- 1Department of Biochemistry, Molecular Biology, Entomology, and Plant Pathology, Mississippi State University, Mississippi State, MS, United States
- 2Department of Plant and Soil Science, Mississippi State University, Mississippi State, MS, United States
- 3Crop Science Research Laboratory, United States Department of Agriculture – Agricultural Research Service (USDA-ARS), Mississippi State, MS, United States
- 4Department of Soil and Crop Sciences, Texas A&M University, College Station, TX, United States
Upland cotton is sensitive to 2,4-dichlorophenoxyacetic acid (2,4-D), and spray drift from adjacent crops can cause yield loss. The identification of potentially 2,4-D-tolerant cotton chromosome substitution (CS) lines with well-characterized tolerance mechanisms could provide a significant step into the development and genetic improvement of upland cotton. Experiments were conducted to understand the possible mechanism of herbicide tolerance in CS-T04-15, CS-T07, and CS-B15sh, 2,4-D herbicide-tolerant cotton CS lines compared with TM-1, the 2,4-D herbicide-susceptible recurrent parent of the CS line. Using [14C]2,4-D, the percent absorption rate and the translocation patterns of the 14C-labeled herbicide at 6 to 48 h after treatment (HAT) were determined. The tolerant cotton CS lines showed 15%–19% [14C]2,4-D uptake, while TM-1 exhibited a reduced uptake of only 1.4% [14C]2,4-D at 24 HAT. Distribution of the absorbed [14C]2,4-D showed that 2%–5% was translocated outside the treated leaf in the tolerant CS lines. In contrast, 77% of the herbicide was translocated above and below the treated leaf in line TM-1. Interestingly, CS-T04-15 showed a restricted movement of 14C below the treated leaf at 6 to 48 HAT, suggesting novel information on the mechanisms of herbicide tolerance to 2,4-D in cotton. This finding is the first report on upland cotton demonstrating a complex differential uptake and translocation associated with herbicide tolerance for [14C]2,4-D in cotton CS lines.
Introduction
Herbicides used in production agriculture are typically synthetic chemicals used to control the growth of undesirable plants. In cotton, herbicides are the most widely used weed management tool because of their speed, flexibility, and low cost. These compounds are grouped into families based on a specific mode of action, including growth regulation, cell membrane disruption, and inhibition of amino acid synthesis, lipid synthesis, photosynthesis, pigment formation, or seedling growth (Gunsolus and Curran, 1999). Since most herbicides are small molecules designed to inhibit specific target sites critical to plant biochemical pathways and physiological processes, understanding the mode of action of herbicide chemistries is essential for developing appropriate weed management strategies in agricultural production systems (Dayan et al., 2010).
The development of 2,4-dichlorophenoxyacetic acid (2,4-D) initiated the revolution of chemical weed control in the 1940s (Peterson et al., 2016). Discovery of this growth-promoting substance's toxic effects on plants and its recognition as a potential chemical warfare agent during World War II prompted the development of 2,4-D as a selective herbicide which targets dicot species (Peterson, 1967). 2,4-D is a synthetic auxin that contains phenoxyalkanoic acid as a major chemical group and mimics the natural auxin in plants, indole-3-acetic acid (IAA) (Song, 2014). Auxin herbicides like 2,4-D can mimic an overdose of IAA, causing massive upregulation of ethylene biosynthesis and abscisic acid (ABA) response (Sterling and Hall, 1997; Grossman, 2003; Perez et al., 2022). Cereal crops are generally tolerant to 2,4-D relative to most dicotyledonous crops and weeds. In sensitive genotypes, the 2,4-D-induced auxinic herbicide syndrome includes leaf epinasty, tissue swelling, stem curling, tissue necrosis, senescence, and plant death (Grossman, 2007; Grossman, 2010). At the vegetative stages, cotton yield losses of up to 49% may occur from 2,4-D particle drift or misapplication exposures, whereas a yield loss of up to 71% is possible due to misapplication at the preflowering squaring stages (Egan et al., 2014). Cotton is not as sensitive to 2,4-D once they start to develop bolls, but can incur yield losses of up to 6% from drift exposures.
Herbicide tolerance in crops grown at a commercial scale is primarily based on transgenic cultivars expressing specific genes for enzymes or proteins that block the action or degrade the active ingredient of the herbicide. The most important example is the glyphosate-tolerant 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) transgene from a soil bacterium that confers resistance to glyphosate in the widely grown transgenic Roundup Ready soybean and cotton cultivars (Delannay et al., 1995; Nida et al., 1996). Transgenic cotton varieties have the tolerance to at least six herbicide modes of action inhibiting PS-II, EPSPS, acetolactate synthase (ALS), glutamine synthetase, hydroxyphenylpyruvate dioxygenase, and synthetic auxins (Vulchi et al., 2022). The commercial production of these herbicide-tolerant cotton varieties in the USA has expanded significantly from 10% in 1997 to 95% in 2019 (ISAAA, 2020). Tolerance to auxin herbicides in broadleaf crop cultivars and weed populations has also been reported (Wright et al., 2010; Preston and Malone, 2014; Robinson et al., 2015; Rey-Caballero et al., 2016; Shergill et al., 2018; Kumar et al., 2019).
Understanding the herbicide tolerance mechanism is central to identifying and developing appropriate weed management control measures based on the response to specific modes of action. The tolerance mechanisms may involve altered absorption or translocation of the herbicide active compound in the plant which leads to reduced herbicide injury. In general, herbicides traverse the cell wall, plasma membrane, and different plant cell organelles to reach their particular site of action and induce toxicity in plants (Sterling, 1994). As early as the 1950s, experiments using carbon-14 [14C]-labeled herbicides, including 2,4-D, were conducted to investigate the translocation, distribution, and metabolism of these compounds in plants (Day, 1952; Barrier and Loomis, 1957; Yamaguchi and Crafts, 1958). The mechanisms of 2,4-D resistance can be inferred from [14C]2,4-D uptake, translocation, and degradation patterns. For example, in 2,4-D-resistant and 2,4-D-susceptible waterhemp (Amaranthus tuberculatus), herbicide uptake and translocation patterns were the same, while an increased metabolism of the compound was observed in 2,4-D-resistant waterhemp (Figueiredo et al., 2018). Similarly, there was no correlation between resistance and reduced translocation of [14C]2,4-D in wild radish (Raphanus raphanistrum L.) (Goggin et al., 2018). In contrast to the above findings, 2,4-D resistance in mustard has been associated with a reduced translocation from the treated leaf (Dang et al., 2018). A resistant ground ivy (Glechoma hederacea), a perennial weed in lawn turf, also demonstrated reduced herbicide uptake (Kohler et al., 2004). Other factors, including temperature regimes, also impacted the efficacy, absorption, and translocation of 2,4-D in both resistant and susceptible types of ragweed species (Ambrosia artemisiifolia and Ambrosia trifida) (Ganie et al., 2017).
Petiolar absorption of [phenyl-14C]-2,4-D has been examined in herbicide-tolerant cotton expressing the tfdA transgene (Laurent et al., 2000). The tfdA transgene isolated from the soil bacterium, Alcaligenes eutrophus (strain JMP134), encodes a 2,4-dichlorophenoxyacetate monooxygenase that degrades 2,4-D herbicide into non-phytotoxic dichlorophenol (DCP) (Streber and Willmitzer, 1989; Lyon et al., 1993). In the transgenic herbicide-tolerant cotton, 2,4-D was degraded to 2,4-dichlorophenol (2,4-DCP), which was then converted into various polar metabolites, including 2,4-DCP-β-О-glucoside and the complex glucosides, 2,4-DCP-(6-О-malonyl)glucoside and 2,4-DCP-(6-О-sulfate)glucoside (Laurent et al., 2000). However, only recently that 2,4-D-tolerant cotton varieties using the Enlist™ technology (Dow Agrosciences, Indianapolis, IN) have been introduced to the commercial landscape. The Enlist™ technology uses an aryloxyalkanoate dioxygenase, AAD-12, from Delftia acidovorans (Wright et al., 2010; Braxton et al., 2017) to degrade 2,4-D into non-phytotoxic compounds, and this transgene technology provides resistance as well to several additional herbicides, including triclopyr and fluroxypyr (Wright et al., 2010). Aside from the AAD transgenes, no other forms of tolerance to auxin herbicides, particularly 2,4-D, have been developed in upland cotton.
The narrow genetic base of cotton limits the genetic improvement of upland cotton varieties. While it is widely recognized in cotton that exotic germplasm contains potentially valuable genes, the exotic gene pools from Gossypium species other than G. hirsutum L. mostly remain untapped, uncharacterized, and underutilized due to the paucity of information about the beneficial alleles in these species and the biological and technical challenges associated with interspecific introgression of valuable traits. Solutions for the weed problems are best addressed through the breeding of herbicide-resistant or herbicide-tolerant varieties. Genetic diversity exists in the relatives of upland cotton, such as the wild accessions of G. hirsutum L. and the tetraploid species, G. barbadense L., G. tomentosum Nuttal ex Seeman, and G. mustelinum Meers ex Watt. Although these wild accessions potentially contain novel genetic solutions to diverse breeding needs, commercial breeding programs generally avoid unadapted germplasm due to their adverse effects on agronomic performance of multiple quality traits. A number of chromosome substitution (CS) lines of upland cotton were jointly developed in the genetic background of inbred TM-1 with introgressions from G. barbadense, G. tomentosum, and G. mustelinum (Saha et al., 2012). Previous studies show that the CS lines are a novel breeding tool for targeted introgression of many valuable traits such as resistance to drought, nematodes, and fusarium wilt, and improved agronomic and fiber traits in upland cotton (Reddy et al., 2020; Ulloa et al., 2020). In our previous work, we identified 2,4-D-tolerant and 2,4-D-susceptible CS lines of G. hirsutum containing specific chromosome introgressions from G. barbadense, G. tomentosum, and/or G. mustelinum from greenhouse and field experiments (Perez et al., 2021). We also confirmed the genetic inheritance of the 2,4-D tolerance trait through progeny test. This paper investigates the tolerance mechanisms associated with absorption and translocation of 2,4-D by comparing the selected 2,4-D-tolerant non-transgenic CS lines with the susceptible and recurrent parent TM-1.
Materials and methods
Plant materials and experimental design
Selected cotton CS lines, including CS-T04-15 (301-8), CS-T07 (206-2), CS-B15sh (31-4), and the susceptible cultivar Texas Marker 1 (TM-1), were used as genetic materials in the study. In previous experiments, we conducted screening of cotton CS lines for tolerance to 2,4-D at 1× field application rate (1.12 kg ae ha−1) under both greenhouse and field conditions (Perez, 2021). Two CS lines, CS-T04-15 and CS-T07, were identified showing tolerance to 2,4-D in both field and greenhouse experiments (Figure 1A). In addition, CS-B15sh showed reduced injury exhibiting some form of tolerance to the herbicide compared to TM-1 (Figure 1C). CS-T07 (206-2), CS-B15sh (31-4) are considered to be largely isogenic to TM-1 and two each other, aside from the substituted chromosomes; CS-T04-15, however, is not expectedly isogenic. The cotton CS line CS-T04-15 has substituted segments of chromosomes 4 and 15 pair, and CS-T07 has a complete pair of chromosome 7 from G. tomentosum in TM-1 genetic background. At the same time, CS-B15sh is homozygous for chromosome 15 in which the short arm comes from G. barbadense. Comparative analysis of the CS lines with their almost isogenic recurrent parent, the inbred TM-1 (G. hirsutum), provided a method to detect and discover important traits with specific substituted chromosome or chromosome segments from the alien species (Saha et al., 2004; Saha et al., 2006).
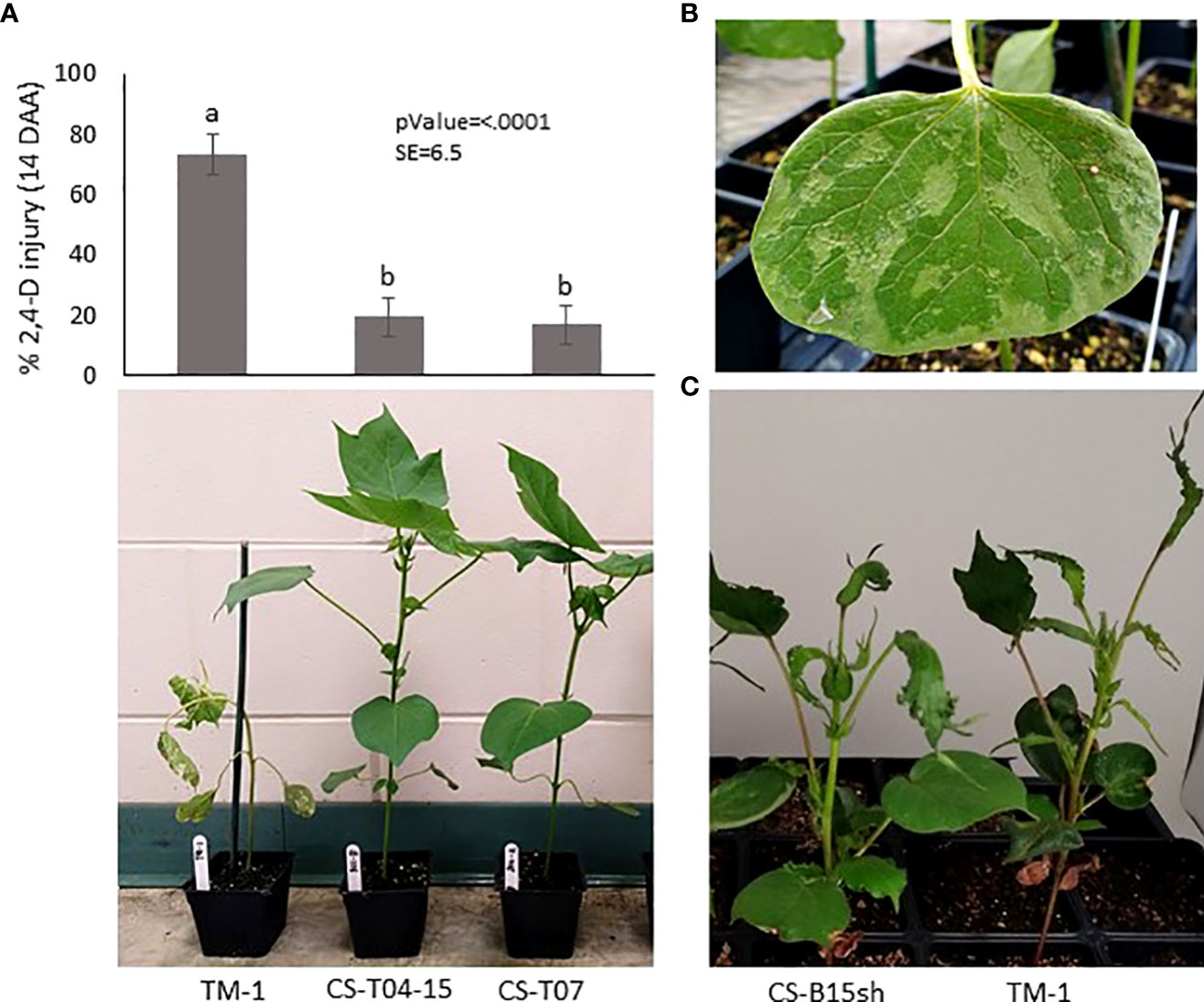
Figure 1 Herbicide injury and symptoms observed in the cotton accessions used in this study. (A) Response to 2,4-D observed on TM-1, CS-T04-15, and CS-T07 treated with 1x rate of 2,4-D in the greenhouse: Top, histogram of 2,4-D injury (mean) observed on 4-leaf stage cotton seedlings at 14 days after application (DAA), with four replications and four plants per replication; Bottom – visible 2,4-D injury of the representative individual plant at 7 DAA; (B) Close-up image of 2,4-D injury on seedling cotyledon of TM-1 after one day of treatment with 1x rate of 2,4-D; (C) Image shows reduced herbicide injury symptoms of CS-B15sh relative to TM-1 after 35 days from application of 0.75× rate of 2,4-D.
Seeds were sown directly into 18-cell Landmark plastic inserts, size 53-cm length and 26-cm width (Landmark Plastic, OH, USA), containing soil (BX PRO-MIX Growing Medium 10280, BWI Companies, Inc., Nash, TX 75569, USA), mixed with 1–2 g of basal fertilizer, (Osmocote Plus, The Scotts Company, Marysville, OH 43041, USA), so that each cell contained one cotton seedling. The experimental design for both uptake and translocation experiments followed a completely randomized design with four replications and one plant per replication. The plants were maintained in the greenhouse from November to December 2020, where the mean temperature and relative humidity were 25°C and 58%, respectively.
Treatment of radiolabeled 2,4-D solution on cotton seedlings
Cotton seedlings used for the [14C]2,4-D treatment were at the four-leaf growth stage. Before the treatment, the plants were transferred to a controlled growth chamber for 3 days of acclimatization to a 12-h photoperiod (07:00 to 19:00), 30°C/25°C day/night temperatures, and 60% relative humidity. A radiolabeled 2,4-D solution was prepared at 0.517 kBq µl−1 working concentration from a commercial stock of 2,4-dichlorophenoxy acetic acid [carboxyl-14C] containing 0.1 mCi ml−1, specific activity of 50 mCi mmol−1, and >98% radiochemical purity (ARC 0722, American Radiolabeled Chemicals, Inc., St. Louis, MO 63146, USA). The working stock of [14C]2,4-D used was prepared using distilled water as a diluent and was maintained at −20°C before the treatment.
For the uptake experiments, [14C]2,4-D was applied to the adaxial surface (upper surface) of the second true leaf of the cotton seedling. A total of 5.17 kBq was applied to each leaf, similar to the procedure described by Johnston et al. (2018), where ten 1-µl spots (total volume of 10 µl per treated leaf) were delivered using a micropipette (P-10, Eppendorf, Hamburg, Germany). The plants were allowed to air dry for 30–60 min on the bench to allow absorption of the radiolabeled herbicide on the treated leaf. The treated plants were moved to and maintained in the growth chamber under the same conditions described above until the collection and processing of tissues for analysis. At 24 h after treatment (HAT), the treated leaves were cut from individual plants and immediately washed in a 50-ml Falcon tube containing 5 ml of 1:1 water:ethanol solution for 60 s with gentle mixing using speed no. 7 (5–35 rpm) on a Roto-Shake Genie (Scientific Industries Inc., Bohemia, NY 11716, USA) to remove non-adsorbed [14C]2,4-D from the leaf surface. The 5-ml wash solutions were mixed with 14 ml of Ultima Gold liquid scintillation (PerkinElmer, Waltham, MA 02451, USA) solution and placed in 20-ml scintillation vials (VWR, Radnor, PA 19087, USA). Residual [14C]2,4-D after 24 h was counted and analyzed using a liquid scintillation counter (Perkin Elmer Tri-Carb 2900TR, Waltham, MA 02451, USA) to determine the percentage absorption of the [14C]2,4-D compound by the plant.
To determine the movement of [14C]2,4-D from the treated leaf to other parts of the plant, tissues were collected from the individual treated plants at 6, 12, 24, and 48 HAT (Johnston et al., 2018). The tissues collected from each plant were divided into three categories: treated leaf, including the petiole (T); above the treated leaf (AT), including all leaf and stem tissues collected from above sample T; and below the treated leaf (BT), including all leaf, cotyledon, and stem tissues below sample T. The collected tissues were individually wrapped in paper wipes (Kimwipes, VWR, Radnor, PA 19087, USA), labeled, and dried overnight (~12 h) in an oven at 60°C–70°C. After drying, the tissues were removed and processed in a biological oxidizer (PerkinElmer Model 307, Waltham, MA 02451, USA) using 0.5 min of combustion time. Radioactivity in terms of disintegrations per minute (DPM) of [14C] in each sample was quantified using a liquid scintillation counter (PerkinElmer Tri-Carb 2900TR, Waltham, MA 02451, USA). The overall method was followed as per the previous report of a similar experiment with weeds investigating how application timing influences the translocation of auxinic herbicides in Palmer amaranth (Johnston et al., 2018).
Data analysis
Percent absorption was calculated as the total [14C]2,4-D in disintegration per minute obtained from leaf washes at 24 HAT divided by the total [14C]2,4-D recovered from all tissue fractions of each plant, including AT, T, and BT tissues. Translocation of the radiolabeled 2,4-D was analyzed by determining the absolute radioactivity detected on each tissue fraction of the cotton lines from 6 to 48 HAT. Percent translocation of [14C]2,4-D was calculated following the formula below:
Analysis of variance and comparison of means using the Tukey–Kramer HSD was applied using the JMP 14 statistical package (SAS Institute, Cary, NC 27513-2414, USA).
Results
The overall absorption of [14C]2,4-D after 24 h, as calculated from the 14C recovered in leaf surface washes, was a modest 13% to 15% among the cotton CS lines, while TM-1 showed uptake of only 3% at 24 HAT, which was significantly lower than the CS lines (Figure 2). Both CS-B15sh and CS-T04-15 showed that 15% and 13% of the applied [14C]2,4-D was absorbed by the plant, respectively.
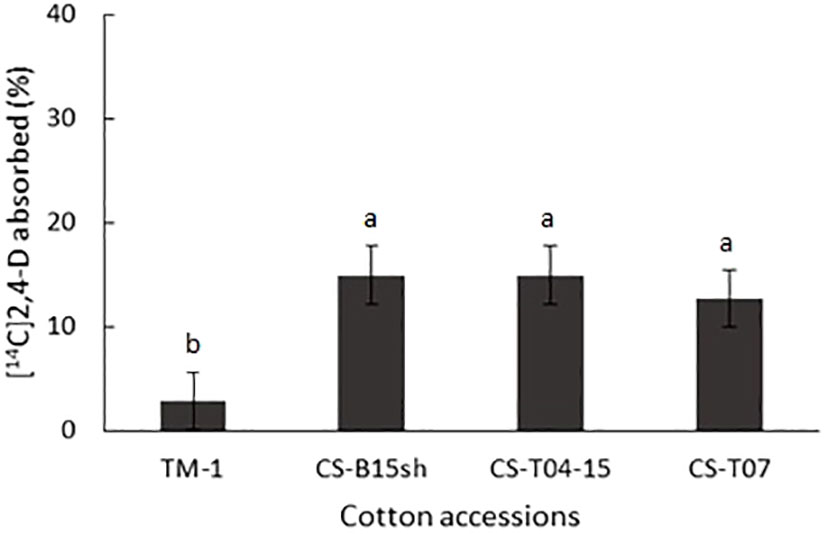
Figure 2 Percent absorption of [14C]2,4-D on cotton seedlings of TM-1, CS-B15sh (31-4), CS-T04-15 (301-8), and CS-T07 (206-2) at 24 h after treatment (HAT). Means are separated with Tukey–Kramer HSD (α = 0.05). Means of the same letter are not significantly different.
The translocation patterns of absorbed [14C]2,4-D among the different tissue fractions after 24 h are presented in Table 1. The data reflect how much [14C]2,4-D was retained in the treated leaf versus the proportion that was translocated into tissues above or below the treated leaf. All of the cotton CS lines exhibited a similar pattern in which 91%–95% of the absorbed [14C]2,4-D remained within the treated leaf tissue. In contrast, only 23% of the radioactivity entering the TM-1 plants remained in the treated leaf fraction (Table 1).

Table 1 Translocation of [14C]2,4-D in the cotton accessions after 24 h of treatment a .
In TM-1 plants, 35% of the herbicide taken into the plants moved into the tissues above the treated leaves, while 42% was transported to the tissues below the treated leaves (Table 1). The movement of 14C above and below the treated leaves exhibited similar patterns in two of the three 2,4-D-tolerant genotypes. In CS-T07, 3% of absorbed [14C]2,4-D was translocated above and below the treated leaf tissue, while in CS-B15sh, 5% of the absorbed [14C]2,4-D was translocated to the tissues above the treated leaves, and 4% was translocated into the tissues below the treated leaves. The CS-T04-15 line displayed a similar pattern for translocation, with 4% of the absorbed label translocated to the tissues above the treated leaf and a slightly lower amount of the herbicide (2%) translocated to the tissues below the treated leaf at 24 HAT (Table 1).
In an attempt to gain insights regarding radiolabeled material movement and herbicide tolerance in the cotton CS lines, we examined the time courses for the appearance of radiolabel in aerial tissues above and below the treated leaves in control and tolerant plants. The four tested lines showed different patterns of radiolabel movement over 48 h (Figure 3). In CS-T04-15, there was no apparent change in radioactivity detected in tissues above the treated leaf from 6 to 12 h, while radiolabel movement increased from 1,068 to 2,408 dpm between 12 and 24 h (Figure 3A). It showed a slight decrease to 2,183 dpm at 48 h, whereas no apparent change in the movement of label into the tissues below the treated leaf was seen over the entire time course. There was no significant difference in radiolabel movement to tissues above and below the treated leaf of CS-T04-15 at 6 to 48 HAT. These data for CS-T04-15 suggest a restricted basipetal movement of [14C]2,4-D into tissues below the treated leaf, which may point to a mechanism of herbicide tolerance distinct from the mechanism(s) underpinning tolerance in CS-T07 and CS-B15sh. A similar trend of radioactivity in both AT and BT tissues was detected in CS-T07 (Figure 3B). There was an increase in radiolabel movement from 6 to 12 h in AT and BT at 1,274 to 1,406 and 504 to 1,341, respectively. After 12 HAT, the radioactivity detected on each sample was reduced to 796 and 866 in AT and BT, respectively, at 48 h. The maximum radioactivity detected was 3,222 at 24 h in CS-T04-15 and 2,747 at 12 h in CS-T07 (Figures 3A, B). In contrast, CS-B15sh showed a steep increase of radiolabel movement over the entire time course with 13,616 being the highest detected at 48 h (Figure 3C). At 6 HAT, the radiolabel was 155 and 521 in the tissues above and below the treated leaf, respectively. From 12 to 48 h, there was a continuous increase of label detected in AT of CS-B15sh at 1,177 to 9,315. Similarly, an increase from 2,061 to 4,301 was detected in the tissues below the treated leaf of CS-B15sh from 12 to 48 h. There was no significant difference in the amount of label detected between tissue samples at 6 to 24 h. The radiolabel detected in AT (9,315) was significantly higher compared to BT (4,301) at 48 h. In TM-1, radiolabel was also increasing in both AT and BT from 6 to 48 h similar to CS-B15sh (Figure 3D). At 6 h, radioactivity detected in AT and BT was 270 and 564, respectively. The label increased to 3,399 and 2,740 at 48 h in AT and BT, respectively. The maximum radioactivity detected in TM-1 was 6,139 at 48 h.
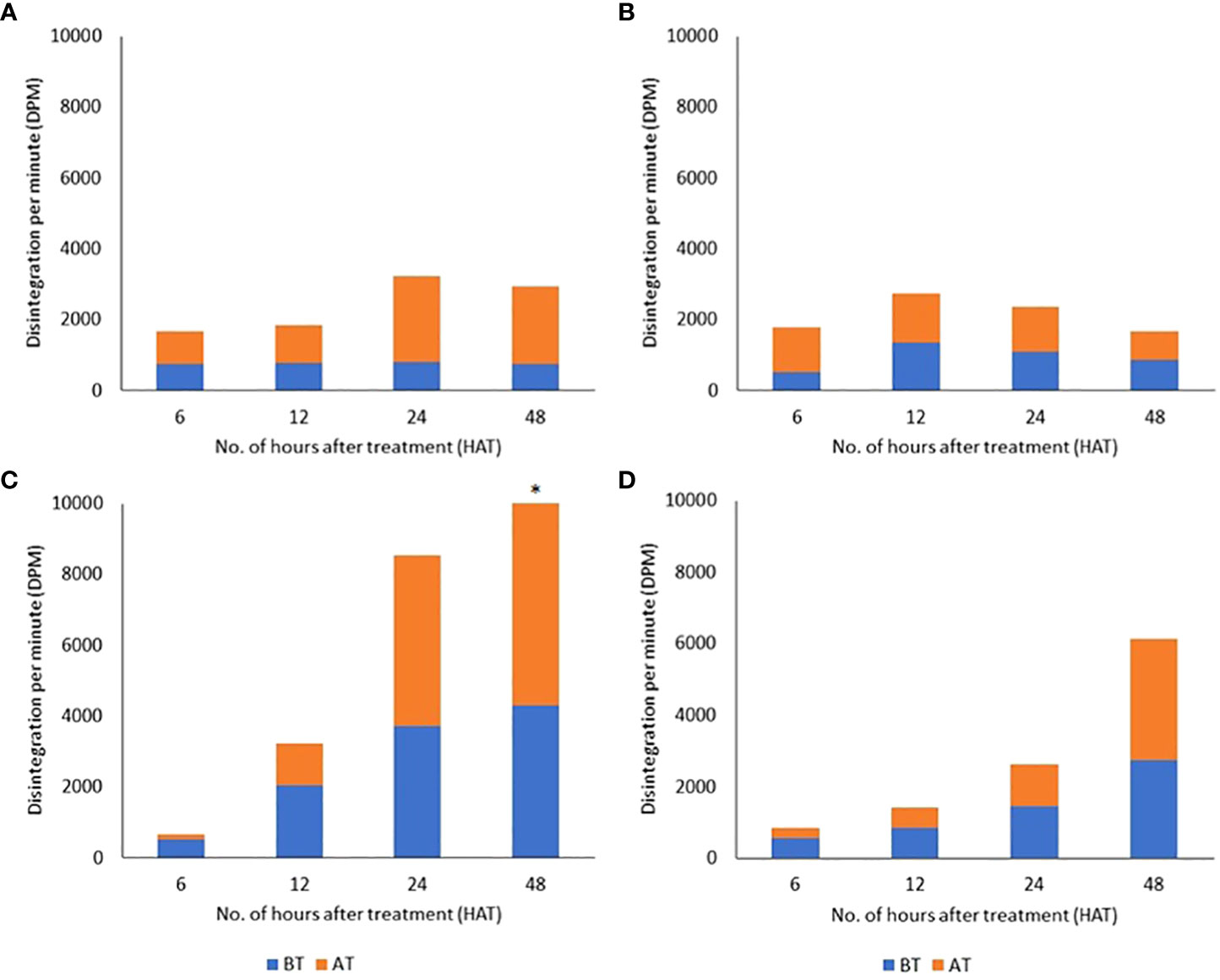
Figure 3 Distributions of [14C]2,4-D detected in tissue fractions above the treated leaf (AT) and below the treated leaf (BT) in cotton seedlings of four genotypes: CS-T04-15 (A), CS-T07 (B), CS-B15sh (C), and TM-1 (D) at 6 to 48 hours after treatment (HAT). Asterisk atop stacked bar indicates that the difference in radioactivity detected between AT and BT tissues was significant.
Discussion
The low absorption rates for 14C-labeled 2,4-D in upland cotton seen in this study were similar to previous reports on 14C uptake in 2,4-D-resistant prickly lettuce treated with [14C]2,4-D (Riar et al., 2011). Our findings of low [14C]2,4-D absorption in cotton contrast with the approximately 50% [14C]2,4-D uptake by 2,4-D-susceptible Palmer amaranth 24 h after applying the same total amount of radiolabel (5.17 kBq) to treated leaves (Johnston et al., 2018). High rates of [14C]2,4-D uptake were also observed in 2,4-D-resistant wild radish with only 2%–4% of the total of 3 kBq [14C]2,4-D remaining on leaf surfaces 24 h after treatment (Goggin et al., 2016). However, the increased uptake of [14C]2,4-D among the cotton CS lines compared with the parental line, TM-1 control, suggests physiological differences in leaf surface components between the introgressed cotton CS lines and TM-1. Our data indicate that reduced absorption of [14C]2,4-D is not associated with the mechanism of 2,4-D tolerance observed in the cotton CS lines. However, it is interesting to note that TM-1 greatly restricts the uptake of [14C]2,4-D with only 1.4% of the radiolabeled herbicide taken up after 24 h compared to the cotton CS lines. The data presented here only account for the distribution of [14C]2,4-D in the aerial tissues of treated plants 24 HAT. It will be interesting to expand these observations to additional time points and belowground tissues. In one of our greenhouse experiments, the adaxial surface of the seedling cotyledon of TM-1 treated with 1× rate exhibited severe leaf abrasions as early as 1 day after herbicide application (Figure 1B). This shows the extreme sensitivity of TM-1 to the 2,4-D herbicide that even a small amount of herbicide penetrating the cuticle is enough to kill the plant. Our previous study showed that CS lines had significant variation in their root, shoot, and leaf morphology (Ayubov et al., 2021). In addition, G. tomentosum, one of the donor parents of CS-T04-15 (301-8) and CS-T07 (206-2), is endemic to Hawaii and shows very distinct morphology from other Gossypium tetraploid species. It has hairy, silvery-green to gray-green, palmately veined leaves, sulfur-yellow corollas without petal spots; forms strongly exerted stigmas; and is devoid of leaf, bracteole, and extra-floral nectaries (Saha et al., 2006). It will be interesting to study in the future if leaf morphology differences, including leaf surface components such as cuticles, wax, and the presence of lysigenous gossypol glands with phenolic compounds, may be factors contributing to the reduced uptake of [14C]2,4-D in cotton species (Johnston et al., 2018; Benedict et al., 2004).
A pattern of reduced translocation of 14C-labeled 2,4-D from the treated leaves was reported for 2,4-D-resistant biotypes of prickly lettuce relative to the susceptible biotype (Riar et al., 2011). Reduced movement of 14C-labeled 2,4-D beyond the treated leaf was also observed in 2,4-D-resistant wild radish (Goggin et al., 2016). These patterns are associated with non-target site resistance mechanisms such as herbicide detoxification and vacuolar sequestration of herbicides or their metabolites, detected in the weeds evolving to become herbicide-resistant under intense selection pressure (Yuan et al., 2007; Delye et al., 2013). Although in a much later study involving 11 resistant populations of wild radish, there was no relationship between resistance and reduced translocation of 2,4-D (Goggin et al., 2018). Among the cotton CS lines, the reduced translocation outside the treated leaf may indicate sequestration of the [14C]2,4-D and any derived metabolites within the treated leaves of the cotton CS lines, but this has not yet been investigated. It is possible that genomic regions in CS-B15sh, CS-T04-15, and CS-T07 introgressed from the donor alien Gossypium species during CS line development could potentially carry genes that oxidize 2,4-D or add different conjugates necessary to inactivate and/or sequester 2,4-D and its derivatives (Delye et al., 2013). The source of CS fragments in CS-T04-15 and CS-T07 is G. tomentosum, a wild allotetraploid Gossypium species native to the Hawaiian area. Although the two lines carry different chromosome introgression segments of chromosomes 4 and 15 for CS-T04-15 and chromosome 7 for CS-T07 (Perez, 2021; Saha et al., 2015), it is interesting to note that both exhibited high tolerance to the herbicide (Figure 1A). Under field conditions, CS-T07 and CS-B15sh also showed heat and drought tolerance as exhibited by enhanced photosynthesis, stomatal conductance, transpiration, and pollen germination compared to TM-1 (Reddy et al., 2020).
On the other hand, the CS line CS-B15sh could potentially carry unique genetic loci from the substitutions on the short arm of chromosome 15 from G. barbadense L. (Stelly et al., 2005; Perez, 2021). Since CS-B15sh is less tolerant than the other two CS lines but more tolerant to 2,4-D than TM-1, we can equally infer that reduced translocation is also responsible for the tolerance in CS-B15sh and its lower tolerance than the other CS lines is due to the fact that it translocates a bit more 2,4-D out of the treated leaf. The tolerance of CS-B15sh to 2,4-D can be due to altered auxin perception within the plant. This study is the first report on 14C translocation in cotton from the treated leaf and beyond (above and below) the treated tissues to understand the potential mechanism of 2,4-D tolerance observed on the selected upland cotton CS lines.
Weeds are the most critical pests in US agriculture based on the consideration of the percentage of hectares treated with herbicides versus the percentage of hectares treated with other pesticides (Fernandez-Cornejo and Jans, 1999). Weed management strategies in cotton often rely primarily on chemical herbicides due to their simplicity, ease of application, efficiency, and cost-effectiveness. The wide application of the 2,4-D herbicide as a sole means of weed control (especially after the introduction of auxinic herbicide-resistant transgenic crops) increases the risk of development of herbicide-resistant weeds. Transgenic crop lines have provided farmers with a cost-effective method to control weeds in the cotton field. However, one of the important disadvantages of using 2,4-D is that wild-type cotton is extremely sensitive to this herbicide, and injury can occur when 2,4-D drifts into neighboring non-transgenic cotton fields. The cotton CS lines tolerant against 2,4-D provide a complementary genetic tool to solve the drift effect problem from 2,4-D in weed management based on non-transgenic cotton cultivars. The genes associated with 2,4-D tolerance in the CS lines could be stacked as a new source of 2,4-D-tolerant genes in a breeding program providing an additional strategy to tackle weed management in cotton. The discovery of unique genetic loci in the cotton CS lines, which have been introgressed from the wild and unadopted genetic resource of cotton germplasm, suggests that the potential herbicide tolerance in the selected cotton CS lines can be transferred by breeding and hybridization in an upland cotton genetic improvement program. Ultimately, the introgression of this useful trait into commercial cotton varieties, protecting non-GM cotton cultivars against off-target drift effects of auxin herbicides applied to neighboring fields, will be a great benefit to cotton farmers.
Conclusion
There is little information available on the physiological mechanism(s) associated with 2,4-D tolerance in cotton. Our results unveiled for the first time that 2,4-D tolerance may be based on reduced translocation, comparing the 2,4-D-tolerant CS lines with the susceptible TM-1 line. This research has, for the first time, 1) provided genetic information on the potential association of different chromosomes or chromosome segments from the alien species of G. barbadense and G. tomentosum with 2,4-D tolerance, 2) revealed that different 2,4-D-tolerant CS lines exhibit different patterns of absorption and translocation, 3) provided a tool to develop natural (non-transgenic) herbicide-tolerant cotton CS lines, and (4) generated insights into genetic 2,4-D tolerance mechanisms in cotton that could connect future possibilities of metabolomics with genomic technologies in developing herbicide-tolerant cotton lines.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
T-MT, LP, JD, and SS designed the experiment. LP and ZY conducted the experiment and data analysis. LP wrote the original version of this paper. JJ and DS provided technical expertise on cotton chromosome substitution lines. All authors have read, contributed to, and approved the manuscript.
Funding
Funding for this project was provided by Cotton Incorporated and the Mississippi Agricultural and Forestry Experiment Station. This material was based on the work that was supported by the National Institute of Food and Agriculture, US Department of Agriculture, Hatch project under accession number 230100.
Acknowledgments
The authors would like to thank Dr. Danica E. Goggins for the critical review of this manuscript. We acknowledge Ms. Evita Lopez for providing the seeds of the cotton lines and for her technical help in the conduct of the experiment.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ayubov M. S., Norov T. M., Saha S., Tseng T. M., Reddy K. R., Jenkins J. N., et al. (2021). Alteration of root and shoot morphologies by interspecific replacement of individual upland cotton chromosome or chromosome segment pairs. Euphytica 217, 1–23. doi: 10.1007/s10681-021-02771-6
Barrier G. E., Loomis W. E. (1957). Absorption and translocation of 2,4-dichlorophenoxyacetic acid and [32P] by leaves. Plant Physiol. 32, 225–231. doi: 10.1104/pp.32.3.225
Benedict C. R., Martin G. S., Liu J., Puckhaber L., Magill C. W. (2004). Terpenoid aldehyde formation and lysigenous gland storage sites in cotton: Variant with mature glands but suppressed levels of terpenoid aldehydes. Phytochemistry 65, 1351–1359. doi: 10.1016/j.phytochem.2004.03.032
Braxton L. B., Richburg J. S., York A. C., Culpepper A. S., Haygood R. A., Lovelace M. L., et al. (2017). Resistance of enlist™ (AAD-12) cotton to glufosinate. Weed Technol. 31, 380–386. doi: 10.1017/wet.2017.14
Dang H. T., Malone J. M., Boutsalis P., Krishnan M., Gill G., Preston C. (2018). Reduced translocation in 2,4-d-resistant oriental mustard populations (Sisymbrium orientale l.) from Australia. Pest Manage. Sci. 74, 1524–1532. doi: 10.1002/ps.4845
Davis D. E., Funderbark H. H., Sansing N. G. (1959). The absorption and translocation of 14C-labeled simazine by corn, cotton, and cucumber. Weeds 7, 300–309. doi: 10.2307/4040338
Day B. E. (1952). The absorption and translocation of 2,4-dichlorophenoxyacetic acid by bean plants. Plant Physiol. 27, 143–152. doi: 10.1104/pp.27.1.143
Dayan F. E., Duke S. O., Grossmann K. (2010). Herbicides as probes in plant biology. Weed Sci. 58, 340–350. doi: 10.1614/WS-09-092.1
Delannay X., Bauman T. T., Beighley D. H., Buettner M. J., Coble H. D., DeFelice M. S., et al. (1995). Yield evaluation of a glyphosate-tolerant soybean line after treatment with glyphosate. Crop Sci. 35, 1461–1467. doi: 10.2135/cropsci1995.0011183X003500050033x
Delye C., Jasieniuk M., Corre V. L. (2013). Deciphering the evolution of herbicide resistance in weeds. Trends Genet. 29, 649–658. doi: 10.1016/j.tig.2013.06.001
Egan J. F., Barlow K. M., Mortensen D. A. (2014). A meta-analysis on the effects of 2, 4-d and dicamba drift on soybean and cotton. Weed Sci. 62, 193–206. doi: 10.1614/WS-D-13-00025.1
Fernandez-Cornejo J., Jans S. (1999). Pest management in U.S. agriculture (Washington DC: US Department of Agriculture).
Figueiredo M. R. A., Leibhart L. J., Reicher Z. J., Tranel P. J., Nissen S. J., Westra P., et al. (2018). Metabolism of 2,4-dichlorophenoxyacetic acid contributes to resistance in a common waterhemp (Amaranthus tuberculatus) population. Pest Manage. Sci. 74, 2356–2362. doi: 10.1002/ps.4811
Ganie Z. A., Jugulam M., Jhala A. J. (2017). Temperature influences efficacy, absorption, and translocation of 2,4-d or glyphosate in glyphosate-resistant and glyphosate-susceptible common ragweed (Ambrosia artemisiifolia) and giant ragweed (Ambrosia trifida). Weed Sci. 65, 588–602. doi: 10.1017/wsc.2017.32
Goggin D. E., Cawthray G. R., Powles S. B. (2016). 2,4-d resistance in wild radish: Reduced herbicide translocation via inhibition of cellular transport. J. Exp. Bot. 67, 3223–3235. doi: 10.1093/jxb/erw120
Goggin D. E., Kaur P., Owen M. J., Powles S. B. (2018). 2, 4-d and dicamba resistance mechanisms in wild radish: subtle, complex and population specific. Ann. Bot. 122, 627–640. doi: 10.1093/aob/mcy097
Grossman K. (2003). ““News from old compounds: The mode of action of auxin herbicides,”,” in Chemistry of crop protection: Progress and prospects in science and regulation. Eds. Voss G., Ramos G. (Weinheim: Wiley-VCH Verlag GmbH), 131–142.
Grossman K. (2007). Auxin herbicide action. Plant Signaling Behav. 2, 421–423. doi: 10.4161/psb.2.5.4417
Grossman K. (2010). Auxin herbicides: Current status of mechanism and mode of action. Pest Manage. Sci. 66, 113–120. doi: 10.1002/ps.1860
Gunsolus J. L., Curran W. S. (1999). Herbicide mode of action and injury symptoms (Minnesota: University of Minnesota Extension Service).
International Service for the Acquisition of Agri-biotech Applications (2020) Crop biotech update. Available at: https://www.isaaa.org/kc/cropbiotechupdate/article/default.asp?ID=18209 (Accessed August 18, 2022).
Johnston C. R., Eure P. M., Grey T. L., Culpepper A. S., Vencill W. K. (2018). Time of application influences translocation of auxinic herbicides in palmer amaranth (Amaranthus palmeri). Weed Sci. 66, 4–14. doi: 10.1017/wsc.2017.36
Kohler E. A., Throssell C. S., Reicher Z. J. (2004). 2,4-D Rate response, absorption, and translocation of two ground Ivy (Glechoma hederacea) populations. Weed Technol. 18, 917–923. doi: 10.1614/WT-03-089R1
Kumar V., Liu R., Boyer G., Stahlman P. W. (2019). Confirmation of 2,4-d resistance and identification of multiple resistance in a Kansas palmer amaranth (Amaranthus palmeri) population. Pest Manage. Sci. 75, 2925–2933. doi: 10.1002/ps.5400
Laurent F., Debrauwer L., Rathahao E., Scalla R. (2000). 2,4-dichlorophenoxyacetic acid metabolism in transgenic tolerant cotton (Gossypium hirsutum). J. Agric. Food Chem. 48, 5307–5311. doi: 10.1021/jf990672c
Lyon B. R., Cousins Y. L., Llewellyn D. J., Dennis E. S. (1993). ). cotton plants transformed with a bacterial degradation gene are protected from accidental spray drift damage by the herbicide 2,4-dichlorophenoxyacetic acid. Transgenic Res. 2, 162–169. doi: 10.1007/BF01972610
Nida D. L., Kolacz K. H., Buehler R. E., Deaton W. R., Schuler W. R., Armstrong T. A., et al. (1996). Glyphosate-tolerant cotton: genetic characterization and protein expression. J. Agric. Food Chem. 44, 1960–1966. doi: 10.1021/jf9505640
Perez L. M. (2021). “The genetics and molecular mechanisms of tolerance to 2,4-dichlorophenoxyacetic acid (2,4-d) in upland cotton (Gossypium hirsutum L.),” (Starkville, MS: Mississippi State University).
Perez L. M., Mauleon R., Arick M. A., Magbanua Z. V., Peterson D. G., Dean J. F. D.. (2022). Transcriptome analysis of the 2,4-dichlorophenoxyacetic acid (2,4-D)-tolerant cotton chromosome substitution line CS-B15sh and its susceptible parental lines G. hirsutum L. cv. Texas Marker-1 and G. barbadense L. cv. Pima 379. Front. Plant Sci. 13, 910369. doi: 10.3389/fpls.2022.910369
Peterson M. A., McMaster S. A., Riechers D. E., Skelton J., Stahlman P. W. (2016). 2,4-d past, present, and future: A review. Weed Technol. 30, 303–345. doi: 10.1614/WT-D-15-00131.1
Preston C., Malone J. M. (2014). Inheritance of resistance to 2,4-d and chlorsulfuron in a multiple-resistant population of Sisymbrium orientale. Pest Manage. Sci. 71, 1523–1528. doi: 10.1002/ps.3956
Reddy K. R., Bheemanahalli R., Saha S., Singh K., Lokhande S. B., Gajanayake B., et al. (2020). High-temperature and drought-resilience traits among interspecific chromosome substitution lines for genetic improvement of upland cotton. Plants 9, 1747. doi: 10.3390/plants9121747
Rey-Caballero J., Menéndez J., Giné-Bordonaba J., Salas M., Alcántara R., Torra J. (2016). Unravelling the resistance mechanisms to 2,4-d (2,4-dichlorophenoxyacetic acid) in corn poppy (Papaver rhoeas). Pestic. Biochem. Physiol. 133, 67–72. doi: 10.1016/j.pestbp.2016.03.002
Riar D. S., Burke I. C., Yenish J. P., Bell J., Gill K. (2011). Inheritance and physiological basis for 2,4-d resistance in prickly lettuce (Lactuca serriola l.). J. Agric. Food Chem. 59, 9417–9423. doi: 10.1021/jf2019616
Robinson A. P., Simpson D. M., Johnson W. G. (2015). Response of aryloxyalkanoate dioxygenase-12 transformed soybean yield components to postemergence 2,4-d. Weed Sci. 63, 242–247. doi: 10.1614/WS-D-14-00036.1
Saha S., Raska D. A., Stelly D. M. (2006). Upland (Gossypium hirsutum L.) x Hawaiian cotton (G. tomentosum nutt. ex seem.) F1 hybrid hypoaneuploid chromosome substitution series. J. Cotton Sci. 10, 263–272.
Saha S., Stelly D. M., Makamov A. K., Ayubov M. S., Raska D., Gutie´rrez O. A., et al. (2015). Molecular confirmation of Gossypium hirsutum chromosome substitution lines. Euphytica journal. doi: 10.1007/s10681-015-1407-2
Saha S., Stelly D. M., Raska D. A., Wu J., Jenkins J. N., McCarty J. C., et al. (2012). “Chromosome substitution lines: concept, development and utilization in the genetic improvement of upland cotton,” in Plant breeding. Ed. Abdurakhmonov I. Y. (Croatia: InTech).
Saha S., Wu J., Jenkins J. N., McCarty J. C., Gutierrez O. A., Stelly D. M., et al. (2004). Effect of chromosome substitutions from Gossypium barbadense l. 3-79 into G. hirsutum l. TM-1 on agronomic and fiber traits. J. Cotton Sci. 8, 162–169.
Shergill L. S., Barlow B. R., Bish M. D., Bradley K. W. (2018). Investigations of 2,4-d and multiple herbicide resistance in a Missouri waterhemp (Amaranthus tuberculatus) population. Weed Sci. 66, 386–394. doi: 10.1017/wsc.2017.82
Song Y. (2014). Insight into the mode of action of 2,4-dichorophenoxyacetic acid (2,4-d) as an herbicide. J. Integr. Plant Biol. 56, 106–113. doi: 10.1111/jipb.12131
Stelly D., Saha S., Raska D., Jenkins J., McCarty J., Gutierrez O. (2005). Registration of 17 upland (Gossypium hirsutum) germplasm lines disomic or different G. barbadense chromosome or arm substitutions. Crop Sci. 45, 2663–2665. doi: 10.2135/cropsci2004.0642
Sterling T. M. (1994). Mechanisms of herbicide absorption across plant membranes and accumulation in plant cells. Weed Sci. 42, 263–276. doi: 10.1017/S0043174500080383
Sterling T. M., Hall J. C. (1997). “Mechanism of action of natural auxins and the auxinic herbicides,” in Herbicide activity: Toxicology, biochemistry, and molecular biology. Eds. Roe R. M., Burton J. D., Kuhr R. J. (Amsterdam: IOS Press), 111–141.
Streber W. R., Willmitzer L. (1989). Transgenic tobacco plants expressing a bacterial detoxifying enzyme are resistant to 2,4-d. Nat. Biotechnol. 7, 811–816. doi: 10.1038/nbt0889-811
Ulloa M., Hutmacher R. B., Schramm T., Ellis M. L., Nichols R., Roberts P. A., et al. (2020). Sources, selection and breeding of fusarium wilt (Fusarium oxysporum f. sp. vasinfectum) race 4 (FOV4) resistance in upland (Gossypium hirsutum L.) cotton. Euphytica 216, 1–18. doi: 10.1007/s10681-020-02643-5
Vulchi R., Bagavathiannan M., Nolte S. A. (2022). History of herbicide-resistant traits in cotton in the U.S. and the importance of integrated weed management for technology stewardship. Plants 11, 1189. doi: 10.3390/plants11091189
Wright T. R., Shan G., Walsh T. A., Lira J. M., Cui C., Song P., et al. (2010). Robust crop resistance to broadleaf and grass herbicides provided by aryloxyalkanoate dioxygenase transgenes. Proc. Natl. Acad. Sci. U.S.A. 107, 20240–20245. doi: 10.1073/pnas.1013154107
Yamaguchi S., Crafts A. S. (1958). Autoradiographic method for studying absorption and translocation of herbicides using C14-labeled compounds. Hilgardia 28, 161–191. doi: 10.3733/hilg.v28n06p161
Keywords: upland cotton, herbicide resistance mechanism, herbicide absorption, herbicide translocation, abiotic stress tolerance, plant breeding, auxin
Citation: Perez LM, Yue Z, Saha S, Dean JFD, Jenkins JN, Stelly DM and Tseng T-M (2022) Absorption and translocation of [14C]2,4-dichlorophenoxyacetic acid in herbicide-tolerant chromosome substitution lines of Gossypium hirsutum L. Front. Agron. 4:936119. doi: 10.3389/fagro.2022.936119
Received: 04 May 2022; Accepted: 30 August 2022;
Published: 06 October 2022.
Edited by:
Joel Torra, Universitat de Lleida, SpainReviewed by:
Marcelo Figueiredo, Duke University, United StatesMuhammad Shahid Iqbal, Cotton Research Institute (CAAS), China
Copyright © 2022 Perez, Yue, Saha, Dean, Jenkins, Stelly and Tseng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Te-Ming Tseng, dC50c2VuZ0Btc3N0YXRlLmVkdQ==
†Present address: Loida M. Perez, Philippine Rice Research Institute, SC Muñoz, Nueva Ecija, Philippines
‡ORCID: Loida M. Perez, orcid.org/0000-0003-4066-2749
Johnie N. Jenkins, orcid.org/0000-0002-3586-8411
David M. Stelly, orcid.org/0000-0002-3468-4119
 Loida M. Perez
Loida M. Perez Ziming Yue2
Ziming Yue2 Sukumar Saha
Sukumar Saha Jeffrey F. D. Dean
Jeffrey F. D. Dean Johnie N. Jenkins
Johnie N. Jenkins David M. Stelly
David M. Stelly Te-Ming Tseng
Te-Ming Tseng