
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Anim. Sci. , 17 January 2025
Sec. Precision Livestock Farming
Volume 5 - 2024 | https://doi.org/10.3389/fanim.2024.1514923
 Naqash Goswami
Naqash Goswami Ifeanyi Solomon Ahamba
Ifeanyi Solomon Ahamba Lionel Kinkpe
Lionel Kinkpe Ali Mujtaba Shah
Ali Mujtaba Shah Ye Xiangyang
Ye Xiangyang Bing Song*
Bing Song* Xianggui Dong
Xianggui Dong Shuhui Wang
Shuhui Wang Zhanjun Ren*
Zhanjun Ren*Rabbit farming plays a crucial role in meeting the global demand for high-quality meat and sustainable agricultural practices. In recent years, significant attention has been directed toward the integration of Genomic-Assisted Selection (GAS) and Precision Nutrition (PN) as synergistic strategies to improve productivity and animal welfare in rabbit production systems. This integration is particularly critical given the intricate interplay between genetic predispositions and nutritional demands in optimizing rabbit farming outcomes. The present review systematically explores the genetic diversity inherent to rabbit breeds, examines the impact of key genetic traits on productivity and welfare metrics, and highlights advancements in genomic research and associated tools within the context of rabbit breeding practices. By examining the importance of personalized nutrition tailored to individual rabbit requirements, as well as nutritional strategies to optimize productivity and longevity. In this regard, we provide essential understanding for farmers and researchers in the rabbit farming industry. Our goal is to provide a new perspective on the significance of this integrated approach and its implications for future practices and research in rabbit production, emphasizing the need for continued efforts to harness the full potential of genomics and nutrition in enhancing rabbit farming practices.
Rabbits are commonly farmed for their meat, skin, or hides, and therefore, concerns about rabbit breeding practices have grown among farmers and consumers over the last decade (Pilarczyk et al., 2020). Rabbit meat is a nutrient-rich product with a higher content of protein and polyunsaturated fatty acids while low levels of carbohydrates, sodium, and cholesterol, as well as energy levels comparable to many types of red meat that are commonly consumed (Siddiqui et al., 2023). Similarly, rabbit skin and hides are sustainable industrial raw materials for the production of high-quality leather commercially (Sikiru et al., 2020). Thereby, rabbit farming is one of the sustainable and profitable agricultural practices due to the higher fecundity, efficient feed conversion, and the ability of rabbits to adapt to a variety of environments as well as the lower initial investment and a small production area required to breed rabbits on the large scale (Wongnaa et al., 2023). However, the efforts to develop genetically superior rabbit breeds are still lacking along with the limited studies on the effect of nutrition, diseases, genetic characteristics, and housing conditions of rabbits to improve the productivity and efficiency of farming. On the whole, the genetic characteristics and nutritional factors of rabbits play a significant role in rabbit farming, particularly in terms of productivity and welfare. For instance, the prolactin receptor gene (PRLR) polymorphisms significantly affected milk production in rabbits in larger liters (Benedek et al., 2023). In addition, genomic selection and crossbreeding reduced mortality while enhancing the growth rates of rabbits ultimately resulting in economical rabbit farming (Assan, 2018). Similarly, nutrition is also a critical factor in rabbit production since rabbits have a complex microbiota in their gastrointestinal tract that plays a critical role in feed digestion, vitamin production, fermentative activity, and stimulation of immune response (Mancini and Paci, 2021). By selecting rabbits with superior genetic characteristics and adopting suitable diets supplemented with additives, farmers can significantly enhance productivity and profitability in rabbit farming.
Nowadays, genomic-assisted selection (GAS) and precision nutrition (PN) approaches are used to enhance the production and farming potential of rabbits as the demand for rabbit meat and sustainable raw materials is growing worldwide. GAS, particularly genomic selection (GS), Marker-assisted Selection (MAS), Genome-wide Association Studies (GWAS), Genomic prediction models, and genotyping and phenotyping, have been widely adopted in animal breeding programs due to its potential to improve selection accuracy, minimize phenotyping, reduce cycle time, and increase genetic gains. GS utilizes genomic-estimated breeding values of individuals obtained from genome-wide markers to choose candidates for the next breeding cycle, making it a powerful approach to improve quantitative traits such as productivity and welfare traits in rabbit farming (Sharma et al., 2024). PN, on the other hand, involves the use of personalized nutrition interventions based on an individual’s unique characteristics and needs. This approach takes into account various factors such as nutrition intake, lifestyle, phenotype, and genotype to achieve personalized and accurate nutrition interventions. PN techniques such as nutrigenomics, metabolomics, microbiota profiling, deep phenotyping, artificial intelligence, and wearable technologies can be used to develop personalized nutrition recommendations that are tailored to an individual’s unique characteristics and needs (Pomar and Remus, 2023). Therefore, by combining GAS and PN approaches, rabbit farming can be optimized to meet the growing demand for rabbit meat and sustainable raw materials worldwide as GAS has the potential to improve selection accuracy and genetic potential, while PN can be used to optimize reproductive performance and health outcomes in rabbits. This integrated approach can lead to improved productivity, sustainability, and welfare in rabbit farming, contributing to a more efficient and responsible production system. This review explores the genetic diversity in rabbit breeds, key genetic traits influencing productivity and welfare, and advances in genetic research and genomic tools in rabbit breeding. The nutritional needs of rabbits for optimal growth and health, the importance of personalized nutrition in meeting individual rabbit requirements, and nutritional strategies to enhance productivity and longevity in rabbits are also discussed. Overall, the review provides a comprehensive overview of the integration of GAS and PN approaches in rabbit farming, highlighting the potential benefits and challenges of this integrated approach for sustainable and efficient rabbit farming.
The genetic diversity of rabbit breeds is crucial for understanding the underlying structure and variation within these breeds, which plays a pivotal role in the selection of distinct physical and physiological traits, such as coat color and structure (Demars et al., 2018; Fontanesi, 2021). Several studies have explored the genetic diversity of indigenous rabbit breeds, such as the Sichuan White, Tianfu Black, Fujian Yellow, and Fujian Black, using advanced techniques like restriction-site-associated DNA sequencing (RAD-seq) and whole-genome resequencing (Ren et al., 2019; Xie et al., 2024). These studies reveal that the Fujian Yellow breed exhibits the highest genetic diversity, followed by the Tianfu Black and Sichuan White breeds, with the principal component analysis (PCA) clearly distinguishing the breeds. Similarly, Xie et al. (2024) found significant genetic diversity across various breeds in relation to key traits, such as coat color, reproductive capacity, disease resistance, and body size, highlighting the importance of understanding these genetic foundations for breeding purposes. Moreover, studies have shown that the integration of genomic selection with genetic diversity assessment can significantly improve traits that are economically important for rabbit farming, such as growth rate, feed conversion ratio, and meat quality (Yang et al., 2020; Piles et al., 2024). Research on the genetic mechanisms underlying beneficial traits, such as disease resistance, larger body size, and adaptability in Indonesian rabbits, also supports the idea that local genetic resources can be key to enhancing productivity and welfare in commercial rabbit farming (Setiaji et al., 2023).
Additionally, genetic studies have focused on optimizing productivity traits such as fertility, longevity, and litter size, which are crucial for enhancing rabbit farming commercially. For example, Ragab et al. (2021) demonstrated that crossbreeding four maternal rabbit lines led to increased longevity, especially in the early reproductive cycles, which is important for the sustainability of rabbit farming. Similarly, Biada et al. (2024) found that genetic differences in gut microbiota diversity were linked to better resistance to diseases and longer lifespans, emphasizing the potential of manipulating the gut microbiome to improve the health and productivity of rabbits. Furthermore, the identification of genes associated with reproductive capacity, such as Cellular Retinoic Acid Binding Protein-1 (CRABP1), could serve as a tool for improving the breeding practices of farmed rabbits (Bao et al., 2024). Ultimately, combining Genetic-Assisted Selection (GAS) with Precision Nutrition (PN) strategies can enable more effective targeting of specific traits to improve both the productivity and welfare of farmed rabbits.
Rabbits require a carefully balanced diet to meet their energy needs for growth, metabolism, reproduction, lactation, and the maintenance of nutrient reserves, with these requirements varying according to environmental factors (e.g., temperature, humidity), vital functions (such as reproduction and lactation), and physiological characteristics like breed, sex, and age. Understanding and addressing these needs is essential to optimize productivity, including meat, fur, and milk production. To determine the specific nutritional requirements of farmed rabbits, several methods are employed, including long-term feeding experiments, calorimetric measurements, comparative slaughter techniques, and non-invasive methods for body composition analysis. These approaches help predict the dietary needs of farmed and domestic rabbits, ensuring high productivity and healthy growth (Xiccato and Trocino, 2020).
To measure the varying energy needs of farmed rabbits, different methods such as long-term feeding experiments, calorimetric measurements, comparative slaughter techniques, and non-destructive methods for body composition measurement are used to predict the required nutrition of farmed or domestic rabbits and ensure productivity. Figure 1 illustrates the pyramid for rabbit food.
A well-balanced diet is crucial for promoting productive traits and maintaining rabbit health. For instance, an optimal fiber-to-starch ratio supports healthy digestive motility, stimulates the gut microbial community, and enhances fermentation. High-quality proteins are essential during growth and fattening stages, while dietary lipids support small intestine health. Essential minerals and vitamins are key for metabolic processes, particularly in growing and reproducing rabbits (Gidenne et al., 2020). Personalized nutrition strategies have been developed to meet the specific needs of rabbits at different growth stages and health conditions. Delgado et al. (2019) highlighted the importance of dietary fiber, particularly soluble fibers, in improving the health of crossbred hybrid rabbits post-weaning. Their study showed that while omega-3 fatty acids did not have significant health benefits, a high-fiber diet reduced mortality by 87% and improved digestibility and immune function.
Similarly, Shao et al. (2021) studied the effects of fat supplementation in rabbit diets, showing that adding 10% lard to the diet increased fat accumulation, serum cholesterol, and thyroid hormone levels, underscoring the importance of controlling fat levels to prevent obesity and other metabolic disorders in farmed rabbits. In another study, Marín-García et al. (2020) explored the impact of genetic selection on the nutritional needs of rabbits, revealing that genetically selected rabbits with higher growth rates (e.g., Line R) required specific protein levels for optimal growth and body composition. These findings emphasize the importance of tailoring diets to genetic traits and growth rates to ensure optimal productivity.
Additionally, Birolo et al. (2022) assessed genotype-diet interactions in two rabbit breeds, Grimaud and Hyla. Their research found that Hyla rabbits responded better to high-energy diets, exhibiting faster growth, improved feed conversion, and better nutrient digestibility, while Grimaud rabbits showed better carcass traits despite different dietary regimens. These results suggest that understanding the genotype-diet relationship is crucial for optimizing nutrition to improve the overall efficiency of rabbit farming.
To further optimize rabbit nutrition, Precision Nutrition (PN) strategies have been developed. These strategies use genetic, environmental, and physiological data to create personalized feeding plans. For instance, de Toro-Martín et al. (2017) discussed various PN approaches, including microbiota manipulation through probiotics and prebiotics, targeted supplementation, and the use of functional feeds to enhance nutrient absorption and feed efficiency. Paës et al. (2020) demonstrated that pre-weaning diets containing prebiotics significantly altered the gut microbiota, improving nutrient absorption and reducing mortality. These studies highlight the potential of PN to improve growth rates, feed efficiency, and overall rabbit health by tailoring nutrition to specific needs.
Understanding and addressing the complex nutritional requirements of rabbits through personalized feeding strategies is key to improving productivity and animal welfare. By considering factors like breed, growth stage, and environmental conditions, these strategies ensure that rabbits receive optimal nutrition for both health and productivity, contributing to more efficient and sustainable rabbit farming.
GAS makes use of both genotypic and phenotypic data of the reference population to predict the genetic traits of individual animals and select the most favorable breeding values to increase the technical and economic efficiency of animal breeding. The basic principle of genomic selection involves using marker data to assess the breeding value of animals without having the information of gene location. The process begins by gathering phenotypic and genotypic data from a reference population. The animals in the reference population are genotyped for SNPs across the entire genome. This data is then used to build a statistical model to estimate how each SNP affects the traits of interest. A predictive equation, known as the genomic estimated breeding value (GEBV), is developed based on this model. The GEBV can then be used to calculate the genomic breeding value of new animals based on their genotypes, even in the absence of reliable phenotypic data (Figure 2). The accuracy of the GEBV depends on factors such as trait heritability, the number of animals in the reference population, and the q parameter. Genomic selection accelerates the pace of genetic gain and improves productivity compared to traditional breeding techniques (Demircioglu, 2024; Ibtisham et al., 2017). In breeding selection programs of rabbits, different criteria are used to select the favorable traits in maternal and paternal lines to enhance the efficiency and production of rabbit farming. For instance, the maternal rabbit lines are selected based on the litter size and weight at weaning reflecting the fertility and the maternal ability of the doe as well as its ability for lactating and nourishing the progeny. On the other hand, the paternal rabbit lines are selected based on the daily weight gain from weaning to slaughtering. However, some multi-purpose farming used to select the rabbit lines with both the reproductive and growth traits to ensure productivity and welfare (García and Argente, 2020). Under the parent category of GAS, several subcategories including GS, MAS, and GWAS used genomic data of rabbit populations to improve rabbit breeding practices and enhance productivity and welfare on a large scale.
This representation showed that The GEBV can then be used to calculate the genomic breeding value of new animals based on their genotypes, even in the absence of reliable phenotypic data.
GS is a potential breeding tool used to reduce the generation gaps, enhance the genetic gain, improve the accuracy and intensity of the selection, and optimize the economic traits of dairy or model animals including rabbits. Though the efficiency of GS for selecting low heritability traits in rabbits is considerable, the genotyping cost in identifying genetic marker information and genomic breeding values is too high to enable genetic predictions in selective breeding programs of rabbits. Therefore, Mancin et al. (2021) employed genotype imputation to identify SNP density across the rabbit genome to ultimately improve the cost-efficiency of GS in rabbits. Thereby, Ning et al. (2022) improved the accuracy of GS for woolen traits in Angora rabbits through single-trait model evaluation using marker density and statistical designs. The efforts were directed to optimize the rabbit breeding pipeline by increasing the number of SNPs across the rabbit genome. A total of 629 Angora rabbits were taken and kept under farming conditions, the wool was harvested at 70, 140, and 210 days of age and the wool characteristics including diameter and length of fine wool, length, and production rate of coarse wool, as well as the weight of sheared wool were measured, genomic DNA was isolated from ear samples, pair-end libraries were constructed and sequenced using low-coverage whole genome sequencing, and marker densities were evaluated to estimate the heritabilities and stable genomic breeding values. The results showed 3.84% genomic coverage and a total of 18,577,154 high-quality SNPs were imputed with an increased accuracy of genetic imputation by 98%, revealing the potential of GS performance for the successful imputation of multi-trait models to boost animal breeding. Subsequently, Li et al. (2024) employed marker density data and a multi-trait genomic best linear unbiased prediction model to improve the GC accuracy for growth and slaughter traits of meat rabbits. A total of 1515 Kangda V line meat rabbits along with 399 litters were studied (1159 were examined for growth and the remaining 798 were slaughtered), the growth traits including feed conversion ratio and weight were recorded at 35, 49, and 70 days of the age, remaining rabbits were slaughtered at the age of 70 days to determine the eviscerated weight, net head weight, kidney weight, and leg weight, genomic DNA was extracted from blood samples and sequenced using low-coverage whole genome sequencing method, and finally the genotypic data was imputed. As a result, a total of 20,125,019 high-quality SNPs were produced with the imputation accuracy greater than 98% using the multi-trait GS model providing a baseline for the optimization of breeding programs of farmed rabbits to boost productivity and welfare. Since SNPs serve as a potential molecular marker for understanding rabbit traits and improving the efficiency of rabbit breeding employing genomic-assisted data, Li et al. (2022) identified a total of 32,144 SNPs within a rabbit genome through genotyping-by-sequencing method, indicating relevant genes associated with the important rabbit traits like fur color and weight, which can be used to improve the GS-based breeding techniques of rabbits on the large scale.
GWAS is a powerful tool used in animal breeding programs to identify genetic markers associated with traits of interest. The basic idea behind GWAS is to scan the entire genome for specific genetic variations called SNPs that are significantly linked to the traits being studied (Degen and Müller, 2023). To conduct a GWAS, a reference population of animals with both genetic marker data and trait measurements is established. These animals are genotyped using advanced sequencing technologies to identify millions of SNPs across their genomes. Statistical models are then used to find SNPs that show strong associations with the traits of interest. The identified SNPs are then used as markers and their estimated effects can then be incorporated into genomic selection models to improve the accuracy of breeding value estimation and accelerate genetic progress in animal breeding programs (El-Sabrout et al., 2020). GWAS uses genetic marker information, without needing to know the exact location of the genes, to estimate the breeding values of animals and select the most favorable individuals for breeding. It can be particularly useful for complex traits that are influenced by many genes. By scanning the entire genome, GWAS can detect associations even for traits with complex genetic architectures. The identified markers can then be used for advanced selection techniques in breeding programs (Sneller et al., 2021; Uffelmann et al., 2021). Sánchez et al. (2020) identified 189 SNPs across the genome of rabbits and found 20 candidate genes associated with feed efficiency and growth performance, both are economically productive traits in rabbit farming, using GWAS which can be manipulated to optimize the breeding values of farmed rabbits and choose the favorable genetic lines as parents of the next generation. Similarly, litter size is also an important reproductive trait of maternal rabbit lines which can be optimized by selecting the rabbit breeds with the excellent combination of functional genes associated with the uterine capacity identified through GWAS as Sosa-Madrid et al. (2020) employed GWAS to find high-density SNPs across 181 does from a control population and investigated the associated genes for characteristics like total number born, number born alive, number born dead, ovulation rate, and implanted embryos of rabbit, and identified the relevant genomic area on chromosome 11 and chromosome 17 closely linked with the reproductive traits of rabbit, providing a baseline for choosing maternal lines with superior reproductive ability during breeding programs. Zhang et al. (2023) identified a total of 91,456 SNPs evenly distributed on 21 autosomes across the rabbit genome using a restriction-site associated DNA sequencing approach and applied GWAS using a mixed linear model to identify functional SNPs containing coat-color associated genes on a particular genomic area. The study investigated the blood samples from the ear of 250 genetically unrelated Rex rabbits with different coat colors, the genomic DNA was extracted, sequenced, and mapped to identify potential SNPs, and then the genomic distribution and association were analyzed. The results showed a single genomic region on chromosome 4 containing the most important set of 24 SNPs in which the coat-color-associated agouti signaling protein (ASIP) gene was found that can help to choose the rabbit lines with the functional gene to improve phenotypic traits among farmed rabbits. Though, the coat color of a rabbit is a trait not linked with the productivity of the animals but important to select fancy rabbit lines with desirable phenotypes. Besides, GWAS has also become a promising approach for improving the meat quality of rabbits by identifying the genetic factors that directly affect the organoleptic properties of meat including fat consistency, shelf life, and nutritional values, improving the productive traits of breeding lines in the next rabbit generations. El Nagar et al. (2023) identified a genomic area across the genome of rabbits containing candidate genes on the rabbit chromosomes 1, 5, and 19 associated with the traits of intramuscular fats, saturated fatty acids, monounsaturated fatty acids, and polyunsaturated fatty acids using GWAS and provided insights into the manipulation of candidate genes to optimize the nutritional content of rabbit meat to ultimately improve the breeding efficiency and welfare. The study also emphasized the fact that the genetic variations of maternal rabbit lines influence the relevant genomic characteristics of progeny associated with the quality of meat and intramuscular fat deposition, suggesting the importance of selecting superior maternal rabbit lines during genomic-based selection programs. Similarly, Liao et al. (2021) identified growth-related genes across the genome of meat rabbit lines using the GWAS approach to enhance the longevity and growth potential of farmed rabbits. Targeting feed conversion ratio and residual feed intake of rabbits, Garreau et al. (2023) employed the GWAS approach and identified17 significant SNPs feed conversion ratio and 111 for residual feed intake with the estimated heritability traits for growth ranging from 0.14 to 0.33, for feed intake and efficiency traits ranged from 0.40 to 0.47, indicating a strong genetic component influencing these traits. The most significant peaks for feed conversion ratio and residual feed intake were located on rabbit chromosome 7 and 18, respectively along with a significant region on chromosome 18 was identified, containing the putative functional candidate gene, GOT1, involved in the metabolism of amino acids and urea, which aligns with the observed improvements in nitrogen balance and reduced nitrogen output in selected rabbit lines. Overall, GWAS used in this study has made significant contributions to rabbit breeding selection programs based on genetic information by identifying genome-wide significant SNPs associated with feed efficiency traits.
MAS is a breeding technique in animals that uses genetic markers to select individuals with desirable traits for breeding. The basic principle of MAS is to identify genetic markers, such as SNPs, that are associated with traits of interest and then use these markers to assist in the selection of breeding animals (Kandel et al., 2023). To implement MAS, genetic markers associated with traits of interest are identified by scanning the entire genome. Once these markers are identified, animals are genotyped to determine their genotypes at the identified markers, and animals with the most favorable marker genotypes, indicating the presence of desirable alleles, are selected as parents for the next generation. The advantage of MAS over traditional phenotypic selection is that it allows for selection at an earlier age, even before the trait is expressed, and can improve the accuracy of selection, especially for traits that are difficult or expensive to measure (de Koning, 2016). Candidate genes for the most important rabbit production and welfare traits including growth hormone genes, insulin-like growth factor 2 gene, myostatin gene, fat mass and obesity-associated genes, and leptin genes related to growth, carcass, and meat quality of rabbits have been identified for MAS to revolutionize the rabbit farming industry (Helal et al., 2022). Ramadan et al. (2020) evaluated the polymorphism of growth hormone genes, insulin-like growth factor 2 genes, and progesterone receptor genes within the genome of Gabali rabbits and studied their association with the productive traits of rabbits such as growth, litter size, and milk production through genomic DNA extraction, PCR amplification, and genotyping to provide functional markers for MAS rabbit breeding. The study findings revealed that the C/T and Del/Del genotypes of the growth hormone gene and insulin-like growth factor 2 gene for growth improvement, the C/T genotype of the growth hormone gene, and the G/G genotype of the progesterone receptor gene for litter size, and the C/C genotype of the growth hormone gene, the A/A genotype of the insulin-like growth factor 2 gene, and the A/A genotype of the progesterone receptor gene were favorable for incorporating these genotypes into MAS programs. Similarly, Safaa et al. (2023) also concluded that the Growth hormone genotypes of rabbits affect the carcass and slaughter traits, the fat mass and obesity-associated SNPs affect the intramuscular fat content, and insulin receptor substrate-1 SNPs influence the drip loss ability of rabbit meat, providing a genetic map to manipulate as markers for genomic selection of rabbit breeds. Whereas, Gencheva et al. (2022) suggested SNPs associated with the growth hormone gene and the related growth hormone receptor responsible for the body weight of rabbits. To provide genetic markers for carcass and meat quality of rabbit, Migdal et al. (2018) analyzed three leptin genes for polymorphism and identified their associations with the complex traits of rabbit carcass and meat quality revealing that the content intramuscular fat, protein, ash, and dry matter of rabbit meat is influenced by the polymorphism in the leptin gene-containing genomic area that can be manipulated while selecting meat rabbits on the basis of genetic markers. However, Wang et al. (2017a) have previously suggested the calpastatin gene as a candidate for the meat quality traits of rabbits in addition to leptin that can be used in a combination to improve the marker-based rabbit selection in the future. Nonetheless, MAS also has some limitations in that it only considers a few markers associated with the traits, while genomic selection (GS) uses the joint merit of all markers across the genome to estimate breeding values. GS has allowed for unprecedented advances in commercial breeding in recent years. Despite these limitations, MAS remains a valuable tool in animal breeding programs, especially when used in combination with other selection methods like GS to improve the accuracy and efficiency of selection.
The following table summarizes key candidate genes associated with important traits in rabbits, including meat and carcass characteristics, growth and body weight (Table 1), disease resistance (Table 2), and coat quality (Table 3). Table 4 further extends these findings by summarizing the integration of GAS and precision nutrition (PN) strategies in rabbit farming. It also indicates the required genotypes for each gene, providing valuable insights for optimizing genomic-assisted selection (GAS) strategies in rabbit breeding.
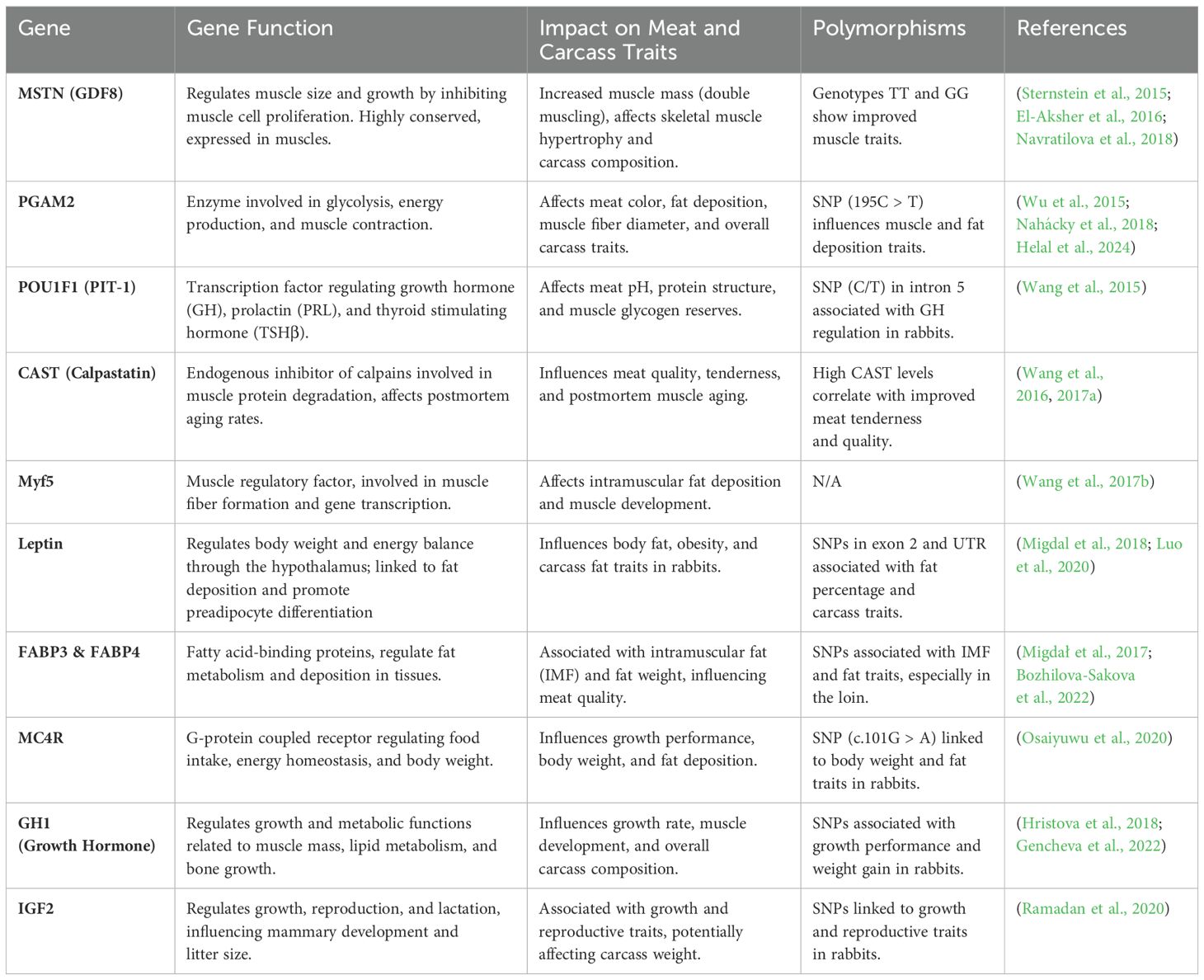
Table 1. Key candidate genes associated with meat, carcass traits, growth, and body weight in rabbits.
Precision nutrition (Figure 3) refers to the determination of nutrient requirements of livestock for protein accretion and maintenance to prevent nutrient deficiencies or excess (Bailey, 2020). Personalized feeding approaches cater to the specific needs of rabbits to improve the growth rate, feed conversion efficiency, and overall animal welfare by optimizing the nutrient utilization (Nutautaitė et al., 2023). Various commercial poultry growth promoters have a positive impact on the final body weight gain of rabbits than the others fed with a general diet. Oyinbo and Shaahu (2020) carried out a feed trial in which twenty unknown breeds of rabbits were fed with four different diet groups containing different poultry growth promoters namely non-inclusion (T1), el-rox (T2), glofast (T3), and maxgrowth (T4) to determine whether the specific diet influence the growth performance, fattening, or carcass characteristics of the rabbit. As a result, the rabbit groups fed with modified diet containing growth promoters (T2, T3, and T4) showed enhanced body weight (1020.40, 1084.00, and 1119.20) as well as individual organ weight than the control group (T1) with the maximum final body weight in a group fed with glofast, indicating that the growth promoter supplementation in rabbit diet is the best way to enhance the rabbit growth and fattening. Singh et al. (2021) also determined the effect of four dietary supplementation groups (t1, t2, t3, and t4) based on the melon seed oil (0%, 0.2%, 0.4%, and 0.6%) on the growth performance and immune response of thirty-six rabbits of mixed breeds and examined the average daily weight gain, average feed intake and conversion ratio, mortality, and antioxidation activities for 12 weeks. The findings turned out to be significant in which the average daily weight gain and average daily feed intake increased with the increase of melon seed oil concentration while the mortality was only recorded in the control group, and also indicated that the precise nutrient supplementation strengthen the immune system of rabbits, promoting survival, productivity, and rabbit welfare. The results were not so different when rabbits were fed with precise diet supplementation regimens based on prebiotic (Abd El-Aziz et al., 2022), organic selenium (Hosny et al., 2020), pepper oil (Abdelnour et al., 2018), n-3 polyunsaturated fatty acids (Castellini et al., 2019), organic selenium and chromium (Amer et al., 2019), and the combination of probiotics and prebiotics (Ayyat et al., 2018).
The integration of Genomic-Assisted Selection (GAS) with Precision Nutrition (PN) offers significant potential to enhance the productivity, feed efficiency, and overall welfare of rabbits. Precision nutrition in rabbits is critical due to their specialized digestive system, which requires a delicate balance of fiber, protein, and energy to ensure optimal health and growth (Pinheiro and Gidenne, 2024). Imbalances in the diet can lead to severe health issues like gastrointestinal stasis (Reponen, 2024), and thus, providing the correct nutrients at precise levels tailored to the genetic profiles of rabbits is paramount for sustainable farming. Recent developments in precision feeding systems further improve this process, particularly when combined with genomic insights that identify rabbits with superior genetic traits such as enhanced growth rates and better feed conversion efficiency.
Genomic-assisted selection allows for the identification of genetic markers that correlate with growth performance and feed efficiency, thereby optimizing breeding programs. For example, studies have shown that genetic traits linked to lean muscle development and efficient nutrient metabolism can be selectively bred for, allowing farmers to focus on improving the genetic base of their herds (Li et al., 2024). Combining GAS with PN allows for the formulation of diets that are fine-tuned to meet the specific nutritional needs of these genetically superior rabbits, which in turn enhances productivity and reduces feed waste. Precision nutrition systems can further support this synergy by customizing dietary regimens based on individual genetic profiles, thus ensuring that rabbits receive the optimal nutrients at every stage of growth.
An important advancement in this area is the development of automated and precise feeding systems that enhance the delivery of tailored nutrition. For example, Mohamad et al. (2023) introduced an automatic rabbit feeding system based on the eight-step George E. Dieter design, capable of distributing pellets across multiple cages at a rate of 400 grams per day. This system ensures each rabbit receives the correct amount of feed based on its specific nutritional needs. Similarly, Kharisma and Sintawati (2023) developed an IoT-based system for managing rabbit breeding that integrates feeding, drinking, temperature control, and environmental monitoring. This system leverages ultrasonic sensors, real-time clocks, and IoT cloud dashboards to adjust feeding and environmental conditions automatically or manually, ensuring optimal conditions for the rabbits’ health and productivity. The synergy between GAS, PN, and automated feeding systems creates a holistic approach to rabbit farming. This integration not only optimizes the use of resources and reduces feed waste but also significantly improves both productivity and animal welfare. Moreover, innovative strategies like feed restriction (FR), particularly post-weaning feed restriction, have proven effective in enhancing feed efficiency and maintaining digestive health in young rabbits. As demonstrated by Ebeid et al. (2022), controlled feed restriction strategies such as modifying the quantity and quality of feed at key developmental stages can promote efficient growth while reducing the need for medication. Similarly, Martignon et al. (2021) showed that fragmented feed distribution (FFD) enhances feed conversion and improves growth rates in growing rabbits, adding further value to precision feeding systems.
These developments in precision feeding, in conjunction with GAS, offer an effective means to ensure that rabbits receive the optimal nutrition for growth and welfare. Furthermore, the increasing use of genomic-assisted selection for identifying traits linked to feed conversion efficiency (FCE) and growth potential (as shown in studies by Li et al., 2024 and Sánchez et al., 2020) allows for a more targeted approach to breeding and feeding strategies. As genetic research continues to advance, and as automated technologies become more prevalent, the potential for fully personalized nutrition systems based on genetic profiles is set to revolutionize rabbit farming. This will make the industry more sustainable, cost-effective, and capable of meeting the growing demands for rabbit products while maintaining high animal welfare standards.
The combination of genomic selection and targeted nutritional techniques has the potential to synergistically improve productivity, health, and welfare in rabbit farming. For example, selecting superior rabbit breeds for gut microbiome resilience, providing prebiotic feeds, and examining the use of locally available, sustainable feed resources like agricultural by-products and macroalgae could also enhance the environmental and economic sustainability of rabbit production (Song et al., 2021). Additionally, rabbit farming as a low-input, climate-smart agriculture strategy holds the promise for enhancing food security and income generation, especially for smallholder farmers in developing regions. Although, the integration of genomics and nutrition presents an innovative approach for optimizing rabbit production, some limitations and challenges are still remained to be addressed. For instance, there is a lack of comprehensive genomic data and the understanding of the genetic basis of important traits in rabbits (Johnson et al., 2024). This makes it difficult to identify beneficial genetic markers linked to desired production traits, such as disease resistance, feed efficiency, and meat quality, and develop effective genomic selection programs. Additionally, accurately measuring and optimizing nutrient requirements, especially for specialized rabbit breeds and production systems, remains a challenge as nutritional needs vary significantly based on factors like age, health status, and environmental conditions (Moto, 2024). The limited availability and high cost of specialized feed ingredients like macroalgae that provide nutritional and health benefits to rabbits also brings a challenge as scaling up production and supply chains for these novel feed resources is difficult (Al-Soufi et al., 2022). Finally, inadequate technical support and extension services to help farmers adopt new genomic and nutritional technologies is a significant limitation. In this regard, improving knowledge transfer to farmers is crucial.
By overcoming these limitations and capitalizing on the potential synergies, integrating genomics and nutrition has promising prospects for enhancing productivity, sustainability, and welfare in rabbit production. Thereby, precision feeding strategies, developed based on individual genetic needs, should be practiced to minimize feed waste and create a more sustainable production system. Similarly, the selection for rabbits with enhanced disease resistance, facilitated by genomic information, must be carried out, leading to a significant decrease in antibiotic usage, benefiting both animal welfare and public health by addressing antibiotic resistance concerns. Finally, genes linked to faster growth rates should be identified to accelerate farm productivity by allowing breeders to select rabbits that reach market weight in short period of time. Overall, integrating genomics and nutrition in rabbit farming offers a promising technique for achieving significant advancements, but requires active research and development efforts to fully realize its potential.
In conclusion, the integration of GAS and PN in rabbit farming is a promising approach to enhance productivity and welfare in a sustainable and efficient manner. Using GAS, farmers can select rabbits with superior genetic traits for improved productivity and welfare. This approach, coupled with PN techniques tailored to individual needs, can lead to personalized nutrition interventions that optimize reproductive performance and health outcomes in rabbits. The importance of this integrated approach lies in its potential to meet the growing demand for rabbit meat and sustainable raw materials worldwide. By enhancing selection accuracy, genetic potential, and nutritional optimization, this approach can lead to improved productivity, sustainability, and welfare in rabbit farming. However, it is essential to address challenges such as the lack of comprehensive genomic data, understanding genetic markers for desired traits, and optimizing nutrient requirements for specialized rabbit breeds. Additionally, efforts should focus on knowledge transfer to farmers, scaling up production of novel feed resources, and providing technical support for adopting new technologies. Overall, integrating GAS and PN in rabbit farming holds great promise for advancing the industry towards more efficient, sustainable, and responsible practices. Continued research and development in this area will be crucial for realizing the full potential of this integrated approach and ensuring the long-term success of rabbit production systems.
NG: Conceptualization, Formal analysis, Investigation, Validation, Writing – original draft, Writing – review & editing. IS: Writing – original draft, Writing – review & editing. LK: Writing – original draft, Writing – review & editing. AM: Writing – original draft, Writing – review & editing. YX: Writing – original draft, Writing – review & editing. BS: Supervision, Visualization, Writing – original draft, Writing – review & editing. XD: Writing – original draft, Writing – review & editing. SW: Writing – original draft. ZR: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The authors express their gratitude to the National Science Foundation of China (No 32102567 and 31972529), the Program for Shaanxi Science & Technology (2022KJXX-13) for financial support.
I acknowledge members of the innovative research team of special economic animals’ group, (Northwest A&F University, Shaanxi, China) for their guidance and encouragement through writing process.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abd El-Aziz A. H., El-Kasrawy N. I., Abd El-Hack M. E., Kamel S. Z., Mahrous U. E., El-Deeb E. M., et al. (2022). Growth, immunity, relative gene expression, carcass traits and economic efficiency of two rabbit breeds fed prebiotic supplemented diets. Anim. Biotechnol. 33, 417–428. doi: 10.1080/10495398.2020.1800485
Abdelnour S., Alagawany M., Abd El-Hack M. E., Sheiha A. M., Saadeldin I. M., Swelum A. A. (2018). Growth, carcass traits, blood hematology, serum metabolites, immunity, and oxidative indices of growing rabbits fed diets supplemented with red or black pepper oils. Animals 8, 168. doi: 10.3390/ani8100168
Abe O., Amusan S., Ozoje M. (2019). Phenotypic correlation among morphometric traits in new zealand white rabbits reared under tropical condition. Nigerian J. Anim. Production 46, 17–22. doi: 10.51791/NJAP.V46I5.222
Al-Soufi S., García J., Muíños A., López-Alonso M. (2022). Marine macroalgae in rabbit nutrition—a valuable feed in sustainable farming. Animals 12, 2346. doi: 10.3390/ani12182346
Amer S. A., Omar A. E., Abd El-Hack M. E. (2019). Effects of selenium-and chromium-enriched diets on growth performance, lipid profile, and mineral concentration in different tissues of growing rabbits. Biol. Trace Element Res. 187, 92–99. doi: 10.1007/s12011-018-1356-4
Assan N. (2018). Factors influencing post-weaning growth and mortality in rabbits. Sci. J. Anim. Sci. 7, 486–492. doi: 10.14196/SJAS.V7I3.2511
Ayyat M. S., Al-Sagheer A. A., Abd El-Latif K. M., Khalil B. A. (2018). Organic selenium, probiotics, and prebiotics effects on growth, blood biochemistry, and carcass traits of growing rabbits during summer and winter seasons. Biol. Trace Element Res. 186, 162–173. doi: 10.1007/s12011-018-1293-2
Bailey C. A. (2020). Precision poultry nutrition and feed formulationAnimal Agriculture. Elsevier, 367–378. doi: 10.1016/B978-0-12-817052-6.00021-5
Ballan M., Bovo S., Schiavo G., Schiavitto M., Negrini R., Fontanesi L. (2022). Genomic diversity and signatures of selection in meat and fancy rabbit breeds based on high-density marker data. Genet. Selection Evol. 54, 3. doi: 10.1186/s12711-022-00696-9
Bao Z., Chen Y., Li J., Cai J., Yang J., Zhai P., et al. (2024). 4D label-free quantitative proteomic analysis identifies CRABP1 as a novel candidate gene for litter size in rabbits. Biol. Reprod., ioae038. doi: 10.1093/biolre/ioae038
Benedek I., Altbäcker V., Zsolnai A., Nagy I., Mezőszentgyörgyi D., Molnár T. (2023). The role of PRLR gene polymorphisms in milk production in European wild rabbit (Oryctolagus cuniculus). Animals 13, 671. doi: 10.3390/ani13040671
Biada I., Ibáñez-Escriche N., Blasco A., Casto-Rebollo C., Santacreu M. A. (2024). Microbiome composition as a potential predictor of longevity in rabbits. Genet. Selection Evol. 56, 25. doi: 10.1186/s12711-024-00895-6
Birolo M., Xiccato G., Bordignon F., Dabbou S., Zuffellato A., Trocino A. (2022). Growth performance, digestive efficiency, and meat quality of two commercial crossbred rabbits fed diets differing in energy and protein levels. Animals 12, 2427. doi: 10.3390/ani12182427
Blasco A., Martínez-Álvaro M., García M.-L., Ibáñez-Escriche N., Argente M.-J. (2017). Selection for environmental variance of litter size in rabbits. Genet. Selection Evol. 49, 1–8. doi: 10.1186/s12711-017-0323-4
Bovo S., Ribani A., Schiavo G., Taurisano V., Bertolini F., Fornasini D., et al. (2024). Genome-wide association studies for diarrhoea outcomes identified genomic regions affecting resistance to a severe enteropathy in suckling rabbits. J. Anim. Breed. Genet. 141, 328–342. doi: 10.1111/jbg.12844
Bozhilova-Sakova M., Viryanski D., Dimitrova I., Ignatova M., Stoykova-Grigorova R. (2022). Genetic factors influencing rabbit breeding. An overview. Bulgarian J. Anim. Husbandry/Životnov Dni Nauki 59.
Castellini C., Mattioli S., Signorini C., Cotozzolo E., Noto D., Moretti E., et al. (2019). Effect of Dietary n-3 Source on Rabbit Male Reproduction. Oxid. Med. Cell. Longevity 2019, 3279670. doi: 10.1155/2019/3279670
Chen S. Y., Zhang W. X., Zhang G. W., Peng J., Zhao X. B., Lai S. J. (2013). Case–control study and mRNA expression analysis reveal the MyD88 gene is associated with digestive disorders in rabbit. Anim. Genet. 44, 703–710. doi: 10.1111/age.12052
Degen B., Müller N. (2023). Advanced marker-assisted selection versus genomic selection in breeding programs. bioRxiv. doi: 10.1101/2023.02.20.529263
de Koning D.-J. (2016). Meuwissen et al. on Genomic Selection. Genetics 203, 5–7. doi: 10.1534/genetics.116.189795
Delgado R., Menoyo D., Abad-Guamán R., Nicodemus N., Carabaño R., García J. (2019). Effect of dietary soluble fibre level and n-6/n-3 fatty acid ratio on digestion and health in growing rabbits. Anim. Feed Sci. Technol. 255, 114222. doi: 10.1016/j.anifeedsci.2019.114222
Demars J., Iannuccelli N., Utzeri V. J., Auvinet G., Riquet J., Fontanesi L., et al. (2018). New insights into the melanophilin (MLPH) gene affecting coat color dilution in rabbits. Genes 9, 430. doi: 10.3390/genes9090430
Demircioglu H. B. (2024). Genomic Selection in Animal BreedingBIO Web of Conferences. EDP Sci., 01069. doi: 10.1051/bioconf/20248501069
de Toro-Martín J., Arsenault B. J., Després J.-P., Vohl M.-C. (2017). Precision nutrition: a review of personalized nutritional approaches for the prevention and management of metabolic syndrome. Nutrients 9, 913. doi: 10.3390/nu9080913
Diribarne M., Mata X., Rivière J., Bouet S., Vaiman A., Chapuis J., et al. (2012). LIPH expression in skin and hair follicles of normal coat and Rex rabbits. PloS One 7, e30073. doi: 10.1371/journal.pone.0030073
Ebeid T. A., Tůmová E., Al-Homidan I. H., Ketta M., Chodová D. J. M. S. (2022). The potential role of feed restriction on productivity, carcass composition, meat quality, and muscle fibre properties of growing rabbits: A review. Meat Science, 191, 108845. doi: 10.1016/j.meatsci.2022.108845
El-Aksher S. H., Sherif H., Khalil M., El-Garhy H. A., Ramadan S. (2016). Polymorphism of progesterone receptor gene in Moshtohor line rabbits and their parental lines using PCR-RFLP technique. Anim. Biotechnol. 25, 25.
El Nagar A. G., Heddi I., Sosa-Madrid B. S., Blasco A., Hernández P., N. I.-E. (2023). Genome-Wide Association Study of Maternal Genetic Effects on Intramuscular Fat and Fatty Acid Composition in Rabbits. Animals 13, 3071. doi: 10.3390/ani13193071
El-Sabrout K., Aggag S., de Souza J. B. F. Jr (2020). Some recent applications of rabbit biotechnology–a review. Anim. Biotechnol. 31, 76–80. doi: 10.1080/10495398.2018.1539005
Fathi M., Abdelsalam M., Al-Homidan I., Abou-Emera O., Rayan G. (2023). Effect of genotype of growing rabbits on productive performance with special reference to residual feed intake at hot temperature. Anim. Bioscience 36, 1067. doi: 10.5713/ab.22.0355
Fatima N., Jia L., Liu B., Li L., Bai L., Wang W., et al. (2023). A homozygous missense mutation in the fibroblast growth factor 5 gene is associated with the long-hair trait in Angora rabbits. BMC Genomics 24, 298. doi: 10.1186/s12864-023-09405-2
Fontanesi L. (2021). Rabbit genetic resources can provide several animal models to explain at the genetic level the diversity of morphological and physiological relevant traits. Appl. Sci. 11, 373. doi: 10.3390/app11010373
Fu L., Yang Z.-J., Chen S.-Y., Wang J., Lai S.-J. (2014). Investigation of JAK1 and STAT3 polymorphisms and their gene–gene interactions in nonspecific digestive disorder of rabbits. Gene 543, 8–14. doi: 10.1016/j.gene.2014.04.017
Fu L., Zhao M.-D., Chen S.-Y., Jia X.-B., Lai S.-J. (2015). Investigation of genetic susceptibility to nonspecific digestive disorder between TYK2, JAK1, and STAT3 genes in rabbits. Livestock Sci. 181, 137–142. doi: 10.1016/j.livsci.2015.08.014
García M.-L., Argente M.-J. (2020). “The genetic improvement in meat rabbits,” in Lagomorpha Charact, vol. 10. BoD – Books on Demand, 2021. p. 144. doi: 10.5772/intechopen.93896
Garreau H., Labrune Y., Chapuis H., Ruesche J., Riquet J., Demars J., et al. (2023). Genome wide association study of growth and feed efficiency traits in rabbits. World Rabbit Sci. 31, 163–169. doi: 10.4995/wrs.2023.18215
Gencheva D. G., Velikov K. P., Veleva P. M. (2022). Association analysis of nucleotide polymorphisms in growth hormone (GH) and its receptor (GHR) with body weight in Californian rabbits. World Rabbit Sci. 30, 95–102. doi: 10.4995/wrs.2022.13127
Gidenne T., Lebas F., Licois D., García J. (2020). Nutrition and feeding strategy: impacts on health statusNutrition of the Rabbit. CAB Int. Wallingford UK, 193–221. doi: 10.1079/9781789241273.0193
Gunia M., David I., Hurtaud J., Maupin M., Gilbert H., Garreau H. (2018). Genetic parameters for resistance to non-specific diseases and production traits measured in challenging and selection environments; application to a rabbit case. Front. Genet. 9. doi: 10.3389/fgene.2018.00467
Helal M., Ahmed M., Ragab M., Ateya A., Sakr S. (2024). Association of single nucleotide polymorphisms in Neuropeptide Y (NPY) and Phosphoglycerate Mutase 2 (PGAM2) genes with growth traits in rabbits. Trop. Anim. Health Production 56, 239. doi: 10.1007/s11250-024-04085-w
Helal M., Hany N., Maged M., Abdelaziz M., Osama N., Younan Y. W., et al. (2022). Candidate genes for marker-assisted selection for growth, carcass and meat quality traits in rabbits. Anim. Biotechnol. 33, 1691–1710. doi: 10.1080/10495398.2021.1908315
Hosny N. S., Hashem N. M., Morsy A. S., Abo-Elezz Z. R. (2020). Effects of organic selenium on the physiological response, blood metabolites, redox status, semen quality, and fertility of rabbit bucks kept under natural heat stress conditions. Front. Veterinary Sci. 7. doi: 10.3389/fvets.2020.00290
Hristova D. G., Tanchev S., Velikov K., Gonchev P., Georgieva S. (2018). Single nucleotide polymorphism of the growth hormone (GH) encoding gene in inbred and outbred domestic rabbits. World Rabbit Sci. 26, 49–55. doi: 10.4995/WRS.2018.7211
Ibtisham F., Zhang L., Xiao M., An L., Ramzan M. B., Nawab A., et al. (2017). Genomic selection and its application in animal breeding. Thai J. Veterinary Med. 47, 301–310. doi: 10.56808/2985-1130.2838
Jia X., Ding P., Chen S., Zhao S., Wang J., Lai S. (2021). Analysis of MC1R, MITF, TYR, TYRP1, and MLPH genes polymorphism in four rabbit breeds with different coat colors. Animals 11, 81. doi: 10.3390/ani11010081
Johnson K. E., Hayes J., Davidson P., Tinago C. B., Anguyo G. (2024). [amp]]lsquo;Never cry for food’: food security, poverty, and recurring themes in news media regarding rabbit farming in East Africa. Renewable Agric. Food Syst. 39, e2. doi: 10.1017/S1742170523000480
Kandel R., Kadariya I. P., Bohara K., Adhikari S. (2023). A review on molecular breeding techniques: Crucial approach in livestock improvement. Arch. Agric. Environ. Sci. 8, 639–651. doi: 10.26832/24566632.2023.0804027
Kharisma A., Sintawati A. (2023). An Automatic Internet of Things-Based System for Rabbit Cage. Ultima Computing: Jurnal Sistem Komputer 15, 52–57. doi: 10.31937/sk.v15i2.3404
Li J., Chen Y., Liu M., Chen Q., Zhou J., Bao G., et al. (2020). Association of Melanophilin (MLPH) gene polymorphism with coat colour in Rex rabbits. World Rabbit Sci. 28, 29–38. doi: 10.4995/wrs.2020.12082
Li Y., Hu S., Wang J., Chen S., Jia X., Lai S. (2018). Molecular cloning, polymorphism, and expression analysis of the LKB1/STK11 gene and its association with non-specific digestive disorder in rabbits. Mol. Cell. Biochem. 449, 127–136. doi: 10.1007/s11010-018-3349-1
Li W., Li W., Song Z., Gao Z., Xie K., Wang Y., et al. (2024). Marker Density and Models to Improve the Accuracy of Genomic Selection for Growth and Slaughter Traits in Meat Rabbits. Genes 15, 454. doi: 10.3390/genes15040454
Li C., Li Y., Zheng J., Guo Z., Mei X., Lei M., et al. (2022). Trait analysis in domestic rabbits (Oryctolagus cuniculus f. domesticus) using SNP markers from genotyping-by-sequencing data. Animals 12, 2052. doi: 10.3390/ani12162052
Liao Y., Wang Z., Glória L. S., Zhang K., Zhang C., Yang R., et al. (2021). Genome-wide association studies for growth curves in meat rabbits through the single-step nonlinear mixed model. Front. Genet. 12. doi: 10.3389/fgene.2021.750939
Liu W.-C., Zeng Y., Chen S.-Y., Jia X.-B., Lai S.-J. (2017). The polymorphism and gene expression of chemokine receptor 6 is associated with digestive disorders in rabbits. Indian J. Anim. Res. 51, 269–274. doi: 10.18805/ijar.11318
Liu Y.-F., Zhang G.-W., Xiao Z.-L., Yang Y., Deng X.-S., Chen S.-Y., et al. (2013). Single nucleotide polymorphisms of NLRP12 gene and association with non-specific digestive disorder in rabbit. Asian-Australasian J. Anim. Sci. 26, 1072. doi: 10.5713/ajas.2013.13087
Lona-Durazo F., Hernandez-Pacheco N., Fan S., Zhang T., Choi J., Kovacs M. A., et al. (2019). Meta-analysis of GWA studies provides new insights on the genetic architecture of skin pigmentation in recently admixed populations. BMC Genet. 20, 1–16. doi: 10.1186/s12863-019-0765-5
Luo G., Hu S., Lai T., Wang J., Wang L., Lai S. (2020). MiR-9-5p promotes rabbit preadipocyte differentiation by suppressing leptin gene expression. Lipids Health Dis. 19, 1–11. doi: 10.1186/s12944-020-01294-8
Mancin E., Sosa-Madrid B. S., Blasco A., Ibáñez-Escriche N. (2021). Genotype imputation to improve the cost-efficiency of genomic selection in rabbits. Animals 11, 803. doi: 10.3390/ani11030803
Mancini S., Paci G. (2021). Probiotics in rabbit farming: Growth performance, health status, and meat quality. Animals 11, 3388. doi: 10.3390/ani11123388
Marín-García P., Ródenas L., Martínez-Paredes E., Cambra-López M., Blas E., Pascual J. (2020). A moderate protein diet does not cover the requirements of growing rabbits with high growth rate. Anim. Feed Sci. Technol. 264, 114495. doi: 10.1016/j.anifeedsci.2020.114495
Martignon M., Burel C., Cauquil L., Combes S., Gidenne T. J. A. (2021). Impact of feed restriction and fragmented feed distribution on performance, intake behaviour and digestion of the growing rabbit. Animal 15, 100270. doi: 10.1016/j.animal.2021.100270
Matics Z., Gerencsér Z., Kasza R., Terhes K., Nagy I., Radnai I., et al. (2021). Effect of ambient temperature on the productive and carcass traits of growing rabbits divergently selected for body fat content. Animal 15, 100096. doi: 10.1016/j.animal.2020.100096
Migdał Ł., Kozioł K., Migdał W., Pałka S., Kmiecik M., Migdał A., et al. (2017). Rabbits Breeding in Poland–Possibility of Implementation of Marker Assisted Selection (Mas) in Breeding Vol. 347 (Institute For Animal Husbandry).
Migdal L., Koziol K., Palka S., Migdal W., Zabek T., Otwinowska-Mindur A., et al. (2018). Mutations in leptin (LEP) gene are associated with carcass and meat quality traits in crossbreed rabbits. Anim. Biotechnol. 29, 153–159. doi: 10.1080/10495398.2017.1347881
Mohamad M. A. H., Yahya M. S., Noor H. M., Jamadi M. H., Janon M. N., Jumari M. Z. I. (2023). A New Design of Automatic Rabbit Feeding System. Int. J. Business Technol. Manage. 5, 1–9.
Molette C., Gilbert H., Larzul C., Balmisse E., Ruesche J., Manse H., et al. (2016). Direct and correlated responses to selection in two lines of rabbits selected for feed efficiency under ad libitum and restricted feeding: II. Carcass and meat quality. J. Anim. Sci. 94, 49–57. doi: 10.2527/jas.2015-9403
Moto E. (2024). Potential, challenges and prospects of rabbit farming in urban and peri-urban areas of Dodoma city, Tanzania. World Rabbit Sci. 32, 31–42. doi: 10.4995/wrs.2024.20238
Nahácky J., Židek R., Gábor M., Miluchová M., Bučko O., Kováčik. A. (2018). MC4R and PGAM2 genes polymorphism association with production traits in rabbit (Oryctolagus cuniculus). J. Microbiology Biotechnol. Food Sci. 7, 537. doi: 10.15414/jmbfs.2018.7.5.493-495
Nasr M. A., Abd-Elhamid T., Hussein M. A. (2017). Growth performance, carcass characteristics, meat quality and muscle amino-acid profile of different rabbits breeds and their crosses. Meat Sci. 134, 150–157. doi: 10.1016/j.meatsci.2017.07.027
Navratilova A., Požgajova M., Trakovicka A., Rafay J. (2018). Rabbit myostatine gene polymorhism (c. 747 + 34C> T and c. 194A> G) as a marker for meat production. J. Cent. Eur. Agric. 19, 948–953. doi: 10.5513/JCEA01/19.4.2367
Ning C., Xie K., Huang J., Di Y., Wang Y., Yang A., et al. (2022). Marker density and statistical model designs to increase accuracy of genomic selection for wool traits in Angora rabbits. Front. Genet. 13. doi: 10.3389/fgene.2022.968712
Nutautaitė M., Racevičiūtė-Stupelienė A., Bliznikas S., Vilienė V. (2023). Enhancement of rabbit meat functionality by replacing traditional feed raw materials with alternative and more sustainable freshwater Cladophora glomerata macroalgal biomass in their diets. Foods 12, 744. doi: 10.3390/foods12040744
Osaiyuwu O., Bolaji U., Adeyinka O., Akinyemi M., Salako A. (2020). Melanocortin 4 receptor (Mc4r) gene polymorphism and its association with body weights of some breeds of rabbit. Nigerian J. Anim. Production 47, 1–7. doi: 10.51791/njap.v47i2.48
Oyinbo E. O., Shaahu D. T. (2020). Growth response and carcass yield of rabbit fed diets containing supplemented poultry growth promoters. Biol. Res. 3, 45–54.
Paës C., Gidenne T., Bébin K., Duperray J., Gohier C., Guené-Grand E., et al. (2020). Early introduction of solid feeds: ingestion level matters more than prebiotic supplementation for shaping gut microbiota. Front. Veterinary Sci. 7. doi: 10.3389/fvets.2020.00261
Pilarczyk B., Tomza-Marciniak A., Pilarczyk R., Januś E., Stanek P., Seremak B., et al. (2020). The effect of the sex, age, and breed of farmed rabbits and the choice of management system on the extensity and intensity of Eimeria infection. Veterinary World 13, 1654. doi: 10.14202/vetworld.2020.1654-1660
Piles M., Mora M., Kyriazakis I., Tusell L., Pascual M., Sánchez J. (2024). Novel phenotypes of feeding and social behaviour and their relationship with individual rabbit growth and feed efficiency. Animal 18, 101090. doi: 10.1016/j.animal.2024.101090
Pinheiro V., Gidenne T. (2024). “Rabbit Nutrition and Feeding,” in Veterinary Care of Farm Rabbits: A Complete Practice Guide to Rabbit Medicine and Production (Cham: Springer International Publishing), 201–234. doi: 10.1007/978-3-031-44542-2_10
Pomar C., Remus A. (2023). Fundamentals, limitations and pitfalls on the development and application of precision nutrition techniques for precision livestock farming. Animal 17, 100763. doi: 10.1016/j.animal.2023.100763
Ragab M., Baselga M., El Nagar A. G., Mínguez C., Pablo Sánchez J. (2021). Genetic analysis of functional longevity in a complete diallel crossing experiment involving four maternal rabbit lines. J. Anim. Breed. Genet. 138, 474–481. doi: 10.1111/jbg.12520
Ramadan S., Manaa E., El-Attrony M., Nagar A. E. (2020). Association of growth hormone (GH), insulin-like growth factor 2 (IGF2) and progesterone receptor (PGR) genes with some productive traits in Gabali rabbits. World Rabbit Sci. 28, 135–144. doi: 10.4995/wrs.2020.12610
Ren A., Du K., Jia X., Yang R., Wang J., Chen S.-Y., et al. (2019). Genetic diversity and population structure of four Chinese rabbit breeds. PloS One 14, e0222503. doi: 10.1371/journal.pone.0222503
Reponen R. A. (2024). The Role of Dietary Carbohydrates in Digestive Disorders of Rabbits. University of Zagreb, Faculty of Veterinary Medicine. Digital Repository of the University of Zagreb. Available online at: https://urn.nsk.hr/urn:nbn:hr:178:140777.
Safaa H. M., Ragab M., Ahmed M., El-Gammal B., Helal M. (2023). Influence of polymorphisms in candidate genes on carcass and meat quality traits in rabbits. PloS One 18, e0294051. doi: 10.1371/journal.pone.0294051
Sánchez J. P., Legarra A., Velasco-Galilea M., Piles M., Sánchez A., Rafel O., et al. (2020). Genome-wide association study for feed efficiency in collective cage-raised rabbits under full and restricted feeding. Anim. Genet. 51, 799–810. doi: 10.1111/age.12988
Setiaji A., Lestari D. A., Pandupuspitasari N. S., Agusetyaningsih I., Khan F. A. (2023). Genetic characteristics of complete mtDNA genome sequence of Indonesian local rabbit (Oryctolagus cuniculus). J. Genet. Eng. Biotechnol. 21, 96. doi: 10.1186/s43141-023-00546-1
Shao J., Wang J., Li Y., Elzo M. A., Tang T., Lai T., et al. (2021). Growth, behavioural, serum biochemical and morphological changes in female rabbits fed high-fat diet. J. Anim. Physiol. Anim. Nutr. 105, 345–353. doi: 10.1111/jpn.13459
Sharma P., Doultani S., Hadiya K., George L., Highland H. (2024). Overview of Marker-assisted Selection in Animal Breeding. J. Adv. Biol. Biotechnol. 27, 303–318. doi: 10.9734/jabb/2024/v27i5790
Siddiqui S. A., Gerini F., Ikram A., Saeed F., Feng X., Chen Y. (2023). Rabbit meat—production, consumption and consumers’ attitudes and behavior. Sustainability 15, 2008. doi: 10.3390/su15032008
Sikiru A. B., Alemede I. C., Arangasamy A., Egena S. S. A., Ijaiya A. T., Makinde J. O. (2020). ) Rabbit: an animal at the nexus of food production and bioscience research for sustainable development in developing countries. Trop. Subtropical Agroecosystems 23. doi: 10.56369/tsaes.2806
Singh A., Alagbe J., Sharma S., Oluwafemi R., Agubosi O. (2021). Effect of dietary supplementation of melon (Citrallus lanatus) seed oil on the growth performance and antioxidant status of growing rabbits. Indonesian J. Innovation Appl. Sci. (IJIAS) 1, 134–143. doi: 10.47540/ijias.v1i2.175
Sneller C., Ignacio C., Ward B., Rutkoski J., Mohammadi M. (2021). Using genomic selection to leverage resources among breeding programs: consortium-based breeding. Agronomy 11, 1555. doi: 10.3390/agronomy11081555
Song J., Zhang J., Xu J., Garcia-Barrio M., Chen Y. E., Yang D. (2021). Genome engineering technologies in rabbits. J. Biomed. Res. 35, 135. doi: 10.7555/JBR.34.20190133
Sosa-Madrid B. S., Santacreu M. A., Blasco A., Fontanesi L., Pena R. N., Ibáñez-Escriche N. (2020). A genomewide association study in divergently selected lines in rabbits reveals novel genomic regions associated with litter size traits. J. Anim. Breed. Genet. 137, 123–138. doi: 10.1111/jbg.12451
Sternstein I., Reissmann M., Maj D., Bieniek J., Brockmann G. A. (2015). A comprehensive linkage map and QTL map for carcass traits in a cross between Giant Grey and New Zealand White rabbits. BMC Genet. 16, 1–12. doi: 10.1186/s12863-015-0168-1
Szendrő K., Szendrő Z., Gerencsér Z., Radnai I., Horn P., Matics Z. (2016). Comparison of productive and carcass traits and economic value of lines selected for different criteria, slaughtered at similar weights. World Rabbit Sci. 24, 15–23. doi: 10.4995/wrs.2016.3684
Uffelmann E., Huang Q. Q., Munung N. S., De Vries J., Okada Y., Martin A. R., et al. (2021). Genome-wide association studies. Nat. Rev. Methods Primers 1, 59. doi: 10.1038/s43586-021-00056-9
Wang J., Elzo M. A., Jia X., Chen S., Lai S. (2017a). Calpastatin gene polymorphism is associated with rabbit meat quality traits. J. Appl. Anim. Res. 45, 342–345. doi: 10.1080/09712119.2016.1191498
Wang J., Elzo M. A., Yan L., Chen S., Jia X., Zhang M., et al. (2016). A single nucleotide polymorphism in CAST gene is associated with meat quality traits in rabbits. Anim. Sci. Papers Rep. 24, 269–278.
Wang J., Hu Y., Elzo M. A., Shi Y., Jia X., Chen S., et al. (2017b). Genetic effect of Myf5 gene in rabbit meat quality traits. J. Genet. 96, 673–679. doi: 10.1007/s12041-017-0822-7
Wang J., Li G., Elzo M. A., Yan L., Chen S., Jia X., et al. (2015). A novel single nucleotide polymorphism of the POU1F1 gene associated with meat quality traits in rabbits. Ann. Anim. Sci. 15, 611–620. doi: 10.1515/aoas-2015-0015
Wang D., Xie K., Wang Y., Hu J., Li W., Yang A., et al. (2022). Cost-effectively dissecting the genetic architecture of complex wool traits in rabbits by low-coverage sequencing. Genet. Selection Evol. 54, 75. doi: 10.1186/s12711-022-00766-y
Wongnaa C. A., Afful-Kwadam K., Asempah M. K., Hagan M. A. S., Awunyo-Vitor D. (2023). Is it profitable and viable to invest in commercialization of rabbit production? Implication on rural enterprise development. Sustain. Technol. Entrepreneurship 2, 100048. doi: 10.1016/j.stae.2023.100048
Wu Z., Chen S., Jia X., Lai S. (2015). Association of a synonymous mutation of the PGAM2 gene and growth traits in rabbits. doi: 10.17221/8079-CJAS
Xiccato G., Trocino A. (2020). Energy and protein metabolism and requirementsNutrition of the Rabbit (CAB International Wallingford UK), 89–125. doi: 10.1079/9781845936693.0083
Xie K., Ning C., Yang A., Zhang Q., Wang D., Fan X. (2024). Resequencing Analyses Revealed Genetic Diversity and Selection Signatures during Rabbit Breeding and Improvement. Genes 15, 433. doi: 10.3390/genes15040433
Yang X., Deng F., Wu Z., Chen S.-Y., Shi Y., Jia X., et al. (2020). A genome-wide association study identifying genetic variants associated with growth, carcass and meat quality traits in rabbits. Animals 10, 1068. doi: 10.3390/ani10061068
Yang Y., Zhang G.-W., Chen S.-Y., Peng J., Lai S.-J. (2013). Polymorphism of NLRP3 gene and association with susceptibility to digestive disorders in rabbit. Asian-Australasian J. Anim. Sci. 26, 455. doi: 10.5713/ajas.2012.12522
Zhang K., Wang G., Wang L., Wen B., Fu X., Liu N., et al. (2023). A genome-wide association study of coat color in Chinese Rex rabbits. Front. Veterinary Sci. 10. doi: 10.3389/fvets.2023.1184764
Keywords: precise nutrition, genomic assisted selection, rabbit, breeding, animal welfare
Citation: Goswami N, Solomon Ahamba I, Kinkpe L, Mujtaba Shah A, Xiangyang Y, Song B, Dong X, Wang S and Ren Z (2025) Enhancing rabbit farming efficiency with integrated genomics and nutritional strategies. Front. Anim. Sci. 5:1514923. doi: 10.3389/fanim.2024.1514923
Received: 21 October 2024; Accepted: 17 December 2024;
Published: 17 January 2025.
Edited by:
Joao Hc Costa, University of Vermont, United StatesReviewed by:
Martina Miluchová, Slovak University of Agriculture, SlovakiaCopyright © 2025 Goswami, Solomon Ahamba, Kinkpe, Mujtaba Shah, Xiangyang, Song, Dong, Wang and Ren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhanjun Ren, cmVuemhhbmp1bkBud2FmdS5lZHUuY24=; Bing Song, YmluZ3NvbmdAbndhZnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.