- 1JHU Applied Physics Laboratory, Laurel, MD, United States
- 2NASA Goddard Space Flight Center, Greenbelt, MD, United States
Analysis of long timespan heliophysics and space physics data or application of machine learning algorithms can require access to petabyte-scale and larger data sets and sufficient computational capacity to process such “big data”. We provide a summary of Python support and performance statistics for the major scientific data formats under consideration for access to heliophysics data in cloud computing environments. The Heliophysics Data Portal lists 21 different formats used in heliophysics and space physics; our study focuses on Python support for the most-used formats of CDF, FITS, and NetCDF4/HDF. In terms of package support, there is no single Python package that supports all of the common heliophysics file types, while NetCDF/HDF5 is the most supported file type. In terms of technical implementation within a cloud environment, we profile file performance in Amazon Web Services (AWS). Effective use of AWS cloud-based storage requires Python libraries designed to read their S3 storage format. In Python, S3-aware libraries exist for CDF, FITS, and NetCDF4/HDF. The existing libraries use different approaches to handling cloud-based data, each with tradeoffs. With these caveats, Python pairs well with AWS’s cloud storage within the current Python ecosystem for existing heliophysics data, and cloud performance in Python is continually improving. We recommend anyone considering cloud use or optimization of data formats for cloud use specifically profile their given data set, as instrument-specific data characteristics have a strong effect on which approach is best for cloud use.
1 Introduction
We summarize our survey and Python performance evaluation of major scientific data formats that are under consideration for access to heliophysics data in cloud computing environments, which provide scaleable high performance computing (HPC) capability alongside very massive data sets. The ability to engage in science for an entire data sets includes the ability to find and categorize thousands of events, to analyze irradiances and field value variations over long time baselines, create forecasting and predictive models, and to explore new science problems that span large dimensionality and long timespans, among others. In addition, the existence of data sets from multiple satellites stored within cloud systems allows for cross-instrument science in multiple domains. We define the terms ‘big data’ to characterize all categories of analysis using data sets larger than can easily be managed on a single desktop machine, and ‘machine learning’ (ML) as a subset of big data analysis that uses machine learning algorithms for engaging in prediction, event finding, or modeling using entire data sets. This paper both captures the current state of the Python landscape with respect to cloud usage, and provides evaluation criteria and a methodology cloud users should consider when deciding how best to make cloud use of their data within Python.
With data sizes in heliophysics ranging from smaller terabyte (TB, 1 TB = 1,000 gigabytes) sets such as with the SuperMAG archive up through petabyte-sized (PB, 1 PB = 1000 TB) sets such as with the Solar Dynamics Observatory, science analysis looking at entire solar cycles and machine learning approaches that require full data access increasing are using cloud-based computing environments to store, access, and analyze big data sets. Our analysis of Python package support will apply for any cloud or HPC environment. In addition, we provide performance statistics for file access specifically within the Amazon Web Services (AWS) cloud environment.
The challenge with science problems that look at entire data sets is the data sets are too large to fit onto a standard laptop or desktop; transferring the data from an archive or virtual observatory to one’s home machine takes prohibitively long; the processing time needed to analyze big data on a single machine is infeasible. Cloud systems are set up to handle big data problems by 1) storing entire instrument data sets in an easy-to-access fashion, 2) moving the compute capability to the data, rather than requiring users to download data to their machine, and 3) allocating multiple CPU processors for analysis tasks so that algorithms can be run on TB or PB of data in finite time. The AWS cloud environment uses AWS servers in what they call their Elastic Cloud Computing (EC2) service and allows storage of yottabytes (YB = 1000 PB) of data on their inexpensive high-capacity Simple Storage Service (S3) object-oriented disk storage. User access can be from user-developed code (such as Jupyter notebooks, Python programs, other programming development tools) and commercial software (such as IDL) running on AWS servers using their Elastic Cloud Computing (EC2) service.
Heliophysics data are available from many archives in a wide variety of formats. For example, the Heliophysics Data Portal1 provides access to mission data in 21 different formats. Fortunately, the bulk of data is primarily encapsulated in only three common formats: CDF, FITS, and NetCDF/HDF5. We also indicate support for Comma-Separate Variable (CSV) files when applicable. There is currently no single dominant format, as each instrument team chooses a format based on their development needs. As a result, a given satellite or instrument community tends to write their analysis tools to work with the subset of data file types that particular team expects; developing generic tools that work with all file types requires extra development time that would take away from science analysis time.
Python is widely used as a language for data analysis in heliophysics and space physics (Hassell et al., 2017; Burrell et al., 2018). There is a movement to create tools that are filetype-independent via the community-created PyHC packages such as SunPy, SpacePy, HAPI, and others, and we support such efforts, but cannot mandate that individual scientists support multiple file types in their personal codes. Likewise, requiring data archives to provide all their data in all three formats would result in 3x storage costs with no proven advantage. We accept that the pragmatic state of the landscape is any cloud analysis project can expect to have to deal with either CDF, FITS, or NetCDF/HDF5. We therefore provide performance statistics comparing these three file types when in use in the AWS S3 environment, and likewise document which file types are supported in current Python packages.
We examined the most prominent and widely-used formats for this report. Our assumptions include that scientists tend to be trained in (and therefore prefer) their existing workflows and may be new to cloud methods. Barriers for cloud use include the need to adopt new data formats, and a potential shift from traditional instrument-provided raw data to derived data sets which have undergone post-processing, potentially including re-binning or resampling. Adoption of cloud requires scientists to potentially switch from their standard files (CDF, FITS, or NetCDF/HDF5) to cloud-optimized file formats, of which the foremost candidate is the Zarr format. The NASA grant proposal ‘ROSES’ opportunities include a current call for preparing ML-ready data sets that are post-processed and downsampled to generate new big data products, therefore scientists will be expected to adapt to cloud-specific data needs over time.
One core issue with cloud data is tool use, and whether the big data sets created in the cloud are accessible only from within the cloud, or if they are also accessible to the scientists’ traditional work computer workflow. To encourage use of big data sets, cloud architects should maintain the ability for scientists to work with the data using tools they are already familiar with, and we argue that user interface development for cloud environments is as important as raw power.
In Earth Observation, similar considerations have been reported. Lynnes et al. (2020) used the criteria of usability, tools, standards, and cost and found ‘no one-size-fits-all’ with long-term archives remaining in their existing stable self-described file formats, and derived multidimensional imagery and array data sets using Cloud-optimized GeoTIFF and Zarr (respectively). We agree, in general, that archives should retain their existing formats while derived ML data sets are free to explore more heliophysics-optimal formats, particularly NetCDF and HDF.
For our specific cloud environment, HelioCloud (NASA/GSFC 2022) is an AWS-based cloud environment and big data analysis cache created by NASA/GSFC Heliophysics to facilitate easy access to cloud capabilities specifically for heliophysics and space physics research while minimizing the learning curve for scientists (who should not need to be cloud experts to use cloud capabilities). We used it for our profiling of file formats in the AWS cloud, and likewise our code is available via the associate HelioCloud github code repository for easy replicability.
In providing a short encyclopedia of primary heliophysics and space physics Python packages, we hope to inform scientists as to which data file formats are accessible for both traditional “home desktop” analysis and big data analysis, as well as hoping that developers will use this information to create better cross-package compatibility. Combined with the file performance statistics in the cloud environment, this paper provides current performance expectations for heliophysics traditional and cloud use in Python so that scientists can choose the appropriate packages and file types for engaging in traditional analysis, big data analysis, and ML science analysis.
For our specific performance statistics for file access speeds in a cloud environment, cloud bulk storage such as AWS S3 is slower within the cloud-based EC2 compute environment compared to the faster (but more expensive) AWS conventional disk storage, called Elastic Block Storage (EBS). Likewise AWS S3 is slower than using your local disk on your local machine as with a traditional workflow. A check of basic filesystem performance found that AWS S3 was 30x slower in accessing files than what a user experiences on a reference machine of a local MacBook Pro, and 95x slower in writing and delete speed versus the same local system. Accepting S3 usage is predicated on understanding that it will be slower than conventional disk access, but allow for larger data sets to be stored at a lower cost. With this compromise in mind, we can look at specific file format performance comparisons.
2 Materials and methods
The materials of cloud file storage are the different file types and the Python packages plus underlying libraries that support those file types. We focus on the community drivers of the need for access to big data sets, and the use of large contiguous data sets for machine-learning (ML) research2. The performance for a presumed ML-ready data set is the base use case for the context of our analysis, as ML problems typically involve codes that must run on the entirely of a given data set. The community has two approaches towards creating and using data sets for ML. The first is to create tools to feed existing instrument data archives into ML pipelines to perform the analysis, while the second is to create a new derived data set by processing, calibrating, curating and collating massive data sets and apply feature extraction, tagging, or other analytic data reduction.
In the tools category, examples include Bobra and Mason’s (2018) published Jupyter Notebooks that use a variety of data sets as input. Schneider et al. (2021) created a Python toolset for generating ML-ready data set from SOHO and SDO into either local FITS files or an HDF5 datacube. In the curated data sets category, Galvez et al. (2019) prepared an ML-ready data set for all three of SDO’s instruments: AIA, HMI, and EVE. Their 6.5 TB curated and collated set is made available via the Stanford Digital Repository as sets of 2–6 GB tar bundles containing compressed Numpy-readable files. Antunes et al. were awarded a 2021 NASA ROSES Tools and Methods proposal to create an ML-ready set for STEREO + SOHO, and a NASA grant proposal program under their ROSES opportunities includes a 2022 call specifically for creating ML-ready data sets.
In terms of technical implementation, AWS S3 storage “supports” all file types, storing them as objects rather than as files within a conventional disk system. Access can be via “copy then use”, where files are copied from AWS S3 to local disk then read by the program, or using libraries that can directly read them from AWS S3 in object format, which we call “S3-aware libraries”. In Python, S3-aware libraries exist for FITS (AstroPy), NetCDF/HDF (netCDF4), HDF5, CDF (MAVENSDC3 libcdf or AstroPy), and Zarr.
Likewise, the three file types are similarly well supported in the Python package environment: both Pandas (Zaitsev et al., 2019) and Xarray libraries have good support for NetCDF and HDF5; Xarray supports Zarr; AstroPy supports FITS and HDF5; SpacePy supports CDF and HDF5; SunPy4 supports FITS, CDF, and NetCDF. We note that NetCDF and HDF5 are often considered identical file formats, and it is not within the scope of this paper to evaluate the compatibility between NetCDF and HDF5 in terms of importing one from the other’s library.
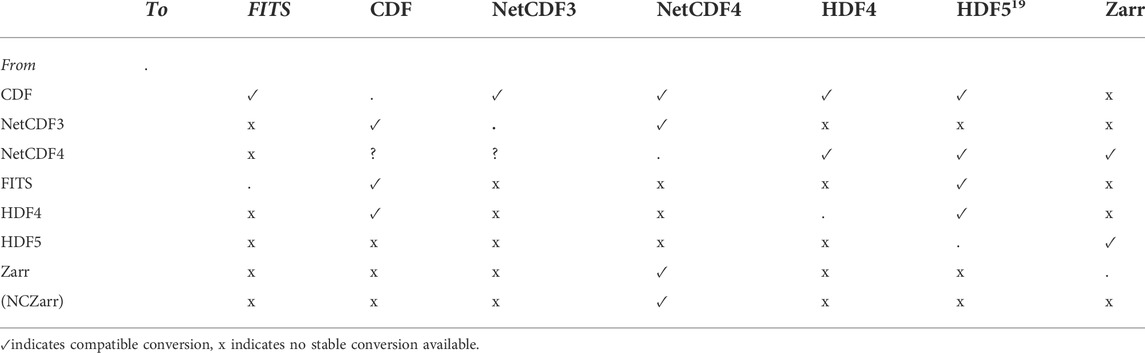
Both the CDF and NetCDF projects provide stand-alone file conversion tools to go between the three file formats. In the CDF ecosystem, there are supported tools for going to or from FITS, NetCDF, and HDF5. For NetCDFs, there are tools to convert to and from the older NetCDF3 and the current NetCDF4/HDF5 format. Converting to HDF5 and Zarr from FITS, CDF and NetCDF5 are problematic. Not all files can be converted, and converting from the richer data types (e.g., HDF5, NetCDF) to the simpler file types (FITS, CDF) is lossy. We did not look deeply at the accuracy or fidelity of metadata for these transformations, as that was outside the scope of our performance evaluations.
In terms of package support, there is no single Python package that supports all the common heliophysics file types, while NetCDF (with cross-support for HDF5) is the most-supported file type. This necessitates that scientists be aware that a given analysis package may or may not support the data file type they require, which presents a barrier to both analysis and code sharing. Assuming scientists choose a package that can read the data format, we assert that Python currently has good support for data access directly from AWS S3 for cloud usage. One concern is performance, as there are three library approaches to handling S3 data:
1) Copying from AWS S3 to local storage, then reading the file locally for analysis: fast, easy, but inefficiently duplicates disk storage
2) Reading from AWS S3 and storing the full data set in memory, then carrying out the analysis: fast but can overwhelm RAM for large/many files
3) Reading from AWS S3 directly in pieces and analyzing the data in smaller chunks: efficient but slow due to S3 limitations and requires your workflow be designed to operate with chunks
Which approach is “best” is very dependent on the specific task at hand. A pipeline processing of small data files, for example, can easily live within memory and gain that speed performance boost, while a machine learning algorithm that must hold vast sets of training data simultaneously will have to do a memory analysis dependent on the project size as well as the capacity of the analysis hardware. Going deeper into these tradeoffs is not within the scope of this paper.
2.1 File types
We converted between file formats using the CDF toolset to explore file size metrics. The tools are provided by NASA and have the straightforward names of netCDF-to-cdf, cdf-to-netCDF, cdf-to-fits and fits-to-cdf (cdf.gsfc.nasa.gov/html/dttools.html). This was the most robust tool set, but as it uses CDF as its intermediate transfer function (for example, doing FITS to NetCDF required fits-to-cdf then cdf-to-netCDF); this may have introduced a bias towards CDF files having the typically smaller file size for identical data.
A key concern in conversion is the preservation and accessibility of metadata. For example, the cdflib library returns this information for an MMS file:
CDF’: PosixPath(“mms1. cdf”), “Version”: “3.6.0”, ‘Encoding”: 6, “Majority”: “Column_major”, “rVariables” [], “zVariables”: [“Epoch”, “mms1_fgm_b_gse_brst_l2”, “mms1_fgm_b_gsm_brst_l2” [etc].
While the same data converted to FITS yields these header fields:
TFIELDS = 12/number of fields in each row.
TTYPE1 = ‘Epoch '/label for field one.
TFORM1 = ‘30A30 '/data format of field: ASCII Character.
TUNIT1 = ‘ns '/physical unit of field.
TTYPE2 = ‘mms1_fgm_b_gse_brst_l2’/label for field two.
TFORM2 = ‘4E '/data format of field: 4-byte REAL.
TUNIT2 = ‘nT '/physical unit of field.
[etc]
The core elements are there (variable names, associated metadata) but interpretation and validation are not seamless. Programs to validate metadata across the conversion would be needed. In terms of accessibility, the difference in file formats require either conversion programs within the Python ingest routine, or a known mapping so that users of one approach (CDF) and library (SunPy) can access the other approach (FITS) and library (AstroPy). Similarly, engaging in analysis within one Python library creates internal data representations (such as SunPy maps) that are not immediately useable within a different Python library (such as with AstroPy and its NDData representations). There is a funded effort already underway to unify several of the core Python heliophysics libraries so that they can share datatypes. Table 1 lists the current cross-file conversion capabilities available for immediate validated bulk conversion between the file formats we discuss.
As a cautionary note, chaining to convert a file (e.g., going from CDF to HDF5 to Zarr) is usually one-way, and not reversible. Conversion from CDF to Zarr is possible via chaining, but not from Zarr to CDF6,7. In general, the multidimensional or more complex data formats (NetCDF, HDF, Zarr) cannot down-convert to the tabular data formats (FITS, some CDF). We also consider that Zarr is not mature enough yet to warrant bulk-converting data archives; if we convert now to Zarr, we might have to re-convert later as the specification evolves.
To dig deeper, in Table 2 we benchmark three different mission data sets: the Magnetic Multiscale Mission (MMS) FGM measurements (CDF-native format), Parker Solar Probe WISPR image data (FITS-native format), and Global Ultraviolet Imager (GUVI) Spectral data (NetCDF-native format). We converted each data set to all three types (FITS, CDF, NetCDF) and measured the resulting file sizes. File sizes upon conversion were somewhat inconsistent, and we do not recommend converting existing archives at this time.
2.2 Support examples in AWS
We provide short code examples for accessing FITS, CDF and NetCDF data within the AWS S3 environment. In all cases we are accessing a presumed AWS S3 object that it in a bucket (S3 directory-equivalent) named “mybucket” with a file name in subdirectories path “mypath” named in some form of “example.*”. These three examples show the similarities and differences in accessing S3 data.
2.2.1 FITS
Access to FITS data is via the astropy and boto3 libraries.
import astropy. io.fits
s3c = boto3. client (‘s3′)
fobj = s3c.get_object (Bucket = 'mybucket',Key = 'mypath/example.fts')
rawdata = fobj ['Body']. read ()
bdata = io. BytesIO(rawdata).
Valid_data = astropy. io.fits.open (bdata).
Valid_header = valid_data [0]. header.
2.2.2 CDF
Access to CDF data is via the MAVENSDC cdflib and boto3 libraries.
import cdflib
Valid_data = cdflib. CDF (‘s3://mybucket/mypath/example.cdf).
2.2.3 NetCDF/HDF5
Access to NetCDF and HDF5 data is via the Python xarray library8,9.
import xarray
filename = “https://s3. yourregion.amazonaws.com/bucketname/mypath/example.h5”.
fgrab = fs. open (s3name).
Valid_data = xarray. open_data set (fgrab).
3 Results
We considered the following issues when examining each file format:
1) Compatibility with existing user code and tools.
2) Documented support by software package and programming lang.
3) Current usage of data format in heliophysics community.
4) Ease of conversion between formats.
5) Data file size.
6) Performance in AWS.
7) Support in multiple languages and support in Python.
3.1 File types
The most common file types in heliophysics and solar physics are currently CDF, FITS, and NetCDF/HDF5. CDF (Common Data Format10) has large file support and allows for compression, and has a large body of existing tools. Language bindings include Python, C, FORTRAN, Java, Perl, C#/Visual Basic, IDL, and Matlab. Many space physics data sets are in CDF format. However, not all Python implementations of CDF are cloud-aware, and it often requires an external C or Fortran library for rapid read speeds.
The FITS (Flexible Image Transport System11) standard is heavily used in solar physics and astronomy and has a large body of support tools. Conversely, as an older format, it is not designed for modern data. Its language bindings include C, FORTRAN, Python, C++, C#,.Net, Pascal, IDL, Java, JavaScript, Perl, Tcl, Matlab, LabVIEW, Mathematica, IGOR Pro, R, Photoshop (plugin), golang, and Swift. In Python, most implementations use the AstroPy fitsio library as a FITS handling, making the read/write usage identical across use cases.
As an evolution of FITS, ASDF (Advanced Scientific Data Format12) was proposed in 2015 (Greenfield et al., 2015) as an evolutionary replacement for the FITS format, which is widely-used in the astronomical community. It has language bindings in Python, C++ (incomplete), and Julia (incomplete). Its advantages include use of JSON, ability to stream data, and ability to be embedded in FITS. However, it is designed as an interchange format rather than an archival format, is not cloud-aware, and the standard is still young and in flux. FITS has also been implemented on top of HDF5 (Price et al., 2015).
NetCDF (Network Common Data Format13) is built on top of the HDF5 format and includes S3 support. HDF (Hierarchical Data Format14) is very wildly used, has a large existing body of tools, and is highly scalable. Language bindings exist for nearly every language. S3 support is built into NetCDF, and available for HDF5 via a virtual file systems layer and read-only. NetCDF is relatively stable while HDF5 is an evolving standard.
Zarr15 is a newer Python-supported cloud-native standard. It is based on NumPy, so a large body of existing code can use it. The Zarr standard is still evolving and is not natively used for existing helio/space physics data sets. Its most common use is currently as an alternative storage format specifically for cloud data.
3.2 File performance in python
Performance of file types on AWS was very dependent on the data type (time series, spectra, image) and instrument specifics. Performance also varies as individual Python libraries are improved and updated over time. For three sample cases (MMS FGM data in NetCDF4, GUVI spectral data in CDF, and WISPR images in FITS), we converted all three into FITS, CDF and NetCDF. We found file sizes varied by factors of 1-5x and S3 access times varied by 1-3x. Library support for chunking (not needing to load an entire file when only parts of it are needed) leads to implementation-specific speed advantages as well, and further library development is recommended.
Within the HelioCloud setup, we compare EC2 access to S3 access for the file types of FITS, CDF, and NetCDF using the Python-native interpreters of AstroPy, netCDF, and cdflib. The AstroPy FITS reader and the cdflib reader used the AWS-supported boto3 S3 library, while NetCDF requires use of the external fs. s3 file system library and the Xarray Python package. We ran this test twice over a 6 month period, during which the Python libraries had improved and produced more consistent results. We present the current state as of the September 2022 version of the libraries. Results are generally valid to within +/-10%, that is, for any given set of 100 runs, the timings will vary across runs but fall within 10% of our results here.
For a given file size, FITS reads were fastest, and CDF reads were slowest. Both FITS and CDF reads were longer as file sizes increased. The NetCDF4 read for the smallest file size took longer than for the two larger file sizes; as AWS S3 requires some overhead to initially access any file, that this indicates NetCDF4 read speeds are fast and the bottleneck is initially accessing S3. Averaged, typical FITS reads were on order of 20 ± 17 MB/s while CDF reads were on the order of 2.2 ± 1.6 MB/s and NetCDF at 24 ± 23 MB/sec. These rates are also on order of 2-3x faster than results from 6 months ago (not pictured), again an indication of library improvements. The wide variance of read times within each file type reinforces that all data conversions and transfers are very instrument-specific and thus broad conclusions over a ‘best’ file format cannot be made without specifically profiling the desired data set.
With the latest version of the libraries, we saw no strong performance preference for accessing a given instrument in its original native format, versus performance from the same data transformed into the other two file formats. As Python library support matures, this suggests that converting datasets to a file format for performance increases is not infeasible. The limiting factor in deciding on such conversions is whether the conversion itself is feasible. For our chosen sets, for example, the MMS FGM instrument was able to convert from CDF to NetCDF4, but the MMS FEEPS instrument was not able to generate a valid NetCDF4. Conversion between file formats requires that the new format support the multidimensionality and internal data organization and therefore we do not advise projects convert to a new format without first performing data validation.
We plot size (in MB) versus scaled read speeds (using average read time from 100 reads) [Figure 1] for three different instruments, each provided for the three different file types (of differing sizes). While the units are ‘MB’ and ‘seconds’, we recommend observing trends rather than documenting specific performance, primarily because in cloud environments the choice of cloud processor will affect read speeds. Therefore absolute values will differ based on hardware, while relative load speeds will remain proportional. In terms of file performance, one would expect a linear trend that, as file sizes increase, file read times also increase; this trend was consistent for FITS and CDF files but NetCDF4 file read times as noted struggled with the small GUVI data file.
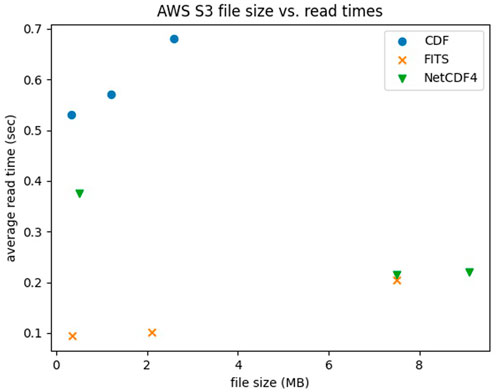
FIGURE 1. Average read speeds (in sec) for three different datasets (MMS, WISPR and GUVI) in each of three file formats (CDF, FITS and NetCDF4).
FITS was the fastest format, a result that surprised us; possibly it is because it is a flat format with little parsing needed, but we hesitate to ascribe a cause without further study. We recommend that library optimizations for S3-aware reads are low-hanging fruit in terms of boosting performance, preferential to reformatting entire data archives. Based on our analysis, we recommend cloud-ready archives do a quick check on their data to see if conversion yields significant (2x storage or >2x S3 speed) gains because of how the gains or losses are very instrument-specific. However, they should not abandon their native formats because those were chosen to support their existing user communities and toolsets, and need to be retained.
Additionally, ML-ready data sets should aim for accessibility and choose a format that allows analysis both within and outside of the Python ecosystem, with HDF5 as the strongest potential future candidate based on the number of languages that support it, support within existing Python libraries such as AstroPy (HDF native) and SunPy (HDF via NetCDF4 support).
3.3 Native python support
Defining the ecosystem of Python package support is always a work in progress, and we capture the current state of the heliophysics-relevant core Python package external file types and internal data types supported. There is a funded effort this year to ensure that SunPy, AstroPy, SciPy and HAPI are able to read each other’s formats. This and related efforts will go a long way towards simplifying the data landscape for Python users.
1) Pandas: CSV, NetCDF4, HDF5, Feather
2) Xarray: CSV, NetCDF3, NetCDF4, HDF5, Zarr, pandas
3) AstroPy [Pandas DataFrames]: CSV, FITS, HDF5
4) SpacePy [SpaceData, Numpy-compatible]: CDF, HDF5
5) SunPy [Map, TimeSeries]: FITS, pandas, AstroPy, J2000, ASDF, CDF, ANA, GenX
6) HelioPy: CDF, astropy, sunpy
7) SciPy: IDL, Matlab, pandas
8) PySat16: NetCDF, xarray, pandas.
We look at Python-specific library support by python data types and python packages. For each package, we summarize the file formats supported and indicate the data structures and basic readers included. In notable cases we include code examples illustrating the data reads. Note that Python natively supports CSV for Lists and Dictionaries. The primary internal data objects across Python heliophysics packages are:
• in AstroPy: Table
• in Numpy: ndarray
• in Pandas: DataFrame
• in SpacePy: SpaceData
• in SunPy: Maps, TimeSeries, NDCube
• in Xarray: xarray or DataFrame
3.4 Python packages
We provide per-package details on the primary Heliophysics Python packages, including their internal data object representations and which files they support, as well as additional relevant details. This information is intended to support users seeking out packages to work with specific files and data, and for developers who are interested in investigating package interoperability.
3.4.1 AstroPy (astropy.org)
The primary AstroPy data objects are Table and CCDData, and Pandas DataTable and DataFrame are supported. Supported file formats are HDF/HDF5, CSV and FITS, as well as JSON and others. The built-in read() and write() functions determine the file types when reading. CCDData can be converted to a Numpy ndarray or an NDData object. AstroPy is designed to support astronomy as well as encouraging interoperability.
3.4.2 Cdaswd library (cdaweb.gsfc.nasa.gov
The cdaswd library supports the SpacePy data model using the NASA CDF C library (requiring a C compiler or pre-built binary package to install), and also supports cdaweb’s netCDF17.
3.4.3 csv (built into python)
CSV files can be read into Python with no additional library needed.
3.4.4 Cdflib (github.com/MAVENSDC/cdflib)
A variant CDF library that does not require the NASA CDF C library, cdflib does require the Python Numpy package.
3.4.5 HAPI, hapi-server.org
The primary HAPI data objects are CSV or JSON data formatted to the HAPI specification. Available Java and Python HAPI server code bases support ingest and streaming of CDF, HDF, and CSV files. The HAPI Python client program uses Numpy arrays as its internal representation. HAPI is a time series download and streaming format specification intended as a common data access API for space science and space weather data.
3.4.6 HDF5 (h5py.org)
The primary HDF5 data objects are dataset (which are like Numpy arrays) and Group (which are like a Python dictionary). The h5py function is used to ingest files.
3.4.7 HelioPy (heliopy.org, no longer supported as of 2022)
The primary HelioPy data object is the SunPy TimeSeries object. HelioPy uses the Python CDFlib and can convert cdf to a Pandas Dataframe.
3.4.8 netCDF4 (unidata.github.io/netcdf4-python/)
The primary netCDF54 data objects are dataset and Variable.
3.4.9 NumPy, numpy.org
The primary NumPy (sometimes written as Numpy) data object is the ndarray. Numpy does not yet have a standard way to deal with metadata. Numpy supports CSV files and data. Many Python packages use ndarrays within their internal data representations.
3.4.10 Pandas (pandas.pydata.org)
The primary Pandas data objects is the DataFrame, and for HDF there is also HDFStore. Pandas supports HDF and CSV files. Pandas does not yet have a standard way to deal with metadata.
3.4.11 PyTables, pytables.org
The primary PyTables object is the Table, and it also supports any NumPy data type. PyTables is built on top of the HDF5 and NumPy libraries.
3.4.12 SciPy: scipy.org
SciPy is a package that can read IDL and Matlab data. It requires the NumPy, SciPy, Matplotlib, IPython, SymPy, and Pandas packages. SciPy provides algorithm and data structure support for a variety of science domains.
3.4.13 SpacePy: spacepy.github.io
The primary SpacePy data object is SpaceData and also has the dmarray class that is equivalent to NumPy arrays plus attributes. Supported file formats include CDF and HDF. SpacePy is NumPy-compatible. Future versions of SpacePy intend to use CDF as the primary internal datatype instead of SpaceData. SpacePy is a data anlysis, modeling and visualization package for space science.
3.4.14 SunPy (docs.sunpy.org)
The primary SunPy data objects are Map and TimeSeries. Supported file formats include FITS, but they recommend using the AstroPy FITS reader as it is more robust. Internal data is primary a wrapper around the Pandas DataFrame and NumPy ndarray. SunPy is an open source solar data analysis environment.
3.4.15 Xarray: xarray.pydata.org
The primary Xarray data object is the xarray, and Pandas DataFrame and NumPy ndarray are also supported. Supported file formats include CDF, NetCDF, HDF5, CSV and Zarr.
3.4.16 PySat (github.com/pysat)
The primary PySat data object is the xarray. Supported file formats include NetCDF3, NetCDF4 and HDF5. PySat is intended to provide an extensible framework for data sources from satellite instruments and constellations.
4 Discussion
Most languages suppx`ort heliophysics core datafile formats, but are not necessarily S3-aware. The 0th-level solution of “copy files from S3 to local storage” is always a fallback option, but we recommend resources be put into adding and improving S3 capabilities in libraries as this is generally not difficult (relies on existing external libraries and drivers) and benefits the community. Python has strong support for the major datafile formats both in S3 and local storage as a language, but no single package supports all the formats. For the major three formats of FITS, CDF, and NetCDF/HDF5, Python does support direct S3 reads, with overall good performance statistics.
For archives looking at reformatting existing data, L1/L2 bulk file convert should be tested, and is not usually recommended because the file size and performance gains are frequently marginal and occasionally detrimental. Analysis, model and ML-ready data set creation (L3/L4 data sets) should look into HDF5 because of its support and cross-compatibility across a variety of languages. As a side note, we notice the ML community L3/L4 data sets are currently a mix of “tools to make ML streams” and “L4 ML-ready data sets”.
Python supports S3 for the major file/data types, but there are idiosyncrasies across packages. In addition, Python + S3 is a dominant cloud architecture, but we note the community wants the user experience and UI to be more like their native tools. For specific Cloud and ML use cases, the trade-off between S3 access methods of “read into memory” (fast, resource-heavy) and “read chunks as needed from S3” (slow but resource-light) means use of High Performance Computing (HPC) tasks with S3 need to be diligent in tracking their disk and memory architecture for their specific task.
While there is no “one size fits all” approach to using S3 for heliophysics, there are also no impediments to using S3 and Cloud within Python in the current state of the art of the Python heliophysics ecosphere18.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://git.mysmce.com/heliocloud/cloud-tutorials/-/blob/master/S3_all_tests.ipynb.
Author contributions
AA and EW carried out the bulk of the analysis, while JB, BT, and JDV provided infrastructure and analysis support.
Funding
This project was supported under the NASA/Goddard Space Flight Center award AD-79106 “Enhancing Science Analysis with Cloud-Based Tools and Frameworks”. This work was enabled by the NASA GSFC HelioCloud environment.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
1https://heliophysicsdata.gsfc.nasa.gov/.
2https://towardsdatascience.com/guide-to-file-formats-for-machine-learning-columnar-training-inferencing-and-the-feature-store-2e0c3d18d4f9.
3https://github.com/MAVENSDC/cdflib.
4https://docs.sunpy.org/en/stable/code_ref/io.html.
5https://discourse.pangeo.io/t/netcdf-to-zarr-best-practices/1119.
6https://www.unidata.ucar.edu/mailing_lists/archives/netcdf-hdf/1992/msg00017.html.
7https://cdf.gsfc.nasa.gov/html/dttools.html.
8http://opendap.ccst.inpe.br/Observations/ARGO/tmp/netCDF4-0.9.8/docs/netCDF4.dataset-class.html.
9https://howto.eurec4a.eu/netcdf_datatypes.html.
11https://fits.gsfc.nasa.gov/.
12https://asdf-standard.readthedocs.io/.
13https://www.unidata.ucar.edu/software/netcdf/.
15https://zarr.readthedocs.io/en/stable/.
16https://github.com/pysat/pysatCDF.
17https://cdaweb.gsfc.nasa.gov/WebServices/REST/py/FAQ.html.
19https://support.hdfgroup.org/products/hdf5_tools/convert-netcdf.html.
20https://fits2hdf.readthedocs.io/en/latest/getting_started.html.
References
Burrell, A. G., Halford, A., Klenzing, J., Stoneback, R. A., Morley, S. K., Annex, A. M., et al. (2018). “Snakes on a spaceship—an overview of Python in heliophysics.” J. Geophys. Res. Space Phys., vol. 123, no. 12, Dec. 2018. DOI.org (Crossref), doi:10.1029/2018JA025877
Greenfield, P., Droettboom, M., and Bray, E. (2015). Asdf: A new data format for astronomy. Astronomy Comput. 12, 240–251. Astronomy and Computing. doi:10.1016/j.ascom.2015.06.004
Hassell, D., Gregory, J., Blower, J., Bryan, N. L, and Karl, E. T. (2017). “A data model of the climate and forecast metadata conventions (CF-1.6) with a software implementation (Cf-Python v2.1).” Geosci. Model Dev., vol. 10, no. 12, Dec.2017, pp. 4619–4646. DOI.org (Crossref), doi:10.5194/gmd-10-4619-2017
Lynnes, C., Quinn, P., Durbin, C., and Shum, D. (2020). Cloud optimized data formats. Comm. Earth Observing Satell. Meet. #4.
NASA/GSFC (2022). The HelioCloud Project [cloud environment] Available at: https://heliocloud.org.
Price, D. C., Barsdell, B., and Greenhill, L. (2015). Hdfits: Porting the FITS data model to HDF5. Astronomy Comput. 12, 212–220. arXiv.org. doi:10.1016/j.ascom.2015.05.001
SPDF Filenaming recommendations. Available at: https://spdf.gsfc.nasa.gov/guidelines/filenaming_recommendations.html.
Zaitsev, I. “The best format to save pandas data.” Medium, 29 Mar. 2019, Available at: https://towardsdatascience.com/the-best-format-to-save-pandas-data-414dca023e0d
Keywords: python (programming language), AWS (amazon web service) cloud services, NetCDF file format, HelioCloud, machine learning, cloud computing
Citation: Antunes AK, Winter E, Vandegriff JD, Thomas BA and Bradford JW (2022) Profiling heliophysics data in the pythonic cloud. Front. Astron. Space Sci. 9:1006839. doi: 10.3389/fspas.2022.1006839
Received: 29 July 2022; Accepted: 26 September 2022;
Published: 21 October 2022.
Edited by:
Angeline G. Burrell, United States Naval Research Laboratory, United StatesReviewed by:
Lei Cai, University of Oulu, FinlandLuke Barnard, University of Reading, United Kingdom
Copyright © 2022 Antunes, Winter, Vandegriff, Thomas and Bradford. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alex K. Antunes, c2FuZHkuYW50dW5lc0BqaHVhcGwuZWR1
 Alex K. Antunes
Alex K. Antunes Eric Winter1
Eric Winter1