
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Anim. Sci., 04 December 2023
Sec. Animal Physiology and Management
Volume 4 - 2023 | https://doi.org/10.3389/fanim.2023.1236582
This article is part of the Research TopicAnimal FibersView all 4 articles
Objective: The goal of this study was to characterize and investigate the effect of single nucleotide polymorphisms (SNPs) or InDels in tyrosinase related protein 1 (TYRP1), dopachrome tautomerase (DCT), and Ras-related protein 38 (RAB38) genes on coat color in South American camelids. TYRP1 is among one of the major regulators of pigmentation in mammals. DCT is a pigment cell-specific gene that plays an important role in the melanin synthesis pathway and the expression of skin color. RAB38 is involved in the transport of tyrosinase (TYR) and TYRP1, which are key enzymes in melanin production.
Methods: Five fleece color tones from 327 alpacas (white, black, brown, gray and light fawn) were investigated to characterize and identify polymorphisms that play a role in coat color. Among the SNPs and Indels identified in the coding region, statistical analysis was performed to determine the association between the polymorphisms and fleece color in camelids, as well as the determination of allelic frequencies.
Results: Twenty two polymorphisms in the coding region, seven of them were nonsynonymous, eleven were synonymous, three located in the 5’UTR region and one deletion in the 3’UTR region were identified in the TYRP1 gene; in the DCT gene were identified twenty two polymorphisms, where five of them were non-synonymous, twelve were synonymous, one polymorphism in the 5’UTR region and four in the 3’UTR region; in adition, in the RAB38 gene six polymorphisms were identified, one was nonsynonymous, two were synonymous, in addition to three polymorphisms in the 3’UTR region were found in the RAB38 gene. There were low to moderate polymorphisms in the populations. Significant differences (P<0.001) were found with color fiber in the alpaca populations studied for TYRP1 and DCT genes.
Domestic camelids are the basis of the family economy of high Andean communities. Among all domestic South American camelids uses, fiber production is one of the most important for breeders and for the industry (Anello et al., 2022), with white alpacas being the most appreciated by the industry due to the ease of dyeing the fiber and causing a favoring of this color, decreasing the population of other natural colors. However, in recent years there has been an increase in demand for natural products that do not harm the environment, with camelids having a wide range of colors ranging from white to black with quality fiber for the international market.
The numerous tonalities of color in animals depend on the pigment produced by melanocytes, which produce two chemically distinct types of melanin pigments, eumelanin and pheomelanin (Le Pape et al., 2008; Cieslak et al., 2011). Variation in coat color results from a complex interaction of the melanins produced, their ratio (eumelanin and pheomelanin), the intensity of pigmentation and the distribution of pigment throughout the hairs (Bhat et al., 2019) and the distribution of these pigments on the body (Cieslak et al., 2011). Three main mechanisms have been identified that define coat color variation; i) melanocyte migration and differentiation ii) melanocyte signaling and regulation and iii) melanin production and transport (Bhat et al., 2019).
Melanin synthesis occurs in specialized pigment cells called melanocytes and is under the catalytic control of several enzymes including Tyrp1 (Feeley et al., 2016). Tyrp1 is responsible for the oxidation of an eumelanin monomer, formed during pigment synthesis, which then allows for the formation of a more stable, black eumelanin (Feeley et al., 2016). Loss-of-function mutations in TYRP1 have been associated with brown phenotypes in dogs (Schmutz et al., 2002; Hrckova Turnova et al., 2017), sheep (Gratten et al., 2007), bears (Puckett et al., 2023), American mink (Cirera et al., 2016) among other species.
Dopachrome tautomerase (DCT) plays an important role in vertebrate melanin pigmentation (Jackson et al., 1992). The TYRP2 protein was identified as the DCT enzyme on chromosome 14 in the mouse (Kroumpouzos et al., 1994) and is known as DCT or TYRP2 (Tsukamoto et al., 1992). Hair melanin content was detected at a higher level in DCT knockout mice than in slaty mutant mice (Guyonneau et al., 2004).
Tyrosinase-related protein 1 (TYRP1) and dopachrome tautomerase (DCT) are involved in melanin production and are therefore likely to be involved in determining coat color (Becker et al., 2015; Paterson et al., 2015). However, the mechanisms by which these genes act to produce coat color are unclear (Peng et al., 2018).
Previous analysis confirms that RAB38 is a melanosomal protein, mutated in the mouse pigmentation mutant, chocolate (cht), and important for tyrosinase-related protein 1 sorting of melanosomal protein tyrosinase (TYRP1) in melanocytes (Loftus et al., 2002). The role of RAB38 in the biogenesis of melanosomes and melanin and the genetic mechanism contribute to hair color variety and intensity in American mink. (Manakhov et al., 2020).
Although the role in brown and black coat coloration of the TYRP1 gene has been evaluated in alpacas (Feeley et al., 2016; Alshanbari et al., 2019), no work has yet been done with the DCT and RAB38 genes in camelids, so the aim of this study was to investigate polymorphisms in these three genes to evaluate their associations with different fleece colors in high Andean camelids, mainly in alpacas.
Three hundred twenty-seven unrelated camelids were studied for TYRP1, DCT and RAB38 genes sequencing; 327 were alpacas (158 white, 88 black, 49 brown, 88 gray and 15 light fawn). Animals were from the Cusco and Puno region, two regions with the highest population of alpacas in Peru. Figure 1 shows some examples of white, brown, black and LF (Light fawn) alpacas. The samples were obtained by venipuncture from the jugular vein into EDTA vacutainer tubes. The blood was frozen at -20 °C until handling at the La Raya research center from Universidad Nacional de San Antonio Abad del Cusco. DNA (deoxyribonucleic acid) was extracted from whole blood of all sampled animals using the Purelink Genomic mini kit (Qiagen) according to the manufacturer’s protocol. DNA quality and quantity were evaluated using the Qubit 4.0 fluorometer (Invitrogen).

Figure 1 Photographs of different coat color. (A) White, (B) LF or light fawn, (C) Black, (D) Brown and (E) Gray.
Primers to amplify the coding region and 5’ and 3’ part were designed with the Ion Ampliseq 6.0.1 designer for Ion Torrent New Generation (Life Technologies, www.ampliseq.com) from the VicPac 2.0.1 reference sequence. A total of 29 Amplicons ranging from 96 bp to 326 bp covered 100% of the target sequence (Table 1S). Primers were supplied in two pools (Forward and reverse), each at a concentration of 100 nM (Life Technologies, Carlsbad, CA, USA). PCR was conducted in a total volume of 20 μL, containing 10 μL QIAGEN Taq PCR Master Mix, 2.4 μL primer mix (0.5 μm end concentration in the PCR), 6.6 μL Milli-Q water and 1 μL of template DNA. A single PCR program was used: denaturation at 95°C for 30 s, 40 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 45 s, and a final extension at 72°C for 5 min. The sequencing library was prepared with the Ion AmpliSeq library kit version 2.0 according to the manufacturer’s protocols. The amplified library was diluted to 10 pM, and 25 μL was used for template preparation according to the Ion PGM Template OT2 200 Kit protocol using the Ion PGM Template OT2 200 kit. The clone amplified library was enriched in Ion OneTouch E.S. Sequencing was performed on an Ion Torrent bulk sequencing machine using the Ion PGM Sequencing 200 v2 kit and an Ion 540 chip. Using Torrent Suite (version 3.6.) variant call format (VCF) files were loaded into Ion Reporter V4.0 (https://ionreporter.lifetechnologies.com/ir/) for variant annotation.
The sequence variants in the 3 genes in each sample were identified using the Torrent Suite Variant Caller (TSVC; v5.8) GATK software tools (version 4.2.2.0; http://www.broadinstitute.org) were used for genotype calling. and the resulting VCF were further annotated using online Ion Reporter software (https://ionreporter.lifetechnologies.com/ir/secure/home.html). The sequences were aligned using the software Bosque 2.0.2 (Ramírez-Flandes and Ulloa, 2008) and Mega 11 (Tamura et al., 2021).
Genotypic and allelic frequencies, as well as the distribution of genotypes among colors were evaluated by Fisher’s exact test and Chi-squared test using the R language (version 4.1.3 http://www.R-project.org). Population polymorphism information content (PIC) was calculated according to Nei’s methods (Chesnokov and Artemyeva, 2015). JMP®, Version 17.1. SAS Institute Inc., Cary, NC, 1989-2023 was also used to perform a contingency analysis containing a mosaic plot, contingency table and complementary statistical tests (Table S2). Contingency analysis allows measuring the interaction between the coat color and genotype variables. DnaSP v6 (Rozas et al., 2017) was used to determine different neutrality tests (Table S3). PopART V1.7 software (Clement et al., 2002) was used to construct a haplotype network for TYRP1 and DCT genes, respectively, where polymorphisms with significant effect were found from four coat colors (White, black, brown and LF).
1664 bp was obtained for the TYRP1 gene, 1624 bp for the DCT gene and 791 bp for the RAB38 gene, corresponding to 100% of the coding region of the three genes and partial regions of the 5’ and 3’UTR region (Figure 2).
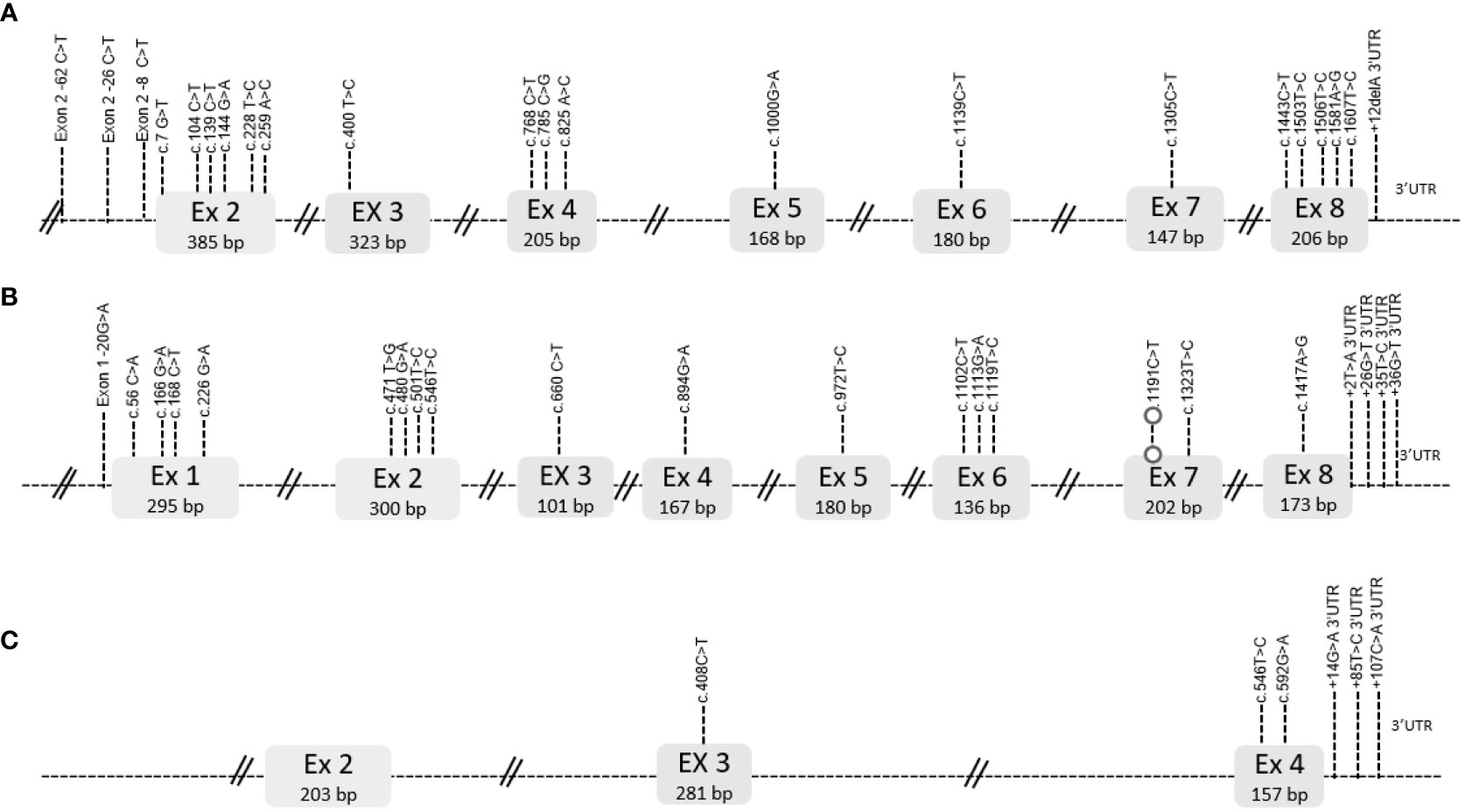
Figure 2 Structure of the TYRP1 (A), DCT (B) and RAB38 (C) genes. The exons are shown in the gray boxes with their size in base pairs (bp), in the case of introns they are shown as dashed lines. The polymorphisms found in each gene are shown according to their exon or 5’ or 3’UTR region.
Thirteen haplotypes were identified for the TYRP1 gene (Figure 3), where haplotypes 1, 4, 5, 7, 9 and 10 are distributed in the four coat colors of the alpacas analyzed, haplotypes 2, 3 and 8 are distributed in three colors and haplotypes 11, 12 and 13 are exclusive to one coat color.
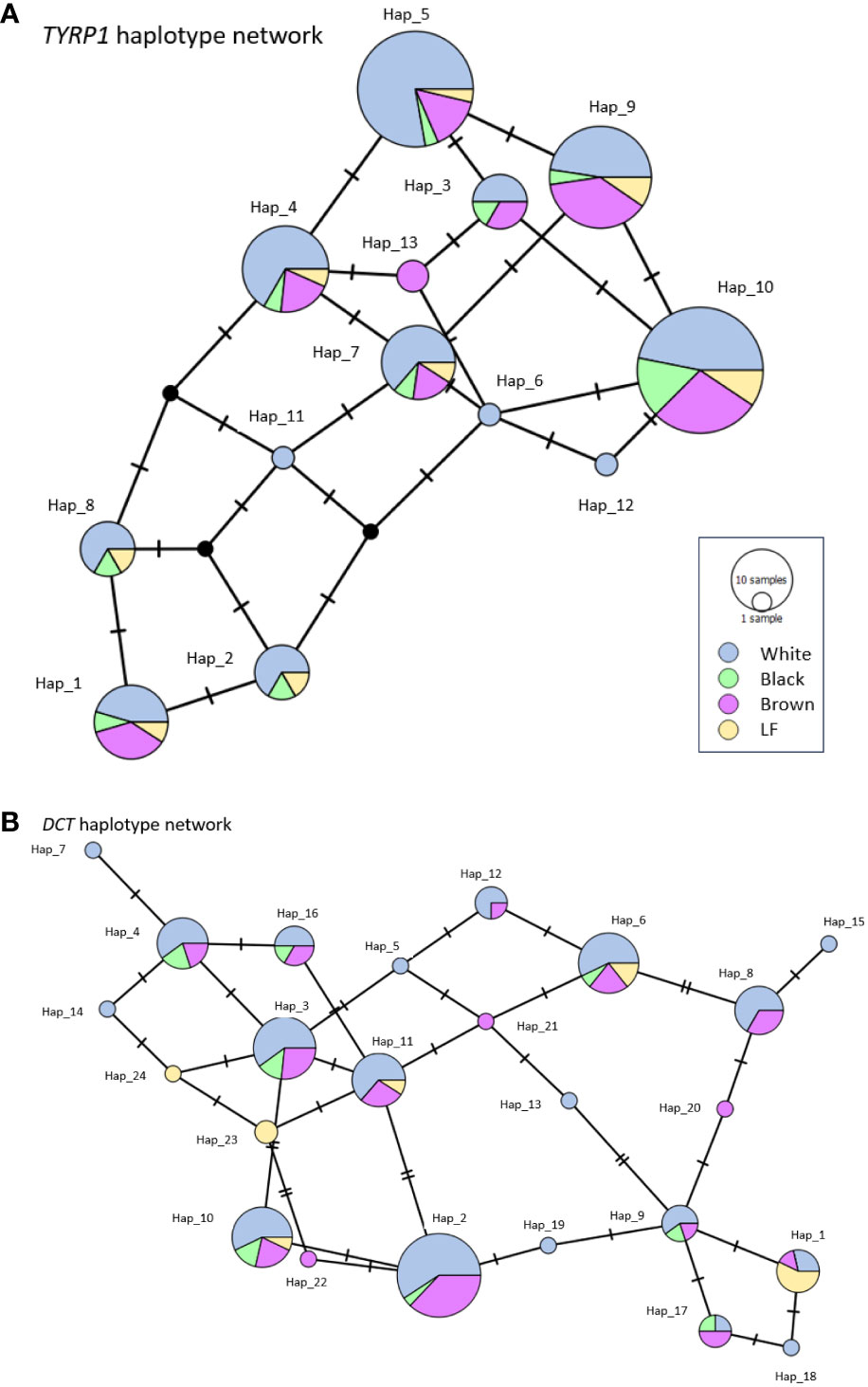
Figure 3 TCS-network of TYRP1 (A) and DCT (B) haplotypes showing all variants identified in alpacas. The size of the circle corresponds to haplotype. Colors represent circulating haplotypes identified in white, black, brown and LF alpacas.
In the case of the DCT gene, 24 haplotypes were identified (Figure 3), where haplotypes 6 and 10 are distributed in the colors analyzed. Haplotypes 1, 2, 3, 4, 9, 11, 16 and 17 are distributed in three colors, haplotypes 8 and 12 in two colors and haplotypes 5, 7, 13, 14 15, 18, 19, 20, 21, 22, 23 and 24 are exclusive for one coat color in the studied alpacas.
Twenty-two polymorphisms were identified (Table S3), twenty-one of SNP type and one deletion in the 3’UTR region. The exons with the highest number of polymorphisms were exon 2 and exon 5 (5 polymorphisms each), detailed in Figure 2. Seven of the polymorphisms were of non-synonymous type. Significant association (Using the value of P<0.001 as a value of greater statistical rigor) was found with 5 polymorphisms (Table 1), one involved amino acid change (c. 7G>T), three were synonymous changes (c.400T>C, c. 1305C>T, c.1443T>C) and one was in the 5’ region (5’c.-26C>T). Likewise, the TYRP1 gene sequence translation revealed an amino acid size of 537 amino acids.
Twenty-two polymorphisms were identified (Table S3), all of SNP type. The exon with the highest number of polymorphisms was exon 1 (4 polymorphisms), detailed in Figure 2. Five of the polymorphisms were of the non-synonymous type. Significant association (P<0.001) was found with 7 polymorphisms, four were synonymous changes (c.168C>T, c. 546T>C, c.660C>, c.894G>A), one was in the 5’ region (5’c.-20G>A) and two were in the 3’region (c.+2T>A, c.+36G>T). Likewise, the DCT gene sequence translation revealed an amino acid size of 519 amino acids.
Six polymorphisms were identified (Table S3) all of SNP type. The exon with the highest number of polymorphisms was exon 4 (2 polymorphisms), detailed in Figure 2. One of the polymorphisms was of non-synonymous type. No significant association (P<0.001) was found with any polymorphism. The RAB38 gene sequence translation revealed an amino acid size of 211 amino acids.
Genotypic frequencies of polymorphisms with significance (P<0.001).
The genotypic and allelic frequencies were analyzed statistically for the polymorphisms of the TYRP1, DCT and RAB38 genes that complied with values of P<0.001. In addition, the PIC values were calculated for white, black and brown alpacas due to the large population analyzed (Tables 2, 3). The information for all the colors analyzed can be found in Table S1 and in Figure S1 including all coat color analyzed.
For the TYRP1 gene, the five SNPs with P values less than 0.001 were found to show a low to moderate PIC value ranging from 0.0303 to 0.3748 (Mean 0.2555). For the -26T>C locus, the most frequent genotype was TT with 82.84%, 62.50% and 87.50% for white, black and brown. The most frequent genotype for the 7G>T locus was GG in the white, black and brown colors with 96.88%, 69.57% and 93.62%, being the TT genotype of null frequency. At the 400T>C locus, the most frequent genotype for white and brown alpacas was TT with 57.27% and 65.85%, and for black alpacas it was TC with 70.59%. At locus 1305C>T, the most frequent genotype was CC for white and brown alpacas with 64.28% and 73.33%, and for black alpacas it was CT with 50%. For locus 1443, the most frequent genotype was TT for white alpacas with 35.83%, for black alpacas it was TC with 68.63% and for brown alpacas it was CC with 52.63%.
For the DCT gene, the seven SNPs with P values less than 0.001 showed a low to moderate PIC value ranging from 0.1453 to 0.4999 (average 0.2636). For the -20G>A locus, the most frequent genotype was GG with 73.18% and 84.44% for white and brown colors in alpacas and, in the case of black alpacas it was AG with 61.54%. The most frequent genotype for the 168G>T locus was CC in white and brown colors with 72.95% and 82.61% and, for black alpacas the CT genotype with 70.27%. At locus 546T>C, the most frequent genotype for white and black alpacas was TC with 49.57% and 71.67%, and for brown alpacas it was TT with 70.73%. For the 660C>T locus, the most frequent genotype was CC for white and brown alpacas with 68.42% and 89.48%, and for black alpacas it was CT with 76.19%. For the 894G>A locus, the most frequent genotype was GG for white and brown alpacas with 72% and 89.13%, for black alpacas it was GA with 60%. At the +2T>A3’UTR locus, the most frequent genotype for white and brown alpacas was TT with 78.72% and 75.61% and for black alpacas it was AT with 68.57%. In case of the +36G>T locus, the most frequent genotype in white and black alpacas was GT with 50.85% and 75.44% and, for brown alpacas it was GG with 62.5%.
In the TYRP1 gene, two SNPs can affect black and white coat color, including c.1305C>T and c.1443T>C in homozygous state for white alpacas and in heterozygous state for black alpacas; as well as three SNPs in the DCT gene, which are 5’c.-20G>A, c.546T>C and 3’c.+36G>T. One SNP can affect black and brown coat (c.400T>C) in heterozygous state for black and homozygous dominant for brown in the TYRP1 gene. Four SNPs can affect the coat color of black alpacas, including c.168C>T, c.660C>T, c.894G>A and 3’c.+2T>A in the DCT gene.
Coat color in domestic camelids is an important characteristic, and in recent years there has been great interest in finding genes responsible for the range of colors present in these animals.
TYRP1, DCT and RAB38 genes play an important role in melanin synthesis. In this work we investigated the variations of the TYRP1, DCT and RAB38 genes in the four South American camelid species, taking large populations of white, black, and brown alpacas, as well as small populations of gray and LF colors, finding a total of 45 SNPs and 1 Indel in these three genes. From statistical analysis, 12 SNP polymorphisms related to coat color in alpacas were obtained, with variable distribution levels in the colors analyzed. These polymorphisms may have a role in color formation in alpacas.
When analyzing TCS-network of TYRP1 and DCT haplotypes, most haplotypes were shared, with no color-specific clusters. At the time of the conquest, when camelids were displaced by sheep, they were not interested in selection by specific colors, but by fleece weight (Marín et al., 2018). The samples obtained were from breeders who manage alpaca selection by coat color. However, the work of selection by defined colors should continue in order to obtain offspring with the same colors as the parents and not vary in their tonality.
Genetic diversity is an important basis for evaluating the status of breeding germplasm resources, and it is the genetic basis for the population’s adaptation to the environment and evolution (Jia et al., 2021). The PIC value in population analysis is important because it reflects genetic diversity. PIC values higher than 0.5 indicate high polymorphism, values between 0.25 and 0.50 moderate polymorphism and values lower than 0.25 indicate low polymorphism (Li et al., 2018). For the genes analyzed, a moderate PIC value (0.25 to 0.50) was shown in 11 loci in black alpacas, 6 loci in white alpacas and 4 loci in brown alpacas. Low PIC values (< 0.25) were observed in 1 loci in black alpacas, 6 loci in white alpacas and 8 loci in brown alpacas. These results indicate that the selection for the colors analyzed in alpacas was different. Traditionally, the industry has preferred white because it can be dyed without bleaching (Anello et al., 2022). In Peru, as a result of high prices for white fiber, there has been a large decrease in colored alpacas, causing a loss of genetic diversity that in the field becomes more complex due to the low price obtained from the sale of colored fiber. Fortunately, in recent years, the demand for natural colors by the textile industry has increased along with the global trend towards the consumption of environmentally friendly products (Anello et al., 2022). The results obtained are useful to consider the conservation of genetic resources of coat color in alpacas due to the diversity found in the animals analyzed from the PIC values.
The TYRP1 gene, which is a member of the tyrosinase gene family, has been implicated mainly in the brown coat color of animals such as sheep (Paris et al., 2019), goats (Becker et al., 2015), birds (Li et al., 2019) where loss of function of the gene occurs. In this study, SNPs c.400T>C, c.1305C>T and c.1443T>C were found to have significant association (p<0.001) with white, black and brown coat color in the TYRP1 gene, where they also had moderate PIC values. The frequency of the TT genotype of the c.400T>C polymorphism was higher in white and brown alpacas, as well as TC in black alpacas. In case of the c.1305C>T polymorphism, the frequency of the CC genotype was higher in white and brown alpacas, CT in black alpacas. Finally, for the c.1443T>C polymorphism, the frequency of the TT genotype was higher in white alpacas, CC in brown alpacas and TC in black alpacas. These results indicate that the three polymorphisms mentioned in the TYRP1 gene could be associated with fiber pigmentation in alpacas, even though they are synonymous and do not involve amino acid changes. Similar studies in black and brown alpacas found no association with dark coat color of the TYRP1 gene (Feeley et al., 2016), despite finding 19 mutations, and sharing 11 mutations with those of the present study, where they indicate that the reduced number of samples could be the cause of not finding association with coat color. In a study conducted in camels (Alshanbari et al., 2019) they found no association with brown color and the analyzed polymorphisms found in intronic regions; however, they indicate that despite the results the TYRP1 gene remains an important candidate for color phenotypes in camels, but Holl et al., 2017 find an important association between KIT gene and with spot in camel.
DCT or TYRP2 is involved in the isomerization of dopachrome to DHICA (Bhat et al., 2019). Mutations in DCT cause drastic decrease in the eumelanin content of melanocytes in mice, unexpectedly increasing the pheomelanin content (Costin et al., 2005). In this study, it was found that SNPs c.168C>T, c.546T>C, c.660C>T, 3’c.+2T>A and 3’c.+36G>T of the DCT gene showed strong significant association (p<0.0001) with white, black, and brown coat color in alpacas, with moderate ICP values. For the c.168C>T polymorphism, the frequency of the GG genotype was higher in white and brown alpacas and TC in black alpacas. In case of the c.546T>C polymorphism, the frequency of the TC genotype was higher in white and black alpacas, TT in brown alpacas. The frequency of the GG genotype of the c.660C >T polymorphism was higher in white and brown alpacas, as well as CT in black alpacas. In case of the 3’c.+2T>A polymorphism, the frequency of the TT genotype was higher in white and brown alpacas, TA in black alpacas. Finally, for the 3’c.+36G>T polymorphism, the frequency of the GT genotype was higher in white and black alpacas and GG in brown alpacas. The results shown indicate that the five polymorphisms found in the DCT gene could be associated with pigmentation in alpacas, and as in the case of the TYRP1 gene analysis are of synonymous type. Studies in goats (Bhat et al., 2019) indicate that the DCT gene significantly regulates dark fiber color, having high expressions in brown skin of Pashmina goats. There are no reported works to date on the analysis of the DCT gene in alpacas, so this work hopefully helps to understand the role of this gene in color phenotypes in alpacas.
The RAB38 gene encodes the member of the Rab family of small G proteins, which is involved in intracellular vesicle trafficking and melanosome biogenesis (Osanai et al., 2005). The RAB38 gene is highly expressed in melanocytes (Loftus et al., 2002). RAB38 is involved in the transport of newly synthesized tyrosinase and Tyrp1, which are key enzymes in melanin production from endosomes in the trans-Golgi network to maturing melanosomes (Bultema et al., 2012; Coppola et al., 2016). Mutations in the RAB38 gene were associated with coat color in chocolate mice (Loftus et al., 2002). The present work reports 6 polymorphisms, which would have no association with coat color in alpacas; however, it is the first time we report the study of the RAB38 gene in alpacas.
It was identified that TYRP1 and DCT genes are related to coat color in alpacas, finding significant association between 3 SNPs for the TYRP1 gene and 5 SNPs for the DCT gene. The loci have low to moderate polymorphism and there are differences in their distribution among the colored alpacas analyzed. The results of this study show the role of genes involved in the alpaca coat color phenotype, which could be used as markers in the selection of preferred fleece tonalities in alpacas.
The data presented in the study are deposited in the Genbank repository, accession number OQ916474 - OQ916486 to TYRP1 and OR405502 - OR405518 to DCT genes.
The animal studies were approved by the Ethics Committee of the Professional School of Veterinary Medicine UNSAAC. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
CM-R obtained funding for the research project. CM-R, CZ-C, and PWB-M developed the ideas. CM-R and CZ-C collected the samples. CM-R performed the analyses. CM-R and PWB-M wrote the manuscript with input from all project members. All authors contributed to the article and approved the submitted version.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors thank the following alpaca breeders for providing samples of their animals, without them this research would not have been possible: Elvis Yucra Jacinto (Fundo Oquemarca), María Barragán de Aedo (CAP Huaycho), Víctor Aedo Barragán (Fundo Teresapampa), Máximo Díaz (Fundo Chaupiwasi) and CICAS La Raya (UNSAAC-Cusco).
The authors declare that the research was conducted in the absence of commercial or financial relationships that could be interpreted as possible conflicts of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fanim.2023.1236582/full#supplementary-material
Alshanbari F., Castaneda C., Juras R., Hillhouse A., Mendoza M. N., Gutiérrez G. A., et al. (2019). Comparative FISH-mapping of MC1R, ASIP, and TYRP1 in new and old world camelids and association analysis with coat color phenotypes in the dromedary (Camelus dromedarius). Front. Genet. 10. doi: 10.3389/fgene.2019.00340
Anello M., Daverio M. S., Di Rocco F. (2022). Genetics of coat color and fiber production traits in llamas and alpacas. Anim. Front. Rev. magazine Anim. Agric. 12 (4), 78–86. doi: 10.1093/af/vfac050
Becker D., Otto M., Ammann P., Keller I., Drögemüller C., Leeb T. (2015). The brown coat colour of Coppernecked goats is associated with a non-synonymous variant at the TYRP1 locus on chromosome 8. Anim. Genet. 46 (1), 50–54. doi: 10.1111/age.12240
Bhat B., Singh A., Iqbal Z., Kaushik J. K., Rao A. R., Ahmad S. M., et al. (2019). Comparative transcriptome analysis reveals the genetic basis of coat color variation in Pashmina goat. Sci. Rep. 9 (1), 6361. doi: 10.1038/s41598-019-42676-y
Bultema J. J., Ambrosio A. L., Burek C. L., Di Pietro S. M. (2012). BLOC-2, AP-3, and AP-1 proteins function in concert with Rab38 and Rab32 proteins to mediate protein trafficking to lysosome-related organelles. J. Biol. Chem. 287 (23), 19550–19563. doi: 10.1074/jbc.M112.351908
Chesnokov Y. V., Artemyeva A. M. (2015). Evaluation of the measure of polymorphism information of genetic diversity. Agric. Biol. 50, 571–578. doi: 10.15389/agrobiology.2015.5.571eng
Cieslak M., Reissmann M., Hofreiter M., Ludwig A. (2011). Colours of domestication. Biol. Rev. Cambridge Philos. Soc. 86 (4), 885–899. doi: 10.1111/j.1469-185X.2011.00177.x
Cirera S., Markakis M. N., Kristiansen T., Vissenberg K., Fredholm M., Christensen K., et al. (2016). A large insertion in intron 2 of the TYRP1 gene associated with American Palomino phenotype in American mink. Mamm. Genome 27 (3-4), 135–143. doi: 10.1007/s00335-016-9620-4
Clement M., Snell Q., Walker P., Posada D., Crandall K. (2002). “TCS: Estimating gene genealogies,” in Parallel and Distributed Pro-cessing Symposium, International Proceedings, Vol. 2. 184.
Coppola U., Annona G., D'Aniello S., Ristoratore F. (2016). Rab32 and Rab38 genes in chordate pigmentation: an evolutionary perspective. BMC evolutionary Biol. 16, 26. doi: 10.1186/s12862-016-0596-1
Costin G. E., Valencia J. C., Wakamatsu K., Ito S., Solano F., Milac A. L., et al. (2005). Mutations in dopachrome tautomerase (Dct) affect eumelanin/pheomelanin synthesis, but do not affect intracellular trafficking of the mutant protein. Biochem. J. 391 (Pt 2), 249–259. doi: 10.1042/BJ20042070
Feeley N. L., Bottomley S., Munyard K. A. (2016). Novel mutations in Vicugna pacos (alpaca) Tyrp1 are not correlated with brown fibre colour phenotypes. Small Ruminant Res. 143, 29–34. doi: 10.1016/j.smallrumres.2016.08.012
Gratten J., Beraldi D., Lowder B. V., McRae A. F., Visscher P. M., Pemberton J. M., et al. (2007). Compelling evidence that a single nucleotide substitution in TYRP1 is responsible for coat-colour polymorphism in a free-living population of Soay sheep. Proc. Biol. Sci. 274 (1610), 619–626. doi: 10.1098/rspb.2006.3762
Guyonneau L., Murisier F., Rossier A., Moulin A., Beermann F. (2004). Melanocytes and pigmentation are affected in dopachrome tautomerase knockout mice. Mol. Cell. Biol. 24 (8), 3396–3403. doi: 10.1128/MCB.24.8.3396-3403.2004
Holl H., Isaza R., Mohamoud Y., Ahmed A., Almathen F., Youcef C., et al. (2017). A frameshift mutation in KIT is associated with white spotting in the Arabian camel. Genes 8 (3), 102. doi: 10.3390/genes8030102
Hrckova Turnova E., Majchrakova Z., Bielikova M., Soltys K., Turna J., Dudas A. (2017). A novel mutation in the TYRP1 gene associated with brown coat colour in the Australian Shepherd Dog Breed. Anim. Genet. 48 (5), 626. doi: 10.1111/age.12563
Jackson I. J., Chambers D. M., Tsukamoto K., Copeland N. G., Gilbert D. J., Jenkins N. A., et al. (1992). A second tyrosinase-related protein, TRP-2, maps to and is mutated at the mouse slaty locus. EMBO J. 11 (2), 527–535. doi: 10.1002/j.1460-2075.1992.tb05083.x
Jia X., Ding P., Chen S., Zhao S., Wang J., Lai S. (2021). Analysis of MC1R, MITF, TYR, TYRP1, and MLPH genes polymorphism in four rabbit breeds with different coat colors. Anim. an Open Access J. MDPI 11 (1), 81. doi: 10.3390/ani11010081
Kroumpouzos G., Urabe K., Kobayashi T., Sakai C., Hearing V. J. (1994). Functional analysis of the slaty gene product (TRP2) as dopachrome tautomerase and the effect of a point mutation on its catalytic function. Biochem. Biophys. Res. Commun. 202 (2), 1060–1068. doi: 10.1006/bbrc.1994.2036
Le Pape E., Wakamatsu K., Ito S., Wolber R., Hearing V. J. (2008). Regulation of eumelanin/pheomelanin synthesis and visible pigmentation in melanocytes by ligands of the melanocortin 1 receptor. Pigment Cell melanoma Res. 21 (4), 477–486. doi: 10.1111/j.1755-148X.2008.00479.x
Li J., Bed'hom B., Marthey S., Valade M., Dureux A., Moroldo M., et al. (2019). A missense mutation in TYRP1 causes the chocolate plumage color in chicken and alters melanosome structure. Pigment Cell melanoma Res. 32 (3), 381–390. doi: 10.1111/pcmr.12753
Li T., Qu J., Wang Y., Chang L., He K., Guo D., et al. (2018). Genetic characterization of inbred lines from Shaan A and B groups for identifying loci associated with maize grain yield. BMC Genet. 19 (1), 63. doi: 10.1186/s12863-018-0669-9
Loftus S. K., Larson D. M., Baxter L. L., Antonellis A., Chen Y., Wu X., et al. (2002). Mutation of melanosome protein RAB38 in chocolate mice. Proc. Natl. Acad. Sci. United States America 99 (7), 4471–4476. doi: 10.1073/pnas.072087599
Manakhov A. D., Mintseva M. Y., Andreev I. A., Uralsky L. I., Andreeva T. V., Trapezov O. V., et al. (2020). Genome analysis of American minks reveals link of mutations in Ras-related protein-38 gene to Moyle brown coat phenotype. Sci. Rep. 10 (1), 15876. doi: 10.1038/s41598-020-72239-5
Marín J. C., Rivera R., Varas V., Cortés J., Agapito A., Chero A., et al. (2018). Genetic variation in coat colour genes MC1R and ASIP provides insights into domestication and management of South American camelids. Front. Genet. 9. doi: 10.3389/fgene.2018.00487
Osanai K., Takahashi K., Nakamura K., Takahashi M., Ishigaki M., Sakuma T., et al. (2005). Expression and characterization of Rab38, a new member of the Rab small G protein family. Biol. Chem. 386 (2), 143–153. doi: 10.1515/BC.2005.018
Paris J. M., Letko A., Häfliger I. M., Ammann P., Flury C., Drögemüller C. (2019). Identification of two TYRP1 loss-of-function alleles in Valais Red sheep. Anim. Genet. 50 (6), 778–782. doi: 10.1111/age.12863
Paterson E. K., Fielder T. J., MacGregor G. R., Ito S., Wakamatsu K., Gillen D. L., et al. (2015). Tyrosinase depletion prevents the maturation of melanosomes in the mouse hair follicle. PloS One 10 (11), e0143702. doi: 10.1371/journal.pone.0143702
Peng Y., Wang Y., Wang R., Geng L., Ma R., Zhang C., et al. (2018). Exploring differentially expressed genes associated with coat color in goat skin using RNA-seq. Can. J. Anim. Sci. 99 (2), 357–366. doi: 10.1139/CJAS-2018-0026
Puckett E. E., Davis I. S., Harper D. C., Wakamatsu K., Battu G., Belant J. L., et al. (2023). Genetic architecture and evolution of color variation in American black bears. Curr. biol: CB 33 (1), 86–97.e10. doi: 10.1016/j.cub.2022.11.042
Ramírez-Flandes S., Ulloa O. (2008). Bosque: integrated phylogenetic analysis software. Bioinf. (Oxford England) 24 (21), 2539–2541. doi: 10.1093/bioinformatics/btn466
Rozas J., Ferrer-Mata A., Sánchez-Del Barrio J. C., Guirao-Rico S., Librado P., Ramos-Onsins S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34 (12), 3299–3302. doi: 10.1093/molbev/msx248
Schmutz S. M., Berryere T. G., Goldfinch A. D. (2002). TYRP1 and MC1R genotypes and their effects on coat color in dogs. Mamm. Genome 13 (7), 380–387. doi: 10.1007/s00335-001-2147-2
Tamura K., Stecher G., Kumar S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38 (7), 3022–3027. doi: 10.1093/molbev/msab120
Keywords: alpaca, TYRP1, DCT, RAB38, SNP, coat color
Citation: Melo-Rojas C, Bravo-Matheus PW, Amaht Araoz C and Zapata-Coacalla C (2023) Identification of polymorphisms in TYRP1, DCT and RAB38 genes and their association with coat color in alpacas. Front. Anim. Sci. 4:1236582. doi: 10.3389/fanim.2023.1236582
Received: 07 June 2023; Accepted: 11 October 2023;
Published: 04 December 2023.
Edited by:
Seyed Abbas Rafat, University of Tabriz, IranReviewed by:
Semir Bechir Suheil Gaouar, University of Tlemcen, AlgeriaCopyright © 2023 Melo-Rojas, Bravo-Matheus, Amaht Araoz and Zapata-Coacalla. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carola Melo-Rojas, Y2Fyb2xhLm1lbG9AdW5zYWFjLmVkdS5wZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.