- 1Instituto Nacional de Tecnología Agropecuaria (INTA), Estación Experimental Agropecuaria (EEA) Marcos Juárez, Córdoba, Argentina
- 2Soil, Water and Crop Management Group, Estación Experimental Agropecuaria (EEA) Oliveros Instituto Nacional de Tecnología Agropecuaria (INTA), Oliveros, Santa Fe, Argentina
- 3Instituto de Agrobiotecnología y Biología Molecular (IABIMO) Conicet-Instituto Nacional de Tecnología Agropecuaria (INTA), Buenos Aires, Argentina
Introduction: The ecosystem services provided by soil microbial communities are critical for the resilience of agroecosystems, ensuring environmental conservation and food security. Long-term experiments comparing contrasting crop rotations are valuable tools for monitoring microbial responses, but they rarely include all crop phases within a single year. Therefore, the long-term agronomic impact may be masked by the immediate effect of the crop evaluated. In this study, we compared different crop rotations based on the Intensification Sequence Index (ISI), which considers the soil occupation time, and analyzed the impact of cover crops and two nitrogen fertilization strategies.
Material and methods: We used an experiment initiated in 2006 with the following crop rotations: Soybean-Soybean, Maize-Wheat/Soybean, and Maize-Soybean-Wheat/Soybean. Soil samples were taken after the harvest of each summer crop phase (i.e., Soybean, Maize, Wheat/Soybean), and the soil prokaryotic community was monitored using 16S rRNA gene sequencing.
Results and discussion: We observed that ISI and crop phase were the main predictors of microbial community composition, explaining 14.7% and 13.0% of the variation, respectively. Nitrogen fertilization had a minor effect (3.12%) and was detected only after maize sampling; cover crops had no significant effect. However, the presence of cover crops showed higher alpha diversity and an increased abundance of Proteobacteria. Maize enriched the abundance of certain taxa of Planctomycetes and Verrucomicrobia, while Soybean increased the abundance of Bacteroidetes and Proteobacteria. Comparatively, Soybean enriched Mucilaginibacter and Geobacter, while Wheat/Soybean enriched Brevundimonas and Roseimicrobium.
Conclusion: Our results demonstrate that crop phase is as important as the long-term legacy of crop rotations in shaping the microbial community and that specific taxa responses are highly dependent on the crop phase surveyed.
1 Introduction
Soil is a highly complex biological system that provides essential ecosystem services, such as habitat and support for life, regulation of climate, nutrients, and water, as well as the production of food, fuel, and fiber (Jónsson and Davíðsdóttir, 2016). The soil’s biological compartment, particularly the microbiome (soil-borne microorganisms), has received considerable attention in recent years (Hermans et al., 2020). Soil microbial communities can improve soil health by storing carbon (C), regulating nutrient cycling, and helping to maintain or restore soil structure (Garnica et al., 2020; Kibblewhite et al., 2008). The soil microbiome can be influenced by biotic and abiotic factors, including soil management and plant identity (Custódio et al., 2022; Chaparro et al., 2012; Hartman et al., 2018). At the same time, soil-plant interactions moderated by soil microbes may affect plant health, including plant growth, plant-pathogens association, and nutrient-use efficiency (Finzi et al., 2014; van der Heijden et al., 2008). Understanding the continuous soil-microbiome-crop linkages is essential to creating sustainable agroecosystems.
Modern agriculture is under pressure to produce more food to meet the future increase in global demand in a scenario of resource scarcity and climate change (Tilman et al., 2001). Argentina is a major global supplier of soybean, maize, and wheat (FAO, 2021). Across the fertile Argentinean pampas, the predominance of soybean (Glycine max L.) monoculture, widespread overuse of pesticides, and conspicuous nutrient exports with grains exceeding N inputs have challenged sustainability in the last twenty years (Mazzilli et al., 2015). Previous reports have demonstrated that unsustainable practices reduce biodiversity with significant losses of endemic microbial species (Figuerola et al., 2012). Such impacts on the soil microbiome can potentially lead to adverse consequences for ecosystem services, crop yields, and overall soil health, thereby impacting global food security (Dubey et al., 2019). Therefore, sustainable practices are necessary to ensure the ecological resilience of agroecosystems (Miner et al., 2020). These practices include the use of no-tillage for reducing soil disturbance, crop rotations that alternate between legumes and cereal crops, the inclusion of cover crops between cash crops, and an optimal reposition of nutrients that are exported with grains from the agroecosystems (Gaudin et al., 2015; Kleijn et al., 2019; Lupwayi et al., 2018; McDaniel et al., 2014; Novelli et al., 2017).
Simultaneously achieving high crop yields while preserving soil health forms the basis of the sustainable intensification (SI) paradigm (Pretty and Bharucha, 2014). This paradigm is supported by ecological principles that advocate more intensive and efficient use of environmental resources, including water, solar radiation, and nutrients, with the ultimate aim of enhancing overall land productivity (Robledo et al., 2024). Agricultural practices within this framework are strategically designed to extend land cover periods and shorten fallow phases. This is achieved through measures such as increasing the frequency of cereals in crop rotation through double cropping or using cover crops (Andrade et al., 2017; Caviglia and Andrade, 2010). The optimization of crop nutrition is achieved by complementing the application of inorganic fertilizers and additional nutrient sources such as leguminous crops or organic manure (Correndo et al., 2021; Enrico et al., 2020; Miner et al., 2020). The main beneficial effects of SI are higher crop yields (Andrade et al., 2017; Cano et al., 2023; Stefan et al., 2021), increased C stocks in stable fractions (Rodríguez et al., 2020; Romaniuk et al., 2018), and reduced greenhouse gas emissions (Piccinetti et al., 2021). However, there is a critical balance between the need to increase the frequency of cereals in crop sequences to enhance SI benefits and a greater reliance on N fertilization, with the deleterious environmental impact of N in cereal-rich agroecosystems.
Sustainable intensification practices affect the soil microbiome directly and indirectly, resulting in divergent ecological responses of bacterial groups (Frene et al., 2022). For example, the increase of living cover and crop diversity can generate a constant input of C via rhizodeposition and favor certain copiotrofic groups, such as Proteobacteria and Bacteroidetes, with an either negative or positive impact on soil C storage and soil health indicators (Agomoh et al., 2020; Frene et al., 2022; Li et al., 2014; Novelli et al., 2011). Actinobacteria, a phylum characterized by several plant-growth-promoting members, can be augmented in more diverse crop rotations with a positive impact on crop productivity (Stefan et al., 2021). Notably, crop species identity can be a major driver of current soil microbial communities due to the chemical composition of plant tissues, and root exudates, as well as crop-specific requirements of agronomical practices (Fang et al., 2022; Fox et al., 2020; Sasse et al., 2018). For example, cereal crops require high inputs of N fertilization and produce large amounts of residues with high C:N ratios, while legumes can partially satisfy N requirement through biological fixation and produce small amounts of residue with low C:N ratios. Acidobacteria might prevail after cereal cultivation, whereas Actinobacteria and Proteobacteria may dominate after legumes (Chamberlain et al., 2020; Feng et al., 2017; Stefan et al., 2021). Also, cereal N fertilization induces changes in soil pH and alters the abundance of key bacterial and archaeal species involved in soil N-cycling, particularly in no-tillage systems (Viso et al., 2024).
Understanding the interconnection between soil microbiome and agronomical practices is imperative for establishing resilient, productive, and sustainable agroecosystems (Wall et al., 2019). The ratio between cereals and legumes will determine the net input of N (Novelli et al., 2011) and both crop rotations (Frene et al., 2022; Nivelle et al., 2016; Pinto et al., 2017), and extra N supplementation (Viso et al., 2024; Zhang et al., 2021) have measurable impacts on the soil microbiome. We then investigate the extent to which N fertilization and the inclusion of cover crops modified prokaryotic communities in crop sequences with different land occupations. However, in crop rotation analysis, it is unclear whether the effects on soil biota are a consequence of the net effect of long-term rotation or the previous crop in a particular rotation (Fox et al., 2020; Ishaq et al., 2017). To avoid this confounding effect, the evaluation of all phases of a rotation (i.e., all previous crops that are part of this rotation are evaluated simultaneously) is recommended so that the response of a particular crop to a given rotation can be compared each year. In this study, we compared the long-term (13 years) effect of sequences with different land occupations, including all the cash crops on the crop rotation. We hypothesized that 1) the crop phase sampling would have a stronger impact on the prokaryotic diversity than the crop rotational history; 2) given a particular crop sequence, the cereal N fertilization and the use of winter cover crops would have a minor effect on the soil microbiome in comparison with the long-term crop rotational management.
2 Materials and methods
2.1 Study site and experimental design
The experiment was performed at INTA Oliveros Research Station (32° 32’ S, 60° 51’ W) Argentina, on a deep, well-drained, carbonates-free Typic Argiudoll (USDA Soil Taxonomy) up to a depth of 240 cm, with a silty loam texture in the surface horizon (clay 209 g kg−1, silt 708 g kg−1, sand 83 g kg−1) and predominant of illite in the clay fraction, pH 6 (1/2.5 soil/water), C.E.C. 19.4 cmolc kg−1. The climate is humid temperate with a mean annual temperature of 17.6°C and a mean annual rainfall of 1042 mm. Rainfall occurs mainly in fall and spring, while the summer months usually present deficits of varying intensity in the agro-climatic balance.
A long-term no-till experiment was established in 2006 to evaluate crop sequences with different intensities of cereal and soybean frequencies. The area has been under cultivation for the last 50 years and under no-tillage from the last 8 years before the beginning of the experiment. The experiment was arranged in a randomized complete block design with three replicates, with each experimental unit being 13 m wide × 50 m long. Treatments consisted of a combination of crop sequences and N fertilization levels in cereals. Each phase, with its respective intensification practice (N fertilization or cover crop treatments), was cultivated concomitantly every year. This distinctive approach sets this study apart from others in the field.
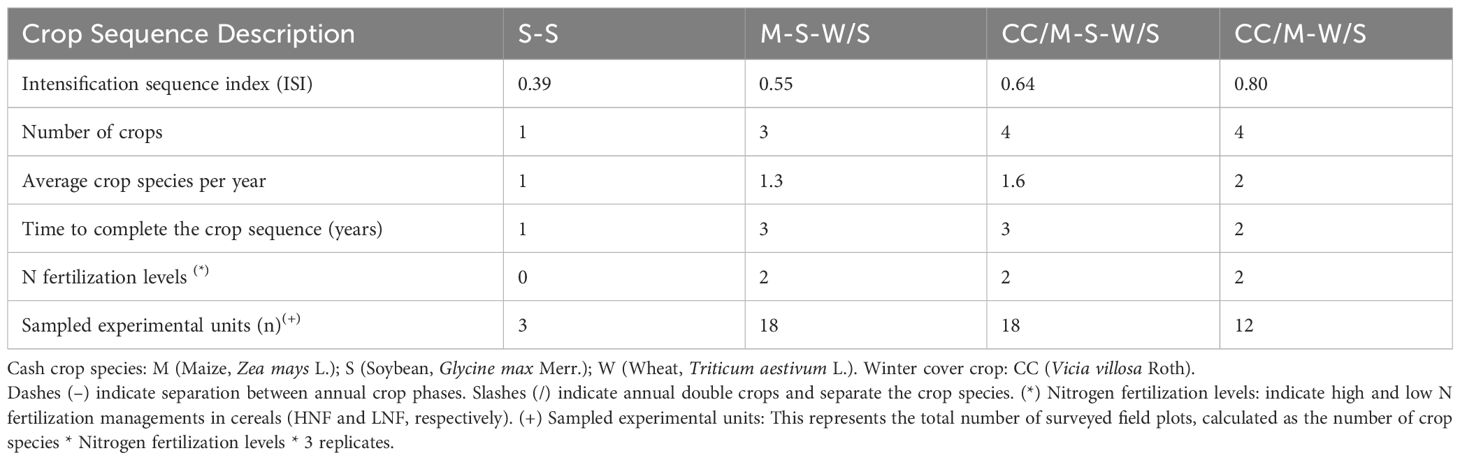
Crop sequences were as follows: (i) soybean monoculture (S-S): ii) Maize-Wheat/Soybean (M-W/S) and iii) Maize-Soybean-Wheat/Soybean (M-S-W/S). In both (ii) and (iii) the inclusion of a winter cover crop (CC) was tested: iv) winter cover crop/Maize-Wheat/Soybean (CC/M-W/S) and v) - maize- winter cover crop/soybean–wheat/soybean (M-CC/S-W/S) and winter cover crop/Maize-Soybean-Wheat/Soybean (CC/M-S-T/S). Winter cover crops consisted of hairy vetch (Vicia villosa) planted before maize and killed at mid-bloom stage, with 3 L ha-1 of glyphosate (48% active principle).
Two N fertilizer rates were evaluated in the sequences that included cereals (wheat or maize). These N fertilizer rates were calculated by measuring soil N content at planting as N-NO3 at a depth of 60 cm, which was subtracted from a threshold of available N (N-NO3 at 60 + fertilizer N) set for two-grain yield goals: a threshold of 160 and 190 kg N ha-1 for grain yields above and below 10000 kg ha-1 in maize (Salvagiotti et al., 2011; Correndo et al., 2021) and a threshold of 92 and 135 kg N ha-1 for grain yields above and below 5000 kg ha-1 in wheat (Salvagiotti et al., 2004; Ferrari et al., 2010). The sequences that received the low and the high N fertilizer rates in cereals are now identified as LNF and HNF, respectively. Therefore, the experiment had in total 4 sequences that included cereals combined with 2 N fertilizer recommendations and a soybean monoculture as a control.
The general management was carried out as usual by farmers in the region for pest and weed control. All phases of each rotation were present every year. At sowing, soybean was inoculated with Bradyrhizobium sp. and fertilized with 70 kg ha−1 of triple super phosphate. Maize and wheat were fertilized at sowing with 80 kg ha−1 of mono ammonium phosphate. Cover crops did not receive fertilization.
The intensification sequence index (ISI), which expresses the relative number of days of the year occupied by crops in a given crop sequence, was calculated as the ratio between the number of days with crops in each crop sequence and the length of the sequence (Caviglia and Andrade, 2010) Taking into account the real-time occupied by crops in each sequence, the following ISIs were established: 0.39, 0.65, 0.55, 0.8, and 0.64 for S-S, M-W/S, M-S-W/S, CC/M-W/S, and M-CC-S-W/S, respectively (Figure 1; Table 1).
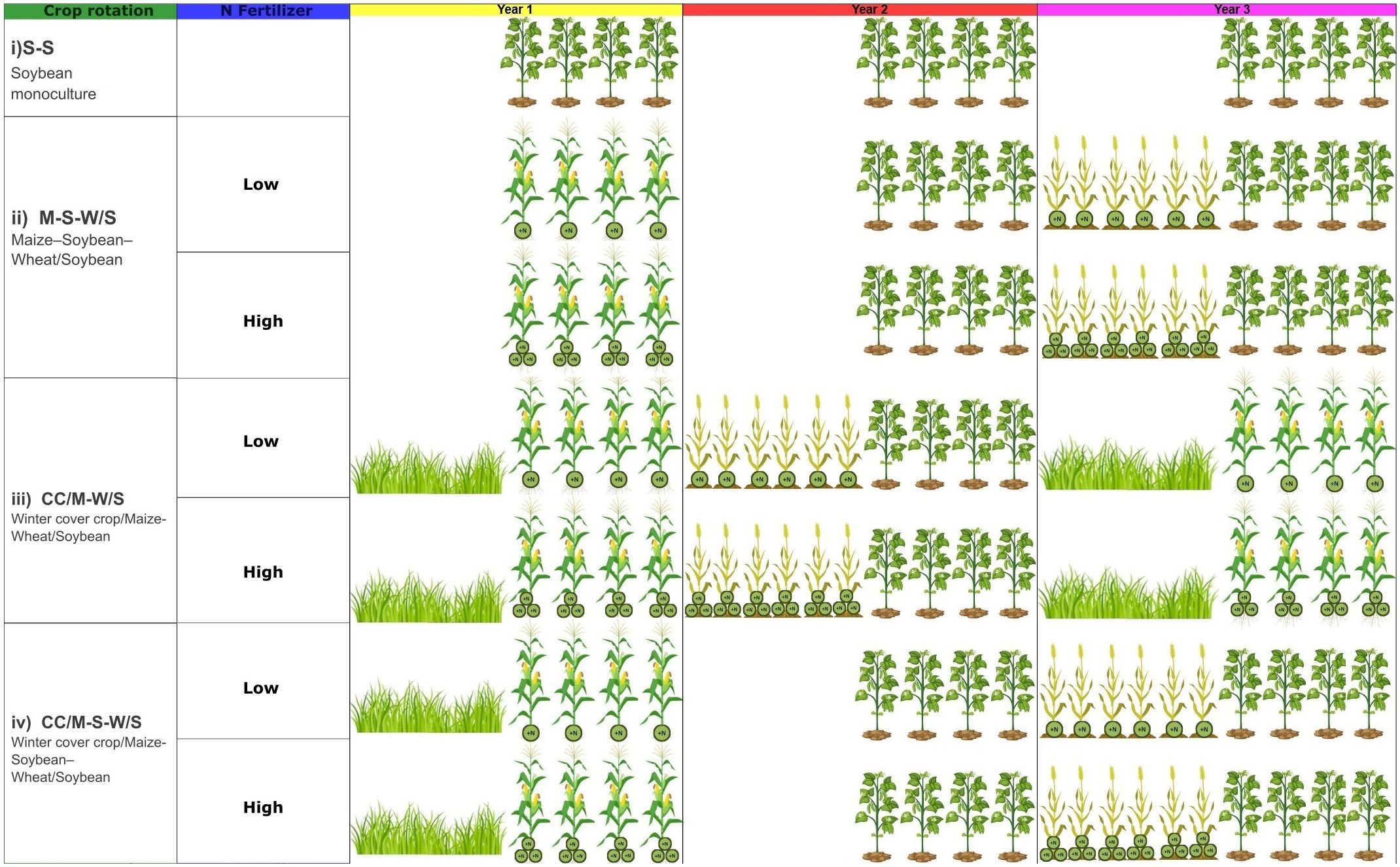
Figure 1. Crop sequence scheme. In the presence of cereals, two levels of nitrogen, low and high, were applied to the crop rotation (LNF and HNF, respectively). The experiment started in 2006, and soil sampling was performed after three complete crop rotational cycles in 2019 after the harvest of summer crops: Maize, Soybean, Wheat/Soybean. Since the longest crop sequence is three years in duration, the graphical scheme spans three years.
2.2 Soil sampling
In May 2019 (after the 2018-19 cropping season ended), soil samples were taken at 0–5 cm depth and replicated three times in each experimental unit. The soil samples were taken from the summer cash crop in each phase of the crop rotations, so after the harvest, all the crops in each rotation were sampled. Therefore, the cumulative effect of each rotation was determined for the summer cash crops (maize, soybean, and soybean from the wheat/soybean double-crop phase) (Figure 1). Hereafter, we use the terms ‘crop phase’ and ‘crop identity’ indistinctly to refer to the three summer cash crops mentioned above. Subsamples were mixed and homogenized in the field, air-dried, and sieved to pass a 2 mm screen. A subset of soil samples was stored at field moisture at -20°C until analysis for microbial measurements.
2.3 DNA extraction and sequencing analysis
DNA Extraction was performed using a Power soil kit (MO BIO Laboratories, Inc. Carlsbad, CA) according to the manufacturer’s instructions. The extracted DNA was sent to the European Molecular Biology Laboratory (EMBL, Heidelberg, Germany) for sequencing in 2019. The V4 region of the 16S rRNA gene, which targets bacterial and archaeal taxa, was amplified using primers 515F (Parada et al., 2016) and 806R (Apprill et al., 2015) and the Illumina MiSeq platform. The raw reads are available at the ENA under the project number PRJEB33480. They are also publicly available at the MGnify website (https://www.ebi.ac.uk/metagenomics/) MGYS00005093.
2.4 Bioinformatics analysis
Bioinformatics processing of reads was performed using QIIME 2 version 2022.2 (Bolyen et al., 2019), Illumina adapters and primers were removed with the ‘cutadapt’ function. Next, denoising processing was performed using DADA2 in the QIIME 2 pipeline (Callahan et al., 2016). Reads with more than 25 score quality and fragments larger than 100 bp were retained and ASVs (amplicon sequence variants) were generated. The taxonomic assignment of each ASV was annotated using the q2-classify-sklearn module using the SILVA database version 13 (Quast et al., 2013), trained for the V4 16S rRNA region, 515F-806R (Bokulich et al., 2018).
2.5 Statistical analysis
Data were analyzed and plotted in R using the stat, agricolae, vegan, phyloseq, DESeq2, and ggplot2 packages (R Development Core Team, 2022). Before microbial data analysis, each sample was rarefied to the minimum number of reads to correct for sampling effort using a subset of bacterial sequences. The Shannon diversity index was estimated using rarefied ASV data. The Poisson regression was used to model the relationship between ISI and Shannon index and the relative abundance of dominant phyla. Distance matrices were created at the ASV level using Bray-Curtis dissimilarity, and Principal Coordinate Analysis (PCoA) ordinations were constructed for these matrices. Treatment effects on β-diversity were determined using permutational analysis of variance (PERMANOVA) with the function adonis (Oksanen et al., 2015). Multiple comparisons using PERMANOVA and Bonferroni P-adjustment were used to determine differences in community composition between contrasts of interest. To identify those taxa associated with a particular crop rotation, we fitted a general linear model (GLM) based on the negative binomial distribution using the DESeq function of the DESeq2 package in R (Love et al., 2014). DESeq2 identified the taxa (ASVs) that responded to each treatment and which taxa were enriched in each treatment. We used phyloseq version 1.34.2 (McMurdie and Holmes, 2013) to visualize the relative abundance of the taxa driving the compositional differences between treatments. To assess the effect of intensification (ISI) on bacterial phyla, we used a GLM based on the Poisson distribution with the plot as a random effect. When the data were overdispersed and the conditional variance was higher than the conditional mean (i.e., phylum abundance) (Bliss, 1953; Ross and Preece, 1985), we performed a negative binomial regression, with the plot as a random effect for each phylum abundance, using the MASS package (Venables and Ripley, 2012).
3 Results
3.1 Whole community analysis in response to crop identity, intensification, and N fertilization
The PERMANOVA analysis showed that ISI (P = 0.001) and crop identity (P = 0.001) were the strongest predictors of the composition of prokaryotic communities, followed by N fertilization (P = 0.028), cover crops were not statistically significant (Supplementary Table 1). There was no significant interaction between the treatments (Supplementary Table 1; Figure 2A). The ISI accounted for 14.7% of the prokaryotic community variation, while the crop identity accounted for 13.0% of the variation. There was no interaction between the ISI and the crop phase. Finally, N fertilization and cover crops accounted for 3.12% and 2.33%, respectively (Supplementary Table 1; Figure 2A). Principal coordinate analysis (PCoA) ordinations of Bray-Curtis dissimilarity for soil prokaryotic community structure indicated that the crop phases were distributed along PCoA axis 2, with both Soybean and Soybean from the Wheat/Soybean double-crop situated on the positive values and Maize located on the negative values of axis 2 (Figure 2B). Notably, it is possible to observe the intensification effect on prokaryotic community composition along PCoA axis 1, with less intensified samples positioned on the negative values and more intensified samples positioned on the positive values (Figure 2B). Non significant association between PCoA axis 1 and ISI was found on Maize, but significant relations were shown on Soybean and Wheat/Soybean phases (Figures 2C–E). In contrast, there was no effect for the cover crops (Supplementary Figure 1A) and N fertilization had a strong effect on the Maize phase but not on the Soybean and Wheat/Soybean phases (Supplementary Figure 1B; Supplementary Table 2).
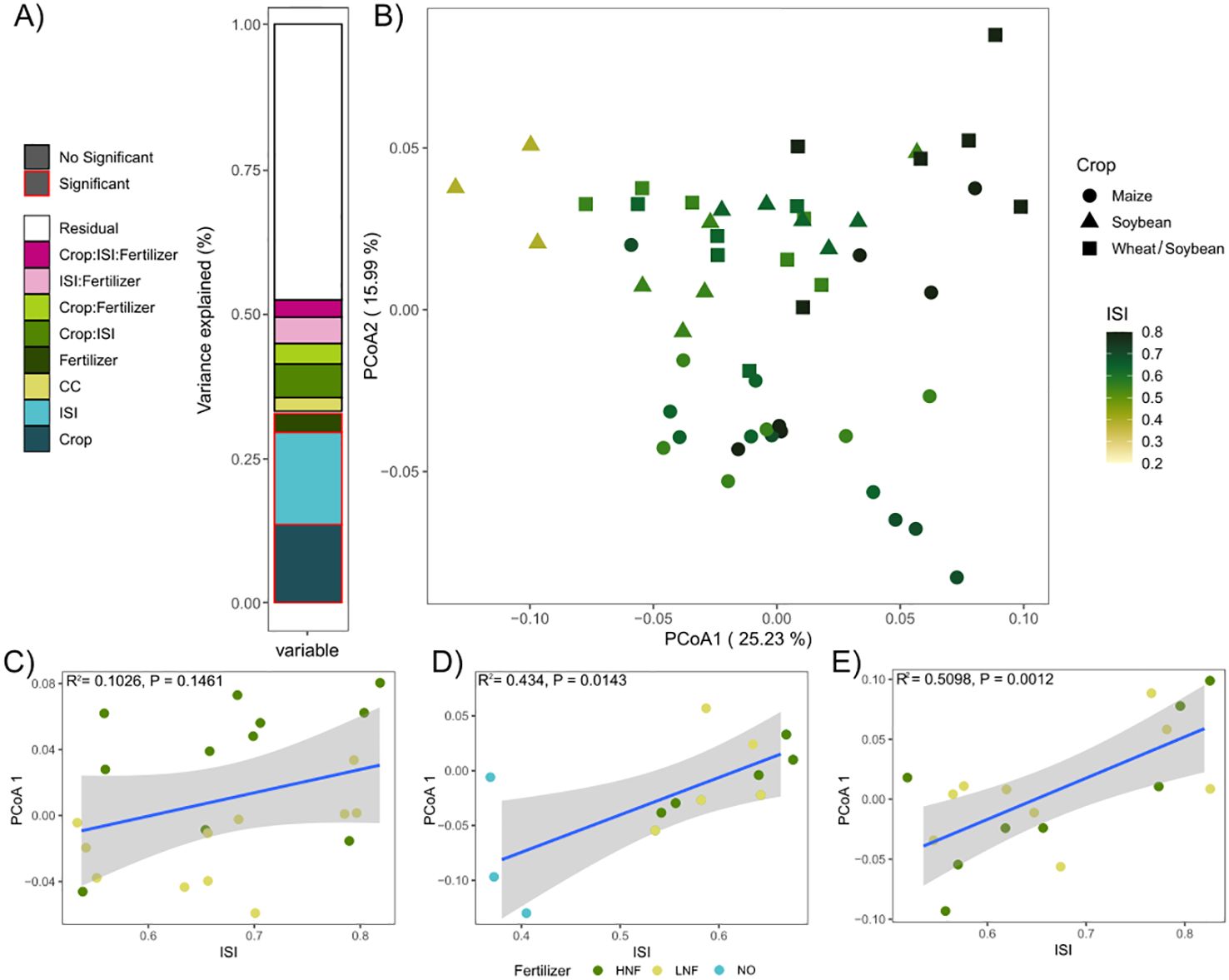
Figure 2. (A) Variance analyzed by PERMANOVA test. (B) Principal Coordinate Analyses (PCoA) ordinations of Bray-Curtis dissimilarity for soil prokaryotic community composition. The PCoA plot shows the distribution pattern of microbial communities among different crops and for Intensification Sequence Index (ISI). Relationship between ISI and PCoA1 axis in different crop phases (C) Maize, (D) Soybean, and (E) Wheat/Soybean; colored by fertilization level.
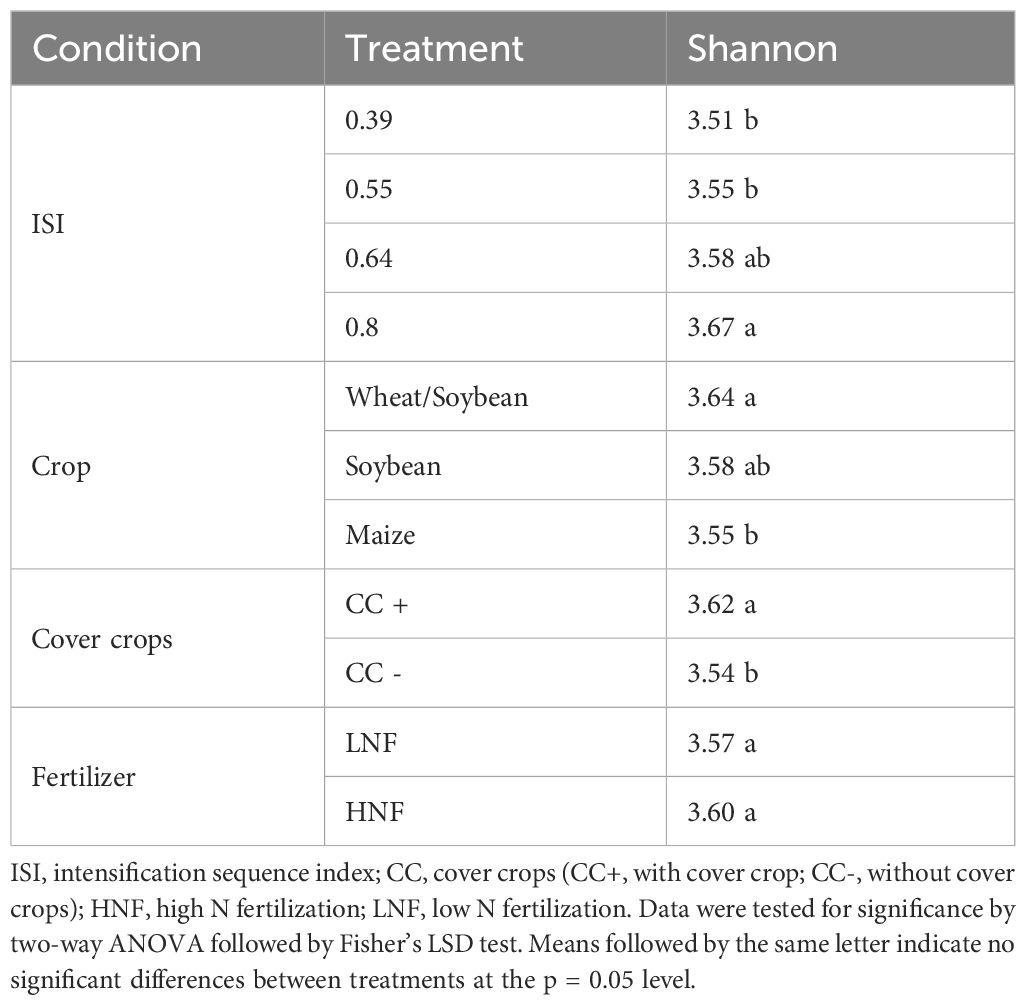
3.2 Prokaryotic α-diversity in response to crop intensification
The α-diversity for soil bacterial/archaeal community based on the Shannon index showed differences for the crop phase (P < 0.05) and cover crops (P < 0.01) (Table 2) but not for N fertilization (P > 0.05) (Table 2). The Wheat/Soybean double crops averaged 3.64 and were significantly higher than Maize. When all rotations were averaged, the inclusion of cover crops increased the α-diversity by 10% in the three crops (Figure 3, inset). Finally, Shannon diversity increased with intensification (ISI) for Maize (P = 0.039) and Wheat/Soybean (P = 0.005) (Figure 3). The linear regression model between the ISI value and the α-diversity showed a significant correlation (R2 = 0.226, P < 0.001) (Figure 3). In particular, we found that ISI value and the α-diversity were significantly correlated in Soybean (R2 = 0.30, P = 0.002) and the Wheat/Soybean double crop (R2 = 0.24, P = 0.002), but not in the Maize crop (R2 = 0.05, P = 0.162) (Figure 3).
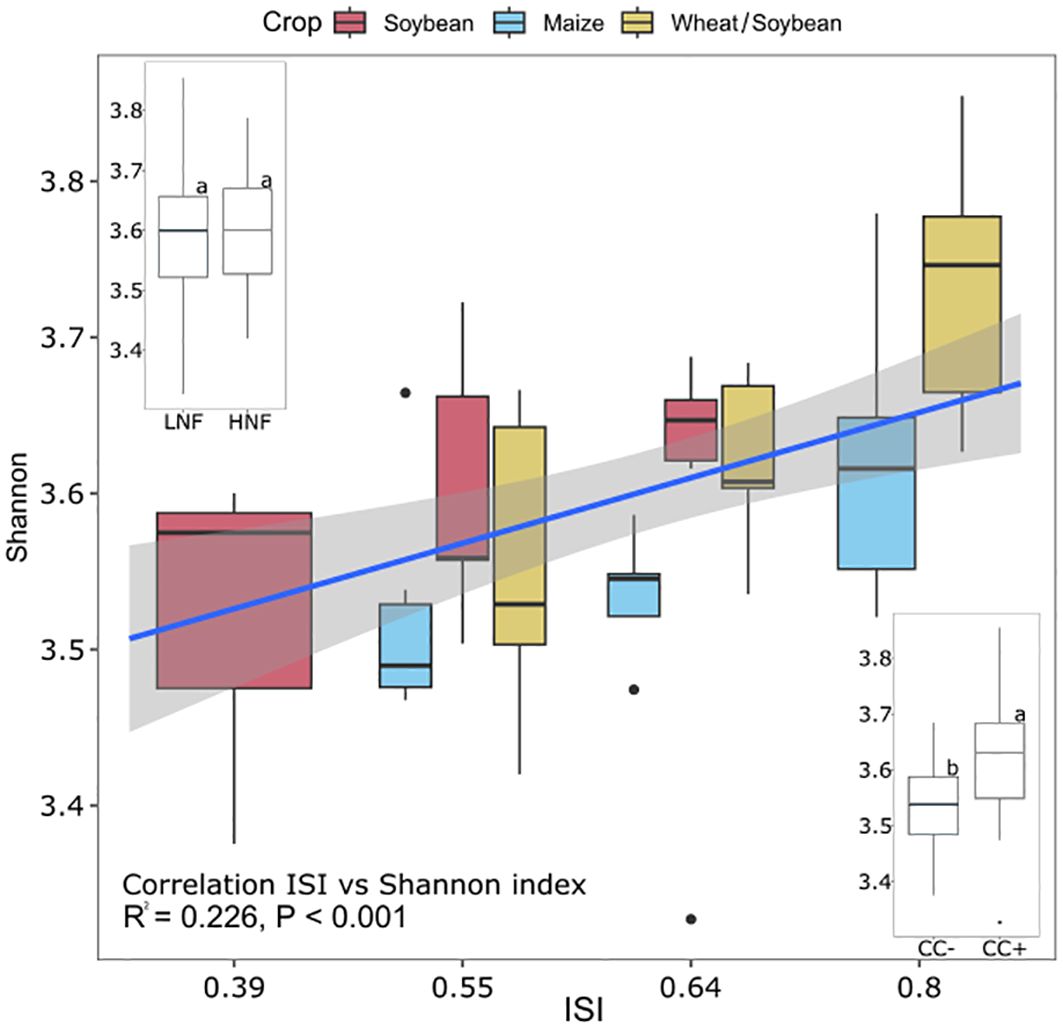
Figure 3. Microbial community alpha-diversity was measured by the Shannon index for each Intensification Sequence Index (ISI), N fertilization, and cover crop. Error bars indicate standard error. The line represents the linear correlation between ISI and Shannon index. Data were tested for significance by GLM ANOVA followed by Fisher’s LSD test. The letters represent significant differences between treatments at the p = 0.05 level.
3.3 Prokaryotic community structure and response to crop intensification and crop sequence
A total of 79 phyla were recovered from this soil. Acidobacteria comprised the majority of the 16S reads recovered (35.75% ± 3.11%), followed by Verrucomicrobia (15.64% ± 2.37%), Planctomycetes (11.57% ± 1.45%), Proteobacteria (10.79% ± 1.36%), and Chloroflexi (8.90% ± 1.14%) (Figure 4). Together, these five phyla accounted for more than 80% of the sequences. Soybean significantly increased the abundance of Acidobacteria (P = 0.037) and Armatimonadetes (P < 0.001), while the sequences containing Soybean and the double-cropped Wheat/Soybean increased the abundance of Bacteroidetes (P < 0.001). In contrast, Maize significantly increased the abundance of Planctomycetes (P = 0.002) and Verrucomicrobia (P < 0.001). Finally, Proteobacteria showed an interaction effect between crop phase and N fertilization, being more abundant in the double-cropped Wheat/Soybean for both cases (P < 0.05). We noted an increase in Chloroflexi, Gemmatimonadetes, and Thraumarchaeota in HNF, while Acidbacteria and Nitrospirae were more abundant in LNF (P < 0.05). Finally, the cover crops significantly increased the abundance of Proteobacteria (P < 0.05) (Figure 4).
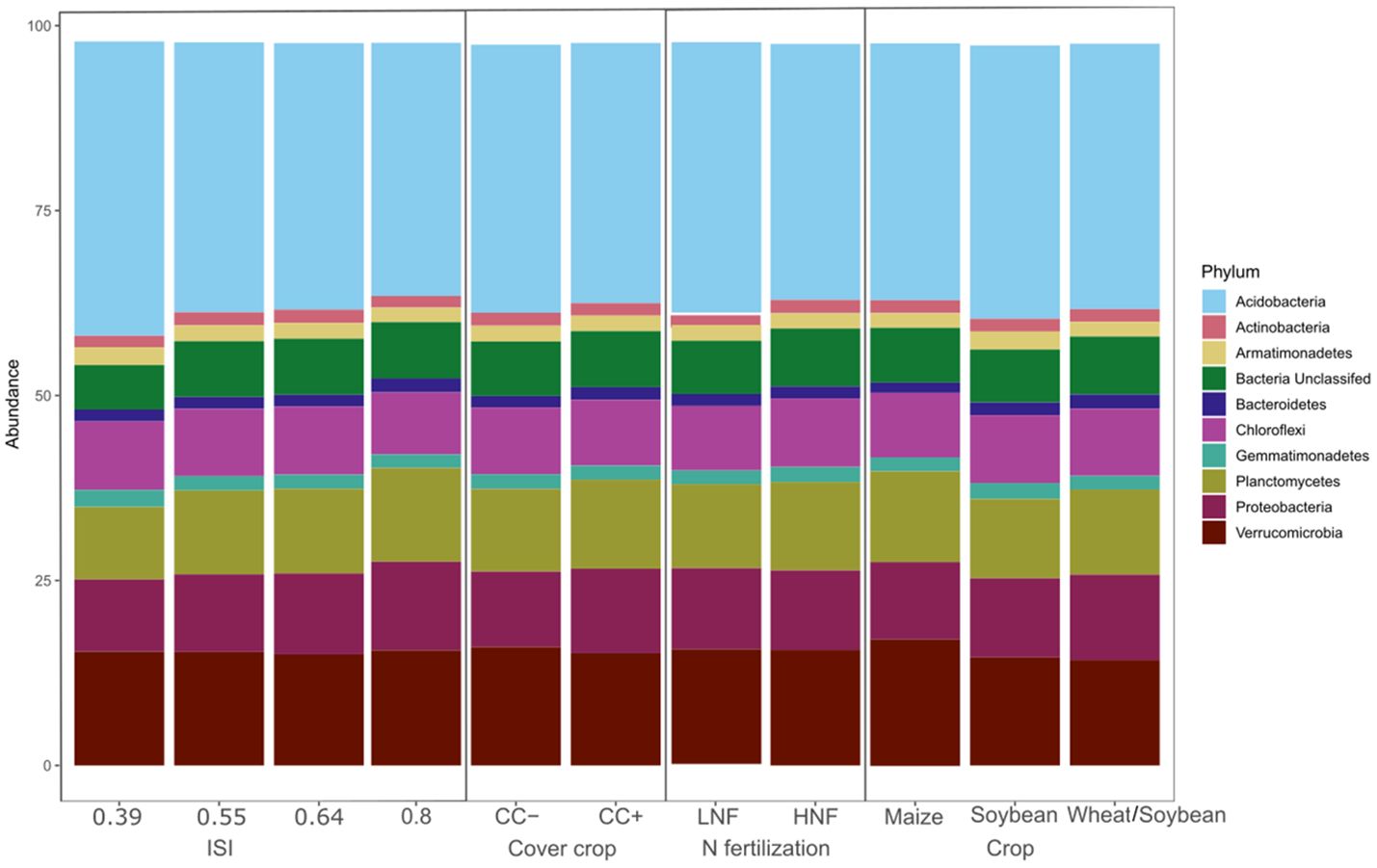
Figure 4. Prokaryotic composition at phylum level in soil with different crop rotations, cover crops, N fertilization, and intensification. Prokaryotic composition at the phylum level (relative abundance) in soil with different Intensification Sequence Index (ISI), cover crop (CC-, CC+) N fertilization (LNF and HNF), crop (Maze, Soybean, Wheat/Soybean).
We analyzed the impact of IRI on different phyla using the Poisson regression model for each rotation and the different phases of each rotation (Figure 5). Planctomycetes (R2 = 0.37, P = 0.0145), Proteobacteria (R2 = 0.51, P = 0.00108), and Bacteroidetes (R2 = 0.36, P = 0.0170) showed a positive relationship with ISI (Figures 5). The Wheat/Soybean double-crop exhibited a high number of responding phylum to the crop intensification, showing a positive correlation with Planctomycetes (R2 = 0.27, P = 0.00168) and Bacteroidetes (R2 = 0.29, P = 0.0296), while in Maize, the abundance of Proteobacteria (R2 = 0.31, P = 0.00677) was positively correlated with ISI. Finally, the abundance of Acidobacteria, Verrucomicrobia, Armatimonadetes, Chloroflexi, Gemmatimonadetes, and Thraumarchaeota correlated negatively, but not significantly, with ISI, while the abundance of Nitrospirae and Firmicutes correlated positively but not significant with ISI.
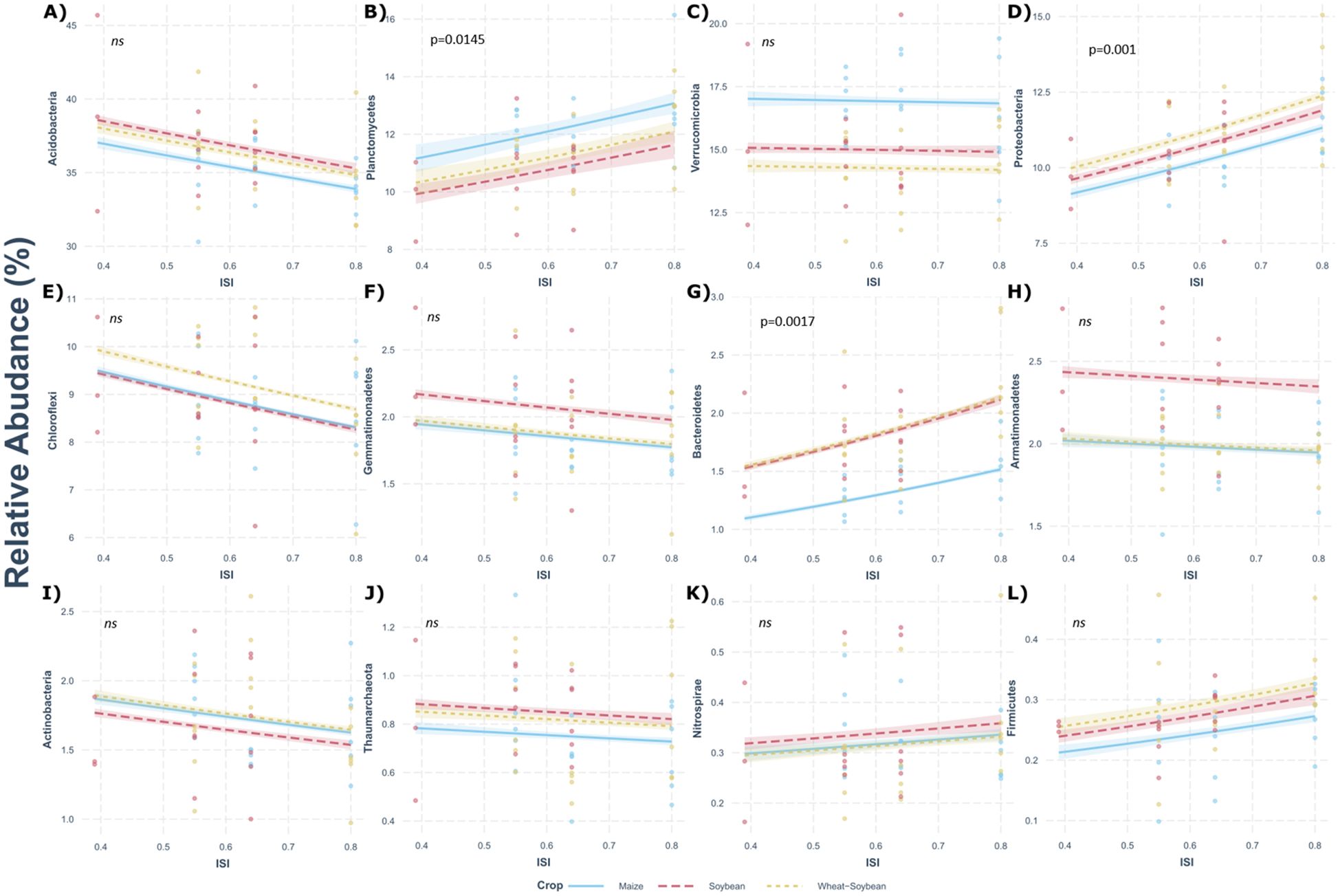
Figure 5. Poisson regression relationship between the relative abundance (%) of dominant phyla for (A) Acidobacteria, (B) Planctomycetes, (C) Verrumicrobia, (D) Proteobacteria, (E) Chloroflexi, (F) Gemmatimonadetes, (G) Bacteroidetes, (H) Armatimonadetes, (I) Actinobacteria, (J) Thaumarchaeota, (K) Nitrospirae, and (L) Firmicutes, and Intensification Sequence Index (ISI). Significance p-value of regression are shown; ns, non-significant.
3.4 Prokaryotic community structure and response to the crop identity
We used DESeq2 differential abundance analysis (alpha = 0.001) to determine which taxa differed in presence and abundance among the soils conditioned by the different plant species included in the different crop rotations. DESeq2 analysis suggested that Maize enriched the abundance of Gemmata, Singulisphaera, and Pirellula (both Planctomycetes) and other unclassified taxa belonging to Planctomycetes (dots below the 0 represent taxa enhanced in Maize) in comparison with Soybean and Wheat/Soybean double crop (Figures 6A, B). The other two taxa enriched in Maize were Acidimicrobiaceae unclassified (phylum: Actinobacteria) and Candidatus Udeobacter (phylum: Verrucomicrobia). In contrast, Soybean enriched the abundance of Flavobacterium and Chryseolinea (both Bacteroidetes) and other Bacteroidetes unclassified (dots above 0 on the plot) (Figures 6A, B). Other taxa increased in Soybean crops were Novosphingobium, Sphingomonadaceae unclassified (phylum: Proteobacteria; class: Alpha-proteobacteria), and Spartobacteria unclassified (Verrucomicrobia) (Figures 6A, B). The comparison between the Soybean and the Wheat/Soybean double crop showed few differential taxa. Soybean enriched Muciliginibacter (phylum: Bacteroidetes) and Geobacter (phylum: Proteobacteria, classes: Delta-proteobacteria) (both above the 0 on the plot), while Wheat/Soybean enriched Brevisfollis and Roseimicrobium (phylum: Verrucomicrobia) (below 0 on the plot) (Figure 6C).
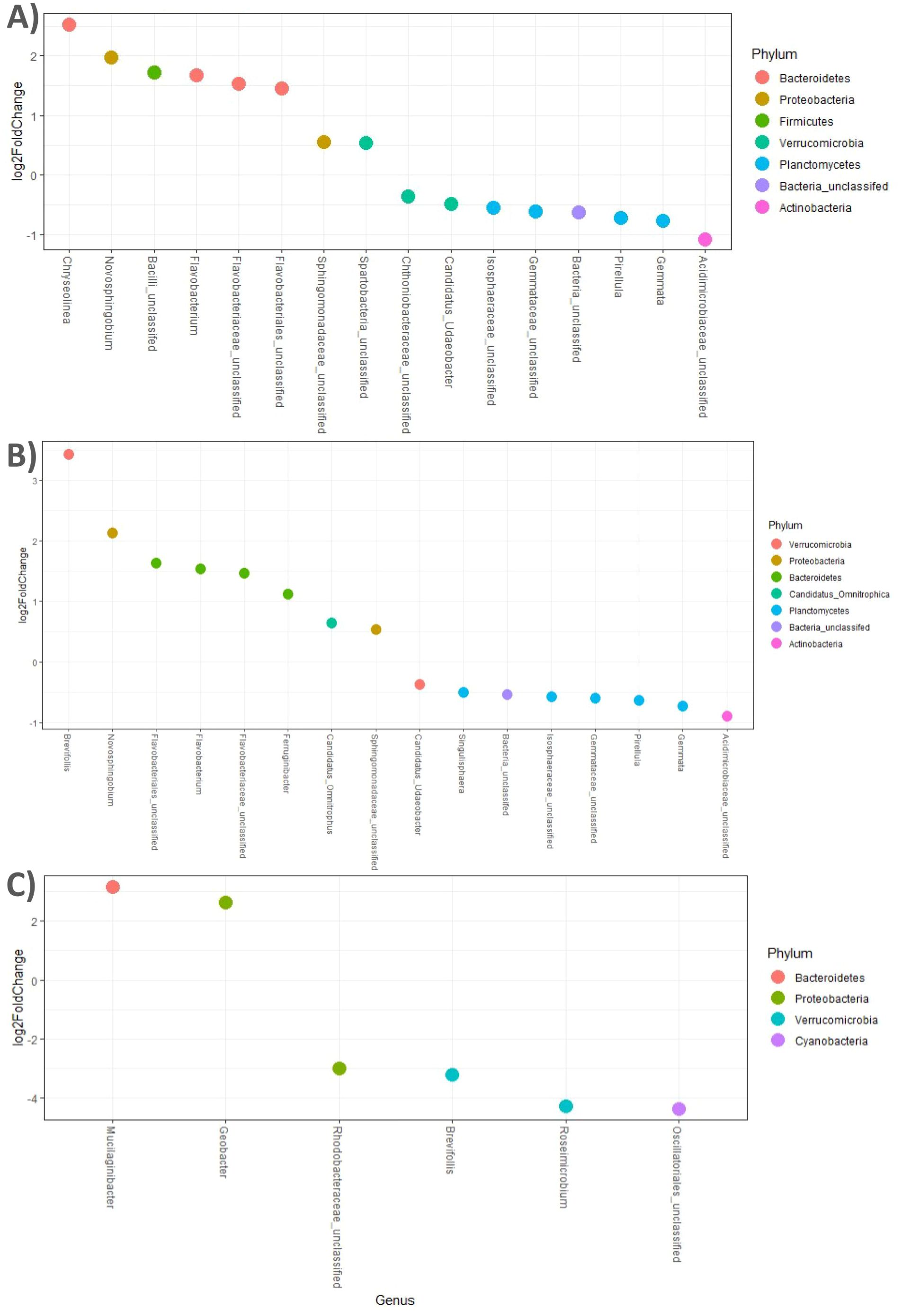
Figure 6. Differential representation of significant abundance ASVs at the genus level between (A) Soybean vs. Maize, (B) Wheat/Soybean vs. Maize, and (C) Soybean vs. Wheat/Soybean crop rotations. Dots represent significantly abundant ASVs, colored by phylum, and are labeled by genus. Multiple dots per genus represent different species of a given genus. Only the probabilities.
4 Discussion
We evaluated the prokaryotic community using high-throughput sequencing of the 16S rRNA gene in soils grown with three cash crops (Wheat, Soybean, and Maize) under contrasting intensification management in a long-term experimental trial. In this study, the immediate effect of the three commercial crops was evaluated simultaneously in each rotation, thus allowing us to evaluate the isolated impact of each phase in the rotation on the microbial communities. Our results showed that both crop phase and crop intensification, as measured by the Intensification Sequence Index (ISI), had a similar impact on the structure of the soil prokaryotic microbiome. Additionally, our results showed a small but significant effect of N fertilization on the prokaryotic community. Meanwhile, cover crops showed a non-significant effect on the community structure but markedly increased α-diversity.
4.1 Impact of crop identity on soil bacterial community
Soil biodiversity is crucial to maintaining and enhancing the ecological functions of the soil (Bender et al., 2016). Bacterial/archaeal richness, measured by the Shannon index, showed significant differences according to crop identity (Ishaq et al., 2017). Several authors showed that wheat increased diversity in comparison with other crops such as cotton, rice, and faba beans (Wei et al., 2019; Granzow et al., 2017). On the contrary, Ashworth et al. (2017) showed that the wheat-soybean sequence had richness values similar to those found in either continuous maize or continuous soybeans.
The prokaryotic community is a crucial component of the soil ecosystem and can be used for evaluating soil and plant health (Sharma et al., 2010). In this study, we found a clear and significant difference in soil prokaryotic community composition among Maize, Soybean, and Wheat/Soybean, as previously reported by Li et al. (2017). This phenomenon may primarily be due to the differential response of microbes to the quantity and quality of root exudates or rhizodeposition caused by the crops (Costa et al., 2006). We found members of Planctomycetes (Gemmata, Singulisphaera, and Pirellula), Actinobacteria (Acidimicrobiaceae), and Verrucomicrobia (Candidatus Udeobacter) associated with maize. Gemmata and Pirellula have been associated with N cycling, especially denitrification (Han et al., 2019). However, Gemmata has been related to wheat residue degradation (Zhou et al., 2018). In contrast, both Soybean and Wheat/Soybean double-crop led to the active increase in Alpha-proteobacteria (Novosphingobium) and Bacteroidetes (Flavobacterium and Chryseolinea), both of which are considered copiotrophs (Fierer et al., 2007; İnceoğlu et al., 2011).
Soybean was mainly enriched in Bacteroidetes, which might be consistent with soybean having more rhizodeposition than maize (Qiao et al., 2017; Mazzilli et al., 2015). soybean rhizodeposits have a lower C/N ratio than those of maize and are rapidly mineralized by microbes, which is likely responsible for the greater shift in the bacterial community (Miao et al., 2021). Bacteroidetes are commonly associated with soils rich in N and highly available nutrients (Liu et al., 2017). For example, the genera Flavobacterium and Chryseobacterium have been associated with plant growth promotion and disease protection (de Sousa Lopes et al., 2021). The Wheat/Soybean double-crop was enriched on Brevifollis and another Verrumicrobia taxa, where this phylum is extremely sensitive to changes in chemical factors related to soil fertility in native forests or grassland (Lupwayi et al., 2020; Stefan et al., 2021).
4.2 Impact of intensification index on soil bacterial community
Agricultural practices under the sustainable intensification paradigm are designed for soil conservation, enhancing crop production and soil health (Bommarco et al., 2013). Previous studies have shown a positive effect of this agricultural management on ecosystemic services, such as yield (Bowles et al., 2020; Gaudin et al., 2015), organic carbon stocks (Tamburini et al., 2020; Romaniuk et al., 2018; Beltrán et al., 2016), pest management (Emery et al., 2021), earthworms and mesofauna abundance (Rodríguez et al., 2020) and nitrous oxide emissions (Piccinetti et al., 2021). Global-scale studies have shown that soil microbiomes, especially soil bacteria, are shaped by many factors (Fierer, 2017). Additionally, Fan et al. (2020) observed significant, biologically meaningful correlations between crop yield and the abundance of specific root-associated microbial taxa and functional genes, highlighting the importance of the prokaryotic community in regulating soil functional potential and plant productivity.
Crop intensification implies an increase in the number of crops per year, and thus, more active roots throughout the year and the amount of C supplied to the soil. Here, the increase in the intensification sequence index (ISI) showed a positive effect on biodiversity, as measured by the Shannon index, in the three crops, mainly Maize and Wheat/Soybeans double-crop. Venter et al. (2017) carried out a meta-analysis that found that approximately 15.1% and 3.4% of microbial richness and diversity, respectively, were increased in the rotational treatments compared to the monocultures. Other studies have shown that management intensification has a significant effect (Bender et al., 2016; Garland et al., 2020) or an inconsistent effect on biodiversity (Stefan et al., 2021). Increasing the number of legume crops every year improves the supply of N due to the biological fixation of N and crop residues (Lal, 2017). Furthermore, Romaniuk et al. (2018) and Beltrán et al. (2016) demonstrated that eight years after the adoption of adequate intensification was sufficient to accumulate more labile C and macronutrients in stable C deposits. Both C and N stocks may contribute to increasing soil bacterial diversity (Garland et al., 2021; Zak et al., 2003).
In this study, we found that the relative abundance of several bacterial and archaeal phyla was significantly changed by the ISI. Overall, some copiotroph bacteria such as Proteobacteria, Firmicutes, Planctomycetes, and Bacteroidetes were consistent with the ecological theory that copiotrophic behavior (or r-strategies) is well adapted to nutrient-rich conditions and positively responds to intensification (Li et al., 2014). Firmicutes and Bacteroidetes can use ammonium or nitrate as their sole N source (Ashworth et al., 2017; Frene et al., 2022). On the contrary, Actinobacteria is a group that has been found in greater proportion in grassland (Liao et al., 2023) and it is negatively affected by soil disturbance (Gou et al., 2024). The changes in the microbial community structure have been associated with aboveground processes linked to crop diversity and composition (Delgado-Baquerizo et al., 2018).
In contrast to copiotroph microorganisms, the relative abundances of oligotrophic groups (e.g., Acidobacteria, Chloroflexi, Gemmatimonadetes, and Armatimonadetes) did not significantly respond to crop intensification. Acidobacteria is a well-known group that thrives in acidic, nutrient-poor soils and does not respond effectively to increased carbon input from additional plant biomass (Fierer et al., 2007; Kalam et al., 2020). Members of Chloroflexi have been identified as pathogenic bacteria (DeBruyn et al., 2011) that cannot respond to crop intensification due to crop rotation’s positive influence on crop health (Custódio et al., 2022). Finally, Gemmatimonadetes and Armatimonadetes have been reported to respond negatively to soil C and P stocks (Gou et al., 2024) and, in the present study both phyla negatively responded to crop intensification. These findings indicate that crop intensification could cause a shift in the life-history strategy.
4.3 Effect of N supply on soil prokaryotic community by fertilizer and cover crops
Our findings revealed a differential effect between N fertilization and cover crops: although N fertilization considerably altered the structure of the prokaryotic community, cover crops increased the α-diversity index. The increment in microbial diversity associated with cover crops has been associated with greater soil enzyme activities and N and P availability, directly influencing soil C and N cycles (Chavarria et al., 2018; Kim et al., 2020; McClelland et al., 2021). Kim et al. (2020) found that cover crops greatly increased the biomass, activity, and variety of soil microbial communities. However, the observed small effect of winter cover crops on soil prokaryotic community composition in our study may be related to the timing of sampling; our samples were collected after the summer cash crop harvest. According to Liu et al. (2021), the influence of cover crops on microbial community composition is determined not only by the identity of the cash and cover crops but also by the collection period. In contrast, N fertilization showed a minor but considerable influence on the structure of the community composition (Li et al., 2018). This effect was mainly observed in maize treatments probably because soybean does not receive any N fertilization (Castle et al., 2021). It has been documented that N fertilization alters the abundance of dominant bacterial phyla (Dai et al., 2018; Nguyen et al., 2018; Viso et al., 2024). Our findings revealed a pronounced decrease in Acidobacteria and Nitrospirae abundance, both of which are known to use an oligotrophic growth strategy that is less favorable in fertilized soils (Legrand et al., 2018; Ramirez et al., 2016).
5 Conclusions
Our findings revealed that both intensification and crop phase influenced the composition of the soil prokaryotic community after thirteen years (2006-2019) of soil management intensification. In addition, cover crops and N fertilizer may have had a limited impact on soil bacterial/archaeal community structure and diversity. The bacterial phylum associated with copiotrophic life forms appeared to benefit from intensification, but certain oligotrophic phylum did not. Although the copiotrophic/oligotrophic hypothesis is simplistic, it provides a basic framework for linking the response of soil prokaryotic communities to crop rotations. There is a clear need for more comprehensible and comparable data about the dynamics of soil microbiomes in agroecosystems, as well as their integration in general models with other physical, chemical, and biological parameters, to better understand the impact of different and alternative agricultural practices. Further studies will determine if this information is linked to crop productivity and soil health indexes, contributing to optimized agricultural management and addressing the problems caused by unsuitable practices.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
JF: Formal analysis, Software, Visualization, Writing – original draft. SB: Conceptualization, Investigation, Methodology, Resources, Writing – review & editing. MM: Investigation, Methodology, Writing – review & editing. JO: Investigation, Methodology, Writing – review & editing. MR: Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing. FS: Conceptualization, Investigation, Methodology, Writing – review & editing. VF: Conceptualization, Funding acquisition, Investigation, Methodology, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the grant “Capacity building for bioinformatics in Latin America-Network” (CABANA-net), funded by the agreement (2022– 316296) between the Chan Zuckerberg Initiative (CZI) and the Universidad de Costa Rica.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2024.1446404/full#supplementary-material
References
Agomoh I. V., Drury C. F., Phillips L. A., Reynolds W. D., Yang X. (2020). Increasing crop diversity in wheat rotations increases yields but decreases soil health. SSSA Jour. 84, 170–181. doi: 10.1002/saj2.20000
Andrade J. F., Poggio S. L., Ermácora M., Satorre E. H. (2017). Land use intensification in the Rolling Pampa, Argentina: Diversifying crop sequences to increase yields and resource use. Eur. Jour. Agron. 82, 1–10. doi: 10.1016/j.eja.2016.09.013
Apprill A., McNally S., Parsons R., Weber L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases the detection of SAR11 bacterioplankton. Aqua. Microb. Ecol. 75, 129–137. doi: 10.3354/ame01753
Ashworth A. J., DeBruyn J. M., Allen F. L., Radosevich M., Owens P. R. (2017). Microbial community structure is affected by cropping sequences and poultry litter under long-term no-tillage. Soil Biol. Biochem. 114, 210–219. doi: 10.1016/j.soilbio.2017.07.019
Beltrán M., Brutti L., Bacigaluppo S., Romaniuk R., Salvagiotti F., Sainz-Rozas H., et al. (2016). Calidad de la material organica y disponibilidad de macro y micronutrientes por la inclusión de trigo como cultivo de cobertura. Cienc Suelo 34, 67–79.
Bender S. F., Wagg C., van der Heijden M. G. (2016). An underground revolution: biodiversity and soil ecological engineering for agricultural sustainability. Trends. Ecol. Evol. 6, 440–452. doi: 10.1016/j.tree.2016.02.016
Bliss C. I. (1953). “Note on the efficient fitting of the negative binomial,” in Fitting the negative binomial distribution to biological data, 9th ed, vol. 9. Ed. Fisher R. A. Biometrics.
Bokulich N. A., Kaehler B. D., Rideout J. R., Dillon M., Bolyen E., Knight R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6, 1–17. doi: 10.1186/s40168-018-0470-z
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, interactive, scalable, and extensible microbiome data science using QIIME 2. Nat. Biotech. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bommarco R., Kleijn D., Potts S. G. (2013). Ecological intensification: harnessing ecosystem services for food security. Trends Ecol. Evol. 28, 230–238. doi: 10.1016/j.tree.2012.10.012
Bowles T. M., Mooshammer M., Socolar Y., Calderón F., Cavigelli M. A., Culman S. W., et al. (2020). Long-term evidence shows that crop-rotation diversification increases agricultural resilience to adverse growing conditions in North America. One Earth 2, 284–293. doi: 10.1016/j.oneear.2020.02.007
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Cano P. B., Cabrini S. M., Peper A. M., Poggio S. L. (2023). Multi-criteria assessment of cropping systems for the sustainable intensification in the Pampas. Agric. Syst. 210, 103723. doi: 10.1016/j.agsy.2023.103723
Castle S. C., Samac D. A., Gutknecht J. L. (2021). Impacts of cover crops and nitrogen fertilization on agricultural soil fungal and bacterial communities. Plant Soil 466, 139–150. doi: 10.1007/s11104-021-04976-z
Caviglia O. P., Andrade F. H. (2010). Sustainable intensification of agriculture in the Argentinean Pampas: capture and use efficiency of environmental resources. Am. J. Plant Sci. Biotechnol. 3, 1–8. doi: 10.1016/j.fcr.2003.10.002
Chamberlain L. A., Bolton M. L., Cox M. S., Suen G., Conley S. P., Ané J. M. (2020). Crop rotation, but not cover crops, influenced soil bacterial community composition in a corn-soybean system in southern Wisconsin. App. Soil Ecol. 154, 103603. doi: 10.1016/j.apsoil.2020.103603
Chaparro J. M., Sheflin A. M., Manter D. K., Vivanco J. M. (2012). Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fertility Soils 48, 489–499. doi: 10.1007/s00374-012-0691-4
Chavarria D. N., Pérez-Brandan C., Serri D. L., Meriles J. M., Restovich S. B., Andriulo A. E., et al. (2018). Response of soil microbial communities to agroecological versus conventional systems of extensive agriculture. Agric. Ecosyst. Environ. 264, 1–8. doi: 10.1016/j.agee.2018.05.008
Correndo A. A., Gutiérrez-Boem F. H., García F. O., Alvarez C., Álvarez C., Angeli A., et al. (2021). Attainable yield and soil texture as drivers of maize response to nitrogen: A synthesis analysis for Argentina. Field Crops Res. 273, 108299. doi: 10.1016/j.fcr.2021.108299
Costa R., Götz M., Mrotzek N., Lottmann J., Berg G., Smalla K. (2006). Effects of site and plant species on rhizosphere community structure as revealed by molecular analysis of microbial guilds. FEMS Microbiol. Ecol. 56, 236–249. doi: 10.1111/j.1574-6941.2005.00026.x
Custódio V., Gonin M., Stabl G., Bakhoum N., Oliveira M. M., Gutjahr C., et al. (2022). Sculpting the soil microbiota. Plant J. 109 (3), 508–522. doi: 10.1111/tpj.15568
Dai Z. M., Su W. Q., Chen H. H., Barberán A., Zhao H. C., Yu M. J., et al. (2018). Long-term nitrogen fertilization decreases bacterial diversity and favors the growth of Actinobacteria and Proteobacteria in agro-ecosystems across the globe. Glob. Change Biol. 24, 3452–3461. doi: 10.1111/gcb.14163
DeBruyn J. M., Nixon L. T., Fawaz M. N., Johnson A. M., Radosevich M. (2011). Global biogeography and quantitative seasonal dynamics of Gemmatimonadetes in soil. App. Environ. Microbi. 77, 6295–6300. doi: 10.1128/AEM.05005-11
Delgado-Baquerizo M., Fry E. L., Eldridge D. J., de Vries F. T., Manning P., Hamonts K. (2018). Plant attributes explain the distribution of soil microbial communities in two contrasting regions of the globe. N. Phytol. 219, 574–587. doi: 10.1111/nph.15161
de Sousa Lopes L., Mendes L. W., Antunes J. E. L., de Souza Oliveira L. M., Melo V. M. M., de Araujo Pereira A. P., et al. (2021). Distinct bacterial community structure and composition along different cowpea producing ecoregions in Northeastern Brazil. Sci. Rep. 11 (1), 831. doi: 10.1038/s41598-020-80840-x
Dubey A., Malla M. A., Khan F., Chowdhary K., Yadav S., Kumar A., et al. (2019). Soil microbiome: a key player for conservation of soil health under changing climate. Biodiv. Conser. 28, 2405–2429. doi: 10.1007/s10531-019-01760-5
Emery S. E., Jonsson M., Silva H., Ribeiro A., Mills N. J. (2021). High agricultural intensity at the landscape scale benefits pests, but low intensity practices at the local scale can mitigate these effects. Agricul. ecosys. Environm. 306, 107199. doi: 10.1016/j.agee.2020.107199
Enrico J. M., Piccinetti C. F., Barraco M. R., Agosti M. B., Eclesia R. P., Salvagiotti F. (2020). Biological nitrogen fixation in field pea and vetch: Response to inoculation and residual effect on maize in the Pampean region. Eur. Jour. Agron. 115, 126016. doi: 10.1016/j.eja.2020.126016
Fan K., Delgado-Baquerizo M., Zhu Y., Chu H. (2020). Crop production correlates with soil multitrophic communities at a large spatial scale. Soil Biol. Biochem. 151, 108047. doi: 10.1016/j.soilbio.2020.108047
Fang Y., Van Zwieten L., Rose M. T., Vasileiadis S., Donner E., Vancov T., et al. (2022). Unraveling microbiomes and functions associated with strategic tillage, stubble, and fertilizer management. Agriculture Ecosyst. Environ. 323, 107686. doi: 10.1016/j.agee.2021.107686
FAO (2021) F. A. O. S. T. A. T. Food and agriculture organization of the United Nations (Rome). Available at: https://www.fao.org/faostat/en/#home (Accessed June 2023).
Feng Y., Guo Z., Zhong L., Zhao F., Zhang J., Lin X. (2017). Balanced fertilization decreases environmental filtering on soil bacterial community assemblage in North China. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02376
Ferrari M., Castellarín J., Rozas H. S., Vivas H., Melchiori R., Gudelj V. (2010). Evaluación de métodos de diagnóstico de fertilidad nitrogenada para el cultivo de trigo en la región pampeana. Agronómicas 46, 10. Available at: http://www.ipni.net/publication/ia-lacs.nsf/0/D2A4C1BA5069500F852579840057F9E4/$FILE/IA%2046.pdf#page=10.
Fierer N., Bradford M. A., Jackson R. B. (2007). Toward an ecological classification of soil bacteria. Ecology 88 (6), 1354–1364. doi: 10.1890/05-1839
Fierer N. (2017). Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590. doi: 10.1038/nrmicro.2017.87
Figuerola E. L. M., Guerrero L. D., Rosa S. M., Simonetti L., Duval M. E., Galantini J. A., et al. (2012). Bacterial indicator of agricultural management for soil under no-till crop production. PloS One 7, e51075. doi: 10.1371/journal.pone.0051075
Finzi A. C., Raymer P. C. L., Giasson M. A., Orwig D. A. (2014). Net primary production and soil respiration in New England hemlock forests affected by the hemlock woolly adelgid. Ecosphere 5, 1–16. doi: 10.1890/ES14-00102.1
Fox A., Lüscher A., Widmer F. (2020). Plant species identity drives soil microbial community structures that persist under a following crop. Ecol. Evol. 10, 8652–8668. doi: 10.1002/ece3.6560
Frene J. P., Figuerola E., Gabbarini L. A., Erijman L., Wall L. G. (2022). Impact of diversification and intensification of crop rotation (DICR) in soil bacterial microbiota in on-farm study after four and seven years. App. Soil Ecol. 179, 104592. doi: 10.1016/j.apsoil.2022.104592
Garland G., Edlinger A., Banerjee S., Degrune F., Garcia-Palacios P., Pescador D. S., et al. (2020). Crop cover is more important than rotational diversity for soil multifunctionality and cereal yields in European cropping systems. Nat. Food 2, 28–37. doi: 10.1038/s43016-020-00210-8
Garnica S., Rosenstein R., Schön M. E. (2020). Belowground fungal community diversity, composition, and ecological functionality associated with winter wheat in conventional and organic agricultural systems. PeerJ 8, e9732. doi: 10.7717/peerj.9732
Gaudin A. C. M., Tolhurst T. N., Ker A. P., Janovicek K., Tortora C., Martin R. C., et al. (2015). Increasing crop diversity mitigates weather variations and improves yield stability. PloS One 10, e0113261. doi: 10.1371/journal.pone.0113261
Gou Z., Wang Y., Cui Z., Yan B., Gao Y., Wu B., et al. (2024). Effects of four-year oilseed flax rotations on the soil bacterial community in a semi-arid agroecosystem. Agronomy 14, 740. doi: 10.3390/agronomy14040740
Granzow S., Kaiser K., Wemheuer B., Pfeiffer B., Daniel R., Vidal S., et al. (2017). The effects of cropping regimes on fungal and bacterial communities of wheat and faba bean in a greenhouse pot experiment differ between plant species and compartment. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00902
Han X. X., Feng J. W., Zhang L., Cui B. H., Zhang J. B., Pan L. J., et al. (2017). Micro-polluted water treatment by biological contact oxidation process: aeration mode and bacteria community analysis. Environ. Eng. Sci. 36, 1491–1502. doi: 10.1089/ees.2019.0198
Hartman K., van der Heijden M. G., Wittwer R. A., Banerjee S., Walser J. C., Schlaeppi K. (2018). Cropping practices manipulate abundance patterns of root and soil microbiome members paving the way to smart farming. Microbiome 6, 1–14. doi: 10.1186/s40168-017-0389-9
Hermans S. M., Buckley H. L., Case B. S., Curran-Cournane F., Taylor M., Lear G. (2020). Using soil bacterial communities to predict physico-chemical variables and soil quality. Microbiome 8 (1), 79. doi: 10.1186/s40168-020-00858-1
İnceoğlu Ö., Al-Soud W. A., Salles J. F., Semenov A. V., van Elsas J. D. (2011). Comparative analysis of bacterial communities in a potato field as determined by pyrosequencing. PloS One 6, e23321. doi: 10.1371/journal.pone.0023321
Ishaq S. L., Johnson S. P., Miller Z. J., Lehnhoff E. A., Olivo S., Yeoman C. J., et al. (2017). Impact of cropping systems, soil inoculum, and plant species identity on soil bacterial community structure. Microb. Ecol. 73, 417–434. doi: 10.1007/s00248-016-0861-2
Jónsson J.Ö.G., Davíðsdóttir B. (2016). Classification and valuation of soil ecosystem services. Agricul. Syst. 145, 24–38. doi: 10.1016/j.agsy.2016.02.010
Kalam S., Basu A., Ahmad I., Sayyed R. Z., El-Enshasy H. A., Dailin D. J., et al. (2020). Recent understanding of soil acidobacteria and their ecological significance: a critical review. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.580024
Kibblewhite M. G., Ritz K., Swift M. J. (2008). Soil health in agricultural systems. Phil. Tran. R. Soc B. 363, 685–701. doi: 10.1098/rstb.2007.2178
Kim N., Zabaloy M., Guan K., Villamil M. (2020). Do cover crops benefit soil microbiome? A meta-analysis of current research. Soil Biol. Biochem. 142, 107701. doi: 10.1016/j.soilbio.2019.107701
Kleijn D., Bommarco R., Fijen T. P. M., Garibaldi L. A., Potts S. G., van der Putten W. H. (2019). Ecological intensification: bridging the gap between science and practice. Trends Ecol. Evol. 34, 154–166. doi: 10.1016/j.tree.2018.11.002
Lal R. (2017). Improving soil health and human protein nutrition by pulses-based cropping systems. Adv. Agron. 145, 167–204. doi: 10.1016/bs.agron.2017.05.003
Legrand F., Picota A., Cobo-Díaza J. F., Carofc M., Chend W., Le Floch G. (2018). Effect of tillage and static abiotic soil properties on microbial diversity. App. Soil Ecol. 132, 135–145. doi: 10.1016/j.apsoil.2018.08.016
Li F., Chen L., Zhang J., Yin J., Huang S. (2017). Bacterial community structure after long-term organic and inorganic fertilization reveals important associations between soil nutrients and specific taxa involved in nutrient transformations. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00187
Li H., Luo L., Tang B., Guo H., Cao Z., Zeng Q., et al. (2018). Dynamic changes in the rhizosphere bacterial community in monoculture and intercropping maize and soybean during various crop growth stages. Res. Square. doi: 10.21203/rs.3.rs-49276/v1
Li X. Z., Rui J. P., Mao Y. J., Yannarell A., Mackie R. (2014). Dynamics of the bacterial community structure in the rhizosphere of a maize cultivar. Soil Biol. Biochem. 68, 392–401. doi: 10.1016/j.soilbio.2013.10.017
Liao J., Dou Y., Yang X., An S. (2023). Soil microbial community and their functional genes during grassland restoration. Jour. Environ. Manage. 325, 116488. doi: 10.1016/j.jenvman.2022.116488
Liu X., Hannula S. E., Li X., Hundscheid M. P. J., Gunnewiek P., Clocchiatti A., et al. (2021). Decomposing cover crops modify root-associated microbiome composition and disease tolerance of cash crop seedlings. Soil Biol. Biochem. 160, 108343. doi: 10.1016/j.soilbio.2021.108343
Liu J., Yu Z., Yao Q., Hu X., Zhang W., Mi G., et al. (2017). Distinct soil bacterial communities in response to the cropping system in a Mollisol of northeast China. Appl. Soil Ecol. 119, 407–416. doi: 10.1016/j.apsoil.2017.07.013
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21. doi: 10.1186/s13059-014-0550-8
Lupwayi N. Z., Fernandez M. R., Kanashiro D. A., Petri R. M. (2020). Profiles of wheat rhizobacterial communities in response to repeated glyphosate applications, crop rotation, and tillage. Can. J. Soil Sci. 2020, 8. doi: 10.1139/cjss-2020-0008
Lupwayi N. Z., May W. E., Kanashiro D. A., Petri R. M. (2018). Soil bacterial community responses to black medic cover crop and fertilizer N under no-till. Appl. Soil Ecol. 124, 95–103. doi: 10.1016/j.apsoil.2017.11.003
Mazzilli S. R., Leamoam A. R., Ernst O. R., Jackson R. B., Pineiro G. (2015). Greater humification of belowground than aboveground biomass carbon intoparticulate soil organic matter in no-till corn and soybean crops. Soil Biol. Biochem. 85, 22–30. doi: 10.1016/j.soilbio.2015.02.014
McClelland S. C., Paustian K., Schipanski M. E. (2021). Management of cover crops in temperate climates influences soil organic carbon stocks: a meta-analysis. Ecol. Appl. 00, e02278. doi: 10.1002/eap.2278
McDaniel M. D., Grandy A. S., Tiemann L. K., Weintraub M. N. (2014). Crop rotation complexity regulates the decomposition of high and low-quality residues. Soil Biol. Biochem. 78, 243–254. doi: 10.1016/j.soilbio.2014.07.027
McMurdie P. J., Holmes S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8, e61217. doi: 10.1371/journal.pone.0061217
Miao S., Qiao Y., Jin J., Wang Y., Tang C. (2021). Greater variation of bacterial community structure in soybean- than maize-grown Mollisol soils in responses to seven-year elevated CO2 and temperature. Sci. Total Environ. 764, 142836. doi: 10.1016/j.scitotenv.2020.142836
Miner G. L., Delgado J. A., Ippolito J. A., Stewart C. E., Manter D. K., Del Grosso S. J., et al. (2020). Assessing manure and inorganic nitrogen fertilization impacts on soil health, crop productivity, and crop quality in a continuous maize agroecosystem. JWSC 75, 481–498. doi: 10.2489/jswc.2020.00148
Nguyen L. T. T., Osanai Y., Lai K., Anderson I. C., Bange M. P., Tissue D. T., et al. (2018). Responses of the soil microbial community to nitrogen fertilizer regimes and historical exposure to extreme weather events: Flooding or prolonged drought. Soil Biol. Biochem. 118, 227–236. doi: 10.1016/j.soilbio.2017.12.016
Nivelle E., Verzeaux J., Habbib H., Kuzyakov Y., Decocq G., Roger D., et al. (2016). Functional response of soil microbial communities to tillage, cover crops and nitrogen fertilization. Appl. Soil Ecol. 108, 147–155. doi: 10.1016/j.apsoil.2016.08.004
Novelli L. E., Caviglia O. P., Melchiori R. J. M. (2011). Impact of soybean cropping frequency on soil carbon storage in Mollisols and Vertisols. Geoderma 167, 254–260. doi: 10.1016/j.geoderma.2011.09.015
Novelli L. E., Caviglia O. P., Piñeiro G. (2017). Increased cropping intensity improves crop residue inputs to the soil and aggregate-associated soil organic carbon stocks. Soil Till. Resea. 165, 128–136. doi: 10.1016/j.still.2016.08.008
Oksanen J., Guillaume B. F., Kindt R. (2015). Vegan: Community Ecology Package (R package version 2.3-0). Available at: http://CRAN.R-project.org/package=vegan (accessed June 2024).
Parada A. E., Needham D. M., Fuhrman J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Piccinetti C. F., Bacigaluppo S., Di Ciocco C. A., De Tellería J. M., Salvagiotti F. (2021). Soybean in rotation with cereals attenuates nitrous oxide emissions as compared with soybean monoculture in the Pampas region. Geoderma 402, 115192. doi: 10.1016/j.geoderma.2021.115192
Pinto P., Long M. E. F., Piñeiro G. (2017). Including cover crops during fallow periods for increasing ecosystem services: Is it possible in croplands of Southern South America? Agri. Ecosys. Environ. 248, 48–57. doi: 10.1016/j.agee.2017.07.028
Pretty J., Bharucha Z. P. (2014). Sustainable intensification in agricultural systems. Ann. Bot. 114, 1571–1596. doi: 10.1093/aob/mcu205
Qiao Y., Miao S., Han X., Yue S., Tang C. (2017). Improving soil nutrient availability increases carbon rhizodeposition under maize and soybean in Mollisols. Sci. Total Environ. 603–604, 416–424. doi: 10.1016/j.scitotenv.2017.06.090
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Opens external link in new window. Nucl. Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Ramirez K. S., Lauber C., Knight R., Bradford M., Fierer N. (2016). Consistent effects of nitrogen fertilization on soil bacterial communities in contrasting systems. Wiley 91, 3463–3470. doi: 10.6084/m9.figshare.c.3303624.v1
R Development Core Team (2014). A Language and Environment for Statistical Computing (Vienna, Austria: R Foundation for Statistical Computing). Available at: http://www.R-project.org/. (Accessed June 2024)
Robledo N. B., Frene J. P., Wall L. G. (2024). Agriculture intensification as a critical step to enhance sustainable productive systems. JWSC 79, 19A–22A. doi: 10.2489/jswc.2024.1230A
Rodríguez M. P., Domínguez A., Moreira Ferroni M., Wall L. G., Bedano J. C. (2020). Diversification and intensification of crop rotations under no-till promote earthworm abundance and biomass. Agronomy 10, 919. doi: 10.3390/agronomy10070919
Romaniuk R., Beltrán M., Brutti L., Costantini A., Bacigaluppo S., Sainz-Rozas H., et al. (2018). Soil organic carbon, macro- and micronutrient changes in soil fractions with different lability in response to crop intensification. Soil Till. Resear. 181, 136–143. doi: 10.1016/j.still.2018.04.014
Ross G. J. S., Preece D. A. (1985). The negative binomial distribution. J. R. Statist. Soc Ser. D. (The Statistician) 34, 323–335. doi: 10.2307/2987659
Salvagiotti F., Castellarín J. M., Ferraguti F. J., Pedrol H. M. (2011). Dosis óptima económica de nitrógeno en maíz según potencial de producción y disponibilidad de nitrógeno en la región pampeana norte. Cien. Suelo 29, 199–212.
Salvagiotti F., Gerster G., Bacigalupo S., Castellarín J., Galarza C., González N., et al. (2004). Residual and fresch effects of phosphorous and sulphur on soybean following wheat. Cien. Suelo 22, 92–101.
Sasse J., Martinoia E., Northen T. (2018). Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci. 23, 25–41. doi: 10.1016/j.tplants.2017.09.00
Sharma S. K., Ramesh A., Sharma M. P., Joshi O. P., Govaerts B., Steenwerth K. L., et al. (2010). “Microbial community structure and diversity as indicators for evaluating soil quality,” in Biodiversity, Biofuels, Agroforestry and Conservation Agriculture (Springer, Berlin/Heidelberg, Germany), 317–358. doi: 10.1007/978-90-481-9513-8_11
Stefan L., Hartmann M., Engbersen N., Six J., Schöb C. (2021). Positive effects of crop diversity on productivity driven by changes in soil microbial composition. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.660749
Tamburini G., Bommarco R., Wanger T. C., Kremen C., van der Heijden M. G. A., Liebman M., et al. (2020). Agricultural diversification promotes multiple ecosystem services without compromising yield. Sci. Adv. 6, eaba1715. doi: 10.1126/sciadv.aba1715
Tilman D., Reich P. B., Knops J., Wedin D., Mielke T., Lehman C. (2001). Diversity and productivity in a long-term grassland experiment. Science 294, 843–845. doi: 10.1126/science.1060391
van der Heijden M. G. A., Bardgett R. D., van Straalen N. M. (2008). The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11, 296–310. doi: 10.1111/j.1461-0248.2007.01139.x
Venables W. N., Ripley B. D. (2012). Modern Applied Statistics with S. (4a ed.). New York, NY: Springer.
Venter Z. S., Scott S. L., Strauss J., Jacobs K., Hawkins H. J. (2017). Increasing crop diversity increased soil microbial activity, nitrogen-sourcing, and crop nitrogen, but not soil microbial diversity. South Afr. J. Plant Soil 34, 371–378. doi: 10.1080/02571862.2017.1317852
Viso N. P., Ortiz J., Maury M., Frene J. P., Iocoli G. A., Lorenzon C., et al. (2024). Long-term maintenance rate fertilisation increases soil bacterial-archaeal community diversity in the subsoil and N-cycling potentials in a humid crop season. Appl. Soil Ecol. 193, 105149. doi: 10.1016/j.apsoil.2023.105149
Wall L. G., Gabbarini L., Ferrari A. E., Frene J. P., Covelli J., Reyna D., et al. (2019). Changes of paradigms in agriculture soil microbiology and new challenges in microbial ecology. Acta Oecol. 95, 68–73. doi: 10.1016/j.actao.2019.02.001
Wei Z., Gu Y., Friman V. P., Kowalchuk G. A., Xu Y., Shen Q., et al. (2019). Initial soil microbiome composition and functioning predetermine future plant health. Sci. Adv. 5, eaaw0759. doi: 10.1126/sciadv.aaw0759
Zak D. R., Holmes W. E., White D. C., Peacock A. D., Tilman. D. (2003). Plant diversity, soil microbial communities, and ecosystem function: are there any links? Ecology 84, 2042–2050. doi: 10.1890/02-0433
Zhang M., Zhang X., Zhang L., Zeng L., Liu Y., Wang X., et al. (2021). The stronger impact of inorganic nitrogen fertilization on soil bacterial comm unity than organic fertilization in short-term conditions. Geoderma 382, 114752. doi: 10.1016/j.geoderma.2020.114752
Keywords: soil microbiome, maize, soybean, wheat, cover crops, N fertilization
Citation: Frene JP, Bacigaluppo S, Maury M, Ortiz J, Rivarola M, Salvagiotti F and Faggioli V (2024) Crop-specific response of soil prokaryotic community to long-term intensification management: the importance of crop phase at sampling. Front. Agron. 6:1446404. doi: 10.3389/fagro.2024.1446404
Received: 09 June 2024; Accepted: 28 November 2024;
Published: 17 December 2024.
Edited by:
Davey Jones, Bangor University, United KingdomReviewed by:
Maria Teresa Ceccherini, University of Florence, ItalyPasquale Napoletano, University of Florence, Italy
Copyright © 2024 Frene, Bacigaluppo, Maury, Ortiz, Rivarola, Salvagiotti and Faggioli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Valeria Faggioli, ZmFnZ2lvbGl2YWxlcmlhQGdtYWlsLmNvbQ==
 Juan P. Frene
Juan P. Frene Silvina Bacigaluppo2
Silvina Bacigaluppo2 Jimena Ortiz
Jimena Ortiz Maximo Rivarola
Maximo Rivarola Fernando Salvagiotti
Fernando Salvagiotti Valeria Faggioli
Valeria Faggioli