- Institute of Bioanalytical Chemistry, Faculty of Chemistry and Mineralogy, Center for Biotechnology and Biomedicine, Leipzig University, Leipzig, Germany
Because of the global spread of multi- and pan-resistant bacteria, there is a need to identify, research, and develop new strategies to combat these pathogens. In a previous proof-of-concept study, we presented an innovative strategy by genetically modifying lytic T7 bacteriophages. We integrated DNA fragments encoding for derivatives of the antimicrobial peptide (AMP) apidaecin into the phage genome to induce the production and release of apidaecin within the T7 infection cycle, thereby also targeting phage-resistant Escherichia coli bacteria. In this follow-up study, we optimized the apidaecin encoding insert to improve the expression of the apidaecin derivative Api805 by adding the secretion signal peptide of the OmpA protein. This prevented the detrimental effects of the peptide on the producing bacterial cell after its production. The integration of two copies of the OmpA-Api805 insert into the phage genome resulted in T7Select-2xOmpA-Api805 phages, which had a partially improved activity in inhibiting phage-resistant E. coli compared to the T7Select phages without insert and with only one copy of the OmpA-Api805 insert. Additionally, we showed that the combinatorial use of the lytic bacteriophage T7Select with the highly active and lytic AMPs CRAMP (cathelicidin-related AMP) and melittin against E. coli made the lysis process of the phage and the peptides more effective and prevented the growth of potentially AMP- and phage-resistant E. coli strains. The integration of DNA sequences derived from CRAMP and melittin into the phage genome resulted in the created T7Select-(M)CRAMP and T7Select-(M)melittin phages, which showed a lysis behavior like the phage without insert and partially inhibited the growth of potentially phage-resistant E. coli strains after the phage-mediated lysis.
1 Introduction
The rise of antibiotic resistance has led to a global public health crisis (O’Neill, 2014). Bacterial infections that were once easily treated with antibiotics are now becoming increasingly difficult to cure due to the emergence of multidrug-resistant bacteria (Nikaido, 2009; van Duin and Paterson, 2016). In 2019, it was estimated that approximately 4.95 million deaths were associated with bacterial antimicrobial resistance, of which 1.27 million deaths were directly caused by the infection (Antimicrobial Resistance Collaborators, 2022). This death toll calls for the exploration of alternative strategies to combat bacterial infections, such as antimicrobial peptides (AMPs), bacteriophages, antisense therapeutics, immunotherapies, vaccines, probiotics, and microbiota-based therapeutics (Kumar et al., 2021; García-Contreras et al., 2022; MacNair et al., 2024).
The potential of AMPs has been recognized due to their broad-spectrum activity against various pathogens (Huan et al., 2020). Among the numerous AMPs identified, CRAMP (cathelicidin-related AMP), melittin, and apidaecin 1b have emerged as promising candidates due to their potent antimicrobial properties. CRAMP (Table 1), produced in mice, is an amphipathic, α-helical peptide that preferentially binds to negatively charged groups of bacterial membranes, thereby damaging them (Gallo et al., 1997). Melittin (Table 1), a component of bee venom, is a linear, water-soluble, cationic, amphipathic peptide (Memariani et al., 2020). It exerts its potent bactericidal activity by acting on the bacterial membrane, causing pore formation and disrupting membrane function, ultimately leading to lysis of the bacterial cells (Fidelio et al., 1984). Apidaecin 1b (GNNRPVYIPQPRPPHPRL), naturally produced by the honey bee (Apis mellifera), and its previously designed derivative Api805 (Table 1) belong to the group of proline-rich AMPs (PrAMPs), which are linear peptides with a high content of proline and arginine (Casteels et al., 1994; Li et al., 2006; Ludwig et al., 2022a). Apidaecins show remarkable efficacy against Gram-negative bacteria (Li et al., 2006), such as Escherichia coli, by entering the cell through the specific transporter SbmA and inhibiting the bacterial 70S ribosome (Krizsan et al., 2014, 2015; Ludwig et al., 2022a).
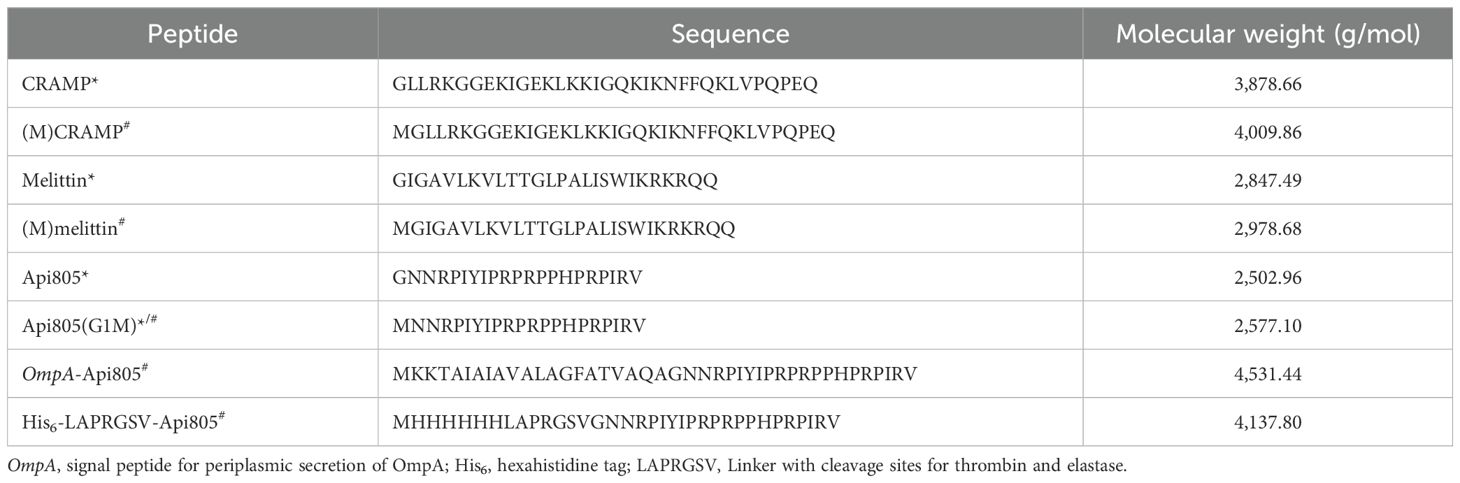
Table 1. Amino acid sequences of peptides synthesized* and/or expressed from coding DNA sequences# in this study.
An alternative approach to targeting bacteria relies on bacteriophages, viruses that infect and destroy bacteria, which have gained increasing attention in recent years as potential therapeutics due to their remarkable specificity and ability to target bacterial strains including multidrug-resistant bacteria (Alqahtani, 2023; Kulshrestha et al., 2024). However, the efficacy of bacteriophages can be limited by several factors, including host-range limitations and the frequent emergence of phage-resistant bacterial strains (Zalewska-Piątek, 2023; Laanto, 2024). To overcome these challenges, synergistic interactions between bacteriophages and other antimicrobial agents have been investigated to enhance their bactericidal activity (Oechslin et al., 2017; Tagliaferri et al., 2019).
Previously, we successfully demonstrated that the combinatorial use of bacteriophage T7Select and apidaecins shows promising synergistic effects by inhibiting the regrowth of phage-resistant strains after phage lysis (Ludwig et al., 2022b). In a proof-of-concept study, we integrated DNA sequences encoding apidaecins into the T7Select phage genome downstream of the gp10 gene encoding the capsid protein to allow expression of apidaecins by the bacterial host (E. coli) during the phage infectious cycle (Ludwig et al., 2022b). With this approach, we aimed to improve the host range limitation of the phage by local production of apidaecin peptides, especially to combat phage-resistant bacterial subpopulations. However, the engineered phages did not show inhibition of phage-resistant bacterial regrowth or improved overall activity compared to the original T7Select phage because the expression levels of apidaecins were too low, even too low to be detected by mass spectrometry (Ludwig et al., 2022b).
In this follow-up study, we report the synergistic effect of CRAMP and melittin with the T7Select phage against E. coli and the construction and characterization of T7Select bacteriophages (Figures 1A, B) engineered to express these membrane-active and lytic AMPs. These AMPs have a different mode of action than the ribosome-inhibiting apidaecins we used previously and therefore should not significantly interfere with their own expression at the 70S ribosome in the host cell. In addition, we aimed to enhance the expression of the apidaecin derivative Api805 by adding the secretion signal peptide of OmpA to the apidaecin sequence (Figures 1A, C), named OmpA-Api805 (Table 1), to prevent its intracellular accumulation with adverse effects on the producing host cell. This should improve peptide expression during bacterial growth and the phage infection cycle by secreting the peptides into the periplasm (Thie et al., 2008; Freudl, 2018; Erkut, 2021). Additionally, we aimed to increase the expression rate by inserting multiple copies of the OmpA-Api805 sequence into the phage genome (Figures 1A, C; Supplementary Figure S1). To avoid replication misalignments (“slippage”) and genetic rearrangements based on direct repeats of homologous DNA sequences (Bzymek and Lovett, 2001) in this case, we altered the codon usage for each OmpA-Api805 copy and intermediate sequences (Supplementary Figure S1). However, this was not possible for the repeated sequences of the T7 promoter and the ribosome-binding site within the insert. Another approach was to attach the Api805 peptide to the viral capsid via a cleavable linker (Table 1; Figures 1A, D).
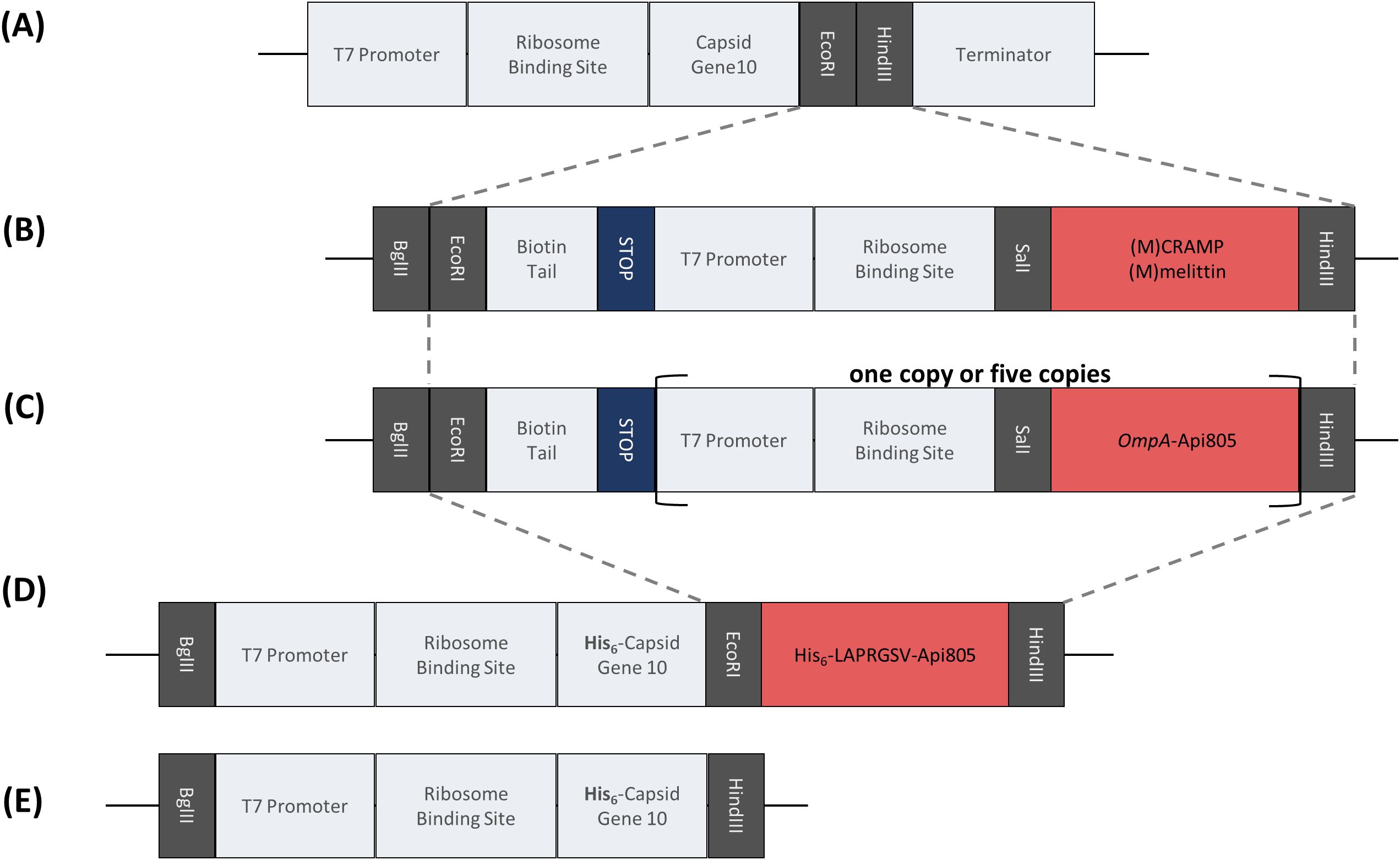
Figure 1. Overview of the peptide coding DNA fragments for integration into the pET28a(+) plasmid and the T7Select phage genome. Gray dashed lines indicate the part to be integrated into the phage genome. Schematic representation of the multiple cloning site in the T7Select phage genome (A). Insert encoding the lytic AMPs (M)CRAMP and (M)melittin (B). Insert encoding the fusion peptide of Api805 with N-terminal addition of the signal peptide of OmpA in one or five copies (C). Insert encoding the direct attachment of Api805 to the viral capsid protein via a linker containing sequences for the hexahistidine tag and cleavage sites for thrombin and elastase (D). Insert of capsid gene 10 for cloning into pET28a(+) for expression in E. coli (E).
2 Materials and methods
2.1 Materials and chemicals
Reagents used are listed in the Supplementary File S1. E. coli and phage strains, plasmids, and primers are listed in Supplementary Table S1. Amino acid sequences of peptides expressed from coding DNA sequences are listed in Table 1. Water was purified using a Purelab Ultra water purification system (electrical resistivity >18.2 kΩ×m; organic content <2 ppb; ELGA LabWater, Celle, Germany).
2.2 Peptide synthesis
Peptides were synthesized on a multiple synthesizer (SYRO2000, MultiSynTech GmbH, Witten, Germany) using the Fmoc/tBu protection strategy, in situ activation with DIC in the presence of HOBt, and Wang resins to obtain the peptides as C-terminal acids. Peptides were cleaved with TFA containing 12.5% (v/v) of a scavenger mixture [ethanedithiol, m-cresol, thioanisole, and water; 1:2:2:2 (by vol)], precipitated with cold diethyl ether, and purified by RP-HPLC on a Jupiter C18-column (21.2 mm ID) using an aqueous acetonitrile gradient in the presence of 0.1% TFA. Peptide purities were determined by RP-HPLC on a Jupiter C18-column (4.6 or 2 mm ID). Monoisotopic masses were confirmed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS; 5800 Proteomic Analyzer; AB Sciex, Darmstadt, Germany).
2.3 Antibacterial activity
Minimum inhibitory concentrations (MICs) were determined at least twice in triplicate using a liquid broth microdilution assay in sterile 96-well plates with a total volume of 100 μL per well. Aqueous peptide solutions were serially diluted twofold in 50 µL of 25% MHBII or LB medium. Overnight cultures were diluted in 25% MHBII or LB medium to 1.5×107 cells per mL and an aliquot of 50 μL was added to each well. The final cell count in each well was 7.5×106 CFU/mL. The plates were incubated at 37°C for 20 ± 2 h. The turbidity of each well was measured at 600 nm. The MIC was defined as the lowest peptide concentration at which the turbidity did not exceed that of the medium alone.
2.4 Plaque assay
For the plaque assay, E. coli Rosetta pLysS was grown to 8×108 CFU/mL in LB medium. Phage lysates were prepared in serial dilutions. An aliquot of the E. coli culture (250 µL) was transferred to molten LB overlay agar (4 mL, 0.75% agar), the diluted phage lysate (100 µL) was added, and the mixture was poured onto solid LB agar plates (1.5% agar). After the upper agar solidified, the plates were inverted and incubated overnight at 37°C. Plaques were counted and phage titers were calculated by multiplying the plaque counts by the dilution factor.
2.5 Phage-peptide synergy assay
E. coli Rosetta pLysS was grown in LB medium to a cell count of 9.2×108 CFU/mL and transferred to a 96-well plate (130 µL/well). PrAMPs (10 µL) were added to a final concentration of 128 µg/mL and serially diluted down to 1 µg/mL. A phage solution (10 µL, 7.5×108 PFU/mL) was added to achieve a final multiplicity of infection (MOI) of 0.0625 and a final volume of 150 µL per well. Optical density at 600 nm (OD600) was recorded in all wells every 15 min after 5 s of medium orbital shaking at a constant temperature of 37°C for a total of 30 h on a PARADIGM™ (Beckman Coulter, Salzburg, Austria).
2.6 DNA constructs for expression and genetic modification of phages
Construct designs of DNA sequences (Figures 1B–E) based on previously reported constructs (Ludwig et al., 2022b) were ordered from Genscript Biotech B.V. (Leiden, The Netherlands). These DNA fragments were cloned into pET28a+ plasmid using BgIII and HindII and transformed into chemically competent E. coli DH5α by heat shock. Plasmids were isolated and purified using the QIAprep® Spin Miniprep Kit (Qiagen, Hilden, Germany). Insertion of the DNA sequences of interest was verified by PCR using the pET28 primer (Supplementary Table S1) and sequencing.
2.7 Peptide and protein expression
The pET28a+ plasmids with different inserts (Supplementary Table S1) were transformed into chemically competent E. coli Rosetta pLysS by heat shock. Bacterial cultures were incubated in LB medium (initial OD600 of 0.05) at 37°C and 200 rpm. When an OD600 of 0.3–0.4 was reached, the culture was split into two and IPTG (final concentration 1 mmol/L) was added to one of the two cultures. Both cultures were incubated at 37°C and 200 rpm for 3 h and the OD600 was recorded hourly. Bacterial cells were harvested (30 min, 4,000 g, 4°C) and analyzed for protein and peptide expression by SDS-PAGE with Coomassie staining. Cells were disrupted using a FastPrep-24™ 5G instrument (MP Biomedicals, Solon, OH, USA) and centrifuged (30 min, 21,000 g, 4°C). The supernatant (soluble protein fraction) and the pellet resuspended in PBS containing urea (4 mol/L, insoluble protein fraction) were analyzed by SDS-PAGE.
2.8 Genetic modification of phages
Genetically modified phages were generated using the T7Select 415 Cloning Kit (Merck Millipore, Burlington, USA) according to the manufacturer’s instructions. The DNA sequences to be inserted (Figures 1B–D) were amplified by PCR using pET28a+ plasmids and pET primers (Supplementary Table S1). The PCR product was digested with the restriction enzymes EcoRI and HindIII (both New England Biolabs, Ipswich, USA) and then ligated into the predigested T7Select vector arms using T4 ligase (New England Biolabs, Ipswich, USA) at 4°C for 16 h. The ligated T7Select genome was added to the provided packaging extract. Packaging was performed for 2 h at room temperature, followed by a plaque assay to obtain infectious particles. Formed plaques were transferred to LB medium (450 µL), incubated at 37°C for 2 h, and chloroform was added (33 µL). For phage propagation and larger lysate production, E. coli Rosetta pLys was grown in LB medium at 37°C to an OD600 ≈ 0.5–1.0 and then 100 µL of undiluted phage lysate was added to 5 mL of bacterial culture. The mixture was then incubated at 37°C until complete lysis, the mixtures were centrifuged (5 min, 5,000 g), sterile filtered (22 μm) in glass culture tubes, and stored at 4°C. Proper insertion was verified by PCR using T7 primers (Supplementary Table S1) and sequencing.
2.9 Time-kill assay
For the time-kill assay, E. coli Rosetta pLysS was grown to 9.2×108 CFU/mL in LB medium and transferred to a clear 96-well plate (130 µL/well). Phage lysate (20 µL, 3.75×108 PFU/mL) was added to achieve a MOI of 0.0625. The outer edge of the 96-well plate, i.e., rows A and H and columns 1 and 12, was filled with 150 µL of sterile medium to reduce edge effects. The plate was incubated at 37°C for at least 30 h and OD600 was measured every 15 min after orbital shaking at medium speed for 5 to 10 s.
2.10 Mass spectrometry
One hundred microliters of bacteria cell culture supernatant, bacteria lysate, or phage lysate were mixed with 1.9 mL of 5% (v/v) acetonitrile in 50 mmol/L ammonium bicarbonate (pH 8.5, ABC solution). A 10-mg weak cation exchange cartridge (Oasis WCX, Waters Corporation, Milford, Massachusetts, USA) was wetted with 1 mL of acetonitrile and equilibrated with 1 mL of ABC solution. Samples were applied in two steps each 1 mL. To wash out unbound material, the cartridge was washed with 1 mL of ABC solution and 1 mL of 5% (v/v) acetonitrile in water with 2% (v/v) formic acid. Peptides were eluted with two steps each 0.3 mL of 60% (v/v) acetonitrile in water with 2% (v/v) formic acid to a low-bind 1.5-mL Eppendorf tube. Forty microliters of the eluate was mixed with 40 µL of 2% (v/v) formic acid in water and transferred to a polypropylene insert (Agilent Technologies, USA). WCX cleaned samples were measured with an M-class UPLC system (Waters, Manchester, UK) coupled online to an ESI-IMS-QTOF instrument (Synapt G2-Si, Waters, Manchester, UK). Sample (5 µL) was injected to Acquity BEH300 C4 trap column (1.7 µm, 180 µm × 20 mm, Waters, Ireland). Trapping was done at a flow rate of 5 µL/min at 20% B, where B was acetonitrile with 0.1% (v/v) formic acid and A was water with 0.1% (v/v) formic acid, for 6 min. The trapped sample was eluted to a BEH300 C4 column (1.7 µm, 100 µm × 100 mm, Waters, Ireland) and equilibrated at 20% B at a flow rate of 1 µL/min with a linear gradient from 20% B to 90% B in 10 min. Re-equilibration before next sample injection was done by 5 min back to 20% B and 14 more minutes at 20% B. Sample ionization was done with a Nanolockspray source (Waters, Manchester, UK) set up with a pre-cut PicoTip® emitter (360 µm OD × 20 µm ID, 10 µm tip 2.5″ length, Waters, Milford, USA) at 3 kV. During the whole measurement every 45 s, the signal of a 100 fmol/µL GluFib solution (m/z 785.843 doubly charged ion signal) was measured for 0.5 s for mass correction. The following were the nano source parameters: 30 V sample cone, 80 V source offset, 100°C source temperature, 20 L/h cone gas, 0.2 bar nano flow gas, and 150 L/h purge gas, with 2 mL/min trap gas (nitrogen, argon for trap, and transfer collision cell), 180 mL/min helium for helium cell, and 90 mL/min IMS cell gas (nitrogen). TriWaveDC settings were 1 V for Trap DC entrance, 2 V for Trap DC bias, −2 V Trap DC, and 0 V Trap DC exit. TriWave IMS settings were 1,000 m/s wave velocity and 40 V wave height. Mass spectra were recorded with MassLynx 4.2 SCN 983 with the following HD-MRM method. Positive polarity was determined with an analyzer in resolution mode from 0 to 30 min for m/z range 50 to 2,000 with a scan rate of 0.5 s. Every 5 s, a radar scan was recorded. Using MRM scan padding, MS/MS was triggered from 10 to 20 min for m/z 570.10, 599.76, 600.40, 606.10, 634.36, 776.30, and 805.67 using transfer collision cell from 18 to 25 V for fragmentation. For data analysis, MassLynx 4.2 SCN983 and Drift Scope 2.9 (Waters, Manchester, UK) were used.
3 Results
3.1 Activity of AMPs and AMP-phage synergy against E. coli
The recently reported apidaecin derivatives Api805 (Ludwig et al., 2022a) and Api805(G1M), also named Api806 (Ludwig et al., 2022b), showed MIC values against E. coli Rosetta DE3 pLysS of 4 and 8 µg/mL in 25% MHBII, but only 128 µg/mL in LB medium (Table 2). A similar medium dependence was observed for the activity of CRAMP and melittin with MICs of 16 µg/mL in 25% MHBII and MICs of 128 and 32 µg/mL in LB medium, respectively. Similar activities were observed for CRAMP, Api805, and Api805(G1M) against the previously isolated T7 phage-resistant E. coli Rosetta DE3 pLysS strain R 2.3 (Ludwig et al., 2022b), whereas melittin was four- and eightfold more active in 25% MHBII and LB, respectively, with MICs of 4 µg/mL in both media (Table 2).
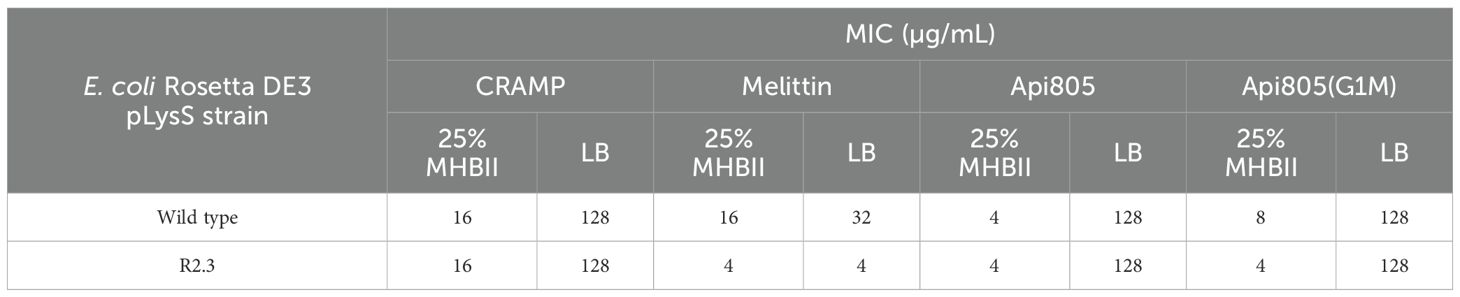
Table 2. MICs of antimicrobial peptides against wild-type E. coli Rosetta DE3 pLysS and the T7Select resistant strain R 2.3 (7.5×106 CFU/mL) cultured in 25% MHB and LB media.
Since ribosome-targeting apidaecin derivatives have been reported to act synergistically with the T7Select phage in a certain concentration range without interfering with phage-mediated lysis, but preventing or delaying regrowth of phage-resistant bacteria (Ludwig et al., 2022b), the effect of the lytic AMPs CRAMP and melittin added to an uninfected and T7Select phage-infected E. coli Rosetta culture was investigated (Figure 2). Uninfected E. coli Rosetta pLys grew in LB medium from an OD600 of ~0.4 (~1.2×108 CFU) measured in a 96-well plate to an OD600 of ~0.55 within 5 h before the OD600 decreased steadily over the next 13 h to ~0.45, which remained stable for the next 12 h. The addition of CRAMP and melittin resulted in different lysis patterns and different regrowth effects after initial lysis, which started at a concentration of 16 µg/mL and further increased with higher AMP concentrations (Figure 2). When incubated with 64 µg/mL CRAMP, the bacteria grew to an OD600 of ~0.5 in the first 3.5 h. The OD600 then decreased steadily over the next 7 h to ~0.35 before increasing to ~0.45 after 30 h of cultivation (Figure 2). Melittin added at the same concentration immediately lysed the bacteria, as indicated by a decrease in OD600 from ~0.4 to ~0.3 in the first 2 h. In the following 20 h, a strong regrowth to an OD600 of ~0.6 was observed, which remained stable for the rest of the cultivation period (Figure 2).
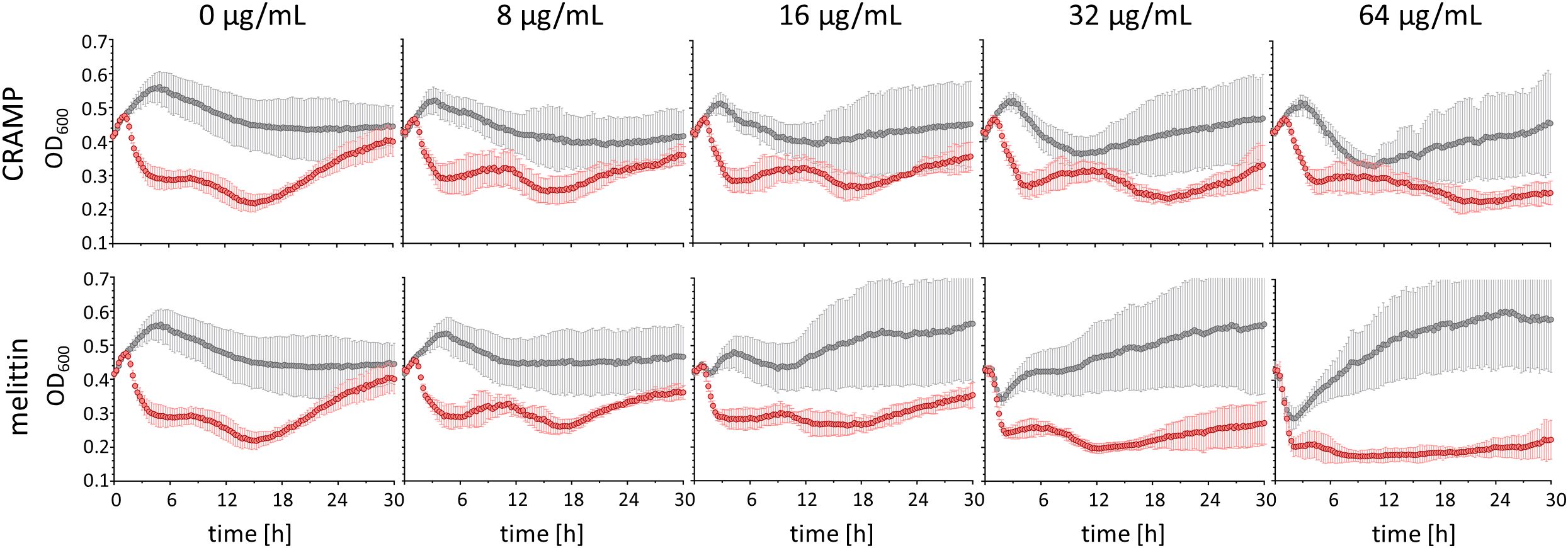
Figure 2. Bacterial growth rates of E. coli Rosetta (t0: ~1.2×108 CFU) uninfected (gray) and infected (red) with Phage T7Select (MOI = 0.0625), and increasing concentrations of the lytic AMPs CRAMP and melittin. Shown are OD600 values recorded every 15 min after 5 s shaking for 30 h. Experiments were performed twice as single determinations and once as duplicates. Error bars indicate the standard deviation of all four replicates.
In the phage-infected E. coli Rosetta pLys cultures, lysis was triggered 1.5 h after phage addition in the absence of AMPs, reducing the OD600 from ~0.5 to ~0.3 in 3 h and further to ~0.2 in the following 10 h. However, regrowth started after approximately 18 h with a steady increase in OD600 to ~0.4 after 30 h of cultivation (Figure 2). The addition of CRAMP did not significantly affect the total lysis phase in the concentration range tested up to 64 µg/mL, but slightly reduced regrowth at 32 µg/mL and completely prevented it at 64 µg/mL over the entire observation period (Figure 2). Melittin addition to a phage-infected E. coli culture induced lysis earlier in a concentration-dependent manner starting at 16 µg/mL and at concentrations of 32 and 64 µg/mL, and the lysis was already completed after 90 min, presumably due to the strong lytic activity of melittin. In addition to a faster lysis, the lysis at 64 µg/mL also appeared to be more effective. As with CRAMP, melittin reduced regrowth at 32 µg/mL and completely inhibited it at 64 µg/mL (Figure 2). It should also be noted that the regrowth induced by the peptides was prevented by the addition of the phage. Thus, CRAMP and melittin appear to be promising candidates for combinatorial use with phages and also for phage engineering, since the lytic activities of phage and peptides were not reduced, but enhanced, and the regrowth of potentially resistant bacteria observed with phage and peptide treatments alone was inhibited.
3.2 Growth effects induced by the expression of apidaecin constructs and lytic AMPs in E. coli
Next, the ability of E. coli to express CRAMP, melittin, Api805, OmpA-Api805, and the fusion protein gene10B-Api805 was tested. Plasmids encoding Api805(G1M), OmpA-Api805, 5xOmpA-Api805, gene10-Api805, (M)CRAMP, and (M)melittin were transformed into E. coli Rosetta pLysS. Bacterial growth was monitored with and without inducing expression from the plasmid using IPTG, and their protein patterns were analyzed by SDS-PAGE (Figure 3). Induced expression of Api805(G1M) significantly reduced bacterial growth compared to the uninduced control with a difference in OD600 values of 0.662 ± 0.017, similar to gene10-Api805 (ΔOD600 = 0.758 ± 0.037), whereas expression of native T7 capsid protein (gene10), OmpA-Api805, and 5xOmpA-Api805 did not reduce bacterial growth (Figure 3A). Induction of (M)CRAMP expression reduced bacterial growth less than Api805(G1M), but still with an ΔOD600 of 0.496 ± 0.076 (Figure 3A). The strongest effect was observed for (M)melittin, which strongly reduced bacterial growth even below the baseline value, indicating a bactericidal effect due to cell lysis upon (M)melittin expression.
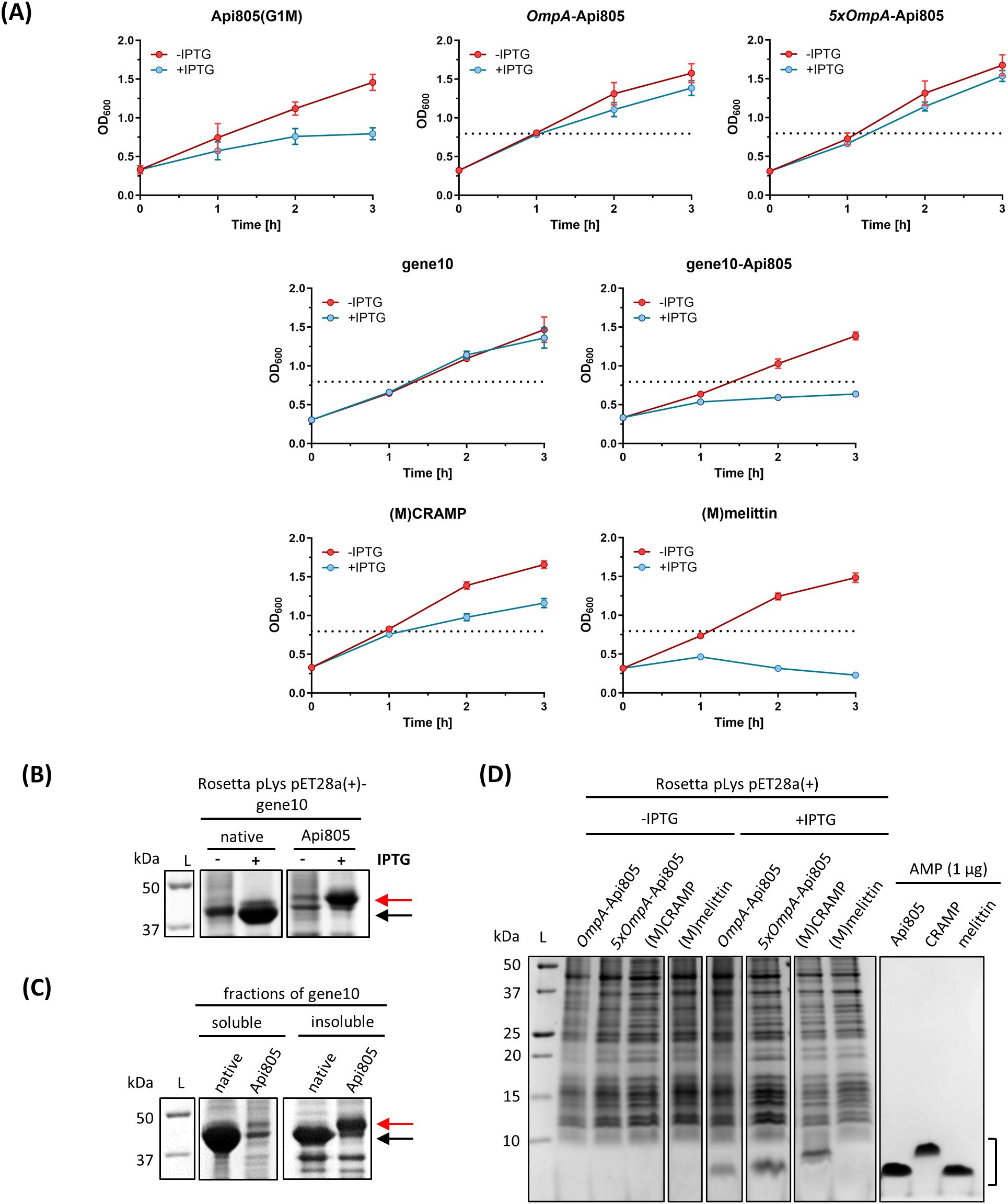
Figure 3. Bacterial growth rates of E. coli Rosetta harboring pET28a(+) vectors containing sequences for AMPs and for gene10 constructs. Growth rates of E. coli cultures were monitored by OD600 values without (red) or with (blue) IPTG-induced peptide/protein expression (A). The horizontal black dotted lines show the growth maximum of Rosetta pLys pET28a(+)-Api805(G1M) (OD600 = 0.795) in the presence of IPTG for comparison. Experiments were performed twice in duplicate. Error bars indicate the standard deviation of all four replicates. Protein/peptide expression in E. coli Rosetta harboring pET28a(+) vectors containing sequences for gene10 constructs and AMPs (B–D). SDS-PAGE (12%) with Coomassie staining of the corresponding E. coli preparations after 3 h of IPTG induction (B) and of the soluble and insoluble fractions after bacterial cell disruption (C). The arrows indicate where the T7 capsid protein gene10 (black) and gene10-Api805 (red) migrate in the gel. SDS-PAGE (16% gel) with Coomassie staining of samples prepared from E. coli cells expressing peptides after 3 h of IPTG induction and of synthetic AMPs (1 µg) for comparison (D). The black bracket indicates the region where the AMPs migrate in the gel.
Both gene10 proteins displayed bands on SDS-PAGE, with the gene10-Api805 band being weaker than the native capsid protein gene10 band (Figure 3B). In contrast to the native capsid, the fusion protein gene10-Api805 was mostly present in insoluble inclusion bodies (Figure 3C). Similar to the previously reported Api805(G1M), also named Api806 (Ludwig et al., 2022b), the Api805 derivatives OmpA-Api805 and 5xOmpA-Api805 were also detected in the cell pellet after 3 h of induction as bands with apparent molecular weights below 10 kDa in SDS-PAGE (Figure 3D). However, the peptides were not detected at the expected molecular weight of ~4.5 kDa, but at an apparent weight of ~2.5 kDa, corresponding to Api805 (Figure 3D; Table 1). Since only Api805 without the OmpA signal peptide and no growth reduction for Rosetta pLys pET28a(+)-OmpA-Api805 and pET28a(+)-5xOmpA-Api805 (Figure 3A) were detectable, we assumed the expression of OmpA-Api805, followed by the natural cleavage of the signal peptide and the secretion of Api805 into the periplasm away from the immediate vicinity of the 70S ribosome, as originally designed. In contrast to (M)CRAMP, no visible band was detectable for (M)melittin in the cell pellet (Figure 3D). Analyzed by mass spectrometry after a WCX-SPE, it was possible to detect (M)melittin mainly in the bacteria culture supernatant and also in the bacterial lysate after lysing the residual cell pellet (Supplementary Figure S2). However, the peptide was not detected in its native variant, but with different modifications. (M)melittin from bacterial lysate was modified at the methionine in position 1 by oxidation (+15.999 Da, Supplementary Figure S2A). (M)melittin identified in bacterial culture supernatant was modified at the methionine like in the lysate (+15.999 Da on methionine); additionally, the tryptophane in position 20 was modified to kynurein (+3.989 Da) and N-formylkynurein (+31.999 Da, Supplementary Figure S2B).
3.3 Generation and activity of AMP-phages
T7Select phage constructs encoding OmpA-Api805, (M)CRAMP, and (M)melittin sequences (Figures 1A–C) resulted in infectious particles (Figure 4A). Analysis of the inserted DNA fragments of the engineered phage genomes by PCR and agarose gel electrophoresis compared to the unmodified T7Select phage showed the correct size confirming the insertion of OmpA-Api805, (M)CRAMP, and (M)melittin (Figure 4B). The phages were genetically stable with respect to the insert, which could be detected and confirmed in all plaques tested even after repeated propagations and in produced phage lysates. Phage lysates of T7Select-OmpA-Api805, T7Select-(M)CRAMP, and T7Select-(M)melittin prepared immediately after phage-mediated lysis were analyzed for AMP production by mass spectrometry after a WCX-SPE. (M)melittin expression was confirmed (Supplementary Figure S4), but as with the bacterial expression samples (Supplementary Figure S2), the peptide was not detected in its native variant, but was modified with a mass shift of +44.990 Da at the methionine (Supplementary Figure S3), which could be due to a carboxylation. However (M)CRAMP, OmpA-Api805, or free Api805 could not be detected by the mass spectrometry method applied. Most likely, (M)CRAMP and Api805 could not be detected due to low amounts, which could be attributed to inefficient expression or loss of the peptides owing to binding to various lysate or medium components during sample preparation. In addition, as with melittin, special modifications could shift the mass of the peptides to quite different m/z values, making it difficult to distinguish between the peptides and all the other components in the phage lysate.
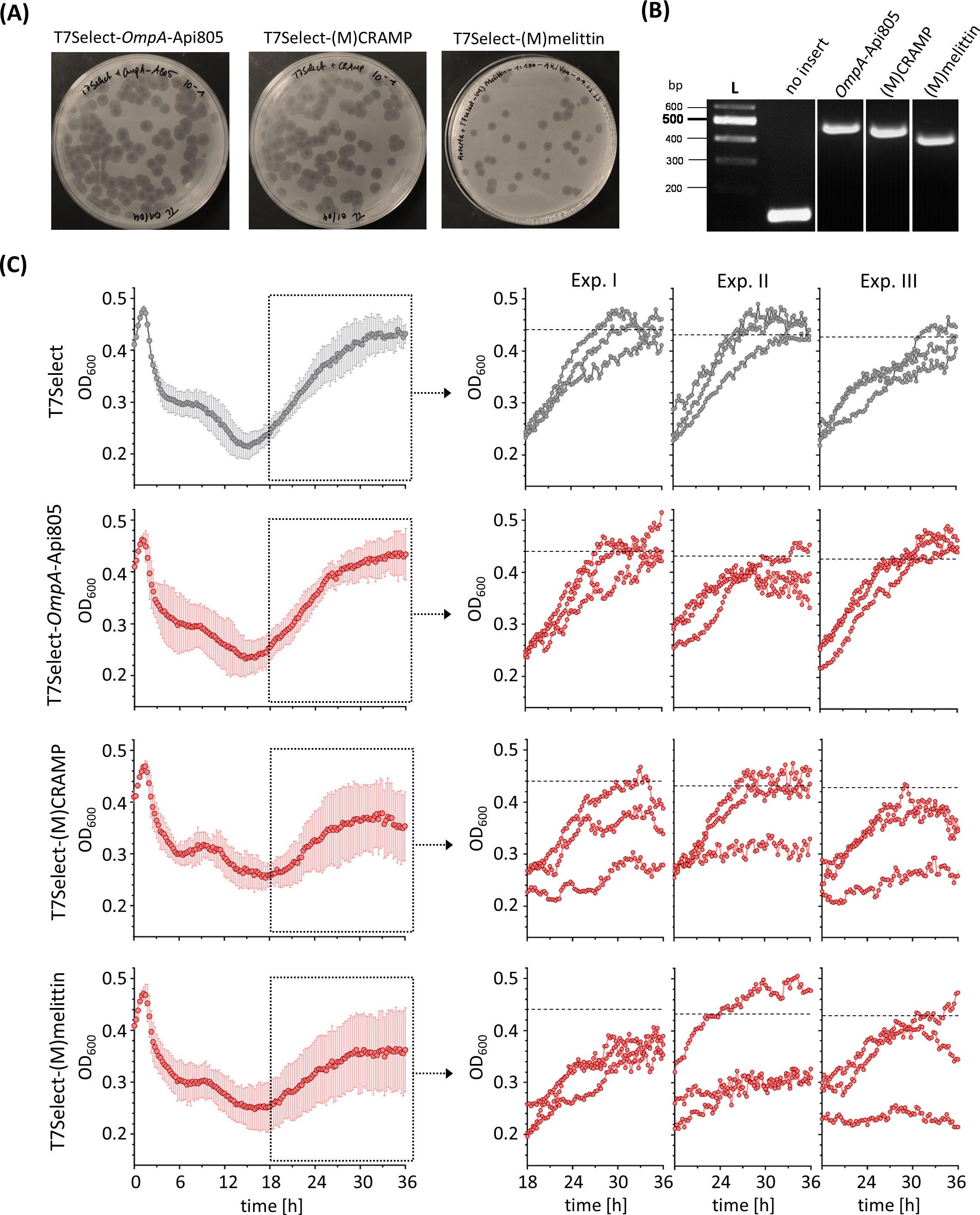
Figure 4. A plaque assay of engineered T7Select phages encoding different AMPs showing the recovery of infectious phages (A). Ethidium bromide-stained agarose gel electrophoresis of DNA fragments amplified by PCR from phage genomes of engineered T7Select phages with T7 fwd and T7 rev primers to confirm the insertion of the AMP inserts OmpA-Api805 (483 bp), (M)CRAMP (462 bp), and (M)melittin (438 bp) (B). Bacterial growth of E. coli Rosetta cultures infected with engineered T7Select phages (MOI 0.0625). Infected cultures with T7Select phage without insert (gray) and with inserts (red) encoding for OmpA-Api805, (M)CRAMP, and (M)melittin (C). Shown are the OD600 values recorded every 15 min after 5 s shaking for 36 h Experiments were performed three times in triplicate. Shown are the curves with error bars (standard deviation) of all nine replicates over the entire cultivation period and the curves of the three individual replicates of each experiment (Exp.) in the regrowth phase, the cultivation period from 18 to 36 h. The horizontal black dotted lines show the mean growth of the culture infected with T7Select after 36 h of the respective experiment for comparison (Exp. 1: OD600 = 0.439 ± 0.021/Exp. 2: OD600 = 0.430 ± 0.008/Exp. 3: OD600 = 0.426 ± 0.017).
The activity of the engineered phages T7Select-OmpA-Api805, T7Select-(M)CRAMP, and T7Select-(M)melittin was determined by a time-kill assay (Figure 4C; Supplementary Figure S4), performed three times in triplicates. The nine replicates of E. coli cultures infected with the control T7Select phage were very uniform and showed no significant variation in growth during the initial lysis and regrowth phases (cultivation phase from 18 to 36 h). The T7Select-OmpA-Api805 phage behaved like the control. The phages with the inserts for (M)CRAMP and (M)melittin also showed similar lysis behavior, but in contrast to the control, the E. coli cultures showed different growth behavior in the regrowth phase (Figure 4C; Supplementary Figure S4). Some replicates behaved similarly to the control, while others showed reduced or even inhibited regrowth, which indicate the presence of phage-induced AMPs.
Integration of the 5xOmpA-Api805 construct (Figures 1A, C) into the phage genome also resulted in infectious particles (Figure 5A). Thirteen plaques of different sizes were picked and the subsequently prepared phage lysates were tested for the insert by PCR and agarose gel electrophoresis (Figure 5A). All lysates except lysate 6 contained phages with a single copy of OmpA-Api805; only lysates 2, 10, 11, and 13 also contained phages with two and four copies of OmpA-Api805. Notably, no lysate was found to contain phages with the full insert of five copies of OmpA-Api805 (Figure 5A). No correlation between plaque size and copy number was observed. Lysate 10 was selected for a subsequent plaque assay (Figure 5B) that attempted to separate phages with different copy numbers of OmpA-Api805. Lysates of 25 plaques were analyzed. While 13 lysates contained mainly phages with only one copy of OmpA-Api805, 11 lysates mainly contained phages with two copies (Figure 5B). One lysate (21) showed faint bands of DNA fragments for one, two, and four copies. Lysate 23 was selected for a subsequent up-scaling propagation and a plaque assay. The phages from lysate 23 were confirmed to be T7Select-2xOmpA-Api805 carrying a genetically stable insert with two copies of OmpA-Api805 (Figures 5C, D), indicating that multiple copies can be incorporated into the phage genome by the chosen strategy. Sequencing of the T7Select-2xOmpA-Api805 genome confirmed the insertion of two copies of OmpA-Api805. Comparison with the 5xOmpA-Api805 sequence revealed a genetic rearrangement of the insert, with the first and fifth copy of the original 5xOmpA-Api805 insert inserted, whereas copies two, three, and four were deleted (Supplementary Figures S1, S5). Compared to the control, the T7Select-2xOmpA-Api805 phage with two copies of OmpA-Api805 showed no differences in the lysis behavior in a time-kill assay. Interestingly, 4 E. coli culture replicates showed reduced and 2 showed delayed regrowth of potentially phage-resistant populations, while the 10 others regrew similarly to the control (Figure 5E). This was overall an improvement over the single-copy OmpA-Api805 phage.
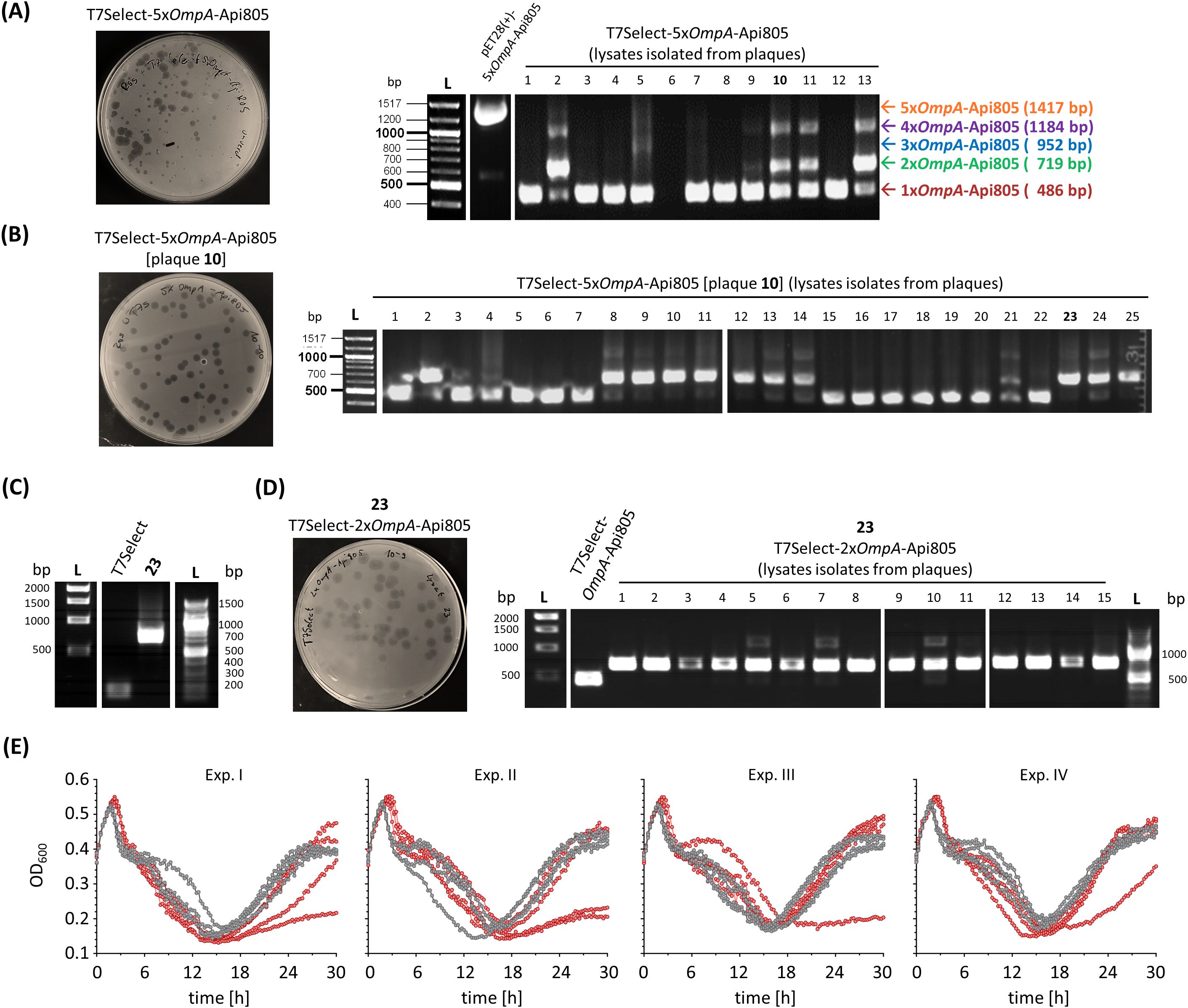
Figure 5. Agar plate after a plaque assay using the generated T7Select-5xOmpA-Api805 phage lysate against E. coli Rosetta and ethidium bromide-stained agarose gels of PCR-amplified DNA fragments from pET28(+)-5xOmpA-Api805 and T7Select-5xOmpA-Api805 phage lysates isolated from 13 different plaques (A). Agar plate after a plaque assay using the T7Select-5xOmpA-Api805 phage lysate isolated from plaque 10 against E. coli Rosetta and ethidium bromide-stained agarose gel of DNA fragments from T7Select-5xOmpA-Api805 [plaque 10] phage lysates isolated from 25 different plaques (B). Ethidium bromide-stained agarose gel of PCR-amplified DNA fragments from T7Select lysate and lysate 23 after an up-scaling propagation, which was confirmed as T7Select-2xOmpA-Api805 phage lysate (C). Agar plate after a plaque assay using the T7Select-2xOmpA-Api805 phage lysate against E. coli Rosetta and ethidium bromide-stained agarose gel of DNA fragments from T7Select-2xOmpA-Api805 phage lysates isolated from 15 different plaques (D). Bacterial growth of E. coli Rosetta cultures infected with T7Select (gray) and T7Select-2xOmpA-Api805 (red) with a MOI of 0.0625 (E). Shown are the OD600 values recorded every 15 min after 10 s shaking for 30 h Experiments were performed four times in quadruplicates. Shown are the curves of the four individual replicates for each phage of each experiment (Exp.) over the entire cultivation.
Cloning of sequences encoding His6-LAPRGSV-Api805 without an additional stop codon, T7 promoter, and ribosome binding site (Figures 1A, D), which should result in direct attachment of the peptide to the capsid, did not yield infectious phages.
4 Discussion
Phages are considered a potential alternative therapeutic strategy to overcome disadvantages of conventional antibiotics. However, their therapeutic applications are limited by significant pharmacological drawbacks. One major limitation is that most phages have a narrow host range targeting only specific bacterial strains (Sulakvelidze et al., 2001; Du et al., 2023). While this is not necessarily a drawback, it hinders their use as broad-spectrum antimicrobial agents. Another challenge is that even phage-susceptible bacteria can rapidly develop resistance, sometimes during the first treatment (Oechslin et al., 2017; Zalewska-Piątek, 2023). Our results consistently confirmed the emergence and proliferation of phage-resistant bacteria. Extending recent research showing a cooperative effect between phages and standard antibiotics or apidaecin-derived PrAMPs (Tagliaferri et al., 2019; Ludwig et al., 2022b), we also observed such dose-dependent synergistic effects for membranolytic CRAMP and melittin in combination with T7Select phages. The lytic activity of either phage or peptide was independent of each other and was not reduced when combined; in some cases, the effects were even enhanced. Regrowth of bacteria resistant or less susceptible to phages or peptides was inhibited when both antibacterial agents were combined. The significantly improved activity of melittin against the T7Select-resistant E. coli strain R 2.3 confirmed an indirect synergistic effect, because the adaptation of the bacterial host to the phage increases the sensitivity to the AMP. Considering previous studies by Qimron et al., Mutalik et al., and Ebbensgaard et al., it is most likely that gene deletions affecting the composition and type of lipopolysaccharide (LPS) structure in the outer membrane trigger both phage resistance and increased susceptibility to AMPs, including melittin (Qimron et al., 2006; Ebbensgaard et al., 2018; Mutalik et al., 2020).
Based on our results, we have developed genetically engineered lytic phages to specifically target phage-susceptible bacteria by inducing CRAMP and melittin expression in the host, which could attack nearby phage-resistant bacteria upon release from the lysed host cell. The partial reduction and inhibition of regrowth by these engineered phages provided significant improvements over the original T7Select phage and phages harboring apidaecin-derived sequences (Ludwig et al., 2022b). Engineering of the phage genome by insertion of sequences encoding CRAMP and melittin was demonstrated and the desired AMP production after phage infection and host lysis could be confirmed for (M)melittin, but not for (M)CRAMP, which does allow, at least for (M)melittin, a direct link between regrowth inhibition and AMP production. Although these effects were not fully reproducible, most likely due to different expression levels of (M)melittin and also (M)CRAMP in the single replicates, they provide a solid base for further development.
The T7Select 415-1b phage display kit and alternative methods have been successfully used to integrate sequences encoding peptides or proteins (Chen et al., 2017; Lemon et al., 2019; Ludwig et al., 2022b; Du et al., 2023; Meile et al., 2023) including antimicrobial agents, such as the anti-biofilm protein dispersin B (Lu and Collins, 2007), biofilm disrupting AMP 1018 (Lemon et al., 2019), the leaderless bacteriocin lacticin Q (Masuda et al., 2021), colicin-like bacteriocins (Du et al., 2023), and apidaecins (Ludwig et al., 2022b). However, phage-mediated expression was only confirmed for superfolder green fluorescent protein (sfGFP), fluorescent protein mCherry, β-galactosidase, and luciferase (Chen et al., 2017; Lemon et al., 2019; Ludwig et al., 2022b; Meile et al., 2023), whereas antimicrobial agents could not be detected after phage lysis, even by sensitive mass spectrometry methods (Lu and Collins, 2007; Lemon et al., 2019; Masuda et al., 2021; Ludwig et al., 2022b), despite improved activities of the engineered phages (Lu and Collins, 2007; Lemon et al., 2019; Masuda et al., 2021; Du et al., 2023). Therefore, based on the current data and to the best of our knowledge, the detection of (M)melittin is the first direct evidence of phage-induced AMP expression.
In contrast to the expression of Api805(G1M), the N-terminal extension of Api805 with the OmpA signal peptide reversed the reduction in bacterial growth rate upon IPTG-induced expression. This was expected since the addition of the OmpA signal peptide should trigger peptide translocation into the periplasm and thus reduce its intracellular concentration and inhibitory effect on the bacterial ribosome. When integrated into the phage genome, this modification had no impact on phage activity. It was also possible to introduce multiple sequences of OmpA-Api805 into the phage genome, although this was genetically instable, ultimately leading to phage genomes carrying two or one encoded OmpA-Api805 sequence. In contrast to the intermediate sequences and the OmpA-Api805 sequences, the sequences of the T7 promoter (TAATACGACTCACTATAGGGAGA) and the ribosome binding site (TTTAACTTTAAGAAGGAGATATACAT) in the 5xOmpA-Api805 insert could not be altered for each copy (Supplementary Figure S1). Thus, there were still homologous sequences in the repeats that could induce deletion of some copies and genetic rearrangement of the insert (Bzymek and Lovett, 2001). However, the incorporation of two copies appears to be genetically stable, which was shown by the isolation of the T7Select-2xOmpA-Api805 phage with partially improved activity compared to the T7Select and T7Select-OmpA-Api805 phage with only one copy of OmpA-Api805. This strategy could be applied to other AMPs, such as CRAMP and melittin, to obtain the desired improvements. Considering the already partially improved activity of the T7Select-(M)CRAMP and T7Select-(M)melittin phages, and the fact that the incorporation of two copies of an AMP into the phage genome appears to be stable and improves the activity, two copies may be sufficient to increase (M)CRAMP and (M)melittin expression and provide more reproducible activity of these phages.
The third approach, the direct attachment of Api805 peptide to the viral capsid via a cleavable linker, did not yield infectious phages. We hypothesize that the attachment of apidaecin reduces the production of the capsid protein below a critical level and that the insolubility, misfolding, or additional high positive charge of the gene10-Api805 construct disrupts the assembly of the T7 capsid shell, preventing the formation of infectious phages. If the high net positive charge of the attached Api805 (+5 under physiological conditions) prevents proper phage formation, this also would most likely apply to other cationic AMPs, including CRAMP and melittin with net charges of +6 and +5, respectively (Ramalingam et al., 1992; Hancock and Diamond, 2000; Islam et al., 2023).
Besides the combinatorial use of bacteriophages and AMPs, the insertion of coding sequences for AMP into the phage genome is an elegant and innovative approach combining these antimicrobial agents. However, further studies are necessary to optimize the reproducibility of the in vitro activity of the AMP-phages presented here and to investigate their effectiveness in an infection model.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
AK: Conceptualization, Data curation, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing, Methodology, Project administration. TL: Writing – original draft, Writing – review & editing, Conceptualization, Data curation, Methodology. DV: Writing – original draft, Writing – review & editing, Conceptualization, Data curation, Methodology.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the European Fund for Regional and Structure Development (EFRE, EU, and Free State of Saxony; Grant Number 100327731), the Federal Ministry of Education and Research (BMBF; Grant Number 16GW0299K), and the University Foundation of Leipzig. The project was co-financed by tax funds on the basis of the budget adopted by the Saxony state parliament.
Acknowledgments
We gratefully acknowledge Ralf Hoffmann for funding support and intensive scientific discussions and thank Daniel Knappe and Gubran Khalil Mohammad for peptide synthesis. We acknowledge support from Leipzig University for Open Access Publishing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frabi.2024.1515874/full#supplementary-material
References
Alqahtani A. (2023). Bacteriophage treatment as an alternative therapy for multidrug-resistant bacteria. Saudi Med. J. 44, 1222–1231. doi: 10.15537/smj.2023.44.12.20230366
Antimicrobial Resistance Collaborators (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet (London England) 399, 629–655. doi: 10.1016/S0140-6736(21)02724-0
Bzymek M., Lovett S. T. (2001). Instability of repetitive DNA sequences: the role of replication in multiple mechanisms. Proc. Natl. Acad. Sci. U S A 98, 8319–8325. doi: 10.1073/pnas.111008398
Casteels P., Romagnolo J., Castle M., Casteels-Josson K., Erdjument-Bromage H., Tempst P. (1994). Biodiversity of apidaecin-type peptide antibiotics. Prospects of manipulating the antibacterial spectrum and combating acquired resistance. J. Biol. Chem. 269, 26107–26115. doi: 10.1016/S0021-9258(18)47165-7
Chen J., Alcaine S. D., Jackson A. A., Rotello V. M., Nugen S. R. (2017). Development of engineered bacteriophages for Escherichia coli detection and high-throughput antibiotic resistance determination. ACS sensors 2, 484–489. doi: 10.1021/acssensors.7b00021
Du J., Meile S., Baggenstos J., Jäggi T., Piffaretti P., Hunold L., et al. (2023). Enhancing bacteriophage therapeutics through in situ production and release of heterologous antimicrobial effectors. Nat. Commun. 14, 4337. doi: 10.1038/s41467-023-39612-0
Ebbensgaard A., Mordhorst H., Aarestrup F. M., Hansen E. B. (2018). The role of outer membrane proteins and lipopolysaccharides for the sensitivity of Escherichia coli to antimicrobial peptides. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02153
Erkut E. (2021). Bacterial signal peptides: structure, optimization, and applications. Eureka 6(1), 12–19. doi: 10.29173/eureka28759
Fidelio G. D., Maggio B., Cumar F. A. (1984). Interaction of myelin basic protein, melittin and bovine serum albumin with gangliosides, sulphatide and neutral glycosphingolipids in mixed monolayers. Chem. Phys. Lipids 35, 231–245. doi: 10.1016/0009-3084(84)90049-5
Freudl R. (2018). Signal peptides for recombinant protein secretion in bacterial expression systems. Microb. Cell Fact 17, 52. doi: 10.1186/s12934-018-0901-3
Gallo R. L., Kim K. J., Bernfield M., Kozak C. A., Zanetti M., Merluzzi L., et al. (1997). Identification of CRAMP, a cathelin-related antimicrobial peptide expressed in the embryonic and adult mouse. J. Biol. Chem. 272, 13088–13093. doi: 10.1074/jbc.272.20.13088
García-Contreras R., Martínez-Vázquez M., González-Pedrajo B., Castillo-Juárez I. (2022). Editorial: alternatives to combat bacterial infections. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.909866
Hancock R. E., Diamond G. (2000). The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 8, 402–410. doi: 10.1016/s0966-842x(00)01823-0
Huan Y., Kong Q., Mou H., Yi H. (2020). Antimicrobial peptides: classification, design, application and research progress in multiple fields. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.582779
Islam M. M., Asif F., Zaman S. U., Arnab M. K. H., Rahman M. M., Hasan M. (2023). Effect of charge on the antimicrobial activity of alpha-helical amphibian antimicrobial peptide. Curr. Res. microbial Sci. 4, 100182. doi: 10.1016/j.crmicr.2023.100182
Krizsan A., Knappe D., Hoffmann R. (2015). Influence of the yjiL-mdtM Gene Cluster on the Antibacterial Activity of Proline-Rich Antimicrobial Peptides Overcoming Escherichia coli Resistance Induced by the Missing SbmA Transporter System. Antimicrobial Agents chemotherapy 59, 5992–5998. doi: 10.1128/AAC.01307-15
Krizsan A., Volke D., Weinert S., Sträter N., Knappe D., Hoffmann R. (2014). Insect-derived proline-rich antimicrobial peptides kill bacteria by inhibiting bacterial protein translation at the 70S ribosome. Angewandte Chemie (International Ed. English) 53, 12236–12239. doi: 10.1002/anie.201407145
Kulshrestha M., Tiwari M., Tiwari V. (2024). Bacteriophage therapy against ESKAPE bacterial pathogens: Current status, strategies, challenges, and future scope. Microbial pathogenesis 186, 106467. doi: 10.1016/j.micpath.2023.106467
Kumar M., Sarma D. K., Shubham S., Kumawat M., Verma V., Nina P. B., et al. (2021). Futuristic non-antibiotic therapies to combat antibiotic resistance: A review. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.609459
Laanto E. (2024). Overcoming bacteriophage resistance in phage therapy. Methods Mol. Biol. (Clifton N.J.) 2738, 401–410. doi: 10.1007/978-1-0716-3549-0_23
Lemon D. J., Kay M. K., Titus J. K., Ford A. A., Chen W., Hamlin N. J., et al. (2019). Construction of a genetically modified T7Select phage system to express the antimicrobial peptide 1018. J. Microbiol. (Seoul Korea) 57, 532–538. doi: 10.1007/s12275-019-8686-6
Li W. F., Ma G. X., Zhou X. X. (2006). Apidaecin-type peptides: biodiversity, structure-function relationships and mode of action. Peptides 27, 2350–2359. doi: 10.1016/j.peptides.2006.03.016
Lu T. K., Collins J. J. (2007). Dispersing biofilms with engineered enzymatic bacteriophage. Proc. Natl. Acad. Sci. U S A 104, 11197–11202. doi: 10.1073/pnas.0704624104
Ludwig T., Hoffmann R., Krizsan A. (2022b). Construction and characterization of T7 bacteriophages Harboring Apidaecin-derived sequences. Curr. Issues Mol. Biol. 44, 2554–2568. doi: 10.3390/cimb44060174
Ludwig T., Krizsan A., Mohammed G. K., Hoffmann R. (2022a). Antimicrobial activity and 70S ribosome binding of apidaecin-derived api805 with increased bacterial uptake rate. Antibiotics (Basel Switzerland) 11, 430. doi: 10.3390/antibiotics11040430
MacNair C. R., Rutherford S. T., Tan M. W. (2024). Alternative therapeutic strategies to treat antibiotic-resistant pathogens. Nat. Rev. Microbiol. 22, 262–275. doi: 10.1038/s41579-023-00993-0
Masuda Y., Kawabata S., Uedoi T., Honjoh K. I., Miyamoto T. (2021). Construction of leaderless-bacteriocin-producing bacteriophage targeting E. coli and neighboring gram-positive pathogens. Microbiol. Spectr. 9, e0014121. doi: 10.1128/Spectrum.00141-21
Meile S., Du J., Staubli S., Grossmann S., Koliwer-Brandl H., Piffaretti P., et al. (2023). Engineered reporter phages for detection of Escherichia coli, Enterococcus, and Klebsiella in urine. Nat. Commun. 14, 4336. doi: 10.1038/s41467-023-39863-x
Memariani H., Memariani M., Moravvej H., Shahidi-Dadras M. (2020). Melittin: a venom-derived peptide with promising anti-viral properties. Eur. J. Clin. Microbiol. Infect. Dis. 39, 5–17. doi: 10.1007/s10096-019-03674-0
Mutalik V. K., Adler B. A., Rishi H. S., Piya D., Zhong C., Koskella B., et al. (2020). High-throughput mapping of the phage resistance landscape in E. coli. PLoS Biol. 18, e3000877. doi: 10.1371/journal.pbio.3000877
Nikaido H. (2009). Multidrug resistance in bacteria. Annu. Rev. Biochem. 78, 119–146. doi: 10.1146/annurev.biochem.78.082907.145923
O’Neill J. (2014). Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations/the Review on Antimicrobial Resistance Chaired by Jum O’Neill. Attribution 4.0 International (CC BY 4.0).
Oechslin F., Piccardi P., Mancini S., Gabard J., Moreillon P., Entenza J. M., et al. (2017). Synergistic interaction between phage therapy and antibiotics clears pseudomonas Aeruginosa infection in endocarditis and reduces virulence. J. Infect. Dis. 215, 703–712. doi: 10.1093/infdis/jiw632
Qimron U., Marintcheva B., Tabor S., Richardson C. C. (2006). Genomewide screens for Escherichia coli genes affecting growth of T7 bacteriophage. Proc. Natl. Acad. Sci. U S A 103, 19039–19044. doi: 10.1073/pnas.0609428103
Ramalingam K., Aimoto S., Bello J. (1992). Conformational studies of anionic melittin analogues: effect of peptide concentration, pH, ionic strength, and temperature–models for protein folding and halophilic proteins. Biopolymers 32, 981–992. doi: 10.1002/bip.360320809
Sulakvelidze A., Alavidze Z., Morris J. G. (2001). Bacteriophage therapy. Antimicrobial Agents chemotherapy 45, 649–659. doi: 10.1128/AAC.45.3.649-659.2001
Tagliaferri T. L., Jansen M., Horz H. P. (2019). Fighting pathogenic bacteria on two fronts: phages and antibiotics as combined strategy. Front. Cell. infection Microbiol. 9. doi: 10.3389/fcimb.2019.00022
Thie H., Schirrmann T., Paschke M., Dübel S., Hust M. (2008). SRP and Sec pathway leader peptides for antibody phage display and antibody fragment production in E. coli. New Biotechnol. 25, 49–54. doi: 10.1016/j.nbt.2008.01.001
van Duin D., Paterson D. L. (2016). Multidrug-resistant bacteria in the community: trends and lessons learned. Infect. Dis. Clinics North America 30, 377–390. doi: 10.1016/j.idc.2016.02.004
Keywords: bacteriophages, antimicrobial peptides (AMP), melittin, CRAMP, apidaecins, synergy, phage engineering, OmpA signal peptide
Citation: Ludwig T, Volke D and Krizsan A (2025) Analysis of engineered T7 bacteriophages containing genetic sequences encoding antimicrobial peptides. Front. Antibiot. 3:1515874. doi: 10.3389/frabi.2024.1515874
Received: 23 October 2024; Accepted: 23 December 2024;
Published: 15 January 2025.
Edited by:
Venkatesan Jayaprakash, Birla Institute of Technology, Mesra, IndiaReviewed by:
Amaç Fatih Tuyun, Istanbul University, TürkiyeGourav Rakshit, Birla Institute of Technology, Mesra, India
Copyright © 2025 Ludwig, Volke and Krizsan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andor Krizsan, YW5kb3Iua3JpenNhbkB1bmktbGVpcHppZy5kZQ==
†These authors have contributed equally to this work
 Tobias Ludwig
Tobias Ludwig Daniela Volke
Daniela Volke Andor Krizsan
Andor Krizsan