- 1Institute of Agricultural Biology and Biotechnology, National Research Council, Lodi, Italy
- 2Department of Veterinary Science, University of Turin, Torino, Italy
- 3Institute of Sciences of Food Production, National Research Council, Milan, Italy
- 4Department of Agricultural and Environmental Sciences - Production, Territory, Agroenergy (DiSAA), University of Milan, Milan, Italy
In dairy ruminants, a diet supplemented with feed rich in unsaturated fatty acids can be an effective medium to increase the health-promoting properties of milk, although their effect on the pathways/genes involved in these processes has not been properly and completely defined to date. To improve our knowledge of the cell’s activity in specific conditions, next-generation RNA-sequencing technology was used to allow whole transcriptome characterization under given conditions. In addition to this, microRNAs (miRNAs) have recently been known as post-transcriptional regulators in fatty acid and cholesterol metabolism by targeting lipid metabolism genes. In this study, to analyze the transcriptome and miRNAs in goat milk after a supplemental diet enriched with linoleic acid (hemp seeds), next-generation RNA-sequencing was used in order to point out the general biological mechanisms underlying the effects related to milk fat metabolism. Ten pluriparous Alpine goats were fed with the same pretreatment diet for 40 days; then, they were arranged to two dietary treatments consisting of control (C) and hemp seed (H)-supplemented diets. Milk samples were collected at 40 (time point = T0) and 140 days of lactation (time point = T1). Milk fatty acid (FA) profiles revealed a significant effect of hemp seeds that determined a strong increment in the preformed FA, causing a reduction in the concentration of de-novo FA. Monounsaturated and polyunsaturated n−3 FAs were increased by hemp treatment, determining a reduction in the n−6/n−3 ratio. After removing milk fats and proteins, RNA was extracted from the milk cells and transcriptomic analysis was conducted using Illumina RNA-sequencing. A total of 3,835 genes were highly differentially expressed (p-value < 0.05, fold change > 1.5, and FDR < 0.05) in the H group. Functional analyses evidenced changes in metabolism, immune, and inflammatory responses. Furthermore, modifications in feeding strategies affected also key transcription factors regulating the expression of several genes involved in milk fat metabolism, such as peroxisome proliferator-activated receptors (PPARs). Moreover, 38 (15 known and 23 novel) differentially expressed miRNAs were uncovered in the H group and their potential functions were also predicted. This study gives the possibility to improve our knowledge of the molecular changes occurring after a hemp seed supplementation in the goat diet and increase our understanding of the relationship between nutrient variation and phenotypic effects.
1 Introduction
Cow, goat, and sheep milk have been recognized to give beneficial effects on humans, and over the years, exhaustive activity has been done on milk production as well as on increasing its nutritional components (Ibeagha-Awemu et al., 2016). Lipid composition, in particular, is one of the most important components of the nutritional quality of goat milk. The different caprine fatty acids (FAs) (short- and medium-chain, saturated, branched, mono- and polyunsaturated, cis and trans, conjugated) are potentially involved as positive or negative predisposing factors for the health of human consumers (Parodi, 1999). For example, it is known that conjugated linoleic acid (CLA) plays a protective function against several human diseases such as cancer and cardiovascular diseases and also an important role in maintaining a healthy gastrointestinal tract and in modulating immune function (Conte et al., 2017). As mammals are not able to produce this kind of FAs, it is necessary to integrate CLA into the diet of animals.
In ruminants, the process for the formation of milk fat is complex and under multifactorial regulation; therefore, there are several strategies including nutrition, seasonal feed changes, and genome variation that can be used to achieve increased milk beneficial components (Hanuš et al., 2018).
For years, the use of diets in which milk FA composition has been manipulated (e.g., soybean oil, corn oil, safflower oil, linseed oil, canola oil, marine algae, and fish oil) represented a good approach for the nutraceutical improvement of milk dairy goats, with a potential positive consequence on human and animal health (Savoini et al., 2019). Many factors, such as the type of fat supplementation and the interaction with the rumen microbiome, influence the FA transfer from diet to milk (Chilliard et al., 2000; Chilliard et al., 2003).
In the last few years, there has been an increasing demand for Cannabis sativa L. (hemp), a wind-pollinated annual plant (Mukherjee et al., 2008) grown for its fibers, oil, and seeds. In particular, the main value of hemp seed resides in its fatty acid composition, consisting of two dietary essential fatty acids, namely, linoleic acid (LA; 18:2ω6) and α-linolenic acid (ALA; 18:3ω3) in the ratio of 2.5–3:1, which have been affirmed to be ideal for human nutrition (Simopoulos, 2008). In addition, metabolites such as γ-linolenic acid (GLA; 18:3ω6) and stearidonic acid (SDA; 18:4ω3), present in hemp seed oil, make the nutritional value of hemp seed superior to other seed oils (Leizer et al., 2000). These metabolites are the precursors for longer chain fatty acid synthesis which are involved in many important biological processes (Guil-Guerrero et al., 2010).
In this regard, hemp seeds might be a good and new source of nutrients for livestock (Russo and Reggiani, 2015; Iannaccone et al., 2019); they are rich in oil and proteins and provide a considerable amount of dietary fiber, vitamins, and minerals. Hemp seeds for livestock nutrition have been mainly used to improve the quantity and quality of production: especially in cattle, positive effects were obtained on beef tissue, while in ewes, after hemp seed feeding, the most affected pathways were correlated to the production of energy (Gibb et al., 2005; Iannaccone et al., 2019).
Many studies have investigated the molecular mechanisms involved in mammary lipogenesis and fat secretion in ruminants induced by changes in the diet, providing further knowledge on the network of genes and transcription regulators driving milk fat synthesis (Bauman et al., 2008; Bionaz and Loor, 2008; Harvatine et al., 2009; Kadegowda et al., 2009; Invernizzi et al., 2010; Mach et al., 2011; Mu et al., 2021).
Next-generation RNA-sequencing technology (RNA-Seq) is used for the analysis of the expression of genes in specific conditions providing deeper knowledge of the transcriptomic events under such conditions. Nowadays, next-generation sequencing (NGS) has become increasingly popular also in nutrigenomics investigations in dairy ruminants, providing, for example, more understanding of the regulatory mechanisms underlying the effect of the supplementation on bovine milk fat synthesis and concomitant increase in milk polyunsaturated fatty acids (PUFAs) of benefit to human health (Ibeagha-Awemu et al., 2016).
In goats, different studies have investigated the effect of a variety of feeding strategies on milk fat composition; however, the physiological basis for this particular response is not yet fully understood. For example, the results of the study of Ollier and coworkers (Ollier et al., 2009) showed that milk yield and composition were affected by whole intact rapeseeds or sunflower oil in high-forage or high-concentrate diets but not the mammary mRNA expression of the key genes involved in lactose (e.g., α-lactalbumin), protein (e.g., β-casein), and lipid metabolism (e.g., lipoprotein, lipase). In addition, transcriptome analysis performed using a bovine oligonucleotide microarray did not provide evidence of treatments inducing significant changes in the expression of specific genes in the mammary gland, suggesting that after the dietary treatment, other factors, such as substrate availability for mammary metabolism, could play an important role in contributing to milk FA responses to changes in caprine diet composition. Also, in the most recent study by Toral et al. (2013) on lactating goats fed sunflower seed oil supplement, the response to the dietary treatments did not seem to be mediated by changes in the mRNA abundance of genes encoding key lipogenic enzymes and the related transcription factors.
On the contrary, diets supplemented with extruded linseed alone or in combination with fish oil in lactating goats showed both to largely change milk FA composition and to have effects on mRNA linked to protein metabolism and transport pathways than to lipid metabolism (Faulconnier et al., 2018).
More recently, the effect of the supplementation with linseed on milk fatty acid composition and gene expression in the mammary gland was evaluated also in dairy ewes. In this case, the linseed supplementation has been shown not only to improve the health and nutritional properties of dairy milk but also to influence the expression of gene networks related to energy balance, RNA binding, circadian rhythm, and metabolism of nutrients (Conte et al., 2021).
MicroRNAs (miRNAs) are a class of small non-coding RNAs 18–25 nt in length that are able to modulate post-transcriptional gene expression through negative regulation or mRNA degradation (Bartel, 2004). In the last few years, thanks to high-throughput sequencing technologies, significant progress has been made in miRNA studies, as evidenced by the 38,589 mature miRNAs available in the miRBase database, release 22.1 (Kozomara et al., 2019).
The miRNAs play an important role in the processes of cell differentiation, biological development, and physiology also in farm animals, including the regulation of muscle development and hypertrophy, adipose tissue growth, oocyte maturation, and early embryonic development (Wang et al., 2013).
Lactating mammary gland miRNAs were characterized in ruminants, such as buffalo (Cai et al., 2017), goat (Mobuchon et al., 2015; Xuan et al., 2020), sheep (Hao et al., 2020; Wang et al., 2021), and cattle (Li et al., 2012; Wang et al., 2016; Billa et al., 2019), highlighting the miRNAs related to pregnancy and the different lactating and non-lactating periods. miRNAs play key roles also in the regulation of milk fat synthesis and catabolism in ruminants (Chen et al., 2018; Dysin et al., 2021). Furthermore, the role of miRNAs as regulators of bovine milk protein and milk fat synthesis affected by the diet was recently reported (Wang et al., 2018).
However, despite the growing number of studies (Lu et al., 2021), the specific mechanism of action of miRNAs remains to be more deeply explored to fully comprehend milk fat metabolism mechanisms in ruminants. In the papers cited above, transcriptome and miRNAs were often studied together, thus characterizing the interaction between mRNA and miRNAs.
In our two previous studies, we have investigated the effect of hemp seeds on milk production, energy and nitrogen balance, methane emissions (Rapetti et al., 2021), and rumen microbiome (Cremonesi et al., 2018) in dairy goats. The objectives of this study were to i) evaluate the effect of a diet enriched with hemp seeds on goat milk composition, ii) identify changes in goat milk transcriptome and miRNA, and (iii) determine the general biological mechanisms induced by the diet.
2 Materials and Methods
2.1 Ethics Statement
The University of Milan Ethics Committee for animal use and care (authorization 88/14, 29 January 2014) approved the animal procedures; experiments were managed in accordance with the guidelines of the Italian law on animal welfare for experimental animals (Legislative Decree 116/92).
2.2 Animals and Diet Treatment
The study was conducted at the Research Center “Cascina Baciocca” at Cornaredo (Milan) of the University of Milan (Italy). The procedures for animal management were reported in our previous papers on the same group of animals (Cremonesi et al., 2018; Rapetti et al., 2021). In particular, for this study, 10 secundiparous Alpine goats previously selected as unaffected by caprine arthritis encephalitis virus (CAEV) and mastitis and with similar phenotypic and genetic traits were randomly divided into two groups with different dietary treatments. The treatments were arranged to evaluate the main effects of a dietary lipid supplementation with hemp seed (35.7% EE on a DM basis), on milk transcriptome and lipid metabolism as follows: control diet (C) and a diet supplemented with 9.4% on DM of hemp seed (H). In order to balance the fat supplementation in the experimental diets, corn flakes (18.6%) and solvent soybean meal (6.6%) were included in the C diet. The diet composition has been reported in detail in Cremonesi et al. (2018).
The animals were fed for 40 days with a transition diet (time point = T0) administered after the calving diet that was used for about the first 10 days of lactation. The transition diet was very similar to the control diet fed in T1, but with a slightly higher forage to concentrate ratio (51/49 vs. 49/51, for T0 and T1, respectively). Thereafter, until 140 ± 7 days of lactation (T1), goats were fed different rations corresponding to different dietary treatments. During the entire experiment, the goats had free access to water and were fed twice daily (08:30 a.m. and 17:30 p.m.).
2.3 Milk Sample Collection
Individual milk samples for milk yield and composition analysis were collected three times during the last week of the transition diet (DIM = 43, on average) and three times during the last week of the experimental diet administration (DIM = 137, on average).
At the two time points (T0 and T1), 2 h later (2H) after morning milking (MM), the milk samples were collected for transcriptomic analysis, based on goat milk somatic cell viability previously described in Turri et al. (2015). Briefly, these analyses have revealed that the percentage of viable cells in milk increased significantly from MM to 2H samples (63.97 ± 2.35a and 77.01 ± 2.25b, respectively). Moreover, the kind of milking did not affect the number of somatic cells (neutrophils: MM = 54.7 ± 2.9, 2H = 55.6 ± 3.5; macrophages: MM = 3.9 ± 0.3, 2H = 4.1 ± 0.4; and epithelial cells: MM = 5.7 ± 0.8, 2H = 6.6 ± 1.0).
For transcriptomic analysis, 100 ml of the milk samples collected from the two half-udders 2 h later after the MM were transferred to falcon tubes and instantly chilled on ice and transferred within 1 h to the molecular laboratory. Then the samples were centrifuged at 750×g at 4°C for 10 min. The fat layer and the supernatant were discarded, and the cell pellet was then resuspended and washed twice in phosphate-buffered saline solution (PBS) pH 7.2 (Invitrogen, Milan, Italy). After centrifugation at 450×g for 10 min, the supernatant was discarded and the pellet was resuspended in 3 ml of TRIzol (Invitrogen, Milan, Italy) and stored at −80°C until the RNA extraction.
2.4 RNA Extraction
Total RNA was extracted from at least 106 of milk somatic cells with TRIzol reagent (Invitrogen, Milan, Italy), following the instructions of the supplier (Invitrogen, Milan, Italy) and then analyzed with the Agilent Bioanalyzer 2100 for quantification and evaluation of quality and purity (only high-quality RNA with RIN greater than 7 and A260/280 ratio greater than 1.8 was used in the transcriptomic analysis).
2.5 Fatty Acid Profile
The fatty acid composition of individual milk samples was determined by gas chromatography (GC) after base-catalyzed transesterification of fat as described in De Noni and Battelli (2008).
2.6 Library Preparation and Sequencing
For RNA-Seq analysis, 1 μg of total RNA was used for library construction. Libraries were generated using the Illumina TruSeq® RNA Sample Preparation v2 Kit (Illumina, Inc., San Diego, CA, USA) according to the manufacturer’s instructions using poly(A) enrichment. Libraries were quality-checked and quantified on Agilent 2200 TapeStation, High Sensitivity D1000 (Agilent, Inc., Santa Clara, CA, USA). The samples were then used for cluster generation and subsequent sequencing on Illumina HiSeq 2000 (Illumina, Inc., San Diego, CA, USA), and 100-base paired-end reads were generated.
For miRNA-Seq analysis, small RNA libraries were generated using the Illumina TruSeq Small RNA Preparation kit (Illumina, Inc., San Diego, CA, USA) according to the manufacturer’s instructions. Libraries were quality-checked and quantified on Agilent 2200 TapeStation, High Sensitivity D1000. A pool of libraries was then size-selected by the Pippin Prep system (Sage Science, MA, USA) to recover the 125- to 167-nt fraction containing mature miRNAs. The pool was then used for cluster generation and subsequent sequencing on Illumina HiSeq 2000 (Illumina, Inc., San Diego, CA, USA), and 50-base single reads were generated.
2.7 Bioinformatic Analysis
For RNA-Seq analysis, raw data were trimmed using Trimmomatic (Bolger et al., 2014). Sequences were aligned to the goat reference genome version 1.0 using STAR_2.3.0 (Dobin et al., 2013). Subsequently, HTSeq-count (Anders et al., 2015) was used to count sequences aligned to each gene. The software package edgeR of Bioconductor (Robinson et al., 2009) was used to estimate differentially expressed genes (DEGs). Cluster analysis was performed with Genesis (Sturn et al., 2002).
For miRNA-Seq analysis, miRNA detection and discovery were carried out with miRDeep2 on Illumina high-quality trimmed sequences. Capra hircus miRNAs available at MirBase (http://www.mirbase.org/) were used to accomplish known miRNA detection on the trimmed sequences. Known miRNAs from related species (sheep, cow, and horse) available at MirBase were also input into miRDeep2 to support the individuation of novel miRNAs.
The miRDeep2 quantifier module was used to quantify expression and retrieve counts for the detected known and novel miRNAs. Differential expression analyses were run with the Bioconductor edgeR package (Sturn et al., 2002). Enrichment analysis of DEGs was performed using pathfindR (Ulgen et al., 2019). Box plot graphics were obtained via the online version of BoxPlotR (http://shiny.chemgrid.org/boxplotr/).
2.8 Statistical Analysis
Data of milk production and composition within each time point were averaged by goat by the MEANS procedure of SAS (SAS version 9.4, SAS Institute, Cary, NC, USA) (SAS Institute Inc, 2013); then, for each time point, the data were analyzed using the generalized linear model procedure of SAS (SAS version 9.4, SAS Institute, Cary, NC, USA) (SAS Institute Inc, 2013) with the following model:
where Y is the dependent variable, µ is the overall mean, Ti is the treatment effect (i = 1, 2), and ei is the residual error. Least squares mean estimates were reported. For all statistical analysis, significance was declared at p <0.05 and trend at p <0.1.
Principal component analysis (PCA) was performed using Genesis version 1.8.1 (Sturn et al., 2002).
2.9 Validation of Gene Expression by Real-Time qPCR
To validate the RNA-Seq data, a subset of genes, randomly selected—such as fatty acid synthetase (FAS); acyl CoA diacylglycerol acyltransferase enzyme (DGAT2); retinoid X receptor alpha (RXRA); retinoic acid receptor alpha (RARA); JunB proto-oncogene, AP-1 transcription factor subunit (JUNB); low-density lipoprotein receptor (LDLR); lipin 2 (LPIN2); interleukin-1 receptor type 2 (IL1R2)—and two housekeeping genes, namely, ribosomal protein S9 (RPS9) and keratin 8 (KRT8), were analyzed. All primers were designed by using the specific sequences and the Primer Express® Software v2.0 (Supplementary Table 1). A pool of RNAs from H and C animals in the two time points (T0 and T1) was retrotranscribed with SuperScript™ II Reverse Transcriptase (Thermo Fisher Scientific, Waltham, MA, USA), following the manufacturer’s instructions. Quantitative real-time polymerase chain reaction was carried out on cDNAs with 7900HT Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA). Reactions were done in 10-µl volumes containing 0.5 µl of each forward primer and 5 µl of each reverse primer, 4 µl of cDNA, and 5 µl of SYBR™ Green PCR Master Mix (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. Negative controls using water in place of the sample were performed alongside each reaction. RT-PCR reaction was run using the cycling parameters of 95°C for 10 min, 40 cycles of 95°C for 15 s, and 60°C for 1 min. Relative expression levels were calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001).
2.10 Functional Annotation and Pathway Analyses of Differentially Expressed Genes
For RNA-Seq, functional analysis of the DEGs was performed using pathfindR, an R package for the comprehensive identification of enriched pathways in omics data (Ulgen et al., 2019). Human homologous differentially expressed miRNAs (DE-miRNAs) were analyzed using miRWalk2.0 to attribute target genes. Finally, functional analysis for miRNA target mRNAs was performed using pathfindR.
3 Results
3.1 Effect of Treatments on Milk Fat/Protein Percentages and Individual Fatty Acid Profiles
No significant differences were observed at T0 in milk yield and milk FA composition, confirming that the two groups of animals were homogeneous in terms of milk quality.
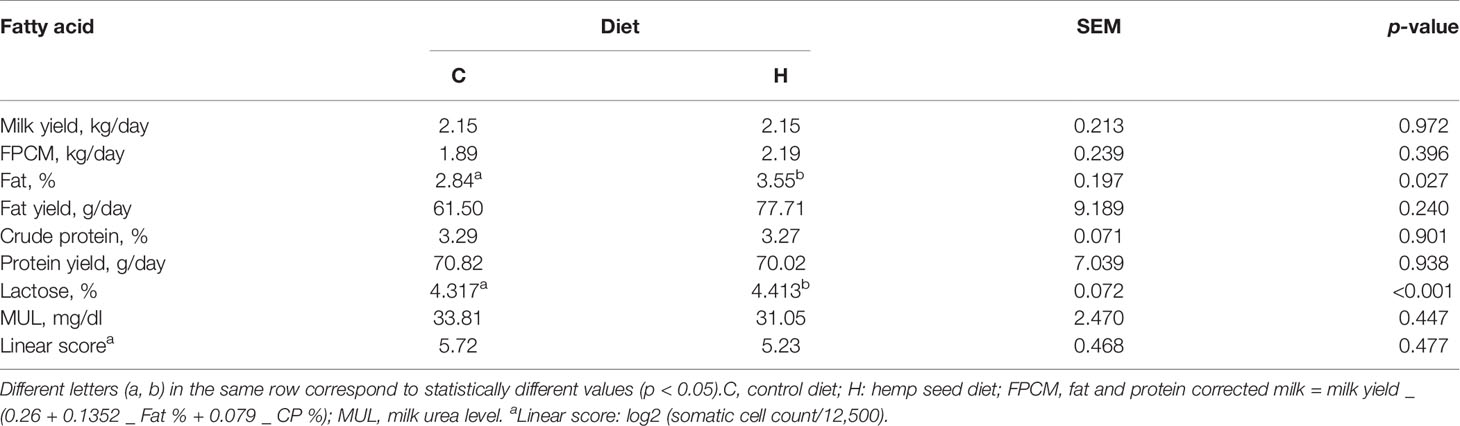
As shown in Table 1, milk production was on average 2.15 kg/day without differences among diets. FPCM, crude protein, protein yield, MUL, and linear score were not affected by the diet.
Milk fat was influenced by dietary lipid supplementation, showing an increment of the fat % (+25%) (p-value = 0.027) in the goats fed the H diet as compared to those fed the C diet.
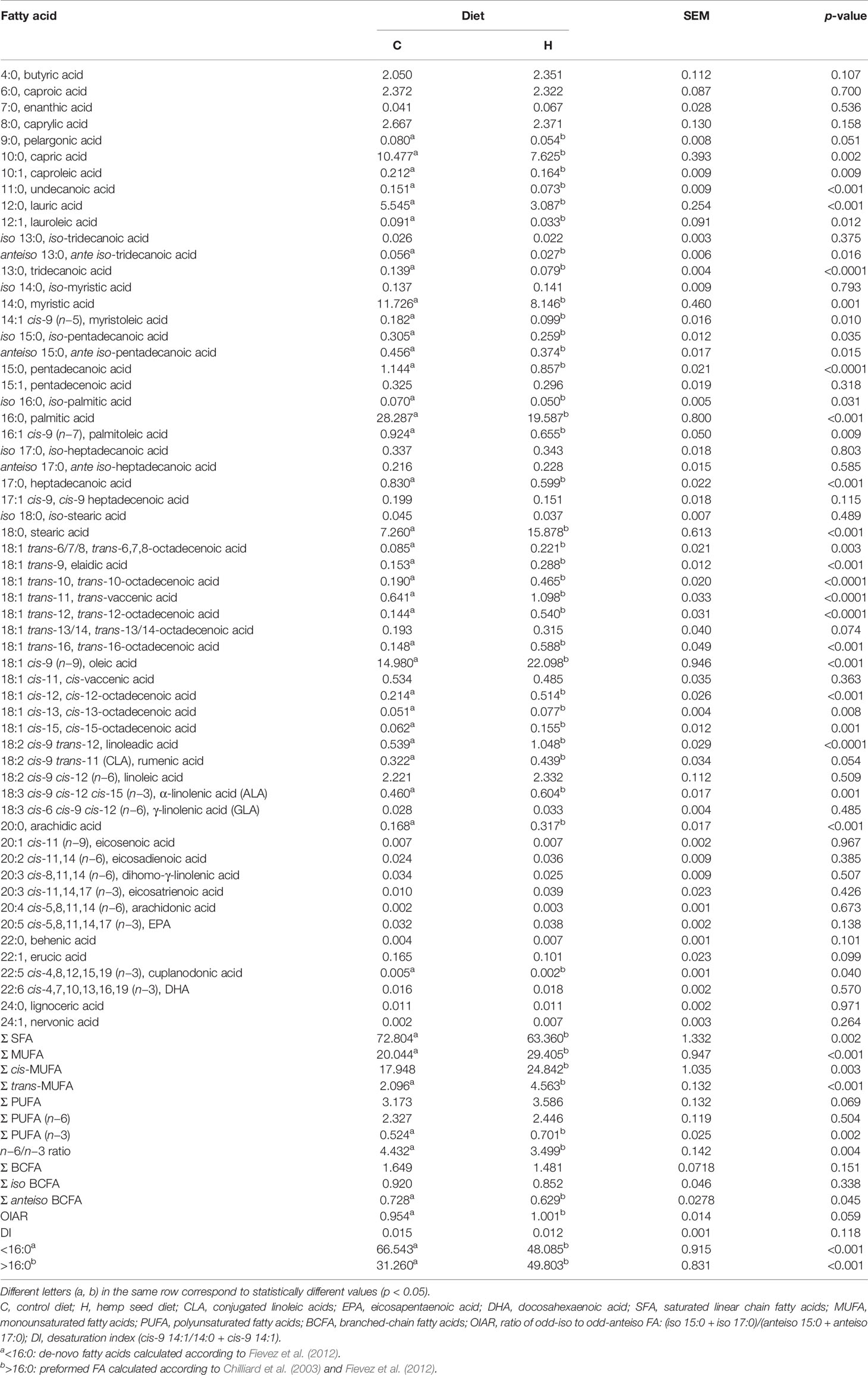
The administration of the two different rations caused the change in the fatty acid profile of milk fat of goats receiving rations supplemented with hemp seed, changing its nutritional and nutraceutical values after diet differentiation (T1) (Table 2). The hemp diet, characterized by higher concentrations of LA and ALA than the control diet (+52% and +28% for LA and ALA, respectively), determined an increase of the most abundant long-chain FA in milk, particularly stearic (+118%) and oleic (+48%) acids. Concerning FA with nutraceutical value, hemp treatment increased the concentration in milk of CLA (0.32% vs. 0.44%, p-value = 0.054) and ALA (0.46% vs. 0.60%, p-value < 0.001). The latter contributed to an increase in PUFA n−3 concentration (+34%), reducing the n−6/n−3 ratio (4.43 vs. 3.50, for C and H, respectively; p-value = 0.004). On the other hand, the milk produced by the goats fed with the H diet resulted in lower de-novo FA concentration, particularly capric (−27%), lauric (−44%), myristic (−31%), and palmitic (−31%) acids.
3.2 RNA-Seq Results
3.2.1 RNA-Seq Output
RNA-Seq was obtained from milk somatic cells (MSCs) isolated from five animals for each diet (C and H) and time point (T0 and T1). An average of 15.3 million counts/sample, generated by RNA-Seq analysis, was mapped against the goat reference genome (CHIR_1.0 GenBank assembly accession: GCA_000317765.1, ftp://ftp.ncbi.nih.gov/genomes/Capra_hircus). The principal component analysis (PCA) clearly separated four out of five animals fed with the hemp diet from the other samples (PC1 = 42.95%) (Figure 1). Other animals from C at T1 were also close to H at T1, likely indicating an effect on transcriptomics occurring during time-course lactation.
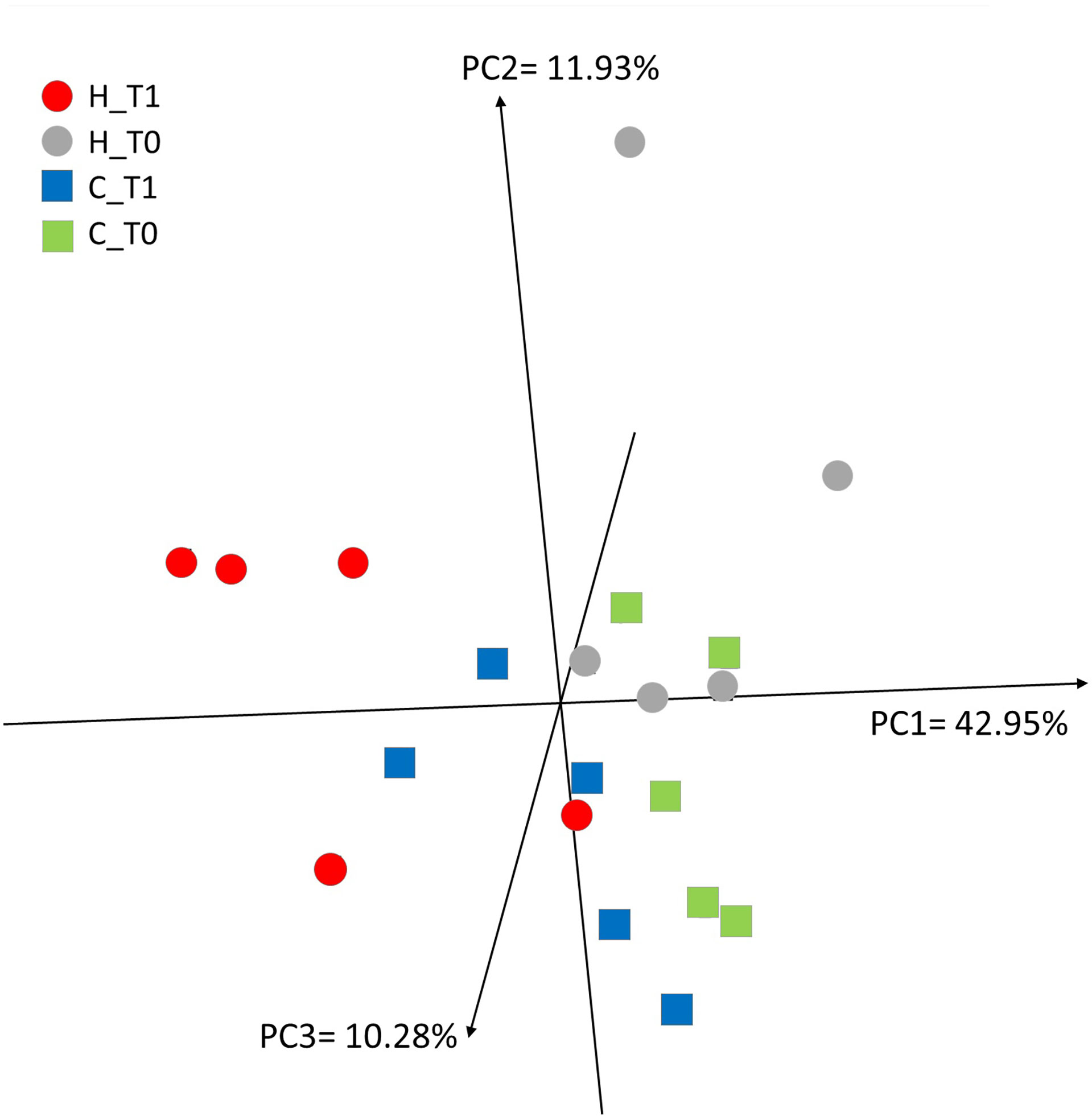
Figure 1 Principal component analysis describing the 13,063 mRNAs expressed in milk somatic cells (MSCs) from the five goats fed with the control (C, square) diet and five goats fed with the hemp seed (H, circle) diet at the two time points (T0 and T1).
When the DEGs were determined between time points, animals given hemp supplementation showed a highly reshaped gene expression [3,835 DEGs in T1 vs. T0, with p-value < 0.05, fold change > 1.5, and false discovery rate (FDR) < 0.05], whereas the control animals showed a limited number of DEGs (374 DEGs in T1 vs. T0, with p-value < 0.05, fold change > 1.5, and FDR < 0.05), probably related to the different animal lactation stages rather than diet (Supplementary Files 1, 2). In fact, most of the DEGs (318 of 374 DEGs) found in the control animals were similarly differentially expressed between the two time points in H animals (Figure 2).
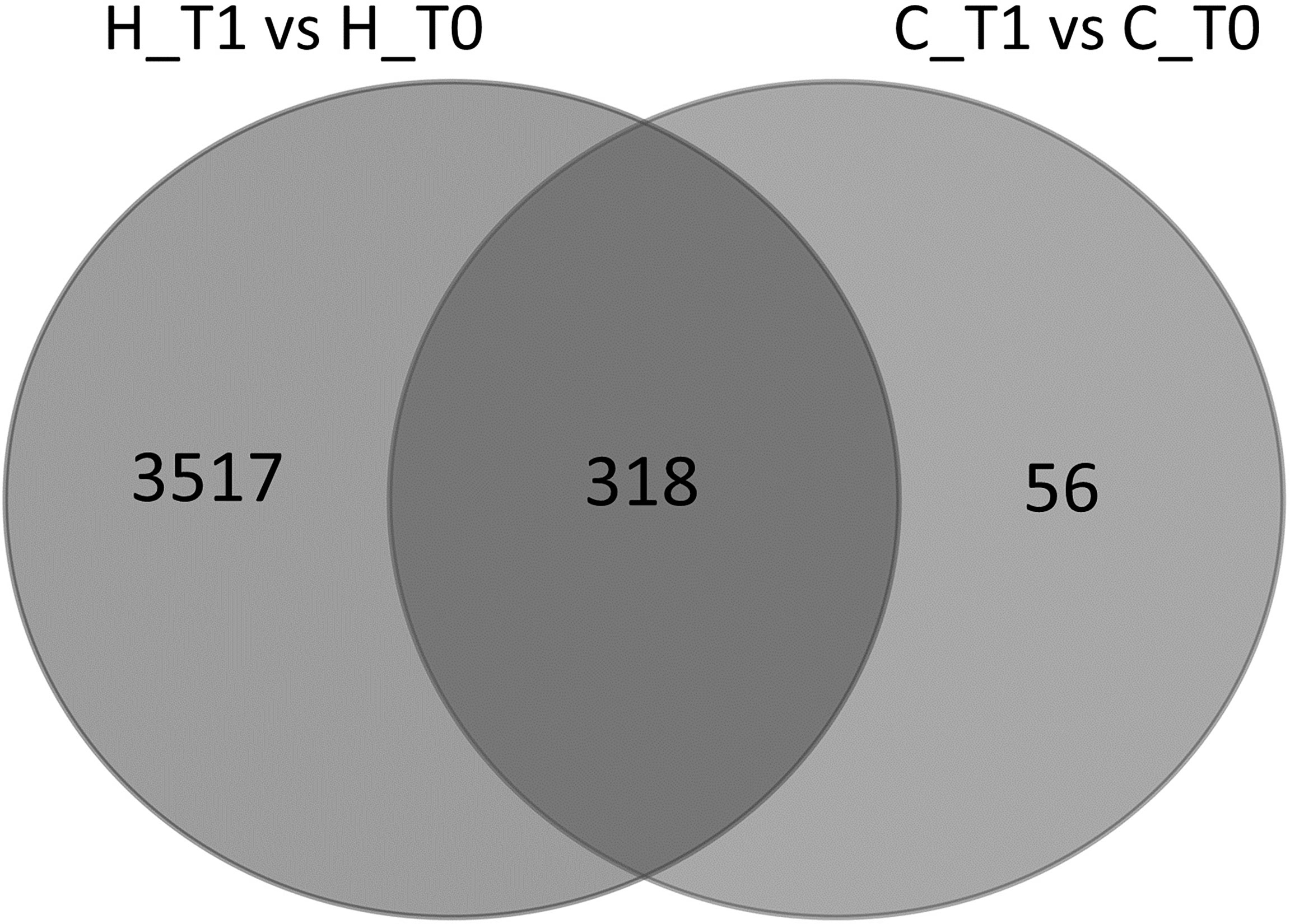
Figure 2 Venn diagram representing the intersection between the list of differentially expressed genes (DEGs) (p-value < 0.05 and FDR < 0.05) in milk somatic cells (MSCs) from goats fed with the hemp seed (H) diet and control (C) diet in the two time points (T0 and T1).
3.2.2 miRNA-Seq Output
An average of 3.5 million counts/sample, generated by miRDeep analysis, was obtained from the comparison of the triplicate of the H and C animals.
In total, 1,175 unique miRNAs were detected in MSCs using miRDeep2. Among these, 406 were known C. hircus miRNAs, 276 were homologous of known miRNAs from other species, and 493 were new candidate miRNAs (Supplementary File 3).
Thirty-eight (15 known and 23 novel, respectively) were DE-miRNAs after diet conversion for the H animals, whereas the control animals showed 17 DE-miRNAs (10 known and 7 novel, respectively) (Supplementary File 4, Table 3).
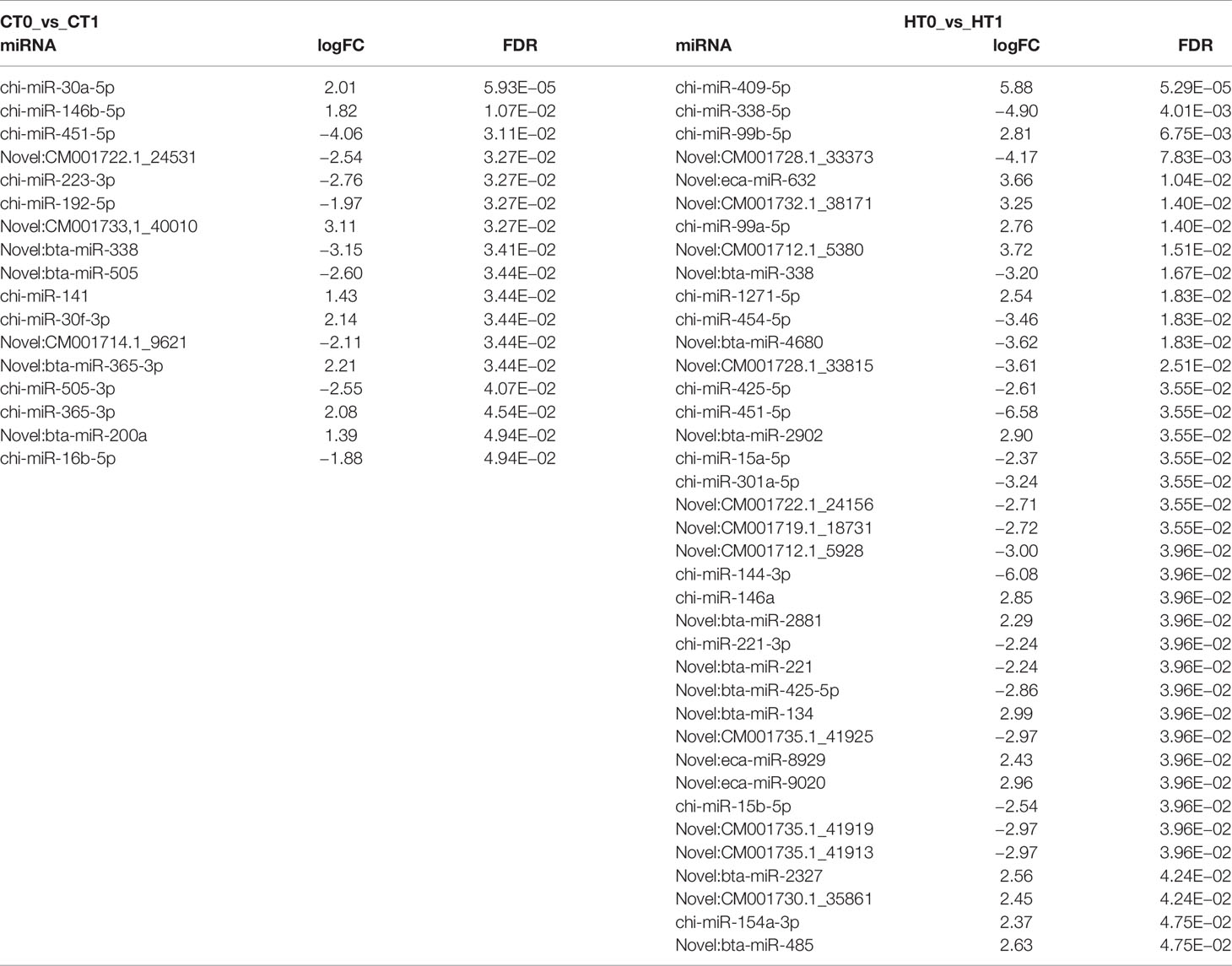
Table 3 Differentially expressed miRNAs (DE-miRNAs) in milk somatic cells (MSCs) from animals given the hemp (H) and control (C) diets in the two time points (T0 and T1).
3.2.3 Functional Categorization of DEGs and Bioinformatic Pathway Analysis
The effect of diet with H supplements versus control animals on biological response was studied to identify functions and pathways that were most affected after 3 months of feeding the experimental diets.
Functional analysis of the 3,835 (H) and 374 (C) DEGs showed 137 and 66 pathways that were significantly enriched from the H and C diets, respectively (Supplementary File 5). The top 15 significant pathways enriched in both diets are shown in Figure 3.
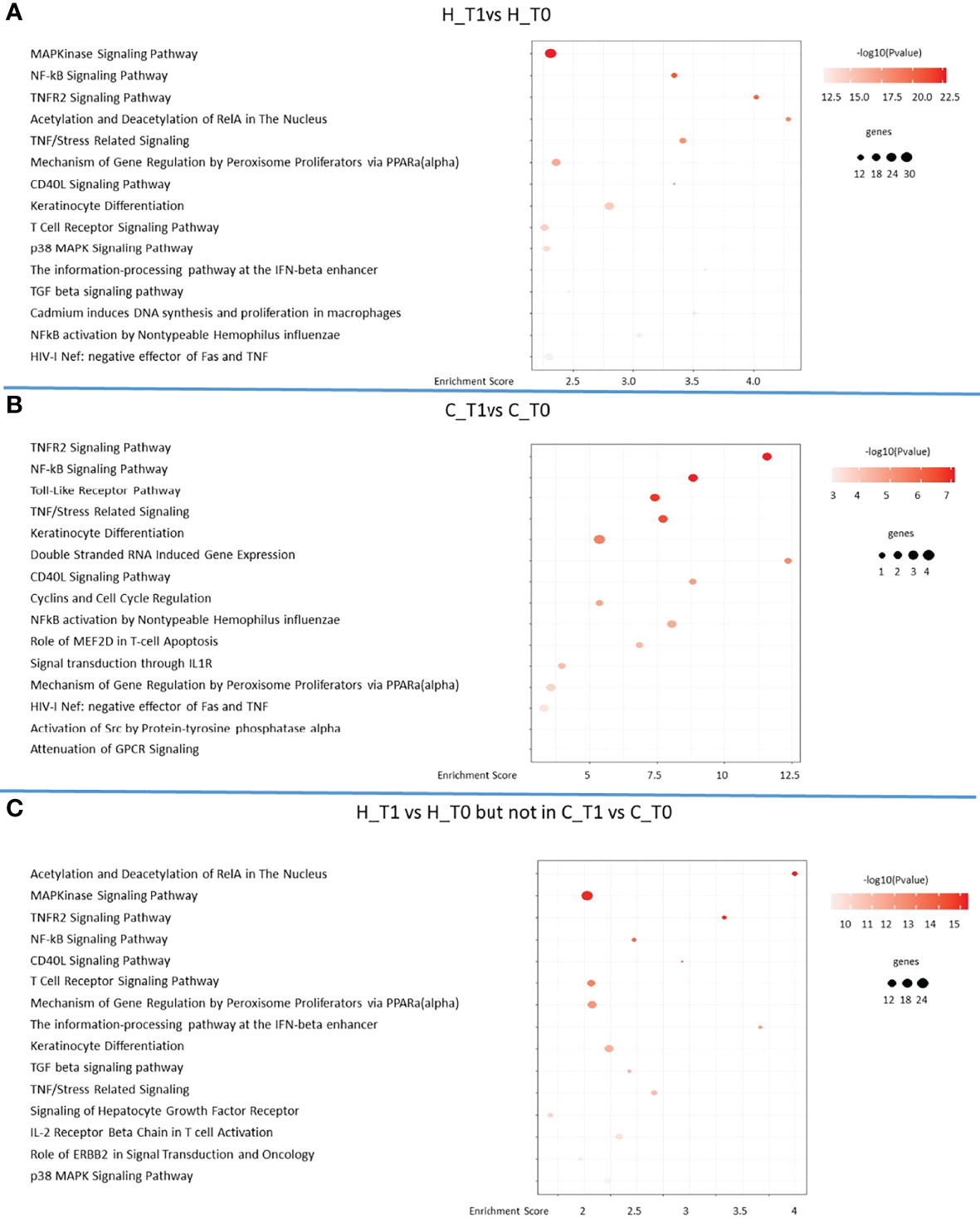
Figure 3 The 15 most significant pathways identified in goats fed with hemp seeds (H) plot (A) and the control (C) group plot (B) in the time course (T0 and T1) and after a comparison between the two diets plot (C).
Fifty-eight significant pathways were together altered in the H and C animals, showing a similar reshaping of their transcriptome during the experimental time course probably related to the progression of lactation, even though the H diet seemed to highly increase the intensity and the number of genes involved in pathway regulation (Supplementary File 5).
The predicted functional landscape of differentially expressed genes in the two groups was prevalently associated with the regulation of immune response (Figure 3, Supplementary File 4). In addition, the peroxisome proliferator-activated receptor (PPAR) signaling was observed to be enriched in the pathway analysis, where the H diet influenced the genes indirectly associated with PPARα signaling. The gene regulation by peroxisome proliferators via the PPAR(alpha) pathway in the H and C animals in the time course (T0 and T1) is shown in detail in Figure 4.
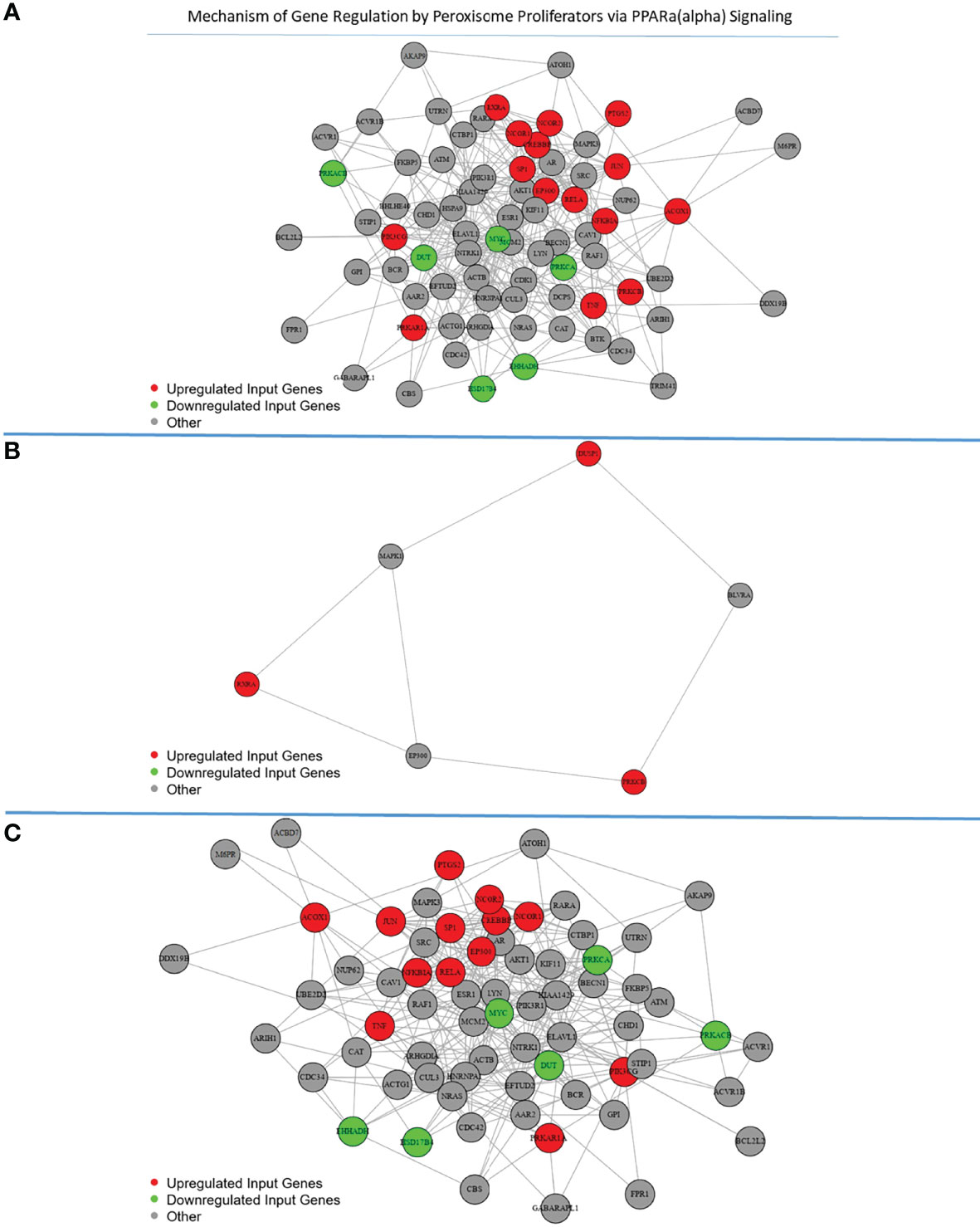
Figure 4 Mechanism of gene regulation by peroxisome proliferators via the PPAR(alpha) pathway: the genes involved in the pathway are reported in red (upregulated) or green (downregulated) for the hemp diet plot (A) and the control diet plot (B) in the time course (T0 and T1) and after a comparison between the two diets plot (C).
In the case of miRNA results, the Gene Ontology (GO) analysis for target genes resulted in the enrichment of immune response pathways for the H diet (Supplementary Table 3, Supplementary File 6). Moreover, in animals given the H diet, the PPAR pathway was highly enriched in the GO analysis, with many putative miRNAs targeting genes regulated by the peroxisome proliferators via PPAR(alpha).
3.3 Real-Time qPCR Validation of RNA-Seq Results
A subset of DEGs (FAS, RXRA, RARA, JUNB, LDLR, PLIN2, IL1R2, and DGAT2) after diet conversion for the H animals was randomly selected and validated by qPCR, using RPS9 and KRT8 as reference genes (Supplementary Table 2).
The supplementation with hemp seeds impacted the transcriptomics of these genes showing a higher expression compared to the control diet (Figure 5A), and real-time qPCR validated the RNA-Seq results, showing also in this case a higher expression in animals fed hemp seeds for all the genes tested (Figure 5B).
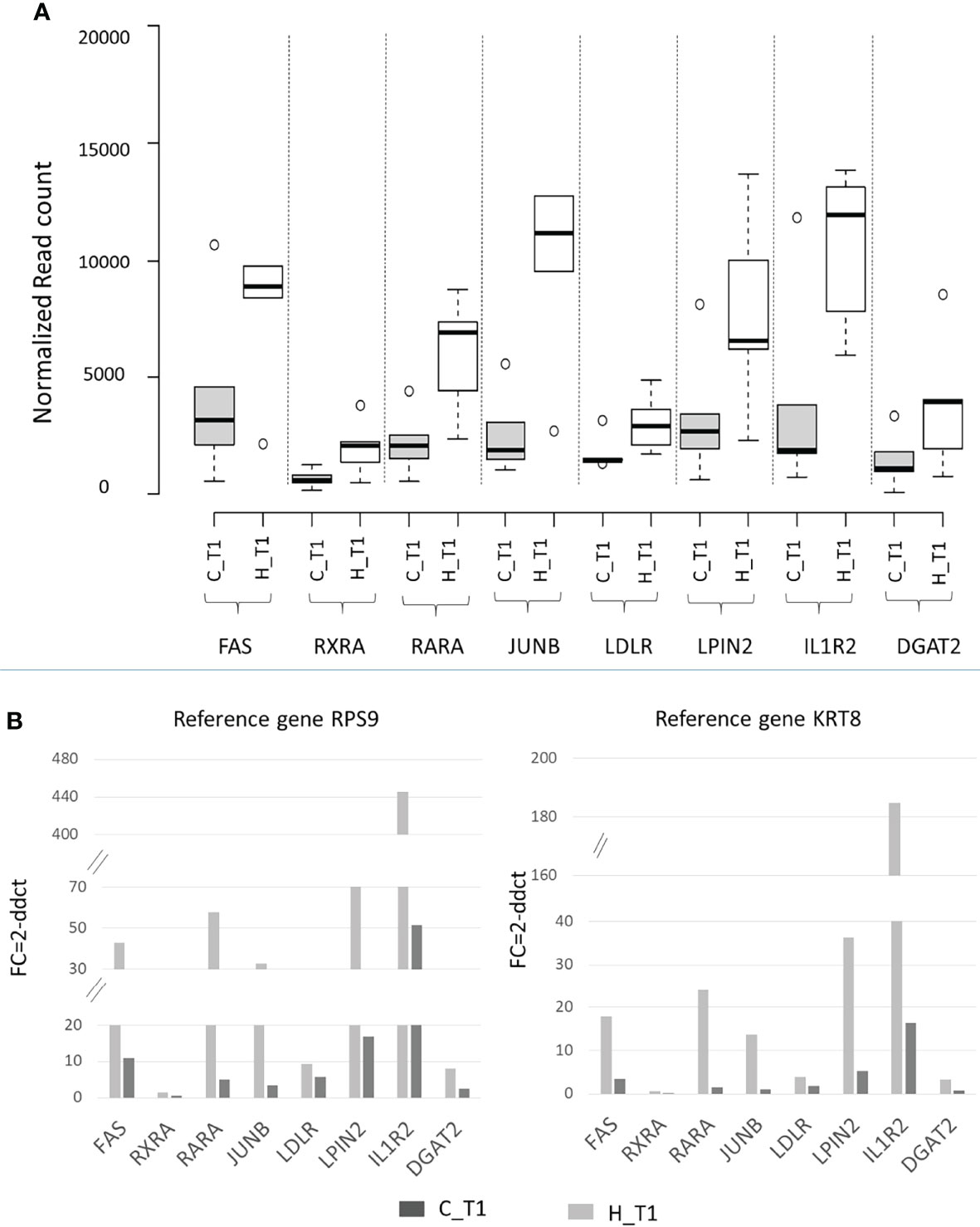
Figure 5 (A) Box plot of RNA expression (normalized read count) and (B) real-time qPCR expression (fold change) for a subset of DEGs (FAS, RXRA, RARA, JUNB, LDLR, PLIN2, IL1R2, and DGAT2) in the hemp (H) and control (C) groups at T1. Ribosomal protein S9 (RPS9) and keratin 8 (KRT8) were used as reference genes.
4 Discussion
The availability of “omics” technologies in nutritional research in ruminants has led to greater knowledge of how nutrients can affect the animal at the cellular level, where nutrients and bioactive compounds in diets can indirectly interact with the genome through mediators as transcription factors (Osorio and Moisa, 2019). Feeding is the most possible approach to produce changes in the milk FA profile of ruminants at the mammary gland level, improving milk quality. In our study, hemp seed supplementation affected milk FA composition, showing higher concentrations of LA, ALA, stearic and oleic acids, and CLA, compared to the non-supplemented group, as reported in Table 2.
Recently, hemp seed supplementation in dairy animal feeding has been described as a way of transferring bioactive substances to milk for human consumption (Bailoni et al., 2021). In agreement with the results obtained by Cozma and coworkers who evaluated the effect of hemp seed oil in Carpathian goats (Cozma et al., 2015), also in our study, the lipid supplementation with hemp seeds did not affect goat milk yield. In addition, these authors found a significant increase in the PUFA concentrations in milk produced by goats supplemented by hemp seed oil, without an effect on n−3 fatty acid contents. It was found that the content of cis-9 and trans-11 CLA increased on average during the experimental period, reaching the peak during the second week of oil supplementation but then decreasing from the third week. In our work, an increase in milk fat of long-chain (>16:0) fatty acids in the H group was observed, especially for CLA, ALA, and PUFA n−3 concentrations, which are beneficial to human health (Cozma et al., 2015; Dixit et al., 2015), without adversely affecting goat performance or health.
As previously described (Al-Khalaifah, 2020), dietary FA supplementations were able to modulate not only metabolic but also inflammatory and immune processes, both through direct action on receptors and transcription factors and through their bioactive oxygenated metabolites, and in-vivo and in-vitro (Agazzi et al., 2004; Thanasak et al., 2004; Bronzo et al., 2010; Lecchi et al., 2011) studies demonstrated, in general, positive effects on immune functions.
The present study pointed out that among the top 15 significant pathways enriched in the H animals, the “MAPK signaling pathway,” “T cell receptor signaling pathway,” “mechanism of gene regulation by peroxisome proliferators via PPAR(alpha),” and “keratinocyte differentiation” were the most represented (Figure 3).
In particular, in “MAPK signaling pathway,” hemp supplementation influenced the expression of 28 genes that were all overexpressed, while only one (MYC) gene was downregulated. MAPK genes are mainly involved in driving cellular responses to a wide range of stimuli, such as mitogens, osmotic stress, heat shock, and pro-inflammatory cytokines (Guo et al., 2016). They control cell functions which comprise apoptosis, mitosis, gene expression, proliferation, differentiation, and cell survival.
The “T cell receptor signaling pathway,” involved in the functioning of T cells and the formation of the immunological synapse, showed 11 upregulated and 5 downregulated genes in H goats, while the “mechanism of gene regulation by peroxisome proliferators via PPAR(alpha)” and “keratinocyte differentiation” pathways revealed 13 and 17 overexpressed and 6 and 1 downregulated genes in the H animals, respectively. The first pathway is involved in the catabolism and homeostasis of the lipids stimulating beta-oxidation of fatty acids, while the second is involved in the regulation of keratinocyte differentiation and the function of the skin barrier.
These four pathways share JUN, FOS, NFKB1, and NFKBIA genes, all overexpressed following hemp diet supplementation. JUN plays a role in cell proliferation, cell survival, and apoptosis, and it has been recently described as involved in lipid accumulation by binding directly to the SREBF1 promoter (Rincon et al., 2001). Moreover, the activation of FOS and JUN occurs as a downstream event of three MAPK signaling pathways (Marion-Letellier et al., 2015) with a key role in signal transduction by gene transcription modulation in the nucleus after changes in the cellular environment.
Interestingly, FAs can target NFKB that regulates key genes (such as NFKB1 and NFKBIA) implicated in inflammatory processes such as cytokines, chemokines, adhesion molecules, and matrix metalloproteinases (DeWille et al., 1979), confirming previously well-described results (Hwang, 1989; Bronzo et al., 2010).
Hemp diet supplementation is also able to modulate the PPARs; these are well-studied receptors in ruminants (Burris et al., 2013), where they are found to be key regulators of lipid and carbohydrate metabolism (Anthos et al., 2021), development, growth, and inflammatory responses (Faulconnier et al., 2018; Anthos et al., 2021).
In goats, PPARs showed their ability to be initialized by dietary fatty acids in different tissues (e.g., liver, skeletal muscle, mammary gland, adipose, immune cells) (Agazzi et al., 2010): in this species, PPARγ has been associated with adipogenic effects in the mammary gland (Coyne et al., 2008), while PPARα has been associated with FA oxidation in the liver during the transition period (Gonzalez-Calvo et al., 2014).
In particular, in our study, the PPAR signaling pathway was modulated by diet supplementation via PPARα. In the animals fed hemp seeds, the genes PIK3CG, NFKBIA, NCOR2, PTGS2, TNF, PRKAR1A, CREBBP, PRKCB, NCOR1, ACOX1, RELA, EP300, RXRA, SP1, and JUN were upregulated, while MYC, HSD17B4, EHHADH, PRKACB, DUT, and PRKCA were downregulated.
As previously described, PPARs are activated by various FAs that bind the ligand-dependent activation function present in the PPAR structure, and thus, they serve as major transcriptional sensors of fatty acids (Wolfrum et al., 2001). According to Wolfrum and coworkers (Wolfrum et al., 2001), the higher concentration of LA and ALA in goats fed with the hemp-supplemented diet makes the PUFAs potent agonists to the PPARα gene. Moreover, a significant upregulation of PPARα gene expression in the liver of goats fed the high ALA diet has been described by Ebrahimi et al. (2015), suggesting that PPARα is the key messenger responsible for the translation of nutritional stresses into changes in hepatic gene expression. In a more recent work on goat mammary cells (Vargas-Bello-Pérez et al., 2019), PPARs revealed an indirect role in PUFA regulation of milk fat-related genes showing their permissive effect for trans10-cis12-CLA and a blocking effect for DHA. In our work, the significant effect of hemp treatment on the increase of PUFA n−3 can explain the upregulation of PPARα, as reported by Hassan et al. (2021). Moreover, the inhibitory effect of PUFA n−6 on PPAR receptor signaling reported by these authors was not observed in our trial because the differences between treatments in terms of PUFA n−6 concentration in milk were very small and not significant. On the other hand, unlike what was previously reported in cows (Ebrahimi et al., 2015), in our experiment, we did not observe any downregulation of lipogenic genes, as demonstrated also by the total amount of de-novo FAs synthesized in the mammary gland that was very similar between treatments. In fact, the differences observed in terms of their concentration were counterbalanced by the higher total milk fat percentage. The results obtained in this study explain only partially the mechanism of action of hemp seeds for FA secretion in goats; the modulation of gene expression may involve a huge number of biochemical processes and genes, as previously reported (Ollier et al., 2009; Conte et al., 2021).
Even epigenetic mechanisms play a significant role as mediators of nutrient–gene interactions in ruminants with implications in ruminant nutrition and physiology that are only now beginning to be discovered. Among the various epigenetic mechanisms, non-coding RNAs and, in particular, microRNAs have been extensively studied in recent years in ruminants, especially in cattle (Wang et al., 2012; Wang et al., 2013; Koufariotis et al., 2015). Furthermore, recent studies have indicated that miRNAs are an important regulator of fat synthesis and lipolysis (Lu et al., 2021).
In our study, we uncovered 38 differentially expressed miRNAs in goats fed the hemp seed diet. Interestingly, as reported in Supplementary Table 3, the pathway analysis of putative miRNA target genes revealed the same pathways related to immunity and lipid metabolism that were significantly enriched by the H diet in the transcriptome analysis, e.g., “MAPK signaling pathway” and “mechanism of gene regulation by peroxisome proliferators via PPAR(alpha),” confirming the role of the identified miRNAs as mediators of nutrient–gene interactions in ruminants.
For instance, in our study, miR-15b-5p, involved in the “mechanism of gene regulation by peroxisome proliferators via PPAR(alpha),” was downregulated by the H diet (Table 3). miR-15b has been described as a regulator of lipid metabolism in caprine mammary epithelial cells, where inhibition of miR-15b expression increased lipid content in mammary epithelial cells through elevation of the lipid synthesis enzyme fatty acid synthetase (FASN), and the overexpression of miR-15b reduced lipid content in mammary epithelial cells with decreasing levels of FASN (Chu et al., 2018).
Another miRNA we found down-expressed in the H animals, miR-454, was recently described as a regulator of triglyceride synthesis in bovine mammary epithelial cells by means of a direct interaction with PPARγ (Zhang et al., 2019). Triglycerides account for 99% of milk fat and play a central role in determining dairy product quality. In the work of Zhang and coworkers (Zhang et al., 2019), both the mRNA and protein levels of PPARγ were negatively correlated with miR-454 expression. Moreover, triglyceride production was inversely correlated with miR-454 expression.
In a study conducted by Mobuchon et al. (2015) on miRNA expressed in lactating goat mammary gland, the genome position analysis showed that 114 caprine miRNAs were located within known bovine quantitative trait locus (QTL) regions for milk production and composition. Among these, mir-1271, located in BTA 7; mir-338, located in BTA 19; and miR-154a, located in BTA 21, were associated with QTL for milk fat percentage and content, whereas miR-425, located in BTA 22, was associated with QTL for milk fat yield. In our work, these four miRNAs were found to be differentially expressed in animals fed the H diet.
Other pathways identified for putative miRNA target genes were related to immunity functions (Table S3).
Accumulating evidence suggests that miRNAs are an important part of the complex regulatory networks that control the processes of immune cells (Dysin et al., 2021). For instance, miR-146a, found in our study to be differentially expressed in animals fed the H diet and involved in many immunity-related pathways, is a well-studied miRNA involved in immune responses initiated by Toll-like receptor signaling pathways and regulates nuclear transcription factor kappa B (NF-κB) activation (Lai et al., 2020). Previous studies had already shown that miR-146a expression in bovine mammary gland tissues was significantly increased in cases of mastitis caused by Staphylococcus aureus, Escherichia coli, or mixed bacterial infection (Wang et al., 2016), supporting the hypothesis that bovine miR-146a regulates the secretion of inflammatory cytokines in bovine mammary epithelial cells (Wang et al., 2017).
Likewise, miR-144, found differentially expressed in animals fed the H diet, has been described to be upregulated in the mammary gland infected with S. aureus and downregulated in the mammary gland infected with E. coli, identifying a total of 31 potential target genes involved in immunity (Luoreng et al., 2018). In our work, we found miR-144 involved in 23 pathways, including EGF signaling, MAPK signaling, and MAPK inactivation of SMRT corepressor pathways.
Moreover, miR-15a, involved in our study in as many as 28 pathways, was described as controlling functions that regulate the inflammation and differentiation of immune cells (Calin et al., 2008; Moon et al., 2014; Chen et al., 2019) as well as mediating the differentiation of adipocytes in goats, thus suggesting also a role in stimulating the synthesis of milk fat (Chen et al., 2016).
In conclusion, hemp seed supplementation in dairy goats increased monounsaturated and polyunsaturated n−3 FAs, determining a reduction in n−6/n−3 ratio. Therefore, feeding goats with hemp seed supplementation increases the functional and nutraceutical properties of milk, making it more useful for human consumption and health.
In addition, this diet caused changes in the genes involved in metabolism, immune, and inflammatory responses. In particular, modifications in feeding strategies affected also key transcription factors regulating the expression of several genes involved in milk fat metabolism, such as PPAR signaling. Finally, the identified differentially expressed miRNAs play a role as mediators of gene–nutrient interactions, acting on the same functional pathways of the differentially expressed genes.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/bioproject/, Accession Number PRJNA851350. Further enquiries can be directed to the corresponding author.
Ethics Statement
The animal procedures were conducted under the approval of the University of Milan Ethics Committee for animal use and care (authorization 88/14, 29 January 2014), in accordance with the guidelines of the Italian law on animal welfare for experimental animals (Legislative Decree 116/92), in force at the time of the trial.
Author Contributions
PC participated in the feeding trial, sample collection, RNA extraction, qPCR, data analysis, data interpretation, and manuscript drafting. EC contributed to RNA-Seq and miRNA-Seq, qPCR, data analysis, data interpretation, and manuscript drafting. FT participated in the feeding trial, sample collection, milk cell analysis, data analysis, data interpretation, and manuscript drafting. BL contributed to bioinformatic analyses. SCh contributed to sample collection, RNA extraction, and qPCR. GB contributed to fatty acid analyses. SCo participated in the feeding trial and sample collection. LR and BC contributed to the study formulation, design and coordination, data interpretation and visualization, and manuscript drafting. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Agreement Regione Lombardia—National Research Council (CNR), Project “FilAgro -Strategie Innovative e sostenibili per la filiera agroalimentare.”
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors wish to thank Christian Mallo for his valuable collaboration in the feeding trial and sample collection during his graduate internship.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fanim.2022.909271/full#supplementary-material
Supplementary File 1 | HTSeq Count.
Supplementary File 2 | List of the differentially expressed genes in Hemp and Control treatment.
Supplementary File 3 | List of miRNAs identified in this study.
Supplementary File 4 | List of the differentially expressed miRNAs in Hemp and Control treatment.
Supplementary File 5 | Functional analysis of the differentially expressed genes in Hemp and Control treatment.
Supplementary File 6 | List of miRNAs predicted target genes and pathways.
Supplementary File 7 | List of mature and novel microRNAs.
References
Agazzi A., Cattaneo D., Dell’Orto V., Moroni P., Bonizzi L., Pasotto D., et al. (2004). Effect of Administration of Fish Oil on Aspects of Cell-Mediated Immune Response in Periparturient Dairy Goats. Small Rumin Res. 55, 77–83. doi: 10.1016/j.smallrumres.2004.02.007
Agazzi A., Invernizzi G., Campagnoli A., Ferroni M., Fanelli A., Cattaneo D., et al. (2010). Effect of Different Dietary Fats on Hepatic Gene Expression in Transition Dairy Goats. Small Rumin Res. 93, 31–40. doi: 10.1016/j.smallrumres.2010.04.027
Al-Khalaifah H. (2020). Modulatory Effect of Dietary Polyunsaturated Fatty Acids on Immunity, Represented by Phagocytic Activity. Front. Vet. Sci. 7. doi: 10.3389/fvets.2020.569939
Anders S., Pyl P. T., Huber W. (2015). HTSeq-A Python Framework to Work With Highthroughput Sequencing Data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Anthos C., Eirini K., Chinmay J., Vassiliki A. B. (2021). The Role of Peroxisome Proliferator-Activated Receptors (PPAR) in Immune Responses. Metabolism 114, 154338. doi: 10.1016/j.metabol.2020.154338
Bailoni L., Bacchin E., Trocino A., Arango S. (2021). Hemp (Cannabis Sativa L.) Seed and Co-Products Inclusion in Diets for Dairy Ruminants: A Review. Animals 11, 856. doi: 10.3390/ani11030856
Bartel D. P. (2004). MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116:281–297. doi: 10.1016/s0092-8674(04)00045-5
Bauman D. E., Perfield J. W., Harvatine K. J., Baumgard L. H. (2008). Regulation of Fat Synthesis by Conjugated Linoleic Acid: Lactation and the Ruminant Model. J. Nutr. 138, 403–409. doi: 10.1093/jn/138.2.403
Billa P. A., Faulconnier Y., Ye T., Chervet M., Le Provost F., Pires J. A. A., et al. (2019). Deep RNA-Seq Reveals Mirnome Differences in Mammary Tissue of Lactating Holstein and Montbéliarde Cows. BMC Genom. 20, 1–11. doi: 10.1186/s12864-019-5987-4
Bionaz M., Loor J. J. (2008). Gene Networks Driving Bovine Milk Fat Synthesis During the Lactation Cycle. BMC Genomics 9, 366. doi: 10.1186/1471-2164-9-366
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic. A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bronzo V., Puricelli M., Agazzi A., Invernizzi G., Ferroni M., Moroni P., et al. (2010). Effects of Protected Fish Oil in the Diet of Periparturient Dairy Goats on Phenotypic Variation in Blood and Milk Leukocytes. Animal 4, 1510–1517. doi: 10.1017/S1751731110000522
Burris T. P., Solt L. A., Wang Y., Crumbley C., Banerjee S., Griffett K., et al. (2013). Nuclear Receptors and Their Selective Pharmacologic Modulators. Pharmacol. Rev. 65, 710–778. doi: 10.1124/pr.112.006833
Cai X., Liu Q., Zhang X., Ren Y., Lei X., Li S., et al. (2017). Identification and Analysis of the Expression of microRNA From Lactating and Nonlactating Mammary Glands of the Chinese Swamp Buffalo. J. Dairy Sci. 100, 1971–1986. doi: 10.3168/jds.2016-11461
Calin G. A., Cimmino A., Fabbri M., Ferracin M., Wojcik S. E., Shimizu M., et al. (2008). MiR-15a and MiR-16-1 Cluster Functions in Human Leukemia. Proc. Natl. Acad. Sci. U.S.A. 105, 5166–5171. doi: 10.1073/pnas.0800121105
Chen Z., Qiu H., Ma L., Luo J., Sun S., Kang K., et al. (2016). MiR-30e-5p and MiR-15a Synergistically Regulate Fatty Acid Metabolism in Goat Mammary Epithelial Cells via LRP6 and YAP1. Int. J. Mol. Sci. 17, 1909. doi: 10.3390/ijms17111909
Chen Z., Shi H., Sun S., Luo J., Zhang W., Hou Y., et al. (2018). MiR-183 Regulates Milk Fat Metabolism via MST1 in Goat Mammary Epithelial Cells. Gene 646, 12–19. doi: 10.1016/j.gene.2017.12.052
Chen Z., Zhou J., Wang X., Zhang Y., Lu X., Fan Y., et al. (2019). Screening Candidate MicroR-15a- IRAK2 Regulatory Pairs for Predicting the Response to Staphylococcus Aureus-Induced Mastitis in Dairy Cows. J. Dairy Res. 86, 425–431. doi: 10.1017/S0022029919000785
Chilliard Y., Ferlay A., Mansbridge R. M., Doreau M. (2000). Ruminant Milk Fat Plasticity: Nutritional Control of Saturated, Polyunsaturated, Trans and Conjugated Fatty Acids. Anim Res. 49, 181–205. doi: 10.1051/animres:2000117
Chilliard Y., Ferlay A., Rouel J., Lamberet G. (2003). A Review of Nutritional and Physiological Factors Affecting Goat Milk Lipid Synthesis and Lipolysis. J. Dairy Sci. 86, 1751–1770. doi: 10.3168/jds.S0022-0302(03)73761-8
Chu M., Zhao Y., Yu S., Hao Y., Zhang P., Feng Y., et al. (2018). miR-15b Negatively Correlates With Lipid Metabolism in Mammary Epithelial Cells. Am. J. Physiol. Cell. Physiol. 314, C43–C52. doi: 10.1152/ajpcell.00115.2017
Conte G., Giordani T., Vangelisti A., Serra A., Pauselli M., Cavallini A., et al. (2021). Transcriptome Adaptation of the Ovine Mammary Gland to Dietary Supplementation of Extruded Linseed. Animals 11, 2707. doi: 10.3390/ani11092707
Conte G., Serra A., Mele M. (2017). Dairy Cow Breeding and Feeding on the Milk Fatty Acid Pattern. In Nutrients Dairy Their Implications Health Dis. Acad. Press CHAPTER 2, 19–41. doi: 10.1016/B978-0-12-809762-5.00002-4
Coyne G. S., Kenny D. A., Childs S., Sreenan J. M., Waters S. M. (2008). Dietary N-3 Polyunsaturated Fatty Acids Alter the Expression of Genes Involved in Prostaglandin Biosynthesis in the Bovine Uterus. Theriogenology 70, 772–782. doi: 10.1016/j.theriogenology.2008.05.048
Cozma A., Andrei S., Pintea A., Miere D., Filip L., Loghin F., et al. (2015). Effect of Hemp Seed Oil Supplementation on Plasma Lipid Profile, Liver Function, Milk Fatty Acid, Cholesterol, and Vitamin A Concentrations in Carpathian Goats. Czech J. Anim Sci. 60, 289–301. doi: 10.17221/8275-CJAS
Cremonesi P., Conte G., Severgnini M., Turri F., Monni A., Capra E., et al. (2018). Evaluation of the Effects of Different Diets on Microbiome Diversity and Fatty Acid Composition of Rumen Liquor in Dairy Goat. Animal 12, 1856–1866. doi: 10.1017/S1751731117003433
De Noni I., Battelli G. (2008). Terpenes and Fatty Acid Profiles of Milk Fat and “Bitto” Cheese as Affected by Transhumance of Cows on Different Mountain Pastures. Food Chem. 109, 299–309. doi: 10.1016/j.foodchem.2007.12.033
DeWille J. W., Fraker P. J., Romsos D. R. (1979). Effects of Essential Fatty Acid Deficiency, and Various Levels of Dietary Polyunsaturated Fatty Acids, on Humoral Immunity in Mice. Nutr. J. 109, 1018–1027. doi: 10.1093/jn/109.6.1018
Dixit S. P., Sivalingam J., Tyagi A. K., Saroha V., Sharma A., Nagda R. K. (2015). Association of Novel SNPs in the Candidate Genes Affecting Caprine Milk Fatty Acids Related to Human Health. Meta Gene 21, 45–56. doi: 10.1016/j.mgene.2015.01.004
Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. (2013). STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Dysin A. P., Barkova O. Y., Pozovnikova M. V. (2021). The Role of microRNAs in the Mammary Gland Development, Health, and Function of Cattle, Goats, and Sheep. Noncoding RNA 7, 78. doi: 10.3390/ncrna7040078
Ebrahimi M., Rajion M. A., Meng G. Y., Farjam A. S., Oskoueian E., Jafari S. (2015). Diet High in α-Linolenic Acid Up-Regulate PPAR-α Gene Expression in the Liver of Goats. Electr. J. Biotechnol. 18, 210–214. doi: 10.1016/j.ejbt.2015.03.009
Faulconnier Y., Bernard L., Boby C., Domagalski J., Chilliard Y., Leroux C. (2018). Extruded Linseed Alone or in Combination With Fish Oil Modifies Mammary Gene Expression Profiles in Lactating Goats. Animal 12, 1564–1575. doi: 10.1017/S1751731117002816
Fievez V., Colman E., Castro-Montoya J.M., Stefanov I., Vlaeminck B. Milk odd- and branched-chain fatty acids as biomarkers of rumen function-An update. Anim. Feed Sci. Technol. (2012) 172, 51–65.
Gibb D. J., Shah M. A., Mir P. S., Mcallister T. A. (2005). Effect of Full-Fat Hemp Seed on Performance and Tissue Fatty Acids of Feedlot Cattle. Can. J. Anim Sci. 85, 223–230. doi: 10.4141/A04-078
Gonzalez-Calvo L., Joy M., Alberti C., Ripoll G., Molino F., Serrano M., et al. (2014). Effect of Finishing Period Length With Alpha-Tocopherol Supplementation on the Expression of Vitamin E-Related Genes in the Muscle and Subcutaneous Fat of Light Lambs. Gene 552, 225–233. doi: 10.1016/j.gene.2014.09.037
Guil-Guerrero J. L., Rincon-Cervera M. A., Venegas-Venegas E. (2010). Gamma-Linolenic and Stearidonic Acids: Purification and Upgrading of C18-PUFA Oils. Eur. J. Lipid Sci. Technol. 112, 1068–1081. doi: 10.1002/ejlt.200900294
Guo J., Fang W., Sun L., Lu Y., Dou L., Huang X., et al. (2016). Reduced miR-200b and miR-200c Expression Contributes to Abnormal Hepatic Lipid Accumulation by Stimulating JUN Expression and Activating the Transcription of Srebp1. Oncotarget 7, 36207–36219. doi: 10.18632/oncotarget.9183
Hanuš O., Samková E., Křížová L., Hasoňová L., Kala R. (2018). Role of Fatty Acids in Milk Fat and the Influence of Selected Factors on Their Variability-A Review. Molecules 23, 1636. doi: 10.3390/molecules23071636
Hao Z., Zhou H., Hickford J. G., Gong H., Wang J., Hu J., et al. (2020). Identification and Characterization of Circular RNA in Lactating Mammary Glands From Two Breeds of Sheep With Different Milk Production Profiles Using RNA-Seq. Genomics 112, 2186–2193. doi: 10.1016/j.ygeno.2019.12.014
Harvatine K. J., Perfield J. W., Bauman D. E. (2009). Expression of Enzymes and Key Regulators of Lipid Synthesis is Upregulated in Adipose Tissue During CLA Induced Milk Fat Depression in Dairy Cows. J. Nutr. 139, 849. doi: 10.3945/jn.108.099994
Hassan F., Nadeem A., Li Z., Javed M., Liu Q., Azhar J., et al. (2021). Role of Peroxisome ProliferatorActivated Receptors (PPARs) in Energy Homeostasis of Dairy Animals: Exploiting Their Modulation Through Nutrigenomic Interventions. Int. J. Mol. Sci. 22, 12463. doi: 10.3390/ijms222212463
Hwang D. (1989). Essential Fatty Acids and Immune Response. FASEB J. 3, 2052–2206. doi: 10.1096/fasebj.3.9.2501132
Iannaccone M., Ianni A., Contaldi F., Esposito S., Martino C., Bennato F., et al. (2019). Whole Blood Transcriptome Analysis in Ewes Fed With Hemp Seed Supplemented Diet. Sci. Rep. 9, 16192. doi: 10.1038/s41598-019-52712-6
Ibeagha-Awemu E. M., Li R., Ammah A. A., Dudemaine P. L., Bissonnette N., Benchaar C., et al. (2016). Transcriptome Adaptation of the Bovine Mammary Gland to Diets Rich in Unsaturated Fatty Acids Shows Greater Impact of Linseed Oil Over Safflower Oil on Gene Expression and Metabolic Pathways. BMC Genom. 17, 104. doi: 10.1186/s12864-016-2423-x
Invernizzi G., Thering B., McGuire M., Savoini G., Loor J. J. (2010). Sustained Upregulation of Stearoyl-CoA Desaturase in Bovine Mammary Tissue With Contrasting Changes in Milk Fat Synthesis and Lipogenic Gene Networks Caused by Lipid Supplements. Funct. Integr. Genomic 10, 561–575. doi: 10.1007/s10142-010-0179-y
Kadegowda A. K., Bionaz M., Piperova L. S., Erdman R. A., Loor J. J. (2009). Peroxisome Proliferator-Activated Receptor-Gamma Activation and Long-Chain Fatty Acids Alter Lipogenic Gene Networks in Bovine Mammary Epithelial Cells to Various Extents. J. Dairy Sci. 92, 4276. doi: 10.3168/jds.2008-1932
Koufariotis L. T., Chen Y. P., Chamberlain A., Vander Jagt C., Hayes B. J. (2015). A Catalogue of Novel Bovine Long Noncoding RNA Across 18 Tissues. PLoS One 10, e0141225. doi: 10.1371/journal.pone.0141225
Kozomara A., Birgaoanu M., Griffiths-Jones S. (2019). Mirbase: From microRNA Sequences to Function. Nucleic Acids Res. 8, D155–D162. doi: 10.1093/nar/gky1141
Lai Y., Lai Y., Rahman M. M., Chen H., Husna A. A., Fujikawa T., et al. (2020). Bovine Milk Transcriptome Analysis Reveals microRNAs and RNU2 Involved in Mastitis. FEBS J. 287, 1899–1918. doi: 10.1111/febs.15114
Lecchi C., Invernizzi G., Agazzi A., Ferroni M., Pisani L. F., Savoini G., et al. (2011). In Vitro Modulation of Caprine Monocyte Immune Functions by 3 Polyunsaturated Fatty Acids. Vet. J. 189, 353–355. doi: 10.1016/j.tvjl.2010.09.001
Leizer C., Ribnicky D., Poulev A., Dushenkov S., Raskin I. (2000). The Composition of Hemp Seed Oil and its Potential as an Important Source of Nutrition. J. Nutraceut Funct. Med. Foods 2, 35–53. doi: 10.1300/J133v02n04_04
Li Z., Liu H., Jin X., Lo L., Liu J. (2012). Expression Profiles of microRNAs From Lactating and non-Lactating Bovine Mammary Glands and Identification of miRNA Related to Lactation. BMC Genomics 13, 731. doi: 10.1186/1471-2164-13-731
Livak K. J., Schmittgen T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)). Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu Q., Chen Z., Ji D., Mao Y., Jiang Q., Yang Z., et al. (2021). Progress on the Regulation of Ruminant Milk Fat by Noncoding RNAs and ceRNAs. Front. Genet. 12. doi: 10.3389/fgene.2021.733925
Luoreng Z. M., Wang X. P., Mei C. G., Zan L. S. (2018). Comparison of microRNA Profiles Between Bovine Mammary Glands Infected With Staphylococcus Aureus and Escherichia Coli. Int. J. Biol. Sci. 14, 87. doi: 10.7150/ijbs.22498
Mach N., Jacobs A. A. A., Kruijt L., van Baal J., Smits M. A. (2011). Alteration of Gene Expression in Mammary Gland Tissue of Dairy Cows in Response to Dietary Unsaturated Fatty Acids. Animal 5, 1217–1230. doi: 10.1017/S1751731111000103
Marion-Letellier R., Savoye G., Ghosh S. (2015). Polyunsaturated Fatty Acids and Inflammation. IUBMB Life 67 (9), 659–667. doi: 10.1002/iub.1428
Mobuchon L., Marthey S., Boussaha M., Le Guillou S., Leroux C., Le Provost F. (2015). Annotation of the Goat Genome Using Next Generation Sequencing of microRNA Expressed by the Lactating Mammary Gland: Comparison of Three Approaches. BMC Genom. 16, 285. doi: 10.1186/s12864-015-1471-y
Moon H. G., Yang J., Zheng Y., Jin Y. (2014). MiR-15a/16 Regulates Macrophage Phagocytosis After Bacterial Infection. J. Immunol. 193, 4558–4567. doi: 10.4049/jimmunol.1401372
Mu T., Hu H., Ma Y., Feng X., Zhang J., Gu Y. (2021). Regulation of Key Genes for Milk Fat Synthesis in Ruminants. Front. Nutr. 8. doi: 10.3389/fnut.2021.765147
Mukherjee A., Roy S. C., De Bera S., Jiang H. E., Li X., Li C. S., et al. (2008). Results of Molecular Analysis of an Archaeological Hemp (Cannabis Sativa L.) DNA Sample From North West China. Genet. Resour. Crop Evol. 55, 481–485. doi: 10.1007/s10722-008-9343-9
Ollier S., Leroux C., de la Foye A., Bernard L., Rouel J., Chilliard Y. (2009). Whole Intact Rapeseeds or Sunflower Oil in High-Forage or High-Concentrate Diets Affects Milk Yield, Milk Composition, and Mammary Gene Expression Profile in Goats. J. Dairy Sci. 92, 5544–5560. doi: 10.3168/jds.2009-2022
Osorio J. S., Moisa S. J. (2019). Gene Regulation in Ruminants: A Nutritional Perspective. Gene Expression and Control. Dairy Sci. Publ. Database 2142. doi: 10.5772/intechopen.82193
Parodi P. W. (1999). Conjugated Linoleic Acid and Other Anticarcinogenic Agents of Bovine Milk Fat. J. Dairy Sci. 82, 1339–1349. doi: 10.3168/jds.S0022-0302(99)75358-0
Rapetti L., Colombini S., Battelli G., Castiglioni B., Turri F., Galassi G., et al. (2021). Effect of Linseeds and Hemp Seeds on Milk Production, Energy and Nitrogen Balance, and Methane Emissions in the Dairy Goat. Animals 11, 2717. doi: 10.3390/ani11092717
Rincon M., Flavell R. A., Davis R. J. (2001). Signal Transduction by MAP Kinases in T Lymphocytes. Oncogene 20, 2490–2497. doi: 10.1038/sj.onc.1204382
Robinson M. D., McCarthy D. J., Smyth G. K. (2009). Edger: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Russo R., Reggiani R. (2015). “Protein Concentration and Amino Acid Profile in Hemp Seed and Flaxseed Meal,” in Eucarpia International Symposium on Protein Crops. Ed. Ron A. (Pontevedra: Spanish Association for Legumes), 193–195.
Savoini G., Omodei Zorini F., Farina G., Agazzi A., Cattaneo D., Invernizzi G. (2019). Effects of Fat Supplementation in Dairy Goats on Lipid Metabolism and Health Status. Animals 9, 917. doi: 10.3390/ani9110917
Simopoulos A. P. (2008). The Importance of the Omega-6/Omega-3 Fatty Acid Ratio in Cardiovascular Disease and Other Chronic Diseases. Exp. Biol. Med. 233, 674–688. doi: 10.3181/0711-MR-311
Sturn A., Quackenbush J., Trajanoski Z. (2002). Genesis: Cluster Analysis of Microarray Data. Bioinformatics 18, 207–220. doi: 10.1093/bioinformatics/18.1.207
Thanasak J., Rutten V. P., Schonewille J. T., Hoek A., Beynen A. C., Noordhuizen J. P., et al. (2004). Effect of a Dietary N-6 Polyunsaturated Fatty Acid Supplement on Distinct Immune Functions of Goats. J. Vet. Med. A Physiol. Pathol. Clin. Med. 51, 1–9. doi: 10.1111/j.1439-0442.2004.00595.x
Toral P. G., Bernard L., Delavaud C., Gruffat D., Leroux C., Chilliard Y. (2013). Effects of Fish Oil and Additional Starch on Tissue Fatty Acid Profile and Lipogenic Gene mRNA Abundance in Lactating Goats Fed a Diet Containing Sunflower-Seed Oil. Animal 7, 948–956. doi: 10.1017/S1751731113000049
Turri F., Castiglioni B., Cremonesi P., Chessa S., Rapetti L., Gliozzi T. M. (2015). Microscopic Differential Cell Counting and Viability Evaluation of Milk Somatic Cells During Lactation in Goat. Ital. J. Anim. Sci. 14, supplement 1, P-123.
Ulgen E., Ozisik O., Sezerman O. U. (2019). Pathfindr: An R Package for Comprehensive Identification of Enriched Pathways in Omics Data Through Active Subnetworks. Front. Genet. 10. doi: 10.3389/fgene.2019.00858
Vargas-Bello-Pérez E., Zhao W., Bionaz M., Luo J., Loor J. J. (2019). Nutrigenomic Effect of Saturated and Unsaturated Long Chain Fatty Acids on Lipid-Related Genes in Goat Mammary Epithelial Cells: What Is the Role of Pparγ? Vet. Sci. 6 (2), 54. doi: 10.3390/vetsci6020054
Wang D., Liang G., Wang B., Sun H., Liu J., et al. (2016). Systematic Micrornaome Profiling Reveals the Roles of microRNAs in Milk Protein Metabolism and Quality: Insights on Low-Quality Forage Utilization. Sci. Rep. 6, 21194. doi: 10.1038/srep21194
Wang X., Gu Z., Jiang H. (2013). MicroRNAs in Farm Animals. Animal 7, 1567–1575. doi: 10.1017/S1751731113001183
Wang J., Hao Z., Hu J., Liu X., Li S., Shen J., et al. (2021). Small RNA Deep Sequencing Reveals the Expressions of microRNAs in Ovine Mammary Gland Development at Peak-Lactation and During the non-Lactating Period. Genomics 113, 637–646. doi: 10.1016/j.ygeno.2020.09.060
Wang X. P., Luoreng Z. M., Zan L. S., Li F., Li N. (2017). Bovine MiR-146a Regulates Inflammatory Cytokines of Bovine Mammary Epithelial Cells via Targeting the TRAF6 Gene. J. Dairy Sci. 100, 7648–7658. doi: 10.3168/jds.2017-12630
Wang X. P., Luoreng Z. M., Zan L. S., Raza S. H. A., Li F., Li N., et al. (2016). Expression Patterns of MiR-146a and MiR-146b in Mastitis Infected Dairy Cattle. Mol. Cell. Probes 30, 342–344. doi: 10.1016/j.mcp.2016.08.004
Wang M., Moisa S., Khan M. J., Wang J., Bu D., Loor J. J. (2012). MicroRNA Expression Patterns in the Bovine Mammary Gland are Affected by Stage of Lactation. J. Dairy Sci. 95 (11), 6529–6535. doi: 10.3168/jds.2012-5748
Wang X., Zhang L., Jin J., Xia A., Wang C., Cui Y., et al. (2018). Comparative Transcriptome Analysis to Investigate the Potential Role of miRNAs in Milk Protein/Fat Quality. Sci. Rep. 19, 6250. doi: 10.1038/s41598-018-24727-y
Wolfrum C., Borrmann C. M., Borchers T., Spener F. (2001). Fatty Acids and Hypolipidemic Drugs Regulate Peroxisome Proliferator-Activated Receptors Alpha - and Gamma-Mediated Gene Expression via Liver Fatty Acid Binding Protein: A Signaling Path to the Nucleus. Proc. Natl. Acad. Sci. U.S.A. 98, 2323–2328. doi: 10.1073/pnas.051619898
Xuan R., Chao T., Wang A., Zhang F., Sun P., Liu S., et al. (2020). Characterization of microRNA Profiles in the Mammary Gland Tissue of Dairy Goats at the Late Lactation, Dry Period and Late Gestation Stages. PLoS One 15, e0234427. doi: 10.1371/journal.pone.0234427
Keywords: goat, hemp seeds, milk, supplemented diet, fatty acids, transcriptome, miRNAs
Citation: Cremonesi P, Capra E, Turri F, Lazzari B, Chessa S, Battelli G, Colombini S, Rapetti L and Castiglioni B (2022) Effect of Diet Enriched With Hemp Seeds on Goat Milk Fatty Acids, Transcriptome, and miRNAs. Front. Anim. Sci. 3:909271. doi: 10.3389/fanim.2022.909271
Received: 31 March 2022; Accepted: 27 May 2022;
Published: 07 July 2022.
Edited by:
Aristide Maggiolino, University of Bari Aldo Moro, ItalyReviewed by:
María Del Carmen López De Las Hazas Mingo, IMDEA Food Institute, SpainVincenzo Landi, University of Bari Aldo Moro, Italy
Jiangjiang Zhu, Southwest Minzu University, China
Copyright © 2022 Cremonesi, Capra, Turri, Lazzari, Chessa, Battelli, Colombini, Rapetti and Castiglioni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bianca Castiglioni, YmlhbmNhLmNhc3RpZ2xpb25pQGliYmEuY25yLml0
 Paola Cremonesi
Paola Cremonesi Emanuele Capra1
Emanuele Capra1 Federica Turri
Federica Turri Barbara Lazzari
Barbara Lazzari Stefania Chessa
Stefania Chessa Giovanna Battelli
Giovanna Battelli Stefania Colombini
Stefania Colombini Bianca Castiglioni
Bianca Castiglioni