
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Agron., 07 December 2021
Sec. Plant-Soil Interactions
Volume 3 - 2021 | https://doi.org/10.3389/fagro.2021.781027
Salicylic acid (SA) and nitric oxide (NO) are key signaling molecules required to activate the plant's innate immunity against abiotic stresses and biotrophic attackers. Stress-induced signaling and accumulation of SA and NO triggers extensive transcriptional reprogramming of defense-related genes, induced biosynthesis of secondary metabolites and anti-microbial compounds, thereby protecting/steering plant growth and immunity. Transcriptional regulation of SA and NO signaling are crucial for fine-tuning important cellular and metabolic functions, thus making plant defense impervious against many pathogens. The development of an impenetrable immune response is often associated with an unavoidable trade-off in the form of active suppression of plant growth and reproduction. Therefore, we highlighted recent advancements and research to unravel transcriptional regulation of SA and NO signaling essential for fulfilling their role as defense signaling molecules. We also emphasized comprehensive knowledge related to transcriptional reprogramming of SA and NO signaling important in strengthening plant growth-immunity trade-off. We also highlighted the progress on SA and NO signaling playing an indispensable role in stimulating plant-microbe interaction to modulate crucial plant functions.
Rapidly changing environmental conditions are the major limiting factor for the growth, development, and productivity of agriculturally important crops significantly affected by various biotic and abiotic stresses (van Butselaar and Van den Ackerveken, 2020). In addition, rapid urbanization/industrialization is the primary cause for reducing agricultural lands and water resources that will limit food availability to humans and livestock in the future (Thurlow et al., 2019). Furthermore, abiotic stresses execute a confronting role in ascertaining crop growth and productivity and influence the dissemination of plant species across different geographical locations (Foyer et al., 2016). Conversely, various anthropogenic activities have significantly altered the climatic conditions that have exacerbated the abiotic stresses on a global scale with increased irregularities in rainfall and unpredictive temperature (Ma et al., 2017). Therefore, fostering new breeds of economically important crops with improved adaptative ability to a diverse range of environmental fluctuations is of utmost requirement (Ma et al., 2017).
Interestingly, one of the most intriguing features of abiotic stresses is the establishment and modulation of signal transduction pathways leading to the activation of diverse defense responses in plants. These defense responses are activated by the complex interaction of transcriptional regulators, genes/proteins, and their cross-talk at physiological, biochemical, and molecular levels (van Butselaar and Van den Ackerveken, 2020). An insight into how these complex biological processes modulate abiotic stress tolerance in plants is still lagging. It is currently being investigated for identifying key mediators/elicitors of abiotic stress tolerance in plants.
Recently, SA and NO have earned an important place in the plant science community as one of the endogenous “mediators/elicitors” which are capable of initiating a wide range of physiological and metabolic responses in plants exposed to stress conditions (Domingos et al., 2015; van Butselaar and Van den Ackerveken, 2020). SA is considered a ubiquitous signaling molecule involved in conferring biotic and abiotic stress tolerance in plants by regulating the expression of critical genes and proteins of the stress defense pathway (Ding and Ding, 2020). Furthermore, several studies have reported that exogenous application of SA strengthens enzymatic and non-enzymatic antioxidants, thereby maintaining reactive oxygen species (ROSs) at the basal level (Koo et al., 2020). ROSs are an unavoidable cause of aerobic metabolisms, which can induce oxidative stress if their stories are not contrived below a threshold level (Mittler, 2017). NO is also an endogenous free radical signaling molecule capable of regulating a wide array of physiological and molecular processes in plants grown in extreme environments (Domingos et al., 2015). Growing shreds of evidence have suggested that NO is a critical player in regulating the growth and productivity of plants exposed to abiotic and biotrophic attackers (Fancy et al., 2017; Corpas and Palma, 2020). NO provokes stress defense response either by acting as an antioxidant or by elicitor of ROS scavenging system, thus stimulating the activities of various antioxidative enzymes and metabolites (Corpas and Palma, 2020). Additionally, multiple studies have delineated that NO can also regulate seed germination, dormancy, fruit ripening, and other physiological processes more effectively than other phytohormones (Sharma et al., 2020b).
To date, several studies have investigated the diverse physiological and biochemical role of SA and NO in plants under various abiotic stress conditions by stimulating ROS scavengers, stress-responsive genes, and complex hormone-mediated signaling events. However, the functional mechanism by which they exert stress defense response is still far from clear. Here, we review an in-depth molecular mechanism for understanding SA and NO-mediated key transcriptional regulators to perceive plant responses under diverse environmental fluctuations. Furthermore, we emphasized the current perceptions regarding SA and NO biosynthesis, transport, signaling, and mode of action/interaction in plants. This review also highlights the underlying methodologies targeting multiple biological processes by mediating crosstalk with other phytohormones/signaling molecules, potential significance, and future perspectives in plant stress biology.
The “sixth” phytohormone SA is a β-hydroxy phenolic acid widely produced in some prokaryotes and crop plants. In plants, it acts as a chemical messenger capable of inflecting many immune responses like extensive transcriptional reprograming, restoring membrane damage, regulating secondary metabolism, and enzyme synthesis to combat stress cues (Zhang and Li, 2019). However, the repercussions of the robust immune system are often associated with the dynamic repression of plant growth and development, often referred to as a “growth-immunity trade-off” (van Butselaar and Van den Ackerveken, 2020). In plants, this “growth-immunity trade-off” is significantly affected either synergistically or antagonistically as a direct effect of phytohormones portraying an intricate balance between growth and immune response upon their accumulation (Zhang and Li, 2019). Raymond F. White (1979) first reported the role of SA in plant immunity, and it was introduced as the “sixth phytohormone” in Raskin (1992). It was said to provide immunity to tobacco plants against tobacco mosaic virus (TMV) by activating Pathogenesis-Related (PR) proteins when applied exogenously (Ding and Ding, 2020). Extensive studies over the past decades have confirmed the immunoregulatory role of SA in plants conferring resistance to plants against several biotic and abiotic stresses overview of which has been depicted in Figure 1 (Janda and Ruelland, 2015; Zheng et al., 2015).
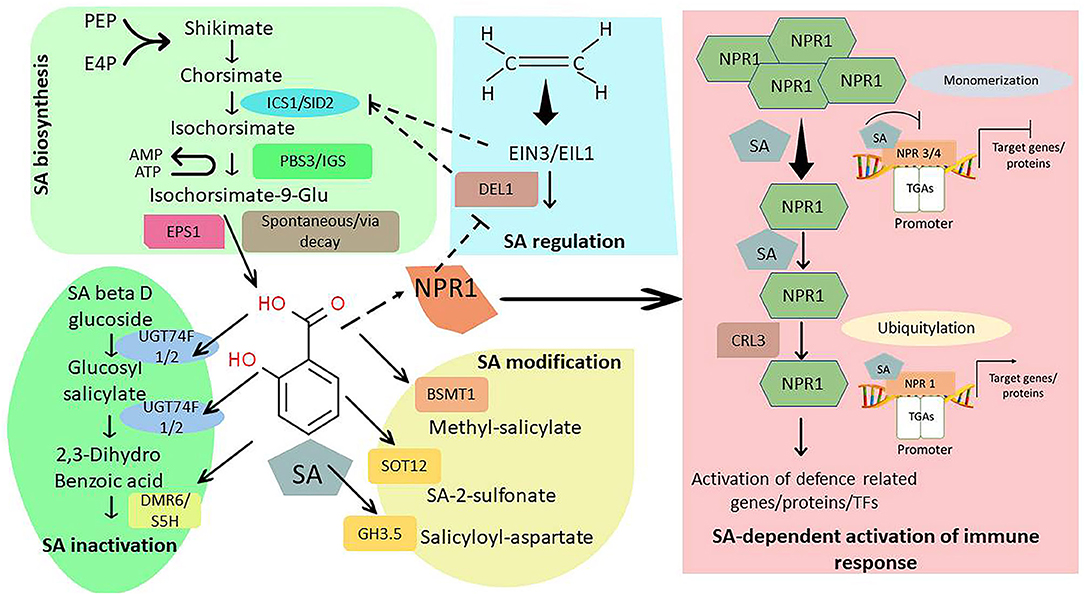
Figure 1. An overview of salicylic acid (SA) synthesis, regulation and signaling in plants. The bioactive SA synthesis starts from shikimate catalyzed by isochorismate synthase (ICS1)/SA induction deficient 2 (SID2). Enhanced disease susceptibility 1 (EDS1) transfer isochorsimate from chloroplast to cytosol which is then converted into isochorsimate-9-glu by AVRPPHB susceptible 3 (PBS3) and isochorismoyl-glutamate synthase (IGS). The isochorsimate-9-glu finally yields SA either spontaneously or by activity of enhanced pseudomonas susceptibility 1 (EPS1). Higher accumulation of ethylene responsive genes such as ethylene insensitive 3 (EIN3) or EIN3 like 1 (EIL1) under the influence of DP-E2F-like1 (DEL1) transcription factor can inhibit transcriptional reprogramming of ICS1 and EDS5 thus inhibiting SA synthesis. However, at higher SA level the activity of EIN3 is repressed by NPR1. Overexpression of UDP-glucosyltransferase 74F1 and F2 (UGT74F1 and F2) along with downy mildew resistant 6/ salicylate-5-hydroxylase (DMR6/S5H) can result in the formation of inactivated SA like SA glucose ester. Similarly, BA/SA carboxyl methyltransferase 1 (BSMT1), sulfotransferase 1 (SOT12) and Gretchen Hagen 3.5 (GH3.5) can modify SA into different conjugates. SA mediated induction of immune response involves monomerization of non-expresser of pathogenic receptor 1 (NPR1) gene which upon higher SA accumulation interact and induce TGA transcription factors that activate 26S proteasome complex leading to ubiquitylation processing stimulated by Cullin-Ring ubiquitin E3 ligase (CRL3).
In plants, SA is synthesized via two specific routes, involving the isochorismate (IC) pathway and phenylalanine ammonia-lyase (PAL) pathway using chorismate as a substrate which is the end product of the shikimate pathway (Janda and Ruelland, 2015). Various studies have confirmed that the genes viz., Enhanced Disease Susceptibility 5 (EDS5), Isochorismate Synthase 1 (ICS1), and avrPphB Susceptible 3 (PBS3) were essentially involved in producing SA via seven-step enzymatic reaction in the chloroplast and cytosol. Upon stress exposure, the genes ICS1 and ICS2 are activated, catalyzing the biosynthesis of IC from chorismate, which is then transported into the cytosol via EDS5 (Torrens-Spence et al., 2019; Ding and Ding, 2020). In prokaryotes, isochorismate lyase (IPL) directly converts IC into pyruvate and SA whereas, in plants, the enzyme Isochorismoyl-Glutamate Synthase (IGS)/PBS3 converts IC into isochorismate-9-glutamate (IC-9-Glu), which is then spontaneously converted into SA and 2-hydroxy-acryloyl-N-glutamate (2HNG). In the PAL pathway, phenylalanine and tyrosine are collectively involved in the biosynthesis of SA using distinct ways (Guilfoyle et al., 2015). The enzyme anthranilate synthase (AS) converts chorismate into tryptophan and at the same time to prephenate by chorismate mutase 1 (CM1), which is then transported into the cytosol by an alternative pathway involving phenylpyruvate aminotransferase (Tanaka et al., 2015). Both cytosolic and plastidial phenylalanine are converted into trans-cinnamic acid by PAL, then converted into SA via ortho-coumaric acid. The trans-cinnamic acid is converted to benzaldehyde and benzoic acid, finally hydroxylated to yield SA (Zhang and Li, 2019).
SA signaling is mediated either by pathogen triggered immunity (PTI), or effectors triggered immunity (ETI) depending upon the nature of abiotic /biotic stress is being perceived by the plant's innate immunity (Huang et al., 2020). In PTI, the relatively conserved molecular signatures of pathogen attacking plants are detected by pattern recognition receptors (PRRs) which are the critical element of plant innate immunity. On the contrary, ETI employs effector resistance proteins, often called nucleotide-binding leucine-rich repeats (NB-LRR), to trigger defense response (Lu et al., 2016). The genes of the ICS biosynthetic pathway are modulated by efficient expression of Systemic Acquired Resistance (SAR) Deficient 1 (SARD1) and Calmodulin Binding Protein 60g (CBP60g), provoking transcriptional regulation of the ICS pathway upon binding to its promoter region (GAAATTTTGG) (Janda and Ruelland, 2015). Additionally, these SARD1 and CBP60g upon accumulation also induce the expression of EDS1, one of the critical genes involved in SA biosynthesis and signaling (Chen et al., 2021). EDS1 is a lipase-like protein which upon interacting with Phytoalexin Deficient 4 (PAD4) or Sequence Associated Gene (SAG101) forms hetero-complexes thus initiating SA signaling (Zheng et al., 2015). The EDS1, PAD4, and SAG101 triad contain lipase-like domains that regulate complex interactions, thereby modulating SA biosynthesis, signaling, and plant innate immune response under different climate extremes (Dongus and Parker, 2021).
Nitric oxide is one of the most acclaimed components of the plant's signaling repertoire that enable plants to thrive under substandard environments (Domingos et al., 2015). In the recent decades, several experimental data have witnessed the multiple roles of NO in germination, reproduction, and strengthening several other physiological and molecular processes in plants (Corpas and Barroso, 2015). However, the exact mechanisms of NO production and its signaling cascade have yet to be explored. Nonetheless, various researchers working in the plant science community have speculated that there are two potential routes for NO biosynthesis in plants (i) reductive pathways and (ii) oxidative pathways (Chamizo-Ampudia et al., 2017). The reductive pathway is the most prominent pathway for NO synthesis in plants mediated via nitrate reductase (NR) enzyme. The NR pathway, located in the plant's cytosol, uses NADPH as a cofactor to reduce nitrate to nitrite, further reducing to NO (Santolini et al., 2017). Researchers have also concluded that under normal growing conditions, the efficiency of NR in NO synthesis is only 1%; nonetheless, it is considered as the essential enzymatic NO production in plants (Gupta et al., 2011). It has also been documented that several post-translational modifications (PTMs) of NR enzymes have significant repercussions on NO biosynthesis under both in-vivo and in-vitro conditions (Corpas and Barroso, 2015). These PTMs such as biotinylation and SUMOylation deactivate NR enzymes by altering magnesium's activity, which replaces aspartate with serine, thus reducing the specificity NR against its substrate, leading to low NO emission.
Additionally, several reports have highlighted the in-vivo synthesis of NO within the plasma membranes via a plasma membrane-bound NR enzyme known as NiNOR, which uses nitrite as substrate generated in an NR-coupled reaction (Domingos et al., 2015; Santolini et al., 2017). Scientists have simulated NO's biosynthesis under in-vitro conditions by exploiting artificial electron donors such as hydrogen sulfide, methyl viologen, cytochrome C and recorded enhanced NO synthesis (Hancock and Whiteman, 2016). Another route of NO biosynthesis in plants is nitrite reduction by a peroxisomal enzyme Xanthine Oxidoreductase (XOR). However, this reactive conversion of nitrite to NO by XOR occurs only under anaerobic conditions using NADPH or xanthine as substrate (Hancock and Whiteman, 2016). Anaerobic synthesis of NO within the peroxisomes could be due to the possible interaction with ROS or reactive nitrogen species (RNS), which has been observed in Pisum sativum and Lupinus albus plants. In the early twentieth century, researchers provide significant insight regarding the existence of an oxidative pathway by which plants synthesize NO by employing an arginine-dependent Nitric Oxide Synthase (NOS)-like biochemical pathway (Gupta et al., 2011). Several studies have shown that the application of exogenous arginine modulated the release of NO in the root tip of Arabidopsis thaliana seedlings by stimulating the synthesis of spermine and spermidine as analyzed by diaminorhodamine-4M fluorescence (Winter et al., 2015). Further, the exogenous application of hydroxylamine to NR deficient tobacco cell lines has resulted in increased NO synthesis; however, the magnitude of production was significantly lower than NO production in the NR pathway (Shivaraj et al., 2020). A diagrammatic representation of NO biosynthesis, signaling, modification, and regulation of immune response in plants has been depicted in Figure 2.
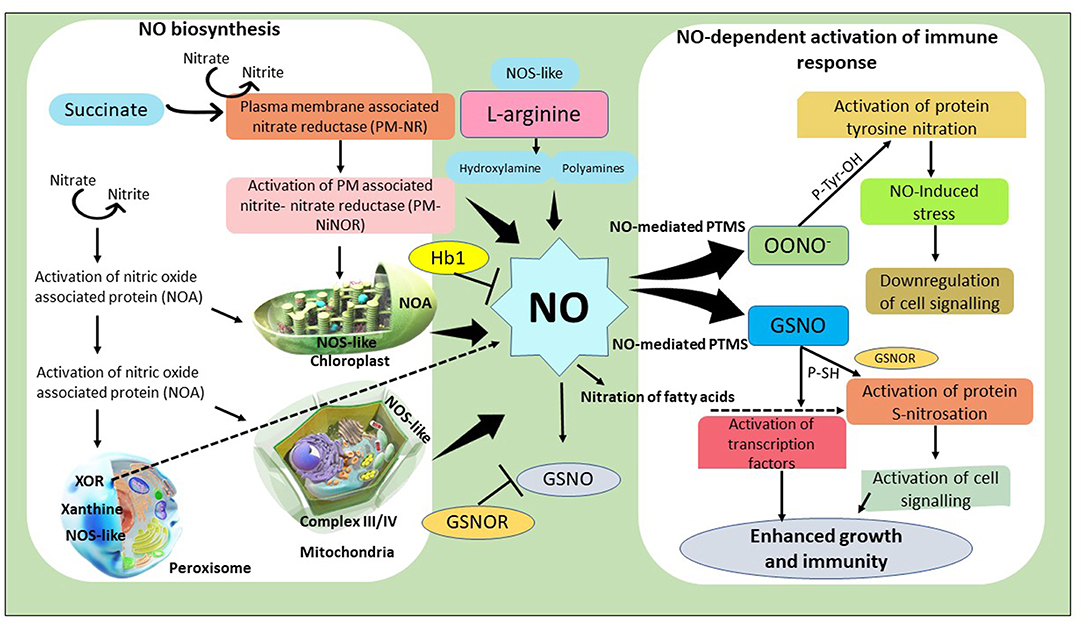
Figure 2. Schematic representation of nitric oxide (NO) biosynthesis in different subcellular compartments in plants detailing about different biochemical reactions involved in biosynthesis and the effect they exert after they are released from respective compartments. In addition, details about different mode of NO production that occur in plants via L-arginine pathway has also been discussed and how the function of NO is affected by other signaling molecules has also been shown. Furthermore, mechanisms involved in the regulation of NO-dependent activation of immune response and activation of other defense responsive genes/proteins which ultimately leads to improve plant growth and development is also shown. PM-NR, Plasma membrane bound nitrate reductase; NiNOR, Plasma membrane bound Nitrite-nitric oxide reductase; NOA, NO associated protein; XOR, Xanthine oxidoreductase; NOS, Nitric oxide synthase; GSNO, S-Nitrosoglutathione; GSNOR, S-Nitrosoglutathione reducatse; PTMs, Post-translational modifications; OONO, peroxynitrite; Hb1, Heamoglobin 1.
In plants, the effect of SA and its influence on plant growth and immune responses have been extensively studied in the model plant Arabidopsis thaliana, which is executed firmly by the positive modulation of Non-expresser Pathogenesis-Related protein gene 1 (NPR1) (Cui et al., 2017). The SA-mediated modulation of growth involves the NPR1 from the cytoplasm to the nucleus, where it interacts with various transcription factors (TFs) signaling genes (Caarls et al., 2015). The TFs like WRKY, No apical meristem/Arabidopsis transcription activation factor/Cup-shaped cotyledon (NAC), Apetella 2/Ethylene responsive factor (AP2/ERF), and Squamosa Binding Proteins (SBP) has been reported to upregulate the expression of genes involved cell wall strengthening, secondary metabolite production and antimicrobial proteins like chitinases and glucanases (Choi et al., 2010). Exogenous application of SA at lower concentrations viz., 0.1–0.5 mM has been shown to enhance the ability of plants to improvise under extreme environments by stimulating the expression of ICS1 and other vital genes (Rivas-San Vicente and Plasencia, 2011). SA has also been known to impact several auxin-mediated cell-to-cell transport processes that strongly affect auxin signaling in the plant's root and shoot (Tamas et al., 2015). The in-depth investigation revealed that SA exerts these growth-promoting responses by fine-tuning the expression of PINOID (PIN) genes that act as auxin carriers and localization (Armengot et al., 2014).
Various studies have indicated that SA alone doesn't have a growth-promoting effect. Instead, it works in conjunction with JA and NO signaling to improve the growth and immunity of plant species. However, the mechanisms by which NO stimulates growth and development in plants are similar to SA, but the functional evidence is still limited. The growth-stimulating effect of NO was first identified in Medicago truncatula. The researchers observed that the NO was directly involved in the regulation of cell division but not for its subsequent progression (Xu et al., 2010). It has also been well-documented that NO is engaged in the differentiation of vascular bundles in various plants by inflecting cell lignifications and apoptosis (Cam et al., 2012).
Additionally, several published reports have delineated that cGMP mediated NO signaling in conjunction with Ca2+ can instigate aerenchyma formation and cell polarity in Zea mays roots and Ceratopteris richardii (Krasylenko et al., 2010). Further, the cytoarchitectural advancement of the roots has also been reported to be dependent upon NO-mediated stimulation of auxin response as exogenously applied NO has been delineated to imitate IAA response to induce de-novo organogenesis in various plants such as C. Sativus and Arabidopsis thaliana (Garcia et al., 2010). NO has also been reported to cause lateral root formation in Solanum lycopersicum upon interacting with Azospirillum. NO exerted this function by stimulating the biosynthesis ROSs which enhanced the production of glutathione that strengthened the symbiotic association of plants with respective microorganisms (Nabi et al., 2019). This gravitropic response mediated by NO has also been confirmed in the roots of Glycine max, where the exogenous application of sodium nitroprusside (SNP) (NO donor) induced differentiation of cortical microtubules, which in turn stimulated lateral and adventitious root formation (Nabi et al., 2019).
The photochemical reactions occurring in the thylakoid membrane of the chloroplast are the primary site for ROS-mediated oxidative damages provoking inequality in the electron flow from oxygen-evolving complex (OEC) to photosystem II (PSII) (Janda et al., 2012). Inhibition/decrease in photosynthetic rate could be attributed to the reduction of Phospholipase D (PLD) activity which may ultimately affect Ribulose Bisphosphate Carboxylase/Oxygenase (RUBISCO) activity (Wani et al., 2017). Recent exploration demonstrated the involvement of SA in regulating PLD activity under stress conditions which sequentially activates phosphatidic acid (PA), thereby modulating vital metabolic processes (Janda et al., 2012). SA-mediated amelioration of photosynthesis and electron transfer by modulating the activity of either PLD or PA has been observed in Arabidopsis thaliana, Brassica napus, and Glycine max under stress conditions (Liu L. et al., 2016; Liu Z. et al., 2016; Wani et al., 2017). SA is believed to activate PLD or PA under stress conditions by triggering transcriptomic changes in the host organisms causing activation of several PLD-dependent genes such as PR-1 and NPR1 (Kalachova et al., 2013). Further, SA is also known to stimulate Phosphatidylinositol-4-Kinase (PI4K), resulting in the formation of Phosphatidylinositol-4-Phosphate (PI4P) and phosphatidylinositol-4-5-bisphosphate, thereby enabling the activity of RUBISCO (Kalachova et al., 2013). In several plants, including Arabidopsis thaliana, both endogenous and exogenous SA (0.5 and 1.0 mM) has modulated the expression of phosphatidylinositol-4-5-bisphosphate under heat stress resulting in enhanced accumulation of PLDs and RUBISCO synthesis (Janda et al., 2015).
Exogenous application of SNP has been reported to rebuild various physiological and biochemical processes in plants, including photosynthesis (Prochazkova et al., 2013). Foliar application of SNP at lower concentrations has stimulated chlorophyll synthesis in several crop plants, including Solanum lycopersicum, Solanum tuberosum, and Lactuca sativa (Prochazkova et al., 2013). Treating plants with a higher concentration of SNP has resulted in the decreased level of photosynthesis enzyme and thus limiting photosynthesis rates in Medicago sativa, Glycine max, and Phaseolus vulgaris (Liu et al., 2014). To decipher the mechanism of how exogenous NO regulate photosynthesis in plants, NO hypersensitive mutants were created and genetically screened in Arabidopsis thaliana (Lozano-Juste and León, 2010). Among all the mutants created, six exhibited mutation on single locus NOX1 (NADPH oxidase 1), a close homolog of CUE1 (Chlorophyll a/b binding unexpressed 1). The CUE1 is responsible for synthesizing chloroplast phosphoenolpyruvate/phosphate translocator, which is involved in various physiological processes and NO production in plants (Lechon et al., 2020). These NOX1 (CUE1) mutants displayed intense disorganization of chloroplast phosphoenolpyruvate/phosphate translocator, which suppressed vital physiological processes, including photosynthesis and electron transport system (Lechon et al., 2020). Additionally, NOX1 (CUE1) mutants exhibited late flowering compared to non-mutant counterparts, as the former have elevated NO levels due to increased synthesis (Yun et al., 2016). The NOX1 (CUE1) mutants generated in other model plants per se., Medicago truncatula and Lotus japonicus, have also shown reduced growth of floral meristem due to downregulation of key gene LFY (LEAFY) and CO (Co-regulator) (Parankusam et al., 2017). Recent findings in Arabidopsis thaliana have shown that the expression of both LFY and CO genes is in the same oscillatory circadian rhythm path as NOX1 and NOS1 (Parankusam et al., 2017). Thus, the above findings strongly indicate NO's involvement in the regulation of photosynthesis, flowering, and various other vital processes in plants.
Researchers have shown that an increase in auxin level or signaling decreases SA signaling and vice-versa (Iglesias et al., 2011; Miura and Tada, 2014). Krizek et al. (2016) further validated this hypothesis, who observed improved photosynthesis and relative water content in auxin-insensitive mutant's arx2-1 and double mutant's aintegumenta-like 6 (ail6) exhibiting increased SA levels. Furthermore, Arabidopsis thaliana plants with altered Wallsarethin 1 (WAT1) protein synthesis improved secondary cell wall and development due to enhanced SA in both roots and shoots compared to auxin level (Miedes et al., 2014). Similarly, cotton plants mutated for WAT1, WAT2, and WAT3 genes showed robust expression of ICS1, resulting in enhanced accumulation of SA under pathogen attack (Tang et al., 2019). Arabidopsis thaliana plants over-expressing SA-inducible DNA binding with One Finger (DOF) protein stimulated OBF Binding Protein (OBP3) TF that significantly improved root and shoot growth by preventing membrane damage and maintaining ion/osmotic homeostasis (Song et al., 2016). SA signaling can also activate the Target of Rapamycin (TOR) signaling pathways, which are highly conserved in plants associated with growth and developmental processes (Schepetilnikov and Ryabova, 2018). TOR signaling pathways have been widely reported to improve ion transition, nutrient transition, and protein synthesis, thus improving the growth and reproduction of crop plants (Schepetilnikov and Ryabova, 2018).
In plants, cGMP has been widely reported to perform a central role in the growth and developmental pathways, including NO signaling (Silveira et al., 2016). Researchers have actively documented NO-mediated cGMP synthesis modulated the expression of defense-related genes including PR-1 and PAL in transgenic tobacco plants under pathogen attack (Charrier et al., 2012; Chun et al., 2012). Recently, studies have also postulated that cGMP and cyclic adenosine 5'-diphosphoribose (cADPR) strongly influence NO synthesis in plants (Sanz et al., 2015). The exogenous application of SNP showed prevention toward membrane damage by regulating the level of thiobarbituric acid reactive substances and plant relative water contents under stress conditions (Rai et al., 2018a,b, 2020a). The cGMP-cADPR-NO triad has been shown to participate in various signaling pathways, including the abscisic acid pathway, which modulates vacuolar ryanodine receptor (RYR) like proteins to stimulate the biosynthesis of membrane proteins (Khan et al., 2014). Additionally, both cGMP and cADPR pathways function synergistically to regulate the NO biosynthesis to activate RYR like proteins and other genes/proteins of the defense pathway.
A large body of literature has documented the role of SA (low concentration) in stimulating growth-related processes and immunity in plants either independently or by interacting/modulating with other phytohormones and signaling molecules. Interestingly, studies have also postulated that most SA regulators do not have a specific DNA/protein motif to stimulate SA biosynthesis. How these regulators regulate SA biosynthesis still seems farfetched (An and Mou, 2011). Which raises a big question how SA-mediated growth-immunity trade-off can be circumvented in a way that makes it possible to breed disease resistance plants without causing a significant reduction of their growth? To answer this question, a forward genetics approach was undertaken by creating loss of function mutants for SA-analog benzothiadiazole non-responsive to bth (nrb) in Arabidopsis thaliana (van Butselaar and Van den Ackerveken, 2020). These SA-analog suppressor mutants showed increased growth but lost their immune response to specific pathogens (Caarls et al., 2017). A similar observation was made for Hydroxycinnamoyl Transferase (HCT)-RNAi sid2-1 and TOR-overexpression lines in Oryza sativa. The SA deficient mutants also exhibited enhanced performance growth and productivity by compromising their innate immune response (van Butselaar and Van den Ackerveken, 2020).
Additionally, studies have also indicated the role of NPR1 in stimulating the growth, productivity, and immunity of rice plants by modulating endogenous SA levels in response to pathogen attacks (Dinler and Tasci, 2020). However, NPR1 independent SA signaling requires activation of WRKY TFs to induce defense response in plants (Gao et al., 2018). The mechanism by which SA activates WRKY TFs is still unclear; however, it has been postulated that the master regulator of SA signaling, i.e., NPR1 undergoes a series of redox-dependent modifications (Li et al., 2020). One such modification involves the activation of thioredoxins which reduces intermolecular disulfide bonds in NPR1, thus causing a conformational change in its monomeric form and activating the expression of WRKY and other TFs (Gao et al., 2018). Another aspect by which SA modulates the growth and yield of crop plants without imperiling immunity is activating the Ideal Plant Architecture 1 (IPA1) gene that encodes squamosa binding protein (Zhang et al., 2020a,b). In rice and Arabidopsis thaliana, site-directed mutagenesis of Os-Sporocyteless 14 (OsSPL14) variant downregulated the expression of miRNA156, which in turn activated the transcription of Isopentenyl Adenine 1D (IPA1D) that improved the immunity, and yield of rice plants against biotic stress (Yin et al., 2019).
Similarly, a large body of evidence has confirmed that S-nitrosoglutathione (GSNO) is considered NO reservoir in plants that can modulate a variety of peptide and protein nitrosylation (Gupta et al., 2011; Corpas et al., 2013; Begara-Morales et al., 2018). This S-nitrosylation, in turn, affects the expression of various downstream genes, TFs, and activity of different intracellular and extracellular enzymes (Corpas et al., 2013). To decipher NO's role in stimulating growth-immunity trade-off, NO deficient mutants were created in Arabidopsis thaliana that showed impaired growth and development and early senescence compared to wild plants. Intriguingly, exogenous NO application to the above mutants markedly improved their growth by regulating the expression of SAG12 and SAG13 compared to their wild counterparts (Shanmugam et al., 2015). The increased accumulation of SAG proteins could be due to the increased nitrosylation of GSNO reductase (GSNOR), leading to enhanced GSNO production (Lindermayr et al., 2010). Again, to test this hypothesis, Antisense RNA-mediated silencing of GSNOR was executed in Arabidopsis thaliana. Their results showed that the lower production of S-nitrosothiols (SNO) was responsible for decreased growth and enhanced susceptibility to pathogenic attack (Bogdan, 2015). Furthermore, in Pisum sativum and Oryza sativa, NO is also known to stimulate growth and yield by regulating the level of methyl-jasmonate (Fancy et al., 2017). Most recently, researchers have confirmed that sequential reduction of NO in Pisum sativum plants showed decreased accumulation of NOS gene, which ultimately resulted in reduced growth and yield (Yadu et al., 2017). Additionally, NO deficient Arabidopsis thaliana mutants created for Nitric Oxide Synthase and Nitric Oxide-Associated 1 (NOS1/NOA1) showed stunted growth and early senescence wild plants as a result of increased accumulation of SAGs (Shen et al., 2013). Recent findings have indicated that NO also acts as a negative regulator of ABA biosynthesis genes such as 9-cis-epoxycarotenoid dioxygenase (NCED9) and a positive regulator of cytokinin biosynthetic genes such as isopentenyl adenine (IPT1) and zeaxanthin reductase (ZR) (Kong et al., 2016). Evidence has also documented the role of both SA and NO in stimulating pollen growth and in flowering by modulating the transcription of Constans (CO), Leafy (LFY), and Flowering Locus C (FLC). Previous findings have reported that NO can regulate the expression of the above genes by activating several TFs, including MADS-box TFs (Falak et al., 2021). These MADS-box TFs orchestrate Ca2+ level, which triggers calmodulin (CaM) protein, thus oscillating growth and natural circadian rhythm in plants under stress conditions (Falak et al., 2021).
Raymond F. White first incepted involvement of SA in plant immunity in 1979, where he demonstrated that the application of acetyl-SA (aspirin) protected Nicotiana tabacum plants against viral infection. Further, in-depth characterization revealed that the SA-mediated accumulation of PR proteins boosted Nicotiana's innate immunity (Fragniere et al., 2011). Later, researchers advocated that plant defense response against pathogen attack is mediated by developing a hypersensitive reaction that triggers the release of SA, thus curbing pathogen penetration and spread (Wani et al., 2017). Controversially, some researchers have postulated that plants over-accumulating a salicylate hydroxylase (NahG) encoding gene negatively regulates SAR and causes SA depletion in plant tissues (Janda and Ruelland, 2015). Likewise, a study conducted by Vos et al. (2015) also confirmed that SA failed to induce insect resistance in plants as an egg laid by an insect on a plant's leaf dramatically accelerated SA accumulation that impaired Jasmonic acid (JA) mediated resistance against pathogens. However, some studies have disapproved the above notion and concluded that insect' egg could stimulate the development of SAR, which helps control microbial populations associated with plant roots, thus boosting plant immunity (Cui et al., 2019). SAR development is often positively regulated by SA-mediated signal perception proteins NPR1 and NPR2 and negatively regulated by NPR3 and NPR4. Recently, Li et al. (2021) reported that priming Vitis vinifera plants with β-aminobutyric acid (BABA) induces the expression of NPR1, NPR3, and Heat Shock Proteins 24 (HSP24), thus providing defense against the necrotrophic fungus Botrytis cinerea.
Similarly, Yildiz et al. (2021) confirmed that the mobile SAR signal induced by treating Arabidopsis thaliana plants with N-hydroxypipecolic acid (NHP) stimulated NPR1 mediated transcriptional reprogramming of Flavin-dependent Monooxygenase 1 (FMO1) and other downstream genes, thus systematically activating an immune response against pathogenic attack. The role of plant natural product NHP in activating immune response has been identified more recently (Yildiz et al., 2021), which is synthesized directly from pipecolic acid catalyzed by FMO1 in a series of transamination/reduction steps mediated by SARD-4 and AGD2-like defense response protein (ALD1). SA-mediated activation of NPR1 has also been shown to stimulate the ubiquitination of several cytoplasmic proteins by forming SA-induced condensates (SINC). These SINCs are rich in stress proteins that trigger ETI by facilitating programmed cell death and SAR (Li et al., 2021). The fundamental role of NPR1 and NPR2 homologs in conferring viral resistance have also been identified and characterized in Cymbidium orchids (Ren et al., 2020), thus indicating SA-induced NPR proteins act as a central hub for modulating the expression of defense-related genes.
NO has also been shown to induce PTI and ETI, thereby stimulating programmed cell death, causing inactivation of pathogenic proteins by increasing the synthesis of cryptogenic and initiating global reprogramming of gene expression (Martínez-Medina et al., 2019a). Researchers have well-corroborated that NO and RNS mediated S-nitrosylation of cysteines residues unequivocally control NO function (Nurnberger and Kemmerling, 2018). In plants, S-Nitrosylation is either catalyzed by thioredoxin or the GSNOR system, both of which strengthen the plant's immune response (Martínez-Medina et al., 2019a). Studies have also confirmed that plants displaying higher GSNOR activity are more resistant to powdery or downy mildew pathogens (Jahnova et al., 2020). In addition, treating potato plants with the inducers of SAR showed a steep increase in GSNOR activity and increased immunity against hemibiotrophic oomycete Phytophthora infestans (Jedelska et al., 2021). Another study has been undertaken to validate the role of GSNOR in imparting pathogenic resistance by generating GSNOR/NOX1 mutant in Arabidopsis thaliana. The mutants showed decreased basal resistance to Pseudomonas syringae and increased S-nitrosothiol levels which could be the main reason behind the abolition of NO-mediated resistance (Xu et al., 2013). RNA interference (RNAi) studies have also confirmed that the higher NO levels have a growth stimulatory effect on a fungal pathogen such as Aspergillus niger, Monilinia fructicola, and Penicillium italicum (Thalineau et al., 2016). Recently, researchers have confirmed the synergistic relation between NO and phytoglobins in plants exposed to pathogenic attacks. They concluded that an increase in NO levels induces the expression of phytoglobins 1 (PHYTGOB1) in tomato roots, thus generating immune system and defense response (Martínez-Medina et al., 2019a). Further, exogenous application of pipecolic acid (Pip) to barley leaves, in turn, activates GSNOR activity leading to the development of SAR, thereby reducing the toxic effect of P. syringae (Lenk et al., 2019).
Temperatures fluctuations, i.e., both high and low temperatures, other than required for the optimum growth of plants, significantly affect cellular and metabolic activities (Ruelland and Zachowski, 2010). Exogenous application of SA either by foliar application, seed soaking, or irrigation has been shown to differentially regulate temperature tolerance in plants by modulating the expression of defense responsive genes (Rai et al., 2018a,b, 2020a,b). Exogenous application of SA (0.5, 1.0, and 2.0 mM) modulated the expression of various antioxidant enzymes, which ultimately resulted in improved physiological and cellular function of Citrus limon plants exposed to chilling stress, Lablab purpureus plants exposed to high-temperature stress, and Solanum lycopersicum plants exposed to heat stress (Siboza et al., 2014; Rai et al., 2018a; Jahan et al., 2019a). Recently, Cingoz and Gurel (2016) reported that exogenous application of SA (0.15 mM) significantly boosted the accumulation of cardenolide, enzymatic and non-enzymatic antioxidants that compellingly inhibited the adverse effect of heat stress on Digitalis trojana plants. Similarly, Zhang et al. (2020b) have also confirmed the stimulatory growth role of SA applied at different concentrations in Capsicum annum plants exposed to high-temperature stress. In another study, Wassie et al. (2020) reported that treating Medicago sativa plants with varying concentrations of SA ameliorates heat-stress-induced oxidative damage by improving growth, photosynthetic efficiency, and modulating defense expression responsive genes.
Likewise, foliar application of NO donor SNP (0.35 and 0.50 mM) enhanced tolerance of Lablab purpureus plants exposed to high-temperature stress by modulating transcriptional reprogramming of stress-responsive gene/enzymes (Rai et al., 2020a,b). Cheng et al. (2018) also reported that treating Populus trichocarpa with hydrogen sulfide enhances NO-mediated nitrosylation of ROS, leading to the decreased activity of GSNOR and improved tolerance to heat stress. Furthermore, integrated transcriptomic and metabolomic analysis of NO-deficient NIA1, 2NOA1-2 mutants of Arabidopsis thaliana confirmed the potential role of NO in attenuating the production of critical secondary metabolites osmoprotectants and stress-responsive proteins under freezing stress (Costa-Broseta et al., 2018). Much research has confirmed NO's involvement in conferring temperature tolerance by activating signaling components/molecules, regulating TFs, and stress-responsive genes/proteins (Parankusam et al., 2017). Various studies have also reported that NO could also act downstream of ROS-mediated signaling, thus preventing the expression of HSP's in plants (Wang et al., 2014). The possible mechanism by which exogenous NO could activate defense response is by enhancing Ca2+ levels, which activates NADPH oxidase enzyme, leading to mitogen-activated protein kinase (MAPK) cascades (Costa-Broseta et al., 2018). Exogenous application of NO to S. lycopersicum seedlings ameliorates chilling-induced oxidative stress by modulating the levels of putrescine and spermidine (Parankusam et al., 2017). Likewise, treating Chrysanthemum morifolium with exogenous NO enhanced SNOs content, which activated the expression of defense responsive genes/proteins, thereby enhancing high-temperature induced oxidative damage (Cheng et al., 2018). Researchers have demonstrated that NO's interaction with SA or H2O2 has significantly modulated the efficiency of the Calvin cycle/ascorbate/glutathione cycle and trehalose accumulation in cucumber and potato plants (Liu et al., 2020; Wu et al., 2020).
Drought-affected plants exhibit prolonged stomatal closure, decreased relative water content, and increased membrane damage that adversely limits their cellular and metabolic activities (Prakash et al., 2021). To maintain optimum growth and adequate resistance against dehydration stress, plants activate many signaling molecules such as SA and NO that help them retain ion/osmotic homeostasis under adverse conditions (Naser Alavi et al., 2014). A study conducted by Sohag et al. (2020) has indicated that hydroponic treatment of SA to rice seedlings significantly enhanced drought stress tolerance by stimulating antioxidant defense-related genes/enzymes and accumulation of osmoprotectants like proline and glycine-betaine. Similarly, Li Q. et al. (2019) generated an overexpression line of tobacco by introducing the salicylic acid-binding protein (SABP) LcSABP gene from Lycium chinense that enhances drought stress tolerance in tobacco plants by improving their physiological and biochemical signaling. Likewise, a combined application of both SA and NO improved vegetative growth and dehydration tolerance in safflower plants by stimulating osmolytes accumulation and induction of defense-related enzymes (Chavoushi et al., 2019). In addition, foliar application of SA at varying concentrations (0.5–2.0 mM) induced transcriptional reprogramming of several TFs such as Dehydration responsive element binding protein (DREB), AP2/ERF, WRKY, and HSPs that significantly contributed toward enhanced immunity of crop plants against drought stress (Lee et al., 2019). SA is an omnipotent activator of several downstream signaling genes that accumulate essential metabolites such as sinapyl alcohol dehydrogenase, cinnamyl alcohol dehydrogenase, and cytochrome p450, improving the growth and reproduction drought of exposed plants (Chavoushi et al., 2019).
Likewise, treating Brassica napus plants with NO significantly improved fresh/dry weights, antioxidant enzymes, and membrane damage, thereby reducing the adverse effect of drought stress (Akram et al., 2018). Pre-treatment of rapeseed with NO (0.5 mM SNP) stimulated both antioxidative defense and glyoxalase system, thereby conferring polyethylene glycol (PEG) induced drought tolerance (Hasanuzzaman et al., 2017). Foliar application of arginine (NO donor) has also been shown to induce short-term drought stress tolerance in wheat seedlings by stimulating expression of methylglyoxal system and antioxidative defense signaling (Hasanuzzaman et al., 2018). Similarly, a large body of literature has corroborated NO's protective role in plants exposed to drought stress by stimulating various signaling cascades such as MAPK, calcium-dependent protein kinase (CDPK), and cyclic guanosine monophosphate (Gayatri et al., 2013). NO has also been implicated in activating defense response at a molecular level by preventing DNA methylation and genome-wide alteration, thereby regulating epigenetic-mediated defense response in plants (Gupta et al., 2020). Exogenous application of SNP also induced the expression of microRNAs (miRNAs) that are potentially involved in regulating epigenetic modification in Medicago sativa plants leading to enhanced growth and development under drought stress (Zhao et al., 2020). Similarly, treating soybean plants with SNP (0.1 mM) stimulates osmolytes accumulation and antioxidant defense responses under drought stress (Rezayian et al., 2020).
Soil salinity alone or in combination with other abiotic stresses significantly affect agricultural production, thus hampering sustainable food production and global food security around the globe (Kim et al., 2018). Strategies like breeding local cultivars with wild relatives, molecular breeding approaches to identify QTLs, or developing transgenic plants are currently being used to enhance their immunity against salt stress (Jayakannan et al., 2015). Exogenous application of SA at lower concentration has been shown to stimulate plant defense response, growth, and productivity by regulating osmolytes and proteins of stress defense pathway such as HSP, DREB, AP2/ERF, and ICS1 (Kim et al., 2018). To study the response of salt stress, SA was exogenously applied (10−8-10−5 M) to glutathione reductase (gr) mutants of Arabidopsis thaliana. Compared to wild-type plants, the gr1 mutant exhibited more redox potential and scavenging activity due to enhanced activity of enzymatic antioxidants and ascorbate-glutathione (ASH-GSH) pools (Csiszar et al., 2018).
Similarly, researchers have functionally characterized the effect of SA in inducing the expression of Jatropha curcas JcWRKY TFs. In transgenic tobacco plants, ectopic expression of JcWRKY enhances salt tolerance in overexpressing tobacco plants by modulating ion homeostasis, photosynthetic efficiency, and antioxidant defense system (Agarwal et al., 2016). Foliar application of SA (100 mg L−1) to leaf and root of tomato seedlings also showed enhanced salinity tolerance. It improved ion/osmotic homeostasis, fresh and dry weights, and increased K and Fe accumulation in leaves of treated plants (Souri and Tohidloo, 2019).
There are contradictory reports regarding the functional role of NO in conferring salt stress tolerance as the effect of NO dramatically varies depending upon concentration applied, plant species, duration, and respective plant parts used for the treatment (Egbichi et al., 2014). Therefore, the molecular mechanism behind NO's role in stimulating plant immune response under salt stress conditions remains elusive (Wang et al., 2015). Nonetheless, researchers of the plant scientific community are on the verge of deciphering the interacting mechanism underlying NO-salt stress response. Recently, the researchers have proposed a hypothesis that it could be due to the enhanced synthesis of polyamines that instigate defense response in plants (Campos et al., 2019). Enhanced accumulation of polyamines in salt exposed plants and improved NO level could be one of the probable molecular mechanisms involved in salt stress tolerance (Hu et al., 2017). Several researchers have documented that exogenous application of SNP improved the growth and immunity of soybean and spinach plants exposed to salt stress by stimulating enzymatic and non-enzymatic antioxidants, osmoprotectants, and G-protein signaling (Egbichi et al., 2014; Wang et al., 2015).
Similarly, Campos et al. (2019) have demonstrated the protective role of exogenous NO in conferring salt stress tolerance in Lactuca sativa plants by ameliorating membrane damage, ion homeostasis, and inducing antioxidants. Exogenous application of NO has been known for triggering innate immunity of Arabidopsis thaliana by stimulating protein Arg methyltransferase 5 (PRMT5). The PRMT5 activity was induced upon NO-mediated S-nitrosylation of Cys-125, which in turn has resulted in the activation of aberrant splicing of stress-related genes causing demethylation in the PRMT5C125S/prmt5-1 plants (Hu et al., 2017). Likewise, rat neuronal NOS (nNOS) overexpression in rice plants under the constitutive expression of ubiquitin promoter (Cai et al., 2015) displayed enhanced salinity tolerance and water holding capacity due to up-accumulation of OsCAT, OsPOX, OsSNAC1, OsLEA3, and OsRD29A genes. A study conducted by Zhou et al. (2016) indicated that overexpression of AtCaM1 and AtCaM4 in transgenic Arabidopsis thaliana stimulated salt stress tolerance by increasing the cellular level of NO, which in turn regulated ion absorption, antioxidant enzymes, and GSNOR1 activity. The combined action of exogenously applied SA and NO stimulated immunity of Vigna angularis plants by regulating growth, photosynthesis, and the ASH-GSH cycle under salt stress conditions (Ahanger et al., 2020).
Heavy metals (HMs) such as cadmium, lead, arsenic, nickel, etc. find their way into the environment due to intensive use of chemical fertilizers and exaggerated industrial wastes, thus reaching alarming levels in the agricultural soils, thereby limiting the growth and productivity of important crops (Sharma et al., 2020b; Rai et al., 2021). Concerning restoring the signal transduction pathway under HM stress, several signaling molecules perse, SA and NO, have come to light robustly targeted to stimulate defense systems in plants (Wani et al., 2017; Sharma et al., 2020a). Foliar spray of SA (0.5 mM) significantly activates the activities of enzymatic and non-enzymatic antioxidants, thereby strengthening the growth and immunity of Sorghum bicolor L. plants exposed to chromium toxicity (Sihag et al., 2019). Likewise, SA and hydrogen sulfide's combined application to Zea mays plants improves Pb tolerance by regulating the free amino acid composition and glutathione content (Zanganeh et al., 2019). Moreover, various studies have corroborated that foliar application of SA activates defense response in HMs stressed plants by regulating the expression of defense responsive genes/proteins (Guo et al., 2018; Li N. et al., 2019; Faraz et al., 2020). Recently, SA-mediated induction of Myeloblastosis (MYB) TF conferred resistance to transgenic rice plants against Cr toxicity by overexpressing OsMYB-R1 TF (Tiwari et al., 2020). Additionally, their RNA-seq data reveals that SA signaling was involved in modulating signaling pathways such as CDPKs, MAPKs, and HSPs activation (Tiwari et al., 2020), thus confirming the complex nature of SA signaling involved in the activation defense response under HMs stress.
Being a multifunctional gaseous molecule, NO plays a pivotal role in modulating plant immunity against HMs both endogenously and exogenously, the effect of which is dependent upon the type HMs, dose, and duration of treatment (Xiong et al., 2010). For example, exogenous NO application stimulated the expression of essential signal transduction pathway genes such as Alternative oxidase 1 (Aox1), which enhanced the adaptability of barley plants against As stress (Shukla et al., 2015; Kolbert et al., 2017). Likewise, exogenous application of NO modulated antioxidant defense system and thiol metabolism, thereby increasing tolerance of rice plants exposed to As stress (Singh et al., 2016). The ameliorative effect of NO on As toxicity could be due to decreased accumulation of As in root and shoot plants by enhancing the expression of AsIII transporter (OsLsi1 and OsLsi2) and GSH/GSSG (oxidized glutathione) levels (Singh et al., 2017). Additionally, exogenous application of SNP has also been implicated in improving the physiological, biochemical, and molecular basis of Ni-stress tolerance in rice plants, and this ameliorative effect of NO was reversed upon the application of 2-4-carboxyphenyl-4,4,5,5-tetramethylimidazoline−1-oxyl-3-oxide (cPTIO) (NO scavenger) (Rizwan et al., 2018).
In contrast, studies have also shown that endogenous production of NO in Pb exposed Arabidopsis thaliana adversely affected its peroxisomal function by decreasing catalase activity up to 50% due to increase NO-mediated PTMs (Corpas and Barroso, 2017). In addition, transcriptome analysis reveals essential insight into NO-mediated signaling in Cd stress-exposed Pleurotus eryngii. The researchers elucidated that NO under HMs stress can activate putative genes like HSPs, oxidoreductases, steroid biosynthesis, and glycerolipid metabolism, thus enhancing plant immunity (Li et al., 2018). Synergistic effect of silicon and exogenous NO compellingly reduced As uptake in Brassica juncea plants, thus promoting its growth and productivity by regulating the activities of CAT, SOD, APX, GR, and GST, membrane damage, and osmolytes (Ahmad et al., 2021). Both exogenous and endogenous NO effectively regulates ion channels/transporters, which are involved in the uptake and extrusion of HMs. However, the actual mechanism behind these remains largely unknown (Terron-Camero et al., 2019). Recent findings have also suggested biosynthesis of NO under HM stress can regulate redox-dependent chromatin remodelings such as histone acetylation/methylation and DNA methylation in plants, thus improving the survival of plants under HMs stress (Ageeva-Kieferle et al., 2019). Further, the researchers also concluded that NO-mediated PTMs such as nitration/S-nitrosylation inhibit histone deacetylation complexes, thus stimulating histone acetylation leading to enhance expression of defense responsive genes (Ageeva-Kieferle et al., 2019).
Plant growth regulators such as SA, NO, Jasmonic acid (JA), Abscisic acid (ABA), and strigolactones can regulate a wide array of plant functions under changing environmental conditions (Klessig et al., 2018). Being active at low concentrations, these phytohormones also play an indispensable role in improving underground communication between plants and beneficial microbes (Mine et al., 2014). Despite their tight spatial synthesis and regulation, they play a vital role in establishing plant-microbe communication, symbiosis, nodulation, and probable function. These interactions are robustly observed in nitrogen-fixing bacteria and arbuscular mycorrhizal (AM fungi), which are associated with almost 70% of total plants present on earth (Foo et al., 2019). In this context, researchers have confirmed the involvement of SA in promoting/modulating interaction between plant and biotrophic microbes (Beckers and Spoel, 2006; Lu, 2009). Exogenous application of SA induces differential response to inhibit nodule formation in alfalfa and vetch, whereas improved nodulation was observed in Lotus japonicus (van Spronsen et al., 2003; Nakagawa and Kawaguchi, 2006). Another study reported modulation of endogenous SA level by overexpressing NahG that significantly improved nodulation in both L. japonicus and Medicago truncatula (Stacey et al., 2006). Similarly, exogenous application of SA inhibited mycorrhizal development in the first week, and after fungal infection, the AM colonization was enhanced with subsequent growth (de Roman et al., 2011). This notion was further confirmed by studying SA-overproducing and NahG deficient tobacco plants exhibiting increased and decreased root colonization followed by fungal infection (Medina et al., 2003). Furthermore, researchers confirmed that specific bacterial taxa influence the SA-based modulation of the root microbiome (Lebeis et al., 2015). Their SynCom experiments concluded that plants treated with exogenous SA showed differential rhizosphere changes where the root isolates were enriched in Flavobacterium sp. 40 and Terracoccus sp. 273 of which abundance growth was found observed in Terracoccus sp. 273 (Lebeis et al., 2015). Martinez-Medina et al. (2017) also reported the antagonistic role of SA over JA in stimulating root colonization of Trichoderma harzianum T-78 in tomato plants. Their study confirmed that SA-dependent elicitation of defense response negatively affected T-78 endophytism, whereas JA-dependent signaling enhanced colonization of T-78. This notion was also confirmed in liverworts where the exogenous application of SA to Marchantia polymorpha enhances its susceptibility against four pathogenic fungal strains Irpex lacteus Marchantia-infectious (MI)1, Phaeophlebiopsis peniophoroides MI2, Bjerkandera adusta MI3, and Bjerkandera adusta MI4 (Matsui et al., 2020). However, treating Marchantia polymorpha with bioactive jasmonate (JA) ameliorated the damage induced by pathogenic fungi, thereby suggesting the existence of antagonistic signaling between SA-JA as observed in seeded plants (Matsui et al., 2020).
NO also regulates plant-fungal mutualistic interactions in a ubiquitously diverse way that often leads to enhanced growth and plant immunity under changing environmental conditions (Mur et al., 2006). This intimate mutualistic relationship between plants and fungi is effectively established, maintained, and regulated by efficient recognition and substantial coordination of their fungal associates to facilitate positive colonization (Mur et al., 2006; Meilhoc et al., 2011). Besides NO's indispensability in stimulating plant immunity against biotic/abiotic stresses (as discussed above), researchers have well-speculated that NO might also play a vital role in establishing and maintaining plant-microbe interactions (Gupta et al., 2011). The role of NO in stimulating plant-mycorrhizal interaction was first incepted by Calcagno et al. (2012). They observed enhanced NO signaling in the roots of M. truncatula plants upon treating them with AM fungus Gigaspora margarita. They concluded that treating plants with AM fungus significantly boosted their nitrate reductase activity, causing an upsurge in the endogenous NO level, strengthening symbiotic establishment (Calcagno et al., 2012). Later, researchers also identified that NO could induce differential regulation of Fix genes from Lotus japonicus (FixLJ), which plays a crucial role in modulating late symbiotic function under oxygen limitation conditions (Defez et al., 2016). Under micro-aerobic conditions, NO phosphorylates FixLJ, which causes transcriptional activation of FixK and NifA genes responsible for activating Fix genes (respiration) and Nif genes for stimulating symbiotic nitrogen fixation (Defez et al., 2016).
Additionally, NO is also responsible for the regulation of either FixK or nitrate reductase (NnR) genes (both of which belong to Fumarate Nitrate Reductase (FNR)/Cyclic AMP Receptor Protein (CRP) gene family) in Sinorhizobium meliloti thus indicating the existence of differential signaling of NO for both the genes (Bueno et al., 2017). Furthermore, NO has been reported to modulate FixLJ-FixK2 regulatory cascades, responsible for activating genes in B. Japonicum required for nitrogen fixation and survival under micro-aerobic conditions (Bueno et al., 2017). Likewise, scientists working on AM fungi have demonstrated that the NO-mediated PTMs are responsible for the perception of diffusible fungal signals by plants upon transcriptional reprogramming of Master Regulator of Cell Cycle Entry and Proliferative Metabolism (MYC) factors that facilitate plant-microbe interaction (Chabaud et al., 2011; Genre et al., 2013). The researchers exploited different bacterial mutants, including S. meliloti, to stringently regulate endogenous levels of NO inside M. truncatula, which activated tyrosine nitration of glutamate synthetase, thereby stimulating the assimilation of ammonia (Blanquet et al., 2015). Researchers have recently documented that treating tomato plants with T. harzianum stimulated NO signaling and triggered transcriptional activation of phytoglobin genes such as PHYTOGB1, PHYTOGB2, and PHYTOGB3 within 4 h of treatment (Martínez-Medina et al., 2019a). In the above study, the authors also confirmed that T-78 mediated NO-burst stimulated the expression of phytoglobin genes that effectively regulated plant immunity and root colonization (Martínez-Medina et al., 2019a). Likewise, in-vitro-grown cultures of Rhizophagus irregularis were used on tomato plants to decipher AM-associated induction of NO signaling/response on the root treated with Fusarium oxysporum (Martínez-Medina et al., 2019b). The researchers reported that an early NO-release was triggered by both the fungi that stimulated the expression of PHYTOGB1 in tomato roots, strengthening plant-fungal mutualistic symbiosis. In contrast, transgenic tomato plants with silenced PHYTOGB1 decreased NO level that conversely altered the plant-microbe interaction (Martínez-Medina et al., 2019b). A hypothetical model describing the potential mechanism by which SA and NO establish plant-microbe interaction leading to subsequent development of immune response is represented in Figure 3.
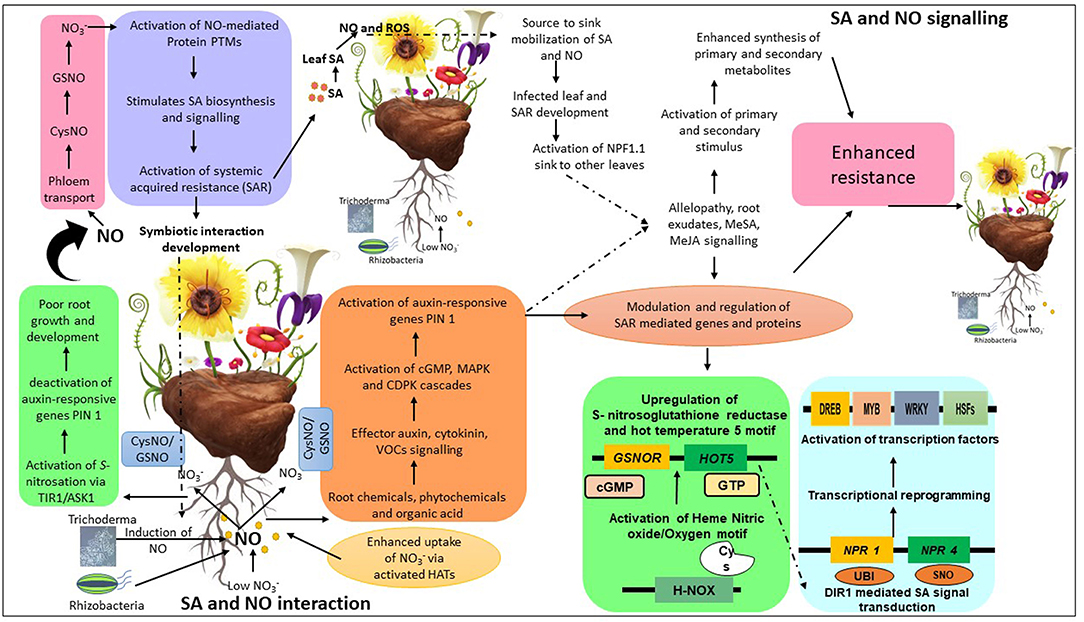
Figure 3. A model depicting interactive mechanisms of salicylic acid (SA)/nitric oxide (NO) and microbes and their repercussions on plant growth and immunity. Both plants and rhizomicrobes stimulate the biosynthesis of various signaling molecules such as SA and NO to establish a mutual relationship. The NO level in root is induced in the presence of beneficial microbes which is then rapidly transported to different part of the plant by activation of histone acetyl transferase (HATs) which in turn activate various signaling cascades involved in the regulation of root growth and development. Whereas, NO mediated post translation modification (NO-PTMs) such as S-nitrosation deactivates the root auxin biosynthesis gene PIN1 resulting in poor root growth. Once NO moves to leaf area via phloem, it interacts and activates SA biosynthesis and signaling. NO-mediated activation of SA leads to the development of systemic acquired resistance (SAR) which results in the stimulation of secondary metabolite synthesis and transcriptional reprogramming of various stress responsive genes/proteins via activation of non-expresser of pathogenesis related protein 1 (NPR1) resulting in enhanced growth and immunity. CysNO, Cysteine nitric oxide; GSNO, S-Nitrosoglutathione; TIR1, Transport inhibitor response 1; ASK1, Kinase associated protein 1; PIN1, Pinnoid1; PTMs, Post-translational modifications; ROS, Reactive oxygen species; MAPK, Mitogen activated protein kinase; CDPK, Calcium dependent protein kinase; VOCs, Volatile organic compounds; H-NOX, Heme nitric oxide; NPF1.1, Nitrate transporter 1/peptide transporter family protein; UBI, Ubiquitination; SNO, S-nitrosation; DREB, Dehydration responsive element binding protein; MYB, Myeloblastosis; HSFs, Heat shock factors.
Perception of an invader leads to transcriptional regulation of signaling molecules such as SA, NO, JA, ABA, polyamines, etc., that play a vital role in regulating downstream genes involved in strengthening defense response (Prakash et al., 2021). Commonly, SA is believed to be most effective against biotrophic pathogens by modulating SAR. At the same time, NO is responsible for activating defense signaling networks, thereby wielding plant response as per the outcome of an activated defense system. Recent evidence has indicated both antagonistic and synergistic relationships between SA and NO in regulating plant responses under stress conditions (Asgher et al., 2017). Likewise, SA and JA-mediated signaling cascades are interdependent and function as complex regulatory networks.
In contrast, ABA has been well-documented to stimulate NO generation in guard cells which is vital for activating ABA-induced stomatal closure (Prakash et al., 2021). Furthermore, polyamines, methylglyoxal, and spermidine can differentially regulate defense-related responses to improve plant growth; however, their actions can be reversed by SA-NO mediated signaling (Prakash et al., 2021). This section highlights critical regulatory mechanisms by which SA and NO interact with other plant growth regulators and how they can together orchestrate virtual signaling networks that enhance plant growth and immunity (Figure 4).
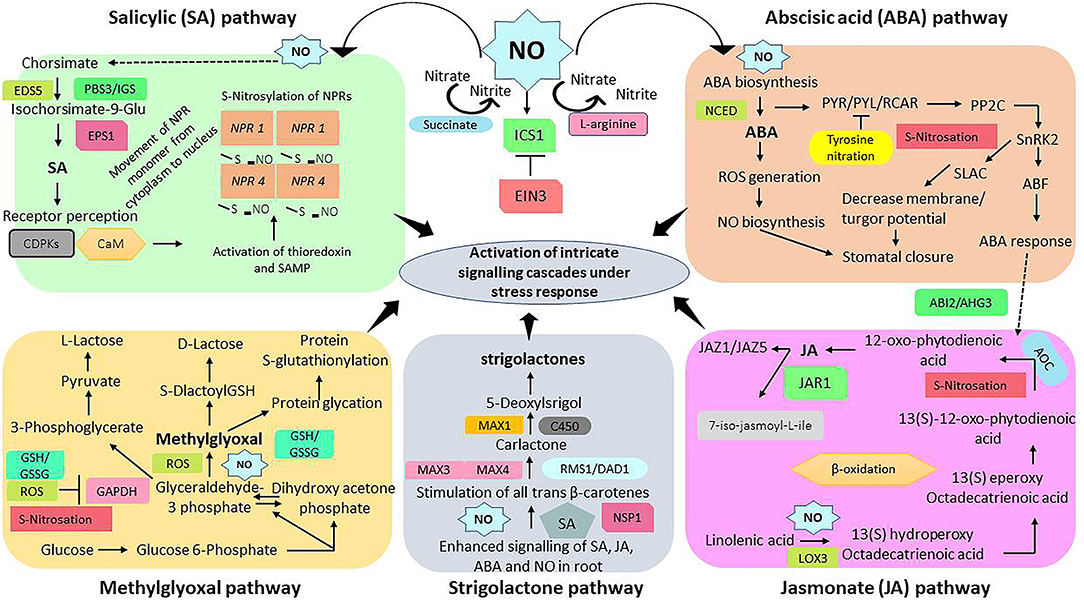
Figure 4. An interactive representation of salicylic acid, nitric oxide, abscisic acid, jasmonic acid, methylglyoxal, and strigolactones signaling cascades. Summary versions of biosynthetic pathways of salicylic acid, nitric oxide, abscisic acid, jasmonic acid, methylglyoxal, and strigolactones and mechanisms of their coordinative functions in modulating defense related genes/enzymes to improve growth and immunity of plants under stress conditions. EDS5, Enhanced disease susceptibility 5; PBS3/IGS, AVRPPHB susceptible 3/isochorismoyl-glutamate synthase; EPS1, Enhanced pseudomonas susceptibility 1; CDPKs, Calcium dependent protein kinases; CaM, Calmodulin; S-NO, S-nitrosation; NPR1, Non-expresser of pathogenesis related protein 1; ICS1, Isochorsimate synthase 1; EIN3, Ethylene insensitive 3; NCED, Nine-cis-epoxycarotenoid dioxygenase; SLAC, Slow anion channel; PP2C, Protein phosphatase 2C; SnRK2, Sucrose nonfermenting 1 (SNF1)-related protein kinase 2; ABF, Abscisic acid responsive factor; LOX3, Lipoxygenase 3; JAR1, Jasmonate resistance 1; AOC, Allene oxide cyclase; ABI2, Abscisic acid insensitive 1; AHG3, Member of PP2C protein family; JAZ1 and 5, Jasmonate ZIM-Domain 1 and 5; NSP1, Nodulation signaling pathway 1; MAX 1,3,4, More auxiliary growth 1, 3, 4; C450, Cytochrome P450; RMS1/DAD1, Carotenoid cleavage dioxygenase family gene; ROS, Reactive oxygen species; GSH/GSSG, Reduced glutathione/oxidized glutathione.
The function of master regulator NPR1 has been extensively implicated in regulating SA/JA mediated redox signaling, which is known to suppress the JA-responsive pathway at the level of initiation of gene transcription (Van der Does et al., 2013; Caarls et al., 2015). Several recent studies have indicated that SA induced monomerization, reduction, and translocation of NPR1 to the nucleus can suppress JA signaling by differentially regulating F-box protein Coronatine-insensitive-Isoleucine 1 (COI1) and Jasmonate ZIM domain (JAZ) repressor proteins (Van der Does et al., 2013; Song et al., 2015). The activated JAZ repressor proteins in the absence of JA get associated with Topless (TPL) repressor protein by novel interactor of JAZ (NINJA) or Histone Deacetylase 6 (HDA6) adaptor protein causing transcriptional inactivation of MYC, Ethylene Insensitive 3 (EIN3), and EIN Like 1 (EIL1) (Vos et al., 2013). However, in the presence of bioactive JA, the master regulator COI1 binds with JA-Ile that initiates proteolytic degradation of JAZ repressor proteins, thereby activating JA-responsive genes (Van der Does et al., 2013). Conversely, researchers have documented SA receptors activated JA signaling to stimulate effectors triggered immunity via the non-canonical pathway (Liu L. et al., 2016). They further indicated that the canonical pathway was established upon SA accumulation sensed by NPR3 and NPR4, thus activating JA signaling by differentially regulating the expression of JAZ repressor proteins and COI1-mediated genes (Liu L. et al., 2016). Additionally, Kloth et al. (2016) performed transcriptome analysis to decipher the role of WRKY22 TF in regulating SA/JA signaling. Their results indicated that ectopic expression of WRKY22 and WRKY 29 induces SA/JA signaling, thereby conferring resistance to Arabidopsis thaliana against the biotrophic pathogen. Similarly, a study conducted by De Vleesschauwer et al. (2016) also corroborated the above notion where the role of DELLA protein Slender Rice 1 (SLR1) stimulated SA/JA-mediated defense signaling, which conferred hemibiotrophic resistance to rice plants. Recently, integrated transcriptome analysis revealed the coordinated role of SA/JA-mediated defense signaling regulated growth and immunity in poplar plants against Melampsora larici-populina, Septoria musiva, and Marssonina brunnea (Luo et al., 2019).
The signaling molecule NO has also been reported to induce JA signaling in plants by activating key JA biosynthesis enzymes such as Allene Oxide Synthase (AOS) and Lipoxygenase 2 (LOX2), thereby triggering a cascade of defense response (Huang et al., 2004). Furthermore, researchers have also well-indicated NO-mediated elicitation of jasmonate biosynthesis stimulated the expression of a wide range of JA-responsive genes such as LOX3, 12-oxophytodienoate reductase 1, 2, and 3 (Mur et al., 2013). Additionally, NO is integrated into JA signaling either by NO-mediated initiation of SA biosynthesis or by activating the ICS1 gene and TGA TFs. Subsequently, SA induces the production of thioredoxins that causes monomerization of NPR1 within the nucleus and, after binding to TGA via ORA59 promoter, suppresses JA signaling (Mur et al., 2013). Another notion is upon infection with necrotrophic pathogen or attacker, enhanced endogenous level of NO induces the expression of LOX3 and 12-oxo-phytodienoic acid (OPDA) reductase (OPR) 1,2, and 3, which causes transcriptional activation of JA biosynthetic genes (Mur et al., 2013). Correspondingly, NO-mediated activation of the ascorbate-glutathione cycle upon exogenous application of JA conferred drought stress tolerance in wheat seedlings, thereby confirming the synergistic interaction of NO/JA signaling in improving plant growth and immunity (Shan et al., 2015). NO-mediated stimulation of jasmonate-dependent basal defense response conferred resistance to tomato plants against root-knot nematode (RKN) by increasing the expression of Protease Inhibitor 2 (Zhou et al., 2015). Studies have also shown that enhanced endogenous levels of NO causes downregulation of JAZ repressor proteins JAZ1 and JAZ10 in the meristematic root regions, thereby improving root architecture and stomatal apparatus in transgenic Arabidopsis thaliana (Barrera-Ortiz et al., 2018; Yastreb et al., 2018). Recently, a study conducted by Avila et al. (2019) confirmed the synergistic role of NO/JA in stimulating defense response in Solanum quitoense plants against F. oxysporum, thereby suggesting the activation of the corresponding signaling pathway by both defense priming compounds.
Elucidation of crosstalk between SA and ABA in stress signaling pathways has become the center of attraction among plant biologists. Increasing evidence has well-recognized ABA stimulates defense response and improves growth and developmental processes in plants (Jiang et al., 2010). Congruently, SA also boosts plant immunity against many biotic/abiotic stresses, thereby positively impacting plant growth and development. However, researchers have demonstrated SA and ABA work antagonistically where SA antagonizes ABA signaling components at both cellulars and transcriptional levels by regulating the activity of Pyrabactin Resistance 1/PYR1-Like Regulatory Component of ABA Receptor (PYR/PYL/RCAR) (Manohar et al., 2017). Further, studies have shown that SA antagonizes ABA signaling by differentially regulating type 2C Protein Phosphatase (PP2C), PP2A, and SnRK2s (Sucrose nonfermenting 1-Related subfamily 2 protein Kinase) which are essential for initiating ABA signaling in plants under stress conditions (Manohar et al., 2017). A large body of literature has revealed the antagonistic relationship between SA and ABA signaling pathway. For example, Meguro and Sato (2014) reported that exogenous application of SA inhibited ABA signaling, affecting shoot growth by downregulating the genes involved in the cell cycle in rice plants.
Similarly, many researchers have also implicated that exogenous application of SA antagonizes ABA-responsive genes by suppressing key ABA signaling components such as ABA1, ABA2, RD29A, thereby suppressing the growth of mutant plants compared to their broad type (Manohar et al., 2017). Correspondingly, several studies have also demonstrated a synergistic relationship between SA/ABA where Horváth et al. (2015) observed upregulation of ABA biosynthesis genes such as Zeaxanthin Epoxidase (SlZEP1), 9-Cis-Epoxycarotenoid Dioxygenase (SlNCED1), and Aldehyde Oxidases (SlAO1 and AlAO2) in SA treated plants exposed to salt stress. Likewise, Saleh et al. (2020) reported that both SA and ABA modulate the expression of various stress-responsive genes and TFs, thereby conferring salt stress tolerance to Vitis vinifera plants. Similarly, co-ordinated signaling of both SA and ABA was found to activate the Ca2+/CPK-dependent mechanism that triggers an immune response in Arabidopsis thaliana plants exposed to a biotrophic pathogen, thereby preventing stomatal closure (Prodhan et al., 2018). Recently, Park et al. (2021) observed that SA-stimulated ROS accumulation in drought exposed plants stimulated ABA signaling genes such as MYC2 and NCED3, activating Ca2+dependent defense response in a distinct phase of drought exposure.
NO metabolism instigates cellular and biochemical reactions to mechanistically regulate ABA homeostasis by modulating ABA-responsive genes such as PYR/PYL/RCAR, PP2C, and SnRK2 (Neill et al., 2002). Additionally, NO-mediated PTMs such as tyrosine nitration and S-nitrosylation have been positively correlated with the regulation of the NCED enzyme, which is central for ABA biosynthesis and NO accumulation (Lozano-Juste and León, 2010). Furthermore, in Arabidopsis thaliana, the cytochrome P450-707A2 (CYP707A2) gene has been well-documented to regulate ABA catabolism and NO biosynthesis, thereby releasing seed dormancy (Lozano-Juste and León, 2010). However, researchers have also corroborated that NO might negatively modulate ABA signaling in guard cells by down-regulating Open Stomata 1 (OST1) and SnRK2 genes via NO-mediated PTMs (Wang et al., 2015). Further, they also concluded that impairment of NO-mediated PTMs in gsnor1-3 mutant exaggerated NO accumulation in guard cells leading to S-nitrosylation of SnRK2 and dysfunction of ABA signaling (Wang et al., 2015). Likewise, Wang et al. (2015) reported that exogenous application of NO breaks seed dormancy and was also influential in alleviating the inhibitory effect of ABA on seed germination by simulating NO-mediated PTMs, which may have resulted in the inactivation of SnRK2. Phytoglobins inactivation in drought-stressed Zea mays plants exhibited low ABA levels that induced cell death in developing embryos. However, exogenous application of ABA reverses this effect causing activation of phytoglobins in Zmpgb1.1 suppressing line with increased accumulation of NO (Kapoor et al., 2018). Conversely, exogenous application of NO and ABA coordinatively regulated various defense-related genes/TFs, thereby conferring PEG-induced drought stress tolerance in Brassica plants (Sahay et al., 2019).
Low molecular weight organic compound polyamines are ubiquitously present in all lifeforms. They can induce a hypersensitive response in plants upon interaction with the pathogenic attacker and stress conditions (Szepesi, 2011). The PAs which are most commonly accumulated in the plant kingdom are putrescine (Put), spermidine (Spd), and spermine (Spm). Several studies have demonstrated that PA biosynthesis and signaling are regulated upon interaction with other phytohormones perse, SA, and NO (Szepesi, 2011). For example, exogenous application of SA modulated PA biosynthesis in maize and tomato plants by increasing the activities of crucial biosynthesis enzymes arginine decarboxylase (ADC) and ornithine decarboxylase (ODC) (Zhang et al., 2011). Likewise, exogenous application of SA and Spm compellingly enhanced the expression of several antioxidant enzymes, metabolites, and defense-related genes that ultimately improved photosynthesis, root/shoot dry weight, and growth of Echinacea purpurea, tomato, maize, oat, and plum plants (Szalai et al., 2016; Takács et al., 2016; Darvizheh et al., 2018, 2019; Canales et al., 2019; Koyuncu et al., 2019). Recently, Pal et al. (2019) also reported that treating wheat dwarf mutant created for reduced height (Rht1 and Rht3) with exogenous polyamine significantly boosted polyamine metabolism, growth, and development of mutant plants by stimulating SA and ABA signaling. Likewise, Liu et al. (2020) also confirmed the above notion where Put was responsible for ROS-mediated activation of SA-signaling, which triggered ETI and SAR in Arabidopsis thaliana by enhancing the accumulation of EDS1 and NPR1 genes in response to pathogen attack.
Recently, NO and PA has emerged as an essential player in regulating growth and defense signaling pathways in plants (Wimalasekera et al., 2011). Studies have concluded that exogenous application of PA-induced NO signaling via unknown mechanisms, thereby stimulating a plethora of physiological and biochemical responses to ameliorate stress-induced oxidative damage in wheat and Arabidopsis thaliana (Diao et al., 2017). Additionally, the concerted action of NO and PA stimulates biosynthesis of various metabolites, antioxidants, and ROS and RNS, thereby regulating plant responses against environmental alterations (Wimalasekera et al., 2011). One of the possible mechanisms by which PA induces NO biosynthesis was deciphered by Agurla et al. (2018), where NO and ROS were over accumulated in response to PA-induced stomatal closure in Arabidopsis thaliana. They identified that PA-induced oxidative burst resulted from enhanced activity of NADPH oxidase and amine oxidase where ROS acted as upstream of NO production by modulating NOS-like activity, thereby completely reversing the inhibitory effect of PA on stomatal closure (Agurla et al., 2018). Similarly, exogenous application of melatonin, a pleiotropic signaling molecule, regulated PA metabolism and NO biosynthesis, and all three in a coordinative manner alleviated heat-induced oxidative damage in tomato seedlings (Jahan et al., 2019b). Likewise, Tailor et al. (2019) also reported that exogenous application of NO significantly enhanced intracellular Spm as evidenced by upregulation of PA biosynthetic enzymes arginine decarboxylase, S-adenosylmethionine decarboxylase, thereby conferring salt tolerance to sunflower seedlings.
Methylglyoxal is a short-oxygenated aldehyde released as a by-product of aerobic metabolisms such as glycolysis and the pentose phosphate pathway (Li, 2016). When synthesized at a higher level, MG can disrupt the array of physiological and biochemical processes in plants leading to loss of critical cellular functions and, in some cases, may induce a detrimental effect on plant survival (Li, 2016). MG detoxification is catalyzed by the glyoxalase (Gly) pathway, i.e., Gly I and Gly II, that efficiently convert MG into non-toxic molecules, reducing its adverse effect on plant growth and development (Li, 2016). Fewer studies have been conducted to analyze crosstalk between SA and MG detoxification signaling in regulating plant growth and immunity against environmental cues (Kaur et al., 2016). A study indicated that SA antagonizes selenium-induced oxidative damage in rice plants by stimulating the activities of enzymatic and non-enzymatic antioxidants along with GlyI and GlyII enzymes that limited the detrimental effect of MG on rice plants (Mostofa et al., 2020). Likewise, exogenous application of SA improves the growth and immunity of Brassica rapa plants under salt stress conditions by regulating the ASH-GSH cycle and MG detoxification system (Kamran et al., 2020). Similarly, SA mediated NO accumulation and signaling counters As-induced toxicity in maize plants by modulating enzymatic and non-enzymatic defense systems and glyoxalase pathway enzymes (Kaya et al., 2020).
NO crosstalk with MG involves the regulatory function of GSH, where increased synthesis of GSH due to enhanced accumulation of NO may be involved differential regulation of MG detoxification system and consequently enhancing oxidative stress tolerance in plants (Mostofa et al., 2018). Increasing evidence has corroborated the above notion. Researchers have observed and linked NO-mediated GSH signaling to play a critical regulatory role in modulating MG and glyoxalase systems in plants (Mostofa et al., 2018). For example, exogenous application of NO enhances antioxidative defense response and methylglyoxal detoxification system in wheat seedlings, thereby improving their growth and development under salinity stress (Hasanuzzaman et al., 2011, 2017). Moreover, Nahar et al. (2016) reported that crosstalk between PA and NO exerted an antagonistic effect on cadmium-induced oxidative stress in mung bean plants by modulating defense-related pathways' critical enzymes glyoxalase defense system.
Furthermore, NO's exogenous application mitigates nickel-induced oxidative damage in eggplants by regulating antioxidant defense system, osmolyte accumulation, and MG detoxification system (Soliman et al., 2019). The study analyzed crosstalk between NO and hydrogen sulfide to stimulate resilience in maize plants exposed to heavy metal stress. The researchers observed that both NO and hydrogen sulfide effectively ameliorated chromium-induced oxidative damage in maize plants by controlling excess ROS generation, MG production, improving membrane integrity, and enzymatic and glyoxalase system (Kharbech et al., 2020).
Strigolactones, a butanolide-class of multifunctional phytohormones derived from the carotenoid catabolism pathway, regulate many plant functions under stress conditions (Torres-Vera et al., 2014). SL is exclusively synthesized in roots and the shoot downstream of carotenoid catabolism, where carlactone is the critical precursor for its biosynthesis (Torres-Vera et al., 2014). Strikingly, researchers have also documented the involvement of SL in regulating nodule formation, mycorrhizal colonization, and mediating plant-microbe interaction (Marzec and Muszynska, 2015). Conversely, Marzec and Muszynska (2015) highlighted the first report for SL's possible involvement in conferring stress tolerance. They observed that SL biosynthesis was enhanced upon pathogen infection, which differentially modulated defense response in Arabidopsis thaliana and O. sativa plants by activating the defense system under the tight control of SA and JA signaling. Later, several researchers indicated that SA acts synergistically to SL and initiates SL biosynthesis by stimulating the expression of critical genes [More Axillary Growth (MAX1, MAX3, and MAX4)] involved in SL production. Various researchers corroborated the above notion. They identified that mutant plants infected with Rhodococcus fascians showed decreased accumulation of SL, causing leafy gall syndrome compared to wild-type plants (Stes et al., 2015).
Interestingly, studies have shown that enhanced expression of MAX2 was observed upon ROS accumulation, thereby inducing oxidative damage in mutant plants compared to wild-type plants where the level of MAX2 was in control (Walter, 2020). Similarly, exogenous application of SA and GR24 (a synthetic analog of SL) stimulated transcriptional activation of SL biosynthetic genes, thereby conferring drought tolerance in wheat cultivars by regulating the antioxidative defense system (Sedaghat et al., 2017, 2020). Moreover, the positive role of SL in stimulating plant defense against fungal pathogens such as Botrytis cinerea, Alternaria alternata, and Meloidogyne incognita has also been demonstrated in SL biosynthetic mutant tomato plants upon interacting with other phytohormones such as ABA, SA, and JA (Aliche et al., 2020).
The interplay between NO and SL was deciphered only recently by Bharti and Bhatla (2015). They treated sunflower seedlings with GR24 that causing a significant reduction in lateral and adventitious root formation, calcium signal, PIN protein distribution, and root NO level. In addition, they also observed a two-fold increase in SL-biosynthetic enzymes in the presence of NO scavenger cPTIO, thus suggesting antagonistic relation between NO and SL. This hypothesis was further confirmed by Manoli et al. (2016). They also observed that exogenous application of NO down-regulates SL-biosynthetic genes by stimulating S-nitrosylation of NO biosynthetic enzymes, thereby activating PIN-dependent auxin regulation and cell differentiation. In this continuation, Sun et al. (2016) demonstrated that NO was the critical signaling mediator in regulating root growth and SL signaling in rice plants grown under nitrogen and phosphorus limiting conditions. They confirmed that SL inhibitor abamine significantly reduced root length in wild-type plants which were reversed by applying NO.
Moreover, the implication of SL and NO in regulating important plant function was also observed by Lv et al. (2018). They confirmed SL signaling played a significant role in stimulating NO and H2O2 production, thereby initiating stomatal closure in the Arabidopsis thaliana in an ABA-dependent manner. They further concluded that SL-related mutants showed an enhanced stomatal opening. In contrast, exogenous application of SL could induce stomatal closure by regulating essential genes of guard cell ABA signaling such as MAX2 and Slow Anion Channel-Associated 1 (SLAC1) (Lv et al., 2018). The interactive role of SL and NO was also recently analyzed by Olah et al. (2020), where they reported that mutant Arabidopsis thaliana plants (max1-1 and max2-1) showed reduced SL synthesis and enhanced NO and S-nitrosothiol levels due to reduced expression of GSNOR protein, thus strengthening root architectural system of Arabidopsis thaliana.
In recent years, technological advancements have provided significant insights into SA and NO signaling that have deepened our understanding of their regulatory role in improving plant growth and metabolism in changing environments. The involvement of SA and NO in mediating plant-microbe interaction has also opened a new realm for plant biologists and microbiologists, allowing them to decipher functional mechanisms by which microbes can be beneficial for improving plant growth and immunity. Nonetheless, a big question still needs an answer: how recent discoveries made regarding SA and NO signaling in plants and their crosstalk with other phytohormones could be successfully exploited to foster agriculture. Researchers have also hypothesized a “triple threat” theory for amalgamating responses of three crucial elements, i.e., plants, their association with a biotrophic pathogen, and environmental cues such as light, temperature, drought, salinity, soil composition, and cold. However, integrating the above information by linking recent breakthroughs regarding SA and NO signaling for improving plant growth, immunity, and ultimately agricultural productivity is still a great challenge to achieve. Therefore, researchers from the plant scientific community need to exercise a stringent work plan focused on establishing a direct link among SA, NO, and plant growth responses for breeding stress-tolerant/resistant plants. Additionally, the research should focus on the systematic use of cutting-edge technologies to help us understand their interactive mechanism regulating plant growth, immunity, and trade-off.
KR conceived the idea and wrote the manuscript and prepared all the figures. KR, NP, NR, SR, and SP-R edited and prepared the final draft of the manuscript. All authors contributed to the article and approved the submitted version.
The authors are also thankful to DST (Department of Science and Technology), DBT-ISLS facility, BHU, Govt. of India (Grant Nos. DST/BHU/PURSE 2017-18 and DST/BHU/FIST 2016-17) for financial support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Agarwal, P., Dabi, M., Sapara, K. K., Joshi, P. S., and Agarwal, P. K. (2016). Ectopic expression of JcWRKY transcription factor confers salinity toslerance via salicylic acid signaling. Front. Plant Sci. 7:1541. doi: 10.3389/fpls.2016.01541
Ageeva-Kieferle, A., Rudolf, E. E., and Lindermayr, C. (2019). Redox-dependent chromatin remodeling: a new function of nitric oxide as architect of chromatin structure in plants. Front. Plant Sci. 10:625. doi: 10.3389/fpls.2019.00625
Agurla, S., Gayatri, G., and Raghavendra, A. S. (2018). Polyamines increase nitric oxide and reactive oxygen species in guard cells of Arabidopsis thaliana during stomatal closure. Protoplasma 255, 153–162. doi: 10.1007/s00709-017-1139-3
Ahanger, M. A., Aziz, U., Alsahli, A. A., Alyemeni, M. N., and Ahmad, P. (2020). Influence of exogenous salicylic acid and nitric oxide on growth, photosynthesis, and ascorbate-glutathione cycle in salt stressed Vigna angularis. Biomolecules 10:42. doi: 10.3390/biom10010042
Ahmad, A., Khan, W. U., Shah, A. A., Yasin, N. A., Naz, S., Ali, A., et al. (2021). Synergistic effects of nitric oxide and silicon on promoting plant growth, oxidative stress tolerance and reduction of arsenic uptake in Brassica juncea. Chemosphere 262:128384. doi: 10.1016/j.chemosphere.2020.128384
Akram, N. A., Iqbal, M., Muhammad, A., Ashraf, M., Al-Qurainy, F., and Shafiq, S. (2018). Aminolevulinic acid and nitric oxide regulate oxidative defense and secondary metabolisms in canola (Brassica napus L.) under drought stress. Protoplasma 255, 163–174. doi: 10.1007/s00709-017-1140-x
Aliche, E. B., Screpanti, C., De Mesmaeker, A., Munnik, T., and Bouwmeester, H. J. (2020). Science and application of strigolactones. New Phytol. 227, 1001–1011. doi: 10.1111/nph.16489
An, C., and Mou, Z. (2011). Salicylic acid and its function in plant immunity. J. Integr. Plant Biol. 53, 412–428. doi: 10.1111/j.1744-7909.2011.01043.x
Armengot, L., Marquès-Bueno, M. M., Soria-Garcia, A., Müller, M., Munné-Bosch, S., and Martínez, M. C. (2014). Functional interplay between protein kinase CK 2 and salicylic acid sustains PIN transcriptional expression and root development. Plant J. 78, 411–423. doi: 10.1111/tpj.12481
Asgher, M., Per, T. S., Masood, A., Fatma, M., Freschi, L., Corpas, F. J., et al. (2017). Nitric oxide signaling and its crosstalk with other plant growth regulators in plant responses to abiotic stress. Environ. Sci. Pollut. Res. 24, 2273–2285. doi: 10.1007/s11356-016-7947-8
Avila, A. C., Ochoa, J., Proaño, K., and Martínez, M. C. (2019). Jasmonic acid and nitric oxide protects naranjilla (Solanum quitoense) against infection by Fusarium oxysporum f. sp. quitoense by eliciting plant defense responses. Physiol. Mol. Plant Pathol. 106, 129–136. doi: 10.1016/j.pmpp.2019.01.002
Barrera-Ortiz, S., Garnica-Vergara, A., Esparza-Reynoso, S., García-Cárdenas, E., Raya-González, J., Ruiz-Herrera, L. F., et al. (2018). Jasmonic acid-ethylene crosstalk via ETHYLENE INSENSITIVE 2 reprograms Arabidopsis root system architecture through nitric oxide accumulation. J. Plant Growth Regul. 37, 438–451. doi: 10.1007/s00344-017-9741-3
Beckers, G. J. M., and Spoel, S. H. (2006). Fine-tuning plant defence signalling: salicylate versus jasmonate. Plant Biol. 8, 1–10. doi: 10.1055/s-2005-872705
Begara-Morales, J. C., Chaki, M., Valderrama, R., Sánchez-Calvo, B., Mata-Pérez, C., Padilla, M. N., et al. (2018). Nitric oxide buffering and conditional nitric oxide release in stress response. J. Exp. Bot. 69, 3425–3438. doi: 10.1093/jxb/ery072
Bharti, N., and Bhatla, S. C. (2015). Nitric oxide mediates strigolactone signaling in auxin and ethylene-sensitive lateral root formation in sunflower seedlings. Plant Signal. Behav. 10:e1054087. doi: 10.1080/15592324.2015.1054087
Blanquet, P., Silva, L., Catrice, O., Bruand, C., Carvalho, H., and Meilhoc, E. (2015). Sinorhizobium meliloti controls nitric oxide–mediated post-translational modification of a Medicago truncatula nodule protein. Mol. Plant Microbe Interact. 28, 1353–1363. doi: 10.1094/MPMI-05-15-0118-R
Bogdan, C. (2015). Nitric oxide synthase in innate and adaptive immunity: an update. Trends Immunol. 36, 161–178. doi: 10.1016/j.it.2015.01.003
Bueno, E., Robles, E. F., Torres, M. J., Krell, T., Bedmar, E. J., Delgado, M. J., et al. (2017). Disparate response to microoxia and nitrogen oxides of the Bradyrhizobium japonicum napEDABC, nirK and norCBQD denitrification genes. Nitric Oxide 68, 137–149. doi: 10.1016/j.niox.2017.02.002
Caarls, L., Pieterse, C. M., and Van Wees, S. (2015). How salicylic acid takes transcriptional control over jasmonic acid signaling. Front. Plant Sci. 6:170. doi: 10.3389/fpls.2015.00170
Caarls L. Van der Does D. Hickman R. Jansen W. Verk M. C. V. Proietti S. (2017) Assessing the role of ETHYLENE RESPONSE FACTOR transcriptional repressors in salicylic acid-mediated suppression of jasmonic acid-responsive genes. Plant Cell Physiol. 58, 266–278. 10.1093/pcp/pcw187.
Cai, W., Liu, W, Wang, W. S., Fu, Z. W., Han, T. T., and Lu, Y. T. (2015). Overexpression of rat neurons nitric oxide synthase in rice enhances drought and salt tolerance. PLoS ONE 10:e0131599. doi: 10.1371/journal.pone.0131599
Calcagno, C., Novero, M., Genre, A., Bonfante, P., and Lanfranco, L. (2012). The exudate from an arbuscular mycorrhizal fungus induces nitric oxide accumulation in Medicago truncatula roots. Mycorrhiza 22, 259–269. doi: 10.1007/s00572-011-0400-4
Cam, Y., Pierre, O., Boncompagni, E., Hérouart, D., Meilhoc, E., and Bruand, C. (2012). Nitric oxide (NO): a key player in the senescence of Medicago truncatula root nodules. New Phytol. 196, 548–560. doi: 10.1111/j.1469-8137.2012.04282.x
Campos, F. V., Oliveira, J. A., Pereira, M. G., and Farnese, F. S. (2019). Nitric oxide and phytohormone interactions in the response of Lactuca sativa to salinity stress. Planta 250, 1475–1489. doi: 10.1007/s00425-019-03236-w
Canales, F. J., Montilla-Bascón, G, Rispail, N., and Prats, E. (2019). Salicylic acid regulates polyamine biosynthesis during drought responses in oat. Plant Signal Behav. 14:e1651183. doi: 10.1080/15592324.2019.1651183
Chabaud, M., Genre, A., Sieberer, B. J., Faccio, A., Fournier, J., Novero, M., et al. (2011). Arbuscular mycorrhizal hyphopodia and germinated spore exudates trigger Ca2+ spiking in the legume and nonlegume root epidermis. New Phytol. 189, 347–355. doi: 10.1111/j.1469-8137.2010.03464.x
Chamizo-Ampudia, A., Sanz-Luque, E., Llamas, A., Galvan, A., and Fernandez, E. (2017). Nitrate reductase regulates plant nitric oxide homeostasis. Trends Plant Sci. 22, 163–174. doi: 10.1016/j.tplants.2016.12.001
Charrier, A., Planchet, E., Cerveau, D., Gimeno-Gilles, C., Verdu, I., Limami, A. M., et al. (2012). Overexpression of a Medicago truncatula stress-associated protein gene (MtSAP1) leads to nitric oxide accumulation and confers osmotic and salt stress tolerance in transgenic tobacco. Planta 236, 567–577. doi: 10.1007/s00425-012-1635-9
Chavoushi, M., Najafi, F., Salimi, A., and Angaji, S. A. (2019). Improvement in drought stress tolerance of safflower during vegetative growth by exogenous application of salicylic acid and sodium nitroprusside. Ind. Crops Prod. 134, 168–176. doi: 10.1016/j.indcrop.2019.03.071
Chen, H., Li, M., Qi, G., Zhao, M., Liu, L., Zhang, J., et al. (2021). Two interacting transcriptional coactivators cooperatively control plant immune responses. bioRxiv. doi: 10.1101/2021.03.21.436112
Cheng, T., Shi, J., Dong, Y., Ma, Y., Peng, Y., Hu, X., et al. (2018). Hydrogen sulfide enhances poplar tolerance to high-temperature stress by increasing S-nitrosoglutathione reductase (GSNOR) activity and reducing reactive oxygen/nitrogen damage. Plant Growth Regul. 84, 11–23. doi: 10.1007/s10725-017-0316-x
Choi, J., Huh, S. U., Kojima, M., Sakakibara, H., Paek, K. H., and Hwang, I. (2010). The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev. Cell 19, 284–295. doi: 10.1016/j.devcel.2010.07.011
Chun, H. J., Park, H. C., Koo, S. C., Lee, J. H., Park, C. Y., Choi, M. S., et al. (2012). Constitutive expression of mammalian nitric oxide synthase in tobacco plants triggers disease resistance to pathogens. Mol. Cells 34, 463–471. doi: 10.1007/s10059-012-0213-0
Cingoz, G. S., and Gurel, E. (2016). Effects of salicylic acid on thermotolerance and cardenolide accumulation under high temperature stress in Digitalis trojana Ivanina. Plant Physiol. Biochem. 105, 145–149. doi: 10.1016/j.plaphy.2016.04.023
Corpas, F. J., Alché, J. D. D., and Barroso, J. B. (2013). Current overview of S-nitrosoglutathione (GSNO) in higher plants. Front. Plant Sci. 4, 126. doi: 10.3389/fpls.2013.00126
Corpas, F. J., and Barroso, J. B. (2015). Nitric oxide from a “green” perspective. Nitric Oxide 45, 15–19. doi: 10.1016/j.niox.2015.01.007
Corpas, F. J., and Barroso, J. B. (2017). Nitric oxide synthase-like activity in higher plants. Nitric Oxide 68, 5–6. doi: 10.1016/j.niox.2016.10.009
Corpas, F. J., and Palma, J. M. (2020). Assessing nitric oxide (NO) in higher plants: an outline. Nitrogen 1, 12–20. doi: 10.3390/nitrogen1010003
Costa-Broseta, Á., Perea-Resa, C., Castillo, M. C., Ruíz, M. F., Salinas, J., and León, J. (2018). Nitric oxide controls constitutive freezing tolerance in Arabidopsis by attenuating the levels of osmoprotectants, stress-related hormones and anthocyanins. Sci. Rep. 8:9268. doi: 10.1038/s41598-018-27668-8
Csiszar, J., Brunner, S., Horváth, E., Bela, K., Ködmön, P., Riyazuddin, R., et al. (2018). Exogenously applied salicylic acid maintains redox homeostasis in salt-stressed Arabidopsis gr1 mutants expressing cytosolic roGFP1. Plant Growth Reg. 86, 181–194. doi: 10.1007/s10725-018-0420-6
Cui, H., Gobbato, E., Kracher, B., Qiu, J., Bautor, J., and Parker, J. E. (2017). A core function of EDS1 with PAD4 is to protect the salicylic acid defense sector in Arabidopsis immunity. New Phytol. 213, 1802–1817. doi: 10.1111/nph.14302
Cui, N., Lu, H., Wang, T., Zhang, W., Kang, L., and Cui, F. (2019). Armet, an aphid effector protein, induces pathogen resistance in plants by promoting the accumulation of salicylic acid. Philos. Trans. R. Soc. B. 374:20180314. doi: 10.1098/rstb.2018.0314
Darvizheh, H., Zahedi, M., Abaszadeh, B., and Razmjoo, J. (2018). Effects of irrigation regime and foliar application of salicylic acid and spermine on the contents of essential oil and caffeic acid derivatives in Echinacea purpurea L. J. Plant. Growth Reg. 37, 1267–1285. doi: 10.1007/s00344-018-9874-z
Darvizheh, H., Zahedi, M., Abbaszadeh, B., and Razmjoo, J. (2019). Changes in some antioxidant enzymes and physiological indices of purple coneflower (Echinacea purpurea L.) in response to water deficit and foliar application of salicylic acid and spermine under field condition. Sci. Hortic. 247, 390–399. doi: 10.1016/j.scienta.2018.12.037
de Roman, M., Fernandez, I., Wyatt, T., Sahrawy, M., Heil, M., and Pozo, M. J. (2011). Elicitation of foliar resistance mechanisms transiently impairs root association with arbuscular mycorrhizal fungi. J. Ecol. 99, 36–45. doi: 10.1111/j.1365-2745.2010.01752.x
De Vleesschauwer, D., Seifi, H. S., Filipe, O., Haeck, A., Huu, S. N., Demeestere, K., et al. (2016). The DELLA protein SLR1 integrates and amplifies salicylic acid-and jasmonic acid-dependent innate immunity in rice. Plant Physiol. 170, 1831–1847. doi: 10.1104/pp.15.01515
Defez, R., Esposito, R., Angelini, C., and Bianco, C. (2016). Overproduction of indole-3-acetic acid in free-living rhizobia induces transcriptional changes resembling those occurring in nodule bacteroids. Mol. Plant Microbe Interact. 29, 484–495. doi: 10.1094/MPMI-01-16-0010-R
Diao, Q., Song, Y., Shi, D., and Qi, H. (2017). Interaction of polyamines, abscisic acid, nitric oxide, and hydrogen peroxide under chilling stress in tomato (Lycopersicon esculentum Mill.) seedlings. Front. Plant Sci. 8:203. doi: 10.3389/fpls.2017.00203
Ding, P., and Ding, Y. (2020). Stories of salicylic acid: a plant defense hormone. Trends Plant Sci. 25, 549–565. doi: 10.1016/j.tplants.2020.01.004
Dinler, B. S., and Tasci, E. (2020). Regulation of NPR1 under salinity and osmotic stress in soybean (Glycine max L.) Leaves. J. Stress Physiol. Biochem. 16:132. Available online at: https://www.proquest.com/scholarly-journals/regulation-npr1-under-salinity-osmotic-stress/docview/2449743031/se-2?accountid=26421
Domingos, P., Prado, A. M., Wong, A., Gehring, C., and Feijo, J. A. (2015). Nitric oxide: a multitasked signaling gas in plants. Mol. Plant 8, 506–520. doi: 10.1016/j.molp.2014.12.010
Dongus, J. A., and Parker, J. E. (2021). EDS1 signalling: at the nexus of intracellular and surface receptor immunity. Curr. Opin. Plant Biol. 62, 102039. doi: 10.1016/j.pbi.2021.102039
Egbichi, I., Keyster, M., and Ludidi, N. (2014). Effect of exogenous application of nitric oxide on salt stress responses of soybean. S. Afr. J. Bot. 90, 131–136. doi: 10.1016/j.sajb.2013.11.002
Falak, N., Imran, Q. M., Hussain, A., and Yun, B. W. (2021). Transcription factors as the “blitzkrieg” of plant defense: a pragmatic view of nitric oxide's role in gene regulation. Int. J. Mol. Sci. 22:522. doi: 10.3390/ijms22020522
Fancy, N. N., Bahlmann, A. K., and Loake, G. J. (2017). Nitric oxide function in plant abiotic stress. Plant Cell Environ. 40, 462–472. doi: 10.1111/pce.12707
Faraz, A., Faizan, M., Sami, F., Siddiqui, H., and Hayat, S. (2020). Supplementation of salicylic acid and citric acid for alleviation of cadmium toxicity to Brassica juncea. J. Plant Growth Regul. 39, 641–655. doi: 10.1007/s00344-019-10007-0
Foo, E., Plett, J. M., Lopez-Raez, J. A., and Reid, D. (2019). The role of plant hormones in plant-microbe symbioses. Front. Plant Sci. 10:1391. doi: 10.3389/fpls.2019.01391
Foyer, C. H., Rasool, B., Davey, J. W., and Hancock, R. D. (2016). Cross-tolerance to biotic and abiotic stresses in plants: a focus on resistance to aphid infestation. J. Exp. Bot. 67, 2025–2037. doi: 10.1093/jxb/erw079
Fragniere, C., Serrano, M., Abou-Mansour, E., Métraux, J. P., and L'Haridon, F. (2011). Salicylic acid and its location in response to biotic and abiotic stress. FEBS Lett. 585, 1847–1852. doi: 10.1016/j.febslet.2011.04.039
Gao, J., Bi, W., Li, H., Wu, J., Yu, X., Liu, D., et al. (2018). WRKY transcription factors associated with NPR1-mediated acquired resistance in barley are potential resources to improve wheat resistance to Puccinia triticina. Front. Plant Sci. 9:1486. doi: 10.3389/fpls.2018.01486
Garcia, M. J., Lucena, C., Romera, F. J., Alcántara, E., and Pérez-Vicente, R. (2010). Ethylene and nitric oxide involvement in the up-regulation of key genes related to iron acquisition and homeostasis in Arabidopsis. J. Exp. Bot. 61, 3885–3899. doi: 10.1093/jxb/erq203
Gayatri, G., Agurla, S., and Raghavendra, A. S. (2013). Nitric oxide in guard cells as an important secondary messenger during stomatal closure. Front. Plant Sci. 4:425. doi: 10.3389/fpls.2013.00425
Genre, A., Chabaud, M., Balzergue, C., Puech-Pagès, V., Novero, M., Rey, T., et al. (2013). Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 198, 190–202. doi: 10.1111/nph.12146
Guilfoyle, T., Hagen, G., Liu, X., Rockett, K. S., Kørner, C. J., and Pajerowska-Mukhtar, K. M. (2015). Salicylic acid signalling: new insights and prospects at a quarter-century milestone. Essays Biochem. 58, 101–113. doi: 10.1042/bse0580101
Guo, J., Zhou, R., Ren, X., Jia, H., Hua, L., Xu, H., et al. (2018). Effects of salicylic acid, Epi-brassinolide and calcium on stress alleviation and Cd accumulation in tomato plants. Ecotox. Environ. Saf. 157, 491–496. doi: 10.1016/j.ecoenv.2018.04.010
Gupta, K. J., Fernie, A. R., Kaiser, W. M., and van Dongen, J. T. (2011). On the origins of nitric oxide. Trends Plant Sci. 16, 160–168. doi: 10.1016/j.tplants.2010.11.007
Gupta, K. J., Kolbert, Z., Durner, J., Lindermayr, C., Corpas, F. J., Brouquisse, R., et al. (2020). Regulating the regulator: nitric oxide control of post-translational modifications. New Phytol. 227, 1319–1325. doi: 10.1111/nph.16622
Hancock, J. T., and Whiteman, M. (2016). Hydrogen sulfide signaling: interactions with nitric oxide and reactive oxygen species. Ann. N. Y. Acad. Sci. 1365, 5–14. doi: 10.1111/nyas.12733
Hasanuzzaman, M., Hossain, M. A., and Fujita, M. (2011). Nitric oxide modulates antioxidant defense and the methylglyoxal detoxification system and reduces salinity-induced damage of wheat seedlings. Plant Biotech. Rep. 5, 353–365. doi: 10.1007/s11816-011-0189-9
Hasanuzzaman, M., Nahar, K., Hossain, M. S., Anee, T. I., Parvin, K., and Fujita, M. (2017). Nitric oxide pretreatment enhances antioxidant defense and glyoxalase systems to confer PEG-induced oxidative stress in rapeseed. J. Plant Interact. 12, 323–331. doi: 10.1080/17429145.2017.1362052
Hasanuzzaman, M., Oku, H., Nahar, K., Bhuyan, M. B., Al Mahmud, J., Baluska, F., et al. (2018). Nitric oxide-induced salt stress tolerance in plants: ROS metabolism, signaling, and molecular interactions. Plant Biotechnol. Rep. 12, 77–92. doi: 10.1007/s11816-018-0480-0
Horváth, E., Csiszár, J., Gallé, Á., Poór, P., Szepesi, Á., and Tari, I. (2015). Hardening with salicylic acid induces concentration-dependent changes in abscisic acid biosynthesis of tomato under salt stress. J. Plant Physiol. 183, 54–63. doi: 10.1016/j.jplph.2015.05.010
Hu, J., Yang, H., Mu, J., Lu, T., Peng, J., Deng, X., et al. (2017). Nitric oxide regulates protein methylation during stress responses in plants. Mol Cell 67, 702–710. doi: 10.1016/j.molcel.2017.06.031
Huang, W., Wang, Y., Li, X., and Zhang, Y. (2020). Biosynthesis and regulation of salicylic acid and N-hydroxypipecolic acid in plant immunity. Mol. Plant 13, 31–41. doi: 10.1016/j.molp.2019.12.008
Huang, X., Stettmaier, K., Michel, C., Hutzler, P., Mueller, M. J., and Durner, J. (2004). Nitric oxide is induced by wounding and influences jasmonic acid signaling in Arabidopsis thaliana. Planta 218, 938–946. doi: 10.1007/s00425-003-1178-1
Iglesias, M. J., Terrile, M. C., and Casalongu,é, C. A. (2011). Auxin and salicylic acid signalings counteract the regulation of adaptive responses to stress. Plant Signal. Behav. 6, 452–454. doi: 10.4161/psb.6.3.14676
Jahan, M. S., Shu, S., Wang, Y., Chen, Z., He, M., Tao, M., et al. (2019a). Melatonin alleviates heat-induced damage of tomato seedlings by balancing redox homeostasis and modulating polyamine and nitric oxide biosynthesis. BMC Plant Biol. 19:414. doi: 10.1186/s12870-019-1992-7
Jahan, M. S., Wang, Y., Shu, S., Zhong, M., Chen, Z., Wu, J., et al. (2019b). Exogenous salicylic acid increases the heat tolerance in tomato (Solanum lycopersicum L) by enhancing photosynthesis efficiency and improving antioxidant defense system through scavenging of reactive oxygen species. Sci. Hort. 247, 421–429. doi: 10.1016/j.scienta.2018.12.047
Jahnova, J., Cinčalová, L., Sedlárová, M., Jedelská, T., Sekaninová, J., Mieslerová, B., et al. (2020). Differential modulation of S-nitrosoglutathione reductase and reactive nitrogen species in wild and cultivated tomato genotypes during development and powdery mildew infection. Plant Physiol. Biochem. 155, 297–310. doi: 10.1016/j.plaphy.2020.06.039
Janda, K., Hideg, É., Szalai, G., Kovács, L., and Janda, T. (2012). Salicylic acid may indirectly influence the photosynthetic electron transport. J. Plant Physiol. 169, 971–978. doi: 10.1016/j.jplph.2012.02.020
Janda, M., and Ruelland, E. (2015). Magical mystery tour: salicylic acid signalling. Environ. Exp. Bot. 114, 117–128. doi: 10.1016/j.envexpbot.2014.07.003
Janda, M., Šašek, V., Chmelarová, H., Andrejch, J., Nováková, M., Hajšlová, J., et al. (2015). Phospholipase D affects translocation of NPR1 to the nucleus in Arabidopsis thaliana. Front. Plant Sci. 6:59. doi: 10.3389/fpls.2015.00059
Jayakannan, M., Bose, J., Babourina, O., Rengel, Z., and Shabala, S. (2015). Salicylic acid in plant salinity stress signalling and tolerance. Plant Growth Regul. 76, 25–40. doi: 10.1007/s10725-015-0028-z
Jedelska, T., Sedlárová, M., Lochman, J., Cinčalová, L., Luhová, L., and Petrivalský, M. (2021). Protein S-nitrosation differentially modulates tomato responses to infection by hemi-biotrophic oomycetes of Phytophthora spp. Hortic. Res. 8, 1–15. doi: 10.1038/s41438-021-00469-3
Jiang, C. J., Shimono, M., Sugano, S., Kojima, M., Yazawa, K., Yoshida, R., et al. (2010). Abscisic acid interacts antagonistically with salicylic acid signaling pathway in rice–Magnaporthe grisea interaction. Mol. Plant Microbe. Interact. 23, 791–798. doi: 10.1094/MPMI-23-6-0791
Kalachova, T., Iakovenko, O., Kretinin, S., and Kravets, V. (2013). Involvement of phospholipase D and NADPH-oxidase in salicylic acid signaling cascade. Plant Physiol. Biochem. 66, 127–133. doi: 10.1016/j.plaphy.2013.02.006
Kamran, M., Xie, K., Sun, J., Wang, D., Shi, C., Lu, Y., et al. (2020). Modulation of growth performance and coordinated induction of ascorbate-glutathione and methylglyoxal detoxification systems by salicylic acid mitigates salt toxicity in choysum (Brassica parachinensis L.). Ecotoxcol. Environ. Saf. 188:109877. doi: 10.1016/j.ecoenv.2019.109877
Kapoor, K., Mira, M. M., Ayele, B. T., Nguyen, T. N., Hill, R. D., and Stasolla, C. (2018). Phytoglobins regulate nitric oxide-dependent abscisic acid synthesis and ethylene-induced program cell death in developing maize somatic embryos. Planta 247, 1277–1291. doi: 10.1007/s00425-018-2862-5
Kaur, C., Sharma, S., Singla-Pareek, S. L., and Sopory, S. K. (2016). Methylglyoxal detoxification in plants: role of glyoxalase pathway. Indian J. Plant Physiol. 21, 377–390. doi: 10.1007/s40502-016-0260-1
Kaya, C., Ashraf, M., Alyemeni, M. N., Corpas, F. J., and Ahmad, P. (2020). Salicylic acid-induced nitric oxide enhances arsenic toxicity tolerance in maize plants by upregulating the ascorbate-glutathione cycle and glyoxalase system. J. Hazard. Mater. 399:123020. doi: 10.1016/j.jhazmat.2020.123020
Khan, M. N., Mohammad, F., Mobin, M., and Saqib, M. A. (2014). “Tolerance of plants to abiotic stress: a role of nitric oxide and calcium”, in Nitric Oxide in Plants: Metabolism and Role in Stress Physiology, ed M. N. Khan (Cham: Springer), 225–242. doi: 10.1007/978-3-319-06710-0_14
Kharbech, O., Sakouhi, L., Massoud, M. B., Mur, L. A. J., Corpas, F. J., Djebali, W., et al. (2020). Nitric oxide and hydrogen sulfide protect plasma membrane integrity and mitigate chromium-induced methylglyoxal toxicity in maize seedlings. Plant Physiol. Biochem. 157, 244–255. doi: 10.1016/j.plaphy.2020.10.017
Kim, Y., Mun, B. G., Khan, A. L., Waqas, M., Kim, H. H., Shahzad, R., et al. (2018). Regulation of reactive oxygen and nitrogen species by salicylic acid in rice plants under salinity stress conditions. PLoS ONE 13:e0192650. doi: 10.1371/journal.pone.0192650
Klessig, D. F., Choi, H. W., and Dempsey, D. M. A. (2018). Systemic acquired resistance and salicylic acid: past, present, and future. Mol. Plant Microbe. Interact. 31, 871–888. doi: 10.1094/MPMI-03-18-0067-CR
Kloth, K. J., Wiegers, G. L., Busscher-Lange, J., Van Haarst, J. C., Kruijer, W., Bouwmeester, H. J., et al. (2016). AtWRKY22 promotes susceptibility to aphids and modulates salicylic acid and jasmonic acid signalling. J. Exp. Bot. 67, 3383–3396. doi: 10.1093/jxb/erw159
Kolbert, Z., Feigl, G., Bord,é, Á., Molnár, Á., and Erdei, L. (2017). Protein tyrosine nitration in plants: present knowledge, computational prediction and future perspectives. Plant Physiol. Biochem. 113, 56–63. doi: 10.1016/j.plaphy.2017.01.028
Kong, X., Wang, T., Li, W., Tang, W., Zhang, D., and Dong, H. (2016). Exogenous nitric oxide delays salt-induced leaf senescence in cotton (Gossypium hirsutum L.). Acta Physiol. Plant 38:61. doi: 10.1007/s11738-016-2079-9
Koo, Y. M., Heo, A. Y., and Choi, H. W. (2020). Salicylic acid as a safe plant protector and growth regulator. Plant Pathol. J. 36:1. doi: 10.5423/PPJ.RW.12.2019.0295
Koyuncu, M. A., Erbas, D., Onursal, C. E., Secmen, T., Guneyli, A., and Uzumcu, S. S. (2019). Postharvest treatments of salicylic acid, oxalic acid and putrescine influences bioactive compounds and quality of pomegranate during controlled atmosphere storage. J. Food Sci. Technol. 56, 350–359. doi: 10.1007/s13197-018-3495-1
Krasylenko, Y. A., Yemets, A. I., and Blume, Y. B. (2010). Functional role of nitric oxide in plants. Russ. J. Plant Physiol. 57, 451–461. doi: 10.1134/S1021443710040011
Krizek, B. A., Bequette, C. J., Xu, K., Blakley, I. C., Fu, Z. Q., Stratmann, J. W., et al. (2016). RNA-Seq links the transcription factors AINTEGUMENTA and AINTEGUMENTA-LIKE6 to cell wall remodeling and plant defense pathways. Plant Physio. 171, 2069–2084. doi: 10.1104/pp.15.01625
Lebeis, S. L., Paredes, S. H., Lundberg, D. S., Breakfield, N., Gehring, J., McDonald, M., et al. (2015). Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349, 860–864. doi: 10.1126/science.aaa8764
Lechon, T., Sanz, L., Sánchez-Vicente, I., and Lorenzo, O. (2020). Nitric oxide overproduction by cue1 mutants differs on developmental stages and growth conditions. Plants 9:1484. doi: 10.3390/plants9111484
Lee, B. R., Zhang, Q., Park, S. H., Islam, M. T., and Kim, T. H. (2019). Salicylic acid improves drought-stress tolerance by regulating the redox status and proline metabolism in Brassica rapa. Hortic. Environ. Biotechnol. 60, 31–40. doi: 10.1007/s13580-018-0099-7
Lenk, M., Wenig, M., Bauer, K., Hug, F., Knappe, C., Lange, B., et al. (2019). Pipecolic acid is induced in barley upon infection and triggers immune responses associated with elevated nitric oxide accumulation. Mol. Plant Microbe. Interact. 32, 1303–1313. doi: 10.1094/MPMI-01-19-0013-R
Li, C., Cao, S., Wang, K., Lei, C., Ji, N., Xu, F., et al. (2021). Heat shock protein (HSP) 24 is involved in the baba-induced resistance to fungal pathogen in postharvest grapes underlying an NPR1-dependent manner. Front. Plant Sci. 12:292. doi: 10.3389/fpls.2021.646147
Li, H., Wu, J., Shang, X., Geng, M., Gao, J., Zhao, S., et al. (2020). WRKY transcription factors shared by BTH-induced resistance and NPR1-mediated acquired resistance improve broad-spectrum disease resistance in wheat. Mol. Plant Microbe. Interact. 33, 433–443. doi: 10.1094/MPMI-09-19-0257-R
Li, N., Han, X., Feng, D., Yuan, D., and Huang, L. J. (2019). Signaling crosstalk between salicylic acid and ethylene/jasmonate in plant defense: do we understand what they are whispering? Int. J. Mol. Sci. 20:671. doi: 10.3390/ijms20030671
Li, Q., Huang, W., Xiong, C., and Zhao, J. (2018). Transcriptome analysis reveals the role of nitric oxide in Pleurotus eryngii responses to Cd2+ stress. Chemosphere 201, 294–302. doi: 10.1016/j.chemosphere.2018.03.011
Li, Q., Wang, G., Guan, C., Yang, D., Wang, Y., Zhang, Y., et al. (2019). Overexpression of LcSABP, an orthologous gene for salicylic acid binding protein 2, enhances drought stress tolerance in transgenic tobacco. Front. Plant Sci. 10:200. doi: 10.3389/fpls.2019.00200
Li, Z. G. (2016). Methylglyoxal and glyoxalase system in plants: old players, new concepts. Bot. Rev. 82, 183–203. doi: 10.1007/s12229-016-9167-9
Lindermayr, C., Sell, S., Müller, B., Leister, D., and Durner, J. (2010). Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell 22, 2894–2907. doi: 10.1105/tpc.109.066464
Liu, C., Atanasov, K. E., Arafaty, N., Murillo, E., Tiburcio, A. F., Zeier, J., et al. (2020). Putrescine elicits ROS-dependent activation of the salicylic acid pathway in Arabidopsis thaliana. Plant Cell Environ. 43, 2755–2768. doi: 10.1111/pce.13874
Liu, L., Sonbol, F. M., Huot, B., Gu, Y., Withers, J., Mwimba, M., et al. (2016). Salicylic acid receptors activate jasmonic acid signalling through a non-canonical pathway to promote effector-triggered immunity. Nat. Commun. 7:13099. doi: 10.1038/ncomms13099
Liu, S., Dong, Y., Xu, L., and Kong, J. (2014). Effects of foliar applications of nitric oxide and salicylic acid on salt-induced changes in photosynthesis and antioxidative metabolism of cotton seedlings. Plant Growth Regul. 73, 67–78. doi: 10.1007/s10725-013-9868-6
Liu, Z., Ding, Y., Wang, F., Ye, Y., and Zhu, C. (2016). Role of salicylic acid in resistance to cadmium stress in plants. Plant Cell Rep. 35, 719–731. doi: 10.1007/s00299-015-1925-3
Lozano-Juste, J., and León, J. (2010). Enhanced abscisic acid-mediated responses in nia1nia2noa1-2 triple mutant impaired in NIA/NR-and AtNOA1-dependent nitric oxide biosynthesis in Arabidopsis. Plant Physiol. 152, 891–903. doi: 10.1104/pp.109.148023
Lu, H. (2009). Dissection of salicylic acid-mediated defense signaling networks. Plant Signal. Behav. 4, 713–717. doi: 10.4161/psb.4.8.9173
Lu, H., Greenberg, J. T., and Holuigue, L. (2016). Salicylic acid signalling networks. Front. Plant Sci. 7, 238. doi: 10.3389/fpls.2016.00238
Luo, J., Xia, W., Cao, P., Xiao, Z. A., Zhang, Y., Liu, M., et al. (2019). Integrated transcriptome analysis reveals plant hormones jasmonic acid and salicylic acid coordinate growth and defense responses upon fungal infection in poplar. Biomolecules 9:12. doi: 10.3390/biom9010012
Lv, S., Zhang, Y., Li, C., Liu, Z., Yang, N., Pan, L., et al. (2018). Strigolactone-triggered stomatal closure requires hydrogen peroxide synthesis and nitric oxide production in an abscisic acid-independent manner. New Phytol. 217, 290–304. doi: 10.1111/nph.14813
Ma, Z, Liu, H., Mi, Z., Zhang, Z., Wang, Y., Xu, W., et al. (2017). Climate warming reduces the temporal stability of plant community biomass production. Nat. Commun. 8:15378. doi: 10.1038/ncomms15378
Manohar, M., Wang, D., Manosalva, P. M., Choi, H. W., Kombrink, E., and Klessig, D. F. (2017). Members of the abscisic acid co-receptor PP 2C protein family mediate salicylic acid–abscisic acid crosstalk. Plant Direct 1:e00020. doi: 10.1002/pld3.20
Manoli, A., Trevisan, S., Voigt, B., Yokawa, K., Baluška, F., and Quaggiotti, S. (2016). Nitric oxide-mediated maize root apex responses to nitrate are regulated by auxin and strigolactones. Front. Plant Sci. 6:1269. doi: 10.3389/fpls.2015.01269
Martinez-Medina, A., Fernandez, I., Lok, G. B., Pozo, M. J., Pieterse, C. M., and Van Wees, S. C. (2017). Shifting from priming of salicylic acid-to jasmonic acid-regulated defences by trichoderma protects tomato against the root knot nematode Meloidogyne incognita. New Phytol. 213, 1363–1377. doi: 10.1111/nph.14251
Martínez-Medina, A., Pescador, L., Fernández, I., Rodríguez-Serrano, M., García, J. M., Romero-Puertas, M. C., et al. (2019b). Nitric oxide and phytoglobin PHYTOGB1 are regulatory elements in the Solanum lycopersicum–Rhizophagus irregularis mycorrhizal symbiosis. New Phytol. 223, 1560–1574. doi: 10.1111/nph.15898
Martínez-Medina, A., Pescador, L., Terrón-Camero, L. C., Pozo, M. J., and Romero-Puertas, M. C. (2019a). Nitric oxide in plant–fungal interactions. J. Exp. Bot. 70, 4489–4503. doi: 10.1093/jxb/erz289
Marzec, M., and Muszynska, A. (2015). In silico analysis of the genes encoding proteins that are involved in the biosynthesis of the RMS/MAX/D pathway revealed new roles of strigolactones in plants. Int. J. Mol. Sci. 16, 6757–6782. doi: 10.3390/ijms16046757
Matsui, H., Iwakawa, H., Hyon, G. S., Yotsui, I., Katou, S., Monte, I., et al. (2020). Isolation of natural fungal pathogens from Marchantia polymorpha reveals antagonism between salicylic acid and jasmonate during liverwort–fungus interactions. Plant Cell Physiol. 61, 265–275. doi: 10.1093/pcp/pcz187
Medina, M. J. H., Gagnon, H., Piché, Y., Ocampo, J. A., Garrido, J. M. G., and Vierheilig, H. (2003). Root colonization by arbuscular mycorrhizal fungi is affected by the salicylic acid content of the plant. Plant Sci. 164, 993–998. doi: 10.1016/S0168-9452(03)00083-9
Meguro, A., and Sato, Y. (2014). Salicylic acid antagonizes abscisic acid inhibition of shoot growth and cell cycle progression in rice. Sci. Rep. 4:4555. doi: 10.1038/srep04555
Meilhoc, E., Boscari, A., Bruand, C., Puppo, A., and Brouquisse, R. (2011). Nitric oxide in legume–rhizobium symbiosis. Plant Sci. 181, 573–581. doi: 10.1016/j.plantsci.2011.04.007
Miedes, E., Vanholme, R., Boerjan, W., and Molina, A. (2014). The role of the secondary cell wall in plant resistance to pathogens. Front Plant Sci. 5:358. doi: 10.3389/fpls.2014.00358
Mine, A., Sato, M., and Tsuda, K. (2014). Toward a systems understanding of plant-microbe interactions. Front. Plant Sci. 5:423. doi: 10.3389/fpls.2014.00423
Miura, K., and Tada, Y. (2014). Regulation of water, salinity, and cold stress responses by salicylic acid. Front Plant Sci. 5:4. doi: 10.3389/fpls.2014.00004
Mostofa, M. G., Ghosh, A., Li, Z. G., Siddiqui, M. N., Fujita, M., and Tran, L. S. P. (2018). Methylglyoxal–a signaling molecule in plant abiotic stress responses. Free Radic. Biol. Med. 122, 96–109. doi: 10.1016/j.freeradbiomed.2018.03.009
Mostofa, M. G., Rahman, M. M., Siddiqui, M. N., Fujita, M., and Tran, L. S. P. (2020). Salicylic acid antagonizes selenium phytotoxicity in rice: selenium homeostasis, oxidative stress metabolism and methylglyoxal detoxification. J. Hazard. Mater. 394:122572. doi: 10.1016/j.jhazmat.2020.122572
Mur, L. A., Carver, T. L., and Prats, E. (2006). NO way to live; the various roles of nitric oxide in plant–pathogen interactions. J. Exp. Bot. 57, 489–505. doi: 10.1093/jxb/erj052
Mur, L. A., Prats, E., Pierre, S., Hall, M. A., and Hebelstrup, K. H. (2013). Integrating nitric oxide into salicylic acid and jasmonic acid/ethylene plant defense pathways. Front. Plant Sci. 4:215. doi: 10.3389/fpls.2013.00215
Nabi, R. B. S., Tayade, R., Hussain, A., Kulkarni, K. P., Imran, Q. M., Mun, B. G., et al. (2019). Nitric oxide regulates plant responses to drought, salinity, and heavy metal stress. Environ. Exp. Bot. 161, 120–133. doi: 10.1016/j.envexpbot.2019.02.003
Nahar, K., Hasanuzzaman, M., Alam, M. M., Rahman, A., Suzuki, T., and Fujita, M. (2016). Polyamine and nitric oxide crosstalk: antagonistic effects on cadmium toxicity in mung bean plants through upregulating the metal detoxification, antioxidant defense and methylglyoxal detoxification systems. Ecotox. Environ. Saf. 126, 245–255. doi: 10.1016/j.ecoenv.2015.12.026
Nakagawa, T., and Kawaguchi, M. (2006). Shoot-applied MeJA suppresses root nodulation in Lotus japonicus. Plant Cell Physiol. 47, 176–180. doi: 10.1093/pcp/pci222
Naser Alavi, S. M., Arvin, M. J., and Manoochehri Kalantari, K. (2014). Salicylic acid and nitric oxide alleviate osmotic stress in wheat (Triticum aestivum L.) seedlings. J. Plant Interact. 9, 683–688. doi: 10.1080/17429145.2014.900120
Neill, S. J., Desikan, R., Clarke, A., and Hancock, J. T. (2002). Nitric oxide is a novel component of abscisic acid signaling in stomatal guard cells. Plant Physiol. 128, 13–16. doi: 10.1104/pp.010707
Nurnberger, T., and Kemmerling, B. (2018). Pathogen-associated molecular patterns (PAMP) and PAMP-triggered immunity. Annu. Plant Rev. 34, 16–47. doi: 10.1002/9781119312994.apr0362
Olah, D., Feigl, G., Molnár, Á., Ördög, A., and Kolbert, Z. (2020). Strigolactones interact with nitric oxide in regulating root system architecture of Arabidopsis thaliana. Front. Plant Sci. 11:1019. doi: 10.3389/fpls.2020.01019
Pal, M., Ivanovska, B., Oláh, T., Tajti, J., Hamow, K. Á., Szalai, G., et al. (2019). Role of polyamines in plant growth regulation of Rht wheat mutants. Plant Physiol. Biochem. 137, 189–202. doi: 10.1016/j.plaphy.2019.02.013
Parankusam, S., Adimulam, S. S., Bhatnagar-Mathur, P., and Sharma, K. K. (2017). Nitric oxide (NO) in plant heat stress tolerance: current knowledge and perspectives. Front. Plant Sci. 8:1582. doi: 10.3389/fpls.2017.01582
Park, S. H., Lee, B. R., Al Mamun, M., Bae, D. W., and Kim, T. H. (2021). Characterization of salicylic acid-and abscisic acid-mediated photosynthesis, Ca2+ and H2O2 accumulation in two distinct phases of drought stress intensity in Brassica napus. Environ. Exp. Bot. 186:104434. doi: 10.1016/j.envexpbot.2021.104434
Prakash, V., Singh, V. P., Tripathi, D. K., Sharma, S., and Corpas, F. J. (2021). Nitric oxide (NO) and salicylic acid (SA): a framework for their relationship in plant development under abiotic stress. Plant Biol. 23, 39–49. doi: 10.1111/plb.13246
Prochazkova, D., Haisel, D., Wilhelmová, N., Pavlíková, D., and Száková, J. (2013). Effects of exogenous nitric oxide on photosynthesis. Photosynthetica 51, 483–489. doi: 10.1007/s11099-013-0053-y
Prodhan, M. Y., Munemasa, S., Nahar, M. N. E. N., Nakamura, Y., and Murata, Y. (2018). Guard cell salicylic acid signaling is integrated into abscisic acid signaling via the Ca2+/CPK-dependent pathway. Plant Physiol. 178, 441–450. doi: 10.1104/pp.18.00321
Rai, K. K., Pandey, N., Meena, R. P., and Rai, S. P. (2021). Biotechnological strategies for enhancing heavy metal tolerance in neglected and underutilized legume crops: A comprehensive review. Ecotox. Environ. Saf. 208:111750. doi: 10.1016/j.ecoenv.2020.111750
Rai, K. K., Pandey, N., and Rai, S. P. (2020a). Salicylic acid and nitric oxide signaling in plant heat stress. Physiol. Plant. 168, 241–255. doi: 10.1111/ppl.12958
Rai, K. K., Rai, N., Aamir, M., Tripathi, D., and Rai, S. P. (2020b). Interactive role of salicylic acid and nitric oxide on transcriptional reprogramming for high temperature tolerance in Lablab purpureus L.: structural and functional insights using computational approaches. J. Biotechnol. 309, 113–130. doi: 10.1016/j.jbiotec.2020.01.001
Rai, K. K., Rai, N., and Rai, S. P. (2018a). Salicylic acid and nitric oxide alleviate high temperature induced oxidative damage in Lablab purpureus L plants by regulating bio-physical processes and DNA methylation. Plant Physiol. Biochem. 128, 72–88. doi: 10.1016/j.plaphy.2018.04.023
Rai, K. K., Rai, N., and Rai, S. P. (2018b). Response of Lablab purpureus L. to high temperature stress and role of exogenous protectants in mitigating high temperature induced oxidative damages. Mol. Biol. Rep. 45, 1375–1395. doi: 10.1007/s11033-018-4301-x
Ren, R., Wei, Y., Ahmad, S., Jin, J., Gao, J., Lu, C., et al. (2020). Identification and characterization of NPR1 and PR1 homologs in Cymbidium orchids in response to multiple hormones, salinity and viral stresses. Int. J. Mol. Sci. 21:1977. doi: 10.3390/ijms21061977
Rezayian, M., Ebrahimzadeh, H., and Niknam, V. (2020). Nitric oxide stimulates antioxidant system and osmotic adjustment in soybean under drought stress. J. Soil. Sci. Plant Nut. 20, 1122–1132. doi: 10.1007/s42729-020-00198-x
Rivas-San Vicente, M., and Plasencia, J. (2011). Salicylic acid beyond defence: its role in plant growth and development. J. Exp. Bot. 62, 3321–3338. doi: 10.1093/jxb/err031
Rizwan, M., Mostofa, M. G., Ahmad, M. Z., Imtiaz, M., Mehmood, S., Adeel, M., et al. (2018). Nitric oxide induces rice tolerance to excessive nickel by regulating nickel uptake, reactive oxygen species detoxification and defense-related gene expression. Chemosphere 191, 23–35. doi: 10.1016/j.chemosphere.2017.09.068
Ruelland, E., and Zachowski, A. (2010). How plants sense temperature. Environ. Exp. Bot. 69, 225–232. doi: 10.1016/j.envexpbot.2010.05.011
Sahay, S., Khan, E., and Gupta, M. (2019). Nitric oxide and abscisic acid protects against PEG-induced drought stress differentially in brassica genotypes by combining the role of stress modulators, markers and antioxidants. Nitric Oxide 89, 81–92. doi: 10.1016/j.niox.2019.05.005
Saleh, B., Alshehadah, E., and Slaman, H. (2020). Abscisic acid (ABA) and salicylic acid (SA) content in relation to transcriptional patterns in grapevine (Vitis vinifera l.) under salt stress. J. Plant Biochem. Physiol. 8:245. doi: 10.35248/2329-9029.20.8.245
Santolini, J., André, F., Jeandroz, S., and Wendehenne, D. (2017). Nitric oxide synthase in plants: where do we stand? Nitric Oxide 63, 30–38. doi: 10.1016/j.niox.2016.09.005
Sanz, L., Albertos, P., Mateos, I., Sánchez-Vicente, I., Lechón, T., Fernández-Marcos, M., et al. (2015). Nitric oxide (NO) and phytohormones crosstalk during early plant development. J. Exp. Bot. 66, 2857–2868. doi: 10.1093/jxb/erv213
Schepetilnikov, M., and Ryabova, L. A. (2018). Recent discoveries on the role of TOR (target of rapamycin) signaling in translation in plants. Plant Physiol. 176, 1095–1105. doi: 10.1104/pp.17.01243
Sedaghat, M., Ferrero, M., Caracci, A., Strnad, M., Novak, O., Schubert, A., et al. (2017). “On the strigolactones-salicylic acid relationship in tomato under drought,” in Joint Congress SIBV-SIGA. (Italy: Proceedings of the Joint Congress SIBV-SIGA, PISA).
Sedaghat, M., Sarvestani, Z. T., Emam, Y., Bidgoli, A. M., and Sorooshzadeh, A. (2020). Foliar-applied gr24 and salicylic acid enhanced wheat drought tolerance. Russ. Plant Physiol. 67, 733–739. doi: 10.1134/S1021443720040159
Shan, C., Zhou, Y., and Liu, M. (2015). Nitric oxide participates in the regulation of the ascorbate-glutathione cycle by exogenous jasmonic acid in the leaves of wheat seedlings under drought stress. Protoplasma 252, 1397–1405. doi: 10.1007/s00709-015-0756-y
Shanmugam, V., Wang, Y. W., Tsednee, M., Karunakaran, K., and Yeh, K. C. (2015). Glutathione plays an essential role in nitric oxide-mediated iron-deficiency signaling and iron-deficiency tolerance in Arabidopsis. Plant J. 84, 464–477. doi: 10.1111/tpj.13011
Sharma, A., Sidhu, G. P. S., Araniti, F., Bali, A. S., Shahzad, B., Tripathi, D. K., et al. (2020a). The role of salicylic acid in plants exposed to heavy metals. Molecules 25:540. doi: 10.3390/molecules25030540
Sharma, A., Soares, C., Sousa, B., Martins, M., Kumar, V., Shahzad, B., et al. (2020b). Nitric oxide-mediated regulation of oxidative stress in plants under metal stress: a review on molecular and biochemical aspects. Physiol. Plant 168, 318–344. doi: 10.1111/ppl.13004
Shen, Q., Wang, Y. T., Tian, H., and Guo, F. Q. (2013). Nitric oxide mediates cytokinin functions in cell proliferation and meristem maintenance in Arabidopsis. Mol. Plant 6, 1214–1225. doi: 10.1093/mp/sss148
Shivaraj, S. M., Vats, S., Bhat, J. A., Dhakte, P., Goyal, V., Khatri, P., et al. (2020). Nitric oxide and hydrogen sulfide crosstalk during heavy metal stress in plants. Physiol. Plant 168, 437–455. doi: 10.1111/ppl.13028
Shukla, P., Singh, S., Dubey, P., Singh, A., and Singh, A. K. (2015). Nitric oxide mediated amelioration of arsenic toxicity which alters the alternative oxidase (Aox1) gene expression in Hordeum vulgare L. Ecotox. Environ. Saf. 120, 59–65. doi: 10.1016/j.ecoenv.2015.05.030
Siboza, X. I., Bertling, I., and Odindo, A. O. (2014). Salicylic acid and methyl jasmonate improve chilling tolerance in cold-stored lemon fruit (Citrus limon). J. Plant Physiol. 171, 1722–1731. doi: 10.1016/j.jplph.2014.05.012
Sihag, S., Brar, B., and Joshi, U. N. (2019). Salicylic acid induces amelioration of chromium toxicity and affects antioxidant enzyme activity in Sorghum bicolor L. Int. J. Phytoremediation 21, 293–304. doi: 10.1080/15226514.2018.1524827
Silveira, N. M., Frungillo, L., Marcos, F. C., Pelegrino, M. T., Miranda, M. T., Seabra, A. B., et al. (2016). Exogenous nitric oxide improves sugarcane growth and photosynthesis under water deficit. Planta 244, 181–190. doi: 10.1007/s00425-016-2501-y
Singh, A. P., Dixit, G., Kumar, A., Mishra, S., Kumar, N., Dixit, S., et al. (2017). A protective role for nitric oxide and salicylic acid for arsenite phytotoxicity in rice (Oryza sativa L.). Plant Physiol. Biochem. 115, 163–173. doi: 10.1016/j.plaphy.2017.02.019
Singh, A. P., Dixit, G., Kumar, A., Mishra, S., Singh, P. K., Dwivedi, S., et al. (2016). Nitric oxide alleviated arsenic toxicity by modulation of antioxidants and thiol metabolism in rice (Oryza sativa L.). Front. Plant Sci. 6, 1272. doi: 10.3389/fpls.2015.01272
Sohag, A. A. M., Tahjib-Ul-Arif, M., Brestic, M., Afrin, S., Sakil, M. A., Hossain, M. T., et al. (2020). Exogenous salicylic acid and hydrogen peroxide attenuate drought stress in rice. Plant Soil Environ. 66, 7–13. doi: 10.17221/472/2019-PSE
Soliman, M., Alhaithloul, H. A., Hakeem, K. R., Alharbi, B. M., El-Esawi, M., and Elkelish, A. (2019). Exogenous nitric oxide mitigates nickel-induced oxidative damage in eggplant by upregulating antioxidants, osmolyte metabolism, and glyoxalase systems. Plants 8:562. doi: 10.3390/plants8120562
Song, A., Gao, T., Li, P., Chen, S., Guan, Z., Wu, D., et al. (2016). Transcriptome-wide identification and expression profiling of the DOF transcription factor gene family in Chrysanthemum morifolium. Front. Plant Sci. 7:199. doi: 10.3389/fpls.2016.00199
Song, G. C., Choi, H. K., and Ryu, C. M. (2015). Gaseous 3-pentanol primes plant immunity against a bacterial speck pathogen, Pseudomonas syringae pv. tomato via salicylic acid and jasmonic acid-dependent signaling pathways in Arabidopsis. Front. Plant Sci. 6:821. doi: 10.3389/fpls.2015.00821
Souri, M. K., and Tohidloo, G. (2019). Effectiveness of different methods of salicylic acid application on growth characteristics of tomato seedlings under salinity. Chem. Biol. Technol. 6, 1–7. doi: 10.1186/s40538-019-0169-9
Stacey, G., McAlvin, C. B., Kim, S. Y., Olivares, J., and Soto, M. J. (2006). Effects of endogenous salicylic acid on nodulation in the model legumes Lotus japonicus and Medicago truncatula. Plant Physiol. 141, 1473–1481. doi: 10.1104/pp.106.080986
Stes, E., Depuydt, S., De Keyser, A., Matthys, C., Audenaert, K., Yoneyama, K., et al. (2015). Strigolactones as an auxiliary hormonal defence mechanism against leafy gall syndrome in Arabidopsis thaliana. J. Exp. Bot. 66, 5123–5134. doi: 10.1093/jxb/erv309
Sun, H., Bi, Y., Tao, J., Huang, S., Hou, M., Xue, R., et al. (2016). Strigolactones are required for nitric oxide to induce root elongation in response to nitrogen and phosphate deficiencies in rice. Plant Cell Environ. 39, 1473–1484. doi: 10.1111/pce.12709
Szalai, G., Pál, M., Árendás, T., and Janda, T. (2016). Priming seed with salicylic acid increases grain yield and modifies polyamine levels in maize. Cereal Res. Commun. 44, 537–548. doi: 10.1556/0806.44.2016.038
Szepesi, Á. (2011). Interaction between salicylic acid and polyamines and their possible roles in tomato hardening processes. Acta Biol. Szeged. 55, 165–166. Available online at: http://abs.bibl.u-szeged.hu/index.php/abs/article/view/2740
Tailor, A., Tandon, R., and Bhatla, S. C. (2019). Nitric oxide modulates polyamine homeostasis in sunflower seedling cotyledons under salt stress. Plant Signal. Behav. 14:1667730. doi: 10.1080/15592324.2019.1667730
Takács, Z., Poór, P., and Tari, I. (2016). Comparison of polyamine metabolism in tomato plants exposed to different concentrations of salicylic acid under light or dark conditions. Plant Physiol. Biochem. 108, 266–278. doi: 10.1016/j.plaphy.2016.07.020
Tamas, L., Mistrík, I., Alemayehu, A., Zelinová, V., Bočová, B., and Huttová, J. (2015). Salicylic acid alleviates cadmium-induced stress responses through the inhibition of Cd-induced auxin-mediated reactive oxygen species production in barley root tips. J. Plant Physiol. 173, 1–8. doi: 10.1016/j.jplph.2014.08.018
Tanaka, S., Han, X., and Kahmann, R. (2015). Microbial effectors target multiple steps in the salicylic acid production and signaling pathway. Front. Plant Sci. 6:349. doi: 10.3389/fpls.2015.00349
Tang, Y., Zhang, Z., Lei, Y., Hu, G., Liu, J., Hao, M., et al. (2019). Cotton WATs modulate SA biosynthesis and local lignin deposition participating in plant resistance against Verticillium dahliae. Front. Plant Sci. 10:526. doi: 10.3389/fpls.2019.00526
Terron-Camero, L. C., Peláez-Vico, M. Á., Del-Val, C., Sandalio, L. M., and Romero-Puertas, M. C. (2019). Role of nitric oxide in plant responses to heavy metal stress: exogenous application versus endogenous production. J. Exp. Bot. 70, 4477–4488. doi: 10.1093/jxb/erz184
Thalineau, E., Truong, H. N., Berger, A., Fournier, C., Boscari, A., Wendehenne, D., et al. (2016). Cross-regulation between N metabolism and nitric oxide (NO) signaling during plant immunity. Front. Plant Sci. 7:472. doi: 10.3389/fpls.2016.00472
Thurlow, J., Dorosh, P., and Davis, B. (2019). Demographic change, agriculture, and rural poverty. Sust. Food Agric. 31–53. doi: 10.1016/B978-0-12-812134-4.00003-0
Tiwari, P., Indoliya, Y., Chauhan, A. S., Singh, P., Singh, P. K., Singh, P. C., et al. (2020). Auxin-salicylic acid cross-talk ameliorates OsMYB–R1 mediated defense towards heavy metal, drought and fungal stress. J. Hazard. Mater. 399:122811. doi: 10.1016/j.jhazmat.2020.122811
Torrens-Spence, M. P., Bobokalonova, A., Carballo, V, Glinkerman, C. M., Pluskal, T., Shen, A., et al. (2019). PBS3 and EPS1 complete salicylic acid biosynthesis from isochorismate in Arabidopsis. Mol. Plant 12, 1577–1586. doi: 10.1016/j.molp.2019.11.005
Torres-Vera, R., García, J. M., Pozo, M. J., and López-Ráez, J. A. (2014). Do strigolactones contribute to plant defence?. Mol. Plant Pathol. 15, 211–216. doi: 10.1111/mpp.12074
van Butselaar, T., and Van den Ackerveken, G. (2020). Salicylic acid steers the growth–immunity tradeoff. Trends Plant Sci. 25, 566–576. doi: 10.1016/j.tplants.2020.02.002
Van der Does, D., Leon-Reyes, A., Koornneef, A., Van Verk, M. C., Rodenburg, N., Pauwels, L., et al. (2013). Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25, 744–761. doi: 10.1105/tpc.112.108548
van Spronsen, P. C., Tak, T., Rood, A. M., van Brussel, A. A., Kijne, J. W., and Boot, K. J. (2003). Salicylic acid inhibits indeterminate-type nodulation but not determinate-type nodulation. Mol. Plant Microbe. Interact. 16, 83–91. doi: 10.1094/MPMI.2003.16.1.83
Vos, I. A., Moritz, L., Pieterse, C. M., and Van Wees, S. (2015). Impact of hormonal crosstalk on plant resistance and fitness under multi-attacker conditions. Front. Plant Sci. 6:639. doi: 10.3389/fpls.2015.00639
Vos, I. A., Pieterse, C. M., and Van Wees, S. C. (2013). Costs and benefits of hormone-regulated plant defences. Plant Pathol. 62, 43–55. doi: 10.1111/ppa.12105
Walter, M. H. (2020). “Strigolactones: nutrient stress hormones affecting plant development and biotic interactions”, in Biology, Chemistry, and Applications of Apocarotenoids, ed S. Ramamoorthy (United States: CRC Press), 189–211. doi: 10.1201/9780429344206-11
Wang, L., Guo, Y., Jia, L., Chu, H., Zhou, S., Chen, K., et al. (2014). Hydrogen peroxide acts upstream of nitric oxide in the heat shock pathway in Arabidopsis seedlings. Plant Physiol. 164, 2184–2196. doi: 10.1104/pp.113.229369
Wang, P., Du, Y., Hou, Y. J., Zhao, Y., Hsu, C. C., Yuan, F., et al. (2015). Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proc. Nat. Acad. Sci. U.S.A. 112, 613–618. doi: 10.1073/pnas.1423481112
Wani, A. B., Chadar, H., Wani, A. H., Singh, S., and Upadhyay, N. (2017). Salicylic acid to decrease plant stress. Environ. Chem. Lett. 15, 101–123. doi: 10.1007/s10311-016-0584-0
Wassie, M., Zhang, W., Zhang, Q., Ji, K., Cao, L., and Chen, L. (2020). Exogenous salicylic acid ameliorates heat stress-induced damages and improves growth and photosynthetic efficiency in alfalfa (Medicago sativa L.). Ecotox. Environ. Saf. 191:110206. doi: 10.1016/j.ecoenv.2020.110206
White, R. F. (1979). Acetylsalicylic acid (aspirin) induces resistance to tobacco mosaic virus in tobacco. Virology 99, 410–412. doi: 10.1016/0042-6822(79)90019-9
Wimalasekera, R., Tebartz, F., and Scherer, G. F. (2011). Polyamines, polyamine oxidases and nitric oxide in development, abiotic and biotic stresses. Plant Sci. 181, 593–603. doi: 10.1016/j.plantsci.2011.04.002
Winter, G., Todd, C. D., Trovato, M., Forlani, G., and Funck, D. (2015). Physiological implications of arginine metabolism in plants. Front. Plant Sci. 6:534. doi: 10.3389/fpls.2015.00534
Wu, P., Xiao, C., Cui, J., Hao, B., Zhang, W., Yang, Z., et al. (2020). Nitric oxide and its interaction with hydrogen peroxide enhance plant tolerance to low temperatures by improving the efficiency of the calvin cycle and the ascorbate–glutathione cycle in cucumber seedlings. J. Plant Growth Regul. 1–19. doi: 10.1007/s00344-020-10242-w
Xiong, J., Fu, G., Tao, L., and Zhu, C. (2010). Roles of nitric oxide in alleviating heavy metal toxicity in plants. Arch. Biochem. Biophys. 497, 13–20. doi: 10.1016/j.abb.2010.02.014
Xu, J., Wang, W., Yin, H., Liu, X., Sun, H., and Mi, Q. (2010). Exogenous nitric oxide improves antioxidative capacity and reduces auxin degradation in roots of Medicago truncatula seedlings under cadmium stress. Plant Soil 326, 321–330. doi: 10.1007/s11104-009-0011-4
Xu, S., Guerra, D., Lee, U., and Vierling, E. (2013). S-nitrosoglutathione reductases are low-copy number, cysteine-rich proteins in plants that control multiple developmental and defense responses in Arabidopsis. Front. Plant Sci. 4:430. doi: 10.3389/fpls.2013.00430
Yadu, S., Dewangan, T. L., Chandrakar, V., and Keshavkant, S. (2017). Imperative roles of salicylic acid and nitric oxide in improving salinity tolerance in Pisum sativum L. Physiol. Mol. Biol. Plant. 23, 43–58. doi: 10.1007/s12298-016-0394-7
Yastreb, T. O., Kolupaev, Y. E., Kokorev, A. I., Horielova, E. I., and Dmitriev, A. P. (2018). Methyl jasmonate and nitric oxide in regulation of the stomatal apparatus of Arabidopsis thaliana. Cytol. Genet. 52, 400–405. doi: 10.3103/S0095452718060129
Yildiz, I., Mantz, M., Hartmann, M., Zeier, T., Kessel, J., Thurow, C., et al. (2021). The mobile SAR signal N-hydroxypipecolic acid induces NPR1-dependent transcriptional reprogramming and immune priming. Plant Physiol. 186, 1679–1705. doi: 10.1093/plphys/kiab166
Yin, H., Hong, G., Li, L., Zhang, X., Kong, Y., Sun, Z., et al. (2019). miR156/SPL9 regulates reactive oxygen species accumulation and immune response in Arabidopsis thaliana. Phytopathology 109, 632–642. doi: 10.1094/PHYTO-08-18-0306-R
Yun, B. W., Skelly, M. J., Yin, M., Yu, M., Mun, B. G., Lee, S. U., et al. (2016). Nitric oxide and S-nitrosoglutathione function additively during plant immunity. New Phytol. 211, 516–526. doi: 10.1111/nph.13903
Zanganeh, R., Jamei, R., and Rahmani, F. (2019). Role of salicylic acid and hydrogen sulfide in promoting lead stress tolerance and regulating free amino acid composition in Zea mays L. Acta Physiol. Plant. 41, 1–9. doi: 10.1007/s11738-019-2892-z
Zhang, H. X., Feng, X. H., Jin, J. H., Khan, A., Guo, W. L., Du, X. H., et al. (2020a). CaSBP11 participates in the defense response of pepper to Phytophthora capsici through regulating the expression of defense-related genes. Int. J. Mol. Sci. 21:9065. doi: 10.3390/ijms21239065
Zhang, X., Shen, L., Li, F., Meng, D., and Sheng, J. (2011). Methyl salicylate-induced arginine catabolism is associated with up-regulation of polyamine and nitric oxide levels and improves chilling tolerance in cherry tomato fruit. J. Agric. Food Chem. 59, 9351–9357. doi: 10.1021/jf201812r
Zhang, Y., and Li, X. (2019). Salicylic acid: biosynthesis, perception, and contributions to plant immunity. Curr. Opin. Plant Biol. 50, 29–36. doi: 10.1016/j.pbi.2019.02.004
Zhang, Z., Lan, M., Han, X., Wu, J., and Wang-Pruski, G. (2020b). Response of ornamental pepper to high-temperature stress and role of exogenous salicylic acid in mitigating high temperature. J. Plant Growth Regul. 39, 133–146. doi: 10.1007/s00344-019-09969-y
Zhao, Y., Ma, W., Wei, X., Long, Y., Zhao, Y., Su, M., et al. (2020). Identification of exogenous nitric oxide-responsive miRNAs from Alfalfa (Medicago sativa L.) under drought stress by high-throughput sequencing. Genes 11:30. doi: 10.3390/genes11010030
Zheng, X. Y., Zhou, M., Yoo, H., Pruneda-Paz, J. L., Spivey, N. W., Kay, S. A., et al. (2015). Spatial and temporal regulation of biosynthesis of the plant immune signal salicylic acid. Proc. Nat. Acad. Sci. U.S.A. 112, 9166–9173. doi: 10.1073/pnas.1511182112
Zhou, J., Jia, F., Shao, S., Zhang, H., Li, G., Xia, X., et al. (2015). Involvement of nitric oxide in the jasmonate-dependent basal defense against root-knot nematode in tomato plants. Front. Plant Sci. 6:193. doi: 10.3389/fpls.2015.00193
Keywords: salicylic acid, nitric oxide, plant immunity, transcriptional regulation, strigolactones
Citation: Rai KK, Pandey N, Rai N, Rai SK and Pandey-Rai S (2021) Salicylic Acid and Nitric Oxide: Insight Into the Transcriptional Regulation of Their Metabolism and Regulatory Functions in Plants. Front. Agron. 3:781027. doi: 10.3389/fagro.2021.781027
Received: 22 September 2021; Accepted: 16 November 2021;
Published: 07 December 2021.
Edited by:
Massimo Zacchini, Research Institute on Terrestrial Ecosystems, National Research Council (CNR), ItalyReviewed by:
Huan Chen, University of South Carolina, United StatesCopyright © 2021 Rai, Pandey, Rai, Rai and Pandey-Rai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shashi Pandey-Rai, c2hhc2hpLmJodWJvdGFueUBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.