- 1Department of Mathematics, Duke University, Durham, NC, United States
- 2Department of Statistics, Stanford University, Stanford, CA, United States
- 3Department of Pediatrics and Rady Children’s Hospital, University of California at San Diego, San Diego, CA, United States
- 4Department of Cellular & Molecular Medicine, Department of Pediatrics, University of California at San Diego, San Diego, CA, United States
Herein we describe new frontiers in mathematical modeling and statistical analysis of oscillatory biomedical signals, motivated by our recent studies of network formation in the human brain during the early stages of life and studies forty years ago on cardiorespiratory patterns during sleep in infants and animal models. The frontiers involve new nonlinear-type time–frequency analysis of signals with multiple oscillatory components, and efficient particle filters for joint state and parameter estimators together with uncertainty quantification in hidden Markov models and empirical Bayes inference.
1 Introduction
The 2017 Nobel Prize in Physiology or Medicine was awarded to Jeffrey Hall and Michael Rosbash of Brandeis University, and Michael Young of Rockefeller University, “for their discoveries of molecular mechanisms controlling the circadian rhythm.” In 1984, they succeeded in isolating the “period gene” (i.e., the gene that controls the circadian rhythm). Hall and Rosbash then went on to “discover PER, the protein encoded by period, accumulated during the night and degraded during the day.” In 1994, Young answered a “tantalizing puzzle” concerning how PER produced in the cytoplasm could reach the cell nucleus where genetic material is located. He discovered a second gene timeless, encoding the TIM protein so that TIM bound to PER can enter the cell nucleus to block the period gene activity. “Such a regulatory feedback mechanism explained how this oscillation of cellular protein levels emerged, but questions lingered,” such as what controlled the frequency of the oscillations. Young identified another gene doubletime encoding the DBT protein that delayed the accumulation of the PER protein. The three laureates identified additional proteins required for the activation of the period gene, as well as for the mechanisms by which light can synchronize the circadian clock.
One of us (Muotri) was PI of a project on “spontaneous network formation” displaying “periodic and regular oscillatory events that were dependent on glutamatergic and GABAergic signaling” during early the brain maturations, for which structural and transcriptional changes “follow fixed developmental programs defined by genetics,” see [1] who also found that “the oscillatory activity transitioned to more spatiotemporally irregular patterns which synchronous network activity resembled features similar to those observed in preterm human EEG.” This project is similar in spirit to the exemplary work of Hall, Rosbash, and Young but the “experimental inaccessibility” of the human brain during the early stages of life pushes mathematical modeling and statistical analysis of the oscillatory signals and events to new frontiers that we present in the next section. We describe in the next paragraph the underlying biomedical background of this project.
One of the major recent realizations, especially in the neurosciences, is that while we can obtain important information from animal studies, there are major differences between humans and animals. This is manifested in many ways, especially in those major clinical trials based on animal findings that did not pan out. Therefore, if we intend to study pathogenesis of disease, treat them, prevent them, or cure diseases across the age spectrum, we need to refocus our scientific approaches and strategies in order to be more efficient and effective. Since embryonic stem cells are often problematic to obtain for ethical reasons, the discovery of being able to re-program somatic cells from humans into induced pluro-potential stem cells (iPSCs, taking these somatic cells back into their “history”) and differentiate them into different relatively mature cell types have opened a major avenue for the scientific community, resulting in the 2012 Nobel Prize in Physiology or Medicine to John Gurdon of Cambridge and Shinya Yamanaka of Kyoto. If these iPSCs are exposed to the right growth factors, they would assemble into the early human brain (brain organoids) by an amazing process of self-organizing the 3-dimensional cellular elements that recapitulate the network, cellular, and membrane properties of neurons and the glia. Many types of organoids such as the kidney, the intestine, the liver, and the lung organoids have been recently developed. These organoids have been particularly useful for studying either normal early human biology or developmental disorders as in neurodevelopmental diseases.
2 Methods
The statistical methods used by Trujillo et al. (2019, pp. 16–19) in their analysis of data on oscillatory signals and events consist of 1) multi-electrode array (MEA) recording and custom analysis, 2) network event analysis that involves detecting spikes (when at least 80% of the maximum spiking values) over the length of the recording when reached at least 1 s away from any other network event, 3) oscillatory spectral power analysis, in which “oscillatory power” is defined as “peaks in the PSD (power spectral density estimated by Peter D. Welch’s method) above the aperiodic
2.1 Time–Frequency Analysis of Signals With Multiple Oscillatory Components
The first author (Wu) has been working on time–frequency analysis (TFA) and its applications to high-frequency biomedical signals in the last ten years. Examples include electrocardiography, electroencephalogram, local field potential, photoplethysmogram (PPG), actinogram, peripheral venous pressure (PVP), arterial blood pressure, phonocardiogram, and airflow respiratory signal, to name several. Usually, these signals are composed of multiple components, each of which reflects the dynamics of a physiological system. The analysis is challenged by the physiological variability that appears in the form of time-varying frequency and amplitude or even time-varying oscillatory pattern that is referred to as the “wave-shape function.” Furthermore, depending on the signal, the “waxing and waning” effect is sometimes inevitable for its components; see [2, Figure 1] for an illustration. Take the widely applied PPG signal as an example, for which [3] has given an introduction to photoplethysmogram (PPG) and its applications “beyond the calculation of arterial oxygen saturation and heart rate.” In addition to the well-known cardiac component reflecting hemodynamic information, PPG may contain the respiratory dynamics as another component. The frequency of the cardiac component (respiratory component, respectively) is impacted by the heart rate variability (breathing rate variability, respectively). Cicone and [4] provide an algorithm to “extract both heart and respiratory rates” from the PPG signal and thereby to analyze their interactions. Such information can be used in conjunction with other biomedical signals reflecting hemodynamics. In particular, PVP is ubiquitous in the hospital environment and a rich source of hemodynamic information [5]. But, it typically has a low signal-to-noise ratio (SNR), and its oscillatory pattern is sensitive to the physiological status, making it much less used in comparison with PPG. Wu et al. [6] have developed new signal processing tools to facilitate its use.
Combining time–frequency analysis (TFA) with statistical analysis, the lack of which in the previous work “presents an opportunity for much future research,” is illustrated in Figure 2 (applied to PPG, fetal ECG, and fetal heart rate variability) of Wu [2] who describes several recent advances in TFA for high-frequency biomedical signals. There are several challenges common to different biomedical signal processing problems. The first is how to estimate the dynamics (e.g., how to quantify the time-varying frequency, amplitude, or wave-shapes) of the signal. The second is to assess signal quality and determine artifacts, distinguishing between physiological and nonphysiological ones. The third is to identify oscillatory components and the fourth is to decompose the signal into constituent components. To address these challenges, several TFA tools have been proposed. In addition to the traditional linear-type time–frequency analysis tools like short-time Fourier transform (STFT), continuous wavelet transform (CWT), and bilinear time-frequency analysis tools [7], several nonlinear-type tools have been developed and applied, including the reassignment method, empirical mode decomposition (EMD), Blaschke decomposition (BKD), adaptive locally iterative filtering (ALIF), sparse time-frequency representation (STFR), synchrosqueezing transform (SST), scattering transform (ST), concentration of frequency and time (ConcFT), de-shape and dynamic diffusion maps [8–14]. The statistical properties of these methods have been relatively unexplored and we are currently investigating them; new methods to handle emerging scientific problems might be developed on the way.
2.2 Efficient Particle Filters for Joint State and Parameter Estimation in HMM
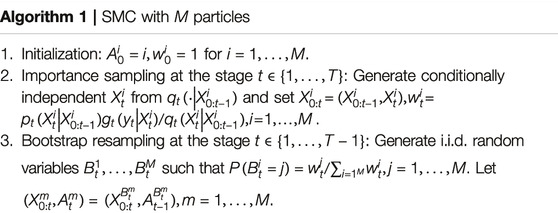
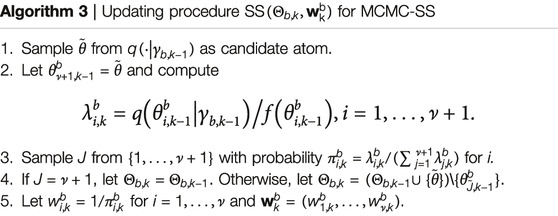
During the past three years, the second author (Lai) has been developing a new Markov Chain Monte Carlo (MCMC) procedure called “MCMC with sequential substitutions” (MCMC-SS) for joint state and parameter estimation in hidden Markov models. The basic idea is to approximate an intractable distribution of interest (or target distribution) by the empirical distribution of N representative atoms, chosen sequentially by an MCMC procedure, so that the empirical distribution approximates the target distribution after a large number of iterations as explained below.
Lai’s work in this area began with the landmark paper of Gordon, Salmond and Smith [15] on the development of sequential Monte Carlo (SMC), also called particle filters, for the estimation of latent states in a hidden Markov model (HMM). Liu’s monograph [16] contains a collection of techniques that have been developed since then, with examples of applications in computational biology and engineering, and Chan and Lai [17] provide a general theory of particle filters. Let
This conditional distribution is often difficult to sample from and the normalizing constant is also difficult to compute for high-dimensional or complicated state spaces, and particle filters use sequential Monte Carlo that involves importance sampling and resampling to circumvent this difficulty. The particle filter computes
By using martingale theory, Chan and Lai [17] provide a comprehensive theory of the SMC estimate
Theorem 1. Under certain Integrability Conditions,
Moreover, letting
Chan and Lai [17, Lemmas 1 and 4] use the following representation of
in which
see Eqs. (3.3) and (3.36) of the study by authors of reference [15] which show that
The assumption of a single fully specified HMM in particle filter is often too restrictive in applications since the model parameters are usually unknown and also need to be estimated sequentially from the observed data. A standard method to estimate unknown parameters is to assume a prior distribution for the unknown parameter vector and to use the Markov chain Monte Carlo (MCMC) to estimate the posterior distribution. Authors of reference [15] carried out this method for time-homogeneous Markov chains
PMCMC uses SMC involving M particles (each of which consists of a sampled parameter and state trajectory) at every iteration k to construct an approximation
in which κ represents an initial burn-in period that is asymptotically negligible as
Theorem 2. Suppose
where
(i) Let
(ii) Let
where
As shown in reference [21], with probability approaching 1 by large k, the candidate atom
for some real-valued function l, which [19] call “potential” in their substantive applications. The advantage of using a zero-mean Gaussian random field reference measure Q is that it is specified by the covariance operator
3 Discussion and Concluding Remarks
Haddad and Lai actually initiated similar research forty years ago when they worked on cardiorespiratory patterns during sleep in a SIDS (Sudden Infant Death Syndrome) project at Columbia University’s Pediatrics Department; see reference [26] which describes the study population consisting of 12 infants “with one or more episodes of aborted SIDS” (four of whom had siblings who died of SIDS), and 19 normal infants, all born full-term except for one aborted SIDS infant born at 37 weeks of gestation. After describing the study design and methods of statistical analysis, the authors of reference [26] presented results on total tidal volume (Vt), respiratory cycle time (Ttot), and increase in Vt/Ttot resulting from 2% increase in CO2 concentration in the sleeping chamber, comparing aborted SIDS to normal infants in both REM (rapid eye movement) and quiet sleep. Because of the inability to induce stress such as loaded breathing as in reference [27, 28], animal models involving sheep, puppies, and dogs were used; see also [29]. In particular, the authors of reference [24] “studied diaphragmatic muscle function during inspiratory flow resistive loaded breathing” in 6 unanesthetized sheep over periods of 6–8 months. Data were collected (baseline) and after application of the loads that were sustained for up to 90 min. Loads were divided into mild (<50 cm H2O
A related direction of our ongoing research is to combine several biomedical signals, which form a multivariate time series, thereby providing a more holographic view of a human subject. For example, in an intensive care unit, PPG can be combined with EEG, EMG, respiratory, and other signals to evaluate a patient’s health status. Wu and his collaborators have applied in [32,33] a combination of ST and EEG channels to study sleep dynamics and an “interpretable machine learning algorithm” to assess consistency of sleep-stage scoring rules across multiple sleep centers. How to utilize available information from multiple centers is a sensor fusion problem. We are currently combining recent advances in sensor fusion with those in TFA to develop integrated statistical analysis of the multivariate time series of multiple biomedical signals.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, and further inquiries can be directed to the corresponding author.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Funding
TL’s research was supported by the National Science Foundation under DMS-1811818. GH’s research was supported by the National Institutes of Health under 1R01HL146530 and 1R21NS111270. AM’s research was supported by the National Institutes of Health under DP2-OD006495.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank TL’s research assistant Chenru Liu at Stanford University for her valuable contributions to the preparation, timely submission, and revision of the paper. We also thank the reviewers whose innovative comments and suggestions have resulted in substantial improvement of the presentation.
References
1. Trujillo, CA, Gao, R, Negraes, PD, Gu, J, Buchanan, J, Preissl, S, et al. Complex Oscillatory Waves Emerging from Cortical Organoids Model Early Human Brain Network Development. Cell Stem Cell (2019). 25(4):558–69. doi:10.1016/j.stem.2019.08.002
2. Wu, H-T. Current State of Nonlinear-type Time-Frequency Analysis and Applications to High-Frequency Biomedical Signals. Curr Opin Syst Biol (2020). 23:8–21. doi:10.1016/j.coisb.2020.07.013
3. Shelley, KH. Photoplethysmography: beyond the Calculation of Arterial Oxygen Saturation and Heart Rate. Anesth Analg (2007). 105:531–6. doi:10.1213/01.ane.0000269512.82836.c9
4. Cicone, A, and Wu, HT. How nonlinear-type time-frequency analysis can help in sensing instantaneous heart rate and instantaneous respiratory rate from photoplethysmography in a reliable way. Front Phy (2017). 8:701.
5. Wardhan, R, and Shelley, K. Peripheral venous pressure waveform. Curr Opin Anesthesiol (2009). 22:814–821.
6. Wu, HT, Alian, A, and Shelley, KH. A new approach to complicated and noisy physiological waveforms analysis: peripheral venous pressure waveform as an example. J Clin Monit Comput (2020). 1–17.
7. Flandrin, P. Wavelet Analysis and its Applications (1999). p. 10.Time-frequency/time-scale Analysis
8. Huang, NE, Shen, Z, Long, RS, Wu, HH, Zheng, Q, et al. The Empirical Mode Decomposition and the hilbert Spectrum for Nonlinear and Non-stationary Time Series Analysis. Proc R Soc Lond A (1998). 454:903–95. doi:10.1098/rspa.1998.0193
10. Daubechies, I, Lu, J, and Wu, H-T. Synchrosqueezed Wavelet Transforms: an Empirical Mode Decomposition-like Tool. Appl Comput Harmonic Anal (2011). 30:243–61. doi:10.1016/j.acha.2010.08.002
11. Daubechies, I, Wang, Y, and Wu, H-t. ConceFT: Concentration of Frequency and Time via a Multitapered Synchrosqueezed Transform. Phil Trans R Soc A (2016). 374:20150193. doi:10.1098/rsta.2015.0193
12. Mallat, S. Group Invariant Scattering. Comm Pure Appl Math (2012). 65:1331–98. doi:10.1002/cpa.21413
13. Lin, C-Y, Su, L, and Wu, H-T. Wave-Shape Function Analysis. J Fourier Anal Appl (2018). 24:451–505. doi:10.1007/s00041-017-9523-0
14. Wang, S-C, Wu, H-T, Huang, P-H, Chang, C-H, Ting, C-K, and Lin, Y-T. Novel Imaging Revealing Inner Dynamics for Cardiovascular Waveform Analysis via Unsupervised Manifold Learning. Anesth Analgesia (2020). 130:1244–54. doi:10.1213/ane.0000000000004738
15. Gordon, NJ, Salmond, DJ, and Smith, AFM. Novel Approach to Nonlinear/non-Gaussian Bayesian State Estimation. IEE Proc F Radar Signal Process UK (1993). 140:107–13. doi:10.1049/ip-f-2.1993.0015
17. Chan, HP, and Lai, TL. A General Theory of Particle Filters in Hidden Markov Models and Some Applications. Ann Stat (2013). 41(6):2877–904. doi:10.1214/13-aos1172
18. Lai, TL Recursive Particle Filters for Joint State and Parameter Estimation in Hidden Markov Models with Multifaceted Applications. In: Proceedings of the 8th ICCM. Boston: International of Press; (2021) in press
19. Chopin, N. A Sequential Particle Filter Method for Static Models. Biometrika (2002). 89(3):539–52. doi:10.1093/biomet/89.3.539
20. Cotter, SL, Roberts, GO, Stuart, AM, and White, D. MCMC Methods for Functions: Modifying Old Algorithms to Make Them Faster. Stat Sci (2013). 28:424–46. doi:10.1214/13-sts421
21. Andrieu, C, Doucet, A, and Holenstein, R. Particle Markov Chain Monte Carlo Methods. J R Stat Soc Ser B (2010). 72(3):269–342. doi:10.1111/j.1467-9868.2009.00736.x
22. Chopin, N, Jacob, PE, and Papaspiliopoulos, O. SMC2: an Efficient Algorithm for Sequential Analysis of State Space Models. J R Stat Soc Ser B (2013). 75(3): 397–426. doi:10.1111/j.1467-9868.2012.01046.x
23. Bezanson, J, Edelman, A, Karpinski, S, and Shah, VB. Julia: A Fresh Approach to Numerical Computing. SIAM Rev (2017). 59(1):65–98. doi:10.1137/141000671
24. Yalamanchili, P, Arshad, U, Mohammed, Z, Garigipati, P, Entschev, P, Kloppenborg, B, et al. ArrayFire - A High Performance Software Library for Parallel Computing with an Easy-To-Use API (2015).
25. Zhu, M. Ph.D. Dissertation in Computer Science. Stanford University (2021). doi:10.1145/3450439.3451855Improving and Accelerating Particle-Based Probabilistic Inference
26. Haddad, GG, Leistner, HL, Lai, TL, and Mellins, RB. Ventilation and Ventilatory Pattern during Sleep in Aborted Sudden Infant Death Syndrome. Pediatr Res (1981). 15(5):879–83. doi:10.1203/00006450-198105000-00011
27. Bazzy, AR, and Haddad, GG. Diaphragmatic Fatigue in Unanesthetized Adult Sheep. J Appl Physiol (1984). 57(1):182–90. doi:10.1152/jappl.1984.57.1.182
28. Haddad, GG, Jeng, HJ, Bazzy, AR, and Lai, TL. Within-breath Electromyographic Changes during Loaded Breathing in Adult Sheep. J Appl Physiol (1986). 61(4):1316–21. doi:10.1152/jappl.1986.61.4.1316
29. Haddad, GG, Jeng, HJ, Lee, SH, and Lai, TL. Rhythmic Variations in R-R Interval during Sleep and Wakefulness in Puppies and Dogs. Am J Physiology-Heart Circulatory Physiol (1984). 247(1):H67–H73. doi:10.1152/ajpheart.1984.247.1.h67
30. Haddad, GG. The Multifaceted Sudden Infant Death Syndrome. Curr Opin Pediatr (1992). 4(3):426–30. doi:10.1097/00008480-199206000-00006
31. Chen, J, Heyse, JF, and Lai, TL. Medical Product Safety Evaluation: Biological Models and Statistical Methods. Boca Raton FL: Chapman & Hall/CRC (2018). doi:10.1201/9781351021982
32. Liu, GR, Lo, YL, Malik, J, Sheu, YC, and Wu, HT. Diffuse to Fuse EEG Spectra–Intrinsic Geometry of Sleep Dynamics for Classification. Biomed Signal Process Control (2019). 55(5):101576.
Keywords: hidden Markov model, time–frequency analysis, oscillatory components, biorhythms, empirical bayes, uncertainty quantification
Citation: Wu H-T, Lai TL, Haddad GG and Muotri A (2021) Oscillatory Biomedical Signals: Frontiers in Mathematical Models and Statistical Analysis. Front. Appl. Math. Stat. 7:689991. doi: 10.3389/fams.2021.689991
Received: 01 April 2021; Accepted: 11 May 2021;
Published: 15 July 2021.
Edited by:
Charles K. Chui, Hong Kong Baptist University, Hong KongReviewed by:
Xiaoming Huo, Georgia Institute of Technology, United StatesHaiyan Cais, University of Missouri–St. Louis, United States
Copyright © 2021 Wu, Lai, Haddad and Muotri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tze Leung Lai, bGFpdEBzdGFuZm9yZC5lZHU=
 Hau-Tieng Wu1
Hau-Tieng Wu1 Gabriel G. Haddad
Gabriel G. Haddad