- 1NIAB East Malling, West Malling, United Kingdom
- 2Tanzania Agricultural Research Institute (TARI) Mikocheni, Dar Es Salaam, Tanzania
- 3Uganda Christian University, Mukono, Uganda
- 4World Vegetable Center, Arusha, Tanzania
- 5NIAB, Park Farm, Cambridge, United Kingdom
African eggplant (Solanum aethiopicum L.) is one of the most common traditional vegetables in Tanzania and Uganda, but its productivity is severely affected by wilt diseases caused by a number of pathogens. Plant stem and root samples were collected in several fields from many neighboring diseased and healthy plants of the Gilo group in Tanzania and from the Shum group in Uganda to identify putative pathogens causing wilt on African eggplants. Through amplicon sequencing of sampled diseased and healthy tissues, we identified putative causal pathogens for the wilt symptoms. Wilting of S. aethiopicum in Uganda is most likely caused by the bacterial pathogen Ralstonia solanacearum whereas, in Tanzania, wilt is most likely caused by the fungal pathogen Macrophomina phaseolina, infecting roots. Infection of stems by Fusarium solani may also contribute to the wilt symptoms in Tanzania. Further artificial inoculation under controlled conditions confirmed that M. phaseolina can cause typical wilting symptoms on S. aethiopcium genotypes. The discovery of different putative causal agents of wilt in the crop demonstrates the need for site specific etiological analysis of wilt before developing and implementing effective control methods. Further research is needed to confirm the results and develop appropriate management measures against specific wilt pathogens.
Introduction
African eggplant (Solanum aethiopicum L.) is one of the common African traditional vegetables grown in Tanzania and Uganda (Abukutsa-Onyango, 2010; Dinssa et al., 2016; Omondi et al., 2016; Ochieng et al., 2018). There are four major types (subgroups) of African eggplant: Gilo, Shum, Kumba, and Aculeatum. Gilo types are grown for their fruits, Shum types for leaves, Kumba types for leaves or fruits, whereas Aculeatum types are ornamental. In Uganda, the Shum group (a leafy type) is highly abundant and commercialized as well as the Gilo group varieties (Sseremba et al., 2018). African eggplant varieties are grown in both rural, urban, and peri-urban areas, and their production is increasingly becoming important in terms of acreage and intensity in terms of investment into inputs, especially in Central Uganda. Their production is becoming a valuable source of income for smallholder farmers, and this has the potential for improving livelihoods.
Low yield of African eggplant achieved in Tanzania is attributed to biotic and abiotic stresses (Mwinuka et al., 2021), including water stress and pathogens. Wilt disease is one of the most important biotic factors limiting the yield potential of African eggplant (Ssekabembe and Odong, 2008). Most research studies, so far, on eggplant wilt disease have been focused on S. melongena (e.g., Bletsos et al., 1997; Lewis Ivey et al., 2007; Dervis et al., 2009; Lebeau et al., 2011; Najar et al., 2011; Li et al., 2017; Rao et al., 2019). Several fungal genera such as Fusarium, Verticillium, Rhizoctonia, and Sclerotium as well as the oomycete Phytophthora can infect S. melongena and cause wilt disease symptoms, resulting in considerable crop losses (Najar et al., 2011). Wilt can also be caused by a bacterial pathogen Ralstonia solanacearum (Nahar et al., 2019; Phukan et al., 2019). Several S. aethiopicum accessions are resistant against one specific wilt causing pathogen F. oxysporum f. sp. Melongenae isolate (Mwaniki et al., 2016). There are no published studies on the causal agent(s) of the wilt disease in field grown S. aethiopicum eggplants.
Given the number of potential pathogens causing wilt symptoms in eggplants of several species, it is important to identify the main pathogen(s) responsible for the wilt disease in any given host species. Timely detection and identification of pathogens is, therefore, important for developing and implementing appropriate disease management strategies in practice. The objective of the current study was to determine putative pathogens of African eggplant wilt in Tanzania and Uganda. We used amplicon-sequencing method to determine putative causative pathogens of the wilt. Two criteria were used to identify putative pathogens based on the current understanding of disease epidemiology: (1) a moderate to high average number of reads per sample and (2) a higher relative abundance in diseased roots since wilting symptoms are assumed to have resulted from infection of roots by soilborne pathogens. This led to the identification of the pathogen Macrophomina phaseolina, which is not normally associated with eggplant wilt disease in Tanzania. Then, we conducted artificial inoculation studies to confirm whether M. phaseolina can cause wilt symptoms in the Gilo type of African eggplants.
Materials and methods
Amplicon sequencing of field diseased and healthy plant samples
Sampling
To minimize the effect of spatial variability in the soil microbial communities, five pairs of immediately neighboring (next to each other within a row) plants—one healthy and one diseased—were randomly picked within each field for sampling rhizosphere soils (soil particles adhered to roots after gentle shaking), and roots and stem tissues (both roots and stems were surface washed with sterile water before DNA extraction). The healthy looking and wilted plants next to each other were uprooted and shaken to remove loose soil. The soil that remained attached to the roots were assumed to represent the rhizosphere and were, therefore, shaken even more vigorously and collected in plastic bags to constitute the rhizosphere samples (any residual root was removed). From the same plant, the fibrous roots without visible soils were also collected in separate bags and washed with sterile water in the lab to obtain root samples. For each pair of plants, there were six samples: two rhizosphere, two roots, and two stems. Six and three farmers’ fields were sampled in Tanzania and Uganda, respectively. In Tanzania, five pairs of diseased/healthy plants were sampled in each field whereas, in Uganda, six pairs were sampled in each field.
In Tanzania, the samples were collected from Arusha, Pwani, and Dar es Salaam administrative regions that are quite different from each other in agroecology. In Arusha region (high altitude and relatively cool agroecology), the samples were collected from World Vegetable Center (WorldVeg) Eastern and Southern Africa Research Station (latitude 3.38°S, longitude 36.81°E), and from two on-farm locations: Kikwe (latitude 3.43°S, longitude 36.85°E) and Sing’isi (latitude 3.37°S, longitude 36.80°E) in March 2019. The samples in Pwani and Dar es Salaam administrative regions (regions characterized by coastal low altitude and high temperature agroecology) were collected from three farms located in Bagamoyo District (latitude 6.2°S, longitude 38.2°E), Kinondoni District (latitude 6.9°S, longitude 39.2°E), and Temeke District (latitude 6.2°S, longitude 39.4°E) in February 2019. In Uganda, samples were obtained in June 2019 in farmers’ fields of the Shum group in Mukono (latitude 0.28°N, longitude 32.30°E); Wakiso (latitude 0.24°N, longitude 32.28°E) and Luwero (latitude 0.49°N, longitude 32.30°E) in the Central Uganda within the Lake Victoria Crescent agroecological zone where most small-scale commercial production of S. aethiopicum is located. This region has a bimodal rainfall pattern with two rainy seasons: one in late March–May and the other, September–November (UBOS, 2013). Distance between sampling sites within each region is at least 35 km.
DNA extraction and sequencing
Samples were freeze dried and dry weight recorded before homogenization with Geno/Grinder 2010 (SPEX CertiPrep) for 2 min at 1500 rpm using 2-ml tubes and two 5-mm steel ball bearings. In Tanzania, DNA from stem and root samples was extracted using a modified cetyltrimethylammonium bromide method as described previously (Allen et al., 2006). For rhizosphere soil samples, DNA was extracted using the Quick-DNA Fecal/Soil Microbe Miniprep Kit from Zymo Research (California, USA) following the manufacturer’s instructions except that DNA was re-suspended in diethylpyrocarbonate-treated water instead of supplied elution buffer. In Uganda, sterile 0.1M phosphate-buffered saline (PBS) buffer was added to the homogenized samples at 1:5 (dry weight [mg]:volume [ml]) ratio. DNA was extracted from 120 ml of PBS resuspended homogenate with DNEasy® Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Pure DNA was eluted from the spin columns in two steps, each using 50 ml of molecular grade water and 1-min incubation before spinning at 14,000 g.
The DNA samples were shipped to Novogene UK (Cambridge, UK) for polymerase chain reaction (PCR), library prep and amplicon sequencing. The target primers were ITS1-1F [ITS1-1F-F: 5′-CTTGGTCATTTAGAGGAAGTAA-3′ (Gardes and Bruns, 1993), ITS2: 5′-GCTGCGTTCTTCATCGATGC-′3 (White et al., 1990)] and 16S V5-V7 [799F: 5′-AACMGGATTAGATACCCKG-3′ (Chelius and Triplett, 2001), 1193R: 5′-ACGTCATCCCCACCTTCC-3′ (Bodenhausen et al., 2013)]. Samples were sequenced on Illumina NovaSeq platform in the 250 nt paired end mode.
Sequencing processing and taxonomy assignment
Sequence data from Tanzania and Uganda samples were analyzed individually. Processing sequences to generating Operational Taxonomic Unit (OTU) counts tables followed previously published pipelines (Papp-Rupar et al., 2022). Sequence processing was performed twice (1) to produce reads for OTU generation and (2) to produce reads for subsequent frequency table generation. Processing for OTU generation used far more stringent filtering to minimize errors. For both processes, sequences with incorrect bases in the barcode or primer region of either forward or reverse reads were discarded. Retained reads were then merged using the UPARSE pipeline (Version 10.0) (Edgar, 2013). For OTU generation, the following settings were used: (1) minimum read length of 250 bases, (2) maximum number of different bases in the merged region of 12 (16S) and 0 (ITS), and (3) minimum merged length of 400 (16S) 185 (ITS). The merged sequences were further filtered for quality with a maximum-expected error threshold of 0.5 (16S) and 0.1 (ITS) per sequence (Edgar and Flyvbjerg, 2015). These differing settings for ITS and 16S reads tended to give equal ratios of usable to discarded reads of ~2:1. Reads were then dereplicated and singletons discarded before generation of OTUs at 97% similarity (UPARSE also removes suspected chimeral sequences). For frequency table generation, reads were merged using the settings already given except for “maximum number of different bases,” which was set to an arbitrarily high number (100) to ensure effectively all reads were merged. No further filtering was performed on these reads. Then, all the unfiltered merged reads were aligned to the OTU representative sequences at the level of 97% similarity to produce an OTU frequency table. To remove host-derived (non-ITS) sequences, OTUs with the representative sequences lacking a recognizable fungal ribosomal small subunit region aligned to hierarchical Markov models available with ITSx (1.1.3) using the HMMER programme (v3.1b2) (Bengtsson-Palme et al., 2013) were removed before further processing. Finally, the SINTAX algorithm (https://www.drive5.com/usearch/manual/sintax_algo.html) was used to assign taxonomic ranks to each OTU with the Unite V8.3 (2021-05-10) fungal database (Kõljalg et al., 2013) and “the RDP training set V18” database for the 16S rRNA gene (Cole et al., 2014). The SINTAX algorithm only resolves bacterial OTUs to the genus level but may resolve fungal OTUs to the species level. Taxonomy assignment confidence was at the 50% level.
Statistical analysis of amplicon data
Only the most abundant OTUs that accounted for 99.0% of the total sequence counts were retained for statistical analysis. The OTU counts data were normalized for library size by the median-of-ratios method implemented in DESeq2 (Love et al., 2014) before statistical analysis. A pair of neighboring healthy/diseased plants was treated as a single block to remove spatial variability; thus, the variation among pairs of neighboring healthy/diseased plants also contained the variation among fields. Sample types (rhizosphere, root, and stem) and diseased status (diseased or healthy) were the other two treatment factors. All statistical analyses were carried out with R 4.1.3 (R Core Team et al., 2019).
Although within-sample (alpha, α) and between-sample (beta, β) diversity indices were analyzed, the present study focused on the differential abundance analysis between diseased and healthy plants in order to identify putative pathogens responsible for wilt symptoms. The rank of α diversity (Shannon and Simpson) indices, calculated with the R vegan 2.3-1 package (Dixon, 2003), were subjected to analysis of variance (ANOVA) to assess the effects of treatment factors via a permutation test. Beta (Bray-Curtis) diversity indices were subjected to permutational multivariate ANOVA (PERMANOVA) to assess the effects of treatment factors (implemented as the Adonis function in the vegan package).
The rationale for identification of putative causal agents was that an increased relative abundance of the potential causal pathogen OTUs will be observed in the diseased samples in comparison to the healthy samples primarily in the rhizosphere/or in the plant roots. DESeq2 analyses were applied to compare relative abundance of individual OTUs between diseased and healthy plants in order to identify putative pathogens. Furthermore, preliminary analysis indicated that there were large interactions between countries and plant disease status, indicating that putative pathogens varied significantly between the two countries. Therefore, the DESeq2 analysis was applied to each sample type (rhizosphere, root, and stem) independently for each country. DESeq2 uses a Wald test to assess for OTUs with significant differential abundance (Anders and Huber, 2010; Love et al., 2014). The p-value was adjusted for multiple testing by the Benjamini-Hochberg method (Benjamini and Hochberg, 1995). In all DESeq2 analyses, the block factor (healthy/diseased plant pairs) was included in the model. For those bacterial OTUs with significant differential abundance between diseased and healthy plants, the OTU representative sequences were searched via BLAST to infer possible taxonomy identities.
Pathogenicity test
Isolate origins and inoculum preparation
DESeq analysis identified M. phaseolina as a putative causal agent of African eggplant (S. aethiopicum) wilt in Tanzania. Although this pathogen was reported to cause root rot of eggplant (S. melongena) in a few specific locations (Khan and Gupta, 1998; Remezani, 2008), it was not commonly known as a pathogen causing the wilt symptoms on African eggplants in Tanzania. Therefore, we conducted pathogenicity studies to confirm whether M. phaseolina can cause wilt symptoms on a few specific African eggplant genotypes from Tanzania. We obtained two M. phaseolina strains, one (PC1/17) isolated from strawberry (Fragaria × ananassa) originating from Egypt and the other (#268866) from CABI (Egham, UK), originated from Phaseolus vulgaris in Sri Lanka for pathogenicity testing. A Fusarium oxysporum strain (PC4/17) from P. vulgaris in the UK was also included in the inoculation.
The two M. phaseolina isolates were sub-cultured in Potato Dextrose Agar (PDA) plates and incubated at 26°C in the dark for 9 days. The plates were blended with 800 ml of water to form mycelial slurry. Viability of inoculum was determined by applying and spreading 100 μl of the inoculum on to PDA plates and assessed after 3 days. F. oxysporum inoculum was prepared in five vials, each containing five mycelium plugs in 80 ml of CMC for 8 days, then the resulting inoculum was filtered and diluted to 500 ml. A haemocytometer was used to estimate the spore concentration.
Plant preparation and inoculation
Three entries (two Gilo S. aethiopicum entry—DB3 and MM1961; and one Shum breeding line E11) were selected for inoculation study, because they represent different use types (fruit and leaf types) and commercial varieties and breeding lines. DB3 is the most popular fruit type African eggplant commercial variety grown in Eastern, Southern, and Western Africa countries. MM1961 is a fruit type advanced breeding line from World Vegetable Center (WorldVeg) in Tanzania, and E11 is a leafy variety of the African eggplant developed by Uganda Christan University. Surface sterilized seeds of the three varieties/lines were pregerminated on pieces of wet filter paper in petri dishes in a growth cabinet set at 27°C daytime (16h) and 21°C at night. After 1 week, germinated seedlings of the same size were selected and potted in a F7 pot (containing John Innes compost No. 2, J. Arthur Bower) individually. Seedlings were grown in a growth cabinet set at 27°C daytime (10h) and 21°C at night for about 4 weeks when plants reached the two to four true leaf stage.
Healthy plants of the same size were selected and split into four groups, each randomly allocated to one of the four treatments that were three isolates and one water control. There were 25 plants of “DB3” per treatment and 12 plants of line E11 per treatment. Seedlings were removed from the pots and the roots were cleaned off compost. The bottom third of the root length was trimmed with scissors. Plants were dipped into the appropriate inoculum suspension (or water for the water control treatment) for 2 min before being re-potted into fresh compost and kept in a growth cabinet.
Assessments and re-isolation
Plants were kept in growth cabinet (27°C daytime, 10h; 21°C at night) post-inoculation and watered every 2–3 days initially and visually monitored for wilting. If no wilting was observed four weeks after inoculation with a specific isolate, water stress was applied to induce wilting: plants were watered every 4–5 days instead of 2–3 days. The experiment was terminated 10 weeks after inoculation.
At termination, wilting plants were taken from the pots, compost removed, and roots washed. Leaves were removed from the plant leaving the stem and roots. Roots and shoots were separated and put into petri dishes where they were surface sterilized by spraying with 70% ethanol, ensuring all the surfaces were covered and left for about 30 s. The roots and shoots were then placed on a piece of sterile filter paper to dry. Once dry, the shoots and roots were cut into 5-mm sections (including the edge of a lesion where obvious), and five sections were placed onto a single PDA plate (one plate per plant). The plates were left to incubate at ambient temperature.
The resulting fungal morphology was compared visually to the original isolates used in inoculation. Then the colonies re-isolated from plants were sequenced and compared to the original isolates. A small piece of mycelium was taken from each colony using a sterile pipette tip, resuspended in 20 μl of extraction solution (E7526, Sigma-Aldrich) incubated at 95°C for 15 min, and then 80 μl of dilution solution (D5688, Sigma-Aldrich) was added to each sample. End point PCR with ITS1 and ITS4 primers (White et al., 1990) was performed in a PTC-220 DYAD™ PCR Engine (BIO-RAD, USA). All reactions were run using 2 μl of DNA extracts in a total volume of 12.5 μl. Each reaction contained 0.05 μl of MolTaq (Molzym, Bremen, Germany), 1.25 μl of MolTaq buffer 10×, 0.25 μl of MgCl2 100 mM, 1.0 μl dNTPs 2.5mM, 1.25 μl of each primer 2 μM and 2.0 μl of 2.0 ng/μl DNA template. PCR conditions were 94°C for 5 min, followed by 30 cycles of 94°C for 1 min, 53°C for 1 min and 72°C for 2 min, with a final extension of 72°C for 7 min. Amplified products were sequenced with LightRun service (Eurofins) using ITS1 and ITS4 primers. Sequences were searched against the Nucleotide BLAST web service search (NCBI) to infer taxonomy and consensus sequences.
Repeat of M. phaseolina strain 268866 inoculations
Due to issues with contamination, it was not possible to definitively compare sequences of the re-isolated and the original strains for M. phaseolina strain 268866, despite being similar morphologically. As such, the pathogenicity test was repeated with just the M. phaseolina strain 268866 as this was the only isolate that had caused wilt symptoms. The repeat test was set up as the first except for the following amendments: Three genotypes were inoculated (12 plants of DB3, 12 plants of E11, and five plants of MM1961). In addition, three plants of each genotype were used in the water control. The repeat test was terminated two weeks after inoculation when majority of inoculated plants had wilted.
Results
Overall sequencing and OTU information
DNA extraction was successful for most of the samples. Table 1 gives the summary of sequences and OTU data.
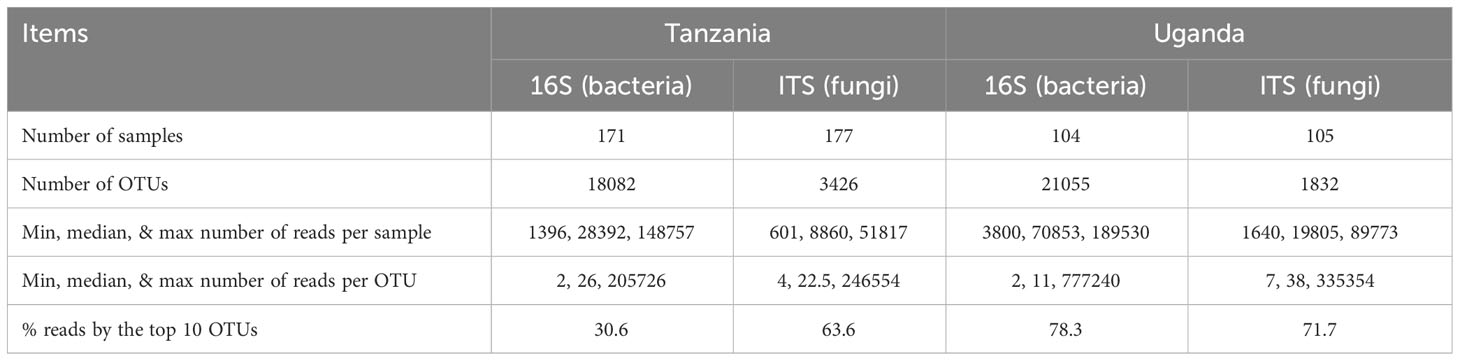
Table 1 Summary of amplicon-sequencing and subsequent bioinformatic processing of sequence reads for those samples from healthy and diseased African eggplant in Tanzania and Uganda.
Bacteria
In total, there were 18,082 and 21,055 bacterial OTUs for Tanzania and Uganda samples, respectively; most OTUs had few reads—median number of reads per OTU was less than 30. The top 10 most abundant OTUs accounted for nearly 31% and 78% of the total reads for Tanzania and Uganda samples, respectively (Table 1). More than 85% of the bacterial OTUs could be assigned to the rank of order at 50% confidence. Nearly all sequences were in the OTUs that could be assigned to the phylum rank; most sequences were from Proteobacteria and Actinobacteria (Figures 1, 2).
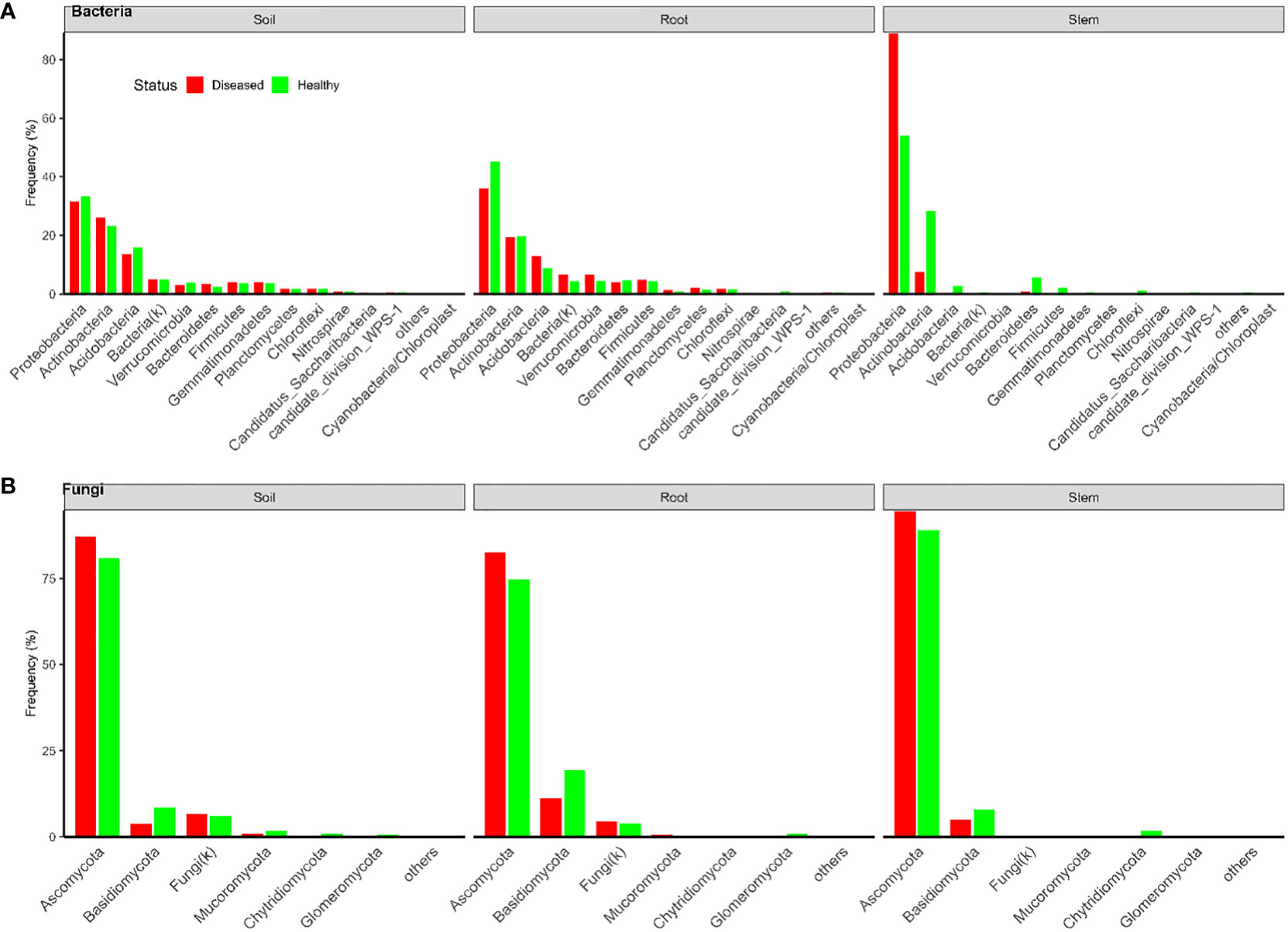
Figure 1 Frequencies of sequences in each bacterial (A) and fungal (B) phylum in three types of tissues of neighboring diseased and healthy eggplants in several fields in Tanzania.
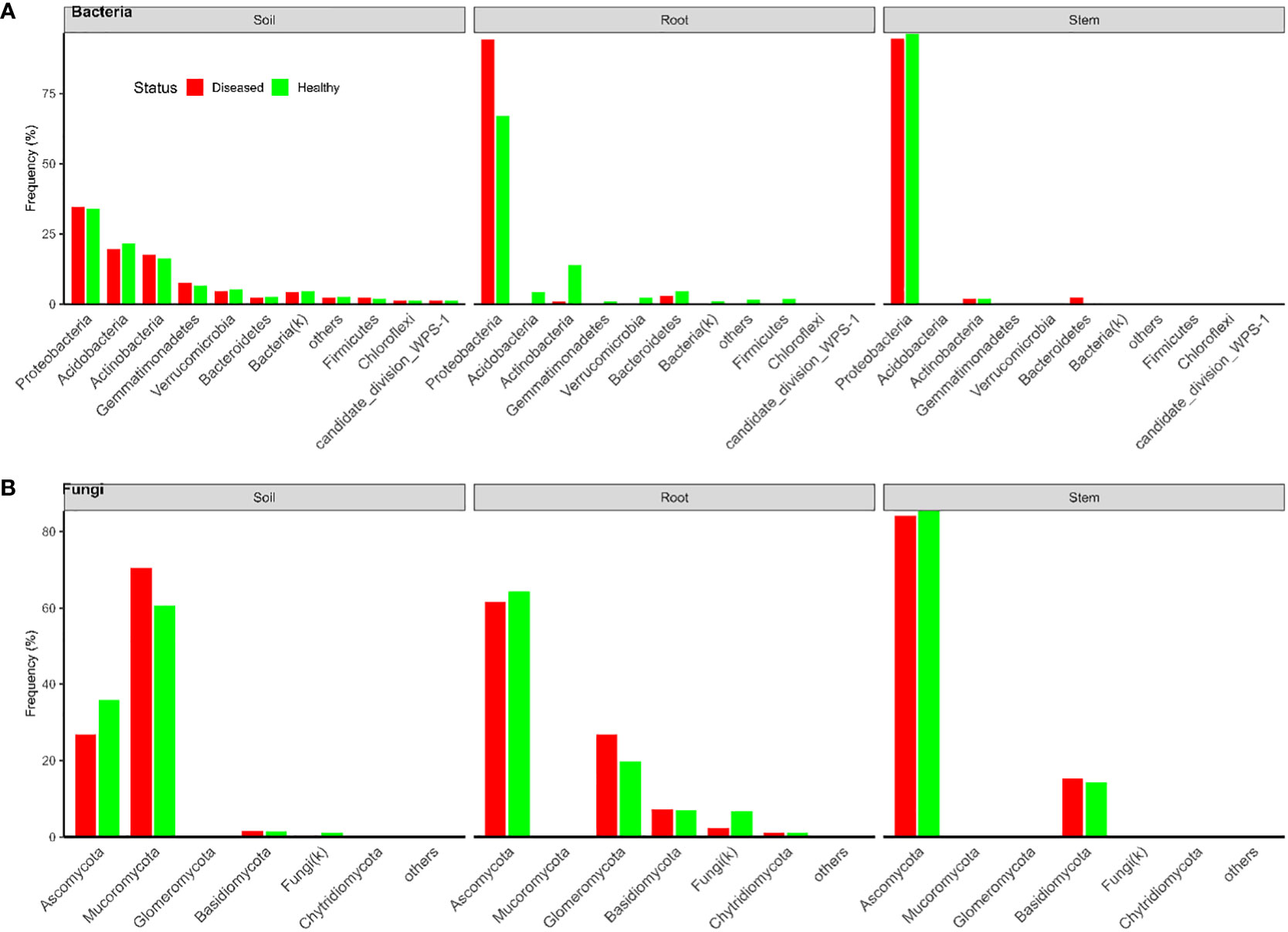
Figure 2 Frequencies of sequences in each bacterial (A) and fungal (B) phylum in three types of tissues of neighboring diseased and healthy eggplants in several fields in Uganda.
Fungi
There were a total of 3,426 and 1,832 fungal OTUs for Tanzania and Uganda samples, respectively; most OTUs had few reads—median number of reads per OTU was less than 40. The top 10 most abundant OTUs accounted for >63% of the total reads (Table 1). About half of the fungal OTUs could not be assigned to the rank of genus at 50% confidence; only about 36% of OTUs were assigned to the species level at 50% confidence. Most of the sequences were in the OTUs that could be assigned to the phylum rank; most of the sequences were from Ascomycota (Figures 1, 2). There were many sequences (>20%) from Glomeromycota from both diseased and healthy root samples in Uganda (Figure 2).
Diversity indices
Bacteria
For bacteria, α diversity indices (Simpson and Shannon) were mostly affected by sample types and followed by spatial location (pairs of plants) (Table 2). Stem samples had the lowest diversity indices whereas rhizosphere had the highest. For Uganda samples, the rhizosphere samples had greater diversity indices than the root samples, compared to the Tanzania samples. Similarly, β diversity was mainly affected by sample types and spatial locations but, to a lesser extent, than the α diversity. There was less clear separation among samples from the three sample types for Tanzania (Figure 3) than for Ugandan (Figure 4) samples. The effect of diseased status was limited but significant, mostly in the form of its interaction with sample types (Table 2).
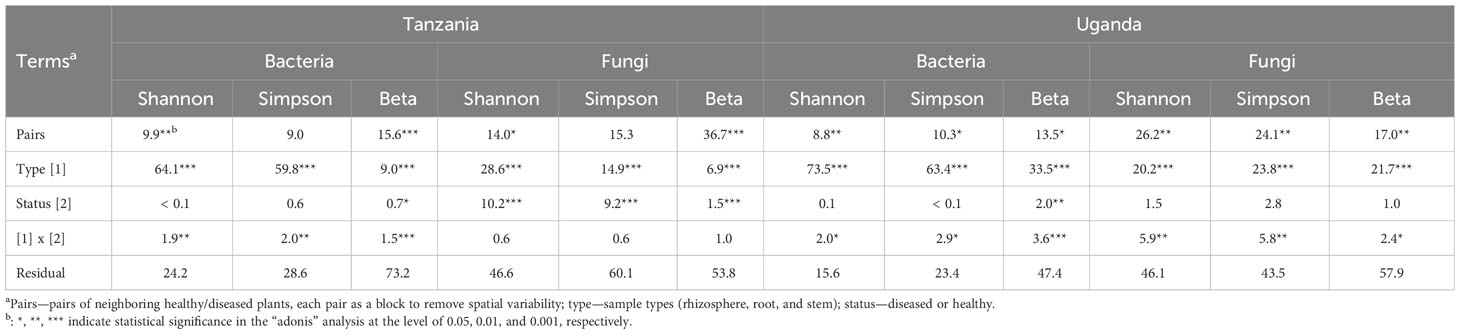
Table 2 Summary of analysis on the contribution (percentage values) of individual experimental terms to the overall variability (sum of squares) in alpha (Shannon and Simpson) and beta diversity indices.
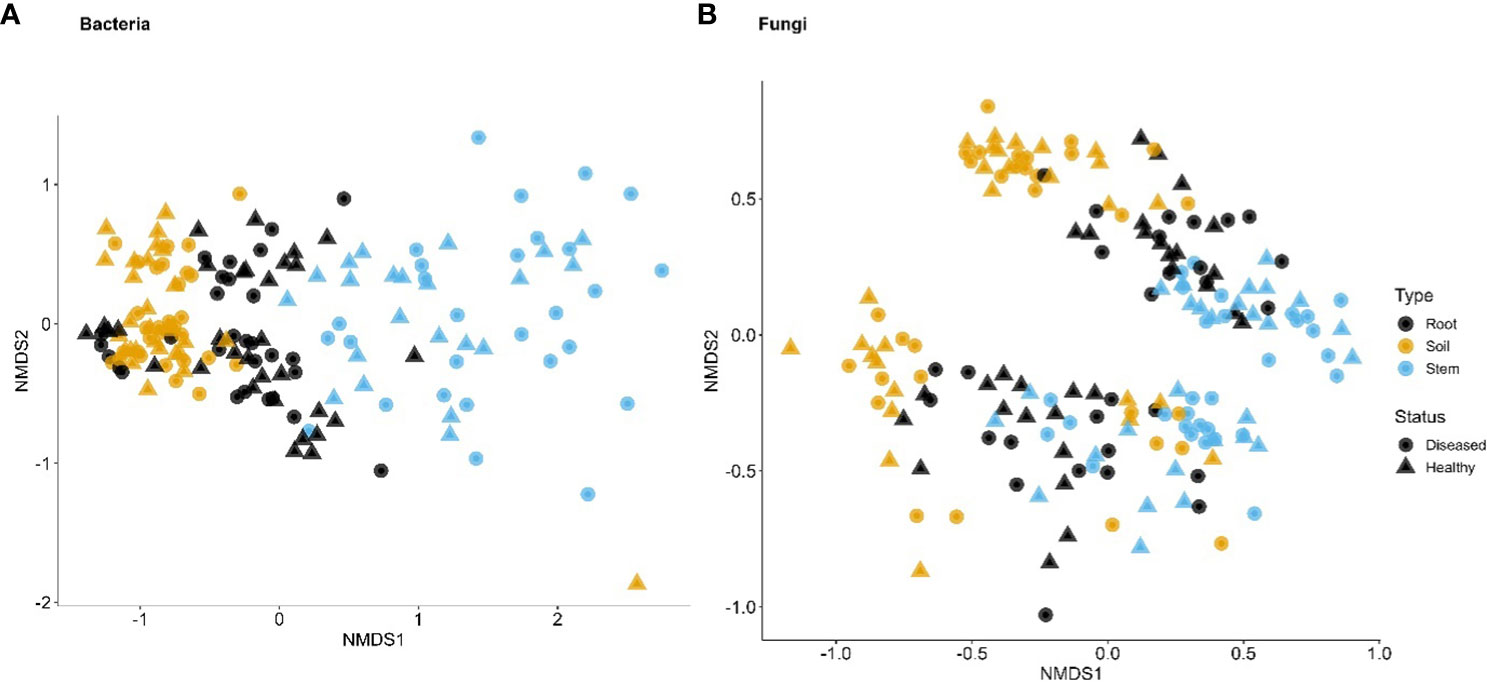
Figure 3 First two dimensions of the non-metric dimensional scaling of Bray–Curtis indices in bacterial (A) and fungal (B) communities in three types of tissues of neighboring diseased and healthy eggplants in a number of fields in Tanzania. The large separation along the second dimension is due to differences among sampling fields.
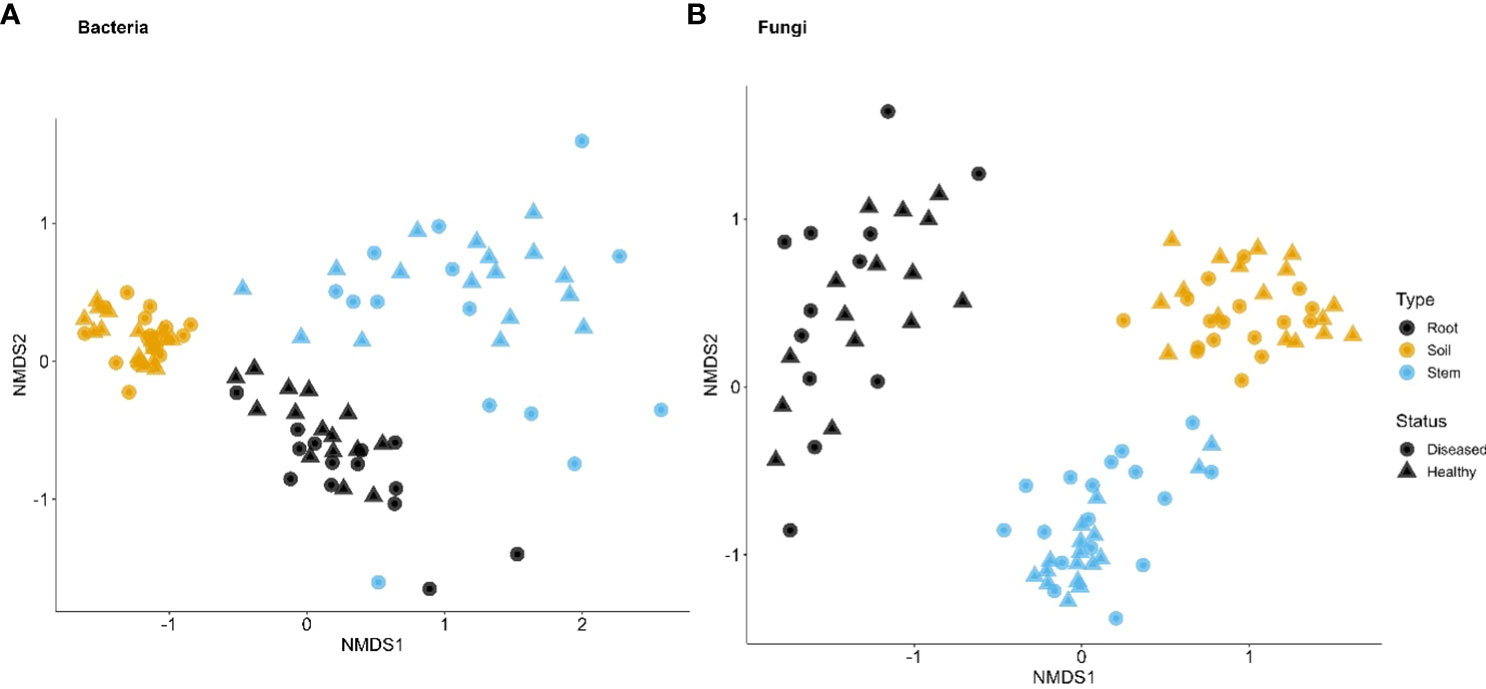
Figure 4 First two dimensions of the non-metric dimensional scaling of Bray–Curtis indices in bacterial (A) and fungal (B) communities in three types of tissues of neighboring diseased and healthy eggplants in a number of fields in Uganda.
Fungi
Although the effect of sample types on the fungal α diversity indices was highly significant, it accounted for far less variability, compared to indices for bacteria (Table 2). The overall trend of the differences among sample types was similar to the bacterial sequences. On the other hand, spatial location accounted for more variability for fungal than for bacterial sequences. Disease status impacted the α diversity of the fungal community far more than the bacterial in one of the Tanzania samples. For β diversity, spatial location was more important in Tanzania than Uganda; in Uganda, both spatial location and sample types were similarly important in affecting β diversity (Figures 3, 4, Table 2). As for bacteria, samples were clearly separated among the three sample types for Uganda (Figure 4).
Differential analysis of OTU-relative abundance
Tanzania – bacteria
For the stem samples, only one of 6064 bacterial OTUs differed significantly in its relative abundance between diseased and healthy plants—an OTU from Enterobacterales (with an average reads number of only 40 per sample) had a much higher abundance in diseased plants (log2 Fold Change = 25.7). None of the 14,935 bacterial OTUs differed in its abundance between diseased and healthy root samples. There were 314 rhizosphere bacterial OTUs, of the total 2,584 OTUs, with differential relative abundance between samples from diseased and healthy plants. Of the 314 OTUs, only four had higher relative abundance in the diseased plants; of the four OTUs, only a Stenotrophomonas OTU had a high average number of reads (= 2,328).
Tanzania – fungi
For the stem samples, five of the 1,120 fungal OTUs differed significantly in their relative abundance between diseased and healthy plants (Table 3). Three of the five OTUs had higher relative abundance in the diseased stems but only one Diaporthe OTU had a very higher number of reads. Another OTU (Candida intermedia) with high reads number had much high abundance on healthy plants. Of the 3,122 rhizosphere fungal OTUs, 22 differed in their relative abundance between samples from diseased and healthy plants, and all had a low number of average reads (Table 3). In the root samples, five of 2,299 OTUs differed in their relative abundance between diseased and healthy roots. However, only one of these OTUs had a moderate number of reads (average = 418) with a much increased relative abundance in the diseased roots—this OTU was identified as M. phaseolina (Table 3). In addition, a F. solani OTU had a higher abundance in diseased plants, but it had much lower reads number (about 8).
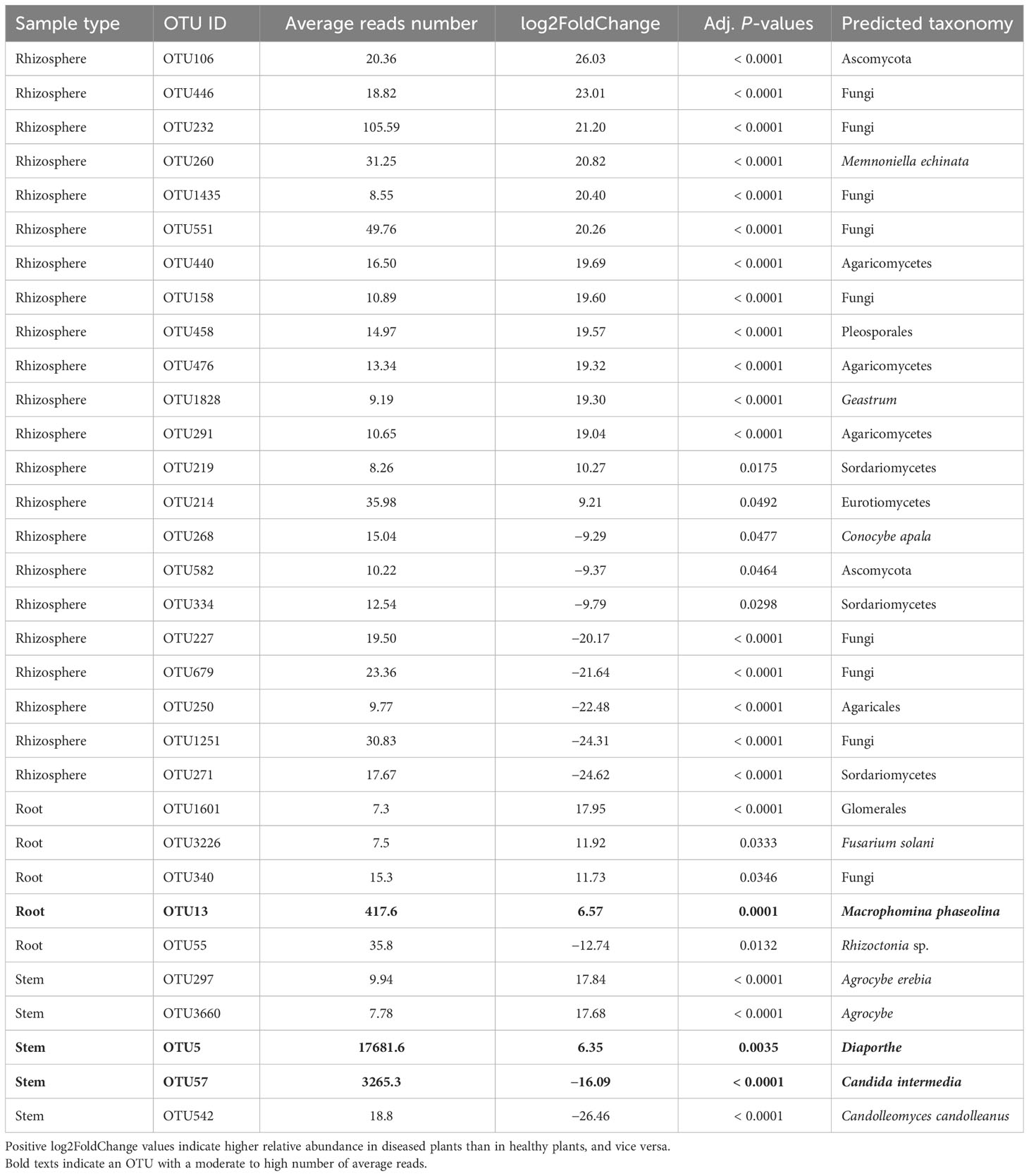
Table 3 Differential analysis summary of fungal taxa groups (OTUs) with differential relative abundance in the rhizosphere, or root, or stem between neighboring wilted and healthy eggplants sampled from three fields in Tanzania.
Uganda – bacteria
For the stem samples, 12 of the 5,592 bacterial OTUs differed significantly in their relative abundance between diseased and healthy plants (Table 4). Seven of the 12 OTUs had higher abundance in the diseased plants, of which only one (Sphingobacterium OTU) had a high number of reads. Four OTUs (one Pantoea and three Pseudomonas) with high reads number had lower abundance in diseased plants (Table 4). For the root samples, diseased and healthy plants differed in the relative abundance of 25 OTUs. Only one of these 25 OTUs had a high number of reads (= 41,980) as well as higher relative abundance in diseased plants; this OTU was identified as Ralstonia (R. solanacearum by further BLAST search). None of the rhizosphere bacterial OTUs differed between samples from diseased and healthy plants.
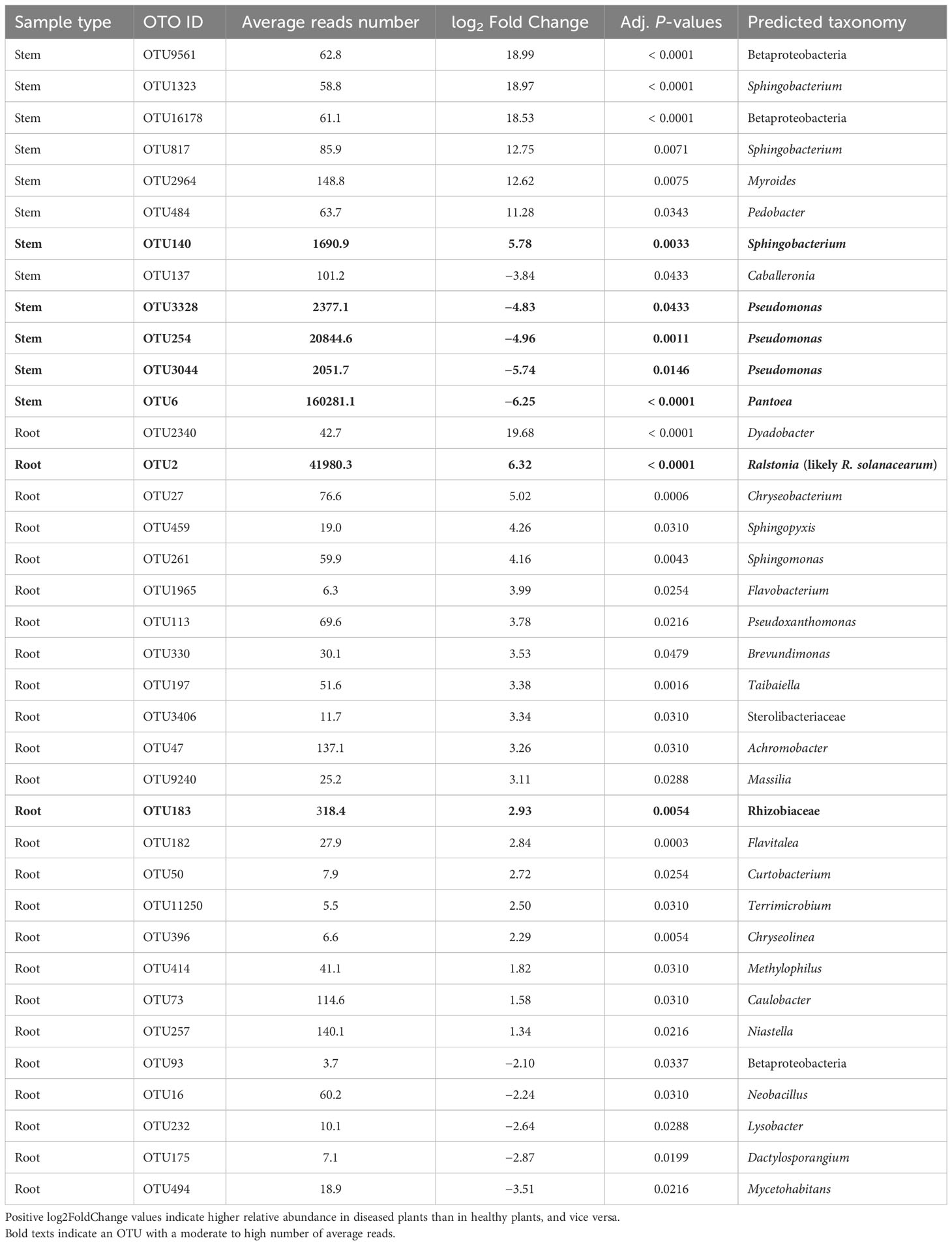
Table 4 Differential analysis summary of bacterial taxa groups (OTUs) with differential relative abundance in the rhizosphere, or root, or stem between neighboring wilted and healthy eggplants sampled from three fields in Uganda.
Uganda – fungi
For the stem samples, only two fungal OTUs differed in the relative abundance between diseased and healthy samples but both had a low average number of reads (Table 5). For root samples, 12 fungal OTUs differed in their relative abundance between diseased and healthy plants, seven of which had higher abundance in diseased plants. However, all these 15 OTUs had a low average number of reads except one Botrytis OTU (average = 386). None of the rhizosphere fungal OTUs differed between samples from diseased and healthy plants.
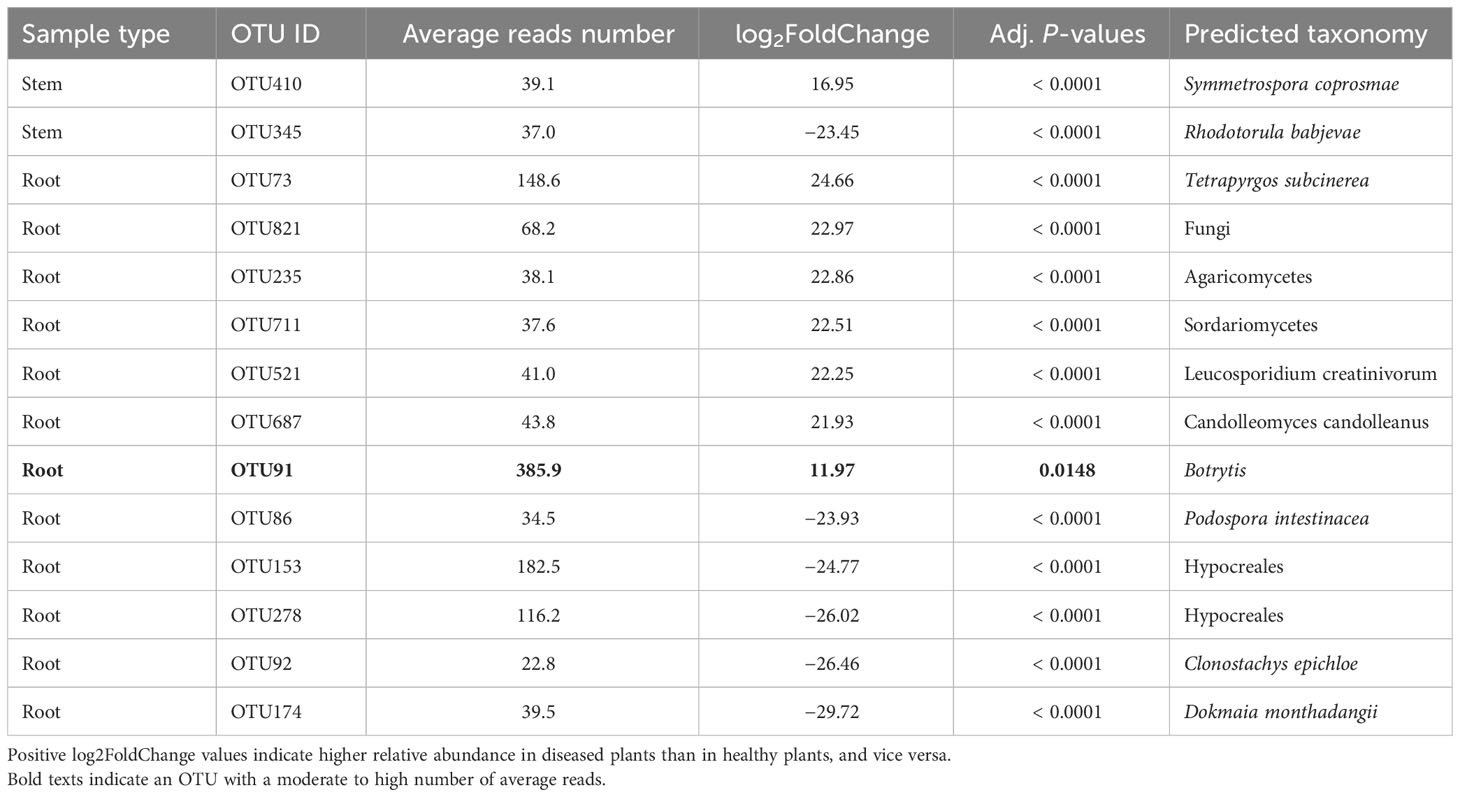
Table 5 Differential analysis summary of fungal taxa groups (OTUs) with differential relative abundance in the rhizosphere, or root, or stem between neighboring wilted and healthy eggplants sampled from three fields in Uganda.
Pathogenicity test
Due to challenges with exchanging plant pathogens or soil samples between countries we identified similar pathogen strains from UK collections to perform pathogenicity tests on S. aethiopicum.
Germination tests on PDA plates confirmed all three isolates used for inoculations were viable. In the first round of inoculations, only the plants that were infected with the M. phaseolina (isolate 268866) showed signs of wilting one week after inoculation. At this stage, wilting plants were sampled for re-isolation as well as healthy water control plants. All 10 wilting DB3 plants sampled and five of the six E11 plants sampled had fungal growth that was morphologically the same as the original isolate used to inoculate (the plate for one E11 plant was contaminated with other fungal growth). The stems sampled from the control (four DB3 and two E11) had growth of endophytic microbes or no growth. Because of subsequent contamination in subculturing, it was not possible to successfully sequence the re-isolated colonies. Non-wilting plants were left to grow for 10 weeks. There was no obvious sign of wilting in plants inoculated with the F. oxysporum isolate or the M. phaseolina PC1/17 isolate, despite drought stress applied—the inoculated plants remained in a similar condition to the control plants. In addition, nine DB3 plants and five E11 plants inoculated with the M. phaseolina (isolate 268866) remained asymptomatic.
In a repeat pathogenicity test with just the M. phaseolina (isolate 268866), two of the 12 DB3 plants, nine of the 12 E11 plants and all five of MM1961 plants showed signs of wilt two weeks after inoculation (Figures 5, 6). The morphology of those colonies re-isolated from stems of wilting plants of all the three genotypes was the same as the original isolate of M. phaseolina (isolate 268866, Figure 7). Sequence results identified the mycelium isolated from all the three S. aethiopicum genotypes to be identical to that of the original isolate.

Figure 5 Three genotypes (DB3, E11 and MM1961) of Solanum aethiopicum plants two weeks after inoculation with the Macrophomina phaseolina isolate 268866. Three control plants of each genotype were dipped in water as opposed to M. phaseolina inoculum.

Figure 6 Phenotypes of plants of three Solanum aethiopicum genotypes inoculated with the Macrophomina phaseolina isolate 268866 or water (control); photo was taken after the compost was washed-off two weeks after inoculation.
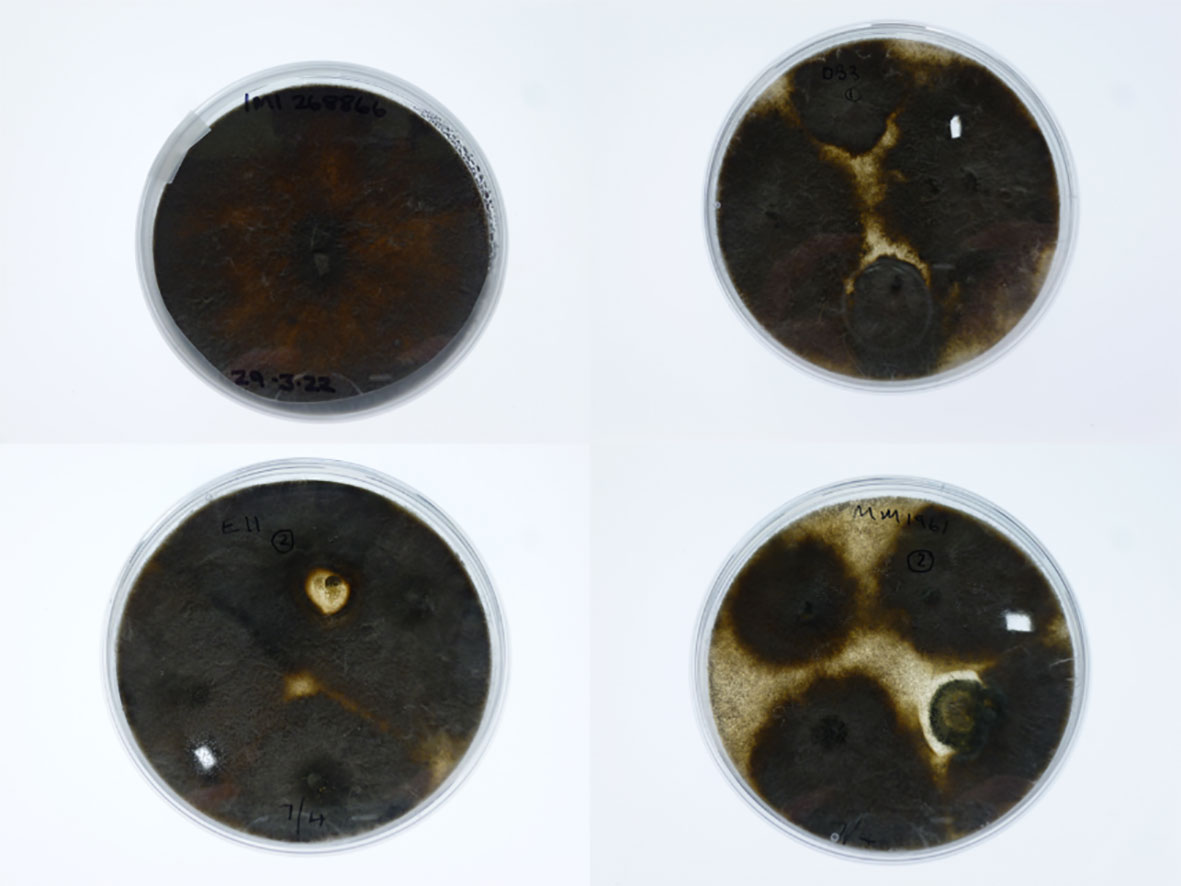
Figure 7 Photos of colony morphology on potato dextrose agar plates: top left—Macrophomina phaseolina Isolate 268866 used for inoculation; top right—fungal colony re-isolated from the stem of an inoculated Solanum aethiopicum DB3 plant; bottom left—fungal colony re-isolated from the stem of an inoculated S. aethiopicum E11 plant; and bottom right—fungal colony re-isolated from the stem of an inoculated S. aethiopicum DMM1961 plant.
Discussion
We identified putative causal pathogens for African eggplant wilt disease based on amplicon sequencing: wilting is most likely to result from a bacterial pathogen (R. solanacearum) in Uganda whereas from a fungal pathogen (M. phaseolina) in Tanzania. Further artificial root inoculation of a number of selected varieties/lines confirmed that M. phaseolina can cause wilting symptoms in African eggplant.
Ralstonia solanacearum, previously known as Pseudomonas solanacearum and Burkholderia solanacearum, can cause bacterial wilt in many plant species (Hayward and Hartman, 1994) and is considered a species complex with considerable variation in host, geographic origin, and pathogenicity properties (Denny, 2006). Its severity also varies with temperature, more severe as temperature increases. It can infect many Solanum species, including tomato (S. lycopersicum) and Thai eggplants (S. melongena) (Akarapisan et al., 2022). There is clear pathogenicity differentiation among R. solanacearum strains from different hosts, which may be considered as evidence for host specialization (Ailloud et al., 2015). Nearly all published research studies on the eggplant wilt disease have been focused on S. melongena. There is considerable amount of genetic diversity of R. solanacearum isolated from S. melongena grown in five provinces of the Philippines (Lewis Ivey et al., 2007). On the other hand, S. melongena genotypes differed in their resistance to a range of R. solanacearum strains (Lebeau et al., 2011), involving both broad-spectrum and strain-specific Quantitative Trait Loci (QTLs) (Salgon et al., 2017). The present study indicated that wilting symptoms on the Shum group of S. aethiopicum in Uganda is most likely, as in S. melongena, to be caused by R. solanacearum. This was not the case for the Gilo group in Tanzania. It is not clear whether such a difference is due to the lack of pathogenic R. solanacearum inoculum in Tanzania fields and/or low level of susceptibility in the Gilo group genotypes. In 2014, ANSES (French Agency for Food, Environmental and Occupational Health and Safety) identified wilt causing R. solanacearum strain Phylotype I sequevar l-31 in samples collected from tomato in Tanzania (Thibault Nordey, per. comm.). Further study is needed to assess the extent of variability of S. aethiopicum resistance/susceptibility to R. solanacearum in relation to the Shum and Gilo groups.
Amplicon sequencing suggested that M. phaseolina is likely a putative pathogen for wilt symptoms on the Gilo group plants in Tanzania. Recently, a new Macrophomina species (M. tecta) has been described (Poudel et al., 2021), which was isolated from the stems of sorghum (Sorghum bicolor) and mungbean (Vigna radiata) plants with charcoal rot symptoms in Australia. Our Macrophomina sequence for the putative wilt pathogen is an exact match for both M. phaseolina and M. tecta (but this new species has not been incorporated into the UNITE database we used [V8.3, 2021-05-10]). It is, therefore, impossible to categorically identify this candidate wilt pathogen based only on the present ITS amplicon sequence. Nevertheless, as we used one archived strain of M. phaseolina for pathogenicity testing, we, thus, still refer to this candidate wilt pathogen OTU as M. phaseolina.
We tried to isolate M. phaseolina from diseased plants in those sampled fields in Tanzania, but only a few fungal strains were isolated from diseased roots of S. aethiopicum in the present study, with the morphology similar to Fusarium spp. (Ruth Minja unpublished) but it was not confirmed molecularly. This may suggest that Fusarium spp. is easier to isolate from diseased tissue than M. phaseolina as the amplicon sequencing data showed the presence of several Fusarium spp. in large abundance. We used two M. phaseolina strains [one originated from Egypt and the other from Sri Lanka] in artificial inoculation studies and demonstrated that M. phaseolina can cause typical wilting symptoms on the eggplants of both the Gilo and Shum groups. M. phaseolina causes damping off, seedling blight, collar rot, stem rot, charcoal rot, basal stem rot, and root rot on many plant species. It can infect over 500 plant species and has a wide geographic distribution (Marquez et al., 2021); large morphological and pathogenic variations exist in M. phaseolina (Iqbal and Mukhtar, 2014). It has been reported to cause root rot of S. melongena (Khan and Gupta, 1998).
It is not possible to exclude the possibility that wilting in the Gilo group plants is caused by Fusarium spp. The most abundant fungal OTU in Tanzania is a Fusarium OTU, most likely from F. solani based on BLAST search; there were other Fusarium OTUs with high counts (possibly F. oxysporum and F. equiseti). These three Fusarium OTUs were more abundant in the stem tissues of diseased plants than in healthy plants (although not statistically significant). In addition to causing wilting in S. melongena (Li et al., 2017), F. solani can also cause stem cankers on S. melongena var. esculentum eggplants (Cerkauskas, 2010). Only one F. solani OTU had a higher abundance in the diseased roots but had low reads number. All other Fusarium OTUs with high reads did not differ significantly in their relative abundance between diseased and healthy roots, suggesting they were unlikely the main agents causing root infections. Three Fusarium species (F. oxysporum, F. solani and F. equiseti) were identified from African eggplant in Tanzania. F. oxysporum and F. equiseti were pathogenic against S. macrocapon (MM 1131) and S. anguivi (N 19) whereas F. solani has previously been shown to be pathogenic against a single S. anguivi genotype (Mwaniki et al., 2011). Wilting in S. melongena can also be caused by F. oxysporum f. sp. melongenae (Safikhani et al., 2013; Rao et al., 2019). When tested against one specific F. oxysporum f. sp. melongenae isolate, a number of S. macrocarpon and S. aethiopicum accessions are resistant whereas S. melongena accessions are overall the most susceptible (Mwaniki et al., 2016). Further research is needed to assess resistance/susceptibility of S. aethiopicum genotypes to these putative fungal pathogens in both the Shum and Gilo groups.
There was a Diaporthe OTU with a high number of reads and more abundant in the stem tissues of diseased plants in Tanzania; Phomopsis vexans is an anamorphic stage of D. vexans. Phomopsis vexans can cause shoot/leaf blight and fruit rot in S. melongena eggplants (Nahar et al., 2019; Bhanushree et al., 2021; Keinath, 2021). Wilt symptoms in those sampled fields in Tanzania may, therefore, have resulted from mixed infections: root infections primarily by M. phaseolina and infection of aboveground tissues by Fusarium spp. and Diaporthe. Roots may be infected simultaneously by several pathogens, such as M. phaseolina and F. solani. Further research is needed to understand which is the primary cause of the wilt disease and/or whether these pathogens can interact to overcome host resistance under field conditions. Once the etiology of the wilt disease on S. aethiopicum is clarified, it will then be possible to develop and implement an integrated approach to manage the disease. This could include the use of beneficial microbes, such as arbuscular mycorrhizal fungi (AMF) and Trichoderma, to improve plant health. The intermediate impact of AMF is the improved resilience to limited water supply, subsequently increasing yield and alleviating the bitter taste of some solanaceous crops, like the leafy S. aethiopicum L., Shum group (Joseph et al., 2022). Trichoderma has mainly been associated with antibiotic remedies and ultimately can improve productivity (Kumar and Ashraf, 2017; Bubici et al., 2019). Interestingly, several Pseudomonas OTUs and one Candida intermedia OTU (with high reads) had a higher relative abundance in the stems of healthy plants in Uganda and Tanzania, respectively. Pseudomonas spp. are known to have biocontrol effect against plant pathogens (Haas and Keel, 2003; Prieto et al., 2009) whereas C. intermedia can produce volatiles that can inhibit Botrytis cinerea (Huang et al., 2011).
There is a Stenotrophomonas OTU with a high number of reads as well as high abundance in rhizosphere of diseased plants in Tanzania. Stenotrophomonas spp. are found throughout the environment, particularly in close association with plant roots, but are not known to infect plants (Ryan et al., 2009). This genus may contain specific strains that have antagonistic effects against plant pathogens (Lottmann and Berg, 1998) or plant growth-promoting effects (Ryan et al., 2009). It is not clear whether there is any biological association of increased relative abundance of this Stenotrophomonas OTU with diseased plants in rhizosphere.
In summary, we have shown that the wilt symptoms of African eggplant (S. aethiopicum) of the Gilo group in Tanzania are most likely due to the fungal pathogens: M. phaseolina infecting roots and F. solani infecting stems. Artificial inoculation demonstrated that M. phaseolina can infect plants of the S. aethiopicum Gilo group. On the other hand, the wilt disease on the S. aethiopicum Shum group plants in Uganda is most likely caused by the bacteria pathogen R. solanacearum. Further research is required to confirm the present results and inference. If these results/inferences are confirmed, then we need to develop and implement pathogen-specific management measures, such as using plant derived (Javaid and Khan, 2022) or synthesized (Zafar et al., 2022) compounds to inhibit M. phaseolina. The difference in putative causal agents between Uganda and Tanzania demonstrates the need for etiological studies before effective control methods can be developed and implemented.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: EBI under the EBI project accession PRJEB59191.
Author contributions
XX: Conceptualization, Formal Analysis, Funding acquisition, Writing – original draft. RM: Investigation, Project administration, Writing – review & editing. EBK: Investigation, Project administration, Writing – review & editing. FD: Data curation, Investigation, Project administration, Writing – review & editing. GD: Formal Analysis, Writing – review & editing. PK: Data curation, Investigation, Writing – review & editing. AK: Data curation, Investigation, Writing – review & editing. EK: Data curation, Investigation, Writing – review & editing. OM: Data curation, Investigation, Writing – review & editing. DM: Data curation, Investigation, Writing – review & editing. HM: Data curation, Investigation, Writing – review & editing. MN: Data curation, Investigation, Writing – review & editing. TP: Data curation, Investigation, Methodology, Writing – review & editing, Project administration. SS: Data curation, Investigation, Writing – review & editing. GS: Data curation, Investigation, Writing – review & editing. ES: Project administration, Supervision, Writing – review & editing. GB: Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The study was funded by UK Research and Innovation’s Biotechnology and Biological Sciences Research Council (BBSRC) through the GCRF SASSA initiative grant number BB/R020655/1.
Acknowledgments
We acknowledge World Vegetable Center and its long-term strategic donors for contributing to the implementation of the study. We thank TARI and Uganda Christian University for administrative and technical support, and thank extension staff and farmers for their assistance in sampling in Tanzania and Uganda.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abukutsa-Onyango M. (2010). Strategic repositioning of African indigenous vegetables in the Horticulture Sector (Entebbe, Uganda: Second Inaugural Lecture of the Jomo Kenyatta University of Agriculture and Technology).
Ailloud F., Lowe T., Cellier G., Roche D., Allen C., Prior P. (2015). Comparative genomic analysis of Ralstonia solanacearum reveals candidate genes for host specificity. BMC Genomics 16, 270. doi: 10.1186/s12864-015-1474-8
Akarapisan A., Kumvinit A., Nontaswatsri C., Puangkrit T., Kositratana W. (2022). Phylotype, sequevar and pathogenicity of Ralstonia solanaceaum species complex from Northern Thailand. J. Phytopathol. 170 (3), 176–184. doi: 10.1111/JPH.13065
Allen G. C., Flores-Vergara M. A., Krasynanski S., Kumar S., Thompson W. F. (2006). A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1 (5), 2320–2325. doi: 10.1038/NPROT.2006.384
Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11 (10), R106. doi: 10.1186/gb-2010-11-10-r106
Bengtsson-Palme J., Ryberg M., Hartmann M., Branco S., Wang Z., Godhe A., et al. (2013). Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 4 (10), 914–919. doi: 10.1111/2041-210x.12073
Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy Stat. Soc. B 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Bhanushree N., Saha P., Tomar B. S., Munshi A. D. (2021). Phomopsis blight in eggplant and strategies to manage through resistance breeding. J. Hortic. Sci. Biotechnol. 97 (1), 34–45. doi: 10.1080/14620316.2021.1966321
Bletsos F. A., Thanassoulopoulos C. C., Roupakias D. G. (1997). Level of resistance to Verticillium dahliae of an interspecific F1 hybrid (Solanum melongena X Solanum torvum). J. Genet. Breed. 51 (1), 69–73.
Bodenhausen N., Horton M. W., Bergelson J. (2013). Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS ONE 8 (2), e56329. doi: 10.1371/journal.pone.0056329
Bubici G., Kaushal M., Prigigallo M. I., Cabanás C. G. L., Mercado-Blanco J. (2019). Biological control agents against Fusarium wilt of banana. Front. Microbiol. 10 (APR). doi: 10.3389/FMICB.2019.00616
Cerkauskas R. F. (2010). Fusarium stem canker of greenhouse eggplant (Solanum melongena var. esculentum) in Ontario. Can. J. Plant Pathol. 30 (4), 614–618. doi: 10.1080/07060660809507562
Chelius M. K., Triplett E. W. (2001). The diversity of archaea and bacteria in association with the roots of Zea mays L. Microbial Ecol. 41 (3), 252–263. doi: 10.1007/s002480000087
Cole J. R., Wang Q., Fish J. A., Chai B., McGarrell D. M., Sun Y., et al. (2014). Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42 (Database issue), D633–D642. doi: 10.1093/nar/gkt1244
Denny T. (2006). “Plant pathogenic Ralstonia species,” in Plant-Associated Bacteria. Ed. Gnanamanickam S. S. (Dordrecht: Springer). doi: 10.1007/978-1-4020-4538-7_16
Dervis S., Yetisir H., Yildirim H., Tok F. M., Kurt S., Karaca F. (2009). Genetic and pathogenic characterization of Verticillium dahliae isolates from eggplant in Turkey. Phytoparasitica 37 (5), 467–476. doi: 10.1007/s12600-009-0061-4
Dinssa F. F., Hanson P., Dubois T., Tenkouano A., Stoilova T., Hughes J. A., et al. (2016). AVRDC – The world vegetable center’s women-oriented improvement and development strategy for traditional African vegetables in sub-Saharan Africa. Eur. J. Hortic. Sci. 81 (2), 91–105. doi: 10.17660/EJHS.2016/81.2.3
Dixon P. (2003). VEGAN, a package of R functions for community ecology. J. Vegetation Sci. 14 (6), 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Edgar R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10 (10), 996–998. doi: 10.1038/nmeth.2604
Edgar R. C., Flyvbjerg H. (2015). Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 31 (21), 3476–3482. doi: 10.1093/bioinformatics/btv401
Gardes M., Bruns T. D. (1993). ITS primers with enhanced specificity for basidiomycetes - application to the identification of mycorrhizae and rusts. Mol. Ecol. 2 (2), 113–118. doi: 10.1111/J.1365-294X.1993.TB00005.X
Haas D., Keel C. (2003). Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu. Rev. Phytopathol. 41, 117–153. doi: 10.1146/annurev.phyto.41.052002.095656
Hayward A. C., Hartman G. L. (1994). “Bacterial wilt: the disease and its causative agent, Pseudomonas solanacearum,” in Bacterial wilt: the disease and its Causative Agent, Pseudomonas solanacearum (Wallingford: CAB International).
Huang R., Li G. Q., Zhang J., Yang L., Che H. J., Jiang D. H., et al. (2011). Control of postharvest Botrytis fruit rot of strawberry by volatile organic compounds of Candida intermedia. Phytopathology 101 (7), 859–869. doi: 10.1094/PHYTO-09-10-0255
Iqbal U., Mukhtar T. (2014). Morphological and pathogenic variability among Macrophomina phaseolina isolates associated with mungbean (Vigna radiata L.) Wilczek from Pakistan. Sci. World J. 2014, 9. doi: 10.1155/2014/950175
Javaid A., Khan I. H. (2022). Chemical profile and antifungal activity of leaf extract of Tabernaemontana divaricata against Macrophomina phaseolina. Plant Prot. 6 (3), 201–208. doi: 10.33804/pp.006.03.4332
Joseph M., Geoffrey S., Tom B., Julian N. M., Brian Justus M., Bishop G. J., et al. (2022). Trader acceptability of African eggplant (Solanum aethiopicum Shum) genotypes and effect of bio-control treatments on consumer sensory acceptability. Int. J. Food Sci. Technol. 57 (9), 6165–6180. doi: 10.1111/IJFS.15977
Keinath A. P. (2021). Differences in susceptibility to Phomopsis blight of seven eggplant cultivars with different fruit types. Plant Health Prog. 23 (1), 57–64. doi: 10.1094/PHP-07-21-0100-RS
Khan M., Gupta J. (1998). Antagonistic efficacy of Trichoderma species against Macrophomina phaseolina on eggplant. J. Plant Dis. Prot. 105, 387–393.
Kõljalg U., Nilsson R. H., Abarenkov K., Tedersoo L., Taylor A. F. S., Bahram M., et al. (2013). Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22 (21), 5271–5277. doi: 10.1111/mec.12481
Kumar M., Ashraf S. (2017). Role of Trichoderma spp. as a Biocontrol Agent of Fungal Plant Pathogens. In: Kumar V., Kumar M., Sharma S., Prasad R. (eds) Probiotics and Plant Health. Singapore: Springer. doi: 10.1007/978-981-10-3473-2_23
Lebeau A., Daunay M. C., Frary A., Palloix A., Wang J. F., Dintinger J., et al. (2011). Bacterial wilt resistance in tomato, pepper, and eggplant: genetic resources respond to diverse strains in the Ralstonia solanacearum species complex. Phytopathology 101 (1), 154–165. doi: 10.1094/PHYTO-02-10-0048
Lewis Ivey M. L., McSpadden Gardener B. B., Opina N., Miller S. A. (2007). Diversity of Ralstonia solanacearum infecting eggplant in the Philippines. Phytopathology 97 (11), 1467–1475. doi: 10.1094/PHYTO-97-11-1467
Li B. J., Li P. L., Li J., Chai A. L., Shi Y. X., Xie X. W. (2017). First report of Fusarium root rot of Solanum melongena caused by Fusarium solani in China. Plant Dis. 101 (11), 1956. doi: 10.1094/PDIS-11-16-1559-PDN
Lottmann J., Berg G. (1998). Interactions between Stenotrophomonas maltophilia and the soilborne pathogen Verticillium dahliae var. longisporum. Bull. OILB/SROP 21 (9), 57–60.
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550. doi: 10.1186/s13059-014-0550-8
Marquez N., Giachero M. L., Declerck S., Ducasse D. A. (2021). Macrophomina phaseolina: general characteristics of pathogenicity and methods of control. Front. Plant Sci. 12. doi: 10.3389/FPLS.2021.634397/BIBTEX
Mwaniki P. K., Abang M., Wagara I. N., Wolukau J. N., Hans-Josef S. (2016). Response of African eggplants to Fusarium spp. and identification of sources of resistance. Afr. J. Biotechnol. 15, 392–400. doi: 10.5897/AJB2015.14874
Mwaniki P. K., Abang M., Wagara I. N., Wolukau J. N., Schroers H.-J., Tenywa J. S., et al. (2011). “Morphology, pathogenicity and molecular identification of Fusarium spp. from wilting eggplants in Tanzania,” in African Crop Science Conference Proceedings, Vol. 10. 217–221.
Mwinuka P. R., Mbilinyi B. P., Mbungu W. B., Mourice S. K., Mahoo H. F., Schmitter P. (2021). Optimizing water and nitrogen application for neglected horticultural species in tropical sub-humid climate areas: A case of African eggplant (Solanum aethiopicum L.). Scientia Hortic. 276, 109756. doi: 10.1016/J.SCIENTA.2020.109756
Nahar N., Islam M. R., Uddin M. M., de Jong P., Struik P. C., Stomph T. J. (2019). Disease management in eggplant (Solanum melongena L.) nurseries also reduces wilt and fruit rot in subsequent plantings: A participatory testing in Bangladesh. Crop Prot. 120, 113–124. doi: 10.1016/J.CROPRO.2019.02.018
Najar A. G., Anwar A., Masoodi L., Khar M. S. (2011). Evaluation of native biocontrol agents against Fusarium solan f. sp. melongenae causing wilt disease of brinjal in Kashmir. J. Phytology 3 (6), 2075–6240.
Ochieng J., Afari-Sefa V., Karanja D., Kessy R., Rajendran S., Samali S. (2018). How promoting consumption of traditional African vegetables affects household nutrition security in Tanzania. Renewable Agric. Food Syst. 33 (2), 105–115. doi: 10.1017/S1742170516000508
Omondi E. O., Debener T., Linde M., Abukutsa-Onyango M., Dinssa F. F., Winkelmann T. (2016). Molecular markers for genetic diversity studies in African leafy vegetables. Adv. Bioscience Biotechnol. 07 (03), 188–197. doi: 10.4236/abb.2016.73017
Papp-Rupar M., Karlstrom A., Passey T., Deakin G., Xu X. (2022). The influence of host genotypes on the endophytes in the leaf scar tissues of apple trees and correlation of the endophytes with apple canker (Neonectria ditissima) development. Phytobiome J 6, 127–138.. doi: 10.1094/PBIOMES-10-21-0061-R
Phukan T., Kabyashree K., Singh R., Sharma P. L., Singh N., Barman A., et al. (2019). Ralstonia solanacearum virulence in eggplant seedlings by the leaf-clip inoculation. Phytopathol. Res. 1 (1), 1–11. doi: 10.1186/S42483-019-0030-X
Poudel B., Shivas R. G., Adorada D. L., Barbetti M. J., Bithell S. L., Kelly L. A., et al. (2021). Hidden diversity of Macrophomina associated with broadacre and horticultural crops in Australia. Eur. J. Plant Pathol. 161 (1), 1–23. doi: 10.1007/S10658-021-02300-0/FIGURES/3
Prieto P., Navarro-Raya C., Valverde-Corredor A., Amyotte S. G., Dobinson K. F., Mercado-Blanco J. (2009). Colonization process of olive tissues by Verticillium dahliae and its in planta interaction with the biocontrol root endophyte Pseudomonas fluorescens PICF7. Microbial Biotechnol. 2 (4), 499–511. doi: 10.1111/j.1751-7915.2009.00105.x
Rao V. G., Kumar B. P., Dhutraj D. N., Apet K. T., Ambadkar C. V., Kumar P., et al. (2019). Characterization and variability of Fusarium oxysporum f. sp. melongenae (Schlecht) Mutuo and Ishigami from wilting eggplants in Marathwada region of Maharashtra. J. Pharmacognosy Phytochem. 8 (5), 1436–1443.
R Core Team, R Core Development Team, R Foundation for Statistical Computing (2019). R: A Language and environment for statistical computing (Vienna, Austria: Austria: R Foundation for Statistical Computing).
Remezani H. (2008). Biological control of root-rot of eggplant caused by Macrophomina phaseolina. American-Eurasian J. Agric. Environ. Sci. 4, 218–220.
Ryan R. P., Monchy S., Cardinale M., Taghavi S., Crossman L., Avison M. B., et al. (2009). The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Rev. Microbiol. 7 (7), 514–525. doi: 10.1038/nrmicro2163
Safikhani N., Morid B., Reza Zamanizadeh H. (2013). First report of Fusarium wilt of eggplant caused by Fusarium oxysporum f. sp. melongenae in Iran. New Dis. Rep. 28 (1), 16–16. doi: 10.5197/J.2044-0588.2013.028.016
Salgon S., Jourda C., Sauvage C., Daunay M. C., Reynaud B., Wicker E., et al. (2017). Eggplant resistance to the Ralstonia solanacearum species complex involves both broad-spectrum and strain-specific quantitative trait loci. Front. Plant Sci. 8. doi: 10.3389/FPLS.2017.00828/FULL
Ssekabembe C. K., Odong T. L. (2008). Division of labour in nakati (Solanum aethiopicum) production in central Uganda. Afr. J. Agric. Res. 3, 400–406.
Sseremba G., Tongoona P., Eleblu J. S. Y., Danquah E. Y., Kaweesi T., Baguma Y., et al. (2018). Stability of Solanum aethiopicum Shum accessions under varied water deficit stress levels and identification of pertinent breeding traits for resistance to water shortage. Euphytica 214 (1), 1–11. doi: 10.1007/S10681-017-2097-8/FIGURES/4
UBOS (2013). 2020 Statistical Abstract Vol. 1 (Kampala, Uganda: Uganda Bureau of Statistics), 38–40.
White T. J., Bruns T. L., Taylor J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogentics,” in A Guide to Molecular Methods and Applications. Eds. Innis M. A., Gelfand D. H., Snisky J. J., White T. J. (New York: Academic Press), 315–322.
Keywords: wilt, amplicon-sequencing, Ralstonia solanacearum, Macrophomina phaseolina, African eggplant
Citation: Xu X, Minja R, Kizito EB, Dinssa F, Deakin G, Kabod PN, Kalala A, Kweka E, Mbwambo O, Mbanzibwa D, Msangi H, Nakanwagi MJ, Passey T, Sentance S, Sseremba G, Stavridou E and Bishop GJ (2024) Amplicon sequencing identified a putative pathogen, Macrophomina phaseolina, causing wilt in African eggplant (Solanum aethiopicum) grown in Tanzania and Uganda. Front. Agron. 5:1300324. doi: 10.3389/fagro.2023.1300324
Received: 23 September 2023; Accepted: 06 December 2023;
Published: 03 January 2024.
Edited by:
Anamika Sharma, Virginia Tech, United StatesReviewed by:
Sudhir Navathe, Agharkar Research Institute, IndiaTariq Mukhtar, Pir Mehr Ali Shah Arid Agriculture University, Pakistan
Copyright © 2024 Xu, Minja, Kizito, Dinssa, Deakin, Kabod, Kalala, Kweka, Mbwambo, Mbanzibwa, Msangi, Nakanwagi, Passey, Sentance, Sseremba, Stavridou and Bishop. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangming Xu, eGlhbmdtaW5nLnh1QG5pYWIuY29t
 Xiangming Xu
Xiangming Xu Ruth Minja2
Ruth Minja2 Elizabeth Balyejusa Kizito
Elizabeth Balyejusa Kizito Fekadu Dinssa
Fekadu Dinssa Greg Deakin
Greg Deakin Pamela Nahamya Kabod
Pamela Nahamya Kabod Deusdedith Mbanzibwa
Deusdedith Mbanzibwa Mildred Julian Nakanwagi
Mildred Julian Nakanwagi Godfrey Sseremba
Godfrey Sseremba Eleftheria Stavridou
Eleftheria Stavridou Gerard J. Bishop
Gerard J. Bishop