- 1Department of Microbiology, Institute of Medical Sciences, Banaras Hindu University, Varanasi, Uttar Pradesh, India
- 2Department of Zoology, Institute of Science, Banaras Hindu University, Varanasi, Uttar Pradesh, India
Introduction: The present study aimed to optimize the doses and schedule of specific bacteriophage cocktails in freshwater fish infections as prophylactic and therapeutic measures.
Methods: The three most active phages against Aeromonas hydrophila (A. hydrophila) (φAHBHU12, φAHBHU16, and φAHBHU19) were characterized phenotypically and genotypically. Intramuscular and water immersion routes were used to calculate the absolute lethal dose of A. hydrophila in Pangasius buchanani. Phage therapy was given simultaneously and after 6, 12, and 24 h of bacterial challenge through intramuscular and water immersion routes.
Results: The prophylactic and early phage administration could save the fish. Furthermore, the dose of intramuscular 1.0 × 104 plaque-forming unit (PFU)/fish and water immersion 1.0 × 106 PFU mL–1 of the phage cocktail was optimal.
Discussion: The efficacy of bacteriophage therapy as preventive or curative measures practical when administered simultaneously or early hours of A. hydrophila infection in aquaculture systems. Phage-based approaches may be used as an alternative to antibiotics in aquaculture to reduce antibiotic use as a part of the “One Health” approach.
1 Introduction
Antibiotic-resistant bacteria are becoming more common as a result of antimicrobial resistance (AMR). The root cause of this problem is the indiscriminate use of antibiotics, including last-resort antibiotics such as colistin, in aquaculture, agriculture, and healthcare (Das et al., 2022). According to projections, AMR will cause 4.95 million deaths in 2019, with 1.3 million deaths directly attributable to treatment-resistant infections (Frei et al., 2023). The Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report 2021 (2021) reported that this figure is expected to rise to 10 million deaths by 2050, and according to the World Bank AMR could result in a 3.8% economic loss by 2050. This demonstrates the urgency with which we must act (O'Neill, 2016). As aquaculture activity expands and intensifies, the problem of microbial infections and the widespread utilization of antibiotics worsens.
Aquaculture is a thriving sector for developing countries in terms of growth and food security. Fisheries are one of India’s fastest-growing industries. Globally, edible fish stocks are declining, which is concerning given that fish is a good source of omega-3 fatty acids and protein (Tahar et al., 2018). According to Yue and Shen (2022), more fish for human consumption are produced by aquaculture than caught in the wild. In 2020, the commercial value of fisheries increased considerably, outpacing the average production rate in the 1990s by more than 60%. This remarkable increase in output is primarily due to the thriving aquaculture industry, which has grown faster than the global population. The report The State of World Fisheries and Aquaculture: Towards Blue Transformation, published in 2022, provides compelling evidence of fisheries’ and aquaculture’s growing importance as sources of food, nutrition, and employment. In 2020, production in these sectors reached a new high of 214 million tons, worth approximately US$424 billion (Of, 2022).
The growth of aquaculture has resulted in high fish mortality rates due to disease outbreaks (Stentiford, 2012). Globally, it is estimated that fish diseases lead to a loss of revenue amounting to US$6 billion annually (Stentiford et al., 2017), and reasons for this may be improper animal care and insufficient methods of protecting against diseases. Poor environmental conditions can lead to increased animal deaths and decreased productivity (Huicab-Pech et al., 2016). Fish production systems globally utilize biocide and antibiotic treatments to prevent infectious pathogens that can lead to diseases (Wanja et al., 2020). These pathogens are frequently present in the aquatic environment where the fish are reared (Kumar et al., 2018).
Aeromonas hydrophila is one of the major bacterial pathogens present in aquatic environments. A. hydrophila is a causative agent of tail and fin rot, hemorrhagic septicemia, also known as motile Aeromonas septicemia (MAS), hemorrhagic septicemia, ulcer disease, and red-sore disease (a disease in freshwater and, to a lesser extent, marine fish). The signs of hemorrhagic septicemia include erosion of the fins, loss of scales, hemorrhages of the gills and vents, abscesses and ulcers, abdominal distension, accumulation of ascitic fluid, anemia, and damage to internal organs and musculature, with generalized liquefaction in the infected fish. In the aquaculture sector, research on phage therapy has already begun, focusing on various pathogenic bacteria (e.g., Vibrio spp., Aeromonas spp.) (Mateus et al., 2014; Laanto et al., 2015; Kalatzis et al., 2016; Hockett and Baltrus, 2017; Duarte et al., 2018; Almeida et al., 2019; Kazimierczak et al., 2019). Research has focused less on phage administration doses and routes and has instead concentrated on the isolation and characterization of virulent phage cocktail formulations.
To reduce the economic loss, bacteriophage treatment of water bodies and fish infections may be one of the potential alternatives to antibiotics under the “One Health” concept. Therefore, the present study planned to evaluate the efficacy of phage therapy in preventing and establishing infections caused by a known fish pathogen, A. hydrophila, in freshwater fish, Pangasius buchanani, in terms of safe doses and timing of phage administration.
2 Material and methods
2.1 Isolation and characterization of A. hydrophila isolates
2.1.1 Isolation and biochemical identification of Aeromonas species
Bacteria were isolated from pond water and diseased fish (12–15 cm in length) near Varanasi, Uttar Pradesh, India. A total of 65 diseased fish were identified by swimming abnormality, pale gills, skin ulceration, and wounds on the body and tail. Specimens for isolation of bacteria were taken from the pond water sample, organs (gills, liver, kidney, ovary, and skin) of diseased fish during postmortem, and swabs from wounds. The culture was performed using blood agar (HiMedia Laboratories, Maharashtra, India) and MacConkey agar (HiMedia Laboratories) and incubated at 37°C overnight. The next day, the plates were examined for growth.
The circular off-gray colonies from blood agar and non-lactose fermenting colonies from MacConkey were subjected to Gram staining and biochemical testing for such things as motility, oxidase production, sugars fermentation, gas production, susceptibility to vibriostatic agent O/129 (decarboxylase testing), and esculin hydrolysis for the identification of A. hydrophila (Samal et al., 2014).
2.1.2 Characterization of the Aeromonas species
Biochemically identified Aeromonas species were subcultured on Mueller–Hinton agar (MHA; HiMedia Laboratories), and then DNA extraction was performed using the phenol–chloroform–isoamyl alcohol (25:24:1) method (Sambrook and Russell, 2006). The isolated genomic DNA was subjected to amplification using 16S rDNA-specific primers targeting genus-specific sequences of Aeromonas species (forward 5′-GGG AGT GCC TTC GGG AAT CAG A-3′ and reverse 5′-TCA CCG CAA CAT TCT GAT TTG-3′) (Hussain et al., 2014). The positive strains for Aeromonas species were further amplified by using A. hydrophila species-specific primers Ahh1 (forward 5′-GCC GAG CGC CCA GAA GGT GAG-3′ and reverse 5′-GAG CGG CTG GAT GCG GTT GT-3′) (Hussain et al., 2014). The master mix (Sigma-Aldrich, St. Louis, MO, USA) consisted of 50 ng of bacterial genomic DNA. The reaction mixture (25 µL) was subjected to 35 cycles on the following program: an initial melting temperature of 95°C for 5 min, with denaturation at 94°C for 45 s, annealing at 59°C for 45 s, and extension at 72°C for 1 s. The final extension was carried out at 72°C for 7 min in a thermal cycler (Bio-Rad Universal Hood II, Hercules, CA, USA). Then, PCR products were visualized on 1% agarose gel electrophoresis.
2.2 Isolation and characterization of A. hydrophila-specific bacteriophages
2.2.1 Bacteriophage isolation and purification
Phages were isolated from the pond, river, and sewage water by using the soft agar (0.8%) overlay method, following the method described by Kutter (2009), with some modifications. A. hydrophila was plated in a lawn culture (1.5 × 108 CFU mL–1) to isolate bacteriophages on MHA and incubated for 6 h to reach the log phase. Water specimens from different sources were collected and treated with 1% chloroform (Sigma-Aldrich) for 10 min and centrifuged three times for 15 min at 10,778 × g. One milliliter of the supernatant was flooded on the 6-h bacterial lawn of the A. hydrophila on an MHA plate and incubated overnight at 37°C. The next day’s lawn was washed with Tris-HCl magnesium gelatin (TMG, pH 7.4) buffer and centrifuged at 10,778 × g for 15 min.
Isolated phages were purified by using plaque counting by applying the 0.8% soft agar overlay method. The single isolated plaque was picked up for further processing (Orlova, 2012). The phage count was increased by lawn and spot methods (Marcó et al., 2012). For purification (toxin-free), the harvested fluid was subjected to membrane dialysis (membrane pore size 20 nm, HiMedia Laboratories) against poly ethylene glycol (PEG) 20% in 2.5 M sodium chloride (NaCl) solution (HiMedia Laboratories) overnight at 4°C and then washed with phosphate buffer saline. The dialysis and washing process was repeated twice (Gangwar et al., 2021).
2.2.2 Bacterial lytic activity of purified phages
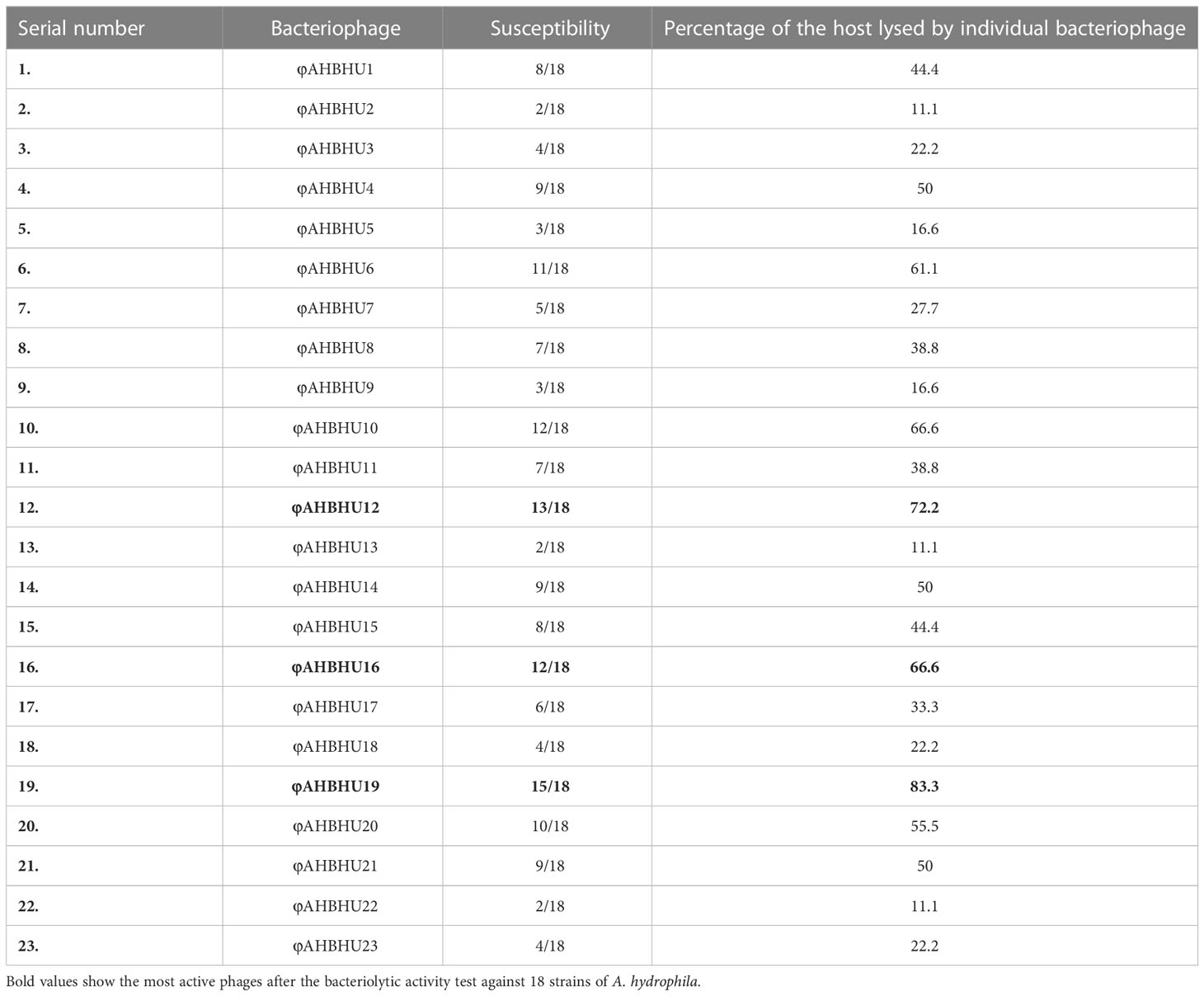
For their bacteriolytic activity, different isolated phages were subjected against different isolates of A. hydrophila. In addition, the lawn culture of A. hydrophila (1.5 × 108 CFU mL–1, 0.5 McFarland) was prepared on MHA. Ten microliters of each phage with the concentration of 1 × 109 PFU mL–1 was spotted on the MHA (Montso et al., 2019). The MHA plates were observed for the lysis of bacteria (clear zone) after incubation at 37°C overnight. Then, the three most active phages (i.e., φAHBHU12, φAHBHU16, and φAHBHU19) were selected for further characterization and experiments.
2.2.3 Host range determination
To see the spectrum of activity of these three phages (φAHBHU12, φAHBHU16, and φAHBHU19), we used different American Type Culture Collection strains of bacteria. The lawn culture of Pseudomonas aeruginosa, Aeromonas sobria, Escherichia coli, Plesiomonas shigelloides, Enterococcus faecalis, Salmonella Typhi, Acinetobacter lwoffii, Enterobacter cloacae, and Staphylococcus aureus (1.5 × 108 CFU mL–1) were made on MHA. The lytic activity of phages against all the bacterial species was determined by spot assay. Spot assay was used to determine the lytic spectrum activity of phage isolates, as described previously.
2.2.4 Effect of different pH and temperature on phage activity
The bacteriophage strains were screened for their lytic activity at different pH ranges (3-12). Bacteriophages (1 × 109 PFU mL–1 final concentrations) were incubated in equal amounts of TMG buffer (1:1) of different pH at 4°C for 2 h. After incubation, the lytic activity of bacteriophages was checked by the spot assay method.
Activities of bacteriophages were screened at different temperatures. For this screening, phages (1mL; 1 × 109 PFU mL–1 final concentrations) were incubated at different temperatures (–80°C, –20°C, 4°C, 28°C, 37°C, 45°C, and 55°C) for 48 h, and then spot assay was performed to check its activity.
2.2.5 Phage morphology
The most virulent phages (i.e., φAHBHU12, φAHBHU16, and φAHBHU19) with titer 1.0 × 1011 PFU mL–1 were filtered through a 0.22-µm syringe filter. The phage suspension was centrifuged at 21,124 × g for 90 min, and the supernatant was decanted. The pellet was washed three times with 0.1 M ammonium acetate (HiMedia Laboratories, pH 7.0). At the final step, the pellet was resuspended in ammonium acetate. The samples were treated with 2% uranyl acetate for negative staining and carbon-coated formvar films and examined under transmission electron microscopy (TALOS, Thermo Scientific, AIIMS, New Delhi, India). The diameter of the head and tail of these phages was determined by ImageJ software.
2.2.6 Restriction digestion analysis
Bacteriophage DNA was extracted and purified using a Norgen Biotek Corp. kit (cat. no 46800). Further phage DNA was subjected to restriction digestion using AluI (10 U µL–1) in accordance with the manufacturer’s instructions (Thermo Fisher Scientific) for 60 min at 37°C in a 20-μL reaction mixture containing 1 μL of DNA (concentration 1 μg μL–1), 2 μL of 10 × buffer, 1 μL of restriction enzyme, and 16 μL of sterile water. The digested products were electrophoresed with 1% agarose gel at 80 V for 1 h. The gel images were captured under ultraviolet light using a gel documentation system (BioRad Universal Hood II). The sizes of the DNA bands were estimated using 1 kb and 100 bp DNA ladders (Invitrogen, Thermo Fisher Scientific).
2.2.7 Genotyping of bacteriophage by enterobacterial repetitive intergenic consensus PCR
The genomic DNA of the phages against A. hydrophila was subjected to enterobacterial repetitive intergenic consensus (ERIC) PCR. The master mix contained 50 ng of phage genomic DNA. The primers were used to amplify phage genomic DNA (forward 5′-ATG TAA GCT CCT GGG GAT TCA-3′ and reverse 5′-AAG TAA GTG ACT GGG GTG AGC G-3′) (Ranjbar et al., 2017). The PCR mixture was subjected to the following program for amplification: melting temperature at 94°C for 7 min with denaturation for 45 s at 92°C, annealing at 31.8°C for 45 s, and extension at 72°C for 1 s. A final extension step at 72°C for 7 min was performed at the end of the 34 cycles. The PCR product was run on 1% agarose gel electrophoresis with 100 bp and 1 kb DNA ladder.
2.3 Fish infection and phage therapy
2.3.1 Experimental fish rearing
The experimental fish, P. buchanani, weighing 12–16 g, 12–15 cm in size, and 6–8 weeks old, were procured from commercial fish farms in Varanasi. The fish were reared in well-aerated 40-L rectangular glass aquariums in the Department of Microbiology, Institute of Medical Sciences, Banaras Hindu University, Varanasi, Uttar Pradesh, India. The permission of the institutional ethics committee was obtained (reference number Dean/2016/CAEC/70/dated 30 March 2017). Before the experiment, the fish were acclimatized for 1 week. Then, the fish were immersed in 0.01% potassium permanganate (HiMedia Laboratories) for 10 min to remove the parasitic infection (Prasad et al., 2011). The aquatic environment of the tank was maintained with dissolved oxygen 8.0 ± 0.5 mg-1, ammonia 0.6 ± 0.05 mg-1, pH 7.2 ± 0.2, and temperature 27°C ± 2°C without chlorination. Each aquarium was aerated with an air pump, and one-third of the water was replaced daily, dead fish were removed, and debris was siphoned from the bottom of the aquarium.
2.3.2 Determination of lethal doses
Two different routes of administration [i.e., intramuscular (IM) and water immersion] were used to establish A. hydrophila infection in the fish.
2.3.2.1 Determination of lethal dose 100 by intramuscular route
A. hydrophila was injected through the IM route in four groups containing 10 fish each group. In each tank (containing 2 L of water), add 0.1% Luria-Bertani broth (Himedia) for the given organic stress. The log phase of bacterial suspension was given to different groups of fish at doses of 8.0 × 102, 8.0 × 103, 8.0 × 104, and 8.0 × 105 CFU/fish. One group was injected with 100µL of 0.85% NaCl as a negative control. Then, we looked for lesions, sickness, and mortality. The lethality was observed for 7 days. After death, the fish were subjected to postmortem examination to ascertain the cause of death. Fish organs (liver, kidney, intestine, and stomach) were cultured on MHA and blood agar media. The whole experiment was repeated three times independently.
2.3.2.2 Determination of lethal dose 100 by water immersion
Forty fish were divided into four groups (10 fish in each group) in 2 L of water with 10 mL of Luria Bertani (LB) broth (HiMedia Laboratories). The first group was not infected. The other three groups were subjected to bacterial inoculation by putting them in a 2-L water tank with different concentrations of A. hydrophila (i.e., 1.0 × 105, 1.0 × 106, and 1.0 × 107 CFU mL–1), and in each tank 10 mL of LB broth was added. The fish were observed for 7 days for any morbidity and mortality (Sarker and Faruk, 2016).
2.3.3 Assessment of the efficacy of the phage cocktail on fish infection
The phage cocktail was used for prophylactic and therapeutic purposes. The fish experiments were set up according to the following plan:
● The first control group was given only 0.85% NaCl.
● The second control group was infected with bacteria only.
● The third control group was given only phage.
All the experiments were repeated three times for robustness of data.
2.3.4 Bacteriophage cocktail given through IM simultaneously, 6, 12, and 24 h after A. hydrophila challenge given through IM
Experimental groups were challenged with 100 μL of A. hydrophila by injecting 8.0 × 105 CFU/fish IM. The bacteriophage cocktail at the doses of 1.0 × 103, 1.0 × 104, 1.0 × 105, 1.0 × 106, 1.0 × 107, and 1.0 × 108 PFU/fish were given IM simultaneously at a different site from that of the bacterial injection.
Each group of 10 fish was placed in the 10-L aquarium. Fish were challenged with 100 μL of A. hydrophila at the dose of 8.0 × 105 CFU/fish through the IM route, and phage cocktails at the quantity of 1.0 × 104 PFU/fish were given simultaneously after 6, 12, and 24 h of bacterial challenge IM. The water was changed daily, as described elsewhere, and the experiment was monitored for 7 days.
2.3.5 Bacteriophage cocktail added into the water simultaneously, 6, 12, and 24 h after A. hydrophila challenge given through IM
Ten groups of P. buchanani were placed in separate aquaria. The fish were challenged with 100μL of A. hydrophila at a concentration 8.0 × 105 CFU/fish through the IM. The phage cocktail at different concentrations (i.e., 1.0 × 104, 1.0 × 105, 1.0 × 106, 1.0 × 107, 1.0 × 108, and 1 × 109 PFU mL–1) was administered through water immersion simultaneously.
The groups, comprising 10 fish each, were challenged with 100 μL of A. hydrophila (8.0 × 105 CFU/fish) through the IM route. However, phage cocktails containing 1.0 × 108 PFU mL–1 were added into the water tank after 6, 12, and 24 of bacterial challenge.
2.3.6 Bacteriophage cocktail added into the water simultaneously, 6, 12, and 24 after A. hydrophila challenge given through water immersion
In water immersion, 1.0 × 107 CFU mL–1 bacterial suspension was added to the 2-L water aquaria with 10 mL of LB broth, and phages cocktails were given simultaneously at a concentration of 1.0 × 103, 1.0 × 104, 1.0 × 105, 1.0 × 106, 1.0 × 107, 1.0 × 108, and 1.0 × 109 PFU mL–1, with 10 mL of LB broth in 2 L by adding in water. Furthermore, the phage cocktail (1.0 × 106 PFU mL–1) was given at 6, 12, and 24 after adding bacteria into the water.
3 Statistical method
One-way ANOVA was applied to check the quality of the data before making different comparisons. When we found that there was a statistically significant difference between the means of three or more independent groups by using an ANOVA test, we used the post hoc test to compare the groups in pairs to obtain the significance levels. Survival at 0 h with survival at 6, 12, and 24 h were compared using t-test. The Student’s t-test was applied to compare the means of paired groups using SPSS package.
4 Results
4.1 Identification of A. hydrophila isolates
A total of 38 Aeromonas spp. were isolated from diseased fish and pond water. From a biochemical and molecular basis, 18 isolates could be confirmed as A. hydrophila, Gram-negative, motile, and oxidase-positive bacteria. It could ferment sugars with gas production and resistance to vibriostatic agent O/129 and hydrolyze esculin hydrolysis. Confirmation of Aeromonas spp. and A. hydrophila was obtained by using primers that are specific to their genus and species. This was determined by the amplicon sizes of 356 bp and 130 bp, respectively (see Figures 1A, B). The AhBHU111 strain of A. hydrophila was used for further experiment.
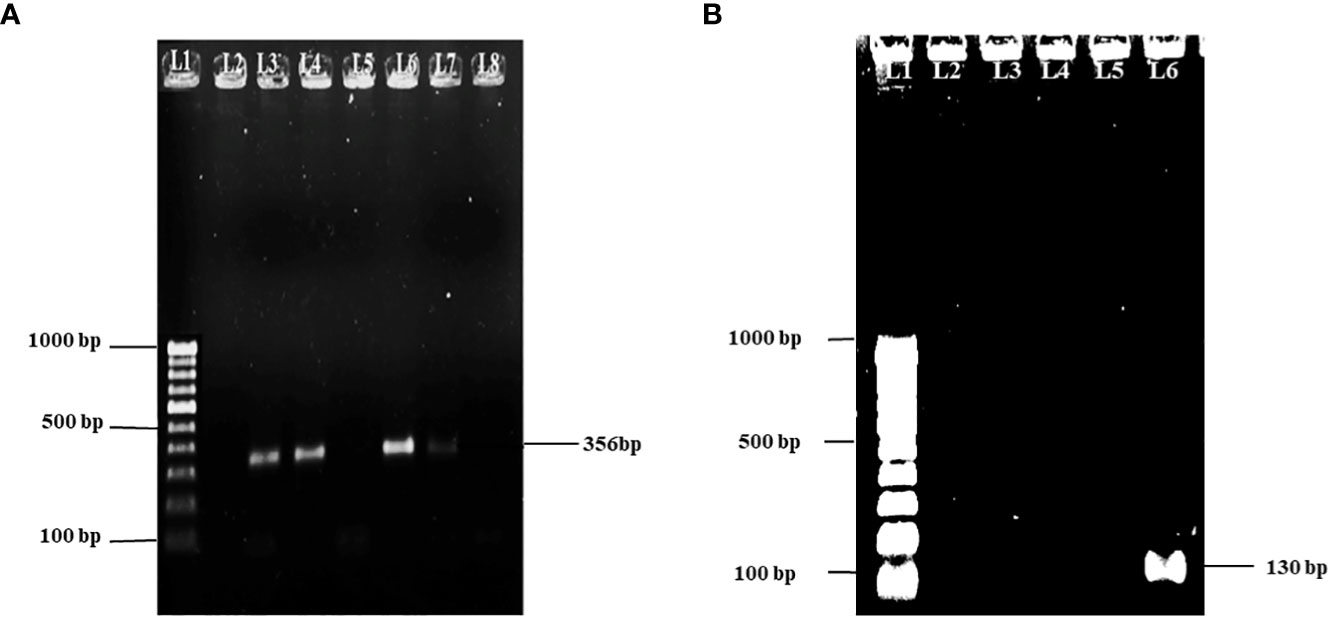
Figure 1 (A) Gel picture showing 100 bp molecular marker in lane 1 and 356 bp amplicon specific for Aeromonas spp. in lanes 3, 4, 6, and 7. (B) Gel picture showing 100 bp molecular marker in lane 1 and 130 bp amplicon specific for A. hydrophila in lane 6.
4.2 Characterization of A. hydrophila-specific bacteriophages
4.2.1 Bacterial lytic activity of isolated bacteriophages
A total of 18 strains of A. hydrophila were tested against 23 bacteriophages by spot assay. In detail, the most virulent bacteriophages, φAHBHU12 (72.2%), φAHBHU16 (66.6%), and φAHBHU19 (83.3%), were used for further experiment (see Table 1).
4.2.2 Host range determination
The most virulent bacteriophages [φAHBHU12 (lysing 72.2%), φAHBHU16 (66.6%), and φAHBHU19 (83.3%)] were selected for further characterization. The three phages could not lyse Pseudomonas aeruginosa, Aeromonas sobria, Escherichia coli, Salmonella Typhi, Acinetobacter lwoffii, Enterobacter cloacae, Plesiomonas shigelloides, Enterococcus faecalis, and Staphylococcus aureus.
4.2.3 Effect of different pH and temperature on phage activity
Good lytic activity of the bacteriophages against A. hydrophila was observed at pH 3–12 (see Figure 2A), and the three phages could survive well at temperatures –80°C, –20°C, 4°C, 28°C, and 37°C. However, φAHBHU12 and φAHBHU19 had satisfactory activity (approximately 50%) at 45°C (see Figure 2B).
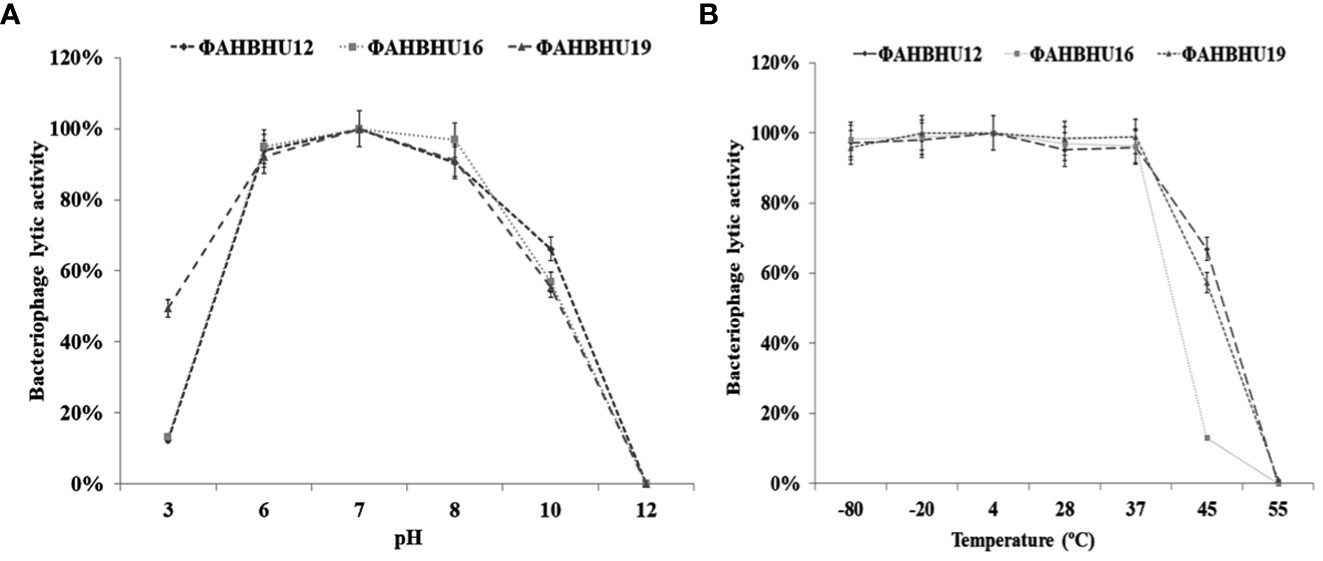
Figure 2 (A) Graph showing effect of pH on Aeromonas hydrophila-specific bacteriophages. (B) Graph showing effect of temperature on A. hydrophila-specific bacteriophages.
4.2.4 Morphological characterization
The bacteriophages φAHBHU12, φAHBHU16, and φAHBHU19 were examined under transmission electron microscopy cryo-TEM (Talos, Thermo Fisher Scientific, Waltham, MA, USA). Based on previous classification (Dion et al., 2020), the φAHBHU12 could be placed in the Podoviridae family, as the diameter of the isometric head was 52.47 nm, and a non-contractile short tail could not be visualized (see Figure 3A). In contrast, bacteriophage φAHBHU16 belongs to the Siphoviridae family, having an icosahedral head (47.82 nm in width) and a long non-contractile tail ranging from 93.55 to 117.30 nm in length (see Figure 3B). The φAHBHU19 could be classified as belonging to the Corticoviridae family, having an icosahedral head (42.17 nm in width) and no tail (see Figure 3C).
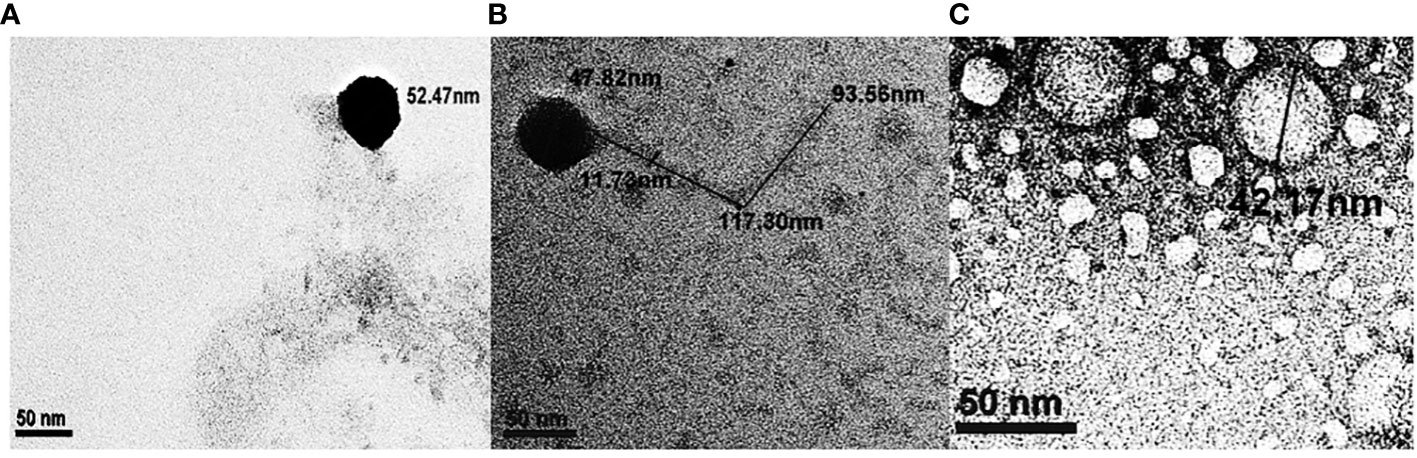
Figure 3 Transmission electron micrograph of bacteriophages, (A) φAHBHU12 belongs to the Podoviridiae family, (B) φAHBHU16 belongs to the Siphoviridae family, and (C) φAHBHU19 belongs to the Corticoviridae family.
4.2.5 The whole-genome fingerprinting by using restriction enzyme digestion (AluI) and enterobacterial repetitive intergenic consensus PCR
The isolated three phages (φAHBHU12, φAHBHU16, and φAHBHU19) were further characterized at the level of whole-genome fingerprinting by restriction digestion and ERIC PCR of their genomic DNA. Different banding patterns were indicated in the restriction digestion with AluI (Figure 4A). Also, in ERIC PCR, distinct band patterns were observed in all three phages (Figure 4B). The results delineate that these three phages have different genetic characteristics.
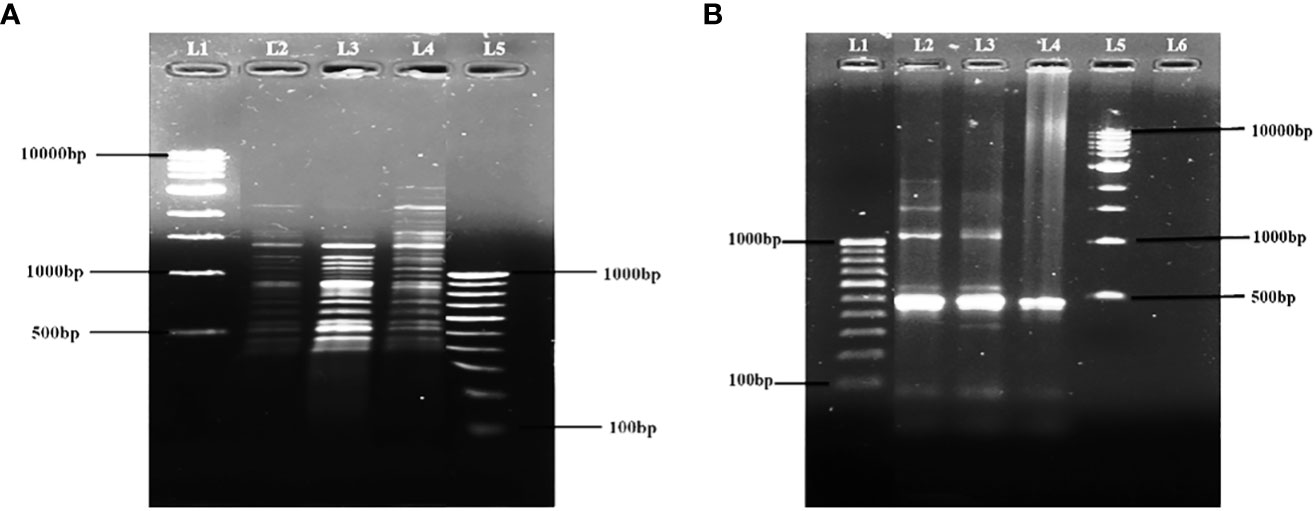
Figure 4 Fingerprinting of φAHBHU12, φAHBHU16, and φAHBHU19 by (A) Restriction digestion with AluI [L1- molecular marker (1 kb), L2- φAHBHU12, L3- φAHBHU16, L4- φAHBHU19, and L5 molecular marker (100 bp)], and (B) ERIC PCR [L1 molecular marker (100 bp), L2- φAHBHU12, L3- φAHBHU16, L4- φAHBHU19, and L5- molecular marker (1 kb marker)].
The isolated potent phages were further characterized at the genomic level using RAPD-PCR and restriction digestion by isolating their genomic DNA. Different banding patterns were observed in the RAPD-PCR analysis (Figure 3A). Also, the restriction digestion with EcoRI shows a distinct band pattern in all three phages (Figure 3B). The results delineate that the isolated potent phages have different genetic makeup.
4.3 Fish infection and phages therapy
4.3.1 Lethal dose 100 of A. hydrophila on intramuscular administration and water immersion
The lethal dose of A. hydrophila killing the P. buchanani fish weighing 12–16 g on intramuscular injection in 7 days was 8.0 × 105CFU/fish. However, the lethal dose through the water immersion route was 1.0 × 107 CFU mL–1 within 7 days when mixed with 10 mL of LB broth containing 2 L of the water tank as organic contamination. Interestingly, no death could be observed in the absence of LB broth (organic matter).
4.3.2 Protection by phage cocktail at different routes of phage administration at different time intervals after A. hydrophila infection
4.3.2.1 Intramuscularly administered varying phage cocktail dose
Figure 5A (IM) shows that when a simultaneous lethal dose of A. hydrophila and different concentrations of phage cocktails were injected through the intramuscular route, 1.0 × 104 PFU/fish could provide 93% protection, which was comparable to the dose of 1.0 × 105 PFU/fish. However, a dose of 1.0 × 106 PFU/fish led to the significantly lower protection of only 43% of the reared fish. When the doses were further reduced to 1.0 × 103 PFU/fish, 57% of the P. buchanani were protected (see Table 2). Better protection (83% and 63%, respectively) could be observed with the intervention at 6 and 12 h at the dose of 1.0 × 105 PFU/fish (see Figure 5B; IM). However, the intervention with phage therapy carried out 24 h after the A. hydrophila challenge resulted in significantly lower protection than when it was carried out simultaneously (see Table 3).
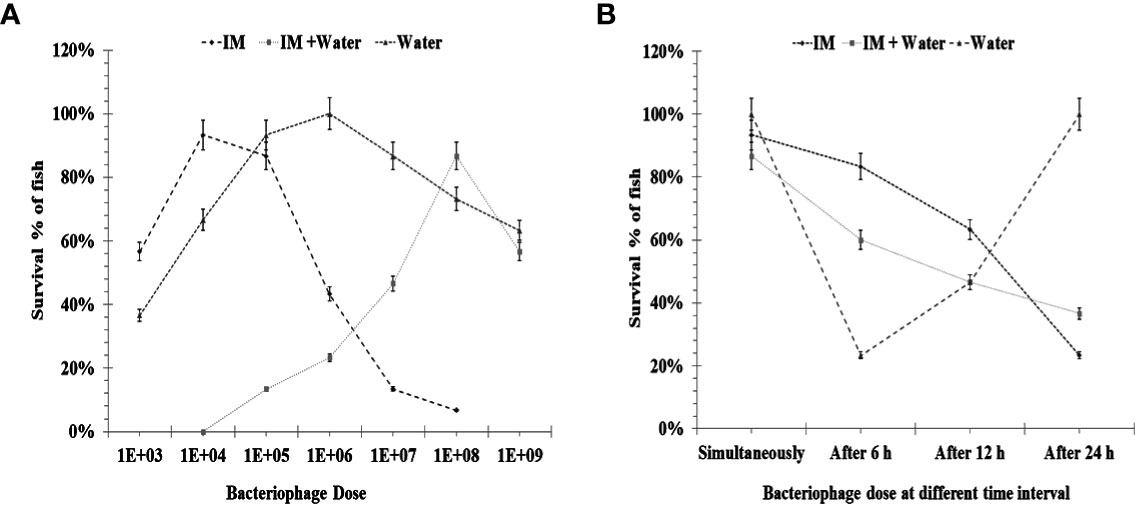
Figure 5 (A) Showing efficacy of phage administration through intramuscular and different doses of bacteriophage when given simultaneously through water immersion. IM: Effect of bacteriophage therapy on Aeromonas hydrophila (8.0 × 105 CFU/fish) infection in Pangasius buchanani. Bacteria and different doses of bacteriophage were given simultaneously through the intramuscular route. IM + water: Showing efficacy of simultaneous addition of different doses of bacteriophage cocktail in water and A. hydrophila (8.0 × 105 CFU/fish) challenge intramuscular in P. buchanani. Water immersion: Effect of bacteriophage therapy on A. hydrophila (1.0 × 107 CFU mL–1) infection in P. buchanani. Bacteria and different doses of bacteriophage were given simultaneously through water immersion. (B) Showing efficacy of phage administration through intramuscular and water immersion at different time intervals after bacterial challenge. IM: Effect of bacteriophage (1.0 × 104PFU/fish) therapy given simultaneously, after 6 h, 12 h, and 24 h of A. hydrophila infection (8.0 × 105CFU/fish) in P. buchanani. Both doses were given through the IM route. IM + water immersion: Effect of bacteriophage (1.0 × 108 PFU mL–1) added into the water (simultaneously, after 6 h, 12 h, and 24 h) with A. hydrophila infection (8.0 × 105CFU/fish) intramuscularly in P. buchanani. Water: Effect of bacteriophage (1.0 × 108 PFU mL–1) added into the water (simultaneously, after 6 h, 12 h, and 24 h) with A. hydrophila infection (8.0 × 105CFU/fish) intramuscularly in P. buchanani..
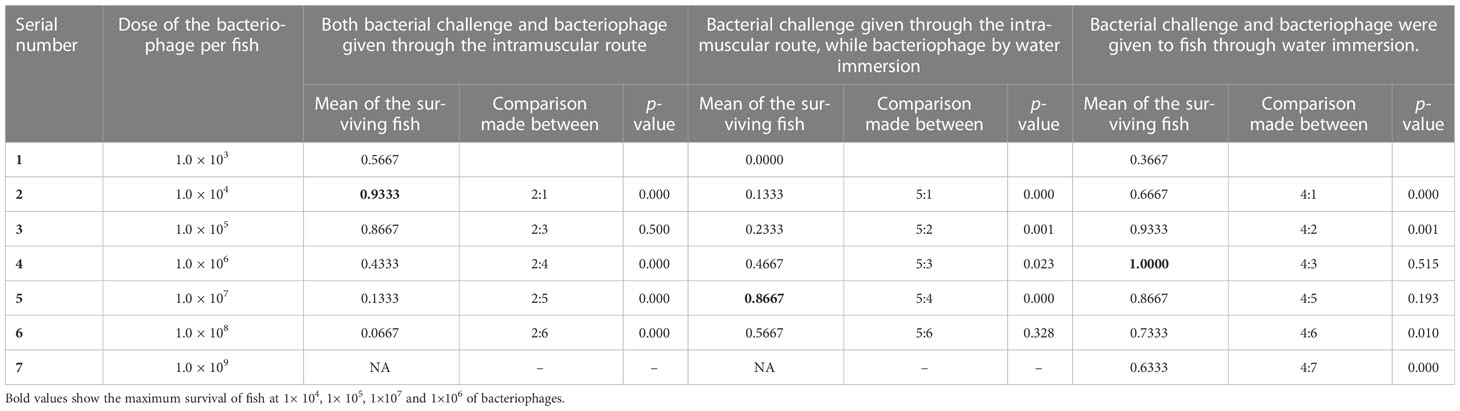
Table 2 Showing efficacy of phage treatment given to the fish by administering simultaneously through intramuscular and water immersion routes at different doses (number of fish in each group = 30).
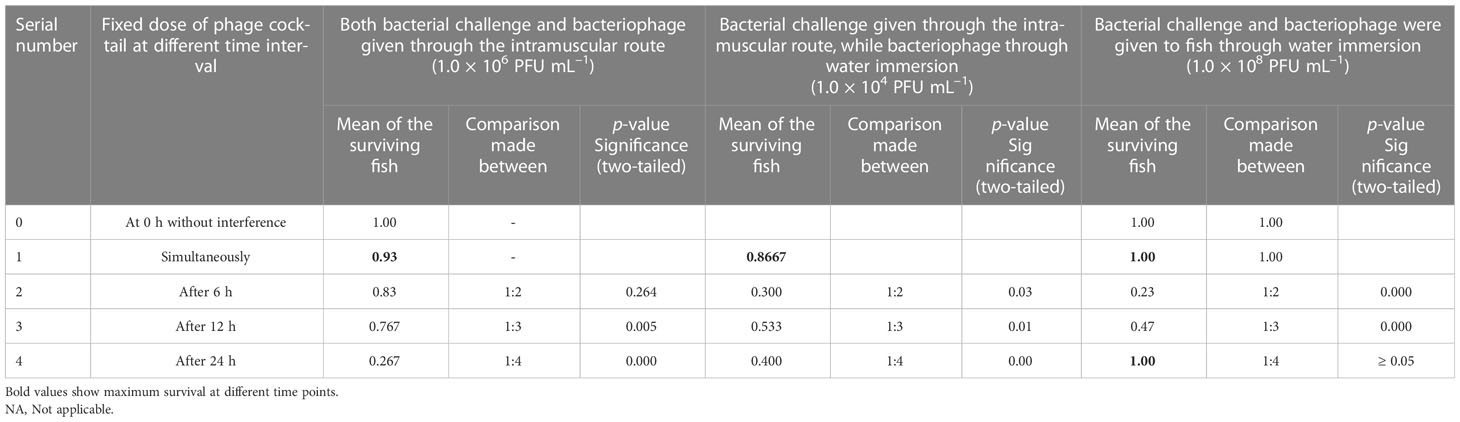
Table 3 Showing efficacy of phage treatment given to the fish by administering simultaneously and at different time intervals after bacterial challenge through intramuscular and water immersion routes at effective doses (number of fish in each group = 30).
4.3.2.2 Route of bacterial dose intramuscular and phages administration challenge through water immersion at different time intervals
The highest protection (87%) through the water immersion route could be observed with the dose of 1.0 × 108 PFU mL–1 in simultaneous addition to aquarium water after IM injection of A. hydrophila 8.0 × 105 CFU/fish. However, comparable protection could also be achieved by mixing the phage cocktail at a concentration of 1.0 × 109 PFU mL–1. However, lowering the phage concentration to 1.0 × 107 PFU mL–1 and 1.0×106 PFUmL-1 resulted in a significant decrease in the protection level to 47% and 23%, respectively (see Figure 5A; IM + water). Interestingly, the phage cocktail at the dose of 1.0 × 108 PFU mL–1 added 6 h after the bacterial challenge provided comparable protection with that given simultaneously at a similar concentration of phage cocktail (see Figure 5B; IM + water).
4.3.2.3 Bacterial dose and varying phage cocktail dose at different time intervals through water immersion
Figure 5A (water) shows that the bacteriophage cocktail concentrations of 1.0 × 105 PFU mL–1 and 1.0 × 106 PFU mL–1 gave comparable protection (93% and 100%, respectively) when both bacterial challenge and phage therapy were given water immersion simultaneously; however, a lower dose of 1.0 × 104 PFU mL–1 gave significantly lower (67%) protection to P. buchanani (see Table 2). However, a 6-h delay in administering a phage cocktail of 1.0 × 106 PFU mL–1 led to only 23% protection, while a delay of 12 h protected 47% of the fish. The other notable finding was that no mortality was observed when the phage cocktail of 1.0 × 106 PFU mL–1 was added to the water 24 h after the P. buchanani infection by A. hydrophila (8.0 × 107 CFU mL–1) (see Figure 5B; water).
5 Discussion
Fisheries and aquaculture production are vital protein sources for human beings, and a growth in aquaculture production is required to meet the high demand for fish and seafood worldwide. However, despite an upsurge in freshwater aquaculture, various microbial infections, including bacteria, pose a big challenge. Therefore, bacteriophage therapy is becoming increasingly popular as a promising alternative to treat/prevent bacterial infection in livestock, including in fish rearing.
However, several conditions must be fulfilled to make the bacteriophages effective in aquaculture. Therefore, the present study aimed to see bacteriophages as preventive/curative tools in aquaculture systems. For this purpose, we reared the P. buchanani fish and determined the lethal dose of A. hydrophilia bacteria by intramuscular and water immersion routes. Furthermore, we evaluated the amount, timing, and mode of delivery of bacteriophages as the prophylactic and therapeutic measures in freshwater aquaculture systems against known fish pathogens (i.e., A. hydrophila).
When P. buchanani was injected intramuscularly, a dose of 8.0 × 105 CFU/fish resulted in the death of all fish within 7 days. However, on simultaneous administration of phage cocktails through the IM route, the doses of 1.0 × 104 and 1.0 × 105 PFU/fish gave the best protection (93% and 87%, respectively). However, decreasing or increasing doses failed to prevent death. A phage cocktail given within the first 6 h protected 83% of the fish. However, high mortality rates (63% and 23%, respectively) could be observed when the injection of the phage cocktail was delayed by 12 and 24 h through the IM route. This observation indicates that the sooner the phage therapy is instituted, the better the results.
In the second experiment, we tried to determine the effect of variation in the route of administration of phages in fish having induced infection by A. hydrophila through the IM route. Interestingly, a higher dose (i.e., 8.0 × 108 PFU mL–1) provided the best protection when the phages were given in water simultaneously. However, the protection rate was significantly reduced when the water immersion phage cocktail was delayed by 24 h in the aquarium containing fish infected intramuscularly.
We induced the lethal infection in the third experiment by adding 1.0 × 107 CFU mL–1 A. hydrophila to the water tank. The lethal dose for P. buchanani could be 1.0 × 107 CFU mL–1 when organic material was added to induce the stress. This observation of organic matter contamination indicates that stress is essential to initiate and establish the infection. We evaluated the different doses of bacteriophage cocktails at different time intervals in the aquaria. Intriguingly, while unprotected fish were dying within 7 days with 1.0 × 107 CFU mL–1, the simultaneous addition of phage cocktail in the water body at concentrations of 1.0 × 105 and 1.0 × 106 PFU mL–1 resulted in 93% and 100% protection, respectively. However, a lower dose of 1.0 × 104 PFU mL–1 could protect only a few of the fish. It is worth noting that when the addition of phage cocktail (1.0 × 106 PFU mL–1) in the water tank was delayed by 6 and 12 h, the protection rates were significantly lower. Surprisingly, when the same dose was delayed by 24 h the mortality was reduced to zero (i.e., 100% protection). Therefore, the amount of bacteriophage cocktail given at a particular time of infection seems extremely important. This varying protection is explained based on the zone phenomenon, an optimum number of bacteriophages in the cocktail leading to sudden lysis of all the bacteria and releasing a considerable amount of endotoxin to which fish succumb. At an early stage of infection, the bacterial count was less, while at a later stage, the bacterial count was higher than the bacteriophages. Even when bacteria are lysed slowly because of their small/large number, tolerable endotoxin is produced. In the latter case, because of a low multiplicity of infection of phages, sudden lysis of the bacteria did not occur, leading to a gradual release of endotoxin. A similar phenomenon has already been reported in treating septicemia with bacteriophages in animal models (Patel et al., 2021; Singh et al., 2022). Consistent with our findings, Zhang et al. (2015) and Karunasagar et al. (2007) have shown the protection of sea cucumber against Vibrio alginolyticus by a spectrum of bacteriophages.
Contrary to this, a few studies have mentioned that lower doses yielded adequate protection, and a difference in treatment efficacy with different quantities of phage cocktails was not reported (Li et al., 2016). In one study, small doses worked because the bacteriophages were self-perpetuating (Lomelí-Ortega and Martínez-Díaz, 2014). A study published in 2017 reported 70% protection when small doses of water immersion administration were carried out shortly after the infection (Wang et al., 2017). Therefore, the phage cocktail given at a particular point in time and the severity of illness may give variable protection. Thus, we can see that prophylactic or early phage therapy provides better protection in freshwater aquaculture systems.
Earlier reports had stated that prophylactic use of bacteriophages 24, 12, and 6 h before bacterial challenge gave significant protection irrespective of the route of administration. Lomelí-Ortega and Martínez-Díaz (2014) have also reported better protection using prophylactic doses and unsatisfactory outcomes when prophylactic use of bacteriophages started 24 h post infection, which agrees with our observation. However, when a phage cocktail was administered in water through immersion, significantly higher (100 to 1,000 times) doses per unit volume were required. A possible explanation for this may be that phages get adsorbed to specific receptors on phage-sensitive bacteria and dead and phage-resistant bacteria. It has already been reported that the immunoglobulin-like domain on the surface of phages makes them more susceptible to getting trapped in the intestinal mucosa (Fraser et al., 2006; Lepage et al., 2008). In addition, the gut is the most diversified, crowded organ of fish and animals, comprising eukaryotic cells, bacteria, fungi, and viruses (Sausset et al., 2020). Therefore, competitive inhibition may also play a role in the efficiency of lysing the target bacteria.
There are several issues to resolve while planning for phage therapy in aquaculture. First, the observations made in this study should not be generalized to all situations in freshwater aquaculture. Lower and repeated doses may reduce mortality when the infection is of longer duration, although this needs to be explored further. Last, if there is an increase in the density of the known bacterial pathogen in the aquaculture system, adding their respective phage cocktail in anticipation at adequate doses as prophylaxis may prevent infection and unnecessary use of antibiotics.
Data availability statement
The original contributions presented in the study are included in the article. Further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by the institutional Ethics Committee for animals permitted the protocol for the study, which was carried out during Dean/2016/CAEC/70 dated 30.03.2017at the Institute of Medical Sciences, Banaras Hindu University, Varanasi, India.
Author contributions
GN, DK, and RC conceived the idea. RK, RY, GN, DK, and RC executed the experimental work and analyzed the data. GN, RK, and DK wrote the initial manuscript. GN, DK, and RC completed the final editing of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by funding from Aquaculture & Fisheries Biotechnology, Department of Biotechnology, Ministry of Science and Technology, Government of India, grant No.BT/PR24773/AAQ/3/863/2017dt. 22/03/2018R/Dev./P-07/671dt.29/05/2018.
Acknowledgments
The authors are also grateful to State Level Viral Research and Diagnostic Laboratory, Department of Microbiology, Institute of Medical Sciences, Banaras Hindu University, for providing an infrastructural facility for the bench work. In addition, we are indebted to the Central Institute of Freshwater Aquaculture, Bhubaneshwar, for providing reference strains. The authors are also grateful to Subhash Lal Karan, Department of Microbiology, Institute of Medical Sciences, Banaras Hindu University, for the statistical analysis of the data. Finally, the authors thank the Sophisticated Analytical Instrumentation Facility, AIIMS Delhi, India, for transmission electron microscopy.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Almeida G. M. F., Mäkelä K., Laanto E., Pulkkinen J., Vielma J., Sundberg L. R. (2019). ‘The fate of bacteriophages in recirculating aquaculture systems (RAS)–towards developing phage therapy for RAS. Antibiotics. 8, 192. doi: 10.3390/ANTIBIOTICS8040192
Briggiler Marcó M., Garneau J. E., Tremblay D., Quiberoni A., Moineau S. (2012). Characterization of two virulent phages of lactobacillus plantarum. Appl. Environ. Microbiol. 78 (24), 8719–8734. doi: 10.1128/AEM.02565-12/ASSET/4F0314A8-98EA-4D23-8712-C1D522716063/ASSETS/GRAPHIC/ZAM9991039300004.JPEG
Das R., Kotra K., Singh P., Loh B., Leptihn S., Bajpai U. (2022). Alternative treatment strategies for secondary bacterial and fungal infections associated with COVID-19. Infect. Dis. Ther. 11 (1), 53–78. doi: 10.1007/s40121-021-00559-8
Dion M. B., Oechslin F., Moineau S. (2020). Phage diversity, genomics and phylogeny. Nat Rev Microbiol. 18 (3), 125–138. doi: 10.1038/s41579-019-0311-5
Duarte J., Pereira C., Moreirinha C., Salvio R., Lopes A., Wang D., et al. (2018). New insights on phage efficacy to control aeromonas salmonicida in aquaculture systems: an in vitro preliminary study. Aquaculture 495, 970–982. doi: 10.1016/J.AQUACULTURE.2018.07.002
Fraser J. S., Yu Z., Maxwell K. L., Davidson A. R. (2006). Ig-like domains on bacteriophages: a tale of promiscuity and deceit. J. Mol. Biol. 359 (2), 496–507. doi: 10.1016/J.JMB.2006.03.043
Frei A., Verderosa A. D., Elliott A. G., Zuegg J., Blaskovich M. A. (2023). Metals to combat antimicrobial resistance. Nat. Rev. Chem. 7, 202–224. doi: 10.1038/s41570-023-00463-4
Gangwar M., Rastogi S., Singh D., Shukla A., Dhameja N., Kumar D., et al. (2021). Study on the effect of oral administration of bacteriophages in Charles foster rats with special reference to immunological and adverse effects. Front. Pharmacol. 12. doi: 10.3389/fphar.2021.615445
Hockett K. L., Baltrus D. A. (2017). Use of the soft-agar overlay technique to screen for bacterially produced inhibitory compounds. J. Vis. Exp. 2017 (119), 1–5. doi: 10.3791/55064
Huicab-Pech Z. G., Landeros-Sánchez C., Castañeda-Chávez M. R., Lango-Reynoso F., López-Collado C. J., Platas Rosado D. E. (2016). Current state of bacteria pathogenicity and their relationship with host and current state of bacteria pathogenicity and their relationship with host and environment in tilapia oreochromis niloticus. J. Aquacult. Res. Dev. 7(5), 1–0. doi: 10.4172/2155-9546.1000428
Hussain I. A., Jeyasekaran G., Shakila R. J., Raj K. T., Jeevithan E. (2014). Detection of hemolytic strains of aeromonas hydrophila and a. sobria along with other aeromonas spp. from fish and fishery products by multiplex PCR. J. Food Sci. Technol. 51 (2), 401–407. doi: 10.1007/S13197-013-1190-9
Kalatzis P. G., Bastías R., Kokkari C., Katharios P. (2016). Isolation and characterization of two lytic bacteriophages, φst2 and φgrn1; phage therapy application for biological control of vibrio alginolyticus in aquaculture live feeds. PloS One 11 (3), 1–18. doi: 10.1371/journal.pone.0151101
Karunasagar I., Shivu M. M., Girisha S. K., Krohne G., Karunasagar I. (2007). Biocontrol of pathogens in shrimp hatcheries using bacteriophages. Aquaculture 268 (1–4), 288–292. doi: 10.1016/J.AQUACULTURE.2007.04.049
Kazimierczak J., Wójcik E. A., Witaszewska J., Guziński A., Górecka E., Stańczyk M., et al. (2019). Complete genome sequences of aeromonas and pseudomonas phages as a supportive tool for development of antibacterial treatment in aquaculture. Virol. J. 16 (1), 1–12. doi: 10.1186/S12985-018-1113-5/TABLES/5
Kumar G., Engle C., Tucker C. (2018). Factors driving aquaculture technology adoption. J. World Aquacult. Soc. 49 (3), 447–76. doi: 10.1111/jwas.12514
Kutter E. (2009). Phage host range and efficiency of plating. Methods Mol. Biol. (Clifton N.J.) 501, 141–149. doi: 10.1007/978-1-60327-164-6_14
Laanto E., Bamford J. K., Ravantti J. J., Sundberg L. R. (2015). The use of phage FCL-2 as an alternative to chemotherapy against columnaris disease in aquaculture. Front. Microbiol. 6:829. doi: 10.3389/FMICB.2015.00829/BIBTEX
Lepage P., Colombet J., Marteau P., Sime-Ngando T., Doré J., Leclerc M. (2008). Dysbiosis in inflammatory bowel disease: a role for bacteriophages? Gut 57 (3), 424–425. doi: 10.1136/GUT.2007.134668
Li Z., Zhang J., Li X., Wang X., Cao Z., Wang L., et al. (2016). Efficiency of a bacteriophage in controlling vibrio infection in the juvenile sea cucumber apostichopus japonicus. Aquaculture 451, 345–352. doi: 10.1016/J.AQUACULTURE.2015.09.024
Lomelí-Ortega C. O., Martínez-Díaz S. F. (2014). Phage therapy against vibrio parahaemolyticus infection in the whiteleg shrimp (Litopenaeus vannamei) larvae. Aquaculture 434, 208–211. doi: 10.1016/J.AQUACULTURE.2014.08.018
Mateus L., Costa L., Silva Y. J., Pereira C., Cunha A., Almeida A. (2014). Efficiency of phage cocktails in the inactivation of vibrio in aquaculture. Aquaculture 424–425, 167–173. doi: 10.1016/J.AQUACULTURE.2014.01.001
Montso P. K., Mlambo V., Ateba C. N. (2019). Characterization of lytic bacteriophages infecting multidrug-resistant shiga toxigenic atypical escherichia coli O177 strains isolated from cattle feces. Front. Public Health 7 (November). doi: 10.3389/fpubh.2019.00355
Of T. H. E. S. (2022). The State of the World series of the Food and Agriculture Organization of the United Nations. World Fish. Aquacult. FAO.
O’Neill J. (2016). Review on antimicrobial resistance: tackling drug-resistant infections globally: final report and recommendations.
Orlova E. V. (2012). Bacteriophages and Their Structural Organisation, Bacteriophages, Kurtboke I. (Ed.). doi: 10.5772/34642
Patel S. R., Pratap C. B., Nath G. (2021). Evaluation of bacteriophage cocktail on septicaemia caused by colistin-resistant acinetobacter baumannii in immunocompromised mice model. Indian J. Med. Res. 154 (1), 141. doi: 10.4103/IJMR.IJMR_2271_18
Prasad Y., Kumar D., Sharma A. K. (2011). Lytic bacteriophages specific to flavobacterium columnare rescue catfish, clarias batrachus (Linn.) from columnaris disease. J. Environ. Biol. 32 (2), 161–168.
Ranjbar R., Tabatabaee A., Behzadi P., Kheiri R. (2017). Enterobacterial repetitive intergenic consensus polymerase chain reaction (ERIC-PCR) genotyping of escherichia coli strains isolated from different animal stool specimens. Iranian J. Pathol. 12 (1), 25–34. doi: 10.30699/ijp.2017.21506
Samal S. K., Das B. K., Pal B. B. (2014). Isolation, biochemical characterization, antibiotic susceptibility study of aeromonas hydrophila isolated from freshwater fish. Int. J. Curr. Microbiol. Appl. Sci. 3 (12), 259–267.
Sambrook J., Russell D. W. (2006). Purification of nucleic acids by extraction with Phenol:Chloroform. Cold Spring Harbor Protoc. 2006 (1), pdb.prot4455. doi: 10.1101/PDB.PROT4455
Sarker J., Faruk M. (2016). Experimental infection of aeromonas hydrophila in pangasius. Progressive Agric. 27 (3), 392–399. doi: 10.3329/pa.v27i3.30836
Sausset R., Petit M. A., Gaboriau-Routhiau V., De Paepe M. (2020). New insights into intestinal phages. Mucosal Immunol. 13 (2), 205–215. doi: 10.1038/s41385-019-0250-5
Singh A., Singh A. N., Rathor N., Chaudhry R., Singh S. K., Nath G. (2022). Evaluation of bacteriophage cocktail on septicemia caused by colistin-resistant klebsiella pneumoniae in mice model. Front. Pharmacol. 13 (February). doi: 10.3389/fphar.2022.778676
Stentiford G. D. (2012). Disease will limit future food supply from the global crustacean fishery and aquaculture sectors. J. Inverterbrat. Patholog. 110 (2), 141–57. doi: 10.1016/j.jip.2012.03.013
Stentiford G. D., Sritunyalucksana K., Flegel T. W., Williams B. A., Withyachumnarnkul B., Itsathitphaisarn O., et al. (2017). New paradigms to help solve the global aquaculture disease crisis. PLoS Pathog. 13 (2), e1006160. doi: 10.1371/journal.ppat.1006160
Tahar A., Kennedy A. M., Fitzgerald R. D., Clifford E., Rowan N. (2018). Longitudinal evaluation of the impact of traditional rainbow trout farming on receiving water quality in Ireland. PeerJ. 6, e5281. doi: 10.7717/peerj.5281
Wang Y., Barton M., Elliott L., Li X., Abraham S., O'Dea M., et al. (2017). Bacteriophage therapy for the control of vibrio harveyi in greenlip abalone (Haliotis laevigata). Aquaculture 473, 251–258. doi: 10.1016/J.AQUACULTURE.2017.01.003
Wanja D. W., Mbuthia P. G., Waruiru R. M., Mwadime J. M., Bebora L. C., Nyaga P. N., et al. (2020). Fish husbandry practices and water quality in central Kenya: potential risk factors for fish mortality and infectious diseases. Vet. Med. Intl. 2020, 6839354. doi: 10.1155/2020/6839354
Yue K., Shen Y. (2022). An overview of disruptive technologies for aquaculture an overview of disruptive technologies for aquaculture. Aquaculture Fisheries 7 (2), 111–120. doi: 10.1016/j.aaf.2021.04.009
Keywords: Aeromonas hydrophila, Pangasius buchanani, bacteriophage therapy, aquaculture, lethal
Citation: Kumari R, Yadav R, Kumar D, Chaube R and Nath G (2023) Evaluation of bacteriophage therapy of Aeromonas hydrophila infection in a freshwater fish, Pangasius buchanani. Front. Aquac. 2:1201466. doi: 10.3389/faquc.2023.1201466
Received: 06 April 2023; Accepted: 25 May 2023;
Published: 20 June 2023.
Edited by:
Beatriz Novoa, Spanish National Research Council (CSIC), SpainReviewed by:
Alberto Falco, Miguel Hernández University of Elche, SpainAlim Isnansetyo, Gadjah Mada University, Indonesia
Copyright © 2023 Kumari, Yadav, Kumar, Chaube and Nath. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gopal Nath, Z29wYWxuYXRoQGdtYWlsLmNvbQ==
†Present address: Ritu Kumari, Department of Botany, Institute of Science, Banaras Hindu University, Varanasi, Uttar Pradesh, India
‡These authors have contributed equally to this work
 Ritu Kumari1†‡
Ritu Kumari1†‡ Deepak Kumar
Deepak Kumar Radha Chaube
Radha Chaube Gopal Nath
Gopal Nath