- 1Center for Dryland Agriculture, Bayero University, Kano, Nigeria
- 2Faculty of Veterinary Medicine, Bayero University, Kano, Nigeria
- 3Faculty of Earth and Environmental Sciences, Bayero University, Kano, Nigeria
- 4Department of Biology and Biotechnology, Worcester Polytechnic Institute, Worcester, MA, United States
- 5Program in Bioinformatics and Computational Biology, Worcester Polytechnic Institute, Worcester, MA, United States
Through enteric fermentation, ruminants convert fibrous biomass into high-quality proteins like meat and milk. In this process however, methanogenic archaea in the ruminant gastrointestinal tract produce methane, a potent greenhouse gas, from the by-products of enteric fermentation: carbon dioxide and hydrogen. Research in ruminant methane mitigation has been extensive, and over the years has resulted in the development of a wide variety of mitigation strategies ranging from cutting our meat consumption, to breeding low emitting cows, to targeting the rumen microbiome. Methods like promotion of reductive acetogenesis, a natural alternative pathway to methanogenesis in the rumen, are at the forefront of rumen microbiome engineering efforts. However, our inability to make acetogenesis a key hydrogen scavenging process in the rumen have limited these manipulation efforts. Herein we comprehensively review these mitigation strategies, with particular emphasis on mechanisms involving the manipulation of rumen acetogenesis. Such manipulation includes the genetic reprogramming of methanogens for reductive acetogenesis. With the advent of CRISPR-Cas genome editing technologies, the potential exists to transform dominant methane-producing archaea, such as Methanobrevibacter ruminantium, into acetate producing organisms. Acetate can, in turn, be utilized by the animal to increase meat and milk production, thus simultaneously reducing emissions and increasing efficiency. The current status and future challenges of these strategies are discussed. We propose that CRISPR offers a promising avenue for sustainable ruminant farming.
1 Methane in global warming and climate change
Greenhouse gases (GHGs) trap heat energy and contribute to global warming, and have been rising dramatically in recent years. Among the greenhouse gases produced by human activities, carbon dioxide (CO2), methane (CH4), and nitrous oxide (N2O) make up the largest proportions, accounting for approximately 75%, 18%, and 4% of the total volume, respectively (Subedi et al., 2022). Although methane ranks second to carbon dioxide in volume, it has a more significant impact on the rate of warming due to its higher global warming potential. Methane warms the earth 86 times more efficiently than carbon dioxide over a 20-year timescale (Cain et al., 2019). However, unlike CO2 which remains in the atmosphere for hundreds of years, methane has a much shorter atmospheric lifetime of 10–12 years (Stavert et al., 2022). Therefore, targeting the emission of CH4 will more immediately slow global warming compared to targeting CO2.
The impact of CH4 includes two major components: climate warming and atmospheric pollution. Methane warms the earth directly by its own radiative forcing (RF) of 0.97 Watts per square meter (W/m2). Oxidation of CH4 in the atmosphere by hydroxyl (OH) radicals or nitrous oxide produces ozone (O3), CO2, and water vapor (Rigby et al., 2017) which add weight to the RF of methane. For instance, when one molecule of CH4 oxidizes in the presence of N2O, it yields an average of 2.7 molecules of ozone, which increases methane’s greenhouse effect by 30% (Collins et al., 2018).
The contribution of methane to atmospheric pollution is primarily driven by ozone, with methane oxidation accounting for approximately 15% of the tropospheric ozone burden (Collins et al., 2018). Ozone poses significant threats to human health, agriculture and ecosystem balance. Globally, about 1 million premature deaths occur annually due to respiratory illnesses resulting from ozone exposure. Half of these deaths are attributable to anthropogenic CH4 emissions (McDuffie et al., 2023). In addition, ozone exposure affects crop growth and productivity. Recently, Mills et al. (2018) reported a loss of 537 million tons of crop yield between the year 2017 and 2019 due to ozone exposure.
The current global emission of methane is approximately 600 teragrams per year (Tg/yr) (Calabrese et al., 2021). This amount is approximately 2.6 times the pre-industrial era emission and accounts for 0.5°C of the current global warming. Generally, research suggests that all significant methane sources are human driven and largely biogenic in nature, i.e., they involve microbial fermentation. Recent estimates suggest emissions from the year 2007 to present are 85% from microbial sources with about half of it coming from the tropics (Basu et al., 2022).
Human derived methane microbial emissions fall under three categories: livestock production (115 Tg CH4/yr), landfills and waste (68 Tg CH4/yr), and rice paddies (30 Tg CH4/yr) (Saunois et al., 2019). Within the livestock sector, emissions from enteric fermentation contribute the largest proportion (85%) amounting to 98 Tg CH4 /yr (Saunois et al., 2020). Cattle account for the majority of enteric fermentation CH4 emissions from livestock worldwide, due to their large population (1.5 billion animals), large rumen size, and digestive characteristics (Malik et al., 2021). On average a beef cow is estimated to emit up to 500 litres (equivalent to 328.5 grams) of methane per day, or nearly 120 kg per animal per year (Figure 1) which exit the animal mainly (95%) through the mouth (Cezimbra et al., 2021) This methane not only harms the environment, but also impacts animal performance as it accounts for about 12%–15% loss of dietary energy (Tapio et al., 2017).
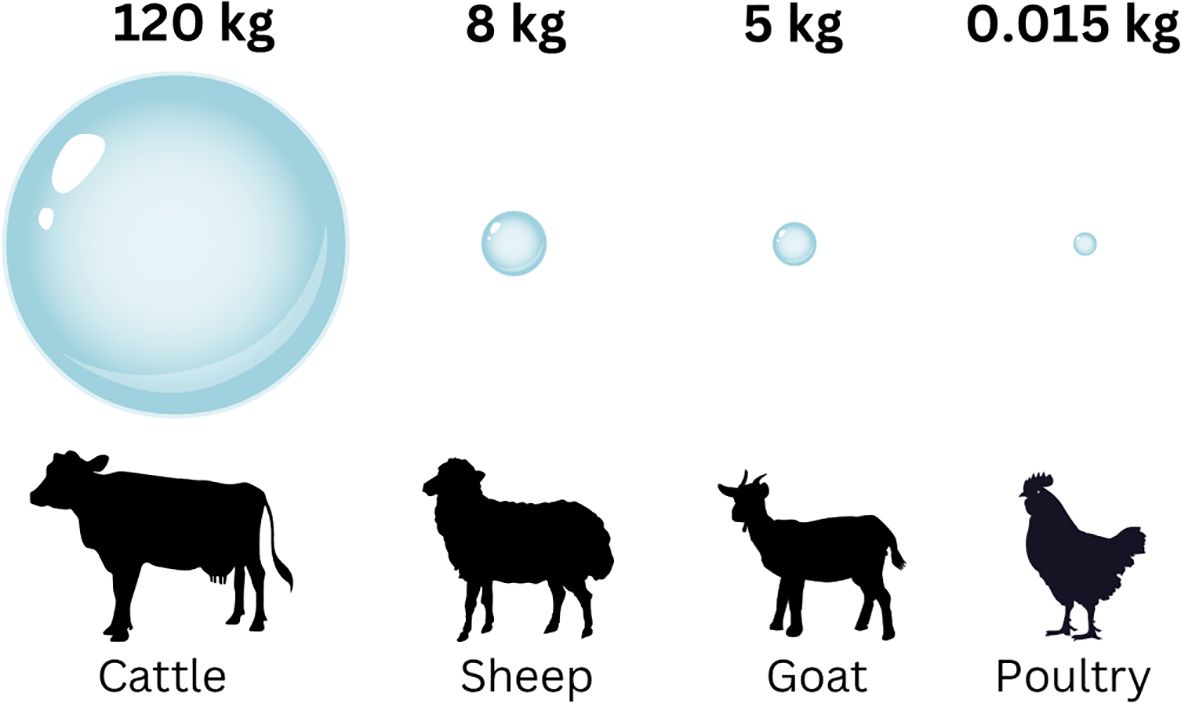
Figure 1. Global yearly emissions of methane by different species. Emissions are shown in kg per animal per year.
As the global population grows and economies strengthen, both consumption and waste production will increase, leading to higher microbial emissions. In the case of the enteric fermentation, by the year 2050 the demand for ruminant products is predicted to increase for meat and milk production by 76% and 63% respectively from their levels in 2010 (FAO, 2018). This extensive production coupled with already existing extreme weather events will exacerbate microbial emissions. For instance, there is strong evidence that a prolonged dry season in the drylands reduces feed quality and causes feed scarcity, both of which reduce digestibility and increase emissions from enteric fermentation (Tadesse and Dereje, 2018). On the other hand, prolonged rain seasons expand ruminant feed sources, which in turn promote overfeeding consequently increasing emissions (Schaefer et al., 2016). Increasing temperatures and additional rainfall creates and expands ideal conditions for methane-emitting archaea in landfills and wetlands (Moomaw et al., 2018).
Methane is therefore an attractive and cost-effective target for climate mitigation strategies given its potency as a GHG and its shorter lifetime in the atmosphere. By targeting methane, visible impact on the climate will be observed within the next few decades. For instance, it is estimated that if emissions from livestock enteric fermentation were entirely halted, the climate would cool by 0.3°C by 2045 (Reisinger et al., 2021).
This review begins by detailing the landscape of broad strategies of methane mitigation in ruminant animals. Then, we expand upon strategies for promoting acetogenesis, which is at the forefront of rumen methane mitigation strategies. We show why acetogenesis is not dominant in the rumen and then report prior efforts to promote acetogenesis. Previous reviews have focused on chemical inhibitors like 3-nitrooxypropanol (3-NOP, Bovaer®) (Kebreab et al., 2023) and macroalgae (Sofyan et al., 2022). Others have focused on nutrition and feed additives (Roques et al., 2024). By revisiting the limited research in acetogenesis, we question the success, persistence, performance, and the fate of rumen enteric fermentation. Finally, we propose hypothetical, sustainable scenarios to maximize the promotion of acetogenesis in the rumen, including how CRISPR-Cas genome editing techniques could be applied to execute the proposals.
2 Broad strategies for reducing methane emission from ruminants
The United Nations Climate Change 26th Conference of Parties (COP26) through the Global Methane Pledge set a goal to decrease agricultural methane emissions by 30% by the year 2030 (Meinshausen et al., 2022). Building on this initiative, the recent COP27 launched an initiative called “fast mitigation sprint”, the goal of which is to focus on big and correctable sources of methane, such as livestock, and to act as quickly as possible (BBC, November 20, 2022).
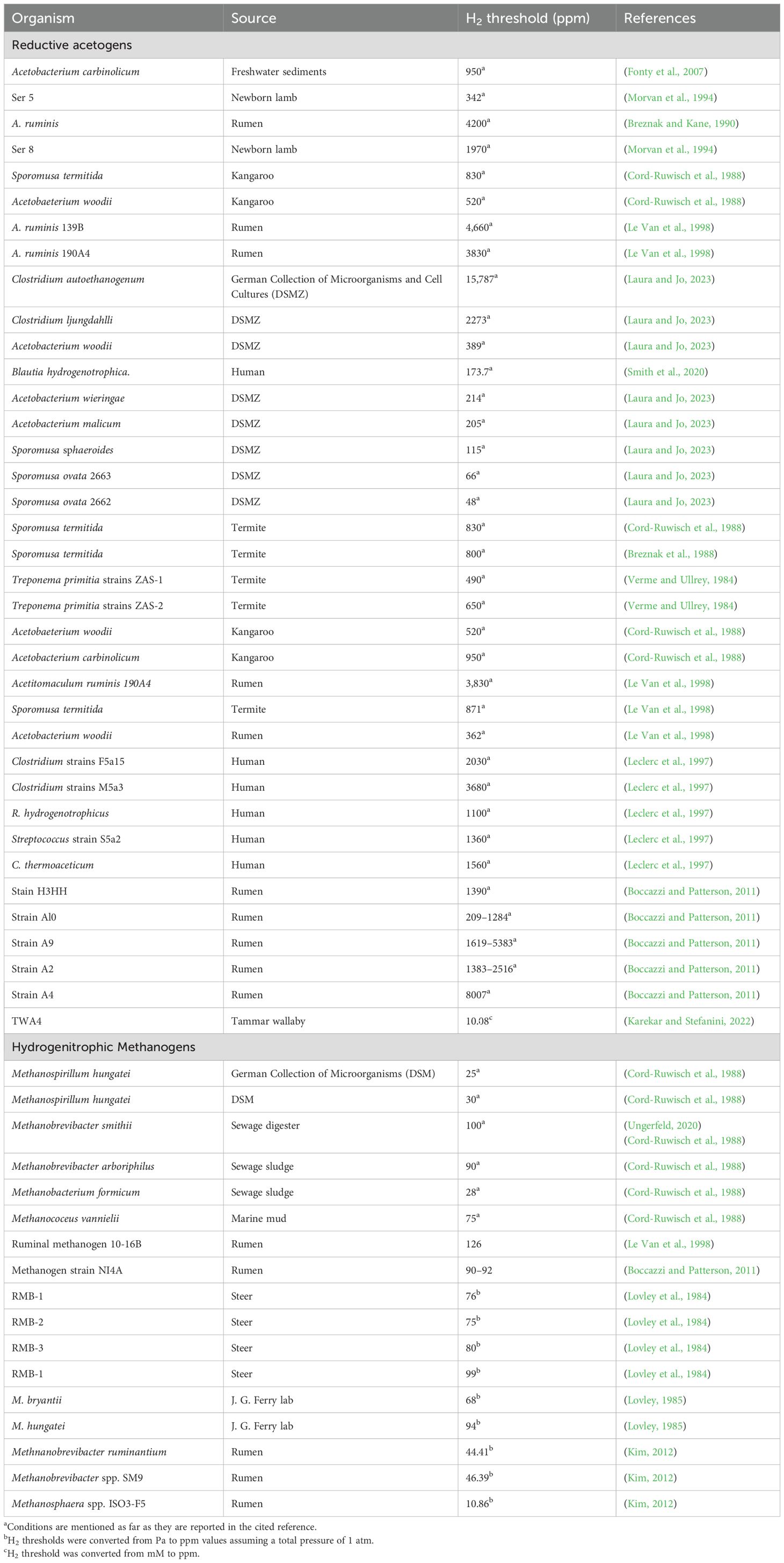
Research on mitigating rumen methane emissions began over 50 years ago. Prior to 2000, studies focused on enhancing the energy utilization efficiency of rumen fermentation to ultimately improve animal productivity (Beauchemin et al., 2020). In the early 2000s, the research focus shifted toward directly reducing rumen methane, resulting in numerous strategies and publications. These methods will be discussed in a historical context below with current research gaps and future research areas highlighted in Table 1.
The initial response from the two most advocacy institutions for reducing livestock emissions, the United Nations Food and Agriculture Organization (FAO) and the Intergovernmental Panel on Climate Change (IPCC), was to advocate for a reduction in global cattle herds by cutting meat consumption (FAO, 2022; IPCC 2022). This approach was supported by several prominent reports, including the Lancet report (Willett et al., 2019) and the Greenpeace report (Greenpeace, 2020), which suggested a 50% reduction in red meat consumption by 2050.
Reduction in meat consumption however, is not practical in numerous ways. First, it conflicts with the World Health Organization (WHO) recommendations to meet some key United Nations Sustainable Development Goals (UNSDGs); e.g., SDG2 on nutritional requirements and food security and SDG1 on poverty reduction. This is especially relevant in the Global South, where animal protein availability is still inadequate (Asano and Biermann, 2019) and consumption is expected to rise as population and average individual incomes increase. Reducing animal numbers in these societies would translate to health problems such as malnutrition, stunted growth, or anemia (Fehér et al., 2020). Second, large-scale reduction in cattle farming would also require substitutes for fertilizers, leather, and pet foods, leading to increased greenhouse gas emissions from synthetic alternatives (Cheng et al., 2022). Third, while repurposing the livestock land for carbon sink through forestation is often proposed, this strategy is not practical where livestock farming dominates, because land use alternatives like crop farming and tree growing are often not feasible (Houzer and Scoones, 2021). Thus, while decreasing cattle farming would reduce GHG emissions, these considerations must be balanced with potential impacts of leaving livestock land vacant, as well as the economic and nutritional needs of cattle farming communities.
2.1 Suggested alternatives to animal proteins
The need to balance human health with environmental concerns has driven the development of alternatives to animal proteins. These alternative proteins can provide necessary nutrition while significantly reducing livestock numbers and emissions. They have existed since the 1960s and include insect protein, plant-based substitutes, cultured meat, and fermentation-derived microbial protein (MP) (Ismail et al., 2020). Each of these categories offers a novel approach to producing protein without relying on conventional livestock farming (Humpenöder et al., 2022).
2.1.1 Insect protein
Insects offer an environmentally sustainable alternative to traditional livestock farming due to their high protein content, minimal methane production, and lower requirements for land, water, and feed compared to cattle. Crickets, for instance, require just 1.7 kilograms of feed to produce 1 kilogram of protein, while cows require 8 kilograms of feed to generate the same amount of beef protein (Moruzzo et al., 2021). Despite this efficiency, edible insects face acceptance barriers. It is worth noting current strategies that aim to enhance the appeal of insect consumption, such as processing insects into powders and adding them to familiar products like protein bars, baked goods, and pasta. These insect-based products have already reached markets in the United States, Canada, and Europe, gradually shifting consumer perceptions and promoting the acceptance of insect-derived foods (Moruzzo et al., 2021).
2.1.2 Plant based protein
Production-wise, plant-based diets have a lower environmental footprint than animal foods (Springmann et al., 2018). Studies have reported that a 70% reduction in GHG emissions and land use could be achieved by shifting our diets to plant-based proteins (Fehér et al., 2020). However, it is important to note that the net environmental benefit is likely to diminish with a complete transition to plant-based products. This is due to: 1) the complex interrelationships in mixed farming systems among feed, fertilizer, and soil quality; and 2) the need for additional land in scaling up plant-based production, which comes at the expense of deforestation and biodiversity decline (Kozicka et al., 2023).
While plant-based production is considered safe, its consumption may lead to more human emissions. Similar to the rumen of animals, humans produce methane from metabolism of methanogenic species (Polag and Keppler, 2019). These methanogens contribute to the production, on average, of 0.35 litres (equivalent to 0.23 grams) of methane per person per day (Polag and Keppler, 2018; Djemai et al., 2021). In 1986, when the human population was 4.7 billion, total human methane production via breath only was estimated to be 0.3 Tg per year (Crutzen et al., 1986). Assuming a direct correlation to population growth, 8 billion humans in 2024 now emit 0.51 Tg per year. For comparison, this amount is significantly lower than emissions from ruminant animals. However, these estimations represent only a fraction (25%) of what humans emit through breath, and excludes the 75% eliminated through flatus, which is difficult to measure and thus underreported. Transition to plant-based diet could significantly increase these numbers, as fiber rich diets produce more methane than protein or fat rich diets (Wilson et al., 2020).
2.1.3 Cultured meat
Cultured meat, or lab meat, is a promising alternative because it bypasses the extravagant length of rearing a whole animal, thus requiring fewer land and water resources (Ismail et al., 2020). However, converting a cell into a steak in the lab is currently very expensive. For instance, in 2013, researchers from Maastricht University spent $2,470,000/kg of production to produce a proof-of-concept cultured meat product (Rubio et al., 2020). At this rate, cultured meat is cost prohibitive as large-scale replacements for beef. Furthermore, consumer concerns persist, with reported fears related to cancer and insufficient regulation (Chriki and Hocquette, 2022).
2.1.4 Microbial protein
Fungi, especially Fusarium venenatum, have a high protein content and an excellent essential amino acid profile, making them a promising source for meeting human protein requirements (Lee et al., 2024). However, this potential can only be realized if a more affordable and eco-friendly energy source replaces the electricity used in bioreactors. Studies have consistently reported that the energy required for mycoprotein production exceeds that of conventional ruminant meat production when comparing land-related and energy-related greenhouse gas emissions (Järviö et al., 2021). For this reason, unless renewable energies like solar and wind become more efficient and cost effective to eventually replace the existing energy system, microbial protein will continue to have a huge environmental impact.
2.2 Reducing emission per kg of animal protein produced
Considering all the challenges of upscaling alternative proteins, it is necessary to explore alternative strategies that can meet emission reduction targets without compromising traditional livestock practices. Proposed approaches can broadly be categorized into two groups: 1) strategies to reduce methane emission per kilogram of animal produced; and 2) strategies to reduce net emissions by targeting microbial fermentation.
Methane intensity refers to enteric methane emitted per unit of animal product produced (g/kg of milk yield or carcass weight) (Beauchemin et al., 2020). Overall, all management practices that enhance animal performance also reduce methane intensity. For example, a high-performing animal that yields more milk and meat may emit higher amounts of methane per animal. But since a smaller proportion of feed is consumed by the animal, emissions intensity is effectively lowered. The idea of methane intensity is highly attractive in low- and medium-income countries as it offers an opportunity to align food security, development objectives, and climate change mitigation goals (FAO, 2019).
2.2.1 Management (husbandry) practices
Advancements in nutrition, disease control, and animal reproduction technologies accelerate animal growth rates, consequently reducing their emissions over the course of their lives. In the United States, for instance, the average annual milk yield in grain-finished cows moved from 1,890 kg to 9,682 kg of milk per cow between 1924 and 2011 and the carbon foot print reduced by 41% between 1944 and 2007 (Capper and Bauman, 2013; Georges et al., 2019). Nutritional management however might introduce competition with human food supply as more land would be needed for grain production to accommodate both human and animal food which could create additional emissions (Ravi Kanth Reddy et al., 2019; Reddy et al., 2019). For this reason, research must focus on identifying nutrients with the potential to reduce emissions without compromising human food systems.
2.2.2 Breeding low emitting animals
Breeding for low emitting animals is possible because the methane production trait has been found to be moderately (0.13–0.35) heritable in cattle and sheep (Black et al., 2021). However, the selection of a stock population is hindered by high expenses of measuring methane (González-Recio et al., 2020). Identification of methane emission proxies could help offset the cost of methane measurements (Lassen and Difford, 2020). But even so, the potential for genetic improvement to reduce methane over 20 years is only 4–8% (Black et al., 2021). This means that lowering global emissions by ruminants is unlikely to be realized within the timeframe of action needed to keep global warming below 1.5°C.
2.2.3 Capture of methane produced by animals and their waste
Many methane emission sources continue to leak methane into the atmosphere despite mitigation efforts, as most strategies are only partially effective (Lidstrom, 2024). Therefore, it has been argued that emission-reduction strategies must be augmented by methane removal from air to slow global warming by 2050 (Warszawski et al., 2021; Nisbet-Jones et al., 2022). Methane, with an energy content of 50.4 MJ/kg, can serve as a potent renewable fuel (Angelidaki et al., 2018). For example, methane from just 12 cows can supply enough gas for daily use by one average household (Crutzen et al., 1986). Additionally, methane can be converted into CO2 and biomass, which can be harvested for use as protein feedstock (Lidstrom, 2024).Traditionally, methane capture has been achieved through manure digesters, though only 5% of methane from animals is emitted through manure, with 95% expelled through eructation. While devices like cow masks have been developed to capture eructed methane, they are costly and raise animal welfare concerns (Kaya and Kaya, 2021).
Methanotrophic bacteria, which oxidize methane, offer a promising alternative. Biofilters using methanotrophs have shown success, though they are more effective at higher methane concentrations located near emission sites (La et al., 2018). Methylotuvimicrobium buryatense 5GB1C is a particularly promising candidate, as it can grow at low methane concentrations (200–1,000 ppm) and demonstrates superior methane consumption rates compared to other strains (He et al., 2023). There remains significant potential for research into methanotrophs that can efficiently remove methane from low-concentration air. For this reason, scientists invariably stress the need to drive down net emissions by targeting rumen microbiota and enteric fermentation.
3 The rumen microbiome
Livestock, particularly ruminants, use low-quality fibrous feedstock and convert it into high-quality livestock products, including meat and milk. This conversion is accomplished through the action of symbiotic microbiota residing in their GI tract (Martínez-Álvaro et al., 2022). These microorganisms ferment feedstock in the rumen to produce short chain fatty acids (SCFAs) which are consumed by the host for its own energy and growth. About 70% of the rumen microbiota pass down the GI tract, where they are themselves digested to further nourish the host with protein, long chain fatty acids, and vitamins (Mizrahi et al., 2021). Concomitant with the nourishing effects, the fermentative action of rumen microbiota produces GHGs, most notably methane.
3.1 Composition of the rumen microbiome
The rumen microbiome is composed of archaea, bacteria, fungi, viruses and protozoa (Wallace et al., 2019). Bacteria are the most numerous and diverse, comprising 95% of rumen microbiota (Pereira et al., 2022). Protozoa are the most abundant by biomass (Solomon et al., 2021) and their diversity and abundance tend to fluctuate more widely across breeds and feed type (Newbold et al., 2015). Numerically, fungi comprise a very small component of the rumen microbiome. However, their efficiency in degrading plant material is remarkable due to their possession of an extensive set of enzymes and rhizoids that penetrate plant structural barriers, subsequently increasing the plant cell surface area for other microbes to colonize (Xue et al., 2020). Viruses are highly abundant and diverse within the rumen, and play a critical role in maintaining the microbial population through intra-ruminal microbial lysis and genetic transfer (Wright and Klieve, 2011). However, rumen viral populations have been the least explored population within the microbiome, primarily due to the challenges in isolation and characterization (Gilbert et al., 2020). The archaeal population in the rumen ranges from 107–109 cells per ml of ruminal content (Wright and Klieve, 2011), and is responsible for all ruminant methane production (Wallace et al., 2019).
As mentioned previously, the microbiota in the rumen interact with one another to perform enteric fermentation, which ultimately benefits their host and fosters their ecological balance. Some bacteria, fungi and protozoa digest plant fiber into sugar monomers, then ferment these products into three major volatile fatty acids: acetate (40–75%), propionate (15–40%), and butyrate (10–20%). These products are then absorbed by the rumen epithelial walls for metabolic use by the animal (Khairunisa et al., 2023). In addition, lactate, ethanol, and succinate are produced as reduced intermediates, then lactate and succinate are further converted to propionate. The products of primary fermentation are passed down the food chain and are ultimately degraded to hydrogen and carbon dioxide (Lyu et al., 2018). Although H2 and CO2 can be expelled from the rumen by eructation (burping), the energy in the mix would be lost to the rumen ecosystem. Consequently, some archaea and some bacteria are able to use the H2 and CO2 for energy conservation and growth and in turn produce methane and/or acetate respectively (Ma et al., 2021).
This complex interdependence between microbiota has been reported in the literature with the interaction between hydrogen producers and hydrogen consumers (Figure 2) repeatedly observed. The interaction between hydrogen producers (Figure 2) and methanogens is the most prevalent because methanogenesis is the key hydrogen sink in the rumen. For instance, this relationship is observed between the rumen fungus Neocallimastix frontalis and methanogen Methanobacterium formicicum (Nakashimada et al., 2000); between the bacterium Ruminococcus and methanogen Methanobrevibacter ruminantium (Henderson et al., 2015); and between protozoans Entodinium, and methanogen Methanobrevibacter thaueri (Xia et al., 2014). The abundance of these hydrogenic organisms has been linked to methane emission and animal production; depleting them from the rumen reduces methane emission but also reduces animal performance, as evidenced by one mitigation strategy called defaunation (Ibrahim et al., 2021). Within this framework, it is important to conduct experiments to explore and understand the metabolic interactions between the two groups of organisms listed in Figure 2. Additionally, investigating the efficiency of hydrogen uptake by organisms listed in Figure 2 could facilitate future development of substitutes for methanogens.
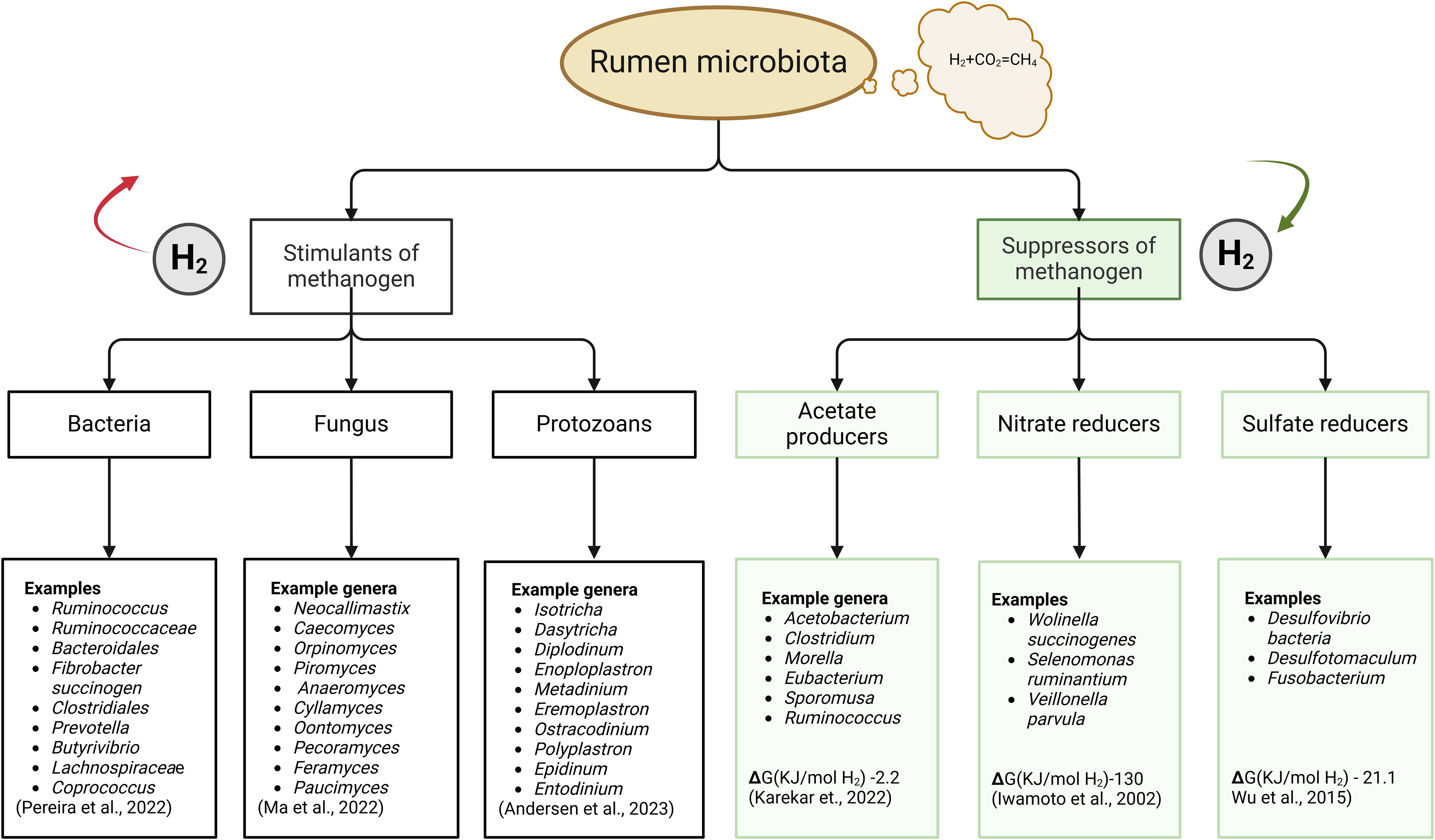
Figure 2. Promising candidates for future research in promoting alternative hydrogen utilizing pathways as a strategy for ruminal methane mitigation. Key hydrogen producers, termed stimulants of methanogens, are shown on the left branch of the diagram; key hydrogen consumers, termed suppressors of methanogens, are shown on the right branch of the diagram.
3.1.1 Factors influencing rumen microbial composition
Generally, the dynamics of the rumen microbiome are largely influenced by three factors: feed (Wilkinson et al., 2020), age of the animal (Martínez-Álvaro et al., 2022) and genetics (Yáñez-Ruiz et al., 2015). Feed type and amount have the most dramatic effect on microbial dynamics and the subsequent effect on methane generation. Poor-quality feed with high fiber content has been significantly linked to diversification of the microbiome and elevated methane emissions as compared to highly digestible, low-fiber feed (Hayek and Garrett, 2018; Vaghar Seyedin et al., 2022). Some experiments in the literature show that animals fed poor-quality feed tend to harbor a high abundance and diversity of hydrogen-producing organisms. Bacteria like Firmicutes and Ruminococcaceae have repeatedly been found to dominate the rumens of high emitters (Wallace et al., 2019; Furman et al., 2020). Due to the nature of the feed, the fermentation process is usually slow, leading to a gradual buildup of hydrogen which supports a greater methanogenic population, ultimately resulting in increased methane production.
On the other hand, Succinivibrionaceae, Quinella spp., and Dialister are commonly found in the rumen of cows that emit extremely low levels of methane (Wallace et al., 2019; O’Hara et al., 2020). These microorganisms are efficient at utilizing hydrogen, resulting in propionate production. It must be noted that when fermentation is fast, which is the case for low-fiber feed, hydrogen builds up in the rumen rapidly (Wang et al., 2016). High hydrogen levels in the rumen shift carbohydrate fermentation toward reactions that consume hydrogen, such as propionate production, to maintain equilibrium (Arndt et al., 2023). It is for this reason that bacteria that are capable of producing lactate and succinate, such as Fibrobacter spp., Kandleria vitulina, Olsenella spp., Prevotella bryantii, and Sharpea azabuensis dominate the rumens of low emitting animals (Wallace et al., 2015). Another hypothesis for low emissions in animals fed a low-fiber diet is that because low-fiber feed is easier to digest, it leads to increased solid waste passage rates that slough off and eliminate the methanogenic population, ultimately resulting in a reduction in total methane production. In one experiment, substituting grass-clover silage with maize silage for dairy cows led to a 15% reduction in methane emissions per kg dry matter intake (Brask-Pedersen et al., 2023).
Age is another factor that affects microbial composition and subsequently methanogenesis. Young animals have less diverse, less abundant, and more dynamic microbiome compositions (Cammack et al., 2018). Their rumen size is small, and they consume few solids that are also easy to digest. As the animal matures, the microbiome stabilizes and becomes more diverse and abundant, thus better supporting methanogenesis. Cattle achieve this stabilization three weeks after birth (Yáñez-Ruiz et al., 2015). Research suggests that beneficial microbial signatures must be imprinted within this sensitive window (first three weeks of life) for the animal to maintain a healthy, diverse, and stable microbiome over time (O’Hara et al., 2020). Thus, the dynamic nature of a young animal’s microbiome presents potential opportunities for manipulation of the microbiome for reduced methane emissions (O’Hara et al., 2020).
Besides feed and age, host genetics is also a main determinant of microbial composition and methane emissions. The composition of the microbiome and the subsequent methane production are influenced by both animal species and breed (Roehe et al., 2016). For example, in a recent exploratory effort to understand the role of animal genetics in microbial composition, researchers discovered a significant association between a region on chromosome 6 with the densities of Euryarchaeota, Actinobacteria and Fibrobacteres. This region encompassed the genes that codes for MEPE, matrix extracellular phosphoglycoprotein (Golder et al., 2018). Despite this achievement, more studies are needed not only to establish these associations, but also determine the extent of the influence and the underlying mechanisms. Overall, rumen microbial composition and the factors that influence their abundance, diversity, and function present opportunities for microbial manipulation, and could serve as potential tools in methane mitigation strategies.
3.2 Targeting the rumen microbiome
3.2.1 Suppression of methanogenesis using chemical inhibitors
Rumen additives ranging from chemical to biological have been used to reduce enteric methane emissions. Historically, the most commonly used chemical additives were antibiotics with ionophore activity to target Gram-positive bacteria, creating a shift in rumen fermentation patterns that are linked with enteric methane suppression. Due to concerns about antibiotic resistance, other inhibitory compounds have recently been developed. In a recent meta-analysis, macroalgae and 3-nitrooxypropanol (3-NOP, Bovaer®) were reported to be the most potent additives with significant methanogen reduction (Kebreab et al., 2023).
The molecular structure of 3-NOP resembles that of methyl-coenzyme M, which is a substrate utilized by coenzyme M reductase (MCR), in the final step of methanogenesis. Molecular docking studies suggest that 3-NOP selectively binds to the active site of MCR and thereby inhibits it. In this process, the nitrate group of 3-NOP is reduced to nitrite, further inactivating MCR (Duin et al., 2016). However, methanogens express a repair system that reactivates MCR (Duin et al., 2016). Therefore, once 3-NOP has been completely metabolized and is absent, CH4 emissions return to their original levels. Studies have shown that supplementing cows with 3-NOP can lead to a reduction in methane emissions on average by up to 30% (Wesemael et al., 2019; Zhang et al., 2020). In vivo experiments with beef (Alemu et al., 2021) and dairy cattle Melgar et al., 2020) demonstrated varying effects depending on dosage. Lower doses (60–75 mg/kg DM) reduced emissions by 16–30%, while higher doses (100–183 mg/kg DM) achieved reductions of 10.6–36.7% (Hodge et al., 2024). In dairy cattle, small amounts (60 mg/kg DM) cut methane production by up to 38%, with higher doses (80 mg/kg DM) reducing emissions by up to 45.1% depending on diet (Van Gastelen et al., 2022). Effects of 3-NOP on animal performance were varied and included: increased hydrogen emissions; reduction in acetate to propionate ratio; reduction in dry matter, organic matter, and energy digestibility; decreased body weight gain; increased milk fat; no change in milk yield or composition; no negative effects on dry matter intake (DMI) (Lileikis et al., 2023). The reasons for these mixed results remain under discussion. Some studies suggest that increased hydrogen accumulation may limit 3-NOP performance. To enhance productivity, 3-NOP can be combined with phloroglucinol, which captures excess hydrogen and generates beneficial metabolites for the host (Martinez-Fernandez et al., 2017). Even though the safety of 3-NOP has been confirmed with regulatory approval in Europe, the U.S., and other countries, questions remain regarding its potential to enhance productivity. Further, there exist economic and logistical challenges, as consistent administration is required to achieve effective methane reduction.
Algae, with their diverse array of properties including antibacterial, antiviral, antioxidant, anti-inflammatory, and anti-carcinogenic effects, have attracted attention in methane mitigation research. Macroalgae, notably Asparagopsis taxiformis, is rich in bromoform. Bromoform is a halomethane compound. It inhibits methanogenesis by binding to vitamin B12, which chemically resembles coenzyme F430, a cofactor required for methanogenesis (Roque et al., 2021). An in vitro study reported that A. taxiformis supplementation at inclusion rates up to 5% of organic matter resulted in methane reduction by 99% without a significant negative impact on volatile fatty acid profiles and organic digestibility (McCauley et al., 2020). Similar to 3-NOP, the effect of A. taxiformis is short lived. To tackle this challenge, current research endeavors focus on devising slow-release formulations and administering them to young animals for a prolonged effect.
3.2.2 Biological control of methanogens by means of phages
Viruses modify the rumen microbial ecosystem via infection, cell lysis, reproduction, and reprograming of microbial metabolism (Gilbert et al., 2020). For this reason, phages can be used as a tool to manipulate the rumen microbial community in a targeted manner, suggesting their potential in methane reduction research (Wolf et al., 2019). Utilizing viruses offers a means to manipulate key components of methanogenesis, such as cellulose-degrading bacteria and methanogenic populations.
Bacterial phages, for instance, can hinder methanogenesis indirectly by targeting hydrogen producers and thereby reducing hydrogen availability. Within this category, phages belonging to families such as Myoviridae, Siphoviridae, Mimiviridae, and Podoviridae (Lobo and Faciola, 2021) have been isolated and are closely associated with dominant rumen bacterial phyla, including Firmicutes and Proteobacteria (Namonyo et al., 2018). Other studies have also discovered phages in the rumen such as, φBrb01 and φBrb02, φSb01, φRa02, and φRa04 that specifically target Bacteroides, Streptococcus and Ruminococcus, respectively (Gilbert et al., 2017).
Methanogen-targeting viruses on the other hand directly suppress methane production by eliminating methanogenic archaea, creating space for other hydrogen-utilizing microorganisms to thrive. Only a limited number of phages specifically targeting methanogens have been investigated thus far. Viral families such as Anaerodiviridae, Leisingerviridae and Speroviridae have been described to infect hosts within the order Methanobacteriales (Wolf et al., 2019). The family Pungoviridae has been found to associate with Methanomicrobiales and family Fervensviridae linked to Methanococcales (Weidenbach et al., 2021). In addition, a dozen proviruses have been found in Methanobacteriales, Methanococcales, Methanosarcinales and Methanonatronarchaeales genomes (Krupovič and Bamford, 2008).
Due to challenges in culturing rumen viruses and overall rumen microorganisms, researchers have shifted their focus to studying viral enzymes. These enzymes can potentially be fused with easily cultivable microorganisms, such as Escherichia coli, to aid in conducting lytic experiments on methanogenic cells. Recent research demonstrated the effectiveness of the lytic enzyme PeiR, sourced from a virus targeting Methanobrevibacter ruminantium, in inhibiting a spectrum of rumen methanogen strains in pure culture (Altermann et al., 2018). In this work, PeiR was found to hydrolyze the pseudomurein cell walls of methanogens. Subsequently, PeiR was utilized to develop bionanoparticles (BNPs) through one-step biosynthesis in E. coli (Altermann et al., 2022). These tailored BNPs were capable of lysing not just the intended methanogenic host archaea, but also a wide range of other rumen methanogen strains leading to a 97% methane reduction for 5 days post-inoculation. Despite these milestones, the use of E. coli as a production host may be limited due to its requirements for oxygen. Similarly, the two-plasmid system requires the application of two different antibiotics and has limited scalability to fed-batch fermentations.
3.2.3 Promotion of alternative consumption of H2 and CO2
Since methanogenesis is influenced by intracellular and intercellular flows of metabolic hydrogen (Ungerfeld, 2020), targeting both its concentration and direction of flow will indirectly influence methanogenesis. Methods to reduce hydrogen availability in the rumen, either by redirecting it to alternative reactions or by lowering the population of hydrogen-producing Gram-positive bacteria, fungi, and protozoa, have been explored (Kelly et al., 2022). One approach involves the use of fats, particularly unsaturated fatty acids. Firstly, unsaturated fatty acids undergo biohydrogenation in the rumen, where specific bacteria add hydrogen to the unsaturated bonds of fatty acids, converting them into more saturated forms. This process consumes hydrogen, thereby reducing the amount available for methanogens to produce methane. Essentially, fats “compete” with methanogens for hydrogen (Yang et al., 2019). Secondly, unsaturated fatty acids can inhibit rumen fermentation, leading to a reduction in hydrogen-producing microorganisms. This includes fermentative bacteria such as Ruminococcus albus, R. flavefaciens, Neocalimastrix spp., and Desulfovibrio (Min et al., 2022), as well as protozoa like Entodinium caudatum (Darabighane et al., 2021) and various fungi (Ibrahim et al., 2021). This inhibition shifts the microbial community in favor of propionate production (Wang et al., 2023). Furthermore, since fats are not fermented in the rumen, they contribute less substrate hydrogen for methanogenesis (Belanche et al., 2020). The methane-suppressing effect of fatty acids, however, is not consistent and largely dependent on the type of fat, concentration, and nutrient composition of the diet fed to the animal. In cattle, a 15% reduction in methane was observed when the diet contained 6% polyunsaturated fatty acids and low fiber. But then 6% of 15 kg of hay (the usual dry matter intake of a cow) equates to almost a kg of oil (0.9 kg) every day for a cow that weighs 500 kg. Given the global vegetable oil shortage, exacerbated by the Russia-Ukraine conflict, more sustainable approaches that avoid competition with the human food supply are critically needed (Mottaleb et al., 2022). Additionally, large-scale oilseed production has significant environmental costs, including high greenhouse gas (GHG) emissions and biodiversity loss. Compared to concentrate feeds production, oilseed production emits nearly equivalent upstream GHG emissions per kilogram of dry matter (1.27 vs 0.70 CO2 equivalents kg dry matter -1) (Arndt et al., 2023).
By contrast, a more useful strategy would be to redirect hydrogen to already existing rumen fermentation pathways like propionate, nitrate, and sulfate, which are nutritionally useful to the animal and its microbial consortium (Min et al., 2022; Tseten et al., 2022). Propionate is the main glucose precursor in ruminants and therefore important to animals with high requirements for glucose such as high producing dairy cows in early lactation (Ungerfeld, 2020). But for the propionate pathway to compete with methanogenesis, animals must be fed either diets rich in starch or precursors of propionate like fumarate, lactate, and succinate. Starch is not only expensive but also its fermentation overwhelms the digestive physiology of cattle leading to a condition known as ruminal acidosis (Elmhadi et al., 2022). In order to avoid the occurrence of lactic acidosis, high-starch diets often include antibiotics such as ionophores to limit bacteria growth. However, the use of antibiotics in livestock production has been restricted worldwide due to concerns about antibiotic resistance. Due to this, researchers have explored the use of propionate-forming bacteria as a strategy to increase propionate production and reduce rumen methane emissions. In an in vitro experiment, (Alazzeh et al., 2012) evaluated the potential for sixteen strains of propionate bacteria in reducing methane emission from concentrate and forage diets. Among them, only six showed significant reduction of methane. Propionibacterium freudenreichii T114, Propionibacterium thoenii T159, and Propionibacterium thoenii ATCC 4874 significantly lowered CH4 production from both substrates, whereas Propionibacterium jensenii T1, Propionibacterium freudenreichii T31, and Propionibacterium freudenreichii T54 only lowered methane production when corn was used as a substrate. Despite these observations, added bacteria were unable to permanently colonize the ruminal cultures and did not change bacterial, archaeal, and protozoal populations, implying that the reduction in methane production was transient.
Unlike the propionate pathway, nitrate reduction (ΔG=−254 kJ/mol H2) versus CO2 reduction to methane (ΔG= −16.9 kJ/mol H2), stands a good chance to compete with methanogenesis. However, there are fewer nitrate reducers in the rumen which restrict the rate of nitrate reduction. Their concentration in the rumen has been reported to range around 103 cells/ml of rumen fluid, whereas the methanogenic population is 109 cells/ml of rumen fluid (Iwamoto et al., 2002). On top of this, the three major nitrate reducers either grow slowly on nitrate and H2 (e.g. Wolinella succinogenes), or reduce nitrate and nitrite at lower rates (e.g. Selenomonas ruminantium and Veillonella parvula) (Iwamoto et al., 2002). To boost nitrate reduction in the rumen, scientists have considered the incorporation of nitrate in animals’ diets. The effect of nitrate supplementation on methane reduction has been reported in many studies. In an in vitro experiment conducted by (Iwamoto et al., 2002), nitrate supplementation to the diet increased the number of nitrate-reducing bacteria such as Wolinella succinogenes and Veillonella parvula. Subsequently, when Wolinella succinogenes was co-cultured with methanogens, methane production decreased from 1.6 mol/L of culture to 0.1 mol/L of culture. Another study reported 16–25% reduction of methane emissions in grams per kilogram of dry matter intake when nitrate was provided at a rate of 21 g NO3−/kg dry matter (DM) (Van Zijderveld et al., 2011). In a more recent experiment, nitrate supplementation reduced in vivo methane production by 31% (Olijhoek et al., 2016).
The methane reduction effect of nitrate has been extensively documented across animal species. Reductions of methane emissions by 12% in beef steers (Alemu et al., 2019; Granja-Salcedo et al., 2019; Feng et al., 2020), 17% in dairy cows (Meller et al., 2019), 26% in sheep (Villar et al., 2019), and 6% in goats (Zhang et al., 2019) have been observed. Despite these huge milestones, application in the field is still restricted because nitrite, which is an intermediate in nitrate reduction, is considered toxic. It causes methemoglobinemia, a condition which decreases the capacity of blood to transport oxygen into tissues, leading to depressed animal performance and even the death of the animal (Lileikis et al., 2023). Some studies speculate the reduction in rumen methanogenesis following nitrate supplementation could largely be due to the toxicity of nitrite on methanogens and other rumen microbiota (Iwamoto et al., 2002). In 2012, the strain NRBB 57 was isolated from the rumen of buffalo, which showed a significant removal of nitrate and nitrite from the medium with a further reduction in methane production (Sakthivel et al., 2012). More studies of this nature are needed to further develop the concept of nitrate supplementation in methane reduction.
Similar to nitrate reduction, sulfate reduction thermodynamics (ΔG= −21.1 kJ/mol H2) can compete with methanogenesis (−16.9 kJ/molH2), but is limited in terms of substrates and organism number (Van Lingen et al., 2016). The abundance of sulfate reducing bacteria in the rumen is generally low, at a concentration of approximately 105 cells/ml, with Desulfovibrio and Desulfotomaculum as the main genera (Lan and Yang, 2019). It has been suggested that increasing sulfate levels in the animal’s diet could boost the competitiveness of sulfate reducing bacteria against methanogens for H2 in the rumen. Few studies have shown a positive effect of sulfate on methanogenesis inhibition. A 16% methane reduction was reported in crossbred lambs following a 2.6% sulfate supplementation (van Zijderveld et al., 2010). A more recent study investigated the effect of supplementing goats with sulfate-reducing bacteria (SRB) along with sulfur (as sodium sulfate) on methane production. SRB was purposely added to minimize the risk of hydrogen sulfide, a toxic end product in the sulfate reduction reaction. Methane (CH4) production (in l/kg of dry matter intake (DMI)) was reduced by 11.8% when sulfur (0.19% DMI) was supplemented along with SRB at the rate 0.5 ml/kg body weight (Uniyal et al., 2023). Therefore, research to identify specific sulfate-reducing bacteria that can oxidize H2S, or a mix of sulfate-reducing bacteria and other rumen microbes that can use H2S, is a promising way to advance research that focus on sulfate reduction as a methane mitigation strategy.
3.2.4 Converting the rumen from methane producer to acetic acid generator
Reductive-acetogenesis, here termed acetogenesis, is a natural hydrogen scavenging reaction in the rumen. Acetate, as the end product of acetogenesis, serves as an excellent energy source for the host animal, and the microbial community within the rumen (Lan and Yang, 2019). There are about 22 diverse genera and over 100 species of acetogens among which the genera of Acetobacterium, Blautia, Clostridium, Morella, Eubacterium, Sporomusa, Ruminococcus and Oxobacter are common in the rumen (Greening et al., 2019; Karekar and Stefanini, 2022). The abundance of rumen acetogens varies widely between animals, from undetectable levels up to 107 cells/ml (Fonty et al., 2007). Acetitomaculum ruminis (A. ruminis) is a notable acetogen in the rumen, producing 2–8 times more acetate than other species (Le Van et al., 1998).
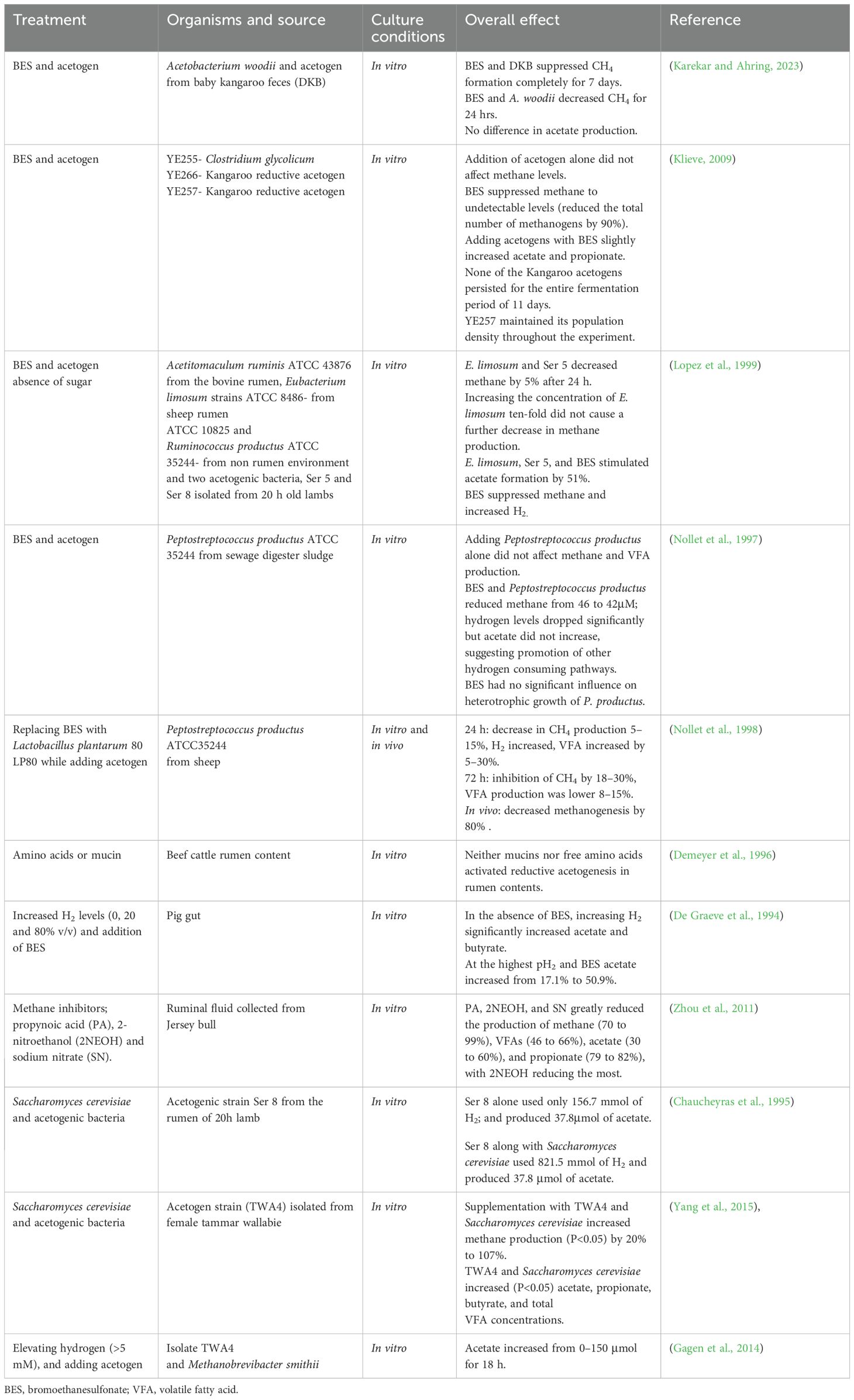
Unlike methanogenesis, acetogenesis is less energetically favorable, with a Gibbs free energy of approximately −95 kJ/mol for reducing 2 moles of CO2 to acetate, compared to −130 kJ/mol for methanogenesis (Kim et al., 2020). Irrespective of this, other anaerobic gut environments like termites (Yang, 2018), kangaroos (Pester and Brune, 2007), wallabies (Leng, 2018), and rumens of young animals (Gagen et al., 2010) use acetogenesis as the dominant hydrogen sink. The dominance of acetogenesis in these other environments is due to the differences in the gut anatomy, physiology and hydrogen concentrations (Hegarty and Gerdes, 1999). Anatomy and physiology can influence both gut metabolism and the types of species that inhabit it. In animals like kangaroos, wallabies and horses, gut anatomy does not support mechanisms such as eructation in ruminants, which is responsible for eliminating gases. To prevent gas buildup that could harm the digestive system, evolution has favored acetogenesis over methanogenesis. It has been reported that methanogenesis is actively prevented in these animals by immune secretions such as antimicrobial peptides, immunoglobulins, innate lymphoid cells and mucin that eliminate or suppress methanogenic archaea and support the growth of acetogenic microbiota (This may have encouraged colonization by diverse acetogen species that are more efficient at capturing hydrogen than those in the rumen (Godwin et al., 2014). Table 2 shows that Sporomusa termitida from termite guts is more efficient at hydrogen uptake (~800 ppm) than A. ruminis from the rumen (3830 ppm). Similarly, acetogens from kangaroos and tammar wallabies also outperform rumen species. Rumen strains A. ruminis 139B and 190A4 exhibit H2 thresholds 30 to 37 times higher than the rumen methanogen strain 10-16B. Overall, six acetogenic strains with notably low H2 thresholds, including strain TWA4, A. tundrae, T. primitia strains ZAS-2 and ZAS-1, E. limosum, and S. sphaeroides, stand out as promising candidates for biotechnological applications (Table 2).
Hydrogen concentration has a central role in the dynamics of acetogenesis during fermentation (Choudhury et al., 2022). High hydrogen concentrations create favorable conditions for acetogenesis, as acetogens have a lower affinity for hydrogen compared to methanogens. In termites, hydrogen levels have been reported to be three times higher than in cattle rumen (Pester and Brune, 2007). This provides a leveling ground for competition with methanogens which naturally thrive in low hydrogen levels because of their high hydrogen affinity (Table 2). The high hydrogen levels in these environments is explained by high abundant protozoa and spirochetes, which are responsible for hydrogen production and also double every 24–48 h, much faster than hydrogenotrophic methanogens (Breznak et al., 1988).
Based on the considerations described above, the question arises: how can we promote acetogenesis in the rumen? Most studies on enhancing rumen acetogenesis have been conducted in vitro, with only one in vivo study (Table 3). Approaches have included adding acetogens Yang et al., 2015; Kim et al., 2018; Pereira et al., 2022), combining methane inhibitors with acetogens (Nollet et al., 1997; Lopez et al., 1999; Gagen et al., 2012), using alternative electron acceptors like caffeic acid (Cord-Ruwisch et al., 1988), and incorporating selective additives such as Saccharomyces cerevisiae. Yang et al., 2015). Overall, adding acetogenic cultures alone in the rumen fluid did not affect acetate or methane production in all systems, even when the concentration of acetogen was increased ten-fold (Lopez et al., 1999). Bromoethanesulfonate (BES) and other methanogenic inhibitors suppressed methanogenesis up to 90% and increased hydrogen levels (Klieve, 2009; Zhou et al., 2011). In one study, increased hydrogen post-BES administration shifted volatile fatty acid concentrations toward propionate production. However, another study showed a methanogenic inhibitor reduced both methane and acetate formation (Zhou et al., 2011). Adding a non-native acetogen strain along with methanogenic inhibitors promoted acetogenesis briefly in some studies. One such report showed that the combination of E. limosum ATCC 8486 and BES stimulated acetate formation by 51% (Lopez et al., 1999). In these studies, methanogenesis resumed during the course of the experiments and all added acetogens perished (Karekar and Ahring, 2023. It is likely the case that these non-native species were unable to establish a niche in the rumen, in addition to the diminishing activity of BES over the course of the experiment. Adding inhibitors while increasing hydrogen levels increased acetate in pig gut (De Graeve et al., 1994). The observation aligns with other findings that collectively suggest that even competent acetogens can only compete with methanogens when hydrogen levels are elevated (Gagen et al., 2014) (Figure 3A). The addition of yeast cells enhanced by more than fivefold the hydrogenotrophic metabolism of the acetogenic strain and its acetate production (Chaucheyras et al., 1995), and yeast stimulate cellulose degradation and elevate hydrogen concentrations (Fonty and Chaucheyras-Durand, 2006). It can thus be concluded that strategies aiming to directly outcompete and displace methanogens with acetogens are unlikely to succeed. However, approaches that promote hydrogen buildup, such as using methane inhibitors or introducing yeast cells, can give acetogens a competitive advantage in the rumen (Figure 3B).
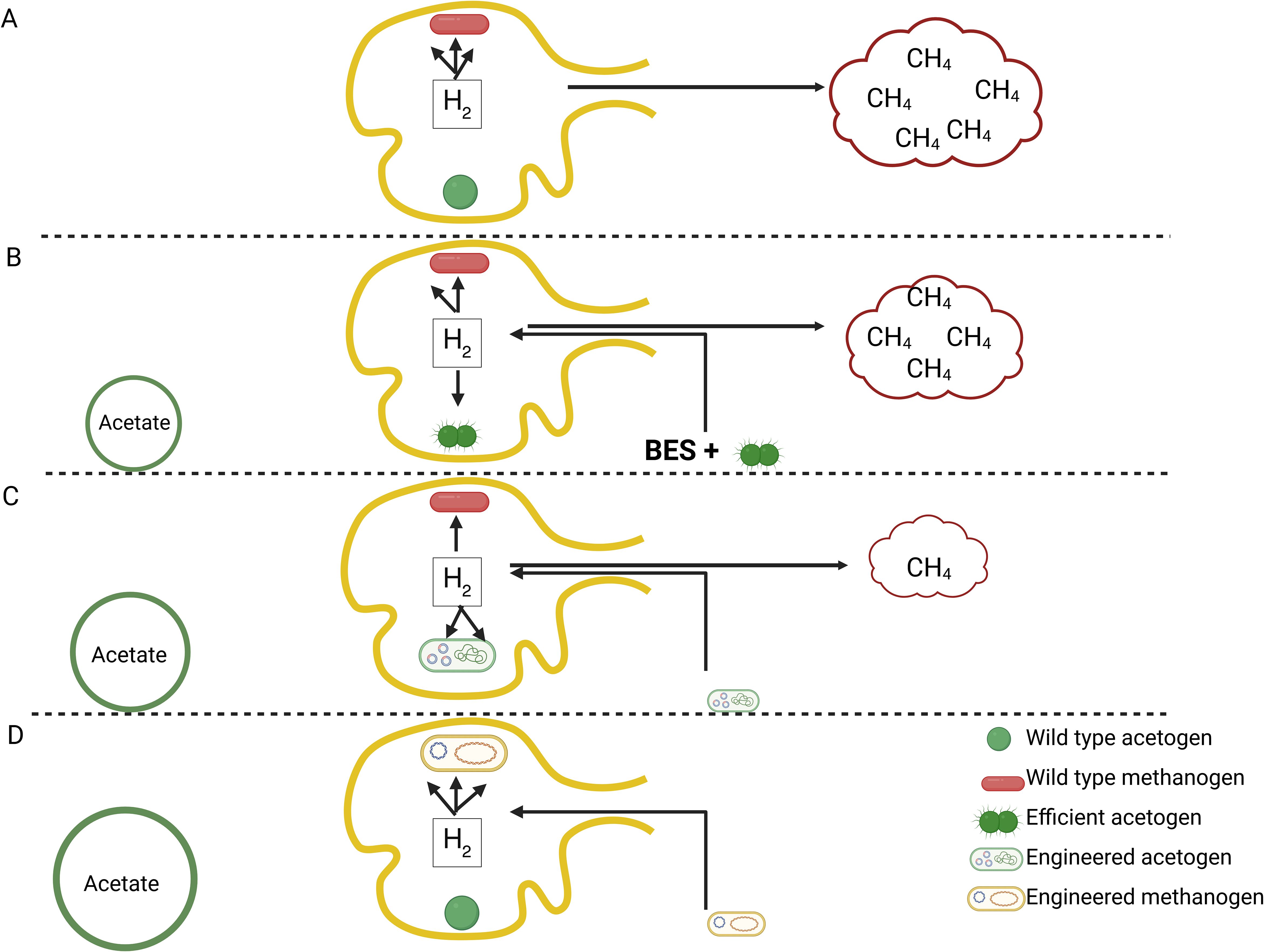
Figure 3. Three hypothetical scenarios for enhancing acetogenesis in the rumen. (A) Existing rumen fermentation. Under normal conditions, methane (CH4) is the main sink of metabolic hydrogen. (B) Current interventions in rumen fermentation, where efficient acetogens from a non-rumen environment are introduced following inhibition of methanogenesis by chemicals like BES. BES creates more available hydrogen for these competent acetogens. Methanogenesis is halted, but only briefly. Methanogenesis resumes once BES expires, or when non-native acetogen species are rejected or fail to establish a niche in the rumen environment. (C) An ideal scenario where native rumen acetogens are enhanced by improving the efficiency of the Wood–Ljungdahl (WL) pathway. (D) A theoretical and highly promising situation in which engineered strains of methanogens permanently replace methane production with acetic acid production.
Learning from all these prior efforts, we now explore hypothetical, sustainable scenarios to maximize the promotion of acetogenesis in the rumen (Figures 3C, D). Increasing hydrogen concentration in the rumen is one way to solve this puzzle. This can be achieved by selecting for microorganisms with high hydrogen production under rumen conditions. But since high hydrogen levels block rumen fermentation, prior to examining this approach, several questions must be addressed (Table 4). For instance, what is the relationship between hydrogen-producing bacteria in the rumen and the methanogens or acetogens? Certain studies indicate a mutualistic relationship where there is a close physical association between hydrogen generators and the methanogens. The question then becomes, is it feasible to selectively disrupt the relationship between H2 producers and methanogens in the rumen? Furthermore, do primary hydrogen generators have any association with acetogens? If not, can we deliberately initiate a syntrophic relationship between them so that hydrogen is more available for acetogens?
The use of efficient acetogens that can compete with methanogens at relatively low hydrogen levels is another promising research area. Highly efficient acetogens from non-rumen environments have been targeted for introduction into the rumen, as demonstrated by Karekar and Ahring (2023). In their study, methanogenesis was suppressed for seven days without a significant change in acetate production. To further advance this line of research, several questions need to be addressed. For example, what could be the collateral damage of the power shift from methanogens to acetogens in a rumen ecosystem that has evolved over millennia to favor domination by methanogens? How challenging would it be to integrate non-native acetogens into the cow rumen? Mimicking the kangaroo gut environment might help, but is it practical or beneficial for animal performance to alter the rumen system so extensively? Furthermore, what happens when hydrogen levels become limiting, which is the case when methanogenic inhibitors like BES are exhausted? While these acetogens are efficient at utilizing hydrogen, they have broad metabolic flexibility and could easily switch to other substrates when hydrogen becomes scarce. Le Van et al. (1998) reported that rumen acetogens prefer substrates other than H2 and CO2. In vivo studies also reveal that replacing methanogens with acetogens reduces hydrogen capture from fermentation by up to 46%, compared to the over 90% efficiency seen in methanogen-dominated rumens (Fonty et al., 2007). This impairs degradative activity of ruminal fermentative bacteria and decreases overall animal performance.
The problems highlighted above can be mitigated by focusing on genetic manipulations. We identify two possible opportunities: 1) genetic engineering native acetogens to increase efficiency of the Wood–Ljungdahl (WL) pathway (Figure 4), or by employing traditional mutant selection approaches used in microbiology. Increasing the copy number of critical genes like Codh and the Acs genes in core rumen acetogens might increase their hydrogen scavenging potential; 2) Methanogens can be custom designed to produce acetate instead of methane (Figure 5). This could be achieved through genetic engineering of methanogens to abate the methane production branch of the WL pathway.

Figure 4. Schematic representation of the WL pathways in acetogens (left) and methanogens (right). Steps restricted to bacteria are marked in bright blue color, whereas those only found in archaea are shown in orange. Dark blue stands for reactions and enzymes observed in both prokaryotic domains. Abbreviations are as follows: Fdh, formate dehydrogenase; Fmd/Fwd, Formylmethanofuran dehydrogenase; Mtr, coenzyme M methyltransferase; Mcr, methyl-coenzyme M reductase; Mer, methyl-ene-H4MPT reductase; Mch, methenyl-H4MPT cyclohydrolase; Ftr, formylmethanofuran:H4MPT formyltransferase; Fch, methenyl-H4F cyclohydrolase; CODH, carbon monoxide dehydrogenase; ACS, acetyl coenzyme A synthase.
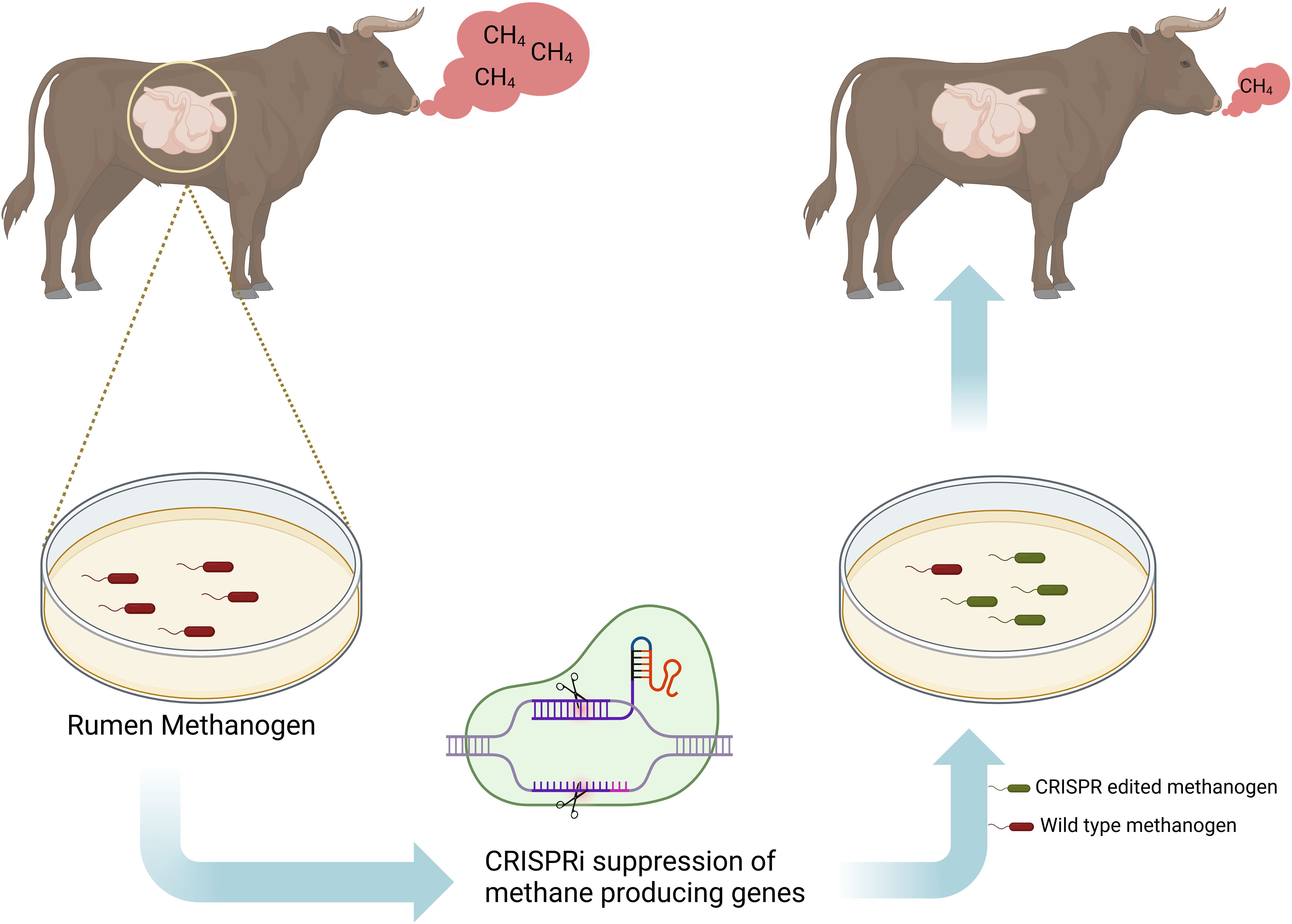
Figure 5. Possible in vitro approach for the gene editing of rumen methanogens. Methanogens would be collected from the rumen and cultured in vitro, at which point methane producing genes are knocked down using CRISPRi. The gene-edited methanogens are then replicated in vitro and reintroduced into the rumen for repopulation. If the edited methanogens can compete effectively within the rumen population, the animal is expected to emit significantly less methane from enteric fermentation. Adapted from Subedi et al. (2022).
3.2.5 Genetic engineering of methanogens to abate methane production
A significant (95%) amount of methane produced in the rumen arises from bioconversion of the products of fermentation (hydrogen and carbon dioxide), which is known as hydrogenotrophic methanogenesis (Mackie et al., 2024). However, there is a natural alternative pathway for bioconversion of CO2 and hydrogen into acetate through the Wood–Ljungdahl (WL) pathway (Figure 4). This pathway is commonly employed by both methanogens and acetogens for carbon fixation and by acetogens also for energy generation (Rosenbaum and Müller, 2021). The production of acetate from CO2 and H2 is here termed primary acetogenesis, whereby CO2 molecules are sequentially reduced under two branches, the carbonyl branch and the methyl branch.
In the methyl branch, CO2 is reduced through a series of sequential hydrogenations reactions to a methyl intermediate (Karekar and Stefanini, 2022). In the carbonyl branch, CO2 is first reduced to carbon monoxide (CO) by means of the enzyme carbon monoxide dehydrogenase (Codh). The CO then is conjugated to the methyl intermediate by means of acetyl-CoA synthase (Acs) to produce acetyl coenzyme A (acetyl-CoA). This acetyl-CoA is subsequently converted to acetate during catabolism or utilized in the synthesis of cell carbon during anabolism (Drake et al., 2013).
If the WL pathway is compared between methanogenic archaea and acetogens it is found that, in archaea, the route that leads to methanogenesis is a metabolic extension and involves two additional enzymes, specifically coenzyme M methyltransferase (Mtr) and methyl-coenzyme M reductase (Mcr), collectively known as the Mtr-Mcr module (Figure 4) (Scheller and Rother, 2022). Thus, there is a clear opportunity to delete or bypass the route that leads to methane production. Acetyl CoA is believed to be an ancient metabolic product, and given that methanogens evolved later than acetogens due to their energy conservation advantage (Martin, 2020), experiments to revert methanogenic archaea to acetogens seem plausible. Therefore, it is theoretically possible to rewire methanogenic metabolism by just diverging all substrate carbon from methanogenesis to flow through acetyl-CoA, and thereby converting methanogens into acetogens. This can be achieved either by spontaneous mutation in an adaptive evolution strategy (i.e. selecting for growth of methanogens in a medium lacking key methanogenesis substrates or with a methanogenesis inhibitor) or through targeted mutations of genes encoding essential enzymes like Mcr, Mtr, and Codh (Figure 4), and several others as highlighted by (Lessner et al., 2006) using advanced technologies like CRISPR-Cas (clustered regularly interspaced short palindromic repeats).
In just a few years since its discovery, CRISPR-Cas-based approaches have propelled forward the area of precision genome engineering. The CRISPR-Cas system is a primitive adaptive immune system found in 40% of bacteria and 90% of archaea (Dhamad and Lessner, 2020). These organisms use CRISPR-Cas systems to defend themselves against viruses. The systems generally have two parts: ‘guide RNA’ molecules that recognize and bind to the targeted viral DNA or RNA, and the Cas enzymes that cut the target. This system can be redirected to delete or add new functions to the cell through direct genome editing. Essentially, the nucleases generate a DNA double-strand break (DSB) at the targeted genome locus, triggering repair by either nonhomologous end joining (NHEJ) or homology-directed repair (HDR). Without a template, NHEJ introduces insertions or deletions that disrupt the target site. With a homologous template, HDR allows precise modifications (Jakočiunas et al., 2016). Although an ever-increasing number of variants of CRISPR-Cas systems have been identified, the first well characterized and widely used system in genome editing is the type II CRISPR-Cas9 system from Streptococcus pyogenes (Jinek et al., 2012).
Beyond DNA modification, adaptations of CRISPR-Cas can also regulate gene expression through CRISPR interference (CRISPRi) and CRISPR activation (CRISPRa) (De Bakker et al., 2022). CRISPRi suppresses gene expression and is particularly valuable for modulating essential genes, where deletion might be lethal (Stachler et al., 2020). Suppression can occur pre-transcriptionally using a catalytically deactivated version of Cas9 (dCas9), where the dCas9–guide RNA complex physically blocks RNA polymerase or transcription factors. Post-transcriptional suppression involves mRNA degradation (Doudna and Charpentier, 2014). For gene activation (CRISPRa), dCas9 is fused to an activator domain, directly or via recruitment domains on the guide RNA, to enhance gene transcription (Bikard et al., 2013).
All these CRISPR-Cas tools can be employed to edit methanogenic archaeal genomes, targeting the genes responsible for methanogenesis. Mcr is a key gene target, which catalyzes the final step of methanogenesis and is found in all methanogens (Beauchemin et al., 2020). Recently, Nayak and Metcalf (2017) deleted Mcr and a heterodisulfide reductase (hdrED) gene using the S. pyogenes-derived CRISPR-Cas9 platform on Methanosarcina acetivorans and showed high efficiency in creating insertions (knock-ins) and deletions (knockouts) through homology directed repair (HDR) without any evidence of off-target mutations. This experiment delivered only five transformants, confirming the essentiality of these genes. Conversely, when deletions were made in non-essential genes responsible for encoding monomethylamine-specific methyltransferases (mtmCB1 and mtmCB2), which facilitate methylamine utilization in the methylotrophic pathway, thousands of transformants were generated (Nayak and Metcalf, 2017). Although the phenotypic behavior of these transformants was not studied, the results suggest that knocking out methyltransferases could be more effective for reducing methane in methylotrophic methanogens, while suppression may be a better approach for essential genes like Mcr and hdrED. Indeed, a simple and efficient CRISPRi system for targeted gene repression has been developed in archaea, successfully achieving a 90% reduction in transcript levels of nitrogen fixation genes in M. acetivorans (Dhamad and Lessner, 2020). Similarly, CRISPRi systems using endogenous type I and type III CRISPR pathways have been implemented in extremophiles Haloferax volcanii and Sulfolobus spp. Testing the effectiveness of CRISPRi systems in downregulating Mcr would be of great interest.
In addition to Mcr, other gene complexes like Mtr and Codh could be targeted. Mtr transfers a methyl group from tetrahydromethanopterin (H4MPT) to coenzyme M (CoM-SH) at the end of the WL pathway (Adam et al., 2022; Figure 4), while Codh plays a central role in CO metabolism (Biester et al., 2022). By overexpressing Codh or inhibiting Mtr, it is possible to promote acetyl-CoA production. In 2004, Rother and Metcalf found that exposing M. acetivorans to varying concentration of CO inhibited Mtr and upregulated Codh activity threefold. In 2006, Lessner et al., further observed an eightfold reduction in Mtr when M. acetivorans was grown on CO as compared to methanol as substrate. Building on these two experiments, Schöne et al. (2022) successfully deconstructed M. acetivorans into an acetogenic archaeon by targeted disruption of Mtr, followed by adaptive evolution. However, since M. acetivorans in the rumen are very few (Nagarajan et al., 2019) and their contribution to methane production is insignificant, extending this work to key emitters like M. ruminantium could be impactful. Unfortunately, hydrogenotrophic methanogens lack the Codh complex (Nagoya et al., 2021), so CRISPR tools could be used to introduce and overexpress Codh in M. ruminantium to explore CO-dependent acetogenesis.
The availability of the genome sequence of M. ruminantium (Leahy et al., 2010) and the recent identification of its operon and functional properties (Bharathi et al., 2020), combined with the extensive understanding of methanogenic biochemical pathways (Deppenmeier, 2002), opens up possibilities for genome editing to mitigate methane emissions in this organism. Advanced tools like DNA-editing all-in-one RNA-guided CRISPR-Cas transposase (DART) and environmental transformation sequencing (ET-seq) now allow for targeted, multi-species editing within complex microbiomes (Rubin et al., 2021). These innovations address key challenges in targeting, delivery, and the survival of edited species. The evidence suggests that when a species of bacteria is isolated from the target environment, engineered, and then returned, their persistence is maintained for longer periods than when non-native species are introduced (Buiatti et al., 2013). However, in situ microbial engineering is still in its infancy; more research must be done before direct editing of the rumen microbial community becomes a reality.
4 Conclusion
Looking forward, we believe engineered biology has the potential to deliver solutions at a scale equivalent to the scale of the climate crisis we are currently facing. The potential to engineer the key methane producer in the rumen of cows is one promising area. If an engineered strain of methanogen is successfully integrated in the microbiome of a cow and subsequently throughout the entire cattle population, a significant and persistent reduction of enteric methane could be achieved. While the technology is promising, there are two critical issues that must be addressed. One, the potential escape of the edited gene which could result in unforeseen consequences for the health of the animal and to the ecosystem at large. Two, the risk of the construct losing its intended function by re-acquiring methanogenic functions from the native populations.
For the former, genetic biocontainment technologies that manage the risk of unintended consequences could be integrated to improve the safety and security of engineered organisms. Approaches that physically confine the cells so that they can be used in the environment without escaping into it have been developed and used. Example, hydrogels have been used to encapsulate E. coli engineered to sense trinitrotoluene (TNT) (Belkin et al., 2017). Techniques have also been developed to create cells that are intact and metabolically active, but cannot replicate and do not transfer DNA to the environment (Fan et al., 2020). Moreover, knocking out an essential biosynthetic gene and exogenously supply the missing metabolite, thereby creating a synthetic auxotroph, is another approach (Gallagher et al., 2015). Therapeutic Lactococcus lactis was controlled by knocking out the thymidylate synthase gene, blocking growth in the gut where thymidine/thymine are rare (Steidler et al., 2003). Despite these advancements, genetic biocontainment remains vulnerable to factors such as cross-feeding, genetic mutations, or unforeseen environmental conditions, all of which must be accounted for to ensure consistent and stable containment (Payne et al., 2024).
The potential of engineered species to re-acquire methanogenic functions from the native population via horizontal gene transfer (HGT) warrants attention, as this would dilute the intended effect of targeted engineering approaches. While HGT is a common process among members in microbial communities, the literature indicates that natural competence and transformation in archaea are relatively rare (Gophna and Altman-Price, 2022). Even archaea that are naturally transformable typically show lower transformation frequencies as compared to bacterial counterparts. Moreover, DNA uptake mechanisms, such as the competence (Com) proteins ComEA and ComEC, have not been identified in archaea, and only a few archaeal viruses have been shown to facilitate HGT. Irrespective of this, several synthetic genetic strategies have recently been proposed to either prevent or penalize the loss of function as well as eliminating cells that bear mutations in DNA sequences of interest. The strategies range from editing the genomes of the host cell for eliminating insertion sequences, to active genetic circuits for detecting underperformance of engineered constructs (Nikel and De Lorenzo, 2021). It is crucial to emphasize the need for extensive research to thoroughly evaluate and quantify 1) the likelihood of horizontal gene transfer and 2) the efficacy of built-in biocontainment mechanisms as safety controls.
Given the rapid advancement in artificial intelligence capabilities, it may be possible to develop predictive tools that can identify ecological risks quickly as they occur, including deleterious genetic rearrangements and horizontal gene transfer. Continued developments in the area of biocontainment will ensure that the engineered organisms remain controllable, stable, and predictable in the environment. While completely eliminating the burden of programmed cells may be unattainable, engineering efforts such as those described here can at least mitigate its impact and allow the development of synthetic biology systems that work efficiently and in better harmony with the natural environment.
Methane is dramatically more potent at warming the planet than carbon dioxide, and responsible for 30% of global temperature rise since the Industrial Revolution. More than half of human-driven methane emissions are microbial in origin. Cattle, through enteric fermentation, are at the top of these emissions, making them a key piece of the climate change puzzle. Some of the proposed strategies such as reducing livestock production do not address the root source of emission. In this review, we holistically examined solutions to the problem of cattle methane emissions, with emphasis on biotechnological solutions related to the rumen microbiome. We argue that applying CRISPR technology to methanogen engineering could result in a permanently low-methane cow. Moreover, such organisms are likely to be scaled to other biogenic sources such as wetlands and rice paddies. We postulate that the WL pathway, especially channeling H2 to acetogenesis, can be a remedy to rumen methane production and provide a pervasive avenue for addressing global methane emission beyond the rumen ecosystem.
While HGT is a common phenomenon, natural competence and transformation in archaea have been reported to be scarce (Gophna and Altman-Price, 2022) and even those archaea that are naturally transformable typically show low transformation frequencies when compared to their bacterial counterparts. Furthermore, DNA uptake mechanisms such us the competence (Com) proteins ie ComEA/ComEC have not been found in archaea and, very few archaeal viruses have also shown to serve as agents of HGT. Despite this, a number of synthetic genetic approaches have been proposed to either prevent or penalize the loss of function as well as eliminating cells that bear mutations in DNA sequences of interest. The strategies range from editing the genomes of the host cell for eliminating insertion sequences, to active genetic circuits for detecting underperformance of engineered constructs (Nikel and De Lorenzo, 2021). However, multiple studies are needed to qualify and quantify these risks. More lab-scale and field-scale studies are needed to characterize 1) the likelihood of horizontal gene transfer and 2) the efficacy of built-in biocontainment mechanisms as safety controls.
Given the rapid advancement in artificial intelligence capabilities, it may be possible to develop predictive tools that can identify ecological risks quickly as they occur, including deleterious genetic rearrangements and horizontal gene transfer. Continued developments in the area of biocontainment will ensure that the engineered organisms remain controllable, stable, and predictable in the environment. It may not be possible to ever eliminate burden when programming cells to do extra tasks; however, engineering efforts such as those described here can at least mitigate its impact and allow the development of synthetic biology systems that work efficiently and in better harmony with the natural environment.
Methane is dramatically more potent at warming the planet than carbon dioxide, and responsible for 30% of global temperature rise since the Industrial Revolution. More than half of human-driven methane emissions are microbial in origin. Cattle, through enteric fermentation, are at the top of these emissions, making them a key piece of the climate change puzzle. Some of the proposed strategies such as reducing livestock production do not address the root source of emission. In this review, we holistically examined solutions to the problem of cattle methane emissions, with emphasis on biotechnological solutions related to the rumen microbiome. We argue that applying CRISPR technology to methanogen engineering could result in a permanently low-methane cow. Moreover, such organisms are likely to be scaled to other biogenic sources such as wetlands and rice paddies. We postulate that the WL pathway, especially channeling H2 to acetogenesis, can be a remedy to rumen methane production and provide a pervasive avenue for addressing global methane emission beyond the rumen ecosystem.
Author contributions
RM: Conceptualization, Visualization, Writing – original draft, Writing – review & editing. AMA: Supervision, Writing – review & editing. KU: Supervision, Writing – review & editing. AA: Supervision, Writing – review & editing. NF: Funding acquisition, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. RM is supported by the Regional Scholarship and Innovation Fund (RSIF) through the Partnership for Skills in Applied Sciences, Engineering and Technology (PASET) program. NF is supported by new faculty start-up funds from Worcester Polytechnic Institute. The funders were not involved in preparation of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adam P. S., Kolyfetis G. E., Bornemann T. L. V., Vorgias C. E., Probst A. J. (2022). Genomic remnants of ancestral methanogenesis and hydrogenotrophy in Archaea drive anaerobic carbon cycling. Sci. Advance 8, eabm9651. doi: 10.1126/sciadv.abm9651
Alazzeh A. Y., Sultana H., Beauchemin K. A., Wang Y., Holo H., Harstad O. M., et al. (2012). Using strains of Propionibacteria to mitigate methane emissions in vitro. Acta Agricult. Scandinavica A: Anim. Sci. 62, 263–272. doi: 10.1080/09064702.2013.773056
Alemu A. W., Pekrul L. K. D., Shreck A. L., Booker C. W., McGinn S. M., Kindermann M., et al. (2021). 3-nitrooxypropanol decreased enteric methane production from growing beef cattle in a commercial feedlot: implications for sustainable beef cattle production. Front. Anim. Sci. 2. doi: 10.3389/fanim.2021.641590
Alemu A. W., Romero-Pérez A., Araujo R. C., Beauchemin K. A. (2019). Effect of encapsulated nitrate and microencapsulated blend of essential oils on growth performance and methane emissions from beef steers fed backgrounding diets. Animals 9, 1–19. doi: 10.3390/ani9010021
Altermann E., Reilly K., Young W., Ronimus R. S., Muetzel S. (2022). Tailored nanoparticles with the potential to reduce ruminant methane emissions. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.816695
Altermann E., Schofield L. R., Ronimus R. S., Beatty A. K., Reilly K. (2018). Inhibition of Rumen methanogens by a novel archaeal lytic enzyme displayed on tailored bionanoparticles. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02378
Angelidaki I., Treu L., Tsapekos P., Luo G., Campanaro S., Wenzel H., et al. (2018). Biogas upgrading and utilization: Current status and perspectives. Biotechnol. Adv. 36, 452–466. doi: 10.1016/j.bioteChadv.2018.01.011
Arndt C., Hristov A. N., Price W. J., McClelland S. C., Pelaez A. M., Cueva S. F., et al. (2023). Full adoption of the most effective strategies to mitigate methane emissions by ruminants can help meet the 1.5°C target by 2030 but not 2050. PNAS 119, 1–10. doi: 10.1073/pnas
Asano Y. M., Biermann G. (2019). Rising adoption and retention of meat-free diets in online recipe data. Nat. Sustainabil. 10, 621–627. doi: 10.1038/s41893-019-0316-0
Basu S., Lan X., Dlugokencky E., Michel S., Schwietzke S., Miller J. B., et al. (2022). Estimating emissions of methane consistent with atmospheric measurements of methane and δ13 C of methane. Atmos. Chem. Phys. 22, 15351–15377. doi: 10.5194/acp-22-15351-2022
Beauchemin K. A., Ungerfeld E. M., Eckard R. J., Wang M. (2020). Review: Fifty years of research on rumen methanogenesis: Lessons learned and future challenges for mitigation. Animal 14, S2–S16. doi: 10.1017/S1751731119003100
Belanche A., Newbold C. J., Morgavi D. P., Bach A., Zweifel B., Yáñez-Ruiz D. R. (2020). A meta-analysis describing the effects of the essential oils blend agolin ruminant on performance, rumen fermentation and methane emissions in dairy cows. Animals 10, 1–15. doi: 10.3390/ani10040620
Belkin S., Yagur-Kroll S., Kabessa Y., Korouma V., Septon T., Anati Y., et al. (2017). Remote detection of buried landmines using a bacterial sensor. Nat. Biotechnol. 35, 308–310. doi: 10.1038/nbt.3791
Bharathi M., Senthil Kumar N., Chellapandi P. (2020). Functional prediction and assignment of methanobrevibacter ruminantium M1 operome using a combined bioinformatics approach. Front. Genet. 11. doi: 10.3389/fgene.2020.593990
Biester A., Marcano-Delgado A. N., Drennan C. L. (2022). Structural insights into microbial one-carbon metabolic enzymes ni–fe–S-dependent carbon monoxide dehydrogenases and acetyl-coA synthases. Biochemistry 61, 2797–2805. doi: 10.1021/acs.biochem.2c00425
Bikard D., Jiang W., Samai P., Hochschild A., Zhang F., Marraffini L. A. (2013). Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 41, 7429–7437. doi: 10.1093/nar/gkt520
Black J. L., Davison T. M., Box I. (2021). Methane emissions from ruminants in Australia: Mitigation potential and applicability of mitigation strategies. Animals 11, 1–20. doi: 10.3390/ani11040951
Boccazzi P., Patterson J. A. (2011). Using hydrogen-limited anaerobic continuous culture to isolate lowhydrogen threshold ruminal acetogenic bacteria. Agric. Food Anal. Bacteriol. 1, 33. Available at: www.AFABjournal.Com.
Brask-Pedersen D. N., Lamminen M., Mogensen L., Hellwing A. L. F., Johansen M., Lund P., et al. (2023). Effect of substituting grass-clover silage with maize silage for dairy cows on nutrient digestibility, rumen metabolism, enteric methane emission and total carbon footprint. Livestock Sci. 274, 105273. doi: 10.1016/j.livsci.2023.105273
Breznak J. A., Kane M. D. (1990). Microbial H 2/CO 2 acetogenesis in animal guts: Nature and nutritional significance. FEMS Microbiol. Lett. 87, 309–314. doi: 10.1111/j.1574-6968.1990.tb04929.x
Breznak J. A., Switzer J. M., Seitz H. J. (1988). Sporomusa termitida sp. Nov., an H2/CO2-utilizing acetogen isolated from termites. Arch. Microbiol. 150, 282–288. doi: 10.1007/BF00407793
Buiatti M., Christou P., Pastore G. (2013). The application of GMOs in agriculture and in food production for a better nutrition: Two different scientific points of view. Genes Nutr. 8, 255–270. doi: 10.1007/s12263-012-0316-4
Cain M., Lynch J., Allen M. R., Fuglestvedt J. S. (2019). Improved calculation of warming-equivalent emissions for short-lived climate pollutants. NPJ Climate Atmos. Sci. 2, 1–7. doi: 10.1038/s41612-019-0086-4
Calabrese S., Garcia A., Wilmoth J. L., Zhang X., Porporato A. (2021). Critical inundation level for methane emissions from wetlands. Environ. Res. Lett. 16, 1–11. doi: 10.1088/1748-9326/abedea
Cammack K. M., Austin K. J., Lamberson W. R., Conant G. C., Cunningham H. C. (2018). Ruminnat nutrition symposium: Tiny but mighty: The role of the rumen microbes in livestock production. J. Anim. Sci. 96, 752–770. doi: 10.1093/jas/skx053
Capper J. L., Bauman D. E. (2013). The role of productivity in improving the environmental sustainability of ruminant production systems. Annu. Rev. Anim. Biosci. 1, 469–489. doi: 10.1146/annurev-animal-031412-103727
Cezimbra I. M., de Albuquerque Nunes P. A., de Souza Filho W., Tischler M. R., Genro T. C. M., Bayer C., et al. (2021). Potential of grazing management to improve beef cattle production and mitigate methane emissions in native grasslands of the Pampa biome. Sci. Total Environ. 780, 1–8. doi: 10.1016/j.scitotenv.2021.146582
Chaucheyras F., Fonty G., Bertin G., Gouet P. (1995). In vitro H2 utilization by a ruminal acetogenic bacterium cultivated alone or in association with an archaea methanogen is stimulated by a probiotic strain of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 61, 3466–3467. doi: 10.1128/aem.61.9.3466-3467.1995
Cheng M., McCarl B., Fei C. (2022). Climate change and livestock production: A literature review. Atmosphere 13, 1–20. doi: 10.3390/atmos13010140
Choudhury P. K., Jena R., Tomar S. K., Puniya A. K. (2022). Reducing enteric methanogenesis through alternate hydrogen sinks in the rumen. Methane 1, 320–341. doi: 10.3390/methane1040024
Chriki S., Hocquette J. (2022). Is “cultured meat” a viable alternative to slaughtering animals and a good comprise between animal welfare and human expectations? Animal Frontiers. 12(1), 35–42. doi: 10.1093/af/vfac002
Collins W. J., Webber C. P., Cox P. M., Huntingford C., Lowe J., Sitch S., et al. (2018). Increased importance of methane reduction for a 1.5 degree target. Environ. Res. Lett. 13, 1–10. doi: 10.1088/1748-9326/aab89c
Cord-Ruwisch R., Seitz H. J., Conrad R. (1988). The capacity of hydrogenotrophic anaerobic bacteria to compete for traces of hydrogen depends on the redox potential of the terminal electron acceptor. Arch. Microbiol. 149, 350–357. doi: 10.1007/BF00411655
Crutzen P. J., Aselmann I., Seiler W. (1986). Methane production by domestic animals, wild ruminants, other herbivorous fauna, and humans. Tellus B: Chem. Phys. Meteorol. 38, 271–284. doi: 10.3402/tellusb.v38i3-4.15135
Darabighane B., Tapio I., Ventto L., Kairenius P., Stefański T., Leskinen H., et al. (2021). Effects of starch level and a mixture of sunflower and fish oils on nutrient intake and digestibility, rumen fermentation, and ruminal methane emissions in dairy cows. Animals 11, 1–19. doi: 10.3390/ani11051310
De Bakker V., Liu X., Bravo A. M., Veening J.-W. (2022). CRISPRi-seq for genome-wide fitness quantification in bacteria. Nat. Protoc. 17, 252–281. doi: 10.1038/s41596-021-00639-6
De Graeve K. G., Grivet J. P., Durand M., Beaumatin P., Cordelet C., Hannequart G., et al. (1994). Competition between reductive acetogenesis and methanogenesis in the pig large-intestinal flora. J. Appl. Microbiol. 76 (1), 55–61. doi: 10.1111/j.1365-2672.1994.tb04415.x
Demeyer D., Fiedler D., De Graeve K. (1996). Attempted induction of reductive acetogenesis into the rumen fermentation in vitro. Reprod. Nutr. Dev. 36, 233–240. doi: 10.1051/rnd:19960301
Deppenmeier U. (2002). The unique biochemistry of methanogenesis. Prog. Nucleic Acid Res. Mol. Biol. 71, 223–283. doi: 10.1016/s0079-6603(02)71045-3
Dhamad A. E., Lessner D. J. (2020). A CRISPRi-dCas9 System for Archaea and Its Use To Examine Gene Function during Nitrogen Fixation by Methanosarcina acetivorans. Appl. Environ. Microbiol. 86 (21), e01402–e01420. doi: 10.1128/AEM.01402-20
Djemai K., Drancourt M., Tidjani Alou M. (2021). Bacteria and methanogens in the human microbiome: A review of syntrophic interactions. Microb. Ecol. 0123456789, 1–19. doi: 10.1007/s00248-021-01796-7
Doudna J. A., Charpentier E. (2014). The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096. doi: 10.1126/science.1258096
Duin E. C., Wagner T., Shima S., Prakash D., Cronin B., Yáñez-Ruiz D. R., et al. (2016). Mode of action uncovered for the specific reduction of methane emissions from ruminants by the small molecule 3-nitrooxypropanol. PNAS 113 (22), 6172–6177. doi: 10.1073/pnas.1600298113
Elmhadi M. E., Ali D. K., Khogali M. K., Wang H. (2022). Subacute ruminal acidosis in dairy herds: Microbiological and nutritional causes, consequences, and prevention strategies. Anim. Nutr. 10, 148–155. doi: 10.1016/j.aninu.2021.12.008
Fan C., Davison P. A., Habgood R., Zeng H., Decker C. M., Gesell Salazar M., et al. (2020). Chromosome-free bacterial cells are safe and programmable platforms for synthetic biology. Proc. Natl. Acad. Sci. 117, 6752–6761. doi: 10.1073/pnas.1918859117
Food and Agricultural Organization (2019). Options for low-emission development in the Tanzania dairy sector: Reducing enteric methane for food security and livelihoods. FAO, Rome, Italy. Retrieved on 11 February 2025 from https://openknowledge.fao.org/server/api/core/bitstreams/8a3892bc-3daa-433e-9520-7ab86480dffd/content.
Food and Agricultural Organization (2018). The state of food security and nutrition in the world: building climate resilience for food security and nutrition (Rome, Italy: FAO). Available at: https://openknowledge.fao.org/server/api/core/bitstreams/f5019ab4-0f6a-47e8-85b9-15473c012d6a/content.
Food and Agricultural Organization (2022). FAO strategy on climate change (Rome, Italy: FAO). Available at: https://openknowledge.fao.org/server/api/core/bitstreams/f6270800-eec7-498f-9887-6d937c4f575a/content.
Fehér A., Gazdecki M., Véha M., Szakály M., Szakály Z. (2020). A comprehensive review of the benefits of and the barriers to the switch to a plant-based diet. Sustainabil. (Switzerland) 12, 1–18. doi: 10.3390/su12104136
Feng X. Y., Dijkstra J., Bannink A., van Gastelen S., France J., Kebreab E. (2020). Antimethanogenic effects of nitrate supplementation in cattle: A meta-analysis. J. Dairy Sci. 103, 11375–11385. doi: 10.3168/jds.2020-18541
Fonty G., Chaucheyras-Durand F. (2006). Effects and modes of action of live yeasts in the rumen. Biologia 61, 741–750. doi: 10.2478/s11756-006-0151-4
Fonty G., Joblin K., Chavarot M., Roux R., Naylor G., Michallon F. (2007). Establishment and development of ruminal hydrogenotrophs in methanogen-free lambs. Appl. Environ. Microbiol. 73, 6391–6403. doi: 10.1128/AEM.00181-07
Furman O., Shenhav L., Sasson G., Kokou F., Honig H., Jacoby S., et al. (2020). Stochasticity constrained by deterministic effects of diet and age drive rumen microbiome assembly dynamics. Nat. Commun. 11, 1–13. doi: 10.1038/s41467-020-15652-8
Gagen E. J., Denman S. E., Padmanabha J., Zadbuke S., Jassim R. A., Morrison M., et al. (2010). Functional gene analysis suggests different acetogen populations in the bovine rumen and tammar wallaby forestomach. Appl. Environ. Microbiol. 76, 7785–7795. doi: 10.1128/AEM.01679-10
Gagen E. J., Mosoni P., Denman S. E., Al Jassim R., McSweeney C. S., Forano E. (2012). Methanogen colonisation does not significantly alter acetogen diversity in lambs isolated 17 h after birth and raised aseptically. Microb. Ecol. 64, 628–640. doi: 10.1007/s00248-012-0024-z
Gagen E. J., Wang J., Padmanabha J., Liu J., De Carvalho I. P. C., Liu J., et al. (2014). Investigation of a new acetogen isolated from an enrichment of the tammar wallaby forestomach. BMC Microbiol. 14, 1–14. doi: 10.1186/s12866-014-0314-3
Gallagher R. R., Patel J. R., Interiano A. L., Rovner A. J., Isaacs F. J. (2015). Multilayered genetic safeguards limit growth of microorganisms to defined environments. Nucleic Acids Res. 43, 1945–1954. doi: 10.1093/nar/gku1378
Georges M., Charlier C., Hayes B. (2019). Harnessing genomic information for livestock improvement. Nat. Rev. Genet. 20, 135–156. doi: 10.1038/s41576-018-0082-2
Gilbert R. A., Townsend E. M., Crew K. S., Hitch T. C. A., Friedersdorff J. C. A., Creevey C. J., et al. (2020). Rumen virus populations: technological advances enhancing current understanding. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00450
Gilbert R. A., Kelly W. J., Altermann E., Leahy S. C., Minchin C., Ouwerkerk D., et al. (2017). Toward understanding phage: Host interactions in the rumen; complete genome sequences of lytic phages infecting rumen bacteria. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02340
Godwin S., Kang A., Gulino L.-M., Manefield M., Gutierrez-Zamora M.-L., Kienzle M., et al. (2014). Investigation of the microbial metabolism of carbon dioxide and hydrogen in the kangaroo foregut by stable isotope probing. ISME J. 8, 1855–1865. doi: 10.1038/ismej.2014.25
Golder H. M., Thomson J. M., Denman S. E., McSweeney C. S., Lean I. J. (2018). Genetic markers are associated with the ruminal microbiome and metabolome in grain and sugar challenged dairy heifers. Front. Genet. 9. doi: 10.3389/fgene.2018.00062
González-Recio O., López-Paredes J., Ouatahar L., Charfeddine N., Ugarte E., Alenda R., et al. (2020). Mitigation of greenhouse gases in dairy cattle via genetic selection: 2. Incorporating methane emissions into the breeding goal. J. Dairy Sci. 103, 7210–7221. doi: 10.3168/jds.2019-17598
Gophna U., Altman-Price N. (2022). Horizontal gene transfer in archaea—From mechanisms to genome evolution. Annu. Rev. Microbiol. 76, 481–502. doi: 10.1146/annurev-micro-040820-124627
Granja-Salcedo Y. T., Fernandes R. M. I., De Araujo R. C., Kishi L. T., Berchielli T. T., De Resende F. D., et al. (2019). Long-term encapsulated nitrate supplementation modulates rumen microbial diversity and rumen fermentation to reduce methane emission in grazing steers. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00614
Greening C., Geier R., Wang C., Woods L. C., Morales S. E., McDonald M. J., et al. (2019). Diverse hydrogen production and consumption pathways influence methane production in ruminants. ISME J. 13, 2617–2632. doi: 10.1038/s41396-019-0464-2
Greenpeace (2020). Farming for Failure: How European animal farming fuels the climate emergency. Brussels, Belgium: Greenpeace 18. Available at: https://www.greenpeace.org/static/planet4-eu-unit-stateless/2020/09/20200922-Greenpeace-report-Farming-for-Failure.pdf
Hayek M. N., Garrett R. D. (2018). Nationwide shift to grass-fed beef requires larger cattle population. Environ. Res. Lett. 13, 1–9. doi: 10.1088/1748-9326/aad401
He L., Groom J. D., Wilson E. H., Fernandez J., Konopka M. C., Beck D. A. C., et al. (2023). A methanotrophic bacterium to enable methane removal for climate mitigation. Proc. Natl. Acad. Sci. 120, e2310046120. doi: 10.1073/pnas.2310046120
Hegarty R. S., Gerdes R. (1999). Hydrogen production and transfer in the rumen. Recent Adv. Anim. Nutr. Aust. 12, 37–44.
Henderson G., Cox F., Ganesh S., Jonker A., Young W., Janssen P. H., et al. (2015). Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 5, 1–15. doi: 10.1038/srep14567
Hodge I., Quille P., O’Connell S. (2024). A review of potential feed additives intended for carbon footprint reduction through methane abatement in dairy cattle. Animals 14, 568. doi: 10.3390/ani14040568
Houzer E., Scoones I. (2021). Are livestock always bad for the planet? Rethinking the protein transition and climate change debate. Brighton: PASTRES, 1–80. doi: 10.19088/STEPS.2021.003
Humpenöder F., Bodirsky B. L., Weindl I., Lotze-campen H., Linder T., Popp A. (2022). Projected environmental benefits of replacing beef with microbial protein. Nature 10, 1–21. doi: 10.1038/s41586-022-04629-w
Ibrahim N. A., Alimon A. R., Yaakub H., Samsudin A. A., Candyrine S. C. L., Wan Mohamed W. N., et al. (2021). Effects of vegetable oil supplementation on rumen fermentation and microbial population in ruminant: A review. Trop. Anim. Health Product. 53, 422. doi: 10.1007/s11250-021-02863-4
IPCC (2018). Global warming of 1.5°C: An IPCC special report on the impacts of global warming of 1.5°C above pre-industrial levels and related global greenhouse gas emission pathways, in the context of strengthening the global response to the threat of climate change, sustainable development, and efforts to eradicate poverty. Eds. Delmotte V. M., Zhai P., Pörtner H.-O., Roberts D., Skea J., Shukla P. R., et al (Cambridge; New York, NY: Cambridge University Press).
IPCC (2022). Climate change 2022: Impacts, adaptation and vulnerability. contribution of working group II to the sixth assessment report of the intergovernmental panel on climate change. Eds. Pörtner H.-O., Roberts D. C., Tignor M., Poloczanska E. S., Mintenbeck K., Alegría A., et al (Cambridge; New York, NY: Cambridge University Press).
Ismail I., Hwang Y. H., Joo S. T. (2020). Meat analog as future food: A review. J. Anim. Sci. Technol. 62, 111–120. doi: 10.5187/jast.2020.62.2.111
Iwamoto M., Asanuma N., Hino T. (2002). Ability of Selenomonas ruminantium, Veillonella parvula, and Wolinella succinogenes to reduce nitrate and nitrite with special reference to the suppression of ruminal methanogenesis. Anaerobe 8, 209–215. doi: 10.1006/anae.2002.0428
Jakočiunas T., Jensen M. K., Keasling J. D. (2016). CRISPR/Cas9 advances engineering of microbial cell factories. Metab. Eng. 34, 44–59. doi: 10.1016/j.ymben.2015.12.003
Järviö N., Maljanen N. L., Kobayashi Y., Ryynänen T., Tuomisto H. L. (2021). An attributional life cycle assessment of microbial protein production: A case study on using hydrogen-oxidizing bacteria. Sci. Total Environ. 776 (145764), 1–12. doi: 10.1016/j.scitotenv.2021.145764
Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., Charpentier E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Karekar S. C., Ahring B. K. (2023). Reducing methane production from rumen cultures by bioaugmentation with homoacetogenic bacteria. Biocatal. Agric. Biotechnol. 47, 102526. doi: 10.1016/j.bcab.2022.102526
Karekar S., Stefanini R. (2022). Homo-acetogens: their metabolism and competitive relationship with hydrogenotrophic methanogens. Microorganisms 10, 1–22. doi: 10.3390/microorganisms10020397
Kaya A., Kaya A. (2021). The effect of some vegetable oils added to dairy calf rations on in vitro feed value and enteric methane production. J. Agric. Product. 2, 1–6. doi: 10.29329/agripro.2021.344.1
Kebreab E., Bannink A., Pressman E. M., Walker N., Karagiannis A., van Gastelen S., et al. (2023). A meta-analysis of effects of 3-nitrooxypropanol on methane production, yield, and intensity in dairy cattle. J. Dairy Sci. 106, 927–936. doi: 10.3168/jds.2022-22211
Kelly W. J., Mackie R. I., Attwood G. T., Janssen P. H., McAllister T. A., Leahy S. C. (2022). Hydrogen and formate production and utilisation in the rumen and the human colon. Anim. Microb. 4, 1–8. doi: 10.1186/s42523-022-00174-z
Khairunisa B. H., Heryakusuma C., Ike K., Mukhopadhyay B., Susanti D. (2023). Evolving understanding of rumen methanogen ecophysiology. Frontiers in Microbiology 14, 1–30. doi: 10.3389/fmicb.2023.1296008
Kim C. C. (2012). Identification of rumen methanogens, characterization of substrate requirements and measurement of hydrogen thresholds. [dissertation/master's thesis]. Palmerston North: Massey University.
Kim S. H., Mamuad L. L., Choi Y. J., Sung H. G., Cho K. K., Lee S. S. (2018). Effects of reductive acetogenic bacteria and lauric acid on in vivo ruminal fermentation, microbial populations, and methane mitigation in Hanwoo steers in South Korea. J. Anim. Sci. 96, 4360–4367. doi: 10.1093/jas/sky266
Kim S.-H., Mamuad L. L., Islam M., Lee S.-S. (2020). Reductive acetogens isolated from ruminants and their effect on in vitro methane mitigation and milk performance in holstein cows. J. Anim. Sci. 62 (1), 1–13. doi: 10.5187/jast.2020.62.1.1
Klieve A. (2009). Using kangaroo bacteria to reduce emissions of methane and increase productivity (Meat & Livestock Australia). Available at: https://era.dpi.qld.gov.au/id/eprint/2360/1/NBP.354_Final_Report.pdf.
Kozicka M., Havlík P., Valin H., Wollenberg E., Deppermann A., Leclère D., et al. (2023). Feeding climate and biodiversity goals with novel plant-based meat and milk alternatives. Nat. Commun. 14, 1–13. doi: 10.1038/s41467-023-40899-2
Krupovič M., Bamford D. H. (2008). Archaeal proviruses TKV4 and MVV extend the PRD1-adenovirus lineage to the phylum Euryarchaeota. Virology 375, 292–300. doi: 10.1016/j.virol.2008.01.043
La H., Hettiaratchi J. P. A., Achari G., Dunfield P. F. (2018). Biofiltration of methane. Biores. Technol. 268, 759–772. doi: 10.1016/j.biortech.2018.07.043
Lan W., Yang C. (2019). Ruminal methane production: Associated microorganisms and the potential of applying hydrogen-utilizing bacteria for mitigation. Sci. Total Environ. 654, 1270–1283. doi: 10.1016/j.scitotenv.2018.11.180
Lassen J., Difford G. F. (2020). Review: Genetic and genomic selection as a methane mitigation strategy in dairy cattle. Animal Int. J. Anim. Biosci. 14, 473–483. doi: 10.1017/S1751731120001561
Laura M., Jo P. (2023). No acetogen is equal: Strongly different H2 thresholds reflect diverse bioenergetics in acetogenic bacteria. Environ. Microbiol. 25 (10), 2032–2040. doi: 10.1111/1462-2920.16429
Leahy S. C., Kelly W. J., Altermann E., Ronimus R. S., Yeoman C. J., Pacheco D. M., et al. (2010). The genome sequence of the rumen methanogen methanobrevibacter ruminantium reveals new possibilities for controlling ruminant methane emissions. PloS One 5, e8926. doi: 10.1371/journal.pone.0008926
Leclerc M., Bernalier A., Donadille G., Lelait M. (1997). H2/CO2Metabolism in acetogenic bacteria isolated from the human colon. Anaerobe 3, 307–315. doi: 10.1006/anae.1997.0117
Lee D., Pan J. H., Kim D., Heo W., Shin E. C., Kim Y. J., et al. (2024). Mycoproteins and their health-promoting properties: Fusarium species and beyond. Compr. Rev. Food Sci. Food Saf. 23 (3), 1541–4337. doi: 10.1111/1541-4337.13365
Leng R. A. (2018). Unravelling methanogenesis in ruminants, horses and kangaroos: The links between gut anatomy, microbial biofilms and host immunity. Anim. Product. Sci. 58, 1175. doi: 10.1071/AN15710
Lessner D. J., Li L., Li Q., Rejtar T., Andreev V. P., Reichlen M., et al. (2006). An unconventional pathway for reduction of CO 2 to methane in CO-grown Methanosarcina acetivorans revealed by proteomics. Proc. Natl. Acad. Sci. 103, 17921–17926. doi: 10.1073/pnas.0608833103
Le Van T. D., Robinson J. A., Ralph J., Greening R. C., Smolenski W. J., Leedle J. A. Z., et al. (1998). Assessment of reductive acetogenesis with indigenous ruminal bacterium populations and acetitomaculum ruminis. Appl. Environ. Microbiol. 64, 3429–3436. doi: 10.1128/AEM.64.9.3429-3436.1998
Lidstrom M. E. (2024). Direct methane removal from air by aerobic methanotrophs. Cold Spring Harbor Perspect. Biol. 16, a041671. doi: 10.1101/cshperspect.a041671
Lileikis T., Nainienė R., Bliznikas S., Uchockis V. (2023). Dietary ruminant enteric methane mitigation strategies: current findings, potential risks and applicability. Animals 13, 2586. doi: 10.3390/ani13162586
Lobo R. R., Faciola A. P. (2021). Ruminal phages – a review. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.763416
Lopez S., McIntosh F. M., Wallace R. J., Newbold C. J. (1999). Effect of adding acetogenic bacteria on methane production by mixed rumen microorganisms. Anim. Feed Sci. Technol. 78, 1–9. doi: 10.1016/S0377-8401(98)00273-9
Lovley D. R. (1985). Minimum threshold for hydrogen metabolism in methanogenic bacteria. Appl. Environ. Microbiol. 49, 1530–1531. doi: 10.1128/aem.49.6.1530-1531.1985
Lovley D. R., Greening R. C., Ferry J. G. (1984). Rapidly growing rumen methanogenic organism that synthesizes coenzyme M and has a high affinity for formate. Appl. Environ. Microbiol. 48, 81–87. doi: 10.1128/aem.48.1.81-87.1984
Lyu Z., Shao N., Akinyemi T., Whitman W. B. (2018). Methanogenesis. Curr. Biol. 28, R727–R732. doi: 10.1016/j.cub.2018.05.021
Ma J., Kooijmans L. M. J., Cho A., Montzka S. A., Glatthor N., Worden J. R., et al. (2021). Inverse modelling of carbonyl sulfide: Implementation, evaluation and implications for the global budget. Atmos. Chem. Phys. 21, 3507–3529. doi: 10.5194/acp-21-3507-2021
Mackie R. I., Kim H., Kim N. K., Cann I. (2024). Hydrogen production and hydrogen utilization in the rumen: Key to mitigating enteric methane production. Anim. Biosci. 37, 323–336. doi: 10.5713/ab.23.0294
Malik P. K., Trivedi S., Mohapatra A., Kolte A. P., Sejian V., Bhatta R., et al. (2021). Comparison of enteric methane yield and diversity of ruminal methanogens in cattle and buffaloes fed on the same diet. PloS One 16, 1–19. doi: 10.1371/journal.pone.0256048
Martin W. F. (2020). Older than genes: the acetyl coA pathway and origins. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00817
Martínez-Álvaro M., Auffret M., Duthie C., Dewhurst R., Cleveland M., Watson M., et al. (2022). Bovine host genome acts on rumen microbiome function linked to methane emissions. Commun. Biol. 5, 350. doi: 10.1038/s42003-022-03293-0
Martinez-Fernandez G., Denman S. E., Cheung J., McSweeney C. S. (2017). Phloroglucinol degradation in the rumen promotes the capture of excess hydrogen generated from methanogenesis inhibition. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01871
McCauley J. I., Labeeuw L., Jaramillo-Madrid A. C., Nguyen L. N., Nghiem L. D., Chaves A. V., et al. (2020). Management of enteric methanogenesis in ruminants by algal-derived feed additives. Curr. pollut. Rep. 6, 188–205. doi: 10.1007/s40726-020-00151-7
McDuffie E. E., Sarofim M. C., Raich W., Jackson M., Roman H., Seltzer K., et al. (2023). The social cost of ozone-related mortality impacts from methane emissions. Earth’s Future 11, e2023EF003853. doi: 10.1029/2023EF003853
Meinshausen M., Lewis J., McGlade C., Gütschow J., Nicholls Z., Burdon R., et al. (2022). Realization of Paris Agreement pledges may limit warming just below 2°C. Nature 604, 304–309. doi: 10.1038/s41586-022-04553-z
Melgar A., Harper M. T., Oh J., Giallongo F., Young M. E., Ott T. L., et al. (2020). Effects of 3-nitrooxypropanol on rumen fermentation, lactational performance, and resumption of ovarian cyclicity in dairy cows. J. Dairy Sci. 103, 410–432. doi: 10.3168/jds.2019-17085
Meller R. A., Wenner B. A., Ashworth J., Gehman A. M., Lakritz J., Firkins J. L. (2019). Potential roles of nitrate and live yeast culture in suppressing methane emission and influencing ruminal fermentation, digestibility, and milk production in lactating Jersey cows. J. Dairy Sci. 102, 6144–6156. doi: 10.3168/jds.2018-16008
Mills G., Sharps K., Simpson D., Pleijel H., Frei M., Burkey K., et al. (2018). Closing the global ozone yield gap: Quantification and cobenefits for multistress tolerance. Glob. Change Biol. 24, 4869–4893. doi: 10.1111/gcb.14381
Min B., Lee S., Jung H., Miller D. N. (2022). Enteric methane emissions and animal performance in dairy and beef cattle production: strategies, opportunities, and impact of reducing emissions. Animals 12, 1–27. doi: 10.3390/ani12080948
Mizrahi I., Wallace R. J., Moraïs S. (2021). The rumen microbiome: Balancing food security and environmental impacts. Nat. Rev. Microbiol. 19, 553–566. doi: 10.1038/s41579-021-00543-6
Moomaw W. R., Chmura G. L., Davies G. T., Finlayson C. M., Middleton B. A., Natali S. M., et al. (2018). Wetlands in a changing climate: science, policy and management. Wetlands 38, 183–205. doi: 10.1007/s13157-018-1023-8
Morvan B., Dore J., Rieu-Lesme F., Foucat L., Fonty G., Gouet P. (1994). Establishment of hydrogen-utilizing bacteria in the rumen of the newborn lamb. FEMS Microbiol. Letters. 117 (3), 249–256. doi: 10.1111/j.1574-6968.1994.tb06775.x
Moruzzo R., Mancini S., Guidi A. (2021). Edible insects and sustainable development goals. Insects 12, 557. doi: 10.3390/insects12060557
Mottaleb K. A., Kruseman G., Snapp S. (2022). Potential impacts of Ukraine-Russia armed conflict on global wheat food security: A quantitative exploration. Global Food Secur. 35, 100659. doi: 10.1016/j.gfs.2022.100659
Muñoz C., Villalobos R., Peralta A. M. T., Morales R., Urrutia N. L., Ungerfeld E. M. (2021). Long-term and carryover effects of supplementation with whole oilseeds on methane emission, milk production and milk fatty acid profile of grazing dairy cows. Animals 11, 1–19. doi: 10.3390/ani11102978
Nagarajan D., Lee D. J., Chang J. S. (2019). Integration of anaerobic digestion and microalgal cultivation for digestate bioremediation and biogas upgrading. Biores. Technol. 290, 1–53. doi: 10.1016/j.biortech.2019.121804
Nagoya M., Kouzuma A., Watanabe K. (2021). Codh/acs-deficient methanogens are prevalent in anaerobic digesters. Microorganisms 9, 1–8. doi: 10.3390/microorganisms9112248
Nakashimada Y., Srinivasan K., Murakami M., Nishio N. (2000). Direct conversion of cellulose to methane by anaerobic fungus Neocallimastix frontalis and defined methanogens. Biotechnol. Lett. 22, 223–227. doi: 10.1023/A:1005666428494
Namonyo S., Wagacha M., Maina S., Wambua L., Agaba M. (2018). A metagenomic study of the rumen virome in domestic caprids. Arch. Virol. 163 (12), 3415–3419. doi: 10.1007/s00705-018-4022-4
Nayak D. D., Metcalf W. W. (2017). Cas9-mediated genome editing in the methanogenic archaeon Methanosarcina acetivorans. Proc. Natl. Acad. Sci. United States America 114, 2976–2981. doi: 10.1073/pnas.1618596114
Newbold C. J., de la Fuente G., Belanche A., Ramos-Morales E., McEwan N. R. (2015). The role of ciliate protozoa in the rumen. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.01313
Nikel P. I., De Lorenzo V. (2021). “Metabolic Engineering for Large-Scale Environmental Bioremediation,” in Metabolic Engineering, 1st ed. Eds. Nielsen J., Stephanopoulos G., Lee S. Y. (Weinheim, Germany: Wiley), 859–890. doi: 10.1002/9783527823468.ch22
Nisbet-Jones P. B. R., Fernandez J. M., Fisher R. E., France J. L., Lowry D., Waltham D. A., et al. (2022). Is the destruction or removal of atmospheric methane a worthwhile option? Philos. Trans. R. Soc. A: Math. Phys. Eng. Sci. 380, 20210108. doi: 10.1098/rsta.2021.0108
Nollet L., Demeyer D., Verstraete W. (1997). Effect of 2-bromoethanesulfonic acid and Peptostreptococcus productus ATCC 35244 addition on stimulation of reductive acetogenesis in the ruminal ecosystem by selective inhibition of methanogenesis. Appl. Environ. Microbiol. 63, 194–200. doi: 10.1128/aem.63.1.194-200.1997
Nollet L., Mbanzamihigo L., Demeyer D., Verstraete W. (1998). Corrigendum to “Effect of the addition of Peptostreptococcus productus ATCC 35244 on reductive acetogenesis in the ruminal ecosystem after inhibition of methanogenesis by cell-free supernatant of Lactobacillus plantarum 80. Anim. Feed Sci. Technol. 74, 189. doi: 10.1016/s0377-8401(98)00193-x
O’Hara E., Kenny D. A., McGovern E., Byrne C. J., McCabe M. S., Guan L. L., et al. (2020). Investigating temporal microbial dynamics in the rumen of beef calves raised on two farms during early life. FEMS Microbiol. Ecol. 96, 1–16. doi: 10.1093/femsec/fiz203
Olijhoek D. W., Hellwing A. L. F., Brask M., Weisbjerg M. R., Højberg O., Larsen M. K., et al. (2016). Effect of dietary nitrate level on enteric methane production, hydrogen emission, rumen fermentation, and nutrient digestibility in dairy cows. J. Dairy Sci. 99, 6191–6205. doi: 10.3168/jds.2015-10691
Payne S., Wick S., Carr P. A., Guido N. J. (2024). A methodology for the assessment and prioritization of genetic biocontainment technologies for engineered microbes. Appl. Biosafe. 29 (2), 1–12. doi: 10.1089/apb.2023.0025
Pereira A. M., de Lurdes Nunes Enes Dapkevicius M., Borba A. E. S. (2022). Alternative pathways for hydrogen sink originated from the ruminal fermentation of carbohydrates: Which microorganisms are involved in lowering methane emission? Anim. Microb. 4, 1–12. doi: 10.1186/s42523-021-00153-w
Pester M., Brune A. (2007). Hydrogen is the central free intermediate during lignocellulose degradation by termite gut symbionts. ISME J. 1, 551–565. doi: 10.1038/ismej.2007.62
Polag D., Keppler F. (2018). Long-term monitoring of breath methane. Sci. Total Environ. 624, 69–77. doi: 10.1016/j.scitotenv.2017.12.097
Polag D., Keppler F. (2019). Global methane emissions from the human body: Past, present and future. Atmos. Environ. 214, 9. doi: 10.1016/j.atmosenv.2019.116823
Ravi Kanth Reddy P., Srinivasa Kumar D., Raghava Rao E., Seshiah V., Sateesh K., Pradeep Kumar Reddy Y., et al. (2019). Assessment of eco-sustainability vis-à-vis zoo-technical attributes of soybean meal (SBM) replacement with varying levels of coated urea in Nellore sheep (Ovis aries). PloS One 14, e0220252. doi: 10.1371/journal.pone.0220252
Reddy P. R. K., Kumar D. S., Rao E. R., Ch. V., Sateesh K., Rao K. A., et al. (2019). Environmental sustainability assessment of tropical dairy buffalo farming vis-a-vis sustainable feed replacement strategy. Sci. Rep. 9, 16745. doi: 10.1038/s41598-019-53378-w
Reisinger A., Clark H., Cowie A. L., Emmet-Booth J., Gonzalez Fischer C., Herrero M., et al. (2021). How necessary and feasible are reductions of methane emissions from livestock to support stringent temperature goals? Philos. Trans. R. Soc. A: Math. Phys. Eng. Sci. 379, 1–18. doi: 10.1098/rsta.2020.0452
Rigby M., Montzka S. A., Prinn R. G., White J. W. C., Young D., O’Doherty S., et al. (2017). Role of atmospheric oxidation in recent methane growth. Proc. Natl. Acad. Sci. 114, 5373–5377. doi: 10.1073/pnas.1616426114
Roehe R., Dewhurst R. J., Duthie C. A., Rooke J. A., McKain N., Ross D. W., et al. (2016). Bovine host genetic variation influences rumen microbial methane production with best selection criterion for low methane emitting and efficiently feed converting hosts based on metagenomic gene abundance. PloS Genet. 12, 1–20. doi: 10.1371/journal.pgen.1005846
Roque B. M., Venegas M., Kinley R. D., De Nys R., Duarte T. L., Yang X., et al. (2021). Red seaweed (Asparagopsis taxiformis) supplementation reduces enteric methane by over 80 percent in beef steers. PloS One 16, 1–20. doi: 10.1371/journal.pone.0247820
Roques S., Martinez-Fernandez G., Ramayo-Caldas Y., Popova M., Denman S., Meale S. J., et al. (2024). Recent advances in enteric methane mitigation and the long road to sustainable ruminant production. Annu. Rev. Anim. Biosci. 12, 321–343. doi: 10.1146/annurev-animal-021022-024931
Rosenbaum F. P., Müller V. (2021). Energy conservation under extreme energy limitation: The role of cytochromes and quinones in acetogenic bacteria. Extremophiles 25, 413–424. doi: 10.1007/s00792-021-01241-0
Rubin B. E., Diamond S., Cress B. F., Crits-Christoph A., Lou Y. C., Borges A. L., et al. (2021). Species- and site-specific genome editing in complex bacterial communities. Nat. Microbiol. 7, 34–47. doi: 10.1038/s41564-021-01014-7
Rubio N. R., Xiang N., Kaplan D. L. (2020). Plant-based and cell-based approaches to meat production. Nat. Commun. 11, 1–11. doi: 10.1038/s41467-020-20061-y
Sakthivel P. C., Kamra D. N., Agarwal N., Chaudhary L. C. (2012). Effect of sodium nitrate and nitrate reducing bacteria on in vitro methane production and fermentation with buffalo rumen liquor. Asian-Australas J Anim Sci. 25 (6), 812–817. doi: 10.5713/ajas.2011.11383
Saunois M., Stavert A. R., Poulter B., Bousquet P., Canadell J. G., Jackson R. B., et al. (2020). The global methane budget 2000–2017. Earth Sys. Sci. Data 12, 1561–1623. doi: 10.5194/essd-12-1561-2020
Saunois M., Stavert A. R., Poulter B., Bousquet P., Josep G., Jackson R. B., et al. (2019). The global methane budget 2000-2017. Earth Sys. Sci. Data 7, 1–136. doi: 10.5194/essd-2019-128
Schaefer H., Fletcher S. E. M., Veidt C., Lassey K. R., Brailsford G. W., Bromley T. M., et al. (2016). A 21st-century shift from fossil-fuel to biogenic methane emissions indicated by 13CH4. Science 352, 80–84. doi: 10.1126/science.aad2705
Scheller S., Rother M. (2022). Deconstructing Methanosarcina acetivorans into an acetogenic archaeon. PNAS 119, 1–7. doi: 10.1073/pnas.2113853119/-/DCSupplemental.Published
Schöne C., Poehlein A., Jehmlich N., Adlung N., Daniel R., Von Bergen M., et al. (2022). Deconstructing Methanosarcina acetivorans into an acetogenic archaeon. Proc. Natl. Acad. Sci. 119, e2113853119. doi: 10.1073/pnas.2113853119
Smith N. W., Shorten P. R., Altermann E., Roy N. C., McNabb W. C. (2020). Mathematical modelling supports the existence of a threshold hydrogen concentration and media-dependent yields in the growth of a reductive acetogen. Biopro. Biosyst. Eng. 43, 885–894. doi: 10.1007/s00449-020-02285-w
Sofyan A., Irawan A., Herdian H., Jasmadi, Harahap M. A., Sakti A. A., et al. (2022). Effects of various macroalgae species on methane production, rumen fermentation, and ruminant production: A meta-analysis from in vitro and in vivo experiments. Anim. Feed Sci. Technol. 294, 115503. doi: 10.1016/j.anifeedsci.2022.115503
Solomon R., Wein T., Levy B., Eshed S., Dror R., Reiss V., et al. (2021). Protozoa populations are ecosystem engineers that shape prokaryotic community structure and function of the rumen microbial ecosystem. bioRxiv 16, 1187–1197. doi: 10.1101/2020.05.15.080218
Springmann M., Clark M., Mason-D’Croz D., Wiebe K., Bodirsky B. L., Lassaletta L., et al. (2018). Options for keeping the food system within environmental limits. Nature 562 (7728), 519–525. doi: 10.1038/s41586-018-0594-0
Stachler A. E., Schwarz T. S., Schreiber S., Marchfelder A. (2020). CRISPRi as an efficient tool for gene repression in archaea. Methods 172, 76–85. doi: 10.1016/j.ymeth.2019.05.023
Stavert A. R., Saunois M., Canadell J. G., Poulter B., Jackson R. B., Regnier P., et al. (2022). Regional trends and drivers of the global methane budget. Global Change Biol. 28, 182–200. doi: 10.1111/gcb.15901
Steidler L., Neirynck S., Huyghebaert N., Snoeck V., Vermeire A., Goddeeris B., et al. (2003). Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nat. Biotechnol. 21, 785–789. doi: 10.1038/nbt840
Subedi U., Kader K., Jayawardhane K. N., Poudel H., Chen G., Acharya S., et al. (2022). The potential of novel gene editing-based approaches in forages and rumen archaea for reducing livestock methane emissions. Agric. (Switzerland) 12, 1–21. doi: 10.3390/agriculture12111780
Tadesse G., Dereje M. (2018). Impact of climate change on smallholder dairy production and coping mechanism in Sub-Saharan Africa-review. Adv. Life Sci. Technol. 65, 41–54. doi: 10.19080/ARTOAJ.2018.16.556000
Tapio I., Snelling T. J., Strozzi F., Wallace R. J. (2017). The ruminal microbiome associated with methane emissions from ruminant livestock. J. Anim. Sci. Biotechnol. 8, 7. doi: 10.1186/s40104-017-0141-0
Tseten T., Sanjorjo R. A., Kwon M., Kim S.-W. (2022). Strategies to mitigate enteric methane emissions from ruminant animals. J. Microbiol. Biotechnol. 32, 269–277. doi: 10.4014/jmb.2202.02019
Ungerfeld E. M. (2020). Metabolic hydrogen flows in rumen fermentation: principles and possibilities of interventions. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00589
Uniyal S., Chaudhary L. C., Kala A., Agarwal N., Chaturvedi V. B. (2023). Effect of supplementing sulphate-reducing bacteria along with sulphur on growth performance, nutrient utilization and methane emission in goats. Trop. Anim. Health Product. 55, 3. doi: 10.1007/s11250-022-03419-w
Vaghar Seyedin S. M., Zeidi A., Chamanehpour E., Nasri M. H. F., Vargas-Bello-Pérez E. (2022). Methane emission: strategies to reduce global warming in relation to animal husbandry units with emphasis on ruminants. Sustainability 14, 16897. doi: 10.3390/su142416897
Van Gastelen S., Dijkstra J., Heck J. M. L., Kindermann M., Klop A., De Mol R., et al. (2022). Methane mitigation potential of 3-nitrooxypropanol in lactating cows is influenced by basal diet composition. J. Dairy Sci. 105, 4064–4082. doi: 10.3168/jds.2021-20782
Van Lingen H. J., Plugge C. M., Fadel J. G., Kebreab E., Bannink A., Dijkstra J. (2016). Thermodynamic driving force of hydrogen on rumen microbial metabolism: A theoretical investigation. PloS One 11, 1–18. doi: 10.1371/journal.pone.0161362
van Zijderveld S. M., Gerrits W. J. J., Apajalahti J. A., Newbold J. R., Dijkstra J., Leng R. A., et al. (2010). Nitrate and sulfate: Effective alternative hydrogen sinks for mitigation of ruminal methane production in sheep. J. Dairy Sci. 93, 5856–5866. doi: 10.3168/jds.2010-3281
Van Zijderveld S. M., Gerrits W. J. J., Dijkstra J., Newbold J. R., Hulshof R. B. A., Perdok H. B. (2011). Persistency of methane mitigation by dietary nitrate supplementation in dairy cows. J. Dairy Sci. 94, 4028–4038. doi: 10.3168/jds.2011-4236
Verme L. J., Ullrey D. E. (1984). Fawn growth. White-Tailed Deer Ecol. Management1 70, 91–118. doi: 10.1128/AEM.70.3.1307
Villar L., Hegarty R., Van Tol M., Godwin I., Nolan J. (2019). Dietary nitrate metabolism and enteric methane mitigation in sheep consuming a protein-deficient diet. Anim. Product. Sci. 60, 232–241. doi: 10.1071/AN18632
Wallace R. J., Rooke J. A., McKain N., Duthie C. A., Hyslop J. J., Ross D. W., et al. (2015). The rumen microbial metagenome associated with high methane production in cattle. BMC Genomics 16 (1), 1–14. doi: 10.1186/s12864-015-2032-0
Wallace R., Sasson G., Garnsworthy P. C., Tapio I., Gregson E., Bani P., et al. (2019). A heritable subset of the core rumen microbiome dictates dairy cow productivity and emissions. Sci. Adv. 5. doi: 10.1126/sciadv.aav8391
Wang M., Wang R., Xie T. Y., Janssen P. H., Sun X. Z., Beauchemin K. A., et al. (2016). Shifts in rumen fermentation and microbiota are associated with dissolved ruminal hydrogen concentrations in lactating dairy cows fed different types of carbohydrates. J. Nutr. 146, 1714–1721. doi: 10.3945/jn.116.232462
Wang K., Xiong B., Zhao X. (2023). Could propionate formation be used to reduce enteric methane emission in ruminants? Sci. Total Environ. 855, 1–12. doi: 10.1016/j.scitotenv.2022.158867
Warszawski L., Kriegler E., Lenton T. M., Gaffney O., Jacob D., Klingenfeld D., et al. (2021). All options, not silver bullets, needed to limit global warming to 1.5°C: A scenario appraisal. Environ. Res. Lett. 16, 064037. doi: 10.1088/1748-9326/abfeec
Weidenbach K., Wolf S., Kupczok A., Kern T., Fischer M. A., Reetz J., et al. (2021). Characterization of blf4, an archaeal lytic virus targeting a member of the methanomicrobiales. Viruses 13, 1934. doi: 10.3390/v13101934
Wesemael D. V., Vandaele L., Ampe B., Cattrysse H., Duval S., Kindermann M., et al. (2019). Reducing enteric methane emissions from dairy cattle: Two ways to supplement 3-nitrooxypropanol. J. Dairy Sci. 102, 1780–1787. doi: 10.3168/jds.2018-14534
Wilkinson T., Korir D., Ogugo M., Stewart R. D., Watson M., Paxton E., et al. (2020). 1200 high-quality metagenome-assembled genomes from the rumen of African cattle and their relevance in the context of sub-optimal feeding. Genome Biol. 21, 1–25. doi: 10.1186/s13059-020-02144-7
Willett W., Rockström J., Loken B., Springmann M., Lang T., Vermeulen S., et al. (2019). Food in the Anthropocene: The EAT–Lancet Commission on healthy diets from sustainable food systems. Lancet 393. doi: 10.1016/S0140-6736(18)31788-4
Williams S. R. O., Hannah M. C., Eckard R. J., Wales W. J., Moate P. J. (2020). Supplementing the diet of dairy cows with fat or tannin reduces methane yield, and additively when fed in combination. Animal 14, s464–s472. doi: 10.1017/S1751731120001032
Wilson A. S., Koller K. R., Ramaboli M. C., Nesengani L. T., Ocvirk S., Chen C., et al. (2020). Diet and the human gut microbiome: an international review. Digest. Dis. Sci. 65, 723–740. doi: 10.1007/s10620-020-06112-w
Wolf S., Fischer M. A., Kupczok A., Reetz J., Kern T., Schmitz R. A., et al. (2019). Characterization of the lytic archaeal virus Drs3 infecting Methanobacterium formicicum. Arch. Virol. 164, 667–674. doi: 10.1007/s00705-018-04120-w
Wright A. G., Klieve A. V. (2011). Does the complexity of the rumen microbial ecology preclude methane mitigation? Anim Feed Sci Technol. 167, 248–253. doi: 10.1016/j.anifeedsci.2011.04.015
Xia Y., Kong Y. H., Seviour R., Forster R. J., Kisidayova S., Mcallister T. A. (2014). Fluorescence in situ hybridization probing of protozoal Entodinium spp. And their methanogenic colonizers in the rumen of cattle fed alfalfa hay or triticale straw. J. Appl. Microbiol. 116, 14–22. doi: 10.1111/jam.12356
Xue M. Y., Sun H. Z., Wu X. H., Liu J. X., Guan L. L. (2020). Multi-omics reveals that the rumen microbiome and its metabolome together with the host metabolome contribute to individualized dairy cow performance. Microbiome 8, 1–19. doi: 10.1186/s40168-020-00819-8
Yáñez-Ruiz D. R., Abecia L., Newbold C. J. (2015). Manipulating rumen microbiome and fermentation through interventions during early life: A review. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.01133
Yang C. (2018). Acetogen communities in the gut of herbivores and their potential role in syngas fermentation. Fermentation 4, 1–17. doi: 10.3390/fermentation4020040
Yang C., Guan L., Liu J., Wang J. (2015). Rumen fermentation and acetogen population changes in response to an exogenous acetogen TWA4 strain and Saccharomyces cerevisiae fermentation product. J. Zhejiang University: Sci. B 16, 709–719. doi: 10.1631/jzus.B1500013
Yang C., McKain N., McCartney C. A., Wallace R. J. (2019). Consequences of inhibiting methanogenesis on the biohydrogenation of fatty acids in bovine ruminal digesta. Anim. Feed Sci. Technol. 254, 114189. doi: 10.1016/j.anifeedsci.2019.05.012
Zhang X. M., Gruninger R. J., Alemu A. W., Wang M., Tan Z. L., Kindermann M., et al. (2020). 3-Nitrooxypropanol supplementation had little effect on fiber degradation and microbial colonization of forage particles when evaluated using the in situ ruminal incubation technique. J. Dairy Sci. 103, 8986–8997. doi: 10.3168/jds.2019-18077
Zhang X., Medrano R. F., Wang M., Beauchemin K. A., Ma Z., Wang R., et al. (2019). Effects of urea plus nitrate pretreated rice straw and corn oil supplementation on fiber digestibility, nitrogen balance, rumen fermentation, microbiota and methane emissions in goats. J. Anim. Sci. Biotechnol. 10, 6. doi: 10.1186/s40104-019-0312-2
Keywords: methane, ruminant, Wood–Ljungdahl pathway, mitigation, CRISPR-Cas
Citation: Mrutu RI, Abdussamad AM, Umar KM, Abdulhamid A and Farny NG (2025) Mitigating methane emissions and promoting acetogenesis in ruminant livestock. Front. Anim. Sci. 6:1489212. doi: 10.3389/fanim.2025.1489212
Received: 02 September 2024; Accepted: 22 January 2025;
Published: 28 February 2025.
Edited by:
Michael D. Flythe, United States Department of Agriculture, United StatesReviewed by:
Ravikanth Reddy Poonooru, University of Missouri, United StatesJourdan Lakes, United States Department of Agriculture (USDA), United States
Copyright © 2025 Mrutu, Abdussamad, Umar, Abdulhamid and Farny. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Natalie G. Farny, bmZhcm55QHdwaS5lZHU=
 Rehema Iddi Mrutu1
Rehema Iddi Mrutu1 Abdussamad Muhammad Abdussamad
Abdussamad Muhammad Abdussamad Kabir Mustapha Umar
Kabir Mustapha Umar Natalie G. Farny
Natalie G. Farny