
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Agron., 03 October 2022
Sec. Disease Management
Volume 4 - 2022 | https://doi.org/10.3389/fagro.2022.984360
This article is part of the Research TopicNew Approaches in the Detection of Plant PathogensView all 4 articles
 Fabiana Stolowicz1
Fabiana Stolowicz1 Luciana Larocca1
Luciana Larocca1 Santiago Werbajh1
Santiago Werbajh1 Yanil Parma1
Yanil Parma1 Carolina Carrillo1
Carolina Carrillo1 Lorena Ogas1
Lorena Ogas1 Juan Pedro Agostini2
Juan Pedro Agostini2 Jonathan Redes2
Jonathan Redes2 Bjorn Welin3
Bjorn Welin3 Atilio Castagnaro4
Atilio Castagnaro4 Adrian Vojnov1*
Adrian Vojnov1*Huanglongbing (HLB) is one of the most devastating diseases in citrus worldwide. The Gram-negative bacterial plant pathogen “Candidatus Liberibacter spp.” is phloem-limited and vectored by citrus psyllids. The species “Candidatus Liberibacter asiaticus” (C.Las) has been detected in Argentina, and its vector has been found in at least nine provinces. Early detection of C.Las is critical for a successful management of HLB disease. Currently, HLB molecular diagnosis is carried out by PCR, nested PCR, real-time PCR, or another combination of these techniques, which require purification of genomic DNA, sophisticated equipment, and highly trained personnel. We have developed a prototype of a sensitive colorimetric kit to detect C.Las based on the specific DNA isothermal amplification of this microorganism. The reaction buffer contains hydroxynaphthol blue (HNB), an indicator dye that turns from violet to blue/light blue when the DNA amplification reaction is positive. Similar sensitivity to visualize a positive reaction was observed between HNB loop-mediated isothermal amplification and agarose gel electrophoresis analysis. The detection of C.Las-infected plants was up to 8 ng of total infected plant genomic DNA, similar to quantitative PCR. A blind validation test of the prototype kit was performed with purified DNA extracted from healthy or C.Las-infected midrib plants. Our kit showed 100% concordance with the results of a gold-standard quantitative PCR technique applied by the Laboratorio de Biología Molecular de EEA Montecarlo. The analysis of samples, without DNA purification to detect C.Las, showed a similar sensitivity to the analysis of the same samples in which C.Las DNA was previously purified.
Huanglongbing (HLB), shown as citrus greening, is the most serious disease of citrus worldwide. Symptoms of HLB can be seen in both leaves (yellow shoots, blotchy mottle leaf and cork) and fruits (smaller, lighter, highly acidic lopsided fruit with green color remaining on the stylar end and aborted seeds) (Etxeberria et al., 2009; Bové, 2014). Once the plant becomes with HLB, there are no curative procedures, and after a few years, the tree dies. Although there are three fastidious α-proteobacteria species of “Candidatus Liberibacter” associated with HLB, namely “Candidatus Liberibacter asiaticus” (C.Las), ”Candidatus Liberibacter americanus”, and ”Candidatus Liberibacter africanus” (Gottwald, 2010), C.Las is the most widespread and economically important.
C.Las is transmitted by infected propagating material (buds or infected plant parts), and in America and Asia, its transmission by the sap-sucking psyllid Diaphorina citri has also been detected (Grafton-Cardwell et al., 2013). This psyllid spreads rapidly in residential and commercial plantings through natural pathways; however, its spread also occurs because of commercial transport of infected plant material (Manjunath et al., 2008; Grafton-Cardwell et al., 2013).
The disease, originally from China, spread from the Asian continent to the rest of the world, showing an exponential advance in the American continent in the last 10 years. In particular, in Argentina, as soon as HLB was detected, the Servicio Nacional de Sanidad y Calidad Agroalimentaria (SENASA) developed and implemented the National HLB Prevention Program to prevent entry and spread within the country. This program focuses on performing permanent monitoring, diagnosis in both plants and psyllids, and eradication of the infected plants. Currently, the major initial detection procedure for C.Las is visual inspection based on disease symptoms in trees. Samples that are suspected to be positive are sent to diagnostic laboratories for secondary analysis. Several methodologies have been developed to detect C.Las in these samples, including serological assays, electronic microscopy, biological assays, DNA probes, PCR, and real-time PCR (Garnier et al., 1987; Kim and Wang, 2009; Gottwald, 2010). Those methods have the drawback of being time-consuming and requiring complex facilities. A successful management of HLB requires an early detection, and the detection of infected insects is critical in preventing the spread of the disease. The most common molecular detection methods in use are conventional PCR with 16S rDNA-based primers as a target of the three Candidatus Liberibacter spp. (Jagoueix et al., 1996; Teixeira et al., 2008; Nageswara-Rao et al., 2013); real-time PCR targeting the 16S rDNA region (Bové, 2006; Li et al., 2006; Pietersen et al., 2010) as well as multiple genetic loci such as hyvI (LasAI) and hyvII (LasAII) in C.Las prophage genes (Zhou et al., 2011; Morgan et al., 2012). A combination of real-time PCR and nested PCR in a single tube to detect the elongation factor Ts gene (Lin et al., 2010), recombinase polymerase amplification (RPA) (Ghosh et al., 2018; Qian et al., 2018), and loop-mediated isothermal amplification (LAMP). LAMP provides an amplification of target DNA under isothermal temperatures (60–65°C), using Bst polymerase with strand displacement activity and a set of four or six primers (Notomi et al., 2000; Nagamine et al., 2002). It has been an advantage of being highly specific, rapid, efficient, and labor-saving (Notomi et al., 2000; Wong et al., 2018). In contrast to both conventional PCR and real-time PCR, a precision thermocycler is not needed because LAMP is performed at a single temperature. LAMP was developed for the detection of C.Las (Okuda et al., 2005; Rigano et al., 2014; Ghosh et al., 2016; Choi et al., 2018), Candidatus Liberibacter solanacearum (Ravindran et al., 2012), Erwinia amylovora (Temple and Johnson, 2011), Tylenchulus semipenetrans (Song et al., 2017), and wheat streak mosaic virus (Lee et al., 2015).
In this study, we report the development of a new prototype colorimetric LAMP kit using hydroxynaphthol blue (HNB). Our results demonstrated that this kit is a reliable, rapid, and cost-effective method for the detection of C.Las in citrus.
Midribs of HLB-infected and healthy leaves of citrus lemon were provided by the Estación Experimental Agropecuaria–INTA Montecarlo, Misiones, Argentina. Midribs were separated from leaf samples and stored submerged in 75% ethanol for −20°C until DNA extraction. Total genomic DNA of the midrib leaves was extracted using a PuriPrep-V Kit protocol (Inbio Highway, Argentina), and the concentration of plant genomic purified DNA was measured with a NanoDrop One Microvolume UV-Vis Spectrophotometer Thermo Fisher Scientific. Alternatively, a simple preparation from 30 mg of midribs was performed using 180 µl NaOH 0.5 N solution, heated for 10 min at 100°C, and neutralized with 120 µl HCL 0.5 N.
The localization of target sequences and the LAMP primer design targeted a hypothetical protein sequence on the gene CLIBASIA_05175, highly specific for C.Las, as reported previously (Rigano et al., 2014). The LAMP assay was performed using a dry thermal block with a 0.5-ml PCR tube holder, incubated at 64°C for 60 min. The final LAMP conditions used were as follows: 40 pmol each of primers Las-FIP and Las-BIP, 5 pmol each of outer primers Las-F3 and Las-B3, 20 pmol each of loop primers Las-LF and Las-LB, 8 units of Bst DNA polymerase large fragment (New England Biolabs), 120 µM HNB (Sigma-Aldrich), 6 mM MgSO4, 1 mM of deoxynucleoside triphosphate (dNTP) mix, 20 mM Tris–HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 0.1% Tween 20, and 1 M betaine, in a final volume of 40 μl including the template.
To compare HNB-LAMP with quantitative PCR (qPCR), 10 µl of 80, 8, 0.8, 0.08, and 0.008 ng of purified genomic DNA of C.Las-infected plants from the same preparation (Inbio Highway, Argentina) was used for both techniques. We used 4 µl of crude plant extracts (without purification) as a template only for direct HNB-LAMP reaction.
HNB colorimetric reaction: Because Mg2+ ion concentration decreases as the LAMP reaction progresses, the LAMP reaction can be quantified by measuring the Mg2+ ion concentration in the reaction solution. Initially, the solution color is violet. A color change is not expected to be observed in negative controls (when DNA is not used or the genomic DNA of a healthy plant is used as a template). When the genomic DNA of C.Las-infected plants or similar is used as the molecular template, amplification happens, and the solution color changes from violet to blue/light blue color (positive control).
To analyze if our HNB-LAMP reaction showed a typical ladder pattern for stem-loop structures, 4 µl of the amplification product was subjected to electrophoresis at 50 V for 50 min on a 1.5% p/v agarose gel, with TAE 1× (Tris-acetate with EDTA) and SYBR Safe 1× (Invitrogen). We then compared the colorimetric results with the expected banding pattern for this reaction.
The qPCR, including primer sequence and program, was carried out following the previously published conditions (Li et al., 2006). Two primers and one TaqMan probe were empirically designed based on the 16S rDNA sequences of C.Las (GenBank Accession No. L22532). The reverse primer 16SrDNA-r 5′CTTGATGGCAACTAGAGGCA-3′, the forward primer 16SrDNA-f 5′-CTTTTCGGAGACCTTTACACA-3′, and the HLB probe 16SrDNA hlb-p 5′-TGCGCTCGTTGCGGGACTTA-3′, labeled at 5´-terminal nucleotide with 6-carboxy-fluorescein (FAM) reporter dye and at 3´-terminal nucleotide with Black Hole Quencher-1 (BHQ-1), are specific for C.Las TaqMan assays.
The qPCR amplifications were performed using a Step One™ real-time PCR system (Applied Biosystems) in a 25-µl final volume. The optimized concentrations were 250 nM each primer forward and reverse, 150 nm target probe, 6.0 mM MgCl2, 250 µM dNTP mix, 1× PCR buffer, and 1 unit PEGASUS Taq DNA polymerase (PB-L, Argentina). The standard amplification protocol was 95°C for 20 s and consisted of 50 cycles at 95°C for 15 s and 58°C for 40 s, followed by 58°C for 30-s reactions. The data were analyzed using the Step One™ software (Version 2.1).
In order to estimate the sensitivity of the HLB prototype kit assay, the genomic DNA of C.Las-infected plants (previously confirmed positive by qPCR at Estación Experimental Agropecuaria–INTA Montecarlo, Misiones, Argentina) was purified by a Puriprep V-Kit (Inbio Highway, Argentina) and serially diluted from 10−1 to 10−5 (80–0.008 ng, respectively). Ten microliters of each dilution, of the same preparation, was used as a template for HNB-LAMP and qPCR. All reactions were carried out in triplicate for HNB-LAMP and qPCR.
The limit of detection of C.Las by HNB-LAMP was defined positive when the color change from violet to blue/light blue occurred in 100% of the replicated samples of each dilution. To determine the limit of the detectable DNA of C.Las by qPCR, the Ct obtained in the different serial dilutions were analyzed. If the ΔCt between two consecutives dilutions were close to the theoretical difference of 3.3 cycles, they were within the dynamics range and considered detectable (Okuda et al., 2005; Garmendia and Vero, 2007; Cuevas-reyes et al., 2016).
To evaluate the performance of HNB-LAMP for HLB diagnosis, the prototype kit was used for in-field validation. Purified DNA samples (16 positive and 14 negative samples) taken from C.Las-infected and non-infected lemon leaves, which were previously analyzed by conventional qPCR, were used to determine the field sensitivity of HNB-LAMP. A contingency table was carried out to compare the results obtained with the HNB-LAMP with those obtained with the qPCR.
We optimized the reaction conditions for a colorimetric, rapid, and simple assay for C.Las detection. HNB, a colorimetric indicator for the titration of Ca2+ and Mg2+ (Goto et al., 2009), was used as an indicator for the LAMP reaction by monitoring the change in the Mg2+ ion concentration. Magnesium ions react with pyrophosphate derived from dNTPs during LAMP reaction. The resulting depletion of magnesium ions in the HNB–Mg2+ interaction induced a color change from violet to blue/light blue, indicating a positive reaction. The prototype kit contains the primers for the LAMP and the dye, both dry in the tubes, and the mixture of reagents and Bst enzyme necessary for the reaction. Figure 1 shows the performance of the colorimetric reaction. The reaction tubes with the samples, a negative control (−) and DNA from infected leaves (+, positive control) before incubation, showed a violet coloration (Figure 1A). After 60 min of incubation, we observed that only the color solution from the positive control, containing DNA from infected leaves (+), changed to light blue color (Figure 1B). These tubes were analyzed by agarose gel electrophoresis, as detailed in the Materials and methods section, showing a clear amplification (ladder-like pattern) only in positive control after incubation (+) whereas no amplification was observed in the negative control (−) (Figure 1C). These results indicated that the visible readout of our prototype kit reaction is consistent to electrophoresis results (Figure 1).
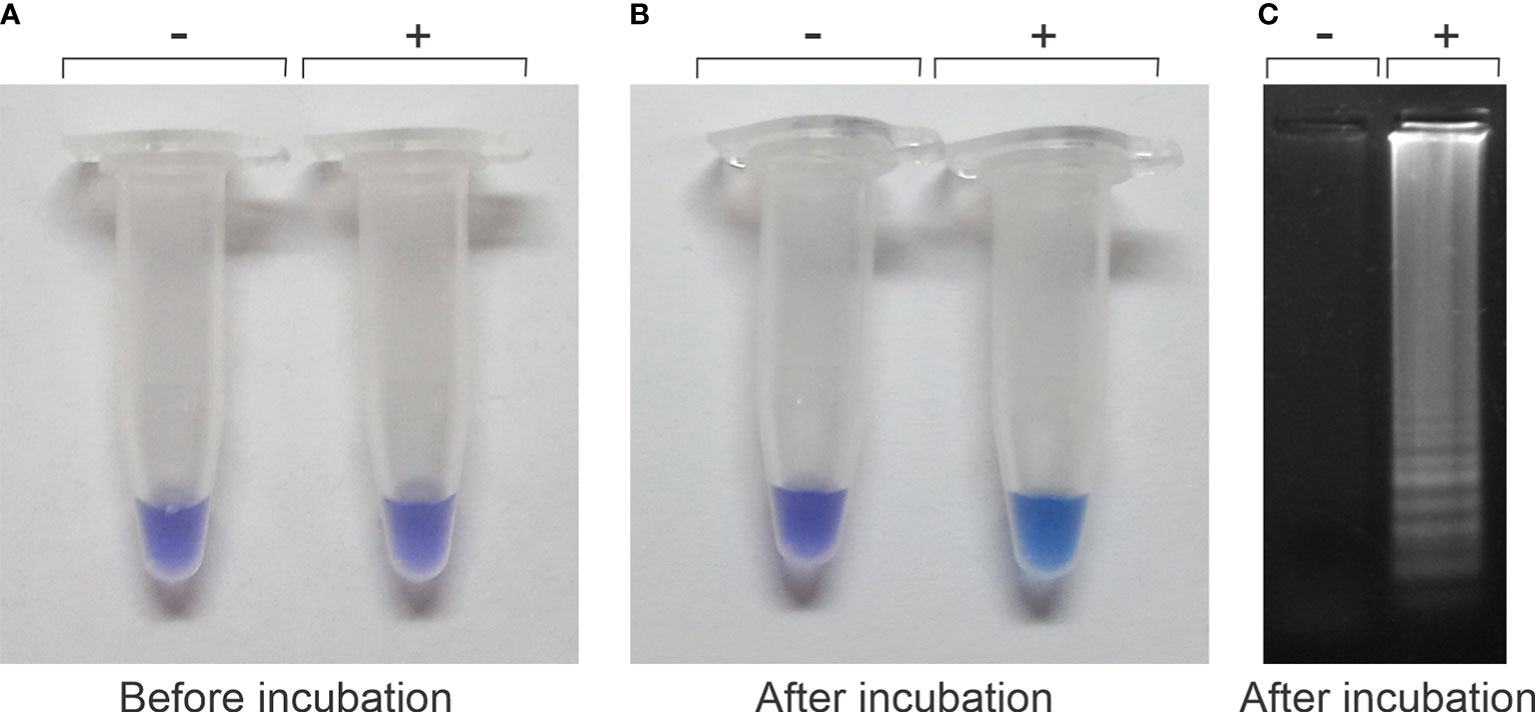
Figure 1 Visualization of color changes from violet to blue/light blue before or after the loop-mediated isothermal amplification (LAMP) reaction. (A) The reaction tubes, a negative and a positive control, before incubation are both violet, indistinguishable from each other. (B) They differ after the incubation period where the positive control (+) changes to light blue, whereas the negative control remains violet (−). (C) Visualization of the LAMP-banding pattern of DNA by agarose gel electrophoresis of the positive reaction observed in panel (B).
Comparative detection sensitivity studies between qPCR and LAMP assay were performed by using purified DNA from C.Las-infected lemon leaves (previously confirmed positive by qPCR at Estación Experimental Agropecuaria–INTA Montecarlo, Misiones, Argentina). The purified DNA was serially diluted up to 10−5 and then used as a template to evaluate the detection limit through both methods. Figure 2 shows the results of the HNB-LAMP assay sensitivity and the confirmation by agarose gel electrophoresis, in parallel with the qPCR, in order to compare each other.
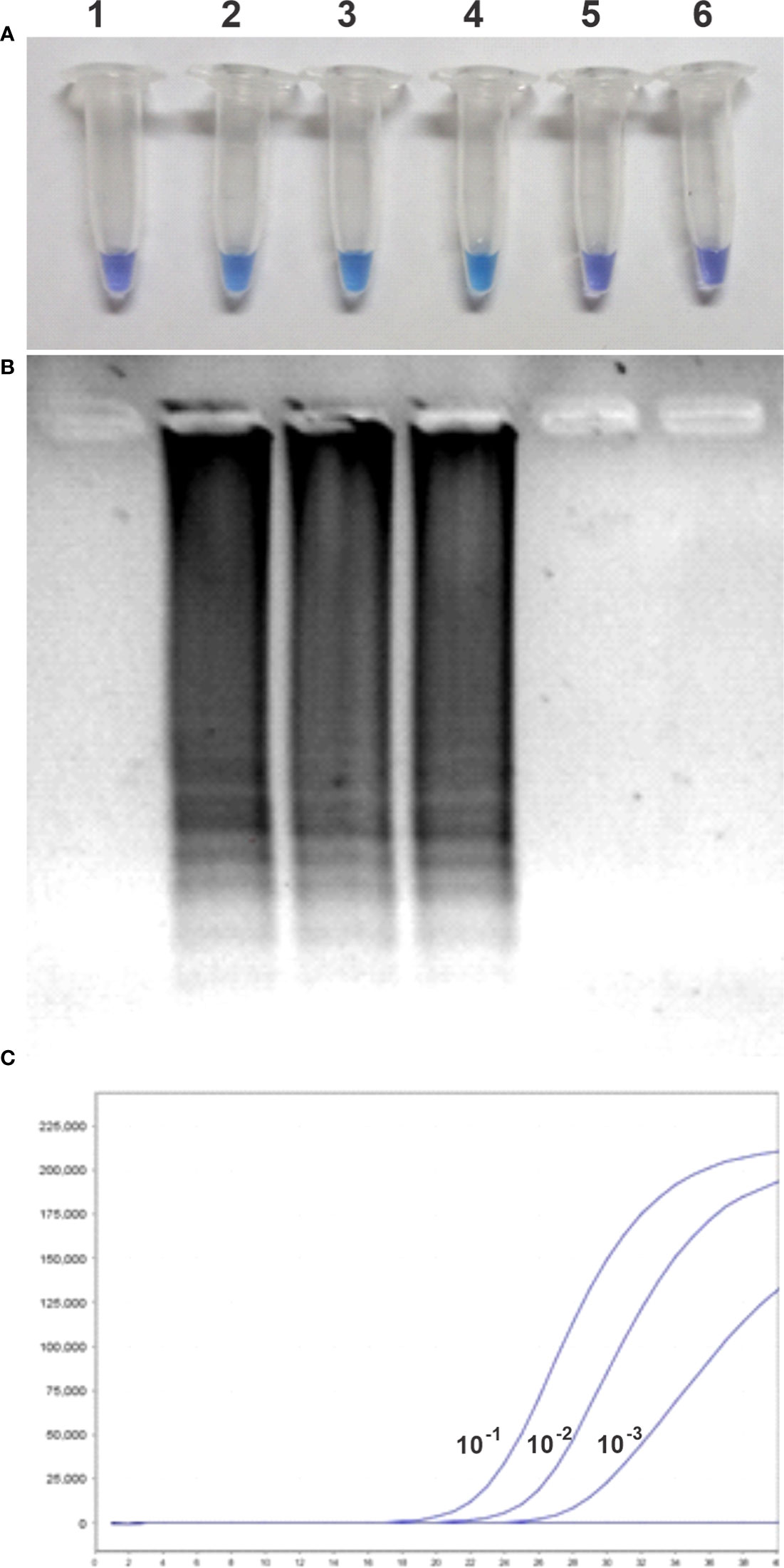
Figure 2 Comparative sensitivities between hydroxynaphthol blue (HNB)–loop-mediated isothermal amplification (LAMP) and qPCR for detecting HLB. (A) Visual examination of LAMP results. (B) Electrophoresis analysis of LAMP products on 1.5% (p/v) agarose gel. (C) Sensitivity of real-time PCR for detecting HLB. Lanes 1–6, negative control and serially diluted genomic DNA (10−1, 10−2, 10−3, 10−4 and 10−5).
The change of solution color from violet to light blue of 100% of the reaction tubes of HNB-LAMP, corresponding to a positive amplification reaction, was observed up to 10−3 dilution of the purified DNA sample (8 ng of total genomics DNA of C.Las-infected plants). The same samples, analyzed by qPCR, showed Cts of 22.91, 26.9, 29.65, 36.75, and 50.00, corresponding to the 10−1, 10−2, 10−3, 10−4 and 10−5 sample dilutions, respectively. Under optimal conditions, the Ct should increase in around 3.3 cycles for every log decrease in the number of DNA molecules. The results showed that the DNA template dilutions of ≥10−4 were out of dynamic range and, therefore, out of technique sensibility. Taken together, these results demonstrate that HNB-LAMP was the same as the sensitivity of qPCR (Figure 2C).
To evaluate the performance of HNB-LAMP for HLB diagnosis, the prototype kit was used for in-field validation. Purified DNA samples (16 positive and 14 negative samples) taken from C.Las-infected and non-infected lemon leaves, which were previously analyzed by conventional qPCR, were used to determine the field sensitivity of HNB-LAMP. The contingency table of the results shows that all the positive and negative results obtained by HNB-LAMP matched with the results obtained by qPCR (Table 1). In other words, no false-negative or false-positive results were raised. The results obtained showed an in-field specificity and sensitivity of 100% (Table 1), indicating that HNB-LAMP assay has a good agreement with the conventional qPCR technique.

Table 1 Sensitivity and specificity of loop-mediated isothermal amplification (LAMP) methods in comparison with qPCR.
We choose 10 midribs of C.Las-infected and healthy leaves of citrus lemon samples (five negatives and five positives) to be analyzed by a direct assay without a DNA purification. Plant extracts were used as a molecular template in an HNB-LAMP analysis (see Materials and methods section for more details). The results shown in Figure 3 are consistent with those previously observed with the same samples with purified DNA (Table 1) and demonstrate that a simpler test, without DNA purification, is feasible to achieve amplification in the samples from C.Las-infected lemon leaves.
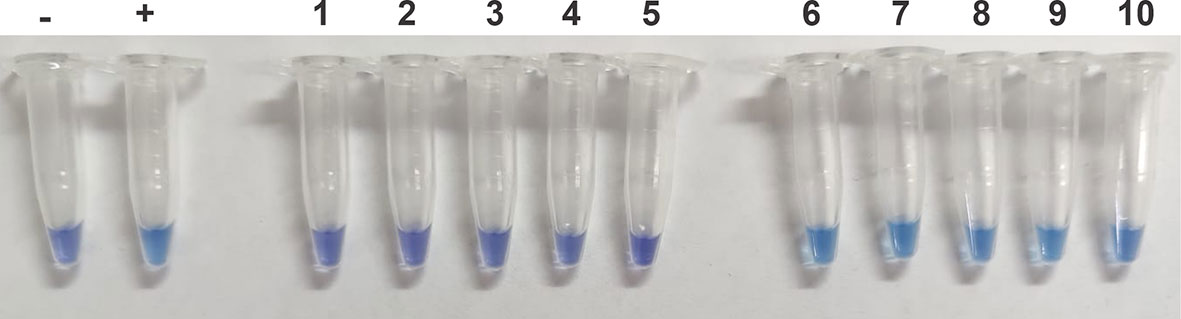
Figure 3 Performance of hydroxynaphthol blue (HNB)–loop-mediated isothermal amplification (LAMP) to detect C.Las in samples without purified DNA. The samples were previously treated with NaOH solutions, heated for 10 min at 100°C, and neutralized with HCl solution. The extract was used as a template for the LAMP reaction. Negative control (−), positive control (+), 1–5 samples from uninfected plants, and 5–10 samples from plants infected with C.Las.
HLB is the most destructive disease of citrus worldwide and, so far, has no cure. It is caused by an intracellular pathogen “Candidatus Liberibacter spp.” whose detection is done by two common methods: visual inspection of symptoms and PCR tests; both methods are accurate but expensive and time-consuming.
With the overall goal of preventing and/or minimizing the spread of HLB, we previously reported a sensitive HLB-LAMP detection method targeting CLIBASIA_05175 (GenBank: ACT57606.1) and using a lateral flow rod by Rigano et al. (2014). In the present study, we developed a new detection kit whose results can be observed without opening the reaction tube cap after incubation. Considering that HNB does not inhibit the amplification reaction, its addition to the detection kit prior to the pathogen detection reaction, the colorimetric change can be observed without opening the reaction tube. This results in a rapid and accurate diagnostic colorimetric kit, which showed superior results to those obtained using the other LAMP kits because it allows for reduced risks of contamination.
Because C.Las cannot be cultured, purified and quantified C.Las genomic DNA was not available for use as a positive control. Therefore, to compare the detection sensitivity between qPCR and LAMP, we used DNA extracted from lemon leaves that were confirmed previously as C.Las-infected by qPCR. The results shown in Figure 2 indicate that the new HNB-LAMP method has similar sensitivity to qPCR when used at 64°C reaction temperature. Finally, to simplify the methods described here, we developed a sample treatment protocol for use in C.Las detection without DNA purification. This new version allows a simpler and more economical detection of the causal agent of HLB disease.
The data presented in the study are deposited in the Figshare repository, accessible at DOI: 10.6084/m9.figshare.21197812.
FS contributed in all the designed experiment work and contributed in performing all of them. LL, SW, CC, and LO contributed in the experimental qPCR experiment. YP contributed in the design of the sample treatment of the new version test avoiding the ADN purification. JA and JR contributed in the samples that were checked, previously, in their lab. BW and AC contributed in coordinating the study and the writing of the manuscript. AV participated in the design of the experiment, participated in the analysis and interpretation of the data, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was carried out with the support of Consejo Nacional de Investigaciones Científicas y Técnicas through a Proyecto Unidades Ejecutoras–2018 ICT Milstein.
FS, LL, CC, BW, AC, and AV are members of CONICET. LO is supported by a ANPCyT fellowship. We thank Dr. Isabel Bianco for the critical reading of the manuscript, Dr. Valeria Conforte for qPCR results analysis, and Dr. Gustavo Leiros for the design of the figures.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Bové J. M. (2006). JOURNAL of PLANT NUTRITION. J. Plant Pathol. 88, 7–37. doi: 10.1080/01904167.2015.1112950
Bové J. M. (2014). Huanglongbing or yellow shoot, a disease of gondwanan origin: Will it destroy citrus worldwide? Phytoparasitica 42, 579–583. doi: 10.1007/s12600-014-0415-4
Choi C. W., Hyun J. W., Hwang R. Y., Powell C. A. (2018). Loop-mediated isothermal amplification assay for detection of candidatus liberibacter asiaticus, a causal agent of citrus huanglongbing. Plant Pathol. J. 34, 499–505. doi: 10.5423/PPJ.FT.10.2018.0212
Cuevas-reyes E., Carrillo-morales M., Treviño-quintanilla L. G. (2016). Oligonucleotides evaluation for measuring gene expression during tomato bacterial wilt. Rev. fitotec. mex [online]. 39, 141–150. doi: 10.35196/rfm.2016.2.141-150
Etxeberria E., Gonzalez P., Achor D., Albrigo G. (2009). Anatomical distribution of abnormally high levels of starch in HLB-affected Valencia orange trees. Physiol. Mol. Plant Pathol. 74, 76–83. doi: 10.1016/J.PMPP.2009.09.004
Garmendia, Vero (2007). Tiempo real 2 . extracción de ADN. Rev. del Lab. Tecnológico del Uruguay, 47–50. doi: 10.26461/06.10
Garnier M., Martin-Gros G., Bové J. M. (1987). Monoclonal antibodies against the bacterial-like organism associated with citrus greening disease. Ann. Inst. Pasteur. Microbiol. 138, 639–650. doi: 10.1016/0769-2609(87)90142-6
Ghosh D. K., Bhose S., Warghane A., Motghare M., Sharma A. K., Dhar A. K., et al. (2016). Loop-mediated isothermal amplification (LAMP) based method for rapid and sensitive detection of ‘Candidatus liberibacter asiaticus’ in citrus and the psyllid vector, diaphorina citri kuwayama. J. Plant Biochem. Biotechnol. 25, 219–223. doi: 10.1007/s13562-015-0332-8
Ghosh D. K., Kokane S. B., Kokane A. D., Warghane A. J., Motghare M. R., Bhose S., et al. (2018). Development of a recombinase polymerase based isothermal amplification combined with lateral flow assay (HLB-RPA-LFA) for rapid detection of “Candidatus liberibacter asiaticus”. PloS One 13:1–23. doi: 10.1371/journal.pone.0208530
Gottwald T. R. (2010). Current epidemiological understanding of citrus huanglongbing. Annu. Rev. Phytopathol. 48, 119–139. doi: 10.1146/ANNUREV-PHYTO-073009-114418
Grafton-Cardwell E. E., Stelinski L. L., Stansly P. A. (2013). Biology and management of Asian citrus psyllid, vector of the huanglongbing pathogens. Annu. Rev. Entomol. 58, 413–432. doi: 10.1146/ANNUREV-ENTO-120811-153542
Jagoueix S., Bové J. M., Garnier M. (1996). PCR detection of the two “Candidatus” liberobacter species associated with greening disease of citrus. Mol. Cell. Probes 10, 43–50. doi: 10.1006/MCPR.1996.0006
Kim J. S., Wang N. (2009). Characterization of copy numbers of 16S rDNA and 16S rRNA of candidatus liberibacter asiaticus and the implication in detection in planta using quantitative PCR. BMC Res. Notes 2:1–4. doi: 10.1186/1756-0500-2-37
Lee S., Kim J. H., Choi J. Y., Jang W. C. (2015). Loop-mediated isothermal amplification assay to rapidly detect wheat streak mosaic virus in quarantined plants. Plant Pathol. J. 31, 438–440. doi: 10.5423/PPJ.NT.06.2015.0110
Li W., Hartung J. S., Levy L. (2006). Quantitative real-time PCR for detection and identification of candidatus liberibacter species associated with citrus huanglongbing. J. Microbiol. Methods 66, 104–115. doi: 10.1016/J.MIMET.2005.10.018
Lin H., Chen C., Doddapaneni H., Duan Y., Civerolo E. L., Bai X., et al. (2010). A new diagnostic system for ultra-sensitive and specific detection and quantification of candidatus liberibacter asiaticus, the bacterium associated with citrus huanglongbing. J. Microbiol. Methods 81, 17–25. doi: 10.1016/J.MIMET.2010.01.014
Manjunath K. L., Halbert S. E., Ramadugu C., Webb S., Lee R. F. (2008). Detection of “Candidatus liberibacter asiaticus” in diaphorina citri and its importance in the management of citrus huanglongbing in Florida. Phytopathology 98, 387–396. doi: 10.1094/PHYTO-98-4-0387
Morgan J. K., Zhou L., Li W., Shatters R. G., Keremane M., Duan Y. P. (2012). Improved real-time PCR detection of “Candidatus liberibacter asiaticus” from citrus and psyllid hosts by targeting the intragenic tandem-repeats of its prophage genes. Mol. Cell. Probes 26, 90–98. doi: 10.1016/J.MCP.2011.12.001
Nagamine K., Hase T., Notomi T. (2002). Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 16, 223–229. doi: 10.1006/mcpr.2002.0415
Nageswara-Rao M., Irey M., Garnsey S. M., Gowda S. (2013). Candidate gene makers for candidatus liberibacter asiaticus for detecting citrus greening disease. J. Biosci. 38, 229–237. doi: 10.1007/s12038-013-9315-x
Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:e63 (I-VII). doi: 10.1093/NAR/28.12.E63
Okuda M., Matsumoto M., Tanaka Y., Subandiyah S., Iwanami T. (2005). Characterization of the tufB-secE-nusG-rplKAJL-rpoB gene cluster of the citrus greening organism and detection by loop-mediated isothermal amplification. Plant Dis. 89, 705–711. doi: 10.1094/PD-89-0705
Pietersen G., Arrebola E., Breytenbach J. H. J., Korsten L., Le Roux H. F., La Grange H., et al. (2010). A survey for “Candidatus liberibacter“ species in south Africa confirms the presence of only “Ca. l. africanus” in commercial citrus. Plant Dis. 94, 244–249. doi: 10.1094/PDIS-94-2-0244
Qian W., Lu Y., Meng Y., Ye Z., Wang L., Wang R., et al. (2018). Field detection of citrus huanglongbing associated with ’ candidatus liberibacter asiaticus’ by recombinese polymerase amplification within 15 min. J. Agric. Food Chem. 66, 5473–5480. doi: 10.1021/acs.jafc.8b01015
Ravindran A., Levy J., Pierson E., Gross D. C. (2012). Development of a loop-mediated isothermal amplification procedure as a sensitive and rapid method for detection of “candidatus liberibacter solanacearum” in potatoes and psyllids. Phytopathology 102, 899–907. doi: 10.1094/PHYTO-03-12-0055-R
Rigano L. A., Malamud F., Orce I. G., Filippone M. P., Marano M. R., Do Amaral A. M., et al. (2014). Rapid and sensitive detection of candidatus liberibacter asiaticus by loop mediated isothermal amplification combined with a lateral flow dipstick. BMC Microbiol. 14:1–9. doi: 10.1186/1471-2180-14-86
Song Z. Q., Cheng J. E., Cheng F. X., Zhang D. Y., Liu Y. (2017). Development and evaluation of loop-mediated isothermal amplification assay for rapid detection of tylenchulus semipenetrans using DNA extracted from soil. Plant Pathol. J. 33, 184–192. doi: 10.5423/PPJ.OA.10.2016.0224
Teixeira D. C., Saillard C., Couture C., Martins E. C., Wulff N. A., Eveillard-Jagoueix S., et al. (2008). Distribution and quantification of candidatus liberibacter americanus, agent of huanglongbing disease of citrus in são paulo state, brasil, in leaves of an affected sweet orange tree as determined by PCR. Mol. Cell. Probes 22, 139–150. doi: 10.1016/J.MCP.2007.12.006
Temple T. N., Johnson K. B. (2011). Evaluation of loop-mediated isothermal amplification for rapid detection of erwinia amylovora on pear and apple fruit flowers. Plant Dis. 95, 423–430. doi: 10.1094/PDIS-09-10-0636
Wong Y. P., Othman S., Lau Y. L., Radu S., Chee H. Y. (2018). Loop-mediated isothermal amplification (LAMP): A versatile technique for detection of micro-organisms. J. Appl. Microbiol. 124, 626–643. doi: 10.1111/jam.13647
Zhou L., Powell C. A., Hoffman M. T., Li W., Fan G., Liu B., et al. (2011). Diversity and plasticity of the intracellular plant pathogen and insect symbiont “Candidatus liberibacter asiaticus“ as revealed by hypervariable prophage genes with intragenic tandem repeats. Appl. Environ. Microbiol. 77, 6663–6673. doi: 10.1128/AEM.05111-11
Keywords: Huanglongbing, citrus, diagnosis, LAMP, Hidroxynaphtol blue, Candidatus liberibacter asiaticus
Citation: Stolowicz F, Larocca L, Werbajh S, Parma Y, Carrillo C, Ogas L, Agostini JP, Redes J, Welin B, Castagnaro A and Vojnov A (2022) A colorimetric, sensitive, rapid, and simple diagnostic kit for the HLB putative causal agent detection. Front. Agron. 4:984360. doi: 10.3389/fagro.2022.984360
Received: 01 July 2022; Accepted: 02 September 2022;
Published: 03 October 2022.
Edited by:
Sara Franco Ortega, University of York, United KingdomReviewed by:
Shimin Fu, Citrus Research Institute, (CAAS), ChinaCopyright © 2022 Stolowicz, Larocca, Werbajh, Parma, Carrillo, Ogas, Agostini, Redes, Welin, Castagnaro and Vojnov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adrian Vojnov, YWF2b2pub3ZAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.