- 1Department of Animal Sciences, Washington State University, Pullman, WA, United States
- 2Washington Animal Disease Diagnostics Laboratory, Pullman, WA, United States
- 3Department of Animal, Veterinary and Food Sciences, University of Idaho, Moscow, ID, United States
Introduction: Reproductive efficiency in dairy cattle bred by artificial insemination (AI) or via embryo transfer (ET) is important as a means of facilitating the rate of genetic improvement by utilizing superior genetics from sires and donor dams. Fertility is especially challenging in primiparous cows as they have recently calved, are still growing, and are lactating. Fertility, measured by the conception rate from the first service (CCR1), is the number of cows that become pregnant as a result of the first AI or ET compared to all cows serviced, whereas times bred (TBRD) is the number of inseminations cows bred by AI needed to achieve a successful pregnancy.
Methods: This study aimed to identify the loci associated with and gene sets enriched for the CCR1 in primiparous cows that were bred by AI (n = 882) or were ET recipients (n = 353), and to identify the loci associated with the TBRD (n= 1,851) in primiparous cows. A genome-wide association analysis and gene set enrichment analysis were performed using single-nucleotide polymorphism (SNP) data for the CCR1 and TBRD in primiparous cows. In cows bred by AI, two loci containing five positional candidate genes were associated with the CCR1 (p < 1 × 10−5) and one gene set with 18 leading-edge genes was enriched (NES ≥3) for the CCR1.
Results and discussion: In cows that were ET recipients, five loci were associated with the CCR1, but no gene sets were enriched for the CCR1. For the second phenotype, five loci with nine positional candidate genes were associated with the TBRD and two gene sets with eight leading-edge genes were enriched for the TBRD. Identifying the loci associated with or gene sets enriched for the CCR1 and TBRD provides an opportunity to better understand embryonic loss in AI-bred and ET-recipient cows and to use this information to select for higher reproductive efficiency.
1 Introduction
Fertility decreases when cattle are under stress, such as the energy demand needed for lactation concurrent with growth in primiparous cows (Bisinotto et al., 2012; Wathes et al., 2007). Dairy cattle have low pregnancy rates, which is likely due to increased inbreeding and selection for production traits at the expense of fertility traits (Bedere et al., 2018; González-Recio et al., 2007; Lucy, 2001). Conception rates at the first breeding have declined from 55% in Holstein heifers to 39% in primiparous cows (CDCB, 2023). Previous studies have shown that fertilization rates in Holsteins are upwards of 90%; indicating that failure of conception is not the cause of infertility (Diskin and Sreenan, 1980; Diskin et al., 2012). The majority (57%) of infertility stems from embryonic loss, which occurs between 0 and 42 days of gestation (Dunne et al., 2000; Inskeep and Dailey, 2005). This reproductive inefficiency results in cows being culled from the herd, and costs producers up to $1,000 per cow due to a higher number of inseminations needed to achieve a pregnancy, and the increased costs of feed and labor incurred as the time between calving increases (Chebel et al., 2004; De Vries, 2006). Since 2010, traits associated with fertility have been added to producer indices to aid in selection and have successfully improved fertility as cow conception rates (CCR) have increased by 4% in the past 14 years (CDCB, 2024).
Fertility traits incorporated into selection indices have not considered different types of breeding services, such as artificial insemination (AI) and embryo transfer (ET). The use of ET has increased in recent years, allowing for genetic progress via the collection of oocytes from females with high genetic merit (Viana, 2023). The genomic selection of elite female donors and highly fertile bulls to be bred by AI is well established, however, research is limited on the selection of ET recipient cows. This area of research is important to discuss as the cost of ET technologies for producers ranges from $250 to $400 per transfer compared to the average cost of $46 for a single AI breeding (Kaniyamattam et al., 2018). Embryonic loss rates in ET have been found to be higher at 21.5% than those of conventional AI at 16.3% (Demetrio et al., 2007). This increase in embryonic loss could be partially due to differences in the selection of ET recipients and how the transferred embryo interacts within the uterine environment of the recipient. Different biological processes have been described in the development of a pregnancy initiated by being bred by AI compared with those in an ET recipient. Studies have suggested an increased rate of blastulation and abnormal placentation in in vitro-produced embryos when compared with AI-bred or natural service embryos (Goossens et al., 2007; Lee et al., 2011; Zhao et al., 2019). Other mammalian models also have identified gene expression differences for embryonic loss associated with in vitro- and in vivo-produced embryos (Assou et al., 2010; Giritharan et al., 2007). Although there are biological differences between pregnancy establishment in AI-bred cows and ET recipients, the loci associated with and the gene sets enriched in successful pregnancy establishment in ET recipients have not been investigated in primiparous cows. Investigation of the primiparous cow conception rate from the first service (CCR1) for AI-bred cows and cows that are ET recipients allows for the identification of regions and genes that differ between these breeding services. Identifying breeding service-specific loci allows for the selection of cattle that may be successfully AI-bred or serve as an ET recipient. The identification of gene sets enriched for CCR1 provides a better understanding of how genes essential to pregnancy establishment interact in the first month after conception.
The number of inseminations required to achieve a successful pregnancy (TRBD), is also an economically important trait. As the TBRD increases, the producer spends $84 per cow (on average) for each additional AI (Lima et al., 2010). TBRD has recently been added as a trait to fertility indices. The average TBRD for Holstein primiparous cows in 2010 was 2.5 and has since decreased to 2.0 with its inclusion into fertility indices (Norman et al., 2020). The Council on Dairy Cattle Breeding reports a similar metric, stating that the average number of inseminations per conception for primiparous cattle was 2.0 in 2022 (CDCB, 2022). No updates for inseminations per conception have been released for 2023 and 2024. The further identification and validation of loci associated with the TBRD in AI-bred cattle could further improve the accuracy of selection and result in a greater rate of genetic progress in fertility and profitability for dairy producers. The objective of this study was to identify the loci associated with and gene sets enriched for the CCR1 and TBRD in cows bred by AI, and the loci associated with and gene sets enriched for the CCR1 in primiparous cows that were ET recipients. Identifying the genomic regions associated or enriched for the CCR1 in AI-bred and ET recipients and the TBRD for AI-bred cows allows for the validation of fertility-related loci and positional candidate and leading-edge genes that can be used to accelerate genetic progress.
2 Materials and methods
2.1 Study animals and phenotype
This study, #6743, was approved by the Washington State University Institutional Animal Care and Use Committee. The study population has been previously described (Kelson et al., 2024) but briefly, and all primiparous cows (n = 5,750) were from one dairy farm in Idaho. Dairy Comp 305 (Valley Agricultural Software, Tulare, CA, USA) breeding and animal health records were evaluated for all the primiparous cattle. Animals with incomplete or missing records (n = 2430) were removed from the study. Exclusion criteria for removing genotyped animals from the study included cattle that were too young to be bred, pregnancy data not yet available, or cattle that had died or were culled prior to breeding. Animals with health events (n = 200), such as mastitis, metabolic issues, or lameness, between 21 days before breeding and 30 days post-breeding were also removed from the study. The exclusion of animals that experienced health events was performed to minimize the environmental effects of infertility and enhance the genetic effects on embryonic loss.
Primiparous cows were bred by AI or received an ET once per estrous cycle. Some primiparous cows were bred exclusively with ET (n = 1,053) or AI (n = 2,067). The animals bred by AI were inseminated following observed estrus, while the embryo transfer recipients had 7-day-old embryos transferred on day 7 of their estrous cycle following a double ovulation synchronization protocol. Pregnancy was determined approximately 30 days post-breeding by rectal palpation and ultrasound. CCR1 for all the primiparous cows was 40% (1235/3120), 42% (882/2067) for the cows bred by AI, and 52% (353/679) for the ET-recipient cows.
The primiparous cows that were only bred by AI were followed to determine their TBRD. The cows that were bred by AI and were ET recipients were removed from the analysis (n = 674). Cows that experienced a pregnancy loss after day 42 of gestation (n = 159) or those with no TBRD data due to culling or age (n = 92) were also removed from the analysis. The removal of cows from the study that experienced a fetal loss eliminated any potential identification of loci that were due to fetal loss rather than embryonic loss. The TBRD ranged from one to ≥ five. A total of 1,851 animals remained for the TBRD analysis (Table 1).
2.2 DNA extraction and genotyping
CLARIFIDE® Plus (Zoetis Precision Animal Health, Parsnippany, NJ, USA) genotypes were obtained with the dairy farm’s approval. Raw genotypes encompassing 45,790 single-nucleotide polymorphisms (SNPs) on average were then imputed to 634,433 SNPs using an in-house reference population of approximately 4,800 US Holsteins that were genotyped on the Illumina (San Diego, CA, USA) BovineHD BeadChip with 777,962 SNPs and an accuracy of >95% (Browning and Browning, 2016; Kiser et al., 2019b). Genotypes of 29,741–53,594 SNPs were shared between the Zoetis and the Illumina BovineHD BeadChip.
2.3 Quality control
Genotypes were imputed using Beagle version 4.1 and animals were subjected to quality control within the SNP and Variation Suite (SVS) software version 8.1 (Golden Helix, Bozeman, MT, USA). No cows were removed for errors in sex or for duplicate samples. SNPs with a call rate < 0.90 (n = 21,462), with a minor allele frequency < 0.01 (n = 109,409), and those that failed Hardy-Weinberg equation testing (p < 10 x 10−100, n = 6,687) were removed. After quality control filtering, 3,120 primiparous cows and 496,891 SNPs remained for the final analysis.
2.4 Genome-wide association analysis
A genome-wide association analysis (GWAA) was performed for the CCR1 in cows bred by AI and cows that were ET recipients using SVS software version 8.1 (Golden Helix, Bozeman, MT, USA). The efficient mixed-model association eXpedited (EMMAX) statistical approach and an identity-by-state matrix was used for the GWAA. There was no pruning of genotypes for linkage disequilibrium prior to the GWAA. The statistical model for EMMAX is expressed as , where y = the vector of observed phenotypic values, X = a matrix containing fixed effects, β = regression coefficients, Z = a matrix containing the observed random effects, u = vector of random effects concerning variants of allele substitutions in the population, and 𝝐 = residual effects (Kang et al., 2010). Due to the unknown inheritance of CCR in cattle, three genotypic models (additive, dominant, and recessive) were analyzed for each phenotype. In the additive model, two minor alleles (aa) result in twice the effect that a single minor allele (Aa) would have on fertility, whereas in the dominant model, the effect of at least one minor allele (Aa/aa) is compared to the effect on the phenotype when there are no minor alleles (AA). For the recessive model, the presence of two minor alleles (aa) is compared to the effect on the phenotype when at least one major allele (Aa/AA) is present. Significance thresholds for unadjusted p-values were based on The Wellcome Trust’s recommendations for the GWAA to determine associations with the CCR1 and TBRD in cows bred by AI and with the CCR1 in cows that were ET recipients (The Wellcome Trust Case Control Consortium, 2007). The threshold for moderate evidence for an association was p < 1 x 10−5 and p < 5 x 10−7 for a strong association.
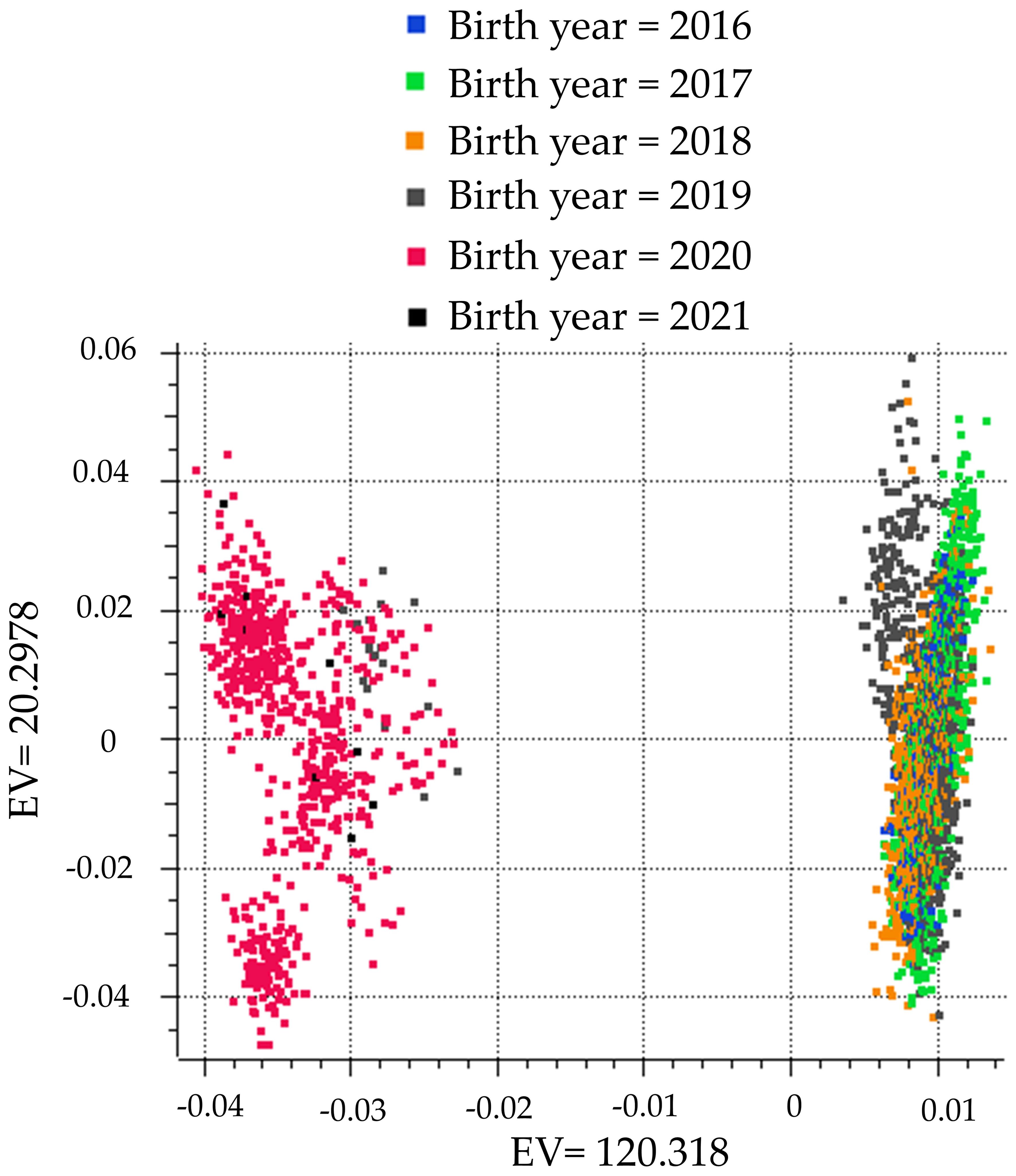
A principal component analysis plot was created using the methods of Halko et al. (2010). The population was divided into two distinct clusters segregated by birth year (Figure 1). To control for the difference in genotypes due to birth year, birth year was used as a covariate in the GWAA. The genomic inflation factor lambda (λGC) was calculated as described by Devlin and Roeder (1999) to assess whether the population stratification was adequately corrected with the use of birth year as a covariate. The λGC for all GWAAs and models ranged from 0.975–1.04. When multiple SNPs were associated with either CCR1 or TBRD on the same chromosome, a linkage disequilibrium threshold of D’ > 0.7 was used to define a locus (Sargolzaei et al., 2008; Weiss and Clark, 2002).
The average haplotype size was calculated in SVS using the method outlined by Gabriel et al., 2002. The average size of a haplotype was 31.1 kb. This haplotype size was used to define the genomic region investigated for the identification of positional candidate genes associated with CCR1 and TBRD. Genes within 31.1 kb of an associated SNP in the 5’ or 3’ directions were identified as positional candidate genes based on the bovine ARS-UCD 1.2 genome assembly.
2.5 Gene set enrichment analysis-single nucleotide polymorphism
A gene set enrichment analysis based on SNPs (GSEA-SNP) was completed using the GenGen software package (Holden et al., 2008). SNPs were mapped to genes within a haplotype based on the ARS-UCD 1.2 genome assembly. A single SNP that was most significantly associated with CCR1 or TBRD within the gene’s haplotype was used to represent each of the 34,450 genes. Gene proxy SNPs were then ranked by their p-values for their association with the CCR1 in AI-bred and ET-recipient cows. Running sum statistics were used to calculate the enrichment scores (ESs) for gene sets based on the SNP ranking. When a gene’s SNP was encountered in the gene set that was highly ranked due to its association with the phenotype, the ES increased. The genes that positively contributed to the ES prior to the peak ES were defined as leading-edge genes. Leading-edge genes are the genes that result in the gene set meeting significance for being enriched for either CCR1 or TBRD. The leading-edge genes are, therefore, the most important genes for the phenotype in the gene set. In contrast, if a gene in the gene set was not associated or ranked, the ES decreased. ES statistics were calculated similarly to Kolmogorov–Smirnov’s weighted statistic (Holden et al., 2008; Charmpi and Ycart, 2015). The GenABEL R package calculated permutated p-values for 10,000 phenotype-based permutation tests for each gene set (Aulchenko et al., 2007). Gene sets were generated from five different databases: Gene Ontology (GO), Reactome (R), BioCarta (M), Kyoto Encyclopedia of Gene and Genomes Pathways (KEGG), and Protein Analysis Through Evolutionary Relationships (PANTER). The ES values were normalized (NES) to account for variances in the number of leading-edge genes present within each gene set. The threshold of significance for the GSEA-SNP was NES ≥ 3.0.
3 Results
3.1 Genome-wide association analysis for cow conception rate from the first service
There were two unique loci and one shared locus associated with the CCR1 in the cattle bred by AI in the additive and dominant models (Figure 2; Table 2). One locus on BTA3 at 117 Mb was associated with CCR1 in the additive (p = 3.91 x 10−7) and dominant models (p = 7.06 x 10−7), and an additional locus on BTA8 at 102 Mb (p = 8.51 x 10−6) was associated with CCR1 in the dominant model. There were five positional candidate genes (ADGB, LOC101906837, LRRFIP1, RAMP1, and RBM44) associated with the CCR1 in cattle bred by AI (Table 2). There were no loci associated with CCR1 in the recessive inheritance model.
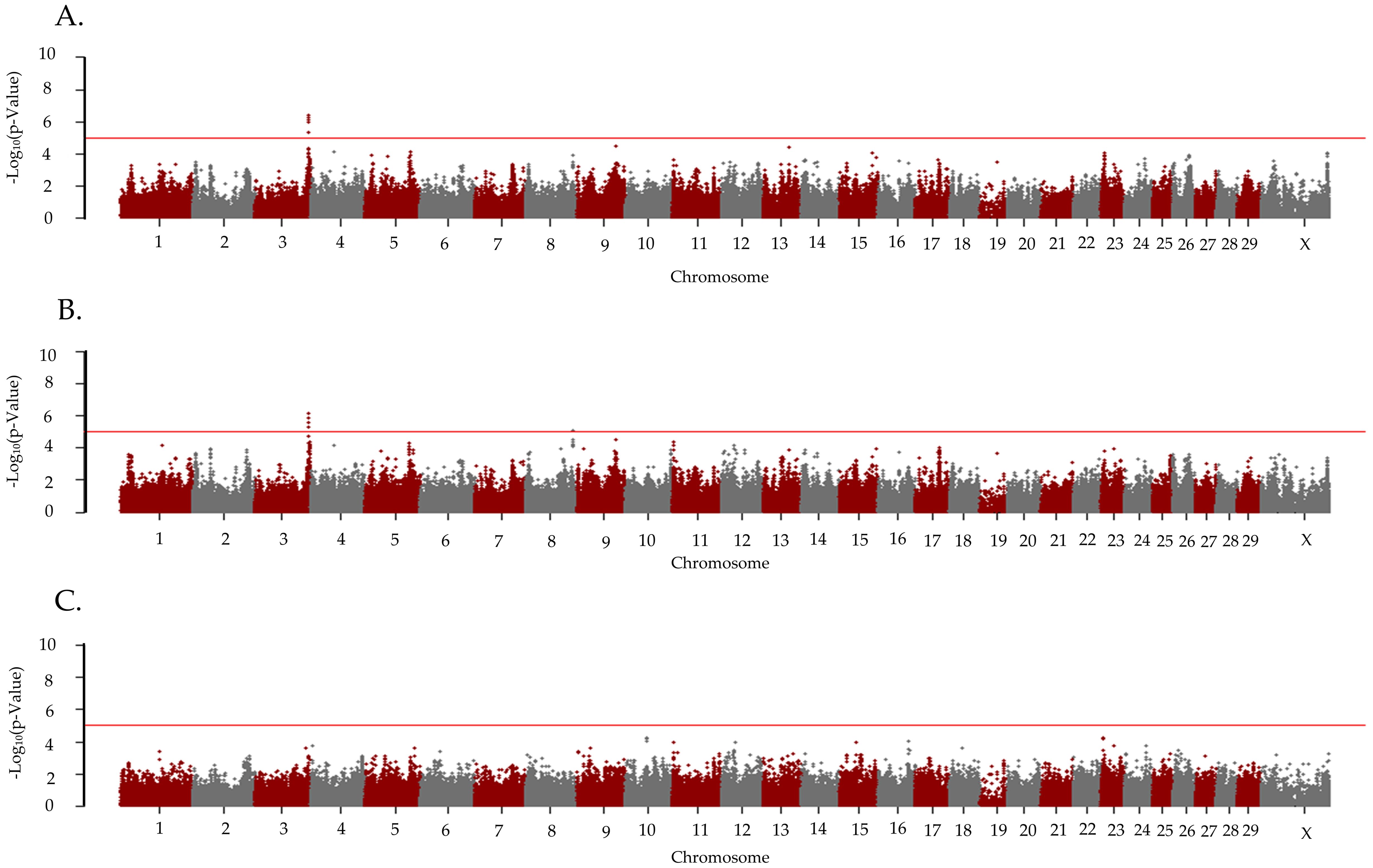
Figure 2. Loci associated with conception due to the first service in primiparous Holstein cows bred by artificial insemination. Manhattan plots are labeled for the (A) additive, (B) dominant, and (C) recessive models. All plots are labeled with the Bos taurus chromosome on the x-axis, and the –log10p-value on the y-axis. Due to all animals being female the Y chromosome is not present on the x-axis. The line indicates the threshold for significance with the red line indicating a p-value < 1x10−5.
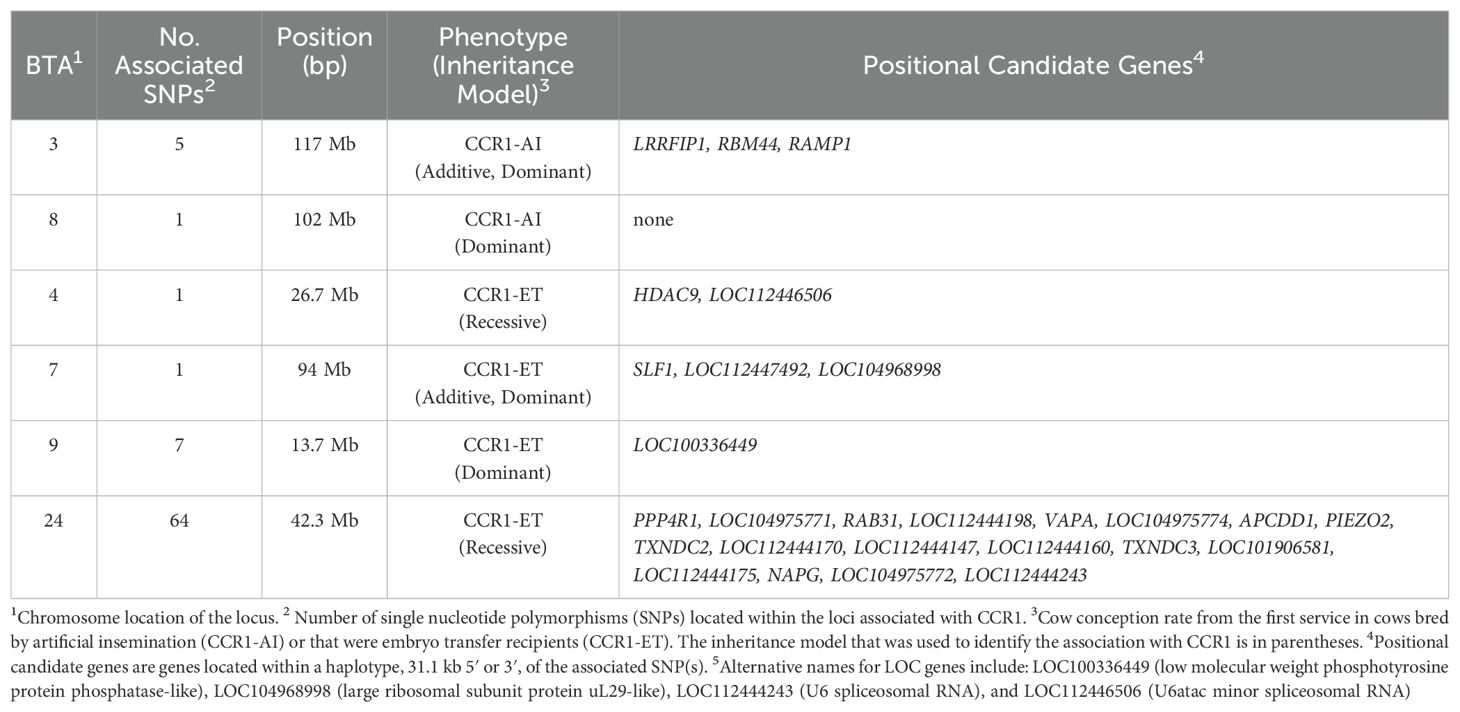
Table 2. Loci and positional candidate genes associated with Holstein primiparous cow conception rate.
The λGC for the CCR1 in the cows that were ET recipients was 0.975 for all the inheritance models. There were five unique loci associated with CCR1 in cows that were ET recipients (Figure 3; Table 2). In the recessive model, two loci were associated with CCR1. The associated locus (P = 8.18 x 10−6) on BTA4 had two positional candidate genes (HDAC9 and LOC112446506) and the associated (p = 1.02 x 10−6) locus on BTA24 held 18 positional candidate genes (Table 2). In the additive inheritance model, a locus on BTA7 at 94 Mb was associated (p = 3.40 x 10−6) with the CCR1 in the ET recipients. This locus was also associated (p = 3.70 x 10−6) in the dominant model and contained three positional candidate genes (SLF1, LOC112447492, and LOC104968998). A second locus on BTA9 at 13 Mb was also associated (p = 3.37x 10−6) with CCR1 in the ET-recipient cows in the dominant model and contained the positional candidate gene LOC100336449. There were no loci that were associated with the CCR1 in the AI-bred cows that were also associated with the CCR1 in the cows that were ET recipients.
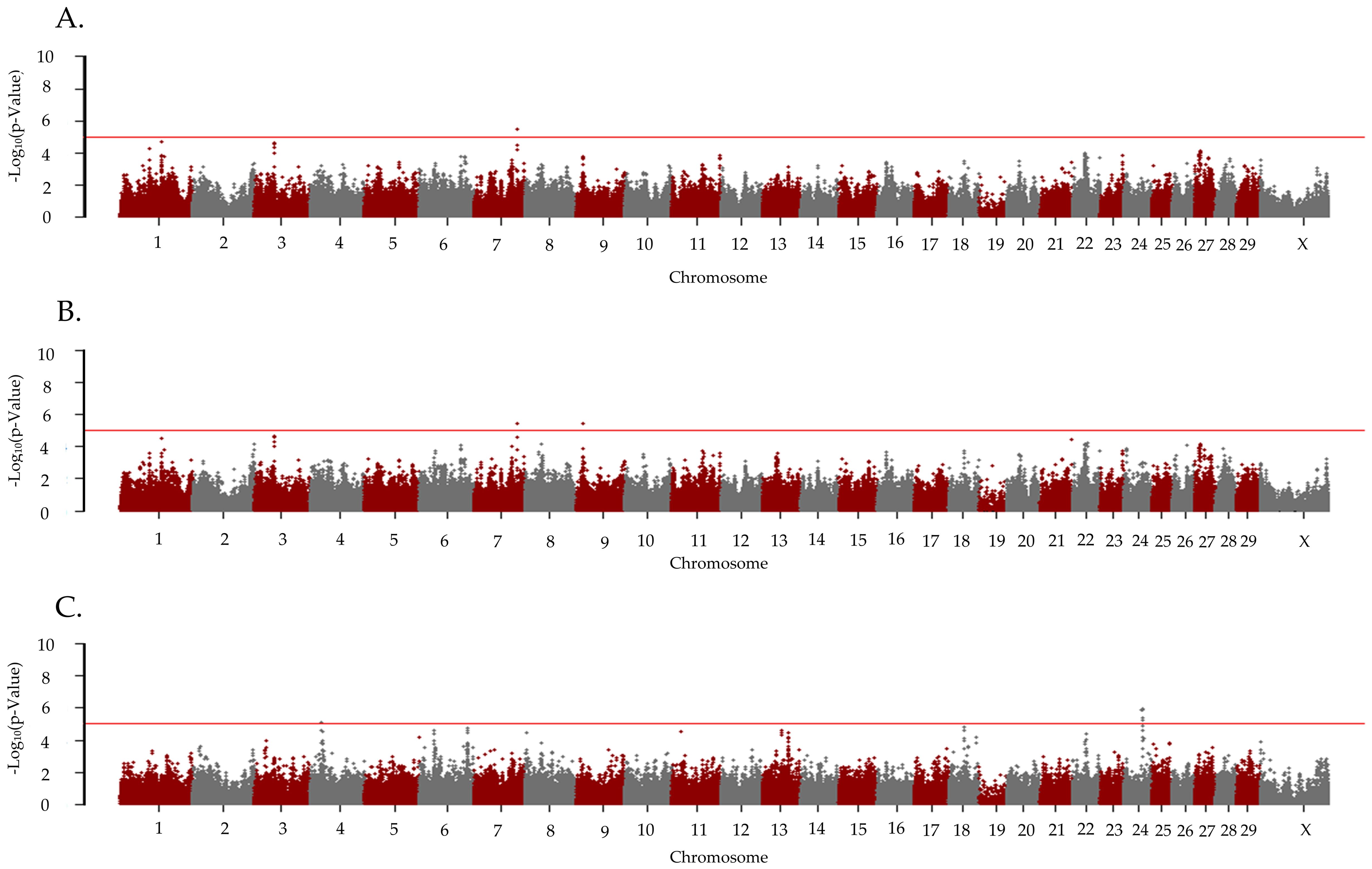
Figure 3. Loci associated with pregnancy due to the first embryo transfer in primiparous Holstein cows. Manhattan plots are labeled for the (A) additive, (B) dominant, and (C) recessive models. All plots are labeled with the Bos taurus chromosome on the x-axis, and the –log10p-value on the y-axis. Due to all animals being female the Y chromosome is not present on the x-axis. The line indicates the threshold for significance with the red line indicating a p-value < 1x10−5.
3.2 Genome-wide association analysis for the number of times bred by artificial insemination prior to a confirmed pregnancy
The λGC for TBRD was 0.99, 0.99, and 1.00 for the additive, dominant, and recessive models, respectively. Five unique loci were associated (p < 1 x 10−5) with the TBRD in the primiparous cows bred by AI (Table 3; Figure 4). For the recessive model, a locus on BTA5 was associated (p = 1.40 x 10−6) with TBRD at 121 Mb with two positional candidate genes (LGR5, and LOC112446921). A locus on BTA9 at 83 Mb was associated (p = 2.84 x 10−6) with TBRD in the additive and dominant models and contained six positional candidate genes (ADGB, LOC101906837, LOC101906979, LOC101907062, LOC512881, and SAMD5). Other loci associated with TBRD in the dominant inheritance model were on BTA3 (p = 9.09 x 10−6) at 117 Mb and BTA10 (P = 9.92 x 10−6) at 99 Mb. Only one positional candidate gene (LRRFIP1) was identified on the locus on BTA3 and four positional candidate genes were identified on BTA10 (Table 3).
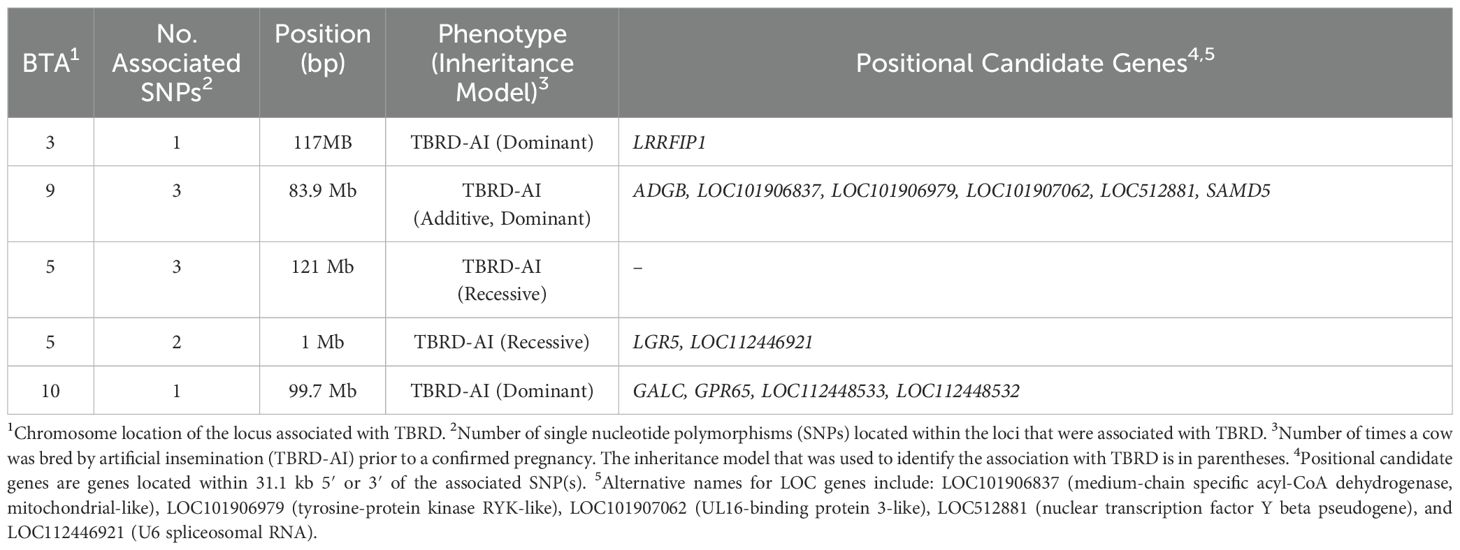
Table 3. Loci and positional candidate genes associated with the number of artificial inseminations required for a successful pregnancy in Holstein primiparous cows.
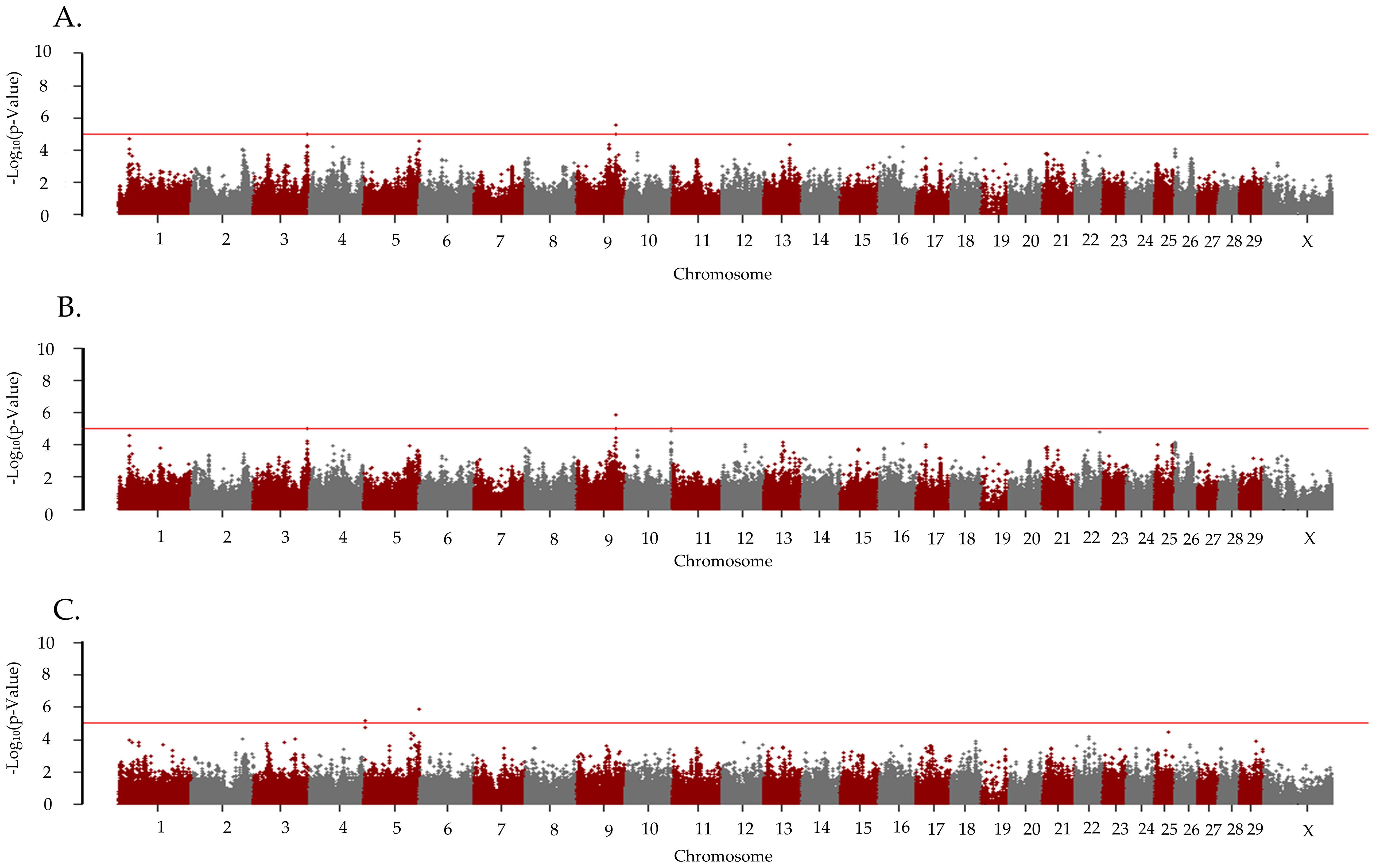
Figure 4. Loci associated with the number of artificial inseminations required for successful pregnancy in Holstein primiparous cattle. Manhattan plots are labeled for the (A) additive, (B) dominant, and (C) recessive models. All plots are labeled with the Bos taurus chromosome on the x-axis, and the –log10p-value on the y-axis. Due to all animals being female the Y chromosome is not present on the x-axis. The line indicates the threshold for significance with the red line indicating a p-value < 1x10−5.
3.3 Gene set enrichment analysis-SNP
One gene set, Pattern Specification Process (GO:0007389) was enriched (NES = 3.0) for the CCR1 in the cows bred by AI. This gene set included 64 genes and 18 leading-edge genes (ASPH, BAX, BMI1, DUSP6, FBXL15, FGF2, GPR161, LFNG, MEF2C, MSX2, NR2F2, OSR1, PKD2, PSEN2, RBPJ, STK4, TGFBR1, and WNT2). No gene sets were enriched for the CCR1 in the ET-recipient cows.
Two gene sets were enriched for TBRD in the cows bred by AI. The first gene set that was enriched (NES=3.40) was Morphogenesis of a Branching Structure (GO:0001763) and consisted of 37 genes and 8 leading-edge genes (BAX, ERMN, FGF2, MSX2, PAK1, PKD2, STK4, and WNT2). The second gene set enriched (NES = 3.17) was Morphogenesis of a Branching Epithelium (GO:0061138) which contained 36 genes and 7 leading-edge genes (BAX, FGF2, MSX2, PAK1, PKD2, STK4, and WNT2). Seven (BAX, FGF2, MSX2, PAK1, PKD2, STK4, and WNT2) of the leading-edge genes enriched for TBRD were found in two or more gene sets. Of these seven genes, four (FGF2, PKD2, TGFBR1 and WNT2) have extensive connections to female fertility and will be discussed in more detail.
4 Discussion
Results from the GWAA and the GSEA-SNP provide a foundation to identify areas of interest in the genome that are associated with embryonic loss to the first service. These analyses provide opportunities to forecast which cattle are more likely to experience embryonic loss to an AI or ET service as well as insights into potential mechanisms of the loss. Variations in the genome that alter positional candidate or leading-edge genes may hinder or cause the overexpression of genes that then influence embryonic survival. Utilizing previous studies focused on genes that were identified as having a role in embryonic loss can help refine which positional candidate genes are potentially important in CCR1 and TBRD phenotypes.
The identification of shared loci in independent populations provides additional support for their importance in embryonic loss in AI or ET. For the determination of shared loci between studies, SNPs reported to be associated with fertility in this study or previous studies were required to be within an average haplotype (± 31 kb) of each other or share the same positional candidate or leading-edge genes.
The locus on BTA8 associated with the CCR1 in AI-bred cows was previously reported to be associated with the CCR1 in an unrelated Holstein population (Kiser et al., 2019a). Similarly, the locus on BTA24 associated with the CCR1 in ET recipient cows was found within a locus associated with the TBRD in a separate Holstein population (Galliou et al., 2020). The locus on BTA24 was also associated with the time in days from calving to first insemination in Nordic Holstein cows (Höglund et al., 2014).
The positional candidate gene associated with the CCR1 in the AI-bred primiparous cows, LRR binding FLII interacting protein 1 (LRRFIP1), is associated with the wingless and Int-1 (WNT) signaling pathway. When LRRF1P1 is silenced or alternatively spliced the interaction with the WNT signaling pathway is distorted, which is thought to impede normal embryonic development (Douchi et al., 2015; Labbé et al., 2017). The second positional candidate gene, receptor activity modifying protein 1 (RAMP1), is associated with calcium receptors as a chaperone protein to route calcium receptors to the cell surface (Yallampalli et al., 2004). The relocation of these receptors is associated with an increase in the progesterone needed for pregnancy establishment, consistent with the elevated RAMP1 levels that are present in pregnant mice (Yallampalli et al., 2004). The positional candidate genes ADGB, LOC101906837, and RBM44 need further investigation to determine their roles in fertility and embryonic loss.
Of the thirteen positional candidate genes associated with the TBRD in the AI-bred primiparous Holstein cows, three (LRRFIP1, GPR65, and LGR5) have previously been identified as being involved in fertility at the embryonic stage. The role of LRRFIP1 has previously been discussed as it was also a positional candidate gene for CCR1. The positional candidate gene on BTA10 is G protein-coupled receptor 65 (GPR65). Overexpression of GPR65 is associated with early pregnancy loss in humans (Mao et al., 2023). However, it is unknown why the overexpression of GRP65 results in reoccurring pregnancy loss. Leucine-rich repeat-containing G protein-coupled receptor 5 (LGR5) on BTA5 is essential for preimplantation development, blastocyst attachment, and the production of progesterone in mice (Sun et al., 2014). Deletion of LGR5 increases the expression of 20𝜶HSD, which is responsible for the metabolism of progesterone within the corpus luteum. In mice, the decrease in progesterone due to a loss of LGR5 and its subsequent increased expression of 20𝜶HSD leads to a premature regression of the corpus luteum and higher instances of embryonic loss (Sun et al., 2014).
There were eight positional candidate genes that were differentially expressed in previous studies. The AI-bred CCR1 positional candidate gene LOC101906837 was differentially expressed in the endometrium of highly fertile open and pregnant heifers (Moraes et al., 2018). Two additional AI-bred CCR1 positional candidate genes, LRRFIP1 and RAMP1, were expressed in the bovine placenta from days 17 to 50 of gestation (Davenport et al., 2023; Supplementary Figure 1). For the positional candidate genes associated with the CCR1 in the cows that were ET recipients, two positional candidate genes, PIEZO2 and PPP4R1, were differentially expressed between the conceptuses of highly fertile and sub-fertile heifers (Moraes et al., 2018). In cows with multiple AI services, the positional candidate gene SAMD5 was found to be differentially expressed between the conceptuses of highly fertile and sub-fertile heifers (Moraes et al., 2018). The AI TBRD positional candidate gene LOC512881 was also differentially expressed between the endometrium of highly fertile and sub-fertile heifers (Moraes et al., 2018).
Three positional candidate genes (VAPA, RAB31, and NAPG) associated with the CCR1 in cows that were ET recipients were leading-edge genes enriched for embryonic loss in a separate Holstein cow population (Neibergs, personal communication). Three genes (GPR65, LGR5, and LOC512881) found to be positional candidate genes for TBRD to AI services, were leading-edge genes enriched for embryonic loss in Holstein heifers in an independent population (Neibergs, personal communication). Finally, GPR65 was negatively associated with embryo implantation in a human study (Mao et al., 2023).
Nine leading-edge genes (ASPH, DUSP6, NR2F2, OSR1, PSEN2, RBPJ, TGFBR1, STK4, and WNT2) enriched for the CCR1 in the AI-bred cows were expressed at different time points during the embryonic period in bovine placentas (Supplementary Figure 1; Davenport et al., 2023). Three leading-edge genes (FGF2, STK4, and WNT2) enriched for the TBRD in the AI-bred cows were expressed in binucleate cells, endothelial cells, endometrial luminal epithelial cells, macrophages, mesenchyme, neutrophils, and uninucleate cells at 17 to 50 days of gestation (Supplementary Figure 1; Davenport et al., 2023).
Four leading-edge genes (ASPH, BAX, PKD2, and TGFBR1) were enriched for the conception rate to the first service in heifers that were ET recipients and the CCR1 in the cows bred by AI (Kelson et al., 2024). PKD2 was also enriched for the TBRD in the primiparous cows bred by AI and heifers were ET recipients. The roles of ASPH and BAX in relation to fertility during the embryonic stage have not been established. These results support that ASPH, BAX, PKD2, and TGFBR1 are important in pregnancy establishment and maintenance during the embryonic period across parities and in AI-bred and ET-recipient cows.
Of the genes enriched for the TBRD in cows bred by AI, four (FGF2, PKD2, TGFBR1, and WNT2) have extensive connections to female fertility. Fibroblast growth factor 2 (FGF2) is associated with in vitro embryo production and is used in embryo culture mediums (Du et al., 2021; Stoecklein et al., 2021; Stoecklein et al., 2022). FGF2 is expressed in the uterus of many mammals prior to implantation and has a role in the successful implantation of an embryo and placental development (Torry et al., 2007; Mondal et al., 2015). In cattle specifically, the effects of FGF2 induce angiogenesis of the embryo both in vitro and in vivo (Krneta et al., 2006; Lin et al., 2008). Inhibition of FGF2 and the formation of new blood vessels is detrimental to embryonic and fetal development and can be attributed to pregnancy loss (Torry et al., 2007; Zygmunt et al., 2003). Without the expansion of the vascular system, the embryo or fetus will suffer from developmental issues or complete pregnancy loss (Torry et al., 2007). Polycystin-2 (PKD2) is a leading-edge gene that was enriched for the CCR1 and TBRD in the AI-bred cows. Expression of PKD2 is highest in the first 5 weeks of development in humans (Chauvet et al., 2002). PKD2 also has a role in the development of placental tissue in mice and is essential for fetal survivability (Garcia-Gonzalez et al., 2010). PKD2 mice knockouts affect placental development, causing decreased vascularization, disorganization of placental smooth muscle, and a buildup of amniotic fluids (Garcia-Gonzalez et al., 2010). The transforming growth factor beta receptor 1 (TGFBR1) uses the WNT pathway as a mediator in cell signaling. Knockouts of TGFBR1 show myometrium abnormalities and a loss of function in WNT signaling. WNT2 is a leading-edge gene that was enriched for the CCR1 in the AI-bred cows and is involved in placental, and embryonic development (Kallapur et al., 1999; Ingman and Robertson, 2002; Chen et al., 2009; Douchi et al., 2015; Labbé et al., 2017). WNT2 increases placental defects such as decreased size, abnormal accumulation of maternal blood, and less efficient establishment of fetal capillaries in mice when mutations of WNT2 lead to complete loss of function (Monkley et al., 1996). In cattle, WNT2 is expressed in the endometrium and in the conceptus during the morula stage (Tríbulo et al., 2017). Additionally, the deletion of TGFBR1 identified it as essential for forming early uterine development (Li et al., 2011; Gao et al., 2014; Monsivais et al., 2017). The identification of TGFB family-related genes and interacting genes such as the WNT pathway across multiple studies suggests their crucial roles in early pregnancy maintenance and high levels of embryonic loss. These tissues and developmental time points are crucial for a successful pregnancy.
Many of the identified genes had direct connections to fertility in cattle or other mammalian species or have been identified as being associated with or enriched for CCR1 or TBRD. However, unique genes and gene sets were also identified as associated with or enriched for CCR1 and TBRD. This is not unexpected as the genetics, management, and environment of different herds are likely to lead to the discovery of unique genes as well as validate the results of previously identified loci, positional candidate genes, gene sets, and leading-edge genes for CCR1 and TBRD. This is why the validation of genomic results is so important in independent cattle populations before adoption for genomic selection. The investigation of loci, positional candidate genes, gene sets, and leading-edge genes for ET-recipient cattle was also expected to identify loci, gene sets, and genes that were shared with the AI-bred cattle but also unique from those associated with the AI-bred cattle. Unique genomic regions were expected as they would reflect the differences in the establishment of pregnancy and development of an embryo in vitro compared to an embryo developed within an AI-bred cow’s uterus.
The results of this study are relevant to the dairy industry as it supports and expands the loci that can be used for genomic selection to improve fertility in Holstein cattle bred by AI and those that are ET recipients. The GWAA provided two loci that were validated for the CCR in the cows bred by AI and one locus that was validated in the primiparous cows for TBRD. The incorporation of newly validated loci for CCR can improve fertility conception rates through genomic selection as it expands the proportion of variance explained for this trait. The GSEA-SNP provided information on the gene networks and biological pathways that are essential for supporting pregnancy during the embryonic period. This information can be used to identify critical biological mechanisms that are failing during embryonic loss through functional genomic and physiology studies. The investigation of the role of the genetics of the ET-recipient cattle in embryonic loss is innovative as the establishment of ET pregnancies differs in some respects from those initiated by AI, as identified by physiological studies (Goossens et al., 2007; Moraes et al., 2018). This study confirms that these differences also extend to the genome of the ET-recipient cow and that fertility can be improved through genomic selection. As more cattle are chosen to remain in the herd as ET recipients, selection for conception rate and TBRD in ET-recipient cattle should be considered.
The limitations of this study include a small sample size for embryo transfer recipients and the inability to validate loci through an independent Holstein population. To combat these limitations, an independent study is being conducted on a different herd to identify loci associated with the CCR and TBRD in ET-recipient cattle.
5 Conclusions
This study investigated the genomic factors associated with the conception rate to the first AI breeding, the rate of pregnancy success to the first ET, and the number of times a cow was bred for the maintenance of pregnancy. There were two loci associated with the CCR1 in cows bred by AI, four loci associated with the CCR1 in cows that were ET recipients, and four loci associated with the TBRD in cows bred by AI. A total of five positional candidate genes were associated with the CCR1 in the AI-bred cows, 23 positional candidate genes associated with the CCR1 in cows that were ET recipients, and 13 positional candidate genes associated with the TBRD in cows bred by AI. The GSEA-SNP resulted in 18 leading-edge genes enriched for the CCR1 in cows that were bred by AI, and 8 leading-edge genes enriched for the TBRD in cows bred by AI.
This study validated two loci, BTA8 associated with the CCR1 in AI-bred cows and BTA24 associated with the TBRD, that were previously identified in Holstein fertility studies (Kiser et al., 2019b; Galliou et al., 2020; Höglund et al., 2014). In cows bred by AI, three positional candidate genes (LOC101906837, LRRFIP1, and RAMP1) were previously associated with a differential expression in fertility cattle studies (Moraes et al., 2018; Davenport et al., 2023). Analysis of the CCR1 in ET-recipient cows resulted in five positional candidate genes (NAPG, PIEZO2, PPP4R1, RAB31, and VAPA) that were supported by their roles in fertility in unrelated populations (Moraes et al., 2018; Neibergs, personal communication). A total of nine leading-edge genes (ASPH, DUSP6, NR2F2, OSR1, PSEN2, RBPJ, TGFBR1, STK4, and WNT2) enriched for the CCR1 in the AI-bred cows were expressed in bovine placentas during embryonic development (Davenport et al., 2023).
This study also supported four positional candidate genes (GPR65, LGR5, LOC512881, and SAMD5) and their function in fertility in unrelated cattle populations for the number of times bred for a successful pregnancy in AI breeding (Neibergs, personal communication; Moraes et al., 2018). Three leading-edge genes (FGF2 STK4, and WNT2) enriched for the number of times bred for a successful pregnancy for AI were supported as genes involved in fertility as they were expressed in the bovine placenta during early gestation (Davenport et al., 2023).
A total of four leading-edge genes were enriched in both heifers and cows in this population (ASPH, BAX, PKD2, and TGFBR1) regardless of the breeding type. Many of the identified genes are associated with the production or regulation of hormones such as progesterone or are connected to placental defects.
In summary, the results of this study identify the loci, positional candidate genes, and leading-edge genes involved in embryonic loss. Embryonic loss poses a challenge for Holstein cattle and dairy producers as it leads to substantial financial loss. Pinpointing the genomic regions linked to embryonic loss during AI or ET services holds promise for producers by incorporating these regions into genomic selection indices. The use of genomic selection for fertility traits such as CCR1 and TBRD has increased since 2010. The inclusion of additional markers that address fertility differences across parities and cattle bred by AI or those that are ET recipients will lead to higher reproductive efficiency.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://doi.org/10.17605/OSF.IO/QYM4E.
Ethics statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Washington State University Institutional Animal Use and Care Committee #6743. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
VK: Data curation, Formal analysis, Investigation, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. JK: Data curation, Methodology, Resources, Software, Validation, Writing – review & editing. KD: Data curation, Investigation, Resources, Visualization, Writing – review & editing. ES: Formal analysis, Validation, Writing – review & editing. BM: Funding acquisition, Investigation, Resources, Validation, Writing – review & editing. HN: Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This project is supported by Agriculture and Food Research Initiative Competitive Grant no. 2023-67015-39524 from the USDA National Institute of Food and Agriculture.
Acknowledgments
The authors would like to recognize the Idaho dairy for their assistance in collecting of records for phenotyping as well as Zoetis for the delivery of the raw genotypes.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fanim.2024.1458088/full#supplementary-material
Supplementary Figure 1 | Leading-edge genes for cow conception rate to first service in cows bred by artificial insemination that were expressed in a bovine placenta single-cell gene study (Davenport et al., 2023). Expression of genes in different placental cell types at days 17 (A), 24 (B), 30 (C), and 50 (D) of gestation. Placental cell types consisted of binucleate cells (BNC), endothelial cells, fetal blood cells, endometrial luminal epithelial cells (LE), macrophages, mesenchyme neutrophils, and uninucleate cells (UNC).
References
Assou S., Boumela I., Haouzi D., Anahory T., Dechaud H., De Vos J., et al. (2010). Dynamic changes in gene expression during human early embryo development: From fundamental aspects to clinical applications. Hum. Reprod. Update 17, 272–290. doi: 10.1093/humupd/dmq036
Aulchenko Y. S., Ripke S., Isaacs A., van Duijn C. M. (2007). Genabel: An R library for genome-wide association analysis. Bioinformatics 23, 1294–1296. doi: 10.1093/bioinformatics/btm108
Bedere N., Cutullic E., Delaby L., Garcia-Launay F., Disenhaus C. (2018). Meta-analysis of the relationships between reproduction, milk yield and body condition score in dairy cows. Livestock Sci. 210, 73–84. doi: 10.1016/j.livsci.2018.01.017
Bisinotto R. S., Greco L. F., Ribeiro E. S., Martinez N., Lima F. S., Staples C. R., et al. (2012). Influences of nutrition and metabolism on fertility of dairy cows. Anim. Reprod. 9, 260–272. Available at: https://animal-reproduction.org/journal/animreprod/article/5b5a6057f7783717068b46e5.
Browning B. L., Browning S. R. (2016). Genotype imputation with millions of reference samples. Am. J. Hum. Genet. 98, 116–126. doi: 10.1016/j.ajhg.2015.11.020
CDCB (2022). Council on Dairy Cattle Breeding. Available online at: https://webconnect.uscdcb.com/#/national-performance-metrics (Accessed April 1, 2024).
CDCB (2023). The Council of Dairy Cattle Breeding Base Means. (4201 Northview Drive Suite 302 Bowie, MD 20716 USA: The Council of Dairy Cattle Breeding). Available at: https://webconnect.uscdcb.com//summary-stats/breed-means-bases-heterosis-inbreeding-regressions.
CDCB (2024). The Council of Dairy Cattle Breeding National Performance Metrics. (4201 Northview Drive Suite 302 Bowie, MD 20716 USA: The Council of Dairy Cattle Breeding). Available at: https://webconnect.uscdcb.com//national-performance-metrics.
Charmpi K., Ycart B. (2015). Weighted Kolmogorov Smirnov testing: An alternative for gene set enrichment analysis. Stat. Appl. Genet. Mol. Biol. 14. doi: 10.1515/sagmb-2014-0077
Chauvet V., Qian F., Boute N., Cai Y., Phakdeekitacharoen B., Onuchic L. F., et al. (2002). Expression of PKD1 and PKD2 transcripts and proteins in human embryo and during normal kidney development. Am. J. Pathol. 160, 973–983. doi: 10.1016/s0002-9440(10)64919-x
Chebel R. C., Santos J. E. P., Reynolds J. P., Cerri R. L. A., Juchem S. O., Overton M. (2004). Factors affecting conception rate after artificial insemination and pregnancy loss in lactating dairy cows. Anim. Reprod. Sci. 84, 239–255. doi: 10.1016/j.anireprosci.2003.12.012
Chen Q., Zhang Y., Lu J., Wang Q., Wang S., Cao Y., et al. (2009). Embryo-uterine cross-talk during implantation: The role of Wnt Signaling. Mol. Hum. Reprod. 15, 215–221. doi: 10.1093/molehr/gap009
Davenport K. M., Ortega M. S., Liu H., O’Neil E. V., Kelleher A. M., Warren W. C., et al. (2023). Single-nuclei RNA sequencing (snrna-seq) uncovers trophoblast cell types and lineages in the mature bovine placenta. Proc. Natl. Acad. Sci. 120. doi: 10.1073/pnas.2221526120
Demetrio D. G. B., Santos R. M., Demetrio C. G. B., Vasconcelos J. L. M. (2007). Factors affecting conception rates following artificial insemination or embryo transfer in lactating holstein cows. J. Dairy Sci. 90, 5073–5082. doi: 10.3168/jds.2007-0223
Devlin B., Roeder K. (1999). Genomic control for association studies. Biometrics 55, 997–1004. doi: 10.1111/j.0006-341x.1999.00997.x
De Vries A. (2006). Economic value of pregnancy in dairy cattle. J. Dairy Sci. 89, 3876–3885. doi: 10.3168/jds.s0022-0302(06)72430-4
Diskin M. G., Parr M. H., Morris D. G. (2012). Embryo death in cattle: An update. Reproduction Fertility Dev. 24, 244. doi: 10.1071/rd11914
Diskin M. G., Sreenan J. M. (1980). Fertilization and embryonic mortality rates in beef heifers after artificial insemination. Reproduction 59, 463–468. doi: 10.1530/jrf.0.0590463
Douchi D., Ohtsuka H., Ariake K., Masuda K., Kawasaki S., Kawaguchi K., et al. (2015). Silencing of LRRFIP1 reverses the epithelial–mesenchymal transition via inhibition of the Wnt/β-catenin signaling pathway. Cancer Lett. 365, 132–140. doi: 10.1016/j.canlet.2015.05.023
Du C., Davis J. S., Chen C., Li Z., Cao Y., Sun H., et al. (2021). FGF2/FGFR signaling promotes cumulus–oocyte complex maturation in vitro. Reproduction 161, 205–214. doi: 10.1530/rep-20-0264
Dunne L. D., Diskin M. G., Sreenan J. M. (2000). Embryo and foetal loss in beef heifers between day 14 of gestation and full term. Anim. Reprod. Sci. 58, 39–44. doi: 10.1016/s0378-4320(99)00088-3
Gabriel S. B., Schaffner S. F., Nguyen H., Moore J. M., Roy J., Blumenstiel B., et al. (2002). The structure of haplotype blocks in the human genome. Science 296, 2225–2229. doi: 10.1126/science.1069424
Galliou J. M., Kiser J. N., Oliver K. F., Seabury C. M., Moraes J. G., Burns G. W., et al. (2020). Identification of loci and pathways associated with Heifer Conception Rate in U.S. Holsteins. Genes 11, 767. doi: 10.3390/genes11070767
Gao Y., Bayless K. J., Li Q. (2014). TGFBR1 is required for mouse myometrial development. Mol. Endocrinol. 28, 380–394. doi: 10.1210/me.2013-1284
Garcia-Gonzalez M. A., Outeda P., Zhou Q., Zhou F., Menezes L. F., Qian F., et al. (2010). PKD1 and Pkd2 are required for normal placental development. PLoS One 5. doi: 10.1371/journal.pone.0012821
Giritharan G., Talbi S., Donjacour A., Di Sebastiano F., Dobson A. T., Rinaudo P. F. (2007). Effect of in vitro fertilization on gene expression and development of mouse preimplantation embryos. Reproduction 134, 63–72. doi: 10.1530/rep-06-0247
González-Recio O., López de Maturana E., Gutiérrez J. P. (2007). Inbreeding depression on female fertility and calving ease in Spanish dairy cattle. J. Dairy Sci. 90, 5744–5752. doi: 10.3168/jds.2007-0203
Goossens K., Van Soom A., Van Poucke M., Vandaele L., Vandesompele J., Van Zeveren A., et al. (2007). Identification and expression analysis of genes associated with bovine blastocyst formation. BMC Dev. Biol. 7. doi: 10.1186/1471-213x-7-64
Halko N., Martinsson P. G., Tropp J. A. (2010). Finding structure with randomness: Probabilistic algorithms for constructing approximate matrix decompositions. SIAM Rev. 53, 217–288. doi: 10.1137/090771806
Höglund J. K., Sahana G., Guldbrandtsen B., Lund M. S. (2014). Validation of associations for female fertility traits in Nordic Holstein, Nordic red and Jersey Dairy Cattle. BMC Genet. 15, 8. doi: 10.1186/1471-2156-15-8
Holden M., Deng S., Wojnowski L., Kulle B. (2008). GSEA-SNP: Applying gene set enrichment analysis to SNP data from genome-wide association studies. Bioinformatics 24, 2784–2785. doi: 10.1093/bioinformatics/btn516
Ingman W. V., Robertson S. A. (2002). Defining the actions of transforming growth factor beta in reproduction. BioEssays 24, 904–914. doi: 10.1002/bies.10155
Inskeep E. K., Dailey R. A. (2005). Embryonic death in cattle. Veterinary Clinics North America: Food Anim. Pract. 21, 437–461. doi: 10.1016/j.cvfa.2005.02.002
Kallapur S., Ormsby I., Doetschman T. (1999). Strain dependency of TGFbeta1 function during embryogenesis. Mol. Reprod. Dev. 52, 341–349. doi: 10.1002/(sici)1098-2795(199904)52:4<341::aid-mrd2<3.0.co;2-n
Kang H. M., Sul J. H., Service S. K., Zaitlen N. A., Kong S., Freimer N. B., et al. (2010). Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42, 348–354. doi: 10.1038/ng.548
Kaniyamattam K., Block J., Hansen P. J., De Vries A. (2018). Economic and genetic performance of various combinations of in vitro-produced embryo transfers and artificial insemination in a dairy herd. J. Dairy Sci. 101, 1540–1553. doi: 10.3168/jds.2017-13475
Kelson V. C., Kiser J. N., Davenport K. M., Suarez E. M., Murdoch B. M., Neibergs H. L. (2024). Identifying regions of the genome associated with conception rate to the first service in holstein heifers bred by artificial insemination and as embryo transfer recipients. Genes 15, 765. doi: 10.3390/genes15060765
Kiser J. N., Clancey E., Moraes J. G., Dalton J., Burns G. W., Spencer T. E., et al. (2019a). Identification of loci associated with conception rate in primiparous Holstein Cows. BMC Genomics 20. doi: 10.1186/s12864-019-6203-2
Kiser J. N., Cornmesser M. A., Blackburn R., McGuirk S. M., Taylor J. F., Seabury C. M., et al. (2019b). Validating loci associated with bovine respiratory disease complex in pre-weaned Holstein Calves. Anim. Genet. 51, 91–94. doi: 10.1111/age.12878
Krneta J., Kroll J., Alves F., Prahst C., Sananbenesi F., Dullin C., et al. (2006). Dissociation of angiogenesis and tumorigenesis in follistatin- and activin-expressing tumors. Cancer Res. 66, 5686–5695. doi: 10.1158/0008-5472.can-05-3821
Labbé P., Faure E., Lecointe S., Le Scouarnec S., Kyndt F., Marrec M., et al. (2017). The alternatively spliced LRRFIP1 isoform-1 is a key regulator of the Wnt/β-catenin transcription pathway. Biochim. Biophys. Acta (BBA) - Mol. Cell Res. 1864, 1142–1152. doi: 10.1016/j.bbamcr.2017.03.008
Lee Y. S., Latham K., Sapienza C., Shore S. K., Litvin J. (2011). Maternal determinants of oocyte and embryo quality. Temple University, Philadelphia, PA. Philadelphia, PA. Available at: https://scholarshare.temple.edu/bitstream/handle/20.500.12613/577/Lee_temple_0225E_10588.pdf?sequence=1&isAllowed=y.
Li Q., Agno J. E., Edson M. A., Nagaraja A. K., Nagashima T., Matzuk M. M. (2011). Transforming growth factor β receptor type 1 is essential for female reproductive tract integrity and function. PLoS Genet. 7. doi: 10.1371/journal.pgen.1002320
Lima F. S., De Vries A., Risco C. A., Santos J. E. P., Thatcher W. W. (2010). Economic comparison of natural service and timed artificial insemination breeding programs in dairy cattle. J. Dairy Sci. 93, 4404–4413. doi: 10.3168/jds.2009-2789
Lin S.-Y., Craythorn R. G., O’Connor A. E., Matzuk M. M., Girling J. E., Morrison J. R., et al. (2008). Female infertility and disrupted angiogenesis are actions of specific follistatin isoforms. Mol. Endocrinol. 22, 415–429. doi: 10.1210/me.2006-0529
Lucy M. C. (2001). Reproductive loss in high-producing dairy cattle: Where will it end? J. Dairy Sci. 84, 1277–1293. doi: 10.3168/jds.s0022-0302(01)70158-0
Mao J., Feng Y., Zheng Y., Gao Y., Zhang L., Sun X., et al. (2023). GPR65 inhibits human trophoblast cell adhesion through upregulation of MYLK and downregulation of fibronectin via camp-erk signaling in a low ph environment. Cell Communication Signaling 21. doi: 10.1186/s12964-023-01249-3
Mondal S., Mor A., Reddy I. J., Nandi S., Parameswaragupta P. (2015). Effect of fibroblast growth factor 2 (FGF2) and insulin transferrin selenium (its) on in vitro maturation, fertilization and embryo development in sheep. Braz. Arch. Biol. Technol. 58, 521–525. doi: 10.1590/s1516-8913201500059
Monkley S. J., Delaney S. J., Pennisi D. J., Christiansen J. H., Wainwright B. J. (1996). Targeted disruption of the WNT2 gene results in placentation defects. Development 122, 3343–3353. doi: 10.1242/dev.122.11.3343
Monsivais D., Matzuk M. M., Pangas S. A. (2017). The TGF-β family in the reproductive tract. Cold Spring Harbor Perspect. Biol. 9. doi: 10.1101/cshperspect.a022251
Moraes J. G., Behura S. K., Geary T. W., Hansen P. J., Neibergs H. L., Spencer T. E. (2018). Uterine influences on conceptus development in fertility-classified animals. Proc. Natl. Acad. Sci. 115. doi: 10.1073/pnas.1721191115
Norman H. D., Guinan F. L., Megonial J. H., Dürr J. W. (2020). Reproductive status of cows in Dairy Herd Improvement programs and bred using artificial insemination, (2020). Available online at: https://queries.uscdcb.com/publish/dhi/current/reproall.html. March 12, 2024.
Sargolzaei M., Schenkel F. S., Jansen G. B., Schaeffer L. R. (2008). Extent of linkage disequilibrium in Holstein cattle in North America. J. Dairy Sci. 91, 2106–2117. doi: 10.3168/jds.2007-0553
Stoecklein K. S., Garcia-Guerra A., Duran B. J., Prather R. S., Ortega M. S. (2022). Actions of FGF2, Lif, and IGF1 on bovine embryo survival and conceptus elongation following slow-rate freezing. Front. Anim. Sci. 3. doi: 10.3389/fanim.2022.1040064
Stoecklein K. S., Ortega M. S., Spate L. D., Murphy C. N., Prather R. S. (2021). Improved cryopreservation of in vitro produced bovine embryos using FGF2, LIF, and IGF1. PLoS One 16. doi: 10.1371/journal.pone.0243727
Sun X., Terakawa J., Clevers H., Barker N., Daikoku T., Dey S. K. (2014). Ovarian LGR5 is critical for successful pregnancy. FASEB J. 28, 2380–2389. doi: 10.1096/fj.13-248344
The Wellcome Trust Case Control Consortium (2007). Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678. doi: 10.1038/nature05911
Torry D. S., Leavenworth J., Chang M., Maheshwari V., Groesch K., Ball E. R., et al. (2007). Angiogenesis in implantation. J. Assisted Reprod. Genet. 24, 303–315. doi: 10.1007/s10815-007-9152-7
Tríbulo P., Moss J. I., Ozawa M., Jiang Z., Tian X., Hansen P. J. (2017). Wnt regulation of embryonic development likely involves pathways independent of nuclear CTNNB1. Reproduction 153, 405–419. doi: 10.1530/rep-16-0610
Viana J. H. M. (2023). Embryo technology newsletter. Available online at: https://www.iets.org/Portals/0/Documents/Public/Committees/DRC/IETS_Data_Retrieval_Report_2022.pdf (Accessed February 2, 2024).
Wathes D. C., Fenwick M., Cheng Z., Bourne N., Llewellyn S., Morris D. G., et al. (2007). Influence of negative energy balance on cyclicity and fertility in the high producing Dairy Cow. Theriogenology 68. doi: 10.1016/j.theriogenology.2007.04.006
Weiss K. M., Clark A. G. (2002). Linkage disequilibrium and the mapping of complex human traits. Trends Genet. 18, 19–24. doi: 10.1016/s0168-9525(01)02550-1
Yallampalli C., Kondapaka S. B., Lanlua P., Wimalawansa S. J., Gangula P. R. R. (2004). Female sex steroid hormones and pregnancy regulate receptors for calcitonin gene-related peptide in rat mesenteric arteries, but not in aorta1. Biol. Reprod. 70, 1055–1062. doi: 10.1095/biolreprod.103.022467
Zhao L., Zheng X., Liu J., Zheng R., Yang R., Wang Y., et al. (2019). PPAR signaling pathway in the first trimester placenta from in vitro fertilization and embryo transfer. Biomedicine Pharmacotherapy 118, 109251. doi: 10.1016/j.biopha.2019.109251
Keywords: cattle, embryonic loss, dairy, loci, conception rate, artificial insemination, embryo transfer (ET)
Citation: Kelson VC, Kiser JN, Davenport KM, Suarez EM, Murdoch BM and Neibergs HL (2024) Genomic regions associated with embryonic loss in primiparous Holstein cows. Front. Anim. Sci. 5:1458088. doi: 10.3389/fanim.2024.1458088
Received: 01 July 2024; Accepted: 20 September 2024;
Published: 31 October 2024.
Edited by:
Gagandeep Kaur, Western University of Health Sciences, United StatesReviewed by:
Taiyong Yu, Northwest A&F University, ChinaJennifer Thomson, Montana State University, United States
Copyright © 2024 Kelson, Kiser, Davenport, Suarez, Murdoch and Neibergs. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Holly L. Neibergs, bmVpYmVyZ3NAd3N1LmVkdQ==
 Victoria C. Kelson
Victoria C. Kelson Jennifer N. Kiser
Jennifer N. Kiser Kimberly M. Davenport
Kimberly M. Davenport Emaly M. Suarez1
Emaly M. Suarez1 Brenda M. Murdoch
Brenda M. Murdoch Holly L. Neibergs
Holly L. Neibergs