- 1Division of Animal Sciences, University of Zagreb Faculty of Agriculture, Zagreb, Croatia
- 2Faculty of Agriculture, University of Zagreb, Zagreb, Croatia
- 3Centre for Livestock Breeding, Croatian Agency for Agriculture and Food, Zagreb, Croatia
African swine fever (ASF) is a highly contagious viral disease affecting domestic and wild pigs, leading to high mortality rates and significant economic losses. Local pig breeds, such as the Black Slavonian pig, are particularly vulnerable due to increased contact with wild boar. This study aimed to assess the genetic diversity parameters of Black Slavonian pigs in Eastern Croatia following a recent ASF outbreak using pedigree-based analyses. Pedigree data comprising 13,306 animals were analyzed, with a reference population of 1,658 individuals from 2018 onward. Genetic diversity parameters, including the average inbreeding coefficient, average relatedness, and effective population size, were estimated under two scenarios: one assuming no ASF-related culling and another excluding animals culled due to ASF. An optimal contribution selection procedure was applied to minimize relatedness within the population, and mating plans were developed for both scenarios to estimate future inbreeding and relatedness. The average inbreeding coefficient was 5.21% in the scenario without ASF-related culling and 4.27% in the scenario with ASF-related culling. Effective population size was 47.10 in the first scenario and 42.94 in the second, indicating a reduction in genetic diversity. Despite the slightly improved genetic diversity parameters in the ASF scenario, the reduced number of mating candidates increased the risk of inbreeding due to a higher likelihood of pairing related individuals. While genetic diversity remained relatively stable, the results suggest that ASF-related culling influenced population structure by removing highly related animals. However, the long-term impact on genetic variability requires further investigation. Future studies incorporating molecular genetic data would enhance the accuracy of relatedness estimation, as pedigree-based analyses may over- or underestimate genetic diversity due to shallow ancestry records.
1 Introduction
African swine fever (ASF) is a highly contagious viral disease that affects domestic pigs and wild suids, such as warthogs, bush pigs, and feral pigs with mortality that approaches nearly 100%. It is caused by the ASF virus (ASFV), a large, double-stranded DNA virus belonging to the family Asfarviridae (Alonso et al., 2018). Today, ASF is a major concern for the global swine industry due to its high mortality rate and the absence of a commercial vaccine. The first known scientific description of the diseases was provided by Mongomery (1921) who provided comprehensive documentation of its clinical signs and epidemiology. ASF was almost forgotten in the western hemisphere, until it occured in Portugal in 1957 and Spain in 1960. After eradicating the disease in Europe in the end of 20th century, in 2014, the first outbreaks were reported in the European Union, affecting wild boar from the Baltic States and Poland. Since then, the disease has spread to other EU countries and neighbouring non-EU countries, and in recent years there have also been outbreaks in Asia, Oceania and some American countries. In 2023, the first occurrence of ASF was recorded in eastern Croatia in the areas of the Slavonia region on the border with Serbia and Bosnia and Herzegovina.
ASF has devastating consequences for the regions where it appears. Besides high mortality, strict control measures include a ban on the transport of live pigs and pork (Commission Implementing Regulation (EU), 2023). A high mortality caused by disease and strict control measures can affect the number of pigs in infected and restricted area. This can be particularly dangerous for local pig breeds (Zhao et al., 2021), which are characterised by a smaller population size where disease outbreaks can have a very strong impact on the number of individuals in the breed.
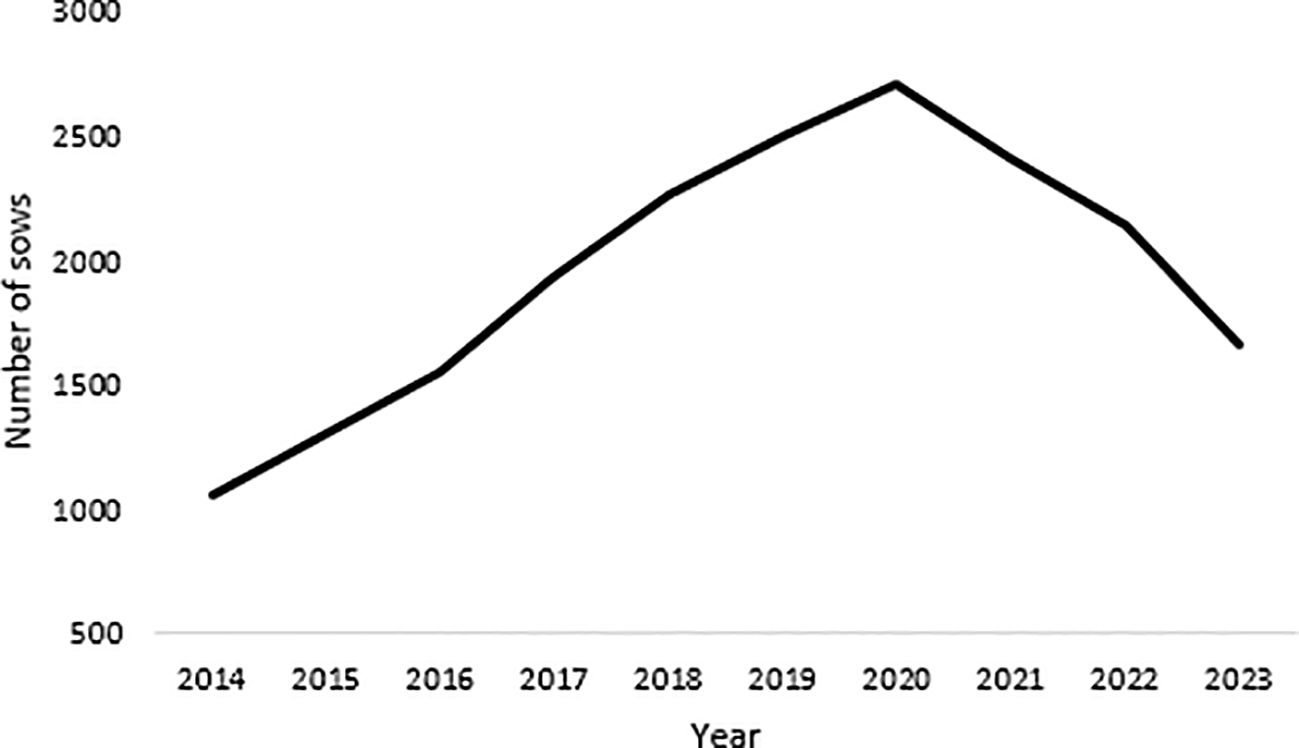
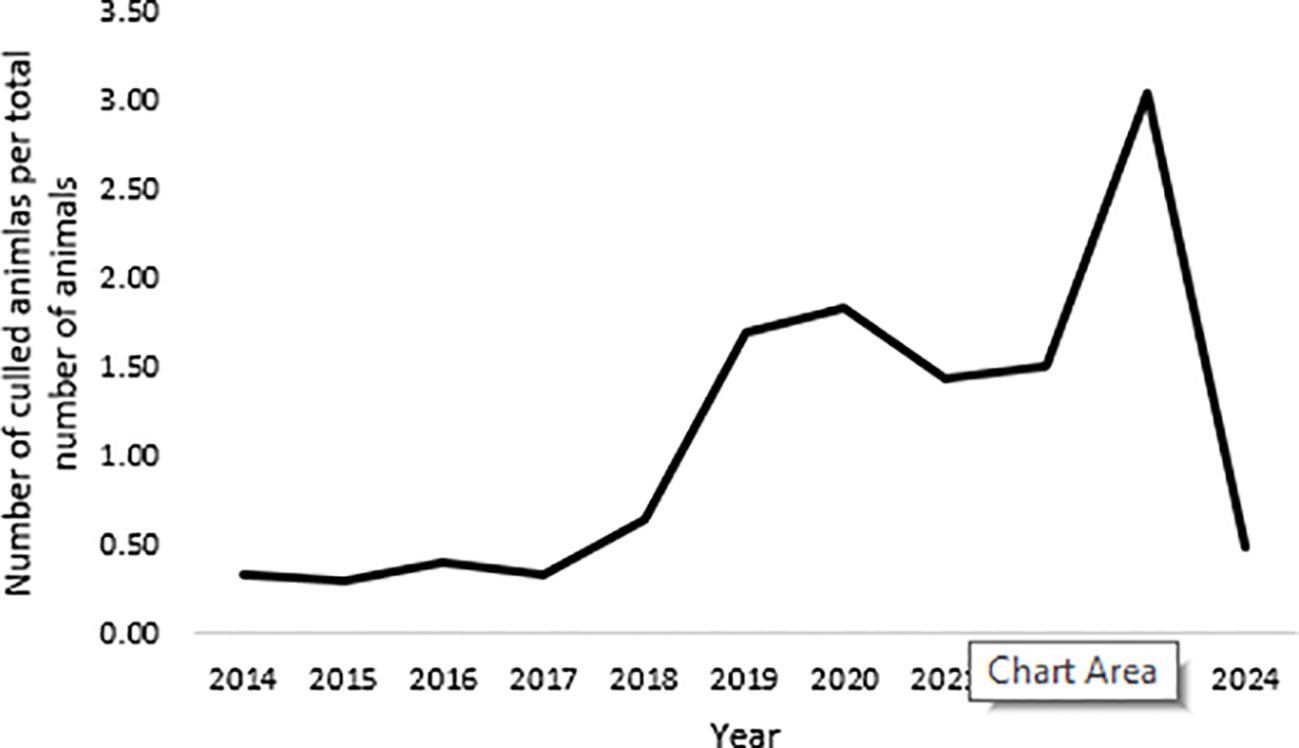
An example of such a breed is the Black Slavonian pig, a Croatian autochthonous black coated pig breed that originated in the 19th century (Figure 1). The breed has modest production characteristics but is known for its very high-quality meat and fat, which is suitable for processing into cured meat products with Protected Geographical Indication (Commission Implementing Regulation (EU), 2017). Originally, breeding area of Black Slavonian pig is Eastern Croatia. However, the breed has spread all over the country in recent years with stable population census (Figure 2). However, the decrease of the population census is recorded even before 2023. The main reasons for this situation are a saturated market and a population size that corresponds to consumer demand. A critical point contributing to this trend is the unfavourable age structure of the breeders. The Black Slavonian pig is mainly bred in rural areas where households are predominantly older. In addition, the low interest of younger generations in pig farming has led to a decline in both the number of farms and the total number of livestock. This trend is also evident in Figure 3, which shows an increase in the number of animals culled after 2017, reflecting similar factors that have contributed to the overall decline in number of animals.
This breed is mostly reared outdoors, making it more exposed to various pathogens, including ASFV. Unfortunately, this became clear in the summer of 2023, when 345 breeding sows and 11 breeding boars died or were culled due to the outbreak of ASF in eastern Slavonia (Figure 4), a region with the largest population of Black Slavonian pigs. Considering that the total number of breeding sows in 2023 was 1,662, one-fifth of the population died or was euthanised due to ASF (Croatian Agency for Food and Agriculture, 2023).
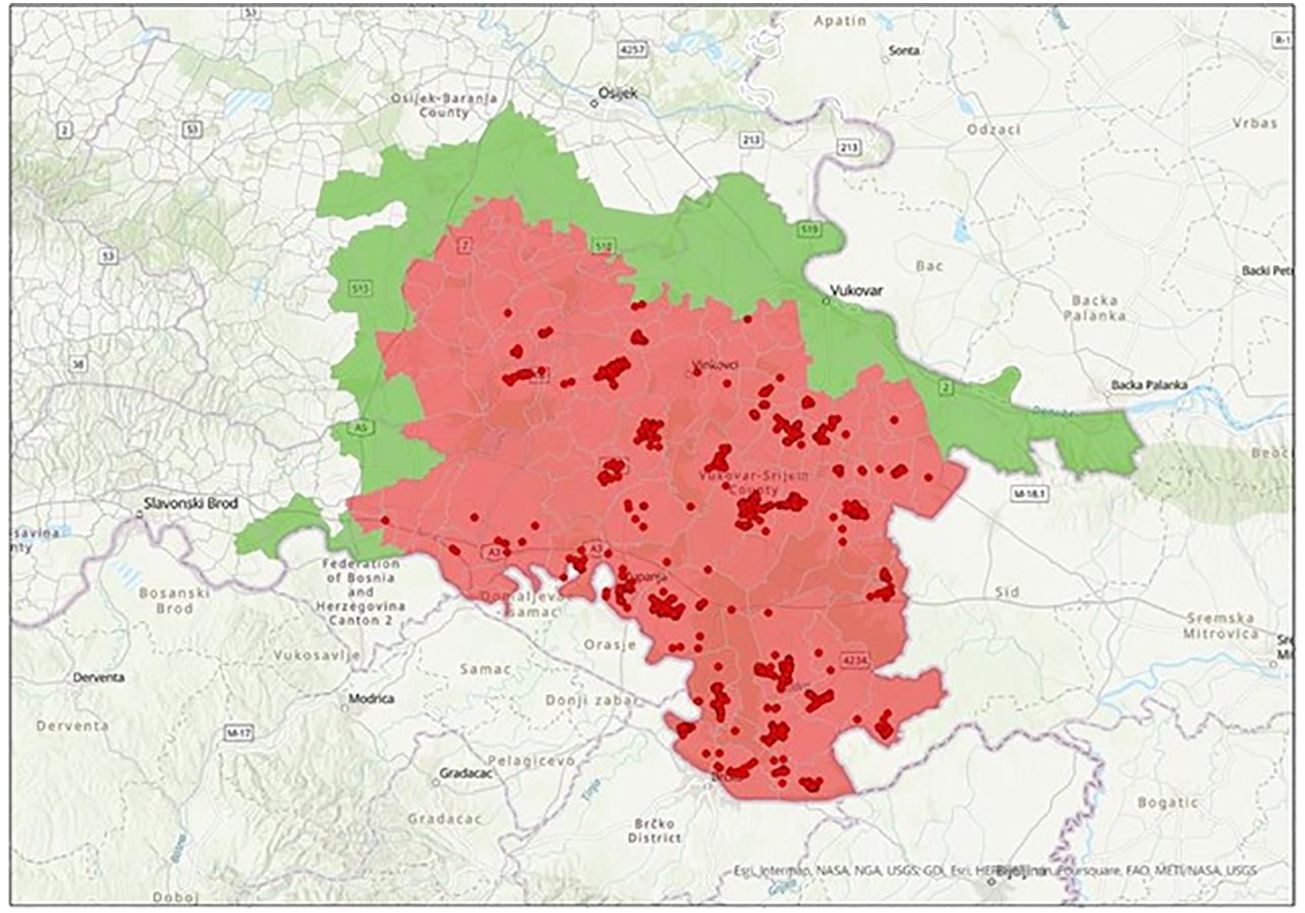
Figure 4. ASF affected area in Slavonia region in Eastern Croatia (red – restricted zone, green - surveillance zone).
While the ecological and economic impacts of ASF are well-documented, its long-term effects on the genetic diversity of domestic local pig breeds remain underexplored. However, there are studies that explored the effect of ASF on the genetic diversity of wild boar Griciuvienė et al. (2021) analysed changes in the genetic structure of Lithuania’s wild boar population after the outbreak of ASF, where the authors find an altered genetic structure between infected and non-infected area. Understanding impacts of ASF on domestic pig populations is crucial for designing effective breeding and conservation strategies. Genetic diversity of small populations is usually impaired, and the main goal of breeding programmes of such populations is based on preserving genetic diversity of population. According to genealogical analyses of the Black Slavonian pig by Škorput et al. (2022) and analysis of genomic information by Zorc et al. (2022), although the population census of the breed has remained stable in recent years, the Black Slavonian pig should be considered as an endangered breed when genetic diversity parameters are taken into account, and preserving its genetic diversity continues to be the primary breeding goal for the population. Given the above, the significant loss of breeding individuals in the population can have a negative impact on genetic diversity of the Black Slavonian pig. The aim of the study was therefore to analyse genetic diversity parameters in the Black Slavonian pig after the ASF epidemic in eastern Croatia using genealogical information and to estimate effects of disease mortality and strict control measures on genetic diversity.
2 Material and methods
2.1 Data
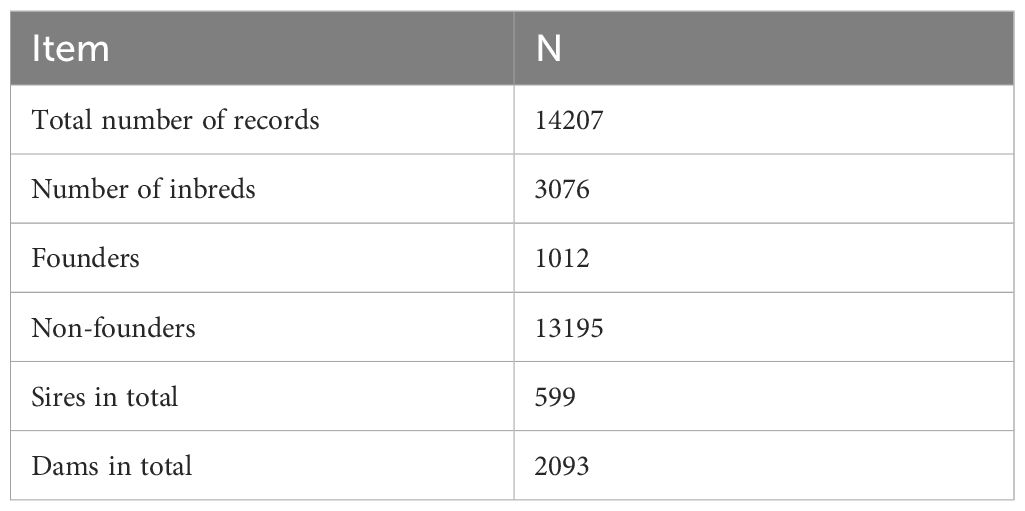
Genealogical information for Black Slavonian pig was provided by Croatian Agency for Agriculture and Food. The pedigree data was available from the start of herdbook recording in 1995. The whole pedigree contained 13 306 animals. However, the reference population was set starting from 2018 with the aim of gaining a better insight into the current genetic diversity of the population contained 1658 individuals. All animals that were culled due to ASF, were coded in the pedigree with the status “positive”. Pedigree structure (Table 1) was determined using CFC software (Sargolzaei et al., 2006). Of the total number of animals in the pedigree, nearly one-fifth were inbred.
2.2 Genetic diversity
To determine the quality and integrity of the pedigree, ENDOG v4.8 software (Gutiérrez and Goyache, 2005) was used. The calculated pedigree quality was evaluated using the average number of maximum generations traced back, the average number of full generations, and number of equivalent generations. The equivalent complete generations is computed as the sum over all known ancestors of the terms computed as the sum of (1/2)n where n is the number of generations separating the individual to each known ancestor (Maignel et al., 1996).
The same software was used to estimate parameters which describe the population structure: the individual and mean inbreeding coefficient (F), defined as the probability that an individual has two identical alleles by descent and calculated using Meuwissen and Luo (1992) algorithm; the average relatedness coefficient (AR) (Goyache et al., 2003; Gutiérrez et al., 2008), defined as the probability that an allele randomly chosen from the whole population in the pedigree belongs to the animal; inbreeding rate (ΔF), and the effective population size (Ne), defined as the number of breeding animals that would lead to the actual increase in inbreeding if they contributed equally to the next generation and usually calculated as follows:
where b is regression coefficient that corresponds to the inbreeding between two generations. Moreover, to obtain reliability of estimation of effective population size, we estimated the effective population size by individual increase in inbreeding (ΔFi) (Gutiérrez et al., 2008), by which we were able to estimate standard error of effective population size:
The average relatedness, the inbreeding coefficient and the effective population size were calculated for two scenarios: in the first scenario, based on the assumption that there was no culling due to ASF parameters were estimated for all available animals. In the second scenario, analysis was performed accounting for animals that were culled due to ASF disease or contact with infected animals.
OptiSel package (Wellmann, 2019) in the R programming environment (R Core Team, 2020) was used to choose available animals in the population and analyse relatedness between mating candidates in the reference population. Moreover, we have created relationship matrix between culled animals. In order to obtain reliable estimation of relatedness between animals, only animals with an equivalent number of generations greater than three were selected as breeding candidates. Adopted optimal contribution selection procedure was used with the aim of minimising relationship between mating candidates in population when creating plans. Moreover, future inbreeding was estimated trough different scenarios accounting for effect of ASF epidemic. The optimisation problems were solved using the CCCP solver from the R package OptiSel. The package contains routines for solving cone constrained convex problems using interior-point methods that are partially ported from Python’s CVXOPT and based on Nesterov–Todd scaling (Vandenberghe, 2010). The solver uses a primal-dual path following algorithms for linear and quadratic cone constrained programming. Mating plans were created for two scenarios:
- In the first scenario average relatedness and future inbreeding were calculated for reference population assuming there was no culling due to ASF epidemic.
- In the second scenario, the same parameters were calculated excluding mating candidates that were culled to ASF epidemic.
- Using all criteria, the total number of mating candidates derived for both scenarios.
3 Results
The trend in the number of culled animals per total number of animals trough years is presented in Figure 2. The sharp increase can be seen in 2023, which coincides with the outbreak of ASF in Croatia and the implementation of strict measures to control ASF. All culls were carried out in restriction zones in eastern Croatia.
Genetic diversity parameters based on genealogical information for the Black Slavonian pig were estimated for a reference population starting from 2018. Population parameters obtained from pedigree (Table 2) indicate impaired genetic diversity of the breed in both scenarios. The average inbreeding coefficient in the referent population was 5.21% in the first scenario and 4.27% in the second scenario, respectively. However, much more informative measures than inbreeding coefficient alone were the changes of average inbreeding coefficient between generations and the effective population size generated from inbreeding rate. Inbreeding rate in both scenarios (1.16% without ASF and 1.06% with ASF, respectively) was higher than 1% and effective population size was lower than 50, which are general FAO recommendation (FAO, 2000) placing Black Slavonian pig, despite stable population size, in breeds with endangered genetic diversity. Moreover, results were obtained for relatively shallow pedigree, which could lead to overestimation of effective population size or underestimation of average relatedness of the population.Differences between the three measures suggest that while some ancestral lines extend deeper (4.51 generations), most individuals lack full generational records (0.47 full generations), which might impact the reliability of pedigree-based genetic analyses. For this reason, the diversity parameters and future scenarios were estimated for individuals with number of equivalent generations higher than three.
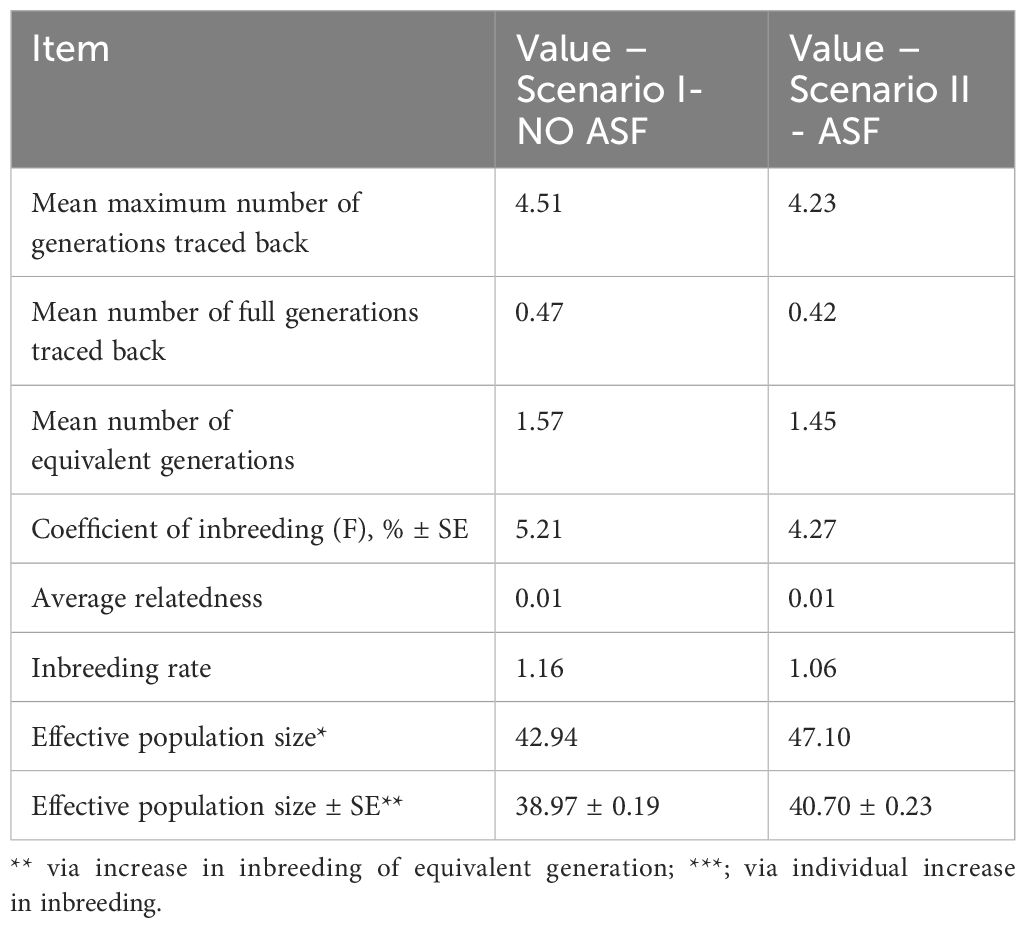
Table 2. Genetic diversity in Black Slavonian pig obtained from pedigree information - *reference population 2018.
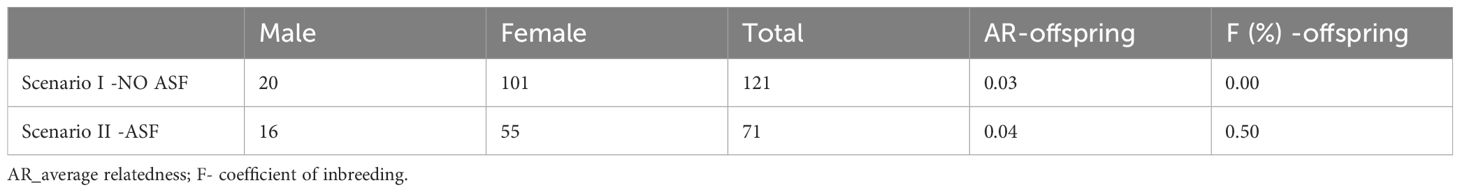
In the first scenario, parameters were estimated including all available mating candidates as well as those culled due to ASF. In the second scenario, parameters were estimated only for animals that remained in the population after ASF occurrence in 2023. Contrary to expectations, the genetic diversity parameters were slightly more favourable after analysing the data in the second scenario. The number of available mating candidates decreased, and less mating candidates were available after the culling of animals and applying strict measures for the control of ASF spreading. Average relatedness between individuals in both scenarios was similar while average relatedness among culled animals was 0.03 and was slightly higher than average relatedness in both scenarios.
The number of mating candidates was significantly reduced after the outbreak of ASF. The number of available boars decreased from 20 to 16 and the number of available sows decreased from 101 to 55. This reduction has not resulted in change in relatedness between mating candidates. Although the analysis of population parameters on the reference population has not shown detrimental effect of ASF on genetic diversity, the reduced number of mating candidates resulted in higher probability of mating between more closely related animals and an increase in the average relatedness of the offspring and consequently results in an increase in inbreeding in the next generation (Table 3).
4 Discussion
Despite the stable number of individuals in the Black Slavonian pig breed in recent decades, the genetic diversity of the population remains one of the main problems for sustainable breeding and existence of the breed, as with most local pig breeds. In most cases, the preservation of genetic diversity in indigenous breeds remains one of the main goals of breeding programs, especially in populations where sustainable conservation is not achieved (Muñoz et al., 2019). Genetic diversity of Black Slavonian pig was analysed in several studies and based on different data type (Gvozdanović et al., 2020; Zorc et al., 2022). All studies to date indicate that despite the relatively large number of individuals in the population, a careful approach to maintaining genetic diversity and management is required, focusing on matings that minimise the increase in inbreeding. Although the mating design within the breeding programme aims to reduce the average relatedness, the diversity parameters retain marginal values. The inbreeding rate exceeds 1%, and the effective populations size remains just below 50, which is a clear indication that inbreeding depression may occur in the population in the future if the decline of the genetic diversity is to continue. For this reason, an analysis of inbreeding depression combining pedigree data should be considered. However, Škorput et al. (2022) showed that although the main breeding goal in the Black Slavonian pig is a conservation of genetic diversity, genetic improvement of economically important traits such as number of piglets born alive or meat quality is possible through the application of the optimal contribution selection frame.
Studies on direct relation between genetic diversity in domestic and ASF are rare. Mujibi et al. (2018) analysed genetic diversity of Kenyan domestic pigs in an area known to be endemic for ASF. Authors highlighted concerns that crossbreeding with commercial breeds, often in response to disease pressures like ASF, threatens the genetic diversity of local pigs. However, authors noticed that a proportion of indigenous pigs have tested ASFV positive but do not present with clinical symptoms of disease indicating some form of tolerance to infection. This fact represents a basis to explore possible genetic determinants underlying tolerance to infection by ASF and prevent the loss of genetic material and possible bottleneck in local pig breeds. Indeed, in a recent study, Qi et al. (2024) investigated the genetic basis for natural resistance to ASF in the local Chinese Xiang pig breed and identified adaptive genes affected by population-stratified genomic structural variants between ASF-susceptible and –resistant pigs.On the contrary, Tsybenko and Vashchenko (2020) reported critical reduction of genealogical lines and strong loss of genetic diversity in of the Mirgorod pig after outbreak of ASF in Ukraine a s a consequence not only of high mortality due to ASF, but also because of strict measure for controlling the disease.
Although a substantial part of the Black Slavonian pig population was affected by ASF, there was no significant decrease in genetic diversity parameters estimated from pedigree. Moreover, the estimated parameters were slightly better (lower inbreeding rate and higher effective population size) in the second scenario that accounted for losses due to ASF. There are several possible explanations for such a situation. First, ASF affected only relatively small geographic areas, and selective mortality disproportionately affected individuals with higher relatedness (e.g. due to shared susceptibility genes), so the surviving population might show lower average relatedness and inbreeding. ASF may have reduced the overall population size but also reduced competition between closely related individuals, leading to a more balanced contribution to reproduction and an increase in effective population size. However, it is crucial to consider whether this effect is sustainable or whether diversity will decline over subsequent generations due to the reduced population size. If breeding is limited to a smaller number of surviving individuals, long-term diversity could still be at risk despite the temporary increase in genetic variation. At the genetic basis, ASF may have caused a population bottleneck, selectively reducing certain genetic lineages while allowing others to persist. If the surviving population retained a more balanced representation of genetic variation compared to the pre-ASF population, this could temporarily result in higher genetic diversity estimates. If individuals with higher heterozygosity or rare alleles had a survival advantage, their genetic contribution to the next generations would be disproportionately higher, leading to an apparent increase in diversity. Additionally, the sudden reduction in population size could lead to a shift in allele frequencies due to genetic drift, amplifying genetic variability in ways not typically observed in stable populations. If breeding is limited to a small number of surviving individuals, long-term diversity could still be at risk despite the temporary increase in genetic variation.
It is common situation that pedigree records of local pig breeds are not fully representative due to missing or incorrect data. Thus, if pedigree data do not fully capture the animals lost due to ASF or if there is incomplete recording of affected individuals, the perceived genetic diversitys might appear artificially better in the ASF scenario. This can be overcome by using molecular information, and the combination of genealogical and molecular data could give a better insight into the status of genetic diversity (Álvarez et al., 2008). However, sampling of animals on farms in infected areas is not encouraging due to strict biosecurity measures and genealogical information remains a method of choice during the epidemic.
Future studies should include contemporary molecular methods for estimating genetic diversity parameters, such as inbreeding determined by identifying runs of homozygosity (ROH) or effective population size estimated from the linkage disequilibrium (LD) analysis. Also, molecular information might be useful in determining relatedness between the remaining animals. In this way, molecular tools can help assess the genetic impact of African swine fever at a finer scale, complement pedigree-based estimates of diversity loss. Moreover, genomic data can be useful for estimating inbreeding depression for economically important traits, such as fertility of fitness. However, due to the strict control measures, sampling pigs is currently not recommended in order to minimise the risk of disease introduction into healthy herds.
The occurrence of ASF in Slavonia in eastern Croatia, the region with the highest number of registered Black Slavonian pigs, resulted with the loss of 20% of herdbook animals. In addition to the direct losses from the culling of infected animals and those that have been in contact with the infected animals, the consequences on the ability to select breeding candidates have also been reduced due to strict measures to control the spread of ASF. As a result, fewer mating candidates were available and the choice for farmers was limited. A complicating factor is that most of the losses occurred in the region with the largest number of Black Slavonian pigs. Škorput et al. (2018) reported a high level of genetic connectedness of herds from infected region with all other herds of Black Slavonian pigs. The herds in the eastern part of the country were pioneer herds in the preservation of the breed and the main source of genetic material for the herds in other regions of the country. The possible detrimental effect of the loss of relatively large number of animals might lead to genetic bottlenecks and a loss of genetic variation. Even though the overall genetic diversity parameters in the population remained stable, the reduced number of mating candidates resulted in a higher likelihood of pairing related individuals, which might result with increased inbreeding rate and a deterioration of genetic diversity parameters in future generations. Despite the reduction in the number of mating candidates, the maintenance of general genetic diversity parameters in the Black Slavonian pig population suggests some level of resilience. However, proactive measures are necessary to sustain this genetic stability. First of all, biosecurity measures that can prevent further outbreaks of ASFV in the population must be conducted strictly. Furthermore, to counteract the loss of genetic diversity, targeted breeding programs that prioritise maximising genetic diversity should be implemented. Introducing less related individuals or utilising molecular markers for selection could help prevent further erosion of genetic diversity. One of the key measures that should be considered is the creation of a protected nucleus of pig breeds aimed at preserving the genetic pool. Moreover, Black Slavonian pigs should be further dispersed to ensure that the occurrence of ASF or other diseases does not have devastating effects on the population’s status and its genetic diversity.
5 Conclusions
Despite the stable number of animals in the population over several years, the estimated parameters indicated low genetic diversity in the Black Slavonian pig population. With the occurrence of ASF in 2023, the genetic diversity of the population was additionally expected to be contracted by culling herdbook boars and sows from the population. Moreover, the implementation of restrictive measures for control of ASF did not allow the exchange of genetic material that origins from restricted areas. The result of the disease outbreak was a reduced number of breeding candidates and available mating combinations. However, we have not observed the significant impact of culling a large number of pigs (approximately one-fifth) from the population on the overall genetic diversity based on genealogical information. Moreover, diversity parameters from pedigree were slightly better when analysis was done with ASF scenario, probably due to the culling of highly related animals. Results are also highly dependent on pedigree completeness that is often a problem in local breeds. Future studies should focus on the use of molecular information that could be useful for determining relatedness between mating candidates to support genealogical information and a more reliable estimation of key parameters of genetic diversity.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation. Requests to access these datasets should be directed to Dubravko Škorput,ZHNrb3JwdXRAYWdyLmhy.
Author contributions
DŠ: Conceptualization, Data curation, Investigation, Methodology, Software, Supervision, Writing – original draft, Writing – review & editing. IS: Conceptualization, Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing. MŠ: Resources, Writing – original draft, Writing – review & editing. DK: Methodology, Supervision, Validation, Writing – original draft, Writing – review & editing. AK: Investigation, Methodology, Writing – original draft, Writing – review & editing. ZL: Supervision, Writing – original draft.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alonso C., Borca M., Dixon L., Revilla Y., Rodriguez F., Escribano J. M., et al. (2018). ICTV virus taxonomy profile: asfarviridae. J. Gen. Virol. 99, 613–614. doi: 10.1099/jgv.0.001049
Álvarez I., Royo L. J., Gutiérrez J. P., Fernández I., Arranz J. J., Goyache F. (2008). Relationship between genealogical and microsatellite information characterizing losses of genetic variability: empirical evidence from the rare Xalda sheep breed. Livest Sci. 115, 80–88. doi: 10.1016/j.livsci.2007.06.009
Commission Implementing Regulation (EU). (2017). 2017/1992 of 6 November 2017 entering a name in the register of protected designations of origin and protected geographical indications (‘Slavonski kulen’/’Slavonski kulin’ (PGI)) (Text with EEA relevance). Off J. Eur. Communities. L288, 2–3.
Commission Implementing Regulation (EU). (2023). 2023/594 of 16 March 2023 laying down special disease control measures for African swine fever and repealing Implementing Regulation (Text with EEA relevance). Off J. Eur. Communities. L79, 65.
Croatian Agency for Food and Agriculture (2023). Annual report (Osijek, Croatia: Croatian Agency for Food and Agriculture).
FAO (2000). Secondary guidelines for development of national farm animal genetic resources management plans: Management of small populations at risk (Rome: Food and Agriculture Organization).
Goyache F., Gutierrez J. P., Fernandez I., Gomez E., Alvarez I., Dıez J., et al. (2003). Using pedigree information to monitor genetic variability of endangered populations: the Xalda sheep breed of Asturias as an example. J. Anim. Breed Genet. 120, 95–103. doi: 10.1046/j.1439-0388.2003.00378.x
Griciuvienė L., Janeliūnas Ž., Jurgelevičius V., Paulauskas A. (2021). The effects of habitat fragmentation on the genetic structure of wild boar (Sus scrofa) population in Lithuania. BMC Genom. Data 22, 53. doi: 10.1186/s12863-021-01008-8
Gutiérrez J. P., Cervantes I., Molina A., Valera M., Goyache F. (2008). Individual increase in inbreeding allows estimating realised effective sizes from pedigrees. Genet. Sel. Evol. 40, 359–378. doi: 10.1186/1297-9686-40-4-359
Gutiérrez J. P., Goyache F. (2005). A note on ENDOG: a computer program for analysing pedigree information. J. Anim. Breed Genet. 122, 172–360. doi: 10.1111/j.1439-0388.2005.00512.x
Gvozdanović K., Škorput D., Djurkin Kušec I., Salajpal K., Kušec G. (2020). Estimation of population differentiation using pedigree and molecular data in Black Slavonian pig. Acta Fytotech. Zootech. 23, 241–249. doi: 10.15414/afz.2020.23.mi-fpap.241-249
Maignel L., Boichard D., Verrier E. (1996). Genetic variability of French dairy breeds estimated from pedigree information. Interbull Bull. 14, 49–54.
Meuwissen T. I., Luo Z. (1992). Computing inbreeding coefficients in large populations. Genet. Sel Evol. 24, 305–313. doi: 10.1186/1297-9686-24-4-305
Mongomery R. E. (1921). On A form of swine fever occurring in British East Africa (Kenya colony). J. Comp. Pathol. Ther. 34, 159–191. doi: 10.1016/S0368-1742(21)80031-4
Mujibi F. D., Okoth E., Cheruiyot E. K., Onzere C., Bishop R. P., Fèvre E. M., et al. (2018). Genetic diversity, breed composition and admixture of Kenyan domestic pigs. PloS One 13, e0190080. doi: 10.1371/journal.pone.0190080
Muñoz M., Bozzi R., Garcia-Casco J., Nuñez Y., Ribani A., Franci A., et al. (2019). Genomic diversity, linkage disequilibrium and selection signatures in European local pig breeds assessed with a high density. Sci. Rep. 9, 13546. doi: 10.1038/s41598-019-49830-6
Qi F., Chen X., Wang J., Niu X., Li S., Huang. S., et al. (2024). Genome-wide characterization of structure variations in the Xiang pig for genetic resistance to African swine fever. Virulence 15, 2382762. doi: 10.1080/21505594.2024.2382762
R Core Team (2020). A language and environment for statistical computing (Vienna (Austria: R Foundation for Statistical Computing).
Sargolzaei M., Iwaisaki H., Colleau J. J. (2006). “CFC: a tool for monitoring genetic diversity,” in Proceedings of the 8th World Congress on Genetics Applied to Livestock Production (CD Comm, Belo Horizonte (Brazil), 27–28.
Škorput D., Špehar M., Luković Z. (2018). Connectedness between contemporary groups in Black Slavonian pig. Liv. Sci. 216, 6–8. doi: 10.1016/j.livsci.2018.06.012
Škorput D., Špehar M., Luković Z. (2022). Managing genetic diversity in pig populations: implications of optimal contribution selection in the Black Slavonian pig. Ital. J. Anim. Sci. 21, 1259–1267. doi: 10.1080/1828051X.2022.2104661
Tsybenko V. H., Vashchenko P. A. (2020). Genealogical analysis of the Mirgorod pig breed before and after outbreak of African swine fever. Veterinary Sci. Technol. Anim. Husbandry Nat. Manage. 5, 216–221. doi: 10.31890/vttp.2020.05.38
Vandenberghe L. (2010). The CVXOPT linear and quadratic cone program solvers. University of California, Los Angeles. Available online at: http://www.seas.ucla.edu/vandenbe/publications/ (Accessed 2022 Jul 22).
Wellmann R. (2019). Optimum contribution selection for animal breeding and conservation: the R package OptiSel. BMC Bioinf. 20, 25. doi: 10.1186/s12859-018-2450-5
Zhao Q., Liu H., Raza Qadri Q., Wang Q., Yuchung P., Guosheng S. (2021). Long-term impact of conventional and optimal contribution conservation methods on genetic diversity and genetic gain in local pig breeds. Heredity 127, 546–553. doi: 10.1038/s41437-021-00484-z
Keywords: pigs, African swine fever, genetic diversity, conservation, inbreeding
Citation: Škorput D, Stupnišek I, Špehar M, Karolyi D, Kaić A and Luković Z (2025) Effect of African swine fever on genetic diversity in Black Slavonian pig. Front. Anim. Sci. 6:1553271. doi: 10.3389/fanim.2025.1553271
Received: 30 December 2024; Accepted: 24 February 2025;
Published: 11 March 2025.
Edited by:
Regina Roessler, University of Kassel, GermanyReviewed by:
Loreta Griciuvienė, Vytautas Magnus University, LithuaniaElżbieta Jadwiga Szymańska, Warsaw University of Life Sciences, Warsaw, Poland
Copyright © 2025 Škorput, Stupnišek, Špehar, Karolyi, Kaić and Luković. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana Kaić, YWthaWNAYWdyLmhy
 Dubravko Škorput
Dubravko Škorput Ivan Stupnišek
Ivan Stupnišek Marija Špehar3
Marija Špehar3 Danijel Karolyi
Danijel Karolyi Ana Kaić
Ana Kaić