
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Agron., 09 August 2022
Sec. Disease Management
Volume 4 - 2022 | https://doi.org/10.3389/fagro.2022.936032
This article is part of the Research TopicInsights in Disease ManagementView all 6 articles
In the last years, the diffusion and implementation of next-generation sequencing and the reduction of costs raised the interest in phytyobiome studies allowing to dissect the ecological interactions regulating the holobiont. Indeed, crop plants are associated with a wide diversity of microorganisms in all their parts. Crop microbiota influences plant phenotype, growth, yield and quality by contributing to plant resistance toward diseases, plant adaptation to abiotic stresses, and plant nutrition. The association between terrestrial plants and microbes developed at least 460 million years ago, as suggested by the fossil evidence of the earliest land plants, indicating the essential role of microbes for plants. Recent studies indicate that plants actively recruit beneficial microorganisms to facilitate their adaptation to environmental conditions. Cultivation methods and disease control measures can influence plant microbiome structure and functions. Both pesticide and biological control agent applications may alter the biodiversity inside the phytobiota and suppress beneficial functions. Nonetheless, to date, the effects of disease control measures on phytobiota and their possible side consequences on plant growth, crop productivity and quality remain a neglected field of study. The present work summarizes the known effects on phytobiota providing evidence about the role of plant microbial community in determining the overall efficacy of the applied control measure and suggests that future studies on plant disease control consider also the microbe-mediated effects on plant fitness.
Plants live in close association with a dynamic phytobiota, consisting of a macrobiota as well as a microbiota, in particular the microbes that inhabit the soil in which plants grow. The importance of soil microbial communities is well recognized for their role in plant performance, including the improvement of plant growth, vigor and fitness traits (Marschner and Rumberger, 2004; Lau and Lennon, 2011; Chaney and Baucom, 2020), as well as nutrient acquisition (Chaparro et al., 2012; Reverchon et al., 2015), associated with the presence of rare taxa (Hol et al., 2010), and the attraction of pollinators, predators and parasites of herbivores (Wagner et al., 2014; Berg et al., 2016; Etemadi et al., 2018). Plants modulate rhizosphere microbiota (Berendsen et al., 2012; Kwak et al., 2018) and selectively recruit beneficial microbes (Park and Ryu, 2021) thanks to signals, including root exudates, volatile compounds (VOCs) and phytohormones, that help to shape the phytobiota by recruiting, repelling and coordinating the interactions (Dicke, 2016). Plant-associated microorganisms form complex networks with other members of the same species as well as with different species, genera, families and even domains of life (Wassermann et al., 2019a). The cooperation between plants and microorganisms, as well as the intra-microbiome interplay, require an intense communication that regulates community assembly (Agler et al., 2016; Venturi and Keel, 2016).
The homeostatic balance between both microbe-microbe and host-microbe interactions is critical for plant health. The perturbation of this balance is often referred to as microbial dysbiosis and it may represent an important mechanism of disease (Martins dos Santos et al., 2010; Sekirov et al., 2010; Kemen, 2014). For example, under high humidity conditions, several Arabidopsis immuno-compromised mutants display leaf-tissue damages related to an altered composition of the phyllosphere microbiota, somewhat similar to the dysbiosis occurring in human inflammatory bowel disease (Chen et al., 2020). Importantly, experiments of bacterial community transplantation showed that the application of a dysbiotic leaf bacterial community to healthy plants results in tissue damage, demonstrating that, in this case, dysbiosis is the cause of the observed symptomatology (Chen et al., 2020). Nevertheless, it is worth mentioning that stress-induced deviation from eubiosis is not always associated with reduced plant performance (Paasch and He, 2021).
The phytobiota protects crops from diseases through different modes of action: i) indirect effects mediated by the activation of plant induced systemic resistance (ISR), a primed resistance against pathogen infections transmitted from roots to plant tissues (Pieterse et al., 2014; Conrath et al., 2015), ii) niche exclusion of pathogens by competition for nutrients and space (Spadaro and Droby, 2016), and/or iii) direct interactions with pathogens through hyperparasitism (invasion and killing of mycelium, spores and resting structures) or antibiosis (production of antimicrobial secondary metabolites) (Raaijmakers and Mazzola, 2012; Ghorbanpour et al., 2018). Finally, some fungal viruses are used to induce hypovirulence (Milgroom and Cortesi, 2004; Double et al., 2018) and microbial antagonists can act via the inactivation of enzymes involved in pathogen infections (Elad, 2000) or the enzymatic degradation of pathogen structures (Köhl et al., 2019).
One easily overlooked, but nonetheless intrinsically important, aspect to consider when studying phytobiota is that pathogenic species are themselves part of all microbiota (Kamada et al., 2013). This is actually a core aspect of epidemiology since the presence of a pathogen does not necessarily lead to infection and disease is caused only under specific inducive conditions (Scholthof, 2007). In the context of dysbiosis, the pathobiota concept, which integrates pathogenic agents within biotic environments, was established and applied to several pathosystems (Baltrus, 2017). The analyses of pathobiota often revealed that pathogens do not operate independently, and multiple pathogens might be involved in severe dysbiosis (Singer, 2010; Lamichhane and Venturi, 2015; Berg et al., 2021). This is the case of Fusarium Head Blight, a highly dynamic disease caused by a complex of different Fusarium species, whose composition is geographically dependent, and each of them can respond differently to the changing climate (Yli-Mattila, 2010; Vaughan et al., 2016). Even if predominant, the highly virulent F. graminearum is only one member of the species complex (Ioos et al., 2005; Xu et al., 2005) and it was recently established that control strategies against F. graminearum are hampered by the presence of the weakly pathogenic F. poae, showing the complexity of developing control strategies against plant diseases caused by multiple pathogens (Tan et al., 2021). A similar scenario has been observed for phytopathogenic bacteria. Pseudomonas syringae pv. actinidiae is always present in syndemic association with Pseudomonas syringae pv. syringae and Pseudomonas viridiflava, two other kiwifruit pathogens, suggesting the establishment of a pathogenic consortium leading to a higher pathogenesis capacity (Purahong et al., 2018). For these reasons, the novel disease control strategies should target the whole phytobiota and the possible pathogenic consortia therein rather than single pathogens to develop suppressive microbial communities able to restrict several pathogens at the same time.
Our current knowledge of communications within the phytobiota is still largely based on interactions involving two or three components, and is frequently measured in controlled conditions. However, a further level of complexity in studying phytobiota relates to eukaryotic organisms that interact with plants, each of them harboring its own internal and external microbiota, influencing the interactions between plants and their microbial communities (Cusano et al., 2011; Khaitov et al., 2015). The intense communication among plant holobiont members and the observations that signals can be co-opted, modified, or even destroyed, by another member of the community reinforce the need for a system-level analysis of communication mechanisms to exploit phytobiota manipulation for crop improvement (Figure 1).
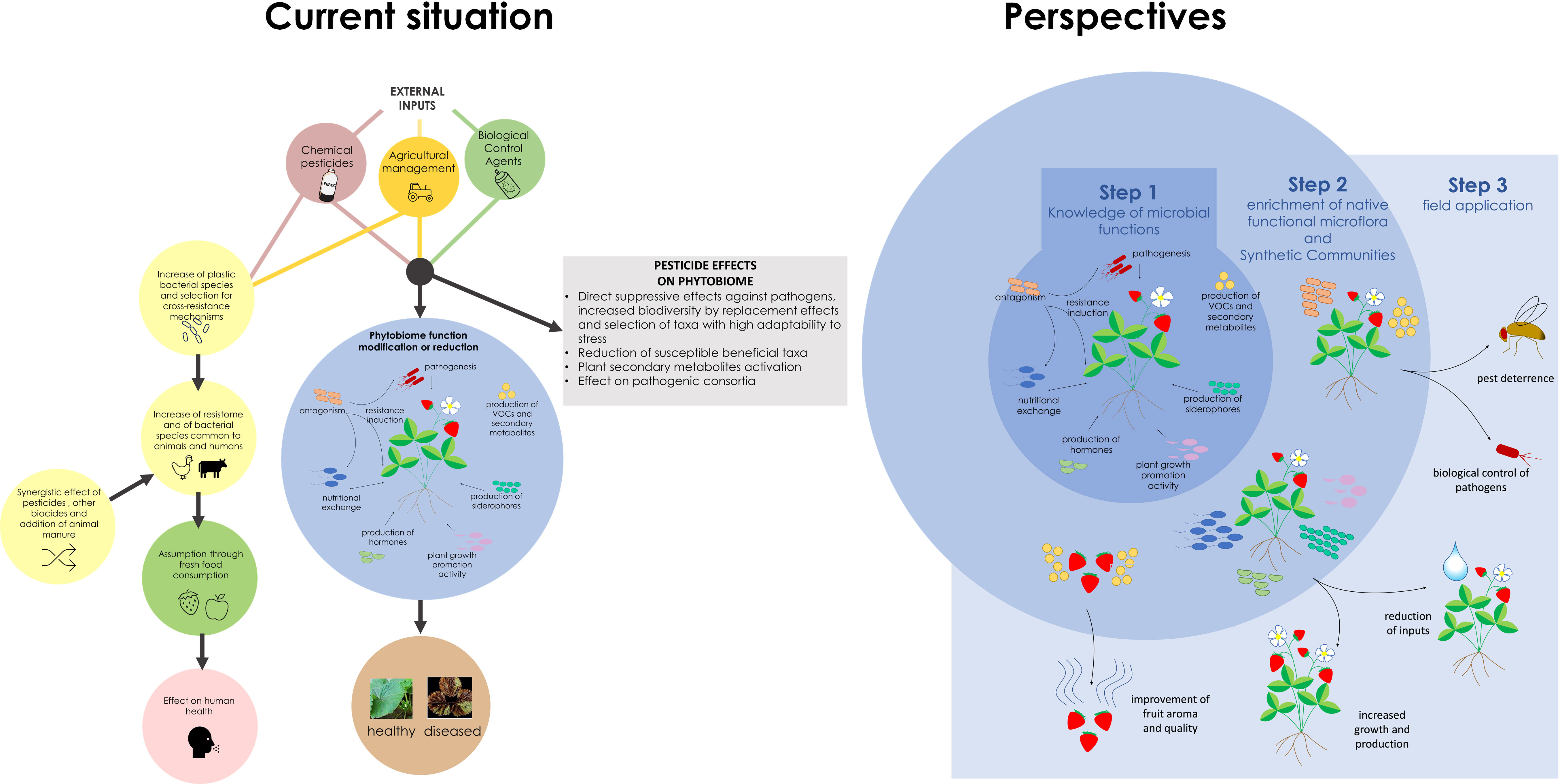
Figure 1 Role of phytobiota and effects of disease control measures on phytobiota (current situation). The panel on future development highlight different strategies to achieve a long-lasting and sustainable disease control approach based on phytobiota manipulation.
Chemical pesticides are extensively used in intensive agriculture and they are still the most effective, reliable and economic tool to minimize losses caused by pests and diseases (Huang et al., 2021a). In this view, pesticides and fertilizers are essential to ensure food security for the ever-growing world population. At global level, more than 2 million tons of pesticides are utilized annually (Sharma et al., 2019). In Europe, the awareness of the risks of pesticides led to the development of policies for their sustainable use (Directive 2009/128/EC) and actions, such as the Farm2ForK Strategy, aiming at a reduction of 50% of pesticide use by 2030. Indeed, in the last decade, pesticide use in the EU was reduced by 10.2% with the lowest total volume (333,500 tones) of sales recorded in 2019. Fungicides, bactericides and herbicides are still the major groups of pesticides currently used (EUROSTAT- European Statistics, 2021). The rise of concern on the effect of pesticides on human and environmental health fostered the research on sustainable alternatives, as well as the extensive study of the direct and indirect effects of their use on non-target organisms and on the ecosystem services they provide (Ramakrishna et al., 2019). Soil microbiota is crucial for ecosystem functions (Karas et al., 2018). Since synthetic fertilizers and pesticides have a long persistence in soil, in the last decade, several studies investigated the effect of pesticides and fertilizers on soil microflora and microbial fertility, which have a direct impact on environmental quality as well as plant health and productivity (Hartmann et al., 2014; Zhu et al., 2016; Huang et al., 2021b). This led the European Food Safety Authority (EFSA) to the inclusion of soil microorganisms in the environmental risk assessment of pesticides (EFSA Panel on Plant Protection Products and their Residues (PPR), 2010; Karas et al., 2018). However, pesticide toxicology assessment is based only on their impact on the N mineralization test (OECD, 2000) and, thus, it does not provide information about key functions of soil microbiota (Karas et al., 2018). Despite the growing body of knowledge on the impact of pesticides on soil microorganisms, standardized methods have not been developed yet and the majority of studies have been performed at a lab-scale or using exposure levels higher than the recommended application doses (Medo et al., 2015; Wang et al., 2016). Soil microbial communities respond to organic and conventional fertilizer inputs by specific community adaptations (Hartmann et al., 2014). This perturbation often leads to an increase in community richness partially related to the higher proportions of microbial groups associated with the degradation of the introduced pesticide (Ramakrishna et al., 2019; Cernava et al., 2019a; Huang et al., 2021a; Huang et al., 2021b).
Concerning the plant-associated microbiota, the effect of xenobiotic pesticides is a neglected field of study, with scarce information about their impact on the structure and dynamic of epiphytic, endophytic and rhizosphere microbial communities (Table 1). Furthermore, no study has been conducted on the interplay of plant-associated microbiota on the uptake, translocation and degradation of pesticides (Vryzas, 2016). Root tip exudates mediated by rhizobacteria could modify the uptake of specific pesticides, while bacterial ligands and enzymes can affect the metabolism and fate of pesticides within crop plants (Vryzas, 2016). Finally, the impact on the phytobiota of transformation products, which may have a higher toxicity than the original pesticide (Wu et al., 2014), has not been elucidated yet. The effect on the phytobiota depends on the pesticide application strategy (i.e. seed coating, pre- or post-emergent crop) and the affinity to systematic translocation. Pre-emergent pesticides are generally applied to the soil. Soil is also the major accumulation site of pesticides from seed coating. In addition to the direct effect on shaping soil microbiota, these practices also define the species abundance and diversity of available beneficial microorganisms that can be recruited by crop plants (Berg, 2009; Cernava et al., 2019a). Karas and colleagues investigated the effect of TCP, 3,5,6-trichloro-2-pyridynol (Chlorpyrifos (CHL), isoproturon (IPU) and tebuconazole (TBZ), which are three pesticides heavily used in Europe and represent the three major pesticide classes (insecticide, herbicide, and fungicide) (Karas et al., 2018). The findings highlighted that functional microbial guilds involved in N and S cycling are the most sensitive endpoints to pesticide exposure. Ammonia-oxidizing microorganisms are the most sensitive and show a consistent response to pesticide exposure, while, in contrast, denitrifying bacteria are stimulated upon exposure to all pesticides. Interestingly, the combined application of pesticides along with biofertilizers may mitigate their negative effects (Huang et al., 2021a; Huang et al., 2021b). In a study conducted on sugar cane, the combination pesticide/biofertilizer enhanced bacterial community diversity compared with control treatments and led to an increase of the expression of the nitrification-related genes pmoA-amoA and the denitrification-related genes nirK and norB (Huang et al., 2021b). However, the type of fertilizer may also have different specific effects. Organic fertilizers generally enhance microbial beneficial functions (Cai et al., 2017; Xiong et al., 2017), whereas synthetic and high nitrogen fertilizers tend to induce dominance effects by stimulating the multiplication of fast-growing microbes (Esperschütz et al., 2007) and to impair soil C/N cycling (Paungfoo-Lonhienne et al., 2015). In this view, to provide a comprehensive elucidation of the effects of a specific pesticide on phytobiota, not only the chemical properties of the pesticide should be considered, but also its interactions with the crop and the other agricultural inputs, including the type of fertilizer and the time of application.
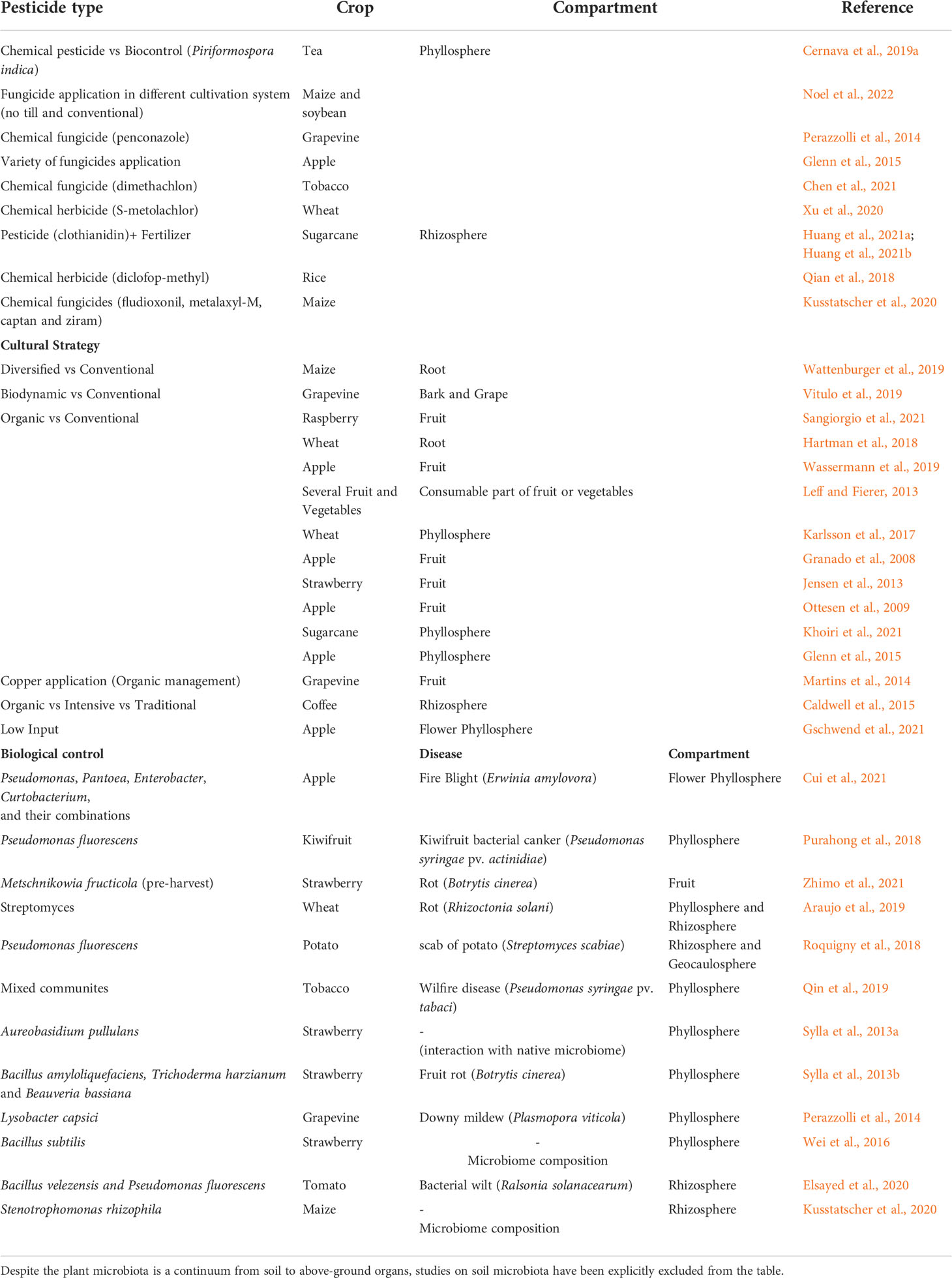
Table 1 List of research works focusing on the influence of chemical and biological pesticide or agricultural management strategies on phytobiota.
In post-emergent application, pesticide sprays target the above-ground plant organs to selectively reduce pathogenic microorganisms in complex communities. In addition to the effects on pathogen epiphytic growth, pesticides also impact the phyllosphere microbial biocoenosis that hosts a variety of microorganisms, thus influencing plant susceptibility to diseases (Purahong et al., 2018), plant secondary metabolism (Sangiorgio et al., 2021) as well as fruit quality and productivity (Sangiorgio et al., 2022a; Sangiorgio et al., 2022b). The phyllosphere is an oligotrophic and dynamic environment subjected to both circadian and seasonal changes of abiotic (e.g. RH, temperature, light, nitrogen deposition) and biotic (e.g. plant exudates and secondary metabolism) conditions. Thus, epiphytic microorganisms evolved several adaptation mechanisms to this ever-changing environment (Bringel and Couée, 2015; Etemadi et al., 2018). Pesticides elicit a distinct shift in rare phyllosphere microbiota (Cernava et al., 2019a; Xu et al., 2020; Chen et al., 2021), with minor or no effect on the core microbiota, generally leading to increased microbial diversity in treated plants when compared to untreated samples (Cernava et al., 2019a; Xu et al., 2020). However, the increase in biodiversity can be detrimental to its functions. Indeed, beneficial bacteria, known for their intrinsic association with plants, are among the most sensitive responders to the employed fungicide (Chen et al., 2021). Moreover, the pesticide-increased biodiversity is associated with replacement effects, where the niche created by the biocidal activity against pathogen is occupied by microbial competitors (Cernava et al., 2019a; Chen et al., 2021) and linked with community expansion related to the selection of those taxa with distinct stress response systems (Cernava et al., 2019a; Xu et al., 2020). For example, in a study conducted on tea leaves, the most abundant bacterial genera in lime sulfur pesticide -specific signatures were assigned to Brevundimonas, Arsenophonus and candidatus Portiera, which also includes species adapted to resist harsh conditions (Cernava et al., 2019a). Similarly, in tobacco plants, the application of the broad-spectrum fungicide N-(3,5-dichlorophenyl) succinimide induces an increase in Gammaproteobacteria over Alphaproteobacteria (Chen et al., 2021). Gammaproteobacteria are common colonizers of plants and are characterized by a fast growth when nutrients are not a limiting factor. To confirm the shift toward taxa with higher adaptability to stress, genes related to the pathways associated with stress responses, biofilm formation and efflux pumps, are increased by pesticide treatment. Both in the soil and the phyllosphere, pesticide-induced changes may also increase the occurrence of potential human pathogens, such as members of the Enterobacteriaceae family, which are common residents of crop phyllosphere and excellent degraders of organic compounds (Brandl, 2006; Teplitski et al., 2011; Erlacher et al., 2015; Cernava et al., 2019b). Due to the commonalities between the adaptation mechanisms to pesticides and the resistance to antimicrobial compounds, such as antibiotics, the pesticide-induced changes in phytobiota can facilitate the development of adaptive mechanisms to multiple antimicrobial resistance, especially in microbes able to degrade the pesticides, which may also be transferred to the human pathogens present in the phytobiota (Kurenbach et al., 2015; Cernava et al., 2019a; Cernava et al., 2019b). For example, glyphosate (N-(phosphonomethyl) glycine) can induce the cross-resistance to antibiotics in Escherichia coli and Salmonella enterica serovar typhimurium (Kurenbach et al., 2015). Methyl salicylate, along with its derivatives sodium salicylate, acetylsalicylate and salicyl alcohol, may facilitate the development of antibiotic resistance through the activation of the mar operon in Escherichia coli (Cohen et al., 1993). Thus, pesticides can increase and diversify the plant resistome, representing an important reservoir of antibiotic resistance genes, especially in fresh consumed vegetables and fruit, therefore posing potential issues to human health (Blair et al., 2015; Cernava et al., 2019b). Finally, the increase in the phyllosphere of taxa able to metabolize pesticides may result in the accumulation in/on fruit and vegetables of intermediate products, which may exert a higher toxicity than the original pesticide (Cycoń and Piotrowska-Seget, 2015).
Although many studies demonstrated that rhizosphere or phyllosphere microbiota, in field conditions, are not greatly affected by the introduced biological control agents (BCAs) (Perazzolli et al., 2014; Wei et al., 2016; Roquigny et al., 2018), other researches, conversely, reported that the application of BCAs significantly change the autochthonous microbial community in terms of both diversity and composition (Schmidt et al., 2014; Qin et al., 2019), which can be neutral or even beneficial for the host plant (Pieterse and Dicke, 2007; Méndez-Bravo et al., 2018). Indeed, microbiota alteration may represent an added value for the plants, thanks to a more diversified community enrichment (Araujo et al., 2019; Cernava, 2021), or through the decrease in the abundance of pathogenic strains, allowing for instance the suppression of decay development during fruit storage (Zhimo et al., 2021). Nevertheless, it cannot be ruled out that the modification of the structure and functions of the microbial community by BCA application can also negatively impact plant fitness and health. Indeed, individual members of a community contribute to the overall stability or instability of the system and, consequently, BCAs, as new community members, must be viewed as potential internal drivers of microbial community assemblage (Hacquard et al., 2015). Moreover, BCAs must present specific features, such as a high adaptability and stress tolerance, a rapid growth and a high competitiveness, providing an advantage to BCAs and leading to a shift in the microbiota related to dominance effects (Viera-Arroyo et al., 2020). Because of the ecological and economic impacts of invasive species on natural sustainability and societal development (Cullen et al., 2008), there is also a concern that BCAs may eventually become a dominating species in crop microbiota, causing permanent damage to local ecosystems, as documented outside the field of plant pathology (Brown and Mathews, 2007; Gaba et al., 2016). From an ecological point of view, biological control may lead to the exclusion of other species in addition to pathogens and, in extreme cases, threaten phytobiota ecological function and resilience (Sanders et al., 2015), although there is no clear evidence of such phenomenon in plant pathology (Barbosa et al., 2017). Moreover, BCAs may become emerging pathogens of local crops or even humans due to host-jumping or other eco-evolutionary events. In this view a crucial role is played by horizontal gene transfer which is facilitated by mobile genetic elements (Frost et al., 2005). For example, Delftia tsuruhatensis strains have been used for biological control and plant growth promotion. However, recently D. tsuruhatensis has also became an emergent human pathogen (Yin et al., 2022). Interestingly, the pan-genome of D. tsuruhatensis shows extensive genetic diversity a high degree of genetic plasticity characterized by diverse mobile genetic elements, massive genomic rearrangement, and horizontal genes suggesting that horizontal gene transfer and purifying selection are important forces in D. tsuruhatensis genetic evolution and adaptation to new hosts (Yin et al., 2022).
BCAs may also contribute to the induction of virulence in autochthonous microbial species, by interfering with the communication system of the pathogens. In a microbial consortium, bacteria may respond to self-like autoinducers from other species, as reported for the olive knot pathogen Pseudomonas savastanoi pv. savastanoi and the harmless endophyte cooperator Erwinia toletana that form a stable community in the olive knot, where they share quorum-sensing signals and cooperate, resulting in a more aggressive disease (Caballo-Ponce et al., 2018). Going further in this concept, Pseudomonas chlororaphis isolates stimulate plant growth through direct pathogen antagonism as well as the induction of systemic resistance in the plant by an array of metabolites under the control of a global regulatory system, the Gac/Rsm regulon (Anderson et al., 2017). Since, in Pseudomonads, this pathway governs the expression of a wide range of genes involved in bacterial fitness, motility, tolerance to stress, biofilm formation and virulence (Latour, 2020), it cannot be excluded that these beneficial bacteria may activate such signaling pathway in pathogenic species present in the phytobiota, leading to an induction of their virulence.
Unraveling the effect of cultivation management on plant native microbiota and the ecological services that they provide is pivotal for the evaluation of the sustainability of the different agricultural practices (Wattenburger et al., 2019). Indeed, the exploitation of plant-associated microorganisms in agricultural systems is one of the main steps toward the implementation of sustainable management strategies (Compant et al., 2019). Generally, cultivation management can be distinguished between organic and conventional. Although the first consists of the exclusion of synthetic pesticides and fertilizers and low or no-tillage, organic farming may differ from conventional management in several ways (Leff and Fierer, 2013), restricting the comparability among different researches (Lupatini et al., 2017). The cultivation method, affecting the nutrient availability and altering the ecological niches, might also alter the composition and structure of the plant microbiota in several agricultural systems (Caldwell et al., 2015; Hartmann et al., 2014; Sangiorgio et al., 2021). However, the observed effects might be complex and time-dependent (Jonason et al., 2011). Additionally, the comparison among different agricultural management strategies can be compromised by the fact that the different practices are often implemented on farms located in similar, but not identical, areas, thus showing diverse edaphic and environmental conditions. Here, we suggest future experiments to aim at field condition homogeneity to better highlight the contribution of the cultivation management in shaping microbial communities. The influence of management practices on the microbial composition and functions of the below-ground microbiota (Ishaq et al., 2017; Karlsson et al., 2017) has been extensively investigated. Most of these studies focus on the effect of fertilizer management, soil management practices (low/no-tillage vs conventional tillage agriculture) and/or the application of soil health treatments on microbiomes (Hartmann et al., 2014; Bill et al., 2021; Ishaq et al., 2017; Lupatini et al., 2017). Additionally, several efforts have been put into the understanding of belowground microbial communities associated with cover crops, which are widely applied as soil improvement and conservation techniques, although most of the studies focus on annual rather than perennial crops (Castellano-Hinojosa and Strauss, 2020). Cover crops may influence soil characteristics, such as pH and temperature, and thus impact soil microbiota composition.
Although organic farming aims at limiting the negative environmental impacts of agriculture (i.e. water eutrophication, biodiversity loss, toxic chemical accumulation), its implementation does not straightforwardly lead to positive consequences, such as the increase of species richness (Karlsson et al., 2017). For example, in organic agriculture, it is a common practice to combine the application of manure and pesticides to the soils, which might increase the content of antibiotic-resistant genes in soil microbiota, potentially leading to the generation of multiple antimicrobial resistance phenotypes (Ramakrishna et al., 2019). Similarly, the application of copper compounds, widely used in organic agriculture for the control of bacterial and fungal diseases, has been found to be negatively correlated with population size, species richness and evenness of grape yeasts and yeast-like fungi (Martins et al., 2014).
Even though the phyllosphere-associated microbiome gained increasing attention in recent years (Remus-Emsermann and Schlechter, 2018), the study of the influence of the cultivation management on the above-ground microbiota is still in its infancy. Different investigations led to very diverse results (Leff and Fierer, 2013), which might depend on the variety of cultivation and disease control practices implemented, on the rates and frequency of treatments and on the target plant of the study. For example, organically-grown apples display a higher bacterial evenness and diversity (Wassermann et al., 2019b), whereas, in raspberry, conventional cultivation promotes bacterial biodiversity on fruit (Sangiorgio et al., 2021). This might be due to the fact that in low-input agriculture, microbial communities are dominated by highly adapted bacterial species able to outcompete generalist colonizers (Sangiorgio et al., 2021).
As we already underlined, besides the description of the taxonomical variations of microbiotas, we suggest to prioritize the comparative analysis of the functionalities, actually or potentially expressed by the microbial communities, respectively, through metatranscriptomic approaches (Sharuddin et al., 2022) or by means of tools such as Tax4fun (Asshauer et al., 2015), Picrust (Douglas et al., 2020) and FAPROTAX (Louca et al., 2016). For example, this approach highlighted that conventional management lowered the abundance of phyllosphere bacterial genes associated with cell motility and energy metabolism, potentially explaining the lower microbial diversity observed in this condition (Khoiri et al., 2021).
Our knowledge of the impact of xenobiotic pesticides on plant phytobiota is still very limited and no information is currently available regarding the effect of the pesticides-induced shift in phytobiota on crop physiological processes and resistance. In the light of the ‘One Health’ paradigm, the pesticide-induced changes in the structure and functions of phytobiota may pose risks to human health, by influencing human microbiota upon consumption of fresh produces, increasing the spread of cross-resistance against different antimicrobials and leading to the accumulation in fruit and vegetables of residues of intermediate products of microbially-metabolized pesticides (Figures 1, 2). Despite the use of BCAs is generally regarded as a sustainable disease control measure, information is still lacking about their effects on phytobiota ecology and their use may pose unexpected threats to plant health (Figures 1, 2). Future BCAs should be selected from the host native phytobiota by dissecting their ecological relations within the microbial community. Moreover, long-term studies should be performed to figure out their impact on plant, environmental and human microbiota. These studies will allow to understand their influence in a new environment, in particular the biological, ecological and evolutionary outcomes that may unbalance the disease triangle in favor of pathogens. Thus, plant microbiota composition and functions should be precisely monitored following BCA application and this may be achieved thanks to Next Generation Sequencing (NGS) methods, such as metagenomics and metatranscriptomics, powerful techniques that should be accompanied by the knowledge of BCA characteristics and mode of action, allowing human intervention to convert a good candidate into a highly effective BCA, as for Rhizobium rhizogenes K84 (Ellis J.G. 2017). Moreover, it is crucial to decipher the microbial communication mechanisms to minimize the risk of possible BCA conversion and to finely tune the ecological interactions within the phytobiota. Overall, a holistic in-depth understanding on BCA-phytobiota interactions will support better timing, formulation and application of BCAs and prevent failures. In that sense, microbiota studies can enhance the efficiency of BCA selection, but also of “helper” strains, which support BCAs in establishment, survival and antagonistic activity in situ. The identification, selection and characterization of native BCAs could overcome the risk related to phytobiota structure modification due to the introduction of exotic microbial strains. These advances will allow to finely manipulate plant and environmental microbiota to develop strategies going beyond both xenobiotic pesticides and traditional biological control applications (Figure 1).
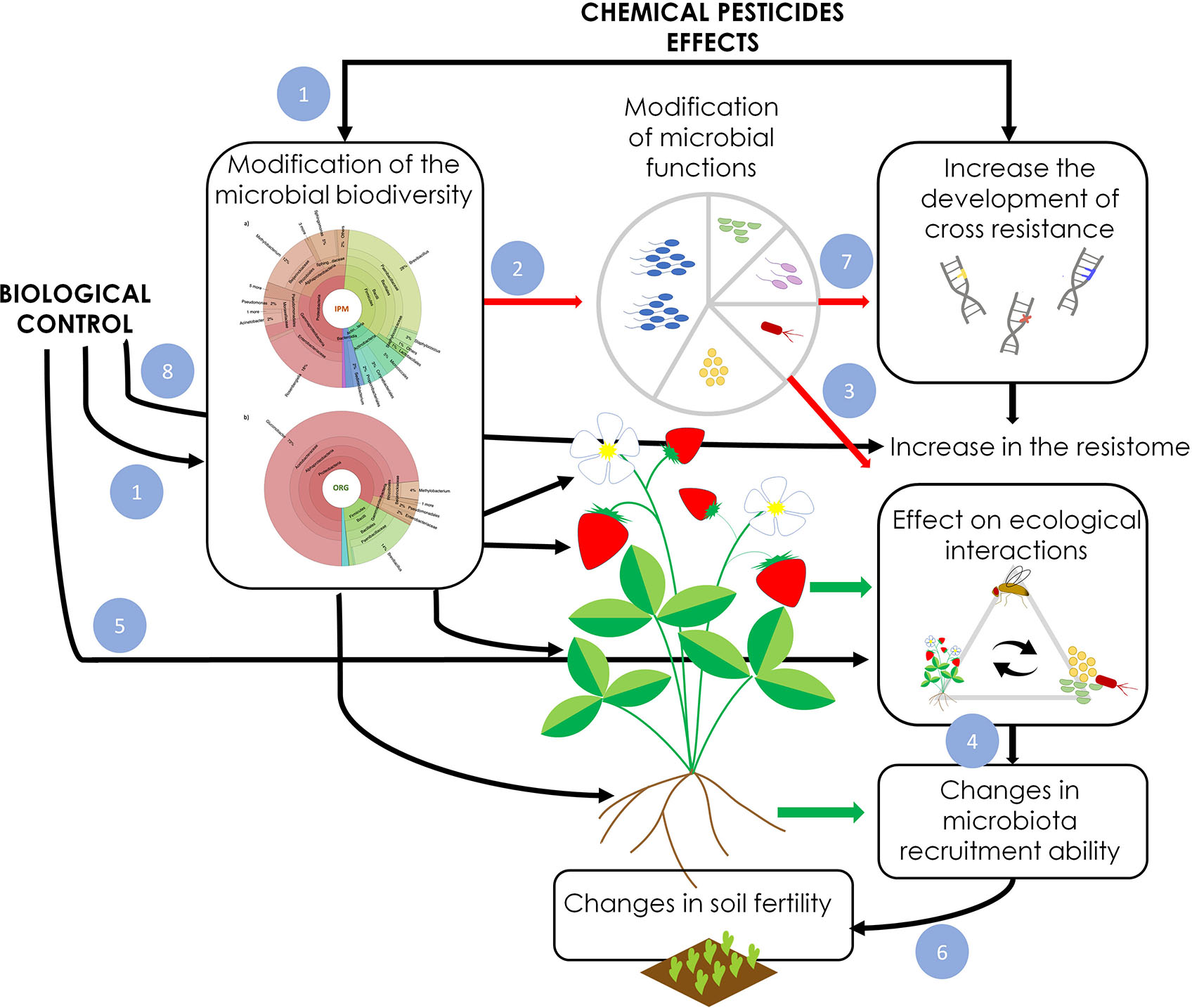
Figure 2 Effects of pesticides and biological control application on the phytobiota. Black and red arrows indicate direct and indirect effects, respectively. The green arrow highlights plant-mediated effects. Both pesticides and biological control agents directly modify (1) the structure of the plant-associated microbial communities of flower, fruit, leaves, root and endosphere. The modifications of phytobiota structure and composition leads to changes (2) in microbial functions which directly affect plant fitness, resistance and phenotype. Moreover, the shifts in microbial functions influence plant ecological interactions (3), including the interplay with pests and pollinators (Liu et al., 2019). Since phytobiota composition and functionality is influenced by the exchange of microbes vectored by other organisms [e.g. pollinators (Keller et al., 2021) and pests (Humphrey and Whiteman, 2020)], pesticides use and biological control influence the plant ability to recruit beneficial microbes (4). These effects are also mediated by the plant which responds to the changes in the phytobioma by producing different signaling molecules (e.g. VOCs, root exudates) (Park and Ryu, 2021). The application of biological control agents directly influences these mechanisms (5). The changes in the phytobiota, the ecological interactions and the recruitment of beneficial microbes affect soil fertility (6), which may further amplify the long-term effects on phytobiota, plant fitness, production and fruit quality. Finally, the pesticide and biological control application may facilitate the selection of broad resistance against antimicrobial compounds (7) which may result in an expansion of the microbial resistome with detrimental effects on human health. Finally, the application of microbial biological control agents, introducing alien microorganisms, may facilitate the horizontal transfer of new genes with the native microbiome directly influencing the expansion of the resistome (8).
Authors equally contributed to the manuscript. EV drafted paragraphs 1, 3 and 5. FS drafted paragraphs 2 and 5. DS paragraph 3 and 4. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aßhauer K. P., Wemheuer B., Daniel R., Meinicke P. (2015). Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data: Fig. 1. Bioinformatics 31, 2882–2884. doi: 10.1093/bioinformatics/btv287
Agler M. T., Ruhe J., Kroll S., Morhenn C., Kim S.-T., Weigel D., et al. (2016). Microbial hub taxa link host and abiotic factors to plant microbiome variation. PloS Biol. 14, e1002352. doi: 10.1371/journal.pbio.1002352
Anderson A. J., Kang B. R., Kim Y. C. (2017). The Gac/Rsm signaling pathway of a biocontrol bacterium, pseudomonas chlororaphis O6. Res. Plant Dis. 23, 212–227. doi: 10.5423/RPD.2017.23.3.212
Araujo R., Dunlap C., Barnett S., Franco C. M. M. (2019). Decoding wheat endosphere–rhizosphere microbiomes in rhizoctonia solani–infested soils challenged by streptomyces biocontrol agents. Front. Plant Sci. 10. doi: 10.3389/FPLS.2019.01038/BIBTEX
Baltrus D. A. (2017). Adaptation, specialization, and coevolution within phytobiomes. Curr. Opin. Plant Biol. 38, 109–116. doi: 10.1016/j.pbi.2017.04.023
Barbosa M., Fernandes G. W., Lewis O. T., Morris R. J. (2017). Experimentally reducing species abundance indirectly affects food web structure and robustness. J. Anim. Ecol. 86, 327–336. doi: 10.1111/1365-2656.12626
Berendsen R. L., Pieterse C. M. J., Bakker P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Berg G. (2009). Plant–microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 84, 11–18. doi: 10.1007/s00253-009-2092-7
Berg G., Kusstatscher P., Abdelfattah A., Cernava T., Smalla K. (2021). Microbiome modulation–toward a better understanding of plant microbiome response to microbial inoculants. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.650610
Berg G., Rybakova D., Grube M., Köberl M. (2016). The plant microbiome explored: implications for experimental botany. J. Exp. Bot. 67, 995–1002. doi: 10.1093/jxb/erv466
Bill M., Chidamba L., Gokul J. K., Labuschagne N., Korsten L. (2021). Bacterial community dynamics and functional profiling of soils from conventional and organic cropping systems 157, 103734. doi: 10.1016/j.apsoil.2020.103734
Blair J. M., Webber M. A., Baylay A. J., Ogbolu D. O., Piddock L. J. (2015). Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42–51. doi: 10.1038/nrmicro3380
Brandl M. T. (2006). Fitness of human enteric pathogens on plants and implications for food safety. Ann. Rev. Phytopathol. 44, 367–392. doi: 10.1146/annurev.phyto.44.070505.143359
Bringel F., Couée I. (2015). Pivotal roles of phyllosphere microorganisms at the interface between plant functioning and atmospheric trace gas dynamics. Front. Microbiol. 6. doi: 10.3389/FMICB.2015.00486
Brown M. W., Mathews C. R. (2007). Conservation biological control of rosy apple aphid, dysaphis plantaginea (Passerini), in Eastern north America. Environ. Entomol. 36, 1131–1139. doi: 10.1603/0046-225X(2007)36[1131:CBCORA]2.0.CO;2
Caballo-Ponce E., Meng X., Uzelac G., Halliday N., Cámara M., Licastro D., et al. (2018). Quorum sensing in pseudomonas savastanoi pv. savastanoi and erwinia toletana: role in virulence and interspecies interactions in the olive knot. Appl. Environ. Microbiol. 84 (18), e00950. doi: 10.1128/AEM.00950-18
Cai F., Pang G., Miao Y., Li R., Li R., Shen Q., et al. (2017). The nutrient preference of plants influences their rhizosphere microbiome. Appl. Soil Ecol. 110, 146–150. doi: 10.1371/JOURNAL.PONE.0106355
Caldwell A. C., Silva L. C.F., da Silva C. C., Ouverney C. C (2015). Prokaryotic diversity in the rhizosphere of organic, intensive, and transitional coffee farms in Brazil. PloS One 10 (6), e0106355. doi: 10.1371/journal.pone.0106355
Castellano-Hinojosa A., Strauss S. L. (2020). Impact of cover crops on the soil microbiome of tree crops. Microorg. 8, 328. doi: 10.3390/MICROORGANISMS8030328
Cernava T. (2021). How microbiome studies could further improve biological control. Biol. Control 160, 104669. doi: 10.1016/j.biocontrol.2021.104669
Cernava T., Chen X., Krug L., Li H., Yang M., Berg G. (2019a). The tea leaf microbiome shows specific responses to chemical pesticides and biocontrol applications. Sci. Total Environ. 667, 33–40. doi: 10.1016/j.scitotenv.2019.02.319
Cernava T., Erlacher A., Soh J., Sensen C. V., Grube M., Berg G. (2019b). Enterobacteriaceae dominate the core microbiome and contribute to the resistome of arugula (Eruca sativa mill.). Microbiome 7, 13. doi: 10.1186/s40168-019-0624-7
Chaney L., Baucom R. S. (2020). The soil microbial community alters patterns of selection on flowering time and fitness-related traits in ipomoea purpurea. Am. J. Bot. 107, 186–194. doi: 10.1002/ajb2.1426
Chaparro J. M., Sheflin A. M., Manter D. K., Vivanco J. M. (2012). Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fertil. Soils 48, 489–499. doi: 10.1007/s00374-012-0691-4
Chen T., Nomura K., Wang X., Sohrabi R., Xu J., Yao L., et al. (2020). A plant genetic network for preventing dysbiosis in the phyllosphere. Nature 580, 653–657. doi: 10.1038/s41586-020-2185-0
Chen X., Wicaksono W. A., Berg G., Cernava T. (2021). Bacterial communities in the plant phyllosphere harbour distinct responders to a broad-spectrum pesticide. Sci. Total Environ. 751, 141799. doi: 10.1016/J.SCITOTENV.2020.141799
Cohen S. P., Levy S. B., Foulds J., Rosner J. L. (1993). Salicylate induction of antibiotic resistance in escherichia coli: activation of the mar operon and a mar-independent pathway. J. Bacteriol 175, 7856–7862. doi: 10.1128/jb.175.24.7856-7862.1993
Compant S., Samad A., Faist H., Sessitsch A. (2019). A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 19, 29–37. doi: 10.1016/j.jare.2019.03.004
Conrath U., Beckers G. J. M., Langenbach C. J. G., Jaskiewicz M. R. (2015). Priming for enhanced defense. Annu. Rev. Phytopathol. 53, 97–119. doi: 10.1146/annurev-phyto-080614-120132
Cui Z., Huntley R. B., Schultes N. P., Steven B., Zeng Q. (2021). Inoculation of stigma-colonizing microbes to apple stigmas alters microbiome structure and reduces the occurrence of fire blight disease. Phytobiomes J. 5 (2), 156–165. doi: 10.1094/PBIOMES-04-20-0035-R
Cullen R., Warner K. D., Jonsson M., Wratten S. D. (2008). Economics and adoption of conservation biological control. Biol. Control 45, 272–280. doi: 10.1016/j.biocontrol.2008.01.016
Cusano A. M., Burlinson P., Deveau A., Vion P., Uroz S., Preston G. M., et al. (2011). Pseudomonas fluorescens BBc6R8 type III secretion mutants no longer promote ectomycorrhizal symbiosis: MHB effect lost in T3SS mutants. Environ. Microbiol. Rep. 3, 203–210. doi: 10.1111/j.1758-2229.2010.00209.x
Cycoń M., Piotrowska-Seget Z. (2015). Biochemical and microbial soil functioning after application of the insecticide imidacloprid. J. Environ. Sci. 27, 147–158. doi: 10.1016/j.jes.2014.05.034
Dicke M. (2016). Plant phenotypic plasticity in the phytobiome: a volatile issue. Curr. Opin. Plant Biol. 32, 17–23. doi: 10.1016/j.pbi.2016.05.004
Double M. L., Jarosz A. M., Fulbright D. W., Davelos Baines A., MacDonald W. L. (2018). Evaluation of two decades of cryphonectria parasitica hypovirus introduction in an american chestnut stand in Wisconsin. Phytopathol. 108, 702–710. doi: 10.1094/PHYTO-10-17-0354-R
Douglas G. M., Maffei V. J., Zaneveld J. R., Yurgel S. N., Brown J. R., Taylor C. M., et al. (2020). PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 38, 685–688. doi: 10.1038/s41587-020-0548-6
EFSA Panel on Plant Protection Products and their Residues (PPR) (2010). Scientific opinion on the development of specific protection goal options for environmental risk assessment of pesticides, in particular in relation to the revision of the guidance documents on aquatic and terrestrial ecotoxicology (SANCO/3268/2001 and SANCO/10329/2002). EFSA J. 8.10, 1821. doi: 10.2903/j.efsa.2010.1821
Elad Y. (2000). Biological control of foliar pathogens by means of trichoderma harzianum and potential modes of action. Crop Prot. 19, 709–714. doi: 10.1016/S0261-2194(00)00094-6
Ellis J. G (2017). Can plant microbiome studies lead to effective biocontrol of plant diseases? Molecular Plant-Microbe Interactions 30 (3), 190–193. doi: 10.1094/MPMI-12-16-0252-CR
Elsayed T. R., Jacquiod S., Nour E. H., Sørensen S. J., Smalla K. (2020). Biocontrol of bacterial wilt disease through complex interaction between tomato plant, antagonists, the indigenous rhizosphere microbiota, and Ralstonia solanacearum. Front. Microbiol. 10, 2835. doi: 10.3389/fmicb.2019.02835
Erlacher A., Cardinale M., Grube M., Berg G. (2015). Biotic stress shifted structure and abundance of enterobacteriaceae in the lettuce microbiome. PloS One 10, e011806. doi: 10.1371/journal.pone.0118068
Esperschütz J., Gattinger A., Mäder P., Schloter M., Fließbach A. (2007). Response of soil microbial biomass and community structures to conventional and organic farming systems under identical crop rotations. FEMS Microbiol. Ecol. 61, 26–37. doi: 10.1111/J.1574-6941.2007.00318.X
Etemadi M., Zuther E., Müller H., Hincha D. K., Berg G. (2018). Ecotype-dependent response of bacterial communities associated with arabidopsis to cold acclimation. Phytobiomes J. 2, 3–13. doi: 10.1094/PBIOMES-04-17-0015-R/ASSET/IMAGES/LARGE/PBIOMES-04-17-0015-R_F7.JPEG
EUROSTAT- European Statistics (2021) Agri-environmental indicator - consumption of pesticides. Available at: https://ec.europa.eu/eurostat/statistics-explained/index.php?title=Agri-environmental_indicator_-_consumption_of_pesticides#Key_messages (Accessed April 29, 2022).
Frost L. S., Leplae R., Summers A. O., Toussaint A. (2005). Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732. doi: 10.1038/nrmicro1235
Gaba S., Gabriel E., Chadœuf J., Bonneu F., Bretagnolle V. (2016). Herbicides do not ensure for higher wheat yield, but eliminate rare plant species. Sci. Rep. 6, 30112. doi: 10.1038/srep30112
Ghorbanpour M., Omidvari M., Abbaszadeh-Dahaji P., Omidvar R., Kariman K. (2018). Mechanisms underlying the protective effects of beneficial fungi against plant diseases. Biol. Control 117, 147–157. doi: 10.1016/j.biocontrol.2017.11.006
Glenn D. M., Bassett C., and Dowd S. E. (2015). Effect of pest management system on ‘Empire’apple leaf phyllosphere populations. Scientia Horticulturae 183, 58–65. doi: 10.1016/j.scienta.2014.12.009
Granado J., Thürig B., Kieffer E., Petrini L., Flieβbach A., Tamm L., et al. (2008). Culturable fungi of stored ‘golden delicious’ apple fruits: a one-season comparison study of organic and integrated production systems in Switzerland. Microbial Ecol. 56 (4), 720–732. doi: 10.1007/s00248-008-9391-x
Gschwend F., Braun-Kiewnick A., Widmer F., Pelludat C.. (2021). Apple blossoms from a swiss orchard with low-input plant protection regime reveal high abundance of potential fire blight antagonists. Phytobiomes J. 5 (2), 145–155. doi: 10.1094/PBIOMES-04-20-0033-R
Hacquard S., Garrido-Oter R., González A., Spaepen S., Ackermann G., Lebeis S., et al. (2015). Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe 17, 603–616. doi: 10.1016/j.chom.2015.04.009
Hartman K., van der Heijden M. G., Wittwer R. A., Banerjee S., Walser J. C., Schlaeppi K, et al (2018). Cropping practices manipulate abundance patterns of root and soil microbiome members paving the way to smart farming. Microbiome 6 (1), 1–14. doi: 10.1038/ismej.2014.210 10.1186/s40168-017-0389-9
Hartmann M., Frey B., Mayer J., Mäder P., Widmer F. (2015). Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 95 9, 1177–1194. doi: 10.1038/ismej.2014.210
Hol W. H. G., de Boer W., Termorshuizen A. J., Meyer K. M., Schneider J. H. M., van Dam N. M., et al. (2010). Reduction of rare soil microbes modifies plant-herbivore interactions. Ecol. Lett. 13, 292–301. doi: 10.1111/j.1461-0248.2009.01424.x
Huang W., Lu Y., Chen L., Sun D., An Y. (2021a). Impact of pesticide/fertilizer mixtures on the rhizosphere microbial community of field-grown sugarcane. Biotech. 11, 1–10. doi: 10.1007/S13205-021-02770-3
Huang W., Sun D., Lu Y., Dai S., Chen L., An Y. (2021b). Effects of pesticide–fertilizer combinations on the rhizosphere microbiome of sugarcane: A preliminary study. Sugar Tech. 23, 571–579. doi: 10.1007/S12355-020-00914-Y
Humphrey P. T., Whiteman N. K. (2020). Insect herbivory reshapes a native leaf microbiome. Nat. Ecol. Evol. 4, 221–229. doi: 10.1038/s41559-019-1085-x
Ioos R., Belhadj A., Menez M., Faure A. (2005). The effects of fungicides on fusarium spp. and microdochium nivale and their associated trichothecene mycotoxins in French naturally-infected cereal grains. J. Crop Prot. 24, 894–902. doi: 10.1016/j.cropro.2005.01.014
Ishaq S. L., Johnson S. P., Miller Z. J., Lehnhoff E. A., Olivo S., Yeoman C. J., et al. (2017). Impact of cropping systems, soil inoculum, and plant species identity on soil bacterial community structure. Microb. Ecol. 73, 417–434. doi: 10.1007/S00248-016-0861-2
Jensen B., Knudsen I. M., Andersen B., Nielsen K. F., Thrane U., Jensen D. F, et al (2013). Characterization of microbial communities and fungal metabolites on field grown strawberries from organic and conventional production. Inter. J. Food Microbiol. 160 (3), 313–322. doi: 10.1016/j.ijfoodmicro.2012.11.005
Jonason D., Andersson G. K. S., Öckinger E., Rundlöf M., Smith H. G., Bengtsson J. (2011). Assessing the effect of the time since transition to organic farming on plants and butterflies. J. Appl. Ecol. 48, 543–550. doi: 10.1111/J.1365-2664.2011.01989.X
Kamada N., Chen G. Y., Inohara N., Núñez G. (2013). Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 14, 685–690. doi: 10.1038/ni.2608
Karas P. A., Baguelin C., Pertile G., Papadopoulou E. S., Nikolaki S., Storck V., et al. (2018). Assessment of the impact of three pesticides on microbial dynamics and functions in a lab-to-field experimental approach. Sci. Total Environ. 637–638, 636–646. doi: 10.1016/J.SCITOTENV.2018.05.073
Karlsson I., Friberg H., Kolseth A. K., Steinberg C., Persson P. (2017). Organic farming increases richness of fungal taxa in the wheat phyllosphere. Mol. Ecol. 26, 3424–3436. doi: 10.1111/MEC.14132
Keller A., McFrederick Q. S., Steffan S., Danforth B. N., Leonhardt S. D. (2021). (More than) hitchhikers through the network: the shared microbiome of bees and flowers. Cur. Opin. Insect Sc. 44, 8–15. doi: 10.1016/j.pbi.2014.04.005
Kemen E. (2014). Microbe–microbe interactions determine oomycete and fungal host colonization. Curr. Op. Plant Biol. 20, 75–81. doi: 10.1016/j.pbi.2014.04.005
Khaitov B., Patiño-Ruiz J. D., Pina T., Schausberger P. (2015). Interrelated effects of mycorrhiza and free-living nitrogen fixers cascade up to aboveground herbivores. Ecol. Evol. 5, 3756–3768. doi: 10.1002/ece3.1654
Khoiri A. N., Cheevadhanarak S., Jirakkakul J., Dulsawat S., Prommeenate P., Tachaleat A., et al. (2021). Comparative metagenomics reveals microbial signatures of sugarcane phyllosphere in organic management. Front. Microbiol. 12. doi: 10.3389/FMICB.2021.623799/BIBTEX
Köhl J., Kolnaar R., Ravensberg W. J. (2019). Mode of action of microbial biological control agents against plant diseases: relevance beyond efficacy. Front. Plant Sci 10, 845. doi: 10.3389/fpls.2019.00845
Kurenbach B., Marjoshi D., Amabile-Cuevas C. F., Ferguson G. C., Godsoe W., Gibson P., et al. (2015). Sublethal exposure to commercial formulations of the herbicides dicamba, 2,4-dichlorophenoxyacetic acid, and glyphosate cause changes in antibiotic susceptibility in escherichia coli and salmonella enterica serovar typhimurium. MBio 6, e00009–e00015. doi: 10.1128/mBio.00009-15
Kusstatscher P., Wicaksono W. A., Thenappan D. P., Adam E., Müller H., Berg G. (2020). Microbiome management by biological and chemical treatments in maize is linked to plant health. Microorganisms 8 (10), 1506. doi: 10.3390/microorganisms8101506
Kwak M.-J., Kong H. G., Choi K., Kwon S.-K., Song J. Y., Lee J., et al. (2018). Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 36, 1100–1109. doi: 10.1038/nbt.4232
Lamichhane J. R., Venturi V. (2015). Synergisms between microbial pathogens in plant disease complexes: a growing trend. Front. Plant Sci. 06. doi: 10.3389/fpls.2015.00385
Latour X. (2020). The evanescent GacS signal. Microorganisms 8, 1746. doi: 10.3390/microorganisms8111746
Lau J. A., Lennon J. T. (2011). Evolutionary ecology of plant–microbe interactions: soil microbial structure alters selection on plant traits. New Phytol. 192, 215–224. doi: 10.1111/j.1469-8137.2011.03790.x
Leff J. W., Fierer N. (2013). Bacterial communities associated with the surfaces of fresh fruits and vegetables. PloS One 8, e59310. doi: 10.1371/journal.pone.0059310
Liu H., Macdonald C. A., Cook J., Anderson I. C., Singh B. K. (2019). An ecological loop: host microbiomes across multitrophic interactions. Trends Ecol. Evol. 34, 1118–1130. doi: 10.1126/science.aaf4507/suppl_file/louca.sm.pdf
Louca S., Parfrey L. W., Doebeli M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science 353 (6305), 1272–1277. doi: 10.1126/science.aaf4507
Lupatini M., Korthals G. W., de Hollander M., Janssens T. K. S., Kuramae E. E. (2017). Soil microbiome is more heterogeneous in organic than in conventional farming system. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.02064
Marschner P., Rumberger A. (2004). Rapid changes in the rhizosphere bacterial community structure during re-colonization of sterilized soil. Biol. Fertil. Soils 40, 1–6. doi: 10.1007/s00374-004-0736-4
Martins dos Santos V., Müller M., de Vos W. M. (2010). Systems biology of the gut: the interplay of food, microbiota and host at the mucosal interface. Curr. Opin. Biotechnol. 21, 539–550. doi: 10.1016/j.copbio.2010.08.003
Martins G., Vallance J., Mercier A., Albertin W., Stamatopoulos P., Rey P., et al. (2014). Influence of the farming system on the epiphytic yeasts and yeast-like fungi colonizing grape berries during the ripening process. Int. J. Food Microbiol. 177, 21–28. doi: 10.1016/j.ijfoodmicro.2014.02.002
Medo J., Maková J., Kovácsová S., Majerčíková K., Javoreková S. (2015). Effect of dursban 480 EC (chlorpyrifos) and talstar 10 EC (bifenthrin) on the physiological and genetic diversity of microorganisms in soil. J. Environ. Sci. Health B. 50, 871–883. doi: 10.1080/03601234.2015.1062659
Méndez-Bravo A., Cortazar-Murillo E. M., Guevara-Avendaño E., Ceballos-Luna O., Rodríguez-Haas B., Kiel-Martínez A. L., et al. (2018). Plant growth-promoting rhizobacteria associated with avocado display antagonistic activity against phytophthora cinnamomi through volatile emissions. PloS One 13, e0194665. doi: 10.1371/journal.pone.0194665
Milgroom M. G., Cortesi P. (2004). Biological control of chestnut blight with hypovirulence: a critical analysis. Annu. Rev. Phytopathol. 42, 311–338. doi: 10.1146/annurev.phyto.42.040803.140325
Noel Z. A., Longley R., Benucci G. M. N., Trail F., Chilvers M. I., Bonito G, et al (2022). Non-target impacts of fungicide disturbance on phyllosphere yeasts in conventional and no-till management. ISME Communications 2 (1), 1–10. doi: 10.1038/s43705-022-00103-w
OECD (2000) OECD guideline for the testing of chemicals - 216, soil microorganisms: Nitrogen transformation test) (Accessed April 29, 2022).
Ottesen A. R., White J. R., Skaltsas D. N., Newell M. J., Walsh C. S. (2009). Impact of organic and conventional management on the phyllosphere microbial ecology of an apple crop. J. Food Protection 72 (11), 2321–2325. doi: 10.4315/0362-028X-72.11.2321
Paasch B. C., He S. Y. (2021). Toward understanding microbiota homeostasis in the plant kingdom. PloS Pathog. 17, e1009472. doi: 10.1371/journal.ppat.1009472
Park Y. S., Ryu C. M. (2021). Understanding plant social networking system: avoiding deleterious microbiota but calling beneficials. Int. J. Mol. Sci. 22 (7), 3319. doi: 10.3390/ijms22073319
Paungfoo-Lonhienne C., Yeoh Y. K., Kasinadhuni N. R. P., Lonhienne T. G. A., Robinson N., Hugenholtz P., et al. (2015). Nitrogen fertilizer dose alters fungal communities in sugarcane soil and rhizosphere. Sci. Rep. 51 5, 1–6. doi: 10.1038/srep08678
Perazzolli M., Antonielli L., Storari M., Puopolo G., Pancher M., Giovannini O., et al. (2014). Resilience of the natural phyllosphere microbiota of the grapevine to chemical and biological pesticides. Appl. Environ. Microbiol. 80, 3585–3596. doi: 10.1128/aem.00415-14/suppl_file/zam999105399so1.pdf
Pieterse C. M. J., Dicke M. (2007). Plant interactions with microbes and insects: from molecular mechanisms to ecology. Trends Plant Sci. 12, 564–569. doi: 10.1016/j.tplants.2007.09.004
Pieterse C. M. J., Zamioudis C., Berendsen R. L., Weller D. M., Van Wees S. C. M., Bakker P. A. H. M. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. doi: 10.1146/annurev-phyto-082712-102340
Purahong W., Orrù L., Donati I., Perpetuini G., Cellini A., Lamontanara A., et al. (2018). Plant microbiome and its link to plant health: host species, organs and pseudomonas syringae pv. actinidiae infection shaping bacterial phyllosphere communities of kiwifruit plants. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01563
Qian H., Zhu Y., Chen S., Jin Y., Lavoie M., Ke M, et al. (2018). Interacting effect of diclofop-methyl on the rice rhizosphere microbiome and denitrification. Pesticide Biochem. Physiol. 146, 90–96. doi: 10.1016/j.pestbp.2018.03.002
Qin C., Tao J., Liu T., Liu Y., Xiao N., Li T., et al. (2019). Responses of phyllosphere microbiota and plant health to application of two different biocontrol agents. AMB Express 9, 1–13. doi: 10.1186/S13568-019-0765-X/FIGURES/6
Raaijmakers J. M., Mazzola M. (2012). Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu. Rev. Phytopathol. 50, 403–424. doi: 10.1146/annurev-phyto-081211-172908
Ramakrishna W., Yadav R., Li K. (2019). Plant growth promoting bacteria in agriculture: Two sides of a coin. Appl. Soil Ecol. 138, 10–18. doi: 10.1016/J.APSOIL.2019.02.019
Remus-Emsermann M. N. P., Schlechter R. O. (2018). Phyllosphere microbiology: at the interface between microbial individuals and the plant host. New Phytol. 218, 1327–1333. doi: 10.1111/NPH.15054
Reverchon F., Bai S. H., Liu X., Blumfield T. J. (2015). Tree plantation systems influence nitrogen retention and the abundance of nitrogen functional genes in the Solomon islands. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.01439
Roquigny R., Novinscak A., Léger G., Marcoux N., Joly D. L., Filion M. (2018). Deciphering the rhizosphere and geocaulosphere microbiomes of potato following inoculation with the biocontrol agent pseudomonas fluorescens strain LBUM223. Phytobiomes J. 2, 92–99. doi: 10.1094/pbiomes-03-18-0013-r/asset/images/large/pbiomes-03-18-0013-r
Sanders D., Kehoe R., van Veen F. J. F. (2015). Experimental evidence for the population-dynamic mechanisms underlying extinction cascades of carnivores. Curr. Biol. 25, 3106–3109. doi: 10.1016/j.cub.2015.10.017
Sangiorgio D., Cellini A., Donati I., Ferrari E., Tanunchai B., Fareed Mohamed Wahdan S., et al. (2022a). Taxonomical and functional composition of strawberry microbiome is genotype-dependent. J. Adv. Res. doi: 10.1016/J.JARE.2022.02.009
Sangiorgio D., Cellini A., Spinelli F., Farneti B., Khomenko I., Muzzi E., et al. (2021). Does organic farming increase raspberry quality, aroma and beneficial bacterial biodiversity? Microorganisms 9, 1617. doi: 10.3390/MICROORGANISMS9081617
Sangiorgio D., Cellini A., Spinelli F., Pastore C., Farneti B., Savioli S., et al. (2022b). Contribution of fruit microbiome to raspberry volatile organic compounds emission. Postharvest Biol. Technol. 183, 111742. doi: 10.1016/J.POSTHARVBIO.2021.111742
Schmidt R., Köberl M., Mostafa A., Ramadan E., Monschein M., Jensen K., et al. (2014). Effects of bacterial inoculants on the indigenous microbiome and secondary metabolites of chamomile plants. Front. Microbiol. 5. doi: 10.3389/fmicb.2014.00064
Scholthof K.-B. G. (2007). The disease triangle: pathogens, the environment and society. Nat. Rev. Microbiol. 5, 152–156. doi: 10.1038/nrmicro1596
Sekirov I., Russell S. L., Antunes L. C. M., Finlay B. B. (2010). Gut microbiota in health and disease. Physiol. Rev. 90, 859–904. doi: 10.1152/physrev.00045.2009
Sharma A., Kumar V., Shahzad B., Tanveer M., Sidhu G. P. S., Handa N., et al. (2019). Worldwide pesticide usage and its impacts on ecosystem. SN Appl. Sci. 1, 1–16. doi: 10.1007/S42452-019-1485-1
Sharuddin S. S., Ramli N., Yusoff M. Z. M., Muhammad N. A. N., Ho L. S., Maeda T. (2022). Advancement of metatranscriptomics towards productive agriculture and sustainable environment: a review. Int. J. Mol. Sci. 23, 3737. doi: 10.3390/ijms23073737
Singer M. (2010). Pathogen-pathogen interaction: a syndemic model of complex biosocial processes in disease. Virulence 1, 10–18. doi: 10.4161/viru.1.1.9933
Spadaro D., Droby S. (2016). Development of biocontrol products for postharvest diseases of fruit: The importance of elucidating the mechanisms of action of yeast antagonists. Trends Food Sci. Technol. 47, 39–49. doi: 10.1016/j.tifs.2015.11.003
Sylla J., Alsanius B. W., Krüger E., Reineke A., Bischoff-Schaefer M., Wohanka W, et al (2013a). Introduction of Aureobasidium pullulans to the phyllosphere of organically grown strawberries with focus on its establishment and interactions with the resident microbiome. Agronomy 3 (4), 704–731. doi: 10.3390/agronomy3040704
Sylla J., Alsanius B. W., Krüger E., Becker D., Wohanka W.. (2013b). In vitro compatibility of microbial agents for simultaneous application to control strawberry powdery mildew (Podosphaera aphanis). Crop Protection 51, 40–47. doi: 10.1016/j.cropro.2013.04.011
Tan J., De Zutter N., De Saeger S., De Boevre M., Tran T. M., van der Lee T., et al. (2021). Presence of the weakly pathogenic fusarium poae in the fusarium head blight disease complex hampers biocontrol and chemical control of the virulent fusarium graminearum pathogen. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.641890
Teplitski M., Warriner K., Bartz J., Schneider K. R. (2011). Untangling metabolic and communication networks: interactions of enterics with phytobacteria and their implications in produce safety. Trends Microbiol. 19, 121–127. doi: 10.1016/j.tim.2010.11.007
Vaughan M., Backhouse D., Ponte E. M. D. (2016). Climate change impacts on the ecology of fusarium graminearum species complex and susceptibility of wheat to fusarium head blight: a review. World Mycotoxin J. 9, 685–700. doi: 10.3920/WMJ2016.2053
Venturi V., Keel C. (2016). Signaling in the rhizosphere. Trends Plant Sci. 21, 187–198. doi: 10.1016/j.tplants.2016.01.005
Viera-Arroyo W. F., Tello-Torres C. M., Martínez-Salinas A. A., Navia-Santillán D. F., Medina-Rivera L. A., Delgado-Párraga A. G., et al. (2020). Biological control: A tool for sustainable agriculture, a point of view of its benefits in Ecuador. J. Selva Andina Biosph. 8, 128–149. doi: 10.36610/j.jsab.2020.080200128x
Vitulo N., Lemos Jr W. J.F., Calgaro M., Confalone M., Felis G. E., Zapparoli G, et al. (2019). Bark and grape microbiome of Vitis vinifera: influence of geographic patterns and agronomic management on bacterial diversity. Front. Microbiol. 9, 3203. doi: 10.3389/fmicb.2018.03203
Vryzas Z. (2016). The plant as metaorganism and research on next-generation systemic pesticides - prospects and challenges. Front. Microbiol. 7. doi: 10.3389/FMICB.2016.01968
Wagner M. R., Lundberg D. S., Coleman-Derr D., Tringe S. G., Dangl J. L., Mitchell-Olds T. (2014). Natural soil microbes alter flowering phenology and the intensity of selection on flowering time in a wild arabidopsis relative. Ecol. Lett. 17, 717–726. doi: 10.1111/ele.12276
Wang C., Wang F., Zhang Q., Liang W. (2016). Individual and combined effects of tebuconazole and carbendazim on soil microbial activity. Eur. J. Soil Biol. 72, 6–13. doi: 10.1016/j.ejsobi.2015.12.005
Wassermann B., Cernava T., Müller H., Berg C., Berg G. (2019a). Seeds of native alpine plants host unique microbial communities embedded in cross-kingdom networks. Microbiome 7, 108. doi: 10.1186/s40168-019-0723-5
Wassermann B., Müller H., Berg G. (2019b). An apple a day: which bacteria do we eat with organic and conventional apples? Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01629
Wattenburger C. J., Halverson L. J., Hofmockel K. S. (2019). Agricultural management affects root-associated microbiome recruitment over maize development. Phytobiomes J. 3, 260–272. doi: 10.1094/PBIOMES-03-19-0016-R/ASSET/IMAGES/LARGE/PBIOMES-03-19-0016-R
Wei F., Hu X., Xu X. (2016). Dispersal of bacillus subtilis and its effect on strawberry phyllosphere microbiota under open field and protection conditions. Sci. Rep. 6, 1–9. doi: 10.1038/srep22611
Wu X., Yin Y., Wang S., Yu Y. (2014). Accumulation of chlorothalonil and its metabolite, 4-hydroxychlorothalonil, in soil after repeated applications and its effects on soil microbial activities under greenhouse conditions. Environ. Sci. pollut. Res. 21, 3452–3459. doi: 10.1007/S11356-013-2318-1
Xiong W., Guo S., Jousset A., Zhao Q., Wu H., Li R., et al. (2017). Bio-fertilizer application induces soil suppressiveness against fusarium wilt disease by reshaping the soil microbiome. Soil Biol. Bioch. 114, 238–247. doi: 10.1007/s10658-005-2446-7
Xu X. M., Parry D. W., Nicholson P., Thomsett M. A., Simpson D., Edwards S. G., et al (2005). Predominance and association of pathogenic fungi causing Fusarium ear blightin wheat in four European countries. CA Cancer J Clin 112 (2), 143–154. doi: 10.1007/s10658-005-2446-7
Xu N., Qu Q., Zhang Z., Yuan W., Cui H., Shen Y., et al. (2020). Effects of residual s-metolachlor in soil on the phyllosphere microbial communities of wheat (Triticum aestivum l.). Sci. Total Environ. 748, 141342. doi: 10.1016/J.SCITOTENV.2020.141342
Yin Z., Liu X., Qian C., Sun L., Pang S., Liu J., et al. (2022). Pan-genome analysis of delftia tsuruhatensis reveals important traits concerning the genetic diversity, pathogenicity, and biotechnological properties of the specie. Microbiol. Spectr. 10, e0207221. doi: 10.1128/spectrum.02072-21
Yli-Mattila T. (2010). Ecology and evolution of toxigenic fusarium species in cereals in northern Europe and Asia. J. Plant Pathol. 92, 7–18. doi: 10.4454/jpp.v92i1.10
Zhimo V. Y., Kumar A., Biasi A., Salim S., Feygenberg O., Toamy M. A., et al. (2021). Compositional shifts in the strawberry fruit microbiome in response to near-harvest application of metschnikowia fructicola, a yeast biocontrol agent. Postharvest Biol. Technol. 175, 111469. doi: 10.1016/J.POSTHARVBIO.2021.111469
Keywords: crop protection, biological control, plant resistome, microbiome, phyllosphere, bacterial signaling, plant pathogen
Citation: Sangiorgio D, Spinelli F and Vandelle E (2022) The unseen effect of pesticides: The impact on phytobiota structure and functions. Front. Agron. 4:936032. doi: 10.3389/fagro.2022.936032
Received: 04 May 2022; Accepted: 15 July 2022;
Published: 09 August 2022.
Edited by:
Davide Spadaro, University of Turin, ItalyReviewed by:
Muneer Ahmad Malla, Dr. Hari Singh Gour University, IndiaCopyright © 2022 Sangiorgio, Spinelli and Vandelle. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francesco Spinelli, ZnJhbmNlc2NvLnNwaW5lbGxpM0B1bmliby5pdA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.