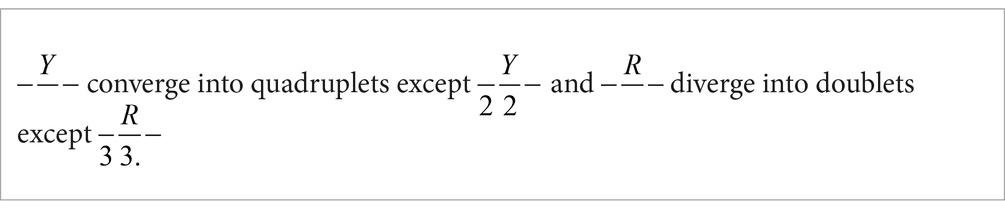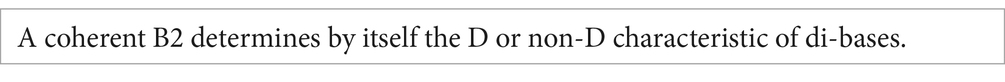- 1Grupo Interdisciplinario de Biología Teórica, Instituto de Neurociencia Cognitiva y Traslacional (INCyT) Universidad Favaloro-INECO-CONICET, Buenos Aires, Argentina
- 2Instituto de Biología Celular y Neurociencia (IBCN) UBA-CONICET, Facultad de Medicina, Universidad de Buenos Aires, Buenos Aires, Argentina
- 3Centro Internacional Franco-Argentino de Ciencias de la Información y de Sistemas (CIFASIS), CONICET, Rosario, Argentina
The present study is devoted to describing the “logic” implicit in the standard genetic code. Bases are considered as physicochemical entities possessing two essential properties: molecular type and number of Hydrogen bonds involved (bases pairing) in the codon-anticodon specific interactions. It is proposed that the codon structure possesses a dual informative function: on the one hand, it determines its discriminating or non-discriminating character, and on the other hand, it determines a specific amino acid. These two aspects constitute the codon global information. Two different sets of rules are introduced to describe these different phenomena. It is established that, depending on the type of base occupying the second position, only two or three of the six codon properties located at defined positions determine the discriminating or non-discriminating behavior. With regard to the amino acid determining function of the codons for different sets of synonymous (singlets, doublets, triplets, quadruplets, or sextets), the number of informative properties integrating the codon and their typical positions characteristically change. Based on the rules presented here, it can be postulated that a codon can be defined as an asymmetric informative entity, whose global informative capacity results from the spatially organized combination of the six properties assigned by the three bases.
1 Introduction
The genetic information required for protein synthesis is encoded in specific DNA segments, the so-called protein-coding genes. The genetic information resides on the specific sequence of four nucleotides possessing each of them a distinct nitrogenous base. The specific sequence of bases determines the specific amino acid (AAs) sequences of each protein. The sequence of DNA bases (exons) specifies the sequence of bases of the primary transcript (pre-mRNA). After processing the primary transcript, the sequence of codons (CD), i.e., a set of three adjacent bases of the mRNA determines the amino acid sequence of proteins. The genetic code is a set of rules that ribosome uses to incorporate AA into a protein using the information of the mRNA (1, 2). These rules are the net result of a complex process that involves several steps of specific molecular matching that takes place during the translation of mRNA into proteins: (a) aminoacyl-tRNA synthetases attach an amino acid to the cognate tRNA (“operational code”), (b) the aminoacyl-tRNA is then used for translation upon binding to mRNA according to (c) the codon-anticodon specific interaction on the ribosome (3–6).
Despite the abundant literature on these molecular interactions, a definitive physical–chemical explanation of the code degeneracy/redundancy is not yet available, and several theoretical models were proposed to explain it. In the present study, these molecular aspects are considered a “black box” and, based on theoretical analyses, explore the possibility of conceiving general formalizations that reliably describe the phenomenon of code degeneracy/redundancy.
The code redundancy refers to the fact that, excluding the 3 stop codons, 61 codons specify 20 amino acids. More specifically, this term refers to the following facts: (a) the anticodon of some tRNA matches more than one codon and (b) some amino acids can be specified by more than one tRNA type having different anticodons (7–11).
The set of codons that specify the same amino acid was classically designed as “synonymous.” As a rule, they share the bases located at the first and second positions. Thus, several authors define a codon as the association between a dinucleotide (those located in the first and second positions) and the nucleotide of the third position (3, 12–15). Instead of the term “di-nucleotide,” the term “di-base” is used in this study.
The set of four codons corresponding to a particular di-base behaves either as one quadruplet (one set of four synonymous) or as two doublets (two sets composed of two synonymous each) or displays an even more complex behavior that will be later described.
In the present analysis, di-bases are characterized as discriminating (D) or non-discriminating (Non-D). A di-base is defined as discriminating when its corresponding four codons diverge into two doublets depending on the molecular type (pyrimidine or purine) of the base in the third position. A di-base is defined as non-discriminating when its corresponding four codons behave as a quadruplet independently of the base in the third position.
The present study aims to define a set of rules that formally describes the D and Non-D character of the different types of di-bases. This study also attempts to characterize the roles played by different bases in assigning such character to di-bases. Given that bases are considered dual physicochemical entities with two essential properties: (a) molecular type (mT) and (b) the number of Hydrogen bonds (nHb) involved in the codon–anticodon interaction, the rules introduced in this study describe how the combination of four properties of a di-base assign discriminating or non-discriminating character to codons. Finally, the study also introduces rules that describe how different combinations of the six codon properties participate in amino acid specifications.
2 The rules of the genetic code degeneracy/redundancy
This section introduces statements that describe the “logic” implicit in the code degeneracy. These rules can be deduced from classical data about (a) the correspondence between codons and amino acids and (b) the occurrence of redundant codons (1, 2, 16).
2.1 Definitions and nomenclature
In the first part, some classical concepts will be recalled, new terms will be defined, and the relationship between them will be established.
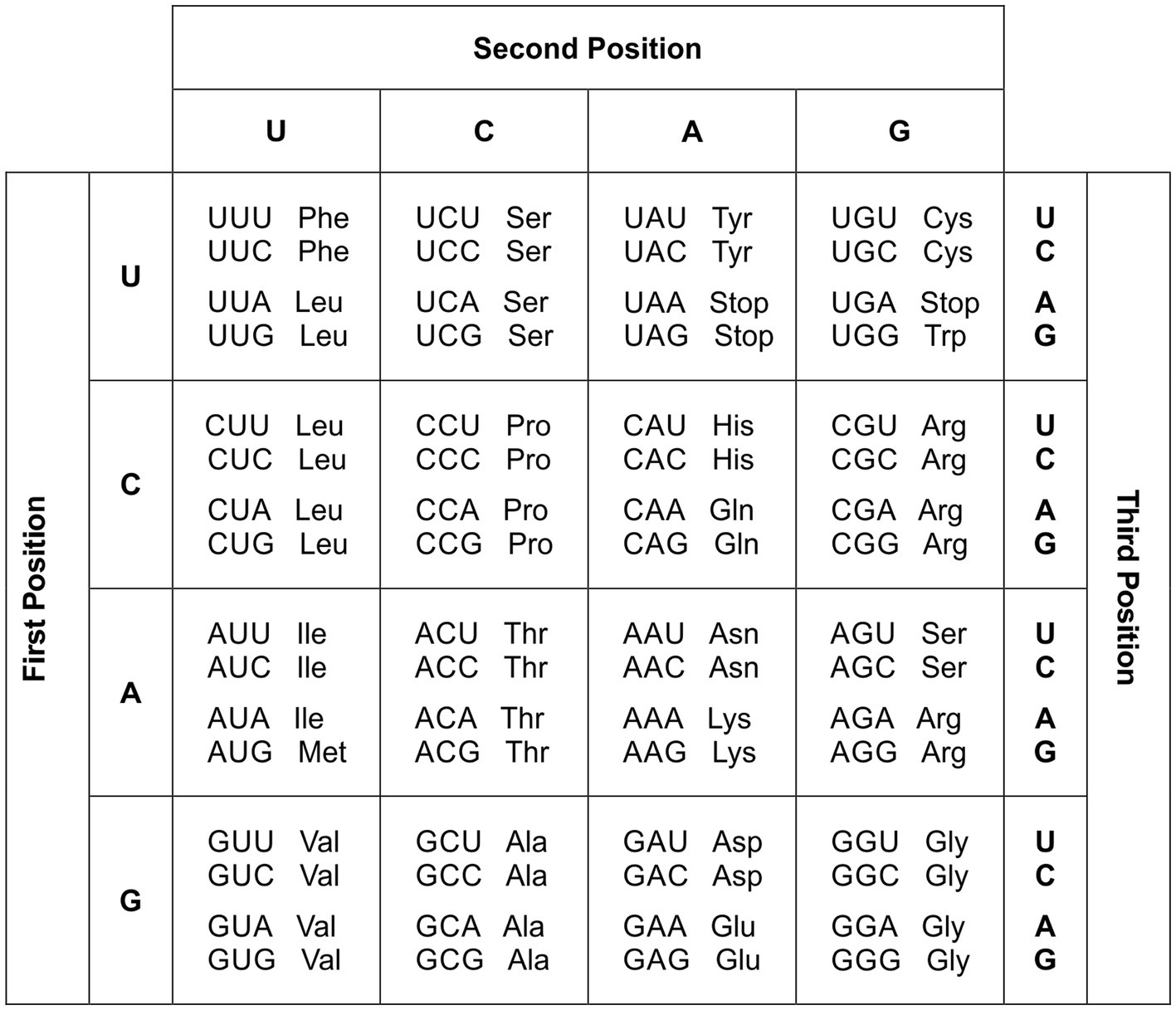
- The codon is the informative unit of the mRNA. It is a sequence of three adjacent bases (B). There are four different bases: uracil (U), cytosine (C), adenine (A), and guanine (G).
- Each codon is denoted by the sequence of three bases (BBB). The base in the first position is denoted as B1 or B− −; in the second position is denoted as B2 or– B –, and in the third position is denoted as B3 or – – B.
- A di-base is denoted as B1B2 or BB–. There are 16 BB–.
- For each di-base BB–, there are four codons given that – –B could be U, C, A, or G.
- A di-base BB– is designated as non-discriminating (Non-D) when the four codons specify a unique amino acid. Then, the four codons of a non-D BB– are synonyms forming a quadruplet.
- A di-base BB– is designated as discriminating (D) when two codons specify a particular amino acid and the other two specify another amino acid. Then, the four codons conform to two groups composed of two synonymous each, i.e., they diverge into two doublets.
- Each base is characterized by two properties: molecular type (mT) and number of H bonds (nHb).
- Two bases, U and C, belong to the pyrimidine type and are denoted as Y. The other two bases, A and G, belong to the purine type and are denoted as R.
- Bases belonging to both molecular types (Y and R) may possess two or three H bonds. This property is denoted as 2 or 3.
- Two bases, U and A, share the property (nHb) 2. The other two, C and G, share the property (nHb) 3.
- Each base may be denoted by indicating its specific pair of properties: Y or R/3 or 2.
- Bases of pyrimidine type are denoted as follows: U is ; C is .
- Bases of purine type are denoted as follows: A is ; G is .
- A base is defined as coherent when its properties (Y or R and 3 or 2) coherently contribute to the discriminating or non-discriminating behavior of the di-base BB– they compose.
- A base is defined as non-coherent when its properties (Y or R and 3 or 2) do not contribute coherently to the discriminating or non-discriminating behavior of the di-base BB– they compose. The notions of coherence and non-coherence will be defined in the following paragraphs.
- A CD can be denoted by specifying the six properties of its corresponding three bases.
Example: The CD UGC is denoted as .
2.2 Rules about the properties molecular type and number of Hydrogen bonds
There are sets of synonymous with different numbers of codons (Figure 1): singlets (unitary set of codons), doublets (set of two synonymous), triplets (set of three synonymous), quadruplets (set of four synonymous), and sextets (set of six synonymous). A triplet can be considered as an association between a doublet and a singlet, and a sextet can be considered as an association between a quadruplet and a doublet.
To make the analysis easier, the four codons corresponding to each BB– will be considered as if they were just quadruplets or doublets. There are, however, two special cases (AU– and UG–) that do not exactly adjust to this definition. As a first approach, these BB– will be considered as being discriminating. In this way, in the simplest diagram (Figure 2), each of the 16 BB– is represented as being discriminating or non-discriminating. In the following sections, the details concerning the special cases for the standard genetic code will be considered step by step, from the simplest picture to the most complex one, and the rules of increasing complexity will then be introduced.
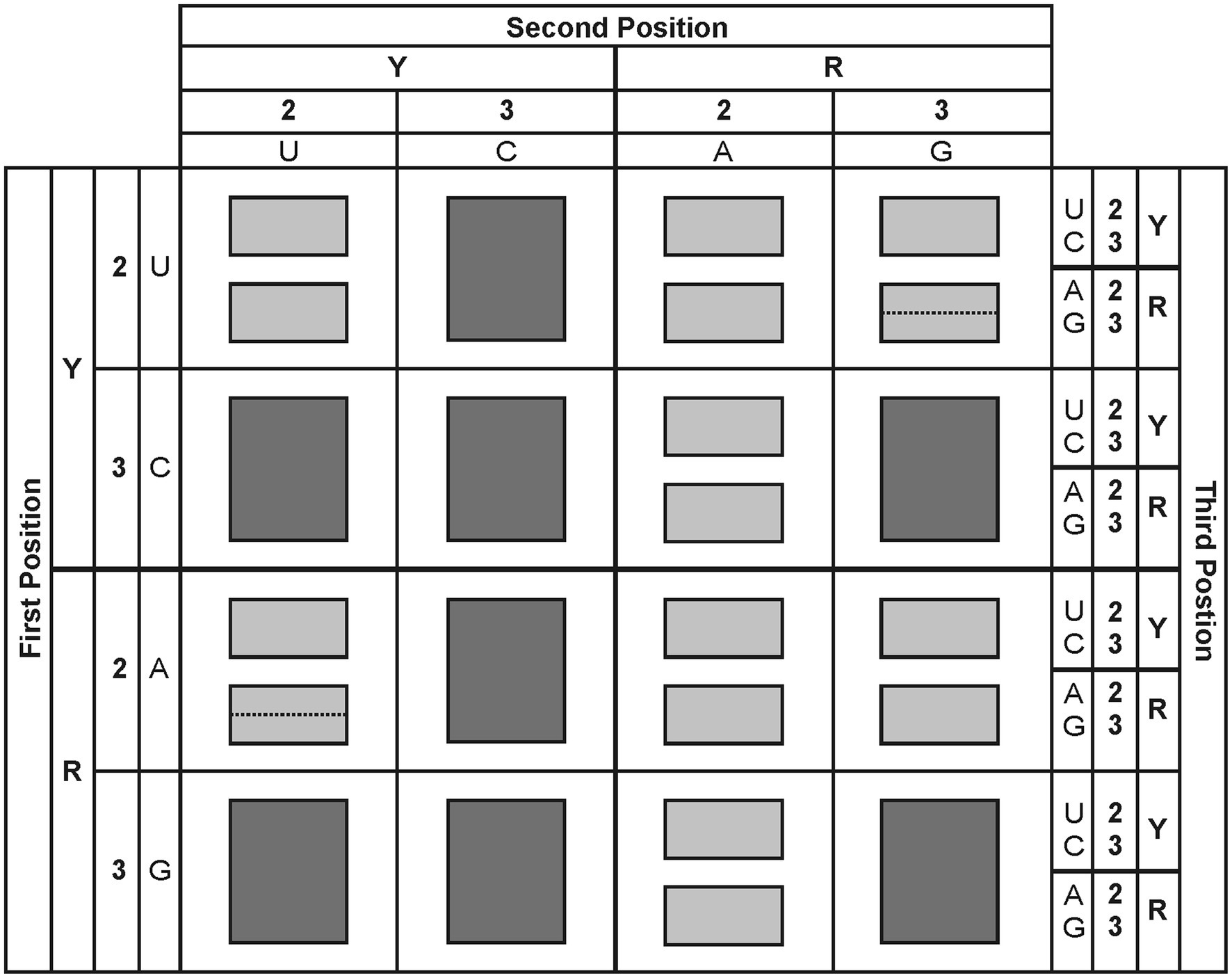
Figure 2. Simplified picture of the genetic code degeneracy. The four codons of each BB are represented as if they were only quadruplets or doublets. Dark gray: quadruplets. Light gray: doublets. Dotted lines indicate special cases of doublets divergence into two singlets. The distribution of the properties Y, R, 2, and 3 is indicated.
2.2.1 The role of bases: adenine and cytosine
With regard to considering the bases A and C, it is shown in Figure 2 as follows:
This rule reveals three essential facts which are as follows:
(1) C assigns a non-D behavior to di-bases. C possesses a non-D characteristic.
(2) A assigns a D behavior to di-bases. A possesses a D characteristic.
Note that BB–, composed of combinations of A and C, such as CA– and AC–, consists of combinations of bases with opposing characteristics. This opposition of characteristics is solved in a way indicating that B1 and B2 (the first and the second positions) display different informative roles.
(3) B2, the second position, has a relevant informative value. In considering those BB– that are combinations of C and A, the discriminating or non-discriminating behavior is determined by B2, i.e., it depends on the characteristic of the base located in the second position.
2.2.2 The role of bases: uracil and guanine
There is no equivalent rule applicable to bases U and G. It will be explained later that these bases are different from A and C with respect to a characteristic defined as coherence.
Regarding BB–, composed of combinations of U and G (a) codons containing the combination GU– converge into a quadruplet, and (b) codons containing the combination UG– diverge into two doublets.
These facts imply that.
(1) The structure –U– assigns a non-D behavior to di-bases. U possesses a non-D characteristic;
(2) The structure –G– assigns a D behavior to di-bases. G possesses a D characteristic.
2.2.3 The role of the property molecular type (Y, R)
Taking into account that C and U share the property Y and that A and G share the property R, from the previous paragraphs can be enunciated another general rule:
2.2.4 The role of the property nHb (2, 3)
C and G are characterized by property 3 and U and A are characterized by property 2. Analyzing the distribution of quadruplets and doublets in connection with properties 2 and 3 (Figure 2), another general rule can be postulated:
A more general enunciation of this rule is as follows:
All these statements are included in the following rule:
2.3 Toward more general rules
The following paragraphs introduce two sets of rules to describe that the codon structure possesses a dual informative function: on the one hand, it determines (a) the discriminating or non-discriminating character, and on the other hand, it determines (b) a specific amino acid. These rules reveal the significance of the spatial position, i.e., the spatial organization, of the properties Y, R, 3, and 2.
2.3.1 The rules of determination of the discriminating or non-discriminating character of the BB–(quadruplets vs. doublets)
The behavior described by Rule 1 is due to the fact that (a) base C is characterized by the coincidence of two non-discriminating properties (Y and 3), and (b) base A is defined by the coincidence of two discriminating properties (R and 2). These conditions are defined as coherence. C and A are coherent bases.
Bases U and G do not fulfill this condition. Base U possesses a discriminating property associated with a non-discriminating one (2 and Y, respectively), while G is characterized by the association of a non-discriminating with a discriminating property (3 and R, respectively). We define this condition as non-coherence. U and G are non-coherent bases.
The relevance of the spatial position is revealed by the fact that all the rules previously enunciated can be integrated into a unique statement:
The specific positions of these three properties within the codon structure can be indicated as follows:
The other three properties of the codon are irrelevant, and they do not possess informative value in this respect.
A derivation of the second law dictates that
From this, two additional enunciations are derived as follows:
In these two conditions, the remaining four properties are informatively irrelevant.
Another corollary of the first law establishes that
2.3.2 Rules about the amino acid-specifying information of the codon
There are BB– whose four codons converge into quadruplets and they specify a single amino acid.
There are BB– whose four codons diverge into doublets that specify two different amino acids.
There are BB– whose four codons diverge into one doublet and two singlets. In some cases, the doublet + one singlet conform a triplet that specifies a single amino acid while the other singlet specify a different amino acid. In other cases, the singlets correspond to the Start or Stop codon.
The following paragraphs show that for each of the above-mentioned conditions, there are different rules describing how a codon specifies a particular amino acid. These rules describe that the “quantity” of information, i.e., the number of informative properties, required for the specification of an amino acid by quadruplets, doublets, and singlets significantly differs. Only when an amino acid is specified by a singlet, the complete set of informative properties of the codon is required.
2.3.2.1 The rule of the specification of amino acids by quadruplets
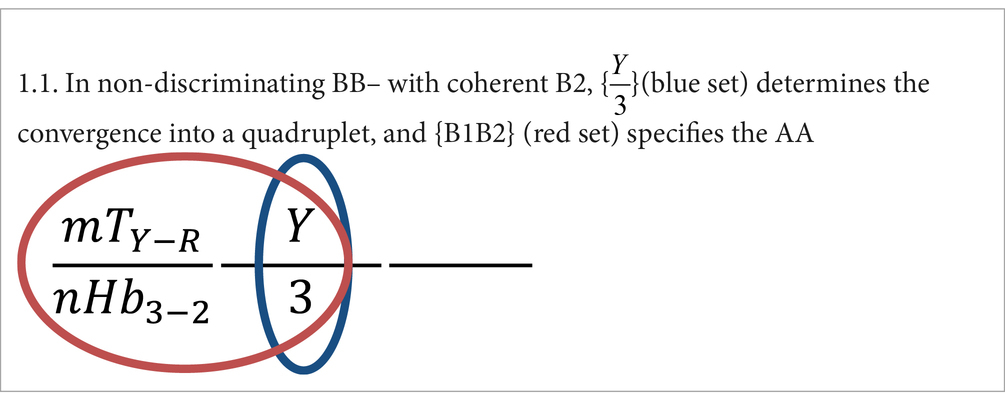
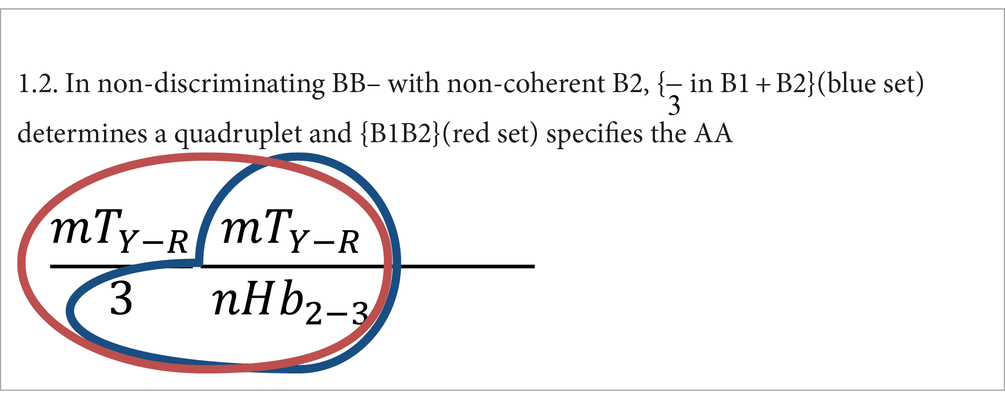
1. In non-discriminating BB– the amino acid specification depends on B2 + B1
2.3.2.2 The rule of the specification of amino acids by doublets
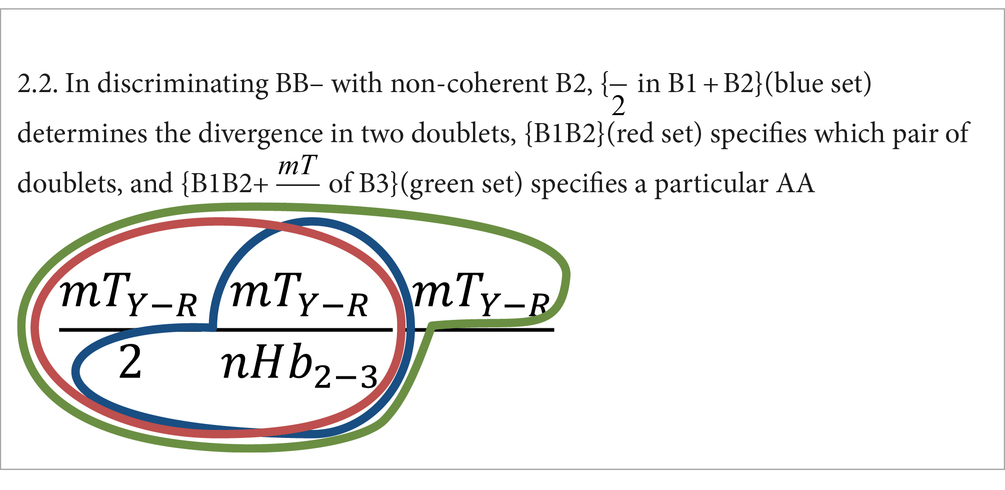
2. In discriminating BB–, the amino acid specification also depends on the property mT ( vs. ) of B3.
2.3.2.3 The rule of amino acid specification by singlets
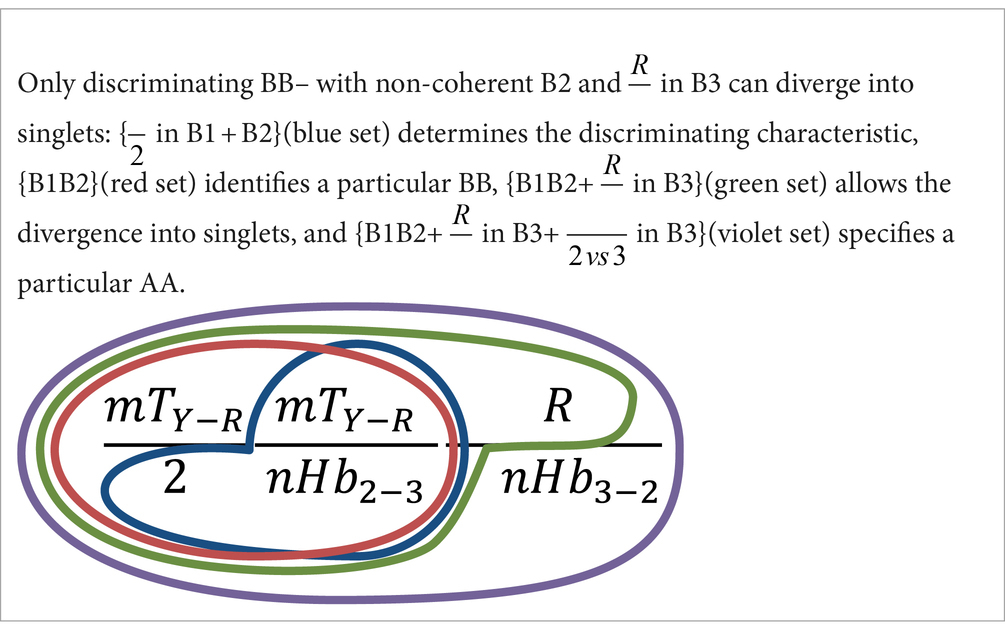
The specification of an amino acid by singlets can be considered special cases in which doublets diverge into two singlets. This condition requires using the six properties of the codon and takes place only when a discriminating BB– possesses a non-coherent B2 and in B3. In these cases, the amino acid specification depends also on the property nHb () of B3.
2.3.2.4 The hierarchy of the informative properties of the codon as revealed by a tree structure
A tree structure was already used to analyze the syntactic structure of the genetic code and investigate the relationship between the codon tree and the hierarchy of the amino acid categorizations (17–19). In this study, a tree structure was used to represent the hierarchy of the properties mT and nHb in relation to their position within the codon, in the determination of the discriminating or non-discriminating character of di-bases and in the specification of amino acids (Figure 3).
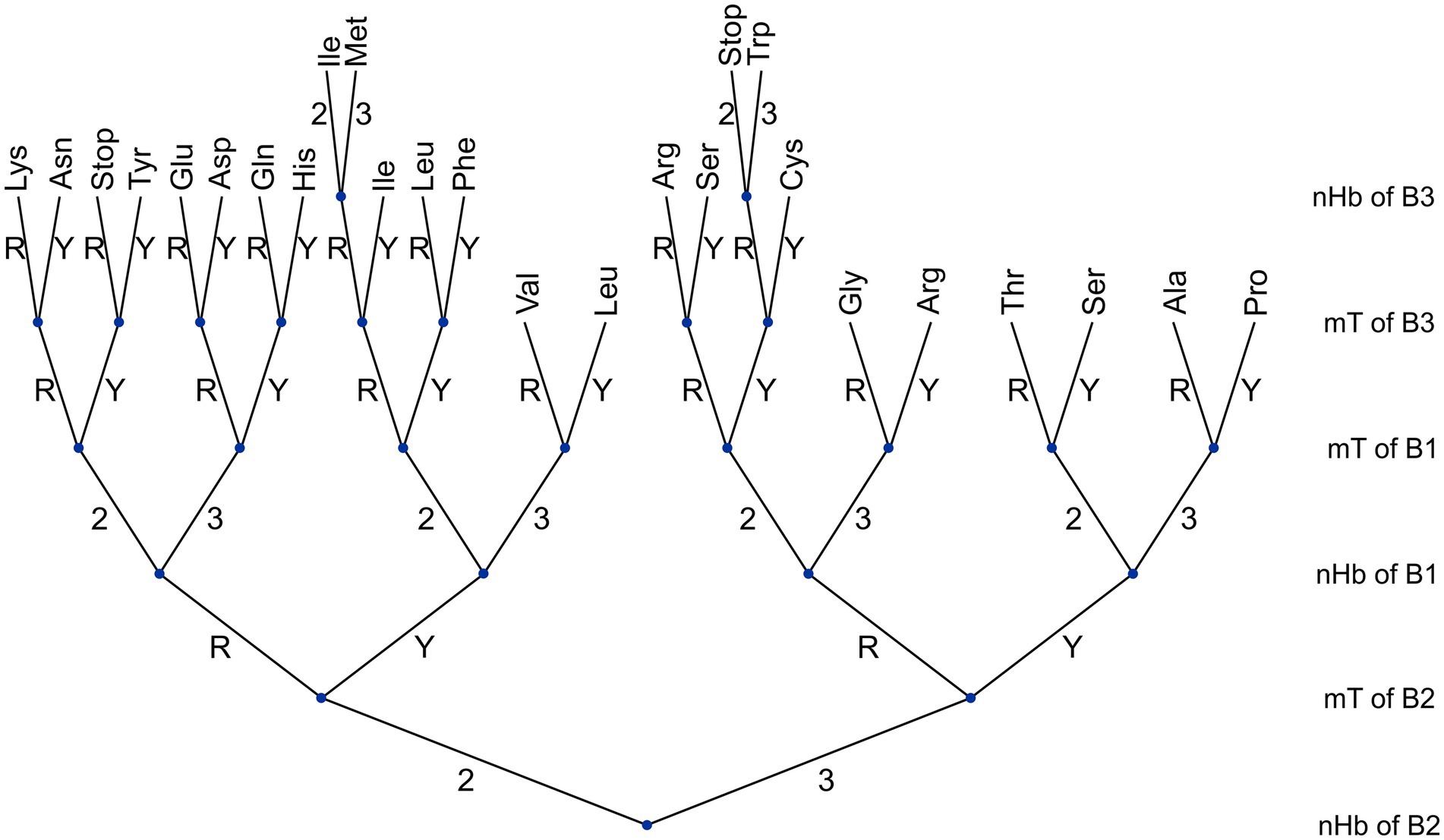
Figure 3. A tree representing the specification of amino acids in terms of the essential properties of the bases.
The induction algorithm ID3 (20) was implemented in order to classify the 64 codons into 21 classes (20 amino acids and stop codons). The six properties of codons were used as attributes.
The height of the tree is determined by the six properties of the three bases. Each node can be thought of as the answer to a question: Is the property discriminating or non-discriminating? With regard to the bifurcation at each node, the branch directed to the right corresponds to non-discriminating and that directed to the left corresponds to discriminating. When all the branches originated at a definite node (a subtree) correspond to the same amino acid, the subtree is replaced by a terminal or “leaf” node. The “morphology” of the tree depends on the order in which the six codon properties are organized in successive nodes.
According to the algorithm, the information gain, i.e., the relative importance of the attributes is expressed by the series: nHb of B2 > mT of B2 > nHb of B1 = mT of B1 > mT of B3 > nHb of B3. Among the different possible trees that maximize information gain, Figure 3 represents one in which the properties of the bases were arranged according to the following decreasing hierarchy: nHb of B2 > mT of B2 > nHb of B1 > mT of B1 > mT of B3 > nHb of B3. This hierarchical order was chosen because it coincided with that expected from the sets of rules introduced in this study. The hierarchy of the different properties depends on their positions within the codon: the maximal hierarchy (the root node) corresponds to the properties of the second position (nHb = mT), the following hierarchy corresponds to the first position (nHb > mT), and the last hierarchy corresponds to the third position (mT > nHb). The tree graphically shows that four, five, and six nodes, i.e., codon properties, are required to specify quadruplets, doublets, and singlets, respectively. It can be noted that this tree reliably reproduces the three rules about amino acid specification described under the title, 2.3.2 Rules about the amino acid-specifying information of the codon.
3 Discussion and concluding remarks
The relevance of nucleotide positions within the codon was proposed and exhaustively analyzed from different perspectives since many years ago. As early as 1960–70, Woese (21, 22) proposed that the origin of a codon assignment, i.e., specification of related amino acids, could have depended on a primitive translation process in which groups of chemically related codons could specify groups of chemically related amino acids, a hypothesis, based on direct physicochemical interactions between nucleic acids and amino acids that was named as “group codon assignments.” In 1966, F. Crick (23) proposed that the interaction between the nucleotide of the 5’end of the anticodon and that of the 3’position of the codon does not respond to the classical base pairing rules, a phenomenon known as the “wobble” hypothesis. Later, Lagerkvist (16, 24) observed that the intensity of the pairing force of the nucleotides located at the first two positions and the purine/pyrimidine nature of the base of the second position are relevant criteria for the categorization of codons into two families of degeneracy. It is interesting that other authors also considered the properties of mT and nHb of bases as criteria for the construction of Boolean algebraic models of degeneracy (25, 26). More recent models of code degeneracy assigned significant importance to the architecture of the anticodon loop of the tRNA and the molecular organization of the ribosome-decoding center (4). Analyses based on cryogenic electron microscopy also highlight the role of the ribosome in establishing the degeneracy (27).
The role of the nHb was also highlighted by Danckwerts and Neubert (28), who showed that when the sum of the nHb in B1B2– is 6, they converge into a quadruplet; however, when the sum is 4, they diverge into doublets. Konjevoda and Štambuk (29) show that when the sum is 5, the mT of B2 defines whether the di-base forms quadruplets or doublets.
Some research works that study the code degeneracy from an evolutionary perspective have attempted to elucidate the possible origin and evolution of the genetic code, in the context of the biology of primitive organisms (protocells), considering two aspects: (a) direct stereochemical interactions between codons and amino acids and (b) the biosynthetic pathways of various amino acids’ synthesis (30–35). Some of these studies were devoted to studying these interactions through molecular dynamics simulations in order to analyze the forces acting between adjacent atoms (36, 37). Other evolutionary analyses have focused on the role natural selection could have played in shaping a robust code, minimizing the impact of mutations and mistranslation on protein structure and function (38–42). This phenomenon is closely associated with codon usage (43–45). The relational model of the structure of the standard genetic code analyzed by Konjevoda and Štambuk (29) is a valuable attempt to unify these and other evolving studies. Other authors emphasize the role of tRNA in the origin and evolution of the genetic code, highlighting the interactions between aminoacyl-tRNA synthetases and different tRNA domains, an operational code of tRNA aminoacylation and a standard tRNA code composed of 46 anticodons (46–52).
The set of rules introduced in this study considers a codon as an entity with a double function. The spatial organization of the six attributes of codons informs about two related, but essentially different, phenomena. Apart from determining a specific amino acid, there is a “logic” that associates the codon composition with its discriminating or non-discriminating character. These two aspects constitute the global information of the codon, and two different sets of rules are needed to describe these different phenomena.
These rules establish that, depending on the type of base (coherent or non-coherent) occupying the second position, only two or three of the six codon properties located at defined positions determine the discriminating or non-discriminating behavior. In addition, the number of properties and also their positions characteristically change when an amino acid is specified by different sized sets of synonymous codons (quadruplets, doublets, and singlets).
It is clear that the second position possesses a relevant function: a coherent B2 per se determines the discriminating or non-discriminating character of codons. However, in the case of codons with non-coherent B2, additional information is required to specify the discriminating vs. non-discriminating character. In considering a codon with a non-coherent B2, the other properties possess different roles depending on their location at B1 or B3: (a) the property nHb of B1 is needed to determine the discriminating vs. non-discriminating character. Thus, the property 2 allows the divergence into two doublets. In this case, the property mT of B1 does not have an informative value; (b) on the other hand, the property mT of B3 is needed to allow the divergence into singlets. In fact, R in B3 allows the divergence while Y does not. Conversely, the property nHb of B3 does not have such an informative value.
On these bases, the present study reinforces the idea that a codon is an asymmetrically organized informative entity since the same informative elements possess different informative roles when they are in different positions, i.e., B1 or B3, with respect to a non-coherent B2.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
MR: Conceptualization, Formal analysis, Methodology, Writing – review & editing. RK: Conceptualization, Formal analysis, Writing – original draft. VF: Conceptualization, Formal analysis, Methodology, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
Some of the text of the present manuscript has previously appeared online in HAL-Inria, Inria’s Research or Technical Reports, [Research Report] RR-5938, INRIA. 2006 ⟨inria-00081981v2⟩. https://inria.hal.science/INRIA-RRRT/inria-00081981v2.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
2. Crick, FHC. The origin of the genetic code. J Mol Biol. (1968) 38:367–79. doi: 10.1016/0022-2836(68)90392-6
3. Grosjean, H, and Westhof, E. An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res. (2016) 44:8020–40. doi: 10.1093/nar/gkw608
4. Lehmann, J, and Libchaber, A. Degeneracy of the genetic code and stability of the base pair at the second position of the anticodon. RNA. (2008) 14:1264–9. doi: 10.1261/rna.1029808
5. Sciarrino, A, and Sorba, P. Codon-anticodon interaction and the genetic code evolution. Biosystems. (2013) 111:175–80. doi: 10.1016/j.biosystems.2013.02.004
6. Woese, CR, Olsen, GJ, Ibba, M, and Söll, D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol Mol Biol Rev. (2000) 64:202–36. doi: 10.1128/MMBR.64.1.202-236.2000
7. Chiusano, ML, Frappat, L, Sciarrino, A, and Sorba, P. Codon usage correlations and crystal basis model of the genetic code. EPL. (2001) 55:287–93. doi: 10.1209/epl/i2001-00411-9
8. Frappat, L, Sciarrino, A, and Sorba, P. A crystal base for the genetic code. Phys Lett A. (1998) 250:214–21. doi: 10.1016/S0375-9601(98)00761-0
9. Frappat, L, Sciarrino, A, and Sorba, P. Symmetry and codon usage correlations in the genetic code. Phys Lett A. (1999) 259:339–48. doi: 10.1016/S0375-9601(99)00418-1
10. Frappat, L, Sciarrino, A, and Sorba, P. Crystallizing the genetic code. J Biol Phys. (2001) 27:1–34. doi: 10.1023/A:1011874407742
11. Lei, L, and Burton, ZF. Evolution of the genetic code. Transcription. (2021) 12:28–53. doi: 10.1080/21541264.2021.1927652
12. Karasev, VA, and Stefanov, VE. Topological nature of the genetic code. J Theor Biol. (2001) 209:303–17. doi: 10.1006/jtbi.2001.2265
13. Gonzalez, DL, Giannerini, S, and Rosa, R. On the origin of degeneracy in the genetic code. Interface Focus. (2019) 9:20190038. doi: 10.1098/rsfs.2019.0038
14. Percudani, R. Restricted wobble rules for eukaryotic genomes. Trends Genet. (2001) 17:133–5. doi: 10.1016/s0168-9525(00)02208-3
15. Pak, D, Kim, Y, and Burton, ZF. Aminoacyl-tRNA synthetase evolution and sectoring of the genetic code. Transcription. (2018) 9:1–15. doi: 10.1080/21541264.2018.1467718
16. Lagerkvist, U. “two out of three”: an alternative method for codon reading. Proc Natl Acad Sci USA. (1978) 75:1759–62. doi: 10.1073/pnas.75.4.1759
17. Jiménez-Montaño, MA. On the syntactic structure of protein sequences and the concept of grammar complexity. Bull Math Biol. (1984) 46:641–59. doi: 10.1007/BF02459508
18. Jiménez-Montaño, MA. On the syntactic structure and redundancy distribution of the genetic code. Biosystems. (1994) 32:11–23. doi: 10.1016/0303-2647(94)90015-9
19. Jiménez-Montaño, MA. The fourfold way of the genetic code. Biosystems. (2009) 98:105–14. doi: 10.1016/j.biosystems.2009.07.006
20. Quinlan, JR. Learning efficient classification procedures In: RS Michalski, JG Carbonell, and TM Mitchell, editors. Machine learning: An artificial intelligence approach : Morgan Kaufmann. Tokyo, New York: Springer-Verlag Berlin Heidelberg (1983). 463–82.
21. Woese, CR, Dugre, DH, Dugre, SA, Kondo, M, and Saxinger, WC. On the fundamental nature and evolution of the genetic code. Cold Spring Harb Symp Quant Biol. (1966) 31:723–36. doi: 10.1101/sqb.1966.031.01.093
22. Woese, CR. Evolution of the genetic code. Naturwissenschaften. (1973) 60:447–59. doi: 10.1007/BF00592854
23. Crick, FH. Codon--anticodon pairing: the wobble hypothesis. J Mol Biol. (1966) 19:548–55. doi: 10.1016/s0022-2836(66)80022-0
24. Lagerkvist, U. Unconventional methods in codon reading. BioEssays. (1986) 4:223–6. doi: 10.1002/bies.950040509
25. Jiménez-Montaño, MA, de la Mora-Basánez, CR, and Pöschel, T. The hypercube structure of the genetic code explains conservative and non-conservative aminoacid substitutions in vivo and in vitro. Biosystems. (1996) 39:117–25. doi: 10.1016/0303-2647(96)01605-x
26. Sánchez, R, Morgado, E, and Grau, R. A genetic code Boolean structure. I. The meaning of Boolean deductions. Bull Math Biol. (2005) 67:1–14. doi: 10.1016/j.bulm.2004.05.005
27. Ye, S, and Lehmann, J. Genetic code degeneracy is established by the decoding center of the ribosome. Nucleic Acids Res. (2022) 50:4113–26. doi: 10.1093/nar/gkac171
28. Danckwerts, HJ, and Neubert, D. Symmetries of genetic code-doublets. J Mol Evol. (1975) 5:327–32. doi: 10.1007/BF01732219
29. Konjevoda, P, and Štambuk, N. Relational model of the standard genetic code. Biosystems. (2021) 210:104529. doi: 10.1016/j.biosystems.2021.104529
30. Hobish, MK, Wickramasinghe, NS, and Ponnamperuma, C. Direct interaction between amino acids and nucleotides as a possible physicochemical basis for the origin of the genetic code. Adv Space Res. (1995) 15:365–82. doi: 10.1016/s0273-1177(99)80108-2
31. Jiménez-Sánchez, A. On the origin and evolution of the genetic code. J Mol Evol. (1995) 41:712–6. doi: 10.1007/BF00173149
32. Saier, MH Jr. Understanding the genetic code. J Bacteriol. (2019) 201:e00091–19. doi: 10.1128/JB.00091-19
33. Taylor, FJ, and Coates, D. The code within the codons. Biosystems. (1989) 22:177–87. doi: 10.1016/0303-2647(89)90059-2
34. Wong, JT. A co-evolution theory of the genetic code. Proc Natl Acad Sci USA. (1975) 72:1909–12. doi: 10.1073/pnas.72.5.1909
35. Wong, JT. Coevolution theory of the genetic code at age thirty. BioEssays. (2005) 27:416–25. doi: 10.1002/bies.20208
36. Halpern, A, Bartsch, LR, Ibrahim, K, Harrison, SA, Ahn, M, Christodoulou, J, et al. Biophysical interactions underpin the emergence of information in the genetic code. Life. (2023) 13:1129. doi: 10.3390/life13051129
37. Harrison, SA, Palmeira, RN, Halpern, A, and Lane, N. A biophysical basis for the emergence of the genetic code in protocells. Biochim Biophys Acta Bioenerg. (2022) 1863:148597. doi: 10.1016/j.bbabio.2022.148597
38. Epstein, CJ. Role of the amino-acid “code” and of selection for conformation in the evolution of proteins. Nature. (1966) 210:25–8. doi: 10.1038/210025a0
39. Fimmel, E, Gumbel, M, Starman, M, and Strüngmann, L. Computational analysis of genetic code variations optimized for the robustness against point mutations with wobble-like effects. Life. (2021) 11:1338. doi: 10.3390/life11121338
40. Freeland, SJ, and Hurst, LD. The genetic code is one in a million. J Mol Evol. (1998) 47:238–48. doi: 10.1007/pl00006381
41. Sonneborn, TM. Degeneracy of the genetic code: extent, nature, and genetic implications In: V Bryson and HJ Vogel, editors. Evolving genes and proteins. New York: Academic Press (1965). 377–97. doi: 10.1016/B978-1-4832-2734-4.50034-6
42. Schwersensky, M, Rooman, M, and Pucci, F. Large-scale in silico mutagenesis experiments reveal optimization of genetic code and codon usage for protein mutational robustness. BMC Biol. (2020) 18:146. doi: 10.1186/s12915-020-00870-9
43. Archetti, M. Selection on codon usage for error minimization at the protein level. J Mol Evol. (2004) 59:400–15. doi: 10.1007/s00239-004-2634-7
44. Janzen, E, Shen, Y, Vázquez-Salazar, A, Liu, Z, Blanco, C, Kenchel, J, et al. Emergent properties as by-products of prebiotic evolution of aminoacylation ribozymes. Nat Commun. (2022) 13:3631. doi: 10.1038/s41467-022-31387-0
45. Radványi, Á, and Kun, Á. The mutational robustness of the genetic code and codon usage in environmental context: a non-Extremophilic preference? Life. (2021) 11:773. doi: 10.3390/life11080773
46. Schimmel, P, Giegé, R, Moras, D, and Yokoyama, S. An operational RNA code for amino acids and possible relationship to genetic code. Proc Natl Acad Sci USA. (1993) 90:8763–8. doi: 10.1073/pnas.90.19.8763
47. Guimarães, RC, Moreira, CH, and de Farias, ST. A self-referential model for the formation of the genetic code. Theory Biosci. (2008) 127:249–70. doi: 10.1007/s12064-008-0043-y
48. Rodin, AS, Szathmáry, E, and Rodin, SN. On origin of genetic code and tRNA before translation. Biol Direct. (2011) 6:14. doi: 10.1186/1745-6150-6-14
49. José, MV, Morgado, ER, Guimarães, RC, Zamudio, GS, de Farías, ST, Bobadilla, JR, et al. Three-dimensional algebraic models of the tRNA code and 12 graphs for representing the amino acids. Life. (2014) 4:341–73. doi: 10.3390/life4030341
50. Lei, L, and Burton, ZF. Evolution of life on earth: tRNA, aminoacyl-tRNA Synthetases and the genetic code. Life. (2020) 10:21. doi: 10.3390/life10030021
51. Farias, ST, and Prosdocimi, F. RNP-world: the ultimate essence of life is a ribonucleoprotein process. Genet Mol Biol. (2022) 45:e20220127. doi: 10.1590/1678-4685-GMB-2022-0127
Keywords: genetic code degeneracy, synonymous codons, hydrogen bond, pyrimidine, purine
Citation: Rapacioli M, Katz R and Flores V (2024) Rules governing the genetic code degeneracy/redundancy and spatial organization of the codon informative properties. Front. Appl. Math. Stat. 10:1340640. doi: 10.3389/fams.2024.1340640
Edited by:
Tidjani Negadi, Oran University 1 Ahmed Ben Bella, AlgeriaReviewed by:
Marco V. José, National Autonomous University of Mexico, MexicoPaško Konjevoda, Rudjer Boskovic Institute, Croatia
Zachary Ardern, Wellcome Sanger Institute (WT), United Kingdom
Copyright © 2024 Rapacioli, Katz and Flores. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Melina Rapacioli, bXJhcGFjaW9saUBmYXZhbG9yby5lZHUuYXI=
 Melina Rapacioli
Melina Rapacioli Ricardo Katz3
Ricardo Katz3 Vladimir Flores
Vladimir Flores