
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Agron., 19 October 2023
Sec. Plant-Soil Interactions
Volume 5 - 2023 | https://doi.org/10.3389/fagro.2023.1216520
Agricultural products such as tea, chocolate, coffee and wine are valued for their sensorial and nutritional qualities. Variation in the growing conditions of a crop can influence the plant’s phenotype, thus it behooves agriculturalists to optimize the conditions on their farms to grow the highest quality product. The set of growing conditions associated with a certain geographic location and its influence on the product’s chemistry is known as terroir. Although terroir plays a significant role in marketing and consumer appreciation as well as product identity and valorization, rarely are the biochemical differences or the factors creating them very well understood. The word derives from the Latin for “land”, suggesting terroir is simply a function of the geographical location where a plant grew, while in its modern usage, terroir is understood to be the result of soil type, climate, landscape, topography, biotic interactions and agricultural practice. Except for fermented food products like wine and chocolate, plant associated microbiomes have been little studied for their contribution to a crop’s terroir; however, modern metagenomics and metabolomics technologies have given scientists the tools to better observe how microbial diversity can impact the chemical variation in plant products. Differences in the microbiomes inhabiting plant organs can change phytochemistry by altering host metabolism, for example increasing the nutrients absorbed by roots that then are deposited in leaves, seeds and fruits. Plant associated microbes can consume plant molecules, removing them from the metabolome, or they can contribute smells and flavors of their own. This review aims to synthesize research into rhizosphere, endosphere, phyllosphere, spermosphere, carposphere, and anthosphere microbiome influences on plant biochemistry and crop derived products, while helping to increase the appreciation that beneficial microbes are able to contribute to agriculture by improving phytochemical quality.
Much of modern plant agriculture is dominated by the quest for higher yield, but in niche or luxury products such as wine and coffee, producers also search for methods to enhance or maintain quality. One of the original concepts surrounding agricultural enhancement of flavor, the word “terroir” derives from 14th century France where it referred to specific tracts of land in the province of Burgundy that consistently produced very good wines (Wilson, 2001). French wine laws for the “Protection of the Place of Origin” or Appellation d’Origine Contrôlée (AOC) continue to ensure a particular terroir by stipulating that the products (including but not limited to wine) are made in a traditional manner with ingredients from specifically classified producers within designated geographical areas. Similar legal/commercial definitions of terroir exist in other countries such as Italy’s Denominazione di Origine Controllata e Garantita, Germany’s Qualitätswein bestimmter Anbaugebiete, Spain’s Denominación de Origen Protegida, Portugal’s Denominação de Origem Controlada, South Africa’s Wine of Origin, Canada’s Vintners Quality Alliance, and the USA’s Appellation of Wine Origin. Outside of wine, the concept of terroir also exists for Longjing hand-roasted green tea grown in the West Lake region in Hangzhou. This tea is considered the most prestigious drink in China, having been consumed by many emperors for hundreds of years and is officially recognized for its terroir with geographical indication labelling (Chan, 2012). Such geographic indications are an important part of agricultural marketing and pricing, for example “Café de Colombia” labelled coffee sells on average for at least three times the price of unlabeled roasted coffee (US$6.99/kg), while “Jamaican Blue Mountain Coffee” sells for over 13 times more (Teuber, 2010).
Besides geographic provenance, a product’s perceived quality can also dramatically increase its price; 1-point increases on the 100-point coffee tasting scale at the annual Cup of Excellence competition result in at least 18% increases in price, while price increases of 15-133% result for coffees ranked in the top four (Traore et al., 2018). Perhaps the most famous example is “Esmeralda Special” the product of the coffee varietal called Geisha grown at Hacienda La Esmeralda in western Panama. Said to contain aromas of jasmine, tangerine and bergamot, it was described by Don Holly, the director of roasting and quality control at Green Mountain Coffee Roasters as “the face of God in a cup”. Esmeralda Special won the competition in 2007 and fetched an auction price of US$374.85/kg (Wilson et al., 2012). More recently another cupping competition winner, a Geisha varietal from the Elida Estate farm in Panama became the most expensive coffee in the world, selling for US$2,268.95 per kilogram (Repanich, 2019). Medal winning wines also increase at least 13% in price (Paroissien and Visser, 2020), while those receiving high critical scores on Wine Spectator’s 100-point scale can fetch up to US$1,000 per 750 mL bottle (Palmer and Chen, 2018). The most expensive wine in the world is probably Screaming Eagle’s 1992 Napa Valley Cabernet Sauvignon which was given a near perfect score of 99 by the world’s most influential wine critic, Robert Parker, and sold for US$83,000 per liter (Batchelor, 2022).
From a commercial point of view, it is desirable to be able to prove that your agricultural product has “good terroir” or is at least biochemically distinct. However, this has traditionally been difficult to do, since the human tongue has difficulty discerning and quantifying subtle variations in chemical diversity or concentration. Scientifically speaking, in order to understand metabolic changes in a product’s flavor, it is also important to identify which chemical compounds are varying in an agricultural product. Fortuitously, advances in chromatography and mass spectroscopy have dramatically improved our ability to identify and quantify the distinctive blends of metabolites within different plant products. Metabolomics now allows the objective characterization of terroir, for example biochemically discriminating between wines produced from Pinot noir grapes grown in two distinct “Grands Crus” appellations (Burgundy, France) separated by less than 2 km (Roullier-Gall et al., 2014). Chemical composition was also able to distinguish between Chardonnay grapes produced at seven different Geographical Indications in Southern Australia (Gambetta et al., 2017). The use of untargeted metabolomics (da Silva Taveira et al., 2014), and classical physio-chemical/sensory analysis of green and roasted beans, has also been shown to permit the discrimination of a terroir between coffee grown in different geographical locations, environmental conditions and agricultural techniques (Scholz et al., 2018). Other examples of studies linking phytochemical profiles to terroir of tea, chocolate, saffron, fruits, medicinal plants and a range of other agricultural products has recently been reviewed (Lucini et al., 2020).
Agriculturalists hoping to increase the quality and price of their products might want to attempt to duplicate or engineer a “good terroir” for their plants, but to do this they need to better understand the factors involved. Besides geographic origin, the modern concept of terroir encompasses other contributors as well, stating that it “refers to an area in which collective knowledge of the interactions between the identifiable physical and biological environment and applied agricultural practices develops, providing distinctive characteristics for the products originating from this area. Terroir thus includes specific soil, topography, climate, landscape characteristics and biodiversity features.” (Castellucci, 2010). Figure 1 summarizes the factors contributing to agricultural terroir. Traditional agricultural practices are one important contributor to terroir and could include any number of things such as tilling v. no-till of the soil, organic v. conventional fertilization, etc. Soil is the most famous factor contributing to terroir, and it can vary in physical structure (eg. sand v. clay v. compost), water retention and nutrient content. An example of a good soil for wine grape production, is one that provides little nitrogen to the vine, causing abiotic stress which increases berry sugar, phenolic, anthocyanin and tannin concentration (Van Leeuwen, 2010). Topography refers to the geographic features (ie. mountains, valleys, lakes, etc.) and elevation of the farm. Although the majority of the world’s foremost wine region, Listrac-Médoc, Bordeaux, France, is just above sea level where warmer conditions help Cabernet Sauvignon and Merlot grapes to flourish, growing grapes at high altitudes (3100 m, Finca Altura Maxima, Salta, Argentina) hits the vines with more ozone and UV radiation, creating fruit with thicker and darker skin, resulting in wines with deeper colors and more robust tannin profiles (Krebiehl, 2023). Landscape refers to the immediate lay of the planted land; for example is the farm flat or sloping and which way is it facing? In wine production, the best landscape for a vineyard is a well-drained mid-slope, where the plants are elevated enough to receive good sunlight while being protected from frost and winds (Krebiehl, 2023). Climate (including solar radiation, precipitation, humidity, temperature and wind) is the dominant factor that controls fruit ripening potential and agricultural product characteristics; it also dictates which crop varieties can be grown where. One example of how climate can impact terroir, is of heavy precipitation during grape berry maturation where it can it can trigger fungal pathogen blooms, dilute fruit sugar and flavor levels, and severely limit both fruit yield and wine quality (Jones, 2018). Biodiversity features are the last factor considered in this definition of terroir, and could include any number of plant-organism interactions, from which species of bee pollinates the flowers, the abundance of pathogens that drift onto leaves as spores, or what kinds of mycorrhizae dominate in the soil. Although there are potentially thousands of plant-microbe interactions occurring on any given farm, the majority of these have been invisible until recent advances in DNA sequencing technology have allowed us to zoom in and observe plant microbiome diversity in its totality. Considering the intimate association that plants maintain with a plethora of bacteria, fungi, viruses, protists and archaea, many scientists now consider them altogether as a holobiont – this is a “close association between different individuals, usually host-microbiota symbioses, that together form anatomical, physiological, immunological or evolutionary units” (Baedke et al., 2020). The microbial portion of this holobiont can be vertically or horizontally transmitted, populating plant tissues inside and out. This microbiome is dynamic and can change as new members arrive or environmental conditions change, favoring particular microbes over others (Bulgarelli et al., 2013). It is interesting to realize that a plant holobiont contains both microbes and host plant which can both be affected by external environmental factors, suggesting a complex web of direct (affecting plant) and indirect (affecting microbiome) influences which contribute to terroir.
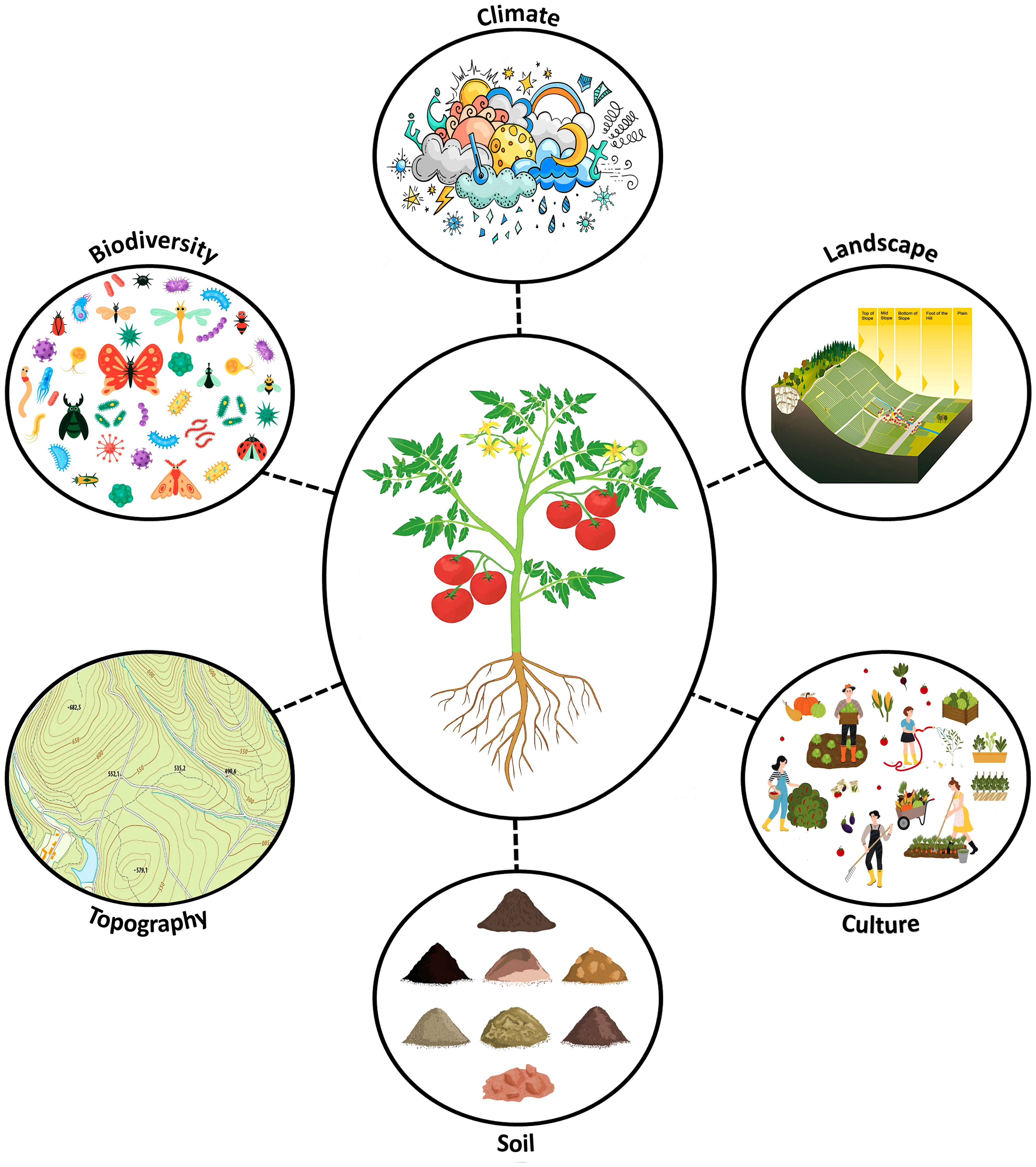
Figure 1 Terroir is an agricultural product’s or crop plant’s phenotype when grown in a defined area and it is shaped by that locality’s characteristics including: applied agricultural practices (culture), soil, topography (elevation), biodiversity features (plant-biotic interactions), climate, and landscape characteristics.
Understanding the ways that microbes contribute to an agricultural product’s chemistry is arguably the final frontier in the study of terroir. Having co-evolved with plants for hundreds of millions of years, microbial endophytes, epiphytes, symbionts and commensals are able to profoundly affect their host’s physiology and metabolism, for example stimulating plant growth through the production of phytohormones, provision of nutrients, enhancing tolerance to abiotic stress, or altering tissue chemistry to help resist pathogen attack (Figure 2) (Johnston-Monje et al., 2019). Usually these plant phenotypes are explained through the activity of one or two particular microbes (eg. nitrogen fixing bacteria in a legume’s nodules), but broad microbiome characteristics like species richness and evenness are also important for structuring communities to provide nutrient cycling and stress/disease resistance to the plant (Bakker et al., 2012). Said another way, “Although specific functions can be attributed to specific microorganisms, it is the total microbiome and its interactions that affect plant health (and terroir).” (Berendsen et al., 2012). Using these new techniques of metagenomics and metabolomics, scientists are beginning to observe novel correlations between plant associated microbiomes to the metabolomes of plants having “good terroir” (O’Banion et al., 2020), especially in grape wine production were the concept of terroir is most strongly developed (Liu et al., 2019; Belda et al., 2020). To be clear, we define microbial terroir as a particular plant associated microbiome (associated with a certain geographic region or not) which can somehow alter the chemical makeup of the plant/agricultural product relative to plants possessing other different microbiomes. This review will highlight research that has begun to prove that microbial populations can impact the chemistry of crop plants and agricultural products. Examples of how single microbes can affect plant chemistry and physiology will also be mentioned to help illustrate the mechanisms involved. Studies of microbial populations in the soil or rhizosphere, carposphere (fruit), endosphere, phyllosphere, spermosphere (seeds) and anthosphere (flowers) influencing plant chemistry, flavor, fragrance, or nutritional content will be presented as evidence supporting the concept of microbial terroir. By understanding how certain microbes or microbiomes can impact plant chemical makeup, agriculturalists may begin to develop practices of microbial terroir engineering to enhance the quality and sale price of their plant products.
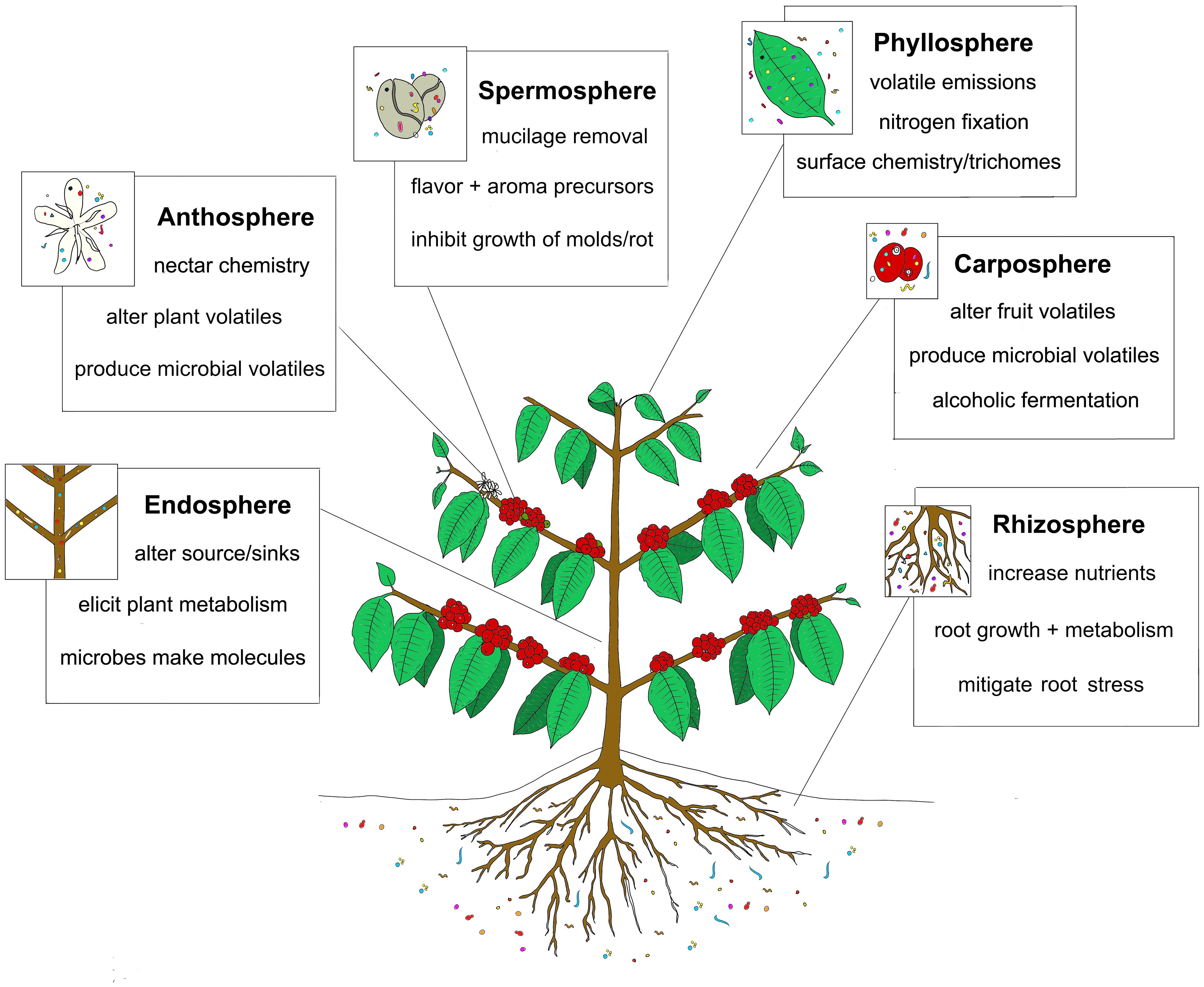
Figure 2 Microbiomes in different plant organs and surfaces contribute to the agricultural terroir which influences the plant metabolome, and product flavor/nutritional value. Microbial populations in the rhizosphere, endosphere, phyllosphere, spermosphere, carposphere, or anthosphere can influence crop chemistry in a number of different ways including microbial production of flavor metabolites, microbial degradation of plant biochemicals and microbial manipulation of plant metabolism.
As the principal factor contributing to classical terroir, good soil is very important for growing healthy and productive plants. Soil is also now known to be the most biodiverse habitat on Earth, with a typical gram of dirt containing 4000 genotypes of bacteria and 2000 different fungi (Decaëns, 2010). Microbiome studies using high throughput sequencing techniques have helped reveal that soil bacterial, protizoan and/or fungal diversity varies in correlation with differences in climate, soil physio-chemistry, agricultural practices and plant species (Bardgett and van der Putten, 2014; Fierer, 2017). Biogeographical variation of soil microbiomes is an important factor spurring exotic plant invasions (Rout and Callaway, 2012), as well as orchid seed germination which depends on the presence of compatible soil symbionts belonging to a Rhizoctonia-like group of fungi (Rasmussen et al., 2015; Voyron et al., 2017), medicinal root saponin content of Panax notoginseng growing in different parts of China (Wei et al., 2020), concentrations of the “impact aroma” molecule rotundone in Shiraz grapes. (Gupta et al., 2019) and the metabolome of Australian Pinot Noir wine (Liu et al., 2020). The influence of mycorrhizae and rhizobacteria on tea productivity and quality has been recently reviewed (Bag et al., 2022).
Comparisons of rhizosphere microbiome to plant metabolome have established a link between the two; this is true of grape (Liu et al., 2019; Zhou et al., 2021), Arabidopsis (Badri et al., 2013), tomato (Escobar Rodríguez et al., 2021) and rice (Deshmukh et al., 2016). With the exception of inoculation studies involving PGPR, mycorrhizae or rhizobia (Van Der Heijden et al., 2008), it is very difficult to know which microbes may have most influenced the plant metabolome (Misra et al., 2017; White, 2020), since very few of these organisms have well known and defined relationships with plants (Wardle et al., 2004). Even less is known about which mechanisms the soil microbes may have used to alter the chemical makeup of the crop plant (Rillig et al., 2018). Soil microbes could translocate to distal organs (ie. enter root and travel to fruit) and there influence the plant metabolome, however we will not explore this topic in our review.
Plants need to absorb 17 different minerals to grow, metabolize and reproduce, including the macronutrients C, H, O, N, P, S, K, Ca, and Mg (Osman, 2013). Conversely, plants can also accumulate toxic compounds such as heavy metals which can be bad for plant growth and human consumption. With little or no saprophytic ability, plant absorption of nutrients depends on soil microbes to digest, breakdown, fix and solubilize compounds in the soil so that the roots can then take up bioavailable minerals. Rhizosphere saprobes will also absorb most of the nutrients they are liberating, forcing the plant to obtain them through symbiosis, to wait for the microbes to die (Kuzyakov and Xu, 2013), or in a recently discovered process called rhizophagy, to ingest and digest the bacteria (White et al., 2018). Alternatively, microbes may also secrete exopolysaccharides, produce biofilms, change the soil pH, chelate, biosorb, precipitate or otherwise lock up soil compounds which may make them unavailable for uptake by the plant. Variation in soil microbial diversity could change nutrient availability for plants in many ways and for example, has been shown to affect litter decomposition by up to 20% (Strickland et al., 2009).
Although nutrients are important to survive, uptake of too much or too little can impact flavor, fragrance and nutritional quality of the agricultural product. In grape production for example, a good soil provides little nitrogen to the vine, causing abiotic stress which reduces plant vigor, berry weight and yield, while increasing berry sugar, phenolic, anthocyanin and tannin concentration – metabolites important in red wine flavor (Van Leeuwen, 2010). Tea leaves grown on phosphorus deficient soil have significant reductions in yield and alterations to their mineral and metabolite content, especially flavonoids, organic acids and amino acids (Ding et al., 2017). Panax notoginseng, called Sanqi in China, is a medicinal plant whose root accumulates higher levels of medically important saponins in soil that is nitrogen and potassium rich (Zhang et al., 2016). Cacao grown on soils containing cadmium tend to accumulate the heavy metal in their seeds, contaminating the resulting chocolate which is banned by some countries if it contains as little as 0.1 mg of Cd/kg (Maddela et al., 2020).
Rhizosphere microbes that alter soil mineral and nutrient availability, could thus affect plant mineral uptake, and thus impact its metabolome. For example, “within-vineyard” enrichment of Alfa- and Gamma-Proteobacteria in soil, correlates with increases of N and S uptake, which in turn correlates with a reduction of berry phenolics, reducing Sangiovese wine quality scores by 19 points (Mocali et al., 2020). Inoculating Tempranillo grapevines with a commercial blend of 5 mycorrhizal fungi (Bioradis Gel by Bioera SLU, Tarragona, Spain) increased phenolic and carotenoid concentrations in leaves, glucose and amino acid content in the fruit (Torres et al., 2019). Similarly, in a pot experiment, inoculation of tea plants with AMF increased N uptake and significantly increased the content of leaf polyphenols, catechuic acid, flavonoids, amino acids, and soluble proteins (Shao et al., 2019). Inoculation of wheat and maize with GFP labelled diazotroph Pseudomonas protegens Pf-5 X940 showed that it localized to root surfaces where it secreted ammonium that increased nitrogen content in both vegetative and reproductive tissues (Fox et al., 2016). The stalk metabolome of bitter and sweet sugarcane varies by 247 different compounds and is highly correlated to the nitrogen concentration and bacterial diversity found in soil (Huang et al., 2021).
Next to N, P is considered the most limiting nutrient for plant growth. Many different soil microbes have been shown to be able to promote plant growth and increase yield by augmenting P availability in soil (Alori et al., 2017). Evidence of soil microbial ability to influence agricultural terroir appears only to exist in wheat and pea whose seeds accumulated significantly greater levels of mineral nutrients or secondary metabolites when inoculated with P uptake enhancing mycorrhizae (Ranjbar Sistani et al., 2020; Shahane et al., 2020; Yadav et al., 2020). Using a series of sensorial tests and an electronic nose, organic wheat inoculated with a commercial blend of mycorrhizae (Micosat F) produced bread that was distinguishable from bread deriving from uninoculated plants (Torri et al., 2013).
Microbe alteration of metal availability in the soil can also affect plant concentrations. Growth promoting and stress resistant rhizobacteria such as certain Pseudomonas, Bacillus, Enterobacter and Burkholderia, are able to reduce cadmium bioavailability in the soil by complexation, biosorption, and precipitation, resulting in soy, wheat, barley, pea, tomato, pumpkin and mustard with lower levels of the toxic metal (Sharma and Archana, 2016). In stark contrast, rhizosphere bacteria (especially Lysobacter, Streptomyces, Agromyces, Nitrospira) of the hyperacumulating plant Arabidopsis halleri appear to be able to help the plant absorb more heavy metals, resulting in 100% more accumulated cadmium and 15% more zinc than plants grown on gamma irradiated soil (Muehe et al., 2015). Similarly, inoculation with certain strains of soil microbe have been shown to be able to “biofortify” seeds by increasing wheat levels of selenium and iron (Yasin et al., 2015). Soil microbiomes and their impact on the chemical and micronutrient content of agricultural plants has been reviewed elsewhere (Cochran, 2017; Kaur et al., 2020; White, 2020).
Some soil microbes are able to alter plant chemistry by changing the way a plant grows and metabolizes. Root growth can be dramatically enhanced by underground bacterial signals which alter plant phytohormone production and gene expression (Ping and Boland, 2004; Vacheron et al., 2013). Various experiments drenched the soil with growth promoting microbes and observed metabolic changes in the plant: Enterobacter and Paenibacillus significantly increased tomato biomass and altered leaf metabolomes (Kalozoumis et al., 2021); Bacillus subtilus GB03 or its cell free biochemical extracts doubled sweet basil weight and increased essential oil accumulation tenfold (Banchio et al., 2009). Similar experiments and results have been seen in tomato (Pérez-Rodriguez et al., 2020), peppermint (Santoro et al., 2011; Santoro et al., 2016), Italian oregano (Banchio et al., 2010), marigold (Cappellari et al., 2013) and lemongrass (Mirzaei et al., 2020). Interestingly, rhizobacterial production of volatile plant growth promoting compounds is itself influenced by environmental conditions, adding a further layer of complexity to the microbial dimension of terroir (Blom et al., 2011). Although mycorrhizae are usually considered to affect plant chemistry by aiding in nutrient uptake, there are also many examples of their ability to stimulate plant growth and metabolism by secreting phytohormones (Rouphael et al., 2015; Avio et al., 2018), resulting in changes to chemistry, nutrients and/or flavours including carotenoids, phenolics, polyphenols, phytoestrogens, flavonoids, antioxidants and essential oils (Ceccarelli et al., 2010; Sbrana et al., 2014; Hart et al., 2015; Battini et al., 2016). Other examples of rhizosphere microbes inducing changes in plant chemistry have been recently reviewed (Qu et al., 2020; Korenblum et al., 2022).
In many cases, soil microbes can change the plant metabolome by simply being detected (Piasecka et al., 2015). Pathogens and symbionts alike can be recognized by their conserved microbe‐associated molecular patterns (MAMPs) or elicitins which trigger above ground responses to mount defenses against pathogens, pests, or abiotic stress, in a process called induced systemic resistance or ISR (Pineda et al., 2017; Rashid and Chung, 2017; Etalo et al., 2018; Vannier et al., 2019; Yu et al., 2019). Detecting elicitors can alert the plant to a nearby pathogen so that it can prepare to defend itself. In grape cell tissue culture, elicitors such as cyclodextrin, and chitosan dramatically increase reserveratrol (molecule with anti-aging, anticarcinogenic, anti-inflammatory and antioxidant properties) production (Jeandet et al., 2014), suggesting that their presence in rhizospheres could have a similar effect. Elicitor mediated plant responses often work through phytohormone signaling, especially jasmonate, methyl jasmonate and salicylic acid; addition of these compounds to plant cell culture can modulate the production of many specialized metabolites (Nabi et al., 2021) suggesting their production (Peñuelas et al., 2014) or indirect modulation (Pineda et al., 2013) by soil microbes could also impact terroir. Inoculation of soil with pure microbial strains has shown that elicitors can influence plant metabolism: grapevines recognize the oomycete Pythium oligandrum by its elicitin oligandrin as it comes in contact with roots, triggering aboveground production of resveratrol (Mohamed et al., 2007); the rhizobacteria Pseudomonas aeruginosa PM12 induces ISR and increases fruit levels of sugars, flavonoids, terpenoids, carotenoids and total phenolics (Fatima et al., 2017) when tomato plants detect the elicitin 3-hydroxy-5-methoxy benzene methanol (Fatima and Anjum, 2017). Further information on the effect of rhizosphere or root microbes and their elicitins on plant metabolism has been recently reviewed (Etalo et al., 2018; Korenblum and Aharoni, 2019).
Variation in soil microbiomes can contribute to different terroirs, however on a farm it is nearly impossible to know which particular microbes or mechanism(s) might have been responsible. Variation in rhizosphere microbiomes correlated with Salvia officialis growth, concentration of essential oils, flavonoids and phenolics (Ghorbanpour et al., 2016); farming history altered the peanut rhizosphere and correlated with changes to plant nutrient metabolism and phytohormone biosynthesis (Li et al., 2019); drenching Arabidopsis roots with different soil microbiomes modulated both shoot biomass and the leaf metabolome, which in turn altered the feeding behavior of Trichoplusia ni caterpillars (Badri et al., 2013); soil fungi influence amino acid concentration in Senecio jacobaea phloem sap, which in turn influenced the feeding behavior of the foliar feeding aphid Aphis jacobaea (Pineda et al., 2017); inoculating B. subtilis Co1-6 and P. polymyxa Mc5Re-14 onto chamomile roots grown in arid Egyptian soil showed an enhancement of the bioactive secondary metabolite apigenin-7-O-glucoside in flowers (Schmidt et al., 2014); microbiota in disease suppressive soil greatly upregulated tomato phenol biosynthesis, lignin deposition, innate immunity and resistance to oxidative stress, leading to higher Fusarium root rot resistance (Chialva et al., 2018). Beta vulgaris and Brassica oleracea were grown on soil missing rare microbes, resulting in higher plant biomass but lower sugar and glucosinalte concentrations (Hol et al., 2010).
Rhizosphere microbiomes can also indirectly influence plant metabolism by mitigating plant stresses. Soil microbes can interrupt plant-soil feedback loops by degrading phytoactive compounds in the rhizosphere, for example bacteria using ACC deaminase to reduce levels of the plant stress hormone ethylene can help roots grow during flooding (Glick, 2014). Phytotoxic compounds in the soil, including plant derived allelochemicals (Misra et al., 2020; Scognamiglio and Schneider, 2020) or human applied herbicides (Zhang et al., 2021; Sun et al., 2022), can stress or kill crops while inducing changes in their metabolic profiles; however, these compounds can be readily degraded by certain soil microbes (Huang et al., 2013). Root diseases caused by microbial pathogens can also trigger production of specialized metabolites in plants, thus for better or for worse (in terms of sensorial or nutritional attributes) soil pathogen biocontrol should be able to preserve the metabolic profile of a healthy plant.
M-tyrosine is an allelopathic chemical secreted by roots of Chewing’s fescue (Festuca rubra) that inhibits growth of other plant species, altering root development, causing leaf chlorosis, retarding growth and reducing levels of amino acids, especially phenylalanine (Zer et al., 2020). By growing Lactuca sativa (lettuce), Phalaris minor and Bambusa arundinacea on sterilized or unsterilized soil that had been amended with M-tyrosine, the soil microbiome was proven to be important in degrading this compound and reducing phenotypic effects on plants (Kaur et al., 2009). Another example of allelopathy is seen in croftonweed (Eupatorium adenophorum) which is native to Central America, but in China, has become a destructive, invasive species that produces two powerful allelochemicals inhibiting the germination and growth of native plants. Soil microbes from different Croftonweed invaded habitats were able to alleviate or eliminate phytotoxicity, whereas microbes present in sand had no effect (Zhu et al., 2011). Tea plants grown in young plantations (2 years old) had significantly greater leaf concentrations of theophylline, total polyphenols, theanine, and total amino acids, than plants grown in soil from older plantations (15 and 30 years old) (Arafat et al., 2020). These metabolic changes in older plantations were explained by soil accumulation of allelochemicals released by leaf litter, reduction of growth promoting genera Sphingomonas, Bacillus, and Prevotella, and increases in soil populations of allelochemical degrading Pseudomonas and Burkholderia. It seems quite plausible then that agricultural plants grown on allelochemical impregnated soils containing different microbial populations would manifest different metabolomes; however, we found no published examples supporting this concept.
Human applied herbicides are meant to take out weeds, but can linger in soil after application and cause stress, yield reductions and metabolic changes to crop plants long after the original intended targets were killed (Fuchs et al., 2021a; Ganugi et al., 2021; Ruuskanen et al., 2022). Glyphosate for example is the most widely used herbicide in the world, which disrupts the shikimate metabolic pathway, blocking aromatic amino acid production and synthesis of downstream metabolites with important implications for sensorial profiles of agricultural products (Fuchs et al., 2021b). Plant metabolomes are very sensitive to herbicides, with rapeseed biochemistry being significantly affected by soil concentrations of glyphosate as low as 5 μM (Petersen et al., 2011). Soil (Zhou et al., 2022) and plant associated (Ke et al., 2021) microbes can also be affected by herbicide presence in the soil, with negative effects reported for rhizobial nodulation in legumes, mycorrhizal associations with roots and of nitrogen metabolism by free living microbes (Meena et al., 2020). Conversely, soil microbiomes are able to degrade or detoxify some herbicides such as glyphosate and atrazine, at different rates (Yale et al., 2017; Zabaloy et al., 2022), suggesting a way that herbicide impregnated soils could differently affect plant metabolomes. In one example of this principle, addition of biochar into fomesafen impregnated soil significantly altered the wheat rhizosphere microbiome and reduced herbicide uptake/toxicity (Meng et al., 2019). Similarly, addition of biochar to atrazine contaminated soil reshapes the soybean rhizosphere microbiome, reducing plant stress, improving nodule formation and increasing plant growth (Zhou et al., 2022). Certain synthetic communities of bacteria (including Firmicutes and Burkholderia) were able to differentially metabolize glyphosate, impacting the growth rates of Arabidopsis (Ramirez-Villacis et al., 2020). Although these case studies prove the concept that microbes can changes soil levels of herbicide and thus impact plant growth, we were unable to find publications explicitly studying this effect in the context of an agricultural plant’s metabolome.
Another way that rhizosphere microbes could contribute to a good terroir is by suppressing pathogens to help maintain the biochemical profile of a healthy plant. Soil pathogens can invade, manipulate (biotrophs) and destroy (necrotrophs) root tissues, inducing a variety of metabolic responses all over the plant (Castro-Moretti et al., 2020). For example, canola roots under attack by Plasmodiophora brassicae (clubroot) defend themselves by producing a variety of antimicrobial metabolites (Pedras et al., 2008). Another example, when the soil transmitted necrotrophic pathogen Ralstonia solanacearum colonizes tomato roots, it induces production of a range of defensive compounds including antimicrobial quinic acids, flavonoids, polyamines (ie. spermidine) and osmoregulatory hexoses (Castro-Moretti et al., 2020). Elevated levels of glycoalkaloids (toxic to humans) are produced in potato tubers trying to resist root rot pathogens like Fusarium sulphureum (Li et al., 2022). In a wonderful example of a soil microbiome preserving a healthy plant metabolome, maize was grown on sterile sand inoculated with soil bacteria that had been filter purified of all protists (predators of bacteria), while some plants were also inoculated with a blend of these bacterial predators: the ciliate (Tetrahymena pyriformis), an amoebae (Acanthamoeba polyphaga) and a flagellate (Cercomonas longicauda). Plants inoculated with protists maintained unstressed metabolic profiles, while plants growing without the influence of the predators experienced significant shifts in their rhizosphere bacterial populations, and accumulated stress induced metabolites (polyols, carbohydrates and phenolic compounds) in their roots and leaves (Kuppardt et al., 2018).
Changes in the diversity of endophytic microbes living within plant tissues like roots or tubers (eg. potato, ginseng, carrot), stems or shoots (eg. celery, cinnamon, asparagus) or leaves (eg. basil, spinache, tea), may also affect their metabolome. Endophytes are derived vertically through the seed (Johnston-Monje et al., 2021b) or via vegetative propagation (Quambusch and Winkelmann, 2018) as well as horizontally from the external environment (Bulgarelli et al., 2013) however no type of transmission is entirely consistent and invariable. The seed transmitted fungal endophyte Epichloë is imperfectly transmitted from 69-99% between different genotypes of host grass (Gagic et al., 2018) and from 0-100% depending on storage conditions; after 18 months at high temperature and humidity, all Epichloë in grass seeds are dead (Hume et al., 2013). Different soil conditions, especially pH, alter the makeup of seed transmitted endophytes colonizing rice plants, which would presumably also have an impact on the plant metabolome (Hardoim et al., 2012). In another example, eradicating the seed transmitted microbiome of metal hyperaccumulating plant Sedum alfredii significantly lowered plant biomass and heavy metal accumulation, while inoculating these axenic plants with certain vertically transmitted endophytes restored the phenotype (Luo et al., 2019). It has long been known that vegetative cuttings and tissue cultured plantlets of crops like potato, grape, strawberry, and cassava, do contain vertically transmitted endophytic bacteria and fungi. Propagation method appears to be able to alter the endophytic microbiome: differently propagated sugar cane contains significantly different populations of endophytic fungi and bacteria (Yang et al., 2023), as do different apple scions grafted onto different apple rootstocks (Liu et al., 2018). There is as of yet no knowledge about the impacts these different microbiomes might have on the plant metabolome; however, it is our opinion that vegetatively propagated endosphere microbiomes represent a subset of the normal seed transmitted microbiome and thus deliver only a fraction of the normal seed microbiome’s phenotypic benefits to these crops. Because tissue culturing attempts to create sterile plantlets, while vegetatively propagated plants are denied sexual reproduction, the resulting propagules may be nearly devoid of their normal vertically transmitted microbiome and likewise nearly devoid of the accompanying phenotypic benefits that seed endophytes would provide. Because many endophytes are thought to be horizontally transmitted (Johnston-Monje et al., 2021a), environmental variation can also influence microbial diversity in the endosphere. This has been shown in grapes, where the majority of bacterial endophytes in roots were observed to originate and vary in relation to the soil microbiome, which in turn was influenced by soil moisture, carbon, temperature, and geographic location (Zarraonaindia et al., 2015). Another study of grape endophytes found that endophytic yeasts in xylem sap originated from soil, which in turn varied from farm to farm and with relation to soil physio-chemical factors (Liu et al., 2020).
An endophyte could most directly contribute to terroir by producing flavorful, nutritious, medicinal or toxic metabolites which remain inside the agricultural product. The most famous seed transmitted endophytes are Epichloë fungi, found in nearly 30% of the Earth’s cool-season grasses – these endophytes produce peramines, lolines, ergot, and indole-diterpene alkaloids which are toxic to insects and mammals (Lee et al., 2021). Another example is the antifungal cancer drug Taxol; found in yew trees at concentrations that can vary by up to 125 times (Soliman et al., 2013), it is synthesized by both the tree and its endophytes like Paraconiothyrium SSM001 which construct Taxol containing endophytic defenses against invading pathogens (Soliman et al., 2015). Similarly, the anticancer drug maytansine is produced by the endophytic microbial community in roots of Putterlickia verrucosa and P. retrospinosa plants (Kusari et al., 2014). Numerous other examples of nutritionally, sensorially and pharmaceutically significant substances produced by endophytes have been reviewed elsewhere (Strobel and Daisy, 2003; Newman and Cragg, 2015; Newman and Cragg, 2020; Tilocca et al., 2020; Mathur and Ulanova, 2022); it is usually unclear how significant their contributions might be in planta.
Endophytes can alter their host plant’s absorption of nutrients by changing the strength of the nutrient sink, for example when leaf endophytes compete for minerals and force the plant to takeup more (Venkateswarlu and Visperas, 1987). Beauveria bassiana for example, stimulates tomato and wheat to uptake more iron (Sánchez-Rodríguez et al., 2015), while Neotyphodium coenophialum increases the concentration of P, Ca, Zn and Cu in P starved tall fescue (Malinowski et al., 2000). Similarly, Epichloë gansuensis-infected grass Achnatherum inebrians enjoyed increased plant biomass, and increased leaf accumulation of C, N, P, metals, tryptophan, threonine, serine, proline, phenylalanine, lysine, L-asparagine, glutamic acid, glutamine, glycine, aspartic acid, arginine, and alanine (Liu et al., 2021b). Inoculating tobacco plants with the dark septate fungus, Acrocalymma vagum, promoted plant growth and reduced heavy metal content in leaves, theoretically resulting in a healthier cigar (Jin et al., 2018). The ability of bacterial endophytes to enhance micro-nutrient accumulation (eg. Zn, Se, Fe) in plants has been reviewed recently (Kaur et al., 2020).
Endophytes inside roots, stems or leaves might also contribute to terroir by influencing or participating in plant metabolism. Endophytic symbiosis between soil transmitted rhizobia and legumes such as common bean is a classic example where nitrogen fixation within the root results in seeds with significantly elevated levels of protein and starch (Massa et al., 2020). In an example of a growth promoting endophyte influencing the plant metabolome, inoculation of grape roots with endophytic Paraburkholderia phytofirmans PsJN, triggered the overexpression of all phenylpropanoid and flavonoid pathway genes, as well as increasing the relative amounts of 32 and 17 different compounds in roots and leaves respectively (Miotto-Vilanova et al., 2019). The root endophyte Piriformospora indica stimulates tomato and Arabidopsis growth by upregulating host production of putrescine; a polyamine so named for smelling like rotting meat (Kundu et al., 2022). Inoculating pot grown apple trees with a blend of 9 different plant growth-promoting bacterial endophytes isolated from wild black cottonwood and willow trees, increased root growth, delayed leaf senescence, and significantly increased fruit mass and soluble sugar content (Rho et al., 2020). Similarly, inoculating grape vines with 8 different fungal endophytes variously increased the content of reducing sugars, flavonoids, phenols, trans-resveratrol and phenylalanine ammonia-lyase in both fruits and leaves (Yang et al., 2016). Inoculating the medicinal plant Artemisia annua with a variety of different bacterial and fungal endophytes, their chemical extracts or elicitins can increase plant growth, stimulate plant defense responses and enhance artemisinin production (Zheng et al., 2021). The bacterial endophytes Bacillus altitudinis and Paenbibacillus polymyxa stimulate growth and medicinal ginsenoside accumulation in roots of ginseng plants (Chu and Bae, 2022). Inoculation of lab grown Catharanthus roseus with leaf endophytes Curvularia sp. CATDLF5 and Choanephora infundibulifera CATDLF6 increased vindoline content by up to 403% although what mechanisms might be involved were unknown (Pandey et al., 2016). Similarly, inoculation and co-culture of tissue cultured grape with different foliar fungal endophytes significantly altered fruit metabolite profiles (Huang et al., 2018). A comparative metabolomics analysis of ryegrass, with or without endophytic Epichloë festucae, found the fungus alters the expression of 30% of the plant’s genes, triggering a dramatic change in plant metabolism; increasing secondary metabolite accumulation (especially flavonoids and anthocyanins), while reducing primary metabolism (Dupont et al., 2015). Not content to passively communicate with the plant, soil bacteria Agrobacterium tumefaciens and A. rhizogenes invade the endosphere and genetically transform plant cells to overproduce auxins and cytokinins resulting in crown gall tumors and hairy roots that produce large amounts of opines; amino acids consumed by the bacteria (Hooykaas, 2000). Many other root endophytes have been documented as being able to alter plant metabolism (Etalo et al., 2018; Korenblum and Aharoni, 2019).
Most studies looking at the influence of an endophytic microbiome on the plant metabolome, establish simple correlations, but do not identify the specific microbes or mechanisms involved. For example in a study of endophytic fungi in different horseradish varieties, 35% of the mycobiome correlated with biochemical variation in the root, but no specific microbes were singled out as responsible (Plaszkó et al., 2022). Increased levels of soil organic matter significantly altered the diversity of bacteria living inside roots of the medicinal plant Echinacea purpurea, which in turn increased the immunostimulant properties of extracts by more than 400% (Haron et al., 2019). A fascinating experiment where specific causal microbes were identified, inoculated Arabidopsis grown on different concentrations of phosphate with a 185-member bacterial synthetic community; shoot levels of orthophosphate correlated negatively with the abundance of the genus Burkholderia (Finkel et al., 2019). This correlation was validated through a dropout inoculation experiment, suggesting that under phosphate starvation, Arabidopsis can’t effectively exclude Burkholderia endophytes, which further sequester the scarce nutrient away from the shoots.
Fruits are enjoyed for their flavor, which consists of mouth perception (sweetness, acidity or bitterness) and aroma, the latter made up of volatile compounds including alcohols, aldehydes, ketones, lactones, terpenoids and apocarotenoids (El Hadi et al., 2013). With an estimated 100 million bacterial cells inside one common apple (Wassermann et al., 2019) and evidence that microbiomes on the surface of fresh fruit and vegetables vary by plant genotype and farming practice (Leff and Fierer, 2013), it is easy to imagine that carposphere microbial terroir might be a thing. Wine grape microbiomes have been variously shown to be influenced by plant cultivar, year, soil, geographic location, vineyard, environmental heterogeneity, agricultural technique and climate (Bokulich et al., 2014; Pinto et al., 2015; Zarraonaindia et al., 2015; Stefanini et al., 2016; Chou et al., 2018). It has also been repeatedly shown that grape microbiome makeup correlates directly with differences in the wine metabolome (Bokulich et al., 2016; Belda et al., 2017; Mezzasalma et al., 2017; Liu et al., 2019). Raspberry metabolomes vary in concentrations of anthocyanidin, acids, ketones, aldehydes, and monoterpenes depending on organic versus IPM cultivation (Sangiorgio et al., 2021). These differences were partially explained by changes in fruit associated populations of volatile chemical emitting Gluconobacter, Sphingomonas, Rosenbergiella, Brevibacillus and Methylobacterium. Experiments by the same author found that surface sterilization of raspberries significantly reduced volatile aldehydes, monoterpenes, norisoprenoids and other aroma-active compounds, although reinoculating the berry with a previously collected surface microbiome was able to partially restore emission of terpenoids (Sangiorgio et al., 2022). Methylobacterium inside strawberries is important in producing the precursors to aromatic furaneol substances (strawberry flavor) such as 2,5-dimethyl-4-hydroxy-2H-furan-3-one (Nasopoulou et al., 2014). When inoculated onto greenhouse grown plants, methylobacteria are able to increase furaneol accumulation in fruit (Verginer et al., 2010b). By metabolically profiling individual bacterial isolates found inside and on the surface of Blaufraenkisch grapes, many of the fruit’s aromas were found to be at least partially attributable to these microbes (Verginer et al., 2010a).
Of course, the most important impact of fruit microbiomes on an agricultural product would be on alcoholic fermentation. Many fruits are fermented to make wine and spirits, although grape wine is by far the most popular (Saranraj et al., 2017). At the end of the 19th century, Louis de Pasteur showed that yeasts on the fruit surface were the main microorganisms responsible for wine fermentation (Barata, 2012). Ninety-three different yeast species have been isolated from grapes growing in 22 countries and vary from plant to plant within a farm, by climatic conditions, by grape variety and by agriculture practices (Liu et al., 2017). Yeasts are responsible for alcoholic fermentation of grapes, converting sugars to carbonyl compounds, esters, acids, acetals, and alcohols, of which ethanol can reach 10-16% final concentration, killing many sensitive microbes which might otherwise influence the quality of the final product (Liu et al., 2017). Although wine is a hostile environment for bacteria (low pH, high ethanol concentration and low nutrients) more than 50 species have been isolated from fruits and must, mostly lactic acid producing bacteria including Lactobacillus, Oenococcus, Pediococcus and Leuconostoc, acetic acid producing bacteria such as Acetobacter and Gluconobacter, and others (Bacillus, Enterococcus, Enterobacter, Pseudomonas, Burkholderia, Serratia, Klebsiella) with less clear oenological functions (Liu et al., 2017). After alcoholic fermentation by yeasts, lactic acid bacteria begin to de-acidifiy the wine by converting citrate and malate to lactate, acetate, diacetyl (buttery flavor), acetoin, 2,3‐butanediol and carbon dioxide; de-acidification is particularly valuable for acidic, cold climate wines such as those made in Canada (Liu, 2002). Malolactic fermentation by bacteria can result in a range of different compounds with flavors described as spicy, floral, oaky, honey, earthy, nutty, roasted, yeasty, sweaty, vanilla, toasty, smoky, bitter and ropy (Liu, 2002). Acetic acid bacteria are considered unfavorable, spoilage organisms as they produce oxidized compounds with unpleasant vinegar-like flavors such as acetaldehyde and acetic acid (Swiegers et al., 2005). Also present on the grape surface are pathogenic and saprophytic fungi which could create off flavors and toxins; these include downy mildew (Plamospara viticola), powdery mildew (Erysiphe necator), grey rot (Botrytis cinerea) and saprophytic moulds (e.g. Cladosporium spp., Aspergillus spp., Penicillium spp.). These fungi do not usually survive during well managed wine fermentations (Barata, 2012).
Geographic influences on vineyard soil microbial diversity are more pronounced for fungi, than for bacteria, suggesting a greater role for these microbes in colonizing the carposphere and contributing to wine terroir during fermentation (Coller et al., 2019). Similarly, studies of spontaneously fermented Cabernet Sauvignon grapes in China (Wei et al., 2022b) and 6 geographically disparate Australian Pinot Noir wines (Liu et al., 2020) found that fruit and must mycobiomes were of primary importance to wine flavor and aroma, with bacterial diversity correlating little with variation in wine chemistry. Studying the microbiomes of over 200 California Cabernet Sauvignon and Chardonnay grape musts and fermentations, found regional microbiota signatures that correlated with wine chemical composition (Bokulich et al., 2016). Although a range of different yeast species could be involved in spontaneous wine fermentation (Capozzi et al., 2015) Saccharomyces cerevisiae is the most common and important, occurring in soil, varying geographically and being transmitted from soil to fruit to fermentation tank (Mandl et al., 2015). Being the dominant microbe in spontaneous wine fermentations, changes in S. cerevisiae strain type or relative abundance can explain the majority of wine flavor differences at a subregional level, when vineyards are separated by less than 12 km (Liu et al., 2021a). Isolating S. cerevisiae from six major New Zealand wine regions and inoculating these into the same Sauvignon Blanc juice resulted in wines with significantly different aromas and flavors (Knight et al., 2015). Controversially, many studies have found instability of S. cerevisiae strains on grapes and in vineyards over time, while others have found that particular strains of S. cerevisiae stably colonize the same winery (building, fermentation tanks, etc) for years, leading to speculation that the latter (known as “winery effect”) is ultimately responsible for most microbial terroir signatures observable in the wine metabolome (Alexandre, 2020). To further confuse the importance of carposphere microbiomes in wine fermentation, most of the world’s wine makers inoculate their grape must with commercial strains of S. cerevisiae, inhibiting and overwhelming most fruit surface microbes, eliminating a potentially important source of chemical uniqueness from their fermentations (Alexandre, 2020).
Caloriewise, the most important agricultural products are the seeds of cereal grasses like maize, rice and wheat which are often ground into flour for making carbohydrate rich bread, tortillas, arepas and noodles. Infection of cereal seeds with fungi can result in bad odor and taste, but also accumulation of dangerous mycotoxins: the fungus Claviceps purpurea (ergot) which infects wheat and rye seeds, produces toxic alkaloids which afflicted tens of thousands of people in Medieval times, causing the disease Saint Anthony’s Fire (Miedaner and Geiger, 2015). Another fungal toxin, deoxynivalenol (also known as vomitoxin) is produced in wheat, barley and corn seeds following infestation by the fungus Fusarium graminearum (Pestka, 2010). Fruit seeds like coffee and cacao are also valued human foods, but they are prized for their flavor and secondary metabolites rather than as a staple source of calories. Coffee and cacao seeds develop within a gelatinous mixture of polysaccharides known as mucilage which is thought to aid in seed dispersal through water and soil, to protect the seed from stomach acids inside animals and to function as a hydrogel, maintaining moisture for germination (Tsai et al., 2021). While some seeds are consumed fresh along with their mucilage (eg. tomato, chia, passion fruit), coffee seeds need to have their mucilage removed before drying to prevent microbial rotting during storage, while cacao seeds need to be fermented before roasting or they will not develop the flavor of chocolate (Schwan and Wheals, 2004; Kadow et al., 2015). To collect and store gel covered seeds for future planting (eg. tomato), mucilage has to be removed before drying and storage. Coffee and cacao are grown in tropical countries, with seed fermentation usually taking place on farm via spontaneous microbiome formation which (depending on the microbial community) can result in the creation of good flavors/aromas/nutrients and the protection against formation of bad ones (Roelofsen, 1958; Lee et al., 2015). The structuring of the seed microbiome appears to depend on the environment in which it was grown: beans (Phaseolus vulgaris) sourced from different farms have distinct microbiomes, suggesting that the majority of the seed microbiome is environmentally derived (Klaedtke et al., 2016). Conversely, a recent study also showed that all plant seeds share a core microbiome (Johnston-Monje et al., 2021b), thus it is still unclear what factors are the most important in shaping the spermosphere microbiome.
Cacao seed fermentation begins by manually opening pods and piling seeds up in tanks/heaps/boxes/platforms/baskets where they are left for 2-10 days to spontaneously ferment, then they are moved into the sun or a mechanical oven to dry for 7-21 days, roasted from 5-120 minutes at 120-150°C and milled to produce chocolate (Lima et al., 2011). The fermentation involves an emergent microbiome that consumes the mucilage while producing acids and alcohols that penetrate the seed, killing the embryo and activating endogenous hydrolytic enzymes. The enzyme catalyzed reactions which form the precursors of chocolate flavor and aroma also reduce bitterness (methylxanthines) and astringency (polyphenols) (Schwan and Wheals, 2004). The typical microbiome in a pile of fermenting cocoa beans is initially dominated by yeasts (producing pectinases, heat, ethanol, carbon dioxide, acids, and volatile compounds), which are then replaced after a few hours by lactic acid bacteria (producing lactic acid, ethanol, acetic acid, glycerol, mannitol, carbon dioxide and volatiles), followed by acetic acid producing bacteria (which oxidize ethanol to acetic acid, producing a lot of heat) and finally by species of Bacillus and molds (which mop up the remaining acetic acid, lactic acid and mannitol, but can sometimes produce off flavors) (Lima et al., 2011). Controlled incubation experiments with sterile beans show that the most important factors to form the chocolate flavor precursors are acid and heat, and not the microbes per se (Kadow et al., 2015). Inoculating sterile beans with a blend of the yeast species Saccharomyces cerevisiae var. chevalieri, the lactic acid-producing Lactobacillus delbrueckii subsp. Lactis and L. plantarum, and the acetic acid bacteria Acetobacter aceti and Gluconobacter oxydans is able to duplicate the quality seen from an average, spontaneous fermentation on a farm (De Vuyst and Leroy, 2020). A number of experiments have shown that through artificial inoculation with carefully selected strains of yeast and bacteria, it is possible to accelerate the fermentation process, reduce the final acid concentration (acidic beans have weak flavor after roasting), increase production of aromas and reduce off flavor production by bacilli and molds at the end of the process (De Vuyst and Leroy, 2020). A variety of sources explain the provenance of the cacao mucilage microbiome, including workers hands, cutting utensils, fruit flies, earphones, leaves, and baskets, however the most important are the surface of cacao pods (Lima et al., 2011). While the terroir of chocolate is marketed at the national level (eg. Hawaiian chocolate is fruity, Venezuelan is nutty, Papua New Guinean is smoky) (Engeseth and Ac Pangan, 2018) and by individual farms (eg. Akesson’s Madagascar estate produces chocolate with hints of raspberries and apricots, while Qantu’s Peruvian bars taste like white wine) (Clark, 2021), the scientific community agrees that cocoa quality/flavor is most influenced by plant genotype and the fermentation microbiome (Hernandez and Granados, 2021). Most of the variability observed in spontaneous cacao bean fermentations is as yet unpredictable – a meta-analysis of 60 articles from Brazil, Ivory Coast and Ghana identified more than 1700 microbes that occur in cacao seed fermentations, but was unable to find a correlation between this variation and geographical location or fermentation method (Taylor et al., 2022).
Coffee seeds are also fermented, primarily to remove the mucilage, producing a naked, green bean that can be effectively dried and stored without microbial rotting. Because the fresh bean contains all the precursors necessary to make standard coffee after roasting, enzymatic or mechanical depulping can be used to remove the mucilage; however, if properly managed, microbial fermentation not only removes the mucilage, but can also improve coffee’s flavor and aroma (Gonzalez-Rios et al., 2007; Haile and Kang, 2019; Elhalis et al., 2023). In wet, mountainous regions like Colombia, Central America and Hawaii, mature coffee cherries are hand-picked (ensuring consistent maturity/quality), mechanically depulped and then wet-fermented for 24–48 h to remove the mucilage (and create numerous novel aromatic compounds) before drying to stop microbial activity (Lee et al., 2015). In dryer, sunnier climates like Brazil and Ethiopia (and representing 90% of world production) cherries are usually machine-harvested (resulting in various levels of maturity/quality), sundried/fermented as whole fruit for 10-25 days, until finally the fruit/mucilage is flaked off (Lee et al., 2015). Other niche methods to remove coffee seed mucilage include passage through the intestines of the Asian palm civet (Kopi Luwak coffee), the intestines of African elephants (black ivory coffee) and the intestines of the Brazilian jacu bird (Lee et al., 2015); while creating a scarce and expensive product that is chemically distinct from traditionally fermented coffees, these are scored poorly by professional coffee tasters who consider them inferior to traditionally fermented beans (Morison, 2020). In any of these methods, beans that are underfermented can result in coffee with bad flavors (such as potato) because residual mucilage on the seed inhibits drying and facilitates growth of molds, while overfermented beans (one of the most cited problems among coffee producers) accumulate propionic and butyric acids that taste like onion or vinegar (Jackels and Jackels, 2005; Haile and Kang, 2019).
Wet fermentations of coffee seeds are usually spontaneously inoculated, employing the microbiome that is naturally assembled on the farm where cherries are harvested; the microbiology of this fermentation method is difficult to control. Although there are many possible inoculum sources in a spontaneous fermentation including soil, leaf litter, fermentation water and workers hands, there is evidence that fruit surfaces transmit the majority of the coffee seed fermentation microbiome (Figure 3) (da Silva Vale et al., 2021). The same publication describes a core coffee seed fermentation microbiome which includes the dominant microbes Gluconobacter, Leuconostoc, Acetobacter (bacteria) and Candida, Pichia, Fusarium, and Hanseniaspora (fungi). Surveying more than 100 studies over the past decades, spontaneous coffee fermentation has been reported to involve a large diversity of microbes including 343 different genera, among these 119 fungi (mostly Aspergillus, Penicillium, Fusarium, Cladosporium, Mucor, and Rhizopus) and 224 bacteria (mostly Bacillus, Leuconostoc, Lactobacillus, Enterobacter, Pseudomonas, Klebsiella, Pantoea, Erwinia, Serratia, Lactococcus and Acinetobacter) (Vaughan et al., 2015; Duong et al., 2020). A typical wet fermentation sees a pH drop from about 5.6 to about 4.6, while glucose is consumed and lactic acid plus ethanol are produced; these fermentation characteristics vary from farm to farm (Jackels and Jackels, 2005). There is evidence from fermentation microbiome deletion experiments (yeasts were eliminated with the antibiotic Natamycin) that yeasts, which are commonly isolated from coffee bean fermentations around the world, are efficient degraders of seed mucilage, produce significant amounts of flavoring compounds (glycerol, isoamyl alcohol, ethanol, acetaldehyde and ethyl acetate), suppress the growth of mycotoxin producing filamentous fungi and significantly improve coffee sensory scores (Elhalis et al., 2020). Lactic acid bacteria are the most abundant prokaryote in coffee fermentations where they produce lactic and acetic acid (providing acidity to coffee), 2,3-butanedione and acetoin (buttery flavor) and ethyl propionate, ethyl acetate, acetaldehyde, phenyl ethanol, and phenylacetaldehyde (fruity and floral notes) (de Melo Pereira et al., 2020). Inoculating seed fermentations with lactic acid bacteria and yeasts can increase the cupping score of the resulting coffee (Cassimiro et al., 2022). Many other bacteria and filamentous fungi can be found at low levels in coffee fermentations, but it is usually unclear what contributions they might make to coffee flavor (Elhalis et al., 2023). Despite not knowing which particular species are involved, variation in the microbiomes of spontaneously fermented coffee beans can lead to differences in coffee flavor: two farms growing Coffea arabica L. var. Acaiá in Minas Gerais, Brazil, both conducted wet fermentation of their beans resulting in distinct fermentation microbiomes and contrasting finished products; one cup with citrus and herbaceous sensory characteristics, while the other also contained nutty flavors (Evangelista et al., 2015). Wet versus dry seed processing results in different fermentation microbiomes (Enterobacteria, lactic acid bacteria and Pichia yeasts versus acetic acid bacteria and non-Pichia yeasts) and different metabolic/flavor profiles (Bruyn et al., 2017).
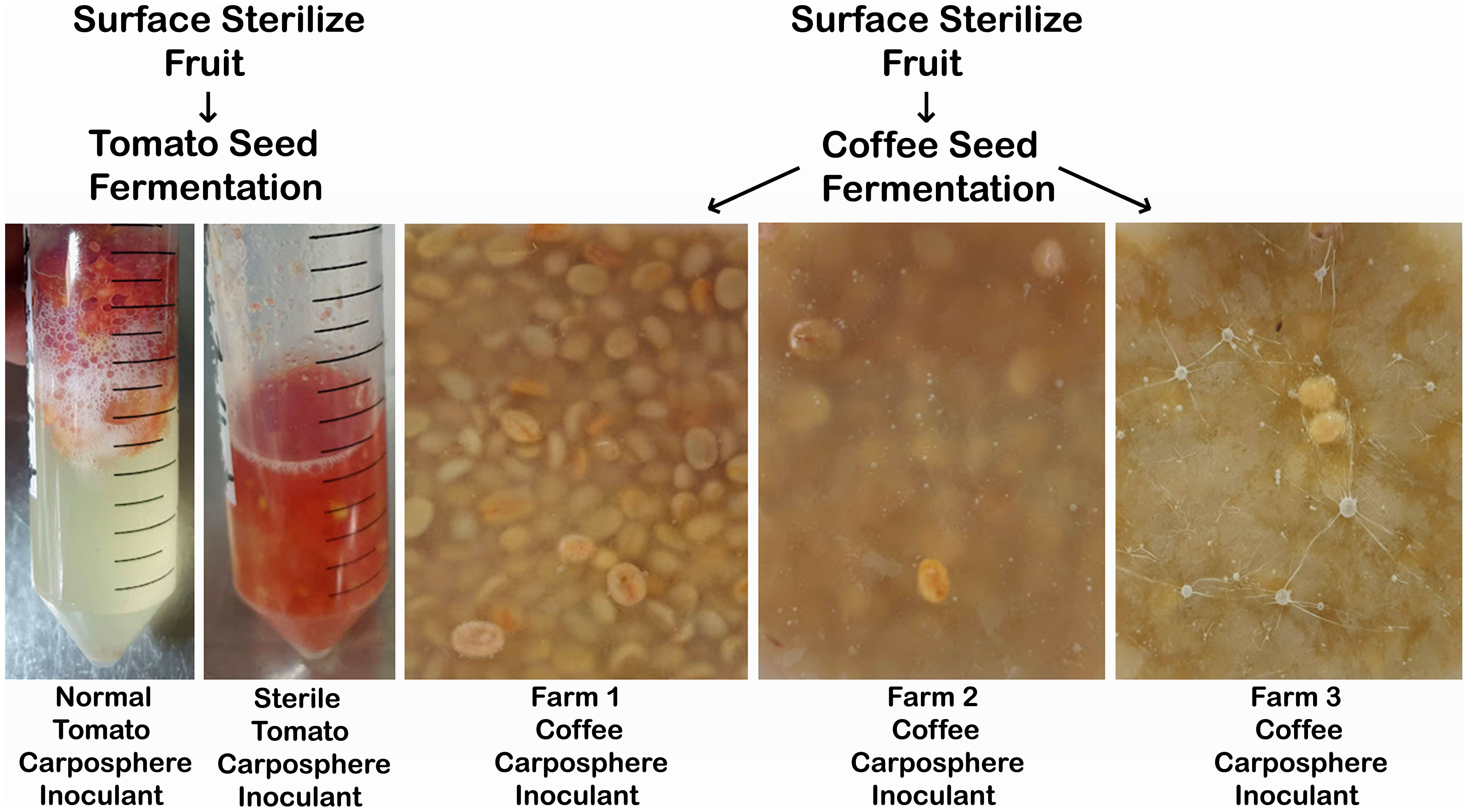
Figure 3 Altering the surface microbiome of a fruit influences the nature of its seed fermentation. Tomatoes and coffee cherries were surface sterilized with two 5 minute washes in 70% ethanol, dissected with sterilized utensils and the seeds extracted. Tomato seeds were then incubated for 72 hours after inoculation with either a normal or sterilized fruit surface microbiome (collected by washing the fruit surface with sterile water). Coffee seeds from surface sterilized fruit were inoculated with surface washes of coffee cherries taken from three different farms and incubated for 72 hours at 25°C (Johnston-Monje, unpublished).
Fermenting green beans a second time is another way that microbes can modify the biochemistry of coffee. In one example, dry, green coffee beans were inoculated with 3 different commercial strains of Saccharomyces and wet fermented for 24 hours before again drying, roasting and conducting chemical and consumer acceptability analysis. The result was a significant increase in antioxidants, polyphenols and flavonoids, but a slightly lower level of consumer acceptance (Kwak et al., 2018). Rhizopus oligosporus is widely used as a starter culture in fermented foods such as soy tempeh and when incubated on dry green coffee beans, it significantly improved the aroma precursors that give rise to coffee fragrance during roasting (pyrazines, thiols, furanones, and guaiacols) (Lee et al., 2016). The company Afineur uses a 5 strain synthetic microbiome to ferment green coffee beans sourced from Guatemala, reducing their acidity and bitterness by consuming catechol, caffeine, hydroxyhydroquinone, acrylamide, trigonelline and pyrogallol, while increasing theophylline, theobromine, vitamin B, inositol and antioxidant content (Martin, 2015; Morison, 2020; Afineur, 2023).
Flowers are plants’ sexual organs and many evolved to attract pollinators; however, humans have also learnt to appreciate them for their beautiful patterns and colors, their fragrance, medicinal value and in some cases, their taste. Roses for example are appreciated for all four – their beauty, their aroma, as soothing teas and as flavors in desserts. Other agriculturally important flowers include chamomile, cannabis, marigold, lavender, jasmine and poppy. Saffron is a colorful spice made from the stigma of Crocus sativus, and has been shown to have both a microbiome (Bhagat et al., 2021) and metabolome that varies by climatic region, agricultural practices and post-harvest processing (Gikas et al., 2021). It has been asserted that “variability in microbial presence, abundance, and composition is a defining feature of the floral microbiome” (Vannette, 2020) suggesting the potential for a variety of different microbiome/metabolome interactions in the anthosphere.
Microbes are present at low levels from the very start of flower formation, but as they bloom and mature, a greater abundance and diversity accumulate (Vannette, 2020). Plant genotype is thought to play a dominant role in shaping both the floral metabolome and microbiome (Wei et al., 2022a), however soil type (Massoni et al., 2021), farm practice (Schaeffer et al., 2023), environmental factors (Johnson et al., 2000; Bartlewicz et al., 2016; Sharaby et al., 2020), and especially visits from pollinators (Ushio et al., 2015), have also been shown to add to or change microbial communities in the flower. Surveying bacterial communities in floral nectar of tobacco trees (Nicotiana glauca) growing all over the world, it was found that increasing temperatures and elevation reduced the diversity and evenness, while geographic separation significantly increased dissimilarity (Sharaby et al., 2020). For over a 100 years it has been known that bees transmit fermentative microbes to flowers, and that floral microbiomes remain free of ascomycete yeasts when pollinators are prevented from visiting, (Vannette, 2020). Microbes on stigmas and in nectar compete for limited resources, meaning that early arrivals claim founder effects, locking up resources quickly and modifying the floral environment to be inhospitable to others; both niches have been found to be species poor and usually dominated by a single type of yeast or bacteria (Vannette, 2020). Petals are richer and more heterogenous in distribution of microbial epiphytes, possessing more of the species typical of a phyllosphere (Hayes et al., 2021).
A flower’s main purpose is to attract pollinators, but they also need to defend themselves from pathogens. In order to both attract and repel, flowers may modulate their chemistry to influence either type of biotic interaction. Some of the volatile organic chemicals produced by flowers are in fact antimicrobial: Arabidopsis thaliana produces sesquiterpene (E)-β-caryophyllene to inhibit the growth of pathogenic Pseudomonas syringae which can damage its stigma (Huang et al., 2012). This volatile and others surge in apple blossoms after infection with fire blight (Erwinia carotovora), repulsing honeybees (Cellini et al., 2019). Microbe free Brassica rapa flowers emit significantly different fragrance profiles after inoculation with floral bacteria, suggesting that the microbes were inducing different secondary metabolite production in the plant, as well as contributing their own (Helletsgruber et al., 2017). Flowers infected with saffron latent virus produce stigmas containing a significantly different metabolome than uninfected plants (Parizad et al., 2019). Female cannabis flowers produce medicinally important phytocannabinoids in their glandular trichomes, where they are thought to help defend against pathogens and herbivory (Taghinasab and Jabaji, 2020; Tanney et al., 2021) – it is as yet unknown whether interaction with anthosphere microbes can impact cannabis flower metabolomes.
While flowers produce fragrance and food rewards to attract pollinators, microbes can alter their quality and quantity by metabolizing plant compounds and secreting some of their own (Vannette, 2020). Nectar is the best studied aspect of flower microbiology, where yeasts and bacteria metabolize sugars, reducing their concentration by up to 60% (Herrera et al., 2008), sometimes producing significant amounts of ethanol (up to 4% of nectar) (Wiens et al., 2008), amino acids, secondary metabolites and other chemical constituents (Lenaerts et al., 2017; Vannette and Fukami, 2018).
Floral microbiomes can influence fragrance by both contributing volatiles or removing compounds being emitted by the flower (Vannette, 2020). In one example, removing the elderberry flower microbiome using 3 broad spectrum antibiotics resulted in a 66% reduction in terpene emission and the loss of β-ocimene, linalool, epoxylinalool and linalool oxide from the blend (Penuelas et al., 2014). Altered floral metabolomes can smell and taste differently to pollinators, who may change their foraging behavior as a result. Ascomycete yeasts in flowers can produce 2-phenylethanol, while many floral bacteria produce acetoin; volatile compounds which can be attractive to pollinators (Vannette, 2020). Presumably following their nose, bumble-bees prefer flower nectar colonized by nectar yeast Metschnikowia reukaufi even though it dramatically reduces sugar concentration (Vannette, 2020).
Leaves are consumed in a variety of different ways including salads (eg. lettuce and spinach), as spices (eg. basil, oregano), for pharmaceuticals (eg. coca, aloe), to make teas (eg. tea, mint) and even to smoke (eg. tobacco). As the primary photosynthesizing organ of a plant, leaves are packed with metabolites involved in light harvesting (ie. carotenoids, antioxidants) and self-defense (eg. alkaloids, waxes). Leaves also absorb carbon dioxide while emitting oxygen, methanol and a variety of volatile compounds (Ameye et al., 2018). The leaf and its encompassing area is called the phyllosphere, which represents the largest biological surface on Earth, containing innumerable microbes whose impact on plant physiology is often unknown (Morris and Kinkel, 2002). Phyllosphere residents are mostly Alphaproteobacteria, Gammaproteobacteria, Bacteroidetes, Actinobacteria and hyperdiverse fungi (Vorholt, 2012). These microbes can come from the seed or the soil, travelling through the vasculature until reaching the leaf. It is also possible that they reach the leaf surface through the air, contact with other plants, water drops or visitation by insects and other organisms. The leaf surface is a relatively inhospitable environment with limited access to water, carbon and nitrogen, and constant exposure to fluctuating temperature, humidity and solar radiation. Phyllosphere microbes could theoretically affect the leaf metabolome by altering plant metabolism, by consuming plant produced biochemicals or by contributing their own metabolites (Bringel and Couée, 2015). Bacteria on a leaf surface can usually be found in the seams between epidermal cells or clustered around trichomes, where they form aggregates of a few thousand cells (Vorholt, 2012). Microbes can also be found colonizing the leaf endophytically and intracellularly, where they can interact with the plant in other ways.
Phyllosphere microbiomes are significantly shaped by plant genotype, so that different species have distinct microbial populations on their leaves (Bringel and Couée, 2015); this makes sense, since that it has been established that the dominant members of the juvenile shoot microbiome are in fact vertically transmitted and not dependent on a commonly available environmental inoculum (Johnston-Monje et al., 2021b). In fact, certain plant phyllosphere mycobiomes can be overwhelmingly made up of species specific seed transmitted microbes, for example Sarocladium zea which can make up 99% of the maize shoot mycoboime, or Penicillium endophytes which can make up 99% of the rice shoot mycobiome (Johnston-Monje et al., 2022). It has been observed in some plants that a significant amount of the phyllosphere microbiome is shared with roots, as various endophytic participants in the rhizophagy cycle can also be found in trichomes and on leaf surfaces (White et al., 2019). Soil could also be an inoculum source for the phyllosphere: about half of all Arabidopsis phyllosphere bacteria have been experimentally estimated to derive from soil (Massoni et al., 2021); soil was concluded to be a key source of grape phyllosphere bacteria (Zarraonaindia et al., 2015); oak seedlings grown on 3 different soils developed distinct populations of phyllosphere bacteria and fungi (Wolfgang et al., 2023). Bacteria and yeasts can be absorbed into roots (Paungfoo-Lonhienne et al., 2010) and a subset of these root microbes are translocated through vascular tissues, xylem and phloem, from roots to shoot meristems (White et al., 2014; Liu et al., 2020; Benito et al., 2022; Micci et al., 2022). Filamentous fungi, such as Fusarium spp., can also enter roots, growing systemically throughout the plant, eventually colonizing shoots and leaves (Lofgren et al., 2018; Rodriguez et al., 2009). Plants do not passively sponge up fungi and deliver them directly to the leaf, apparently filtering them and only allowing certain species through; Bromus tectorum or maize grown on microbially distinct soils nevertheless develop analogous phyllosphere mycobiomes (Nebert, 2018; Ricks and Koide, 2019). Bacteria that travel through vasculature and arrive at nutrient rich shoot meristems can often be observed forming biofilms on the surfaces of developing leaves and intracellularly colonizing dividing meristematic cells (White et al., 2014). As bacterially colonized plant cells mature and differentiate into epidermis or trichomes, they begin to produce superoxides while intracellular endophytes secrete ethylene and reduced forms of nitrogen (ie. ammonia, nitric oxide) which serve as antioxidants (Chang et al., 2021). This nutrient exchange process in plant cells has been termed the ‘nutrient trap mechanism’ because once begun, bacteria cannot cease nitrogen secretion without being damaged by superoxides (Chang et al., 2021). A range of plants from sphagnum moss to tomatoes have been observed to have trichomes, nodules or epidermal cells manifesting this endophytic nutrient trap (Kox et al., 2020; Micci et al., 2022). Of the structures involved in this association, glandular trichomes are especially relevant to mention – they have long been known to contain a diversity of terpenoids, phenolics, antioxidants and other secondary metabolites thought to be involved in defense (Feng et al., 2021). Trichomes have now also been shown to be sites of significant nitrogen fixation by intracellular bacteria (Micci et al., 2022). Bacteria are seen to emerge from the lateral pits or ruptures in trichome walls, accumulating at the bases of the trichome and spreading out onto the leaf surface, thus contributing to the phylloplane microbial community and explaining how soil or seed derived endophytes might make it all the way there (Micci et al., 2022).
Although many phyllosphere microbes come from the seed or soil, leaf surfaces also become colonized by bacteria and fungi that vector through the air, splash from other plant surfaces and are brought in by insects or other organisms (Gong and Xin, 2021). Established leaf microbiomes can be altered by their proximity to other plants, temperature, precipitation, exposure to sand storms and foliar pesticide application (Bringel and Couée, 2015; Cernava et al., 2019). Some of these environmentally transmitted, leaf surface colonizers are the shadow yeasts (Sporobolomyces spp.) that arrive as airborne spores to colonize the endosphere and phyllosphere of many plants (Ling et al., 2020). Many other fungi such as Alternaria spp. and Cladosporium spp., are also known to colonize leaf surfaces (Bao et al., 2022). These fungi are typically epiphytic, but can sometimes enter plants as endophytes and later transition into saprophytes as leaves begin to senesce (Rodriguez et al., 2009).
However they arrive, phyllosphere microbes can impact the leaf metabolome by influencing plant metabolism, by consuming plant produced biochemicals or by contributing their own metabolites. Using quantitative nuclear magnetic resonance and imaging high-resolution mass spectrometry to visualize Arabidopsis leaf surface metabolites, colonization with organoheterotroph Sphingomonas melonis or the phytopathogen Pseudomonas syringae pv. tomato dramatically altered carbohydrate production, arginine metabolism and phytoalexin biosynthesis (Ryffel et al., 2016). In grasses, Ascomycete fungal endophytes of the genus Epichloë grow on and within shoots where they increase plant production of phenolics and other antioxidants (White and Torres, 2010; Xia et al., 2018) while secreting their own alkaloids (peramines, lolines, lolitrems and ergolines); reducing palatability and impacting the feeding behavior of herbivores (Bharadwaj et al., 2020). Leaf endophytes of the genus Paenibacillus isolated from leaves of Leucojum aestivum produce a range of alkaloids (e.g., galanthamine, lycorine, ismine, lycoramine, haemanthamine, tazettine, galanthine, homolycorine, and hippeastrine) that were characteristic of the plant (Ptak et al., 2022); reinoculating these bacteria back into axenic plants increased their alkaloid, chlorophyll and carotenoid content. Foliar spraying of tea (Camellia sinensis) with Bacillus amyloliquefaciens reduced polyphenol, catechin, and caffeine levels, while increasing amino acids and theanin in harvested leaves (Huang et al., 2022). Removing the Sambucus nigra phyllosphere microbiome using antibiotics reduced the phyllosphere concentration of lactate, citraconic acid, acetyl-CoA, isoleucine, and several secondary metabolites including terpenes and phenols (Gargallo-Garriga et al., 2016). A similar experiment with Brassica nigra found that using antibiotics to remove bacteria did not change the emission of volatile compounds from the leaf; however, applying fungicides to remove fungi triggered emission of four new compounds (cyclohexanone, cyclohexyl cyanide and 2 unknown) suggesting the microbes had previously been consuming the molecules (Saunier et al., 2020). In experiments using Korean perilla (Perilla frutescens), the reduction of phyllosphere microbes by rigorous seed surface sterilization resulted in plants producing markedly less mint oils and totally lacking aroma, compared to control seedlings bearing the full microbiome (Linsey Park and James White, unpublished). Glandular trichomes are found on leaves of many dicots, e.g., hemp (Cannabis sativa), tomato (Solanum lycopersicum) and mints (family Lamiaceae), and they contain terpenoids, phenolics and oils (Schilmiller et al., 2008). We believe that because at least some of these compounds are important in fostering nitrogen fixation by endophytic microbes, that their production by the plant is tightly correlated with the presence, diversity and activity of the phyllosphere microbiome.
Plants are holobionts that host a great diversity of microbes within their tissues and on their surfaces. These microbes can influence the plant to change its metabolism, they can intercept, consume and alter plant metabolites and they can secrete metabolites of their own into the phytosphere. If these microbial populations vary (ie. by location, by environment, by agronomic practice) so too their impact on the plant metabolome can vary. In the rhizosphere, microbial populations can help the root acquire and absorb nutrients, they can secrete compounds which induce the plant to alter its metabolism, or they can modify the soil to mitigate biotic or abiotic root stress, maintaining an unstressed metabolic state in the plant. Inside the plant, endophytes can produce agronomically desirable molecules, they can induce the plant to increase its production of desirable metabolites, and they can alter source-sink relationships to make the plant increase nutrient absorption or redistribute molecule storage. In the phyllosphere, microbes can modify volatile emissions and produce their own metabolites, while inside trichomes they participate in a nutrient trap where they produce reduced forms of nitrogen and the plant produces superoxides, terpenes, phenolics and lipids. Microbes in flowers can alter floral scent by emitting volatile compounds themselves or inducing the plant to change what it is releasing. Anthosphere microbes can also influence nectar chemistry, often by consuming sugars therein. In fruits, the most metabolically important microbes are found on the surface where they either emit volatile compounds themselves or they induce plant metabolism to create them. Microbes on the fruit surface also seem to be the most important members of the spontaneous fermentation microbiome which is important for making wine and fermenting coffee/cacao seeds to remove the mucilage, while providing chemical precursors which increase quality of the product. The impact of microbes on the flavors, aromas, nutritional or medicinal value of agricultural products is often studied under controlled conditions, for example by killing particular strains with antibiotics or by inoculating new microbes onto the plant and watching for changes in the plant metabolome. In reality, most agricultural production occurs under the unpredictable conditions of a farm where proof of microbial influence on the plant metabolome usually can’t get any better than observing a correlation. In a lab or in a controlled factory environment where plant microbiomes can be washed away and replaced (ie. washing grapes and inoculating them with alcoholic yeasts), terroir engineering may become increasingly possible, but out in the field with unpredictable environmental variation and biotic interactions, achieving an optimal microbial terroir may continue to depend on a blend of the environmental stability, producer’s savoir faire and good luck.
DJ-M conceived of, designed and wrote most of the paper, made Figures 1 and 3, and also conducted the seed fermentations pictured in Figure 3. LV reviewed literature and wrote part of the sections on seed and fruit microbiomes. JLM reviewed literature and drew Figure 2. JW reviewed literature and wrote the section on phyllospheres. All authors contributed to the article and approved the submitted version.
The writing of this review was funded by The University of the Valley. Publication fees provided by the Max Planck Society.
We are grateful for open access funding provided by the Max Planck Society. Thank you to the reviewers for their thoughtful comments which helped make this a much better paper.
The authors declare that the research was conducted in theabsence of any commercial or financial relationships that could beconstrued as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Afineur (2023) Afineur: We team up with microbes for smarter food production. Available at: https://www.afineur.com/ (Accessed 2023/2/2).
Alexandre H. (2020). Wine yeast terroir: separating the wheat from the chaff—for an open debate. Microorganisms 8 (5), 787. doi: 10.3390/microorganisms8050787
Alori E. T., Glick B. R., Babalola O. O. (2017). Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front. Microbiol. 8 (971). doi: 10.3389/fmicb.2017.00971
Ameye M., Allmann S., Verwaeren J., Smagghe G., Haesaert G., Schuurink R. C., et al. (2018). Green leaf volatile production by plants: a meta-analysis. New Phytol. 220 (3), 666–683. doi: 10.1111/nph.14671
Arafat Y., Ud Din I., Tayyab M., Jiang Y., Chen T., Cai Z., et al. (2020). Soil sickness in aged tea plantation is associated with a shift in microbial communities as a result of plant polyphenol accumulation in the tea gardens. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00601
Avio L., Turrini A., Giovannetti M., Sbrana C. (2018). Designing the ideotype mycorrhizal symbionts for the production of healthy food. Front. Plant Sci. 9 (1089). doi: 10.3389/fpls.2018.01089
Badri D. V., Zolla G., Bakker M. G., Manter D. K., Vivanco J. M. (2013). Potential impact of soil microbiomes on the leaf metabolome and on herbivore feeding behavior. New Phytol. 198 (1), 264–273. doi: 10.1111/nph.12124
Baedke J., Fábregas-Tejeda A., Nieves Delgado A. (2020). The holobiont concept before Margulis. J. Exp. Zoology Part B: Mol. Dev. Evol. 334 (3), 149–155. doi: 10.1002/jez.b.22931
Bag S., Mondal A., Banik A. (2022). Exploring tea (Camellia sinensis) microbiome: Insights into the functional characteristics and their impact on tea growth promotion. Microbiological Res. 254, 126890. doi: 10.1016/j.micres.2021.126890
Bakker M. G., Manter D. K., Sheflin A. M., Weir T. L., Vivanco J. M. (2012). Harnessing the rhizosphere microbiome through plant breeding and agricultural management. Plant Soil 360 (1), 1–13. doi: 10.1007/s11104-012-1361-x
Banchio E., Bogino P. C., Santoro M., Torres L., Zygadlo J., Giordano W. (2010). Systemic induction of monoterpene biosynthesis in Origanum× majoricum by soil bacteria. J. Agric. Food Chem. 58 (1), 650–654. doi: 10.1021/jf9030629
Banchio E., Xie X., Zhang H., Pare P. W. (2009). Soil bacteria elevate essential oil accumulation and emissions in sweet basil. J. Agric. Food Chem. 57 (2), 653–657. doi: 10.1021/jf8020305
Bao L., Sun B., Liu J., Zhang S., Xu N., Zhang X., et al. (2022). Leaf-Associated Epiphytic Fungi of Gingko biloba, Pinus bungeana and Sabina chinensis Exhibit Delicate Seasonal Variations. J. Fungi 8 (6), 631. doi: 10.3390/jof8060631
Barata A. (2012). The microbial ecology of wine grape berries. Int. J. Food Microbiol. 153 (3), 243–259. doi: 10.1016/j.ijfoodmicro.2011.11.025
Bardgett R. D., van der Putten W. H. (2014). Belowground biodiversity and ecosystem functioning. Nature 515 (7528), 505–511. doi: 10.1038/nature13855
Bartlewicz J., Lievens B., Honnay O., Jacquemyn H. (2016). Microbial diversity in the floral nectar of Linaria vulgaris along an urbanization gradient. BMC Ecol. 16 (1), 1–11. doi: 10.1186/s12898-016-0072-1
Batchelor M. (2022). “The world’s most expensive wines are a testimony of survival,” in The CEO magazine. Sydney, Australia.
Battini F., Turrini A., Quartacci M., Malorgio F., Sgherri C., Mariotti L., et al. (2016). Dual inoculation with AMF and associated bacteria improves nutraceutical value of sweet basil grown under commercial conditions. Agrochimica 60 (2), 81–99. doi: 10.12871/0021857201623
Belda I., Gobbi A., Ruiz J., de Celis M., Ortiz-Álvarez R., Acedo A., et al. (2020). “Microbiomics to define wine terroir,” in Reference Module in Food Science (Elsevier).
Belda I., ZarraonaIndia I., Perisin M., Palacios A., Acedo A. (2017). From vineyard soil to wine fermentation: microbiome approximations to explain the “terroir” Concept. Front. Microbiol. 8 (821). doi: 10.3389/fmicb.2017.00821
Benito P., Carro L., Bacigalupe R., Ortúzar M., Trujillo M. E. (2022). From roots to leaves: the capacity of Micromonospora to colonize different legume tissues. Phytobiomes J. 6 (1), 35–44. doi: 10.1094/PBIOMES-02-21-0015-R
Berendsen R. L., Pieterse C. M., Bakker P. A. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17 (8), 478–486. doi: 10.1016/j.tplants.2012.04.001
Bhagat N., Sharma S., Ambardar S., Raj S., Trakroo D., Horacek M., et al. (2021). Microbiome fingerprint as biomarker for geographical origin and heredity in crocus sativus: A feasibility study. Front. Sustain. Food Syst. 5. doi: 10.3389/fsufs.2021.688393
Bharadwaj R., Jagadeesan H., Kumar S., Ramalingam S. (2020). Molecular mechanisms in grass-Epichloë interactions: towards endophyte driven farming to improve plant fitness and immunity. World J. Microbiol. Biotechnol. 36, 1–28. doi: 10.1007/s11274-020-02868-5
Blom D., Fabbri C., Connor E. C., Schiestl F. P., Klauser D. R., Boller T., et al. (2011). Production of plant growth modulating volatiles is widespread among rhizosphere bacteria and strongly depends on culture conditions. Environ. Microbiol. 13 (11), 3047–3058. doi: 10.1111/j.1462-2920.2011.02582.x
Bokulich N. A., Collins T. S., Masarweh C., Allen G., Heymann H., Ebeler S. E., et al. (2016). Associations among wine grape microbiome, metabolome, and fermentation behavior suggest microbial contribution to regional wine characteristics. mBio 7 (3), e00631–e00616. doi: 10.1128/mBio.00631-16
Bokulich N. A., Thorngate J. H., Richardson P. M., Mills D. A. (2014). Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. 111 (1), E139. doi: 10.1073/pnas.1317377110
Bringel F., Couée I. (2015). Pivotal roles of phyllosphere microorganisms at the interface between plant functioning and atmospheric trace gas dynamics. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00486
Bruyn F. D., Zhang S. J., Pothakos V., Torres J., Lambot C., Moroni A. V., et al. (2017). Exploring the impacts of postharvest processing on the microbiota and metabolite profiles during green coffee bean production. Appl. Environ. Microbiol. 83 (1), e02398–e02316. doi: 10.1128/AEM.02398-16
Bulgarelli D., Schlaeppi K., Spaepen S., Van Themaat E. V. L., Schulze-Lefert P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838. doi: 10.1146/annurev-arplant-050312-120106
Capozzi V., Garofalo C., Chiriatti M. A., Grieco F., Spano G. (2015). Microbial terroir and food innovation: The case of yeast biodiversity in wine. Microbiological Res. 181, 75–83. doi: 10.1016/j.micres.2015.10.005
Cappellari L., Santoro M. V., Nievas F., Giordano W., Banchio E. (2013). Increase of secondary metabolite content in marigold by inoculation with plant growth-promoting rhizobacteria. Appl. Soil Ecol. 70, 16–22. doi: 10.1016/j.apsoil.2013.04.001
Cassimiro D., Batista N. N., Fonseca H. C., Naves J. A. O., Dias D. R., Schwan R. F. (2022). Coinoculation of lactic acid bacteria and yeasts increases the quality of wet fermented Arabica coffee. Int. J. Food Microbiol. 369, 109627. doi: 10.1016/j.ijfoodmicro.2022.109627
Castellucci F. (2010). Resolution OIV/VITI 333/2010 - DEFINITION OF VITIVINICULTURAL “TERROIR” (Tbilisi, Georgia: International Organization of Vine and Wine (OIV).
Castro-Moretti F. R., Gentzel I. N., Mackey D., Alonso A. P. (2020). Metabolomics as an emerging tool for the study of plant–pathogen interactions. Metabolites 10 (2), 52. doi: 10.3390/metabo10020052
Ceccarelli N., Curadi M., Martelloni L., Sbrana C., Picciarelli P., Giovannetti M. (2010). Mycorrhizal colonization impacts on phenolic content and antioxidant properties of artichoke leaves and flower heads two years after field transplant. Plant Soil 335 (1-2), 311–323. doi: 10.1007/s11104-010-0417-z
Cellini A., Giacomuzzi V., Donati I., Farneti B., Rodriguez-Estrada M. T., Savioli S., et al (2019). Pathogen-induced changes in floral scent may increase honeybee-mediated dispersal of Erwinia amylovora. The ISME Journal 13 (4), 847–859. doi: 10.1038/s41396-018-0319-2
Cernava T., Chen X., Krug L., Li H., Yang M., Berg G. (2019). The tea leaf microbiome shows specific responses to chemical pesticides and biocontrol applications. Sci. Total Environ. 667, 33–40. doi: 10.1016/j.scitotenv.2019.02.319
Chan S. C. (2012). “Terroir and green tea in China: the case of meijiawu dragon well (Longjing) tea,” in Geographical Indications and International Agricultural Trade: The Challenge for Asia. Eds. Augustin-Jean L., Ilbert H., Saavedra-Rivano N. (London: Palgrave Macmillan UK), 226–238.
Chang X., Kingsley K. L., White J. F. (2021). Chemical interactions at the interface of plant root hair cells and intracellular bacteria. Microorganisms 9 (5), 1041. doi: 10.3390/microorganisms9051041
Chialva M., Salvioli di Fossalunga A., Daghino S., Ghignone S., Bagnaresi P., Chiapello M., et al. (2018). Native soils with their microbiotas elicit a state of alert in tomato plants. New Phytol. 220 (4), 1296–1308. doi: 10.1111/nph.15014
Chou M.-Y., Heuvel J. V., Bell T. H., Panke-Buisse K., Kao-Kniffin J. (2018). Vineyard under-vine floor management alters soil microbial composition, while the fruit microbiome shows no corresponding shifts. Sci. Rep. 8 (1), 11039. doi: 10.1038/s41598-018-29346-1
Chu L. L., Bae H. (2022). Bacterial endophytes from ginseng and their biotechnological application. J. Ginseng Res. 46 (1), 1–10. doi: 10.1016/j.jgr.2021.04.004
Clark C. (2021). “Chocolate’s secret ingredient is fermenting microbes,” in Scientific American (New York, USA: Springer Nature).
Cochran A. T. (2017). “Selenium and the plant microbiome,” in Selenium in plants: Molecular, Physiological, Ecological and Evolutionary Aspects. Eds. Pilon-Smits E. A. H., Winkel L. H. E., Lin Z.-Q. (Cham: Springer International Publishing), 109–121.
Coller E., Cestaro A., Zanzotti R., Bertoldi D., Pindo M., Larger S., et al. (2019). Microbiome of vineyard soils is shaped by geography and management. Microbiome 7 (1), 140. doi: 10.1186/s40168-019-0758-7
da Silva Taveira J. H., Borém F. M., Figueiredo L. P., Reis N., Franca A. S., Harding S. A., et al. (2014). Potential markers of coffee genotypes grown in different Brazilian regions: A metabolomics approach. Food Res. Int. 61, 75–82. doi: 10.1016/j.foodres.2014.02.048
da Silva Vale A., de Melo Pereira G. V., de Carvalho Neto D. P., Sorto R. D., Goés-Neto A., Kato R., et al. (2021). Facility-specific ‘house’ microbiome ensures the maintenance of functional microbial communities into coffee beans fermentation: implications for source tracking. Environ. Microbiol. Rep. 13 (4), 470–481. doi: 10.1111/1758-2229.12921
Decaëns T. (2010). Macroecological patterns in soil communities. Global Ecol. Biogeography 19 (3), 287–302. doi: 10.1111/j.1466-8238.2009.00517.x
de Melo Pereira G. V., da Silva Vale A., de Carvalho Neto D. P., Muynarsk E. S. M., Soccol V. T., Soccol C. R. (2020). Lactic acid bacteria: what coffee industry should know? Curr. Opin. Food Sci. 31, 1–8. doi: 10.1016/j.cofs.2019.07.004
Deshmukh Y., Khare P., Patra D. (2016). Rhizobacteria elevate principal basmati aroma compound accumulation in rice variety. Rhizosphere 1, 53–57. doi: 10.1016/j.rhisph.2016.07.001
De Vuyst L., Leroy F. (2020). Functional role of yeasts, lactic acid bacteria and acetic acid bacteria in cocoa fermentation processes. FEMS Microbiol. Rev. 44 (4), 432–453. doi: 10.1093/femsre/fuaa014
Ding Z., Jia S., Wang Y., Xiao J., Zhang Y. (2017). Phosphate stresses affect ionome and metabolome in tea plants. Plant Physiol. Biochem. 120, 30–39. doi: 10.1016/j.plaphy.2017.09.007
Duong B., Marraccini P., Maeght J.-L., Vaast P., Lebrun M., Duponnois R. (2020). Coffee microbiota and its potential use in sustainable crop management. A review. Front. Sustain. Food Syst. 4. doi: 10.3389/fsufs.2020.607935
Dupont P. Y., Eaton C. J., Wargent J. J., Fechtner S., Solomon P., Schmid J., et al. (2015). Fungal endophyte infection of ryegrass reprograms host metabolism and alters development. New Phytol. 208 (4), 1227–1240. doi: 10.1111/nph.13614
El Hadi M. A., Zhang F. J., Wu F. F., Zhou C. H., Tao J. (2013). Advances in fruit aroma volatile research. Molecules 18 (7), 8200–8229. doi: 10.3390/molecules18078200
Elhalis H., Cox J., Frank D., Zhao J. (2020). The crucial role of yeasts in the wet fermentation of coffee beans and quality. Int. J. Food Microbiol. 333, 108796. doi: 10.1016/j.ijfoodmicro.2020.108796
Elhalis H., Cox J., Zhao J. (2023). Coffee fermentation: Expedition from traditional to controlled process and perspectives for industrialization. Appl. Food Res. 3 (1), 100253. doi: 10.1016/j.afres.2022.100253
Engeseth N. J., Ac Pangan M. F. (2018). Current context on chocolate flavor development — a review. Curr. Opin. Food Sci. 21, 84–91. doi: 10.1016/j.cofs.2018.07.002
Escobar Rodríguez C., Novak J., Buchholz F., Uetz P., Bragagna L., Gumze M., et al. (2021). The bacterial microbiome of the tomato fruit is highly dependent on the cultivation approach and correlates with flavor chemistry. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.775722
Etalo D. W., Jeon J.-S., Raaijmakers J. M. (2018). Modulation of plant chemistry by beneficial root microbiota. Natural product Rep. 35 (5), 398–409. doi: 10.1039/C7NP00057J
Evangelista S. R., Miguel M., Silva C. F., Pinheiro A. C. M., Schwan R. F. (2015). Microbiological diversity associated with the spontaneous wet method of coffee fermentation. Int. J. Food Microbiol. 210, 102–112. doi: 10.1016/j.ijfoodmicro.2015.06.008
Fatima S., Anjum T. (2017). Identification of a Potential ISR Determinant from Pseudomonas aeruginosa PM12 against Fusarium Wilt in Tomato. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00848
Fatima S., Anjum T., Hussain R., Ali B. (2017). PGPR mediated bio-fortification of tomato fruit metabolites with nutritional and pharmacological importance. Pakistan J. Biotechnol. 14 (1), 17–21-17–21. Available at: https://pjbt.org/index.php/pjbt/article/view/723
Feng Z., Bartholomew E. S., Liu Z., Cui Y., Dong Y., Li S., et al. (2021). Glandular trichomes: new focus on horticultural crops. Horticulture Res. 8, 158. doi: 10.1038/s41438-021-00592-1
Fierer N. (2017). Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15 (10), 579. doi: 10.1038/nrmicro.2017.87
Finkel O. M., Salas-González I., Castrillo G., Spaepen S., Law T. F., Teixeira P. J. P. L., et al. (2019). The effects of soil phosphorus content on plant microbiota are driven by the plant phosphate starvation response. PloS Biol. 17 (11), e3000534. doi: 10.1371/journal.pbio.3000534
Fox A. R., Soto G., Valverde C., Russo D., Lagares A. Jr., Zorreguieta Á., et al. (2016). Major cereal crops benefit from biological nitrogen fixation when inoculated with the nitrogen-fixing bacterium Pseudomonas protegens Pf-5 X940. Environ. Microbiol. 18 (10), 3522–3534. doi: 10.1111/1462-2920.13376
Fuchs B., Laihonen M., Muola A., Saikkonen K., Dobrev P. I., Vankova R., et al. (2021a). A glyphosate-based herbicide in soil differentially affects hormonal homeostasis and performance of non-target crop plants. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.787958
Fuchs B., Saikkonen K., Helander M. (2021b). Glyphosate-modulated biosynthesis driving plant defense and species interactions. Trends Plant Sci. 26 (4), 312–323. doi: 10.1016/j.tplants.2020.11.004
Gagic M., Faville M. J., Zhang W., Forester N. T., Rolston M. P., Johnson R. D., et al. (2018). Seed transmission of Epichloë endophytes in Lolium perenne is heavily influenced by host genetics. Front. Plant Sci. 9, 1580. doi: 10.3389/fpls.2018.01580
Gambetta J. M., Cozzolino D., Bastian S. E., Jeffery D. W. (2017). Exploring the effects of geographical origin on the chemical composition and quality grading of Vitis vinifera L. cv. Chardonnay grapes. Molecules 22 (2), 218. doi: 10.3390/molecules22020218
Ganugi P., Miras-Moreno B., Garcia-Perez P., Lucini L., Trevisan M. (2021). Concealed metabolic reprogramming induced by different herbicides in tomato. Plant Sci. 303, 110727. doi: 10.1016/j.plantsci.2020.110727
Gargallo-Garriga A., Sardans J., Pérez-Trujillo M., Guenther A., Llusià J., Rico L., et al. (2016). Shifts in plant foliar and floral metabolomes in response to the suppression of the associated microbiota. BMC Plant Biol. 16 (1), 78. doi: 10.1186/s12870-016-0767-7
Ghorbanpour M., Hatami M., Kariman K., Abbaszadeh Dahaji P. (2016). Phytochemical variations and enhanced efficiency of antioxidant and antimicrobial ingredients in salvia officinalis as inoculated with different rhizobacteria. Chem. Biodiversity 13 (3), 319–330. doi: 10.1002/cbdv.201500082
Gikas E., Koulakiotis N. S., Tsarbopoulos A. (2021). Phytochemical differentiation of saffron (Crocus sativus L.) by high resolution mass spectrometry metabolomic studies. Molecules 26 (8), 2180. doi: 10.3390/molecules26082180
Glick B. R. (2014). Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiological Res. 169 (1), 30–39. doi: 10.1016/j.micres.2013.09.009
Gong T., Xin X.-F. (2021). Phyllosphere microbiota: Community dynamics and its interaction with plant hosts. J. Integr. Plant Biol. 63 (2), 297–304. doi: 10.1111/jipb.13060
Gonzalez-Rios O., Suarez-Quiroz M. L., Boulanger R., Barel M., Guyot B., Guiraud J.-P., et al. (2007). Impact of “ecological” post-harvest processing on the volatile fraction of coffee beans: I. Green coffee. J. Food Composition Anal. 20 (3), 289–296. doi: 10.1016/j.jfca.2006.07.009
Gupta V. V. S. R., Bramley R. G. V., Greenfield P., Yu J., Herderich M. J. (2019). Vineyard soil microbiome composition related to rotundone concentration in Australian cool climate ‘Peppery’ Shiraz grapes. Front. Microbiol. 10 (1607). doi: 10.3389/fmicb.2019.01607
Haile M., Kang W. H. (2019). The role of microbes in coffee fermentation and their impact on coffee quality. J. Food Qual. 2019, 4836709. doi: 10.1155/2019/4836709
Hardoim P. R., Hardoim C. C., Van Overbeek L. S., Van Elsas J. D. (2012). Dynamics of seed-borne rice endophytes on early plant growth stages. PloS One 7 (2), e30438. doi: 10.1371/journal.pone.0030438
Haron M. H., Tyler H. L., Chandra S., Moraes R. M., Jackson C. R., Pugh N. D., et al. (2019). Plant microbiome-dependent immune enhancing action of EChinacea purpurea is enhanced by soil organic matter content. Sci. Rep. 9 (1), 136. doi: 10.1038/s41598-018-36907-x
Hart M., Ehret D. L., Krumbein A., Leung C., Murch S., Turi C., et al. (2015). Inoculation with arbuscular mycorrhizal fungi improves the nutritional value of tomatoes. Mycorrhiza 25 (5), 359–376. doi: 10.1007/s00572-014-0617-0
Hayes R. A., Rebolleda-Gómez M., Butela K., Cabo L. F., Cullen N., Kaufmann N., et al. (2021). Spatially explicit depiction of a floral epiphytic bacterial community reveals role for environmental filtering within petals. MicrobiologyOpen 10 (1), e1158. doi: 10.1002/mbo3.1158
Helletsgruber C., Dötterl S., Ruprecht U., Junker R. R. (2017). Epiphytic bacteria alter floral scent emissions. J. Chem. Ecol. 43 (11), 1073–1077. doi: 10.1007/s10886-017-0898-9
Hernandez C. E., Granados L. (2021). Quality differentiation of cocoa beans: implications for geographical indications. J. Sci. Food Agric. 101 (10), 3993–4002. doi: 10.1002/jsfa.11077
Herrera C. M., García I. M., Pérez R. (2008). Invisible floral larcenies: microbial communities degrade floral nectar of bumble bee-pollinated plants. Ecology 89 (9), 2369–2376. doi: 10.1890/08-0241.1
Hol W. G., De Boer W., Termorshuizen A. J., Meyer K. M., Schneider J. H., Van Dam N. M., et al. (2010). Reduction of rare soil microbes modifies plant–herbivore interactions. Ecol. Lett. 13 (3), 292–301. doi: 10.1111/j.1461-0248.2009.01424.x
Hooykaas P. (2000). “Agrobacterium, a natural metabolic engineer of plants,” in Metabolic engineering of plant secondary metabolism. Alfermann R.V.A.W. eds. (Dordrecht, Netherlands: Springer Dordrecht), 51–67.
Huang M., Sanchez-Moreiras A. M., Abel C., Sohrabi R., Lee S., Gershenzon J., et al. (2012). The major volatile organic compound emitted from Arabidopsis thaliana flowers, the sesquiterpene (E)-β-caryophyllene, is a defense against a bacterial pathogen. New Phytol. 193 (4), 997–1008. doi: 10.1111/j.1469-8137.2011.04001.x
Huang L.-F., Song L.-X., Xia X.-J., Mao W.-H., Shi K., Zhou Y.-H., et al. (2013). Plant-soil feedbacks and soil sickness: from mechanisms to application in agriculture. J. Chem. Ecol. 39 (2), 232–242. doi: 10.1007/s10886-013-0244-9
Huang W., Sun D., Chen L., An Y. (2021). Integrative analysis of the microbiome and metabolome in understanding the causes of sugarcane bitterness. Sci. Rep. 11 (1), 6024. doi: 10.1038/s41598-021-85433-w
Huang X., Tang Q., Li Q., Lin H., Li J., Zhu M., et al. (2022). Integrative analysis of transcriptome and metabolome reveals the mechanism of foliar application of Bacillus amyloliquefaciens to improve summer tea quality (Camellia sinensis). Plant Physiol. Biochem. 185, 302–313. doi: 10.1016/j.plaphy.2022.06.016
Huang L.-H., Yuan M.-Q., Ao X.-J., Ren A.-Y., Zhang H.-B., Yang M.-Z. (2018). Endophytic fungi specifically introduce novel metabolites into grape flesh cells in vitro. PloS One 13 (5), e0196996. doi: 10.1371/journal.pone.0196996
Hume D., Card S., Rolston M. (2013). “Effects of storage conditions on endophyte and seed viability in pasture grasses,” in: Revitalising Grasslands to Sustain our Communities: Proceedings, 22nd International Grassland Congress, 15-19 September, 2013, Sydney, Australia: New South Wales Department of Primary Industry, 405–408.
Jackels S. C., Jackels C. F. (2005). Characterization of the coffee mucilage fermentation process using chemical indicators: A field study in Nicaragua. J. Food Sci. 70 (5), C321–C325. doi: 10.1111/j.1365-2621.2005.tb09960.x
Jeandet P., Clément C., Courot E. (2014). Resveratrol production at large scale using plant cell suspensions. Eng. Life Sci. 14 (6), 622–632. doi: 10.1002/elsc.201400022
Jin H.-Q., Liu H.-B., Xie Y.-Y., Zhang Y.-G., Xu Q.-Q., Mao L.-J., et al. (2018). Effect of the dark septate endophytic fungus Acrocalymma vagum on heavy metal content in tobacco leaves. Symbiosis 74 (2), 89–95. doi: 10.1007/s13199-017-0485-4
Johnson K. B., Stockwell V. O., Sawyer T. L., Sugar D. (2000). Assessment of environmental factors influencing growth and spread of pantoea agglomerans on and among blossoms of pear and apple. Phytopathology® 90 (11), 1285–1294. doi: 10.1094/phyto.2000.90.11.1285
Johnston-Monje D., Arévalo A. L., Bolaños A. C. (2021a). “Friends in low places: Soil derived microbial inoculants for biostimulation and biocontrol in crop production,” in Microbiome Stimulants for Crops (Elsevier), 15–31.
Johnston-Monje D., Castillo-Avila D. K., Raizada M. N., Becerra Lopez-Lavalle L. A. (2019). Paying the Rent: How Endophytic Microorganisms Help Plant Hosts Obtain Nutrients (Pergamon: Elsevier).
Johnston-Monje D., Gutiérrez J. P., Becerra Lopez-Lavalle L. A. (2022). Stochastic inoculum, biotic filtering and species-specific seed transmission shape the rare microbiome of plants. Life 12 (9), 1372. doi: 10.3390/life12091372
Johnston-Monje D., Gutiérrez J. P., Lopez-Lavalle L. A. B. (2021b). Seed-transmitted bacteria and fungi dominate juvenile plant microbiomes. Front. Microbiol. 12 (2945). doi: 10.3389/fmicb.2021.737616
Jones G. V. (2018). The climate component of terroir. Elements: Int. Magazine Mineralogy Geochemistry Petrology 14 (3), 167–172. doi: 10.2138/gselements.14.3.167
Kadow D., Niemenak N., Rohn S., Lieberei R. (2015). Fermentation-like incubation of cocoa seeds (Theobroma cacao L.) – Reconstruction and guidance of the fermentation process. LWT - Food Sci. Technol. 62 (1, Part 1), 357–361. doi: 10.1016/j.lwt.2015.01.015
Kalozoumis P. G., Savvas D., Aliferis K. A., Ntatsi G., Marakis G., Simou E., et al. (2021). Impact of plant growth-promoting rhizobacteria inoculation and grafting on tolerance of tomato to combined water and nutrient stress assessed via metabolomics analysis. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.670236
Kaur H., Kaur R., Kaur S., Baldwin I. T., Inderjit (2009). Taking ecological function seriously: soil microbial communities can obviate allelopathic effects of released metabolites. PloS One 4 (3), e4700. doi: 10.1371/journal.pone.0004700
Kaur T., Rana K. L., Kour D., Sheikh I., Yadav N., Yadav A., et al. (2020). “Microbe-mediated biofortification for micronutrients: present status and future challenges,” in Trends of microbial biotechnology for sustainable agriculture and biomedicine systems: perspectives for human health (Amsterdam: Elsevier), 1–17.
Ke M., Ye Y., Li Y., Zhou Z., Xu N., Feng L., et al. (2021). Leaf metabolic influence of glyphosate and nanotubes on the Arabidopsis thaliana phyllosphere. J. Environ. Sci. 106, 66–75. doi: 10.1016/j.jes.2021.01.002
Klaedtke S., Jacques M.-A., Raggi L., Préveaux A., Bonneau S., Negri V., et al. (2016). Terroir is a key driver of seed-associated microbial assemblages. Environ. Microbiol. 18 (6), 1792–1804. doi: 10.1111/1462-2920.12977
Knight S., Klaere S., Fedrizzi B., Goddard M. R. (2015). Regional microbial signatures positively correlate with differential wine phenotypes: evidence for a microbial aspect to terroir. Sci. Rep. 5 (1), 14233. doi: 10.1038/srep14233
Korenblum E., Aharoni A. (2019). Phytobiome metabolism: beneficial soil microbes steer crop plants' secondary metabolism. Pest Manage. Sci. 75 (9), 2378–2384. doi: 10.1002/ps.5440
Korenblum E., Massalha H., Aharoni A. (2022). Plant–microbe interactions in the rhizosphere via a circular metabolic economy. Plant Cell 4 (9), 3168–3182. doi: 10.1093/plcell/koac163
Kox M. A., van den Elzen E., Lamers L. P., Jetten M. S., van Kessel M. A. (2020). Microbial nitrogen fixation and methane oxidation are strongly enhanced by light in Sphagnum mosses. Amb Express 10 (1), 1–11. doi: 10.1186/s13568-020-00994-9
Krebiehl M. W. (2023). “The differences between high- and low-elevation wine,” in Wine Enthusiast (New York, USA: Wine Enthusiast Companies).
Kundu A., Mishra S., Kundu P., Jogawat A., Vadassery J. (2022). Piriformospora indica recruits host-derived putrescine for growth promotion in plants. Plant Physiol. 188 (4), 2289–2307. doi: 10.1093/plphys/kiab536
Kuppardt A., Fester T., Härtig C., Chatzinotas A. (2018). Rhizosphere protists change metabolite profiles in zea mays. Front. Microbiol. 9 (857). doi: 10.3389/fmicb.2018.00857
Kusari S, Lamshöft M, Kusari P, Gottfried S, Zühlke S, Louven K, et al. (2014). Endophytes are hidden producers of maytansine in Putterlickia roots. J. Natural Products 77 (12), 2577–2584. doi: 10.1021/np500219a
Kuzyakov Y., Xu X. (2013). Competition between roots and microorganisms for nitrogen: mechanisms and ecological relevance. New Phytol. 198 (3), 656–669. doi: 10.1111/nph.12235
Kwak H. S., Jeong Y., Kim M. (2018). Effect of yeast fermentation of green coffee beans on antioxidant activity and consumer acceptability. J. Food Qual. 2018, 5967130. doi: 10.1155/2018/5967130
Lee L. W., Cheong M. W., Curran P., Yu B., Liu S. Q. (2015). Coffee fermentation and flavor – An intricate and delicate relationship. Food Chem. 185, 182–191. doi: 10.1016/j.foodchem.2015.03.124
Lee L. W., Cheong M. W., Curran P., Yu B., Liu S. Q. (2016). Modulation of coffee aroma via the fermentation of green coffee beans with Rhizopus oligosporus: I. Green coffee. Food Chem. 211, 916–924. doi: 10.1016/j.foodchem.2016.05.076
Lee K., Missaoui A., Mahmud K., Presley H., Lonnee M. (2021). Interaction between grasses and epichloë Endophytes and its significance to biotic and abiotic stress tolerance and the rhizosphere. Microorganisms 9 (11), 2186. doi: 10.3390/microorganisms9112186
Leff J. W., Fierer N. (2013). Bacterial communities associated with the surfaces of fresh fruits and vegetables. PloS One 8 (3), e59310. doi: 10.1371/journal.pone.0059310
Lenaerts M., Goelen T., Paulussen C., Herrera-Malaver B., Steensels J., Van den Ende W., et al. (2017). Nectar bacteria affect life history of a generalist aphid parasitoid by altering nectar chemistry. Funct. Ecol. 31 (11), 2061–2069. doi: 10.1111/1365-2435.12933
Li X., Jousset A., de Boer W., Carrión V. J., Zhang T., Wang X., et al. (2019). Legacy of land use history determines reprogramming of plant physiology by soil microbiome. ISME J. 13 (3), 738–751. doi: 10.1038/s41396-018-0300-0
Li H., Li M., Fan Y., Liu Y., Qin S. (2022). Antifungal activity of potato glycoalkaloids and its potential to control severity of dry rot caused by Fusarium sulphureum. Crop Sci 63 (2), 801–811. doi: 10.1002/csc2.20874
Lima L. J., Almeida M. H., Nout M. R., Zwietering M. H. (2011). Theobroma cacao L.,”The food of the Gods”: quality determinants of commercial cocoa beans, with particular reference to the impact of fermentation. Crit. Rev. Food Sci. Nutr. 51 (8), 731–761. doi: 10.1080/10408391003799913
Ling L., Tu Y., Ma W., Feng S., Yang C., Zhao Y., et al. (2020). A potentially important resource: endophytic yeasts. World J. Microbiol. Biotechnol. 36, 1–7. doi: 10.1007/s11274-020-02889-0
Liu S. Q. (2002). Malolactic fermentation in wine – beyond deacidification. J. Appl. Microbiol. 92 (4), 589–601. doi: 10.1046/j.1365-2672.2002.01589.x
Liu J., Abdelfattah A., Norelli J., Burchard E., Schena L., Droby S., et al. (2018). Apple endophytic microbiota of different rootstock/scion combinations suggests a genotype-specific influence. Microbiome 6 (1), 18. doi: 10.1186/s40168-018-0403-x
Liu D., Chen Q., Zhang P., Chen D., Howell K. S. (2020). The fungal microbiome is an important component of vineyard ecosystems and correlates with regional distinctiveness of wine. mSphere 5 (4), e00534-20. doi: 10.1128/mSphere.00534-20
Liu Y., Hou W., Jin J., Christensen M. J., Gu L., Cheng C., et al. (2021b). Epichloë gansuensis Increases the Tolerance of Achnatherum inebrians to Low-P Stress by Modulating Amino Acids Metabolism and Phosphorus Utilization Efficiency. J. Fungi 7 (5), 390. doi: 10.3390/jof7050390
Liu D., Legras J.-L., Zhang P., Chen D., Howell K. (2021a). Diversity and dynamics of fungi during spontaneous fermentations and association with unique aroma profiles in wine. Int. J. Food Microbiol. 338, 108983. doi: 10.1016/j.ijfoodmicro.2020.108983
Liu Y., Rousseaux S., Tourdot-Maréchal R., Sadoudi M., Gougeon R., Schmitt-Kopplin P., et al. (2017). Wine microbiome: A dynamic world of microbial interactions. Crit. Rev. Food Sci. Nutr. 57 (4), 856–873. doi: 10.1080/10408398.2014.983591
Liu D., Zhang P., Chen D., Howell K. (2019). From the vineyard to the winery: how microbial ecology drives regional distinctiveness of wine. Front. Microbiol. 10 (2679). doi: 10.3389/fmicb.2019.02679
Lofgren L.A., LeBlanc N.R., Certano A.K., Nachtigall J., LaBine K.M., Riddle J., et al (2018). Fusarium graminearum: pathogen or endophyte of North American grasses? Phytologist 217 (3), 1203–1212.
Lucini L., Rocchetti G., Trevisan M. (2020). Extending the concept of terroir from grapes to other agricultural commodities: an overview. Curr. Opin. Food Sci. 31, 88–95. doi: 10.1016/j.cofs.2020.03.007
Luo J., Tao Q., Jupa R., Liu Y., Wu K., Song Y., et al. (2019). Role of vertical transmission of shoot endophytes in root-associated microbiome assembly and heavy metal hyperaccumulation in Sedum alfredii. Environ. Sci. Technol. 53 (12), 6954–6963. doi: 10.1021/acs.est.9b01093
Maddela N. R., Kakarla D., García L. C., Chakraborty S., Venkateswarlu K., Megharaj M. (2020). Cocoa-laden cadmium threatens human health and cacao economy: A critical view. Sci. Total Environ. 720, 137645. doi: 10.1016/j.scitotenv.2020.137645
Malinowski D. P., Alloush G. A., Belesky D. P. (2000). Leaf endophyte Neotyphodium coenophialum modifies mineral uptake in tall fescue. Plant Soil 227 (1), 115–126. doi: 10.1023/A:1026518828237
Mandl K., Schieck J., Silhavy-Richter K., Alexander P., Schneider V., Schmidt H. (2015). Vines take up yeasts from soil and transport them through the vine to the stem and skins of grapes. Ithaka J. 2015, 349–355.
Massa N., Cesaro P., Todeschini V., Capraro J., Scarafoni A., Cantamessa S., et al. (2020). Selected autochthonous rhizobia, applied in combination with AM fungi, improve seed quality of common bean cultivated in reduced fertilization condition. Appl. Soil Ecol. 148, 103507. doi: 10.1016/j.apsoil.2020.103507
Massoni J., Bortfeld-Miller M., Widmer A., Vorholt J. A. (2021). Capacity of soil bacteria to reach the phyllosphere and convergence of floral communities despite soil microbiota variation. Proc. Natl. Acad. Sci. 118 (41), e2100150118. doi: 10.1073/pnas.2100150118
Mathur V., Ulanova D. (2022). Microbial metabolites beneficial to plant hosts across ecosystems. Microbial Ecol. 86 (1), 25–48. doi: 10.1007/s00248-022-02073-x
Meena R. S., Kumar S., Datta R., Lal R., Vijayakumar V., Brtnicky M., et al. (2020). Impact of agrochemicals on soil microbiota and management: A review. Land 9 (2), 34. doi: 10.3390/land9020034
Meng L., Sun T., Li M., Saleem M., Zhang Q., Wang C. (2019). Soil-applied biochar increases microbial diversity and wheat plant performance under herbicide fomesafen stress. Ecotoxicology Environ. Saf. 171, 75–83. doi: 10.1016/j.ecoenv.2018.12.065
Mezzasalma V., Sandionigi A., Bruni I., Bruno A., Lovicu G., Casiraghi M., et al. (2017). Grape microbiome as a reliable and persistent signature of field origin and environmental conditions in Cannonau wine production. PloS One 12 (9), e0184615. doi: 10.1371/journal.pone.0184615
Micci A., Zhang Q., Chang X., Kingsley K., Park L., Chiaranunt P., et al. (2022). Histochemical evidence for nitrogen-transfer endosymbiosis in non-photosynthetic cells of leaves and inflorescence bracts of angiosperms. Biol. (Basel) 11 (6), 876. doi: 10.3390/biology11060876
Miedaner T., Geiger H. H. (2015). Biology, genetics, and management of ergot (claviceps spp.) in rye, sorghum, and pearl millet. Toxins 7 (3), 659–678.
Miotto-Vilanova L., Courteaux B., Padilla R., Rabenoelina F., Jacquard C., Clément C., et al. (2019). Impact of paraburkholderia phytofirmans psJN on grapevine phenolic metabolism. Int. J. Mol. Sci. 20 (22), 5775. doi: 10.3390/ijms20225775
Mirzaei M., Ladan Moghadam A., Hakimi L., Danaee E. (2020). Plant growth promoting rhizobacteria (PGPR) improve plant growth, antioxidant capacity, and essential oil properties of lemongrass (Cymbopogon citratus) under water stress. Plant Physiol. 10 (2), 3155–3166. doi: 10.30495/IJPP.2020.672574
Misra B. B., Das V., Landi M., Abenavoli M. R., Araniti F. (2020). Short-term effects of the allelochemical umbelliferone on Triticum durum L. metabolism through GC–MS based untargeted metabolomics. Plant Sci. 298, 110548. doi: 10.1016/j.plantsci.2020.110548
Misra S., Pandey S., Dixit V., Mishra S. K., Khan M. H., Agarwal L., et al. (2017). “Soil microbiome for enhanced crop productivity,” in Mining of Microbial Wealth and MetaGenomics. Eds. Kalia V. C., Shouche Y., Purohit H. J., Rahi P. (Singapore: Springer Singapore), 227–247.
Mocali S., Kuramae E. E., Kowalchuk G. A., Fornasier F., Priori S. (2020). Microbial functional diversity in vineyard soils: sulfur metabolism and links with grapevine plants and wine quality. Front. Environ. Sci 8, 75. doi: 10.3389/fenvs.2020.00075
Mohamed N., Lherminier J., Farmer M.-J., Fromentin J., Béno N., Houot V., et al. (2007). Defense responses in grapevine leaves against Botrytis cinerea induced by application of a Pythium oligandrum strain or its elicitin, oligandrin, to roots. Phytopathology 97 (5), 611–620. doi: 10.1094/PHYTO-97-5-0611
Morison A. (2020). Rhizo coffee a novel fermented coffee product. MSc, Victoria University of Wellington.
Morris C. E., Kinkel L. (2002). “Fifty years of phyllosphere microbiology: significant contributions to research in related fields,” in Phyllosphere microbiology, eds. Lindow S.E.H.-P., Lindow E. I., Elliott V. J. (St. Paul, Minnesota, USA: APS Press), 365–375.
Muehe E. M., Weigold P., Adaktylou I. J., Planer-Friedrich B., Kraemer U., Kappler A., et al. (2015). Rhizosphere Microbial Community Composition Affects Cadmium and Zinc Uptake by the Metal-Hyperaccumulating Plant <span class="named-content genus-species" id="named-content-1">Arabidopsis halleri. Appl. Environ. Microbiol. 81 (6), 2173–2181. doi: 10.1128/aem.03359-14
Nabi N., Singh S., Saffeullah P. (2021). Responses of in vitro cell cultures to elicitation: regulatory role of jasmonic acid and methyl jasmonate: a review. In Vitro Cell. Dev. Biol. - Plant 57 (3), 341–355. doi: 10.1007/s11627-020-10140-6
Nasopoulou C., Pohjanen J., Koskimäki J. J., Zabetakis I., Pirttilä A. M. (2014). Localization of strawberry (Fragaria x ananassa) and Methylobacterium extorquens genes of strawberry flavor biosynthesis in strawberry tissue by in situ hybridization. J. Plant Physiol. 171 (13), 1099–1105. doi: 10.1016/j.jplph.2014.03.018
Nebert L. D. (2018). On Germs and Germination: Uncovering the Hidden Ecology of Seedborne Bacteria and Fungi in Open-Pollinated Maize (Oregon: University of Oregon).
Newman D. J., Cragg G. M. (2015). Endophytic and epiphytic microbes as “sources” of bioactive agents. Front. Chem. 3. doi: 10.3389/fchem.2015.00034
Newman D. J., Cragg G. M. (2020). Plant endophytes and epiphytes: Burgeoning sources of known and “unknown” cytotoxic and antibiotic agents? Planta Med. 86 (13/14), 891–905. doi: 10.1055/a-1095-1111
O’Banion B. S., O’Neal L., Alexandre G., Lebeis S. L. (2020). Bridging the gap between single-strain and community-level plant-microbe chemical interactions. Mol. Plant-Microbe Interactions® 33 (2), 124–134. doi: 10.1094/mpmi-04-19-0115-cr
Osman K. T. (2013). “Plant nutrients and soil fertility management,” in Soils: Principles, Properties and Management (Dordrecht: Springer Netherlands), 129–159.
Palmer J., Chen B. (2018). Wineinformatics: regression on the grade and price of wines through their sensory attributes. Fermentation 4 (4), 84. doi: 10.3390/fermentation4040084
Pandey S. S., Singh S., Babu C. V., Shanker K., Srivastava N., Shukla A. K., et al. (2016). Fungal endophytes of Catharanthus roseus enhance vindoline content by modulating structural and regulatory genes related to terpenoid indole alkaloid biosynthesis. Sci. Rep. 6 (1), 26583. doi: 10.1038/srep26583
Parizad S., Dizadji A., Habibi M. K., Winter S., Kalantari S., Movi S., et al. (2019). The effects of geographical origin and virus infection on the saffron (Crocus sativus L.) quality. Food Chem. 295, 387–394. doi: 10.1016/j.foodchem.2019.05.116
Paroissien E., Visser M. (2020). The causal impact of medals on wine producers' Prices and the gains from participating in contests. Am. J. Agric. Economics 102 (4), 1135–1153. doi: 10.1002/ajae.12037
Paungfoo-Lonhienne C., Rentsch D., Robatzek S., Webb R. I., Sagulenko E., Näsholm T., et al. (2010). Turning the table: plants consume microbes as a source of nutrients. PloS One 5 (7), e11915. doi: 10.1371/journal.pone.0011915
Pedras M. S. C., Zheng Q.-A., Strelkov S. (2008). Metabolic changes in roots of the oilseed canola infected with the biotroph plasmodiophora brassicae: phytoalexins and phytoanticipins. J. Agric. Food Chem. 56 (21), 9949–9961. doi: 10.1021/jf802192f
Peñuelas J., Asensio D., Tholl D., Wenke K., Rosenkranz M., Piechulla B., et al. (2014). Biogenic volatile emissions from the soil. Plant Cell Environ. 37 (8), 1866–1891. doi: 10.1111/pce.12340
Penuelas J., Farré-Armengol G., Llusia J., Gargallo-Garriga A., Rico L., Sardans J., et al. (2014). Removal of floral microbiota reduces floral terpene emissions. Sci. Rep. 4 (1), 6727. doi: 10.1038/srep06727
Pérez-Rodriguez M. M., Pontin M., Lipinski V., Bottini R., Piccoli P., Cohen A. C. (2020). Pseudomonas fluorescens and Azospirillum brasilense Increase Yield and Fruit Quality of Tomato Under Field Conditions. J. Soil Sci. Plant Nutr. 20 (4), 1614–1624. doi: 10.1007/s42729-020-00233-x
Pestka J. J. (2010). Deoxynivalenol: mechanisms of action, human exposure, and toxicological relevance. Arch. Toxicol. 84 (9), 663–679. doi: 10.1007/s00204-010-0579-8
Petersen I. L., Tomasi G., Sørensen H., Boll E. S., Hansen H. C. B., Christensen J. H. (2011). The use of environmental metabolomics to determine glyphosate level of exposure in rapeseed (Brassica napus L.) seedlings. Environ. pollut. 159 (10), 3071–3077. doi: 10.1016/j.envpol.2011.04.005
Piasecka A., Jedrzejczak-Rey N., Bednarek P. (2015). Secondary metabolites in plant innate immunity: conserved function of divergent chemicals. New Phytol. 206 (3), 948–964. doi: 10.1111/nph.13325
Pineda A., Kaplan I., Bezemer T. M. (2017). Steering soil microbiomes to suppress aboveground insect pests. Trends Plant Sci. 22 (9), 770–778. doi: 10.1016/j.tplants.2017.07.002
Pineda A., Soler R., Weldegergis B. T., Shimwela M. M., Van Loon J. J. A., Dicke M. (2013). Non-pathogenic rhizobacteria interfere with the attraction of parasitoids to aphid-induced plant volatiles via jasmonic acid signalling. Plant Cell Environ. 36 (2), 393–404. doi: 10.1111/j.1365-3040.2012.02581.x
Ping L., Boland W. (2004). Signals from the underground: bacterial volatiles promote growth in Arabidopsis. Trends Plant Sci. 9 (6), 263–266. doi: 10.1016/j.tplants.2004.04.008
Pinto C., Pinho D., Cardoso R., Custódio V., Fernandes J., Sousa S., et al. (2015). Wine fermentation microbiome: a landscape from different Portuguese wine appellations. Front. Microbiol. 6 (905). doi: 10.3389/fmicb.2015.00905
Plaszkó T., Szűcs Z., Cziáky Z., Ács-Szabó L., Csoma H., Géczi L., et al. (2022). Correlations between the metabolome and the endophytic fungal metagenome suggests importance of various metabolite classes in community assembly in horseradish (Armoracia rusticana, Brassicaceae) roots. Front. Plant Sci. 13, 921008. doi: 10.3389/fpls.2022.921008
Ptak A., Morańska E., Warchoł M., Gurgul A., Skrzypek E., Dziurka M., et al. (2022). Endophytic bacteria from in vitro culture of Leucojum aestivum L. @ a new source of galanthamine and elicitor of alkaloid biosynthesis. Sci. Rep. 12 (1), 13700. doi: 10.1038/s41598-022-17992-5
Qu Q., Zhang Z., Peijnenburg W., Liu W., Lu T., Hu B., et al. (2020). Rhizosphere microbiome assembly and its impact on plant growth. J. Agric. Food Chem. 68 (18), 5024–5038. doi: 10.1021/acs.jafc.0c00073
Quambusch M., Winkelmann T. (2018). “Bacterial endophytes in plant tissue culture: Mode of action, detection, and control,” Plant Cell culture Protoc. eds. Loyola-Vargas V.M., Ochoa-Alejo N.. eds. (New York, NY, USA: Humana Press), 69–88. doi: 10.1007/978-1-4939-8594-4_4
Ramirez-Villacis D. X., Finkel O. M., Salas-González I., Fitzpatrick C. R., Dangl J. L., Jones C. D., et al. (2020). Root microbiome modulates plant growth promotion induced by low doses of glyphosate. mSphere 5 (4), e00484–e00420. doi: 10.1128/mSphere.00484-20
Ranjbar Sistani N., Desalegn G., Kaul H.-P., Wienkoop S. (2020). Seed metabolism and pathogen resistance enhancement in pisum sativum during colonization of arbuscular mycorrhizal fungi: an integrative metabolomics-proteomics approach. Front. Plant Sci. 11 (872). doi: 10.3389/fpls.2020.00872
Rashid M. H.-O., Chung Y. R. (2017). Induction of systemic resistance against insect herbivores in plants by beneficial soil microbes. Front. Plant Sci. 8 (1816). doi: 10.3389/fpls.2017.01816
Rasmussen H. N., Dixon K. W., Jersáková J., Těšitelová T. (2015). Germination and seedling establishment in orchids: a complex of requirements. Ann. Bot. 116 (3), 391–402. doi: 10.1093/aob/mcv087
Repanich J. (2019) The World Record for Most Expensive Coffee Sold Was Just Shattered. Available at: https://robbreport.com/food-drink/dining/world-record-coffee-sold-Panama-gesha-elida-estate-2859497/ (Accessed 2020/7/25 2020).
Rho H., Van Epps V., Kim S.-H., Doty S. L. (2020). Endophytes increased fruit quality with higher soluble sugar production in honeycrisp apple (Malus pumila). Microorganisms 8 (5), 699. doi: 10.3390/microorganisms8050699
Ricks K. D., Koide R. T. (2019). Biotic filtering of endophytic fungal communities in Bromus tectorum. Oecologia 189 (4), 993–1003. doi: 10.1007/s00442-019-04388-y
Rillig M. C., Lehmann A., Lehmann J., Camenzind T., Rauh C. (2018). Soil biodiversity effects from field to fork. Trends Plant Sci. 23 (1), 17–24. doi: 10.1016/j.tplants.2017.10.003
Rodriguez R., White J. Jr., Arnold A., Redman a. (2009). Fungal endophytes: diversity and functional roles. New Phytol. 182 (2), 314–330. doi: 10.1111/j.1469-8137.2009.02773.x
Roelofsen P. A. (1958). “Fermentation, drying, and storage of cacao beans,” in Advances in Food Research. Eds. Mrak E. M., Stewart G. F. (Elsevier), 225–296.
Roullier-Gall C., Lucio M., Noret L., Schmitt-Kopplin P., Gougeon R. D. (2014). How subtle is the “Terroir” Effect? Chemistry-related signatures of two “Climats de bourgogne”. PloS One 9 (5), e97615. doi: 10.1371/journal.pone.0097615
Rouphael Y., Franken P., Schneider C., Schwarz D., Giovannetti M., Agnolucci M., et al. (2015). Arbuscular mycorrhizal fungi act as biostimulants in horticultural crops. Scientia Hortic. 196, 91–108. doi: 10.1016/j.scienta.2015.09.002
Rout M. E., Callaway R. M. (2012). Interactions between exotic invasive plants and soil microbes in the rhizosphere suggest that 'everything is not everywhere'. Ann. Bot. 110 (2), 213–222. doi: 10.1093/aob/mcs061
Ruuskanen S., Fuchs B., Nissinen R., Puigbò P., Rainio M., Saikkonen K., et al. (2022). Ecosystem consequences of herbicides: the role of microbiome. Trends Ecol. Evol 38 (1), 35–43. doi: 10.1016/j.tree.2022.09.009
Ryffel F., Helfrich E. J. N., Kiefer P., Peyriga L., Portais J.-C., Piel J., et al. (2016). Metabolic footprint of epiphytic bacteria on Arabidopsis thaliana leaves. ISME J. 10 (3), 632–643. doi: 10.1038/ismej.2015.141
Sánchez-Rodríguez A. R., Del Campillo M. C., Quesada-Moraga E. (2015). Beauveria bassiana: An entomopathogenic fungus alleviates Fe chlorosis symptoms in plants grown on calcareous substrates. Scientia Hortic. 197, 193–202. doi: 10.1016/j.scienta.2015.09.029
Sangiorgio D., Cellini A., Spinelli F., Farneti B., Khomenko I., Muzzi E., et al. (2021). Does organic farming increase raspberry quality, aroma and beneficial bacterial biodiversity? Microorganisms 9 (8), 1617. doi: 10.3390/microorganisms9081617
Sangiorgio D., Cellini A., Spinelli F., Pastore C., Farneti B., Savioli S., et al. (2022). Contribution of fruit microbiome to raspberry volatile organic compounds emission. Postharvest Biol. Technol. 183, 111742. doi: 10.1016/j.postharvbio.2021.111742
Santoro M. V., Bogino P. C., Nocelli N., Cappellari L., Giordano W. F., Banchio E. (2016). Analysis of plant growth-promoting effects of fluorescent pseudomonas strains isolated from mentha piperita rhizosphere and effects of their volatile organic compounds on essential oil composition. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01085
Santoro M. V., Zygadlo J., Giordano W., Banchio E. (2011). Volatile organic compounds from rhizobacteria increase biosynthesis of essential oils and growth parameters in peppermint (Mentha piperita). Plant Physiol. Biochem. 49 (10), 1177–1182. doi: 10.1016/j.plaphy.2011.07.016
Saranraj P., Sivasakthivelan P., Naveen M. (2017). Fermentation of fruit wine and its quality analysis: A review. Aust. J. Sci. Technol. 1 (2), 85–97.
Saunier A., Mpamah P., Biasi C., Blande J. D. (2020). Microorganisms in the phylloplane modulate the BVOC emissions of Brassica nigra leaves. Plant Signaling Behav. 15 (3), 1728468. doi: 10.1080/15592324.2020.1728468
Sbrana C., Avio L., Giovannetti M. (2014). Beneficial mycorrhizal symbionts affecting the production of health-promoting phytochemicals. ELECTROPHORESIS 35 (11), 1535–1546. doi: 10.1002/elps.201300568
Schaeffer R. N., Crowder D. W., Illán J. G., Beck J. J., Fukami T., Williams N. M., et al. (2023). Disease management during bloom affects the floral microbiome but not pollination in a mass-flowering crop. J. Appl. Ecol. 60 (1), 64–76. doi: 10.1111/1365-2664.14320
Schilmiller A. L., Last R. L., Pichersky E. (2008). Harnessing plant trichome biochemistry for the production of useful compounds. Plant J. 54 (4), 702–711. doi: 10.1111/j.1365-313X.2008.03432.x
Schmidt R., Köberl M., Mostafa A., Ramadan E. M., Monschein M., Jensen K. B., et al. (2014). Effects of bacterial inoculants on the indigenous microbiome and secondary metabolites of chamomile plants. Front. Microbiol. 5. doi: 10.3389/fmicb.2014.00064
Scholz M., Kitzberger C. S. G., Prudencio S. H., Silva R. S. (2018). The typicity of coffees from different terroirs determined by groups of physico-chemical and sensory variables and multiple factor analysis. Food Res. Int. 114, 72–80. doi: 10.1016/j.foodres.2018.07.058
Schwan R. F., Wheals A. E. (2004). The microbiology of cocoa fermentation and its role in chocolate quality. Crit. Rev. Food Sci. Nutr. 44 (4), 205–221. doi: 10.1080/10408690490464104
Scognamiglio M., Schneider B. (2020). Identification of potential allelochemicals from donor plants and their synergistic effects on the metabolome of Aegilops geniculata. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.01046
Shahane A. A., Shivay Y. S., Prasanna R. (2020). Enhancing phosphorus and iron nutrition of wheat through crop establishment techniques and microbial inoculations in conjunction with fertilization. Soil Sci. Plant Nutr. 66 (5), 763–771. doi: 10.1080/00380768.2020.1799692
Shao Y.-D., Zhang D.-J., Hu X.-C., Wu Q.-S., Jiang C.-J., Gao X.-B., et al. (2019). Arbuscular mycorrhiza improves leaf food quality of tea plants. Notulae Botanicae Horti Agrobotanici Cluj-Napoca 47 (3), 608–614. doi: 10.15835/nbha47311434
Sharaby Y., Rodríguez-Martínez S., Lalzar M., Halpern M., Izhaki I. (2020). Geographic partitioning or environmental selection: What governs the global distribution of bacterial communities inhabiting floral nectar? Sci. Total Environ. 749, 142305. doi: 10.1016/j.scitotenv.2020.142305
Sharma R. K., Archana G. (2016). Cadmium minimization in food crops by cadmium resistant plant growth promoting rhizobacteria. Appl. Soil Ecol. 107, 66–78. doi: 10.1016/j.apsoil.2016.05.009
Soliman S., Greenwood J. S., Bombarely A., Mueller L. A., Tsao R., Mosser D., et al. (2015). An endophyte constructs fungicide-containing extracellular barriers for its host plant. Curr. Biol. 25 (19), 2570–2576. doi: 10.1016/j.cub.2015.08.027
Soliman S. S. M., Trobacher C. P., Tsao R., Greenwood J. S., Raizada M. N. (2013). A fungal endophyte induces transcription of genes encoding a redundant fungicide pathway in its host plant. BMC Plant Biol. 13 (1), 93. doi: 10.1186/1471-2229-13-93
Stefanini I., Albanese D., Cavazza A., Franciosi E., De Filippo C., Donati C., et al. (2016). Dynamic changes in microbiota and mycobiota during spontaneous ‘Vino Santo Trentino’ fermentation. Microbial Biotechnol. 9 (2), 195–208. doi: 10.1111/1751-7915.12337
Strickland M. S., Lauber C., Fierer N., Bradford M. A. (2009). Testing the functional significance of microbial community composition. Ecology 90 (2), 441–451. doi: 10.1890/08-0296.1
Strobel G., Daisy B. (2003). Bioprospecting for microbial endophytes and their natural products. Microbiol. Mol. Biol. Rev. 67 (4), 491–502. doi: 10.1128/MMBR.67.4.491-502.2003
Sun L., Liu L., Wang Y., Feng Y., Yang W., Wang D., et al. (2022). Integration of metabolomics and transcriptomics for investigating the tolerance of foxtail millet (Setaria italica) to atrazine stress. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.890550
Swiegers J. H., Bartowsky E. J., Henschke P. A., Pretorius I. S. (2005). Yeast and bacterial modulation of wine aroma and flavour. Aust. J. Grape Wine Res. 11 (2), 139–173. doi: 10.1111/j.1755-0238.2005.tb00285.x
Taghinasab M., Jabaji S. (2020). Cannabis microbiome and the role of endophytes in modulating the production of secondary metabolites: an overview. Microorganisms 8 (3), 355. doi: 10.3390/microorganisms8030355
Tanney C. A. S., Backer R., Geitmann A., Smith D. L. (2021). Cannabis glandular trichomes: A cellular metabolite factory. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.721986
Taylor A. J., Cardenas-Torres E., Miller M. J., Zhao S. D., Engeseth N. J. (2022). Microbes associated with spontaneous cacao fermentations - A systematic review and meta-analysis. Curr. Res. Food Sci. 5, 1452–1464. doi: 10.1016/j.crfs.2022.08.008
Teuber R. (2010). Geographical indications of origin as a tool of product differentiation: The case of coffee. J. Int. Food Agribusiness Marketing 22 (3-4), 277–298. doi: 10.1080/08974431003641612
Tilocca B., Cao A., Migheli Q. (2020). Scent of a killer: microbial volatilome and its role in the biological control of plant pathogens. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00041
Torres N., Hilbert G., Antolín M. C., Goicoechea N. (2019). Aminoacids and flavonoids profiling in tempranillo berries can be modulated by the arbuscular mycorrhizal fungi. Plants 8 (10), 400. doi: 10.3390/plants8100400
Torri L., Migliorini P., Masoero G. (2013). Sensory test vs. electronic nose and/or image analysis of whole bread produced with old and modern wheat varieties adjuvanted by means of the mycorrhizal factor. Food Res. Int. 54 (2), 1400–1408. doi: 10.1016/j.foodres.2013.09.045
Traore T. M., Wilson N. L., Fields D. (2018). What explains specialty coffee quality scores and prices: a case study from the cup of excellence program. J. Agric. Appl. Economics 50 (3), 349–368. doi: 10.1017/aae.2018.5
Tsai A. Y.-L., McGee R., Dean G. H., Haughn G. W., Sawa S. (2021). Seed mucilage: biological functions and potential applications in biotechnology. Plant Cell Physiol. 62 (12), 1847–1857. doi: 10.1093/pcp/pcab099
Ushio M., Yamasaki E., Takasu H., Nagano A. J., Fujinaga S., Honjo M. N., et al. (2015). Microbial communities on flower surfaces act as signatures of pollinator visitation. Sci. Rep. 5 (1), 8695. doi: 10.1038/srep08695
Vacheron J., Desbrosses G., Bouffaud M.-L., Touraine B., Moënne-Loccoz Y., Muller D., et al. (2013). Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 4 (356). doi: 10.3389/fpls.2013.00356
Van Der Heijden M. G. A., Bardgett R. D., Van Straalen N. M. (2008). The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11 (3), 296–310. doi: 10.1111/j.1461-0248.2007.01139.x
Van Leeuwen C. (2010). “Terroir: the effect of the physical environment on vine growth, grape ripening and wine sensory attributes,” in Managing wine quality (Cambridge, UK: Woodhead Publishing Limited), 273–315.
Vannette R. L. (2020). The floral microbiome: plant, pollinator, and microbial perspectives. Annu. Rev. Ecology Evolution Systematics 51, 363–386. doi: 10.1146/annurev-ecolsys-011720-013401
Vannette R. L., Fukami T. (2018). Contrasting effects of yeasts and bacteria on floral nectar traits. Ann. Bot. 121 (7), 1343–1349. doi: 10.1093/aob/mcy032
Vannier N., Agler M., Hacquard S. (2019). Microbiota-mediated disease resistance in plants. PloS Pathog. 15 (6), e1007740–e1007740. doi: 10.1371/journal.ppat.1007740
Vaughan M. J., Mitchell T., Gardener B. B. M. (2015). What's inside that seed we brew? A new approach to mining the coffee microbiome. Appl. Environ. Microbiol. 81 (19), 6518–6527. doi: 10.1128/AEM.01933-15
Venkateswarlu B., Visperas R. M. (1987). Source-sink relationships in crop plants. IRRI Research Paper Series 125, 1–20.
Verginer M., Leitner E., Berg G. (2010a). Production of volatile metabolites by grape-associated microorganisms. J. Agric. Food Chem. 58 (14), 8344–8350. doi: 10.1021/jf100393w
Verginer M., Siegmund B., Cardinale M., Müller H., Choi Y., Míguez C. B., et al. (2010b). Monitoring the plant epiphyte Methylobacterium extorquens DSM 21961 by real-time PCR and its influence on the strawberry flavor. FEMS Microbiol. Ecol. 74 (1), 136–145. doi: 10.1111/j.1574-6941.2010.00942.x
Vorholt J. A. (2012). Microbial life in the phyllosphere. Nat. Rev. Microbiol. 10 (12), 828–840. doi: 10.1038/nrmicro2910
Voyron S., Ercole E., Ghignone S., Perotto S., Girlanda M. (2017). Fine-scale spatial distribution of orchid mycorrhizal fungi in the soil of host-rich grasslands. New Phytol. 213 (3), 1428–1439. doi: 10.1111/nph.14286
Wardle D. A., Bardgett R. D., Klironomos J. N., Setälä H., van der Putten W. H., Wall D. H. (2004). Ecological linkages between aboveground and belowground biota. Science 304 (5677), 1629–1633. doi: 10.1126/science.1094875
Wassermann B., Müller H., Berg G. (2019). An apple a day: which bacteria do we eat with organic and conventional apples? Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01629
Wei R., Ding Y., Chen N., Wang L., Gao F., Zhang L., et al. (2022b). Diversity and dynamics of microbial communities during spontaneous fermentation of Cabernet Sauvignon (Vitis vinifera L.) from different regions of China and their relationship with the volatile components in the wine. Food Res. Int. 156, 111372. doi: 10.1016/j.foodres.2022.111372
Wei G., Li M., Zhang G., Chen Z., Wei F., Jiao S., et al. (2020). Rhizosphere microbiome is associated with yield and medical value of the perennial panax notoginseng. Research Square, PPR188287. doi: 10.21203/rs.3.rs-41495/v1
Wei N., Whyle R. L., Ashman T.-L., Jamieson M. A. (2022a). Genotypic variation in floral volatiles influences floral microbiome more strongly than interactions with herbivores and mycorrhizae in strawberries. Horticulture Res. 9, uhab005. doi: 10.1093/hr/uhab005
White R. E. (2020). The value of soil knowledge in understanding wine terroir. Front. Environ. Sci. 8 (12). doi: 10.3389/fenvs.2020.00012
White J. F., Kingsley K. L., Verma S. K., Kowalski K. P. (2018). Rhizophagy cycle: an oxidative process in plants for nutrient extraction from symbiotic microbes. Microorganisms 6 (3), 95. doi: 10.3390/microorganisms6030095
White J. F., Kingsley K. L., Zhang Q., Verma R., Obi N., Dvinskikh S., et al. (2019). Endophytic microbes and their potential applications in crop management. Pest Manage. Sci. 75 (10), 2558–2565. doi: 10.1002/ps.5527
White J. F. Jr., Torres M. S. (2010). Is plant endophyte-mediated defensive mutualism the result of oxidative stress protection? Physiologia Plantarum 138 (4), 440–446. doi: 10.1111/j.1399-3054.2009.01332.x
White J. F. Jr., Torres M. S., Somu M. P., Johnson H., Irizarry I., Chen Q., et al. (2014). Hydrogen peroxide staining to visualize intracellular bacterial infections of seedling root cells. Microscopy Res. Technique 77 (8), 566–573. doi: 10.1002/jemt.22375
Wiens F., Zitzmann A., Lachance M.-A., Yegles M., Pragst F., Wurst F. M., et al. (2008). Chronic intake of fermented floral nectar by wild treeshrews. Proc. Natl. Acad. Sci. 105 (30), 10426–10431. doi: 10.1073/pnas.0801628105
Wilson J. E. (2001). Geology and wine 4. The origin and odyssey of terroir. Geosci. Canada 28 (3), 139–141.
Wilson B. R., Conley J. F., Harris T. M., Lafone F. (2012). New terrains of taste: Spatial analysis of price premiums for single origin coffees in Central America. Appl. Geogr. 35 (1), 499–507. doi: 10.1016/j.apgeog.2012.10.004
Wolfgang A., Tack A. J., Berg G., Abdelfattah A. (2023). Reciprocal influence of soil, phyllosphere and aphid microbiomes. Research Square. doi: 10.21203/rs.3.rs-2651152/v1
Xia C., Li N., Zhang Y., Li C., Zhang X., Nan Z. (2018). Role of Epichloë endophytes in defense responses of cool-season grasses to pathogens: A review. Plant Dis. 102 (11), 2061–2073. doi: 10.1094/PDIS-05-18-0762-FE
Yadav R., Ror P., Rathore P., Ramakrishna W. (2020). Bacteria from native soil in combination with arbuscular mycorrhizal fungi augment wheat yield and biofortification. Plant Physiol. Biochem. 150, 222–233. doi: 10.1016/j.plaphy.2020.02.039
Yale R. L., Sapp M., Sinclair C. J., Moir J. W. B. (2017). Microbial changes linked to the accelerated degradation of the herbicide atrazine in a range of temperate soils. Environ. Sci. pollut. Res. 24 (8), 7359–7374. doi: 10.1007/s11356-017-8377-y
Yang D., Lin X., Wei Y., Li Z., Zhang H., Liang T., et al. (2023). Can endophytic microbial compositions in cane roots be shaped by different propagation methods. PloS One 18 (8), e0290167. doi: 10.1371/journal.pone.0290167
Yang M.-Z., Ma M.-D., Yuan M.-Q., Huang Z.-Y., Yang W.-X., Zhang H.-B., et al. (2016). Fungal endophytes as a metabolic fine-tuning regulator for wine grape. PloS One 11 (9), e0163186. doi: 10.1371/journal.pone.0163186
Yasin M., El-Mehdawi A. F., Anwar A., Pilon-Smits E. A., Faisal M. (2015). Microbial-enhanced selenium and iron biofortification of wheat (Triticum aestivum L.)-applications in phytoremediation and biofortification. Int. J. phytoremediation 17 (4), 341–347. doi: 10.1080/15226514.2014.922920
Yu K., Pieterse C. M. J., Bakker P. A. H. M., Berendsen R. L. (2019). Beneficial microbes going underground of root immunity. Plant Cell Environ. 42 (10), 2860–2870. doi: 10.1111/pce.13632
Zabaloy M. C., Allegrini M., Hernandez Guijarro K., Behrends Kraemer F., Morrás H., Erijman L. (2022). Microbiomes and glyphosate biodegradation in edaphic and aquatic environments: recent issues and trends. World J. Microbiol. Biotechnol. 38 (6), 98. doi: 10.1007/s11274-022-03281-w
Zarraonaindia I., Owens S. M., Weisenhorn P., West K., Hampton-Marcell J., Lax S., et al. (2015). The soil microbiome influences grapevine-associated microbiota. MBio 6 (2), e02527-14. doi: 10.1128/mBio.02527-14
Zer H., Mizrahi H., Malchenko N., Avin-Wittenberg T., Klipcan L., Ostersetzer-Biran O. (2020). The phytotoxicity of meta-tyrosine is associated with altered phenylalanine metabolism and misincorporation of this non-proteinogenic phe-analog to the plant's proteome. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00140
Zhang H.-Z., Liu D.-H., Zhang D.-K., Wang Y.-H., Li G., Yan G.-L., et al. (2016). Quality Assessment of Panax notoginseng from Different Regions through the Analysis of Marker Chemicals, Biological Potency and Ecological Factors. PloS One 11 (10), e0164384–e0164384. doi: 10.1371/journal.pone.0164384
Zhang Q., Ye Y., Qu Q., Yu Y., Jin M., Lu T., et al. (2021). Enantioselective metabolomic modulations in Arabidopsis thaliana leaf induced by the herbicide dichlorprop. Sci. Total Environ. 797, 149015. doi: 10.1016/j.scitotenv.2021.149015
Zheng L. P., Li X. P., Zhou L. L., Wang J. W. (2021). Endophytes in Artemisia annua L.: new potential regulators for plant growth and artemisinin biosynthesis. Plant Growth Regul. 95 (3), 293–313. doi: 10.1007/s10725-021-00751-3
Zhou J., Cavagnaro T. R., De Bei R., Nelson T. M., Stephen J. R., Metcalfe A., et al. (2021). Wine terroir and the soil bacteria: an amplicon sequencing–based assessment of the barossa valley and its sub-regions. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.597944
Zhou C., Cheng H., Wu Y., Zhang J., Li D., Pan C. (2022). Bensulfuron-methyl, terbutylazine, and 2,4-D butylate disturb plant growth and resistance by deteriorating rhizosphere environment and plant secondary metabolism in wheat seedlings. J. Agric. Food Chem. 70 (40), 12796–12806. doi: 10.1021/acs.jafc.2c03126
Keywords: plant microbiome, plant metabolome, terroir, microbial terroir, fermentation, rhizosphere, endophytes, carposphere
Citation: Johnston-Monje D, Vergara LI, Lopez-Mejia J and White JF (2023) Plant microbiomes as contributors to agricultural terroir. Front. Agron. 5:1216520. doi: 10.3389/fagro.2023.1216520
Received: 04 May 2023; Accepted: 03 October 2023;
Published: 19 October 2023.
Edited by:
Davey Jones, Bangor University, United KingdomReviewed by:
Sophie Trouvelot, Université de Bourgogne, FranceCopyright © 2023 Johnston-Monje, Vergara, Lopez-Mejia and White. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David Johnston-Monje, ZGFtb2pvbW9AZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.