- 1Department of Biology, University of Massachusetts Boston, Boston, MA, United States
- 2Department of Chemistry, Villanova University, Villanova, PA, United States
- 3Department of Biology, Texas State University, San Marcos, TX, United States
The emerging fungal pathogen Batrachochytrium salamandrivorans (Bsal) threatens the diversity of amphibians, particularly in North America where it is projected to invade. Amphibian skin defenses include a mucosal layer containing microorganisms that can potentially modulate host response to pathogens such as Bsal. In this study, we focused on the composition of the skin microbiome across life stages of spotted salamanders (Ambystoma maculatum). We also evaluated the stress hormone corticosterone and skin microbiome response to inoculations with Bsal and probiotics at both the larval and juvenile developmental stages, and the response to different environmental conditions. Results indicated that both bacterial and fungal communities found on the skin significantly differed in structure and diversity between life stages of A. maculatum. Exposure to three different probiotics (Bacillus thuringiensis, Chryseobacterium rhizoplanae, and Penicillium sp.) and Bsal evoked shifts in the microbiome of larvae and juveniles, and the metabolite profile of the larval mucosal layer of A. maculatum. Despite changes in the microbiome, all tested probiotics and Bsal were unable to persist on the skin. Larval bacterial microbiomes shifted in response to Bsal and B. thuringiensis with no significant impacts on antifungal function or bacteria richness, however fungi strongly responded to Bsal and B. thuringiensis application. This indicates that developmental shifts in the microbiome can be initiated by microbial applications such as B. thuringiensis, a widely used mosquito larvicide. Overall, experimental results indicate that life stage, growth and development, and environmental conditions appeared to be the main factors driving changes in the amphibian skin microbiome and potential anti-Batrachochytrium function.
Introduction
Batrachochytrium salamandrivorans (Bsal) is an emerging fungal pathogen affecting amphibians, primarily salamanders (Martel et al., 2013; Gray et al., 2015), which has the potential to spread worldwide. It is one of two fungi, along with Batrachochytrium dendrobatidis (Bd), known to cause the disease chytridiomycosis in amphibians (reviewed in Woodhams et al., 2018). Clinical signs of disease include lethargy, weight loss, loss of righting ability, skin sloughing or lesions, and eventually dehydration, a systemic inflammatory response, and electrolyte imbalances leading to cardiac arrest and death (Sheley et al., 2023). Infectious zoospores invade keratinized epithelial cells and proliferate, moving from host to host through direct contact or through environmental substrates like soil or water. Bsal has yet to be detected in North America (Waddle et al., 2020), however, projections of the pathogen, including its presence in the pet trade and capacity to persist in soils (McGrath-Blaser et al., 2024), reveal that invasion may be inevitable (Gray et al., 2015). Bsal has already led to rapid declines of fire salamanders (Salamandra salamandra) in Europe (Lötters et al., 2020), while other species appear to be largely unaffected (Martel et al., 2013). The link between fungal pathogens and amphibian skin, including resident microorganisms with antipathogen function (Bates et al., 2019), suggests the incorporation of the microbiome into disease management should be prioritized.
The mucus layer surrounding the skin of amphibians has several components including skin defense peptides, alkaloids, toxins, and a diverse community of microorganisms producing secondary metabolites (reviewed in Woodhams et al., 2023). Many of these components can contribute to pathogen defense. For example, host-derived antimicrobial peptides and symbiotic skin microbes can inhibit the growth of fungal pathogens (Le Sage et al., 2021). Skin microbiomes have received considerable attention due to their potential use in probiotic strategies mitigating chytridiomycosis (Bletz et al., 2013). There are several factors that may influence the skin microbiome of amphibians including body microhabitats, environment (Kueneman et al., 2017), host species or population, life stage, and pathogen exposure (reviewed in Becker et al., 2023). Metamorphosis alone has a substantial role in restructuring the microbial skin community of frogs during which changes in immune function also occur (Humphries et al., 2022), a notion recently demonstrated in newts (Hartmann et al., 2023) and fully aquatic salamanders (Martínez-Ugalde et al., 2022).
Amphibian-associated probiotics represent one potential mitigation strategy against Bd or Bsal infection (Kueneman et al., 2016; Bletz et al., 2018). The application of beneficial bacteria found on amphibians, such as Janthinobacterium lividum, has been effective against Bd when applied to the skin of some species of frogs (Harris et al., 2009). However, in experiments where J. lividum was applied to the skin of Panamanian golden frogs, Atelopus zeteki, J. lividum did not persist on the skin for an extended period and did not improve survival (Becker et al., 2012), nor did genetically modifying an existing symbiont to produce the antifungal compound violacein (Becker et al., 2021). It is likely that using probiotics that are native to species’ habitats, or exposure earlier in development (e.g., Pereira et al., 2018), will provide improved results and persistence on the skin. Furthermore, Kearns et al. (2017) suggested that Bd-inhibitory fungal probiotics might be an effective alternative to bacterial probiotics, and a more natural approach compared to chemical disinfectants applied to multiple individuals simultaneously (Thumsová et al., 2024). Bd-inhibitory fungal probiotics exhibited only a minor stress response, did not inhibit host defense peptides, and were more prevalent on the skin of captive-bred frogs compared to Bd-inhibitory bacteria (Kearns et al., 2017).
In this study, the roles of the bacterial and fungal skin microbiome and probiotics were evaluated in disease dynamics by investigating the spotted salamander, Ambystoma maculatum, and its response to Bsal exposure. We first analyzed the A. maculatum microbiome over different life stages using amplicon sequencing, hypothesizing that both captive conditions and life stage would impact microbial community structure. We next performed two probiotic and Bsal exposure experiments on larval and juvenile salamanders to determine the effect of life stage on pathogen defense and probiotic application. We hypothesized that the potential probiotics would be able to persist on the skin of spotted salamanders and provide protection against Bsal, if applied early in development, or that strong colonization resistance may indicate a disease-resistant phenotype. Overall, our study showed significant shifts in the spotted salamander microbiome due to life stage, probiotic application, and pathogen exposure with potential implications for immune function.
Methods
Surface microbiomes across life stages
We aimed to determine how the surface microbiome differed among the environment (water at field oviposition site), eggs, larvae, juveniles, and adult stages of spotted salamanders, Ambystoma maculatum, located in the Blue Hills Reservation in Milton, Massachusetts, USA (42.214 N, -71.087 W). Microbiome samples were collected from four pond water swabs, 12 egg swabs in the field (3 swabs from 4 unique clutches), 5 captive larvae (1 per tank containing several larvae), 12 individually housed captive post-metamorphic juveniles, and 13 adults in the field (Figure 1). To collect samples, egg masses or salamanders at different life-history stages were lifted with freshly gloved hands and rinsed with sterile water to remove debris and transient microbes before swabbing. Each digit and lateral strokes of the dorsal and ventral regions were collected for 30 swab strokes per animal. Swabs were kept on ice and stored at –20°C until DNA extraction. As discussed below, we note that life stages with similar origins (field sampled or sampled in captivity) are most comparable because both life stage and captive housing can impact the amphibian skin microbiome (e.g., Bates et al., 2019, meta-analysis in Kueneman et al., 2022; Burgess and Bishop, 2023).
Figure 1. Timeline of spotted salamander field sampling and exposure experiments presented in this study.
Metagenomic DNA was extracted using the IBI gMax DNA kit (IBI Scientific, Dubuque, IA, USA) with the addition of lysozyme. We characterized the bacterial communities through PCR by amplifying the 16S rRNA gene using the primer pair 515F/806R following Earth Microbiome Project protocols (Caporaso et al., 2012; Parada et al., 2016; Thompson et al., 2017). Briefly, each sample was amplified in 25μl duplicate reactions with the following chemistry: 11μl of PCR-grade water, 10μl of PCR master mix (5 PRIME HotMasterMix, Quantabio, Beverly, MA, USA), 1μl of forward and reverse primers, and 2μl of undiluted DNA template. Reactions were carried out with the following conditions: 94°C for 3m, followed by 35 cycles of 94°for 45s, 50°C for 60s, and 72°C for 90s, with a final extension at 72°C for 10m. Fungal PCR was processed identically, except for the addition of 3μl of undiluted DNA template per reaction and the use of the ITS1f/ITS2 primer pair (Bokulich and Mills, 2013). For both genes, PCR product was verified via electrophoresis and each sample was pooled prior to purification using the ThermoFisher Scientific GeneJET PCR Purification Kit (ThermoFisher Scientific, Waltham, MA, USA). Samples were analyzed for DNA concentration using a Nanodrop (Thermo Scientific NanoDrop 2000/Spectrophotometer) and pooled in equimolar amounts. Sequencing was performed on an Illumina MiSeq using a v2 300 cycle kit at the University of Massachusetts Boston.
Larval exposure experiment
Three egg clutches were collected at Blue Hills Reservation in Milton, Massachusetts in March 2016 and animals were reared in the lab and co-housed by treatment (see below) according to the methods in (Barnhart et al., 2020). Larvae (n = 36) were divided into three treatment groups (control, Bsal, and probiotic; n= 12 per treatment) to test for effects of treatment on the microbiome. The bacterial probiotic Bacillus thuringiensis was chosen based on its use as a common mosquito control agent around ponds and other amphibian habitats (Northeast Massachusetts Mosquito Control and Wetlands Management District, https://www.nemassmosquito.org/public-education/pages/larvicide; Environmental Protection Agency, https://www.epa.gov/mosquitocontrol/bti-mosquito-control, accessed 14 February 2023; but see Allgeier et al., 2019 and Land et al., 2023 for potential food web impacts) as well as evidence of significant growth inhibition of Bd and Bsal in preliminary experiments (Supplementary Figure S1). Briefly, Bd (strain JEL 423) and Bsal, were cultured on 1% tryptone agar for a period of 5-7 days to stimulate zoospore production. Following incubation, the plates were gently flooded with 1% tryptone solution, and the resultant liquid was filtered through 0.45 µm pore size filters to exclude zoosporangia. The enumeration of zoospores was performed with a hemocytometer and we standardized cellular density to 5 x 106 zoospores ml-1. To assess the inhibitory effects on Bd growth we inoculated 96-well plates with 50 µl of Bd zoospores and an equivalent volume of bacterial supernatant. In control wells, heat-killed Bd zoospores were used alongside wells devoid of zoospores, while a positive control featured 50 µl of 1% tryptone medium. The growth dynamics of Bd were quantitatively assessed at 480 nm on days 0, 3, 5, and 7. Percent inhibition of Bd/Bsal growth in the presence of metabolites was calculated by evaluating the slope of optical density over the 7-day incubation period and we compared this to the average growth slope observed in the positive control wells.
According to calculations based on effective concentrations for mosquito larvicide use, one gram of B. thuringiensis (coated corn-cob pellets, VectoBac WDG) was added to 5 L of artificial pond water and mixed by stirring. The spotted salamander larvae were each placed in 500 ml of the B. thuringiensis solution for 90 minutes. To investigate effects of Bsal exposure, Bsal was collected by flooding Petri plates with 3-6 ml of water with or without thyroid hormone added as in Barnhart et al. (2020). Thyroid hormone is known to induce gene expression in Bd, including induction of subtilisin-like serine protease and other potential virulence factors (Thekkiniath et al., 2015). Bsal was filtered and zoospores were counted using a hemocytometer. Larvae were exposed to Bsal at 5x103 zoospores/ml in 200 ml tanks for 24 hours. Metagenomic DNA was extracted with the IBI gMax kit from a subset of control (n=5), Bsal exposed (n=7), and B. thuringiensis exposed (n=8) individuals and processed for microbiome sequencing as described above.
Juvenile exposure experiment
In a second experiment, we raised spotted salamander eggs collected in March 2016 from the Blue Hills Reservation to the juvenile stage. Animals were co-housed by treatment (see below) in terrariums (2L capacity mouse cages) with 2.5 cm of soil (Atlanta Botanical Garden Mix, Josh’s Frogs, Owasso, MI, USA), sphagnum moss, springtails (Josh’s Frogs), and fed with crickets on a bi-weekly basis. Terrariums were moistened with sterile artificial pond water as needed.
Ninety-five juveniles were divided into one of nine groups and exposed to different treatments, Bsal loads, or probiotic application: (1) control, (2) a stressed ‘agitated’ group, (3) low Bsal dose (5x103 zoospores), (4) high Bsal dose (1.75x106 zoospores), (5) repeated exposure (n=3 exposures) to 5x103 Bsal zoospores, (6) high Bsal (1.75x106 zoospores) with thyroid hormone, (7) B. thuringiensis application, (8) Chryseobacterium rhizoplanae application, (9) Penicillium sp. application. The three microbial strains were chosen based upon their ability to inhibit Bsal (Supplementary Figures S1, S2; Kearns et al., 2017 for Penicillium sp.).
Bsal zoospores were processed as described above in the larval experiment. Chryseobacterium rhizoplanae and B. thuringiensis were cultured for three days in 1% tryptone broth and colony forming units (CFUs) were quantified with dilution plates. Bacteria were diluted in sterile water to a desired concentration of 1.15 x 105 CFUs mL-1 for B. thuringiensis and 1.1 x 107 CFUs ml -1 for C. rhizoplanae. One mL of bacterial solution was added to each salamander exposure bath. Penicillium sp. was cultured for two days on Sabouraud dextrose agar. Two plates were then flooded with 3mL of sterile water and a cell scraper was used to disturb the surface of the agar to collect fungal cells. Animals were exposed to all treatments for 90 minutes. Salamanders were sampled for their microbiome as described above at 16- and 30-days post-exposure to a treatment.
For one bacterial treatment (C. rhizoplanae) and controls, skin associated metabolites were examined at 36 days after exposure to test for metabolomic differences. This isolate was chosen due to the widespread nature of the genus Chryseobacterium on amphibian skin (Kueneman et al., 2019) as well as the isolate’s ability to inhibit both Bd and Bsal (Supplementary Figure S2). Metabolites were extracted from swabs and processed using a HPLC fingerprinting method following Umile et al. (2014). We performed a non-parametric PERMANOVA to assess the effects of treatment on metabolite profiles. In addition, we calculated Kendall correlations with metabolites and sOTU abundance.
To evaluate the stress response of salamanders, we measured corticosterone release rates similarly to Charbonnier et al. (2018). Briefly, juveniles were placed individually in plastic cups with 30 mL of sterile artificial pond water for 1 hour between 0900–1300 h to minimize the effects of circadian rhythms. Samples were collected on the day of exposure (day 0, one-hour post-exposure) and on day 30. Mass was measured with each collection of water-borne hormones to control differences in corticosterone release rates based on size. Corticosterone release rates were measured in units of pg g -1 hr-1. Significant differences in corticosterone were evaluated with an ANOVA.
Microbiome sequence processing and data analysis
All sequencing data was quality filtered and demultiplexed using QIIME2 (version 2022.2; Bolyen et al., 2019). Sequences were clustered into sub-operational taxonomic units (sOTUs) using deblur (Amir et al., 2017). Samples were rarefied to the lowest sampling depth (4,700-5,600 sequences per sample depending on the experiment) to maintain sample diversity and retain the most samples. QIIME2 was used to calculate alpha diversity (sOTU richness and phylogenetic diversity) and beta diversity (UniFrac, Lozupone and Knight, 2005). We chose to use unweighted UniFrac to assay overall shifts in community membership due to treatment, however it is of note that for all experiments there was no significant difference in similarity matrices as determined by a mantel test. Taxonomy was assigned using Greengenes (v. 13.8; McDonald et al., 2013) for bacteria and the UNITE database (v. 9, Abarenkov et al., 2010) for fungi. Subsequent analyses were performed in R. We omitted fungal data from eggs due to insufficient data.
We used generalized linear models to compare alpha diversity and a non-parametric PERMANOVA (Dixon, 2003) to compare community composition. We calculated the mean proportion of putative anti-Batrachochytrium sOTUs by matching sOTUs to the 2023 version of the Antifungal Isolates Database (Woodhams et al., 2015; https://github.com/AmphiBac/AmphiBac-Database) with vsearch (Rognes et al., 2016). Utilization of the Antifungal Isolates Database allowed for prediction of the relative effectiveness of the bacterial microbiome to inhibit Batrachochytrium fungi. A beta regression was performed to model differences in the proportion of putative anti-Batrachochytrium bacteria across the various treatments (glmmTMB). Finally, we used a Kruskal-Wallis test with an FDR-corrected p-value to assess bacterial/fungal genera that respond to Bsal and probiotic application relative to control samples.
Results
Skin microbiomes across life stages
The diversity, composition, and community structure of skin bacterial communities on spotted salamanders differed among life stages (Figure 2). sOTU richness significantly varied among the pond water and life stages (GLM, c2 = 72.313, p < 0.001; Figure 2A). Larvae had lower richness compared to pond water (emmeans, t = 4.916, p < 0.001), eggs (t = 5.420, p < 0.001), juveniles (t = 4.463, p < 0.001), and adults (t = 4.510, p < 0.001), and eggs had higher richness than juveniles (t = 3.656, p = 0.006) and adults (t = 3.654, p = 0.006, Figure 2A). The proportion putative anti-Batrachochytrium bacteria also varied across life stages (glmmTMB Beta Regression: AntiF: c2 = 73.653, p < 0.001; Figure 2C). Pond water had a higher proportion compared to the larvae (emmeans: t = 3.195, p = 0.022), but a lower proportion compared to juveniles (t = 3.009, P = 0.035) and adults (t = 2.900, p = 0.045). Eggs and larvae both had lower putative anti-Batrachochytrium proportions compared to juveniles (Eggs: t = 5.966, p < 0.001; Larvae: t = 6.136, p < 0.001) and adults (Eggs: t = 5.872, p < 0.001; Larvae: t = 6.063, p < 0.001).
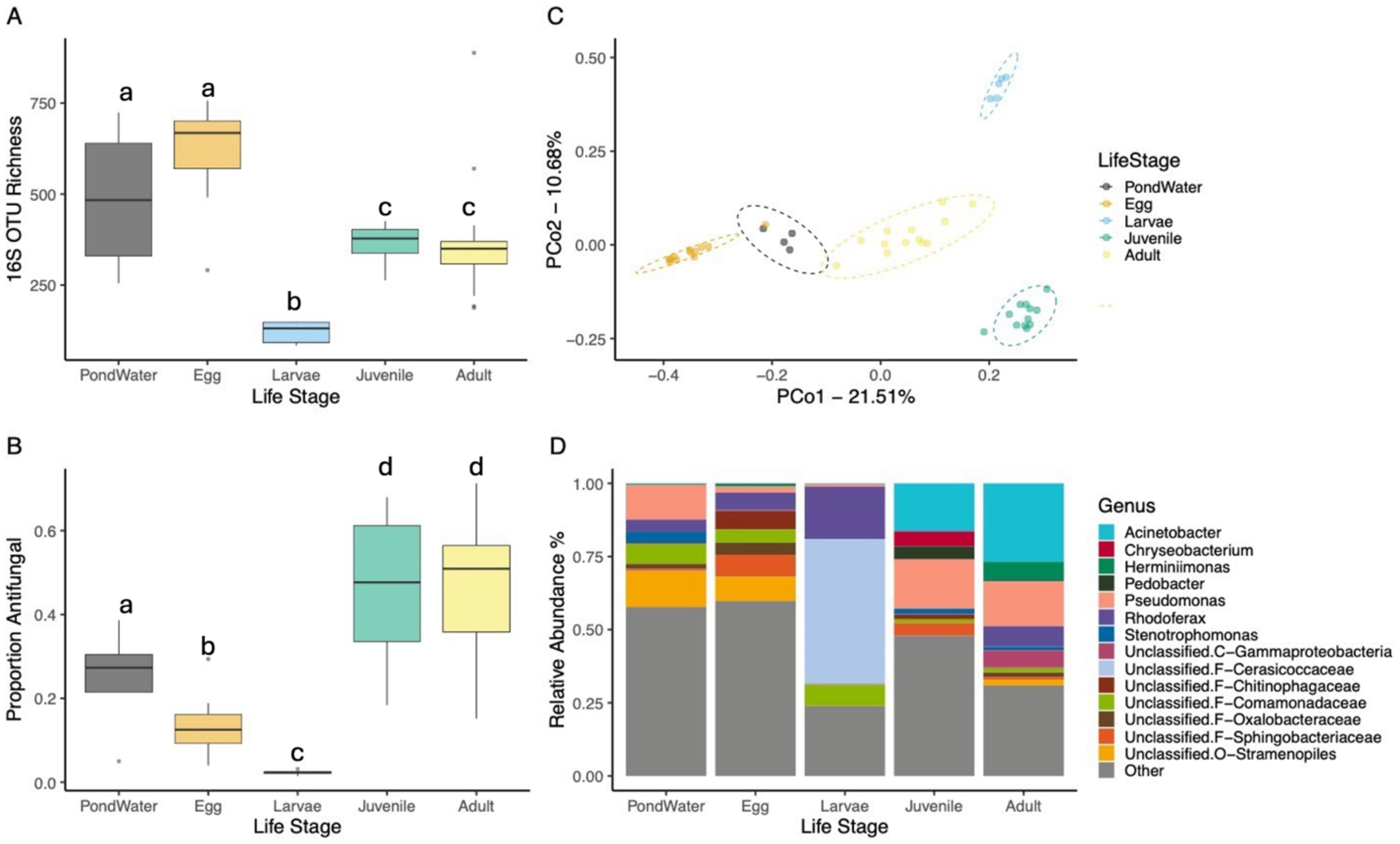
Figure 2. Spotted salamander bacterial communities across different life stages. Adults, eggs, and pond water were sampled in the field and the larvae and juveniles were control animals from the larval and juvenile I lab experiments. (A) sOTU richness, (B) proportion of anti-Batrachochytrium sp. bacteria, (C) principal coordinates analysis based on unweighted UniFrac similarity, and (D) stacked bar-chart of the 14 most abundant bacterial genera. Letters in (A, B) indicate categories that are significantly different.
The four life stages also significantly differed in community composition (PERMANOVA, F4,41 = 4.012, R2 = 0.28, all pairwise p < 0.05; Figure 2C, Supplementary Table S1). Bacterial taxonomic composition (Figure 2D) across all life stages except for larvae was dominated by members of the Proteobacteria. The surface bacteria on eggs were largely composed of bacteria from the orders Burkholderiales, Sphingobacteriales, and Psuedomonadales. The skin bacteria on larvae were dominated by the phylum Verrucomicrobia (family: Cerasicoccaceae), with a smaller presence of the families Comamonadaceae (Phylum: Proteobacteria) and Pseudomonadaceae (Phylum: Proteobacteria). Pond water samples consisted mainly of Burkholderiales, and Pseudomonadales.
As with the bacterial community, the fungal community differed among life stages and between amphibian and the environment (Figure 3). While sOTU richness (Figure 3A) did not significantly vary between life stages or when compared to pond water communities (GLM, all p > 0.05), community composition did, however (F=20.518, R2 = 61.2%, p<0.001; Figure 3B; Supplementary Table S1). The taxonomic identity of communities (Figure 3C) also differed among life stages with all life stages having membership from the classes Dothideomycetes, Eurotiomycetes, Lecanoromycetes, and Leotiomycetes. Juveniles had the most unique taxonomic composition with a sizable abundance of the class Pezizomycotina, a group rare on other life stages.
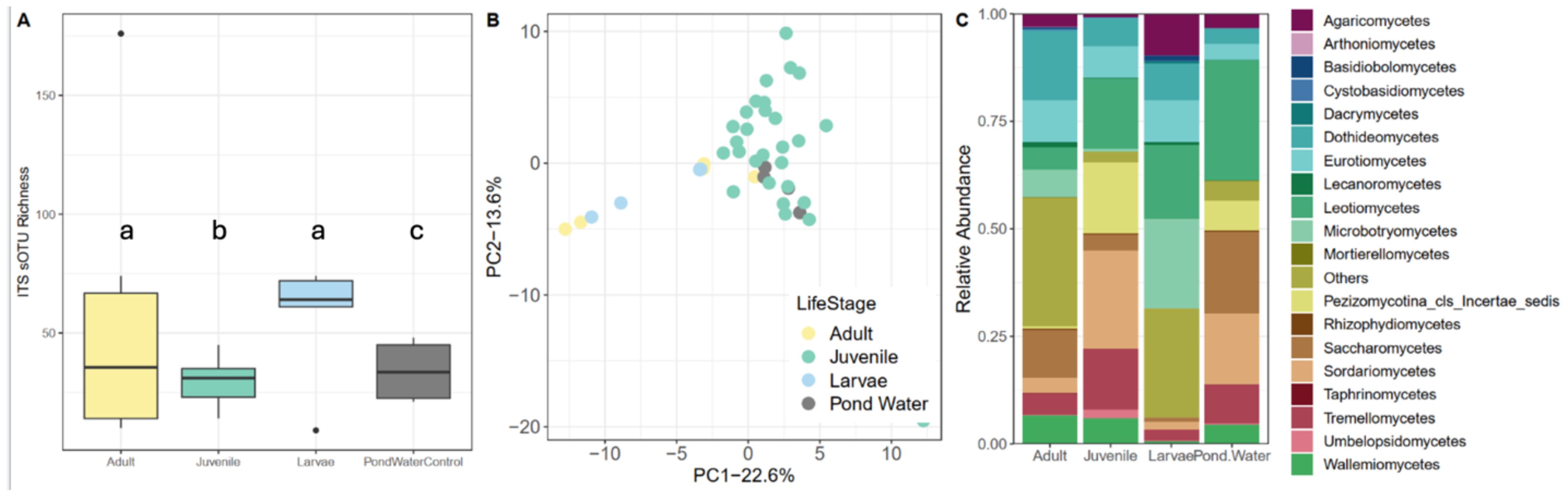
Figure 3. Spotted salamander fungal communities across different life stages. Adults and pond water were sampled in the field and the larvae and juveniles were control animals from the larval and juvenile lab experiments. (A) sOTU richness, (B) principal coordinates analysis based on unweighted UniFrac similarity, and (C) stacked bar-chart of the relative abundance of the most abundance fungal classes. Letters in (A) indicate categories that are significantly different.
Response of larval skin microbiome to Bsal and probiotic application
Larval bacterial skin microbiome sOTU richness did not significantly differ among control, Bsal, and B. thuringiensis probiotic treatment (GLM; all p > 0.05; Figure 4A). The proportion of anti-Batrachochytrium bacteria did not differ significantly among the sample treatments and larvae showed putative anti-Batrachochytrium bacteria made up an average of 21.7 ± 8.3% of the bacterial community on the skin (glmmTMB Beta Regression: AntiF: c2 = 2.229, p = 0.328; Figure 4B). There were significant differences in community composition between the three treatments tested, (PERMANOVA, F2,17 = 1.761, R2 = 15.608%, p < 0.001; Figure 4C, Supplementary Table S2). Unlike bacterial communities, a significant difference was observed in sOTU fungal richness (c2 = 3.32 p = 0.0021) following probiotic application and Bsal exposure (Figure 5A). Community composition shifted in response to treatment (PERMANOVA, F=2.97, R2 = 31.31%, p=0.016, Figure 5B, Supplementary Table S2) as well. Finally, the probiotic B. thuringiensis was not detected through the 16S rRNA gene sequencing data suggesting it could not persist on the skin.
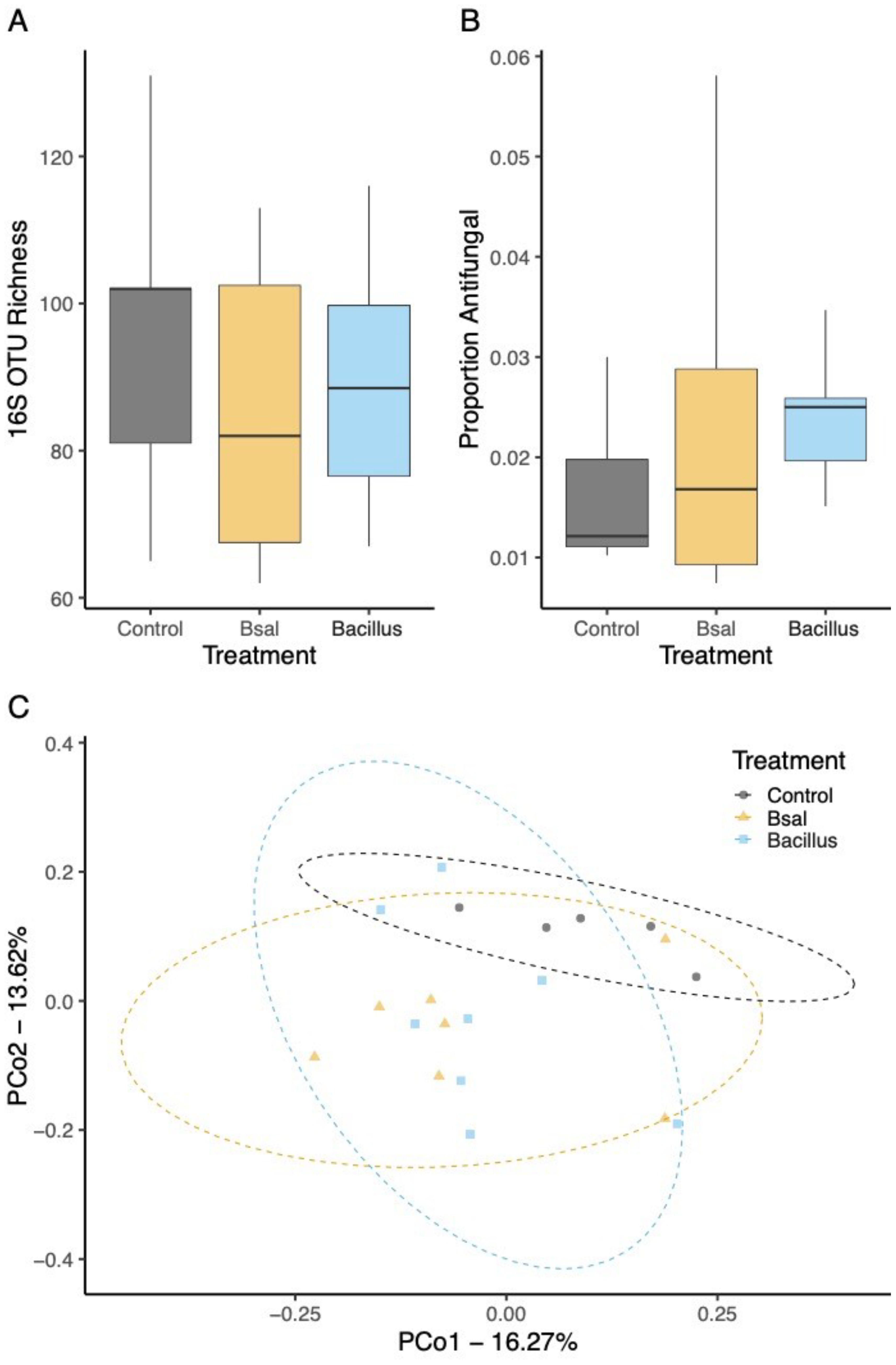
Figure 4. Bacterial communities of larval spotted salamanders 35 days post exposure to Batrachochytrium salamandrivorans or Bacillus. (A) sOTU richness, (B) proportion of anti-Batrachochytrium bacteria, and (C) principal coordinates based on Unweighted UniFrac similarity.
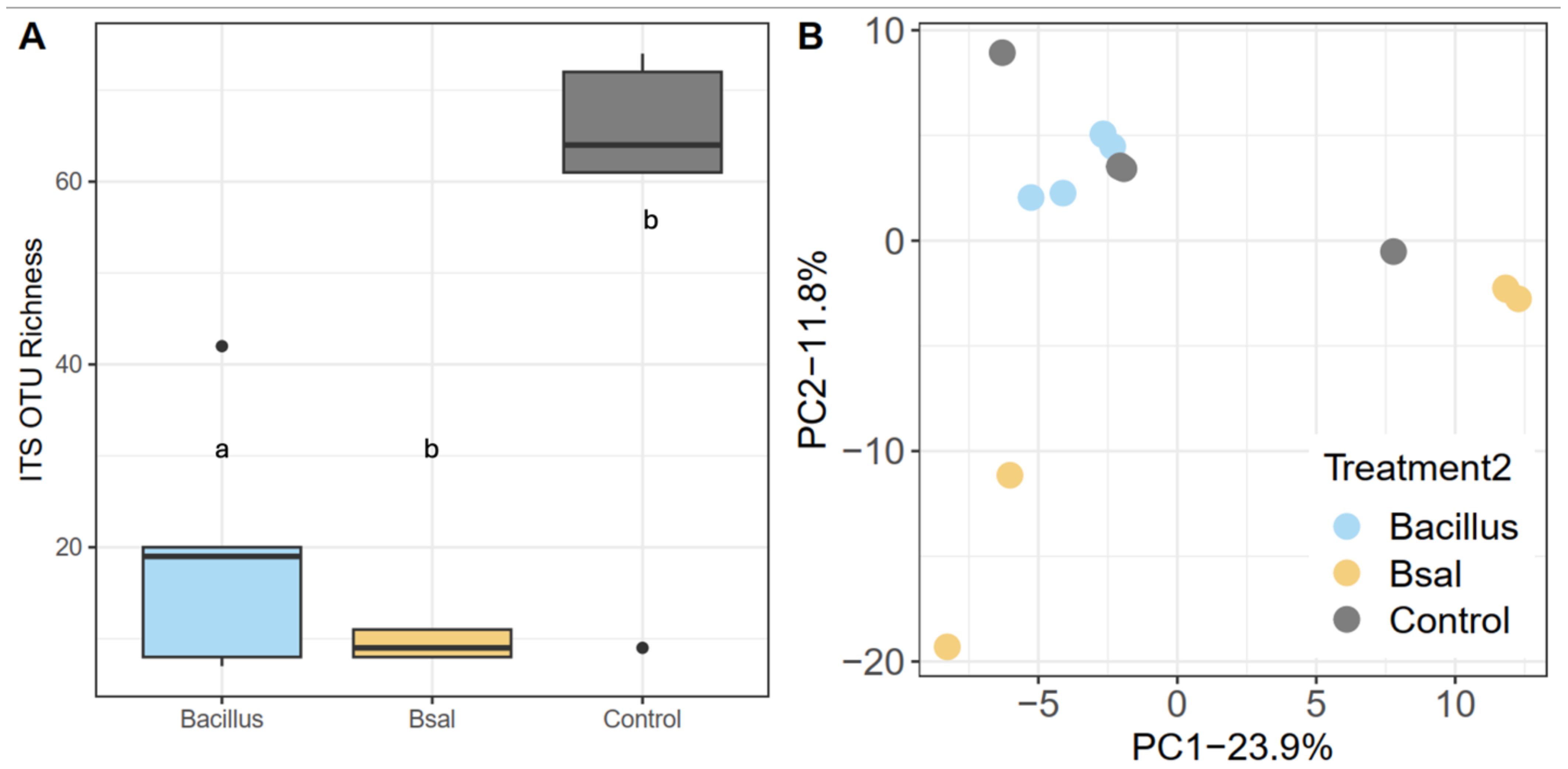
Figure 5. Fungal communities of larval spotted salamanders 35 days post exposure to Batrachochytrium salmandrivorans. (A) sOTU richness and (B) principal coordinates based on unweighted UniFrac similarity. Letters in (A) indicate categories that are significantly different.
Response of juvenile skin microbiome to Bsal and probiotic application
The juvenile microbiome differed in bacterial phylogenetic diversity which varied with the interaction between treatment and sample day (LMER: PD: c2 = 16.142, p = 0.040; Supplementary Figure S3). Both the agitated and Bsal-repeated treatments on day 16 had lower diversity than all other treatments – except for the C. rhizoplanae treatment sampled on day 30 (p < 0.05 for all Bsal-repeated comparisons). The Bsal-thyroid treatment group sampled on day 16 also had lower diversity compared to the high Bsal, low Bsal, and Bsal-thyroid treatment groups sampled on day 30 (all p < 0.05 for Bsal-thyroid comparisons). While the proportion of putative anti-Batrachochytrium reads was not significantly affected by treatments applied to juveniles at days 16 or 30 post-exposure (glmmTMB Beta Regression: Treatment*SampleDay: c2 = 15.202, p = 0.055; SampleDay: c2 = 24.695, p < 0.001; Supplementary Figure S3), on day 16, salamanders had higher putative anti-Batrachochytrium function than on day 30, potentially indicating an acute response (mean % ± SD day 16: 62.0 ± 11.1, day 30: 47.2 ± 12.7; emmeans, t = 11.632, p < 0.001).
Bacterial community composition significantly differed based on both between treatment and time (F17,159 = 1.411, R2 = 13.325%, p < 0.001; Supplementary Figure S3, Supplementary Table S4). Skin-associated metabolites of juvenile salamanders raised under control conditions and one bacterial treatment (C. rhizoplanae) were examined at 36 days post-probiotic application. Beta diversity analysis indicated significant differences (p=0.01, R2 = 19.9%) among treatments (Supplementary Figure S4). While we did not identify their structures, metabolites from all salamander skin swab samples were tested for correlation with sOTU abundances (Supplementary Figure S5). A heat map (Supplementary Figure S5) shows several taxa of bacteria and a single fungal sOTU that correlated with specific metabolites (6.18 minutes) from the HPLC-MS analysis. In general, sOTUs correlated with specific metabolites, while compounds eluting at 13.22 and 18.12 minutes were correlated with several sOTUs. Lastly, despite shifts in the communities, the probiotics did not persist on the skin of the juveniles at a higher level compared to the controls, with C. rhizoplanae absent from skin swab sequencing data (Supplementary Table S3; p > 0.05).
Fungal richness (Supplementary Figure S6) typically did not shift in response to probiotic application and Bsal exposure. The only significant shift in fungal diversity due to treatment was the application of C. rhizoplanae, which significantly decreased overall diversity between 16- and 30-days post application (Welch’s t-test, p<0.001). Fungal community composition shifted as well in response to probiotic and Bsal application (Supplementary Figure S6). All applications experienced a significant shift in community structure when comparing 16- and 30-days post application (PERMANOVA, R2 = 43.1%, p < 0.001; Supplementary Table S4).
At one hour after initial exposure to treatments, the high Bsal and Penicillium sp. exposed juveniles displayed elevated corticosterone release rates compared to all other treatments (ANOVA, p < 0.001, Supplementary Figure S7A). Thirty days after exposure, there were no significant differences between treatments (ANOVA, p = 0.203, Supplementary Figure S7B). There was no significant correlation between corticosterone release rates and mass change (ANOVA, Treatment F = 1.670, p = 0.161); however, salamanders in treatments that showed a significant elevation of corticosterone release rates (Bsal and Penicillium sp.) also exhibited significantly diminished growth (F = 2.658, p = 0.010), reducing proportional growth from 35% to 22% over 8 weeks (Supplementary Figure S8).
Taxonomic response to Bsal and probiotic application
In larval salamanders, we noted a increase in the genus Rhodoferax and the family Cerasicoccaceae in response to both Bsal and Bacillus application (Supplementary Figure S9A). This corresponded with a significant decline in the abundance in Fluvivola and Pseudomonas. In juvenile spotted salamanders, the genus Pseudomonas decreased in abundance in all treatments relative to control animals. This also led to a net increase in the abundance taxa from the genus Acinetobacter and Chryseobacterium (Supplementary Figure S9A). In larval salamanders, several fungal genra responded to both Bsal and Bacillus application (Supplementary Figure S9A). Bsal exposed salamanders saw a large increase in taxa that could not be identified at the phylum level while Bacillus exposed salamanders saw an increase in the yeast Rhodotorula as well the genera Cladosporum, Debaryomyces, and Trichoderma. Probiotic and Bsal treated juvenile salamanders (Supplementary Figure S9B) saw an increase in the genera Humicola, Candidia, and Cladosporum, with a net decrease in Apotrichum and Trichoderma.
Discussion
Elucidating the protective benefit of microbiomes and identifying beneficial pathogen species or compositions has been suggested as a potential avenue to help mitigate or prevent Batrachochytrium fungal infections (Bletz et al., 2013). In this study, we aimed to understand the role of host development, pathogen challenge, and probiotic application on the microbiome of spotted salamanders. Our results demonstrated a shift in both bacterial and fungal community composition in response to developmental life stage. Similar to our results, Kueneman et al. (2014) found lower overall bacterial richness on Rana cascadae tadpoles compared to adults, a similar change documented by others as well (Davis et al., 2017, Prest et al., 2018; Martínez-Ugalde et al., 2022; Hartmann et al., 2023). Amphibians experience costs due to metamorphosis that may also be indicated by shifts in skin bacterial diversity (Humphries et al., 2022). This variation in bacterial diversity is likely driven by a wide range of factors including life history traits, environment, and changes in skin chemistry (pH, mucin content, etc.). Indeed, Burgess and Bishop (2023) found that as embryos of spotted salamanders develop, bacterial diversity tended to decrease. Spotted salamanders in this experiment were reared through development in captivity, which may have altered their microbiome’s developmental trajectory (Kueneman et al., 2022). In addition to community composition, our results indicate significant shifts in potential beneficial taxa (Figures 2, 4) during development. Young spotted salamanders (eggs/larvae) had the lowest abundance of putative anti-Batrachochytrium bacteria relative to juvenile and adult salamanders, suggesting that as the host’s own immune system develops (Humphries et al., 2022) the microbiome and its protective capacity develops as well. This relationship holds for larvae that transition to juveniles in captivity, as well as between eggs and adults sampled from the same field site. It also mirrors the succession of bacterial community diversity during development of another amphibian, boreal toads, Anaxyrus boreas (Prest et al., 2018). The greater impact on the microbiome (richness/composition) of Bsal treated with thyroid compared to untreated Bsal may indicate gene expression changes in Bsal (Thekkiniath et al., 2015) that can directly impact the microbiome and are triggered by developmental signals. The developmental window of host association with the microbiome can impact later life physiology and immune function and thus disease dynamics (Warne et al., 2019).
While the bacterial communities of spotted salamanders shifted with life stage, taxonomic groups such as Burkholderiales, Sphingobacterales, and Pseudomonadales were the most common, as demonstrated previously in a global metanalysis (Kueneman et al., 2019). Previous work on the internal microbiome of spotted salamander eggs demonstrated less diverse microbiomes (Bishop et al., 2021), and while there were overlaps in taxonomic groups (e.g., Burkholderiales), the inside of the egg relative to the surface appears to be more selective for microbial taxa. This result mirrors previous work on the plant seed microbiome (Nelson, 2018). Interestingly, the skin bacteria on larvae were dominated by the phylum Verrucomicrobia (Family: Cerasicoccaceae), a group common on red-backed salamanders, Plethodon cinereus (Loudon et al., 2014) and American bullfrogs, Rana catesbeiana (Kueneman et al., 2019). This group remains uncultivated from amphibians as does its function and role in host defense. Studies on the fungal microbiome are significantly less common than those evaluating bacterial communities. Compared to previous work in poison arrow frogs (Kearns et al., 2017), the spotted salamander fungal communities shared membership from the Dothideomycetes and Leotiomycetes. Fungi associated with red-backed salamanders (Barnes et al., 2021) and Boreal toads (Alexiev et al., 2021) too showed similar fungal groups (Basidiomycota, Ascomycota, and Basidiobolomycota) suggesting they may be important symbionts for amphibians. How fungi contribute to host function in amphibians is unclear and an important future avenue for microbiome research.
The spotted salamanders investigated in this study were not susceptible to Bsal after exposure at various life stages (larvae and juveniles) to the fungal pathogen under standard laboratory conditions, and across a range of exposure doses (Barnhart et al., 2020). Analysis of the stress hormone corticosterone (Supplementary Figure S7) showed elevated levels in salamanders exposed to Bsal, which signifies a shift in energy balance to facilitate coping with a stressor, either behaviorally or physiologically (Barnhart et al., 2020). Here, we also found increased corticosterone release rates in juvenile spotted salamanders exposed to Penicillium sp., but not probiotic bacteria. While infection by Batrachochytrium sp. fungi can shift the microbiome of amphibians (Bates et al., 2019; Jani et al., 2021), the lack of infection post-exposure could indicate a physiological stress response that would have cascading effects on the skin, likely influencing the observed shift in the microbiome. A similar effect was recently observed in tropical frogs (Neely et al., 2023). In both larvae and juvenile spotted salamanders, community composition shifted with Bsal exposure. In all treatments of juveniles, higher putative antifungal function and lower microbial richness was observed between day 16 and day 30. While we did not test the initial microbiome of these treatments, the consistency in a response to treatment suggests that probiotics and Bsal consistently impact the amphibian microbiome. Only C. rhizoplanae treated juveniles maintained stable antifungal function and richness through time. Bsal did not persistently infect any salamanders at any stage (Barnhart et al., 2020). Our data suggest that the host and microbiome responded to the addition of bacteria (at least Chryseobacterium) with an adaptive response including reduced diversity and a shift toward members that may outcompete the probiotics (Woodhams et al., 2023).
All probiotic applications (Bacillus, Chryseobacterium, and Penicillium) in this study shifted the microbiome of larvae and juveniles, and of putative anti-Batrachochytrium bacteria in juveniles. In larvae, the proportion of putative anti-Batrachochytrium bacteria was marginally higher after treatment with Bacillus thuringiensis, a finding reflecting that critical developmental windows may occur early in amphibian development (Knutie et al., 2017; Warne et al., 2017, Warne et al., 2019). Interestingly, despite the lack of persistence of Chryseobacterium on juvenile salamanders, we detected a net shift in skin-associated metabolites. This, when paired with shifts in community composition, suggests that probiotics don’t need to persist to modulate the function of amphibian skin, and could lead to shifts in microbial secondary metabolites that influence amphibian defense or development through mechanisms such as insulin-like signaling pathways (Dallas and Warne, 2023). However, given the limited scope of our metabolomic data, more information is needed to further elucidate this relationship.
Analysis of the presence of probiotics revealed only the fungus, Penicillium, was able to persist despite an overall decrease in abundance following application (Supplementary Table S3). Our results are consistent with previous work that has commonly demonstrated a difficulty in applying probiotics that persist on the skin for extended periods of time (e.g., Woodhams et al., 2020; Becker et al., 2021), which may be indicative of a natural tendency to prevent dysbiosis. There was also a difference in proportional change in mass when comparing the Penicillium sp. treatment to all other treatments: Penicillium sp.-treated juveniles gained less mass overall, perhaps due to the initial elevation of corticosterone release rates and allocation of energy resources elsewhere. Corticosterone release rates did not differ significantly among treatments at 30-days post exposure. In a study using similar methods, Kearns et al. (2017) found that midwife toads, Alytes obstetricans, did not have elevated corticosterone release rates when exposed to fungal probiotics (Penicillium sp.) compared to bacterial probiotics. These results suggest selection of fungal over bacterial probiotic candidates may be beneficial to the host given the apparent Batrachochytrium-inhibitory effects of amphibian-associated fungi (Kearns et al., 2017). Symbiotic groups of microbial cultures may be the next step in treatment research because of the synergistic effects of microbes interacting in communities (Piovia-Scott et al., 2017).
In conclusion, this study demonstrated a shift in the microbiome of spotted salamanders in response to conditions of development, pathogen exposure, and probiotic application. While it is apparent that the amphibian microbiome is dynamic and plays a key role in host protection, maintenance, and putative anti-pathogen function, further investigation is required in various species and disease models. Disease mitigation strategies are vital to conserve amphibian species in the likely event that Bsal continues to spread internationally. Results from this investigation suggest that spotted salamanders may resist Bsal chytridiomycosis in part because of a diverse skin microbiome with inherent putative anti-Batrachochytrium function. It may be prudent to test threatened amphibian species for these traits, and if not present, to apply probiotics during a microbiome reorganization phase such as pre-hatching (Warne et al., 2019), during which the treatment may have the greatest effect.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI Sequence Read Archive, accession PRJNA1158746.
Ethics statement
The animal study was approved by University of Massachusetts Boston IACUC Board. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
KB-M: Writing – review & editing, Writing – original draft. BL: Writing – review & editing, Writing – original draft. PK: Writing – review & editing, Writing – original draft. RA: Writing – review & editing, Writing – original draft. RW: Writing – review & editing, Writing – original draft. MB: Writing – review & editing, Writing – original draft. SA: Writing – review & editing, Writing – original draft. AP: Writing – review & editing, Writing – original draft. AT-P: Writing – review & editing, Writing – original draft. CG: Writing – review & editing, Writing – original draft. KS: Writing – review & editing, Writing – original draft. TU: Writing – review & editing, Writing – original draft. KM: Writing – review & editing, Writing – original draft. DW: Writing – review & editing, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R25GM076321. Funding was also provided in part by the Wildlife Without Borders- Amphibians in Decline program (F15AP00968 to DW), the US National Science Foundation (IOS-1845634 to DW, BII 2120084, and DGE 1249946, Integrative Graduate Education and Research Traineeship (IGERT): Coasts and Communities – Natural and Human Systems in Urbanizing Environments), the Nancy Goranson Endowment Fund, and the UMass Boston Biology Department.
Acknowledgments
The authors thank Alberto Campos, Fanghemei Zhang (Jessica), Kathleen Conroy, Diego Aparicio, Jo-Hanna Azzara, Terence Cook, Sendy Lamour, Vivian Le, Andrea Lu, Modi Maitri, Jessie Ngandjui, Bryan Peguero, Natan Pirete, Kiloni Quiles-Franco, Rayshawn Reece, Marina Teixeira, and Steven Tran. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R25GM076321. Funding was also provided in part by the Wildlife Without Borders- Amphibians in Decline program (F15AP00968 to DW), the US National Science Foundation (IOS-1845634 to DW, BII 2120084, and DGE 1249946, Integrative Graduate Education and Research Traineeship (IGERT): Coasts and Communities – Natural and Human Systems in Urbanizing Environments), the Nancy Goranson Endowment Fund, and the UMass Boston Biology Department.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/famrs.2024.1425570/full#supplementary-material
References
Abarenkov K., Nilsson R.H., Larsson K.-H., Alexander I. J., Eberhardt U., Erland S., et al. (2010). The UNITE database for molecular identification of fungi–recent updates and future perspectives. New Phytol. 186, 281–285. doi: 10.1111/j.1469-8137.2009.03160.x
Alexiev A., Chen M. Y., McKenzie V. J. (2021). Identifying fungal-host associations in an amphibian host system. PloS One 16, e0256328. doi: 10.1371/journal.pone.0256328
Allgeier S., Friedrich A., Brühl C. A. (2019). Mosquito control based on bacillus thuringiensis Israelensis (Bti) interrupts artificial wetland food chains. Sci. Total Environ. 686, 1173–1184. doi: 10.1016/j.scitotenv.2019.05.358
Amir A., McDonald D., Navas-Molina J. A., Kopylova E., Morton J. T., Xu Z. Z., et al. (2017). Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2. doi: 10.1128/msystems.00191-16
Barnes E. M., Kutos S., Naghshineh N., Mesko M., You Q., Lewis J. D. (2021). Assembly of the amphibian microbiome is influenced by the effects of land-use change on environmental reservoirs. Environ. Microbiol. 23, 4595–4611. doi: 10.1111/1462-2920.15653
Barnhart K., Bletz M. C., LaBumbard B., Tokash-Peters A., Gabor C. R., Woodhams D. C. (2020). Batrachochytrium salamandrivorans elicits acute stress response in spotted salamanders but not infection or mortality. Anim. Conserv. 23, 533–546. doi: 10.1111/acv.12565
Bates K. A., Shelton J. M.G., Mercier V. L., Hopkins K. P., Harrison X. A., Petrovan S. O., et al. (2019). Captivity and infection by the fungal pathogen batrachochytrium salamandrivorans perturb the amphibian skin microbiome. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01834
Becker C.G., Greenspan S. E., Martins R. A., Lyra M. L., Prist P., Metzger J. P., et al. (2023). Habitat split as a driver of disease in amphibians. Biol. Rev. 98, 727–746. doi: 10.1111/brv.12927
Becker M. H., Brophy J. A.N., Barrett K., Bronikowski E., Evans M., Glassey E., et al. (2021). Genetically modifying skin microbe to produce violacein and augmenting microbiome did not defend Panamanian golden frogs from disease. ISME Commun. 1, 57. doi: 10.1038/s43705-021-00044-w
Becker C. G., Rodriguez D., Longo A. V., Talaba A. L., Zamudio K. R. (2012). Disease risk in temperate amphibian populations is higher at closed-canopy sites. PloS One 7, e48205.
Bishop C., Jurga E., Graham L. (2021). Patterns of bacterial diversity in embryonic capsules of the spotted salamander ambystoma maculatum: an expanding view of a symbiosis. FEMS Microbiol. Ecol. 97, fiab128. doi: 10.1093/femsec/fiab128
Bletz M. C., Kelly M., Sabino-Pinto J., Bales E., Praet S. V., Bert W., et al. (2018). Disruption of skin microbiota contributes to salamander disease. Proc. R. Soc. B: Biol. Sci. 285, 20180758. doi: 10.1098/rspb.2018.0758
Bletz M. C., Loudon A. H., Becker M. H., Bell S. C., Woodhams D. C., Minbiole K. P.C., et al. (2013). Mitigating amphibian chytridiomycosis with bioaugmentation: characteristics of effective probiotics and strategies for their selection and use. Ecol. Lett. 16, 807–820. doi: 10.1111/ele.12099
Bokulich N. A., Mills D. A. (2013). Improved selection of internal transcribed spacer-specific primers enables quantitative, ultra-high-throughput profiling of fungal communities. Appl. Environ. Microbiol. 79, 2519–2526. doi: 10.1128/AEM.03870-12
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Author correction: reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 1091–1091. doi: 10.1038/s41587-019-0252-6
Burgess W. L., Bishop C. D. (2023). Bacterial diversity in egg capsular fluid of the spotted salamander ambystoma maculatum decreases with embryonic development. Microb. Ecol. 86, 1789–1798. doi: 10.1007/s00248-023-02228-4
Caporaso J.G., Lauber C. L., Walters W. A., Berg-Lyons D., Huntley J., Fierer N., et al. (2012). Ultra-high-throughput microbial community analysis on the illumina hiSeq and miSeq platforms. he ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Charbonnier J. F., Pearlmutter J., Vonesh J. R., Gabor C. R., Forsburg Z. R., Grayson, K. L., et al. (2018). Cross-life stage efects of aquatic larval density and terrestrial moisture on growth and corticosterone in the spotted salamander. Diversity 10, 68.
Dallas J., Warne R. W. (2023). Heat hardening of a larval amphibian is dependent on acclimation period and temperature. J. Exp. Zoology Part A: Ecol. Integr. Physiol. 33S, 339–345.
Davis L. R., Bigler L., Woodhams D. C. (2017). Developmental trajectories of amphibian microbiota: response to bacterial therapy depends on initial community structure. Environ. Microbiol. 19, 1502–1517. doi: 10.1111/1462-2920.13707
Dixon P. (2003). VEGAN, a package of R functions for community ecology. J. Veget. Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Gray M. J., Lewis J. P., Nanjappa, Klocke B., Pasmans F., Martel A. N., et al. (2015). Batrachochytrium salamandrivorans: the north american response and a call for action. PloS Pathog. 11, e1005251.
Harris R. N., Lauer A., Simon M. A., Banning J. L., Alford R. A. (2009). Addition of antifungal skin bacteria to salamanders ameliorates the effects of chytridiomycosis. Dis. Aquat. Organisms 83, 11–16.
Hartmann A. M., McGrath-Blaser S. E., Colón-Piñeiro Z., Longo A. V. (2023). Ontogeny drives shifts in skin bacterial communities in facultatively paedomorphic salamanders. Microbiology 169, 1399. doi: 10.1099/mic.0.001399
Humphries J. E., Lanctôt C. M., Robert J., McCallum H. I., Newell D. A., Grogan L. F. (2022). Do immune system changes at metamorphosis predict vulnerability to chytridiomycosis? Dev Comp Immunol. 136, 104510. doi: 10.1016/j.dci.2022.104510
Jani A. J., Bushell J., Arisdakessian C.G., Belcaid M., Boiano D. M., Brown C., et al. (2021). The amphibian microbiome exhibits poor resilience following pathogen-induced disturbance. ISME J. 15, 1628–1640. doi: 10.1038/s41396-020-00875-w
Kearns J., Fischer S., Fernández-Beaskoetxea S., Gabor C. R., Bosch J., Bowen J. L. C., et al. (2017). Fight fungi with fungi: antifungal properties of the amphibian mycobiome. Front. Microbiol. 8, 2494.
Knutie S. A., Wilkinson C. L., Kohl K. D., Rohr J. R. (2017). Early-life disruption of amphibian microbiota decreases later-life resistance to parasites. Nat. Commun. 8, 86. doi: 10.1038/s41467-017-00119-0
Kueneman J. G., Bletz M. C., Becker M., Gratwicke B., Garcés O. A., Hertz A., et al. (2022). Effects of captivity and rewilding on amphibian skin microbiomes. Biol. Conserv. 271, 109576. doi: 10.1016/j.biocon.2022.109576
Kueneman J. G., Bletz M. C., McKenzie V. J., Becker C. G., Joseph M. B., Abarca J. G., et al. (2019). Community richness of amphibian skin bacteria correlates with bioclimate at the global scale. Nat. Ecol. Evol. 3 , 381–389.
Kueneman J. G., Parfrey L. W., Woodhams D. C., Archer H. M., Knight R., McKenzie V. J. (2014). The amphibian skin-associated microbiome across species, space and life history stages. Mol. Ecol. 23, 1238–1250.
Kueneman J. G., Weiss S., McKenzie V. J. (2017). Composition of micro-eukaryotes on the skin of the cascades frog (Rana cascadae) and patterns of correlation between skin microbes and batrachochytrium dendrobatidis. Front. Microbiol. 8, 2350.
Kueneman J. G., Woodhams D. C., Harris R., Holly M., Rob Knight A., McKenzie V. J. (2016). Probiotic Treatment Restores Protection against Lethal Fungal Infection Lost during Amphibian Captivity. Proc. R. Soc. B: Biol. Sci. 283, 20161553. doi: 10.1098/rspb.2016.1553
Land M., Bundschuh M., Richard J., Hopkins B. P., McKie B. G. (2023). Effects of mosquito control using the microbial agent bacillus thuringiensis Israelensis (Bti) on aquatic and terrestrial ecosystems: A systematic review. Environ. Evidence 12, 26. doi: 10.1186/s13750-023-00319-w
Le Sage E. H., LaBumbard B. C., Reinert L. K., Miller B. T., Richards-Zawacki C. L., Woodhams D. C., et al. (2021). Preparatory immunity: seasonality of mucosal skin defences and batrachochytrium infections in southern leopard frogs. J. Anim. Ecol. 90, 542–554. doi: 10.1111/1365-2656.13386
Lötters S., Wagner N., Albaladejo G., Böning, Dalbeck L., Düssel H., et al. (2020). The amphibian pathogen batrachochytrium salamandrivorans in the hotspot of its european invasive range: past- present-future. Salamandra 56.
Loudon A. H., Woodhams D. C., Parfrey L. W., Archer H., Knight R., McKenzie V., et al. (2014). Microbial community dynamics and effect of environmental microbial reservoirs on red-backed salamanders (Plethodon cinereus). ISME J. 8, 830–840. doi: 10.1038/ismej.2013.200
Lozupone C., Knight R.. (2005). UniFrac: A new phylogenetic method for comparing microbial communities. ISME J. 71, 8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005
Martel A., Spitzen-van der Sluijs A., Blooi M., Bert W., Ducatelle R., Fisher M. C., et al. (2013). Batrachochytrium salamandrivorans sp. Nov. Causes lethal chytridiomycosis in amphibians. Proc. Natl. Acad. Sci. 110, 15325–15329. doi: 10.1073/pnas.1307356110
Martínez-Ugalde E., Ávila-Akerberg V., González Martínez T. M., Vázquez Trejo M., Hernández D. Z., Anaya-Morales S. L., et al. (2022). The skin microbiota of the axolotl ambystoma altamirani is highly influenced by metamorphosis and seasonality but not by pathogen infection. Anim. Microb. 4, 63. doi: 10.1186/s42523-022-00215-7
McDonald D., Price M. N., Goodrich J., Nawrocki E. P., DeSantis T. Z., Probst A., et al. (2013). An improved greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea | The ISME journal. Available online at: https://www.nature.com/articles/ismej2011139 (Accessed February 23, 2024).
McGrath-Blaser S. E., McGathey N., Pardon A., Hartmann A. M., Longo A. V. (2024). Invasibility of a North American soil ecosystem to amphibian-killing fungal pathogens. Proc. R. Soc. B: Biol. Sci. 291, 20232658. doi: 10.1098/rspb.2023.2658
Neely W. J., Martins R. A., Mendonça da Silva C. M., Ferreira da Silva T., Fleck L. E., Whetstone R. D., et al. (2023). Linking microbiome and stress hormone responses in wild tropical treefrogs across continuous and fragmented forests. Commun. Biol. 6, 1–15. doi: 10.1038/s42003-023-05600-9
Nelson E. B. (2018). The seed microbiome: origins, interactions, and impacts. Plant Soil 422, 7–34. doi: 10.1007/s11104-017-3289-7
Parada A. E., Needham D. M., Fuhrman J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Pereira S. A., Jeronimo G. T., Marchiori N. C., Oliveira H. M., Jesus G. F. A., Schmidt E. C., et al. (2018). Tadpoles fed supplemented diet with probiotic bacterium isolated from the intestinal tract of bullfrog lithobates catesbeianus: haematology, cell activity and electron microscopy. Microb. Pathogen. 114, 255–635. doi: 10.1016/j.micpath.2017.11.033
Piovia-Scott J., Rejmanek D., Woodhams D. C., Worth S.J., Kenny H., McKenzie V., et al. (2017). Greater species richness of bacterial skin symbionts better suppresses the amphibian fungal pathogen batrachochytrium dendrobatidis. Microb. Ecol. 74, 217–226. doi: 10.1007/s00248-016-0916-4
Prest T. L., Kimball A. K., Kueneman J. G., McKenzie V. J. (2018). Host-associated bacterial community succession during amphibian development. Mol. Ecol. 27, 1992–2006. doi: 10.1111/mec.14507
Rognes T., Flouri T., Nichols B., Quince C., Mahé F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584.
Sheley W. C., Gray M. J., Wilber M. Q., Cray C., Davis Carter E., Miller D. L. (2023). Electrolyte imbalances and dehydration play a key role in batrachochytrium salamandrivorans chytridiomycosis. Front. Vet. Sci. 9, 1055153. doi: 10.3389/fvets.2022.1055153
Thekkiniath J., Zabet-Moghaddam M., Kottapalli K. R., Pasham M. R., Francisco S. S., Francisco M. S. (2015). Quantitative proteomics of an amphibian pathogen, batrachochytrium dendrobatidis, following exposure to thyroid hormone. PloS One 10, e0123637. doi: 10.1371/journal.pone.0123637
Thompson L. R., Sanders J. G., McDonald D., Amir A., Ladau J., Locey K. J., et al. (2017). A communal catalogue reveals earth’s multiscale microbial diversity. Nature 551, 457–463. doi: 10.1038/nature24621
Thumsová B., González-Miras E., Rubio Á., Granados I., Bates K. A., Bosch J. (2024). Chemical disinfection as a simple and reliable method to control the amphibian chytrid fungus at breeding points of endangered amphibians. Sci. Rep. 14, 5151. doi: 10.1038/s41598-024-55946-1
Umile T. P., McLaughlin P. J., Johnson K. R., Honarvar S., Blackman A. L., Burzynski E. A., et al. (2014). Nonlethal amphibian skin swabbing of cutaneous natural products for HPLC fingerprinting. Anal. Methods 6, 3277–3284. doi: 10.1039/C4AY00566J
Waddle J.H., Grear D. A., Mosher B. A., Campbell Grant E. H., Adams M. J., Backlin A. R., et al. (2020). Batrachochytrium salamandrivorans (Bsal) not detected in an intensive survey of wild North American amphibians. Sci. Rep. 10, 13012. doi: 10.1038/s41598-020-69486-x
Warne R. W., Kirschman L., Zeglin L. (2019). Manipulation of Gut Microbiota during Critical Developmental Windows Affects Host Physiological Performance and Disease Susceptibility across Ontogeny. J. Anim. Ecol. 88, 845–856. doi: 10.1111/1365-2656.12973
Warne R. W., Kirschman L., Zeglin L. (2017). Manipulation of gut microbiota reveals shifting community structure shaped by host developmental windows in amphibian larvae. Integr. Comp. Biol. 57, 786–794.
Woodhams D. C., Alford R. A., Antwis R. E., Archer H., Becker M. H., Belden L. K., et al. (2015). Antifungal isolates database of amphibian skin-associated bacteria and function against emerging fungal pathogens: ecological archives E096-059. Ecology 96, 595–595. doi: 10.1890/14-1837.1
Woodhams D. C., Barnhart K. L., Bletz M. C., Campos A. J., Ganem S. J., Hertz A., et al. (2018). “Batrachochytrium: Biology and Management of Amphibian Chytridiomycosis,” in Encyclopedia of Life Sciences (Hoboken, NJ: John Wiley & Sons, Ltd), 1–18. doi: 10.1002/9780470015902.a0027207
Woodhams D. C., McCartney J., Walke J. B., Whetstone R. (2023). The adaptive microbiome hypothesis and immune interactions in amphibian mucus. Dev. Comp. Immunol. 145, 104690.
Keywords: amphibian, Bacillus thuringiensis, Batrachochytrium salamandrivorans, Chryseobacterium rhizoplanae, chytridiomycosis, corticosterone, disease ecology
Citation: Barnhart-McCarty K, LaBumbard B, Kearns PJ, Ahsan R, Whetstone R, Bletz M, AlKhalifa SE, Poltronetti A, Tokash-Peters A, Gabor CR, Schliep K, Umile TP, Minbiole K and Woodhams DC (2024) Micromanagement: conditions influencing antipathogen function of the skin microbiome in spotted salamanders, Ambystoma maculatum. Front. Amphib. Reptile Sci. 2:1425570. doi: 10.3389/famrs.2024.1425570
Received: 30 April 2024; Accepted: 30 September 2024;
Published: 22 October 2024.
Edited by:
Joana Sabino Pinto, University of Groningen, NetherlandsReviewed by:
Arik Hartmann, University of Florida, United StatesAndrea J. Jani, University of Hawaii at Manoa, United States
Copyright © 2024 Barnhart-McCarty, LaBumbard, Kearns, Ahsan, Whetstone, Bletz, AlKhalifa, Poltronetti, Tokash-Peters, Gabor, Schliep, Umile, Minbiole and Woodhams. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patrick J. Kearns, cGF0cmljay5rZWFybnNAdW1iLmVkdQ==; Douglas C. Woodhams, ZHdvb2RoYW1zQGdtYWlsLmNvbQ==
†Present address: Klaus Schliep, Graz University of Technology, Institute of Molecular Biotechnology, Graz, AustriaMolly Bletz, Ecosystem Science and Management, The Penn State University, State College, University Park, PA, United States
 Kelly Barnhart-McCarty1
Kelly Barnhart-McCarty1 Patrick J. Kearns
Patrick J. Kearns Ross Whetstone
Ross Whetstone Molly Bletz
Molly Bletz Caitlin R. Gabor
Caitlin R. Gabor Klaus Schliep
Klaus Schliep Thomas P. Umile
Thomas P. Umile Kevin Minbiole
Kevin Minbiole Douglas C. Woodhams
Douglas C. Woodhams