- 1Gulf Coast Research and Education Center, Institute of Food and Agriculture Science, University of Florida, Wimauma, FL, United States
- 2Department of Plant Pathology, University of Florida, Gainesville, FL, United States
- 3Horticulture Science Department, University of Florida, Gainesville, FL, United States
Phytophthora crown rot (PhCR) caused by Phytophthora cactorum is one of the most damaging soilborne diseases of strawberry in the United States and worldwide. Limitations on fumigants such as methyl bromide have led to deterioration in the control of P. cactorum in recent years. The development of resistant varieties is a critical component of the strategy for combatting this soilborne disease. Here, we review the biology of the pathogen, molecular mechanisms of infection, and management of PhCR in strawberry. Recent genomics advances in octoploid strawberry breeding have been able to gain a deeper insight into the genetic architecture of resistance to PhCR and identified the genetic resistance sources for the improvement of strawberry varieties against the pathogen. Quantitative trait loci (QTL) associated with PhCR resistance have been identified and introgressed for breeding of PhCR resistance in cultivated octoploid strawberry (F. × ananassa). Further characterizing candidate genes and mechanisms of resistance could facilitate incorporating the resistance genes into commercial varieties through genomics-assisted breeding, respectively. In this review, we address important recent advances and progress for genomics-assisted breeding for the resistance to PhCR and the potential use of CRISPR gene editing in cultivated strawberry.
Introduction
Strawberry is one of the important soft fruit crops popular for its color and taste. Besides its attractive color, strawberry has various health benefits such as high in vitamins, minerals, antioxidants, flavonoids, and phenolic acids (Giampieri et al., 2015). The cultivated strawberry (Fragaria × ananassa) is an allo-octoploid (2n = 8x = 56) and interspecific hybrid originated from a cross between wild octoploid species F. chiloensis and F. virginiana (Darrow, 1966; Edger et al., 2019). In 2017, the United States produced 1.6 billion pounds (726 million kg) of strawberries, with a total value of approximately $3.5 billion (NASS, 2017). The yield and quality of strawberry fruit are affected by various environmental and biological factors. The common strawberry diseases in Florida include charcoal rot (Macrophomina phaseolina), Colletotrichum crown rot (Colletotrichum gloeosporioides), Botrytis fruit rot (Botrytis cinerea), Phytophthora crown rot (Phytophthora cactorum), anthracnose fruit rot (Colletotrichum acutatum), powdery mildew (Podosphaera apahanis), leather rot (P. cactorum and P. nicotianae), Phomopsis leaf blight and soft rot (Phomopsis obscurans), stem end rot and leaf blotch (Gnomonia comari), and angular leaf spot (Xanthomonas fragariae) (Oliveira and Peres, 2020).
Among various diseases, economic losses due to root and crown rots are estimated about $150 million per year in the United States (Samtani et al., 2019). In the Florida industry itself, it is estimated that yield loss due to soilborne diseases costs $15 million in revenue each year, despite annual pre-plant soil fumigation (personal communication with Florida Strawberry Growers Association). Phytophthora crown rot (PhCR) disease caused by P. cactorum is one of the most important soilborne diseases of strawberry in the United States and worldwide, causing up to 40% production losses (Stensvand et al., 1999). This pathogen was first reported in Germany in 1952 (Deutschmann, 1954) and has been problematic in strawberry production worldwide (Schafleitner et al., 2013).
Limitations on fumigants have imposed challenges for the control of P. cactorum in recent years. Commercial cultivars vary widely in their genetic resistance to this pathogen, with the most resistant varieties providing high levels of disease control (Mangandi et al., 2017). Thus, the development of resistant cultivars will be a critical component of the strategy for managing this disease in an era of limited fumigants and increases in demand for organic strawberries. Also, soil fumigation before planting is often ineffective in controlling this disease in fruit production fields because the main inoculum source is infected nursery stock (Browne and Bhat, 2011; Baggio et al., 2021). Although various practices together with chemical pesticides are used to manage PhCR, understanding the mechanism of disease resistance and developing disease-resistant cultivars is the best strategy to protect against the pathogen.
Several breeding efforts have been made in the past to identify the genetic resource of resistance for breeding in both diploid and octoploid strawberries (Eikemo et al., 2003; Shaw et al., 2006; Shaw et al., 2008; Eikemo and Stensvand, 2015). The resistance locus Resistance to Phytophthora cactorum 1 (RPc-1) has been identified in F. vesca (Davik et al., 2015). In the octoploid cultivated strawberry, a major resistance locus, FaRPc2, located in a linkage group 7D confers resistance against P. cactorum (Mangandi et al., 2017). Other quantitative trait loci (QTL), FaRPc6C, FaRPc6D, and FaRPc7D, were also identified in a biparental mapping population for P. cactorum resistance in cultivated strawberry (Nellist et al., 2019). FaRPc2 and FaRPc7D are located in the same genomic region, indicating the same resistance QTL. Pathogen populations can be constitutively diversified in response to resistance genes in plants and high levels of diversity and population structure are observed in the potato late blight pathogen, P. infestans (Wang et al., 2017). It would be important to understand the population genetic structure of pathogens for crafting durable resistance and addressing the potential breakdown of resistance genes in field conditions. Currently, there is no information available about the pathogenic diversity of P. cactorum in U.S. strawberry fruiting fields and plant nurseries. It is not uncommon to observe pathotypes of other Phytophthora species in soybean and potato, as these pathogens have rapidly adapted to overcome resistance genes (Witek et al., 2016; Rojas et al., 2017). In addition, a recent study reported that another Phytophthora species, P. nicotianae, which is closely related to P. cactorum, can cause crown rot of strawberry in the United States (Marin et al., 2018). Unfortunately, little is known about how P. nicotianae compares to P. cactorum with respect to host resistance, especially for FaRPc2, which is the major resistance source for strawberry varieties in the United States. The spatial and temporal deployment of resistance genes of FaRPc2 combined with knowledge of pathogen populations would be a major factor determining broad-spectrum resistance to PhCR in cultivated strawberry.
This review addresses the current knowledge of P. cactorum causing crown rot disease and its management, and recent genomic approaches for the breeding of PhCR resistance in cultivated strawberry.
The genus Phytophthora and P. cactorum causing Phytophthora crown rot disease in strawberry
The genus Phytophthora became widely known among plant pathologists after the late blight disease of potato, caused by Phytophthora infestans, in the United States, in 1843, and in Europe, in 1845 (Bourke, 1991; Peterson et al., 1992). The pathogen destroyed Ireland’s potato fields during 1845–1846, leading the country to mass starvation with sociological and economic impacts (Large, 1940). The genus was first described by de Bary, in 1876 (Erwin and Ribeiro, 2005). Within this genus, several species are described, some with a narrow host range while others are pathogenic to more than 900 plant species, such as P. infestans and P. cinnamomi, respectively (Zentmyer, 1980). Moreover, adaptability, such as hybridization and effective dispersal, have enabled Phytophthora species to shift hosts, colonize new niches, and become more aggressive and invasive pathogens (Brasier et al., 1999; Goss et al., 2011; Man in ‘t Veld et al., 2012; Husson et al., 2015).
Taxonomically, the species P. cactorum is within the kingdom Stramenopila, phylum Heterokontaphyta, class Peronosporomycetes, order Peronosporales, and family Peronosporaceae. This hemibiotrophic oomycete pathogen has been reported for the first time from rotting cacti (Cereus giganteus and Melocactus nigrotomentosus) by Lebert and Cohn (1870) in Czechoslovakia. Several synonyms are listed by Erwin and Ribeiro (2005), but P. cactorum is the valid name nowadays. Although it occurs worldwide, it is mostly found in temperate regions causing root and collar rots, fruit rots, cankers, leaf blight, wilts, and seedling blights in various hosts (Erwin and Ribeiro, 2005). This pathogen can cause disease in more than 200 plant species from 150 different genera and 60 families (Tucker, 1933; Nienhaus, 1960). In strawberry, P. cactorum was first reported to cause leather rot on fruit (Rose, 1924), then crown rot, in 1952 (Deutschmann, 1954). Since then, crown rot of strawberry caused by this Phytophthora species has been detected in most of the United States, Europe, and parts of Asia and Africa.
P. cactorum is a homothallic species producing oospores on several natural media (Gallegly and Hong, 2008). Oogonia features are smooth-walled and hyaline (19–38 µm diameter); antheridia are paragynous and often attached close to the oogonial stalk; oospores could be plerotic or aplerotic within a range of 20–26 µm in diameter. Sporangia are papillate, normally borne terminally, with shape ranging from ellipsoidal to spherical (31.4 ± 4.8 × 26.4 ± 4.0 µm) (Oudemans and Coffey, 1991; Erwin and Ribeiro, 2005). Sporangiophores are often simple or in a close or lax sympodium with clustered sporangia (Erwin and Ribeiro, 2005; Abad et al., 2022). Chlamydospores are not very often produced, but when present, they are terminal or intercalary and globose (17–55 µm in diameter). Colonies have no distinct pattern in V8 agar, potato dextrose agar, and malt extract agar with cardinal temperatures of 4°C for minimum, 24°C for optimum, and 30°C for maximum growth (Abad et al., 2022).
P. cactorum shares the Clade 1a of the Phytophthora phylogenetic tree based on the internal transcribed spacer (ITS) region with P. idaei, P. pseudotsugae, and P. hedraiandra (Abad et al., 2022). Its distinguishable features from the other species are the production of caducous sporangia with short pedicels (less than 40 mm) and paragynous antheridia (Erwin and Ribeiro, 2005). Besides morphology, BLASTs of the ITS and cytochrome c oxidase subunit 1 (COI) regions are often used for Phytophthora species identification (Robideau et al., 2011). However, a method for routine and rapid detection of P. cactorum was created based on high-resolution melting (HRM) assay, which facilitates correct diagnosis, the study of their host ranges and distributions, and accelerates management decisions (Ratti et al., 2019).
Morphology of sporangia and oogonia can slightly vary among P. cactorum isolates from different hosts, but due to subtle variations, these characteristics are not enough for isolate differentiation (Hantula et al., 2000). However, some molecular markers can distinguish among isolates from different plant species (Cooke et al., 1996; Hantula et al., 1997; Lilja et al., 1998). Variations can also be observed among isolates from the same host infecting different tissues, such as isolates infecting strawberry crown and fruit, causing crown rot and leather rot, respectively. For a long time, it has been believed that crown rot could only be caused by strains isolated from strawberry crowns or from soil, whereas isolates from other hosts and strawberry fruit could produce leather rot (Seemüller and Schmidle, 1979; Harris and Stickels, 1981; Hantula et al., 2000). Although crown rot is likely caused by a genetically distinct pathotype of P. cactorum, isolates have the ability to cause leather rot on strawberry; however, not all isolates from leather rot have the ability to cause crown rot (Eikemo et al., 2004; Nellist et al., 2021). Additionally, genetic separation among isolates of P. cactorum in North America and Europe was not obtained with UPGMA analysis; however, host specificity among the strains was observed. Contrastingly, AFLP markers showed that leather rot isolates were different from crown rot isolates during the analysis of 44 isolates of P. cactorum from strawberry and other hosts (Eikemo et al., 2004). Besides P. cactorum, other species can also be associated with causing crown-rot-like symptoms. Phytophthora fragariae causes red stele (red coloration in the core of the root), which can be significant in some strawberry-growing regions leading to severe stunting and mortality. Other species of Phytophthora have also been occasionally reported in strawberry, such as P. citricola, P. citrophthora, P. eryptogea, P. bishii (= bisheria), P. megasperma, P. fragariaefolia, and P. nicotianae (Jeffers and Scott 1953; Meszka and Michalecka 2016; Marin et al., 2018; Farr and Rossman, 2022).
Biology of Phytophthora cactorum on strawberry plants
Disease and symptoms
PhCR of strawberry can be caused by several Phytophthora species; however, P. cactorum is the most important. Early symptoms are characterized by young leaves turning bluish-green, which eventually begin to wilt. Due to disease development and crown collapse, the whole plant wilts and dies within days (Peres and Baggio, 2019). When the diseased plant is removed from the ground, the crown likely breaks at the upper end. Dark reddish-brown discoloration of crowns starting at the upper or bottom part and disintegration of the vascular tissue are characteristic symptoms of this disease (Figure 1), which could be confused with symptoms caused by other crown rot pathogens, such as C. gloeosporioides and M. phaseolina. Depending on the environmental factors, the number of crowns within the plant, and cultivar tolerance, the rotting process of the crown may stop leading to recovery or stunting of the plant (Maas, 1998).
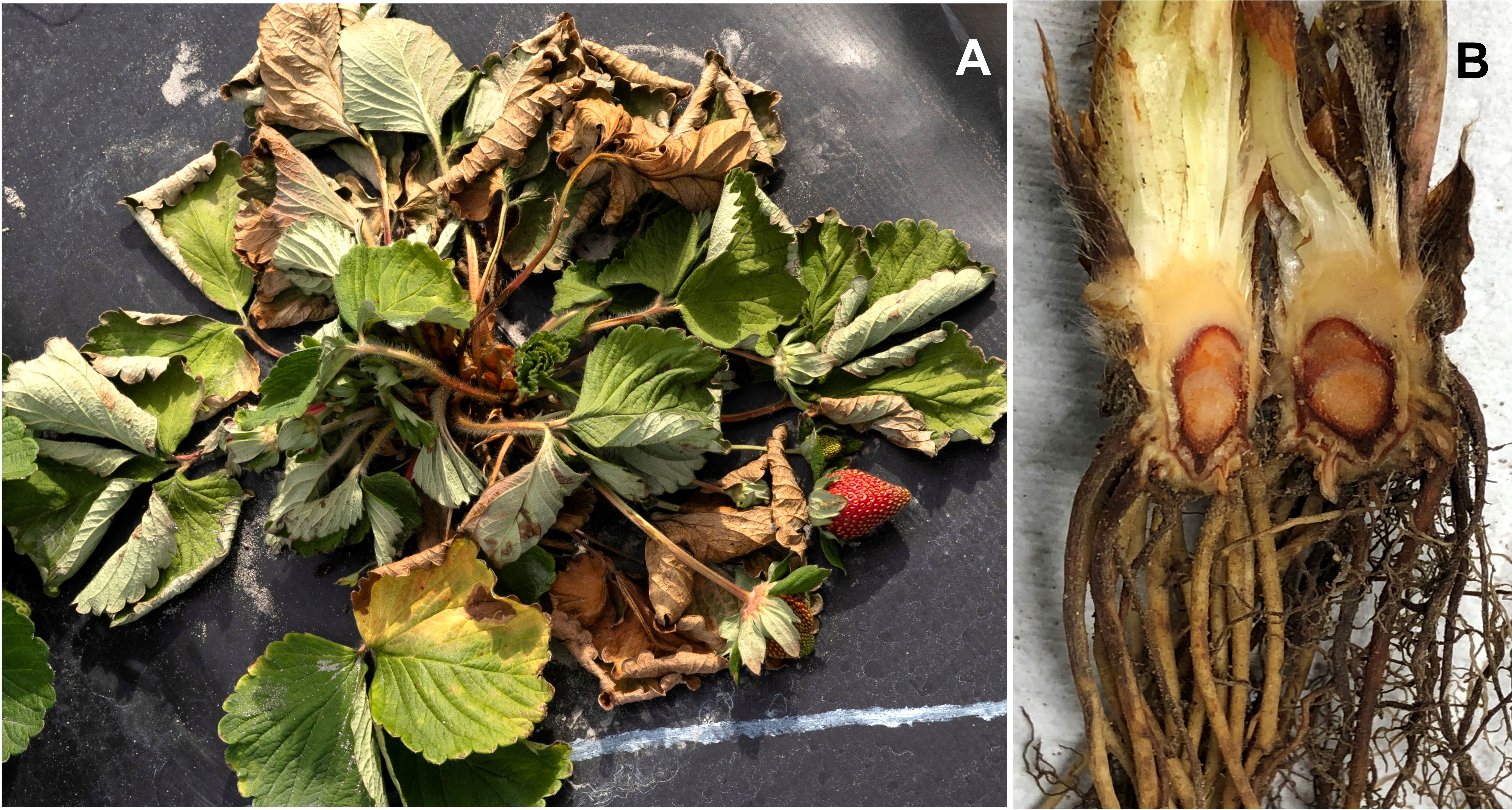
Figure 1 Symptoms of Phytophthora crown rot disease in strawberry. (A) Strawberry plant wilting, and (B) crown rot symptoms caused by Phytophthora cactorum.
Pathogen biology, epidemiology, and disease cycle
Pathogen infection and disease development require specific factors and, if not fulfilled, infections may remain latent, such as in cold-stored plants. Warm weather and abundant wetness trigger disease occurrence and development (Maas, 1998). In strawberry annual production systems in open fields that adopt overhead irrigation after transplanting to promote plant establishment, these conditions are common, leading to outbreaks in the early season of diseases caused by oomycetes (Marin et al., 2019; Baggio et al., 2021). Strawberries are more susceptible at transplanting, and 45 to 60 days later, which correspond to periods when the plant is metabolically more active due to rapid juvenile growth, root development, and fruit production (Marin et al., 2022a). Constant availability of new root growth is a likely cause of the rapid development of damaging Phytophthora populations under favorable conditions. Although cultivar resistance is genetically determined, digging immature plants from the nursery could lead to outbreaks in fruit production fields. Moreover, plants under stress are more prone to disease development, such as frigo plants transplanted during warm weather (Lederer and Seemuller, 1992).
The disease cycle may slightly change depending on the strawberry production system (Figure 2). For the perennial production system and nursery fields, due to the constant presence of strawberry plants, the source of inoculum could be oospores that are produced from sexual reproduction between antheridium and oogonium and survived in the soil or plant debris (Srivastava et al., 2020). This survival structure, under conducive conditions, germinates and produces sporangium that contains zoospores. Either the germinated sporangium or the flagellated zoospores can infect the plants, usually through wounds during transplanting. Zoospores move in the water, discard flagella, and synthesize a cell wall, forming a cyst. The cyst germinates and enters host tissue through natural openings like stomata or penetration of host epidermal cells that occurs with the help of an appressorium-like swollen tube (Hohl and Stössel, 1976). Specifically for nursery production, cold-stored plant material (frigo plants) could also serve as the major source of inoculum (Pettitt and Pegg, 1994). The expression of symptoms is linked to the time of planting, since during winter, the pathogen reduces its activity due to low temperatures, delaying disease progress (Maas, 1998). For annual strawberry production, where plants are usually cultivated during the winter, the pathogen might not survive between production seasons, during summer. In Florida, through nurseries monitoring and studies aiming to access the Phytophthora survival over seasons, the major source of inoculum is likely transplants harboring quiescent infection from nurseries (Baggio et al., 2021); however, this scenario could be different for strawberry growing regions lacking such information. The strawberry propagation system in nursery fields varies, but it is common that at least the third and fourth generations of transplants are multiplied in open fields, being subjected to pathogen infection (Rahman et al., 2015). A similar situation could happen at the nursery level with the exchange of plant stock materials within and among nurseries. After introduction of the pathogen in a field, under wet and favorable conditions, zoospores are produced by infected plants and can disseminate at short distances through free water present in poorly drained soil and penetrate strawberry plants. However, these secondary infections are more likely to occur at the nursery level or in perennial production systems, where the density of plants is higher and plants are maintained in the fields for longer periods, resulting in multiple disease cycles within one crop cycle, characterizing a polycyclic disease. Conversely, in annual production systems, where secondary infections are rare during the same crop cycle, the disease has a monocyclic pattern of development.
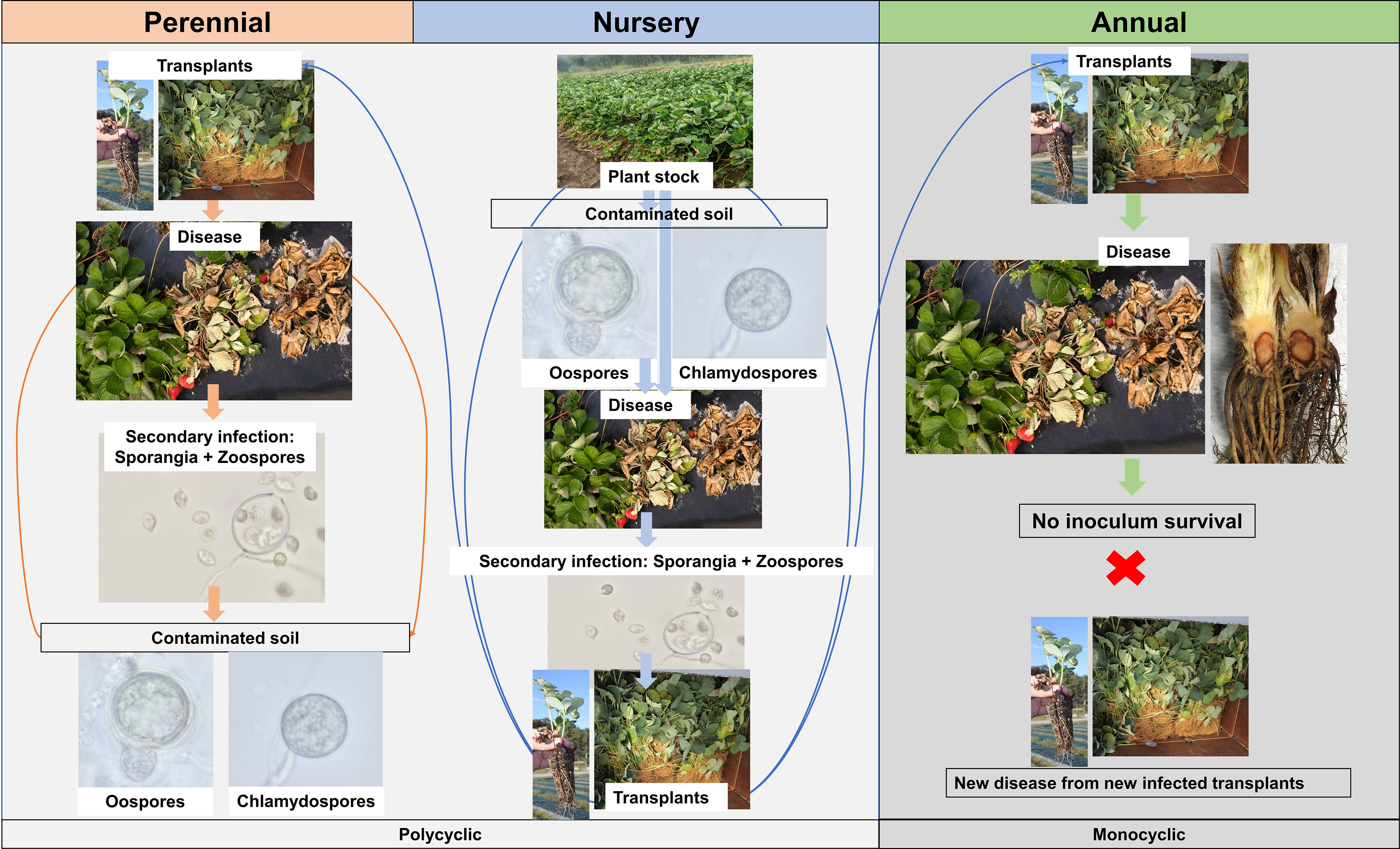
Figure 2 Disease cycle of Phytophthora crown rot caused by Phytophthora cactorum according to the strawberry production system. For the annual production system, the scheme was drawn based on the findings reported by Baggio et al. (2021).
Disease management—cultural control
PhCR is usually managed with a combination of chemical and non-chemical methods. Cultural control methods can vary depending on the strawberry region and production system adopted. Studies carried out in Florida, where the annual production system is adopted, show that the pathogen inoculum likely does not survive in the soil and asymptomatic nursery transplants harboring latent infections are considered the major source of inoculum (Baggio et al., 2021). However, the inoculum survival could greatly vary according to weather and soil type among different strawberry growing regions; therefore, more studies aiming to monitor the inoculum survival over time in other areas are needed, such as the one carried out by Baggio et al. (2021). Foremost, the acquisition of free and clean plant stock is detrimental to avoid the introduction of the pathogen and the start of an epidemic. In perennial production systems, where the pathogen can survive in the soil and plant debris, practices that aim to eliminate or reduce the inoculum present in the soil are usually adopted. Among these methods, soil fumigation and soil solarization have been reported to be effective in controlling Phytophthora species. Solarization is carried out after the beds are formed and can be effective if weather conditions are ideal (30–45 days of hot weather that promotes soil temperatures of at least 50°C). Also, steam disinfestation could be used as an alternative to chemical fumigation, since raising the soil temperature to 70°C for 30 min to a depth of 0 to 25 cm effectively reduced soilborne diseases (Fennimore and Goodhue, 2016).
Regardless of the production system, PhCR incidence is usually lower in sites with appropriate soil drainage. Improved soil drainage is usually achieved with properly prepared and tilled soil, preventing the formation of soil layers that are impervious to water. The pathogen could also be introduced in a disease-free area from infested areas through contaminated irrigation water. Therefore, the adoption of appropriate irrigation practices, such as reducing overhead irrigation to avoid free water availability and monitoring water from reservoirs for pathogen occurrence, could avoid the introduction and minimize the dissemination of the pathogen in a field. The adoption of plastic-mulched, raised beds also contributes to disease management. Besides permitting the use of drip irrigation systems, in which low amounts of water are provided to the plants, it improves soil drainage and allows the application of soil fumigants, which can help with inoculum reduction in soils infested with Phytophthora species.
To manage diseases in the plant stock materials at the nursery level, thermotherapy of asymptomatic nursery transplants infected with P. cactorum could be a good alternative to manage pathogen populations in plant stock. Studies have shown that exposing transplants to aerated steam in a closed chamber at 37°C for 1 h, followed by 44°C for 4 h, was effective in reducing P. cactorum, including mefenoxam-resistant populations and disease incidence in production fields (Baggio et al., 2021). The other chemical alternative would be the use of biological products; however, their efficacy may vary across different seasons if environmental conditions are not always conducive to the biological agent (Hautsalo et al., 2016). Some products containing Streptomyces spp., Bacillus subtilis strain AFS032321, and Pseudomonas chlororaphis strain AFS009 were tested at field conditions in Florida and had a positive effect on reducing PhCR (Mertely et al., 2020; Baggio et al., 2021). However, more studies are needed to really access their efficacy across multiple seasons in Florida and different strawberry growing regions.
The adoption of tolerant or less-susceptible cultivars is a desirable strategy and a sustainable method in managing PhCR. Among some of the FL and CA cultivars, ‘Sweet Charlie’, “Florida Radiance’, ‘Florida 127’ Sensation™, and ‘Florida Brilliance’ are susceptible, whereas ‘Strawberry Festival’, ‘Florida Elyana’, ‘Florida Beauty’, ‘Fronteras’, ‘Merced’, ‘Albion’, ‘San Andreas’, and ‘Portola’ are considered tolerant (Seijo et al., 2020; Marin et al., 2022a). Unfortunately, most cultivars are bred and selected based on fruit yield and quality and not on tolerance to PhCR. Therefore, it falls on breeding programs for the development and deployment of new cultivars with pathogen resistance, fruit quality, and competitive yield.
Disease management—chemical control
In areas where the pathogen may survive in the soil, soil fumigation can provide adequate control of the disease. Nursery and fruit production growers have always relied on soil fumigation using methyl bromide due to its high effectiveness in controlling soilborne pathogens (Santos et al., 2006). However, its phase out in 2013 in the United States and being banned in the EU from 2010 led producers to transition to alternative broad-spectrum fumigants, such as chloropicrin, 1,3-dichloropropene, metam sodium, dazomet, allyl-isothiocyanates, methyl iodide, dimethyl disulfide, and ethanedinitrile, which are usually not as effective (Baggio et al., 2020; Holmes et al., 2020). Limitation of the product movement throughout the soil profile due to its chemical features may jeopardize the reduction of pathogen population in the soil. Regardless of the production system, fungicide applications during transplanting and fruit production are still the predominant control method and, basically, only metalaxyl or mefenoxam and potassium phosphite products are available and registered for strawberry production in the United States (Peres and Baggio, 2019). However, due to the emergence of mefenoxam-resistant isolates and the failure of phosphite products in controlling P. nicotianae, management of PhCR has become even more challenging (Marin and Peres, 2021; Marin et al., 2021). Moreover, even resistance to quinone outside inhibitor fungicides has been reported in P. cactorum and P. nicotianae in Florida (Marin et al., 2022b). Thus, there is a demand for further research to find more effective alternative broad-spectrum fumigants and/or more efficient application methods, fungicides with new modes of action, and alternative biorational products. To improve the management options, alternative oomyticides were investigated and potential products were found; however, they are still not registered for strawberry (Marin and Peres, 2021). Moreover, disease management must be based on integrating the adoption of tolerant cultivars, cultural practices, and chemical applications to minimize the limitations of each control method.
Molecular mechanisms for the pathogen infection in strawberry
Various fungal pathogens use plant cell-wall degrading enzymes like CAZymes and mechanical force to break those barriers to infect and extract nutrients from the host cell. A total of 696 transcripts encoding CAZymes targeting cellulose, hemi-cellulose, and pectin were identified in the genome of P. cactorum 10300 (Armitage et al., 2018). Plant cell wall degrading enzymes, protease inhibitors, and phytotoxins of the PcF Toxin family are the effectors deployed by Phytophthora spp. during the infection of the host (Blackman et al., 2014). The delivery of various effector proteins (apoplastic or cytoplasmic) into the host tissue by a virulent pathogen targets plant immune system-related proteins (Dou and Zhou, 2012) and helps them to defeat plant immunity and initiate infection (Dangl et al., 2013). These effectors are secreted into the apoplast or host cells with the help of haustoria (Petre and Kamoun, 2014). In most of the Phytophthora species, a conserved RXLR motif at the N terminus is contained in one of the important classes of cytoplasmic effectors (Jiang et al., 2008). Various avirulence proteins (RXLR cytoplasmic effectors) like PiAvr3a, PiAvr2, PiAvr4, and PiAvrB1b1 has been localized to haustoria showing it as a site of secretion and delivery into host cells (Birch et al., 2006; van Poppel, 2009; Gilroy et al., 2011). The presence of infection-induced haustorium membrane protein 1 (PiHmp1) is important for infection in potato by P. infestans (Avrova et al., 2008). The importance of this protein for infection is highlighted with transient silencing of PiHmp1 that prevented infection to the host tissue.
Transcriptomic profiling of three different life cycle stages (mycelia, zoospores, and germinating cysts with germ tubes) of P. cactorum revealed the induction of numerous RXLR and Nep1-like proteins (NLPs) and the downregulation of the majority of Crinklers (CRNs) genes in germinating cysts with germ tube stage (Chen et al., 2018). Since cyst with the germ tube stage is an important pre-infection stage, the upregulated genes play a major role during the infection to host tissue. Moreover, gene expression patterns before and after the infection by P. cactorum identified various RXLR effector genes (Chen et al., 2011). Transcriptomic analysis of P. cactorum revealed a gene encoding the small cysteine-rich secretory protein SCR96 to be a virulence factor and important for oxidative stress tolerance (Chen et al., 2014). The comparison of transcriptomes of two P. cactorum isolates Pc407 (high virulence) and Pc440 (low virulence) obtained from Fragaria vesca revealed the high expression of a gene encoding a GAF sensor protein in the high virulence isolate Pc407 that is potentially involved in membrane trafficking processes (Taljoma et al., 2019). Lipid catabolism and elicitins were more highly expressed in Pc407 than in Pc440 at 48 h post-inoculation. The members of the glycosyl hydrolase family 1 involved in the metabolism of glycosylated secondary metabolites were expressed more in Pc407 than in Pc440. In addition, the more virulent isolate Pc407 depends on a smaller set of key RXLR effectors while infecting the host tissue whereas the less virulent isolate Pc440 relies on more RXLR effectors. We propose that P. cactorum infection in strawberry would involve a cascade of enzymes and effectors to infect plants similar to the infection mechanism used by other Phytophthora species.
Genome sequencing of P. cactorum and its application to control Phytophthora crown rot
The exploitation of DNA sequencing helped in learning more about the genetics of Phytophthora, species differentiation, phylogeny construction, and grouping into clades (Armitage et al., 2021). The availability of isolate sequences would help to identify similarities and differences in addition to showing if any gene or DNA region is identical (or nearly identical), in which case the isolates belong to the same species. P. cactorum infects a wide range of plants including ornamentals, trees, and fruit crops. The whole genome sequencing of P. cactorum from Panax species revealed its genome size to be 121.5 Mb using third-generation single-molecule real-time (SMRT) sequencing technology (Yang et al., 2018). The genome is highly heterozygous and is the second largest genome within the Phytophthora genus. On comparing the genome of P. cactorum with other Phytophthora spp., it was revealed that P. parasitica, P. infestans, and P. capsici had the closest relationships (highly syntenic). The extensive expansion of genes encoding detoxification enzymes and carbohydrate-active enzymes (CAZymes) showed comparative analyses of Phytophthora genomes. Also, due to the wide host range, P. cactorum had higher utilization or detoxification ability against ginsenosides (a group of defense compounds synthesized by Panax species) compared to P. sojae. A total of 901 and 282 genes were predicted to putatively encode CAZY enzymes in P. cactorum from Panax and strawberry, respectively (Armitage et al., 2018). Also, tolerance to fungicides in P. cactorum involved many proteins in the detoxification metabolic pathway, revealed by proteomic analysis (Mei et al., 2014). The identification of peroxidase (POD), glutathione S-transferases (GST), cytochrome P450 (CYPs), methyltransferase (MTR), dehydrogenase, ATP-binding cassette (ABC) transporter families, and major facilitator superfamily (MFS) in P. cactorum genome supports the ability of the pathogen to detoxify secreted ginsenosides (Yang et al., 2018).
Utilizing de novo prediction and homology-based comparison, the genome of P. cactorum was found to have 56.7 Mb repetitive elements, which constitutes 46.7% of the assembled genome (Yang et al., 2018). Of the 46.7% repetitive elements in the genome, 45.3% include transposable elements (TEs), 20.3% of which include long terminal repeats (LTR). The majority of unique genes in P. cactorum were related to defense response, the cell cycle, interaction between organisms, regulation of the cell cycle, TOP signaling pathway, and peptidyl-amino acid modification. The genome sequence of P. cactorum isolate 10300 isolated from infected strawberry tissue generated a 59.3-Mb genome assembly with a de novo assembly using ABySS (Armitage et al., 2018). The genome size of P. cactorum from strawberry is smaller compared to the genome of P. cactorum from Panax species (121.5 Mb). Also, the percentage of repetitive elements is 18% in the P. cactorum genome from strawberry versus 46.7% in the genome from Panax. A total of 23,884 genes encoding 24,189 proteins were predicted from its genome, and the number of genes that encoded putatively secreted proteins was 2,234. Although the numbers of genes predicted might be variable within Phytophthora species, the greatest number of complete single copy core eukaryotic genes (CEGs) was obtained in P. cactorum gene models, indicating a good representation of gene space within gene models (Armitage et al., 2018).
The predicted genes revealed a plethora of genes encoding putative secreted effectors containing about 200 RxLR domain-containing effectors, 77 CRNs, and apoplastic effectors such as phytotoxins (PcF proteins) and necrosis inducing proteins that play a significant role in infecting the host tissue. Genome sequencing and comparative analysis of 18 isolates of P. cactorum revealed evidence for host specialization (Armitage et al., 2021). Isolates causing PhCR in strawberry are globally similar whereas greater diversity was observed for apple-infecting isolates. Discrete lineages and non-recombining clades were observed within P. cactorum for specialized strawberry PhCR and apple-specific isolates. Additionally, gain and loss of effector complements were observed in P. cactorum phylogeny presenting determinants of host boundaries. Although the genome sequencing and comparative analysis from different research highlight the role of RxLR effectors, CRNs, and apoplastic effectors, not much has been done to characterize their individual role in infecting strawberry. Additionally, understanding the host specificity of these effectors would allow us to target them for resistance breeding in strawberry to PhCR.
In the future, further genome sequencing of P. cactorum isolates and Phytophthora spp. would allow for a better understanding of the molecular mechanism of infection via comparative genomic analysis. It is crucial to identify and understand the effectors involved during infection to understand the molecular disease mechanism. Also, the durability and functionality of the disease resistance depend on the effector protein secreted by plant pathogens, and these isolate-specific effectors could be targeted for resistance breeding in strawberry. Furthermore, the availability of genome sequences for different isolates of P. cactorum would help in early detection and surveillance for a new isolate if they evolve in the future and to keep track of different isolates infecting strawberry. The comparative analysis of all the isolates of P. cactorum infecting strawberry would additionally provide information about conserved regions across all the isolates. This conserved regions across isolates might serve as the target region in the pathogen genome for resistance breeding against P. cactorum.
QTL discovery for the resistance to Phytophthora crown rot in strawberry
The majority of cultivars grown worldwide are susceptible to P. cactorum, although high level of resistance was observed in a few cultivated strawberries and numerous diploid strawberry accessions (Parikka, 2003; Eikemo et al., 2010). The diploid wild strawberry F. vesca ‘Hawaill 4’ genotype showed resistance against P. cactorum. A major locus, RPc-1 (Resistance to P. cactorum 1), identified from F. vesca explained 74.4% of the phenotypic variation for resistance to PhCR (Davik et al., 2015; Toljamo et al., 2016). In addition to the diploid strawberry, different sources of resistance against P. cactorum are available in the octoploid strawberry (Eikemo et al., 2003; Shaw et al., 2006; Shaw et al., 2008; Mangandi et al., 2017; Nellist et al., 2019). The testing of 26 cultivars and five selections for resistance to P. cactorum crown rot revealed that the genetic background greatly affects the degree of resistance and susceptibility to P. cactorum (Eikemo et al., 2003). In another study, 186 genotypes from the University of California strawberry breeding program were evaluated for the resistance to P. cactorum and identified that the phenotypic variation of 60.6% for the PhCR resistance was due to the different genetic backgrounds among the genotypes (Shaw et al., 2006). The maximum resistance was observed for ‘Albion’, ‘Aromas’, and ‘Camino Real’, moderate resistance for ‘Camarosa’, and minimum resistance for ‘Diamante’ and ‘Ventana’ (Shaw et al., 2006). Conventional and biotechnological approaches could be used to transfer the source of resistance to susceptible cultivars.
In the strawberry breeding program of the University of Florida, a major locus conferring resistance to PhCR referring to FaRPc2 was identified using a pedigree-based analysis in complex, multiparental population sets (Mangandi et al., 2017). The resistance locus FaRPc2 was mapped to linkage group 7D and accounts for most of the genetic variation existing in the breeding population tested for the PhCR resistance. Within the FaRPc2 locus, it was found that two major haplotypes H2 (FaRPc2-H2) and H3 (FaRPc2-H3) are highly associated with resistance against P. cactorum. The two resistant haplotypes may be originated from different resistance sources, and it is possible that two distinct functional resistance alleles are present at FaRPc2. The FaRPc2-H3 genotype is more prevalent in commercial strawberry varieties in the United States, while FaRPc2-H2 seems to be more frequently present in European accessions (Noh et al., 2018). However, very little is known about molecular mechanisms of resistance and candidate genes in the FaRPc2 region. To achieve the long-term goal of achieving durable resistance against P. cactorum, it would be important to combine multiple resistance genes and/or alleles. Additionally, three QTLs (FaRPc6C, FaRPc6D, and FaRPc7D) were reported in European germplasm on chromosomes 6-2, 6-4, and 7-3 using biparental population (‘Emily’ × ‘Fenella’) inoculated in a greenhouse (Nellist et al., 2019). Three QTLs explained about 37% of the phenotypic variation (Nellist et al., 2019). The significant SNP markers for FaRPc7D were located within the same QTL region of FaRPc2. Biparental mapping population could detect locus-determining small genetic variability because it includes less genetic and allelic diversity from fewer parents. However, the use of multi-parental populations enabled the exploration of allelic diversity and increased the accuracy of QTL detection (Mangandi et al., 2017). Thus, QTLs identified using a multifamily population would be effective in diverse genetic backgrounds, whereas QTLs identified using biparental population might be less effective for wide genetic backgrounds.
Genomics-assisted breeding approaches for the resistance against P. cactorum
The application of genomics in strawberry breeding has lagged compared to other major crops because of its highly heterozygous allo-octoploid genome, and limited genomic resources in the past. However, the recent availability of reference genome of octoploid strawberry from 'Camarosa', SNP genotyping array (FanaSNP), and high-throughput DNA tests enabled the use of genomic approaches in strawberry breeding (Bassil et al., 2015; Edger et al., 2019; Whitaker et al., 2020; Oh et al., 2020; Liu et al., 2021). In addition, the recent haplotype-phased genomes from University of California variety 'Royal Royce' and University of Florida elite breeding line 'FL15.89-25' shed light on advanced genomics research in strawberry (Hardigan et al., 2021; Fan et al., 2022). Several disease resistance traits are controlled by major loci such as FaRPc2, FaRCa1, and FaRCg1 (Mangandi et al., 2017; Salinas et al., 2019; Chandra et al., 2021). In some cases, complex traits may not be controlled by a few loci, and the application of genome-wide prediction would be an alternative strategy for improving traits of interest (Whitaker et al., 2020; Tapia et al., 2022). Genome-wide prediction in strawberry had been used in the past for parent selection of positive yield and quality traits (Petrasch et al., 2022). This approach could be useful for predicting breeding values without phenotypic information (Whitaker et al., 2020).
The development of genetically resistant varieties against P. cactorum is the most reliable and long-lasting method to protect strawberry plants from the pathogen. The screening and identification of a durable source of resistance are essential to transfer the source into a susceptible variety. The classical breeding for resistance involved the transfer of R genes from one germplasm to another. Breeding efforts for P. cactorum resistance started in 1992, and mostly focused on testing methods for susceptibility in mature plants (Simpson et al., 1994). Genomic tools have been used to develop molecular markers tightly associated with candidate R genes in the selection of parents and predicting inheritance pattern in progenies for early selection, thus reducing the costs associated with field trials (Akino et al., 2014; Peace, 2017). The development of the first 90K Axiom single nucleotide polymorphism (SNP) array enabled the identification of QTLs associated with the trait of interest to develop molecular markers (Bassil et al., 2015). A compendium of strawberry DNA tests was created and subsequently available to assist the breeders with the molecular markers available for various traits in strawberry (Oh et al., 2020). These DNA tests would help breeders to exploit the genetic resources available in their breeding program to breed better cultivars.
Rapid DNA extraction and high-resolution melting analysis were used for selection at the FaRPc2 loci conferring Phytophthora crown and root rot resistance in the strawberry breeding program at the University of Florida in 2015 (Lee et al., 2016). In addition, the development of high-throughput molecular markers and fine-mapping of FaRPc2 has been described in the octoploid strawberry (Noh et al., 2018). The high-resolution SNP molecular markers were developed for H2 and H3 haplotypes responsible for resistance to P. cactorum on strawberry. A total of seven markers for H2 (FaRPc2-H2) and four for H3 (FaRPc2-H3) resistance haplotype were tested for the University of Florida strawberry breeding population and other varieties from different breeding programs, respectively (Noh et al., 2018). All the markers were successfully utilized to breed new resistant varieties via marker-assisted selection.
Recently, the availability of reference genome and sequencing technologies helped to identify R genes associated with disease resistance in strawberry (Edger et al., 2019; Barbey et al., 2019; Shirasawa et al., 2021). The comparison of R genes present in cultivated strawberry with their diploid ancestors helped to understand their presence and retention during the transition from diploid to octoploid, and expression pattern (Barbey et al., 2019). The diploid ancestral relatives of the modern cultivated strawberry could possess genetic resources for disease resistance that could be introgressed to susceptible varieties.
Candidate genes and molecular mechanisms for the resistance against P. cactorum in strawberry
The R gene-related transcripts including receptor-like kinases, nucleotide-binding site leucine-rich repeat (NBS-LRR), and toll/interleukin-1 receptor domain-containing proteins were highly upregulated in the resistant genotype of F. vesca compared to the susceptible genotypes (Gogoi et al., 2019). A PCR-based approach (NBS profiling) identified 23 distinct resistant gene analog fragments of NBS-LRR from a resistant genotype of F. vesca (Chen et al., 2016). One of the RGAs, RGA109, was characterized as a putative NBS-LRR R protein. RPc-1 locus (resistant to P. cactorum) included one NB-LRR protein that was upregulated in resistant genotypes after the pathogen infection. Similarly, potential resistant genes were also identified in RPc-1 locus from root transcriptomic analysis of F. vesca (Toljamo et al., 2016).
Transcriptomic analysis of F. vesca ‘Hawaill 4’ roots infected with P. cactorum revealed potential candidate genes within the QTL region, RPc1, responsible for defense (Davik et al., 2015; Toljamo et al., 2016). They identified 69 defense-related genes within RPc1; 7 were significantly downregulated, and 19 were upregulated in response to the P. cactorum inoculation. Several genes such as NBS-LRR, L-type-lectin-RKLs, and receptor-like protein (RLP) with a leucine-rich-repeat were highly expressed in inoculated plants. The L-type-lectin-RKLs are involved in Phytophthora resistance, while LRR-RLPs are associated with various disease resistance in plants (Bouwmeester et al., 2011; Zipfel, 2014; Wang et al., 2015). A phenylalanine ammonia-lyase (PAL) encoding gene in RPc1 was upregulated after inoculation of P. cactorum. The PAL-dependent phenylpropanoid pathway is responsible for salicylic acid synthesis, which is an important hormone signaling pathway associated with fungal pathogen defense responses (Yuan et al., 2009). In addition, WRKY transcription factors located in RPc1 were significantly upregulated in response to P. cactorum inoculation. The transcriptomic analysis of F. vesca roots after inoculation with P. cactorum provides the possible molecular mechanisms for resistance associated with the pathogen in strawberry. All these RNA-seq transcriptome studies were carried out in diploid F. vesca, but none of the gene expression data in response to P. cactorum have been reported in the octoploid strawberry.
A number of candidate genes had been identified in plants for Phytophthora resistance; however, only limited work has been achieved in the octoploid cultivated strawberry. Analysis of transcriptome data was conducted for the resistance to C. gloeosporioides in octoploid strawberry (Wang et al., 2017; Chandra et al., 2021). Chandra et al. (2021) identified several candidate genes for the resistance to Colletotrichum crown rot and developed functional markers linked to the major locus FaRCg1 conferring resistance against C. gloeosporioides. It is important to identify candidate genes and incorporate them into elite breeding materials for developing new resistant cultivars. Expression QTL (eQTL) analysis could also effectively discover gene associations with target traits of interests in strawberry (Barbey et al., 2019). This genomics approach successfully characterized important genes associated with fruit flavors and pathways.
Molecular mechanisms of defense against P. cactorum have not been understood in strawberry, although the defense mechanism of various horticulturally important plants against Phytophthora spp. is available. The defense system can be categorized as preformed and induced depending on how they respond to the attacking pathogen (Jones and Dangl, 2006). The preformed defense system responds to a different class of pathogens in a similar way. It includes cell wall, waxy epidermal layer, bark, and cuticle that act as the first line of defense used by plants against the pathogens. The induced defense system responds to the virulence factors or elicitors of the pathogen directly or effecting host targets to defend (Van Loon et al., 2006). The first line of defense in strawberry could include cuticle, cell wall, trichomes, and leaf veins against various pathogens such as B. cinerea, C. acutatum, Tetranychus utricae, and Xantomonas fragariae, respectively (Gooding, 1976; Barritt, 1980; Guidarelli et al., 2011; Steinite and Levinsh, 2003; Kennedy and King, 1962). Biochemical barriers present in various parts of strawberries acts as a defense against various pathogens. The secondary metabolites including glucosinolates, sulfur compounds, phenolics, and saponins form a biological barrier acting at early stage of infection (Kliebenstein, 2004). Salicyclic acid, lignin, and chlorogenic acid are products of the phenylpropanoid pathway and serve as barriers for pathogens in strawberry. A biochemical compound similar to phytoalexin acts to make strawberries resistant to P. fragariae (Mussell and Staples, 1971), whereas the presence of antifungal compounds in the strawberry leaves makes it resistant to Colletotrichum fragariae. The preformed compound had antifungal activity 15 times more in the moderately resistant cultivar 'Sweet Charlie' than in the susceptible cultivar ‘Chandler’ (Terry, 2002).
Pattern recognition receptors (PRRs) in the plant cell membrane expose their pathogen/microbe/damage-associated molecular pattern (PAMP/MAMP/DAMP) recognition domains into the apoplast that recognizes conserved oomycete PAMPs and trigger PAMP-triggered immunity (PTI) (Fawke et al., 2015). The recognition of pathogen’s effectors entering the host cells mediated by intercellular disease resistance proteins (R genes) led to effector-triggered immunity (ETI). PTI is generally measured or marked based on ion fluxes, mitogen-activated protein kinase (MAPK) cascades, production of reactive oxygen species (ROS), cell wall reinforcement, pH alkalinization, callose deposition, and defense gene activation mostly regulated by WRKY transcription factors whereas ETI causes visible controlled cell death known as hypersensitive response (HR) (Ingle et al., 2006). Figure 3 illustrates the potential role of the PAMP- and DAMP-mediated defense pathway against P. cactorum. Numerous DAMPs had been identified in plants; however, their roles in plant immunity and defense are not fully understood. Plants could monitor the cuticle integrity and cell walls through DAMPs and activate defense responses. For example, Arabidopsis leaves treated with cutinase or pathogen expressing cutinase gene display strong resistance against necrotrophic fungus Botrytis cinerea (Chassot et al., 2007). Oligogalacturonic acid responses were enhanced with overexpression of wall associated kinase 1 (WAK1) and increased resistance to B. cinerea (Brutus et al., 2010). Similarly, xyloglucan oligosaccharides act as DAMP and induce immune responses like MAPK activation, callose deposition, and immune gene expression in grapevine and Arabidopsis for resistance against B. cinerea and Hyaloperonospora arabidopsidis (Claverie et al., 2018). Additionally, extracellular amino acid histidine acts as DAMP and induces resistance against soilborne bacterial pathogen Ralstonia solanacearum and B. cinerea in Arabidopsis and tomato, respectively, through activation of the ethylene signaling pathway (Seo et al., 2016). Moreover, the genus Phytophthora induces necrosis and immune response in several plant species using cellulose binding elicitor lectin (CBEL). A receptor-like kinase BAK1 (general regulator of basal immunity) is required for CBEL-induced defense responses and respiratory burst oxidase homologue (RBOH)-mediated production of reactive oxygen burst (Larroque et al., 2013). The result showed that a BAK1- and RBOH-dependent CBEL-triggered immunity is important for Phytophthora resistance, whereas susceptibility to Phytophthora does not correlate with natural quantitative variation of basal immunity triggered by conserved elicitors such as CBEL.
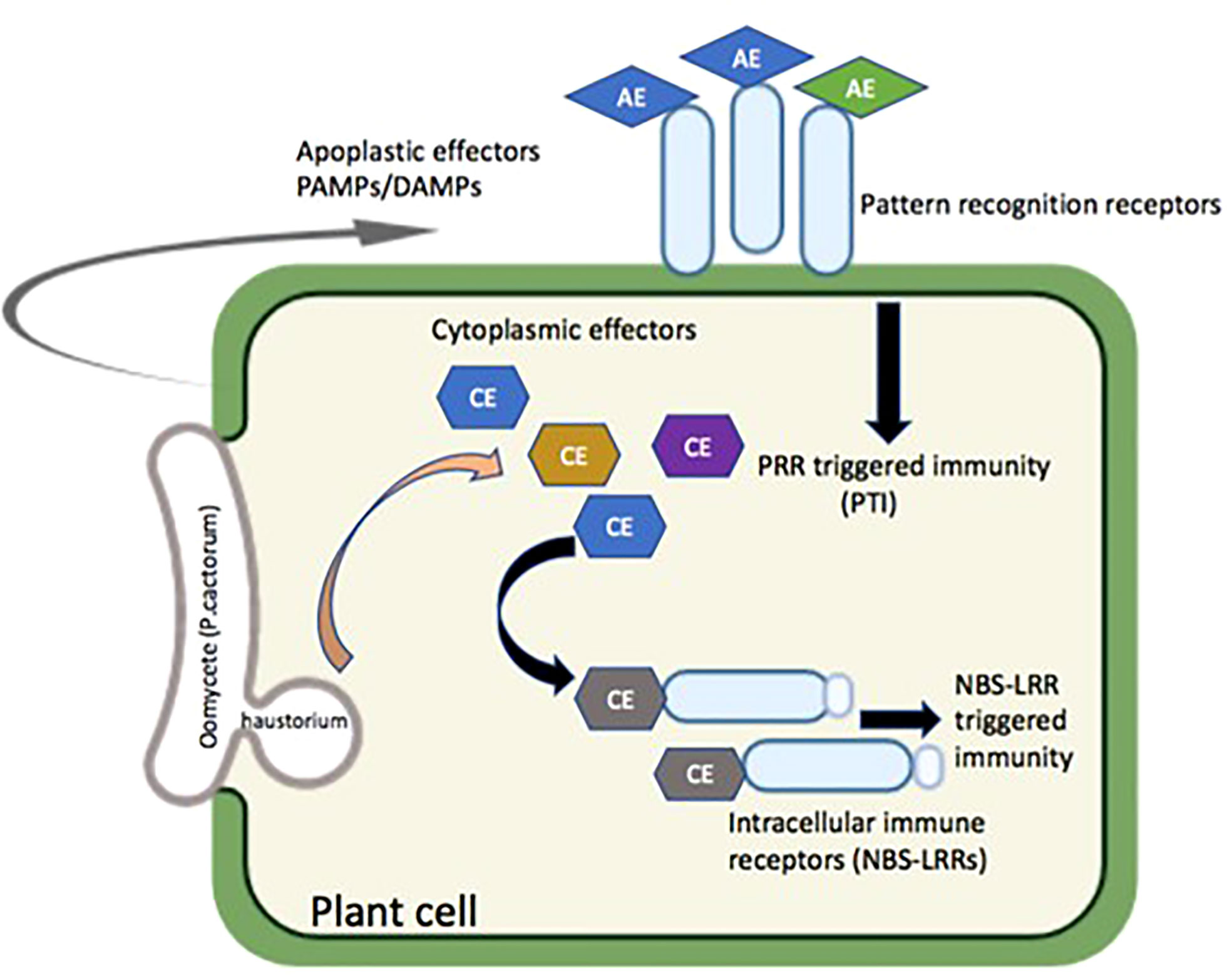
Figure 3 Possible mode of action for the resistance against P. cactorum in strawberry. PAMPs, pathogen-associated molecular patterns; DAMPs, damage-associated molecular patterns; AE, apoplast effectors; CE, cytoplasmic effectors; PRR, pattern recognition receptors; NBS-LRRs, nucleotide binding site-leucine rich repeats.
Potential application of CRISPR gene editing for the PhCR resistance in strawberry
The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9 system has been widely applied for targeted gene editing in humans, animals, and plants (Yang et al., 2013; Shalem et al., 2014; Zhang et al., 2014). In plants, genome editing can be used not only for studying gene functions but also in developing new germplasm by inducing mutations or modifying target genes. The method of gene editing has a great potential to enhance the breeding process for improving fruit quality and disease resistance in strawberry (Abdelrahman et al., 2018; Yang et al., 2013; Ito et al., 2015). In traditional breeding, the linkage drag is often challenging for the introgression of new resistance traits from wild varieties into elite cultivars. Because certain resistance genes could be genetically linked to loci associated with agronomically important traits such as yield and plant development (Lewis et al., 2007; Miah et al., 2013; Summers and Brown, 2013), modifying target genes precisely using CRISPR-guided mutation could be the effective way to resolve this issue of linkage drag.
The technique of CRISPR/Cas9 has been applied to improve cultivars for disease resistance against soilborne pathogens in plants (Table 1). Mutations in Non-expressor of Pathogenesis-Related 3 (TcNPR3) gene induced by transient expression of CRISPR/Cas9 lead to enhanced defense response to P. tropicalis in Theobroma cacao (Fister et al., 2018). TcNPR3 mutagenesis by CRISPR/Cas9 reduced lesion sizes in CRISPR/Cas9-treated leaf tissues and elevated the expression of pathogenesis-related (PR) genes (Fister et al., 2018). In another study, CRISPR/Cas9-induced mutation in Downy Mildew Resistance 6 (DMR6) enhances defense response to broad-spectrum plant pathogens (Xanthomonas spp., Pseudomonas syringae, and P. capsica) in tomato (de Toledo Thomazella et al., 2016). SlDMR6-1 mutagenesis by CRISPR/Cas9 delayed disease progression of P. capsica and the disease symptom was less severe in SlDMR6-1 edited plants (de Toledo Thomazella et al., 2016). Furthermore, CRISPR/Cas9-mediated gene editing could precisely repair mutations on target genes by homologous recombination (HR). Single-nucleotide polymorphisms and small InDel mutations of resistance genes could be potential candidates for CRISPR/Cas9-mediated HR applications. With more knowledge on host–pathogen interactions for PhCR resistance, we would have more feasible options for CRISPR-guided resistance breeding to PhCR in strawberry.
CRISPR/Cas9-mediated target mutagenesis studies have recently been reported in octoploid strawberry (Xing et al., 2018; Martín-Pizarro et al., 2019; Wilson et al., 2019). Transient assay of CRISPR/Cas9 introduced targeted mutations was performed with FvMYB10 (R2R3 MYB transcription factor 10) and FvCHS (chalcone synthase) genes involved in fruit color development. Agrobacterium-mediated transformation was utilized for the stable integration of CRISPR/Cas9 construct targeting PDS (phytoene desaturase). In addition, the CRISPR/Cas9 gene-editing system was first evaluated for the functional change of gene using Tomato MADS box gene6 (TM6) in the octoploid cultivated strawberry. The tm6 mutant lines exhibited morphological abnormalities in anthers and pollen grains (Martín-Pizarro et al., 2019).
Because of the high complexity of polyploid genome of strawberry, designing a single guide RNA (sgRNA) specific to a target gene is necessary to reduce any off-target effects. The 5’-end 20 nucleotide sequences in sgRNA determine target specificity and potential off-targets. Particularly, 8–12 nucleotides proximal to PAM (seed sequence) have a crucial role in the target gene recognition and cleavage (Jiang et al., 2013). A number of bioinformatic tools are currently available for designing sgRNA in different plant species (Table 2). CRISPOR, CRISPRdirect, CRISPR-P, CRISPR RGEN Tools, and CHOPCHOP provide a genome-wide survey of specific sgRNAs in diploid (F. vesca) and octoploid strawberry (F. ×ananassa). Moreover, CRISPR RGEN Tools and PolyOligo provide designing sgRNA on diploid and octoploid strawberry genomes. The recent haplotype phased genome of octoploid strawberry, FaRR1 (cv. Royal Royce), is now available in CHOPCHOP.
Conclusion
The recent genomic advances in both P. cactorum and strawberry have been greatly valuable for the resistance breeding of PhCR in octoploid cultivated strawberry. Because of the availability of high-quality chromosome-scale reference genomes of octoploid strawberry, it could be possible to discover more genes and rare alleles associated with the resistance to multiple pathogens and other important breeding characteristics. Furthermore, genomics research for characterizing gene functions will accelerate to develop gene-specific markers and facilitate the effectiveness of marker-assisted selection in development of new disease-resistant varieties. In new approaches of disease resistance breeding, one of the most exciting prospects would be to discover susceptibility (S) genes using an octoploid strawberry pangenome and utilize them with CRISPR/Cas9-guided mutation tools including a DNA-free gene editing platform suitable for generation of breeding accessions and varieties.
Author contributions
SS, MM, NP, and SL developed ideas and prepared the manuscript. SS, MM, ML, and JB wrote the manuscript, and all the remaining authors contributed to the final version of the manuscript.
Funding
This work was supported by the Florida Strawberry Growers Association, and Specialty Crops Research Initiative grant no. 2017-51181-26833 from the USDA National Institute of Food and Agriculture.
Conflict of interest
Author JB completed this manuscript at University of Florida and is now employed by Syngenta Crop Protection, LLC.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abad ZG, Burgess T, Redford AJ, Bienapfl JC, Mathew R, Srivastava SK, et al. (2022). IDphy: molecular and morphological identification of phytophthora based on the types, in Plant Dis. doi: 10.1094/PDIS-02-22-0448-FE
Abdelrahman M., Al-Sadi A. M., Pour-Aboughadareh A., Burritt D. J., Tran L. S. P. (2018). Genome editing using CRISPR/Cas9–targeted mutagenesis: An opportunity for yield improvements of crop plants grown under environmental stresses. Plant Physiol. Biochem. 131, 31–36.
Akino S., Takemoto D., Hosaka K. (2014). Phytophthora infestans: a review of past and current studies on potato late blight. J. Gen. Plant Pathol. 80 (1), 24–37. doi: 10.1007/s10327-013-0495-x
Armitage A. D., Lysøe E., Nellist C. F., Lewis L. A., Cano L. M., Harrison R. J., et al. (2018). Bioinformatic characterisation of the effector repertoire of the strawberry pathogen Phytophthora cactorum. PloS One 13 (10), e0202305. doi: 10.1371/journal.pone.0202305
Armitage A., Nellist C., Bates H., Sobczyk M., Luberti M., Lewis L., et al. (2021). Comparative analysis of host-associated variation in Phytophthora cactorum. Front. Microbiol. 12.
Avrova A. O., Boevink P. C., Young V., Grenville-Briggs L. J., Van West P., Birch P. R., et al. (2008). A novel Phytophthora infestans haustorium-specific membrane protein is required for infection of potato. Cell. Microbiol. 10 (11), 2271–2284. doi: 10.1111/j.1462-5822.2008.01206.x
Baggio J. S., Marin M. V., Peres N. A. (2021). Phytophthora crown rot of Florida strawberry: inoculum sources and thermotherapy of transplants for disease management. Plant Dis. (First Look) 105, 3496–3502. doi: 10.1094/PDIS-11-20-2476-RE
Baggio J. S., Ruschel R. G., Noling J., Peres N. A. (2020). Physical, cultural, and chemical alternatives for integrated management of charcoal rot of strawberry. Plant Dis 105, 295–304. doi: 10.1094/PDIS-04-20-0917-RE
Barbey C. R., Lee S., Verma S., Bird K. A., Yocca A. E., Edger P. P., et al. (2019). Disease resistance genetics and genomics in octoploid strawberry. G3: Genes Genomes Genet. 9 (10), 3315–3332. doi: 10.1534/g3.119.400597
Bassil N. V., Davis T. M., Zhang H., Ficklin S., Mittmann M., Webster T., et al. (2015). Development and preliminary evaluation of a 90 K axiom® SNP array for the allo-octoploid cultivated strawberry Fragaria× ananassa. BMC Genomics 16 (1), 155. doi: 10.1186/s12864-015-1310-1
Barritt B. H. (1980). Resistance of strawberry clones to botrytis fruit rot. J. Amer. Soc Hortic. Sci. 105, 160–164.
Birch P. R., Rehmany A. P., Pritchard L., Kamoun S., Beynon J. L. (2006). Trafficking arms: oomycete effectors enter host plant cells. Trends Microbiol. 14 (1), 8–11. doi: 10.1016/j.tim.2005.11.007
Blackman L. M., Cullerne D. P., Hardham A. R. (2014). Bioinformatic characterisation of genes encoding cell wall degrading enzymes in the phytophthora parasitica genome. BMC Genomics 15 (1), 785. doi: 10.1186/1471-2164-15-785
Bouwmeester K., De Sain M., Weide R., Gouget A., Klamer S., Canut H., et al. (2011). The lectin receptor kinase LecRK-i. 9 is a novel phytophthora resistance component and a potential host target for a RXLR effector. PloS Pathog. 7 (3), e1001327.
Bourke A. (1991). Potato late blight in Europe in 1845: the scientific controversy. Phytophthora (New York) pp, 12–24.
Brasier C. M., Cooke D. E. L., Duncan J. M. (1999). Origin of a new phytophthora pathogen through interspecific hybridization. Proc. Natl. Acad. Sci. 96, 5878–5883. doi: 10.1073/pnas.96.10.5878
Brauer E. K., Balcerzak M., Rocheleau H., Leung W., Schernthaner J., Subramaniam R., et al. (2020). Genome editing of a deoxynivalenol-induced transcription factor confers resistance to fusarium graminearum in wheat. Mol. Plant-Microbe Interactions® 33, 553–560. doi: 10.1094/MPMI-11-19-0332-R
Brutus A., Sicilia F., Macone A., Cervone F., De Lorenzo G. (2010). A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. 107 (20), 9452–9457.
Chandra S., Oh Y., Han H., Salinas N., Anciro A., Whitaker V. M., et al. (2021). Comparative transcriptome analysis to identify candidate genes for FaRCg1 conferring resistance against colletotrichum gloeosporioides in cultivated strawberry (Fragaria× ananassa). Front. Genet. 12.
Chassot C., Nawrath C., Métraux J. P. (2007). Cuticular defects lead to full immunity to a major plant pathogen. Plant J. 49 (6), 972–980.
Claverie J., Balacey S., Lemaître-Guillier C., Brulé D., Chiltz A., Granet L., et al. (2018). The cell wall-derived xyloglucan is a new DAMP triggering plant immunity in Vitis vinifera and Arabidopsis thaliana. Front. Plant Sci. 9, 1725.
Chen X. R., Brurberg M. B., Elameen A., Klemsdal S. S., Martinussen I. (2016). Expression of resistance gene analogs in woodland strawberry (Fragaria vesca) during infection with Phytophthora cactorum. Mol. Genet. Genomics 291 (5), 1967–1978. doi: 10.1007/s00438-016-1232-x
Chen X. R., Huang S. X., Zhang Y., Sheng G. L., Zhang B. Y., Li Q. Y., et al. (2018). Transcription profiling and identification of infection-related genes in phytophthora cactorum. Mol. Genet. Genomics 293 (2), 541–555. doi: 10.1007/s00438-017-1400-7
Chen X., Klemsdal S. S., Brurberg M. B. (2011). Identification and analysis of Phytophthora cactorum genes up-regulated during cyst germination and strawberry infection. Curr. Genet. 57 (5), 297. doi: 10.1007/s00294-011-0348-0
Chen X. R., Zhang B. Y., Xing Y. P., Li Q. Y., Li Y. P., Tong Y. H., et al. (2014). Transcriptomic analysis of the phytopathogenic oomycete Phytophthora cactorum provides insights into infection-related effectors. BMC Genomics 15 (1), 980. doi: 10.1186/1471-2164-15-980
Cooke D. E. L., Kennedy D. M., Guy D. C., Russell J., Unkles S. E., Duncan J. M. (1996). Relatedness of group I species of phytophthora as assessed by randomly amplified polymorphic DNA (RAPDs) and sequences of ribosomal DNA. Mycological Res. 100, 297–303. doi: 10.1016/S0953-7562(96)80158-4
Dangl J. L., Horvath D. M., Staskawicz B. J. (2013). Pivoting the plant immune system from dissection to deployment. Science 341 (6147), 746–751.
Darrow G. M. (1966). The strawberry: History, breeding, and physiology. New York: Holt, Rinehart and Winston.
Davik J., Eikemo H., Brurberg M. B., Sargent D. J. (2015). Mapping of the RPc-1 locus for Phytophthora cactorum resistance in Fragaria vesca. Mol. Breed. 35 (11), 211. doi: 10.1007/s11032-015-0405-3
de Toledo Thomazella D. P., Brail Q., Dahlbeck D., Staskawicz B. J. (2016). CRISPR-Cas9 mediated mutagenesis of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. BioRxiv 064824. doi: 10.1101/064824
Deutschmann V. F. (1954). Eine wurzelfa¨ule an erdbeeren, hervorgerufen durch Phytophthora cactorum (Leb et Cohn) schroet. Nachrichtenblatt Des. Deutschen Pflanzenschutzdienst 6, 7–9.
Dou D., Zhou J. M. (2012). Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe 12 (4), 484–495. doi: 10.1016/j.chom.2012.09.003
Edger P. P., Poorten T. J., VanBuren R., Hardigan M. A., Colle M., McKain M. R., et al. (2019). Origin and evolution of the octoploid strawberry genome. Nat. Genet. 51 (3), 541–547. doi: 10.1038/s41588-019-0356-4
Eikemo H., Brurberg M. B., Davik J. (2010). Resistance to Phytophthora cactorum in diploid fragaria species. HortScience 45 (2), 193–197. doi: 10.21273/HORTSCI.45.2.193
Eikemo H., Klemsdal S. S., Riisberg I., Bonants P., Stensvand A., Tronsmo A. M. (2004). Genetic variation between Phytophthora cactorum isolates differing in their ability to cause crown rot in strawberry. Mycological Res. 108 (3), 317–324. doi: 10.1017/S0953756204009244
Eikemo H., Stensvand A. (2015). Resistance of strawberry genotypes to leather rot and crown rot caused by phytophthora cactorum. Eur. J. Plant Pathol. 143 (2), 407–413. doi: 10.1007/s10658-015-0685-9
Eikemo H., Stensvand A., Davik J., Tronsmo A. M. (2003). Resistance to crown rot (Phytophthora cactorum) in strawberry cultivars and in offspring from crosses between cultivars differing in susceptibility to the disease. Ann. Appl. Biol. 142 (1), 83–89. doi: 10.1111/j.1744-7348.2003.tb00232.x
Erwin D. C., Ribeiro O. K. (2005). Phytophthora diseases worldwide. (St. Paul, MN: American Phytopathological Society).
Farr D. F., Rossman A. Y. (2022) Fungal databases, U.S. national fungus collections, ARS, USDA. retrieved June 1, 2022. Available at: https://nt.ars-grin.gov/fungaldatabases/.
Fan Z., Tieman D. M., Knapp S. J., Zerbe P., Famula R., Barbey C. R., et al. (2022). A multi-omics framework reveals strawberry flavor genes and their regulatory elements. doi: 10.1111/nph.18416
Fawke S., Doumane M., Schornack S. (2015). Oomycete interactions with plants: infection strategies and resistance principles. Microbiol. Mol. Biol. Rev. 79 (3), 263–280.
Fennimore S. A., Goodhue R. E. (2016). Soil Disinfestation with Steam: A Review of Economics, Engineering, and Soil Pest Control in California Strawberry. Int. J. Fruit Sci. 16:sup1, 71–83. doi: 10.1080/15538362.2016.1195312
Fister A. S., Landherr L., Maximova S. N., Guiltinan M. J. (2018). Transient expression of CRISPR/Cas9 machinery targeting TcNPR3 enhances defense response in theobroma cacao. Front. Plant Sci. 9 (March). doi: 10.3389/fpls.2018.00268
Gallegly M. E., Hong C. (2008). Phytophthora: identifying species by morphology and DNA fingerprints Vol. 158 (St. Paul, MN: American Phytopathological Society (APS Press).
Giampieri F., Forbes-Hernandez T. Y., Gasparrini M., Alvarez-Suarez J. M., Afrin S., Bompadre S., et al. (2015). Strawberry as a health promoter: an evidence based review. Food Funct. 6 (5), 1386–1398. doi: 10.1039/C5FO00147A
Gilroy E. M., Breen S., Whisson S. C., Squires J., Hein I., Kaczmarek M., et al. (2011). Presence/absence, differential expression and sequence polymorphisms between PiAVR2 and PiAVR2-like in phytophthora infestans determine virulence on R2 plants. New Phytol. 191 (3), 763–776. doi: 10.1111/j.1469-8137.2011.03736.x
Gogoi A., Lysøe E., Davik J., Eikemo H., Stensvand A., Brurberg M. B. (2019). “RNA-Seq transcriptome analysis reveals candidate genes involved in crown-rot (Phytophthora cactorum) resistance in strawberry (Fragaria vesca),” in 2019 IS-MPMI XVIII congress (Scotland: ISMPMI).
Goss E. M., Cardenas M. E., Myers K., Forbes G. A., Fry W. E., Restrepo S., et al. (2011). The plant pathogen phytophthora andina emerged via hybridization of an unknown phytophthora species and the Irish potato famine pathogen, p. infestans. PloS One 6, e24543. doi: 10.1371/journal.pone.0024543
Gooding H. (1976). Resistance to mechanical injury and assessment of shelf life in fruits of strawberry (Fragaria × ananassa). Hortic. Res. 16, 71–82.
Guidarelli M., Carbone F., Mourgues F., Perrotta G., Rosati C., Bertolini P., et al. (2011). Colletotrichum acutatum interactions with unripe and ripe strawberry fruits and differential responses at histological and transcriptional levels. Plant Pathol. 60, 685–697. doi: 10.1111/j.1365-3059.2010.02423.x
Hantula J., Lilja A., Nuorteva H., Parikka P., Werres S. (2000). Pathogenicity, morphology and genetic variation of Phytophthora cactorum from strawberry, apple, rhododendron, and silver birch. Mycological Res. 104, 1062–1068. doi: 10.1017/S0953756200002999
Hantula J., Lilja A., Parikka P. (1997). Genetic variation and host specificity of Phytophthora cactorum isolated in Europe. Mycological Res. 101, 565–572. doi: 10.1017/S0953756296002900
Hardigan M. A., Feldmann M. J., Pincot D. D., Famula R. A., Vachev M. V., Madera M. A., et al. (2021). Blueprint for phasing and assembling the genomes of heterozygous polyploids: Application to the octoploid genome of strawberry.
Hardigan M. A., Feldmann M.J., Pincot D. D. A., Famula R.A., Vachev M. V., Madera M.A., et al. (2021). Blueprint for Phasing and Assembling the Genomes of Heterozygous Polyploids: Application to the Octoploid Genome of Strawberry. bioRxiv 11. doi: 10.1101/2021.11.03.467115
Harris D. C., Stickels J. E. (1981). Crown rot (Phytophthora cactorum) in glasshouse-grown strawberries at East malling research station, England, UK. Plant Pathol. 30, 205–212. doi: 10.1111/j.1365-3059.1981.tb01258.x
Hautsalo J., Vestberg M., Parikka P., Kukkonen S., Karhu S., Tahvonen R. (2016). Biological control of strawberry crown rot is substrate dependent phenomenon. J. Berry Res. 6, 65–79. doi: 10.3233/JBR-150110
Hohl H. R., Stössel P. (1976). Host–parasite interfaces in a resistant and a susceptible cultivar of solanum tuberosum inoculated with phytophthora infestans: tuber tissue. Can. J. Bot. 54 (9), 900–912. doi: 10.1139/b76-094
Holmes G. J., Mansouripour S. M., Hewavitharana S. (2020). Strawberries at the crossroads: Management of soilborne diseases in California without methyl bromide. Phytopathology 110, 620 956–968. doi: 10.1094/PHYTO-11-19-0406-IA
Husson C., Aguayo J., Revellin C., Frey P., Ioos R., Marçais B. (2015). Evidence for homoploid speciation in Phytophthora alni supports taxonomic reclassification in this species complex. Fungal Genet. Biol. 77, 12–21. doi: 10.1016/j.fgb.2015.02.013
Ingle R. A., Carstens M., Denby K. J. (2006). PAMP recognition and the plant–pathogen arms race. Bioessays 28 (9), 880–889.
Ito Y., Nishizawa-Yokoi A., Endo M., Mikami M., Toki S. (2015). CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem. Biophys. Res. Commun. 467 (1), 76–82. doi: 10.1016/j.bbrc.2015.09.117
Jeffers W. F., Scott D. H. (1953). “Red stele disease of strawberry,” in Yearbook of Agriculture, 760–765.
Jiang W., Bikard D., Cox D., Zhang F., Marraffini L. A. (2013). RNA-Guided editing of bacterial genomes using CRISPR-cas systems. Nat. Biotechnol. 31 (3), 233–239. doi: 10.1038/nbt.2508
Jiang R. H., Tripathy S., Govers F., Tyler B. M. (2008). RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc. Natl. Acad. Sci. 105 (12), 4874–4879. doi: 10.1073/pnas.0709303105
Kennedy B. W., King T. H. (1962). Angular leaf spot of strawberry caused by Xanthomonas fragariae sp. nov. Phytopathol. 52, 873–875.
KLIEBENSTEIN D. J. (2004). Secondary metabolites and plant/environment interactions: a view through Arabidopsis thaliana tinged glasses. Plant Cell Environ. 27, 675–684. doi: 10.1111/j.1365-3040.2004.01180.x
Larroque M., Belmas E., Martinez T., Vergnes S., Ladouce N., Lafitte C., et al. (2013). Pathogen-associated molecular pattern-triggered immunity and resistance to the root pathogen Phytophthora parasitica in Arabidopsis. J. Exp. Bot. 64 (12), 3615–3625.
Lebert H., Cohn F. (1870). Über die fäule der cactusstämme (On the rot of cactus stems). Beitr. Biol. Pflanz 1, 51–57.
Lederer W., Seemuller E. (1992). Investigations on the predisposition of strawberry plants to crown rot (Phytophthora cactorum). J. Plant Dis. Prot. 99 225, 233.
Lee S., Noh Y. H., Roach J. A., Mangandi J., Verma S., Whitaker V. M., et al. (2016). A high-throughput genotyping system combining rapid DNA extraction and high-resolution melting analysis in allo-octoploid strawberry. In VIII Int. Strawberry Symposium 1156, 89–94.
Lee S., Whitaker V. M., Hutton S. F. (2016). Mini review: potential applications of non-host resistance for crop improvement. Front. Plant Sci. 7, 997. doi: 10.3389/fpls.2016.00997
Lewis R. S., Linger L. R., Wolff M. F., Wernsman E. A. (2007). The negative influence of N-mediated TMV resistance on yield in tobacco: linkage drag versus pleiotropy. Theor. Appl. Genet. 115 (2), 169–178.
Lilja A., Karjalainen R., Parikka P., Kammiovirta K., Nuorteva H. (1998). Pathogenicity and genetic variation of Phytophthora cactorum from silver birch and strawberry. Eur. J. Plant Pathol. 104, 529–535. doi: 10.1023/A:1008644804415
Liu T., Li M., Liu Z., Ai X., Li Y. (2021). Reannotation of the cultivated strawberry genome and establishment of a strawberry genome database. Horticulture Res. 8 (1), 1–9. doi: 10.1038/s41438-021-00476-4
Maas J. L. (1998). Compendium of strawberry diseases USDA. Belsville: APS. doi: 10.1094/9780890546178
Mangandi J., Verma S., Osorio L., Peres N. A., van de Weg E., Whitaker V. M. (2017). Pedigree-based analysis in a multiparental population of octoploid strawberry reveals QTL alleles conferring resistance to phytophthora cactorum. G3: Genes Genomes Genet. 7 (6), 1707–1719. doi: 10.1534/g3.117.042119
Man in ‘t Veld W. A., Rosendahl C. H. M., Hong C. (2012). Phytophthora x serendipita sp. nov. and p. xpelgrandis, two destructive pathogens generated by natural hybridization. Mycologia 104, 1390–1396. doi: 10.3852/11-272
Marin M. V., Peres N. A. (2021). Improving the toolbox to manage phytophthora diseases of strawberry: searching for chemical alternatives. Plant Health Progress 22, 294–299. doi: 10.1094/PHP-02-21-0034-FI
Marin M. V., Seijo T. E., Baggio J. S., Whitaker V. M., Peres N. A. (2022a). Resistance of strawberry cultivars and the effects of plant ontogenesis on Phytophthora cactorum and P. nicotianae causing crown rot. Plant Dis. (First look). doi: 10.1094/PDIS-01-22-0203-RE
Marin M. V., Seijo T., Mertely J., Peres N. A. (2019). First report of crown rot caused by phytopythium helicoides on strawberry in the americas. Plant Dis. 103, 2696–2696. doi: 10.1094/PDIS-03-19-0658-PDN
Marin M. V., Seijo T., Oliveira M. S., Zuchelli E., Mertely J., Peres N. A. (2018). First report of phytophthora nicotianae causing crown rot of strawberry in the united states. Plant Dis. 102, 1463–1463. doi: 10.1094/PDIS-08-17-1333-PDN
Marin M. V., Seijo T., Zuchelli E., Peres N. A. (2021). Resistance to mefenoxam of Phytophthora cactorum and P. nicotianae causing crown and leather rot in Florida strawberry. Plant Dis. 105, 3490–3495. doi: 10.1094/PDIS-11-20-2474-RE
Marin M. V., Seijo T. E., Zuchelli E., Peres N. A. (2022b). Detection and characterization of quinone outside inhibitor-resistant Phytophthora cactorum and p. nicotianae causing leather rot in Florida strawberry. Plant Dis. 106, 1203–1208.
Martín-Pizarro C., Triviño J. C., Posé D. (2019). Functional analysis of the TM6 MADS-box gene in the octoploid strawberry by CRISPR/Cas9-directed mutagenesis. J. Exp. Bot. 70 (3), 949–961. doi: 10.1093/jxb/ery400
Mei X., Yang M., Ding X., Bi Y., Chen L., Deng W., et al. (2014). Proteomic analysis of zoxamide-induced changes in phytophthora cactorum. Pesticide Biochem. Physiol. 113, 31–39. doi: 10.1016/j.pestbp.2014.06.004
Mertely J., Marin M., Martin R., Peres N. A. (2020). Evaluation of biological products for the control of phytophthora crown rot in annual strawberry, 2018-2019. Plant Dis. Manage. Rep. 14, PF003.
Meszka B., Michalecka ,. M. (2016). Identification of Phytophthora spp. isolated from plants and soil samples on strawberry plantations in Poland. J. Plant Dis. Prot. 123, 29–36.
Miah G., Rafii M. Y., Ismail M. R., Puteh A. B., Rahim H. A., Asfaliza R., et al. (2013). Blast resistance in rice: a review of conventional breeding to molecular approaches. Mol. Biol. Rep. 40 (3), 2369–2388. doi: 10.1007/s11033-012-2318-0
Mussell H. W., Staples R. C. (1971). Phytoalexin-like compounds apparently involved in strawberry resistance to Phytophthora fragariae. Phytopathology 61, 515–517.
Nellist C. F., Armitage A. D., Bates H. J., Sobczyk M. K., Luberti M., Lewis L. A., et al. (2021). Comparative analysis of host-associated variation in Phytophthora cactorum. Front. Microbiol. 12, 679936. doi: 10.3389/fmicb.2021.679936
Nellist C. F., Vickerstaff R. J., Sobczyk M. K., Marina-Montes C., Wilson F. M., Simpson D. W., et al. (2019). Quantitative trait loci controlling Phytophthora cactorum resistance in the cultivated octoploid strawberry (Fragaria× ananassa). Horticulture Res. 6 (1), 1–14.
Nienhaus F. (1960). Das wirtsspektrum von Phytophthora cactorum (Leb. et Cohn) schroet. (The Host Spectr. Phytophthora cactorum [Leb Et Cohn] Schroet.) Phytopathol. Z. 38, 33–68.
Noh Y. H., Oh Y., Mangandi J., Verma S., Zurn J. D., Lu Y. T., et al. (2018). High-throughput marker assays for FaRPc2-mediated resistance to phytophthora crown rot in octoploid strawberry. Mol. Breed. 38 (8), 104. doi: 10.1007/s11032-018-0861-7
Oh Y., Chandra S., Lee S. (2020). Development of subgenome-specific markers for FaRXf1 conferring resistance to bacterial angular leaf spot in allo-octoploid strawberry. Int. J. Fruit Sci., 1–13. doi: 10.1080/15538362.2019.1709116
Oliveira M. S., Peres N. A. (2020). Common strawberry diseases in Florida: PP354, 11/2019. EDIS 2020 (2). doi: 10.32473/edis-pp354-2020
Oudemans P., Coffey M. D. (1991). Isozyme comparison within and among worldwide sources of three morphologically distinct species of phytophthora. Mycol. Res. 95, 19–30. doi: 10.1016/S0953-7562(09)81358-0
Peace C. P. (2017). DNA-Informed breeding of rosaceous crops: promises, progress and prospects. Horticulture Res. 4 (1), 1–13. doi: 10.1038/hortres.2017.6
Parikka P. (2003). Susceptibility of strawberry varieties to crown rot (Phytophthora cactorum) in greenhouse tests. Acta Hortic. 626, 183–189.
Peterson P. D. Jr., Campbell C. L., Griffith C. S. (1992). James E. teschemacher and the cause and management of potato blight in the united states. Plant Dis. 76, 754–756.
Petrasch S., Mesquida-Pesci S. D., Pincot D. D., Feldmann M. J., López C. M., Famula R., et al. (2022). Genomic prediction of strawberry resistance to postharvest fruit decay caused by the fungal pathogen botrytis cinerea. G3 12 (1), jkab378. doi: 10.1093/g3journal/jkab378
Petre B., Kamoun S. (2014). How do filamentous pathogens deliver effector proteins into plant cells? PloS Biol. 12 (2), e1001801. doi: 10.1371/journal.pbio.1001801
Pettitt T. R., Pegg G. F. (1994). Sources of crown rot (Phytophthora cactorum) infection in strawberry and the effect of cold storage on susceptibility to the disease. Ann. Appl. Biol. 125 (2), 279–292. doi: 10.1111/j.1744-7348.1994.tb04969.x
Pröbsting M., Schenke D., Hossain R., Häder C., Thurau T., Wighardt L., et al. (2020). Loss of function of CRT1a (calreticulin) reduces plant susceptibility to verticillium longisporum in both arabidopsis thaliana and oilseed rape (Brassica napus). Plant Biotechnol. J. 18, 2328–2344. doi: 10.1111/pbi.13394
Rahman M., Ojiambo P., Louws F. (2015). Initial inoculum and spatial dispersal of colletotrichum gloeosporioides, the causal agent of strawberry anthracnose crown rot. Plant Dis. 99, 80–86. doi: 10.1094/PDIS-02-13-0144-RE
Ratti M. F., Farrer R. A., Cano L. M., Faedda R., Goss E. M. (2019). Evaluation of high-resolution melting for rapid differentiation of phytophthora hybrids and their parental species. Plant Dis. 103, 2295–2304. doi: 10.1094/PDIS-12-18-2291-RE
Robideau G. P., De Cock A. W., Coffey M. D., Voglmayr H., Brouwer H., Bala K., et al. (2011). DNA Barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Mol. Ecol. Resour. 11, 1002–1011. doi: 10.1111/j.1755-0998.2011.03041.x
Rojas J. A., Miles T. D., Coffey M. D., Martin F. N., Chilvers M. I. (2017). Development and application of qPCR and RPA genus-and species-specific detection of phytophthora sojae and p. sansomeana root rot pathogens of soybean. Plant Dis. 101 (7), 1171–1181.
Salinas N., Verma S., Peres N., Whitaker V. M. (2019). FaRCa1: a major subgenome-specific locus conferring resistance to colletotrichum acutatum in strawberry. Theor. Appl. Genet. 132 (4), 1109–1120. doi: 10.1007/s00122-018-3263-7
Samtani J. B., Rom C. R., Friedrich H., Fennimore S. A., Finn C. E., Petran A., et al. (2019). The status and future of the strawberry industry in the united states. HortTechnology 29 (1), 11–24. doi: 10.21273/HORTTECH04135-18
Santos B. M., Gilreath J. P., López -Aranda J. M., Miranda L., Soria C., Medina J. J., et al. (2006). Performance of methyl bromide alternatives for strawberry production in Florida and Spain. Proc. Fla. State Hortic. Soc 119.
Schafleitner S., Bonnet A., Pedeprat N., Rocca D., Chartier P., Denoyes B. (2013). Genetic variation of resistance of the cultivated strawberry to crown rot caused by phytophthora cactorum. J. Berry Res. 3 (2), 79–91. doi: 10.3233/JBR-130052
Seemüller E., Schmidle A. (1979). Einfluß der herkunft von phytophthora cactorum-isolaten auf ihre virulenz an apfelrinde, erdbeerfrüchten und erdbeerfru¨chten. Phytopathologische Z. 94, 218–225. doi: 10.1111/j.1439-0434.1979.tb01553.x
Seijo T., Marin M., Whitaker V. M., Peres N. A. (2020). Evaluation of strawberry cultivars and advanced breeding selections for resistance to Phytophthora crown rot caused by Phytophthora cactoru-2020. Plant Dis. Manage. Rep. 14, PF086.
Seo S., Nakaho K., Hong S. W., Takahashi H., Shigemori H., Mitsuhara I. (2016). L-Histidine induces resistance in plants to the bacterial pathogen Ralstonia solanacearum partially through the activation of ethylene signaling. Plant Cell Physiol. 57 (9), 1932–1942.
Shalem O., Sanjana N. E., Hartenian E., Shi X., Scott D. A., Mikkelsen T. S., et al. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343 (6166), 84 LP–84 87. doi: 10.1126/science.1247005
Shaw D. V., Hansen J., Browne G. T. (2006). Genotypic variation for resistance to Phytophthora cactorum in a California strawberry breeding population. J. Am. Soc. Hortic. Sci. 131 (5), 687–690. doi: 10.21273/JASHS.131.5.687
Shaw D. V., Hansen J., Browne G. T., Shaw S. M. (2008). Components of genetic variation for resistance of strawberry to Phytophthora cactorum estimated using segregating seedling populations and their parent genotypes. Plant Pathol. 57 (2), 210–215. doi: 10.1111/j.1365-3059.2007.01773.x
Shirasawa K., Hirakawa H., Nakayama S., Sasamoto S., Tsuruoka H., Minami C., et al. (2021). A chromosome-scale strawberry genome assembly of a Japanese variety, reikou. bioRxiv. doi: 10.1101/2021.04.23.441065
Simpson D. W., Bell J. A., Harris D. C. (1994). “Breeding for resistance to fungal diseases in strawberry,” in Progress in temperate fruit breeding (Dordrecht: Springer), 63–66.
Srivastava J., Singh A., Sharma R. (2020). Diseases of apples and their management. Dis. Fruits Vegetable Crops: Recent Manage. Approaches.
Steinite I., Levinsh G. (2003). Possible role of trichomes in resistance of strawberry cultivars against spider mite. Acta Univ. Latviensis 662, 59–65.
Stensvand A., Herrero M. L., Talgø V. (1999). Crown rot caused by Phytophthora cactorum in Norwegian strawberry production. EPPO Bull. 29 (1-2), 155–158. doi: 10.1111/j.1365-2338.1999.tb00809.x
Summers R. W., Brown J. K. M. (2013). Constraints on breeding for disease resistance in commercially competitive wheat cultivars. Plant Pathol. 62 (S1), 115–121. doi: 10.1111/ppa.12165
Tapia R., Abd-Elrahman A., Osorio L., Whitaker V. M., Lee S. (2022). Combining canopy reflectance spectrometry and genome-wide prediction to increase response to selection for powdery mildew resistance in cultivated strawberry. J. Exp. Botany. doi: 10.1093/jxb/erac136
Terry L. A. (2002). “Natural disease resistance in strawberry fruit and Geraldton waxflower flowers,” (United Kingdom: Cranfield University).
Toljamo A., Blande D., Kärenlampi S., Kokko H. (2016). Reprogramming of strawberry (Fragaria vesca) root transcriptome in response to phytophthora cactorum. PloS One 11 (8), e0161078. doi: 10.1371/journal.pone.0161078
Toljamo A., Blande D., Munawar M., Kärenlampi S. O., Kokko H. (2019). Expression of the GAF Sensor, Carbohydrate-Active Enzymes, Elicitins, and RXLRs Differs Markedly Between Two Phytophthora cactorum Isolates. Phytopathology. 109 (5), 726–735. doi: 10.1094/PHYTO-04-18-0136-R
Tucker C. M. (1933). Distribution of the genus phytophthora. Univ. Mo. Agric. Exp. Stn. Res. Bull. 184, 80.
Van Loon L. C., Rep M., Pieterse C. M. J. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162.
van Poppel P. M. (2009). The phytophthora infestans avirulence gene PiaAvr4 and its potato counterpart R4.
Wang Y., Cordewener J. H., America A. H., Shan W., Bouwmeester K., Govers F. (2015). Arabidopsis lectin receptor kinases LecRK-IX. 1 and LecRK-IX. 2 are functional analogs in regulating phytophthora resistance and plant cell death. Mol. Plant-Microbe Interact. 28 (9), 1032–1048.
Wang F., Zhang F., Chen M., Liu Z., Zhang Z., Fu J., et al. (2017). Comparative transcriptomics reveals differential gene expression related to colletotrichum gloeosporioides resistance in the octoploid strawberry. Front. Plant Sci. 8, 779. doi: 10.3389/fpls.2017.00779
Whitaker V. M., Knapp S. J., Hardigan M. A., Edger P. P., Slovin J. P., Bassil N. V., et al. (2020). A roadmap for research in octoploid strawberry. Horticulture Res. 7 (1), 1–17. doi: 10.1038/s41438-020-0252-1
Wilson F. M., Harrison K., Armitage A. D., Simkin A. J., Harrison R. J. (2019). CRISPR/Cas9-mediated mutagenesis of phytoene desaturase in diploid and octoploid strawberry. Plant Methods 15 (1), 1–13. doi: 10.1186/s13007-019-0428-6
Witek K., Lin X., Karki H. S., Jupe F., Witek A. I., Steuernagel B., et al. (2016). A complex resistance locus in Solanum americanum recognizes a conserved phytophthora effector. Nat. Plants 7 (2), 198–208.
Xing S., Jia M., Wei L., Mao W., Abbasi U. A., Zhao Y., et al. (2018). CRISPR/Cas9-introduced single and multiple mutagenesis in strawberry. J. Genet. Genomics 45 (12), 685–687. doi: 10.1016/j.jgg.2018.04.006
Yang M., Duan S., Mei X., Huang H., Chen W., Liu Y., et al. (2018). The Phytophthora cactorum genome provides insights into the adaptation to host defense compounds and fungicides. Sci. Rep. 8 (1), 1–11. doi: 10.1038/s41598-018-24939-2
Yang H., Wang H., Shivalila C. S., Cheng A. W., Shi L., Jaenisch R. (2013). One-step generation of mice carrying reporter and conditional alleles by CRISPR/cas-mediated genome engineering. Cell 154 (6), 1370. doi: 10.1016/j.cell.2013.08.022
Yuan Y., Chung J. D., Fu X., Johnson V. E., Ranjan P., Booth S. L., et al. (2009). Alternative splicing and gene duplication differentially shaped the regulation of isochorismate synthase in populus and arabidopsis. Proc. Natl. Acad. Sci. 106 (51), 22020–22025. doi: 10.1073/pnas.0906869106
Zentmyer G. A. (1980). Phytophthora cinnamomic and the disease it causes. monogr. no. 10. Am. Phytopathological Society St. Paul Minm. 96, 1–96.
Zhang L., Huang X., He C., Zhang Q. Y., Zou X., Duan K., et al. (2018). Novel fungal pathogenicity and leaf defense strategies are revealed by simultaneous transcriptome analysis of Colletotrichum fructicola and strawberry infected by this fungus. Front. Plant Sci. 9, 434. doi: 10.3389/fpls.2018.00434
Zhang M., Liu Q., Yang X., Xu J., Liu G., Yao X., et al. (2020). CRISPR/Cas9-mediated mutagenesis of Clpsk1 in watermelon to confer resistance to Fusarium oxysporum f.sp. niveum. Plant Cell Rep. 39, 589–595. doi: 10.1007/s00299-020-02516-0
Zhang H., Zhang J., Wei P., Zhang B., Gou F., Feng Z., et al. (2014). The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12 (6), 797–807. doi: 10.1111/pbi.12200
Keywords: DNA marker, marker-assisted selection (MAS), genome assembly, disease resistance (R) genes, CRISPR genome editing, octoploid reference genome
Citation: Sharma S, Marin MV, Lee MB, Baggio JS, Peres NA and Lee S (2022) Genomic approaches for improving resistance to Phytophthora crown rot caused by P. cactorum in strawberry (Fragaria × ananassa). Front. Agron. 4:941111. doi: 10.3389/fagro.2022.941111
Received: 11 May 2022; Accepted: 21 July 2022;
Published: 22 August 2022.
Edited by:
Jonathan Spencer West, Rothamsted Research, United KingdomReviewed by:
Yunpeng Cao, Chinese Academy of Sciences (CAS), ChinaHåvard Eikemo, Norwegian Institute of Bioeconomy Research (NIBIO), Norway
Copyright © 2022 Sharma, Marin, Lee, Baggio, Peres and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seonghee Lee, c2VvbmdoZWUxMDVAdWZsLmVkdQ==
†Present addresses: Sadikshya Sharma, Department of Plant Science, University of California, Davis, Davis, CA, United States
Juliana S. Baggio, Syngenta Crop Protection, Vero Beach, FL, United States
‡These authors have contributed equally to this work
 Sadikshya Sharma
Sadikshya Sharma Marcus V. Marin
Marcus V. Marin Juliana S. Baggio1†
Juliana S. Baggio1† Natalia A. Peres
Natalia A. Peres Seonghee Lee
Seonghee Lee
