- 1Department of Mathematics and Applied Mathematics, University of Johannesburg, Johannesburg, South Africa
- 2Health, Nutrition, and Population Unit, The World Bank, Harare, Zimbabwe
- 3Faculty of Engineering and the Built Environment, Data Science Across Disciplines, University of Johannesburg, Johannesburg, South Africa
Introduction: The utility of non-contact technologies for screening infectious diseases such as COVID-19 can be enhanced by improving the underlying Artificial Intelligence (AI) models and integrating them into data visualization frameworks. AI models that are a fusion of different Machine Learning (ML) models where one has leveraged the different positive attributes of these models have the potential to perform better in detecting infectious diseases such as COVID-19. Furthermore, integrating other patient data such as clinical, socio-demographic, economic and environmental variables with the image data (e.g., chest X-rays) can enhance the detection capacity of these models.
Methods: In this study, we explore the use of chest X-ray data in training an optimized hybrid AI model based on a real-world dataset with limited sample size to screen patients with COVID-19. We develop a hybrid Convolutional Neural Network (CNN) and Random Forest (RF) model based on image features extracted through a CNN and EfficientNet B0 Transfer Learning Model and applied to an RF classifier. Our approach includes an intermediate step of using the RF's wrapper function, the Boruta Algorithm, to select important variable features and further reduce the number of features prior to using the RF model.
Results and discussion: The new model obtained an accuracy and recall of 96% for both and outperformed the base CNN model and four other experimental models that combined transfer learning and alternative options for dimensionality reduction. The performance of the model fares closely to relatively similar models previously developed, which were trained on large datasets drawn from different country contexts. The performance of the model is very close to that of the “gold standard” PCR tests, which demonstrates the potential for use of this approach to efficiently scale-up surveillance and screening capacities in resource limited settings.
1. Introduction
1.1. Background
In Zimbabwe, the detection of COVID-19 has been central to the public health emergency response to the COVID-19 pandemic. This has allowed for the early identification of individuals that are likely to have the virus and placing them in quarantine or isolation to minimize the onward spread of the virus. Globally, countries have invested in ensuring the early identification of suspected and confirmed cases through testing. Countries such as China and the United States that were most affected also invested in testing and screening of cases as a major response strategy [1, 2].
Given the high costs and feasibility of the main approaches for detecting cases, countries have typically adopted a targeted approach for screening with a focus on presenting symptoms and likelihood of exposure based on travel history. The “gold standard” test for diagnosing COVID-19 is the Reverse Transcript Polymerase Chain Reaction (RT-PCR) test due to its high sensitivity. Czumbel et al. [3] noted, across several studies in their meta-analysis, that the RT-PCR tests based on nasopharyngeal swab tests have a sensitivity of 98% (95% CI 89%-100%) in previously confirmed COVID-19 infected patients. The use of nasopharyngeal test swabs is very common globally including Sub-Saharan Africa (SSA).
Several countries, especially those in SSA, have faced challenges in meeting reasonable COVID-19 testing coverage. This is due to a combination of global shortages, pricing and often intensive logistical requirements associated with conducting these tests at scale. Global data from Our World in Data (OWID, 2022) shows a positive correlation between GDP and testing coverage. The low testing coverage, compounded by the delays in the provision of test results, limits the extent to which the strategy for testing and detection of COVID-19 can effectively be applied as a mechanism for containing the spread of the virus in these settings.
In recent years, notable progress has been made in the potential use of Artificial Intelligence (AI) techniques in screening and diagnosis for COVID-19. Two well-documented means of COVID-19 infection diagnosis using AI have employed the analysis of either X-ray [4–8] or CT scan images [9–12]. The standard need of a radiologist could be side-stepped should AI-based image processing be adopted. In fact, it has been shown that COVID-19 can be diagnosed with even higher accuracy than the RT-PCR based test [13]. Therefore, diagnosis using CT scans may help to overcome the sub-optimal sensitivity of PCR tests [14] and eliminate the need for specialist doctors, who are often not available in rural areas.
Despite the promising opportunities for the use of AI in the detection of COVID-19, there has been limited use of detection models in real world applications. Those developed are mainly from pooled open source datasets established for experimentation and with very few based on smaller datasets drawn from a localized context [15, 16].
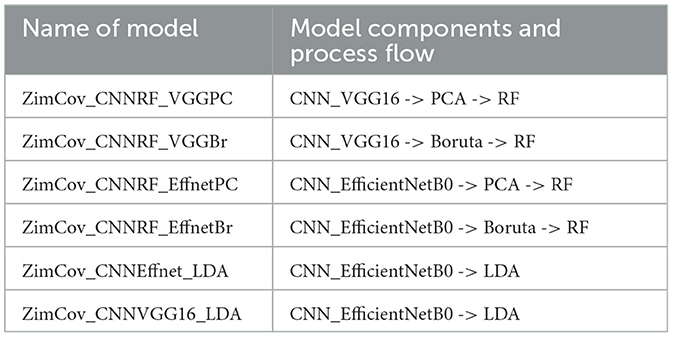
In this paper, we test the applicability of Deep Learning (DL) methods on a small dataset from Zimbabwe. We explore ways to improve the performance of the models through a combination of techniques that include a hybrid DL and Machine Learning (ML) model, image augmentation and image enhancement as well as introduction of newly engineered features derived from unsupervised learning. We propose a new model, ZimCov_CNNRF, that fuses these elements. The performance of the new model is assessed against the base CNN model and the different iterations of the hybrid model.
1.2. Related work
The paper by Shen et al. [17] highlights the remarkable results that have been observed in the application of DL for medical image analysis, largely due to the technique's ability to draw and learn hierarchical feature representations from the image data. Similar to other literature, their paper notes that DL methods require large training data sets for them to be highly effective. Brigato and Iocchi [18] provide a closer look at DL with small datasets. They note that whilst ML models have a high likelihood to overfit the training dataset, CNNs are generally highly resistant to overfitting but model complexity should be considered. They argue that small nets can perform better than bigger complex models in scenarios with smaller training samples.
The use of transfer learning as a key starting point has gained traction in the last years and several real-world applications have utilized transfer learning and DL algorithms for pattern recognition and classification tasks [19]. To complement deep transfer learning, data augmentation has been increasingly used to address the challenges of limited training data sets. It has been observed that data augmentation is effective at improving model performance as it prevents overfitting by modifying small datasets to possess the characteristics of big data [20]. Most recently, hybrid models that combine the classic CNN architecture and the ML-based Random Forest classifier [21] have demonstrated very good performance in image classification accuracy, obtaining 99% compared to a standard CNN model with 94% accuracy [22]. The AI model that combines a CNN architecture and RF algorithm is also in line with the work of Kwak et al. [23], who showed that such a methodology is effective when the dataset is not large. A key feature of the procedure is the use of CNN's automatic feature extraction capability, and the subsequent use of the selected features in the RF classifier. In the absence of feature extraction, the RF would potentially suffer from the curse of dimensionality as they are structured to perform better with fewer number of features relative to the number of observations.
The study by Ghose et al. [24] shows that transfer learning-driven approaches can identify COVID-19 patients from normal individuals with high accuracy using X-ray and CT scans. The proposed method can achieve 99.59% accuracy for X-rays and 99.95% for CT scan images. Iteratively pruned deep learning model ensembles have also been demonstrated for detecting pulmonary manifestations of COVID-19 with chest X-rays [25]. Abiyev and Ma'aitah [26] discuss the use of CNNs for the detection of chest diseases such as chronic obstructive pulmonary disease, pneumonia, asthma, tuberculosis, and lung diseases. They demonstrate the feasibility of classifying chest pathologies in chest X-rays using conventional and deep learning approaches. The architecture and design principle of CNNs are presented and compared with back-propagation neural networks (BPNNs) with supervised learning and competitive neural networks (CpNNs) with unsupervised learning. Their study demonstrates that the use of deep convolutional neural networks in the detection of chest diseases shows promise. The comparative results suggest that CNNs are more accurate and efficient than other approaches such as BPNNs and CpNNs. Research by Polsinelli et al. [27] proposes a light convolutional neural network based on SqueezeNet for the efficient discrimination of COVID-19 CT images with respect to other community-acquired pneumonia and/or healthy CT images. Akinyelu and Blignaut [28] introduce DL-based solutions for COVID-19 diagnosis CT images and 12 cutting-edge DL pre-trained models. All the models are trained on 9,000 COVID-19 samples and 5,000 normal images, which is higher than the COVID-19 images used in most studies. DenseNet121 and VGG16 achieved the best sensitivity, while InceptionV3 and InceptionResNetV2 achieved the best specificity. Varela-Santos and Melin [29] present a new approach to classify COVID-19 based on its manifestation on chest X-rays using texture features and neural networks. The authors conducted experiments using supervised learning models to accurately classify medical images from COVID-19 patients and images of other lung-related diseases. They used image texture feature descriptors, feed-forward and convolutional neural networks to create a baseline for future development of a system that can automatically detect COVID-19 based on its manifestation on chest X-rays and CT images of the lungs. The hybrid approach of combining deep learning (DL) and machine learning (ML) models proved to be effective in accurately classifying the medical images.
The above research articles demonstrate the effectiveness of deep learning models in detecting COVID-19 from medical imaging, including chest X-rays and CT scans. These advancements provide strong justification for the proposed hybrid model to detect COVID-19. However, each of the studies has limitations in terms of dataset size for some, modality-specific knowledge transfer, and computational efficiency. To overcome these limitations and improve the accuracy of COVID-19 diagnosis, a hybrid model that combines the strengths of multiple models could be a promising solution. Hybrid models could leverage the modality-specific knowledge transfer approach, the lightweight CNN architecture, and the use of multiple pre-trained models and large datasets where available though not the case in resource limited settings. Additionally, hybrid models could employ iterative model pruning and ensemble learning techniques to improve performance and reduce computational complexity.
We are not aware of any published work on these advancements in hybrid AI models to detect COVID-19 based on a locally trained dataset in Zimbabwe. The closest reference to similar work in Zimbabwe is an analysis based on Kaggle's open-source data to train a model to screen COVID-19 infected patients versus healthy patients [30]. Their CNN model achieved 99% accuracy.
The development of hybrid AI models for detecting COVID-19 from medical images has the potential to revolutionize diagnosis and treatment in resource-limited settings. The evidence shows that by combining various approaches, hybrid models could improve the accuracy of COVID-19 diagnosis, reduce the risk of false negatives, and enable faster and more efficient diagnosis. Furthermore, the combination of transfer learning, data augmentation, and other deep learning techniques with machine learning algorithms like Random Forests can improve the accuracy and efficiency of medical image analysis, even in settings with limited training data. Hybrid AI models can help clinicians to quickly and accurately diagnose COVID-19 patients, leading to more effective treatment and control of the disease.
2. Methods
2.1. Image data and pre-processing
We analyzed chest X-ray data obtained from a private radiology center in Zimbabwe. A total of 984 chest X-rays were extracted in the Digital Imaging and Communications of Medicine (DICOM) format, an international standard for storing, processing, printing, exporting and displaying medical image data. The R statistical computing software was used to export images into storage folders based on the classes (COVID and non-COVID) and in all our analyses. The export function was configured to store images in PNG format of size 224 × 224 pixels. In addition to the image storage, a separate export function was used to extract and save image metadata in a tabular format and this included data including age, sex and image characteristics.
Given that the original COVID status of patients were based on the radiologist's reports, we validated the COVID status of patients using laboratory data with confirmed PCR test results for the reference period prior to categorizing the image datasets into COVID positive and negative class folders. We used fuzzy matching to link the separate databases using unique identifier string variables. The approach entailed quantifying the dissimilarity between two corresponding unique identifiers from the image database and the laboratory database.
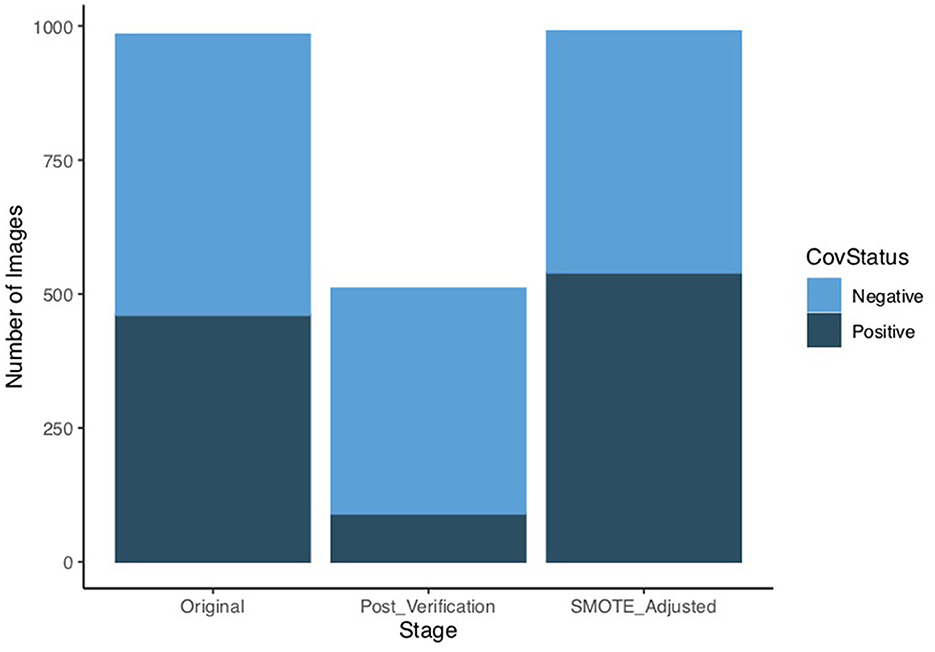
A total of 90 images were confirmed to belong to COVID-19 positive patients based on PCR positive laboratory results and a total of 420 images were retained as COVID-19 negative. The derived sample size post verification introduced class imbalance as the COVID-19 cases represent only 18% of the total 510 images eligible for further analysis. Johnson and Khoshgoftaar [31] note that the class of interest, which in our case is the COVID-19 positive group, is often the minority group having a much smaller sample size. Training prediction models on imbalanced datasets is challenging and often leads to poor performance [32].
2.2. Class imbalance
The verification of classes, i.e. the COVID-19 status, resulted in a significant drop in the size of the dataset from 984 eligible images to only 510 images. As only 90 images remained for the positive class, there was need to address the class imbalance as the key metrics in this analysis, particularly the accuracy, would be unreliable. This would compound the already inherent risk of over-fitting due to the small size of the dataset.
For this analysis we used the Synthetic Minority Oversampling Technique (SMOTE) [33] to generate synthetic samples from the minority COVID-19 positive class. This oversampling technique makes use of an algorithm that generates new data from features' spaces and interpolation. The approach of randomly oversampling helps to address the challenge of overfitting and based on the calibration also helps to overcome the class imbalance. The use of SMOTE enabled us to use a dataset of 990 images that were relatively balanced across the two classes as shown in Figure 1.
2.3. Data augmentation and enhancement
We used data augmentation to address the small size of our image dataset and this entailed using a data generator that reproduced the images in our existing dataset by making minor alterations such as flip, rotations or zoom. The generator for data augmentation was applied on the training sample whilst a standard generator that only re-scaled the images was used for validation and test datasets as per standard practice. A 60%:20%:20% split rule was applied to separate the sample across train, validate and test respectively.
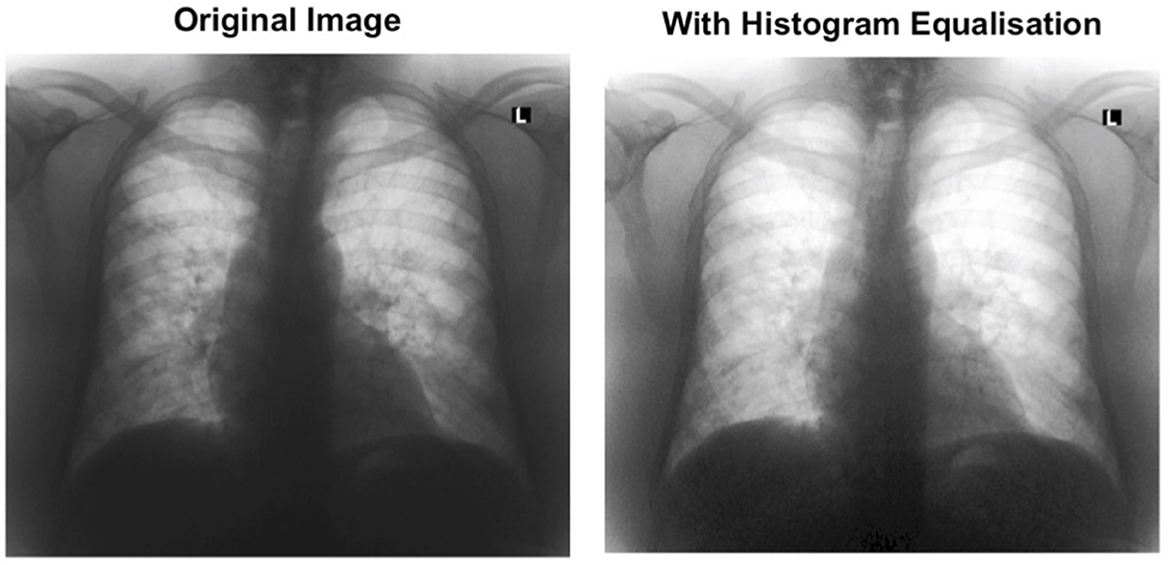
We used the histogram equalization technique [34] to adjust the varying intensities of the images and enhance contrast in a standardized way. Enhancing the image contrast is achieved through spreading the most frequent pixel intensity values and therefore allowing areas in the image having low contrast to gain higher contrast. This approach is typically used when images are either too dark or washed out due to a lack of contrast, balancing out the shadows and highlights in an image. Figure 2 shows a sample image from the dataset in its original form and after enhancement with histogram equalization. For the purposes of this experiment, we used the enhanced image as one of the options to assess its added value in model performance.
2.4. Proposed model architecture and process
Transfer learning has allowed for use of pre-trained CNNs to classify a different problem than the one it was originally trained for. The VGG16, developed by Simonyan and Zisserman [35], is a 16-layer transfer learning architecture that has previously been noted to have significantly improved model performance relative to a basic CNN even with image augmentation [36]. Another family of transfer learning models, EfficientNets, have also been identified as having better accuracy and efficiency than the basic CNNs [37].
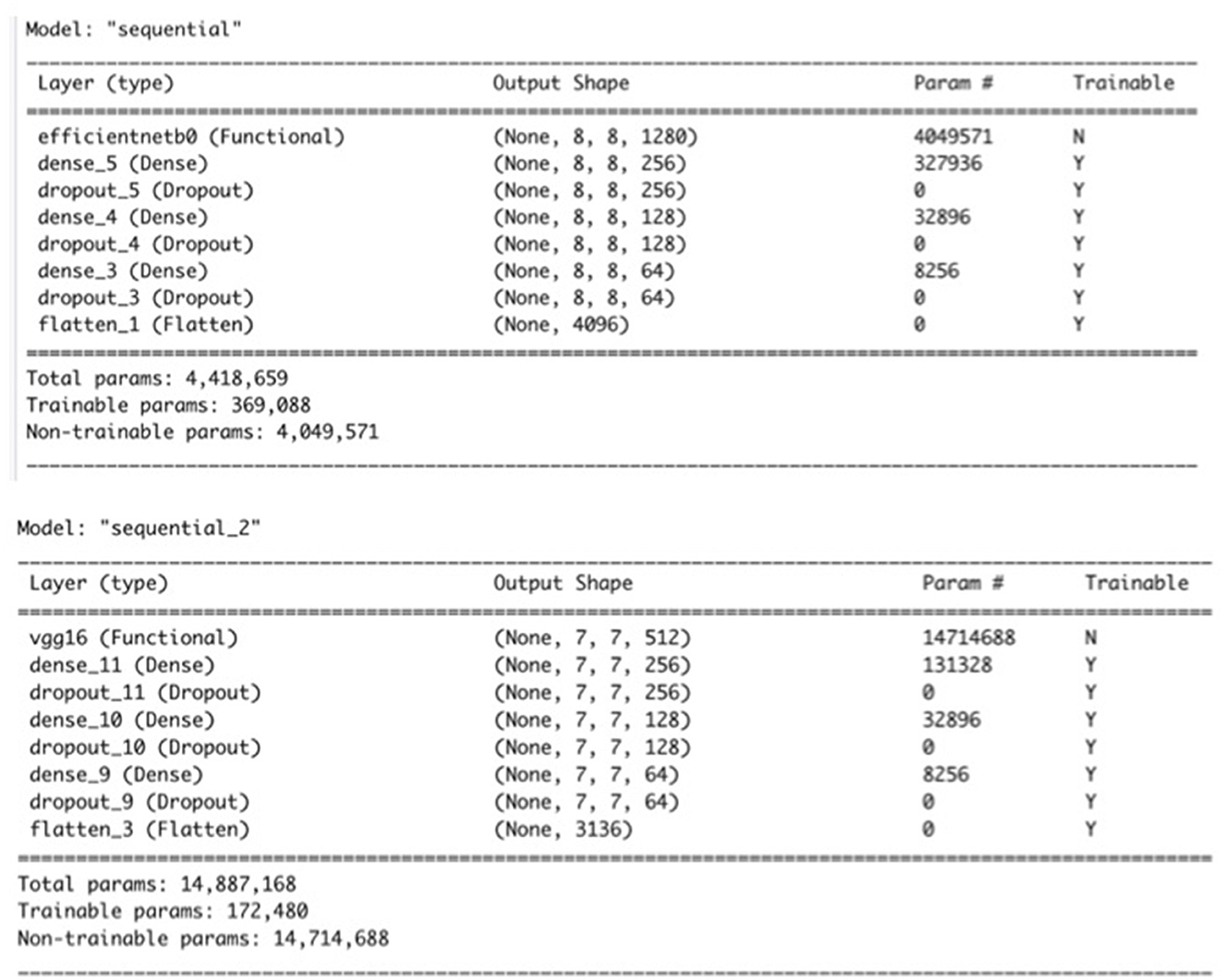
The proposed architecture of the base CNN model of the ZimCov_CNNRF, which we used to extract features from the image dataset consists of pre-trained layers of either the VGG16 or EfficientNetB0 models with frozen weights.
The base CNN model has a sequential architecture consisting of three hidden layers. The first layer is a dense layer with 256 units and a Rectified Linear Unit (ReLU) activation function, followed by a dropout layer with a rate of 0.4. The second layer is another dense layer with 128 units and a ReLU activation function, followed by another dropout layer with a rate of 0.4. The third layer is a dense layer with 64 units and a ReLU activation function, followed by a dropout layer with a rate of 0.4. The fourth layer is a flatten layer that flattens the input to a 1D array, followed by another dropout layer with a rate of 0.4. Finally, the last layer is a dense layer with two [2] units and a softmax activation function. The model takes as input images of size 224 × 224 and is trained using binary cross-entropy as the loss function, Stochastic Gradient Descent (SGD) as the optimizer with a learning rate of 0.01, a decay rate of 1e-6, and momentum of 0.9 with Nesterov momentum, and three evaluation metrics: accuracy, area under the ROC curve (AUC), and recall.
The transfer learning models use the foundational EfficientNet-B0 and VGG16 architectures pre-trained on the ImageNet dataset. The first step is to load the pre-trained EfficientNet-B0 or VGG16 model with the application_efficientnet_b0() or application_vgg16() function. We set the weights parameter to ‘imagenet' to load the pre-trained weights. The include_top parameter is set to FALSE to exclude the final classification layer, which will be replaced by a new set of dense layers. The input_shape parameter is set to (224, 224, 3) to match the size of the input images. Next, we froze the pre-trained weights in the EfficientNet-B0 and VGG16 models by calling the freeze_weights() function. The keras_model_sequential() function is then called to create a sequential model. We added the pre-trained EfficientNet-B0 model to the sequential model using the pipe (%>%) operator. Similar to the CNN Model, the model is compiled with the binary cross-entropy loss function, Adam optimizer with a learning rate of 0.001, and several metrics such as accuracy, AUC, recall, and precision. The model can then be trained on a dataset of binary classification tasks. The pre-trained weights in the EfficientNet-B0 model allow the model to perform well even with a limited amount of training data. For the proposed model, instead of compilation, the model is completed with the flattening of the layer after the third dense layer, after which the relevant features are extracted. Figure 3 presents a summary of the two models.
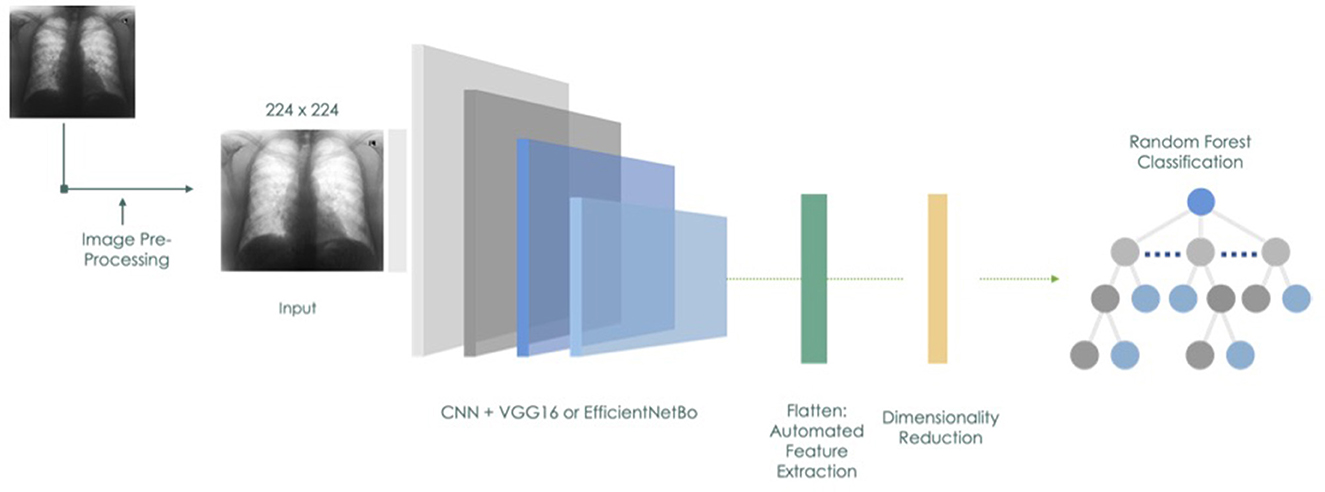
We experimented in reducing the number of extracted features through either Principal Component Analysis (PCA) or the Boruta Algorithm [38]. The Boruta is a wrapper algorithm based on the RF that has robust capabilities in feature selection as it creates a classification model based on shadow and original attributes to assess their importance in prediction. We applied the RF classifier on the reduced set of variables. We extended our experiments to compare the CNN and RF models to EfficientNetB0 and VGG16 CNNs with Linear Discriminant Analysis (LDA) considering the potential of the latter to further enhance the performance by reducing the dimensionality of the learned features and improving the separability of different classes. Figure 4 illustrates the model design and process.
2.5. Evaluating model performance - overview of metrics
For this study, we used Accuracy, Precision, Recall, F1 score and area under curve (AUC) to assess performance of the models. Accuracy is a commonly used metric as it describes how a model performs across all classes. It is a ratio based on correct predictions vs. total number of predictions. This metric becomes less useful when the classes have a different magnitude of importance. For example, in our case, correctly predicting that an individual is COVID-19 positive is of much more importance, given the extensive risk involved in obtaining an incorrect result, which could lead to the continued spread of the disease. Precision on the other hand describes how reliable the model is in classifying the samples as Positive. Recall measures the ability of a given model to correctly detect Positive samples. In general, a higher recall indicates that the model has detected more positive samples. Given the use case of COVID-19 in this analysis, the Recall was of prominent interest to us as the goal of a good model in this case would be to correctly detect all positive samples and the misclassification of negative samples as positive would have minimal impact on society or public health as opposed to positives classified as negatives. Another metric, the F1 Score introduces a balance between Precision and Recall as it combines the precision and recall metrics relative to the positive class. Another metric, the area under curve (AUC) score reflects the model's ability to distinguish between different classes and a higher score depicts better performance. The following formulae summarize the computation of the ratios. The AUC is derived from calculating the area under the Receiver Operating Characteristic (ROC) Curve, a graphical plot that is used to assess performance of binary classifiers and has gained popularity in diagnostic radiology and imaging systems in past decades [39]. The following formulae summarize the computation of the ratios.
where TP = True Positives, TN = True Negatives, FP = False Positives, FN = False Negatives.
3. Findings
In this section we present the findings of experiments conducted with six different models, four of which are derived from our proposed approach. In addition to the base CNN and the CNN_EfficientNetB0, we developed models based on variations in use of the well-known traditional models (EfficientNetB0 vs VGG16) and also applied either Principal Component Analysis or the Boruta Algorithm to reduce dimensionality of the datasets prior to application of RF. Table 1 provides a summary of the four variations of the ZimCov_CNNRF. We compare the original CNN model with the hybrid models to assess changes in model performance. The scenarios presented in the analysis include these variations in the steps toward building the hybrid models, particularly with regards to reducing dimensionality prior to application of the RF.
3.1. Original CNN model and transfer learning models
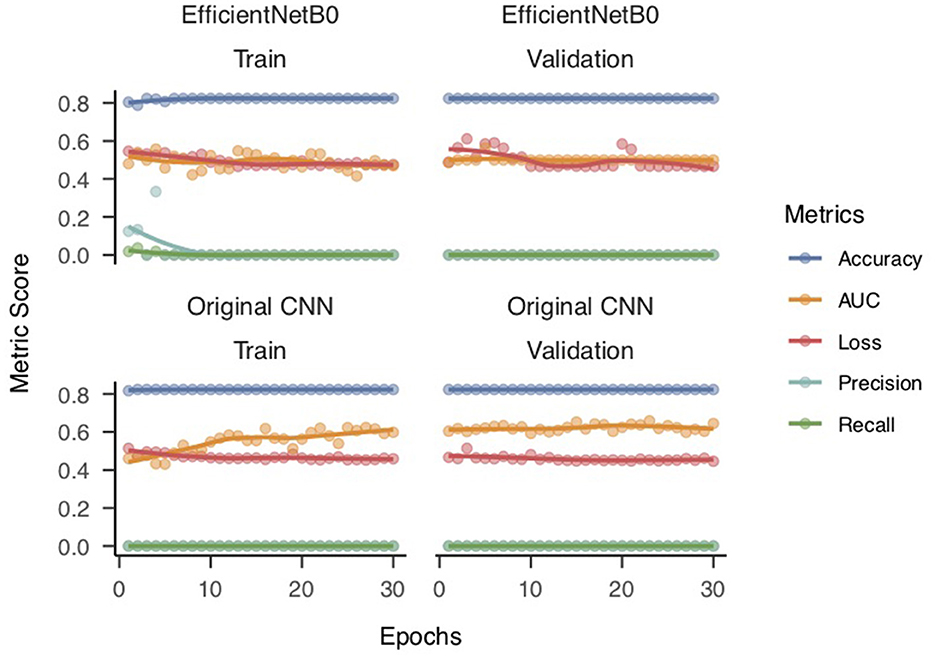
Both the base CNN model (without transfer learning) and the transfer learning based CNN-EfficientNetB0 model obtained an accuracy of 82% after 30 epochs of training but nearly 0% in precision and recall. Figure 5 shows that although both train and validation accuracy estimates performed above 80% for the two models, the precision and recall were at zero, which effectively pulled down the AUC metric. This is consistent with the imbalanced nature of the dataset.
The results imply that the model is predicting all instances as negative (no presence of the target class), and hence, it has zero true positives and high false negatives, leading to zero precision and recall values. Moreover, the model's validation loss does not decrease significantly (from 0.55 for Epoch 1 to 0.48 for Epoch 30), and the validation accuracy does not increase (from 0.48 or Epoch 1 to 0.47 for Epoch 30), which is a clear sign of overfitting. Overfitting occurs when the model starts to memorize the training data instead of learning the underlying patterns. Consequently, the model fails to generalize to new, unseen data. We utilized the SMOTE technique to address the problem of overfitting in our machine learning model. SMOTE works by creating synthetic samples of the minority class by interpolating new instances between existing minority class samples. This helps to balance the class distribution, which in turn reduces the likelihood of the model overfitting on the majority class. We applied the SMOTE technique to our dataset by oversampling the minority class to achieve a more balanced distribution between the two classes. This allowed the model to learn from more representative samples of the minority class and better generalize to unseen data. By using SMOTE, we were able to effectively address the overfitting problem and improve the performance of our model on both the training and testing datasets. The SMOTE-based adjustments to balance the dataset was only applied to the data sets of extracted features for subsequent modeling with RF.
3.2. CNN random forest hybrid model performance
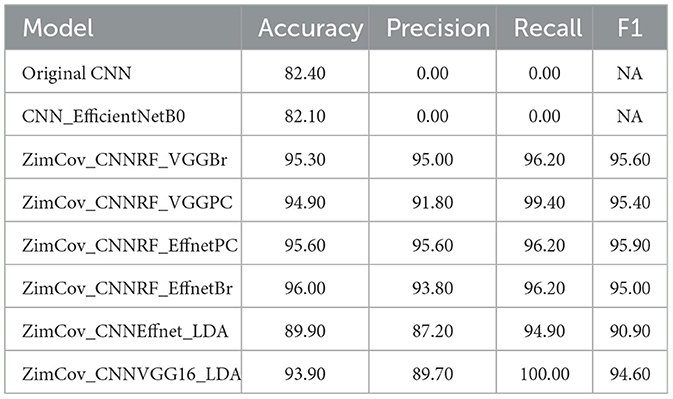
The ZimCov_CNNRF models exhibited improved predictive capacity as the accuracy, precision, recall and F1 increased to above 91% for all variations. The ZimCov_CNNRF_VGGPC had the best recall at 99.4% followed by the two EfficientNetB0 based models, ZimCov_CNNRF_EffnetBr and ZimCov_CNNRF_EffnetPC at 96.2%. Both latter models obtained the highest accuracy scores (96%) whilst the ZimCov_CNNRF_EffnetBr attained the highest F1 Score of 96%.
The EfficientNetB0 with LDA (ZimCov_CNNEffnet_LDA) attained 89.9% Accuracy and 94.9% Recall whilst the VGG16 with LDA (ZimCov_CNNVGG16_LDA) had 93.9% Accuracy and 100% Recall. A summary of the performance of these models is included in Table 2.
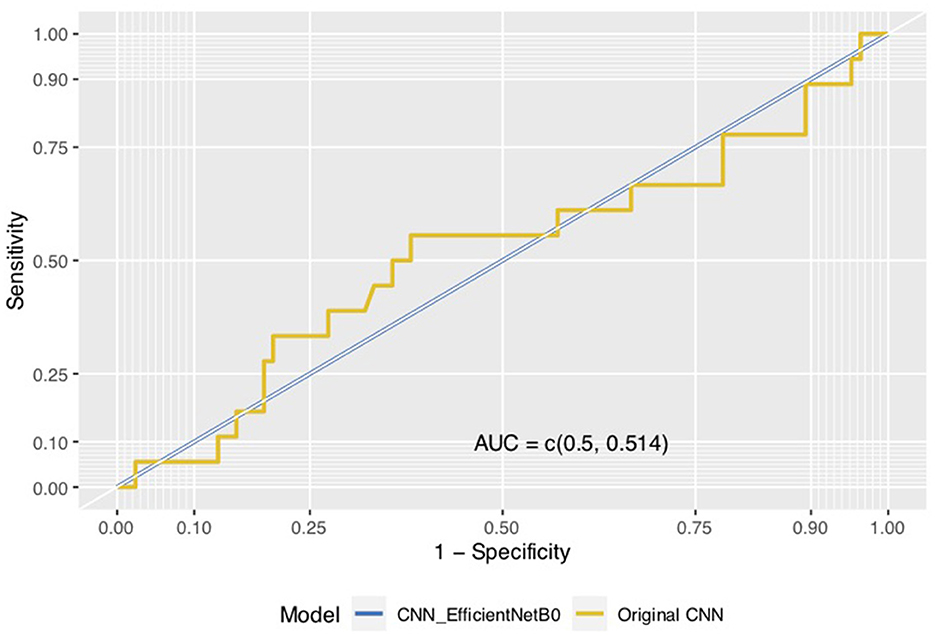
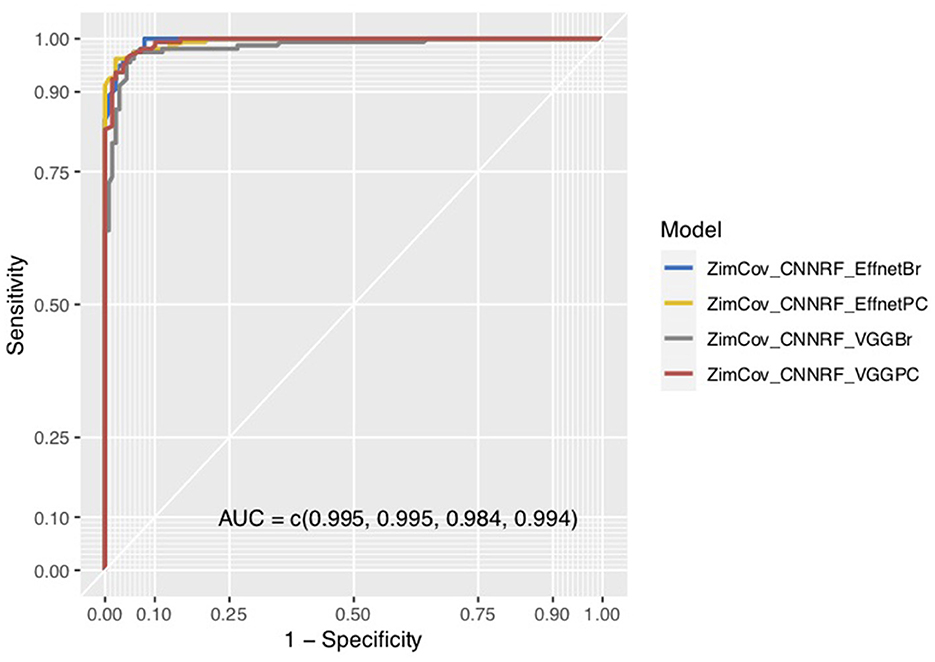
We visualized model performance using ROC curves illustrating the AUC scores. Figures 6, 7 show that the AUC scores increased from 0.5 for the CNN models to a minimum of 0.98 for the hybrid ZimCov_CNNRF models. The curves for the original CNN and the CNN_EfficientNetBo transfer learning models shown in Figure 6 are very close to the 45-degree diagonal line, which indicates the low accuracy of these models. Our proposed models, however, have curves further away from the diagonal line and the estimated area under these curves together with the accuracy measures show that the proposed models have better performance compared to the original CNN models (as seen in Figure 7). The performance of the ZimCov_CNNRF is comparable to that of PCR tests that have 98% accuracy in COVID-19 detection [3].
4. Discussion
CNNs have reliably outperformed other deep learning and machine learning models in their accuracy in image detection for classification of diseases [40]. The deep learning models have, however, typically relied on large datasets with good quality and often curated images for good performance. Whilst Random Forests have proved to be equally robust in handling classification problems, their utility in computer vision has been limited due to the curse of dimensionality arising from the large volume of pixel features defining the images. Our experimental analysis relied on a hybrid model in which the CNN is used to provide the first level of feature extraction, complemented by either additional feature extraction through the Boruta algorithm or dimensionality reduction using Principal Component Analysis; thereafter we implement RF as the final classifier. The hybrid ZimCov_CNNRF models based on images with enhanced quality and adjusted sample size using SMOTE performed better that the CNN model even after inclusion of transfer learning based on the traditional classic models, in this case VGG16 and EfficientNetB0. We observed that the performance of the hybrid models matched those typically obtained in similar analysis undertaken for large publicly obtainable data sets [41].
Our findings show that a stepwise procedure incorporating pre-processing techniques for image enhancement, sample size adjustment using SMOTE, and inclusion of a dimensionality reduction layer enhances performance of CNN deep learning models for detection COVID-19 in a relatively small chest X-ray image data set. The potential value add of a hybrid model with EfficientNetB0 or VGG16 and dimensionality reduction stages are demonstrated through the model performance including for the LDA. By leveraging the learned features from the CNN and the dimensionality reduction capabilities of Boruta, PCA and LDA, the hybrid models can achieve higher accuracy and generalization performance on these tasks compared to either models individually. The advantages of the proposed method are as follows:
• The CNN's feature extraction property proves to be more efficient in feature extraction and enhances generalizability due to its ability to handle a large spectrum of features. The forward and backward propagation algorithm characteristic of the CNN allows it to automatically learn and extract features in convolutional layers much more efficiently [42].
• The application of PCA or Boruta algorithms introduces an important layer of dimensionality reduction as the number of CNN extracted features remains disproportionately high when applying CNN's automatic feature extraction property on a small sample of images. This is important as the performance of the RF is typically affected by the number of features used in the decision trees.
• The RF, as an ensemble of decision trees for classification, enhances the reliability of the prediction model. The RF procedure introduces additional randomness as it grows several trees and searches for the best features during the splitting of a random subset of features, which improves accuracy and lessens variance whilst handling the problem of overfitting.
• The use of image augmentation and sample size adjustments allows for practical use in real world clinical settings where image data is typically few and not curated.
The performance of the ZimCov_CNNRF models is close to relatively similar approaches [41]. Our analysis demonstrated that the hybrid model based on either the EfficientNetB0 or the VGG16 with Random Forest and PCA dimensionality reduction outperformed other comparator models in detecting COVID-19, achieving an accuracy above 95% and recall of up to 99%. This result is consistent with some recent studies that also utilized hybrid models combining CNNs with other algorithms to detect COVID-19. For instance, a study by Zivkovic et al. [43] reported an accuracy of 99% using a hybrid model combining a CNN and an XGBoost classifier. Another study by Ahlawat and Choudhary [44] achieved an accuracy of 99% using a hybrid model that combined a CNN with a support vector machine (SVM) classifier whilst Ozturk et al. [45] achieved an accuracy of 98.08% (binary classification) for their darkNet model of an image detection system (You Only Look Once - YOLO). However, our results are higher than some other studies that utilized only CNN models for COVID-19 detection and that have relatively small size datasets. For example, a study by Apostolopoulos and Mpesiana [46] reported an accuracy of 93.48% using a VGG19 based CNN model for binary classification. Sahinbas and Catak [47] developed CNN transfer learning models using ResNet, DenseNet, InceptionV3, VGG16 and VGG19 foundational architectures to detect COVID-19 from X-ray images with a training dataset of 50 COVID-19 positive and 50 COVID-19 negative patients. VGG16 outperformed the other models with an accuracy of 80%.
Overall, our study demonstrated the superiority of the hybrid model based on EfficientnetB0 with Random Forest in detecting COVID-19, with a higher accuracy compared to other studies that utilized only CNN models or other hybrid models. The work presented here is of interest since, unlike these previous works, we are limited to a small data set obtained from Zimbabwe. The performance of the model, falling only nearly two percent below the accuracy of the “gold standard” PCR tests demonstrates the potential for use of this approach to scale surveillance and screening capacities in resource limited settings. The proposed model and approach offers several practical advantages in resource-limited settings:
• Firstly, these models can be trained on relatively small datasets, as demonstrated in our study, which makes them suitable for settings where access to large datasets may be limited. This can potentially increase the availability of COVID screening tools even in remote rural clinics where specialist medical practitioners such as radiologists are in short supply.
• Moreover, hybrid models combining CNNs with other algorithms can provide higher accuracy in COVID detection compared to models utilizing only CNNs or other algorithms. This can improve the reliability of COVID screening tools in resource-limited settings where there may be a shortage of trained medical personnel.
• In Zimbabwe, three critical considerations that underscore the importance of this method and similar technologies include the numerical inadequacy of specialist health professionals including radiologists given high staff attrition, the high cost of accessing specialist medical services and the absence of structured medical imaging information in public health institutions. A deliberate policy is needed, which will support an investment in best practice approaches for storing and managing medical image data complemented by systemic use of the proposed models as clinical decision support systems for the few radiologists in service. This could go a long way toward facilitating efficiency gains and equity in the provision of screening and diagnostic services.
The proposed approach also has some limitations and potential disadvantages that are discussed below:
• The feature extraction process based on the CNNs without transfer learning can result in a significantly high number of features that cannot be used in subsequent steps with reliable accuracy even after application of dimensionality reduction techniques. The CNN feature extraction procedure is therefore dependent on the use of appropriate transfer learning.
• Data for the experimental analysis was obtained from one institution and it may be useful to validate with data from other institutions in Zimbabwe.
• These models require access to high-quality medical imaging equipment, which may not be available in all settings. Additionally, the development and maintenance of these models may require significant resources and expertise, which may not be available in all resource-limited settings.
5. Conclusion
A novel classification model for diagnosing COVID-19 patients based on a hybrid CNN and RF architecture was presented. The use of RF in the classification of an image-based dataset was made possible due to the representation capability of CNNs supported by the dimensionality reduction capabilities of the Boruta algorithm and PCA. Our method achieved significantly improved accuracy, precision and recall relative to a standard CNN model even after application of transfer learning. The transformation of image data into a tabular format will allow for potential integration of other individual patient characteristics. The steps toward prediction i.e. image quality enhancement, dimensionality reduction via PCA or Boruta and finally the use of a RF for prediction can be fully automated, which allows for integration of this model into digital applications for non-invasive detection of COVID-19. The use of local data, which typically has size limitations, could provide useful insights into how routinely collected medical image data could be integrated into a structured health care management and public health emergency response. Furthermore, these models can be implemented in public health settings with limited resources and can help eliminate the need for specialist doctors, who are often not available particularly in rural areas. Our method can be replicated for other chest X-ray image-based datasets.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving human participants were reviewed and approved by the Medical Research Council of Zimbabwe (Approval no: MRCZ/A/3004). Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author contributions
CS: conceptualization, writing—original draft preparation, writing—review and editing. CH, FN, and MV: writing—review and editing and supervision. All authors contributed to the article and approved the submitted version.
Acknowledgments
The authors are grateful to Godfrey Chatora (MD, FRCR), Sharon Ngururu (MSc) for implementation support, the learning partnership and guidance on medical imaging data, documentation and systems.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Pulia MS, O'Brien TP, Hou PC, Schuman A, Sambursky R. Multi-tiered screening and diagnosis strategy for COVID-19: a model for sustainable testing capacity in response to pandemic. Annal Med. (2020) 52:207–14. doi: 10.1080/07853890.2020.1763449
2. Chen Q, Rodewald L, Lai S, Gao GF. Rapid and sustained containment of covid-19 is achievable and worthwhile: implications for pandemic response. BMJ. (2021) 4:e066169. doi: 10.1136/BMJ-2021-066169
3. Czumbel LM, Kiss S, Farkas N, Mandel I, Hegyi A, Nagy Á, et al. Saliva as a candidate for COVID-19 diagnostic testing: a meta-analysis. Front Med. (2020) 7:465. doi: 10.3389/fmed.2020.00465
4. Wang L, Lin ZQ, Wong A. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci Rep. (2020) 10:19549. doi: 10.1038/s41598-020-76550-z
5. Wong HYF, Lam HYS, Fong AHT, Leung ST, Chin TWY, Lo CSY, et al. Frequency and distribution of chest radiographic findings in patients positive for COVID-19. Radiology. (2020) 296:E72–8. doi: 10.1148/radiol.2020201160
6. Narin A, Kaya C, Pamuk Z. Automatic detection of coronavirus disease (COVID-19) using x-ray images and deep convolutional neural networks. Pattern Anal Appl. (2021) 24:1207–20. doi: 10.1007/s10044-021-00984-y
7. Zhang R, Tie X, Qi Z, Bevins NB, Zhang C, Griner D, et al. Diagnosis of coronavirus disease 2019 pneumonia by using chest radiography: value of artificial intelligence. Radiology. (2021) 298:E88–97. doi: 10.1148/radiol.2020202944
8. Deshpande G, Batliner A, Schuller BW. AI-Based human audio processing for COVID-19: a comprehensive overview. Pattern Recognit. (2022) 122:108289. doi: 10.1016/j.patcog.2021.108289
9. Li X, Zeng W, Li X, Chen H, Shi L, Li X, et al. CT imaging changes of corona virus disease 2019(COVID-19): a multi-center study in Southwest China. J Transl Med. (2020) 18:154. doi: 10.1186/s12967-020-02324-w
10. Han X, Fan Y, Alwalid O, Li N, Jia X, Yuan M, et al. Six-month follow-up chest CT findings after severe COVID-19 Pneumonia. Radiology. (2021) 299:E177–86. doi: 10.1148/radiol.2021203153
11. Xu Z, Zhao L, Yang G, Ren Y, Wu J, Xia Y, et al. Severity assessment of COVID-19 using a CT-based radiomics model. Stem Cells Int. (2021) 2021:2263469. doi: 10.1155/2021/2263469
12. Zhao C, Xu Y, He Z, Tang J, Zhang Y, Han J, et al. Lung segmentation and automatic detection of COVID-19 using radiomic features from chest CT images. Pattern Recognit. (2021) 119:108071. doi: 10.1016/j.patcog.2021.108071
13. Kassania SH, Kassanib PH, Wesolowskic MJ, Schneidera KA, Detersa R. Automatic detection of coronavirus disease (COVID-19) in X-ray and CT images: a machine learning based approach. Biocybernetics Biomed Eng. (2021) 41:867–79. doi: 10.1016/j.bbe.2021.05.013
14. Gietema HA, Zelis N, Nobel JM, Lambriks LJG, van Alphen LB, Oude Lashof AML, et al. CT in relation to RT-PCR in diagnosing COVID-19 in The Netherlands: a prospective study. PLoS One. (2020) 15:e0235844. doi: 10.1371/journal.pone.0235844
15. Yamashita R, Nishio M, Do RKG, Togashi K. Convolutional neural networks: an overview and application in radiology. Insights Imag. (2018) 9:611–29. doi: 10.1007/s13244-018-0639-9
16. D'souza RN, Huang PY, Yeh FC. Structural analysis and optimization of convolutional neural networks with a small sample size. Sci Rep. (2020) 10:834. doi: 10.1038/s41598-020-57866-2
17. Shen D, Wu G, Suk HI. Deep learning in medical image analysis. Ann Rev Biomed Eng. (2017) 19:221–48. doi: 10.1146/annurev-bioeng-071516-044442
19. Gupta J, Pathak S, Kumar G. Deep learning (CNN) and transfer learning: a review. J Phys Conf Series. (2022) 2273:012029. doi: 10.1088/1742-6596/2273/1/012029
20. Shorten C, Khoshgoftaar TM. A survey on image data augmentation for deep learning. J Big Data. (2019) 6:60. doi: 10.1186/s40537-019-0197-0
22. Khozeimeh F, Sharifrazi D, Izadi NH, Joloudari JH, Shoeibi A, Alizadehsani R, et al. RF-CNN-F: random forest with convolutional neural network features for coronary artery disease diagnosis based on cardiac magnetic resonance. Sci Rep. (2022) 12:11178. doi: 10.1038/s41598-022-15374-5
23. Kwak GH, Park C, Lee K, Na S, Ahn H, Park NW, et al. Potential of Hybrid CNN-RF model for early crop mapping with limited input data. Remote Sensing. (2021) 13:1629. doi: 10.3390/rs13091629
24. Ghose P, Alavi M, Tabassum M, Ashraf Uddin M, Biswas M, Mahbub K, Zhao Z. Detecting COVID-19 infection status from chest x-ray and CT scan via single transfer learning-driven approach. Front Genet. (2022) 13:980338. doi: 10.3389/fgene.2022.980338
25. Rajaraman S, Siegelman J, Alderson PO, Folio LS, Folio LR, Antani SK, et al. Iteratively pruned deep learning ensembles for COVID-19 detection in chest x-rays. IEEE Access. (2020) 8:115041–50. doi: 10.1109/ACCESS.2020.3003810
26. Abiyev RH, Ma'aitah MKS. Deep convolutional neural networks for chest diseases detection. J Healthcare Eng. (2018) 2018:1–11. doi: 10.1155/2018/4168538
27. Polsinelli M, Cinque L, Placidi G. A light CNN for detecting COVID-19 from CT scans of the chest. Pattern Recognit Letters. (2020) 140:95–100. doi: 10.1016/j.patrec.2020.10.001
28. Akinyelu AA, Blignaut P. COVID-19 diagnosis using deep learning neural networks applied to CT images. Front Art Int. (2022) 5:919672. doi: 10.3389/frai.2022.919672
29. Varela-Santos S, Melin P. A new approach for classifying coronavirus COVID-19 based on its manifestation on chest X-rays using texture features and neural networks. Inf Sci. (2021) 545:403–14. doi: 10.1016/j.ins.2020.09.041
30. Techzim. Zim Duo Working on AI Based COVID-19 Diagnostic System. (2021). Available online at: https://www.techzim.co.zw/2021/01/zim-duo-working-on-ai-based-covid-19-diagnostic-system/ (accessed March 1, 2021).
31. Johnson JM, Khoshgoftaar TM. Survey on deep learning with class imbalance. J Big Data. (2019) 6:27. doi: 10.1186/s40537-019-0192-5
32. Bauder RA, Khoshgoftaar TM, Hasanin T. 2018 17th IEEE International Conference on Machine Learning and Applications (ICMLA). 785–790. Orlando, FL: IEEE (2018).
33. Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: synthetic minority over-sampling technique. J Artif Int Res. (2002) 16:321–57. doi: 10.1613/jair.953
34. Cheng HD, Shi XJ. A simple and effective histogram equalization approach to image enhancement. Digit Signal Process. (2004) 14:158–70. doi: 10.1016/j.dsp.2003.07.002
35. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. (2014). doi: 10.48550/ARXIV.1409.1556
36. Tammina S. Transfer learning using VGG-16 with deep convolutional neural network for classifying images. IJSRP. (2019) 9:9420. doi: 10.29322/IJSRP.9.10.2019.p9420
37. Tan M, Le QV. EfficientNet: Rethinking model scaling for convolutional neural networks. (2020). Retrieved from: http://arxiv.org/abs/1905.11946
38. Kursa MB, Jankowski A, Rudnicki WR. Boruta a system for feature selection. Fund Inf. (2010) 101:271–85. doi: 10.3233/FI-2010-288
39. Hajian-Tilaki K. Receiver operating characteristic (roc) curve analysis for medical diagnostic test evaluation. Caspian J Int Med. (2013) 4:627–35.
40. Lan T, Hu H, Jiang C, Yang G, Zhao Z. A comparative study of decision tree, random forest, and convolutional neural network for spread-F identification. Adv Space Res. (2020) 65:2052–61. doi: 10.1016/j.asr.2020.01.036
41. Mostafiz R, Uddin MS, Alam NA, Mahfuz Reza M, Rahman MM. Covid-19 detection in chest X-ray through random forest classifier using a hybridization of deep CNN and DWT optimized features. J King Comput Inf Sci. (2022) 34:3226–35. doi: 10.1016/j.jksuci.2020.12.010
42. Liu Y, Pu H, Sun DW. Efficient extraction of deep image features using convolutional neural network (CNN) for applications in detecting and analysing complex food matrices. Trends Food Sci Technol. (2021) 113:193–204. doi: 10.1016/j.tifs.2021.04.042
43. Zivkovic M, Bacanin N, Antonijevic M, Nikolic B, Kvascev G, Marjanovic M, et al. Hybrid CNN and XGBoost model tuned by modified arithmetic optimization algorithm for COVID-19 early diagnostics from X-ray images. Electronics. (2022) 11:3798. doi: 10.3390/electronics11223798
44. Ahlawat S, Choudhary A. Hybrid CNN-SVM classifier for handwritten digit recognition. Proc Comput Sci. (2020) 167:2554–60. doi: 10.1016/j.procs.2020.03.309
45. Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Rajendra Acharya U, et al. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. (2020) 121:103792. doi: 10.1016/j.compbiomed.2020.103792
46. Apostolopoulos ID, Mpesiana TA. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. (2020) 43:635–40. doi: 10.1007/s13246-020-00865-4
Keywords: Artificial Intelligence, machine learning, medical imaging, computer vision, hybrid model
Citation: Sisimayi C, Harley C, Nyabadza F and Visaya MV (2023) AI-enabled case detection model for infectious disease outbreaks in resource-limited settings. Front. Appl. Math. Stat. 9:1133349. doi: 10.3389/fams.2023.1133349
Received: 28 December 2022; Accepted: 16 May 2023;
Published: 08 June 2023.
Edited by:
Aurelio A. de los Reyes V., University of the Philippines Diliman, PhilippinesReviewed by:
Rahib Abiyev, Near East University, CyprusEbenezer Olaniyi, Mississippi State University, United States
Copyright © 2023 Sisimayi, Harley, Nyabadza and Visaya. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chenjerai Sisimayi, Y2hlbmplcmFpLnNpc2ltYXlpQGdtYWlsLmNvbQ==; Y3Npc2ltYXlpQHdvcmxkYmFuay5vcmc=
 Chenjerai Sisimayi
Chenjerai Sisimayi Charis Harley3
Charis Harley3 Farai Nyabadza
Farai Nyabadza