
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Amphib. Reptile Sci., 05 September 2023
Sec. Conservation
Volume 1 - 2023 | https://doi.org/10.3389/famrs.2023.1215723
This article is part of the Research TopicFrom Landscape Modifications to Pathogen Infections: Are Threats to Amphibians the Same in All Biomes?View all 7 articles
 Sarah E. Ball1*
Sarah E. Ball1* Mollie Gupta1
Mollie Gupta1 Sarah J. Aldridge1
Sarah J. Aldridge1 Bryony E. Allen1
Bryony E. Allen1 Sally C. Faulkner2
Sally C. Faulkner2 Gonzalo Oteo-García1
Gonzalo Oteo-García1 Richard A. Griffiths3
Richard A. Griffiths3 Peter Hill4
Peter Hill4 Nicola J. Morris5
Nicola J. Morris5 Michael C. A. Stevens2
Michael C. A. Stevens2 John Wilkinson4
John Wilkinson4 Trenton W. J. Garner1
Trenton W. J. Garner1The alpine newt Ichthyosaura alpestris has achieved a widespread distribution as a non-native (alien) species in Britain since its initial introduction over a century ago, but the patterns of its release and subsequent dispersal have never yet been collectively analysed. We employed a multi-disciplinary combination of methods, using geographic profiling to estimate the likely number and locations of introductions, and mitochondrial DNA polymorphisms to investigate the likely geographic source of primary introductions, including the potential role of the pet trade. In parallel we used population genetic analysis and coalescence-based modelling to infer the demographics and directionality of dispersal from founding populations. Our results show that alpine newts have been released at multiple sites. We found a close resemblance between patterns of mtDNA haplotypes in the pet trade and those of established alpine newt populations, suggesting a relationship between trade, releases, and dispersal. Results from demographic modelling using Approximate Bayesian Computation are also consistent with multiple independent introductions with limited local dispersal, and additionally suggest that releases may occur from intermediate sources, such as captive populations. Our results support the hypothesis that deliberate human activity is largely responsible for both introductions of alpine newts into the UK and their wider dispersal post-introduction. The likely involvement of the international pet trade highlights the risk that ongoing releases of I. alpestris may expose native species to pathogens, whether pre-existing or novel.
Alien invasive species are considered one of the pre-eminent threats to agriculture, fisheries and horticulture, as well as to biodiversity (Wilcove et al., 1998; Mack et al., 2000; Gurevitch and Padilla, 2004). Invasions are at an all-time high (Hulm, 2009; Seebens et al., 2017; Pyšek et al., 2020), showing no sign of slowing as the number of first records of established alien species world-wide continues to increase (Seebens et al., 2017). This has generated an unprecedented “mass invasion event” (Ricciardi, 2007), on a background of increasing globalisation (Amano et al., 2016; Capinha et al., 2017; Hulme, 2021). Legislation and strategies designed to reduce the impact of invasive alien species have been primarily focused on the prevention of introductions across international borders, with early control of introduced alien species (Miller et al., 2006). Control measures for alien species that have already achieved widespread national distributions present logistic and financial challenges (Leung et al., 2002; Renault et al., 2021).
The term alien encompasses all species subsisting outside their native range as the result of human activity (IUCN Guidelines, 2000; CBD Secretariat, 2002). To be further categorized as invasive, the species must have expanded significantly beyond the site of introduction (Valéry et al., 2008; Blackburn et al., 2011), or have a negative impact in its non-native range (IUCN Guidelines, 2000; CBD Secretariat, 2002). However, only a small proportion of alien species show unequivocal progression along the continuum from introduction to invasion (Williamson and Fitter, 1996; Blackburn et al., 2011; Chapple et al., 2012; Daly et al., 2023). This may be context-dependent; a species may be invasive in some non-native settings, but not in others. The factors underlying such differences may provide insight into routes and processes of invasion (Zenni and Nuñez, 2013). Anthropogenic factors may continue to influence range expansion even after an introduction has become established (Ewart et al., 2019). The invasive cane toad (Rhinella marinus) in Australia (Lever, 2001) exploits roads to achieve rapid range expansion (Seabrook and Dettmann, 1996; Brown et al., 2006). Widespread distribution may also arise from repeated introductions, as well as by post-introduction translocation; the discontinuous distribution of the invasive alien North American bullfrog (Lithobates catesbeianus) in southwest France was likely facilitated through translocations by humans (Ficetola et al., 2007).
In this study, we investigate pathways of introduction and dispersal of the alpine newt Ichthyosaura alpestris (formerly Triturus or Mesotriton alpestris) in the UK. I. alpestris is one of the most broadly distributed amphibians in mainland Europe (Sillero et al., 2014; Speybroeck et al., 2020). It does not occur naturally in the UK, where it has long been held in private collections, often as an exotic pond ornamental (Lever, 1977). The oldest documented self-sustaining non-captive population of I. alpestris in England dates to the 1920s, in a garden neighbouring an aquatic nursery (Lever, 1977). Since then, breeding colonies of alpine newts have been recorded at multiple locations (Bell and Bell, 1995; Beebee, 2007; Bond and Haycock, 2008; Wilkinson, 2016). Where the source is known, these have often followed the deliberate or accidental release of newts from captivity. Despite this century-long history of colonisation, a detrimental impact on native biodiversity has not been demonstrated, and I. alpestris is not currently considered invasive in the UK (Roy et al., 2014).
We use geographic profiling (geoprofiling) to estimate the likely number and locations of introductions or translocations of I. alpestris at a national scale, based on records of sightings and ecological surveys. This method, which uses the geographical distribution of known occurrences to infer likely sources (Le Comber and Stevenson, 2012), has previously been applied to investigate the origin and spread of other alien species, including invasive algae Caulerpa species (Papini et al., 2013), a fruit fly pest Drosophila suzukii (Cini et al., 2014), and a geophyte plant Oxalis pes-caprae (Papini et al., 2017). To investigate the provenance of primary introductions, including the potential role of the pet trade, we study geographical patterns of mitochondrial DNA (mtDNA) polymorphisms. In parallel we use microsatellite markers for population genetic analysis and coalescence-based modelling with approximate Bayesian computation (ABC), to infer the demographics and directionality of dispersal from founding populations.
We interpret our findings in comparison with other examples of the introduction of I. alpestris outside its native range with differing invasion outcomes (Zenni and Nuñez, 2013), to address the question of why I. alpestris is not invasive in the UK, despite its long-standing naturalisation and widespread distribution, and whether there are conditions under which this situation might change. We underscore the need for an understanding of the ecology of alien species, especially relating to movement and dispersal (Simberloff, 2003; Wilson et al., 2009; Pittman et al., 2014; Cayuela et al., 2020) in addressing these questions. This approach can help to predict the trajectory of an already established non-native species, and potentially inform management decisions that encompass the human contribution to different components of the invasion process.
We used geographic profiling to estimate the number of spatial clusters associated with I. alpestris in the UK, based on point-pattern data of observations of adult newts. We applied the Dirichlet-Process-Mixture (DPM) model which allows identification of multiple sources, originally developed by Verity et al. (2014), and extended by Faulkner et al. (2016). In brief, in the first step, the model allocates each location to one of a finite number of assumed known source locations, based on distance. This generates clusters of observations allocated to the same source which are then used to re-estimate the central location of the potential sources. The original allocations are discarded, and each datapoint is allocated to the re-estimated source locations in a repeat of the first step. The Markov chain Monte Carlo (MCMC) method is used to alternate between these steps until convergence, assessed by the Gelman-Rubin method (Gelman et al., 2004). The posterior distribution of source locations is then used to produce the geoprofile, a ranked probability surface overlaid on the map of the study area. The higher the location on the surface, the more likely it is to be a point of introduction. Dispersal distance is defined in the model by the parameter sigma, representing the standard deviation of the bivariate normal distribution that makes up each cluster. Sigma describes the dispersal distance from a central source, where 39% of the data is expected to be seen within one standard deviation of a point of introduction, 87% within two and 99% within three (Faulkner et al., 2016).
We used a dataset of 84 spatial records of unique sighting locations, dating from the 1920s to 2016. These were based on data compiled for the GB Non-Native Species Secretariat, which showed adult alpine newts to be established at more than 40 sites in mainland UK (Wilkinson, 2016). We augmented these data with additional records, notified by experienced ecological recorders or published in an ecological report or survey. The dataset is provided in Supplementary Material. Precise location details are withheld to protect privacy of sites, as many are related to private property.
We implemented the DPM model in R (R Core Team, 2014) using a modification of Rgeoprofile (https://github.com/bobverity/Rgeoprofile) (Verity et al., 2014). We set the number of burn-in iterations to 10,000, sampling iterations to 500,000, and the number of MCMC chains to five, allowing sigma to be fitted by the model. Further details of model settings are as in Verity et al. (2014). To test the model’s performance, we assessed its efficiency in identifying known points of human-mediated introduction in three regions, Canterbury, Edinburgh and Leeds. In brief, the test for search efficiency in finding known points of introduction is obtained through a source’s hit score percentage, a metric which is calculated by dividing the area searched before finding the source by the total search area. A smaller hit score thus signifies a more efficient search.
Adult newts were collected from 8 field sites in 5 regions spanning mainland UK (Table 1; Figure 1). Sampling locations were not identical to datapoints of the geoprofiling dataset. For some sites there was pre-existing information on I. alpestris introductions, for others the origin of the newts was unknown. To investigate the amphibian pet trade as a potential source, we purchased captive-bred newts from a UK-based exotic pet shop, and from a UK online site trading in amphibian and reptilian exotics. Full location details are withheld to protect the privacy of sites and retail outlets. Under the Wildlife and Countryside Act (1981) it is illegal to release alpine newts into the wild in the UK. Newts sourced from UK field locations and retail outlets were therefore euthanized in accordance with current regulations [Animals (Scientific Procedures) Act 1986 Amendment Regulations (2012)]. Euthanized newts and tissue samples were preserved in 100% ethanol. To enrich the phylogenetic representation of I. alpestris from its native range in North-Western Europe, tissue specimens from newts captured in the wild in Belgium, France, Switzerland and The Netherlands, were generously provided by colleagues listed in Acknowledgements.
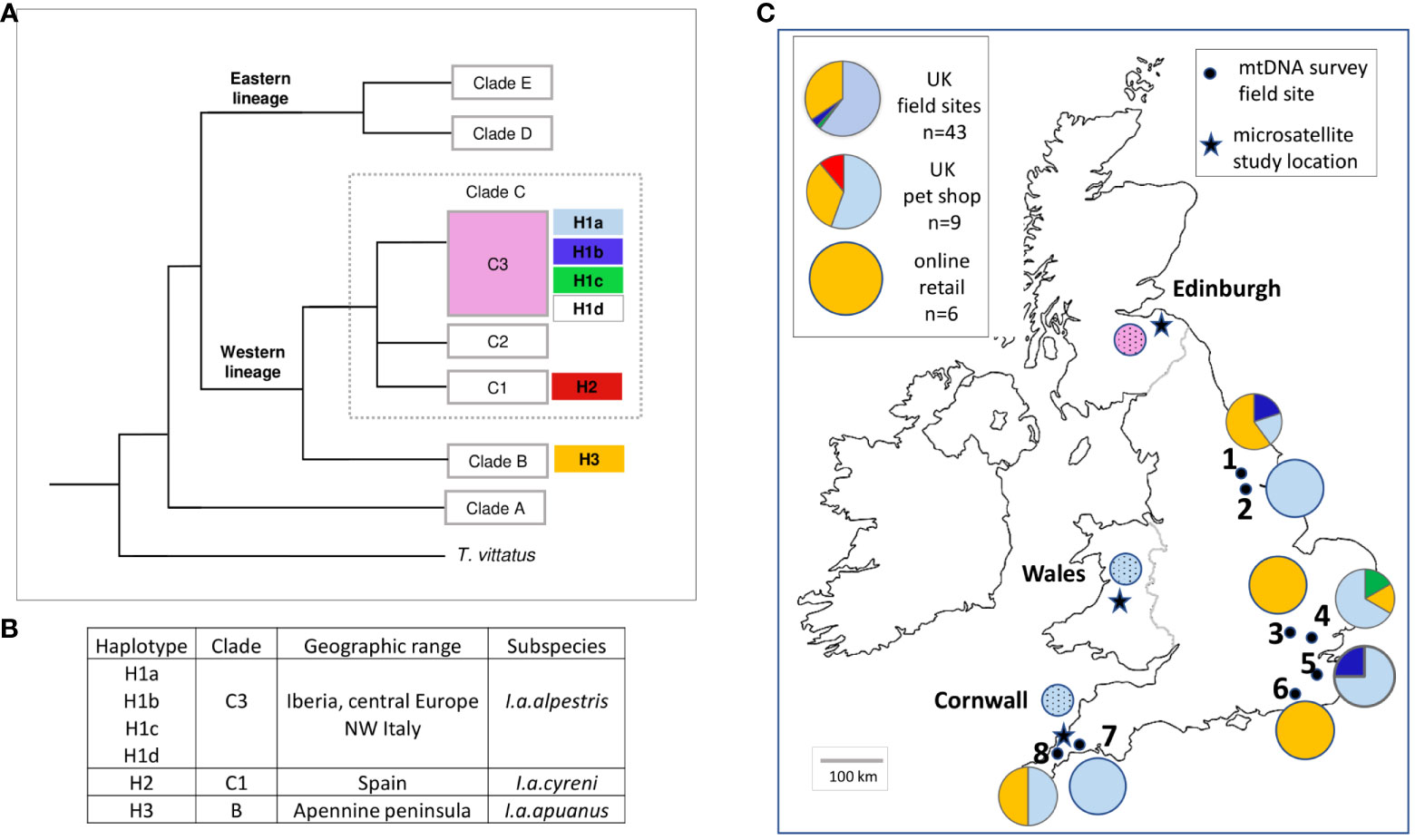
Figure 1 Survey of I. alpestris mtDNA haplotypes in the UK. (A) Phylogeographic relationships of I. alpestris in the UK. Outline phylogenetic tree based on clades described in Sotiropoulos et al. (2007). Haplotypes identified in the UK-wide study are shown next to the likely clade of origin. For Edinburgh samples, where only cytb sequences were obtained, the broad clade of likely origin is indicated (pink). (B) Association between mtDNA haplotypes and I. alpestris subspecies. Table showing the geographic range of the different haplotypes within the native range of I. alpestris, and the subspecies associated with the clades represented in the UK populations and pet trade samples (Sotiropoulos et al., 2007). (C) Distribution of mtDNA haplotypes within UK. Pie charts illustrating the broad geographical distribution of mtDNA haplotypes across the UK, using the same colour scheme as (A). Numbering for field study sites as in Table 1. Pies for microsatellite study regions are stippled. Inset: results for the relative proportions of haplotypes across the UK field study sites, and pet trade samples. Haplotype H1d was not found in any of the UK or pet trade samples.
We extracted DNA from 3 mm toe or tail clips using a DNeasy Blood and Tissue Kit (Qiagen). We amplified mtDNA 16S RNA using primers 16Sar and 16Sbr from Palumbi et al. (1991), and cytochrome b (cytb) using primers L14841 and H15149 from Kocher et al. (1989), loci selected for homology with a phylogeographic study of I. alpestris in Europe reported by Sotiropoulos et al. (2007). DNA was amplified in HotStarTaq Plus or TypeIt Mastermix (Qiagen), with 5μmol/L forward and reverse primer, with 30-40 cycles of 94°C 30 sec, 57°C 90 sec, 72°C 90 sec, final extension 72°C 10 min. Sanger sequencing in each orientation was done by GATC Biotech (Zurich) or Beckman Coulter Genomics, using the PCR primers.
We aligned merged forward and reverse sequences from study samples with homologous I. alpestris sequences accessed from GenBank (Benson et al., 2013), derived from Sotiropoulos et al. (2007), using ClustalW in MEGA 6.0 (Tamura et al., 2013), trimming ends by eye. Trimmed consensus 16S and cytb sequences were concatenated using R v3.1 (R Core Team, 2014) to generate a final 708 bp 16S/cytb sequence. To construct a phylogenetic tree, we aligned concatenated sequences from the UK field study, captive bred newts, and the additional European samples, with published homologous I. alpestris sequences (Sotiropoulos et al., 2007). We used this alignment to construct a maximum likelihood phylogenetic tree in MEGA 6.0, with 10,000 bootstraps, using the Kimura 2-parameter model (Kimura, 1980), identified in MEGA as the most appropriate nucleotide substitution model. Homologous sequences from Triturus vittatus were included as an outgroup.
To investigate local patterns of introduction and spread, we sampled more intensively at three locations, which differed with respect to the area covered, the type of landscape and the likely time since introduction. In two of the regions in our study, Cornwall and Wales, the samples we used for genetic analysis had already been collected from ponds in ecological surveys. We adopted a similar sampling strategy for the Edinburgh (Scotland) sites for consistency. A breeding pond may be considered a panmictic unit in the population genetics of newts (Jehle et al., 2005), facilitating coalescence modeling of demographic history, genetic differentiation between populations, and estimation of intrapopulation genetic diversity, although limiting the application of spatial genetic tests (Prunier et al., 2013). In addition, this approach allows comparison with published studies on newts, in which pond-based sampling is frequently used. In Edinburgh (Scotland), the sites were in urban landscape over an area of approximately 100km2, with inter-pond distances ranging from 30m to ~15km, including one pond on a golf course, one pool in a disused quarry, and two closely situated ponds in parkland. Newts were captured specifically for this study, from the same series of ponds as in the geographic profiling study. Alpine newts are known to have been released from superfluous breeding stock at the local university in the 1950s–1970s, predating legislation against release into the wild. In Cornwall (England) the sites comprised three garden ponds, separated by <1km, in an area where alpine newts are known locally to have been present for 3–30 years, and a fourth natural pond ~13km distant across mixed urban and rural landscape, where newts were first reported in 2015. In Ceredigion (Wales) the sites comprised three closely situated semi-rural ponds, where the presence of alpine newts was first noted in 2014. The sites in Cornwall and Wales were not identical to those sampled for the mtDNA study. Details of each location are summarised in Table 2, with maps in Figure 2.
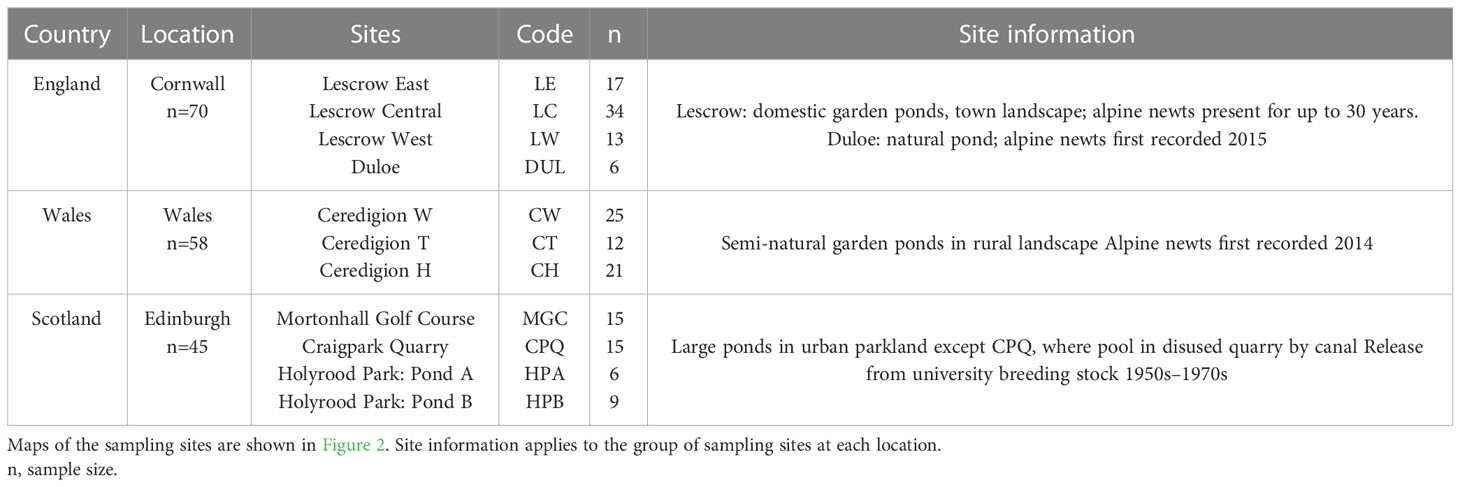
Table 2 Details of case study locations for investigation of local patterns of introduction and spread.
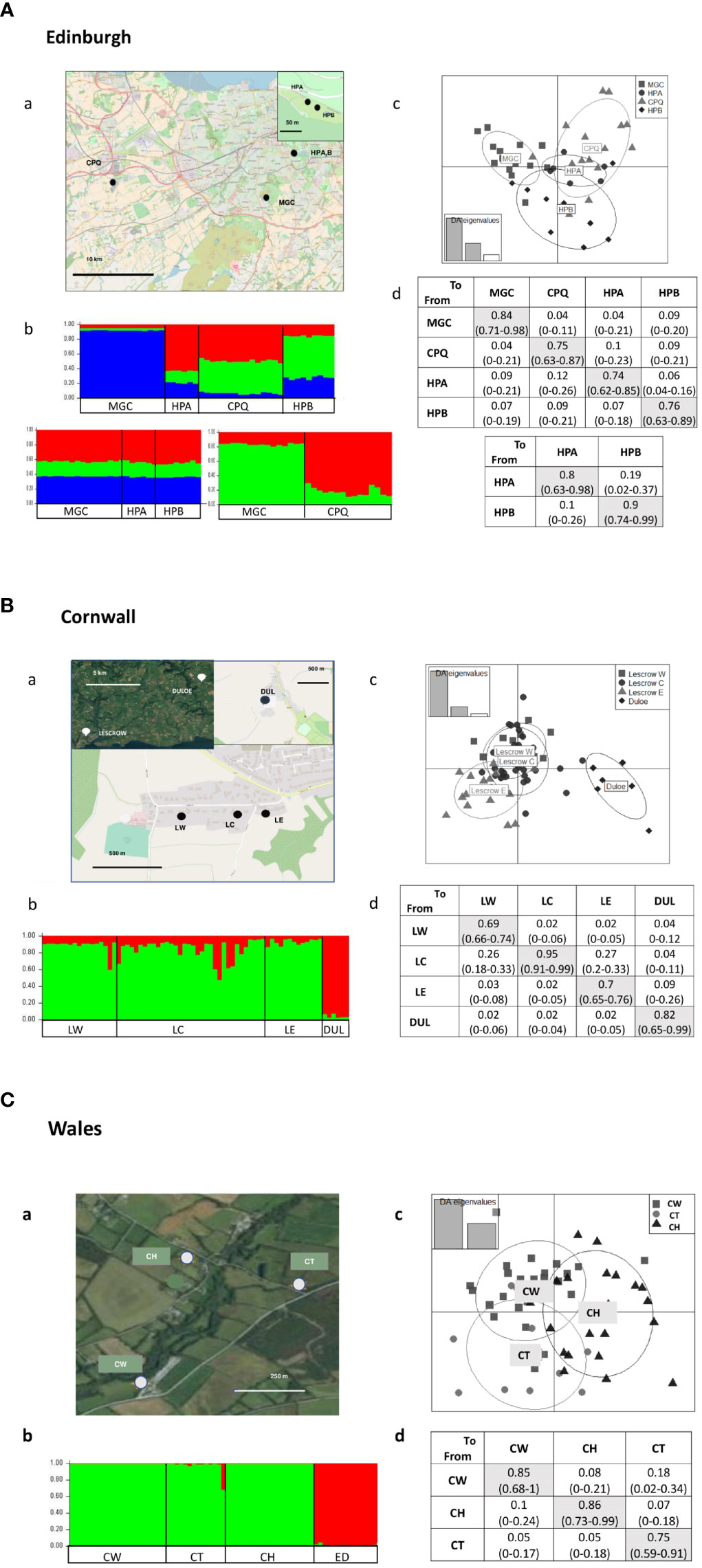
Figure 2 Maps and genetic differentiation between sites in microsatellite study locations [(A) Edinburgh, (B) Cornwall, (C) Wales]. (a) Map of study locations showing relative positions of study sites. Note the scale differs between the study locations and inset maps. (b) STRUCTURE bar charts showing proportional membership coefficients of individuals to each of the inferred clusters, grouped according to their study population. Each colour represents a different cluster. Edinburgh: samples show differentiation between MGC and the other sites, but lack of differentiation between CPQ, HPA and HPB (K=3). When MGC and CPQ are analysed separately there is clear differentiation (K=2). When MGC, HPA and HPB are analysed together, there is no differentiation (K=3) Cornwall: there is differentiation between DUL and the other sites, but no clear pattern between the LE, LC and LW sites (K=2). Wales: no differentiation between sites. Samples from Edinburgh used as PCR cross-location controls are included in analysis (ED) (K=2). (c) DAPC scatterplots showing individuals and inertia ellipses according to their sampling site. Patterns of differentiation are consistent with STRUCTURE results. (d) Matrix of migration rates between inferred in BayesAss, showing posterior mean estimates (95% CI). Left column shows donor site, top row shows recipient site. The diagonal cells (shaded) show inferred proportion of non-migrants. Results are also shown for Edinburgh when data for HPA and HPB are analysed separately (lower panel), illustrating the effect of likely homoplasy on inferred migration when all sites including CPQ are analysed together (upper). When DUL was excluded from analysis in the Cornwall location, there was no effect on the patterns of migration between the LW, LC and LE sites (not shown).
DNA extraction was as for the mtDNA study. Three sets of microsatellite primers were amplified in multiplex (CopTa1; CopTa2; CopTa4), (CopTa6; CopTa11) (Prunier et al., 2012) and (Ta1Ca1; Ta2Caga3) (Garner et al., 2003). CopTa3, CopTa8 and CopTa12 (Prunier et al., 2012) were amplified in simplex. PCR was performed in HotStarTaq Plus or Multiplex Mastermix (Qiagen), with 5μmol/L unlabelled reverse primer and 5μmol/L fluorophore-labelled forward primer (Applied Biosystems), in 40–45 cycles of 94°C 30 sec, 60°C 90 sec, 72°C 90 sec, final extension 72°C 10 min. Amplified products were resolved by capillary electrophoresis on a 3130xl Genetic Analyser (Applied Biosystems) with a LIZ-500 size standard (Applied Biosystems). Template-negative controls and replicates were included in each plate, including samples across different case study locations, to ensure consistency of results. Failed or equivocal reactions were repeated in simplex. Alleles were scored manually, using Peak Scanner 1.0 software (Applied Biosystems).
We analysed amplicon data in Micro-Checker v 2.2.0.3 (Van Oosterhout et al., 2004) for genotyping errors and null alleles, using Bonferroni correction, and in pegas (Paradis, 2010), implemented in R, to test for Hardy-Weinberg equilibrium, and to exclude linkage disequilibrium. We used FSTAT V2.9.3 (Goudet, 2001) to calculate summary statistics (FST, FIS, allele richness, observed and expected heterozygosity) and to compare summary statistics between regions. To estimate the current effective breeding size of the sampled populations we used a full likelihood sibship assignment method (Wang, 2009), implemented in COLONY 2.0.6.3 (Jones and Wang, 2010), assuming random mating and both male and female polygamy, with three medium runs based on a medium sibship prior, not updating allele frequency, and with assumed error rate 0.001. Confidence intervals were obtained by bootstrapping.
We used STRUCTURE V2.3.4 (Pritchard et al., 2000) to infer genetic clustering within each case study region. As sampling was unbalanced across sites, we set the initial α to 1/n, where n=number of sample locations, to improve the accuracy of individual assignments to subpopulations (Wang, 2017). We used the LOCPRIOR setting, to incorporate sampling location (Hubisz et al., 2009), a burn-in of 100,000 iterations, and 500,000 MCMC replications, for 10 independent replicate runs of K, for K=1–6. We used mean log-likelihood values for each value of K and Delta K to determine the optimal value of K at each case study location (Evanno et al., 2005). These were obtained using Structure Harvester (Earl and von Holdt, 2012), or the function evannomethodstructure in the package pophelper (Francis, 2017), implemented in R. To infer clustering, we additionally used discriminant analysis of principal components (DAPC) (Jombart et al., 2010) in adegenet version 2.0.1 (Jombart, 2008), implemented in R. This method uses multivariate analysis both to maximise variation between clusters, or subpopulations, and to minimise variation within clusters.
We used BayesAss v1.3 (Wilson and Rannala, 2003; Faubet et al., 2007) to investigate recent migration rates between populations in each region. This Bayesian approach does not assume symmetric migration rates (Beerli, 1998) and is applicable to nonstationary populations which are not in Hardy-Weinberg or genetic equilibrium (Wilson and Rannala, 2003), in contrast to FST-derived statistics (Whitlock and McCauley, 1999; Neigel, 2002; Meirmans and Hedrick, 2011). BayesAss uses multi-locus genotypes as probabilistic indicators of source populations, to produce estimates of recent or contemporary dispersal rates for each sampling site (Wilson and Rannala, 2003; Pearse and Crandall, 2004). BayesAss was run with 3 million MCMC iterations, with a burn-in of 100,000. Delta values were adjusted to ensure that 30–60% of the total changes were accepted, as recommended in the software manual. A minimum of three independent runs initialised with different starting seeds was carried out for each set to confirm concordance of posterior mean parameter estimates. 95% confidence intervals (CI) for migration rates were estimated as mean ±1.96 standard deviations.
We used ABC implemented in DIYABC version 2.1.0 (Cornuet et al., 2014) to model patterns of introduction and spread in the three case study locations. The underlying process of ABC is to simulate datasets for scenarios based on observed genetic data, drawing parameters from prior distributions (Beaumont et al., 2002; Cornuet et al., 2008), informed by available biological and demographic data. The simulated datasets with summary statistics closest to those of the observed dataset are then used to identify the model scenario that best fits the data, and to infer parameters for the supported model. ABC cannot distinguish between competing models where the expected values for the summary statistics do not differ (Wakeley, 2003). We therefore adopted the approach of sequential comparison of competing scenarios to address specific questions, and then to generate a final model in which to infer posterior parameters for the model. Competing scenarios were designed to be of similar complexity, with prior distributions of equivalent breadth. We used default settings for the microsatellite mutation priors, based on the generalized stepwise model (Di Rienzo et al., 1994; Estoup et al., 2002), as there are no specific data to inform mutation parameters for I. alpestris; all loci were treated as a single group. We used the same ranges of priors for all three regions, with foundation bottleneck duration fixed at 5 generations, and founder size from 2–50. A minimum of 105 simulations was performed per scenario. Scenarios were compared using linear discriminant analysis of summary statistics with logistic regression analysis (Fagundes et al., 2007; Beaumont, 2008), to estimate posterior probabilities with 95% confidence intervals for each competing scenario (Cornuet et al., 2008). Parameters of the model scenarios were estimated from the posterior parameter distributions of the 1% simulated datasets with summary statistics closest to those of the observed dataset. Precision and bias of posterior parameters were assessed using 1000 pseudo-observed datasets with parameter values drawn from the posterior distribution for the relevant scenario. Models were evaluated for goodness of fit and potential discrepancies (Gelman et al., 2004) using the model checking function in DIYABC, with the inclusion of summary statistics not used for the original selection of simulated datasets (Cornuet et al., 2010). Further details of ABC methods including prior settings and summary statistics used for simulation and for model checking are detailed in Supplementary Material.
A total of 38 spatial clusters yielded the largest marginal likelihood for the number of realised sources in the DPM model (Figure 3), equivalent to ~45% of the 84 records in the spatial dataset having resulted from independent introductions. Hit scores of known sources in Canterbury, Edinburgh and Leeds showed that each known source was found more efficiently than by a random, uninformed search. Newt dispersal was estimated by the DPM model to be 1.08 km for the UK-wide data, and 0.78 km, 0.49 km and 1.26 km for Canterbury, Edinburgh and Leeds respectively (detailed in Supplementary Material). In each case sigma was fitted by the model with no prior constraints.
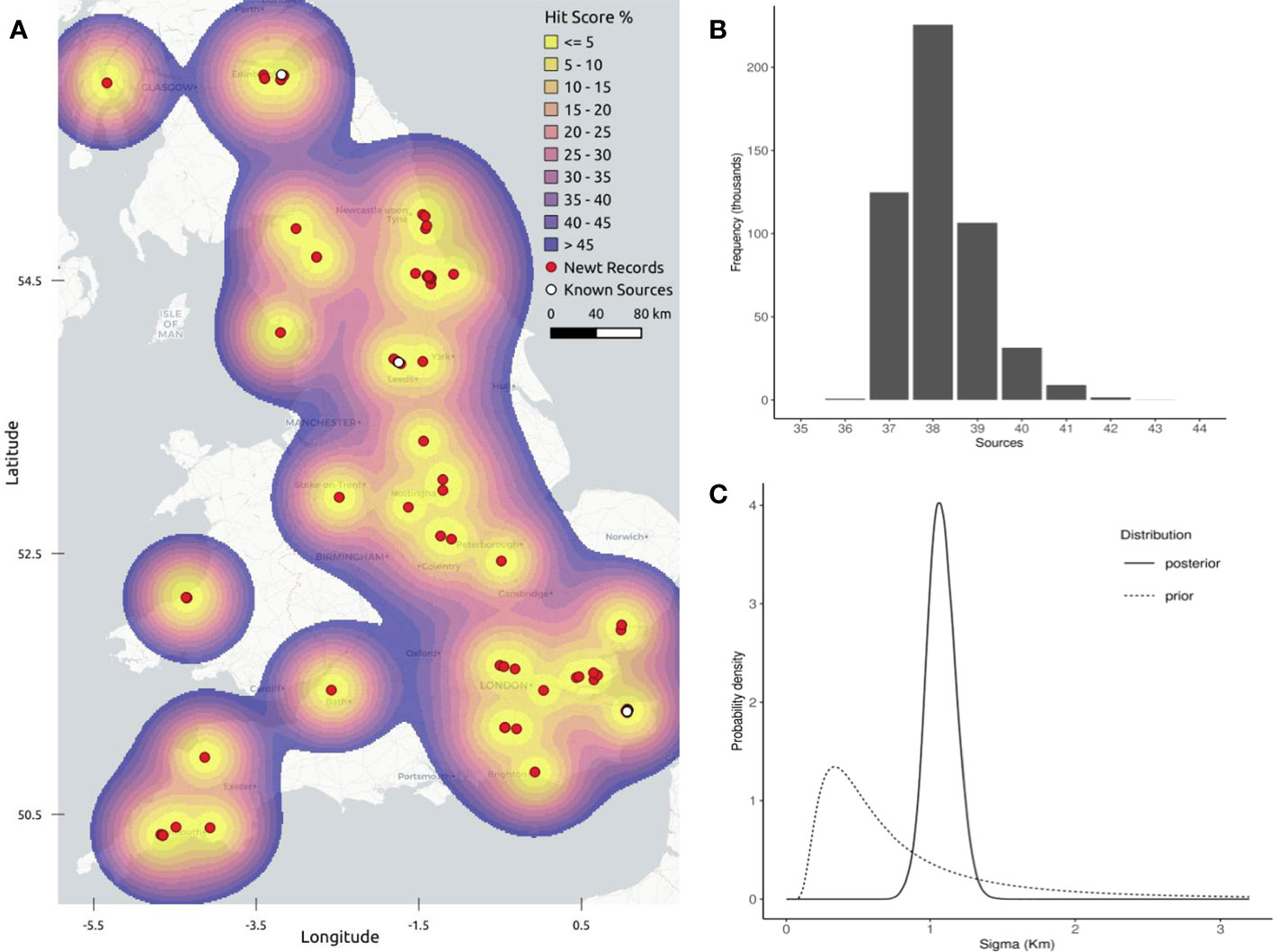
Figure 3 Geographic profiling for UK-wide data: multiple points of introduction. (A) The top 50% of the geographic profile produced by the DPM model analysing UK-wide data points (shown in red). Areas in yellow indicate those locations most likely to contain a source of newt dispersal, indicating the existence of multiple source locations. This map was created using the Carto Light No Labels layer via QGIS.org (2021). (B) The marginal likelihood for the number of source populations, showing the greatest support for 38 spatial clusters associated with the UK-wide data. (C) Prior and posterior distributions describing newt dispersal (sigma). The mean posterior density on newt dispersal, obtained by the DPM model, was estimated to be ~1.08km.
Results are presented in Figure 1. We identified six 16S/cytb mtDNA haplotypes in a total of 68 study sequences (UK field study n=43, captive bred newts n=15, European samples n=10). The phylogenetic tree generated from an alignment including published homologous I. alpestris sequences (n=59) closely resembles that of Sotiropoulos et al. (2007), with division into Eastern and Western lineages, and the same strongly supported clades.
Four haplotypes were represented in the UK survey field site samples, associated with two distinct clades of the Western lineage of I. alpestris. The commonest haplotype, H1a, was found in 26 individuals from 6 study sites. Haplotype H1b was observed in 2 individuals, one from a northern England site, one from the south. A novel haplotype, H1c, was found in a single individual from southeast England. Haplotype H3 was found in 14 individuals from 6 study sites. Five sites had more than one haplotype (2 haplotypes at 3 study sites, 4 haplotypes at 2 sites). All six samples from the online pet shop were of haplotype H3. Three haplotypes were represented in 9 samples from the UK pet shop, most frequently H1a or H3, and a single individual with haplotype H2 (not found in any of the UK field site samples). We identified a total of two haplotypes in 10 samples from the European mainland. Six samples had the H1a haplotype (France n=1, Switzerland n=1, Netherlands n=3, Belgium n=1). Four samples from Belgium had a closely related haplotype, H1d, which was not found in any of the UK field site samples.
All individuals from the Wales and Cornwall microsatellite study sites shared the common H1a (clade C3) haplotype. The cytb sequence of all individuals from the four Edinburgh sites was consistent with a clade 3 origin. However, amplification of 16S was unsuccessful, precluding identification of the full 16S/cytb haplotype. As these clusters of sites had been sampled more intensively than the mtDNA field study sites, we did not include these results in calculations of the distribution frequency of mtDNA haplotypes.
There was no evidence for linkage disequilibrium or allele drop out. Replicate controls gave consistent results. Micro-checker results for one locus (CopTa3) were suggestive of a null allele in the Cornwall study location. CopTa3 was therefore excluded from analysis for Cornwall, but retained for other locations, where there was no evidence for a null allele. Analysis included 45 samples from 4 sites in Edinburgh genotyped at 6 loci, 58 samples from 4 sites in Wales genotyped at 10 loci, and 70 samples from 4 sites in Cornwall genotyped at 8 loci.
Summary statistics for Edinburgh, Cornwall and Wales, derived from the full range of loci for each study location, are shown in Table 3, together with published results from I. alpestris in its native range in Switzerland (Emaresi et al., 2011) and in the Iberian Peninsula (including an allochthonous population in central Spain (Peňalara) (Palomar et al., 2017). These results show high FIS values for Cornwall and Wales, indicative of inbreeding. There was lower allele richness in all three UK regions than in the Spain or Switzerland published studies, despite other indices of diversity being comparable, with the exception of possibly lower genetic diversity (HS) results in the Wales study region. However, both published studies had used microsatellite panels which were only partially overlapping with our study, thus precluding statistical comparison.
To correct for the different number of loci used between study groups and thus to allow statistical comparison, we compared UK study locations using results from the six microsatellite loci common to all three (Table 4). These confirmed significantly lower allele richness and genetic diversity (Hs), and significantly higher FIS in Wales than in Edinburgh (p< 0.01). Hs and FIS, but not allele richness, differed between Cornwall and Edinburgh (p<0.05). This confirmed that the differences between the sites were not the result of the smaller number of loci for the Edinburgh study, although the use of 6 rather than 10 loci did result in a lower value for allele richness and expected heterozygosity in the Wales sites. There was no significant difference between the Wales and Cornwall locations for any of the summary statistics.
In Edinburgh (Figure 2A) STRUCTURE analysis of all four sites indicated K = 2 to be most probable (not shown) with individuals sampled from MGC assigned to one cluster and the remaining individuals to the other. MGC also separates from the other sample locations in the first axis of variation in the DAPC scatterplot. The second axis separates HPB from the other sample sites. As ABC modeling revealed the likely occurrence of homoplasy between CPQ and HPA (detailed below), STRUCTURE was rerun with exclusion of different combinations of CPQ HPA, and HPB. Differentiation between MGC and CPQ was most pronounced with the exclusion of both HPA and HPB. When CPQ was excluded, there was no evidence in STRUCTURE of differentiation between MGC, HPA and HPB. In Cornwall (Figure 2B) individuals from Duloe were genetically differentiated from the other three sites, as indicated by STRUCTURE and DAPC analysis. In Wales (Figure 2C) STRUCTURE analysis of the Welsh sampling sites indicated that K=1 best described the data, consistent with panmixia. The DAPC results similarly show that these sites cluster with a higher degree of overlap than that observed in Cornwall or Edinburgh.
Figure 2 shows inferred posterior migration rates between sampling sites for each region. For the Edinburgh sites the inferred proportion of non-migrants ranged from 0.74–0.84. The inferred migration rates between sites varied from mean 0.04 (95%CI 0–0.12) to 0.12 (0–0.26). The highest inferred migration was between CPQ and HPA. We repeated BayesAss analysis of HPA and HPB in isolation, to remove the confounding influence of likely homoplasy with CPQ. This reduced the inferred proportion of non-migrants for both HPA and HPB, with a higher level of bidirectional migration between the two sites. For the Welsh sites the proportion of non-migrants was 0.75–0.86, with a similar range of inferred migration rates. However, there was apparent asymmetry of migration, being 0.18 (0.02–0.34) from CW to CT, but only 0.05 (0–0.17) in the opposite direction, but with overlapping confidence intervals. Asymmetry was more pronounced in the Cornwall samples, in which there was a high inferred migration from LC to each of LW and LE, 0.26 (0.18–0.33) and 0.27 (0.20–0.33) respectively, with very low rates of migration in the other direction in both cases, with no overlap in confidence intervals. This was reflected in the proportion of inferred non-migrants, which was 0.95 (0.91–0.99) for LC, against 0.69 (0.66–0.74) for LW and 0.70 (0.65–0.78) for LE.
We based initial comparisons on the assumption that each site in the same region originated from a common source population, and that primary introduction events were likely to have been associated with a foundation bottleneck. For sites without support for a primary foundation bottleneck, we considered the possibility of secondary spread from a site of primary introduction, or from an additional unsampled population. Details of scenarios and posterior probabilities in sequential comparisons are given in Supplementary Material. Final working models are shown in Figure 4.
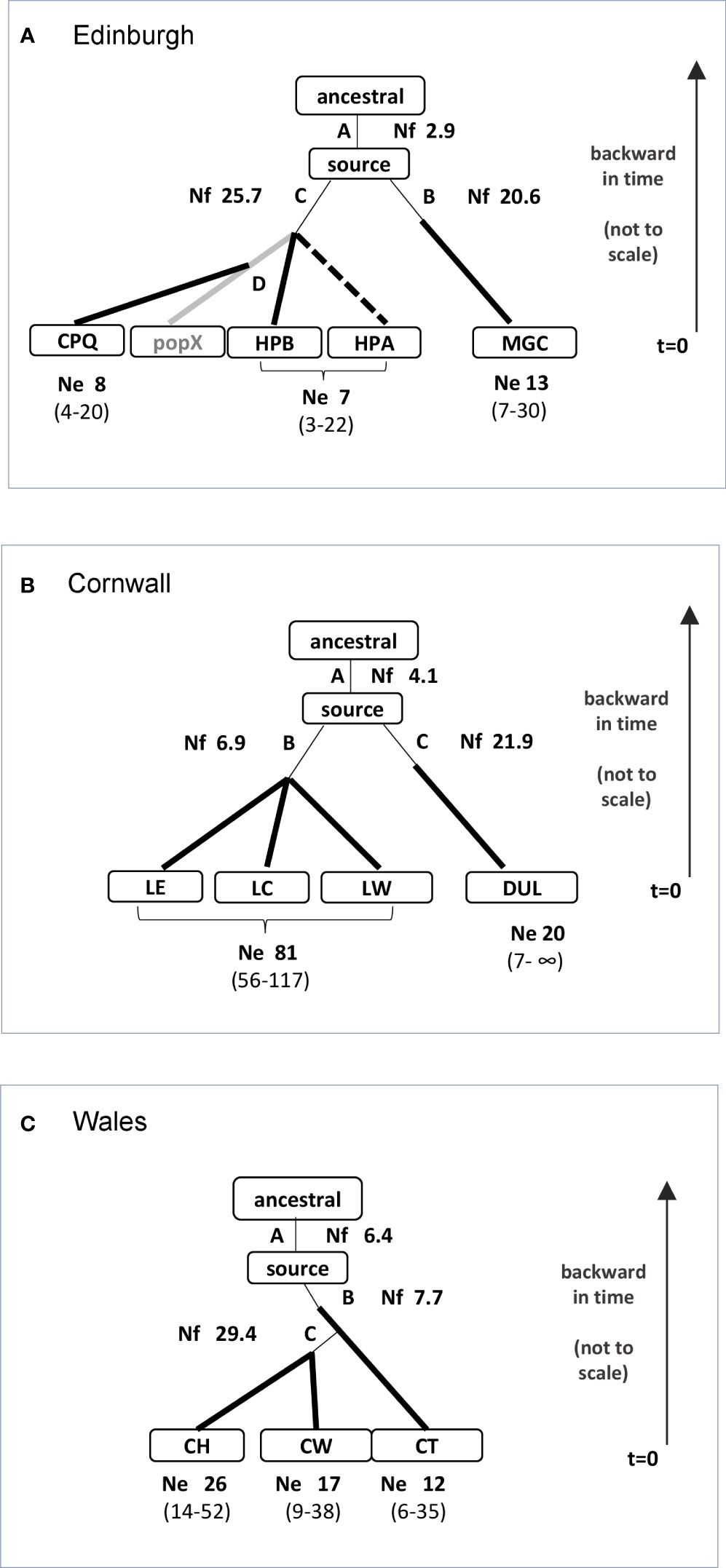
Figure 4 Supported ABC scenarios. Annotated schematic models for introduction events from ancestral and source populations, with modal values for foundation bottlenecks derived from posterior parameter distributions For the supported model in all three locations, the common source population is itself derived with a foundation bottleneck from an ancestral population. Nf: founder size (mode); Ne: effective population size with 95% CI (shown for pooled populations where model does not differentiate). (A) Edinburgh. The ABC model indicates a primary introduction to MGC from the source population, with a foundation bottleneck. HPB and an unsampled “ghost” population (pop X, shown in grey) are derived from the source population, with a common foundation bottleneck. The CPQ population is derived from the unsampled population. The HPA population is added as a dotted line; although it was not included in the comparison scenarios, additional model testing supported a common origin with HPB. relationship between sites in model may have been influenced by presence homoplasy. (B) Cornwall. The ABC model indicates two main introductions from a common source population, one to Duloe, and the other to the Lescrow cluster of ponds (LW, LC, LE). Each introduction is associated with a foundation bottleneck. (C) Wales. The ABC model indicates a primary introduction to CT from the source population, with a foundation bottleneck. CW and CH are derived from the CT population, with a common foundation bottleneck.
Edinburgh (Figure 4A): The distribution of sampling sites in Edinburgh is more scattered than the other two regions. Two ponds, HPA and HPB, are very closely adjacent, MGC and CPQ being more distant. Initial scenario comparisons generated conflicting results with respect to the relative topology of the HPA and CPQ sites, and model checking showed the Fst summary statistic between the two sites to be a consistent outlier (<0.001). This may reflect homoplasy between the sites, in which alleles may be identical by state but not by inheritance, interfering with coalescent modeling (Estoup et al., 2002). In subsequent comparisons we therefore excluded the HPA population, retaining the CPQ site, which is potentially more informative due to its more distant geographical location and higher number of samples. This resolved the problem of conflicting support for competing scenarios. There was strong support for more than one primary introduction from a common source population, one being to the MGC with a foundation bottleneck, and the other giving rise to both HPB and CPQ. There was strong support for at least one unsampled “ghost” population being involved in the origin of the CPQ and HPB populations, although on the data available it was not possible to further define their relationship. In the final working model, we assumed two primary introductions from a common source population, one into MGC, and another into both HPB and an unsampled population, with secondary spread from the unsampled population into CPQ. However, although the model is consistent with STRUCTURE and DAPC clustering, with excellent goodness-of-fit on model checking when HPA is excluded, the possibility of homoplasy also occurring between CPQ and HPB means that the origins of CPQ relative to MGC and HPB should also be interpreted with caution.
Cornwall (Figure 4B): Four sites were included in analysis, of which the three Lescrow ponds are closely clustered, and Duloe is situated ~13 km away, separated by potential urban barriers. The supported model incorporates two main introductions, each with a foundation bottleneck from a common source population, one to Duloe, the other to the Lescrow ponds. This scenario is consistent with results from STRUCTURE and DAPC, in which Duloe clusters independently from the other sites. It was not possible to determine whether any one pond of the Lescrow cluster was the site of primary introduction.
Wales (Figure 4C): Three sites were included in ABC analysis, all situated within a 1 km radius in rural landscape. We reasoned that a single introductory event was likely, given the proximity of the sampled ponds, and their likely panmixia. Comparison of scenarios of competing topology identified CT as the likely site of primary introduction, arising with a foundation bottleneck from the source population, with subsequent secondary spread to the other two sites.
For all three study locations, there was support in ABC for an intermediary ancestral population from which the source population had been derived, again with a foundation bottleneck.
We used the posterior parameter distributions of the final working model scenario for each study region to derive estimates for the size of introductory bottlenecks. In DIYABC, the founder size (Nf) is the effective population size averaged over the duration of the bottleneck. This was arbitrarily fixed at 5 generations, with a uniform prior of [2,50] for Nf in all model scenarios. Posterior distribution curves for Nf in supported models show a modal pattern in comparison with the flat plots for the prior distribution, indicating the influence of genetic data (plots shown in Supplementary Material). We therefore used posterior modal values as an estimate of founder numbers for scenarios (Figure 4). Mean and median values, which are more influenced by prior ranges, are given in Supplementary Material.
Based on the posterior distribution modal values, the estimated founder number for the introductory events for the Wales ponds is 7.7, and for the Lescrow cluster of ponds in Cornwall 6.9. Estimated founder sizes for introductions from the source population are higher for Edinburgh, and for the Duloe pond in Cornwall (Figure 4). These figures cannot be taken to indicate the exact founder size, but do suggest that a colony can become established from the introduction of a small number of individuals. Estimates for bottleneck sizes between ancestral and source populations in model scenarios are also low, but more difficult to evaluate in the absence of information about the processes and number of intermediary populations involved.
Estimates of Ne for the relevant populations are also shown in Figure 4, calculated using the full likelihood sibship assignment method, with confidence intervals derived by bootstrapping.
It was not possible in this study to derive meaningful posterior parameter estimates with respect to timing of demographic events. The relation between the inferred number of generations and chronological time is a function of the mutation rate of the genetic markers used (Kimura, 1968), and the generation time, reflecting the time separating the birth of parents from the birth of their offspring. Although the microsatellite mutation rate observed in the eastern tiger salamander Ambystoma tigrinum tigrinum is consistent with the default prior we used in ABC modeling, there may be log magnitude variation between loci (Bulut et al., 2009). The time to female sexual maturity, around three years in I. alpestris (Griffiths and Teunis, 1996; Miaud et al., 2000), can be used as an approximator of generation time (Lande, 1982), although this will be affected by life history trait variation (Miaud et al., 2000), and by the presence of overlapping generations. Posterior parameters can still be used to infer relative timings within a scenario, and calibrated to known timings, but not between scenarios where different microsatellite panels have been used. For the Edinburgh study, calibration of time parameters is enabled by the information that the university legally released alpine newts from their captive colony in the 1950s and 1970s into ponds in Edinburgh, specifically naming the MGC site (Scottish Golf Environment Group). However, the presence of homoplasy complicates extrapolation to other Edinburgh sites. For Cornwall, the inferred timing of introduction events is consistent with a more recent record of newts at the Duloe site (first recorded 2015) in comparison with the LE/LC/LW cluster of ponds (up to 30 years). The more recent posterior estimates in Wales are consistent with first records from 2014.
Strikingly, the sites where naturalised colonies of alpine newts have been recorded in the UK are almost without exception in human-dominated landscape, often privately owned garden or farm ponds (Bell and Bell, 1995; Beebee, 2007; Humphreys et al., 2011; Wilkinson, 2016). Available records are largely based on reported sightings rather than a systematic and comprehensive survey, and are therefore likely to underestimate the total number of established colonies, especially in rural areas. However, I. alpestris mtDNA was not detected by eDNA metabarcoding in any of the natural UK ponds tested in a study of the indigenous great crested newt Triturus cristatus (Rees et al., 2014; Biggs et al., 2015; Harper et al., 2018), in keeping with alpine newts being rare in a truly wild setting.
Our geoprofiling results indicate that a significant proportion of non-captive colonies of I. alpestris in the UK are likely have arisen from independent introductions. The efficiency of geoprofiling in identifying known points of introduction in the test regions in our study illustrates the potential power of this method. However, in the absence of a systematic survey we were unable to fully exploit this potential, and the proportion inferred in our study (38 sources inferred for 84 spatial records) can only be an approximation. Nonetheless, the combination of the geoprofiling findings and the human-dominated locations of established colonies strongly suggests multiple independent introductions, resulting from the local release or escape of newts from a domestic setting. Interestingly, in a recent geoprofiling study, the distribution of another ornamental escapee in the UK, the non-native ring-necked parakeet Psittacula krameria, was also attributed to multiple local introductions, rather than to spread from a few sites (Heald et al., 2020). Papini et al. (2017) similarly implicated public or private gardens and plant dealers as multiple sources of introduction and spread in their geoprofiling study of an invasive geophyte Oxalis pes-caprae (Papini et al., 2017). In future studies, geoprofiling could be a valuable tool in the study of the spatial distribution of alpine newts between known breeding populations using eDNA sampling of all potential breeding ponds in a defined area, to generate both presence and absence data. This would be a useful adjunct to spatial sampling for genetic studies (Prunier et al., 2013), as used by Papini et al. (2017).
The variety of mtDNA haplotypes identified in established populations of I. alpestris in Britain confirms their origin from multiple sources within the native range of the species. The likely origin of naturalised alpine newts from captive collections is further supported by the close resemblance between mtDNA haplotypes found in newts sourced from the pet trade and those from established alpine newt populations. Commonly found haplotypes correspond to I. alpestris lineages noted for their bright colouration and distinct markings, despite their limited distribution in their native range (Sotiropoulos et al., 2007). While the earliest introductions by primary collectors were probably collected by enthusiasts directly from the species’ native range (Lever, 1977), animals sourced from the various wholesale, retail and online providers of non-native amphibians currently operating in the UK are more likely to derive from commercial breeding stock (Krysko et al., 2011; Tapley et al., 2011; Carpenter et al., 2014; Kraus, 2015; Mohanty and Measey, 2019). The resolution of the mtDNA markers we used in our study is too low to discriminate between direct introductions from the native range and escapes/translocations from captive or established invasive populations located within or outside the UK. This could be improved with mitogenomic and genomic sequencing, also potentially useful to track individual animals from known sources, including trade.
The occurrence of multiple independent introductions is also supported by our finding of genetically distinct alpine newt populations in two of the three study regions selected for microsatellite genetic testing. This is likely to reflect historic genetic differences between source populations rather than post-introduction divergence of isolated populations (Murphy et al., 2008; Landguth et al., 2010; Cayuela et al., 2018), given the short timescale relative to the life history of I. alpestris (Griffiths and Teunis, 1996; Miaud et al., 2000). The finding of genetically distinct populations in Edinburgh was unexpected, as we had initially assumed they had all originated from surplus university breeding stock, as documented for the MGC site (Scottish Golf Environment Group). The CPQ site in Edinburgh is a pool in a disused quarry close to the city canal, in contrast to the parkland setting of the other sites, which may reflect illicit dumping of unwanted animals (Copp et al., 2005; Stringham and Lockwood, 2018). The unsampled source population for CPQ in the supported ABC model could represent an intermediary captive population, possibly from the pet trade, or an additional unsampled naturalised population, the canal acting as a dispersal corridor for unassisted spread (Anderson et al., 2015).
There was a low migration rate between the MGC and CPQ populations, which are separated by 12.6km. Inferred migration was also low between MGC and the HPA/HPB sites, separated by 4km. By contrast, BayesAss results indicated moderate rates of reciprocal migration between the HPA and HPB ponds, which are less than 50m apart, but also a high proportion of inferred non-migrant individuals, consistent with breeding pond philopatry in alpine newts (Joly and Miaud, 1989; Sinsch et al., 2003). These results are consistent with estimates of dispersal by the DPM model in our geoprofiling study, ranging from 0.49–1.26km, and with published studies of newt movement ecology. For example, capture-mark-recapture studies of I. alpestris are consistent with high levels of movement between ponds up to 1.8km apart (Schmidt et al., 2006), while Ribeiro et al. (2011) found 1km to be the distance threshold between ponds in a species richness study of amphibians in Spain.
We found striking asymmetry of migration between the Lescrow group of ponds in Cornwall with a very high inferred rate from one pond to each of the other ponds, with negligible flow in the alternate direction. A similar but less pronounced asymmetry was seen with the Wales ponds. Asymmetry can result from source-sink dynamics (Murphy et al., 2010), or from a pronounced recent bottleneck (Sundqvist et al., 2016). It is likely that the latter explanation applies in this context, Lescrow and Wales ponds having only been recently populated, in each case from a common source population, and with a low founder number estimated from ABC modeling. Patterns generated by colonisation of previously unoccupied sites are unlikely to be at equilibrium, and may be more complex to interpret than isolation by distance (Orsini et al., 2013). It will be very interesting to monitor the evolution of genetic patterns in these study ponds, and to further study dispersal pathways by spatial genetic sampling (Prunier et al., 2013).
The concentration of alpine newt populations in human-dominated landscape in the UK is most likely to reflect their site of initial introduction, with limited dispersal due to the nature of their surroundings. There is nothing to suggest that I. alpestris is a synanthrope which benefits from continuing proximity to populated areas, in contrast to the common myna Sturnus tristis, whose limited dispersal outside townscape after more than a century of colonisation in Australia may reflect a positive preference for urban environments (Sol et al., 2012; Old et al., 2014; Ewart et al., 2019). In addition, captive alpine newts are most likely to be released into the ponds where they establish self-sustaining colonies, rather than into terrestrial habitat. This may not only bypass any selection pressure for the good dispersal abilities often attributed to founding individuals (Williamson and Fitter, 1996; Denoël et al., 2018; Renault et al., 2021; Daly et al., 2023), but also restrict further dispersal. The garden pond can be considered a “discrete and specialised” amphibian habitat (Beebee, 1985), relatively inaccessible to native newts without human intervention (Beebee, 2014). The boundaries that limit the access of native amphibians to garden ponds (Humphreys et al., 2011) are likely to similarly limit the outward spread of alpine newts.
The dispersal of newts to new breeding ponds in their native range is also strongly influenced by landscape connectivity (Pittman et al., 2014). In its native range in Switzerland, forest acts as a dispersal corridor for I. alpestris, and urban landscape as a dispersal barrier (Emaresi et al., 2011). Genetic studies of newts in their native setting show genetic differentiation between populations in fragmented habitat, but genetic homogeneity in continuous habitat (Noël et al., 2007; Beninde et al., 2021). Suburban and urban landscapes are thus likely to limit the unaided dispersal ability of I. alpestris (Hamer and McDonnel, 2008; Emaresi et al., 2011). An equivalent importance of landscape connectivity in invasive spread is supported by the comparison of the limited spread of alpine newts in the UK with the rapid expansion of an allochthonous introduction of I. alpestris cyreni in the Peñalara massif in central Spain (Lope and Cuadrado, 1985; Bosch and Martínez-Solano, 2003; Palomar et al., 2017). Although south of the natural range of the species, this is in protected montane habitat, with a close-set matrix of natural ponds and the availability of terrestrial refugia (Palomar et al., 2017). By contrast, the unsuitable Mediterranean climate explains the very limited dispersal of an I. alpestris breeding population in Larzac in the south of France, outside its natural range, despite the presence of potentially accessible neighbouring ponds (Denoël, 2005), possibly exacerbated by the captive origin of the Larzac population (Carrete and Tella, 2008; Sol and Maspons, 2016). Other factors may also contribute to the invasion success of the Peñalara population, including its direct origin from a natural population, and retained genetic diversity, although possibly offset by inbreeding (Palomar et al., 2017). Biotic factors may also be relevant in the limited expansion of breeding colonies of I. alpestris in the UK, including reduced genetic diversity and likely captive origin (Carrete and Tella, 2008). We also found evidence for inbreeding in the Wales and Cornwall study regions.
Overcoming barriers to dispersal, whether by initial release or secondary translocation into more connected habitat, could allow alpine newts to become invasive sensu Blackburn et al. (2011). A change in distribution would also increase the risk of contact between I. alpestris and native taxa, and thus the potential risk of a detrimental impact. For example, an overlap of breeding range increases the risk of hybridisation between introduced and indigenous species (Crispo et al., 2011), illustrated by the extensive genomic introgression between the introduced non-native Triturus carnifex and the threatened native crested newt T. cristatus in Switzerland (Dufresnes et al., 2016). However, in its native range in mainland Europe the alpine newt is sympatric with all three newt species that are native to the UK, the great crested newt T. cristatus, the smooth newt Lissotriton vulgaris (formerly T. vulgaris) and the palmate newt L. helveticus (formerly T. helveticus) (Speybroeck et al., 2020). The presence of multiple newt taxa, including I. alpestris, has been noted in assemblages at the same breeding ponds (Fasola and Canova, 1992; Covaciu-Marcov et al., 2010; Speybroeck et al., 2016). It is therefore possible that I. alpestris could have little direct negative impact in the event of contact with native newt species. The transmission of disease from introduced alpine newts probably represents a greater risk, discussed further below.
A lag phase between introduction and invasion (Crooks, 2005) may be ended by a novel invader, as well as a change in the established alien population. Genetic investigations have confirmed historical accounts that the invasion of Australia by the alien European rabbit Oryctolagus cuniculus was triggered by the release of rabbits of wild ancestry, rather than the repeated introduction of animals of domestic origin (Alves et al., 2022). With respect to the alpine newt, novel variants could arise pre-introduction from hybridisation caused by the lack of segregation of genetic subtypes in commercial stock. For example, batches of water frogs obtained through a licensed retailer have been shown to include both Marsh frogs (Pelophylax ridibundus) and Anatolian water frogs (P.cf. bedriagae) (Holsbeek et al., 2008). The rapid spread of the invasive rabbit is still likely to have been facilitated by pre-existing network of “sleeper” domestic rabbit populations, with potential parallels with the already widespread distribution of the alpine newt in the UK.
The non-captive populations of I. alpestris in our study most closely correspond with category B3 or C3 in the framework of alien species proposed by Blackburn et al. (2011), being self-sustaining but situated in human-associated landscape within urban confines. However, they represent only a portion of the total I. alpestris in the UK, which also includes animals held captive within the pet trade or privately owned within UK households. This captive reservoir is the likely immediate source population for naturalized alpine newts. The potential risk posed by release of captive non-native species is well-recognized (CBD Secretariat, 2010; Tingley et al., 2015). Despite this, legislation controlling release of alpine newts in the UK clearly provides insufficient protection, given that releases are ongoing. There also remains a grey area with respect to the legal status of newts kept in privately owned garden ponds, unless effectively contained. Deliberate releases of animals into the wild are more likely to be motivated by well-intentioned attempts at “liberation”, coupled with ignorance of the law rather than malicious contravention (Stringham and Lockwood, 2018). Moorhouse et al. (2017) found that the purchase of an exotic pet was less likely when the customer was given information about legality and zoonotic risk. Unfortunately, such information is currently absent or difficult to access on internet pet trade sites.
The number of reported alpine newt locations has continued to rise over the past decade (Harrower et al., 2020; Allain and Lynn, 2021). Higher public awareness, for example in response to requests to report the presence of alien amphibian species (Amphibian and Reptile Conservation), may have played a part, but the magnitude of the increase and lack of plateau suggests that the trend is genuine. It also mirrors the world-wide increase in records of invasive alien species world-wide (Hulm, 2009; Seebens et al., 2017; Pyšek et al., 2020), and is likely to reflect the relative ease of buying exotic pets from internet sources in a global market (Amano et al., 2016; Capinha et al., 2017; Hulme, 2021). An increased density of populations increases propagule pressure (Blackburn et al., 2015), and the risk of secondary contact as discussed above, but the major cause for concern is the continuing risk imposed by ongoing introductions from the exotic pet trade. The risk of disease transmission is the most immediate and concerning (Fisher and Garner, 2007). The international amphibian trade is already implicated in the global spread of pathogens including Batrachochytrium dendrobatidis, B. salamandrivorans and ranaviruses (Fisher and Garner, 2007; Schloegel et al., 2009; Martel et al., 2014). Schloegel et al. (2009) have reported a high prevalence of B. dendrobatidis and ranavirus in recently imported live market frogs at three major US ports of entry. There is also a high level of B. dendrobatidis in imported amphibians in Japan, with a pathogen haplotype over-represented in international trade samples in comparison with locally bred sources (Tamukai et al., 2014). Connected populations of I. alpestris have been implicated as an immune reservoir in the spread of B. salamandrivorans in Germany (Beninde et al., 2021). B. salamandrivorans has not been identified in the wild in the UK (Cunningham et al., 2019) but has been found in private amphibian collections (Fitzpatrick et al., 2018), and infection represents an ongoing risk (Winchester, 2015). New high resolution mitogenomic and genomic markers will be valuable in both disease surveillance, and in barcoding and tracking global trade introductions (Rane et al., 2023).
In this study we provide evidence that the widespread distribution of non-captive populations of I. alpestris in the UK has arisen from repeated human-mediated introductions, most probably from a domestic reservoir of captive animals. While the alpine newt in the UK is not invasive there is no justification for attempted eradication, which would be expensive and logistically difficult, as well as being ultimately futile unless the elusive supply line of primary introductions and subsequent release from captivity can be effectively controlled. Eradication might still be feasible on the neighbouring island of Ireland, where non-native alpine newts have been sighted, but where their distribution is not yet apparently widespread (Meehan, 2013). The eradication of a locally invasive introduced population of I. alpestris in New Zealand was successful because it was confined, early and intense (Arntzen et al., 2016; Sanson, 2017). Extant non-captive populations of I. alpestris may eventually become integrated into native biodiversity (Trigger et al., 2008; Clavero, 2014; Schlaepfer, 2018), but any complacency arising from the apparent lack of an invasive phenotype in long-established populations of I. alpestris in mainland Britain would be misplaced in face of continuing introductions. The potential threat posed by I. alpestris in the UK is ultimately driven by the global amphibian trade, which should be the priority target for control measures.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
The animal study was reviewed and approved by ZSL Ethics Committee for Animal Research.
All authors contributed to the study conception and design. Collection of the distribution data and selection of sampling sites were performed by RG, PH, NM, JW and TG. Geoprofiling analysis was performed by SF and MS. Material preparation, data collection and analysis were performed by MG, SA, BA, GO-G and SB. The first draft of the manuscript was written by SB and MG. All authors commented on previous versions of the manuscript, and read and approved the final manuscript.
This study was supported by core funding by Research England to the Institute of Zoology, Zoological Society of London. Additional funding was from postgraduate students’ project bench fees, in partnership with University College London, Imperial College London and Queen Mary University of London.
The geoprofiling study could not have been done without the knowledge, expertise, and enthusiasm of the late Dr Steven LeComber at QMUL who pioneered the use of this method in conservation. Mark Stevenson, Dr LeComber’s PhD student at QMUL, was responsible for the geoprofiling groundwork underlying the study in this paper. We thank colleagues in mainland Europe for samples from alpine newts in their native range: Sergé Bogaerts and Annamarieke Spitzen (Reptile, Amphibian and Fish Conservation Netherlands (RAVON), The Netherlands); Frank Pasmans (Ghent University, Belgium); Benedikt Schmidt (University of Zürich, Switzerland); Claude Miaud (University of Savoie, France). We are also grateful to Sarah King, who assisted with sampling in the Edinburgh study region. We would also like to thank reviewers for their insightful comments and valuable advice on improving this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author TG declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/famrs.2023.1215723/full#supplementary-material
Supplementary Datasheet 1 | Spatial dataset.
Supplementary Datasheet 2 | Geoprofiling additional information.
Supplementary Datasheet 3 | ABC additional information.
Supplementary Datasheet 4 | Genetic data: spreadsheet of genotypes.
Allain S. J., Lynn V. J. (2021). Distribution of the alpine newt Ichthyosaura alpestris in Great Britain updated using social media. Herpetol. Bull. 158, 28–31. doi: 10.33256/hb158.2831
Alves J. M., Carneiro M., Day J. P., Welch J. J., Duckworth J. A., Cox T. E., et al. (2022). A single introduction of wild rabbits triggered the biological invasion of Australia. Proc. Natl. Acad. Sci. U.S.A. 119, e2122734119. doi: 10.1073/pnas.2122734119
Amano T., Coverdale R., Peh K. S. H. (2016). The importance of globalisation in driving the introduction and establishment of alien species in Europe. Ecography 39, 1118–1128. doi: 10.1111/ecog.01893
Amphibian and Reptile Conservation UK (ARC). Report Your Sightings. Available at: https://www.arc-trust.org/Pages/Category/non-natives (Accessed 29 June 2023).
Anderson N. L., Paszkowski C. A., Hood G. A. (2015). Linking aquatic and terrestrial environments: can beaver canals serve as movement corridors for pond-breeding amphibians? Anim. Conserv. 18, 287–294. doi: 10.1111/acv.12170
Arntzen J. W., King T. M., Denoël M., Martínez-Solano I., Wallis G. P. (2016). Provenance of Ichthyosaura alpestris (Caudata: Salamandridae) introductions to France and New Zealand assessed by mitochondrial DNA analysis. Herpetol. J. 26, 49–56.
Beaumont M. A. (2008). “Joint determination of topology, divergence time, and immigration in population trees,” in Simulations, Genetics and Human Prehistory. Eds. Matsumura S., Forster P., Renfrew C (McDonald Institute Monographs, University of Cambridge), 135–154.
Beaumont M. A., Zhang W., Balding D. J. (2002). Approximate Bayesian computation in population genetics. Genetics 162, 2025–2035. doi: 10.1093/genetics/162.4.2025
Beebee T. J. C. (1985). Discriminant analysis of amphibian habitat determinants in south-east England. Amphib-reptil. 6, 35–43. doi: 10.1163/156853885X00164
Beebee T. (2014). Reptiles and amphibians of a village in Somerset, England. Herpetol. Bull. 128, 16–19.
Beerli P. (1998). “Estimation of migration rates and population sizes in geographically structured populations”, in Advances in Molecular Ecology; NATO-ASI Workshop Series. Ed Carvalho G. (Amsterdam: IOS Press) 306, 39–54.
Bell B., Bell A. P. (1995). 2nd World Congress of Herpetology: Distribution of the introduced alpine newt Triturus alpestris and of native Triturus species in north Shropshire, England. Aust. J. Ecol. 20, 367–375. doi: 10.1111/j.1442-9993.1995.tb00552.x
Beninde J., Keltsch F., Veith M., Hochkirch A., Wagner N. (2021). Connectivity of Alpine newt populations (Ichthyosaura alpestris) exacerbates the risk of Batrachochytrium salamandrivorans outbreaks in European fire salamanders (Salamandra salamandra). Conserv. Genet. 22, 653–659. doi: 10.1007/s10592-021-01377-8
Benson D. A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D. J., Ostell J., et al. (2013). GenBank. Nucleic Acids Res. 41, D36–D42. doi: 10.1093/nar/gks1195
Biggs J., Ewald N., Valentini A., Gaboriaud C., Dejean T., Griffiths R. A., et al. (2015). Using eDNA to develop a national citizen science-based monitoring programme for the great crested newt (Triturus cristatus). Biol. Conserv. 183, 19–28. doi: 10.1016/j.biocon.2014.11.029
Blackburn T. M., Lockwood J. L., Cassey P. (2015). The influence of numbers on invasion success. Mol. Ecol. 24, 1942–1953. doi: 10.1111/mec.13075
Blackburn T. M., Pyšek P., Bacher S., Carlton J. T., Duncan R. P., Jarošik V., et al. (2011). A proposed unified framework for biological invasions. Trends Ecol. Evol. 26, 333–339. doi: 10.1016/j.tree.2011.03.023
Bosch J., Martínez-Solano I. (2003). Factors influencing occupancy of breeding ponds in a montane amphibian assemblage. J. Herpetol. 37, 410–413. doi: 10.1670/0022-1511(2003)037[0410:FIOOBP]2.0.CO;2
Brown G. P., Phillips B. L., Webb J. K., Shine R. (2006). Toad on the road: use of roads as dispersal corridors by cane toads (Bufo marinus) at an invasion front in tropical Australia. Biol. Conserv. 133, 88–94. doi: 10.1016/j.biocon.2006.05.020
Bulut Z., McCormick C. R., Gopurenko D., Williams R. N., Bos D. H., Dewoody J. A. (2009). Microsatellite mutation rates in the eastern tiger salamander (Ambystoma tigrinum tigrinum) differ 10-fold across loci. Genetica 136, 501–504. doi: 10.1007/s10709-008-9341-z
Capinha C., Seebens H., Cassey P., García-Díaz P., Lenzner B., Mang T., et al. (2017). Diversity, biogeography and the global flows of alien amphibians and reptiles. Divers. Distrib. 23, 1313–1322. doi: 10.1111/ddi.12617
Carpenter A. I., Andreone F., Moore R. D., Griffiths R. A. (2014). A review of the international trade in amphibians: the types, levels and dynamics of trade in CITES listed species. Oryx 48, 565–574. doi: 10.1017/S0030605312001627
Carrete M., Tella J. (2008). Wild-bird trade and exotic invasions: a new link of conservation concern? Front. Ecol. Environ. 6, 207–211. doi: 10.1890/070075
Cayuela H., Rougemont Q., Prunier J. G., Moore J. S., Clober J., Besnard A., et al. (2018). Demographic and genetic approaches to study dispersal in wild animal populations: A methodological review. Mol. Ecol. 27, 3976–4010. doi: 10.1111/mec.14848
Cayuela H., Valenzuela-Sánchez A., Teulier L., Martínez-Solano Í., Léna J. P., Merilä J., et al. (2020). Determinants and consequences of dispersal in vertebrates with complex life cycles: a review of pond-breeding amphibians. Q. Rev. Biol. 95, 1–36. doi: 10.1086/707862
CBD Secretariat. (2002) Decision VI/23: Alien species that threaten ecosystems, habitats and species. Document UNEP/CBD/COP/6/23 (Montreal, Canada: Secretariat of the Convention on Biological Diversity Secretariat).
CBD Secretariat (2010). Pets, Aquarium, and Terrarium Species: Best Practices for Addressing Risks to Biodiversity (Montreal, Canada: Secretariat of the Convention on Biological Diversity).
Chapple D. G., Simmonds S. M., Wong B. B. (2012). Can behavioral and personality traits influence the success of unintentional species introductions? Trends Ecol. Evol. 27, 57–64.
Cini A., Anfora G., Escudero-Colomar L. A., Grassi A., Santosuosso U., Seljak G., et al. (2014). Tracking the invasion of the alien fruit pest Drosophila suzukii in Europe. J. Pest Sci. 87, 559–566. doi: 10.1007/s10340-014-0617-z
Clavero M. (2014). Shifting baselines and the conservation of non-native species. Conserv. Biol. 28, 1434–1436. doi: 10.1111/cobi.12266
Copp G. H., Wesley K. J., Vilizzi L. (2005). Pathways of ornamental and aquarium fish introductions into urban ponds of Epping Forest (London, England): the human vector. J. Appl. Ichthyol. 21, 263–274. doi: 10.1111/j.1439-0426.2005.00673.x
Cornuet J. M., Pudlo P., Veyssier J., Dehne-Garcia A., Gautier M., Leblois R., et al. (2014). DIYABC v2. 0: A software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 30, 1187–1189. doi: 10.1093/bioinformatics/btt763
Cornuet J. M., Ravigné V., Estoup A. (2010). Inference on population history and model checking using DNA sequence and microsatellite data with the software DIYABC (v1. 0). BMC Bioinf. 11, 401. doi: 10.1186/1471-2105-11-401
Cornuet J. M., Santos F., Beaumont M. A., Robert C. P., Marin J. M., Balding D. J., et al. (2008). Inferring population history with DIY ABC: A user- friendly approach to approximate Bayesian computation. Bioinformatics 24, 2713–2719. doi: 10.1093/bioinformatics/btn514
Covaciu-Marcov S. D., Cicort-Lucaciu A. S., Mitrea I., Sas I., Caus A. V., Cupsa D. (2010). Feeding of three syntopic newt species (Triturus cristatus, Mesotriton alpestris and Lissotriton vulgaris) from Western ROmania. North Western. J. Zool. 6, 95–108.
Crispo E., Moore J. S., Lee-Yaw J. A., Gray S. M., Haller B. C. (2011). Broken barriers: Human-induced changes to gene flow and introgression in animals: An examination of the ways in which humans increase genetic exchange among populations and species and the consequences for biodiversity. BioEssays 33, 508–518. doi: 10.1002/bies.201000154
Crooks J. A. (2005). Lag times and exotic species: The ecology and management of biological invasions in slow-motion. Ecoscience 12, 316–329. doi: 10.2980/i1195-6860-12-3-316.1
Cunningham A. A., Smith F., McKinley T. J., Perkins M. W., Fitzpatrick L. D., Wright O. N., et al. (2019). Apparent absence of Batrachochytrium salamandrivorans in wild urodeles in the United Kingdom. Sci. Rep. 9, 2831. doi: 10.1038/s41598-019-39338-4
Daly E. Z., Chabrerie O., Massol F., Facon B., Hess M. C., Tasiemski ,. A., et al. (2023). A synthesis of biological invasion hypotheses associated with the introduction–naturalisation–invasion continuum. Oikos 2023, 5. doi: 10.1111/oik.09645
Denoël M. (2005). Persistance et dispersion d’une population introduite de triton alpestre (Triturus alpestris) dans les causses du Larzac (sud de la France). Rev. d’Ecol. (La Terre La Vie). 60, 139–148. doi: 10.3406/revec.2005.1252
Denoël M., Dalleur S., Langrand E., Besnard A., Cayuela H. (2018). Dispersal and alternative breeding site fidelity strategies in an amphibian. Ecography 41, 1543–1555. doi: 10.1111/ecog.03296
Di Rienzo A., Peterson A. C., Garza J. C., Valdes A. M., Slatkin M., Freimer N. B. (1994). Mutational processes of simple sequence repeat loci in human populations. Proc. Natl. Acad. Sci. U. S. A. 91, 3166–3170. doi: 10.1073/pnas.91.8.3166
Dufresnes C., Pellet J., Bettinelli-Riccardi S., Thiébaud J., Perrin N., Fumagalli L. (2016). Massive genetic introgression in threatened northern crested newts (Triturus cristatus) by an invasive congener (T. carnifex) in Western Switzerland. Conserv. Genet. 17, 839–846. doi: 10.1007/s10592-016-0825-6
Earl D. A., von Holdt B. M. (2012). STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Emaresi G., Pellet J., Dubey S., Hirzel A. H., Fumagalli L. (2011). Landscape genetics of the Alpine newt (Mesotriton alpestris) inferred from a strip-based approach. Conserv. Genet. 12, 41–50. doi: 10.1007/s10592-009-9985-y
Estoup A., Jarne P., Cornuet J. M. (2002). Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Mol. Ecol. 11, 1591–1604. doi: 10.1046/j.1365-294X.2002.01576.x
Evanno G., Regnaut S., Goudet J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Ewart K. M., Griffin A. S., Johnson R. N., Kark S., Magory Cohen T., Lo N., et al. (2019). Two speed invasion: assisted and intrinsic dispersal of common mynas over 150 years of colonization. J. Biogeogr. 46, 45–57. doi: 10.1111/jbi.13473
Fagundes N. J. R., Ray N., Beaumont M., Neuenschwander S., Salzano F. M., Bonatto S. L., et al. (2007). Statistical evaluation of alternative models of human evolution. Proc. Natl. Acad. Sci. U. S. A. 104, 17614–17619. doi: 10.1073/pnas.0708280104
Fasola M., Canova L. (1992). Feeding habits of Triturus vulgaris, T. cristatus and T. alpestris (Amphibia, Urodela) in the northern Apennines (Italy). Ital. J. Zool. 59, 273–280. doi: 10.1080/11250009209386682
Faubet P., Waples R. S., Gaggiotti O. E. (2007). Evaluating the performance of a multilocus Bayesian method for the estimation of migration rates. Mol. Ecol. 16, 1149–1166. doi: 10.1111/j.1365-294X.2007.03218.x
Faulkner S. C., Verity R., Roberts D., Roy S. S., Robertson P. A., Stevenson M. D., et al. (2016). Using geographic profiling to compare the value of sightings vs trap data in a biological invasion. Divers. Distrib. 23, 104–112. doi: 10.1111/ddi.12498
Ficetola G. F., Coïc C., Detaint M., Berroneau M., Lorvelec O., Miaud C. (2007). Pattern of distribution of the American bullfrog Rana catesbeiana in Europe. Biol. Invasions. 9, 767–772. doi: 10.1007/s10530-006-9080-y
Fisher M. C., Garner T. W. J. (2007). The relationship between the emergence of Batrachochytrium dendrobatidis, the international trade in amphibians and introduced amphibian species. Fungal Biol. Rev. 21, 2–9. doi: 10.1016/j.fbr.2007.02.002
Fitzpatrick L. D., Pasmans F., Martel A., Cunningham A. A. (2018). Epidemiological tracing of Batrachochytrium salamandrivorans identifies widespread infection and associated mortalities in private amphibian collections. Sci. Rep. 8, 1–10. doi: 10.1038/s41598-018-31800-z
Francis R. M. (2017). pophelper: An r package and web app to analyse and visualize population structure. Mol. Ecol. Resour. 17, 27–32. doi: 10.1111/1755-0998.12509
Garner T. W. J., Schmidt B. R., Hoec k P., Van Buskirk J. (2003). Di- and tetranucleotide microsatellite markers for the Alpine newt (Triturus alpestris): Characterization and cross-priming in five congeners. Mol. Ecol. Notes 3, 186–188. doi: 10.1046/j.1471-8286.2003.00394.x
Gelman A., Carlin J. B., Stern H. S., Rubin D. B. (2004). Bayesian data analysis, 2nd edition (Boca Raton, FL: Chapman and Hall/CRC).
Goudet J. (2001). FSTAT, version 2.9. 3. A program to estimate and test gene diversities and fixation indices (Lausanne, Switzerland: Lausanne University). Available at: http://www.unil.ch/izea/softwares/fstat.html.
Gurevitch J., Padilla D. K. (2004). Are invasive species a major cause of extinctions? Trends Ecol. Evol. 19, 470–474. doi: 10.1016/j.tree.2004.07.005
Hamer A. J., McDonnel l. (2008). Amphibian ecology and conservation in the urbanising world: A review. Biol. Conserv. 141, 2432–2449. doi: 10.1016/j.biocon.2008.07.020
Harper L. R., Lawson Handley L., Hahn C., Boonham N., Rees H. C., Gough K. C., et al. (2018). Needle in a haystack? A comparison of eDNA metabarcoding and targeted qPCR for detection of the great crested newt (Triturus cristatus). Ecol. Evol. 8, 6330–6341. doi: 10.1002/ece3.4013
Harrower C. A., Rorke S. L., Roy H. E. (2020). Joint Nature Conservation Committee B6. Pressure from invasive species. Technical document September 2020. Available at: https://jncc.gov.uk/ukbi-B6.
Heald O. J. N., Fraticelli C., Cox S. E., Stevens M. C. A., Faulkner S. C., Blackburn T. M., et al. (2020). Understanding the origins of the ring-necked parakeet in the UK. J. Zool. 312, 1–11. doi: 10.1111/jzo.12753
Holsbeek G., Mergeay J., Hotz H., Plötner J., Volckaert F. A. M., De Meester L. (2008). A cryptic invasion within an invasion and widespread introgression in the European water frog complex: consequences of uncontrolled commercial trade and weak international legislation. Mol. Ecol. 17, 5023–5035. doi: 10.1111/j.1365-294X.2008.03984.x
Hubisz M. J., Falush D., Stephens M., Pritchard J. K. (2009). Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 9, 1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x
Hulm P. E. (2009). Trade, transport and trouble: managing invasive species pathways in an era of globalization. J. Appl. Ecol. 46, 10–18. doi: 10.1111/j.1365-2664.2008.01600.x
Hulme P. E. (2021). Unwelcome exchange: International trade as a direct and indirect driver of biological invasions worldwide. One Earth 4, 666–679. doi: 10.1016/j.oneear.2021.04.015
Humphreys E., Toms M., Newson S., Baker J., Wormald K. (2011). “An examination of reptile and amphibian populations in gardens, the factors influencing garden use and the role of a “Citizen science” approach for monitoring their populations within this habitat,” in British Trust for Ornithology Research Report, 572. Available at: https://www.bto.org/sites/default/files/shared_documents/publications/research-reports/2010/rr572.pdf.
IUCN Guidelines. (2000). Guidelines for the prevention of biodiversity loss caused by alien invasive species. Approved by IUCN Council Feb 2000. Available at: https://portals.iucn.org/library/efiles/documents/Rep-2000-052.pdf.
Jehle R., Burke T., Arntzen J. W. (2005). Delineating fine-scale genetic units in amphibians: Probing the primacy of ponds. Conserv Genet 6, 227–234. doi: 10.1007/s10592-004-7832-8
Joly P., Miaud C. (1989). Fidelity to the breeding site in the alpine newt Triturus alpestris. Behav. Processes 19, 47–56.
Jombart T. (2008). adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405. doi: 10.1093/bioinformatics/btn129
Jombart T., Devillard S., Balloux F. (2010). Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 11, 1–15. doi: 10.1186/1471-2156-11-94
Jones O. R., Wang J. (2010). COLONY: A program for parentage and sibship inference from multilocus genotype data. Mol. Ecol. Resour. 10, 551–555. doi: 10.1111/j.1755-0998.2009.02787.x
Kimura M. (1968). Evolutionary rate at the molecular level. Nature 217, 624–626. doi: 10.1038/217624a0
Kimura M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Kocher T. D., Thomas W. K., Meyer A., Edwards S. V., Paabo S., Villablance F. X., et al. (1989). Dynamics of mitochondrial DNA evolution in animals: Amplification and sequencing with conserved primers. Proc. Natl. Acad. Sci. U. S. A. 86, 6196–6200. doi: 10.1073/pnas.86.16.6196
Kraus F. (2015). Impacts from invasive reptiles and amphibians. Annu. Rev. Ecol. Evol. Syst. 46, 75–97. doi: 10.1146/annurev-ecolsys-112414-054450
Krysko K. L., Burgess J. P., Rochford M. R., Gillette C. R., Cueva D., Enge K. M., et al. (2011). Verified non-indigenous amphibians and reptiles in Florida from 1863 through 2010: outlining the invasion process and identifying invasion pathways and stages. Zootaxa 3028, 1–64. doi: 10.11646/zootaxa.3028.1.1
Lande R. (1982). A quantitative genetic theory of life history evolution. Ecology 63, 607–615. doi: 10.2307/1936778
Landguth E. L., Cushman S. A., Schwartz M. K., McKelvey K. S., Murphy M., Luikart G. (2010). Quantifying the lag time to detect barriers in landscape genetics. Mol. Ecol. 19, 4179–4191. doi: 10.1111/j.1365-294X.2010.04808.x
Le Comber S. C., Stevenson M. D. (2012). From Jack the Ripper to epidemiology and ecology. Trends Ecol. Evol. 27, 307–308. doi: 10.1016/j.tree.2012.03.004
Leung B., Lodge D. M., Finnoff D., Shogren J. F., Lewis M. A., Lamberti G. (2002). An ounce of prevention or a pound of cure: bioeconomic risk analysis of invasive species. Proc. R. Soc. London. Ser. B: Biol. Sci. 269, 2407–2413. doi: 10.1098/rspb.2002.2179
Lever C. (2001). The cane toad: the history and ecology of a successful colonist (Otley: Westbury Academic and Scientific Pub).
Lope M. J., Cuadrado J. A. (1985). Nota sobre la presencia del tritoín alpino (Triturus alpestris) en el centro de la Peniínsula Ibeírica. Donãna. Acta Vertebrata. 12, 317–318.
Mack R. N., Simberloff D., Lonsdale W. M., Evans H., Clout M., Bazzaz F. A. (2000). Biotic invasions: causes, epidemiology, global consequences, and control. Ecol. Appl. 10, 689–710. doi: 10.1890/1051-0761(2000)010[0689:BICEGC]2.0.CO;2
Martel A., Blooi M., Adriaensen C., Van Rooij P., Beukema W., Fisher M. C., et al. (2014). Recent introduction of a chytrid fungus endangers Western Palearctic salamanders. Science 346, 630–631. doi: 10.1126/science.1258268
Meehan S. T. (2013). IWT National Smooth Newt Survey 2013 report (Dublin, Ireland). Available at: https://iwt.ie/wp-content/uploads/2017/09/Newt-Survey-2013.pdf.
Meirmans P. G., Hedrick P. W. (2011). Assessing population structure: FST and related measures. Mol. Ecol. Resour. 11, 5–18. doi: 10.1111/j.1755-0998.2010.02927.x
Miaud C., Guyetant R., Faber H. (2000). Age, size, and growth of the alpine newt, Triturus alpestris (Urodela: Salamandridae), at high altitude and a review of life-history trait variation throughout its range. Herpetologica 56, 135–144. Available at: http://www.jstor.org/stable/3893265.
Miller C., Kettunen M., Shine C. (2006). “Scope options for EU action on invasive alien species (IAS),” in Final report for the European Commission (Brussels, Belgium: Institute for European Environmental Policy (IEEP), (Brussels, Belgium).
Mohanty N. P., Measey J. (2019). The global pet trade in amphibians: species traits, taxonomic bias, and future directions. Biodiversi. Conserv. 28, 3915–3923. doi: 10.1007/s10531-019-01857-x
Moorhouse T. P., Balaskas M., D'Cruze N. C., Macdonald D. W. (2017). Information could reduce consumer demand for exotic pets. Conserv. Lett. 10, 337–345. doi: 10.1111/conl.12270
Murphy M. A., Dezzani R., Pilliod D. S., Storfer A. (2010). Landscape genetics of high mountain frog metapopulations. Mol. Ecol. 19, 3634–3649. doi: 10.1111/j.1365-294X.2010.04723.x
Murphy M. A., Evans J. S., Cushman S. A., Storfer A. (2008). Representing genetic variation as continuous surfaces: an approach for identifying spatial dependency in landscape genetic studies. Ecography 31, 685–697. doi: 10.1111/j.1600-0587.2008.05428.x
Noël S., Ouellet M., Galois P., Lapointe F. J. (2007). Impact of urban fragmentation on the genetic structure of the eastern red-backed salamander. Conserv. Genet. 8, 599–606. doi: 10.1007/s10592-006-9202-1
Old J. M., Spencer R. J., Wolfenden J. (2014). The common myna (Sturnus tristis) in urban, rural and semi-rural areas in Greater Sydney and its surrounds. Emu 114, 241–248. doi: 10.1071/MU13029
Orsini L., Vanoverbeke J., Swillen I., Mergeay J., De Meester L. (2013). Drivers of population genetic differentiation in the wild: isolation by dispersal limitation, isolation by adaptation and isolation by colonization. Mol. Ecol. 22, 5983–5999. doi: 10.1111/mec.12561
Palomar G., Vörö s J., Bosch J. (2017). Tracking the introduction history of Ichthyosaura alpestris in a protected area of Central Spain. Conserv. Genet. 18, 867–876. doi: 10.1007/s10592-017-0934-x
Palumbi S., Martin A., ROmano S., McMillan W. O., Stice L., Grabowski G. (1991). The simple fool’s guide to PCR, version 2.0 (Honolulu: University of Hawaii).
Papini A., Mosti S., Santosuosso U. (2013). Tracking the origin of the invading Caulerpa (Caulerpales, Chlorophyta) with Geographic Profiling, a criminological technique for a killer alga. Biol. Invasions. 15, 1613–1621. doi: 10.1007/s10530-012-0396-5
Papini A., Signorin i., Foggi B., Della Giovampaola E., Ongaro L., Vivona L., et al. (2017). History vs. legend: Retracing invasion and spread of Oxalis pes-caprae L. in Europe and the Mediterranean area. PloS One 12, e0190237. doi: 10.1371/journal.pone.0190237
Paradis E. (2010). Pegas: An R package for population genetics with an integrated-modular approach. Bioinformatics 26, 419–420. doi: 10.1093/bioinformatics/btp696
Pearse D. E., Crandall K. A. (2004). Beyond FST: analysis of population genetic data for conservation. Conserv. Genet. 5, 585–602. doi: 10.1007/s10592-003-1863-4
Pittman S. E., Osbourn M. S., Semlitsch R. D. (2014). Movement ecology of amphibians: A missing component for understanding population declines. Biol. Conserv. 169, 44–53. doi: 10.1016/j.biocon.2013.10.020
Pritchard J. K., Stephens M., Donnelly P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1111/j.1471-8286.2007.01758.x
Prunier J. G., Kaufmann B., Fenet S., Picard D., Pompanon F., Joly P., et al. (2013). Optimizing the trade-off between spatial and genetic sampling efforts in patchy populations: Towards a better assessment of functional connectivity using an individual-based sampling scheme. Mol. Ecol. 22, 5516–5530. doi: 10.1111/mec.12499
Prunier J., Kaufmann B., Grolet O., Picard D., Pompanon F., Joly P. (2012). Skin swabbing as a new efficient DNA sampling technique in amphibians, and 14 new microsatellite markers in the alpine newt (Ichthyosaura alpestris). Mol. Ecol. Resour. 12, 524–531. doi: 10.1111/j.1755–0998.2012.03116.x
Pyšek P., Hulme P. E., Bacher S., Blackburn T. M., Carlton J. T., Dawson W., et al. (2020). Scientists' warning on invasive alien species. Biol. Rev. 95, 1511–1534. doi: 10.1111/brv.12627
QGIS.org (2021). QGIS Geographic Information System (QGIS Association). Available at: http://www.qgis.org.
Rane R., Walsh T. K., Lenancker P., Gock A., Dao T. H., Nguyen V. L., et al. (2023). Complex multiple introductions drive fall armyworm invasions into Asia and Australia. Sci. Rep. 13, 1–18. doi: 10.1038/s41598-023-27501-x
R Core Team. (2014). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing). Available at: https://www.R-project.org/.
Rees H. C., Bishop K., Middleditch D. J., Patmore J. R. M., Maddison B. C., Gough K. C. (2014). The application of eDNA for monitoring of the Great Crested Newt in the UK. Ecol. Evol. 4, 4023–4032. doi: 10.1002/ece3.1272
Renault D., Manfrini E., Leroy B., Diagne C., Ballesteros-Mejia L., Angulo E., et al. (2021). Biological invasions in France: Alarming costs and even more alarming knowledge gaps. NeoBiota 67, 191–224. doi: 10.3897/neobiota.67.59134
Ribeiro R., Carretero M. A., Sillero N., Alarcos G., Ortiz-Santaliestra M., Lizana M., et al. (2011). The pond network: can structural connectivity reflect on (amphibian) biodiversity patterns? Landscape Ecol. 26, 673–682. doi: 10.1007/s10980-011-9592-4
Ricciardi A. (2007). Are modern biological invasions an unprecedented form of global change? Conserv. Biol. 21, 329–336. doi: 10.1111/j.1523-1739.2006.00615.x
Roy H. E., Peyton J., Aldridge D. C., Bantock T., Blackburn T. M., Britton R., et al. (2014). Horizon scanning for invasive alien species with the potential to threaten biodiversity in Great Britain. Global Change Biol. 20, 3859–3871. doi: 10.1111/gcb.12603
Sanson L. (2017). Update on European Alpine Newt Incursion Management Programme (New Zealand: Department of Conservation). Available at: https://blog.doc.govt.nz/2017/04/28/d-g-direct-april/ (Accessed 10 October 2019).
Schlaepfer M. A. (2018). Do non-native species contribute to biodiversity? PloS Biol. 16, e2005568. doi: 10.1371/journal.pbio.2005568
Schloegel L. M., Picco A. M., Kilpatrick A. M., Davies A. J., Hyatt A. D., Daszak P. (2009). Magnitude of the US trade in amphibians and presence of Batrachochytrium dendrobatidis and ranavirus infection in imported North American bullfrogs (Rana catesbeiana). Biol. Conserv. 142, 1420–1426. doi: 10.1016/j.biocon.2009.02.007
Schmidt P., Weddeling K., Thomas M., Rottscheidt R., Tarkhnishvili D. N., Hachtel M. (2006). Dispersal of Triturus alpestris and T. vulgaris in agricultural landscapes – comparing estimates from allozyme markers and capture-mark-recapture analysis. Proc. 13th. Congress. Societas. Europaea. Herpetol. 143, 139–143. doi: 10.1016/B978-012369442-3/50182-3
Scottish Golf Environment Group. Nature: Pond Management Mortonhall Golf Club. Available at: https://www.sgeg.org.uk/documents/Publications (Accessed 12 February 2019).
Seabrook W. A., Dettmann E. B. (1996). Roads as activity corridors for cane toads in Australia. J. Wildlife. Manage. 60, 363–368. doi: 10.2307/3802236
Seebens H., Blackburn T. M., Dyer E. E., Genovesi P., Hulme P. E., Jeschke J. M., et al. (2017). No saturation in the accumulation of alien species worldwide. Nat. Commun. 8, 14435. doi: 10.1038/ncomms14435
Sillero N., Campos J., Bonardi A., Corti C., Creemers R., Crochet P.-A., et al. (2014). Updated distribution and biogeography of amphibians and reptiles of Europe. Amphibia-Reptilia 35, 1–31. doi: 10.1163/15685381-00002935
Simberloff D. (2003). How much information on population biology is needed to manage introduced species? Conserv. Biol. 17, 83–92. doi: 10.1046/j.1523-1739.2003.02028.x
Sinsch U. L., Lang V. E., Wiemer R. A., Wirtz S. (2003). Dynamik einer Kammmolch-Metapopulation (Triturus cristatus) auf militärischem Übungsgelände (Schmittenhöhe, Koblenz): 2. Saisonale Variation der Bestände in zwei Laichgewässern. Z. für. Feldherpetol. 10, 211–227.
Sol D., Bartomeus I., Griffin A. S. (2012). The paradox of invasion in birds: Competitive superiority or ecological opportunism? Oecologia 169, 553–564. doi: 10.1007/s00442-011-2203-x
Sol D., Maspons J. (2016). “Life history, behaviour and invasion success,” in Biological Invasions and Animal Behaviour. Eds. Weir J. S., Sol D. (Cambridge, UK: Cambridge University Press), 63–81.
Sotiropoulos K., Eleftherakos K., Džukić G., Kalezić M. L., Legakis A., Polymeni R. M. (2007). Phylogeny and biogeography of the alpine newt Mesotriton alpestris (Salamandridae, Caudata), inferred from mtDNA sequences. Mol. Phylogenet. Evol. 45, 211–226. doi: 10.1016/j.ympev.2007.03.012
Speybroeck J., Beukema W., Bok B., van der Voort J. (2016). Field guide to the amphibians and reptiles of Britain and Europe. (London: Bloomsbury Publishing).
Speybroeck J., Beukema W., Dufresnes C., Fritz U., Jablonski D., Lymberakis P., et al. (2020). Species list of the European herpetofauna – 2020 update by the Taxonomic Committee of the Societas Europaea Herpetologica. Amphibia-Reptilia 41, 139–189. doi: 10.1163/15685381-bja10010
Stringham O. C., Lockwood J. L. (2018). Pet problems: biological and economic factors that influence the release of alien reptiles and amphibians by pet owners. J. Appl. Ecol. 55, 2632–2640. doi: 10.1111/1365-2664.13237
Sundqvist L., Keenan K., Zackrisson M., Prodöhl P., Kleinhans D. (2016). Directional genetic differentiation and relative migration. Ecol. Evol. 6, 3461–3475. doi: 10.1002/ece3.2096
Tamukai K., Une Y., Tominaga A., Suzuki K., Goka K. (2014). Batrachochytrium dendrobatidis prevalence and haplotypes in domestic and imported pet amphibians in Japan. Dis. Aquat. Organisms. 109, 165–175. doi: 10.3354/dao02732
Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tapley B., Griffiths R. A., Bride I. (2011). Dynamics of the trade in reptiles and amphibians within the United Kingdom over a ten-year period. Herpetol. J. 21, 27–34.
Tingley R., Weeks A. R., Smart A. S., van Rooyen A. R., Woolnough A. P., McCarthy M. A. (2015). European newts establish in Australia, marking the arrival of a new amphibian order. Biol Invasions 17, 31–37. doi: 10.1007/s10530-014-0716-z
Trigger D., Mulcock J., Gaynor A., Toussaint Y. (2008). Ecological restoration, cultural preferences and the negotiation of ‘nativeness’ in Australia. Geoforum 39, 1273–1283. doi: 10.1016/j.geoforum.2007.05.010
Valéry L., Fritz H., Lefeuvre J. C., Simberloff D. (2008). In search of a real definition of the biological invasion phenomenon itself. Biol. Invasions. 10, 1345–1351. doi: 10.1007/s10530-007-9209-7
Van Oosterhout C., Hutchinson W. F., Wills D. P. M., Shipley P. (2004). Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 4, 535–538. doi: 10.1111/j.1471-8286.2004.00684.x
Verity R., Stevenson M. D., Rossmo D. K., Nichols R. A., Le Comber S. C. (2014). Spatial targeting of infectious disease control: Identifying multiple, unknown sources. Methods Ecol. Evol. 5, 647–655. doi: 10.1111/2041-210X.12190
Wakeley J. (2003). “Inferences about the structure and history of populations: coalescents and intraspecific phylogeography,” in The Evolution of Population Biology. Eds. Singh R., Uyenoyama M. (Cambridge, UK: Cambridge University Press), 193–215.
Wang J. (2009). A new method for estimating effective population sizes from a single sample of multilocus genotypes. Mol. Ecol. 18, 2148–2164. doi: 10.1111/j.1365-294X.2009.04175.x
Wang J. (2017). The computer program structure for assigning individuals to populations: easy to use but easier to misuse. Mol. Ecol. Resour. 17, 981–990. doi: 10.1111/1755-0998.12650
Whitlock M. C., McCauley D. E. (1999). Indirect measures of gene flow and migration: FST≠ 1/(4Nm+ 1). Heredity 82, 117–125. doi: 10.1038/sj.hdy.6884960
Wilcove D. S., Rothstein D., Dubow J., Phillips A., Losos E. (1998). Quantifying threats to imperiled species in the United States. BioScience 48, 607–615. doi: 10.2307/1313420
Wilkinson J. (2016) Alpine Newt, Mesotriton alpestris GB Non-Native Species Secretariat Information Portal nonnativespecies org/factsheet species Id 2215. (Accessed 22 December 2020).
Williamson M. H., Fitter A. (1996). The characters of successful invaders. Biol. Conserv. 78, 163–170. doi: 10.1016/0006-3207(96)00025-0
Wilson J. R., Dormontt E. E., Prentis P. J., Lowe A. J., Richardson D. M. (2009). Something in the way you move: dispersal pathways affect invasion success. Trends Ecol. Evol. 24, 136–144. doi: 10.1016/j.tree.2008.10.007
Wilson G. A., Rannala B. (2003). Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163, 1177–1191. doi: 10.1093/genetics/163.3.1177
Winchester D. (2015). Risk assessment summary sheet for the Alpine Newt (Ichthyosaura alpestris). Available at: http://www.nonnativespecies.org/inde (Accessed June 2020).
Keywords: invasive alien species, ecological genetics, invasion routes, geoprofiling, dispersal, global amphibian trade
Citation: Ball SE, Gupta M, Aldridge SJ, Allen BE, Faulkner SC, Oteo-García G, Griffiths RA, Hill P, Morris NJ, Stevens MCA, Wilkinson J and Garner TWJ (2023) Multiple introductions and human-aided dispersal of the UK’s most widespread non-native amphibian. Front. Amphib. Reptile Sci. 1:1215723. doi: 10.3389/famrs.2023.1215723
Received: 02 May 2023; Accepted: 24 July 2023;
Published: 05 September 2023.
Edited by:
Cynthia A. Paszkowski, University of Alberta, CanadaReviewed by:
David Lesbarreres, Environment and Climate Change Canada (ECCC), CanadaCopyright © 2023 Ball, Gupta, Aldridge, Allen, Faulkner, Oteo-García, Griffiths, Hill, Morris, Stevens, Wilkinson and Garner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sarah E. Ball, U2FyYWguQmFsbEBpb3ouYWMudWs=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.