
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Agron. , 10 January 2022
Sec. Disease Management
Volume 3 - 2021 | https://doi.org/10.3389/fagro.2021.807112
Plant-parasitic nematodes represent a substantial constraint on global food security by reducing the yield potential of all major crops. The soybean cyst nematode (SCN) (Heterodera glycines Ichinohe) is widely distributed across important soybean production areas of the U.S., being the major soybean yield-limiting factor, especially in the Midwestern U.S. Double cropped (DC) soybean is commonly planted following winter wheat. We previously reported double-cropping soybean fields with reduced SCN counts compared to fallow at both R1 growth stage (beginning of flowering) (−31.8%) and after soybean harvest (−32.7%). To test if higher counts of beneficial and SCN antagonistic microorganisms could be correlated with the suppression of SCN in fields previously planted with wheat, three field locations with noted SCN suppression were selected for a metagenomics study. Ten subplots were selected (5 wheat and 5 fallow pre-soybean) from each location. A total of 90 soil samples were selected: 3 fields ×2 treatments × 3 timepoints × 5 replications. Three DNA markers targeted distinct microbial groups: bacteria (16S V4-V5), fungi (ITS2), and Fusarium (tef1). Amplicons were sequenced using an Illumina MiSeq platform (300 bp paired-end). Sequencing datasets were processed in R using the DADA2 pipeline. Fungal populations were affected by location in all sampling periods and differed significantly between DC and fallow plots at soybean planting and after harvest (P < 0.001). Several enriched fungal and bacterial taxa in wheat plots, including Mortierella, Exophiala, Conocybe, Rhizobacter spp., and others, were previously reported to parasitize SCN and other plant-parasitic nematodes, suggesting a potential role of beneficial microbes in suppression of SCN in soybean fields double-cropped with wheat.
Plant-parasitic nematodes are a threat to many crop production systems worldwide, representing a substantial constraint on global food security. Annual crop yield losses linked to nematode parasitism average 12.3% worldwide, reaching ~$157 billion, notably in low technology production zones, exacerbating poverty, food insecurity, and malnutrition (Singh et al., 2015; Bernard et al., 2017; Coyne et al., 2018). Besides direct losses tied to nematodes, these organisms may be part of disease complexes, often causing root system wounds, and allowing other pathogens to infect plants (Xing and Westphal, 2006; Coyne et al., 2018). In the U.S., the soybean cyst nematode (SCN) (Heterodera glycines Ichinohe) is widely disseminated in all major soybean production areas (Tylka and Marett, 2021). SCN is a significant soybean yield-limiting factor, especially in the Midwestern U.S. (Bradley et al., 2021), and yield losses caused by SCN can reach 60% for susceptible cultivars (Hershman, 2014). Moreover, SCN may cause up to 30% of losses without noticeable aboveground symptoms (Mueller et al., 2016), requiring root inspection for proper assessment of pathogen incidence. Several cropping practices are recommended to manage SCN, including using resistant varieties, crop rotation, weed management, and applying nematicides and biocontrol agents as seed treatments (Mueller et al., 2016; Rocha et al., 2021a).
Double cropping (DC) is described as producing more than one crop on the same portion of land in a single growing season (Gesch and Johnson, 2015). DC soybean is frequently planted following the harvest of winter wheat in mid-to-late June (Nafziger, 2009). Double cropping has been more successful in the southern portion of Illinois, due to more favorable weather conditions allowing earlier wheat harvest and soybean planting, while warmer Fall weather provides winter crops conditions to grow for an extended window before being exposed to freezing temperatures (Nafziger, 2009). Knowing that farmers double-crop soybean with wheat in Southern Illinois, with several research reports indicating wheat as a potential suppressor SCN populations, we previously reported soybean fields double-cropped with wheat having reduced SCN counts compared to fallow strips at the R1 growth stage (beginning of flowering) (−31.8%) and after soybean harvest (−32.7%) (Fehr et al., 1971; Rocha et al., 2021b). Similarly, additional research points to the suppression effects of wheat on SCN populations, with authors hypothesizing that a composite of factors is involved. Environmental aspects, the influence of wheat stubble, wheat root exudates, and mechanical interference with host recognition by SCN are proposed to contribute to reduced SCN population densities where wheat preceded soybean (Baird and Bernard, 1984; Koenning, 1991; Hershman and Bachi, 1995; Long and Todd, 2001; Warnke et al., 2006; Wight et al., 2011; Bernard, 2018). However, results from the current study and prior literature suggest that the suppressive effects of wheat on SCN are not entirely observed at soybean planting, indicating that wheat production alone may not be responsible for all SCN suppression. Broder and Wagner (1988) reported an array of fungi groups present in decomposing wheat residue. Noteworthily, bacterial and fungal species are regularly found associated with SCN cysts, with numerous reports in the literature implicating changes in soil microbial communities in suppressing nematode populations in production fields (Tyler et al., 1987; Nour et al., 2003; Song et al., 2016). Other studies report discrepancies in the rhizosphere microbial communities of healthy vs. diseased roots (Mendes et al., 2011; Wolfgang et al., 2019).
In the soil, microbial communities are linked with critical biological functions. They facilitate nutrient intake, nitrogen fixation, limit pathogen pressure, and secrete secondary metabolites, as plants and microbes evolved biochemical mechanisms to communicate with each other (Chaparro et al., 2012; De-la-Peña and Loyola-Vargas, 2014; Meena et al., 2017). Such beneficial organisms include plant growth-promoting rhizobacteria (PGPR), endophytic and arbuscular mycorrhiza (AMF), rhizospheric bacteria, endophytic fungi, actinomycetes, and other microorganisms with mutualistic or non-symbiotic relationships with plants (Meena et al., 2017; Dubey et al., 2020). These interactions often include specific microorganisms functioning in concert with the overall soil microbiome to shape plant health, disease suppression, and crop productivity (Chaparro et al., 2012). Recent progress in several sequencing-based molecular techniques brought a new perspective vis-à-vis the broadness of microbial diversity and powerful tools to elucidate some of the ecological principles driving these communities, their functions, and implications on plant health (Brown and Tiedje, 2011; Kielak et al., 2016). In many instances, these new techniques can be implemented to interpret patterns in microbial diversity and how they correspond with disease suppression in production soils (Peralta et al., 2018). Considering that growing wheat as a winter crop contributes to reduced SCN populations in the succeeding growing season and research suggests microbial community composition as a potential suppression factor, the objectives of this research are to: (1) describe fungal, bacterial, and Fusarium profiles from DC fields previously described to have SCN suppression; (2) identify differentially abundant taxonomic groups across treatments; (3) pinpoint potential microbial groups related to the suppression of SCN; (4) provide further evidence in support of the adoption of wheat in DC systems to manage SCN populations in soybean production fields.
Field experiments were conducted from 2017 to 2018 to assess the effect of wheat production on SCN population densities in DC soybean fields. Research plots were established in row crop fields located on commercial farms in Illinois, allowing to better simulate environmental conditions experienced by growers. Field sites were selected based on an SCN survey conducted by the Illinois Soybean Association (ISA) in 2017, where population densities were determined in 22 fields in 7 counties.
From this survey, nine fields were selected for field experiments, including three locations each for low, moderate, and high initial SCN population densities (Rocha et al., 2021b; Supplementary Figure 1). At each location, the experimental design consisted of two treatments (WT, winter wheat; FL, fallow) with 3 replications. Treatments were assigned to strips (9.14 m wide × 182.9 m long), with 3 strips allocated for winter wheat and 3 strips remaining in fallow over winter. Each strip was subdivided into 3 subplots (9.14 m wide × 61.0 m long). Thus, the study consisted of 18 subplots amounting to 1.1 hectares per location (Supplementary Figure 2). Wheat was planted in Fall 2017 and was terminated with herbicides 2 weeks after emergence to establish and maintain fallow strips. Herbicides were applied to both fallow and wheat fields in the fall and spring to maintain weed free plots and prevent SCN reproduction on volunteer soybeans and weeds, as over 100 weed species are reported to be potential hosts of SCN (Rocha et al., 2021a). Soybean was planted in all subplots following the wheat harvest. Additional field trial information is available in Rocha et al. (2021b).
The population densities of SCN were determined at four intervals: (1) wheat establishment (November 2017); (2) post-wheat harvest and before soybean planting (June 2018); (3) soybean R1 growth stage (August 2018); (4) after soybean harvest (October 2018). From each location, a total of 18 soil samples were collected (2 treatments × 3 replicates × 5 subplots per replicate) for each of the four sampling intervals.
Field locations with contrasting differences in SCN egg densities between wheat and fallow plots were selected for the current metagenomic study. All samples were ranked by the ratio of final [Pf—at soybean harvest (Oct. 2018)] to initial [Pi—at soybean planting (Jun. 2018)] SCN population density. Locations with more distinguished differences in population ratios among WT and FL plots over this period were selected. In these locations, SCN population densities had a sharp increase in FL plots but were stable in WT. These included fields three (Nashville), six (Towers), and eight (Oakdale), all located in Washington County, Illinois. From the original 18 samples collected from each field location (nine WT; nine FL), a total of 10 were selected: five with the largest reductions in SCN counts (five WT) and five with the sharpest increases in SCN counts (five FL). Location descriptions including counts throughout field trials are listed in Supplementary Table 1. For each plot selected, soil samples from 3-time points were included: post-wheat/pre-soybean, mid-soybean (R1 or beginning of flowering), and post-soybean. A total of 90 soil samples were included in the current study: 3 fields × 2 treatments (wheat and fallow) × 3 timepoints (pre-, mid-, and post-soybean) × 5 replications.
Soil samples were collected from each plot at each of the four pre-determined time intervals using a cylindrical soil probe. Each sample was a composite of 20 soil cores (2.5-cm-diam. × 20-cm-deep) collected in a zig-zag pattern, bulked in a bucket, sieved through a 6,350-μm-pore mesh, and kept in a cooler for transport. Subsequent soil samples were collected approximately from the same area at each time point, as collection points were geo-referenced to ensure a 0.30 m-foot distance from the preceding sampling location. This was done to circumvent potential effects of disturbed soil on microbial communities in the surrounding soil.
Total DNA was extracted from soil samples using the PowerLyzer PowerSoil DNA extraction kit (MO BIO Laboratories Inc., Carlsbad, CA, USA) following the manufacturer's suggestions. Final DNA concentrations and ratios (A260/A280 and A260/A230) were estimated utilizing a Nanophotometer P-30 (Implen, Munich, Germany), while quality was verified using 1% agarose gel electrophoresis. DNA samples were stored at −20°C pending downstream applications.
Three DNA markers were used to cover three distinct microbial groups: bacteria, fungi, and Fusarium species. Primers 515FB/926R targeted the 16S ribosomal RNA gene V4-V5 region (400–500 bp amplicon) for bacteria (Walters et al., 2016). For fungi, the internal transcribed spacer (ITS2) was covered using primers ITS86F/ITS4R, returning amplicons of ~370 bp (De Beeck et al., 2014). Finally, the translation elongation factor-1 alpha (tef1) gene was included to provide better resolution when speciating Fusarium species, as this genus comprises species pathogenic to both soybean and wheat, and the ITS region is often uninformative at the species level due to conserved regions across species (O'Donnell et al., 2015). The tef1 factor was first amplified using primers EF1 and EF2 (O'Donnell et al., 1998). Primers Fa150 and Ra-2 were then used to amplify a partial region of the translation elongation factor-1 alpha (tef1) gene (Cobo-Diaz et al., 2019). PCR products were cleaned with the QIAquick® PCR purification kit (QIAGEN, Germantown, MD, USA).
DNA library preparation and sequencing were executed by the Integrated Microbiome Resource (IMR) facility (Dalhousie University—Halifax, Nova Scotia, Canada). Samples were paired-end sequenced with an Illumina MiSeq platform using v3 chemistry (2 × 300 bp).
The sequencing dataset was processed in R (v4.0.2) (R Core Team, 2020) through RStudio (v1.3.1073) (RStudio Team, 2020) employing the DADA2 pipeline (v1.16) (Callahan et al., 2016). Primer trimming was initially performed using CutAdapt (v2.10) (Martin, 2011). Subsequent analyses followed steps suggested by the DADA2 pipeline, with minor modifications for each marker (16S, ITS2, and tef1). Briefly, reads were trimmed and filtered to exclude low-quality sequences, followed by a merging step with at least 12 bases of overlap. Chimeras were deleted using the consensus approach.
For bacteria, taxonomy was assigned using the SILVA 138 ribosomal RNA (rRNA) database (Quast et al., 2013). For ITS2, the UNITE ITS (v8.2) database was utilized to assign fungal taxonomy (Abarenkov et al., 2020). In the case of tef1, a custom database was developed using NCBI entries, following parameters outlined by Boutigny et al. (2019).
After taxonomy and abundance tables were generated, ASV abundance matrices and metadata files were uploaded to the Microbiome analyzer (https://www.microbiomeanalyst.ca/), a web-based platform providing comprehensive statistical, visual, and meta-analysis of microbiome datasets, running the MicrobiomeAnalystR package (Dhariwal et al., 2017; Chong et al., 2020) via R (v4.0.2). An initial low filter was applied, keeping features with 4 counts in at least 10% of samples. Subsequently, a 10% low variance filter was used. The counts matrix was normalized using total sum scaling (TSS) (Paulson et al., 2013).
The following analyses were performed for all markers (16S, ITS2, and tef1): Alpha diversity was estimated using Shannon's diversity index followed by either a T-test (winter option) or an F-Test (location). Subsequently, β-diversity was calculated using the Bray–Curtis similarity index from log-transformed data [log (x + 1)]. Factors included location and winter option. The significance of factors was checked using multivariate permutational analysis of variance (PERMANOVA). Major variance components of bacterial, fungal, and tef1 were plotted using Principal Coordinate Analysis (PCoA) using the Bray-Curtis index as the distance method. The relative abundance of fungal and bacterial phyla among winter option and sampling was tested using the Kruskal–Wallis test (P = 0.05).
Linear discriminant analysis (LDA) Effective Size (LEfSe) was performed to identify features at the genus and species levels most likely to explain differences among fallow and wheat fields (Segata et al., 2011). The LEfSe algorithm employs a non-parametric factorial Kruskal-Wallis (KW) sum-rank test to identify features (genera or species) with significant differential abundance, followed by Linear Discriminant Analysis to estimate the effect size of each differentially abundant feature. Features were considered significant at a p-value of 0.05 and with an effect size threshold of 2 (on a log10 scale). Detailed results with respective p-values and LDA scores are presented in Supplementary Tables 2–4 for bacteria, fungi, and Fusarium, respectively.
Finally, a co-occurrence network was created via MicrobiomeAnalyst using the Sparse Correlations for Compositional data (SparCC) as the distance measure of taxa between features (Friedman and Alm, 2012). The SparCC algorithm recognizes correlations considering that microbiomes are sparse compositional datasets (many taxa are not correlated with one another) by employing a sparse network and log-ratio transformed data. The algorithm performs iterations to find correlations that stand out of background correlations. Bacteria, fungi, and Fusarium datasets were merged, with the relative abundance of Fusarium being divided by the percentage of ITS sequences assigned to the Fusarium genus, as proposed by Cobo-Díaz et al. (2019), to bring further resolution and normalize the abundance of Fusarium species within fungi. Correlations were then drawn between taxa and SCN egg counts, which were obtained from the same soil samples used for this metagenomic study. SCN count data was added to already TSS normalized metagenomics data. Analyses were performed at both mid- and post-soybean, since SCN suppression was observed in wheat fields at these time points. Correlations were performed at genus and species levels, using 200 permutations, with cutoffs of P < 0.05 for significance and >0.50 for correlation. Detailed results for the analyses at the genus and species levels are presented in Supplementary Table 5.
Demultiplexed raw sequence datasets are available via the National Center for Biotechnology Information (NCBI) Sequence Read Archive accession PRJNA735240.
An output of 7,738,883 16S reads was acquired from high-throughput sequencing, culminating in 6,791,110 reads after filtering and processing. A total of 6,110,266 reads were available after low variance and low count filtering, averaging 67,892 reads per sample. 16S rarefaction curves exhibited a plateau in species richness after 25,000 reads (Supplementary Figure 3), indicating the sequencing depth was sufficient to cover the majority of bacterial taxa. The most abundant bacterial phylum across all sampling times was Proteobacteria, with relative abundance increasing from pre-soybean sampling (FL: 25.05%; WT: 26.69%) compared to mid-soybean (FL: 41.35%; WT: 39.10%), with a later decrease at post-soybean (FL: 23.76%; WT: 24.53%) (Table 1). Although compositions were not statistically different between winter options, wheat plots displayed significant higher counts of Proteobacteria (26.69%), Acidobacteria (22.98%), and Planctomycetota (8.59%) at pre-soybean compared to fallow plots, which had 25.05, 21.13, and 7.57% of Proteobacteria, Acidobacteria, and Planctomycetota, respectively (Table 1). Conversely, fallow plots had greater counts of Firmicutes compared to wheat plots at both pre-soybean (FL: 9.74%; WT: 7.03%) and post-soybean (FL: 10.96%; WT: 8.54%). Interestingly, counts of Firmicutes were significantly higher in wheat plots at mid-soybean (6.49%) compared to fallow plots (5.88%) (Table 1). Proteobacteria and Verrucomicrobiota were the only groups with higher relative abundance at mid-soybean compared to the other sampling periods.
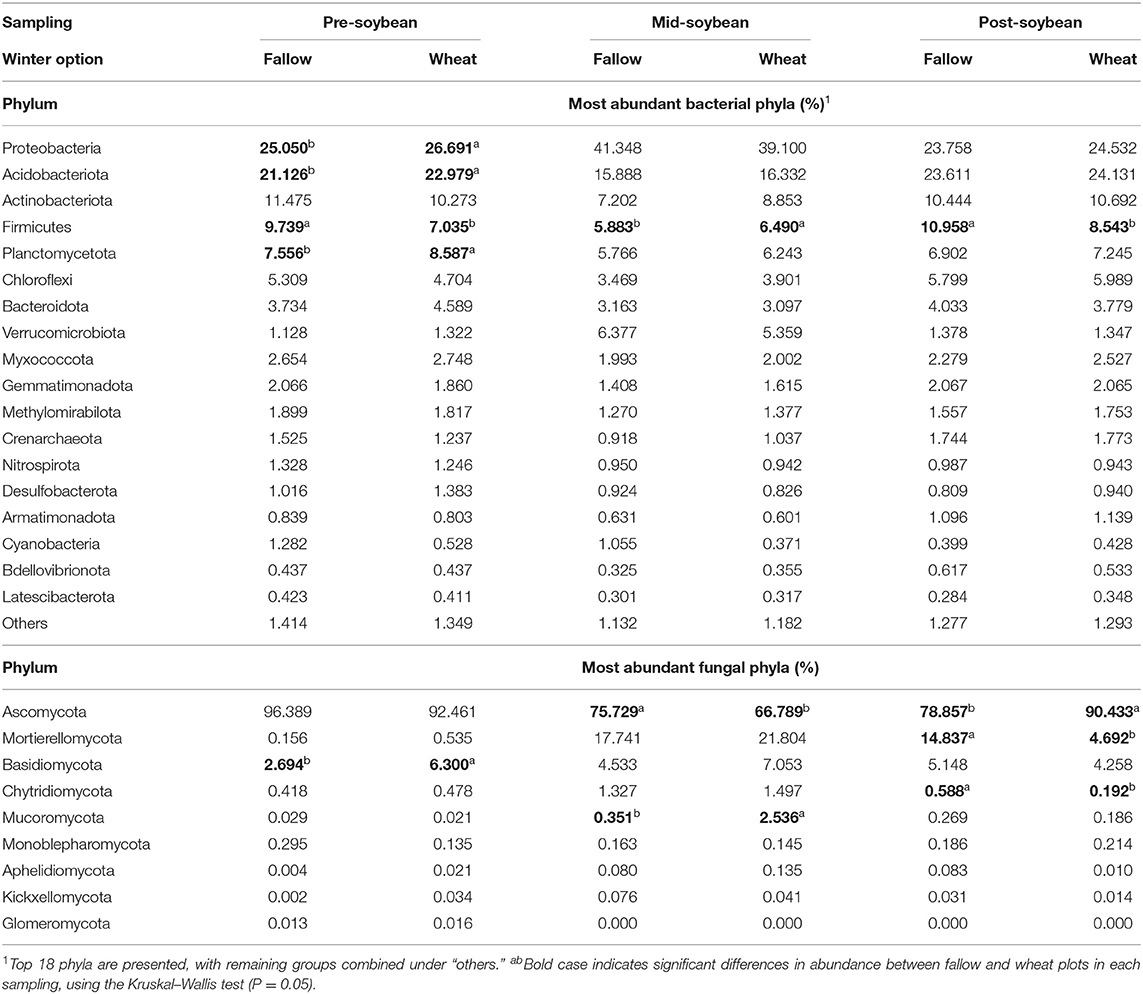
Table 1. The relative abundance of fungal and bacterial phyla affected by winter option and sampling.
For ITS2, a total of 3,468,232 fungal sequences were obtained from high-throughput sequencing, resulting in 2,062,499 reads after filtering and processing. After low count and low variance filtering, 2,058,190 reads were included in downstream analyses, averaging 26,387 reads per sample. The rarefaction curves denoted that the fungal richness did not increase after 15,000 sequences (Supplementary Figure 4), showing that sequencing coverage was sufficient to capture the majority of fungal taxa. The most abundant fungal phyla are presented in Table 1. Overall, Ascomycetes comprised the majority of fungal ASVs for all sampling times, even though variations were observed with greater counts at pre-soybean sampling (FL: 96.39%; WT: 92.46%), a drop at mid-soybean (FL: 75.73%; WT: 66.79%) and another relative increase at soybean harvest (FL: 78.86%; WT: 90.43%) (Table 1). Basidiomycota relative abundance was greater in wheat plots at pre-soybean (WT: 6.30%; FL: 2.69%). At mid-soybean, the abundance of Ascomycetes was significantly higher in fallow plots compared to wheat plots, whereas wheat plots had significantly higher counts of Mucoromycota (WT: 2.54%; FL: 0.35%). A trend of higher counts of Mortierellomycota and Basidiomycota was also observed in wheat plots compared to fallow plots at mid-soybean. At post-soybean, Ascomycota (WT: 90.43%; FL: 78.86%) abundance was higher in WT plots, while Mortierellomycota (WT: 4.69%; FL:14.84%) and Chytridiomycota (WT: 0.59%; FL: 0.19%) counts were higher in fallow vs. wheat plots (Table 1).
Alpha diversity was estimated using Shannon's diversity index, considering winter option (fallow or wheat) and location as factors. Overall, field location had a stronger influence on bacterial richness within samples in comparison to winter option (Table 2). Bacterial alpha diversity was comparable between winter option in all samplings: pre-soybean (T = 0.26206; P = 0.79529), mid-soybean (T = −0.15330; P = 0.87943), and post-soybean (T = −0.93780; P = 0.35659; Figure 1A and Table 2). Across field locations, bacteria alpha diversity was found to be similar at pre-soybean (F = 2.30620; P = 0.11895) and mid-soybean (F = 1.85410; P = 0.17672), but different in post-soybean samples (F = 12.523; P < 0.001; Figure 1B and Table 2).
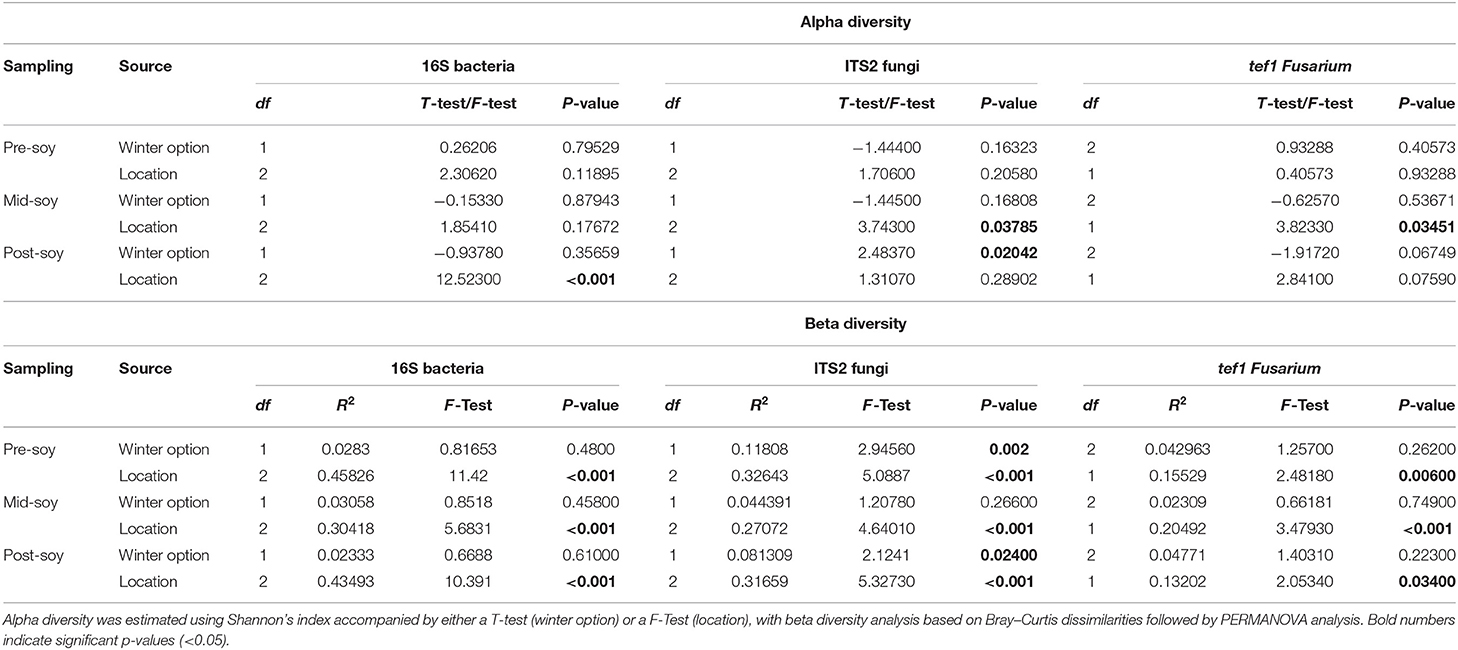
Table 2. Alpha and beta diversity analyses examining the effect of winter option (fallow or wheat) and location (Nashville, Oakdale, and Towers) on soil bacterial and fungal communities.
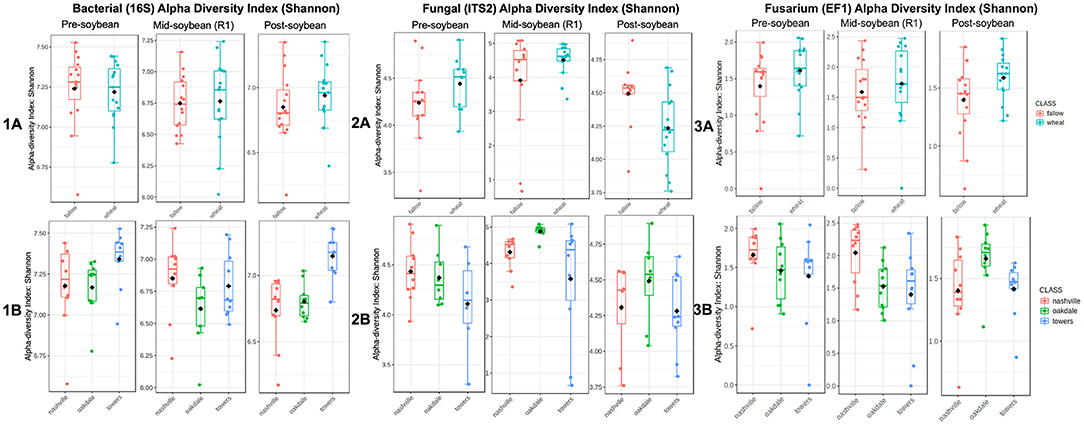
Figure 1. Comparison of bacterial (1), fungal (2), and Fusarium (3) communities' alpha diversities in each winter option (A) (fallow or wheat) and location (B) (Nashville, Oakdale, and Towers). Diversity was measured using Shannon's index based on sequencing data.
In the case of fungal communities, alpha diversity did not vary between winter option at pre-soybean (T = −1.4440; P = 0.16323) or mid-soybean (T = −1.4450; P = 0.16808), but diversity was higher in fallow plots at post-soybean (T = 2.48370; P = 0.02042; Figure 2A and Table 2). Among field locations, alpha diversity was significantly different at mid-soybean (F = 3.743; P = 0.03785), but similar at both pre-soybean (F = 1.706; P = 0.2058) and post-soybean (F = 1.3107; P = 0.28902; Figure 2B and Table 2).
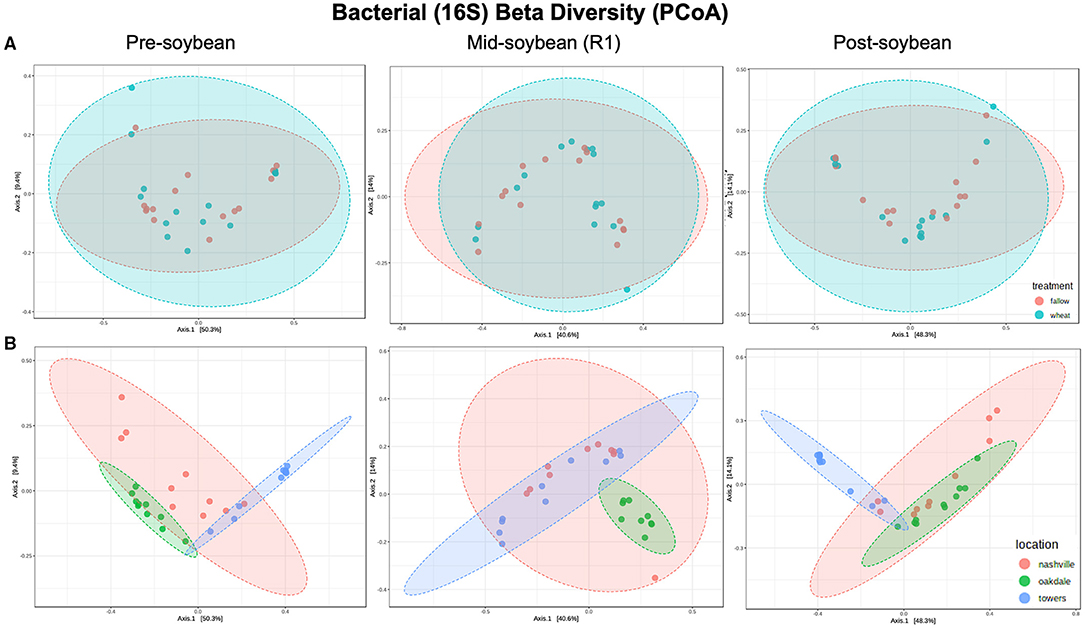
Figure 2. Principal coordinates analysis (PCoA) based on Bray-Curtis distances comparing the composition of bacterial communities across treatment (A) (fallow or wheat) and location (B) (Nashville, Oakdale, and Towers).
For Fusarium (tef1), Shannon's index suggests no influence of winter option on alpha diversity in all sampling times: pre-soybean (T = 0.93288; P = 0.40573), mid-soybean (T = −0.6257; P = 0.53671), and post-soybean (T = −1.9172; P = 0.06749; Figure 3A and Table 2). When comparing alpha diversity of Fusarium communities among field location, data points to differences at mid-soybean (F = 3.8233; P = 0.03541), but similar outcomes for pre- (F = 0.40573; P = 0.93288) and post-soybean (F = 2.8410; P = 0.07590) samples (Figure 3B and Table 2).
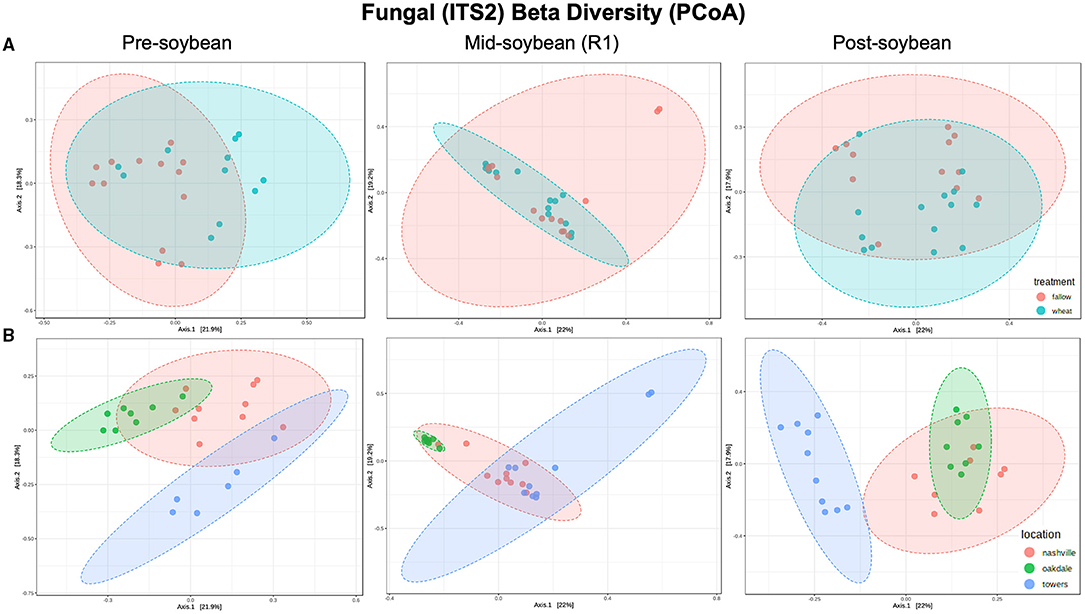
Figure 3. Principal coordinates analysis (PCoA) based on Bray-Curtis distances comparing the composition of bacterial communities across treatment (A) (fallow or wheat) and location (B) (Nashville, Oakdale, and Towers).
Beta diversity was estimated using the Bray–Curtis similarity index as the distance method from log-transformed data [log (x + 1)]. Field location, but not winter option, was the primary driver of the variation in bacterial communities (Table 2). Bacterial composition profiles were comparable when comparing wheat and fallow in all sampling intervals: pre- (R2 = 0.0283; P = 0.48), mid- (R2 = 0.03058; P = 0.458), and post-soybean (R2 = 0.02333; P = 0.610; Figure 2A and Table 2). An effect of field location on bacterial communities was detected in all samplings: pre- (R2 = 0.45826; P < 0.001), mid- (R2 = 0.30418; P < 0.001), and post-soybean samplings (R2 = 0.43493; P < 0.001; Figure 2B and Table 2).
Although the overall composition of bacterial communities was similar between wheat and fallow plots, the Bray-Curtis index indicates wheat altering the composition patterns of fungal communities. Differences were noted at pre- (R2 = 0.11808; P = 0.002) and post-soybean (R2 = 0.081309; P = 0.024), with comparable profiles at mid-soybean (R2 = 0.044391; P = 0.266; Figure 3A and Table 2). Similar to bacterial communities, the fungal beta diversity was strongly affected by location during pre- (R2 = 0.32643; P < 0.001), mid- (R2 = 0.27072; P < 0.001), and post-soybean (R2 = 0.31659; P < 0.001) samplings (Figure 3B and Table 2).
For Fusarium (tef1), no effect of winter option on beta diversity was observed: pre- (R2 = 0.042963; P = 0.26200), mid- (R2 = 0.02309; P = 0.74900), and post-soybean (R2 = 0.04771; P=0.22300; Figure 4A and Table 2). Across field locations, the composition of Fusarium communities was significantly different at pre- (R2 = 0.15529; P < 0.001), mid- (R2 = 0.20492; P < 0.001) and post-soybean (R2 = 0.13202; P = 0.03400; Figure 4B and Table 2).
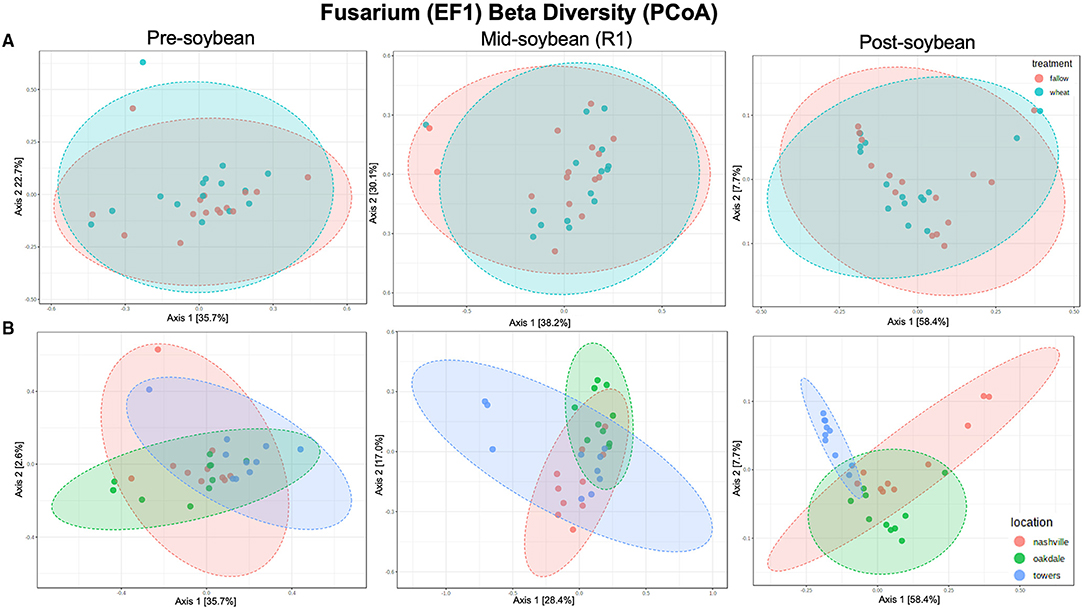
Figure 4. Principal coordinates analysis (PCoA) based on Bray-Curtis distances comparing the composition of Fusarium communities across treatment (A) (fallow or wheat) and location (B) (Nashville, Oakdale, and Towers).
The overall composition of bacterial communities was analyzed at both family and genus levels. Differences at the family level were predominantly due to sampling times, with similar profiles when analyzing samples across winter options (Figure 5A). For instance, at mid-soybean, an overall rise in Comamonadaceae and Chtoniobacteraceae was observed, but patterns were comparable among the wheat and fallow fields (Figure 5A).
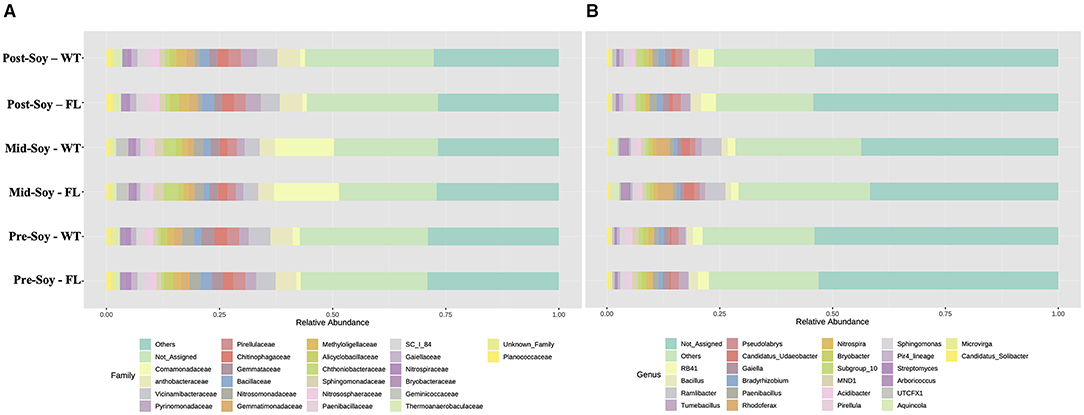
Figure 5. Relative abundance of bacterial communities (16S) sunder different winter options (fallow of wheat) and sampling times (pre, mid and post-soybean). (A) Family; (B) genera.
Similar to the family-level analysis, the overall composition of bacterial communities at the genus level displayed a similar profile between winter options (Figure 5B). In fact, the abundance of Rhodoferax, Ramlibacter, Arboricoccus, and Aquincola increased at mid-soybean, but counts were similar across treatment at each sampling. The following LEfSe analyzes provide a more in-depth assessment showing a significant association of specific genera with wheat plots throughout the soybean growing season.
Although the overall composition of bacterial communities was similar among the wheat and fallow plots, as indicated by the previously described beta diversity analysis, several genera of bacteria were found to be significantly enriched in wheat plots when the LEfSe method was used. At pre-soybean, a sum of 62 bacterial genera was significantly different between fallow and wheat plots, 19 enriched in fallow and 43 in wheat plots (Supplementary Table 2). Genera with highest LDA scores in wheat plots included Citrifermentans (LDA = 4.08; P < 0.001), Acidibacter (LDA = 3.99; P = 0.012), Terrimonas (LDA = 3.95; P <0.026), Ellin6067 (LDA = 3.99; P < 0.001), and Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium (LDA = 3.77; P = 0.036; Figure 6 and Supplementary Table 2).
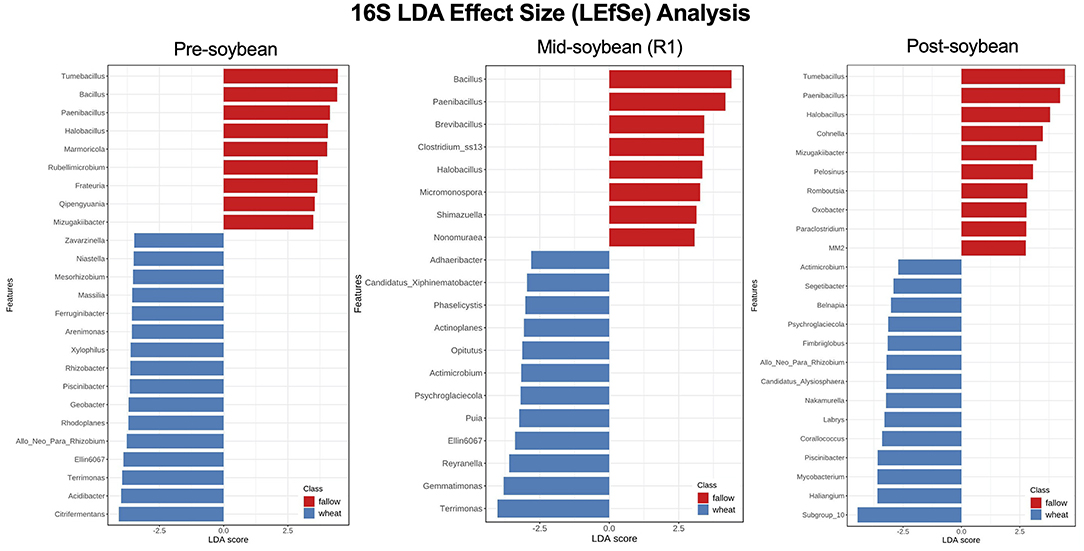
Figure 6. Top 25 bacterial genera most likely to explain differences between winter options (fallow or wheat) at pre-, mid-, and post-soybean, using the LEfSe method. Features are ranked by their LDA scores, with blue/red bars indicating features enriched in wheat/fallow plots (P < 0.05). Supplementary Table 2 includes all analyzed genera and their respective P-values.
Throughout later season samplings, the number of enriched genera across winter options dropped to 20 at mid soybean (FL: 8; WT: 12) and to 24 at soybean harvest (fallow: 10; wheat: 14), suggesting that bacterial communities returned to equilibrium at these sampling dates and that wheat may have only a short-term effect on soil bacterial communities (Supplementary Table 2). At mid-season, Terrimonas (LDA = 4.02; P = 0.023), Gemmatimonas (LDA = 3.80; P = 0.008), Reyranella (LDA = 3.6; P = 0.018), Ellin6067 (LDA = 3.39; P = 0.045), and Puia (LDA = 3.24; P = 0.0.32) had higher LDA scores in wheat fields (Figure 6). At post-soybean, the top 5 LDA scores were attributed to Subgroup_10 (LDA = 4.45; P = 0.029), Haliangium (LDA = 3.6; P = 0.040), Mycobacterium (LDA = 3.6; P = 0.021), Piscinibacter (LDA = 3.59; P = 0.029), and Corallococcus (LDA = 3.4; P = 0.023; Figure 6). While not significant, a trend of higher counts of Bradyrhizobium, a soybean nitrogen-fixing genus, was observed in wheat plots compared to fallow plots at pre- (LDA = 4.18; P = 0.11) and mid-soybean (LDA = 4.26; P = 0.061; Supplementary Table 2).
After considering the relative abundance of fungal taxa at both family and genus levels, the overall fungal family profiles differed significantly between sampling times (Figure 7A), with some differences also noted among winter option within each sampling time. At pre-soybean, Mortiriellaceae was only detected in wheat plots and, although the overall abundance of this family increased at mid-soybean, wheat plots had greater relative abundance in comparison to fallow. Interestingly, after soybean harvest, Mortiriellaceae abundance in fallow plots was higher than in wheat (Figure 7A). Plectosphaerellaceae abundance followed a comparable pattern observed for Mortiriellaceae. Nectriaceae, which comprises several plant pathogenic species (e.g., Fusarium), initially had higher relative abundance in wheat plots, but after mid-soybean, fallow plots displayed slightly higher Nectriacea abundance (Figure 7A). While analyzing individual genera associated with the winter option, several of these fungal groups were significantly enriched (Figure 7B), with further details and p-values available in Supplementary Table 3.
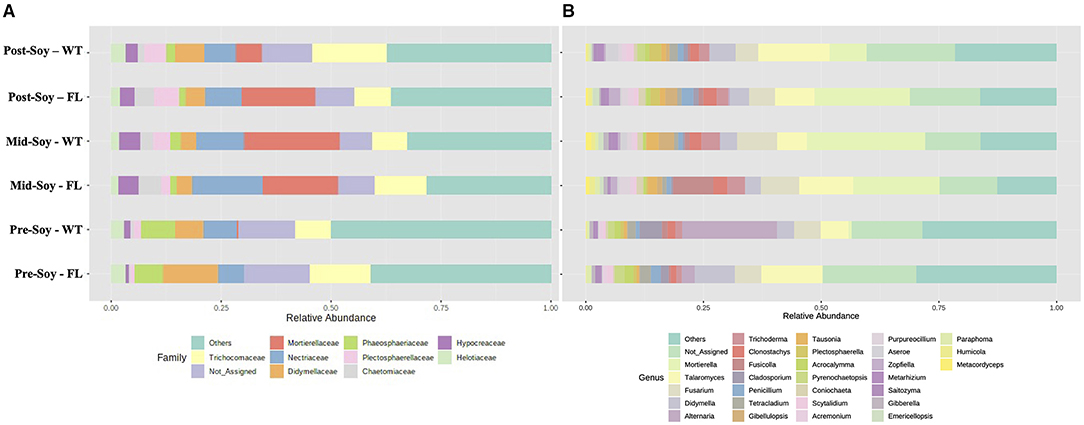
Figure 7. Relative abundance of fungal communities (ITS2) sunder different winter options (fallow of wheat) and sampling times (pre, mid, and post-soybean). (A) Top 30 fungal families; (B) top 10 fungal genera.
Using LEfSe, from a total of 27 fungal genera with significantly different noted abundances between fallow and wheat plots, 10 were enriched in fallow and 17 in wheat plots at pre-soybean. Fungal genera with highest LDA scores in wheat plots at pre-soybean included Alternaria (LDA = 5.71; P = 0.001), Conocybe (LDA: 5.07; P = 0.012), Cladosporium (LDA: 5.04; P = 0.004), Parastagonospora (LDA = 4.85; P = 0.003), and Mortierella (LDA: 4.28, P = 0.029; Figure 8 and Supplementary Table 3). Although Alternaria was identified in both wheat and fallow plots at soybean planting, the relative abundance of this genus was significantly higher in WT fields. These results could be linked to the fact that Alternaria comprises a genus with wheat pathogenic species, including Alternaria triticina, the causal agent of leaf blight of wheat. A trend of higher counts of Trichoderma in wheat plots was observed at pre-soybean (LDA: 4.39; P = 0.111; Supplementary Table 3).
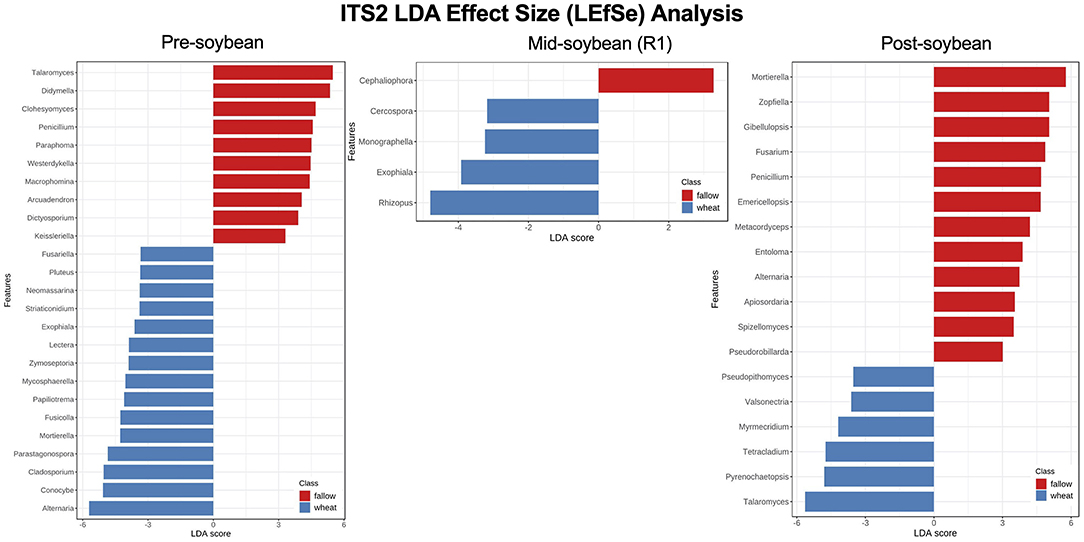
Figure 8. Top 25 fungal genera most likely to explain differences between winter options (fallow or wheat) at pre-, mid-, and post-soybean, using the LEfSe method. Features are ranked by their LDA scores, with blue/red bars indicating features enriched in wheat/fallow plots (p < 0.05). Supplementary Table 3 includes all analyzed genera and their respective P-values.
The number of expressive features identified using LEfSe dropped to four at mid-soybean (fallow: 1; wheat: 3) and increased to 18 at harvest (fallow: 12; wheat: 6). Rhizopus (LDA = 4.8; P = 0.002), Exophiala (LDA = 3.92; P = 0.048), Monographella (LDA = 3.24, P = 0.039), and Cercospora (LDA = 3.18, P = 0.023) were enriched in wheat plots at mid-soybean, while Talaromyces (LDA = 5.64; P < 0.001), Pyrenochaetopsis (LDA = 4.8; P < 0.001), Tetracladium (LDA = 4.74; P = 0.035), Myrmecridium (LDA = 4.19; P = 0.003), and Pseudopithomyces (LDA = 3.53; P = 0.035) were enriched in wheat plots at soybean harvest (Figure 8 and Supplementary Table 3). LEfSe analysis at the species level revealed higher counts of the potential biological control agents Mortierella elongata (LDA = 4.98; P = 0.048), M. exigua (LDA = 4.66; P = 0.027), and M. gamsii (LDA = 3.69; P = 0.012) in wheat plots at mid-soybean (Supplementary Table 3).
Similar to bacterial and fungal communities, the abundance of Fusarium communities was influenced by both sampling and winter options. Although beta-diversity was not significant, some Fusarium species were significantly enriched in wheat plots (Supplementary Table 4). At pre-soybean, F. graminearum (LDA = 5.62; P < 0.001), F. merismoides (LDA = 5.1; P = 0.0406), and F. tricinctum (LDA = 4.32; P = 0.005) were significantly enriched in wheat plots (Supplementary Table 4).
At mid-soybean, F. ipomoeae (Fusarium incarnatum-equiseti species complex—FIESC) abundance was greater in fallow plots (LDA = 5.68; P = 0.015), while F. graminearum counts were higher in wheat plots at soybean harvest (LDA = 4.94; P = 0.022; Supplementary Table 4). After soybean harvest, even though the abundance of wheat pathogenic Fusarium species (F. graminearum and F. armeniacum) was lower compared to the abundance at soybean planting, numbers in wheat plots trended higher compared to fallow (Figure 9 and Supplementary Table 4). Fusarium solani, a soybean pathogenic species, was enriched in fallow plots at post-soybean (LDA = 4.55; P = 0.024).
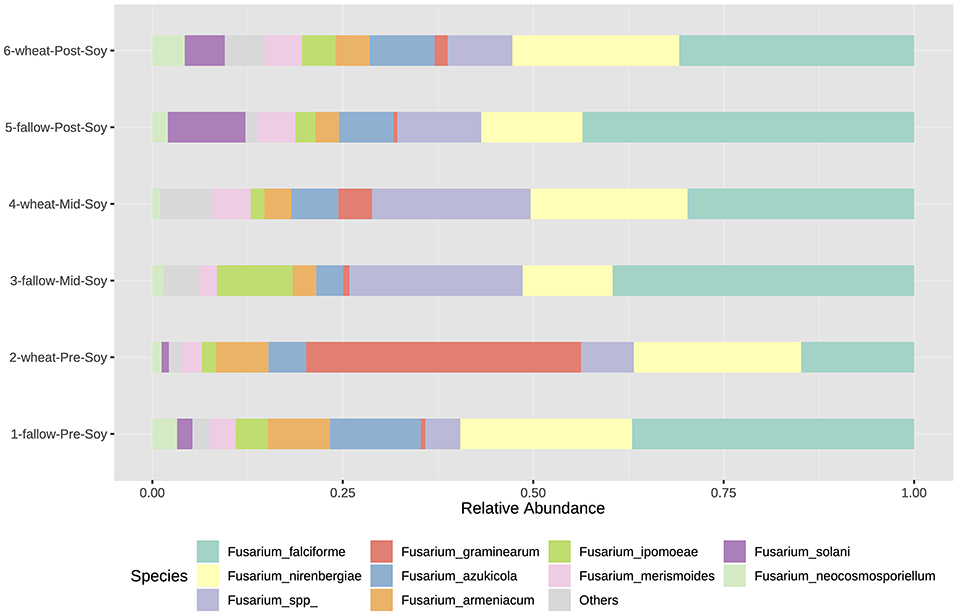
Figure 9. Relative abundance of Fusarium species (tef1) under different winter options (fallow of wheat) and sampling times (pre, mid, and post-soybean).
The co-occurrence network analysis using samples from mid-soybean season revealed 11 positive and 2 negative significant correlations (p < 0.05 and R2 > 0.50) between fungal and bacterial genera and SCN egg counts, and 5 positive correlations at the species level. Negative correlations included Ktedonobacter (R = −0.5322; P = 0.005) and Pullulanibacillus (R = −0.5594; P = 0.005). Looking at post-soybean harvest, 82 (35 negative, 47 positive) significant interactions were observed at the genus level, while 46 (30 negative, 16 positive) were detected at the species level. Noteworthy taxa negatively correlated with SCN eggs counts included Acrocalymma spp. (R = −0.6834; P = 0.005), Clonostachys rosea (R = −0.8769; P = 0.005), Mortierella alpina (R = −0.7914; P = 0.005), Metarhizium anisopliae (R = −0.5299; P = 0.0149), and Purpureocillium lilacinum (R = −0.6458; P = 0.005). Further details, correlation scores and p-values are available in Supplementary Table 5.
Field experiments were conducted with standard crop production practices in Illinois to evaluate the effect of wheat production on SCN population densities. SCN egg counts from double-cropping soybean plots were reduced compared to fallow plots at both R1 stage (−31.8%) and after soybean harvest (−32.7%) (Rocha et al., 2021b). In the present research, we hypothesized that shifts in soil microbial communities are tied to the suppression of SCN in these systems.
To study the potential impact of soil microbial communities on SCN, we initially determined the alpha and beta diversity of bacterial and fungal communities in soil samples. While alpha diversity refers to the species richness in a sample, beta diversity analysis indicates the degree of variation in community composition across samples or environments, in our case winter option or location (Whittaker, 1960). The alpha diversity analysis indicated no effect of the winter option on either bacterial or fungal richness within samples, while substantial differences in richness were associated to field location. However, the composition of fungal communities was significantly distinct across fallow and wheat samples. Collectively, our alpha and beta results propose that planting wheat over winter has a more prominent effect on specific taxa rather than on the overall composition of microbial communities in the tested soil samples. Similarly, in a recent study, the inclusion of wheat straw had slight to no impact on either bacterial or fungal richness. However, differences in microbial communities of watermelon rhizosphere were reported to be enhanced along with a subsequent reduction in Fusarium wilt pressure (Tang et al., 2020). Broder and Wagner (1988) reported lower bacterial populations recovered from decomposing wheat residue compared to residue from soybean and corn, reflecting reduced wheat residue decomposition levels compared to the other materials. Although bacterial communities may rapidly change due to wheat residue decomposition, fungal communities' responses may last longer (Tardy et al., 2015). While Penton et al. (2014) reported 917 fungal genera in a study analyzing the microbial community structure of suppressive and non-suppressive soils, the majority of differences across environments were tied to fewer than 40 genera, including some endophytic species and mycoparasites. Slight variations in microbial profiles triggered by winter wheat may be the basis of SCN suppression in soybean fields double cropped with wheat.
The primary driver of bacterial communities in the present research was field location, as the overall beta diversity did not vary across wheat and fallow plots. Even though the overall community composition was similar across these plots, some bacterial groups were enriched in wheat plots. Several bacterial genera enhanced in DC plots, including Rhizobacter, Shinella, Variovax, Ideonella, Gemmatimonas, Actinoplanes, Phaselicystis, and Candidatus, have been previously reported in nematode suppressive soils (Topalovic et al., 2020). Other genera are major constituents of the SCN microbiome, including Chthoniobacter, Halangium, Massilia, Mesorhizobium, Niastella, and Rhizobacter (Hu et al., 2019), suggesting that some of the soil bacteria enhanced by wheat production may also colonize SCN cysts, potentially affecting SCN population densities.
Regarding fungal communities, several genera observed more prominently in wheat plots are documented to be pathogenic or associated with wheat, including Alternaria species (Ramires et al., 2018), Fusarium spp., Mycosphaerella, Monographella, Neoascochyta (Golzar et al., 2019), Zymoseptoria, Parastagonospora, and Pseudopithomyces spp. (Perelló et al., 2017). Having a susceptible host present (wheat) offers an environment for the increase of these pathogenic species. Although Alternaria includes several plant-pathogenic species, research has shown its potential to reduce SCN egg counts in greenhouse (Haarith et al., 2021). Some of the Fusarium spp. that were enriched in wheat plots (F. graminearum and F. tricinctum) are known wheat pathogens, causing wheat head blight (Cowger et al., 2020) and F. merismoides causing seedling diseases (Liu et al., 2013). As some of these species are also pathogenic to corn, and a typical crop rotation in a corn/soybean production field would be corn (year 1), wheat/soybean (year 1/year 2), and corn (year 3), wheat could potentially increase these pathogenic species in the following corn season. Greater counts of saprophytic fungi (Pluteus, Rhizopus, Pseudopithomyces, and others) throughout the season could be linked to higher amounts of decaying wheat straw in the soil (Broder and Wagner, 1988; Fukasawa et al., 2011; Tardy et al., 2015; Wanasinghe et al., 2018).
In addition to wheat pathogenic and saprophytic fungal groups, several fungal genera previously documented to be engaged in the suppression of plant-parasitic nematode populations were enriched in wheat plots, such as Conocybe, Cladosporium, Acrocalymma, Exophiala, Motierella, and others. Conocybe spp. produce toxin droplets able to immobilize and kill soil-borne nematodes (Hutchison et al., 1996). Cladosporium and Exophiala, endophytic fungal species, were reported to activate systemic defensive responses in pine trees, reducing the incidence of the pine wood nematode (Bursaphelenchus xylophilus) (Chu et al., 2019). Similarly, Exophialla spp. reduced SCN egg density in a greenhouse assay evaluating potential biological control agents (BCA) of SCN (Haarith et al., 2021). Research indicates that this species can penetrate through SCN not only via natural openings, but also via cyst walls (Chen and Dickson, 1996). Mortierella alpina and other Mortierella species have likewise been recognized to dominate decomposing wheat residue (Tardy et al., 2015) and to inhibit SCN egg hatching (Meyer et al., 2004). The enhanced abundance of these species was also correlated with SCN egg density in continuous soybean fields (Strom et al., 2020). Mortierella was also reported to lower egg hatching and trap nematodes via hyphal adhesion, penetration of the nematode's cuticle, and the subsequent digestion of its cellular content (AL-Shammari et al., 2013; DiLegge et al., 2019). Moreover, Mortierella is noted to be antagonistic to other plant-parasitic nematodes, including Meloidogyne spp. (AL-Shammari et al., 2013; DiLegge et al., 2019; Topalovic et al., 2020). In wheat, Mortierella was negatively correlated with reproduction indexes of the cereal cyst nematode, Heterodera avenae (Qiu et al., 2020). Likewise, this fungal group has been reported to be part of the microbiome colonizing SCN cysts, with Mortierella counts enhanced at mid-season soybean (Hu et al., 2018). In our study, reduced SCN population densities were observed during an analogous time window when soybean was planted following wheat strips. This suggests that these antagonists may contribute to the suppressive effect of wheat on SCN population densities.
Additional fungal species previously reported as potential BCAs were negatively correlated with SCN abundance, including Metarhizium anisopliae and Purpureocillium lilacinum. M. anisopliae has been reported to trigger 78.5% juvenile mortality in Meloidogyne incognita in vitro (Youssef et al., 2020). Similarly, a M. anisopliae isolate collected from a suppressive soil parasitized up to 47.15% of Heterodera avenae (cereal cyst nematode) juveniles in vitro (Ghayedi and Abdollahi, 2013). Purpureocillium isolates are known nematode parasites, as some isolates are the base of some commercial biological products. P. lilacinum may also live as a saprophyte, thriving in a wide range of organic materials, but studies have demonstrated that P. lilacinum can rapidly infect and colonize nematode eggs, even disrupting cuticle layers of juveniles inside infected eggs (Stirling, 2014).
Multiple mechanisms are often associated with the lower disease pressure induced by soil microorganisms, including competition for space and resources, mycoparasitism, direct antibiosis, and the activation of induced systemic resistance (ISR) in plants, enhancing thus their defense against a broad range of pathogenic organisms (Pozo and Azcón-Aguilar, 2007; Jung et al., 2012; Selosse et al., 2014; Pimentel et al., 2020; Topalovic et al., 2020). Xu et al. (2015) reported an increase in soil enzymatic activity and microbial biomass associated with altered microbial communities in the watermelon rhizosphere when wheat was grown as a companion crop, which was suggested to be linked to Fusarium wilt suppression. By adding another crop over the winter in double cropping soybean, there is a potential for an escalation in the richness of soil microbial communities (Peralta et al., 2018), as research reveals that plants can shape the soil microbiome via the secretion of root exudates. In fact, the adaptation of plants to withstand multiple abiotic and biotic stresses is partly credited to the long-term co-evolution of plants and associated beneficial microorganisms (Dubey et al., 2020). Broder and Wagner (1988) reported Aspergillus, Fusarium, Penicillium, Pithomyces, Rhizoctonia, Rhizopus, and Trichoderma as major fungal genera present in decomposing wheat residue. Hu et al. (2017) pointed to fungi and bacteria parasitizing cysts as possible biological factors associated with SCN suppressive soils, which are characterized by low disease pressure even though both a virulent pathogen (SCN) and a susceptible host (soybean) are present.
One of the challenges of the adoption of biological control strategies to manage plant-pathogenic nematodes is that practices meant to conserve or enhance suppression of nematodes may not be effective across field sites, since they may rely on the presence of native antagonists (Timper, 2014). This may explain why differences in results are listed in the literature when wheat is used as a suppressor of SCN. To decrease this variation across environments, antagonists could be potentially introduced to field sites in conjunction with conservation practices to enhance the stability, effectiveness, and extent of disease suppression effects (Timper, 2014). Ultimately, specific antagonists must be isolated and further studied, as these key microorganisms may offer new management tools, cutting down costs, and promoting more sustainable disease management practices. Additionally, the effect of wheat suppressing SCN may become more pronounced after multiple growing seasons. Similar studies had indicated that, several years were needed before observing a significant impact of practices such as crop rotation or the use of cover crops on soil nematode communities. Long and Todd (2001) reported reductions in SCN population densities 3–4 years after the establishment of a double-cropping system. Similarly, Tyler et al. (1987) discussed that long-term tillage affected SCN population densities only 3 or 4 years after the establishment of their cropping system. Soil ecosystems may require attaining some degree of equilibrium before suppressiveness is noted, which implies that shifts in soil microbial communities due to cultural practices are incremental (Yeates et al., 1999; Venter et al., 2016; Grabau et al., 2017).
In the current study, metagenomic analysis reveals substantial contrasts in fungal communities across wheat and fallow fields throughout the soybean growing season, with a number of fungal and bacterial taxa being enriched in wheat fields. Several enriched groups in fields double cropped with wheat were previously described to parasitize SCN cysts and eggs, including Mortierella, Exophiala, Conocybe, Rhizobacter spp., and others, suggesting that microbial antagonists may be linked to the suppressive effects of wheat on SCN in DC soybean. Further research is needed to characterize these bacterial and fungal organisms in the soybean rhizosphere and SCN cysts to confirm their role in SCN suppression. To our knowledge, this is the first study employing next-generation sequence to explore the soil microbiome of soybean double cropped with winter wheat. In addition to correlations of microbial taxa with SCN, this work provides an extended wheat/fallow co-occurrence network analysis of bacterial and fungal taxa, which could be valuable for researchers working with soil microbial dynamics and ecology.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: NCBI [accession: PRJNA735240].
LR conducted sample processing, DNA extraction and quality check, analyzed the data, and wrote the manuscript with assistance from AF and JB. LR designed figures, formatted, and submitted manuscript. All authors have read and agreed to the published version of the manuscript.
This project was partially funded by the Illinois Soybean Association Checkoff Program.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
This work was part of first author's Ph.D. dissertation. We are thankful for the Illinois Soybean Association Checkoff Program for funding this research and for the assistance from SIU researchers, undergraduate, and graduate students.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2021.807112/full#supplementary-material
Abarenkov, K., Zirk, A., Piirmann, T., Pöhönen, R., Ivanov, F., Nilsson, R. H., et al. (2020). UNITE General FASTA Release for Fungi. Version 04.02.2020. Oslo: UNITE Community. doi: 10.15156/BIO/786368
AL-Shammari, T. A., Bahkali, A. H., Elgorban, A. M., El-Kahky, M. T., and Al-Sum, B. A. (2013). The use of Trichoderma longibrachiatum and Mortierella alpina against root-knot nematode, Meloidogyne javanica on tomato. J. Pure Appl. Microbiol. 7, 199–207. Available online at: https://microbiologyjournal.org/the-use-of-trichoderma-longibrachiatum-and-mortierella-alpina-against-root-knot-nematode-meloidogyne-javanica-on-tomato/
Baird, S. M., and Bernard, E. C. (1984). Nematode population and community dynamics in soybean-wheat cropping and tillage regimes. J. Nematol. 16, 379–389.
Bernard, E. C.. (2018). “Plant parasitic nematodes of Tennessee and Kentucky,” in Plant Parasitic Nematodes in Sustainable Agriculture of North America. Sustainability in Plant and Crop Protection, eds S. Subbotin and J. Chitambar (Cham: Springer), 305–325. doi: 10.1007/978-3-319-99588-5_12
Bernard, G. C., Egnin, M., and Bonsi, C. (2017). “The impact of plant-parasitic nematodes on agriculture and methods of control,” in Nematology-Concepts, Diagnosis and Control, eds M. M. Shah and M. Mahamoo (London: IntechOpen). 121–151. doi: 10.5772/intechopen.68958
Boutigny, A. L., Gautier, A., Basler, R., Dauthieux, F., Leite, S., Valade, R., et al. (2019). Metabarcoding targeting the EF1 alpha region to assess Fusarium diversity on cereals. PLoS ONE 14:e0207988. doi: 10.1371/journal.pone.0207988
Bradley, C. A., Allen, T., Sisson, A. J., Bergstrom, G. C., Bissonnette, K. M., Bond, J. P., et al. (2021). Soybean yield loss estimates due to diseases in the United States and Ontario, Canada from 2015-2019. Plant Health Prog. doi: 10.1094/PHP-01-21-0013-RS
Broder, M. W., and Wagner, G. H. (1988). Microbial colonization and decomposition of corn, wheat, and soybean residue. Soil Sci. Soc. Am. J. 52, 112–117. doi: 10.2136/sssaj1988.03615995005200010020x
Brown, C., and Tiedje, J. M. (2011). “Metagenomics: the paths forward,” in Handjournal of Molecular Microbial Ecology II: Metagenomics in Different Habitats, ed F. J. Bruijn (Hoboken, NJ: John Wiley and Sons), 579–588. doi: 10.1002/9781118010549.ch54
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Chaparro, J. M., Sheflin, A. M., Manter, D. K., and Vivanco, J. M. (2012). Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fertil. Soils 48, 489–499. doi: 10.1007/s00374-012-0691-4
Chen, S. Y., and Dickson, D. W. (1996). Fungal penetration of the cyst wall of Heterodera glycines. Phytopathology 86, 319–327. doi: 10.1094/Phyto-86-319
Chong, J., Liu, P., Zhou, G. Y., and Xia, J. G. (2020). Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protocols 15, 799–821. doi: 10.1038/s41596-019-0264-1
Chu, H. L., Wang, C. Y., Li, Z. M., Wang, H. H., Xiao, Y. G., Chen, J., et al. (2019). The dark septate endophytes and ectomycorrhizal fungi effect on pinus tabulaeformis carr. seedling growth and their potential effects to pine wilt disease resistance. Forests 10:140. doi: 10.3390/f10020140
Cobo-Diaz, J. F., Baroncelli, R., Le Floch, G., and Picot, A. (2019). A novel metabarcoding approach to investigate Fusarium species composition in soil and plant samples. FEMS Microbiol. Ecol. 95:fiz084. doi: 10.1093/femsec/fiz084
Cobo-Díaz, J. F., Baroncelli, R., Le Floch, G., and Picot, A. (2019). Combined metabarcoding and co-occurrence network analysis to profile the bacterial, fungal and Fusarium communities and their interactions in maize stalks. Front. Microbiol. 10:261. doi: 10.3389/fmicb.2019.00261
Cowger, C., Ward, T. J., Nilsson, K., Arellano, C., McCormick, S. P., and Busman, M. (2020). Regional and field-specific differences in Fusarium species and mycotoxins associated with blighted North Carolina wheat. Int. J. Food Microbiol. 323:108594. doi: 10.1016/j.ijfoodmicro.2020.108594
Coyne, D. L., Cortada, L., Dalzell, J. J., Claudius-Cole, A. O., Haukeland, S., Luambano, N., et al. (2018). Plant-parasitic nematodes and food security in Sub-Saharan Africa. Annu. Rev. Phytopathol. 56, 381–403. doi: 10.1146/annurev-phyto-080417-045833
De Beeck, M. O., Lievens, B., Busschaert, P., Declerck, S., Vangronsveld, J., and Colpaert, J. V. (2014). Comparison and validation of some ITS primer pairs useful for fungal metabarcoding studies. PLoS ONE 9:e97629. doi: 10.1371/journal.pone.0097629
De-la-Peña, C., and Loyola-Vargas, V. M. (2014). Biotic interactions in the rhizosphere: a diverse cooperative enterprise for plant productivity. Plant Physiol. 166, 701–719. doi: 10.1104/pp.114.241810
Dhariwal, A., Chong, J., Habib, S., King, I. L., Agellon, L. B., and Xia, J. (2017). MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 45, W180–W188. doi: 10.1093/nar/gkx295
DiLegge, M. J., Manter, D. K., and Vivanco, J. M. (2019). A novel approach to determine generalist nematophagous microbes reveals Mortierella globalpina as a new biocontrol agent against Meloidogyne spp. nematodes. Sci. Rep. 9:7521. doi: 10.1038/s41598-019-44010-y
Dubey, R. K., Tripathi, V., Prabha, R., Chaurasia, R., Singh, D. P., Rao, C. S., et al. (2020). “Belowground microbial communities: key players for soil and environmental sustainability,” in Unravelling the Soil Microbiome. Springer Briefs in Environmental Science, eds R. K. Dubey, V. Tripathi, R. Prabha, R. Chaurasia, D. P. Singh, C. S. Rao, et al. (Cham: Springer), 5–22. doi: 10.1007/978-3-030-15516-2_2
Fehr, W. R., Caviness, C. E., Burmood, D. T., and Pennington, J. S. (1971). Stage of development descriptions for soybeans, Glycine max (L.) Merrill. Crop Sci. 11, 929–931. doi: 10.2135/cropsci1971.0011183X001100060051x
Friedman, J., and Alm, E. J. (2012). Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 8:e1002687. doi: 10.1371/journal.pcbi.1002687
Fukasawa, Y., Osono, T., and Takeda, H. (2011). Wood decomposing abilities of diverse lignicolous fungi on nondecayed and decayed beech wood. Mycologia 103, 474–482. doi: 10.3852/10-246
Gesch, R. W., and Johnson, J. M. -F. (2015). Water use in camelina–soybean dual cropping systems. Agron. J. 107, 1098–1104. doi: 10.2134/agronj14.0626
Ghayedi, S., and Abdollahi, M. (2013). Biocontrol potential of Metarhizium anisopliae (Hypocreales: Clavicipitaceae), isolated from suppressive soils of the Boyer-Ahmad region, Iran, against J2s of Heterodera avenae. J. Plant Protect. Res. 53, 165–171. doi: 10.2478/jppr-2013-0025
Golzar, H., Thomas, G., Jayasena, K. W., Wright, D., Wang, C., and Kehoe, M. (2019). Neoascochyta species cause leaf scorch on wheat in Australia. Australas. Plant Dis. Notes 14,:1. doi: 10.1007/s13314-018-0332-3
Grabau, Z. J., Maung, Z. T. Z., Noyes, D. C., Baas, D. G., Werling, B. P., Brainard, D. C., et al. (2017). Effects of cover crops on Pratylenchus penetrans and the nematode community in carrot production. J. Nematol. 49, 114–123. doi: 10.21307/jofnem-2017-051
Haarith, D., Kim, D. G., Chen, S. Y., and Bushley, K. E. (2021). Growth chamber and greenhouse screening of promising in vitro fungal biological control candidates for the soybean cyst nematode (Heterodera glycines). Biol. Control 160:104635. doi: 10.1016/j.biocontrol.2021.104635
Hershman, D., and Bachi, P. (1995). Effect of wheat residue and tillage on Heterodera glycines and yield of doublecrop soybean in Kentucky. Plant Dis. 79, 631–633. doi: 10.1094/PD-79-0631
Hershman, D. E.. (2014). 2015 Soybean Cyst Nematode (SCN) Management Recommendations for Kentucky. Lexington, KY: University of Kentucky Extension.
Hu, W., Strom, N., Haarith, D., Chen, S., and Bushley, K. E. (2018). Mycobiome of cysts of the soybean cyst nematode under long term crop rotation. Front. Microbiol. 9:386. doi: 10.3389/fmicb.2018.00386
Hu, W., Strom, N. B., Haarith, D., Chen, S., and Bushley, K. E. (2019). Seasonal variation and crop sequences shape the structure of bacterial communities in cysts of soybean cyst nematode. Front. Microbiol. 10:2671. doi: 10.3389/fmicb.2019.02671
Hu, W. M., Samac, D. A., Liu, X. Z., and Chen, S. Y. (2017). Microbial communities in the cysts of soybean cyst nematode affected by tillage and biocide in a suppressive soil. Appl. Soil Ecol. 119, 396–406. doi: 10.1016/j.apsoil.2017.07.018
Hutchison, L. J., Madzia, S. E., and Barron, G. L. (1996). The presence and antifeedant function of toxin producing secretory cells on hyphae of the lawn-inhabiting agaric Conocybe lactea. Can. J. Bot. 74, 431–434. doi: 10.1139/b96-053
Jung, S. C., Martinez-Medina, A., Lopez-Raez, J. A., and Pozo, M. J. (2012). Mycorrhiza-induced resistance and priming of plant defenses. J. Chem. Ecol. 38, 651–664. doi: 10.1007/s10886-012-0134-6
Kielak, A. M., Barreto, C. C., Kowalchuk, G. A., van Veen, J. A., and Kuramae, E. E. (2016). The ecology of Acidobacteria: moving beyond genes and genomes. Front. Microbiol. 7:744. doi: 10.3389/fmicb.2016.00744
Koenning, S. R.. (1991). Effects of wheat and soybean planting date on Heterodera glycines population dynamics and soybean yield with conventional tillage. Plant Dis. 75, 301–304. doi: 10.1094/PD-75-0301
Liu, B., Giesler, L. J., Jackson-Ziems, T. A., Wegulo, S. N., and Harveson, R. M. (2013). “Major Fusarium diseases on corn, wheat, and soybeans in Nebraska,” in 2013 Crop Production Clinic Proceedings (Lincoln, NE: UNL Extension), 143–145.
Long, J., and Todd, T. (2001). Effect of crop rotation and cultivar resistance on seed yield and the soybean cyst nematode in full-season and double-cropped soybean. Crop Sci. 41, 1137–1143. doi: 10.2135/cropsci2001.4141137x
Martin, M.. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12. doi: 10.14806/ej.17.1.200
Meena, M., Swapnil, P., Zehra, A., Aamir, M., Dubey, M. K., Goutam, J., et al. (2017). “Beneficial microbes for disease suppression and plant growth promotion,” in Plant-Microbe Interactions in Agro-Ecological Perspectives, eds D. P., Singh, H. B., Singh, and R. Prabha (Singapore: Springer), 395–432. doi: 10.1007/978-981-10-6593-4_16
Mendes, R., Kruijt, M., de Bruijn, I., Dekkers, E., van der Voort, M., Schneider, J. H., et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100. doi: 10.1126/science.1203980
Meyer, S. L. F., Huettel, R. N., Liu, X. Z., Humber, R. A., Juba, J., and Nitao, J. K. (2004). Activity of fungal culture filtrates against soybean cyst nematode and root-knot nematode egg hatch and juvenile motility. Nematology 6, 23–32. doi: 10.1163/156854104323072883
Mueller, D., Wise, K., Sisson, A., Smith, D., Sikora, E., Bradley, C., et al. (2016). A Farmer's Guide to Soybean Diseases. St. Paul, MN: APS Press. doi: 10.1094/9780890545157
Nafziger, E.. (2009). “Soybean,” in Illinois Agronomy Handjournal, ed U. O. I. Extension (Urbana, IL: University of Illinois Cooperative Extension Service), 27–36.
Nour, S. M., Lawrence, J. R., Zhu, H., Swerhone, G. D., Welsh, M., Welacky, T. W., et al. (2003). Bacteria associated with cysts of the soybean cyst nematode (Heterodera glycines). Appl. Environ. Microbiol. 69, 607–615. doi: 10.1128/AEM.69.1.607-615.2003
O'Donnell, K., Kistler, H. C., Cigelnik, E., and Ploetz, R. C. (1998). Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. PNAS 5, 2044–2049. doi: 10.1073/pnas.95.5.2044
O'Donnell, K., Ward, T. J., Robert, V. A., Crous, P. W., Geiser, D. M., and Kang, S. (2015). DNA sequence-based identification of Fusarium: current status and future directions. Phytoparasitica 43, 583–595. doi: 10.1007/s12600-015-0484-z
Paulson, J. N., Stine, O. C., Bravo, H. C., and Pop, M. (2013). Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 10, 1200–1202. doi: 10.1038/nmeth.2658
Penton, C. R., Gupta, V. V., Tiedje, J. M., Neate, S. M., Ophel-Keller, K., Gillings, M., et al. (2014). Fungal community structure in disease suppressive soils assessed by 28S LSU gene sequencing. PLoS ONE 9:e93893. doi: 10.1371/journal.pone.0093893
Peralta, A. L., Sun, Y., McDaniel, M. D., and Lennon, J. T. (2018). Crop rotational diversity increases disease suppressive capacity of soil microbiomes. Ecosphere 9:e02235. doi: 10.1002/ecs2.2235
Perelló, A., Aulicino, M., Stenglein, S. A., Labuda, R., and Moreno, M. V. (2017). Pseudopithomyces chartarum associated with wheat seeds in Argentina, pathogenicity and evaluation of toxigenic ability. Eur. J. Plant Pathol. 148, 491–496. doi: 10.1007/s10658-016-1093-5
Pimentel, M. F., Arnao, E., Warner, A. J., Subedi, A., Rocha, L. F., Srour, A., et al. (2020). Trichoderma isolates inhibit Fusarium virguliforme growth, reduce root rot, and induce defense-related genes on soybean seedlings. Plant Dis. 104, 1949–1959. doi: 10.1094/PDIS-08-19-1676-RE
Pozo, M. J., and Azcón-Aguilar, C. (2007). Unraveling mycorrhiza-induced resistance. Curr. Opin. Plant Biol.10, 393–398. doi: 10.1016/j.pbi.2007.05.004
Qiu, W., Su, H., Yan, L., Ji, K., Liu, Q., and Jian, H. (2020). Organic fertilization assembles fungal communities of wheat rhizosphere soil and suppresses the population growth of Heterodera avenae in the field. Front. Plant Sci. 11:1225. doi: 10.3389/fpls.2020.01225
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
R Core Team (2020). R: A Language and Environment for Statistical Computing. Vienna: Austria: R Foundation for Statistical Computing. Available online at: https://www.R-project.org/ (accessed February 15, 2021).
Ramires, F. A., Masiello, M., Somma, S., Villani, A., Susca, A., Logrieco, A. F., et al. (2018). Phylogeny and mycotoxin characterization of Alternaria species isolated from wheat grown in Tuscany, Italy. Toxins 10:472. doi: 10.3390/toxins10110472
Rocha, L. F., Gage, K. L., Pimentel, M. F., Bond, J. P., and Fakhoury, A. M. (2021a). Weeds hosting the soybean cyst nematode (Heterodera glycines Ichinohe): management implications in agroecological systems. Agronomy 11:146. doi: 10.3390/agronomy11010146
Rocha, L. F., Pimentel, M. F., Bailey, J., Wyciskalla, T., Davidson, D., Fakhoury, A. M., et al. (2021b). Impact of wheat on soybean cyst nematode population density in double-cropping soybean production. Front. Plant Sci. 12:640714. doi: 10.3389/fpls.2021.640714
RStudio Team (2020). RStudio: Integrated Development for R. Boston, MA: RStudio, PBC. Available online at: http://www.rstudio.com/ (accessed February 15, 2021).
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Selosse, M.-A., Bessis, A., and Pozo, M. J. (2014). Microbial priming of plant and animal immunity: symbionts as developmental signals. Trends Microbiol. 22, 607–613. doi: 10.1016/j.tim.2014.07.003
Singh, S., Singh, B., and Singh, A. (2015). Nematodes: a threat to sustainability of agriculture. Proc. Environ. Sci. 29, 215–216. doi: 10.1016/j.proenv.2015.07.270
Song, J., Li, S. X., Xu, Y. L., Wei, W., Yao, Q., and Pan, F. J. (2016). Diversity of parasitic fungi from soybean cyst nematode associated with long-term continuous cropping of soybean in black soil. Acta Agric. Scand. B Soil Plant Sci. 66, 432–442. doi: 10.1080/09064710.2016.1158862
Stirling, G. R.. (2014). Biological Control of Plant-Parasitic Nematodes: Soil Ecosystem Management in Sustainable Agriculture. Oxfordshire: CABI. doi: 10.1079/9781780644158.0000
Strom, N., Hu, W. M., Haarith, D., Chen, S. Y., and Bushley, K. (2020). Interactions between soil properties, fungal communities, the soybean cyst nematode, and crop yield under continuous corn and soybean monoculture. Appl. Soil Ecol. 147:103388. doi: 10.1016/j.apsoil.2019.103388
Tang, L. L., Xia, Y., Fan, C., Kou, J. M., Wu, F. Z., Li, W. H., et al. (2020). Control of Fusarium wilt by wheat straw is associated with microbial network changes in watermelon rhizosphere. Sci. Rep. 10:12736. doi: 10.1038/s41598-020-69623-6
Tardy, V., Chabbi, A., Charrier, X., de Berranger, C., Reignier, T., Dequiedt, S., et al. (2015). Land use history shifts in situ fungal and bacterial successions following wheat straw input into the soil. PLoS ONE 10:e0130672. doi: 10.1371/journal.pone.0130672
Timper, P.. (2014). Conserving and enhancing biological control of nematodes. J. Nematol. 46, 75–89. Available online at: https://journals.flvc.org/jon/article/view/83284
Topalovic, O., Hussain, M., and Heuer, H. (2020). Plants and associated soil microbiota cooperatively suppress plant-parasitic nematodes. Front. Microbiol. 11:313. doi: 10.3389/fmicb.2020.00313
Tyler, D., Chambers, A., and Young, L. (1987). No-tillage effects on population dynamics of soybean cyst nematode. J. Agron. 79, 799–802. doi: 10.2134/agronj1987.00021962007900050008x
Tylka, G. L., and Marett, C. C. (2021). Known distribution of the soybean cyst nematode, Heterodera glycines, in the United States and Canada in 2020. Plant Health Progr. 22, 72–74. doi: 10.1094/PHP-10-20-0094-BR
Venter, Z. S., Jacobs, K., and Hawkins, H.-J. (2016). The impact of crop rotation on soil microbial diversity: a meta-analysis. Pedobiologia 59, 215–223. doi: 10.1016/j.pedobi.2016.04.001
Walters, W., Hyde, E. R., Berg-Lyons, D., Ackermann, G., Humphrey, G., Parada, A., et al. (2016). Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1:e00009–15. doi: 10.1128/mSystems.00009-15
Wanasinghe, D. N., Phukhamsakda, C., Hyde, K. D., Jeewon, R., Lee, H. B., Jones, E. B. G., et al. (2018). Fungal diversity notes 709-839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers. 89, 1–236. doi: 10.1007/s13225-018-0395-7
Warnke, S., Chen, S., Wyse, D. L., Johnson, G., and Porter, P. M. (2006). Effect of rotation crops on Heterodera glycines population density in a greenhouse screening study. J. Nematol. 38, 391–398. Available online at: https://journals.flvc.org/jon/article/view/67670
Whittaker, R. H.. (1960). Vegetation of the Siskiyou Mountains, Oregon and California. Ecol. Monogr. 30, 280–338. doi: 10.2307/1948435
Wight, J. P., Allen, F. L., Donald, P. A., Tyler, D. D., and Saxton, A. M. (2011). Impact of crop rotation and bio-covers on soybean cyst nematode. Plant Health Progr. 12, 1–9. doi: 10.1094/PHP-2010-0930-01-RS
Wolfgang, A., Taffner, J., Guimarães, R. A., Coyne, D., and Berg, G. (2019). Novel strategies for soil-borne diseases: exploiting the microbiome and volatile-based mechanisms towards controlling Meloidogyne-based disease complexes. Front. Microbiol. 10:1296. doi: 10.3389/fmicb.2019.01296
Xing, L., and Westphal, A. (2006). Interaction of Fusarium solani f. sp. glycines and Heterodera glycines in sudden death syndrome of soybean. Phytopathology 96, 763–770. doi: 10.1094/PHYTO-96-0763
Xu, W. H., Wang, Z., and Wu, F. (2015). The effect of D123 wheat as a companion crop on soil enzyme activities, microbial biomass and microbial communities in the rhizosphere of watermelon. Fron. Microbiol. 6:899. doi: 10.3389/fmicb.2015.00899
Yeates, G. W., Wardle, D. A., and Watson, R. N. (1999). Responses of soil nematode populations, community structure, diversity and temporal variability to agricultural intensification over a seven-year period. Soil Biol. Biochem. 31, 1721–1733. doi: 10.1016/S0038-0717(99)00091-7
Youssef, M. M., El-Nagdi, W. M., and Lotfy, D. E. (2020). Evaluation of the fungal activity of Beauveria bassiana, Metarhizium anisopliae and Paecilomyces lilacinus as biocontrol agents against root-knot nematode, Meloidogyne incognita on cowpea. Bull. Natl. Res. Cent. 44:112. doi: 10.1186/s42269-020-00367-z
Keywords: Heterodera glycines, metagenomics, suppressive soils, soil microbiome, plant-parasitic nematodes, soybean diseases, biological control
Citation: Rocha LF, Bond JP and Fakhoury AM (2022) Wheat Production Alters Soil Microbial Profiles and Enhances Beneficial Microbes in Double-Cropping Soybean. Front. Agron. 3:807112. doi: 10.3389/fagro.2021.807112
Received: 01 November 2021; Accepted: 06 December 2021;
Published: 10 January 2022.
Edited by:
Giovanni Bubici, Institute for Sustainable Plant Protection, National Research Council (CNR), ItalyReviewed by:
Ahmed Elhady, Julius Kühn-Institute, GermanyCopyright © 2022 Rocha, Bond and Fakhoury. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Leonardo F. Rocha, bGVvbmFyZG8ucm9jaGFAc2l1LmVkdQ==; Ahmad M. Fakhoury, YW1mYWtob3VAc2l1LmVkdQ==; Jason P. Bond, amJvbmRAc2l1LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.