- 1Ruakura Research Centre, The New Zealand Institute for Plant and Food Research Limited, Hamilton, New Zealand
- 2Mount Albert Research Centre, The New Zealand Institute for Plant and Food Research Limited, Auckland, New Zealand
- 3Te Puke Research Centre, The New Zealand Institute for Plant and Food Research Limited, Te Puke, New Zealand
Acibenzolar-S-methyl (ASM), the active ingredient in Actigard® (Syngenta), is a plant defence elicitor used for the management of bacterial canker caused by Pseudomonas syringae pv. actinidiae (Psa) in kiwifruit. We examined changes in gene expression and phytohormone content in two kiwifruit cultivars, ‘Hayward’ (Actinidia chinensis var. deliciosa) and ‘Zesy002’ (A. chinensis var. chinensis), following ASM application. In glasshouse studies, ASM-induced resistance to stem inoculation with Psa was accompanied by upregulation of salicylic acid (SA) defence pathway genes (PR1, PR2, and PR5) in stems and leaves. The expression of PR1 in ASM-treated plants increased by c. 5-fold in ‘Hayward’ and by over 15-fold in ‘Zesy002,’ relative to the untreated controls, whereas PR2 and PR5 increased by up to 4-fold in both cultivars. Orchard studies over three seasons confirmed the utility of PR1, PR2, and PR5 for monitoring ASM-induced responses in mature vines. PR1 and PR5 were more strongly induced by ASM than PR2 and gene upregulation in ‘Hayward,’ was accompanied by a reduction in Psa leaf spotting; no such relationship was measurable for ‘Zesy002’ because leaf spot symptoms are rarely expressed. In the third season, six additional gene candidates, BAD, Gluc2, Class IV Chit, EDS1A, NPR3, and NIMIN2, were responsive to ASM in ‘Hayward’ and ‘Zesy002.’ Gene upregulation was transient, with expression levels increasing by 1 d after ASM application and declining to control levels between 7 and 14 days. Moreover, the amplitude of gene upregulation depended on leaf developmental stage and was greater in the first true leaf and the youngest mature leaves than in immature leaves along the same shoot. Phytohormone content did not show a repeatable response pattern to ASM in potted plants or in vines possibly as a consequence of their wider role in regulating plant growth and mediating environmental responses. In conclusion, this study demonstrates that defence gene expression can be used to monitor responsiveness to ASM in two genetically distinct mature kiwifruit cultivars (‘Hayward’ and ‘Zesy002’) under orchard conditions. The use of defence marker genes could be of broader utility across kiwifruit species and could be used to guide ASM application schedules in the orchard.
Introduction
Acibenzolar-S-methyl (ASM) is the active ingredient in the commercially available elicitor product Actigard®/Bion® (Syngenta Crop Protection, Basel, Switzerland) and is arguably the best known and best characterised plant defence elicitor (Gorlach et al., 1996; Vallad and Goodman, 2004; Faize and Faize, 2018). ASM is used in kiwifruit orchards to protect kiwifruit against infection by Pseudomonas syringae pv. actinidiae (Psa), the most important pathogen of kiwifruit around the world (Vanneste, 2012). Five Psa biovars can be distinguished within the pathovar actinidiae but the recent global outbreak was caused only by strains belonging to biovar 3 (Chapman et al., 2012; McCann et al., 2013; Vanneste et al., 2013). The ability of Psa to survive as an epiphyte and to infect through natural openings and wounds (e.g., stomata, broken trichomes, lenticels, and leaf abscission scars) (Donati et al., 2020) makes it difficult to control. Resistance to Psa in kiwifruit is mediated by the salicylic acid (SA)-responsive defence pathway (Cellini et al., 2014; Wang et al., 2017, 2018; Wurms K. et al., 2017; Wurms K. V. et al., 2017; Michelotti et al., 2018; Song et al., 2019), and ASM, a functional mimic of SA (Tripathi et al., 2019), has been shown to enhance resistance to Psa infection (Reglinski et al., 2013; Cellini et al., 2014; De Jong et al., 2019).
Plants orchestrate their response to different pests and pathogens via interconnected and often antagonistic hormonal signalling pathways and gene regulatory networks (Berens et al., 2017; Chen et al., 2020). In general, resistance to biotrophs is mediated primarily via the SA-responsive pathway, whilst resistance to necrotrophs is directed by the jasmonic acid (JA)-responsive pathway (Pieterse et al., 2012). The intracellular accumulation of SA is important for establishment of systemic acquired resistance (SAR) because it alters cellular redox thus resulting in the nuclear migration of the regulatory protein Non-expressor of Pathogenesis-Related 1 (NPR1) and the subsequent transcription of pathogenesis related (PR) genes (Mou et al., 2003). Moreover, NPR3 and NPR4 have been reported to regulate NPR1-dependent gene expression in a SA concentration dependent manner (Ding et al., 2018; Backer et al., 2019). ASM has been shown to operate at the site of or downstream of SA accumulation (Tripathi et al., 2010) and to induce transcriptional changes typically associated with SA-mediated SAR including the expression of classical SAR marker genes pathogenesis-related 1 (PR1), PR2, and PR5 (Friedrich et al., 1996; Lawton et al., 1996; Durrant and Dong, 2004; Tripathi et al., 2010). Defence genes can be induced directly by ASM or may be “primed” for enhanced expression upon subsequent pathogenic attack (Tripathi et al., 2019).
Advances in our understanding of defence elicitation has been accompanied by heightened interest in the practical implementation of induced resistance for plant disease management (Reglinski et al., 2014). However, plant defence elicitors have shown variable field efficacy and greater understanding of factors affecting elicitor responsiveness in field conditions is lacking. To date, studies on ASM-induced kiwifruit defence responses to Psa have been carried out in tightly controlled environments and generally on small whole plants or detached plant parts (Wang et al., 2017, 2018; Wurms K. et al., 2017; Wurms K. V. et al., 2017; Michelotti et al., 2018; De Jong et al., 2019; Song et al., 2019). This study spanned the laboratory to the field and investigated the use of biochemical (phytohormones) and molecular (defence genes) markers of defence elicitation in kiwifruit to monitor ASM-induced responses over time in potted kiwifruit plants in controlled glasshouse conditions and in mature vines in the highly dynamic orchard environment.
The response to ASM was examined in the green fleshed kiwifruit cultivar Actinidia chinensis var. deliciosa ‘Hayward’ and in the yellow fleshed cultivar A. chinensis var. chinensis ‘Zesy002,’ also known as Gold3. Those two cultivars are the economically most important cultivars in New Zealand and show a different level of resistance to Psa (Vanneste, 2017). In contrast to ‘Hayward,’ leaf spots caused by Psa are only rarely detected on ‘Zesy002’; often the first visible symptoms on this cultivar are shoot or cane dieback, symptoms which are relatively rare on ‘Hayward’ (Vanneste, 2017). The ability to monitor the onset of ASM-induced defence responses in kiwifruit in the orchard has potential to better inform its use in disease management.
Materials and Methods
Glasshouse Trials
Plant Material
Clonal Actinidia chinensis var. deliciosa ‘Hayward’ and A. chinensis var. chinensis ‘Zesy002’ tissue cultured plantlets (Multiflora, Auckland, New Zealand) were individually transplanted from an agar growth medium in sealed plastic tubs to 0.5-L planter bags filled to two thirds with Daltons™ GB mix (Daltons, Matamata, New Zealand) and topped up with a 50:50 ratio mix of potting mix and perlite. The plantlets were placed in a glasshouse under clear plastic tents to maintain humidity and with supplementary heating for the first 2 weeks of growth. Plantlets were further grown in normal glasshouse conditions (15–24°C, 14 h day length) to approximately 30 cm tall, with at least three to four fully expanded leaves when used for experiments. A flood and drain system was used once daily to water the plants.
Treatment With Actigard
Actigard® (acibenzolar-S-methyl [ASM], Syngenta, Auckland, New Zealand) was prepared in de-ionised water at a concentration of 0.2 g/L and applied to potted kiwifruit plants a using a handheld mist sprayer. Untreated control plants were sprayed with sterile de-ionised water (SDW). After treatment, plants were kept in the glasshouse for 6 days until transfer to a quarantine glasshouse for inoculation studies. Pathogen inoculation was performed 1 week after ASM and SDW treatment as described below.
Inoculum Preparation
Pseudomonas syringae pv. actinidiae biovar 3 (Psa), strain 10627, (Vanneste et al., 2013) was grown on King's B medium (King et al., 1954), and incubated at 28°C for at least 48 h. Bacterial colonies were re-suspended in sterile de-ionised water, and then the cell density was estimated using a NovoSpec™ spectrophotometer (Amersham Biosciences, Auckland, New Zealand) (OD = 600 nm). The bacterial suspension was adjusted to give a final concentration of c. 1 × 108 colony forming units per mL(CFU/mL), this was confirmed by plating 1/10th dilutions on King's B medium and counting the number of colonies after 48 h incubation at 28°C.
Plants in the glasshouse trials were inoculated by a single stab to the stem with a sterile needle to a depth of 2–3 mm, at a single point midway between the leaf petiole of the third and fourth unfurled leaf, followed immediately by pipetting 2 μL of Psa suspension into the wound. For mock inoculations, sterile de-ionised water was pipetted instead of inoculum. Inoculated plants were placed into high-humidity tents (>90%) for 48 h, after which the relative humidity was reduced by opening vents on top of the tents. The ambient temperature in the glasshouse ranged from 16 to 28°C and the temperature inside the tents ranged from 20 to 30°C.
Disease Assessments
Disease severity was assessed at between 13 and 28 days post inoculation by measuring the length (mm) of the water soaked lesion extending from the stem inoculation point. There were 12 plants per treatment in the ‘Hayward’ experiment and 20 plants per treatment in the ‘Zesy002’ experiment. A subset of the mock inoculated untreated controls (n = 6, Hayward; n = 8 ‘Zesy002’) were also assessed to confirm that symptom expression was associated with Psa inoculation.
Tissue Sampling for Phytohormone and Gene Expression Analysis
Leaf and stem tissue samples were taken immediately before Psa inoculation (T = 0) and at 3, 6, 24, and 48 h after inoculation. The upper leaf sampled was the fully expanded mature leaf immediately above the inoculation point. The mid-ribs of the leaves were removed before the remainder of the tissue was taken. Stem tissue for gene expression analysis was taken by cutting through the stem 0.5 cm above and below the inoculation point. A 1-cm stem section directly above this, was taken for phytohormone analysis. All tissue was snap frozen in liquid nitrogen and then stored at −80°C. There were four replicates (individual plants) per time point for each treatment. All four replicates of upper leaf and stem samples were processed for phytohormone analysis whilst three of the four replicates were processed for gene expression analysis.
Orchard Trials
Orchard trials were conducted on ‘Hayward’ and ‘Zesy002’ vines at Ruakura Research Orchard (Hamilton, New Zealand) in 2016, 2017, and 2018. Vines were pergola trained with a single vine per bay. Actigard (0.2 g/L in tap water, 1,000 L/ha) was applied using a 5-L Maxi Plus pressurised sprayer fitted with a hollow cone nozzle. There were six single-vine replicates per treatment in the ‘Hayward’ trial in 2016 and seven single-vine replicates in 2017 and 2018. For ‘Zesy002,’ there were nine single-vine replicates in 2016 and five two-vine replicates in 2017 and 2018. The treatment application dates and leaf tissue sampling schedule are outlined in Table 1.
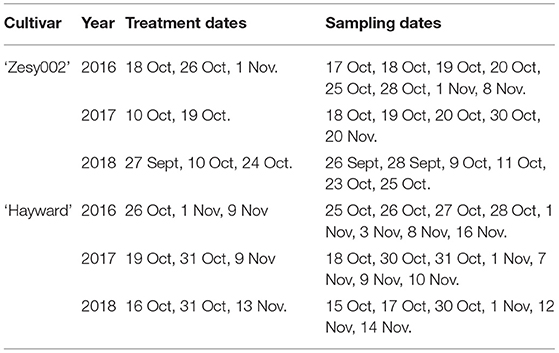
Table 1. Treatment dates and sampling dates for the orchard trials conducted on Actinidia chinensis var deliciosa ‘Hayward’ and A. chinensis var chinensis ‘Zesy002’ in 2016, 2017, and 2018.
Disease Assessment
The incidence and severity of Psa leaf spot necrosis in ‘Hayward’ vines was measured by visually estimating the percentage area of leaf necrosis on each of 100 leaves per replicate in 2016 and 2017, and 150 leaves per replicate in 2018. Assessment dates were 14 Nov, 2016, 10 Nov, 2017, and 16 Nov 2018. Psa leaf spotting is not strongly expressed in ‘Zesy002’ and therefore no disease data was recorded.
Leaf Sampling for Phytohormone and Gene Expression Analysis
The first true leaf was sampled from four fruiting shoots per vine. Four discs (12 mm diameter) were cut from each leaf using a cork borer and the samples were pooled by replicate vine before being snap frozen in liquid nitrogen and stored at −80°C. For phytohormone analysis in ‘Hayward’ there were six replicates in 2016 and seven in 2017 and 2018. For gene expression there were six replicates in 2016, four in 2017, and five in 2018. For ‘Zesy002’ there were nine replicates in 2016 and five replicates in 2017 and 2018 for both phytohormone and gene expression analysis. In 2018, additional leaf samples were taken at 1 d after spray 3 in order to examine effects of leaf maturity on responsiveness to ASM. Three developmentally different leaves were sampled along the length of a fruiting shoot; the first true leaf, the youngest fully expanded leaf and a newly emerged leaf. These represent leaves from the base, the middle and the tip of the growing shoot. There were four shoots sampled per replicate vine (n = 5) and leaves were pooled by leaf type.
Phytohormone Analysis
The samples from the potted plant experiments (100 mg fresh weight) and from the 2016 orchard trials (25 mg freeze dried tissue) were each extracted with 1 mL of chilled (4°C) 80:20 acetonitrile:water to which internal standards had been added (2.5 ng [2H4] SA, 12.5 ng [2H5] JA, and 6.25 ng [2H6] ABA). The samples were shaken for 30 min at 4°C on a flat-bed orbital shaker, extracted overnight at −20°C and then centrifuged at 13,000 g for 10 min at 4°C. An 800-μL aliquot of each sample was transferred to a 96-well deep plate and evaporated to dryness in a refrigerated CentriVap vacuum concentrator at −4°C (Labconco, Kansas City, MO, USA). Samples were reconstituted in 200 μL of chilled (4°C) 80:20 acetonitrile:water and then made basic with the addition of 1.8 mL of 0.67% ammonium hydroxide (aq) prior to SPE clean-up on a SOLA SAX 96-well plate (10 mg/2 mL; Thermo Scientific, CA, USA). Plates were activated using 2x 1.5 mL acetonitrile and equilibrated using 2x 1.5 mL 0.67% ammonium hydroxide (aq). After equilibration, samples were loaded and washed with 1.5 mL of 0.67% ammonium hydroxide (aq) followed by 1.3 mL of methanol. Acidic plant hormones were eluted with 600 μL 3.77% formic acid in acetonitrile and evaporated to dryness in a refrigerated centrivap vacuum concentrator at −4°C. Samples were reconstituted in 200 μL of 10:90 acetonitrile:water for analysis by Liquid Chromatography Mass Spectrometry (LCMS). The 2017 and 2018 orchard samples were extracted using a graphitised carbon solid phase extraction (SPE)-multiwell plate clean-up instead of an anion exchange SPE-multiwell plate to improve the retention and recovery of more polar phytohormones such as salicylic acid glycoside (SAG) and 12-hydroxyjasmonic acid (12-OH JA). The samples, 25 mg freeze-dried ground tissue (DW), were weighed into 1.5-mL Eppendorf tubes on dry ice, and to each was added 1 mL chilled (4°C) extraction solvent (acetonitrile + 0.01% TFA), labelled internal standard mix [2.5 ng [2H4] SA, 25 ng [2H5] JA, 6.4 ng [2H6] ABA, 0.6 ng [2H10] JA-ile (2018 season only)] and 0.8 g stainless steel beads 0.9–2 mm (Next Advance Inc., NY, USA). Samples were prepared as described by Bulley et al. (2021) and were reconstituted in 200 μL of 10:90 acetonitrile:water for analysis by LC-MS. Reagents used for LCMS are described in Supplementary Table 1.
LCMS Analysis
LC-MS/MS experiments were performed on a 5500 QTrap triple quadrupole/linear ion trap (QqLIT) mass spectrometer equipped with a TurboIon-Spray™ interface (AB Sciex, ON, Canada) coupled to an Ultimate 3000 UHPLC (Dionex, CA, USA). Plant hormones were separated on a Poroshell 120 SB-C18 2.7 μm 2.1 × 150 mm ID column (Agilent Technologies, CA, USA) maintained at 60°C. Solvents were (A) water + 0.1% formic acid and (B) acetonitrile + 0.1% formic acid and the flow rate was 400 μL min-1. The initial mobile phase, 2% B, was held for 3 min before ramping linearly to 16% B at 3.5 min, then to 100% B at 7 min and holding at 100% B until 8 min before resetting to the original conditions. Injection size was 10 μL. MS data were acquired in the negative mode using a multiple reaction monitoring (MRM) method with optimised Q1 and Q3 transitions for each of the analysed acidic plant hormones (Supplementary Table 2). Other operating parameters were as follows: dwell time, 10 ms; ionspray voltage, −4,500 V; temperature, 600°C; curtain gas, 45 psi; ion source gas 1, 60 psi; ion source gas 2, 60 psi. All data were analysed and processed using Analyst version 1.6.2 and MultiQuant version 3.0 software packages. Concentrations were calculated on the basis of the peak area for the endogenous compounds relative to those determined for the internal standards.
Gene Expression Analysis
Real-Time Quantitative PCR (qPCR)
Tissue samples from the potted plants in the glasshouse and the orchard vines in 2016 and 2017 were ground by mortar and pestle using liquid nitrogen. All replicates were processed from the glasshouse trials, six replicates from the 2016 orchard trial and four replicates from the 2017 orchard trial. Total RNA was extracted from ~100 mg of ground tissue using the Spectrum Plant Total RNA Kit (Sigma-Aldrich, Auckland, New Zealand) following the supplier's recommendations. RNA concentration and purity were determined using a NanoDrop 200c spectrophotometer (Thermo Scientific, Waltham, MA, USA). To remove any contaminant genomic DNA (gDNA), 1 μg of RNA was treated using a Quanta PerfeCTa DNase 1 kit (DNature, Gisborne, New Zealand) according to the manufacturer's instructions. Absence of gDNA in the samples was checked by qPCR, before first strand cDNA was synthesised from 1 μg of DNAse-treated RNA using a Quanta qScript cDNA SuperMix kit (DNature, Gisborne, New Zealand) according to the manufacturer's recommendations.
The resultant cDNA was diluted before use in qPCR reactions. The optimal cDNA dilution factor was determined by testing different cDNA dilutions on a subset of samples ranging from low to high enzyme activity, and selecting the dilution that produced a doubling of DNA at each cycle, regardless of the amount of enzyme activity. For the ‘Hayward’ and ‘Zesy002’ glasshouse trials, the optimal dilution was 25-fold in nuclease-free water, and for the ‘Hayward’ orchard trials the dilution factor was 10-fold.
Reference genes (RGs) were selected based on previous studies (Wurms et al., 2011; Cellini et al., 2014; Petriccione et al., 2015; Wurms K. V. et al., 2017): actin; ubiquitin-conjugating enzyme (UBC1); protein phosphatase 2A (PP2A), elongation factor (EF); glyceraldehyde 3-phosphate dehydrogenase (GAPDH); 40S ribosomal protein (40S); and β-tubulin. To select the most stably expressed reference genes under the specific conditions of each experiment, a randomised subset of samples from each experiment was tested by qPCR against these RGs, and the two most stable RGs used for normalisation in each trial were selected using geNorm software, version 3.4. The most stably-expressed RGs were: 40s and UBC1 for the ‘Hayward’ glasshouse trial; GAPDH and UBC1 for the ‘Zesy002’ glasshouse trial; actin and UBC1 for the ‘Hayward’ orchard trial 2016; and 40S and PP2A for the ‘Hayward’ orchard trial 2017. A gene expression normalisation factor (N) for each sample, calculated using geNorm and based on the geometric mean of the RGs, was then used for calculation of the relative expression of each gene of interest (GoI), as described by Vandesompele et al. (2002).
GoIs were chosen based on their involvement in induced kiwifruit resistance to Psa as shown by other qPCR studies, proteomic- and transcriptomic-studies and/or because they were reliable markers of different hormonal pathways (Table 2).
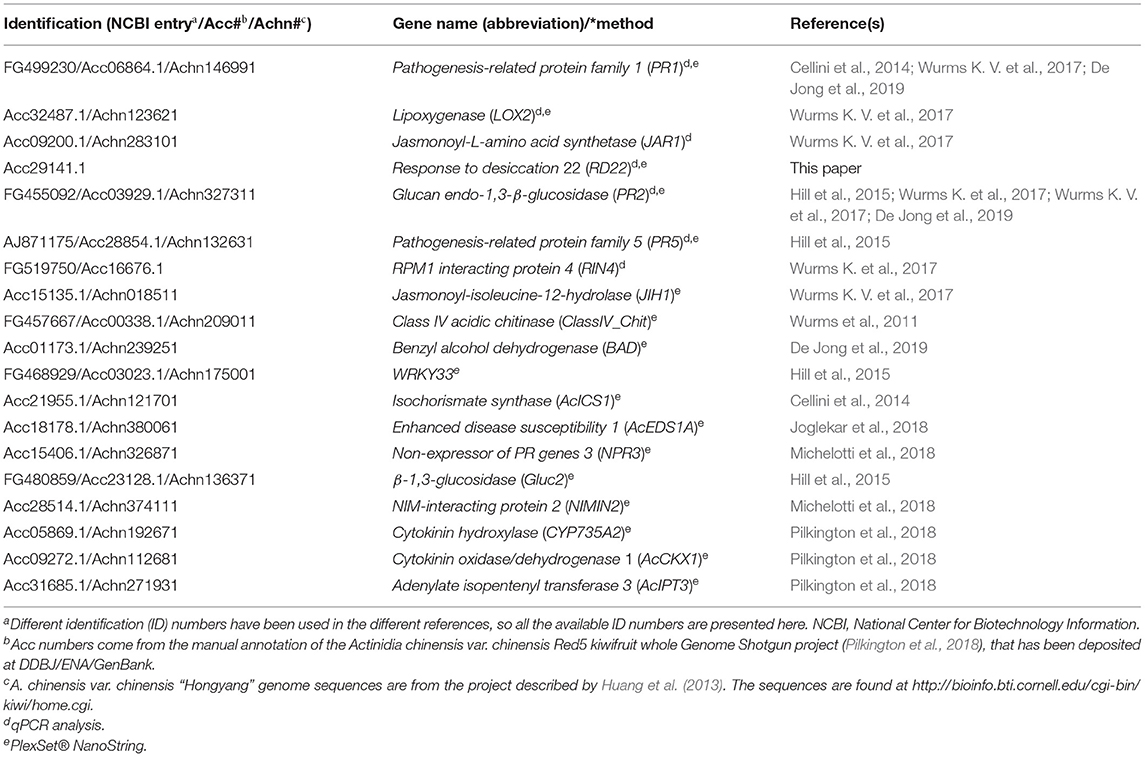
Table 2. Genes of interest for analysis of defence expression by quantitative polymerase chain reaction (qPCR) and by PlexSet® NanoString.
The primers for RG and GoI were designed in-house using Primer3 software (The Whitehead Institute, Cambridge, MA, USA) and were synthesised by Invitrogen (Auckland, New Zealand), except for: actin and PR1, which came from the work of Cellini et al. (2014); PP2A, which was from Nardozza et al. (2015); and LOX2 (Erin Stroud, Plant & Food Research, unpublished data). Primer sequences are shown in Supplementary Table 3. Only primer pairs with efficiencies of 80% or greater were used in the experiment.
Quantitative PCR analysis was performed in triplicate, in a 10 μL reaction volume containing 1 μL of cDNA, 1 μL each of forward and reverse primers (10 μM), 2 μL nuclease-free water and 5 μL of Light Cycler® 480 SYBR Green 1 Master Mix (Roche Diagnostics GmbH, Mannheim, Germany), on a 2-plex Qiagen RotorGene™ 3000. The relative quantification thermal cycling conditions were: denaturation at 95°C for 10 min, followed by 40 cycles of 15 s denaturation at 95°C, 15 s annealing at a different optimised temperature between 55 and 60°C for each primer set, and 20 s extension at 72°C. Inter-run variability was controlled by including a complete set of treatments on each plate, but a separate run for each biological replicate, which were then averaged. Melting curve analysis (60–95°C at 1°C increments with 5 s between each step) was performed after the final qPCR cycle to validate amplicon specificity. Non-template controls were also included in each qPCR run to assess the purity of the reagents.
The relative expression of each GoI was normalised relative to the RGs. The significance of differences between treatments was determined by analysis of variance (ANOVA) (see below).
Orchard Trials—‘Zesy002’ (2016, 2017)
Total RNA was isolated and gDNA removed as described previously for the glasshouse trials. Absence of DNA in the samples was checked by qPCR using primers specific for one of the genes used as a reference gene: GAPDH, actin or 40S rRNA. If no amplicon was obtained, or if the Cq value was over 35, first strand cDNA was synthesised using qScript cDNA SuperMix (Quanta BioSciences) according to the manufacturer's recommendations.
Dilutions of pooled samples were used to find a cDNA concentration which gave a Cq value around 20 when using one of the three RGs. Amplification efficiency was obtained directly from the machine Eco Real-Time PCR System (Illumina) or was calculated as the slope of the regression between the log values and the average Cq values for a range of dilutions.
All qPCR reactions were done in duplicate using PerfeCTa SYBR® Green FastMix (Quanta bio) in an Eco Real-Time PCR System (Illumina). The three genes GAPDH, actin and 40S rRNA gene were analysed as potential RGs for each of the experiments, with 40S rRNA and GAPDH being the two most stable RGs in all cases. Gene expression was determined as described for the glasshouse trials.
Gene Expression Analysis by PlexSet® NanoString
The most stable RGs and the best differentially expressed GoIs from the qPCR experiments were also used for NanoString (Table 2). Additional GoIs were selected on the basis of results from NanoString experiments on the kiwifruit response to Psa, and to represent as many different defence response pathways and temporal stages of defence as possible (Table 2). Oligomer design for the RG and GoI probes was carried out by NanoString Technologies Inc. (Seattle, WA, United States of America), and the oligomers were designed to be highly specific to the target sequence. The oligomer probes were synthesised by Integrated DNA Technologies Private Limited (IDT, Singapore) and comprise a reporter sequence, a capture sequence, plus probes A + B for 24 gene targets (including six reference genes) that hybridise to the target sequences of interest (Supplementary Table 4), forming a tripartite complex.
Sample Preparation
The total RNA from 100 mg samples of ground kiwifruit tissue was extracted, and the concentration and purity were assessed, as described above for qPCR analysis. The RNA samples were sent on dry ice to Grafton Clinical Genomics (University of Auckland, Auckland, New Zealand), where RNA quality and concentrations were checked using a PipeJet® Nanodispenser (BioFluidix GmbH, Freiburg, Germany) and then working aliquots of 50 ng/μL were prepared with nuclease free water and stored at –80°C.
Titration Analysis to Determine the Optimal RNA Input (ng)
To determine the optimum RNA input for the PlexSet, a titration was run using three samples: two samples with expected high expression and one “Mix” sample (a pool of all samples to act as a calibrator for normalisation across different sets of barcodes). Each sample was run at different total RNA input amounts using the titration-24 kits. After an overnight hybridisation of 18 h at 67°C, total barcode counts were calculated and graphed on a scatter plot. The trend line equation was used to determine the optimal RNA input (ng) for 150,000 total counts, this being the optimum level as per NanoString recommendations (Nanostring Technologies Inc, 2017). The resulting optimal RNA input (1,300 ng) was used in the full PlexSet run.
PlexSet Run
For the PlexSet run, the 15 μL reaction volume in each well-contained 5 μL hybridisation buffer, 0.5 μL working probe A (0.6 nM each of Probe As), 0.5 μL working probe B (3 nM each of Probe Bs), 2 μL of the appropriate Plexset (A-H), and 7 μL of sample RNA at 185.7 ng/μL (optimal RNA input was determined by titration analysis). The plate was hybridised in the thermocycler at 67°C for 19 h.
Data Analysis
The results were analysed using the nSolver® software (version 4.0) provided by NanoString Technologies Inc (Seattle, WA, USA). Normalised data were then statistically analysed, as described for qPCR results.
Statistical Analysis
Glasshouse Trials
Separate analyses were carried out for ‘Hayward’ and ‘Zesy002’ cultivars, and for the two sources of plant tissue, stem and upper leaf. Analysis of variance (ANOVA) was carried out for phytohormone content and for gene expression for Treatment, Inoculation and Time. The blocking factor was replicate (four for hormones, three for gene expression).
The data for all the variables were log-transformed, except for ‘Zesy002’/Stem JAR1 and LOX2 which were square root transformed, to ensure normality and homogeneity to meet the assumptions of ANOVA.
Disease assessment data were analysed as a randomised block design (RBD) by ANOVA, with data transformation when necessary to satisfy the assumptions of ANOVA (normal distribution and homogeneity of variance) and means separation by Fisher's Least Significant Difference (P ≤ 0.05).
All analyses were all conducted using GenStat 20th edition (VSN International Limited, United Kingdom (https://genstat.kb.vsni.co.uk/).
Orchard Trials
Separate analyses were carried out for ‘Hayward’ and ‘Zesy002’ cultivars, and for each of the 3 years. The data for all the variables were log-transformed when required to meet the assumptions of ANOVA of normality and homogeneity. The analyses were conducted using GenStat 20th edition with the exception of the phytohormone content data for 2016 and 2017 which were analysed in GenStat 17th edition (VSN International).
Linear mixed models (via ReML) was employed to analyse effects of ASM and time on phytohormone content in 2016 and 2017. ANOVA was carried out to determine the effects of ASM and time on gene expression. For ‘Hayward,’ there were six replicates in 2016 and four in 2017 and for ‘Zesy002’ there were nine replicates in 2016 and five replicates in 2017. In 2018, ANOVA was carried out to determine effect of ASM and time on phytohormone content and gene expression. There were seven replicates for ‘Hayward’ and five for ‘Zesy002.’ Contrast factors were used to compare ASM with the control at each time. ANOVA was also carried out for gene expression and phytohormone content treatment (ASM vs. Control), and leaf type (first true leaf, youngest mature leaf, and immature leaf) after spray 3 only.
Disease assessment data for ‘Hayward’ were assessed using hierarchical generalised linear models to test the incidence and severity of leaf necrosis for the effect of treatment (Control vs. ASM), separately for each year. The random effect was replicate. The analysis was carried out in Genstat 21st edition.
Results
ASM Induces Resistance to Psa Stem Infection and Alters Phytohormone and Gene Expression Patterns in Potted Kiwifruit
Untreated ‘Hayward’ plants developed larger stem lesions than ‘Zesy002’ plants following inoculation with Psa (Table 3). Spray application with ASM, 1 week before stem inoculation, increased resistance to Psa infection as expressed by smaller stem lesions in ASM plants than in controls; lesion length was reduced by ~60% in ‘Hayward’ and by ~45% in ‘Zesy002’ (Table 3). Tissue from the stem inoculation site and the adjacent leaf were analysed to compare changes in phytohormone content and defence gene expression between control and ASM treated plants after inoculation with Psa. The concentrations of ABA, SA, JA, and/or JA-ile were significantly affected by ASM and, in ‘Hayward’ in particular, changed over time (Supplementary Figures 1, 2, and Supplementary Table 5). However, there was no significant interaction between ASM and Time and therefore data showing the effect of ASM alone is presented in Table 4. ASM induced an increase in ABA concentration in both cultivars (Table 4) with the greatest and most consistent response seen in ‘Hayward’ leaves where ABA increased by up to 4-fold compared with controls (Supplementary Figure 1). ASM also induced a reduction in JA but an increase in JA-ile in ‘Hayward’ leaves, and a reduction in SA in ‘Hayward’ stems. In ‘Zesy002,’ JA in stems was higher in ASM-treated plants than in the control (Supplementary Figure 2).
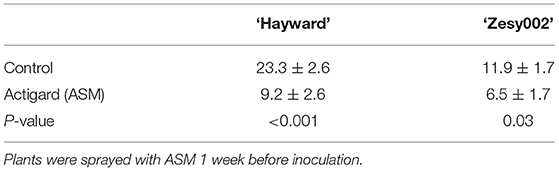
Table 3. Stem lesion length ±SE (mm) in Actinidia chinensis var. deliciosa ‘Hayward’ and A. chinensis var. chinensis ‘Zesy002’ at 21 days after inoculation with Pseudomonas syringae pv. actinidiae.
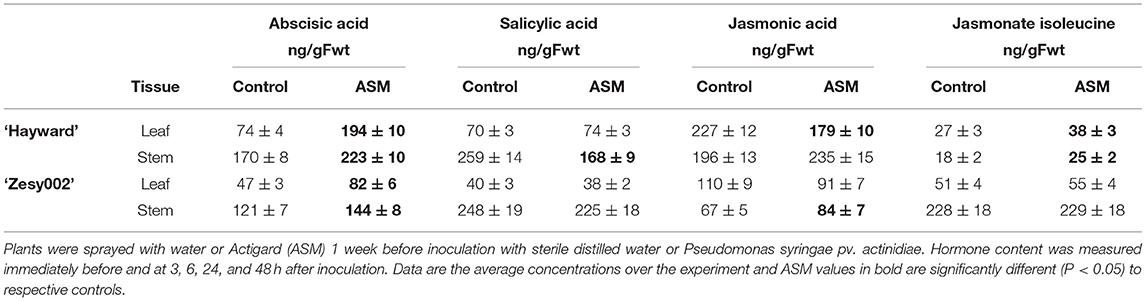
Table 4. Effect of Actigard (ASM) on the concentrations (ng/g Fwt) of salicylic acid (SA), abscisic acid (ABA), jasmonic acid (JA), and jasmonate isoleucine (JA-ile) at the stem inoculation site and in the adjacent leaf in Actinidia chinensis var. deliciosa ‘Hayward’ and A. chinensis var. chinensis ‘Zesy002’ potted plants.
The expression patterns for seven genes (PR1, PR2, PR5, JAR1, LOX2, RD22, and RIN4) were evaluated. The expression of PR1, PR2, and PR5 were the most strongly upregulated by ASM in both cultivars, compared with untreated controls (Figure 1). One week after ASM treatment (T0) PR1 expression was ~5-fold higher in ‘Hayward’ and over 15-fold higher in ‘Zesy002’ in ASM-treated plants than in untreated controls whilst PR5 was 4-fold higher in ‘Hayward’ stems and 3-fold and 5-fold higher, respectively, in the leaves and stems of ‘Zesy002.’ The upregulation of PR2 by ASM at T0, relative to controls, was significant only in ‘Zesy002’ stems (4-fold increase). After inoculation, the expression of PR1 and PR5 increased by up to 20-fold, and PR2 by up to 6-fold in the leaves of ASM-treated plants, when compared with controls. Moreover, gene upregulation was independent of the inoculum (i.e., SDW and Psa) suggesting that ASM may have “primed” the plants to enable faster response to stem wounding itself and/or the associated environmental change whereby inoculated plants were placed under high humidity to favour infection. None of the other genes showed a strong or repeatable response to ASM (Supplementary Figures 3, 4).
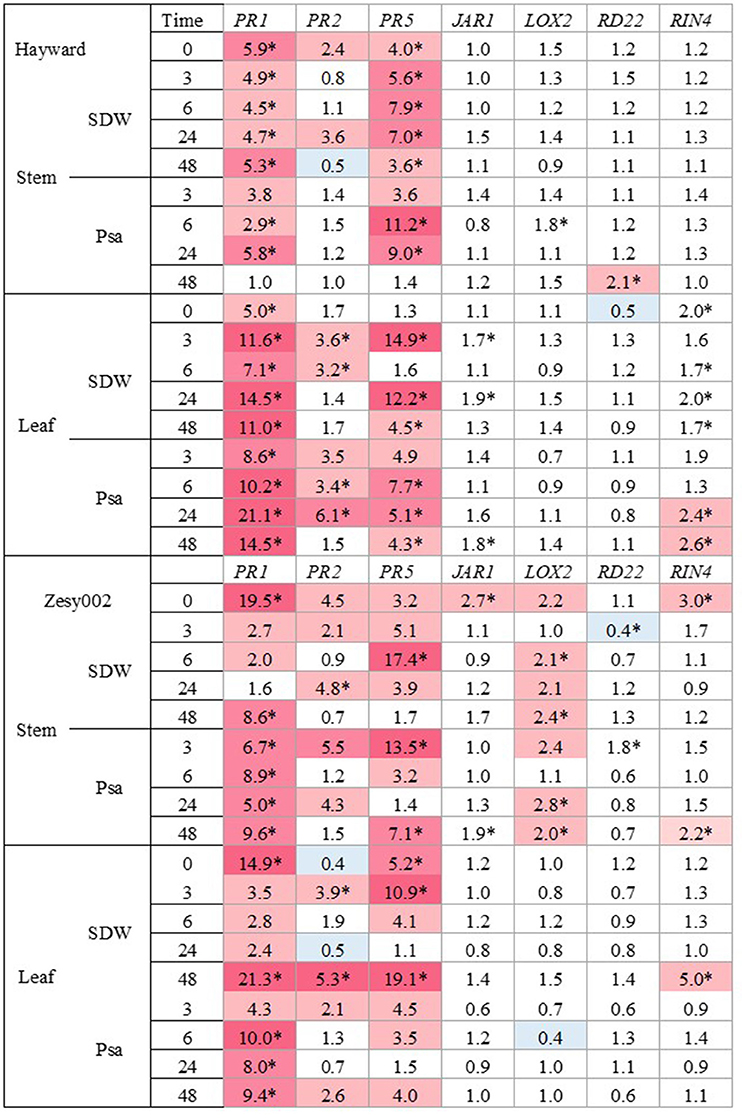
Figure 1. Fold-change in gene expression induced by Actigard at the stem inoculation site and in the adjacent leaf in glasshouse-grown Actinidia chinensis var. deliciosa ‘Hayward’ plants and A. chinensis var. chinensis ‘Zesy002’ plants before (T = 0 h) and at 3, 6, 24, and 48 h after inoculation with sterile distilled water (SDW) or a suspension of Pseudomonas syringae pv. actinidiae (Psa). Plants were sprayed with water (Con) or with 0.2 g/L Actigard (ASM) 1 week before inoculation. Gene expression was quantified by quantitative PCR (qPCR) using 40S rRNA (40S) and ubiquitin-conjugating enzyme (UBC1) as the reference genes. The analysis was performed at the log2 scale and the heat map shows the relative ratio of transcript levels between Actigard-treated plants and the control. Significant differences at individual time points are identified with an asterisk (p < 0.05).
Pre-flowering Applications of ASM Reduce Leaf Spotting Caused by Psa in ‘Hayward’ Vines
Spray application of ASM during the pre-flowering period resulted in a significant reduction in the incidence and severity of leaf spotting caused by Psa in ‘Hayward’ vines in the orchard (Table 5). The incidence of leaf spotting was reduced by ~40% and the severity of infection (% leaf area affected) by ~50% in each year of the study. No measurable leaf spotting was observed in ‘Zesy002’ (data not shown).
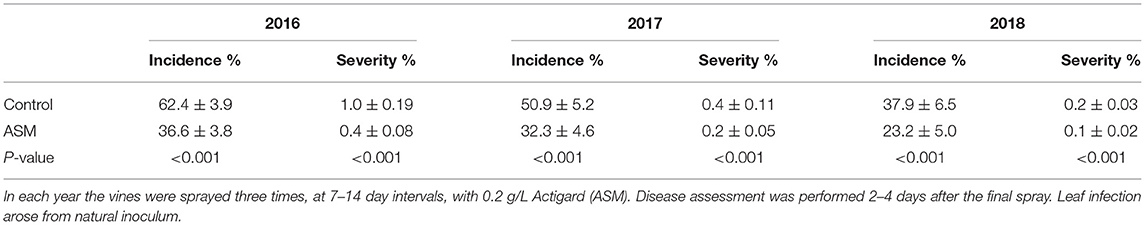
Table 5. Incidence and severity of leaf spotting caused by Pseudomonas syringae pv. actinidiae in mature Actinidia chinensis var. deliciosa ‘Hayward’ vines at Ruakura Research Orchard.
Phytohormones Are Differentially Affected by ASM in ‘Hayward’ and ‘Zesy002’ Vines
Treatment with ASM did not significantly affect leaf hormone content in ‘Hayward’ vines in 2016 (Supplementary Figure 5 and Supplementary Table 6) or 2017 (Supplementary Figure 6 and Supplementary Table 6). Leaf phytohormone content differed significantly over time, suggesting a developmental response, but there was no significant ASM x time interaction (Supplementary Table 6). In 2018, no significant difference in the phytohormonal content of the first true leaf was found between the control and ASM–treated vines at 1 day after the last of three ASM sprays (Supplementary Tables 6, 7).
In ‘Zesy002’ vines, leaf concentrations of SA and ABA differed significantly between ASM–treated vines and the control in 2016 (Supplementary Table 8). SA was lower in ASM-treated vines than in the control at 7 d after spray 1 (p < 0.05), 2 d after spray 2 (p < 0.01), and 7 d after spray 3 (p < 0.01; Supplementary Figure 7). Conversely, ABA concentration was significantly greater (p < 0.001) in leaves from ASM vines than in controls at 1 d and 2 d after treatment (Supplementary Figure 7). Thereafter, ABA concentration did not differ significantly between ASM and the control. In 2017, there was no significant difference in hormone concentrations between ASM and control vines at any of the sampling time points (Supplementary Figure 8 and Supplementary Table 8). In 2018, ASM significantly affected the concentrations of ABA and SAG (Supplementary Table 8); ABA was greater (p < 0.05) in ASM vines than in the control at 1 d after spray 1 and SAG was significantly lower (p < 0.05) in ASM vines than in the control at 13 d after spray 1 (Supplementary Figure 9).
ASM Induces Transient Changes in Gene Expression in ‘Hayward’ and ‘Zesy002’
Pre-flowering spray applications with ASM resulted in significant upregulation of PR1, PR2, and PR5 in leaves in ‘Hayward’ and ‘Zesy002’ vines in 2016 (Figure 2) and 2017 (Figure 3). In 2016, the strongest upregulation occurred at 2 d after the first of three ASM applications in ‘Hayward’ and at 6 d after spray 2 in Zesy002; expression of PR1 and PR5 increased by up to 5- and 8-fold, respectively in ‘Zesy002’ and by 4- and 2-fold, respectively in ‘Hayward.’ PR2 increased by up to 3-fold in ‘Zesy002’ and up to 4.6-fold in ‘Hayward.’ In 2017, three pre-flowering ASM sprays were applied in ‘Hayward’ but only two sprays in ‘Zesy002’ because of adverse weather conditions during spring. Relative to controls the expression of PR1 and PR5 in ASM-treated vines was increased by up to 5- and 3.5-fold, respectively in ‘Zesy002’ and by 3- and 2-fold, respectively in ‘Hayward’ (Figure 3). PR2 increased by up to 3-fold in ‘Zesy002’ and 4-fold in ‘Hayward’ (Figure 3).
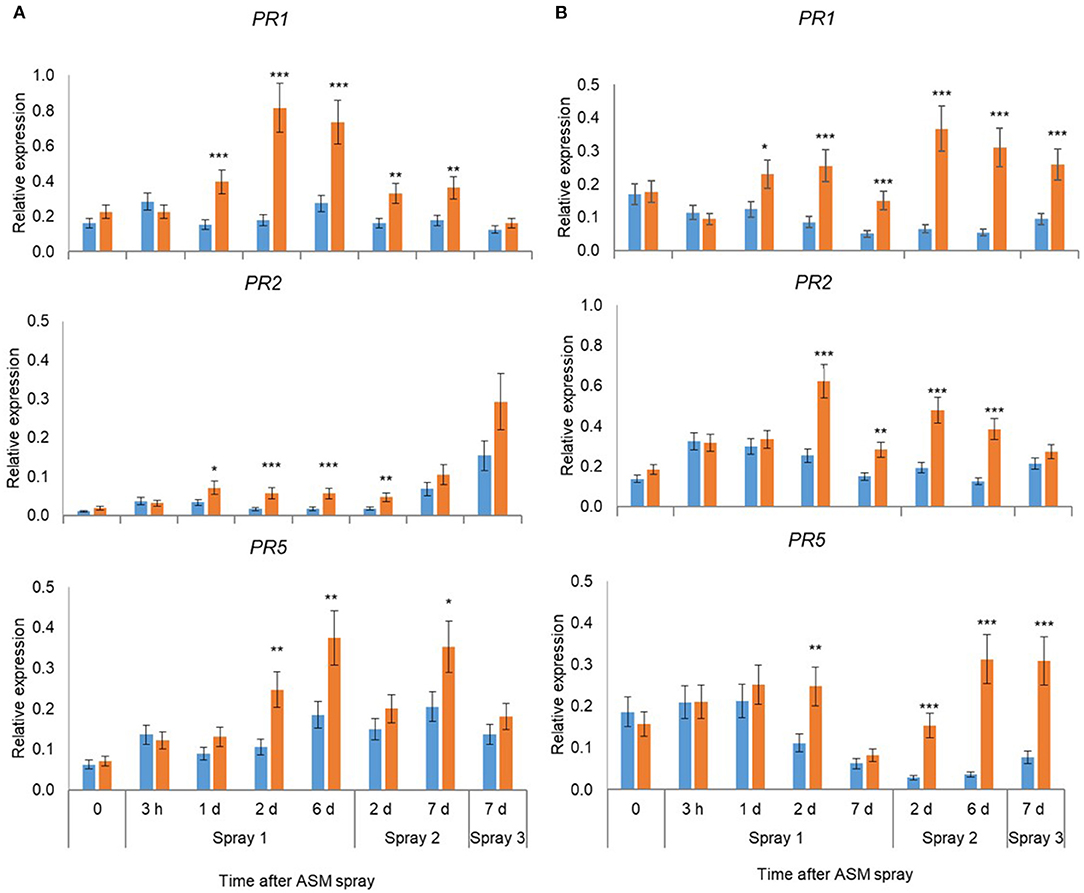
Figure 2. Expression of PR1, PR2, and PR5 in leaves of Actinidia chinensis var. deliciosa ‘Hayward’ (A) and A. chinensis var. chinensis ‘Zesy002’ vines (B) at different times after Actigard (ASM) application during spring in 2016. Actigard (ASM) was sprayed on ‘Zesy002’ on 18 October, 26 October, and 1 November, and on ‘Hayward’ on 26 October, 1 November, and 9 November. Gene expression was quantified by qPCR using actin and ubiquitin-conjugating enzyme as the reference genes for ‘Hayward,’ and 40S rRNA and GAPDH as the reference genes for ‘Zesy002.’ Data were log2 transformed for analysis, but are presented as back-transformed means. Asterisks above bars indicate significant treatment differences, where * <0.05, ** <0.01, and *** <0.001.
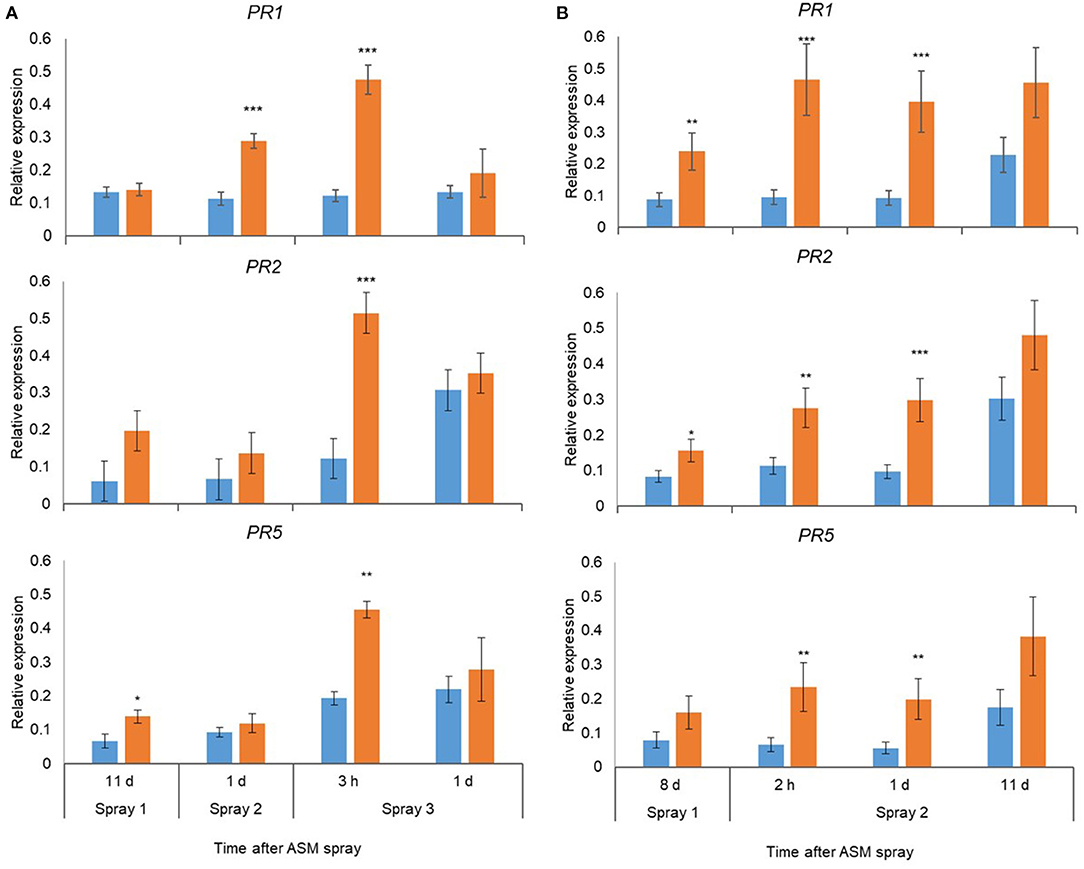
Figure 3. Expression of PR1, PR2, and PR5 in leaves of Actinidia chinensis var. deliciosa ‘Hayward’ (A) and A. chinensis var. chinensis ‘Zesy002’ vines (B) at different times after Actigard (ASM) application during spring in 2017. Actigard (ASM) was sprayed on ‘Zesy002’ on 10 October, and 19 October, and on ‘Hayward’ on 19 October, 31 October, and 9 November. Gene expression was quantified by qPCR using 40S rRNA (40S) and protein phosphatase 2A as the reference genes for ‘Hayward,’ and 40S and GAPDH as the reference gene for ‘Zesy002.’ Data were log2 transformed for analysis, but are presented as back-transformed means. Asterisks above bars indicate significant treatment differences, where * <0.05, ** <0.01, and *** <0.001.
In 2018, the expression of 18 candidate marker genes was investigated, including regulatory proteins (AcEDS1A, AP2_ERF2, NIMIN2, NPR3, WRKY33), enzymes involved with hormone biosynthesis and homeostasis (BAD, AcICS1, LOX2, AtCKX1, CYP735A2, JIH1) and pathogenesis-related proteins (PR1, Gluc2, PR5, PR2, ClassIV Chitinase). The expression of all gene candidates except JIH1 in ‘Hayward’ and JIH1 and CYP7352A in ‘Zesy002’ changed significantly over time, with 10 genes in ‘Hayward’ and 11 genes in ‘Zesy002’ affected by ASM (Table 6). As in 2016 and 2017, the expression of PR1, PR2, and PR5 was significantly upregulated after treatment with ASM in both cultivars (Figure 4). The expression of PR1 in both cultivars and PR2 in ‘Zesy002’ depended on the interaction between time and ASM (Table 6) and was upregulated by 2- to 3-fold after each spray application. Similarly, the interaction between time and ASM affected the expression of AcEDSA1, BAD, NIMIN2, NPR3 in both cultivars (Figure 5) as well as an additional five genes in ‘Zesy002’ (AcICS1, AP2ERF2, ClassIV Chit, Gluc2, and JIH1; Table 6 and Supplementary Figure 10). The most highly upregulated gene in both cultivars was NIMIN2 which was increased by between 2- and 8-fold in ‘Hayward’ and 4- and 10-fold in ‘Zesy002’ relative to respective untreated controls. In general, the effect of ASM on gene expression was transient, with expression increasing by 1 d after treatment and then declining to control levels after 12/13 d (Figures 4, 5 and Supplementary Figure 10, Supplementary Tables 9, 10).
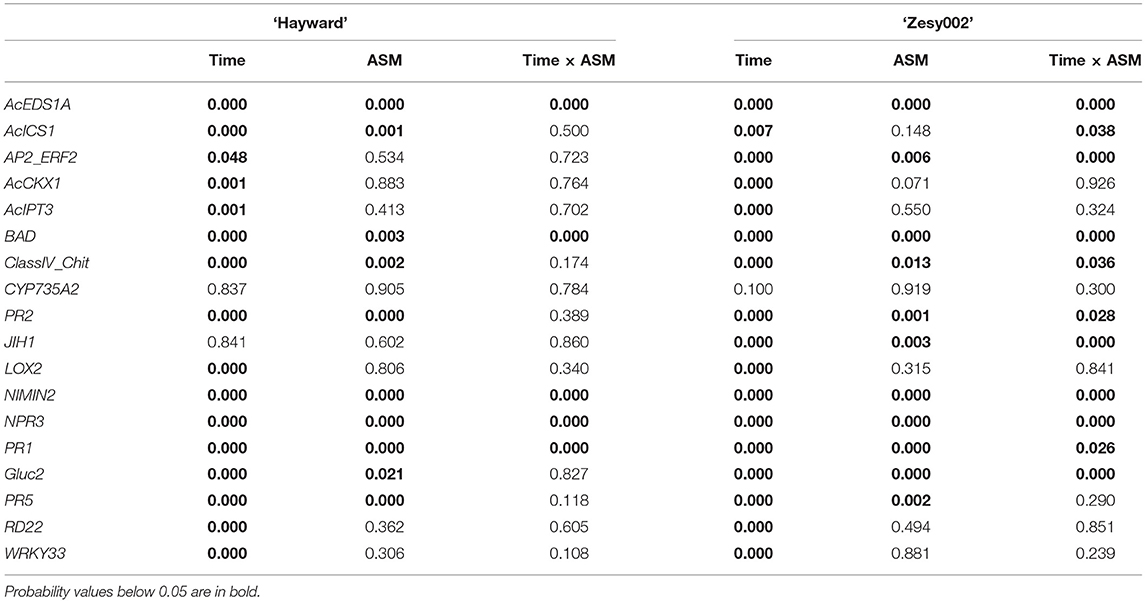
Table 6. P-values from the statistical analyses to determine the effect of Actigard (ASM) application and time on gene expression in leaves of Actinidia chinensis var. deliciosa ‘Hayward’ and A. chinensis var. chinensis ‘Zesy002’ kiwifruit vines in spring in 2018.
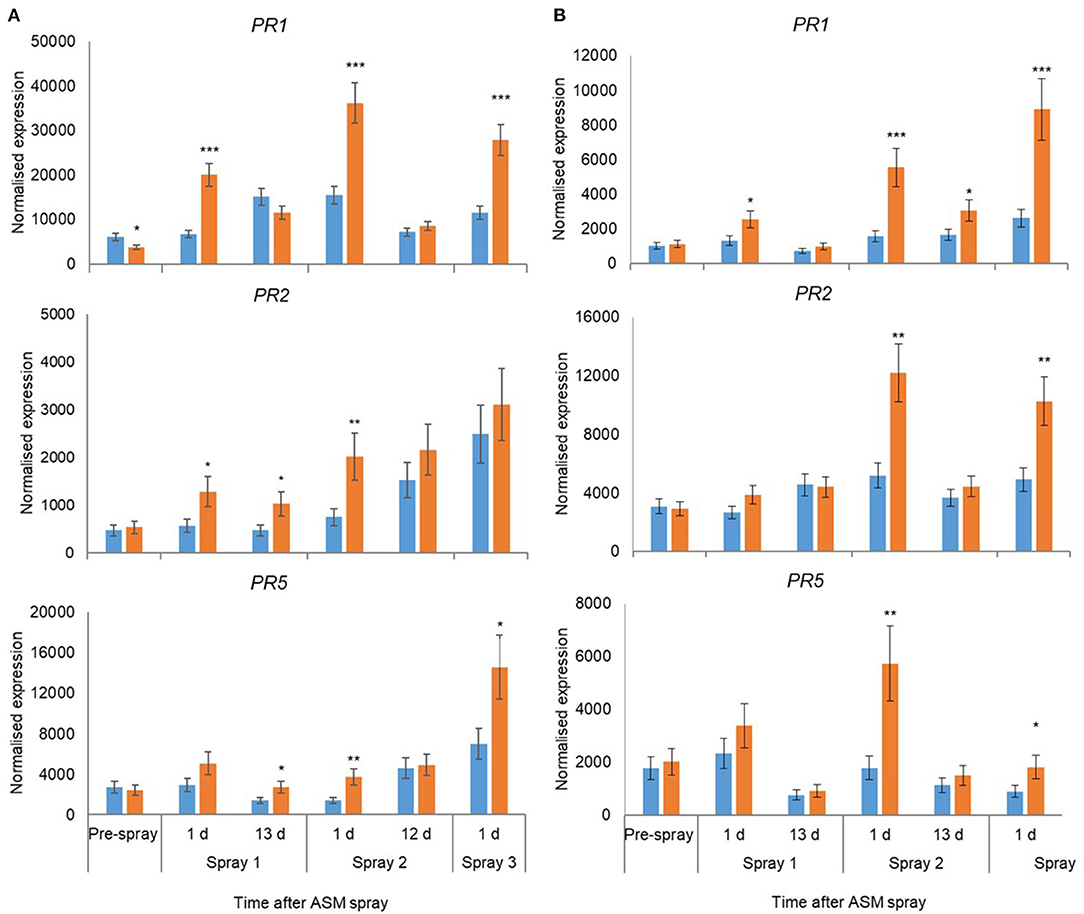
Figure 4. Expression of PR1, PR2, and PR5 in leaves of Actinidia chinensis var. deliciosa ‘Hayward’ (A) and A. chinensis var. chinensis ‘Zesy002’ (B) vines at different times after Actigard (ASM) application during spring in 2018. Actigard (ASM) was sprayed on ‘Zesy002’ on 27 September, 10 October, and 24 October, and on ‘Hayward’ on 16 October, 31 October, and 13 November. Gene expression was measured by PlexSet® NanoString where the Y-axis represents physical counts of the number of molecules expressed. Data were normalised against six references genes—40s rRNA, actin, elongation factor, glyceraldehyde 3-phosphate dehydrogenase, protein phosphatase 2A, and ubiquitin-conjugating enzyme. Data were log2 transformed for analysis, but are presented as back-transformed means. Asterisks above bars indicate significant treatment differences, where * <0.05, ** <0.01, and *** <0.001.
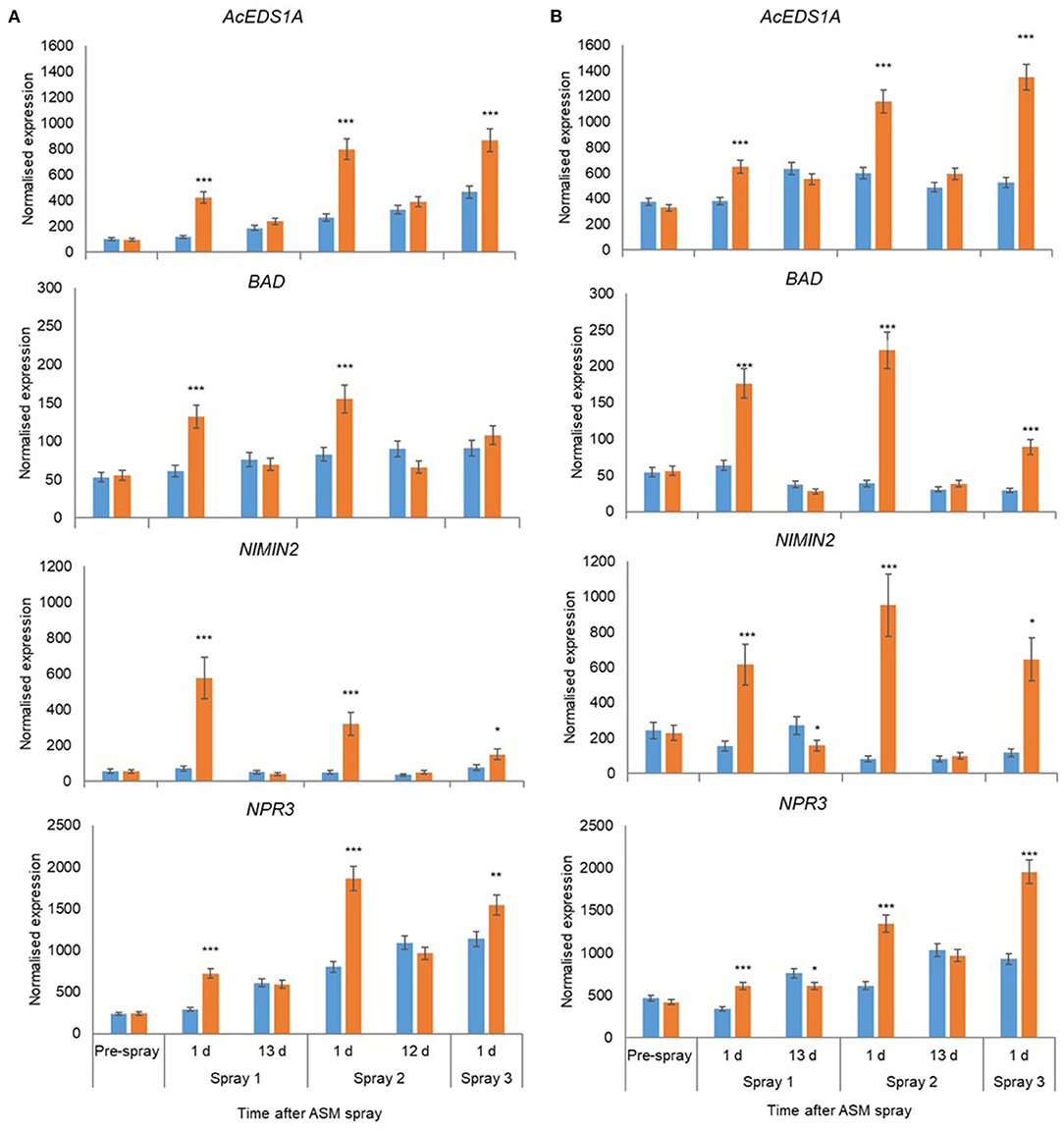
Figure 5. Expression of AcEDS1A, BAD, NIMIN2, and NPR3 in Actinidia chinensis var. deliciosa ‘Hayward’ (A) and A. chinensis var. chinensis ‘Zesy002’ (B) in spring 2018, as measured by PlexSet® NanoString, and the Y-axis represents physical counts of the number of molecules expressed. Actigard (ASM) was sprayed on ‘Zesy002’ on 27 September, 10 October, and 24 October, and on ‘Hayward’ on 16 October, 31 October, and 13 November. Data were normalised against six references genes—40s rRNA, actin, elongation factor, glyceraldehyde 3-phosphate dehydrogenase, protein phosphatase 2A, and ubiquitin-conjugating enzyme. Data were log2 transformed for analysis, but are presented as back-transformed means. Asterisks above bars indicate significant treatment differences, where * <0.05, ** <0.01, and *** <0.001.
Leaf Developmental Stage Affects Responsiveness to ASM
Leaf developmental stage during the pre-flowering period affected basal gene expression and responsiveness to ASM (Supplementary Tables 11, 12). Treatment with ASM induced the upregulation of PR1, PR2, PR5, Gluc2, EDSA1, NIMIN2, NPR3, and BAD in both cultivars; the amplitude of the upregulation depended on leaf developmental stage and was greater in the first true leaf and the youngest mature leaves than in immature leaves (Figures 6, 7). Indeed, in ‘Hayward’ ASM did not affect gene expression in immature leaves whilst in ‘Zesy00’ only AcEDS1A and PR2 were significantly upregulated by ASM. Leaf developmental stage also affected phytohormone levels (Supplementary Table 13) but, in contrast to gene expression, phytohormone levels were generally highest in the immature leaves (Supplementary Table 14). In ‘Hayward’ treatment with ASM resulted in a significant reduction in SA in the immature leaf and a reduction in SAG in the youngest mature and the immature leaf (Supplementary Figure 11). ASM did not affect phytohormone concentrations in ‘Zesy002’ (Supplementary Table 13).
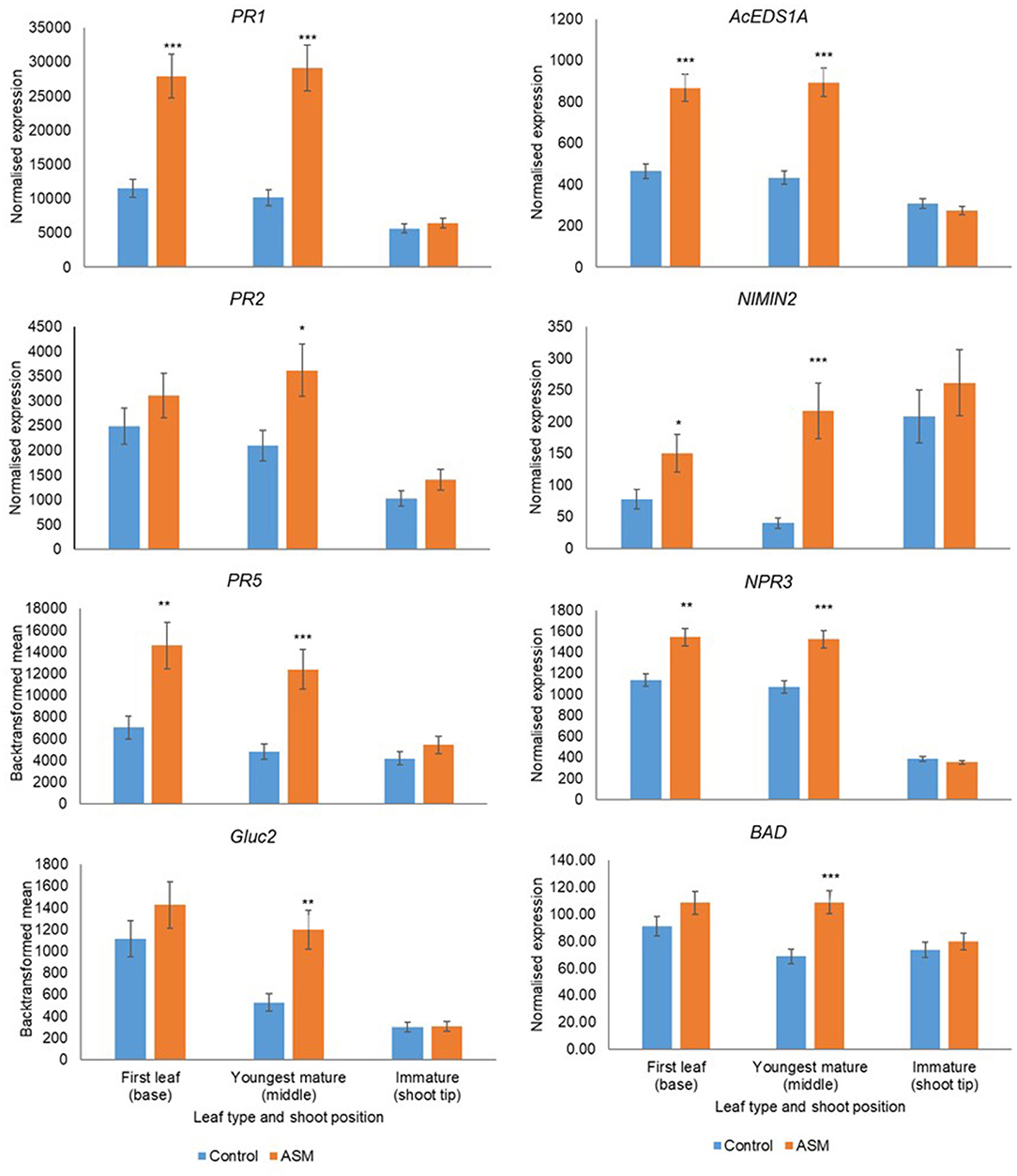
Figure 6. Expression of PR1, PR2, PR5, AcEDS1A, NIMIN2, NPR3, and BAD in Actinidia chinensis var deliciosa 'Hayward' leaves of differing maturity in spring 2018, as measured by PlexSet® NanoString, and the Y-axis represents physical counts of the number of molecules expressed. Actigard (ASM) was sprayed on 16 October, 31 October, and 13 November and leaves were sampled on 14 November. Data were normalised against six reference genes—40s rRNA, actin, elongation factor, glyceraldehyde 3-phosphate dehydrogenase, protein phosphatase 2A, and ubiquitin-conjugating enzyme. Data were log2 transformed for analysis, but are presented as back-transformed means. Asterisks above bars indicate significant treatment differences, where * <0.05, ** <0.01, and *** <0.001.
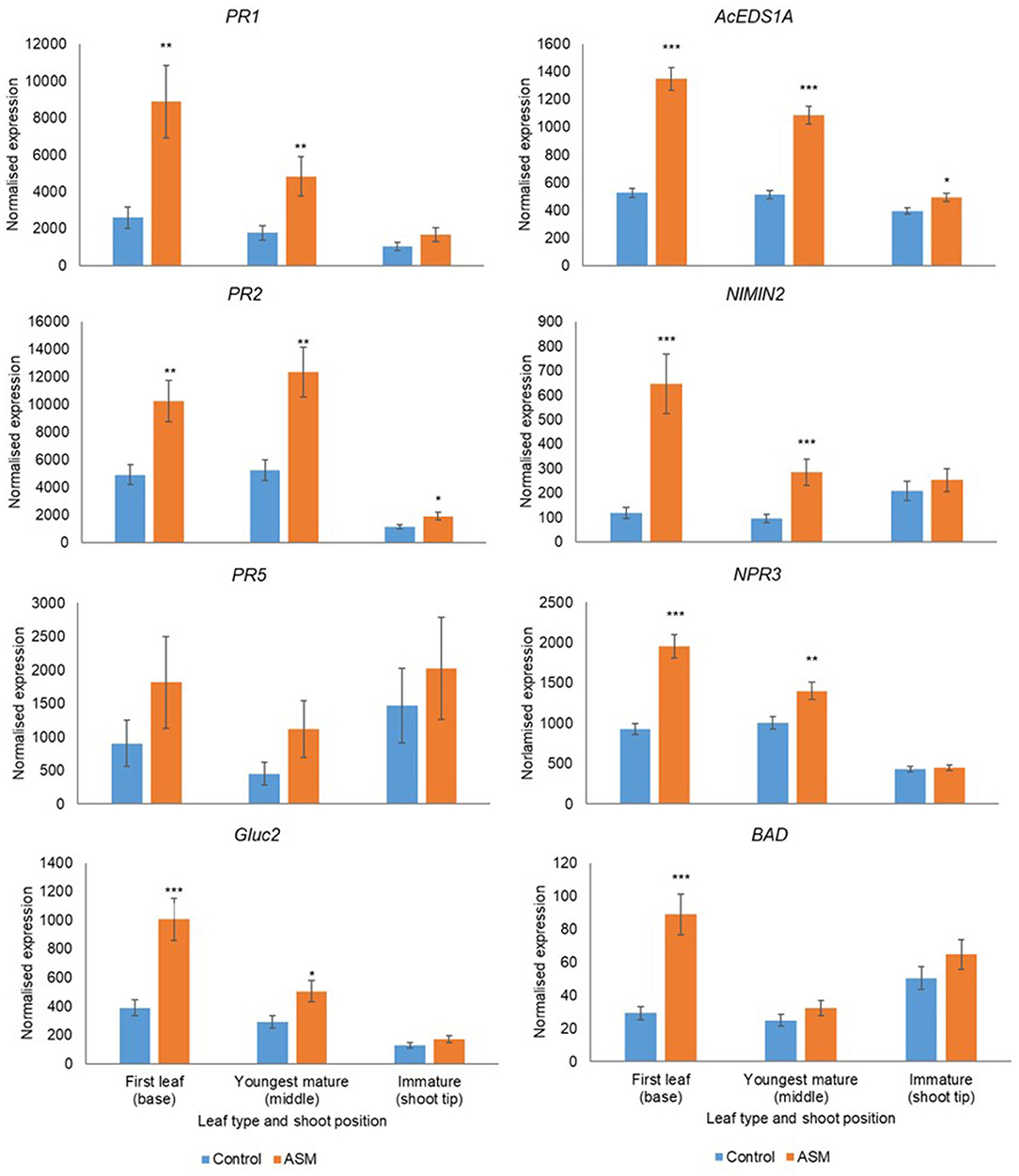
Figure 7. Expression of PR1, PR2, PR5, AcEDS1A, NIMIN2, NPR3, and BAD in Actinidia chinensis var chinensis ‘Zesy002’ leaves of differing maturity in spring 2018, as measured by PlexSet® NanoString, and the Y-axis represents physical counts of the number of molecules expressed. Actigard (ASM) was sprayed on 27 September, 10 October, and 24 October and leaves were sampled on 25 October. Data were normalised against six reference genes—40s rRNA, actin, elongation factor, glyceraldehyde 3-phosphate dehydrogenase, protein phosphatase 2A, and ubiquitin-conjugating enzyme. Data were log2 transformed for analysis, but are presented as back-transformed means. Asterisks above bars indicate significant treatment differences, where * <0.05, ** <0.01, and *** <0.001.
Discussion
Numerous studies have described the use of molecular markers to monitor induced resistance to pests and pathogens in laboratory and glasshouse environments, but relatively few have demonstrated their use to monitor defence elicitation on mature plants in the field (Acimovic et al., 2015; Johnson and Temple, 2016; Bellée et al., 2018; Hu et al., 2018). This study examined changes in phytohormone level and in gene expression in potted ‘Hayward’ and ‘Zesy002’ kiwifruit plants and in mature orchard vines after treatment with the commercial elicitor Actigard [a.i. acibenzolar-S-methyl (ASM)]. Actigard is used for the management of Psa in kiwifruit orchards and its active ingredient, ASM, is a functional mimic of SA and has been shown to activate kiwifruit resistance to Psa by activation of the SA-responsive defence pathway (Cellini et al., 2014; Wurms K. et al., 2017; Michelotti et al., 2018). Accordingly we found that ASM enhanced resistance against Psa in potted kiwifruit and induced the expression of defence genes associated with the SA-pathway, in particular PR1, PR2, and PR5. Moreover, the same three genes as well as other SA markers, BAD, AcEDS1A, NPR3, and NIMIN2, were upregulated in the leaves of orchard grown kiwifruit vines following treatment with ASM. Interestingly, the amplitude of the gene upregulation depended on leaf developmental stage thus demonstrating the potential utility of these markers as a tool to monitor the ASM-induced defence response in field conditions. Conversely, phytohormonal levels were highly variable and did not exhibit consistent response patterns to ASM. This variability, particularly in the orchard, may be a consequence of the highly dynamic role played by phytohormones in regulating plant development and their responses to multiple biotic and abiotic environmental triggers (Verma et al., 2016).
The induction of resistance to Psa in potted kiwifruit by ASM has been shown to correlate with increased expression of these of SA-pathway markers PR1, PR2, and PR5 (Cellini et al., 2014; Beatrice et al., 2017; Wurms K. V. et al., 2017; Michelotti et al., 2018; De Jong et al., 2019). This study confirms and extends these findings by employing these genes to monitor the onset of the ASM-induced defence response in mature kiwifruit in an orchard environment over three consecutive seasons. The same vines were used each year to minimise inter-vine and inter-orchard variations. Nevertheless, there remain many more confounding factors in the orchard environment than in the more tightly controlled environments used for potted plant trials and the consistency of the response of these three genes in the orchard validates their utility as markers, particularly so when contrasted with the hormonal response patterns. PR1, PR2, and PR5 were significantly over-expressed (up to 7-fold increases in expression) in ASM-treated vines compared with the controls in each orchard trial. In each of the three seasons, gene upregulation was transient, with expression levels tending to return to control levels by 11–14 days post treatment. However, this was somewhat confounded by the different spray intervals in the trials, and further studies with more extensive sampling regimes are required to more accurately profile the temporal patterns of gene upregulation after ASM application. The expression profiles of PR1, PR2, and PR5 tended to vary between cultivars and over time with no evidence of a strong cultivar dependent response. Although different RGs were used in the analysis of some qPCR experiments, the conclusions were validated by the Nanostring experiment in 2018 in which all the different RGs were used simultaneously for normalisation.
In the potted plant trials, inoculation with Psa was performed by stabbing the stem. This differs from the Cellini et al. (2014) study where plants were spray inoculated with Psa. Nevertheless, similar results were found with ASM-induced resistance to Psa being accompanied by increased expression of PR1, PR2, and PR5. ASM has been shown to induce resistance by direct activation of gene expression and by augmenting subsequent expression induced by a secondary stimuli, a phenomenon known as priming (Kohler et al., 2002; Beckers and Conrath, 2007). In the potted plant studies there was evidence of both direct activation (before inoculation) and priming (post inoculation) of PR1, PR5, and PR2 in the ASM treated plants. In most cases the priming response was observed in both SDW and Psa inoculated plants thus suggesting that priming was associated with the wound response and not with the pathogen per se. However, the upregulation of PR1 and PR5 was greater in ‘Hayward’ stems at 24 h after inoculation in Psa inoculated plants than in plants inoculated with SDW. The GoIs tended to be more strongly expressed in untreated ‘Hayward’ plants than in their ‘Zesy002’ counterparts and yet ‘Hayward’ was more susceptible to Psa than ‘Zesy002’ as evidenced by the development of larger stem lesions. However, gene upregulation by ASM was generally greater in ‘Zesy002’ than in ‘Hayward,’ relative to the controls. For example, PR1 expression increased by 5-fold in response to ASM in ‘Hayward’ leaves and stems compared with >15-fold increases in ‘Zesy002’ counterparts. One hypothesis is that the greater responsiveness of ‘Zesy002’ to ASM, when compared with ‘Hayward,’ rather than the constitutive expression levels, may, in part, account for its more effective resistance to Psa inoculation.
The modes of action of PR1, PR2, and PR5 may, in part, explain their utility as markers of ASM-induced resistance and their importance in the kiwifruit-Psa interaction. ASM is known to be effective against Psa (Collina et al., 2016) and operates as an SA-mimic in a signalling cascade that culminates in the establishment of SAR (Friedrich et al., 1996; Tripathi et al., 2010). PR1 in particular is regarded an essential player in activation of SAR (Vlot et al., 2009) and upregulation of PR1 by ASM has been shown to be associated with onset of resistance to Psa in potted kiwifruit plants (Cellini et al., 2014). PR1 was also shown to inhibit pathogen growth via sequestration of sterol (Gamir et al., 2017). This is a broad spectrum defence mechanism, which is especially effective against sterol auxotrophs, including most bacteria, because they must obtain sterols from their environment, and sterols play vital roles in regulating membrane fluidity and permeability (Kazan and Gardiner, 2017). The PR2 family include the β-glucosidases [E.C.3.2.1.21] and β-1,3-glucanases that can convert inert stored conjugates of phytohormones and antimicrobial secondary metabolites into more biologically active forms by the removal of sugar residues (Morant et al., 2008). The deployment of the bioactive molecule via enzymatic sugar hydrolysis rather than de novo synthesis may be important in the kiwifruit-Psa interaction. PR5 proteins, also known as thaumatin-like proteins (TLP), are induced by a range of biotic and abiotic stresses (Liu et al., 2010) and have been shown to exhibit antimicrobial activity by affecting fungal cell wall integrity. The involvement of PR5 in defence against bacterial pathogens is less well-understood but may involve membrane permeabilisation. A basic form of a TLP has shown direct antimicrobial activity against a P. syringae pathovar of soybean (Liu et al., 2016). A different member of the PR5 family was found not to respond to ASM treatment on another A. chinensis var. chinensis cultivar (Cellini et al., 2014).
In commercial orchards in New Zealand, ASM is applied during the pre-flowering period to protect vines against infection by Psa. In the current study, pre-flowering applications of ASM resulted in the upregulation of PR1, PR2, and PR5 expression in the leaves of ‘Hayward’ and ‘Zesy002’ vines. Gene upregulation in ‘Hayward’ was accompanied by a reduction in Psa leaf spotting whereas no such relationship was measurable for ‘Zesy002’ because leaf spot symptoms are rarely expressed in this cultivar (Vanneste, 2017). There was evidence of differential expression of PR1, PR2, and PR5 between the two cultivars across the 3 years of the study, with PR1 and PR5 tending to be greater in ‘Hayward’ than ‘Zesy002’ and the reverse being the case for PR2. In 2018, the constitutive expression of PR1 in untreated controls was, on average, approximately 5-fold greater in ‘Hayward’ than in ‘Zesy002.’ However, ASM induced a 2- to 3-fold upregulation in PR1 in both cultivars meaning that the constitutive expression level of PR1 in ‘Hayward’ tended to be greater than the ASM-induced level in ‘Zesy002.’ Differences between constitutive and ASM inducible expression were also observed for PR5 (‘Hayward’ > ‘Zesy002’) and PR2 (‘Zesy002’ > ‘Hayward’) but these were less marked than with PR1. Taken together these data suggest that PR1, PR2, and PR5 are useful markers for ASM-induced responses in kiwifruit vines but their absolute expression levels may not necessarily correlate with relative levels of Psa resistance in these two cultivars as measured by leaf spot symptom expression. Interestingly, this differs with a study in apple where defence induction (28 genes) by ASM in five genotypes correlated with their basal susceptibility to apple scab i.e., the greatest induction by ASM was observed in the most resistant genotype (Marolleau et al., 2017). The same study also reported that two successive ASM applications to apple seedlings provided greater protection against leaf infection by Erwinia amylovara than a single spray. In the current study, the reliance on natural infection and the lack of leaf spot symptoms in ‘Zesy002’ meant that no equivalent measure could be made. Nevertheless, based on gene upregulation there was no evidence of that successive applications of ASM had a cumulative effect on defence induction in kiwifruit vines.
A larger group of host genes was investigated in the orchard in 2018. Of these additional genes, NIMIN2, NPR3, AcEDS1A, and BAD were significantly upregulated by ASM in both cultivars. Gene upregulation was transient after each of three sprays with a strong upregulation 1 d after ASM treatment and then a return to control levels by 12/13 days post treatment and before the next application. The expression levels of AcEDS1A and BAD were similar in both cultivars whereas NIMIN2 and NPR3 were generally greater in ‘Zesy002’ than in ‘Hayward’ for both ASM-treated and control plants. These genes are associated with SA defence pathway with EDS1A playing a role in the regulation of SA accumulation whilst NPR3 actively binds SA and interacts with NIMIN2 to promote the transcription of defence-related genes including PR genes (Chen et al., 2020) (Hermann et al., 2013). It is not known if NPR3 binds the ASM in a similar manner to SA. The role benzyl alcohol dehydrogenase (BAD) in SA defence activation is less well understood, however, upregulation of the same BAD isoform was by ASM and/or a yeast biocontrol agent was shown to correlate with resistance to Psa in ‘Hayward’ kiwifruit in glasshouse studies (De Jong et al., 2019). In other studies BAD has been associated with localised detoxification of fungal toxins at pathogen infections sites (Guillen et al., 1998; Colrat et al., 1999).
ClassIV_Chit (PR3 family) and Gluc2 (PR2 family) were also significantly upregulated by ASM in both cultivars, but did not show a significant Time x ASM interaction. These genes encode for chitinase and glucanases, respectively, and are more commonly normally associated defence against fungal pathogens (Lawrence et al., 2000). However, and up-regulation of the same ClassIV_Chit isoform correlated with ASM-induced reduction of Psa leaf infection in glasshouse ‘Hayward’ plants (De Jong et al., 2019). Markers of the JA and ABA pathways (LOX2, JAR1, and RD22), did not correlate with ASM-induced resistance to Psa in the potted trial, nor did expression show statistically significant or consistent trends that could be related to ASM treatment or cultivar in mature vines. This agrees with other studies reporting that ASM-induced resistance to Psa predominantly involves the SA pathway (Cellini et al., 2014; Michelotti et al., 2018).
The frequency of elicitor application is a crucial consideration for field implementation and particularly so because of potential trade-offs associated with the diversion of finite resources into defence at the expense of growth (Walters and Heil, 2007). Repeat application of elicitors such as ASM has been associated with a decrease in yield in cantaloupe and tomatoes (Kunwar et al., 2017; Egel et al., 2018). Moreover, we found that three applications of ASM at weekly intervals inhibited the growth of potted ‘Zesy002’ plants and observed a possible relationship between pre-flowering ASM sprays and retardation of fruit growth in ‘Zesy002’ vines (Tony Reglinski, Plant & Food Research, unpublished data). In year three of the current study, markers of the cytokinin (CK) pathway (IPT3, CKX1, and CYP735A2) were included because the CK pathway is commonly associated with growth regulation (Schaller et al., 2014) and more recently has been proposed to play a role in plant immunity (Albrecht and Argueso, 2017). Despite being applied at 14 d intervals rather than 21 d, as in commercial orchards, there was no significant effect of ASM on the expression of the CK markers in ‘Hayward’ or ‘Zesy002’ vines, suggesting there was unlikely to be an adverse effect on kiwifruit growth.
Age-related resistance is a commonly observed phenomenon across plant species and several studies have reported that leaves become more resistant to pathogen attack with increasing age (Develey-Rivière and Galiana, 2007; Hu and Yang, 2019). However, far less is understood about the effect of leaf age on elicitor responsiveness and induced resistance. In vine crops like kiwifruit there is a long period of leaf development before flowering (Salinero et al., 2009) and so the leaf canopy during the pre-flowering spray period comprises leaves at varying stages of development. This is one of the periods of greatest risk to leaf infection by Psa (Vanneste, 2017). In the third season of the current study it was shown that kiwifruit leaf development affected responsiveness to ASM and that key genes in the SA-pathway (PR1, NIMIN2, and NPR3) were strongly upregulated in mature leaves but not in the immature leaves in both cultivars. This is an important result and suggests that ASM will not provide full protection to emerging leaves in the leaf canopy from infection by Psa during the “high risk” spring period. Similar “age-related” elicitor responsiveness was reported in potted grapevines where defence activation by laminarin was found to be greater in mature leaves than in younger leaves (Steimetz et al., 2012). More recently, Verly et al. (2020) who reported that the induction of PR1 and PR5 by Bion (a.i. benzothiadiazole, syn. ASM) in Arabidopsis depended not only on developmental stage but also on nitrogen nutrition. Collectively, these studies demonstrate the importance of understanding how interactions between vine phenology and agronomic conditions may limit elicitor responsiveness and defence inducibility in the orchard. This knowledge can facilitate more effective and smarter use of elicitors, for example in the current study the age-dependent elicitor response data would justify the integration of ASM with a directly antimicrobial companion spray to protect immature tissues. Phytohormones were generally not reliable as markers of the ASM-induced response in ‘Hayward’ and ‘Zesy002’ kiwifruit. In glasshouse studies, ASM induced a significant increase in ABA content in stems and leaves of both cultivars, whereas in the orchard there was generally no significant effect of ASM on ABA. There was a differential cultivar response to ASM with respect to SA content in the glasshouse, with ASM causing a significant reduction in SA content in ‘Hayward’ but not in ‘Zesy002.’ In the orchard, SA and SAG content in leaves tended to be lower in ASM-treated vines for both cultivars but this was generally not significantly different to the control. However, ASM has been shown to operate down-stream of SA accumulation in the defence signaling cascade (Friedrich et al., 1996; Tripathi et al., 2010) and therefore may not be expected to directly affect SA and SAG levels in kiwifruit. The lack of a consistent pattern in hormonal response in this study does not negate their importance as regulators of the host defence response but does demonstrate the difficulty of using phytohormones as markers of defence given the complex dynamics of hormonal signaling networks (Berens et al., 2017). Hormones regulate numerous developmental and physiological processes in plants as well as adaptive responses to environmental cues (Wasternack and Hause, 2013) and so differentiating their role in defence-related responses amidst their involvement regulating other stress responses may be too difficult in the orchard environment. More detailed analysis of the hormone response in leaves under controlled conditions may be required to identify patterns that correspond with defence elicitation. Moreover, because phytohormone content varies between plant species it is possible that hormonal ratios at local levels rather than absolute concentrations drive the defence response.
Conclusion
In conclusion, this is the first report describing the use of defence genes to monitor the onset of the ASM-induced defence response in ‘Hayward’ and ‘Zesy002’ kiwifruit in orchard conditions. Genes that were used successfully in glasshouse studies (PR1, PR2, and PR5) to investigate ASM-induced resistance to Psa in potted plants were also responsive to ASM applications in mature orchard vines. Moreover, additional genes associated with the SA-mediated defence pathway, BAD, AcEDS1A, NPR3, and NIMIN2, were shown to be upregulated by ASM treatment. Responsiveness to ASM in orchard vines depended on leaf developmental stage and was transient, with gene expression increasing by 24 h after treatment before returning to untreated control levels after 11–14 days. In contrast, phytohormonal levels in orchard vines exhibited no repeatable response pattern to ASM, possibly reflecting their wider role in the regulation of plant growth and the response to other environmental cues. The successful demonstration of these markers in an orchard environment can open up opportunities to deepen our knowledge of factors affecting elicitor field efficacy and so guide their practical implementation. For example, may is be possible to determine the compatibility of ASM with different agricultural inputs based on the gene upregulation rather than on efficacy alone? Moreover, studies to map spatial and temporal components of defence induction across a vine and to compare effects of vine phenology on elicitor responsiveness would also advance our understanding on potential limitations of elicitors as crop protectants.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author/s.
Author Contributions
TR, KW, and JV contributed to conception, design, and execution of the study. CM and MA performed the statistical analysis. TR and KW wrote the first draft of the manuscript. JC and AAC wrote sections of the manuscript. JV provided editorial input and contributed to manuscript revision. AAC, JY, JO, DC, DJ, TT, JT, and MS contributed significant technical input to the study. All authors contributed to the article and approved the submitted version.
Conflict of Interest
All authors were employed by company The New Zealand Institute for Plant and Food Research Limited.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Thank to Trisha Periera, Hannah Jonson, and Rachelle Anderson (all of Plant & Food Research) for assistance with tissue collection. Thanks also to the New Zealand Ministry of Business, Innovation and Employment (MBIE) (contract C11X1205) and Zespri International Limited for financial support (contract VI1602), Jason Copedo of Grafton Clinical Genomics, Auckland, New Zealand, for carrying out the NanoString assays, Erin Stroud of Plant & Food Research for supplying the LOX2 primers for qPCR, and Drs. Erik Rikkerink, Joanna Bowen and Jay Jayaraman for reviewing the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2021.831172/full#supplementary-material
References
Acimovic, S. G., Zeng, Q., Mcghee, G. C., Sundin, G. W., and Wise, J. C. (2015). Control of fire blight (Erwinia amylovora) on apple trees with trunk-injected plant resistance inducers and antibiotics and assessment of induction of pathogenesis-related protein genes. Front. Plant Sci. 6:16. doi: 10.3389/fpls.2015.00016
Albrecht, T., and Argueso, C. T. (2017). Should I fight or should I grow now? The role of cytokinins in plant growth and immunity and in the growth-defence trade-off. Ann. Bot. 119, 725–735. doi: 10.1093/aob/mcw211
Backer, R., Naidoo, S., and Van Den Berg, N. (2019). The Nonexpressor of Pathogenesis-Related genes 1 (NPR1) and related family: mechanistic insights in plant disease resistance. Front. Plant Sci. 10:102. doi: 10.3389/fpls.2019.00102
Beatrice, C., Linthorst, J. M. H., Cinzia, F., and Luca, R. (2017). Enhancement of PR1 and PR5 gene expressions by chitosan treatment in kiwifruit plants inoculated with Pseudomonas syringae pv. actinidiae. Eur. J. Plant Pathol. 148, 163–179. doi: 10.1007/s10658-016-1080-x
Beckers, G. J., and Conrath, U. (2007). Priming for stress resistance: from the lab to the field. Curr. Opin. Plant Biol. 10, 425–431. doi: 10.1016/j.pbi.2007.06.002
Bellée, A., Cluzet, S., Dufour, M.-C., Mérillon, J.-M., and Corio-Costet, M.-F. (2018). Comparison of the impact of two molecules on plant defense and on efficacy against Botrytis cinerea in the vineyard: a plant defense inducer (Benzothiadiazole) and a fungicide (Pyrimethanil). J. Agric. Food Chem. 66, 3338–3350. doi: 10.1021/acs.jafc.7b05725
Berens, M. L., Berry, H. M., Mine, A., Argueso, C. T., and Tsuda, K. (2017). Evolution of hormone signaling networks in plant defense. Annu. Rev. Phytopathol. 55, 401–425. doi: 10.1146/annurev-phyto-080516-035544
Bulley, S. M., Cooney, J. M., and Laing, W. (2021). Elevating ascorbate in Arabidopsis stimulates the production of abscisic acid, phaseic acid, and to a lesser auxin (IAA) and jasmonates, resulting in increased expression of DHAR1 and multiple transcription factors associated with abiotic stress tolerance. Int. J. Mol. Sci. 23:6743. doi: 10.3390/ijms22136743
Cellini, A., Fiorentini, L., Buriani, G., Yu, J., Donati, I., Cornish, D. A., et al. (2014). Elicitors of the salicylic acid pathway reduce incidence of bacterial canker of kiwifruit caused by Pseudomonas syringae pv. actinidae. Ann. Appl. Biol. 165, 441–453. doi: 10.1111/aab.12150
Chapman, J. R., Taylor, R. K., Weir, B. S., Romberg, M. K., Vanneste, J. L., Luck, J., et al. (2012). Phylogenetic relationships among global populations of Pseudomonas syringae pv. actinidiae. Phytopathology 102, 1034–1044. doi: 10.1094/phyto-03-12-0064-r
Chen, J., Clinton, M., Qi, G., Wang, D., Liu, F., and Fu, Z. Q. (2020). Reprogramming and remodeling: transcriptional and epigenetic regulation of salicylic acid-mediated plant defense. J. Exp. Bot. 71, 5256–5268. doi: 10.1093/jxb/eraa072
Collina, M., Donati, I., Bertacchini, E., Brunelli, A., and Spinelli, F. (2016). Greenhouse assays on the control of the bacterial canker of kiwifruit (Pseudomonas syringae pv. actinidiae). J. Berry Res. 6, 407–415. doi: 10.3233/jbr-160128
Colrat, S., Deswarte, C., Latche, A., Klaebe, A., Bouzayen, M., Fallot, J., et al. (1999). Enzymatic detoxification of eutypine, a toxin from Eutypa lata, by Vitis vinifera cells: partial purification of an NADPH-dependent aldehyde reductase. Planta 207, 544–550. doi: 10.1007/s004250050516
De Jong, H., Reglinski, T., Elmer, P. A. G., Wurms, K., Vanneste, J. L., Guo, L. F., et al. (2019). Integrated use of Aureobasidium pullulans strain CG163 and acibenzolar-S-methyl for management of bacterial canker in kiwifruit. Plants 8:287. doi: 10.3390/plants8080287
Develey-Rivière, M.-P., and Galiana, E. (2007). Resistance to pathogens and host developmental stage: a multifaceted relationship within the plant kingdom. New Phytol. 175, 405–416. doi: 10.1111/j.1469-8137.2007.02130.x
Ding, Y. L., Sun, T. J., Ao, K., Peng, Y. J., Zhang, Y. X., Li, X., et al. (2018). Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 173, 1454–1467. doi: 10.1016/j.cell.2018.03.044
Donati, I., Cellini, A., Sangiorgio, D., Vanneste, J. L., Scortichini, M., Balestra, G. M., et al. (2020). Pseudomonas syringae pv. actinidiae: ecology, infection dynamics and disease epidemiology. Microb. Ecol. 80, 81–102. doi: 10.1007/s00248-019-01459-8
Durrant, W. E., and Dong, X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209. doi: 10.1146/annurev.phyto.42.040803.140421
Egel, D. S., Kleczewski, N. M., Mumtaz, F., and Foster, R. (2018). Acibenzolar-S-methyl is associated with yield reduction when used for managing bacterial wilt (Erwinia tracheiphila) in cantaloupe. Crop Prot. 109, 136–141. doi: 10.1016/j.cropro.2018.03.012
Faize, L., and Faize, M. (2018). Functional analogues of salicylic acid and their use in crop protection. Agronomy-Basel 8:5. doi: 10.3390/agronomy8010005
Friedrich, L., Lawton, K., Ruess, W., Masner, P., Specker, N., Rella, M. G., et al. (1996). A benzothiadiazole derivative induces systemic acquired resistance in tobacco. Plant J. 10, 61–70. doi: 10.1046/j.1365-313X.1996.10010061.x
Gamir, J., Darwiche, R., Van't Hof, P., Choudhary, V., Stumpe, M., Schneiter, R., et al. (2017). The sterol-binding activity of Pathogenesis-Related protein 1 reveals the mode of action of an antimicrobial protein. Plant J. 89, 502–509. doi: 10.1111/tpj.13398
Gorlach, J., Volrath, S., Knaufbeiter, G., Hengy, G., Beckhove, U., Kogel, K. H., et al. (1996). Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell 8, 629–643. doi: 10.1105/tpc.8.4.629
Guillen, P., Guis, M., Martinez-Reina, G., Colrat, S., Dalmayrac, S., Deswarte, C., et al. (1998). A novel NADPH-dependent aldehyde reductase gene from Vigna radiata confers resistance to the grapevine fungal toxin eutypine. Plant J. 16, 335–343. doi: 10.1046/j.1365-313x.1998.00303.x
Hermann, M., Maier, F., Masroor, A., Hirth, S., Pfitzner, A. J. P., and Pfitzner, U. M. (2013). The Arabidopsis NIMIN proteins affect NPR1 differentially. Front. Plant Sci. 4:88. doi: 10.3389/fpls.2013.00088
Hill, M. G., Wurms, K. V., Davy, M. W., Gould, E., Allan, A., Mauchline, N. A., et al. (2015). Transcriptome analysis of kiwifruit (Actinidia chinensis) bark in response to armoured scale insect (Hemiberlesia lataniae) feeding. PLoS ONE 10:e141664. doi: 10.1371/journal.pone.0141664
Hu, J., Jiang, J., and Wang, N. (2018). Control of citrus huanglongbing via trunk injection of plant defense activators and antibiotics. Phytopathology 108, 186–195. doi: 10.1094/phyto-05-17-0175-r
Hu, L., and Yang, L. (2019). Time to fight: molecular mechanisms of age-related resistance. Phytopathology 109, 1500–1508. doi: 10.1094/phyto-11-18-0443-rvw
Huang, S. X., Ding, J., Deng, D. J., Tang, W., Sun, H. H., Liu, D. Y., et al. (2013). Draft genome of the kiwifruit Actinidia chinensis. Nat. Commun. 4:9. doi: 10.1038/ncomms3640
Joglekar, S., Suliman, M., Bartsch, M., Halder, V., Maintz, J., Bautor, J., et al. (2018). Chemical activation of EDS1/PAD4 signaling leading to pathogen resistance in Arabidopsis. Plant Cell Physiol. 59, 1592–1607. doi: 10.1093/pcp/pcy106
Johnson, K. B., and Temple, T. N. (2016). Comparison of methods of Acibenzolar-S-Methyl application for post-infection fire blight suppression in pear and apple. Plant Dis. 100, 1125–1131. doi: 10.1094/pdis-09-15-1062-re
Kazan, K., and Gardiner, D. M. (2017). Targeting pathogen sterols: defence and counterdefence? PLoS Pathog. 13:e1006297. doi: 10.1371/journal.ppat.1006297
King, E. O., Ward, M. K., and Raney, D. E. (1954). Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44, 301–307.
Kohler, A., Schwindling, S., and Conrath, U. (2002). Benzothiadiazole-induced priming for potentiated responses to pathogen infection, wounding, and infiltration of water into leaves requires the NPR1/NIM1 gene in Arabidopsis. Plant Physiol. 128, 1046–1056. doi: 10.1104/pp.010744
Kunwar, S., Paret, M. L., Freeman, J. H., Ritchie, L., Olson, S. M., Colee, J., et al. (2017). Foliar applications of Acibenzolar-S-Methyl negatively affect the yield of grafted tomatoes in fields infested with Ralstonia solanacearum. Plant Dis. 101, 890–894. doi: 10.1094/pdis-03-16-0331-re
Lawrence, C. B., Singh, N. P., Qiu, J. S., Gardner, R. G., and Tuzun, S. (2000). Constitutive hydrolytic enzymes are associated with polygenic resistance of tomato to Alternaria solani and may function as an elicitor release mechanism. Physiol. Mol. Plant Pathol. 57, 211–220. doi: 10.1006/pmpp.2000.0298
Lawton, K. A., Friedrich, L., Hunt, M., Weymann, K., Delaney, T., Kessmann, H., et al. (1996). Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J. 10, 71–82. doi: 10.1046/j.1365-313X.1996.10010071.x
Liu, C., Cheng, F. F., Sun, Y. G., Ma, H. Y., and Yang, X. Q. (2016). Structure-function relationship of a novel PR-5 protein with antimicrobial activity from soy hulls. J. Agric. Food Chem. 64, 948–959. doi: 10.1021/acs.jafc.5b04771
Liu, J. J., Sturrock, R., and Ekramoddoullah, A. K. M. (2010). The superfamily of thaumatin-like proteins: its origin, evolution, and expression towards biological function. Plant Cell Rep. 29, 419–436. doi: 10.1007/s00299-010-0826-8
Marolleau, B., Gaucher, M., Heintz, C., Degrave, A., Warneys, R., Orain, G., et al. (2017). When a plant resistance inducer leaves the lab for the field: integrating ASM and into routine apple protection practices. Front. Plant Sci. 8:1938. doi: 10.3389/fpls.2017.01938
McCann, H. C., Rikkerink, E. H. A., Bertels, F., Fiers, M., Lu, A., Rees-George, J., et al. (2013). Genomic analysis of the kiwifruit pathogen Pseudomonas syringae pv. actinidiae provides insight into the origins of an emergent plant disease. PLoS Pathogens 9:e1003503. doi: 10.1371/journal.ppat.1003503
Michelotti, V., Lamontanara, A., Buriani, G., Orru, L., Cellini, A., Donati, I., et al. (2018). Comparative transcriptome analysis of the interaction between Actinidia chinensis var. chinensis and Pseudomonas syringae pv. actinidiae in absence and presence of acibenzolar-S-methyl. BMC Genomics 19:585 doi: 10.1186/s12864-018-4967-4
Morant, A. V., Jørgensen, K., Jørgensen, C., Paquette, S. M., Sánchez-Pérez, R., Møller, B. L., et al. (2008). β-Glucosidases as detonators of plant chemical defense. Phytochemistry 69, 1795–1813. doi: 10.1016/j.phytochem.2008.03.006
Mou, Z., Fan, W., and Dong, X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113, 935–944. doi: 10.1016/s0092-8674(03)00429-x
Nanostring Technologies Inc (2017). Manual 10044-02. All About PlexSet Technology Data Analysis in nSolver Software. Nanostring Technologies Inc.
Nardozza, S., Martinez-Sanchez, M., Curtis, C., Datson, P. M., and Montefiori, M. (2015). “Screening Actinidia germplasm for different levels of tolerance, or resistance, to Psa (Pseudomonas syringae pv. actinidiae),” in VIII International Symposium on Kiwifruit, eds H. Huang and Q. Zhang (Leuven: Int Soc Horticultural Science), 351–355.
Petriccione, M., Mastrobuoni, F., Zampella, L., and Scortichini, M. (2015). Reference gene selection for normalization of RT-qPCR gene expression data from Actinidia deliciosa leaves infected with Pseudomonas syringae pv. actinidiae. Sci. Rep. 5:16961. doi: 10.1038/srep16961
Pieterse, C. M. J., Van Der Does, D., Zamioudis, C., Leon-Reyes, A., and Van Wees, S. C. M. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. doi: 10.1146/annurev-cellbio-092910-154055
Pilkington, S. M., Crowhurst, R., Hilario, E., Nardozza, S., Fraser, L., Peng, Y. Y., et al. (2018). A manually annotated Actinidia chinensis var. chinensis (kiwifruit) genome highlights the challenges associated with draft genomes and gene prediction in plants. BMC Genomics 19:19. doi: 10.1186/s12864-018-4656-3
Reglinski, T., Dann, E., and Deverall, B. (2014). “Implementation of induced resistance for crop protection,” in Induced Resistance for Plant Defense, eds D. R. Walters, A. C. Newton, and G. D. Lyon (Oxford: Wiley-Blackwell). doi: 10.1002/9781118371848.ch12
Reglinski, T., Vanneste, J. L., Wurms, K., Gould, E., Spinelli, F., and Rikkerink, E. (2013). Using fundamental knowledge of induced resistance to develop control strategies for bacterial canker of kiwifruit caused by Pseudomonas syringae pv. actinidiae. Front. Plant Sci. 4:24. doi: 10.3389/fpls.2013.00024
Salinero, M. C., Vela, P., and Sainz, M. J. (2009). Phenological growth stages of kiwifruit (Actinidia deliciosa ‘Hayward’). Sci. Hortic. 121, 27–31. doi: 10.1016/j.scienta.2009.01.013
Schaller, G. E., Street, I. H., and Kieber, J. J. (2014). Cytokinin and the cell cycle. Curr. Opin. Plant Biol. 21, 7–15. doi: 10.1016/j.pbi.2014.05.015
Song, Y., Sun, L., Lin, M., Chen, J., Qi, X., Hu, C., et al. (2019). Comparative transcriptome analysis of resistant and susceptible kiwifruits in response to Pseudomonas syringae pv. actinidiae during early infection. PLoS ONE 14:e0211913. doi: 10.1371/journal.pone.0211913
Steimetz, E., Trouvelot, S., Gindro, K., Bordier, A., Poinssot, B., Adrian, M., and Daire, X. (2012). Influence of leaf age on induced resistance in grapevine against Plasmopara viticola. Physiol. Mol. Plant Pathol. 79, 89–96. doi: 10.1016/j.pmpp.2012.05.004
Tripathi, D., Jiang, Y.-L., and Kumar, D. (2010). SABP2, a methyl salicylate esterase is required for the systemic acquired resistance induced by acibenzolar-S-methyl in plants. FEBS Lett. 584, 3458–3463. doi: 10.1016/j.febslet.2010.06.046
Tripathi, D., Raikhy, G., and Kumar, D. (2019). Chemical elicitors of systemic acquired resistance - salicylic acid and its functional analogs. Curr. Plant Biol. 17, 48–59. doi: 10.1016/j.cpb.2019.03.002
Vallad, G. E., and Goodman, R. M. (2004). Systemic acquired resistance and induced systemic resistance in conventional agriculture. Crop Sci. 44, 1920–1934. doi: 10.2135/cropsci2004.1920
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:1–11. Available online at: http://genomebiology.com/2002/3/7/research/0034
Vanneste, J. L.. (2012). Pseudomonas syringae pv. actinidiae (Psa): a threat to the New Zealand and global kiwifruit industry. N. Z. J. Crop Hortic. Sci. 40, 265–267. doi: 10.1080/01140671.2012.736084
Vanneste, J. L.. (2017). The scientific, economic, and social impacts of the New Zealand outbreak of bacterial canker of kiwifruit (Pseudomonas syringae pv. actinidiae). Annu. Rev. Phytopathol. 55, 377–399. doi: 10.1146/annurev-phyto-080516-035530
Vanneste, J. L., Yu, J., Cornish, D. A., Tanner, D. J., Windner, R., Chapman, J. R., et al. (2013). Identification, virulence and distribution of two biovars of Pseudomonas syringae pv. actinidiae in New Zealand. Plant Dis. 97, 708–719. doi: 10.1094/pdis-07-12-0700-re
Verly, C., Djoman, A. C. R., Rigault, M., Giraud, F., Rajjou, L., Saint-Macary, M. E., et al. (2020). Plant defense stimulator mediated defense activation is affected by nitrate fertilization and developmental stage in Arabidopsis thaliana. Front. Plant Sci. 11:583. doi: 10.3389/fpls.2020.00583
Verma, V., Ravindran, P., and Kumar, P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16:86. doi: 10.1186/s12870-016-0771-y
Vlot, A. C., Dempsey, D. A., and Klessig, D. F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
Walters, D., and Heil, M. (2007). Costs and trade-offs associated with induced resistance. Physiol. Mol. Plant Pathol. 71, 3–17. doi: 10.1016/j.pmpp.2007.09.008
Wang, T., Wang, G., Jia, Z. H., Pan, D. L., Zhang, J. Y., and Guo, Z. R. (2018). Transcriptome analysis of kiwifruit in response to Pseudomonas syringae pv. actinidiae Infection. Int. J. Mol. Sci. 19:373. doi: 10.3390/ijms19020373
Wang, Z., Liu, Y., Li, L., Li, D., Zhang, Q., Guo, Y., et al. (2017). Whole transcriptome sequencing of Pseudomonas syringae pv. actinidiae-infected kiwifruit plants reveals species-specific interaction between long non-coding RNA and coding genes. Sci. Rep. 7:4910. doi: 10.1038/s41598-017-05377-y
Wasternack, C., and Hause, B. (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. Ann. Bot. 111, 1021–1058. doi: 10.1093/aob/mct067
Wurms, K., Cui, W., Ah Chee, A., Rees-George, J., Bublin, M., and Breiteneder, H. (2011). Down regulation of putative defence-associated transcripts correlates with ripe rot symptoms on kiwifruit (Actinidia chinensis). J. Phytopathol. 159, 435–442. doi: 10.1111/j.1439-0434.2011.01786.x
Wurms, K., Gould, E., Ah Chee, A., Taylor, J., Curran, B., and Reglinski, T. (2017). Elicitor induction of defence genes and reduction of bacterial canker in kiwifruit. N. Z. Plant Protect. 70, 272–284. doi: 10.30843/nzpp.2017.70.61
Keywords: elicitor, phytohormone, pathogenesis-related protein, β-1, 3-glucosidase, induced resistance
Citation: Reglinski T, Wurms K, Vanneste J, Ah Chee A, Yu J, Oldham J, Cornish D, Cooney J, Jensen D, Trower T, Taylor J, Schipper M, Alavi M and McKenzie C (2022) Transient Changes in Defence Gene Expression and Phytohormone Content Induced by Acibenzolar-S-Methyl in Glasshouse and Orchard Grown Kiwifruit. Front. Agron. 3:831172. doi: 10.3389/fagro.2021.831172
Received: 08 December 2021; Accepted: 30 December 2021;
Published: 27 January 2022.
Edited by:
Davide Spadaro, University of Turin, ItalyReviewed by:
Giorgio Gambino, Institute for Sustainable Plant Protection, National Research Council (CNR), ItalyPedro Laborda, Nantong University, China
Copyright © 2022 Reglinski, Wurms, Vanneste, Ah Chee, Yu, Oldham, Cornish, Cooney, Jensen, Trower, Taylor, Schipper, Alavi and McKenzie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tony Reglinski, dG9ueS5yZWdsaW5za2lAcGxhbnRhbmRmb29kLmNvLm56
 Tony Reglinski
Tony Reglinski Kirstin Wurms
Kirstin Wurms Joel Vanneste
Joel Vanneste Annette Ah Chee
Annette Ah Chee Janet Yu1
Janet Yu1 Janine Cooney
Janine Cooney Joseph Taylor
Joseph Taylor Magan Schipper
Magan Schipper Catherine McKenzie
Catherine McKenzie