
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Astron. Space Sci., 06 January 2023
Sec. Astrobiology
Volume 9 - 2022 | https://doi.org/10.3389/fspas.2022.1040633
 Riccardo Pagliarello1,2
Riccardo Pagliarello1,2 Elisabetta Bennici1
Elisabetta Bennici1 Alessia Cemmi3
Alessia Cemmi3 Ilaria Di Sarcina3
Ilaria Di Sarcina3 Cornelis Spelt4
Cornelis Spelt4 Luca Nardi1
Luca Nardi1 Antonella Del Fiore5
Antonella Del Fiore5 Patrizia De Rossi6
Patrizia De Rossi6 Francesca Paolini7,8
Francesca Paolini7,8 Ronald Koes4
Ronald Koes4 Francesca Quattrocchio4
Francesca Quattrocchio4 Eugenio Benvenuto1
Eugenio Benvenuto1 Silvia Massa1*
Silvia Massa1*Introduction: Methods for production of fresh, health food are needed in view of long-term, deep-space manned missions. To this end, crops tailored for better performance under non-terrestrial conditions may be obtained by the exploitation of biochemical patterns related to specialized metabolites known to confer protection against environmental challenges and to be beneficial to human health.
Methods: In this work, for the first time, MicroTom plants have been engineered specifically for agrospace applications to express PhAN4, a MYB-like transcription factor able to regulate the biosynthesis of anthocyanins that influence tomato genes possibly involved in agrospace-relevant functions.
Results: PhAN4 engineering underpinned the genetic background of the dwarf tomato MicroTom while maintaining yield and photosynthetic capacity. PhAN4 expression resulted in the accumulation of anthocyanins and polyphenols, a differential carotenoid profile, increased antioxidant scavenging capacities of fruits compared to the original genotype. Improved ability to counteract ROS generation and to preserve plant protein folding after ex-vivo gamma irradiation was observed.
Discussion: These results highlights that the manipulation of specific metabolic pathways is a promising approach to design novel candidate varieties for agrospace applications.
Humans will venture within the Solar System, mostly heading to Moon and Mars. Lunar and Martian manned missions will be complicated by logistics, extended duration and radiation concerns, among other factors. As man moves into outer space, plants will follow, allowing the setup of Bioregenerative Life Support Systems able to provide necessary raw materials and their recycle (Fu et al., 2016).
Higher plants cultivation in outer space outposts will provide fresh food and will be a source of value-added molecules, as healthy supplements to astronauts’ diet, otherwise strictly dependent on dehydrated initial food payloads (McNulty et al., 2021).
The design of plant ideotypes able to cope with non-terrestrial conditions is mandatory to overcome hindrances to cultivation in space. Pro-oxidant stimuli related to cosmic radiation are among the most prominent challenges for space agriculture (De Pascale et al., 2021; Mortimer and Gilliham, 2022). Acting directly and indirectly to delay oxidative damage, plant enzymatic players and metabolites activating endogenous antioxidant defense systems may help plant growth in space environments. In parallel, generating crops able to counteract overproduction of free radicals induced by harmful ionizing radiation is crucial for human survival in space, as well, and opens the way to the ideal “antioxidant space fresh food” (Gómez et al., 2021). Indeed, the oral intake of currently available, clinically-tested chemical radioprotectants (i.e., thiols, aminothiols, thidiazoles and benzothiazoles) is limited due to toxicity (Copp et al., 2013). Therefore, there is great interest in the development of plants able to accumulate natural antioxidants that may be introduced in astronauts’ diet (Lourenço et al., 2019).
The design of plants suitable to these ends may be achieved by refinement of specialized metabolite pathways that plants activate to counteract oxidative stress. Indeed, the early land plants were forced to evolve in the mechanisms of self-preservation in conditions of primordial Earth, characterized by scarce habitability, in which the first defensive phenolics fulfilled multiple roles in the pre-adaptive development (Graham et al., 2004). Novel genotypes boosting biosynthetic pathways for the production of specialized metabolites can be generated by manipulation of their regulators (Wang et al., 2016).
Plants of the Solanaceae family are considered among the most interesting genotype sources to obtain ideotypes for agrospace applications and have already provided insights into biological processes related to space stimuli exposure (Kim et al., 2004; Nechitailo and Jinying, 2005; Colla et al., 2007; Cheng et al., 2010; De Micco et al., 2014; Dueck et al., 2016; Villani et al., 2017; Arena et al., 2019; Desiderio et al., 2019; Massa et al., 2022). Tomato (Solanum lycopersicum L.) is one of the most cultivated crops worldwide and its fruits are the largest dietary source of lycopene, ascorbic acid, alpha-lipoic acid, choline, folic acid and lutein (Mirdehghan and Valero, 2017). Despite the richness in valuable compounds known for their nutraceutical properties, accumulation of flavonoids in tomato fruits is sub-optimal and, with some exception, anthocyanins, widely known for their potent antioxidant properties, are not accumulated (Mes et al., 2008). Anthocyanins have increasing applications in the food sector and there is growing interest in the design of food crops with improved levels and composition of these anti-oxidant nutraceuticals for “agrospace” applications (Gómez et al., 2021). Over the last decades, attempts to overproduce anthocyanins in tomato have been carried on by both biotechnological approaches and conventional breeding, reporting success in anthocyanins accumulation in fruits and other organs (Bovy et al., 2002; Mathews et al., 2003; Butelli et al., 2008; Povero et al., 2011; Schreiber et al., 2012; Kiferle et al., 2015; Su et al., 2016; Blando et al., 2019; Jian et al., 2019; Colanero et al., 2020). By breeding, tomato lines have been generated by combining the dominant Atv allele with Aft or Abg. As examples, the double mutant Aft/Aft atv/atv, due to anthocyanin accumulation, determined intensely pigmented tomato accumulating anthocyanidins by .12% (Mes et al., 2008) and .28% (Povero et al., 2011) in fruit epidermis. The “Indigo Rose” cultivar containing the Aft and atv loci, bear purple fruit with anthocyanin detected in the fruit peel (Ooe et al., 2016). More recently, a conventional breeding approach aimed at combining different alleles enhancing anthocyanin production in cultivated tomato, produced the Aft/Aft atv/atv combination named “Sun Black” with remarkable phenotype of a deep purple fruit pigmentation (Mazzucato et al., 2013; Blando et al., 2019). In addition to breeding, genetic engineering has been used to determine anthocyanins biosynthesis in tomato. Particularly, genes encoding R2R3-MYB, bHLH and WDR factors have been used to regulate anthocyanins biosynthesis, due to their activity in combination within the MBWW transcription complex (Ramsay and Glover, 2005; Zhang et al., 2014). SlAN11 is a WD40 protein able to promote anthocyanin expression in tomato transgenic lines, physically interacting with bHLH but not with other MYB transcription factors (Gao et al., 2018). The combined expression of a MYB and a bHLH regulators of the anthocyanin pathway from Antirrhinum majus (Delila and Rosea1, respectively) under a fruit-specific promoter, resulted in the production of anthocyanins in tomato fruits peel and flesh (Butelli et al., 2008). Studies have demonstrated that engineering approaches implying the sole use of MYB factors, particularly those belonging to the SG6 clade, are sufficient to restore the biosynthesis of anthocyanins by promotion of the transcription of their bHLH partners and, therefore, by reconstituting the MBWW (Mehrtens et al., 2005; Takos et al., 2006; Zhang et al., 2019). As an example, the ANT1 gene from a tomato wild relative (S. chilense), highly homologous to the Antirrhinum Rosea, induces purple spotting on the epidermis of tomatoes (Mathews et al., 2003; Schreiber et al., 2012). The 35S promoter-driven expression of either the Solanum lycopersicum ANT1 or AN2, has been shown to induce anthocyanins production in the flesh and peel of the fruits and in different organs of tomato plants (Kiferle et al., 2015). Upon overexpression of SlAN2, together with anthocyanins accumulation in fruits, flower organs and vegetative parts, an enhancement of the emission of volatile molecules contributing to the aroma of fruits was found, as well (Jian et al., 2019). In addition, SlAN2 has been related to variation of levels of specialized metabolites other than anthocyanins, and of fruit softening (Meng et al., 2015). These findings shed light to the use of MYB transcription factors of the SG6 clade to affect multiple pathways and to subsequently regulate various, sometimes unrelated, processes in tomato (Stracke et al., 2001; Zimmermann et al., 2004; Zhang et al., 2019). Among such processes, a transcriptomic analysis conducted in a tomato hairy root model, gave a clue that this approach may regulate traits related to resistance to oxidative and other abiotic stresses (Massa et al., 2022).
The Anthocyanin4 gene of P. hybrida (PhAN4) is an SG6 member of a small family of genes encoding very similar MYBs involved in the induction of anthocyanins accumulation in different plant parts (Povero, 2011). PhAN4 has been shown to affect multiple suitable pathways in tomato, referring not only to anthocyanins and other flavonoids accumulation, but also to possibly favorable agronomic features among which plant response to biotic and abiotic stress, ROS detoxification and ROS signaling (Massa et al., 2022).
In this study, MicroTom plants engineered to express PhAN4 were obtained and characterized showing equivalence or improvement of specific agronomical traits that may be beneficial for agrospace applications, compared to wild type. Engineering conferred peculiar metabolite composition and ex-vivo improved capacity to counteract generation of ROS, oxidation and misfolding of soluble proteins upon a potent pro-oxidant stimulus such as exposition to high dose gamma rays, compared to wild type.
Plant engineering has the potential to provide manufacturing capacity to tackle space environmental stresses harmful to both humans and plants. Our initial evaluations reinforce the concept that this strategy may tap into the potential of plants to serve as high-value product sources to meet the agronomical and diet needs of future space manned missions.
Solanum lycopersicum L. miniature-dwarf-determinate cv. MicroTom (Scott and Harbaugh, 1989) was used as starting and control material for genetic transformation. Anthocyanin4 (AN4) cDNA (GenBank: HQ428105.1) from petals of Petunia hybrida cultivar Violet 30 (PhAN4) was amplified with primers containing attB-1 and -2 recombination sites (Supplementary Table S1). Recombination of PCR product and pDONR221 P1-P2 (RU Ghent) produced the entry clone. The resulting pEntry construct was recombined with pKGW.0 (RU Ghent; www.psb.ugent.be/gateway/) using LR clonase, obtaining the 35S:AN4 construct. For plant transformation, tomato seeds were surface-sterilised by immersion into a 2% (v/v) solution of sodium hypochlorite for 15 min and rinsed four times in sterile distilled water. Seeds were placed on 25 mL of half-strength MS medium (Duchefa Biochemie; Harlem, Netherlands) (pH 5.7) containing 15 g/L sucrose and incubated for 7 days at 24°C for germination. Single colonies of Agrobacterium harbouring the 35S:AN4 vector were grown in 5 mL of LB liquid medium with 200 mM acetosyringone overnight. Cotyledon and seedling stems (hypocotils) pieces were cut from tomato seedlings and soaked for 30 or more min in Agrobacterium liquid culture (OD600 = .5), and then placed on the co-cultivation medium consisting of MS containing 20 g/L glucose, 1 mM MES, .75 mg/L trans-zeatin, 1.0 mg/L IAA, and 200 mM acetosyringone, pH 5.7, and incubated at 24°C in the dark. After 2 days, the tissues were transferred to the induction medium (same composition of the co-cultivation medium without acetosyringone, but plus 300 mg/L cefotaxime and 100 mg/L kanamycin). Tissues were sub-cultured every 21 days until shoot formation, incubated at 24°C under a 16 h light photoperiod. Shoots were then transferred to the elongation medium (same composition of the induction medium with .1 mg/L trans-zeatin and .05 mg/L IAA) and incubated as above. Shoots were harvested and transferred to the rooting medium consisting of MS containing 20 g/L glucose, 1 mM MES, .2 mg/L IBA and 400 mg/L carbenicillin.
Standard PCR assays were performed on genomic DNA of primary tranformants (T0) extracted with NucleoSpin Plant II Kit (Macherey-Nagel; Duren, Germany) with primers specific for PhAN4 (Supplementary Table S1). Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen; Valencia, CA, United States) and then treated with amplification grade DNaseI (Invitrogen; Waltham, Massachusetts, United States). cDNA was synthesized using the iScript™ cDNA Synthesis Kit (BioRad Laboratories Inc., Milan, Italy) and used as template for real time-PCR analysis (Kapa SYBR Fast 2x qPCR Master Mix; KAPA Biosystems, Milan, Italy) in an iCycler iQ detection system (BioRad Laboratories Inc., Milan, Italy). The actin 41 gene was used as reference gene. Relative gene expression levels were obtained using the 2−ΔΔCt formula (Livak and Schmittgen, 2001). Samples were amplified at 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing, and extension at 60°C for 30 s.
Plants were grown in a containment greenhouse biosafety level 2 (ENEA Casaccia Research Center, Rome) in hydroponic conditions. Plants were cultivated in mesh pots containing a mix of horticultural perlite and vermiculite in a 1:1 ratio with nutrient solution (1 g/L; Idrofill base, K-Adriatica, Rovigo, Italy), maintained at pH 5.5–6.5 and electrical conductivity 2.2–2.5 dSm−1. Plants received natural light integrated with artificial light (blue LED 70 μmol m−2 s−1, white LED 70 μmol m−2 s−1, red LED 160 μmol m−2 s−1) (PRO 650 lamps, LumiGrow, California, United States) 16/8 h day/night, at a temperature of 25/16°C day/night.
TLA technology was performed on genomic DNA (gDNA) extracted from mature leaves of T3 generation of two primary transformants, namely AN4-M “Magenta” and AN4-P “Purple” plants. The gDNA was extracted using the NucleoSpin Plant II kit (Macherey-Nagel; Duren, Germany), following the user manual and quantified using the Qubit 4TM Fluorometer (ThermoFisher Scientific, Massachussetts, United States). Samples were processed by Cergentis (B.V., Utrecht, Netherlands) (De Vree et al., 2014). Briefly, two primer sets (1–2) were designed on the transgene (Supplementary Table S2) and used in individual TLA amplification. PCR products were purified, the library was prepped using the Illumina Nextera flex protocol and then sequenced on an Illumina sequencer (Illumina Inc., California, United States). The obtained reads were mapped using BWA-SW version .7.15-r1140 with the settings bwasw -b 7 (Li and Durbin, 2010). The NGS reads were aligned to the transgene (TG) and the tomato genome (GCF_000188115.4_SL3.0). Integration sites were detected based on coverage peak(s) in the genome and on the identification of fusion-reads between the TG sequence and host genome.
Plants (20 plants per group) were phenotypically characterized using the following biometric parameters: plant height (by digital calliper), overall leaf area (ImageJ version 1.8.0, National Institute of Health, New York, United States), number of leaves per plant, number of flowers per plant, number of fruits per plant, fruit weight at the ripe stage, equatorial diameter of fruits (by digital calliper) and number of seeds. The photosynthetic maximum photochemical efficiency (Fv/Fm) and the maximum fluorescence yield (Y_Fm) were evaluated as plant stress indices using the Plant Stress Kit (PSK; Opti-sciences, Hudson, United States). Three fully developed leaves from each replicate were clumped with leaf clips for a 30 min dark adaptation before fluorescence measurements. The maximum photochemical efficiency Fv/Fm was calculated on the basis of the equation: Fv/Fm = (Fm − F0)/Fm, where F0 = initial fluorescence, Fm = maximal fluorescence, and Fv = variable fluorescence (Fm/F0) (Baker and Rosenqvist, 2004). Dry matter content was measured at the harvest by weighting six plants/genotype before and after drying at 85°C until no weight variation was recorded.
Three mg (dried weight) of ripe fruits triplicates were ground and resuspended in 2 mL of MeOH 70%, vortexed, shook in Mixer Mill (MM) for 60 min at 20 Hz frequency and gently mixed at room temperature. Samples were then centrifuged at 12,000 g for 10 min. The supernatant was recovered and diluted 1:2 in MeOH 70%. Total anthocyanins content was measured by spectrophotometry (Shimadzu, Duisburg, Germany) following the pH-differential method as described in Brito et al. (2014) and using the extinction coefficient of the most abundant anthocyanin (petunidin-3-(p-coumaroyl)-rutinoside-5-glucoside) (Sigma-Aldrich, Missouri, United States).
Freeze-dried, ground tomato fruits (30 mg) triplicates were extracted with .6 mL EtOH 80% (v/v), shaken in a water bath at 80°C for 40 min and then centrifuged at 3,000 rpm for 15 min. The supernatant was filtered through polytetrafluoroethylene membrane (.45 μm) and stored at −20°C. The antioxidant capacity of the ethanolic extracts was spectrophotometrically tested by DPPH (1,1-diphenyl-2-picrylhyydrazyl) (Li et al., 2010), with some modifications. Briefly, .25 mL of diluted ethanolic extract was added to 2.9 mL of .06 mM DPPH working solution. The mixture was shaken and allowed to stand at room temperature, in the dark, for 30 min. Absorbance was measured at 515 nm (UV-VIS spectrophotometer Shimadzu, Duisburg, Germany). The inhibition of free radical DPPH was expressed as DPPH scavenging effect (% inhibition) I% = {(A0 –A1)/A0*100}, where A1 and A0 are the absorbance values of blank and of tested samples, respectively. Trolox (vitamin E equivalent Antioxidant) was used as a reference standard.
Irradiation tests were performed on ripe fruits-deriving lyophilized material using a pool-type irradiation plant equipped with a 60Co gamma source in a high volume (7 m × 6 m × 3.9 m) shielded cell (Fusion and Nuclear Safety Technologies Department, ENEA, Rome, Italy). The source emits radiations consisting of two gamma photons with a mean energy of 1.25 MeV (Baccaro et al., 2019). Fricke dosimetric system was employed for the determination of the absorbed dose during the irradiation tests. Samples were irradiated at room temperature with .5 Gy (dose rate 5.6 Gy/h) or 2 kGy (dose rate of 1.8 kGywater/h). High dose Gamma rays (2 kGy) was used to investigate the ability to counteract the generation of reactive oxygen species upon potent ROS induction. .5 Gy absorbed dose was used to evaluate the stability of ripe fruits major compounds by UV-Vis spectrophotometry upon the limiting dose for astronauts and cosmonauts based on the recommendations of the International Commission on Radiological Protection. Not irradiated sample sets were used as references. Each set consisted of two replicates.
ROS basal and induced level of accumulation and their species were analysed in wild type and transgenic dried biomass sample sets by ESR Spectroscopy. For each ESR analysis, 8 ± .1 mg of lyophilized powder were split into two PT-Capillaries (NOX-A.8.1-PT NOXYGEN, Holland) inserted in a conventional quartz sample tube (o.d./i.d. of 4/3 mm) closed by a plastic lid. Irradiated samples were analyzed straight after the end of irradiation and ESR signals and were normalized to the sample mass. ESR measurements were acquired using an ESR e-scan spectrometer (Bruker, Massachusetts, United States) operating in the X-band frequency (9.4 GHz) with a field modulation frequency of 86 kHz and modulation amplitude of 5.152 G. ESR spectra were recorded at a central magnetic field of 3,466 G with a sweep width of 160 G, microwave power of .14 mW, microwave frequency of 9.75 GHz. The reported ESR spectra were derived from the accumulation of four scans. Bruker WinEPR data processing software was used for data elaboration.
Tomato fruit total soluble proteins were investigated for maintenance of folding upon high-dose gamma irradiation. Fruit samples were finely ground in liquid nitrogen with mortar and pestle, resuspended and homogenized in phosphate-buffered saline (PBS, 1:3 w/v) containing a protease inhibitor cocktail (Complete, Roche, Basilea, Switzerland) to extract soluble proteins avoiding degradation due to the extraction procedure. Photoluminescence emission spectra of extracts were determined before and straight after 2 kGy absorbed dose of both AN4 and control dried biomass sample sets. Two replicates per set were poured into quartz cuvettes with an optical path length of 1 cm (104F-QS, Hellma Analytics, Munich, Germany). Emission spectra were recorded using the Spectrometer FS 5 (Edinburgh Instruments, Livingstone, United Kingdom) in the range 300–800 nm with 280 nm excitation wavelength. The recorded spectra were normalized and corrected for background scattering (reference: extraction buffer).
Information on polyphenols and carotenoids level under .5 Gy absorbed dose was accomplished on the corresponding ripe fruit-derived extracts. Polyphenols extraction was conducted as already described (Serrano-Díaz et al., 2014) with some adjustments. Briefly, 30 mg of dry lyophilized fruits were extracted with 1.5 mL of water:HCl (100:1, v/v) under shaking for 1 h at 500 rpm (40°C). Carotenoids were extracted as already described (Fikselová et al., 2008): 25 mg of lyophilized biomass were extracted using 100 µL of isopropanol under shaking for 1 h at 500 rpm (60°C). Extracts were centrifuged at 3,500 rpm for 5 min to remove debries. Supernatants were examined at 250–680 nm, by a UV-spectrophotometer (BioSpectrometer® kinetic, Eppendorf, Hamburg, Germany).
GraphPad Prism version 8 (GraphPad Software, San Diego, CA, United States) was used for all graphical and statistical data processing. Data obtained were subjected to ordinary one-way ANOVA with multiple comparisons and to Tukey’s post hoc test as a statistical hypothesis testing. Significant differences were shown as asterisks, where *p-value ≤.05, **p-value ≤.01 and ***p-value ≤.001 were indicated on respective graphs.
The PhAN4 cDNA under the control of the constitutive 35SCaMV promoter was inserted in Solanum lycopersicum cv. MicroTom (Supplementary Figure S1). Eighteen primary transformants were screened for the presence of the transgene by PCR analysis of genomic DNA (Supplementary Figure S2). The third (T3) generation progeny of three primary transformants harbouring one copy of PhAn4, namely AN4-M (homozygous), AN4-P1 (homozygous) and AN4-P2 (hemizygous) was investigated in detail. These plants showed a different colour phenotype, with low (AN4-P1), medium (AN4-M) or strong (AN4-P2) pigmentation corresponding to statistically significant differential expression of PhAN4 in fruits and leaves (Figure 1).
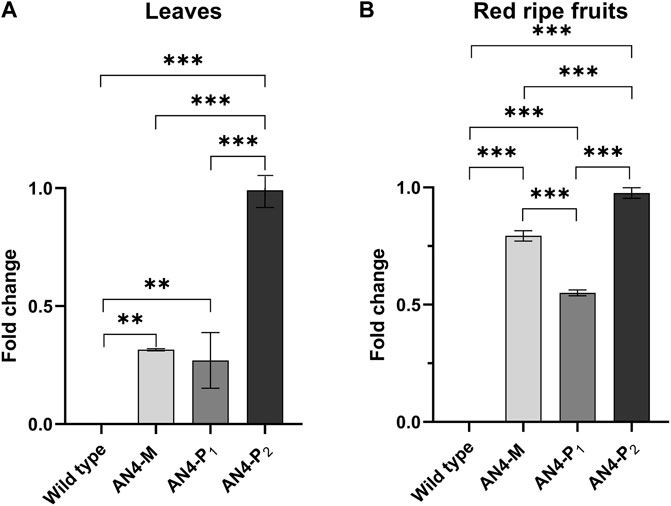
FIGURE 1. PhAN4 gene expression analysis conducted on red ripe fruits (A) and leaves (B) of wild type, AN4-M and AN4-P plants. Statistical relevance: *: p < .05; **: p < .001; ***: p < .0001.
AN4-P1 and AN4-P2, deriving from the same primary transformant, had integrated PhAN4 in chromosome 12, in position 23,824,716–23,824,827. In AN4-M, PhAN4 resulted to be inserted in chromosome 8, in position 11,072,987–11,073,027 (Supplementary Figure S3). The corresponding regions of Solanum lycopersicum cultivar Heinz 1706 chromosome 12 Sl3.0 (NC_015449.3) and chromosome 8 (NC_015445.3) revealed non-relevant coding regions within 600,000 bp upstream and downstream the integration sites in both chromosome 12 and 8, indicating that the integration events did not either interrupted or relapsed into possibly relevant loci and known gene functions of the MicroTom genome.
The AN4-M and the AN4-P2 T3 plants, due to the most favourable features related to anthocyanins accumulation in fruits, were further characterized. These plants developed normally during vegetative growth. Life-cycle traits, such as the duration of vegetative state (i.e., time from sowing to flower development) and time from sowing to fruit development resulted to be 40.5 ± 3.5 days and 55.5 ± 3.5 days for all the genotypes, respectively (Supplementary Figure S4) (Meissner et al., 1997).
The vegetative traits of engineered plants resulted significantly different compared to wild type. Morphometric analysis, indeed, showed that both AN4-M and, more markedly, AN4-P2 plants were significantly smaller-sized than wild type. The reduced habitus was correlated to a decrease of the following parameters: height, number of leaves and leaf area (Figure 2). On the contrary, yield traits (i.e., number of flowers, number of fruits, diameter and weight of fruits) were unchanged in transgenic plants compared to wild type, with the only exception of the production of seeds, that was significantly higher in AN4-M plants compared to both wild type and AN4-P2 (Figure 2).
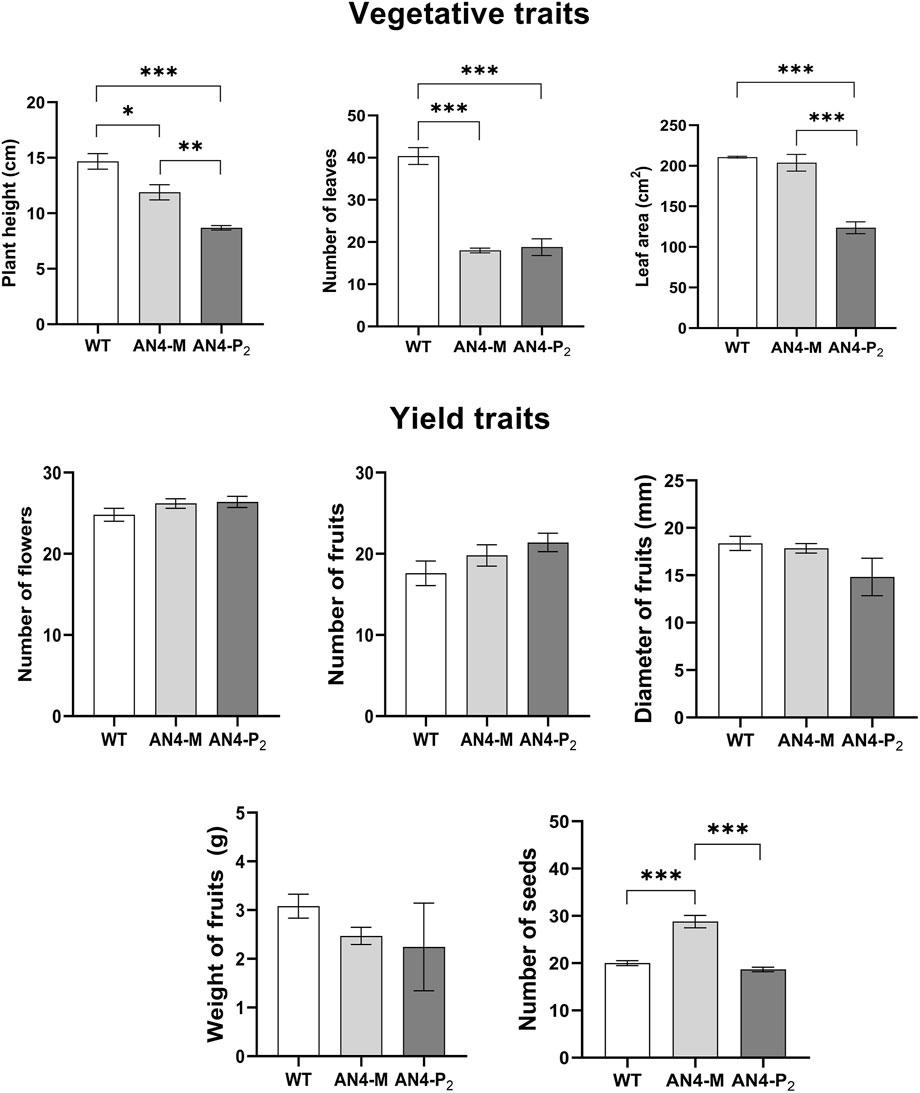
FIGURE 2. Compared morphometric analysis among wild type, AN4-P2 and AN4-M plants. Means ± SE (n = 5 replicates). Statistical relevance: *: p < .05; **: p < .001; ***: p < .0001.
Despite reduced crown, the photosynthetic maximum photochemical efficiency (Fv/Fm) and the maximum fluorescent yield (Y_Fm) resulted in the range (.7/.9) described for healthy leaves (Figure 3) for all the three genotypes. In addition, as an indicator of plant resource strategy between growth and efficient conservation of resources, the AN4-M plants demonstrated the highest dry matter content (Table 1).
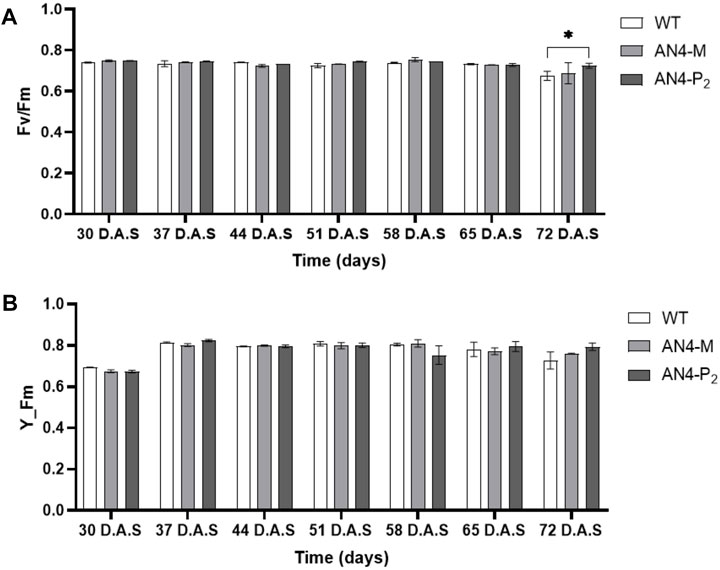
FIGURE 3. Analysis of the Fv/Fm (A) and Y_Fm (B) indices of wild type, AN4-M and AN4-P2 MicroTom along the observation period. Statistical relevance: *: p < .05; **: p < .001; ***: p < .0001. D.A.S.
During development, a purple pigmentation was visible in seeds and, right after sowing, in roots, hypocotyls and cotyledons of both AN4-M and AN4-P2. Plants of the AN4-M line, were phenotypically indistinguishable from AN4-P2 until 3 weeks after sowing, when stems, leaves, stamens, pistil, petals, and calix developed peculiar purple pigmentation depending on the genotype, (Figure 4). Peel and, at a lesser extent, pulp of ripe fruits showed a darker pigmentation compared to wild type, as well, and pigmentation varied along ripening stages. After an intense onset in peel at the immature green stage, the pigmentation extended to placenta and then to mesocarp and endocarp (Figure 4). At maturity, the fruits of AN4-M and AN4-P2 showed a different colour phenotype, with medium (AN4-M) or strong (AN4-P2) pigmentation corresponding to differential accumulation of anthocyanins.
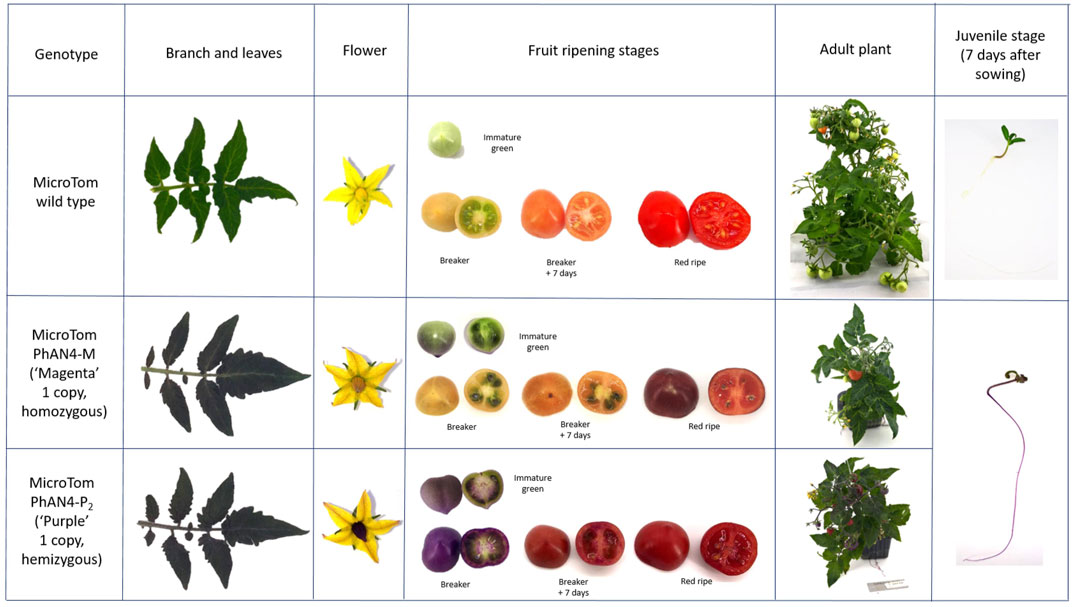
FIGURE 4. Genotype-specific phenotypes of T3 generation MicroTom expressing PhAN4 under the control of the CaMV35S promoter. Phenotypic analysis of wild-type, homozygous AN4-M line and hemizygous AN4-P2 plants. Branches, leaves, flowers, fruits (immature green, breaker, breaker +7 days and red ripe stages) and related cross-section, whole plants and juvenile stages.
Accumulation of anthocyanins, virtually absent in wild type, increased through the ripening stages of AN4-M and AN4-P2 fruits until the breaker + 7 days stage, when both showed the highest anthocyanin concentration (125 ± 6 µg/gfresh weight and 335 ± 17 µg/gfresh weight, respectively). At red ripening, total anthocyanins decreased in both AN4-P2 (177.5 ± 8.9 µg/gfresh weight) and AN4-M (70 ± 4 µg/gfresh weight) (Figure 5A).
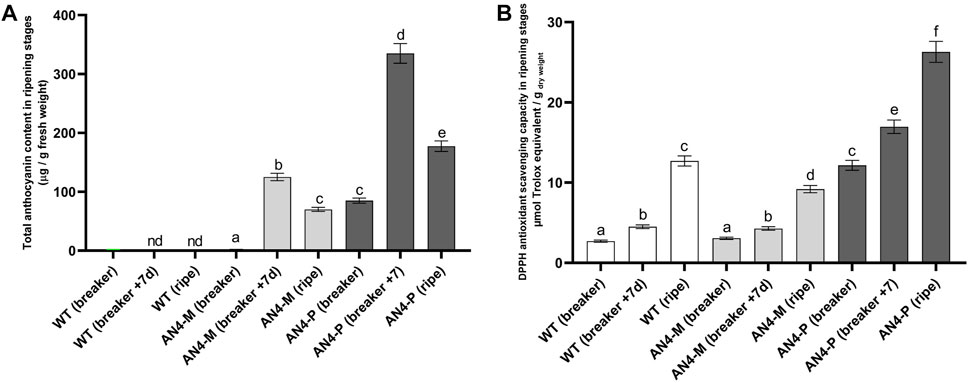
FIGURE 5. Total anthocyanin content in wild type and engineered tomato fruits (A). The amount of anthocyanins is expressed in µg/g of fresh weight. Each analysis consisted of triplicate measurements of each sample and data were averaged over the three measurements. Analysis of antioxidant activity (DPPH) in tomato fruits from wild type and AN4 plants (B). Results are expressed as µg Trolox equivalents/g dry weight (DW). A univariate statistical analysis based on Student’s test and one way-ANOVA (p ≤ .05) was carried out.
Despite the decrease in the anthocyanins concentration at ripening, the DPPH antioxidant scavenging capacity of the AN4-P2 fruits resulted to be significantly higher than wild type along all the stages, including red ripe. The DPPH of AN4-M fruits was not statistically different from wild type, but recorded a statistically relevant decrease at red ripe stage (Figure 5B).
High dose acute gamma radiation, known to efficiently generate peroxyl radicals, was used as a tool to further investigate whether ripe fruits of AN4-M and AN4-P2 plants may better counterbalance ROS generation compared to wild type. Electron Spin Resonance (ESR) allowed to assess the type and the amount of free radicals generated in fruits upon gamma irradiation (Figure 6). MicroTom irradiated fruits, independently from genotype, exhibited a composite ESR spectrum, due to the complexity of the biological matrix of the tomato fruit. In each genotype, irradiation produced two side peaks (doublet) spaced about ± 3 mT from each other (g = 2.014 and g = 2.020), whose hyperfine constants were found to be the same as that of irradiated cellulose and OH-induced oxidation of carbohydrates, which includes the formation of peroxyl radical intermediates (Aleksieva et al., 2009; Yordanov and Aleksieva, 2009; Faure et al., 2014). The ESR maximum intensity of radicals accumulation after irradiation showed a significantly lower intensity of the radical singlet (g = 2.020) in AN4-M compared to wild type (Δh = −470.16), whereas radicals referring to the g = 2.014 singlet accumulated at a slightly higher level in AN4-M (Δh = 162.46) (Figure 6A). The ESR spectra and the maximum intensity (h) reached by radicals’ accumulation upon 2 kGy irradiation for AN4-P2 and wild type are substantially equivalent in correspondence of radical referring to the g = 2.020 (Δh = −232.96), whereas they differ for the g = 2.014 singlet with AN4-P2 showing a significantly lower level of radicals accumulated after irradiation compared to wild type (Δh = 20.57) (Figure 6B). The value calculated for the overall area under the curve (a.u.c.) defined by the doublet g = 2.014/g = 2.020 is similar in AN4-M and AN4-P2 (a.u.c.AN4-M = 7,717; a.u.c.AN4-P2 = 7,716), whereas it is lower compared to wild type (a.u.c.wt = 9,717). Hence, by exposure to an equal dose of radiation, AN4 fruits have higher radical scavenging ability compared to wild type, resulting in lower levels of radicals accumulated.
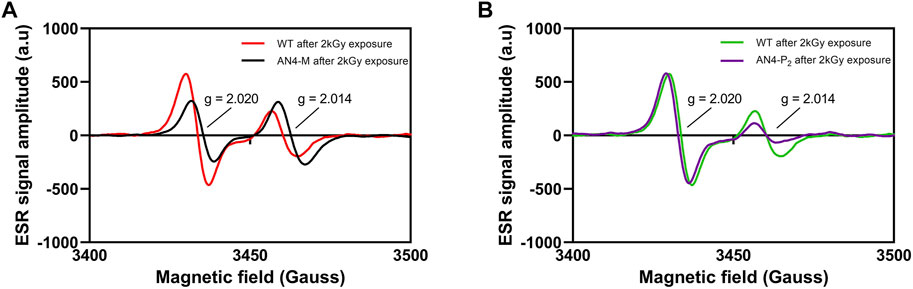
FIGURE 6. ESR spectra of wild type, AN4-M (A) and AN4-P2 fruits (B) at 2 kGy absorbed dose (dose rate = 1.8 kGy/h). The intensity of each signal is expressed as the peak-to-peak height normalized for mass unit and by subtracting the intensities of the signals before irradiation.
Photoluminescence was used to determine the resistance to misfolding and to oxidation of total soluble proteins of MicroTom fruits after high dose acute gamma irradiation (Figure 7). The emission spectra of not irradiated soluble protein samples, characterized by a peak corresponding to tryptophan (Hilaire et al., 2017; Yang et al., 2017), were similar in wild type and AN4-M and AN4-P2 fruits. The tryptophan peak, that is known to be highly dependent on polarity and/or local environment (Vivian and Callis, 2001), shifted at 391 nm in AN4-P2 extracts. High dose gamma irradiation did not cause any detectable decrease in fluorescence emission intensity of the spectra, independently of genotype, demonstrating a general stability of MicroTom fruit soluble proteins tertiary and quaternary structure upon this stress. Nevertheless, a blue shift phenomenon was observed in the spectra of all the three genotypes, being more relevant in wild type (−21 nm) and AN4-P2 (−16 nm) than in AN4-M (−9 nm), and revealing that the side chain indole of tryptophan of proteins of the first two genotypes may be more exposed due to misfolding and more prone to undergo oxidation than AN4-M. In wild type and AN4-M a shoulder appears (306–310 nm) in the emission maximum, mainly referring to tyrosine. In AN4-P2 irradiated proteins, tryptophan derivatives such as kynurenine and 3-hydroxykynurenine (439 and 470 nm) are formed (Daly et al., 2009; Gakamsky et al., 2017).
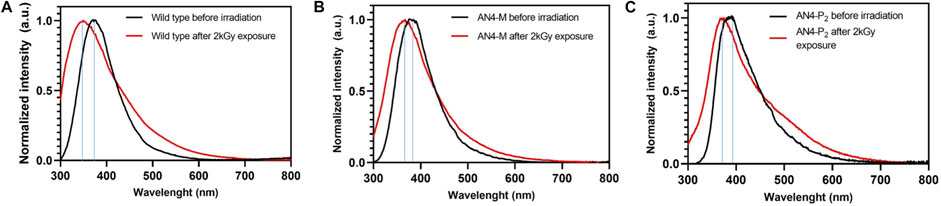
FIGURE 7. Photoluminescence emission spectra in normal conditions and after 2 kGy exposure of wild type (A), AN4-M (B), and AN4-P2 (C).
In absence of irradiation challenge, irrespective of genotype, UV-Vis spectra of polyphenol extracts of ripe fruits show main peaks at 254 and 265 nm, generally referred as isoflavones (Vacek et al., 2008), followed by two shoulders at 282 i.e., flavan-3-ols (Silva et al., 2012) and 319 nm i.e., flavones (Mabry et al., 1970). Anthocyanins and anthocyanins associated with phenolic acids also produce peaks around 280 nm and 320 nm, respectively (Solìs-Oviedo and De La Cruz Pech-Canul, 2019), contributing to the profile of AN4-M and AN4-P2 genotypes (Figure 8). Anthocyanins result in an additional characteristic peak at 530 nm (Vivar-Quintana et al., 2002; da Silva et al., 2007; Fedenko et al., 2017), which, as expected, is observed only in transgenic fruits (especially in AN4-P2, as expected). Despite the correspondence in UV-Vis peaks among genotypes, the intensity of each peak is higher for AN4-P2 followed by AN4-M and wild type, indicating an overall higher content of all the corresponding class of compounds compared to wild type. Gamma irradiation of fruits determined a significant decrease of the intensities of all the peaks referring to polyphenols in all the genotypes. Despite the generalized loss of absorbance, the percentage decrease of peaks intensities is lower for the AN4-M fruits than wild type and the overall intensity of peaks remains higher in both AN4-P2 and AN4-M fruits than in wild type (Figure 8).
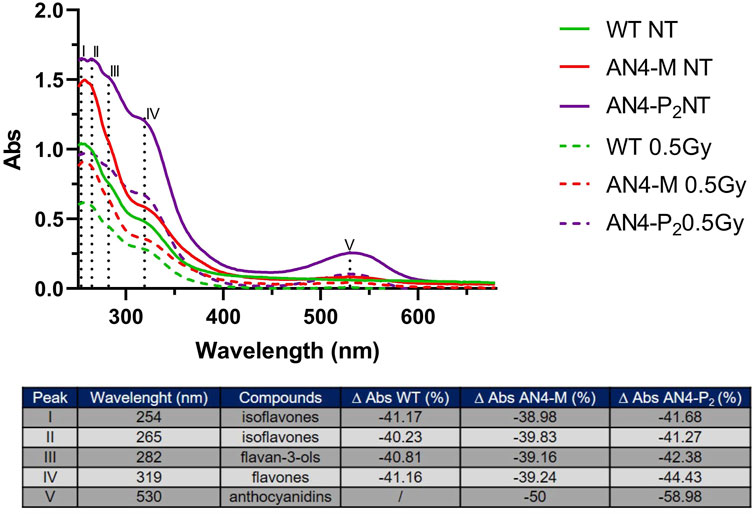
FIGURE 8. UV-VIS spectra of polyphenols extracts of ripe fruits before and after .5 kGy absorbed dose. The table reports the peaks and the corresponding compounds. The ∆ Abs (%) was calculated considering the Abs after and before the irradiation for wild type, AN4-M and AN4-P2 extracts.
In absence of irradiation challenge, carotenoids extracts of all ripe fruits independently of genotype revealed two main peaks or shoulders at 289 (i.e., phytoene) and 302 nm (phytoene and lycopene) followed by four peaks or shoulders of decreasing absorbance at 323 (cis-xanthins), 335 (15-cis-β-cryptoxanthin,13-cis-β-carotene), 353 (phytofluene), and 370 nm (13-cis-astaxanthin) (Figure 9) (Silva et al., 2014). The latter four peaks in AN4-M fruits have similar intensities among each other but lower compared to AN4-P2 and wild type. Thereafter, spectra return comparable for the three genotypes, with a group of four peaks at 409 (sigma-carotene), 432 (9- and 13-cis-violaxanthins), 452 (all-trans-xanthins, all-trans-β-carotene, lutein, neozeaxanthin) and 477 nm (trans-lycopene, neolycopene A) with similar absorbance within each genotype, followed by a shoulder at 509 nm (myxol glycosides) (Silva et al., 2014). The overall intensity of peaks is higher for AN4-P2 and wild type between 280 and 370 nm, followed by AN4-M, indicating a higher content of the class of compounds referring to peaks I-VI in AN4-P2 and wild type compared to AN4-M ripe fruits. From 400 to 550 nm, the intensities of the VII-XI peaks are higher for wild type than engineered fruits. After gamma irradiation, a significant decrease of the intensities of all the peaks for all the genotypes was observed, especially for wild type. Despite this generalized loss of absorbance, the percentage decrease of peaks intensities was higher for wild type than engineered fruits, with the only exception of peaks VII and VIII in AN4-P2 (Figure 9). The AN4-M overall peak intensities resulted the highest among the three genotypes.
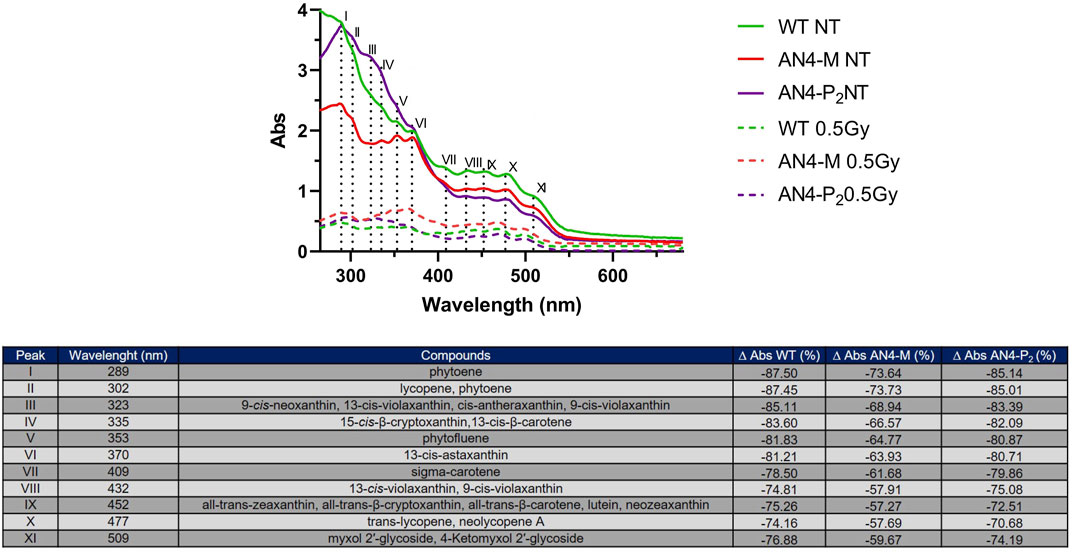
FIGURE 9. UV-VIS spectra of carotenoid extracts of ripe fruits before and after .5 kGy absorbed dose. The table reports the peaks and the corresponding compounds. The ∆ Abs (%) was calculated considering the Abs after and before the irradiation for wild type, AN4-M and AN4-P2 extracts.
There is rising interest in the development of biofortified crops that may cope with outer space conditions thanks to the accumulation of natural antioxidants and that may, in parallel, be usefully introduced in the astronauts’ diet in future deep-space manned missions (Liu et al., 2021). Among the specialized metabolites providing protection against environmental challenges, polyphenols and anthocyanins are well-known for their potent anti-oxidant properties. These compounds are, indeed, candidate protectants against cosmic radiation for humans during spaceflight (Gómez et al., 2021) and seems to be particularly effective if administered in their original phyto-complex. As an example, a diet supplemented with polyphenols-rich dried plums completely prevented bone loss caused by simulated space radiation in mice effectively reducing expression of genes related to both pro-osteoclastogenic and antioxidant responses, whereas artificial antioxidant mixtures failed (Schreurs et al., 2016).
Undoubtedly, currently available tomatoes varieties generated through conventional improvement programs to induce anthocyanins bioaccumulation, are to be considered as a valuable source of antioxidants. Nevertheless, these varieties are generally cultivar with indeterminate growth, and it must be taken in consideration that the ideotypes aiming at agrospace applications need to meet specific minimum requirements for cultivation in space, among which short life cycle, determinate growth with small size and improved traits related to survival in harsh conditions and in the artificial, confined environments like those that are currently foreseen to support life of crews in future deep-space manned missions (Dueck et al., 2016). Nowadays, four determinate, dwarf tomato cultivars have been proposed for cultivation in space, namely, Mohamed, Red Robin, Sweet N′ Neat and MicroTom (Dueck et al., 2016; Spencer et al., 2019). Among them, MicroTom represents the cultivar that has been most comprehensively studied in space (Nechitailo and Jinying, 2005) or in space-simulated conditions (Colla et al., 2007; De Micco et al., 2014; Arena et al., 2019). Therefore, in this work, the tomato cv MicroTom was specifically chosen to be engineered with the aim of a “space evolution” of tomato.
MicroTom was engineered to express the SG6 anthocyanin R2R3-MYB transcription factor PhAN4 from P. hybrida. In a MicroTom hairy roots culture-based model, PhAN4 had already demonstrated to promote not only the biosynthesis of anthocyanins, but also a transcriptome reprogramming of the tomato cell, determining a positive regulation of genes correlated to cell response to biotic, abiotic, and redox stimuli (Massa et al., 2022). Clearly, these features may be very beneficial for a tomato whole plant intended to agrospace applications.
The homozygous AN4-M and the hemizygous AN4-P2 plants were chosen for characterization, due to fertility, single transgene copy number and no interruption of relevant gene sequences upon transformation. In addition, the hemizygous AN4-P2 plants showed the most favourable features related to anthocyanins accumulation compared to both wild type and homozygous lines. The higher accumulation of anthocyanins in hemizygous plants is in accordance with findings obtained for a fruit-specific anthocyanin-related, purple phenotype induced by Delila and Rosea1 (Del/Ros1) transgenes in hemyzigous MicroTom transformants (Butelli et al., 2008). The Del/Ros1 purple phenotype was, thereafter, usefully transferred into other tomato genetic backgrounds by crossing to obtain gene pyramiding (Butelli et al., 2021) and this approach might be taken in consideration for the MicroTom AN4-P2 phenotype, as well.
Life-cycle and yield traits were positively or were not influenced by the PhAN4 transgene. As an indicator of a plant species’ resource use strategy (i.e., its position in a fundamental trade-off between rapid assimilation and growth at one extreme, and efficient conservation of resources within tissues at the other), the dry matter content of these plants pointed to the AN4-M genotype as the most efficient (Vaieretti et al., 2007). In addition, as a pleiotropic effect, engineered plants showed an even more miniaturized habitus compared to wild type. This feature may be explained considering the secondary metabolite-oriented production driven by PhAN4 at the expense of primary metabolism. Further miniaturization, combined with the overall conserved productivity of the PhAN4-engineered plants, are clearly beneficial features in view of the confined and spatially limited environments designed for agrospace (Paradiso et al., 2014).
It is known that anthocyanins and polyphenols act as ROS-scavenging compounds that participate to both non-enzymatic and enzymatic antioxidant mechanisms (Ullah et al., 2019). The over-accumulation of these specialized metabolites in AN4 fruits may explain the enhanced antioxidant capacity of AN4-P2 compared to wild type along the ripening stages. In addition, the improved antioxidant features of the AN4-P2 engineered fruits at red ripe stage, despite the decrease of anthocyanins content, confirms the additional contribution of isoflavones, flavones and flavan-3-ols to antioxidant capacity, as shown by UV-Vis spectra. Anthocyanin degradation and related discoloration is a phenomenon known as “fading.” In maturing fruits, ROS are formed and fading is thought to protect other cellular components from related damages (Passeri et al., 2016).
In addition to polyphenols, the accumulation of carotenoids, generally reaching maximum level at the ripe stage, may be important in improving the anti-oxidant features of PhAN4-engineered ripe fruits, and reveal a possible cross talk between the anthocyanin and the carotenoid pathways, leading to improvement of either carotenoid levels or stability. Indeed, according to UV-Vis spectroscopy, red ripe AN4-P2 fruits show higher absorbance for peaks related to 9-cis-neoxanthin, 13-cis-violaxanthin, cis-antheraxanthin, 9-cis-violaxanthin, 15-cis-β-cryptoxanthin, 13-cis-β-carotene and phytofluene compared to both wild type and AN4-M. Transcriptomic analysis of PhAN4-expressing tomato hairy roots has already revealed the down-regulation of violaxanthin de-epoxidase (VDE) and carotenoid isomerase (CrtISO) (Massa et al., 2022). The downregulation of these genes usually leads to the accumulation of more bioavailable forms of carotenoids than zeaxanthin and lycopene, canonical products of their activity (Zhou et al., 2022). Since VDE normally uses violaxanthin and antheraxanthin as precursors to produce zeaxanthin, the higher intensity of peak III, that indeed refers to these compounds, in carotenoid extracts of AN4-P2 fruits compared to wild type, may be an indication that VDE down-regulation may occur in transformed whole plants, as well. The lower intensity of peak IX (zeaxanthin) in carotenoid extracts from AN4-P2 fruits compared to wild type, may also point to this direction. On the other hand, CrtISO usually converts tetra-cis-lycopene to lycopene. The lower intensity of peak X (trans-lycopene and neolycopene A) in carotenoid extracts of AN4-P2 fruits compared to wild type, may also corroborate the hypothesis. Metabolomic and transcriptomic analyses are currently ongoing to confirm these hypotheses in plants.
Despite gamma radiation is only one component of cosmic rays, the higher Δ Abs % of carotenoids compared to polyphenols after radiation challenge independently of genotype, may point to a higher resistance of polyphenols rather than carotenoids to radiation damage and to their possible major contribution in preserving plant tissues in space settings from accumulation of reactive species. Indeed flavonoids in general are known for their resistance to ionizing radiation (Alcaraz et al., 2021). In a scenario where plants are subjected to the limiting dose for astronauts of .5 Gy, as defined by the International Commission on Radiological Protection, the UV-Vis spectra would point to AN4-M fruits as the most promising (ICRP, 2012).
Both transgenic and conventionally bred tomato that have been previously generated to achieve anthocyanins biosynthesis, show higher levels of these compounds compared to the AN4 ripe fruits (Supplementary Table S3). A part of these works reports on the related antioxidant capacity, as well. These latter works demonstrate that the restored anthocyanins biosynthesis determines a significant improvement of tomato antioxidant capacity (Butelli et al., 2008; Li et al., 2011; Ooe et al., 2016; Blando et al., 2019). Due to the difference in detection methods (Supplementary Table S3), it is difficult to make a solid comparison between these works and the present study in terms of antioxidant capacity. Our aim was primarily to characterize the response of the AN4 tomato under a potent radiative pro-oxidant stimulus. ESR spectroscopy, a leading method for evaluating food matrices (Aleksieva and Yordanov, 2018), revealed that, in tomato fruits, high dose gamma radiation mainly induces cellulose radicals, as expected for a plant matrix (Duliu and Bercu, 2017), and showed that AN4 fruits metabolites have a higher free radical scavenging activity compared to wild type ripe fruits. This trait may be very beneficial both in terms of food consumption and in terms of plant resistance to pro-oxidant stimuli. The discrepancy between the DPPH and ESR analyses of the AN4-M ripe fruits (i.e., lower DPPH value, but higher a.u.c. calculated by ESR, compared to wild type) can be explained by the different redox potential of the DPPH radical used in the DPPH assay compared to the hydroxyl radical species generated by gamma irradiation (Munteanu et al., 2021). The improved antioxidant capacity of engineered AN4-P2 ripe fruits compared to wild type and AN4-M, may also point to the possible additional contribution of ROS-scavenging enzymatic players such as glutathione peroxidase, catalase, and superoxide dismutase. Interestingly, it has been demonstrated that anthocyanins interact with these enzymes during scavenging processes (Salehi et al., 2020). Furthermore, transcriptomic analysis of tomato PhAN4-expressing hairy roots has revealed that PhAN4 upregulates plastidial thioredoxin Y2, that is a player of ROS detoxification involved in the redox signalling network of tomato (Massa et al., 2022). Preliminary transcriptomic evaluation on AN4-P2 ripe fruits seems to corroborate this hypothesis (manuscript in preparation).
The failure of a protein to fold correctly, that can occur in space due to the presence ionizing radiation, can lead to serious dysfunctions (Ecroyd and Carver, 2008). Since tryptophan and tyrosine residues reflect the folding state of a protein (Yang et al., 2017), we used the photoluminescence analysis to determine the loss of tryptophan fluorescence excited at 280 nm in samples. The better response of PhAN4-engineered fruits to the gamma radiative stimulus, may indicate that, since anthocyanins are not specifically extracted by procedures leading to crude soluble protein extracts, this effect may be correlated, again, to a regulation operated by PhAN4 on ROS-scavenging enzymatic players.
Further research is being carried on to evaluate the effect of different radiation-related scenarios on PhAN4-engineered plants in vivo, and logistical challenges required to set up reliable ground-based chronic exposure of plants to the various components of cosmic radiation are being tackled. This work represents the first effort to specifically engineer tomato for agrospace purposes exploiting biotechnological approaches with the aim to optimize plant performances for future long-term space exploration.
The survival of human beings in outer space will depend on the knowledge acquired in different fields, together with the capacity to exploit higher plants to supply fresh health food, and to possibly serve as bioreactors for in situ production of value-added molecules. Carefully planned medicine supply will be necessary for astronauts in future outer space outposts. This task may be partially fulfilled by plant space agriculture, as well. Recent literature highlights, indeed, in-flight instability of drug formulations, many of which would be completely degraded by the end of the proposed first Mars mission duration. (Menezes et al., 2015; Blue et al., 2019).
Viewing plants as factories of value-added food and molecules will definitely help to counteract the space adverse conditions for survival of both plants and humans and will expand the life support capabilities of plants in space.
Due to the nature of the data generated in this research, no repositories are applicable. The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary Materials. Raw data supporting results are available from the corresponding author, [SM], upon reasonable request.
EUB, FQ and SM planned and designed the project. CS performed plant transformation and regeneration of primary transformants. SM and RP performed biometric, fluorometric and PCR screening of plants, sample preparation for all subsequent analyses, as well as wrote the paper and prepared figures. LN set-up hydroponic cultivation. ELB handled growth and maintenance of plants and contributed to sample preparation. AC and IDS performed gamma irradiation experiments, ESR, UV-VIS, and photoluminescence analysis and contributed to writing the manuscript. FP collaborated in the qRT-PCR set-up and analysis. ADF and PDR performed anthocyanins quantification and the Trolox assay. RK and FQ supervised plant transformation. EUB, RK and FQ reviewed the manuscript. All authors approved the submitted version.
This work was supported by the ENEA/ASI (Italian Space Agency) BIOExTREME (Grant ASI n. 2014-007-R.0), HORTSPACE (Grant ASI n. 2017-11-H.0), REBUS (Grant ASI n. 2019-4-U.0) Projects.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fspas.2022.1040633/full#supplementary-material
Alcaraz, M., Olivares, A., Achel, D. G., García-Gamuz, J. A., Castillo, J., and Alcaraz-Saura, M. (2021). Genoprotective effect of some flavonoids against genotoxic damage induced by X-rays in vivo: Relationship between structure and activity. Antioxidants (Basel, Switz). 11. doi:10.3390/antiox11010094
Aleksieva, K., Georgieva, L., Tzvetkova, E., and Yordanov, N. D. (2009). EPR study on tomatoes before and after gamma-irradiation. Radiat. Phys. Chem. 78, 823–825. doi:10.1016/j.radphyschem.2009.05.013
Aleksieva, K. I., and Yordanov, N. D. (2018). Various approaches in epr identification of gamma-irradiated plant foodstuffs: A review. Food Res. Int. 105, 1019–1028. doi:10.1016/j.foodres.2017.11.072
Arena, C., Vitale, E., Mele, B. H., Cataletto, P. R., Turano, M., Simoniello, P., et al. (2019). Suitability of Solanum lycopersicum L. ‘ microtom ’ for growth in bioregenerative life support systems: Exploring the effect of high-LET ionising radiation on photosynthesis, leaf structure and fruit traits. Plant Biol. J. 21, 615–626. doi:10.1111/plb.12952
Baccaro, S., Cemmi, A., and SarcinaDi, I. (2019). Calliope 60Co gamma irradiation facility for space qualification at ENEA-Casaccia research centre, 94–100. doi:10.15406/paij.2019.03.00164
Baker, N. R., and Rosenqvist, E. (2004). Applications of chlorophyll fluorescence can improve crop production strategies: An examination of future possibilities. J. Exp. Bot. 55, 1607–1621. doi:10.1093/jxb/erh196
Blando, F., Berland, H., Maiorano, G., Durante, M., Mazzucato, A., Picarella, M. E., et al. (2019). Nutraceutical characterization of anthocyanin-rich fruits produced by “Sun Black” tomato line. Front. Nutr. 6, 133. doi:10.3389/fnut.2019.00133
Blue, R. S., Bayuse, T. M., Daniels, V. R., Wotring, V. E., Suresh, R., Mulcahy, R. A., et al. (2019). Supplying a pharmacy for NASA exploration spaceflight: Challenges and current understanding. NPJ Microgravity 5, 14. doi:10.1038/s41526-019-0075-2
Bovy, A., De Vos, R., Kemper, M., Schijlen, E., Almenar Pertejo, M., Muir, S., et al. (2002). High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1. Plant Cell 14, 2509–2526. doi:10.1105/tpc.004218
Brito, A., Areche, C., Sepúlveda, B., Kennelly, E. J., and Simirgiotis, M. J. (2014). Anthocyanin characterization, total phenolic quantification and antioxidant features of some chilean edible berry extracts. Molecules 19, 10936–10955. doi:10.3390/molecules190810936
Butelli, E., Bulling, K., Hill, L., and Martin, C. (2021). Beyond purple tomatoes: Combined strategies targeting anthocyanins to generate crimson, magenta, and indigo fruit. Horticulturae 7, 327–414. doi:10.3390/horticulturae7090327
Butelli, E., Titta, L., Giorgio, M., Mock, H. P., Matros, A., Peterek, S., et al. (2008). Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 26, 1301–1308. doi:10.1038/nbt.1506
Cheng, L., Yang, H., Lin, B., Wang, Y., Li, W., Wang, D., et al. (2010). Effect of gamma-ray radiation on physiological, morphological characters and chromosome aberrations of minitubers in Solanum tuberosum L. Int. J. Radiat. Biol. 86, 791–799. doi:10.3109/09553002.2010.484478
Colanero, S., Perata, P., and Gonzali, S. (2020). What’s behind purple tomatoes? Insight into the mechanisms of anthocyanin synthesis in tomato fruits. Plant Physiol. 182, 1841–1853. doi:10.1104/pp.19.01530
Colla, G., Rouphael, Y., Cardarelli, M., Mazzucato, A., and Olimpieri, I. (2007). Growth, yield and reproduction of dwarf tomato grown under simulated microgravity conditions. Plant Biosyst. 141, 75–81. doi:10.1080/11263500601153735
Copp, R. R., Peebles, D. D., Soref, C. M., and Fahl, W. E. (2013). Radioprotective efficacy and toxicity of a new family of aminothiol analogs. Int. J. Radiat. Biol. 89, 485–492. doi:10.3109/09553002.2013.770579
da Silva, F. L., Escribano-Bailón, M. T., Pérez Alonso, J. J., Rivas-Gonzalo, J. C., and Santos-Buelga, C. (2007). Anthocyanin pigments in strawberry. LWT - Food Sci. Technol. 40, 374–382. doi:10.1016/j.lwt.2005.09.018
da Silva Souza, M. A., Peres, L. E. P., Freschi, J. R., Purgatto, E., Lajolo, F. M., and Hassimotto, N. M. A. (2020). Changes in flavonoid and carotenoid profiles alter volatile organic compounds in purple and orange cherry tomatoes obtained by allele introgression. J. Sci. Food Agric. 100, 1662–1670. doi:10.1002/jsfa.10180
Daly, S., Bianchini, R., Polefka, T., Jumbelic, L., and Jachowicz, J. (2009). Fluorescence and coloration of grey hair. Int. J. Cosmet. Sci. 31, 347–359. doi:10.1111/j.1468-2494.2009.00500.x
De Micco, V., Paradiso, R., Aronne, G., Pascale, S. D., Quarto, M., and Arena, C. (2014). Leaf anatomy and photochemical behaviour of Solanum lycopersicum L. Plants from seeds irradiated with low-LET ionising radiation. Sci. World J. 2014, 1–13. doi:10.1155/2014/428141
De Pascale, S., Arena, C., Aronne, G., De Micco, V., Pannico, A., Paradiso, R., et al. (2021). Biology and crop production in space environments: Challenges and opportunities. Life Sci. Sp. Res. 29, 30–37. doi:10.1016/j.lssr.2021.02.005
De Vree, P. J. P., De Wit, E., Yilmaz, M., Van De Heijning, M., Klous, P., Verstegen, M. J. A. M., et al. (2014). Targeted sequencing by proximity ligation for comprehensive variant detection and local haplotyping. Nat. Biotechnol. 32, 1019–1025. doi:10.1038/nbt.2959
Desiderio, A., Salzano, A. M., Scaloni, A., Massa, S., Pimpinella, M., De Coste, V., et al. (2019). Effects of simulated space radiations on the tomato root proteome. Front. Plant Sci. 10, 1334. doi:10.3389/fpls.2019.01334
Dueck, T., Kempkes, F., Meinen, E., and Stanghellini, C. (2016). “Choosing crops for cultivation in space,” in 46th int. Conf. Environ. Syst. 1–9.
Duliu, O. G., and Bercu, V. (2017). ESR investigation of the free radicals in irradiated foods. Electron spin Reson. food Sci. 2017, 17–32. Academic Press. doi:10.1016/B978-0-12-805428-4.00002-7
Ecroyd, H., and Carver, J. A. (2008). Unraveling the mysteries of protein folding and misfolding. IUBMB life 60 (12), 769–774. doi:10.1002/iub.117
Faure, A. M., Sánchez-Ferrer, A., Zabara, A., Andersen, M. L., and Nyström, L. (2014). Modulating the structural properties of β-d-glucan degradation products by alternative reaction pathways. Carbohydr. Polym. 99, 679–686. doi:10.1016/j.carbpol.2013.08.022
Fedenko, V. S., Shemet, S. A., and Landi, M. (2017). UV–vis spectroscopy and colorimetric models for detecting anthocyanin-metal complexes in plants: An overview of in vitro and in vivo techniques. J. Plant Physiol. 212, 13–28. doi:10.1016/j.jplph.2017.02.001
Fikselová, M., Šilhár, S., Mareček, J., and Frančáková, H. (2008). Extraction of carrot (Daucus carota L.) carotenes under different conditions. Czech J. Food Sci. 26, 268–274. doi:10.17221/9/2008-cjfs
Fu, Y., Li, L., Xie, B., Dong, C., Wang, M., Jia, B., et al. (2016). How to establish a bioregenerative life support system for long-term crewed missions to the moon or Mars. Astrobiology 16, 925–936. doi:10.1089/ast.2016.1477
Gakamsky, A., Duncan, R. R., Howarth, N. M., Dhillon, B., Buttenschön, K. K., Daly, D. J., et al. (2017). Tryptophan and non-tryptophan fluorescence of the eye lens proteins provides diagnostics of cataract at the molecular level. Sci. Rep. 7, 40375–40415. doi:10.1038/srep40375
Gao, Y., Liu, J., Chen, Y., Tang, H., Wang, Y., He, Y., et al. (2018). Tomato SlAN11 regulates flavonoid biosynthesis and seed dormancy by interaction with bHLH proteins but not with MYB proteins. Hortic. Res. 5, 27. doi:10.1038/s41438-018-0032-3
Gómez, X., Sanon, S., Zambrano, K., Asquel, S., Bassantes, M., Morales, J. E., et al. (2021). Key points for the development of antioxidant cocktails to prevent cellular stress and damage caused by reactive oxygen species (ROS) during manned space missions. npj Microgravity 7, 35–19. doi:10.1038/s41526-021-00162-8
Graham, L. E., Kodner, R. B., Fisher, M. M., Graham, J. M., Wilcox, L. W., Hackney, J. M., et al. (2004). Early land plant adaptations to terrestrial stress: A focus on phenolics. Evol. Plant Physiol., 155–169. doi:10.1016/B978-012339552-8/50010-X
Hilaire, M. R., Ahmed, I. A., Lin, C. W., Jo, H., DeGrado, W. F., and Gai, F. (2017). Blue fluorescent amino acid for biological spectroscopy and microscopy. Proc. Natl. Acad. Sci. U. S. A. 114, 6005–6009. doi:10.1073/pnas.1705586114
ICRP (2012). ICRP statement on tissue reactions/early and late effects of radiation in normal tissues and organs – threshold doses for tissue reactions in a radiation protection context.
Jian, W., Cao, H., Yuan, S., Liu, Y., Lu, J., Lu, W., et al. (2019). SlMYB75, an MYB-type transcription factor, promotes anthocyanin accumulation and enhances volatile aroma production in tomato fruits. Hortic. Res. 6, 22. doi:10.1038/s41438-018-0098-y
Kiferle, C., Fantini, E., Bassolino, L., Povero, G., Spelt, C., Buti, S., et al. (2015). Tomato R2R3-MYB proteins SlANT1 and SlAN2: Same protein activity, different roles. PLoS One 10, 01363655–e136420. doi:10.1371/journal.pone.0136365
Kim, J. H., Baek, M. H., Byung, Y. C., Seung, G. W., and Kim, J. S. (2004). Alterations in the photosynthetic pigments and antioxidant machineries of red pepper (Capsicum annuum L.) seedlings from gamma-irradiated seeds. J. Plant Biol. 47, 314–321. doi:10.1007/BF03030546
Li, D., Li, X., and Ding, X. (2010). Composition and antioxidative properties of the flavonoid-rich fractions from tartary buckwheat grains. Food Sci. Biotechnol. 19, 711–716. doi:10.1007/s10068-010-0100-4
Li, H., Deng, Z., Liu, R., Young, J. C., Zhu, H., Loewen, S., et al. (2011). Characterization of phytochemicals and antioxidant activities of a purple tomato (Solanum lycopersicum L.). J. Agric. Food Chem. 59, 11803–11811. doi:10.1021/jf202364v
Li, H., and Durbin, R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595. doi:10.1093/bioinformatics/btp698
Liu, Y., Xie, G., Yang, Q., and Ren, M. (2021). Biotechnological development of plants for space agriculture. Nat. Commun. 12 (1), 5998–6003. doi:10.1038/S41467-021-26238-3
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Lourenço, S. C., Moldão-Martins, M., and Alves, V. D. (2019). Antioxidants of natural plant origins: From sources to food industry applications. Molecules 24 (22), 4132. doi:10.3390/molecules24224132
Mabry, T. J., Markham, K. R., and Thomas, M. B. (1970). “The ultraviolet spectra of flavones and flavonols,” in The systematic identification of flavonoids (Berlin, Heidelberg: Springer), 41–164.
Massa, S., Pagliarello, R., Cemmi, A., Di Sarcina, I., Bombarely, A., Demurtas, O. C., et al. (2022). Modifying anthocyanins biosynthesis in tomato hairy roots: A test bed for plant resistance to ionizing radiation and antioxidant properties in space. Front. Plant Sci. 13, 830931. doi:10.3389/fpls.2022.830931
Mathews, H., Clendennen, S. K., Caldwell, C. G., Liu, X. L., Connors, K., Matheis, N., et al. (2003). Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 15, 1689–1703. doi:10.1105/tpc.012963
Mazzucato, A., Willems, D., Bernini, R., Picarella, M. E., Santangelo, E., Ruiu, F., et al. (2013). Novel phenotypes related to the breeding of purple-fruited tomatoes and effect of peel extracts on human cancer cell proliferation. Plant Physiol. biochem. 72, 125–133. doi:10.1016/J.PLAPHY.2013.05.012
McNulty, M. J., Xiong, Y., Yates, K., Karuppanan, K., Hilzinger, J. M., Berliner, A. J., et al. (2021). Molecular pharming to support human life on the moon, Mars, and beyond. Crit. Rev. Biotechnol. 41, 849–864. doi:10.1080/07388551.2021.1888070
Mehrtens, F., Kranz, H., Bednarek, P., and Weisshaar, B. (2005). The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 138, 1083–1096. doi:10.1104/pp.104.058032
Meissner, R., Jacobson, Y., Melamed, S., Levyatuv, S., Shalev, G., Ashri, A., et al. (1997). A new model system for tomato genetics. Plant J. 12, 1465–1472. doi:10.1046/j.1365-313x.1997.12061465.x
Menezes, A. A., Cumbers, J., Hogan, J. A., and Arkin, A. P. (2015). Towards synthetic biological approaches to resource utilization on space missions. J. R. Soc. Interface 12, 20140715. doi:10.1098/rsif.2014.0715
Meng, X., Yang, D., Li, X., Zhao, S., Sui, N., and Meng, Q. (2015). Physiological changes in fruit ripening caused by overexpression of tomato SlAN2, an R2R3-MYB factor. Plant Physiol. biochem. 89, 24–30. doi:10.1016/j.plaphy.2015.02.005
Mes, P. J., Boches, P., Myers, J. R., and Durst, R. (2008). Characterization of tomatoes expressing anthocyanin in the fruit. J. Am. Soc. Hortic. Sci. 133, 262–269. doi:10.21273/jashs.133.2.262
Mirdehghan, S. H., and Valero, D. (2017). Bioactive compounds in tomato fruit and its antioxidant activity as affected by incorporation of aloe, eugenol, and thymol in fruit package during storage. Int. J. Food Prop. 20, 1–9. doi:10.1080/10942912.2016.1223128
Mortimer, J. C., and Gilliham, M. (2022). SpaceHort: Redesigning plants to support space exploration and on-earth sustainability. Curr. Opin. Biotechnol. 73, 246–252. doi:10.1016/j.copbio.2021.08.018
Munteanu, I. G., Apetrei, C., and Hadjipavlou-Litina, D. (2021). Analytical methods used in determining antioxidant activity: A review. Int. J. Mol. Sci. 22 (7), 3380. doi:10.3390/ijms22073380
Nechitailo, G. S., Jinying, L., Huai, X., Yi, P., Chongqin, T., and Min, L. (2005). Influence of long term exposure to space flight on tomato seeds. Adv. Space Res. 36 (7), 1329–1333. doi:10.1016/j.asr.2005.06.043
Ooe, E., Ogawa, K., Horiuchi, T., Tada, H., Murase, H., Tsuruma, K., et al. (2016). Analysis and characterization of anthocyanins and carotenoids in Japanese blue tomato. Biosci. Biotechnol. Biochem. 80, 341–349. doi:10.1080/09168451.2015.1091715
Paradiso, R., De Micco, V., Buonomo, R., Aronne, G., Barbieri, G., and De Pascale, S. (2014). Soilless cultivation of soybean for bioregenerative life-support systems: A literature review and the experience of the MELiSSA project – food characterisation phase I. Plant Biol. 16, 69–78. doi:10.1111/plb.12056
Passeri, V., Koes, R., and Quattrocchio, F. M. (2016). New challenges for the design of high value plant products: Stabilization of anthocyanins in plant vacuoles. Front. Plant Sci. 7, 153. doi:10.3389/fpls.2016.00153
Povero, G., Gonzali, S., Bassolino, L., Mazzucato, A., and Perata, P. (2011). Transcriptional analysis in high-anthocyanin tomatoes reveals synergistic effect of Aft and atv genes. J. Plant Physiol. 168, 270–279. doi:10.1016/j.jplph.2010.07.022
Povero, G. (2011). “Physiological and genetic control of anthocyanin pigmentation in different species,” in Developmental Genetics; AIMMS. NBN urn:nbn:nl:ui:31-1871/26381.
Ramsay, N. A., and Glover, B. J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10, 63–70. doi:10.1016/j.tplants.2004.12.011
Salehi, B., Sharifi-Rad, J., Cappellini, F., Reiner, Z., Zorzan, D., Imran, M., et al. (2020). The therapeutic potential of anthocyanins: Current approaches based on their molecular mechanism of action. Front. Pharmacol. 11, 1300–1320. doi:10.3389/fphar.2020.01300
Schreiber, G., Reuveni, M., Evenor, D., Oren-Shamir, M., Ovadia, R., Sapir-Mir, M., et al. (2012). ANTHOCYANIN1 from Solanum chilense is more efficient in accumulating anthocyanin metabolites than its Solanum lycopersicum counterpart in association with the ANTHOCYANIN FRUIT phenotype of tomato. Theor. Appl. Genet. 124, 295–307. doi:10.1007/s00122-011-1705-6
Schreurs, A. S., Shirazi-Fard, Y., Shahnazari, M., Alwood, J. S., Truong, T. A., Tahimic, C. G. T., et al. (2016). Dried plum diet protects from bone loss caused by ionizing radiation. Sci. Rep. 6, 21343–21411. doi:10.1038/srep21343
Scott, J. W., and Harbaugh, B. K. (1989). Micro-tom: A miniature dwarf tomato. Circular–University of Florida. Agricultural Experiment Stations Num 370, 6.
Serrano-Díaz, J., Sánchez, A. M., Martínez-Tomé, M., Winterhalter, P., and Alonso, G. L. (2014). Flavonoid determination in the quality control of floral bioresidues from Crocus sativus L. J. Agric. Food Chem. 62, 3125–3133. doi:10.1021/jf4057023
Silva, M. A., Ky, I., Jourdes, M., and Teissedre, P. L. (2012). Rapid and simple method for the quantification of flavan-3-ols in wine. Eur. Food Res. Technol. 234, 361–365. doi:10.1007/s00217-011-1628-0
Silva, N. A. D., Rodrigues, E., Mercadante, A. Z., and De Rosso, V. V. (2014). Phenolic compounds and carotenoids from four fruits native from the Brazilian Atlantic forest. J. Agric. Food Chem. 62, 5072–5084. doi:10.1021/jf501211p
Solìs-Oviedo, R. L., and De La Cruz Pech-Canul, Á. (2019). Frontiers and New trends in the science of fermented food and beverages. London, United Kingdom: BoD–Books on Demand. doi:10.5772/intechopen.73404
Spencer, L., Hummerick, M., Stutte, G., Sirmons, T., Graham, T., Massa, G., et al. (2019). “Dwarf tomato and pepper cultivars for space crops,” in 49th international conference on environmental systems.
Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. doi:10.1016/S1369-5266(00)00199-0
Su, X., Xu, J., Rhodes, D., Shen, Y., Song, W., Katz, B., et al. (2016). Identification and quantification of anthocyanins in transgenic purple tomato. Food Chem. 202, 184–188. doi:10.1016/j.foodchem.2016.01.128
Takos, A. M., Jaffé, F. W., Jacob, S. R., Bogs, J., Robinson, S. P., and Walker, A. R. (2006). Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142, 1216–1232. doi:10.1104/pp.106.088104
Ullah, R., Khan, M., Shah, S. A., Saeed, K., and Kim, M. O. (2019). Natural antioxidant anthocyanins—A hidden therapeutic candidate in metabolic disorders with major focus in neurodegeneration. Nutrients 11 (6), 1195. doi:10.3390/NU11061195
Vacek, J., Klejdus, B., Lojková, L., and Kubán, V. (2008). Current trends in isolation, separation, determination and identification of isoflavones: A review. J. Sep. Sci. 31, 2054–2067. doi:10.1002/jssc.200700569
Vaieretti, M. V., Díaz, S., Vile, D., and Garnier, E. (2007). Two measurement methods of leaf dry matter content produce similar results in a broad range of species. Ann. Bot. 99, 955–958. doi:10.1093/aob/mcm022
Villani, M. E., Massa, S., Lopresto, V., Pinto, R., Salzano, A. M., Scaloni, A., et al. (2017). Effects of high-intensity static magnetic fields on a root-based bioreactor system for space applications. Life Sci. Sp. Res. 15, 79–87. doi:10.1016/j.lssr.2017.09.002
Vivar-Quintana, A. M., Santos-Buelga, C., and Rivas-Gonzalo, J. C. (2002). Anthocyanin-derived pigments and colour of red wines. Anal. Chim. Acta 458, 147–155. doi:10.1016/S0003-2670(01)01619-1
Vivian, J. T., and Callis, P. R. (2001). Mechanisms of tryptophan fluorescence shifts in proteins. Biophys. J. 80, 2093–2109. doi:10.1016/S0006-3495(01)76183-8
Wang, H., Wang, H., Shao, H., and Tang, X. (2016). Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front. Plant Sci. 7, 67. doi:10.3389/fpls.2016.00067
Yang, H., Xiao, X., Zhao, X., and Wu, Y. (2017). “Intrinsic fluorescence spectra of tryptophan, tyrosine and phenyloalanine,” in Selected papers of the Chinese society for optical engineering conferences held october and november 2016, 10255, 1199–1206. SPIE. doi:10.2991/icadme-15.2015.46
Yordanov, N. D., and Aleksieva, K. (2009). Preparation and applicability of fresh fruit samples for the identification of radiation treatment by EPR. Radiat. Phys. Chem. 78, 213–216. doi:10.1016/j.radphyschem.2008.10.003
Zhang, H., Koes, R., Shang, H., Fu, Z., Wang, L., Dong, X., et al. (2019). Identification and functional analysis of three new anthocyanin R2R3-MYB genes in Petunia. Plant Direct 3, 001144–e213. doi:10.1002/pld3.114
Zhang, Y., Butelli, E., Alseekh, S., Tohge, T., Rallapalli, G., Luo, J., et al. (2015). Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat. Commun. 6, 8635–8711. doi:10.1038/ncomms9635
Zhang, Y., Butelli, E., and andMartin, C. (2014). Engineering anthocyanin biosynthesis in plants. Curr. Opin. Plant Biol. 19, 81–90. doi:10.1016/j.pbi.2014.05.011
Zhou, X., Rao, S., Wrightstone, E., Sun, T., Lui, A. C. W., Welsch, R., et al. (2022). Phytoene synthase: The key rate-limiting enzyme of carotenoid biosynthesis in plants. Front. Plant Sci. 13, 884720–884729. doi:10.3389/fpls.2022.884720
Keywords: MicroTom, agrospace, biofortification, anthocyanins, antioxidant properties
Citation: Pagliarello R, Bennici E, Cemmi A, Di Sarcina I, Spelt C, Nardi L, Del Fiore A, De Rossi P, Paolini F, Koes R, Quattrocchio F, Benvenuto E and Massa S (2023) Designing a novel tomato ideotype for future cultivation in space manned missions. Front. Astron. Space Sci. 9:1040633. doi: 10.3389/fspas.2022.1040633
Received: 09 September 2022; Accepted: 21 December 2022;
Published: 06 January 2023.
Edited by:
Lyle Whyte, McGill University, CanadaReviewed by:
Neftali Ochoa-Alejo, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional, MexicoCopyright © 2023 Pagliarello, Bennici, Cemmi, Di Sarcina, Spelt, Nardi, Del Fiore, De Rossi, Paolini, Koes, Quattrocchio, Benvenuto and Massa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Silvia Massa, c2lsdmlhLm1hc3NhQGVuZWEuaXQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.