacterium Extorquens AM1. J. Bacteriol. 192 (7), 1813. doi:10.1128/JB.01166-09
Ormeño-Orrillo, E., and Martínez-Romero, E. (2019). A Genomotaxonomy View of the Bradyrhizobium Genus. Front. Microbiol. 10 (1334). doi:10.3389/fmicb.2019.01334
Osinski, G. R., Cockell, C. S., Pontefract, A., and Sapers, H. M. (2020). The Role of Meteorite Impacts in the Origin of Life. Astrobiology 20 (9), 1121–1149. doi:10.1089/ast.2019.2203
Palumbo, A. M., Head, J. W., and Wordsworth, R. D. (2018). Late Noachian Icy Highlands Climate Model: Exploring the Possibility of Transient Melting and Fluvial/lacustrine Activity through Peak Annual and Seasonal Temperatures. Icarus 300, 261–286. doi:10.1016/j.icarus.2017.09.007
Peña-Ocaña, B. A., Ovando-Ovando, C. I., Puente-Sánchez, F., Tamames, J., Servín-Garcidueñas, L. E., González-Toril, E., et al. (2022). Metagenomic and Metabolic Analyses of Poly-Extreme Microbiome from an Active Crater Volcano lake. Environ. Res. 203, 111862. doi:10.1016/j.envres.2021.111862
Phillips, R. J., Zuber, M. T., Solomon, S. C., Golombek, M. P., Jakosky, B. M., Banerdt, W. B., et al. (2001). Ancient Geodynamics and Global-Scale Hydrology on Mars. Science 291 (5513), 2587. doi:10.1126/science.1058701
Prokofeva, M. I., Kublanov, I. V., Nercessian, O., Tourova, T. P., Kolganova, T. V., Lebedinsky, A. V., et al. (2005). Cultivated Anaerobic Acidophilic/acidotolerant Thermophiles from Terrestrial and Deep-Sea Hydrothermal Habitats. Extremophiles 9 (6), 437–448. doi:10.1007/s00792-005-0461-4
Prosser, J. T., and Carr, M. J. (1987). Poás Volcano, Costa Rica: Geology of the summit Region and Spatial and Temporal Variations Among the Most Recent Lavas. J. Volcanology Geothermal Res. 33 (1), 131–146. doi:10.1016/0377-0273(87)90057-6
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 41 (D1), D590–D596. doi:10.1093/nar/gks1219
Rensing, C., Pribyl, T., and Nies, D. H. (1997). New Functions for the Three Subunits of the CzcCBA Cation-Proton Antiporter. J. Bacteriol. 179 (22), 6871. doi:10.1128/jb.179.22.6871-6879.1997
Robbins, S. J., Achille, G. D., and Hynek, B. M. (2011). The Volcanic History of Mars: High-Resolution Crater-Based Studies of the Calderas of 20 Volcanoes. Icarus 211 (2), 1179–1203. doi:10.1016/j.icarus.2010.11.012
Robbins, S. J., Hynek, B. M., Lillis, R. J., and Bottke, W. F. (2013). Large Impact Crater Histories of Mars: The Effect of Different Model Crater Age Techniques. Icarus 225 (1), 173–184. doi:10.1016/j.icarus.2013.03.019
Rodríguez, A., and van Bergen, M. J. (2017). Superficial Alteration Mineralogy in Active Volcanic Systems: An Example of Poás Volcano, Costa Rica. J. Volcanology Geothermal Res. 346, 54–80. doi:10.1016/j.jvolgeores.2017.04.006
Rohwerder, T., Gehrke, T., Kinzler, K., and Sand, W. (2003). Bioleaching Review Part A. Appl. Microbiol. Biotechnol. 63 (3), 239–248. doi:10.1007/s00253-003-1448-7
Rohwerder, T., and Sand, W. (2003). The Sulfane Sulfur of Persulfides Is the Actual Substrate of the Sulfur-Oxidizing Enzymes from Acidithiobacillus and Acidiphilium Spp. Microbiology 149 (7), 1699–1710. doi:10.1099/mic.0.26212-0
Rotto, S., and Tanaka, K. L. (1995). Geologic/geomorphologic Map of the Chryse Planitia Region of Mars. IMAP.
Rowe, G. L., Brantley, S. L., Fernandez, M., Fernandez, J. F., Borgia, A., and Barquero, J. (1992a). Fluid-volcano Interaction in an Active Stratovolcano: the Crater lake System of Poás Volcano, Costa Rica. J. Volcanology Geothermal Res. 49 (1), 23–51. doi:10.1016/0377-0273(92)90003-V
Rowe, G. L., Ohsawa, S., Takano, B., Brantley, S. L., Fernandez, J. F., and Barquero, J. (1992b). Using Crater Lake Chemistry to Predict Volcanic Activity at Poás Volcano, Costa Rica. Bull. Volcanology 54 (6), 494–503. doi:10.1007/BF00301395
Ruff, S. W., Farmer, J. D., Calvin, W. M., Herkenhoff, K. E., Johnson, J. R., Morris, R. V., et al. (2011). Characteristics, Distribution, Origin, and Significance of Opaline Silica Observed by the Spirit Rover in Gusev Crater, Mars. Journal of Geophysical Research: Planets 116(E7). doi:10.1029/2010JE003767
Rymer, H., and Brown, G. (1989). Gravity Changes as a Precursor to Volcanic Eruption at Poás Volcano, Costa Rica. Nature 342 (6252), 902–905. doi:10.1038/342902a0
Rymer, H., Cassidy, J., Locke, C. A., Barboza, M. V., Barquero, J., Brenes, J., et al. (2000). Geophysical Studies of the Recent 15-year Eruptive Cycle at Poás Volcano, Costa Rica. J. Volcanology Geothermal Res. 97 (1), 425–442. doi:10.1016/S0377-0273(99)00166-3
Salvage, R. O., Avard, G., de Moor, J. M., Pacheco, J. F., Brenes Marin, J., Cascante, M., et al. (2018). Renewed Explosive Phreatomagmatic Activity at Poás Volcano, Costa Rica in April 2017. Front. Earth Sci. 6, 160. doi:10.3389/feart.2018.00160
San Martin-Uriz, P., Gomez Manuel, J., Arcas, A., Bargiela, R., and Amils, R. (2011). Draft Genome Sequence of the Electricigen Acidiphilium Sp. Strain PM (DSM 24941). J. Bacteriol. 193 (19), 5585–5586. doi:10.1128/JB.05386-11
Sanford, W. E., Konikow, L. F., Rowe, G. L., and Brantley, S. L. (1995). Groundwater Transport of Crater-lake Brine at Poa´s Volcano, Costa Rica. J. Volcanology Geothermal Res. 64 (3), 269–293. doi:10.1016/0377-0273(94)00080-Z
Schippers, A. (2007). “Microorganisms Involved in Bioleaching and Nucleic Acid-Based Molecular Methods for Their Identification and Quantification,” in Microbial Processing of Metal Sulfides. Editors E. R. Donati, and W. Sand. (DordrechtNetherlands: Springer), 3–33.:
Schmidt, M. E., Farrand, W. H., Johnson, J. R., Schröder, C., Hurowitz, J. A., McCoy, T. J., et al. (2009). Spectral, Mineralogical, and Geochemical Variations across Home Plate, Gusev Crater, Mars Indicate High and Low Temperature Alteration. Earth Planet. Sci. Lett. 281 (3), 258–266. doi:10.1016/j.epsl.2009.02.030
Schneider, K., Peyraud, R., Kiefer, P., Christen, P., Delmotte, N., Massou, S., et al. (2012). The Ethylmalonyl-CoA Pathway Is Used in Place of the Glyoxylate Cycle by Methylobacterium Extorquens AM1 during Growth on Acetate. J. Biol. Chem. 287 (1), 757–766. doi:10.1074/jbc.M111.305219
Schulze-Makuch, D., Dohm, J. M., Fan, C., Fairén, A. G., Rodriguez, J. A. P., Baker, V. R., et al. (2007). Exploration of Hydrothermal Targets on Mars. Icarus 189 (2), 308–324. doi:10.1016/j.icarus.2007.02.007
Seto, M., Noguchi, K., and Cappellen, P. V. (2019). Potential for Aerobic Methanotrophic Metabolism on Mars. Astrobiology 19 (10), 1187–1195. doi:10.1089/ast.2018.1943
Skok, J. R., Mustard, J. F., Ehlmann, B. L., Milliken, R. E., and Murchie, S. L. (2010). Silica Deposits in the Nili Patera Caldera on the Syrtis Major Volcanic Complex on Mars. Nat. Geosci. 3 (12), 838–841. doi:10.1038/ngeo990
Solomon, S. C., Aharonson, O., Aurnou, J. M., Banerdt, W. B., Carr, M. H., Dombard, A. J., et al. (2005). New Perspectives on Ancient Mars. Science 307 (5713), 1214. doi:10.1126/science.1101812
Squyres, S. W., Aharonson, O., Clark, B. C., Cohen, B. A., Crumpler, L., de Souza, P. A., et al. (2007). Pyroclastic Activity at Home Plate in Gusev Crater, Mars. Science 316 (5825), 738. doi:10.1126/science.1139045
Squyres, S. W., Arvidson, R. E., Ruff, S., Gellert, R., Morris, R. V., Ming, D. W., et al. (2008). Detection of Silica-Rich Deposits on Mars. Science 320 (5879), 1063. doi:10.1126/science.1155429
Stamenković, V., Ward, L. M., Mischna, M., and Fischer, W. W. (2018). O2 Solubility in Martian Near-Surface Environments and Implications for Aerobic Life. Nat. Geosci. 11 (12), 905–909. doi:10.1038/s41561-018-0243-0
Tamames, J., and Puente-Sánchez, F. (2019). SqueezeMeta, A Highly Portable, Fully Automatic Metagenomic Analysis Pipeline. Front. Microbiol. 9 (3349). doi:10.3389/fmicb.2018.03349
Tanaka, K. L., Dohm, J. M., Lias, J. H., and Hare, T. M. (1998). Erosional Valleys in the Thaumasia Region of Mars: Hydrothermal and Seismic Origins. J. Geophys. Res. E: Planets 103 (E13), 31407–31419. doi:10.1029/98je01599
Thollot, P., Mangold, N., Ansan, V., Le Mouélic, S., Milliken, R. E., Bishop, J. L., et al. (2012). Most Mars Minerals in a Nutshell: Various Alteration Phases Formed in a Single Environment in Noctis Labyrinthus. Journal of Geophysical Research. Planets 117 (E11). doi:10.1029/2011JE004028
Tibazarwa, C., Wuertz, S., Mergeay, M., Wyns, L., and van der Lelie, D. (2000). Regulation of the Cnr Cobalt and Nickel Resistance Determinant of Ralstonia Eutropha (Alcaligenes Eutrophus) CH34. J. Bacteriol. 182 (5), 1399. doi:10.1128/JB.182.5.1399-1409.2000
Trainer, M. G., Wong, M. H., McConnochie, T. H., Franz, H. B., Atreya, S. K., Conrad, P. G., et al. (2019). Seasonal Variations in Atmospheric Composition as Measured in Gale Crater, Mars. J. Geophys. Res. Planets 124 (11), 3000–3024. doi:10.1029/2019JE006175
Ullrich, S. R., Poehlein, A., Voget, S., Hoppert, M., Daniel, R., Leimbach, A., et al. (2015). Permanent Draft Genome Sequence of Acidiphilium Sp. JA12-A1. Stand. Genomic Sci. 10 (1), 56. doi:10.1186/s40793-015-0040-y
Vago, J. L., Coates, A. J., Jaumann, R., Korablev, O., Ciarletti, V., Mitrofanov, I., et al. (2018). “Chapter 12-Searching for Traces of Life with the ExoMars Rover,” in From Habitability to Life on Mars. Editors N. A. Cabrol, and E. A. Grin (Elsevier), 309–347. doi:10.1016/b978-0-12-809935-3.00011-6
Wächtershäuser, G. (2006). From Volcanic Origins of Chemoautotrophic Life to Bacteria, Archaea and Eukarya. Philosophical Transactions of the Royal Society of London. Series B. Biol. Sci. 361 (1474), 1787–1808. doi:10.1098/rstb.2006.1904
Walter, M. R., and Des Marais, D. J. (1993). Preservation of Biological Information in Thermal Spring Deposits: Developing a Strategy for the Search for Fossil Life on Mars. Icarus 101 (1), 129–143. doi:10.1006/icar.1993.1011
Wang, J., Hynek, B. M., et al. (2020). “Temporal and Metagenomic Studies of an Extreme Martian Analog Ecosystem: Poás Volcano, Costa Rica,” in 51st Lunar and Planetary Science Conference, Houston, LPI (Lunar and Planetary Institute) Abstract 1774.
Wang, Q., Cen, Z., and Zhao, J. (2015). The Survival Mechanisms of Thermophiles at High Temperatures: An Angle of Omics. Physiology 30 (2), 97–106. doi:10.1152/physiol.00066.2013
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 73 (16), 5261. doi:10.1128/AEM.00062-07
Weiss, M. C., Sousa, F. L., Mrnjavac, N., Neukirchen, S., Roettger, M., Nelson-Sathi, S., et al. (2016). The Physiology and Habitat of the Last Universal Common Ancestor. Nat. Microbiol. 1 (9), 16116. doi:10.1038/nmicrobiol.2016.116
Weitz, C. M., Bishop, J. L., Thollot, P., Mangold, N., and Roach, L. H. (2011). Diverse Mineralogies in Two Troughs of Noctis Labyrinthus, Mars. Geology 39 (10), 899–902. doi:10.1130/G32045.1
Wichlacz, P. L., Unz, R. F., and Langworthy, T. A. (1986). Acidiphilium Angustum Sp. nov., Acidiphilium Facilis Sp. nov., and Acidiphilium Rubrum Sp. nov.: Acidophilic Heterotrophic Bacteria Isolated from Acidic Coal Mine Drainage. Int. J. Syst. Evol. Microbiol. 36 (2), 197–201. doi:10.1099/00207713-36-2-197
Wilson, S. A., Morgan, A. M., Howard, A. D., and Grant, J. A. (2021). The Global Distribution of Craters with Alluvial Fans and Deltas on Mars. Geophys. Res. Lett. 48 (4), e2020GL091653. doi:10.1029/2020GL091653
Yamauchi, M., and Yamada, Y. (1981). Glycolate Pathway and Serine Synthesis in Relation to CO2 Concentration in Tomato Leaves. Plant Cel Physiol. 22 (8), 1489–1498. doi:10.1093/oxfordjournals.pcp.a076302
Yen, A. S., Morris, R. V., Clark, B. C., Gellert, R., Knudson, A. T., Squyres, S., et al. (2008). Hydrothermal Processes at Gusev Crater: An Evaluation of Paso Robles Class Soils. Journal of Geophysical Research. Planets 113 (E6). doi:10.1029/2007JE002978
Yen, A. S., Morris, R. V., Ming, D. W., Schwenzer, S. P., Sutter, B., Vaniman, D. T., et al. (2021). Formation of Tridymite and Evidence for a Hydrothermal History at Gale Crater, Mars. J. Geophys. Res. Planets 126 (3), e2020JE006569. doi:10.1029/2020JE006569
Keywords: mars, astrobiology, acid-sulfate hydrothermal systems, extremophiles, metagenomic sequencing, acidiphilium bacteria
Citation: Wang JL, Dragone NB, Avard G and Hynek BM (2022) Microbial Survival in an Extreme Martian Analog Ecosystem: Poás Volcano, Costa Rica. Front. Astron. Space Sci. 9:817900. doi: 10.3389/fspas.2022.817900
Received: 18 November 2021; Accepted: 03 January 2022;
Published: 28 January 2022.
Edited by:
Maggie C. Y. Lau Vetter, Institute of Deep-Sea Science and Engineering (CAS), ChinaReviewed by:
Rachel Lee Harris, Harvard University, United StatesCopyright © 2022 Wang, Dragone, Avard and Hynek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Justin L. Wang, anVzdGluLndhbmdAY29sb3JhZG8uZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.