
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Anim. Sci. , 29 October 2021
Sec. Animal Nutrition
Volume 2 - 2021 | https://doi.org/10.3389/fanim.2021.761930
This article is part of the Research Topic The Relationship of Animal Health and Management to Food Safety View all 6 articles
Greater consumer demand for all natural, antibiotic-free poultry products has led to an increase in pastured poultry operations. Given the increased level of environmental interaction, and the potential increase in exposure to foodborne pathogens, a greater understanding of the prevalence and diversity of Salmonella populations inherent within pastured poultry flocks. To achieve this, 42 pastured poultry flocks from 11 farms were sampled using a farm-to-fork strategy and Salmonella was isolated and characterized through pre-harvest (feces, soil) to post-harvest (ceca, whole carcass rinse) to the final product (whole carcass rinse) the consumer would purchase. Salmonella was isolated from 353 of a total of 2,305 samples, representing an overall prevalence of 18.1%. By far the most prevalent serotype was Kentucky (72.7% of all isolates), with < 16% of all Salmonella representing a top serotype of concern for human health according to the CDC. Even though these flocks were raised antibiotic-free, Salmonella isolates exhibited resistances to a variety of antibiotics, with the two most common resistances being toward tetracycline and streptomycin (68.8 and 64.4% of all isolates, respectively); however, almost 98% of the multidrug resistant isolates were serotype Kentucky. Salmonella prevalence and diversity (both in terms of serotypes and antibiotic resistance profiles) were related more to the farm location than to the type of sample from which the Salmonella was isolated from along the farm-to-fork continuum. Based on these data, while Salmonella prevalence was similar to that from conventional poultry operations, serotypes of lesser concern to human health (Kentucky, Indiana) tended to fill the ecological niche for Salmonella species throughout the farm-to-fork continuum in these pastured poultry flocks. The diversity of these Salmonella populations tended to be farm specific, indicating the need for more tailored intervention strategies to continue to enhance the safety of these products.
Salmonella is a persistent cause of foodborne illness with potential for resurgence because it can colonize many food production environments (Silva et al., 2014; Magossi et al., 2019). Poultry and poultry products are strongly linked to ~50% of reported cases of salmonellosis (Gu et al., 2015). Uncooked poultry meat, especially comminuted or mechanically separated meat, continues to be an important source of Salmonella infections according to the USDA-Food Safety Inspection Service (USDA-FSIS) (USDA-FSIS, 2020a). From 1999 to 2008, 33% of the 621 known-source outbreaks caused by Salmonella were attributed to contaminated poultry products, and contaminated poultry products caused ~336,000 illnesses, ~5,000 hospitalizations, and ~150 deaths annually (Batz et al., 2012). Different serotypes vary in their association with poultry and foodborne disease (Gould et al., 2013; Crim et al., 2014), and poultry products are considered a primary reservoir of salmonellosis for adults and people living in urban areas (Mughini-Gras et al., 2014).
Recently, increased demand for antibiotic-free, “natural” products has pushed consumers toward the organic food market (Dimitri and Oberholtzer, 2009; Reisch et al., 2013). This has impacted the poultry industry, where broiler meat harvested from alternative poultry farming production facilities, such as organic and pastured, have increased in demand (Van Loo et al., 2011; Rothrock et al., 2016). Organic poultry farms are characterized by farms that rear birds without the use of antibiotics and allow the birds access to the outside, while pastured poultry operations require moveable pens/housing that are moved daily to fresh pasture. The microbial safety of conventionally-raised poultry and their products throughout the entire farm-to-fork continuum has been studied in the US (Rodriguez et al., 2006; Pitesky et al., 2012), but organic/pastured production studies related to food safety issues are limited. A comparison between alternative production practices and conventional practices are problematic because of vast differences inherent in these production systems (Van Loo et al., 2012), but some recent studies have worked to statistically compare the two types of farms (Siemon et al., 2007; Alali et al., 2010; Peng et al., 2016; Kassem et al., 2017). Siemon et al. (2007) and Alali et al. (2010) both found that Salmonella contamination of fecal matter and bird feed was significantly lower in organic farms. However, there is no scientific consensus since others have shown no difference (Bailey and Cosby, 2005; Lund, 2006; Lestari et al., 2009), or that conventional practices yielded significantly lower Salmonella levels (Cui et al., 2005). Therefore, more work needs to be done to better understand the ecology of Salmonella in these emerging poultry management systems, especially since they have been shown to be a member of the pastured broiler core microbiome along the farm-to-fork continuum (Rothrock et al., 2019b).
An additional topic of interest is that organic and pastured poultry broilers are both reared antibiotic-free, which could potentially affect the antimicrobial resistance (AR) profile and antibiotic resistance genes (ARG) of the Salmonella populations along the farm-to-fork continuum. There have been conflicting results from studies comparing the abundance of AR foodborne pathogens and ARG in antibiotic-free vs. conventional broiler farms (Sapkota et al., 2011; Zhang et al., 2011; Millman et al., 2013; Garcia and Teixeira, 2017; Davis et al., 2018; Rovira et al., 2019; Pesciaroli et al., 2020). Given the recent decline in clinically important antibiotics due to the rapid appearance of resistant foodborne pathogen strains (Alanis, 2005; Smith and Coast, 2013), it is imperative that the AR potential of Salmonella inherent within alternative poultry production system is better studied and understood.
Therefore, the objective of this study was to better understand Salmonella prevalence and diversity along the pastured poultry farm-to-fork continuum, and characterize those isolates based on the AR profiles to assess the baseline Salmonella AR inherent within these antibiotic-free management systems. From 42 pastured broiler flocks covering eleven farms in the southeastern US, Salmonella was isolated from pre-harvest (feces, pasture soil) and post-harvest (ceca, end of processing and final product whole carcass rinses) samples. Isolates were serotyped and characterized using the National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS) protocols (CDC, 2015) to determine the AR profiles, as well as the prevalence of multidrug resistance (MDR) among these isolates. This information will provide vital Salmonella ecology data (prevalence, diversity, AR) from these types of alternative, antibiotic-free poultry management systems to not only help improve on-farm poultry food safety, but also the growing number of customers that are consuming these poultry products.
Forty-two flocks from eleven pastured poultry farms located in the southeastern U.S. were sampled between 2014 and 2017. The major broiler management parameters for each farm within the study can be seen in Table 1. All broilers flocks were considered all-natural or certified all-natural, pasture-raised using moveable pens and temporary fencing, and never had any antibiotics administered to them during their grow out, nor were they used historically on the farms.
The following samples were collected along the “farm-to-fork” continuum for each flock: (i) feces, (ii) pasture soil, (iii) cecal content at processing, (iv) whole carcass rinse (WCR) directly after processing, and (v) final product WCR after the carcass has been stored at the correct temperature and correct time for each farm. All samples were collected in the field, and returned to the lab in a cooler packed in ice. Pre-harvest samples (feces and soil) were collected from the pasture where the flock was currently residing at the time of sampling, and these sampling occurred 3 times during grow-out: (i) within a few days of being placed on the pasture (Start), (ii) halfway through their time on pasture (Mid), and (iii) on the day which the flock was processed (End). At each sampling time, the pasture area was divided into five separate sections, and five subsamples in each section were pooled into a single sample for each section (five total fecal and five total soil samples were collected on each sampling day). The total volume of sample collected for each field sample was at least 25 g. For the post-harvest processing and final product samples (ceca, WCR), five pooled samples were collected for each sample type, with each pooled sample containing ceca or WCR from five carcasses.
To prepare the environmental samples for homogenization, three g (feces, soil) or five ceca were combined within filtered stomacher bags (Seward Laboratories Systems, Inc., West Sussex, UK), and diluted 1:3 using 10 mmol L−1 phosphate-buffered saline (PBS). For the WCR, 100 ml of 10 mmol L−1 PBS were added to each carcass within the storage bag, and the bags were vigorously shaken for 60 s. Five WCRs were pooled into a single filtered stomaching bag, and this was repeated five times (n = 25 carcasses). No further dilution in 10 mmol L−1 PBS was required for the WCR samples. All samples were homogenized for 60 s and these homogenates were used for all downstream cultural isolations.
As a pre-enrichment step, the stomached homogenates remained in the filtered stomacher bags and incubated overnight at 35°C. Two different enrichments broths were used to isolate Salmonella spp. from these environmental samples: Tetrathionate (TT; Becton-Dickinson, Sparks, MD) broth and Rappaport-Vassiliadis (RV; Becton Dickinson) media. After overnight incubation at 42°C in both of these enrichment broths, one loopful from each enrichment broth was spread on two different differential media: Brilliant Green Sulfa with novobiocin (BGS; Becton Dickinson) agar and xylose lysine tergitol-4 (XLT-4; Becton Dickinson) agar. These plates were incubated overnight at 35°C, and on each plate, three Salmonella–like colonies per subsample were picked and confirmed using triple sugar iron agar (TSI; Becton-Dickinson) and lysine iron agar fermentation (LIA; Becton-Dickinson) using an incubation period of 18–24 h at 35°C. Final confirmation of suspect TSI/LIA isolates was performed using Salmonella polyvalent O antiserum agglutination (Becton-Dickinson), using manufacturer's specifications. Positive salmonellae were serogrouped using individual Salmonella poly O antisera for O groups A through I, following the Kauffman-White scheme (Popoff and Le Minor, 1997).
All recovered Salmonella isolates were serotyped using the dkgB-linked intergenic space region (dkgB-ISR) PCR method (Guard et al., 2012). Single colonies for each isolate were grown in 10 ml of brain heart infusion (BHI) broth (Difco BD, Franklin Lakes, NJ) at 37°C for 16 h. Bacterial cells were pelleted in a Sorvall RC5B Plus centrifuge at 5,000 × g for 15 min in a Sorvall Super-lite SLA 600TC rotor. The DNA from all Salmonella isolates were extracted and purified using the PureLink Genomic DNA Mini Kit (Invitrogen, Grand Island, NY). Spectrometer readings of DNA samples were obtained using a NanoDrop 1000 (ThermoScientific, Wilmington, DE) to assure OD260/280 ratios are >1.7 and that DNA concentrations are above 20 ng ml−1. The PCR protocol and primers targeting the dkgB-linked ISR region (including the entire 5S ribosomal gene) have been described previously (Morales et al., 2006). To determine serotype, amplicon sequence trimmed to the aforementioned ISR region were aligned to reference sequences deposited at NCBI by DNASTAR Lasergene SeqMan Version 8.0.2 using default project assembling parameters except as follows: minimum match percentage 100, minimum sequence length 100. Only perfect matches can be used to call serotype. ISR reference sequences that define serotype have GenBank accession numbers JN105119-JN105125 and JN092293-JN092328.
Recovered isolates were sub-cultured on Blood Agar Plates (BAP) overnight at 36 ± 1°C, twice sequentially. One to two colonies were used to inoculate five mL of demineralized water to achieve a 0.5 McFarland equivalent using the Sensititre nephelometer (ThermoScientific, TREK Diagnostics, Inc., Cleveland, OH). Following vortexing, 10 μL of the cell suspension was transferred to 11 mL of Sensititre Cation adjusted Mueller-Hinton Broth with TES, followed by thorough vortexing. Fifty microliters of the inoculum was transferred to each well of the Sensititre® NARMS Gram-Negative Format CMV3AGNF plate (Trek Diagnostic Systems). These AST plates contained varying concentrations of the following antimicrobials: cefoxitin, azithromycin, chloramphenicol, tetracycline, ceftriaxone, amoxicillin/clavulanic acid (2:1), ciprofloxacin, gentamicin, nalidixic acid, ceftiofur, sulfisoxazole, trimethoprim/sulfamethoxazole, ampicillin, and streptomycin. Plates were sealed and incubated at 36±1°C for 24 h. Quality control strains E. coli ATCC 25922, Staphylococcus aureus ATCC 29213, Enterococcus faecalis ATCC 29212, and Pseudomonas aeruginosa ATCC 27853 were included in susceptibility tests as controls (Clinical and Laboratory Standards Institute, 2010). The published NARMS protocol and breakpoints were used for characterization and antibiotic resistance determination for each isolate (CDC, 2015).
A generalized linear model with a binomial distributed outcome and a log link function was used to determine if there are significant differences in Salmonella prevalence (presence/absence) and the presence of multidrug resistant Salmonella (≥ 3 antibiotics) between farms and sample types. The significance of the model was established using a likelihood ratio test (R function ANOVA with argument test set to “Chisq”). Multiple comparisons of means, i.e., regression coefficients for farms and sample types were done using the multcomp package in R. The mcp function was used to specify linear hypotheses and the glht function was used to make Tukey contrasts. Statistical analyses were performed using R (v4.0.3) and a p < 0.05 was used for all analyses to determine significance.
Overall, 353 of the 2,305 samples (18.1%) were positive for Salmonella, which is similar to the prevalence of Salmonella found in other studies focusing on pastured or free-range broiler flocks (16–31%) (Bailey and Cosby, 2005; Siemon et al., 2007; Melendez et al., 2010; Scheinberg et al., 2013; Shi et al., 2019). While this prevalence in this study is higher than what was reported for Salmonella from conventionally reared poultry in 2014 (3.7%) (USDA-FSIS, 2020b), several direct comparison studies have found that Salmonella prevalence was lower in pastured vs. conventional systems (Lestari et al., 2009; Alali et al., 2010; Van Loo et al., 2012). Table 2 outlines the breakdown of Salmonella prevalence based on either sample type or farm of origin. Pre-harvest Salmonella prevalence (15.2 and 11.5% for feces and soil, respectively) and post-harvest cecal samples (12.9%) were significantly lower (p < 0.001) than processing WCR Salmonella prevalence (28.5%), with final product prevalence (17.8%) almost being significantly lower than processing WCR prevalence as well (p = 0.0502). Farm of origin had a significant effect on Salmonella prevalence (p < 0.001), with farms being categorized into 4 groups based on their Salmonella prevalence: >21% (A, H, I, M); 13–16% (B, E); 2–7% (C, D, K); and 0% (J, L). In general, the farms where flocks were sampled over multiple years (A, B, E, I) tended to have higher prevalence than those flock(s) sampled only within a single year (C, D, J, L), which speaks to the temporal variability of Salmonella on pastured farms. This variation in Salmonella prevalence between farms was not unexpected, considering the variability of environmental and management conditions pastured flocks are exposed to have been shown to effect pathogen concentrations, as previously reviewed (Rothrock et al., 2019a; Shi et al., 2019).
Sixteen isolates were not able to be recovered from frozen stocks, hence the reduced number of isolates seen between the prevalence (353) and diversity (337) analyses. In total, fifteen different serotypes were identified among the 337 Salmonella isolates with nine serotypes being isolated in more than one sample (Table 3). The dominant serotype recovered was Kentucky, representing 72.7% of all the isolates, with Indiana (9.5%) and Infantis (5.9%) representing the other most prevalent serotypes. All other recovered serotypes represented ≤ 3% of the Salmonella recovered throughout the study. The USDA-FSIS has identified Kentucky as the dominant Salmonella serotype in US poultry production consistently, with it representing an ever increasing percentage of all Salmonella recovered from 1998 (26.7%) to 2014 (60.1%) (USDA-FSIS, 2020b). Kentucky has also been previously shown to be the most dominant Salmonella serotype in pastured flocks, representing 53–95% of all serotypes recovered (Melendez et al., 2010; Rothrock et al., 2016). While there is a concern for the safety of pastured raised poultry products due to the less stringent/controlled management systems and increased environmental exposure of the flocks, it should be noted that of all the recovered Salmonella, only 15.7% (53/337 isolates) represented the top 32 Salmonella serotypes that the CDC considered of concern for human health (CDC, 2013).
When looking at serotype diversity on a sample type level (Figure 1A), the distribution of Salmonella was fairly consistent. Kentucky represented a higher percentage of the post-harvest isolates (81.2%) as compared to the pre-harvest environment (68.3%). The serotype richness of Salmonella populations (e.g., number of serotypes recovered) decreased from the pre-harvest (10 and 7 for feces and soil, respectively) to post-harvest (5 and 8 for the ceca and processing WCR, respectively) to the final product WCR (3). It should be noted that there is a spike in Salmonella richness in the processing WCR, half of those serotypes represent only a single isolate for the entire study. Not only did the Salmonella richness decrease throughout the farm-to-fork continuum, so did the prevalence of Salmonella serotypes that are of the greatest concern to human health (Infantis, Enteritidis, Seftenberg, Braenderup/Cholareasuis, Muenchen, 1,2,[5],12:I:-, Javiana, Schwarzengrund, Mbandaka/Typhimurim, and Heidelberg) (CDC, 2013). While these serotypes represented 22.7% of all Salmonella isolated from the fecal samples, they constituted only 12.5% of the processing WCR rinse isolates and only 2.6% of the Salmonella recovered from the final product WCR. In fact, Infantis was the only Salmonella serotype of greatest concern to human health according to the CDC recovered from the final products, and represents only a single isolate from the 230 final product WCR samples in the study.
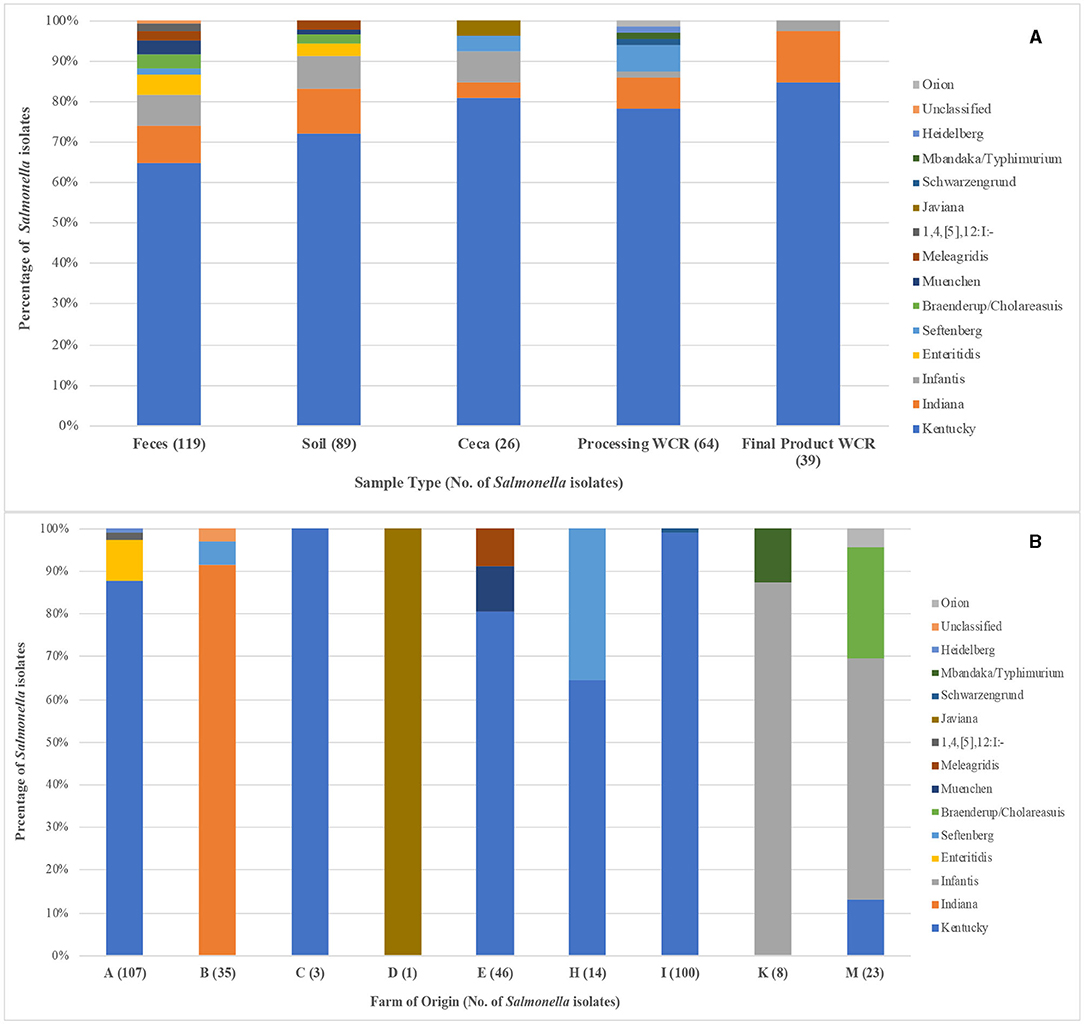
Figure 1. Salmonella serotype diversity of the 337 isolates recovered from pastured poultry flocks as determined by the dkgB-ISR PCR method. (A) Distribution of Salmonella serotypes based on sample type from which the Salmonella was isolated. (B) Distribution of Salmonella serotypes based on the farm from which the Salmonella was isolated. For both graphs, the number in parentheses next to the sample type/farm indicates the number of isolates for that sample type/farm.
As seen in the overall Salmonella prevalence data described above, farm of origin had a direct effect on serotype diversity (Figure 1B). Salmonella was not recovered from flocks from two of the farms (J, L); therefore, only nine farms are analyzed here. Of the fifteen serotypes identified in this study, twelve of them were not isolated from more than one farm. While serotype Indiana was the second most prevalent Salmonella recovered in the study across all sample types (9.5% of isolated Salmonella), it was only isolated from a single farm (B), highlighting how influential farm location was on Salmonella diversity. The three multi-farm serotypes were Kentucky (A, C, E, H, I, M), Infantis (K, M), and Seftenberg (B, H). Kentucky was the dominant serotype on 5 of the 6 farms they were isolated from, ranging from 13.0 to 100%, while Infantis was the dominant serotype recovered from both farms it was found on (87.5 and 56.5% on farms K and M, respectively). Of interest related to the Infantis isolates is the fact they were only recovered from flocks that were sampled in the final year of the study (2017). The significance of Infantis has increased in recent years, with salmonellosis cases attributed to Infantis increased by 165.8% from 2006 to 2016 (CDC, 2018), its prevalence among Salmonella isolated from poultry products increased by 483.6% from 1998 to 2014 (USDA-FSIS, 2020b). Additionally, it was responsible for a multi-state, 129-case outbreak associated with raw poultry products in 2018 that resulted in 25 hospitalizations and 1 death (CDC, 2019). The emergence of Infantis in these final year flocks processed months before the multistate outbreak occurred may indicate that Infantis was present in both conventional and pastured poultry management systems simultaneously, but a greater genetic characterization of the Infantis from this study would need to be performed.
While currently discontinued poultry industry, the prophylactic use antibiotics during rearing provided selective environmental pressure resulting in the proliferation of antibiotic resistance within the poultry-associated microbiome, including pathogens such as Salmonella. Considering all of the flocks in the present study were raised with antibiotic-free/medication-free feed and were not exposed to any exogenous sources of antibiotics pre- or post-harvest, determining the AR profiles of the recovered Salmonella would provide valuable baseline AR data on poultry-related Salmonella. Even within an antibiotic-free management system, several antibiotic resistances were expressed in the recovered Salmonella (Table 4). This was not unexpected, since previous work has shown that S. enterica serotypes to have low level natural resistance to several antibiotics, including tetracycline, macrolides/azithromycin, and streptomycin (Stock and Wiedemann, 2000). Around two-thirds of all Salmonella isolates were resistant to STR (64.4%) and TET (68.8%). Resistance toward STR and TET have been previously observed in pastured poultry management systems, ranging from <40% (Griggs et al., 2006; Alali et al., 2010) to 75–90% (Melendez et al., 2010; Rothrock et al., 2016) of the recovered Salmonella isolates in those studies. The next set of expressed resistances (26–27% of all isolates) comprised three antibiotic classes: b-lactam/b-lactamase inhibitors (AMO), cephems (FOX, TIO, AXO), and penicillins (AMP). While similar levels of AMO and AMP resistance have been observed in previous studies (Griggs et al., 2006; Lestari et al., 2009; Alali et al., 2010), very few described Salmonella isolates with resistance to any of the cephem class of antibiotics (Siemon et al., 2007; Rothrock et al., 2016). Of all of the antibiotics tested against using the gram negative NARMS panel, ciprofloxacin was the only antibiotic that no Salmonella isolates were resistant to, while resistances were found for six other antibiotics in one (FIS, CHL, NAL) or two (GEN, SXT, AZI) isolates.
When comparing the Salmonella AR profiles for the seven antibiotics with the most resistances based on sample type (Figure 2), a similar trend was seen for all sample types. While the resistance rates varied among sample types, in general resistance to STR/TET (52.5–80.8%) was higher than to the AMO/AMP/FOX/TIO/AXO group (7.7–32.7%), as was seen with the serotype diversity above, Salmonella AR profile diversity was much more distinct at the farm level (Figure 3). Three farms were excluded from this analysis since either no Salmonella were recovered (J, L), or all Salmonella that were recovered were pan-susceptible (K). When comparing farms where more than a single flock was followed, two major trends became evident. First, most of the STR/TET resistance came from Salmonella isolated from 3 of the 4 farms where at least 5 flocks were followed (A, E, I). Secondly, the AMO/AMP/FOX/TIO/AXO group resistances were predominantly only found on 2 farms (A, H). One potential cause of this resistance group is carriage by the blaCMY−2gene, which confers resistance to several antibiotic classes, including penicillins (AMP), b-lactam/b-lactamase inhibitors (AMO), and cephems (FOX/TIO/AXO) (Jacoby and Munoz-Price, 2005), and has been found to confer MDR in Salmonella and other bacteria (Miriagou et al., 2004; Mataseje et al., 2009). Preliminary whole genome sequencing analysis of a few of these isolates shows the presence of the blaCMY−2gene (data not shown), supporting this hypothesis. Interestingly, farm A relocated to a new location during the study, and all flocks from the new location were labeled under Farm H after a full year passed between samplings. Therefore, the farmer, management system, and farm equipment were the same, but the physical farm location was completely different (including soil type, composition of pasture, and climate). It has been previously demonstrated that flock management could significantly impact the farm environment (Sossidou et al., 2011; Sánchez-Casanova et al., 2020), so potentially the physical management structures (e.g., housing, farm equipment) have to potential to not only shape new farm environments physically, but also biologically in terms of being potential vectors for pathogens expressing specific AR profiles, or harboring plasmids containing specific ARGs.
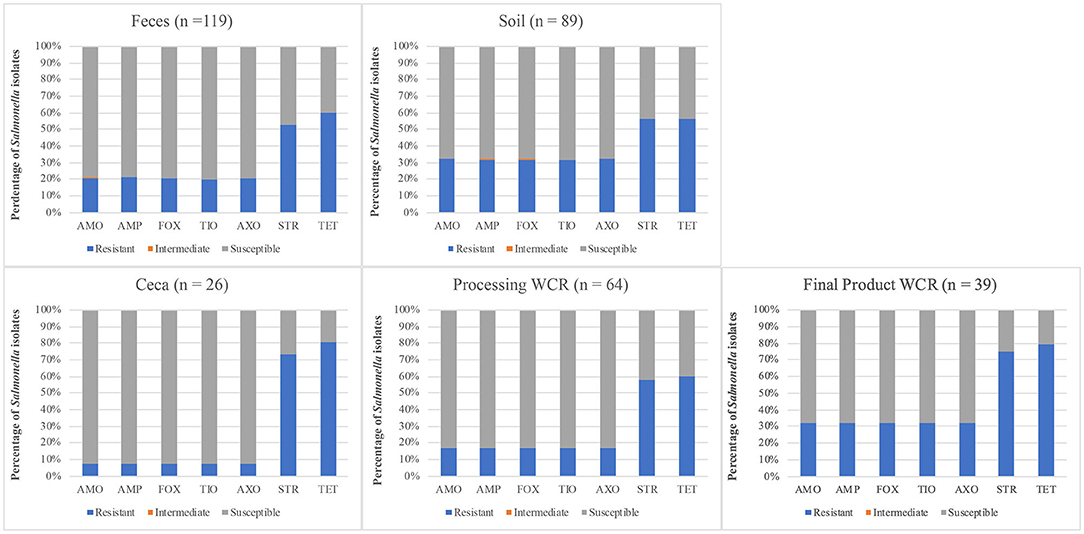
Figure 2. Distribution of antibiotic resistances using the CDC-NARMS method and breakpoints for Salmonella isolated from pastured poultry flocks based on the sample type from which the Salmonella was isolated. The antibiotics include amoxicillin-clavulanic acid (AMO), ampicillin (AMP), cefoxitin (FOX), ceftiofur (TIO), ceftriaxone (AXO), streptomycin (STR), and tetracycline (TET).
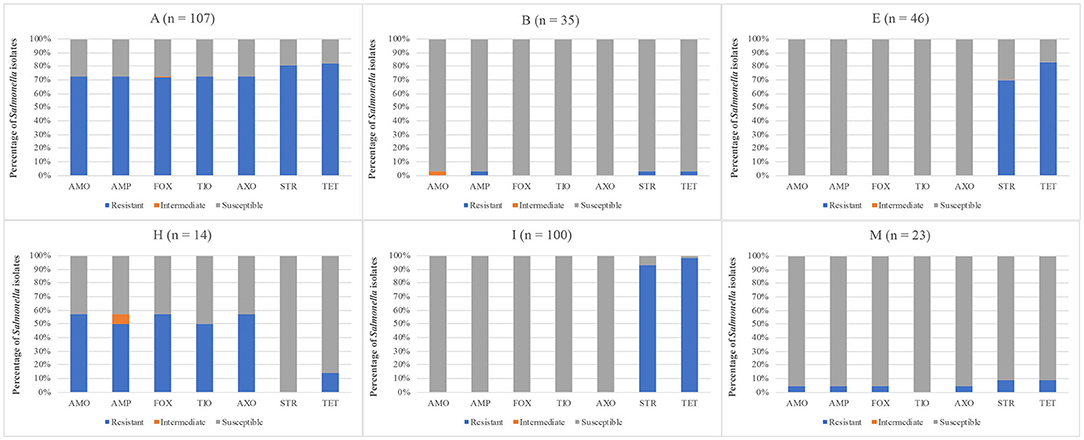
Figure 3. Distribution of antibiotic resistances using the CDC-NARMS method and breakpoints for Salmonella isolated from pastured poultry flocks based on the farm from which the Salmonella was isolated. The antibiotics include amoxicillin-clavulanic acid (AMO), ampicillin (AMP), cefoxitin (FOX), ceftiofur (TIO), ceftriaxone (AXO), streptomycin (STR), and tetracycline (TET).
Even within these antibiotic-free management systems, multidrug resistant (MDR) Salmonella, isolates exhibiting three of more resistances, were prevalent (Table 5). Overall, 27.3% (92/337) of the Salmonella isolates were considered MDR, with 82.6% (76/92) of the MDR isolates exhibiting resistances to seven antibiotics. This MDR rate is in line with what NARMS has found in conventional poultry samples from 2104 to 2017 (8.3–24.5%) (FDA, 2021). Aside from cecal samples (7.7%), the MDR rate across sample types was consistent (20.3–38.2%), and there were no significant differences between the MDR prevalences for any pairwise comparison, although cecal and soil prevalences were nearly significant (p = 0.0615). Conversely, when characterizing MDR Salmonella based on farm of origin, the MDR rates ranged from 0.0% (D, E, I, K) to 100% (C), although 87.0% (80/92) of the MDR Salmonella were recovered from farm A. It should be noted that in terms of serotypes of public health concern, of the 92 MDR Salmonella isolates from this study, 90 (97.8%) were Kentucky, with only a single MDR isolate representing a CDC top 32 serotype (Braenderup/Cholerasuis). Since Kentucky has been shown to be genetically more similar to common environmental E. coli than human pathogenic Salmonella serotypes (Morales et al., 2006), the results from this study demonstrate that Kentucky represents more of a biological vector to transfer MDR to the more pathogenic Salmonella serovars, than an actual threat to public health themselves.
While Salmonella was recovered from pre-harvest, post-harvest, and final product samples from these pastured poultry farms at prevalence rates similar to those seen from conventional poultry operations, only ~16% of all of the Salmonella isolates were serotypes of greatest concern to human health (CDC, 2013). The prevalence of these human health-related serotypes decreased along the farm-to-fork continuum (<3% of Salmonella recovered from the final product samples), indicating that the increased environmental exposure in these flocks did not result in a major health risk to the consumers of these products. Even though these pastured flocks were raised antibiotic-free, resistances to several antibiotic classes were expressed consistently among the Salmonella isolates, although >98% of the MDR isolates were serotyped as Kentucky. Only a single MDR Salmonella serotype of human health importance was isolated (~0.04% of all samples tested in this study), further indicating the relative safety of these pastured poultry products to the consumer. Additionally, these data suggest that Salmonella diversity is driven largely by the farm of origin, rather than the type of sample from which the Salmonella was isolated within the production cycle for that farm. Therefore, while general Salmonella intervention strategies may be successful in pastured poultry systems, due to the diversity in management options based on the need of and availability to these farmers, more targeted understanding of the environmental and management variables on these farms is vital.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material. Further inquiries regarding the data can be directed to the corresponding author/s.
Ethical review and approval was not required for the animal study because while we sampled from active pastured poultry farms, we did not manage the production or processing of the birds. We only collected the pre-harvest samples (soil, feces) after the broilers were moved on the pasture, and the post-harvest samples (ceca, whole carcass rinses) were collected after the farmer or processing facility culled the broilers. Written informed consent was obtained from the owners for the participation of their animals in this study.
MR designed the experiment, oversaw the processing of the samples, analyzed the results, and wrote the manuscript. JG performed the serotyping and edited the manuscript. AO assisted in the analysis of the antibiotic resistance data, performed the statistical analyses, and edited the manuscript. All authors contributed to the article and approved the submitted version.
These investigations were supported by the Agricultural Research Service, USDA CRIS Projects Genetic Analysis of Poultry-Associated Salmonella enterica to Identify and Characterize Properties and Markers Associated with Egg-Borne Transmission of Illness #6040-32000-007-00 and Molecular Approaches for the Characterization of Foodborne Pathogens in Poultry #6612-32000-059-00.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank Katelyn Griffin, Ariella Perry, Hellen Noh, Sarah Khorramzadeh, and Manju Amin for assistance in sample acquisition and pathogen isolation and Laura Lee Rutherford, Cheryl Gresham-Pearson, and Latoya Wiggins for performing the NARMS panels.
AR, antimicrobial resistant; ARG, antibiotic resistance genes; MDR, multidrug resistant; WCR, whole carcass rinse.
Alali, W. Q., Thakur, S., Berghaus, R. D., Martin, M. P., and Gebreyes, W. A. (2010). Prevalence and distribution of Salmonella in organic and conventional broiler poultry farms. Foodborne Pathog. Dis. 7, 1363–1371. doi: 10.1089/fpd.2010.0566
Alanis, A. J. (2005). Resistance to antibiotics: are we in the post-antibiotic era? Arch. Med. Res. 36, 697–705. doi: 10.1016/j.arcmed.2005.06.009
Bailey, J. S., and Cosby, D. E. (2005). Salmonella prevalence in free-range and certified organic chickens. J. Food Prot. 68, 2451–2453. doi: 10.4315/0362-028X-68.11.2451
Batz, M. B., Hoffmann, S., and Morris, J. G. Jr. (2012). Ranking the disease burden of 14 pathogens in food sources in the United States using attribution data from outbreak investigations and expert elicitation. J. Food Prot. 75, 1278–1291. doi: 10.4315/0362-028X.JFP-11-418
CDC (2013). An Atlas of Salmonella in the United States, 1968–2011. Available online at: https://www.cdc.gov/salmonella/pdf/salmonella-atlas-508c.pdf (accessed: June 15, 2021).
CDC (2015). National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS). Available online at: http://www.cdc.gov/narms
CDC (2018). National Enteric Disease Surviellance: Salmonella Annual Report 2016. Available online at: https://www.cdc.gov/nationalsurveillance/pdfs/2016-Salmonella-report-508.pdf (accessed: June 15, 2021).
CDC (2019). Outbreak of Multidrug-Resistant Salmonella Infections Linked to Raw Chicken Products. Available online at: https://www.cdc.gov/salmonella/infantis-10-18/index.html (accessed: June 15, 2021).
Clinical and Laboratory Standards Institute (2010). Performance Standards for Antimicrobial Susceptibility Testing; 20th Informational Supplement. CLSI M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA.
Crim, S. M., Iwamoto, M., Huang, J. Y., Griffin, P. M., Gilliss, D., Cronquist, A. B., et al. (2014). Incidence and trends of infection with pathogens transmitted commonly through food–Foodborne Diseases Active Surveillance Network, 10 U.S. sites, 2006-2013. MMWR Morb. Mortal. Wkly. Rep. 63, 328–332.
Cui, S., Ge, B., Zheng, J., and Meng, J. (2005). Prevalence and antimicrobial resistance of campylobacter spp. and Salmonella serovars in organic chickens from Maryland retail stores. Appl. Environ. Microbiol.71, 4108–4111. doi: 10.1128/AEM.71.7.4108-4111.2005
Davis, G. S., Waits, K., Nordstrom, L., Grande, H., Weaver, B., Papp, K., et al. (2018). Antibiotic-resistant Escherichia coli from retail poultry meat with different antibiotic use claims. BMC Microbiol. 18:174. doi: 10.1186/s12866-018-1322-5
Dimitri, C., and Oberholtzer, L. (2009). Marketing US organic foods: Recent Trends From Farms to Consumers. DIANE Publishing.
FDA (2021). NARMS Now. US Department of Health and Human Services. Available online at: https://www.fda.gov/animal-veterinary/national-antimicrobial-resistance-monitoring-system/narms-now-integrated-data (accessed: July 1, 2021).
Garcia, J. M., and Teixeira, P. (2017). Organic versus conventional food: a comparison regarding food safety. Food Rev. Int. 33, 424–446. doi: 10.1080/87559129.2016.1196490
Gould, L. H., Walsh, K. A., Vieira, A. R., Herman, K., Williams, I. T., Hall, A. J., et al. (2013). Surveillance for foodborne disease outbreaks - United States, 1998-2008. MMWR. Surveill. Summ. 62, 1–34.
Griggs, J. P., Bender, J. B., and Jacob, J. P. (2006). Microbial safety of chickens raised without antibiotics. J. Appl. Poult. Res. 15, 475–482. doi: 10.1093/japr/15.3.475
Gu, W., Vieira, A. R., Hoekstra, R. M., Griffin, P. M., and Cole, D. (2015). Use of random forest to estimate population attributable fractions from a case-control study of Salmonella enterica serotype Enteritidis infections. Epidemiol. Infect. 143, 2786–2794. doi: 10.1017/S095026881500014X
Guard, J., Sanchez-Ingunza, R., Morales, C., Stewart, T., Liljebjelke, K., Kessel, J., et al. (2012). Comparison of dkgB-linked intergenic sequence ribotyping to DNA microarray hybridization for assigning serotype to Salmonella enterica. FEMS Microbiol. Lett. 337, 61–72. doi: 10.1111/1574-6968.12010
Jacoby, G. A., and Munoz-Price, L. S. (2005). The new beta-lactamases. N. Engl. J. Med. 352, 380–391. doi: 10.1056/NEJMra041359
Kassem, I. I., Kehinde, O., Kumar, A., and Rajashekara, G. (2017). Antimicrobial-resistant Campylobacter in organically and conventionally raised layer chickens. Foodborne Pathog. Dis. 14, 29–34. doi: 10.1089/fpd.2016.2161
Lestari, S. I., Han, F., Wang, F., and Ge, B. (2009). Prevalence and antimicrobial resistance of Salmonella serovars in conventional and organic chickens from Louisiana retail stores. J. Food Prot. 72, 1165–1172. doi: 10.4315/0362-028X-72.6.1165
Lund, V. (2006). Natural living - a precondition for animal welfare in organic farming. Livest. Prod. Sci. 100, 71–83. doi: 10.1016/j.livprodsci.2005.08.005
Magossi, G., Cernicchiaro, N., Dritz, S., Houser, T., Woodworth, J., Jones, C., et al. (2019). Evaluation of Salmonella presence in selected United States feed mills. Microbiol. Open 8:e00711. doi: 10.1002/mbo3.711
Mataseje, L. F., Xiao, J., Kost, S., Ng, L. K., Doré, K., and Mulvey, M. R. (2009). Characterization of Canadian cefoxitin-resistant non-typhoidal Salmonella isolates, 2005-06. J. Antimicrob. Chemother. 64, 723–730. doi: 10.1093/jac/dkp249
Melendez, S. N., Hanning, I., Han, J., Nayak, R., Clement, A. R., Wooming, A., et al. (2010). Salmonella enterica isolates from pasture-raised poultry exhibit antimicrobial resistance and class I integrons. J. Appl. Microbiol. 109, 1957–1966. doi: 10.1111/j.1365-2672.2010.04825.x
Millman, J. M., Waits, K., Grande, H., Marks, A. R., Marks, J. C., Price, L. B., et al. (2013). Prevalence of antibiotic-resistant E. coli in retail chicken: comparing conventional, organic, kosher, and raised without antibiotics. F1000Res. 2:155. doi: 10.12688/f1000research.2-155.v2
Miriagou, V., Tassios, P. T., Legakis, N. J., and Tzouvelekis, L. S. (2004). Expanded-spectrum cephalosporin resistance in non-typhoid Salmonella. Int. J. Antimicrob. Agents 23, 547–555. doi: 10.1016/j.ijantimicag.2004.03.006
Morales, C. A., Gast, R., and Guard-Bouldin, J. (2006). Linkage of avian and reproductive tract tropism with sequence divergence adjacent to the 5S ribosomal subunit rrfH of Salmonella enterica. FEMS Microbiol. Lett. 264, 48–58. doi: 10.1111/j.1574-6968.2006.00432.x
Mughini-Gras, L., Enserink, R., Friesema, I., Heck, M., Van Duynhoven, Y., and Van Pelt, W. (2014). Risk factors for human salmonellosis originating from pigs, cattle, broiler chickens and egg laying hens: a combined case-control and source attribution analysis. PLoS ONE 9:e87933. doi: 10.1371/journal.pone.0087933
Peng, M., Salaheen, S., Almario, J. A., Tesfaye, B., Buchanan, R., and Biswas, D. (2016). Prevalence and antibiotic resistance pattern of Salmonella serovars in integrated crop-livestock farms and their products sold in local markets. Environ. Microbiol. 18, 1654–1665. doi: 10.1111/1462-2920.13265
Pesciaroli, M., Magistrali, C. F., Filippini, G., Epifanio, E. M., Lovito, C., Marchi, L., et al. (2020). Antibiotic-resistant commensal Escherichia coli are less frequently isolated from poultry raised using non-conventional management systems than from conventional broiler. Int. J. Food Microbiol. 314:108391. doi: 10.1016/j.ijfoodmicro.2019.108391
Pitesky, M., Charlton, B., Bland, M., and Rolfe, D. (2012). Surveillance of Salmonella Enteritidis in layer houses: a retrospective comparison of the Food and Drug Administration's Egg Safety Rule (2010–2011) and the California Egg Quality Assurance Program (2007–2011). Avian Dis. 57, 51–56, 56. doi: 10.1637/10281-061312-Reg.1
Popoff, M., and Le Minor, L. (1997). “Antigenic formulas of the Salmonella serovars,” in 7th World Health Organization Collaborating Centre for Reference and Research on Salmonella. Paris: Pasteur Institute.
Reisch, L., Eberle, U., and Lorek, S. (2013). Sustainable food consumption: an overview of contemporary issues and policies. Sustain. Sci. Pract. Pol. 9, 7–25. doi: 10.1080/15487733.2013.11908111
Rodriguez, A., Pangloli, P., Richards, H. A., Mount, J. R., and Draughon, F. A. (2006). Prevalence of Salmonella in diverse environmental farm samples. J. Food Prot. 69, 2576–2580. doi: 10.4315/0362-028X-69.11.2576
Rothrock, M. J Jr., Hiett, K. L., Guard, J. Y., and Jackson, C.R. (2016). Antibiotic resistance patterns of major zoonotic pathogens from all-natural, antibiotic-free, pasture-raised broiler flocks in the southeastern United States. J. Environ. Qual. 45, 593–603. doi: 10.2134/jeq2015.07.0366
Rothrock, M. J., Locatelli, A., Feye, K. M., Caudill, A. J., Guard, J., Hiett, K., et al. (2019b). A microbiomic analysis of a pasture-raised broiler flock elucidates foodborne pathogen ecology along the farm-to-fork continuum. Front Vet. Sci. 6:260. doi: 10.3389/fvets.2019.00260
Rothrock, M. J. Jr., Gibson, K. E., Micciche, A. C., and Ricke, S. C. (2019a). Pastured poultry production in the United States: strategies to balance system sustainability and environmental impact. Front. Sustain. Food. Sys. 3:74. doi: 10.3389/fsufs.2019.00074
Rovira, P., Mcallister, T., Lakin, S. M., Cook, S. R., Doster, E., Noyes, N. R., et al. (2019). Characterization of the microbial resistome in conventional and “raised without antibiotics Beef and dairy production systems. Front. Microbiol. 1010:1980. doi: 10.3389/fmicb.2019.01980
Sánchez-Casanova, R., Sarmiento-Franco, L., Phillips, C. J. C., and Zulkifli, I. (2020). Do free-range systems have potential to improve broiler welfare in the tropics? World's Poultry Sci. J. 76, 34–48. doi: 10.1080/00439339.2020.1707389
Sapkota, A. R., Hulet, R. M., Zhang, G., Mcdermott, P., Kinney, E. L., Schwab, K. J., et al. (2011). Lower prevalence of antibiotic-resistant Enterococci on U.S. conventional poultry farms that transitioned to organic practices. Environ. Health Perspect. 119, 1622–1628. doi: 10.1289/ehp.1003350
Scheinberg, J., Doores, S., and Cutter, C. N. (2013). A microbiological comparison of poultry products obtained from farmers' markets and supermarkets in Pennsylvania. J. Food Safety 33, 259–264. doi: 10.1111/jfs.12047
Shi, Z., Rothrock, M. J J.r, and Ricke, S.C. (2019). Applications of microbiome analyses in alternative poultry broiler production systems. Front. Vet. Sci. 6:157. doi: 10.3389/fvets.2019.00157
Siemon, C. E., Bahnson, P. B., and Gebreyes, W. A. (2007). Comparative investigation of prevalence and antimicrobial resistance of Salmonella between pasture and conventionally reared poultry. Avian Dis. 51, 112–117. doi: 10.1637/0005-2086(2007)051[0112:CIOPAA]2.0.CO;2
Silva, C., Calva, E., and Maloy, S. (2014). One health and food-borne disease: salmonella transmission between humans, animals, and plants. Microbiol Spectr 2, OH-0020–2013. doi: 10.1128/9781555818432.ch9
Smith, R., and Coast, J. (2013). The true cost of antimicrobial resistance. Br. Med. J. 346:fl493. doi: 10.1136/bmj.f1493
Sossidou, E. N., Dal Bosco, A., Elson, H. A., and Fontes, C. M. G. A. (2011). Pasture-based systems for poultry production: implications and perspectives. World's Poult. Sci. J. 67, 47–58. doi: 10.1017/S0043933911000043
Stock, I., and Wiedemann, B. (2000). Natural antibiotic susceptibility of Salmonella enterica strains. Int. J. Antimicrob. Agents 16, 211–217. doi: 10.1016/S.0924-8579(00)00204-1
USDA-FSIS (2020a). Sampling Results for FSIS Regulated Products. Washington, DC: US Department of Agriculture (USDA). Available online at: https://www.fsis.usda.gov/wps/portal/fsis/topics/data-collection-and-reports/microbiology/sampling-project-results (accessed May 11, 2020).
USDA-FSIS (2020b). Serotypes Profile of Salmonella Isolates from Meat and Poultry Productions January 1998 through December 2014. Available online at: https://www.fsis.usda.gov/sites/default/files/media_file/2020-10/Salmonella-Serotype-Annual-2014.pdf (accessed June 15, 2021).
Van Loo, E. J., Alali, W., and Ricke, S. C. (2012). Food safety and organic meats. Annu. Rev. Food Sci. Technol. 3, 203–225. doi: 10.1146/annurev-food-022811-101158
Van Loo, E. J., Caputo, V., Nayga, R. M. Jr, Meullenet, J.-F., and Ricke, S. C. (2011). Consumers' willingness to pay for organic chicken breast: evidence from choice experiment. Food Qual. Prefer. 22, 603–613. doi: 10.1016/j.foodqual.2011.02.003
Zhang, J. Y., Massow, A., Stanley, M., Papariella, M., Chen, X., Kraft, B., et al. (2011). Contamination rates and antimicrobial resistance in Enterococcus spp., Escherichia coli, and Salmonella isolated from “No Antibiotics Added”-labeled chicken products. Foodborne Pathog. Dis. 8, 1147–1152. doi: 10.1089/fpd.2011.0852
Keywords: Salmonella, farm-to-fork, pastured poultry, antibiotic resistance, serotyping
Citation: Rothrock MJ Jr, Guard JY and Oladeinde A (2021) Salmonella Diversity Along the Farm-to-Fork Continuum of Pastured Poultry Flocks in the Southeastern United States. Front. Anim. Sci. 2:761930. doi: 10.3389/fanim.2021.761930
Received: 20 August 2021; Accepted: 20 September 2021;
Published: 29 October 2021.
Edited by:
Rand Broadway, Agricultural Research Service (USDA), United StatesReviewed by:
Sara D. Lawhon, Texas A&M University, United StatesCopyright © 2021 Rothrock, Guard and Oladeinde. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael J. Rothrock Jr., bWljaGFlbC5yb3Rocm9ja0B1c2RhLmdvdg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.