
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Agron., 02 April 2025
Sec. Disease Management
Volume 7 - 2025 | https://doi.org/10.3389/fagro.2025.1569408
Introduction: Flavescence dorée (FD) is a European quarantine disease of grapevine with high economic impact. The Flavescence dorée phytoplasmas (FDp) are epidemically spread by the leafhopper Scaphoideus titanus, which has been introduced to Europe from North America. The FDp is of European origin and is widely spread in symptomless alder trees. FD outbreaks were related to three different genetic variants of FDp: MAP-FD1, MAP-FD2 and MAP-FD3. A multitude of non-epidemic genotypes exist in mixed infections in alder trees. Germany is so far free of FD but the possibility of the transmission of MAP-FD2 phytoplasmas from alder to grapevine by the invasive secondary vector Orientus ishidae has been demonstrated experimentally and under natural conditions. Thus, FDp-infected alder represent a risk for FD outbreaks.
Methods: Geodata-based analyses were employed to identify potential alder sites adjacent to vineyards in the major wine-growing regions of Germany in Rhineland-Palatinate and Baden-Wuerttemberg. Webmaps for Plant Protection Services were established. In addition, molecular screening tools for the detection of MAP-FD1 and MAP-FD2 phytoplasmas in mixed infections in alders were developed.
Results and Discussion: 1089 alder samples from 171 sites in Rhineland-Palatinate, 556 alder samples from 104 sites in Baden-Wuerttemberg and 141 samples from 22 sites in Franconia were examined. Almost all samples (98%) were infected with 16SrV phytoplasmas. The distribution of MAP-FD2 phytoplasmas varied greatly among the different regions: no infections were found in the Mosel-Saar-Ruwer region while along the Rhine valley from Rheinhessen to Baden infection rates around 70% were observed. Northern and Southern regions of Baden-Wuerttemberg as well as Franconia had lower infection rates ranging from 15% to 40%. Insect captures in the different regions showed that O. ishidae is present in all regions but infection rates varied indicating that this invasive species is responsible for the spread of MAP-FD2 phytoplasma in alder in Germany. Only few infections with MAP-FD1 genotypes were found. Specific primers for the epidemic map genotype M54 revealed no infection in the alder trees.
Flavescence dorée (FD) is a quarantine disease of grapevine that leads to important economic losses in several southern European countries (Chuche and Thiéry, 2014; Tramontini et al., 2020). It is associated with the Flavescence dorée phytoplasma (FDp), a member of the elm yellows 16SrV group. FD phytoplasmas belong to the two different taxonomic subgroups 16SrV-C and 16SrV-D (Angelini et al., 2001). FD is epidemically transmitted from vine to vine by the leafhopper Scaphoideus titanus, which has been introduced in the 1950ies from North America to Europe (reviewed by Gonella et al., 2024). Only recently, it became evident that the phytoplasma is of European origin and is widespread in alders in Western Europe and in Clematis vitalba in Eastern Europe (Angelini et al., 2004; Malembic-Maher et al., 2020). The FD phytoplasma can be transmitted occasionally from alder to grapevine by the invasive leafhopper Orientus ishidae (Jarausch et al., 2023). This scenario was described for the first time in Germany in 2020 (Jarausch et al., 2021), which so far has been regarded as FD-free. The infected grapevine was promptly uprooted and no FD outbreak occurred since S. titanus was not present in Germany at this time. This situation, however, changed in 2024 when larger populations of S. titanus were discovered in South-western Germany (Askani et al., 2024; Jarausch et al., 2025a).
FD phytoplasmas can be further divided in different clusters based on the epidemiological informative housekeeping gene map (Arnaud et al., 2007; Malembic-Maher et al., 2020). Up to now, 173 different map genotypes have been distinguished (Malembic-Maher et al., 2020; Krstić et al., 2022; Marcone et al., 2024; Jarausch et al., 2025b). Clusters MAP-FD1, MAP-FD2 and MAP-FD3 are related to different FD outbreaks in different geographic regions. MAP-FD1 and FD2 genotypes are present in France and Switzerland while MAP-FD3 genotypes are reported from Southeastern European countries (Malembic-Maher et al., 2020). All three clusters occur in the countries in between, e.g. Italy, Hungary, Croatia (Malembic-Maher et al., 2020; Plavec et al., 2019). The dominant map genotypes in the different clusters are M50 (FD1), M38 and M54 (FD2) and M51 (FD3) (Malembic-Maher et al., 2020). M50, M51 and M54 are widespread in FD epidemics while M38 is also often reported from reservoir plants like alder or alternative insect vectors (Malembic-Maher et al., 2020). Several other map genotypes related to these three clusters differ only by one or two nucleotides from the reference sequence. Most of them have been reported only once or in low number (Malembic-Maher et al., 2020; Krstić et al., 2022). Map genotyping has been applied as a tool to study alternative epidemiological cycles of FD including reservoir plants and alternative insect vectors. In Switzerland and Northern Italy, M50 was dominant or widespread in reservoir plants like alder while M38 was rare (Rizzoli et al., 2021; Rigamonti et al., 2023). By contrast in Germany, M38 was the dominant genotype found in alternative vectors like Orientus ishidae (Jarausch et al., 2023). This genotype could be transmitted by O. ishidae from alder to broadbean and from there further to grapevine by S. titanus. By this, it could be confirmed that FDp were present in naturally infected alder in Germany (Malembic-Maher et al., 2020).
The majority of map genotypes has only been found in alder (Malembic-Maher et al., 2020; Marcone et al., 2024) and has been referred to as Alder Yellows (AldY). These genotypes are transmitted from alder to alder by the Macropsinae Oncopsis alni (Maixner and Reinert, 1999; Maixner et al., 2000; Malembic-Maher et al., 2020). Occasionally, these map genotypes can be transmitted by O. alni to grapevine leading to Palatinate Grapevine Yellows (PGY) that exhibit in grapevine the same symptoms as FD. These PGY map genotypes are not transmissible by S. titanus and, thus, cannot lead to FD outbreaks.
The epidemiological significance of the map genotypes was supported by their correlation to the genetic diversity of two genes encoding for adhesion related proteins (vmpA and vmpB) involved in insect vector transmission (Malembic-Maher et al., 2020). MAP-FD1, FD2 and FD3 genotypes were correlated to vectotypes II and III, which are transmitted by Deltocephalinae vectors like S. titanus, O. ishidae, Allygus spp. or the planthopper Dictyophara europea that transmits MAP-FD3 from Clematis to grapevine. By contrast, PGY-genotypes transmitted by Macropsinae vectors like O. alni belong to the vectotype I (Malembic-Maher et al., 2020).
AldY phytoplasmas are widespread in German alders although infected alder trees do not show any symptoms. All trees examined in a natural habitat without viticulture (Spreewald, Brandenburg) and most of the trees in the viticultural regions of Palatinate and Moselle were found to be infected (Maixner and Reinert, 1999; Holz et al., 2015). While the map genotyping of the samples from the Spreewald revealed mainly PGY genotypes and no MAP-FD2 genotypes (Holz et al., 2015), the field captures of Allygus spp. and O. ishidae from alder trees in the Palatinate were almost exclusively infected with the MAP-FD2 genotype M38 (Jarausch et al., 2023). Whereas Allygus modestus and Allygus mixtus are native species, O. ishidae is an invasive species of Asian origin, which has been introduced in Europe from 1998 onwards (Gaffuri et al., 2011 and references therein; EPPO, 2015). Since 2002, this species rapidly spreads within Germany (EPPO, 2015). As O. ishidae readily transmits M38 from alder to alder test plants (Jarausch et al., 2023), its contribution to the spread of MAP-FD2 phytoplasmas between alders remains unclear.
FD is regulated as quarantine pest in the European Union (EU, 2019/2072) including the mandatory control of S. titanus populations by insecticide treatments and the uprooting of infected vines for containment of the disease. Furthermore, a systematic monitoring of FD is required also in FD-free regions like Germany. This monitoring has to focus on vineyards near alder stands, which have a higher risk of natural FD infection. A geodata based analysis was performed in this study to identify alder trees in the vicinity of vineyards on a large scale in the major viticultural regions of Germany. Furthermore, molecular tools were developed to analyze the distribution of FD phytoplasmas in naturally infected alder trees and to estimate the potential risk of FD phytoplasma transmission to adjacent grapevines.
Alder trees (Alnus glutinosa, Alnus incana) are not specified in any geodata sets. They have to be identified according to their specific properties: alders are trees with dark foliage that grow at the borders of rivers, streams, lakes or wet grounds. All geodata were provided by the Ordnance Surveys of Baden-Wuerttemberg (Landesamt für Geoinformation und Landentwicklung) and Rhineland-Palatinate (Landesamt für Vermessung und Geobasisinformation RLP).
Firstly, towering vegetation was deduced from the geodata to find trees. The basic parameters were height above ground (to find towering structures) and photosynthesis (to exclude other towering objects, e.g., roof tops). As primary data basis, a normalized digital surface model (nDSM) and digital 4-channel orthophotos (DOPs) were used. The orthophotos served as starting point for the calculation of the normalized difference vegetation index (NDVI), which represents the absorption and reflection properties of chlorophyll-containing plants. The nDSM was calculated by subtracting a digital terrain model (DTM) from a digital surface model (DSM).
Trimble eCognition (version 10.4, Trimble Germany GmbH, Munich, Germany) was used for the object based image analysis (Blaschke, 2010). Individual pixels of each DOP-tile (2 by 2 km) were divided into towering objects and ground objects using a “multi-threshold segmentation”. Subsequently, towering objects were separated into vegetation and non-vegetation objects using the NDVI. In an additional step, individual trees, bushes and smaller groups of trees could be identified using a “watershed segmentation algorithm”. This algorithm uses local intensity minima as starting points (seed objects) and expands them through adjacent pixels with increasing intensity values, until they collide with the object boundaries of neighboring seed object. In this case the resulting polygons reflected individual trees, bushes and smaller groups of trees in larger bodies of towering vegetation (e.g., forests). To gain the largest possible data base and amount of impactful parameters for subsequent machine learning steps, these polygons were attributed with a large array of data. In addition to the DOP channel values red, green, blue and near infrared (NIR), NDVI, BAI (Bare Area Index), Wetness-index, slope, nDSM and the gray level co-occurrence matrices (GLCM) Contrast and Dissimilarity were chosen as the most impactful parameters, each with the statistical values: maximum, mean and standard deviation.
Utilizing ArcGIS Pro (ESRI, 2024), the objects were examined according to their proximity to bodies of water (flowing waters and standing waters from ATKIS). All objects with a distance of more than 25 m from water were dropped. The remaining polygons were converted into points, that could subsequently be used as input objects for the machine learning algorithm.
To determine the probability of certain points as alder sites, a Random Forest (RF) algorithm (Breiman, 2001) was run through R scripts (R Core Team, 2024). Through a multitude of different decision trees, each with their different decision levels and branches, the points and their attributed information were divided into different classes: alder trees and non-alder trees.
Samples of alder trees were taken in most of the sites where alders were detected in the various wine-growing regions of Rhineland-Palatinate (Supplementary Table S1) and Baden-Wuerttemberg (Supplementary Table S2). In addition, alder sites were identified and sampled in the wine-growing region of Franconia in Bavaria. A mountain region in the German Alps (Allgäu) was sampled as a control region. The sampling was conducted between 2016 and 2024. One branch per alder tree was analyzed.
Adult Cicadellidae leafhoppers were collected from alder trees in the various wine-growing regions of Rhineland-Palatinate, Baden-Wuerttemberg and Franconia as well as in the control region of Allgäu by sweep-netting in July and August in the years 2016–2024. Specimens of the 16SrV phytoplasma vectors Allygus spp., O. ishidae and O. alni were picked with a mouth aspirator and stored at −20°C prior identification and molecular testing. Dead insects were identified at species level based on morphological characteristics as described in Jarausch et al. (2023).
Total nucleic acids (TNA) were extracted from phloem preparations of the alder branches with a CTAB-based method according to Jarausch et al. (2011). 16SrV phytoplasmas were detected in the alder extracts with elm yellows group-specific primers fAY/rEY (Marcone et al., 1996). Amplification was achieved with TEMPase Mastermix (VWR International GmbH, Darmstadt, Germany). After initial denaturation for 15 min at 95°C, 40 cycles of 15 sec 95°C, 15 sec 63°C and 30 sec 72°C were conducted. Aliquots of the PCR products were analyzed by 2% agarose gel electrophoresis.
The map marker gene sequences M1–M121 described in Malembic-Maher et al. (2020; Supplementary Table S1) were used to select different primer pairs. Sequences were aligned with MEGA 7 (Kumar et al., 2016) using the MUSCLE algorithm. Primers were selected manually with respect to the different objectives: universal map primers MAP U1/MAP U2 exhibited 100% identity with all 121 map genotypes and have already been used in Jarausch et al. (2023), primers M38 for/M38 rev were selected to amplify only MAP-FD2 genotypes while primers M50 for/M50 rev were selected for amplification of MAP-FD1 genotypes. To distinguish the epidemic map genotypes M38 and M54, a M54-specific reverse primer M54 rev was selected, which was used in combination with forward primer M38 for. The primer sequences are given in Table 1.
All primer pairs were designed for real-time PCR amplification with a SYBR Green™ assay. The reaction set up used 0.22 µM of each primer in 1x TEMPase Mastermix (VWR International GmbH, Darmstadt, Germany) with 3 mM Mg2+ and SYBR Green™ I (Lumiprobe GmbH, Hannover, Germany) diluted to 1:66000 in 20 µl reactions with 1 µl TNA extract. Cycle conditions were 15 min at 95°C for hot start followed by 40 cycles of 15 sec 95°C, 15 sec annealing and 30 sec 72°C. After the final elongation step of 4 min at 72°C, a melting curve analysis ranging from 50°C to 90°C was performed. The annealing temperature was specific for each primer pair: 54°C for MAP U1/MAP U2, 63°C for M38 for/M38 rev, 58°C for M38 for/M54 rev and 64°C for M50 for/M50 rev. All PCR reactions were performed either in a Chromo4 Real-Time PCR detector or a CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories GmbH, Feldkirchen, Germany). Data were analyzed with Opticon Monitor3 or CFX Maestro software (Bio-Rad Laboratories GmbH, Feldkirchen, Germany), respectively.
The secY-map gene marker of 16SrV phytoplasmas was amplified with nested PCR primers according to Arnaud et al. (2007). The PCR conditions were adjusted in the first PCR for 20 cycles with primers FD9-F5 and MAP-R1 and in the second PCR for 35 cycles with primers FD9-F6 and MAP-R2 to 15 sec 95°C, 20 sec of 52°C and 1 min 66°C each. The PCR products were purified with Macherey&Nagel PCR clean up kit (Macherey&Nagel, Düren, Germany) and sequenced via Sanger sequencing by Eurofins Genomics (Eurofins Genomics, Ebersberg, Germany). Cloning of FD9-F6/MAP-R2 PCR products was achieved by T/A-cloning in the pUC-vector derivate pT-PCR by standard methods using blue/white-shift selection. White colonies were either directly analyzed via plasmid preparation or by colony PCR. Plasmid DNA was purified with Qiagen Miniprep-kit (Qiagen GmbH, Hilden, Germany). For colony PCR, a master plate of white colonies was prepared and established colonies were transferred with sterile toothpicks to a PCR plate containing 200 µl per well. Cells were broken with the boiling method (5 min, 95°C) and 1 µl per colony extract was transferred to the subsequent PCR reaction. Screening for the MAP-FD2 marker was done with qPCR M38for/M38rev as described above. Plasmid DNA was purified from positive colonies of the master plate as above. Sanger sequencing of the plasmid DNA was performed with standard M13 primers by Eurofins Genomics (Eurofins Genomics, Ebersberg, Germany). Assembling of raw sequences and multiple alignments were done with DNASTAR Lasergene (DNASTAR inc., Madison, USA). Phylogenetic reconstructions using maximum likelihood were performed by MEGA7 (Kumar et al., 2016) with randomized bootstrapping evaluation.
A M38-infected Alnus test plant of the transmission trials reported by Jarausch et al. (2023) was used to establish an in vitro culture of M38-infected alder. Young growing shoots of a greenhouse-maintained alder plant was used for culture initiation as described by Ciccotti et al. (2008). In vitro shoots were maintained and subcultured on Murashige & Skoog medium with 4.44 µM BAP (6-benzylaminopurine) (Duchefa Biochemie, Haarlem, Netherlands). The infected in vitro plants served as homogenous biological control available throughout the year.
The analysis identified potential sites with alder trees near vineyards for all wine-growing areas in Rhineland-Palatinate and Baden-Wuerttemberg. Within the scope of this project, these were the only federal states that provided access to the necessary input geodata.
In 2016, a first training set of reference points for alder and non-alder sites was obtained by field inspections in the Palatinate region. Based on this training data, RF algorithms were established to classify sites with tree vegetation at the border of running and standing waters near to vineyards. Based on this first geodata map, ground truth data for reference points of alder and non-alder trees were obtained in as many as possible different sites in wine-growing regions of Rhineland-Palatinate between 2016 and 2021 and in Baden-Wuerttemberg between 2017 and 2024.
As shown in Supplementary Table S1, 2547 reference points were obtained for the seven wine-growing regions in Rhineland-Palatinate. Respective 6085 reference points for nine wine-growing regions in Baden and seven wine-growing regions in Wuerttemberg are given in Supplementary Table S2. The machine learning analysis using these ground truth data yielded a classification accuracy for alder and non-alder trees of 0.8357. The confusion matrix shows that 86% of non-alder trees were correctly classified and that 74% of alder trees were accurately classified. The ROC (Receiver Operating Characteristic) analysis in Figure 1 confirms this result by showing a high sensitivity (true positive rate) and a good specificity (false positive rate).
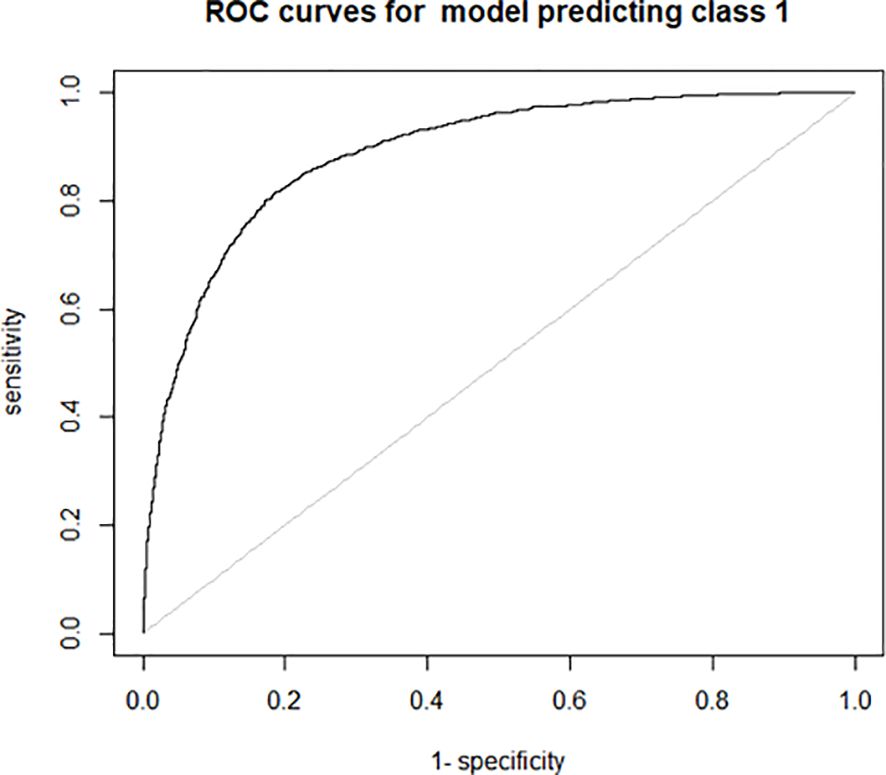
Figure 1. ROC curve for the performance of the Random Forest Classifier to predict alder and non-alder trees.
Based on these results, webmaps were provided to the Plant Protection Services in Rhineland-Palatinate and Baden-Wuerttemberg to identify vineyards near potential alder trees that exhibit a risk for a natural spread of FDp from alder to grapevine. Two levels of probability (70% and 85%) for sites with alder trees are given and vineyards in a flight distance of potential insect vectors of up to 200 m are highlighted.
The webmap for Rhineland-Palatinate (https://risikokarte-fd-rlp.agroscience.de) is password protected but the respective map of Baden-Wuerttemberg is open access and is openly accessible under https://phytomo-bw.agroscience.de/. The reference points for alder in Baden-Wuerttemberg are shown in this map.
First, a universal map gene detection primer pair for 16SrV phytoplasmas was developed to detect all known map genotypes. Tests against different 16Sr group phytoplasmas (16SrI, 16SrIII, 16SrX, 16SrXII) showed that the primer pair MAP U1/MAP U2 is specific for the detection of 16SrV elm yellows group phytoplasmas (Table 2). It detected various strains of ‘Flavesence dorée’ phytoplasmas, Palatinate Grapevine Yellows phytoplasma (PGY), Alder Yellows phytoplasma as well as ‘Candidatus Phytoplasma ulmi’. The sensitivity was tested with 1513 phytoplasma-infected alder samples obtained in this study. The direct standard PCR with elm yellows group-specific primers fAY/rEY reacted positive with 98.9% of the samples while the real-time PCR assay with primers MAP U1/MAP U2 confirmed 16SrV phytoplasma infection in 99.9% of the samples. Thus, the new primers can also be used as detection primers for 16SrV phytoplasmas.
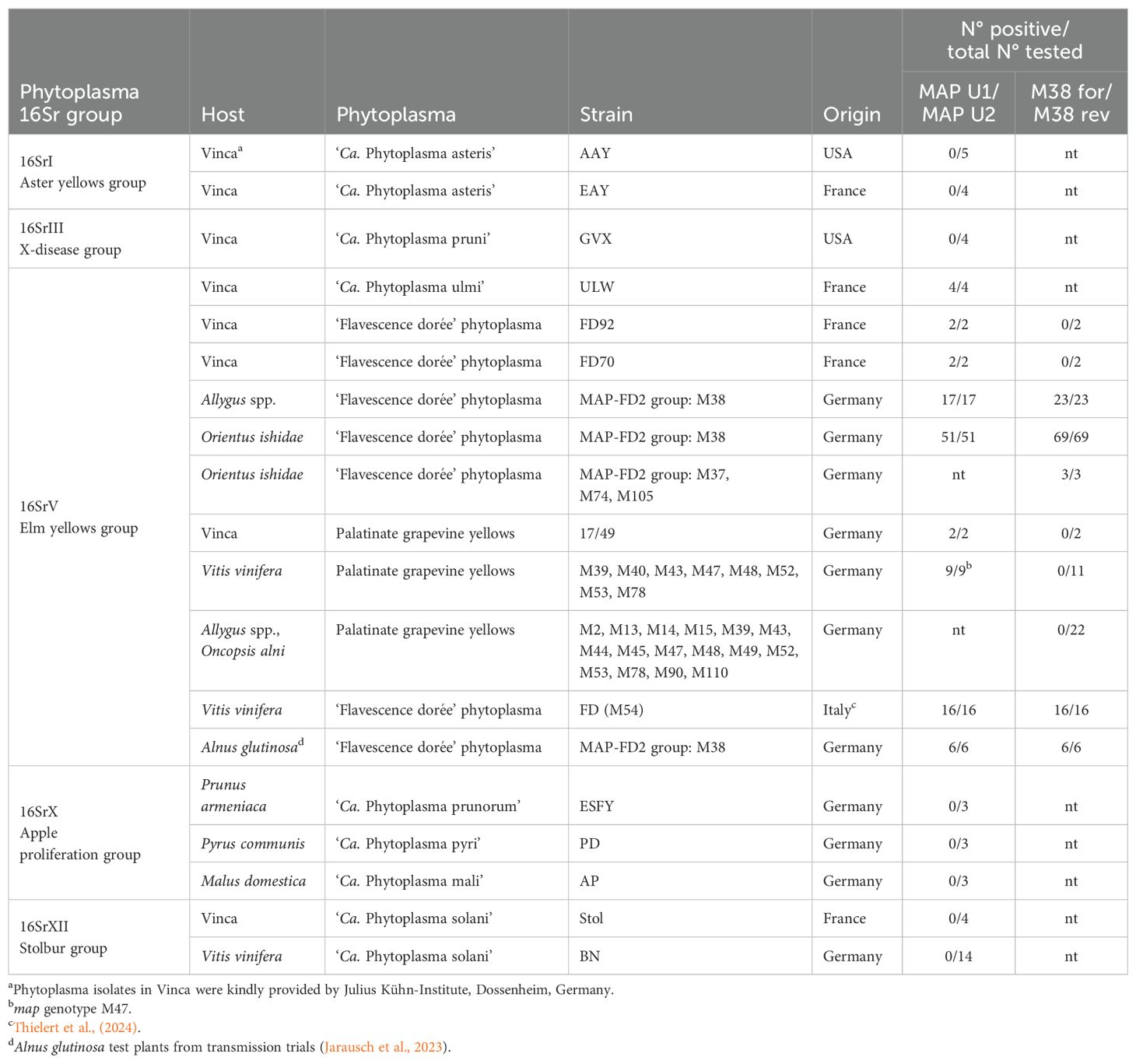
Table 2. Specificity test of universal map primers MAP U1/MAP U2 and MAP-FD2-specific primers M38 for/M38 rev against different 16Sr group reference strains and different 16SrV group phytoplasmas and map genotypes.
Actually, 173 different map genotypes are defined. The list of Malembic-Maher et al. (2020) has been updated by new genotypes published by Krstić et al. (2022); Kogej Zwitter et al. (2023); Radonjić et al. (2023) and Marcone et al. (2024). The list with reference accession numbers can be found in Supplementary Table S3. The universal map primers MAP U1/MAP U2 exhibited 100% identity to 163 map genotypes and had only one mismatch with 10 map genotypes (Table 3). Nine of these genotypes have been found only once or twice in alder or O. alni (Malembic-Maher et al., 2020). The genotype M47, however, has been identified regularly in alder, O. alni and in PGY-infected grapevine. PCR tests of nine M47-infected grapevine samples showed that these map genotypes were also detected by the MAP U1/MAP U2 primers (Table 2).
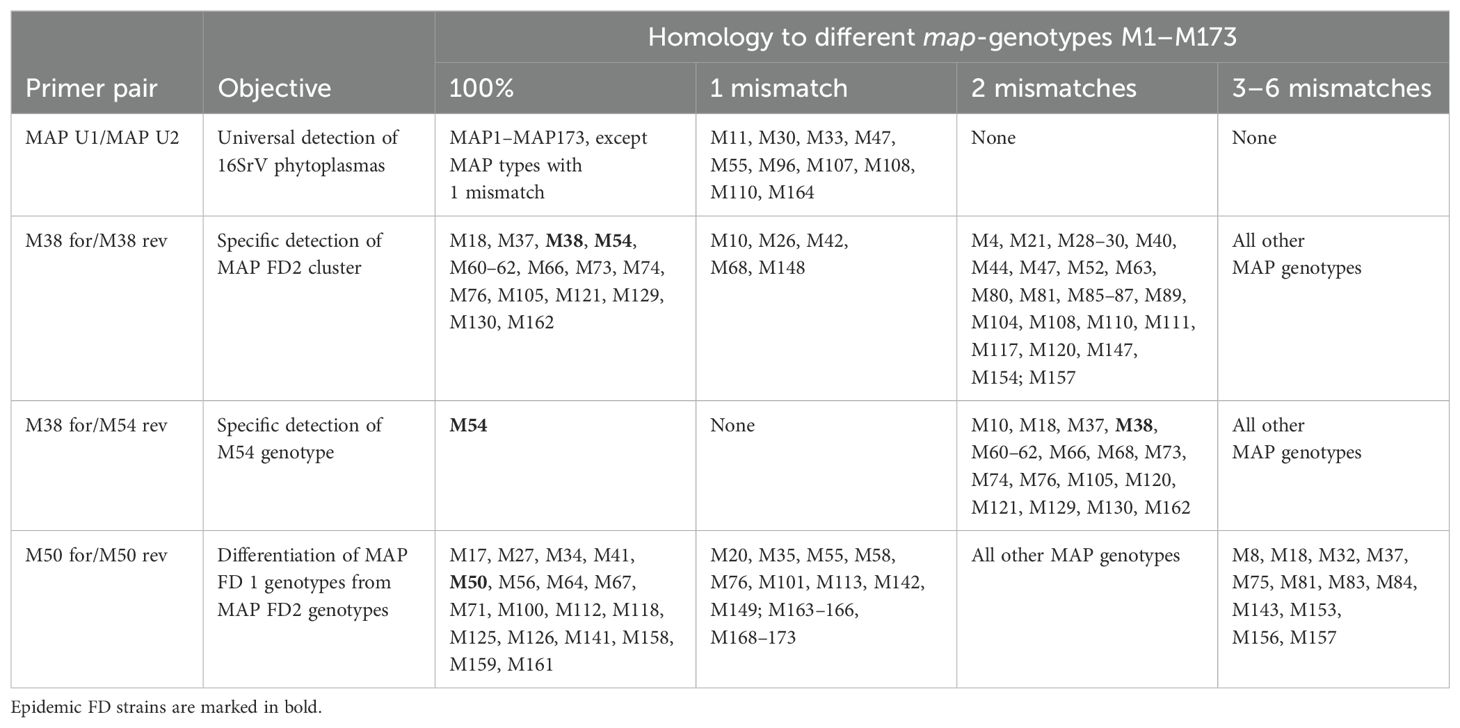
Table 3. Homologies of the primer sequences developed in this study to map-genotypes M1–M173 (Supplementary Table S3).
As M38 was the dominant MAP-FD2 genotype that has been found in grapevine as well as in alder and Deltocephalinae vectors Allygus spp. and O. ishidae (Malembic-Maher et al., 2020), specific primers for this genotypes have been developed. As shown in Table 3, the primer pair M38 for/M38 rev has 100% sequence identity to most of the MAP-FD2 cluster genotypes including the epidemic ‘Flavescence dorée’ strain M54. The primers exhibit one mismatch to seven other genotypes that have been attributed to the MAP-FD2 cluster (Krstić et al., 2022). The only map genotypes that were not detected are M89 and M155, which have been recently found in Vitis vinifera (Krstić et al., 2022). The specificity of the primers has also been tested with PGY-infected V. vinifera samples as well as with AldY-infected Allygus spp. and O. alni samples. As shown in Table 2, no reaction occurred with these genotypes while the M38 genotype was readily detected in all samples of alder, Allygus spp. and O. ishidae. In addition, the M54 genotype was detected in all FD-infected grapevine samples in Italy. O. ishidae individuals infected with MAP-FD2 genotypes M37, M74 and M105 also reacted positive in M38 for/M38 rev real-time PCR assay. Thus, the primers M38 for/M38 rev can be considered specific for MAP-FD2 genotypes.
M54 is the dominant MAP-FD2 genotype in FD epidemics in France. It was therefore important to know, if this genotype is present in German alders. A M54-specific primer M54 rev was designed that enabled a distinction between M38 and M54. Applied in combination with forward primer M38 for, this primer pair did not react with M38-infected alder or M50 (strain FD70) and M53 (PGY) infected Vinca test plants. As shown in Table 3, this primer pair has 100% sequence identity only with genotype M54 and can be used to identify this genotype. It has been successfully applied to M54-infected grapevine samples from Italy, which were also tested positive with M38 for/M38 rev primers (Table 2).
MAP-FD1 genotypes have been identified only very rarely in samples of O. alni in Germany or the adjacent Alsace region in France (Malembic-Maher et al., 2020). However, it cannot be excluded that they are spread in German alders and represent a risk for FD-outbreaks. Therefore, primers were developed that allow detection of MAP-FD1 genotypes. The MAP-FD1 cluster is more heterogeneous as the MAP-FD2 cluster with reference genotype M50 being responsible for major FD epidemics. Further genotypes M27, M34, M112 and M158 have been found in grapevine (Malembic-Maher et al., 2020; Krstić et al., 2022; Kogej Zwitter et al., 2023). Primer pair M50 for/M50 rev was selected to detect these and 13 further MAP-FD1 genotypes with 100% sequence identity. Some other more or less distantly related map genotypes have only one mismatch with the primers (Table 3). Among these is a group of samples (M163–M173) from alder trees in Southern Italy (Marcone et al., 2024). The primer pair detected readily genotype M50 (strain FD70) in infected Vinca samples but did not react with genotypes M38 (in alder), M54 (in grapevine) and M53 (in Vinca).
Naturally infected alder trees harbor a multitude of different 16SrV phytoplasma isolates with several different map genotypes. The majority belongs to AldY- or PGY-group phytoplasmas (Malembic-Maher et al., 2020). Therefore, specific primers are needed to detect MAP-FD1 and MAP-FD2 phytoplasmas in mixed infections in alder. The application of MAP-FD2 primers M38 for/M38 rev had to be a compromise between sensitivity (detection of low concentrations of M38 genotype) and specificity (no cross reaction to other map genotypes). Therefore, a normalization with the universal primers MAP U1/MAP U2 was done. Homogenously M38-infected micropropagated alder plants were used as biological controls. Sixteen samples were tested with primers MAP U1/MAP U2 and M38 for/M38 rev in parallel and the ratios of the obtained Cq-values M38/MAP were made. In total, 51 runs were conducted and the mean ratio was 1.05 ± 0.04. Thus, both primer pairs performed equally well and a ratio of 1 could be attributed to a homogenous infection with genotype M38. This value could be confirmed by the analysis of 17 M38-infected Allygus spp. samples, 51 M38-infected O. ishidae samples and 16 M54-infected grapevine samples (Table 2). The mean ratio of these tests was 1.07 ± 0.05.
For the definition of a threshold ratio up to which a MAP-FD2 detection in a sample with mixed infection is reliable, map genes of infected alder were amplified according to Arnaud et al. (2007) and PCR products were either directly sequenced or cloned. In total, sequences of 46 samples were obtained and three groups of samples were separated for data analysis: 17 samples with a M38/MAP ratio between 1.0 and 1.14, 28 samples with a ratio between 1.15 and 1.40 and one sample with a higher ratio value above 1.40. M38 sequences were detected in 94% of samples with a ratio up to 1.14. Mixed infections were found in all samples with a ratio between 1.15 and 1.40 but a M38 sequence could be confirmed in 53% of the samples. Cloning of the map PCR products of some samples further showed that only one out of fifteen clones had the M38 sequence. In order to obtain more data for samples with M38/MAP ratios above 1.40, map PCR products of five samples with ratios between 1.46 and 1.87 were cloned and a total of 49 clones was analyzed by colony PCR with primers M38 for/M38 rev. No positive reaction was obtained with the 49 clones. By contrast, clones that were obtained from alder samples with a M38/MAP ratio around 1 tested strongly positive in colony PCR with the primers M38 for/M38 rev. Sequence analysis of some positive clones confirmed the infection with map genotype M38. These results indicate that samples with M38/MAP ratios above 1.40 have to be considered negative for MAP-FD2 genotypes. This assay was applied to all alder samples of the different regions. Applying a cut-off at Cq 35 for M38 for/M38 rev PCR, 98% of the samples with ratios M38/MAP between 1.00 and 1.40 were positive. Thus, the ratio of 1.40 was used as threshold for a positive detection of MAP-FD2 phytoplasmas. In consequence, the same upper threshold ratio of 1.40 was applied for the test of mixed infections with MAP-FD1-specific primers M50 for/M50 rev without further experimental confirmation.
Samples of alder were taken wherever possible as part of the field inspections to find alder sites near vineyards in as many different regions as possible. Beforehand, it was checked whether the test of a single branch was sufficient for reliable detection of 16SrV phytoplasmas. For this purpose, 3–5 individual branches were examined from each of 12 infected alders. Phytoplasmas were detected in all samples. For this reason, only one branch per tree was tested during monitoring in Rhineland-Palatinate, Baden-Wuerttemberg and Franconia. An average of five trees were examined per site.
All samples were first tested with primers fAY/rEY for infection with 16SrV phytoplasmas. All positive samples were then tested with primers M38 for/M38 rev and MAP U1/MAP U2 for an infection with MAP-FD2 phytoplasmas. All samples with a M38/MAP ratio up to 1.40 were considered positive for MAP-FD2 phytoplasmas. A subset of negative samples was further tested with primers M50 for/M50 rev for an infection with MAP-FD1 phytoplasmas. Only samples with a M50/MAP ratio up to 1.40 were regarded as positive.
In Rhineland-Palatinate, 1089 samples originating from 171 different sites were analyzed as shown in Table 4. In total, 97% of alders were infected with 16SrV phytoplasmas, of which 55% harbored MAP-FD2 phytoplasmas. However, the MAP-FD2 infections were distributed very differently in the wine-growing regions: almost no MAP-FD2 phytoplasmas were detected in the northwestern regions Ahr, Mosel, Saar, Ruwer and Mittelrhein whereas in the southern parts of the Rhine valley MAP-FD2-infection rates increased from 49% in Nahe to 86% in Rheinhessen and 76% and 68% in northern and southern Palatinate, respectively. Only three MAP-FD1-infections were detected in the 375 MAP-FD2-negative samples tested. No infection with M54 was found in 460 MAP-FD2-positive samples.
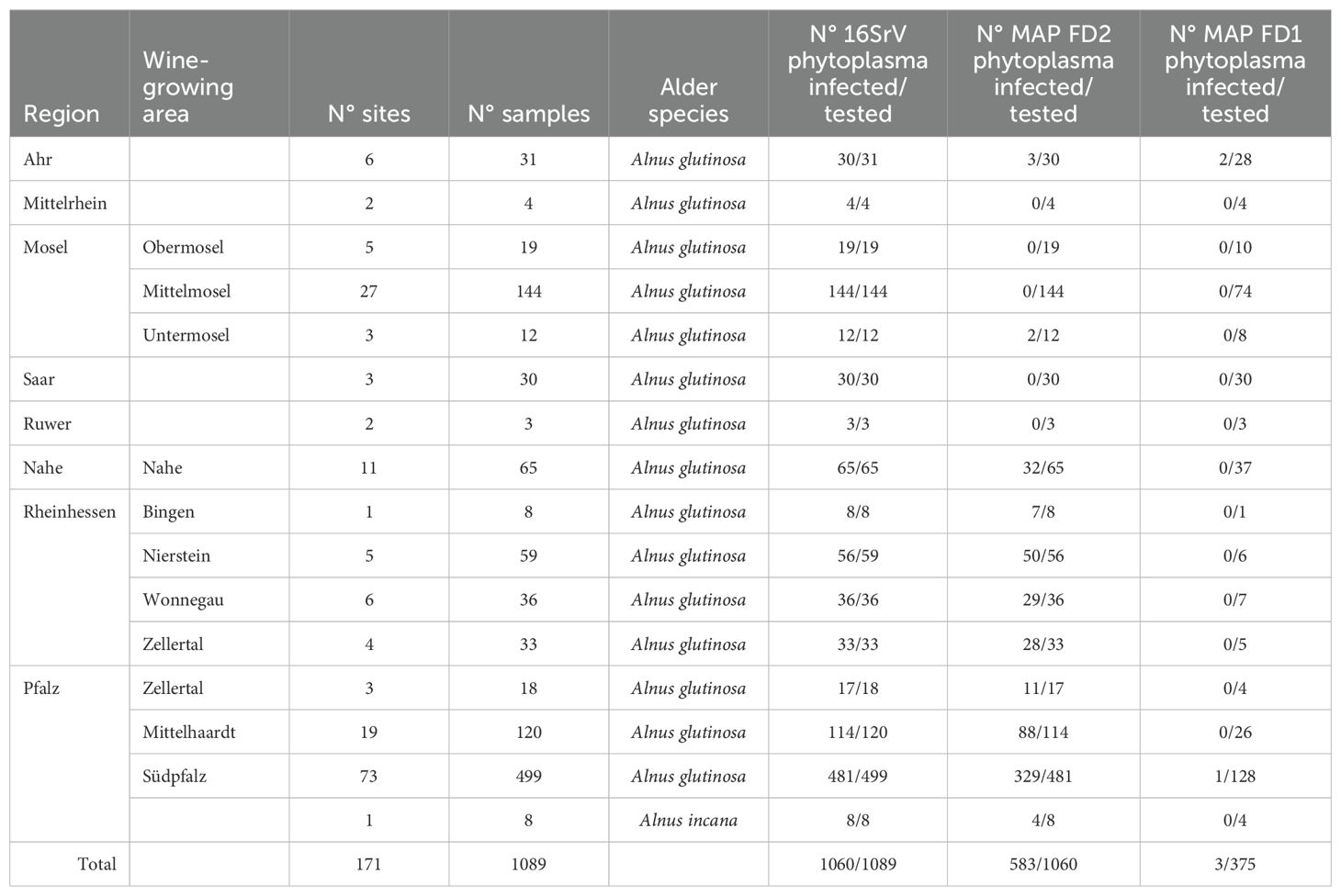
Table 4. Summary of the results obtained with alder samples from different sites in Rhineland-Palatinate for the detection of 16SrV phytoplasmas, MAP-FD2 and MAP-FD1 phytoplasmas using primers fAY/rEY, M38 for/M38 rev and M50 for/M50 rev, respectively.
In Baden-Wuerttemberg, 556 samples originating from 104 different sites were analyzed as shown in Table 5. In total, 98% of alders were infected with 16SrV phytoplasmas, of which 61% harbored MAP-FD2 phytoplasmas. Again, the MAP-FD2 infections were distributed differently in the wine-growing regions: they were high in the Rhine valley in regions neighboring Rheinhessen and Palatinate (Badische Bergstrasse: 78%; Kraichgau: 73%; northern Ortenau: 64%; central and southern Ortenau: 81%) and low in far northern (Tauberfranken: 30%) or far southern (Bodensee: 42%) wine growing regions. The central wine growing regions of Wuerttemberg had high (Hohenlohe and Kochertal: 75% and 71%, respectively) to intermediate (Heilbronner Land, Württembergisch Unterland/Enztal and Remstal 59%, 52% and 63%, respectively) infection rates. The southern wine growing regions of Baden (Kaiserstuhl: 45%, Markgräfler Land: 51%) exhibited likewise intermediate rates of infection with MAP-FD2 phytoplasmas. Among the 276 negative samples tested, 12 infections with MAP-FD1 phytoplasmas were detected across the different regions (Table 5). No infection with M54 was found in 306 MAP-FD2-positive samples.
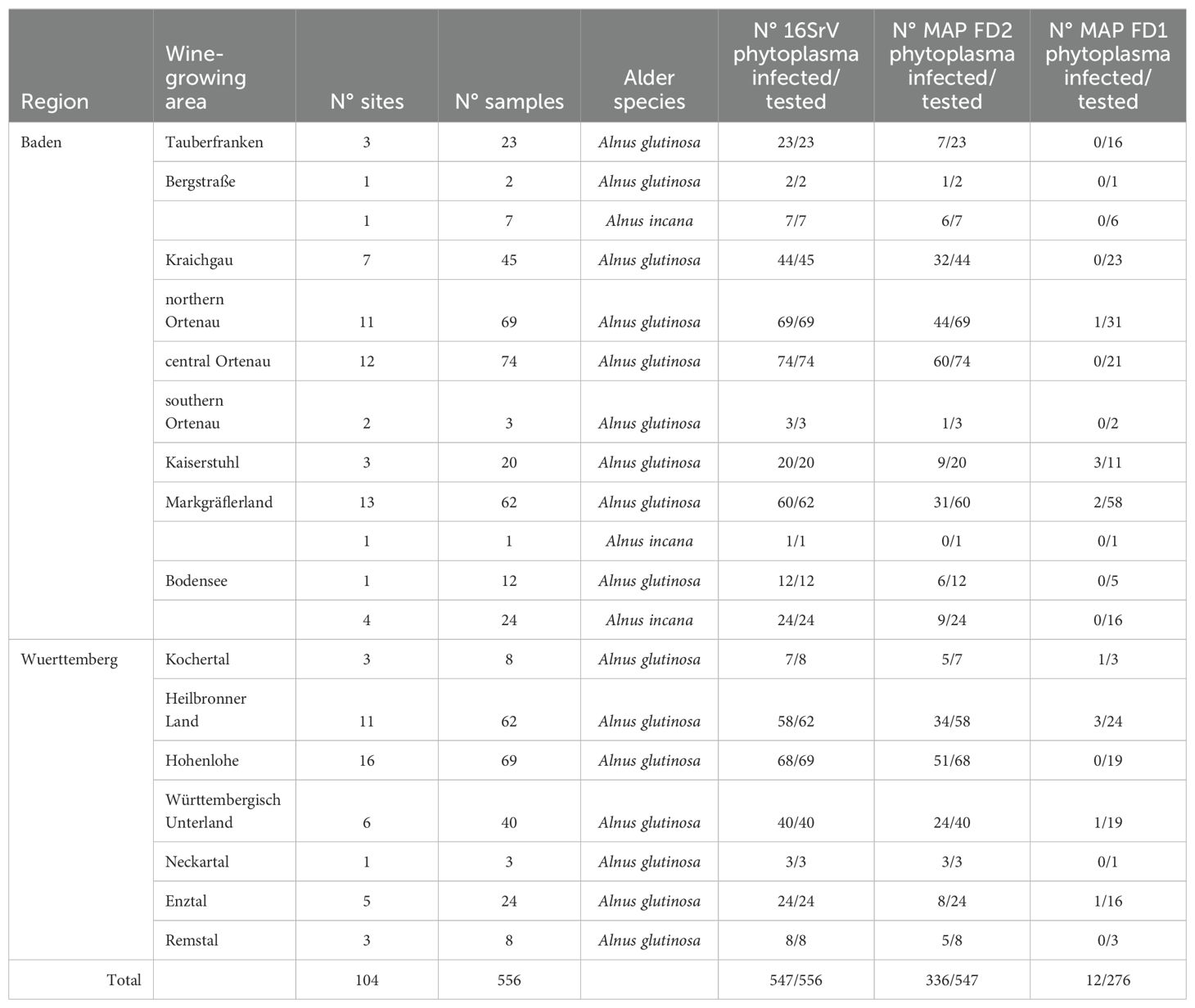
Table 5. Summary of the results obtained with alder samples from different sites in Baden-Wuerttemberg for the detection of 16SrV phytoplasmas, MAP-FD2 and MAP-FD1 phytoplasmas using primers fAY/rEY, M38 for/M38 rev and M50 for/M50 rev, respectively.
Alder samples in the Bavarian wine growing region of Franconia were sampled without support of a geodata analysis. Thus, sampling sites were concentrated along the river Main and in additional known regions with alder stands. In total, 141 samples from 22 different sites were tested (Table 6). As in the other regions, 97% of the samples tested positive for 16SrV phytoplasmas but only 15% were infected with MAP-FD2 phytoplasmas. No infection with MAP-FD1 phytoplasmas was detected in 90 MAP-FD2-negative samples. Fourteen MAP-FD2-positive samples were tested for M54-infection with negative result.
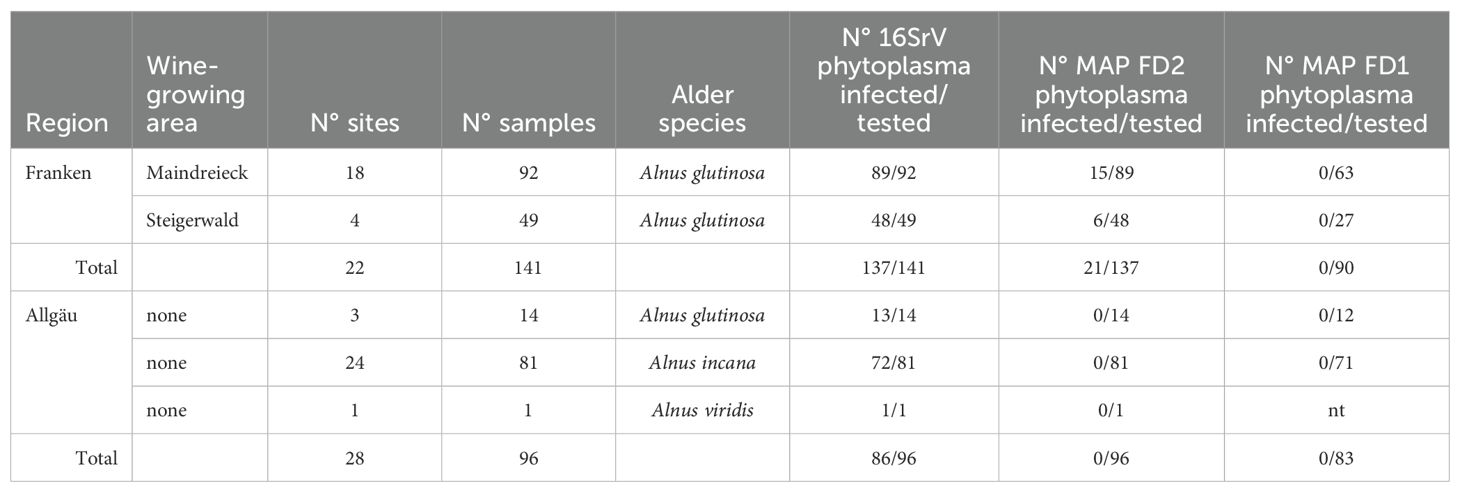
Table 6. Summary of the results obtained with alder samples from different sites in Bavaria for the detection of 16SrV phytoplasmas, MAP-FD2 and MAP-FD1 phytoplasmas using primers fAY/rEY, M38 for/M38 rev and M50 for/M50 rev, respectively.
The distribution of MAP-FD2 phytoplasmas in German wine growing regions of Rhineland-Palatinate, Baden-Wuerttemberg and Franconia is visualized in Figure 2. All raw data of the sampling sites and the test results are available in Supplementary Table S4.
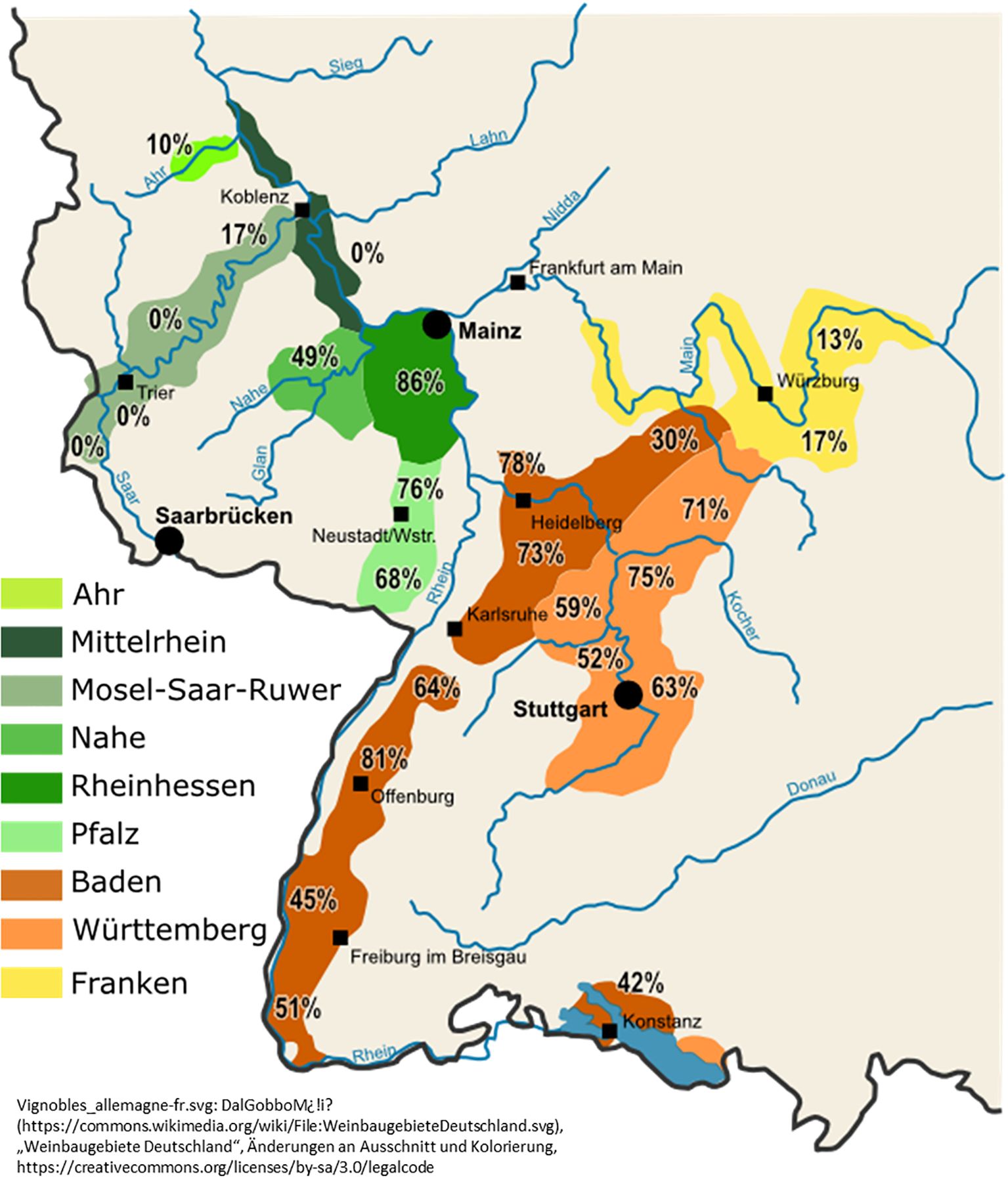
Figure 2. Distribution of MAP-FD2 phytoplasmas in different wine growing regions of Rhineland-Palatinate, Baden-Wuerttemberg and Franconia.
A mountainous region in the Bavarian Alps (Allgäu) without any wine growing was analyzed as control region for the spread of MAP-FD2 and MAP-FD1 phytoplasmas. No FDp-infection was discovered in 96 samples from 28 different sites although 16SrV phytoplasmas were detected in 90% of the samples (Table 6).
From previous work (Malembic-Maher et al., 2020; Jarausch et al., 2023) it was known that the Deltocephalinae O. ishidae and Allygus spp. can be infected to high percentages with the MAP-FD2 genotype M38. Both species can transmit this genotype to Alnus test plants and at least O. ishidae also form alder to grapevine. As O. ishidae is an invasive species in Germany, insect captures were conducted in the different wine growing regions along with alder sampling. Due to time constraints for sampling many sites in a short period of time, it was not possible to systematically monitor the populations. However, the data in Table 7 provide an initial indication of the distribution of O. ishidae in the various regions. The indigenous species Allygus spp. and O. alni were sampled as controls. Individuals of O. ishidae were captured in most of the wine growing regions where a substantial number of samplings could be conducted. Interestingly, O. ishidae was not found on alders in Mosel and Allgäu, two regions were no M38-infections in alder could be recorded.
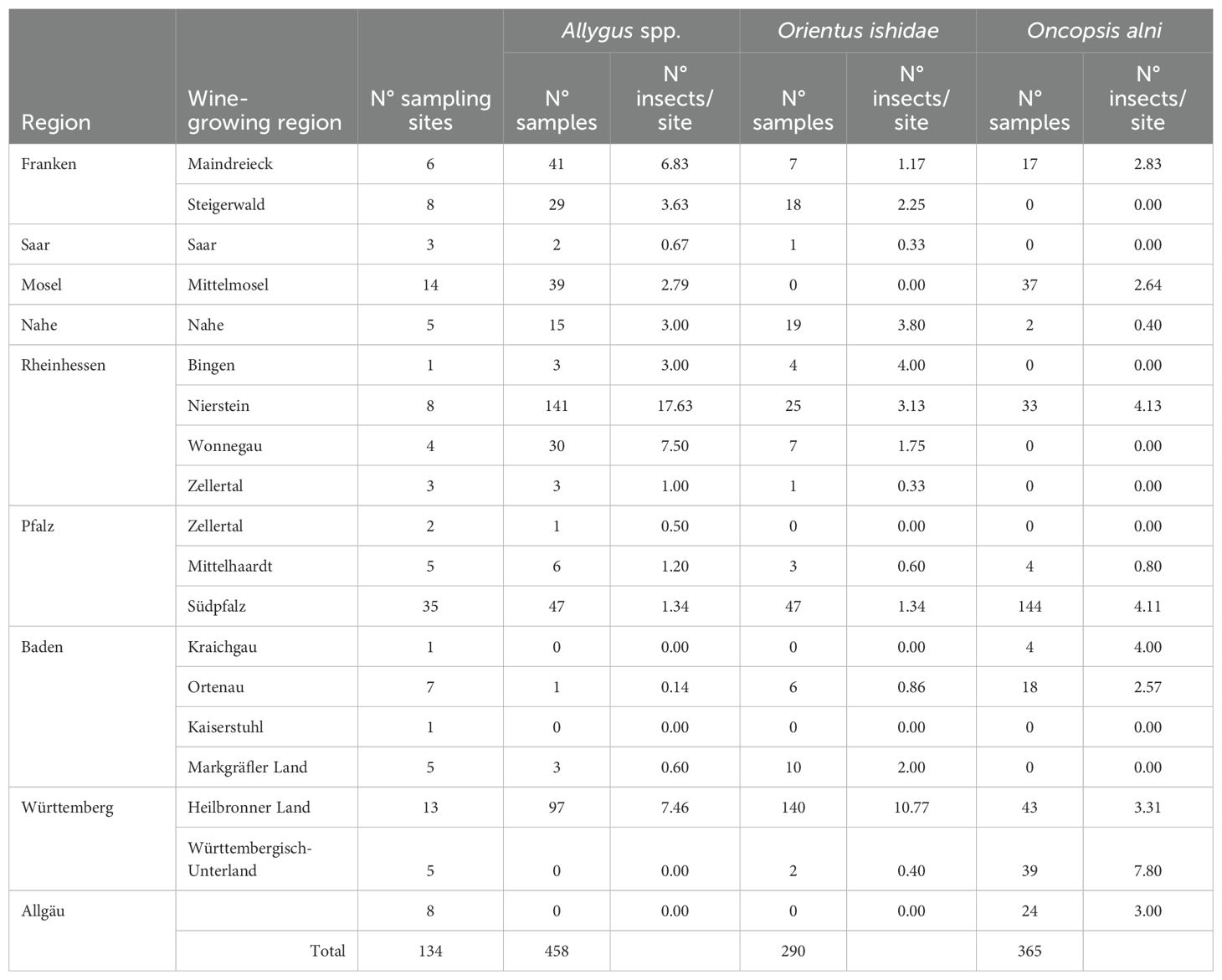
Table 7. Summary of insect captures of 16SrV phytoplasma vectors in different wine-growing regions between 2016 and 2024.
Data on the infection rate of the captured insects and the molecular analysis of the detected phytoplasmas are provided in a separate publication of Jarausch et al. (2025b). Real-time PCR with primers MAP38 for/MAP38 rev confirmed M38-infections in 24 individuals of O. ishidae and 53 individuals of Allygus spp.
So far, Germany has been considered free of Flavescence dorée. However, German wine-growing areas in Baden-Wuerttemberg and Rhineland-Palatinate are on the border of the spread of FD from south and west. It is well documented that Flavescence dorée phytoplasmas are of European origin and are present in reservoir plants like alder (Malembic-Maher et al., 2020). It was recently experimentally demonstrated that the polyphagous leafhopper Orientus ishidae is able to transmit the FDp from naturally infected alder to grapevine (Jarausch et al., 2023). Under natural conditions, this scenario has already been detected in the German wine-growing region of Rheinhessen and in the Alsace region of France, which is close to the wine-growing regions of Baden in Germany (Jarausch et al., 2021; Auriol et al., 2023). These events were not associated with subsequent FD outbreaks as the FD vector S. titanus was not present. However, this situation changed in 2024 when relatively high populations of S. titanus were first discovered in South Baden (Askani et al., 2024; Jarausch et al., 2025a) entailing a drastically increased risk of FD outbreaks in Germany. Previous studies have shown that alders in Germany were infected to a high degree with 16SrV phytoplasmas (Maixner and Reinert, 1999; Holz et al., 2015) which were occasionally transmitted to grapevine by the leafhopper Oncopsis alni provocating the non-epidemic Palatinate Grapevine Yellows disease (Maixner et al., 2000). Since 2016, risk analyses for FD outbreaks starting from infected alder trees were performed in Germany. One part was the search for alternative vectors, which transmit the FDp from alder to grapevine. This research by Jarausch et al. (2023) revealed that a high percentage of field-captured O. ishidae individuals were infected with MAP-FD2 genotype M38. The result signified that M38 must be widespread in German alders. However, data of the distribution of M38 in German alders were missing until we started our investigations in 2016 and found the first putative M38-infections in alders in the Palatinate region (Jarausch et al., 2019). These data are included in the present study. Therefore, the objective of our study was first to identify potential alder sites near vineyards in all major wine-growing regions of Germany and second to elucidate the distribution of FDp in these alders. Taken together, both information should give an actual view of the risk for natural FD outbreaks in Germany, if S. titanus spreads over the territory.
For the realization of the first aspect, geodata analyses were employed to establish web maps of all potential alder sites near vineyards for the entire viticultural regions of Rhineland-Palatinate and Baden-Wuerttemberg. The challenge was to identify alders among towering vegetation. This could be achieved by applying machine learning techniques to the datasets of more than 8,600 reference points. The accuracy of classification of alder and non-alder sites reached 84% as alder sites share common features like proximity to water, soil moisture and dark foliage. The analysis revealed that alder sites in Germany are widespread and occur mostly in flat growing areas and are absent on slopes. The high amount of alder sites near vineyards is probably a specificity of viticultural regions north of the Alps and may represent an increased risk for natural FD spread from these reservoir plants (Auriol et al., 2023). Different reservoir plants of FDp are reported from FD-affected countries south of the Alps, e.g. hazelnut (Coryllus avelana), hornbeam (Carpinus betulus) and willow (Salix sp.) (Casati et al., 2017; Lessio et al., 2019; Mehle et al., 2019). As the flight distance from alternative FDp vectors such as O. ishidae from alder trees or other reservoir plants into the vineyard is limited (Lessio et al., 2016; Casati et al., 2017; Jermini et al., 2019; Rizzoli et al., 2021), we assumed that only vineyards in a distance from up to 200 m from potential alder trees have an increased risk of FDp transmission. As an outcome of this study, we recommend to focus the FD monitoring in Germany on these vineyards. This can reduce the area to be monitored by 83%.
It is well known that naturally infected alders are colonized by a multitude of different 16SrV phytoplasmas (Malembic et al., 2007). Therefore, a real-time PCR based screening assay had to be developed to analyze the large number of samples. It was expected from previous data (Jarausch et al., 2019, 2023) that M38 would be the major FDp that spreads in German alders. Specific nucleotide signatures for M38 and related MAP-FD2 phytoplasmas in the map gene were used to select the specific primers M38 for/M38 rev. In silico analyses and PCR tests with reference samples showed that the primers are specific for MAP-FD2 phytoplasmas. However, a certain background within Cq values between 35 and 40 was obtained with extracts from naturally infected alder. This could be due either to low concentrations of MAP-FD2 phytoplasmas or cross-reactions with other 16SrV phytoplasmas. A normalization with newly developed universal primers for the map gene of 16SrV phytoplasmas was applied to solve this problem. In addition, our data for the analysis of all field-collected samples demonstrate the potential of these primers for 16SrV phytoplasma diagnosis.
The application of the MAP-FD2 screening assay to all alder samples showed that these phytoplasmas are widespread in German alders in the vicinity of vineyards. Sequence data from a high number of infected O. ishidae and Allygus spp. captured on alder trees in different German regions yielded only M38 as MAP-FD2 genotype (Jarausch et al., 2023, 2025b). Thus, it is reasonable that the MAP-FD2 genotypes detected by the assay in the German alder trees belong to the map genotype M38. This finding is in contrast to reports from southern Switzerland and northwestern Italy where the MAP-FD1 genotype M50 is the major FDp detected in reservoir plants like alder as well as in O. ishidae (Rizzoli et al., 2021; Rigamonti et al., 2023). This indicates that the epidemiological cycle of FDp spread among alders is different in Germany compared to regions south of the Alps. This hypothesis is supported by similar data for the Alsace region in the Upper Rhine valley (Auriol et al., 2023) where M38 was found in all infected O. ishidae as well as in alder.
The M38 for/M38 rev assay equally well detects the map genotype M54, which is responsible for major FD outbreaks in France (Malembic-Maher et al., 2020). It has also spread to Swiss (Rizzoli et al., 2021) and Northern Italian vineyards (Rigamonti et al., 2023). While M54 is widespread in grapevine and in S. titanus it has not been reported from reservoir plants in the studies of Malembic-Maher et al. (2020) or Rizzoli et al. (2021). Only Rigamonti et al. (2023) detected M54 occasionally in various reservoir plants like Ailanthus altissima and Corylus avellana but not in alder. We therefore developed a specific primer M54 rev to distinguish M38 and M54. The real-time PCR assay M38 for/M54 rev is specific for M54 and detects no other known map genotype. It readily detected M54 in FD-infected grapevine samples from Northern Italy and might be applied in epidemiological studies. Here, we tested a subset of MAP-FD2-positive alder extracts for the presence of M54 in German alder trees. No single infection among 780 samples from all regions could be found. This further indicates that M54 is a very specific map genotype well adapted to the epidemiological cycle S. titanus–grapevine (Rigamonti et al., 2023).
The MAP-FD1 genotype M50 was only very rarely reported from Germany or the adjacent Alsace region in France. Malembic-Maher et al. (2020) list three infections in the alder leafhopper O. alni indicating a presence of this genotype in German alders. Therefore, MAP-FD1 detection primers M50 for/M50 rev were selected in the available map gene sequences that enabled also a clear distinction from MAP-FD2 genotypes. In silico analyses showed that these primers have 100% identity to all major MAP-FD1 genotypes including those which have been found in FD outbreaks: M27, M34, M50, M112 and M158 (Malembic-Maher et al., 2020; Krstić et al., 2022; Kogej Zwitter et al., 2023). A subset of alder samples from the different German regions that was negative for MAP-FD2 genotypes was analyzed with these primers. Only a few samples tested positive for MAP-FD1 genotypes. This result confirms the data for MAP-FD2 genotypes that the FDp epidemiology in German alder trees differs from countries south of the Alps where MAP-FD1 genotype M50 is prevalent in reservoir plants.
The distribution of MAP-FD2 genotypes is very heterogeneous in the various wine-growing regions (see Figure 2). Especially in the northern and northwestern viticultural areas of Ahr, Mosel-Saar-Ruwer and Mittelrhein, no or only very few infections with FDp (5 samples in total) were found. Similarly, only low FDp infestations were found in the north-eastern wine-growing area of Franconia. In contrast, very high levels of FDp infection (64–86%) are concentrated in viticultural areas along the rivers Rhine, Neckar and Kocher. Data for insect vectors in the Palatinate (Pfalz) region showed that Allygus spp. and O. ishidae were infected with 16SrV phytoplasmas to about 40% (Jarausch et al., 2019). These infected individuals carried the M38 genotype in 80% of Allygus spp. and 98% of O. ishidae. In particular, O. ishidae is a highly efficient vector of M38 to alder test plants (Malembic-Maher et al., 2020; Jarausch et al., 2023). Analyses with quantitative real-time PCR indicated that O. ishidae acquires the FDp by larval feeding on infected alder and emerging adults are already highly infective. The adults have a high phytoplasma load and remain infective throughout the entire flight season (Jarausch et al., 2023). O. ishidae was detected in Germany in different geographic places already in 2002 (Nickel and Remane, 2003). As no MAP-FD2 phytoplasmas have been found in alder in the Palatinate region before the invasion of O. ishidae (Maixner and Reinert, 1999), it can be assumed that the spread of M38 is related to the spread of O. ishidae. This species is also in other countries considered as risk for a FD spread as several reports showed that O. ishidae can be highly infected with MAP-FD1 and FD2 phytoplasmas (Mehle et al., 2010; Casati et al., 2017; Malembic-Maher et al., 2020; Rizzoli et al., 2021). That is why we have randomly captured insects in the studied wine-growing regions. The data provide indication that O. ishidae is present on alder in most of the regions. However, it was not found on alder in the Mosel region and in the control region of Allgäu, two regions were no M38-infections in alder could be recorded. All infected individuals of O. ishidae in all other wine-growing regions carried the M38 genotype while Allygus spp. were mainly infected with PGY genotypes (Jarausch et al., 2025b). This supports the hypothesis that the invasive spread of O. ishidae in German alder trees resulted in a widespread distribution of M38 genotypes in alders. This increases the risk for FDp transmission from alder to grapevine, especially in regions with a high MAP-FD2 prevalence as depicted in Figure 2.
Based on our results, thorough monitoring of FD-infested grapevines should now be carried out, especially in regions with high abundance of FDp in alders and high populations of O. ishidae. This could clarify the true importance of O. ishidae as a vector of FDp under natural conditions.
16SrV phytoplasma infections are commonly reported from black alder (Alnus glutinosa) (Malembic-Maher et al., 2020), which was also the dominant species in Germany. However, grey alder (Alnus incana) is the prevailing species in certain regions like Bodensee and Allgäu. This species was just as heavily infected with 16SrV phytoplasmas as A. glutinosa. Infected A. incana were also reported from Montenegro (Radonjić et al., 2013). Similar results for Italian alder (A. cordata) were recently reported by Marcone et al. (2024). The 16SrV phytoplasma-infection of a shrub of green alder (A. viridis) in a mountainous region of the Allgäu Alps (Table 6) is probably the first report for this species. Marjanovic et al. (2019) tested this species in Serbian mountains but could not confirm an infection. Interestingly, from northern regions it is reported that infected alder trees are highly loaded with 16SrV phytoplasmas but rarely show symptoms (Maixner and Reinert, 1999; Holz et al., 2015; Malembic-Maher et al., 2020) while symptom expression is frequently recorded from studies in southern countries (Cvrković et al., 2008; Radonjić et al., 2013; Atanasova et al., 2014; Marjanovic et al., 2019).
Our study has shown that FDp are widespread in alders near vineyards in Germany. It is almost exclusively the MAP-FD2 type M38, which can be transmitted from O. ishidae to grapevine and may be responsible for FD outbreaks. Our and other data suggest that the invasive species O. ishidae is responsible for the important spread of M38 in German alders. This leads to an increased risk of transmission of FDp to grapevine – especially as the FD vector S. titanus has now reached Germany. The data further suggest a different epidemiological situation in Germany and adjacent regions like Alsace with a wide distribution of alders and the spread of a specific MAP-FD2 type, M38. As part of our work, web-based risk maps were established that allow targeted monitoring of vineyards in the vicinity of alders in Rhineland-Palatinate and Baden-Wuerttemberg. Our data on the distribution of FDp in the various wine-growing regions also allow the responsible Plant Protection Services to focus on the most endangered areas.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
WJ: Conceptualization, Methodology, Project administration, Writing – review & editing. MR: Investigation, Methodology, Writing – original draft. TS: Data curation, Formal Analysis, Visualization, Writing – original draft. DG: Methodology, Software, Writing – original draft. BJ: Resources, Writing – review & editing. MT: Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This research was partly supported by funding from the European Regional Development Fund as part of the INTERREG V InvaProtect project Sustainable protection of plants against invasive pests, by funds of the Fonds für die Entwicklung ländlicher Räume in Rheinland-Pfalz, and by funds of the Federal Ministry of Food and Agriculture (BMEL) based on a decision of the Parliament of the Federal Republic of Germany via the Federal Office for Agriculture and Food (BLE) under the innovation support programme, project PhytoMo FKZ 2818718D19.
We gratefully acknowledge the contribution of Franziska Bischoff for the first field inspections and providing the first training data. We thank Sandra Biancu and Sanela Kugler for field collections and laboratory work with insect samples and Nora Schwind for maintaining micropropagated alder infected with genotype M38.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2025.1569408/full#supplementary-material
Angelini E., Clair D., Borgo M., Bertaccini A., Boudon-Padieu E. (2001). Flavescence dorée in France and Italy—Occurrence of closely related phytoplasma isolates and their near relationships to Palatinate grapevine yellows and an alder yellows phytoplasma. Vitis 40, 79–86. doi: 10.5073/vitis.2001.40.79-86
Angelini E., Squizzato F., Lucchetta G., Borgo M. (2004). Detection of a phytoplasma associated with grapevine Flavescence dorée in Clematis vitalba. Eur. J. Plant Pathol. 110, 193–201. doi: 10.1023/B:EJPP.0000015361.95661.37
Arnaud G., Malembic-Maher S., Salar P., Bonnet P., Maixner M., Marcone C., et al. (2007). Multilocus sequence typing confirms the close genetic interrelatedness of three distinct flavescence dorée phytoplasma strain clusters and group 16SrV phytoplasmas infecting grapevine and alder in Europe. Appl. Environ. Microbiol. 73, 4001–4010. doi: 10.1128/AEM.02323-06
Askani L., Zimmermann O., Zimmermann C., Rinke F., Jarausch B., Hoffmann C., et al. (2024). First report of Scaphoideus titanus (Hemiptera: Cicadellidae) in Germany (Baden-Württemberg). EPPO Bull. 2024, 1–3. doi: 10.1111/epp.13053
Atanasova B., Spasov D., Jakovljević M., Jović J., Krstić O., Mitrović M., et al. (2014). First report of alder yellows phytoplasma associated with common alder (Alnus glutinosa) in the Republic of Macedonia. Plant Dis. 98, 1268–1268. doi: 10.1094/PDIS-03-14-0315-PDN
Auriol A., Salar P., Pedemay S., Lusseau T., Desque D., Lacaze D., et al. (2023). Origin of isolated cases of “flavescence dorée” in North-East of France: search for reservoir plants and insect vectors in semi-natural habitats near vineyards. Phytopathog. Mollicutes 13, 45–46. doi: 10.5958/2249-4677.2023.00023.3
Blaschke T. (2010). Object based image analysis for remote sensing. ISPRS J. Photogramm. Remote Sens. 65, 2–16. doi: 10.1016/j.isprsjprs.2009.06.004
Casati P., Jermini M., Quaglino F., Corbani G., Schaerer S., Passera A., et al. (2017). New insights on flavescence dorée phytoplasma ecology in the vineyard agro-ecosystem in southern Switzerland. Ann. Appl. Biol. 171, 37–51. doi: 10.1111/aab.12359
Chuche J., Thiéry D. (2014). Biology and ecology of the Flavescence dorée vector Scaphoideus titanus: A review. Agron. Sustain. Dev. 34, 381–403. doi: 10.1007/s13593-014-0208-7
Ciccotti A. M., Bisognin C., Battocletti I., Salvadori A., Herdemertens M., Jarausch W. (2008). Micropropagation of apple proliferation-resistant apomictic Malus sieboldii genotypes. Agron. Res. 6, 445–458.
Cvrković T., Jović J., Mitrović M., Petrović A., Krnjajić S., Malembic-Maher S., et al. (2008). First report of alder yellows phytoplasma on common alder (Alnus glutinosa) in Serbia. Plant Pathol. 57, 773. doi: 10.1111/j.1365-3059.2008.01880.x
EPPO (2015). Orientus ishidae: A potential phytoplasma vector spreading in the EPPO region. EPPO Rep. Serv. 5, 8–10.
ESRI (2024). ArcGIS Pro (Version 3.4.0) (Redlands, CA: Environmental Systems Research Institute). Available at: https://www.esri.com/de-de/arcgis/products/arcgis-pro/. (Accesed August 30, 2024).
Gaffuri F., Sacchi S., Cavagna B. (2011). First detection of the mosaic leafhopper, Orientus ishidae, in northern Italian vineyards infected by the flavescence dorée phytoplasma. New Dis. Rep. 24, 22. doi: 10.5197/j.2044-0588.2011.024.022
Gonella E., Benelli G., Arricau-Bouvery N., Bosco D., Duso C., Dietrich C., et al. (2024). Scaphoideus titanus up-to-the-minute: biology, ecology, and role as a vector. Entomol. Generalis 44, 481–496. doi: 10.1127/entomologia/2023/2597
Holz S., Duduk B., Büttner C., Kube M. (2015). Genetic variability of Alder yellows phytoplasma in Alnus glutinosa in its natural Spreewald habitat. For. Pathol. 46, 11–21. doi: 10.1111/efp.2016.46.issue-1
Jarausch B., Askani L., Kugler S., Jarausch W., Fuchs R., Zimmermann O., et al. (2025a). A new invader in Germany: Scaphoideus titanus, the vector of “flavescence dorée” in grapevine. Phytopathog. Mollicutes 15, 43–44. doi: 10.5958/2249-4677.2025.00026.6
Jarausch B., Biancu S., Kugler S., Jarausch W., Maixner M. (2025b). Monitoring of alternative insect vectors of Flavescence dorée-related phytoplasmas in different wine-growing regions in Germany. Phytopathog. Mollicutes 15, 45–46. doi: 10.5958/2249-4677.2025.00027.2
Jarausch B., Biancu S., Kugler S., Wetzel T., Baumann M., Winterhagen P., et al. (2021). First report of Flavescence dorée -related phytoplasma in a productive vineyard in Germany. Plant Dis. 105, 3285. doi: 10.1094/PDIS-02-21-0330-PDN
Jarausch B., Biancu S., Lang F., Desqué D., Salar P., Jarausch W., et al. (2019). Study of the epidemiology of Flavescence dorée (FD)-related phytoplasmas and potential vectors in a FD-free area. Phytopathog. Mollicutes 9, 59–60. doi: 10.5958/2249-4677.2019.00030.6
Jarausch B., Markheiser A., Jarausch W., Biancu S., Kugler S., Runne M., et al. (2023). Risk assessment for the spread of Flavescence dorée-related phytoplasmas from alder to grapevine by alternative insect vectors in Germany. Microorganisms 11, 2766. doi: 10.3390/microorganisms11112766
Jarausch B., Schwind N., Fuchs A., Jarausch W. (2011). Characteristics of the spread of apple proliferation by its vector Cacopsylla picta. Phytopathology 101, 1471–1480. doi: 10.1094/PHYTO-01-11-0012
Jermini M., Conedera M., Rizzoli A., Belgeri E., Filippin L., Angelini E. (2019). Potential role of Orientus ishidae in the “flavescence dorée” epidemics in Ticino, Switzerland. Phytopathog. Mollicutes 9, 67–68. doi: 10.5958/2249-4677.2019.00034.3
Kogej Zwitter Z., Seljak G., Jakomin T., Brodarič J., Vučurović A., Pedemay S., et al. (2023). Epidemiology of flavescence dorée and hazelnut decline in Slovenia: geographical distribution and genetic diversity of the associated 16SrV phytoplasmas. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1217425
Krstić O., Cvrković T., Marinković S., Jakovljević M., Mitrović M., Toševski I., et al. (2022). Genetic diversity of flavescence dorée phytoplasmas in vineyards of Serbia: from the widespread occurrence of autochthonous Map-M51 to the emergence of endemic map-FD2 (Vectotype II) and new map-FD3 (Vectotype III) epidemic genotypes. Agronomy 12, 448. doi: 10.3390/agronomy12020448
Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lessio F., Bocca F., Alma A. (2019). Development, spatial distribution, and presence on grapevine of nymphs of Orientus ishidae (Hemiptera: cicadellidae), a new vector of flavescence dorée phytoplasmas. J. Econ. Entomol. 112, 2558–2564. doi: 10.1093/jee/toz177
Lessio F., Picciau L., Gonella E., Mandrioli M., Tota F., Alma A. (2016). The mosaic leafhopper orientus ishidae: host plants, spatial distribution, infectivity, and transmission of 16SrV phytoplasmas to vines. Bull. Insectol. 69, 277–289.
Maixner M., Reinert W. (1999). Oncopsis alni (Schrank) (Auchenorrhyncha: Cicadellidae) as a vector of the alder yellows phytoplasma of Alnus glutinosa (L.) Gaertn. Eur. J. Plant Pathol. 105, 87–94. doi: 10.1023/A:1008602327715
Maixner M., Reinert W., Darimont H. (2000). Transmission of grapevine yellows by Oncopsis alni (Schrank). Vitis 39, 83–84. doi: 10.5073/vitis.2000.39.83-84
Malembic S., Salar P., Vergnes D., Foissac X. (2007). Detection and diversity of “flavescence dorée”-related phytoplasmas in alders surrounding infected vineyards in Aquitaine (France). Bull. Insectol. 60, 329–330.
Malembic-Maher S., Desque D., Khalil D., Salar P., Danet J. L., Duret M.-S., et al. (2020). When a Palearctic bacterium meets a Nearctic insect vector: genetic and ecological insights into the emergence of the grapevine flavescence dorée epidemics in Europe. PloS Pathog. 16, e 1007967. doi: 10.1371/journal.ppat.1007967
Marcone C., Pierro R., Palmieri C. (2024). Occurrence, impact, and multilocus sequence analysis of alder yellows phytoplasma infecting common alder and italian alder in Southern Italy. Microorganisms 12, 1140. doi: 10.3390/microorganisms12061140
Marcone C., Ragozzino A., Schneider B., Lauer U., Smart C. D., Seemüller E. (1996). Genetic characterization and classification of two phytoplasmas associated with Spartium Witches’-Broom disease. Plant Dis. 80, 365–371. doi: 10.1094/PD-80-0365
Marjanovic M., Stepanovic J., Rekanovic E., Kube M., Duduk B. (2019). Alder yellows phytoplasmas in Alnus species in Serbia. Phytopathog. Mollicutes 9, 57–58. doi: 10.5958/2249-4677.2019.00029.X
Mehle N., Jakoš N., Mešl M., Miklavc J., Matko B., Rot M., et al. (2019). Phytoplasmas associated with declining of hazelnut (Corylus avellana) in Slovenia. Eur. J. Plant Pathol. 155, 1117–1132. doi: 10.1007/s10658-019-01839-3
Mehle N., Seljak G., Rupar M., Ravnikar M., Dermastia M. (2010). The first detection of a phytoplasma from the 16SrV (Elm yellows) group in the mosaic leafhopper Orientus ishidae. New Dis. Rep. 22, 11–11. doi: 10.5197/j.2044-0588.2010.022.011
Nickel H., Remane R. (2003). Verzeichnis der Zikaden (Auchenorrhyncha) der Bundesländer Deutschlands. Entomofauna Germanica 6, 130–154.
Plavec J., Budinšćak Ž., Križanac I., Škorić D., Foissac X., Šeruga Musić M. (2019). Multilocus sequence typing reveals the presence of three distinct Flavescence dorée phytoplasma genetic clusters in Croatian vineyards. Plant Pathol. 68, 18–30. doi: 10.1111/ppa.2019.68.issue-1
Radonjić S., Hrnčić S., Krstić O., Cvrković T., Mitrović M., Jović J., et al. (2013). First report of alder yellows phytoplasma infecting common and grey alder (Alnus glutinosa and A. incana) in Montenegro. Plant Dis. 97, 686–686. doi: 10.1094/PDIS-11-12-1087-PDN
Radonjić S., Krstić O., Cvrković T., Hrnčić S., Marinković S., Mitrović M., et al. (2023). The first report on the occurrence of Flavescence dorée phytoplasma affecting grapevine in vineyards of Montenegro and an overview of epidemic genotypes in natural plant reservoirs. J. Plant Pathol. 105, 419–427. doi: 10.1007/s42161-023-01318-z
R Core Team (2024). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing). Available at: https://www.R-project.org/. (Accesed August 30, 2024).
Rigamonti I. E., Salvetti M., Girgenti P., Bianco P. A., Quaglino F. (2023). Investigation on Flavescence dorée in North-Western Italy identifies Map-M54 (16SrV-D/Map-FD2) as the only phytoplasma genotype in Vitis vinifera L. and reveals the presence of new putative reservoir plants. Biology 12, 1216. doi: 10.3390/biology12091216
Rizzoli A., Belgeri E., Jermini M., Conedera M., Filippin L., Angelini E. (2021). Alnus glutinosa and Orientus ishidae (Matsumura 1902) share phytoplasma genotypes linked to the ‘Flavescence dorée’ epidemics. J. Appl. Entomol. 145, 1015–1028. doi: 10.1111/jen.12933
Thielert B., Menz P., Götte G., Runne M., Michel M., Wagner S., et al. (2024). PhenoTruckAI: on-Site Hyperspectral Measurement for Distinction of Quarantine Grapevine Disease “flavescence dorée” and non-Quarantine Disease “bois noir” in a Mobile Laboratory. INFORMATIK 2024, 1253–1260. doi: 10.18420/inf2024_110
Keywords: Alnus glutinosa, Flavescence dorée, geodata analysis, map gene, Orientus ishidae, PCR screening assay, phytoplasma, risk assessment
Citation: Jarausch W, Runne M, Schell T, Guerniche D, Jarausch B and Trapp M (2025) Analysis of the spread of Flavescence dorée-related phytoplasmas in naturally infected alder in Germany based on molecular and geodata. Front. Agron. 7:1569408. doi: 10.3389/fagro.2025.1569408
Received: 31 January 2025; Accepted: 18 March 2025;
Published: 02 April 2025.
Edited by:
David Ezra, Agricultural Research Organization (ARO), IsraelReviewed by:
Domenico Bosco, University of Turin, ItalyCopyright © 2025 Jarausch, Runne, Schell, Guerniche, Jarausch and Trapp. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wolfgang Jarausch, d29sZmdhbmcuamFyYXVzY2hAYWdyb3NjaWVuY2UucmxwLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.