- 1Environmental Genomics and Systems Biology Research Group, Institute for Natural Resource Sciences, Zurich University of Applied Sciences (ZHAW), Wädenswil, Switzerland
- 2Molecular Plant Breeding, Institute of Agricultural Sciences, ETH Zurich, Zurich, Switzerland
- 3Plant Sciences Unit, Flanders Research Institute for Agriculture, Fisheries and Food (ILVO), Merelbeke, Belgium
- 4Bioinformatics and Systems Biology, Justus-Liebig University Giessen, Giessen, Germany
The seven pathovars of Xanthomonas hortorum and Xanthomonas hydrangeae, referred to as the X. hortorum – X. hydrangeae species complex, cause disease on a multitude of plants, including crops, ornamental and wild plants. Cross-pathogenicity was proven for some of the strains within this species complex. It is thus important to have highly specific and fast diagnostics methods for members of the X. hortorum – X. hydrangeae species complex. A comparative genomic analysis was conducted for representative members within the complex to identify singletons for use as genomic targets for the assays. Seven loop-mediated isothermal amplification (LAMP) diagnostics assays were developed for the detection of six clades within the X. hortorum – X. hydrangeae species complex, in addition to one assay specific for the entire species complex. Primer sets were tested on a set of 62 reference strains. The primer sets amplified their respective targets within 15 minutes. Based on the reference set, all assays had a sensitivity, specificity, and efficiency of 100%. The assays were used on a validation set of 60 strains. According to the LAMP results, out of the 60 strains, 39 strains were assigned to one of the clades within the complex, 9 were assigned to the complex but to yet undefined clades within the complex, and 12 strains were previously misclassified as X. hortorum since their genomic DNA did not yield amplification with any of the assays. The seven genome-based assays are promising for use as diagnostic tools for various members within the X. hortorum – X. hydrangeae species complex, and for assigning new and historical isolates to this complex.
Introduction
Diagnostic methods are an essential part of integrated pest management programs, especially for the control of bacterial spot diseases (Agrios, 2005; Janse, 2005) such as the ones caused by Xanthomonas hortorum and Xanthomonas hydrangeae (Dia et al., 2021; Dia et al., 2022). Robust, field-deployable methods, with minimal sample preparation and post-amplification handling, are valuable tools for in-field or point-of-entry molecular detection of plant pathogens (Donoso and Valenzuela, 2018). Field-deployable methods, such as loop-mediated isothermal amplification (LAMP) and recombinase polymerase amplification (RPA) are effective methods that are of interest in diagnostic procedures (Donoso and Valenzuela, 2018; Catara et al., 2021). These two isothermal methods rely on strand displacement by the polymerase, and exponentially amplify the target regions at a constant temperature (Notomi et al., 2000; Piepenburg et al., 2006). Results can be detected visually by turbidity or color change (Mori et al., 2004; Tomita et al., 2008), or at end-point amplification by gel electrophoresis, or alternatively in real-time by measuring fluorescence (Bühlmann et al., 2013; Gétaz et al., 2017; Ruinelli et al., 2017). Both LAMP and RPA have been previously used for the detection of plant pathogens, including those within the X. hortorum – X. hydrangeae species complex, such as X. hortorum pathovars (pvs.) gardneri, carotae and pelargonii (Temple and Johnson, 2009; Temple et al., 2013; Strayer-Scherer et al., 2019; Stehlíková et al., 2020; Dia et al., 2021).
The X. hortorum – X. hydrangeae species complex currently comprises seven pathovars of X. hortorum (Morinière et al., 2020) and the recently described novel species X. hydrangeae (Dia et al., 2021). The species X. hortorum includes the devastating bacterial pathogen of geranium (X. hortorum pv. pelargonii) (Munnecke, 1954; Nameth et al., 1999), the internationally regulated seed-borne pathogen affecting carrot (X. hortorum pv. carotae) (Scott and Dung, 2020), the pathogen present in most lettuce-growing regions (X. hortorum pv. vitians) (Sahin, 1997; Morinière et al., 2020), and the pathogen which, in addition to three other Xanthomonas pathogens, causes bacterial spot of tomato and pepper (X. hortorum pv. gardneri) (Jones et al., 2004; Osdaghi et al., 2021). The three remaining pathovars of X. hortorum (pvs. taraxaci, hederae and cynarae) cause, respectively, bacterial spot and/or bacterial blight on dandelion (Niederhauser, 1943), ivy (Arnaud, 1920; White, 1934) and artichoke (Ridé, 1956; Trébaol et al., 2000; Timilsina et al., 2019). X. hydrangeae causes bacterial leaf spot on Hydrangea (Dia et al., 2021). Strains within the species complex affect 65 plant species spread over 15 botanical families (Dia et al., 2022).
The goal of this study was to develop specific and rapid genomics-based LAMP assays targeting six X. hortorum – X. hydrangeae clades, in addition to one assay detecting the totality of the complex. Assay metrics (e.g., sensitivity, specificity, and efficiency) were assessed based on the genomic DNA of reference strains (n = 62). The assays were used to differentiate various xanthomonads (e.g., unclassified or with an uncertain classification) obtained from culture collections and diagnostic laboratories. Furthermore, two assays, targeting the species complex and X. hydrangeae, were used to diagnose X. hydrangeae on Hydrangea spp. with typical leaf spots, showing the direct application of the developed assay.
Materials and Methods
Bacterial Strains, Bacterial Boiled Cells, Bacterial Genomic DNA, and Plant Extracts Preparation
Bacterial strains were stored in a 1:1 (v:v) NYGB and 100% glycerol at -80°C, and were revived from glycerol stocks. Strains were cultivated on nutrient-yeast glycerol agar (NYGA, 5 g L−1 peptone, 3 g L−1 yeast extract, 20 g L−1 glycerol and 15 g L−1 agar) or in NYG broth overnight or for two days at 28°C (with shaking at 200 rpm). Cells grown overnight in NYGB were used to prepare boiled cells or to extract genomic DNA.
Boiled cells were prepared by transferring one loop of fresh bacterial cells into 1 ml of ddH2O, followed by boiling at 98°C for 15 minutes and dilution at 1:100 with ddH2O. To obtain high-quality DNA, bacterial genomic DNA was extracted from overnight cultures with the NucleoSpin tissue kit (Macherey-Nagel, Düren, Germany) according to the instructions of the manufacturer. The genomic DNA was checked using a Q5000 spectrophotometer (Quawell, San Jose, CA). Genomic DNA amounts were normalized to 1 ng µl−1 based on spectrophotometer values, and the normalized concentration of random samples was checked using Quant-iT PicoGreen double-stranded DNA quantification assay (Thermo Fisher Scientific, Waltham, MA).
Plant material was prepared as follows: two fresh 1 cm-diameter punches from leaves of Hedera helix, Taraxacum officinale, Lactuca sativa, Pelargonium sp. and Hydrangea macrophylla, and 50 mg ( ± 3 mg) of Daucus carota seed were placed in tubes containing steel beads (OptiGene Ltd, Horsham, United Kingdom). For spiking the template with plant extracts, the plant material was bead-beaten using the plant lysis kit according to the manufacturer’s protocol (OptiGene Ltd). The prepared plant material was flash-frozen in liquid nitrogen and stored at -80°C for subsequent use.
Genome-Informed Target Identification
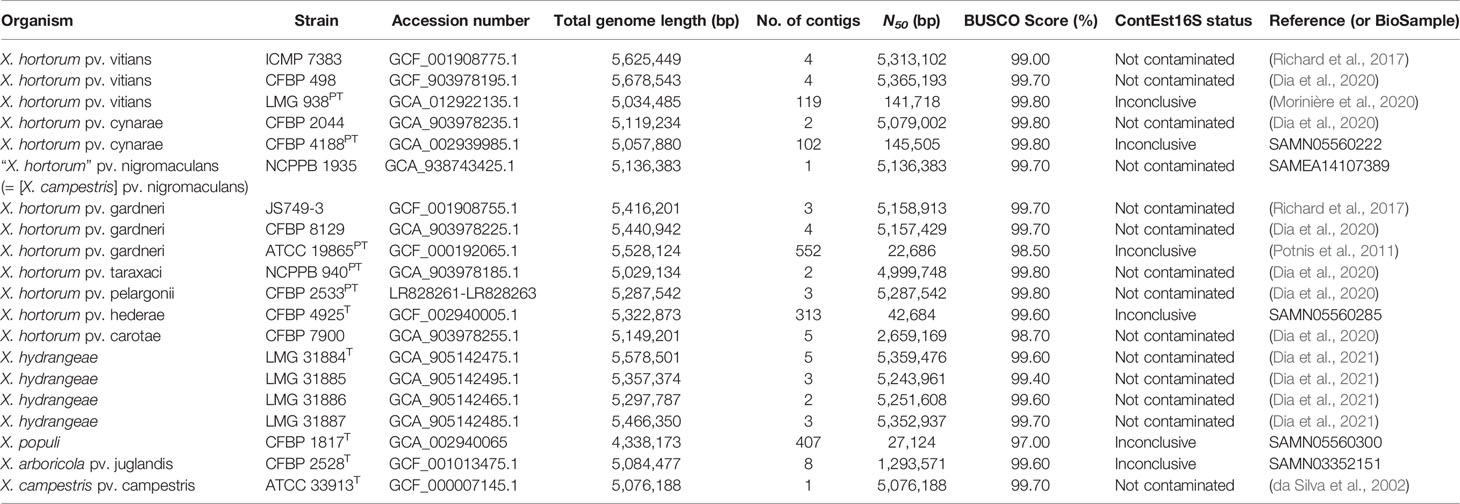
Target identification was based on the whole-genome phylogeny of the X. hortorum – X. hydrangeae species complex (Dia et al., 2021). Genomes of 17 X. hortorum – X. hydrangeae species complex strains and three outgroup strains (Xanthomonas arboricola pv. juglandis CFBP 2528T, Xanthomonas populi CFBP 1817T and Xanthomonas campestris pv. campestris ATCC 33913T) (Table 1) were submitted to the comparative genomics platform EDGAR v3.0 (Dieckmann et al., 2021). Genome completeness was analyzed based on 1,152 core genes using BUSCO v5.2.2 (Manni et al., 2021) using the xanthomonadales_odb10 (2020-03-06), the mode “genome” and dependencies hmmsearch v3.1 and prodigal v2.6.3. BUSCO scores were all ≥ 97%. Potential genome contamination was assessed using ContEst16S (Lee et al., 2017). The 16S rRNA sequences were intact for all genomes except for five draft genomes, for which the integrity check was inconclusive as only one 16S rRNA copy was found.
The phylogeny of the 20 genomes was built in the EDGAR platform, based on a core genome of 2,345 genes per genome. There was a total of 831,037 amino acid residues per genome. Each of the proteins was independently aligned with MUSCLE (Edgar, 2004), and the resulting alignments were then concatenated and used to construct the whole-genome Maximum-Likelihood phylogenetic tree. The generated newick tree was then visualized using MEGA X v10.0.5 (Kumar et al., 2018). For assays developed based on two or more genomes, the core genome of the target strains was searched for candidate chromosomal singletons against all other strains in the EDGAR project. For assays developed based on only one genome, the totality of the genome was used to search for singletons. An overview of the workflow is presented in Figure 1.
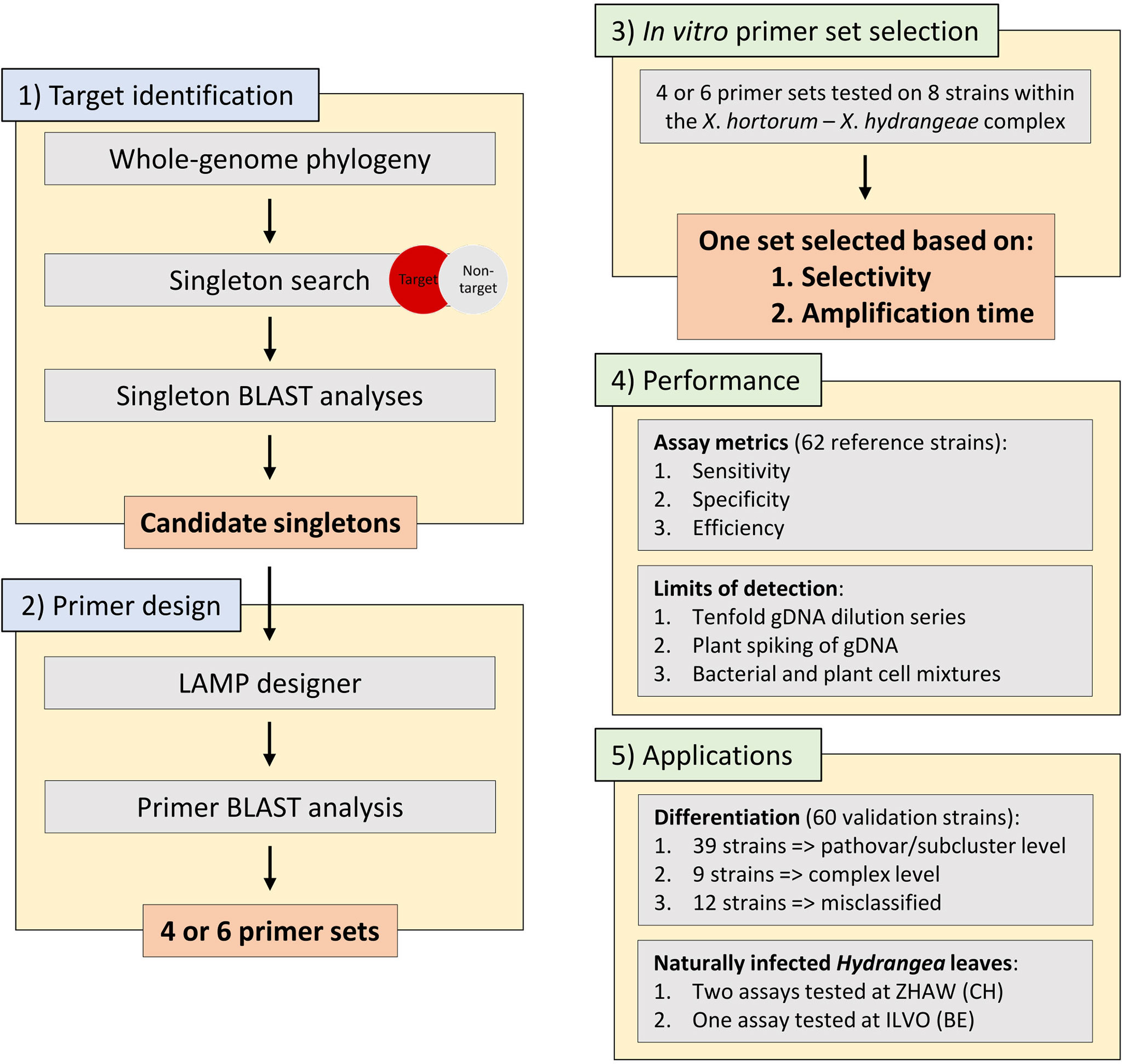
Figure 1 The workflow used within this work starting from 1) target identification using a singleton whole genome phylogeny approach, 2) subsequent LAMP primer design, 3) an in vitro primer set selection based on eight representative strains within the Xanthomonas hortorum – Xanthomonas hydrangeae species complex, 4) performance testing of the seven assays on 62 reference strains including assays metrics (e.g., sensitivity, specificity, and efficiency) and limits of detection (e.g., tenfold genomic DNA dilution series and spiking of genomic DNA with plant material) and, 5) the two main applications of the seven assays. gDNA, genomic DNA.
Since EDGAR uses a bidirectional best BLASTp hits approach for calculating singletons, gene duplication and paralogs are treated as singletons, if unidentical (Blom et al., 2016). The in silico specificity of each singleton was thus further verified using a BLASTn v2.7.1 analysis (Camacho et al., 2009) against a local database of the 20 genomes used for the genome-informed target identification including Xanthomonas spp. genomes sequenced at ZHAW (Table 1), and against the nucleotide nr/nt (Xanthomonadales, taxid:135614) NCBI collection (accessed February 26, 2020). To refine results, the singleton specificity was further checked using the whole-genome shotgun contigs (WGS) database (Xanthomonadales, taxid:135614) at NCBI (accessed on February 26, 2020), a crucial step in genomics-based assay development as the nucleotide and the WGS databases contain different types of genome sequence data.
To ensure that specific singletons are not erroneously discarded, the identity of the genomes with significant hits to the singletons of the various assays was verified by calculating average nucleotide identity (ANI) values using fastANI v1.1 (Jain et al., 2018) between the genome sequences. Singletons were considered specific and retained for primer design if BLASTn did not yield significant hits besides the respective target organism(s), if the genes were not located in the close vicinity (~10 kb on each side) of potentially functional transposases, if the genes were located on the chromosome (in complete genomes) or on contigs tentatively assigned as chromosomal, and if they were present in only one copy.
LAMP Primer Design, Synthesis, and Selection
Primer sets (outer primers F3/B3, loop primers LoopF/LoopB and internal primers FIP/BIP) were designed using LAMP Designer v1.16 (Premier Biosoft, San Francisco, CA). Default reaction conditions and primer parameters were used. Depending on the number of specific singletons for the various assays (Supplementary Table 1), primer sets were designed for either the same or different singleton(s) (Supplementary Table 2). Primer sets without a secondary structure, with an increased stability at primer ends and an optimal distance between ends of F2 and B2 sequences, were selected for further analysis.
In silico specificity of the outer primers and their amplicons was analyzed using primer-BLAST and BLASTn, respectively, using databases as previously described (accessed February 28, 2020). Primer sets fulfilling both specificity criteria were selected for in vitro analysis. Based on LAMP primer design recommendations, “TTTT” linkers were added for the internal primers (Notomi et al., 2000). For each assay, four primer sets were synthesized (Microsynth AG, Balgach, Switzerland), except for the assay targeting X. hortorum pv. hederae, for which six primer sets were prepared.
To select the optimal LAMP primer set per target group, the selectivity of the four or six primer sets (Supplementary Table 2) was first tested on a sub-selection of eight representative strains within the X. hortorum – X. hydrangeae species complex using genomic DNA or boiled cells. For each assay, one primer set was then selected for further analysis based on two criteria: selective amplification of the genomic DNA of the target organism(s) within the X. hortorum – X. hydrangeae species complex, and, given fulfilment of first criterion, shortest amplification time. Finally, selectivity of assays was then confirmed on the normalized genomic DNA of strains within the reference group, and the assays were used to differentiate strains within the validation set. The sequences of the retained primer sets are reported in Supplementary Table 3. The locus tags, annotation and contig accession numbers of the retained singletons are described in Supplementary Table 4.
General LAMP Experiment and Assay Conditions
The LAMP reaction composition for a single sample is reported in Supplementary Table 5. In all experiments, ddH2O served as no template control (NTC). All measurements were carried on a Light Cycler 480 Real-Time machine (Roche Diagnostics, Rotkreuz, Switzerland) in white 96-well plates (Roche). The target temperature was 64°C, ramp rate was 2.2°C s-1 and amplifications ran for 55 min. Samples with ambiguous amplifications (i.e., melting curve not in expected range, unexpected amplification) were repeated in triplicates. Melting curve analysis was conducted for 5 min from 64°C to 95°C at 0.01°C s-1; melting temperatures of the assays are reported in Supplementary Table 3.
Assay Metrics of LAMP Assays
Assay metrics were based on the normalized genomic DNA of the reference strains and were calculated as follows (Blaser et al., 2018): the assay sensitivity (true-positive rate) was calculated using formula [1], while specificity (true-negative rate) was calculated using formula [2], and assay efficiency (correct assay results) with formula [3],
[1]
[2]
[3]
in which NTP represents the number of true positive samples, NTN the number of true negatives, NFP the number of false positives and NFN the number of false negatives.
Limits of Detection of DNA- and Crude Bacterial Cell-Based Assays
The limits of detection (LoD) of the DNA-template based assays were tested with tenfold dilutions series from 1 ng µl-1 to 1 fg µl-1 of representative target strains, with and without spiking plant extract in experimental triplicates (Supplementary Table 6). In addition, the effect of non-host plant extract was tested on normalized genomic DNA in experimental duplicates. Genomic copy equivalents were calculated using the DNA amount (in ng), a template of average size 5.5 Mb and average weight of a base-pair of 650 Daltons (Supplementary Table 7).
For the crude bacterial cell-template based assays, representative strains were grown overnight on NYGA at 28°C. Single colonies were picked and transferred in sterile 0.8% KCl, and the OD was adjusted to OD600 ≈ 0.5 (corresponding to around 108 CFU ml-1). Then, ten-fold serial dilutions of 105, 103, 102, 101 and 100 CFU ml-1 were prepared and used for testing the performance of crude bacterial cell-based assays.
In steel ball bearings tubes, 1 ml of bacterial suspension and 1 ml of lysis buffer (OptiGene Ltd) were pipetted to the previously added plant extracts, and the bacterial-plant mixture was then vigorously bead-beaten for 2 min. Twenty µl of the mixture was further diluted in 1 ml dilution buffer (OptiGene Ltd), and 2.5 µl of the diluted mixture was used as template for the LAMP reaction. Measurements were taken from biological and experimental duplicates (Supplementary Table 8).
Performance of Two Assays on Naturally Infected Hydrangea
In July 2021, the Plant Diagnostic Center at ILVO (Merelbeke, Belgium) received Hydrangea arborescens cv. ‘Annabelle’ plants exhibiting characteristic Xanthomonas-leaf spots from a retail nursery in Belgium. LAMP assays were done on the naturally infected leaves at ILVO (only with the X. hydrangeae assay) and at Zurich University of Applied Sciences (ZHAW, Wädenswil, Switzerland) (with the X. hydrangeae and species complex assays) by the same protocol. Briefly, leaf-discs of 10 mm in diameter were cut out from the symptomatic leaves, vigorously bead-beaten and further processed as described above except that the LAMP reactions at the ILVO laboratory were done on a Genie II device (OptiGene Ltd).
Results
Bacterial Strains
The 122 bacterial strains used in this study comprised 62 reference strains (Table 2 and Supplementary Table 9), and a validation set of 60 xanthomonad strains unclassified or doubtfully classified by culture collections or diagnostic laboratories (Supplementary Table 9). The bacterial strains were isolated from over 50 plant species, in different years (1942 to 2021) and 30 countries. Strains were obtained from various international culture collections and national diagnostic laboratories as indicated in Supplementary Table 9. The reference group included 24 strains from the X. hortorum – X. hydrangeae species complex, for which whole genomes were available (Table 1). These genomes were included in the development of the primer sets. The remaining strains were chosen to reflect the broad diversity within the Xanthomonas genus, in addition to six pseudomonads. The validation set comprised 60 strains for which no whole genome sequences were available. The partial gyrB gene of 38 strains within the validation set was sequenced (data not shown).
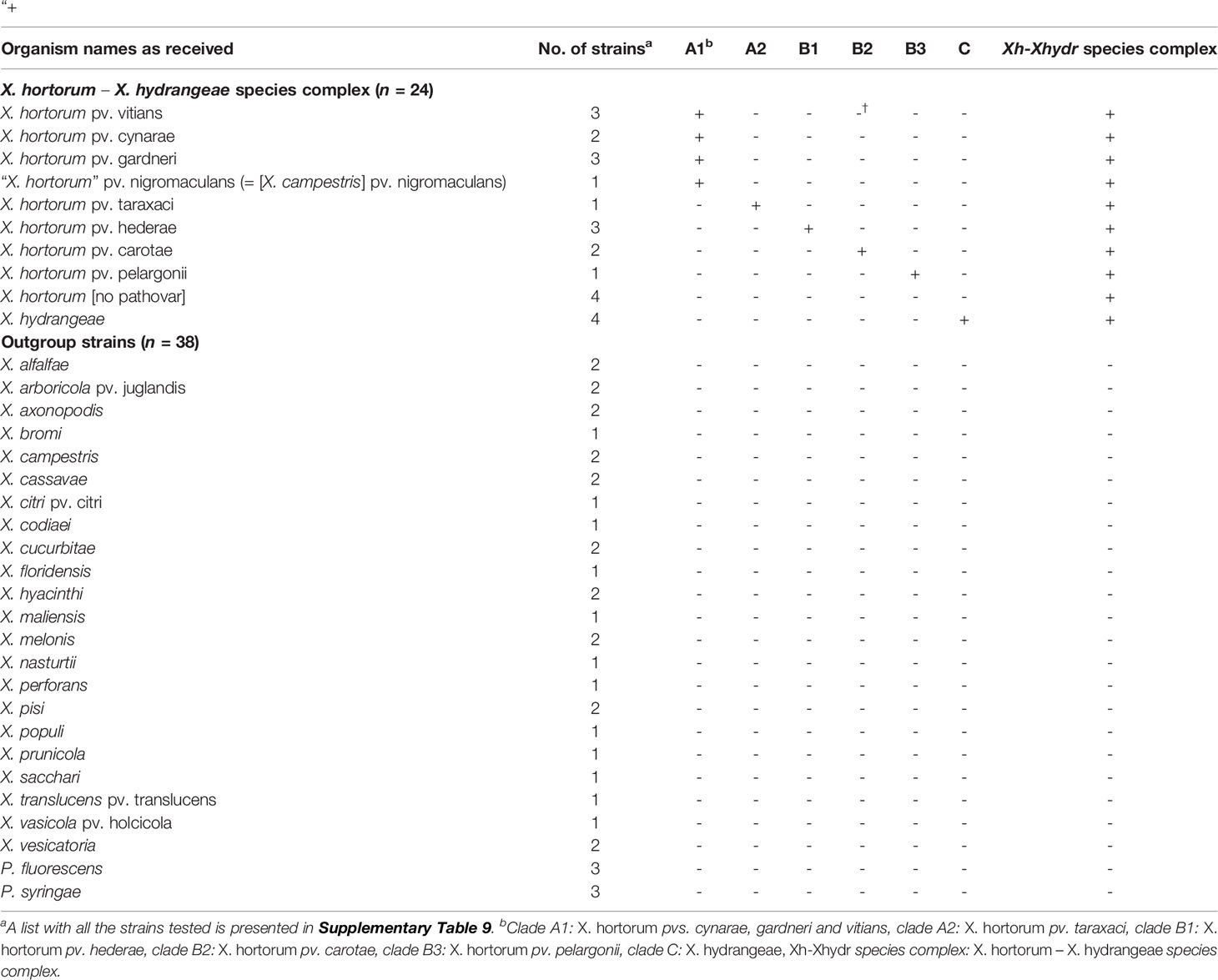
Table 2 Reference strains used in this study and results of the LAMP assays based on their normalized genomic DNA. Selective amplification is marked with a sign “+”, and “-” denotes absence of amplification.
Seven LAMP Assays Developed
The recently published whole-genome phylogeny of the X. hortorum – X. hydrangeae species complex revealed three major clades (Figure 2): 1) clade A, encompassing X. hortorum pvs. gardneri, cynarae, vitians (clade A1) and pv. taraxaci (clade A2); 2) clade B, including X. hortorum pv. hederae (clade B1), pv. carotae (clade B2) and pv. pelargonii (clade B3); and 3) X. hydrangeae (clade C) (Dia et al., 2021). These clades served as a base for finding candidate and specific singletons (Supplementary Table 1), the latter being used as LAMP assay targets. Seven LAMP assays were thus developed, one for the overarching X. hortorum – X. hydrangeae species complex and six other assays, each targeting a separate clade within the species complex (Figure 2). More information on assay development (e.g., candidate and specific singletons, candidate primer sets and selected singletons) is provided as Supplementary Text 1.

Figure 2 Whole-genome Maximum-Likelihood core phylogeny of the Xanthomonas hortorum – Xanthomonas hydrangeae species complex. The tree shows the various clades within the species complex (grey arrow): clades A1 (X. hortorum pvs. gardneri, cynarae, vitians and [X. hortorum] pv. nigromaculans, grey square) and A2 (X. hortorum pv. taraxaci, blue circle), clades B1(X. hortorum pv. hederae, green circle), B2 (X. hortorum pv. carotae, red circle) and B3 (X. hortorum pv. pelargonii, yellow circle), and clade C (X. hydrangeae, grey diamond) in addition to three outgroup strains. Accession numbers of the 20 genomes used in this study are noted in Table 1.
Assay Metrics of LAMP Assays
The sensitivity, specificity, and performance of all seven assays was 100% (Table 3). Of the X. hortorum – X. hydrangeae strains within the reference group, the genomic DNA of 20 strains was selectively amplified with two assays: 1) the overarching species complex assay and, 2) one clade-specific assay. The genomic DNA of four Xanthomonas sp. strains, namely GBBC 1934, SB 3701, SB 3727 and Xc407, yielded an amplification with the assay targeting the species complex only (Supplementary Table 9). No amplification was observed for the genomic DNA of the 38 strains not belonging to the X. hortorum – X. hydrangeae species complex (Table 2 and Supplementary Table 9).
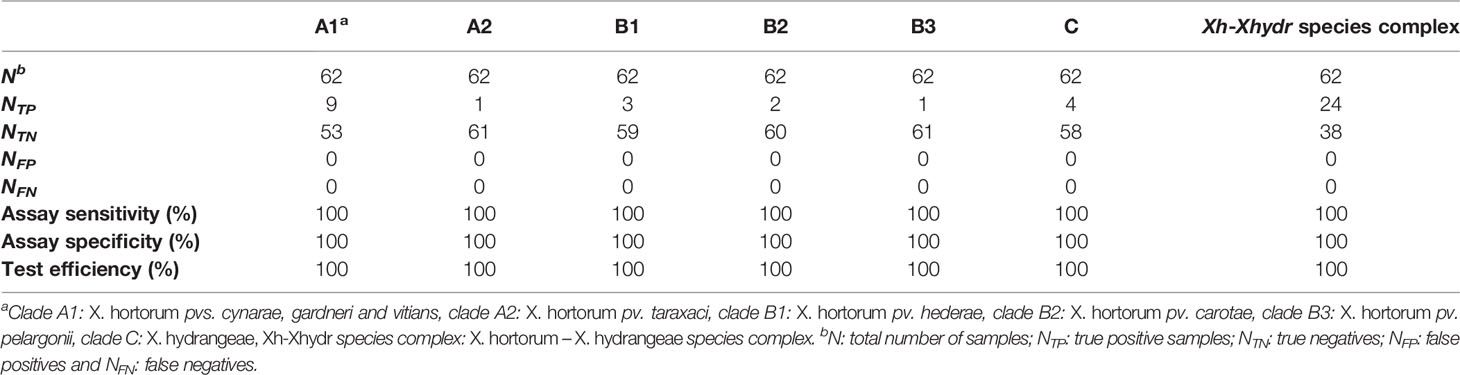
Table 3 Specificity, sensitivity, and efficiency of the LAMP assays based on the genomic DNA of the 62 reference strains.
Limits of Detection of Bacterial Genomic DNA- and Crude Bacterial Cell-Template Based Assays
All assays yielded a positive reaction with bacterial genomic DNA quantities down to at least 2.5 pg per reaction (Table 4), equivalent to 421 genome-copies (Supplementary Table 7). The consistent LoD varied between the assays. Without plant extract spiking, the consistent LoD was 2.5 pg per reaction for the assays targeting clade A1 and the species complex, and down to 250 fg per reaction for the other assays. Spiking of the reaction mix with plant extracts decreased the consistent LoD of assays targeting clades A1, A2, B3 and C tenfold, after spiking with plant extracts from lettuce, dandelion, pelargonium, or hydrangea. The consistent LoD of the assays targeting clades B1, B2 and the species complex were not affected by the addition of plant extracts.
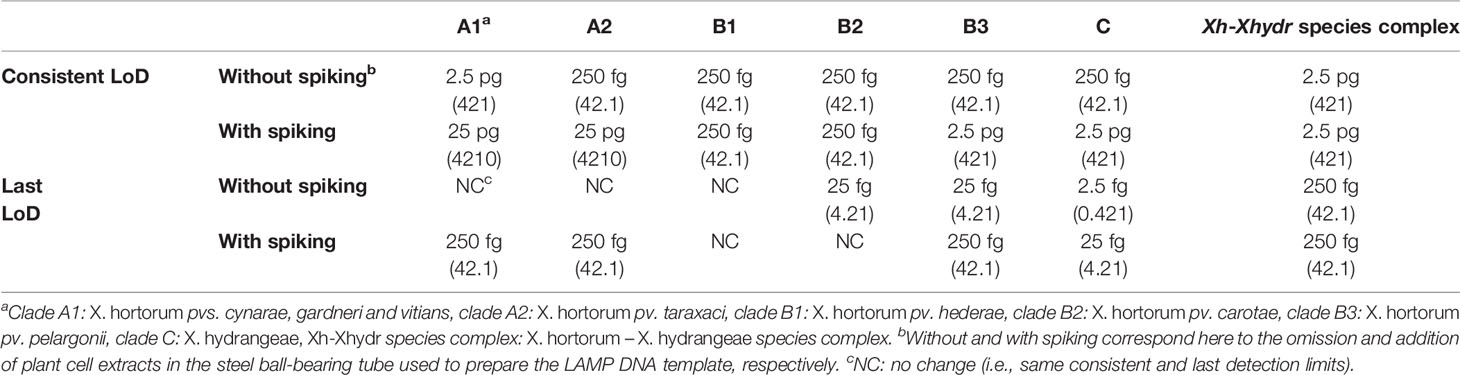
Table 4 Summary of the consistent and last limits of detection (LoD) of genomic DNA-based assays. Genome-copy equivalents are reported between parentheses.
When using boiled bacterial cells, all assays detected at least about 1,000 cells µl-1 of LAMP reaction mix (equivalent to 1 × 106 cells ml-1) (Table 5 and Supplementary Table 8). Other assays, like the ones targeting clades A2 and B2, were also able to consistently detect 100 cells µl-1 of LAMP reaction mix (equivalent to 1 × 105 cells ml-1). Representative amplifications by the assay targeting the species complex are shown in Figures 3A–D.

Table 5 Summary of the consistent and last limits of detection (LoD) of cell-based assays, reported in number of bacteria in 1 µl of LAMP reaction.
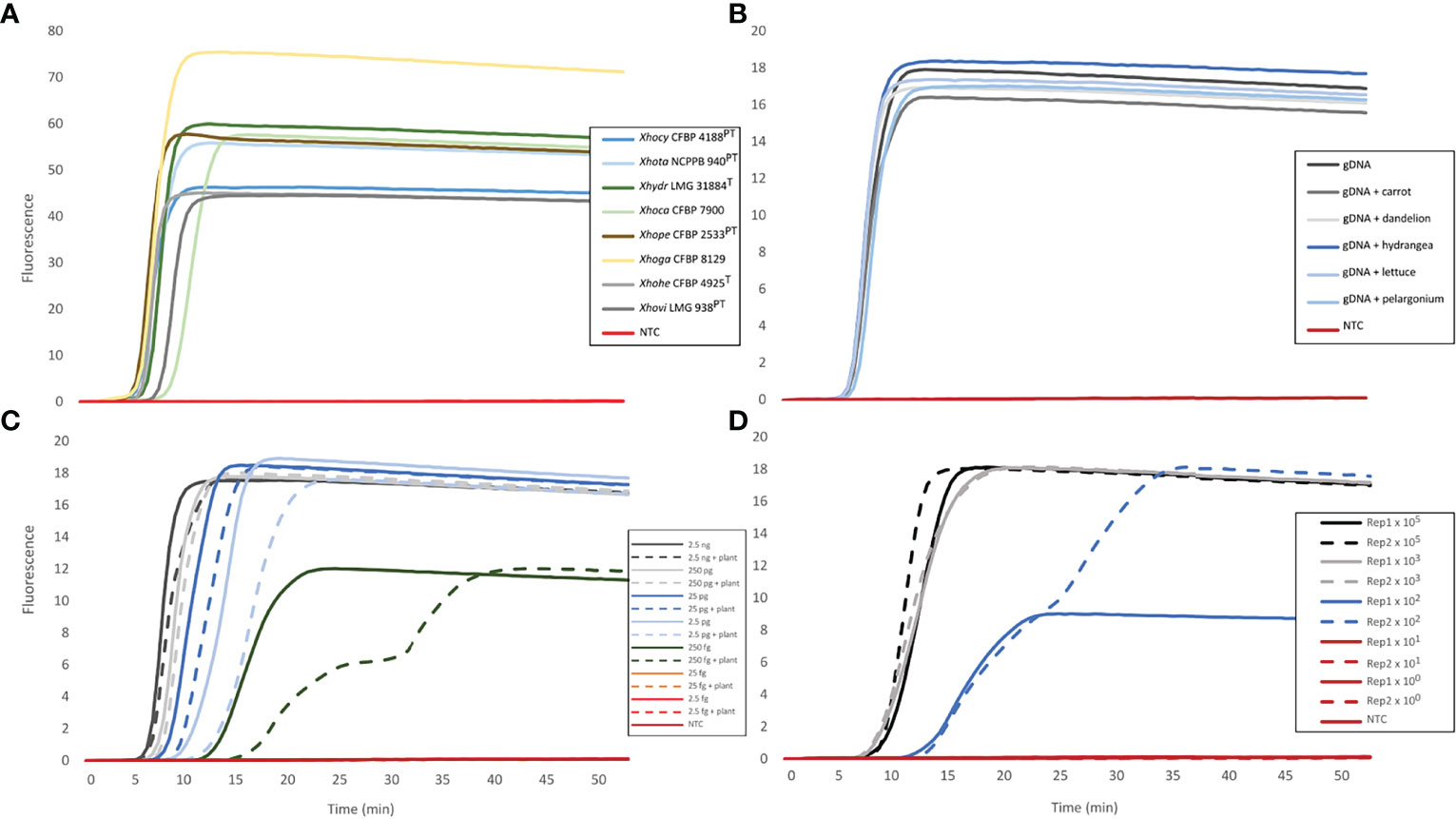
Figure 3 Representative amplifications of the assay targeting the Xanthomonas hortorum– Xanthomonas hydrangeae complex. The amplifications were obtained with: (A) the genomic DNA of the eight reference strains for which abbreviations are as follows: X. hortorum pvs. cynarae (Xhocy), gardneri (Xhoga), taraxaci (Xhota), pelargonii (Xhope), carotae (Xhoca), vitians (Xhovi) and hederae (Xhohe), in addition to X. hydrangeae (Xhydr), (B) the spiked genomic DNA (gDNA) of X. hortorum pv. hederae strain CFBP 4925T with non-host plant extracts (i.e., carrot, dandelion, hydrangea, lettuce or pelargonium), (C) a 10-fold genomic DNA dilution series of X. hortorum pv. hederae strain CFBP 4925T, with and without ivy extract spiking, and (D) crude bacterial (X. hortorum pv. hederae strain CFBP 4925T) and ivy mixture in two biological replicates (Rep1 and Rep2).
Practical Applications of the LAMP Assays
To examine the applicability of the assays, the seven assays were tested on a validation set of 60 unclassified strains or strains with an uncertain classification received as Xanthomonas spp. or X. hortorum from culture collections or diagnostic laboratories (Supplementary Table 9). The genomic DNA of 39 strains of this validation set yielded an amplification with the assay targeting the species complex, and with an assay targeting one of the six clades, assigning them to one of the defined clades. The genomic DNA of nine strains were only amplified with the assay targeting the complex, suggesting that these strains belong to the X. hortorum – X. hydrangeae species complex, but not to any of the defined clades. Genomic DNA of 12 strains did not yield an amplification with any of the assays (Supplementary Table 9) and thus do not belong to the complex, according to the LAMP results. One of the 12 strains, strain NCB 1082, could have yielded false negative signals for the assays targeting A1 and the species complex. The partial gyrB sequence of this strain clustered with various strains of X. hortorum pvs. gardneri and vitians (clade A1) (data not shown).
Two of the LAMP assays were used to identify X. hydrangeae from naturally infected Hydrangea leaves. Specific and rapid amplifications from all tested samples were obtained with the LAMP assay targeting clade C (X. hydrangeae) in both laboratories at ZHAW and ILVO. Furthermore, the LAMP assay targeting the complex was also tested on the infected leaves at ZHAW and showed positive results as well.
Discussion
Assay Development
Assays were originally planned for each separate pathovar within clade A, in addition to an assay encompassing the totality of clade A. However, no suitable singletons were found between representative genomes of X. hortorum pvs. gardneri, cynarae, vitians and taraxaci. Out of the four pathovars within that clade, suitable singletons were only found for X. hortorum pv. taraxaci. Thus, two assays were developed: one targeting specifically clade A1 containing X. hortorum pvs. gardneri, vitians and cynarae and one targeting clade A2 specific for X. hortorum pv. taraxaci. This can be explained by the very high genetic relatedness between strains within the clade A1. The ANI values between strains of X. hortorum pvs. cynarae, gardneri and vitians were above 98%, whereas ANI values between X. hortorum pv. taraxaci (clade A2) and clade A1 strains were below 98% (Morinière et al., 2020). The threshold for finding singletons acceptable as genomic targets is thus related to the ANI values, and the higher the ANI value, the more challenging finding a suitable singleton.
Limits of Detections
Insufficient information is available concerning the LoD of the existing molecular diagnostic assays targeting single pathovars of X. hortorum. Out of 18 PCR-based and isothermal amplification studies present in the literature, only six reported LoD values (Dia et al., 2022) and these ranged from 2 pg to 200 fg for DNA experiments, whereas bacterial cell-based LoD values were between 5 × 104 CFU ml-1 and 5 × 106 CFU ml-1.
The consistent LoD of the DNA-template based assays developed in this study is comparable to LAMP and PCR assays previously published for detecting X. hortorum pv. gardneri (Stehlíková et al., 2020) and X. hortorum pv. carotae (primer set 9B) (Meng et al., 2004). On the other hand, the LAMP assays developed in this study are more sensitive than the PCR/Multiplex-PCR assays targeting X. hortorum pv. gardneri for which LoD was estimated to be 50 pg μl-1 (Araújo et al., 2012). The sensitivity of our assays, however, is lower than two previously published assays (conventional PCR and qPCR), targeting X. hortorum pv. carotae (primer set 3S) (Meng et al., 2004) and X. hortorum pv. pelargonii (Farahani and Taghavi, 2016). The greater sensitivity of qPCR when compared to LAMP has also been previously reported (Bühlmann et al., 2013). The assays in the current study on boiled bacterial cells had a consistent detection limit in the same order of magnitude as previously published studies (Araújo et al., 2012; Strayer et al., 2016; Strayer-Scherer et al., 2019).
LAMP Assays to Identify X. hortorum – X. hydrangeae Strains
All seven assays were highly sensitive and specific, irrespective of their taxon-level of detection. LAMP assays were able to assign reference strains, initially classified as Xanthomonas sp. based on partial sequence analysis of gyrB, to a level lower than the species complex (Table 2). The genomic DNA of those strains resulted in amplification with two assays (i.e., the assay targeting the whole complex and a single assay targeting one of the six clades). For example, the genomic DNA of strains GBBC 1967 and GBBC 950, both isolated from Hedera helix, was amplified by the assay targeting the whole complex and the one targeting X. hortorum pv. hederae (clade B1), assigning them to X. hortorum pv. hederae. This was further confirmed using whole-genome sequence data (data not shown).
Several strains from both the reference and validation set were accurately identified as belonging to the X. hortorum – X. hydrangeae species complex but their genomic DNA did not yield amplification by any of the six other assays targeting various clades. Based on LAMP results, this would suggest that they potentially belong to new clades within the X. hortorum – X. hydrangeae species complex. Some of these strains were isolated from plants that were not previously reported as host plants within X. hortorum (Dia et al., 2022). This suggests that several novel lineages may exist within the X. hortorum – X. hydrangeae species complex requiring further investigation. In case that novel lineages are confirmed, and depending on their relevance to plant health, the development of new assays targeting these novel lineages might be required.
Furthermore, results from the LAMP assays on the validation set also showed that several strains had been misassigned to the species complex by the diagnostic laboratories that provided the strains, as their genomic DNA did not yield amplification with any of the LAMP assays. More information regarding those strains is presented in Supplementary Text 2 and Supplementary Figure 1.
Performance of Two Assays on Naturally Infected Hydrangea Plants
The performance of diagnostic assays on field samples is a crucial metric of applicability as a diagnostic method. We previously had tested the LAMP assays to detect X. hydrangeae in artificially inoculated plants (Dia et al., 2021), using a 48-hour growth step. In this study, two LAMP assays were tested directly on extracts of naturally infected Hydrangea leaves. The time required from receiving the infected leaves to LAMP results was two hours, and it can be further lowered by using a shorter amplification time. No bacterial growth step was needed as leaf spot macerate was diluted and directly used as LAMP reaction template. The assays identified the causal agent of bacterial leaf spot of Hydrangea as a member of clade C (X. hydrangeae). The first assay targeting the whole X. hortorum – X. hydrangeae species complex provided additional certainty to the diagnosis results.
The LAMP assay targeting clade C was used at ILVO with the same protocol used at ZHAW except that it was performed on a portable machine. The same conclusion was drawn identifying X. hydrangeae as the causative agent of the bacterial leaf spot on hydrangea. This validation and transferability step is extremely important in diagnostics (EPPO, 2021), further attesting to the robustness of the developed assay. Additionally, the field-deployable capacity of the assays developed here was also demonstrated, a very valuable characteristic within plant pathogen diagnostics.
Even though the detection limits of some of the assays decreased when spiked with plant extracts, the results obtained from the two LAMP assays on naturally infected Hydrangea demonstrate the applicability of the assays on plants with an on-going infection with X. hortorum – X. hydrangeae species complex strains. The performance of the assays in latent phases of infections and on other hosts remains to be assessed. This is of particular relevance as some X. hortorum members can survive on plant surfaces as epiphytes (Toussaint et al., 2012) and as the presence of non-pathogenic Xanthomonas species (Vauterin et al., 1996; Cesbron et al., 2015) cannot be excluded.
Concluding Remarks
The LAMP assays developed in this study can be used for multiple purposes, both in silico and in the laboratory. In silico, the specific singletons can be used to identify suspected strains within the X. hortorum – X. hydrangeae species complex from their genome sequences and, potentially assign them to one of the defined clades without detailed analysis of the genomes. This is invaluable given the ever-changing pathogen taxonomy, not always quickly adopted in diagnostic or research laboratories, and the prevalent mischaracterization and misnaming of strains and genomes.
The LAMP assays can be used in field settings by using macerated and diluted symptomatic tissue as LAMP template. The LAMP assays can also be used in a laboratory setting, for example, to (re)assign strains within bacterial collections or to identify X. hortorum – X. hydrangeae strains when tested on extracted genomic DNA or crude boiled bacterial cells. In this study, we have shown that the LAMP assays were able to assign previously unassigned strains to clades within the X. hortorum – X. hydrangeae species complex. Some strains could be assigned to the species complex but not to a lower taxonomical level. These strains could represent novel groups within the complex and require further phylogenic analysis. The combination of various taxon detection levels (i.e., species to pathovar level) provided a higher degree of discriminative power, in addition to minimal sample preparation and post-amplification handling (e.g., no gel electrophoresis step), higher tolerance to inhibitors present in crude plant samples and with field-deployable capacities.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
ND, TS, and JP conceptualized the study and analyzed the data. ND and JP created the methodology, and curated the data with the assistance from JB. ND, BC, TS, and JP were involved in the investigations. JB and JP helped with software. ND, BC, TS, and JP were involved in the validation. ND, TS, and JP contributed to the data visualization. TS and JP contributed to project administration and supervision. ND, TS, and JP prepared the original draft with assistance from BC and JB for review and editing. All the authors revised the final version of the manuscript, while JP acted as the corresponding author. All authors contributed to the article and approved the submitted version.
Funding
ND was supported by grant no. IZCOZ0_177064 from the Swiss National Science Foundation (SNSF). TS and JP were supported by the Department of Life Sciences and Facility Management of the Zurich University of Applied Sciences (ZHAW) in Wädenswil. The EDGAR platform is funded by the BMBF grant FKZ 031A533 within the de.NBI network. The open access article processing charges of this publication were funded by SNSF.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank Nicola Rhyner (ZHAW) and Cinzia Vanmalderghem (ILVO) for their assistance in the laboratory, Marilena Palmisano (ZHAW) for her technical advice. We thank Andrew Aspin (Fera Science Ltd., UK), Dr. Eliška Hakalová (Mendel University in Brno, CZ), Dr. Tanja Dreo and Dr. Manca Pirc (both National Institute of Biology, SI), and Prof. Svetoslav Bobev (Agricultural University-Plovdiv, Bulgaria) for providing strains used in this study. The authors would also like to thank the HPC team of the School for Life Sciences and Facility Management at ZHAW for providing computing resources and support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2022.898778/full#supplementary-material
References
Araújo E. R., Costa J. R., Ferreira M. A. S. V., Quezado-Duval A. M. (2012). Simultaneous Detection and Identification of the Xanthomonas Species Complex Associated With Tomato Bacterial Spot Using Species-Specific Primers and Multiplex PCR. J. Appl. Microbiol. 113, 1479–1490. doi: 10.1111/j.1365-2672.2012.05431.x
Arnaud G. (1920). Une maladie bactérienne du lierre (Hedera helix L.). Comptes Rendus Hebdomadaires des Séances de l’Académie des Sciences 171, 121-122.
Blaser S., Diem H., Felten A., Gueuning M., Andreou M., Boonham N., et al. (2018). From Laboratory to Point of Entry: Development and Implementation of a Loop-Mediated Isothermal Amplification (LAMP)-Based Genetic Identification System to Prevent Introduction of Quarantine Insect Species. Pest Manage. Sci. 74, 1504–1512. doi: 10.1002/ps.4866
Blom J., Kreis J., Spänig S., Juhre T., Bertelli C., Ernst C., et al. (2016). EDGAR 2.0: An Enhanced Software Platform for Comparative Gene Content Analyses. Nucleic Acids Res. 44, W22–W28. doi: 10.1093/nar/gkw255
Bühlmann A., Pothier J. F., Tomlinson J. A., Frey J. E., Boonham N., Smits T. H. M., et al. (2013). Genomics-Informed Design of Loop-Mediated Isothermal Amplification for Detection of Phytopathogenic Xanthomonas Arboricola Pv. Pruni at the Intraspecific Level. Plant Pathol. 62, 475–484. doi: 10.1111/j.1365-3059.2012.02654.x
Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K, et al. (2009). BLAST+: Architecture and Applications. BMC Bioinformatics 10, 421. doi: 10.1186/1471-2105-10-421
Catara V., Cubero J., Pothier J. F., Bosis E., Bragard C., Đermić E., et al. (2021). Trends in Molecular Diagnosis and Diversity Studies for Phytosanitary Regulated Xanthomonas. Microorganisms 9, 862. doi: 10.3390/microorganisms9040862
Cesbron S., Briand M., Essakhi S., Gironde S., Boureau T., Manceau C., et al. (2015). Comparative Genomics of Pathogenic and Nonpathogenic Strains of Xanthomonas arboricola Unveil Molecular and Evolutionary Events Linked to Pathoadaptation. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.01126
da Silva A.C.R., Ferro J.A., Reinach F.C., Farah C.S., Furlan L.R., Quaggio R.B., et al. (2002). Comparison of the Genomes of Two Xanthomonas Pathogens With Differing Host Specificities. Nature 417, 459–463. doi: 10.1038/417459a
Dia N. C., Morinière L., Cottyn B., Bernal E., Jacobs J. M., Koebnik R., et al. (2022). Xanthomonas hortorum – Beyond Gardens: Current Taxonomy, Genomics, and Virulence Repertoires. Mol. Plant Pathol. 23, 597–621. doi: 10.1111/mpp.13185
Dia N. C., Van Vaerenbergh J., Van Malderghem C., Blom J., Smits T. H. M., Cottyn B., et al. (2021). Xanthomonas hydrangeae Sp. Nov., A Novel Plant Pathogen Isolated From Hydrangea arborescens. Int. J. System. Evol. Microbiol. 71, 5163. doi: 10.1099/ijsem.0.005163
Dia N.C., Rezzonico F., Smits T.H.M., Pothier J.F. (2020). Complete or High-Quality Draft Genome Sequences of Six Xanthomonas Hortorum Strains Sequenced With Short- and Long-Read Technologies. Microbiology Resource Announcements 9, e00828-00820. doi: 10.1128/MRA.00828-20.
Dieckmann M. A., Beyvers S., Nkouamedjo-Fankep R. C., Hanel P. H. G., Jelonek L., Blom J., et al. (2021). EDGAR3.0: Comparative Genomics and Phylogenomics on a Scalable Infrastructure. Nucleic Acids Res. 49, W185–W192. doi: 10.1093/nar/gkab341
Donoso A., Valenzuela S. (2018). In-Field Molecular Diagnosis of Plant Pathogens: Recent Trends and Future Perspectives. Plant Pathol. 67, 1451–1461. doi: 10.1111/ppa.12859
Edgar R. C. (2004). MUSCLE: Multiple Sequence Alignment With High Accuracy and High Throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Farahani A. S., Taghavi S. M. (2016). Sensitive and Specific Detection of Xanthomonas hortorum Pv. pelargonii in Geranium by Real-Time PCR. J. Plant Protect. Res. 56, 265–269. doi: 10.1515/jppr-2016-0039
Gétaz M., Bühlmann A., Schneeberger P. H. H., Van Malderghem C., Duffy B., Maes M., et al. (2017). A Diagnostic Tool for Improved Detection of Xanthomonas fragariae Using a Rapid and Highly Specific LAMP Assay Designed With Comparative Genomics. Plant Pathol. 66, 1094–1102. doi: 10.1111/ppa.12665
Jain C., Rodriguez-R L. M., Phillippy A. M., Konstantinidis K. T., Aluru S. (2018). High Throughput ANI Analysis of 90K Prokaryotic Genomes Reveals Clear Species Boundaries. Nat. Commun. 9, 1–8. doi: 10.1038/s41467-018-07641-9
Jones J. B., Lacy G. H., Bouzar H., Stall R. E., Schaad N. W. (2004). Reclassification of the Xanthomonads Associated With Bacterial Spot Disease of Tomato and Pepper. System. Appl. Microbiol. 27, 755–762. doi: 10.1078/0723202042369884
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis Across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lee I., Chalita M., Ha S.-M., Na S.-I., Yoon S.-H., Chun J. (2017). ContEst16S: An Algorithm That Identifies Contaminated Prokaryotic Genomes Using 16S RNA Gene Sequences. Int. J. Syst. Evol. Microbiol. 67, 2053–2057. doi: 10.1099/ijsem.0.001872
Manni M., Berkeley M. R., Seppey M., Simão F. A., Zdobnov E. M. (2021). BUSCO Update: Novel and Streamlined Workflows Along With Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol. Biol. Evol. 38, 4647–4654. doi: 10.1093/molbev/msab199
Meng X. Q., Umesh K. C., Davis R. M., Gilbertson R. L. (2004). Development of PCR-Based Assays for Detecting Xanthomonas campestris pv. carotae, the Carrot Bacterial Leaf Blight Pathogen, From Different Substrates. Plant Dis. 88, 1226–1234. doi: 10.1094/PDIS.2004.88.11.1226
Mori Y., Kitao M., Tomita N., Notomi T. (2004). Real-Time Turbidimetry of LAMP Reaction for Quantifying Template DNA. J. Biochem. Biophys. Methods 59, 145–157. doi: 10.1016/j.jbbm.2003.12.005
Morinière L., Burlet A., Rosenthal E. R., Nesme X., Portier P., Bull C. T., et al. (2020). Clarifying the Taxonomy of the Causal Agent of Bacterial Leaf Spot of Lettuce Through a Polyphasic Approach Reveals That Xanthomonas cynarae Trébaol Et Al. 2000 Emend. Timilsina Et Al. 2019 is a Later Heterotypic Synonym of Xanthomonas hortorum Vauterin Et Al. 1995. System. Appl. Microbiol. 43, 126087. doi: 10.1016/j.syapm.2020.126087
Nameth S. T., Daughtrey M. L., Moorman G. W., Sulzinski M. A. (1999). Bacterial Blight of Geranium: A History of Diagnostic Challenges. Plant Dis. 83, 204–212. doi: 10.1094/PDIS.1999.83.3.204
Niederhauser J. S. (1943). A Bacterial Leaf Spot and Blight of the Russian Dandelion. Phytopathology 33, 959–961.
Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., et al. (2000). Loop-Mediated Isothermal Amplification of DNA. Nucleic Acids Res. 28, E63. doi: 10.1093/nar/28.12.e63
Osdaghi E., Sharma A., Goss E., Abrahamian P., Newberry E., Potnis N., et al. (2021). A Centenary for Bacterial Spot of Tomato and Pepper. Mol. Plant Pathol. 22, 1500–1519. doi: 10.1111/mpp.13125
Piepenburg O., Williams C. H., Stemple D. L., Armes N. A. (2006). DNA Detection Using Recombination Proteins. PloS Biol. 4, e204. doi: 10.1371/journal.pbio.0040204
Potnis N., Krasileva K., Chow V., Almeida N., Patil P., Ryan R., et al. (2011). Comparative Genomics Reveals Diversity Among Xanthomonads Infecting Tomato and Pepper. BMC Genomics 12, 146.
Richard D., Boyer C., Lefeuvre P., Canteros B.I., Beni-Madhu S., Portier P., et al. (2017). Complete Genome Sequences of Six Copper-Resistant Xanthomonas Strains Causing Bacterial Spot of Solaneous Plants, Belonging to X. Gardneri, X. Euvesicatoria, and X. Vesicatoria, Using Long-Read Technology. Genome Announcements 5, e01693-01616. doi: 10.1128/genomeA.01693-16
Ridé M. (1956). Sur une maladie nouvelle de l’artichaut (Cynara scolymus L.). Comptes Rendus Hebdomadaires des Séances de l’Académie des Sciences 243, 174-177.
Ruinelli M., Schneeberger P. H. H., Ferrante P., Bühlmann A., Scortichini M., Vanneste J. L., et al. (2017). Comparative Genomics-Informed Design of Two LAMP Assays for Detection of the Kiwifruit Pathogen Pseudomonas syringae pv. actinidiae and Discrimination of Isolates Belonging to the Pandemic Biovar 3. Plant Pathol. 66, 140–149. doi: 10.1111/ppa.12551
Sahin F. (1997). Detection, Identification and Characterization of Strains of Xanthomonas campestris Pv. Vesicatoria by Traditional and Molecular Methods, and Resistance in capsicum Species to Xanthomonas campestris pv. vesicatoria Pepper Race 6 (Columbus (OH: The Ohio State University).
Scott J. C., Dung J. K. S. (2020). Distribution of Xanthomonas hortorum pv. carotae Populations in Naturally Infested Carrot Seed Lots. Plant Dis. 104, 2144–2148. doi: 10.1094/PDIS-12-19-2674-RE
Stehlíková D., Beran P., Cohen S. P., Čurn V. (2020). Development of Real-Time and Colorimetric Loop Mediated Isothermal Amplification Assay for Detection of Xanthomonas gardneri. Microorganisms 8, 1301. doi: 10.3390/microorganisms8091301
Strayer A., Jeyaprakash A., Minsavage G. V., Timilsina S., Vallad G. E., Jones J. B., et al. (2016). A Multiplex Real-Time PCR Assay Differentiates Four Xanthomonas Species Associated With Bacterial Spot of Tomato. Plant Dis. 100, 1660–1668. doi: 10.1094/PDIS-09-15-1085-RE
Strayer-Scherer A., Jones J. B., Paret M. L. (2019). Recombinase Polymerase Amplification Assay for Field Detection of Tomato Bacterial Spot Pathogens. Phytopathology 109, 690–700. doi: 10.1094/PHYTO-03-18-0101-R
Temple T. N., du Toit L. J., Derie M. L., Johnson K. B. (2013). Quantitative Molecular Detection of Xanthomonas hortorum pv. carotae in Carrot Seed Before and After Hot-Water Treatment. Plant Dis. 97, 1585–1592. doi: 10.1094/PDIS-03-13-0262-RE
Temple T. N., Johnson K. B. (2009). Detection of Xanthomonas hortorum pv. carotae on and in Carrot With Loop-Mediated Isothermal Amplification (LAMP). Phytopathology 99, S186.
Timilsina S., Kara S., Jacques M. A., Potnis N., Minsavage G. V., Vallad G. E., et al. (2019). Reclassification of Xanthomonas gardneri (Ex Šutič 1957) Jones Et Al. 2006 as a Later Heterotypic Synonym of Xanthomonas cynarae Trébaol Et Al. 2000 and Description of x. cynarae Pv. cynarae and x. cynarae pv. gardneri Based on Whole Genome Analyses. Int. J. System. Evol. Microbiol. 69, 343–349. doi: 10.1099/ijsem.0.003104
Tomita N., Mori Y., Kanda H., Notomi T. (2008). Loop-Mediated Isothermal Amplification (LAMP) of Gene Sequences and Simple Visual Detection of Products. Nat. Protoc. 3, 877–882. doi: 10.1038/nprot.2008.57
Toussaint V., Benoit D. L., Carisse O. (2012). Potential of Weed Species to Serve as a Reservoir for Xanthomonas campestris pv. vitians, the Causal Agent of Bacterial Leaf Spot of Lettuce. Crop Prot. 41, 64–70. doi: 10.1016/j.cropro.2012.05.018
Trébaol G., Gardan L., Manceau C., Tanguy J. L., Tirilly Y., Boury S. (2000). Genomic and Phenotypic Characterization of Xanthomonas cynarae Sp. Nov., a New Species That Causes Bacterial Bract Spot of Artichoke (cynara scolymus L.). Int. J. System. Evol. Microbiol. 50, 1471–1478. doi: 10.1099/00207713-50-4-1471
Vauterin L., Yang P., Alvarez A., Takikawa Y., Roth D. A., Vidaver A. K., et al. (1996). Identification of non-Pathogenic Xanthomonas Strains Associated With Plants. System. Appl. Microbiol. 19, 96–105. doi: 10.1016/S0723-2020(96)80016-6
Keywords: singletons, bacterial leaf spot, diagnostic methods, natural infection, hydrangea
Citation: Dia NC, Cottyn B, Blom J, Smits THM and Pothier JF (2022) Differentiation of the Xanthomonas hortorum – Xanthomonas hydrangeae Species Complex Using Sensitive and Selective LAMP Assays. Front. Agron. 4:898778. doi: 10.3389/fagro.2022.898778
Received: 17 March 2022; Accepted: 05 May 2022;
Published: 01 June 2022.
Edited by:
Sara Franco Ortega, University of York, United KingdomReviewed by:
Sanjeet Kumar, Max Planck Institute for the Science of Human History (MPI-SHH), GermanyDavid J. Studholme, University of Exeter, United Kingdom
Copyright © 2022 Dia, Cottyn, Blom, Smits and Pothier. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joël F. Pothier, am9lbC5wb3RoaWVyQHpoYXcuY2g=
 Nay C. Dia
Nay C. Dia Bart Cottyn
Bart Cottyn Jochen Blom
Jochen Blom Theo H. M. Smits
Theo H. M. Smits Joël F. Pothier
Joël F. Pothier