- 1Department of Animal and Poultry Sciences, Virginia Tech, Blacksburg, VA, United States
- 2Virginia Bioinformatics Institute, Virginia Tech, Blacksburg, VA, United States
- 3Department of Animal Sciences, Purdue University, West Lafayette, IN, United States
The ratio of concentrate to forage within diets is known to alter rumen microbial profiles, but comparatively less information is available on the effect of differing sources of individual nutrients on the microbiome. The objective of this study was to investigate rumen microbial responses to diets composed of protein and fiber sources expected to vary in nutrient degradability. The responses of interest included relative abundances of bacterial taxa as well as estimations of community richness and diversity. Ten ruminally cannulated wethers (Suffolk, Dorset, or Suffolk × Dorset) received four diet treatments consisting of either beet pulp or timothy hay and soybean meal (SBM) or heat-treated soybean meal (HSBM) in a partially replicated 4 × 4 Latin square experiment for 21 days. Timothy hay and beet pulp were expected to provide differing rumen degradabilities of neutral detergent fiber (NDF) while the soybean meals were expected to provide differing rumen degradabilities of crude protein (CP). Solid and liquid samples of rumen contents were collected for microbial DNA isolation and Next-Generation sequencing. Numerous rumen bacterial population shifts were observed due to change in fiber source, with increased abundances (P < 0.05) of fibrolytic populations associated with timothy hay diets compared with beet pulp diets. Conversely, populations of the pectin-degrading genera, Treponema and Lachnospira, increased on the beet pulp treatment (P = 0.015 and P = 0.0049, respectively). Limited impact on bacterial taxa was observed between diets differing in protein source. The Paraprevotellaceae genus YRC22 was observed to increase in abundance on HSBM diets (P = 0.023) and the phylum Spirochaetes tended to be more abundant on SBM than HSBM diets (P = 0.071). Beet pulp decreased rumen bacterial diversity (P = 0.0027) and tended to decrease bacterial species richness (P = 0.051) compared to timothy hay. Our results serve to further underscore the sensitivity of rumen microbes to changes in their preferred substrates, particularly of those associated with fiber degradation.
Introduction
The rumen ecosystem is composed of diverse and dynamic populations of microorganisms, including bacteria, fungi, and protozoa, that are responsible for the vast majority of feed digestion by the host animal (Hungate et al., 1964; Krause et al., 2013; Jewell et al., 2015). The energy harvested from volatile fatty acids (VFA) produced through microbial fermentation of plant material is estimated at 70% of the total metabolic energy used by the ruminant (Bergman, 1990). In addition, 60–85% of amino acids reaching the small intestine are supplied by rumen microbial protein synthesis (Storm et al., 1983). The rumen microbiome therefore serves as an important intermediate between diet and animal performance (Mullins et al., 2013; Gleason and White, 2018).
High-throughput or Next-Generation sequencing technologies, such as Illumina, have propelled investigations of the rumen microbiome forward with their ability to rapidly and economically sequence the bacterial 16S rRNA gene. Along with bioinformatics techniques and pipelines, Next-Generation sequencing has allowed for detailed characterization of the microbiome, which has greatly enhanced our understanding of this complex ecosystem (Lima et al., 2015). Common targets of investigation include relative abundances of the major bacterial taxa, in addition to estimates of species richness and diversity. Variations in these measurements have been linked to variations in livestock production traits, including milk yield and components (Indugu et al., 2017; Wu et al., 2021), average daily gain (Min et al., 2019), and feed efficiency (McLoughlin et al., 2020).
Previous research has demonstrated that the microbial community can be influenced by diet forage-to-concentrate ratio (de Menezes et al., 2011; Carberry et al., 2012; Petri et al., 2013) and by alterations in amounts of specific concentrate ingredients (Callaway et al., 2010; Petri et al., 2014). However, little information on the effects of differing nutrient degradabilities is currently available. Therefore, our objective was to examine how the rumen microbiome responds to supplies of protein and fiber sources possessing differing degradability profiles of crude protein (CP) and neutral detergent fiber (NDF), respectively.
Materials and Methods
Animals, Experimental Design, and Treatments
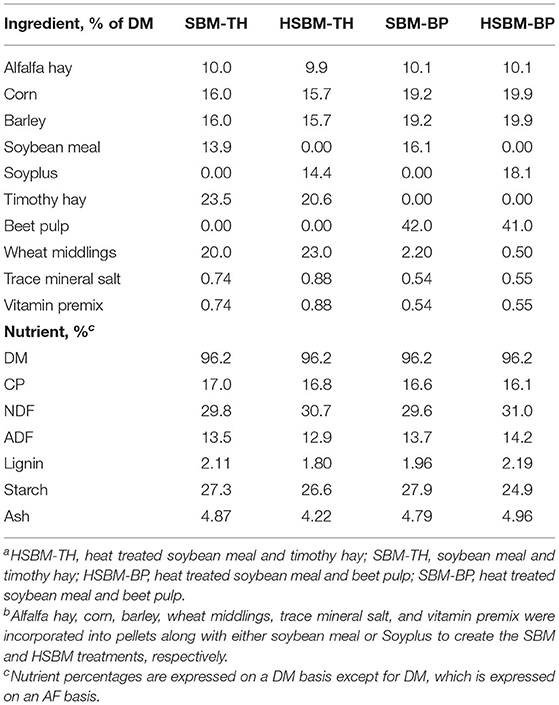
All animal use and procedures were approved by the Virginia Tech Institutional Animal Care and Use Committee (Protocol #18-096). Ten ruminally cannulated commercial wethers (Suffolk, Dorset, or Suffolk × Dorset) were housed in individual stalls at the Smithfield Farm, Virginia Tech, Blacksburg, VA. All wethers were consuming a basic forage diet prior to study enrollment. Wethers were ~1.5 years of age and weighed an average of 62.1 ± 6.6 kg at trial commencement. Wethers were assigned to treatments in a partially replicated 4 × 4 Latin square so that treatment groups were balanced for initial body weight. Treatments were assigned using a 2 × 2 factorial approach and included feedstuffs intended to supply varying rumen degradabilities of CP and NDF. Soybean meal (SBM) and heat-treated soybean meal (HSBM) were utilized as the CP sources with high and low rumen degradabilities, respectively (NRC, 2016). These meals were pelleted with alfalfa, corn, barley, wheat middlings, trace mineral salt, and a sheep vitamin premix (Table 1). Pelleted beet pulp (BP) and long timothy hay (TH) represented the NDF sources, with BP expected to undergo a faster rate of fiber degradation compared to TH (Van Soest et al., 1991; DePeters et al., 1997). Diets were prepared by combining the appropriate protein pellet with the appropriate fiber source to create the 4 treatments: highly degradable CP plus lowly degradable NDF (SBM-TH), highly degradable CP plus highly degradable NDF (SBM-BP), lowly degradable CP plus lowly degradable NDF (HSBM-TH), and lowly degradable CP plus highly degradable NDF (HSBM-BP). Each animal consumed each of the 4 diets for 21 days. Gradual diet adaptation occurred during the first 3 days (with animals receiving 25, 50, and 75% of their ration as the new diet on each day, respectively) and 100% of the diet treatment was offered for the remaining 18 days. Upon the completion of a period, diet adaptation for the next period began on the following day. Animals were fed once daily at 0800 h. Clean, fresh water was available at all times.
Sample Collection and DNA Isolation
Fluid and solid samples of rumen contents from all animals were collected at 0730 h on day 16 of each period. The remaining days of the period through d 21 were devoted to investigations into rumen fermentation responses (Gleason et al., unpublished). The early timing of rumen sampling in relation to feeding was chosen so as to not interfere with other experimental activities that were occurring around the same time (ruminal infusions and bolusing, etc., intended for the separate publication mentioned). Contents were collected via the cannula from the dorsal sac after slight hand mixing and strained through 1 layer of gauze to separate the fluid and solid fractions. Samples were stored in cryovials at −80°C until total DNA extraction. Extraction was performed on 0.5 g of a liquid or solid sample using a QIAamp DNA stool minikit (Qiagen, Valencia, CA) following the manufacturer's protocol. Sample DNA concentrations and 260/280 ratios were obtained using a spectrophotometer (Epoch2 Microplate Reader, Biotek, Winooski, VT). Extracted DNA was stored at −80°C until further processing.
PCR Amplification, Next-Generation Sequencing, and Bioinformatic Analysis
The universal primers 515F and 926R were chosen to amplify the V4–V5 region of the 16S rRNA gene following the Earth Microbiome Project protocol (https://www.earthmicrobiome.org/). The PCR reaction mixture included 13.0 μL of PCR-grade water, 1.0 μL of template DNA, 10 μM of each primer, and 10.0 μL of 5PRIME HotMasterMix (2 × ) (Quantabio, Beverly, MA). Samples were amplified in duplicate under the following thermocycler conditions: 94°C for 3 min for initial denaturing, then 35 cycles of 94°C for 45 s, 50°C for 60 s, and 72°C for 90 s. A final elongation step occurred at 72°C for 10 min followed by a hold at 4°C. After pooling duplicates, all amplicons were visualized on a 2% agarose gel and quantitated on a Qubit fluorometer (Fisher Scientific, Hampton, NH). Normalization was performed based on Qubit results, and amplicons were purified on a Pippin Prep (Sage Science, Beverly, MA) targeting a 520 bp range. Quantitative PCR was then run on the pool. Amplicons were loaded at 9.5 pM and sequenced using the MiSeq v3 600-cycle kit on the Miseq platform (Illumina, Inc., San Diego, CA). All sequences were submitted to the NCBI Sequence Read Archive (accession# PRJNA753122). Sequence analyses were conducted using the QIIME 2 bioinformatics platform version 2019.10 (Bolyen et al., 2019) following the protocol of Estaki et al. (2020). Sequence quality filtering and denoising was performed using Deblur (Amir et al., 2017). Sequences were then clustered into OTUs and taxonomic identities assigned at a 97% identity cut-off using a naive Bayes classifier with the Greengenes database as a reference (DeSantis et al., 2006). Relative abundances of taxa were obtained by dividing the reads assigned to a given taxon by the total number of reads present. Richness and diversity estimates were obtained using the Core Diversity command.
Statistical Analysis
Results were analyzed using the nlme package (Pinheiro et al., 2020) in R version 3.6.1 (R Core Team, 2019). Response variables included percent relative abundances of bacterial taxa at the phylum, family, and genus levels, number of observed OTUs, and Shannon diversity index value. Response variables were analyzed using the linear model:
where μ represents the overall mean, αi is the effect of the ith CP source, βj is the effect of the jth NDF source, αβij is the interaction of CP source i and NDF source j, ck represents the random effect of animal k, dl represents the random effect of period l, and eijkl is the residual error associated with CP source i, NDF source j, animal k, and period l. Compound Symmetry, Unstructured, and 1st Order Autoregressive residual error variance structures were compared for each response variable and the Akaike information criterion (AIC) was used to assess model quality (Hurvich and Tsai, 1989). The model with the lowest AIC value was subjected to ANOVA and least square means were determined. Statistical significance was considered when P < 0.05 and a tendency considered when 0.05 ≤ P < 0.10.
Results
Demultiplexed sequence counts totaled 14,851,796 reads with a median length of 300 nucleotides and an average of 198,024 sequences per sample. The predominant bacterial phylum identified was Bacteroidetes, ranging between 73.1 and 78.5% relative abundance across treatment means in rumen fluid (Table 2) and between 52.4 and 74.5% in rumen solids (Table 3). Other phyla representing at least 1% mean relative abundance included Firmicutes, Fibrobacteres, Proteobacteria, Spirochaetes, Tenericutes, and Cyanobacteria in both fractions. Populations of Fibrobacteres and Spirochaetes were considerably more pronounced in the solid fraction than in fluid (Figures 1A,B), which is consistent with previous observations (de Menezes et al., 2011). Their representatives at the family level (Figures 2A,B) and genus level (Figures 3A,B) also reflected these differences. A number of taxonomic lines were found to display variation in response to the different diet treatments. These belonged largely to the predominant phyla identified, including Bacteroidetes, Firmicutes, Spirochaetes, Tenericutes, and Verrucomicrobia. Increases in relative abundance were mainly associated with the TH treatment and decreases with the BP treatment, with few exceptions.
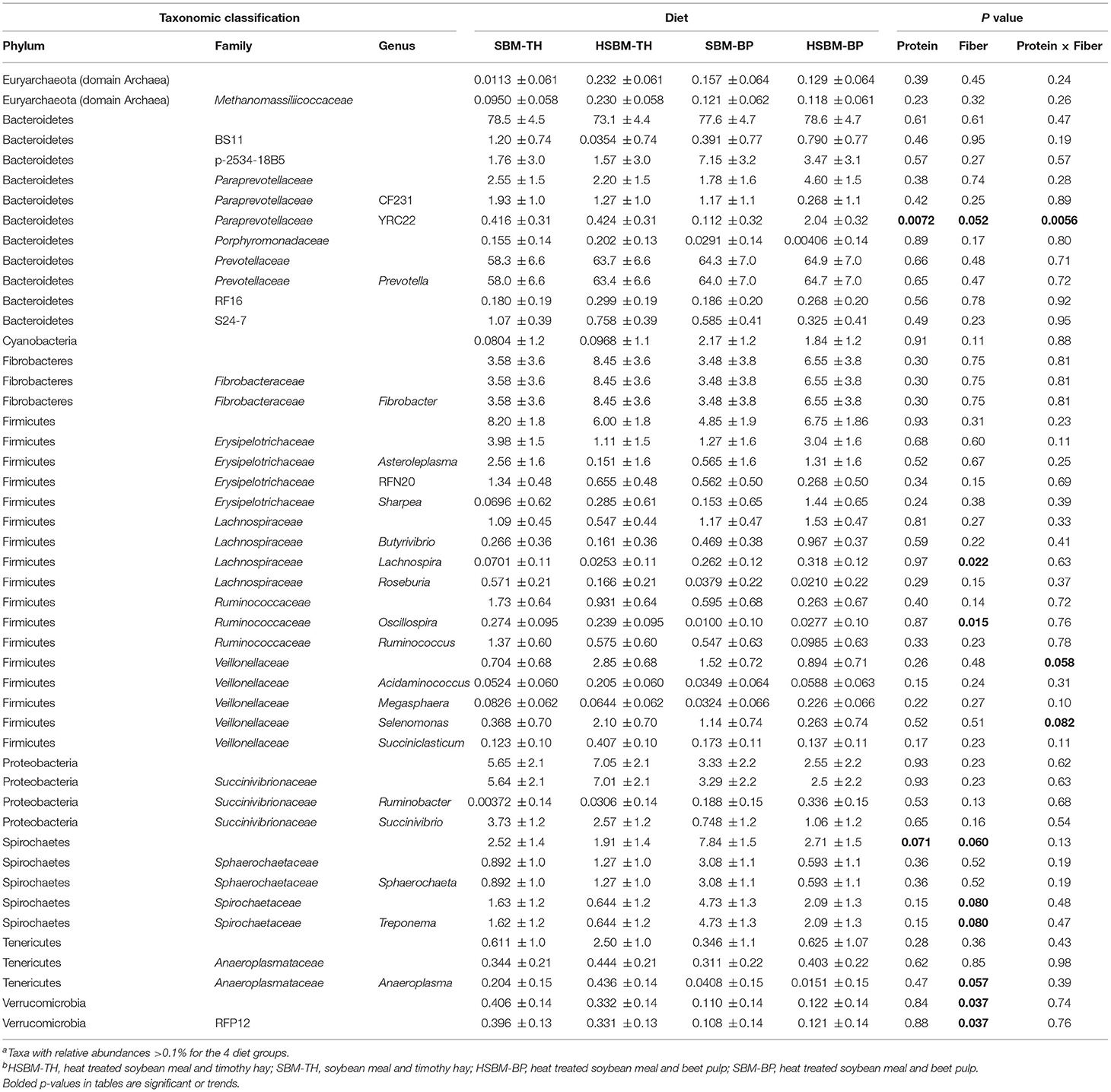
Table 2. LS means ± SE for percent relative abundances of bacterial taxa identified in the rumen fluid fraction as differentiated by diet along with P values for the effects of protein source, fiber source, and the interaction of protein and fiber sourcea,b.
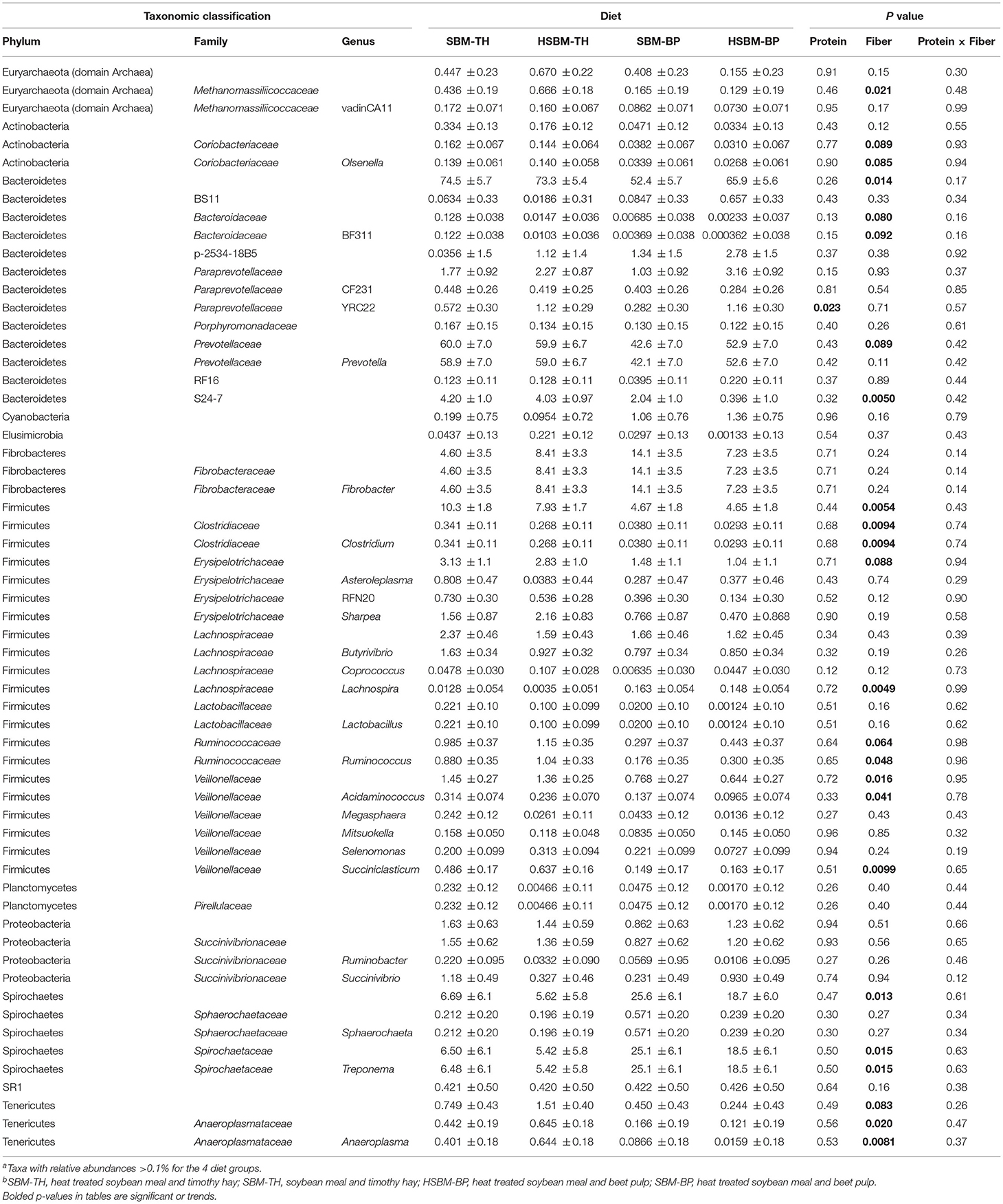
Table 3. LS means ± SE for percent relative abundances of bacterial taxa identified in the rumen solid fraction as differentiated by diet along with P values for the effects of protein source, fiber source, and the interaction of protein and fiber sourcea,b.
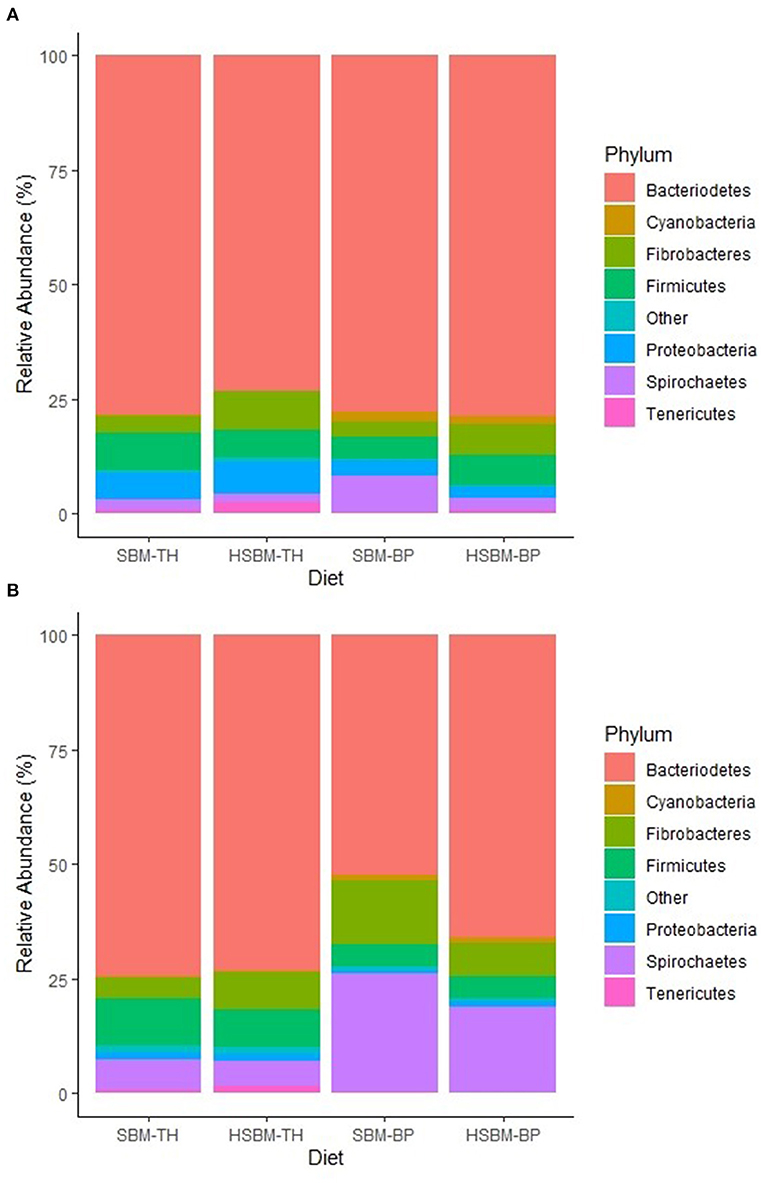
Figure 1. (A) Phylum-level composition of bacteria identified in the rumen fluid fraction as differentiated by diet. The category “Other” is comprised of taxa with relative abundances < 1.0% for all diets. (B) Phylum-level composition of bacteria identified in the rumen solid fraction as differentiated by diet. The category “Other” is comprised of taxa with relative abundances < 1.0% for all diets.
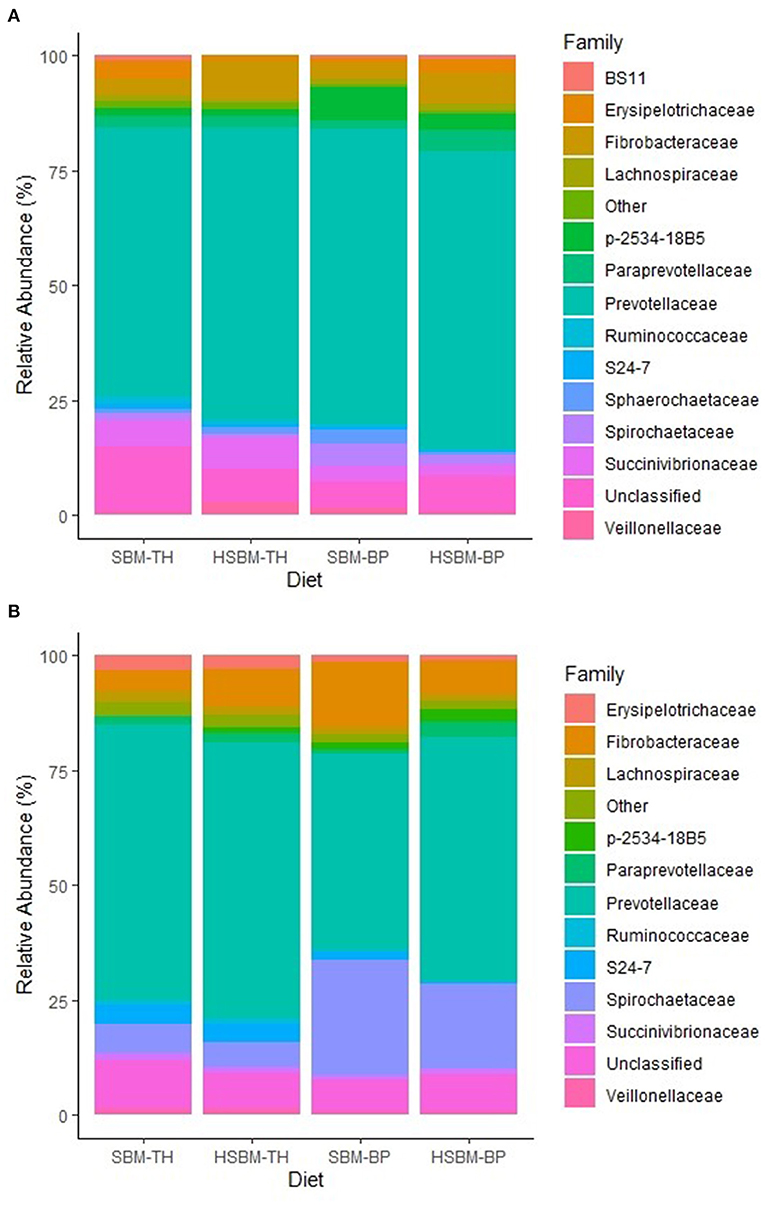
Figure 2. (A) Family-level composition of bacteria identified in the rumen fluid fraction as differentiated by diet. The category “Other” is comprised of taxa with relative abundances < 1.0% for all diets. (B) Family-level composition of bacteria identified in the rumen solid fraction as differentiated by diet. The category “Other” is comprised of taxa with relative abundances < 1.0% for all diets.
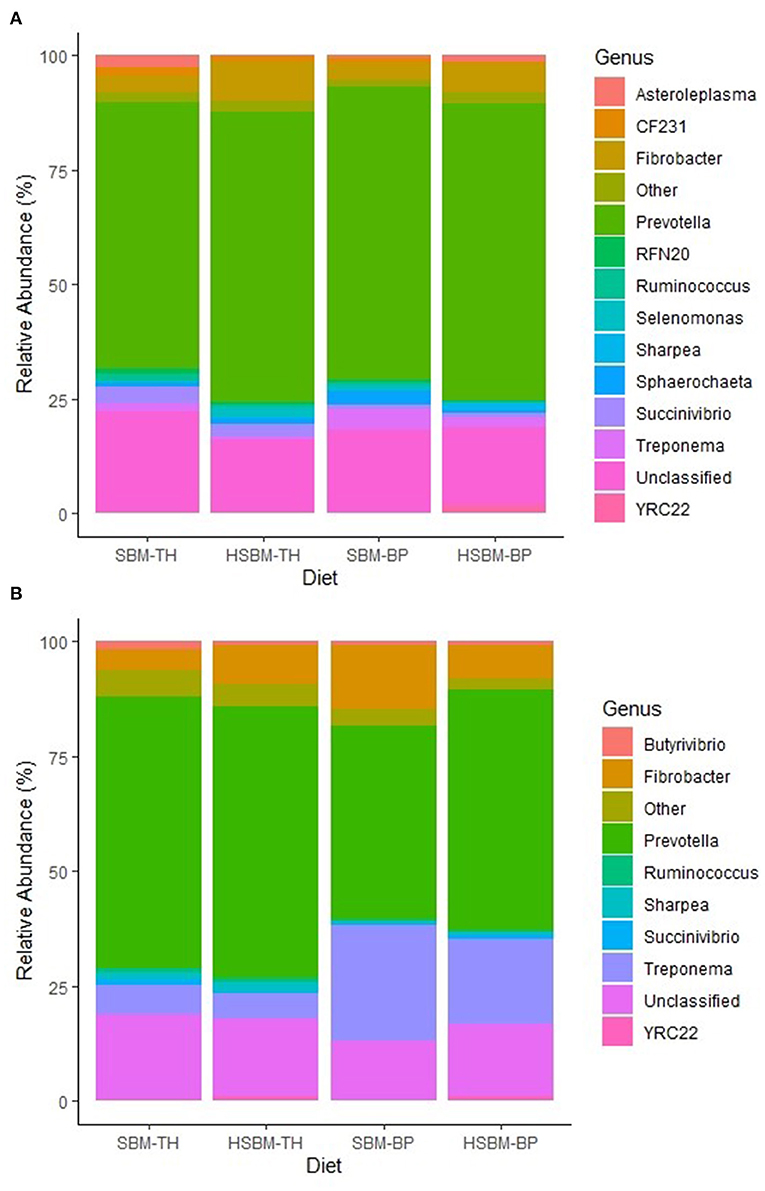
Figure 3. (A) Genus-level composition of bacteria identified in the rumen fluid fraction as differentiated by diet. The category “Other” is comprised of taxa with relative abundances < 1.0% for all diets. (B) Genus-level composition of bacteria identified in the rumen solid fraction as differentiated by diet. The category “Other” is comprised of taxa with relative abundances < 1.0% for all diets.
Fiber Effects on Bacterial Composition
Bacteroidetes and its family S24-7 increased (P = 0.014 and P = 0.0050, respectively; Table 3) in animals receiving the TH treatment compared to those on the BP treatment. Two additional Bacteroidetes families, Bacteroidaceae and Prevotellaceae, tended to be more prominent in response to the TH treatment (P = 0.089 and P = 0.080, respectively; Table 3) as well as two genera, BF311 (P = 0.092; Table 3) and YRC22 (P = 0.052; Table 2). Similarly, Firmicutes was more abundant on the TH treatment than on BP (P = 0.0054; Table 3), as were its families Clostridiaceae and Veillonellaceae (P = 0.0094 and P = 0.016, respectively; Table 3). Erysipelotrichaceae and Ruminococcaceae populations tended to be less abundant on BP as well (P = 0.088 and P = 0.064, respectively; Table 3). The Firmicutes genera Acidaminococcus, Clostridium, Oscillospira, Ruminococcus, and Succiniclasticum decreased in abundance (P < 0.05; Tables 2, 3) in response to BP treatment. Population decreases associated with BP were also observed in Verrucomicrobia and its family RFP12 (P = 0.037 and P = 0.037, respectively; Table 2) and in the Tenericutes family Anaeroplasmataceae and its genus Anaeroplasma (P = 0.020 and P = 0.0081, respectively; Table 3). Tenericutes itself, along with the family Coriobacteriaceae, and Coriobacteriaceae's genus Olsenella tended to be less abundant on BP compared to the TH treatment (P = 0.083, P = 0.089, and P = 0.085, respectively; Table 3). Methanomassiliicoccaceae also exhibited a population decline when BP diets were fed (P = 0.021; Table 3). The phylum Spirochaetes increased in abundance on the BP treatment compared to TH (P = 0.013; Table 3). The family Spirochaetaceae and genus Treponema reflected this effect as well (P = 0.015). The Firmicutes genus Lachnospira was also observed to increase on the BP treatment (P = 0.0049; Table 3).
Protein Effects on Bacterial Composition
Few changes in bacterial community composition were observed when protein source was varied, contrasting with the numerous population shifts associated with differences in fiber source. The phylum Spirochaetes tended to increase on SBM compared to HSBM (P = 0.071; Table 2). The Paraprevotellaceae genus YRC22 was observed to increase in abundance on HSBM diets, and this effect was consistent both in the fluid fraction (P = 0.0072, Table 2) and solid fraction (P = 0.023; Table 3).
Richness and Diversity Responses
Richness and diversity of the rumen microbiome varied with fiber source, but not protein source. Shannon diversity of the solid fraction samples was greater in animals consuming TH compared to BP (P = 0.0027; Table 4). The number of OTUs identified tended to increase on TH diets compared to BP diets (P = 0.051; Table 4).
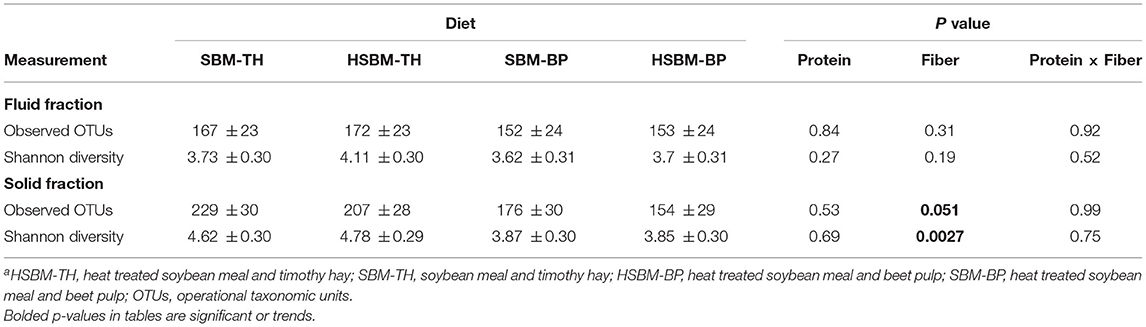
Table 4. LS means ± SE for alpha diversity metrics of the bacterial community identified in the rumen fluid and solid fractions as differentiated by diet along with P values for the effects of protein source, fiber source, and the interaction of protein and fiber sourcea.
Discussion
Bacterial Population Shifts Associated With Fiber Source
The majority of microbial variation due to fiber source was identified in the solid ruminal fraction compared to the fluid fraction. This was unsurprising given that fibrolytic activity would be most prominent in the fiber mat of the rumen. Fibrolytic activities are abundant within the Bacteroidetes phylum (Terrapon et al., 2015). Because Bacteroidetes is generally the most prominent phylum in the rumen and cellulose is the main energy source of ruminants (Ensminger et al., 1990), it is logical that this bacterial group would be sensitive to changes in the fiber substrate provided. The responses to alterations in fiber source demonstrated by Bacteroidetes, its families S24-7, Bacteroidaceae and Prevotellaceae, and its genera BF311 and YRC22 that we observed are consistent with this logic. Bacteroidaceae, Prevotellaceae and S24-7 are known to be involved in digestion of fiber (Lan et al., 2006; Ivarsson et al., 2014; Gamage et al., 2018). Even though the Bacteroidaceae genus BF311's substrate preferences have yet to be fully characterized (Bi et al., 2018), observations of increased abundance in goats when hay was fed also support an involvement in forage fiber degradation (Zhang et al., 2018). The same experiment reported similar behavior of YRC22 (Zhang et al., 2018). Our observations of fiber effects on Bacteroidetes and its family S24-7, as well as tendencies for Prevotellaceae, Bacteroidaceae, BF311, and YRC22 to be affected by fiber source may reflect their known (or probable) functional roles and indicate sensitivity to changes in their substrate.
Like Bacteroidetes, the Firmicutes phylum was more abundant on the TH treatment than on BP and the population shifts that we observed in its members are logical given what is currently known about their substrate preferences. Ruminococcus and Clostridium, both Firmicutes genera, have been established as prominent fiber digesters in the rumen (Preston and Leng, 1987; Chesson and Forsberg, 1997). Oscillospira and Succiniclasticum are also cellulolytic and have been shown to increase as the dietary forage-to-concentrate ratio increases (Mackie et al., 2003; Han et al., 2019). Additionally, fiber has been shown to stimulate Acidaminococcus populations in vitro (Gong et al., 2019). Although we only observed a tendency of the family Erysipelotrichaceae to increase on TH, previous research has demonstrated greater prominence of this family in cattle consuming hay as a forage source compared to corn silage or grass silage (Deusch et al., 2017). Our results serve to further underscore the sensitivity of these taxa to changes in their fiber substrate.
Our observations of the responses of Actinobacteria, Tenericutes, and Verrucomicrobia members to changes in fiber source parallel similar findings in the literature. The Verrucomicrobia family RFP12 has been shown to decrease in abundance as dietary forage content is decreased (Plaizier et al., 2017). Anaeroplasma, a Tenericutes genus, has been correlated with fiber digestion (Niu et al., 2015), and Olsenella, an Actinobacteria genus, has been suggested to participate prominently in the degradation of ryegrass (Huws et al., 2016). Our results indicate that these taxa are more strongly associated with the degradation of cellulose and hemicellulose in forages than with the degradation of soluble fiber in beet pulp. The behavior of Methanomassiliicoccaceae in response to forage or fiber content appears to be less clear. The sole Archaeal family in our report, Methanomassiliicoccaceae has been reported to decrease in abundance when dietary forage level was reduced (Zhu et al., 2018), but has also been observed to increase as forage fiber was replaced with wheat bran and soy hulls as non-forage fiber sources (Wang et al., 2018), in direct contradiction to our findings. Additional work is necessary to fully characterize the effects of forage fiber vs. non-forage fiber on Methanomassiliicoccaceae.
Contrasting with the previous taxa mentioned above, the phylum Spirochaetes was approximately 3 to 4 times more abundant in animals on the BP treatment compared to those receiving TH. The effect was demonstrated by its family Spirochaetaceae and genus Treponema as well. The Firmicutes genus Lachnospira was the only other taxonomic group displaying a population increase in association with the BP treatment rather than a decrease. This response is most likely due to the high pectin content of beet pulp (Codling and Woodman, 1929). Treponema and Lachnospira are both known to be involved in pectin degradation (Wojciechowicz and Ziołecki, 1979; Kasperowicz, 1994). Previous research has also demonstrated increased abundances of these genera in response to stimulation by pectin (Liu et al., 2015; Bang et al., 2018), and according to Liu et al. (2015), ruminal Treponema populations were “remarkably supported” by the provision of pectin. The positive responses we observed in these taxa provide further evidence of their sensitivity to pectin content and broadly underscore the significance of substrate availability in influencing microbial population shifts.
Numerous investigations have evaluated the effects of hay inclusion on the rumen microbial community within the context of comparing different forage-to-concentrate ratios (Fernando et al., 2010; Hook et al., 2011; Klevenhusen et al., 2017; Zhang et al., 2017). The recurring theme throughout these studies is that fibrolytic bacterial populations increase when forage content is increased, in agreement with our observations. Comparatively less research attention, however, has been dedicated to rumen microbial changes on beet pulp-based diets and to our knowledge, no studies have utilized BP as a replacement for forage in the diet and compared differences in the microbial profiles. Studies have instead focused on using BP as a high-fiber/low-starch energy source to replace starchy concentrate feeds (Zhao et al., 2013; Münnich et al., 2018). Nonetheless, these studies have still observed reductions in fiber-degrading bacterial populations in response to BP treatment, which is consistent with our findings (Zhao et al., 2013; Münnich et al., 2018). These results suggest that the optimal type of fiber substrate likely differs between various bacterial populations involved in its breakdown and that fibrolytic species may not be supported as adequately by non-forage fiber sources. This highlights the need for future ruminant nutrition work to move away from broad classification of nutrients and toward characterizing specific substrate supplies.
Bacterial Population Shifts Associated With Protein Source
In contrast to the microbial responses associated with fiber type, very little microbial variation was evident when protein source was varied. Major taxa associated with ruminal protein degradation, including Butyrivibrio, Prevotella, and Ruminobacter (Mackie et al., 2001; McDonald et al., 2011) displayed similar relative abundances across diet treatments, appearing to indicate no sensitivity to the provision of heat-treated vs. non-treated SBM. The sole population shift that we observed was that of the Paraprevotellaceae genus YRC22, which increased in abundance on the HSBM treatment compared to SBM. A tendency for the Spirochaetes phylum to increase in abundance on SBM was also noted. These responses may be evidence of involvement in protein metabolism. Some members of Spirochaetes have been observed to produce enzymes with roles in peptide degradation, including trypsin, chymotrypsin, and arylamidases (Nordhoff et al., 2005; Newbrook et al., 2017). Because our observed shift in Spirochaetes was quite minimal, however, more research is necessary to clarify the sensitivity of this phylum to changes in protein source. YRC22 has been described as a minor genus with relatively unknown function (Zhu et al., 2018). Previous research has identified a strong correlation between YRC22 abundance and concentrations of certain amino acids in rumen fluid, leading investigators to conclude this genus may be involved in their metabolism to some degree (Hua et al., 2017). The sensitivity of YRC22 to changes in protein source that we observed provides additional evidence of a potential link with the processing or utilization of protein and, along with its response to our fiber treatments, underscores the need for further investigation of this genus.
As explained above, our protein treatments were represented by SBM and heated SBM. Treating SBM with heat leads to denaturation of proteins, which may decrease their solubility and slow their ruminal degradation rate (Russell et al., 1992). Decreased ruminal availability of SBM protein due to heat treatment is well documented in both cattle (Plegge et al., 1985; Ljøkjel et al., 2000; Borucki Castro et al., 2007) and sheep (Glimp et al., 1967; Demjanec et al., 1995), but very little research has investigated the impacts on rumen microbial populations. Plegge et al. (1985) reported that heat treatment of SBM did not impact rumen bacterial protein synthesis efficiency or bacterial nitrogen flow, but no information of the effects on the microbial populations themselves was available. Investigators evaluating the substitution of SBM with dried distillers grains with solubles (DDGS) observed no rumen bacterial population changes in cattle (Castillo-Lopez et al., 2017) or sheep (Shen et al., 2020). Although this protein source substitution differed from our experiment, it bears the similarity that DDGS possesses a higher proportion of rumen-undegradable protein (RUP) to rumen-degradable protein (RDP) compared to SBM (NRC, 2016). The results of these studies and ours indicate that the rumen microbiome is resistant to change in terms of population dynamics due to heat treating SBM or substituting it with another protein feed. Additionally, the high amount of CP present in our diets made it unlikely that the animals were deficient in RDP. This provides another reason for the lack of sensitivity to protein source that we observed.
Overall, our investigation was unable to detect widespread change in bacterial population dynamics due to protein source. A similar observation was made by Gong et al. (2019), who reported that gut microbial growth patterns displayed a weaker correlation with protein than with carbohydrates. Aside from the likely absence of RDP-deficient conditions in our study, the limited overall effect of protein source on the rumen microbial community compared to fiber source may be due to the fact that plant fiber is the main nutrient source that the rumen microbiome has evolved to process and that vast numbers of rumen bacterial taxa are directly involved in fibrolytic activities (Hobson and Stewart, 1997). Additionally, rumen bacteria have the capability of utilizing recycled urea as a source of nitrogen for synthesizing their own microbial proteins (McDonald et al., 2011). It is therefore possible that the microbiome is comparatively less sensitive to changes in dietary protein solubility than it is to variations in fiber characteristics.
Microbial Richness, Diversity, and Links With Productive Efficiency
While unchanged by protein source, Shannon diversity of the microbiome was observed to decrease in response to the BP treatment compared to the TH treatment. Similarly, the number of identified OTUs tended to be lower on the BP treatment. The number of OTUs is an indication of species richness of the microbiome whereas Shannon diversity is a measure of species representation. Previous reports have found that richness and diversity may be sensitive to diet forage-to-concentrate ratio, with richness and diversity greatest on forage diets and lowest at high levels of concentrate feeding (Petri et al., 2013; Zhang et al., 2017). Our results indicate that the type of fiber source, rather than just fiber concentrations, may also influence these metrics because we balanced our experimental diets to have similar concentrations of NDF. The decline in Shannon diversity and the tendency for the number of OTUs to decline with BP treatment also appears to mirror the bacterial population decreases associated with the BP diets discussed above.
Lower levels of richness and diversity have been linked with increased feed efficiency and reduced methane production in cattle (Shabat et al., 2016). According to Shabat et al. (2016), efficient rumen microbiomes are composed of a smaller number of more dominant taxa that utilize a narrower range of metabolic pathways and produce metabolites that are more closely aligned with the energetic needs of the host. Less efficient microbiomes can be characterized by more diverse populations that employ a wide range of metabolic pathways, some of them resulting in greater energy loss, such as through methane production (Shabat et al., 2016). Methane represents wasted dietary energy for the animal as it cannot be absorbed and also contributes to agricultural greenhouse gas emissions (Johnson and Ward, 1996). The reduction in abundance of the methanogen family Methanomassiliicoccaceae that we observed may be further evidence that substituting TH with BP could potentially shift the microbiome in a more energetically efficient direction. This family is poorly characterized, however, and additional research is necessary to clarify its contribution to ruminant methane production (Huang et al., 2016). Two bacterial taxa that were also observed to decrease in abundance on BP diets, Clostridium and Succiniclasticum, have been previously linked with poor feed efficiency in growing steers (Hernandez-Sanabria et al., 2012; Myer et al., 2015). Because we utilized mature wethers in our experiment and measuring emissions was outside our study scope, further work will be needed to confirm if the taxonomic shifts we observed would result in improved growth efficiency and lower methane production.
Conclusions
In this study, we sought to explore rumen microbial responses to differing dietary sources of fiber and protein and, in doing so, further the understanding of the complex relationship between the rumen microbiome and diet. The responses we observed indicate that nutrient source, rather than simply nutrient concentration, exerts a marked impact on bacterial population dynamics in the ovine rumen microbiome. Additional research is required to determine if the differing nutrient sources utilized in our study and their subsequent influence on the rumen microbiome may translate into biologically and economically relevant alterations in livestock performance.
Data Availability Statement
The datasets generated for this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: PRJNA753122).
Ethics Statement
The animal study was reviewed and approved by Virginia Tech Institutional Animal Care and Use Committee.
Author Contributions
CG was responsible for animal monitoring, sample collection, data analysis, and manuscript preparation. RS conducted all bioinformatics analyses. LB assisted with experiment execution and contributed to manuscript editing. RW served as the study's principal investigator, securing necessary funds and determining study design. RW also contributed substantially to manuscript editing. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by funding from the U.S. Department of Agriculture—National Institute of Food and Agriculture (2017-05943).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Amir, A., McDonald, D., Navas-Molina, J. A., Kopylova, E., Morton, J. T., Zech Xu, Z., et al. (2017). Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2, e00191–e00116. doi: 10.1128/mSystems.00191-16
Bang, S.-J., Kim, G., Lim, M. Y., Song, J.-E., Jung, H. D., Kum, S. J., et al. (2018). The influence of in vitro pectin fermentation on the human fecal microbiome. AMB Exp. 8:98. doi: 10.1186/s13568-018-0629-9
Bergman, E. N. (1990). Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 70, 567–590. doi: 10.1152/physrev.1990.70.2.567
Bi, Y., Zeng, S., Zhang, R., Diao, Q., and Tu, Y. (2018). Effects of dietary energy levels on rumen bacterial community composition in Holstein heifers under the same forage to concentrate ratio condition. BMC Microbiol. 18:69. doi: 10.1186/s12866-018-1213-9
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Borucki Castro, S. I., Phillip, L. E., Lapierre, H., Jardon, P. W., and Berthiaume, R. (2007). Ruminal degradability and intestinal digestibility of protein and amino acids in treated soybean meal products. J. Dairy Sci. 90, 810–822. doi: 10.3168/jds.S0022-0302(07)71565-5
Callaway, T. R., Dowd, S. E., Edrington, T. S., Anderson, R. C., Krueger, N., Bauer, N., et al. (2010). Evaluation of bacterial diversity in the rumen and feces of cattle fed different levels of dried distillers grains plus solubles using bacterial tag-encoded FLX amplicon pyrosequencing. J. Anim. Sci. 88, 3977–3983. doi: 10.2527/jas.2010-2900
Carberry, C. A., Kenny, D. A., Han, S., McCabe, M. S., and Waters, S. M. (2012). Effect of phenotypic residual feed intake and dietary forage content on the rumen microbial community of beef cattle. Appl. Environ. Microbiol. 78, 4949–4958. doi: 10.1128/AEM.07759-11
Castillo-Lopez, E., Jenkins, C. J. R., Aluthge, N. D., Tom, W., Kononoff, P. J., et al. (2017). The effect of regular or reduced-fat distillers grains with solubles on rumen methanogenesis and the rumen bacterial community. J. Appl. Microbiol. 123, 1381–1395. doi: 10.1111/jam.13583
Chesson, A., and Forsberg, C. W. (1997). “Polysaccharide degradation by rumen microorganisms,” in The Rumen Microbial Ecosystem (London: Chapman and Hall), 329–381. doi: 10.1007/978-94-009-1453-7_8
Codling, A. J., and Woodman, H. E. (1929). Sugar-beet pulp as a source of pectin. J. Agric. Sci. 19, 701–714. doi: 10.1017/S0021859600013411
de Menezes, A. B., Lewis, E., O'Donovan, M., O'Neill, B. F., Clipson, N., and Doyle, E. M. (2011). Microbiome analysis of dairy cows fed pasture or total mixed ration diets. FEMS Microbiol. Ecol. 78, 256–265. doi: 10.1111/j.1574-6941.2011.01151.x
Demjanec, B., Merchen, N. R., Cremin, J. D., Aldrich, C. G., and Berger, L. L. (1995). Effect of roasting on site and extent of digestion of soybean meal by sheep: I. Digestion of nitrogen and amino acids. J. Anim. Sci. 73, 824–834. doi: 10.2527/1995.733824x
DePeters, E. J., Fadel, J. G., and Arosemena, A. (1997). Digestion kinetics of neutral detergent fiber and chemical composition within some selected by-product feedstuffs. Anim. Feed Sci. Technol. 67, 127–140. doi: 10.1016/0377-8401(96)01145-5
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/AEM.03006-05
Deusch, S., Camarinha-Silva, A., Conrad, J., Beifuss, U., Rodehutscord, M., and Seifert, J. (2017). A structural and functional elucidation of the rumen microbiome influenced by various diets and microenvironments. Front. Microbiol. 8:1605. doi: 10.3389/fmicb.2017.01605
Ensminger, M. E., Oldfield, J. E., and Heinemann, W. W. (1990). Feeds and Nutrition. 2nd ed. Clovis, CA: The Ensminger Publishing Company.
Estaki, M., Jiang, L., Bokulich, N. A., McDonald, D., González, A., Kosciolek, T., et al. (2020). QIIME 2 enables comprehensive end-to-end analysis of diverse microbiome data and comparative studies with publicly available data. Curr. Protoc. Bioinform. 70:e100. doi: 10.1002/cpbi.100
Fernando, S. C., Purvis, H. T., Najar, F. Z., Sukharnikov, L. O., Krehbiel, C. R., Nagaraja, T. G., et al. (2010). Rumen microbial population dynamics during adaptation to a high-grain diet. Appl. Environ. Microbiol. 76, 7482–7490. doi: 10.1128/AEM.00388-10
Gamage, H. K. A. H., Tetu, S. G., Chong, R. W. W., Bucio-Noble, D., Rosewarne, C. P., et al. (2018). Fiber supplements derived from sugarcane stem, wheat dextrin and Psyllium husk have different in vitro effects on the human gut microbiota. Front. Microbiol. 9:1618. doi: 10.3389/fmicb.2018.01618
Gleason, C. B., and White, R. R. (2018). Variation in animal performance explained by the rumen microbiome or by diet composition. J. Anim. Sci. 96, 4658–4673. doi: 10.1093/jas/sky332
Glimp, H. A., Karr, M. R., Little, C. O., Woolfolk, P. G., Mitchell, G. E., and Hudson, L. W. (1967). Effect of reducing soybean protein solubility by dry heat on the protein utilization of young lambs. J. Anim. Sci. 26, 858–861. doi: 10.2527/jas1967.264858x
Gong, L., Chi, H., Wang, J., Zhang, H., and Sun, B. (2019). In vitro fermentabilities of whole wheat as compared with refined wheat in different cultivars. J. Funct. Foods. 52, 505–515. doi: 10.1016/j.jff.2018.11.027
Han, X., Li, B., Wang, X., Chen, Y., and Yang, Y. (2019). Effect of dietary concentrate to forage ratios on ruminal bacterial and anaerobic fungal populations of cashmere goats. Anaerobe 59, 118–125. doi: 10.1016/j.anaerobe.2019.06.010
Hernandez-Sanabria, E., Goonewardene, L. A., Wang, Z., Durunna, O. N., and Moore, S. S. (2012). Impact of feed efficiency and diet on adaptive variations in the bacterial community in the rumen fluid of cattle. Appl. Environ. Microbiol. 78, 1203–1214. doi: 10.1128/AEM.05114-11
Hobson, P. N., and Stewart, C. S. (1997). The Rumen Microbial Ecosystem. London: Chapman and Hall. doi: 10.1007/978-94-009-1453-7
Hook, S. E., Steele, M. A., Northwood, K. S., Dijkstra, J., France, J., Wright, A. D. G., et al. (2011). Impact of subacute ruminal acidosis (SARA) adaptation and recovery on the density and diversity of bacteria in the rumen of dairy cows. FEMS Microbiol. Ecol. 78, 275–284. doi: 10.1111/j.1574-6941.2011.01154.x
Hua, C., Tian, J., Tian, P., Cong, R., Luo, Y., Geng, Y., et al. (2017). Feeding a high concentration diet induces unhealthy alterations in the composition and metabolism of ruminal microbiota and host response in a goat model. Front. Microbiol. 8:138. doi: 10.3389/fmicb.2017.00138
Huang, X. D., Martinez-Fernandez, G., Padmanabha, J., Long, R., Denman, S. E., and McSweeney, C. S. (2016). Methanogen diversity in indigenous and introduced ruminant species on the Tibetan Plateau. Archaea. 2016, 1–10. doi: 10.1155/2016/5916067
Hungate, R. E., Bryant, M. P., and Mah, R. A. (1964). The rumen bacteria and protozoa. Annu. Rev. Microbiol. 18, 131–166. doi: 10.1146/annurev.mi.18.100164.001023
Hurvich, C. M., and Tsai, C.-L. (1989). Regression and time series model selection in small samples. Biometrika 76, 297–307. doi: 10.1093/biomet/76.2.297
Huws, S. A., Edwards, J. E., Creevey, C. J., Rees Stevens, P., Lin, W., Girdwood, S. E., et al. (2016). Temporal dynamics of the metabolically active rumen bacteria colonizing fresh perennial ryegrass. FEMS Microbiol. Ecol. 92:fiv137. doi: 10.1093/femsec/fiv137
Indugu, N., Vecchiarelli, B., Baker, L. D., Ferguson, J. D., Vanamala, J. K., Pitta, D. W., et al. (2017). Comparison of rumen bacterial communities in dairy herds of different production. BMC Microbiol. 17:190. doi: 10.1186/s12866-017-1098-z
Ivarsson, E., Roos, S., Liu, H. Y., and Lindberg, J. E. (2014). Fermentable non-starch polysaccharides increases the abundance of Bacteroides–Prevotella–Porphyromonas in ileal microbial community of growing pigs. Animal 8, 1777–1787. doi: 10.1017/S1751731114001827
Jewell, K. A., McCormick, C. A., Odt, C. L., Weimer, P. J., and Suen, G. (2015). Ruminal bacterial community composition in dairy cows is dynamic over the course of two lactations and correlates with feed efficiency. Appl. Environ. Microbiol. 81, 4697–4710. doi: 10.1128/AEM.00720-15
Johnson, D. E., and Ward, G. M. (1996). Estimates of animal methane emissions. Environ. Monit. Assess. 42, 133–141. doi: 10.1007/BF00394046
Kasperowicz, A. (1994). Comparison of utilization of pectins from various sources by pure cultures of pectinolytic rumen bacteria and mixed cultures of rumen microorganisms. Acta Microbiol. Pol. 43, 47–56.
Klevenhusen, F., Petri, R. M., Kleefisch, M.-T., Khiaosa-ard, R., Metzler-Zebeli, B. U., and Zebeli, Q. (2017). Changes in fibre-adherent and fluid-associated microbial communities and fermentation profiles in the rumen of cattle fed diets differing in hay quality and concentrate amount. FEMS Microbiol. Ecol. 93. doi: 10.1093/femsec/fix100
Krause, D. O., Nagaraja, T. G., Wright, A. D. G., and Callaway, T. R. (2013). Board-invited review: Rumen microbiology: leading the way in microbial ecology. J. Anim. Sci. 91, 331–341. doi: 10.2527/jas.2012-5567
Lan, N. P. T., Sakamoto, M., Sakata, S., and Benno, Y. (2006). Bacteroides barnesiae sp. nov., Bacteroides salanitronis sp. nov. and Bacteroides gallinarum sp. nov., isolated from chicken caecum. Int. J. Syst. Evol. Microbiol. 56, 2853–2859. doi: 10.1099/ijs.0.64517-0
Lima, F. S., Oikonomou, G., Lima, S. F., Bicalho, M. L., Ganda, E. K., de Oliveira Filho, J. C., et al. (2015). Prepartum and postpartum rumen fluid microbiomes: characterization and correlation with production traits in dairy cows. Appl. Environ. Microbiol. 81, 1327–1337. doi: 10.1128/AEM.03138-14
Liu, J., Pu, Y.-Y., Xie, Q., Wang, J.-K., and Liu, J.-X. (2015). Pectin induces an in vitro rumen microbial population shift attributed to the pectinolytic Treponema group. Curr. Microbiol. 70, 67–74. doi: 10.1007/s00284-014-0672-y
Ljøkjel, K., Harstad, O. M., and Skrede, A. (2000). Effect of heat treatment of soybean meal and fish meal on amino acid digestibility in mink and dairy cows. Anim. Feed Sci. Technol. 84, 83–95. doi: 10.1016/S0377-8401(00)00104-8
Mackie, R. I., Aminov, R. I., Hu, W., Klieve, A. V., Ouwerkerk, D., Sundset, M. A., et al. (2003). Ecology of uncultivated Oscillospira species in the rumen of cattle, sheep, and reindeer as assessed by microscopy and molecular approaches. Appl. Environ. Microbiol. 69, 6808–6815. doi: 10.1128/AEM.69.11.6808-6815.2003
Mackie, R. I., McSweeney, C. S., and Aminov, R. I. (2001). “Rumen,” in Encyclopedia of Life Sciences, ed John Wiley and Sons, Ltd. (Chichester: John Wiley and Sons, Ltd), a0000404. doi: 10.1038/npg.els.0000404
McDonald, P., Greenhalgh, J. F. D., Morgan, C. A., Edwards, R., and Sinclair, L. (2011). Animal Nutrition. 7th edn. New York, NY: Pearson.
McLoughlin, S., Spillane, C., Claffey, N., Smith, P. E., O'Rourke, T., Diskin, M. G., et al. (2020). Rumen microbiome composition is altered in sheep divergent in feed efficiency. Front. Microbiol. 11:1981. doi: 10.3389/fmicb.2020.01981
Min, B. R., Gurung, N., Shange, R., and Solaiman, S. (2019). Potential role of rumen microbiota in altering average daily gain and feed efficiency in meat goats fed simple and mixed pastures using bacterial tag-encoded FLX amplicon pyrosequencing. J. Anim. Sci. 97:3523–3534. doi: 10.1093/jas/skz193
Mullins, C. R., Mamedova, L. K., Carpenter, A. J., Ying, Y., Allen, M. S., Yoon, I., et al. (2013). Analysis of rumen microbial populations in lactating dairy cattle fed diets varying in carbohydrate profiles and Saccharomyces cerevisiae fermentation product. J. Dairy Sci. 96, 5872–5881. doi: 10.3168/jds.2013-6775
Münnich, M., Khol-Parisini, A., Klevenhusen, F., Metzler-Zebeli, B. U., and Zebeli, Q. (2018). Graded replacement of maize grain with molassed sugar beet pulp modulated ruminal microbial community and fermentation profile in vitro: sugar beet pulp, rumen microbiota and fermentation in vitro. J. Sci. Food Agric. 98, 991–997. doi: 10.1002/jsfa.8547
Myer, P. R., Smith, T. P., Wells, J. E., Kuehn, L. A., and Freetly, H. C. (2015). Rumen microbiome from steers differing in feed efficiency. PLoS ONE 10:e0129174. doi: 10.1371/journal.pone.0129174
Newbrook, K., Staton, G. J., Clegg, S. R., Birtles, R. J., Carter, S. D., and Evans, N. J. (2017). Treponema ruminis sp. nov., a spirochaete isolated from the bovine rumen. Int. J. Syst. Evol. Microbiol. 67, 1349–1354. doi: 10.1099/ijsem.0.001812
Niu, Q., Li, P., Hao, S., Zhang, Y., Kim, S. W., Li, H., et al. (2015). Dynamic distribution of the gut microbiota and the relationship with apparent crude fiber digestibility and growth stages in pigs. Sci. Rep. 5:9938. doi: 10.1038/srep09938
Nordhoff, M., Taras, D., Macha, M., Tedin, K., Busse, H.-J., and Wieler, L. H. (2005). Treponema berlinense sp. nov. and Treponema porcinum sp. nov., novel spirochaetes isolated from porcine faeces. Int. J. Syst. Evol. Microbiol. 55, 1675–1680. doi: 10.1099/ijs.0.63388-0
NRC (2016). Nutrient Requirements of Beef Cattle. 8th edn. Washington, DC: National Academies Press.
Petri, R. M., Mapiye, C., Dugan, M. E., and McAllister, T. A. (2014). Subcutaneous adipose fatty acid profiles and related rumen bacterial populations of steers fed red clover or grass hay diets containing flax or sunflower-seed. PLoS ONE 9:e104167. doi: 10.1371/journal.pone.0104167
Petri, R. M., Schwaiger, T., Penner, G. B., Beauchemin, K. A., Forster, R. J., McKinnon, J. J., et al. (2013). Characterization of the core rumen microbiome in cattle during transition from forage to concentrate as well as during and after an acidotic challenge. PLoS ONE 8:e83424. doi: 10.1371/journal.pone.0083424
Pinheiro, J., Bates, D., DebRoy, S., Sarkar, D., and R Core Team. (2020). nlme: Linear and Nonlinear Mixed Effects Models. Vienna: R Foundation for Statistical Computing. Available online at: https://CRAN.R-project.org/package=nlme (accessed October 01, 2020).
Plaizier, J. C., Li, S., Danscher, A. M., Derakshani, H., Andersen, P. H., and Khafipour, E. (2017). Changes in microbiota in rumen digesta and feces due to a grain-based subacute ruminal acidosis (SARA) challenge. Microb. Ecol. 74, 485–495. doi: 10.1007/s00248-017-0940-z
Plegge, S. D., Berger, L. L., and Fahey, G. C. (1985). Effect of roasting temperature on the proportion of soybean meal nitrogen escaping degradation in the rumen. J. Anim. Sci. 61, 1211–1218. doi: 10.2527/jas1985.6151211x
Preston, T. R., and Leng, R. A. (1987). Matching Ruminant Production Systems with Available Resources in the Tropics and Sub-tropics. Armidale, N.S.W: Penambul Books.
R Core Team (2019). A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing.
Russell, J. B., O'Connor, J. D., Fox, D. G., Van Soest, P. J., and Sniffen, C. J. (1992). A net carbohydrate and protein system for evaluating cattle diets: I. Ruminal fermentation. J. Anim. Sci. 70, 3551–3561. doi: 10.2527/1992.70113551x
Shabat, B. S. K., Sasson, G., Doron-Faigenboim, A., Durman, T., Yaacoby, S., Berg Miller, M. E., et al. (2016). Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 10, 2958–2972. doi: 10.1038/ismej.2016.62
Shen, J., Li, Z., Yu, Z., and Zhu, W. (2020). Effects of dietary replacement of soybean meal with dried distillers grains with solubles on the microbiota occupying different ecological niches in the rumen of growing Hu lambs. J. Anim. Sci. Biotechnol. 11:93. doi: 10.1186/s40104-020-00499-2
Storm, E., Ørskov, E. R., and Smart, R. (1983). The nutritive value of rumen micro-organisms in ruminants: 2. The apparent digestibility and net utilization of microbial N for growing lambs. Br. J. Nutr. 50, 471–478. doi: 10.1079/BJN19830114
Terrapon, N., Lombard, V., Gilbert, H. J., and Henrissat, B. (2015). Automatic prediction of polysaccharide utilization loci in Bacteroidetes species. Bioinformatics 31, 647–655. doi: 10.1093/bioinformatics/btu716
Van Soest, P. J., Robertson, J. B., and Lewis, B. A. (1991). Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74, 3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2
Wang, K., Nan, X., Chu, K., Tong, J., Yang, L., Zheng, S., et al. (2018). Shifts of hydrogen metabolism from methanogenesis to propionate production in response to replacement of forage fiber with non-forage fiber sources in diets in vitro. Front. Microbiol. 9:2764. doi: 10.3389/fmicb.2018.02764
Wojciechowicz, M., and Ziołecki, A. (1979). Pectinolytic enzymes of large rumen treponemes. Appl. Environ. Microbiol. 37, 136–142. doi: 10.1128/aem.37.1.136-142.1979
Wu, X., Huang, S., Huang, J., Peng, P., Liu, Y., Han, B., et al. (2021). Identification of the potential role of the rumen microbiome in milk protein and fat synthesis in dairy cows using metagenomic sequencing. Animals. 11:1247. doi: 10.3390/ani11051247
Zhang, J., Shi, H., Wang, Y., Li, S., Cao, Z., Ji, S., et al. (2017). Effect of dietary forage to concentrate ratios on dynamic profile changes and interactions of ruminal microbiota and metabolites in Holstein heifers. Front. Microbiol. 8:2206. doi: 10.3389/fmicb.2017.02206
Zhang, R. Y., Jin, W., Feng, P. F., Liu, J. H., and Mao, S. Y. (2018). High-grain diet feeding altered the composition and functions of the rumen bacterial community and caused the damage to the laminar tissues of goats. Animal 12, 2511–2520. doi: 10.1017/S175173111800040X
Zhao, X. H., Liu, C. J., Liu, Y., Li, C. Y., and Yao, J. H. (2013). Effects of replacing dietary starch with neutral detergent-soluble fibre on ruminal fermentation, microbial synthesis and populations of ruminal cellulolytic bacteria using the rumen simulation technique (RUSITEC). J. Anim. Physiol. Anim. Nutr. 97, 1161–1169. doi: 10.1111/jpn.12025
Keywords: microbiome, next-generation sequencing, nutrients, rumen, sheep
Citation: Gleason CB, Settlage RE, Beckett LM and White RR (2021) Characterizing Effects of Ingredients Differing in Ruminally Degradable Protein and Fiber Supplies on the Ovine Rumen Microbiome Using Next-Generation Sequencing. Front. Anim. Sci. 2:745848. doi: 10.3389/fanim.2021.745848
Received: 22 July 2021; Accepted: 03 September 2021;
Published: 08 October 2021.
Edited by:
James Levi Klotz, Agricultural Research Service (USDA), United StatesReviewed by:
Suban Foiklang, Maejo University, ThailandAhmed Elolimy, University of Arkansas for Medical Sciences, United States
Copyright © 2021 Gleason, Settlage, Beckett and White. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robin R. White, cnJ3aGl0ZUB2dC5lZHU=
 Claire B. Gleason
Claire B. Gleason Robert E. Settlage
Robert E. Settlage Linda M. Beckett
Linda M. Beckett Robin R. White
Robin R. White