- 1College of Animal Science and Technology, Hebei Agricultural University, Baoding, China
- 2Baoding Xingrui Agriculture and Animal Husbandry Development Co., Ltd., Baoding, China
- 3Hebei Animal Husbandry and Veterinary Research Institute, Baoding, China
- 4College of Animal Science and Technology, China Agricultural University, Beijing, China
Eggshell pores can be subdivided into micrometer-scale gas pores, submicroscopic bubble pores, and nanoscale mesopores. All are important indicators of eggshell quality ensuring gas exchange between the inside and outside of the eggshell and preventing invasion by external bacteria. Although previous studies on eggshell pores focused on gas pores, recent studies have shown that bubble pores may play an more important role in regulating gas exchange. In order to investigated the relationship between gas and bubble pores and the mechanisms of genetic regulation. In this study, 40-week-old Brown-Egg Dwarf Layers (DWL) eggs were selected, and the quantity of gas pores (QGP), quantity of mammillary (QM), and quantity of bubble pores (QBP), area sum of bubble pores (ASBP), and other bubble pore-related indexes were determined. The correlation between each index was calculated, and genome-wide association analysis (GWAS) was performed based on whole genome resequencing (WGR). The results showed that the CVs of QGP and QM were 15.69% and 15.49%, respectively, and the CVs of the related pore indices, such as QBP and ASBP, were 29.22%-44.82%. The correlation coefficient between QGP and QM was 0.59 (P< 0.01), and there was no correlation between QGP, QM, and the bubble pore-related indicators (P > 0.05). These results above suggest that the gas and bubble pores may be two independent pore systems. A total of 32 single nucleotide polymorphisms (SNPs) associated with the suggestively significant level of bubble pore correlation indexes were detected in GWAS, and the corresponding genes were ANXA10, CDH10, AADAT, RXFP1, FNIP2, DDX60, PCDH10, RAPGEF2, FSTL5 and SPOCK3. KEGG enrichment analysis showed that these genes were mainly expressed in the calcium ion binding pathway, indicating that the genes and pathways may play a regulatory role in forming bubble pores during eggshell calcification. This study provides a basis for revealing the genetic regulatory mechanism of eggshell pores and a reference and direction for further improvement in eggshell quality.
Introduction
Eggshells contain approximately 95.0% calcium carbonate, 3.5% organic matrix (Nys et al., 2004), and small amounts of magnesium carbonate and calcium phosphate (Rama et al., 2007), which are formed in the uterus of chickens. The biomineralization process involves transforming amorphous calcium carbonate to calcite crystals (Li et al., 2018). Eggshells can be divided into mammillary layers (ML), palisade layers (PL), and vertical crystalline layers (VCL) based on differences in the morphology of the calcite crystals (Perrott et al., 1981). These mineralized layers contain different types of pores (Sara et al., 2021), which are classified into three categories based on size: gas pores at the micro-scale, bubble pores at the sub-micrometer scale, and nanoscale mesopores (Zhou et al., 2011). Among them, the most extensive and intensive studies have been conducted on gas pores, with an average pore size of 10-20 µm, and an average density of 1.35/mm2, and are approximately twice as numerous at the blunt end as sharp end (Riley et al., 2014). Most penetrate through all mineralized layers of the eggshell (Sara et al., 2021). There are relatively few studies on bubble pores. Riley et al (Riley et al., 2014 visualized the size of bubble pores by X-ray micro-computed tomography, and concluded that most of the bubble pores had a pore size of approximately 250 nm, and the total void occupied approximately 1/3 of the total volume of the mineralized layer. With regard to the distribution pattern, the quantity of bubble pores (QBP) was relatively uniformly distributed at both ends of the eggshell. Still, there were large differences between the different mineralized layers, with PL having the most, followed by ML, with VCL having the least (Arzate-Vázquez et al., 2019). Mesopores are interstitial voids located between the VCL and cuticle, with dimensions of less than 10 nm (Zhou et al., 2011). The formation of gas pores was mainly caused by incomplete fusion of the mammillary interstices and columnar extension of the PL during the early stages of eggshell formation (Tullett, 1975). However, the cause of bubble pore formation has not been reported, whereas similarly structured reticulated pore systems have been reported in eggshell studies of other species, such as rhea, emu, and various birds of prey (Tyler, 1957; Board and Tulett, 1975), suggesting that bubble pores are a widespread type of pore.
Eggshells and their special pore structures combine during the incubation of laying hen embryos to ensure gas exchange between the inside and outside of the shell (Arzate-Vázquez et al., 2019) and to prevent the invasion of external bacteria into the embryo (Olivier et al., 2008a). Gas pores were long considered to dominate eggshell gas exchange (Tullett, 1984) jointly; however, Zhou et al (Zhou et al., 2011 showed that bubble pores mainly determined the rate of eggshell gas conduction through statistical calculations of bubble pore size and gas conduction experiments. At the physical level, bacterial invasion was inhibited by, the outermost epidermal layer (Sparks and Board, 1984; Chavez et al., 2002) and nano-sized mesopores, and the VCL (Olivier et al., 2008b; Zhou et al., 2011). These inhibit microorganism and bacterial invasion by decreasing the number of pores in the eggshell. At the chemical level, C-type lysozymes in the cuticle are solubilized and released during microbial contamination to minimize damage (Olivier et al., 2008a). Ovocalyxin-36 in eggshell membranes dissolves mammillary calcium reserves to provide antimicrobial protection (Gautron et al., 2007).
Avian eggshell formation is influenced by various environmental, nutritional, and genetic factors (Roberts, 2004). No eggshell gas or bubble pore QTL has been identified in published articles or multiple databases. However, many traits of eggshell quality are largely determined by genetic factors. For example, in an F2 population of hens derived from crosses between the standard breed White Leghorn (WL) and the Chinese indigenous strain of Dongxiang (DX) chickens, both eggshell thickness (0.21 to 0.31) and strength (0.20 to 0.27) were moderately heritable at 32-72 weeks of age, and ITPR2, PIK3C2G, and NCAPG were the three most promising loci for eggshell quality (Sun et al., 2015). The quantity of mammillary (QM) had low heritability (0.19) in the 66-week-old population of F2 hens, and searched for the KNDC1 gene, which may regulate QM, in a genome-wide association analysis (GWAS) study (Duan et al., 2016). The single nucleotide polymorphism (SNP)-based heritabilities of crystal total integral intensity and degree of orientation were 0.23 and 0.06, respectively, and searched for six genes in GWAS, such as PLCZ1, ABCC9, and ITPR2, which may be the key genes for the total crystal integral intensity (Li et al., 2021).
Whole genome resequencing (WGR) is a highly efficient method for screening genes regulated by complex traits (Parveen et al., 2020), and the first published genome sequence of Red Junglefowl by Hillier et al (Hillier et al., 2004 greatly contributed to the progress of multifaceted research at the genome level of the domestic chicken. Whole genome resequencing technology has been widely applied to chicken performance enhancement and important advances have been made. For example, Rubin et al (Rubin et al., 2010 resequenced the genomes of chickens representing eight different domestic chicken populations and the Red Junglefowl, identifying more than 7million SNPs and nearly 1300 segmental base deletions, upon which to hypothesize the evolutionary direction of broilers and egg-laying chickens. Wang et al (Wang et al., 2015 found that AATF, CYBB, FOXM1, and BCDO2 were involved in the ROS process; thus, in the regulation of Ca2+ concentration and hypoxia response, revealing the regulatory mechanism of high-altitude hypoxia acclimatization in Tibetan chickens through whole-genome sequencing of Tibetan chickens, village chickens, pheasants and Red Junglefowl.
Since pores form during the calcification of eggshells, it is likely that genetic factors may influence their formation. Therefore, this study aimed to analyze the genome-wide association of Brown-Egg Dwarf Layer (DWL) eggshell pores using WGR technology to reveal the genetic structure of eggshell pores and identify candidate gene loci that could provide a reference for the genetic regulation of eggshell biomineralization.
Materials and methods
Experimental hens
Approximately 1,000 healthy, 40-week-old DWL hens of genotype ZdwW (Ning, 2004) from Hebei Rongde Poultry Breeding Co., Ltd. (Hengshui, China) were selected for the experiment. During the experiment, the pure line was bred until the 7th generation. From 18 weeks of age to the end of the experiment, all laying hens were kept in the same fully enclosed chicken house, individual single cages, under the same light program (16L:8D), and feed and water were maintained during the experiment. On the experiment day when the hens were 40 weeks old, 80 hens were randomly selected to lay eggs on the same day (unbroken, clean and smooth eggs), and the eggs were collected in a one-chicken-one-egg fashion to be used in the next step of the experiment.
Eggshell sample preparation
The eggs were broken at the middle end, the contents were poured out, and the egg whites attached to the eggshells were washed with deionized water. Eggshell pieces of approximately 1.0 × 1.0 cm2 were taken from the blunt, middle, and sharp ends for the quantity of gas pores (QGP) and QM measurement. Another three pieces of eggshell were taken for bubble pore-related index measurement. The QGP and QM measurements were performed: boiled eggshells were placed in 1% NaOH solution (Mreda Technology Co Ltd., Beijing, China) for 15 min. An optical microscope (RX-45B1, RuiXian Optical Instrument Co Ltd., Dongguan, China) was used to observe the gas pores and mammilla to confirm that the fibrous membrane on the inner surface of the eggshell was removed and that the mammilla could be observed. The eggshells were washed with deionized water and soaked in 1% HCl solution (Nanjing Chemical Reagent Co Ltd., Nanjing, China) for 30 s to enlarge the gas pores. The inner surface of the eggshell was washed again with deionized water and left to dry. The eggshells were stained with 0.1% methylene blue (Solarbio Science & Technology Co Ltd., Beijing, China) and allowed to stand for 10 min. After the penetration of methylene blue into the outer surface of the eggshell, an optical microscope was used to observe the QGP per unit area (0.5 × 0.5 cm2) of the outer eggshell surface (Mehlum et al., 1987).
Eggshells sized 0.3 × 0.3 cm2 were taken from the three sections of the eggshells, and a cross-section of the eggshells was measured for bubble pores. The eggshell samples were placed on 12.5 mm diameter sample trays (Rigorous Technology Co Ltd., Shenzhen, China) using electrically conductive adhesive tape (Precise Trading Co Ltd., Shenzhen, China). Gold was sprayed thrice for 15 s each using an ion-sputtering instrument (Sputter Coater 108, Cressington Scientific Instruments Ltd., Watford, UK). Subsequently, the bubble pores in the middle of the three layers of the eggshell, namely the ML, PL, and VCL, were photographed using a scanning electron microscope (Prisma E, Thermo Scientific., Massachusetts, USA) at 10,000 ×. Photoshop (Photoshop CC 2018, Adobe Systems Corp., San Jose, CA, USA), Ipwin32 (version 1.41, National Institutes of Health, MD), and ImageJ-Win64 (version 6.0, Media Cybernetics Corp., Silver Spring, MD) were used to aligned QBP, as shown in Figure 1. The average area of bubble pores (AABP), area sum of bubble pores (ASBP), total perimeter of bubble pores (TPBP), area ratio of bubble pores (ARBE), and the detailed procedure was based on the methods of Wang et al (Wang et al., 2017). To photograph the inner surface of the eggshell at 200 × condition QM, statistical methods were used as described by Duan et al (Duan et al., 2016).
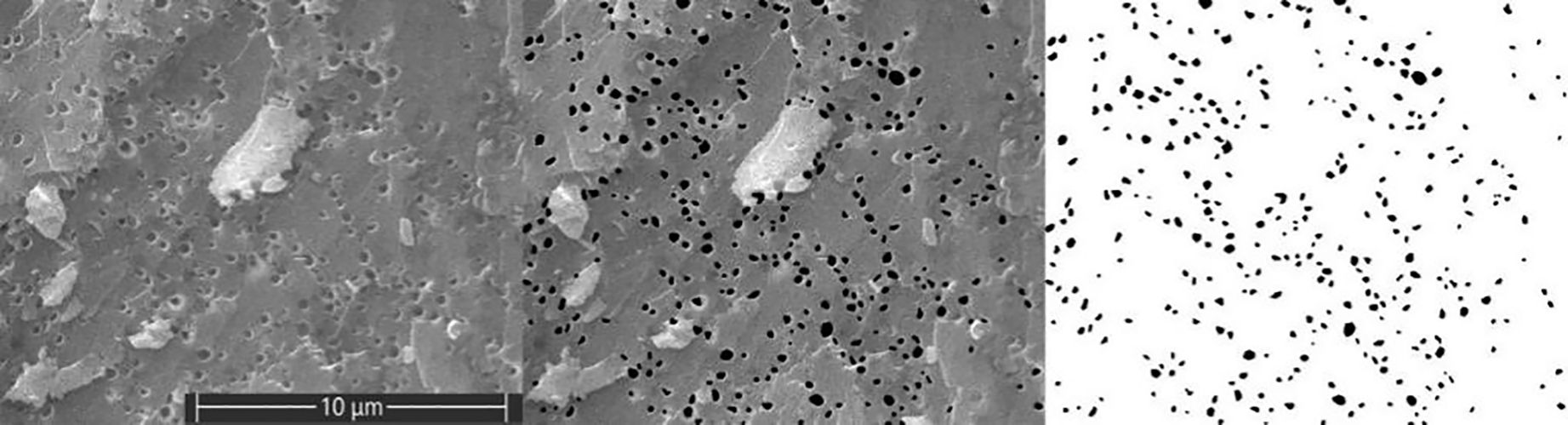
Figure 1. Measurements of bubble pores from cross-sectional of eggshell. From left to right, the first image shows the bubble pores at 10,000 × SEM, the second image is a Photoshopped image to increase the color contrast between the bubble pores and the surrounding eggshell area, and the third image shows the bubble pores measured using Ipwin-32 software.
DNA extraction and inspection
Blood was collected from 80 DWL corresponding to eggshell pore indexes, DNA was extracted using a genomic DNA extraction kit (Tiangen Biotech Co Ltd., Beijing, China), and the quality of the samples was checked using a 721G spectrophotometer (INESA Analytical Instrument Co. Ltd., Shanghai, China) to ensure that the OD260/OD280 values of the samples were within the range of 1.8-2.0.
Whole genome resequencing
After DNA shearing, adapter ligation, PCR product selection and other operations to construct a 300-350 bp DNA library, we then sequenced the blood samples using the Illumina HiSeq PE150 platform (San Diego, CA, USA). The raw reads were filtered using the Fastp v0.23.1 (Chen et al., 2018) quality control filtering criteria, which were as follows: (1) removal of splice sequences; (2) removal of reads with N (non-AGCT) bases greater than or equal to 5; (3) sliding window with 4 bases as the size of windows, removing the average base mass value less than 20; and (4) reads with lengths less than 75 bp or average base mass value less than 15bp. The clean reads obtained were subsequently randomly aligned to the Red Junglefowl GRCg6a version of the reference genome using BWA v0.7.17 (Bolger et al., 2014), with an average alignment rate of 99.58% and an average alignment depth of 11.74 ×. The results were reordered according to the chromosome order using SAMtools v1.10 (Li et al., 2009), Picard’s MarkDuplicates module was used to remove duplicate reads (https://broadinstitute.github.io/picard/), and analysis results of the aligned genome results, such as the GC content were obtained by Qualimap v2.3 (Okonechnikov et al., 2015). The sequencing data were all within the normal range. GATK v4.0.12 (Aaron et al., 2010) was screened for SNPs across the genome in the following steps: (1) the filtering parameters were QD< 2.0 || MQ< 40.0 || FS > 60.0 || SOR > 3.0 || MQRankSum< -12.5 || ReadPosRankSum< -8.0; (2) more than 20% of the individuals were of the deletion genotype (./.) are removed; (3) SNP loci are generally dichotomous genotypes, so we filter out loci with more than two completely different genotypes; (4) Minor allele frequency (MAF) is the proportion of alleles with a low number of occurrences in the whole population, and we remove SNP loci with an MAF less than 0.05. Finally, we obtained a total of 9,084,960 SNP loci. Gene annotation results were obtained by ANNOVAR (Wang et al., 2010) calculation. Resequenced clean reads were deposited in the National Center for Biotechnology Information BioProject database (https://www.ncbi.nlm.nih.gov/bioproject) under accession number PRJNA1016067.
Phenotype analysis
Phenotypic correlates of pore traits, such as QGP, QM, and QBP, were analyzed using SPSS (IBM SPSS Statistics version 26.0, Armonk, NY), using Pearson correlation analysis, with correlation significance thresholds set at P< 0.05, and extreme significance thresholds set at P< 0.01.
Population structure
Genome-wide SNPs were filtered to eliminate false-positive statistical results because of population stratification using PLINK v1.9 (Purcell et al., 2007) with the parameter –indep–pairwise 50 5 0.5, and 970,235 SNPs that were not tightly linked were selected. The identical by state (IBS) matrix was calculated using Plink, and then neighbor-joining in PHYLIP v3.69 (Plotree and Plotgram, 1989) was used to construct phylogenetic trees for all samples. Subsequently, PCA analysis was performed using Eigensoft v7.2.1 (Price et al., 2006) to extract principal components for mapping.
Genome-wide association analysis
A mixed linear model (MLM) was used for the association analysis of eggshell pore traits. To increase the accuracy of the results, the population genetic structure and individual kinship matrix were added to the model. The following model was used:
y: phenotypic traits, X: indicator matrix for fixed effects, α: estimated parameters for fixed effects, Z: indicator matrix for SNPs, β: effect of SNPs; W: indicator matrix for random effects, μ: the additive polygenic effect (0, GVg), with G the genomic kinship matrix, and the additive effect variance Vg; e: entourage residuals, obeying e ~ (0, δe2).
The Kinship matrix was calculated using EMMAX v0.94.1 (Zhou and Stephens, 2012), and the first four PCA components were selected as covariates to correlate the large number of variance loci with the experimental objective traits. Manhattan plots and quantile-quantile (Q-Q) plots of the GWAS results were plotted using the “gap” package in R v4.1.3 (Zhou and Stephens, 2012). The genome-wide suggestive and significant P-values were 1.1 × 10-7 (1/9,084,960) and 5.5 × 10-9 (0.05/9,084,960), respectively, determined by Bonferroni correction.
Functional annotation and linkage disequilibrium analysis
Based on the significant SNP loci for each trait, Linkage Disequilibrium (LD) analysis was performed using the solid spine algorithm by Haploview v4.2 (Barrett et al., 2005) to detect regions that may be linked to the significant loci. Genes within 50 kb upstream and downstream of the potentially linked loci were collected based on the Ensembl chicken genome annotation file and analyzed for significant SNPs for gene GO annotation and KEGG enrichment using Metascape (Zhou et al., 2019).
Results and discussion
Phenotypic characteristics
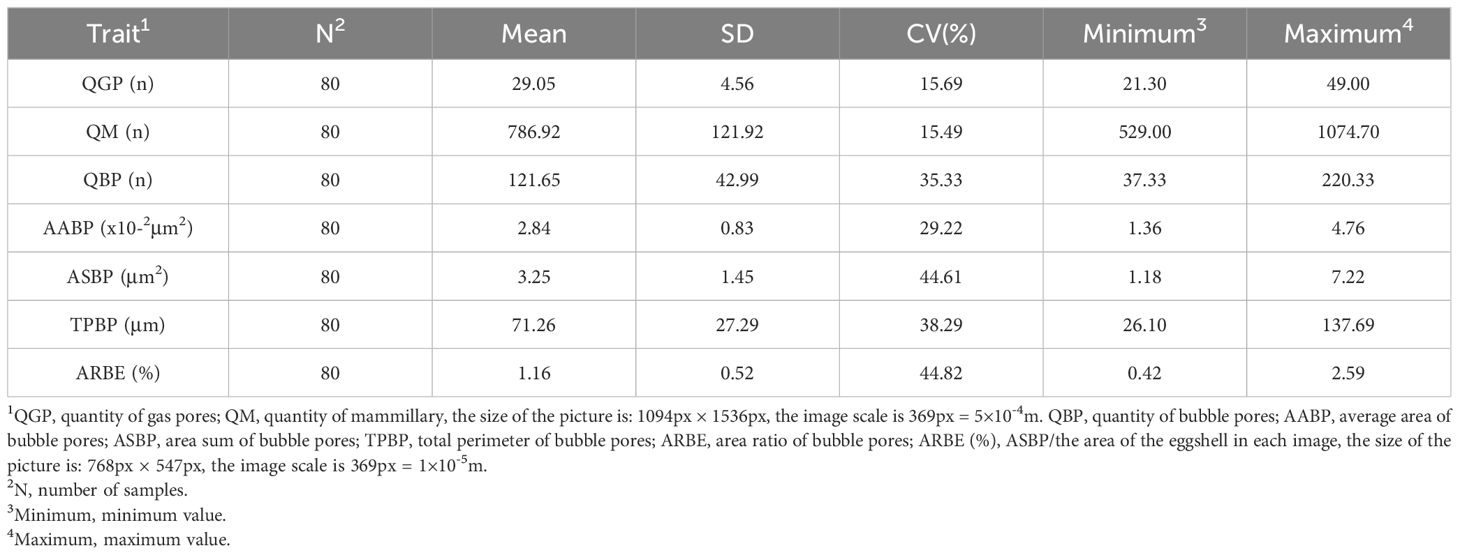
Descriptive results of the eggshell pore-related metrics are shown in Table 1. The CV values of QGP and QM were 15.69% and 15.49%, respectively, indicating that the degree of trait dispersion was low. The overall value was low, which is consistent with the QGP in the 42-week-old WL population in a previous study by Wang et al (Wang et al., 2017 and with the QM in the 45-week-old Lohmann population in the study by Cristina et al (Benavides-Reyes et al., 2021). Furthermore, QGP mean values were generally consistent with the 72-week-old all pink-shell-laying hens populations of Lin et al (Lin et al., 2023), and the QM mean values were generally consistent with the results of a 30-week-old Hyline variety brown population in the study by Parket et al (Park and Sohn, 2018), indicating that measurements of this type of indicator are relatively stable. The mean value of QBP in this experiment was lower than the QBP measurements of the WL population eggshell palisade layer in Wang et al (Wang et al., 2017). Usually, the QBP of ML and VCL is significantly lower than that of PL (Arzate-Vázquez et al., 2019), whereas the QBP in this study was the mean value of ML, PL, and VCL; thus, it also lowered the measurements of the corresponding ASBP, ARBE, and TPBP, which also explained why the QBP, AABP, and other bubble pore-related metrics 29.22-44.82% of the data variability was greater relative to QGP and QM.
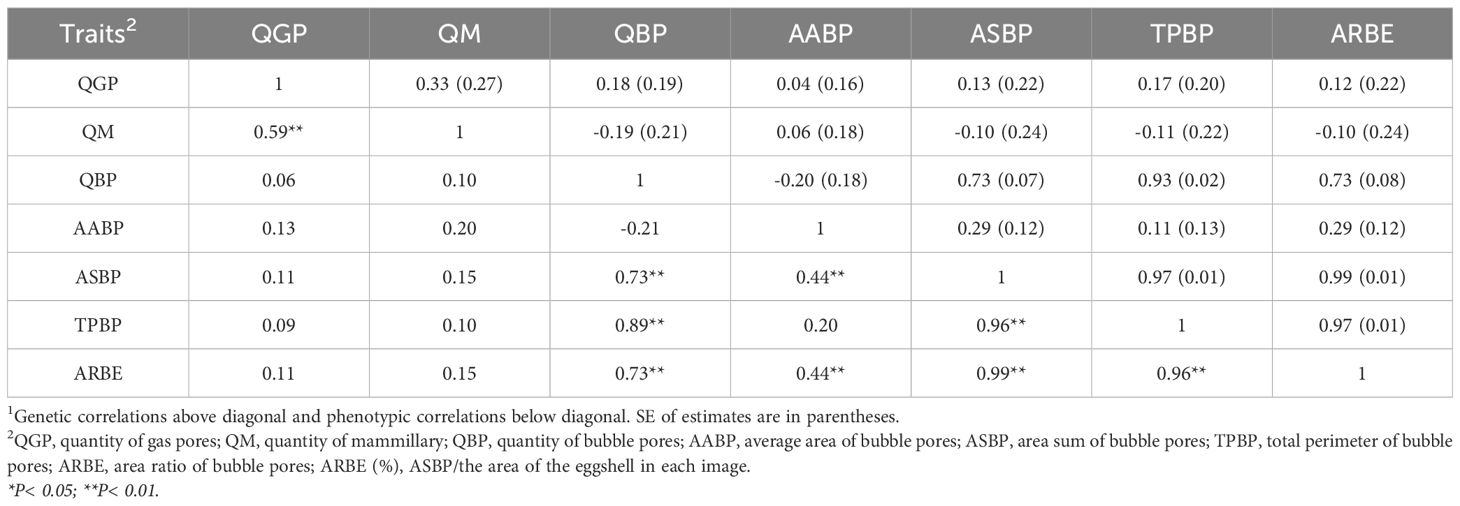
The phenotypic correlations of eggshell pore-related indicators are shown in Table 2. The genetic correlation between QGP and QM was (0.33) and the phenotypic correlation coefficient between QGP and QM was 0.59 (P< 0.01), which was consistent with the positive correlation between the two indexes (0.918) in Tullett’s studies (Tullett, 1975; Tullett, 1984). During eggshell formation, the inward gaps among the mammillary units correspond to the junction of the bottom of the mammilla and the PL to form gas pore channels; thus, an increase in QM per unit area increases the effective QGP, improving the ability to exchange gas between the inside and outside of the eggshell. There was no phenotypic correlation among QGP, QM, and QBP, AABP, and other bubble pore-related indicators(P > 0.05), and the genetic correlations between both QGP and QM and the indicators related to the bubble pores were low at (0.04-0.18) and (-0.19-0.06), respectively. Combined with the characteristics that gas pores are distributed throughout the eggshell mineralized layer (diameter between 10-20 µm) (Zhou et al., 2011) and bubble pores are diffusely distributed throughout the eggshell mineralized layer (diameter of about 0.25 µm) (Riley et al., 2014), it is speculated that the QGP and the QBP may be two separate pore systems. which suggests that the formation mechanisms of gas and bubble pores may differ completely. The phenotypic correlation coefficients between QBP and ASBP, TPBP and ARBE were 0.73-0.89 (P< 0.01) and the genetic correlation coefficients were (0.73-0.93), while the phenotypic correlation coefficients between AABP and ASBP and ARBE were both 0.44 (P< 0. 01) and genetic correlation coefficients were 0.29. However, there was no correlation between QBP and AABP, which indicated that QBP and AABP were independent, and that the variation in eggshell ASBP was more predominant because of the variation of QBP and that in AABP plays a secondary role.
Whole genome resequencing and population structure
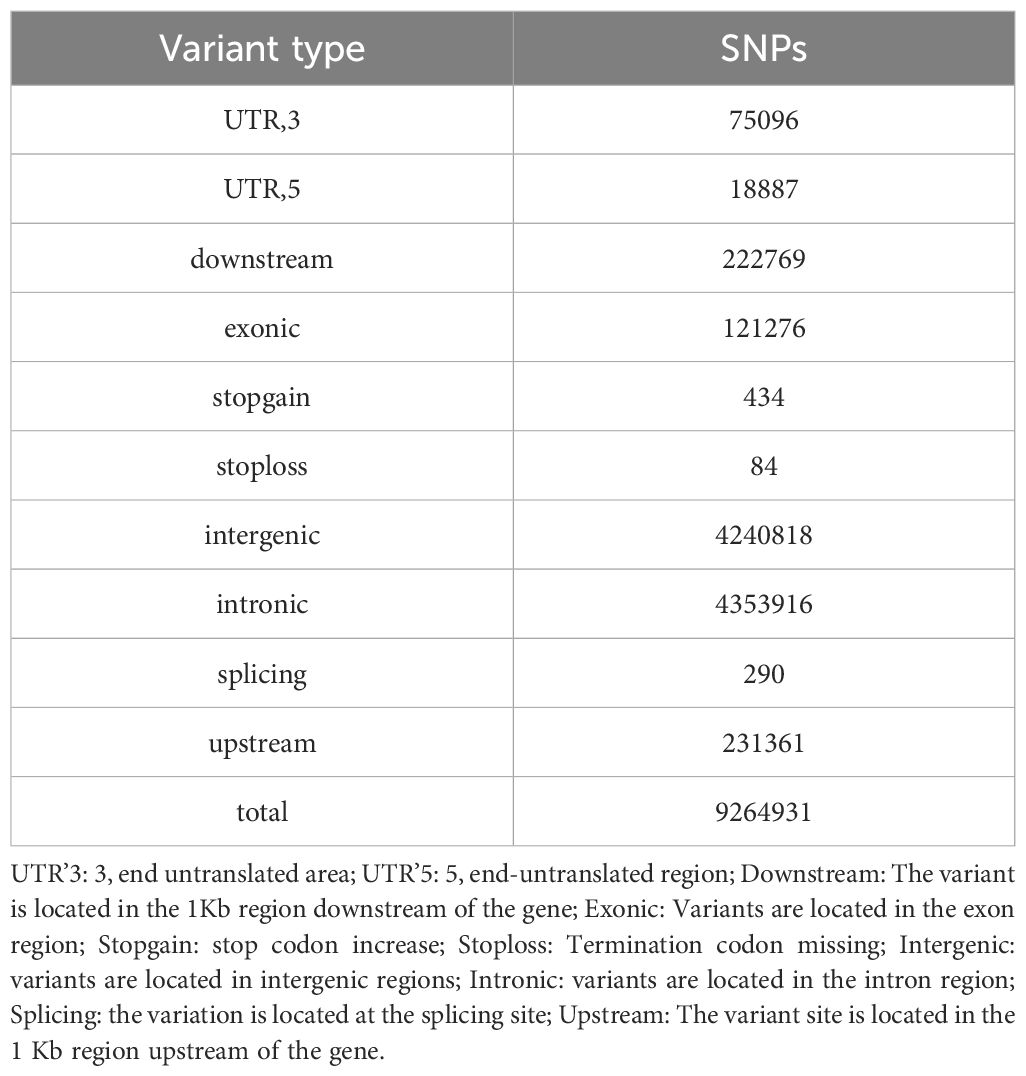
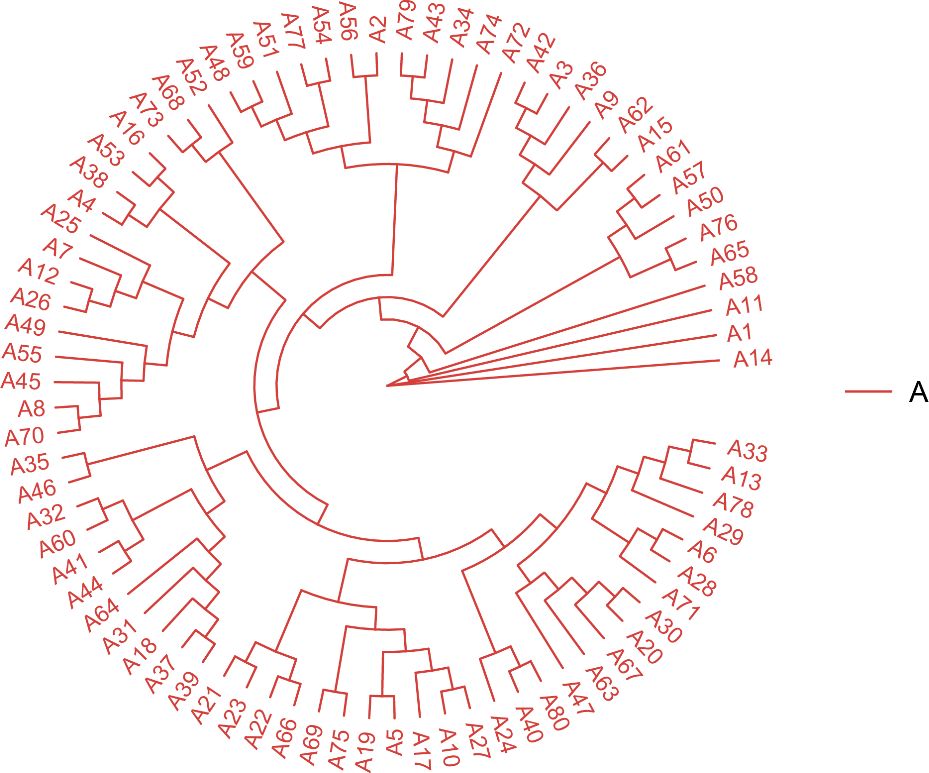
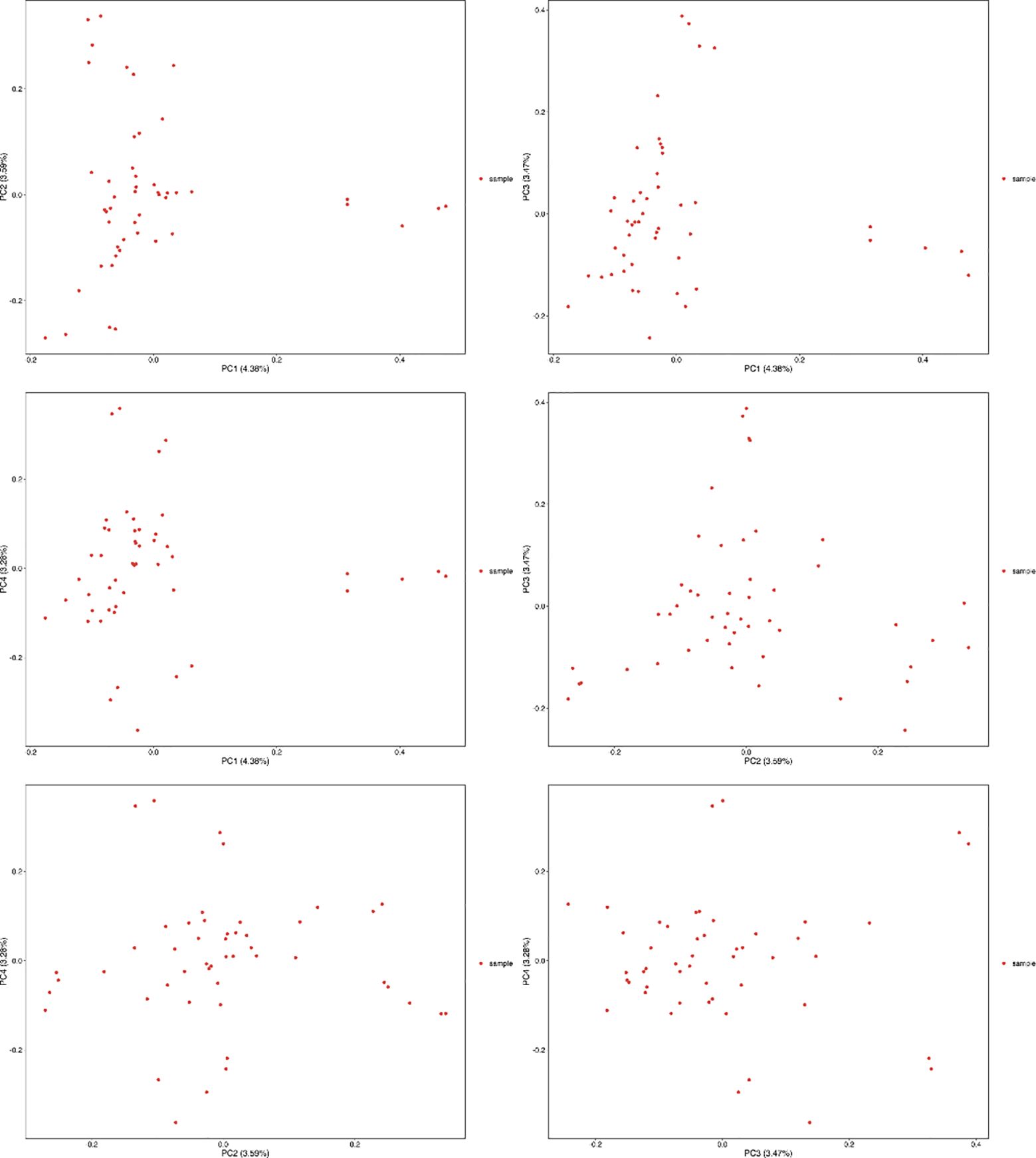
The resequencing results are shown in Table 3, where a total of 1020.16 Gb raw reads were obtained after sequencing and a total of 1017.04 Gb clean reads were obtained after quality control. Population SNPs specific annotation distribution is shown in Table 4, and a total of 9,264,931 SNP sites were obtained. The phylogenetic tree for all individuals is shown in Figure 2 which showed that there may not be significant genetic distances present in the populations. PCA plotted the first four principal components for all individuals as shown in Figure 3, where the first four principal components contributed 4.38%, 3.59%, 3.47%, and 3.28% of the genetic variance, respectively. The results indicated no obvious subpopulation differentiation within the group, and the populations were more tightly clustered. The results of the phylogenetic tree of population and principal component analysis analyses coincided, suggesting that they came from the same group and were suitable for the subsequent GWA analysis.
Genome-wide association study
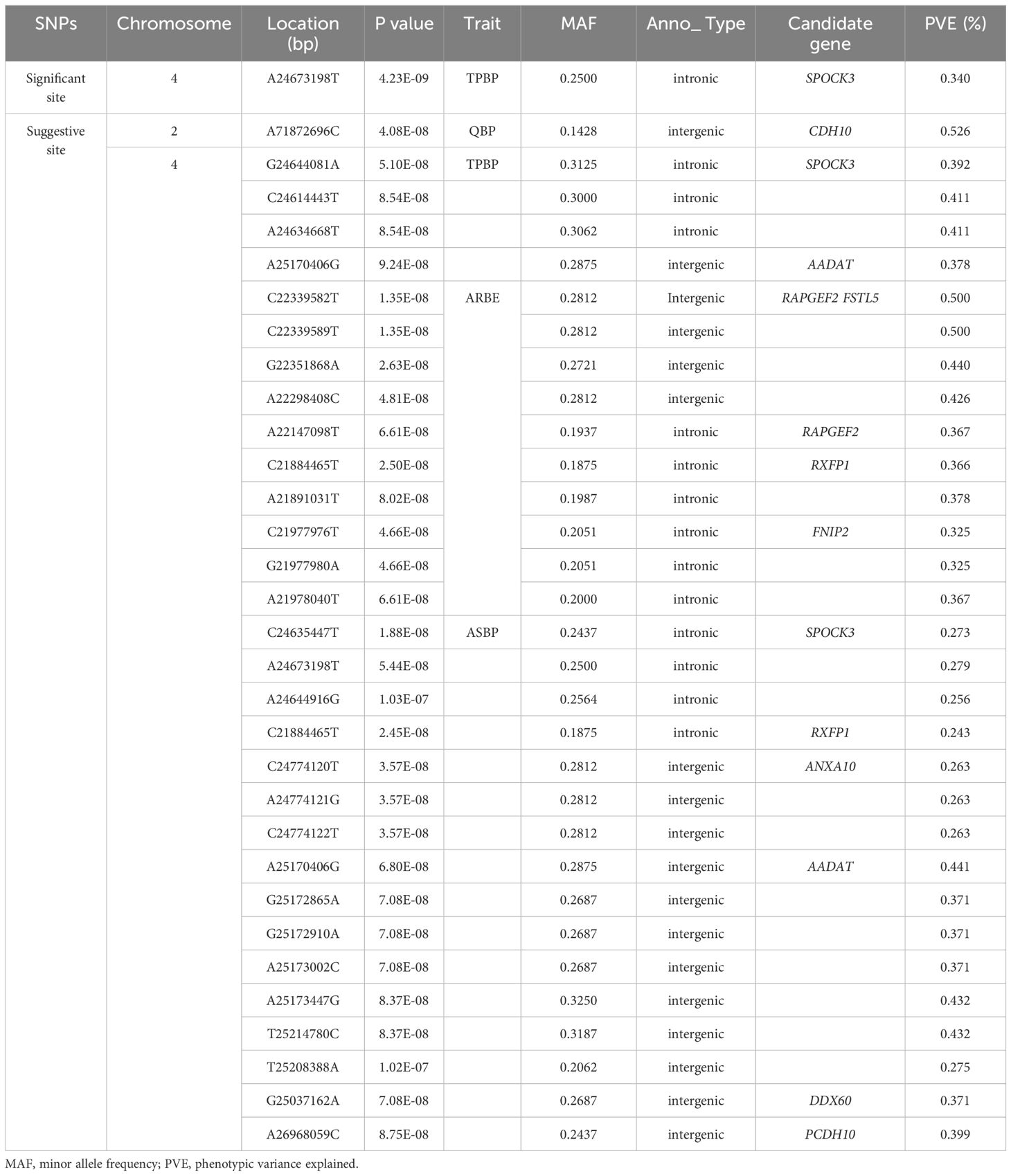
Manhattan and Q-Q plots of the 7 eggshell pore-related metrics are shown in Figure 4, and a short list of significant SNPs detected by the GWAS is shown in Table 5. In this experiment, GWAS detected 31 SNPs above the threshold of suggestive significance on chromosome 4 (GGA4), including ASBP (Olivier et al., 2008b), TPBP (Sara et al., 2021), and ARBE (Tyler, 1957), and one SNP in TPBP above the threshold of significance. Detection of one QBP-associated SNP was above the suggested threshold for GGA2. The GWAS in our study did not identify significant SNPs in the QM and QGP metrics, which may be caused by the small phenotypic variance of QM and QGP and Duan et al (Duan et al., 2016 estimated the genetic parameter of QM to be 0.19, which indicates low heritability. This suggests that the mechanism of QM formation is controlled by microefficiency genes, making it difficult to detect SNPs, as in the case of QGP.
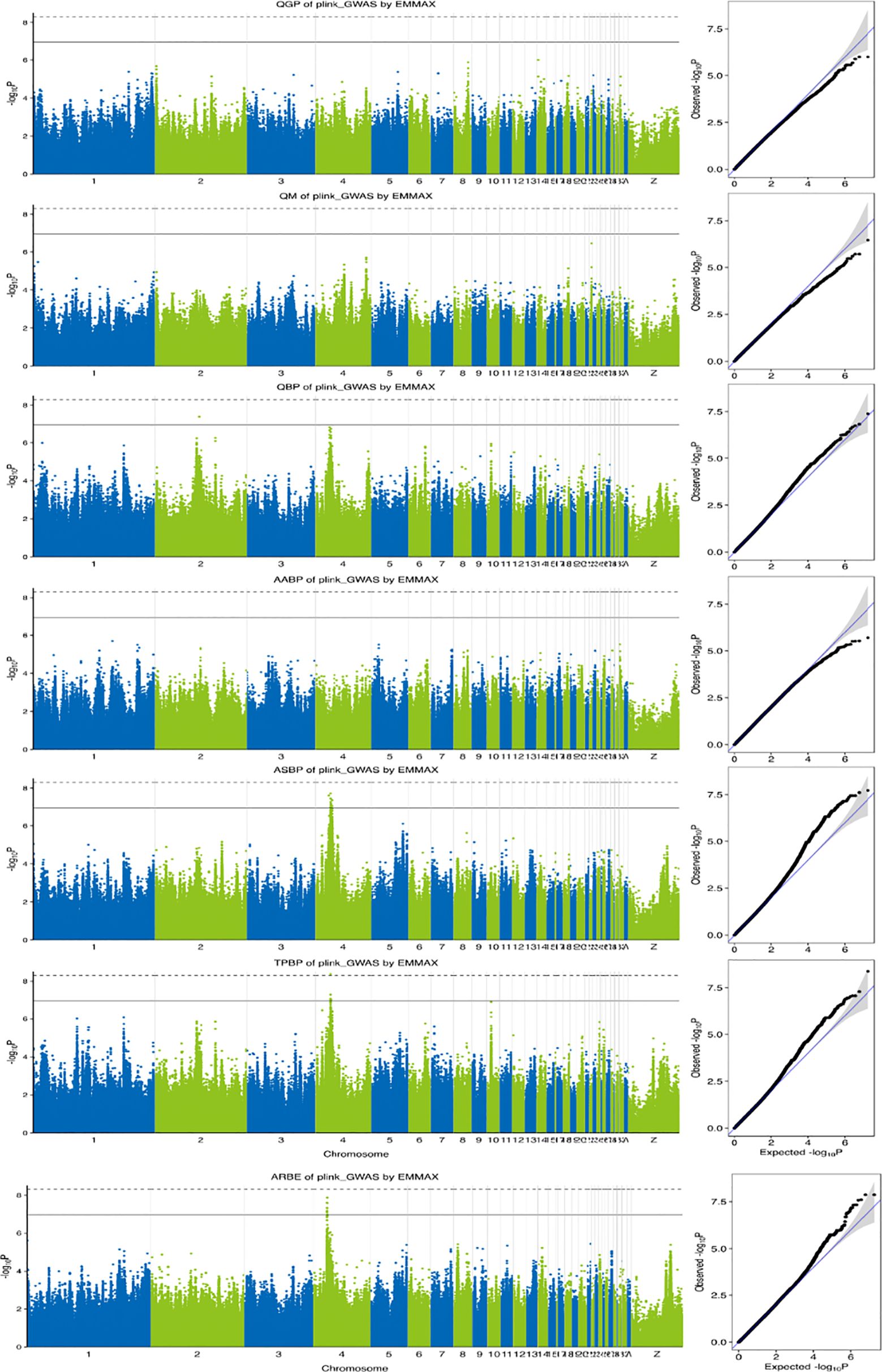
Figure 4. Manhattan plots and quantile–quantile plots of the observed P-values for pore-related indicators. The Manhattan plots indicate -log10 (observed P-values) for genome-wide SNPs (y-axis) plotted against their respective positions on each chromosome (x-axis). And the horizontal solid and dashed lines corresponding to suggestive (1.1×10−7) and significant (5.5×10−9) thresholds, respectively. For quantile-quantile plots, the x-axis shows the expected -log10-transformed P-values, and the y-axis represents the observed -log10-transformed P-values. QGP, quantity of gas pores; QM, quantity of mammillary; QBP, quantity of bubble pores; AABP, average area of bubble pores; ASBP, area sum of bubble pores; TPBP, total perimeter of bubble pores; ARBE, area ratio of bubble pores.
For QBP, only one suggestive significant SNP was found and located in the cadherin 10 (CDH10) intergenic region on GGA2. CDH10 was one of the first adhesion molecules described (Takeichi, 1990), and calmodulin molecules interact with each other through the formation of motifs in a highly flexible manner at their tips, which plays an important role in the treatment of cancers (Cavallaro and Christofori, 2004). Therefore, this gene may be an important indicator of QBP.
For TPBP, a total of five SNPs above the threshold of suggestive significance were detected on GGA4, four SNPs were located on the intronics of SPARC (osteonectin), cwcv, and kazal-like domain proteoglycan 3 (SPOCK3). One was detected above the significance level with a P value of 4.23E-09, and one SNP was located in the intergenic region of aminoadipate aminotransferase (AADAT). SPOCK3 belongs to the SPOCK family of highly conserved, multi-structural domain extracellular matrix glycoproteins, which are proteins that regulate the development of the central nervous system and include three basic structures: the FS structural domain, EC structural domain, and TY structural domain, whereas the EC structural domain has a strong affinity for calcium ions (Hartmann and Maurer, 2001). Although no relevant studies have been conducted on poultry, it may also be an important candidate gene as a TPBP indicator. AADAT was first identified in the rat liver (Nakatani et al., 1970) and where it degrades lysine (Higashino et al., 1971). In contrast, 70-75% of eggshell membranes required for precipitation attachment during eggshell calcification are other proteins and glycoproteins containing lysine-derived crosslinks (Leach, 1982; Zhao and Chi, 2009), so AADAT may also be an important candidate gene for TPBP.
For ASBP, 16 suggestive significant SNPs were detected on GGA4, among which the candidate genes SPOCK3 and AADAT were also detected; one SNP site each was found to be the same as that detected by the TPBP index. AADAT may influence lysine and eggshell membrane formation during eggshell formation, whereas the EC structural domain of SPOCK3 affects the binding of calcium ions during eggshell formation. Together, these regulate eggshell formation, thereby regulating the overall TPBP and ASBP metrics of the bubble pores. In addition, the candidate genes identified in ASBP were relaxin family peptide receptor 1 (RXFP1), Annexin A10 (ANXA10), DExD/H-box helicase 60 (DDX60), and protocadherin 10 (PCDH10). Relaxin is systemically and endocronically circulated in pregnant females (Bathgate et al., 2013), and the axial pathway between RXFP1 and relaxin formation has been identified (Jane and Espey, 1973; Bennett, 2009), and plays a role in promoting the growth and softening of the reproductive tract of pregnant females (Hyung-Yul et al., 2005). It has not been studied in avian species and may be an important candidate for ASBP indicators. ANXA10 belongs to the family of membrane-bound proteins, plays a role in apoptosis and calcium signaling (Mussunoor and Murray, 2008), is a prognostic biomarker and suppressor of hepatocellular carcinoma (Zhang et al., 2023), may be related to calcification during ovulation in laying hens and is an important candidate gene for ASBP. DDX60 is a DEAD-box RNA-deconjugating enzyme that promotes the RIG-I-like receptor-mediated signaling pathway and plays an important role in antiviral immunity (Moeko et al., 2011). It is also considered an important candidate gene. PCDH10 belongs to the same family of calmodulins as CDH10 and is found in the central portion of the visual system of chicken embryos during hatching. PCDH10 and CDH10 are partially expressed in the central part of the visual system during chick embryo hatching (Becker and Redies, 2003), are essential for neurodevelopment, and may be important candidate genes.
For ARBE, 10 suggestive significant SNPs were located on GGA4, and one SNP identical to the RXFP1 SNP locus of a candidate gene identified during association analysis with ASBP. The remaining candidate genes included rap guanine nucleotide exchange factor 2 (RAPGEF2), Follistatin like 5 (FSTL5), and folliculin interacting protein 2 (FNIP2). RapGEF2 is one of the many guanine nucleotide exchange factors (GEFs) that specifically activate Rap1 (Ras-proximate-1), plays a role in signaling pathways that control a variety of processes, including cell adhesion (Boettner and Aelst, 2009), and RapGEF2 been found to have an important positive effect on embryonic hematopoiesis in studies in mice (Ande et al., 2010). FSTL5 is an extracellular matrix-secreted protein involved in cell migration, proliferation, differentiation and organ development (Emanuel et al., 2009). It has been implicated in human studies as a possible new avenue for treating of hepatocellular carcinoma (Zhang et al., 2020). The liver plays an important role in egg production in laying hens, and the fat synthesized in the liver is transported via the bloodstream to the adipose tissue for storage or to the ovaries for egg production, and the detected FNIP2 gene has been found to have a positive role in lipid metabolism in chickens (Guo et al., 2021). We therefore suggest that the FNIP2 gene may be in some way linked to the liver influencing ovulation and thus pore traits in the eggshell. Therefore, RXFP1, RapGEF2, FSTL5, and FNIP2 may be interconnected and jointly regulate the production and supply of blood and fat, all of which may be important candidate genes.
GO and KEGG analyses
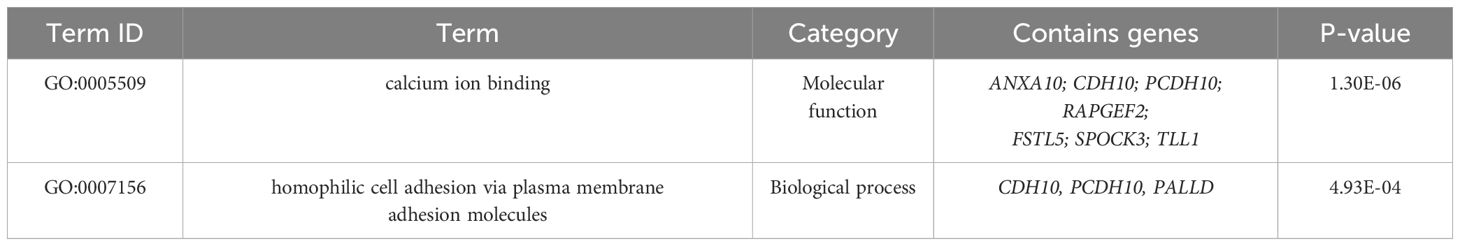
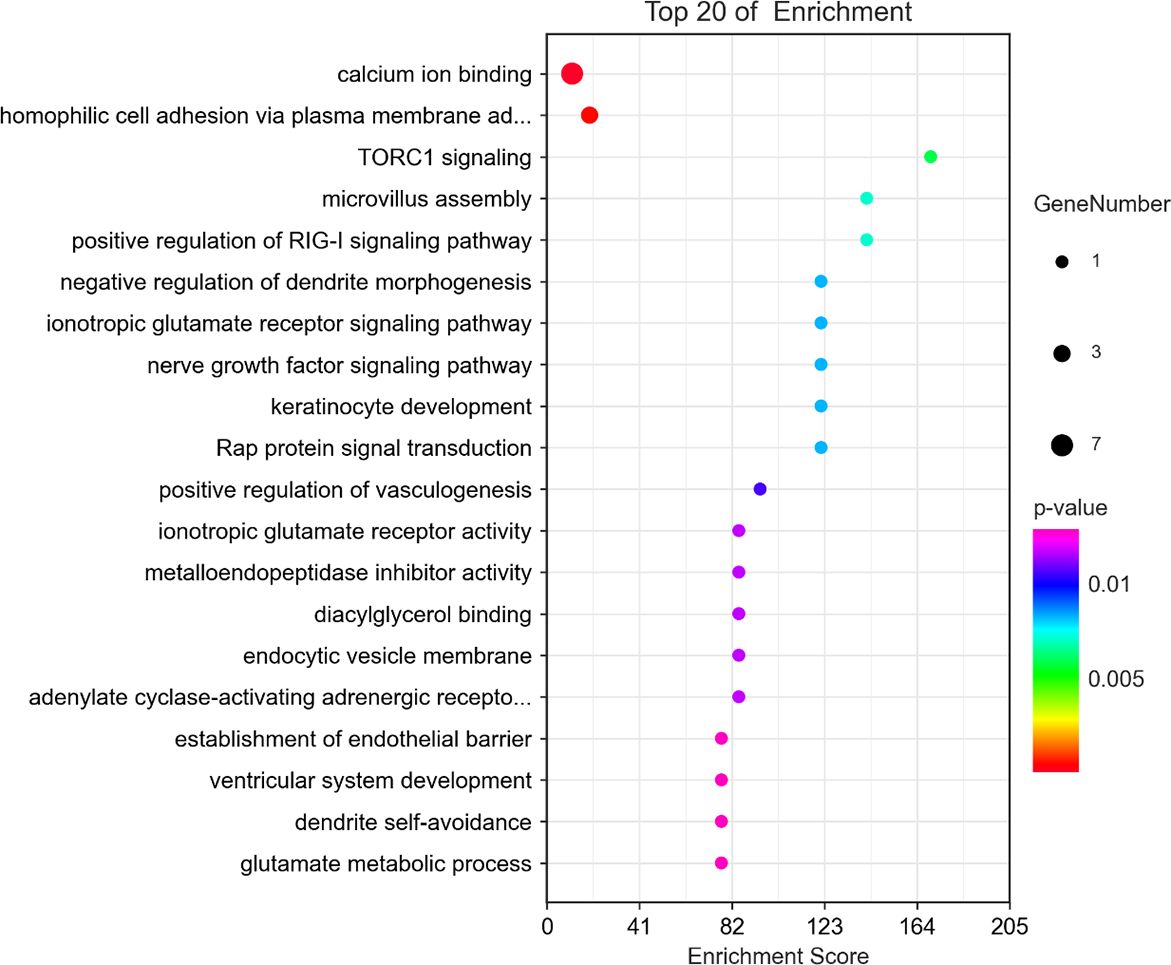
We performed GO and KEGG analyses on 32 significant SNPs in 23 genes in the anterior and posterior 50 kb regions. The GO analysis results are shown in Figure 5. GO terms were classified into three categories: biological processes (BP), cellular components (CC) and molecular functions (MF). There was a total of 35 GO terms, of which 17 were included in BP, five in MF, and 13 in CC; the top 10 terms of the three categories with P values are shown in Figure 6. There was one significant term for both MF and BP. The results of the KEGG pathway analysis are shown in Table 6 and Figure 7, which indicated that pore-associated metrics were related to the calcium ion binding pathway and homophilic cell adhesion via the plasma membrane adhesion molecule pathway.
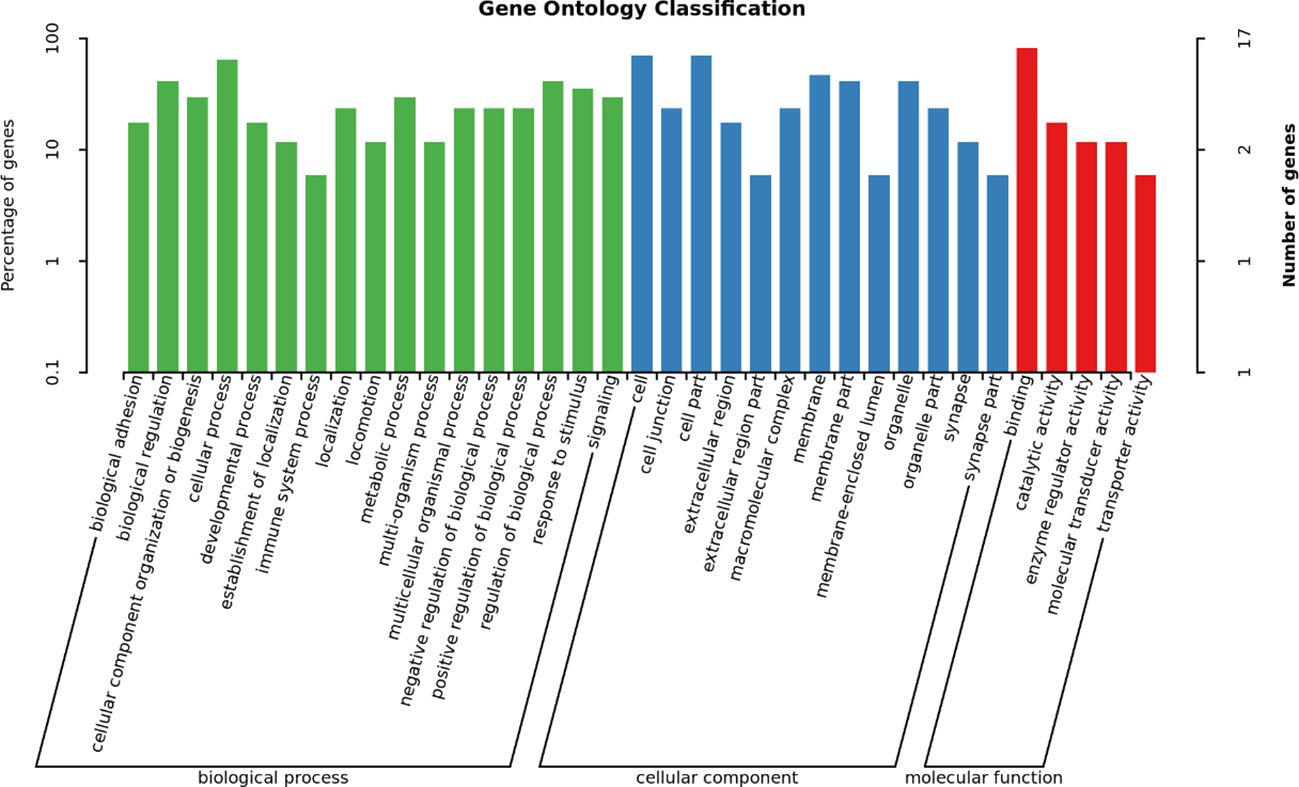
Figure 5. GO enrichment analysis of DEGs in molecular function, cellular component, and biological process GO, Gene Ontology.
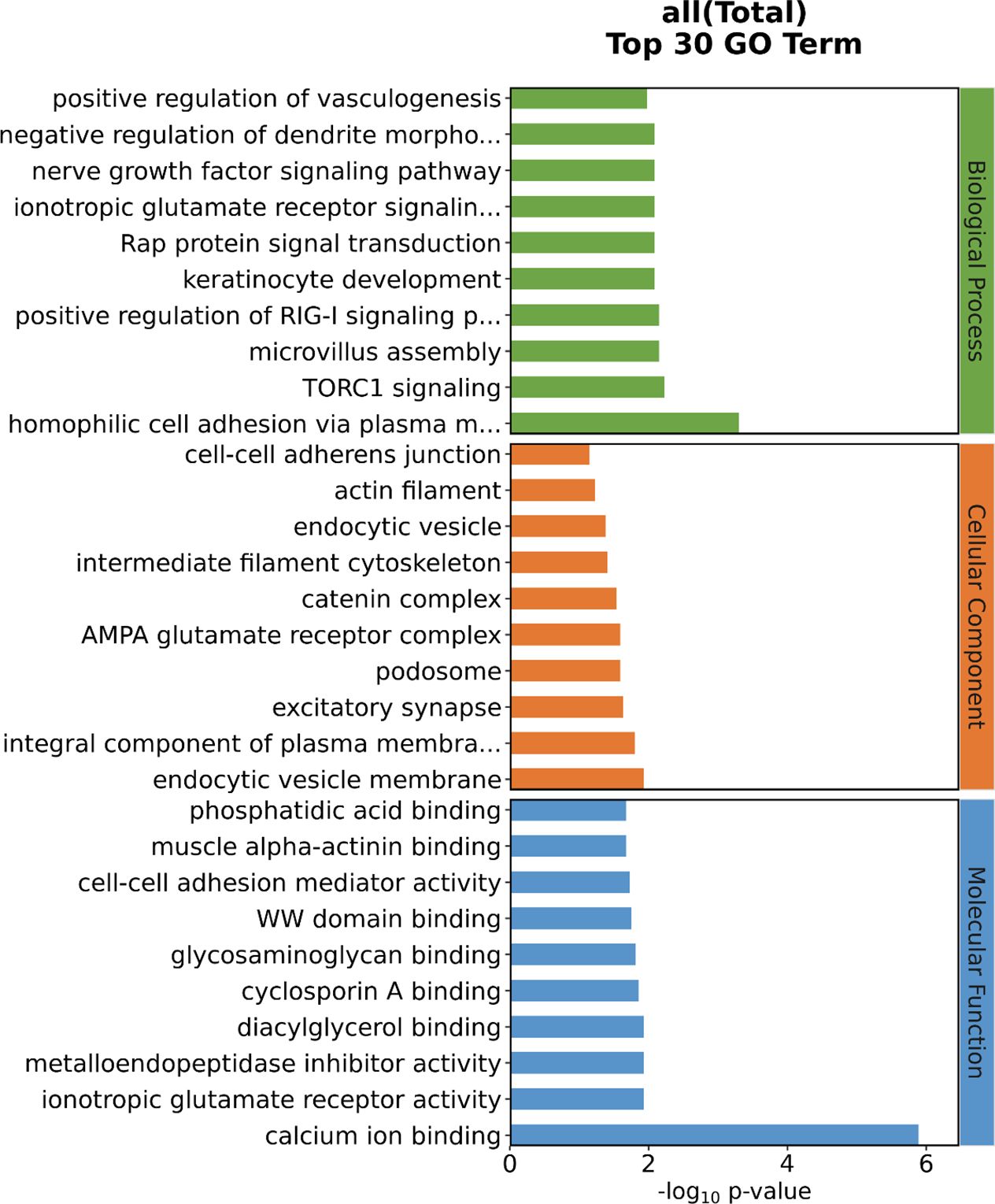
Figure 6. Top 10 GO enrichment analysis of DEGs in molecular function, cellular component, and biological process. GO, Gene Ontology.
In this experiment, seven and three genes were identified in the calcium-binding and neutrophil adhesion pathways via the plasma membrane adhesion molecules, respectively, ANXA10, CDH10, PCDH10, RAPGEF2, FSTL5, SPOCK3, TLL1, CDH10, PCDH10, and PALLD. Combined with the annotated functions of the SNPs detected in the GWAS, there are related to cytosolic calcium adhesion, hepatic regulation, or ovulation. Large amounts of calcium ions, carbonate ions, and matrix proteins are required during eggshell mineralization in laying hens. The source of these substances, and the mineralization process occurs in the uterine fluid, which is a cell-free environment. The uterus does not store either of these ions and must be continuously replenished with the blood. Ca2+ is transported intracellularly from the plasma to glandular cells via the calcium ion pathway, TRPV6, and a calcium-binding protein conjugated to CALB1 (Striem and Bar, 1991). When Ca2+ arrives at the basal lamina of the glandular cells of the uterine lumen, it is transported via Ca2+/H+ ion-exchange channels (ATP2B1, ATP2B2) and Na+/Ca2+ ion exchange channels (SLC8A1, SLC8A3) into the uterine fluid (Vincent et al., 2012), thus ensuring that eggshell calcification proceeds smoothly. The detected SNPs are likely involved in and regulated by these processes. Therefore, all of them may be candidate genes related to eggshell pore traits.
LD analysis
For GGA2, the region where CDH10 is located was analyzed for LD, the results are shown in Figure 8. The 71.871 to 71.876 Mb (5.28 Kb) region is all LD, with 2-71872593, 2-71871719, 2-71873875, 2-71873987, 2-71874359, 2-71875440, 2-71875875 being strong LD. However, only one point is significant in Figure 8, indicating that this SNP is independent, followed by an examination of the MAF for this SNP, which is 0.142857, suggesting that it may play a separate regulatory role for QBP and needs to be focused on. Only one LD analysis was selected for the analysis of significant SNPs in GGA4; the information is shown in Figure 9. The specific regulatory position 4-24774120 of ANXN10 is included in 24.774-24.774 (0.5kb), which has a strong LD with the other two loci 4-24774121 and 4-24774122 detected by this GWAS. The MAFs of the three SNPs were all 0.28125, and the LDs with the other non-significant SNPs in the region were very high. The length of this section of the 0.5kb SNP sequence is influential to ASBP.
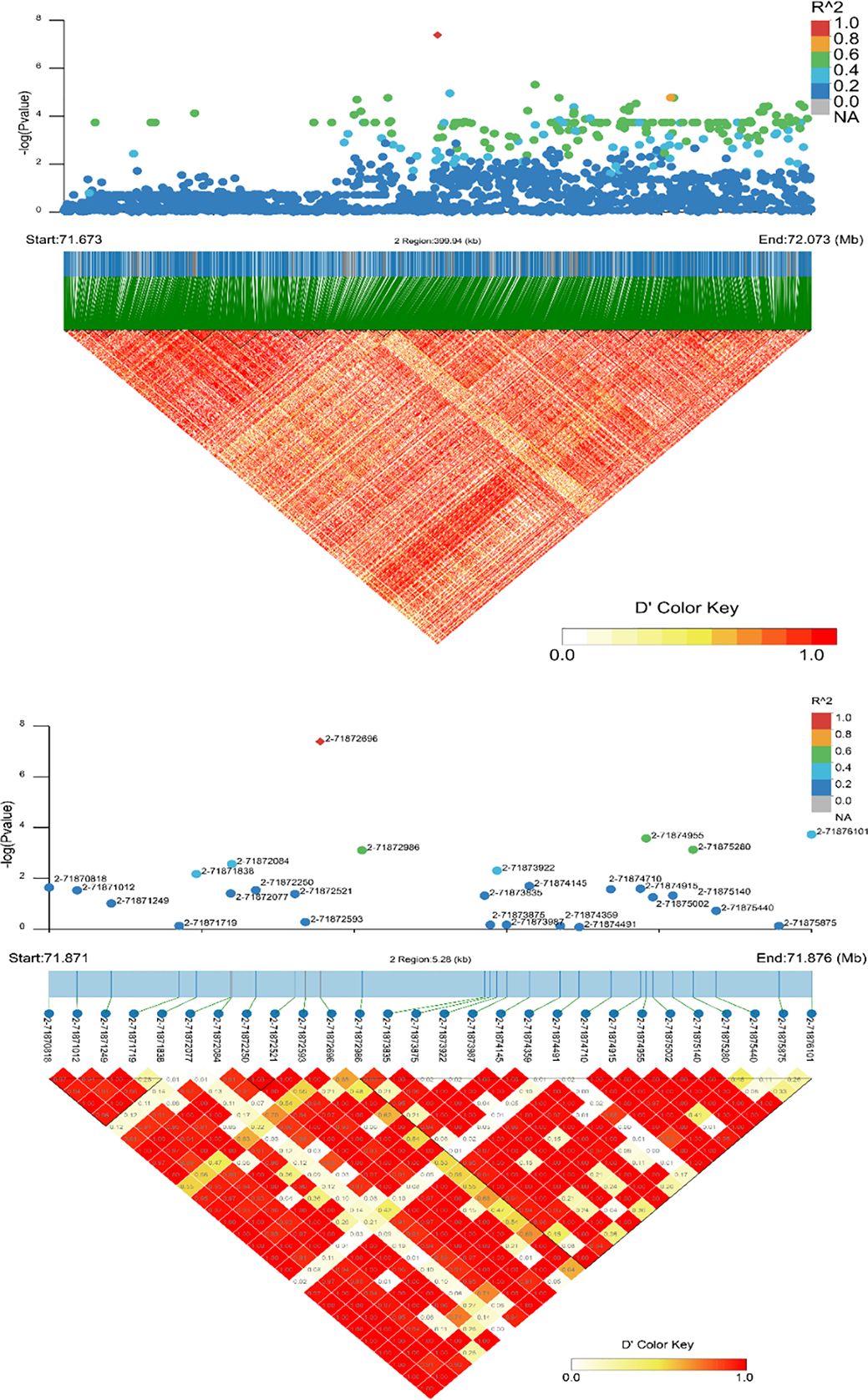
Figure 8. Linkage disequilibrium (LD) analysis of loci in the significant region associated with CDH10. The strong LD block is defined as D’≥0.8.
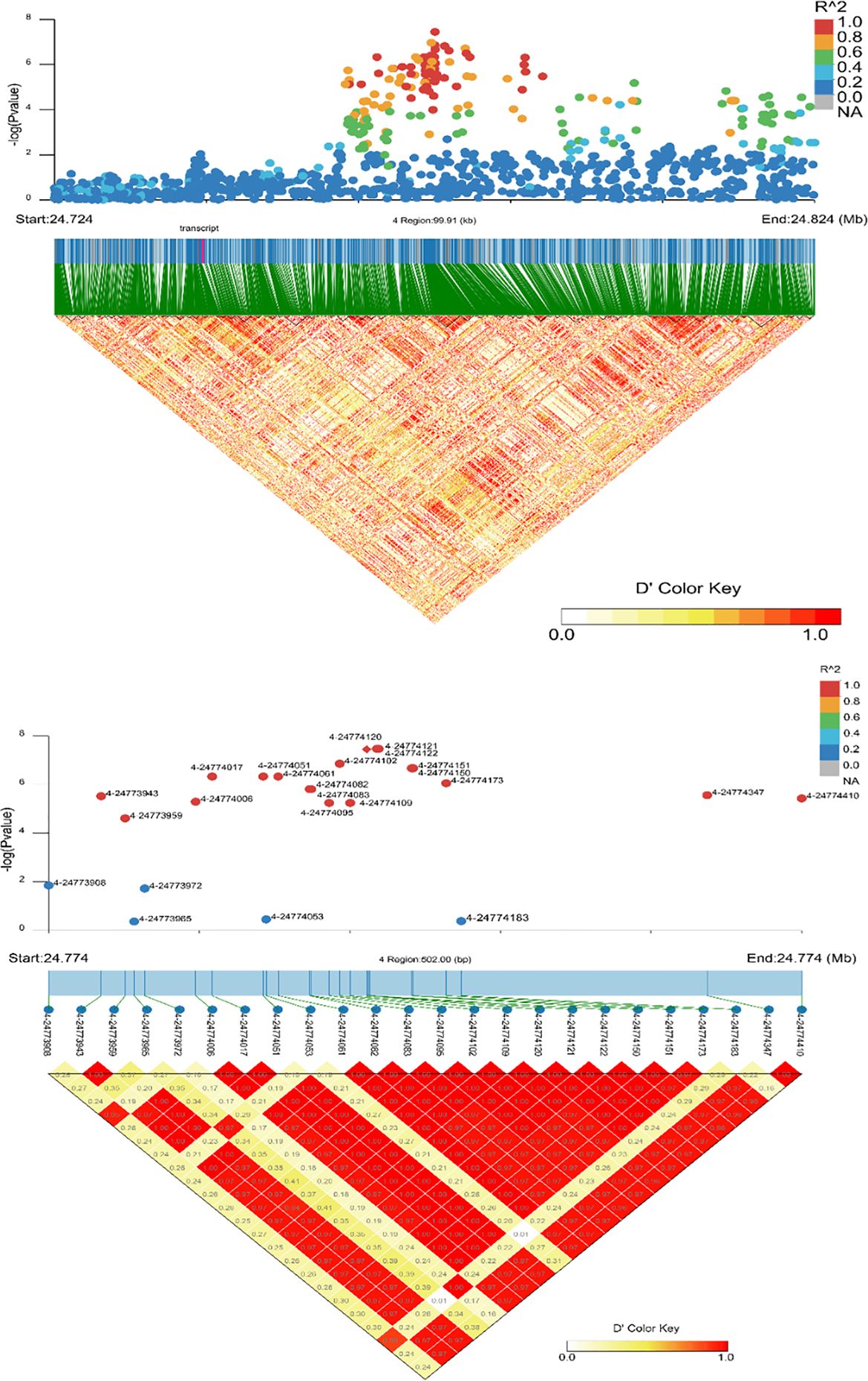
Figure 9. Linkage disequilibrium (LD) analysis of loci in the significant region associated with ANXA10. The strong LD block is defined as D’≥0.8.
Summary
This study explored the relationship between gas and bubble pores indicators for the first time. The CV values of bubble pore-related indicators were all larger than those of QGP and QM, and the bubble pore-related indicators were not correlated with QGP and QM (P > 0.05). This suggests that the gas and bubble pores may not belong to the same pore system, and different mechanisms may form them.
This study was also the first to screen genes related to gas and bubble pores by GWAS and to analyze the genetic mechanism of gas and bubble pores preliminarily. GWAS detected 32 SNPs associated with eggshell bubble pores, and the related genes were mainly located in GGA4, which, combined with KEGG enrichment analysis, was tightly linked to the calcium ion-binding pathway and could may play a regulatory role in the formation of bubble pores during eggshell calcification. The SNPs associated with QGP and QM were not detected in the GWAS. This could have occurred because of the difference in genetic mechanisms or may be regulated by micro-effector genes. This requires further evaluation by genetic parameter estimation.
In this study, we conducted a preliminary exploration of eggshell pore indices. Still, the mechanism of the gene-related loci we searched for is unclear, and the relationship between pore-related indices and eggshell quality has not been investigated. Therefore, further experiments are needed for deeper investigation. Our findings revealed the genetic basis of pore space in the eggshell ultrastructure and provided a favorable theoretical basis for the genetic regulation of eggshell biomineralization.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA1016067.
Ethics statement
The animal study was approved by Hebei Agricultural University, China. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
JZ: Conceptualization, Data curation, Writing – original draft, Writing – review & editing. YC: Methodology, Writing – review & editing. LS: Validation, Visualization, Writing – review & editing. YW: Software, Validation, Writing – review & editing. XZ: Resources, Validation, Writing – review & editing. RZ: Project administration, Writing – review & editing. HC: Funding acquisition, Resources, Visualization, Writing – review & editing. HL: Data curation, Resources, Writing – review & editing. ZN: Resources, Visualization, Writing – review & editing. DW: Conceptualization, Funding acquisition, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The work was supported by the Modern Agricultural Industry Technology System of Layerindustry Innovation Team in Hebei (HBCT2024260204), the National Natural Science Foundation of China (31902141), Natural Science Foundation for The Excellent Youth of Hebei Province, China (C2023204186), Chicken Modern Seed Industry Science and Technology Innovation Team (21326303D).
Acknowledgments
The authors would like to thank Hebei Rongde Poultry breeding Co., Ltd. for its support to this experimental.
Conflict of interest
Author XY was employed by the company Baoding Xingrui Agriculture and Animal Husbandry Development Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aaron M. K., Matthew H., Eric B., Andrey S., Kristian C., Andrew K., et al. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Ande S., Orri G. K., Xiu C., Vincenzo C., Lino T., KJ R., et al. (2010). RapGEF2 is essential for embryonic hematopoiesis but dispensable for adult hematopoiesis. Blood 116, 2921–2931. doi: 10.1182/blood-2010-01-262964
Arzate-Vázquez I., Méndez-Méndez J. V., Flores-Johnson E. A., Nicolás-Bermúdez J., Chanona-Pérez J. J., Santiago-Cortés E. (2019). Study of the porosity of calcified chicken eggshell using atomic force microscopy and image processing. Micron 118, 50–57. doi: 10.1016/j.micron.2018.12.008
Barrett J. C., Fry B., Maller J., J.Daly M. (2005). Haploview: analysis and visualization of LD and haplotype maps. Bioinf. (Oxford England) 21, 263–265. doi: 10.1093/bioinformatics/bth457
Bathgate R. D., Halls M. L., Westhuizen E. T., Callander G. E., Kocan M., Summers R. J. (2013). Relaxin family peptides and their receptors. Physiol. Rev. 93, 405–480. doi: 10.1152/physrev.00001.2012
Becker T., Redies C. (2003). Internal structure of the nucleus rotundus revealed by mapping cadherin expression in the embryonic chicken visual system. J. Comp. Neurol. 467, 536–548. doi: 10.1002/cne.10954
Benavides-Reyes C., Folegatti E., Dominguez-Gasca N., Litta G., Sanchez-Rodriguez E., Rodriguez-Navarro A. B., et al. (2021). Research Note: Changes in eggshell quality and microstructure related to hen age during a production cycle. Poult Sci. 100, 101287. doi: 10.1016/j.psj.2021.101287
Bennett R. G. (2009). Relaxin and its role in the development and treatment of fibrosis. Transl. Res. 154, 1–6. doi: 10.1016/j.trsl.2009.03.007
Board R. G., Tulett S. G. (1975). The pore arrangement in the emu (Dromaius novæhollandiæ) eggshell as shown by plastic models. J. Microsc 103, 281–284. doi: 10.1111/j.1365-2818.1975.tb03906.x
Boettner B., Aelst L. V. (2009). Control of cell adhesion dynamics by Rap1 signaling. Curr. Opin. Cell Biol. 21, 684–693. doi: 10.1016/j.ceb.2009.06.004
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinf (Oxford England) 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Cavallaro U., Christofori G. (2004). Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat. Rev. Cancer 4, 118–132. doi: 10.1038/nrc1276
Chavez C., Knape K. D., Coufal C. D., Carey J. B. (2002). Reduction of eggshell aerobic plate counts by ultraviolet irradiation. Poult Sci. 81, 1132–1135. doi: 10.1093/ps/81.8.1132
Chen S. F., Zhou Y. Q., Chen Y. R., Gu J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinf (Oxford England) 34, 884–890. doi: 10.1093/bioinformatics/bty560
Duan Z. Y., Sun C. J., Shen M. M., Wang K. H., Yang N., Zheng J. X., et al. (2016). Genetic architecture dissection by genome-wide association analysis reveals avian eggshell ultrastructure traits. Sci. Rep. 6, 28836. doi: 10.1038/srep28836
Emanuel K., Deniz Ö. R., Thomas M., Walter B., Michael G. (2009). Activins and follistatins: Emerging roles in liver physiology and cancer. World J. Hepatol. 1, 17. doi: 10.4254/wjh.v1.i1.17
Gautron J., Murayama E., Vignal A., Morisson M., Mckee M. D., Réhault S., et al. (2007). Cloning of ovocalyxin-36, a novel chicken eggshell protein related to lipopolysaccharide-binding proteins, bactericidal permeability-increasing proteins, and plunc family proteins. J. Biol. Chem. 282, 5273–5286. doi: 10.1074/jbc.M610294200
Guo L. J., Chao X. H., Huang W. I., Li Z. H., Luan K., Ye M., et al. (2021). Whole transcriptome analysis reveals a potential regulatory mechanism of LncRNA-FNIP2/miR-24-3p/FNIP2 axis in chicken adipogenesis&13. Front. Cell Dev. Biol. 9. doi: 10.3389/fcell.2021.653798
Hartmann U., Maurer P. (2001). Proteoglycans in the nervous system — the quest for functional roles in vivo. Matrix Biol. 20, 23–35. doi: 10.1016/s0945-053x(00)00137-2
Higashino K., Fujioka M., Yamamura Y. (1971). The conversion of L-lysine to saccharopine and alpha-aminoadipate in mouse. Arch. Biochem. Biophys. 142, 606–614. doi: 10.1016/0003-9861(71)90525-X
Hillier L. W., Miller W., Birney E., Warren W., Hardison R. C., Ponting C. P., et al. (2004). Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nat: Int. Wkly J. Sci. 432, 695–716. doi: 10.1038/nature03154
Hyung-Yul L., Zhao S. P., Fields P. A., Sherwood O. D. (2005). The extent to which relaxin promotes proliferation and inhibits apoptosis of cervical epithelial and stromal cells is greatest during late pregnancy in rats. Endocrinol 146, 511–518. doi: 10.1210/en.2004-0796
Jane C. H., Espey L. L. (1973). Utilization of the relaxed symphysis pubis of Guinea pigs for clues to the mechanism of ovulation. Endocrinol 93, 1441–1445. doi: 10.1210/endo-93-6-1441
Leach R. M. (1982). Biochemistry of the organic matrix of the eggshell. Poult Sci. 61, 2040–2047. doi: 10.3382/ps.0612040
Li H. M., Bob H., Alec W., Tim F., Jue R., Nils H., et al. (2009). The sequence alignment/map format and SAMtools. Bioinf (Oxford England) 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li Q. L., Duan Z. Y., Sun C. J., Zheng J. X., Xu G. Y., Yang N. (2021). Genetic variations for the eggshell crystal structure revealed by genome-wide association study in chickens. BMC Genomics 22, 786. doi: 10.1186/s12864-021-08103-1
Li Y., Li Y., Liu S., Tang Y., Mo B., Liao H. (2018). New zonal structure and transition of the membrane to mammillae in the eggshell of chicken Gallus domesticus. J. Struct. Biol. 203, 162–169. doi: 10.1016/j.jsb.2018.04.006
Lin X., Shi X. F., Xu G. Y., Zheng J. X. (2023). Research Note: Three-dimensional microstructural basis for chicken eggshell translucency. Poult Sci. 102, 103149. doi: 10.1016/j.psj.2023.103149
Mehlum F., Rahn H., Bech C., Haftorn S. (1987). Interrelationships between egg dimensions, pore numbers, incubation time, and adult body mass in Procellariiformes with special reference to the Antarctic petrel Thalassoica Antarctica. Polar Res. 5, 53–58. doi: 10.1111/j.1751-8369.1987.tb00355.x
Moeko M., Hiroyuki O., Misako M., Tsukasa S. (2011). DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Mol. Cell Biol. 31, 3802–3819. doi: 10.1128/MCB.01368-10
Mussunoor S., Murray G. I. (2008). The role of annexins in tumour development and progression. J. Pathol. 216, 131–140. doi: 10.1002/path.2400
Nakatani Y., Fujioka M., Higashino K. (1970). Alpha-aminoadipate aminotransferase of rat liver mitochondria. Biochim. Biophys. Acta 198, 219–228. doi: 10.1016/0005-2744(70)90054-9
Ning Z. H. (2004). Breeding of new supporting lines for grain-saving small laying hens and research on supporting breeding technology [D] (Beijing, China: China Agric. Univ.).
Nys Y., Gautron J., Garcia-Ruiz J. M., Hincke M. T. (2004). Avian eggshell mineralization: biochemical and functional characterization of matrix proteins. C. R. Palevol. 3, 549–562. doi: 10.1016/j.crpv.2004.08.002
Okonechnikov K., Conesa A., García-Alcalde F. (2015). Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinf. (Oxford England) 32, 292–294. doi: 10.1093/bioinformatics/btv566
Olivier W. L., Picman J., Hincke M. T. (2008a). Antimicrobial activity of the Anseriform outer eggshell and cuticle. Comp. Biochem. Physiol. Part B. 149, 640–649. doi: 10.1016/j.cbpb.2008.01.001
Olivier W. L., Picman J., Hincke M. T. (2008b). Antimicrobial activity of cuticle and outer eggshell protein extracts from three species of domestic birds. Br. Poult Sci. 49, 133–143. doi: 10.1080/00071668408454866
Park J.-A., Sohn S.-H. (2018). The influence of hen aging on eggshell ultrastructure and shell mineral components. Korean J. Food Sci. Anim. Resour. 38, 1080–1091. doi: 10.5851/kosfa.2018.e41
Parveen A., Jackson C. D., Dey S., Tarrant K., Anthony N., Rhoads D. D. (2020). Identification and validation of quantitative trait loci for ascites syndrome in broiler chickens using whole genome resequencing. BMC Genet. 21, 54. doi: 10.1186/s12863-020-00859-x
Perrott H. R., Scott V. D., Board R. G. (1981). Crystal orientation in the shell of the domestic fowl: an electron diffraction study. Calcif Tissue Int. 33, 119–124. doi: 10.1007/BF02409423
Plotree D., Plotgram D. (1989). Phylip-phylogeny inference package (version 3.2). Cladistics 5, 163–166. doi: 10.1086/416571
Price A. L., Patterson N. J., Plenge R. M., Weinblatt M. E., Shadick N. A., Reich D. (2006). Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909. doi: 10.1038/ng1847
Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. R., Bender D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
Rama K. D. S., Siddharthan A., Seshadri S. K., Sampath K. T. S. (2007). A novel route for synthesis of nanocrystalline hydroxyapatite from eggshell waste. J. Mater Sci: Mater Med. 18, 1735–1743. doi: 10.1007/s10856-007-3069-7
Riley A., Sturrock C. J., Mooney S. J., Luck M. R. (2014). Quantification of eggshell microstructure using X-ray micro computed tomography. Br. Poult Sci. 55, 311–320. doi: 10.1080/00071668.2014.924093
Roberts J. R. (2004). Factors affecting egg internal quality and egg shell quality in laying hens. Poultry Sci. 41, 161–177. doi: 10.2141/jpsa.41.161
Rubin C. J., Zody M. C., Eriksson. J., Meadows. J. R. S., Sherwood E., Webster M. T., et al. (2010). Whole-genome resequencing reveals loci under selection during chicken domestication. Nat 464, 587–591. doi: 10.1038/nature08832
Sara E. O., Chin K., Sertich J. J. W., Varricchio D. J., Choi S., Rifkin J. (2021). Tiny, ornamented eggs and eggshell from the Upper Cretaceous of Utah represent a new ootaxon with theropod affinities. Sci. Rep. 11, 10021. doi: 10.1038/s41598-021-89472-1
Sparks N., Board R. G. (1984). Cuticle, shell porosity and water uptake through hens’ eggshells. Br. Poult Sci. 25, 267–276. doi: 10.1080/00071668408454866
Striem S., Bar A. (1991). Modulation of quail intestinal and egg shell gland calbindin (Mr 28,000) gene expression by vitamin D3, 1,25-dihydroxyvitamin D3 and egg laying. Mol. Cell Endocrinol. 75, 169–177. doi: 10.1016/0303-7207(91)90232-H
Sun C. J., Qu L., Yi G. Q., Yuan J. W., Duan Z. Y., Shen M. M., et al. (2015). Genome-wide association study revealed a promising region and candidate genes for eggshell quality in an F2 resource population. BMC Genomics 16, 565. doi: 10.1186/s12864-015-1795-7
Takeichi M. (1990). Cadherins: a molecular family important in selective cell-cell adhesion. Annu. Rev. Biochem. 59, 237–252. doi: 10.1146/annurev.bi.59.070190.001321
Tullett S. G. (1975). Regulation of avian eggshell porosity. J. Zool 177, 339–348. doi: 10.1016/0300-9629(84)90083-5
Tullett S. G. (1984). The porosity of avian eggshells. Comp. Biochem. Physiol Part A: Physiol. 78, 5–13. doi: 10.1016/0300-9629(84)90083-5
Tyler C. (1957). Some chemical, physical and structural Properties of moa egg shells. J. Polynesian Soc. 66, 110–130. doi: 10.1080/03014220709510542
Vincent J., Brionne A., Gautron J., Nys Y. (2012). Identification of uterine ion transporters for mineralisation precursors of the avian eggshell. BMC Physiol. 12, 10. doi: 10.1186/1472-6793-12-10
Wang D. H., Li Y. J., Liu L., Liu J. S., Bao M., Yang N., et al. (2017). Traits of eggshells and shell membranes of translucent eggs. Poult Sci. 96, 351–358. doi: 10.3382/ps/pew328
Wang K., Li M. Y., Hakonarson H. (2010). ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164. doi: 10.1093/nar/gkq603
Wang M. M., Li Y., Peng M. S., Zhong L., Wang Z. J., Li Q. Y., et al. (2015). Genomic analyses reveal potential independent adaptation to high altitude in Tibetan chickens. Mol. Biol. Evol. 32, 1880–1889. doi: 10.1093/molbev/msv071
Zhang C. H., Peng L. L., Gu H. T., Wang J. J., Wang Y. X., Xu Z. Q. (2023). ANXA10 is a prognostic biomarker and suppressor of hepatocellular carcinoma: a bioinformatics analysis and experimental validation. Sci. Rep. 13, 1583. doi: 10.1038/s41598-023-28527-x
Zhang D. Y., Lei J. S., Sun W. L., Wang D. D., Lu Z. (2020). Follistatin Like 5 (FSTL5) inhibits epithelial to mesenchymal transition in hepatocellular carcinoma. Chin. Med. J. 133, oul1978–804. doi: 10.1097/CM9.0000000000000847
Zhao Y., Chi Y. (2009). Characterization of collagen from eggshell membrane. Biotechnol 8, 254–258. doi: 10.3923/biotech.2009.254.258
Zhou J., Wang S. T., Nie F. Q., Feng L., Zhu G. S., Jiang L. (2011). Elaborate architecture of the hierarchical hen’s eggshell. Nano Res. 4, 171–179. doi: 10.1007/s12274-010-0067-8
Zhou X., Stephens M. (2012). Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 44, 821–824. doi: 10.1038/ng.2310
Keywords: hen, GWAS, bubble pore, SNP, GGA4
Citation: Zhang J-J, Chen Y-F, Shi L, Wang Y-T, Zhao X-Y, Zhou R-Y, Chen H, Liu H-G, Ning Z-H and Wang D-H (2024) Genome-wide association analysis of eggshell pore traits based on whole genome resequencing. Front. Anim. Sci. 5:1469859. doi: 10.3389/fanim.2024.1469859
Received: 24 July 2024; Accepted: 21 November 2024;
Published: 06 December 2024.
Edited by:
Birendra Mishra, University of Hawaii at Manoa, United StatesReviewed by:
Ning Gao, Hunan Agricultural University, ChinaJingwei Yuan, Chinese Academy of Agricultural Sciences, China
Copyright © 2024 Zhang, Chen, Shi, Wang, Zhao, Zhou, Chen, Liu, Ning and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: De-He Wang, dGhlY29uY2VydGV2ZW50QGNhdS5lZHUuY24=
 Jun-Jie Zhang1
Jun-Jie Zhang1 Yi-Fan Chen
Yi-Fan Chen Rong-Yan Zhou
Rong-Yan Zhou De-He Wang
De-He Wang