- 1Biogenix Labs, G42 Healthcare, Abu Dhabi, United Arab Emirates
- 2Research, G42 Healthcare, Abu Dhabi, United Arab Emirates
- 3Insights Research Organization and Solutions (IROS), Abu Dhabi, United Arab Emirates
- 4United Arab Emirates University, Al-Ain, United Arab Emirates
- 5Khalifa University, Abu Dhabi, United Arab Emirates
Introduction: The COVID-19 pandemic has led to the rapid development and launch of several commercial RT-PCR-based assays for identification of SARS-CoV-2. However, there is need for peer-reviewed evaluation of these assays that can support their clinical performance. In this study, we, therefore, conduct an in-house evaluation of the automated Cobas 68000 RT-PCR assay in detecting SARS-CoV-2 infections using different pooling techniques.
Methods: An observational study is conducted to evaluate the clinical performance of the Cobas 6800 SARS-CoV-2 assay in comparison with the Labgun Exofast RT-PCR kit, using both pooled and non-pooled sample techniques. A total of 300 nasopharyngeal swab samples, 40 known positive samples and 260 negative samples, are used for pooling, while the performance is evaluated in three different sample pool sizes of 4, 5, and 6.
Results: The sensitivity and specificity of the Cobas 6,800 was 100% when compared to the comparable assay. The sample pooling technique showed that specificity was 100% in all pool sizes and the sensitivity varied from 95% in the 6-pooled sample to 100% in both the 5- and 4-pooled samples. The lower limit of detection was verified as 25 copies/ml for un-pooled samples, and, therefore, the limit of detection was 100, 125, and 150 copies/ml for the 4, 5, and 6 sample pools, respectively. Strong correlation was observed between the Ct values of the target genes of both assays.
Conclusion: Cobas 6800 RT-PCR assay is a reliable platform for qualitative and rapid detection of SARS-CoV-2 and can be effectively utilized for pooling of samples with highly efficient performance when disease prevalence is lower.
Introduction
As of September 2022, the COVID-19 pandemic had led to more than 614 million laboratory-confirmed cases that were positive for SARS-CoV-2 (WHO, 2022). The rising number of cases has burdened diagnostic facilities around the world, as laboratory diagnosis of COVID-19 is necessary to identify infections, monitor progress, and effect contact tracing. Apart from diagnosis, population-wide screening is carried out to prevent community spread of the disease. A negative RT-PCR test result for SARS-CoV-2 virus is a requirement for travel and for access to public areas as a part of pandemic management guidelines. These public health measures and large-scale screenings have brought new challenges to screening methods including the need to catch up with increasing demands for tests requiring less sophistication and expertise, and a faster turnaround time.
The reverse transcription real-time PCR (RT-PCR) is the most commonly recommended test for laboratory diagnosis of SARS-CoV-2, and it helps detect at least two genes from the target genes of SARS-CoV-2. The target genes of SARS-CoV-2 for diagnostic purposes include the specific structural spike protein (S), the envelope (E), the nucleocapsid (N) genes, the nonstructural RNA-dependent RNA polymerase (RdRp), and the replicase open reading frame (ORF) 1a/b, ORF 1 b-nsp14 (Abduljalil, 2020; Sharma et al., 2021). Thus far, the RT-PCR remains the most feasible, suitable, and reliable diagnostic test for identifying SARS-CoV-2 infections (Chung et al., 2021). However, there are some limitations with this method as the RT-PCR platform requires expertise and facilities with appropriate infrastructure. Distinct primers and probes are required for every target, which limits prompt scaling up for other nucleic acid targets (Aziz et al., 2021). Some of the other concerns about RT-PCR platforms are that the sensitivity is affected by sampling errors, the viral load of the sample, and the lack of differentiation between live and inactive viral fragments (Asif et al., 2021).
To accommodate the huge demand, there is increasing need to develop efficient automated solutions and effective sample pooling strategies. Sample pooling has been a vital strategy for testing large sample volumes where extracts from a random number of samples are combined into a single tube for analysis. This strategy is highly advantageous when disease prevalence is low. Studies on the feasibility and the accuracy of sample pooling techniques have shown the strategy to be an effective approach for wide-scale population screening (Sahajpal et al., 2020). The method has proven cost-effective in mass screening, and with optimization of pool size, the pooling strategy can help with substantially reducing cost and turnaround time without compromising sensitivity (Cherif et al., 2020; Hogan et al., 2020). Despite the advantages of pooling strategies, sample pooling can be very challenging, especially when the pool is larger. Deconvoluting larger pools can be challenging and time-consuming; therefore, optimizing pool size based on the positivity rate and the technique used becomes crucial (Mishra et al., 2020; Mahmoud et al., 2021).
There is immense growth and expansion in the commercial SARS-CoV-2 RT-PCR kits used in fully automated systems. Automated solutions for molecular diagnostics can help handle the demand as large numbers of samples can be processed in a very short turnaround time with minimal hands-on required and can be scaled to keep pace with the increasing demands (Cobb et al., 2017). This current pandemic has led to rapid development and launch of several commercial RT-PCR-based assays, and evaluation of these assays is mandated. Nevertheless, it is necessary to evaluate the analytical performance of any diagnostic test to understand the capabilities and limitations of that test. The Cobas 68000 RT-PCR test is a fully enclosed automated system with minimal manual interaction and high throughput. Therefore, in this study, we conduct an in-house evaluation of the automated Cobas 68,000 RT-PCR assay for detecting SARS-CoV-2 infections using different pooling techniques.
Materials and methods
Study setting
The study was approved by the review board at the Department of Health (DOH), Abu Dhabi, UAE. Informed consent was waived by the review committee.
An observational study was conducted to evaluate the diagnostic accuracy of the Cobas 6800 SARS-CoV-2 assay. The nasopharyngeal samples used in this study were collected from 300 individuals referred for COVID-19 testing. All the samples were tested individually for the presence of SARS-CoV-2 immediately after collection from our laboratory collection center using our laboratory-standardized protocol.
The clinical performance of the Cobas 6800 SARS-CoV-2 assay was compared with that of the Labgun Exofast RT-PCR kit using a total of random 300 nasal swab samples, collected as a part of the routine SARS-CoV-2 screening in our laboratory during the months of August 2021 to January 2022.
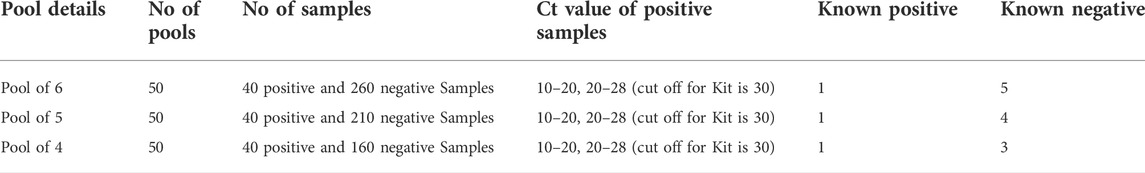
All 300 samples were individually tested, and 40 known positive samples and 260 negative nasopharyngeal swab samples were used for this study. The 40 individual positive specimens had Ct values between 10.0 and 28.0, including a subset of low viral load positive samples (19 out of 40 samples) with Ct values between 20.0 and 28.0. These samples were used for pooling, and 50 pools were created for each pool size of 4, 5, and 6. The pools were tested with one positive sample in each batch, and positive samples with both high and low viral loads were used.
Instruments and reagents
• Cobas 6800 real-time RT-PCR system (Manufacturer: Roche; serial number 2409);
• Line Gene 9,600 (Manufacturer: Bioer; Serial Number: BYQ6.613E-540410) used for qualitative detection of nucleic acids from SARS-CoV-2;
• Automated Sample Preparation System: MGISP-960 (Serial Number: 30030900200035);
• Roche Kit (Lot number: G21065);
• MGI Extraction Kit (Lot Number: H0062);
• LabGun COVID-19 RT-PCR reagent (Lot Number: 6220200423);
• Control material used for LOD (AccuPlex): 0505-0168.
Sample preparation
The nasopharyngeal samples utilized for this study were collected in our laboratory from individuals who underwent testing for SARS-COV-2 infections and were transported using viral transport medium (VTM); 0.6 ml of UTM-RT aliquots were transferred into secondary tubes and loaded on the Cobas 6800 m system.
Sample pooling
A uniquely labeled secondary tube was assigned for each pool. Samples to be pooled were denoted with the identification label of the pooling tube via the sample tracking system, as per the manufacturer’s recommendations for pooling the samples. One positive sample with five negative samples were pooled for the 6-sample pools, and one positive and four negatives and one positive and three negative samples were pooled for the 5- and 4-pool samples, respectively. A biological safety cabinet was used to ensure safety while pooling samples, one pool at a time. Samples from the same pool were pooled using a calibrated micro-pipettor with a fresh pipette tip for each sample. Complete mixing of each pool sample was achieved through pipetting up and down, ensuring no bubbles, foam, or aerosols formed in the tube. Pooled samples were then processed in the Cobas 6800 system by first logging into the system’s user interface and initiating processing by importing the samples’ tracking sheet in each pool. After this, the system’s reagents and consumables were refilled as prompted by the system, and the sample pools were loaded. The run was initiated on the user interface after the batch was full. Results and reports were then exported and reviewed for qualitative analysis of COVID-19, in terms of which the Cobas 6800 system automatically detected the SARS-CoV-2, for each processed sample and control, displaying target results for samples, as well as test validity and overall results for controls.
Cobas 6800 reverse transcription real-time PCR assay
The Cobas 6800 SARS-CoV-2 is based on a fully automated system performing nucleic acid extraction and purification followed by PCR amplification and detection. It works on the principle of selective amplification of target nucleic acid from the sample, achieved by the use of target-specific forward and reverse primers for the ORF1 a/b non-structural region (Target 1) that is unique to SARS-CoV-2. Additionally, there is a conserved region in the structural protein envelope E-gene (Target 2), which will also detect the SARS-CoV-2 virus. Selective amplification of RNA internal control is achieved by the use of non-competitive sequence-specific forward and reverse primers that have no homology with the coronavirus genome. A thermostable DNA polymerase enzyme is used for amplification. The coronavirus and RNA internal control detection probes are each labeled with unique fluorescent dyes that act as reporters. In this study, testing was conducted on pooled samples with pool sizes of 4, 5, and 6 samples, along with one positive and negative control. Positive results are reported when the ORF1ab gene is detected with a positive or negative result of the E gene, and when the E gene is positive with a negative ORF1ab gene, it is reported as presumptive positive. Whenever a positive or a presumptive positive result is detected, the entire batch of pooled samples is run individually to identify the positive samples (FDA, 2021).
Comparison: The Labgun Exofast reverse transcription real-time PCR kit
The LabGun Exofast COVID-19 RT-PCR Kit is a real-time test designed to detect RNA from the SARS-CoV-2 in nasopharyngeal, oropharyngeal, anterior nasal, and mid-turbinate nasal swabs and sputum samples. The SARS-CoV-2 primer/probe set is designed to detect the RdRp gene and the N gene of the SARS-CoV-2 virus. This comparable system is an FDA-approved, well-developed, standardized, in-house-validated protocol for detection of SARS CoV-2 from nasopharyngeal swabs. Nucleic acids are isolated and purified from nasopharyngeal samples using an MGI extraction kit. The purified nucleic acid is directly amplified using the LabGunTMCOVID-19 RT-PCR kit on the Bioer real-time PCR detection system targeting the RdRp and the N gene. Positive results are reported when the RdRp gene is detected with a positive or negative result of the N gene. When the N gene is positive with a negative RdRp gene, it is reported presumptive positive, and the sample is repeated on another testing platform (FDA, 2020).
The LabGun Exofast COVID-19 RT-PCR assay was chosen for comparison because this assay has a similar limit of detection as Cobas 6800. The turnaround time (TAT) is 40 min, and it has been well-tested, verified, and validated in house, having been in routine use in our laboratory since January 2021. It is currently considered the reference standard in our laboratory for SARS-CoV-2 detection.
The performance of the Cobas 6800-RT-PCR assay for identifying SARS-CoV-2 virus was evaluated against the comparable Labgun using 40 positive and 260 negative samples.
Statistical analysis
The performance of the Cobas 6800-RT-PCR assay was assessed by calculating the sensitivity, specificity, and agreement of results using Cohen’s kappa coefficient of variance (CV) to assess the inter-assay precision, the limit of detection (LOD), the carry-over test to calculate the error limit, and Pearson’s correlation between the Ct values of the target genes of both assays.
Results
The diagnostic accuracy of the Cobas 6800-RT-PCR assay was tested by using both the pooled and the non-pooled sample techniques. A total of 300 nasal swab samples, 40 positive samples and 260 negative samples, were used for the pooling technique assessment.
Pooled samples
Sample pooling was conducted, and a total of 50 pools with pool sizes of 6, 5, and 4 samples were compared using both platforms. Details of the pooling are shown in Table 1.
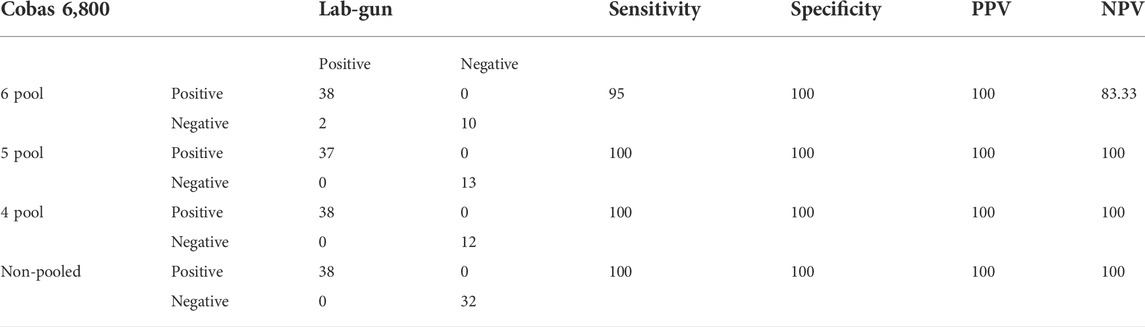
The sensitivity and specificity of the Cobas 6800 RT-PCR assay with a pool size of 6 was found to be 95% and 100%, respectively, with a Cohen’s kappa agreement of 96%. When pool sizes of 5 and 4 samples were evaluated, the sensitivity and specificity was 100% compared to the reference test in our laboratory. Cohen’s kappa agreement was 100% for both the 5- and 4-sample pool evaluations (Table 2).
Non-pooled samples
Seventy samples were run on both platforms: 38 samples turned out positive and 32 samples turned out negative on both platforms, which clearly demonstrates a sensitivity and specificity of 100% (Table 2).
Inter-assay precision
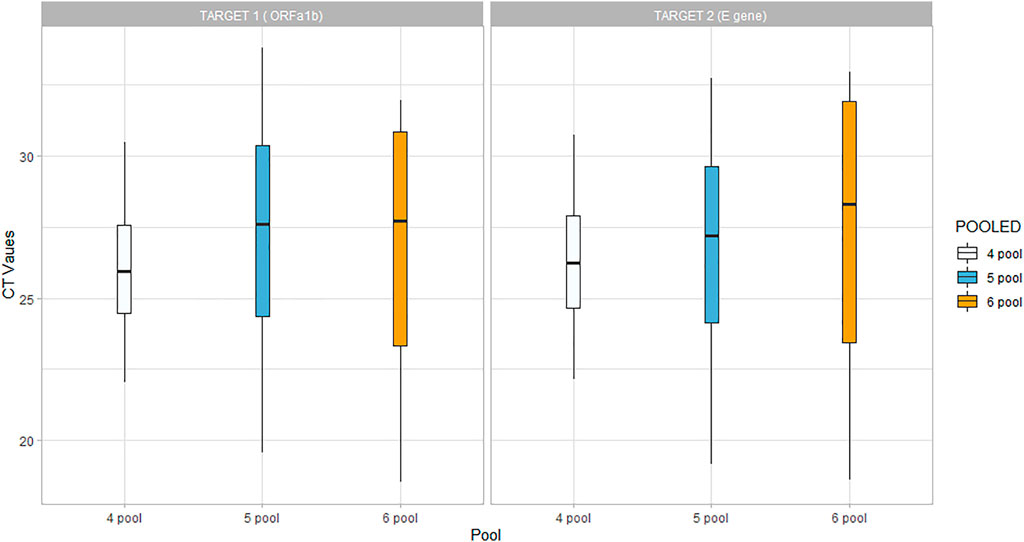
Inter-assay precision was determined using the coefficient of variance CV (%), while sample pools with one internal control were repeated, and the Ct values were determined for the same sample pool on 5 separate days. The inter-assay precision was determined for the 6-, 5-, and 4-pooled samples, and the CV% varied between 1.5% and 4%. The maximum deviation in the Ct values ranged from 0.23 to 2.21 (Figure 1).
Correlation between ct values of Target 1 and comparative RNA-dependent RNA polymerase gene
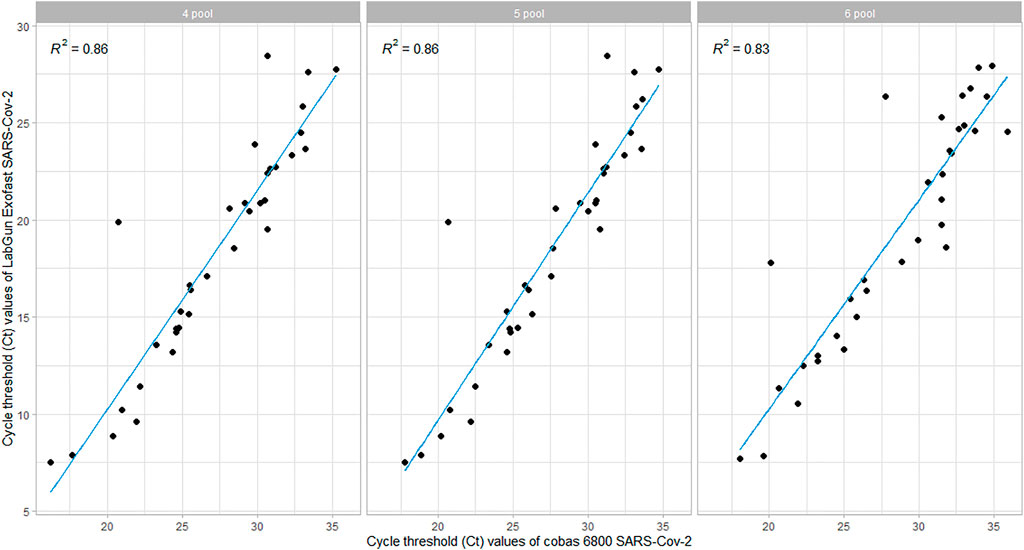
The Ct values of Target-1 (ORF1ab) were compared with the Ct values of the comparable assay (targeting the viral RdRp). The linear relationship between the Ct values was evaluated using Pearson’s correlation, and we found that there was a strong correlation when tested with all three different sample pool sizes. However, the correlation coefficient was found to be lower in the 6-pooled sample compared to the 4- and 5-pooled samples (Figure 2).
Limit of detection
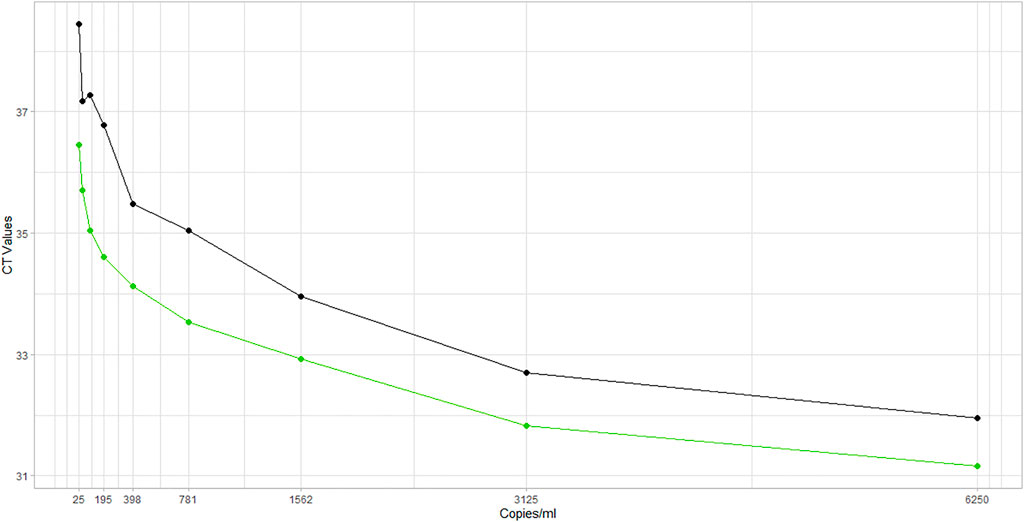
Analytical sensitivity is determined by performing limit of detection experiments which determine the lowest concentration of SARS-CoV-2 detected, at which level, approximately 95% of all true positive samples tested positive. The LOD was determined using the Cobas 6800-RT-PCR assay. The AccuPlex SARS-CoV-2 Verification Panel–Full Genome was used, which contains different concentrations of positive reference material (non-replicative recombinant viruses), to enable evaluation of test performance at multiple points across the assay range. A vial concentration of 100,000 copies/ml was used for analysis after serial dilutions to reach a lower limit of 25 copies/ml. The LOD was established by estimating the Ct values from nine different dilutions of the control sample, ranging from 6,250 copies/ml to 25 copies per ml. The lower limit of detection was verified as 25 copies/ml as per the manufacturer’s claim; hence, the limit of detection is expected to be 100, 125, and 150 copies/ml for the 4-, 5-, and 6-sample pools, respectively (Figure 3).
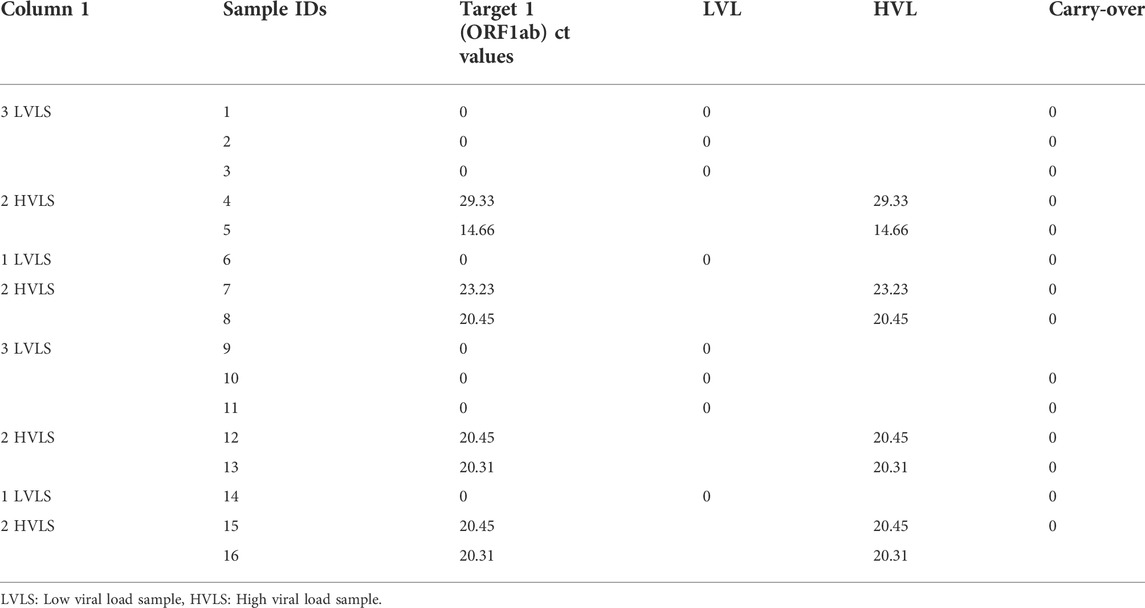
Carry-over test
RT-PCR was performed on eight high positive samples and eight confirmed negative samples, measured by analyzing high and low viral concentration samples in the sequence as per Table 3. Standard deviation (SD) of the low–low results was calculated, and the error limit was three times the SD. The mean Ct value was found to be the same for both high and low viral load specimens with a 0% carry-over.
Discussion
The COVID-19 pandemic has increased the global demand for laboratory testing services, including test reagents, sampling devices, laboratory instruments, and personal protective equipment. At the same time, the restriction on movement and travel has led to a massive disruption of supply chains around the world. As a result, there is scarcity of resources to meet the need of the hour for newer assays with faster turnaround time that can accommodate techniques like sample pooling to meet the diagnostic demands. Automation is the most efficient way of utilizing skilled manpower and is cost-effective. Unlike other platforms that use varied nucleic acid binding techniques optimized for preferential isolation of nucleic acids, the Cobas 6800 uses a uniform, universal sample preparation process to isolate, purify, and extract the nucleic acids in the sample processing module (Cobb et al., 2017).
Sensitivity and specificity
This study found that the sensitivity and specificity of the Cobas 6800 was 100% compared to those of the standard assay. The performance of the Cobas 6800 when using pooled samples showed that specificity was 100%, while sensitivity varied from 95% in the 6-pooled sample to 100% in both the 5- and 4-pooled samples. These results agree with other validation study reports (Nörz et al., 2020; Poljak et al., 2020; Kogoj et al., 2021).
From the 6-pool sample strategy, discordant results were identified among the low viral load samples with Ct values >27. Among the low viral load samples that were detected in the Labgun RT-PCR kit with Ct > 25, the E target was the only gene that was detected with the Cobas 6800 assay. Similar observation was also reported in another evaluation of Cobas 6800, reiterating the manufacturer’s claim of higher sensitivity to the E gene compared to the ORF1ab gene (Wirden et al., 2020).
Limit of detection
This study evaluated the manufacturer’s claim of a lower limit of detection of 25 copies/ml for Target 1 and 32 copies/ml for Target 2 (FDA, 2021). Similar results were found in this study where nine different dilutions, ranging from 25 copies/ml to 6,250 copies/ml were tested, and 25 copies/ml was found to be the lowest detectable concentration for both the target genes. Other studies have also supported this claim that the Cobas 6,800 has a lower limit of detection for SARS-COV-2 detection and that the Cobas E gene is more sensitive among the various other target genes in low viral load samples (Pujadas et al., 2020; Lee et al., 2021).
Anti-interference
The Cobas 6800 assay uses the RNA internal control (IC)–AmpErase enzyme (uracil N-glycosylase) in the PCR mix, which can enzymatically remove the PCR amplicon from a previous reaction without degrading naive DNA (Cobb et al., 2017). This study also demonstrated that there was no carry-over observed between samples and the error limit was calculated to be 0. False positive results may occur if carry-over of samples is not adequately controlled during sample handling and processing.
The IC is tested with every individual pool of samples during sample processing to monitor the success of sample preparation and the PCR amplification process. This helps identify the samples containing interfering agents that could affect nucleic acid isolation and PCR amplification. In addition to the IC, the test utilizes an external positive and a negative control with each run to rule out inhibition or contamination (FDA, 2021).
Stability and reproducibility
The Cobas 6800 RT-PCR assay showed very good assay reproducibility. The samples were subjected to testing on 5 consecutive days using 6-pooled, 5-pooled and 4-pooled samples. The coefficients of variation (CV%) were between 1.5% and 4%, which is within the acceptable range of 5% in reference to our laboratory standards. The study also observed a strong correlation between the Ct values of the target genes of both the assays, which is also supported by other studies that demonstrate a strong correlation between the SARS-CoV-2-specific targets obtained by the Cobas and comparative assays (Poljak et al., 2020).
Pooling of samples
The pooling technique run on the Cobas 6800 was highly efficient when the positivity rate was low. During the months of August 2021 to December 2021, the average positivity rate was around 0.3%–0.1% in our laboratory. The pooling technique was proving more cost effective and quicker with faster TAT when the positivity rate was low. However, after the emergence of the new Omicron variant in the latter half of December 2021, the average daily cases reported in the UAE increased, which in turn increased the slide positivity rates to up to 8% during the month of January 2022. When the slide positivity rate was high, the pooling method led to an average positive repeat percentage of 27%, and therefore the technique was not cost-effective when the positivity rate increased. This is supported by other studies showing that pooling techniques are effective only when the prevalence rate is low (Abdalhamid et al., 2020; Aragón‐Caqueo et al., 2020; Mahmoud et al., 2021).
The Cobas 6800 assay uses primers designed to detect the ORF1 a/b genomic region, and studies have shown that ORF1ab, RdRp primers have better analytical performance in identifying the SARS-CoV2-RNA (Mollaei et al., 2020). This allows the Cobas 6800 to be more efficient in detecting SARS-CoV-2 infections.
Critical analysis
The Cobas 6800 has high sensitivity and specificity and is highly efficient in the sample pooling technique, even when the sample pool has as many as six samples. It has a lower limit of detection for SARS-COV-2 detection of 25 copies/ml for un-pooled samples and has an inbuilt carry-over contamination control system preventing carry-over between samples and interference from contaminants. However, when selecting a SARS-CoV-2 RNA assay, the clinical performances of sensitivity and specificity alone are not themselves sufficient. There are other parameters that play a paramount role in selection, especially when employed in large-scale testing. Sample throughput, turnaround time (TAT), and the accommodation of techniques such as sample pooling, hands-on time, ease of method, availability of reagents, and cost of testing are considered equally when it comes to large-scale testing.
The Cobas 6800 is a fully automated platform with a turnaround time (TAT) of 2 hours. However, the techniques of pooling require significant time spent in the preparation of sample pools. The barcodes of pooled samples are scanned and captured in the system, and after the pooling of samples into the secondary tubes, secondary barcodes are created. The system then matches the secondary barcodes to the related primary sample barcodes. For a single run of five pooled samples, 480 samples need to be pooled, and this preparation, including sample barcoding and aliquoting, alone requires an average of 2 hours. Therefore, the total TAT to run 480 samples is about 5 h. By contrast, the Labgun Exofast RT-PCR kit has a TAT of 40 min to run 96 samples. However, the initial sample barcode scanning and aliquoting, followed by nucleic acid extraction and purification with the MGI extraction kit, requires an additional 90 min. This shows the Cobas 6800 RT-PCR platform is still time-efficient when compared to the Labgun Exofast RT-PCR kit.
The Cobas 6800 is an expensive test compared to the Labgun Exofast RT-PCR kit, as the price of the cartridge is higher. However, the pooling techniques employed greatly reduce the per capita cost of the test. Furthermore, the automation process of the Cobas 6800 platform reduces the cost of labor as it eliminates the need for skilled personnel, whereas the Labgun requires additional personnel with specific skills required to work at different work stations, such as nucleic acid isolation and extraction, and to run the PCR assays. Hence, when the sample load is high, the Cobas 6800 platform has proven more cost-effective than the Labgun.
Our study has some limitations in that it does not include clinical data on severity to correlate with sensitivity, which could have given further insight into the performance of the automated Cobas 6800 platform.
In conclusion, the overall performance of the Cobas 6800 is superior to the standard assay for detection of SARS-COV-2 from an operational standpoint. The automated system significantly improves work flow and processes large numbers of samples with shorter turnaround time. This study concludes that the Cobas 6800 RT-PCR assay is a reliable platform for qualitative and rapid detection of SARS-CoV-2 and can be effectively utilized for pooling of samples with highly efficient performance when disease prevalence is lower.
Data availability statement
The raw data supporting the conclusion of this article will be made available by the authors, on approval of request.
Ethics statement
The studies involving human participants were reviewed and approved by the Ethics review board at the Department of Health (DOH), Abu Dhabi, UAE. The ethics committee waived the requirement for written informed consent for participation.
Author contributions
SM: conception, design, data acquisition, analysis of data, interpretation of data, and substantial revision of the manuscript; SG: conception, design, data acquisition, analysis of data, interpretation of data, and drafting and substantial revision of the manuscript; PR: data acquisition, analysis of data, interpretation of data, and substantial revision of the manuscript; FC: analysis of data, interpretation of data, graphical representation of data, and substantial revision of the manuscript; HA: interpretation of data and substantial revision of the manuscript and WZ: interpretation of data and substantial revision of the manuscript.
Acknowledgments
We thank all lab personnel and the skilled technicians at the Biogenix lab, who contributed to this study.
Conflict of interest
Authors SM, SG, PR, FC, HA, and WZ were employed by G42 Healthcare. Authors SG, FC, HA, and WZ were employed by Insights Research Organization and Solutions (IROS).
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdalhamid, B., Bilder, C. R., McCutchen, E. L., Hinrichs, S. H., Koepsell, S. A., and Iwen, P. C. (2020). Assessment of specimen pooling to conserve SARS CoV-2 testing resources. Am. J. Clin. Pathol. 153 (6), 715–718. doi:10.1093/ajcp/aqaa064
Abduljalil, J. (2020). Laboratory diagnosis of SARS-CoV-2: available approaches and limitations. New Microbes New Infect. 36, 100713. doi:10.1016/j.nmni.2020.100713
Aragón‐Caqueo, D., Fernández‐Salinas, J., and Laroze, D. (2020). Optimization of group size in pool testing strategy for SARS‐CoV‐2: A simple mathematical model. J. Med. Virol. 92 (10), 1988–1994. doi:10.1002/jmv.25929
Asif, M., Xu, Y., Xiao, F., and Sun, Y. (2021). Diagnosis of COVID-19, vitality of emerging technologies and preventive measures. Chem. Eng. J. 423, 130189. doi:10.1016/j.cej.2021.130189
Aziz, A., Asif, M., Ashraf, G., Farooq, U., Yang, Q., and Wang, S. (2021). Trends in biosensing platforms for SARS-CoV-2 detection: a critical appraisal against standard detection tools. Curr. Opin. Colloid Interface Sci. 52, 101418. doi:10.1016/j.cocis.2021.101418
Cherif, A., Grobe, N., Wang, X., and Kotanko, P. (2020). Simulation of pool testing to identify patients with coronavirus disease 2019 under conditions of limited test availability. JAMA Netw. Open 3 (6), e2013075. doi:10.1001/jamanetworkopen.2020.13075
Chung, Y. S., Lee, N. J., Woo, S. H., Kim, J. M., Kim, H. M., Jo, H. J., et al. (2021). Validation of real-time RT-PCR for detection of SARS-CoV-2 in the early stages of the COVID-19 outbreak in the Republic of Korea. Sci. Rep. 11 (1), 14817–14818. doi:10.1038/s41598-021-94196-3
Cobb, B., Simon, C. O., Stramer, S. L., Body, B., Mitchell, P. S., Reisch, N., et al. (2017). The Cobas® 6800/8800 system: a new era of automation in molecular diagnostics. Expert Rev. Mol. diagn. 17 (2), 167–180. doi:10.1080/14737159.2017.1275962
FDA 2020. LabGunTM COVID-19 RT-PCR kit user’s manual. Available at: https://www.fda.gov/media/137483/download.
FDA 2021. Qualitative assay for use on the Cobas 6800/8800 Systems. Available at: https://www.fda.gov/media/136049/download.
Hogan, C. A., Sahoo, M. K., and Pinsky, B. A. (2020). Sample pooling as a strategy to detect community transmission of SARS-CoV-2. JAMA 323, 1967–1969. doi:10.1001/jama.2020.5445
Kogoj, R., Kmetic, P., Valencak, A. O., Komlos, K. F., Seme, K., Sagadin, M., et al. (2021). Real-life head-to-head comparison of performance of two high-throughput automated assays for the detection of SARS-CoV-2 RNA in nasopharyngeal swabs: The alinity m and Cobas 6800 SARS-CoV-2 assays. J. Mol. Diagn. 23, 920–928. doi:10.1016/j.jmoldx.2021.05.003
Lee, C. K., Tham, J. W., Png, S., Chai, C. N., Ng, S. C., Tan, E. J., et al. (2021). Clinical performance of Roche Cobas 6800, luminex ARIES, MiRXES fortitude kit 2.1, altona RealStar, and applied biosystems TaqPath for SARS‐CoV‐2 detection in nasopharyngeal swabs. J. Med. virology 93 (7), 4603–4607. doi:10.1002/jmv.26940
Mahmoud, S. A., Ibrahim, E., Thakre, B., Teddy, J. G., Raheja, P., Ganesan, S., et al. (2021). Evaluation of pooling of samples for testing SARS-CoV-2 for mass screening of COVID-19. BMC Infect. Dis. 21 (1), 360–369. doi:10.1186/s12879-021-06061-3
Mishra, B., Behera, B., Mohanty, M., Ravindra, A., and Ranjan, J. (2020). Challenges and issues of SARS-CoV-2 pool testing. Lancet. Infect. Dis. 20 (11), 1233. doi:10.1016/S1473-3099(20)30463-1
Mollaei, H. R., Afshar, A. A., Kalantar-Neyestanaki, D., Fazlalipour, M., and Aflatoonian, B. (2020). Comparison five primer sets from different genome region of COVID-19 for detection of virus infection by conventional RT-PCR. Iran. J. Microbiol. 12 (3), 185–193.
Nörz, D., Frontzek, A., Eigner, U., Oestereich, L., Wichmann, D., Kluge, S., et al. (2020). Pushing beyond specifications: Evaluation of linearity and clinical performance of the Cobas 6800/8800 SARS-CoV-2 RT-PCR assay for reliable quantification in blood and other materials outside recommendations. J. Clin. Virol. 132, 104650. doi:10.1016/j.jcv.2020.104650
Poljak, M., Korva, M., Knap Gašper, N., Fujs Komloš, K., Sagadin, M., Uršič, T., et al. (2020). Clinical evaluation of the Cobas SARS-CoV-2 test and a diagnostic platform switch during 48 hours in the midst of the COVID-19 pandemic. J. Clin. Microbiol. 58 (6), 005999-20. doi:10.1128/JCM.00599-20
Pujadas, E., Ibeh, N., Hernandez, M. M., Waluszko, A., Sidorenko, T., Flores, V., et al. (2020). Comparison of SARS‐CoV‐2 detection from nasopharyngeal swab samples by the Roche Cobas 6800 SARS‐CoV‐2 test and a laboratory‐developed real‐time RT‐PCR test. J. Med. Virol. 92 (9), 1695–1698. doi:10.1002/jmv.25988
Sahajpal, N. S., Mondal, A. K., Njau, A., Ananth, S., Jones, K., Ahluwalia, P. K., et al. (2020). Proposal of RT-PCR–based mass population screening for severe acute respiratory syndrome coronavirus 2 (coronavirus disease 2019). J. Mol. Diagn. 22 (10), 1294–1299. doi:10.1016/j.jmoldx.2020.07.001
Sharma, K., Agarwala, P., Gandhi, D., Mathias, A., Singh, P., Sharma, S., et al. (2021). Correction: comparative analysis of various clinical specimens in detection of SARS-CoV-2 using rRT-PCR in new and follow up cases of COVID-19 infection: Quest for the best choice. Plos one 16 (6), e0253355. doi:10.1371/journal.pone.0253355
WHO 2022 WHO coronavirus (COVID-19) dashboard. Available at: https://covid19.who.int/(Accessed 11 March 2022).
Keywords: RT-PCR, pooling, Cobas, SARS-CoV-2, COVID-19
Citation: Mahmoud S, Ganesan S, Raheja P, Cantarutti F, Ateia H and Zaher W (2022) Diagnostic accuracy of the Cobas 6800 RT-PCR assay for detection of SARS-CoV-2 RNA. Front. Anal. Sci. 2:1030701. doi: 10.3389/frans.2022.1030701
Received: 29 August 2022; Accepted: 13 October 2022;
Published: 01 November 2022.
Edited by:
Liang Qiao, Fudan University, ChinaReviewed by:
Ashis Kumar Mondal, Augusta University, United StatesMuhammad Asif, Wuhan Institute of Technology, China
Copyright © 2022 Mahmoud, Ganesan, Raheja, Cantarutti, Ateia and Zaher. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sally Mahmoud, c2FsbHkuYWJkdWxsYUBnNDIuYWk=
 Sally Mahmoud1*
Sally Mahmoud1* Subhashini Ganesan
Subhashini Ganesan Flavia Cantarutti
Flavia Cantarutti Walid Zaher
Walid Zaher