
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Anim. Sci. , 01 June 2022
Sec. Animal Physiology and Management
Volume 3 - 2022 | https://doi.org/10.3389/fanim.2022.898808
This article is part of the Research Topic Embryonic Development, Pregnancy Establishment and Loss View all 9 articles
 M. Sofia Ortega1*
M. Sofia Ortega1* Jason A. Rizo1
Jason A. Rizo1 Jessica N. Drum1
Jessica N. Drum1 Eleanore V. O'Neil1
Eleanore V. O'Neil1 Ky G. Pohler2
Ky G. Pohler2 Karl Kerns1,3
Karl Kerns1,3 Amanda Schmelze1
Amanda Schmelze1 Jonathan Green1
Jonathan Green1 Thomas E. Spencer1*
Thomas E. Spencer1*The mechanisms regulating early stages of placentation and trophectoderm differentiation in the ruminant conceptus remain poorly understood. Here we present a model of trophectoderm (TE) differentiation in vitro from outgrowths of individual in vitro derived embryos. Cell outgrowths expressed markers of mononucleate (MNC) and binucleate (BNC) TE cells. The percentage of BNC ranged from 14 to 39% in individual outgrowths as determined by flow cytometry. Pregnancy-associated glycoproteins (PAGs), produced by BNC, were measured in culture media on days 35 to 54. Continuous secretion of PAGs was observed and indicative of BNC functionality. Gene expression was evaluated in 20 embryo cell outgrowths derived from two different sires. Expression of HAND1, which is involved in TE differentiation, and CSH2, a BNC-specific gene, was altered in cell outgrowths between the two sires tested. Single-cell RNA-seq analysis of day 40 TE cell outgrowths revealed 11 distinct cell populations, with specific clusters genes involved in TE lineage specification, proliferation, and differentiation. In addition, whole -RNAseq analysis was performed in day 35 and 40 TE cell outgrowths and confirmed sustained expression of genes expressed by BNC, such as CSH2 and some PAGs. The developed in vitro bovine embryo outgrowth culture found evidence for MNC and BNC differentiation and continuous production of PAGs, recapitulating key features of early bovine placenta development. This model can be used to understand the developmental biology of TE cells, provide insights into paternal influences on TE differentiation, and impact our understanding of early pregnancy loss in cattle.
After hatching from the zona pellucida by day 9 post-fertilization, the bovine blastocyst grows into an ovoid shape by day 13 and starts the process of elongation (Brooks et al., 2014; Spencer et al., 2016). The elongating conceptus secretes interferon tau (IFNT), a trophectoderm (TE)-specific gene product that acts as the maternal recognition signal in ruminants (Spencer and Bazer, 2004; Spencer et al., 2007; Roberts et al., 2008). Around day 19, the TE begins to adhere to the luminal epithelium of the endometrium and initiates placentation (Spencer et al., 2007). Placentation involves TE proliferation and differentiation of mononucleate TE cells (MNC) into binucleate cells (BNC), which occurs around day 24 of pregnancy (Greenstein et al., 1958). Trophectoderm BNCs are considered crucial for the formation of placentomes and thus vital for fetal and placental growth (Wooding, 1992). Further, BNC secrete specific pregnancy-associated glycoproteins (PAGs) that can be detected in maternal circulation and used to diagnose pregnancy and monitor placental function (Green et al., 2005; Pohler et al., 2013, 2015). Of note, pregnancies of low fertility sires exhibit increased loss between days 19 and 30 (Ortega et al., 2018). Also, PAG production by the placenta is sire dependent, and low PAG concentrations in the maternal circulation by day 30 of gestation are indicative of embryonic mortality (Franco et al., 2018).
The mechanisms regulating these early stages of placentation in ruminants remain poorly understood, and it is difficult to monitor these processes effectively in vivo or mimic them in vitro. To date, several in vitro culture systems have been developed to study TE differentiation and development that are dependent (Talbot et al., 2000; Bai et al., 2011; Pillai et al., 2019) or independent (Shimada et al., 2001; Hashizume et al., 2006) of a monolayer of feeder cells. The objective here was to develop and evaluate an in vitro model of MNC and BNC differentiation using in vitro derived embryos that was independent of feeder cells and could be used to evaluate sire effects on early placenta development.
Embryo media and supplements were prepared as previously described (Ortega et al., 2017b), and bovine embryo production was performed using standard techniques (Ortega et al., 2018, 2020). Cumulus-oocytes complexed (COCs) were obtained from a single company that provides abattoir-derived COCs from Bos taurus crosses (no breed specific). COCs with at least three layers of compact cumulus cells and homogeneous cytoplasm were placed in groups of 50 into 2 ml glass sterile vials containing 1 ml of oocyte maturation medium equilibrated with air containing 5% (v/v) CO2 covered with mineral oil. Tubes with COC were shipped overnight in a portable incubator (Minitube USA Inc., Verona, WI, USA) at 38.5°C to the University of Missouri. For all experiments, up to three Holstein sires were used. The percentage of putative zygotes that cleaved was determined on day 3 (day 0 = day of insemination), and the number of embryos that reached the blastocyst stage was recorded on day 8 of development. Following IETS grading guidelines (Stringfellow and Seidel, 1998), expanded blastocysts (stage 7), with compact inner cell mass and clear blastocoel (Grade 1) were transferred to a new culture plate with equilibrated bovine synthetic oviduct fluid (SOF-BE2). Blastocysts were supplemented with 10% (v/v) FBS and cultured for 48 h to promote hatching from zona pellucida.
Four reproductive tracts from post-pubertal, non-pregnant cows were collected at slaughter and processed at room temperature (RT). Endometrial stromal fibroblast cells were isolated and cultured as previously described with modifications (Fitzgerald et al., 2019). Briefly, uteri were disinfected with 70% ethanol and rinsed with DMEM/F12 (ThermoFisher Scientific, Valencia, CA) supplemented with 1% penicillin/streptomycin (Sigma, St. Louis, MO) to remove blood and debris. Intercaruncular regions were dissected into strips, transferred to a petri dish, and finely minced with scissors. Approximately 5 cm3 of minced tissue was incubated in 20 mL of digestion solution [DMEM/F12, 0.4 mg/mL Collagenase V (Sigma), 1.25 U/mL Dispase II (Sigma), and 1% antibiotic-antimycotic (Thermofisher)] for 1 h at 38.5°C with agitation. Following digestion, supernatants were filtered through a 40-μm cell strainer, and 20 mL of culture medium [DMEM/F12, 1% antibiotic-antimycotic, 10% (v/v) FBS] was added to stop enzymatic digestion. The filtered suspension was then centrifuged at 500 × g for 5 min at RT. The stromal fibroblast cell pellet was suspended in 3 mL of culture medium, transferred to 75 cm2 culture flasks containing 15 mL of equilibrated culture medium, and maintained at 38.5°C in a humidified 5% CO2 environment. After 6 h, the medium was removed to prevent epithelial cell attachment, and fresh warm medium was added to stromal cells that remained adhered to the flask. Culture medium was changed every 48 h until cells reached 95% confluency, all cells used were between passage 0 and 3. To obtain stroma fibroblast-conditioned medium, medium was changed and collected after 24 h of culture, for up to 4 times per batch of cells. Conditioned medium was combined, filtered (0.2 μm), and stored at 4°C until use.
At day 10, individual hatched blastocysts were transferred to either Collagen IV-coated 24-well plates (Corning, NY, USA) or 8-chamber slides (Ibidi, Martinsried, Planegg, Germany) with TE growth medium [1:1 fresh medium (DMEM/F-12, 10% FBS, 1% antibiotic/antimycotic) and stromal cell conditioned medium] (Hashizume et al., 2006). The initial volume of medium was 400 or 250 μl for wells and chambers, respectively. Cultures were maintained at 38.5°C in a humidified 5% CO2 environment. Attachment was assessed on days 12, 14, 16, 18 and 20 for all embryos by gently swirling the culture plate while assessed using an inverted microscope. Fifty percent of the medium was removed and replaced every 4 days until TE outgrowths covered 50% of the well surface. The medium was then changed every 3 days. Media from 24-well plates was collected for PAG analysis.
Approximately 1 x 106 cells from four TE cell outgrowths were stained in a volume of 500 μL for 15 min with 1:500 H33342. Image-based flow cryometric (IBFC) data acquisition was performed following previous methodology (Kerns et al., 2018). Using a FlowSight image-based flow cytometer (FS) fitted with a 20X microscope objective (numerical aperture of 0.9) with an imaging rate of up to 2,000 events per second. The flow-core diameter was 10 μm and the speed was 66 mm per second. Raw image data were acquired using INSPIRE® software. To produce the highest resolution, the camera setting was at 1.0 μm per pixel of the charged-coupled device. In INSPIRE® FS data acquisition software, two bright-field channels were collected (channels 1 and 9), one side scatter (SSC; channel 6), and one H33342 (channel 7) per cell with a minimum of 1,000 cells collected. The following laser and power settings were used: 405 nm (to excite H33342): 5.0 mW and 785 nM SSC laser: 10.0 mW. First, cells were gated to select those only in focus, utilizing the Gradient RMS calculation of brightfield images in channel 1. Next, single, as opposed to multiple, cells were gated for by plotting the area of M01 (brightfield mask) vs. the aspect ratio of M01 (brightfield mask). A histogram for H33342 intensity was generated to visualize the data.
On day 30 of development, TE cell outgrowths, cultured in 8-chamber slides, were washed 3 times with warmed PBS and then fixed in cold acetone for 15 min at −20°C. After fixation, slides were air dried for 1 min at RT. Cells were then washed twice with PBS with 2 mg/mL of polyvinylpyrrolidone (PBS/PVP) and permeabilized using PBS with 0.5% (v/v) Triton X-100 for 1 h at RT. Cells were washed again with PBS/PVP and blocked with 10% normal goat serum (Life Technologies, ThermoFisher) for 1 h at RT. Cytokeratin 8 antibody (Rat TROMA-I; DSHB, University of Iowa) was added at a 1:200 dilution in blocking buffer (10% normal goat serum), and cells were incubated for 1 h at RT. Cells were washed twice with PBS/PVP and then incubated for 1 h at RT with secondary Goat anti-Rat IgG conjugated with Alexa Fluor-488 (ThermoFisher) at a 1:400 dilution in blocking buffer. Following incubation, cells were washed twice with PBS/PVP and counterstained with Hoescht 33342 (2.5 μg/ml in ddH2O) for 10 min in the dark. Cells were then rinsed twice with Milli-Q water and covered with SlowFade Gold antifade reagent (ThermoFisher, S36936). Images of three TE cell outgrowths were acquired by confocal microscopy (Leica TCS SP8 MP) and analyzed using ImageJ version V. 1.6 (National Institutes of Health, Bethesda, MD).
To evaluate the presence of BNC, TE outgrowths from three different embryos that reached 80% confluency (around day 24), were mechanically disrupted, and passaged into new collagen IV-coated plates (Corning, NY, USA). After 6 days in culture, cells were collected and pooled to determine the presence of genes known to be expressed in the trophectoderm, including GATA Binding Protein 3 (GATA3) (Bai et al., 2011) and Heart and Neural Crest Derivatives Expressed 1 (HAND1) (Scott et al., 2000), Interferon Tau (IFNT2) (Roberts et al., 2008), placental lactogen (CSH2) (Ushizawa et al., 2007), and Pregnancy Associated Glycoprotein 1 (PAG1) (Ushizawa et al., 2007) by end-point PCR. Primers details are in Supplementary Table 1. Primers for PAG1 and the housekeeping gene Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) were designed and validated as previously described (Ortega et al., 2017a). Primers for the rest of the genes that were previously published (Shimada et al., 2001; Bai et al., 2011), were optimized for the annealing temperature to produce a single distinct PCR product of known size (visualized in an agarose gel), and amplicon sequence was validated through Sanger Sequencing before use. A day 20 conceptus and chorioallantois of a day 30 pregnancy were used as positive controls, and bovine fetal fibroblasts (BFF) were used as negative controls. Total RNA was isolated using Direct-zolTM RNA miniprep kit (Zymo Research, Irvine, CA) according to manufacturer's protocol. Each PCR reaction consisted of 2 μl of cDNA sample, 2 μl of 10X PCR buffer, 1.6 μl of dNTPs mix (0.4 mM each dNTP), 0.4 μl of each primer (10 μM primer solution), 0.25 μl of Hot Start Taq polymerase (Takara Bio, Mountain View, CA), and 14.35 μl of nuclease-free water for a total volume reaction of 20 μl. Amplification conditions were: 95°C for 2 min; 40 cycles at 95°C for 10 sec; 55°C for 30 sec; and 72°C for 1 min followed by 72°C for 10 min. PCR products were separated and visualized in a 1.5% (wt/vol) agarose gel. A total of 20 cell outgrowths were evaluated.
Expression of MNC and BNC markers was quantified by qPCR using specific primers (Supplementary Table 1) validated following described procedures (Ortega et al., 2017a). A total of 20 TE cells outgrowths from two different Holstein sires (10 cells outgrowths/sire) were analyzed. RNA was isolated using Direct-Zol™ RNA miniprep kit (Zymo Research), while reversed transcription and qPCR analysis were performed as previously described (Ortega et al., 2018, 2020). Data were normalized to the geometric mean of two housekeeping genes [GAPDH and Succinate Dehydrogenase Complex Flavoprotein Subunit A (SDHA)] and expressed as fold change relative to housekeeping genes. Gene expression data was analyzed using the GLM procedure of SAS v9.4 (SAS Institute Inc., Cary, NC).
Bovine PAGs concentration from TE cell culture media were detected using an ELISA assay as previously described (Green et al., 2005; Pohler et al., 2015, 2016). The assay sensitivity was 0.2 ng/mL, while intra- and inter-assay coefficients of variation was <10%, respectively. PAGs were evaluated from 30 individual wells among three sires (1 well = 1 embryo, 10 embryos/sire) at days 35, 40, 45, 50, and 54 of development (day 0 = day insemination). Data was tested for normality using the UNIVARIATE procedure and the Shapiro-Wilk test of SAS v 9.4 (SAS Institute Inc.). Differences in PAGs among sires per day of development were determined using a mixed model with a repeated measures statement. The model included sire, day of development, and their interaction. A repeated statement was included with the variable embryo within bull as the repeated subject. The LSMEANS statement with a Tukey-Kramer adjustment was used to determine the means of sire by day of development interaction.
Single-cell RNA-seq analysis was performed using TE outgrowths from three different embryos from one sire at day 40 of culture. TE cells were first mechanically suspended in culture medium and then incubated in a digestion solution [DMEM/F-12, 0.4 mg/mL DNase 1 (Sigma), 0.5 mg/mL collagenase IV (Sigma), and 0.125% (v/v) Trypsin] for 8 to 10 min. Following digestion, cells were filtered through a 40-μm strainer and 1% FBS was added to the filtrate to stop enzymatic digestion. The cell suspension was centrifuged at 300 x g for 5 min. To wash the cells, the pellet was resuspended in 3 mL of culture medium and centrifuged at 100 x g for 5 min, this step was repeated 2 times. After washing, cells were resuspended in base medium and filtered through a 10 μm strainer. Cells were washed as described above. After washing, cells were resuspended in 1 mL of PBS/BSA [PBS supplemented with 0.4% BSA fraction V (Sigma)]. Cells were then counted, and viability was assessed using a hemocytometer and a Trypan blue stain (Thermofisher), and only samples with cell viability above 80% were analyzed. Lastly, cells were resuspended to a concentration of 1,000 viable cells/μL in PBS/BSA. Droplet generation of single cells and library preparation was performed using a 10X Chromium system following manufacturer's protocol at the University of Missouri DNA Core Facility. Bases with high quality (Q > 30) were estimated to be of the unique molecular identity counts. Libraries were sequenced on an Illumina NextSeq sequencer with a target of 25,000 reads per cell. The base call files were processed using Cell Ranger (v. 3.0.1), the proprietary pipeline for single-cell sequence analysis by 10X Genomics. FASTQ files were mapped to bovine reference genome ARS-UCD1.2 using the STAR aligner (Rosen et al., 2020). The Cell Ranger pipeline was also used to perform background filtration and generate feature-barcode matrices as previously described (Fitzgerald et al., 2019). Pre-processing workflow and quality control was performed using the R package “Seurat” following procedures described elsewhere (Butler et al., 2018; Stuart and Satija, 2019). Cells with low gene expression (<200), and high expression of mitochondrial origin (above 15%) were removed from further analysis. After quality control, there were on average 3,147 cells per sample that were used for single cell RNA-seq analysis. After normalization and integration of samples, clusters of cells with similar gene expression profiles were identified using the non-linear dimensional reduction UMAP. In addition, markers that define each cluster were identified using the “FindAllMarkers” function of the Seurat package. As an additional cutoff, only genes that were expressed in at least 50% of the cells of each cluster were considered to find cluster markers.
Whole TE transcriptome analysis was performed in a total of 8 TE outgrowths from a single sire, four at day 35 of and four at day 40 of culture, respectively. High-throughput sequencing was done at the University of Missouri Genomics Technology Core. Libraries were constructed following the manufacturer's protocol with reagents supplied in Illumina's TruSeq mRNA stranded sample preparation kit. The sample concentration was determined by Qubit fluorometer (Invitrogen) using the Qubit HS RNA assay kit, and the RNA integrity was checked using the Fragment Analyzer automated electrophoresis system. Briefly, the poly-A containing mRNA is purified from total RNA (1 ug), RNA is fragmented, double-stranded cDNA is generated from fragmented RNA, and the index containing adapters are ligated to the ends. The amplified cDNA constructs were purified by addition of Axyprep Mag PCR Clean-up beads. The final construct of each purified library was evaluated using the Fragment Analyzer automated electrophoresis system, quantified with the Qubit fluorometer using the Qubit HS dsDNA assay kit, and diluted according to Illumina's standard sequencing protocol for sequencing on the NovaSeq 6000.The quality of files was checked with the FastQC tool (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Illumina adapters were trimmed and reads with fewer than 10 nucleotides were discarded. After quality control, reads were mapped to the bovine reference genome (Bos_taurus.ARS-UCD1.2) using the Hisat2 (Kim et al., 2015). Read counts were determined using the program FeatureCounts (Liao et al., 2014). Differentially expressed genes (DEGs) between sample groups were determined by edgeR-robust (Zhou et al., 2014), using a false discovery rate (FDR) <0.05 was used as a threshold for statistical significance.
Following a published model (Hashizume et al., 2006) with modifications, outgrowths of TE cell monolayers were established from individual bovine embryos by prolonged culture using bovine uterine endometrial stroma fibroblast cell conditioned medium. Embryos attached to a type IV collagen matrix between days 14 and 16, and TE outgrowth was observed by day 22 (Figure 1). Histologically, BNC were observed by day 30 (Figures 1, 2A). Image-based flow cytometry was used to quantify DNA content in cells and provide evidence for BNC differentiation (Figure 2B). BNC percentages ranged from 14 to 39% across the four outgrowths analyzed. Expression of the MNC gene markers GATA3, IFNT2 and HAND1, as well as the BNC gene markers CSH2 and PAG1 were observed in in vitro derived TE (IVT) cells (Figure 2C).
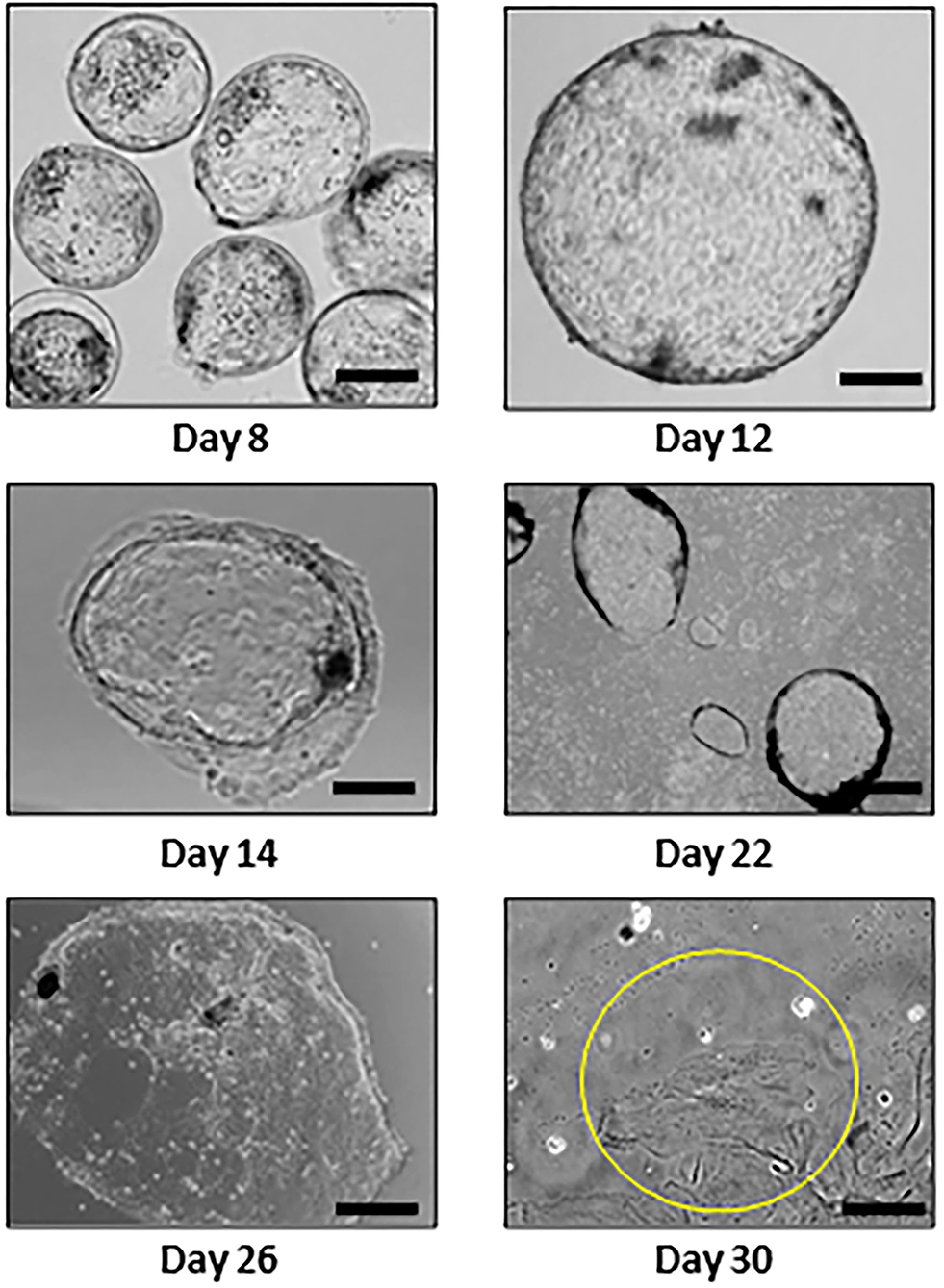
Figure 1. Individual embryos and trophectoderm outgrowths during 30 days of in vitro culture (day 0 = insemination). The cells circled in yellow in the day 30 outgrowth are binucleated. Bar = 100 μm.
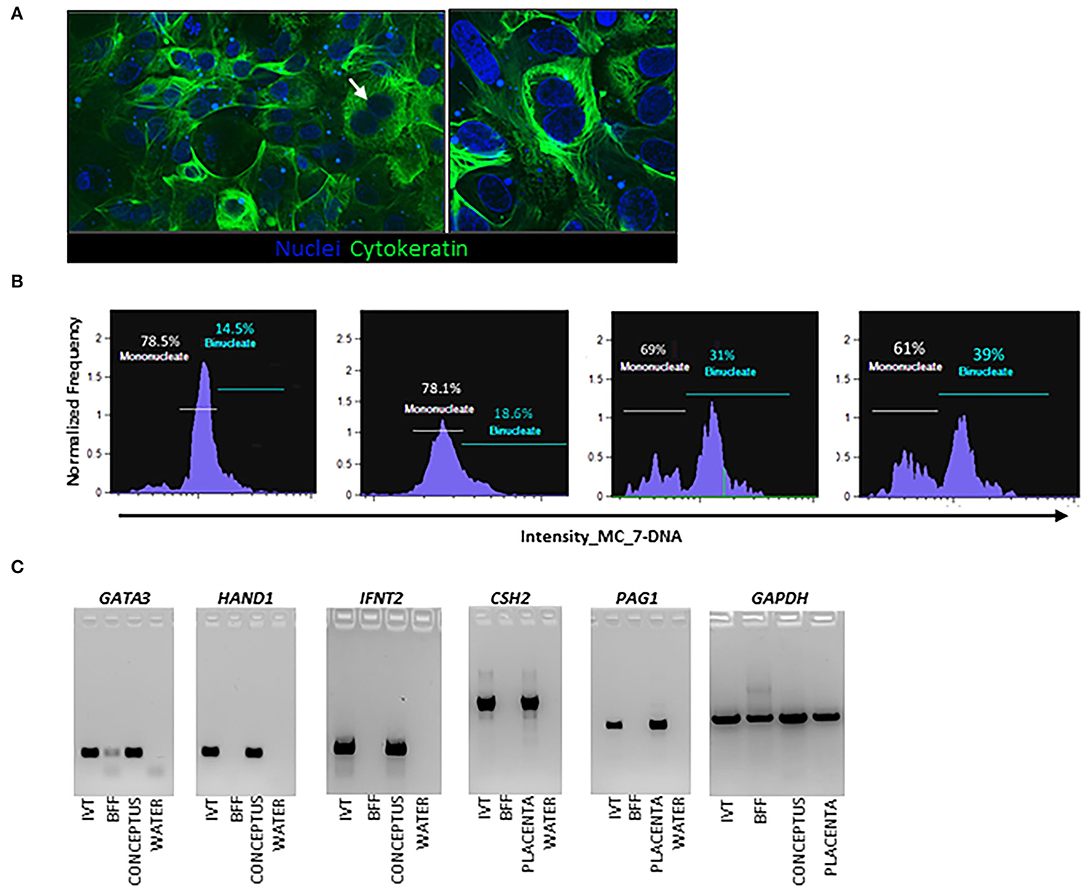
Figure 2. Characterization of in vitro derived trophectoderm (IVT) cells from bovine embryo outgrowths. (A) Cytokeratin analysis in day 30 embryo outgrowth cultures. Cytokeratin is green with nuclei-stained blue using Hoechst dye, a total of three samples were analyzed. (B) Flow cytometry analysis, a total of four independent samples were analyzed. (C) Endpoint PCR analysis. IVT are embryo outgrowths on day 30 of culture (pool of three different embryo outgrowths). Positive controls are a day 20 conceptus and day 30 bovine placenta. Negative controls are bovine fibroblasts (BFF), and reaction control was water.
To determine male influences on trophectoderm differentiation, expression of MNC and BNC genes and secretion of PAGs by were evaluated. For gene expression analysis, 20 independent embryo cell outgrowths from two different sires (n = 10 cell outgrowths per sire) were evaluated at day 30 of culture. Expression of GATA2, GATA3, and IFNT2 was not different (P > 0.10) among the two sires (Figure 3). Trophoblast Kunitz domain protein 1 (TKDP1) was increased (P < 0.05) in cells from sire A compared to sire B. In contrast, expression of HAND1 and CSH2 was increased (P = 0.005) in sire B than sire A, whereas PAG1 was not different (P = 0.10).
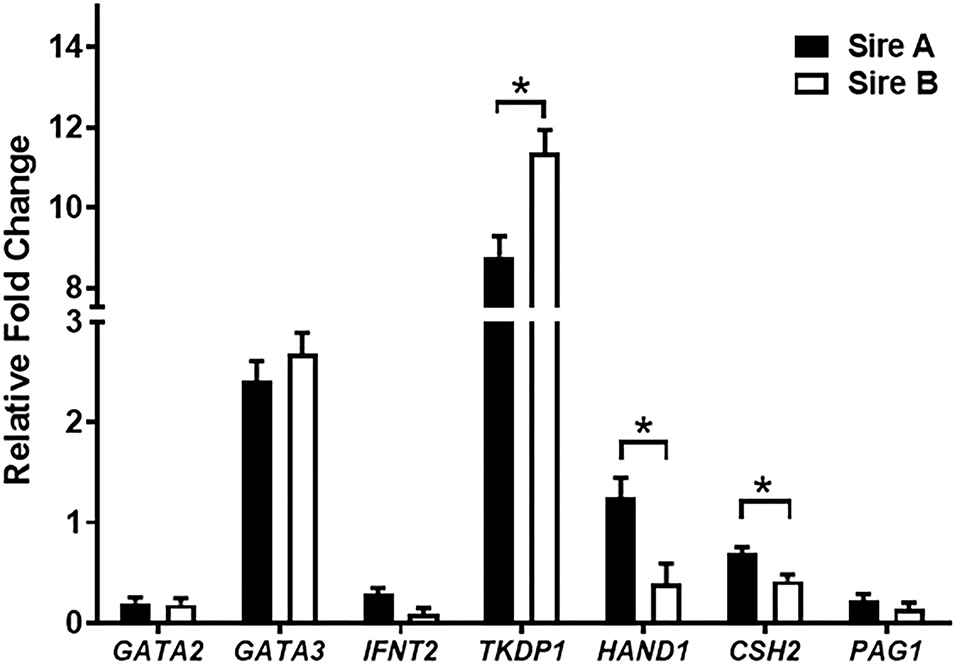
Figure 3. Gene expression in trophectoderm cell outgrowths on day 30. A total of 20 cell outgrowths were analyzed (10 outgrowths/sire). Values are presented as LSMEANS ± SEM. The * denotes difference (P < 0.05) between sires.
In a separate experiment, cell outgrowths from three different sires were assayed for the secretion of PAGs (n = 10 cell outgrowths per sire). There was a bull effect (P = 0.02) on PAG secretion, but not a day of culture (P = 0.20) or interaction effect of bull and day of culture (P = 0.47). When comparing individual bulls in a post-hoc analysis, the amount of PAGs in media of TE cultures from sire 3 were higher (P < 0.01) than Sire 2 on days 35 and 40 (P = 0.0007) of culture. By day 45, Sire 1 had higher (P < 0.01) levels of PAG than Sire 3. By day 50–54 PAG release decreased in all samples relative to day 35 (Figure 4).

Figure 4. Pregnancy associated glycoproteins (PAGs) in cell culture media. A total of 30 cell outgrowths (10 outgrowths per sire) were evaluated in an ELISA specific for BNC-secreted PAGs. Bars marked with an * above represents differences (P < 0.05) between sires.
Single-cell RNA-seq analysis was performed in three different TE outgrowths from one sire at day 40 of culture. A total of 11 cell populations (Clusters) were identified (Figure 5A). The number of representative genes per cluster ranged from 54 to 405 (Supplementary Table 2). Among representative genes per cluster, ribosomal genes (RPL10, RPL12, RPS3-23) were present in cluster 1, genes involved in ubiquitin pathway and angiogenesis marked cluster 2, and TE differentiation genes such as HAND1, CITED1, and TKDP1 denoted cluster 7. Genes associated with growth and remodeling such as JAG1 and EPCAM were representative of cluster 11. Cluster 3 had increased expression of TE markers including TKDP1 and the ancient PAGs PAG8 and PAG12. Genes associated with TE stem cells and lineage specification such as SOX15 and KLF4 were found in Cluster 4. TE differentiation markers CITED1, PLAC8, and HAND1 were abundant in Cluster 7. Except for PAG1, modern PAGs (PAG7, PAG9, and PAG17) were not found. In addition, all genes expressed in each cluster can be found in the Supplementary Data Sheet 1.
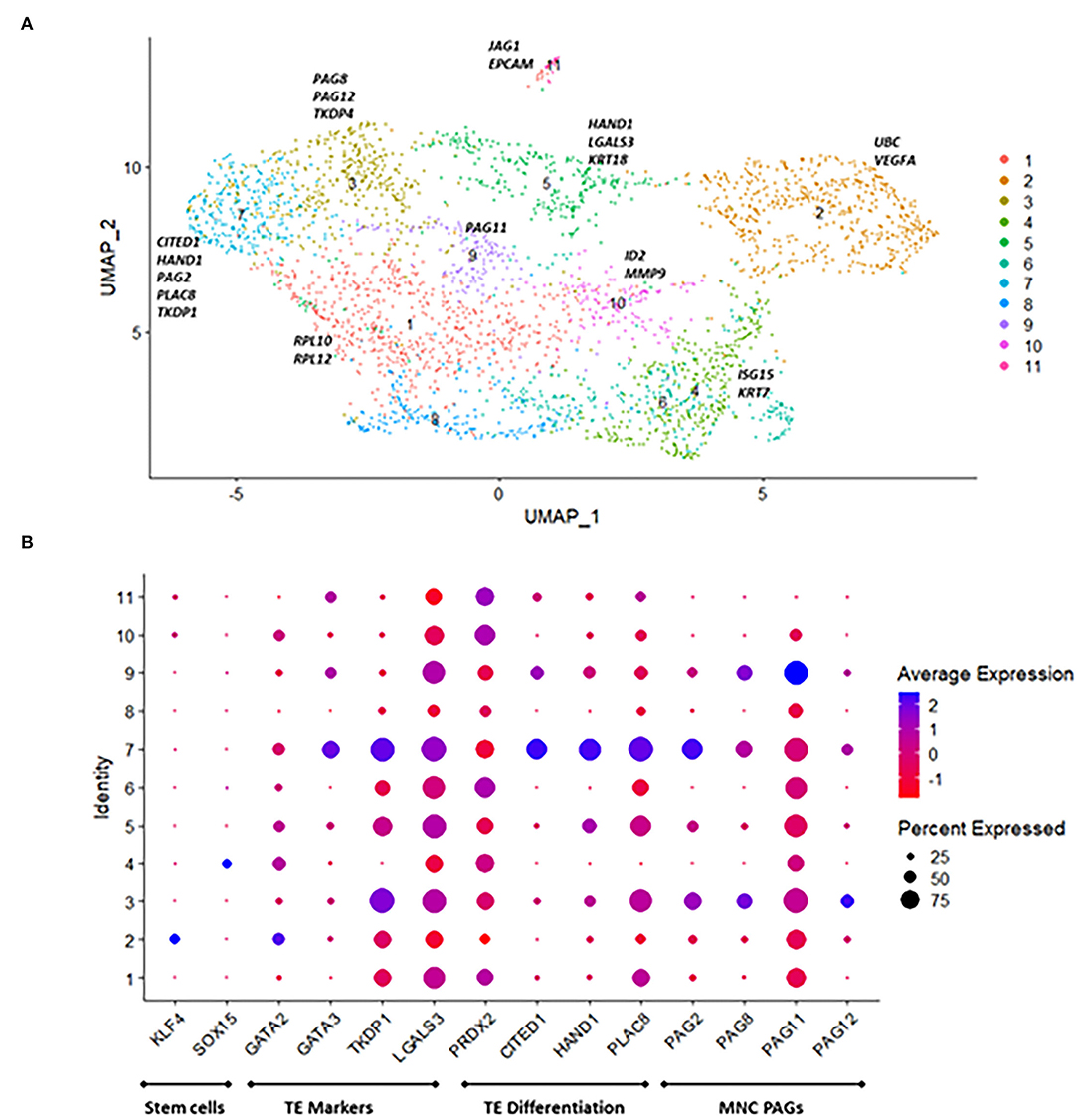
Figure 5. Single-RNA seq profiling of in vitro derived trophectoderm (IVT) outgrowths on day 40. (A) UMAP plot revealing 11 cell populations based on marker gene analysis. (B) DotPlot representing genes involved in trophectoderm specification, differentiation, and PAGs. Dot Size indicates the percentage of cells in the cluster expressing the gene, and shading represents level of expression (higher expression = blue shade lower expression = red shade).
To further corroborate the presence of MNC and BNC markers in the TE outgrowths, whole transcriptome analysis was performed in a total of 8 TE outgrowths at days 35 and 40 of culture (Supplementary Table 3). In all samples, there was expression of genes specifically expressed by MNC such as GATA3, HAND1, PAG11, PAG12 and PAG2 (Figure 6A; Supplementary Table 2), and of genes specifically expressed by BNC such as: CSH2, PAG1, PAG3, PAG4, PAG5, PAG7, PAG9, PAG10, PAG16, PAG17, PAG18, PAG20 and PAG21 (Figure 6B; Supplementary Table 3).
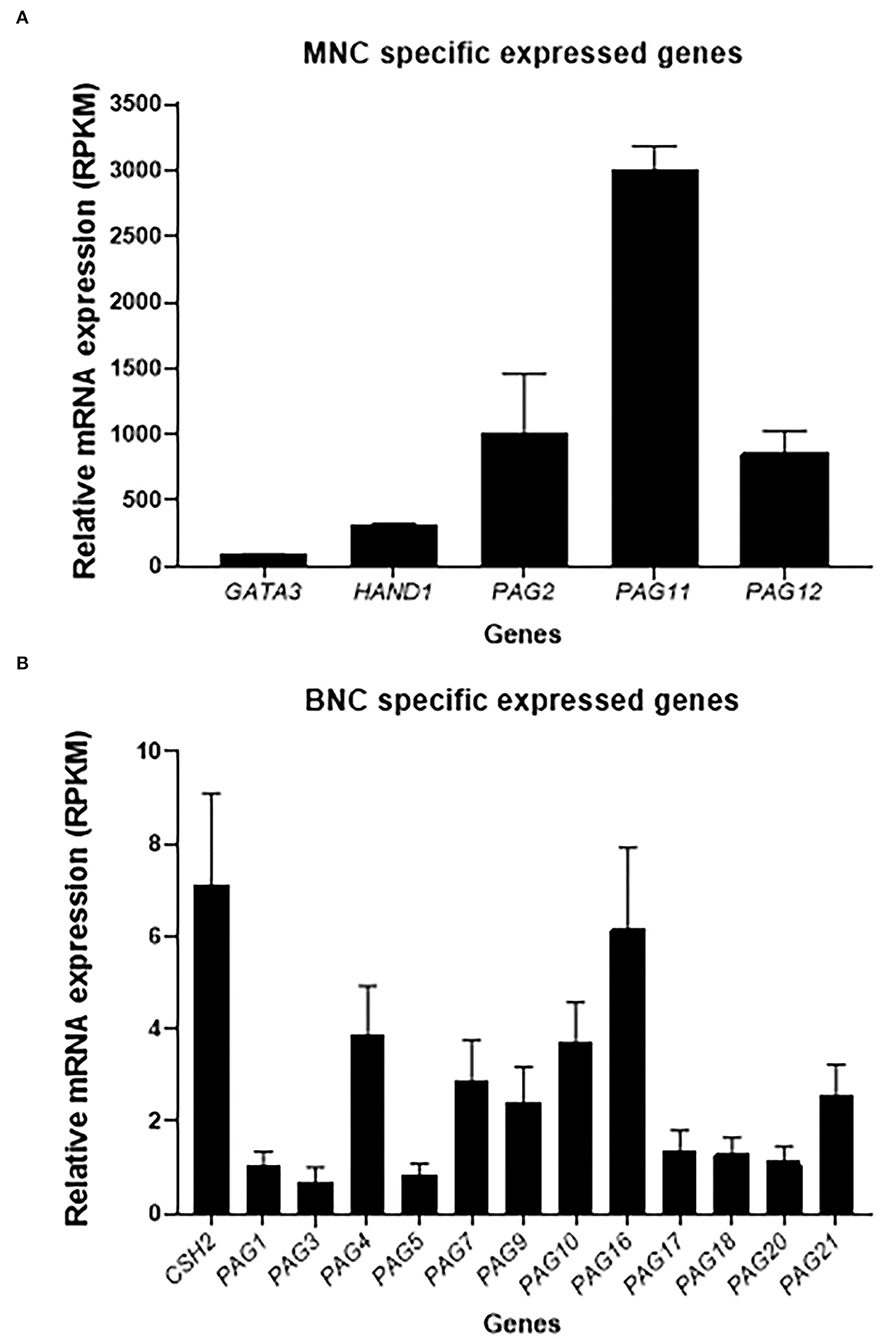
Figure 6. Gene expression of TE marker genes through transcriptome analysis. A total of eight outgrowths at day 35 and 41 were analyzed. (A) MNC specific genes expressed TE outgrowths. (B) BNC specific genes expressed in the TE outgrowths. Values are presented as RPKM ± SEM.
Here we evaluated a model for bovine TE differentiation derived from single IVP blastocysts. Trophectoderm outgrowths have been established in the past from in vitro embryos using feeder cells (Talbot et al., 2000; Bai et al., 2011; Ramos-Ibeas et al., 2014), Matrigel (Yang et al., 2011), or type I collagen (Shimada et al., 2001; Hashizume et al., 2006). For this work, the extracellular matrix was changed to type IV collagen to promote cell adhesion and proliferation given its abundance in the endometrium and particularly in the basal lamina supporting epithelial cells (Yamada et al., 2002). Further, Type IV collagen is increased in the endometrium and trophoblast during early pregnancy (Oefner et al., 2015) and used in the derivation of trophoblast stem cells in vitro (Okae et al., 2018).
When evaluated by endpoint and quantitative PCR, expression of MNC marker genes IFNT2, GATA3, and TKDP1 was observed in the cells of the embryo outgrowths. IFNT2 is expressed and secreted by elongating conceptus of cattle (Spencer and Bazer, 2004), and GATA3 expression is restricted to the trophectoderm and acts in parallel pathways to CDX2 to induce expression of trophectoderm lineage genes (Ralston et al., 2010; Bai et al., 2011). TKDP1 expression is increased at the time of differentiation of MNC to BNC, suggesting a potential role in this process (MacLean II et al., 2003; Smith et al., 2010). Expression of CSH2 and PAG1, classical markers of BNC in vivo (Xie et al., 1997; Green et al., 2005; Spencer et al., 2007), was also observed. The mixed population of MNC and BNC cells is like the one observed in the developing conceptus during early placentation. Interestingly, continuous PAG secretion from these cells into the culture media was observed and corroborated by PAGs expression on the whole TE transcriptome data in vivo, PAGs are secreted by BNC in the placenta, can be detected in maternal circulation as early as day 22 of pregnancy, and serve as a marker to monitor pregnancy (Wallace et al., 2015; Wijma et al., 2016; Filho et al., 2020). There is evidence that lower circulating PAGs can be a predictor of embryonic mortality and influence by sire (Pohler et al., 2016).
Here, expression of MNC and BNC markers was influenced by sire, as indicated by the decreased expression of HAND1 and CSH2 in trophectoderm cells from one of the two tested sires. Given that Hand1 is involved in the differentiation of giant cells in the mouse placenta (Scott et al., 2000), and HAND1-positive cells secrete CSH2 in cattle, it is possible that the lower expression of HAND1 indicates decreased ability of the TE to differentiate into BNC. As TE cells can be successfully generated from individual embryos, the model presented here could be a useful tool to identify sires prone to high embryonic mortality based on their ability to secrete PAGs in vitro.
Furthermore, there were individual differences in PAG concentrations depending upon sire. In vivo, PAGs are secreted by BNC in the placenta, can be detected in maternal circulation as early as day 22 of pregnancy, and serve as a marker to monitor pregnancy (Wallace et al., 2015; Wijma et al., 2016; Filho et al., 2020). There is evidence that lower circulating PAGs can be a predictor of embryonic mortality (Pohler et al., 2016), and similar to what we observed in the present study, PAGs concentrations are affected by the sire (Franco et al., 2018). Furthermore, secretion of PAGs by TE cells was higher at day 35 followed by a decrease by day 40 and a second increase in secretion by day 50 (wave pattern), which is a similar to the pattern of secretion of PAGs in vivo (Pohler et al., 2013). As TE cells can be successfully generated from individual embryos, the model presented here could be a useful tool to identify sires prone to high embryonic mortality based on their ability to secrete PAGs in vitro.
Single-cell transcriptome analysis revealed distinct populations of cells in the embryo outgrowths. Expression of genes associated with trophectoderm stem cells and lineage specification such as KLF4 and SOX15 were highly expressed in a small population of cells in Clusters 2 and 4, respectively. KLF4, is a pluripotency-related gene found in induced TE cell lines in cattle (Kawaguchi et al., 2016). In the mouse, Sox15 drives the transcriptional activity of Hand1, which is involved in trophoblast giant cell differentiation (Scott et al., 2000; Yamada et al., 2006). Likewise, genes involved in TE lineage specification in human, mouse, and bovine GATA2 and GATA3 (Roberts et al., 2004; Cheng and Handwerger, 2005; Bai et al., 2011; Schiffmacher and Keefer, 2013) were present in Cluster 2 and 7, respectively. LGALS3, is abundant in the elongating conceptus (Cammas et al., 2005; Baldwin, 2019) and expressed in all clusters in this dataset. Galectin 3 binding protein (LGALS3BP) was also expressed in all cells in this dataset, and it has been shown to be increased in immune cells of pregnant heifers during early pregnancy (Rocha et al., 2020). Galectins contribute to cell-to-cell adhesion and control immune cell activities (Chaney et al., 2022), but their exact role in early pregnancy remains unclear.
CITED1, HAND1 and PLAC8 were abundant in the same cluster. CITED1 and HAND1 are involved in trophectoderm differentiation in the mouse (Scott et al., 2000; Rodriguez et al., 2004), and increased in conceptuses from high fertility sires (Ortega et al., 2018). In the human, PLAC8 is involved in TE differentiation and regulates the migration of extravillous trophoblast cells (Chang et al., 2018). The ancient PAGs 2, 8 and 12 were represented in two clusters, while PAG11 was expressed across all cells. These PAGs are mostly expressed in mononucleated trophoblast cells (Telugu et al., 2009, 2010; Wallace et al., 2015). The exact function of PAGs is still unclear, but there is speculation about their role in facilitating adhesion between trophoblast and endometrial epithelium (Wallace et al., 2015). The fact that several markers (Figure 5) are present in various clusters with different levels of expression could indicate some transition in the function of the cells in these clusters.
All the samples used for single-cell analysis were selected after measuring PAG proteins in the cell culture supernatant, to ensure that functional BNC were present. The lack of expression of BNC markers' such as PAG1 and CSH2 could have been the result of two scenarios: physical exclusion of BNC in the straining steps during sample preparation due to the larger size of BNC, or low number of cells and reads per cell in the single-cell RNA-seq resulting in failure to capture the expression of BNC. Moving forward, one option to determine single cell transcriptome analysis in multinucleated large cells could be the use of in situ (Vickovic et al., 2019).
Nevertheless, BNC markers genes and proteins were detected in these cells through other analyses in this work. Whole TE transcriptome, confirmed not only the expression of BNC markers detected by Endpoint PCR, like PAG1 and CSH2, but other markers for BNC such as the other members of PAGs family (PAG1, PAG3, PAG4, PAG5, PAG7, PAG9, PAG10, PAG16, PAG17, PAG18, PAG20 and PAG21) (Green et al., 2000; Polei et al., 2020). Some of the PAGs that are mostly secreted by MNC, such as PAG2, PAG11 and PAG12 (Green et al., 2000; Polei et al., 2020) were also found the expressed in the whole TE transcriptome, confirming the mixed population of MNC and BNC in the culture system.
In conclusion, we have developed and present an improved model to derive TE cells in vitro from embryo outgrowths. The trophectoderm cells undergo differentiation into BNC and display continuous secretion of PAGs, which are BNC-specific proteins, similar to what is found during early bovine pregnancy in vivo. Furthermore, these data corroborate previous evidence reporting that bovine placentation is influenced by the sire (Pohler et al., 2013; Ortega et al., 2018). Even when the uterus is necessary for conceptus elongation (Spencer et al., 2016), this model allows for the study of trophectoderm development and can help in understanding the biology of these cells, as well as provide insights on how male-driven factors regulate trophectoderm differentiation and contribute to pregnancy loss during early placentation in the bovine.
The data discussed in this publication are accessible through GEO Series accession number GSE200216 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE200216).
MO, JR, JD, EO'N, KP, KK, AS, JG, and TS participated in study execution, analysis, manuscript drafting, and critical discussion. All authors contributed to the article and approved the submitted version.
This work was supported by Agriculture and Food Research Initiative Competitive Grant 2019-67015-28998, and Postdoctoral Fellowship Award 2019-67012-34005 from the USDA National Institute of Food and Agriculture.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank Dr. Pramod Dhakal for his assistance with immunolocalization procedure, Dr. Frank Baker and Dr. Alexander Jurkevich from the University of Missouri Molecular Cytology Core for their training and assistance with the confocal microscope. Acknowledgments are also extended to Dr. Bo Harstine from Select Sires, Inc. (Plain City, Ohio) for donation of bovine semen used throughout the project.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fanim.2022.898808/full#supplementary-material
Bai, H., Sakurai, T., Someya, Y., Konno, T., Ideta, A., Aoyagi, Y., et al. (2011). Regulation of trophoblast-specific factors by GATA2 and GATA3 in bovine trophoblast CT-1 cells. J. Reprod. Dev. 57, 518–525. doi: 10.1262/jrd.10-186K
Baldwin, H. L.. (2019). Investigation of Bovine Conceptus Galectins and the Effect of Bovine Conceptus Secretory Proteins on the Endometrial Epithelial Transcriptome - ProQuest. Available online at: https://www.proquest.com/openview/85072ff160d13e0754b9770c2ff22cda/1?pq-origsite=gscholar&cbl=18750&diss=y (accessed March 6, 2022).
Brooks, K., Burns, G., and Spencer, T. E. (2014). Conceptus elongation in ruminants: roles of progesterone, prostaglandin, interferon tau and cortisol. J. Anim. Sci. Biotechnol. 5, 53. doi: 10.1186/2049-1891-5-53
Butler, A., Hoffman, P., Smibert, P., Papalexi, E., and Satija, R. (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420. doi: 10.1038/nbt.4096
Cammas, L., Reinaud, P., Dubois, O., Bordas, N., Germain, G., and Charpigny, G. (2005). Identification of differentially regulated genes during elongation and early implantation in the ovine trophoblast using complementary DNA array screening1. Biol. Reprod. 72, 960–967. doi: 10.1095/biolreprod.104.034801
Chaney, H. L., Grose, L. F., LaBarbara, J. M., Sirk, A. W., Blancke, A. M., Sánchez, J. M., et al. (2022). Galectin-1 induces gene and protein expression related to maternal-conceptus immune tolerance in bovine endometrium. Biol. Reprod. 106, 487–502. doi: 10.1093/biolre/ioab215
Chang, W.-L., Liu, Y.-W., Dang, Y.-L., Jiang, X.-X., Xu, H., Huang, X., et al. (2018). PLAC8, a new marker for human interstitial extravillous trophoblast cells, promotes their invasion and migration. Development 145, dev148932. doi: 10.1242/dev.148932
Cheng, Y.-H., and Handwerger, S. (2005). A placenta-specific enhancer of the human syncytin gene1. Biol. Reprod. 73, 500–509. doi: 10.1095/biolreprod.105.039941
Filho, R. V. O., Franco, G. A., Reese, S. T., Dantas, F. G., Fontes, P. L. P., Cooke, R. F., et al. (2020). Using pregnancy associated glycoproteins (PAG) for pregnancy detection at day 24 of gestation in beef cattle. Theriogenology 141, 128–133. doi: 10.1016/j.theriogenology.2019.09.014
Fitzgerald, H. C., Dhakal, P., Behura, S. K., Schust, D. J., and Spencer, T. E. (2019). Self-renewing endometrial epithelial organoids of the human uterus. Proc. Natl. Acad. Sci. U. S. A. 116, 23132–23142. doi: 10.1073/pnas.1915389116
Franco, G. A., Peres, R. F. G., Martins, C. F. G., Reese, S. T., Vasconcelos, J. L. M., and Pohler, K. G. (2018). Sire contribution to pregnancy loss and pregnancy-associated glycoprotein production in Nelore cows. J. Anim. Sci. 96, 632–640. doi: 10.1093/jas/sky015
Green, J. A., Parks, T. E., Avalle, M. P., Telugu, B. P., McLain, A. L., Peterson, A. J., et al. (2005). The establishment of an ELISA for the detection of pregnancy-associated glycoproteins (PAGs) in the serum of pregnant cows and heifers. Theriogenology 63, 1481–1503. doi: 10.1016/j.theriogenology.2004.07.011
Green, J. A., Xie, S., Quan, X., Bao, B., Gan, X., Mathialagan, N., et al. (2000). Pregnancy-associated bovine and ovine glycoproteins exhibit spatially and temporally distinct expression patterns during pregnancy1. Biol. Reprod. 62, 1624–1631. doi: 10.1095/biolreprod62.6.1624
Greenstein, J. S., Murray, R. W., and Foley, R. C. (1958). Observations on the morphogenesis and histochemistry of the bovine pre-attachment placenta between 16 and 33 days of gestation. Anat. Rec 132, 321–341. doi: 10.1002/ar.1091320308
Hashizume, K., Shimada, A., Nakano, H., and Takahashi, T. (2006). “Bovine Trophoblast Cell Culture Systems,” in Placenta and Trophoblast: Methods and Protocols Volume 1 Methods in Molecular MedicineTM, eds M. J. Soares and J. S. Hunt (Totowa, NJ: Humana Press), 179–188.
Kawaguchi, T., Cho, D., Hayashi, M., Tsukiyama, T., Kimura, K., Matsuyama, S., et al. (2016). Derivation of induced trophoblast cell lines in cattle by doxycycline-inducible piggyBac vectors. PLOS ONE 11, e0167550. doi: 10.1371/journal.pone.0167550
Kerns, K., Zigo, M., Drobnis, E. Z., Sutovsky, M., and Sutovsky, P. (2018). Zinc ion flux during mammalian sperm capacitation. Nat. Commun. 9, 2061. doi: 10.1038/s41467-018-04523-y
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
MacLean, J. A. II., Chakrabarty, A., Xie, S., Bixby, J. A., Roberts, R. M., et al. (2003). Family of Kunitz proteins from trophoblast: expression of the trophoblast Kunitz domain proteins (TKDP) in cattle and sheep. Mol. Reprod. Dev. 65, 30–40. doi: 10.1002/mrd.10262
Oefner, C. M., Sharkey, A., Gardner, L., Critchley, H., Oyen, M., and Moffett, A. (2015). Collagen type IV at the fetal–maternal interface. Placenta 36, 59–68. doi: 10.1016/j.placenta.2014.10.012
Okae, H., Toh, H., Sato, T., Hiura, H., Takahashi, S., Shirane, K., et al. (2018). Derivation of human trophoblast stem cells. Cell Stem Cell 22, 50–63.e6. doi: 10.1016/j.stem.2017.11.004
Ortega, M. S., Kelleher, A. M., O'Neil, E., Benne, J., Cecil, R., and Spencer, T. E. (2020). NANOG is required to form the epiblast and maintain pluripotency in the bovine embryo. Mol. Reprod. Dev. 87, 152–160. doi: 10.1002/mrd.23304
Ortega, M. S., Kurian, J. J., McKenna, R., and Hansen, P. J. (2017a). Characteristics of candidate genes associated with embryonic development in the cow: evidence for a role for WBP1 in development to the blastocyst stage. PLoS ONE 12, e0178041. doi: 10.1371/journal.pone.0178041
Ortega, M. S., Moraes, J. G. N., Patterson, D. J., Smith, M. F., Behura, S. K., Poock, S., et al. (2018). Influences of sire conception rate on pregnancy establishment in dairy cattle. Biol. Reprod. 99, 1244–1254. doi: 10.1093/biolre/ioy141
Ortega, M. S., Wohlgemuth, S., Tribulo, P., Siqueira, L. G., Cole, J. B., and Hansen, P. J. (2017b). A single nucleotide polymorphism in COQ9 affects mitochondrial and ovarian function and fertility in Holstein cows. Biol. Reprod. 96, 652–663. doi: 10.1093/biolre/iox004
Pillai, V. V., Siqueira, L. G., Das, M., Kei, T. G., Tu, L. N., Herren, A. W., et al. (2019). Physiological profile of undifferentiated bovine blastocyst-derived trophoblasts. Biol. Open 8, bio03793, 1–12. doi: 10.1242/bio.037937
Pohler, K. G., Geary, T. W., Johnson, C. L., Atkins, J. A., Jinks, E. M., Busch, D. C., et al. (2013). Circulating bovine pregnancy associated glycoproteins are associated with late embryonic/fetal survival but not ovulatory follicle size in suckled beef cows1. J. Anim. Sci. 91, 4158–4167. doi: 10.2527/jas.2013-6348
Pohler, K. G., Green, J. A., Geary, T. W., Peres, R. F. G., Pereira, M. H. C., Vasconcelos, J. L. M., et al. (2015). “Predicting embryo presence and viability,” in Regulation of Implantation and Establishment of Pregnancy in Mammals: Tribute to 45 Year Anniversary of Roger V. Short's “Maternal Recognition of Pregnancy” Advances in Anatomy, Embryology and Cell Biology, eds R. D. Geisert and F. W. Bazer (Cham: Springer International Publishing), 253–270. doi: 10.1007/978-3-319-15856-3_13
Pohler, K. G., Peres, R. F. G., Green, J. A., Graff, H., Martins, T., Vasconcelos, J. L. M., et al. (2016). Use of bovine pregnancy-associated glycoproteins to predict late embryonic mortality in postpartum Nelore beef cows. Theriogenology 85, 1652–1659. doi: 10.1016/j.theriogenology.2016.01.026
Polei, M., Günther, J., Koczan, D., and Fürbass, R. (2020). Trophoblast cell differentiation in the bovine placenta: differentially expressed genes between uninucleate trophoblast cells and trophoblast giant cells are involved in the composition and remodeling of the extracellular matrix and O-glycan biosynthesis. BMC Mol. Cell Biol. 21, 1. doi: 10.1186/s12860-020-0246-8
Ralston, A., Cox, B. J., Nishioka, N., Sasaki, H., Chea, E., Rugg-Gunn, P., et al. (2010). Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development 137, 395–403. doi: 10.1242/dev.038828
Ramos-Ibeas, P., Calle, A., Pericuesta, E., Laguna-Barraza, R., Moros-Mora, R., Lopera-Vásquez, R., et al. (2014). An efficient system to establish biopsy-derived trophoblastic cell lines from bovine embryos1. Biol. Reprod. 91, 1–10. doi: 10.1095/biolreprod.114.118430
Roberts, R. M., Chen, Y., Ezashi, T., and Walker, A. M. (2008). Interferons and the maternal–conceptus dialog in mammals. Semin. Cell Dev. Biol. 19, 170–177. doi: 10.1016/j.semcdb.2007.10.007
Roberts, R. M., Ezashi, T., and Das, P. (2004). Trophoblast gene expression: transcription factors in the specification of early trophoblast. Reprod. Biol. Endocrinol. 2, 47. doi: 10.1186/1477-7827-2-47
Rocha, C. C., da Silva Andrade, S. C., de Melo, G. D., Motta, I. G., Coutinho, L. L., Gonella-Diaza, A. M., et al. (2020). Early pregnancy-induced transcripts in peripheral blood immune cells in Bos indicus heifers. Sci. Rep. 10, 13733. doi: 10.1038/s41598-020-70616-8
Rodriguez, T. A., Sparrow, D. B., Scott, A. N., Withington, S. L., Preis, J. I., Michalicek, J., et al. (2004). Cited1 is required in trophoblasts for placental development and for embryo growth and survival. Mol. Cell. Biol. 24, 228–244. doi: 10.1128/MCB.24.1.228-244.2004
Rosen, B. D., Bickhart, D. M., Schnabel, R. D., Koren, S., Elsik, C. G., Tseng, E., et al. (2020). De novo assembly of the cattle reference genome with single-molecule sequencing. GigaScience 9, giaa021. doi: 10.1093/gigascience/giaa021
Schiffmacher, A. T., and Keefer, C. L. (2013). CDX2 regulates multiple trophoblast genes in bovine trophectoderm CT-1 cells. Mol. Reprod. Dev. 80, 826–839. doi: 10.1002/mrd.22212
Scott, I. C., Anson-Cartwright, L., Riley, P., Reda, D., and Cross, J. C. (2000). The HAND1 basic helix-loop-helix transcription factor regulates trophoblast differentiation via multiple mechanisms. Mol. Cell. Biol. 20, 530–541. doi: 10.1128/MCB.20.2.530-541.2000
Shimada, A., Nakano, H., Takahashi, T., Imai, K., and Hashizume, K. (2001). Isolation and characterization of a bovine blastocyst-derived trophoblastic cell line, BT-1: development of a culture system in the absence of feeder cell. Placenta 22, 652–662. doi: 10.1053/plac.2001.0702
Smith, C. S., Berg, D. K., Berg, M., and Pfeffer, P. L. (2010). Nuclear transfer-specific defects are not apparent during the second week of embryogenesis in cattle. Cell. Reprogram. 12, 699–707. doi: 10.1089/cell.2010.0040
Spencer, T. E., and Bazer, F. W. (2004). Conceptus signals for establishment and maintenance of pregnancy. Reprod. Biol. Endocrinol. 2, 49. doi: 10.1186/1477-7827-2-49
Spencer, T. E., Forde, N., and Lonergan, P. (2016). Insights into conceptus elongation and establishment of pregnancy in ruminants. Reprod. Fertil. Dev. 29, 84–100. doi: 10.1071/RD16359
Spencer, T. E., Johnson, G. A., Bazer, F. W., and Burghardt, R. C. (2007). Fetal-maternal interactions during the establishment of pregnancy in ruminants. Soc. Reprod. Fertil. Suppl. 64, 379–396. doi: 10.5661/RDR-VI-379
Stringfellow, D. A., and Seidel, S. M. (1998). Manual of the International Embryo Transfer Society (IETS), 4th Edn. Champaign, IL: International Embryo Transfer Society.
Stuart, T., and Satija, R. (2019). Integrative single-cell analysis. Nat. Rev. Genet. 20, 257–272. doi: 10.1038/s41576-019-0093-7
Talbot, N. C., Caperna, T. J., Edwards, J. L., Garrett, W., Wells, K. D., and Ealy, A. D. (2000). Bovine blastocyst-derived trophectoderm and endoderm cell cultures: interferon tau and transferrin expression as respective in vitro markers. Biol. Reprod. 62, 235–247. doi: 10.1095/biolreprod62.2.235
Telugu, B. P. V., Walker, A. M., and Green, J. A. (2009). Characterization of the bovine pregnancy-associated glycoprotein gene family – analysis of gene sequences, regulatory regions within the promoter and expression of selected genes. BMC Genomics 10, 185. doi: 10.1186/1471-2164-10-185
Telugu, B. P. V. L., Palmier, M. O., Doren, S. R. V., and Green, J. A. (2010). An examination of the proteolytic activity for bovine pregnancy-associated glycoproteins 2 and 12. Biol. Chem. 391, 259–270. doi: 10.1515/bc.2010.016
Ushizawa, K., Takahashi, T., Hosoe, M., Ishiwata, H., Kaneyama, K., Kizaki, K., et al. (2007). Global gene expression analysis and regulation of the principal genes expressed in bovine placenta in relation to the transcription factor AP-2 family. Reprod. Biol. Endocrinol. 5, 17. doi: 10.1186/1477-7827-5-17
Vickovic, S., Eraslan, G., Salmén, F., Klughammer, J., Stenbeck, L., Schapiro, D., et al. (2019). High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods 16, 987–990. doi: 10.1038/s41592-019-0548-y
Wallace, R. M., Pohler, K. G., Smith, M. F., and Green, J. A. (2015). Placental PAGs: gene origins, expression patterns, and use as markers of pregnancy. Reproduction 149, R115–R126. doi: 10.1530/REP-14-0485
Wijma, R., Stangaferro, M. L., Kamat, M. M., Vasudevan, S., Ott, T. L., and Giordano, J. O. (2016). Embryo mortality around the period of maintenance of the corpus luteum causes alterations to the ovarian function of lactating dairy cows 1. Biol. Reprod. 95, 1–14. doi: 10.1095/biolreprod.116.142075
Wooding, F. B. P.. (1992). The synepitheliochorial placenta of ruminants: binucleate cell fusions and hormone production. Placenta 13, 101–113. doi: 10.1016/0143-4004(92)90025-O
Xie, S., Green, J., Bao, B., Beckers, J.-F., Valdez, K. E., Hakami, L., et al. (1997). Multiple Pregnancy-associated glycoproteins are secreted by day 100 ovine placental tissue. Biol. Reprod. 57, 1384–1393. doi: 10.1095/biolreprod57.6.1384
Yamada, K., Kanda, H., Tanaka, S., Takamatsu, N., Shiba, T., and Ito, M. (2006). Sox15 enhances trophoblast giant cell differentiation induced by Hand1 in mouse placenta. Differentiation 74, 212–221. doi: 10.1111/j.1432-0436.2006.00070.x
Yamada, O., Todoroki, J., Takahashi, T., and Hashizume, K. (2002). The dynamic expression of extracellular matrix in the bovine endometrium at implantation. J. Vet. Med. Sci. 64, 207–214. doi: 10.1292/jvms.64.207
Yang, Q. E., Fields, S. D., Zhang, K., Ozawa, M., Johnson, S. E., and Ealy, A. D. (2011). Fibroblast growth factor 2 promotes primitive endoderm development in bovine blastocyst outgrowths1. Biol. Reprod. 85, 946–953. doi: 10.1095/biolreprod.111.093203
Keywords: trophectoderm, pregnancy-associated glycoproteins, sire, in vitro fertilization, binucleate cells, mononucleate cells
Citation: Ortega MS, Rizo JA, Drum JN, O'Neil EV, Pohler KG, Kerns K, Schmelze A, Green J and Spencer TE (2022) Development of an Improved in vitro Model of Bovine Trophectoderm Differentiation. Front. Anim. Sci. 3:898808. doi: 10.3389/fanim.2022.898808
Received: 17 March 2022; Accepted: 19 April 2022;
Published: 01 June 2022.
Edited by:
Carlo C. Lazado, Norwegian Institute of Food, Fisheries and Aquaculture Research (Nofima), NorwayReviewed by:
Kazuya Kusama, Tokyo University of Pharmacy and Life Sciences, JapanCopyright © 2022 Ortega, Rizo, Drum, O'Neil, Pohler, Kerns, Schmelze, Green and Spencer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: M. Sofia Ortega, b3J0ZWdhb2JhbmRvbUBtaXNzb3VyaS5lZHU=; Thomas E. Spencer, c3BlbmNlcnRlQG1pc3NvdXJpLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.