
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Agron., 10 February 2022
Sec. Disease Management
Volume 4 - 2022 | https://doi.org/10.3389/fagro.2022.836360
This article is part of the Research TopicCocoa Diseases and their ManagementView all 5 articles
 Angra Paula Bomfim Rego1
Angra Paula Bomfim Rego1 Irma Yuliana Mora-Ocampo1
Irma Yuliana Mora-Ocampo1 Carlos Priminho Pirovani1
Carlos Priminho Pirovani1 Edna Dora Martins Newman Luz2†
Edna Dora Martins Newman Luz2† Ronan Xavier Corrêa1*†
Ronan Xavier Corrêa1*†Species of the genus Phytophthora cause black-pod rot, which is the disease responsible for the largest losses in cocoa production in the world. The species Phytophthora palmivora affects cacao tree cultivation in all producing countries. However, proteomic level studies of the Theobroma cacao–P. palmivora interaction are incipient. Thus, the aim of this study was to analyze this pathosystem through comparative proteomics and systems biology analyses. The proteins were extracted from leaves of T. cacao PA 150 (resistant) and SIC 23 (susceptible) clones 48 h after inoculation with P. palmivora using inoculation with sterile distilled water as controls. There were differences in the protein profile between the control and inoculated treatments of both clones. Thirty-seven distinct proteins were identified on 88 spots of the PA 150 treatments, and 39 distinct proteins were identified on 120 spots of the SIC 23 treatments. The metabolisms of ATP, carbohydrates, and nitrogen compounds had higher percentages of proteins with increased accumulation after inoculation in both clones. Systems biology analysis demonstrated that the networks contain a higher number of proteins in the clusters corresponding to processes of photosynthesis and glucose metabolism, suggesting that they are the most affected by the infection. In addition, lipoxygenase (LOX), 2-methylene-furan-3-one reductase-like, and co-chaperonin CPN20 proteins and a probable CC-NBS-LRR protein may be involved in resistance to black-pod disease caused by P. palmivora.
Theobroma cacao L. beans are the raw material for chocolate production, with an international trade valued at US $103 billion per year. However, losses caused by diseases affect up to 38% of the annual global cocoa harvest (Marelli et al., 2019). The most economically important cocoa tree disease black-pod rot is caused by Phytophthora species that infect the fruits, being responsible for the loss of 20–25% of cocoa production, which is approximately 700,000 metric tons on a global scale (Adeniyi, 2019). Phytophthora megakarya (Brasier and Griffin) occurs only in countries of West and Central Africa and is considered a significant pathogen only in cacao, whereas Phytophthora palmivora (E. J. Butler) is present in all cocoa producing countries and has a wide range of hosts (Ali et al., 2017; Perrine-Walker, 2020).
Phytophthora palmivora is a hemibiotrophic Oomycete capable of infecting more than 200 species of plants (Perrine-Walker, 2020) with the capacity to infect leaves, branches, fruits, stalk, and roots. The symptoms appear in infected fruits around 4 days after penetration of the germ tube in the mesocarp and in the biotrophic phase establishment. During the necrotrophic phase, secondary hyphae kill the host cell, a brown lesion develops, and the fruit is no longer suitable for harvesting (Bowers et al., 2001; Perrine-Walker, 2020).
Black-pod rot can be controlled using cultivation practices such as removal of infected fruits and pods and phytosanitary pruning of the cacao tree, associated with chemical, genetic, and biological methods to ensure integrated disease control management in cocoa tree plantations (Macagnan et al., 2006; Zhang and Motilal, 2016). The use of resistant varieties is a more effective alternative for disease control (Barreto et al., 2015), although many cocoa cultivars are susceptible to this disease.
Studies on the Phytophthora–T.cacao interaction have focused on anatomic/histological differences, enzymes, genes, and metabolites of varying influence on resistance to black-pod rot (Bailey et al., 2016; Gu et al., 2020). However, there are few studies specifically on the P. palmivora–T.cacao interaction. One such study evaluated 262 cacao genotypes regarding genetic resistance and it was found that resistance to black-pod rot is polygenic in cross-breeds involving resistant and susceptible genotypes (Dantas Neto et al., 2005) and oligogenic in cultivars of average resistance (Barreto et al., 2015). In a transcriptomic analysis, the expression of 5,264 cocoa genes was altered after P. palmivora infection, including subsets of genes involved in biosynthesis of phenylpropanoid, biosynthesis and action of ethylene and jasmonic acid (JA), plant defense signal transduction, and endocytosis, which were induced in response to infection (Ali et al., 2017). Furthermore, a large subset of genes that codify putative proteins related to pathogenesis (PR) also showed differential expression in response to infection (Ali et al., 2017).
However, biological issues can only be addressed at protein level due to different levels of genic regulation and post-translational modifications (Quirino et al., 2010). This type of analysis provides information on the molecular mechanisms of resistance in plants (Geddes et al., 2008).
Some aspects of the T. cacao–Moniliophthora perniciosa pathosystem have already been addressed through proteomic analysis (Almeida et al., 2017; dos Santos et al., 2020; Mares et al., 2020). Proteins from the leaf water wash (LWW) of CCN51 genotype were analyzed by 2D-SDS/PAGE followed by tandem mass spectrometry, and 42 proteins (28 from the cocoa and 14 from bacteria) were identified, including proteins related to defense and synthesis of defense metabolites and involved in nucleic acid metabolism (Almeida et al., 2017). The changes in protein expression in basidiospores of the fungus M. perniciosa in response to the LWW of two contrasting cacao varieties for resistance to witches' broom disease were described in a proteomic analyses (Mares et al., 2020). The proteomic analysis was performed by the 2D-PAGE technique combined with mass spectrometry (MS) revealed proteins associated with energy (ATP synthase) and protein (BiP) metabolism, whose accumulation was reduced by basidiospores germinated in leaf wash from “Catongo” cacao. Furthermore, proteins involved in virulence were identified along with fungal resistance to polyketide cyclase, glycoside hydrolase, multidrug transporter protein (SFM), and proteins related to oxidative stress and fermentation, such as catalase A and alcohol dehydrogenase (ADH) (Mares et al., 2020). On the other hand, in a 2D gel-based approach followed by MS analysis comparing the response of contrasting cacao genotypes for resistance to M. perniciosa, the resistant genotype showed expressed proteins associated with stress and defense, while the susceptible genotype pathogenesis related proteins (PRs), oxidative stress regulation related proteins, and trypsin inhibitors were repressed (dos Santos et al., 2020). Nevertheless, despite the large economic impact caused by black pod rot, protein level studies of the response of T. cacao to infection by P. palmivora are incipient. Thus, this study aims to analyze the P. palmivora–T. cacao interaction through a comparative proteomic and systems biology approach, using T. cacao PA 150 (resistant) and SIC 23 (susceptible) clones inoculated with P. palmivora to uncover the possible defense mechanisms of the plant.
Fully expanded and physiologically mature leaves from T. cacao clones PA 150, resistant, and SIC 23 susceptible to black-pod rot were collected from plants of the active germplasm bank of the Cacao Research Center—CEPEC/Executive Commission of the Cacao Tree Plantation Plan—CEPLAC. So, 10 adult 20-year-old plants were used to obtain leaves from each cacao clone. The P. palmivora isolate was obtained from the culture collection Arnaldo Medeiros of the CEPEC (Luz et al., 2008). The inoculations were performed on leaf discs according to the methodology developed by Nyassé et al. (1995), with modifications to the aliquot volume to 20 μl of the zoospore suspension of P. palmivora at a concentration of 3 × 105 zoospores/ml.
The total protein extracts from leaves of the control and inoculated treatments after 48 h were obtained using phenol extraction followed by precipitation with ammonium acetate 0.1 mol L−1 in methanol, as described by Pirovani et al. (2008). A total of 1.0 g of leaf tissue, previously macerated in liquid nitrogen and in the presence of polyvinylpyrrolidone antioxidant (PVPP), was used for each treatment, using a procedure based on successive washes, associated with steps of sonication. In the second extraction stage, phenols and dense SDS were used. The precipitated proteins were resuspended in rehydration buffer, composed by 8 mol L−1 urea, 2% chaps, 2% IPG buffer 3–10, bromophenol blue 0.002%. The proteins were quantified using 2D-Quant (GE HealthCare) kit, following the manufacturer's instructions.
First dimension electrophoresis was carried out in strips in the non-linear (NL) 3–10 pH band (Amersham Biosciences, Immobiline Dry-Strip). Total protein (500 μg) was homogenized in rehydration buffer containing Dithiothreitol (DTT), at a concentration of 50 mmol L−1 and 0.5% ampholytes, totaling 250 μl. The strips were focalized using EttanIPGhor3 (GE Healthcare) equipment, with Ettan IPGhor3 software, in accordance with a rehydration time of 12 h at 20°C and running conditions of 500 Vh for 1 h, 1,000 Vh for 1 h 04, 8,000 Vh for 2 h 30, and 8,000 Vh for 40 min. After isoelectric focalization of the strips, they were incubated for 15 min with equilibrium buffer (Urea at 6 M, Tris-HCl (75 mmol L−1, pH 8.8), glycerol at 30%, SDS at 2%, and bromophenol blue at 0.002%), containing DTT 10 mg ml−1; for a further 15-min period in iodoacetamide at 25 mg ml−1 (Buffer Tris-HCl 50 mM, pH 8.8, urea 6 M, glycerol at 30%, SDS at 2%, iodoacetamide at 1%, trace of bromophenol blue); and, finally, for 15 more min in running buffer 1X (Tris at 0.25 mol L−1, glycine at 1.92 mol L−1, SDS at 1%, pH 8.5). The strips were placed on vertical SDS-PAGE gel at 12.5%. The second dimension (2-DE) was carried out on a Ruby SE600 (GE Healthcare) system: 15 mA/gel for 15 min, 40 mA/gel for 30 min, and, finally, 50 mA/gel for 5 h, for each strip, at a constant temperature of 11°C. All the 2-D gel separations were repeated three times for each treatment. After electrophoresis, the proteins were visualized with 0.08% Coomassie Blue G-250 (Neuhoff et al., 1988). The gels were colored and discolored for 7 days under constant agitation. After this period, the gels were maintained in acetic acid at 7%. The gel images were obtained through a LabScanner (Amersham Bioscience) and analyzed using Image Master 2D Platinum 7.0 software (GE Healthcare). The gels were reproduced in triplicate for each sample in order to increase reproducibility of the analysis. The control samples were compared to samples inoculated with P. palmivora for each cacao clone. The analysis of spots differentially accumulated between the variety of treatments was based on ANOVA calculation, where values of p ≤ 0.05 and spots with variations of intensity (fold) >1.5 were considered (Supplementary Tables 1, 2).
The spots of interest were excised from 2-DE gel using a scalpel and proteolytic digestion was carried out according to Shevchenko et al. (2007). The solution containing the proteolytic was fragmented through reverse phase chromatography in a nanoAcquity UPLC (WATERS) attached to the Q-Tof micro mass spectrometer (Waters), as per Silva et al. (2013). The obtained spectra were analyzed using ProteinLynx v2.3 software and compared with the T. cacao genome database of NCBI, using the MASCOT MS/MS IonSearch tool (www.matrixscience.com; Supplementary Tables 3, 4; Argout et al., 2008). The criteria used for research were trypsin enzyme digestion, carbamidomethyl (Cys) with fixed modification, and oxidation (Met), as variable modification; a maximum of one loss of the cleavage site; and ±0.3 Da for the peptide tolerance error and 0.1 Da for fragmented ions error (Silva et al., 2013; Villela-Dias et al., 2014). The generated FASTA sequences were analyzed using Blast2GO software (http://www.blast2go.com), which provides important information, such as ontology, functions, biological processes, and cellular localization.
To obtain information on the protein–protein interactions based on the proteomic profiles of T. cacao, homologous proteins in Arabidopsis thaliana were sought out. To achieve this objective, all the identified proteins with their FASTA sequences were processed using STRING 11.0 (http://string-db.org) software. The proteins were individually analyzed with the following parameters: meaning of network edges: confidence; active interaction sources: textmining, experiments, databases, co-expression, neighborhood, gene fusion, and co-occurrence; minimum required interaction score: high confidence (0.700) more than 50 interactions, significance level of 0.7; max number of interactors to show: first and second shell: no more than 50 interactions. The file for each network was downloaded in TSV format and the files were subsequently merged and analyzed using Cytoscape software version 3.7.1. The properties of modularity and centrality (betweenness and node degree; Supplementary Tables 5, 6) of the network were calculated through the igraph package of the RStudio statistical tool (RStudio Team, 2019). An analysis of genic ontology enrichment was carried out for each cluster (Supplementary Tables 7, 8) through the BiNGO plugin version 3.0.3 (Shannon, 2003).
There were differences in the protein profile between the treatment inoculated with P. palmivora zoospores and the control treatment (placebo), both for clone PA 150 (resistant) and for clone SIC 23 (susceptible) (Figure 1). A total of 392 spots were identified in the inoculated treatment of the clone PA 150, whereas 326 spots were identified in the control treatment. Of these, 143 were identified exclusively in the inoculated treatment and 77 exclusively in the control treatment, while 249 spots were common to both treatments. Of these, 30 were differentially accumulated (fold >1.5; 0.05 ANOVA).
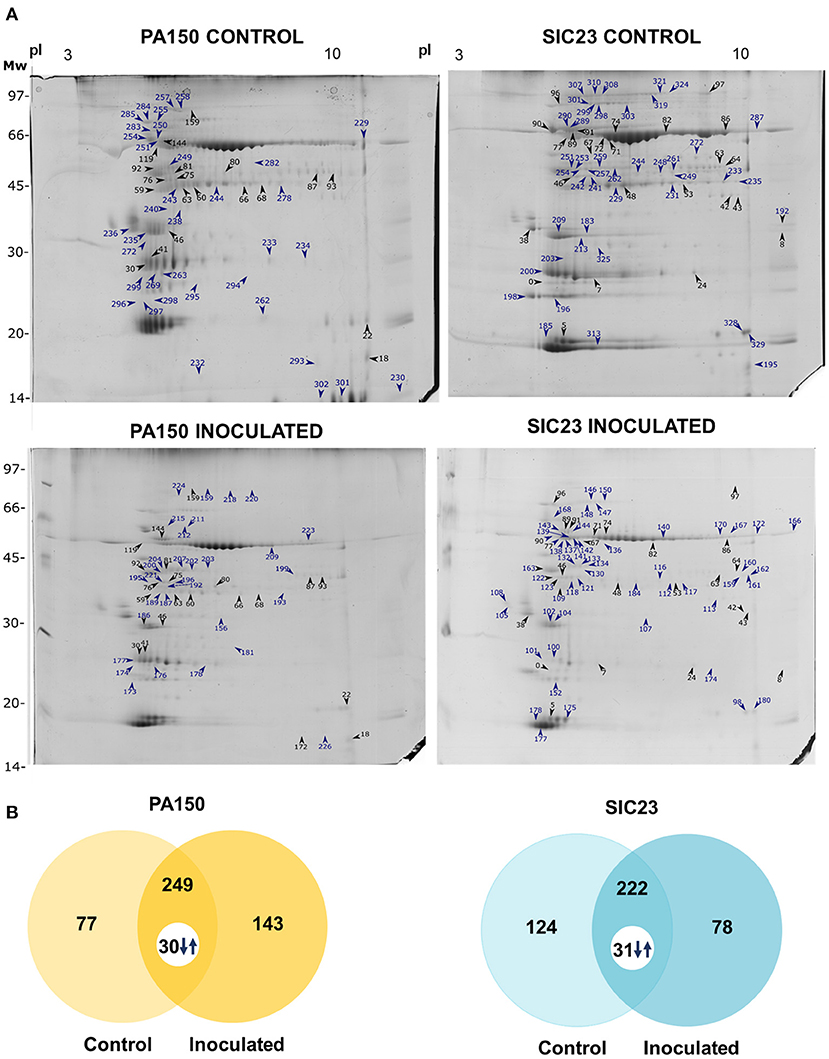
Figure 1. Electrophoretic profiles of cacao leaves inoculated and not inoculated with P. palmivora. (A) SDS-PAGE-2D representative of the inoculated and control treatments of each clone. The spots marked with arrows and numbers are those from which the proteins in Tables 1, 2 were identified. Blue arrows and numbers indicate the spots detected exclusively in each treatment; black arrows and numbers indicate common spots that are differentially accumulated; (B) Venn diagrams representing the spots detected in resistant (PA 150) and susceptible (SIC 23) clones in the control and inoculated treatments. The numbers accompanied by arrows are the spots common to both treatments that had a significant difference in accumulation considering the ANOVA p < 0.05 and fold >1.5 parameters.
In turn, 300 spots were identified in the inoculated treatment of the clone SIC 23, and 346 spots were identified in the control treatment. Of these, 78 were identified exclusively in the inoculated treatment and 124 were detected exclusively in the control treatment, while 222 spots were common to both treatments. Among the spots in common, 31 were differentially accumulated (fold >1.5; p < 0.05 ANOVA).
Finally, 37 distinct proteins were identified on 88 spots of the PA 150 treatments (Table 1) and 39 distinct proteins were identified on 120 spots of the SIC 23 treatments (Table 2).
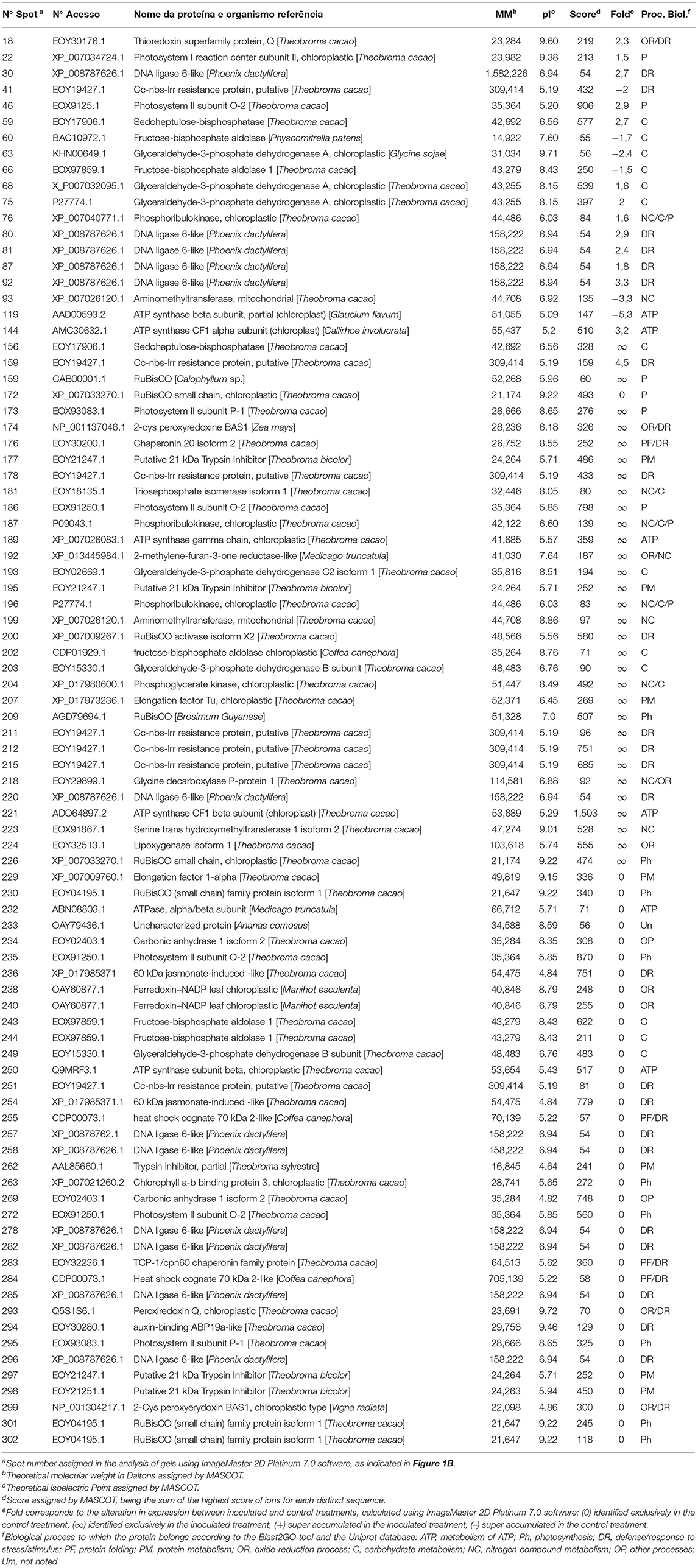
Table 1. Proteins identified through mass spectrometry in leaves of the PA 150 resistant clone of T. cacao inoculated and not inoculated with P. palmivora.
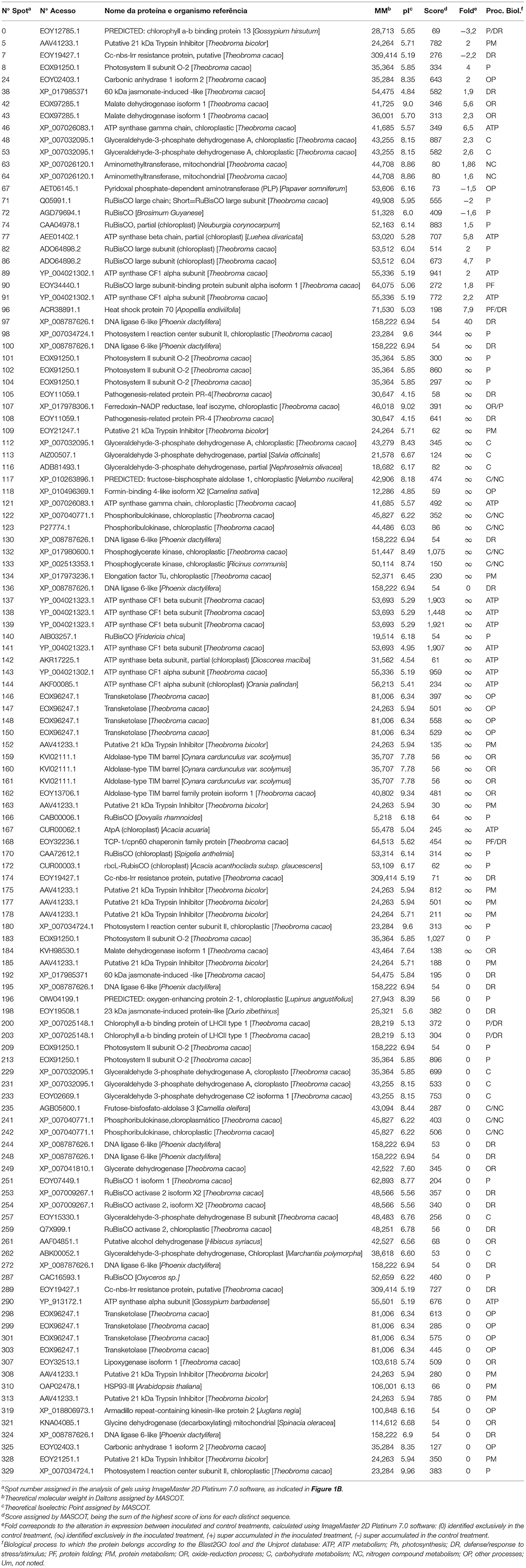
Table 2. Proteins identified through mass spectrometry in leaves of the SIC 23 susceptible clone of T. cacao inoculated and not inoculated with P. palmivora.
The identified proteins were organized into 10 biological processes (Figure 2). It was observed that some proteins belonged to more than one process (Tables 1, 2). The graph in Figure 2 shows the percentage of proteins that had reduced or increased accumulation, in each process, at 48 hai in the PA 150 (Figure 2A) and SIC 23 (Figure 2B) clones. Proteins identified on more than one spot with different accumulation were added both to up-accumulated and down-accumulated proteins.
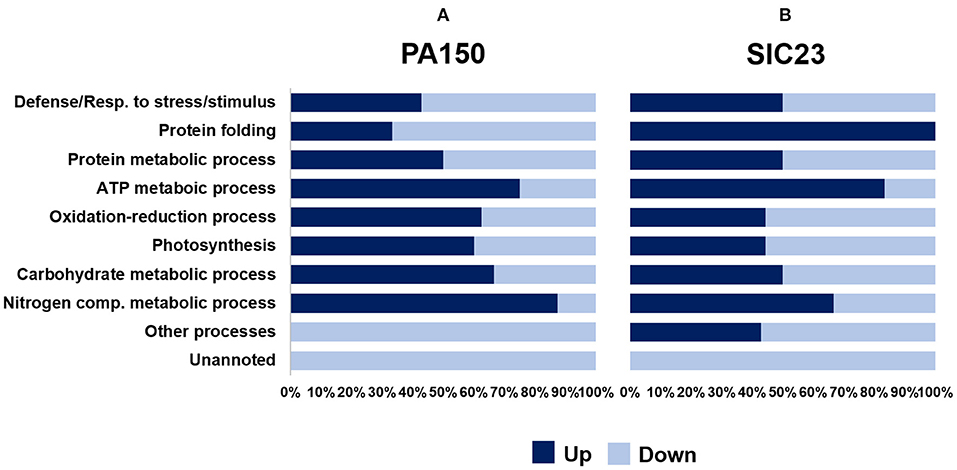
Figure 2. Classification by category of proteins identified in cacao leaves inoculated with P. palmivora. The percentage of proteins identified with increased accumulation (dark blue) and with decreased accumulation (light blue) in leaf discs from (A) resistant clone PA 150 and from (B) susceptible clone SIC 23, 48 hai with oomycete P. palmivora, in comparison to their controls.
In the resistant genotype (Figure 2A), the processes that had a higher percentage of proteins with increased accumulation after inoculation with P. palmivora were the metabolic process of ATP, the oxide-reduction process, photosynthesis, and the metabolic process of carbohydrates and nitrogen compounds. On the other hand, the processes that had a higher percentage of proteins with reduced accumulation during infection were defense or response to stress or stimulus, protein folding, other processes, and not noted. The metabolic process of proteins was maintained at the same percentage of proteins with increased accumulation and reduced accumulation.
In the susceptible clone (Figure 2B), the processes that had a higher percentage of proteins with increased accumulation at 48 hai were protein folding, the metabolic process of ATP, and nitrogen compound metabolism. Those that had a higher percentage of proteins with reduced accumulation were the oxide-reduction process, photosynthesis, other processes, and not noted. However, the processes of defense or response to the stimulus, protein metabolism, and carbohydrate metabolism maintained the same percentage.
The interaction network corresponds to the homologous proteins identified in the PA 150 (Figure 3A) clone resulting in 1,339 knots, 28,131 connectors, and 14 clusters, to which biological functions were attributed, such as response to wounds, lipid transport, photosynthesis, biosynthesis of cuticular hydrocarbons, and protein folding, among others (Supplementary Table 7). In the same network, there was a total of 177 bottleneck proteins and 515 protein hubs (Supplementary Table 5).
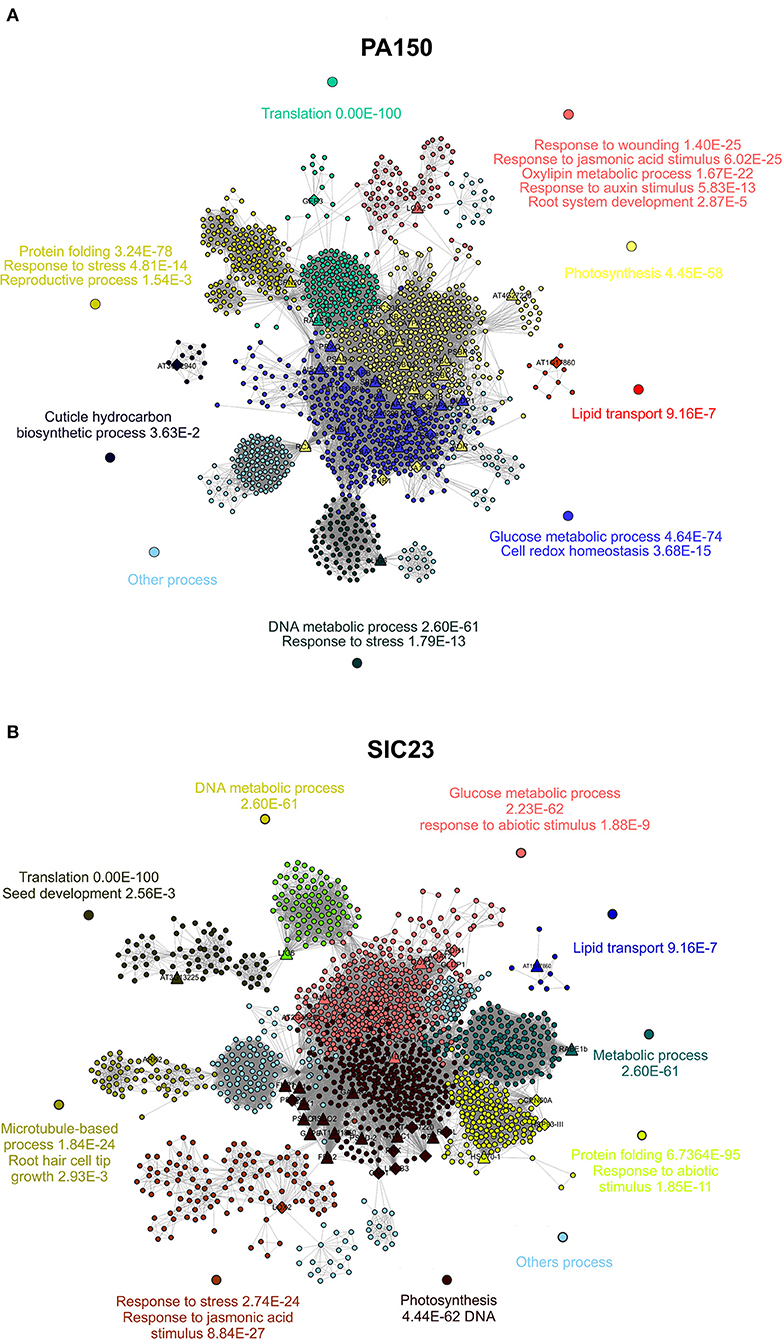
Figure 3. Protein interaction networks of A. thaliana. Interaction network of the proteins of A. thaliana homologous to those identified in leaves from (A) resistant clone PA 150 and (B) susceptible clone T. cacao, 48 hai with P. palmivora. Each knot represents a protein; the larger knots in triangle shape represent proteins homologous to those identified in T. cacao as up-accumulated and the larger knots in diamond shape represent the proteins homologous to those identified in T. cacao as down-accumulated. The networks were divided into clusters, each represented by a color. At least one biological function with its p-value was attributed to each cluster.
In turn, the network corresponding to the proteins homologous to those identified in the SIC 23 clone (Figure 3B) presented a total of 1,466 knots, 27,198 connectors, and 15 clusters, which corresponded to biological functions such as response to stress, microtubule-based process, and photosynthesis, among others (Supplementary Table 8). In this network, 201 proteins were bottleneck and there were 573 hubs (Supplementary Table 6).
The black-pod rot disease, caused by species of the genus Phytophthora, is responsible for great losses in global cacao production. Here, responses of two contrasting clones for P. palmivora resistance were compared at proteomic level. Proteins related to photosynthesis, glucose metabolism, response to stress and oxidation–reduction process were identified (Figure 1). Through the different patterns of protein accumulation, differences in molecular responses between susceptible and resistant genotypes were elucidated, such as declining cellular energy, balance in the production and detoxification of ROS, control of cytotoxic metabolites, production of JA, and participation of a protein encoded by a resistance gene (R).
Ribulose bisphosphate carboxylase/oxygenase (rubisco) protein, was identified on seven spots in the PA 150 clone (Table 1); four with reduced accumulation (spots 172, 230, 301, and 302) and three with increased accumulation (spots 159, 209, and 226). In turn, the same protein was identified on 12 spots in the SIC 23 clone (Table 2); eight with increased accumulation (spots 74, 82, 86, 90, 140, 166, 170, and 172) and four with reduced accumulation (spots 71, 72, 251, and 287). This protein is one of the main enzymes in the second phase of photosynthesis. Interestingly, RuBisCO Activase (RCA), which is required for rubisco to function properly, showed reduced accumulation in the susceptible clone (spots 253, 254, and 259) and increased accumulation in PA 150 (spot 200). However, it required ATP to perform its activase function (Portis et al., 2007; Carmo-Silva and Salvucci, 2011). Thus, six spots corresponding to ATP synthase subunits were identified in the resistant clone, of which three showed an increased accumulation (Table 1; spots 144, 189, and 221) and three showed reduced accumulation (spots 119, 250, and 232), whereas the susceptible clone had 14 spots identified as corresponding to ATP synthase subunits, of which 13 had increased accumulation (Table 2; spots 46, 77, 89, 91, 121,137–139, 141–144, and 167).
Therefore, in the susceptible clone, where RCA had reduced accumulation, there was a higher number of spots corresponding to rubisco and ATPase with increased accumulation, in contrast to the resistant clone. As a result, we can propose the following scenario: a decrease in the quantity of RCA in the SIC 23 clone resulted in deficient rubisco activity, which leads to an imbalance in the Calvin cycle, and, as a consequence, a decrease in sugars production in the cell. As a result, energy production would be reduced. Thus, there should have been cellular compensation for the energy loss, with increased expression of rubisco and ATP synthase.
RuBisCO Activase is one of the most important proteins within the interaction network related to homologous proteins identified in both clones (Figure 3) since its values of betweenness and node degree are close to the maximum values (Supplementary Tables 5, 6). Therefore, it is considered a protein with an important role in regulation and signaling within the organism (Verli, 2014).
The photosystems I and II subunits were identified on different spots (Tables 1, 2; Figure 2), which had different accumulation in both clones 48 hai. It is well-known that these reaction centers are the primary source of reactive oxygen species (ROS) (Asada, 2006); a decrease in photosystem expression causes an increase in ROS production (Oukarroum et al., 2015). Thus, the different expression patterns of the subunits of PSI and PSDII observed in the PA 150 and SIC 23 clones may be involved in varying levels of ROS production.
Furthermore, it was observed that the chlorophyll a-b binding protein, also called light-harvesting chlorophyll a/b-binding (LHCB), had reduced accumulation in both clones (Table 1, spot 263; Table 2, spots 0, 200, and 203). This protein absorbs solar light and drives the transport of photosynthetic electrons (Jansson, 1994; Liu et al., 2013). Moreover, LHCBs are positively involved in ABA signaling by modulating ROS homeostasis (Xu et al., 2012). Therefore, a decrease in the expression of LHCB protein in both clones suggests a decrease in the ABA pathway, promoting the accumulation of H2O2 (Asselbergh et al., 2008). In that regard, the co-chaperonin CPN20 was identified in the PA 150 clone as exclusive (spot 176). This protein negatively regulates the signaling of ABA (Zhang et al., 2013), reinforcing the theory that ABA pathway is repressed, and is responsible for the activation of iron superoxide dismutase (FeSODs; FSDs) enzymes, whose function is to dismutase superoxide (O−) in H2O2 (Kuo et al., 2013a,b).
Moreover, a 2-cys peroxyredoxine BAS1, an enzyme involved in the regulation of plastidial H2O2 concentration (Awad et al., 2015), was exclusively identified in the PA 150 clone (Table 1; spot 174). A 2-cys peroxyredoxine was also identified with increased accumulation in the resistant clone of the pathosystem T. cacao–M. perniciosa (dos Santos et al., 2020). This evidence suggests that there is a more efficient ROS production and detoxification system against infection with P. palmivora.
The metabolism of carbohydrates positively regulates the expression of genes related to defense (Rojas et al., 2014). In this regard, it was observed that the triosephosphate isomerase enzyme (TPI) (Table 1; spot 181) had increased accumulation in the PA 150 clone after inoculation, which is an indication of glycolytic pathway increase (Valcu et al., 2009). In addition, TPI assists in the control of methylglyoxal levels, a cytotoxic metabolite and glycolysis by-product, which is accumulated in plants under stress and which participates as a molecular signaler (Kaur et al., 2015).
In turn, the glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) protein, in its cytosolic form (GAPC), intermediates in both energy metabolism and signal transference for long-term adjustment and protection against redox imbalance (Schneider et al., 2018). In the pathosystem T. cacao–M. perniciosa, two GAPC isoforms were identified with increased accumulation in the resistant clone at 72 hai (dos Santos et al., 2020). Interestingly, a GAPC had differentiated accumulation between the clones since it was super accumulated in the PA 150 clone (spot 193) and had reduced accumulation in the SIC 23 clone (spot 233), which may influence the resistance of T. cacao to P. palmivora. This is also represented in the interaction network of the resistant clone (Figure 3A; Supplementary Table 7), in which the cluster where the GAPC isoforms meet had assigned processes for glucose metabolism and cellular redox homeostasis.
The fact that the clusters with a greater number of proteins were those related to photosynthesis and metabolism of sugar (Supplementary Tables 5, 6) suggests that these biological functions were the most affected during P. palmivora infection. Network analysis demonstrates that the proteins homologous to those identified in SIC 23 with reduced accumulation have greater influence on the clusters corresponding to processes of photosynthesis and metabolism of sugars in comparison to the proteins homologous to those identified with increased accumulation, based on node degree value. The opposite occurs in the PA 150 clone, where the proteins homologous to those identified with increased accumulation have a higher node degree value. This may translate into an increase in the resistant clone and a decrease in the susceptible clone for these biological processes.
A phosphoribulokinase (PRK) was found to be differentially expressed (Tables 1, 2). In the inoculated PA 150 clone, all of the spots corresponding to PRK (spots 76, 187, and 196) had increased accumulation. However, in the SIC 23 clone, the PRK protein varied in accumulation between the different spots, with two of them (spots 122 and 123) showing increased accumulation and the other two (spots 241 and 242) showing decreased accumulation after inoculation. This protein was identified as repressed in susceptible black pepper inoculated with Phytophthora capsici (Mahadevan et al., 2016) and is involved in the defense response to bacteria in Arabidopsis (Jones et al., 2006). Therefore, this difference in the accumulation pattern may be involved in resistance in the PA 150 clone. Furthermore, the PRK is regulated by thioredoxin (Feierabend, 2005), which was identified in two spots in the inoculated PA 150 clone (Table 1); one with increased accumulation (spot 18) and the other with reduced accumulation (spot 293).
Lipoxygenase (LOX) was identified as up-accumulated in the PA 150 clone and down-accumulated in the SIC 23 clone (Table 1, spot 224; Table 2, spot 307). In tobacco plants inoculated with Phytophthora parasitica nicotianae, it was observed that LOX gene transcripts increased and their activity was increased in the initial stages of infection in resistant plants (Veronesi et al., 1996). The LOX pathway results in the production of taumatin, JA, oxylipins, and volatile aldehydes that play an important role in wound healing, synthesis of antimicrobial substances, and membrane damage during the hypersensitive response (HR) (Thakur and Udayashankar, 2019). Jasmonic acid enhances superoxide production and activates genes encoding proteins involved in defense responses (Vasyukova and Ozeretskovskaya, 2009; Karpets et al., 2014), increases resistance to Phytophthora infestans in tomatoes and potatoes (Cohen et al., 1993) and the defense measured by this hormone is crucial to the resistance of peppers to P. capsici (Ueeda et al., 2005).
After inoculation, 2-methylene-furan-3-one reductase-like (enone oxidoreductase) was found to increase accumulation in the resistant genotype. This protein is induced by ripening and inhibited by auxin in strawberries (Schiefner et al., 2013). It was also found only in the Prunus persica clone resistant to the fungus Taphrina deformans (Goldy et al., 2017). Furthermore, the protein is involved in the biosynthesis of furaneol, which is a volatile compound (Raab et al., 2006) that may be involved in defense signaling (Peinado-Guevara et al., 2017). The homolog of this protein in the network is AOR, a NADPH dependent oxidoreductase on, that aids in detoxifying stromal reactive carbonyls produced under oxidative stress (Yamauchi et al., 2011, 2012).
A probable CC-NBS-LRR protein was identified in both clones, albeit with different accumulation patterns. In the SIC 23 clone, three spots (Table 2; spots 7, 174, and 289) were identified corresponding to this protein, whereby only one of them had increased accumulation. In the PA 150 clone, seven spots were identified corresponding to the same protein (Table 1); five had increased accumulation (spots 159, 178, 211, 212, and 215) and two (spots 251 and 41) reduced accumulation.
The CC-NBS-LRR proteins are codified by one of the genes identified as genes of plant resistance (R) to diseases. Their structure contains an N-terminal coiled-coil (CC) domain, a nucleotide binding site (NBS) domain, and a C-terminal leucine-rich repeat (LRR) domain. The CC-NBS-LRR proteins recognize products derived from specific pathogens and initiate a resistance response that generally includes a type of cell death known as hypersensitivity response (HR) (Moffett et al., 2002). It has already been observed that the expression of this protein confers resistance to P. infestans on currant tomatoes (Zhang et al., 2014) and positive regulation was discovered for various NBS-LRR genes that suppress P. infestans infection in Solanum pinnatisectum (Gu et al., 2020). The cacao genome contains several copies corresponding to the genes that encodes CC-NBS-LRR isoforms (https://cocoa-genome-hub.southgreen.fr). Therefore, the accumulation of the different spots correspondent to this protein in PA 150 suggests that this clone can express more isoforms of this protein than the susceptible clone, giving it an advantage in its defense mechanism against P. palmivora.
Leaves from PA 150 (resistant) and SIC 23 (susceptible) clones of T. cacao inoculated with P. palmivora present differences in the protein profile when compared with their respective controls (mock-inoculated). These differences revealed evidence of the defense mechanisms presented in each clone (Figure 4).
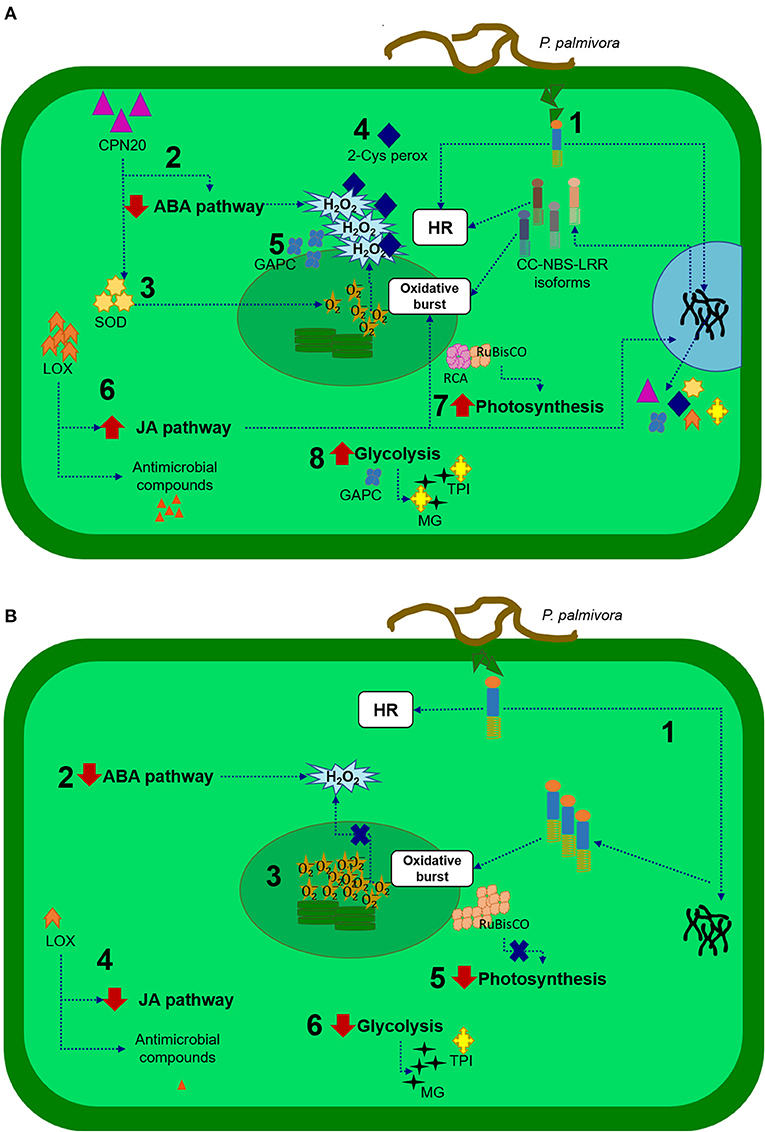
Figure 4. Representative scheme of the defense responses of the cacao tree to P. palmivora. In the resistant clone (A): (1) A CC-NBS-LRR protein recognizes products derived from the pathogen, initiating a response that includes hypersensitive response (HR) and signaling for expression of isoforms and other proteins that help in defense responses; (2) CPN20 protein negatively regulates the signaling of ABA, promoting H2O2 accumulation; (3) CPN20 also activates superoxide dismutase (SOD) enzymes that dismutate superoxide in H2O2 and O2; (4) 2-cys peroxiredoxin regulates H2O2 concentration; (5) A cytosolic glyceraldehyde-3-phosphate-dehydrogenase (GAPC) protect against redox imbalance; (6) A lipoxygenase (LOX) protein activates jasmonic acid (JA) production, which enhances superoxide production and activates genes encoding proteins involved in defense responses; (7) RuBisCO activase (RCA) is increased, assisting in the activity of RuBisCO increasing photosynthesis and (8) Active glycolysis, where the triosephosphate isomerase (TPI) enzyme assists in the control of methylglyoxal levels (MG), a cytotoxic by-product of glycolysis. In the susceptible clone (B): (1) A CC-NBS-LRR protein recognizes products derived from the pathogen. This isoform could initiate responses like HR, but there would be no expression of other isoforms; (2) ABA pathway reduced allows the accumulation of H2O2; (3) In CPN20, 2-Cys peroxiredoxin or GAPC accumulation could lead to imbalance redox; (4) Decrease in LOX accumulation causes a decrease in the production of jasmonic acid and antimicrobial compounds; (5) RuBisco not activated can affect photosynthesis; (6) Glycolysis pathway is reduced, and production of TPI could be insufficient to protect against cytotoxic compounds.
According to the expression pattern of various identified proteins, processes of photosynthesis, and metabolism of sugars increase in the resistant clone and decrease in the susceptible clone and were apparently the most affected processes during P. palmivora infection.
RuBisCO Activase is thought to be a protein that plays an important role in regulation and signaling both clones defense responses. The reduction in the quantity of RCA in the SIC 23 clone suggests deficient rubisco activity, and, therefore, energy decline in the cell. Thus, increased accumulation of rubisco and ATP synthase is being attempted to compensate for this energy loss.
In the resistant clone, there was evidence of better management of ROS production and regulation and in the control of cytotoxic by-products of glycolysis, and protection against redox imbalances, reduction in the ABA pathway, and, therefore, accumulation of H2O2. In addition, the increased accumulation of the LOX protein suggests production of JA, oxylipins, and volatile aldehydes important in wound healing, synthesis of antimicrobial substances, and membrane damage during the HR. It has already been demonstrated that JA is involved in the resistance of plants to Phytophthora species. In turn, an oxidoreductase may contribute to detoxifying compounds produced under oxidative stress, and a probable CC-NBS-LRR protein recognizes products derived from the pathogen, initiating a response that includes a HR.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
AR, EL, and RC contributed to the conception and design of the study. AR and IM-O organized the database and performed the statistical analysis. AR wrote the first draft of the manuscript. CP supervised the proteomics analysis. EL and RX supervised the phytopathological experiments and obtained financial support for the project. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was supported by the Conselho Nacional de Ciência e Tecnologia – CNPq, a Brazilian Federal Government agency (grant number 309841/2015-1 and 308959/2019-1); and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – CAPES, Bahian State Government (Scholarship number 481074/2013-9 e 307915/2012-3 granted to AR).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors are grateful to Comissão Executiva do Plano da Lavoura Cacaueira — CEPLAC, for their assistance in preparing Phytophthora zoospores for cocoa inoculation, and Universidade Estadual de Santa Cruz — UESC, for providing the facilities in Greenhouse and Center of Biotechnology and Genetics CBG.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2022.836360/full#supplementary-material
Adeniyi, D. (2019). “Diversity of cacao pathogens and impact on yield and global production,” in Theobroma Cacao - Deploying Science for Sustainability of Global Cocoa Economy, ed P. Aikpokpodion (London: IntechOpen). doi: 10.5772/intechopen.81993 Available online at: https://www.intechopen.com/chapters/65131
Ali, S. S., Shao, J., Lary, D. J., Strem, M. D., Meinhardt, L. W., and Bailey, B. A. (2017). Phytophthora megakarya and P. palmivora, causal agents of black pod rot, induce similar plant defense responses late during infection of susceptible cacao pods. Front. Plant Sci. 8, 169. doi: 10.3389/fpls.2017.00169
Almeida, D. S. M., Gramacho, K. P., Cardoso, T. H. S., Micheli, F., Alvim, F. C., and Pirovani, C. P. (2017). Cacao phylloplane: the first battlefield against Moniliophthora perniciosa, which causes witches' broom disease. Phytopathology 107, 864–871. doi: 10.1094/PHYTO-06-16-0226-R
Argout, X., Fouet, O., Wincker, P., Gramacho, K., Legavre, T., Sabau, X., et al. (2008). Towards the understanding of the cocoa transcriptome: production and analysis of an exhaustive dataset of ESTs of Theobroma cacao L. generated from various tissues and under various conditions. BMC Genomics. 9, 512. doi: 10.1186/1471-2164-9-512
Asada, K. (2006). Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 141, 391–396. doi: 10.1104/pp.106.082040
Asselbergh, B., De Vleesschauwer, D., and Höfte, M. (2008). Global switches and fine-tuning—ABA modulates plant pathogen defense. Mol. Plant Microbe Interact. 21, 709–719. doi: 10.1094/MPMI-21-6-0709
Awad, J., Stotz, H. U., Fekete, A., Krischke, M., Engert, C., Havaux, M., et al. (2015). 2-Cysteine peroxiredoxins and thylakoid ascorbate peroxidase create a water-water cycle that is essential to protect the photosynthetic apparatus under high light stress conditions. Plant Physiol. 167, 1592–1603. doi: 10.1104/pp.114.255356
Bailey, B. A., Ali, S. S., Akrofi, A. Y., and Meinhardt, L. W. (2016). “Phytophthora megakarya, a causal agent of black pod rot in Africa,” in eds B. A. Bailey and L. W. Meinhardt, Cacao Diseases (Cham: Springer International Publishing), 267–303. doi: 10.1007/978-3-319-24789-2_8
Barreto, M. A., Santos, J. C. S., Corrêa, R. X., Luz, E. D. M. N., Marelli, J., and Souza, A. P. (2015). Detection of genetic resistance to cocoa black pod disease caused by three Phytophthora species. Euphytica 206, 677–687. doi: 10.1007/s10681-015-1490-4
Bowers, J. H., Bailey, B. A., Hebbar, P. K., Sanogo, S., and Lumsden, R. D. (2001). The impact of plant diseases on world chocolate production. Plant Health Prog. 2, 12. doi: 10.1094/PHP-2001-0709-01-RV
Carmo-Silva, A. E., and Salvucci, M. E. (2011). The activity of Rubisco's molecular chaperone, rubisco activase, in leaf extracts. Photosynth Res. 108, 143–155. doi: 10.1007/s11120-011-9667-8
Cohen, Y., Gisi, U., and Niderman, T. (1993). Local and systemic protection against Phytophthora infestans induced in potato and tomato plants by jasmonic acid and jasmonic methyl ester. Phytopathology 83, 1054–1062. doi: 10.1094/Phyto-83-1054
Dantas Neto, A., Corrêa, R. X., Monteiro, W. R., Luz, E. D. M. N., Gramacho, K. P., and Lopes, U.V. (2005) Caracterização de uma população de cacaueiro para mapeamento de genes de resistência à vassoura-de-bruxa e podridão parda. Fitopatol. Bras. 30, 380–386. doi: 10.1590/S0100-41582005000400007
dos Santos, E. C., Pirovani, C. P., Correa, S. C., Micheli, F., and Gramacho, K. P. (2020). The pathogen Moniliophthora perniciosa promotes differential proteomic modulation of cacao genotypes with contrasting resistance to witches' broom disease. BMC Plant Biol. 20, 1. doi: 10.1186/s12870-019-2170-7
Feierabend, J. (2005). “Catalases in plants: molecular and functional properties and role in stress defence,” in Antioxidants and Reactive Oxygen Species in Plants, ed N. Smirnoff (Oxford: Blackwell Publishing Ltd.), 101–130.
Geddes, J., Eudes, F., Laroche, A., and Selinger, L. B. (2008). Differential expression of proteins in response to the interaction between the pathogen Fusarium graminearum and its host, Hordeum vulgare. Proteomics 8, 545–554. doi: 10.1002/pmic.200700115
Goldy, C., Svetaz, L. A., Bustamante, C. A., Allegrini, M., Valentini, G. H., Drincovich, M. F., et al. (2017). Comparative proteomic and metabolomic studies between Prunus persica genotypes resistant and susceptible to Taphrina deformans suggest a molecular basis of resistance. Plant Physiol. Biochem. 118, 245–255. doi: 10.1016/j.plaphy.2017.06.022
Gu, B., Cao, X., Zhou, X., Chen, Z., Wang, Q., Liu, W., et al. (2020). The histological, effectoromic, and transcriptomic analyses of Solanum pinnatisectum reveal an upregulation of multiple NBS-LRR genes suppressing Phytophthora infestans infection. Int. J.Mol.Sci. 21:9,3211. doi: 10.3390/ijms21093211
Jansson, S. (1994). The light-harvesting chlorophyll ab-binding proteins. Biochim Biophys. Acta Bioenerg. 1184, 1–19. doi: 10.1016/0005-2728(94)90148-1
Jones, A. M. E., Thomas, V., Bennett, M. H., Mansfield, J., and Grant, M. (2006). Modifications to the Arabidopsis defense proteome occur prior to significant transcriptional change in response to inoculation with Pseudomonas syringae. Plant Physiol. 142, 1603–1620. doi: 10.1104/pp.106.086231
Karpets, Y. V., Kolupaev, Y. E., Lugovaya, A. A., and Oboznyi, A. I. (2014). Effect of jasmonic acid on the pro-/antioxidant system of wheat coleoptiles as related to hyperthermia tolerance. Russ. J. Plant Physiol. 61, 339–346. doi: 10.1134/S102144371402006X
Kaur, C., Sharma, S., Singla-Pareek, S. L., and Sopory, S. K. (2015). “Methylglyoxal, triose phosphate isomerase, and glyoxalase pathway: implications in abiotic stress and signaling in plants,” in Elucidation of Abiotic Stress Signaling in Plants (New York, NY Springer), 347–366. doi: 10.1007/978-1-4939-2211-6_13
Kuo, W. Y., Huang, C. H., and Jinn, T. L. (2013b). Chaperonin 20 might be an iron chaperone for superoxide dismutase in activating iron superoxide dismutase (FeSOD). Plant Signal. Behav. 8, 2. doi: 10.4161/psb.23074
Kuo, W. Y., Huang, C. H., Liu, A. C., Cheng, C. P., Li, S. H., Chang, W. C., et al. (2013a). CHAPERONIN 20 mediates iron superoxide dismutase (FeSOD) activity independent of its co-chaperonin role in Arabidopsis chloroplasts. New Phytol. 197, 99–110. doi: 10.1111/j.1469-8137.2012.04369.x
Liu, R., Xu, Y.-H., Jiang, S.-C., Lu, K., Lu, Y.-F., Feng, X.-J., et al. (2013). Light-harvesting chlorophyll a/b-binding proteins, positively involved in abscisic acid signalling, require a transcription repressor, WRKY40, to balance their function. J. Exp. Bot. 64, 5443–5456. doi: 10.1093/jxb/ert307
Luz, E. D. M. N., Silva, S. D. V. M., Bezerra, J. L., Souza, J. T., de Santos, A. F., and dos. (2008). Glossário Ilustrado de Phytophthora: Técnicas Especiais para o Estudo de Oomicetos. Itabuna, BA: Fapesb.
Macagnan, D., Romeiro, R. D. S, de Souza, J. T., and Pomella, A. W. V. (2006). Isolation of actinomycetes and endospore-forming bacteria from the cacao pod surface and their antagonistic activity against the witches' broom and black pod pathogens. Phytoparasitica 34, 122–132. doi: 10.1007/BF02981312
Mahadevan, C., Krishnan, A., Saraswathy, G. G., Surendran, A., Jaleel, A., and Sakuntala, M. (2016). Transcriptome- assisted label-free quantitative proteomics analysis reveals novel insights into Piper nigrum—Phytophthora capsici phytopathosystem. Front. Plant Sci. 7, 785. doi: 10.3389/fpls.2016.00785
Marelli, J.-P., Guest, D. I., Bailey, B. A., Evans, H. C., Brown, J. K., Junaid, M., et al. (2019). Chocolate under threat from old and new cacao diseases. Phytopathology 109, 1331–1343. doi: 10.1094/phyto-12-18-0477-rvw
Mares, J. H., Gramacho, K. P., Santana, J. O., Oliveira de Souza, A., Alvim, F. C., and Pirovani, C. P. (2020). Hydrosoluble phylloplane components of Theobroma cacao modulate the metabolism of Moniliophthora perniciosa spores during germination. Fungal Biol. 124, 73–81. doi: 10.1016/j.funbio.2019.11.008
Moffett, P., Farnham, G., Peart, J., and Baulcombe, D. C. (2002). Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 21, 4511–4519. doi: 10.1093/emboj/cdf453
Neuhoff, V., Arold, N., Taube, D., and Ehrhardt, W. (1988). Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9, 255–262. doi: 10.1002/elps.1150090603
Nyassé, S., Cilas, C., Herail, C., and Blaha, G. (1995). Leaf inoculation as an early screening test for cocoa (Theobroma cacao L.) resistance to Phytophthora black pod disease. Crop Prot. 14, 657–663. doi: 10.1016/0261-2194(95)00054-2
Oukarroum, A., Bussotti, F., Goltsev, V., and Kalaji, H. M. (2015). Correlation between reactive oxygen species production and photochemistry of photosystems I and II in Lemna gibba L. plants under salt stress. Environ. Exp. Bot. 109, 80–88. doi: 10.1016/j.envexpbot.2014.08.005
Peinado-Guevara, L. I., López-Meyer, M., López-Valenzuela, J. A., Maldonado-Mendoza, I. E., Galindo-Flores, H., Campista-León, S., et al. (2017). Comparative proteomic analysis of leaf tissue from tomato plants colonized with Rhizophagus irregularis. Symbiosis 73, 93–106. doi: 10.1007/s13199-016-0470-3
Perrine-Walker, F. (2020). Phytophthora palmivora–cocoa interaction. J. Fungi. 6, 167. doi: 10.3390/jof6030167
Pirovani, C. P., Carvalho, H. A. S., Machado, R. C. R., Gomes, D. S., Alvim, F. C., Pomella, A. W. V., et al. (2008). Protein extraction for proteome analysis from cacao leaves and meristems, organs infected by Moniliophthora perniciosa, the causal agent of the witches' broom disease. Electrophoresis 29, 2391–2401. doi: 10.1002/elps.200700743
Portis, A. R., Li, C., Wang, D., and Salvucci, M. E. (2007). Regulation of rubisco activase and its interaction with Rubisco. J. Exp. Bot. 59, 1597–1604. doi: 10.1093/jxb/erm240
Quirino, B. F., Candido, E. S., Campos, P. F., Franco, O. L., and Krüger, R. H. (2010). Proteomic approaches to study plant–pathogen interactions. Phytochemistry 71, 351–362. doi: 10.1016/j.phytochem.2009.11.005
Raab, T., López-Ráez, J. A., Klein, D., Caballero, J. L., Moyano, E., Schwab, W., et al. (2006). FaQR, required for the biosynthesis of the strawberry flavor compound 4-hydroxy-2,5-dimethyl-3(2H)-furanone, encodes an enone oxidoreductase. Plant Cell 18, 1023–1037. doi: 10.1105/tpc.105.039784
Rojas, C. M., Senthil-Kumar, M., Tzin, V., and Mysore, K. S. (2014). Regulation of primary plant metabolism during plant–pathogen interactions and its contribution to plant defense. Front. Plant Sci. 5, 17. doi: 10.3389/fpls.2014.00017
RStudio Team (2019). RStudio: Integrated Development for R (1.2.5033). RStudio, Inc. Available online at: http://www.rstudio.com/ (accessed July 7, 2018).
Schiefner, A., Sinz, Q., Neumaier, I., Schwab, W., and Skerra, A. (2013). Structural basis for the enzymatic formation of the key strawberry flavor compound 4-hydroxy-2,5-dimethyl-3(2 H)-furanone. J. Biol. Chem. 288, 16815–16826. doi: 10.1074/jbc.M113.453852
Schneider, M., Knuesting, J., Birkholz, O., Heinisch, J. J., and Scheibe, R. (2018). Cytosolic GAPDH as a redox-dependent regulator of energy metabolism. BMC Plant Biol. 18, 184. doi: 10.1186/s12870-018-1390-6
Shannon, P. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shevchenko, A., Tomas, H., Havlis, J., Olsen, J. V., and Mann, M. (2007). In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860. doi: 10.1038/nprot.2006.468
Silva, F. A. C., Pirovani, C. P., Menezes, S., Pungartnik, C., Santiago, A. S., Costa, M. G. C., et al. (2013). Proteomic response of Moniliophthora perniciosa exposed to pathogenesis-related protein-10 from Theobroma cacao. Genet. Mol. Res. 12, 4855–4868. doi: 10.4238/2013.October.22.5
Thakur, M., and Udayashankar, A. C. (2019). “Lipoxygenases and their function in plant innate mechanism,” in Bioactive Molecules in Plant Defense, eds S. Jogaiah and M. Abdelrahman (Cham: Springer International Publishing), 133–143. doi: 10.1007/978-3-030-27165-7_8
Ueeda, M., Kubota, M., and Nishi, K. (2005). Contribution of jasmonic acid to resistance against Phytophthora blight in Capsicum annuum cv. SCM334. Physiol. Mol. Plant Pathol. 67, 149–154. doi: 10.1016/j.pmpp.2005.12.002
Valcu, C.-M., Junqueira, M., Shevchenko, A., and Schlink, K. (2009). Comparative proteomic analysis of responses to pathogen infection and wounding in Fagus sylvatica. J. Proteome Res. 8, 4077–4091. doi: 10.1021/pr900456c
Vasyukova, N. I., and Ozeretskovskaya, O. L. (2009). Jasmonate-dependent defense signaling in plant tissues. Russ. J. Plant Physiol. 56, 581–590. doi: 10.1134/S102144370905001X
Verli, H. (2014). (Org.) “Biologia de sistemas,” in Bioinformática da Biologia à Flexibilidade Molecular, 1st Edn. (Sociedade Brasileira de Bioquímica e Biologia Molecular-SBBq), 282. Available online at: https://www.ufrgs.br/bioinfo/ebook/ (accessed July 7, 2018).
Veronesi, C., Rickauer, M., Fournier, J., Pouenat, M. L., and Esquerre-Tugaye, M. T. (1996). Lipoxygenase gene expression in the tobacco–Phytophthora parasitica nicotianae interaction. Plant Physiol. 112, 997–1004. doi: 10.1104/pp.112.3.997
Villela-Dias, C., Camillo, L. R., de Oliveira, G. A. P., Sena, J. A. L., Santiago, A. S., de Sousa, S. T. P., et al. (2014). Nep1-like protein from Moniliophthora perniciosa induces a rapid proteome and metabolome reprogramming in cells of Nicotiana benthamiana. Physiol. Plant. 150, 1–17. doi: 10.1111/ppl.12061
Xu, Y.-H., Liu, R., Yan, L., Liu, Z.-Q., Jiang, S.-C., Shen, Y.-Y., et al. (2012). Light-harvesting chlorophyll a/b-binding proteins are required for stomatal response to abscisic acid in Arabidopsis. J. Exp. Bot. 63, 1095–1106. doi: 10.1093/jxb/err315
Yamauchi, Y., Hasegawa, A., Mizutani, M., and Sugimoto, Y. (2012). Chloroplastic NADPH-dependent alkenal/one oxidoreductase contributes to the detoxification of reactive carbonyls produced under oxidative stress. FEBS Lett. 586, 1208–1213. doi: 10.1016/j.febslet.2012.03.013
Yamauchi, Y., Hasegawa, A., Taninaka, A., Mizutani, M., and Sugimoto, Y. (2011). NADPH-dependent reductases involved in the detoxification of reactive carbonyls in plants. J. Biol. Chem. 286, 6999–7009. doi: 10.1074/jbc.M110.202226
Zhang, C., Liu, L., Wang, X., Vossen, J., Li, G., Li, T., et al. (2014). The Ph-3 gene from Solanum pimpinellifolium encodes CC-NBS-LRR protein conferring resistance to Phytophthora infestans. Theor. Appl. Genet. 127, 1353–1364. doi: 10.1007/s00122-014-2303-1
Zhang, D., and Motilal, L. (2016). “Origin, dispersal, and current global distribution of cacao genetic diversity,” in Cacao Diseases (Cham: Springer International Publishing), 3–31. doi: 10.1007/978-3-319-24789-2_1
Keywords: black-pod disease, Oomycete, genetics, phytopathology, plant improvement, plant–pathogen interaction, proteomics, systems biology
Citation: Rego APB, Mora-Ocampo IY, Pirovani CP, Luz EDMN and Corrêa RX (2022) Protein Level Defense Responses of Theobroma cacao Interaction With Phytophthora palmivora. Front. Agron. 4:836360. doi: 10.3389/fagro.2022.836360
Received: 15 December 2021; Accepted: 18 January 2022;
Published: 10 February 2022.
Edited by:
Yuxuan Hou, China National Rice Research Institute, Chinese Academy of Agricultural Sciences (CAAS), ChinaCopyright © 2022 Rego, Mora-Ocampo, Pirovani, Luz and Corrêa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ronan Xavier Corrêa, cm9uYW54Y0B1ZXNjLmJy
†These authors share senior authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.