- 1Key Laboratory for Agro-Ecological Processes in Subtropical Region, National Engineering Laboratory for Pollution Control and Waste Utilization in Livestock and Poultry Production, Hunan Provincial Key Laboratory of Animal Nutritional Physiology and Metabolic Process, Institute of Subtropical Agriculture, Chinese Academy of Sciences, Changsha, China
- 2College of Animal Science and Technology, Hunan Agricultural University, Changsha, China
- 3College of Modern Agriculture, University of Chinese Academy of Sciences, Beijing, China
Here, the effects of non-protein nitrogen sources on fermentation parameters and microbial diversity were explored using three fistula goats as rumen fluid donors. The experiments involved six fermenters in a replicated 3 × 3 Latin square design with three dietary non-protein sources [ammonium chloride (A), biuret (B), and glutamine (G)] as treatment factors. A dual-flow continuous culture fermentation system was used. Microbial protein content in group B was significantly lower than that in the other two groups (P < 0.05). Ammonia nitrogen concentration significantly differed among the three groups (P < 0.01), following the order of G > A > B group. The acetate-to-propionate ratio in group G was significantly lower than that in the other two groups (P < 0.01). At the phylum level, the relative abundances of Cyanobacteria, Elusimicrobia, and Armatimonadetes were the highest in group G, being significantly higher than those in group B (P < 0.05). At the genus level, the relative abundance of Ruminococcus_1 was significantly higher in group A than in group B (P < 0.05). Overall, glutamine shifted the fermentation pathway from acetate to propionate, and the lower microbial crude protein content and relative abundances of the major fiber-degrading bacteria Ruminococcus_1 and protein-degrading bacteria Prevotellaceae_UCG-001 in group B indicate that biuret is not suitable as a dietary non-protein nitrogen source.
1 Introduction
The number of ruminants has dramatically increased in recent years, resulting in evident increases in the import of high-quality forage and protein feed and, ultimately, the production costs and product prices in China. Therefore, new protein feed resources must be developed and the utilization efficiency of dietary protein must be improved in the ruminant husbandry industry. A large number of bacteria in the rumen primarily derive nutrients from nitrogen; therefore, appropriate supplementation of nitrogen, including non-protein nitrogen (NPN), is imperative for ruminants (Dayani et al., 2010). Despite the long history of research, NPN remains a hotspot in the field of ruminant nutrition. Belasco (1954) and Henderickx (1967) demonstrated the high efficiency of ammonium succinate in promoting microbial protein synthesis. Further experiments revealed that organic ammonium salts could provide nitrogen for bacterial growth and cellulose digestion, and ammonium sulfate and acetamide could enhance microbial protein synthesis (Henderickx and Demeyer, 1967). Rumen microorganisms produce urease for degrading NPN to NH3 and CO2 and synthesizing amino acids and then utilize the produced amino acids, peptides, and nitrogen, along with adenosine triphosphate (ATP), for growth, finally supplying microbial proteins to the host animal (Dinic´ et al., 2013). The positive effects of NPN utilization in ruminants are well-known.
In dairy buffaloes, coated urea supplementation improved milk production and fat content by augmenting rumen function through enhanced microbial protein turnover and increased fiber degradation (Aquino et al., 2014; Nadeem et al., 2014). Chegeni et al. (2013) found that polymer-coated urea could be used as a substitute to soybean meal for sheep fed high levels of maize straw. As such, sufficient energy was available in the rumen, and the slower degradation rate of polymer-coated urea was more suitable for microbial growth and could supply sufficient nitrogen for microbial protein synthesis. Abdel et al. (2013) observed that daily body weight gain was significantly higher in lambs fed a urea-treated diet, which was attributed to the improved palatability and digestibility of nutrients and their enhanced digestion and passage rates from the rumen (Mahr-un-Nisa et al., 2004; Singh, 2014). During grass withering, molasses–urea blocks provide NPN to rumen microorganisms, increasing microbial protein concentration and promoting microbial growth and reproduction (Li et al., 2017). Corte et al. (2018) showed that in Nellore steer, dietary urea or slow-release urea supplementation improved the efficiency of microbial protein synthesis, with a significantly higher efficiency observed with slow-release urea than with urea. Specifically, slow-release urea degrades slowly in the rumen, and the synchronization between fermentable energy and degradable nitrogen in the rumen maximizes microbial growth and promotes microbial protein synthesis. Dietary nitrogen sources and degradation patterns affect microbial protein synthesis and utilization by rumen microorganisms. Dietary urea and coated urea supplementation positively affects the animal body, and the superior effect of coated urea to that of urea has been demonstrated in many studies. In the rumen, the microbial growth rate is slower than the urea degradation rate, and rapid ammonia release results in inefficient utilization of nitrogen and wastage of nitrogen sources (Xin et al., 2010).
To date, there has been extensive research on urea as an NPN source. Although urea positively affects rumen fermentation parameters and microbial growth in ruminants, it degrades rapidly, rendering its prompt use by microorganisms difficult and leading to the wastage of nitrogen sources. Therefore, further studies are warranted to explore more suitable nitrogen sources as substitutes for protein feed. In the present study, we used the artificial rumen method to examine the effects of different NPN sources [ammonium chloride (A), biuret (B), and glutamine (G)] on rumen fermentation characteristics and microbial diversity. Our results provide novel information for elucidating the effects of NPN sources on rumen fermentation and microbial reproduction and guiding the formulation of ruminant diets in practice.
2 Materials and Methods
2.1 Ethics
All experiments were approved by and conducted in accordance with the animal care guidelines of the Animal Care Committee of the Institute of Subtropical Agriculture, Chinese Academy of Sciences.
2.2 Substrates and In Vitro Incubation
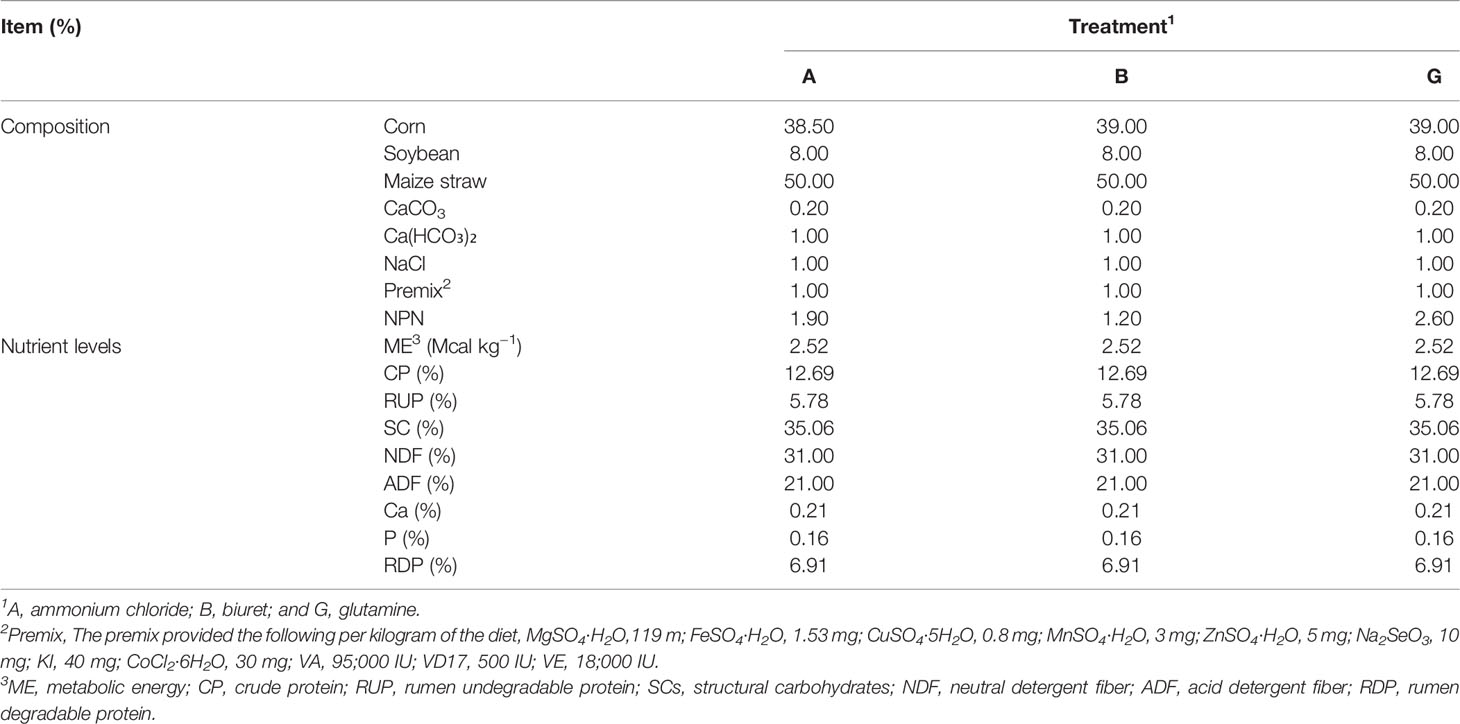
Three types of fermentation substrates were used for in vitro incubation: (1) a basic diet containing ammonium chloride (A), (2) a basic diet containing biuret (B), and (3) a basic diet containing glutamine (G). Maize stover of “Kexiangtian 1” (locally breed) was selected as forage. The diets were dried at 65°C, ground, and stored in air-tight bags. Ammonium chloride, biuret, and glutamine were purchased from the China National Medicines Co. Ltd. (Beijing, China). The composition and nutrient levels of basic experimental diets are presented in Table 1.
2.3 Animals and Diets
Three ruminally fistulated Xiangdong black goats (a local breed in Hunan Province, China) with an average body weight of 25.0 ± 2.5 kg (mean ± SD) were used in the experiment. The goats were fed a basic diet comprising a mixture of corn–soybean meal concentrate and maize stover (50:50). The diet was provided twice a day as two meals at 08:00 and 17:00 h, and water was provided ad libitum.
2.3.1 RUSITEC Incubation
Rumen contents of three Xiangdong black goats were collected before morning feeding, pooled, strained through four layers of cheesecloth and brought to the laboratory in a thermo-insulated container prefilled with CO2. A fermentation tank with a nominal volume of 1,400 ml was inoculated with 500 ml of McDougall’s buffer (1948) formula configuration and 500 ml of strained rumen fluid. In addition, approximately 20 g of substrate was weighed into the tank under a stream of CO2 at 39.5 ± 0.5°C. The dual-flow continuous culture fermentation system is described in detail by Shen (2012). The RUSITEC system comprised six fermentation vessels allocated to three groups (two vessels per group). Each vessel within a group was assigned to one of the two rotation treatments over three periods (runs), following a balanced 3 × 3 Latin square design. Each period (run) lasted 8 days, including 5 days for acclimatization and 3 days for sample collection. Each tank was connected to a pressure sensor and a pH meter. Data were automatically recorded in real time using a computer. Artificial saliva was infused to maintain a liquid passage rate of 6%·h−1, and approximately 1,400 ml effluent flew out of the fermenters per day, leading to liquid turnover once a day. The overflow liquid and small digested feed particles from each fermentation vessel were collected in a water-cooled glass container at a constant temperature of 4°C maintained with circulating cold water. When fermentation started, 40 g of the experimental diet was added to each fermentation vessel daily in two equal portions at 08:00 and 20:00 h. Nitrogen gas supply was started before feeding and immediately stopped after feeding.
2.3.2 Sample Collection
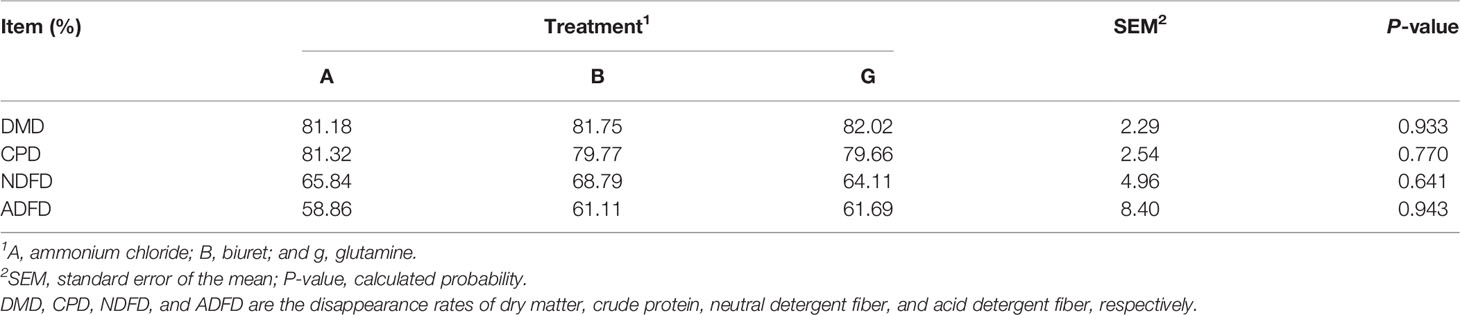
All samples for the three phases were collected at 08:00 and 20:00 h before feeding on 3 consecutive days (days 6 to 8). Fermentation liquid (5 ml) was collected from each tank for protozoa counts. Approximately 2 ml of the liquid effluent was collected and centrifuged at 15,000 ×g for 10 min at 4°C. The supernatant was acidified using 0.15 ml of 25% (w/v) metaphosphoric acid for the analysis of volatile fatty acids (VFAs) and ammonia (NH3-N) and stored at −20°C until sample analysis. Subsequently, the sample bottles were shaken thoroughly to ensure that the representative portions of the liquid and particle fractions could be obtained. Then, 2 and 10 ml of microbial samples were collected, immediately frozen with liquid N2, and stored at −80°C for subsequent DNA extraction and microbial protein determination, respectively. Finally, the residual fermented liquid was filtered into a nylon bag; dried at 65°C for 8 h; weighed; and ground to determine the rate of disappearance of dry matter (DMD), crude protein (CPD), neutral detergent fiber (NDFD), and acid detergent fiber (ADFD) in the substrate.
2.4 Chemical Analysis
In all feed samples, DM (method 945.15), CP (method 954.01), NDF, and ADF (method 973.18) contents were analyzed following published methodologies (Williams et al., 1990). The pH of the fermentation tank was automatically detected by the pH electrode installed in the tank and entered into the computer.
The individual VFA concentrations in the supernatant were measured using gas chromatography (HP5890, Agilent 5890; Agilent, Santa Clara, CA) according to the method described by Chen et al. (2017). The NH3-N concentration was determined on the basis of the ultraviolet absorption value recorded at 700 nm using a spectrophotometer (UV-2300, Shimadzu, Japan), according to the method described by Wang et al. (2016). Microbial crude protein (MCP) production was estimated according to the purine method described by Hu (2005).
2.5 Protozoa Counts
Protozoa counts were obtained according to the method described by Wu (2017). Using staining microscopy, 5 ml of rumen fluid and 10 ml of methyl green formalin solution were collected, mixed well, and stained for 24 h. A disposable pipette was used to remove the stained solution. Then, the suspension was swiftly dropped onto a blood cell counting plate until it covered the entire counting chamber. The liquid-filled counting plate was gently enclosed with a glass coverslip. Finally, cell count was determined under an optical microscope.
In the selected plate count area of 1 mm2, five squares each at the four corners and in the middle, totaling 25 squares, were analyzed. The average count was calculated as follows:
where X is the total number of protozoa of five squares, D is the fold-dilution of rumen fluid, and 1,000 is the coefficient for the conversion of 1 mm3 square volume to 1 ml.
2.6 DNA Extraction and Sequencing
Microbial DNA was extracted using a bacterial DNA kit (Omega Bio-Tek Inc., Norcross, GA, USA), according to the manufacturer’s protocol. The extracted DNA samples were detected by 1% agarose gel electrophoresis and spectrophotometry (Nanodrop 2000, America) (260/280 nm density ratio) for quality check. The V3–4 hypervariable region of the bacterial 16S rRNA gene was amplified using primers 338F (ACTCCTACGGGAGGCAGCAG) and 806R (GGACTACHVGGGTWTCTAAT) (Mumbi et al., 2015). For each sample, a 10-digit barcode sequence was added at the 5′-end of the forward and reverse primers (Allwegene Technology Inc., Beijing). Polymerase chain reaction (PCR) (LightCycler 480II, Germany) was performed on a MasterCycler Gradient (Eppendorf, Germany) using a 25-μl reaction system containing 12.5 μl of KAPA 2G Robust Hot Start Ready Mix, 1 µl of forward primer (5 µM), 1 µl of reverse primer (5 µM), 5 µl of DNA (total template quantity = 30 ng), and 5.5 µl of H2O. The reaction conditions were as follows: 95°C for 5 min, followed by 28 cycles at 95°C for 45 s, 55°C for 50 s, and 72°C for 45 s and final extension at 72°C for 10 min. The PCR products were purified using the QIAquick Gel Extraction Kit (QIAGEN, Germany), quantified using real-time PCR, and sequenced on the MiSeq platform at Allwegene Technology Inc., Beijing.
2.7 Sequence Data Analyses
Raw data were removed from consideration if they were shorter than 200 bp, had a low-quality score (≤20), contained ambiguous bases, or did not exactly match the primer sequences and barcode tags. Qualified reads were separated using sample-specific barcode sequences and trimmed using the Illumina Analysis Pipeline version 2.6. The dataset was then analyzed using QIIME. The sequences were clustered into operational taxonomic units (OTUs) at 97% similarity (Edgar, 2013). The Ribosomal Database Project (RDP) tool was used to classify all sequences into different taxonomic groups (Cole et al., 2009).
The evolutionary distances between microbial communities in each sample were calculated using the TAYC coefficient and represented as an unweighted pair group method with arithmetic mean (UPGMA) clustering tree describing the dissimilarity (1 − similarity) between multiple samples (Jiang et al., 2013).
2.8 Statistical Analysis
The statistical model included dietary treatments (n = 3) as the fixed effects, treatment replicates (n = 6) as a repeated measure, and period (n = 3) and fermentation tank (n = 6) as the random effects. Data were subjected to single factor analysis in SPSS 19.1.0. Significance was declared at P < 0.05, and 0.05 ≤ P ≤ 0.10 indicated a significant trend.
3 Results
3.1 Nutrient Disappearance Rate
DMD, CPD, NDFD, and ADFD were not affected (P > 0.05) by NPN sources (Table 2). However, DMD and ADFD were numerically higher (P > 0.05) but CPD and NDFD were lower (P > 0.05) in group G than those in the other groups.
3.2 Rumen Fermentation Parameters
3.2.1 Protozoa Count
NPN sources (Table 3) did not affect protozoa counts (P > 0.05). However, protozoa count in group G was, respectively, 17.74% and 11.57% higher than that in groups A and B.
3.2.2 Fermentation Parameters
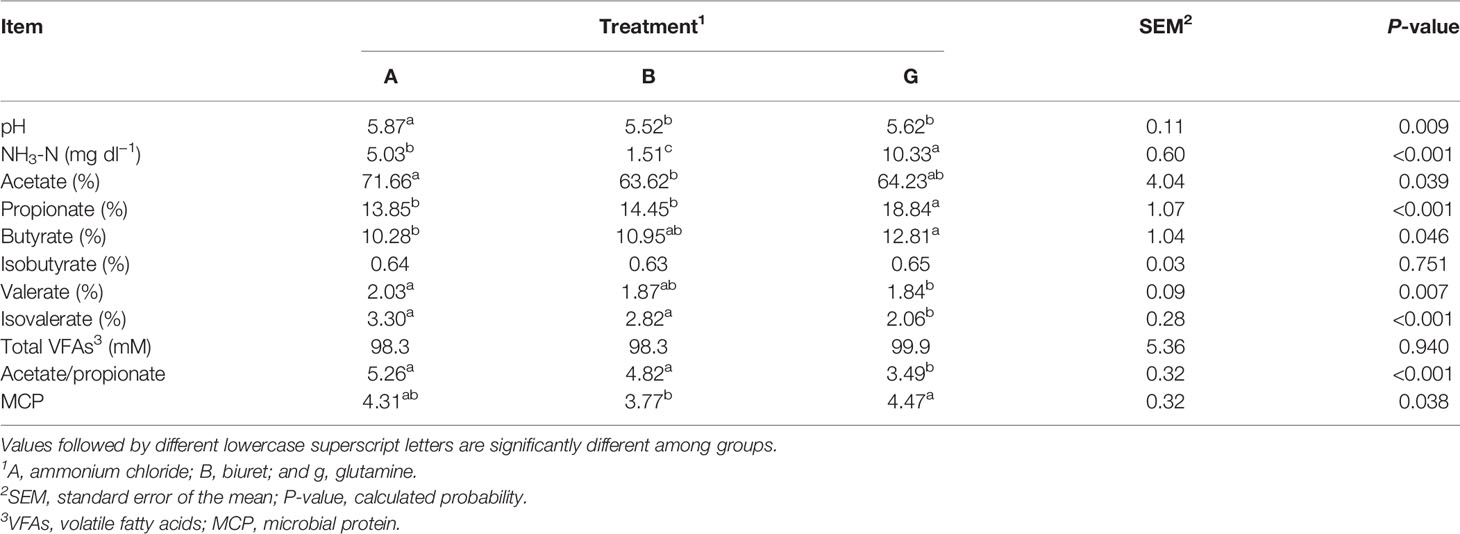
The results are presented in Table 4. pH in group A was significantly higher (P < 0.01) than that in the other two groups. NH3-N concentration was the highest in group G, being, respectively, 2.05 and 6.84 (P < 0.01) times higher than that in groups A and B. The total VFA content was not significantly different among the three groups (P > 0.05). Acetate proportion in group A (71.66%) was significantly higher (P < 0.05) than that in group B. Butyrate proportion in group G was (12.81%) was significantly higher (P < 0.05) than that in group A. Propionate proportion in group G was significantly higher than that in the other two groups (P < 0.01). Valerate proportion in group A was significantly higher (P < 0.05) than that in group G. Isovaleric acid proportion and acetate-to-propionate ratio in group G were significantly lower (P < 0.01) than those in the other two groups. MCP content in group G (18.57%) higher (P < 0.05) than that in group B.
3.3 Rumen Microbial Diversity
3.3.1 Microbial Richness and Diversity
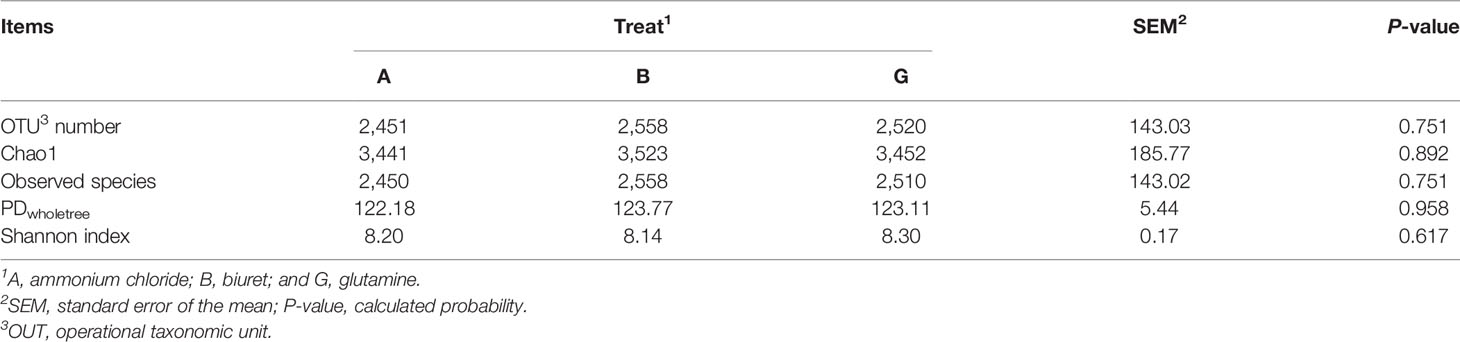
As shown in Table 5, the alpha diversity indices were not significantly different (P > 0.05) among the three groups. OTU number and microbial richness were relatively higher but microbial community diversity was numerically lower (P > 0.05) in group B than that in the other two groups.
3.3.2 Structural Analysis of Rumen Microbial Colonies
As indicated in Table 6, at the phylum level, the relative abundances of Cyanobacteria and Elusimicrobia were the highest in group G (169% and 168%, respectively), being 41.24% and 95.37% (P = 0.002) higher than those in groups A and B, respectively. The relative abundances of Planctomycetes and Armatimonadetes in group G were, respectively, 83.38% (P = 0.018) and 247% (P = 0.039) higher than those in group B group. The relative abundance of SR1_Absconditabacteria was higher in group B than in the other two groups (P < 0.05). There were no differences in the abundances of other bacteria among the three groups (P > 0.05).
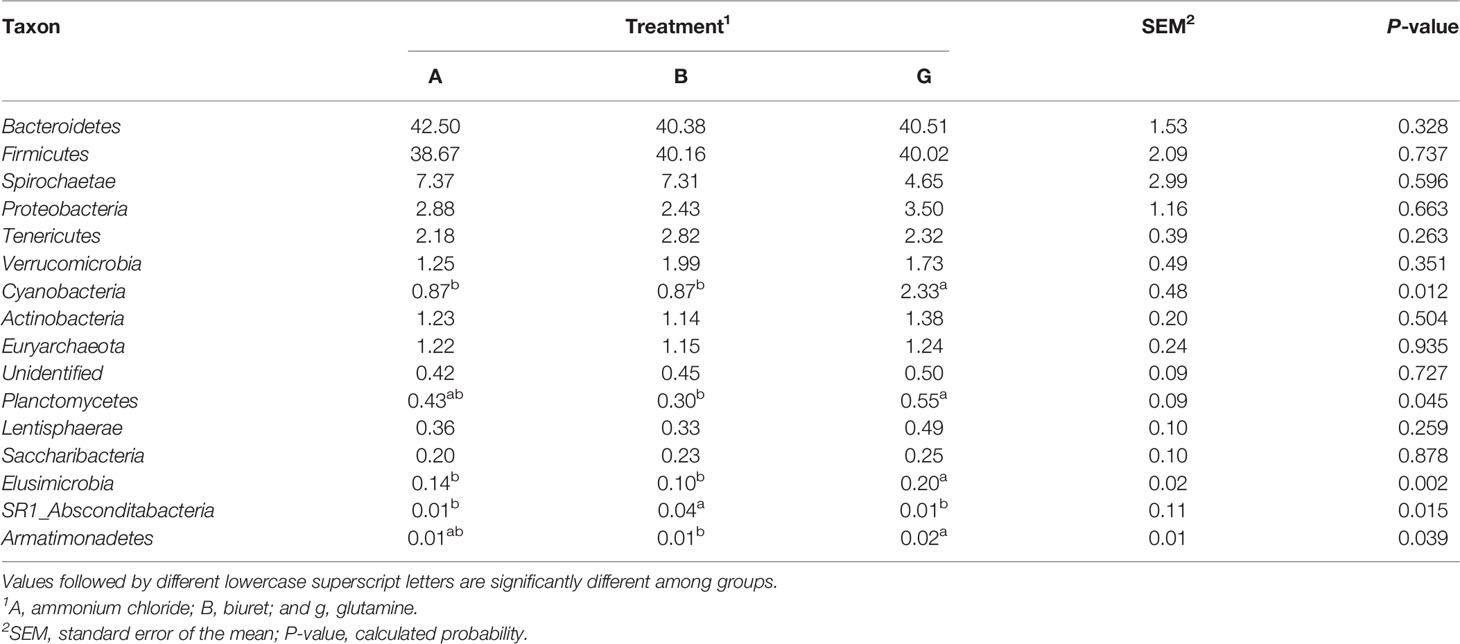
Table 6 Relative abundance (%) of the 16 most abundant bacteria at the phylum level as affected by dietary non-protein nitrogen sources.
Relative abundances of the top 20 bacteria at the genus level, as affected by different NPN sources, are shown in Table 7. Within Bacteroidetes, the relative abundance of Rikenellaceae_RC9_gut_group was the highest in group B (7.4727%), being significantly higher (P < 0.05) than that in group G. The relative abundances of SP3-e08 and Prevotellaceae_UCG-001 in were the highest in group A, although the values were not significantly different (P > 0.05) from those in groups B and G. Within Firmicutes, the relative abundance of Butyrivibrio_2 was higher (P < 0.05) in group B than in the other two groups, whereas the relative abundance of Ruminococcus_1 was significantly higher in group A (P < 0.05) than in group B.
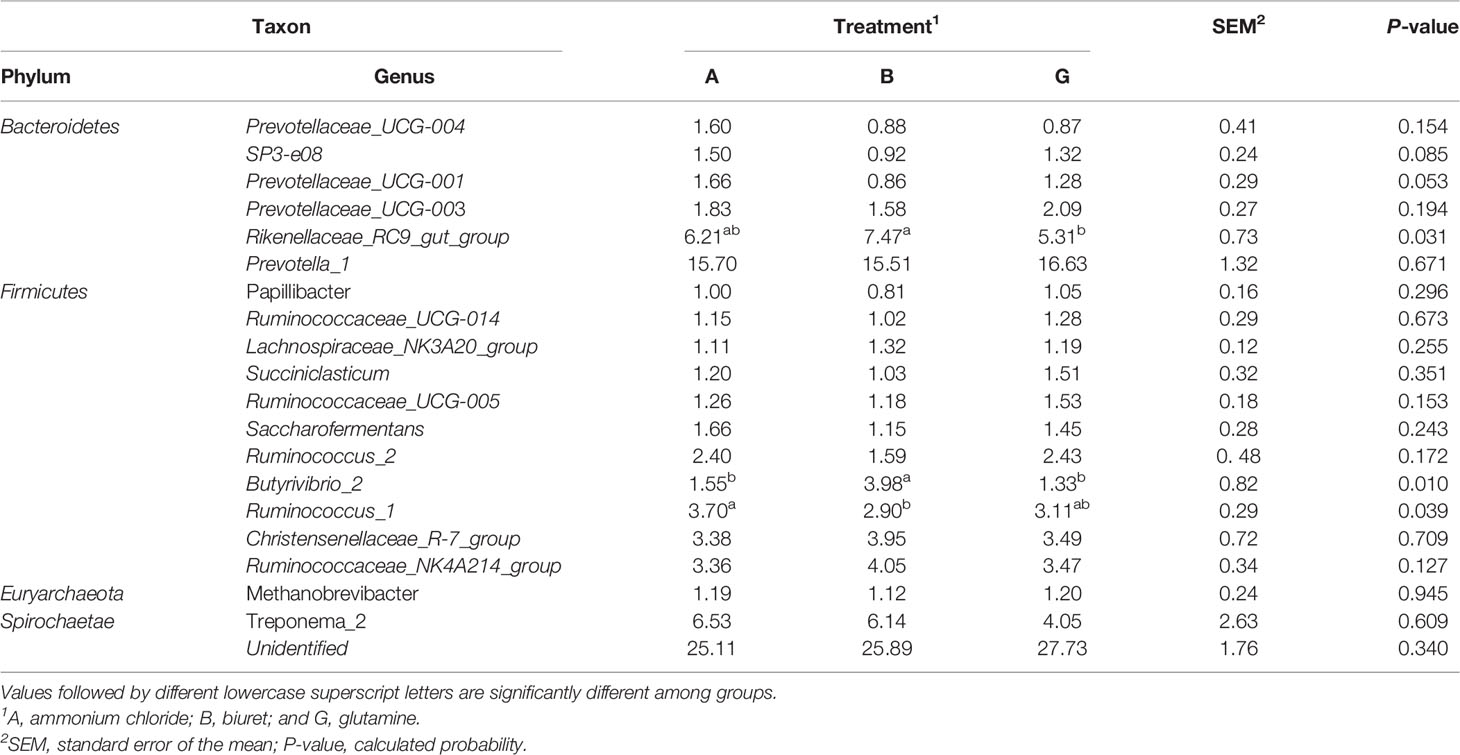
Table 7 Relative abundance (%) of the most abundant bacteria at the genus level affected by dietary non-protein nitrogen sources.
4 Discussion
4.1 Effects of NPN Sources on the Disappearance Rates of Nutrients
DM and NDF are important indicators of dietary utilization during ruminal fermentation. Increasing NPN level in in vitro cultures of rumen inocula enhanced DMD (Muthia et al., 2021). Further, NPN source affected DMD; specifically, biuret-straw and urea-straw significantly improved DMD, because starch, as a carbohydrate, readily provides energy for rumen fermentation (Muthia et al., 2021). However, the present experiment showed that DMD, CPD, ADFD, and NDFD did not differ among the treatments. In supplemented diets, nitrogen and energy are available simultaneously and in proportions required by rumen microbes, thus promoting diet digestion (Muthia et al., 2021). Although increased dietary energy levels significantly enhance apparent digestibility, the dietary levels of the three treatments were equal in terms of nitrogen and energy supply in the present study. This may explain the lack of significant differences in apparent digestibility among the experimental groups.
4.2 Effects of NPN Sources on Rumen Fermentation Parameters
pH reflects the overall state of rumen fermentation; however, it is easily affected by various factors, and a relatively stable pH value is considered beneficial to animal health and feed utilization (Bao et al., 2011). In the present experiment, the pH of approximately 5.5–6.0 maintained the normal state of the fermentation tank. In a study by Wang et al. (2010), treatments including ammonium chloride in different molecular forms of nitrogen showed the highest pH values. Similarly, in the present experiment, ammonium chloride showed the highest pH value. Ammonium chloride is a highly water-soluble ionic compound. Ammonium radicals and chloride ions are ionized in water; thus, ammonia gas accumulation elevates pH. In vitro results showed that the NH3-N concentration in group G was the highest among the three treatments. Protozoa exhibit a stronger deamination activity than bacteria. Protozoa are the net ammonia-producing microorganisms but cannot use ammonia to synthesize bacterial proteins (Huntington and Joan, 2021). Therefore, in group G, which showed a higher protozoa count, NH3-N concentration in the rumen was higher than that in groups A and B. In addition, glutamine and ammonium chloride could be directly utilized by microorganisms, and ammonium ions can dissociate, increasing the NH3-N concentration (Geisseler et al., 2011). These results are also consistent with the lowest NH3-N concentration in group B of the present experiment. Low ruminal NH3-N concentrations (<2.94 mM) can limit microbial growth and ruminal fermentation (Wang et al., 2018), thereby reducing the MCP content. These results were consistent with the lowest MCP in group B. NH3-N acts as the raw material for MCP synthesis and can directly reflect the production of microbial proteins. Simultaneously, in the present experiment, SP3-e08 and Prevotellaceae_UCG-001, which degrade carbohydrates, were more abundant in group A than in group G. As the synthesis efficiency of MCP is affected by many factors, carbohydrates in the feed can be fermented by microorganisms, providing energy and affecting the efficiency of microbial protein synthesis (Feng et al., 1993; Xie et al., 2019).
VFAs are the final products of rumen fermentation and provide energy to the body. In the present study, total VFAs did not differ among the treatments, due perhaps to the equal nitrogen and energy levels and similar DM degradability of the three treatments. Specifically, group G showed the highest propionate and butyrate levels, which may result from the highest NH3-N concentration in this group. As such, increased ruminal NH3-N concentrations improve energy production from straw (Lee et al., 1985). Furthermore, the molar ratio of acetate to propionate is an important measure of microbial fermentation. In the present experiment, the lowest acetate-to-propionate ratio was recorded in group G, probably because the propionate content in this group was significantly higher than that in groups A and B. In a study on lactating dairy cows, a linear decrease in isobutyrate with increase ammonium chloride concentration was observed, although it did not significantly affect other VFA. However, these results are inconsistent with our findings of the highest valerate and isovalerate proportion in group A. In dairy cows that are fed a diet based on maize stover as the only source of roughage, rumen valerate and isovalerate concentrations increase significantly (Weng, 2013). Increased pH, enhanced affinity of rumen microorganisms to fiber, and augmented ATP production affect VFA generation (Strobel and Russell, 1986). Furthermore, the type of roughage may affect valerate and isovalerate production (Yue et al., 2009; Ponce et al., 2016). In the present experiment, maize stover was added as roughage to the experimental diets. Nevertheless, the acetate-to-propionate ratio was the highest in group A, which tended toward acetate fermentation, changed the rumen fermentation mode, and increased the fiber content during fermentation. These results are consistent with the highest relative abundance of Ruminococcus_1, a major fiber-degrading bacteria in the rumen. Thus, greater maize stover fermentation in the rumen increased valerate and isovalerate proportion in group A compared with those in the other groups. Therefore, roughage utilization affects VFA generation, regardless of the concentrate-to-roughage ratio.
4.3 Effects of NPN Sources on Rumen Microbial Diversity
Ruminal microbiome is a complex and important natural fermentation system, which has long been a focus of ruminant research (Abecia et al., 2014). Rumen microorganisms are always in dynamic equilibrium. Dietary factors are the key influences, and the corresponding rumen microorganisms also affect host metabolism and nutrition (Sommer and Ckhed, 2013). Fibers and carbohydrates in the diet are degraded and fermented by ruminal microorganisms to synthesize MCPs and provide nutrition to the host animals. Short-chain fatty acids produced during anaerobic fermentation can meet 70% of the daily energy requirements of animals (Kelsea et al., 2015; Shabat et al., 2016). In addition, 50% of the ruminant protein demand is derived from ruminal microorganisms (Yeoman and White, 2015). Since the structural composition of the colony could not be deduced based on alpha diversity indices, we further analyzed the differences in bacterial abundances among the three groups. Jami and Mizrahi (2012) characterized the core microbiome of bovine rumen and identified Bacteroidetes, Firmicutes, and Spirochaetae as the top three phyla, consistent with the findings of the present study. The dominant microflora was not affected by diet, although the addition of different nitrogen sources affected the relative abundances of some rare rumen bacteria. Specifically, the relative abundances of Cyanobacteria, Planctomycetes, Elusimicrobia, and Armatimonadetes were the highest in group G and significantly higher than those in the B group. This may be because the NH3-N concentration in group B was very low, which likely limited the growth of microorganisms and reduced their activity. Surprisingly, the abundance of SR1_Absconditabacteria was highest in group B, with the lowest microbial protein and NH3-N concentration concentrations. Sun et al. (2019) reported that 45% low-concentration diets increased the relative abundance of SR1_Absconditabacteria on at the phylum level compared with 55% high-concentration diets. SR1_Absconditabacteria may not rely primarily on nitrogen sources for nutrition in the rumen; therefore, its activity was not restricted even at low NH3-N and protein levels. At the genus level, the relative abundances of Rikenellaceae_RC9_gut_group and Prevotella_1 were the highest in all groups, indicating that these genera play pivotal roles in rumen NPN metabolism. Prevotellaceae_UCG-001 is one of the major protein-degrading bacteria in the rumen (Avgustin et al., 1997). In the present study, the relative abundances of Prevotellaceae_UCG-001 and SP3-e08 were the lowest in group B, which is consistent with the lower ruminal MCP content in this group. However, the relative abundance of Rikenellaceae_RC9_gut_group was the highest in group B and the lowest in group G. Biuret is a urea derivative, and the mechanism through which rumen microorganisms convert urea to bacterial proteins differs from that through which they convert other NPN sources (Balasubramanian et al., 2013). Biuret cannot be directly degraded by microorganisms, but it can be catalyzed by ureases secreted by microorganisms (Strope et al., 2011). Moreover, the solubility of biuret is low, its degradation rate of is slow, and it remains in the rumen for a longer time, which may be conducive to the growth of Rikenellaceae_RC9_gut_group and alter the activity of other microorganisms (Ali, 2007). Butyrivibrio_2 is a fiber-degrading bacterium, and its relative abundance was the highest in group B. Based on the results of apparent indicators of nutrient disappearance rates, NDFD was the highest in group B. This may explain the higher microbial activity of Butyrivibrio_2 in group B. Griffith et al. (2016) reported that ammoniated barley straw feed is more suitable for fiber degradation by rumen microbes. Moreover, a higher relative abundance of Ruminococcus_1 was observed in group A, possibly because ammonium chloride was more easily used by these microbes, which promoted their proliferation. Finally, the relative abundance of Unidentified microbes was 25%, whose phyla and functions were unclear. Further studies on these microbes are warranted to better understand the metabolic mechanisms of NPN in the rumen.
In conclusion, the relative abundances of the major fiber-degrading bacteria Ruminococcus_1 and protein-degrading bacteria SP3-e08 and Prevotellaceae_UCG-001 were lower in group B than in the other groups. Group G showed significantly increased the concentrations of propionate, NH3-N, and microbial proteins, which decreased the acetate-to-propionate ratio and appeared to shift the fermentation pathways from acetate to propionate. Thus, glutamine is more suitable than biuret as a dietary source of non-protein nitrogen.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at https://www.ncbi.nlm.nih.gov/, PRJNA774940.
Ethics Statement
The animal study was reviewed and approved by The experiments were conducted according to the animal care guidelines of the Animal Care Committee, Institute of Subtropical Agriculture, Chinese Academy of Sciences.
Author Contributions
Animal experimentation: JZ and AR; data analysis: JZ and AR; data curation: JZ; writing—original draft: JZ; writing—review and editing: CZ, JJ, and JZ; funding acquisition: CZ. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Ministry of Science and Technology of China (2018YFD0501903), the National Natural Science Foundation of China (31372342, 3177131431, and 31730092), the Youth Innovation Team Project of ISA, the Chinese Academy of Sciences (2017QNCXTD_ZCS), Chinese Academy of Sciences (XDA27040306), and Science and Technology Department of Tibet (XZ202101ZD003N, XZDZKJ-2021-01-09, and XZ202101ZY0008N-KT01-Z03).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors express their heartfelt thanks to Professor CS Zhou for useful suggestions during paper writing. The authors also thank Dr. L. Chen and A. Ren for academic help. The authors thank Professors WJ Shen for providing the dual-flow rumen fermentation system.
References
Abecia L., Ramos-Morales E., Mart Nez-Fernandez G., Arco A., Mart N-Garc A A. I., Newbold C. J., et al. (2014). Feeding Management in Early Life Influences Microbial Colonisation and Fermentation in the Rumen of Newborn Goat Kids. Anim. Prod. Sci. 9, 1449–1454. doi: 10.1071/AN14337
Abdel Hameed A. A., Fedel E.L., Seed A.M. (2013). Growth Performance and Rumen Fermentation of Lambs Fed Untreated or Urea Treated Groundnut Hull with Different Protein Sources. J. Anim. Prod. Adv. 3 (3), 86—96.
Ali C. S. (2007). In Vitro Utilization of NPN Sources by Increasing Levels of Corn Starch in Straw-Based Diets. Pak. Vet. J. 27, 95–101.
Aquino D. L., Rosario M. V. D., Vergarra K. F. (2014). Effect of Augmented Feeding With Bypass Amino Acids and Slow-Release non-Protein Nitrogen Supplements on Milk Peak, Lactation Persistency, Milk Quality and Post-Partum Reproductive Performance of Brazilian Buffaloes. Buffalo. Bulletin. 32, 961–965. doi: 10.4081/ijas.2013.e85
Avgustin G., Wallace R. J., Flint H. J. (1997). Phenotypic Diversity Among Ruminal Isolates of Prevotella Ruminicola: Proposal of Prevotella Brevis Sp. Nov. Prevotella Bryantii Sp. Nov. And Prevotella Albensis Sp. Nov. And Redefinition of Prevotella Ruminicola. Int. J. Syst. Evol. Mic. 47 (2), 284–288. doi: 10.1099/00207713-47-2-284
Balasubramanian A., Durairajpandian V., Elumalai S., Mathivanan N., Munirajan A. K., Ponnuraj K. (2013). Structural and Functional Studies on Urease From Pigeon Pea (Cajanus Cajan). Int. J. Biol. Macromol. 58, 301–309. doi: 10.1016/j.ijbiomac.2013.04.055
Bao L., Huang Q., Chang L., Zhou J., Lu H. (2011). Screening and Characterization of a Cellulase With Endocellulase and Exocellulase Activity From Yak Rumen Metagenome. J. Mol. Catal. B-Enzym. 73, 104–110. doi: 10.1016/j.molcatb.2011.08.006
Belasco I. J. (1954). New Nitrogen Feed Compounds for Ruminants. A Laboratory Evaluation. J. Anim. Sci. 13, 601–610. doi: 10.2134/jas1954.133601x
Bondarenko G., Dvorska J. (2015). Non-protein. Nitro. (OPTIGEN™) High. Product. Dairy. Cow.: Ukrain. Exp. 205, 10–14.
Chegeni A., Li Y. L., Deng K. D., Jiang C. G., Diao Q. Y. (2013). Effect of Dietary Polymer-Coated Urea and Sodium Bentonite on Digestibility, Rumen Fermentation, and Microbial Protein Yield in Sheep Fed High Levels of Corn Stalk. Live. Sci. 157, 141–150. doi: 10.1016/j.livsci.2013.07.001
Chen L., Ren A., Zhou C., Tan Z. (2017). Effects of Lactobacillus Acidophilus Supplementation for Improving In Vitro Rumen Fermentation Characteristics of Cereal Straws. Ital. J. Anim. Sci. 16, 52–60. doi: 10.1080/1828051X.2016.1262753
Cole J. R., Wang Q., Cardenas E., Fish J., Chai B., Farris R. J., et al. (2009). The Ribosomal Database Project: Improved Alignments and New Tools for rRNA Analysis. Nucleic. Acids Res. 37, D141–D145. doi: 10.1093/nar/gkn879
Corte R. R., Brito F. O., Leme P. R., Pereira A., Freitas J. E., Renn F. P., et al. (2018). The Effects of Partial Substitution of Soybean With Urea or Slow-Release Urea on Finishing Performance, Meat Quality, and Digestion Parameters of Nellore Steers. Anim. Prod. Sci. 58 (12), 2242–2248. doi: 10.1071/AN16609
Dayani O., Tahmasbi R., Sabetpay M. R. (2010). Effect of Feeding Dietary Treated Wheat Straw With Urea and Whey on Fattening Lambs Performance. J. Anim. Vet. Adv 9 (21), 265–271. doi: 10.3923/javaa.2010.2763.2767
Dinic´ B., Terzic´ D., Blagojevic´ M., Markovic´ J., Lugic´ Z., Stanisavljevic´ R., et al. (2013). Effect of Addition of NPN Substances and Inoculants on Fermentation Process and Nutritive Value of Corn Silage. Acta Agr. Ser. 18, 11–21.
Edgar R. C. (2013). UPARSE: Highly Accurate OTU Sequences From Microbial Amplicon Reads. Nat. Methods 10, 996–998. doi: 10.1038/NMETH.2604
Feng P., Hoover W. H., Miller T. K., Blauwiekel R. (1993). Interactions of Fiber and Nonstructural Carbohydrates on Lactation and Ruminal Function. J. Dairy. Sci. 76, 1324–1333. doi: 10.3168/jds.S0022-0302(93)77463-9
Geisseler D., Horwath W. R., Joergensen R. G., Ludwig B. (2011). Pathways of Nitrogen Utilization by Soil Microorganisms – A Review. Soil Biol. Biochem. 42, 2058–2067. doi: 10.1016/j.soilbio.2010.08.021
Griffith C. L., Ribeiro G. O. Jr., Oba M., McAllister T. A., Beaucheminl K. A. (2016). Fermentation of Ammonia Fiber Expansion Treated and Untreated Barley Straw in a Rumen Simulation Technique Using Rumen Inoculum From Cattle With Slow Versus Fast Rate of Fiber Disappearance. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01839
Henderickx H. K. (1967). Urea as a Protein Supplement || Chapter 3-The Effectiveness of Urea and Other Non-Protein Nitrogen Compounds in Ruminant Feeding 73–82. doi: 10.1016/b978-0-08-012078-2.50007-6
Henderickx H. K., Demeyer D. I. (1967). Ammonia Production by Rumen Microbes In Vitro. Sci. Nat-Heidelberg. 54, 369–370. doi: 10.1007/BF00636642
Hu W. L. (2005). Study on the Effect of Saponins on Rumen Fermentation, Methane Production and Animal Performance (Hangzhou: Zhejiang University), 50–52.
Huntington G. B., Joan E. (2021). 352 A Century and a Half of Nitrogen and Protein Metabolism in Ruminants. J. Anim. Sci 99, 196. doi: 10.1093/jas/skab235.354
Jami E., Mizrahi I. (2012). Composition and Similarity of Bovine Rumen Microbiota Across Individual. Anim. PloS One 7, e33306. doi: 10.1371/journal.pone.0033306
Jiang X. T., Peng X., Deng G. H., Sheng H. F., Wang Y., Zhou H. W., et al. (2013). Illumina Sequencing of 16S rRNA Tag Revealed Spatial Variations of Bacterial Communities in a Mangrove Wetland. Microb. Ecol. 66, 96–104. doi: 10.1007/s00248-013-0238-8
Kelsea A., Jewell, Caroline A., McCormick, Christine L., Odt, et al. (2015). Ruminal Bacterial Community Composition in Dairy Cows Is Dynamic Over the Course of Two Lactations and Correlates With Feed Efficiency. Appl. Environ. Microb. 81, 4697–4710. doi: 10.1128/AEM.00720-15
Lee G. J., Hennessy D. W., Williamson P. J., Nolan J. V., Leng R. A. (1985). Responses to Protein Meal Supplements by Lactating Beef Cattle Given a Low-Quality Pasture Hay. Aust. J. Agr. Res. 36, 729–741. doi: 10.1071/AR9850729
Li C., Xue A., Zhao Q. (2017). 686 Effect of Licking Molasses–Urea Block on Weight Gain, Rumen Fermentation, and the Main Microbe Populations of Grazing Sheep During Grass Withering Period. J. Anim. Sci 95 (suppl4), 335. doi: 10.2527/asasann.2017.686
Mahr-un-Nisa Sarwar, M., Khan M. A. (2004). Influence of Ad Libitum Feeding of Urea-Treated Wheat Straw With or Without Corn Steep Liquor on Intake, in Situ Digestion Kinetics, Nitrogen Metabolism, and Nutrient Digestion in Nili-Ravi Buffalo Bulls. Crop Pasture Sci. 55, 229–236. doi: 10.1071/AR02236
McDougall E. J. (1948). Studies on Ruminant Saliva. 1. The Composition and Output of Sheep's Saliva. Biochem. J. 43, 99–108. doi: 10.1042/bj0430099
Mumbi M. P., Eissa N., Noah B. C., Ehsan K., Jean-Eric G., Heimesaat M. M. (2015). Antepartum Antibiotic Treatment Increases Offspring Susceptibility to Experimental Colitis: A Role of the Gut Microbiota. Plos. One 25, 1–22. doi: 10.1371/journal.pone.0142536
Muthia D., Ridla M., Laconi E. B., Ridwan R., Jayanegara A. (2021). Effects of Ensiling, Urea Treatment and Autoclaving on Nutritive Value and in-Vitro Rumen Fermentation of Rice Straw. Adv. Anim. Vete. Sci. 9 (5), 655–661. doi: 10.17582/journal.aavs/2021/9.5.655.661
Nadeem M. S., Pasha T. N., Jabbar M. A., Javed K., Ditta Y. A. (2014). Effect of Different non-Protein Nitrogen (NPN) Sources on Performance of Lactating Nili-Ravi Buffaloes. Int. Livestock. Nutr. Conf. 24, 1–4.
Ponce C. H., Schutz J. S., Smith D. R., Galyean M. L. (2016). Effects of Wet Distillers Grains With Solubles in Diets Based on Steam-Flaked Corn Containing Moderate or High Roughage Levels on Nutrient Digestibility in Feedlot Cattle and In Vitro Fermentation. Prof. Anim. Ent. 32, 580–590. doi: 10.15232/pas.2016-01521
Shabat S. K. B., Sasson G., Doron-Faigenboim A., Durman T., Yaacoby S., Berg Miller M. E., et al. (2016). Specific Microbiome-Dependent Mechanisms Underlie the Energy Harvest Efficiency of Ruminants. Isme. J. 10, 2958–2972. doi: 10.1038/ismej.2016.62
Shen W. J. (2012). Development and Operation Parameters of a New Dual Outflow Rumen Simulation System (Changsha, China: Hunan Agricultural University).
Singh K. (2014). Biotechnological Approaches on Conversion of Agrocellulosic Residues and Dairy Wastes Into Useful End-Products. Indian. J. Anim. Sci. 36, 1159–1162. doi: 10.1002/sia.1865
Sommer F. B., Ckhed F. (2013). The Gut Microbiota–Masters of Host Development and Physiology. Nat. Rev. Microbiol. 11, 227–238. doi: 10.1038/nrmicro2974
Strobel H. J., Russell J. B. (1986). Effect of pH and Energy Spilling on Bacterial Protein Synthesis by Carbohydrate-Limited Cultures of Mixed Rumen Bacteria. J. Dairy. Sci. 69, 2941–2947. doi: 10.3168/jds.S0022-0302(86)80750-0
Strope P. K., Nickerson K. W., Harris S. D., Moriyama E. N. (2011). Molecular Evolution of Urea Amidolyase and Urea Carboxylase in Fungi. Bmc. Evol. Biol. 11, 1–15. doi: 10.1186/1471-2148-11-80
Sun F. Y., Zhao Y. G., Xue F. G., Hua D. K., Du C. M., Jiang L. S., et al. (2019). Effects of Kelp Powder on Rumen Fermentation Parameters and Microflora Structure of Dairy Cows Fed a High Concentrate Diet. Chin. J. Anim. Nutr. 31, 2842–2853.
Wang M. Z., Yu L. H., Wang H. R., Li G. X., Cao H. C. (2010). Effects of Different Molecular Nitrogen Sources on Rumen Microbial Fermentation and Protein Synthesis. China J. Anim. Husb. 46, 20–24.
Wang P., Zhao S., Nan X., Jin D., Wang J. (2018). Influence of Hydrolysis Rate of Urea on Ruminal Bacterial Diversity Level and Cellulolytic Bacteria Abundance In Vitro. Peer. J. 6, e5475. doi: 10.7717/peerj.5475
Weng X. X. (2013). Studies on Rumen Fermentation and VFA Absorption Characteristics and Related Gene Expression of Dairy Cows Fed Different Diets (Lanzhou, China: Gansu Agricultural University), 1–89.
Williams S., Williams S., Firestndone D., Williams S.N., WilliamsScott M. (201990). Official Methods of Analyses of the Association of Official Analytical Chemists.
Wu M. N. (2017). Optimization of Some Operating Parameters of a Novel Dual-Outflow Continuous Culture Rumen Simulation System (Changsha, China: Hunan Agricultural University), 1–53.
Xie Y., Wu Z., Wang D., Liu J. (2019). Nitrogen Partitioning and Microbial Protein Synthesis in Lactating Dairy Cows With Different Phenotypic Residual Feed Intake. J. Anim. Sci. Biotech. 10, 2–8. doi: 10.1186/s40104-019-0356-3
Xin H. S., Schaefer D. M., Liu Q. P., Axe D. E., Meng Q. X. (2010). Effects of Polyurethane Coated Urea Supplement on In Vitro Ruminal Fermentation, Ammonia Release Dynamics and Lactating Performance of Holstein Dairy Cows Fed a Steam-Flaked Corn-Based Diet. Asian Austral J. Anim. 23, 491–500. doi: 10.5713/ajas.2010.90153
Yeoman C. J., White B. A. (2015). Gastrointestinal Tract Microbiota and Probiotics in Production Animals. Annu. Rev. Anim. B. 2, 469–486. doi: 10.1146/annurev-animal-022513-114149
Yue Q., Yang H. J., Cao Y. C., Zhang D. F., Jiang Y. H., Wang J. Q. (2009). Feruloyl and Acetyl Esterase Production of an Anaerobic Rumen Fungus Neocallimastix Sp. YQ2 Effected by Glucose and Soluble Nitrogen Supplementations and its Potential in the Hydrolysis of Fibrous Feedstuffs. Anim. Feed. Tech. 153, 263–277. doi: 10.1016/j.anifeedsci.2009.06.019
Keywords: non-protein nitrogen, artificial rumen, ammonia nitrogen, protozoa, microbial diversity
Citation: Zhu J, Ren A, Jiao J, Shen W, Yang L, Zhou C and Tan Z (2022) Effects of Non-Protein Nitrogen Sources on In Vitro Rumen Fermentation Characteristics and Microbial Diversity. Front. Anim. Sci. 3:891898. doi: 10.3389/fanim.2022.891898
Received: 08 March 2022; Accepted: 05 May 2022;
Published: 10 June 2022.
Edited by:
Georgios Theodorou, Agricultural University of Athens, GreeceReviewed by:
Kampanat Phesatcha, Nakhon Phanom University, ThailandIrida Palamidi, Agricultural University of Athens, Greece
Copyright © 2022 Zhu, Ren, Jiao, Shen, Yang, Zhou and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuanshe Zhou, emNzQGlzYS5hYy5jbg==
 Jinjia Zhu
Jinjia Zhu Ao Ren1,2
Ao Ren1,2