- 1Department of Anesthesiology and Perioperative Medicine, University of Alabama at Birmingham, Birmingham, AL, United States
- 2Department of Pharmacology, Ascension St. Vincent’s, Birmingham, AL, United States
Antimicrobial resistance in the intensive care unit is an ongoing global healthcare concern associated with high mortality and morbidity rates and high healthcare costs. Select groups of bacterial pathogens express different mechanisms of antimicrobial resistance. Clinicians face challenges in managing patients with multidrug-resistant bacteria in the form of a limited pool of available antibiotics, slow and potentially inaccurate conventional diagnostic microbial modalities, mimicry of non-infective conditions with infective syndromes, and the confounding of the clinical picture of organ dysfunction associated with sepsis with postoperative surgical complications such as hemorrhage and fluid shifts. Potential remedies for antimicrobial resistance include specific surveillance, adequate and systematic antibiotic stewardship, use of pharmacokinetic and pharmacodynamic techniques of therapy, and antimicrobial monitoring and adequate employment of infection control policies. Novel techniques of combating antimicrobial resistance include the use of aerosolized antibiotics for lung infections, the restoration of gut microflora using fecal transplantation, and orally administered probiotics. Newer antibiotics are urgently needed as part of the armamentarium against multidrug-resistant bacteria. In this review we discuss mechanisms and patterns of microbial resistance in a select group of drug-resistant bacteria, and preventive and remedial measures for combating antibiotic resistance in the critically ill.
Introduction
Antimicrobial resistance in the ICU (intensive care unit) is a growing healthcare concern. The Centers for Disease Control and Prevention (CDC) estimates that more than 2.8 million antibiotic-resistant infections occur each year, with more than 35,000 deaths as a result (Despotovic et al., 2020). Continued exposure to antibiotics is one of the most important factors, if not the most important factor, in the development of antimicrobial resistance (Bell et al., 2014). In the ICU, infections due to multidrug-resistant (MDR) bacteria lead to poor clinical outcomes, prolonged ICU and hospital stays, high mortality rates, and higher healthcare costs (Campion and Scully, 2018).
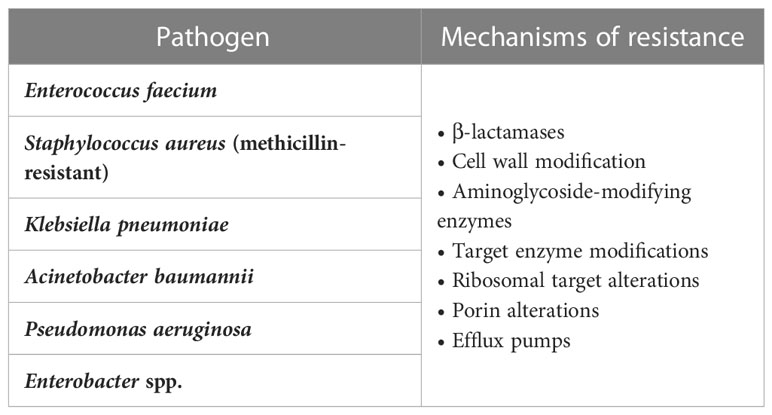
A set of nosocomial pathogens, colloquially termed ESKAPE organisms, are of particular interest because of their propensity for antimicrobial resistance; these organisms are Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species (Rice, 2008). ICU providers face challenges in diagnosing and treating MDR bacteria in the form of differentiating colonization from infection, mimicry of sepsis by non-infective syndromes such as heart failure and inflammatory conditions, the slowness and inaccuracy of current diagnostic modalities of bacterial infections in terms of Gram stains and microbial cultures, and the confounding of signs of organ system dysfunction in sepsis by postoperative complications such as hypotension and hemorrhage (Lam et al., 2018; Kisat and Zarzaur, 2022). Although previous reports have focused on specific genres of either bacteria or antibiotic resistance, this report provides a more comprehensive approach to the problem of antimicrobial resistance in a sample of Gram-positive and Gram-negative bacteria. In this report we will discuss risk factors and mechanisms of antimicrobial resistance with a focus on a group of resistant bacteria frequently encountered in the ICU. We will also provide an update on current and future preventive and therapeutic measures to manage antimicrobial resistance.
Risk factors for acquiring MDR bacteria in the ICU
Risk factors for acquiring MDR bacteria in the ICU can be classified as patient, epidemiological, or geographic factors. Patient factors include an age of more than 60 years; diabetes mellitus; immunosuppression; a history of use of third-generation cephalosporins, vancomycin, or corticosteroids; and chronic liver or kidney disease. Epidemiological factors include duration of hospitalization, use of invasive catheters, hemodialysis, and high colonization pressure in the community. Geographical factors include travel from one geographic area with a high prevalence of MDR to another area (Burillo et al., 2019). Several scoring systems have been developed to predict, and stratify by risk, patients who are prone to become infected by MDR with variable sensitivity, specificity, and predictive values (Vasudevan et al., 2014; Teysseyre et al., 2019).
Types and mechanisms of antimicrobial resistance
Antimicrobial resistance can be broadly categorized into two distinct groups: natural and acquired. Acquired resistance can be a consequence of repeated exposure to antibiotic therapy, leading to gene mutations within a population of bacteria. It can also come about through a phenomenon called horizontal gene transfer (HGT). Indeed, HGT is the single-most important determinant of high-level antibiotic resistance in ICUs around the world and is directly correlated to antibiotic exposure (Baditoiu et al., 2017). Although it is true that genetic mutations impart resistance to a single class of antibiotics, it would take an innumerable amount of time for enough mutations to accumulate and produce resistance to multiple classes within the same organism. HGT bypasses this mechanism via the exchange of genetic material (i.e., DNA) encoding various resistance genes, giving rise to multidrug-resistant (MDR) organisms. ESKAPE organisms are a notable set of nosocomial pathogens because of their ability to evade and adapt to antimicrobial exposure through various modes of resistance (De Oliveira et al., 2020; Prajapati et al., 2021).
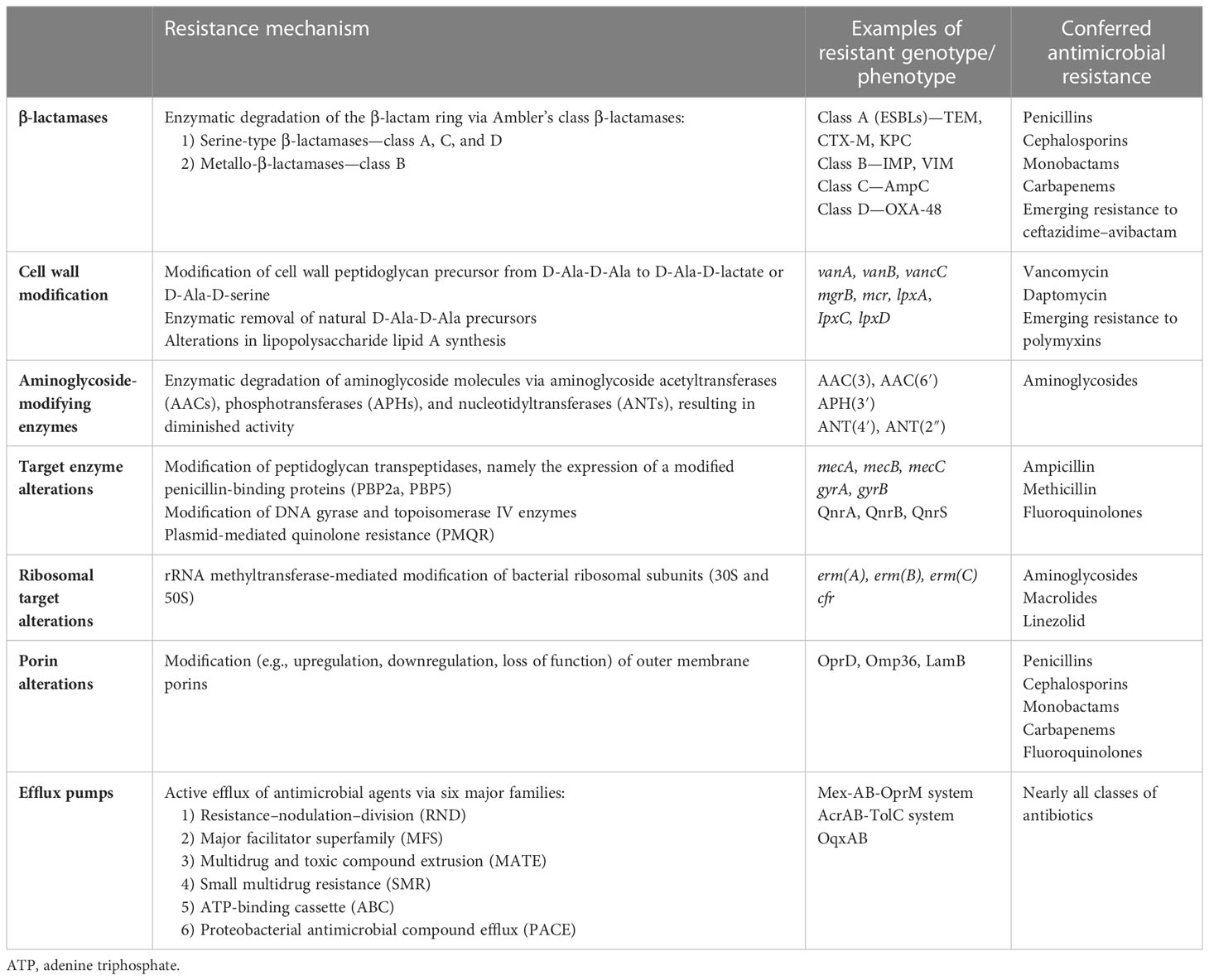
Natural resistance is defined as a genetically encoded trait that is shared by the entire population of a species regardless of antibiotic exposure. For example, vancomycin is a large glycopeptide antibiotic with activity against Gram-positive organisms that inhibits cell wall synthesis by binding the terminal d-Ala, d-Ala of the peptidoglycan precursor.9 Gram-negative bacteria are naturally resistant to the effects of vancomycin owing to the limited permeability of their outer cell wall to this large molecule (https://paperpile.com/c/MbHZBP/CZmw). Another well-documented example is the propensity of members of the genus Enterococcus to have a naturally occurring high level of cephalosporin resistance due in part to an expression of low-affinity penicillin-binding proteins (PBPs), notably PBP5 (Rice et al., 2009). In addition, natural resistance may also manifest itself as a spontaneous genetic mutation, granting resistance to an organism that is usually susceptible to a particular agent. For example, Escherichia coli has been shown to acquire resistance to quinolones through mutations in the quinolone resistance-determining regions of the gyrA and parC genes (Zeng et al., 2020) (Figure 1).
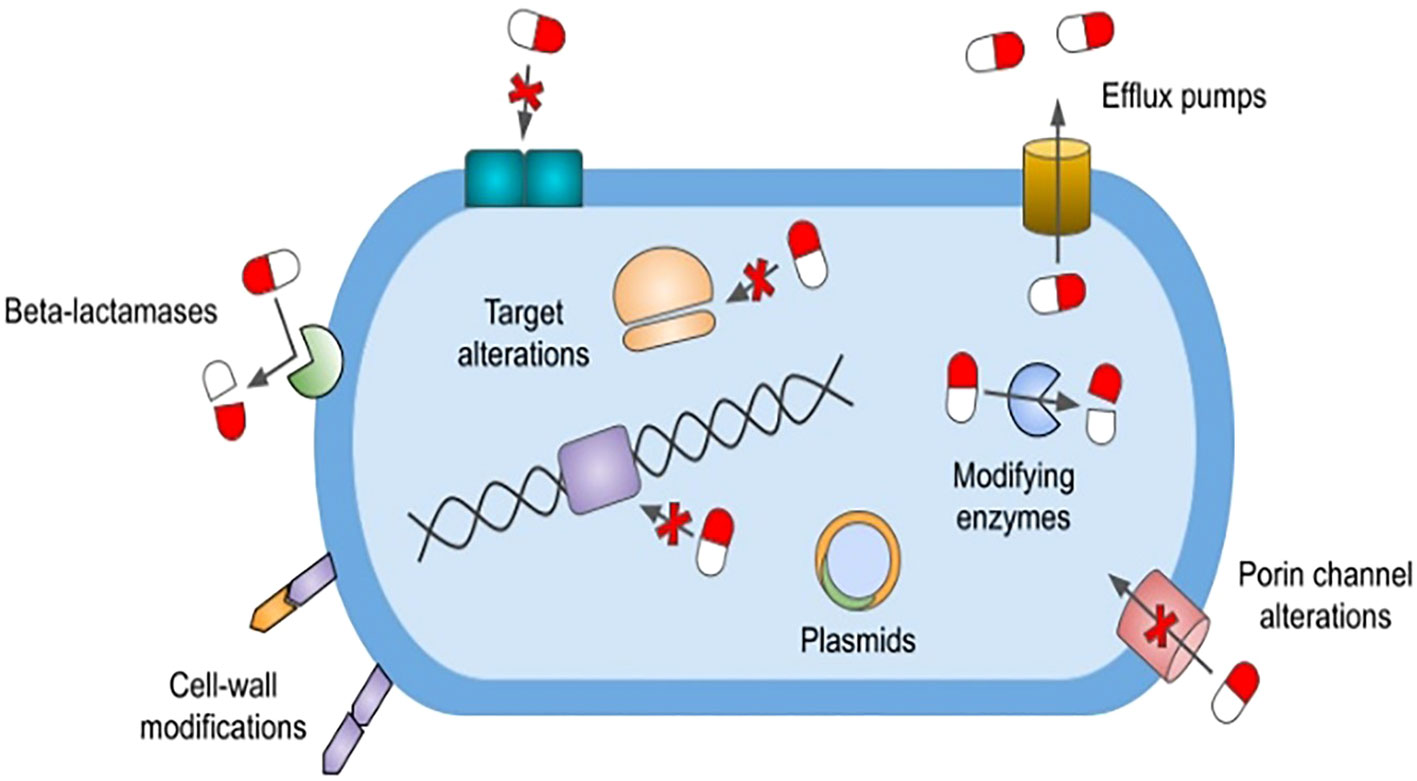
Figure 1 Mechanisms of antimicrobial resistance. The figure depicts various antimicrobial resistance strategies utilized by ESKAPE pathogens. β-lactamases confer various levels of resistance to β-lactam antibiotics and are classified as Amber’s class A, B, C, and D. Cell wall modifications alter the synthesis of lipopolysaccharide lipid A or peptidoglycan precursors, thereby preventing the inhibition of cell wall synthesis. Bacteria can also develop antimicrobial resistance by altering target binding sites via genetic mutations. These alterations include modification to penicillin-binding proteins, alterations to DNA gyrase and topoisomerase IV enzymes, and methyltransferase-mediated modifications to the 30S and 50S ribosomal subunits. Outer membrane porin channel alterations prohibit entry of various antimicrobials into the cell. The expression of transmembrane efflux pumps, classified into six major families, allows bacteria to actively remove nearly all classes of antibiotics from their cytoplasm. Plasmids are small mobile genetic elements composed of extrachromosomal DNA. They can spread resistance genes between bacteria of the same or different species, with the plasmid-mediated quinolone resistance gene being one such example. *ESKAPE pathogens: Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species.
Examples of drug-resistant bacteria
1. Enterococcus faecium isolates can be pathogenic and have a tendency to be highly resistant to multiple classes of antibiotics, including β-lactams (especially cephalosporins), aminoglycosides, and folate reductase inhibitors (i.e., sulfamethoxazole/trimethoprim) (Rosselli Del Turco et al., 2021). An appreciable increase in ampicillin-resistant Enterococcus spp. within U.S. hospitals was noted to occur in the late 1980s and was mainly driven by the widespread use of penicillin in the preceding decades.19 The predominant mechanism of resistance was found to be the development of low-affinity PBPs, specifically PBP5 (Leclercq et al., 1988). Around the same time period, the use of vancomycin was on the rise because of an increase in methicillin-resistant Staphylococcus aureus (MRSA) isolates (D'Agata et al., 2009). Vancomycin-resistant Enterococcus (VRE) isolates were first reported in England in 1988, followed by rapid identification in other parts of Europe and the United States. Resistance to vancomycin is conferred by the vanA, vanB, and vanC genes, which are found within various species of Enterococcus. The inducible vanA and vanB transposons have the propensity for horizontal transfer between various Enterococcus species, whereas the vanC gene is an intrinsic resistance mechanism found in Enterococcus gallinarum and Enterococcus casseliflavus (Arthur and Courvalin, 1993). Although the incidence of VRE infections between 2012 and 2017 decreased, the CDC expects the VRE burden to reach 54,500 cases and lead to 5,400 deaths annually (Reyes et al., 2016).
2. Staphylococcus aureus is of particular concern owing to its ability to develop methicillin resistance via acquisition of the staphylococcal cassette chromosome mec, which carries the resistance gene mecA (Hardy et al., 2004). Since the discovery of MRSA in the 1960s it has been a pathogen of interest for both community- and hospital-acquired infections. A 2005 report by Kuehnert et al. showed that there were 291,542 hospital discharges with S. aureus-related-infection diagnoses between 1999 and 2000. They also reported the overall rate of methicillin resistance for S. aureus to be 43.2% (Kuehnert et al., 2005). More recently, a French study looked at the impact of antibiotic exposure and selection of MRSA in hospital settings. They found that a more prolonged antibiotic exposure in ICUs than in general wards promoted the dissemination of MRSA in the hospital (Patry et al., 2008). In addition, other studies have corroborated the finding that excessive antibiotic exposure promotes the emergence of MRSA infections (Kardas-Sloma et al., 2011). Despite the high utilization rates of antibiotics in ICUs across the nation, a 2019 CDC report reported a 74% drop in the incidence of hospital-acquired MRSA bloodstream infections between 2005 and 2016, with an estimated 119,247 cases of S. aureus bloodstream infections and 19,832 associated deaths in 2017 (Wunderink et al., 2020).
3. Klebsiella pneumoniae is a commensal organism, normally found in the human intestine, and an opportunistic pathogen that can cause a range of infections. The 2019 CDC antimicrobial resistance threat report lists carbapenem-resistant Enterobacterales (CRE) as an urgent threat and extended-spectrum β-lactamase (ESBL)-producing Enterobacterales as a serious threat, with 13,100 estimated cases and 1,100 estimated deaths in hospitalized patients and 197,400 estimated cases and 9,100 estimated deaths in hospitalized patients, respectively, that year (CDC 2022). K. pneumoniae can express a multitude of resistance genes rendering a wide range of β-lactams, including penicillins, broad-spectrum cephalosporins, and carbapenems, ineffective in its treatment. Resistance to penicillins and early-generation cephalosporins can arise from mutations of genes encoding TEM-1, TEM-2, or SHV-1, whereas resistance to third-generation cephalosporins is encoded by CTX-M, OXA, or AmpC β-lactamase (Paterson, 2006). Infections with ESBL-producing organisms place a strain on the healthcare system and are associated with mortality rates ranging between 3.7% and 22.1% (Gutierrez-Gutierrez et al., 2016). A report published in 2014 looked at healthcare-associated infections in 11,282 patients from 183 hospitals. The most frequent infections were found to be pneumonia (21.8%), surgical site infections (21.8%), gastrointestinal infections (17.1%), urinary tract infections (12.9%), and bloodstream infections (9.9%), with the leading causative organisms being Clostridium difficile (12.1%), S. aureus (10.7%), Klebsiella (9.9%), and Escherichia coli (9.3%) (Magill et al., 2014).
4. Acinetobacter baumannii is a Gram-negative opportunistic bacterium, commonly found in soil and water samples, and is a CDC pathogen that poses an urgent threat, with an estimated 8,500 infections and 700 estimated deaths per year (CDC 2022). It can contaminate and survive on the surface of the skin and medical devices, such as ventilator tubing, respirators, and arterial line pressure monitoring devices, for up to 27 days (Jawad et al., 1998). Acinetobacter infections are common in ICUs and can lead to prolonged exposure to antibiotics, increased ICU length of stay, and mortality rates ranging from 13% to 30% (Vincent et al., 2009)(Mathai et al., 2012)(Garnacho-Montero et al., 2015)(Hidron et al., 2008) (Garnacho-Montero et al., 2015). What makes Acinetobacter infections particularly dangerous and difficult to treat is their propensity for rapid and high-level development of resistance to most available antibiotics, including “last-resort” agents such as tigecycline, colistin, and polymyxins (Lee et al., 2017). A. baumannii possesses multiple intrinsic and extrinsic mechanisms of resistance that include a wide range of β-lactamases (e.g., TEM-1, CTX-M, OXA, and class B metallo-β-lactamases), defects in cell wall permeability porin channels, expression of multidrug efflux pumps, and alteration of target sites (PBPs and mutations in DNA gyrase). These mechanisms have to led to the isolation of MDR (resistance to three or more classes of antibiotics), extensively drug-resistant (XDR; MDR plus resistance to carbapenems), and pan-drug-resistant (PDR; XDR plus resistance to polymyxins) bacteria (Breijyeh et al., 2020). New treatment strategies must adapt to developing resistances, whether through the use of “old-school” agents like polymyxin, combination therapies with colistin and carbapenems, or the development of new therapeutic agents such as eravacycline and cefiderocol (Falcone et al., 2021).
5. Pseudomonas aeruginosa is a familiar nosocomial pathogen often found in ICUs across the USA. Although it has been isolated from normal gastrointestinal flora, it may also cause community-acquired and nosocomial infections in immunocompromised hosts (Chopra et al., 2008). Multidrug resistant Pseudomonas is a global health concern and a serious threat according to the CDC, with an estimated 32,600 cases and 2,700 deaths among hospitalized patients in 2017 (CDC 2022). Resistance to multiple classes of antibiotics is attained via intrinsic or acquired mechanisms, including the expression of β-lactamases (e.g., TEM, SHV, CTX-M, OXA, MBLs, and AmpC), target binding site alteration (e.g., 16S ribosomal RNA mutations, aminoglycoside-modifying enzymes, and gyraA and parC mutations), loss of surface porins such as OprD (conferring carbapenem resistance), and overexpression of efflux pumps capable of actively transporting antibiotics out of the cell (Fang et al., 2014). In addition to the mechanisms listed above, Pseudomonas has the ability to survive on devices found in healthcare settings (e.g., ventilators, urinary tract devices, and central line kits) through the formation of biofilm and persister cells (Hong et al., 2015). Prolonged exposure to anti-pseudomonal antibiotics has been reported to promote the selection and proliferation of MDR Pseudomonas and lead to high rates of colonization and prolonged duration of therapy in ICUs around the country (Raman et al., 2018).
6. Enterobacter is a genus of Gram-negative bacteria known to cause drug-resistant nosocomial infections. It has been isolated from the respiratory and urinary tracts, surgical wounds, cardiac and bone tissues, and blood samples of patients residing in ICUs. According to the Surveillance and Control of Pathogens of Epidemiologic Importance (SCOPE) report, Enterobacter was found to cause 4.7% of all ICU infections in US hospitals (Wisplinghoff et al., 2004). Particular attention is being paid to Enterobacter aerogenes and Enterobacter cloacae for their propensity to develop resistance to multiple antibiotics. All Enterobacter are naturally resistant to ampicillin, amoxicillin-clavulanic acid, and cefoxitin via the production of β-lactamases (Wisplinghoff et al., 2004). Moreover, they naturally express the AmpC β-lactamase, which is further inducible by exposure to broad-spectrum cephalosporins and has been found integrated into a large plasmid element capable of horizontal gene transmission (Lee et al., 2003). In addition, resistance to aminoglycosides and fluoroquinolones is acquired via intrinsic mutations or plasmid-encoded drug-modifying enzymes (Mezzatesta et al., 2012). Finally, more recent data show the emergence and spread of hospital-associated carbapenem-resistant E. cloacae across the USA, as well as the emergence of pan-drug-resistant strains, with resistance to colistin, around the world (Diene et al., 2013).
A summary of the mechanisms of MDR to antibiotics is presented in Tables 1 and 2.
Prevention and treatment of antimicrobial resistance
Antibiotic stewardship
Antimicrobial stewardship (AS) refers to an organized program designed to monitor, improve, and measure the responsible use of antibiotics in the ICU. It has emerged to overcome the risks of the inappropriate and inadequate use of antibiotics in the ICU. Inappropriate use of antibiotics describes the very early use of antibiotics with a too-broad spectrum, leading to an increased risk of resistant bacteria. Inadequate use of antibiotics describes the very early use of antibiotics that do not cover suspected organisms (Zilberberg et al., 2014).
Essentials of AS entail the following:
1. Facility-specific guidelines based on national guidelines and local antibiograms.
2. Restrictive programs in which a release of an antibiotic requires approval by an AS team member. This approval is based on cost, spectrum, and risk of side effects from the antibiotic of interest.
3. A persuasive program [also called prospective audit and feedback (PAF)] in which feedback in the form of modification or discontinuation of an antibiotic is provided by an AS team member on antibiotics that are prescribed in a special ICU.
4. Automatic stop orders in which there is a set date after which the prescribed antibiotics require an additional order to extend the duration of the prescription.
5. An antibiotic “time out” in which there is a prompt to review the prescribed antibiotics at a certain time point after initiation.
6. Rapid microbial identification using rapid diagnostic testing, such as real-time polymerase chain reaction (rtPCR), for the diagnosis of the genes of resistant bacteria such as MRSA from positive cultures.
7. Systematic de-escalation of antibiotics based on the kinetics of certain biomarkers, the most studied of which is procalcitonin.
8. Embedding a member of an AS team in ICU rounds.
9. Implementation of alerts in prescription software to guide clinicians at different stages of antibiotic prescription (Doernberg and Chambers, 2017).
Antibiograms
An antibiogram is a snapshot of microbial susceptibility to various antibiotics reported over time. It can provide useful insights into current resistance patterns and past and future trends, and help guide empiric antibiotic treatment. Antibiograms are commonly encountered in a table format that categorizes bacteria as Gram positive or Gram negative and breaks them down further by individual species. Susceptibility rates to commonly used antibiotics are reported as percentages. Depending on facility capabilities, antibiograms can drill down into specific susceptibilities based on patient location, such as ICUs, and sites of infections, such as blood, sputum, or urine. Susceptibility patterns can also have intrahospital variability. A study by Kaufman et al. compared hospital-wide susceptibility data with isolates from surgical intensive care unit (SICU) patients. The two most commonly isolated organisms were S. aureus and P. aeruginosa. Surprisingly, the susceptibility patterns were significantly different between the hospital-wide and SICU data (Kaufman et al., 1998). Another study by Chang et al. (2023) evaluated the effect of empiric antibiotics on mortality in patients with bloodstream infections in the emergency department (ED). They found that empiric antibiotic selection based on a local antibiogram was associated with a reduced mortality rate and a shorter length of stay in the ICU (Chang et al., 2023).
Antibiograms are a useful tool in the clinician’s arsenal to help prevent the emergence of antimicrobial resistance, but they also have some limitations. First, antibiogram reporting is binary, indicating whether or not a specific isolate is susceptible, and does not account for minimum inhibitory concentrations (MICs). The Clinical and Laboratory Standards Institute (CLSI) publishes an annual report with MICs for various bacterial pathogens. According to the 2023 CLSI report, S. aureus with a vancomycin MIC of ≤ 2 µg/ml is considered susceptible (“M100” n.d.). Consequently, every S. aureus isolate with that MIC will be reported as susceptible on an antibiogram. Despite the fact that an antibiogram is technically accurate, it is recommended to avoid using vancomycin to treat S. aureus isolates with an MIC of 2 µg/mL or greater (Rybak et al., 2009). Second, antibiograms may provide false information regarding the timing of cultures, which may lead to erroneous diagnosis of time-dependent classifications, such as community-acquired pneumonia (CAP) compared with hospital-acquired pneumonia (HAP). This in turn may lead to inappropriate use of antibiotics, as the treatment for CAP and HAP is different and aims at covering different pathogens based on indication.
In summary, antibiograms are an important tool in helping ICU practitioners select appropriate empiric antibiotic regimens that will provide a broad coverage and simultaneously help curtail the development of antimicrobial resistance. They also come with important limitations and are meant to be used as an aid and not a replacement for clinical judgment or multidisciplinary collaboration with members of the ICU team, including physicians, mid-level practitioners, and pharmacists.
Surveillance
Surveillance entails monitoring trends in the susceptibilities of microorganisms isolated in one or more ICUs within a hospital, identifying patients colonized or infected with MDR bacteria, and monitoring trends in the use of antibiotics. Practices vary in terms of the population of patients to undergo surveillance, the laboratory assays used to perform surveillance, the ICUs put under surveillance, and the microorganisms surveilled. In addition to the time and expense of routine universal ICU surveillance, there is mixed evidence on whether or not it truly guides initial antibiotic therapy because of its low sensitivity (Hayon et al., 2002; Sanders et al., 2008).
Pharmacokinetic/pharmacodynamic optimization
Alterations in renal function and flow, ranging from augmented renal clearance to acute kidney injury (AKI) and failure requiring the initiation of renal replacement therapy, have strong implications for antibiotics that are renally cleared, such as β-lactams. Patients with augmented renal clearance may need an increased initial dosage or infusion of β-lactam antibiotics to achieve adequate tissue levels. On the other hand, patients with AKI may need renal dose adjustments. An aggressive pharmacodynamic (PK) approach, though not a standard of care at present, may be needed for novel β-lactam antibiotics to reduce resistance (Gatti and Pea, 2021).
Adequate infection control measures
Adequate infection control measures in the form of standard precautions (hand hygiene, gloves, gowns, and eye protection) in addition to transmission (i.e., droplet, airborne, or contact)-based precautions were developed to reduce the transmission of resistant microorganisms from one site to another. Adherence to isolation precautions has been conclusively shown to reduce the transmission of MRSA and extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae (Bassetti et al., 2015).
Adequate source control
Source control is the removal of the source of infection by the drainage of pus or inflammatory material, or debridement of necrotic tissue, and is especially important in surgical patients. Achieving early source control is associated with a reduction in antibiotic duration and resistance. Timing of source control should be guided by the severity of infection, source of infection, and hemodynamic stability of the patient (Kisat and Zarzaur, 2022). Successful signs of source control include, but are not limited to, the resolution of clinical signs of infection, such as fever and leukocytosis, the amelioration of tenderness, radiological resolution in the form of reduction in the size of infected fluid, and the disappearance of previously recognized fistulas or abscesses and a reduction in drainage from tubes and drains (Solomkin et al., 2013).
Conclusion
Antibiotic resistance remains an ongoing challenge for patients admitted to the ICU. The imbalance between limited antibiotic pools on one side and rapidly evolving microbial resistance on the other warrants attention and the implementation of more rigorous preventive strategies, such as rapid diagnostic testing, AS policies, strict infection control measures, and PK/PD drug dosing and monitoring. Novel areas of research, such as studying the interactions of antibiotics and gut microbiota using metagenomic techniques, the protection and restoration of the commensal gut ecosystem via fecal transplantation or orally administered probiotics (Ruppe et al., 2018), and the novel aerosolized routes for administration of antibiotics for lung infections (Montgomery et al., 2014), are urgently needed in the current battle against antimicrobial resistance.
Author contributions
AZ developed the concept and the manuscript design, and wrote and edited the final manuscript draft. RK and DS participated in manuscript design and wrote the initial manuscript draft. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Arthur M., Courvalin P. (1993). Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob. Agents Chemother. 37 (8), 1563–1571. doi: 10.1128/AAC.37.8.1563
Baditoiu L., Axente C., Lungeanu D., Muntean D., Horhat F., Moldovan R., et al. (2017). Intensive care antibiotic consumption and resistance patterns: a cross-correlation analysis. Ann. Clin. Microbiol. Antimicrob. 16 (1), 71. doi: 10.1186/s12941-017-0251-8
Bassetti M., De Waele J. J., Eggimann P., Garnacho-Montero J., Kahlmeter G., Menichetti F., et al. (2015). Preventive and therapeutic strategies in critically ill patients with highly resistant bacteria. Intensive Care Med. 41 (5), 776–795. doi: 10.1007/s00134-015-3719-z
Bell B. G., Schellevis F., Stobberingh E., Goossens H., Pringle M. (2014). A systematic review and meta-analysis of the effects of antibiotic consumption on antibiotic resistance. BMC Infect. Dis. 14, 13. doi: 10.1186/1471-2334-14-13
Breijyeh Z., Jubeh B., Karaman R. (2020). Resistance of gram-negative bacteria to current antibacterial agents and approaches to resolve it. Molecules 25 (6). doi: 10.3390/molecules25061340
Burillo A., Munoz P., Bouza E. (2019). Risk stratification for multidrug-resistant gram-negative infections in ICU patients. Curr. Opin. Infect. Dis. 32 (6), 626–637. doi: 10.1097/QCO.0000000000000599
Campion M., Scully G. (2018). Antibiotic use in the intensive care unit: Optimization and de-escalation. J. Intensive Care Med. 33 (12), 647–655. doi: 10.1177/0885066618762747
Chang C. M., Hsieh M. S., Yang C. J., How C. K., Chen P. C., Meng Y. H. (2023). Effects of empiric antibiotic treatment based on hospital cumulative antibiograms in patients with bacteraemic sepsis: a retrospective cohort study. Clin. Microbiol. Infect. doi: 10.1016/j.cmi.2023.01.004
Chopra I., Schofield C., Everett M., O'Neill A., Miller K., Wilcox M., et al. (2008). Treatment of health-care-associated infections caused by gram-negative bacteria: a consensus statement. Lancet Infect. Dis. 8 (2), 133–139. doi: 10.1016/S1473-3099(08)70018-5
Lee C. R., Park M., KS P., IK B., YB K., et al. (2017). Biology of acinetobacter baumannii: Pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front. Cell Infect. Microbiol. 7. doi: 10.3389/fcimb.2017.00055
D'Agata E. M., Webb G. F., Horn M. A., Moellering R. C. Jr., Ruan S. (2009). Modeling the invasion of community-acquired methicillin-resistant staphylococcus aureus into hospitals. Clin. Infect. Dis. 48 (3), 274–284. doi: 10.1086/595844
De Oliveira D. M. P., Forde B. M., Kidd T. J., Harris P. N. A., Schembri M. A., Beatson S. A., et al. (2020). Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 33 (3). doi: 10.1128/CMR.00181-19
Despotovic A., Milosevic B, Milosevic I, Mitrovic N, Cirkovic A, Jovanovic S, et al. (2020). Hospital-acquired infections in the adult intensive care unit-epidemiology, antimicrobial resistance patterns, and risk factors for acquisition and mortality. Am. J. Infect. Control 48 (10), 1211–1215. doi: 10.1016/j.ajic.2020.01.009
Diene S. M., Merhej V., Henry M., El Filali A., Roux V., Robert C., et al. (2013). The rhizome of the multidrug-resistant enterobacter aerogenes genome reveals how new "killer bugs" are created because of a sympatric lifestyle. Mol. Biol. Evol. 30 (2), 369–383. doi: 10.1093/molbev/mss236
Doernberg S. B., Chambers H. F. (2017). Antimicrobial stewardship approaches in the intensive care unit. Infect. Dis. Clin. North Am. 31 (3), 513–534. doi: 10.1016/j.idc.2017.05.002
Falcone M., Tiseo G., Nicastro M., Leonildi A., Vecchione A., Casella C., et al. (2021). Cefiderocol as rescue therapy for acinetobacter baumannii and other carbapenem-resistant gram-negative infections in intensive care unit patients. Clin. Infect. Dis. 72 (11), 2021–2024. doi: 10.1093/cid/ciaa1410
Fang Z. L., Zhang L. Y., Huang Y. M., Qing Y., Cao K. Y., Tian G. B., et al. (2014). OprD mutations and inactivation in imipenem-resistant pseudomonas aeruginosa isolates from china. Infect. Genet. Evol. 21, 124–128. doi: 10.1016/j.meegid.2013.10.027
Garnacho-Montero J., Dimopoulos G., Poulakou G., Akova M., Cisneros J. M., De Waele J., et al. (2015). Task force on management and prevention of acinetobacter baumannii infections in the ICU. Intensive Care Med. 41 (12), 2057–2075. doi: 10.1007/s00134-015-4079-4
Gatti M., Pea F. (2021). Pharmacokinetic/pharmacodynamic target attainment in critically ill renal patients on antimicrobial usage: focus on novel beta-lactams and beta lactams/beta-lactamase inhibitors. Expert Rev. Clin. Pharmacol. 14 (5), 583–599. doi: 10.1080/17512433.2021.1901574
Gutierrez-Gutierrez B., Perez-Galera S., Salamanca E., de Cueto M., Calbo E., Almirante B., et al. (2016). A multinational, preregistered cohort study of beta-Lactam/beta-Lactamase inhibitor combinations for treatment of bloodstream infections due to extended-Spectrum-beta-Lactamase-Producing enterobacteriaceae. Antimicrob. Agents Chemother. 60 (7), 4159–4169. doi: 10.1128/AAC.00365-16
Hardy K. J., Hawkey P. M., Gao F., Oppenheim B. A. (2004). Methicillin resistant staphylococcus aureus in the critically ill. Br. J. Anaesth. 92 (1), 121–130. doi: 10.1093/bja/aeh008
Hayon J., Figliolini C., Combes A., Trouillet J. L., Kassis N., Dombret M. C., et al. (2002). Role of serial routine microbiologic culture results in the initial management of ventilator-associated pneumonia. Am. J. Respir. Crit. Care Med. 165 (1), 41–46. doi: 10.1164/ajrccm.165.1.2105077
Hong D. J., Bae I. K., Jang I. H., Jeong S. H., Kang H. K., Lee K. (2015). Epidemiology and characteristics of metallo-beta-Lactamase-Producing pseudomonas aeruginosa. Infect. Chemother. 47 (2), 81–97. doi: 10.3947/ic.2015.47.2.81
Jawad A., Seifert H., Snelling A. M., Heritage J., Hawkey P. M. (1998). Survival of acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J. Clin. Microbiol. 36 (7), 1938–1941. doi: 10.1128/JCM.36.7.1938-1941.1998
Kardas-Sloma L., Boelle P. Y., Opatowski L., Brun-Buisson C., Guillemot D., Temime L. (2011). Impact of antibiotic exposure patterns on selection of community-associated methicillin-resistant staphylococcus aureus in hospital settings. Antimicrob. Agents Chemother. 55 (10), 4888–4895. doi: 10.1128/AAC.01626-10
Kaufman D., Haas C. E., Edinger R., Hollick G. (1998). Antibiotic susceptibility in the surgical intensive care unit compared with the hospital-wide antibiogram. Arch. Surg. 133 (10), 1041–1045. doi: 10.1001/archsurg.133.10.1041
Kisat M., Zarzaur B. (2022). Antibiotic therapy in the intensive care unit. Surg. Clin. North Am. 102 (1), 159–167. doi: 10.1016/j.suc.2021.09.007
Kuehnert M. J., Hill H. A., Kupronis B. A., Tokars J. I., Solomon S. L., Jernigan D. B. (2005). Methicillin-resistant-Staphylococcus aureus hospitalizations, united states. Emerg. Infect. Dis. 11 (6), 868–872. doi: 10.3201/eid1106.040831
Lam S. W., Bauer S. R., Fowler R., Duggal A. (2018). Systematic review and meta-analysis of procalcitonin-guidance versus usual care for antimicrobial management in critically ill patients: Focus on subgroups based on antibiotic initiation, cessation, or mixed strategies. Crit. Care Med. 46 (5), 684–690. doi: 10.1097/CCM.0000000000002953
Leclercq R., Derlot E., Duval J., Courvalin P. (1988). Plasmid-mediated resistance to vancomycin and teicoplanin in enterococcus faecium. N Engl. J. Med. 319 (3), 157–161. doi: 10.1056/NEJM198807213190307
Lee S. H., Jeong S. H., Park Y. M. (2003). Characterization of blaCMY-10 a novel, plasmid-encoded AmpC-type beta-lactamase gene in a clinical isolate of enterobacter aerogenes. J. Appl. Microbiol. 95 (4), 744–752. doi: 10.1046/j.1365-2672.2003.02040.x
Magill S. S., Edwards J. R., Bamberg W., Beldavs Z. G., Dumyati G., Kainer M. A., et al. (2014). Multistate point-prevalence survey of health care-associated infections. N Engl. J. Med. 370 (13), 1198–1208. doi: 10.1056/NEJMoa1306801
Mezzatesta M. L., Gona F., Stefani S. (2012). Enterobacter cloacae complex: clinical impact and emerging antibiotic resistance. Future Microbiol. 7 (7), 887–902. doi: 10.2217/fmb.12.61
Montgomery A. B., Vallance S., Abuan T., Tservistas M., Davies A. (2014). A randomized double-blind placebo-controlled dose-escalation phase 1 study of aerosolized amikacin and fosfomycin delivered via the PARI investigational eFlow(R) inline nebulizer system in mechanically ventilated patients. J. Aerosol Med. Pulm Drug Deliv. 27 (6), 441–448. doi: 10.1089/jamp.2013.1100
Paterson D. L. (2006). Resistance in gram-negative bacteria: enterobacteriaceae. Am. J. Med. 119 (6 Suppl 1), S20–S28. doi: 10.1016/j.amjmed.2006.03.013
Patry I., Leroy J., Henon T., Talon D., Hoen B., Bertrand X. (2008). [Evaluation of antibiotic prescription in a french university hospital]. Med. Mal Infect. 38 (7), 378–382. doi: 10.1016/j.medmal.2008.03.009
Prajapati J. D., Kleinekathofer U., Winterhalter M. (2021). How to enter a bacterium: Bacterial porins and the permeation of antibiotics. Chem. Rev. 121 (9), 5158–5192. doi: 10.1021/acs.chemrev.0c01213
Raman G., Avendano E. E., Chan J., Merchant S., Puzniak L. (2018). Risk factors for hospitalized patients with resistant or multidrug-resistant pseudomonas aeruginosa infections: a systematic review and meta-analysis. Antimicrob. Resist. Infect. Control. 7, 79. doi: 10.1186/s13756-018-0370-9
Reyes K., Bardossy A. C., Zervos M. (2016). Vancomycin-resistant enterococci: Epidemiology, infection prevention, and control. Infect. Dis. Clin. North Am. 30 (4), 953–965. doi: 10.1016/j.idc.2016.07.009
Rice L. B. (2008). Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J. Infect. Dis. 197 (8), 1079–1081. doi: 10.1086/533452
Rice L. B., Carias L. L., Rudin S., Hutton R., Marshall S., Hassan M., et al. (2009). Role of class a penicillin-binding proteins in the expression of beta-lactam resistance in enterococcus faecium. J. Bacteriol. 191 (11), 3649–3656. doi: 10.1128/JB.01834-08
Rosselli Del Turco E., Bartoletti M., Dahl A., Cervera C., Pericas J. M. (2021). How do i manage a patient with enterococcal bacteraemia? Clin. Microbiol. Infect. 27 (3), 364–371. doi: 10.1016/j.cmi.2020.10.029
Ruppe E., Burdet C., Grall N., de Lastours V., Lescure F. X., Andremont A., et al. (2018). Impact of antibiotics on the intestinal microbiota needs to be re-defined to optimize antibiotic usage. Clin. Microbiol. Infect. 24 (1), 3–5. doi: 10.1016/j.cmi.2017.09.017
Rybak M. J., Lomaestro B. M., Rotschafer J. C., Moellering R. C., Craig W. A., Billeter M., et al. (2009). Vancomycin therapeutic guidelines: a summary of consensus recommendations from the infectious diseases society of america, the american society of health-system pharmacists, and the society of infectious diseases pharmacists. Clin. Infect. Dis. 49 (3), 325–327. doi: 10.1086/600877
Sanders K. M., Adhikari N. K., Friedrich J. O., Day A., Jiang X., Heyland D., et al. (2008). Previous cultures are not clinically useful for guiding empiric antibiotics in suspected ventilator-associated pneumonia: secondary analysis from a randomized trial. J. Crit. Care 23 (1), 58–63. doi: 10.1016/j.jcrc.2008.01.009
Solomkin J. S., Ristagno R. L., Das A. F., Cone J. B., Wilson S. E., Rotstein O. D., et al. (2013). Source control review in clinical trials of anti-infective agents in complicated intra-abdominal infections. Clin. Infect. Dis. 56 (12), 1765–1773. doi: 10.1093/cid/cit128
Teysseyre L., Ferdynus C., Miltgen G., Lair T., Aujoulat T., Lugagne N., et al. (2019). Derivation and validation of a simple score to predict the presence of bacteria requiring carbapenem treatment in ICU-acquired bloodstream infection and pneumonia: CarbaSCORE. Antimicrob. Resist. Infect. Control. 8, 78. doi: 10.1186/s13756-019-0529-z
Vasudevan A., Mukhopadhyay A., Li J., Yuen E. G., Tambyah P. A. (2014). A prediction tool for nosocomial multi-drug resistant gram-negative bacilli infections in critically ill patients - prospective observational study. BMC Infect. Dis. 14, 615. doi: 10.1186/s12879-014-0615-z
Wisplinghoff H., Bischoff T., Tallent S. M., Seifert H., Wenzel R. P., Edmond M. B. (2004). Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39 (3), 309–317. doi: 10.1086/421946
Wunderink R. G., Srinivasan A., Barie P. S., Chastre J., Dela Cruz C. S., Douglas I. S., et al. (2020). Antibiotic stewardship in the intensive care unit. an official american thoracic society workshop report in collaboration with the AACN, CHEST, CDC, and SCCM. Ann. Am. Thorac. Soc 17 (5), 531–540. doi: 10.1513/AnnalsATS.202003-188ST
Zeng L., Zhang J., Li C., Fu Y., Zhao Y., Wang Y., et al. (2020). The determination of gyrA and parC mutations and the prevalence of plasmid-mediated quinolone resistance genes in carbapenem resistant klebsiella pneumonia ST11 and ST76 strains isolated from patients in heilongjiang province, china. Infect. Genet. Evol. 82, 104319. doi: 10.1016/j.meegid.2020.104319
Keywords: MRSA, antibiotic resistance, antibiotic stewardship, ESBL, multidrug-resistant bacteria
Citation: Karukappadath RM, Sirbu D and Zaky A (2023) Drug-resistant bacteria in the critically ill: patterns and mechanisms of resistance and potential remedies. Front. Antibiot. 2:1145190. doi: 10.3389/frabi.2023.1145190
Received: 21 January 2023; Accepted: 06 June 2023;
Published: 30 June 2023.
Edited by:
Girish B. Nair, Beaumont Health, United StatesReviewed by:
Bahareh Hajikhani, Shahid Beheshti University of Medical Sciences, IranParth Shah, William Beaumont Hospital, United States
Copyright © 2023 Karukappadath, Sirbu and Zaky. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ahmed Zaky, azaky@uabmc.edu
 Riaz M. Karukappadath
Riaz M. Karukappadath