- 1Department of Crop Sciences, Tshwane University of Technology, Pretoria, South Africa
- 2Department of Crop Science, University for Development Studies, Tamale, Ghana
- 3Chemistry Department, Tshwane University of Technology, Pretoria, South Africa
To evaluate cowpea genotypic differences in response to inoculation with Bradyrhizobium sp. strains BR 3267 and CB756 in the field, 15 genotypes were assessed for N2 fixation using 15N natural abundance and assays of xylem N solutes (ureides, nitrates, and amino-N). Carbon isotope discrimination (δ13C) was used as a surrogate for water use efficiency and grain yield determined at maturity. Within each location, the tested cowpeas elicited contrasting responses to inoculation with either Bradyrhizobium strains. For example, inoculating Apagbaala and IT90K-277-2 with strain BR 3267 doubled N-fixed and grain yield at Nyankpala when compared to inoculation of the same genotypes with strain CB756, whereas the inoculation of Padituya with strain CB756 increased those same parameters relative to non-inoculation. Similar contrasting results were also obtained at Savelugu and Gbalahi, where bacterial inoculation with each strain resulted in up to a five-fold increase in N-fixed and a four-fold increase in grain yield depending on the cowpea/inoculant combinations used and the planting location. The main effect of genotype showed that the percent N derived from N2 fixation (%Ndfa) by the test cowpeas ranged between 74.4 and 89.8%, 11.7 and 50.5%, and 71.9 and 90.3% at Nyankpala, Savelugu, and Gbalahi, respectively. The genotypes grown at Savelugu generally exhibited low %Ndfa which was augmented by greater soil N uptake leading to grain yield increases, a finding supported by significant positive correlations when N-fixed and N content were each plotted against grain yield within locations. The inverse relationship between symbiosis and soil N uptake was also supported by negative correlations between xylem nitrate concentration and relative ureide N abundance (RU-N), and between xylem nitrate concentration and %Ndfa measured from 15N natural abundance. Some high yielding genotypes also elicited greater water use efficiency (δ13C), a trait that could be exploited for increased cowpea production in water limiting environments. This study demonstrates the presence of genotypic differences in cowpea response to inoculation in the field and contributes to the literature regarding the factors influencing legume inoculation response in the tropics.
Introduction
Grain legumes are highly valued because of their nutritional and health benefits in addition to their contribution to soil fertility through symbiotic relationships with diverse rhizobia in the soil (Foyer et al., 2016). Moreover, their inclusion in cropping systems reduces chemical fertilizer usage and greenhouse gas emissions, both of which contribute to global warming (De Ron et al., 2017). In Africa, cowpea (Vigna unguiculata L. Walp.) is widely cultivated for its edible leaves and grain (Ehlers and Hall, 1997; Xiong et al., 2016) which are rich in protein (23.9–34.1%), carbohydrate (64%), and essential S-containing amino acids, such as lysine, which is often lacking in cereals (Iqbal et al., 2006; Hall, 2012; Santos et al., 2012; Ddamulira and Santos, 2015; Shevkani et al., 2015). Furthermore, cowpea has considerable tolerance to drought, a trait that makes it a preferred crop in low rainfall areas and an integral component of low-input farming systems where its N2-fixing activities supply N to subsequent non-legumes planted in rotation (Dakora and Keya, 1997; Peoples et al., 2009). Because of the low C/N ratio of legumes like cowpea, their residues have faster rates of decomposition, which makes them important in conservation agriculture where they also help in C sequestration and improved soil microbial activities (Guopeng et al., 2020).
Despite the nutritional and agronomic importance of cowpea in Africa, average grain yield of the crop is low (160–810 kg.ha−1) across the continent due to poor soil fertility, pests and diseases, and the cultivation of low yielding landraces by farmers (Tittonell and Giller, 2013; De Ron et al., 2017). For example, average cowpea yields of 171, 220, 362, and 417 kg.ha−1 have been reported in major producing countries like Niger, Mali, Senegal and Nigeria, respectively (Hall, 2012). Nevertheless, a greater proportion of the world's cowpea comes from Africa, where Nigeria and Niger together produced 67% of the global output in 2019 (FAO, 2021). In Ghana, up to 2,440 kg.ha−1 of grain yield is attainable with recommended agronomic practices. Yet, the yield gap is further widened due to much lower yields recorded by most farmers compared to the potential yield of 3,500 kg.ha−1 (MoFA, 2010, 2016). Despite the low yields however, cowpea production in Ghana is still more than its consumption, suggesting that the country could increase its net export of the crop by bridging the yield gap through improved agronomic practices and selection for drought tolerance and increased symbiosis in low N soils (MoFA, 2010; Belane and Dakora, 2011; Boddey et al., 2017).
Several studies with different cowpea genotypes in the field have reported up 96% symbiotic dependence, suggesting that the N2-fixing ability of the crop could be tapped for its increased production in the N limited soils that characterize most parts of Africa (Belane and Dakora, 2009, 2010; Belane et al., 2011). When planted in soils that contain large numbers of ineffective rhizobia, however, legumes such as cowpea are often inoculated with superior N2-fixing rhizobial strains to optimize the symbiosis (Ulzen et al., 2016). Nevertheless, inoculation failures often occur when the introduced strain has poor competitive ability for nodule occupancy compared to the already adapted native rhizobial population (Catroux et al., 2001; Herridge et al., 2002). Aside the adverse effects of environmental factors, negative inoculation response in the fields can also result from poor quality of inoculants, suggesting the need to address existing lapses in quality control (Herrmann and Lesueur, 2013). To enhance inoculation success in the field, seed inoculation is often adopted to promote the colonization of nodules by the introduced strains for improved plant growth (Lopes et al., 2021). Moreover, in the formulation of inoculants, the choice of a suitable carrier can enhance rhizobial survival by protecting them against adverse soil factors (Herrmann and Lesueur, 2013). Although previous studies have shown the presence of effective cowpea rhizobia in African soils (Hall, 2004), recent studies have observed inoculation success in Ghana, Mozambique, Tanzania, and Brazil that led to effective nodulation and significant grain yield increases (Martins et al., 2003; Nyoki and Ndakidemi, 2014; Ulzen et al., 2016; Boddey et al., 2017; Kyei-Boahen et al., 2017). However, despite the widespread cultivation of diverse cowpea germplasm in Africa, the few reports regarding the inoculation response of the crop are based on a limited number of genotypes (Ulzen et al., 2016; Boddey et al., 2017; Kyei-Boahen et al., 2017). Furthermore, most breeder-released cowpea varieties, breeding lines, and landraces have not been adequately characterized in in terms of their symbiotic performance, especially in relation to Bradyrhizobium inoculation response, an aspect much needed for varietal choice, recommendation, and breeding purposes aimed at improving the cowpea-rhizobia symbiosis. An evaluation of possible cowpea genotypic differences in inoculation-response in the field is therefore required to guide the selection of cowpea/rhizobia combinations that optimize symbiosis for cheaper and sustainable farming systems (Ulzen et al., 2016).
As with legumes in the tribe Phaseoli, cowpea exports its fixed N products in root nodules to the aerial parts as ureides (allantoin and allantoic acid) through the xylem stream (Baral et al., 2016). Therefore, the concentration of ureides relative to other xylem N solutes (i.e., nitrates and amino acids) is often used as a measure of the N sources being assimilated by the legume (Herridge et al., 1990). Furthermore, the 15N natural abundance method has been widely used to quantify N2 fixation by grain legumes, including cowpea (Belane and Dakora, 2010, 2011), soybean (Mapope and Dakora, 2016), and Bambara groundnut (Mohale et al., 2014), under field conditions. The joint application of such different techniques is expected to yield a more reliable assessment of legume N2 fixation.
Although cowpea is widely reported for its drought tolerance via several mechanisms, which include stomatal closure and drought escape (Itani et al., 1992), not much information is documented on the water relations of most cultivated varieties in Ghana and elsewhere. With climate change however, information on drought tolerance is needed to select climate-smart cowpea genotypes to bridge the existing yield gap of the crop (Vadez et al., 2012). Carbon isotope discrimination in C3 plants during photosynthesis is known to correlate with water use efficiency, and therefore allows for the use of the former as a reliable surrogate for assessing plant water use efficiency (Farquhar et al., 1989). Plants subjected to water limiting conditions show higher water use efficiency via reduced discrimination against 13CO2, leading to higher δ13C values. Conversely, under adequate water supply, plants exhibit lower δ13C values due to increased discrimination against 13CO2 (Cernusak et al., 2013). Consequently, carbon isotope discrimination has been successfully used to estimate plant water relations in several grain legumes under field conditions (Mohale et al., 2014; Mokgehle et al., 2014; Mapope and Dakora, 2016).
This study evaluated 15 cowpea genotypes, including breeding lines and a local landrace, for genotypic differences in response to two Bradyrhizobium inoculants at three locations in the Northern region of Ghana in relation to symbiotic performance and grain yield. The 15N natural abundance technique and colorimetric assays of xylem N solutes were used to assess N2 fixation while the 13C isotopic technique was used as a surrogate for plant water use efficiency.
Materials and Methods
Experimental Site Description
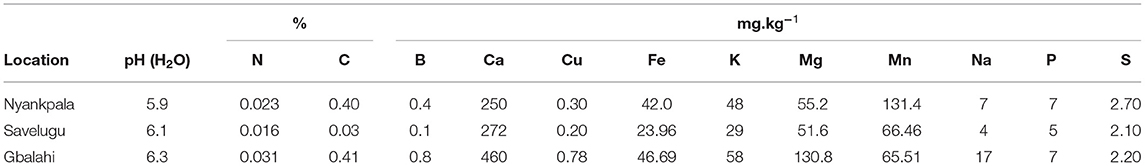
The experiments were conducted at three locations (Nyankpala, Savelugu and Gbalahi) in the Northern region of Ghana. Nyankpala is located on latitude 9.404 and longitude −0.982, while Gbalahi is located on latitude 9.437 and longitude −0.734. Savelugu is located on latitude 9.569 and longitude −0.830. The field at Nyankpala had been fallowed for over 5 years while the Savelugu and Gbalahi locations were respectively planted with okra and maize in the year prior to this experiment. Prior to experimental layout and planting, soil samples were cored from several points in each field, bulked, and sub-sampled for analysis of pH (H2O), %C (Walkley and Black, 1934), P (Bray and Kurtz, 1945), B, Ca, Cu, Fe, K, Mg, Mn, Na, S (ammonium acetate method), and total N (using N digest).
The Northern Region has a unimodal rainfall of 800–1,200 mm which is distributed between April and October each year (Owusu and Waylen, 2013). The total annual rainfall during the study year (in 2013) was 1081.5 and 1009.7 mm at Nyankpala and Savelugu, respectively. Rainfall data at Gbalahi (857.3 mm) was collected from January to October 2013. The total rainfall from planting until plant sampling (early podding stage of cowpea) was 481 mm at Nyankpala, 471 mm at Savelugu, and 305 mm at Gbalahi. The average weekly distribution of rainfall at the three locations are illustrated in Supplementary Figure 1.
Source of Seeds, Experimental Design, and Planting
The 15 cowpea genotypes used in this study included breeder-released varieties (Apagbaala, Asetenapa, Bawutawuta, Marfotuya, Nhyira, Padituya, Songotra, Soronko, and Zayura), breeding lines (SARVx-09-001, SARVx-09-002, SARVx-09-003, and SARVx-09-004, IT90K-277-2), and a local landrace (Omandaw) sourced from CSIR-Savanna Agricultural Research Institute (CSIR-SARI) in Tamale and the Crops Research Institute (CRI) in Kumasi, both in Ghana. The local landrace is adapted to the savanna agroecology of Ghana where it is widely cultivated by rural farmers. The study was a factorial experiment involving the 15 cowpea genotypes together with three levels of inoculation (Bradyrhizobium sp. BR 3267, Bradyrhizobium sp. CB756 and control/non-inoculation). Thus, there were a total of 45 treatment combinations, which were laid in a randomized complete block design with four replications at each location. Planting was done on July 2, 2013 in Nyankpala, July 10, 2013 in Savelugu, and on July 16, 2013 in Gbalahi. Seeds were sown in plots measuring 2.4 × 2 m at a spacing of 60 cm between rows and 20 cm between plants, with 1 m path between plots and 1.5 m between blocks. Weeding was done using a hand hoe.
Inoculation of Seeds With Bradyrhizobium Strains
Seed inoculation was done in the shade using 10 g peat-based inoculant per kg seed which yielded 108 rhizobial cells.g−1 of inoculant. The seeds were coated with stimulin (a gel sticker) to aid attachment of the inoculant to the seed. To avoid cross contamination, uninoculated/control plots were planted first, followed by inoculated treatments.
Sampling and Processing of Plants
Plants were sampled at early podding at 54 days after planting (DAP) in Nyankpala and Savelugu, and at 55 DAP in Gbalahi. For this, five plants were randomly dug up from each plot in all experiments and separated into shoots, roots, and nodules. The number of nodules per plant was counted and recorded. The shoots and nodules were separately oven-dried (65°C for 48 h) and weighed to determine shoot dry matter and nodule dry matter, respectively. The shoots were then finely ground (0.50 mm sieve) for 15N and 13C isotopic analysis. Shoots of non-legume reference plants were concurrently sampled in each location and similarly processed for 15N isotopic analysis.
15N/14N Isotopic Analysis on Plant Shoots
Weighed ground shoot samples (2.0–2.5 mg) of both legume and reference plants were prepared, and each sample fed into a Carlo Erba NA1500 elemental analyzer (Fisons Instruments SpA, Strada, Rivoltana, Italy) coupled to a Finnigan MAT252 mass spectrometer (Fisons Instrument SpA, Strada, Rivoltana, Italy) via conflo II open-split device to measure 15N/14N isotopic composition, as described by Mapope and Dakora (2016). The 15N natural abundance of the test plants were expressed as δ notation and is the per mille deviation of the 15N natural abundance of the sample from that of atmospheric N2 (0.36637 atom % 15N). An internal standard of Nasturtium spp. was included after every five runs. The δ15N of shoot samples was calculated as (Unkovich et al., 2008):
where the 15N/14Nsample is the abundance ratio of 15N and 14N in the sample and 15N/14Natm is the abundance ratio of 15N and 14N in the atmosphere.
Shoot N
The %N of shoot samples were obtained directly from the mass spectrometer, and shoot N content was measured as the product of shoot dry matter and %N (Pausch et al., 1996).
Percent N Derived From Fixation (%Ndfa) and N-Fixed
Shoot %Ndfa of cowpea was calculated as (Shearer and Kohl, 1986):
where δ15Nref is the 15N natural abundance of reference plant, δ15Nleg is the 15N natural abundance of legume plant, and the B value is the 15N natural abundance of the shoot of legume completely dependent on N2 fixation for its N nutrition. The B value (−1.95‰) used in this study was earlier determined by inoculating cowpea genotype Padituya with Bradyrhizobium sp. TUTVUSA28 which was isolated from cowpea nodules in South Africa (Mohammed et al. unpublished data). The mean δ15N of all reference plants at each location was determined and used to calculate the %Ndfa of cowpea genotypes per location.
The N-fixed by cowpea was calculated as (Maskey et al., 2001):
The soil N uptake by legume plants was calculated as the shoot N content less N-fixed in shoot.
Shoot 13C/12C Isotopic Analysis
The ground shoot samples of cowpea were similarly subjected to 13C/12C isotopic analysis, as described for 15N/14N. The 13C natural abundance (δ13C) was calculated as (Farquhar et al., 1989):
where 13C/12Csample is the isotopic ratio of the sample and 13C/12Cstandard is the isotopic ratio of Pee Dee Belemnite (PDB), a universally accepted standard from Belemnite Pee Dee limestone formation (Craig, 1957).
Collection of Xylem Sap
Xylem sap was collected from the same plants that were sampled and processed for 15N and 13C isotopic analysis. Five plants from each plot were decapitated at the crown level, and root-bleeding xylem sap were collected into 2 ml Eppendorf tubes using clean Pasteur glass pipettes. The xylem samples were stored at −20°C until analysis for N solutes (ureide-N, nitrate-N, and amino-N).
Measurement of Xylem N Solutes
Ureide-N in Xylem Sap
The concentration of ureides in xylem sap was colorimetrically determined, as described by Young and Conway (1942) or Unkovich et al. (2008). Varying concentrations of allantoin (0.1, 0.02, 0.04, 0.1, and 0.15 mM) were prepared as standards for estimating ureides in xylem sap, after colorimetric assay (Young and Conway, 1942).
Nitrate-N in Xylem Sap
Nitrate-N in xylem sap was determined using the salicylic acid method (Cataldo et al., 1975), as described by Unkovich et al. (2008). Again, varying concentrations of nitrate (0, 1.25, 2.5, 5, 10, and 15 mM) were used as standards to construct curves for extrapolating nitrate values from xylem sap.
Amino-N Determination
Amino-N in xylem sap was measured using the ninhydrin method (Yemm and Cocking, 1955; Unkovich et al., 2008). As with ureide and nitrate-N determinations, a standard curve was prepared from assaying different concentrations of amino acid (0, 0.1, 0.2, 0.4, and 1 mM).
Relative Ureide Nitrogen (RU-N)
The relative ureide-N in xylem sap or relative ureide abundance was calculated as (Herridge and Peoples, 1990):
Where a, b, and c are the molar concentrations of ureides (ureides contain four nitrogen atoms per molecule), nitrate-N, and amino-N, respectively (Herridge et al., 1990).
Determination of Grain Yield
At maturity, 20 plants were harvested from each plot for determination of grain yield. The pods were detached from plants, air-dried to 15% moisture content, and threshed to obtain seeds. The seeds were weighed, and grain yield was expressed per hectare based on plant population.
Statistical Analysis
All data gathered were tested for normality and then subjected to ANOVA using Statistica data analysis software, version 10.0 (Statsoft Inc, 2011). A two-way ANOVA was used to compare the means of genotype and inoculation within each location while a three-way ANOVA was used to compare the performance of genotypes between locations. Where treatment means showed significant differences, Duncan's Multiple Range Test (DMRT) was used to separate them at p ≤ 0.05. Correlation and regression analyses were used to assess functional relationship between measured parameters.
Results
Soil Chemical Properties
Pre-planting soil samples from Nyankpala, Savelugu, and Gbalahi recorded pH (H2O) of 5.9, 6.1, and 6.3, respectively (Table 1). The concentrations of various nutrient elements in the soils at the three test locations are presented in Table 1. Both soil N and organic carbon contents were much lower in the soils at Savelugu when compared to values recorded at Nyankpala and Gbalahi.
Plant Growth, Symbiotic Parameters, δ13C, and Grain Yield of Cowpea
Main Effect of Cowpea Genotype
A two-way ANOVA of data within locations revealed significant effect (p ≤ 0.05) of genotype on nodule dry matter, shoot biomass, δ15N, %Ndfa, N-fixed, soil N uptake, C/N ratio, δ13C, and grain yield of cowpea in the three test locations (Tables 2–4).
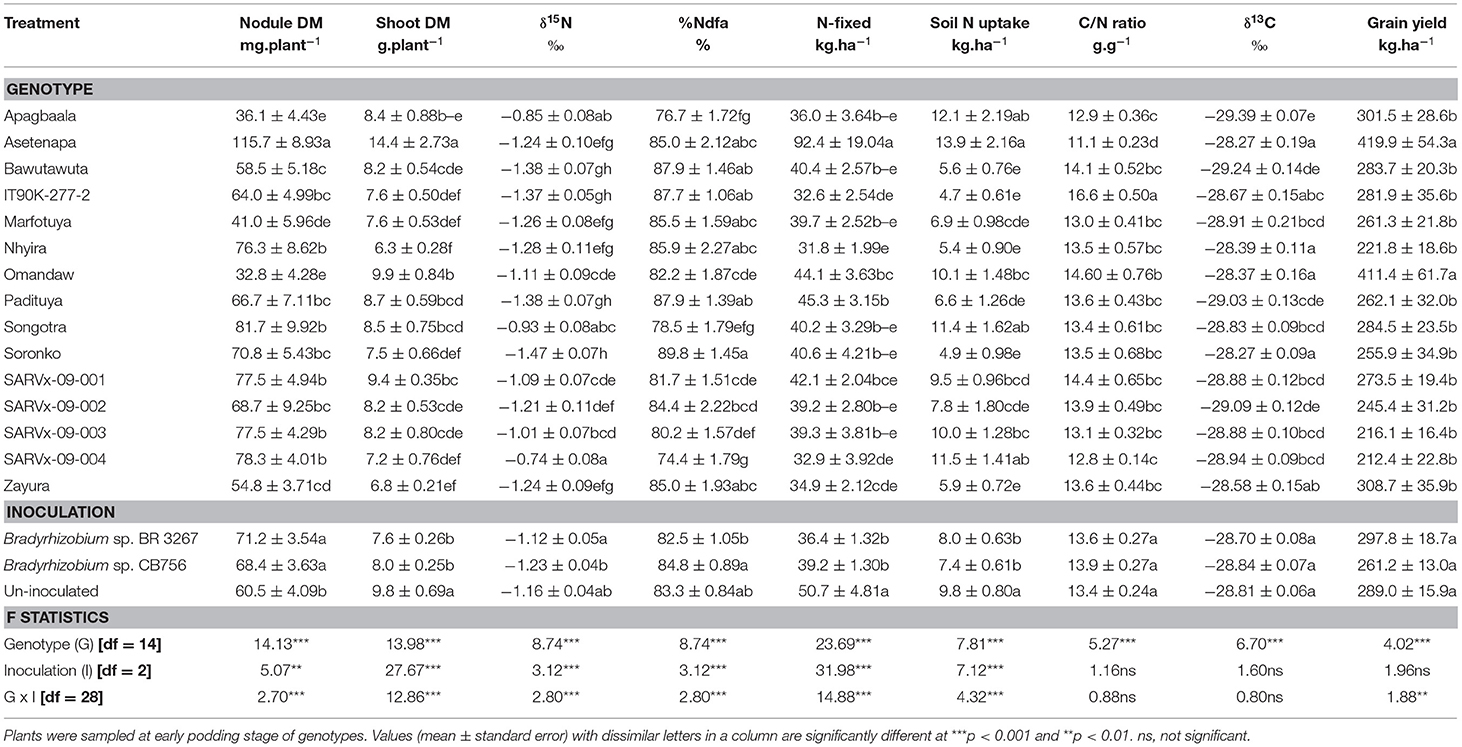
Table 2. Nodulation, shoot dry matter (DM), symbiotic parameters, soil N uptake, δ13C, and grain yield of 15 cowpea genotypes inoculated with two Bradyrhizobium strains and planted in Nyankpala in 2013.
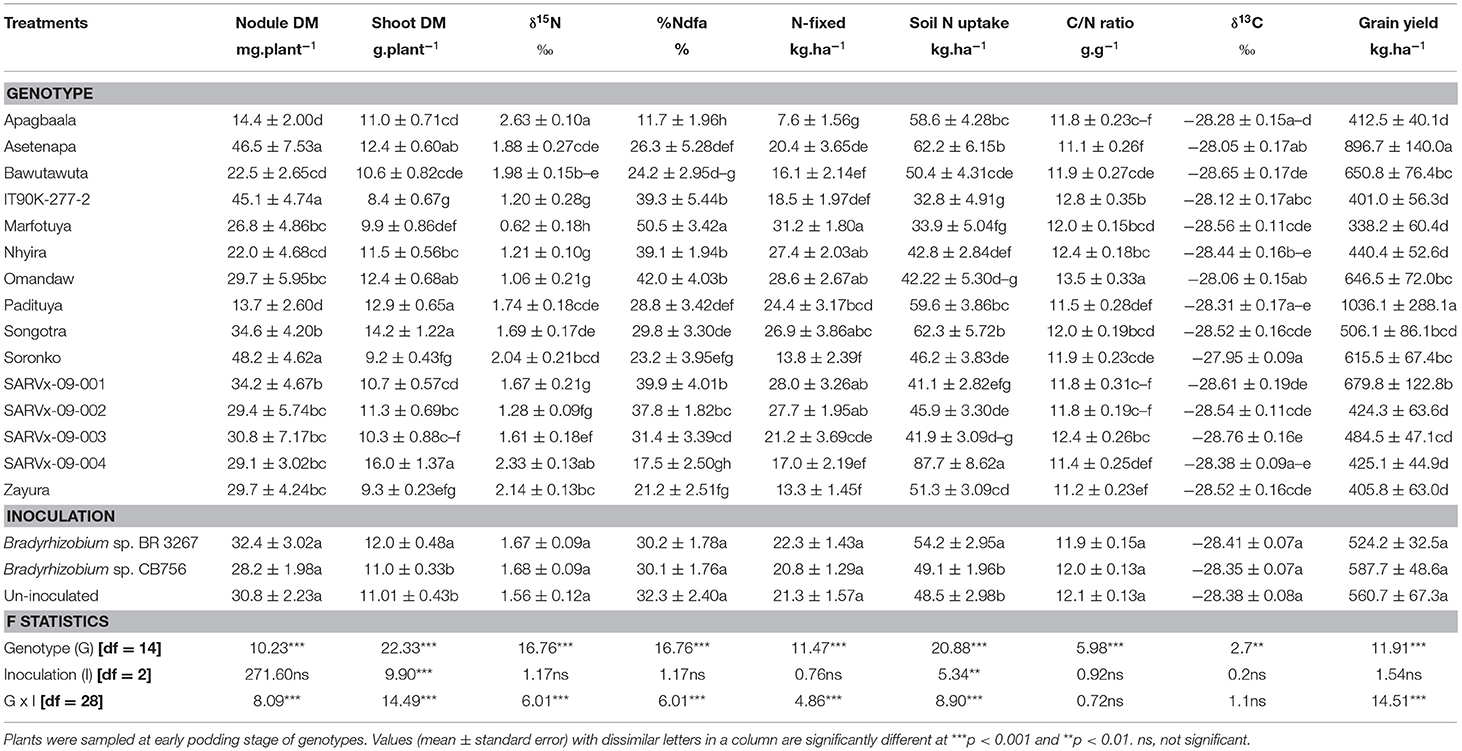
Table 3. Nodulation, shoot dry matter (DM), symbiotic parameters, soil N uptake, δ13C, and grain yield of 15 cowpea genotypes inoculated with two Bradyrhizobium strains and planted in Savelugu in 2013.
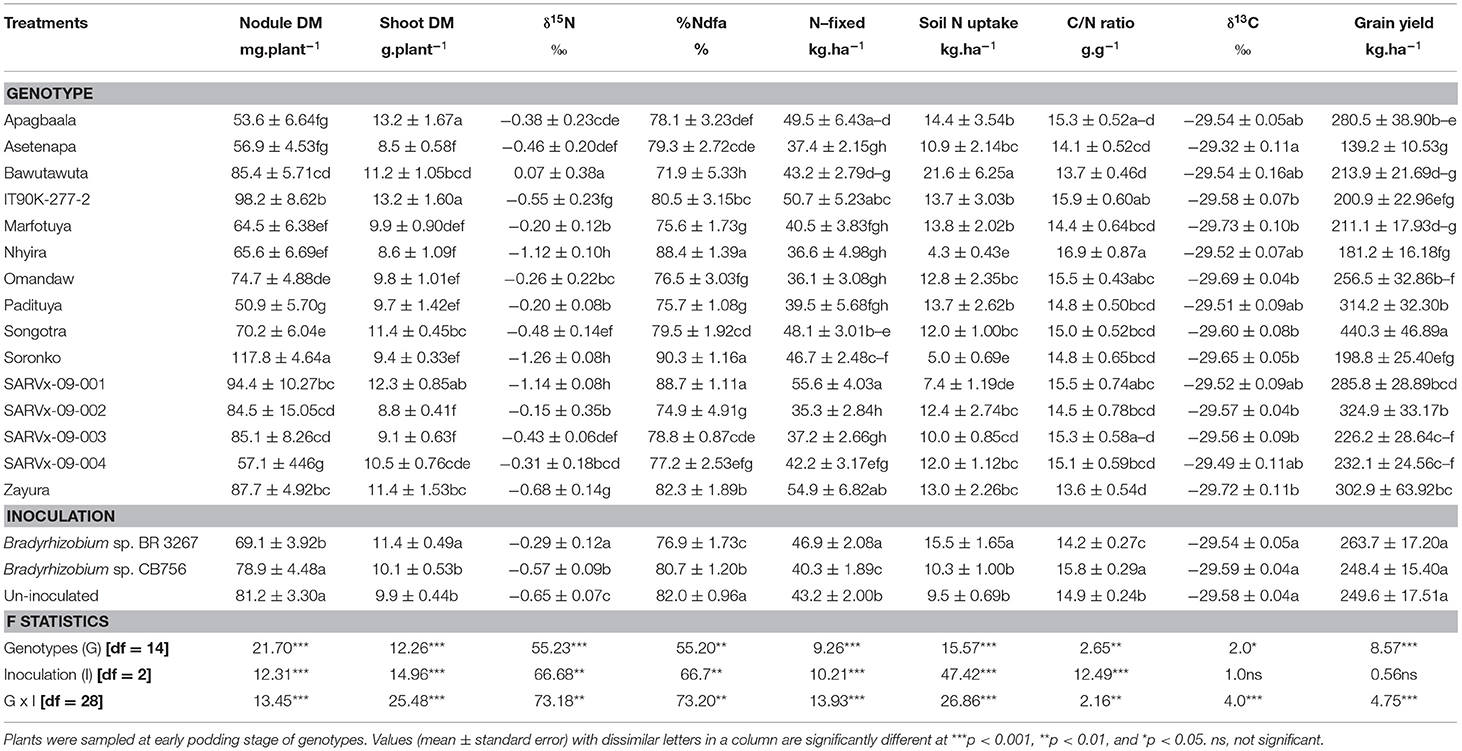
Table 4. Nodulation, shoot dry matter (DM), symbiotic parameters, soil N uptake, δ13C, and grain yield of 15 cowpea genotypes inoculated with two Bradyrhizobium strains and planted in Gbalahi in 2013.
From the analysis of main effects in Nyankpala, genotype Asetenapa recorded the highest nodule mass (115.7 mg.plant−1), followed by genotypes Songotra, Nhyira, SARVx-09-001, SARVx-09-003, and SARVx-09-004 which had relatively lower but similar nodule mass ranging between 76.33 and 81.67 mg.plant−1 (Table 2). The least nodule mass in Nyankpala was recorded in genotypes Apagbaala, Omandaw, and Marfotuya (Table 2).
The genotype Asetenapa, which produced the highest nodule mass, also exhibited the highest shoot biomass, followed by Omandaw and SARVx-09-0001 (Table 2). The least shoot biomass in Nyankpala was produced by genotypes Nhyira and Zayura and (Table 2).
Genotype Soronko recorded the least δ15N in Nyankpala followed by Asetenapa, Bawutawuta, IT90K-277-2, Marfotuya, Nhyira, Padituya, and Zayura, also with lower δ15N values when compared to the other genotypes at the location (Table 2). The genotype SARVx-09-004, Apagbaala, and Songotra had the highest δ15N values in Nyankpala. As expected, %Ndfa was greater in the genotypes which had lower δ15N, with values ranging between 85.02% in genotypes Asetenapa and Zayura and 89.78% in Soronko. Due to their greater δ15N values, genotypes SARVx-09-0004, Apagbaala, and Songotra recorded relatively lower %Ndfa (74.4, 76.76, and 78.50%, respectively) in Nyankpala (Table 2).
The amount of N-fixed was highest in genotype Asetenapa (92.42 kg.ha−1) followed by Padituya (45.26 kg.ha−1) and Omandaw (44.05 kg.ha−1). Despite their greater %Ndfa however, genotypes IT90K-277-2 and Nhyira recorded lower N-fixed due to their reduced shoot biomass, whilst genotype SARVx-09-004 coupled reduced shoot biomass with lower %Ndfa and N-fixed (Table 2). The genotype Asetenapa, which recorded the highest N-fixed at Nyankpala, also recorded the highest soil N uptake at the location, whilst Apagbaala, Songotra, and SARVx-09-004 recorded relatively lower N-fixed but greater soil N uptake (Table 2). Soil N uptake was least in genotypes Bawutawuta, IT90K-277-2, Nhyira, Soronko, and Zayura, all of which had high %Ndfa. Despite the high %Ndfa by IT90K-277-2 however, the genotype together with Omandaw recorded high C/N ratio at Nyankpala due to their reduced %N (Table 6) when compared to other genotypes at the location. The least C/N ratio was recorded in genotype Asetenapa followed by Apagbaala and SARVx-09-004 (Table 2).
Further, the genotypes Asetenapa, Nhyira, Omandaw, Soronko, and Zayura recorded higher shoot δ13C values in Nyankpala than the other cowpea genotypes (Table 2). In contrast, genotype Apagbaala, together with Bawutawuta and SARVx-09-002, elicited lower δ13C values when compared to the other genotypes in Nyankpala (Table 2). Despite the observed high %Ndfa by genotypes in Nyankpala, their grain yield was generally low, with the highest values being recorded in genotypes Asetenapa (419.9 kg.ha−1) and Omandaw (411.4 kg.ha−1) which also exhibited greater δ13C values. Grain yield was, however, lower but similar between and among the remaining cowpea genotypes, with values ranging between 212.4 and 308.7 kg.ha−1 (Table 2).
In the Savelugu location, there was a significant effect (p ≤ .05) of cowpea genotype on nodulation, shoot biomass, symbiotic parameters, soil N uptake, δ13C, and grain yield (Table 3). Here, the genotypes Asetenapa, IT90K-277-2, and Soronko, together with Padituya and SARVx-09-001, recorded higher nodule mass when compared to the other genotypes. The least nodule mass at Savelugu was recorded in genotypes Apagbaala and Padituya, followed by Bawutawuta and Nhyira (Table 3).
The genotypes Asetenapa and Songotra, which had high nodule mass and genotypes Omandaw and SARVx-09-004, recorded greater shoot biomass in Savelugu. However, Padituya had high shoot biomass despite reduced nodule biomass in the Savelugu location (Table 3). In contrast, genotypes IT90K-277-2, Soronko, and Zayura exhibited much lower shoot biomass in Savelugu despite their relatively high nodule biomass at the location (Table 3).
Further, shoot δ15N values were positive for all cowpea genotypes planted in Savelugu (Table 3). Here, Marfotuya recorded the least δ15N value (+0.62‰), with genotypes, such as IT90K-277-2, Nhyira, Omandaw, SARVx-09-001, and SARVx-09-002, also exhibiting much lower δ15N values (+1.06 to +1.67‰) when compared to other genotypes at the location (Table 3). As a result, %Ndfa was highest in Marfotuya (50.5%), followed by the other genotypes which had lower δ15N values in that order, with values ranging from 37.8 to 42.0% (Table 4). Because of their high δ15N values, %Ndfa was much lower in genotypes Apagbaala (11.7%), SARVx-09-004 (17.5%), Soronko (23.2%), and Zayura (21.2%) (Table 3).
The highest N-fixed was recorded in genotype Marfotuya (31.18 kg.ha−1) followed by Nhyira, Omandaw, SARVx-09-001, SARVx-09-002, and then Songotra with values ranging between 26.91 and 28.60 kg.ha−1 (Table 3). Genotype Apagbaala, together with Zayura, SARVx-09-004, Soronko, and Bawutawuta, recorded markedly lower N-fixed values (7.58–17.02 kg.ha−1) (Table 3). Soil N uptake was highest in genotype SARVx-09-004 due to its higher δ15N and lower %Ndfa values. Other genotypes with high soil N uptake in Savelugu include Asetenapa and Songotra. However, soil N uptake was lower in genotypes Bawutawuta, IT90K-277-2, and SARVx-09-001 which all recorded relatively high %Ndfa in Savelugu (Table 3). Shoot C/N ratio was highest in genotype Omandaw (13.5 g.g−1) followed by IT90K-277-2 (12.8 g.g−1), and least in genotypes Asetenapa and Zayura. Other genotypes with low C/N ratio include Apagbaala, Padituya, SARVx-09-001, SARVx-09-002, and SARVx-09-004 with values ranging between 11.2 and 11.8 g.g−1 (Table 3).
At Savelugu, the cowpea genotype Soronko recorded the highest water use efficiency measured as shoot δ13C (−27.95‰), followed by Asetenapa and Omandaw all with high δ13C at Nyankpala (Table 3). The least δ13C was recorded in genotype SARVx-09-004 (−28.38‰) followed by Bawutawuta (28.65‰) and SARVx-09-001 (28.61‰). The highest grain yield in Savelugu (1036.1 kg.ha−1) was recorded in genotype Padituya followed by Asetenapa (896.7 kg.ha−1), Omandaw (646.5 kg.ha−1), and Soronko (615.5 kg.ha−1) which all recorded high δ13C at the location (Table 3). Cowpea genotype SARVx-09-004 also recorded relatively high grain yield (679.8 kg.ha−1) despite much lower δ13C value (Table 3). The least grain yield at Savelugu was recorded in genotypes Apagbaala, IT90K-277-2, Marfotuya, Nhyira, SARVx-09-002, SARVx-09-004, and Zayura with values between 338.4 and 440.4 kg.ha−1 (Table 3).
At Gbalahi, there was a marked effect of genotype on nodule mass, shoot biomass, symbiotic parameters, soil N uptake, δ13C, and grain yield of cowpea (Table 4). The genotype Soronko recorded the highest nodule mass followed by IT90K-277-2, SARVx-09-001, and Zayura, also, with high nodule dry matter (Table 4). The least nodule mass was recorded in genotypes SARVx-09-004 and Padituya. Other genotypes with lower nodule mass at Gbalahi include Apagbaala, Asetenapa, Marfotuya, and Nhyira (Table 4).
The genotypes Apagbaala and IT90K-277-2 recorded the highest shoot biomass at Gbalahi followed by SARVx-09-001 and Zayura also with high shoot biomass. The least shoot biomass was recorded in genotypes Asetenapa, Nhyira, SARVx-09-002, SARVx-09-003, and then followed by Omandaw and Padituya (Table 4).
Genotypes Nhyira, Soronko, and SARVx-09-001 recorded the lowest δ15N values followed by Zayura and IT90K-277-2 also with lower δ15N (Table 4). The highest δ15N value was recorded in genotype Bawutawuta followed by Marfotuya, Padituya, and SARVx-09-002 which also exhibited high δ15N values (Table 4). As a result, %Ndfa was highest in genotypes Nhyira (88.4%), Soronko (90.3%), and SARVx-09-001 (88.7%), followed by Zayura (82.3%) and IT90K-277-2 (80.5%). Genotype Bawutawuta recorded the least %Ndfa in Gbalahi, followed by Marfotuya, Padituya, and SARVx-09-002, all of which had high δ15N values (Table 4). The amount of N-fixed was highest in genotype SARVx-09-001, followed by Zayura and IT90K-277-2, while the least N-fixed was recorded in genotype SARVx-09-002 followed by Omandaw, Nhyira, Padituya, Marfotuya, and SARVx-09-003 also with low N-fixed values (Table 4). Genotype Bawutawuta, together with Apagbaala, IT90K-277-2, Marfotuya, and Padituya recorded greater soil N uptake while Nhyira, Soronko, and SARVx-09-001 recorded lower soil N uptake. Shoot C/N ratio ranged between 13.56 and 16.89 g.g−1 in Gbalahi, with genotypes Nhyira, IT90K-277-2, and Omandaw recording higher C/N ratio in the location while Bawutawuta and Zayura recorded the least values (Table 4).
Shoot δ13C ranged from −29.73‰ in genotype Marfotuya to −29.32‰ in Asetenapa at Gbalahi (Table 4). The highest grain yield (440.3 kg.ha−1) in Gbalahi was recorded in genotype Songotra followed by SARVx-09-002 (324.9 kg.ha−1) and Padituya (314.2 kg.ha−1), while Asetenapa recorded the least grain yield at the location (139.2 kg.ha−1). Other genotypes with lower grain yield at Gbalahi include Nhyira, Soronko, Bawutawuta, and Marfotuya (Table 4).
Main Effect of Inoculation
The main effect of inoculation was significant (p ≤ .05) for nodule dry matter, shoot dry matter, δ15N, %Ndfa, N-fixed, and soil N uptake at Nyankpala (Table 2). Although inoculation with either Bradyrhizobium strains increased dry nodule mass over non-inoculation at Nyankpala, shoot biomass was lowered when compared to uninoculated plots (Table 2). Together with non-inoculation, bacterial inoculation with strain CB756 decreased overall shoot δ15N, resulting in greater %Ndfa when compared to inoculation with strain BR 3267 (Table 2). As observed with shoot biomass, non-inoculation increased overall N-fixed and soil N uptake in Nyankpala when compared to plots inoculated with either Bradyrhizobium strains (Table 2). However, overall shoot C/N ratio, δ13C, and grain yield were unaltered by bradyrhizobial inoculation at Nyankpala (Table 2).
At Savelugu, the overall plant nodulation and other symbiotic parameters, along with δ13C and grain yield of cowpea, were unaltered by bradyrhizobial inoculation (Table 3). However, the main effect of inoculation was significant for shoot biomass and soil N uptake in Savelugu (Table 3). Overall, seed inoculation with Bradyrhizobium sp. strain BR 3267 increased shoot biomass and soil N uptake when compared to non-inoculation or inoculation with strain CB756 (Table 3).
At Gbalahi, the main effect of inoculation was significant for nodule mass, shoot biomass, all symbiotic parameters, and soil N uptake (Table 4). Here, bacterial inoculation with strain BR 3267 increased shoot biomass despite reduced nodule mass compared to non-inoculation or inoculation with strain CB756. However, δ15N was least with non-inoculation followed by inoculation with strain CB756. As a result, %Ndfa was highest with non-inoculation, followed by inoculation with strain CB756, and least with strain BR 3267 inoculation (Table 4). Bacterial inoculation with strain BR 3267 increased soil N uptake and decreased C/N ratio compared to other treatments. Here, overall δ13C and grain yield were unaltered by inoculation (Table 4).
Genotype × Inoculation Interaction Effect
There was a marked effect of genotype × inoculation interaction on nodule dry matter, shoot biomass, δ15N, N-fixed, soil N uptake, N content, and grain yield across the three test locations (Tables 2–4). However, results are described for shoot biomass, N-fixed, N content, and grain yield for cowpea genotypes that showed consistent patterns of interactions for those parameters within the test locations (Figures 1–3).
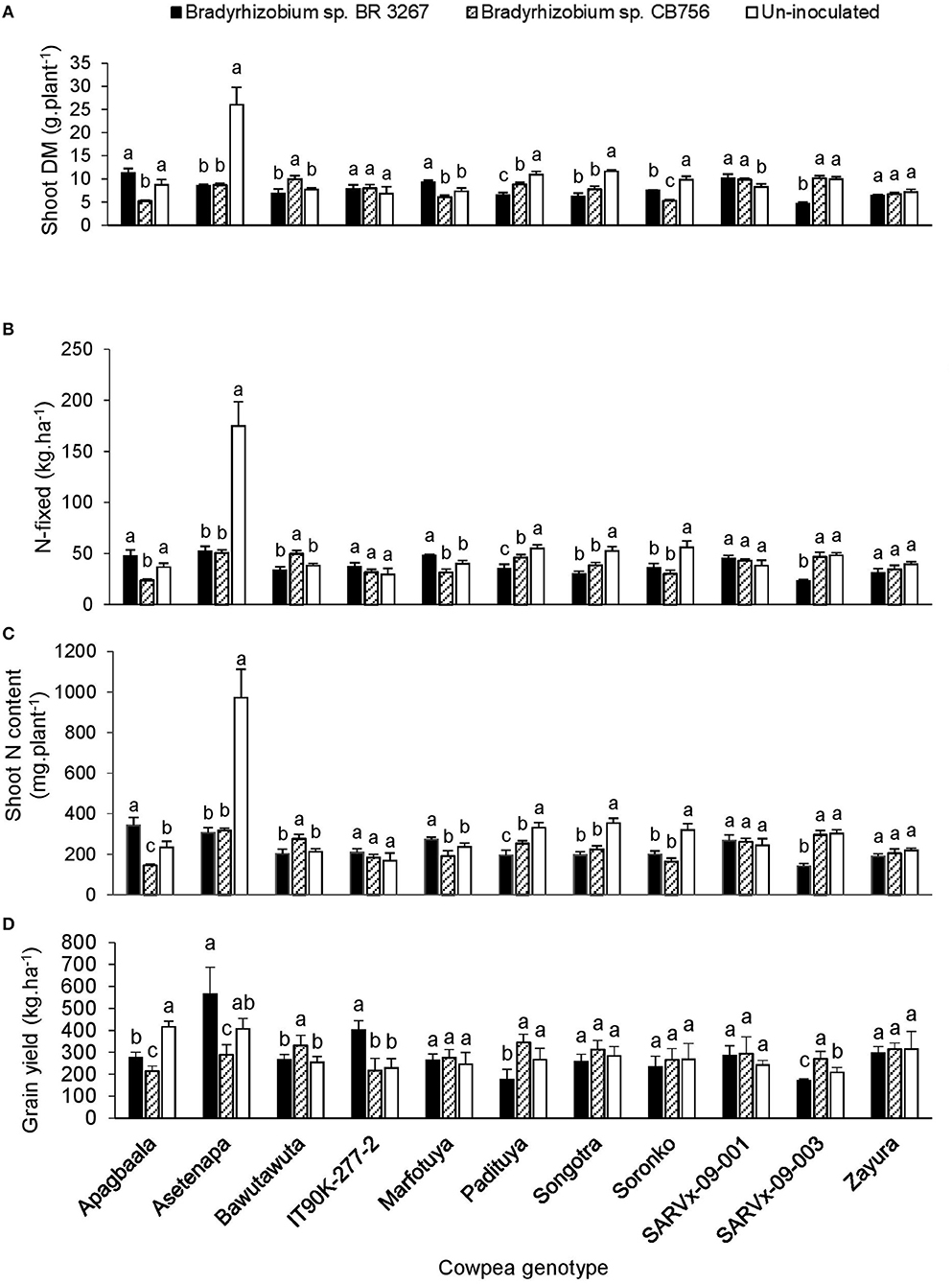
Figure 1. The interactive effect of genotype x inoculation on (A) shoot dry matter, (B) N-fixed, (C) N content, and (D) grain yield of 11 cowpea genotypes planted in Nyankpala in 2013. For each genotype, bars with dissimilar letters are significantly different at p ≤ 0.05. Error bars represent standard errors.
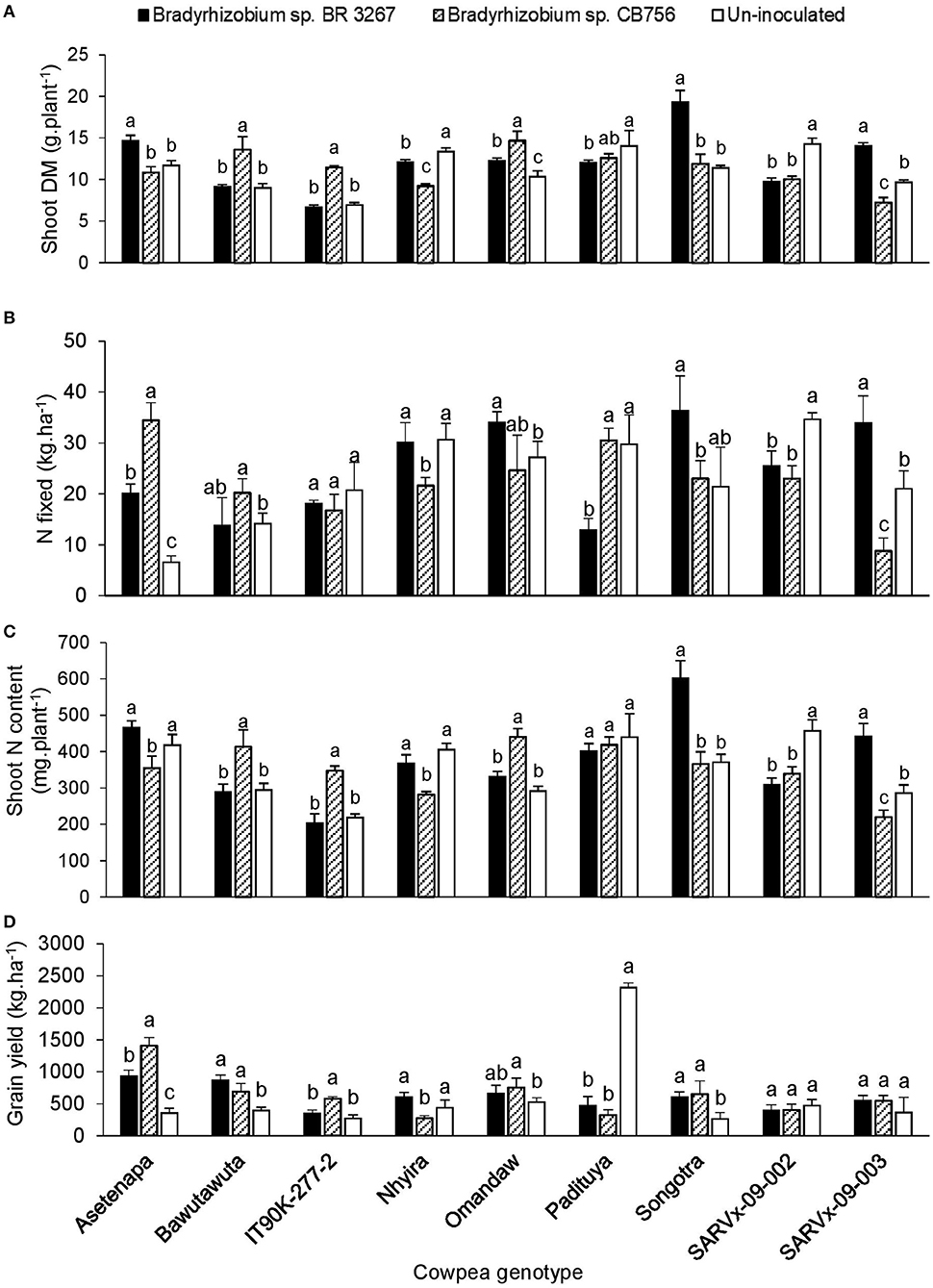
Figure 2. The interactive effect of genotype x inoculation on (A) shoot dry matter, (B) N-fixed, (C) N content, and (D) grain yield of 9 cowpea genotypes planted in Savelugu in 2013. For each genotype, bars with dissimilar letters are significantly different at p ≤ 0.05. Error bars represent standard errors.
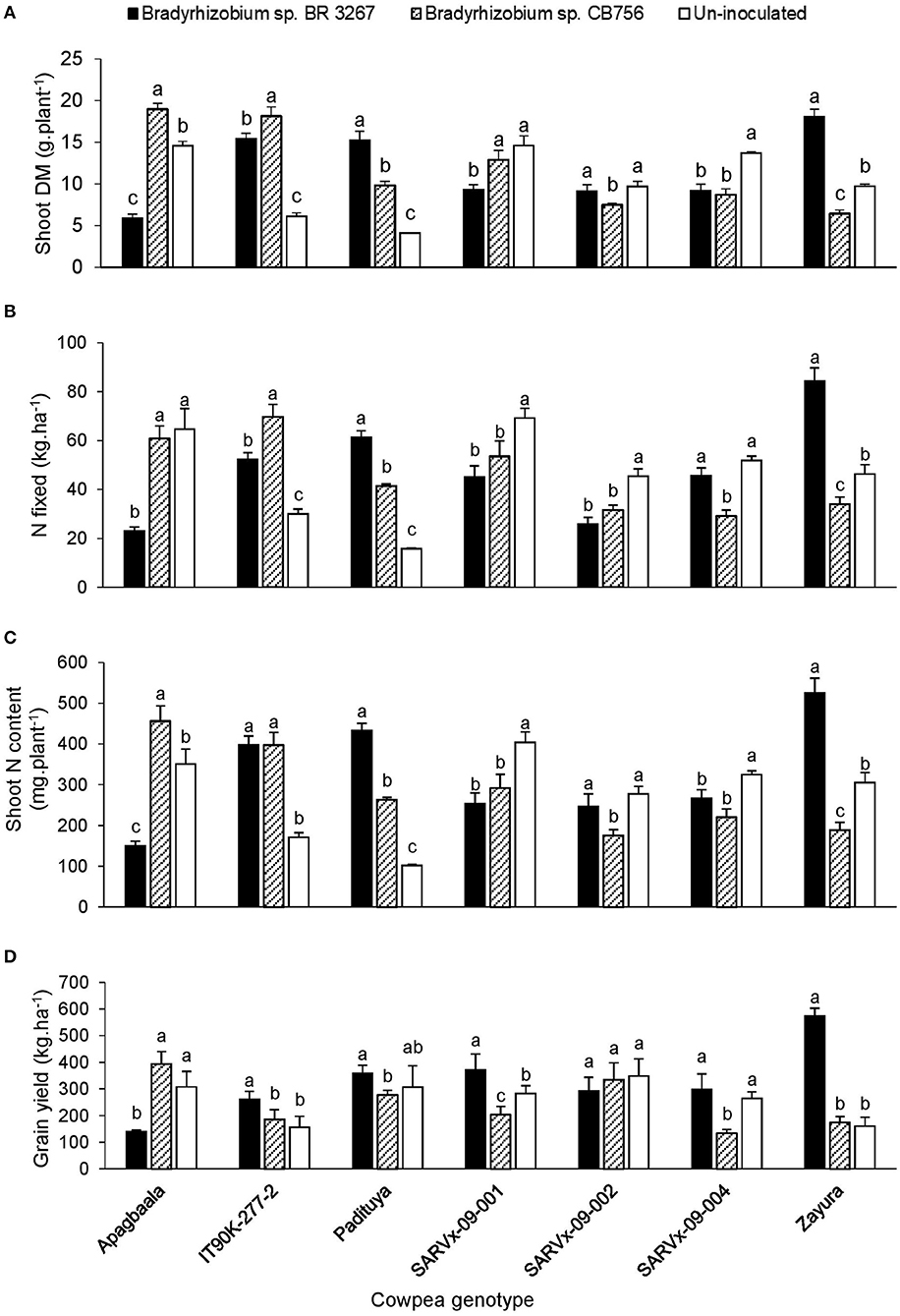
Figure 3. The interactive effect of genotype x inoculation on (A) shoot dry matter, (B) N-fixed, (C) N content, and (D) grain yield of 7 cowpea genotypes planted in Gbalahi in 2013. For each genotype, bars with dissimilar letters are significantly different at p ≤ 0.05. Error bars represent standard errors.
In Nyankpala, inoculating genotype Apagbaala with Bradyrhizobium sp. strain BR 3267 doubled N-fixed and slightly increased the parameter in IT90K-277-2 when compared to inoculation with strain CB756 (Figures 1A–D). On the other hand, bacterial inoculation of Bawutawuta, Padituya, and SARVx-09-003 with strain CB756 significantly increased shoot biomass, resulting in a 32–200% increase in the amounts of N-fixed and shoot N accumulation when compared to inoculation with strain BR 3267 (Figure 1). Consequently, grain yield was increased by 1.3- to 2-fold in those cowpea/inoculant strain combinations in Nyankpala (Figure 1). In contrast, non-inoculation of Apagbaala, Asetenapa, Padituya, and SARVx-09-003 in Nyankpala each increased N-fixed by 1.5- to 3.4-fold (36.5–55.0 kg.ha−1), and also resulted in greater shoot N accumulation. As a result, grain yield was increased by 120–194% in those control plants when compared to inoculation with strain CB756 in the case of Apagbaala and Asetenapa, and relative to strain BR 3267 in the case of Padituya and SARVx-09-003 (Figure 1). Furthermore, although non-inoculation of genotypes Songotra and Soronko resulted in greater shoot biomass, N-fixed, and N accumulation when compared to inoculation with either strain, the corresponding increases in grain yield were not significant (Figure 1). Similarly, bacterial inoculation of Soronko with strain BR 3267 only increased grain yield slightly despite significant increases in shoot biomass, N-fixed, and N accumulation (Figure 1).
At Savelugu, although N2 fixation was generally low in plants, bacterial inoculation of Asetenapa with either strain BR 3267 or CB756 resulted in a 3- to 5-fold increase in N-fixed, and, respectively, increased grain yield by 2.6-fold (932 kg.ha−1) and 4-fold (1403 kg.ha−1) (Figure 2). On the other hand, inoculating genotype Bawutawuta with strain CB756 in Savelugu increased shoot biomass, N-fixed (by 143%), shoot N accumulation, and, consequently, grain yield by 175% (Figure 2). Furthermore, bacterial inoculation of genotypes Nhyira, Omandaw, and Songotra with strain BR 3267 increased N-fixed by 126–157% and grain yield by up to 2-fold when compared to non-inoculation in the case of Omandaw and Songotra, and relative to strain CB756 in the case of Nhyira (Figure 2). Although inoculating IT90K-277-2 with Bradyrhizobium sp. CB756 resulted in lower N-fixed, the shoot biomass and N accumulation were greater due to higher soil N uptake, which resulted in up to two-fold increase in grain yield when compared to non-inoculation or inoculation with strain BR 3267. Here, non-inoculation increased N-fixed in genotype Nhyira (30.6 kg.ha−1) and doubled it in Padituya when compared to inoculation with strains CB756 and BR 3267, respectively. Consequently, grain yield was also increased by 61% in uninoculated plants of Nhyira (440.7 kg.ha−1) relative to plants inoculated with strain CB756, and up to 5 to 7-fold in Padituya (2318.7 kg.ha−1) relative to inoculation with either strain (Figure 2).
In Gbalahi, the genotype x inoculation interaction was significant for several parameters, including shoot biomass, N-fixed, N content, and grain yield (Table 5). Here, up to a 2.5-fold increase in N-fixed (84.5 kg.ha−1) and 3.6-fold increase in grain yield (573.7 kg.ha−1) was obtained by inoculating IT90K-277-2, SARVx-09-004 and Zayura with Bradyrhizobium sp. BR 3267, and Apagbaala with strain CB756. Consequently, grain yield was increased by 57–149% in those cowpea-inoculant strain combinations (Figure 3). On the other hand, non-inoculation increased N-fixed by 2.8-fold and 78% in genotypes Apagbaala (64.7 kg.ha−1) and SARVx-09-004 (51.8 kg.ha−1) relative to inoculation of those genotypes with Bradyrhizobium sp. BR 3267 or Bradyrhizobium sp. CB756, respectively in Gbalahi. Moreover, non-inoculation also resulted in a 2-fold and 97% grain yield increase in genotypes Apagbaala and SARVx-09-004 relative to inoculation of those genotypes with Bradyrhizobium sp. strain BR 3267 or CB756, respectively (Figure 3).
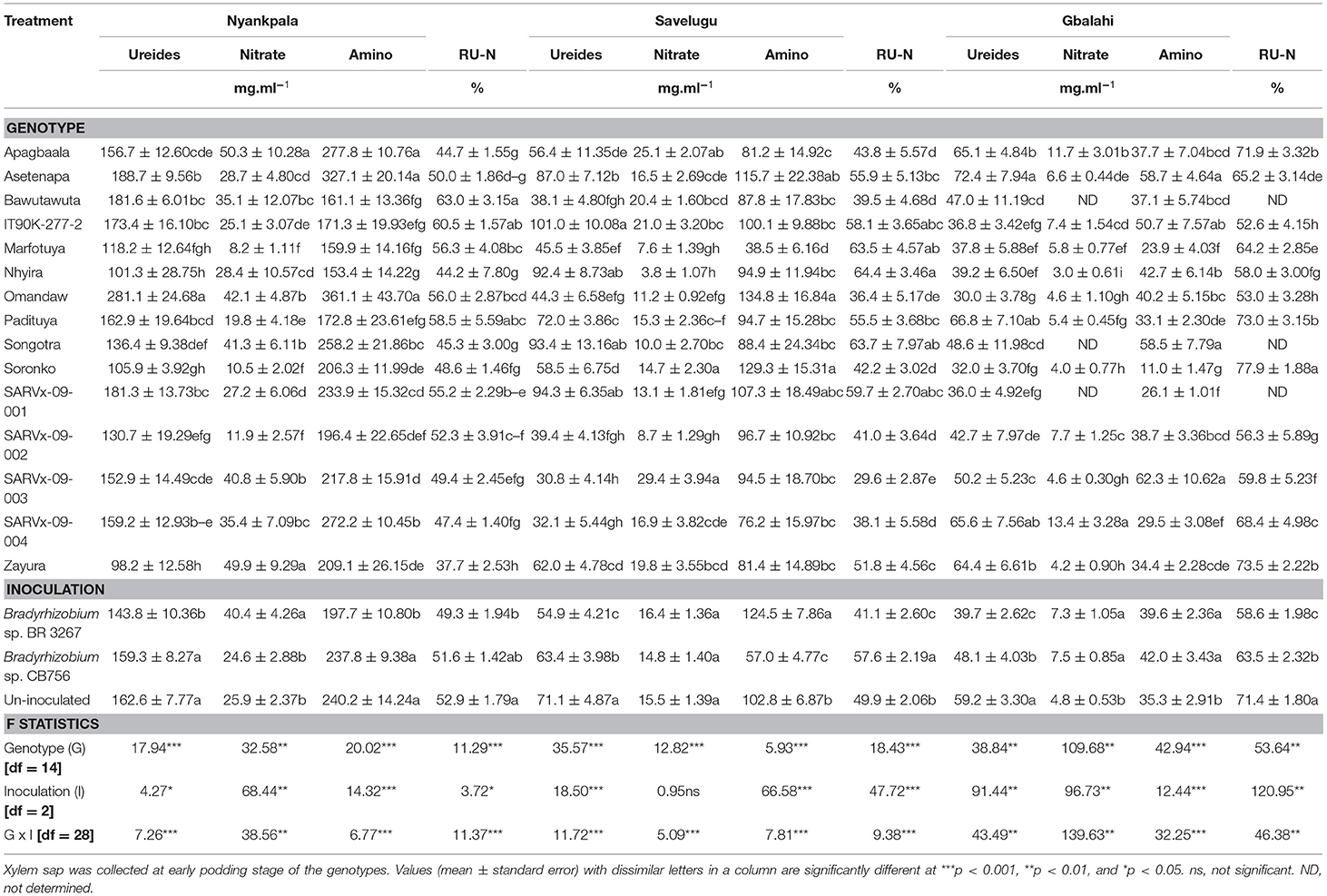
Table 5. The concentrations of ureides, nitrate, amino-N, and Relative ureide-N in xylem sap of cowpea genotypes inoculated with two Bradyrhizobium strains and planted in Nyankpala, Savelugu and Gbalahi in 2013.
Main Effects of Genotype and Cropping System on N Solutes in Xylem Sap
Within each location, there was a significant effect of genotype on the concentration of ureides, nitrate, and amino acids in the xylem sap of the test cowpeas (Table 5).
At Nyankpala, genotypes Omandaw and Asetenapa had greater concentration of ureides in xylem sap while Nhyira, Soronko, and Zayura recorded the least concentrations of ureides (Table 5). Other cowpea genotypes with high concentration of ureides in xylem sap in Nyankpala include Bawutawuta, IT90K-277-2, and SARVx-09-001 (Table 5). The genotypes Apagbaala, Songotra, SARVx-09-003, and Zayura coupled lower ureides in xylem sap with greater concentrations of nitrate. However, the concentrations of ureides and nitrates were both high in the sap of genotype Omandaw (Table 5). Moreover, the concentration of amino acids was greater in genotypes Apagbaala, Asetenapa, Omandaw, and SARVx-09-004 but least in Bawutawuta, Nhyira, and Padituya (Table 5). With high %Ndfa values from 15N natural abundance, the cowpea genotypes Bawutawuta, IT90K-277-2, and Padituya also recorded higher RU-N in xylem sap at Nyankpala. In contrast, genotypes Apagbaala, Nhyira, Songotra, and Zayura recorded much lower RU-N at Nyankpala, although Nhyira and Zayura had high %Ndfa at the location based on 15N natural abundance (Tables 2, 5).
The main effect of inoculation was also significant for the concentrations of ureides, nitrate, amino acids, and RU-N in xylem sap of the test cowpeas at Nyankpala (Table 5). Here, as observed with the 15N natural abundance, non-inoculation, together with bradyrhizobial inoculation with strain CB75, increased xylem ureides and amino acids, but decreased nitrate concentration, resulting in greater RU-N when compared to inoculation with strain BR 3267 (Table 5).
In Savelugu, there was a significant effect (p ≤ 0.05) of genotype on the concentrations of ureides, nitrate, and amino acids and RU-N in the xylem sap of cowpeas planted at Savelugu (Table 5). Here, genotype IT90K-277-2 recorded the highest ureide concentration in xylem sap despite having lower N-fixed measured from 15N natural abundance (Tables 3, 5). However, the genotypes Nhyira, Songotra, and SARVx-09-001 exhibited high ureide levels and relatively lower nitrate concentrations, which resulted in greater RU-N and %Ndfa measured from 15N natural abundance (Tables 3, 5). The least concentration of ureides was recorded in the xylem sap of SARVx-09-003 followed by SARVx-09-002, SARVx-09-004, Marfotuya, Bawutawuta, and Omandaw which also had low ureides in xylem sap (Table 3). With low ureide concentration, the genotypes Soronko, SARVx-09-003, and Apagbaala recorded the highest level of nitrate in xylem sap (Table 3). Moreover, the genotypes Omandaw and Soronko recorded the highest concentration of amino acids in xylem sap at Savelugu, followed by Asetenapa and SARVx-09-001 also with high concentrations of amino acids (Table 3). The least RU-N was recorded in genotype SARVx-09-003 followed by Omandaw, SARVx-09-004, Bawutawuta, SARVx-09-002, and Apagbaala in that order (Table 3). Other genotypes which coupled high RU-N with relatively high %Ndfa from 15N natural abundance include Marfotuya and IT90K-277-2 (Tables 3, 5).
The main effect of inoculation at Savelugu was also significant for xylem ureides, amino acid concentrations, and RU-N but not for nitrates (Table 3). Overall, non-inoculation or bacterial inoculation with Bradyrhizobium sp. strain CB756 each increased xylem ureide concentration when compared to inoculation with strain BR 3267 (Table 3). The concentration of nitrate in the xylem sap was however unaltered by bradyrhizobial inoculation at Savelugu. However, the concentration of amino acids was greater in plots inoculated with Bradyrhizobium sp. BR 3267 followed by the uninoculated plots, and least in plots inoculated with strain CB756. Non-inoculation and bradyrhizobial inoculation with strain CB756 each increased RU-N when compared to inoculation with strain BR 3267 at Savelugu although %Ndfa from 15N natural abundance was unaltered by the inoculation treatment (Table 3).
In Gbalahi, there was a marked effect of genotype on the concentrations of ureides, nitrate, amino acids, and RU-N in xylem sap of cowpea (Table 5). Here, genotype Asetenapa recorded the highest concentration of ureides in xylem sap followed by Padituya and SARVx-09-004. Other genotypes with high ureide levels in the location include Apagbaala and Zayura. The least concentration of ureide was recorded in genotype Omandaw followed by Soronko, IT90K-277-2, Marfotuya, and Nhyira also with low xylem ureide concentration (Table 5). With high ureides however, genotypes Apagbaala and SARVx-09-004 also recorded higher concentration of nitrate in xylem sap. Together with genotypes Apagbaala, Soronko, and Zayura which recorded high levels of ureides, Padituya also recorded greater RU-N despite having reduced concentration of ureides in xylem sap (Table 5). Moreover, genotypes Soronko and Zayura recorded high RU-N as well as greater %Ndfa measured using 15N natural abundance (Table 5).
In Gbalahi, there was a marked effect of inoculation on the concentration of ureides, nitrate, amino acids, and RU-N in xylem sap (Table 5). Overall, the highest concentration of ureides in xylem sap in Gbalahi was obtained by non-inoculation followed by bacterial inoculation with strain CB756. Non-inoculation also reduced overall nitrate and amino acids in xylem sap when compared to inoculation with either Bradyrhizobium strains in Gbalahi. Here, together with non-inoculation, bacterial inoculation with strain CB756 each increased RU-N and %Ndfa measured from 15N natural abundance when compared to inoculation with strain BR 3267 (Tables 4, 5).
N and C Assimilation in Cowpea Genotypes
The 15 cowpea genotypes markedly varied in their shoot N and C concentrations (%) and N and C contents (mg.plant−1) in Nyankpala, Savelugu, and Gbalahi (Table 6).
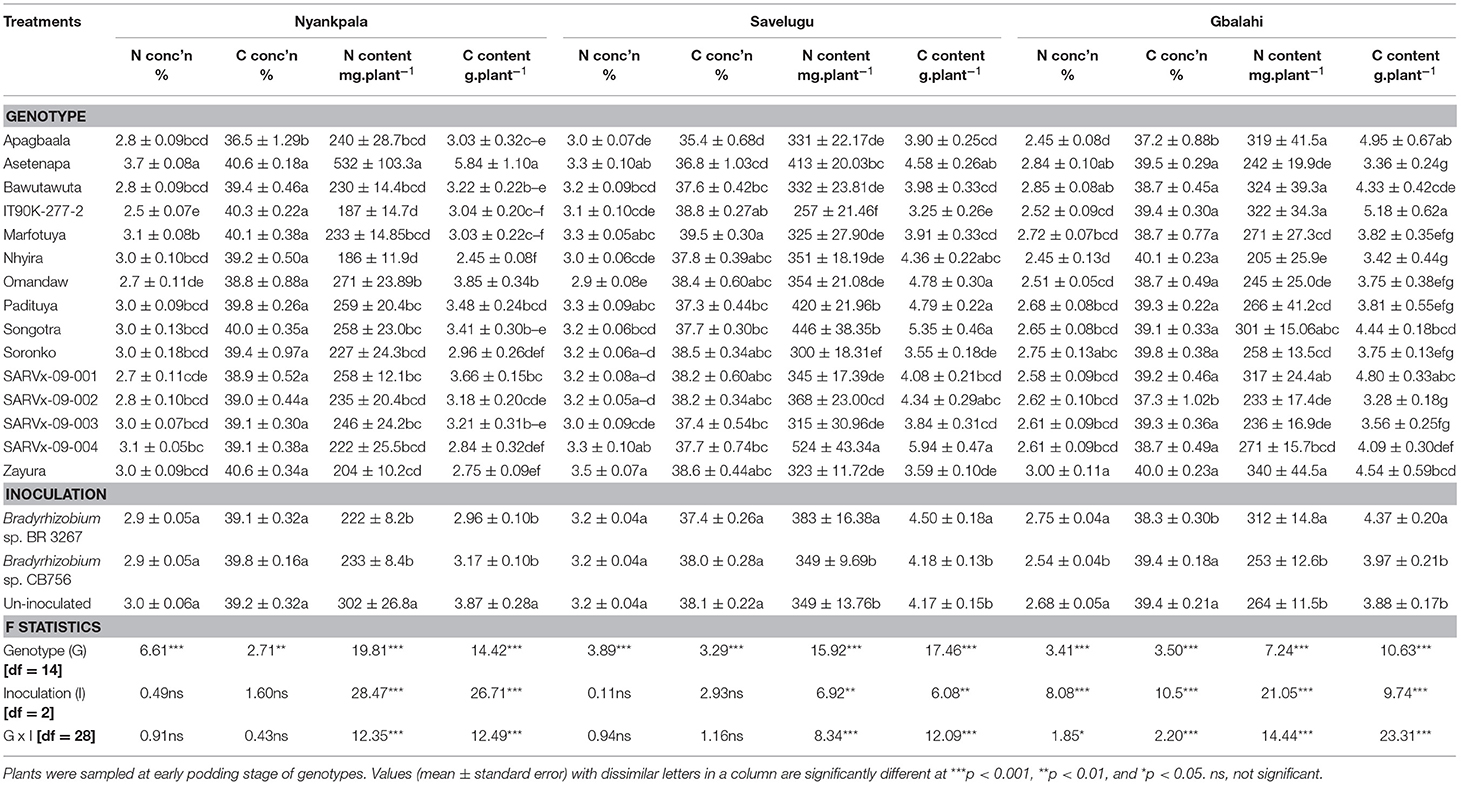
Table 6. Shoot N and C assimilation in 15 cowpea genotypes inoculated with two Bradyrhizobium strains and planted in Nyankpala, Savelugu, and Gbalahi in 2013.
In Nyankpala, the genotype Asetenapa recorded the highest %N followed by Marfotuya and SARVx-09-004 also with high %N. On the other hand, genotypes IT90K-277-2 and Omandaw recorded the least %N at the location (Table 6). Further, genotype Apagbaala recorded the least %C (36.5%) in Nyankpala, while the remaining genotypes exhibited markedly greater but similar %C values (38.8–40.6%) in that location (Table 6). With high %N and %C, the genotype Asetenapa coupled higher shoot N accumulation with greater C content in Nyankpala. Other genotypes with high N and C contents in Nyankpala include Omandaw and SARVx-09-001 (Table 6).
The main effect of inoculation was significant for N and C contents in Nyankpala, with non-inoculation resulting in greater shoot N and C accumulation when compared to inoculation with either Bradyrhizobium strains at the location. However, the overall %N and %C were unaltered by inoculation (Table 6).
In Savelugu, there was a significant effect of genotype on %N, %C, and N and C contents (mg.plant−1) (Table 6). Here, %N was highest in genotype Zayura followed by Asetenapa, SARVx-09-004, and then Marfotuya and Padituya, but least in Omandaw and Apagbaala (Table 6). Genotype Marfotuya recorded the highest %C in Savelugu, followed by IT90K-277-2, Omandaw, Soronko, SARVx-09-001, SARVx-09-002, and Zayura also with high %C. The genotypes SARVx-09-004, Songotra, Padituya, and Asetenapa exhibited high N and C contents in Savelugu while Omandaw recorded higher C content but lower N content (Table 6). On the other hand, genotypes such as Apagbaala and Soronko had lower N and C contents in Savelugu (Table 6).
The main effect of inoculation was also significant for N and C contents in Savelugu but not %N and %C. Here, bacterial inoculation with strain BR 3267 increased overall N and C contents when compared to non-inoculation or inoculation with strain CB756 in Savelugu (Table 6).
At Gbalahi, the genotypes showed marked variations in %N, %C, N, and C contents (Table 6). The genotype Zayura had the highest %N in shoot followed by Asetenapa and Bawutawuta also with high %N. In contrast, Apagbaala and Nhyira recorded the least %N at Gbalahi (Table 6). The least %C was recorded in genotypes Apagbaala (37.2%) and Nhyira (37.3%) while the remaining genotypes recorded higher but similar %C ranging between 38.7 and 40.1%. In Gbalahi, genotypes Apagbaala, IT90K-277-2, and SARVx-09-001 coupled high N contents with greater C content. On the other hand, genotypes Bawutawuta and Zayura recorded relatively lower C contents despite greater N content in Gbalahi. The least N and C contents at Gbalahi were recorded in genotypes SARVx-9-002, SARVx-09-003, Nhyira, Omandaw, and Asetenapa (Table 6).
The main effect of Bradyrhizobium inoculation markedly influenced %N, %C, as well as N and C contents in Gbalahi (Table 6). Here, together with non-inoculation, bacterial inoculation with strain BR 3267 increased overall %N compared to inoculation with strain CB756. On the other hand, non-inoculation or inoculation with strain CB756 each increased %C when compared to inoculation with strain BR 3267. However, bacterial inoculation with strain BR 3267 increased shoot N and C contents at Gbalahi when compared to non-inoculation or inoculation with strain CB756 at the location (Table 6).
Three-Way ANOVA Comparison of Parameters among Locations
Plant Growth, Symbiotic Parameters, δ13C, and Grain Yield
A three-way ANOVA revealed marked effect of planting location on nodule mass, shoot biomass, symbiotic parameters, soil N uptake, δ13C, and grain yield of the cowpea genotypes (Supplementary Table 2). Overall, nodule mass of cowpea was highest in Gbalahi, followed by Nyankpala, and least in Savelugu. However, overall shoot biomass was highest in Savelugu despite reduced nodulation by plants at the location followed by plants in Gbalahi (Supplementary Table 2).
The least δ15N was recorded by plants in Nyankpala followed by those at Gbalahi while plants in Savelugu recorded the highest overall δ15N (Supplementary Table 2). As a result, average %Ndfa and N-fixed were both higher in plants in Nyankpala and Gbalahi when compared to the plants in Savelugu (Supplementary Table 2). With higher δ15N and lower %Ndfa, plants in Savelugu recorded the highest soil N uptake while those in Nyankpala recorded the least. The overall C/N ratio was highest in plants in Gbalahi followed by those in Nyankpala, and least in plants in Savelugu (Supplementary Table 2). On the other hand, %N was highest in plants in Savelugu, whilst %C was least in the plants at the same location. The shoot N and C contents of cowpea closely mirrored patterns in their δ13C values across locations, with the highest values being recorded in Savelugu (Supplementary Table 2). Overall, average δ13C values of cowpea was highest in Savelugu followed by Gbalahi, and least in the plants grown in Nyankpala. With greater δ13C, plants in Savelugu also recorded the highest grain yield while those in Gbalahi recorded the least (Supplementary Table 2).
Ureides, Nitrate, Amino-N, and Relative Ureide-N in Xylem Sap
The concentration of xylem N solutes (ureides, nitrate and amino-N) and RU-N of the cowpea genotypes markedly varied between the test locations (Supplementary Table 3). The concentrations of ureides, nitrate, and amino-N in xylem sap were highest in plants grown in Nyankpala followed by those in Savelugu, and least in Gbalahi. However, RU-N was highest in plants in Gbalahi followed by those in Nyankpala due to relatively lower concentrations of nitrate and amino-N in xylem sap of cowpea in the former location (Supplementary Table 3). The plants grown in Savelugu coupled low RU-N with the least overall %Ndfa measured from 15N natural abundance (Supplementary Table 3).
Correlation and Regression Analysis
Correlation and regression analyses revealed significant functional relationships between several parameters (Figures 4–7). There were significant negative correlations between xylem nitrate-N and RU-N in all three locations, and xylem nitrate-N and %Ndfa measured from 15N natural abundance at Nyankpala and Savelugu (Figures 4A–F). %Ndfa and soil N uptake also showed negative correlation. Further, there were significant positive correlations between shoot biomass and N-fixed, soil N uptake, and shoot N content in all test locations (Figures 5A–I). Grain yield also showed significant positive correlations with N-fixed and shoot N content in all three locations (Figures 6A–F) while %N and %C were also positively correlated in Nyankpala and Savelugu. As to be expected, shoot biomass and C content correlated positively in all three locations (Figures 7A–E).
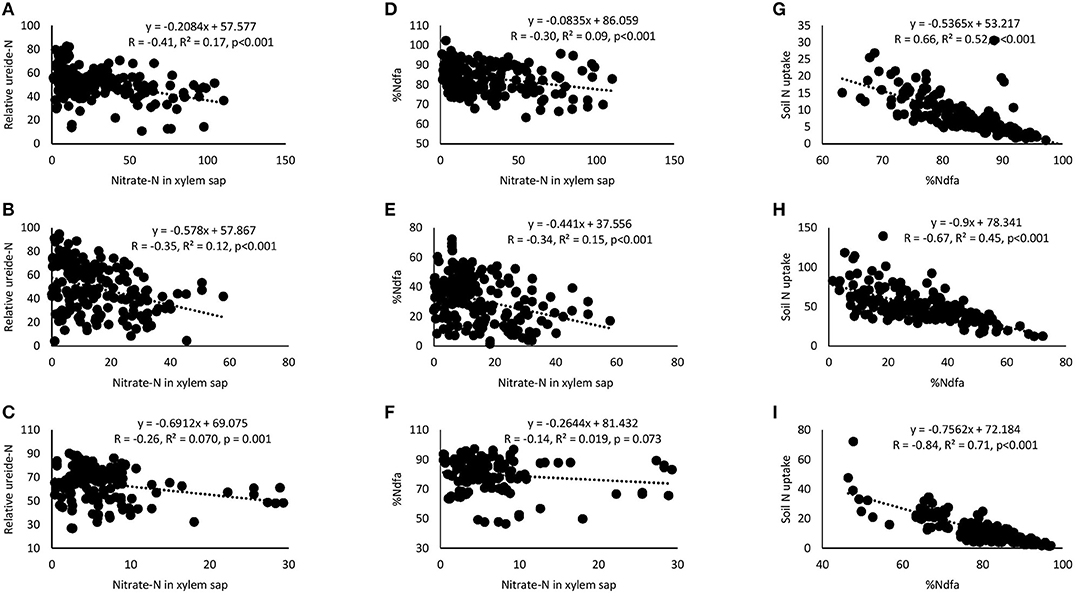
Figure 4. Correlation and regression analysis between xylem nitrate-N and relative ureide-N (RU-N) of 15 cowpea genotypes planted in (A) Nyankpala, (B) Savelugu, and (C) Gbalahi, and between xylem nitrate-N and %Ndfa at (D) Nyankpala, (E) Savelugu, and (F) Gbalahi, and between %Ndfa and soil N uptake in (G) Nyankpala, (H) Savelugu, and (I) Gbalahi.
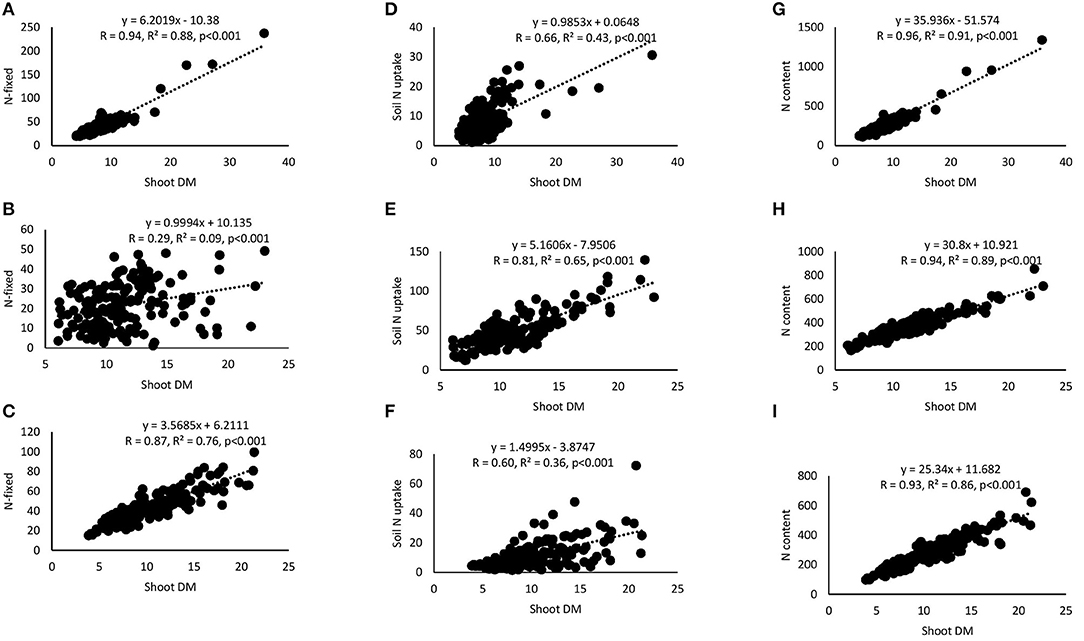
Figure 5. Correlation and regression analysis between shoot dry matter (DM) and N-fixed by 15 cowpea genotypes planted in (A) Nyankpala, (B) Savelugu, and (C) Gbalahi, and between shoot DM and soil N uptake at (D) Nyankpala, (E) Savelugu, and (F) Gbalahi, and between shoot DM and N content at (G) Nyankpala, (H) Savelugu, and (I) Gbalahi.
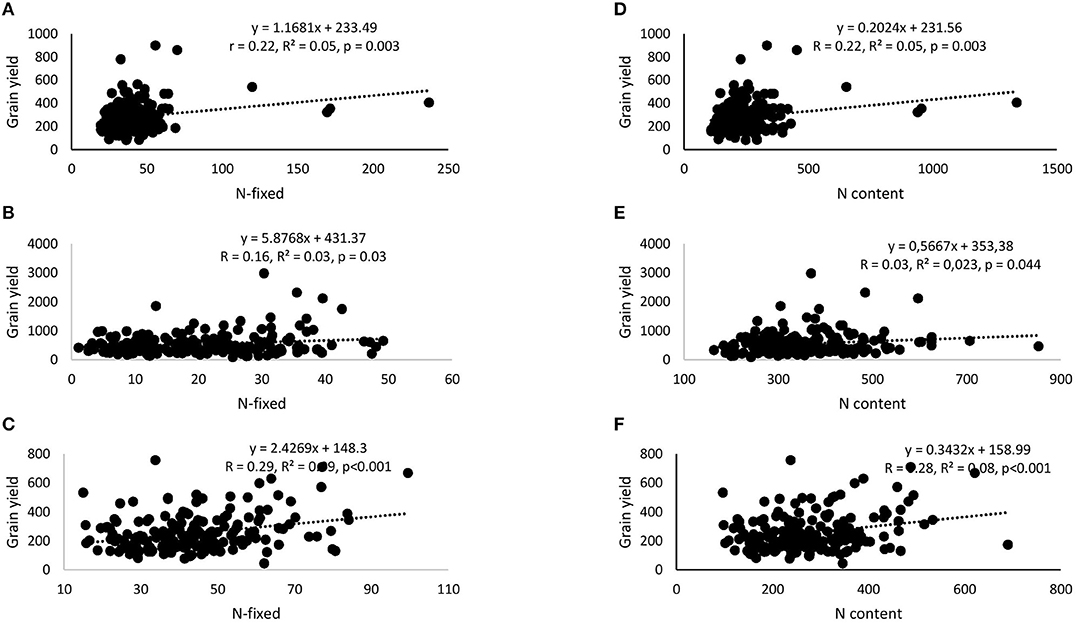
Figure 6. Correlation and regression analysis between N-fixed and grain yield of 15 cowpea genotypes planted in (A) Nyankpala, (B) Savelugu, and (C) Gbalahi, and between shoot N content and grain yield in (D) Nyankpala, (E) Savelugu, and (F) Gbalahi.
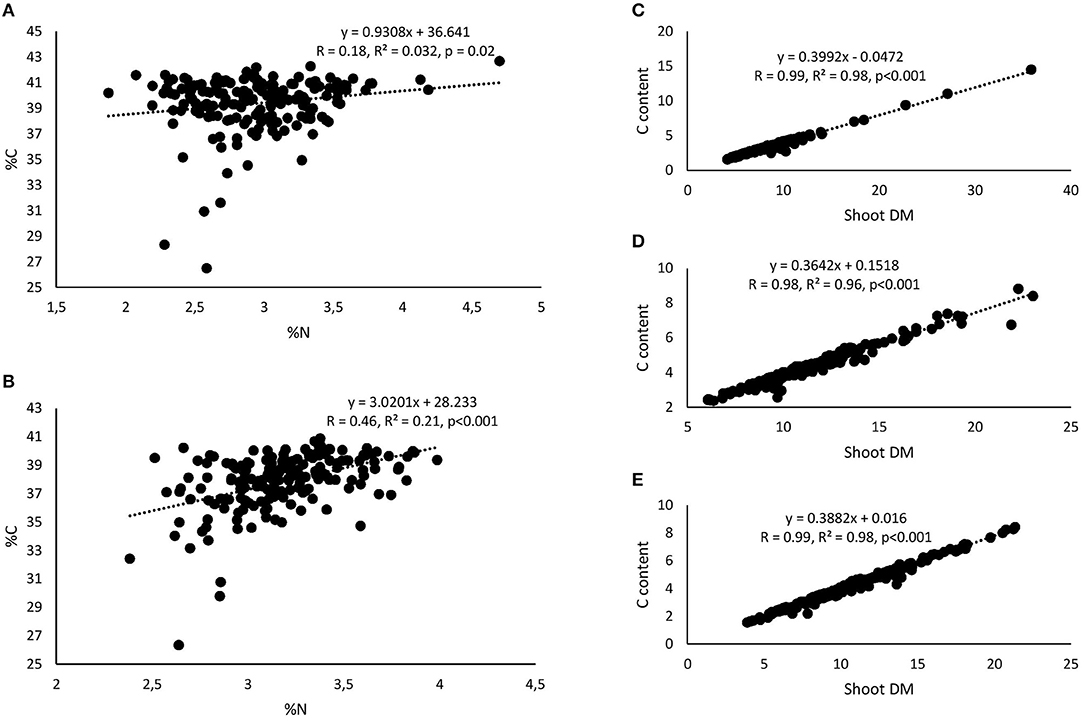
Figure 7. Correlation and regression analysis between shoot %N and %C of 15 cowpea genotypes planted in (A) Nyankpala and (B) Savelugu and between shoot dry matter (DM) and C content at (C) Nyankpala, (D) Savelugu, and (E) Gbalahi.
Discussion
Cowpea Genotypic Differences in Growth, Symbiotic N2 Fixation and Grain Yield
Although several interaction effects were significant within test locations, analysis of the main effects also revealed marked variations in nodulation, plant growth, and symbiotic performance of the test cowpeas, a finding similar to previous reports among cowpea germplasm grown in contrasting environments in Africa (Belane and Dakora, 2010; Belane et al., 2014). Although greater nodulation was expected to promote symbiosis and plant growth, instances of higher nodulation were not always accompanied by increases in plant growth and symbiotic performance by the test cowpeas. For example, even though increased nodulation in genotype Asetenapa promoted greater biomass accumulation at Nyankpala, other genotypes, including Nhyira, Songotra, and SARVx-09-001, elicited reduced plant growth and N-fixed despite higher nodulation at the location. Similar trends between cowpea nodulation, plant growth, and N2-fixing parameters were also observed in Savelugu and Gbalahi, clearly suggesting that the extent of nodulation alone did not always explain plant growth and legume N2 fixation in this study. These results agree with previous reports that rather than the number of nodules, it is the N2-fixing efficiency of the rhizobial microsymbionts which is a key determinant of legume symbiotic performance (Westhoek et al., 2017; Mohammed et al., 2018).
As expected, the cowpea genotypes in this study varied in their symbiotic dependence, with %Ndfa ranging between 74.4 and 89.8% in Nyankpala, 11.7 and 50.5% in Savelugu and 71.9 and 90.3% in Gbalahi. Similarly, previous reports have shown that some genotypes can derive up to 96% of their N requirements from the atmosphere (Belane and Dakora, 2010). The observed high symbiotic dependence by cowpeas in this study partly accounts for the wider adaptation of the crop to nutrient depleted soils in which other crops may fail to survive (Belane and Dakora, 2010; Hall, 2012). The low symbiotic dependence by most cowpeas in Savelugu resulting from relatively high soil N (0.16% N) in the location was augmented by greater soil N uptake leading to grain yield increases, a finding supported by significant positive correlations when N-fixed and N content were each plotted against shoot biomass and grain yield within the test locations. The observed soil N inhibition of symbiosis (Mbah and Dakora, 2017) was also evidenced by significant negative correlations between xylem nitrate-N and RU-N, xylem nitrate-N and %Ndfa measured from 15N natural abundance, and between %Ndfa and soil N uptake in all test locations (Figures 4A–I).
With only few exceptions however, the xylem N solute measure of symbiosis in individual cowpea genotypes did not always follow the same pattern as that measured using the 15N natural abundance method, an observation attributed to the instantaneous nature of the ureides technique in estimating legume symbiotic dependence (Unkovich et al., 2008). Nevertheless, the overall symbiotic dependence measured from the ureides (RU-N) and 15N natural abundance (%Ndfa) were both least in plants grown in Savelugu compared to those in Nyankpala and Gbalahi, indicating the usefulness of both techniques in estimating legume N2 fixation.
Grain yield of cowpea in this study ranged between 212.4 and 419.9 kg.ha−1 in Nyankpala, 338.2 and 1036.1 kg.ha−1 in Savelugu, and 139.2 and 440.3 kg.ha−1 in Gbalahi. Despite the much lower %Ndfa and N-fixed by the test cowpeas grown at Savelugu, those genotypes augmented their low symbiotic N with increased soil N uptake, leading to higher grain yields. The complementary roles of symbiosis and soil N uptake in promoting the growth and yield of the test cowpeas was evidenced by significant positive correlations when N-fixed and N content were each plotted against shoot biomass and grain yield (Figures 6A–F). The genotypes Asetenapa and Omandaw (local landrace) respectively recorded higher grain yields in Nyankpala (419.9 and 411.4 kg.ha−1) and Savelugu (896.7 and 646.5 kg.ha−1), suggesting possible wider adaptation of those genotypes to the different test environments. The highest grain yield in Savelugu (1036.1 kg.ha−1) was however recorded in genotype Padituya which also recorded relatively high grain yield (314.2 kg.ha−1) in Gbalahi when compared to other genotypes in the location. The fact that most cowpeas in Gbalahi had relatively lower grain yields despite deriving up to 90% of N from symbiosis was unexpected. However, other location specific factors including lower rainfall/soil moisture or its distribution (Supplementary Figure 1) might have contributed to the observed lower grain yield at the location.
Genotypic Variations in N and C Assimilation, and Water Use Efficiency (δ13C) in Field-Grown Cowpea
Despite the widely reported drought tolerance of cowpea (Hall, 2012), most cultivated varieties are yet to be assessed for their water relations in the field. This study revealed marked differences in the N and C concentrations of the test cowpea genotypes. For example, except for Apagbaala in Nyankpala, and together with SARVx-09-002 in Gbalahi which recorded much lower %C in shoots (36.5–37.6%), the remaining genotypes had higher but similar shoot %C (38.7–40.6%) (Table 3). Moreover, the fact that genotype Asetenapa in Nyankpala, and Marfotuya, Padituya, SARVx-09-004, and Zayura in Savelugu, each coupled greater %N with relatively higher %C in those locations suggest that increased C gain via photosynthesis stimulated N nutrition (Rogers et al., 2006). The functional link between N and C nutrition via photosynthesis was previously reported in field grown cowpeas (Belane and Dakora, 2011), and was further supported by positive correlations between %N and %C in this study (Figures 7A,B). Given the rising atmospheric CO2 levels with climate change, incorporating legume genotypes eliciting greater N and C accumulation in cropping systems could help in C sequestration, and therefore reduce greenhouse gas emissions. Moreover, the typically low C/N ratio (11.1–16.9 g.g−1) recorded for cowpeas across the three locations is characteristic of N2-fixing legumes, and partly accounts for their ease of decomposition which makes them valuable in conservation agriculture where they improve C sequestration and soil microbial activities (Jarecki and Lal, 2003).
Aside their potential N contribution to cropping systems, the test cowpeas also showed marked variation in plant water use efficiency (δ13C) within the test locations. Interestingly, the cowpea genotypes Soronko, Asetenapa, Nhyira, Omandaw, and IT90K-277-2 elicited greater water use efficiency in both Nyankpala (−28.67 to −28.27‰) and Savelugu (−28.12 to −28.12‰). Further, the fact that genotypes Asetenapa and Omandaw in Nyankpala together with Padituya and Soronko in Savelugu coupled greater water use efficiency (δ13C) with relatively higher grain yields in the respective locations are traits that could be exploited for crop improvement programs aimed at screening for enhanced plant-water relations and grain yield. The robustness of carbon isotope discrimination in estimating plant water relations as observed in this study has been reported in other field-grown legumes (Mohale et al., 2014; Mapope and Dakora, 2016).
Effect of Planting Location (Environment) on Cowpea Symbiotic N Nutrition, N and C Assimilation and Water Use Efficiency (δ13C)
Although genotypic differences in plant growth and symbiotic relations exist among cowpea germplasm, these physiological traits are also influenced by environmental differences including soil and climate factors (Belane and Dakora, 2009; Nyemba and Dakora, 2010; Belane et al., 2011). Consequently, plant growth and symbiotic performance of the test cowpeas in this study was markedly influenced by planting location. For example, overall plant growth was greater in Savelugu despite reduced nodulation and symbiosis at the location due to high soil N, whereas cowpea plants in Gbalahi coupled greater nodulation with relatively higher plant growth when compared to those grown in Nyankpala. However, overall symbiotic dependence (%Ndfa) and N-fixed were much higher in Nyankpala and Gbalahi due to reduced δ15N resulting from effective symbiosis in those locations when compared to Savelugu. Since cowpea exports fixed N as ureides (Baral et al., 2016), xylem N solutes (ureide, nitrate and amino-N) were also used as a measure of symbiotic dependence (Unkovich et al., 2008). The concentrations of all xylem N solutes were greater in Nyankpala followed by Savelugu, and least in plants grown in Gbalahi (Supplementary Table 3). As observed with %Ndfa from 15N natural abundance, the relative ureide-N abundance was higher in Nyankpala and Gbalahi, and again, least in Savelugu, an indication that both techniques are robust in assessing legume N2 fixation. The observed lower symbiotic dependence by cowpeas in Savelugu was probably due to its suppression by endogenous soil N in the location, which resulted in greater soil N uptake to augment the shortfall in N requirement, a finding supported by significant negative correlations between %Ndfa and soil N uptake, xylem nitrates and RU-N, and between xylem nitrates and %Ndfa measured from 15N natural abundance (Figure 4). Despite the reduced symbiotic dependence (%Ndfa) by plants in Savelugu, C/N ratio was much lower in those plants due to greater %N and shoot N accumulation resulting increased soil N uptake (Supplementary Table 2). The lower grain yield recorded by plants in Gbalahi despite higher symbiotic performance was probably due to reduced rainfall or soil moisture at the location during the pod filling stage (Supplementary Figure 1).
Variable Response of Cowpea to Inoculation With Bradyrhizobium Strains in the Field
To test for possible cowpea genotypic differences in response to inoculation with Bradyrhizobium sp. strains BR 3267 and CB756, 15 cowpea genotypes were assessed for N2 fixation and grain yield at three locations in the Northern Region of Ghana. The test cowpeas were found to elicit contrasting responses to inoculation with either Bradyrhizobium strains within and between locations in relation to nodulation, shoot growth, symbiotic performance, and grain yield. For brevity, selected interaction effects within test locations are discussed for genotypes that showed consistent patterns in relation to most symbiotic parameters. Bacterial inoculation of cowpea respectively increased N-fixed and grain yield by up to 5- and 4-fold, depending on genotype-inoculant strain combinations and planting location. For example, inoculating genotype Apagbaala with Bradyrhizobium sp. BR 3267 in Nyankpala doubled N-fixed (47.7 kg.ha−1), and slightly increased it in IT90K-277-2 when compared to inoculation with strain CB756. On the other hand, bacterial inoculation of Bawutawuta, Padituya, and SARVx-09-003 with strain CB756 significantly increased shoot biomass, resulting in 32 – 200% increases in the amounts of N-fixed and shoot N accumulation when compared to inoculation of the same genotype with strain BR 3267 (Figure 2). Consequently, grain yield was increased by 1.3- to 2-fold in those cowpea-inoculant strain combinations in Nyankpala (Figure 1). In contrast, non-inoculation of Apagbaala, Asetenapa, Padituya, and SARVx-09-003 in Nyankpala each increased N-fixed by 1.5- to 3.4-fold (36.5–55.0 kg.ha−1), resulting in greater shoot N accumulation and 1.2- to 2-fold increase in grain yield in those cowpea-inoculant combinations (Figure 1), a finding that confirms the presence of effective cowpea-compatible rhizobia in the studied soils. Although this is the first report of inoculation response in genotypes Apagbaala and Asetenapa, the increased N2 fixation and grain yield in uninoculated Padituya is in contrast with an earlier study that found up to 57% increase in grain yield of the genotype due to inoculation with Bradyrhizobium sp. strain BR 3267 (Boddey et al., 2017). The discrepancy in inoculation response could be attributed to differences in the study locations together with prevailing climatic conditions, a finding consistent with earlier reports which also found inoculation failures in cowpea in the same growing season due to differences in planting location (Ulzen et al., 2016). Although N2 fixation was generally low in plants in Savelugu, bacterial inoculation of Asetenapa with either strain BR 3267 or CB756 resulted in 3 to 5-fold increase in N-fixed, and, respectively, increased grain yield by 2.6- (932 kg.ha−1) and 4-fold (1403 kg.ha−1). On the other hand, inoculating Bawutawuta with strain CB756 at Savelugu increased shoot biomass, N-fixed (by 143%), shoot N accumulation, and, consequently, grain yield by 175% (Figure 2). However, non-inoculation increased N-fixed in genotype Nhyira (30.6 kg.ha−1) and doubled it in Padituya when compared to inoculation with Bradyrhizobium sp. strain CB756 or BR 3267, respectively. As a result, grain yield was increased by 61% (440.7 kg.ha−1) in uninoculated Nhyira relative to inoculation with Bradyrhizobium sp. CB756, and up to 5 to 7-fold in uninoculated Padituya (2318.7 kg.ha−1) relative to inoculation with either Bradyrhizobium strains (Figure 2). At Gbalahi, up to a 2.5-fold increase in N-fixed (84.5 kg.ha−1) and 3.6-fold increase in grain yield (573.7 kg.ha−1) was obtained by inoculating IT90K-277-2, SARVx-09-004, and Zayura with Bradyrhizobium sp. strain BR 3267 and Apagbaala with strain CB756. Here, non-inoculation respectively increased N-fixed by 2.5-fold and 78% in genotypes Apagbaala (60.8 kg.ha−1) and SARVx-09-004 (51.8 kg.ha−1) in Gbalahi, and resulted in grain yield increases of 2-fold and 97% relative to inoculation with strain BR 3267 and CB756 at the location. These findings are consistent with an earlier study which reported up to 38% increase in grain yield of cowpea resulting from inoculation with Bradyrhizobium sp. BR 3267 relative to non-inoculation although the same inoculant failed to increase the parameter in another location (Ulzen et al., 2016).
Clearly, cowpea inoculation response in this study was markedly influenced by the cowpea-inoculant strain combinations used and the planting location, a finding consistent with previous reports that inoculation success is influenced by the compatibility between crop genotype and inoculant strain, location-dependent factors, such as the population and N2-fixing efficiency of native rhizobia, and the adaptation and competitive ability of introduced strains against the prevailing soil microbiome (Catroux et al., 2001). The fact that some cowpea genotypes, including Apagbaala, Asetenapa, and Padituya in Nyankpala, Padituya in Savelugu, and Apagbaala and SARVx-09-004 in Gbalahi, formed effective symbiosis with native rhizobia in the respective locations leading to enhanced plant growth and grain yield indicates the presence of effective indigenous cowpea-compatible rhizobia in the studied soils (Figures 2, 3). These findings also agree with a recent report by Mohammed et al. (2018) who found the presence of highly effective rhizobial symbionts responsible for cowpea nodulation in the studied locations. Although his study is the first report of inoculation response in most of the test cowpea genotypes, Padituya was earlier found to respond to inoculation with Bradyrhizobium sp. BR 3267, leading to improved grain yield over non-inoculation (Ulzen et al., 2016). The observed interactions within the test locations in this study further reveal the complexity of the factors influencing legume inoculation response in the field (Catroux et al., 2001), which therefore suggests a need to study the functional diversity of cowpea nodule microsymbionts alongside their adaptive traits such as tolerance to biotic and abiotic stresses that often occur under field conditions.
Conclusion
The test cowpeas exhibited variable nodulation, plant growth, and symbiotic performance across the test environments. The fact that cowpeas in this study could obtain up to 90% of their N requirement from symbiosis stresses their survival advantage in the N limited soils in most parts of Africa. Although endogenous soil N suppressed cowpea symbiotic dependence in Savelugu, increased soil N uptake augmented the shortfall leading to grain yield increases. Grain yields of up to 1,036 kg.ha−1 was recorded without any inorganic inputs, suggesting that the cowpea-rhizobia symbiosis could be exploited for increased yield. The fact that genotypes such as Asetenapa, Omandaw, Soronko, and Padituya coupled greater δ13C with higher grain yield is a desirable trait that could be harnessed for increased production of the crop under water limiting conditions, and for breeding programs to improve plant-water relations. The fact that inoculation response was markedly influenced by genotype-inoculant strain combination and location effects suggest that the introduction of inoculant strains into new areas require field testing with widely grown varieties to assess their feasibility.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
MM conducted field experiments, analyzed and interpreted data, and prepared the manuscript. GM took part in xylem N solute analyses. ES helped in field experiments. FD funded and coordinated the research and helped in manuscript preparation. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors appreciate support from Dr Benjamin D. K. Ahiabor of the CSIR-Savanna Agricultural Research Institute at Nyankpala, Ghana for the supply of inoculant and support during field experimentation. We are grateful to the South African Research Chair in Agrochemurgy and Plant Symbioses, the National Research Foundation, and the Tshwane University of Technology (TUT) for financial support to FD's research, and for a competitive TUT Doctoral Bursary to MM.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2022.764070/full#supplementary-material
References
Baral, B., Teixeira da Silva, J. A., and Izaguirre-Mayoral, M. L. (2016). Early signaling, synthesis, transport and metabolism of ureides. J. Plant Physiol. 193, 97–109. doi: 10.1016/j.jplph.2016.01.013
Belane, A. K., Asiwe, J., and Dakora, F. D. (2011). Assessment of N2 fixation in 32 cowpea (Vigna unguiculata L. Walp) genotypes grown in the field at Taung in South Africa, using 15N natural abundance. Afric. J. Biotechnol. 10, 11450–11458. doi: 10.5897/AJB11.674
Belane, A. K., and Dakora, F. D. (2009). Measurement of N, fixation in 30 cowpea (Vigna unguiculata L. Walp.) genotypes under field conditions in Ghana, using the 15N natural abundance technique. Symbiosis 48, 47–56. doi: 10.1007/BF03179984
Belane, A. K., and Dakora, F. D. (2010). Symbiotic N2 fixation in 30 field-grown cowpea (Vigna unguiculata L. Walp.) genotypes in the Upper West Region of Ghana measured using 15N natural abundance. Biol. Fertil. Soils 46, 191–198. doi: 10.1007/s00374-009-0415-6
Belane, A. K., and Dakora, F. D. (2011). Photosynthesis, symbiotic N and C accumulation in leaves of 30 nodulated cowpea genotypes grown in the field at Wa in the Guinea savanna of Ghana. Field Crops Res. 124, 279–287. doi: 10.1016/j.fcr.2011.07.014
Belane, A. K., Pule-Meulenberg, F., Makhubedu, T. I., and Dakora, F. D. (2014). Nitrogen fixation and symbiosis-induced accumulation of mineral nutrients by cowpea (Vigna unguiculata L. Walp.). Crop Pasture Sci. 65, 250–258. doi: 10.1071/CP13283
Boddey, R. M., Fosu, M., Atakora, W. K., Miranda, C. H. B., Boddey, L. H., Guimaraes, A. P., et al. (2017). Cowpea (Vigna unguiculata) crops in Africa can respond to inoculation with Rhizobium. Exp. Agric. 53, 578–587. doi: 10.1017/S0014479716000594
Bray, R. H., and Kurtz, L. T. (1945). Determination of total, organic, and available forms of phosphorus in soils. Soil Sci. 59, 39–46. doi: 10.1097/00010694-194501000-00006
Cataldo, D., Haroon, M., Schrader, L. E., and Youngs, V. L. (1975). Rapid colorimetric determination of nitrate in plant tissue by nitration of salicylic acid. Commun. Soil Sci. Plant Anal. 6, 71–80
Catroux, G., Hartmann, A., and Revellin, C. (2001). Trends in rhizobial inoculant production and use. Plant Soil 230, 21–30. doi: 10.1023/A:1004777115628
Cernusak, L. A., Ubierna, N., Winter, K., Holtum, J. A. M., Marshall, J. D., and Farquhar, G. D. (2013). Environmental and physiological determinants of carbon isotope discrimination in terrestrial plants. New Phytol. 200, 950–965. doi: 10.1111/nph.12423
Craig, H. (1957). Isotopic standards for carbon and oxygen and correction factors for mass-spectrometric analysis of carbon dioxide. Geochim. Cosmochim. Acta 12, 133–149. doi: 10.1016/0016-7037(57)90024-8
Dakora, F. D., and Keya, S. O. (1997). Contribution of legume nitrogen fixation to sustainable Agriculture in Sub-Saharan Africa. Soil Biol. Biochem. 29, 809–817. doi: 10.1016/S0038-0717(96)00225-8
Ddamulira, G., and Santos, C. (2015). Grain yield and protein content of Brazilian cowpea genotypes under diverse ugandan environments. Am. J. Plant Sci. 6, 2074. doi: 10.4236/ajps.2015.613208
De Ron, A. M., Sparvoli, F., Pueyo, J. J., and Bazile, D. (2017). Editorial: Protein crops: food and feed for the future. Front. Plant Sci. 8, 105. doi: 10.3389/fpls.2017.00105
Ehlers, J. D., and Hall, A. E. (1997). Cowpea (Vigna unguiculata L. Walp.). Field Crop. Res. 53, 187–204. doi: 10.1016/S0378-4290(97)00031-2
FAO (2021). Available online at: http://www.fao.org/faostat/en/#data/QCL/visualize. (accessed Aug 24, 2021).
Farquhar, G. D., Ehleringer, J. R., and Hubick, K. T. (1989). Carbon isotope discrimination and photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40, 503–537. doi: 10.1146/annurev.pp.40.060189.002443
Foyer, C. H., Lam, H.-M., Nguyen, H. T., Siddique, K. H. M., Varshney, R. K., Colmer, T. D., et al. (2016). Neglecting legumes has compromised human health and sustainable food production. Nat. Plants 2, 16112. doi: 10.1038/nplants.2016.112
Guopeng, Z., Weidong, C., Jinshun, B., Changxu, X. U., Naohua, Z., Songjuan, G., et al. (2020). Co-incorporation of rice straw and leguminous green manure can increase soil available nitrogen (N) and reduce carbon and N losses: an incubation study. Pedosphere, 30, 661–670. doi: 10.1016/S1002-0160(19)60845-3
Hall, A. E. (2004). Breeding for adaptation to drought and heat in cowpea. Eur. J. Agron. 21, 447–454. doi: 10.1016/j.eja.2004.07.005
Hall, A. E. (2012). Phenotyping cowpeas for adaptation to drought. Front. Physiol. 3, 155. doi: 10.3389/fphys.2012.00155
Herridge, D., Gemell, G., and Hartley, E. (2002). Legume inoculants and quality control. ACIAR Proc. 109e, 105–115. Available online at: https://www.researchgate.net/profile/David-Herridge/publication/242302221_Legume_Inoculants_and_Quality_Control/links/0a85e52e8327d092e2000000/Legume-Inoculants-and-Quality-Control.pdf (accessed January 19, 2022).
Herridge, D. F., Bergersen, F. J., and Peoples, M. B. (1990). Measurement of nitrogen fixation by soybean in the field using the ureide and natural 15N abundance methods. Plant Physiol. 93, 708–16. doi: 10.1104/pp.93.2.708
Herridge, D. F., and Peoples, M. B. (1990). Ureide assay for measuring nitrogen fixation by nodulated soybean calibrated by N methods. Plant Physiol. 93, 495–503. doi: 10.1104/pp.93.2.495
Herrmann, L., and Lesueur, D. (2013). Challenges of formulation and quality of biofertilizers for successful inoculation. Appl. Microbiol. Biotechnol. 97, 8859–8873. doi: 10.1007/s00253-013-5228-8
Iqbal, A., Khalil, I., Ateeq, N., and Khan, M. S. (2006). Nutritional quality of important food legumes. Food Chem. 97, 331–335. doi: 10.1016/j.foodchem.2005.05.011
Itani, J., Utsunomiya, N., and Shigenaga, S. (1992). Drought tolerance in cowpea: comparative study on water and photosynthesis among cowpea, common bean and greengram plants under water stress conditions. Japanese J. Trop. Agric. 36, 269–274.
Jarecki, M. K., and Lal, R. (2003). Crop management for soil carbon sequestration. Crit. Rev. Plant. Sci. 22, 471–502. doi: 10.1080/713608318
Kyei-Boahen, S., Savala, C. E. N., Chikoye, D., and Abaidoo, R. (2017). Growth and yield responses of cowpea to inoculation and phosphorus fertilization in different environments. Front. Plant Sci. 8, 646. doi: 10.3389/fpls.2017.00646
Lopes, M. J. D. S., Dias-Filho, M. B., and Gurgel, E. S. C. (2021). Successful plant growth-promoting microbes: inoculation methods and abiotic factors. Front. Sustain. Food Syst. 5, 48. doi: 10.3389/fsufs.2021.606454
Mapope, N., and Dakora, F. D. (2016). N2 fixation, carbon accumulation, and plant water relations in soybean (Glycine max L. Merrill) varieties sampled from farmers' fields in South Africa, measured using 15N and 13C natural abundance. Agric. Ecosyst. Environ. 221, 174–186. doi: 10.1016/j.agee.2016.01.023
Martins, L. M. V., Xavier, G. R., Rangel, F. W., Ribeiro, J. R. A., Neves, M. C. P., Morgado, L. B., et al. (2003). Contribution of biological nitrogen fixation to cowpea: a strategy for improving grain yield in the semi-arid region of Brazil. Biol. Fertil. Soils 38, 333–339. doi: 10.1007/s00374-003-0668-4
Maskey, S. L., Bhattarai, S., Peoples, M. B., and Herridge, D. F. (2001). On-farm measurements of nitrogen fixation by winter and summer legumes in the hill and terai regions of nepal. F. Crop. Res. 70, 209–221. doi: 10.1016/S0378-4290(01)00140-X
Mbah, G. C., and Dakora, F. D. (2017). Nitrate inhibition of N2 fixation and its effect on micronutrient accumulation in shoots of soybean (Glycine max L. Merr.), Bambara groundnut (Vigna subterranea L. Vedc) and Kersting's groundnut (Macrotyloma geocarpum Harms.). Symbiosis 2, 1–12. doi: 10.1007/s13199-017-0531-2
MoFA (2010). Agriculure in Ghana: Facts and figures. Ministry of Food and Agriculture (MoFA)—Statistics, Research and Information Directorate (SRID). Available online at: http://gis4agricgh.net/POLICIES/AGRICULTURE-IN-GHANA-FF-2010.pdf (accessed January 19, 2022).
MoFA (2016). Agriculture in Ghana, Facts and Figures. Ministry of Food and Agriculture (MoFA)—Statistics, Research and Information Directorate (SRID). Available online at: https://www.scribd.com/document/471493223/Agriculture-in-Ghana-Facts-and-Figures-2015-pdf (accessed January 19, 2022).
Mohale, K. C., Belane, A. K., and Dakora, F. D. (2014). Symbiotic N nutrition, C assimilation, and plant water use efficiency in Bambara groundnut (Vigna subterranea L. Verdc) grown in farmers' fields in South Africa, measured using 15N and 13C natural abundance. Biol. Fertil. Soils 50, 307–319. doi: 10.1007/s00374-013-0841-3
Mohammed, M., Jaiswal, S. K., and Dakora, F. D. (2018). Distribution and correlation between phylogeny and functional traits of cowpea (Vigna unguiculata L. Walp.)-nodulating microsymbionts from Ghana and South Africa. Sci. Rep. 8, 18006. doi: 10.1038/s41598-018-36324-0
Mokgehle, S. N., Dakora, F. D., and Mathews, C. (2014). Variation in N2 fixation and N contribution by 25 groundnut (Arachis hypogaea L.) varieties grown in different agro-ecologies, measured using 15N natural abundance. Agric. Ecosyst. Environ. 195, 161–172. doi: 10.1016/j.agee.2014.05.014
Nyemba, R. C., and Dakora, F. D. (2010). Evaluating N2 fixation by food grain legumes in farmers' fields in three agro-ecological zones of Zambia, using 15N natural abundance. Biol. Fertil. Soils 46, 461–470. doi: 10.1007/s00374-010-0451-2
Nyoki, D., and Ndakidemi, P. A. (2014). Influence of Bradyrhizobium japonicum and phosphorus on micronutrient uptake in cowpea. A case study of Zinc (Zn), Iron (Fe), Copper (Cu) and Manganese (Mn). Am. J. Plant Sci. 5, 427–435. doi: 10.4236/ajps.2014.54056
Owusu, K., and Waylen, P. R. (2013). The changing rainy season climatology of mid-Ghana. Theor. Appl. Climatol. 112, 419–430. doi: 10.1007/s00704-012-0736-5
Pausch, R. C., Mulchi, C. L., Lee, E. H., and Meisinger, J. J. (1996). Use of 13C and 15N isotopes to investigate O3 effects on C and N metabolism in soybeans. Part II. Nitrogen uptake, fixation, and partitioning. Agric. Ecosyst. Environmen 60, 61–69. https://doi.org/10.1016/S0167-8809(96)01062-6
Peoples, M. B., Brockwell, J., Herridge, D. F., Rochester, I. J., Alves, B. J. R., Urquiaga, S., et al. (2009). The contributions of nitrogen-fixing crop legumes to the productivity of agricultural systems. Symbiosis 48, 1–17. doi: 10.1007/BF03179980
Rogers, A., Gibon, Y., Stitt, M., Morgan, P. B., Bernacchi, C. J., Ort, D. R., et al. (2006). Increased C availability at elevated carbon dioxide concentration improves N assimilation in a legume. Plant Cell Environ. 29, 1651–1658. doi: 10.1111/j.1365-3040.2006.01549.x
Santos, C. A. F., Da Costa, D. C. C., Da Silva, W. R., and Boiteux, L. S. (2012). Genetic analysis of total seed protein content in two cowpea crosses. Crop Sci. 52, 2501–2506. doi: 10.2135/cropsci2011.12.0632
Shearer, G., and Kohl, D. H. (1986). N2-fixation in field settings: estimations based on natural 15N abundance. Aust. J. Plant Physiol. 13, 699–756. doi: 10.1071/PP9860699c
Shevkani, K., Kaur, A., Kumar, S., and Singh, N. (2015). Cowpea protein isolates: Functional properties and application in gluten-free rice muffins. LWT Food Sci. Technol. 63, 927–933. doi: 10.1016/j.lwt.2015.04.058
Statsoft Inc (2011). Statistica (data analysis software system). version 10. Available online at: www.statsoft.com (accessed January 19, 2022).
Tittonell, P., and Giller, K. E. (2013). When yield gaps are poverty traps: the paradigm of ecological intensification in African smallholder agriculture. F. Crop. Res. 143, 76–90. doi: 10.1016/j.fcr.2012.10.007
Ulzen, J., Abaidoo, R. C., Mensah, N. E., Masso, C., and Abdel Gadir, A. H. (2016). Bradyrhizobium inoculants enhance grain yields of soybean and cowpea in Northern Ghana. Front. Plant Sci. 7, 1770. doi: 10.3389/fpls.2016.01770
Unkovich, M., Herridge, D., Peoples, M., Cadisch, G., Boddey, B., Giller, K., et al. (2008). Measuring Plant-Associated Nitrogen Fixation in Agricultural Systems. Canberra, Australia: Australian Centre for International Agricultural Research. Available online at: https://www.aciar.gov.au/publication/books-and-manuals/measuring-plant-associated-nitrogen-fixation-agricultural-systems. (accessed January 19, 2022).
Vadez, V., Berger, J. D., Warkentin, T., Asseng, S., Ratnakumar, P., Rao, K. P. C., et al. (2012). Adaptation of grain legumes to climate change: a review. Agron. Sustain. Dev. 32, 31–44. doi: 10.1007/s13593-011-0020-6
Walkley, A., and Black, I. A. (1934). An examination of the degtjareff method for determining soil organic matter, and a proposed modification of the chromic acid titration method. Soil Sci. 37, 29–38. doi: 10.1097/00010694-193401000-00003
Westhoek, A., Field, E., Rehling, F., Mulley, G., Webb, I., Poole, P. S., et al. (2017). Policing the legume-Rhizobium symbiosis: a critical test of partner choice. Sci. Rep. 7, 1–10. doi: 10.1038/s41598-017-01634-2
Xiong, H., Shi, A., Mou, B., Qin, J., Motes, D., Lu, W., et al. (2016). Genetic diversity and population structure of cowpea (Vigna unguiculata L. Walp). PLoS ONE 11:e0160941. doi: 10.1371/journal.pone.0160941
Yemm, E., and Cocking, R. R. (1955). The determination of amino-acids with ninhydrin. Analyst 80, 209–204. doi: 10.1039/AN9558000209
Keywords: symbiosis, ureides, N-fixed, water use efficiency, δ13C
Citation: Mohammed M, Mbah GC, Sowley ENK and Dakora FD (2022) Cowpea Genotypic Variations in N2 Fixation, Water Use Efficiency (δ13C), and Grain Yield in Response to Bradyrhizobium Inoculation in the Field, Measured Using Xylem N Solutes, 15N, and 13C Natural Abundance. Front. Agron. 4:764070. doi: 10.3389/fagro.2022.764070
Received: 24 August 2021; Accepted: 03 January 2022;
Published: 10 February 2022.
Edited by:
Mohamed Farissi, Université Sultan Moulay Slimane, MoroccoReviewed by:
Yaosheng Wang, Chinese Academy of Agricultural Sciences (CAAS), ChinaMohammed Mouradi, Université Sultan Moulay Slimane, Morocco
Copyright © 2022 Mohammed, Mbah, Sowley and Dakora. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Felix D. Dakora, RGFrb3JhRkRAdHV0LmFjLnph
 Mustapha Mohammed
Mustapha Mohammed Glory C. Mbah
Glory C. Mbah Elias N. K. Sowley2
Elias N. K. Sowley2 Felix D. Dakora
Felix D. Dakora