
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Anim. Sci., 16 September 2021
Sec. Animal Physiology and Management
Volume 2 - 2021 | https://doi.org/10.3389/fanim.2021.650347
 Kaixuan Guo1,2†
Kaixuan Guo1,2† Yue Zhao1,2†
Yue Zhao1,2† Luqing Cui1,2
Luqing Cui1,2 Zhengzheng Cao1,2
Zhengzheng Cao1,2 Fan Zhang1,2
Fan Zhang1,2 Xiangru Wang1
Xiangru Wang1 Jiawei Feng1,2
Jiawei Feng1,2 Menghong Dai1,2*
Menghong Dai1,2*Bacterial resistance is a complex scientific issue. To manage this issue, we need to deeply understand the influencing factors and mechanisms. Based on the background of livestock husbandry, this paper reviews the factors that affect the acquisition of bacterial resistance. Meanwhile, the resistance mechanism is also discussed. “Survival of the fittest” is the result of genetic plasticity of bacterial pathogens, which brings about specific response, such as producing adaptive mutation, gaining genetic material or changing gene expression. To a large extent, bacterial populations acquire resistance genes directly caused by the selective pressure of antibiotics. However, mobile resistance genes may be co-selected by other existing substances (such as heavy metals and biocides) without direct selection pressure from antibiotics. This is because the same mobile genetic elements as antibiotic resistance genes can be co-located by the resistance determinants of some of these compounds. Furthermore, environmental factors are a source of resistance gene acquisition. Here, we describe some of the key measures that should be taken to mitigate the risk of antibiotic resistance. We call on the relevant governments or organizations around the world to formulate and improve the monitoring policies of antibiotic resistance, strengthen the supervision, strengthen the international cooperation and exchange, and curb the emergence and spread of drug-resistant strains.
The emergence and spread of bacterial pathogen resistance is a natural response to the selection pressure of antibiotics (Witte et al., 1999). The latest report shows that the total global sales of antimicrobial agents for livestock in 2017 was 93,309 tons, and sales are expected to increase by 11.5% to 1,04,079 tons in 2030 (Tiseo et al., 2020). The application of antibiotics in animals has three main purposes: the treatment of pathogen infection (therapeutic use), the prevention of animal disease outbreak (prevention) and the promotion of growth (Gould, 2016; Roth et al., 2019). Antibiotic resistant bacteria associated with animals are easily transmitted to humans through the food chain, and spread in the environment through animal feces (Van Boeckel et al., 2015; Manyi-Loh et al., 2018). Therefore, when animals are infected by bacterial pathogens, most countries advocate antimicrobial treatment. This treatment can effectively control the spread of disease and reduce the excretion of pathogens from animals. In addition to treating infections, antibiotics are also used in animals in many developing countries and a few developed countries as a preventive measure to prevent outbreaks of disease, even if the animals are healthy and there are no known infectious diseases in the feeding environment (Prescott, 2017; Collineau et al., 2018). Finally, the use of antibiotics as growth promoters (AGPs) in animal husbandry in developing countries has been controversial. For example, in the review (Sharma et al., 2016), the studies reported the types of antibiotics registered as feed additives for livestock (Hobman and Crossman, 2015) and poultry in the United States, Canada, the European Union and Australia to promote their healthy growth. After antibiotics are fed to farm animals in low or sub-dose concentrations, 75–90% will be lost to the environment in the form of urine and feces (Lekshmi et al., 2017). But the concentration of sub-therapeutic antibiotics in some tissues or organs (such as intestines) will lead to non-specific mutation of bacteria, which may promote the development of bacterial resistance (Cogliani et al., 2011). In addition, the low concentrations of heavy metals in the intestines and feces are often in close contact with bacteria. Usually, some heavy metals are mixed in feed as nutritional additives, because they may be necessary elements to maintain various physiological functions of animal body (Hejna et al., 2018). However, some heavy metals with no established biological functions are considered as pollutants. In recent years, bacteria have developed drug resistance under the influence of heavy metals (especially copper and zinc). Different from the short half-life and biodegradability of antibiotics, heavy metals are not biodegradable in various environments with relatively low concentrations (lower than their minimal inhibitory concentration), and their effects are extensive and lasting (Luo et al., 2014; Waseem et al., 2014; Gumpu et al., 2015; Zhao et al., 2015). All in all, microorganisms can be subjected to long-term selective pressure by metals. Some data show that there is a significant correlation between heavy metal resistance genes and antibiotic and disinfectant resistance genes in food borne Salmonella (Yang et al., 2016; Zhang et al., 2016; Deng et al., 2018). In livestock husbandry, biocides are widely used in environmental surface because of their disinfection effect and the treatment of wounds. Biocides used in diverse applications eventually enter the environment. There is no doubt that fungicides exert selective pressure on microorganisms, even at low concentrations (Ons et al., 2020). The mechanisms leading to resistant bacterial populations from antibiotic uses, heavy metals uses, and exposure to biocides have all gained a widespread attention. Furthermore, relevant experiments have proved that bacteria can be induced or selected to produce adaptability when they are exposed to these non-antibiotic antibiotics, thereby reducing sensitivity to one or more antibiotics (Wales and Davies, 2015). For example, after exposure to phenolic fungicides or sub-inhibitory concentrations of triclosan, the frequency of mutations in antibiotic resistance of Salmonella increases (Randall et al., 2004). Pathogens will also be transmitted through certain ways, such as, from animals to humans through food chain (Gupta et al., 2020; Lei et al., 2020). Additionally, bacteria can also acquire exogenous resistance genes through other factors in the environment, such as flies (Zhao et al., 2019). The sources of selective pressure and the routes of drug resistance transmission are shown in Figure 1. Vertical gene transfer (VGT), a process of cell growth (Seoane et al., 2011), and horizontal gene transfer (HGT), the transfer of DNA by mobile plasmids between genera (Li et al., 2019a), coexisted in the natural environments (Qiu et al., 2018). Under the pressure of long-term selection, the inseparable connection among human, animal, environment and food may be an important factor to induce drug resistance and cause vertical or horizontal transmission of drug-resistant factors.
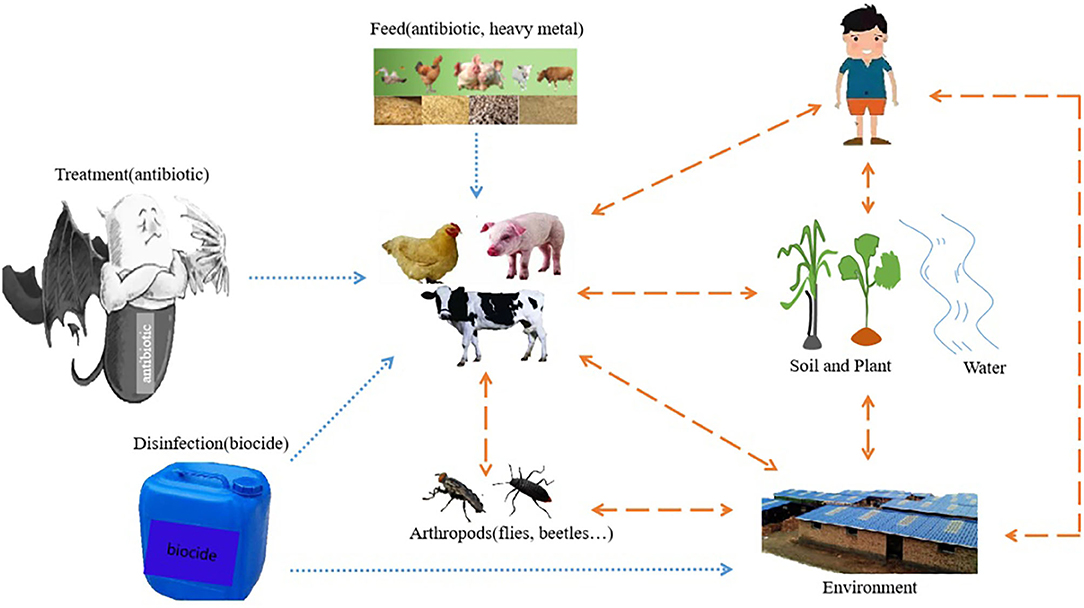
Figure 1. The selective pressure source and drug resistance transmission pathway of bacteria resistance. The blue one-way arrow indicates the sources of selective pressure, including antibiotics used to treat or prevent diseases, feeds containing feed additives such as antibiotics and heavy metals, and biocides used for disinfection in places such as livestock houses or the environment. The Yellow two-way arrow indicates the route of transmission of drug resistance. It includes two-way transmission among human, animal, plant, biological media, water and livestock environment. The dot arrow represents the source of selective pressure on the bacteria; the dash arrow represents the route of resistance genes transmission; the solid arrow represents both the dot and the dash arrow.
Antibiotic resistance is a global problem and poses a major threat to human and animal health. Driven by conditions such as selective pressure of antibacterial drugs, spontaneous mutation, recombination and horizontal gene transfer, the unreasonable use of antibiotics has led to the increasing resistance of bacteria to antibiotics. The resistance of bacteria to antibacterial drugs is caused by genetic variation of different mechanisms. The increase in human health risks associated with antimicrobial resistance has prompted us to study the development mechanism of drug resistance, and in-depth exploration of the factors related to the spread of resistance genes in order to take necessary measures to reduce the growing threat. The resistance mechanisms of bacteria to antibiotics, heavy metals and biocides are introduced as follow.
Genetic plasticity is a prominent feature of bacteria, which will enable bacteria to respond to various threats in the environment, including the existence of antibiotic molecules that may endanger their survival. The main bactericidal mechanisms of antibiotics are: hindering the synthesis of cell wall, inhibiting nucleic acid and protein synthesis, affecting bacterial metabolism and the function of cell membrane. From the perspective of evolution, bacteria mainly adapt to the “attack” of antibiotics through two genetic strategies (Munita and Arias, 2016). (i) In general, gene mutation is associated with the mechanism of action of compounds and (ii) horizontal gene transfer (HGT) is used to obtain exogenous DNA encoding the determinants of resistance. Traditionally, bacteria obtain external genetic material through three main methods: transformation, transduction, and combination. Emergence of resistance often involves conjugation. Conjugation is likely to occur in the gastrointestinal tract of animals receiving antibiotic treatment, which involves cell-to-cell contact, and is a relatively effective method of gene transfer. As a common rule, mobile genetic elements (MGEs) are used as carriers to share valuable genetic information. Plasmids and transposons are the most important MGEs, which play an important role in the development and spread of drug resistance in clinical related microorganisms. In addition, integron is one of the most effective mechanisms for accumulating antibiotic resistance genes, and it is also one of the main driving forces of bacterial evolution. It is a site-specific recombination system that can capture mobile gene cassettes carrying resistance genes and provide an effective mechanism to ensure the expression of new genes. In particular, it is worth noting that carrying plasmids usually results in huge adaptation costs, which means that if the bacteria carrying plasmids want to survive in the population, the cost of plasmids needs to be balanced by the selection pressure brought by antibiotics, for example. In other words, the existence of antibiotics is conducive to the maintenance of plasmid carrying resistance gene. The effects, the use of different types of antibiotics on the generation, spread of bacterial drug resistance and maintenance of drug resistant plasmids will be described in detail in Antibiotics.
Metallic oxide played a role in retarding or declining bacterial growth through the generation of active oxygen (Qiu et al., 2018). Copper and zinc compounds are widely used in animal husbandry as feed additives (Yazdankhah et al., 2014). The use of copper compounds causes copper to enter the environment and causes copper pollution in the environment, which leads to the production of copper-resistant genes by many microorganisms (Yazdankhah et al., 2014). The resistance mechanism to copper is basically non-ATP enzyme excretion mode, and its copper resistance system mainly includes cut system and pco system, as well as cusCFBA efflux system in copper oxidase CueO. Copper-resistant plasmids are often endowed with the gene family pco (Tetaz and Luke, 1983; Yang et al., 2020). Some bacteria have strong adaptability to zinc and can automatically change heavy metals from highly toxic state to low toxic state or non-toxic state through biotransformation or physiological metabolic activities. For instance, E.coli evolved a genetic system of tolerance to heavy metal zinc, showing resistance to heavy metal zinc (Grass et al., 2002). So far, it has been found that E. coli has four systems to keep intracellular Zn2+ in balance, namely znuABC, zupT, zntA and CDF system. ZnuABC and zupT systems are responsible for absorbing Zn2+, while zntA and CDF systems are responsible for pumping Zn2+ out. Studies have shown that (Rensing et al., 1997) the gene zntA, which is usually integrated into the chromosome of bacteria, can encode Zn(II) export protein and is responsible for the specific resistance to Zn(II), Pb(II) and Cd(II) in E. coli (Yang et al., 2020). In prokaryotes, the main mechanism of reducing accumulation is the active efflux of cations (Nies, 1992). Up to now, all bacterial cation efflux systems are encoded and induced by plasmids. In gram positive multi metal resistant Staphylococcus aureus, membrane bound cadA protein (a P-type ATPase) catalyzes the efflux of Cd2+ (and possibly Zn2+). The second protein (CADC) is necessary for complete drug resistance, while the third protein (Cadr) is considered to be the decisive factor in regulating drug resistance. The proteins necessary for CO2+, Zn2+ and Cd2+ efflux (czca, czcb and czcc) and the determinants of regulatory drug resistance (czcd) are all encoded by the determinants of Czc from gram negative multi heavy metal resistant bacteria Alcaligenes eutrophus. The efflux of chromate catalyzed by the ChrA efflux pump leads to chromate resistance (Juhnke et al., 2002). Two determinants of chromate resistance genes were found in this bacterium. That is, the chr(1) [genes chrI, chrB(1), chrA(1), chrC, chrE, chrF(1)] on the known plasmid pMOL28 and the chr(2) on the chromosome [genes chrB(2), chrA(2), chrF(2)]. The gene rcnA commonly found in E. coli is specifically induced by nickel or cobalt and confers resistance to nickel and cobalt based on the efflux system (Rodrigue et al., 2005). Additionally, the research on the effect of heavy metal on bacterial resistance is mainly as a selective pressure to induce antibiotic resistance and promote the spread of resistance genes. The effects of the presence of heavy metal on the generation, spread of bacterial drug resistance will be described in detail in Heavy metals.
The academic definition of bacterial resistance to biocides are: (i) a bacterial strain that is not killed by a biocide concentration to which the majority of the bacterial species are susceptible and (ii) bacterial cells in a culture that survive biocide exposure that kills the majority of the bacterial population in that culture (Maillard, 2018). Biocides usually put pressure on bacterial cells and cause the bacteria to die (Maillard, 2018). The main bactericidal mechanism is destroying the integrity and permeability of the cell membrane, leading to leakage of intracellular material and DNA degradation (Lin et al., 2019; Yi et al., 2019). It can also affect DNA replication through protein denaturation (Chindera et al., 2016) or inhibit biofilm formation (Wang et al., 2018; Halicki et al., 2020). In response, bacteria express several mechanisms to defend against the adverse effects caused by biocides. These mechanisms include the ability of bacteria to repair damage, which can sufficiently reduce the concentration of biocides so that they no longer cause damage to bacterial cells (Maillard, 2018). Therefore, The resistance mechanisms to biocides include decreasing the concentration of biocides in bacteria (reducing biocide penetration; efflux pumps; enzymatic degradation), changing physiology and metabolism and producing mutations, which has been described in detail in a review (Maillard, 2018). Due to selective pressure of disinfectant, mutations or differential gene expression can significantly contribute to disinfectant resistance (Samantha Mc Carlie, 2020); the mechanism of acquired resistance also includes the effective transfer of mobile genetic elements that can carry single or multiple resistance determinants (Samantha Mc Carlie, 2020). Efflux of biocides is also a major mechanism (Russell, 2001; Mohsen and Brown, 2017). Many biocides can induce adaptive multiple antibiotic resistance phenotypes. This may be due to the increased expression of efflux pump and decreased synthesis of outer membrane porin, or both (Molnar et al., 2015). The effects of the present of biocides on the generation, spread of bacterial resistance will be described in detail in Biocides.
Since the 1940s, antibiotics have been widely used in livestock and poultry production as a kind of medicine with the functions of medical treatment, and have played a huge role in promoting the development of animal husbandry (Kumar et al., 2019). When antibiotics are used to treat the infection of pathogenic bacteria, the premise of bactericidal effect is to reach the effective drug concentration and action time, which is concentration and time-dependent on the action of bacteria. When antibiotics are used for treatment, prevention and feed additive to promote growth, they often expose to the pathogenic bacteria and the common bacteria that are usually in the sub-lethal concentration. Due to the selective pressure of antibiotics, both the pathogenic bacteria and the common bacteria develop resistance. The resistant bacteria and their resistance genes will spread horizontally between animals, environment and humans, thus posing a serious threat to human health. Therefore, relevant national governments or institutions need to improve regulatory measures and strengthen supervision to ensure the healthy, stable and sustainable development of the animal husbandry industry. The main antibiotics commonly used in clinic are β-lactam, tetracycline, aminoglycoside, chloramphenical, quinolone, sulfonamides and polypeptide. As most countries in the world banned the use of multiple antibiotics as ways to prevent animal diseases or as feed additives, the resistance of bacteria to some antibiotics has declined. For example, the European Commission (EC) issued a decree to ban the use of antibiotics as growth promoters in feed since 2006 (Huyghebaert et al., 2011). Subsequent investigations showed that antibiotic resistance is 6–20% lower in places where the use of antibiotics as growth promoters is prohibited than in places where is no policy (Huyghebaert et al., 2011). In 2015, the 68th World Health Assembly passed the Global Antimicrobial Resistance Action Plan (Laxminarayan et al., 2020). All member states recognized the serious threat that antibiotics pose to human health and advocated specific actions to solve this problem. Since 2015, the implementation of the U.S. Food and Drug Administration (FDA) Veterinary Feed Directive has reduced the sales of antibiotics as growth promoters (Doshi et al., 2020). In the United States, where fluoroquinolone drugs are banned, the drug resistance rate does not exceed 5%, while in Brazil, China, and the European Union, where the drug is registered, the drug resistance rate is not <40 % on average (Roth et al., 2019). In 2016, India decided to ban unreasonable use of drugs and promised to ban colistin as a growth promoter in order to implement better antibacterial management (Laxminarayan et al., 2020). In recent years, the Chinese government has successively published a list of antibiotics banned in agriculture, and has achieved remarkable results in reducing and replacing the use of antibiotics (Xu et al., 2020). Although in the short term, these antibiotic uses appeared to be beneficial, in fact this practice not only caused to bacterial resistance more easily, but may even induce super resistant bacteria (Kumar et al., 2019). There have been many reports on the effect of low level antibiotics on bacterial resistance, as shown in Table 1.
Heavy metal elements are important components of a series of bioactive substances such as enzymes, hormones and vitamins in human and animal bodies. They participate in the composition of various enzymes and coenzymes, and play an important role in regulating the acid-base balance of body fluids, maintaining the osmotic pressure balance between cells and blood, and promoting the activities of digestive enzymes. It was found that heavy metals such as copper, zinc, chromium, nickel, lead and cadmium were widely used as feed additives in some intensive dairy farming in China (Cannatelli et al., 2014). However, the heavy metal elements may lead to resistance, which is associated with excessive use of Cu and Zn as feed additive. High levels of zinc oxide are also often used as feed additives. This feed contains calcium and iron, which can not only maintain the production of laying hens, but also prevent layers from contracting Salmonella (Li et al., 2019b). However, the presence of heavy metals provides a selective pressure on bacteria. Their stability and persistence may encourage bacteria in food and animals to develop resistance to heavy metals when heavy metals reach a critical concentration. For example, zinc rich supplements in pig feed have chosen more zinc tolerant E. coli (Noh et al., 2017). It is worth noting that bacteria may develop antibiotic resistance at sublethal concentrations of heavy metals, promote plasmid-mediated horizontal transfer of antibiotic resistance genes, and drive the abundance of ARGs (Sharma et al., 2016). Some metals increase the frequency of bacterial resistance through genetic mutations under long-term exposure to sub-MIC levels (Noh et al., 2017). For example, the MIC of bacteria to ciprofloxacin increased by 24.8, 15.6 and 3.9 times respectively after being induced to be mutated at the sublethal levels of Ag(I), Cu(II) and Zn(II) (Johanns et al., 2019). These resistant mutants also exhibited multi-drug resistance. For example, Zn(II) evolved ciprofloxacin-resistant mutants, and at the same time showed resistance to chloramphenicol and tetracycline (Maillard, 2005). In a pig isolate, it was identified that a plasmid carrying a gene named tcrB on a conjugated plasmid made Enterococcus facialis copper resistant (Maillard, 2005). Studies have shown that the use of copper selects new resistance genes. For example, copper-resistant Salmonella can be isolated from pig feed supplemented with copper (Aarestrup et al., 2002; Li et al., 2019c). Cu resistance genes pcoA was found in multi-resistant Enterobacteriaceae with blaNDM−1 or blaCTX−M−15 genes, and these genes were mostly located on plasmids (Medardus et al., 2014). The resistance of high-zinc feed may increase due to the correlation between zinc tolerance and antimicrobial resistance. It has recently been found that the copy number of tetracycline and sulfonamide resistance genes is increased in the intestines of weaned piglets fed a high diet of zinc oxide (Poole, 2017). The zinc concentration in liquid pig manure and phenotypic antimicrobial resistance in E. coli have been linked in recent publications in Germany (Yang et al., 2018). And the level of pharmacological zinc feeding is related to the increase in the prevalence and persistence of methicillin-resistant S. aureus (Vahjen et al., 2015). However, the research found that there was no correlation between phenotypic zinc resistance and multiple antimicrobial resistance in piglet fecal E. coli isolates (Hölzel et al., 2012). Observations showed that the MIC of E. coli isolated from animals on zinc or copper did not change when exposed to high concentrations of zinc.
Biocides generally refer to all chemicals such as disinfectants and preservatives, fungicides, algaecides, rodenticides and insecticides (Hölzel et al., 2012; Slifierz et al., 2015b) and phages and bacteria and other biological agents used to kill pathogenic microorganisms (Slifierz et al., 2015b). In a sense, biocides are a very diverse group of chemicals. Nowadays, chemical biocides are widely used in consumer goods, water, food industry, commodity manufacturing industry, pharmaceutical industry, health care and veterinary departments, as well as oil and gas industry. The universal application reflects the versatility of biocide products in environmental disinfection, product preservation and anti-corrosion (Ghazisaeedi et al., 2020). The biocides used for disinfection and antisepsis mainly include: Alcohols (ethanol, isopropanol); Aldehydes (glutaraldehyde, formaldehyde); Anilides (triclocarban); Biguanides (chlorhexidine, alexidine, polymeric, biguanides); Bisphenols (triclosan, hexachlorophene); Diamideines (propamidine, dibromopropamidine) (Paul et al., 2019). In addition, disinfectants based on quaternary ammonium compounds (QAC) are stable, have low cytotoxicity, and are non-corrosive to farm equipment. Therefore, it has become the most commonly used disinfectant in industry and animal production. The effect of biocides on bacterial resistance is shown in Table 2. Although there are many reports that microbial resistance will be affected with the use of fungicides, there are different views in the field of research. The use of disinfectants in the poultry and swine industry and their contribution to the antibiotic and disinfectant susceptibility of E. coli strains obtained after cleaning and disinfection were studied. They found that all E. coli strains were sensitive to formaldehyde, benzalkonium chloride, acetic acid and hydrogen peroxide at moderate concentrations, indicating that anti disinfectants were not selected in the actual use of disinfectants (Maillard, 2005). Randal and others discussed the adaptability and spread of multi-drug resistant (MAR) Salmonella and Salmonella typhimurium in the intestines of chickens under the choice of farm disinfectant. Their research shows that the resistant strains screened by farm disinfectants will not be preferentially selected because of the use of disinfectant, and their transmission ability is weak, and will not cause more harm to chickens than other Salmonella (McDonnell and Russell, 1999). A large veterinary institution in Australia has detected the pathogenic bacterium Enterobacter homachi in the hand washing pool of ICU in a hospital. Despite two subsequent attempts to disinfect it with biocides, the microorganism still survived for several weeks (McDonnell and Russell, 1999). However, the level of disinfectant tolerance can be slightly increased by the disinfectant mutants of Salmonella typhimurium (Maertens et al., 2019). In conclusion, improper disinfection program combined with the use of antibiotics may be still at the risk of leading to a higher level of bacterial resistance.
Arthropods, such as flies, beetles, cockroaches and so on, are common in livestock units. The persistent fecal substrate, thermal insulation materials and accumulated dust provide a suitable living environment for them. They have a special ability to transfer pathogens and resistance genes from different environmental levels. Flies carrying Salmonella with the same serotype and subtype as livestock and poultry can usually be caught around livestock and poultry farms that are positive for Salmonella (Hancock et al., 1998; Olsen and Hammack, 2000; Davies and Breslin, 2003; Seier-Petersen et al., 2014). Clostridium perfringens can also often be isolated from flies and beetles around infected broiler flocks (Hancock et al., 1998; Olsen and Hammack, 2000; Davies and Breslin, 2003; Kinde et al., 2005; Seier-Petersen et al., 2014). Salmonella reported to be isolated from mantis samples from poultry farms in the Middle East (Craven et al., 2001) and in a hatchery and poultry feed mill in the USA (Fathpour et al., 2003). Cockroaches are closely related to people and animals. They have been repeatedly accused of being vectors of food poisoning bacteria (Kopanic et al., 1994). Bacterial pathogens can live on their body surface and digestive tract and spread through direct transfer or during reflux and defecation (Ostrolenk and Welch, 1942). From the above description, we can know flies may be a health indicator of the prevalence of antimicrobial resistance (Schaumburg et al., 2016; Onwugamba et al., 2018). This view was previously confirmed by Zurek and Ghosh (Zurek and Ghosh, 2014). Flies entering the broiler house increase the risk of colonization of multi-drug resistant E. coli in broiler chickens (Solà-Ginés et al., 2015; Poudel et al., 2019). However, no similar transfer studies have been found in other arthropods on the farm.
Bacterial resistance is also affected by stress factors. The resistance of diseased livestock and poultry is regulated by nerves, body fluids and endocrine to a certain extent. Under the influence of various stress factors, such as climate upheaval (supercooling or overheating), excessive humidity, strong light, poor ventilation, group transfer, the normal physiological functions of the body will be seriously disordered, and the body's constitution will be reduced, and the resistance will be reduced, and the drug resistance will produce. Studies have shown that some components of the immune system have broad-spectrum antibacterial activity (Andersson et al., 2016). Under long-term action, bacteria can develop resistance to these broad-spectrum antibacterial substances. When the veterinarian uses antibiotics to treat diseased animals, the pathogenic bacteria population will be exposed to sublethal levels of antibiotics. Once some pathogenic bacteria survive, they also may become resistant to antibiotics and spread horizontally or vertically (Andersson et al., 2016). Therefore, to create a good environment for livestock and poultry, reduce the incidence of drug resistance, measures such as reducing stress factors, strengthening feeding management, purifying and disinfecting pens should be taken.
The causes of antibiotic resistance are complex. In animal husbandry, bacterial resistance to antibiotics is caused by many factors, such as medication procedure, management system, breeding environment, selection pressure and so on (Manyi-Loh et al., 2018). Generally, drug resistance is caused by the combination of environmental selection and gene mutation (Manyi-Loh et al., 2018). Long term use of antibiotics in domestic animals may lead to drug resistance in both human and animals. In order to reduce the incidence of bacterial drug resistance, it is necessary to avoid creation of the settings that beneficial for resistance genes persistence in bacterial communities, reduce dispersal routes for resistant bacteria to microbiome, limit the selection pressure for resistant pathogens and strengthen feeding management. Therefore, it is urgent to take appropriate measures to alleviate the generation and spread of drug-resistant strains according to the actual situation.
Ensure that antibiotics are used correctly in humans and animals (van den Bogaard and Stobberingh, 1999). In order to prevent and control the occurrence of epidemic diseases, some farms add antibiotics to feed and drinking water, and frequently use the same kind of antibiotics, so that long-term drug use leads to bacterial resistance to antibiotics. Secondly, not changing the antibiotics in time or increasing the dosage of drugs are also one of the important reasons for the increase of drug-resistant strains. In order to solve this problem, we can consider changing the way of drug use (Friedman et al., 2007). If we use interval or shuttle drug, the frequency of drug resistance will be greatly reduced. Interval medication refers to the use of a drug in the process, not continuous use, can be divided into batches and doses. Shuttle drug use mainly refers to the use of different drugs interspersed, or in different seasons, different years, so as to reduce drug resistance. In addition, the combination of many antibiotics has obvious synergistic effect. For example, the combination of penicillin and aminoglycoside antibiotics, which is the most common in clinic, not only broadens the antibacterial spectrum, but also reduces the drug dosage and plays the role of the best efficacy.
Prevention of bacterial infection and transmission. “There is a correlation between the use of antibiotics in livestock and poultry breeding and the infection of drug-resistant bacteria in humans and livestock,” which is still further aggravating (Chen et al., 2018). Improving public and personal health conditions to prevent the generation and spread of pathogens is a crucial measure to solve this problem. The fewer people and livestock are infected, the less antibiotics are needed and the slower resistance develops. Developing countries should focus on improving infrastructure and providing more clean water and sanitation. The focus of other countries should be to reduce the infection of pathogenic bacteria and prevent the emergence of super resistant bacteria.
Develop new antibiotics or find alternative therapies (Cox and Wright, 2013). After the emergence of metallolactamase NDM-1, the only way to treat it is to use polymyxin as the “last line of defense.” Due to its high toxicity, polymyxin has always been a kind of antibiotic that is not easily used by human beings. Therefore, the development of a new generation of antibiotics has become an urgent task in the world. Chemical synthesis and natural product research are two main methods for human beings to discover antibiotics. Compared with chemical synthesis, most of the antibiotics used by human beings are derived from natural products, such as β–lactam antibiotics, vancomycin and polymyxin, which are the “last line of defense” against bacterial infection (Okano et al., 2017). However, due to the complex structure of natural products and low synthesis efficiency, the production line of new antibiotics is not optimistic, and alternative solutions are needed (Rossiter et al., 2017). One possible alternative is vaccination. In fact, despite the rapid emergence of antibiotic resistance, vaccines can allow infections to be controlled more sustainably (Tagliabue and Rappuoli, 2018).
Participate in international cooperation to reduce the risk of drug resistance transmission. Drug resistance is not a problem that can be solved by a single country, or even by a region. In fact, the world has taken intervention measures from many aspects to deal with the challenges brought by antibiotic resistance (Laxminarayan et al., 2013). For example, the European Medical Agency (EMEA) is mainly responsible for the evaluation, monitoring and early warning of EU drugs. There is a special group to carry out specific work, including the Scientific Advisory Group on Antimicrobials (SAGAM), which makes recommendations to the Committee for Medicinal Products for Veterinary Use (CVMP) on the approval and use of veterinary antibiotics, especially the development of guidelines on antimicrobial resistance. The Federation of Veterinarians of Europe (FVE) is a veterinary organization from 35 European countries, including four veterinary groups: industry veterinarians, public health veterinarians, education, research and enterprise veterinarians, and state veterinarians. In 1999, FVE established an ad hoc group on antimicrobial resistance and formulated guidelines for the use of antimicrobial agents in animals to reduce the occurrence and spread of drug resistance. However, due to a lack of access to investment, infrastructure and modern equipment some developing countries struggle to adequately implement strategies available to wealthier nations. It can be seen that, to a large extent, there is a lack of coordinated global action, especially at the national and international political levels. At present, countries in the world are more interconnected and interdependent than ever before. Especially in the field of international health care and health, we should advocate the consciousness of “a community of shared future for mankind.”
To improve the monitoring of antibiotics for human and animal use. To this end, the world should first make efforts to monitor and quantify antibiotic resistance in the environment. This requires countries to provide adequate support for the construction of advanced drug resistance monitoring networks and biological risk assessment systems. In 2014, the World Health Organization (WHO) for the first time issued a global antibiotic resistance monitoring report, mainly reports the 7 kinds of common pathogens of antimicrobial resistance of international concern. The World Organization for Animal Health (OIE) launched its first annual data collection on antimicrobial use in animals in 2015, published the first edition of its Annual Report on Antimicrobial Use in Animals in 2016 and the second report in 2017 (Tornimbene et al., 2018). It sorted out the use of veterinary antibiotics in 146 countries and regions from 2013 to 2016. Although these reports may be considered to have limitations on the effectiveness of the impact due to mixed factors such as obsolescence, national attributes and information bias. These reports still provide valuable reference for the global sharing of information on the development trend of bacterial resistance, and also bring new hope for the global joint response and solution of antibiotic resistance.
Strengthen the propaganda, education and training of antibiotics. Enhance global awareness of antibiotic resistance, disseminate awareness and appropriate information about the risks and consequences of rapidly developing antibiotic resistance. Some countries are looking for alternatives to antibiotics in animal husbandry in order to reduce the use of antibiotics on farms and ensure that the economic benefits of the farm will be further improved under the guidance of the policy (Prescott, 2017; Antão and Wagner-Ahlfs, 2018). It is very important to solve this global problem that everyone in the agricultural sector, public sector, health sector, drug administration department and the public at different levels should treat the drug resistance problem of animal origin bacteria as the master. In a word, it is not an overnight thing to solve the problem of antibiotic resistance. The whole world needs an united front to formulate long-term plans and goals, work together in the same boat, and contribute to a better future for the world.
This paper reviews the influencing factors and mechanisms of bacterial resistance, and puts forward some possible suggestions on how to alleviate the emergence and spread of bacterial resistance. In livestock and its environment, animals are both recipients and providers of selective stress, and ultimately bacteria are survived in a sub-lethal level of antimicrobial environment. Under the selective pressure from low levels of antibiotics, heavy metals and biocides, bacteria produce corresponding resistance. And this pressure can promote the inter-bacterial spread of resistance genes. Most notably, non-antibiotic antibacterial agents can induce or select bacterial adaptation, thereby reducing sensitivity to one or more antibiotics. Apart from antibiotics, heavy metals and biocides, bacterial resistance is also affected by arthropods in the environment. Flies carrying drug-resistant bacteria and resistant genes can transmit them to food animals and bacteria in their environment, and they have the risk of being transmitted to human beings through the food chain, which poses a threat to public health. Furthermore, from the physiological point of view of animals, stress can lead to physiological dysfunction of animals, which indirectly leads to bacterial resistance. Briefly, many factors directly or indirectly cause bacterial resistance. Therefore, countries or alliances around the world should combine the monitoring of animal-derived bacterial resistance and the application of veterinary drug supervision platforms to improve the scientific management of veterinary antimicrobials, effectively promote the reduction of veterinary antimicrobials, and ensure production safety in the global aquaculture industry, Livestock and poultry product quality safety, public health safety and ecological environment safety.
KG, YZ, and MD contributed conception and design of the study. KG, YZ, LC, and MD investigated and surveyed literatures. YZ wrote the first draft of the manuscript. KG wrote the second draft of the manuscript. KG, YZ, LC, ZC, FZ, XW, JF, and MD revised the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was supported by the National Key R&D Program of China, grant number 2017YFC1600100, the Fundamental Research Funds for the Central Universities, grant number 2662017JC034, the Applied Basic Research Programs of Wuhan, grant number 2017020201010228 to MD, and the Earmarked Fund for China Agriculture Research System, grant number CARS-35 to XW.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank the National Key R&D Program of China, the Fundamental Research Funds for the Central Universities, Applied Basic Research Programs of Wuhan and the Earmarked Fund for China Agriculture Research System for funding this study.
Aarestrup, F. M., Hasman, H., Jensen, L. B., Moreno, M., Herrero, I. A., Domínguez, L., et al. (2002). tcrB, a gene conferring transferable copper resistance in Enterococcus faecium: occurrence, transferability, and linkage to macrolide and glycopeptide resistance. Appl. Environ. Microbiol. 8, 4127–4129. doi: 10.1016/j.metabol.2009.05.029
Andersson, D. I., Hughes, D., and Kubicek-Sutherland, J. Z. (2016). Mechanisms and consequences of bacterial resistance to antimicrobial peptides. Drug Resist. Update 26, 43–57. doi: 10.1016/j.drup.2016.04.002
Antão, E., and Wagner-Ahlfs, C. (2018). Antibiotikaresistenz. Bundesgesundheitsblatt - Gesundheitsforschung - Gesundheitsschutz 61, 499–506. doi: 10.1007/s00103-018-2726-y
Bhattacharya, G., Dey, D., Das, S., and Banerjee, A. (2017). Exposure to sub-inhibitory concentrations of gentamicin, ciprofloxacin and cefotaxime induces multidrug resistance and reactive oxygen species generation in meticillin-sensitive Staphylococcus aureus. J. Med. Microbiol. 66, 762–769. doi: 10.1099/jmm.0.000492
Cannatelli, A., Di Pilato, V., Giani, T., Arena, F., Ambretti, S., Gaibani, P., et al. (2014). In vivo evolution to colistin resistance by PmrB sensor kinase mutation in KPC-producing Klebsiella pneumoniae is associated with low-dosage colistin treatment. Antimicrob. Agents Chemother. 58, 4399–4403. doi: 10.1128/AAC.02555-14
Chen, Q., Gong, X., Zheng, F., Ji, G., Li, S., Stipkovits, L., et al. (2018). Interplay between the phenotype and genotype, and efflux pumps in drug-resistant strains of Riemerella anatipestifer. Front. Microbiol. 9:2136. doi: 10.3389/fmicb.2018.02136
Chen, Y., Sun, J., Liao, X., Shao, Y., Li, L., Fang, L., et al. (2016). Impact of enrofloxacin and florfenicol therapy on the spread of OqxAB gene and intestinal microbiota in chickens. Vet. Microbiol. 192, 1–9. doi: 10.1016/j.vetmic.2016.05.014
Chindera, K., Mahato, M., Sharma, A. K., Horsley, H., Kloc-Muniak, K., Kamaruzzaman, N. F., et al. (2016). The antimicrobial polymer PHMB enters cells and selectively condenses bacterial chromosomes. Sci. Rep. 6:23121. doi: 10.1038/srep23121
Cogliani, C., Goossens, H., and Greko, C. (2011). Restricting antimicrobial use in food animals: lessons from Europe. Features 6, 274–279. doi: 10.1128/microbe.6.274.1
Collineau, L., Bougeard, S., Backhans, A., Dewulf, J., Emanuelson, U., Grosse, B. E., et al. (2018). Application of multiblock modelling to identify key drivers for antimicrobial use in pig production in four European countries. Epidemiol. Infect. 146, 1003–1014. doi: 10.1017/S0950268818000742
Cottell, A., Denyer, S. P., Hanlon, G. W., Ochs, D., and Maillard, J. Y. (2009). Triclosan-tolerant bacteria: changes in susceptibility to antibiotics. J. Hosp. Infect. 72, 71–76. doi: 10.1016/j.jhin.2009.01.014
Cox, G., and Wright, G. D. (2013). Intrinsic antibiotic resistance: mechanisms, origins, challenges and solutions. Int. J. Med. Microbiol. 303, 287–292. doi: 10.1016/j.ijmm.2013.02.009
Craven, S. E., Stern, N. J., Bailey, J. S., and Cox, N. A. (2001). Incidence of Clostridium perfringens in broiler chickens and theirenvironment during production and processing. Avian Dis. 45, 887–896. doi: 10.2307/1592868
Curiao, T., Marchi, E., Viti, C., Oggioni, M. R., Baquero, F., Martinez, J. L., et al. (2015). Polymorphic variation in susceptibility and metabolism of triclosan-resistant mutants of Escherichia coli and Klebsiella pneumoniae clinical strains obtained after exposure to biocides and antibiotics. Antimicrob. Agents Chemother. 59, 3413–3423. doi: 10.1128/AAC.00187-15
Davies, R., and Breslin, M. (2003). Observations on Salmonella contamination of commercial laying farms before and after cleaning and disinfection. Vet. Rec. 10, 283–287. doi: 10.1136/vr.152.10.283
Deng, W., Quan, Y., Yang, S., Guo, L., Zhang, X., Liu, S., et al. (2018). Antibiotic resistance in Salmonella from retail foods of animal origin and its association with disinfectant and heavy metal resistance. Microb. Drug Resist. 24, 782–791. doi: 10.1089/mdr.2017.0127
Doshi, P., Morten, C. J., and Herder, M. (2020). Integrated drug reviews at the US food and drug administration-reply. JAMA Intern. Med. 180, 1261–1262. doi: 10.1001/jamainternmed.2020.1978
Fathpour, H., Emtiazi, G., and Ghasemi, E. (2003). Cockroaches as reservoirs and vectors of drug resistant Salmonella spp. Fresen Environ. Bull. 12, 724–727. doi: 10.1016/S1382-6689(02)00123-0
Friedman, D. B., Kanwat, C. P., Headrick, M. L., Patterson, N. J., Neely, J. C., and Smith, L. U. (2007). Importance of prudent antibiotic use on dairy farms in South Carolina: a pilot project on farmers' knowledge, attitudes and practices. Zoonoses Public Health 54, 366–375. doi: 10.1111/j.1863-2378.2007.01077.x
Ge, B., Domesle, K. J., Yang, Q., Young, S. R., Rice-Trujillo, C. L., Bodeis Jones, S. M., et al. (2017). Effects of low concentrations of erythromycin, penicillin, and virginiamycin on bacterial resistance development in vitro. Sci. Rep. UK 1:11017. doi: 10.1038/s41598-017-09593-4
Ghazisaeedi, F., Ciesinski, L., Bednorz, C., Johanns, V., Pieper, L., Tedin, K., et al. (2020). Phenotypic zinc resistance does not correlate with antimicrobial multi-resistance in fecal E. coli isolates of piglets. Gut. Pathog. 12:4. doi: 10.1186/s13099-019-0342-5
Gould, K. (2016). Antibiotics: from prehistory to the present day. J. Antimicrob. Chemoth. 71, 572–575. doi: 10.1093/jac/dkv484
Grass, G., Wong, M. D., Rosen, B. P., Smith, R. L., and Rensing, C. (2002). ZupT is a Zn(II) uptake system in Escherichia coli. J. Bacteriol. 184, 864–866. doi: 10.1128/JB.184.3.864-866.2002
Gullberg, E., Albrecht, L. M., Karlsson, C., Sandegren, L., and Andersson, D. I. (2014). Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. Mbio 5, e1914–e1918. doi: 10.1128/mBio.01918-14
Gullberg, E., Cao, S., Berg, O. G., Ilbäck, C., Sandegren, L., Hughes, D., et al. (2011). Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 7:e1002158. doi: 10.1371/journal.ppat.1002158
Gumpu, M. B., Sethuraman, S., Krishnan, U. M., and Rayappan, J. B. B. (2015). A review on detection of heavy metal ions in water – An electrochemical approach. Sens. Actuators. B. Chem. 213, 515–533. doi: 10.1016/j.snb.2015.02.122
Gupta, A., Bansal, M., Wagle, B., Sun, X., Rath, N., Donoghue, A., et al. (2020). Sodium butyrate reduces Salmonella enteritidis infection of chicken enterocytes and expression of inflammatory host genes in vitro. Front. Microbiol. 11:553670. doi: 10.3389/fmicb.2020.553670
Halicki, P., Radin, V., von Groll, A., Nora, M. V., Pinheiro, A. C., Da, S. P., et al. (2020). Antibiofilm potential of arenecarbaldehyde 2-pyridinylhydrazone derivatives against acinetobacter baumannii. Microb. Drug Resist. 26, 1429–1436. doi: 10.1089/mdr.2019.0185
Han, Y., Zhou, Z. C., Zhu, L., Wei, Y. Y., Feng, W. Q., Xu, L., et al. (2019). The impact and mechanism of quaternary ammonium compounds on the transmission of antibiotic resistance genes. Environ. Sci. Pollut. Res. Int. 26, 28352–28360. doi: 10.1007/s11356-019-05673-2
Hancock, D. D., Besser, T. E., Rice, D. H., Ebel, E. D., Herriott, D. E., and Carpenter, L. V. (1998). Multiple sources of Escherichia coli O157 in feedlots and dairy farms in the Northwestern USA. Prev. Vet. Med. 35, 11–19. doi: 10.1016/S0167-5877(98)00050-6
Hejna, M., Gottardo, D., Baldi, A., Dell Orto, V., Cheli, F., Zaninelli, M., et al. (2018). Review: nutritional ecology of heavy metals. Animal 12, 2156–2170. doi: 10.1017/S175173111700355X
Hobman, J. L., and Crossman, L. C. (2015). Bacterial antimicrobial metal ion resistance. J. Med. Microbiol. 64, 471–497. doi: 10.1099/jmm.0.023036-0
Hölzel, C. S., Müller, C., Harms, K. S., Mikolajewski, S., Schäfer, S., Schwaiger, K., et al. (2012). Heavy metals in liquid pig manure in light of bacterial antimicrobial resistance. Environ. Res. 113, 21–27. doi: 10.1016/j.envres.2012.01.002
Huyghebaert, G., Ducatelle, R., and Immerseel, F. V. (2011). An update on alternatives to antimicrobial growth promoters for broilers. Vet. J. 187, 182–188. doi: 10.1016/j.tvjl.2010.03.003
Johanns, V. C., Ghazisaeedi, F., Epping, L., Semmler, T., Lübke-Becker, A., Pfeifer, Y., et al. (2019). Effects of a four-week high-dosage zinc oxide supplemented diet on commensal Escherichia coli of weaned pigs. Front. Microbiol. 10:2734. doi: 10.3389/fmicb.2019.02734
Juhnke, S., Peitzsch, N., Hübener, N., Grosse, C., and Nies, D. H. (2002). New genes involved in chromate resistance in Ralstonia metallidurans strain CH34. Arch. Microbiol. 1, 15–25. doi: 10.1007/s00203-002-0492-5
Jutkina, J., Marathe, N. P., Flach, C. F., and Larsson, D. G. J. (2018). Antibiotics and common antibacterial biocides stimulate horizontal transfer of resistance at low concentrations. Sci. Total Environ. 616–617, 172–178. doi: 10.1016/j.scitotenv.2017.10.312
Kamathewatta, K., Bushell, R., Rafa, F., Browning, G., Billman-Jacobe, H., and Marenda, M. (2020). Colonization of a hand washing sink in a veterinary hospital by an Enterobacter hormaechei strain carrying multiple resistances to high importance antimicrobials. Antimicrob. Resist. Infect. Control 9:163. doi: 10.1186/s13756-020-00828-0
Karatzas, K. A. G., Webber, M. A., Jorgensen, F., Woodward, M. J., Piddock, L. J. V., and Humphrey, T. J. (2007). Prolonged treatment of Salmonella enterica serovar Typhimurium with commercial disinfectants selects for multiple antibiotic resistance, increased efflux and reduced invasiveness. J. Antimicrob. Chemother. 60, 947–955. doi: 10.1093/jac/dkm314
Kimon, A. G. K., Randall, L. P., Webber, M., Laura, J. V. P., Humphrey, T. J., Woodward, M. J., et al. (2008). Phenotypic and proteomic characterization of multiply antibiotic-resistant variants of Salmonella enterica Serovar Typhimurium selected following exposure to disinfectants. Appl. Environ. Microb. 74, 1508–1516. doi: 10.1128/AEM.01931-07
Kinde, H., Castellan, D. M., Kerr, D., Campbell, J., Breitmeyer, R., and Ardans, A. (2005). Longitudinal monitoring of two commercial layer flocks and their environments for Salmonella enterica serovar enteritidis and other Salmonellae. Avian Dis. 49, 189–194. doi: 10.1637/7228-062704R
Kopanic, R. J., Sheldon, B. W., and Wright, C. G. (1994). Cockroaches as vectors of Salmonella: laboratory and field trials. J. Food Prot. 57, 125–135. doi: 10.4315/0362-028X-57.2.125
Kumar, M., Jaiswal, S., Sodhi, K. K., Shree, P., Singh, D. K., Agrawal, P. K., et al. (2019). Antibiotics bioremediation: perspectives on its ecotoxicity and resistance. Environ. Int. 124, 448–461. doi: 10.1016/j.envint.2018.12.065
Laxminarayan, R., Duse, A., Wattal, C., Zaidi, A. K., Wertheim, H. F., Sumpradit, N., et al. (2013). Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 13, 1057–1098. doi: 10.1016/S1473-3099(13)70318-9
Laxminarayan, R., Van Boeckel, T., Frost, I., Kariuki, S., Khan, E. A., Limmathurotsakul, D., et al. (2020). The lancet infectious diseases commission on antimicrobial resistance: 6 years later. Lancet Infect. Dis. 20, e51–e60. doi: 10.1016/S1473-3099(20)30003-7
Lei, C. W., Zhang, Y., Kang, Z. Z., Kong, L. H., Tang, Y. Z., Zhang, A. Y., et al. (2020). Vertical transmission of Salmonella Enteritidis with heterogeneous antimicrobial resistance from breeding chickens to commercial chickens in China. Vet. Microbiol. 240:108538. doi: 10.1016/j.vetmic.2019.108538
Lekshmi, M., Ammini, P., Kumar, S., and Varela, M. F. (2017). The food production environment and the development of antimicrobial resistance in human pathogens of animal origin. Microorganisms 5:11. doi: 10.3390/microorganisms5010011
Li, B., Qiu, Y., Song, Y., Lin, H., and Yin, H. (2019a). Dissecting horizontal and vertical gene transfer of antibiotic resistance plasmid in bacterial community using microfluidics. Environ. Int. 131:105007. doi: 10.1016/j.envint.2019.105007
Li, J., Xu, Y., Wang, L., and Li, F. (2019b). Heavy metal occurrence and risk assessment in dairy feeds and manures from the typical intensive dairy farms in China. Environ. Sci. Pollut. R. 26, 6348–6358. doi: 10.1007/s11356-019-04125-1
Li, X., Gu, A. Z., Zhang, Y., Xie, B., Li, D., and Chen, J. (2019c). Sub-lethal concentrations of heavy metals induce antibiotic resistance via mutagenesis. J. Hazard Mater. 369, 9–16. doi: 10.1016/j.jhazmat.2019.02.006
Lin, D., Chen, K., Xie, M., Ye, L., Chan, E. W., and Chen, S. (2017). Effect of ceftiofur and enrofloxacin on E. coli sub-population in pig gastrointestinal tract. J. Glob. Antimicrob. Resist. 10, 126–130. doi: 10.1016/j.jgar.2017.05.010
Lin, W., Yuan, D., Deng, Z., Niu, B., and Chen, Q. (2019). The cellular and molecular mechanism of glutaraldehyde-didecyldimethylammonium bromide as a disinfectant against Candida albicans. J. Appl. Microbiol. 126, 102–112. doi: 10.1111/jam.14142
Luo, X. S., Ip, C. C. M., Li, W., Tao, S., and Li, X. D. (2014). Spatial–temporal variations, sources, and transport of airborne inhalable metals (PM10) in urban and rural areas of northern China. Atmos. Chem. Phys. Discuss. 14, 13133–13165. doi: 10.5194/acpd-14-13133-2014-supplement
Maertens, H., De Reu, K., Meyer, E., Van Coillie, E., and Dewulf, J. (2019). Limited association between disinfectant use and either antibiotic or disinfectant susceptibility of Escherichia coli in both poultry and pig husbandry. BMC Vet. Res. 15:310. doi: 10.1186/s12917-019-2044-0
Maillard, J. (2018). Resistance of Bacteria to Biocides. Microbiol. Spectr. 6:ARBA-0006-2017. doi: 10.1128/microbiolspec.ARBA-0006-2017
Maillard, J. Y. (2005). Antimicrobial biocides in the healthcare environment: efficacy, usage, policies, and perceived problems. Ther. Clin. Risk Manage. 1, 307–320.
Manyi-Loh, C., Mamphweli, S., Meyer, E., and Okoh, A. (2018). Antibiotic use in agriculture and its consequential resistance in environmental sources: potential public health implications. Molecules 23:795. doi: 10.3390/molecules23040795
Mavri, A., Ribič, U., and MoŽina, S. S. (2016). The biocide and antibiotic resistance in Campylobacter jejuni and Campylobacter coli. Food Eng. Ser. 4, 887–896. doi: 10.1007/978-3-319-24040-4_15
McDonnell, G., and Russell, A. D. (1999). Antiseptics and disinfectants: activity, action, and resistance. Clin. Microbiol. Rev. 12, 147–179. doi: 10.1128/CMR.12.1.147
Medardus, J. J., Molla, B. Z., Nicol, M., Morrow, W. M., Rajala-Schultz, P. J., Kazwala, R., et al. (2014). In-feed use of heavy metal micronutrients in U.S. swine production systems and its role in persistence of multidrug-resistant Salmonellae. Appl. Environ. Microb. 80, 2317–2325. doi: 10.1128/AEM.04283-13
Mohsen, C., and Brown, M. H. (2017). The role played by drug efflux pumps in bacterial multidrug resistance. Biochemistry 61, 127–139. doi: 10.1042/EBC20160064
Molnar, D., Piotrowski, M., Schultz, D., and Wagner, D. (2015). Sublethal exposure to commercial formulations of the herbicides dicamba, 2,4-dichlorophenoxyacetic acid, and glyphosate cause changes in antibiotic susceptibility in escherichia coli and salmonella enterica serovar typhimurium. Mbio 6, 13–30. doi: 10.1128/mBio.00009-15
Munita, J. M., and Arias, C. A. (2016). Mechanisms of antibiotic resistance. Microbiol. Spectr. 4:VMBF-0016-2015. doi: 10.1128/microbiolspec.VMBF-0016-2015
Nhung, N., Thuy, C., Trung, N., Campbell, J., Baker, S., Thwaites, G., et al. (2015). Induction of antimicrobial resistance in Escherichia coli and non-typhoidal Salmonella strains after adaptation to disinfectant commonly used on farms in Vietnam. Antibiotics 4, 480–494. doi: 10.3390/antibiotics4040480
Nies, D. H. (1992). Resistance to cadmium, cobalt, zinc, and nickel in microbes. Plasmid 27, 17–28. doi: 10.1016/0147-619X(92)90003-S
Noh, H., Kim, H., Heo, S. J., Cho, H. H., and Koh, H. B. (2017). Guanosine 5′-monophosphate-chelated calcium and iron feed additives maintains egg production and prevents Salmonella Gallinarum in experimentally infected layers. J. Vet. Sci. 18:291. doi: 10.4142/jvs.2017.18.3.291
Okano, A., Isley, N. A., and Boger, D. L. (2017). Total syntheses of vancomycin-related glycopeptide antibiotics and key analogues. Chem. Rev. 117, 11952–11993. doi: 10.1021/acs.chemrev.6b00820
Olsen, A. R., and Hammack, T. S. (2000). Isolation of Salmonella spp. from the housefly, Musca domestica L., and the dump fly, Hydrotaea aenescens (Wiedemann) (Diptera: Muscidae), at caged-layer houses. J. Food Protect. 63:958. doi: 10.4315/0362-028X-63.7.958
Ons, L., Bylemans, D., Thevissen, K., and Cammue, B. P. A. (2020). Combining biocontrol agents with chemical fungicides for integrated plant fungal disease control. Microorganisms 8:1930. doi: 10.3390/microorganisms8121930
Onwugamba, F. C., Fitzgerald, J. R., Rochon, K., Guardabassi, L., Alabi, A., Kühne, S., et al. (2018). The role of ‘filth flies' in the spread of antimicrobial resistance. Travel Med. Infect. Dis. 22, 8–17. doi: 10.1016/j.tmaid.2018.02.007
Ostrolenk, M., and Welch, H. (1942). The house fly as a vector of food poisoning organisms in food producing establishments. Am. J. Public Health Nations Health 32, 487–494. doi: 10.2105/AJPH.32.5.487
Paul, D., Chakraborty, R., and Mandal, S. M. (2019). Biocides and health-care agents are more than just antibiotics: inducing cross to co-resistance in microbes. Ecotox Environ. Safe 174, 601–610. doi: 10.1016/j.ecoenv.2019.02.083
Poole, K. (2017). At the nexus of antibiotics and metals: the impact of Cu and Zn on antibiotic activity and resistance. Trends Microbiol. 25, 820–832. doi: 10.1016/j.tim.2017.04.010
Poudel, A., Hathcock, T., Butaye, P., Kang, Y., Price, S., Macklin, K., et al. (2019). Multidrug-Resistant Escherichia coli, Klebsiella pneumoniae and Staphylococcus spp. in houseflies and blowflies from farms and their environmental settings. Int. J. Environ. Res. Public Health 16:3583. doi: 10.3390/ijerph16193583
Prescott, J. F. (2017). History and current use of antimicrobial drugs in veterinary medicine. Microbiol. Spectr. 5:ARBA-0002-2017. doi: 10.1128/microbiolspec.ARBA-0002-2017
Qiu, Y., Zhang, J., Li, B., Wen, X., Liang, P., and Huang, X. (2018). A novel microfluidic system enables visualization and analysis of antibiotic resistance gene transfer to activated sludge bacteria in biofilm. Sci. Total Environ. 642, 582–590. doi: 10.1016/j.scitotenv.2018.06.012
Randall, L. P., Bagnall, M. C., Karatzas, K. A., Coldham, N. C., Piddock, L. J. V., and Woodward, M. J. (2007a). Fitness and dissemination of disinfectant-selected multiple-antibiotic-resistant (MAR) strains of Salmonella enterica serovar Typhimurium in chickens. J. Antimicrob. Chemother. 61, 156–162. doi: 10.1093/jac/dkm415
Randall, L. P., Cooles, S. W., Coldham, N. G., Penuela, E. G., Mott, A. C., Woodward, M. J., et al. (2007b). Commonly used farm disinfectants can select for mutant Salmonella enterica serovar Typhimurium with decreased susceptibility to biocides and antibiotics without compromising virulence. J. Antimicrob. Chemother. 60, 1273–1280. doi: 10.1093/jac/dkm359
Randall, L. P., Cooles, S. W., Piddock, L. J. V., and Woodward, M. J. (2004). Effect of triclosan or a phenolic farm disinfectant on the selection of antibiotic-resistant Salmonella enterica. J. Antimicrob. Chemoth. 54, 621–627. doi: 10.1093/jac/dkh376
Rebekah, N. (2011). Whitehead TWO. Exposure of Salmonella enterica Serovar Typhimurium to high level biocide challenge can select multidrug resistant mutants in a single step. PLoS ONE 7:e22833. doi: 10.1371/journal.pone.0022833
Rensing, C., Mitra, B., and Rosen, B. P. (1997). The zntA gene of Escherichia coli encodes a Zn(II)-translocating P-type ATPase. Proc. Natl. Acad. Sci. U. S. A. 94, 14326–14331. doi: 10.1073/pnas.94.26.14326
Rodrigue, A., Effantin, G., and Mandrand-Berthelot, M. (2005). Identification of rcnA (yohM), a nickel and cobalt resistance gene in Escherichia coli. J. Bacteriol. 8, 2912–2916. doi: 10.1128/JB.187.8.2912-2916.2005
Rossiter, S. E., Fletcher, M. H., and Wuest, W. M. (2017). Natural products as platforms to overcome antibiotic resistance. Chem. Rev. 117, 12415–12474. doi: 10.1021/acs.chemrev.7b00283
Roth, N., Kasbohrer, A., Mayrhofer, S., Zitz, U., Hofacre, C., and Domig, K. J. (2019). The application of antibiotics in broiler production and the resulting antibiotic resistance in Escherichia coli: a global overview. Poult. Sci. 98, 1791–1804. doi: 10.3382/ps/pey539
Russell, A. D. (2001). Mechanisms of bacterial insusceptibility to biocides. Am. J. Infect. Control. 29, 259–261. doi: 10.1067/mic.2001.115671
Samantha Mc Carlie, C. E. B. R. (2020). Molecular basis of bacterial disinfectant resistance. Drug Resist. Update 48:100672. doi: 10.1016/j.drup.2019.100672
Schaumburg, F., Onwugamba, F. C., Akulenko, R., Peters, G., Mellmann, A., Köck, R., et al. (2016). A geospatial analysis of flies and the spread of antimicrobial resistant bacteria. Int. J. Med. Microbiol. 306, 566–571. doi: 10.1016/j.ijmm.2016.06.002
Schuurmans, J. M., van Hijum, S. A. F. T., Piet, J. R., Händel, N., Smelt, J., Brul, S., et al. (2014). Effect of growth rate and selection pressure on rates of transfer of an antibiotic resistance plasmid between E. coli strains. Plasmid 72, 1–8. doi: 10.1016/j.plasmid.2014.01.002
Seier-Petersen, M. A., Jasni, A., Aarestrup, F. M., Vigre, H., Mullany, P., Roberts, A. P., et al. (2014). Effect of subinhibitory concentrations of four commonly used biocides on the conjugative transfer of Tn916 in Bacillus subtilis. J. Antimicrob. Chemother. 69, 343–348. doi: 10.1093/jac/dkt370
Seoane, J., Yankelevich, T., Dechesne, A., Merkey, B., Sternberg, C., and Smets, B. F. (2011). An individual-based approach to explain plasmid invasion in bacterial populations. Fems Microbiol. Ecol. 75, 17–27. doi: 10.1111/j.1574-6941.2010.00994.x
Sharma, V. K., Johnson, N., Cizmas, L., McDonald, T. J., and Kim, H. (2016). A review of the influence of treatment strategies on antibiotic resistant bacteria and antibiotic resistance genes. Chemosphere 150, 702–714. doi: 10.1016/j.chemosphere.2015.12.084
Sheridan, À., Lenahan, M., Duffy, G., Fanning, S., and Burgess, C. (2012). The potential for biocide tolerance in Escherichia coli and its impact on the response to food processing stresses. Food Control 26, 98–106. doi: 10.1016/j.foodcont.2012.01.018
Shun-Mei, E., Zeng, J., Yuan, H., Lu, Y., Cai, R., and Chen, C. (2018). Sub-inhibitory concentrations of fluoroquinolones increase conjugation frequency. Microb. Pathog. 114, 57–62. doi: 10.1016/j.micpath.2017.11.036
Slifierz, M. J., Friendship, R., and Weese, J. S. (2015b). Zinc oxide therapy increases prevalence and persistence of methicillin-resistant Staphylococcus aureus in pigs: a randomized controlled trial. Zoonoses Public Health 62, 301–308. doi: 10.1111/zph.12150
Slifierz, M. J., Friendship, R. M., and Weese, J. S. (2015a). Methicillin-resistant Staphylococcus aureus in commercial swine herds is associated with disinfectant and zinc usage. Appl. Environ. Microb. 81, 2690–2695. doi: 10.1128/AEM.00036-15
Solà-Ginés, M., González-López, J. J., Cameron-Veas, K., Piedra-Carrasco, N., Cerdà-Cuéllar, M., and Migura-Garcia, L. (2015). Houseflies (Musca domestica) as vectors for extended-spectrum β-Lactamase-producing Escherichia coli on Spanish broiler farms. Appl. Environ. Microb. 81, 3604–3611. doi: 10.1128/AEM.04252-14
Tagliabue, A., and Rappuoli, R. (2018). Changing priorities in vaccinology: antibiotic resistance moving to the top. Front. Immunol. 9:1068. doi: 10.3389/fimmu.2018.01068
Tetaz, T. J., and Luke, R. K. (1983). Plasmid-controlled resistance to copper in Escherichia coli. J. Bacteriol. 154, 1263–1268. doi: 10.1128/jb.154.3.1263-1268.1983
Tiseo, K., Huber, L., Gilbert, M., Robinson, T. P., and Van Boeckel, T. P. (2020). Global trends in antimicrobial use in food animals from 2017 to 2030. Antibiotics (Basel) 9:918. doi: 10.3390/antibiotics9120918
Tornimbene, B., Eremin, S., Escher, M., Griskeviciene, J., Manglani, S., and Pessoa-Silva, C. L. (2018). WHO global antimicrobial resistance surveillance system early implementation 2016–17. Lancet Infect. Dis. 18, 241–242. doi: 10.1016/S1473-3099(18)30060-4
Vahjen, W., Pietruszyńska, D., Starke, I. C., and Zentek, J. (2015). High dietary zinc supplementation increases the occurrence of tetracycline and sulfonamide resistance genes in the intestine of weaned pigs. Gut. Pathog. 7:23. doi: 10.1186/s13099-015-0071-3
Van Boeckel, T. P., Brower, C., Gilbert, M., Grenfell, B. T., Levin, S. A., Robinson, T. P., et al. (2015). Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. U. S. A. 112, 5649–5654. doi: 10.1073/pnas.1503141112
van den Bogaard, A. E., and Stobberingh, E. E. (1999). Antibiotic usage in animals: impact on bacterial resistance and public health. Drugs 58, 589–607. doi: 10.2165/00003495-199958040-00002
Wales, A., and Davies, R. (2015). Co-selection of resistance to antibiotics, biocides and heavy metals, and its relevance to foodborne pathogens. Antibiotics 4, 567–604. doi: 10.3390/antibiotics4040567
Wang, Y., Wang, Y., Sun, L., Grenier, D., and Yi, L. (2018). Streptococcus suis biofilm: regulation, drug-resistance mechanisms, and disinfection strategies. Appl. Microbiol. Biotechnol. 102, 9121–9129. doi: 10.1007/s00253-018-9356-z
Waseem, A., Arshad, J., Iqbal, F., Sajjad, A., Mehmood, Z., and Murtaza, G. (2014). Pollution status of Pakistan: a retrospective review on heavy metal contamination of water, soil, and vegetables. Biomed. Res. Int. 2014, 1–29. doi: 10.1155/2014/813206
Wistrand-Yuen, E., Knopp, M., Hjort, K., Koskiniemi, S., Berg, O. G., and Andersson, D. I. (2018). Evolution of high-level resistance during low-level antibiotic exposure. Nat. Commun. 9:1599. doi: 10.1038/s41467-018-04059-1
Witte, W., Klare, I., and Werner, G. (1999). Selective pressure by antibiotics as feed additives. Infection 27(Suppl. 2), S35–S38. doi: 10.1007/BF02561669
Xu, J., Sangthong, R., McNeil, E., Tang, R., and Chongsuvivatwong, V. (2020). Antibiotic use in chicken farms in northwestern China. Antimicrob. Resist. Infect. Control 9:10. doi: 10.1186/s13756-019-0672-6
Yang, Q. E., Agouri, S. R., Tyrrell, J. M., and Walsh, T. R. (2018). heavy metal resistance genes are associated with blaNDM-1- and blaCTX-M-15-carrying Enterobacteriaceae. Antimicrob. Agents Chemother. 62, e2617–e2642. doi: 10.1128/AAC.02642-17
Yang, S., Deng, W., Liu, S., Yu, X., Mustafa, G. R., Chen, S., et al. (2020). Presence of heavy metal resistance genes in Escherichia coli and Salmonella isolates and analysis of resistance gene structure in E. coli E308. J. Glob. Antimicrob. Resist. 21, 420–426. doi: 10.1016/j.jgar.2020.01.009
Yang, S., Wu, G., Long, M., Deng, W., Wang, H., and Zou, L. (2016). Antibiotic and disinfectant resistance of Salmonella isolated from egg production chains. Yi Chuan 38, 948–956. doi: 10.16288/j.yczz.16-185
Yazdankhah, S., Rudi, K., and Bernhoft, A. (2014). Zinc and copper in animal feed - development of resistance and co-resistance to antimicrobial agents in bacteria of animal origin. Microb. Ecol. Health Dis. 25:25862. doi: 10.3402/mehd.v25.25862
Yi, J., Zhang, Y., Lin, W., Niu, B., and Chen, Q. (2019). Effect of polyhexamethylene biguanide functionalized silver nanoparticles on the growth of Staphylococcus aureus. Fems Microbiol. Lett. 366:fnz036. doi: 10.1093/femsle/fnz036
Zhang, A., He, X., Meng, Y., Guo, L., Long, M., Yu, H., et al. (2016). Antibiotic and disinfectant resistance of Escherichia coli isolated from retail meats in Sichuan, China. Microb. Drug Resist. (Larchmont, N.Y.) 22, 80–87. doi: 10.1089/mdr.2015.0061
Zhang, P. Y., Xu, P. P., Xia, Z. J., Wang, J., Xiong, J., and Li, Y. Z. (2013). Combined treatment with the antibiotics kanamycin and streptomycin promotes the conjugation of Escherichia coli. FEMS Microbiol. Lett. 348, 149–156. doi: 10.1111/1574-6968.12282
Zhao, F. J., Ma, Y., Zhu, Y., Tang, Z., and McGrath, S. P. (2015). Soil contamination in China: current status and mitigation strategies. Environ. Sci. Technol. 49, 750–759. doi: 10.1021/es5047099
Zhao, S., Wang, C. L., Chang, S. K., Tsai, Y. L., and Chou, C. H. (2019). Characterization of Escherichia coli isolated from day-old chicken fluff in Taiwanese hatcheries. Avian. Dis. 63, 9–16. doi: 10.1637/11935-072318-Reg.1
Keywords: acquired resistance, antibiotic, heavy metal, biocide, resistance mechanism, mitigation measures
Citation: Guo K, Zhao Y, Cui L, Cao Z, Zhang F, Wang X, Feng J and Dai M (2021) The Influencing Factors of Bacterial Resistance Related to Livestock Farm: Sources and Mechanisms. Front. Anim. Sci. 2:650347. doi: 10.3389/fanim.2021.650347
Received: 07 April 2021; Accepted: 23 August 2021;
Published: 16 September 2021.
Edited by:
Pasquale De Palo, University of Bari Aldo Moro, ItalyReviewed by:
Matthew Whitney Jorgensen, Agricultural Research Service, USDA, United StatesCopyright © 2021 Guo, Zhao, Cui, Cao, Zhang, Wang, Feng and Dai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Menghong Dai, ZGFpbWVuZ2hvbmdAbWFpbC5oemF1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.