
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Anal. Sci. , 03 January 2025
Sec. Omics
Volume 4 - 2024 | https://doi.org/10.3389/frans.2024.1512573
This article is part of the Research Topic Thought Leaders in Analytical Science Research View all 12 articles
Lysine carbamylation is a non-enzymatic protein post-translational modification (PTM) that plays important roles in regulating enzymatic activity and the pathogenesis of diseases such as atherosclerosis, rheumatoid arthritis, and uremia. The progress of understanding the roles of carbamylation in biological systems has been delayed due to lack of systematic assays to study its functions. To aggravate this scenario, carbamylation is a major artifact in proteomics analysis given that urea, which is used during sample preparation, induces carbamylation. In addition, anti-acetyllysine antibodies co-purify carbamylated and acetylated peptides. In a recent paper, we leveraged co-purification with anti-acetyllysine antibodies to develop a method for analyzing carbamylated proteomes. In this perspective article, we discuss how this method may be applied to characterize the physiological functions of carbamylation in humans and other biological models, as well as the utility of establishing novel disease biomarkers.
Post-translational modifications (PTMs) of proteins expand the repertoire properties of the 21 genetically coded amino acids. These modifications are critical for regulating protein function, activity, localization, interaction, and turnover. To date, there are over 400 known PTMs (Li et al., 2022), but most of the knowledge is concentrated around a few modifications, such as phosphorylation, glycosylation, acetylation, and ubiquitination. Carbamylation is a PTM that occurs via nonenzymatic modification of the amine groups of protein N-terminus or lysine side chains. Lysine carbamylation has been identified as an important regulator of disease development and thus serves as a biomarker. Mechanistically, carbamylation regulates protein/enzyme activity, gene expression, cell signaling and metabolism (Figure 1A). Mass spectrometry-based proteomic analysis provides an instrumental resource for discovering novel roles of protein PTMs, including carbamylation in biological systems. However, protein carbamylation is a major artifact in proteomics sample preparation, representing a major challenge. In this perspective article, we describe recent developments in proteomics analysis of protein carbamylation as well as opportunities and needs to advance the field.

Figure 1. Lysine carbamylation. (A) Biological functions of protein carbamylation. (B) Mechanism of protein carbamylation and decarbamylation. MPO; Myeloperoxidase. (C) A role of carbamylation on ornithine transcarbamylase in the urea cycle. Kcx-307; carbamylated lysine-307 (D) Structure of carbamyl-lysine and related molecules. The structural differences are highlighted by the colors.
The current mechanisms for known to induce lysine carbamylation are non-enzymatic reaction of cyanate, isocyanate or carbamoyl-phosphate that adds a carbamoyl moiety (–CONH2) forming Nε’-carbamyl-lysine (also known as carbamoyl-lysine or homocitrulline) (Figure 1B). Cyanate and isocyanate are formed by upstream degradation of urea or by oxidation of thiocyanate by myeloperoxidase (MPO) (Figure 1B) (Verbrugge et al., 2015). In the case of carbamoyl-phosphate, this is a natural metabolite of the organism, being an intermediate of the urea cycle and the de novo pyrimidine biosynthesis pathway (Joshi et al., 2015; Matsumoto et al., 2019; Chen et al., 2023). Carbamylation activates ornithine transcarbamylase (Hu et al., 2023), the first step of the urea cycle, by transferring the carbamyl moiety from carbamoyl phosphate to ornithine to produce citrulline (Figure 1C). Citrulline is converted to arginine by argininosuccinate lyase, which is consequently cleaved by arginase to release urea and recycle ornithine (Figure 1C) (Matsumoto et al., 2019). The released urea is excreted into urine to detoxify the body from nitrogen (Matsumoto et al., 2019). This mechanism is regulated by sirtuins, specifically, SIRT4 (Hu et al., 2023). SIRT4 removes carbamylation from ornithine transcarbamylase, reducing its activity (Figure 1C) (Hu et al., 2023). However, in amino acid excess (the main source of nitrogen in the body), SIRT4 expression is downregulated to increase urea production and excretion (Hu et al., 2023). In humans, the sirtuin family comprises of 7 enzymes that cleave multiple acyl groups from lysine residues, including acetylation, lactylation, and malonylation (Bheda et al., 2016). Therefore, it is possible that other members of this family also have decarbamylase activity. Sirtuins are part of a much larger group of proteins named histone deacetylases (HDACs) (Park and Kim, 2020); yet the decarbamylase activity of other HDACs has not been investigated. Therefore, the current literature suggests that lysine carbamylation occurs through non-enzymatic mechanisms, while its removal being catalyzed by enzymes, such as SIRT4. However, it is possible that other enzymes, particularly within HDAC families, may also exhibit decarbamylase activity.
The use of various buffers, detergents, and salts for efficient cell lysis and protein extraction is fundamental in proteomics. Urea, a common denaturing agent, was first used in protein denaturation in the early 20th century (Mirsky and Pauling, 1936) and has been used for proteomics since the 1990s (Henzel et al., 1993; Shevchenko et al., 1996). Urea effectively denatures and solubilizes proteins; however, it also induces carbamylation, an artifact that can mislead protein analysis results. Carbamylation occurs when urea decomposes into cyanate in aqueous solutions, which then reacts with N-terminal amino groups and the side chains of lysines (Figure 1B) (Marier and Rose, 1964; Kollipara and Zahedi, 2013). This artifact not only alters carbamylation levels in target proteins but also disrupts quantitative chemical labeling (e.g., tandem mass tags), blocks trypsin digestion, and modifies peptide charge, mass, and retention time (Stanfill et al., 2018). Moreover, artifactual carbamylated peptides negatively affect the enrichment of lysine-acetylated peptides during immuno-purification by competing for binding with anti-acetylated lysine antibodies. This cross-reaction is due to the structural similarities between carbamyl- and acetyl-lysines (Figure 1D) (Martinez-Val et al., 2017). Several strategies can mitigate artifact carbamylation, such as adjusting temperature, pH, urea concentration, and incubation time. Freshly prepared urea and room temperature sample preparation significantly reduces carbamylation since urea decomposes faster into cyanate at higher temperatures (Wisniewski et al., 2009). Detergents like sodium deoxycholate (SDC) can be used as an alternative to urea to avoid protein carbamylation. SDC improves protein denaturation and solubilization, increasing the number of detected proteins and enhancing spectra quality (Lau and Othman, 2019; Lin et al., 2008; Masuda et al., 2008; León et al., 2013; Shahinuzzaman et al., 2020). Using SDC limits carbamylation to endogenous levels, with studies showing over a 67% reduction in carbamylated lysine residues compared to urea-based buffers (Martinez-Val et al., 2017; Lau and Othman, 2019). Recently, we used SDC to avoid artifactual carbamylation and leveraged the copurification of carbamylated peptides using anti-acetylated lysine antibodies (You et al., 2023). We tested the approach to study the role of carbamylation in inflammation using a macrophage cell line treated with bacterial lipopolysaccharide (LPS). Quantitative analysis revealed 2,378 endogenously carbamylated peptides, 360 of which were regulated by LPS. Functional-enrichment analysis indicates that these proteins are involved in diverse cellular processes from metabolism to signaling pathways. We also found that carbamylation regulates ubiquitination machinery (You et al., 2023). Overall, the use of non-carbamylating denaturing agents, such as SDC, prevents the formation of artifactual carbamylation and the use of anti-acetyl-lysine antibodies enables a comprehensive analysis of the carbamylated proteome. This opens opportunities to discover new roles for protein carbamylation, evidenced by our discovery of carbamylation regulation of ubiquitination machinery.
As mentioned above for ornithine transcarbamylase, lysine carbamylation is a regulator of enzymatic activity. Carbamylation is also a component of enzyme catalytic sites. For instance, ureases bear a carbamylated lysine along with multiple histidines, and an aspartate in their catalytic site, which coordinate two metal ions (Proshlyakov et al., 2021). Most of the known ureases have two nickel ions but others have two iron ions, which induce the cleave and release of ammonia from urea (Figure 2A) (Proshlyakov et al., 2021). Similarly, most of the members of the cyclic amidohydrolase family have a carbamylated lysine in their catalytic site that coordinates metal ion binding (Guan et al., 2021; Huang, 2020; Huang et al., 2023). This family includes allantoinase, dihydroorotase, dihydropyrimidinase, hydantoinase, and imidase (Figure 2B) (Guan et al., 2021; Huang, 2020; Huang et al., 2023). For instance, in the case of Escherichia coli allantoinase, the catalytic site is composed of four histidines, one aspartate and one carbamylated lysine that coordinate two zinc ions (Figure 2C) (Huang et al., 2023). Therefore, lysine carbamylation is an important structural component of the catalytic site of the cyclic amidohydrolase family, helping in their activity of opening or closing the rings of their substrates (Guan et al., 2021; Huang, 2020; Huang et al., 2023). It might not be a coincidence that the cyclic aminohydrolases have similar mechanism than urease, their substrates are all ureides (Figure 2B). It is possible that elevated urea concentrations induce carbamylation, leading to the activation of these enzymes.
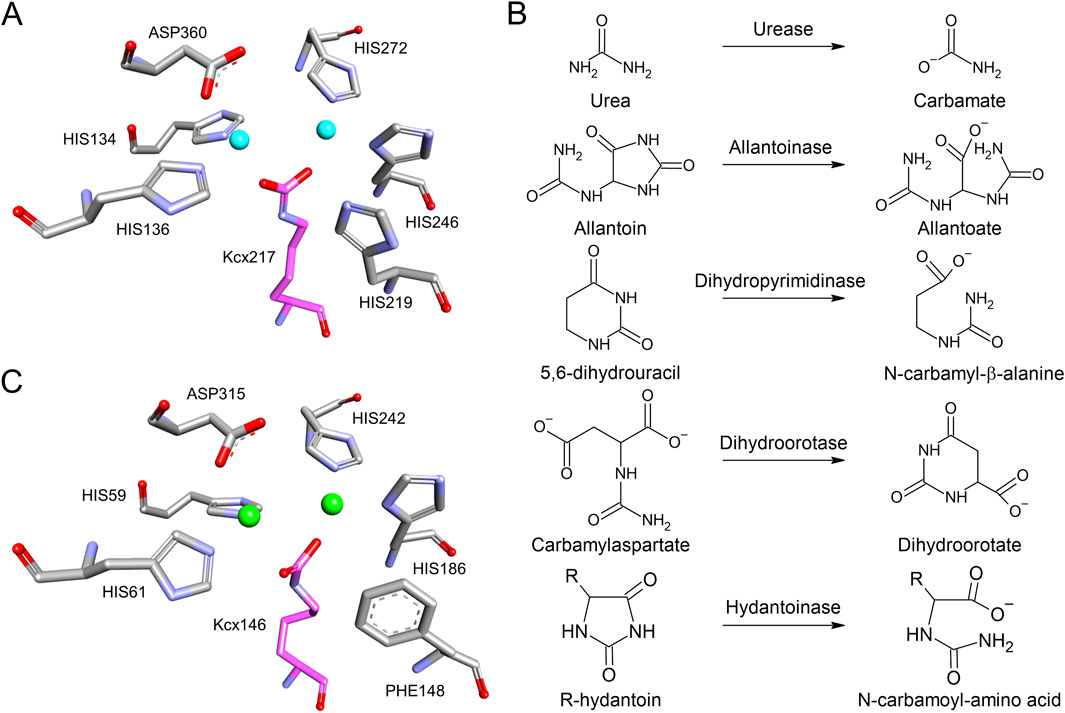
Figure 2. Carbamylated lysine in enzyme catalytic sites. (A) The catalytic site of urease from K. aerogenes (PDB 1FWJ). Nikel ions are presented by cyan spheres. (B) Substrates and products of urease and the cyclic amidohydrolase family, which have carbamylated lysine in their catalytic sites. (C) The catalytic site of E. coli allantoinase (PDB 8HFD). Zinc ions are presented by green spheres. Carbamyl-lysine is colored by magenta.
Another mechanism by which carbamylation regulates enzymatic activity is by modifying substrates and preventing them to be used or recognized by the enzymes. For example, carbamylation of ubiquitin blocks the formation of polyubiquitin chains (Pawloski et al., 2022). This can have detrimental effects on cells, as polyubiquitination regulates a variety of cellular processes, including targeting proteins for degradation in the proteasome. Indeed, carbamylated proteins are not degraded in the proteasome and accumulate in cells (Desmons et al., 2019). Carbamylation also affects the de-polymerization of polyubiquitin chains (You et al., 2023). We have recently shown that carbamylation of M1-linked diubiquitin blocks it from being cleaved by the deubiquitinase OTULIN (You et al., 2023). In summary, carbamylation has important roles in regulating enzymatic activity. The advances in carbamylated proteomics analysis will enable to further study the role of carbamylation in other enzymes and proteins with other functions.
In vivo carbamylation can be mediated by myeloperoxidase, a critical inducer of neutrophile extracellular trap (NET) formation, and by urea-derived cyanate (Figure 1B). MPO-mediated protein carbamylation is implicated in the pathogenesis of numerous diseases such as atherosclerosis, rheumatoid arthritis, periodontitis, and chronic kidney disease (Wang et al., 2007; Shi et al., 2011; Gorisse et al., 2016). MPO-induced carbamylation of lipoproteins is implicated in the pathogenesis of atherosclerosis (Wang et al., 2007). MPO is enriched in atherosclerotic lesions (Hazen, 2004) and has been shown to carbamylate low-density lipoprotein (LDL) and cardioprotective high density lipoprotein (HDL), resulting in proatherosclerotic conversion of HDL (Denimal, 2023). Carbamylated HDL is elevated in both type 1 (T1D) and type 2 diabetes (T2D), where it correlates to patient cardiovascular risk in T2D and glycemic control in T1D (Denimal, 2023). Carbamylated LDL is also elevated in T2D prior to kidney dysfunction, where it has been shown to directly induce epithelial cell death, suggestive of a central role of carbamylation in the concurrent pathogenesis of T2D, atherosclerosis, and chronic kidney disease (Shiu et al., 2014; Ok et al., 2005). Carbamylated LDL promotes thrombus formation, oxidative stress, and endothelial nitric oxide synthase uncoupling (Denimal, 2023). Smoking promotes MPO-mediated carbamylation and is thought to be a critical mediator of smoking-induced atherosclerosis (Wang et al., 2007). Given de-carbamylation is dependent on enzyme availability and often modifies proteins with long half-lives, including extracellular matrix proteins, carbamylation has been proposed as a biomarker of aging as its tissue accumulation is inversely associated with lifespan (Gorisse et al., 2016). Such associations of this PTM with the pathogenesis of numerous diseases make the development of methods to study carbamylation critical. While we have outlined several lines of evidence, suggesting the overall biological significance of carbamylation, further developments enabling high throughput analysis of global protein carbamylation is needed to characterize its biological significance, regulation, and function.
Given anti-citrullinated protein antibodies (ACPAs) are well-established biomarkers of rheumatoid arthritis (RA), antibodies to other PTMs, such as carbamylated proteins have also been explored as disease biomarkers. Citrullination and carbamylation result in the structurally similar PTMs citrulline and carbamyl-lysine, respectively (Figure 1D). While less well-explored, anti-carbamylated protein (anti-CarP) antibodies are detected in nearly half of patients with RA and may serve as useful biomarkers to predict both preclinical disease and disease progression to bone erosion (Shi et al., 2011; O'Neil et al., 2020). Such PTMs accumulate over time and are especially long-lived in the joint given the slow turnover of extracellular matrix proteins in the articular cartilage of synovial joints. In transgenic mice overexpressing the humanized RA MHC II HLADRB1* 04:01, NETs externalize carbamylated peptides and induce anti-carbamylated antibody responses (O'Neil et al., 2020). However, protein carbamylation in the inflamed human joint has not been thoroughly investigated (McHugh, 2023). Several other studies have suggested a role of carbamylated proteins in autoimmunity. Ex vivo cell culture models have shown that carbamylation of vimentin induces T-cell reactivity as indicated by IFNγ production (Choudhury et al., 2023). Carbamylated aldolase and cytokeratin display increased MHC I binding affinity compared to non-modified proteins (Shah et al., 2023). It appears that not all individuals with RA that are ACPA-positive display anti-CarP antibodies. In systemic lupus erythematosus (SLE), anti-CarP antibodies correlate with markers of systemic inflammation and arthralgia (Li et al., 2020). A consequence of many autoimmune diseases includes progressive aging phenotypes, such as early-onset of age-related muscle wasting (sarcopenia) and cardiometabolic disease, both of which target striated muscle. However, the mechanisms leading to such phenotypes are mainly described as chronic inflammation-related and lack well-defined targets aside from anti-inflammatory therapeutics primarily targeting direct disease activity (Casanova-Vallve et al., 2020; Mehta et al., 2023). Tissue-specific neutrophil activation and NET formation are implicated in cardiovascular and sarcopenic phenotypes that are increased in autoimmune diseases (Ma, 2021; Nie et al., 2024). For example, skeletal muscle IL-8 abundance, an activator of neutrophil NET formation, correlates to disease activity in RA patients (Huffman et al., 2017). In patients with heart failure with dilated cardiomyopathy, cardiac NET abundance correlates with cardiac dysfunction and clinical outcomes (Ichimura et al., 2024). Together, these data suggest a potential role of carbamylation in mediating the pathogenesis and severity of both cardiometabolic and autoimmune disease phenotypes and highlight the vast clinical utility of studying protein carbamylation as a molecular regulator and target in human diseases.
Co-purification of acetylated and carbamylated peptides with anti-acetyl-lysine antibodies facilitated our method development for analyzing carbamylated proteomes (You et al., 2023). This method also provides a future opportunity to integrate multiple PTMs in proteomic analyses as carbamylation and acetylation data can be obtained simultaneously. However, the similar masses of lysine acetylation (+42.0103 Da) and carbamylation (+43.00543 Da) represents a challenge for reliable peptide identification. This is especially relevant when comparing carbamylation to the 13C isotope of acetylation +43.0137 Da, only an 8 mDa difference. Isotopic correction, mass recalibration, and narrow mass error tolerances during peptide identification can reduce misidentification (You et al., 2023). To improve identification reliability, peptides can be validated by the presence of characteristic fragments in the tandem mass spectrum. For instance, the fragmentation of acetyl-lysines generates a fragment at m/z 126.1 (Nakayasu et al., 2014). This fragment is present in 98% of the acetylated peptide tandem mass spectra (Trelle and Jensen, 2008), but absent in tandem mass spectra of carbamylated peptides (Guo et al., 2018). A diagnostic fragment for carbamylation has not been described yet, but as more data becomes available, it opens the opportunity of performing systematic studies on the fragmentation patterns (Geiszler et al., 2023). This can improve the annotation of spectra from carbamylated peptides.
The development of specific affinity purification methods for carbamylated peptides provides the opportunity to reduce misidentification and may even improve proteome coverage as the sample complexity would be reduced. This is a challenging issue because the antibodies should have a broad specificity to enrich carbamylated peptides, but at the same time should distinguish carbamylation from acetylation and citrullination. We have previously tested one anti-carbamylation antibody conjugated to agarose beads (Cayman), but it was not ideal for proteomics analysis and only yielded identification of 128 carbamylated peptides (unpublished observations). Another point to consider is that carbamylated antibodies display significant cross-reactivity with citrullinated peptides. For instance, Sahlström et al. tested 12 anti-citrullination monoclonal antibodies and observed half (6) to display significant cross-reactivity to protein carbamylation (Sahlstrom et al., 2020). Therefore, there is a need to systematically test anti-carbamylation antibody specificity and performance to develop proteomics assays.
As mentioned above, the method developed for analyzing carbamylation has already enabled the discovery of new functions for this PTM. However, there are remaining technical needs for the development of reliable carbamylated peptide identification and enrichment methods.
Increasing carbamylated proteome coverage will open opportunities to study the role of this PTM in metabolic regulation and cell signaling. The current literature demonstrates that carbamylation regulates nitrogen metabolism, including the urea cycle and metabolism of ureides. The fact that carbamylation is non-enzymatic, makes us believe that it may result in lysine modifications in catalytic or co-factor-binding sites of enzymes. Such modifications could then activate the enzymes, such as ureases and cyclic amidohydrolases, or inhibit enzymes that require the free primary amine groups of the lysine residues. We have identified similar mechanism in which acetylation modifies and subsequently inhibits enzymes involved in the central carbon metabolism in bacteria (Nakayasu et al., 2017).
Similar to acetylation, carbamylation is implicated in many cell-signaling processes as well. For instance, recognition of carbamylated TLR5 by anti-carbamylated protein autoantibodies induces receptor activation and pro-inflammatory signaling. Another example is the suppression of mTOR signaling by carbamylation (Wang et al., 2019). In chronic kidney disease, high urea levels are associated with depression by inducing the carbamylation of mTOR, which leads to loss of neuronal synapses (Wang et al., 2019). In-depth carbamylated proteome analysis integrated with other PTMs will be instrumental for determining new roles for carbamylation in cell signaling. A multi-PTM approach facilitates comprehensive characterization of differential pathway regulation by different PTMs. Our current method analyzes carbamylation and acetylation simultaneously, and additional PTMs can be easily obtained by sequential purification on the same sample, spurring additional mechanistic insight. In our previous work, we combined carbamylation, acetylation, and phosphorylation to identify a function of carbamylation in regulating ubiquitination machinery (You et al., 2023). In addition to multi-omics integration, structural analysis is instrumental for identifying important carbamylated modification sites. Modifications on catalytic or cofactor-binding sites, as well in interacting regions and activation loops on proteins can provide key functional information for PTMs. We have used such approach to study the role of PTMs in bacteria and to discover how carbamylation on ubiquitin regulates the activity of the deubiquitinase OTULIN (You et al., 2023; Nakayasu et al., 2017; Feid et al., 2022; Walukiewicz et al., 2024). In summary, we believe that analysis of carbamylated proteomes integrated with other omics and structural analyses will have major impact in discovery new roles for carbamylation in regulating the cell metabolism and signaling.
Histones are among the most carbamylated proteins. In neutrophils, MPO produces cyanate that induces histone carbamylation (O'Neil et al., 2020). Histone carbamylation is also induced by carbamoyl-phosphate produced by carbamoyl-phosphate synthase located in the nucleus of the cell (Joshi et al., 2015). Similar to acetylation, carbamylation also promotes transcriptional activation (Joshi et al., 2015). However, it is not known whether acetylation and carbamylation share the same transcription regulators or how the dynamics between these two modifications regulate transcription. Similar to how mass spectrometry contributes to understanding the role of acetylation and other PTMs on histones, it will likely provide insight into the functional role of carbamylation on gene regulation. Mass spectrometry analysis of carbamylated histone proteins can provide a detailed view of the functional role of PTMs in disease pathogenesis and treatment. The combination of these PTM signatures with chromatin immunoprecipitation with sequencing (ChIP-seq), assay for transposase-accessible chromatin with sequencing (ATAC-seq), and RNA sequencing should illustrate transcriptional network regulation by carbamylation. In addition, affinity purification-mass spectrometry (AP-MS) can be used to discover factors that bind to carbamylated proteins, facilitating the identification of transcription factors that are regulated by carbamylation. Furthermore, mass spectrometry can be used to discover decarbamylases, which may play a role in downregulating gene expression. Overall, mass spectrometry will be instrumental in determining the roles of carbamylation in the regulation of gene expression.
Current and future advances in protein carbamylation analysis mass spectrometry will also have an impact on biomarker discovery and validation. As discussed above, carbamylation is involved in a variety of conditions, making carbamylated proteins promising biomarkers. In biomarker studies, it is often crucial to prepare and analyze hundreds to thousands of samples. A recently developed automated platform to enrich for acetylated peptides can be applied to carbamylation analysis in the future (Gritsenko et al., 2024). This automated enrichment platform with the increased speed of the new generations of LC-MS, allows for the analysis of dozens to hundreds of samples a day. Therefore, the current technology is ready for carrying out large scale biomarker studies of protein carbamylation.
Lysine carbamylation has a variety of roles in human physiology and disease pathogenesis. However, progress in comprehensively understanding the role and mechanisms of protein carbamylation in health and disease has been slow given it is a major artifact in proteomic analyses. Development of workflows to perform carbamylation/acetylation proteomics opens many opportunities to study the roles of carbamylation in health and in disease. Further optimization of proteomic analyses along with multi-PTM integration will further advance our understanding of the functional role of protein carbamylation.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
YY: Conceptualization, Visualization, Writing–original draft, Writing–review and editing. GM: Conceptualization, Visualization, Writing–original draft, Writing–review and editing. EN: Conceptualization, Funding acquisition, Visualization, Writing–original draft, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Catalyst Award from the Human Islet Research Network (HIRN) (to EN) (via U24 DK104162) and by National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grants R01 DK138335 (to EN) and U01 DK127505 (to EN).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
The authors declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Bheda, P., Jing, H., Wolberger, C., and Lin, H. (2016). The substrate specificity of sirtuins. Annu. Rev. Biochem. 85, 405–429. doi:10.1146/annurev-biochem-060815-014537
Casanova-Vallve, N., Constantin-Teodosiu, D., Filer, A., Hardy, R. S., Greenhaff, P. L., and Chapman, V. (2020). Skeletal muscle dysregulation in rheumatoid arthritis: metabolic and molecular markers in a rodent model and patients. PLoS One 15, e0235702. doi:10.1371/journal.pone.0235702
Chen, J., Yang, S., Li, Y., Ziwen, X., Zhang, P., Song, Q., et al. (2023). De novo nucleotide biosynthetic pathway and cancer. Genes Dis. 10, 2331–2338. doi:10.1016/j.gendis.2022.04.018
Choudhury, R. H., Daniels, I., Vaghela, P., Atabani, S., Kirk, T., Symonds, P., et al. (2023). Immune responses to citrullinated and homocitrullinated peptides in healthy donors are not restricted to the HLA SE shared allele and can be selected into the memory pool. Immunology 169, 467–486. doi:10.1111/imm.13645
Denimal, D. (2023). Carbamylated lipoproteins in diabetes. World J. Diabetes 14, 159–169. doi:10.4239/wjd.v14.i3.159
Desmons, A., Okwieka, A., Doue, M., Gorisse, L., Vuiblet, V., Pietrement, C., et al. (2019). Proteasome-dependent degradation of intracellular carbamylated proteins. Aging (Albany NY) 11, 3624–3638. doi:10.18632/aging.102002
Feid, S. C., Walukiewicz, H. E., Wang, X., Nakayasu, E. S., Rao, C. V., and Wolfe, A. J. (2022). Regulation of translation by lysine acetylation in Escherichia coli. mBio 13, e0122422. doi:10.1128/mbio.01224-22
Geiszler, D. J., Polasky, D. A., Yu, F., and Nesvizhskii, A. I. (2023). Detecting diagnostic features in MS/MS spectra of post-translationally modified peptides. Nat. Commun. 14, 4132. doi:10.1038/s41467-023-39828-0
Gorisse, L., Pietrement, C., Vuiblet, V., Schmelzer, C. E., Kohler, M., Duca, L., et al. (2016). Protein carbamylation is a hallmark of aging. Proc. Natl. Acad. Sci. U. S. A. 113, 1191–1196. doi:10.1073/pnas.1517096113
Gritsenko, M. A., Tsai, C. F., Kim, H., and Liu, T. (2024). Automated immunoprecipitation workflow for comprehensive acetylome analysis. Methods Mol. Biol. 2823, 173–191. doi:10.1007/978-1-0716-3922-1_12
Guan, H. H., Huang, Y. H., Lin, E. S., Chen, C. J., and Huang, C. Y. (2021). Structural analysis of Saccharomyces cerevisiae dihydroorotase reveals molecular insights into the tetramerization mechanism. Molecules 26, 7249. doi:10.3390/molecules26237249
Guo, C., Guo, X., Zhao, L., Chen, D., Wang, J., and Sun, J. (2018). Optimization of carbamylation conditions and study on the effects on the product ions of carbamylation and dual modification of the peptide by Q-TOF MS. Eur. J. Mass Spectrom. 24, 384–396. doi:10.1177/1469066718788665
Hazen, S. L. (2004). Myeloperoxidase and plaque vulnerability. Arterioscler. Thromb. Vasc. Biol. 24, 1143–1146. doi:10.1161/01.ATV.0000135267.82813.52
Henzel, W. J., Billeci, T. M., Stults, J. T., Wong, S. C., Grimley, C., and Watanabe, C. (1993). Identifying proteins from two-dimensional gels by molecular mass searching of peptide fragments in protein sequence databases. Proc. Natl. Acad. Sci. U. S. A. 90, 5011–5015. doi:10.1073/pnas.90.11.5011
Hu, S. H., Feng, Y. Y., Yang, Y. X., Ma, H. D., Zhou, S. X., Qiao, Y. N., et al. (2023). Amino acids downregulate SIRT4 to detoxify ammonia through the urea cycle. Nat. Metab. 5, 626–641. doi:10.1038/s42255-023-00784-0
Huang, C. Y. (2020). Structure, catalytic mechanism, posttranslational lysine carbamylation, and inhibition of dihydropyrimidinases. Adv. Protein Chem. Struct. Biol. 122, 63–96. doi:10.1016/bs.apcsb.2020.05.002
Huang, Y. H., Yang, P. C., Lin, E. S., Ho, Y. Y., Peng, W. F., Lu, H. P., et al. (2023). Crystal structure of allantoinase from Escherichia coli BL21: a molecular insight into a role of the active site loops in catalysis. Molecules 28, 827. doi:10.3390/molecules28020827
Huffman, K. M., Jessee, R., Andonian, B., Davis, B. N., Narowski, R., Huebner, J. L., et al. (2017). Molecular alterations in skeletal muscle in rheumatoid arthritis are related to disease activity, physical inactivity, and disability. Arthritis Res. Ther. 19, 12. doi:10.1186/s13075-016-1215-7
Ichimura, S., Misaka, T., Ogawara, R., Tomita, Y., Anzai, F., Sato, Y., et al. (2024). Neutrophil extracellular traps in myocardial tissue drive cardiac dysfunction and adverse outcomes in patients with heart failure with dilated cardiomyopathy. Circ. Heart Fail 17, e011057. doi:10.1161/CIRCHEARTFAILURE.123.011057
Joshi, A. D., Mustafa, M. G., Lichti, C. F., and Elferink, C. J. (2015). Homocitrullination is a novel histone H1 epigenetic mark dependent on aryl hydrocarbon receptor recruitment of carbamoyl phosphate synthase 1. J. Biol. Chem. 290, 27767–27778. doi:10.1074/jbc.M115.678144
Kollipara, L., and Zahedi, R. P. (2013). Protein carbamylation: in vivo modification or in vitro artefact? Proteomics 13, 941–944. doi:10.1002/pmic.201200452
Lau, B. Y. C., and Othman, A. (2019). Evaluation of sodium deoxycholate as solubilization buffer for oil palm proteomics analysis. PLOS ONE 14, e0221052. doi:10.1371/journal.pone.0221052
León, I. R., Schwämmle, V., Jensen, O. N., and Sprenger, R. R. (2013). Quantitative assessment of in-solution digestion efficiency identifies optimal protocols for unbiased protein analysis. Mol. and Cell. Proteomics 12, 2992–3005. doi:10.1074/mcp.M112.025585
Li, Y., Jia, R., Liu, Y., Tang, S., Ma, X., Shi, L., et al. (2020). Antibodies against carbamylated vimentin exist in systemic lupus erythematosus and correlate with disease activity. Lupus 29, 239–247. doi:10.1177/0961203319897127
Li, Z. Y., Li, S. F., Luo, M. Q., Jhong, J. H., Li, W. S., Yao, L. T., et al. (2022). dbPTM in 2022: an updated database for exploring regulatory networks and functional associations of protein post-translational modifications. Nucleic Acids Res. 50, D471–D479. doi:10.1093/nar/gkab1017
Lin, Y., Zhou, J., Bi, D., Chen, P., Wang, X. C., and Liang, S. P. (2008). Sodium-deoxycholate-assisted tryptic digestion and identification of proteolytically resistant proteins. Anal. Biochem. 377, 259–266. doi:10.1016/j.ab.2008.03.009
Ma, Y. (2021). Role of neutrophils in cardiac injury and repair following myocardial infarction. Cells 10, 1676. doi:10.3390/cells10071676
Marier, J. R., and Rose, D. (1964). Determination of cyanate, and a study of its accumulation in aqueous solutions of urea. Anal. Biochem. 7, 304–314. doi:10.1016/0003-2697(64)90135-6
Martinez-Val, A., Garcia, F., Ximénez-Embún, P., Martínez Teresa-Calleja, A., Ibarz, N., Ruppen, I., et al. (2017). Urea artifacts interfere with immuno-purification of lysine acetylation. J. Proteome Res. 16, 1061–1068. doi:10.1021/acs.jproteome.6b00463
Masuda, T., Tomita, M., and Ishihama, Y. (2008). Phase transfer surfactant-aided trypsin digestion for membrane proteome analysis. J. Proteome Res. 7, 731–740. doi:10.1021/pr700658q
Matsumoto, S., Haberle, J., Kido, J., Mitsubuchi, H., Endo, F., and Nakamura, K. (2019). Urea cycle disorders-update. J. Hum. Genet. 64, 833–847. doi:10.1038/s10038-019-0614-4
McHugh, J. (2023). Carbamylated NETs promote bone erosion in RA. Nat. Rev. Rheumatol. 19, 193. doi:10.1038/s41584-023-00938-0
Mehta, P. K., Levit, R. D., Wood, M. J., Aggarwal, N., O'Donoghue, M. L., Lim, S. S., et al. (2023). Chronic rheumatologic disorders and cardiovascular disease risk in women. Am. Heart J. Plus 27, 100267. doi:10.1016/j.ahjo.2023.100267
Mirsky, A. E., and Pauling, L. (1936). On the structure of native, denatured, and coagulated proteins. Proc. Natl. Acad. Sci. 22, 439–447. doi:10.1073/pnas.22.7.439
Nakayasu, E. S., Burnet, M. C., Walukiewicz, H. E., Wilkins, C. S., Shukla, A. K., Brooks, S., et al. (2017). Ancient regulatory role of lysine acetylation in central metabolism. mBio 8, e01894. doi:10.1128/mBio.01894-17
Nakayasu, E. S., Wu, S., Sydor, M. A., Shukla, A. K., Weitz, K. K., Moore, R. J., et al. (2014). A method to determine lysine acetylation stoichiometries. Int. J. Proteomics 2014, 730725. doi:10.1155/2014/730725
Nie, H., Liu, Y., Zeng, X., and Chen, M. (2024). Neutrophil-to-lymphocyte ratio is associated with sarcopenia risk in overweight maintenance hemodialysis patients. Sci. Rep. 14, 3669. doi:10.1038/s41598-024-54056-2
Ok, E., Basnakian, A. G., Apostolov, E. O., Barri, Y. M., and Shah, S. V. (2005). Carbamylated low-density lipoprotein induces death of endothelial cells: a link to atherosclerosis in patients with kidney disease. Kidney Int. 68, 173–178. doi:10.1111/j.1523-1755.2005.00391.x
O'Neil, L. J., Barrera-Vargas, A., Sandoval-Heglund, D., Merayo-Chalico, J., Aguirre-Aguilar, E., Aponte, A. M., et al. (2020). Neutrophil-mediated carbamylation promotes articular damage in rheumatoid arthritis. Sci. Adv. 6, eabd2688. doi:10.1126/sciadv.abd2688
Park, S. Y., and Kim, J. S. (2020). A short guide to histone deacetylases including recent progress on class II enzymes. Exp. Mol. Med. 52, 204–212. doi:10.1038/s12276-020-0382-4
Pawloski, W., Komiyama, T., Kougentakis, C., Majumdar, A., and Fushman, D. (2022). Site-specific detection and characterization of ubiquitin carbamylation. Biochemistry 61, 712–721. doi:10.1021/acs.biochem.2c00085
Proshlyakov, D. A., Farrugia, M. A., Proshlyakov, Y. D., and Hausinger, R. P. (2021). Iron-Containing ureases. Coord. Chem. Rev. 448, 214190. doi:10.1016/j.ccr.2021.214190
Sahlstrom, P., Hansson, M., Steen, J., Amara, K., Titcombe, P. J., Forsstrom, B., et al. (2020). Different hierarchies of anti-modified protein autoantibody reactivities in rheumatoid arthritis. Arthritis Rheumatol. 72, 1643–1657. doi:10.1002/art.41385
Shah, S., Cook, K. W., Symonds, P., Weisser, J., Skinner, A., Al Omari, A., et al. (2023). Vaccination with post-translational modified, homocitrullinated peptides induces CD8 T-cell responses that mediate antitumor immunity. J. Immunother. Cancer 11, e006966. doi:10.1136/jitc-2023-006966
Shahinuzzaman, A. D. A., Chakrabarty, J. K., Fang, Z. X., Smith, D., Kamal, A. M., and Chowdhury, S. M. (2020). Improved in-solution trypsin digestion method for methanol-chloroform precipitated cellular proteomics sample. J. Sep. Sci. 43, 2125–2132. doi:10.1002/jssc.201901273
Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858. doi:10.1021/ac950914h
Shi, J., Knevel, R., Suwannalai, P., van der Linden, M. P., Janssen, G. M., van Veelen, P. A., et al. (2011). Autoantibodies recognizing carbamylated proteins are present in sera of patients with rheumatoid arthritis and predict joint damage. Proc. Natl. Acad. Sci. U. S. A. 108, 17372–17377. doi:10.1073/pnas.1114465108
Shiu, S. W., Xiao, S. M., Wong, Y., Chow, W. S., Lam, K. S., and Tan, K. C. (2014). Carbamylation of LDL and its relationship with myeloperoxidase in type 2 diabetes mellitus. Clin. Sci. (Lond) 126, 175–181. doi:10.1042/CS20130369
Stanfill, B. A., Nakayasu, E. S., Bramer, L. M., Thompson, A. M., Ansong, C. K., Clauss, T. R., et al. (2018). Quality control analysis in real-time (QC-ART): a tool for real-time quality control assessment of mass spectrometry-based proteomics data. Mol. Cell Proteomics 17, 1824–1836. doi:10.1074/mcp.RA118.000648
Trelle, M. B., and Jensen, O. N. (2008). Utility of immonium ions for assignment of epsilon-N-acetyllysine-containing peptides by tandem mass spectrometry. Anal. Chem. 80, 3422–3430. doi:10.1021/ac800005n
Verbrugge, F. H., Tang, W. H., and Hazen, S. L. (2015). Protein carbamylation and cardiovascular disease. Kidney Int. 88, 474–478. doi:10.1038/ki.2015.166
Walukiewicz, H. E., Farris, Y., Burnet, M. C., Feid, S. C., You, Y., Kim, H., et al. (2024). Ancient regulatory role of lysine acetylation in central metabolism. mBio 8, e01894. doi:10.1128/mBio.01894-17
Wang, H., Huang, B., Wang, W., Li, J., Chen, Y., Flynn, T., et al. (2019). High urea induces depression and LTP impairment through mTOR signalling suppression caused by carbamylation. EBioMedicine 48, 478–490. doi:10.1016/j.ebiom.2019.09.049
Wang, Z., Nicholls, S. J., Rodriguez, E. R., Kummu, O., Horkko, S., Barnard, J., et al. (2007). Protein carbamylation links inflammation, smoking, uremia and atherogenesis. Nat. Med. 13, 1176–1184. doi:10.1038/nm1637
Wisniewski, J. R., Zougman, A., Nagaraj, N., and Mann, M. (2009). Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362. doi:10.1038/nmeth.1322
Keywords: carbamylation, post-translational modification, proteomics, function, disease
Citation: You Y, Many G and Nakayasu ES (2025) Protein carbamylation and proteomics: from artifacts to elucidation of biological functions. Front. Anal. Sci. 4:1512573. doi: 10.3389/frans.2024.1512573
Received: 16 October 2024; Accepted: 06 December 2024;
Published: 03 January 2025.
Edited by:
Makusu Tsutsui, Osaka University, JapanReviewed by:
Liam McDonnell, Fondazione Pisana per la Scienza Onlus, ItalyCopyright © 2025 You, Many and Nakayasu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ernesto S. Nakayasu, ZXJuZXN0by5uYWtheWFzdUBwbm5sLmdvdg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.