- Instituto de Pesquisas de Produtos Naturais Walter Mors, Centro de Ciências da Saúde, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil
Ornithine decarboxylase (ODC) catalyzes the decarboxylation of ornithine to produce putrescine, the first step in the metabolism of polyamines (putrescine, spermidine, and spermine), which are essential growth factors in eukaryotic cells. ODC is active as a homodimer and depends on pyridoxal 5′-phosphate (PLP) as a cofactor. An increase in the concentration of polyamines has been associated with carcinogenesis. Therefore, there is much interest in identifying inhibitors of this pathway as potential chemotherapeutic and chemopreventive agents. The best-known inhibitor of mammalian ODC is α-difluoromethylornithine (DFMO), a highly selective compound that alkylates Cys-360 (a residue of the ODC active site). Although DFMO was initially developed for the treatment of cancer, the World Health Organization recommends its use in combination with nifurtimox for the treatment of human African trypanosomiasis. Considering the importance of ODC as a promising target for the treatment of various types of cancer and other infectious diseases, choosing the right method for screening potential inhibitors can help to accelerate the discovery of new drugs. Several methods for the determination of ODC activity are found in the literature. Among these, we can mention analysis with radioactive markers, colorimetric assays using auxiliary enzymes to detect CO2 or H2O2 release, chromatographic separations with putrescine derivatization, mass spectrometry, and spectroscopic techniques. In this review, the main analysis methods used will be described, highlighting their advantages and disadvantages, as well as identifying the most promising methods for high-throughput screening.
1 Introduction
Ornithine decarboxylase (ODC, EC 4.1.1.17) is an enzyme that limits the rate of the polyamine biosynthetic pathway. Polyamines play an important role in the cellular homeostasis of most living organisms, and ODC has been reported in several species (Miller-Fleming et al., 2015; Bateman et al., 2021). ODC, a pyridoxal 5′-phosphate (PLP) dependent enzyme, catalyzes the decarboxylation of l-ornithine to putrescine via Schiff base formation (Figure 1). Putrescine is the precursor of spermidine and spermine in the first step of polyamine biosynthesis (Michael, 2016). Elevated expression of ODC, as well as abnormally high levels of polyamines, are correlated with the occurrence of many diseases, including cancer and Alzheimer’s (Gerner and Meyskens, 2004; Mäkitie et al., 2010; Miller-Fleming et al., 2015; Somani et al., 2017; Igarashi and Kashiwagi, 2019; Holbert et al., 2022; Ivanov and Khomutov, 2022). Consequently, ODC is considered an oncogenic enzyme and a target for the treatment of several diseases (McCann and Pegg, 1992; Marton and Pegg, 1995; Mukhopadhyay and Madhubala, 1995; Casero and Marton, 2007; Smithson et al., 2010a; Colotti and Ilari, 2011; Chakraborty et al., 2013; Somani et al., 2017; Perdeh et al., 2020; Sweeney, 2020; Khan et al., 2021). ODC is a homodimer in a dynamic equilibrium between the active dimeric form and the inactive monomeric form (Solano et al., 1985; Mitchell et al., 1988; Mitchell and Chen, 1990; Coleman et al., 1994; Chai et al., 2020). The two identical active sites in the homodimer are formed at the dimer interface by residues from the N-terminal domain (Lys69) of one subunit and the C-terminal domain (Cys360) of the other (Tobias and Kahana, 1994; Chai et al., 2020). Lys69 was biochemically identified as the covalent binding site for PLP, while Cys360 is involved in covalent adduct formation with DFMO (α-difluoromethylomithine) (Poulin et al., 1992; Coleman et al., 1994; Grishin et al., 1999). In addition, other important interactions at the binding site have also been described, such as Glu274, which stabilizes the positive charge on the pyridine nitrogen of the PLP, increasing the ability to withdraw electrons from the ring, and Arg277, which is necessary for the binding of the high affinity of PLP through interaction with the cofactor 5′-phosphate (Figure 1) (Osterman et al., 1995, 1997; Brooks and Phillips, 1997).
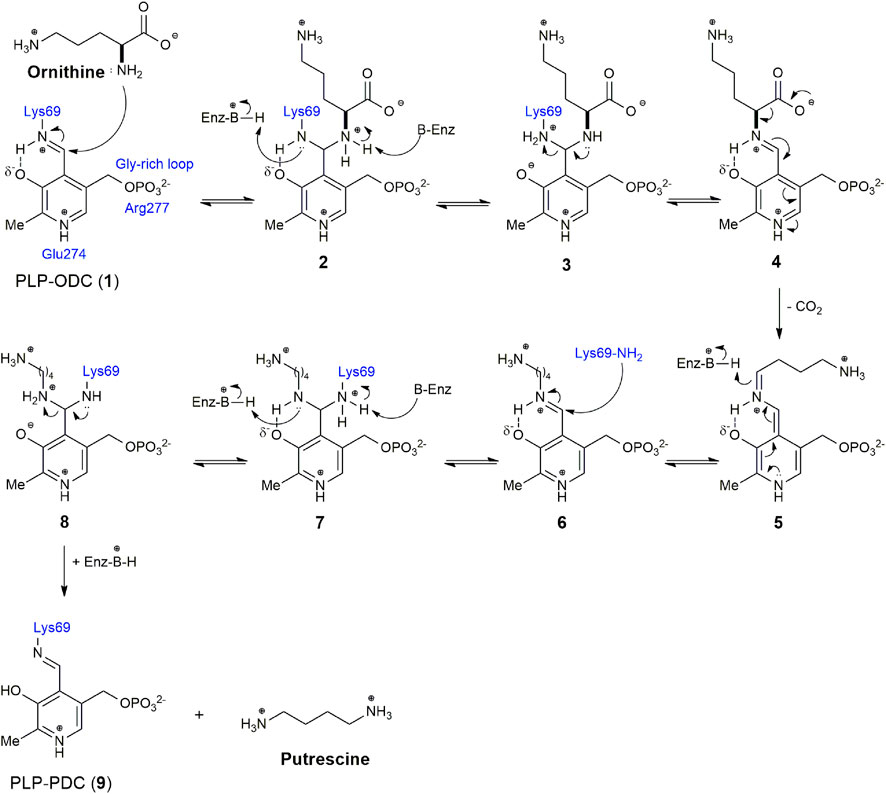
FIGURE 1. Reaction mechanism of Ornithine decarboxylase (ODC) with the pyridoxal 5′-phosphate (PLP) cofactor.
Due to its importance in several cellular functions and wide distribution in several species, since the 1970s there has been a great therapeutic interest in the inhibition of ODC, with the consequent depletion of polyamine biosynthesis (Mamont et al., 1978; Metcalf et al., 1978). According to the mechanism of the ODC reaction shown in Figure 1, the enzyme and its reaction can be inhibited by cofactor (PLP), substrate (ornithine), or product (putrescine) analogues. α-difluoromethylornithine (DFMO), an ornithine analogue, is the most important and best-known inhibitor of ODC. DFMO, marketed as Eflornithine, belongs to the WHO Model List of Essential Medicines to treat sleeping sickness (WHO 2022). Although it is not effective for the treatment of cancer alone, it has been widely used in combination with other drugs. It is in several clinical trials as an adjuvant or chemopreventive agent and appears to be highly useful in the treatment of neuroblastoma (Kim et al., 2017; Alhosin et al., 2020; Schultz et al., 2021). In addition to the cofactor, substrate, and ODC reaction product analogues, other classes of inhibitors (benzothiazoles, indoles, bisbiguanide, and dithioamidine) with different binding modes were identified, indicating allosteric sites in the enzyme. The identification of these new classes of inhibitors was made from a high-throughput enzymatic screening with 316,114 different molecules, demonstrating the importance of this methodology for the identification of more selective and potent inhibitors for ODC, with improved pharmacokinetic properties (Smithson et al., 2010a). There are several methods in the literature to assess ODC activity. The most used methods are based on the detection of radiolabeled CO2 or radiolabeled DMFO linked to ODC, on the capture of CO2 for the conversion of NADH into NAD with the use of Phosphoenolpyruvate Carboxylase (PEPC) and Malate Dehydrogenase (MDH) enzymes, on the chemiluminescence or colorimetric detection of H2O2 produced by putrescine oxidation by amine oxidases, and in derivatization for analysis by high-performance liquid chromatography (HPLC) or mass spectrometry (MS) and fluorescence. However, we did not find a compilation of existing methods in the literature, which makes it difficult to choose the most appropriate method to assess ODC activity and screening for inhibitors. Therefore, we will make a brief description of the main methods found (Table 1), seeking, whenever possible, to present their advantages and disadvantages and the possibility of application in high-throughput screening (HTS).
2 Methods to assess ODC activity and inhibition
2.1 Radiolabeling
The measurement of 14CO2 release from [14C]carboxyl-labeled amino acids is widely used as a method of assay for decarboxylase activity (Kobayashi, 1963). For in vitro ODC activity, the usual procedure is to incubate the enzyme sample in a medium with EDTA, sodium/potassium phosphate buffer, pyridoxal phosphate, and the [1–14C]-l-ornithine in a vial sealed with the cap containing the paper discs impregnated with hyamine/sodium hydroxide to capture the release of radiolabeled carbon dioxide. The enzymatic reaction is stopped by the addition of sulfuric/citric acid. After an additional incubation period to complete 14CO2 absorption, the caps are removed and transferred to a scintillation vial containing scintillation fluid. Measurements of disintegrations per minute are made in a liquid scintillation spectrometer. The specific ODC activity is expressed as nmol CO2 released/min/mg protein (Jones et al., 1972; Beaven et al., 1978; Smith and Marshall, 1988; Henley et al., 1996; Vanisree and Shyamaladevi, 2006; Schultz et al., 2020).
Another method to monitor the ODC activity is by using α-[5–14C]Difluoromethylornithine, an irreversible ODC inhibitor that forms a covalent bond with the enzyme. For in vitro ODC activity, mouse tissues are treated with α-[5–14C]DFMO and incubated in the enzymatic reaction medium. After precipitation of protein by addition of acid and centrifugation, the pellet is dissolved in hydroxide sodium solution, mixed with scintillation fluid, and has the radioactivity counted in a liquid scintillation spectrometer (Seely et al., 1982). For in vivo controlling, the labeled ODC can be monitored by autoradiographic examination of mouse kidneys or livers after administration of α-[5–14C]DFMO (Pegg et al., 1982; Pösö and Pegg, 1983; Zagon et al., 1984).
2.2 CO2 consumption by coupled enzymes (PEPC-MDH)
The most used method for the detection of CO2 produced by decarboxylases, such as ODC, makes use of CO2 consumption through reactions with the enzymes Phosphoenolpyruvate Decarboxylase (PEPC) and Malate Dehydrogenase (MDH), known as PEPC-MDH (Figure 2). From the CO2 produced by the decarboxylase reaction, PEPC catalyzes the condensation of bicarbonate with phosphoenol pyruvate to form oxaloacetate. In the next step, MDH (using NADH as a cofactor) reduces oxaloacetate to form malate and NAD+. NADH absorbs light at 340 nm, but NAD+ does not. The presence of CO2 in the reaction system leads to a decrease in light absorbance, used to assess the production of CO2 (Scriven et al., 1988). These reactions can be performed using commercial kits, standardized for CO2 detection (Smithson et al., 2010b; Liao et al., 2015; Chai et al., 2020). This assay allows the monitoring of the reaction in real-time and is considered by some authors as a good candidate for adaptation to HTS (Smithson et al., 2010b; Chai et al., 2020). Smithson et al. (Smithson et al., 2010b), optimized this assay using a commercial system for the detection of bicarbonate, carried out the experiments in N2 atmosphere, performed the analyzes in an automated system in 384-well microplates, and optimized all the conditions reactions to evaluate 3,600 compounds. This method was used to screen a library of 316,114 compounds as inhibitors of T. brucei ODC. New classes of ODC inhibitors were identified that probably bind to new non-active sites of the enzyme, demonstrating the importance of this type of screening for the identification and development of new inhibitors (Smithson et al., 2010a).
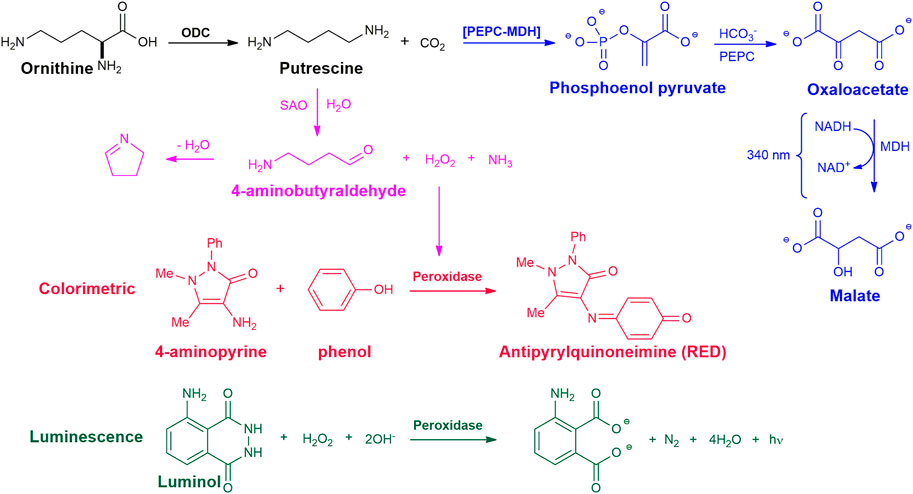
FIGURE 2. Chemical reactions for the enzymatic assay of ODC activity. Blue - detection of CO2 formation, with Phosphoenolpyruvate Decarboxylase (PEPC) and Malate Dehydrogenase (MDH) enzymes. Magenta - detection of putrescine formation with soyabean amine oxidase (SAO) and horseradish peroxidase enzymes; Red - colorimetric analysis, Green - luminescence.
2.3 Putrescine oxidation: H2O2 released
2.3.1 Colorimetric
This method is based on the oxidation of putrescine by soyabean amine oxidase (SAO), does not require polyamine purification, and can be performed in a 96-well plate for analysis in a plate reader. In the ODC reaction, the concentration of putrescine formed is determined by the amount of H2O2 generated by the reaction with SAO. Oxidation of 1 mol of putrescine by SAO generates 1 mol of H2O2 which reacts with 4-aminoantipyrine and phenol catalyzed by horseradish peroxidase to produce a colored complex that absorbs at 505 nm (Figure 2). Negative control can be done by replacing protein or substrate with buffer in the ODC enzyme reaction mix (Badolo et al., 1999; Das et al., 2015). In these assays, the soy amine oxidase enzyme was isolated from a soy plant and used in crude (Das et al., 2015) or purified form (Badolo et al., 1999), which increases the test performance by one more step. Furthermore, the possibility of test compounds acting as inhibitors of one of the additional enzymes should always be considered in HTS assays, which could lead to false positives for ODC.
2.3.2 Luminescence
The luminescence-based method was developed for the determination of total polyamines in biological samples (Fagerström et al., 1984) and adapted to determine ODC activity in cell extracts (Wang and Bachrach, 2000). This method consists in the oxidation of putrescine with a diamine oxidase, producing H2O2. The quantity of H2O2 formed is determined by chemiluminescence from the reaction with luminol and a peroxidase. Cell extracts are incubated with l-ornithine and placed on phosphocellulose paper strips. After drying, the contaminants are removed by washing the paper with ammonium hydroxide. To elute the putrescine, the paper is placed in a buffer solution containing magnesium sulfate and kept under agitation. After elution, the solution containing the putrescine is oxidized by a diamine oxidase generating H2O2. The hydrogen peroxide formed is subjected to a reaction with luminol and a peroxidase (Figure 2). This simple analytical method has the sensitivity of conventional assays based on the use of radioactive l-ornithine (Wang and Bachrach, 2000). Despite the relative simplicity and sensitivity of detecting putrescine on a picomolar scale, this method requires an initial step of extraction from paper, the value measured to produce putrescine may be interfered with by other polyamines and depends on other enzymes.
2.4 Adducts of putrescine with trinitrophenyl groups
Luqman and colleagues (Luqman et al., 2013) developed a colorimetric method to determine the formation of putrescine by ODC. This method is based on the solubility differences between substrates and product adducts and was developed aiming at the use of low-cost reagents and usual laboratory equipment. It is an endpoint method based on the reaction of picrylsulfonic acid (2,4,6 trinitrobenzenesulfonic acid, TNBS) with amino groups at alkaline pH forming colored trinitrophenyl (TNP) groups, resulting in water-soluble TNP-ornithine-TNP and adducts of TNP-putrescine-TNP soluble in 1-pentanol, which can be separated by phase extraction. The absorbance of the upper layer, containing TNP-putrescine-TNP, should be read at 426 nm. The difference in absorbance between the sample with and without enzyme will be related to the amount of putrescine formed, which must be compared with a standard putrescine plot for quantification. For the inhibition assay, a control set without the inhibitor needs to be compared to samples with the inhibitor. Despite being an inexpensive method and being applied by several authors (Nilam et al., 2017; El-Sayed et al., 2019; Villa-Ruano et al., 2019; Chai et al., 2020), it is not suitable for high-throughput screening with many samples. The method is time-consuming and laborious due to the need for several vortex mixing and centrifugation steps, in addition to the many transfers between new sets of tubes. To ensure the correct TNP-putrescine-TNP formation reaction and extractions, small aliquots must be pipetted many times correctly, giving many sources of errors that depend heavily on the analyst’s skills.
2.5 HPLC assays
Due to its great resolution, specificity, and increased sensibility, HPLC assays are among the most reliable methods for the detection of polyamines. The major drawbacks are the cost, long analysis time, and the need for a laborious pretreatment of the sample. Several methods describe the detection and analysis of polyamines through HPLC analysis, but only a few have been applied to monitor ODC activity. In general, the enzymatic reaction is terminated by the addition of HClO4, before centrifugation for protein precipitation. The amine-containing supernatant is then used for derivatization and analysis.
This pretreatment and a classical derivatization procedure using NaOH and benzoyl chloride was recently performed by Liu and coworkers for the screening of ODC inhibitors (Chai et al., 2020). The dansylation of the diamines can also be performed using the amine-containing supernatant, saturated sodium carbonate solution, and dansyl chloride. After derivatization, a pre-purification of the mixture using a Bond-Elut C18 cartridge was performed before HPLC analysis (Kabra et al., 1986; Legaz et al., 2001). o-Phthalaldehyde (OPA) can also be used for amine derivatization from the mixture of the neutralized supernatant, OPA, a borate buffer, and mercaptoethanol for reagent stabilization. A fluorescence detector with standard filters for OPA was used for detection (Kvannes and Flatmark, 1987; Bito et al., 2019). Tritium-labeled ornithine was used for the detection of ODC activity, obtained directly from the murine embryonic submandibular gland. After complete disruption of the sample in phosphate buffer (pH 7.3), followed by centrifugation, analysis was performed by HPLC equipped with a scintillation counter (Beeman and Rossomando, 1989). Other recent derivatizations include carbamoylation with isobutyl chloroformate (Ah Byun et al., 2008), derivatization with diethyl ethoxymethylenemalonate (del Rio et al., 2018), and the use of commercially available derivatization kits (Xiao et al., 2009).
2.6 MS assays
Given the importance of detecting very low concentrations (traces) of polyamines in biological fluids, many chromatographic methods have been used, mainly based on HPLC separation with fluorometric detection (Yoshida et al., 2004), GC separation (Li et al., 2008), and HPLC with MS detection (Ducros et al., 2009; Liu et al., 2011). The detection by HPLC/Q-TOF MS of benzoylated polyamines allowed the determination of very low concentrations of polyamines in urine (Liu et al., 2011). Mass spectrometry without chromatographic separation was used by Gaboriou and colleagues to detect and quantify biologically relevant polyamines after dansylation. In the Atmospheric Pressure Chemical Ionization (APCI) mass spectra, the positive ions for each dansylated polyamine were generated after optimization by flow injection analysis (FIA). FIA together with MS detection by monitoring selected ions has greatly increased the sensitivity of polyamine detection. The method proved to be linear over a wide range of polyamine concentrations allowing detection in the fmols range. The method was considered fast, efficient, economical, reproducible, and simple enough to allow its routine application (Gaboriau et al., 2003). Subsequently, this method was used to detect the putrescine formed in vitro during the decarboxylation of l-ornithine by ODC present in cell extracts of a mouse hepatoma cell line, allowing detection of ODC activity as low as 0.05 nmol/mg (Gaboriau et al., 2005).
2.7 Circular dichroism
l-ornithine is chiral and when decarboxylated by ODC it is converted to putrescine, achiral. This difference in chirality makes it possible to use circular dichroism to monitor ODC reactions. As an amino acid, l-ornithine generates a positive signal at 201 nm in the CD spectrum due to the interaction of the carboxylate group with chiral Cα. This signal is dependent on the concentration of l-ornithine, with a linear decay of its intensity along the ODC reaction. The monitoring of ODC reactions by CD is very simple, requiring only buffer (phosphate or tris–less interference in the spectrum), substrate, PLP, and enzyme. However, it has the limitation that several organic compounds absorb around 201 nm and cannot be used with cell extracts or mixtures of compounds (Brooks and Phillips, 1996). In addition, it cannot be analyzed on microplates, which makes it difficult to perform many analyses simultaneously.
2.8 Fluorescence assays
A fluorescence-based assay to continuously monitor ODC activity was developed based on the principle of the tandem supramolecular enzyme assay. Tandem assays rely on pairs of reporter compounds with different affinities for a macrocyclic receptor and a fluorescent dye. This difference in macrocycle affinity causes a change in the spectroscopic properties of the fluorescent dye. In tandem enzymatic assays, the macrocycle receptor interacts with the substrate or enzymatic reaction product, leading to a displacement of the fluorescent dye. The change in fluorescence to be detected will depend on the difference in affinity of the substrate or product for the macrocycle compared to the fluorescent dye (Ghale and Nau, 2014). In the assays carried out by Nilam et al. (Nilam et al., 2017) trans-4-[4-(dimethylamino)styryl]-1-methylpyridinium iodide (DSMI) was used as a fluorescent dye and cucurbit [6]uril (CB6) as the macrocyclic receptor (Nau et al., 2009; Li et al., 2012). l-ornithine has little affinity for the macrocycle, so DSMI is preferentially encapsulated by the receptor (CB6) and is strongly fluorescent. With the conversion of l-ornithine into putrescine by the ODC, the putrescine, with greater affinity for the macrocycle, will be encapsulated, continuously releasing the DSMI, with a decrease in the fluorescence intensity with the course of the reaction. This assay can be adapted for reading on microplates and allowed the determination of the enzymatic kinetic parameters of the ODC, using the known inhibitors DFMO and (−)-epigallocatechin (EGCG) as positive controls (Nilam et al., 2017).
3 Perspective for HTS
Despite the importance of ODC as a therapeutic target, the identification of new inhibitors from the screening of large libraries of compounds is very limited by the difficulty in assaying ODC activity in a high-throughput manner. This has been a major challenge for decarboxylases, as both the substrate and the product are not directly detected by spectroscopic techniques, needing derivatization or conversion processes into spectrophotometrically detectable complexes. Among the assays presented, only the PEPC-MDH and the fluorescence assay proved to be suitable for HTS due to their biological relevance, sensitivity, robustness, and economic viability (Blay et al., 2020). However, just in the PEPC-MDH the experimental and automation conditions were evaluated and optimized to meet the criteria for a high-quality HTS. Given the difficulties of HTS assays for ODC based on enzymatic reactions, a promising alternative would be to perform HTS assays based on ligand-enzyme processes (Bergsdorf and Ottl, 2010), employing libraries of compounds or molecular fragments (Erlanson, 2012) − which have shown to be very promising in the last few decades− developing several compounds that are in clinical phase trials and some drugs on the market (Erlanson et al., 2016).
Author contributions
LT Planned the organization of the manuscript, wrote and revised BS Wrote topic about HPLC JS Wrote topic about Adducts of putrescine with trinitrophenyl groups FF Wrote and revised the manuscript.
Funding
National Council for Scientific and Technological Development (CNPQ): LT—Research Fellowship—317008/2021-8, JS undergraduate research fellowships. Carlos Chagas Filho Foundation for Research Support of the State of Rio de Janeiro (FAPERJ): LT research project- E26/010.001718/2019, 260003/011346/2021, 260003/015693/2021.
Acknowledgments
The authors would like to thank CNPq, CAPES, and FAPERJ for financial support. JS is grateful to CNPq for undergraduate research fellowships.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ah Byun, J., Lee, S. H., Jung, B. H., Choi, M. H., Moon, M. H., and Chung, B. C. (2008). Analysis of polyamines as carbamoyl derivatives in urine and serum by liquid chromatography-tandem mass spectrometry. Biomed. Chromatogr. 22, 73–80. doi:10.1002/bmc.898
Alhosin, M., Razvi, S. S. I., Sheikh, R. A., Khan, J. A., Zamzami, M. A., and Choudhry, H. (2020). Thymoquinone and difluoromethylornithine (DFMO) synergistically induce apoptosis of human acute T lymphoblastic leukemia jurkat cells through the modulation of epigenetic pathways. Technol. Cancer Res. Treat. 19, 1533033820947489. doi:10.1177/1533033820947489
Badolo, L., Berlaimont, V., Helson-Cambier, M., Hanocq, M., and Dubois, J. (1999). Simple and rapid enzymatic assay of ornithine decarboxylase activity. Talanta 48, 127–134. doi:10.1016/S0039-9140(98)00228-8
Bateman, A., Martin, M. J., Orchard, S., Magrane, M., Agivetova, R., Ahmad, S., et al. (2021). UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 49, D480–D489. doi:10.1093/NAR/GKAA1100
Beaven, M. A., Wilcox, G., and Terpstra, G. K. (1978). A microprocedure for the measurement of 14CO2 release from [14C]carboxyl-labeled amino acids. Anal. Biochem. 84, 638–641. doi:10.1016/0003-2697(78)90089-1
Beeman, C. S., and Rossomando, E. F. (1989). Assay of ornithine decarboxylase activity by reversed-phase high-performance liquid chromatography. J. Chromatogr. 496, 101–110. doi:10.1016/S0378-4347(00)82556-6
Bergsdorf, C., and Ottl, J. (2010). Affinity-based screening techniques: Their impact and benefit to increase the number of high quality leads. Expert Opin. Drug Discov. 5, 1095–1107. doi:10.1517/17460441.2010.524641
Bito, T., Okamoto, N., Otsuka, K., Yabuta, Y., Arima, J., Kawano, T., et al. (2019). Involvement of spermidine in the reduced lifespan of caenorhabditis elegans during Vitamin B12 deficiency. Metabolites 9, E192. doi:10.3390/metabo9090192
Blay, V., Tolani, B., Ho, S. P., and Arkin, M. R. (2020). High-throughput screening: today’s biochemical and cell-based approaches. Drug Discov. Today 25, 1807–1821. doi:10.1016/J.DRUDIS.2020.07.024
Brooks, H. B., and Phillips, M. A. (1997). Characterization of the reaction mechanism for Trypanosoma brucei ornithine decarboxylase by multiwavelength stopped-flow spectroscopy. Biochemistry 36, 15147–15155. doi:10.1021/bi971652b
Brooks, H. B., and Phillips, M. A. (1996). Circular dichroism assay for decarboxylation of optically pure amino acids: Application to ornithine decarboxylase. Anal. Biochem. 238, 191–194. doi:10.1006/ABIO.1996.0274
Casero, R. A., and Marton, L. J. (2007). Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat. Rev. Drug Discov. 6, 373–390. doi:10.1038/NRD2243
Chai, X., Zhan, J., Pan, J., He, M., Li, B., Wang, J., et al. (2020). The rational discovery of multipurpose inhibitors of the ornithine decarboxylase. FASEB J. 34, 10907–12921. doi:10.1096/FJ.202001222R
Chakraborty, D., Saravanan, P., Patra, S., and Dubey, V. K. (2013). Studies on ornithine decarboxylase of Leishmania donovani: Structure modeling and inhibitor docking. Med. Chem. Res. 22, 466–478. doi:10.1007/s00044-012-0035-9
Coleman, C. S., Stanley, B. A., Viswanath, R., and Pegg, A. E. (1994). Rapid exchange of subunits of mammalian ornithine decarboxylase. J. Biol. Chem. 269, 3155–3158. doi:10.1016/S0021-9258(17)41842-4
Colotti, G., and Ilari, A. (2011). Polyamine metabolism in leishmania: From arginine to trypanothione. Amino Acids 40, 269–285. doi:10.1007/S00726-010-0630-3
Das, M., Kumar, R., and Dubey, V. (2015). Ornithine decarboxylase of Leishmania donovani: Biochemical properties and possible role of N-terminal extension. Protein Pept. Lett. 22, 130–136. doi:10.2174/0929866521666140616115357
del Rio, B., Alvarez-Sieiro, P., Redruello, B., Martin, M. C., Fernandez, M., Ladero, V., et al. (2018). Lactobacillus rossiae strain isolated from sourdough produces putrescine from arginine. Sci. Rep. 8, 3989. doi:10.1038/s41598-018-22309-6
Ducros, V., Ruffieux, D., Belva-Besnet, H., de Fraipont, F., Berger, F., and Favier, A. (2009). Determination of dansylated polyamines in red blood cells by liquid chromatography–tandem mass spectrometry. Anal. Biochem. 390, 46–51. doi:10.1016/J.AB.2009.04.007
El-Sayed, A. S. A., George, N. M., Yassin, M. A., Alaidaroos, B. A., Bolbol, A. A., Mohamed, M. S., et al. (2019). Purification and characterization of ornithine decarboxylase from Aspergillus terreus; kinetics of inhibition by various inhibitors. Molecules 24, E2756. doi:10.3390/MOLECULES24152756
Erlanson, D. A., Fesik, S. W., Hubbard, R. E., Jahnke, W., and Jhoti, H. (2016). Twenty years on: The impact of fragments on drug discovery. Nat. Rev. Drug Discov. 15, 605–619. doi:10.1038/nrd.2016.109
Erlanson, D. A. (2012). Introduction to fragment-based drug discovery. Top. Curr. Chem. 317, 1–32. doi:10.1007/128_2011_180
Fagerström, R., Seppänen, P., and Jänne, J. (1984). A rapid chemiluminescence-based method for the determination of total polyamines in biological samples. Clin. Chim. Acta. 143, 45–50. doi:10.1016/0009-8981(84)90036-6
Gaboriau, F., Havouis, R., Groussard, K., Moulinoux, J. P., and Lescoat, G. (2005). Measurement of ornithine decarboxylase activity in cell extracts using mass spectrometry detection of dansylated putrescine. Anal. Biochem. 341, 385–387. doi:10.1016/J.AB.2005.01.026
Gaboriau, F., Havouis, R., Moulinoux, J. P., and Delcros, J. G. (2003). Atmospheric pressure chemical ionization-mass spectrometry method to improve the determination of dansylated polyamines. Anal. Biochem. 318, 212–220. doi:10.1016/S0003-2697(03)00214-8
Gerner, E. W., and Meyskens, F. L. (2004). Polyamines and cancer: Old molecules, new understanding. Nat. Rev. Cancer 4, 781–792. doi:10.1038/nrc1454
Ghale, G., and Nau, W. M. (2014). Dynamically analyte-responsive macrocyclic host-fluorophore systems. Acc. Chem. Res. 47, 2150–2159. doi:10.1021/AR500116D/ASSET/IMAGES/LARGE/AR-2014-00116D
Grishin, N. v., Osterman, A. L., Brooks, H. B., Phillips, M. A., and Goldsmith, E. J. (1999). X-ray structure of ornithine decarboxylase from Trypanosoma brucei: The native structure and the structure in complex with α difluoromethylornithine. Biochemistry 38, 15174–15184. doi:10.1021/bi9915115
Henley, C. M., Muszynski, C., Cherian, L., and Robertson, C. S. (1996). Activation of ornithine decarboxylase and accumulation of putrescine after traumatic brain injury. J. Neurotrauma 13, 487–496. doi:10.1089/NEU.1996.13.487
Holbert, C. E., Cullen, M. T., Casero, R. A., and Stewart, T. M. (2022). Polyamines in cancer: Integrating organismal metabolism and antitumour immunity. Nat. Rev. Cancer 22, 467–480. doi:10.1038/s41568-022-00473-2
Igarashi, K., and Kashiwagi, K. (2019). The functional role of polyamines in eukaryotic cells. Int. J. Biochem. Cell Biol. 107, 104–115. doi:10.1016/J.BIOCEL.2018.12.012
Ivanov, A. v., and Khomutov, A. R. (2022). Biogenic polyamines and related metabolites. Biomolecules 12, 14. doi:10.3390/BIOM12010014
Jones, R. D., Hampton, J. K., and Preslock, J. P. (1972). Ornithine decarboxylase in mouse placenta assayed by a paper disc method for 14CO2 capture. Anal. Biochem. 49, 147–154. doi:10.1016/0003-2697(72)90252-7
Kabra, P. M., Lee, H. K., Lubich, W. P., and Marton, L. J. (1986). Solid-phase extraction and determination of dansyl derivatives of unconjugated and acetylated polyamines by reversed-phase liquid chromatography: Improved separation systems for polyamines in cerebrospinal fluid, urine and tissue. J. Chromatogr. 380, 19–32. doi:10.1016/S0378-4347(00)83621-X
Khan, A., Gamble, L. D., Upton, D. H., Ung, C., Yu, D. M. T., Ehteda, A., et al. (2021). Dual targeting of polyamine synthesis and uptake in diffuse intrinsic pontine gliomas. Nat. Commun. 12, 1–13. doi:10.1038/s41467-021-20896-z
Kim, H. I., Schultz, C. R., Buras, A. L., Friedman, E., Fedorko, A., Seamon, L., et al. (2017). Ornithine decarboxylase as a therapeutic target for endometrial cancer. PLoS One 12, 1–18. doi:10.1371/journal.pone.0189044
Kobayashi, Y. (1963). Determination of histidine decarboxylase activity by liquid scintillation counting of C-14-O2. Anal. Biochem. 5, 284–290. doi:10.1016/0003-2697(63)90080-0
Kvannes, J., and Flatmark, T. (1987). Rapid and sensitive assay of ornithine decarboxylase activity by high-performance liquid chromatography of the o-phthalaldehyde derivative of putrescine. J. Chromatogr. 419, 291–295. doi:10.1016/0378-4347(87)80289-X
Legaz, M. E., Fontaniella, B., de Armas, R., and Vicente, C. (2001). Determination by high performance liquid chromatography of ornithine and lysine decaboxylases in sugar cane juices. Chromatographia 53 (1), S260–S265. doi:10.1007/BF02490339
Li, L., Hara, K., Liu, J., Yu, Y., Gao, L., Wang, Y., et al. (2008). Rapid and simultaneous determination of hair polyamines as N-heptafluorobutyryl derivatives by gas chromatography-mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 876, 257–260. doi:10.1016/j.jchromb.2008.10.054
Li, Z., Sun, S., Liu, F., Pang, Y., Fan, J., Song, F., et al. (2012). Large fluorescence enhancement of a hemicyanine by supramolecular interaction with cucurbit[6]uril and its application as resettable logic gates. Dyes Pigments 93, 1401–1407. doi:10.1016/J.DYEPIG.2011.10.005
Liao, C., Wang, Y., Tan, X., Sun, L., and Liu, S. (2015). Discovery of novel inhibitors of human S-adenosylmethionine decarboxylase based on in silico high-throughput screening and a non-radioactive enzymatic assay. Sci. Rep. 5, 10754. doi:10.1038/SREP10754
Liu, R., Jia, Y., Cheng, W., Ling, J., Liu, L., Bi, K., et al. (2011). Determination of polyamines in human urine by precolumn derivatization with benzoyl chloride and high-performance liquid chromatography coupled with Q-time-of-flight mass spectrometry. Talanta 83, 751–756. doi:10.1016/J.TALANTA.2010.10.039
Luqman, S., Luqman, S., Masood, N., Srivastava, S., and Dubey, V. (2013). A modified spectrophotometric and methodical approach to find novel inhibitors of ornithine decarboxylase enzyme: A path through the maze. Protoc. Exch. 4, 1–18. doi:10.1038/PROTEX.2013.045
Mäkitie, L. T., Kanerva, K., Polvikoski, T., Paetau, A., and Andersson, L. C. (2010). Brain neurons express ornithine decarboxylase‐activating antizyme inhibitor 2 with accumulation in alzheimer’s disease. Brain Pathol. 20, 571–580. doi:10.1111/J.1750-3639.2009.00334.X
Mamont, P. S., Duchesne, M. C., Grove, J., and Bey, P. (1978). Anti-proliferative properties of DL-α-difluoromethyl ornithine in cultured cells. A consequence of the irreversible inhibition of ornithine decarboxylase. Biochem. Biophys. Res. Commun. 81, 58–66. doi:10.1016/0006-291X(78)91630-3
Marton, L. J., and Pegg, A. E. (1995). Polyamines as targets for therapeutic intervention. Annu. Rev. Pharmacol. Toxicol. 35, 55–91. doi:10.1146/ANNUREV.PA.35.040195.000415
McCann, P. P., and Pegg, A. E. (1992). Ornithine decarboxylase as an enzyme target for therapy. Pharmacol. Ther. 54, 195–215. doi:10.1016/0163-7258(92)90032-U
Metcalf, B. W., Bey, P., Danzin, C., Jung, M. J., Casara, P., and Vevert, J. P. (1978). Catalytic irreversible inhibition of mammalian ornithine decarboxylase (E.C.4.1.1.17) by substrate and product analogs. J. Am. Chem. Soc. 100, 2551–2553. doi:10.1021/JA00476A050
Michael, A. J. (2016). Biosynthesis of polyamines and polyamine-containing molecules. Biochem. J. 473, 2315–2329. doi:10.1042/BCJ20160185
Miller-Fleming, L., Olin-Sandoval, V., Campbell, K., and Ralser, M. (2015). Remaining mysteries of molecular biology: The role of polyamines in the cell. J. Mol. Biol. 427, 3389–3406. doi:10.1016/J.JMB.2015.06.020
Mitchell, J. L. A., and Chen, H. J. (1990). Conformational changes in ornithine decarboxylase enable recognition by antizyme. Biochim. Biophys. Acta 1037, 115–121. doi:10.1016/0167-4838(90)90109-S
Mitchell, J. L. A., Rynning, M. D., Chen, H. J., and Hicks, M. F. (1988). Interrelation between the charge isoforms of mammalian ornithine decarboxylase. Arch. Biochem. Biophys. 260, 585–594. doi:10.1016/0003-9861(88)90485-7
Mukhopadhyay, R., and Madhubala, R. (1995). Leishmania donovani: Cellular control of ornithine decarboxylase in promastigotes. Int. J. Biochem. Cell Biol. 27, 947–952. doi:10.1016/1357-2725(95)00053-R
Nau, W. M., Ghale, G., Hennig, A., Bakirci, H., and Bailey, D. M. (2009). Substrate-selective supramolecular tandem assays: Monitoring enzyme inhibition of arginase and diamine oxidase by fluorescent dye displacement from calixarene and cucurbituril macrocycles. J. Am. Chem. Soc. 131, 11558–11570. doi:10.1021/ja904165c
Nilam, M., Gribbon, P., Reinshagen, J., Cordts, K., Schwedhelm, E., Nau, W. M., et al. (2017). A label-free continuous fluorescence-based assay for monitoring ornithine decarboxylase activity with a synthetic putrescine receptor. SLAS Discov. 22, 906–914. doi:10.1177/2472555216689288
Osterman, A. L., Brooks, H. B., Rizo, J., and Phillips, M. A. (1997). Role of Arg-277 in the binding of pyridoxal 5’-phosphate to Trypanosoma brucei ornithine decarboxylase. Biochemistry 36, 4558–4567. doi:10.1021/BI962916H/ASSET/IMAGES/LARGE/BI962916HF00007.JPEG
Osterman, A. L., Kinch, L. N., Grishin, N. v., and Phillips, M. A. (1995). Acidic residues important for substrate binding and cofactor reactivity in eukaryotic ornithine decarboxylase identified by alanine scanning mutagenesis. J. Biol. Chem. 270, 11797–11802. doi:10.1074/JBC.270.20.11797
Pegg, A. E., Seely, J., and Zagon, I. S. (1982). Autoradiographic identification of ornithine decarboxylase in mouse kidney by means of α-[5-14C]difluoromethylornithine. Sci. (1979) 217, 68–70. doi:10.1126/science.6806900
Perdeh, J., Berioso, B., Love, Q., LoGiudice, N., Le, T. L., Harrelson, J. P., et al. (2020). Critical functions of the polyamine putrescine for proliferation and viability of Leishmania donovani parasites. Amino Acids 52, 261–274. doi:10.1007/s00726-019-02736-z
Pösö, H., and Pegg, A. E. (1983). Measurement of the amount of ornithine decarboxylase in Saccharomyces cerevisiae and Saccharomyces uvarum by using alpha-[5-14C]difluoromethylornithine. Biochim. Biophys. Acta 747, 209–214. doi:10.1016/0167-4838(83)90099-7
Poulin, R., Lu, L., Ackermann, B., Bey, P., and Pegg, A. E. (1992). Mechanism of the irreversible inactivation of mouse ornithine decarboxylase by alpha-difluoromethylornithine. Characterization of sequences at the inhibitor and coenzyme binding sites. J. Biol. Chem. 267, 150–158. doi:10.1016/S0021-9258(18)48472-4
Schultz, C. R., Gruhlke, M. C. H., Slusarenko, A. J., and Bachmann, A. S. (2020). Allicin, a potent new ornithine decarboxylase inhibitor in neuroblastoma cells. J. Nat. Prod. 83, 2518–2527. doi:10.1021/acs.jnatprod.0c00613
Schultz, C. R., Swanson, M. A., Dowling, T. C., and Bachmann, A. S. (2021). Probenecid increases renal retention and antitumor activity of DFMO in neuroblastoma. Cancer Chemother. Pharmacol. 88, 607–617. doi:10.1007/s00280-021-04309-y
Scriven, F., Wlasichuk, K. B., and Palcic, M. M. (1988). A continual spectrophotometric assay for amino acid decarboxylases. Anal. Biochem. 170, 367–371. doi:10.1016/0003-2697(88)90644-6
Seely, J. E., Poso, H., and Pegg, A. E. (1982). Measurement of the number of ornithine decarboxylase molecules in rat and mouse tissues under various physiological conditions by binding of radiolabelled alpha-difluoromethylornithine. Biochem. J. 206, 311–318. doi:10.1042/BJ2060311
Smith, T. A., and Marshall, J. H. A. (1988). The oxidative decarboxylation of ornithine by extracts of higher plants. Phytochemistry 27, 703–710. doi:10.1016/0031-9422(88)84079-2
Smithson, D. C., Lee, J., Shelat, A. A., Phillips, M. A., and Guy, R. K. (2010a). Discovery of potent and selective inhibitors of Trypanosoma brucei ornithine decarboxylase. J. Biol. Chem. 285, 16771–16781. doi:10.1074/jbc.M109.081588
Smithson, D. C., Shelat, A. A., Baldwin, J., Phillips, M. A., and Guy, R. K. (2010b). Optimization of a non-radioactive high-throughput assay for decarboxylase enzymes. Assay. Drug Dev. Technol. 8, 175–185. doi:10.1089/ADT.2009.0249
Solano, F., Peñafiel, R., Solano, M. E., and Lozano, J. A. (1985). Equilibrium between active and inactive forms of rat liver ornithine decarboxylase mediated by L-ornithine and salts. FEBS Lett. 190, 324–328. doi:10.1016/0014-5793(85)81311-9
Somani, R. R., Rai, P. R., and Kandpile, P. S. (2017). Ornithine decarboxylase inhibition: A strategy to combat various diseases. Mini Rev. Med. Chem. 18, 1008–1021. doi:10.2174/1389557517666170927130526
Sweeney, C. (2020). Targeting the polyamine pathway—“a means” to overcome chemoresistance in triple-negative breast cancer. J. Biol. Chem. 295, 6278–6279. doi:10.1074/jbc.H120.013736
Tobias, K. E., and Kahana, C. (1994). Formation of Functional Cross-Species Heterodimers of Ornithine Decarboxylase* * the two active sites in ornithine decarboxylase (ODC) are formed at the dimer interface with Lys-69 and Cys-360 contributing to each active site from opposite monomers. Biochemistry 33, 13662–13667. Available at: https://pubs.acs.org/sharingguidelines (Accessed May 11, 2022).
Vanisree, A. J., and Shyamaladevi, C. S. (2006). The effect of N-acetylcysteine in combination with vitamin C on the activity of ornithine decarboxylase of lung carcinoma cells - in vitro. Life Sci. 79, 654–659. doi:10.1016/j.lfs.2006.02.009
Villa-Ruano, N., Pacheco-Hernández, Y., Zárate-Reyes, J. A., Cruz-Durán, R., and Lozoya-Gloria, E. (2019). Volatile composition and biological activities of the leaf essential oil from zanthoxylum limoncello grown in oaxaca, méxico. Chem. Biodivers. 16, e1800498. doi:10.1002/CBDV.201800498
Wang, Y., and Bachrach, U. (2000). A luminescence-based test for determining ornithine decarboxylase activity. Anal. Biochem. 287, 299–302. doi:10.1006/ABIO.2000.4873
WHO (2022), WHO includes combination of eflornithine and nifurtimox in its Essential List of Medicines for the treatment of human African trypanosomiasis. Available at: https://www.who.int/news/item/08-05-2009-who-includes-combination-of-eflornithine-and-nifurtimox-in-its-essential-list-of-medicines-for-the-treatment-of-human-african-trypanosomiasis [Accessed August 2, 2022].
Xiao, Y., McCloskey, D. E., and Phillips, M. A. (2009). RNA Interference-mediated silencing of ornithine decarboxylase and spermidine synthase genes in trypanosoma brucei provides insight into regulation of polyamine biosynthesis. Eukaryot. Cell 8, 747–755. doi:10.1128/EC.00047-09
Yoshida, H., Harada, H., Nakano, Y., Nohta, H., Ishida, J., and Yamaguchi, M. (2004). Liquid chromatographic determination of polyamines in human urine based on intramolecular excimer-forming fluorescence derivatization using 4-(1-pyrene)butanoyl chloride. Biomed. Chromatogr. 18, 687–693. doi:10.1002/BMC.377
Keywords: radiolabeling, CO2 release, putrescine oxidation, TNBS colorimetric assay, HPLC, mass spectrometry, spectroscopic assays
Citation: Tinoco LW, da Silva Santos BM, Soares JMdS and Finelli FG (2022) Analytical assays to evaluate enzymatic activity and screening of inhibitors for ornithine decarboxylase. Front. Anal. Sci. 2:1018080. doi: 10.3389/frans.2022.1018080
Received: 12 August 2022; Accepted: 22 September 2022;
Published: 10 October 2022.
Edited by:
Quezia B. Cass, Federal University of São Carlos, BrazilReviewed by:
Surabhi Chandra, University of Nebraska at Kearney, United StatesRobert Stephen Phillips, University of Georgia, United States
Copyright © 2022 Tinoco, da Silva Santos, Soares and Finelli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luzineide W. Tinoco, bHd0aW5vY29AaXBwbi51ZnJqLmJy
 Luzineide W. Tinoco
Luzineide W. Tinoco Bruno Maia da Silva Santos
Bruno Maia da Silva Santos Jhones Matheus da Silva Soares
Jhones Matheus da Silva Soares Fernanda Gadini Finelli
Fernanda Gadini Finelli