- 1Department of Agriculture, Food, and Resource Sciences, University of Maryland Eastern Shore, Princess Anne, MD, United States
- 2Department of Computer Science and Informatics, Uva Wellassa University, Badulla, Sri Lanka
Salmonella is a foodborne pathogenic bacterium that causes salmonellosis worldwide. Also, Salmonella is considered a serious problem for food safety and public health. Several antimicrobial classes including aminoglycosides, tetracyclines, phenols, and β-Lactams are used to treat Salmonella infections. Antibiotics have been prescribed for decades to treat infections caused by bacteria in human and animal healthcare. However, intensive use of antibiotics resulted in antibiotic resistance (AR) among several foodborne bacteria including Salmonella. Furthermore, multi-drug resistance (MDR) of Salmonella has increased dramatically. In addition to MDR Salmonella, extensively drug resistant (XDR) as well as pan drug resistant (PDR) Salmonella were reported globally. Therefore, increasing AR is becoming a serious universal public health crisis. Salmonella developed many mechanisms to ensure its survival against antimicrobials. The most prominent defense mechanisms against these antibiotics include enzymatic inactivation, expelling drugs from the cell through efflux pumps, altering the structure of drugs, and changing or protecting the targets of drugs. Additionally, the formation of biofilms and plasmid-mediated AR by Salmonella, enhancing its resistance to various antibiotics, making it a challenging pathogen in both healthcare and food industry settings. This review focuses exclusively on providing a detailed overview of the mechanisms of AR in Salmonella.
1 Introduction
Salmonella is a member of the Enterobacteriaceae family, a facultative anaerobic, Gram-negative, rod-shaped bacterium that causes the foodborne disease salmonellosis (Andersen et al., 2015; Punchihewage-Don et al., 2022, 2024). The Salmonella genus primarily comprises two species: Salmonella bongori and Salmonella enterica. S. enterica is categorized into six subspecies including Salmonella enterica subsp. arizonae, diarizonae, enterica, indica, houtenae, and salamae. These subspecies are further divided into serogroups based on the immunoreactivity of cell surface structures, the O (the outermost layer of the bacteria’s surface) and H (thin, thread-like structures found in the flagella) antigens (CDC, 2011, 2022; Ryan et al., 2017; Ferrari et al., 2019; Popa and Papa, 2021). Based on the immunoreactivity of cell surface structures of Salmonella, over 2,500 different serovars have been identified to date (WHO, 2018).
Broadly, Salmonella can be further categorized into typhoidal and non-typhoidal types. Typhoidal Salmonella, which includes Salmonella enterica serovar Typhi, Sendai, and Paratyphi A, B, C, are host-adapted to humans and cause systemic and sometimes life-threatening infections such as typhoid and paratyphoid fever, depending on the serovar that invades human cells (Gal-Mor et al., 2014; Smith et al., 2016; Punchihewage-Don et al., 2021; Schultz et al., 2021; Plumb et al., 2023). In contrast, non-typhoidal Salmonella, such as Salmonella enterica serovar Enteritidis and Typhimurium, typically cause gastroenteritis and can infect a wide range of hosts. Some serovars are host-specific (e.g., S. Dublin in cattle and S. Choleraesuis in pigs), while many serovars can infect a variety of hosts worldwide, causing disease in both humans and animals (WHO, 2018; CDC, 2022). Non-typhoidal Salmonella directly causes mild, severe, or life-threatening foodborne poisoning in humans (WHO, 2018). Notably, Salmonella infections are transmitted through a wide range of contaminated products such as raw/uncooked or undercooked meat and poultry or poultry products, fresh fruits and vegetables as well as unpasteurized milk and other dairy products (FDA, 2020; Punchihewage-Don et al., 2021).
The rapid development in antibiotic resistance (AR) among pathogenic Salmonella strains in recent years has had a significant impact (Punchihewage-Don et al., 2023). This is because antibiotic-resistant Salmonella is directly linked to a rise in human deaths, extended hospital stays, and increased treatment costs due to the failure of therapies (Jajere, 2019). According to the Centers for Disease Control and Prevention (CDC), each year in the United States, Salmonella is linked to roughly 1.35 million cases of infection, leads to 26,500 hospital admissions, and results in 420 deaths (CDC, 2023). Furthermore, among these cases, the CDC estimates that about 212,500 are due to drug-resistant non-typhoidal Salmonella each year, leading to roughly 70 deaths. Additionally, these infections incur a significant financial burden, amounting to an estimated $400 million in direct medical costs annually (CDC, 2019, 2023). Diarrhea, fever, and stomach cramps are the common symptoms of salmonellosis. Antibiotics are used as a treatment in severe cases of salmonellosis (CDC, 2023). A broad spectrum of antibiotics such as β-lactams, aminoglycosides, tetracyclines, quinolones, cephalosporins, and trimethoprim-sulfamethoxazole are used to combat Salmonella. The improper and overuse of antibiotics contributes to the development of AR in pathogenic bacteria (Akinyemi and Ajoseh, 2017; Millan, 2018). Moreover, the reason for the emergence of AR is that bacteria change their response to these antibiotics (WHO, 2023).
Antibiotics help to prevent and fight against infections caused by bacteria such as salmonellosis, pneumonia, tuberculosis, gonorrhea, etc. (WHO, 2023). Overuse of antibiotics has resulted in the development of pathogenic bacteria that are resistant to antibiotics, including multi-drug resistant (MDR) strains. The reasons for AR for not only the overuse and misuse of antibiotics. Several other reasons can confer AR, such as poor infection prevention and control. Individuals, policymakers, health professionals, the healthcare industry, and the agricultural sector are responsible for minimizing the influence and limiting the spread of AR (WHO, 2023). Some strains of Salmonella have developed resistance to several classes of antibiotics such as aminoglycosides, β-lactam antibiotics, chloramphenicol, quinolones, tetracyclines, sulphonamides, and trimethoprim (Urban-Chmiel et al., 2022). Table 1 describes the summary of the Salmonella AR mechanisms.
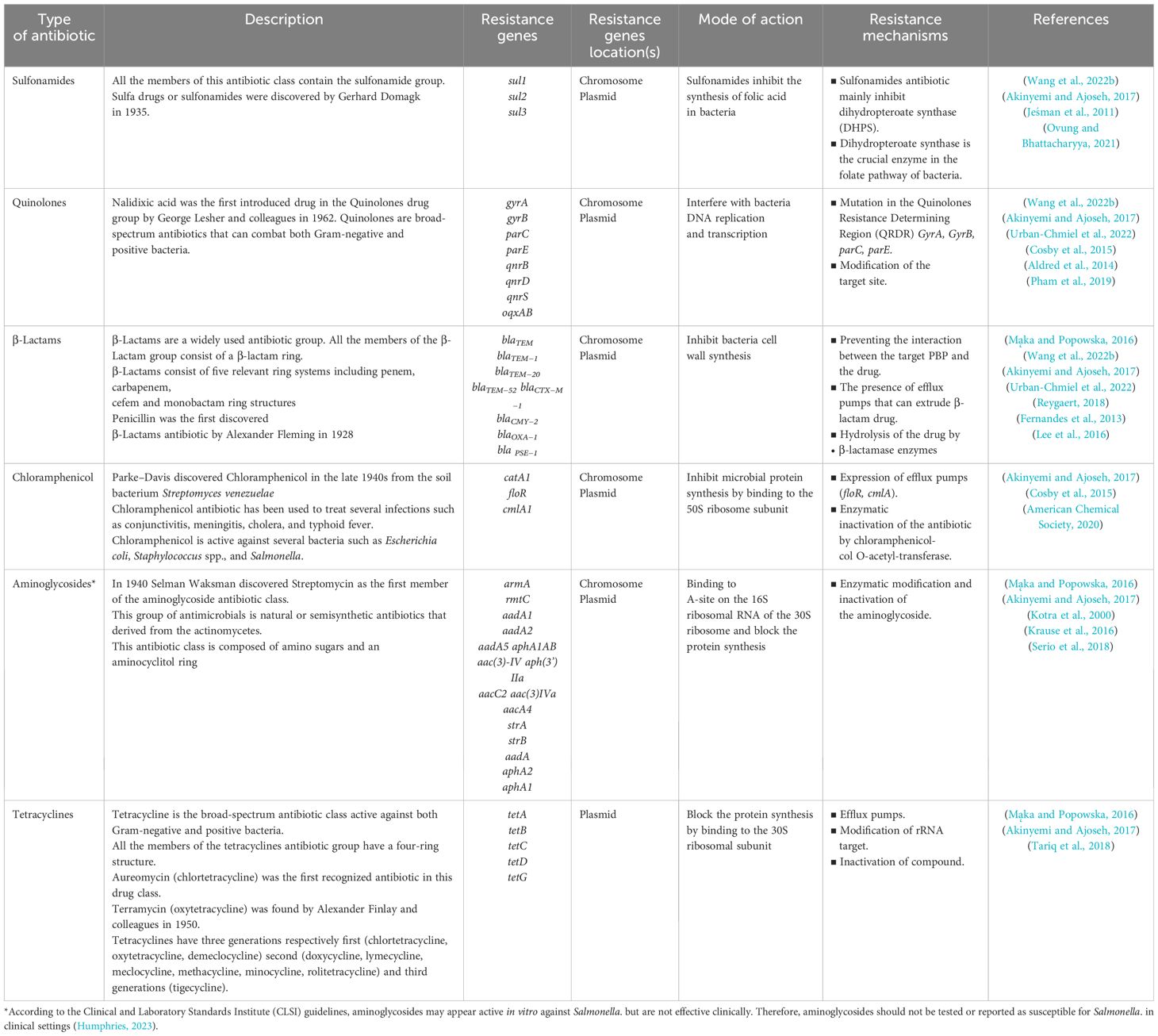
Table 1. Brief categorization of clinically important antibiotics, resistance genes, resistance mechanisms and mode of action of Salmonella.
There are two major molecular mechanisms involved in AR: intrinsic resistance and acquired resistance. Intrinsic resistance, also known as natural or innate resistance, is a natural property of certain bacterial species that makes them resistant to specific antimicrobial agents. This type of resistance is genetically predetermined and persistent, as it is an inherent feature of the organism’s genome (Tanwar et al., 2014; Abebe et al., 2016; Reygaert, 2018). Intrinsic resistance is a universal trait within a bacterial species, independent of previous antibiotic exposure, not related to horizontal gene transfer, and controlled by chromosomal genes (Cox and Wright, 2013). Mechanisms of intrinsic resistance include producing enzymes that neutralize antimicrobial compounds, structural barriers that prevent compounds from reaching target sites, reduced permeability of the bacterial outer membrane, natural activity of efflux pumps, and the lack of appropriate target sites for antibiotics. Bacteria with intrinsic resistance often employ these mechanisms to withstand the effects of antimicrobial agents (Cox and Wright, 2013; Olivares Pacheco et al., 2013; Abebe et al., 2016; Reygaert, 2018).
In contrast, acquired resistance refers to the ability of bacteria to become resistant to antibiotics through genetic changes. This type of resistance develops in previously susceptible bacteria and can occur through genetic mutations or acquisition of resistance genes from other resistant bacteria (Reygaert, 2018; Mancuso et al., 2021; Chin et al., 2023; Harris et al., 2023). Mechanisms of acquired resistance include modification of antibiotic target sites, production of enzymes that inactivate antibiotics, and enhanced efflux of antibiotics from bacterial cells. Acquired resistance can spread through horizontal gene transfer involving processes like transformation, transposition, and conjugation (Culyba et al., 2015; Abebe et al., 2016; Reygaert, 2018; Mancuso et al., 2021; Harris et al., 2023). Both single-drug resistant and MDR pathogenic microorganisms developed resistance mechanisms as their surveillance strategy (Andersen et al., 2015).
Notably, over the years, most Salmonella have evolved resistance mechanisms to numerous antibiotics (Akinyemi and Ajoseh, 2017; CDC, 2019). As a result, AR in Salmonella is a rising problem in the food industry as well as the health sector (Mąka and Popowska, 2016; Punchihewage-Don et al., 2022, 2024). AR has been identified in both typhoidal and non-typhoidal Salmonella serovars (Baugh, 2014; CDC, 2019; Punchihewage-Don et al., 2024). Moreover, MDR Salmonella increased its prevalence by combating clinically essential antibiotics such as fluoroquinolones and third-generation cephalosporins (CDC, 2019; Jajere, 2019; Punchihewage-Don et al., 2022). Several AR mechanisms are encoded in the genome of Salmonella such as active drug efflux pumps, decreased membrane permeability, enzymatic inactivation of the antibiotics, target site modification, plasmid-mediated resistance, and biofilm formation. These mechanisms allow Salmonella cells to expel antibiotics and reduce the antibiotic concentration within the cytoplasm. In addition, these mechanisms are capable of conferring MDR (Shrestha et al., 2015).
Despite extensive knowledge of AR mechanisms in bacteria, a significant gap exists in understanding emerging resistance patterns and the role of novel efflux pump families like PACE and AbgT in Salmonella. This review addresses this gap by analyzing various AR mechanisms, including these new efflux pumps. By synthesizing recent findings, we aim to provide a detailed overview of the AR mechanisms exclusively for Salmonella.
2 Drug efflux
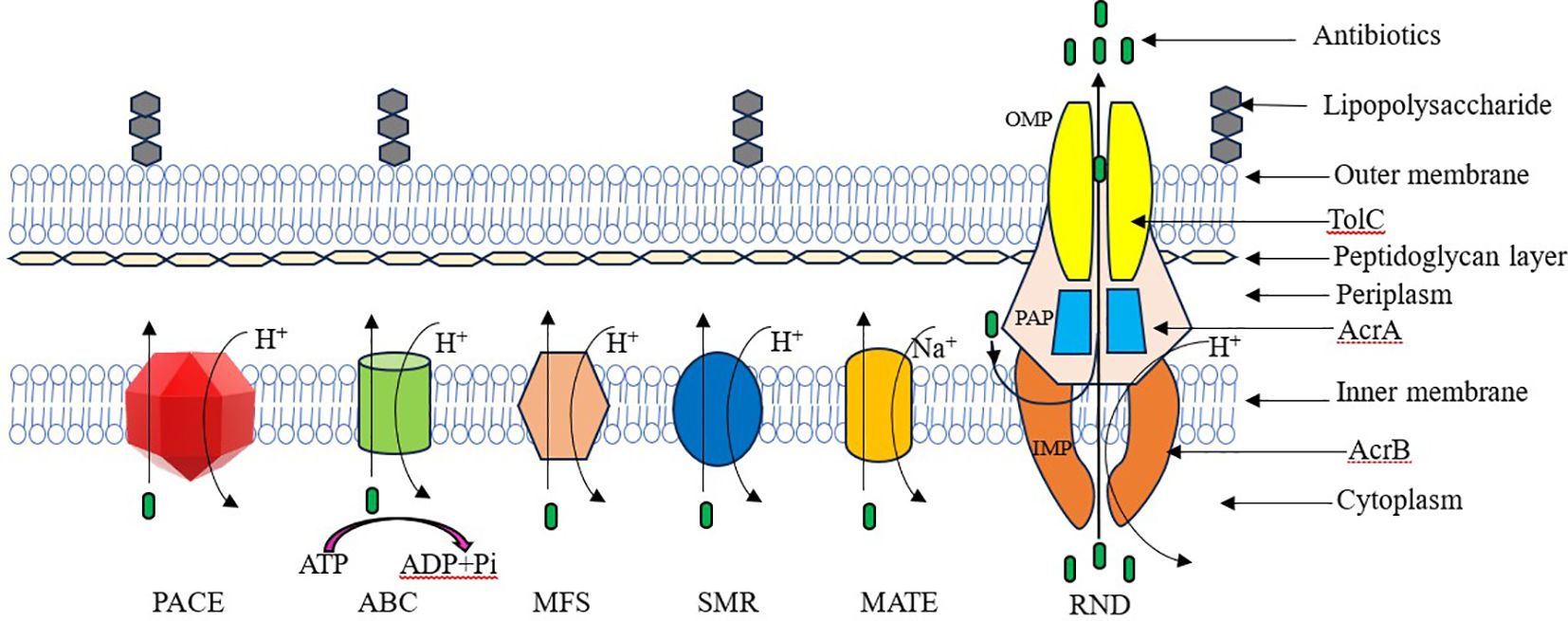
Antibiotic efflux is a widely used AR mechanism among bacteria. This mechanism allows bacteria to extrude antibiotics from their intracellular environment to the extracellular environment (Noto, 2022). Salmonella utilizes different types of efflux pumps. Those drug efflux systems are divided into five main families depending on their energy source and structure (Figure 1). They are ATP-binding cassette (ABC) family, small multi-drug resistance (SMR) family, multi-drug and toxic compound extrusion (MATE) family, major facilitator superfamily (MFS), and resistance nodulation cell division (RND) family (Reygaert, 2018; Nishino et al., 2021; Gaurav et al., 2023; Duffey et al., 2024).
Efflux pumps can be categorized by their energy sources into primary and secondary active transporters. Primary active transporters, such as members of the ABC family, obtain energy through the hydrolysis of ATP. In contrast, secondary active transporters, including members of the MATE, MFS, RND, and SMR families, utilize energy derived from hydrogen ions (H+) or the electrochemical gradient of sodium ions (Na+), generated by the proton motive force (Sharma et al., 2019).
Salmonella encodes at least eleven MDR pumps from each family except the SMR family (Bogomolnaya et al., 2013). S. Typhimurium harbor functional drug efflux pump systems that belong to four main efflux pump system families. These efflux pump systems belong to the following families: the MFS family (including EmrAB and MdfA), the ABC family (MacAB), the MATE family (MdtK), and the RND family (AcrAB, AcrD, AcrEF, MdtABC, MdsAB) (Andersen et al., 2015). According to the latest studies, two new bacterial energy efflux pump families have been discovered. These novel drug efflux systems are the proteobacterial antimicrobial compound efflux (PACE) superfamily and the p-aminobenzoyl-glutamate transporter (AbgT) family (Hassan et al., 2018).
2.1 Resistance nodulation cell division family
Resistance nodulation cell division (RND) family effluxes are more effective for the extrusion of a wide range of antibiotics such as chloramphenicol, novobiocin, tetracyclines, some β-lactams, fusidic acid and fluoroquinolones, detergents, bile salts, metals and biocides (Alenazy, 2022b). The RND family is crucial to MDR in Salmonella (Shrestha et al., 2015). The three major proteins are involved in forming the tripartite efflux system in RND family efflux pumps. They are an inner-membrane protein (IMP), an outer-membrane protein (OMP), and a periplasmic adapter protein (PAP) (Alenazy, 2022b). There are five RND multi-drug efflux systems belonging to Salmonella. They are AcrAB, AcrD, AcrEF, MdtABC, and MdsABC efflux pumps (Yamasaki et al., 2011). The AcrAB-TolC efflux system is the common and extensively studied drug efflux pump system in Salmonella. The AcrAB drug efflux pump system of S. Typhimurium is located on the chromosome and it is encoded by the acrAB genes. The AcrAB-TolC system comprises a tripartite efflux pump in Salmonella. Furthermore, the AcrAB-TolC efflux pump forms three protein subunits. They are AcrB (inner membrane transporter protein), AcrA (periplasmic adaptor protein), and TolC (the outer membrane porin protein) (Baugh, 2014).
The tripartite AcrAB-TolC drug efflux pump system of S. Typhimurium vigorously excludes a wide range of antibiotics such as chloramphenicol, tetracycline, ciprofloxacin, acriflavine, fusidic acid, novobiocin, erythromycin, rifampin, and β-lactam. Fernandes et al. (2013) elaborated on the significance of the Salmonella AcrAB-TolC system in the pathogenesis of the bacterium (Fernandes et al., 2013). The AcrD efflux pump of Salmonella actively extrudes kanamycin, tobramycin, gentamicin, and amikacin. The AcrEF efflux system in Salmonella is homologous and functionally similar to the AcrAB system, and together with TolC, forms the AcrEF-TolC tripartite multi-drug efflux system that complements the AcrAB-TolC system’s function. In Salmonella, the AcrEF-TolC tripartite multi-drug efflux system is composed of three components. They are AcrE (membrane fusion protein), AcrF (inner membrane transporter), and TolC (outer membrane channel protein). The member of AcrEF efflux in family RND resistance to ciprofloxacin, tetracycline, nalidixic acid, chloramphenicol, and triclosan. The MdsABC efflux pump is found only in Salmonella (Shrestha et al., 2015).
Inner membrane RND-type transporter (MdsB), periplasmic membrane-fusion protein (MdsA), and outer membrane protein (MdsC) are the major three components of the Salmonella MdsABC tripartite efflux pump (Song et al., 2015). Salmonella MdsABC efflux pump resistant to novobiocin, deoxycholate, some β-lactams, copper and zinc (Blair et al., 2014). MdtABC drug efflux systems of Salmonella have two different transporters called MdtB and MdtC. Both of them are co-transcribed along with MdtA (a membrane fusion protein) in the same operon. In Salmonella, MdtABC efflux pumps resistance to antibiotics such as β-lactams, novobiocin, and bile salts. Furthermore, the MdtABC efflux pump detoxified the cell from copper, zinc, and tungstate (Shrestha et al., 2015).
2.2 Small multi-drug resistance family
The small multi-drug resistance (SMR) family is the smallest efflux transporter family among the other four families. This family of efflux transporters is restricted to prokaryotic cells (Higgins, 2007). The SMR family obtains energy from the proton-motive force (H+) (Reygaert, 2018). SMR transporters confer resistance to a diverse number of quaternary ammonium compounds and lipophilic cations like benzalkonium, cetyltrimethylammonium bromide (CTAB), cetylpyridinium chloride (CTPC), methyl viologen, and tetraphenylphosphonium (TPP). In addition, SMR efflux pumps confer resistance to antibiotics such as β-lactams, cephalosporins, dihydrofolate inhibitors, and aminoglycosides (Shrestha et al., 2015).
2.3 ATP-binding cassette family
The ATP-binding cassette (ABC) family of drug efflux pumps derives its energy from ATP hydrolysis to remove drugs from the intracellular environment (Kabra et al., 2019). The ABC family is known as the primary active transporters because it utilizes ATP as its energy source to extrude substances out of the cell. MacAB-TolC is one of the known drug efflux pump systems in Salmonella. MacAB-TolC can form tripartite complex. MacB functions as the inner membrane pump protein (efflux pump). Periplasmic protein and the outer membrane channel of the Salmonella MacAB-TolC tripartite efflux pump are respectively MacA and TolC. In Salmonella, MacAB-TolC Efflux system resistance to the macrolides such as erythromycin, and azithromycin (Baugh, 2014). In Salmonella, the PhoPQ regulatory system, which is a two-component system, controls the expression of the MacAB pump (Nishino et al., 2006, 2009). Specifically, PhoP, a component of the PhoPQ system, binds to the macAB ABC transporter and represses its activity (Nishino et al., 2006). This regulatory mechanism is crucial in controlling the virulence of Salmonella (Kato and Groisman, 2008). Furthermore, the functions of the PhoPQ system in Salmonella are multifaceted, encompassing various key physiological processes. These include the regulation of Mg2+ homeostasis, providing resistance against antimicrobial peptides, and enabling growth in acidic environments with a low pH level (Groisman et al., 2021). Moreover, the MacAB drug efflux system, a member of the ABC family in Salmonella, serves as a critical defense mechanism, safeguarding the bacteria from oxidative stress. This efflux pump is notably activated in response to exposure to hydrogen peroxide (H2O2). Such induction facilitates the survival of Salmonella under conditions where peroxide is present, highlighting the pump’s crucial role in the bacterial defense mechanism against oxidative damage (Bogomolnaya et al., 2013).
2.4 Major facilitator superfamily
The major facilitator superfamily (MFS) is generally introduced as the largest family of secondary active solute transporters (Kumar et al., 2013). MFS transporters have broad substrate specificity, such as ions, amino acids, carbohydrates, lipids, nucleosides, and other small molecules (Madej, 2014). The EmrAB efflux pump, a member of the MFS family, significantly contributes to the internal drug resistance of Salmonella. EmrAB and MdfA are the most described efflux pumps in Salmonella. The EmrAB efflux pump comprises the membrane fusion protein EmrA and functions as a multi-drug efflux system, expelling substrates such as novobiocin, nalidixic acid, and sodium deoxycholate from the bacterial cell. The MdfA efflux pump in S. Typhimurium acts as a single cytoplasmic efflux protein, conferring resistance to norfloxacin, chloramphenicol, tetracycline, and doxorubicin (Baugh, 2014; Andersen et al., 2015).
2.5 Multi-drug and toxic compound extrusion
The multi-drug and toxic compound extrusion (MATE) efflux family acquires energy by using the Na+ gradient (Reygaert, 2018). The MdtK efflux pump is a member of the MATE-type drug efflux pump in Salmonella. MdtK efflux pump is substrate-specific and excludes antibiotics such as fluoroquinolones, cation drugs, and aminoglycosides from the cells (Alenazy, 2022a). MdtK efflux pump of Salmonella confers resistance to antibiotics such as norfloxacin, doxorubicin, and acriflavine (Shrestha et al., 2015).
2.6 Proteobacterial antimicrobial compound efflux superfamily
Recently identified, the proteobacterial antimicrobial compound efflux (PACE) superfamily with AceI from Acinetobacter baumannii as the prototype. The PACE bacterial drug efflux transport proteins (AceI) are encoded by genes that are discovered in the genomes of numerous pathogenic bacteria species such as Pseudomonas, Klebsiella, Enterobacter, Salmonella, and Burkholderia species (Huang et al., 2022). The PACE multi-drug transporters are secondary active transporters that utilize the proton motive force to transport biocides such as benzalkonium, diqualinium, acriflavin, proflavin, and chlorhexidine (Hassan et al., 2018; Zhao et al., 2022; Gaurav et al., 2023; Ahmad et al., 2024). PACE family members are particularly common among proteobacteria. There are no findings so far that PACE efflux systems exist in archaea and eukaryotes (Zhao et al., 2022).
2.7 p-Aminobenzoyl-glutamate transporter family
The AbgT family of transporters is crucial for bacterial folate synthesis, essential for growth, and acts as a drug efflux pump, facilitating resistance to various sulfa drugs. The AbgT family uses the proton motive force for transporting biocides (Delmar and Yu, 2016). This family includes two primary members, YdaH and MtrF, who play key roles in these biological processes (Shome et al., 2021). Both efflux systems YdaH and MtrF vigorously mediate the bacterial resistance against sulfonamide antimetabolite drugs. AbgT‐type efflux proteins have been discovered in both Gram-negative bacteria (S. enterica, E. coli, N. gonorrhoeae, A. borkumensis) and Gram-positive bacteria including Staphylococcus aureus and Streptomyces coelicolor. Also, these efflux proteins can be found in eukaryotes including the yeast Saccharomyces arboricola (Delmar and Yu, 2016). However, further research is necessary to understand the prevalence and specific functions of the AbgT transporter family in Salmonella. This knowledge gap highlights the need for more in-depth studies to elucidate how these transporters operate and contribute to the bacterial life cycle and drug resistance mechanisms in Salmonella.
Table 2 provides a comparison of the newly discovered PACE and AbgT efflux pump families with the established efflux pump families (ABC, SMR, MATE, MFS, and RND), highlighting differences in energy utilization, substrate specificity, and structural properties.
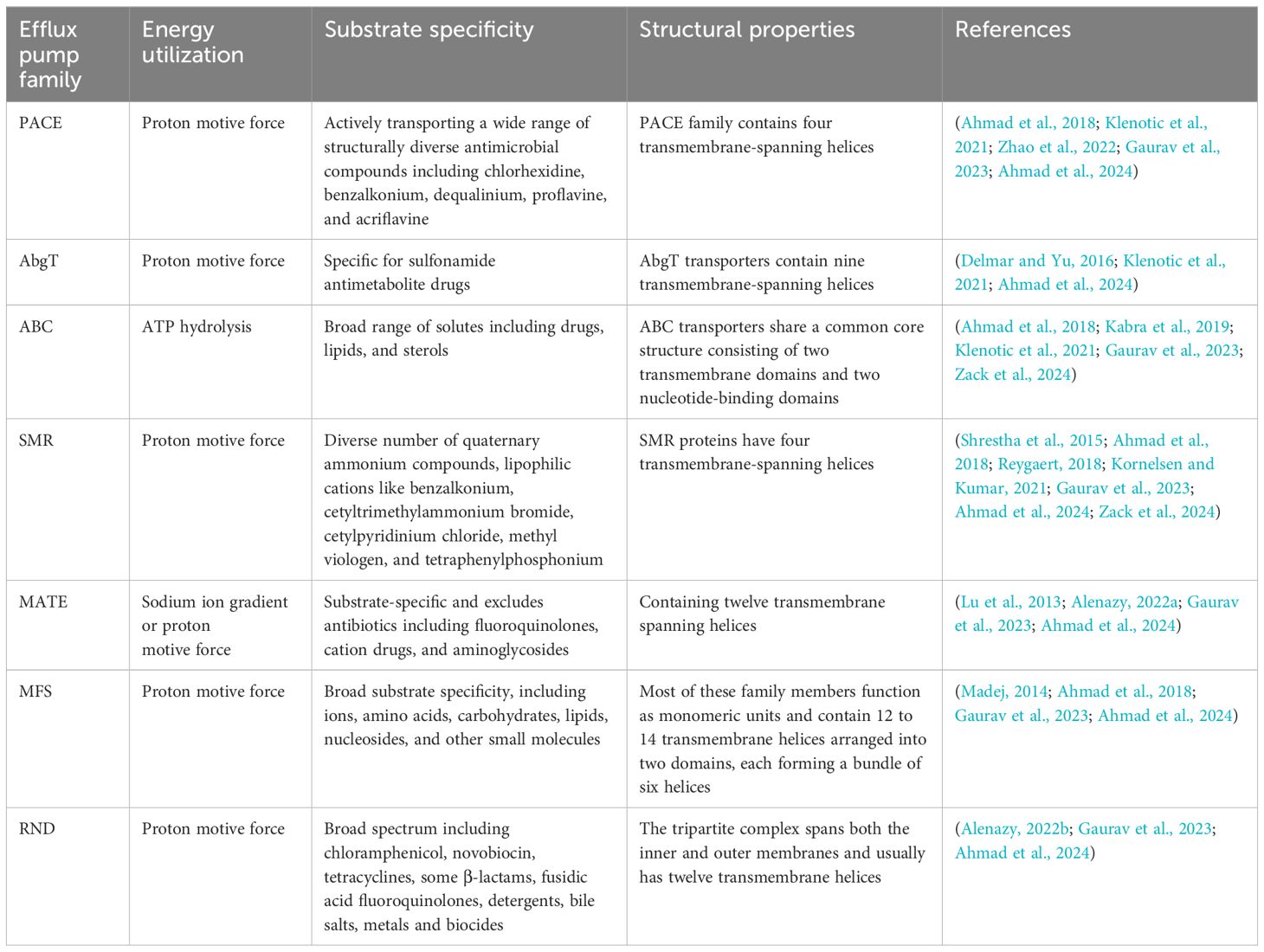
Table 2. Comparison of efflux pump families in Salmonella: energy utilization, substrate specificity, and structural properties.
3 Altered membrane permeability
Reduced permeability is a well-known AR mechanism among Salmonella (Akinyemi and Ajoseh, 2017). There are two mechanisms involved in reducing antibiotic permeability in Gram-negative bacteria including Salmonella. They are alterations of the outer membrane lipid barrier and porin-mediated permeability. Structural and functional arrangements of Salmonella’s lipopolysaccharide (LPS) layer provide a barrier to some molecules including the antibiotics (Sirisena, 2000; Reygaert, 2018). Furthermore, having a hydrophobic lipid bilayer with pore-forming proteins of specific size-exclusion properties leads the external membrane to act as a penetration barrier (Delcour, 2009).
The cell wall of Salmonella is mainly composed of three different layers. They are the outer membrane, the peptidoglycan layer, and the inner membrane (Rycroft, 2000). The external membrane of Salmonella is an asymmetrical, highly complex lipid bilayer. Its inner layer forms the phospholipids and its outer layer forms the lipopolysaccharides. The lipopolysaccharide molecule is amphipathic. It includes both hydrophobic and hydrophilic components on the same molecule. The LPS molecules consist of three regions: a hydrophobic anchor lipid A, a core oligosaccharide with several anionic moieties, and the O-antigen (Rycroft, 2000). Salmonella modifies its own LPS as a mechanism to resist antimicrobial agents such as antimicrobial peptides. PhoPQ and PmrA/B are two pairs of component systems that allow modifications in the LPS of Salmonella. The genes crucial for LPS modification in Salmonella, such as pmrD, pmrC, pmrG, the pmrH-M operon, and pmrE, fall under the regulatory control of the two regulatory component systems PhoPQ and PmrAB (Devi, 2011).
In Gram-negative bacteria, the phosphorylation of lipid A and core sugars contributes to the anionic properties of the cell surface. As a strategy, Gram-negative bacteria like Salmonella are used to reduce the negative charge of the cell exterior surface through the addition of positive charges into lipid A. This process manipulates the decreasing affinity for antimicrobial peptides. The lipid A segment of LPS undergoes three structural modifications, such as the addition of palmitate, the addition of phosphoethanolamine, and the attachment of aminoarabinose (Abdi et al., 2019). For instance, in S. typhimurium, the addition of the amine-containing sugar aminoarabinose to the lipid A phosphate group confers resistance to polymyxin B (Band and Weiss, 2015).
Lipid A acylation is a crucial mechanism by which Salmonella resists antimicrobial peptides. This process enhances the bacterium’s ability to reduce the fluidity of its outer membrane by increasing its hydrophobicity. In Salmonella, palmitate chains are added to lipid A to form a hepta-acylated lipid A, a process facilitated by the outer membrane enzyme PagP, which is regulated by the PhoPQ system. The hepta-acylated structure of lipid A is significant because it inhibits the incorporation of cationic antimicrobial peptides, thereby protecting the bacteria from these antimicrobial agents (Li et al., 2013; Boll et al., 2015; Matamouros and Miller, 2015; Saha et al., 2022). In addition, a study revealed that Salmonella lipopolysaccharide modifications prevent the outer membrane penetration of novobiocin (Nobre et al., 2015).
Porins are protein channels that reside in the outer membrane of Gram-negative bacteria, such as Escherichia coli and Salmonella. These channels are crucial for the passive transport of small molecules, including nutrients and certain antibiotics, across the outer membrane (Galdiero et al., 2012; Choi and Lee, 2019; Ude et al., 2021). Generally, porins permit the passive transport of hydrophilic molecules with a molecular weight of less than 600 Da. In addition, bacterial porins facilitate the entry of nutrients and the excretion of waste and toxic compounds from the bacterial cell environment. The major porins of S. Typhimurium are OmpF, OmpC, PhoE and OmpD (Ipinza et al., 2014). Two major porin-based mechanisms can be identified in Gram-negative bacteria including Salmonella, E. coli, P. aeruginosa, Klebsiella pneumoniae. The two mechanisms involve modifications to the outer membrane porins, encompassing their loss, reduction, or alteration, along with specific mutations that directly lead to changes in porin functionality (Andersen et al., 2015). Members of the family Enterobacteriaceae are well-known examples of reducing the number of porins as their resistance mechanism against antibiotics. Sometimes, bacteria completely stop producing certain porins (Reygaert, 2018). In Salmonella, OmpC and OmpF porins allow β-lactams to enter the bacterial cell and reach their penicillin-binding protein targets. It has been reported that a reduction in either OmpF or OmpC porin amount leads to an increase in the resistance to β-lactams including ampicillin, cefoxitin, and other cephalosporins (Cosby et al., 2015). In addition, the deficiency of OmpC porin in S. Typhimurium can lead to carbapenem resistance (Vergalli et al., 2020). Moreover, a deficiency of OmpF porin expression has been reported in some quinolone-resistant Salmonella strains (Al Gallas and Aissa, 2017).
4 Inactivation of drugs
Drug inactivation is one of the well-studied antibiotic defensive mechanisms. This mechanism enables the destruction or inactivation of the antibiotic compounds by enzymatic hydrolysis. The β-lactamase enzyme is the most typical example of an enzymatic hydrolysis process (Noto, 2022). Enterobacteriaceae family including Salmonella attain resistance to β-lactam by synthesizing β-lactamase or extended-spectrum β-lactamase (Akinyemi and Ajoseh, 2017). The main mechanism of the β-lactamases enzyme is to inactivate the β-lactam antibiotic by hydrolyzing a specific site in the β-lactam ring structure. This process leads to cleaves the β-lactam ring. Furthermore, the open-ring structure of antibiotics is unable to bind their target penicillin-binding protein (PBP) (Reygaert, 2018; Tooke et al., 2019).
Extended-spectrum β-lactamases (ESBLs) are enzymes that confer broad-spectrum AR. Many of these ESBLs are plasmid-mediated enzymes and help bacteria resist a wide range of β-lactams (Bradford, 2001; Naas et al., 2008). ESBLs have been predominantly detected in the Enterobacteriaceae family including Klebsiella, Escherichia coli, Salmonella, Enterobacter, Proteus, Citrobacter, Serratia and Providencia (Teklu et al., 2019). ESBLs can inactivate broad-spectrum cephalosporins, penicillins, and aztreonam. Additionally, β-lactamase inhibitors such as sulbactam, clavulanic acid, and tazobactam are highly effective against ESBLs (Rahman et al., 2018). Salmonella uses enzymatic inactivation mechanisms to resist chloramphenicol and aminoglycosides. Plasmid-mediated resistance genes in the Salmonella genome lead to the production of enzymes such as phosphotransferases and chloramphenicol acetyltransferases, which inactivate chloramphenicol (Guerra et al., 2000). Moreover, Salmonella inactivates aminoglycosides through the production of aminoglycoside-modifying enzymes (AME) such as adenyltransferases, acetyltransferases, and phosphotransferases (Noto, 2022; Thacharodi and Lamont, 2022).
5 Antibiotic target site modification
In this AR mechanism, bacteria can hinder the effectiveness of antibiotics by obstructing their binding sites. Bacteria employ several strategies to change the target sites such as target protection, point mutations in the genes coding the binding site, and replacement or avoidance of the original binding sites (Munita and Arias, 2016; Schaenzer and Wright, 2020). The mutations that occurred in the DNA gyrase and topoisomerase IV are the best examples to discuss under this topic. Salmonella develops resistance to fluoroquinolones and quinolones through mutations in DNA gyrase and topoisomerase IV, resulting in structural changes to these enzymes. Also, it allows for the reduction or complete elimination of the antibiotic’s ability to attach to these target sites (Reygaert, 2018; Herrera-Sánchez et al., 2021). DNA gyrase is identified as the primary target of quinolones in salmonellae (Kongsoi et al., 2015). Both enzymes are large and complex, each consisting of two subunit pairs: GyrA (a 97 kDa protein encoded by the gyrA gene) and GyrB (a 90 kDa protein encoded by the gyrB gene). ParC (75 kDa) and ParE (70 kDa) are named as the corresponding subunits of topoisomerase IV. DNA gyrase has the ability to add negative supercoils into DNA molecules and can also abolish both positive and negative supercoils. Additionally, it can catenate and decatenate closed circular molecules. DNA topoisomerase IV can eliminate both positive and negative supercoils as well but plays a better role in decatenation compared to DNA gyrase. Both enzymes collaborate in the replication, transcription, recombination, and repair of DNA (Jacoby, 2005).
Quinolones target the bacterial enzymes DNA gyrase and topoisomerase IV, which are essential for DNA replication. Quinolones inhibit the enzymes’ ability to re-ligate the DNA strands after inducing double-strand breaks, ultimately leading to the accumulation of DNA breaks and bacterial cell death (Drlica and Zhao, 1997; Fàbrega et al., 2009; Aldred et al., 2014; Kongsoi et al., 2015; Hooper and Jacoby, 2016; Pham et al., 2019). Quinolone resistance initiates with the chromosomal mutations in Quinolone Resistance Determinant Regions (QRDR) of DNA gyrase and topoisomerase IV (Li et al., 2018). Furthermore, the major mechanism of Salmonella resistance to quinolones is mutations that change the target site of the antibiotics with DNA gyrase. In Salmonella, mutations occurred in the specific region (QRDR) of the gyrA gene, i.e., between amino acids 67 and 106 (Souza et al., 2011). In Salmonella, point mutations in the QRDR of the gyrA gene could be adequate to mediate resistance to non-fluorine quinolones such as nalidixic acid and decrease sensitivity to ciprofloxacin. Therefore, additional mutations are needed for a higher level of fluoroquinolone resistance (Souza et al., 2011; Li et al., 2018). Mutation sites within the QRDRs of the gyrA gene are positioned at amino acid positions Serine-83 (Ser-83) and Aspartate-87 (Asp-87). The most common amino acid substitutions in nalidixic acid-resistant strains are Leu, Thr, Phe, Tyr, or Ala at Ser-83, and Gly, Lys, Asn, or Tyr at Asp-87. In cases of high-level resistance, double mutations at both positions 83 and 87 in the gyrA gene have been identified in clinical isolates of S. Typhimurium. These mutations are known to significantly alter the DNA gyrase enzyme, leading to reduced binding of fluoroquinolone antibiotics and thus contributing to high-level resistance. Additionally, Salmonella strains can also exhibit amino acid substitution mutations at other positions, such as Ala-67 (to Pro), Gly-81 (to Ser, Asp, Cys, or His), and Leu-98 (to Val) (Li et al., 2018).
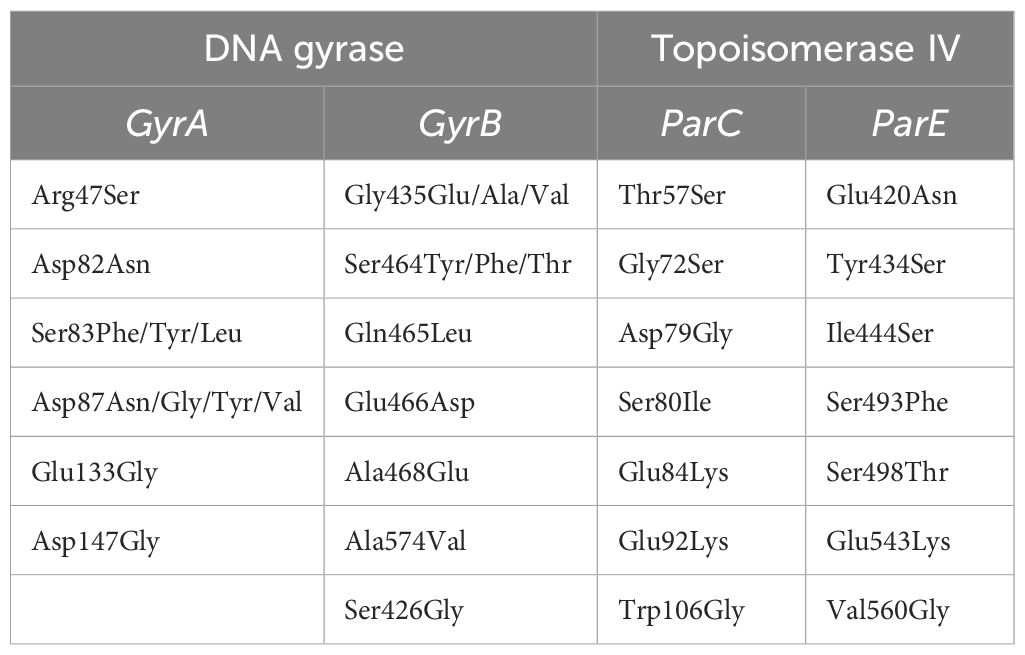
Souza et al. (2011) reported that 105 Salmonella strains (94 epidemic and 11 of poultry origin) were resistant to nalidixic acid out of a total of 123 Salmonella strains (Souza et al., 2011). They aimed to evaluate mutations in the QRDR of the gyrA gene related to resistance to the nalidixic acid and decreased susceptibility to ciprofloxacin using allele-specific PCR and restriction fragment length polymorphism (AS-PCR-RFLP). The study found a high incidence of mutations, particularly at codons that code for Asp-87 and Ser-83, which are associated with quinolone resistance. The research concluded that a need for judicious use of quinolones to treat Salmonella infections due to the potential for resistance development. Diverse mutations have also been discovered in the QRDRs of DNA gyrase (GyrA and GyrB) and topoisomerase IV (ParC and ParE) of typhoidal Salmonella. These mutations are mentioned in Table 3 (Shaheen et al., 2021). A recent study has identified novel mutations within the QRDR of DNA gyrase and topoisomerase IV in Salmonella isolates from Jiangsu Province, China. These findings reveal new insights into the mechanisms of quinolone resistance in Salmonella. This research reported seven novel mutations in the GyrB (S426G), ParC (D79G), and ParE (S498T, E543K, V560G, I444S, Y434S) expanding our understanding of the genetic diversity underlying quinolone resistance (Qian et al., 2020). Research on chromosomal alterations in the QRDRs of DNA gyrase and topoisomerase IV in Salmonella has provided valuable insights into the emergence and dissemination of quinolone resistance. By identifying specific mutation patterns and understanding their impact on resistance, these studies contribute to improved diagnostic methods, treatment approaches, and surveillance efforts. Ultimately, this knowledge supports the development of targeted strategies to combat quinolone-resistant Salmonella.
6 Plasmid-mediated resistance
In this mechanism, bacteria transfer AR genes which are carried on plasmids (Millan, 2018). Plasmids are small, extrachromosomal circular DNA molecules primarily found in bacteria, but they also exist in some eukaryotes. Plasmids replicate independently, and the genes they carry provide genetic advantages to bacteria, such as AR. Furthermore, plasmids facilitate the horizontal transfer of resistance genes between pathogenic bacteria through conjugation (Bennett, 2008; National Human Genome Research Institute, 2014).
Resistance plasmids contain one or more AR genes, and the majority of them have the ability to transfer between bacteria through conjugation (Bennett, 2008). AR plasmids comprise single or multiple AR genes that can develop an MDR phenotype (Nikaido, 2009). Plasmids carry AR genes in bacteria, conferring resistance to various antibiotics. These plasmids, which possessed AR genes, are known as R factors or resistance plasmids. These R factors are categorized into specific groups known as incompatibility groups (Inc groups) (Rycroft, 2000). Plasmid incompatibility refers to the inability of two plasmids to persist simultaneously within the same cell line (Singh et al., 2022). Plasmids from various incompatibility groups have been connected to various AR genes in Salmonella and other bacteria (Carattoli, 2003). Specifically, IncA/C plasmids isolated from Salmonella carry genes that exhibit resistance to various antibiotics, including aminoglycosides, β-lactams, chloramphenicol, sulfisoxazole, tetracyclines, and trimethoprim (McMillan et al., 2019).
Most AR genes in Salmonella are frequently found on plasmids. Salmonella contains multiple large conjugative plasmids carrying AR genes, conferring resistance to antibiotics such as β-lactams, tetracyclines, aminoglycosides, and quinolones (Millan, 2018). The conjugation of plasmids carrying AR mechanisms has contributed to the universal dissemination of AR genes within the Enterobacteriaceae family, including Salmonella. These plasmids have the capacity to confer resistance to various antibiotics, including β-lactamases (CMY, DHA, GES, LAP, NDM, SHV, TEM), extended-spectrum β-lactamases (CTX, VEB), metallo β-lactamases (IMP), carbapenemases (KPC, VIM), quinolone resistance (Qep, Qnr), aminoglycoside resistance (AAC, Arm, RmtB), tetracycline resistance (Tet), sulfonamide resistance (Sul), and colistin resistance (Huddleston, 2014).
The resistance genes like blaSHV, blaTEM, blaCTX, blaCMY, and blaOXA encoded by Salmonella plasmids are responsible for the ESBL plasmid-mediated resistance (Mąka and Popowska, 2016). Salmonella primarily employs an enzymatic inactivation mechanism as its main defense strategy to resist ESBLs. Furthermore, plasmid-mediated resistance has played a significant role in conferring resistance to ESBLs among Salmonella. These plasmids often carry genes like blaCMY-2 and blaCTX-M-3, which are responsible for resistance and can be shared among bacterial organisms, regardless of bacterial species. This has contributed to the widespread occurrence of ESBL resistance (Su and Chiu, 2007).
Quinolones are widely used to treat salmonellosis in both human and veterinary medicine (Pribul et al., 2017). However, their continuous application over the years has resulted in an increase in resistance (Jacoby et al., 2014). Bacteria exhibit three main mechanisms to resist quinolones: protection of the target from the antibiotics through plasmid-mediated genes, modification of antibiotics, and reducing intracytoplasmic quinolone concentration through efflux pumps (Jacoby et al., 2014; Ruiz, 2019; Usman and Hamza, 2022). Notably, the main three defensive mechanisms for Plasmid-Mediated Quinolone Resistance (PMQR) have been revealed since 1988 (Jacoby et al., 2014; Ruiz, 2019). In brief, PMQR involves genes such as qnrA, qnrB, qnrC, qnrD, qnrS, and qnrVC, which produce proteins from the pentapeptide repeat family. These proteins protect DNA gyrase and topoisomerase IV against quinolone inhibition. Typically, qnr genes are found within sul1-type integrons and are often associated with mobilizing or transposable elements within plasmids (Lee et al., 2021). The next defensive mechanism is acetylation of quinolones. The aac(6′)-Ib-cr gene encodes an enzyme known as aminoglycoside acetyltransferase, which has the ability to confer resistance simultaneously against both aminoglycoside antibiotics and quinolones/fluoroquinolone antibiotics (Lee et al., 2021; Gharbi et al., 2023). In addition to aac(6′)-Ib-cr gene, the newly described plasmid-mediated phosphorylase gene (crpP) was responsible for inactivating ciprofloxacin with the reaction of aminoglycoside phosphotransferase. This gene was detected in pUM505 plasmid, isolated from a clinical Pseudomonas aeruginosa isolate (Chávez-Jacobo et al., 2018; Ruiz, 2019; Lee et al., 2021). However, additional research is required to confirm the presence of crpP gene in the Salmonella genome. The third resistance mechanism is the depletion of the intracellular concentration of quinolones via efflux pumps. Plasmid genes can also lead to resistance by promoting the production of drug efflux systems, such as QepAB and OqxAB. These efflux pumps help remove quinolones from the bacterial cell, thereby enhancing quinolone and fluoroquinolone resistance (Jacoby et al., 2014; Lee et al., 2021). These resistance mechanisms collaboratively contribute to the challenge of combating quinolone resistance in bacterial infections.
A study conducted in Brazil aimed to investigate the occurrence of PMQR in Salmonella and its correlation with susceptibility to fluoroquinolones. The study collected a total of 129 samples from diverse sources, including animal-derived food, environmental samples, animals, and humans. Among the isolated samples, S. Typhimurium and S. Enteritidis were identified. Interestingly, these isolates displayed resistance to a range of fluoroquinolones, including enrofloxacin, ciprofloxacin, ofloxacin, levofloxacin, and nalidixic acid. Among the isolates, qnr genes were detected in 15 instances, with 8 carrying qnrS, 6 carrying qnrB, and 1 carrying qnrD. Additionally, the aac(6′)-Ib gene was identified in 23 isolates. Furthermore, the integron gene was identified in 67 of the isolates (Pribul et al., 2017).
A recent study examined the potential presence of PMQR genes within Salmonella enterica (Lee et al., 2021). The study involved collecting Salmonella enterica samples from patients diagnosed with salmonellosis between 2016 to 2019 in South Korea. Among the 208 clinical isolates of Salmonella obtained from humans, thirty-four strains exhibited reduced susceptibility to fluoroquinolones. Within this subset, 22 Salmonella strains were found to carry single PMQR genes, including qnrA, qnrB, or qnrS, while three Salmonella strains were observed to harbor two PMQR genes, specifically qnrS and aac(6’)-Ib-cr, or qnrA and qnrB (Lee et al., 2021).
7 Creation of biofilm barriers
Bacterial biofilms can be described as clusters of bacteria that stick to a surface and are enclosed by a self-produced matrix (Vestby et al., 2020). Salmonella biofilm extracellular matrix is mainly composed of cellulose, biofilm-associated protein, O-antigen capsule, curli protein (amyloid fimbriae), and extracellular DNA (Daxin, 2016). In Salmonella, curli proteins are thin hair-like filaments encoded by csgBAC and csgDEFG operons. The primary roles of curli proteins include facilitating surface adhesion, promoting cell aggregation, and formation of biofilm. Additionally, curli proteins are involved in mediating the adhesion and invasion of host cells, and they are also effective in activating the host’s inflammatory response (Steenackers et al., 2012; Baugh, 2014; Daxin, 2016). In Salmonella biofilms, cellulose, a key exopolysaccharide made up of β-1-4-D-glucose units, is synthesized by two operons, bcsABZD and bcsEFG (Daxin, 2016).
The formation of biofilms in Salmonella increases its chances of survival in hostile environments and facilitates resistance against antimicrobial compounds (Trampari et al., 2021). The formation of Salmonella biofilms facilitates contributing to its persistence in particular surface areas (Sakarikou et al., 2020). These biofilms can develop on both living and non-living surfaces (Daxin, 2016). For instance, Salmonella biofilms can be found on biotic surfaces such as epithelial cells within a host, including human cholesterol gallstones, and the chicken intestinal epithelium (Castelijn, 2013). Salmonella biofilms facilitate Salmonella to survive in several conditions such as elevated temperatures, malnourishment conditions, acidic pH, diverse atmospheres, and antimicrobials (Ćwiek et al., 2019). Moreover, bacterial biofilms play an important role in antimicrobial agents through a variety of mechanisms (Cadena et al., 2019). Biofilm production is recognized as one of the best AR mechanisms. Several mechanisms of biofilm structure contribute to antimicrobial resistance. The activation of efflux pumps, limited penetration of antibiotics through the biofilm polysaccharide matrix, quorum-sensing, physiological alterations due to slow growth rates and starvation process, and persisted bacterial cells-like mechanisms allowed biofilms to resist the antimicrobials (Andersen et al., 2015). Furthermore, bacterial cells in biofilms exhibit greater AR than planktonic cells due to their several mechanisms (Ćwiek et al., 2019). Moreover, Salmonella forms biofilms on food products, which cause foodborne illnesses, and are challenging to eliminate due to their higher resistance to antibiotics compared to planktonic cells (Alenazy, 2022a). Both typhoid and non-typhoid Salmonella were able to perform biofilms. Usually, non-typhoidal Salmonella biofilms cause problems in several industries such as veterinary and medical settings. As well as biofilms of S. Typhi can be identified on gallstones during infection (Baugh, 2014). The formation of biofilms is important to Salmonella for their growth and survival in the environment. Salmonella covered by biofilm exhibits increased resistance to chemical agents such as antibiotics and disinfectants, as well as to physical stresses. Also, the creation of biofilms enhanced Salmonella virulence (Daxin, 2016). Salmonella biofilms are resistant to several antibiotics such as ampicillin, ciprofloxacin, gentamicin, tetracycline, or third-generation cephalosporins such as ceftriaxone and cefotaxime (Ćwiek et al., 2019).
One of the key mechanisms through which Salmonella biofilms develop AR is efflux. Among the multi-drug efflux pumps in Salmonella, the AcrAB–TolC efflux system is well-characterized. The MDR efflux pumps found in Salmonella are classified into four distinct drug efflux families: five in the RND family, two in the MFS family, one in the MATE family, and one in the ABC family. The inactivation of any multi-drug efflux pumps reduces biofilm production in Salmonella Typhimurium (Baugh, 2014). Additionally, the presence of mutants in S. Typhimurium, such as tolC, acrB, acrD, acrEF, mdtABC, mdsABC, emrAB, mdfA, mdtK, and macAB, can hinder biofilm formation because Salmonella strains possessing these mutants lack MDR efflux systems (Baugh, 2014; Daxin, 2016). Nonetheless, a study reported that changing efflux function decreases the formation of Salmonella biofilms (Trampari et al., 2021). The AcrD efflux pump is important to Salmonella biofilm formation. Mutations in AcrD significantly reduce both biofilm formation and the expression of key biofilm proteins encoded by csgBD (Buckner et al., 2016). In S. Typhimurium, tolC and acrB mutants cannot form a functional biofilm and suppress the transcription of both curli operons (csgBAC and csgDEFG) (Baugh, 2014).
The extracellular matrix within microbial biofilms serves a critical function, such as providing structural support, facilitating communication between cells, and protecting the bacterial community by blocking antibiotics from penetrating the bacteria (Vestby et al., 2020; Singh et al., 2022; Prinzi and Rohde, 2023). Salmonella biofilms enhance the likelihood of persistence in hostile environments by facilitating defiance against antimicrobial compounds. This occurs because of their structure and the composition of their extracellular matrix (Trampari et al., 2021). A study demonstrated that Salmonella biofilms show resistance to ciprofloxacin due to the formation of biofilm structure (González et al., 2018). In addition, Salmonella exhibits tolerance to triclosan, a widely used biocide against bacteria, including Salmonella (Tabak et al., 2007). This tolerance is attributed to the limited diffusion of triclosan through the extracellular matrix formed by Salmonella as part of its biofilm formation. Moreover, extracellular DNA (eDNA) within Salmonella biofilms enhances AR to specific antibiotics by altering the outer membrane’s magnesium ion concentration. DNA, being an anionic molecule, can chelate cations like Mg2+. Notably, Mg2+ restriction in S. Typhimurium acts as an environmental signal, triggering the induction of the PhoPQ and PmrAB two-component systems, which subsequently promote AR mechanisms (Dincer et al., 2020).
8 Conclusion and future perspectives
In conclusion, this review highlights the complex nature of AR in Salmonella, emphasizing the various resistance mechanisms that include drug efflux, target site modification, permeability alterations, and biofilm formation. The extensive and improper use of antibiotics in both clinical and agricultural settings has considerably contributed to the acceleration of resistance traits among various Salmonella serovars. The rising MDR Salmonella presents an alarming public health challenge with significant implications for treatment efficacy and infection control.
Efforts to combat this issue must be multi-pronged, emphasizing the development of novel antimicrobial agents, such as neoglycosides (Labby and Garneau-Tsodikova, 2013), and the use of antibiotic adjuvants (Wright, 2016). Strategies should also include the combination of multiple antibiotics (Parmanik et al., 2022; Wang et al., 2022a; Lobertti et al., 2024), the creation of enzyme specific inhibitors such as novel β-lactamase inhibitors, AEM inhibitors to overcome aminoglycoside resistance (Wang et al., 2022a), and chloramphenicol acetyltransferase inhibitors to combat chloramphenicol resistance. Moreover, efflux pump inhibitors, including berberine and palmatine (Morita et al., 2016; Aghayan et al., 2017; Li and Ge, 2023), should be employed. The use of nanoparticles as drug delivery systems and the incorporation of plant-derived metabolites (Parmanik et al., 2022) are promising approaches. Furthermore, the judicious use of existing antibiotics, enhanced surveillance for resistance patterns, and rigorous infection prevention strategies are essential components of this comprehensive approach.
Additionally, public health education on antibiotic stewardship and the importance of comprehensive hygiene practices in food preparation are critical in curbing the spread of these resistant pathogens. Future research should focus on innovative approaches to overcome bacterial defensive mechanisms, such as the development of new therapeutic molecules, the use of bacteriophage therapy, the use of competitive exclusion to suppress pathogenic bacteria in the food environment, genetic modification of surrogate bacteria using gene editing tools such as CRISPR-Cas techniques to reduce AR Salmonella and the exploration of vaccine potentials. To combat the threat of AR Salmonella and ensure proper treatment for those infected, coordinated and collaborative actions across the globe are essential.
Author contributions
AP: Conceptualization, Software, Visualization, Writing – original draft, Writing – review & editing. PR: Conceptualization, Data curation, Software, Visualization, Writing – original draft, Writing – review & editing. SP: Funding acquisition, Resources, Supervision, Writing – review & editing.
Funding
The authors declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by USDA Evans-Allen funds (Grant No: 4-451621) and from the School of Graduate Studies of the University of Maryland Eastern Shore (UMES).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdi M., Mirkalantari S., Amirmozafari N. (2019). Bacterial resistance to antimicrobial peptides. J. Pept. Sci. 25, e3210. doi: 10.1002/psc.3210
Abebe E., Tegegne B., Tibebu S. (2016). A review on molecular mechanisms of bacterial resistance to antibiotics. EJAS 8, 301–310. doi: 10.5829/idosi.ejas.2016.8.5.1143
Aghayan S. S., Kalalian Mogadam H., Fazli M., Darban-Sarokhalil D., Khoramrooz S. S., Jabalameli F., et al. (2017). The effects of berberine and palmatine on efflux pumps inhibition with different gene patterns in Pseudomonas aeruginosa isolated from burn infections. Avicenna. J. Med. Biotechnol. 9, 2–7.
Ahmad I., Ali M., Ali R., Nawaz N., Patching S. G. (2024). Multidrug efflux protein families in bacteria: ABC, RND, MFS, SMR, MATE, PACE, AbgT (An Update). Biochemistry. 1–32. doi: 10.13140/RG.2.2.32729.12640
Ahmad I., Nawaz N., Ur Rahman S., Sajid A., Khan F., Khan S. B., et al. (2018). Multifaceted roles of efflux proteins in prokaryotes. Asian. J. Biol. Sci. 7, 1–7.
Akinyemi K. O., Ajoseh S. O. (2017). “Factors contributing to the emergence and spread of antibiotics resistance in Salmonella species,” in Current topics in Salmonella and salmonellosis. Ed. Mihai M. (IntechOpen, Rijeka), Ch. 6.
Aldred K. J., Kerns R. J., Osheroff N. (2014). Mechanism of quinolone action and resistance. Biochem. 53, 1565–1574. doi: 10.1021/bi5000564
Alenazy R. (2022a). Antibiotic resistance in Salmonella: Targeting multidrug resistance by understanding efflux pumps, regulators and the inhibitors. J. King. Saud. Univ. Sci. 34, 102275. doi: 10.1016/j.jksus.2022.102275
Alenazy R. (2022b). Drug efflux pump inhibitors: A promising approach to counter multidrug resistance in Gram-negative pathogens by targeting AcrB protein from AcrAB-TolC multidrug efflux pump from Escherichia coli. Biol. 11, 1328. doi: 10.3390/biology11091328
Al Gallas N., Aissa R. B. (2017). Fluoroquinolones resistance Salmonella: state of knowledge. Adv. Biotechnol. Microbiol. 7, 0095–0097. doi: 10.19080/AIBM.2017.07.555717
American Chemical Society. (2020). Chloramphenicol. Available online at: https://www.acs.org/molecule-of-the-week/archive/c/chloramphenicol.html (Accessed August 17, 2023).
Andersen J. L., He G.-X., Kakarla P., Ranjana K. C., Kumar S., Lakra W. S., et al. (2015). Multidrug efflux pumps from Enterobacteriaceae, Vibrio cholerae and Staphylococcus aureus bacterial food pathogens. Int. J. Environ. Res. Public. Health 12, 1487–1547. doi: 10.3390/ijerph120201487
Band V. I., Weiss D. S. (2015). Mechanisms of antimicrobial peptide resistance in Gram-negative bacteria. Antibiotics 4, 18–41. doi: 10.3390/antibiotics4010018
Baugh S. (2014). The role of multidrug efflux pumps in biofilm formation of Salmonella enterica serovar Typhimurium (Birmingham, England: University of Birmingham).
Bennett P. M. (2008). Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. Br. J. Pharmacol. 153, 347–357. doi: 10.1038/sj.bjp.0707607
Blair J. M. A., Smith H. E., Ricci V., Lawler A. J., Thompson L. J., Piddock L. J. V. (2014). Expression of homologous RND efflux pump genes is dependent upon AcrB expression: implications for efflux and virulence inhibitor design. J. Antimicrob. Chemother. 70, 424–431. doi: 10.1093/jac/dku380
Bogomolnaya L. M., Andrews K. D., Talamantes M., Maple A., Ragoza Y., Vazquez-Torres A., et al. (2013). The ABC-type efflux pump MacAB protects Salmonella enterica serovar Typhimurium from oxidative stress. mBio 4, e00630–e00613. doi: 10.1128/mBio.00630-13
Boll J. M., Tucker A. T., Klein D. R., Beltran A. M., Brodbelt J. S., Davies B. W., et al. (2015). Reinforcing lipid A acylation on the cell surface of Acinetobacter baumannii promotes cationic antimicrobial peptide resistance and desiccation survival. mBio 6, 1–11. doi: 10.1128/mbio.00478-15
Bradford P. A. (2001). Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14, 933–951. doi: 10.1128/cmr.14.4.933-951.2001
Buckner M. M., Blair J. M., La Ragione R. M., Newcombe J., Dwyer D. J., Ivens A., et al. (2016). Beyond antimicrobial resistance: Evidence for a distinct role of the AcrD efflux pump in Salmonella biology. mBio 7, 1–10. doi: 10.1128/mBio.01916-16
Cadena M., Kelman T., Marco M. L., Pitesky M. (2019). Understanding antimicrobial resistance (AMR) profiles of Salmonella biofilm and planktonic bacteria challenged with disinfectants commonly used during poultry processing. Foods 8, 1–16. doi: 10.3390/foods8070275
Carattoli A. (2003). Plasmid-mediated antimicrobial resistance in Salmonella enterica. Curr. Issues. Mol. Biol. 5, 113–122. doi: 10.21775/cimb.005.113
Castelijn G. A. A. (2013). Salmonella Typhimurium biofilms (Wageningen, Netherlands: Wageningen University).
CDC. (2011). National Salmonella surveillance overview. Available online at: https://www.cdc.gov/nationalsurveillance/salmonella-surveillance.html (Accessed March 28, 2023).
CDC. (2019). Antibiotic resistance threats in the United States. Available online at: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf (Accessed January 17 2023).
CDC. (2022). Serotypes and the importance of serotyping Salmonella. Available online at: https://www.cdc.gov/salmonella/reportspubs/salmonella-atlas/serotyping-importance.html (Accessed August 12, 2023).
CDC. (2023). Salmonella . Available online at: https://www.cdc.gov/salmonella/index.html (Accessed June 19 2023).
Chávez-Jacobo V. M., Hernández-Ramírez K. C., Romo-Rodríguez P., Pérez-Gallardo R. V., Campos-García J., Gutiérrez-Corona J. F., et al. (2018). CrpP is a novel ciprofloxacin-modifying enzyme encoded by the Pseudomonas aeruginosa pUM505 plasmid. Antimicrob. Agents. Chemother. 62, 1–11. doi: 10.1128/aac.02629-17
Chin K. W., Michelle Tiong H. L., Luang-In V., Ma N. L. (2023). An overview of antibiotic and antibiotic resistance. Environ. Adv. 11, 100331. doi: 10.1016/j.envadv.2022.100331
Choi U., Lee C.-R. (2019). Distinct roles of outer membrane porins in antibiotic resistance and membrane integrity in Escherichia coli. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00953
Cosby D. E., Cox N. A., Harrison M. A., Wilson J. L., Buhr R. J., Fedorka-Cray P. J. (2015). Salmonella and antimicrobial resistance in broilers: A review. J. Appl. Poult. Res. 24, 408–426. doi: 10.3382/japr/pfv038
Cox G., Wright G. D. (2013). Intrinsic antibiotic resistance: Mechanisms, origins, challenges and solutions. Int. J. Med. Microbiol. 303, 287–292. doi: 10.1016/j.ijmm.2013.02.009
Culyba M. J., Mo C. Y., Kohli R. M. (2015). Targets for combating the evolution of acquired antibiotic resistance. Biochem. 54, 3573–3582. doi: 10.1021/acs.biochem.5b00109
Ćwiek K., Bugla-Płoskońska G. B., Wieliczko A. (2019). Salmonella biofilm development: Structure and significance. Adv. Hyg. Exp. Med. 73, 937–943. doi: 10.5604/01.3001.0013.7866
Daxin P. (2016). “Biofilm formation of salmonella,” in Microbial Biofilms. Eds. Dharumadurai D., Nooruddin T. (IntechOpen, Rijeka), Ch. 12.
Delcour A. H. (2009). Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta 1794, 808–816. doi: 10.1016/j.bbapap.2008.11.005
Delmar J. A., Yu E. W. (2016). The AbgT family: A novel class of antimetabolite transporters. Protein. Sci. 25, 322–337. doi: 10.1002/pro.2820
Devi V. (2011). Antimicrobial peptides and Salmonella pathogenesis. Doctoral dissertation, Indian Institute of Science). Available online at: https://etd.iisc.ac.in/handle/2005/1049.
Dincer S., Fatima Masume U., Anil D. (2020). “Antibiotic resistance in biofilm,” in Bacterial Biofilms. Eds. Dincer S., Melis Sümengen Ö., Afet A. (IntechOpen, Rijeka), Ch. 9.
Drlica K., Zhao X. (1997). DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61, 377–392. doi: 10.1128/mmbr.61.3.377-392.1997
Duffey M., Jumde R. P., da Costa R. M. A., Ropponen H. K., Blasco B., Piddock L. J. V. (2024). Extending the potency and lifespan of antibiotics: inhibitors of gram-negative bacterial efflux pumps. ACS. Infect. Dis. 10, 1458–1482. doi: 10.1021/acsinfecdis.4c00091
Fàbrega A., Madurga S., Giralt E., Vila J. (2009). Mechanism of action of and resistance to quinolones. Microb. Biotechnol. 2, 40–61. doi: 10.1111/j.1751-7915.2008.00063.x
FDA. (2020). Get the facts about Salmonella. U.S. Food and Drug Administration (U.S. Food and Drug Administration). Available online at: https://www.fda.gov/animal-veterinary/animal-health-literacy/get-facts-about-salmonella (Accessed January 17 2023).
Fernandes R., Amador P., Prudêncio C. (2013). [amp]]beta;-Lactams: chemical structure, mode of action and mechanisms of resistance. Rev. Res. Med. Microbiol. 24, 7–17. doi: 10.1097/MRM.0b013e3283587727
Ferrari R. G., Rosario D. K. A., Cunha-Neto A., Mano S. B., Figueiredo E. E. S., Conte-Junior C. A. (2019). Worldwide epidemiology of Salmonella serovars in animal-based foods: a Meta-analysis. Appl. Environ. Microbiol. 85, e00591–e00519. doi: 10.1128/AEM.00591-19
Galdiero S., Falanga A., Cantisani M., Tarallo R., Della Pepa M. E., D'Oriano V., et al. (2012). Microbe-host interactions: structure and role of Gram-negative bacterial porins. Curr. Protein. Pept. Sci. 13, 843–854. doi: 10.2174/138920312804871120
Gal-Mor O., Boyle E. C., Grassl G. A. (2014). Same species, different diseases: how and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Front. Microbiol. 5. doi: 10.3389/fmicb.2014.00391
Gaurav A., Bakht P., Saini M., Pandey S., Pathania R. (2023). Role of bacterial efflux pumps in antibiotic resistance, virulence, and strategies to discover novel efflux pump inhibitors. Microbiol. 169, 1–13. doi: 10.1099/mic.0.001333
Gharbi M., Abbas M. A. S., Hamrouni S., Maaroufi A. (2023). First report of aac(6)-Ib and aac(6)-Ib-cr variant genes associated with mutations in gyrA encoded fluoroquinolone resistance in avian Campylobacter coli strains collected in Tunisia. Int. J. Mol. Sci. 24, 16116. doi: 10.3390/ijms242216116
González J. F., Alberts H., Lee J., Doolittle L., Gunn J. S. (2018). Biofilm formation protects Salmonella from the antibiotic ciprofloxacin in vitro and in vivo in the mouse model of chronic carriage. Sci. Rep. 8, 222. doi: 10.1038/s41598-017-18516-2
Groisman E. A., Duprey A., Choi J. (2021). How the PhoP/PhoQ system controls virulence and Mg(2+) homeostasis: Lessons in signal transduction, pathogenesis, physiology, and evolution. Microbiol. Mol. Biol. 85, e0017620. doi: 10.1128/mmbr.00176-20
Guerra B., Soto S., Cal S., Mendoza M. C. (2000). Antimicrobial resistance and spread of class 1 integrons among Salmonella serotypes. Antimicrob. Agents. Chemother. 44, 2166–2169. doi: 10.1128/aac.44.8.2166-2169.2000
Harris M., Fasolino T., Ivankovic D., Davis N. J., Brownlee N. (2023). Genetic factors that contribute to antibiotic resistance through intrinsic and acquired bacterial genes in urinary tract infections. Microorganisms 11, 1407. doi: 10.3390/microorganisms11061407
Hassan K. A., Liu Q., Elbourne L. D. H., Ahmad I., Sharples D., Naidu V., et al. (2018). Pacing across the membrane: the novel PACE family of efflux pumps is widespread in Gram-negative pathogens. Res. Microbiol. 169, 450–454. doi: 10.1016/j.resmic.2018.01.001
Herrera-Sánchez M. P., Castro-Vargas R. E., Fandiño-de-Rubio L. C., Rodríguez-Hernández R., Rondón-Barragán I. S. (2021). Molecular identification of fluoroquinolone resistance in Salmonella spp. isolated from broiler farms and human samples obtained from two regions in Colombia. Vet. World. 14, 1767–1773. doi: 10.14202/vetworld.2021.1767-1773
Higgins C. F. (2007). Multiple molecular mechanisms for multidrug resistance transporters. Nature 446, 749–757. doi: 10.1038/nature05630
Hooper D. C., Jacoby G. A. (2016). Topoisomerase inhibitors: fluoroquinolone mechanisms of action and resistance. Cold. Spring. Harb. Perspect. Med. 6, 1–21. doi: 10.1101/cshperspect.a025320
Huang L., Wu C., Gao H., Xu C., Dai M., Huang L., et al. (2022). Bacterial multidrug efflux pumps at the frontline of antimicrobial resistance: An overview. Antibiotics 11, 1–18. doi: 10.3390/antibiotics11040520
Huddleston J. R. (2014). Horizontal gene transfer in the human gastrointestinal tract: potential spread of antibiotic resistance genes. Infect. Drug Resist. 7, 167–176. doi: 10.2147/idr.S48820
Humphries R. M. (2023). AST News Update June 2023: New! CLSI M100-Ed33: Updated Aminoglycoside Breakpoints for Enterobacterales and Pseudomonas aeruginosa (Clinical and Laboratory Standards Institute). Available online at: https://clsi.org/about/blog/ast-news-update-june-2023-new-clsi-m100-ed33-updated-aminoglycoside-breakpoints-for-enterobacterales-and-pseudomonas-aeruginosa/ (Accessed June 09, 2024).
Ipinza F., Collao B., Monsalva D., Bustamante V. H., Luraschi R., Alegría-Arcos M., et al. (2014). Participation of the Salmonella OmpD porin in the infection of RAW264.7 macrophages and BALB/c mice. PloS One 9, 1–11. doi: 10.1371/journal.pone.0111062
Jacoby G. A. (2005). Mechanisms of resistance to quinolones. Clin. Infect. Dis. 41 Suppl 2, S120–S126. doi: 10.1086/428052
Jacoby G. A., Strahilevitz J., Hooper D. C. (2014). Plasmid-mediated quinolone resistance. Microbiol. Spectr. 2, 1–24. doi: 10.1128/microbiolspec.PLAS-0006-2013
Jajere S. M. (2019). A review of Salmonella enterica with particular focus on the pathogenicity and virulence factors, host specificity and antimicrobial resistance including multidrug resistance. Vet. World. 12, 504–521. doi: 10.14202/vetworld.2019.504-521
Jeśman C., Młudzik A., Cybulska M. (2011). History of antibiotics and sulphonamides discoveries. Pol. Merkur. Lekarski 30, 320–322.
Kabra R., Chauhan N., Kumar A., Ingale P., Singh S. (2019). Efflux pumps and antimicrobial resistance: paradoxical components in systems genomics. Prog. Biophys. Mol. Biol. 141, 15–24. doi: 10.1016/j.pbiomolbio.2018.07.008
Kato A., Groisman E. A. (2008). The PhoQ/PhoP regulatory network of Salmonella enterica. Adv. Exp. Med. Biol. 631, 7–21. doi: 10.1007/978-0-387-78885-2_2
Klenotic P. A., Moseng M. A., Morgan C. E., Yu E. W. (2021). Structural and functional diversity of resistance-nodulation-cell division transporters. Chem. Rev. 121, 5378–5416. doi: 10.1021/acs.chemrev.0c00621
Kongsoi S., Yokoyama K., Suprasert A., Utrarachkij F., Nakajima C., Suthienkul O., et al. (2015). Characterization of Salmonella Typhimurium DNA gyrase as a target of quinolones. Drug Test. Anal. 7, 714–720. doi: 10.1002/dta.1744
Kornelsen V., Kumar A. (2021). Update on multidrug resistance efflux pumps in Acinetobacter spp. Antimicrob. Agents. Chemother. 65, e0051421. doi: 10.1128/aac.00514-21
Kotra L. P., Haddad J., Mobashery S. (2000). Aminoglycosides: perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicrob. Agents. Chemother. 44, 3249–3256. doi: 10.1128/aac.44.12.3249-3256.2000
Krause K. M., Serio A. W., Kane T. R., Connolly L. E. (2016). Aminoglycosides: an overview. Cold. Spring. Harb. Perspect. Med. 6. doi: 10.1101/cshperspect.a027029
Kumar S., Floyd J. T., He G., Varela M. F. (2013). Bacterial antimicrobial efflux pumps of the MFS and MATE transporter families: a review. Recent Res. Dev. Antimicrob. Agents. Chemother. 7, 1–21.
Labby K. J., Garneau-Tsodikova S. (2013). Strategies to overcome the action of aminoglycoside-modifying enzymes for treating resistant bacterial infections. Future Med. Chem. 5, 1285–1309. doi: 10.4155/fmc.13.80
Lee D., Das S., Dawson N. L., Dobrijevic D., Ward J., Orengo C. (2016). Novel computational protocols for functionally classifying and characterising serine Beta-Lactamases. PloS Comput. Biol. 12, e1004926. doi: 10.1371/journal.pcbi.1004926
Lee S., Park N., Yun S., Hur E., Song J., Lee H., et al. (2021). Presence of plasmid-mediated quinolone resistance (PMQR) genes in non-typhoidal Salmonella strains with reduced susceptibility to fluoroquinolones isolated from human salmonellosis in Gyeonggi-do, South Korea from 2016 to 2019. Gut. Pathog. 13, 35. doi: 10.1186/s13099-021-00431-7
Li Y., Ge X. (2023). Role of berberine as a potential efflux pump inhibitor against MdfA from Escherichia coli: in vitro and in silico studies. Microbiol. Spectr. 11, e03324–e03322. doi: 10.1128/spectrum.03324-22
Li J., Haihong H., Abdul S., Heying Z., Zonghui Y. (2018). “Fluoroquinolone resistance in Salmonella: mechanisms, fitness, and virulence,” in Salmonella. Ed. Maria Teresa M. (IntechOpen, Rijeka), Ch. 6.
Li Y., Wang Z., Chen J., Ernst R. K., Wang X. (2013). Influence of lipid A acylation pattern on membrane permeability and innate immune stimulation. Mar. Drugs 11, 3197–3208. doi: 10.3390/md11093197
Lobertti C. A., Gizzi F. O., Magni C., Rial A., Chabalgoity J. A., Yim L., et al. (2024). Enhancing colistin efficacy against Salmonella infections with a quinazoline-based dual therapeutic strategy. Sci. Rep. 14, 5148. doi: 10.1038/s41598-024-55793-0
Lu M., Symersky J., Radchenko M., Koide A., Guo Y., Nie R., et al. (2013). Structures of a Na+-coupled, substrate-bound MATE multidrug transporter. PNAS 110, 2099–2104. doi: 10.1073/pnas.1219901110
Madej M. G. (2014). Function, structure, and evolution of the major facilitator superfamily: The LacY manifesto. Adv. Biol. 2014, 1–20. doi: 10.1155/2014/523591
Mąka Ł., Popowska M. (2016). Antimicrobial resistance of Salmonella spp. isolated from food. Rocz. Panstw. Zakl. Hig. 67, 343–358.
Mancuso G., Midiri A., Gerace E., Biondo C. (2021). Bacterial antibiotic resistance: the most critical pathogens. Pathogens 10, 1310. doi: 10.3390/pathogens10101310
Matamouros S., Miller S. I. (2015). S. Typhimurium strategies to resist killing by cationic antimicrobial peptides. BBA 1848, 3021–3025. doi: 10.1016/j.bbamem.2015.01.013
McMillan E. A., Gupta S. K., Williams L. E., Jové T., Hiott L. M., Woodley T. A., et al. (2019). Antimicrobial resistance genes, cassettes, and plasmids present in Salmonella enterica associated with United States food animals. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00832
Millan A. S. (2018). Evolution of plasmid-mediated antibiotic resistance in the clinical context. Trends Microbiol. 26, 978–985. doi: 10.1016/j.tim.2018.06.007
Morita Y., Nakashima K.-i., Nishino K., Kotani K., Tomida J., Inoue M., et al. (2016). Berberine is a novel type efflux inhibitor which attenuates the MexXY-mediated aminoglycoside resistance in Pseudomonas aeruginosa. Front.s Microbiol. 7. doi: 10.3389/fmicb.2016.01223
Munita J. M., Arias C. A. (2016). Mechanisms of antibiotic resistance. Microbiol. Spectr. 4, 1–24. doi: 10.1128/microbiolspec.VMBF-0016-2015
Naas T., Poirel L., Nordmann P. (2008). Minor extended-spectrum beta-lactamases. Clin. Microbiol. Infect. 14 Suppl 1, 42–52. doi: 10.1111/j.1469-0691.2007.01861.x
National Human Genome Research Institute (2014).Plasmid/plasmids. Available online at: https://www.nature.com/scitable/definition/plasmid-plasmids-28/ (Accessed August 16, 2023).
Nikaido H. (2009). Multidrug resistance in bacteria. Annu. Rev. Biochem. 78, 119–146. doi: 10.1146/annurev.biochem.78.082907.145923
Nishino K., Latifi T., Groisman E. A. (2006). Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 59, 126–141. doi: 10.1111/j.1365-2958.2005.04940.x
Nishino K., Nikaido E., Yamaguchi A. (2009). Regulation and physiological function of multidrug efflux pumps in Escherichia coli and Salmonella. BBA 1794, 834–843. doi: 10.1016/j.bbapap.2009.02.002
Nishino K., Yamasaki S., Nakashima R., Zwama M., Hayashi-Nishino M. (2021). Function and inhibitory mechanisms of multidrug efflux pumps. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.737288
Nobre T. M., Martynowycz M. W., Andreev K., Kuzmenko I., Nikaido H., Gidalevitz D. (2015). Modification of Salmonella lipopolysaccharides prevents the outer membrane penetration of novobiocin. Biophys. J. 109, 2537–2545. doi: 10.1016/j.bpj.2015.10.013
Noto H. (2022). Review on antibiotic resistant Salmonella in animals: public health and economic importance. JVMAC 1, 1–10.
Olivares Pacheco J. A., Bernardini A., Garcia-Leon G., Corona F., Sanchez M. B., Martinez J. L. (2013). The intrinsic resistome of bacterial pathogens. Fronti. Microbiol. 4, 1–15. doi: 10.3389/fmicb.2013.00103
Ovung A., Bhattacharyya J. (2021). Sulfonamide drugs: structure, antibacterial property, toxicity, and biophysical interactions. Biophys. Rev. 13, 259–272. doi: 10.1007/s12551-021-00795-9
Parmanik A., Das S., Kar B., Bose A., Dwivedi G. R., Pandey M. M. (2022). Current treatment strategies against multidrug-resistant bacteria: A Review. Curr. Microbiol. 79, 388. doi: 10.1007/s00284-022-03061-7
Pham T. D. M., Ziora Z. M., Blaskovich M. A. T. (2019). Quinolone antibiotics. Medchemcomm. 10, 1719–1739. doi: 10.1039/c9md00120d
Plumb I., Fields P., Bruce B., CDC (2023). Salmonellosis, Nontyphoidal; CDC Yellow Book 2024. Available online at: https://wwwnc.cdc.gov/travel/yellowbook/2024/infections-diseases/salmonellosis-nontyphoidal (Accessed August 22, 2023).
Popa G. L., Papa M. I. (2021). Salmonella spp. infection - a continuous threat worldwide. Germs 11, 88–96. doi: 10.18683/germs.2021.1244
Pribul B. R., Festivo M. L., Rodrigues M. S., Costa R. G., Rodrigues E., de Souza M. M. S., et al. (2017). Characteristics of quinolone resistance in Salmonella spp. isolates from the food chain in Brazil. Fronti. Microbiol. 8, 1–7. doi: 10.3389/fmicb.2017.00299
Prinzi A., Rohde R. (2023).The role of bacterial biofilms in antimicrobial resistance. American society for microbiology. Available online at: https://asm.org/articles/2023/march/the-role-of-bacterial-biofilms-in-antimicrobial-re (Accessed August 06, 2024).
Punchihewage-Don A. J., Hasan N. A., Rashed S. M., Parveen S. (2023). Microbiome analysis of organic and conventional chickens processed using whole carcass enrichment and rinse methods. J. Food Prot. 86, 100176. doi: 10.1016/j.jfp.2023.100176
Punchihewage-Don A. J., Hawkins J., Adnan A. M., Hashem F., Parveen S. (2022). The outbreaks and prevalence of antimicrobial resistant Salmonella in poultry in the United States: An overview. Heliyon. 8, 1–11. doi: 10.1016/j.heliyon.2022.e11571
Punchihewage-Don A. J., Parveen S., Schwarz J., Hamill L., Nindo C., Hall P., et al. (2021). Efficacy and quality attributes of antimicrobial agent application via a commercial electrostatic spray cabinet to inactivate Salmonella on chicken thigh meat. J. Food Prot. 84, 2221–2228. doi: 10.4315/JFP-21-206
Punchihewage-Don A. J., Schwarz J., Diria A., Bowers J., Parveen S. (2024). Prevalence and antibiotic resistance of Salmonella in organic and non-organic chickens on the Eastern Shore of Maryland, USA. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1272892
Qian H., Cheng S., Liu G., Tan Z., Dong C., Bao J., et al. (2020). Discovery of seven novel mutations of gyrB, parC and parE in Salmonella Typhi and Paratyphi strains from Jiangsu Province of China. Sci. Rep. 10, 7359. doi: 10.1038/s41598-020-64346-0
Rahman S. U., Ali T., Ali I., Khan N. A., Han B., Gao J. (2018). The growing genetic and functional diversity of extended spectrum beta-lactamases. Biomed. Res. Int. 2018, 9519718. doi: 10.1155/2018/9519718
Reygaert W. C. (2018). An overview of the antimicrobial resistance mechanisms of bacteria. AIMS. Microbiol. 4, 482–501. doi: 10.3934/microbiol.2018.3.482
Ruiz J. (2019). Transferable mechanisms of quinolone resistance from 1998 onward. CMR 32, 1–59. doi: 10.1128/CMR.00007-19
Ryan M. P., O’Dwyer J., Adley C. C. (2017). Evaluation of the complex nomenclature of the clinically and veterinary significant pathogen Salmonella. BioMed. Res. Int. 2017, 3782182. doi: 10.1155/2017/3782182
Rycroft A. N. (2000). “Structure, function and synthesis of surface polysaccharides in Salmonella,” in Salmonella in Domestic Animals. Ed. Wray C. (CABI Publishing, Wallingford, United Kingdom), 19–33.
Saha S., Pupo E., Zariri A., van der Ley P. (2022). Lipid A heterogeneity and its role in the host interactions with pathogenic and commensal bacteria. Microlife 3, uqac011. doi: 10.1093/femsml/uqac011
Sakarikou C., Kostoglou D., Simões M., Giaouris E. (2020). Exploitation of plant extracts and phytochemicals against resistant Salmonella spp. in biofilms. Food. Res. Int. 128, 108806. doi: 10.1016/j.foodres.2019.108806
Schaenzer A. J., Wright G. D. (2020). Antibiotic resistance by enzymatic modification of antibiotic targets. TMM 26, 768–782. doi: 10.1016/j.molmed.2020.05.001
Schultz B. M., Melo-Gonzalez F., Salazar G. A., Porto B. N., Riedel C. A., Kalergis A. M., et al. (2021). New insights on the early interaction between typhoid and non-typhoid Salmonella serovars and the host cells. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.647044
Serio A. W., Keepers T., Andrews L., Krause K. M. (2018). Aminoglycoside revival: review of a historically important class of antimicrobials undergoing rejuvenation. EcoSal. Plus. 8, 1–20. doi: 10.1128/ecosalplus.ESP-0002-2018
Shaheen A., Tariq A., Iqbal M., Mirza O., Haque A., Walz T., et al. (2021). Mutational diversity in the quinolone resistance-determining regions of type-II topoisomerases of Salmonella serovars. Antibiotics 10, 1–26. doi: 10.3390/antibiotics10121455
Sharma A., Gupta V. K., Pathania R. (2019). Efflux pump inhibitors for bacterial pathogens: From bench to bedside. Indian. J. Med. Res. 149, 129–145. doi: 10.4103/ijmr.IJMR_2079_17
Shome S., Sankar K., Jernigan R. L. (2021). Simulated drug efflux for the AbgT family of membrane transporters. J. Chem. Inf. Model. 61, 5673–5681. doi: 10.1021/acs.jcim.1c00516
Shrestha U., Ranaweera I., Kumar S., Kc R., Kakarla P., Lakra W. S., et al. (2015). “Multidrug resistance efflux pumps of Salmonella enterica,” in Salmonella: prevalence, risk factors and treatment options. Ed. Hackett C. B. (Nova Science Publishers, Inc, New York), 1–30.
Singh A., Shahid M., Singh P. G., Khan H. M. (2022). “Mobile genetic elements,” in Beta-Lactam resistance in Gram-negative bacteria: threats and challenges. Eds. Shahid M., Singh A., Sami H. (Springer Nature, Singapore), 141–153.
Sirisena D. M. (2000). Outer membrane permeability of lipopolysaccharide deficient mutants of Salmonella Typhimurium. JNSF 28, 89–100. doi: 10.4038/jnsfsr.v28i2.2678
Smith S. I., Seriki A., Ajayi A. (2016). Typhoidal and non-typhoidal Salmonella infections in Africa. Eur. J. Clin. Microbiol. Infect. Dis. 35, 1913–1922. doi: 10.1007/s10096-016-2760-3
Song S., Lee B., Yeom J. H., Hwang S., Kang I., Cho J. C., et al. (2015). MdsABC-Mediated pathway for pathogenicity in Salmonella enterica serovar Typhimurium. Infect. Immun. 83, 4266–4276. doi: 10.1128/iai.00653-15
Souza R. B. D., Magnani M., Ferrari R. G., Kottwitz L. B., Sartori D., Tognim M. C., et al. (2011). Detection of quinolone-resistance mutations in Salmonella spp. strains of epidemic and poultry origin. Braz. J. Microbiol. 42, 211–215. doi: 10.1590/s1517-83822011000100026
Steenackers H., Hermans K., Vanderleyden J., De Keersmaecker S. C. (2012). Salmonella biofilms: an overview on occurrence, structure, regulation and eradication. Food. Res. Int. 45, 502–531. doi: 10.1016/j.foodres.2011.01.038
Su L. H., Chiu C. H. (2007). Salmonella: clinical importance and evolution of nomenclature. Change Gung. Med. J. 30, 210–219.
Tabak M., Scher K., Hartog E., Romling U., Matthews K. R., Chikindas M. L., et al. (2007). Effect of triclosan on Salmonella Typhimurium at different growth stages and in biofilms. FEMS. Microbiol. Lett. 267, 200–206. doi: 10.1111/j.1574-6968.2006.00547.x
Tanwar J., Das S., Fatima Z., Hameed S. (2014). Multidrug resistance: an emerging crisis. Interdiscip. Perspect. Infect. Dis. 2014, 541340. doi: 10.1155/2014/541340
Tariq S., Rizvi S. F. A., Anwar U. (2018). Tetracycline: classification, structure activity relationship and mechanism of action as a theranostic agent for infectious lesions-A mini review. BJSTR 7, 5787–5796. doi: 10.26717/BJSTR.2018.07.001475
Teklu D. S., Negeri A. A., Legese M. H., Bedada T. L., Woldemariam H. K., Tullu K. D. (2019). Extended-spectrum beta-lactamase production and multi-drug resistance among Enterobacteriaceae isolated in Addis Ababa, Ethiopia. Antimicrob. Resist. Infect. Control. 8, 39. doi: 10.1186/s13756-019-0488-4
Thacharodi A., Lamont I. L. (2022). Aminoglycoside-modifying enzymes are sufficient to make Pseudomonas aeruginosa clinically resistant to key antibiotics. Antibiotics 11, 884. doi: 10.3390/antibiotics11070884
Tooke C. L., Hinchliffe P., Bragginton E. C., Colenso C. K., Hirvonen V. H. A., Takebayashi Y., et al. (2019). [amp]]beta;-Lactamases and β-lactamase inhibitors in the 21st century. J. Mol. Biol. 431, 3472–3500. doi: 10.1016/j.jmb.2019.04.002
Trampari E., Holden E. R., Wickham G. J., Ravi A., Martins L. O., Savva G. M., et al. (2021). Exposure of Salmonella biofilms to antibiotic concentrations rapidly selects resistance with collateral tradeoffs. NPJ Biofilms Microbiomes. 7, 3. doi: 10.1038/s41522-020-00178-0
Ude J., Tripathi V., Buyck J. M., Söderholm S., Cunrath O., Fanous J., et al. (2021). Outer membrane permeability: Antimicrobials and diverse nutrients bypass porins in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 118, 1–8. doi: 10.1073/pnas.2107644118
Urban-Chmiel R., Marek A., Stępień-Pyśniak D., Wieczorek K., Dec M., Nowaczek A., et al. (2022). Antibiotic resistance in bacteria-A review. Antibiotics 11, 1–40. doi: 10.3390/antibiotics11081079
Usman J., Hamza A. (2022). Review on the emergence of quinolone resistance against Salmonella Typhi in Nigeria. Int. J. Sci. Glob. Sustain. 8, 10–10.
Vergalli J., Bodrenko I. V., Masi M., Moynié L., Acosta-Gutiérrez S., Naismith J. H., et al. (2020). Porins and small-molecule translocation across the outer membrane of Gram-negative bacteria. Nat. Rev. Microbiol. 18, 164–176. doi: 10.1038/s41579-019-0294-2
Vestby L. K., Grønseth T., Simm R., Nesse L. L. (2020). Bacterial biofilm and its role in the pathogenesis of disease. Antibiotics 9, 1–29. doi: 10.3390/antibiotics9020059
Wang Y., Ge H., Wei X., Zhao X. (2022b). Research progress on antibiotic resistance of Salmonella. Food Qual. Saf. 6, 1–10. doi: 10.1093/fqsafe/fyac035
Wang N., Luo J., Deng F., Huang Y., Zhou H. (2022a). Antibiotic combination therapy: a strategy to overcome bacterial resistance to aminoglycoside antibiotics. Front. Pharmacol. 13. doi: 10.3389/fphar.2022.839808
WHO. (2018). Salmonella (non-typhoidal) (World Health Organization). Available online at: https://www.who.int/news-room/fact-sheets/detail/Salmonella-(non-typhoidal) (Accessed April 24, 2022).
WHO. (2023). Antibiotic resistance. Available online at: https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance (Accessed August 6, 2024).
Wright G. D. (2016). Antibiotic adjuvants: Rescuing antibiotics from resistance. Trends Microbiol. 24, 862–871. doi: 10.1016/j.tim.2016.06.009
Yamasaki S., Nagasawa S., Hayashi-Nishino M., Yamaguchi A., Nishino K. (2011). AcrA dependency of the AcrD efflux pump in Salmonella enterica serovar Typhimurium. J. Antibiot. 64, 433–437. doi: 10.1038/ja.2011.28
Zack K. M., Sorenson T., Joshi S. G. (2024). Types and mechanisms of efflux pump systems and the potential of efflux pump inhibitors in the restoration of antimicrobial susceptibility, with a special reference to Acinetobacter baumannii. Pathogens 13, 197. doi: 10.3390/pathogens13030197
Keywords: Salmonella mechanisms, antibiotic resistance, multi-drug resistance, efflux pumps, biofilm formation, plasmid-mediated resistance, antibiotic target modification, drug inactivation
Citation: Punchihewage-Don AJ, Ranaweera PN and Parveen S (2024) Defense mechanisms of Salmonella against antibiotics: a review. Front. Antibiot. 3:1448796. doi: 10.3389/frabi.2024.1448796
Received: 13 June 2024; Accepted: 26 August 2024;
Published: 17 September 2024.
Edited by:
Abid Ali, Texas A&M University, United StatesReviewed by:
Moiz Ashraf Ansari, Texas A&M University, United StatesHaris Saeed, University of Southern California, United States
Sanath H. Kumar, Central Institute of Fisheries Education (ICAR), India
Copyright © 2024 Punchihewage-Don, Ranaweera and Parveen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anuradha Jeewantha Punchihewage-Don, amFwdW5jaGloZXdhZ2Vkb25AdW1lcy5lZHU=
 Anuradha Jeewantha Punchihewage-Don
Anuradha Jeewantha Punchihewage-Don Priyanka Nilmini Ranaweera
Priyanka Nilmini Ranaweera Salina Parveen
Salina Parveen