- Department of Animal Sciences, Institute of Food and Agricultural Sciences, University of Florida, Gainesville, FL, United States
Introduction: Dietary fatty acids play a pivotal role in energy metabolism and various physical functions. However, the molecular mechanisms underlying the absorption of long-chain fatty acids (LCFA) in ruminant animals remain poorly understood. To address this gap, we investigated the occurrence and distribution of genes and proteins previously characterized in nonruminant models within the gastrointestinal tract of cattle.
Methods: Tissue samples from the rumen, descending duodenum, proximal jejunum, medial jejunum, distal jejunum, and distal ileum were collected at slaughter from eight Holstein steers. RT‒PCR and capillary electrophoretic-based chemiluminescence were used to determine the mRNA and protein levels of genes and proteins involved in the uptake, activation, intracellular trafficking, re-esterification, and lipoprotein assembly of LCFA.
Results and discussion: Measurable mRNA levels were found for 38 genes involved in the intestinal absorption of LCFA in all examined sections of the bovine gastrointestinal tract. The mRNA abundances of all genes were greater in the intestinal sections than in the rumen sections (p < 0.05). At the protein level, the expression of nine key transporters and enzymes, namely, CD36, SLC27A4, ACSL1, FABP1, FABP2, DGAT1, MTP, Apo-AI, and ACAT2, was detected in most of the intestinal sections. The distribution profile of these proteins in the small intestine suggested a prominent role for protein-mediated absorption of LCFA in the proximal and distal intestine in cattle. The low expression levels of most proteins in the rumen suggest little absorption of LCFA at this location in cattle. Overall, the findings from this study support the presence, in cattle, of genes and proteins involved in the intestinal absorption of LCFA described in models of nonruminants. Identifying the proteins involved in the absorption of LCFA in the small intestine is the first step in understanding how the expression of these proteins influences the capacity of the small intestine to absorb fatty acids in cattle.
1 Introduction
Dietary fats serve as a primary energy source for mammalian cells, supplying approximately 2.25 times more energy than protein or carbohydrates. In addition to their critical role in energy metabolism, fatty acids, the building blocks of fats, also serve various important physiological functions. For instance, they act as structural components within biological membranes, serve as precursors for hormones and eicosanoids, and contribute to intracellular signaling as well as transcriptional and posttranslational gene regulation (Kazantzis and Stahl, 2012; Wang et al., 2013). It is important to note that not all fatty acids can be synthesized by mammalian cells, and they must be obtained from the diet (Kaur et al., 2014). Consequently, understanding fatty acid absorption is important for maximizing nutrient utilization, enabling critical physiological processes, and optimizing energy metabolism.
The anatomy of the gastrointestinal tract differs significantly between ruminant and nonruminant animals, resulting in distinct effects on nutrient digestion and metabolism. Ruminant animals such as cattle and sheep possess a distinctive gastrointestinal tract consisting of four stomach compartments − the rumen, reticulum, omasum, and abomasum − in addition to the small and large intestine. This contrasts with nonruminant animals such as humans, rodents, and swine, whose gastrointestinal tract encompasses the stomach and intestines. In nonruminants, triglycerides constitute the primary class of fats within the diet and remain mostly unaltered as they reach the small intestine. Once in the small intestine, these triglycerides undergo hydrolysis into free fatty acids and mono-diacylglycerols before their uptake. In ruminants, dietary triglycerides, phospholipids, and galactolipids undergo extensive microbial hydrolysis and biohydrogenation in the rumen (Harrison and Leat, 1975). Therefore, fatty acids flowing to the small intestine are predominantly free saturated long-chain fatty acids (LCFA), which are believed to be absorbed exclusively through the small intestine (Doreau and Chilliard, 1997).
Ruminant and nonruminant animals exhibit substantial differences in the total tract digestibility of fatty acids. For example, digestibility in swine typically ranges from 80 to 90%, whereas in dairy cows, it falls within the range of 67 to 75% (Boerman et al., 2015; Daley et al., 2020). The variation in digestibility may arise from differences in the mechanisms of fatty acid absorption. The widespread practice of supplementing fats in ruminant diets, especially for dairy cows, emphasizes the need for a detailed understanding of the absorptive process. Such comprehension could not only enhance nutrient utilization but also contribute to the development of strategies promoting sustainable production systems and economic viability.
Although it was previously thought that fatty acids passively diffuse across cell membranes (Hamilton et al., 2002), it is now recognized that the absorption of fatty acids, particularly LCFAs, involves a more complex and intricate process. The absorption of LCFA involves specific proteins and a sequence of steps, including fatty acid uptake, activation, intracellular trafficking, re-esterification, and lipoprotein synthesis (Abumrad et al., 1998; Schwenk et al., 2010). Most, if not all, of the transporters and enzymes involved in the intestinal absorption of fatty acids have been characterized using rodents and human cell lines as models (Ockner and Manning, 1974; Stahl et al., 1999). Current knowledge about the molecular mechanisms of fatty acid absorption in ruminants is limited. The available information relies solely on gene expression and fails to encompass all sections of the gastrointestinal tract (Chao et al., 2017). Therefore, this descriptive study was designed with the objective of investigating two initial basic questions: 1) Do genes and proteins associated with the absorption of LCFA in nonruminant models also manifest in cattle? 2) What is the distribution profile of these genes and proteins within the gastrointestinal tract of cattle? We hypothesize that genes and proteins responsible for LCFA absorption, as described in nonruminant models, will be expressed in cattle. Additionally, we postulate that variations in their distribution along the gastrointestinal tract of cattle are due to inherent differences in fat digestion.
2 Materials and methods
2.1 Tissue harvesting and processing
Tissue samples were collected from eight Holstein steers at the slaughterhouse of the University of Florida Department of Animal Sciences. The animals had an average body weight of 490 ± 32 kg and grazed on Bahiagrass pasture while receiving a mineral supplement for 40 days prior to slaughter. The animals were fasted for approximately 12 hours before harvest, stunned by captive bolt, killed by exsanguination, and immediately eviscerated. Samples were taken from the rumen sacs (cranial, ventral, caudoventral, dorsal, and caudodorsal), descending duodenum (15-30 cm distal to the pylorus), proximal jejunum (30 cm distal to the end of duodenum), medial jejunum (at the midpoint between the end of the duodenum and the ileocecal fold), distal jejunum (50 cm before the ileocecal fold), and distal ileum (15 cm before the ileocecal junction). Excised segments were rinsed extensively with phosphate-buffered saline to remove any digesta residues. Epithelial tissue was dissected from muscle layers and connective tissue using forceps, and samples of epithelial tissue (approximately 2 g) from each gastrointestinal section were transferred to cryo-tubes, snap frozen in liquid nitrogen, and stored at −80°C until analysis.
2.2 Laboratory analyses
2.2.1 RNA isolation, cDNA synthesis, and RT−PCR
Total RNA was isolated from 50 mg of epithelial tissue using the miRNeasy kit (Cat. no. 7217084, Qiagen) following the manufacturers’ protocols. Briefly, genomic DNA was removed via an on-column method using DNaseI digestion mix (Cat. no. 79254, Qiagen), and RNA was eluted with 50 µL of nuclease-free water. RNA purity and integrity were assessed using a UV−Vis spectrophotometer (NanoDrop ND-1000, Thermo Scientific) and an automated electrophoresis system (4200 TapeStation, Agilent), respectively. All the samples exhibited a purity with an A260/A280 ratio above 2.0, and their RNA integrity number (RIN) was equal to or greater than 7.8. The RNA was diluted to 100 ng/μL with DNase/RNase-free water for cDNA synthesis. Samples of diluted RNA of the five rumen sacs were pooled equally by volume within each animal before cDNA synthesis. cDNA was synthesized using a commercial kit (Cat. no. AB14534A, Thermo Scientific) following the manufacturer’s protocol, and the reaction was performed in a thermal cycler at 42°C for 30 min, followed by 95°C for 2 min. Thirty-eight primers of genes involved in the intestinal absorption of LCFAs were designed using the NCBI database and aligned against the NCBI database through BLASTN to determine primer compatibility with already annotated sequences of the corresponding genes in both databases. Primers were validated by evaluating the amplification efficiency and met the MIQE guidelines of r2 > 0.98 and an efficiency of 90–110% (Bustin et al., 2009).
Gene expression across the cattle gastrointestinal tissues was analyzed by SYBR Green quantitative real-time PCR using an RT−PCR cycler (CFX96, Bio-Rad) and a three-step protocol following the procedure outlined by Dickson et al. (2020) with modifications to the reaction temperature conditions. These modifications included an initial denaturation step at 95°C for 20 seconds, followed by 40 cycles of denaturation at 95°C for 5 seconds, annealing at 60°C for 10 seconds, and a final extension at 72°C for 20 seconds. The samples were run in duplicate, and three internal control genes (β-actin, GAPDH, and RPS9) were used to normalize the data. Internal control genes were stably expressed across all tissues. Normalized relative quantities (fold change) for genes were calculated according to Pfaffl (2001). The expression values for each tissue within the animal were normalized against the values of the duodenum of the corresponding animal. Details about primer sequence and real-time PCR performance can be found in the Supplementary Tables S1 and S2.
2.2.2 Protein expression
Tissue samples were suspended in a protein extraction reagent (Cat. no. 87787; Fisher Scientific) containing a protease and phosphatase inhibiting cocktail (Cat. no. 78442; Fisher Scientific). Subsequently, the samples were homogenized (Tissue-Tearor 985370, BioSpec Products), gently mixed on a rocker for 1 hour at 4°C, and centrifuged at 5,000 rpm for 20 min at 4°C. The supernatant was collected and used to determine the total protein concentration by employing a Pierce BCA protein assay kit (Cat. no. 23227; Thermo Scientific). Tissue samples for each gastrointestinal section were pooled from all animals and stored at –80°C until analysis. Protein expression of the long-chain fatty acid transport protein 4 (SLC27A4), fatty acid translocase (CD36), fatty acid binding protein, liver (FABP1), fatty acid binding protein, intestinal (FABP2), fatty acid binding protein, heart (FABP3), acyl-CoA synthetase long chain family 1 (ACSL1), microsomal triglyceride transfer protein (MTP), acyl-CoA-cholesterol acyltransferase 2 (ACAT2), and apolipoprotein A-I (Apo-AI) was determined through capillary size-based chemiluminescence using a Simple Western Protein Analyzer (Abby, ProteinSimple). The procedures were performed according to the manufacturer’s recommendations using a 12-230 kDa Separation Module (Cat. no. SM-W004; ProteinSimple). Briefly, tissue lysates were diluted to the desired concentration using sample buffer supplied with the separation module. Subsequently, they were combined with dithiothreitol and the fluorescent master mix and heated at 95°C for 5 min. The denatured samples, blocking reagent (antibody diluent 2 or milk free), primary antibodies (in antibody diluent), streptavidin horseradish peroxidase, secondary antibodies, and chemiluminescent substrates were added to a 25-well plate. Afterwards, the plate and a capillary cartridge were inserted into the instrument, and the following instrument settings were utilized: stacking and separation at 475 volts for 30 min, blocking for 5 min, immunoprobing with primary and secondary antibodies for 30 min each, and luminol/peroxide chemiluminescence detection for approximately 30 min. The optimal exposure time for the measurement of chemiluminescent signals of each primary antibody was determined by analyzing the data at different exposure times to identify substrate depletion. The antibodies were used at saturation concentrations. To discriminate low target protein signals from background noise, chemiluminescence signals were quality-checked using the following criteria: the peak signal-to-noise ratio had to be equal to or greater than 10, and the baseline (expressed as a percentage of the peak area) had to be less than 20% (Beekman et al., 2018). Chemiluminescence peaks of target proteins were normalized to within-sample β-actin expression. Normalized data are expressed as arbitrary units of chemiluminescence (AU). Electropherograms for individual proteins are included in the Supplementary Figure S1. Information about the antibodies and controls used are included in the Supplementary Table S3.
2.3 Statistical analysis
The data were analyzed using the MIXED procedure of SAS 9.4 (SAS Institute) according to the following model:
where Yij is the variable of interest, µ is the overall mean, ai is the random effect of animal (i = 1–8), Mj is the fixed effect of tissue, and eij is the residual error. The data were log-2 transformed before the analysis and then log-back transformed afterwards. The normality of the residuals was checked with normal probability and box plots, and the homogeneity of variances was determined with plots of the residuals and predicted values. The data are presented as least-squares means with standard errors of the mean. Significance was declared at p≤ 0.05.
3 Results
3.1 Expression of genes involved in long-chain fatty acid uptake
The mRNA expression of SLC27A gene family members 1 to 6, CD36, and SRB1 was greater in the small intestine than in the rumen (p< 0.01, Figure 1). Among the intestinal sections, SLC27A1 exhibited greater expression in the duodenum than in the ileum (p< 0.01). The mRNA levels of SLC27A2 were greater in the duodenum, proximal and distal jejunum, and ileum than in the medial jejunum (p< 0.01). SLC27A3 and SLC27A4 were more highly expressed in the distal jejunum than in the proximal and medial jejunum (p< 0.01). The SLC27A5 mRNA abundance was greater in the duodenum than in the medial and distal jejunum and ileum (p< 0.01). The expression of SLC27A6 was greater in the duodenum than in the jejunal sections (p< 0.01). The CD36 levels were similar among the small intestine sections (p< 0.01). The mRNA level of SRB1 was lower in the medial jejunum than in the other intestinal sections (p< 0.01).
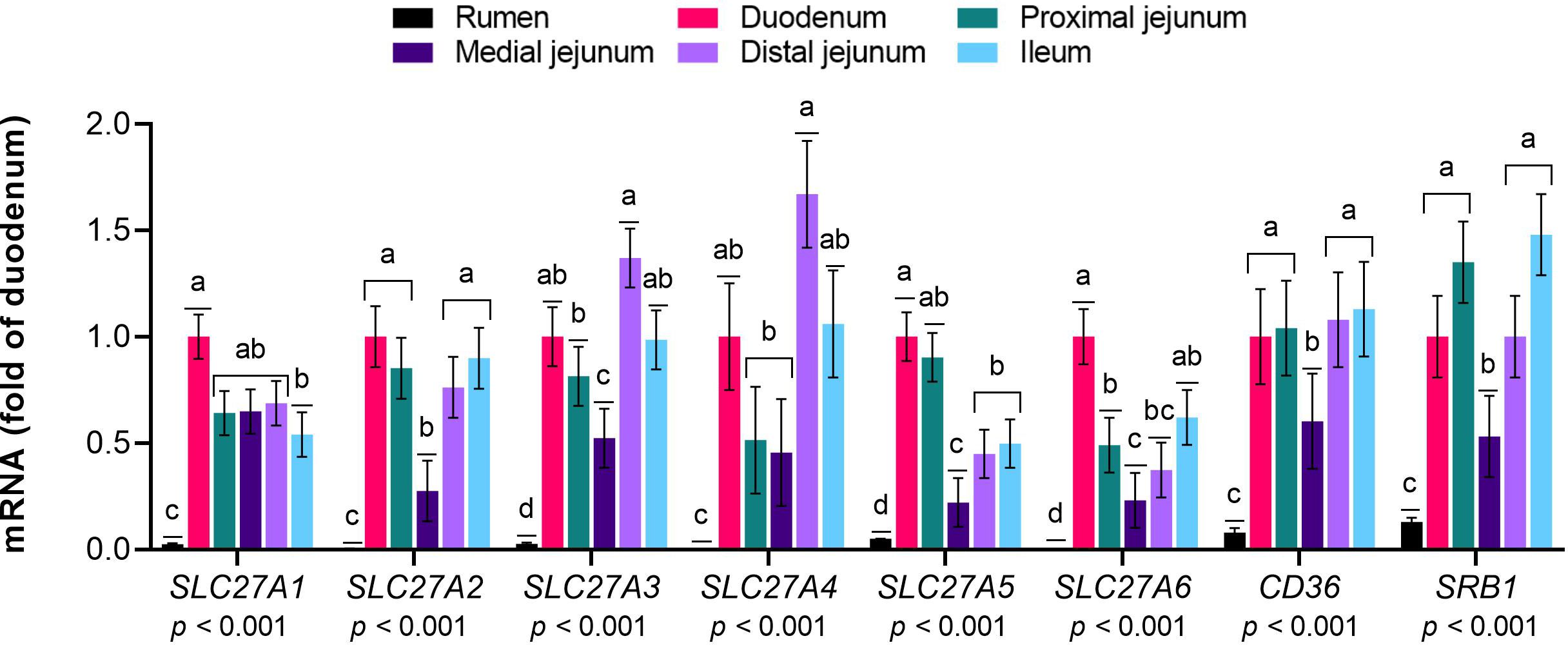
Figure 1 Expression of genes responsible for the uptake of long-chain fatty acids (LCFA) in the gastrointestinal tract of cattle. Relative expression (fold change in the duodenum) of genes involved in the uptake of LCFA in the rumen, duodenum, proximal jejunum, medial jejunum, distal jejunum, and ileum of cattle. The values are the means ± SEM. For the tissue effect, means without a common letter differ (p< 0.05).
3.2 Expression of genes involved in intracellular trafficking and activation of long-chain fatty acids
The mRNA expression levels of FABP family members 2 to 5 and DBI (Figure 2) and ACSL family members 1, 3, 4, 5 and 6 (Figure 3) were greater in the intestinal sections than in the rumen (p≤ 0.01). Among the small intestine sections, FABP1 was predominantly expressed in the duodenum, while the FABP2 levels were greater in the duodenum and ileum than in the other sections (p< 0.01). The mRNA levels of FABP3 were greater in the duodenum, proximal jejunum and ileum than in the medial jejunum (p< 0.01). The expression of FABP4 was greater in the duodenum and proximal jejunum than in the ileum and medial jejunum (p< 0.01). FABP5 expression was greater in the ileum than in the medial jejunum (p< 0.01). The mRNA abundance of FABP6 was extremely low throughout the gastrointestinal tract, with slightly greater levels in the duodenum than in the other intestinal sections (p= 0.01). The expression of ACSL1 and ACSL5 was lower in the medial jejunum than in the other intestinal sections (p< 0.01). DBI, ACSL3, ACSL4, and ACSL6 were expressed at similar levels across the intestinal sections (p< 0.01).
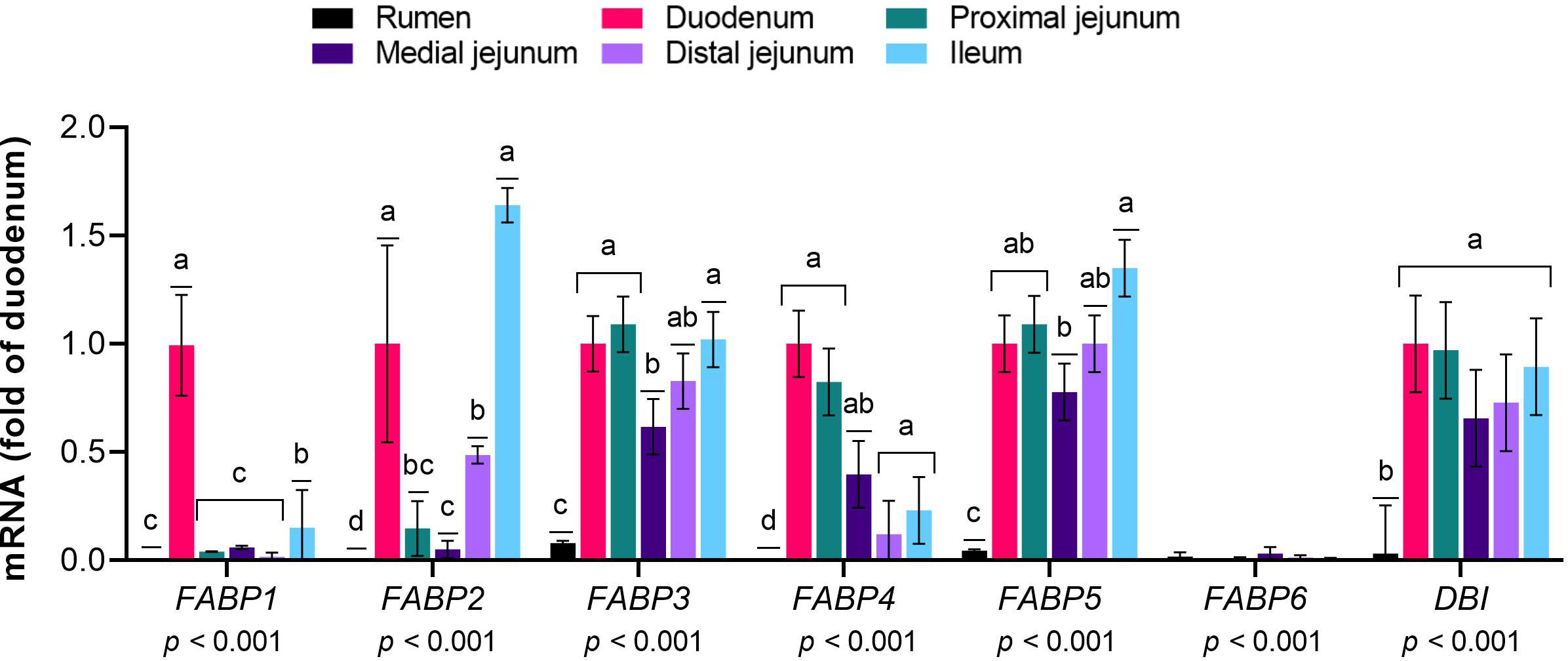
Figure 2 Expression of genes responsible for the intracellular trafficking of long-chain fatty acids (LCFA) in the gastrointestinal tract of cattle. Relative expression (fold change in the duodenum) of genes involved in the intracellular trafficking of LCFA in the rumen, duodenum, proximal jejunum, medial jejunum, distal jejunum, and ileum of cattle. The values are the means ± SEM. For the tissue effect, means without a common letter differ (p< 0.05).
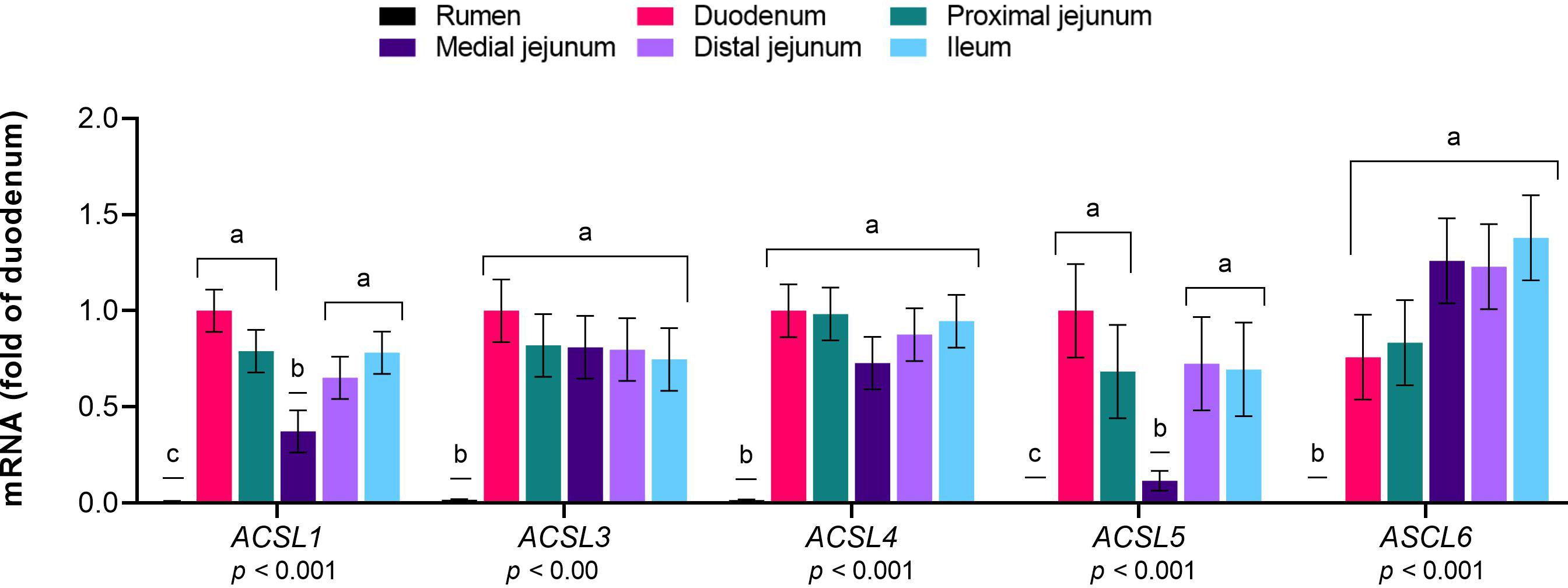
Figure 3 Expression of genes responsible for the activation of long-chain fatty acids (LCFA) in the gastrointestinal tract of cattle. Relative expression (fold change in the duodenum) of genes involved in the activation of LCFA to fatty acyl-CoA in the rumen, duodenum, proximal jejunum, medial jejunum, distal jejunum, and ileum of cattle. The values are the means ± SEM. For the tissue effect, means without a common letter differ (p< 0.05).
3.3 Expression of genes involved in re-esterification of long-chain fatty acids
The expression levels of GPAT1, GPAT3, AGPAT family members 1, 2, 3, and 5, LPIN 1 to 3, and DGAT 1 and 2 were greater in the intestinal sections than in the rumen (p≤ 0.01, Figure 4). Within the intestinal segments, GPAT1 and GPAT3 were more highly expressed in the ileum than in the duodenum (p< 0.01). AGPAT1 was expressed at higher levels in the ileum than in the medial jejunum (p< 0.01), while AGPAT2 was expressed at constant levels in intestinal sections (p= 0.01). AGPAT 3 and 5 exhibited greater mRNA abundance in the ileum than in the duodenum (p< 0.01). LPIN1 expression was greater in the ileum than in the duodenum and medial jejunum (p< 0.01), and LPIN2 levels were greater in the ileum than in the jejunal sections (p< 0.01). LPIN3 exhibited greater expression in the duodenum than in the jejunal sections (p< 0.01). DGAT1 mRNA levels were lower in the medial jejunum than in the other intestinal sections (p< 0.01), and DGAT2 showed similar expression in the small intestine (p< 0.01).
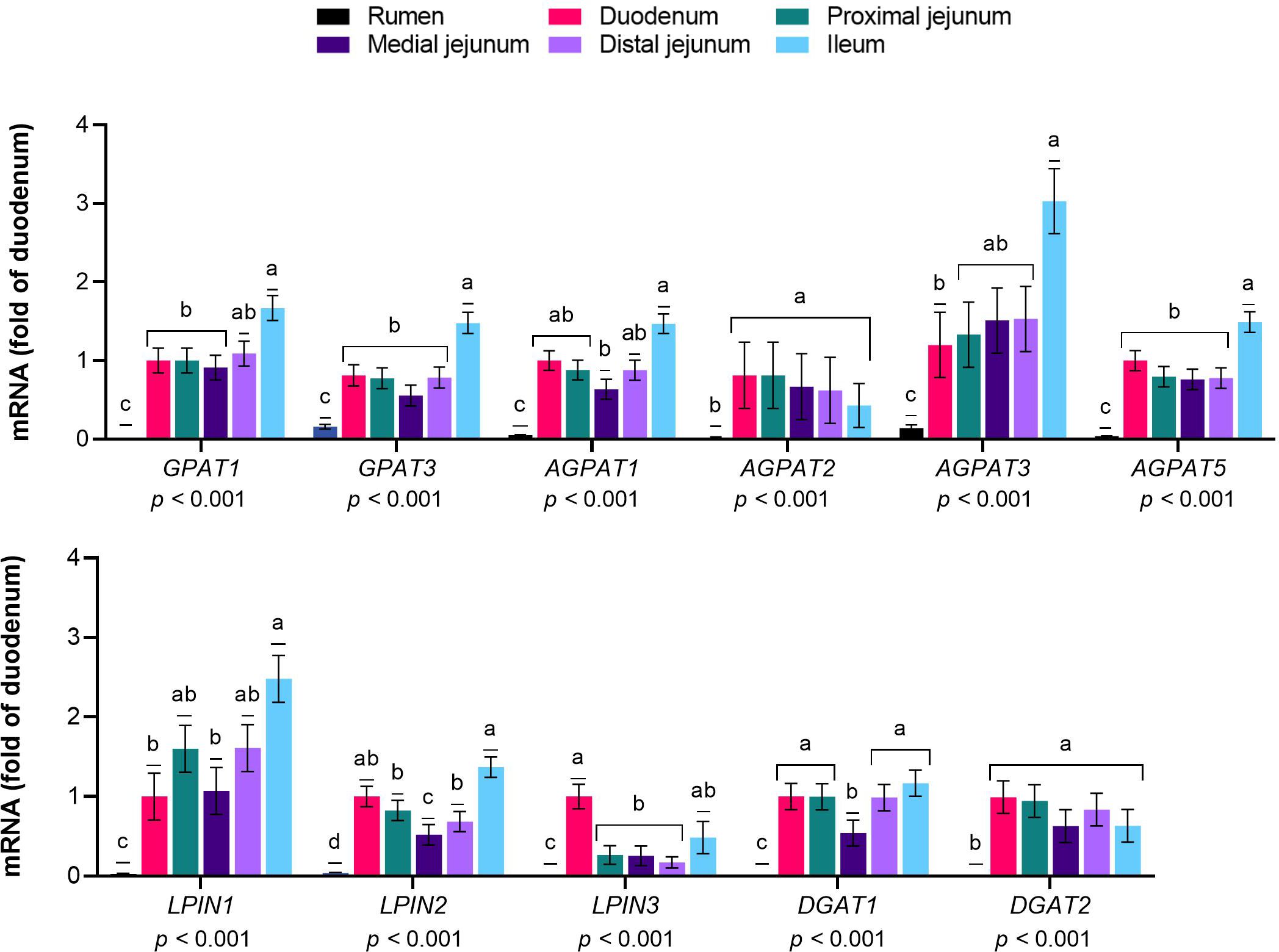
Figure 4 Expression of genes responsible for the re-esterification of long-chain fatty acids (LCFA) in the gastrointestinal tract of cattle. Relative expression (fold change in the duodenum) of genes involved in the re-esterification of LCFA into triglycerides/phospholipids in the rumen, duodenum, proximal jejunum, medial jejunum, distal jejunum, and ileum of cattle. The values are the means ± SEM. For the tissue effect, means without a common letter differ (p< 0.05).
3.4 Expression of genes involved in lipoprotein synthesis
The mRNA expression of MTP, APO family members A1, A4, C3, and D, and ACAT2 was greater in the intestinal sections than in the rumen (p< 0.05, Figure 5). Within the small intestine, MTP expression was lower in the medial jejunum than in the other sections (p< 0.01). APOA1 levels were similar among the intestinal sections (p< 0.01), and APOA4 and APOC3 were more highly expressed in the ileum than in the medial jejunum (p< 0.01 and p= 0.02,, respectively). No differences were observed in the APOB levels between the evaluated sections (p = 0.20). APOD was expressed at similar levels in the intestinal sections (p< 0.01), and ACAT2 was more highly expressed in the duodenum than in the jejunum (p< 0.01).
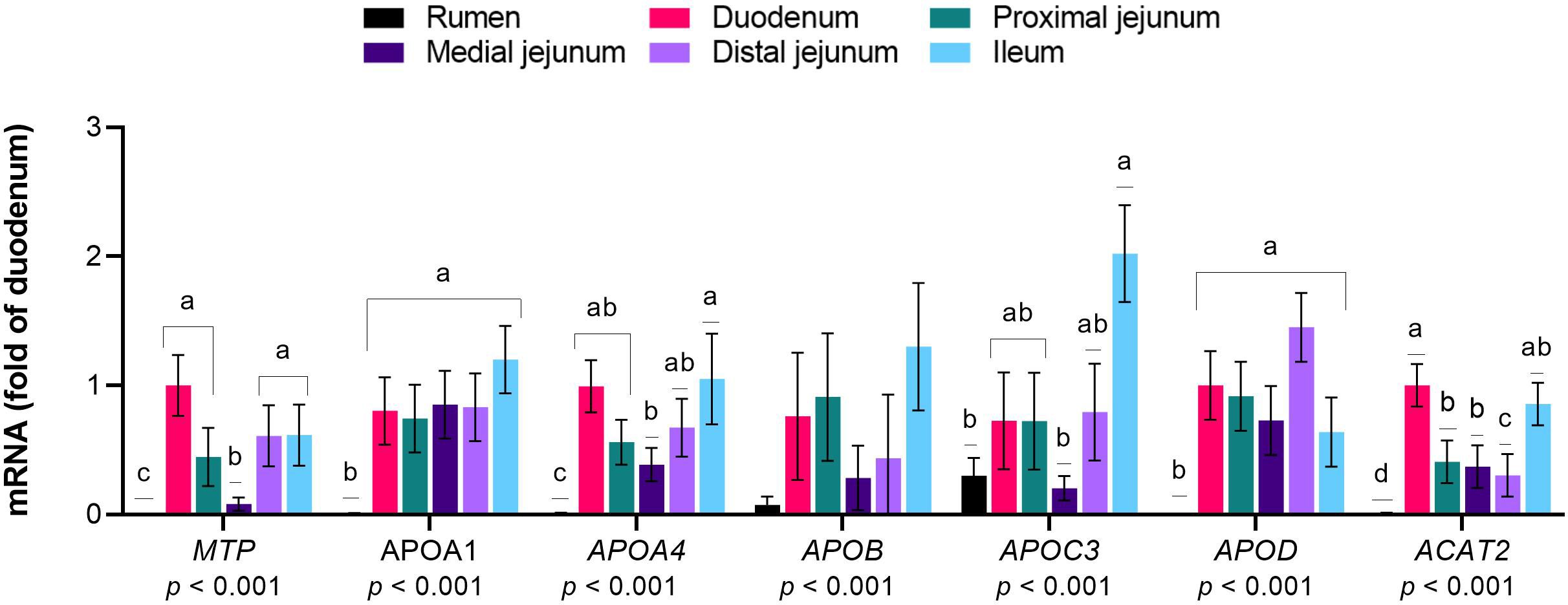
Figure 5 Expression of genes responsible for the synthesis of lipoproteins from long-chain fatty acids (LCFA) in the gastrointestinal tract of cattle. Relative expression (fold change in the duodenum) of genes involved in the incorporation of LCFA into lipoproteins in the rumen, duodenum, proximal jejunum, medial jejunum, distal jejunum, and ileum of cattle. The values are the means ± SEM. For the tissue effect, means without a common letter differ (p< 0.05).
3.5 Protein expression
To further determine whether the expression of specific genes observed via real-time PCR analysis was reflected at the protein level, we performed capillary electrophoretic-based chemiluminescence in the same sections of the cattle gastrointestinal tract. We focused this analysis on key transporters and enzymes implicated in the main steps of the LCFA absorption process according to models from studies in nonruminants. As explained in the methods section, samples were pooled from all animals by tissue for this analysis; thus, statistical comparisons among sections were not possible. An immunoreactive signal against CD36 was detected in the rumen and proximal and distal jejunum (Figure 6). SLC27A4 was expressed longitudinally throughout all examined intestinal sections. The protein expression of ACSL1 was detected across all jejunal sections and the ileum. For FABP1, very strong signals were identified across all small intestine sections. FABP2 was expressed at low levels in all small intestine sections. MTP was expressed at relatively low levels in the duodenum, proximal and medial jejunum, and ileum, with no signal in the rumen or distal jejunum. For DGAT1 and Apo-AI, immunoreactive signals were detected in all examined sections.
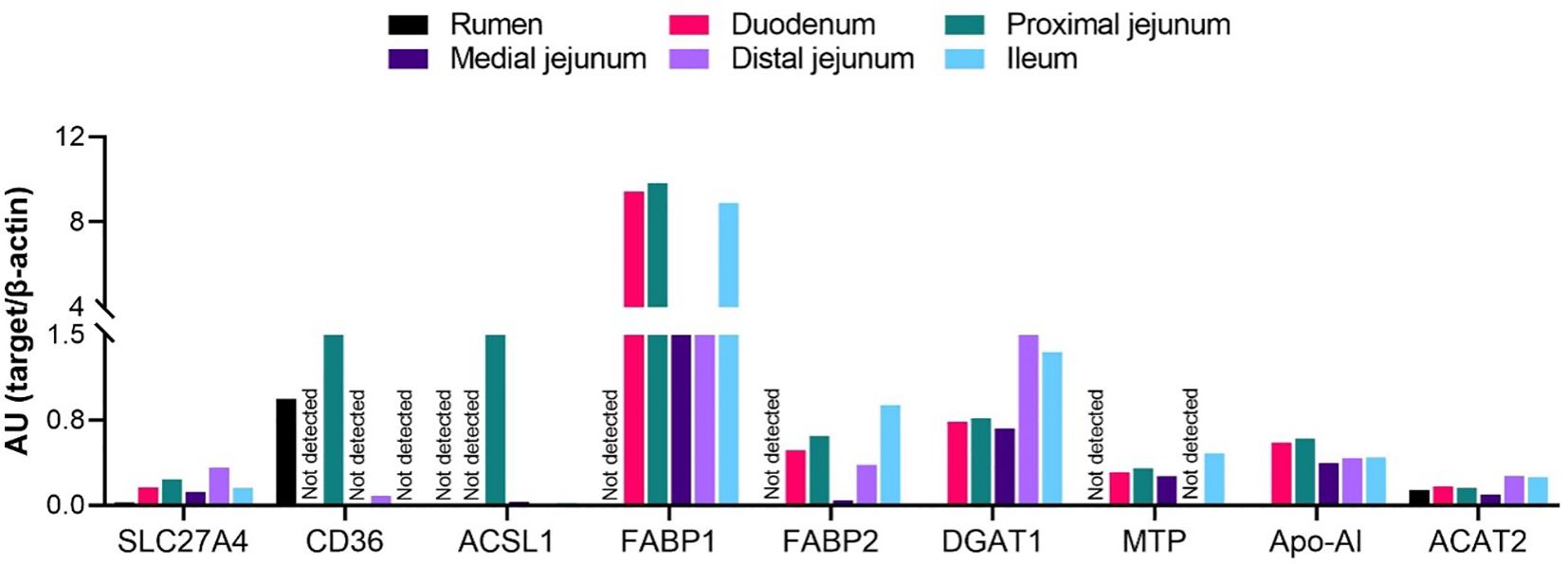
Figure 6 Expression of proteins involved in the absorption of long-chain fatty acids in the gastrointestinal tract of cattle. Protein expression of SLC27A4, CD36, ACSL1, FABP1-2, DGAT1, MTP, Apo-AI, and ACAT2 in the rumen, duodenum, proximal jejunum, medial jejunum, distal jejunum, and ileum of cattle. Protein expression data were normalized to the expression of β-actin and are expressed as arbitrary units (AU) of chemiluminescence. Statistical comparisons among tissues were not possible due to n=1.
4 Discussion
The central role of the intestine in regulating whole-body lipid metabolism is underscored by its ability to absorb luminal fatty acids (Dedousis et al., 2022). Consequently, the significance of intestinal mechanisms for fatty acid absorption becomes evident in this context. The present study offers a report on molecular mechanisms associated with absorption of LCFA in ruminants. First, we showed that the gastrointestinal tract of cattle expresses genes involved in all the main steps of the intestinal absorption of LCFA described in nonruminant models, confirming our hypothesis. Except for the rumen, whose expression levels were minimal for nearly the totality of genes, all intestinal sections expressed the whole set of genes investigated. In sequence, we showed that the expression of key genes implicated in intestinal LCFA absorption translates to the protein level in the gastrointestinal tract of cattle, strengthening the notion that ruminants and nonruminants share similarities in protein-mediated fatty acid absorption processes. However, the distribution profile of most proteins along the small intestine did not parallel that observed in nonruminants, indicating species-specific expression profiles.
The intestinal uptake of LCFA comprises their entry from the lumen into enterocytes (Wang et al., 2013). Studies conducted in ruminants fitted with intestinal cannulas demonstrated that the greatest disappearance of free LCFA, as well as of triglycerides and phospholipids, occurs in the medial to distal jejunum, and by the time the digesta reaches the ileum, the assimilation of these fatty acids is almost complete (Lennox and Garton, 1968). Our RT−PCR results revealed that for most uptake genes, the mRNA levels in the ileum were similar to those in the duodenum and proximal jejunum. Similarly, at the protein level, the expression levels of SCL27A4 and FABP1 and 2 were comparable between these sections, suggesting that the distal intestine plays an important role in the uptake of LCFA in ruminants. In rats, under physiological conditions, fat is primarily taken up in the jejunum, but at high dietary fat concentrations, the ileum is called upon to take up greater amounts of fat (Booth et al., 1961). It is plausible that the physiological importance of genes and proteins involved in the uptake of LCFA in the distal region of the small intestine is related to a similar function in cattle. Recent evidence in nonruminants supports two protein-mediated putative pathways for the uptake of LCFA: 1) CD36 alone or in association with enterocyte FABP accepts LCFA at the brush border membrane, transporting them to the inner side of enterocytes, and 2) LCFA are transported by SLC27A4 (Lagakos et al., 2011; Wang et al., 2013). We detected the CD36 protein in the proximal (at high levels) and distal (very low levels) portions of the jejunum and at relatively high levels in the rumen. The presence of CD36 at higher levels in the proximal jejunum is consistent with published data in humans and rodents reporting CD36 functions in fat uptake to be localized more proximally in the intestine (Lobo et al., 2001; Nassir et al., 2007). The CD36 exhibits high affinity for LCFA; however, its Km for fatty acid uptake by the enterocytes is very low, indicating that it will be rapidly saturated (Nassir et al., 2007). The functional significance of the presence of protein CD36 in the rumen is unknown, but because it is a transmembrane protein with broad ligand specificity, its function in this organ may not necessarily relate to lipid absorption or metabolism. Previous studies have identified high levels of CD36 mRNA in the rumen of cattle (Kong et al., 2016) and dairy goats (Cao et al., 2016). A role of nutrition in the regulation of CD36 has been reported, as its protein levels were upregulated in the small intestine of pigs fed a high-fat diet (33% oleic acid and 25% palmitic acid) compared to those in a control low-fat group (Wang et al., 2023). In contrast, the intraduodenal infusion of oleic acid resulted in a rapid and marked downregulation of CD36 mRNA levels, and a dramatic reduction in oleic acid uptake by isolated enterocytes in rats (Chen et al., 2001) suggesting a role of CD36 in the uptake of this LCFA. However, the substrate preference of CD36 for LCFA remains elusive and should be the focus of future research.
The SLC27A family comprises six proteins that mediate the uptake of long- and very-long-chain (≥ C22) fatty acids (Stahl, 2004). Among them, SLC27A4 has been shown to predominate in the small intestine in mice, with higher expression levels in the jejunum and ileum (Anderson and Stahl, 2013). Our results revealed mRNAs for SLC27A 1 to 6 across all examined intestinal sections, similar to what was observed at the protein level, in which SLC27A4 was identified throughout the small intestine. In mice, SLC27A4 is expressed only on the apical side of enterocytes, indicating a role for this protein in the uptake of luminal LCFA rather than its import from the blood in nonruminants (Stahl et al., 1999). This might be associated with the low expression levels of SLC27A4 in our study since the animals were withheld from food for 12 h before harvest, and the luminal uptake of fatty acids was likely low. In enterocytes isolated from mice, SLC27A4 showed a strong substrate preference for palmitic acid and, to a lesser extent, for oleic and linolenic acid (Stahl et al., 1999). Additionally, the incubation of enterocytes with increasing concentrations of an SLC27A4 antisense oligonucleotide − which restricts the expression of SLC27A4 − resulted in proportionate decreases in the uptake of palmitic and oleic acid in mice, confirming a major role of SLC27A4 in the uptake of these LCFAs (Stahl et al., 1999; Zhou et al., 2010). In addition to CD36 and SLC27A4, FABP have also been implicated in the uptake of LCFA. Enterocytes contain high levels of two members of this family of proteins with high affinity for LCFA: liver-FABP and intestinal-FABP (FABP1 and FABP2, respectively) (Storch and Thumser, 2010). FABP1 expression is maximal at a more proximal location and almost absent in the ileum in mice, whereas FABP2 is essentially found in the ileum (Gordon et al., 1985). Our RT−PCR analysis detected mRNAs for FABP1−6 across all examined intestinal sections, similar to the results observed at the protein level for FABP1 and FABP2. The protein expression of FABP1 throughout the intestine was substantially greater than that of FABP2, but the distribution profile revealed that FABP 1-2 was expressed at comparable levels in the proximal and distal intestine sections. These findings are consistent with previous data in cattle (Hayashi et al., 2013) and confirm that enterocyte FABP are also located in the distal intestine in cows, unlike in nonruminants. Enterocytes are unique in that they are exposed to fatty acids on their apical (lumen) and basolateral (blood) surfaces and metabolize them differently depending upon the entry site in the cells (Alpers et al., 2000). FABP1-2 are localized throughout the enterocyte cytosol but participate in specific pathways of intestinal lipid metabolism (Lagakos et al., 2011). FABP1 seems to bind and target LCFA for oxidation, regardless of their entry point into enterocytes, while FABP2 mainly binds LCFA on the apical side, directing them to triglyceride synthesis (Alpers et al., 2000; Lagakos et al., 2011). This might be attributed to the high expression levels of FABP1 relative to those of FABP2 in the present research, given that the animals were in a fasted state at harvest. FABP 1-2 bind stearic acid with greater affinity than oleic and linolenic acid and palmitic acid with much lower affinity (Nemecz et al., 1991; Smathers and Petersen, 2011).
Once at the inner side of the membrane, the processing of nonesterified free fatty acids by the enterocyte is initiated by esterification to CoA to form fatty acyl-CoA. This reaction, called activation, traps fatty acids within the cell and enables an otherwise nonreactive fatty acid to be used in fatty acid synthesis and oxidation and phospholipid synthesis (Watkins, 1997). Enzymes that catalyze this reaction in LCFA belong mainly to the acyl-CoA synthetase family specific for long chains (ACSL, members 1, 3, 4, 5, and 6), although some members of the SLC27A family (including SLC27A4) also display long-chain acyl-CoA synthetase activity (Anderson and Stahl, 2013). In this study, mRNAs encoding all five ACSL were detected throughout the examined intestinal segments. At the protein level, ACSL1 was expressed at relatively high levels in the proximal jejunum but at relatively low levels in the medial and distal intestinal sections. In ruminants, ACSL1 was shown to be the most abundant ACSL isoform in the mammary tissue of dairy cattle and was also detected in the duodenum of sheep (Bionaz and Loor, 2008; Chao et al., 2017). Conversely, in rodents, ACLS5 is the major intestinal synthetase, while ACSL1 is expressed at very low levels (Suzuki et al., 1990; Meller et al., 2013). The newly synthesized fatty acyl-CoAs that are not immediately targeted to biosynthetic or catabolic pathways are bound to the acyl-CoA binding protein (DBI) (Knudsen et al., 2000). In the present study, mRNA expression of this gene was detected in every examined intestinal region. DBI binds long-chain acyl-CoA esters with high affinity and acts along with FABP and ACAT2 as a reservoir, lowering the concentration of free cytosolic acyl-CoA (Knudsen et al., 2000).
In eukaryotes, fatty acyl-CoA is rapidly re-esterified to triglycerides during the postprandial period through two major pathways, the de novo glycerol phosphate pathway and the monoacylglycerol pathway (Coleman and Lee, 2004). Monoacylglycerol is the predominant pathway in the small intestine of humans and nonruminants (Yu et al., 2018); however, because ruminants absorb essentially no monoacylglycerols (Bauchart, 1993) we focused our RT−PCR analysis only on genes of the glycerol phosphate pathway. In this pathway, glycerol-3-phosphate acyltransferases (GPAT) catalyze the initial reaction by transferring a fatty acid from acyl-CoA to glycerol-3-phosphate, generating lysophosphatidic acid, which is further acylated at the sn-2 position of the glycerol backbone to form phosphatidic acid via the action of the 1-acyl glycerol-3-phosphate acyltransferase (AGPAT) family. At this point, phosphatidic acid can be dephosphorylated by phosphatidic acid phosphatases (also known as lipin) to form diacylglycerol or be directed to phospholipid synthesis. Diacylglycerol receives one last fatty acid from acyl-CoA to produce triglycerides through the action of acyl-CoA diacylglycerol acyltransferases (DGAT) (Yamashita et al., 2014). The detection of mRNAs encoding genes involved in all steps of this pathway and of the protein DGAT1 in the present study indicates its physiological importance in ruminants, with a prominent role in the distal intestine, since most genes and the protein DGAT1 were expressed most distally. These findings point to differences with respect to the nonruminant model, in which the expression of enzymes of the de novo glycerol phosphate pathway is highest in the proximal intestine (Cases et al., 1998; Khatun et al., 2016). GPAT3 (microsomal GPAT) is the rate-limiting enzyme in the glycerol phosphate pathway due to its low specific activity and uses C18:1-CoA as the preferred substrate compared to C16:0-CoA, C14:0-CoA, and C18:0-CoA. The levels of GPAT3 were slightly lower than those of GPAT1 in our study, which is likely associated with the fasting state at harvest, since GPAT1 selectively assigns acyl-CoA to β-oxidation or triglycerides synthesis (Yu et al., 2018). DGAT, which are coexpressed with DGAT1 and DGAT2, catalyze the only irreversible step in triglycerides synthesis and show a preference for C18:1-CoA over C16:0-CoA (Cases et al., 2001).
Triglycerides and phospholipids formed in enterocytes are available for lipoprotein or fat droplet synthesis. For lipoproteins to assemble, substrates need to be transferred from the membrane to the cisternae of the endoplasmic reticulum. The microsomal triglyceride transfers large subunit (MTP, 97 kDa) protein transfers triglycerides, cholesteryl esters, and phospholipids, especially in the synthesis of ApoB-containing lipoproteins (Hussain et al., 2012). These lipoproteins, which include chylomicrons, low-density lipoproteins (LDL) and very-low density proteins (VLDL), are the main lipid transport vehicles in nonruminants (Sirwi and Hussain, 2018). The relatively low MTP protein levels found in the present research might be attributed to the fact that, unlike nonruminants, bovine lipoproteins are mainly not ApoB-based. In dairy cattle, HDL accounts for the majority of plasma lipoproteins (~80%), with lower amounts of LDL and VLDL (Bauchart, 1993; Duran et al., 2021; Behling-Kelly and Wong, 2022). Cholesteryl esters and phospholipids represent approximately 90% of the lipid content in bovine high-density lipoproteins (HDL), and Apo-AI is the core structural protein, although other apoproteins are associated with these particles, including Apo A-IV and C-III (Raphael et al., 1973; Feingold, 2000). Cholesteryl esters, triglycerides, and phospholipids are incorporated into LDL and VLDL (Raphael et al., 1973). The protein expression of Apo A-I was comparable throughout all intestinal sections in the present study, and at the gene level, the mRNA levels of A-IV, B, C-III, and D were also detected. Our results revealed that the protein ACAT2 was expressed in parallel with the protein Apo-AI in all the examined sections. Notably, the amount of ACAT2 in the rumen was comparable to that in the small intestine, although the functional significance of this expression is unknown. ACAT2 is the main enzyme responsible for esterification and the incorporation of cholesterol into lipoproteins in enterocytes in nonruminants.
It is very important to highlight that, although all main proteins involved in the intestinal absorption of LCFA described in models for nonruminants were detected in cattle in this study, the exact function of these proteins in the molecular mechanisms of intestinal LCFA absorption and how they are regulated under normal physiological conditions in ruminants remains largely unknown.
5 Conclusion
The findings from this study indicate the presence of genes and proteins involved in the intestinal absorption of LCFA in cattle described in nonruminant models. The protein distribution profile points toward a prominent role of the proximal and distal sections of the small intestine in protein-mediated absorption of LCFA in cattle. Future investigations are needed to address the influence of individual fatty acids and their intestinal flow level on the expression of proteins involved in the intestinal absorption of LCFA and how the expression levels of these proteins influence the capacity of the small intestine to absorb fatty acids in ruminants.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because samples were collected after animals were slaughtered.
Author contributions
FH: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. FB: Conceptualization, Data curation, Formal analysis, Methodology, Project administration, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors would like to express their gratitude to Daniel de Oliveira, Bruno Emanuel Barreta, and Dérick Cantarelli Rosler, all from the University of Florida, for their assistance in collecting the samples, and to Dr. Milerky Perdomo for her assistance with the RT-PCR.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial and financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fanim.2024.1435257/full#supplementary-material
References
Abumrad N., Harmon C., Ibrahimi A. (1998). Membrane transport of long-chain fatty acids: evidence for a facilitated process. J. Lipid Res. 39, 2309–2318. doi: 10.1016/S0022-2275(20)33310-1
Alpers D. H., Bass N. M., Engle M. J., Deschryver-Kecskemeti K. (2000). Intestinal fatty acid binding protein may favor differential apical fatty acid binding in the intestine. Biochim. Biophys. Acta 1483, 352–362. doi: 10.1016/S1388-1981(99)00200-0
Anderson C. M., Stahl A. (2013). SLC27 fatty acid transport proteins. Mol. Aspects Med. 34, 516–528. doi: 10.1016/j.mam.2012.07.010
Bauchart D. (1993). Lipid absorption and transport in ruminants. J. Dairy Sci. 76, 3864–3881. doi: 10.3168/jds.S0022-0302(93)77728-0
Beekman C., Janson A. A., Baghat A., Van Deutekom J. C., Datson N. A. (2018). Use of capillary Western immunoassay (Wes) for quantification of dystrophin levels in skeletal muscle of healthy controls and individuals with Becker and Duchenne muscular dystrophy. PloS One 13, e0195850. doi: 10.1371/journal.pone.0195850
Behling-Kelly E., Wong C. (2022). Agarose gel electrophoresis determination of bovine lipoproteins compared with a wet chemistry method. JDS Commun. 3, 373–376. doi: 10.3168/jdsc.2022-0223
Bionaz M., Loor J. J. (2008). ACSL1, AGPAT6, FABP3, LPIN1, and SLC27A6 are the most abundant isoforms in bovine mammary tissue and their expression is affected by stage of lactation. J. Nutri 138, 1019–1024. doi: 10.1093/jn/138.6.1019
Boerman J. P., Firkins J. L., St-Pierre N. R., Lock A. L. (2015). Intestinal digestibility of long-chain fatty acids in lactating dairy cows: A meta-analysis and meta-regression. J. Dairy Sci. 98, 8889–8903. doi: 10.3168/jds.2015-9592
Booth C. C., Read A. E., Jones E. (1961). Studies on the site of fat absorption: 1. The sites of absorption of increasing doses of I-labelled triolein in the rat. Gut 2, 23–31. doi: 10.1136/gut.2.1.23
Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Cao D., Luo J., Chen D., Xu H., Shi H., Jing X., et al. (2016). CD36 regulates lipopolysaccharide-induced signaling pathways and mediates the internalization of Escherichia coli in cooperation with TLR4 in goat mammary gland epithelial cells. Sci Rep 6, 23132–23144. doi: 10.1038/srep23132
Cases S., Smith S. J., Zheng Y. W., Myers H. M., Lear S. R., Sande E., et al. (1998). Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc. Natl. Acad. Sci. U.S.A. 95, 13018–13023. doi: 10.1073/pnas.95.22.13018
Cases S., Stone S. J., Zhou P., Yen E., Tow B., Lardizabal K. D., et al. (2001). Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. J. Biol. Chem. 276, 38870–38876. doi: 10.1074/jbc.M106219200
Chao T., Wang G., Ji Z., Liu Z., Hou L., Wang J., et al. (2017). Transcriptome analysis of three sheep intestinal regions reveals key pathways and hub regulatory genes of large intestinal lipid metabolism. Sci. Rep. 7, 5345. doi: 10.1038/s41598-017-05551-2
Chen M., Yang Y., Braunstein E., Georgeson K. E., Harmon C. M. (2001). Gut expression and regulation of FAT/CD36: possible role in fatty acid transport in rat enterocytes. Am. J. Physiol. Endocrinol. Metabol. 281, E916–E923. doi: 10.1152/ajpendo.2001.281.5.E916
Coleman R. A., Lee D. P. (2004). Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 43, 134–176. doi: 10.1016/S0163-7827(03)00051-1
Daley V. L., Armentano L. E., Kononoff P. J., Hanigan M. D. (2020). Modeling fatty acids for dairy cattle: Models to predict total fatty acid concentration and fatty acid digestion of feedstuffs. J. Dairy Sci. 103, 6982–6999. doi: 10.3168/jds.2019-17407
Dedousis N., Teng L., Kanshana J. S., Kohan A. B. (2022). A single-day mouse mesenteric lymph surgery in mice: an updated approach to study dietary lipid absorption, chylomicron secretion, and lymphocyte dynamics. J. Lipid Res. 63, 1–14. doi: 10.1016/j.jlr.2022.100284
Dickson M. J., Piersanti R. L., Ramirez-Hernandez R., De Oliveira E. B., Bishop J. V., Hansen T. R., et al. (2020). Experimentally induced endometritis impairs the developmental capacity of bovine oocytes. Biol. Repro. 103, 508–520. doi: 10.1093/biolre/ioaa069
Doreau M., Chilliard Y. (1997). Digestion and metabolism of dietary fat in farm animals. Brit J. Nutr. 78 Suppl 1, S15–S35. doi: 10.1079/BJN19970132
Duran M. J., Kannampuzha-Francis J., Nydam D., Behling-Kelly E. (2021). Characterization of particle size distribution of plasma lipoproteins in dairy cattle using high-resolution polyacrylamide electrophoresis. Front. Anim. Sci. 2. doi: 10.3389/fanim.2021.678085
Feingold K. R. (2000). “Introduction to Lipids and Lipoproteins,” in Endotext. Eds. Feingold K. R., Anawalt B., Blackman M. R., Boyce A., Chrousos G., Corpas E., De Herder W. W., Dhatariya K., Dungan K., Hofland J., Kalra S., Kaltsas G., Kapoor N., Koch C., Kopp P., Korbonits M., Kovacs C. S., Kuohung W., Laferrère B., Levy M., Mcgee E. A., Mclachlan R., New M., Purnell J., Sahay R., Shah A. S., Singer F., Sperling M. A., Stratakis C. A., Trence D. L., Wilson D. P. (MDText.com, Inc, South Dartmouth (MA). Copyright © 2000-2023, MDText.com, Inc.
Gordon J. I., Elshourbagy N., Lowe J. B., Liao W. S., Alpers D. H., Taylor J. M. (1985). Tissue specific expression and developmental regulation of two genes coding for rat fatty acid binding proteins. J. Biol. Chem. 260, 1995–1998. doi: 10.1016/S0021-9258(18)89501-1
Hamilton J. A., Kamp F., Guo W. (2002). Mechanism of cellular uptake of long-chain fatty acids: Do we need cellular proteins? Mol. Cell Biochem. 239, 17–23. doi: 10.1023/A:1020542220599
Harrison F. A., Leat W. M. F. (1975). Digestion and absorption of lipids in non-ruminant and ruminant animals: a comparison. Proc. Nutri Soc. 34, 203–210. doi: 10.1079/PNS19750040
Hayashi H., Maruyama S., Fukuoka M., Kozakai T., Nakajima K., Onaga T., et al. (2013). Fatty acid-binding protein expression in the gastrointestinal tract of calves and cows. Anim. Sci. J. 84, 35–41. doi: 10.1111/j.1740-0929.2012.01038.x
Hussain M. M., Rava P., Walsh M., Rana M., Iqbal J. (2012). Multiple functions of microsomal triglyceride transfer protein. Nutri Metabol. 9, 14. doi: 10.1186/1743-7075-9-14
Kaur N., Chugh V., Gupta A. K. (2014). Essential fatty acids as functional components of foods- a review. J. Food Sci. Technol. 51, 2289–2303. doi: 10.1007/s13197-012-0677-0
Kazantzis M., Stahl A. (2012). Fatty acid transport proteins, implications in physiology and disease. Biochim. Biophys. Acta 1821, 852–857. doi: 10.1016/j.bbalip.2011.09.010
Khatun I., Clark R. W., Vera N. B., Kou K., Erion D. M., Coskran T., et al. (2016). Characterization of a novel intestinal glycerol-3-phosphate acyltransferase pathway and its role in lipid homeostasis. J. Biol. Chem. 291, 2602–2615. doi: 10.1074/jbc.M115.683359
Knudsen J., Neergaard T. B. F., Gaigg B., Jensen M. V., Hansen J. K. (2000). Role of acyl-CoA binding protein in acyl-CoA metabolism and acyl-CoA–mediated cell signaling. J. Nutri 130, 294S–298S. doi: 10.1093/jn/130.2.294S
Kong R. S. G., Liang G., Chen Y., Stothard P., Guan L. L. (2016). Transcriptome profiling of the rumen epithelium of beef cattle differing in residual feed intake. BMC Genomics 17, 592. doi: 10.1186/s12864-016-2935-4
Lagakos W. S., Gajda A. M., Agellon L., Binas B., Choi V., Mandap B., et al. (2011). Different functions of intestinal and liver-type fatty acid-binding proteins in intestine and in whole body energy homeostasis. Am. J. Physiol. Gastrointest. Liver Physiol. 300, G803–G814. doi: 10.1152/ajpgi.00229.2010
Lennox A. M., Garton G. A. (1968). The absorption of long-chain fatty acids from the small intestine of the sheep. Brit J. Nutri 22, 247–254. doi: 10.1079/BJN19680030
Lobo M. V., Huerta L., Ruiz-Velasco N., Teixeiro E., de la Cueva P., Celdrán A., et al. (2001). Localization of the lipid receptors CD36 and CLA-1/SR-BI in the human gastrointestinal tract: towards the identification of receptors mediating the intestinal absorption of dietary lipids. J. Histochem. Cytochem. 49, 1253–1260. doi: 10.1177/002215540104901007
Meller N., Morgan M. E., Wong W. P. S., Altemus J. B., Sehayek E. (2013). Targeting of Acyl-CoA synthetase 5 decreases jejunal fatty acid activation with no effect on dietary long-chain fatty acid absorption. Lipids Health Dis. 12, 88. doi: 10.1186/1476-511X-12-88
Nassir F., Wilson B., Han X., Gross R. W., Abumrad N. A. (2007). CD36 is important for fatty acid and cholesterol uptake by the proximal but not distal intestine. J. Biol. Chem. 282, 19493–19501. doi: 10.1074/jbc.M703330200
Nemecz G., Hubbell T., Jefferson J. R., Lowe J. B., Schroeder F. (1991). Interaction of fatty acids with recombinant rat intestinal and liver fatty acid-binding proteins. Arch. Biochem. Biophys. 286, 300–309. doi: 10.1016/0003-9861(91)90044-J
Ockner R. K., Manning J. A. (1974). Fatty acid-binding protein in small intestine. Identification, isolation, and evidence for its role in cellular fatty acid transport. J. Clin. Invest. 54, 326–338. doi: 10.1172/JCI107768
Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45. doi: 10.1093/nar/29.9.e45
Raphael B. C., Dimick P. S., Puppione D. L. (1973). Lipid characterization of bovine serum lipoproteins throughout gestation and lactation. J. Dairy Sci. 56, 1025–1032. doi: 10.3168/jds.S0022-0302(73)85300-7
Schwenk R. W., Holloway G. P., Luiken J. J., Bonen A., Glatz J. F. (2010). Fatty acid transport across the cell membrane: regulation by fatty acid transporters. Prostaglandins Leukot. Essent. Fatty Acids 82, 149–154. doi: 10.1016/j.plefa.2010.02.029
Sirwi A., Hussain M. M. (2018). Lipid transfer proteins in the assembly of apoB-containing lipoproteins. J. Lipid Res. 59, 1094–1102. doi: 10.1194/jlr.R083451
Smathers R. L., Petersen D. R. (2011). The human fatty acid-binding protein family: Evolutionary divergences and functions. Hum. Genomics 5, 170. doi: 10.1186/1479-7364-5-3-170
Stahl A. (2004). A current review of fatty acid transport proteins (SLC27). Pflügers Archiv 447, 722–727. doi: 10.1007/s00424-003-1106-z
Stahl A., Hirsch D. J., Gimeno R. E., Punreddy S., Ge P., Watson N., et al. (1999). Identification of the major intestinal fatty acid transport protein. Mol. Cell 4, 299–308. doi: 10.1016/S1097-2765(00)80332-9
Storch J., Thumser A. E. (2010). Tissue-specific functions in the fatty acid-binding protein family. J. Biol. Chem. 285, 32679–32683. doi: 10.1074/jbc.R110.135210
Suzuki H., Kawarabayasi Y., Kondo J., Abe T., Nishikawa K., Kimura S., et al. (1990). Structure and regulation of rat long-chain acyl-CoA synthetase. J. Biol. Chem. 265, 8681–8685. doi: 10.1016/S0021-9258(19)38942-2
Wang K., She Y., Mangat R., Makarowski A., Roy B. C., Bruce H. L., et al. (2023). Preferential deposition of dairy derived fatty acids in muscle tissue is partially due to the upregulation of CD36 in a low-birth-weight swine model. J. Anim. Sci. 101, 1–13. doi: 10.1093/jas/skad113
Wang T. Y., Liu M., Portincasa P., Wang D. Q. (2013). New insights into the molecular mechanism of intestinal fatty acid absorption. Eur. J. Clin. Invest. 43, 1203–1223. doi: 10.1111/eci.12161
Watkins P. A. (1997). Fatty acid activation. Prog. Lipid Res. 36, 55–83. doi: 10.1016/S0163-7827(97)00004-0
Yamashita A., Hayashi Y., Matsumoto N., Nemoto-Sasaki Y., Oka S., Tanikawa T., et al. (2014). Glycerophosphate/acylglycerophosphate acyltransferases. Biology 3, 801–830. doi: 10.3390/biology3040801
Yu J., Loh K., Song Z.-Y., Yang H.-Q., Zhang Y., Lin S. (2018). Update on glycerol-3-phosphate acyltransferases: the roles in the development of insulin resistance. Nutri Diabetes 8, 34. doi: 10.1038/s41387-018-0045-x
Keywords: intestine, rumen, ruminants, transport, uptake
Citation: Hentz F and Batistel F (2024) Characterization of genes and proteins involved in the absorption of long-chain fatty acids in the gastrointestinal tract of cattle. Front. Anim. Sci. 5:1435257. doi: 10.3389/fanim.2024.1435257
Received: 20 May 2024; Accepted: 17 July 2024;
Published: 01 August 2024.
Edited by:
James Levi Klotz, United States Department of Agriculture, United StatesReviewed by:
Ronald Joseph Trotta, University of Kentucky, United StatesVincenzo Lopreiato, University of Messina, Italy
Copyright © 2024 Hentz and Batistel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fernanda Batistel, ZmVybmFuZGFiYXRpc3RlbEB1ZmwuZWR1
 Fernanda Hentz
Fernanda Hentz Fernanda Batistel
Fernanda Batistel